Abstract
Liquid–liquid phase separation (LLPS) is a novel principle for explaining the precise spatial and temporal regulation in living cells. LLPS compartmentalizes proteins and nucleic acids into micron-scale, liquid-like, membraneless bodies with specific functions, which were recently termed biomolecular condensates. Biomolecular condensates are executors underlying the intracellular spatiotemporal coordination of various biological activities, including chromatin organization, genomic stability, DNA damage response and repair, transcription, and signal transduction. Dysregulation of these cellular processes is a key event in the initiation and/or evolution of cancer, and emerging evidence has linked the formation and regulation of LLPS to malignant transformations in tumor biology. In this review, we comprehensively summarize the detailed mechanisms of biomolecular condensate formation and biophysical function and review the recent major advances toward elucidating the multiple mechanisms involved in cancer cell pathology driven by aberrant LLPS. In addition, we discuss the therapeutic perspectives of LLPS in cancer research and the most recently developed drug candidates targeting LLPS modulation that can be used to combat tumorigenesis.
Subject terms: Cancer genetics, Epigenomics
Introduction
In the densely packed cellular space, the coordination of complex biochemical reactions in a spatial and temporal manner is important for performing biological function.1 Disruption of the precise spatiotemporal regulation can result in dysregulation of diverse cellular processes, including transcription,2 genomic integrity,3 chromatin organization,4 RNA processing5 and intracellular signaling.6,7 These processes are conceptual mechanisms underlying the complex hallmarks of cancer, including the loss of control over cell growth and proliferation, resistance to cell death, and metabolic reprogramming.8 Therefore, it is necessary to maintain normal spatiotemporal control in cells.
A possible cellular strategy for coordinating these reactions involves the formation of different compartments, where the density of the reaction components at a specified intracellular location can be adjusted.9 In fact, enzymatic reaction components are usually packaged in distinct subcellular compartments.9 For instance, classical organelles with a defined stoichiometry, such as the nucleus, Golgi apparatus, mitochondria and others, are lipid bilayer membrane-bounded compartments that make internal compounds inaccessible to almost all biomolecules and extracompartmental properties.
Notably, eukaryotic cells also achieve subcellular compartmentalization by forming a variety of nonstoichiometric biomolecular condensates without membranous structures,10 including promyelocytic leukemia (PML) protein bodies, nucleoli, paraspeckles, and Cajal bodies in the nucleus and stress granules (SGs), signaling puncta, and processing bodies (P bodies) found in the cytoplasm.11–13 Biomolecular condensates are composed of weak, multivalent interactions between macromolecules9 (for example, proteins and nucleic acids). In addition to the abovementioned punctate membraneless bodies, other subcellular structures also share similar physical properties, including heterochromatin14 and membrane receptor clusters at the cell membrane.15 The majority of these condensates exchange subunits rapidly with their surroundings within seconds or minutes.16–18
Increasing lines of evidence suggest that biomolecular condensates are reversibly and dynamically assembled via liquid–liquid phase separation (LLPS).11 LLPS is a physiological process that spontaneously drives the separation of a homogeneous solution of constituents into two or more coexisting phases: a dilute phase and a dense phase.19 Additionally, many studies have demonstrated that biomolecular condensates can be transformed into materials in different states, such as viscous liquids, gels, and even solid aggregates.20–22 The material features of biomolecular condensates are pivotal to distinct functions; for example, biocondensates form biochemical reaction centers, signaling hubs and the supporting architecture. Although cells have developed a variety of mechanisms to guarantee well-controlled LLPS, aberrant forms of phase separation are causatively related to many of the dysregulated cellular processes in cancer.20 In Table 1, we summarize the names, genes, localizations, structures and functions of 25 molecules related to the oncogenic mechanisms related to LLPS. For instance, the LLPS of NUP98-HOXA9 contributes to the formation of a broad super-enhancer (SE)-like binding pattern that potentiates the transcriptional activation of leukemogenic genes.23 Furthermore, it has been estimated that most cell signaling proteins, and even a large number of cancer-related proteins, have long intrinsically disordered regions (IDRs), which play critical roles in promoting LLPS.24
Table 1.
Tumor-associated protein molecules that have been discovered to undergo LLPS
| Protein | Organelle/Biomolecular condensates | Localization | Function | Role of phase separation in tumor |
|---|---|---|---|---|
| 53BP1 | Nuclear body | Nucleus |
1. Respond to DNA damage.126 2. Assists the tumor suppressor effect of p53.131 |
Hyper-accumulation of 53BP1 on chromatin and enhanced LLPS compromise cell survival in cancer cells.199 |
| BRD4 | Enhanceosome; nuclear body; nuclear bodies that occur at super enhancers in mESCs | Nucleus114 |
1. Epigenetic and transcriptional regulation.310,311 2. Respond to DNA damage.312 |
The involvement of BRD4 in SEs plays a role in oncogene transcriptional addiction and cancer cell survival.312 |
| CBX2 | PcG protein complex; euchromatin; heterochromatin; PcG chromatin condensates313 | Nucleus |
1. Chromatin remodeling and modification of histones.314 2. Promote the proliferation, invasion and migration of cancer cells.315 |
PcG protein complex can compact chromatin,314 and CBX2 is particularly upregulated in various cancers.316 |
| DAXX | Nuclear body; nuclear protein granule; SPOP/DAXX body | Nucleus and cytoplasm |
1. Transcriptional regulation. 2. DNA repair, and respond in viral infection. 3. Impact apoptosis and cell signaling. |
SPOP/DAXX bodies formed via LLPS is important in inducing cancer cell apoptosis.97,128 |
| SPOP | Nuclear body; nuclear protein granule; SPOP/DAXX body | Nucleus and cytoplasm |
1. An E3 ubiquitin ligase substrate binding subunit of the proteasome complex that has both oncogenic and tumor-suppressive function in human cancers. 2. SPOP is frequently mutated in different cancers.214,225,226,317 |
Cancer-associated mutations of SPOP disrupt LLPS, and result in reduced protein ubiquitination.215 |
| EWS | Nuclear protein granule318 | Nucleus, cytoplasm and plasma membrane |
1. Histone modification. 2. DNA methylation.319 3. Fuse with transcription factor ELI.320 |
EWS-FLI1 specifically recruit BAF complex to activate target genes via binding to tumor-specific enhancers in Ewing sarcoma.163 |
| HNRNPA1, HNRPA1 | Cytoplasmic stress granule (SG) | Nucleus and cytoplasm |
1. Forming SGs.104 2. RNA splicing.321 |
SGs participate in apoptosis, immune modulation, and signaling pathways.322 SG assembly is aberrantly elevated in human cancers.323 |
| HP1a | Heterochromatin116 | Nucleus |
1. Promote the formation of heterochromatin and plays a role in gene silencing. |
Normal packaging and organization of heterochromatin is often compromised in cancer.327 |
| MED1 | Enhanceosome; nuclear body; nuclear bodies that occur at super enhancers in mESCs | Nucleus |
1. A coactivator that involved in cancer-related transcriptional regulation and dysregulation.328–330 2. Regulate autophagy.331 |
1. MED1 overexpression is associated with drug resisitance.332 2. SEs mediate transcriptional addiction.333 |
| NONO | Paraspeckle | Nucleus |
1. It is required for mRNA splicing, DNA unwinding, transcriptional regulation, nuclear retention of defective RNA and DNA repair.334 2. Induce cellular senescence. 3. Regulator of RNA:DNA hybrid related telomere instability.335–337 |
Paraspeckles influence the tumor stability to develop drugs resistance.338 |
| SFPQ | Paraspeckle | Nucleus and cytoplasm |
1. Regulate transcriptional activity, mRNA processing and splicing. 2. Regulator of RNA:DNA hybrid related telomere instability. 3. High SFPQ expression level in liver cancer is associated with cisplatin resistance.337,339–341 4. DNA repair. |
Paraspeckles influence the tumor stability to develop drugs resistance.338 |
| NPM1 | Nucleolus; granular component342 | Nucleus |
1. Cooperate with MYC to induce transcription of target proteins, thereby regulating the proliferation of normal cells and cancer cells. 2. It is mutated in acute myeloid leukemia. 3. NPM1 silencing cells play a role in migration and invasion ability.343–347 |
Larger nucleolar size and number are hallmarks of various cancers.348 Deregulation of nucleolar functions is correlated with tumorigenesis.349 |
| NUP98 | Nuclear pore central transport channel; selective hydrogel-like meshwork formed by FG-nucleoporins in nuclear pore central channel | Nucleus |
1. Chromosomal translocations, changes in protein expression levels, and single point mutations. |
NUP98-HOXA9 formed via LLPS induces leukemic transformation.23 |
| PML | PML body | Nucleus and cytoplasm |
1. Mediator of multiple apoptotic pathways. 2. A tumor suppressor. 3. Transcriptional regulation. 4. Regulate growth and invasion of cancer differentially.352–354 |
Disruption of PML bodies drives initiation of acute promyelocytic leukemia.355,356 |
| YTHDF1, DF1 | P-body; cytoplasmic stress granule;357 neuronal ribonucleoprotein granule | Cytoplasm |
1. Higher expression in tumors than normal tissue in human cancers. 2. Regulate immune response and antigen processing and presentation.358 3. mRNA binding, processing and degradation.359 4. Regulate tumorigenicity and stem cell-like activity in cancer cells via Wnt/β-catenin pathway.360 5. Overexpression of YTHDF1 promotes breast cancer progression.361 |
1. P-body-based regulation of mRNA metabolism plays an important role in cancer development and progression.258 2. SG assembly is upregulated in cancer.323 |
| YTHDF2, DF2 | P-body; cytoplasmic stress granule; neuronal ribonucleoprotein granule | Nucleus and cytoplasm |
1. mRNA binding, processing, stability and degradation. 2. YTHDF2 SUMOylation is important in post-transcriptional gene expression regulation and cancer progression.362–365 |
1. P-body-based regulation of mRNA metabolism plays an important role in cancer development and progression.258 2. SG assembly is upregulated in cancer.323 |
| YTHDF3, DF3 | P-body; cytoplasmic stress granule; neuronal ribonucleoprotein granule | Cytoplasm |
1. A key player in YAP signaling. 2. Translational regulation. 3. Higher expression in tumors than normal tissue in human cancers. 4. Overexpression of YTHDF1 and YTHDF3 promotes breast cancer progression.359,361,366,367 |
1. P-body-based regulation of mRNA metabolism plays an important role in cancer development and progression.258 2. SG assembly is upregulated in cancer.323 |
| TAF15 | Nuclear protein granule | Nucleus and cytoplasm |
1. mRNA binding, stabilization and regulation. 2. Stress response and DNA repair.56 3. Translational control. 4. mRNA and protein levels of TAF15 are upregulated in liposarcoma.368–370 5. TAF15 overexpression is associated with poor prognosis in patients NSCLC patients.371 |
Aberrant gene transcription through loci-specific phase separation, which contribute to oncogenic transformation ability in relevant cancers.372 |
| P-TEFb | 7SK snRNP complex, super elongation complex (SEC) | Nucleus | 1. Elongation control of cellular transcription.373 | SEC is a target for the mixed lineage leukemia (MLL) protein to activate the MLL target genes expression and promote leukemogenesis.374 |
| OCT-4 | MED1 droplets at SEs | Nucleus and cytoplasm | 1. Cancer stem cells maintain expression of Oct4.375 | SEs mediate transcriptional addiction in diverse cancers.333 |
| YAP | YAP-TEAD complex/ YAP-TAZ-TEAD complex | Nucleus and cytoplasm |
1. Transcriptional coactivator. 2. Effector of the Hippo signaling cascade. 3. Induce cancer stem cell attributes, proliferation, chemoresistance, and metastasis.366,376 |
YAP-TEAD complex is not only hyperactivated, but also confers a strong oncogenic activity in tumor tissues.377,378 |
| TAZ | YAP-TAZ-TEAD complex | Nucleus, cytoplasm and plasma membrane |
1. Transcriptional coactivator. 2. Effector of the Hippo signaling cascade. 3. Induce cancer stem cell attributes, proliferation, chemoresistance, and metastasis.366,376,379 |
YAP-TAZ-TEAD complex is not only hyperactivated, but also confers a strong oncogenic activity in tumor tissues.377,378 |
| DDX3 | Cytoplasmic stress granule | Nucleus, cytoplasm and plasma membrane |
1. ATP-dependent RNA helicase involved in DNA repair.380 2. Transcriptional regulation. 3. Initiate and regulate translation.381 4. Involved in stress and inflammatory responses.382 |
Cancer-associated mutations of DDX3X cause SG hyper-assembly and translation impairment.383 |
| HSF1 | HSF1 nuclear stress bodies (HSF1 foci) | Nucleus |
1. Transcriptional regulator of chaperones. 2. Cancer cell invasion, proliferation, and metabolism.384 |
HSF1 foci are preferentially located in cancer cells of primary human tumors.384 |
| ENL | Nuclear protein granule | Nucleus | 1. Frequently fuse with the MLL protein and resultant fusion proteins function as oncogenic drivers in acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL).385 | ENL LLPS enhance recruitment of SEC and drive transcription.386,387 |
In this review, we describe recently obtained insights and findings on the formation, dynamics, regulation, and function of LLPS and highlight the various effects of biomolecular condensates on cell biology. Our review specifically focuses on the dysregulated LLPS that occurs in aberrant cellular processes associated with carcinogenesis, including chromatin organization, epigenetics, oncogenic transcription, aberrant signaling pathways, and telomere lengthening mediated by LLPS. Clinical evidence for the occurrence of abnormal LLPS processes in cancer patients has also been extensively collected, and this information provides a substantial basis for the importance of LLPS in tumor biology. For example, Meng et al. observed phase-separated droplets formed by Merlin (NF2) in dissected samples from vestibular schwannoma patients.25 In addition, emerging evidence suggests that biomolecular condensates affect the pharmacodynamic properties of antineoplastic medicines; therefore, regulating the LLPS process could be a potential strategy for novel cancer therapies. Therefore, we evaluate the great potential of effectively regulating LLPS in anticancer therapy and propose perspectives on condensates that might contribute to future investigations in oncology.
Liquid–liquid phase separation
LLPS is a decent explanation for the formation of multiple membraneless structures in cells.26 The profound exploration of membraneless organelles began with the discovery of P granules in Caenorhabditis elegans in 2009. The P granules are liquid-like structures composed of proteins and RNAs, which can flow, fuse, deform and fission under shear force.27 Following the study of P granules, other intracellular substructures, such as the nucleoli, SGs, and paraspeckles, have also been revealed to be formed by LLPS and are enriched in RNA-binding proteins (RBPs) and RNAs.28,29
LLPS is thought to be triggered by weak, multivalent interactions between proteins and nucleic acids13 rather than covalent, high-affinity interactions. These weak interactions also play a pivotal role in concentrating components at discrete cellular sites,30 which is very important to ensure the accurate spatiotemporal regulation of normal cell biological activities. Below, we summarize the thermodynamic conditions necessary to trigger LLPS and various interactions between proteins and nucleic acids that promote LLPS.
Thermodynamic conditions of LLPS
If we consider the cytoplasm or nucleoplasm as a macromolecular solution, the surface of macromolecules would exhibit weak, transient, nonspecific interactions with each other and with the solvent.31 These low-affinity interactions tend to dissolve the molecules and evolve the entire system into a well-mixed state (entropy).31 The solubility of macromolecules is dominated by the balance of weak interactions between macromolecules and macromolecules versus macromolecules and solvent.32 As the concentration of macromolecules in the solution is increased to the solubility limit—the threshold concentration—the interactions between the macromolecules will become stronger than the interactions between the macromolecules and the solvent, and as a result, this solution will gain propensity to LLPS.33 The threshold concentration depends on several biophysical parameters, such as the salt concentration, temperature, and other ions.34 Therefore, the concentration dependence suggests that the threshold concentrations are hallmarks of LLPS.19
Multivalency driving LLPS
The macromolecules undergoing LLPS can be classified into scaffolds and clients.35 Scaffold molecules are characterized by multivalent proteins or nucleic acids that are indispensable for triggering LLPS.36 Higher valency enables the formation of larger oligomers or polymers at lower saturation.37 Multivalency can be achieved by proteins containing multiple folded domains, IDRs,28 and nucleic acid chains.38 The client proteins are molecules with low valency that are recruited by the scaffold and partition into condensate structures. Clients cannot themselves undergo LLPS, and their recruitment level is determined by the scaffold stoichiometry.35
Compared with the very-low-affinity interactions determining solubility, the multivalent interactions between macromolecules show higher affinity and high stereospecificity and enable assembly into large oligomers or polymers.39
Multiple folded domains that promote LLPS
LLPS in living cells was initially investigated based on the multivalent interactions among Nephrin, Nck and neural Wiskott-Aldrich Syndrome protein (N-WASP). The phosphotyrosines (pTyrs) in Nephrin can each bind the SH2 domain of Nck, and Nck possesses three SH3 domains that can bind the ~six proline-rich motifs (PRMs) of N-WASP (as shown in Fig. 1a).37 When the adhesion receptor Nephrin is attached to the lipid bilayer, the three-component system (Nephrin/Nck/N-WASP) can also undergo LLPS and form liquid-like clusters on the membrane surface.40 Similarly, the signaling proteins governing the organization of actin in T cells can also assemble into membrane puncta in response to T-cell receptor (TCR) activation.15
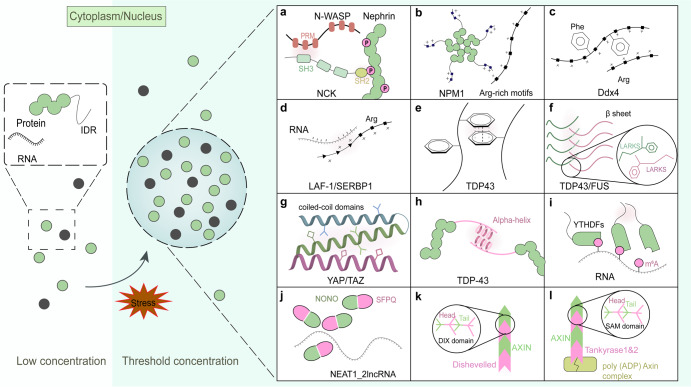
Fig. 1.
Summary of various types of interactions that promote the occurrence and maintenance of LLPS. a Nephrin contains three phosphotyrosines (pTyrs) motifs, which bind the SH2 domain of Nck, and Nck possesses three SH3 domains that can bind the ~six proline-rich motifs (PRMs) of N-WASP. b The unphosphorylated form of the Npm1 monomer oligomerizes into pentamers through its N-terminus (Npm-N) and binds to proteins with Arg-rich linear motifs. c In Ddx4, Phe and Arg motifs in the intrinsically disordered regions (IDRs) drive LLPS. d In LAF-1 and SERBP1 proteins, the positively charged Arg/Gly-rich (RGG/RG) domain binds to negatively charged RNA and effectively promote LLPS. e In TDP43, the pi-pi interactions in IDRs facilitates LLPS. f Low-complexity amyloid-like reversible kinked segments (LARKS) in TDP43, and FUS proteins would give rise to phase separation via mediating reversible amyloid-like interactions. g Hydrophobic interactions existing in coiled-coil domains are essential for phase separation. h Local α-helix in the C-terminus of the TDP-43 protein enables TDP-43 self-connections and facilitates LLPS. i LLPS of YTHDF1 can be enhanced by the mRNAs with multiple m6A residues. j NEAT1_2 lncRNA subdomains selectively bind NONO/SFPQ proteins and dynamically oligomerize. k In the Wnt signaling cascade, Disheveled contacts the DIX domain of the AXIN protein in a head-to-tail manner and transduce Wnt signals into the nucleus. l SAM domains of tankyrase protein form dynamic puncta via head-to-tail polymerization with AXIN, promoting Wnt signaling
Another example is Nucleophosmin (Npm1), a highly abundant protein that contributes to the maintenance of genome stability and regulation of the p53 tumor suppressor pathway and has been implicated in mediating the LLPS of nucleolar granule components.41–43 The Npm1 monomer encodes only a 2-valent interaction partner, which is insufficient for triggering phase separation. However, the unphosphorylated form of the Npm1 monomer oligomerizes into pentamers through its N-terminus (Npm-N) and binds to proteins with Arg-rich linear motifs. The oligomerizing process effectively increases the valency to 10, which is sufficiently high to mediate phase separation (Fig. 1b).44,45 This excellent example proves the importance of multivalency between interacting motifs for controlling LLPS. Similarly, the speckled POZ protein (SPOP), a tumor suppressor, is a cullin-3-RING ubiquitin ligase (CRL3) substrate adapter that can self-assemble into higher-order polymerized forms and localize to nuclear speckles.46
IDRs promote LLPS
The IDRs in proteins are another method for gaining multivalency and driving LLPS. IDRs lack a stable tertiary structure, which allows access to a wider conformational space and enables the formation of three-dimensional networks of protein molecules.47 These regions have low amino acid sequence complexity and comprise only a limited set of amino acid types, such as Gly, Ser, and Gln, and aromatic residues, including Phe and Tyr.9,48,49 Recent evidence suggests that the aromatic residues in IDRs are particularly important for the promoting effect. In Ddx4, the cation-pi interactions between Phe and Arg motifs have been proven to be significant for driving LLPS (Fig. 1c).50 Analogously, the Tyr residues in other RBPs, such as fused in sarcoma (FUS),51 hnRNPA152 and BuGZ,53 can also promote the process of phase separation in vitro and/or in cells. The mutation of Tyr residues in BuGZ53 and FUS54 significantly blocks the formation of phase-separated liquid droplets.
Additionally, the electrostatic interactions (salt bridges) between opposing charge residues contribute to the promotion of LLPS. In the Caenorhabditis elegans protein LAF-1, the positively charged Arg/Gly-rich (RGG/RG) domain binds to negatively charged RNA and effectively promotes the formation of P granules (Fig. 1d).55 The same interaction pattern can also be observed in nucleation events at DNA damage response (DDR) sites, where negatively charged poly(ADP-ribose) (PAR) can rapidly recruit positively charged proteins containing IDRs and cause liquid demixing upon DNA damage (Fig. 2).56 Moreover, IDRs can drive LLPS through other weak interactions, such as pi-pi interactions mediated by aromatic residues (Fig. 1e),57 reversible amyloid-like interactions occurred in TDP43, and FUS proteins (Fig. 1f),58,59 hydrophobic interactions triggered by coiled-coil domains (Fig. 1g),60–62 along with dipolar interactions (data was not shown).63
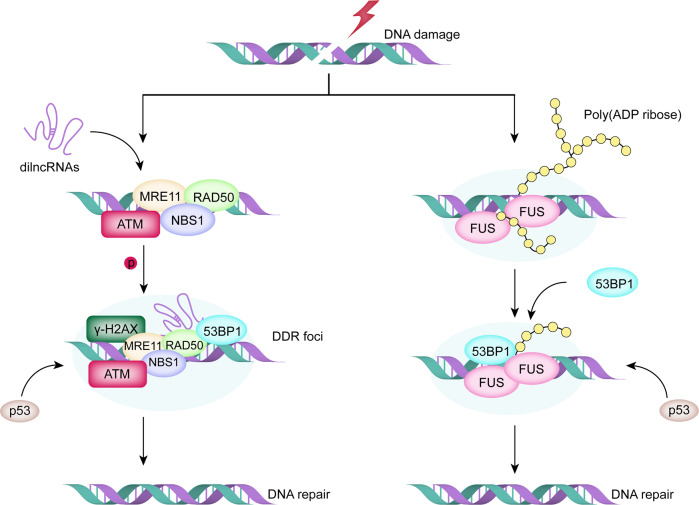
Fig. 2.
LLPS supports various DNA damage response (DDR) mediator to form DNA repair foci in different pathways. (Left) MRE11–RAD50–NBS1 complexes bind to the exposed DNA damage sites to initiate the pathway. Subsequently, the damage-induced lncRNAs (dilncRNAs) and P53-binding protein 1 (53BP1) are recruited to the DDR site to promote the formation of DNA damage repair foci via LLPS. (Right) Nucleation of PAR with FUS and 53BP1 at DDR sites forms liquid-like compartments and facilitates subsequent signaling and repair
In addition, the short stretches of amino acids in IDRs can form localized structures and promote self-interaction. For instance, the IDRs located in the C-terminus of the TDP-43 protein contain a structural domain that forms a local α-helix, which enables TDP-43 self-connections and facilitates LLPS (Fig. 1h). Mutations affecting the structure or intermolecular contacts in the TDP-43 alpha-helix and/or nearby regions can disrupt multivalent interactions and result in altered phase separation.64
RNAs modulate phase behavior
As mentioned above, RNAs are found in membraneless condensates19,20,65 and have been shown to promote LLPS.65 RNAs can not only drive LLPS via electrostatic interactions, but repetitive intermolecular base pairing can also achieve multivalency and thus drive the formation of clusters in vitro and in vivo.65 For example, RNAs added to SERPINE1 mRNA-binding protein 1 (SERBP1) medium effectively induce LLPS by interacting with the RG/RGG-rich domains of SERBP1 (Fig. 1d). Furthermore, fluorescence signals are more rapidly recovered in the presence of RNA, which suggests that RNA at a certain concentration (0.05 mg/ml) (measurement of the RNA sequence 5’-GCGCGGG-3’) makes SERBP1 droplets more dynamic and fluid.66 However, the experiments conducted by Burke and Janke suggest that FUS monomers can interact with RNA to initiate the construction of fibrillar FUS condensates, but a higher quantity of RNA dissolves FUS condensates.51
N6-methyladenosine (m6A) is the most frequent nucleotide modification of mRNA67,68 and is necessary for various cellular and physiological processes. The expression level of the correlated proteins is frequently increased in a variety of human cancers.69,70 In diverse RNP granules, including SGs, keratin granules, and P-bodies, the m6A-binding protein YTHDFs in the cytoplasm undergo spontaneous LLPS in vitro and in extracted cells, and the process can be clearly enhanced by the addition of mRNAs containing multiple m6A residues (Fig. 1i).68 Polymethylated mRNAs may function as scaffolds for proteins with multivalency in combination with YTHDF proteins through their IDRs, resulting in LLPS.68
In addition, a great number of long noncoding RNAs (lncRNAs) are localized on chromatin and often form an RNA cloud in a particular nuclear area to regulate gene expression. ncRNAs are major components of several membraneless structures, such as the nucleolus and paraspeckles,54,71 the formation of which is frequently dysregulated in cancers. Numerous findings have recently demonstrated that NEAT1_2 lncRNA subdomains can selectively attach to NONO/SFPQ proteins, and these proteins can dynamically oligomerize and recruit additional proteins through LLPS to facilitate the assembly of paraspeckles (Fig. 1j).72
New mechanism to multivalency—head-to-tail polymerization
In contrast to IDRs, there remain stably structured domains in proteins that can attain multivalency by improving local condensation.73 The DIX and SAM domains are two distinct domains capable of spontaneously assembling dynamic head-to-tail polymers, condense into filaments and are then crosslinked to form three-dimensional condensates.74,75 The DIX domain was discovered in the Wnt signaling cascade, Disheveled,76 which can contact the DIX domain of the AXIN protein in a head-to-tail manner and transduce Wnt signals into the nucleus (Fig. 1k).77 Analogously, the SAM domains of tankyrase protein can also form dynamic puncta via head-to-tail polymerization, promoting Wnt signaling by binding to and ribosylating the poly (ADP) AXIN destruction complex (Fig. 1l).78 The difference between the DIX and SAM domain interactions is that the affinities of SAM domains are higher than those of DIX domain-mediated interactions.74
Physiological regulation of LLPS
Any factor that influences the properties of the condensate component affects the condensate structure, dissolution, viscoelasticity, and other physicochemical features that influence their functions,12,38 such as protein concentration, posttranslational modifications (PTMs) or environmental elements, providing a vigorous mechanism for the regulation of cells.79
Biophysical parameters
The concentrations of biomolecular components are essential factors in condensate formation and dissolution. Expanding the volume of nucleoli by placing them into a hypotonic solution can cause reversible solubilization of PML bodies and nucleoli as the matrix is diluted.80 A variety of pathways influence the concentration, such as influencing biosynthesis, degradation, transportation, and localization.38
Because of the importance of thermodynamics, for some of the molecules that reach their dissolution limit, even a slight disturbance to physical parameters, such as a concentration or temperature chance, can induce rapid phase changes. For instance, a change of 1 °C may lead to BUgZ, DDX4, or FUS droplets condensing or dissolving.9 The prion-like domains of these proteins can sense pressure regulated by the environment, which in turn influences the solubility and phase behavior of the protein.81 For example, TDP43 and FUS cluster to SGs under physiological stress conditions such as heat and oxidative stress.82,83
Other biophysical elements, such as the salt concentration in the milieu66 and the addition of PEG3000 and glycerol, can also effectively regulate LLPS.66
Posttranslational modifications
Emerging evidence suggests that PTMs, which include phosphorylation, acetylation, arginine (Arg)-methylation, and SUMOylation, play pivotal roles in regulating phase separation.84,85 The downstream reactions of cells to various stimuli are often influenced by PTM-triggered signaling.86 PTMs induce a wide range of effects on the structural properties of intrinsically disordered proteins and potentially drive complete state changes among different states, such as intrinsically disordered states, folded states, dispersed monomeric, and phase-separated states.87 In LLPS, PTMs can alter the physicochemical characteristics of the regulated amino acids in scaffold proteins, such as by changing their valency, electric charge, or volume.35 PTMs can also influence the interactive conditions to affect phase separation, such as by directly diminishing or enhancing the multivalent interactions between macromolecules, recruiting certain macromolecules into the condensate or excluding macromolecules from the condensate.84 For instance, the Arg residues of RGG/RG motifs in FUS are largely modified by the deposition of asymmetric dimethyl groups by protein arginine methyltransferase (PRMT) 1 or 8,88–90 which in turn decreases the LLPS rate of FUS and enhances condensate dynamics.91 An overabundance of Arg methylation mediated by PRMT1 increases the rate of cytosolic FUS and SG partitioning in response to oxidative stress.82 Additionally, Arg methylation reduces hnRNPA2 phase separation and destabilizes the Ddx4 droplets by diminishing Arg-aromatic(pi) interactions.50,92
In addition, another PTM called SUMOylation involves the covalent attachment of small ubiquitin-like modifiers (SUMOs) that modulate cellular processes in the nucleus.93 Lys residues in disordered regions have been found to be the preferred target of SUMOylation, different from other common Lys PTMs.94 SUMOylation in PMLs contributes significantly to the formation of the PML nuclear body, whereas de-SUMOylation can lead to a constituent protein being released and nuclear bodies being separated during mitosis.50,95 In addition, death domain-associated protein (DAXX) possesses highly conserved SUMO-interacting motifs,96 which are needed for DAXX linkage with SUMOylated PML oncogenic domains, and its expression is upregulated in multiple cancers.97 DAXX can bind to SUMOylated SMAD4 and suppress SMAD4-mediated transcription.27 SMAD4 is activated downstream of the cellular effects of TGF-β, which induces apoptosis and prevents proliferation, and the loss of SMAD4 expression potentiates tumorigenesis,93,98 which suggests the potential significance of DAXX in carcinogenesis. In addition, SUMOylation of SERBP1 is thought to be a trigger for glioblastoma multiform progression because aberrant SUMOylation pathways may result in cancer progression.99
Furthermore, it has been demonstrated that in response to DNA damage, the N-terminus of FUS is phosphorylated by DNA-dependent protein kinase (DNA-PK), and this phosphorylation leads to FUS translocation from the nucleus to the cytoplasm. The translocation is mediated by the phosphorylation of serine or threonine residues on the N-terminus of FUS by DNA-PK, but the exact mechanism remains uncertain.100 Additionally, Ding et al. revealed that the phosphorylation of Ser61 occurs specifically at the Ser61 site of FUS, which can effectively disrupt the intra- and intermolecular interactions that maintain pathological aggregation in cells.101 Therefore, we believe that diverse PTMs can be an effective approach for regulating the process of phase separation and possibly affect oncogenic processes.
ATP and phase separation
The role of ATP in LLPS has thus far been subject to considerable uncertainty. First, ATP prevents RNA/proteosome assembly while maintaining protein solubility.102,103 A high concentration of ATP inhibits the tendency of IDRs in granular components to assemble into stable amyloid fibrils.104,105 Brangwynne et al. also found that the liquid-like properties in the spherical state of the nucleolus depend on ATP, which suggests that the nonspherical nucleolar profile may indicate changes in metabolism.106 However, a range of evidence confirms that ATP promotes the formation of nuclear condensates.107 Therefore, the exact role played by ATP in LLPS remains difficult to discern, but we look forward to more ideas from future researchers.
Chaperones and phase separation
Molecular chaperones are essential components of the quality control system in cells to sustain protein homeostasis (proteostasis) and thus avoid aberrant folding and aggregation.108 A wide spectrum of molecular chaperones has been proven to undergo LLPS, including heat shock protein 27 (Hsp27), class I and II Hsp40, Hsp70, and Hsp90, and most of chaperones have been found to be incorporated into SGs.109,110 Liu et al. demonstrated that small Hsp27 prevents LLPS of FUS protein by interfering with intra- and intermolecular transient interactions of the low-complexity domain in FUS. However, the phosphorylation of Hsp27 induced by cellular stress conditions, including sodium arsenite, heat shock, or oxidative stress, reduces its inhibitory function.110 Analogously, in neuron terminals, transportin also functions as a chaperone of FUS proteins, inhibiting the phase separation process and SG partitioning.111
In patients with fibrolamellar carcinoma, loss of myristylation and gain of Hsp70 binding by oncogenic DnaJB1-PKAcat are responsible for abolishing RIα LLPS. The DnaJB1-PKAcat fusion oncogene has been detected in nearly all fibrolamellar carcinoma patients, and the inhibition of RIα LLPS results in disruption of cAMP compartmentation and deregulation of the cAMP/PKA signaling pathway, which consequently leads to tumorigenesis.112 Therefore, the above-described example illustrates the importance of normal phase separation to the organism and the pathogenic potentiality of molecular chaperones in the regulation of LLPS.
Functions of biomolecular condensates
LLPS is involved in multiple biological processes in cells, such as chromatin architecture, DNA damage repair, transcriptional regulation, intracellular signaling, and protein degradation.113 These activities take place throughout all types of cells, and abnormalities in these processes are central events in the study of tumor biology. Therefore, we reasoned that LLPS might contribute to tumor pathogenesis and tumor progression.
LLPS in chromatin organization
LLPS of chromatin-associated factors can promote the organization of the chromatin structure to regulate transcription.14,114,115 Gibson et al. found that reconstituted chromatin undergoes LLPS both in vitro and in cells.4 Recent studies suggest that heterochromatin protein 1 (HP1) can form liquid-like droplets via phase separation.14,116 Heterochromatin is a tightly packaged chromatin structure that is an important component of eukaryotic genome sequences and is pivotal for the normal organization of chromosomes, genome integrity, DNA replication,117 transposon silencing and gene expression.118 A prominent characteristic of heterochromatin is trimethylation of Lys 9 on histone H2 (H3K9me2) and H3K9me3.119 Some findings have revealed that LLPS specifically takes place at transcriptionally active regions of DNA with lower chromatin density. Shin et al. revealed that phase-separated liquid condensates are preferentially formed at chromatic regions with low density and mechanically push out nontargeted chromatin. Thus, distant targeted genomic loci can be mechanically pulled together and restructured through the fusion of droplets.114 The causative linkage between LLPS and chromatin compression has been demonstrated in an increasing number of studies.120 DNA demethylation in heterochromatin has been linked to chromosome translocations in different types of cancer, such as breast, lymphoid, and endothelial tract cancer.121,122 Additionally, reduced hypermethylation of heterochromatin has been highly correlated with human cancer progression and metastasis.123,124
LLPS in DNA damage repair
The DDR is responsible for safeguarding genomic integrity and stability, whereas defects in the DDR may result in oncogenic mutations.125 In response to DNA damage, LLPS supports the process of various DDR mediators forming repair compartments in two different ways (Fig. 2).126 Upon DNA damage, MRE11–RAD50–NBS1 complexes can recognize and bind to the exposed DNA damage sites and recruit damage-induced lncRNAs (dilncRNAs). dilncRNAs then attract the tumor suppressor P53-binding protein 1 (53BP1) to the DDR site and promote the formation of DNA damage repair foci via LLPS (Fig. 2 left).127 This method of repairing DNA double-strand breaks is very common in cancer cells and results in the avoidance of apoptosis.128
The earliest cellular response to DNA lesions in another pathway starts with the nucleation of PAR with multiple intrinsically disordered proteins, including FUS, at DDR sites.127,129 FUS is precisely directed to DNA damage sites by long and branched PAR chain formation.130 After the PAR signal terminates, 53BP1 gains access to the DNA damage sites, forms liquid-like compartments and facilitates subsequent signaling and repair (Fig. 2 right).131
LLPS in transcriptional regulation
As a critical step in gene expression, dysregulated gene transcription can initiate the uncontrolled proliferation of cancer cells.132 Interestingly, emerging evidence demonstrates that LLPS plays an important role in the progression of transcription and RNA processing.133,134 For example, the C-terminal domain (CTD) of the RPB1 subunit of human RNA polymerase (Pol) II is composed of a highly repetitive, unstructured protein domain of low complexity.135 The FUS, Ewing sarcoma (EWS), and TAF15 genes can form a liquid-like phase-separated state, directly bind the CTD of RNA Pol II and activate transcription.51,136 Furthermore, recent evidence demonstrates that transcription factors (TFs), RNA Pol II, chromatin regulators and various coactivators aggregate into condensates (called SEs)137 via phase separation at those highly transcribed genes.138
In addition, strong evidence demonstrates that the transcriptional coactivator Yes-associated protein (YAP)139 and the transcriptional effector PDZ-binding motif (TAZ)140 compartmentalize the transcriptional cofactors and coactivators to facilitate the expression of target genes by LLPS.62 In lung cancer, the YAP-generated condensates can further form YAP/TEAD/SRC-1 compartments by interacting with SRC-1 and extensively improve YAP transcription (as shown in Fig. 3e).141 It is well known that YAP and TAZ are downstream effectors of the Hippo pathway—a tumor suppressor signaling pathway that is important for biological activities such as immune regulation, epithelial homeostasis, and tissue regeneration.142 Therefore, aberrant YAP/TAZ-mediated transcriptional condensates may contribute to cancer-related pathophysiology.
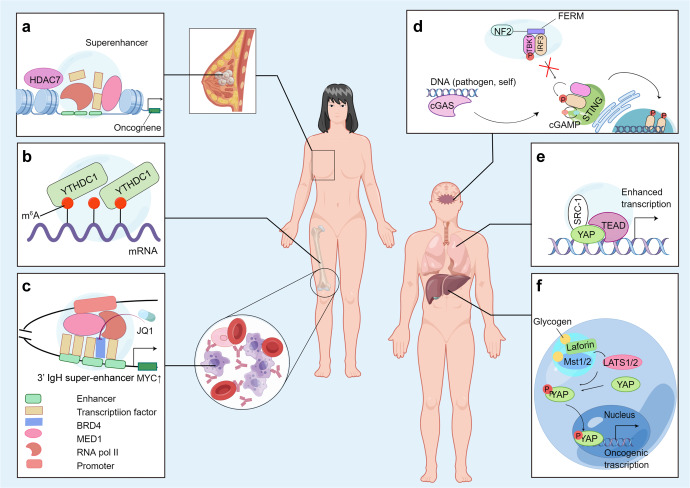
Fig. 3.
The roles of phase separation in various cancers. (By Figdraw.). a In stem-like breast cancer cell model, the histone deacetylase HDAC7 binds near the transcriptional start site and to SEs of various oncogenes. b YTHDC1 undergoes LLPS via binding with m6A-mRNA, and the number of resulting nuclear condensates (nYACs) is greatly increased in acute myeloid leukemia (AML) cells. c In multiple myeloma (MM) cells, the 3’ IgH super-enhancer (SE) inserts near the MYC locus, driving the upregulation of MYC expression. d Mutant FERM domain of NF2 form phase-separated condensates with IRF3 and abrogates the antitumor immunity initiated by STING. e In lung cancer, the liquid-like YAP/TEAD/SRC-1 compartments in nucleus can broadly upregulate YAP transcription. f Glycogen accumulation and phase separation lead to the formation of Laforin-Mst1/2 complex, thus activate oncogenic YAP signaling
Furthermore, intracellular phase-separated liquid-like structures regulate the localization and processing of mRNA.143,144 For example, AKAP95, a nuclear protein that participates in RNA splicing, generates liquid-like phase-separated condensates in vitro and in cells.145 AKAP95 is frequently overexpressed in human breast cancer, and the liquid condensates possess the abilities to support tumorigenesis with proper liquidity and dynamicity,145 which indicates that modifying the properties of biomolecular condensates can potentially target LLPS and provide useful ideas for cancer therapy. The mechanisms of RNA distribution and processing in cells are important for subsequent protein localization and function, which implicates the multistep nature of gene expression regulation.146
LLPS and intracellular signaling
Various signaling transduction pathways have been implicated in the regulation of cell proliferation, differentiation, metabolism, angiogenesis, apoptosis and senescence.147,148 Emerging evidence suggests the importance of phase separation in orchestrating signaling pathways through the compartmentalization of significant factors.149 For example, when TCR phosphorylation is triggered, the downstream signaling proteins also form liquid-like clusters spontaneously via LLPS, which results in the promotion of signal outputs.15
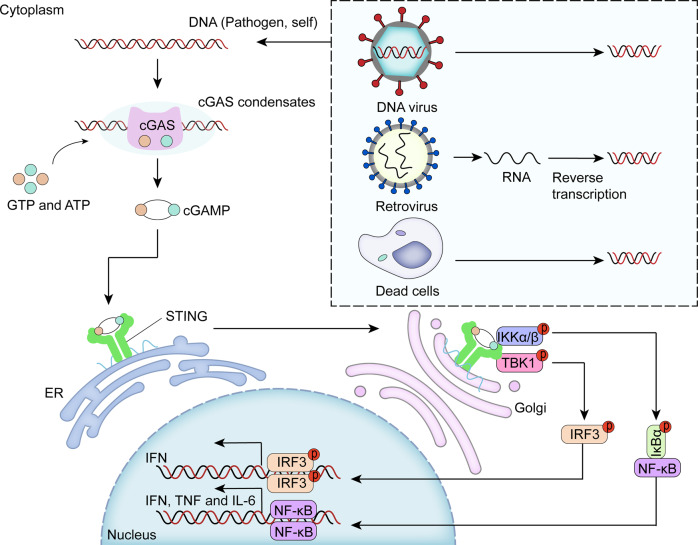
Furthermore, the cellular enzyme cyclic GMP-AMP synthase (cGAS) functions as a direct DNA sensor and produces the secondary messenger cyclic GMP-AMP (cGAMP), which has an innate immune function (Fig. 4).150 Interactions with DNA induce phase separation of cGAS and promote the production of cGAMP, which interacts with the receptor stimulator of interferon genes (STING), activates downstream type I interferon and NF-κB signaling151 and facilitates innate immunity.150 Therefore, normal and properly regulated LLPS is essential for the human body to maintain normal immune signaling pathways, and if the LLPS process is aberrantly changed, it is likely to cause pathological consequences, such as cancer.
Fig. 4.
Phase separation of enzyme cyclic GMP-AMP synthase (cGAS) contributes to innate immunity. The cellular enzyme cGAS directly induces phase separation via binding with DNA and produces cGAMP. cGAMP interacts with STING and activates downstream innate immunity
LLPS and autophagy
Phase separation is also involved in autophagy.20 Autophagosome formation involves the process of LLPS at specific sites, and LLPS can positively regulate autophagy activity through different mechanisms.152 Specifically, phase separation plays a role in modulating TORC1 activity.153 TORC1 is a Ser/Thr kinase complex that regulates multiple cellular processes and cellular metabolism in response to nutrient availability in the milieu.154 The inhibition of TORC1 signaling can induce autophagy, and dysregulated TORC1 and autophagy have been demonstrated to be correlated with tumorigenesis.155 Mechanistically, LLPS of yeast Pbp1 in combination with a cellular redox state can lead to TORC1 downregulation and consequently promote autophagy.155 In addition, LLPS plays a role in forming aminopeptidase Ape1 condensates, which are trafficked by double-membrane-bound Cvt vesicles and thereby enable selective engulfment.156
Furthermore, biomolecular condensates can control protein quality by not undergoing autophagic degradation.157 For example, stress-sensitive proteins in the nucleus misfold under pressure and then aggregate into the nucleolus after being in a nucleoplasmic dispersion state. After combining with nucleolar proteins through LLPS, these misfolded proteins are protected from irreversible aggregation,158 which facilitates refolding during recovery from stress.
In summary, LLPS is important to a variety of cellular processes, as explained herein. Therefore, we will discuss the possible oncogenic effects of various aberrant LLPS and the states of LLPS processes in tumor cells of different cancer patients.
Aberrant LLPS in cancer
As described previously, biomolecular condensates are involved in the control and regulation of various cellular biological processes, and the constituent macromolecules of biomolecular condensates are affected by genetic abnormalities in various cancers, which suggests that condensates are significant for unraveling the carcinogenesis process and prompting new advancements in cancer therapy. Malignant cells acquire genomic mutations that influence various biological processes mediated by LLPS during tumorigenesis, including chromatin changes, transcription, DNA damage repair, and tumor suppression. These mutations may result in aberrant cellular activity, such as driving unlimited proliferation and replicative immortality, angiogenesis, cancer cell evasion from growth suppressors, resistance to death, invasion and metastasis.3,132 Furthermore, recent studies have demonstrated that aberrant LLPS can influence epigenetic regulation, which is also associated with the onset and progression of cancers. Therefore, we suspect that LLPS may provide a useful framework for understanding the emergence and progression of diverse cancers and for ultimately finding appropriate treatments.
The deregulation of LLPS in chromatin organization contributes to tumorigenesis
A high frequency of mutations in genes encoding the elements modulating chromatin architectures has been found in numerous cancers.159–161 Genetic alterations of the components of phase-separated droplets regulating chromatin organization have also been found in recent studies.162 For instance, the chromatin remodeling complex BRG1/BRM-associated factor (BAF) is recruited by the EWS-FLI1 chimeric protein to activate oncogenic gene expression in Ewing sarcoma.163 The BAF complex mediates chromatin remodeling in an ATP-dependent manner, and cells with mutated BAF subunits show damaged chromatin structures and are unable to express multiple genes.164 The IDRs in EWSR1 effectively mediate strong interactions with FLI1 and form liquid-like compartments, which is significant for the tumorigenicity of Ewing sarcoma,163,165 and the genes encoding BAF complex subunits are also frequently mutated in different cancers.166 To develop therapeutic agents for Ewing sarcoma, Martin et al. extensively summarized various agents and factors affecting EWS-FLI1 activity, such as hypoxia, miRNAs, and antibodies against the IGF-1/IGF-1R pathway.165
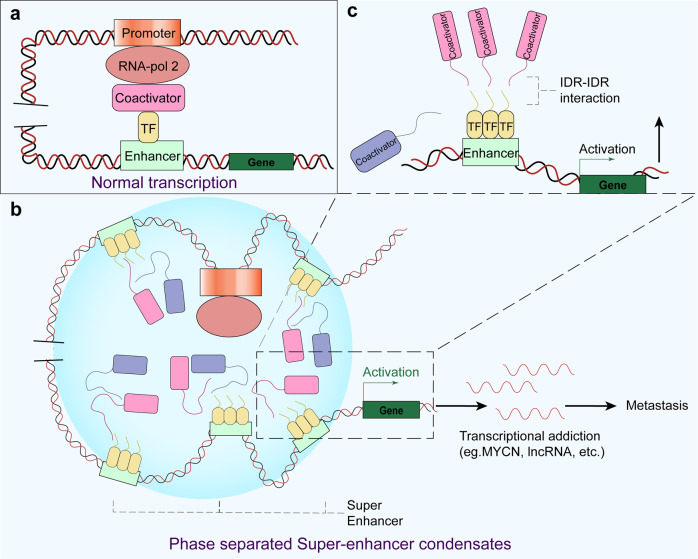
SEs drive oncogenic transcription
Abnormal gene transcription driven by mutations in the genetic and epigenetic landscape contributes to the initiation of uncontrollable growth and proliferation of cancer cells.132 Different from normal transcriptional pathway (Fig. 5a), the activation of prominent oncogenes and other genes related to tumor pathogenesis is possibly induced through SEs, which comprise several hundred clusters of enhancer elements, TFs and enhancer-related modifications that regulate gene transcription important to different cell types (Fig. 5b, c).137,167 These genes are particularly sensitive to the disruption of oncogenic signaling pathways.168 Emerging evidence suggests that TFs containing IDRs, transcription coactivators and RNA pol II form phase-separated condensates at SEs169 and that the protein and nucleic acid components of SEs are subject to phosphorylation; SEs bind proteins based on their phosphorylation status.137
Fig. 5.
Components and processes in normal transcription and SE-mediated transcriptional addiction of genes in cancer. a The normal transcription of a gene involves proper interactions among the promoter, RNA pol 2, coactivators, TFs and enhancers. b The SE condensates form a liquid-like complex via LLPS and may result in the transcriptional addiction of certain genes. c Detailed illustration of the IDR-IDR interaction that promotes SE formation
Researchers have discovered that Epstein–Barr virus (EBV) nuclear antigen 2 (EBNA2) and its coactivator EBNALP undergo LLPS mediated by their IDRs at active SEs by interacting with numerous TFs, which promotes their own transcription.170 EBV is one of the most significant human tumor viruses and is correlated with various cancers, including nasopharyngeal carcinoma, gastric cancers, and Hodgkin lymphoma.171–175 Malignant cells may acquire SEs via diverse mechanisms, including genomic rearrangements, focal amplifications of SEs commonly associated with other genes, and highly asymmetric loading of oncogenic TFs.176–178 SE acquisition through these mechanisms is related to a large number of oncogenes that are being currently studied.176,179,180 Notably, the protein products of these genes are important for controlling the identity, growth and proliferation of cells.167
Recently reported lines of evidence indicate that the IDRs of signaling factors in the Wnt/β-catenin pathway are components of condensates that form at SEs in a Wnt-inducible manner.181 Moreover, as the pivotal component of the Wnt pathway, β-catenin contributes to the hyperactivation of Wnt signaling via abnormal phase separation induced by IDR-IDR interactions at SEs;182,183 this hyperactivation of Wnt signaling is one of the early events in the carcinogenesis of diverse cancers. For instance, almost all colorectal cancers (CRCs) exhibit a hyperactivated Wnt pathway,184,185 and the suppression of Wnt signaling can significantly restrict CRC initiation and extend survival.186 However, in the absence of Wnt ligands, the multiprotein destruction complex suppresses Wnt signaling and requires the activation of downregulated pathways.187 The destruction complex comprises the tumor suppressor APC, the scaffold AXIN, and two kinases—GSK3 and CK1. Recent studies have demonstrated that AXIN and APC undergo LLPS, which is critical for their function in inhibiting the Wnt/β-catenin pathway.187,188 Hyperactivation of the Wnt pathway caused by APC mutations is prevalent in a large portion of human CRCs, and these mutants play a pivotal role in the initiation of tumorigenesis by influencing cell differentiation and facilitating rapid proliferation.189,190 The above-described examples strongly demonstrate that the regulation of Wnt signaling pathway hyperactivation can be performed in two directions: inhibiting the phase separation induced by Wnt signaling or promoting the LLPS of the destruction complex.
The TF MYC, which has potent cell growth- and proliferation-promoting metabolic activities, is unleashed by genetic and epigenetic dysregulation in cancer.191 Mutations or translocations of overactivated enhancers adjacent to the MYC gene are correlated with the pathogenesis of malignancy.192 In numerous cancer cells, excessively large SEs have been observed in the gene desert near the MYC gene but are rarely found in their healthy counterparts.167 For instance, in multiple myeloma (MM), malignant cells often contain a translocation in which the 3′ IgH SE is inserted near the MYC locus, which drives upregulation of MYC expression167,193 (Fig. 3c). In another example, the SE-enriched transcriptional coactivators BRD4 and MED1 are phase-separated into condensates at SEs, which leads to an effectively compartmentalized and assembled transcriptional apparatus.134 Indeed, some evidence suggests that BRD4 might mediate transcriptional addiction to the MYC oncogene.132 The intervention of MM tumor cell proliferation with the BET-bromodomain inhibitor JQ1 causes a specific reduction in BRD4 at SEs, which results in disruption of MYC transcription elongation.193,194 Moreover, BRD4 knockdown can induce cell apoptosis and inhibit the growth of MYCN (a member of the MYC family)-amplified neuroblastomas.195 Zhang et al. showed that the focal amplification of SEs in the 3’ direction relative to MYC in lung adenocarcinoma and endometrial cancer are physically connected to the MYC promoter and are correlated with MYC gene overexpression.178 SEs not only activate the transcription of protein-coding genes but also regulate the transcription and maturation of noncoding genes, such as miRNAs196 and lncRNAs.197 Studies have also observed reduced activity of SEs in certain tumor cells. That is, SEs activated in the process of cell carcinogenesis are often related to cancer-promoting miRNAs, whereas inactivated SEs mainly regulate the production of cancer-suppressing miRNAs.196
In summary, we realize that SEs can be novel biomarkers useful for the discovery of cancer-specific pathology, and these findings contribute to a deeper understanding of cancer biology, diagnosis, and therapy.167
The deregulated LLPS of tumor suppressors contributes to tumorigenesis
One of the hallmark capabilities of cancer is the evasion of tumor suppressors to limit cell growth and proliferation.3 Tumorigenesis is actively inhibited by a spectrum of tumor suppressors, including p53, TGF-β, RB1 and PTEN, which regulate tumor immunology and immune integrity.198
p53
Among these suppressors, p53 appears to be extremely crucial because it responds to multiple stress signals by coordinating distinct cellular activities, such as permanent and impermanent cell cycle arrest, apoptosis and cell senescence, which are all correlated with tumor suppression.199–203 p53 is a cellular stress sensor that suppresses tumorigenesis via transcriptional activation,204 and 53BP1 helps to stabilize and enhance p53 gene expression.199
However, p53 is mutated in more than 50% of all human cancers205 and is functionally inactivated by mutational, viral, or cellular patterns in most types of cancer.206 A large proportion of mutant p53 species are highly overexpressed in cancer cells, and some p53 mutations exert negative dominance effects through coaggregation, hetero-oligomerization, and prion-like aggregation with mutant or normal p53 protein. Tumor-associated stress is identified as a strong inducer of p53 aggregation in cell lines.207–212 Patients harboring these mutants may have poor clinical outcomes,212 which makes these proteins promising therapeutic targets. Increasing studies have attempted to develop methods to inhibit the activity of mutant p53s or to re-establish some wild-type functions that are very promising for cancer therapy.213
SPOP
The tumor suppressor gene SPOP has gained much attention recently because of its mutation in various cancers.214 SPOP forms phase-separated, membraneless clusters in nuclear speckles, and these droplets play central roles in suppressing the tumorigenesis of multiple human malignancies, such as gastric, liver, and prostate cancers.46,128 The SPOP droplets function as CRL3 substrate adapters that attract oncogenic substrates for ubiquitination and subsequent proteasomal degradation by a ligase.128 Both SPOP self-association and SPOP interactions with substrates can enhance LLPS.215–217 When the SPOP substrate protein DAXX is coexpressed with SPOP, another type of liquid-like droplet—SPOP/DAXX bodies—can be formed via LLPS, leading to the ubiquitination of DAXX and thus reducing the DAXX level.218 DAXX maintains the survival of cancer cells by downregulating the transcription of various tumor suppressors, including the TF p5397 and SMAD4.93 DAXX degradation by SPOP effectively induces cancer cell apoptosis and degrades potential therapeutic targets.219–221 Moreover, mutations of SPOP are frequently observed in solid tumors, such as breast,222 endometrial,223 gastric224 and prostate cancers,224,225 and are associated with early events in tumorigenesis.218,226 The consequences of oncogenic mutations in SPOP activities include DAXX recruitment and LLPS disruption, which largely prevents the formation of SPOP/DAXX bodies and thus results in the accumulation of a large number of DAXX proteins.
TGF-β
Another tumor suppressor protein, TGF-β, induces growth arrest of cancer cells and repression of the c-MYC proto-oncogene at the early phase.227 However, in later stages of malignancy, TGF-β initiates cell invasion and modulates the microenvironment to benefit cancer cell growth.228 The interaction of the TGF-β/SMAD and Wnt pathways plays a crucial role in cellular biology, and the opposite function of TGF-β and Wnt is significant for the joint regulation of bone genesis and resorption.229 During bone metastasis, the TGF-β pathway maintains disseminated tumor cell dormancy in bone or osteolytic outgrowth.229,230 Recently, Esposito et al. demonstrated that TGF-β induces phase separation of the bone metastasis-promoting protein DACT1, which represses the Wnt signaling pathway by sequestering the Wnt pathway activator casein kinase 2. Eliminating the IDRs in the DACT1 protein effectively abolishes its capability to generate liquid-like condensates and the effect of suppression on Wnt signaling.229 These findings elucidate the mechanism of cancer bone metastasis and encourage future research toward the inhibition of DACT1 condensate formation.
Aberrant LLPS interferes with antitumor signaling pathways
As mentioned before, appropriately regulated LLPS can function as a significant hub to promote signal outputs to modulate cellular activities. However, aberrant LLPS is triggered via oncogenic mutations and consequently disturbs the signaling pathways.
Merlin (NF2/schwannomin), another tumor suppressor protein, integrates and regulates intracellular signaling pathways (including the Hippo signaling pathway) and the extracellular matrix and promotes innate immunity against cancer.25,231 However, genetic inactivation and mutations of NF2 can be found in a wide range of malignancies, including CRC, schwannomas, type 2 neurofibromatosis, and skin tumors.232 Recently, Meng et al. demonstrated that a mutant FERM domain of NF2 potently suppresses the cGAS-STING signaling pathway by forming phase-separated condensates with IRF3 and impedes the antitumor immunity initiated by STING (Fig. 3d).25 Clinically, NF2-IRF3 condensates can be observed in surgically resected samples from bilateral vestibular schwannomas.233 Therefore, at least in NF2-related malignancies, we can attempt to inhibit the formation of intracellular membraneless structures composed of NF2 to restore the antitumor immunity of the Hippo pathway and cGAS-STING pathway.
In addition, we used to believe that LLPS can only occur in proteins and nucleic acids, but intriguingly, recent studies have shown that other molecules can also generate biomolecular condensates through LLPS and exhibit their own cancer-causing effects. Liu et al. found that the accumulation of glycogen is frequently detected in tumor cells to support increased glucose consumption for tumor growth. The accumulated glycogen can undergo LLPS, leading to the formation of the Laforin-Mst1/2 complex in liver tumors, as shown in Fig. 3f. The Mst1/2 (two Hpo homologs) kinases are significant components of the Hippo pathway in regulating immune systems and cell proliferation via a tumor suppression mechanism.234 However, the membraneless Laforin-Mst1/2 complex robustly sequesters the Hippo kinases Mst1/2 and abolishes their repression of oncogenic YAP signaling. Thus, the elimination of glycogen storage can potentially abrogate liver growth and cancer initiation.235
Furthermore, a nonreceptor protein tyrosine phosphatase encoded by PTPN11, SHP2, plays an essential role in MAPK signal transduction and organism development.236 Disease-associated mutant SHP2 can undergo LLPS and recruit condensates, leading to RAS-MAPK signaling hyperactivation and dysregulation, which is crucial in tumorigenesis events.237,238 Researchers have hypothesized that the robust hyperactivation of mutant SHP2 compared with wild-type SHP2 is due to conformational transition. SHP2 allosteric inhibitors effectively inhibit the phase separation of SHP2 mutants and abrogate the capabilities gained by mutated SHP2 droplets, which strongly supports this opinion.237
LLPS of RNA and RBPs in tumorigenesis
In RNA-containing condensates, RBPs often recognize their RNA targets in a specific manner,239 and LLPS of RNA with RBPs is facilitated by the distinct properties of the RNA,240 such as its identity, length, structure, modifications and expression level. Paraspeckle, a cancer-related biomolecular condensate discovered within the past 20 years,241 is a nuclear body constructed via a specific lncRNA—NEAT1_2 RNA—that docks at transcription starting sites of active genes.72 A capture hybridization analysis of RNA target experiments demonstrated that NEAT1 RNA is crosslinked to the transcription start sites of active genes in a breast cancer cell line, but the evidence is insufficient to prove that LLPS drives NEAT_2 RNA activity because the probes do not target NEAT1_2 specifically.242 In addition, the NEAT1 gene, including the region that encodes NEAT1_2, has been found to be mutated in biopsied samples collected from liver cancer patients,243 and hypoxia can trigger the upregulated transcription of NEAT1_2 via hypoxia-inducible factors, which results in a critical upsurge of paraspeckles in breast cancer cell lines.244 However, under certain conditions, paraspeckles appear to play tumor-suppressive roles.245
Moreover, a study conducted in 2021 showed that the phosphatidic acid-binding lncRNA small nucleolar RNA host gene 9 (SNHG9) facilitates LLPS of LATS1—one of the key kinases of the Hippo pathway—to promote oncogenic YAP signaling.246 Clinically, SNHG9 expression was positively correlated with YAP and Ki-67 expression and breast cancer progression.246 The role played by RNA in tumorigenesis has been extensively explored by researchers, and we believe that LLPS is an important complement in the causal relationship between multiple RNAs and tumorigenesis.
Epigenetic dysregulation mediated by LLPS in cancer
Many studies in the field of cancer have concentrated on determinants of genetic alterations that inhibit or facilitate the acquisition of cancer phenotypes. However, a growing body of evidence suggests that destroying certain epigenetic processes can also exert a major impact on cancer development.247,248 Epigenetic modifications refer to changes through which cells exhibit a distinct profile of gene expression in identical DNA sequences without irreversibly changing the genetic information.249 Among epigenetic modifications involved in regulatory functions, histone modifications, DNA or RNA methylation, chromosome condensation,116,250 and lncRNA molecule expression251 have been reported to play significant roles in the genesis, progression and metastasis of cancers.252
Recent evidence also suggests that EBNA2 undergoes LLPS that reorganizes the host chromatin topology. The N-terminus of EBNA2 mediates the reorganization of chromatin topology via LLPS to induce accessible chromatin domains (ACDs). The CTD of EBNA2 is significant in the epigenetic regulation of host gene expression by recruiting histone acetylase p300 to ACDs, which then mediates the acetylation of certain regions in histone H3K27.253,254 Moreover, a study performed in 2019 revealed that histone modifications are essential for maintaining cancer stem cells in human breast cancer. In this study, researchers formed a stem-like breast cancer cell model and observed that the histone deacetylase HDAC7 binds near the transcriptional start site and to SEs of various oncogenes (Fig. 3a) and results in the activation of SE-associated oncogenes.255
The most common internal modification of mRNAs, m6A, is significantly correlated with gene expression in various cancers.256 Recent studies have shown that mRNA modification by multiple m6A modifications can function as a multivalent scaffold, promote distinct binding with cytosolic YTHDF proteins and facilitate the LLPS process, which results in the formation of various RNP granules, including P bodies and SGs.68,257 P bodies are responsible for RNA decay and storage, and different components of P bodies play different roles in tumorigenesis.258 Emerging evidence also demonstrates that abnormalities in the expression and/or activity of SG components contribute to drug resistance and the tumorigenesis of diverse cancers, including CRC,259 pancreatic cancer,260 and leukemia.261 Furthermore, YTHDC1 in the nucleus can undergo LLPS by binding with m6A-mRNA (Fig. 3b), and the number of resulting nuclear condensates (nYACs) is greatly increased in acute myeloid leukemia cells.262 nYACs protect leukemia-promoting mRNAs from degradation263 and maintain cells in an undifferentiated state, which is significant for cell survival and leukemia maintenance.262
Therefore, deregulated epigenetic changes may result in tumor cell adaptation and resistance to anticancer therapy, which remain profound challenges to therapeutic intervention,264 and targeting the mechanisms of LLPS is a new direction that may lead to elegant strategies for overcoming the complicated situations in cancer therapy.
LLPS as a strategy for the alternative lengthening of telomeres
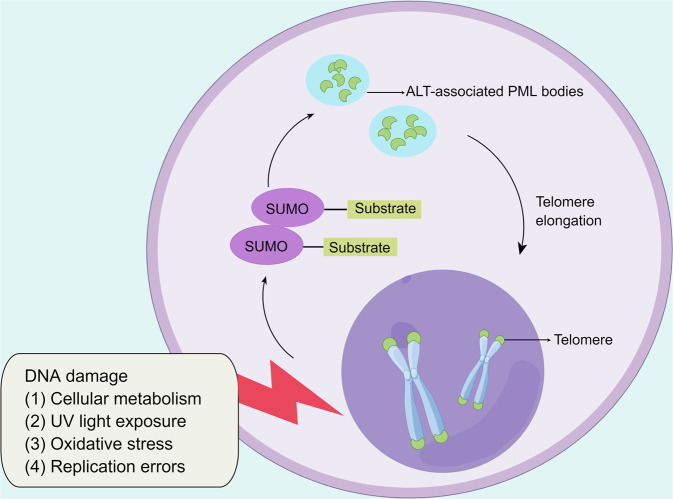
Telomeres are nucleoprotein structures formed by a repetitive nucleotide sequence (TTAGGG)n that constitutes a “cap structure” at the end of a chromosome, which results in the preservation of genome stability.265–267 Telomeres are parts of constitutive heterochromatic regions and are enriched with di- and trimethylated heterochromatin histones,268 namely, H3K9me3 and H4K20me3, and characterized by HP1 binding.269,270 During eukaryotic cell replication, telomeres continuously shorten, which ultimately leads to cell senescence and apoptosis. In tumorigenesis, tumor cells almost universally acquire telomere DNA maintenance mechanisms (TMMs) that prevent activation of the DDR, which results in counteracting telomere shortening. Two types of TMMs have been recently well characterized: telomerase and alternative lengthening of telomeres (ALTs). In fact, most human tumors exhibit telomerase-based TMM. However, after the development of drugs targeting telomerase, some cancer cells can still escape death, highlighting the less frequently engaged ALT pathway.267,271,272 The hallmark of an ALT-related cancer is excessive telomere clustering in PML bodies, which are known as ALT-associated PML bodies (APBs) and are observed as large bright telomere foci.273 In response to DNA damage, liquid-like APB formation can be triggered via poly (SUMO)-poly SUMO interaction motif-mediated LLPS (Fig. 6).274 The mechanism of telomere lengthening has been shown to be correlated with mitotic DNA synthesis in APB-like foci,275,276 and a large quantity of aggregated telomeres at these foci can facilitate mitotic DNA synthesis-mediated ALT.277 Some evidence supports the notion that the telomere length is positively associated with the risk of malignancy, such as melanoma, B-cell lymphoma and chronic lymphocytic leukemia.278–280
Fig. 6.
LLPS is a strategy for the alternative lengthening of telomeres. (By Figdraw.). In ALT-related cancer, excessive telomere clustering in PML bodies (ALT-associated PML bodies) can be triggered via poly (SUMO)-poly SUMO interaction motif-mediated LLPS
Prospects of applying LLPS in cancer treatment
Because LLPS can affect tumorigenesis through different pathways, practical strategies for treating these cancer-associated proteins and their upstream/downstream signaling remain to be developed. For the formation mechanisms of biomolecular condensates, we can sustain normal LLPS by controlling the concentration of related proteins/nucleic acids, specifically disrupting the phase separation process, partitioning of cancer drugs in biomolecular condensates, and modifying LLPS by interfering with PTMs, among other strategies.
Normalizing the protein concentration
Membrane-less condensates assembled from phase-separated proteins are collections of molecules at high concentrations in specific locations, which means that changes in the concentration of relevant molecules can have an impact on the formation and size of LLPS-formed droplets and thus control their function.134,281 Therefore, normalizing the protein contents by inducing up- or downregulation of their expression might be a pivotal and valid method for regulating phase separation.
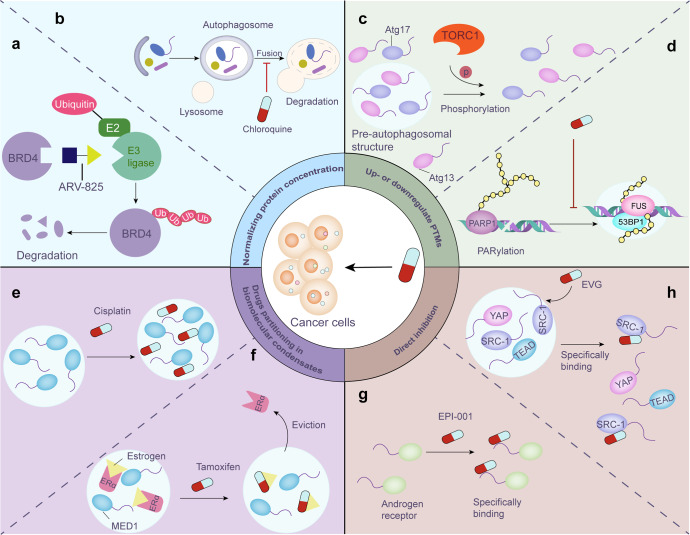
To downregulate the expression of target proteins, an emerging technology referred to as proteolytic targeting chimera (PROTAC) technology can link target proteins to E3 ligases and thus induce their precise degradation in the cell.282 PROTACs exploit the cell’s own protein destruction mechanism to clear specific target proteins from cells. Lu et al. designed a heterobifunctional PROTAC, ARV-825, which can lead to efficient and prolonged degradation of BRD4 in BL cell lines by recruiting BRD4 to the E3 ubiquitin ligase cereblon (Fig. 7a)283 and thus downregulating the expression of MYC.284 In addition, autophagy‐targeting chimera technology is an advanced approach that selectively eliminates target proteins via an autophagy‐dependent pathway.282,285
Fig. 7.
Future prospects of cancer treatment via regulating LLPS. a ARV-825, a novel PROTAC, efficiently degrades BRD4 protein in BL cell lines by linking BRD4 to E3 ubiquitin ligase. b Chloroquine primarily blocks autophagy by impairing autophagosome-lysosomal fusion, thereby upregulate target protein level. c Phosphorylation mediated by activated TORC1 significantly inhibits LLPS and impairs autophagosome prestructure (PAS) formation. d Inhibition of PARylation prevents the formation of DNA damage repair foci. e Cisplatin preferentially concentrates in biomolecule condensates, which can help improve drug efficacy. f Tamoxifen efficiently expels ERα from MED1 condensates via specifically partitioning into these condensates. g The small-molecule compound EPI-001 selectively binds to the IDR of the androgen receptor, thereby slowing the progression of castration-resistant prostate cancer. h EVG can directly target the SRC-1/YAP/TEAD droplets to restrict cancer cell growth in a YAP-dependent manner
In contrast, to upregulate certain protein contents, preventing their degradation may be a viable option, and in this context, proteasome inhibitors and lysosomal inhibitors have been tested in many trials.286,287 For example, chloroquine, an FDA-approved drug that blocks autophagy primarily by impairing autophagosome-lysosomal fusion, has been shown in clinical trials to enhance the potential of combination anticancer therapies by sensitizing tumor cells (Fig. 7b).287
However, considering the normal physiological function of the target proteins in different signaling pathways, the specific up- or downregulation of the expression of distinct proteins might be harmful to normal biological activities. Therefore, we need to be careful about the possible side effects when regulating protein expression levels. Detailed clinical trials should be conducted to anticipate side effects and prevent them as much as possible before applying these treatments.
Induction/inhibition of posttranslational modifications to influence LLPS
PTMs are vital modulators of condensation and affect the properties of membraneless compartments; for example, phosphorylation and methylation of the C-terminal LCD in the fragile X protein, which causes mental retardation, induces contradictory influences on in vitro translation regulation: phosphorylation promotes phase separation and leads to translation inhibition,288 and methylation decreases the propensity for LLPS.289 Under high nutrient availability, LLPS mediated by the interaction between the autophagy‐related proteins Atg13 and Atg17 is inhibited by phosphorylation via activated TORC1 and then impairs pre‐autophagosomal structure formation (Fig. 7c).290 PARylation is another reversible PTM process that is mediated by the catalysis of PAR polymerase (PARP).291 The inhibition of PARP1 prevents DNA damage repair foci formation and jeopardizes the process of DDR (Fig. 7d).130
Drug concentrations in biomolecular condensates
Interestingly, researchers have found that some antineoplastic drugs can be selectively concentrated in specific condensates via physicochemical interactions, which may exert curative effects or induce drug resistance in cancer.292,293 For example, cisplatin is a widely used antineoplastic agent that partitions selectively with a partition coefficient of 600 in transcriptional condensates where SE DNA can be platinated (Fig. 7e).294 This finding reveals the potential of increasing drug target engagement by partitioning the therapeutic components into condensates.
Tamoxifen is another antineoplastic drug that is important for the treatment of estrogen receptor (ER)-positive breast cancer. ERα selectively concentrates into MED1 condensates in an estrogen-dependent manner and is evicted from the condensate by tamoxifen, which also preferentially partitions into transcriptional condensates and competes for binding to estrogen (Fig. 7f). The overexpression of MED1 results in the volume expansion of transcriptional condensates, which results in dilution of the concentrations of tamoxifen in the condensates and counteraction of the efficacy on ERα eviction from the condensate.294
Both of these examples provide strong evidence indicating that specific compartmentalization and the concentration of small-molecule cancer therapeutics in condensates can impact drug pharmacodynamics and interventions on phase‐separated complexes and may therefore be efficient for targeting undruggable molecules.
Drugs directly interacting with biomolecular condensates
Similar to previous expectations, some evidence suggests that certain drugs interact directly with condensates; that is, the application of certain drugs can specifically prevent the formation of condensates that contribute to disease pathology.294 Notably, IDRs were preliminarily thought to be undruggable due to their conformational heterogeneity and dynamicity.295 However, various recent studies have challenged the identification of different strategies to bind IDRs. For instance, EPI‐001 is a small‐molecule compound that attenuates the progression of castration-resistant prostate cancer by selectively binding to IDRs of the androgen receptor (Fig. 7g).296 Moreover, researchers have also analogously demonstrated that the anticancer adjuvant melatonin is capable of inhibiting the intrinsically disordered N-terminal region of prion-mediated phase separation in cancer, which results in the amelioration of multidrug resistance.297
Elvitegravir (EVG) was originally developed to treat HIV infection and can potently suppress cancer metastasis by directly targeting the m6A methyltransferase METTL3.298 EVG can effectively target the SRC-1/YAP/TEAD droplets to restrict cancer cell growth in a YAP-dependent manner by specifically disrupting LLPS of SRC-1 (Fig. 7h).141 Recent research has also shown that allosteric regulators may influence LLPS because allosteric sites in intrinsically disordered proteins can be managed to enhance signaling interactions between various mechanistic components.299
In addition, interfering with RNA to influence LLPS can be a potential regulatory method. For instance, the RNA helicase DDX3X plays a role in mediating the maturation and disassembly of SGs, and Dhh1 accelerates the aggregation of PBs by linking to Pat1.300,301 Furthermore, the molecules that bind to proteins in LLPS, including molecular chaperones and ligands, are important regulatory components.183,302
Concluding remarks and future perspectives
Recent studies have revealed the significance of coordinated actions of biomolecular condensates in orchestrating diverse cellular processes. The driving forces of LLPS are the multivalent interactions between macromolecules accomplished via multiple modular domains, IDRs, and nucleic acid chains. Cutting-edge research on phase separation has successfully reconstituted biomolecular condensates in vitro that simulate the biological features and functions of biomolecular liquid-like droplets in vivo.37 Although the IDRs lack a stable 3-dimensional structure, which makes the drug discovery process more challenging, a variety of websites can be used to predict the IDRs in diverse proteins and provide increasingly comprehensive messages of intrinsically disordered proteins.303–305 Small-molecule inhibitors directly targeting IDRs have been discovered recently.306 For instance, EPI-002 is the first drug tested in a clinical trial that directly binds to the IDR of androgen and shows signs of efficacy in castration-resistant prostate cancer patients.307 The analog of EPI-002, EPI-7386, exhibits improved pharmacokinetics and metabolic stability but has never been tested clinically.307
When normal LLPS is disrupted by genetic or epigenetic mutations, aberrant biomolecular condensates may be involved in tumorigenesis because of their role in dysregulated chromosome organization, signal transduction, and transcriptional dysregulation and consequently facilitate the development of cancer. Ming et al. demonstrated that cancer cells might be more sensitive and more addictive to LLPS, which suggests the therapeutic potential of LLPS in cancer.308 The application of 1,6-hexanediol to pancreatic cancer cells can significantly abrogate the LLPS process and thereby downregulate the expression of the MYC oncogene.308
Because LLPS is involved in many cancer mechanisms, various studies have regulated the phase separation process by determining the concentration of related proteins/nucleic acids, directly targeting components that undergo phase separation, or modifying LLPS by interfering with PTMs. In addition, the specific partitioning of antineoplastic drugs in subcellular condensates is also important for drug efficacy because the concentrations of active ingredients can be extremely high in these condensates. According to this characteristic action, we can detect the distribution of anticancer drugs in cells or by linking anticancer drugs to molecules that can specifically aggregate in liquid droplets such that they can directly act on the carcinogenic targets in subcellular condensates.
However, despite the promise of a therapeutic target through LLPS interference, it must be remembered that IDRs are widely distributed in the human body. More than 30% of the regions in the proteome are disordered,309 which suggests that we still face enormous challenges in determining the specificity of drug action. We encourage the development of more cell models and animal models to explore the development of anticancer drugs that modulate LLPS, and we hope that more clinical evidence will confirm our conjecture. We believe that joint explorations conducted by cancer researchers, cell biologists, and biophysicists can uncover the mystery of cancer phase separation and can identify more tumor treatment strategies related to phase separation.
Acknowledgements
This study was jointly supported by the National Natural Science Foundation of China (U21A20374 and 82072698), Shanghai Municipal Science and Technology Major Project (21JC1401500), Scientific Innovation Project of Shanghai Education Committee (2019-01-07-00-07-E00057), Clinical Research Plan of Shanghai Hospital Development Center (SHDC2020CR1006A), and Xuhui District Artificial Intelligence Medical Hospital Cooperation Project (2021-011).
Author contributions
X.T., R.T., W.W., J.X., Y.Z., X.Y., and S.S. contributed to the writing and revising the paper. X.Y. and S.S. supervised the review. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xuhui Tong, Rong Tang
Contributor Information
Xianjun Yu, Email: yuxianjun@fudanpci.org.
Si Shi, Email: shisi@fudanpci.org.
References
- 1.Zhang H, et al. RNA controls PolyQ protein phase transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti S, Dormann D. Liquid-liquid phase separation in disease. Annu Rev. Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Gibson BA, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.e421. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang E, Aifantis I. RNA splicing and cancer. Trends Cancer. 2020;6:631–644. doi: 10.1016/j.trecan.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Calses PC, Crawford JJ, Lill JR, Dey A. Hippo pathway in cancer: aberrant regulation and therapeutic opportunities. Trends Cancer. 2019;5:297–307. doi: 10.1016/j.trecan.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Li L, et al. Notch signaling pathway networks in cancer metastasis: a new target for cancer therapy. Med. Oncol. 2017;34:180. doi: 10.1007/s12032-017-1039-6. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, et al. Emerging roles of liquid-liquid phase separation in cancer: from protein aggregation to immune-associated signaling. Front. Cell Dev. Biol. 2021;9:631486. doi: 10.3389/fcell.2021.631486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taniue K, Akimitsu N. Aberrant phase separation and cancer. FEBS J. 2022;289:17–39. doi: 10.1111/febs.15765. [DOI] [PubMed] [Google Scholar]
- 12.Jiang S, et al. Protein phase separation and its role in tumorigenesis. Elife. 2020;9:e60264. doi: 10.7554/eLife.60264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nozawa RS, et al. Nuclear microenvironment in cancer: control through liquid-liquid phase separation. Cancer Sci. 2020;111:3155–3163. doi: 10.1111/cas.14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su X, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pak CW, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dundr M, et al. In vivo kinetics of Cajal body components. J. Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidtkamp-Peters S, et al. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 19.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci. 2020;63:953–985. doi: 10.1007/s11427-020-1702-x. [DOI] [PubMed] [Google Scholar]
- 21.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 22.Wegmann S, et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37:e98049. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn JH, et al. Phase separation drives aberrant chromatin looping and cancer development. Nature. 2021;595:591–595. doi: 10.1038/s41586-021-03662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng F, et al. Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol. Cell. 2021;81:4147–4164 e4147. doi: 10.1016/j.molcel.2021.07.040. [DOI] [PubMed] [Google Scholar]
- 26.Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer. 2022;22:239–252. doi: 10.1038/s41568-022-00444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 28.Boeynaems S, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchan JR. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 32.Flory PJ, Krigbaum WR. Thermodynamics of high polymer solutions. Annu. Rev. Phys. Chem. 1951;2:383–402. doi: 10.1146/annurev.pc.02.100151.002123. [DOI] [Google Scholar]
- 33.Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cable J, et al. Phase separation in biology and disease-a symposium report. Ann. N. Y Acad. Sci. 2019;1452:3–11. doi: 10.1111/nyas.14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banani SF, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boija A, Klein IA, Young RA. Biomolecular condensates and cancer. Cancer Cell. 2021;39:174–192. doi: 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockmayer WH. Molecular distribution in condensation polymers. J. Polym. Sci. 1952;9:69–71. doi: 10.1002/pol.1952.120090106. [DOI] [Google Scholar]
- 40.Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014;3:e04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindstrom MS. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem. Res. Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colombo E, Alcalay M, Pelicci PG. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30:2595–2609. doi: 10.1038/onc.2010.646. [DOI] [PubMed] [Google Scholar]
- 44.Mitrea DM, et al. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl Acad. Sci. USA. 2014;111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitrea DM, et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marzahn MR, et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016;35:1254–1275. doi: 10.15252/embj.201593169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr. Opin. Struct. Biol. 2021;67:41–50. doi: 10.1016/j.sbi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017;292:19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699 e616. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang S, et al. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang H, et al. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elbaum-Garfinkle S, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altmeyer M, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat. Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt HB, Barreau A, Rohatgi R. Phase separation-deficient TDP43 remains functional in splicing. Nat. Commun. 2019;10:4890. doi: 10.1038/s41467-019-12740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes MP, Goldschmidt L, Eisenberg DS. Prevalence and species distribution of the low-complexity, amyloid-like, reversible, kinked segment structural motif in amyloid-like fibrils. J. Biol. Chem. 2021;297:101194. doi: 10.1016/j.jbc.2021.101194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes MP, et al. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, et al. Pressure and temperature phase diagram for liquid-liquid phase separation of the RNA-binding protein fused in sarcoma. J. Phys. Chem. B. 2021;125:6821–6829. doi: 10.1021/acs.jpcb.1c01451. [DOI] [PubMed] [Google Scholar]
- 61.Yu M, et al. Interferon-gamma induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell. 2021;81:1216–1230 e1219. doi: 10.1016/j.molcel.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Lu Y, et al. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020;22:453–464. doi: 10.1038/s41556-020-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zbinden A, Perez-Berlanga M, De Rossi P, Polymenidou M. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev. Cell. 2020;55:45–68. doi: 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 64.Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baudin A, et al. Structural characterization of the RNA-binding protein SERBP1 reveals intrinsic disorder and atypical RNA binding modes. Front Mol. Biosci. 2021;8:744707. doi: 10.3389/fmolb.2021.744707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ries RJ, et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lan Q, et al. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 71.Han TW, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 72.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 73.Pancsa R, Schad E, Tantos A, Tompa P. Emergent functions of proteins in non-stoichiometric supramolecular assemblies. Biochim. Biophys. Acta Proteins Proteom. 2019;1867:970–979. doi: 10.1016/j.bbapap.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 74.Bienz M. Head-to-tail polymerization in the assembly of biomolecular condensates. Cell. 2020;182:799–811. doi: 10.1016/j.cell.2020.07.037. [DOI] [PubMed] [Google Scholar]
- 75.Bienz M. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem. Sci. 2014;39:487–495. doi: 10.1016/j.tibs.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 76.Schwarz-Romond T, Merrifield C, Nichols BJ, Bienz M. The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J. Cell Sci. 2005;118:5269–5277. doi: 10.1242/jcs.02646. [DOI] [PubMed] [Google Scholar]
- 77.Gammons M, Bienz M. Multiprotein complexes governing Wnt signal transduction. Curr. Opin. Cell Biol. 2018;51:42–49. doi: 10.1016/j.ceb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 78.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 79.Chong PA, Vernon RM, Forman-Kay JD. RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 2018;430:4650–4665. doi: 10.1016/j.jmb.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 80.Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J. Struct. Biol. 2004;146:281–290. doi: 10.1016/j.jsb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Franzmann TM, Alberti S. Protein phase separation as a stress survival strategy. Cold Spring Harb. Perspect. Biol. 2019;11:a034058. doi: 10.1101/cshperspect.a034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamaguchi A, Kitajo K. The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS ONE. 2012;7:e49267. doi: 10.1371/journal.pone.0049267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gal J, et al. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol. Aging. 2011;32:2323 e2327–2323.e2340. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hofweber M, Dormann D. Friend or foe-post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019;294:7137–7150. doi: 10.1074/jbc.TM118.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanikawa C, et al. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 2018;22:1473–1483. doi: 10.1016/j.celrep.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 86.Monahan Z, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bah A, Forman-Kay JD. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 89.Scaramuzzino C, et al. Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS ONE. 2013;8:e61576. doi: 10.1371/journal.pone.0061576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wooderchak WL, et al. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry. 2008;47:9456–9466. doi: 10.1021/bi800984s. [DOI] [PubMed] [Google Scholar]
- 91.Hofweber M, et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–719.e713. doi: 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 92.Ryan VH, et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell. 2018;69:465–479.e467. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang CC, et al. Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J. Biol. Chem. 2005;280:10164–10173. doi: 10.1074/jbc.M409161200. [DOI] [PubMed] [Google Scholar]
- 94.Hendriks IA, et al. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017;24:325–336. doi: 10.1038/nsmb.3366. [DOI] [PubMed] [Google Scholar]
- 95.Ishov AM, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin DY, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 97.Mahmud I, Liao D. DAXX in cancer: phenomena, processes, mechanisms and regulation. Nucleic Acids Res. 2019;47:7734–7752. doi: 10.1093/nar/gkz634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fox BM, et al. SUMOylation in glioblastoma: a novel therapeutic target. Int J. Mol. Sci. 2019;20:1853. doi: 10.3390/ijms20081853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deng Q, et al. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 2014;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ding X, et al. Amyloid-forming segment induces aggregation of FUS-LC domain from phase separation modulated by site-specific phosphorylation. J. Mol. Biol. 2020;432:467–483. doi: 10.1016/j.jmb.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 102.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 103.Rice AM, Rosen MK. ATP controls the crowd. Science. 2017;356:701–702. doi: 10.1126/science.aan4223. [DOI] [PubMed] [Google Scholar]
- 104.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shakya A, Park S, Rana N, King JT. Liquid-liquid phase separation of histone proteins in cells: role in chromatin organization. Biophys. J. 2020;118:753–764. doi: 10.1016/j.bpj.2019.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 109.Gu J, et al. Hsp40 proteins phase separate to chaperone the assembly and maintenance of membraneless organelles. Proc. Natl Acad. Sci. USA. 2020;117:31123–31133. doi: 10.1073/pnas.2002437117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Z, et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 2020;27:363–372. doi: 10.1038/s41594-020-0399-3. [DOI] [PubMed] [Google Scholar]
- 111.Qamar S, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell. 2018;173:720–734.e715. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang JZ, et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell. 2020;182:1531–1544 e1515. doi: 10.1016/j.cell.2020.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 114.Shin Y, et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175:1481–1491.e1413. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Quinodoz SA, et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–757 e724. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Larson AG, et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singh PB, Belyakin SN, Laktionov PP. Biology and physics of heterochromatin-like domains/complexes. Cells. 2020;9:1881. doi: 10.3390/cells9081881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Narlikar GJ. Phase-separation in chromatin organization. J. Biosci. 2020;45:5. doi: 10.1007/s12038-019-9978-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rupa DS, Hasegawa L, Eastmond DA. Detection of chromosomal breakage in the 1cen-1q12 region of interphase human lymphocytes using multicolor fluorescence in situ hybridization with tandem DNA probes. Cancer Res. 1995;55:640–645. [PubMed] [Google Scholar]
- 122.Wong N, et al. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am. J. Pathol. 2001;159:465–471. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Norwood LE, et al. A requirement for dimerization of HP1Hsalpha in suppression of breast cancer invasion. J. Biol. Chem. 2006;281:18668–18676. doi: 10.1074/jbc.M512454200. [DOI] [PubMed] [Google Scholar]
- 124.Ding L, Kleer CG. Enhancer of Zeste 2 as a marker of preneoplastic progression in the breast. Cancer Res. 2006;66:9352–9355. doi: 10.1158/0008-5472.CAN-06-2384. [DOI] [PubMed] [Google Scholar]
- 125.Cuella-Martin R, et al. 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol. Cell. 2016;64:51–64. doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mirza-Aghazadeh-Attari M, et al. 53BP1: a key player of DNA damage response with critical functions in cancer. DNA Repair. 2019;73:110–119. doi: 10.1016/j.dnarep.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 127.Pessina F, et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019;21:1286–1299. doi: 10.1038/s41556-019-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cai D, Liu Z, Lippincott-Schwartz J. Biomolecular condensates and their links to cancer progression. Trends Biochem. Sci. 2021;46:535–549. doi: 10.1016/j.tibs.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 129.Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44:993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Singatulina AS, et al. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019;27:1809–1821.e1805. doi: 10.1016/j.celrep.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 131.Kilic S, et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu H, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318–323. doi: 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sabari BR, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harlen KM, Churchman LS. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017;18:263–273. doi: 10.1038/nrm.2017.10. [DOI] [PubMed] [Google Scholar]
- 136.Kwon I, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hnisz D, et al. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 139.Cai D, et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019;21:1578–1589. doi: 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shreberk-Shaked M, Oren M. New insights into YAP/TAZ nucleo-cytoplasmic shuttling: new cancer therapeutic opportunities? Mol. Oncol. 2019;13:1335–1341. doi: 10.1002/1878-0261.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu G, et al. Pharmacological inhibition of SRC-1 phase separation suppresses YAP oncogenic transcription activity. Cell Res. 2021;31:1028–1031. doi: 10.1038/s41422-021-00504-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Disco. 2020;19:480–494. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Langdon EM, Gladfelter AS. A new lens for RNA localization: liquid-liquid phase separation. Annu. Rev. Microbiol. 2018;72:255–271. doi: 10.1146/annurev-micro-090817-062814. [DOI] [PubMed] [Google Scholar]
- 144.Hu J, et al. AKAP95 regulates splicing through scaffolding RNAs and RNA processing factors. Nat. Commun. 2016;7:13347. doi: 10.1038/ncomms13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li W, et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol. 2020;22:960–972. doi: 10.1038/s41556-020-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lecuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 147.Adjei AA, Hidalgo M. Intracellular signal transduction pathway proteins as targets for cancer therapy. J. Clin. Oncol. 2005;23:5386–5403. doi: 10.1200/JCO.2005.23.648. [DOI] [PubMed] [Google Scholar]
- 148.Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020;9:2308. doi: 10.3390/cells9102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Su Q, Mehta S, Zhang J. Liquid-liquid phase separation: orchestrating cell signaling through time and space. Mol. Cell. 2021;81:4137–4146. doi: 10.1016/j.molcel.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ablasser A, et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen D, et al. Inositol polyphosphate multikinase inhibits liquid-liquid phase separation of TFEB to negatively regulate autophagy activity. Dev. Cell. 2020;55:588–602 e587. doi: 10.1016/j.devcel.2020.10.010. [DOI] [PubMed] [Google Scholar]
- 153.Noda NN, Wang Z, Zhang H. Liquid-liquid phase separation in autophagy. J. Cell Biol. 2020;219:e202004062. doi: 10.1083/jcb.202004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 155.Yang YS, et al. Yeast Ataxin-2 forms an intracellular condensate required for the inhibition of TORC1 signaling during respiratory growth. Cell. 2019;177:697–710 e617. doi: 10.1016/j.cell.2019.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yamasaki A, et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol. Cell. 2020;77:1163–1175.e1169. doi: 10.1016/j.molcel.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 157.Rekulapally P, Suresh SN. Nucleolus: a protein quality control compartment. Trends Biochem. Sci. 2019;44:993–995. doi: 10.1016/j.tibs.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 158.Frottin F, et al. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 159.Kemp CJ, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Debaugny RE, Skok JA. CTCF and CTCFL in cancer. Curr. Opin. Genet. Dev. 2020;61:44–52. doi: 10.1016/j.gde.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang B, et al. Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target Ther. 2021;6:290. doi: 10.1038/s41392-021-00678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boulay G, et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell. 2017;171:163–178.e119. doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 2016;6:a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Herrero-Martin D, et al. Factors affecting EWS-FLI1 activity in Ewing’s Sarcoma. Sarcoma. 2011;2011:352580. doi: 10.1155/2011/352580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cho WK, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Peng Q, et al. Phase separation of Epstein-Barr Virus EBNA2 and its coactivator EBNALP controls gene expression. J. Virol. 2020;94:e01771–19. doi: 10.1128/JVI.01771-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Portis T, Dyck P, Longnecker R. Epstein-Barr Virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;102:4166–4178. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- 172.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cao Y. EBV based cancer prevention and therapy in nasopharyngeal carcinoma. NPJ Precis Oncol. 2017;1:10. doi: 10.1038/s41698-017-0018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tu C, et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole-genome sequencing. Carcinogenesis. 2018;39:1517–1528. doi: 10.1093/carcin/bgy108. [DOI] [PubMed] [Google Scholar]
- 175.Taylor GS, et al. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 2015;33:787–821. doi: 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 176.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Affer M, et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014;28:1725–1735. doi: 10.1038/leu.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zamudio AV, et al. Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell. 2019;76:753–766 e756. doi: 10.1016/j.molcel.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang Y, Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020;13:165. doi: 10.1186/s13045-020-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Boija A, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855.e1816. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schatoff EM, Leach BI, Dow LE. Wnt signaling and colorectal cancer. Curr. Colorectal Cancer Rep. 2017;13:101–110. doi: 10.1007/s11888-017-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 186.Tran TQ, et al. Alpha-Ketoglutarate attenuates Wnt signaling and drives differentiation in colorectal cancer. Nat. Cancer. 2020;1:345–358. doi: 10.1038/s43018-020-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schaefer KN, Peifer M. Wnt/Beta-catenin signaling regulation and a role for biomolecular condensates. Dev. Cell. 2019;48:429–444. doi: 10.1016/j.devcel.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shi Q, Kang K, Chen YG. Liquid-liquid phase separation drives the beta-catenin destruction complex formation. Bioessays. 2021;43:e2100138. doi: 10.1002/bies.202100138. [DOI] [PubMed] [Google Scholar]
- 189.Schepers A, Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb. Perspect. Biol. 2012;4:a007989. doi: 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fearon ER. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 191.Stine ZE, et al. MYC, metabolism, and cancer. Cancer Disco. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang X, Cairns MJ, Yan J. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 2019;47:11481–11496. doi: 10.1093/nar/gkz1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Disco. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative MicroRNA network analysis. Cell. 2017;168:1000–1014 e1015. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xie JJ, et al. Super-enhancer-driven long non-coding RNA LINC01503, regulated by TP63, is over-expressed and oncogenic in squamous cell carcinoma. Gastroenterology. 2018;154:2137–2151.e2131. doi: 10.1053/j.gastro.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 198.Munoz-Fontela C, Mandinova A, Aaronson SA, Lee SW. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat. Rev. Immunol. 2016;16:741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ghodke I, et al. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. Mol. Cell. 2021;81:2596–2610.e2597. doi: 10.1016/j.molcel.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 201.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 202.Hu W, Feng Z, Levine AJ. The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer. 2012;3:199–208. doi: 10.1177/1947601912454734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 205.Zhou X, Hao Q, Lu H. Mutant p53 in cancer therapy-the barrier or the path. J. Mol. Cell Biol. 2019;11:293–305. doi: 10.1093/jmcb/mjy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang XW, Harris CC. p53 tumor-suppressor gene: clues to molecular carcinogenesis. J. Cell Physiol. 1997;173:247–255. doi: 10.1002/(SICI)1097-4652(199711)173:2<247::AID-JCP30>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 207.Rangel LP, Costa DC, Vieira TC, Silva JL. The aggregation of mutant p53 produces prion-like properties in cancer. Prion. 2014;8:75–84. doi: 10.4161/pri.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ano Bom AP, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J. Biol. Chem. 2012;287:28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Xu J, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 210.Higashimoto Y, et al. Unfolding, aggregation, and amyloid formation by the tetramerization domain from mutant p53 associated with lung cancer. Biochemistry. 2006;45:1608–1619. doi: 10.1021/bi051192j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ishimaru D, et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42:9022–9027. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- 212.De Smet F, et al. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J. Pathol. 2017;242:24–38. doi: 10.1002/path.4872. [DOI] [PubMed] [Google Scholar]
- 213.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Clark A, Burleson M. SPOP and cancer: a systematic review. Am. J. Cancer Res. 2020;10:704–726. [PMC free article] [PubMed] [Google Scholar]
- 215.Bouchard JJ, et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell. 2018;72:19–36.e18. doi: 10.1016/j.molcel.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Geng C, et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36:4767–4777. doi: 10.1038/onc.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dai X, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 2017;23:1063–1071. doi: 10.1038/nm.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kwon JE, et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 2006;281:12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- 220.An J, et al. Truncated ERG oncoproteins from TMPRSS2-ERG fusions are resistant to SPOP-mediated proteasome degradation. Mol. Cell. 2015;59:904–916. doi: 10.1016/j.molcel.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 221.Zhang Z, et al. ERG the modulates Warburg effect and tumor progression in cervical cancer. Biochem. Biophys. Res. Commun. 2020;522:191–197. doi: 10.1016/j.bbrc.2019.11.079. [DOI] [PubMed] [Google Scholar]
- 222.Kim B, et al. Breast cancer metastasis suppressor 1 (BRMS1) is destabilized by the Cul3-SPOP E3 ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 2011;415:720–726. doi: 10.1016/j.bbrc.2011.10.154. [DOI] [PubMed] [Google Scholar]
- 223.Le Gallo M, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kim MS, et al. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS. 2013;121:626–633. doi: 10.1111/apm.12030. [DOI] [PubMed] [Google Scholar]
- 225.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Song Y, et al. The emerging role of SPOP protein in tumorigenesis and cancer therapy. Mol. Cancer. 2020;19:2. doi: 10.1186/s12943-019-1124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Drabsch Y, ten Dijke P. TGF-beta signaling in breast cancer cell invasion and bone metastasis. J. Mammary Gland Biol. Neoplasia. 2011;16:97–108. doi: 10.1007/s10911-011-9217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Esposito M, et al. TGF-beta-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat. Cell Biol. 2021;23:257–267. doi: 10.1038/s41556-021-00641-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bragado P, et al. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat. Cell Biol. 2013;15:1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Stamenkovic I, Yu Q. Merlin, a “magic” linker between extracellular cues and intracellular signaling pathways that regulate cell motility, proliferation, and survival. Curr. Protein Pept. Sci. 2010;11:471–484. doi: 10.2174/138920310791824011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Asthagiri AR, et al. Neurofibromatosis type 2. Lancet. 2009;373:1974–1986. doi: 10.1016/S0140-6736(09)60259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Petrilli AM, Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35:537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang Y, et al. Hippo kinases MST1/2 regulate immune cell functions in cancer, infection, and autoimmune diseases. Crit. Rev. Eukaryot. Gene Expr. 2020;30:427–442. doi: 10.1615/CritRevEukaryotGeneExpr.2020035775. [DOI] [PubMed] [Google Scholar]
- 235.Liu Q, et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell. 2021;184:5559–5576.e5519. doi: 10.1016/j.cell.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 236.Tajan M, et al. SHP2 sails from physiology to pathology. Eur. J. Med. Genet. 2015;58:509–525. doi: 10.1016/j.ejmg.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 237.Zhu G, et al. Phase separation of disease-associated SHP2 mutants underlies MAPK hyperactivation. Cell. 2020;183:490–502.e418. doi: 10.1016/j.cell.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets. 2012;16:103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Polishchuk M, et al. A combined sequence and structure based method for discovering enriched motifs in RNA from in vivo binding data. Methods. 2017;118-119:73–81. doi: 10.1016/j.ymeth.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 240.Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021;22:183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Fox AH, et al. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/S0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 242.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fujimoto A, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 244.Choudhry H, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mello SS, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Li RH, et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021;31:1088–1105. doi: 10.1038/s41422-021-00530-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lieberthal JG, Kaminsky M, Parkhurst CN, Tanese N. The role of YY1 in reduced HP1alpha gene expression in invasive human breast cancer cells. Breast Cancer Res. 2009;11:R42. doi: 10.1186/bcr2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 249.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wang L, et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell. 2019;76:646–659.e646. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 251.Ntziachristos P, Mullenders J, Trimarchi T, Aifantis I. Mechanisms of epigenetic regulation of leukemia onset and progression. Adv. Immunol. 2013;117:1–38. doi: 10.1016/B978-0-12-410524-9.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Park JW, Han JW. Targeting epigenetics for cancer therapy. Arch. Pharm. Res. 2019;42:159–170. doi: 10.1007/s12272-019-01126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yang Y, et al. Phase separation of Epstein-Barr virus EBNA2 protein reorganizes chromatin topology for epigenetic regulation. Commun. Biol. 2021;4:967. doi: 10.1038/s42003-021-02501-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kempkes B, Ling PD. EBNA2 and its coactivator EBNA-LP. Curr. Top. Microbiol. Immunol. 2015;391:35–59. doi: 10.1007/978-3-319-22834-1_2. [DOI] [PubMed] [Google Scholar]
- 255.Caslini C, et al. HDAC7 regulates histone 3 lysine 27 acetylation and transcriptional activity at super-enhancer-associated genes in breast cancer stem cells. Oncogene. 2019;38:6599–6614. doi: 10.1038/s41388-019-0897-0. [DOI] [PubMed] [Google Scholar]
- 256.Ma S, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Gao Y, et al. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 2019;29:767–769. doi: 10.1038/s41422-019-0210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nsengimana B, et al. Processing body (P-body) and its mediators in cancer. Mol. Cell Biochem. 2022;477:1217–1238. doi: 10.1007/s11010-022-04359-7. [DOI] [PubMed] [Google Scholar]
- 259.Legrand N, Dixon DA, Sobolewski C. Stress granules in colorectal cancer: current knowledge and potential therapeutic applications. World J. Gastroenterol. 2020;26:5223–5247. doi: 10.3748/wjg.v26.i35.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Grabocka E, Bar-Sagi D. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell. 2016;167:1803–1813.e1812. doi: 10.1016/j.cell.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Podszywalow-Bartnicka P, et al. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle. 2014;13:3727–3741. doi: 10.4161/15384101.2014.965013. [DOI] [PubMed] [Google Scholar]
- 262.Cheng Y, et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39:958–972 e958. doi: 10.1016/j.ccell.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zlotorynski E. m(6)A nuclear condensates support AML. Nat. Rev. Mol. Cell Biol. 2021;22:442. doi: 10.1038/s41580-021-00385-3. [DOI] [PubMed] [Google Scholar]
- 264.Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer. 2020;20:743–756. doi: 10.1038/s41568-020-00302-4. [DOI] [PubMed] [Google Scholar]
- 265.Chakravarti D, LaBella KA, DePinho RA. Telomeres: history, health, and hallmarks of aging. Cell. 2021;184:306–322. doi: 10.1016/j.cell.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Moyzis RK, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Bryan TM, et al. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Garcia-Cao M, et al. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 269.Gonzalo S, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 270.Montero JJ, et al. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018;9:1548. doi: 10.1038/s41467-018-03916-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.De Vitis M, Berardinelli F, Sgura A. Telomere length maintenance in cancer: at the crossroad between telomerase and alternative lengthening of telomeres (ALT) Int. J. Mol. Sci. 2018;19:606. doi: 10.3390/ijms19020606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 273.Zhang JM, et al. Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. Mol. Cell. 2021;81:1027–1042 e1024. doi: 10.1016/j.molcel.2020.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhang H, et al. Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell. 2020;31:2048–2056. doi: 10.1091/mbc.E19-10-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ozer O, Bhowmick R, Liu Y, Hickson ID. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget. 2018;9:15836–15846. doi: 10.18632/oncotarget.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Min J, Wright WE, Shay JW. Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol. Cell Biol. 2017;37:e00226–17. doi: 10.1128/MCB.00226-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Min J, Wright WE, Shay JW. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019;33:814–827. doi: 10.1101/gad.324905.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Machiela MJ, et al. Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: a report from the female lung cancer consortium in Asia. Int J. Cancer. 2015;137:311–319. doi: 10.1002/ijc.29393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ojha J, et al. Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol. Biomark. Prev. 2016;25:1043–1049. doi: 10.1158/1055-9965.EPI-15-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.McNally EJ, Luncsford PJ, Armanios M. Long telomeres and cancer risk: the price of cellular immortality. J. Clin. Invest. 2019;129:3474–3481. doi: 10.1172/JCI120851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Yang Y, et al. Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nat. Commun. 2019;10:3759. doi: 10.1038/s41467-019-11671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zou Y, Ma D, Wang Y. The PROTAC technology in drug development. Cell Biochem. Funct. 2019;37:21–30. doi: 10.1002/cbf.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lu J, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Liao X, et al. ARV-825 demonstrates antitumor activity in gastric cancer via MYC-targets and G2M-checkpoint signaling pathways. Front. Oncol. 2021;11:753119. doi: 10.3389/fonc.2021.753119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Takahashi D, et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell. 2019;76:797–810 e710. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 286.Nunes AT, Annunziata CM. Proteasome inhibitors: structure and function. Semin. Oncol. 2017;44:377–380. doi: 10.1053/j.seminoncol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Mauthe M, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Tsang B, et al. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl Acad. Sci. USA. 2019;116:4218–4227. doi: 10.1073/pnas.1814385116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010;19:1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hawkins WD, Klionsky DJ. A separation that’s for the best: coming together at the PAS. Cell Res. 2020;30:372–373. doi: 10.1038/s41422-020-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Luo YY, Wu JJ, Li YM. Regulation of liquid-liquid phase separation with focus on post-translational modifications. Chem. Commun. 2021;57:13275–13287. doi: 10.1039/D1CC05266G. [DOI] [PubMed] [Google Scholar]
- 292.Girdhar A, et al. Computational insights into mechanism of AIM4-mediated inhibition of aggregation of TDP-43 protein implicated in ALS and evidence for in vitro inhibition of liquid-liquid phase separation (LLPS) of TDP-43(2C)-A315T by AIM4. Int. J. Biol. Macromol. 2020;147:117–130. doi: 10.1016/j.ijbiomac.2020.01.032. [DOI] [PubMed] [Google Scholar]
- 293.Strzyz P. Drugs enter a liquid phase. Nat. Rev. Mol. Cell Biol. 2020;21:419. doi: 10.1038/s41580-020-0268-2. [DOI] [PubMed] [Google Scholar]
- 294.Klein IA, et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Iconaru LI, et al. Discovery of small molecules that inhibit the disordered protein, p27(Kip1) Sci. Rep. 2015;5:15686. doi: 10.1038/srep15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.De Mol E, et al. EPI-001, a compound active against castration-resistant prostate cancer, targets transactivation unit 5 of the androgen receptor. ACS Chem. Biol. 2016;11:2499–2505. doi: 10.1021/acschembio.6b00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Loh D, Reiter RJ. Melatonin: regulation of prion protein phase separation in cancer multidrug resistance. Molecules. 2022;27:705. doi: 10.3390/molecules27030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Liao, L. et al. Anti-HIV drug elvitegravir suppresses cancer metastasis via increased proteasomal degradation of m6A methyltransferase METTL3. Cancer Res.10.1158/0008-5472.CAN-21-4124 (2022). [DOI] [PubMed]
- 299.Rehman AU, Rahman MU, Arshad T, Chen HF. Allosteric modulation of intrinsically disordered proteins. Adv. Exp. Med. Biol. 2019;1163:335–357. doi: 10.1007/978-981-13-8719-7_14. [DOI] [PubMed] [Google Scholar]
- 300.Saito M, et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019;15:51–61. doi: 10.1038/s41589-018-0180-7. [DOI] [PubMed] [Google Scholar]
- 301.Sachdev R, et al. Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. Elife. 2019;8:e41415. doi: 10.7554/eLife.41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Mateju D, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Piovesan D, et al. MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res. 2021;49:D361–D367. doi: 10.1093/nar/gkaa1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Punta M, Simon I, Dosztanyi Z. Prediction and analysis of intrinsically disordered proteins. Methods Mol. Biol. 2015;1261:35–59. doi: 10.1007/978-1-4939-2230-7_3. [DOI] [PubMed] [Google Scholar]
- 305.Wu Z, Hu G, Oldfield CJ, Kurgan L. Prediction of intrinsic disorder with quality assessment using QUARTER. Methods Mol. Biol. 2020;2165:83–101. doi: 10.1007/978-1-0716-0708-4_5. [DOI] [PubMed] [Google Scholar]
- 306.Iconaru LI, et al. Small molecule sequestration of the intrinsically disordered protein, p27(Kip1), within soluble oligomers. J. Mol. Biol. 2021;433:167120. doi: 10.1016/j.jmb.2021.167120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sadar MD. Discovery of drugs that directly target the intrinsically disordered region of the androgen receptor. Expert Opin. Drug Disco. 2020;15:551–560. doi: 10.1080/17460441.2020.1732920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Ming Y, et al. Targeting liquid-liquid phase separation in pancreatic cancer. Transl. Cancer Res. 2019;8:96–103. doi: 10.21037/tcr.2019.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Ruff KM, Pappu RV. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 2021;433:167208. doi: 10.1016/j.jmb.2021.167208. [DOI] [PubMed] [Google Scholar]
- 310.Sabari BR, Dall’Agnese A, Young RA. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 2020;45:961–977. doi: 10.1016/j.tibs.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: going beyond transcriptional regulation. Mol. Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Plys AJ, et al. Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33:799–813. doi: 10.1101/gad.326488.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tatavosian R, et al. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019;294:1451–1463. doi: 10.1074/jbc.RA118.006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zeng M, Li B, Yang L, Guan Q. CBX2 depletion inhibits the proliferation, invasion and migration of gastric cancer cells by inactivating the YAP/beta-catenin pathway. Mol. Med. Rep. 2021;23:137. doi: 10.3892/mmr.2020.11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Wang S, et al. A potent, selective CBX2 chromodomain ligand and its cellular activity during prostate cancer neuroendocrine differentiation. Chembiochem. 2021;22:2335–2344. doi: 10.1002/cbic.202100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wang Z, et al. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat. Rev. Urol. 2020;17:339–350. doi: 10.1038/s41585-020-0314-z. [DOI] [PubMed] [Google Scholar]
- 318.Chong S, et al. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell. 2022;82:2084–2097.e5. doi: 10.1016/j.molcel.2022.04.007. [DOI] [PubMed] [Google Scholar]
- 319.Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J. Mol. Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 320.Chong S, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Markmiller S, et al. Persistent mRNA localization defects and cell death in ALS neurons caused by transient cellular stress. Cell Rep. 2021;36:109685. doi: 10.1016/j.celrep.2021.109685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wang F, et al. Targeting stress granules: a novel therapeutic strategy for human diseases. Pharm. Res. 2020;161:105143. doi: 10.1016/j.phrs.2020.105143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Shi Q, et al. Prostate Cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by upregulating Caprin1-dependent stress granule assembly. Mol. Cancer. 2019;18:170. doi: 10.1186/s12943-019-1096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Chang CW, Shen YC, Yan SJ. HP1a-mediated heterochromatin formation inhibits high dietary sugar-induced tumor progression. Cell Death Dis. 2021;12:1130. doi: 10.1038/s41419-021-04414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Liu Y, Zhang D. HP1a/KDM4A is involved in the autoregulatory loop of the oncogene gene c-Jun. Epigenetics. 2015;10:453–459. doi: 10.1080/15592294.2015.1048059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Strom AR, et al. HP1alpha is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. Elife. 2021;10:e63972. doi: 10.7554/eLife.63972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Carone DM, Lawrence JB. Heterochromatin instability in cancer: from the Barr body to satellites and the nuclear periphery. Semin. Cancer Biol. 2013;23:99–108. doi: 10.1016/j.semcancer.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lee YL, et al. Mediator subunit MED1 is required for E2A-PBX1-mediated oncogenic transcription and leukemic cell growth. Proc. Natl Acad. Sci. USA. 2021;118:e1922864118. doi: 10.1073/pnas.1922864118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Rasool RU, et al. CDK7 inhibition suppresses castration-resistant prostate cancer through MED1 inactivation. Cancer Disco. 2019;9:1538–1555. doi: 10.1158/2159-8290.CD-19-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Guo Q, et al. miR-374a-5p inhibits non-small cell lung cancer cell proliferation and migration via targeting NCK1. Exp. Ther. Med. 2021;22:943. doi: 10.3892/etm.2021.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Zhou J, et al. MED1 mediator subunit is a key regulator of hepatic autophagy and lipid metabolism. Autophagy. 2021;17:4043–4061. doi: 10.1080/15548627.2021.1899691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Nagalingam A, et al. Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis. 2012;33:918–930. doi: 10.1093/carcin/bgs105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Thandapani P. Super-enhancers in cancer. Pharm. Ther. 2019;199:129–138. doi: 10.1016/j.pharmthera.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 334.Iino K, et al. RNA-binding protein NONO promotes breast cancer proliferation by post-transcriptional regulation of SKP2 and E2F8. Cancer Sci. 2020;111:148–159. doi: 10.1111/cas.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Huang CJ, et al. Altered stoichiometry and nuclear delocalization of NonO and PSF promote cellular senescence. Aging. 2016;8:3356–3374. doi: 10.18632/aging.101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Feng P, et al. NONO and tumorigenesis: more than splicing. J. Cell Mol. Med. 2020;24:4368–4376. doi: 10.1111/jcmm.15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Petti E, et al. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019;10:1001. doi: 10.1038/s41467-019-08863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Pisani G, Baron B. NEAT1 and paraspeckles in cancer development and chemoresistance. Noncoding RNA. 2020;6:43. doi: 10.3390/ncrna6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Meng Y, et al. LncRNA-422 suppresses the proliferation and growth of colorectal cancer cells by targeting SFPQ. Clin. Transl. Med. 2022;12:e664. doi: 10.1002/ctm2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Ru Y, et al. NEAT1_2-SFPQ axis mediates cisplatin resistance in liver cancer cells in vitro. OncoTargets Ther. 2018;11:5695–5702. doi: 10.2147/OTT.S163774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Rajesh C, Baker DK, Pierce AJ, Pittman DL. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Mitrea DM, et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 2018;9:842. doi: 10.1038/s41467-018-03255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Kim JY, Cho YE, Park JH. The nucleolar protein GLTSCR2 is an upstream negative regulator of the oncogenic nucleophosmin-MYC axis. Am. J. Pathol. 2015;185:2061–2068. doi: 10.1016/j.ajpath.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 344.Falini B, et al. Diagnostic and therapeutic pitfalls in NPM1-mutated AML: notes from the field. Leukemia. 2021;35:3113–3126. doi: 10.1038/s41375-021-01222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Falini B, Brunetti L, Martelli MP. How I diagnose and treat NPM1-mutated AML. Blood. 2021;137:589–599. doi: 10.1182/blood.2020008211. [DOI] [PubMed] [Google Scholar]
- 346.Luo C, et al. The role of NPM1 in the invasion and migration of drug resistant bladder cancer. Urol. J. 2021;18:452–459. doi: 10.22037/uj.v18i.6087. [DOI] [PubMed] [Google Scholar]
- 347.Qin G, et al. NPM1 upregulates the transcription of PD-L1 and suppresses T cell activity in triple-negative breast cancer. Nat. Commun. 2020;11:1669. doi: 10.1038/s41467-020-15364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Orsolic I, et al. The relationship between the nucleolus and cancer: current evidence and emerging paradigms. Semin. Cancer Biol. 2016;37-38:36–50. doi: 10.1016/j.semcancer.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 349.Roche B, Arcangioli B, Martienssen R. New roles for dicer in the nucleolus and its relevance to cancer. Cell Cycle. 2017;16:1643–1653. doi: 10.1080/15384101.2017.1361568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Simon DN, Rout MP. Cancer and the nuclear pore complex. Adv. Exp. Med. Biol. 2014;773:285–307. doi: 10.1007/978-1-4899-8032-8_13. [DOI] [PubMed] [Google Scholar]
- 351.Capitanio JS, Montpetit B, Wozniak RW. Human Nup98 regulates the localization and activity of DExH/D-box helicase DHX9. Elife. 2017;6:e18825. doi: 10.7554/eLife.18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Li Y, et al. PML nuclear body biogenesis, carcinogenesis, and targeted therapy. Trends Cancer. 2020;6:889–906. doi: 10.1016/j.trecan.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 353.Tampakaki M, et al. PML differentially regulates growth and invasion in brain cancer. Int. J. Mol. Sci. 2021;22:6289. doi: 10.3390/ijms22126289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.El-Asmi F, Chelbi-Alix MK. [PML isoforms and TGF-beta response] Med. Sci. 2020;36:50–56. doi: 10.1051/medsci/2019269. [DOI] [PubMed] [Google Scholar]
- 355.di Masi A, et al. PML nuclear body disruption impairs DNA double-strand break sensing and repair in APL. Cell Death Dis. 2016;7:e2308. doi: 10.1038/cddis.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.de The H, Pandolfi PP, Chen Z. Acute promyelocytic leukemia: a paradigm for oncoprotein-targeted cure. Cancer Cell. 2017;32:552–560. doi: 10.1016/j.ccell.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 357.Fu Y, Zhuang X. m(6)A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 2020;16:955–963. doi: 10.1038/s41589-020-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Hu J, et al. YTHDF1 is a potential pan-cancer biomarker for prognosis and immunotherapy. Front. Oncol. 2021;11:607224. doi: 10.3389/fonc.2021.607224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582–1595 e1518. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Bai Y, et al. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front. Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am. J. Cancer Res. 2020;10:2546–2554. [PMC free article] [PubMed] [Google Scholar]
- 362.Hou G, et al. SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 2021;49:2859–2877. doi: 10.1093/nar/gkab065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Dixit D, et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Disco. 2021;11:480–499. doi: 10.1158/2159-8290.CD-20-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Li J, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer. 2020;19:152. doi: 10.1186/s12943-020-01267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Einstein JM, et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell. 2021;81:3048–3064 e3049. doi: 10.1016/j.molcel.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Ni W, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Chang G, et al. YTHDF3 induces the translation of m(6)A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell. 2020;38:857–871 e857. doi: 10.1016/j.ccell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Pan L, et al. TRPM2-AS promotes cancer cell proliferation through control of TAF15. Int. J. Biochem. Cell Biol. 2020;120:105683. doi: 10.1016/j.biocel.2019.105683. [DOI] [PubMed] [Google Scholar]
- 369.Andersson MK, et al. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Spitzer JI, et al. mRNA and protein levels of FUS, EWSR1, and TAF15 are upregulated in liposarcoma. Genes Chromosomes Cancer. 2011;50:338–347. doi: 10.1002/gcc.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Singh AK, Kapoor V, Thotala D, Hallahan DE. TAF15 contributes to the radiation-inducible stress response in cancer. Oncotarget. 2020;11:2647–2659. doi: 10.18632/oncotarget.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Zuo L, et al. Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. Nat. Commun. 2021;12:1491. doi: 10.1038/s41467-021-21690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.He N, Zhou Q. New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC) J. Neuroimmune Pharm. 2011;6:260–268. doi: 10.1007/s11481-011-9267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Trosko JE. From adult stem cells to cancer stem cells: Oct-4 Gene, cell-cell communication, and hormones during tumor promotion. Ann. N. Y. Acad. Sci. 2006;1089:36–58. doi: 10.1196/annals.1386.018. [DOI] [PubMed] [Google Scholar]
- 376.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Qiao Y, et al. RUNX3 is a novel negative regulator of oncogenic TEAD-YAP complex in gastric cancer. Oncogene. 2016;35:2664–2674. doi: 10.1038/onc.2015.338. [DOI] [PubMed] [Google Scholar]
- 378.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Moya IM, et al. Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science. 2019;366:1029–1034. doi: 10.1126/science.aaw9886. [DOI] [PubMed] [Google Scholar]
- 380.Cargill MJ, Morales A, Ravishankar S, Warren EH. RNA helicase, DDX3X, is actively recruited to sites of DNA damage in live cells. DNA Repair. 2021;103:103137. doi: 10.1016/j.dnarep.2021.103137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Shen H, et al. Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol. Cell. 2022;S1097-2765:00385–00389. doi: 10.1016/j.molcel.2022.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Samir P, Kanneganti TD. DDX3X sits at the crossroads of liquid-liquid and prionoid phase transitions arbitrating life and death cell fate decisions in stressed cells. DNA Cell Biol. 2020;39:1091–1095. doi: 10.1089/dna.2020.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Valentin-Vega YA, et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 2016;6:25996. doi: 10.1038/srep25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Gaglia G, et al. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 2020;22:151–158. doi: 10.1038/s41556-019-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Deshpande AJ, Bradner J, Armstrong SA. Chromatin modifications as therapeutic targets in MLL-rearranged leukemia. Trends Immunol. 2012;33:563–570. doi: 10.1016/j.it.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Kabra A, Bushweller J. The intrinsically disordered proteins MLLT3 (AF9) and MLLT1 (ENL)—multimodal transcriptional switches with roles in normal hematopoiesis, MLL fusion leukemia, and kidney cancer. J. Mol. Biol. 2022;434:167117. doi: 10.1016/j.jmb.2021.167117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Guo C, et al. ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Sci. Adv. 2020;6:eaay4858. doi: 10.1126/sciadv.aay4858. [DOI] [PMC free article] [PubMed] [Google Scholar]