Abstract
The nucleotide context surrounding stop codons significantly affects the efficiency of translation termination. In eukaryotes, various 3′ contexts that are unfavorable for translation termination have been described; however, the exact molecular mechanism that mediates their effects remains unknown. In this study, we used a reconstituted mammalian translation system to examine the efficiency of stop codons in different contexts, including several previously described weak 3′ stop codon contexts. We developed an approach to estimate the level of stop codon readthrough in the absence of eukaryotic release factors (eRFs). In this system, the stop codon is recognized by the suppressor or near-cognate tRNAs. We observed that in the absence of eRFs, readthrough occurs in a 3′ nucleotide context-dependent manner, and the main factors determining readthrough efficiency were the type of stop codon and the sequence of the 3′ nucleotides. Moreover, the efficiency of translation termination in weak 3′ contexts was almost equal to that in the tested standard context. Therefore, the ability of eRFs to recognize stop codons and induce peptide release is not affected by mRNA context. We propose that ribosomes or other participants of the elongation cycle can independently recognize certain contexts and increase the readthrough of stop codons. Thus, the efficiency of translation termination is regulated by the 3′ nucleotide context following the stop codon and depends on the concentrations of eRFs and suppressor/near-cognate tRNAs.
Keywords: stop codon context, readthrough, eRF1, eRF3, translation termination, ribosome
Abbreviations: eEF, eukaryotic elongation factor; eIFs, eukaryotic translation initiation factors; eRFs, eukaryotic release factors; HS, high salt; IC, initiation complex; LS, low salt; preTC, pretermination complex; PTC, premature termination codons; TMV, tobacco mosaic virus
Protein synthesis is completed when the stop codon (UAA, UAG, or UGA) occupies the ribosomal A site, where the eukaryotic release factors (eRFs) decode it. In eukaryotes, the tRNA-mimicking factor eRF1 recognizes all three stop codons and promotes the release of the synthesized peptide from the peptidyl-transferase center. It is stimulated by eRF3, which resembles the elongation factor 1A (eEF1A) (1, 2, 3, 4). Remarkably, positional and conformational similarities exist between the termination eRF1–eRF3 complex and elongation aa tRNA–eEF1 complexes (5, 6). Stop codon recognition is implemented by the conserved TASNIKS and YxCxxxF motifs of the N-domain of eRF1 (7, 8). The lysine of the NIKS motif can be hydroxylated, which improves termination efficiency (9). cryo-EM structures of mammalian ribosomal complexes containing a stop codon at the A site have shown that binding of eRF1 leads to changes in mRNA configuration so that the fourth nucleotide following the three bases of the stop codon is pulled into the A site (10, 11, 12). This configuration differs from the shape of sense codons recognized by tRNAs and implicates a complex three-dimensional interplay of eRF1 and 18S rRNA. This also explains the strong impact of the identity of the 3′ nucleotide downstream of the stop codon on termination (13).
However, in some instances, the amino acid is incorporated into the nascent polypeptide chain instead of proper translation termination. Such an event is a result of stop codon suppression or readthrough, when the stop codon in the ribosomal A site is interpreted as a sense codon and is recognized by near-cognate tRNAs instead of eRF1. The basal level of stop codon readthrough commonly has a frequency of < 0.1% (14), although in some cases, the level of readthrough was shown to be higher than 10% (15, 16). Phylogenetic analysis of the 12 Drosophila species revealed more than 280 conserved stop codon readthroughs. This was confirmed by ribosome profiling analysis, which indicated that readthrough was a relatively common event (17, 18). Subsequent studies have identified readthrough in fungi (19), and numerous studies have described readthrough for a large number of transcripts in mammals (15, 16, 18, 20, 21, 22, 23). In addition, premature stop codons also sustain a sufficient percentage of readthrough (<1%) (14), which is essential for disease therapeutics, occurring as a result of nonsense mutations. These data demonstrate the importance of readthrough events, which can be considered not only as an error during the termination process but also as an important regulatory mechanism.
It has been shown that the nucleotide context of stop codons significantly affects the readthrough level in different groups of eukaryotes (24, 25, 26). The nearest 5′ and 3′ nucleotides of stop codons can decrease and increase the translation termination efficiency (27). Previously, the strongest influence on translation termination was demonstrated for +4 nucleotides immediately following the stop codon (13, 28, 29, 30, 31, 32, 33). Few studies investigating the influence of the 3′ context have suggested that terminating signals include six nucleotides (31, 32). Purines are preferred over pyrimidines in eukaryotic genomes (33, 34). Additionally, nucleotide distribution up to +9 in Saccharomyces cerevisiae and most likely in all eukaryotes is not random. It was demonstrated that positions +4, +5, +6, +8, and +9 were the key, and the +7 position did not have any effect (35). The most effective suppression motif, CAA UUA entirely conforms to the 3′ context of stop codon UAG of tobacco mosaic virus (TMV). In mammalian cells, Cridge et al. (36) affirmed the high impact on the readthrough of +4 and +8 nucleotides independently of the type of stop codon, and +5 and +6 positions determined the increase or decrease in readthrough depending on the stop codon and +4 nucleotides.
There is evidence that different factors can influence readthrough levels in cooperation with the stop codon context. It has been shown that the eukaryotic translation initiation factor eIF3 increases readthrough in weak termination contexts, possibly promoting the incorporation of near-cognate tRNAs (37). The posttranslational hydroxylation of prolyl in ribosomal protein Rps23 of the 40S subunit can also modulate termination accuracy in a context-dependent manner (38). However, the exact mechanism underlying the effect of the 3′ stop codon context on translation termination remains unknown. According to our previous study (39), there is no apparent connection between nucleotide frequencies in the 3′ stop codon context and their effect on peptide-release efficiency. We investigated the effects of several 3′ stop codon contexts on readthrough and peptide release in the reconstituted translation termination system and revealed the molecular mechanism of this process.
Results
Model system to study stop codon readthrough
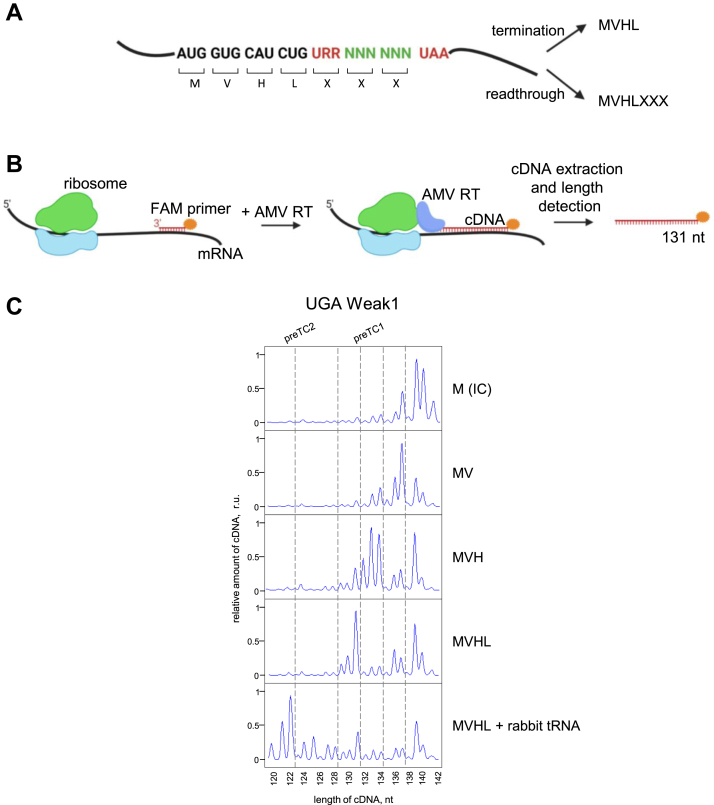
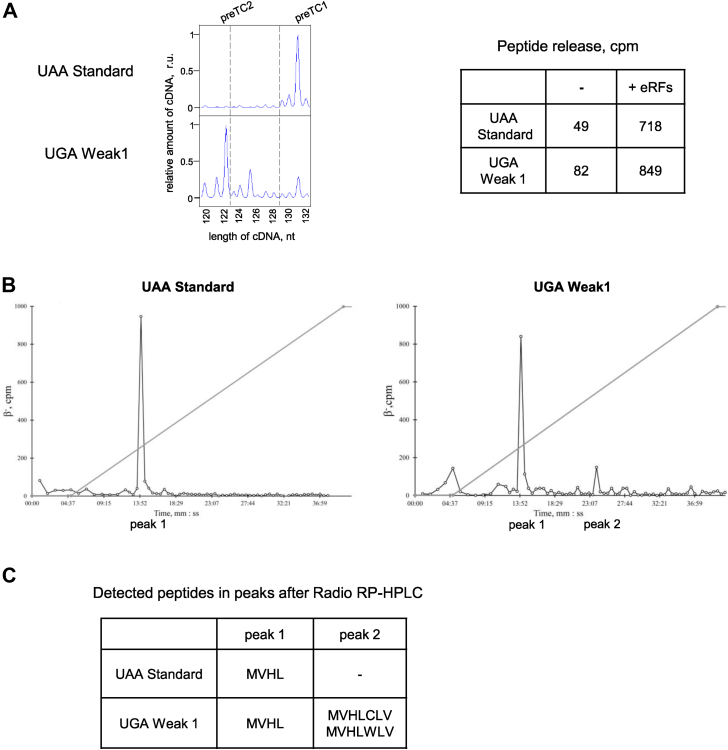
To study the mechanism underlying stop codon readthrough, we constructed model mRNAs containing two stop codons separated by hexanucleotide sequences (Fig. 1A). After the first stop codon (UAA, UAG, or UGA), we inserted several hexanucleotide sequences reported to be preferable for readthrough (15, 29, 40). We chose the most frequent stop codon, UAA, to be the second stop codon to exclude secondary readthrough and estimate the level of the first stop codon readthrough. The 3′ context of the second stop codon was A-rich AAG CUU, which ensured efficient translation termination according to our data (39). After termination at the first stop codon, we obtained the MVHL tetrapeptide, and after readthrough, we obtained the heptapeptide MVHLXXX (Fig. 1A).
Figure 1.
An approach for determining the level of stop codon readthrough in the reconstituted eukaryotic translation system.A, the scheme of the dual-stop mRNA used for quantification of the level of the stop codon readthrough. The mRNA contains an ORF encoding the MVHL tetrapeptide and ending with the first stop codon (UAA, UAG, or UGA). The ORF is followed by a 3՛ context (6 nt) followed by the second stop codon (UAA). When readthrough on the first stop codon occurs, MVHLXXX heptapeptide is synthetized. Created with BioRender.com. B, the scheme of the toe-printing assay used to detect ribosome position on the mRNA. AMV reverse transcriptase synthesizes cDNA on the mRNA template before colliding with the ribosome. The length of cDNA corresponds to position of the ribosome. Created with BioRender.com. C, toe-printing analysis of the ribosomal complexes assembled at the dual-stop mRNAs containing UGA Weak1 context. Different a.a. tRNAs (V, H, L) were added to the initiation complex. An addition of the rabbit tRNA leads to readthrough of the first stop codon and appearance of the preTC2. IC, initiation complex; preTC1 and preTC2, pretermination complexes at the first and second stop codons. a.a. aminoacylated; cDNA, complementary DNA; preTC, pretermination complex.
To estimate the stop codon readthrough efficiency, we performed a fluorescent toe-printing assay of the ribosomal complexes assembled at the model mRNA. Fluorescent toe-printing is based on a reverse transcription reaction with fluorescently labeled primers that anneal downstream of the ribosomal complex (Fig. 1B). The position of the bound ribosome on the mRNA was then determined by the length of the fragment which extended the primer during reverse transcription.
The consensus sequence CUAG, following the UGA stop codon, was previously shown to stimulate readthrough in a few mammalian genes (15). Based on this sequence, we designed a model mRNA with a CUA GUA (Weak1) context located between the UGA and UAA stop codons. We reconstituted translation initiation 48S and 80S complexes on the mRNA (Fig. 1C) in the presence of the initiator methionyl-tRNA using an in vitro translation system (3). We then added Val-tRNAVal and translation elongation factors eEF1 and eEF2 in the presence of GTP to the initiation complex (IC). We obtained an elongation complex with dipeptidyl-tRNA (MV-tRNA) located at the P site. The addition of His-tRNAHis moved the ribosome to the next codon, and we obtained MVH-tRNA at the P site. The final addition of Leu-tRNALeu caused the formation of pretermination complex 1 (preTC1) with MVHL-tRNA at the P-site and UGA stop codon at the A site.
Total rabbit aminoacylated (a.a.) tRNA moved preTC1 through the first stop codon to the second stop codon, giving the appearance of a +9 nucleotide peak (Fig. 1C), indicating the formation of preTC2. Intermediate peaks corresponding to elongation complexes that stopped at the CUA and GUA codons were also detected. Therefore, in the absence of eRFs, stop codon could be recognized by suppressor or near-cognate tRNAs at least in the specific 3′ nucleotide context. We suggest that the stop codon readthrough efficiency is determined by the presence of such tRNAs in the cell. In this experiment rabbit tRNAs and rabbit ribosomes were used (Fig. 1C). To determine the effect of tRNAs from different organisms on readthrough, we compared the effects of calf, rabbit, and yeast total tRNA (Fig. S1). It appeared that all tested total tRNA induced UGA readthrough. This indicates that they contain a sufficient amount of suppressor or near-cognate tRNA to recognize the UGA stop codon. However, the calf tRNA was less active than the others which means that it contained smaller amounts of appropriate tRNA. It is noteworthy that the second stop codon UAA in the strong A-rich 3′ context (39) was not recognized by any of the tested preparations of tRNA. Therefore, different stop codons have different readthrough potentials.
Factors influencing stop codon readthrough efficiency
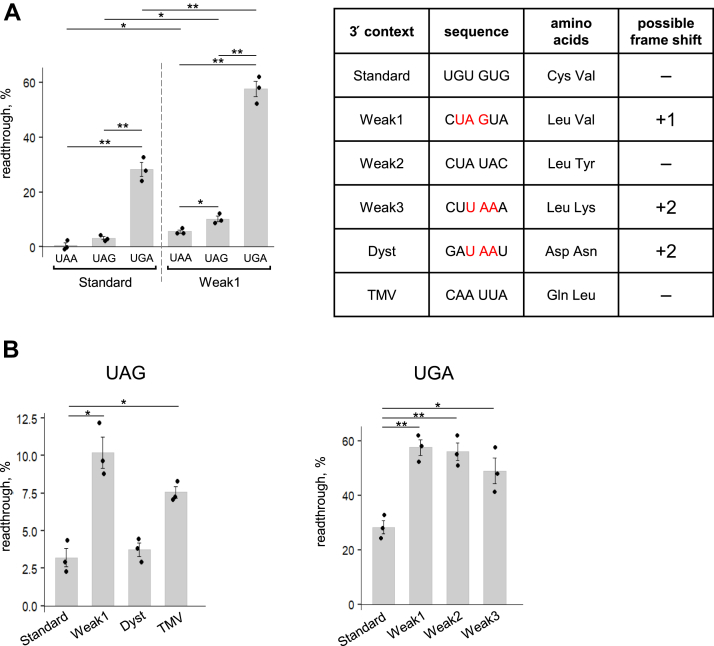
To determine the factors affecting the efficiency of stop codon readthrough in the absence of eRFs, we compared how all three stop codons in the Standard and Weak1 3′ contexts were decoded (Fig. 2A, row data are presented in Fig. S2). The 3ʹ context UGU GUG was chosen as the Standard. This context has been used in all our previous studies in the reconstituted mammalian translation system (3, 39, 41, 42, 43, 44, 45, 46, 47). The frequencies of triplets from this sequence in the human genome calculated earlier (1–1.8%) were close to that for random triplet NNN 1/64 = 1.56% (39). Therefore, this sequence can be used as a control sequence.
Figure 2.
Stop codon readthrough on different stop codons and 3′ contexts in the absence of release factors.A, readthrough efficiency on all three stop codons in the Standard and Weak1 contexts. Characteristics of the 3′ contexts are shown in the table. B, readthrough efficiency on the UAG and UGA stop codons in the Standard, Weak1, Dyst, TMV, Weak2, and Weak3 contexts. For the Standard and Weak1 contexts, the data are duplicated from A. Individual measurements are shown as dots, mean ± SEM are shown on histogram. n = 3, ∗p ≤ 0.05 and ∗∗p ≤ 0.01. Statistically insignificant differences are not shown. TMV, tobacco mosaic virus.
To exclude mistakes in all cases where we observed the appearance of intermediate peaks between the first and second stop codons, which indicated a lack of complementary tRNA, we determined the efficiency of readthrough by summarizing all the peaks appearing after the first stop codon. Thus, the effect of deficiency of the corresponding tRNAs in the calculations was reduced.
For stop codons in the Standard context, readthrough varied from 0.5% to 28% (Table 1 and Fig. 2A). We observed that UAA was the strongest stop codon and UGA was the weakest. We determine the weakness of the stop codon based on the readthrough frequency. In the Standard context, the readthrough efficiency of these codons differed by a factor of 55 (Table 1). Stop codons in the Weak1 context demonstrated the same readthrough dependence: UAA was the strongest and UGA was the weakest stop codon. However, the overall readthrough rate was higher, ranging from 5% for UAA to 57% for UGA. Therefore, the readthrough level of these stop codons differed by a factor of 10 in the Weak1 context (Table 1). We revealed that the efficiency of stop codon readthrough fits into an exponential curve for both Standard and Weak1 contexts. Thus, it is a property of the stop codons themselves, regardless of the 3′ context. It is difficult to determine the cause of such an exponential dependence; obviously, it reflects a much more efficient decoding of the UGA stop codon by the suppressor or near-cognate tRNAs.
Table 1.
Ratio of readthrough efficiency
| mRNA | Mean ± SEM | Ratio to |
||||
|---|---|---|---|---|---|---|
| UAA | UAG | UGA | Weak1 | Standard | ||
| UAA Standard | 0.5 ± 1.0 | – | 0.16 | 0.02 | 0.09 | – |
| UAG Standard | 3.2 ± 0.6 | 6.32 | – | 0.11 | 0.31 | – |
| UGA Standard | 28.3 ± 2.5 | 55.68 | 8.81 | – | 0.49 | – |
| UAA Weak1 | 5.6 ± 0.6 | – | 0.55 | 0.10 | – | 11.01 |
| UAG Weak1 | 10.2 ± 1.0 | 1.83 | – | 0.18 | – | 3.18 |
| UGA Weak1 | 57.5 ± 2.8 | 10.29 | 5.63 | – | – | 2.03 |
| UAG Dyst | 3.7 ± 0.4 | – | – | – | – | 1.17 |
| UAG TMV | 7.6 ± 0.4 | – | – | – | – | 2.36 |
| UGA Weak2 | 56.0 ± 3.2 | – | – | – | – | 1.98 |
| UGA Weak3 | 48.9 ± 4.7 | – | – | – | – | 1.73 |
Regarding the influence of the sequence of the 3′ stop codon context, we also found significant differences in readthrough efficiency for the same codons in different contexts (Fig. 2A and Table 1). However, in this case, the difference was smaller than that between different stop codons in the same context. Thus, the readthrough of the UAA stop codon in the Standard context was 10 times worse than that in the Weak1 context, and the readthrough of the UGA stop codon was 2 times worse. Thus, the main factors determining readthrough efficiency are the type of stop codon and the sequence of the 3′ context.
The influence of the type of stop codon on readthrough is likely determined by the availability of suppressor or near-cognate tRNAs in the cell and by the ability of tRNA to recognize stop codons successfully. Thus, UAA and UAG codons, in addition to suppressor tRNAs, can be recognized by glutamine or tyrosine tRNA, whereas the UGA codon can be recognized by cysteine and tryptophan tRNAs (48, 49). It can be assumed that tryptophan tRNA, paired with the UGG codon, efficiently recognizes the UGA stop codon, since the first two nucleotides in these codons coincide, and there is a purine in the third position of both codons. Conversely, in the cysteine and tyrosine codons, where the first two nucleotides (UG and UA, respectively) match the stop codons, pyrimidine (C or U) is in the third position, which conformationally differs from the purine of the stop codon. In the glutamine codons CAA and CAG, nucleotides in the second and third positions match the UAA and UAG stop codons, but in the first position, there is a C. Since the first nucleotide of the codon is important for accurate pairing with the anticodon of tRNA, pairs of stop codons and glutamine tRNA are rarely formed in the ribosome. We believe that the similarity of the UGA stop codon with the tryptophan (UGG) codon determines a high percentage of its readthrough in any 3′ context (Table 1).
To ensure that readthrough differed at the same stop codons in the absence of eRFs, we explored the potential of the other 3′ contexts. For this purpose, we cloned four additional weak 3′ contexts following the UGA and UAG stop codons (Fig. 2A): CUA UAC (Weak2) and CUU AAA (Weak3) contexts, containing a probable weak CU dinucleotide, CAA UUA (TMV) context from the TMV (29), and GAU AAU (Dyst) context, which corresponds to the sequence followed the nonsense mutation 651d in the human dystrophin transcript variant Dp4271 (40). These contexts have previously been described as weak only for UAG or UGA stop codons. The effects of Weak1, Dyst, and TMV contexts were compared on the UAG stop codon, and the effects of Weak1, Weak2, and Weak3 on the UGA stop codon (Figs. 2B and S2). We revealed that the readthrough was more efficient on the UAG in the Weak1 context (10%) and in the TMV context (7.5%). In dystrophin context, the readthrough of the UAG did not differ from the Standard context (Fig. 2B and Table 1). On the UGA stop codon, all three contexts induced a higher level of readthrough than the Standard context (Fig. 2B and Table 1).
Thus, we revealed that the readthrough on the same stop codon in different 3′ contexts proceeded differently (Fig. 2B). This dependence has two possible explanations: (i) presence or absence of tRNA complementary to a certain 3′ context (strong/weak context, respectively) and (ii) recognition of certain 3′ contexts by other participants of the elongation cycle (ribosome or eEFs). The possible effect of tRNA availability on readthrough thus requires consideration. When complementary to the 3′ context, tRNA is absent, one round of elongation with near-cognate tRNA passing through the stop codon and accumulation of the ribosomal complex on the next sense codon should be observed. In our experiments, we did not observe this pattern of toe-printing when comparing the different contexts (Fig. S2). For example, on the UAA stop codon in the Standard context, there is practically no readthrough, but the intermediate peaks between the first and second stop codons completely coincide with the peaks of the Weak1 context, which gives 10 times more efficient readthrough. In addition, for the UGA stop codon, the intermediate peaks on the Standard context, giving 28% readthrough, and on the Weak1 context, giving 57% readthrough, are the same. These observations allowed us to conclude that the 3′ context is recognized by the ribosome or eEFs, and its effect on the stop codon readthrough does not depend on the presence of the tRNAs complementary to the 3′ context.
3′ stop codon contexts do not affect translation termination efficiency
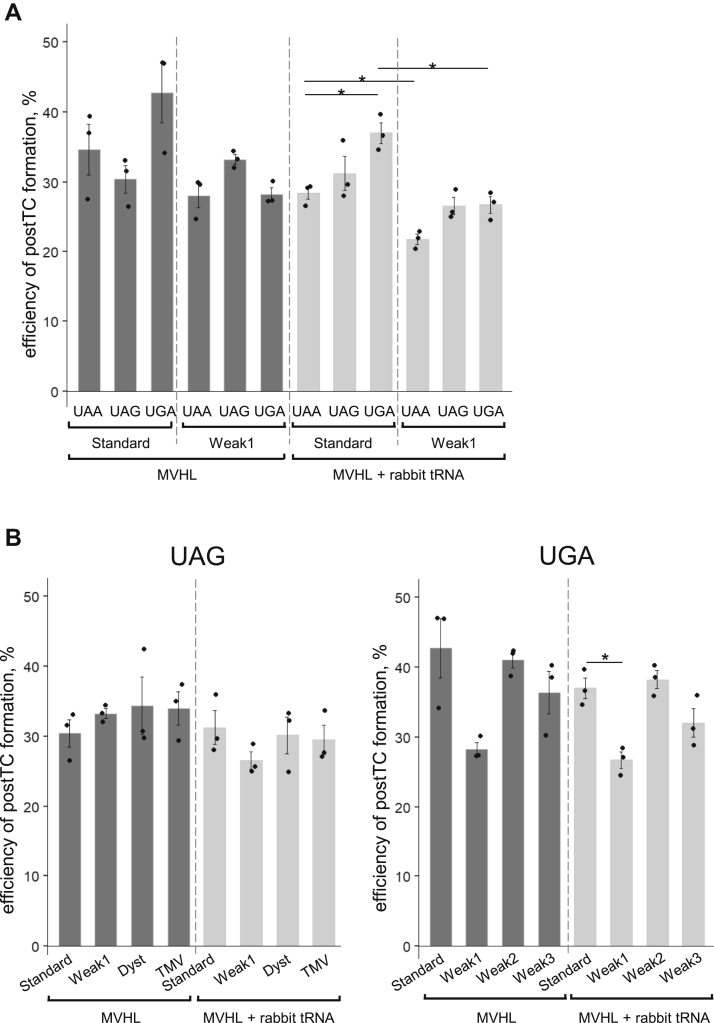
To determine how the chosen weak 3′ contexts affected translation termination, a complex of human release factors, eRF1-eRF3a-GTP, was added to preTC1s assembled in the presence of individual M, V, H, and L tRNAs (Figs. 3 and S3). During stop codon recognition by eRF1 and eRF3, the ribosome protects additional nucleotides on the mRNA, which can be detected in toe-printing assays as a one- or two-nucleotide forward shift of the ribosomal complex (3, 10, 11, 50). The preTC1 was paused at the first stop codon, and the addition of eRFs to this complex led to the appearance of a +2 peak corresponding to the post-termination complex (postTC) (Fig. S3). Translation termination efficiency was estimated based on the appearance of the postTC peak.
Figure 3.
Translation termination induced by eRF1–eRF3a complex on different stop codon contexts.A, postTC formation efficiency on all three stop codons in the Standard and Weak1 contexts in the absence and presence of rabbit tRNA. B, postTC formation efficiency on the UAG and UGA stop codons in the Standard, Weak1, Dyst, TMV, Weak2, and Weak3 contexts. For the Standard and Weak1 contexts, the data are duplicated from A. Individual measurements are shown as dots, mean ± SEM are shown on histogram. n = 3, ∗p ≤ 0.05, ∗∗p ≤ 0.01. Statistically insignificant differences are not shown. eRFs, eukaryotic release factors; TMV, tobacco mosaic virus; postTC, post-termination complex.
In the absence of competition with suppressor or near-cognate tRNAs, eRF1 recognized all stop codons with the same efficiency (visible differences are not significant). We did not find any dependence of translation termination efficiency on either the type of stop codon or the 3′ context (Fig. 3). However, when rabbit tRNA was added to the translation, we observed a slight but significant decrease in the efficiency of postTC peak formation on the UAA and UGA codons in the Weak1 context (Fig. 3A). No decrease in the postTC peak was observed for stop codons in other contexts (Fig. 3). We interpret this result as very efficient binding of near-cognate tRNA to the stop codon in the Weak1 context, so the readthrough can compete with translation termination.
Moreover, when rabbit tRNA and eRFs were present in the translation, toe-print peaks corresponding to preTC2 at the second stop codon did not form (Fig. S4). This indicates that eRFs completely suppressed the readthrough of the first stop codon. We can conclude that translation termination is a much more efficient process than stop codon readthrough. The complex of release factors successfully competes with near-cognate tRNAs for binding to stop codons, regardless of the type of stop codon and the 3′ context. Therefore, eRFs ensure high efficiency of translation termination in all 3′ contexts.
Influence of translation initiation factors on the stop codon readthrough
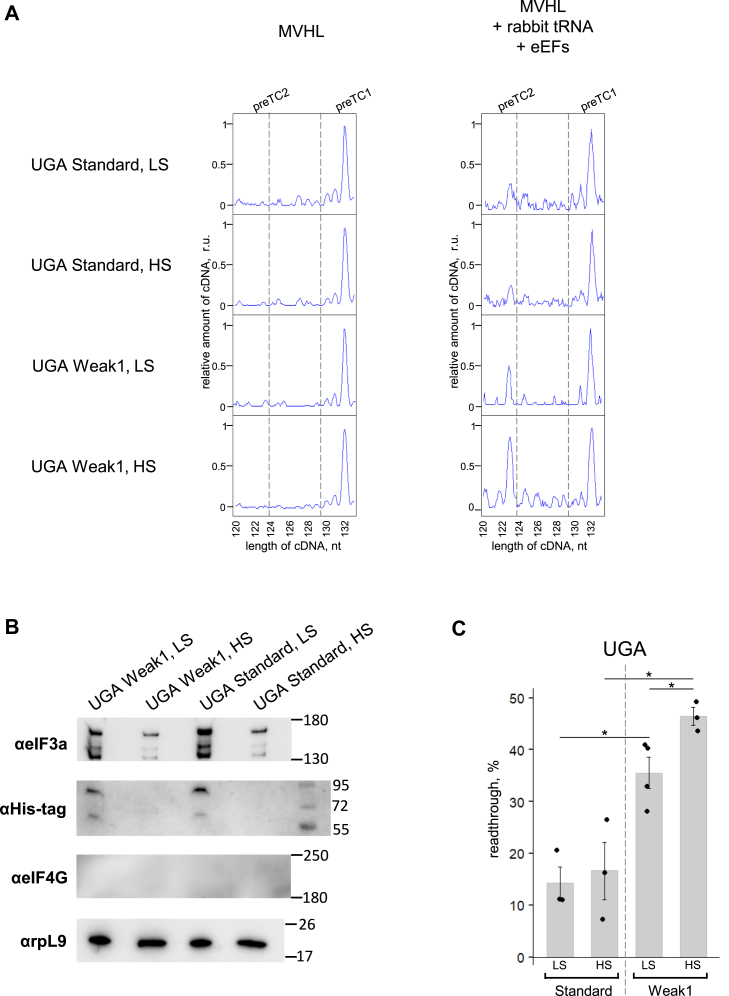
We recently showed that several eIFs, including eIF3 and eIF3j, stimulate translation termination (45). As we performed previous experiments on readthrough and termination in the reconstituted in vitro system in the presence of all translation factors, the influence of eIFs on these processes should be excluded. We performed sucrose gradient centrifugation of two preTCs assembled on the UGA stop codon after elongation with individual M, V, H, and L tRNAs in the Standard and Weak1 contexts in parallel (Fig. 4A). To control the composition of the resulting complexes, Western blotting was performed with antibodies specific to eIF3a, eIF4G, and His-tag, as all recombinant proteins used in the reconstructed system had a His-tag (Fig. 4B). Western blotting showed that under low-salt (LS) conditions, significant amounts of eIF3 remained in the preTC(LS), which was also shown in our previous study (45). After high-salt (HS) centrifugation, we observed a significant decrease of eIF3 in the preTC(HS). eIF4G is a scaffold protein that can contaminate ribosomes and native proteins. This protein was not detected by antibodies in any of the examined preTCs. Anti-His-tag antibodies revealed some amounts of recombinant eIFs in the preTC(LS), which were completely lost in the preTC(HS). Electrophoresis with recombinant eIF4B and eIF5 showed that these factors have the same mobility in PAAG (Fig. S5). As a control, we stained preTCs with antibodies specific to ribosomal protein L9 and demonstrated that the amount remained unchanged in all complexes.
Figure 4.
Readthrough efficiency dependence on the presence of eIFs.A, toe-printing of the purified preTC1 assembled on the UGA stop codon in the Standard and Weak1 contexts in high or low concentration of the salt. Readthrough efficiency was measured on the preTC1. Elongation factors and an excess of a.a. rabbit tRNA were added to induce readthrough on the first stop codon. B, Western blot analysis of the purified preTC1s with antibodies raised against eIF3a, His-tag, eIF4G, rpL9. C, readthrough efficiency on the UGA stop codon in the Standard and Weak1 contexts. Individual measurements are shown as dots, mean ± SEM are shown on histogram. n = 3 (4 for Weak1 LC), ∗p ≤ 0.05. Statistically insignificant differences are not shown. a.a. aminoacylated; eIFs, eukaryotic translation initiation factors; preTC1, pretermination complex 1.
After the addition of eEFs and rabbit tRNA, we analyzed the stop codon readthrough in purified preTCs (Fig. 4A). We found that readthrough occurred on both pure preTC(HS) and preTC(LS), containing eIF3, eIF5, and eIF4B (Fig. 4, A and C). We observed that the presence of eIFs in the preTC(LS) slightly decreased the readthrough in the Weak1 context. Interestingly, in the presence of all translation components, readthrough on the UGA stop codon was approximately 30% in the Standard context and 60% in the Weak1 context (Fig. 2); however, on the pure preTC1s, we observed 15% in the Standard context and 35 to 45% in the Weak1 context (Fig. 4). The overall level of readthrough decreased, but the ratio of readthrough in different 3′ contexts was the same. Thus, in a pure system, the readthrough was two times lower, but it still proceeded effectively in the presence of only rabbit tRNA and eEFs. We propose that eIFs are somehow involved in the process of tRNA binding to stop codons in this in vitro system, but they are not 3′ context-recognizing participants. Since the ratio of levels of readthrough was preserved, it can be confidently stated that the 3′ context of the stop codon is recognized by one of the participants of the elongation cycle (ribosome or eEFs) and no additional protein factors are involved in this process.
Mechanism of stop codon readthrough
It should be noted that Weak1, Weak3, and Dyst contexts contain stop codons that can be recognized when the reading frame is shifted forward by one or two nucleotides (Table in Fig. 2). Our data showed that readthrough occurred without frameshifts (Fig. S2). Thus, during stop codon readthrough, the reading frame is preserved, which indicates the usual elongation reaction and precise interaction between nucleotides of the stop codon and anticodon of near-cognate tRNAs.
Retention of the reading frame and synthesis of the corresponding peptides during readthrough were confirmed through analysis of the synthesized peptides (Fig. 5). PreTCs labeled with radioactive methionine were assembled on UAA Standard and UGA Weak1 mRNAs. They were then purified by centrifugation in a 10 to 30% sucrose gradient. After measuring the radioactivity in the fractions and obtaining the results of toe-printing, the fraction with the maximum signal of radioactivity was selected from each gradient, in which the presence of preTC was confirmed according to the results of toe-printing (Fig. 5A). Next, to perform termination and peptide release, eRF1 and eRF3a were added to the complexes. We observed the release of radiolabeled peptides in both preTCs (Table in Fig. 5A).
Figure 5.
Detection of the synthesized peptides during stop codon readthrough.А, toe-printing analysis of the purified preTCs assembled on the UAA Standard and UGA Weak1 contexts. Hydrolysis of peptidyl-tRNA induced by the addition of eRF1 and eRF3a to the preTCs. B, radio RP-HPLC analysis of released peptides after incubation with eRF1 and eRF3a. C, peptides detected by MALDI-TOF MS in the peak 1 and peak 2 obtained in radio RP-HPLC. c.p.m., counts per minute; eRFs, eukaryotic release factors; r.u., relative units; preTC, pretermination complex.
eRFs were also added to the remaining volume of the preTC (130 μl), and peptides released during translation termination were purified from the high-molecular-weight components by centrifugation through Ultracel-10K. The fraction of low molecular weight components was analyzed using reverse phase HPLC. As a result, a radioactive HPLC profile was obtained for both the samples (Fig. 5B). The profile of the Standard context showed only one distinct peak (peak 1), whereas the Weak1 context profile showed two radioactive peaks (peak 1 and peak 2). Next, the fractions corresponding to peaks 1 and 2 were analyzed by MALDI-TOF MS (Fig. 5C; raw data are shown in Figs. S6 and S7). As a result, we detected the tetrapeptide MVHL (in the peak 1) and two heptapeptides MVHLWLV and MVHLСLV (in the peak 2) on the UGA Weak1 template and did not detect additional peptides except for MVHL (in the peak 1) on the UAA Standard template. Thus, during stop codon readthrough, common translation elongation occurs and extended peptides are formed.
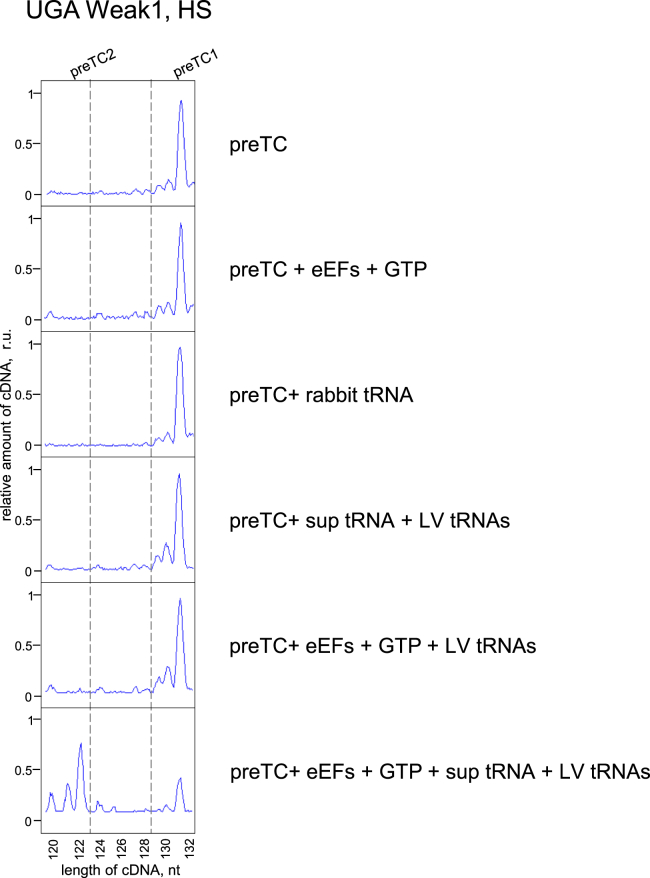
Based on these data and the results obtained on purified preTСs, we hypothesized that the ribosome, eEFs, and suppressor or near-cognate tRNA are the main participants in the process of stop codon readthrough. To verify this, we performed experiments with the individual L and V tRNAs, complementary to the codons of the Weak1 context, and synthetic Ser-tRNAsup on the preTC(HS) (Fig. 6). After the addition of eEFs or rabbit tRNA to preTC(HS), we did not observe any readthrough; however, a 35% readthrough was observed when both components were added (Fig. 4C). Thus, all components of translation elongation are required for readthrough, which is a result of stop codon recognition by a suitable tRNA. This was confirmed by the addition to preTC(HS) of purified synthetic L and V tRNA and suppressor tRNA in the presence of eEFs. The suppressor tRNA recognizes the stop codon, the following codons are recognized by L and V tRNA, and preTC2 is formed (Fig. 6). Thus, necessary and sufficient participants of the stop codon readthrough are stop codons in the correct 3′ context, the ribosome, suppressor or near-cognate tRNAs, and eEFs.
Figure 6.
Stop codon readthrough reconstituted from the individual components. Example of toe-printing analysis of the purified preTC1s after addition of different combination of eEFs, sup tRNA, and L, V tRNAs. PreTC1 was assembled on the UGA stop codon in the Weak1 context in the high salt concentration. ru, relative units. eEF, eukaryotic elongation factor; preTC1, pretermination complex 1; preTC2, pretermination complex 2.
Discussion
We conducted a study on the effect of the 3′ region of the stop codon on the efficiency of its readthrough and translation termination. Surprisingly, this study revealed that the “weak” contexts did not significantly suppress translation termination, while they induced stop codon readthrough entirely at the elongation stage (Figs. 2 and 6). Therefore, the ability of eRFs to recognize stop codons and cause peptide release is not affected by mRNA context. Thus, the efficiency of translation termination is based on equilibrium between the amount and affinity to the stop codon of eRFs and suppressor/near-cognate tRNAs.
During experiments on purified preTCs, we showed that the necessary and sufficient translational components for readthrough were the ribosome with the positioned stop codon in a certain context, suppressor or near-cognate tRNAs, and eEFs (Fig. 6). This is another round of elongation, which takes place if the stop codon in the ribosome is preferable for recognition by tRNA conformation. This was confirmed through experiments on the preservation of the reading frame and synthesis of the corresponding peptides (Fig. 5). It is noteworthy that eEFs and tRNAs are equally available to all mRNAs; therefore, the question remains on how the cell determines which stop codon will be read during readthrough. From our data, the determining factor for triggering stop codon readthrough is the recognition of a favorable 3′ context by the participants of elongation cycle (ribosome or eEFs).
These data are in agreement with previous studies that discussed the mechanism by which the 3′ context impacts readthrough. Previously, destabilization of the ribosomal secondary structure owing to the interaction between rRNA and mRNA was proposed, which led to an increased probability of the stop codon binding with tRNAs than with eRF1. Two regions of S. cerevisiae rRNA, potentially capable of pairing with the 3′ context on mRNA, were revealed (32). Computational analyses of 3′ UTRs from 14 eukaryotic species revealed 18S rRNA complementarity with the first 50 nucleotides of the 3′ UTRs, which form an evolutionarily conserved pattern localized around the ribosomal mRNA entry channel (51). Another study conducted on mammalian cells also proposed that translation termination efficiency is influenced by interactions of the ribosome with six mRNA nucleotides downstream of the stop codon, occupying the entry channel (36). Interestingly, in that study, it was shown that among weak contexts, only the UGA CUA sequence did not respond to increased cellular levels of eRF1, although reducing the eRF1 level through siRNAs dramatically increased the overall readthrough efficiency. This agrees with our data, which shows that weak contexts induce high levels of readthrough in the absence of eRFs, which is practically undetectable in the presence of eRFs (Figs. 2 and 3). Additionally, recent studies have implicated interactions between mRNA and ribosomal proteins in accounting for context effects in start codon selection (52, 53). It is possible that similar interactions can also occur during recognition of the 3′ context of the stop codon. Although we do not have structural data demonstrating the interaction of the stop codon 3′ context with the ribosome, our biochemical experimental data indicate that such an interaction may exist. In experiments with the preTC purified in the sucrose gradient, the stop codon readthrough depended on the mRNA 3′ context (Fig. 4). In this system, there are only three participants: preTC, a.a. tRNAs, and eEFs. Thus, one of the three participants in this reaction recognized the 3′ context. It cannot be a tRNA, complementary to the stop codon, since it does not have any structural features required for this recognition. Therefore, they are either ribosomes or eEFs. Considering the data on the recognition of the context of start codons and the absence of any data on the interaction of eEFs with the mRNA, we concluded that the weak stop codon contexts are more likely recognized by the ribosome.
This result is interesting from the perspective of the mechanism of the readthrough of premature termination codons (PTCs). It is noteworthy that the local concentrations of eRFs in various regions of the translated mRNA are different. At the stop codon of the main reading frame, eRFs are recruited and activated by the poly(A)-binding protein PABP, eIF3j, and other factors (42, 45, 46, 47). The stop codons of the upstream ORFs are recognized by eRFs recruited by eIF3j and eIF3 (45). However, the local concentration of eRFs near the PTC is low because it lacks determinants (poly(A) tail and 5′ end of mRNA) for the binding of protein loading and activating eRFs. This creates favorable conditions for stop codon readthrough, since at a reduced concentration of eRFs, suppressor or near-cognate tRNAs can compete with them more successfully. Thus, PTC readthrough should be more efficient than the readthrough of native stop codons. According to previous studies, the vast majority of 3′ stop codon contexts are neutral for the ribosome, that is, the ribosome is unable to recognize them. However, nonsense mutations occur randomly; therefore, the context of PTC is also random and unable to interact with the ribosome. Consequently, an increased level of PTC readthrough is not determined by the interaction of the 3′ context with the ribosome but depends only on the local concentration of eRFs, which is undoubtedly a cellular strategy to deal with emerging nonsense mutations.
In conclusion, it is noteworthy that the insensitivity of eRF1 to the 3′ context allows the ribosome to finish translation in the majority of the 3′ stop codon contexts. Such an approach is reasonable because any random mutation in this region leads to the immediate cessation of protein synthesis and wasting of cell resources. However, in some instances, a signal impairing translation termination is required. This function is realized by a small number of specific sequences capable of interacting with the ribosome.
Experimental procedures
Construction of model mRNAs
MVHL-stop mRNAs containing one of the three stop codons (UAA, UAG, or UGA) were described previously (3, 41). To obtain plasmid with the desired first stop codon, 3′ stop codon context and with the second UAA stop codon, an amplified fragment of pET28-MVHL plasmid (with the Bgl and mutant primers listed in the Supplementary Data Table S1 and S2) was cloned into original pET28-MVHL using Bgl II and Hind III restriction sites. To obtain mRNA, we amplified fragments of the obtained plasmids using Bgl and globin_rev primers. PCR product was transcribed by the T7 RiboMAX Express Large Scale RNA Production System kit (Promega) according to the manufacturer's protocol. mRNAs were purified sequentially by isolation in acidic phenol, precipitation with 3 M LiCl, and gel filtration on NAP-5 (Cytiva). The resulting mRNAs contained four CAA repeats, 5′UTR of 52 nt, an MVHL coding sequence ending with the first stop codon, 3′ context of 6 nt, second stop codon (UAA), and 3′UTR of 420 nt.
Construction and aminoacylation of individual tRNAs
To obtain individual tRNAs, we linearized by BstOI (Promega) the pUC18 plasmid, in which the human tRNA sequences were inserted. The suppressor tRNA was obtained from the serine tRNA sequence, in which the anticodon was changed to the UCA complementary to the UGA stop codon. Sup tRNA was aminoacylated with serine. The linearized fragment (in a final concentration of 0.1 g/l) was transcribed with the 8 u/μl T7 RNA polymerase (Thermofisher) in the presence of TBx1 buffer supplemented with 4 mM ATP, 4 mM UTP, 4 mM CTP, 4 mM GTP, 20 mM MgCl2, 10 mM DTT, 2% PEG 8000, 0.68 u/μl Ribolock (Thermofisher) on 37 °C for 4 h. Then, tRNAs were purified sequentially by isolation in acidic phenol and gel filtration on NAP-5 (Cytiva). Aminoacylation and isolation of the aminoacyl-tRNA synthetases were performed on the basis of the method described (54). For aminoacylation, a mixture containing 10 μM of tRNA, 0.5 g/l aminoacyl-tRNA synthetases from the S100 RRL fraction, 50 μM of the corresponding amino acid, 40 mM Tris–HCl pH 7.5, 15 mM MgCl2, 2 mM DTT, 10 mM ATP, 1 mM CTP, 0.5 u/μl Ribolock (Thermofisher) was incubated at 37 °C for 7 min. Then, a.a. tRNAs were purified sequentially by acidic phenol isolation and gel filtration on NAP-5 (Cytiva).
Ribosomal subunits and translation factors
40S and 60S ribosomal subunits, eEF1, and eEF2 were purified from a RRL cell lysate, and eIF2 and eIF3 were purified from a HeLa cell lysate, as described previously (3). Human translation factors eIF1, eIF1A, eIF4A, eIF4B, ΔeIF4G, ΔeIF5B, eIF5, eRF1 were produced as recombinant proteins in Escherichia coli strain BL21 with subsequent protein purification using Ni-NTA agarose and ion-exchange chromatography (3). Human full-sized eukaryotic release factor eRF3a (GSPT1), kindly provided by Dr Christiane Schaffitzel, was expressed in insect cell line Sf21 with baculovirus EMBacY from a MultiBac expression system and then purified as described previously (42).
PreTC1 assembly, stop codon readthrough, and translation termination
Ribosome complexes were assembled in vitro as previously described with modifications (3). ICs were assembled at 4 °C in 10 μl contained 0.375 pmol mRNA, 3 pmol Met-tRNAiMet, 1 pmol each of 40S and 60S ribosomal subunits, 5 pmol eIF2, 1 pmol eIF3, 7 pmol eIF4A, and 2 pmol each of ΔeIF4G, eIF4B, eIF1, eIF1A, eIF5, and ΔeIF5B, supplemented with buffer composed of 20 mM Tris acetate, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT, 0.4 U/μl Ribolock (Thermofisher), 1 mM ATP, 0.25 mM spermidine, and 0.2 mM GTP. The reaction mixture was kept at 37 °C for 15 min to allow ribosomal–mRNA complex formation. Peptide elongation (preTC1 assembly) was performed by the addition of 3 pmol a.a. Val, His and Leu tRNAs, 3 pmol eEF1, and 0.5 pmol eEF2 to the IC and was incubated for 15 min at 37 °C. Stop codon readthrough was performed by the addition of 120 pmol of a.a. total tRNA (rabbit, calf, or yeast), during elongation. Translation termination was performed by the addition to 10 μl sample containing preTC1 of 0.17 pmol eRF1 and eRF3a. During the competition termination-readthrough experiments, 120 pmol a.a. rabbit tRNA was added. The reaction mixture was incubated at 37 °C for 10 min. Ten microliter samples of each complexes were subsequently subjected to a toe-print assay.
Purified preTCs assembly
Either [35S]-labeled or unlabeled eukaryotic preTCs on MVHL-UAA_Standard, MVHL-UGA_Standard, and MVHL-UGA_Weak1 mRNAs were assembled and purified as previously described (3). Briefly, ICs were assembled in a 500-μl solution containing 37 pmol of mRNA, 200 pmol Met-tRNAiMet or [35S]-labeled Met-tRNAiMet, 90 pmol 40S and 60S ribosomal subunits, 200 pmol eIF2, 90 pmol eIF3, and 125 pmol of eIF4A, ΔeIF4G, eIF4B, eIF1, eIF1A, eIF5, ΔeIF5B supplemented with buffer A (25 mM Tris–HCl (pH 7.5), 50 mM KOAc, 2.5 mM MgCl2, 2 mM DTT, 0.3 U/μl Ribolock (Thermofisher), 1 mM ATP, 0.25 mM spermidine, and 0.2 mM GTP). The reaction mixture was maintained at 37 °C for 15 min to allow the ribosomal–mRNA complex formation. Peptide elongation was performed by addition of 150 pmol individual a.a. V, H, and L tRNAs (to obtain pure preTC1) or 6000 pmol total rabbit tRNA (to obtain radiolabeled peptides after readthrough), 200 pmol eEF1, and 50 pmol eEF2 to the IC, followed by incubation for another 15 min at 37 °C. The ribosomal complexes were centrifuged in a Beckman SW55 rotor for 95 min at 4 °C and 50,000 rpm in a linear sucrose density gradient (10–30%, w/w) prepared in buffer A containing 5 mM MgCl2 or in buffer A containing 5 mM MgCl2 and 300 mM KOAc for preTC1(HS) complexes. After centrifugation, 13 lower gradient fractions of 150 μl were collected, and fractions corresponding to the preTC1 were detected by a toe-printing assay and by the presence of [35S]-Met. The preTC fractions were combined and diluted 3-fold with buffer A containing 1.25 mM MgCl2 (to a final concentration of 2.5 mM Mg2+) and used in the peptide-release assay or for toe-printing analysis. Additionally, 10 μl of each preTC1(HS) and preTC1(LS) were analyzed by Western blotting using antibodies against eIF3a (Abcam 86146), eIF4GI (Cell Signaling 2858), His-tag (Miltenyi Biotec 130-092-785), and rpL9 (Abcam 182556).
Readthrough efficiency analysis on purified preTCs
Purified preTC(HS) and preTC(LS) on mRNAs with Standard or Weak1 contexts were used to determine readthrough efficiency in the presence of total tRNA or individual a.a. V, L, and sup tRNAs. For analysis, 30 μl aliquots containing 0.015 pmol preTCs were incubated at 37 °C for 15 min with 3 pmol eEF1, 0.5 pmol eEF2, and 120 pmol a.a. total tRNA or individual a.a. V, L, and sup tRNAs in the presence of 0.2 mM GTP. Samples were analyzed using toe-printing.
Toe-printing analysis
Toe-printing analyses were performed as described (50). The samples with preTCs or postTC were incubated with AMV reverse transcriptase and a 5′-FAM-labeled primer complementary to the 3′ UTR sequence. Complementary DNAs were separated by electrophoresis on an Syntol LLC Nanophor 05 Genetic Analyser (Syntol, LLC) using SD-450 size standard (Syntol, LLC) and PDMA-6 polymer (Syntol, LLC) by manufacturer protocol with injection time 30 s. The efficiency of readthrough was calculated using rfu signals for ribosomal complexes in the formula with subtraction of the mean signal in the absence of a.a. total tRNA. The efficiency of postTC formation was calculated as described (50) in the formula with subtraction of the mean signal in the absence of eRFs.
Statistical analysis
All experiments were carried out in at least 3 replicates (the specific replicates are shown as dots). All data are presented as mean ± SEM. An unpaired two-tailed t test was used to compare mean values between two groups. For multiple comparisons, the Holm correction of p value was used (55). The difference was considered significant when p value was less than 0.05 (∗) or 0.01 (∗∗).
p values were calculated using t_test (rstatix library) and p.adjust (stats library) functions in R.
Radio RP-HPLC and MALDI-TOF Mass Spectrometry of peptides
One hundred fifty microliters of radiolabeled preTCs, assembled on MVHL UAA Standard mRNA (40,000 c.p.m.) and MVHL UGA Weak1 mRNA (45,500 c.p.m.), were incubated with 70 pmol eRF1 and 10 pmol eRF3a with 0.2 mM GTP and 0.2 mM MgCl2 at 37 °C for 15 min.
Ribosomes, proteins and tRNA from 10 μl of samples were pelleted before and after peptide release with ice-cold 5% trichloroacetic acid and centrifuged at 14,000g at 4 °C for 15 min. The amount of released [35S]-peptides was determined by scintillation counting of supernatant using a liquid scintillation counter Tri-Carb 4810 (PerkinElmer Life Sciences). The rest of the samples were subjected to centrifugation through Ultracel – 10K centrifugal filters at 14,000g at 4 °C for 20 min. Flowthrough fractions with addition of 20 μg of equimolar unlabeled control peptides MVHL, MVHLCLV, and MVHLWLV were applied to Radio RP-HPLC.
Radio RP-HPLC of peptides was performed on a Gilson HPLC instrument, using Phenomenex Luna C18(2) column (5 μm, 100Å, 250 × 4.6 mm) and post column detection on a liquid scintillation counter Tri-Carb 4810 (PerkinElmer Life Sciences) using the Ultima-Gold scintillation cocktail (PerkinElmer). RP-HPLC separation was performed at a flow rate of 0.7 ml/min using a gradient of water (A)-acetonitrile (B), both (A) and (B) solvents contained 0.5% of propionic acid (v/v). The gradient profile was as follows: 0 to 5 min, 0% (B); 5 to 40 min, 0 to 100% (B); 40 to 45 min, 100% (B); 45 to 50 min 100 to 0% (B); and 50 to 55 min, 0% (B). Fractions were collected every 30 s (350 μl) and the elution profile of the [35S]-labeled peptides was monitored by scintillation counting of 50 μl aliquots (each aliquot was mixed with 450 μl of scintillation cocktail). The residual 300 μl of a fraction (if its radioactivity > 100 cpm (signal/noise >5)) were lyophilized on CentriVap Benchtop Vacuum Concentrator at 30 °C, dissolved in 20 μl of acetonitrile and analyzed by MALDI-TOF Mass Spectrometry.
The samples were spotted with matrix solution (10 mg/ml 2,5-Dihydroxybenzoic acid in 50% ACN/0.3% TFA) on the ground steel MALDI-TOF target and analyzed with MALDI-TOF MS. Spectra were acquired on Ultraflex II MALDI-TOF/TOF (Bruker Daltonics) in the positive reflector mode. Each spectrum was acquired from 500 laser shots. The monoisotopic masses registered in the spectra correspond to synthetic witness peptides.
Data availability
The data supporting the findings of this study are contained in the article and supporting information. All the source data generated for this study are available from the corresponding author (Dr Elena Alkalaeva; alkalaeva@eimb.ru) upon reasonable request.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest regarding the content of this article.
Acknowledgments
The study of the effect of the stop codon context on the efficiency of translation termination was supported by the Russian Science Foundation (Grant No. 19-74-10078). Mass spectrometry studies were performed using the "Human Proteome" resource center equipment within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021–2030) (№122030100168-2). We are grateful to Ludmila Frolova for providing us with plasmids encoding release factors, Tatyana Pestova and Christopher Hellen for providing us with plasmids encoding initiation factors, and Christiane Schaffitzel for providing us with plasmids encoding eRF3a. cDNA fragment analyses were performed by the center of the collective use "Genome" of EIMB RAS. We are thankful to the Centre for Precision Genome Editing and Genetic Technologies for Biomedicine for access to the facilities necessary for this study.
Author contributions
N. B., E. S., D. V. Y., I. Y. T., A. S., and T. E. investigation; N. B. and T. E. visualization; T. E. and E. A. writing–review and editing; E. A. supervision; E. A. conceptualization; E. A. writing–original draft.
Edited by Karin Musier-Forsyth
Contributor Information
Tatiana Egorova, Email: tatvladegorova@gmail.com.
Elena Alkalaeva, Email: alkalaeva@eimb.ru.
Supporting information
References
- 1.Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stansfield I., Jones K.M., Ter-Avanesyan M.D., Tuite5 M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In Vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura Y., Ito K., Ehrenberg M. Mimicry grasps reality in translation termination. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]
- 5.Voorhees R.M., Schmeing T.M., Kelley A.C., Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.des Georges A., Hashem Y., Unbehaun A., Grassucci R.A., Taylor D., Hellen C.U.T., et al. Structure of the mammalian ribosomal pre-termination complex associated with eRF1•eRF3•GDPNP. Nucl. Acids Res. 2014;42:3409–3418. doi: 10.1093/nar/gkt1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K., Frolova L., Seit-Nebi A., Karamyshev A., Kisselev L., Nakamura Y. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng T., Yamamoto A., Wilkins S.E., Sokolova E., Yates L.A., Münzel M., et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell. 2014;53:645–654. doi: 10.1016/j.molcel.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matheisl S., Berninghausen O., Becker T., Beckmann R. Structure of a human translation termination complex. Nucl. Acids Res. 2015;43:8615–8626. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao S., Murray J., Brown A., Taunton J., Ramakrishnan V., Hegde R.S. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell. 2016;167:1229–1240.e15. doi: 10.1016/j.cell.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeling K.M., Xue X., Gunn G., Bedwell D.M. Therapeutics based on stop codon readthrough. Annu. Rev. Genomics Hum. Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., et al. Evidence of efficient stop codon readthrough in four mammalian genes. Nucl. Acids Res. 2014;42:8928–8938. doi: 10.1093/nar/gku608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gärtner J., et al. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife. 2014;3 doi: 10.7554/eLife.03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jungreis I., Lin M.F., Spokony R., Chan C.S., Negre N., Victorsen A., et al. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21:2096–2113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013;2 doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitag J., Ast J., Bölker M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature. 2012;485:522–525. doi: 10.1038/nature11051. [DOI] [PubMed] [Google Scholar]
- 20.Chittum H.S., Lane W.S., Carlson B.A., Roller P.P., Lung F.D.T., Lee B.J., et al. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi Y., Hayashi A., Campagnoni C.W., Kimura A., Inuzuka T., Baba H. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J. Biol. Chem. 2012;287:17765–17776. doi: 10.1074/jbc.M111.314468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eswarappa S.M., Potdar A.A., Koch W.J., Fan Y., Vasu K., Lindner D., et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014;157:1605–1618. doi: 10.1016/j.cell.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loughran G., Jungreis I., Tzani I., Power M., Dmitriev R.I., Ivanov I.P., et al. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018;293:4434–4444. doi: 10.1074/jbc.M117.818526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brar G.A. Beyond the triplet code: context cues transform translation. Cell. 2016;167:1681–1692. doi: 10.1016/j.cell.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes – the impact of RNA sequence. RNA Biol. 2015;12:950–958. doi: 10.1080/15476286.2015.1068497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baranov P.V., Atkins J.F., Yordanova M.M. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat. Rev. Genet. 2015;16:517–529. doi: 10.1038/nrg3963. [DOI] [PubMed] [Google Scholar]
- 27.Bertram G., Innes S., Minella O., Richardson J., Stansfield I. Endless possibilities: translation termination and stop codon recognition. Microbiology. 2001;147:255–269. doi: 10.1099/00221287-147-2-255. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen W.T., Curran J.F. Effects of the nucleotide 3??? To an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J. Mol. Biol. 1991;219:231–241. doi: 10.1016/0022-2836(91)90564-m. [DOI] [PubMed] [Google Scholar]
- 29.Skuzeski J.M., Nichols L.M., Gesteland R.F., Atkins J.F. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- 30.Li G., Rice C.M. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol. 1993;67:5062–5067. doi: 10.1128/jvi.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole E.S., Major L.L., Mannering S.A., Tate W.P. Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucl. Acids Res. 1998;26:954–960. doi: 10.1093/nar/26.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namy O., Hatin I., Rousset J.P. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavener D.R., Ray S.C. Eukaryotic start and stop translation sites. Nucl. Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cridge A.G., Major L.L., Mahagaonkar A.A., Poole E.S., Isaksson L.A., Tate W.P. Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucl. Acids Res. 2006;34:1959–1973. doi: 10.1093/nar/gkl074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams I., Richardson J., Starkey A., Stansfield I. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucl. Acids Res. 2004;32:6605–6616. doi: 10.1093/nar/gkh1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cridge A.G., Crowe-McAuliffe C., Mathew S.F., Tate W.P. Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucl. Acids Res. 2018;46:1927–1944. doi: 10.1093/nar/gkx1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beznosková P., Wagner S., Jansen M.E., Von Der Haar T., Valášek L.S. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucl. Acids Res. 2015;43:5099–5111. doi: 10.1093/nar/gkv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loenarz C., Sekirnik R., Thalhammer A., Ge W., Spivakovsky E., Mackeen M.M., et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sokolova E.E., Vlasov P.K., Egorova T.V., Shuvalov A.V., Alkalaeva E.Z. The influence of A/G composition of 3’ stop codon contexts on translation termination efficiency in eukaryotes. Mol. Biol. 2020;54:739–748. doi: 10.31857/S0026898420050080. [DOI] [PubMed] [Google Scholar]
- 40.Bidou L., Hatin I., Perez N., Allamand V., Panthier J., Rousset J. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 2004;11:619–627. doi: 10.1038/sj.gt.3302211. [DOI] [PubMed] [Google Scholar]
- 41.Alkalaeva E., Eliseev B., Ambrogelly A., Vlasov P., Kondrashov F.A., Gundllapalli S., et al. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009;583:3455–3460. doi: 10.1016/j.febslet.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov A., Mikhailova T., Eliseev B., Yeramala L., Sokolova E., Susorov D., et al. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucl. Acids Res. 2016;44:7766–7776. doi: 10.1093/nar/gkw635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kryuchkova P., Grishin A., Eliseev B., Karyagina A., Frolova L., Alkalaeva E. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucl. Acids Res. 2013;41:4573–4586. doi: 10.1093/nar/gkt113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Susorov D., Mikhailova T., Ivanov A., Sokolova E., Alkalaeva E. Stabilization of eukaryotic ribosomal termination complexes by deacylated tRNA. Nucl. Acids Res. 2015;43:3332–3343. doi: 10.1093/nar/gkv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egorova T., Biziaev N., Shuvalov A., Sokolova E., Mukba S., Evmenov K., et al. eIF3j facilitates loading of release factors into the ribosome. Nucl. Acids Res. 2021;49:11181–11196. doi: 10.1093/nar/gkab854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuvalova E., Egorova T., Ivanov A., Shuvalov A., Biziaev N., Mukba S., et al. Discovery of a novel role of tumor suppressor PDCD4 in stimulation of translation termination. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov A., Shuvalova E., Egorova T., Shuvalov A., Sokolova E., Bizyaev N., et al. Polyadenylate-binding protein-interacting proteins PAIP1 and PAIP2 affect translation termination. J. Biol. Chem. 2019;294:8630–8639. doi: 10.1074/jbc.RA118.006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beznosková P., Bidou L., Namy O., Valášek L.S. Increased expression of tryptophan and tyrosine tRNAs elevates stop codon readthrough of reporter systems in human cell lines. Nucl. Acids Res. 2021;49:5202–5215. doi: 10.1093/nar/gkab315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beznosková P., Gunišová S., Valášek L.S. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016;22:456–466. doi: 10.1261/rna.054452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egorova T., Sokolova E., Shuvalova E., Matrosova V., Shuvalov A., Alkalaeva E. Fluorescent toeprinting to study the dynamics of ribosomal complexes. Methods. 2019;162–163:54–59. doi: 10.1016/j.ymeth.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Pánek J., Kolář M., Herrmannová A., Valášek L.S. A systematic computational analysis of the rRNA–3′ UTR sequence complementarity suggests a regulatory mechanism influencing post-termination events in metazoan translation. RNA. 2016;22:957–967. doi: 10.1261/rna.056119.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong J., Aitken C.E., Thakur A., Shin B.-S., Lorsch J.R., Hinnebusch A.G. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2126–E2135. doi: 10.1073/pnas.1620569114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong J., Hinnebusch A.G. uS5/Rps2 residues at the 40S ribosome entry channel enhance initiation at suboptimal start codons in vivo. Genetics. 2022;220 doi: 10.1093/genetics/iyab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pestova T.V., Hellen C.U.T. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are contained in the article and supporting information. All the source data generated for this study are available from the corresponding author (Dr Elena Alkalaeva; alkalaeva@eimb.ru) upon reasonable request.