Abstract
Cancer cells undergo diverse metabolic adaptations to meet the energetic demands imposed by dysregulated growth and proliferation. Assessing metabolism in intact tumors allows the investigator to observe the combined metabolic effects of numerous cancer cell-intrinsic and -extrinsic factors that cannot be fully captured in culture models. We have developed methods to use stable isotope-labeled nutrients (e.g., [13C]glucose) to probe metabolic activity within intact tumors in vivo, in mice and humans. In these methods, the labeled nutrient is introduced to the circulation through an intravenous catheter prior to surgical resection of the tumor and adjacent nonmalignant tissue. Metabolism within these tissues during the infusion transfers the isotope label into metabolic intermediates from pathways supplied by the infused nutrient. Extracting metabolites from surgical specimens and analyzing their isotope labeling patterns provides information about metabolism in the tissue. We provide detailed information about this technique, from introduction of the labeled tracer through data analysis and interpretation, including streamlined approaches to quantify isotope labeling in informative metabolites extracted from tissue samples. We focus on infusions with [13C]glucose and the application of mass spectrometry to assess isotope labeling in intermediates from central metabolic pathways, including glycolysis, the tricarboxylic acid cycle and nonessential amino acid synthesis. We outline practical considerations to apply these methods to human subjects undergoing surgical resections of solid tumors. We also discuss the method’s versatility and consider the relative advantages and limitations of alternative approaches to introduce the tracer, harvest the tissue and analyze the data.
Introduction
Proper control of metabolism is required for normal tissue function and health. Alterations in metabolism contribute to many diseases, and importantly, many of these diseases can be treated by reversing the perturbations or identifying ways to compensate for them. In cancer, perturbed metabolism is a phenotypic hallmark as transformed cells modify key pathways (e.g., energy formation, macromolecular synthesis and redox homeostasis) in order to support malignant levels of growth. Several diagnostic modalities and therapies exploit the perturbed metabolic state of tumors, emphasizing the clinical value in understanding metabolic reprogramming1.
A great deal of research in cancer metabolism has been performed in ex vivo model systems. With the increasing application of metabolic flux analysis and metabolomics to cancer research, cell culture models have been the most prominent experimental systems over the last few decades. These systems allow for precise experimental control and have been highly effective at identifying regulatory mechanisms governing metabolic flux2. Their major limitation is that they do not fully recapture all of the processes that impact metabolism in bona fide tumors, particularly cell-extrinsic factors, including the heterogeneous mix of cell types, variable nutrient availability, disordered mechanical forces and systemic metabolic influences of the host. With the realization that these processes profoundly impact the metabolic properties of malignant cells, there has been an increasing demand for versatile, informative methods to study tumor metabolism in vivo.
Metabolic tracing in vivo
Metabolic tracing with stable isotopes has been used for years to assess metabolite turnover, metabolic fluxes and disease-associated metabolic perturbations in humans and animal models. This technique involves introducing a nutrient labeled with a stable isotope—that is, an isotope that does not undergo radioactive decay but can be recognized by mass spectrometry (MS) or nuclear magnetic resonance (NMR) spectroscopy—into the subject. Routes of introduction may be enteral (e.g., via eating or drinking) or parenteral (intravenous or intraperitoneal injection). 13C is commonly used to track carbon fates from glucose and other nutrients. Transfer of the isotope label from the nutrient tracer into downstream metabolites occurs as a result of metabolic activity in tissues. Comparing metabolite labeling patterns between the blood and the tissue or between tumor and adjacent nonmalignant tissue can report tissue-specific metabolic activities. Metabolomics and stable isotope tracing provide complementary views of the metabolic phenotype, with metabolomics providing a broad view of steady-state metabolite abundance and isotope tracing adding a component of dynamic metabolite turnover. Advancements in the resolving power and sensitivity of MS systems have increased the variety of metabolites that can be analyzed for abundance and labeling. Other techniques can be integrated into the workflow of isotope-tracing experiments to assign metabolic activities to particular cell types or even organelles within the tissue.
In this protocol, we describe methods using stable isotopes to study cancer metabolism in mice and patients. The approach is informative and versatile, capable of studying tumors at many anatomic locations and straightforward to combine with drug treatments, dietary modifications and other parameters. We discuss experimental considerations, including the choice of tracer, method of delivery, metabolite extraction, MS and data analysis.
Development of the protocol
We initially developed this protocol to analyze human tumor metabolism through the intravenous infusion of labeled nutrients (e.g., [13C]glucose) in patients. The methods described in this protocol have been applied to a variety of tumor types in patients, including glioblastoma3, lung cancer4,5, renal cell carcinoma6 and solid extracranial malignancies in children7. Similar approaches have been translated to mouse models of cancer in our laboratory and many others8-12.
Delivery of the tracer
In human cancer metabolism studies, isotopes have been delivered intravenously as a simple bolus13 or by continuous infusion, with or without an initial bolus. Each approach introduces 13C-labeled metabolites into the circulation, resulting in the labeling of downstream metabolic pathways. However, important differences between the methods should be considered.
Metabolite labeling in tumors depends on multiple factors, including enrichment of the precursor pool, metabolism in other organs, and the time required to establish complex labeling patterns. Advantages of the bolus-only method include ease of delivery, with the tracer usually delivered as a short intravenous dose in the preoperative holding room. The lack of a continuous infusion also reduces the total amount of the tracer required, and therefore reduces the cost. Bolus administration of isotope-labeled nutrients can provide high-quality data about metabolic activity in human tumors13,14. The relative limitations of bolus-only administration include lower overall 13C enrichment in target metabolites and non-steady-state labeling dynamics4. These factors can reduce the richness of the data yield, particularly for labeling patterns that require an extended time to develop, such as successive turns of the tricarboxylic acid (TCA) cycle.
Continuous administration of the tracer addresses these issues, thereby improving some aspects of computational modeling of the data. A challenge associated with persistent delivery through continuous label infusion is the impact of metabolism in tissues other than the tumor, which introduces labeled intermediates into the circulation over time. These labeled intermediates need to be considered as potential sources of label into the tumor.
Optimizing extent of labeling
We have also used continuous isotope infusions without the initial bolus component to assess cancer metabolism. In the glioma patients assessed by this method, steady-state labeling in the plasma was not achieved within a practical time frame, and NMR analysis of tumor metabolites was limited by low 13C enrichment3. Extending the infusion period would likely address these limitations, but we have found that providing the isotope as an initial bolus followed by an infusion of 2–3 h is both practical for human surgical studies and capable of producing high-quality labeling data. In a small number of surgical cases in non-small cell lung cancer (NSCLC), the infusion of [13C]glucose extended up to 6–10 h. In these rare cases, we did not observe an appreciable difference in the labeling of central carbon metabolites (e.g., glycolytic metabolites and TCA cycle intermediates) when compared with our standard 3 h infusion protocol. This finding suggests that isotopic steady state is approached within 3 h in these infusions, at least in central carbon metabolism. Note that considerable label transfer into lipids and other macromolecules may take longer than the time necessary to approach steady state in precursors from the TCA cycle and other central pathways. Our limited analysis of tumor lipids has not revealed labeling from [13C]glucose in lung cancer.
Other methods of tracer delivery
Isotope tracers can also be administrated via the diet, eliminating the complications of anesthesia or surgical procedures for delivery15,16. The duration of delivery can extend from hours to days, thus allowing for isotopic enrichment of metabolites synthesized de novo, including macromolecules such as nucleotides. An advantage of this method is the physiological route of tracer entry. Potential challenges include inconsistencies in feeding, although published studies have reported acceptable consistency among mice subjected to these methods. The prolonged exposure to labeled nutrients may complicate some aspects of the data analysis and interpretation, as the label is transferred to a large number of metabolic intermediates over time, and these intermediates supply many different pathways.
Physiological responses to 13C infusions
In developing these methods, we optimized multiple parameters that influenced the extent of labeling and physiological responses. The effects of the isotope label appear to be negligible in our experience, but the nutrients themselves do induce metabolic effects, as expected. These effects could become pathological if the infusion causes persistent, supraphysiological levels of the nutrient in the circulation, so the rate of infusion must be tailored to minimize these risks. Brisk [13C]glucose infusion may induce physiological insulin release by the pancreas, and hyperglycemia could occur if the infusion rate is too high. Although our protocols allow hyperglycemic episodes to be treated with exogenous insulin administered by the anesthesiologist, this is expected to have undesirable effects on glucose metabolism in the tumor and elsewhere in the body. We therefore aim to avoid even transient hyperglycemia. During [13C]lactate infusions, we monitor patients for metabolic acidosis by measuring pH, bicarbonate and partial pressure of CO2 in the blood. Lactate is metabolized rapidly, and we have not observed episodes of metabolic acidosis, although a few patients had respiratory acidosis during the procedure (i.e., low pH and high partial pressure of CO2 without a reduction in bicarbonate). This condition is easily distinguished from metabolic acisosis, is unrelated to the infusion, and typically resolves rapidly by increasing the rate of mechanical ventilation5.
Other physiological consequences should be considered for each new nutrient. For example, glutamine is a major inter-organ shuttle of nitrogen, so infusing [13C]glutamine introduces nitrogen that ultimately needs to be converted to urea in the liver. Caution should be used in patients with liver disease. Furthermore, the spontaneous deamidation of [13C]glutamine in solution can result in a substantial amount of ammonia in the stock, and this could be dangerous to patients with compromised hepatic function. We measure free ammonia in stocks of [13C]glutamine prior to infusion into patients.
Integration into surgical workflows
We designed the protocol to be minimally disruptive to the standard surgical workflow during tumor resections or biopsies in patients. Ensuring that the priming bolus has been completed and the continuous infusion has started before the patient is moved to surgery is an important step. This eliminates the need for the research team to be at the bedside in the operating room, reducing any interruption of workflow during surgical preparation. Blood draws for isotope analysis are performed in concert with the anesthesia team, and can often coincide with standard-of-care blood draws.
Once the surgeon has removed the tumor, if possible, the samples for metabolic analysis are chosen together with the surgeon and delivered to the research team before the specimen is sent to pathology. Tumor fragments acquired at the end of the infusion are immediately frozen in the operating room, and later subjected to metabolite extraction and MS to determine how the tracer supplies carbon to central metabolic pathways, particularly glycolysis, the TCA cycle, the pentose phosphate pathway and amino acid synthesis. In cases where the tumor margin is of critical importance, the samples must be collected under the supervision of a pathologist in the operating room.
Applications of the method
This approach has been applied to numerous tumor types in numerous tissues. The discussion below draws heavily from our experiences in cancer, but the general method can also be applied to other contexts in which diseased tissues are sampled by surgery or biopsy, particularly in tissues with rapid metabolite turnover. These could include disorders of the liver or kidney, skeletal myopathies, cardiomyopathies, diabetes and other conditions. Isotope infusions have also long been used to assess physiological metabolic changes, including changes induced by exercise17-19.
Depending on the size of the tumor, several tumor regions can be sampled, allowing for the examination of intratumoral metabolic heterogeneity. However, a limiting factor of these experiments is that the analysis has typically been performed in bulk tissue samples, consisting of a mixture of cell types. While many tumors are primarily composed of cancer cells, essentially all contain nonmalignant cells as well (endothelial cells, fibroblasts, immune cells), and in some tumors these outnumber the cancer cells. Histopathological examination on adjacent fragments can help assess the relative proportions of tumor cells compared with nonmalignant stromal cells.
A major technical challenge at present is to develop ways to reliably assess the discrete metabolic activities of different cell types within a complex microenvironment. A recent study in mice demonstrated the proof of principle that specific cell populations can be sorted and studied from a bulk sample. In this example, mice were infused with 13C-glucose, with and without infection by Listeria. T cells were then enriched from bulk spleen tissue and analyzed by MS to identify infection-specific aspects of T-cell metabolism20. In principle similar methods could be applied to other cell types.
Experimental design
Overview
The instructions are divided into three procedures. Procedure 1 describes the preparation of clinical samples, starting with assembling the team and the candidate patients. Procedure 2 describes infusion in mice. Procedure 3 describes the MS analysis.
Advanced planning, flexibility and communication
Unexpected deviations from the planned timing of these studies occur commonly in the operating room because of the complexity of surgeries in patients. It is important to maintain a seamless flow of communication between the clinical and research team and, above all, for the research team to ‘go with the flow’, keeping the patient’s safety and clinical needs as the top priority. Last-minute scheduling changes and alterations of the surgical approach based on real-time observations in the operating room can derail the study if communication is ineffective. Patients may also decide to withdraw consent for any of a number of reasons, and it is valuable to receive this information prior to preparing the [13C]-labeled material for infusion. Other pitfalls to keep in mind:
Unexpected delays may result in a cessation of [13C] infusion prior to removal of the tumor unless the team knows about this and prepares additional material
Alternatively, some surgeries are unexpectedly short, making it difficult to reach a persistent state of labeling. For procedures expected to be short, it is advisable (if possible) to initiate the infusion earlier in the preoperative holding period
Some medications, particularly corticosteroids, can induce hyperglycemia and make it difficult to achieve adequate labeling in the circulating glucose pool
Prolonged devascularization of the tumor and adjacent tissue may cause artifacts in metabolite labeling. In our experience, 13C labeling is relatively durable after devascularization7, but it is important to minimize this time as much as possible while not interfering with the surgical standard of care. Biopsies in nondevascularized tissues likely provide a less perturbed view of metabolism, but we have observed similar labeling between tumors analyzed by biopsy versus resection in NSCLC5
While every effort should be made to freeze tumor samples in liquid nitrogen immediately after removing from the patient, the complexity of the surgery, needs of the clinical pathologist and other issues make delays unavoidable in some cases. Deviations from the standard workflow should be documented and considered when analyzing the data
To minimize the contributions of labeled metabolites from blood, the samples should be rinsed briefly in ice-cold saline before freezing, as the presence of large amounts of blood in a tissue sample can alter the measurement of metabolite levels or labeling patterns. Samples with obvious hemorrhage should be excluded
Clinical infusions
The labeled nutrients are prepared before surgery under standard high-risk compounding within the United States Pharmacopeia guidelines. Under these guidelines, the labeled nutrients can be stored up to 30 h at ambient temperature, but are typically prepared no more than 12 h before surgery. We have provided specific details for the infusion of glucose. If you use a different nutrient, then refer to Table 1 for advice. If your nutrient of interest is not in this table, then it is important to consider factors such as its physiological concentration to help choose a priming dose and rate of infusion. Communication with the clinical pharmacy team about this and other factors helps ensure safe delivery of the labeled nutrient.
Table 1 ∣.
Calculations for 13C infusions in human patients
| Metabolite | MWt | Stock | Bolus | Infusion (calculated for 150 min) |
|---|---|---|---|---|
| [U-13C]glucose | 186.1 g/mol | 133 g/L (715 mmol/L) | 4.3 mmol/min × 10 mins (43 mmol = 8 g) | 0.36 mmol/min (54 mmol = 10 g) |
| Na [U-13C]lactate | 115.05 g/mol | 40.3 g/L (350 mmol/L) | 30 μmol/kg/min × 5 min (for 75 kg: 1.29 g) | 10 μmol/kg/min (for 75 kg: 12.9 g) |
| Na [U-13C]acetate | 84.02 g/mol | 30–37 g/L (357–440 mmol/L) | 36 μmol/kg/min × 5 min (for 75 kg: 1.13 g) | 18 μmol/kg/min (for 75 kg: 17.0 g) |
The amount of tissue required for analysis depends on the the sensitivity of the analytical platform used. We have obtained adequate signal with as little as ~5 mg of tissue on gas chromatography–mass spectrometry (GC-MS), and other MS platforms offer even better sensitivity for metabolite detection. We recommend a pilot experiment with the relevant tissue to determine the amount of material necessary to detect metabolites of interest. These preliminary experiments can be performed using an analogous tissue sample from mice if the clinical sample is limited. Otherwise, samples can be measured by weight or protein content to track the amount of sample necessary to obtain high-quality data on the mass spectrometer.
We recommend analyzing samples from various positions within the tumor and the surrounding tissue. Tissue sampling can be planned in advance with the help of clinical imaging such as 18FDG-PET to identify regions of heterogeneous glucose uptake, or proximity to anatomical landmarks clearly visible on MRI (e.g., blood vessels, bronchi or other structures). In general, larger tumors increase the likelihood of regional metabolic heterogeneity. Nevertheless, we have observed large tumors with remarkably similar metabolic characteristics from different sampling locations, and overall that different regions of the same tumor are more similar to each other than regions acquired from different tumors4. Conversely, we have sometimes observed metabolic heterogeneity in tumors as small as 1 cm in diameter.
[13C] infusions in mouse models of cancer
This part of the protocol describes the use of intravenous isotope infusions to assess tumor metabolism in mice, using methods similar in design to those described for patients. We focus on xenograft-bearing NOD.CB17-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice21, although the approach is versatile and can be used in other models, including genetically modified mice with autochthonous tumors. We place a catheter in the lateral tail vein while the mice are anesthetized and continuously infuse with the isotope-labeled nutrient at concentrations and rates described below. Our use of anesthesia is designed to mimic the surgical conditions in human subjects. Infusions can also be performed in nonanesthetized mice using jugular vein catheters tunneled subcutaneously to exit behind the neck22,23. These are particularly useful for prolonged infusions and experiments studying the brain, where general anesthesia is undesirable.
In subcutaneous xenografts, tumor size is an important consideration when analyzing the metabolome, and consistency in tumor size may improve reproducibility11. Regional heterogeneity in metabolite abundance in large subcutaneous tumors has been demonstrated to influence gene expression and other aspects of tumor biology24. In our experience, tumors well below or above 1 cm in diameter may display differences in total metabolite enrichment even if the isotopologue distribution is conserved.
The doses in Table 2 were chosen from published literature, concentrations of each nutrient in the circulation and empirical observations of nutrient enrichment during infusion experiments. In cases where we were unable to reference published infusions in mice, we optimized infusion parameters to achieve 30–40% enrichment in the circulation. If you need to try a different metabolite, a similar procedure of optimization can be performed. It is also important to consider the physiological responses and measurements of the administered nutrient. Multiple animal models (e.g., rat, pig) can also be used for these infusions. Each model will likely require optimization, as nutrient turnover rates may differ considerably.
Table 2 ∣.
13C nutrient measurements for mouse infusions
| Labeled nutrient | Dose (g/kg) | Mouse weight (g) | 13C amount (mg) | Total volume (μL) |
|---|---|---|---|---|
| Glucose | 2.48 | 25 | 62 | 750 |
| Lactate | 1.44 | 25 | 36 | 750 |
| Glutamine | 1.73 | 25 | 43 | 1,500 |
The infusion parameters described in Step 17 in Procedure 2 were determined by similar principles. We initially mimicked the glucose infusion parameters for other labeled nutrients. During these optimization experiments, we noted that delivering lactate rapidly over a 1 min bolus was poorly tolerated by the mice, so we lowered the priming rate to deliver the bolus over 10 min.
It is important to practice the tissue harvesting procedure; in our experience, this leads to facile and rapid sample handling and improves reproducibility of the metabolite quality and yield. Metabolites vary widely in their stability. The concentrations of some metabolites, e.g., change only slightly in the 10 min after removal from the mouse. We have also found that the degree and nature of the isotope labeling is also stable7.
However, some metabolites—for example, samples containing adenine (adenosine, S-adenosyl methionine, etc.)—are highly labile, and their degradation produces adenine, making it difficult to analyze labeling patterns. Furthermore, if it is important to quantify the abundance of any metabolite, it is even more important to freeze the sample as quickly as possible, as delays will create numerous artifacts.
To improve reproducibility in measurement by MS, strategies such as spiked-in controls can be used to assess efficiency and quantify metabolites of interest. These internal controls (e.g., d27 myristic acid, norvaline, etc.) can be introduced during processing of the tissue sample. If particular metabolites are essential to the hypothesis being tested, it is advantageous to test the lability of those metabolites separately. This can be done by testing standards for degradation over time.
Metabolite analysis by MS
Several MS platforms can be used to assess isotope labeling. Multiple excellent protocols are available for gas chromatography/MS25,26, liquid chromatography/tandem MS27,28 and high-resolution MS29. For amino acids and intermediates from the TCA cycle and other central pathways, analysis with a single quadrupole mass spectrometer (e.g., GC-MS) is a straightforward and reliable approach, but requires derivatization (e.g., by tert-butyldimethylsilyl) of the metabolites.
Gas chromatography/MS.
Gas chromatography/MS is a reliable, well-established means of measuring isotopically labeled compounds from central metabolic pathways. Limitations of GC-MS include that it requires that metabolites be volatilized into gas phase for analysis, usually with the aid of a separate derivatization step. Single quadrupole mass spectrometers are also generally less sensitive than the high-resolution platforms described below, although this is rarely an issue for abundant metabolites from central pathways.
Liquid chromatography–high-resolution mass spectrometry.
Liquid chromatography–high-resolution mass spectrometry offers enhanced sensitivity, eliminates the need for derivatization to volatilize metabolites, and provides a much more accurate assessment of metabolite masses. Together, these differences broaden the scope of labeled metabolites that can be detected and discriminate multiple isotope labels simultaneously.
Equipment.
Multiple high-resolution mass spectrometer (HRMS) systems can be used. In this example, we describe settings for a ThermoScientific Q-Exactive. We recommend coupling the HRMS system to an ultra-high-performance liquid chromatography (UHPLC) module capable of constant flow rates with low variability. The system must be stable to low-concentration salts and moderately basic pH.
Expertise needed to implement this workflow
Preparation of 13C nutrients for patient infusion should be performed by a clinical pharmacist
Placement of intravenous lines in the patient should be performed by a registered nurse or medical doctor
Acquisition and interpretation of 13C labeling as described here requires access to mass spectrometers and analytical expertise
Materials
Reagents
GC/MS derivitatization
N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide, >97% (Sigma-Aldrich, 394882)
Methoxyamine hydrochloride (Sigma Aldrich, 226904)
Pyridine >99.9% (Sigma Aldrich, 270407)
Human infusion
[13C] labeled nutrient, certified as sterile and pyrogen-free, e.g., [U-13C]glucose, >99% atom (Cambridge Isotope Laboratories, CLM 1396-MPT-PK) (see ‘Reagent setup’ for more detail)
Liquid nitrogen (Airgas, cat. no. NI NF240LT22)
Mouse infusion
[U-13C]glucose, >99% atom (Cambridge Isotope Laboratories, CLM-1396. MilliporeSigma, cat. no. 389374)
[U-13C]lactate, >99% atom (Cambridge Isotope Laboratories, CLM-1579. MilliporeSigma, cat. no. 485926)
[3-13C]lactate, >99% atom (Cambridge Isotope Laboratories, CLM-1578. MilliporeSigma, cat. no. 490040)
[U-13C]glutamine, >99% atom (Cambridge Isotope Laboratories, CLM-1822 (alternative 13C, 15N, 2H molecules can be purchased for customized studies))
Ketamine/xylazine, 30 mg/mL (ARC Drug Services)
Ketamine, 100 mg/mL (ARC Drug Services)
Isotonic saline solution (Baxter, cat. no. 2F7123)
Metabolite extraction
Tissue ruptor (Qiagen, cat. no. 9001271)
Tissue ruptor probes (Qiagen, cat. no. 990890)
Pellet pestles (Fisher, cat. no. 2-141-364)
Centrifuge or microcentrifuge operating at 4 °C
HPLC grade methanol (Fisher Scientific, cat. no. A456-4)
HPLC grade acetonitrile (VWR Scientific, cat. no. BJAH015-4)
HPLC grade water (VWR Scientific, cat. no. BJ365-4)
Equipment
Human infusion
4 mL polyethylene sample vials (Fisher Scientific, cat. no. 03-338-1E)
EDTA blood tubes (Fisher Scientific, cat. no. 02-687-107)
Blood glucometer (Bayer, cat. no. 9545C)
Weighing dishes (Fisher Scientific, cat. no. S67091A)
5 mL syringes
Scalpel
Mouse equipment
Infusion pump, New Era Pump Systems (Fisher Scientific, cat. no. NC9722205)
Heating pads (Fisher, cat. no. 04-777-144)
Tuberculin syringe, Monoject, 1, 3, 5 mL regular tip (Cardinal Health, cat. no. 1180100555)
Butterfly needle, Terumo Winged Infusion Set, 27 G (Terumo, cat no. 22-289-912)
Micro-Renathane tubing (Braintree Scientific, cat. no. MRE-025)
Tissue tape (3M, cat no. 29501)
Microcapillary tubes (Fisher, cat. no. 22-362-566)
25 G needle tips (BD, cat. no. 305122)
Insulin syringe with permanently attached needle (Fisher, cat. no. 14-826-79)
1.5 mL polypropylene microcentrifUge tubes (Fisher Scientific, cat. no. 05-408-129)
Liquid nitrogen dewer (Fisher Scientific, 11-670-4B)
Reagent setup
Tracers to assess glycolysis and TCA cycle metabolism in humans
Prepare these as described in Table 1. These are prepared under standard high-risk compounding within the United States Pharmacopeia guidelines before surgery. Under these guidelines, the labeled nutrients can be stored up to 30 h at ambient temperature, but are typically prepared up to 12 h before surgery.
Equipment setup
GC-MS
We use an Agilent 7890 gas chromatograph networked to an Agilent 5977b mass selective detector with electron impact ionization. This is set up as presented in the table below.
| Gas chromatography | Carrier gas: UHP helium (99.995%). Nitrogen gas can also be used |
| Mass selective detector | Electron impact ionization −70 eV. Quadrupole temperature 180 °C. Source temperature 250 °C |
| Scan parameters | Range 50–1,000 m/z. Scan speed 562 u/s. Frequency 1.6 scans/s. Cycle time 630.57 ms. Step size 0.1 m/z. Solvent delay 9 min |
| Injection and chromatography | The inlet is run in split mode (10:1) at 250 °C. Septum purge flow: 3 mL/min at 8.2 psi.Column flow of 1.0 mL/min |
| Oven temperatures | 60 °C for 1 min. Ramp from 60 °C to 320 °C (10 °C/min). End: hold at 320 °C for 10 min |
Note: autotune the mass spectrometer. Ensure that H2O is <5%, and check for leaks. EM volts should be <2,500.
LC-MS
We use a Thermo Scientific Q Exactive-HF-X hybrid orbitrap system, coupled to a Vanquish UHPLC. We run a hydrophilic interaction chromatography (HILIC) method, with a Millipore ZIC-pHILIC column (5 μm, 2.1 × 150 mm). We separate metabolites on this column using a binary solvent system of 10 mM ammonium acetate in water, pH 9.8 (solvent A) and acetonitrile (solvent B) with a constant flow rate of 0.25 mL/min. For gradient separation, the column was equilibrated with 90% solvent B. After injection, the gradient proceeded as presented in the table below.
| Time | Gradient | %B |
|---|---|---|
| 0–15 | Linear ramp | 90–30 |
| 15–18 | Isocratic | 30 |
| 18–19 | Linear ramp | 30–90 |
| 19–27 | Column regeneration, isocratic | 90 |
▲CRITICAL When using a HILIC column, never flow 100% of either mobile phase onto the column after equilibration. HILIC chromatography relies on the establishment of aqueous and organic microenvironments within the stationary phase for adequate separation.
Procedure 1: clinical infusions
! CAUTION All procedures must be approved by the institutional committee that oversees research on human subjects, e.g., the institutional review board (IRB). Essential considerations during this process include administration of 13C nutrients, patient confidentiality and adverse event monitoring. It is often helpful to discuss the study with an IRB representative before drafting the protocol.
Background preparation
- Assemble the team. Constructing the right team for these studies is essential. The specific personnel needs will depend on the hypotheses to be tested, but our teams have included the following:
- The clinicians who will identify study subjects (often but not always surgeons)
- The surgeon who will perform the biopsy or resection
- Study coordinator and/or study nurse
- Clinical pharmacist (prepares the isotope-labeled material for infusion)
- Anesthesiologist (often responsible for fluid infusions and sometimes drawing blood samples)
- Radiologist (reviews presurgical images to help plan tissue sampling)
- Pathologist (assists in postsurgical analysis of tissue specimens)
- Research team member (receives samples in the operating room and performs analyses)
Identify potential study subjects, and obtain written consent. Surgical candidates who are eligible for isotope infusions are identified by the clinical team, usually at a presurgical clinic appointment. These potential study subjects are contacted by the research team, and written consent is obtained by a study coordinator. The eligibility and potential contraindications depend on the study details but may include underlying medical conditions that complicate the infusions (e.g., glucose infusions are contraindicated for diabetes) or an inability to participate in presurgical imaging (e.g., if the patient has metal-containing stents, this precludes MRI). Once consent has been obtained, the coordinator notifies the personnel involved in the study.
- Hold a multidisciplinary meeting to address the key factors for successful infusion.
- If the study involves medical imaging to help guide tissue sampling, the images should be reviewed in consultation with the radiologist and surgeon 1–2 d before the procedure
- The expected duration of the surgical procedures should be discussed to ensure that enough [13C] labeled nutrient is prepared. This also allows the research team to monitor for any unexpected delays or complications that arise. In this event, the research team needs to communicate with clinical pharmacy to prepare additional [13C] to prolong the infusion
- It is helpful to involve the pharmacist as early as possible in discussions about how to handle the labeled nutrients, because there are special considerations related to their use for human infusion
Perform pathogen/pyrogen testing of materials for infusion. [13C]-labeled nutrients should be certified as sterile and pyrogen-free by the commercial supplier. We have found it useful to test again for endotoxin contamination after the material is prepared into aliquots and shipped to us. Our practice has been to test at least one aliquot per lot using a commercial endotoxin assay, e.g., GenScript, L00350. We do not revalidate sterility of the material, as the reconstituted nutrient will be filtered by the pharmacist prior to use (see below).
Day of surgery
Preparation of [13C]-labeled nutrients ● Timing ~60 min
-
5The material to be infused is delivered to the clinical pharmacy team, who prepares the infusion bag. This is the procedure that we use for [13C]glucose:
- Dilute the sterile and pyrogen-free [13C]glucose in sterile water, to create a stock solution of 0.133 g/mL
-
Pass this stock through a 0.2 μm filter directly into an infusion bag. Generally, the infusion bag is prepared the morning of the surgery, but can also be prepared prior to surgery and stored at 4 °C. Our pharmacists prefer to use the material within 24 h of filter sterilizationThe volume of the bag and duration of infusion are important considerations. We aim to continue the infusion for at least 2–3 h, as this approach produces a prolonged period of steady-state labeling of the precursor in the plasma and abundant labeling in metabolites from the human tumor types we have studied. Following this reasoning and the table in ‘Reagent setup’, a [13C]glucose bag to last 4 h would be prepared by mixing 24 g of glucose in 180 mL. Infusions ranging from 45 min to well over 4 h have provided informative data on our patients.
Patient preparation
▲CRITICAL The research nurses obtain the bag for [13C] infusion from the pharmacy and prepare the patient for the study.
-
6
Place a peripheral intravenous (PIV) catheter while the patient is in the preoperative holding area. This PIV will be used to infuse the labeled nutrient. Ensure that there is a separate catheter for blood sampling to determine 13C enrichment in the circulation. An arterial catheter can be used for this purpose if one is required by the surgical team; otherwise another PIV can be used, but this second PIV needs to be in a different limb that the one used for the infusion to avoid artifactually high levels of 13C enrichment. Note that these studies usually require placement of one PIV beyond the vascular access needed by the surgical team, and this should be explained to the patient in the consent form.
-
7
Draw an aliquot of ~3 mL of blood prior to injecting the bolus, and store in an EDTA tube on ice. For this and all subsequent blood draws, total blood glucose levels are immediately measured with a glucometer to monitor the patient for hyperglycemia, and these values are recorded. The remaining blood sample is stored on ice, until it is processed in the laboratory for MS analysis, or placed in storage at −80 °C.
-
8
Cross-check the label on the infusion bag with the patient identification prior to the infusion. Any additional documentation required by the clinical study, including the additional PIV and infusion of [13C], can be entered into the medical record at this time.
[13C] Nutrient administration
▲CRITICAL These steps are performed by a medical professional, such as a dedicated reseach nurse. The aim is to have the patient under continuous infusion before being transferred to the operating room.
-
9
(See Fig. 1 for overview.) Start the infusion before the patient is transferred to the operating room, to minimize distraction of the anesthesia and surgical teams.
-
10
Deliver a bolus of [13C]glucose at a rate of 6 mL/min for 10 min, which delivers 8 g of glucose.
-
11
Reduce the infusion rate to 0.5 mL/min (or 4 g glucose per hour) for the duration of the surgery.
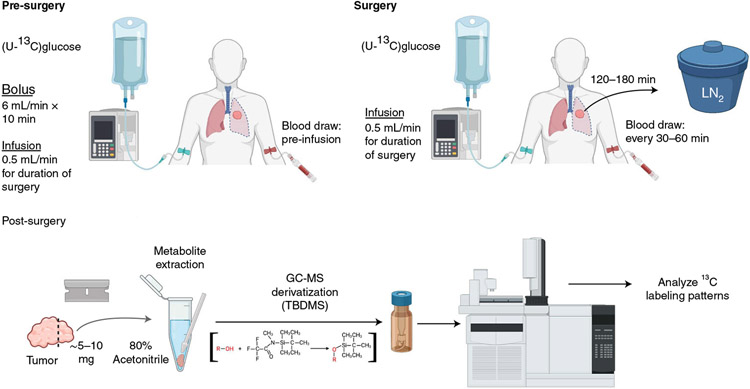
Fig. 1 ∣. Patient infusion workflow.
GC-MS, gas-chromatography-mass spectrometry; LN2, liquid nitrogen; MOX, methoxyamine; TBDMS, tert-butyldimethylsilyl.
During surgery ● Timing typically between 1 and 4 h
-
12
Obtain blood during the infusion. These blood samples will be used to measure the abundance and labeling of the infused nutrient in the circulation. The process for obtaining these samples depends, in part, on whether vascular access is already an established part of the surgical protocol. In the case of thoracic surgeries, most patients have an arterial line placed by the anesthesia team, and blood can be sampled from this line during the surgery. The research team coordinates with anesthesia to draw ~3 mL of blood approximately every 30–60 min. This timing can vary as care is taken not to interrupt the interaction between the anesthesia and surgical teams. If no arterial line is placed, blood draws from other sites can be used. In pediatric patients, where implantation of multiple PIVs can be difficult, blood draws by finger stick can be used. These samples provide sufficient blood for glucometer measurements and [13C] analysis.
-
13
Monitor the patient. Infusion monitoring occurs throughout the procedure, and communication between the research and anesthesia teams is critical. In our experience, supraphysiological glucose levels are rare in nondiabetic patients. Our clinical protocols are written to allow the anesthesia team to administer insulin to reverse hyperglycemia if blood glucose levels exceed 220 mg/dL. If this were to occur, the glucose infusion would be discontinued immediately. The research team also needs to monitor progress of the surgery to make sure that there is enough [13C]glucose for the duration of the procedure. If additional glucose is needed, the research team communicates with clinical pharmacy to prepare an additional [13C] infusion bag.
-
14
Document surgical details. Record the duration of the infusion, when blood supply to the tumor is interrupted (e.g., pulmonary artery ligation during resection of lung tumors), and when the tissue is removed from the body.
-
15
Sample collection is done by the surgeon, as follows: after removing the tumor, quickly remove small samples from various regions of tumor and adjacent tissue as planned from presurgical imaging, and deliver these specimens to the research team.
▲CRITICAL STEP We aim to use at least 5 mg of tissue for GC/MS analysis of isotope labeling. If sufficient tissue is available, it is advisable to analyze three independent fragments to assess metabolic heterogeneity, which can be substantial in solid tumors4,5.
-
16
The research team immediately freezes the samples in liquid nitrogen. Note the time between removal from the body and freezing.
■PAUSE POINT Collected tissues can be stored indefinitely at −80 °C. Blood samples can be centrifuged to remove red blood cells, and stored at −80 °C.
Procedure 2: mouse infusion
Mouse infusion ● Timing ~1–5 h
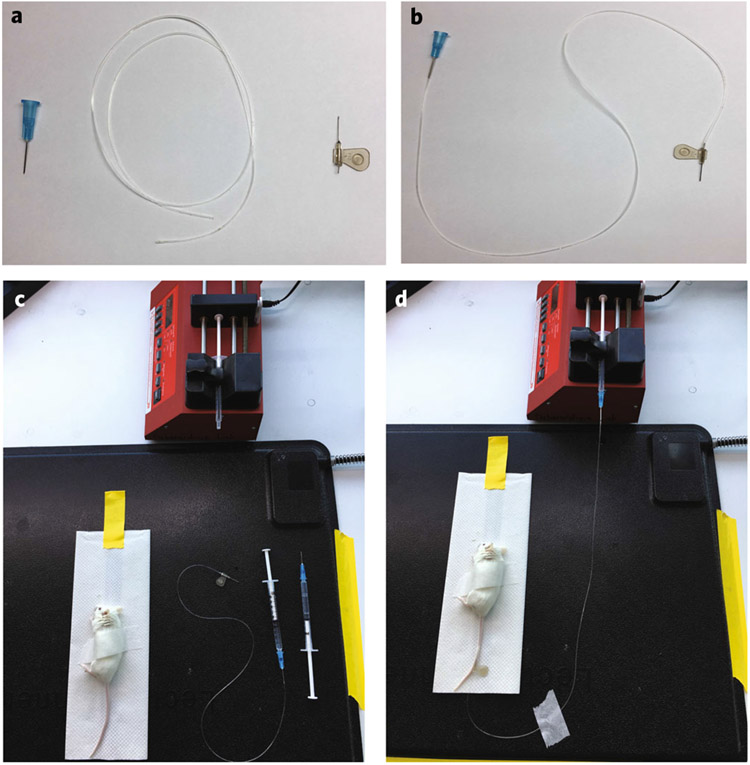
(See Fig. 2 for overall workflow.) Assemble the infusion needle (Fig. 3a,b).
Weigh the mice.
Calculate the volume of [13C] solution necessary to infuse the mice. Prepare the necessary dose following the guidelines in Table 2.
Calculate the amount of ketamine/xylazine necessary for the initial dose weight (100 μL/25 g mouse).
Calculate the ketamine necessary for maintenance doses (1/3 to 1/4 volume of initial dose, per hour of infusion).
Label all collection tubes.
Warm the mice by placing a heating pad under the cage (37 °C). Place a thermometer in the cage to monitor temperature.
Prepare the infusion pump settings. Check that the syringe diameter and rate are correctly calibrated for the infusion (see Step 17 below).
Remove air bubbles from the [13C] solution-filled syringe.
Load the [13C] syringe onto the infusion pump.
Anesthetize the mouse by intraperitoneal injection of ketamine/xylazine.
-
Monitor reflexes at least every 30 min by pinching the paw. Additionally, monitor signs of arousal, including an increase in the respiratory rate or movement of the whiskers.
! CAUTION Sensitivity to ketamine/xylazine can be dependent upon the age, size, strain and overall health of the mouse. In our experience, metastasis confers an additional sensitivity to ketamine in NSG mice, compared with NSG mice with a solitary subcutaneous tumor. After the initial dose, maintenance doses of ketamine (1/3 or 1/4 of initial dose) can be applied every 45–60 min of infusion.
Position the mouse on its side, lateral tail vein up (Fig. 3c). Hang the tail over the side of the bench to increase the venous blood volume. Immobilize the mouse by taping across the hips to a paper towel. Ensure accessibility to the paws (so that it easy to pinch them to monitor reflexes) and abdomen (to enable ketamine injections).
Curve the mouse tail over your finger to insert the catheter (bevel up) at an angle near perpendicular to your finger. Puncture the skin with the needle tip. Adjust the angle so the needle is parallel to the vein. Advance the needle. If the needle is in the vein, it should advance smoothly. If the vein is punctured in the process, move towards the base of the tail and try again.
- Check for proper needle placement using the following techniques:
- Unhook the saline-filled syringe from the needle. This creates a change in pressure, causing blood to flow into tubing if the needle is in position. If unsuccessful (e.g., no blood flow), fill the opened needle cap with saline before reattaching to the syringe. This prevents the introduction of air bubbles into the infusion line
- Alternatively, inject a small volume of saline to test the needle placement. If there is no resistance during injection, the vein will flush without swelling or blanching of the tail, confirming correct placement. Avoid flushing excess saline. If the needle is not correctly placed, injection of excess saline will cause edema in the tail and make needle placement more difficult.
Once successful, detach the saline syringe from the hub of the needle. Fill this hub with saline to prevent air bubbles. Attach it to the syringe on the infusion apparatus. Lift the tape holding the paper towel on the heating pad, and move the mouse closer to the pump (Fig. 3d). Connect the tubing to the infusion pump.
- Begin the infusion using the parameters shown below.
Labeled nutrient Syringe
Infusion parameters
Volume (mL) Diameter (mm) Bolus (μL/min) Infusion (μL/min) Glucose 1 4.699 125 (1 min) 2.5 (180 min) Lactate 1 4.699 15 (10 min) 2.0 (180 min) Glutamine 3 8.585 150 (1 min) 2.5 (300 min)
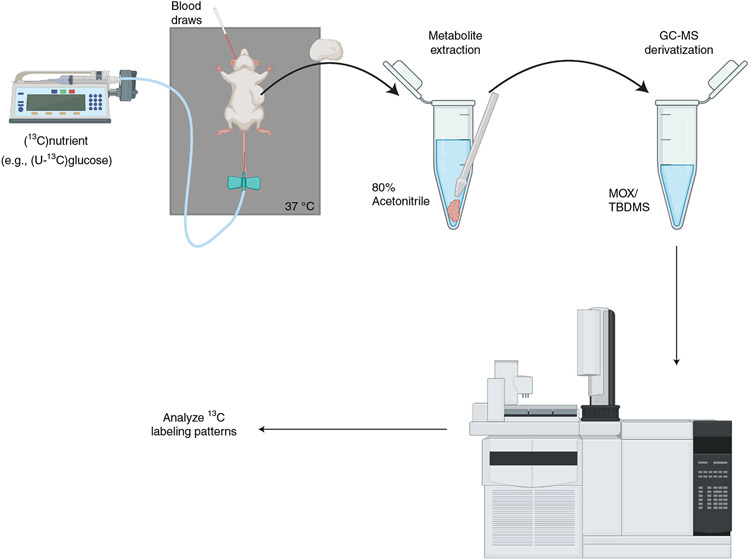
Fig. 2 ∣. Mouse infusion workflow.
An automated pump infuses the [13C] nutrient into the mice via a butterfly catheter inserted into the tail vein. At the end of infusion, the tumor is dissected and a sample is homogenized in 80% acetonitrile. The homogenized sample is dried and derivatized by MOX/TBDMS before injection onto GC-MS.
Fig. 3 ∣. Mouse infusion apparatus.
a, Butterfly infusion needle and tubing. b, Assembled version of a. c, Apparatus used in mouse infusions. The [13C] syringe is loaded into the infusion pump, the mouse is secured to a paper towel on a heating pad and a saline-filled syringe is attached to the butterfly needle. d, Image of an active infusion. The infusion tubing is secured to the heating pad, and the mouse is accessible for both blood draws and ketamine maintenance dosing.
Blood sampling by retro-orbital blood draw
▲CRITICAL Blood can be drawn from mice using several methods, but some of these are impractical during isotope infusions. As this is a terminal procedure with anesthetized mice, we have found retro-orbital draws to be most useful for isotope infusions as they are consistent, can be performed multiple times during the infusion and do not disrupt the vasculature near the site of [13C] administration.
-
18
Place the capillary tube behind the eye, ~4 mm deep (often indicated by a mark or line on the capillary tube). Gently spin the capillary tube between your fingers to draw blood.
-
19
Once the sample is acquired, immediately compress the eye to stop bleeding and clean the area.
-
20
Use a 200 μL pipette to flush the blood from the capillary tube into a collection tube.
-
21
Store on ice during the procedure, or spin to separate blood and plasma.
■PAUSE POINT Samples can be stored at −80 °C indefinitely.
Tissue harvesting in the mouse
Bulk tumor tissue
-
22
Euthanize the mouse by cervical dislocation.
-
23
Spray the mouse with 70% ethanol.
-
24
Harvest tissues with tweezers/scissors, carefully removing contaminating tissues (i.e., skin, muscle).
-
25
Flash freeze in liquid nitrogen. Samples can be stored at −80 °C indefinitely.
▲CRITICAL STEP Metabolite lability is an important consideration in the timing from sample excision to freezing.
Procedure 3: downstream analysis
▲CRITICAL Various processing methods (metabolite extraction, cell isolation) and analytical methods (GC-MS, LC-MS) can be applied to samples obtained from mice or from patients. Our standard methods are summarized below.
Sample processing
From tissue
Place snap-frozen tissue samples on dry ice.
Use a clean scalpel and forceps to cut a small piece of tissue (weighing 5–25 mg). Larger pieces can be cut into smaller fragments to facilitate dissociation.
- Dissociate the material either manually (option A) or mechanically (option B).
- Manual dissociation method
- Place tissue fragment in a labeled, clean 1.5 mL Eppendorf containing 400 μL 80% acetonitrile on ice.
- Grind the tissue using a pestle until fully dissociated.
- Mechanical dissociation method
- Place the fragment in a labeled, clean 50 mL conical tube containing 400 μL 80% acetonitrile, on ice.
- Grind the tissue using a motorized grinder (e.g., bead or probe-based). Intermittent grinding can be used to minimize the amount of heat generated. Most tumors dissociate easily, while fibrous or elastic tissues (e.g., lung) require extended grinding.
Lyse the samples by freeze–thaw cycles. Transfer samples between liquid nitrogen and ice three times.
Spin for 10 min at 10,000g (4 °C) to remove debris.
Transfer the supernatant to a new tube.
Perform protein quantification on both the pellet and the supernatant. The pellet can be resuspended in a cell lysis buffer (e.g., RIPA), and both the pellet and supernatant can be quantified by a bicinchoninic acid assay. Protein quantification of the pellet can be used to normalize samples, while testing the concentration of protein in the supernatant can prevent saturating the signal on the mass spectrometer. Multiple methods of normalizing metabolite abundance can be used, including weight of wet tissue, weight of lyophilized sample, protein abundance, etc. In our experience, normalizing to protein concentration in the pellet has led to reproducible data between samples.
-
Dilute the supernatant to a final concentration that does not saturate the mass spectrometer. We dilute to 0.1 μg protein/μL with 80% acetonitrile.
■ PAUSE POINT Store at −80 °C until injection into the mass spectrometer.
From plasma
-
9
Centrifuge 50 μL of plasma at 14,000g (10 min, 4 °C) to remove any contaminating cells.
-
10
Transfer the supernatant to a new tube.
-
11
Dilute the supernatant into 80% acetonitrile. 10 μL into 990 μL of 80% acetonitrile is used for LC-MS analysis, and 50 μL into 450 μL is used for GC-MS analysis.
-
12
Shake/vortex to mix.
-
13
Spin for 10 min at 10,000g (4 °C).
-
14
Transfer supernatant to new tube.
■PAUSE POINT Store at −80 °C until further preparation for MS.
Mass spectrometry
-
15At this stage, the frozen samples from Steps 8 and 14 can be analyzed by MS. We present our methods for GC-MS and LC-MS analysis.
- GC-MS sample preparation
-
Dry down the extracted supernatant by an air or N2 stream, or speed vacuum centrifugation. Note: dried samples can be stored indefinitely at −80 °C until processed for GC-MS.! CAUTION Some metabolites, including those with reactive thiols, may be oxidized by the O2 present in air. Using N2 rather than air is preferable for analyzing these metabolites.
-
Prepare methoxyamine HCl (methoxyamine in pyridine (10 mg/mL)).! CAUTION Derivatization solvents should be used in a chemical fume hood.
- Add 40 μL of methoxyamine to each dried sample.
- Vortex for 5 s. Spin (10,000 rpm, 30 s) to remove any remaining debris.
-
Transfer supernatant to a GC-MS vial with a glass insert. Cap the GC-MS vial.▲CRITICAL STEP GC-MS vial inserts are used so that less liquid needs to be present in the vial for successful autosampler injection. This is crucial for injecting the sample at a sufficiently high concentration.
- Heat for 10 min at 70 °C.
- Add 80 μL of MTBSFA + 1% TMCS.
- Vortex for 5 s.
- Heat for 60 min at 70 °C.
- Add samples to GC-MS for injection. Samples can remain at ambient temperatures for up to 48 h. Alternatively, samples can be stored at −80 °C for an indefinite period of time. Critical samples can be stored at −80 °C immediately after injection, until the data have been processed and deemed satisfactory.
-
GC-MS acquisition
Check that the GC-MS is ready as described in ‘Equipment setup’.
Start the autosampler.
-
(B)HRMS analysis
- Check that the HILIC-MS system is set up as described in ‘Equipment setup’.
- Thaw the samples, and prepare aliquots of ~100 μL into LC-MS vials with insert.
- Acquire mass spectra from isotopically labeled samples by HRMS full scan (precursor ion only). Utilize method switching between positive and negative polarities at a resolving power of 120,000 full width at half maximum and an automatic gain control (AGC) target of 1,000,000. All data are acquired with the profile spectral acquisition setting and a standard mass range of 80–1,200 Da.
- Use unlabeled samples (e.g., non-infused mouse tissue, or blood samples collected before infusion) to generate data-dependent spectra. This enables high-confidence identification of traced metabolites.
- Acquire product ion spectra from unlabeled, T0 specimens using a data-dependent tandem MS method in a single polarity. Product ion spectra are acquired with a resolving power of 15,000 full width at half maximum without a fixed mass range. The AGC target value is set to 200,000 with a maximum injection time of 150 ms. Data-dependent parameters were set to acquire the top ten ions with a dynamic exclusion of 30 s and a mass tolerance of 5 ppm. Isotope exclusion is enabled and a stepped normalized collision energy applied with values of 20, 35 and 50. The minimum AGC target is set to 8,000.
-
Prepare the samples in the autosampler, and analyze the experimental samples, controls and standards. We recommend injecting samples in sets intermixed with injections of quality control samples throughout the run.The blank injection consists of 80% acetonitrile. The check standard is is a mix of proline, phenylalanine, succinate and 2-hydroxyglutarate at 10 nM concentrations. This injection is used to monitor instrument function after calibrations or column changes, as well as to assess instrument sensitivity. Quality control standards are standards used to monitor changes in the equipment with time. This could be a plasma sample, for example. We recommend making a single solution that contains small, equal aliquots from all the samples that are to be analyzed in the experiment. We use the following injection format:
- Blank
- Check standard (two injections: one data-dependent MS/MS (ddMS/MS) positive, one ddMS/MS negative)
- Quality control (three injections: one ddMS/MS positive, one ddMS/MS negative, one MS1 only polarity switch)
- 20 experimental samples (10 h of run time)
- Blank
- Fresh quality control (two injections: one ddMS/MS positive, one ddMS/MS negative)
- Repeat the ‘Blank, 20 samples, QC’ pattern until all samples are analyzed, then
- Blank
- Fresh quality control (two injections: one ddMS/MS positive, one ddMS/MS negative)
- Blank
-
End sequence▲CRITICAL STEP Quality controls and blank samples are important internal controls within and between MS runs. The measured ions can contain contaminants from processing, dirty solvents, adducts and other artifacts, which can in part be accounted for by injecting blank samples. Quality control samples (i.e., freshly prepared serum extracts, or controlled tissue extracts) are used to monitor changes in metabolite measurements throughout a run, and between batches. These controls are important to monitor instrument performance and successfully identify metabolites.
Analysis of isotope enrichment
-
16Several software programs exist to analyze MS data for peak quality, signal-to-noise ratio, isotopologue resolution and other factors. Examples include Tracefinder from ThermoScientific; El-Maven, an opensource freeware from Elucidata; Chemstation or Profinder from Agilent, etc. The specific software and its capabilities may depend on the MS systems being used. Example steps for how to do this are provided for GC-MS and HRMS (options A and B, respectively).
- GCMS peak analysis.
- Use unlabeled, derivatized metabolite standards to generate an in-house library of mass spectra. These are used as references for the experimental metabolite peaks.
- Match the metabolite ion distribution and retention time of each metabolite peak to make sure that they have been identified correctly.
- Calculate the abundance of each relevant metabolite ion.
- Correct these data for naturally abundant isotopes, as the tert-butyldimethylsilyl derivatization adds a significant number of atoms that need to be accounted for (see ‘Isotope corrections’ section below).
- HRMS peak analysis.
- Analyze the metabolites qualitatively using a spectral browser (i.e., ThermoScientific FreeStyle or QualBrowser), to verify three specific aspects of data quality:
- Is the m/z of the monoisotopic ion within 5 ppm of the theoretical m/z predicted by the chemical formula?
- Are the m/z values of metabolite product ions within 10 ppm of known metabolite production ions reported in spectral libraries such as mzCloud or product ion spectra obtained from a purified standard?
- Are the retention times of metabolites within 5% of the retention time of a purified standard analyzed with the same chromatographic method?
-
Compare the results in the data files with an in-house compound library containing the theoretical masses of isotopically labeled metabolites.▲CRITICAL STEP When making a custom library of isotopologues, the m/z of all labeled species must be calculated to the fourth decimal place since the data are being searched with a 5 ppm tolerance. We recommend the following mass differences between nuclei for calculating m/z values for isotopologue libraries.
Nucleus Delta per nucleus 12C to 13C 1.003355 1H to 2H 1.006276 14N to 15N 0.997035 - Use quantitative software to automatically identify and integrate peaks from the compound library at a 5 ppm tolerance.
- Carefully review all automated integrations and software-identified retention times. Only isotopologues with identical retention times should be integrated and used to calculate fractional enrichment. The retention times of isotopologues should be within 1% of each other, and the monoisotopic mass should be confirmed using the criteria described in steps i–iii) (Fig. 4).
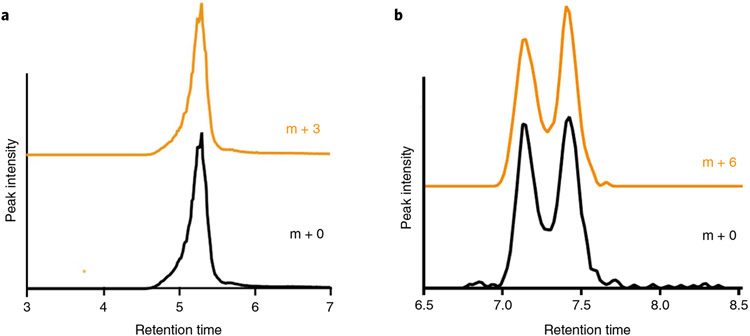
Fig. 4 ∣. Confirmation of [13C] peaks in HRMS.
a,b, The extracted ion chromatograms (XICs) of lactate and [U-13C] lactate (m + 0 and m + 3, respectively) (a), and glucose and [U-13C]glucose (m + 0 and m + 6, respectively) (b), are overlaid. Comparison of the glucose XIC with a chemically purified standard confirms the correct peak is identified. To calculate fractional enrichment, only the overlapping peaks in each XIC are integrated. In the case of glucose, we resolve two structural isomers of the sugar in our HILIC chromatography (confirmed with chemical standards), so both peaks in the bimodal distribution between 7 and 7.5 min are integrated to calculate fractional enrichment.
Isotope corrections
-
17
The isotopes used for labeling also occur in nature—albeit at a much lower level (13C is naturally abundant at 1.1% of all carbon). To determine the experimental 13C enrichment occurring as a consequence of infusion with a labeled nutrient, a mathematical correction must be performed to account for naturally occurring 13C. Isotope correction requires the chemical formula of the ion measured, which is especially critical in derivatized samples where the analysis needs to account for atoms not associated with the native metabolite but introduced through derivatization. There are various computational methods for performing this correction, many of which are freely available. One example adapted by our laboratory uses a customized R script, which can be found at the GitHub repository (https://github.com/wencgu/nac). The script was written by adapting the AccuCor algorithm29. Other examples of isotope correction programs are Metran30, IsoCor31 and others.
▲CRITICAL STEP When reviewing labeled data on a HRMS instrument, ensure that all isotopologues are fully resolved (baseline separation between m/z peaks in the Full MS1 spectrum) from other naturally abundant isotopes. This resolution can be determined either through proprietary software sold with the instruments, or though freely available software (e.g., El-Maven). Naturally abundant isotopes are present in MS1 data at predictable rates but can be resolved from your isotopologues of interest on a high-resolution instrument. For example, if tracing 15N glutamine in the HRMS method described here, the peak is baseline resolved from the naturally abundant 13C peak and the m/z differences are >5 ppm. Therefore, the 15N peak does not need to be corrected for naturally abundant 13C signal, but only naturally abundant 15N signal.
Troubleshooting
Mouse infusions
Low isotope enrichment
Low enrichment can occur for several reasons: (1) check tubing and syringe connections periodically for leaks during infusion, as poor delivery of the labeled nutrient will reduce isotopic enrichment in downstream metabolites; (2) verify that the infusion settings on the pump (e.g., tube diameter, infusion rate) were entered correctly; (3) double-check the infusion solution calculations for appropriate concentrations. If testing a new metabolite infusion, consider increasing the bolus or continuous infusion.
Difficulty with needle placement in the tail vein
Replace the butterfly catheter between experiments. Dull needles decrease the success of tail-vein insertion. Before infusion, check tubing and syringe connections for blockage or clogs. A blocked needle prevents the observation of blood flow needed to ensure successful needle placement, and can hinder infusion.
Mass spectrometry
Signal intensity
Low signal may arise from a dirty instrument or insufficient nutrient abundance.
Dirty chromatograms
If the instrument is dirty, first remove and clean the sweep cone and ion transfer tube in the mass spectrometer. The sweep cone can be cleaned with a mild abrasive like sialic acid, and the ion transfer tube can be sonicated in 25% formic acid. Additionally, the source needle may be dirty. Look for residue at the end of the needle, and change if necessary.
Poor yield
If an extraction did not yield enough material, more sample can be injected onto the column. We have found that up to 20 μL of 80% acetonitrile can be injected without dramatic changes in chromatographic performance of analytes.
Mass accuracy
Isotopic peaks may not be properly identified if the instrument is out of calibration. To avoid this issue, calibration should be performed at least weekly. Additionally, the ion optics or ion focusing may be out of calibration. We recommend a full instrument evaluation with CalMix every 7 d. Finally, if the column is overloaded, mass accuracy will be affected from space charging in the orbitrap. If this happens, reduce the injection volume or dilute the sample extract in the autosampler vial.
Mass range on LC-MS
Setting the mass range low-end to m/z 80 is a compromise that minimizes low-end mass dilution while tracing central carbon metabolism comprehensively. However, if the user wishes to trace into smaller metabolites like glycine, glyoxal and methylglyoxal, the mass range of the scan event must be lowered accordingly. If the mass range is lowered below 60, the large amount of acetate in the mobile phase (m/z 59 in negative mode) will influence total ion counts and dilute signal from metabolites of interest.
Timing
Procedure 1: clinical infusion
Background preparation: timing depends on multiple factors (e.g., length of time for IRB review and approval can vary greatly)
Pathogen and pyrogen testing of labeled nutrient: ~1 h. This testing occurs when the shipment arrives from the commercial supplier, and generally takes 1 h to complete. Samples need to be tested before delivery to clinical pharmacy
Identification of potential subjects: timing varies. Suitable subjects for infusion studies are identified by the clinical team, and sent to the research coordinator for consent. In general, these patients are identified and consented several weeks before surgery, but we have occasionally studied patients consented just 1–2 d prior to the surgery
Multidisciplinary meeting: 0.5 h. Occurs after the patient has undergone clinical imaging, and before the day of surgery
Preparation of [13C]-labeled nutrients: ~1 h. The labeled nutrients generally take 1 h to prepare, and the preparation should be completed at least 1 h before the scheduled start of a surgery
Patient preparation: ~1 h: the placement of an additional PIV line and the time-zero blood draw are performed ~1 h before surgery. Placement of the IV and the blood draw take ~5 min
Nutrient administration: timing varies. The bolus is timed to finished 5–15 min before the patient is moved from the preoperative rooms to the operating room. The total duration of infusion is dependent upon the length of surgery, which is influenced by a variety of factors on the day of surgery that may be difficult to predict
Procedure 2: mouse infusion
Needle preparation: 0.5 h: the infusion needle can be prepared any time before the mouse infusion
Preparation of the labeled nutrient: ~0.5 h
Mouse preparation: ~0.5 h
Tail-vein cannulation: timing varies with experience. Successful needle insertion into the tail vein can take 2–3 min per mouse by an experienced user. Inexperienced users may take significantly longer
Labeled nutrient infusion: timing varies based on nutrient and experimental conditions
Procedure 3: downstream analysis
Sample preparation: timing varies according to the number of samples to be processed. Tissue lysis requires ~2–3 min per sample. Freeze–thaw cycles can take 30 min, and centrifugation will take 10–15 min
Determining protein concentration: 0.5 h
GC-MS drying down samples: 3–4 h. Alternatively, samples can be dried overnight in a speedvac
GC-MS: ~45 min per sample
LC-MS: ~30 min per sample
Anticipated results
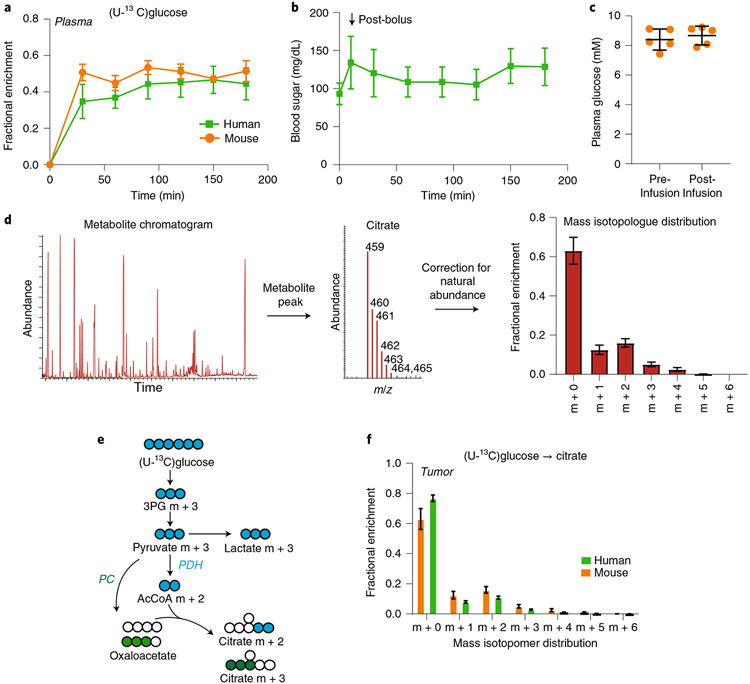
The method described here enables the investigation of metabolic processes in vivo, particularly in tumors and adjacent nonmalignant tissues. The technique achieves similar levels of enrichment in plasma glucose when infusing [13C]glucose into humans or mice (Fig. 5a), and does not significantly alter circulating glucose levels (Fig. 5b,c). These infusions also produce multiple labeled forms of central carbon metabolites in tumors, dependent upon several factors, including individual enzymes and TCA cycle activity.
Fig. 5 ∣. Data from [U-13C]glucose infusions in patients and mice.
a, Time course of plasma glucose enrichment during [U-13C]glucose infusions in patients and mice. b, Plasma glucose concentration in patients during [13C]glucose infusions. See Supplementary Dataset. c, Plasma glucose concentration in mice before and after [U-13C]glucose infusion. d, Workflow of data analysis. The chromatogram is analyzed for peaks of interest. Next, the peak for citrate is identified, and the abundances of citrate masses (459–465) are recorded. The values are then corrected for natural abundance to measure the mass isotopologue distribution of citrate. e, Schematic of labeling downstream of [U-13C]glucose, highlighting the expected effects of PDH or PC. f, Mass isotopologues of citrate from human NSCLC and mice bearing melanoma PDXs after infusion with [U-13C]glucose. Data are expressed as average and standard deviation. m/z, mass-to-charge ratio; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase.
The overall flow of analysis is to examine the chromatogram of the sample and identify peaks corresponding to metabolites of interest (e.g., citrate in the illustrated example). The ions of interest are quantified and corrected for natural abundance to generate the mass isotopologue distribution of the labeled metabolite (Fig. 5d). In addition to overall metabolite labeling, the pattern of isotopologues provides important information about metabolic activity. Citrate m + 2 is usually generated when pyruvate dehydrogenase (PDH) converts fully labeled pyruvate (m + 3) to fully labeled acetyl-CoA (AcCoA, m + 2), and this AcCoA is condensed with an unlabeled oxaloacetate by citrate synthase. Similarly, citrate m + 3 arises from the combination of oxaloacetate m + 3 generated from pyruvate m + 3 by pyruvate carboxylase (PC) and an unlabeled AcCoA (Fig. 5e). Citrate m + 1 may be generated by TCA cycle turnover when AcCoA enrichment is low4 and/or by refixation of 13CO2 by carboxylase reactions, including PC. In both tumor-bearing mice and patient tumors, the tumors contain detectable levels of citrate m + 1, m + 2, m + 3, etc., indicating that the method reports multiple routes of glucose metabolism (Fig. 5f).
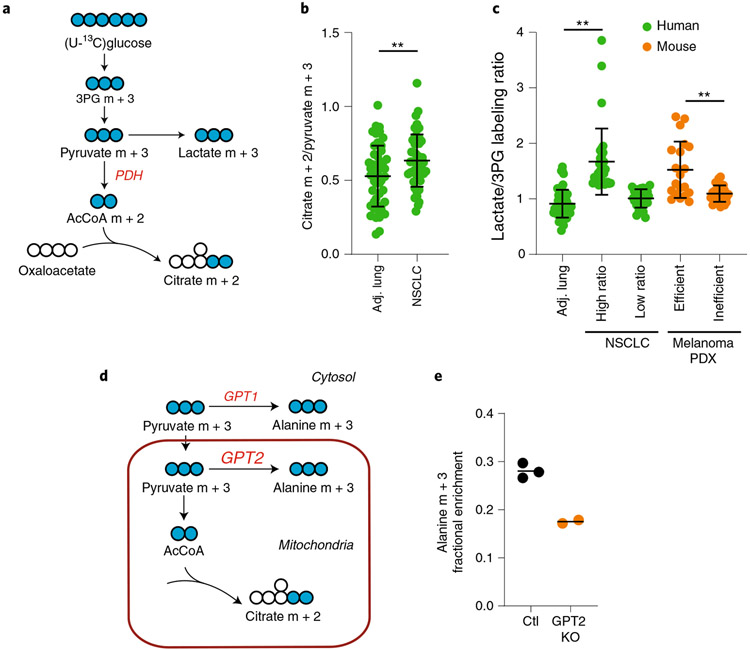
Comparing the relative abundance of different labeled metabolites can also provide information about particular metabolic activities within the tissue. For example, after infusions with [U-13C] glucose, the ratio of citrate m + 2 to pyruvate m + 3 labeling is used as a surrogate of PDH (Fig. 6a). Data from NSCLC patients reveal that tumors and lungs contain ratios ≥0.5, indicating that both tissues contain substantial PDH activity (Fig. 6b). The ratio is modestly elevated in the tumors, suggesting an activation of PDH in NSCLC.
Fig. 6 ∣. Examining metabolic features among tumor and tissue types.
a, Schematic of labeling downstream of [U-13C]glucose. b, Citrate m + 2/pyruvate m + 3 ratios in healthy lung and tumors. **P < 0.01 (Welch’s t-test). c, Lactate/3PG labeling ratios in patients and xenograft-bearing mice. Human data: **P < 0.001, Kruskal–Wallis test, Dunn’s post-hoc. PDX data: **P < 0.01 Mann–Whitney test. d, Schematic of alanine labeling from 13C-pyruvate via either of two isoforms of GPT (GPT1 or GPT2). e, Alanine labeling after infusion of [U-13C]glucose into mice with control (Ctl) xenografts or xenografts lacking GPT2 (GPT2 KO). 3PG, 3-phosphoglycerate; PDH, pyruvate dehydrogenase; PC, pyruvate carboxylase; GPT, glutamate pyruvate transaminase; NSCLC, non-small cell lung cancer; PDX, patient-derived xenograft. Data are expressed as average and standard deviation.
Other comparisons of metabolite enrichment can provide information about nutrient uptake5,21. 3-Phosphoglycerate (3PG) is a glycolytic metabolite that is typically not produced from lactate in nongluconeogenic tissues. Therefore, after infusion with [13C]glucose, comparing the labeling ratio between 3PG and lactate provides information about the source of these labeled species. Because both 3PG and lactate can be produced from glucose, if lactate labeling is equal to or lower than labeling in 3PG, the data are consistent with production of 13C-lactate from 13C-glucose (proceeding through 13C-3PG) in the tissue. On the other hand, if lactate labeling exceeds 3PG labeling, one possible explanation is import of 13C-lactate from the bloodstream; this labeled lactate could arise from conversion of 13C-glucose to 13C-lactate by other tissues, followed by secretion of 13C-lactate into the bloodstream. Some human NSCLCs and melanoma patient-derived xenografts (PDXs) have quite high lactate/3PG labeling ratios (Fig. 6c), and in the melanoma PDXs this feature correlates with metastatic efficiency5,21.
These methods can be used to test hypotheses about metabolic activities within tumors. For example, many tissues including tumors express multiple isoforms of enzymes localized to different subcellular compartments. Glutamate pyruvate transaminase (GPT) converts pyruvate into alanine using an amino group derived from glutamate. GPT1 is cytosolic, while GPT2 is mitochondrial (Fig. 6d). To test whether GPT2 contributes to alanine metabolism in NSCLC cells, we created a CRISPR-mediated knockout of this enzyme and implanted the cells into mice to establish subcutaneous xenografts, then infused with [U-13C]glucose. GPT2 deficiency markedly reduced the appearance of labeled alanine in the tumors, indicating that GPT2 contributes to alanine metabolism in these tumors (Fig. 6e).
Further considerations
We have highlighted a few examples of how infusion with [U-13C]glucose can be used to test hypotheses about central carbon metabolism in tumors. However, the type and timing of a labeled-nutrient infusion depends on the pathway of interest. Many other commercially available tracers can be utilized to interrogate other pathways. These include alternatively labeled forms of glucose that may be preferable to assess particular aspects of central metabolism. For example, [3,4-13C]glucose provides a specific means to assess PC activity. This tracer produces two molecules of [1-13C]pyruvate through glycolysis. PDH releases this 13C from pyruvate as 13CO2, such that the label will be absent from TCA cycle intermediates resulting from this reaction. On the other hand, PC converts [1-13C] pyruvate into labeled oxaloacetate, resulting in 13C-labeled TCA cycle intermediates. Other isotopes can be used to assess pathways beyond carbon metabolism. For example, [15N]glutamine is used to evaluate transamination reactions, amide transfer reactions, glutathione biosynthesis, the urea cycle and many other pathways.
Thoughtful analysis of isotope enrichment data as described above can provide a wealth of information about metabolism within tissues. Complementing this initial, qualitative examination of labeling data with computational tools can provide a fuller picture of the metabolic network. For example, any of a variety of modeling software packages, including INCA32, tcaSIM33, Metran34 and 13CFLUX235, allow the user to fit the experimental data into a metabolic network model of the user’s design. Relative fluxes throughout the network can be estimated by fixing one reaction, for example the reaction catalyzed by citrate synthase, to an arbitrary value of 1.0 and then calculating ranges for other fluxes around the network that fit the experimental labeling data. These approaches do not produce absolute flux values, but they can be used to detect differences in relative activities and to test whether empirical data can be explained by (i.e., ‘fit’) a modeled network. Iterating parameters within the model (e.g., the direction of a particular reaction or transporter) allows the user to examine whether a particular change improves or worsens the data fit4,5. This exercise helps the user develop hypotheses about metabolism within the tissue, and these hypotheses can then be tested in experimental systems. Another useful application of metabolic network models is to simulate the labeling features predicted to be produced from a particular tracer entering a modeled network, before the infusion is performed. This provides a framework to help choose the optimal tracer, plan the timing of the infusion and anticipate the label distribution.
Labeling data can be analyzed further by incorporating additional methods to calculate absolute fluxes within the tissue23,36-38. Quantitative analyses like these are assisted by constraining the range of possible fluxes by accounting for total metabolite exchanges into and out of the tissue of interest, for example by cannulating the arterial supply and venous drainage of an organ to measure metabolite content in each37. This approach is challenging when vascular access is impractical or impossible (e.g., in tumors with multiple routes of venous drainage). Nevertheless, there has been increasing success in using such approaches, together with multiple [13C]tracers, multicompartment modeling and physiological perturbations such as dietary modulation, to comprehensively assess nutrient metabolism in mammalian systems36.
Supplementary Material
Acknowledgements
R.J.D. is a Howard Hughes Medical Institute (HHMI) Investigator, the Robert L. Moody, Sr. Faculty Scholar at UT Southwestern and Joel B. Steinberg, M.D. Distinguished Chair in Pediatrics. S.J.M. is an HHMI Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the Kathryn and Gene Bishop Distinguished Chair in Pediatric Research, the director of the Hamon Laboratory for Stem Cells and Cancer, and a Cancer Prevention and Research Institute of Texas Scholar. The research was supported by the Cancer Prevention and Research Institute of Texas (RP170114 and RP180778), the National Institutes of Health (R35 CA220449; U01 CA228608) and the Robert A. Welch Foundation (I-1733). A.T. was supported by the Leopoldina Fellowship (LPDS 2016-16) from the German National Academy of Sciences and the Fritz Thyssen Foundation. B.F. is supported by the National Institutes of Health (K99/R00 CA237724-01A1). Figures were generated using BioRender.com.
Appendix
Related links
Key data used in this protocol
- Faubert B et al. Cell 171, 358–371 (2017): 10.1016/j.cell.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdogan A et al. Nature 577, 115–120 (2020): 10.1038/s41586-019-1847-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT. et al. Cell 164, 681–694 (2016): 10.1016/j.cell.2015.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
The software can be found at https://github.com/wencgu/nac.
Competing interests
R.J.D. is an adviser for Agios Pharmaceuticals and Vida Ventures, and a founder and adviser for Atavistik Bio. S.J.M. is an adviser for Frequency Therapeutics and Protein Fluidics.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-021-00605-2.
Data availability
Previously unpublished data are included in the Supplementary Information. The remaining supporting data can be found in refs. 4,5,21.
References
- 1.Faubert B & DeBerardinis RJ Analyzing tumor metabolism in vivo. Annu. Rev. Cancer Biol 1, 99–117 (2016). [Google Scholar]
- 2.Buescher JM et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol 34, 189–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher EA et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 25, 1234–1244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hensley CT et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faubert B et al. Lactate metabolism in human lung tumors. Cell 171, 358–371 e359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courtney KD et al. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 28, 793–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston K et al. Isotope tracing reveals glycolysis and oxidative metabolism in childhood tumors of multiple histologies. Med 2, 395–410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oizel K et al. Glutamine uptake and utilization of human mesenchymal glioblastoma in orthotopic mouse model. Cancer Metab. 8, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Momcilovic M et al. The GSK3 signaling axis regulates adaptive glutamine metabolism in lung squamous cell carcinoma. Cancer Cell 33, 905–921 e905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson SM et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X et al. The abundance of metabolites related to protein methylation correlates with the metastatic capacity of human melanoma xenografts. Sci. Adv 3, eaao5268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grinde MT et al. Glutamine to proline conversion is associated with response to glutaminase inhibition in breast cancer. Breast Cancer Res. 21, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan TW et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 8, 41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellers K et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin. Invest 125, 687–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun RC et al. Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nat. Commun 8, 1646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang C, Chen L & Rabinowitz JD Metabolomics and isotope tracing. Cell 173, 822–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roussel R, Carlier PG, Robert JJ, Velho G & Bloch G 13C/31P NMR studies of glucose transport in human skeletal muscle. Proc. Natl Acad. Sci. USA 95, 1313–1318 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romijn JA, Coyle EF, Sidossis LS, Rosenblatt J & Wolfe RR Substrate metabolism during different exercise intensities in endurance-trained women. J. Appl. Physiol 88, 1707–1714 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Coggan AR, Kohrt WM, Spina RJ, Bier DM & Holloszy JO Endurance training decreases plasma glucose turnover and oxidation during moderate-intensity exercise in men. J. Appl. Physiol 68, 990–996 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Ma EH et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8(+) T cells. Immunity 51, 856–870 e855 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Tasdogan A et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marin-Valencia I et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui S et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan M et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol 18, 1090–1101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J et al. 13C isotope-assisted methods for quantifying glutamine metabolism in cancer cells. Methods Enzymol. 542, 369–389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long CP & Antoniewicz MR High-resolution (13)C metabolic flux analysis. Nat. Protoc 14, 2856–2877 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Yuan M et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat. Protoc 14, 313–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broekaert D & Fendt SM Measuring in vivo tissue metabolism using (13)C glucose infusions in mice. Methods Mol. Biol 1862, 67–82 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Su X, Lu W & Rabinowitz JD Metabolite spectral accuracy on orbitraps. Anal. Chem 89, 5940–5948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoniewicz MR, Kelleher JK & Stephanopoulos G Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng 9, 68–86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millard P, Letisse F, Sokol S & Portais JC IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Young JD INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alger JR, Sherry AD & Malloy CR tcaSIM: a simulation program for optimal design of (13)C tracer experiments for analysis of metabolic flux by NMR and mass spectroscopy. Curr. Metabolomics 6, 176–187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo H, Antoniewicz MR, Stephanopoulos G & Kelleher JK Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J. Biol. Chem 283, 20621–20627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weitzel M et al. 13CFLUX2-high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics 29, 143–145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui S et al. Quantitative fluxomics of circulating metabolites. Cell Metab. 32, 676–688 e674 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang C et al. Metabolite exchange between mammalian organs quantified in pigs. Cell Metab. 30, 594–606 e593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, Dai Z, Cooper DE, Kirsch DG & Locasale JW Quantitative analysis of the physiological contributions of glucose to the TCA cycle. Cell Metab. 32, 619–628 e621 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously unpublished data are included in the Supplementary Information. The remaining supporting data can be found in refs. 4,5,21.