Abstract
Mutations in the global regulatory genes gacS and gacA render Pseudomonas syringae pv. syringae strain B728a completely nonpathogenic in foliar infiltration assays on bean plants. It had been previously demonstrated that gac genes regulate alginate production in Pseudomonas species, while other published work indicated that alginate is involved in the pathogenic interaction of P. syringae on bean plants. Together, these results suggested that the effects of gacS and gacA mutations on virulence in B728a might stem directly from a role in regulating alginate. In this report, we confirm a role for gac genes in both algD expression and alginate production in B728a. However, B728a mutants completely devoid of detectable alginate were as virulent as the wild-type strain in our assay. Thus, factors other than, or in addition to, a deficiency of alginate must be involved in the lack of pathogenicity observed with gacS and gacA mutants.
Alginate production has long been studied in the genus Pseudomonas due to the link between cystic fibrosis symptoms in humans and the isolation of mucoid Pseudomonas aeruginosa from patient lungs (2). Since the production of extracellular polysaccharide had also been implicated in the virulence of several phytopathogenic bacteria such as Erwinia stewartii (3) and Ralstonia solanacearum (10, 22), the strong foundation of the molecular genetics of the alginate biosynthetic pathway in P. aeruginosa (23) made a natural starting point for studies in closely related plant pathogens such as P. syringae that were also known to produce alginate (6). The alginate biosynthetic genes in P. syringae are highly conserved compared to the P. aeruginosa biosynthetic cluster (5, 7, 17). A role for alginate production in pathogenic interactions of P. syringae pathovars on their plant hosts had been suggested (4, 21, 27). Mutant screens in another plant pathogen, Pseudomonas viridiflava, had demonstrated a link between the gacS-gacA regulon in that organism and the production of alginate and pathogenicity (15). These results were reinforced by the finding that algD, a biosynthetic gene central to alginate production, requires gacS for its efficient expression in Azotobacter vinelandii (1). The gacS-gacA two-component system is widely distributed in gram-negative bacteria and regulates diverse gene systems involved with moderating the bacterial interaction with the extracellular milieu. The two members of this gene pair, either singly or together, have been implicated in the expression of a wide variety of phenotypes in a number of bacterial genera. Of particular significance have been results that showed that gacS and gacA play a role in regulating virulence factors in animal pathogens such as Salmonella spp. (9) and Vibrio cholerae (26). In P. syringae pv. syringae B728a, a causal agent of bacterial brown spot of the snap bean, gacS and gacA have been shown to regulate bacterial swarming, the production of syringomycin, protease, and N-acyl-l-homoserine lactone, in addition to the pathogenicity defects originally described (12; see also reference 13). However, none of these individual phenotypes have been directly related to the effects of gac mutations on pathogenicity. The requirement for gacS and gacA in alginate production (1, 14) and the reported involvement of alginate in pathogenic interactions on bean plants (27) made it seem possible that alginate was a significant contributing factor to the lesion-minus phenotype exhibited by gacS and gacA mutants of B728a. Here we report that this is probably not the case.
An ongoing project in our laboratory uses the reporter transposon TnlacZ to directly search for genes that are affected for expression in a B728a gacA mutant background (15; T. G. Kinscherf, J. J. Holmstadt, E. M. Ostertag, A. K. Savage, C. A. Hinkley, T. Kitten, J. L. McEvoy, and D. K. Willis, unpublished results). During the course of this work, two transposon insertions were isolated, cloned, and characterized as being in the algD gene. This locus encodes GDP-mannose dehydrogenase (2) and is the first gene in the alginate biosynthetic operon of P. syringae pv. syringae (17). Partial sequence analysis indicated appropriately high similarities to Pseudomonas algD genes already in the GenBank database. A cosmid containing the TnlacZ insertion in algD was mated into strains B728a (wild type), BGACΩ1 (gacA), and BSAL1 (salA). Potential kanamycin-resistant, tetracycline-sensitive chromosomal exchanges were isolated in all three backgrounds and were checked by Southern blotting for the recombinational inheritance of the TnlacZ mutation (data not shown).
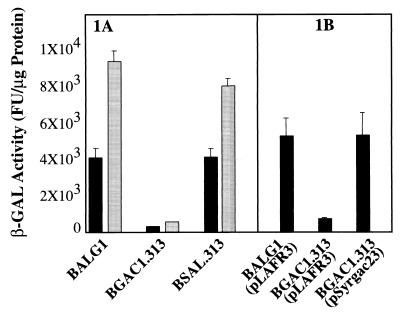
B728a was tested over a range of sorbitol and NaCl concentrations on mannitol-glutamic acid-yeast extract (MGY) plates at 28°C, and maximal alginate production (as evidenced by visual mucoidy) occurred in the presence of 0.6 M sorbitol (data not shown), in agreement with the previously published data (17). In contrast, the algD exchange mutant BALG1 was nonmucoid and did not produce detectable alginate under these same conditions (Table 1). Mucoidy was restored to BALG1 by the introduction of plasmid pSK2 containing the alginate biosynthetic cluster from the P. syringae pv. syringae strain FF5 (17), indicating that the insertion in algD was causal to the nonmucoid phenotype. Maximal expression of the chromosomal TnlacZ reporter in strain BALG1 also occurred at 0.6 M sorbitol in MGY liquid medium (data not shown). Figure 1A shows the expression of the chromosomal algD1::TnlacZ reporter in various genetic backgrounds (wild type, gacA2::Ω, and salA1::Ω) in the presence or absence of 0.6 M sorbitol. The addition of 0.6 M sorbitol to the medium resulted in an approximately twofold induction of algD expression from all strains tested regardless of the mutational background. This twofold effect of sorbitol was independent of the presence or absence of the gacA gene (Fig. 1A), although the lack of an intact gacA gene by itself caused a dramatic reduction of expression (12.7- to 16.7-fold) within both of the gacA mutant reporter strains tested. Expression of the algD reporter was restored completely by introduction of the gacA gene on a plasmid (Fig. 1B). Table 1 shows that neither the gacS mutant NPS3136 nor the gacA mutant BGACΩ1 produces alginate, confirming the regulation of the alginate pathway by this two-component regulator. The salA gene is a regulator of antibiotic production and virulence in B728a that is dependent upon gacS and gacA for its expression (13). The salA mutant BSAL1 produced normal levels of alginate (Table 1), and a mutation in salA did not significantly affect either the basal expression of the algD reporter or the twofold sorbitol induction of expression (Fig. 1). In addition, the alginate production of the acyl-homoserine lactone-deficient mutant BHSL (12) was not affected (Table 1). This indicates that alginate production lies in a separate branch of the gac regulon from salA. We also tested a collection of field strains of P. syringae pv. syringae and their respective gacS mutants (19) and, in all cases, alginate production (as judged by mucoidy on MGY medium containing sorbitol) was found to be gacS dependent (data not shown).
TABLE 1.
Alginate production by P. syringae pv. syringae B728a and various mutant strains
| Strain | Description | Alginate productiona (μg/mg of total protein)b |
|---|---|---|
| B728a | Wild type | + (495) |
| NPS3136 | gacS1::Tn5 mutant of B728a (25) | − (<20) |
| BGACΩ1 | gacA2::Ω mutant of B728a; the Ω cassette (18) was cloned into the gacA gene and exchanged into the chromosome of B728a (Kitten et al.)c | − (<20) |
| BALG1 | algD1::TnlacZ insertion exchanged into B728a | − (<20) |
| BALG1(pSK2) | Alginate biosynthetic region (17) mated into BALG1 | + (442) |
| BHSL | ahlI::lacZ mutant of B728a (12) | + (372) |
| BSAL1 | salA1::Ω mutant of B728a (13) | + (588) |
| BGAC1.313 | algD1::TnlacZ reporter exchange into BGACΩ1 | − (ND) |
| BGAC2.313 | Independent algD::TnlacZ reporter exchange into BGACΩ1 | − (ND) |
| BSAL.313 | algD::TnlacZ reporter exchange into BSAL1 | − (ND) |
| B728a(pLAFR3) | B728a containing plasmid vehicle pLAFR3 (24); used as a wild-type plasmid-containing control | + (ND) |
| BALG1(pLAFR3) | BALG1 containing plasmid vehicle pLAFR3 (24); used as a plasmid-containing control | − (ND) |
| BGAC1.313(pLAFR3) | BGAC1.313 containing plasmid vehicle pLAFR3 (24); used as a plasmid-containing control | − (ND) |
| BGAC1.313(pSyrgac23) | BGAC1.313 containing a wild-type gacA gene in pLAFR3 to yield plasmid pSyrgac23 (20) | − (ND) |
The plus or minus sign indicates whether mucoidy was produced on MGY (17) plus 0.6 M sorbitol plates.
Alginate was quantitated using a modified version of a previously described assay (16); the standard errors were ±120, ±78, ±66, and ±70 for B728a, BALG1(pSK2), BHSL, and BSAL1, respectively. Other strains tested too low for meaningful standard error or were not done (ND).
T. Kitten, E. M. Ostertag, T. G. Kinscherf, and D. K. Willis, unpublished data.
FIG. 1.
algD TnlacZ reporter activity in B728a mutants. Six individual β-galactosidase assays were performed (FluorReporter lacZ/Galactosidase Quantitation Kit F-2905; Molecular Probes) with each culture of strains grown in MGY (17) for 16 h at ambient temperature with aeration. Shown is the mean β-galactosidase activity as fluorescence units (FU)/microgram of total cellular protein for the mean of two repetitions of the experiment. The error bars represent the standard errors of the means. (A) Expression of the reporter in algD in various genetic backgrounds without (black column) or with (gray column) 0.6 M sorbitol added to the MGY medium. The β-galactosidase activity from BGAC1.313 was 322 (±21) FU without or 566 (±23) FU with sorbitol in the medium. The independent exchange mutant BGAC2.313 gave 310 (±27) FU and 563 (±23) FU, respectively (data not shown). (B) Restoration of algD expression by wild-type gacA on a plasmid (pSyrgac23). Strains were grown in MGY containing 0.6 M sorbitol and 10 μg of tetracycline per ml. The β-galactosidase activity from BGAC1.313(pLAFR3) was 677 (±99) FU. The cosmid pLAFR3 was used as the vector control for pSyrgac23.
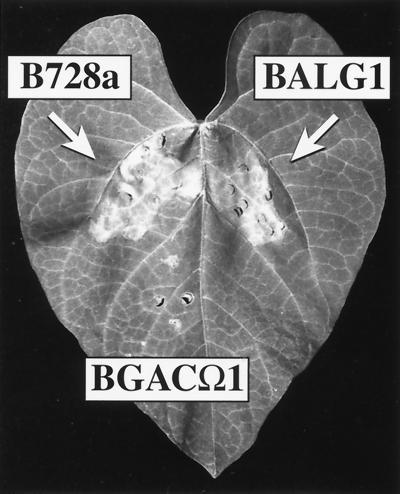
The effect of TnlacZ insertions in algD on the virulence of B728a on the snap bean (Phaseolus vulgaris) was tested in the same bean leaf infiltration assay we used to define the nonpathogenic phenotype of gacS and gacA mutants (Fig. 2). Primary bean leaves were inoculated with our standard assay range of cell concentrations, i.e., 107, 106, and 105 CFU/ml. No difference in the virulence level was observed between B728a and BALG1 in three experiments, with lesion manifestation occurring with both strains at all of the tested inoculum levels. When the gacA mutant BGACΩ1 was inoculated along with the other strains in the same experiment, no disease symptoms were observed for that strain, as always (Fig. 2). Bacterial growth following infiltration of primary bean leaves was assayed using B728a(pLAFR3), BALG1(pLAFR3), and the alginate-restored mutant BALG1(pSK2) in a leaf infiltration growth assay (25). The in planta population growth of the three strains was essentially identical over a period of 3 days (data not shown), at which time the level of necrosis made further collection of leaf disks problematic with all three strains.
FIG. 2.
Lesion formation by B728a and mutant derivatives on bean leaves. Bacterial suspensions were made from King's medium B plates grown for 2 days at 28°C. Bacteria were suspended in water and diluted to 106 CFU per ml in 10 mM phosphate buffer (pH 7.2). Bacteria were infiltrated locally into the leaves of 14-day-old bean plants. The photograph was taken 3 days postinoculation using a Kodak DCS420 digital camera. The arrows indicate the pathogenic reaction caused by infiltration of B728a or BALG1. The small circular wounds within the area infiltrated by BGACΩ1 were caused by the infiltration process.
The data presented here clearly support the inference from earlier work (1, 14) that gacS and gacA regulation of alginate production is general among pseudomonads and probably among other gram-negative bacteria. The profound effects of gacA mutations on the expression of algD would appear to be sufficient to explain the requirement for functional gacS and gacA genes in alginate production. The algD gene is the first gene in the P. syringae biosynthetic cluster and encodes GDP-mannose dehydrogenase, an essential synthetic enzyme in the alginate pathway (17). However, it is important not to oversimplify the situation, since it is clear that multiple levels of regulation are involved in this system. While sorbitol had only relatively small effects on algD expression, it was still absolutely required for the manifestation of mucoidy on plates. This suggests that osmolarity might be affecting the expression of other genes involved in alginate production. The algT gene has been shown to be osmoregulated and encodes a sigma factor (ς22) that is required for alginate production (11). The conditional expression of this sigma factor would have the capacity to affect multiple loci within the biosynthetic cluster, and it may be the cumulative effects of this regulation that produces the requirement for osmotic stress that we observed. Previous studies measuring expression of an algD reporter on a plasmid reported significantly greater effects for sorbitol induction, possibly reflecting differences in plasmid copy number (17). We found that a chromosomal location for the reporter was critical for the accurate determination of algD expression. Our preliminary tests using the algD reporter on a plasmid in P. syringae resulted in only a threefold gac effect on expression and no significant induction by sorbitol (data not shown). This effectively masked the separation of the gac and sorbitol effects that our chromosomal reporter demonstrated, with the twofold sorbitol induction occurring independently of the presence or absence of an intact gacA gene. The environmental signal(s) to which the gacS-gacA system responds remains unknown, and our results indicate that it is neither sorbitol nor the osmotic effects of sorbitol that provide this signal. This finding was reinforced by other experiments that showed that the expression of a salA reporter fusion, previously demonstrated to be strongly regulated by gacA (13), was also unaffected by 0.6 M sorbitol (data not shown).
The lack of effect by a salA mutation on either algD expression or alginate production demonstrates that alginate production lies in the salA-independent branch of the gacS-gacA regulon. The salA gene was originally isolated as a copy number suppressor of gacS phenotypes in B728a and was subsequently found to encode a regulator that was dependent on the presence of intact gacS and gacA genes for its expression (13). Phenotypes associated with salA mutations in B728a are defects in antibiotic production and severe attenuation of pathogenicity in laboratory assays. These represent a subset of known gacS and gacA phenotypes in B728a, and thus, salA effectively defines the branch point in the gac regulon leading to the manifestation of plant disease. From this perspective it is not too surprising that alginate production, being in the salA-independent part of the regulon, does not appear to play a major role in virulence. Repeated infiltration experiments failed to demonstrate deficiencies in lesion forming ability by the algD mutant relative to the wild type (Fig. 2). Previous work reported by other investigators described a small decrease in lesion number in growth chamber experiments with a P. syringae mutant affected in the algL gene (27), as well as the absence of satellite lesions in infiltrated leaves. We have never observed satellite lesions around sites infiltrated with B728a (19), nor have others that regularly perform bean infiltration assays with P. syringae pv. syringae (S. Hirano, personal communication). The leaf infiltration assay that is used in our laboratory is very reproducible and has shown a high degree of correlation with results from large-scale field studies (8; S. S. Hirano and C. D. Upper, unpublished data). This is the method that we have routinely used to define the pathogenicity phenotype in gacS and gacA mutants of B728a (13, 20, 25). Thus, the virulence exhibited by algD mutants in this assay would appear to make it unlikely that the gacS-gacA-mediated loss of alginate production is responsible for the complete lack of pathogenicity exhibited by gacS and gacA mutants (Fig. 2). It remains to be determined if defects in alginate production contribute to a loss of fitness under field conditions such as has been demonstrated with a gacS mutant (8). This possibility is currently under investigation.
Acknowledgments
We thank Susan Hirano and Chris Upper for helpful suggestions on the manuscript.
This work was briefly supported by NSF grant MCB-9419023 to D. K. Willis, and J. J. Holmstadt was supported by the NSF/DOE/USDA Arabidopsis Training Grant at UW—Madison.
REFERENCES
- 1.Castaneda M, Guzman J, Moreno S, Espin G. The GacS sensor kinase regulates alginate and poly-beta-hydroxybutyrate production in Azotobacter vinelandii. J Bacteriol. 2000;182:2624–2628. doi: 10.1128/jb.182.9.2624-2628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deretic V, Gill J F, Chakrabarty A M. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987;15:4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolph P J, Majerczak D R, Coplin D L. Characterization of a gene cluster for exopolysaccharide biosynthesis and virulence in Erwinia stewartii. J Bacteriol. 1988;170:865–871. doi: 10.1128/jb.170.2.865-871.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fett W F, Dunn M F. Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol. 1989;89:5–9. doi: 10.1104/pp.89.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fett W F, Wijey C, Lifson E R. Occurrence of alginate gene sequences among members of the pseudomonad rRNA homology groups I–IV. FEMS Microbiol Lett. 1992;78:151–157. doi: 10.1016/0378-1097(92)90017-i. [DOI] [PubMed] [Google Scholar]
- 6.Fett W M, Osman S F, Fishman M L, Siebles T S., III Alginate production by plant-pathogenic pseudomonads. Appl Environ Microbiol. 1986;52:466–473. doi: 10.1128/aem.52.3.466-473.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fialho A M, Zielinski N A, Fett W F, Chakrabarty A M, Berry A. Distribution of alginate gene sequences in the Pseudomonas rRNA homology group I-Azomonas-Azotobacter lineage of superfamily B procaryotes. Appl Environ Microbiol. 1990;56:436–443. doi: 10.1128/aem.56.2.436-443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano S S, Ostertag E M, Savage S A, Baker L S, Willis D K, Upper C D. Contribution of the regulatory gene lemA to field fitness of Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1997;63:4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 10.Kao C C, Barlow E, Sequeira L. Extracellular polysaccharide is required for wild-type virulence of Pseudomonas solanacearum. J Bacteriol. 1992;174:1068–1071. doi: 10.1128/jb.174.3.1068-1071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keith L M, Bender C L. AlgT (ς22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J Bacteriol. 1999;181:7176–7184. doi: 10.1128/jb.181.23.7176-7184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinscherf T G, Willis D K. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone gene ahlI. J Bacteriol. 1999;181:4133–4136. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly-identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–930. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 14.Liao C-H, McCallus D, Fett W. Molecular characterization of two gene loci required for production of the key pathogenicity factor pectate lyase in Pseudomonas viridiflava. Mol Plant-Microbe Interact. 1994;7:391–400. doi: 10.1094/mpmi-7-0391. [DOI] [PubMed] [Google Scholar]
- 15.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May T B, Chakrabarty A M. Isolation and assay of Pseudomonas aeruginosa alginate. Methods Enzymol. 1994;235:295–304. doi: 10.1016/0076-6879(94)35148-1. [DOI] [PubMed] [Google Scholar]
- 17.Peñaloza-Vázquez A, Kidambi S P, Chakrabarty A M, Bender C L. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J Bacteriol. 1997;179:4464–4472. doi: 10.1128/jb.179.14.4464-4472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 19.Rich J J, Hirano S S, Willis D K. Pathovar-specific requirement for the Pseudomonas syringae lemA gene in disease lesion formation. Appl Environ Microbiol. 1992;58:1440–1446. doi: 10.1128/aem.58.5.1440-1446.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph K W E, Gross M, Ebrahim-Nesbat F, Nollenburg M, Zomorodian A, Wydra K, et al. The role of extracellular polysaccharide as virulence factors for phytopathogenic pseudomonads and xanthomonads. In: Kado C I, Crosa J H, editors. Molecular mechanisms of bacterial virulence. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 357–378. [Google Scholar]
- 22.Saile E, McGarvey J A, Schell M A, Denny T P. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 1997;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 23.Shankar S, Ye R W, Schlictman D, Chakrabarty A M. Exopolysaccharide alginate synthesis in Pseudomonas aeruginosa: enzymology and regulation of gene expression. Adv Enzymol Relat Areas Mol Biol. 1995;70:221–255. doi: 10.1002/9780470123164.ch4. [DOI] [PubMed] [Google Scholar]
- 24.Staskawicz B J, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol Plant-Microbe Interact. 1990;3:149–156. [Google Scholar]
- 26.Wong S M, Carroll P A, Rahme L G, Ausubel F M, Calderwood S B. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect Immun. 1998;66:5854–5861. doi: 10.1128/iai.66.12.5854-5861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Peñaloza-Vázquez A, Chakrabarty A M, Bender C L. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol Microbiol. 1999;33:712–720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]