Abstract
Main conclusion
Plant responds to Agrobacterium via three-layered immunity that determines its susceptibility or resistance to Agrobacterium infection.
Abstract
Agrobacterium tumefaciens is a soil-borne Gram-negative bacterium that causes crown gall disease in plants. The remarkable feat of interkingdom gene transfer has been extensively utilised in plant biotechnology to transform plant as well as non-host systems. In the past two decades, the molecular mode of the pathogenesis of A. tumefaciens has been extensively studied. Agrobacterium has also been utilised as a premier model to understand the defence response of plants during plant–Agrobacterium interaction. Nonetheless, the threat of Agrobacterium-mediated crown gall disease persists and is associated with a huge loss of plant vigour in agriculture. Understanding the molecular dialogues between these two interkingdom species might provide a cure for crown gall disease. Plants respond to A. tumefaciens by mounting a three-layered immune response, which is manipulated by Agrobacterium via its virulence effector proteins. Comparative studies on plant defence proteins versus the counter-defence of Agrobacterium have shed light on plant susceptibility and tolerance. It is possible to manipulate a plant’s immune system to overcome the crown gall disease and increase its competence via A. tumefaciens-mediated transformation. This review summarises the recent advances in the molecular mode of Agrobacterium pathogenesis as well as the three-layered immune response of plants against Agrobacterium infection.
Keywords: Agrobacterium tumefaciens, Plant–Agrobacterium interaction, Plant immunity
Introduction
Agrobacterium tumefaciens is a soil-borne, Gram-negative bacterium that infects and causes tumours, called crown gall disease, in a variety of plant species. More than a century ago, Agrobacterium was isolated from a crown gall tumour by two plant pathologists Smith and Townsend (Smith and Townsend 1907). There were originally three biovars of pathogenic Agrobacteria based upon the host range and manner of pathogenic response in the host. Biovar I includes A. tumefaciens, biovar II includes A. rhizogenes and biovar III includes A. vitis (Slater et al. 2009). However, research on the taxonomic classification and position of Agrobacterium is still ongoing (Gan and Savka 2018; Ormeno-Orrillo et al. 2015). Among all biovars, biovar I, Agrobacterium (C58), was the first whose genome was sequenced and made available in the database (https://www.ncbi.nlm.nih.gov/genome), where it was renamed as A. fabrum. Furthermore, Shams et al. used another synonym for Agrobacterium, i.e. Rhizobium radiobacter (Shams et al. 2013). For the convenience of the readers, the older name, i.e. A. tumefaciens will be used in this review because it is predominantly used in ongoing research and review articles.
There are two distinct lifestyles of Agrobacterium in nature: one is free-living, saprophytic and non-pathogenic and the other one is pathogenic, based on plant tissue as its ecological niche instead of soil (Meyer et al. 2018). In bacteria, the change between these two lifestyles is coordinated with the change in the gene expression pattern resulting from the perception of environmental cues (Duprey et al. 2014; Valentini et al. 2018). Agrobacterium begins its pathogenic lifestyle when it perceives signals from wounded plant cells. There is a large tumour-inducing (Ti) plasmid in Agrobacterium, which confers pathogenicity. All Ti plasmids contain at least four gene clusters or operons. These operons possess different functions. For instance, the Vir operon contains all the virulence genes that get activated during the pathogenic process, the repABC operon maintains the replication and separation of the Ti plasmid, the tra operon facilitates the conjugation of DNA and the trb operon synthesises the secretion system required when transmitting pTi from one to another bacterium (Wetzel et al. 2015). The virulence protein processes the T-DNA region of the Ti plasmid in response to an environmental signal. During processing, only 25 base pair direct repeats at the left and right borders of the T-DNA are processed. The T-strand, along with several virulence proteins, enters the host cytoplasm, travels towards the nucleus and is integrated into the plant genome (Nester 2015; Gelvin 2017). As T-DNA harbours auxin (iaaM, iaaH), cytokinin (ipt) and opine synthesis genes, it causes hormonal imbalances in plants and results in malignant growth. Opine synthesis genes lead to the production of opines, which serve as a source of nutrition for Agrobacterium and create a new ecological niche for it (Lacroix and Citovsky 2013). Recently, Agrobacterium fitness gene has been identified, which constitutes 3–8% of its total genes and is important for its competitive survival in plants (Torres et al. 2022).
Agrobacterium ranks third among the most pathogenic bacteria, next only to Pseudomonas syringae pathovars and Ralstonia solanacearum (Mansfield et al. 2012), which severely affect plant growth and vigour during crown gall disease. Agrobacterium has been known for several years as a plant pathogen. Studies have reported that the native T-DNA of Agrobacterium can be replaced with any gene of interest and that it can transform plants without causing tumours (Fraley et al. 1983; Caplan et al. 2019).
The unintended plant transformation activity of Agrobacterium makes it not only an important plant pathogen but also a potent biotechnological tool. Recent studies have also shown that Agrobacterium can transform non-host plants (Song et al. 2019) as well as non-plant systems, such as yeast (Bundock et al. 1995), fungi (De Groot et al. 1998; Li et al. 2017) and even human cells (Lacroix et al. 2006; Lacroix and Citovsky 2018).
Moreover, Agrobacterium has been utilised for the production of pharmaceutical proteins in plants (Kopertekh and Schiemann 2019). While Agrobacterium is extensively used in the field of plant biotechnology and serves as a model for plant–pathogen interaction, molecular mode of pathogenesis, etc., some plants are recalcitrant to transformation. The recalcitrance depends on the type of Agrobacterium used, the type of plant and the explants (Tzfira and Citovsky 2006). Moreover, the extensive use of Agrobacterium in biotechnological industries overshadowed the adverse effect of its natural action, namely crown gall disease. Hence, it is important to understand the innate immune response of plants and the downstream defence activation during plant–Agrobacterium interaction.
In nature, plants and pathogens coevolved over time. Plants have evolved a complex and versatile immune response that detects the pathogen with a wide array of receptors. Simultaneously, pathogens have managed to escape from the plant immune system with the help of effector proteins. The innate immune system of a plant is triggered during pathogen attack. The plant does not recognise the whole pathogen but instead detects its signature molecular pattern. In case of pathogens, these molecular patterns are called ‘pathogen-associated molecular pattern (PAMP)’. On the contrary, in the case of non-pathogens, the molecular signals are termed ‘microbe-associated molecular pattern (MAMP)’ (Ausubel 2005). These PAMPs are a part of the general elicitor and are present in a vast group of pathogenic bacteria that are evolutionary stable and essential for the pathogenesis of the microbe. Besides PAMP, damage-associated molecular pattern (DAMP), a product of host cellular damage after pathogen invasion, also triggers the immune response. Plants are able to detect these extracellular and intracellular milieus using a cell surface receptor named pattern recognition receptor (PRR). In this way, the plant immune system serves as a surveillance system for detecting these signals during pathogen attack (Cook et al. 2015; Gust et al. 2017). The recognition of PAMP by PRR is the first level of defence and is referred to as PAMP-triggered immunity (PTI). PTI is a determinant of the plant resistance at an initial level that affects both basal and non-host resistance. Basal resistance denotes the resistance of susceptible plants after getting infected with the adapted pathogen. However, when a plant develops resistance to a non-adapted pathogen, either pathogenic or non-pathogenic, it is called non-host resistance (Couto and Zipfel 2016; Tang et al. 2017). As soon as PAMP perception and PTI activation occur, the downstream signalling event commences immediately and blocks the infection process at an early stage. The downstream signalling events include the reactive oxygen species (ROS) burst and the activation of different kinases, such as the mitogen-activated protein kinase (MAPK) cascade that phosphorylates the defence-related genes (Noman et al. 2019). To overcome PTI, the pathogen makes plant cells more susceptible to infection using numerous virulence effector proteins. This process of pathogen counteraction is called effector triggered susceptibility (ETS). A plant’s compatibility with the pathogen is further determined by the second level of defence, which is called effector triggered immunity (ETI). ETI causes a hypersensitive reaction involving localised cell death of the host cells to prevent infection propagation (Janda et al. 2019). This review summarises the latest research on the molecular mechanism of T-DNA transfer in plant cells, with a focus on understanding the plant defence mechanisms and Agrobacterium counter-defence during plant–Agrobacterium interaction.
Molecular mechanism of Agrobacterium-mediated pathogenesis
Agrobacterium is used as a model organism to study plant–pathogen interactions. The pathogenesis of Agrobacterium has been studied extensively at the bacterial level. Despite this study, it is unclear how plants respond to this process. Only a small number of observations have been reported regarding host factors and their importance in Agrobacterium pathogenesis. These observations confirm that Agrobacterium, apart from its virulence protein, utilises the host cellular machinery for T-DNA cytoplasmic trafficking, nuclear import and its integration into the plant genome (Gelvin 2003; Citovsky et al. 2004; Michielse et al. 2004; Li and Pan 2017; Yang et al. 2017). As a pathogen, Agrobacterium has been extensively studied. Its pathogenesis begins with the activation of its virulence genes after being stimulated by a chemical signal released by the plant cell (Gelvin 2017). The entire process is quite complex and has already been examined extensively in many articles. Therefore, this review discusses only the most recent advancements. There are four steps in the infection process: (1) release of chemical signals and the onset of pathogenic lifestyle of Agrobacterium, (2) activation and induction of the virulence gene, (3) generation of the T-complex and its cytoplasmic trafficking and nuclear import and (4) integration of T-DNA into the plant genome and its expression.
Release of chemical signal and onset of pathogenic lifestyle of Agrobacterium
When plants are subjected to biotic or abiotic stresses, the very first response is oxidative burst, which is followed by the production of phenolic compounds and other secondary metabolites (Baker et al. 2020). Acetosyringone (AS), a phenolic metabolite of plants, is most effective (Guo et al. 2017). AS is synthesised via the phenylpropanoid pathway (Maury et al. 2010), and phenylpropanoids are involved in plant defence (Fraser and Chapple 2011). However, Agrobacterium somehow utilises this defence signal to initiate infection. Recent studies on Agrobacterium have identified an antibiotic-resistant RND-type efflux pump called the MexE/MexF/AmeC pump, which enhances the concentration of the inducer required for the induction of virulence genes (Binns and Zhao 2020). Agrobacterium becomes chemotactic and travels towards the wound site once it finds an appropriate signal (Guo et al. 2017). CheW proteins of Agrobacterium are implicated in bridging CheA kinase and chemoreceptor, thus forming a core chemoreceptor complex that facilitates chemotaxis (Huang et al. 2018). When the process is successful, Agrobacterium attaches to the plant cell with the help of binding and attachment proteins (encoded by ChvA, ChvB, PscA and Att) (Tzfira and Citovsky 2002; Cangelosi et al. 2007).
Activation and induction of the virulence gene
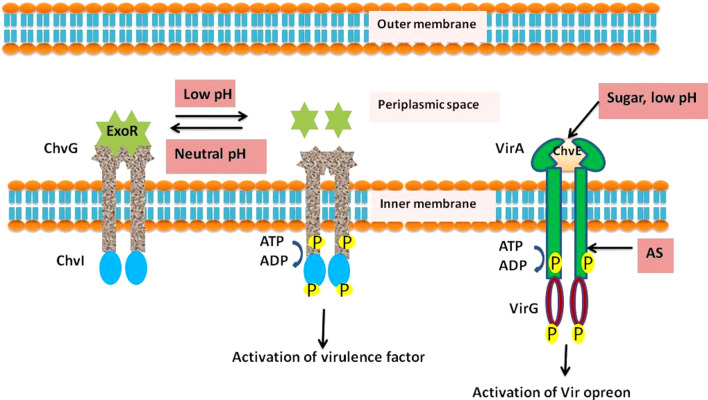
The rhizosphere must contain three factors to activate and induce the virulence system of Agrobacterium, namely, low pH, sugar and phenolic compounds (Lacroix and Citovsky 2013). Agrobacterium’s virulence system is triggered by low pH and sugar, but these factors are not necessary to induce the virulence genes. Phenolic compounds, such as and hydroxyacetosyringone, induce the virulence genes. Virulence property of an Agrobacterium is determined by its chromosomal virulence genes (chv) and Ti plasmid virulence genes (vir genes). ExoR, a periplasmic regulator, detects low pH and activates the virulence process. Under neutral pH, ExoR interacts with the chromosome-based two-component regulatory system ChvG. However, under acidic pH condition, ExoR gets cleaved by periplasmic proteases and frees ChvG, which gets autophosphorylated and transfers its phosphate group to the response regulator ChvI (Heckel et al. 2014; Wu et al. 2012; Yuan et al. 2008; Subramoni et al. 2014). ChvI, in turn, activates the other virulence genes and also T6SS (Yuan et al. 2008; Wu et al. 2012), and the Ti plasmid-based virulence protein, VirG (Li et al. 2002; Yuan et al. 2008). It is noteworthy that low pH not only activates the ExoR-ChvG-ChvI regulatory system but also suppresses the plant’s defence response. Stable low pH affects the distribution of Ca+2 ions and suppresses the expression of the marker defence-related genes NDR1/HIN1-like 10 (NHL10) and FLG22-induced receptor-like kinase 1 (FRK-1), thereby enhancing the susceptibility of Agrobacterium-mediated gene transfer (Wang et al. 2018a). Although VirG transcription is activated by the ExoR-ChvG-ChvI cascade, VirG induction requires the phosphorylation of its Asp 52 residue by VirA sensor kinase after detecting the phenolic signal. Additionally, sugar from the plant exudates acts as a signalling molecule for virulence activation and is detected by other chromosome-based virulence proteins, such as ChvE (Cangelosi et al. 1990). Interestingly, low pH increases the affinity of ChvE to sugar (Hu et al. 2013). Additionally, low pH promotes the interaction of the ChvE protein with VirA at its periplasmic domain, which results in VirA protein activation (Gao and Lynn 2005; Nair et al. 2011). The host range of A. tumefaciens is reportedly determined by the interaction between ChvE and VirA (He et al. 2009; Hu et al. 2013). VirA/VirG is a plasmid-based regulatory system in Agrobacterium that is essential for its virulence (Lin et al. 2014; Wise and Binns 2016). In the two-component regulatory system VirA/VirG, VirA acts as sensor histidine kinase, whereas VirG acts as a response regulator. When AS is detected by VirA, it gets autophosphorylated and, in turn, phosphorylates the Asp 52 residue of VirG response regulator, which is located in the cytoplasm of Agrobacterium. In the Ti plasmid, phosphorylated VirG binds to the 12 bp Vir box region, which is located upstream of the transcriptional start site of the virulence gene (Subramoni et al. 2014). As a result, the vir genes, which are distributed in 11 operons, are activated. The vir genes include virA, virB,virC, virD, virE, virF, virG, virH, virK, virL and virM. The virB operon encodes most proteins, 11 in total, whereas the virA and virG operons always exhibit low levels of expression (Nester 2015) so that they can sense the extracellular stimuli. The model for signal perception is depicted in Fig. 1. The single-stranded T-strand is generated from the Ti plasmid via nicking at the right and left borders by VirD2 and VirD1. Additionally, the generation of multiple copies of T-strand and its conjugative transfer are maintained by VirC1 and VirC2 proteins (Atmakuri et al. 2007). These four virulence effector proteins contribute to efficient T-strand generation, and VirD2 remains associated with the right border of T-strand. The T-strand–VirD2 nucleoprotein complex, along with several virulence effector proteins, such as VirE2, VirF, VirE3 and VirD5, is then translocated to the host cell via the type 4 secretion system (T4SS). T4SS consists of the cell envelope spanning transporter and the extracellular pilus and is synthesised from VirD4 and VirB1-B11 proteins (Li and Christie 2018). Agrobacterium and plant cells are physically attached to each other with the aid of the VirB2 protein of Agrobacterium and the cell membrane proteins of the plant. To detect the membrane protein involved in attachment to the bacterium, Hwang used yeast two-hybrid assays to screen out the interacting partner of VirB2 and observed that the Arabidopsis reticulon-like (RTNL) proteins, AtRTNLB1, AtRTNLB2 and AtRTNLB4, interacted with the VirB2 protein (Hwang and Gelvin 2004).
Fig. 1.
Model for signal perception in Agrobacterium. Three factors, low pH, sugar and phenolic compound (Acetosyringone:AS) are sensed by ChvG/ChvI, ChvE and VirA/VirG regulatory systems, respectively. Under neutral pH, ExoR remain bounded to ChvG and make them inactive which get activated under low pH by proteolytic cleavage and dissociation of ExoR. This allow the ChvG for autophosphorylation and transfer phosphate group to ChvI response regulator which then activate VirG. Sugar and low pH induces the ChvE for binding and activation of VirA. Simultaneously, AS perceived by cytoplasmic domain of VirA which allow the autophosphorylation of VirA. VirA phosphorylates VirG and induce it for activation of Vir operon for activation of other virulence proteins
VirD4 is a part of the transmembrane domain and may use ATP to transfer the T-strand to the pilus. VirD4 contains a C-terminal glutamine-rich conserved region that enables the recognition of the ‘VirD2-T-strand’ complex as a substrate for translocation in plants (Das 2020). VirE2 enters the host cell via clathrin-mediated endocytosis (Li and Pan 2017); however, the rest of the mechanism is yet to be elucidated.
Generation of the T-complex and its cytoplasmic trafficking and nuclear import
VirE2 is a single-stranded DNA binding (SSB) protein. VirE2 binds to the T-strand and forms a right-handed, cord-like structure inside the host cell, thus forming the T-complex (Abu-Arish et al. 2004). VirE2 has also been proposed to protect the T-strand from host nucleolytic degradation (Citovsky et al. 1989). Both VirE2 and VirD2 contain nuclear localization signals (NLSs), which import the T-strand into the nuclei. The VirD2 and VirE2 proteins use the host importins and VIP1 and VIP2 to create a ‘super T-complex’ that allows T-DNA nuclear import (Gelvin 2010, 2012; Guo et al. 2009, 2019; Shi et al. 2014). Prior to the nuclear import, the T-strand must, however, be trafficked in the host cytoplasm, which is largely accomplished by the VirE2 protein. To enter the nucleus, VirE2 gets associated with the F-actin network and the endoplasmic reticulum. Furthermore, VirE2 utilises the host myosin XI-K system for trafficking (Tu et al. 2018; Yang et al. 2017).
Integration of the T-DNA into the plant genome and its expression
When the T-DNA enters the host nucleus, it must first be stripped of its associated Vir and host proteins so that it can be integrated into the host genome. Agrobacterium VirF is similar to the F-box protein of the host, which is utilised to strip off the virulence proteins. VirF employs the host ubiquitin/proteasomal activity to degrade these proteins. VirF, however, is prone to proteasomal degradation in the host cell, and another virulence protein, VirD5, protects VirF inside the cell (Wang et al. 2014, 2018b). Interestingly, the Agrobacterium VirF mutant strain did not alter the transformation susceptibility in Arabidopsis or Nicotiana, which suggests that some other factor is involved in this step too. The host F-box protein has been identified and named as VIP1 binding F-box protein (VBF4). This protein targets VIP1 and VirE2 for proteasomal degradation via the S-PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1)-CULLIN1 (CUL1)-Fbox protein (SCFVBF) pathway (Zaltsman et al. 2010, 2013). It can, therefore, be said that although Agrobacterium utilises the host’s cellular machinery, it also possesses a backup strategy that involves the virulence protein.
It has been suggested that transcriptionally active regions of the genome that contain a high proportion of A = T sequences are the most likely sites for the integration of T-DNAs (Bourras et al. 2015). Later, the sequence of T-DNA/plant junction was analysed in plants without antibiotic selection, and the pattern of T-DNA integration was found to be random (Shilo et al. 2017). These junction sites are typically the double-stranded DNA repair sites in the plant genome (Kleinboelting et al. 2015; Gelvin 2017). Although the T-DNA integration process resembles a DNA repair process, such as non-homologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ), the integration does not entail the same proteins involved in these repair processes (van Attikum et al. 2003; Park et al. 2015).
The T-strand enters the nucleus as a single-stranded DNA, but whether the T-DNA integrates into the host genome as a single or double-stranded form is still under investigation. However, the double-stranded T-DNA integration is comparatively more favourable (Kleinboelting et al. 2015). In 2016, it was shown that the T-DNA is integrated as a double-stranded break at the microhomologous site in the genome via the annealing and repair process. In a study by van Kregten et al., it was proven that the mutation of DNA polymerase θ gene in Arabidopsis inhibits its stable transformation but not its transient transformation via Agrobacterium. The group suggested that DNA polymerase θ initiates the first step in T-DNA integration (Van Kregten et al. 2016). In contrast, Nishizawa-Yokoi et al. proposed that T-DNA integration occurs via multiple redundant pathways and that it might involve some other unknown pathway (Nishizawa-Yokoi et al. 2021).
A T-DNA sequence with eukaryotic promoter elements, such as the TATA box, CAAT box and polyadenylation signal, is expressed in eukaryotic cells (Zhang et al. 2015). This feature indicates the eukaryotic origin of the T-DNA fragment. T-DNA also exhibits microhomologies with eukaryotic promoter core elements at its 3′ end, which signifies its likelihood of integration at promoter sites (Bourras et al. 2012). The model for Agrobacterium-mediated T-DNA transfer is illustrated in Fig. 2A.
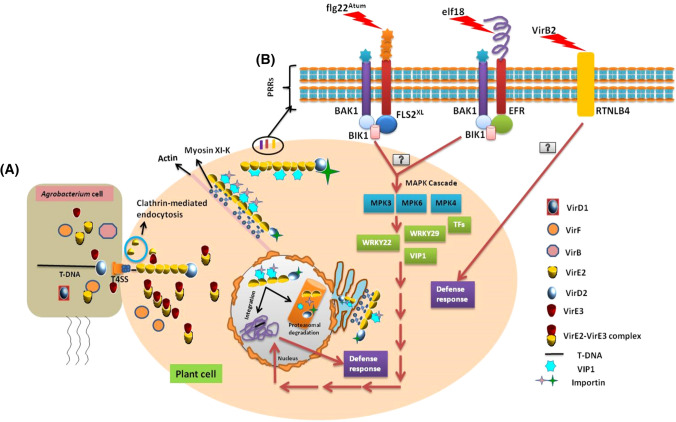
Fig. 2.
Agrobacterium-mediated T-DNA transfer and plant innate immunity. A Model for Agrobacterium-mediated T-DNA transfer: T-DNA and other virulence proteins getting out from Agrobacterium via T4SS. VirE2 enters in plant cell via clathrin-mediated endocytosis. T-complex associates with several host protein and trafficked via myosin XI-K system towards nucleus. After entering to the nucleus, T-DNA get stripped off with the help of VirF and then integrated to the genome. Other virulence factors is degraded by proteasomal degradation, B Model for PAPMs perception and plant immune response during Agrobacterium infection: flg22Atum and elf18 PAMPs of Agrobacterium recognised by cell surface PRRs, FLS2XL and EFR, respectively. VirB2 peptide also recognised by RTNLB4. The downstream MAP-Kinase cascade activation of FLS2XL and EFR induces several transcription factors which then enters to the nucleus and activates defence response
Defence response of the plant during plant–Agrobacterium interaction
Similar to the innate immune system of the animals, plants also have their defence system to fight against pathogen attack. When a wound occurs, plants secrete metabolites, such as H ions, phenolics and carbohydrates, to heal the cell damage at the wound site (Lacroix and Citovsky 2013). During wound formation, plant exudates are the first level of check to prevent Agrobacterium infection. For instance, unlike the crown gall-susceptible dicot plants, maize seedlings secrete chemicals that block the growth of Agrobacterium. Chemicals such as DIMBOA (2,4-dihydroxy7-methoxy-2H-1,4-benzixazin-3(4H)-one) and MDIBOA (2-hydroxy-4,7-dimethoxybenzoxazin-3-one) inhibit the growth of Agrobacterium (Zhang et al. 2000). Hence, the non-natural host of Agrobacterium is incapable of inducing virulence, thereby protecting it from crown gall disease. This review discusses the defence response of plants against Agrobacterium, which starts with the perception of pathogen signals, and then moves on to disease development.
Pathogen signal perception and innate plant immunity
Plants respond to elicitors, such as flagellin, elongation factor-thermo unstable (EF-Tu) and lipopolysaccharide, which trigger innate immune response during pathogen attack (Janda et al. 2019). The plant PRRs comprise two types of membrane receptors, namely, receptor-like kinases (RLKs) and receptor-like proteins (RLPs) (Couto and Zipfel 2016; Boutrot and Zipfel 2017). Both RLKs and RLPs possess extracellular and transmembrane domains; however, while RLKs contain an intracellular kinase domain, RLPs do not (Saijo et al. 2018). An Arabidopsis thaliana PRR that has been well characterised so far is flagellin sensing 2 (FLS2), which possesses leucine-rich repeats (LRRs) as a ligand-binding motif and belongs to the RLK family (Chinchilla et al 2007). FLS2 recognises the 22-amino-acid long peptide of flagellin (monomer of flagella), flg22. The perception of flg22 via FLS2 stimulates plant immune response by modulating protein activity and increasing reactive oxygen species (ROS) accumulation and phytohormone synthesis, such as salicylic acid (SA) and jasmonic acid (JA) (Bigeard et al. 2015). This step is followed by the activation of defence-related genes as well as callose deposition at the cell wall to strengthen the wall composition and prevent pathogen ingress into the plant (Muthamilarasan and Prasad 2013; Janda et al. 2019).
Some bacterial pathogens escape FLS2 immunodetection by modifying the flg22 epitope. In A. tumefaciens, for instance, flg22 is modified into flg22Atum, which is undetectable by the FLS2 immunoreceptor (Felix et al. 1999; Trdá et al. 2014). flg22Atum differs from flg22 in that half of the 22 amino acid residues are modified. As a part of the plant–pathogen co-evolution effort, the flg22Atum receptor from the cell culture of Vitis riparia was identified and named FLS2XL (FLS2 with eXtended Ligand recognition). FLS2XL had an additional 16-amino-acid extension, which might differ from FLS2 in the ligand recognition process (Fürst et al. 2020). In comparison with FLS2 (VrFLS2), FLS2XL can bind flg22Atum with a higher affinity than flg22. FLS2XL ligand accommodation in its extracellular domain differs from that of FLS2 in that the former accommodates the ligand with lesser steric hindrance. To understand the perception at the domain level, chimeric receptors with the extracellular LRR domain of FLS2XL and the cytoplasmic domain of VrFLS2 were generated, thus resulting in a chimeric FLS2XL (c-FLS2XL). The c-FLS2XL showed that 12–18 LRRs of FLS2XL were crucial for flg22Atum immunodetection; nevertheless, the other LRRs also provided sensitivity for flg22Atum epitope detection. Nicotiana plant expressing c-FLS2XL was found to be resistant to tumour formation after Agrobacterium infection. In contrast, the plant expressing FLS2 was susceptible to crown gall disease (Fürst et al. 2020). In addition to FLS2, the EF-Tu receptor (EFR) has been identified in Arabidopsis for its potential role in Agrobacterium infection. EFR belongs to the LRR-RLK type of PRR because FLS2 and EF-Tu from Agrobacterium work as eliciting ligands. EF-Tu is a highly conserved and abundant bacterial protein that plays a role in the protein translation process. EFR recognises the N-terminal 18 amino acid residues in EF-Tu, named elf18, which serves as a ligand epitope. In addition, EFR activates the downstream defence response, which is not identical but similar to FLS2 (Zipfel et al. 2006; Wan et al. 2019). Unlike FLS2 which is found in almost all higher plants, EFR is only found in the Brassicaceae family of plants. However, the transgenic expression of EFR in other groups of plants, such as rice (Lu et al. 2015; Schwessinger et al. 2015), wheat (Hj et al. 2015) tomato, Nicotiana and Medicago (Lacombe et al. 2010; Pfeilmeier et al. 2019), provides tolerance against pathogenic bacteria. By generating efr mutant Arabidopsis plants, it was confirmed that this pathway does not perceive elf18 and enables the infection of the plant by Agrobacterium (Zipfel et al. 2006). The manner in which EF-Tu gets exposed to the outer membrane osf Agrobacterium is still under research, but it has been shown that EF-Tu is secreted to the outer membranes by other pathogens, such as Xanthomonas campestris and Erwinia chrysanthemi (Watt et al. 2005; Kazemi‐Pour et al. 2004). The EF-Tu protein from Acinetobacter baumannii is associated with outer membrane vesicle (OMV), cell surface and fibronectin (Dallo et al. 2012). Based on these findings, it appears that EF-Tu is transported to the outer membrane by vesicle trafficking, but the precise mechanism is yet to be elucidated. It is noteworthy that the downstream signalling of both EFR and FLS2 generates the same kind of defence response during Agrobacterium infection in Arabidopsis. Also, the combination of EF-Tu and flg22 fails to induce synergistic defence responses but exerts the same impact on MAP-kinase transduction cascade (Zipfel et al. 2006; Dafny-Yelin et al. 2008). Because the cytoplasmic fragment of FLS2XL is similar to FLS2, it can be speculated that FLS2XL is likely to exhibit a signalling akin to that of FLS2 during Agrobacterium infection. Additionally, RTNLB4, a membrane associated protein that interacts with VirB2 in the T-pilus plays a possible role in defence during Agrobacterium infection (Hwang and Gelvin 2004). Upon infection with the elf18 peptide of Agrobacterium, plants with abnormal levels of RTNLB4, either overexpression or mutant lines, showed hampered immunity. Furthermore, the rtnlb4 mutant plants were more resistant to Agrobacterium infection than the wild-type plant, suggesting its probable role in addition to defence. Agrobacterium VirE2 utilises cytoplasmic trafficking to enter the nucleus. RTNLB4 is found on the plasma membrane and endoplasmic reticulum and is involved in intracellular trafficking. Therefore, RTNLB4 is supposedly involved in the intracellular transmission of T-DNA (Huang and Hwang 2020). Furthermore, RAB8A, 8B and 8D interact with several RTNLB proteins and participate in the Agrobacterium infection process (Huang et al 2021).
Because the VirB2 protein interacts with the RTNLB4 membrane protein, it was tested whether the VirB2 peptide acted as PAMP, similar to elf18 and flg22. Two peptide regions of VirB2, S111-T58 and I63-I80, were found to alleviate plant defence response, and the residues of the VirB2 peptide from Q-48 to V-101 might be involved in plant–Agrobacterium interaction. In addition, both, elf18 and VirB2 have been shown to activate the early defence-related genes, namely, MPK3, MPK6, WRKY22, WRKY29, FRK1 and PR1, during Agrobacterium infection (Huang and Hwang 2020).
MAPK signalling
As a response to PAMPs, PTI activates the MAPK signalling pathway in several ways, including protein phosphorylation, ROS burst and transcriptional reprogramming of defence genes (Boutrot and Zipfel 2017). A previously held theory suggested that PTI induced an ROS burst that acted upstream of the MAPK signalling cascade, but later it became clear that PTI signalling was split into two distinct pathways, one triggering MAPK signalling and the other triggering ROS burst. The β subunit of G-protein (AGB1) associates with NADPH-oxidase and contributes to ROS generation. The EFR-mediated PAMP perception of Agrobacterium during infection is associated with the AGB1-mediated ROS burst. The EFR-AGB1 downstream signalling hampers Agrobacterium infection in Arabidopsis (Xu et al. 2014; Ishikawa 2009). Somatic embryogenesis receptor kinase (SERK)3/brassinosteroid (BR)-associated kinase (BAK)1 is a key co-receptor that recognises PAMP via FLS2. In plants, BAK1 activates the MAPK signal transduction cascade. BAK1 has previously been reported to be activated during brassinosteroid hormone regulation, which, in turn, activates the brassinosteroid-insensitive 1 (BRI1) receptor. During innate immunity too, BAK1 interacts with BRI1 and then activates the MAPK signalling cascade (Heese et al. 2007; Bigeard et al. 2015). This cascade is composed of three signalling modules, namely, MAPKKKs, MAPKKs and MAPKs. Arabidopsis encodes 60 MAPKKKs, 20 MAPKKs and 10 MAPKs, which suggests that a single MAPK can activate multiple MAPKKs, which, in turn, can activate multiple MAPKKKs. In Arabidopsis, four MAPKs, namely, MPK3, MPK6, MPK4 and MPK11, have been found to be responsive to pathogen infection (Bigeard et al. 2015).
Defence responsive genes
A PTI response activates the MAP-kinase signal transduction cascade, which further activates the transcription factors such as WRKY33 and VIP1, thus leading to the activation of several defence-related proteins. An in-depth analysis of the interaction between Ageratum and Agrobacterium has been performed to determine how plants respond to Agrobacterium infection (Ditt et al. 2001). Ditt et al. showed that plants expressed defence responsive genes after 24–48 h of Agrobacterium infection, which mostly include PR protein, NtPRp27 from tobacco, defence responsive proteins of phenylpropanoid pathway, cytochrome P450 monooxygenase from Arabidopsis and disease-resistant protein, Xa21, from rice (Ditt et al. 2001, 2006). Also, Veena et al. demonstrated that the defence responsive genes are differentially expressed during the early hours (3–6 h) of Agrobacterium infection in plants, which include glutathione-S-transferase, PR genes and SARs (Veena et al. 2003).
In addition to triggering the innate immune response of plants, Agrobacterium hijacks the host machinery during infection. Arabidopsis VirE2 interacting protein 1 (VIP1), a bZIP transcription factor, is activated by phosphorylation via MAPK (MPK3), which further activates the pathogenesis-related 1 (PR1) promoter containing defence genes (Djamei et al. 2007). VIP1 binds to the VIP1 responsive element (VRE; ACNGCT) of the PR1 promoter (Pitzschke et al. 2009). VIP1 is localised in the cytoplasm under non-stress conditions; however, upon infection with Agrobacterium, it gets phosphorylated at the Ser79 residue (via MPK3) and relocalises to the nucleus to activate the defence genes (Djamei et al. 2007). VirE2, an Agrobacterium effector protein, utilises the nuclear import activity of VIP1 to transport T-DNA into the host nucleus (Tzfira et al. 2001). In another study, it was found that VirE2 binds to VIP1 and decreases its level inside the cell, which, in turn, lowers the level of defence response during Agrobacterium infection. (Shi et al. 2014). According to Lapham et al., VirE2 localised in the cytoplasm modulates plant RNAs and genes to promote transformation (Lapham et al. 2021).
Plant transcription factor VFP4 (VirF binding protein) and its downstream gene ATL31 have both been found to be activated in response to Agrobacterium infection, and the overexpression of the gene renders the plant resistant to infection. VFP4 differentially regulates the defence responsive genes, including antibacterial genes; thus, VFP4 provides another defence layer against Agrobacterium. The bacterium detects VFP4 via VirF effector protein, which is then processed for proteasomal degradation via the SCFVirF pathway (García-Cano et al. 2018). Besides Arabidopsis VIP1, HvVIP1 from barley has also been identified to be activated during Agrobacterium infection. The HvVIP1 protein contains a conserved bZIP domain and exhibits a positive correlation with barley’s PR genes, such as HvPR1, HvPR4 and HvPR10. Apart from PR1 activation, HvVIP1 also activates one of the MAP-kinase members, HvMPK1 (El Sarraf et al. 2019). It is possible that HvVIP1 confers Agrobacterium resistance in barley. A VIP1 from Populus trichocarpa, PtVIP1, also serves against pathogen invasion by activating the PR1 gene (Wang et al. 2019). Recently, a PR 10 gene from the Agrobacterium-recalcitrant plant Hypericum perforatum has been identified and named as phenolic oxidative coupling protein (Hyp-1). This defence gene has been found to hinder Agrobacterium infection in Tobacco (Hou et al. 2020). During infection, Hyp-1 transcripts get upregulated and play an important role in plant defence (Karppinen et al. 2016). It has been suggested that Hyp-1 confers tolerance to Agrobacterium by downregulating auxin signalling pathway genes, such as NtaTIR1 and NtaAFR8. During pathogen attack, Hyp-1 induces the expression of MiR160, which targets the ARF10, ARF16 and ARF17 transcripts of the auxin signalling cascade and downregulates their expression (Hou et al. 2020; Pinweha et al. 2015; Wójcik et al. 2017). Dunoyer found low levels of small RNAs associated with the iaaM (tryptophan 2 oxygenase) and ags (agropine synthase) genes of T-DNA in Nicotiana tabaccum after 3 days of Agrobacterium infiltration. Additionally, RNAi-deficient plants were susceptible to Agrobacterium, suggesting that RNA silencing may provide a defence against Agrobacterium.
However, small interfering RNA (siRNA) has been detected during the initial days of infection in the tissues; later, the anti-silencing state is maintained by Agrobacterium, which inhibits the synthesis of siRNA. Thus, again, Agrobacterium takes over the plant defence system and leads to successful infection (Dunoyer et al. 2006). Recently, we have identified a tau class GST in rice, GSTU5, which interacts with the VirE2 protein of Agrobacterium and hinders its single-stranded DNA binding (SSB) property. The GSTU5-knockdown lines in rice were more susceptible to Agrobacterium infection in comparison with its overexpressing lines, thus alluding the probable role of GSTU5 as a defence gene in rice (Tiwari et al. 2022). Therefore, it can be said that plants have a three-layered immunity against Agrobacterium. In the first layer, PRRs, such as FLS2XL, FLS2, EFR and RTNLB4, play a role (further confirmation is required for RTNLB4). The second layer of plant immunity is related to signal integration, which involves MAP-kinase defence-related genes, especially MPK3, MPK4, MPK6 and MPK11. The last layer of immune response involves defence induction and amplification of defence-related transcription factors, such as VIP1, WRKY22 and WRKY29, which activate the defence-related genes. Thus, during plant–Agrobacterium interaction, disease susceptibility and resistance are dependent upon the layered immunity of plants and the virulence effector proteins of Agrobacterium. A model for PAMP perception and plant immune response during Agrobacterium infection is given in Fig. 2B.
A list of genes that act during Agrobacterium infection in different plants is presented in Table 1.
Table 1.
A list of plant genes act during Agrobacterium infection
| Plant genes | Specific role | Plant | Reference |
|---|---|---|---|
| Flagellin sensing 2 (FLS2) | Perceives flg22 of Agrobacterium | Arabidopsis thaliana | Chinchilla et al. (2007) |
| FLS2 with eXtended Ligand recognition (FLS2XL) | Perceives flg22Atum of Agrobacterium | Vitis riparia | Fürst et al. (2020) |
| Elongation factor-Thermo unstable (EF-Tu) receptor (EFR) | Perceives 18 aa residues in EF-Tu named elf18 of Agrobacterium | Arabidopsis thaliana | Zipfel et al. (2006) |
| RETICULON-LIKE4 (RTNLB4) | Perceives elf18 peptide of Agrobacterium | Arabidopsis thaliana | Hwang and Gelvin (2004) |
| NtPRp27 | Defence gene | Nicotiana tabacum | Ditt et al. (2001); Ditt et al. (2006) |
| Cytochrome P450 monooxygenase | Defence gene | Arabidopsis thaliana | Ditt et al. (2001); Ditt et al. (2006) |
| Xa21 | Defence gene | Oryza sativa | Ditt et al. (2001); Ditt et al. (2006) |
| VirE2 interacting protein (VIP1) | Activates defence genes | Arabidopsis thaliana | Djamei et al. (2007) |
| HvVIP1 | Activates PR1 gene(HvPR1, HvPR4, and HvPR10) | Hordeum vulgare | El Sarraf et al. (2019) |
| PtVIP1 | Activates the PR1 gene | Populus trichocarpa | Wang et al. (2019) |
| Phenolic oxidative coupling protein (Hyp-1) | Defence gene | Nicotiana tabacum | Hou et al. (2020); Hou et al. (2020); Pinweha et al. (2015); Wójcik et al. (2017) |
| OsGSTU5 | Defence gene | Oryza sativa | Tiwari et al. (2022) |
| MPK3, MPK6 | MAP-kinase defence genes | Arabidopsis thaliana | Huang and Hwang (2020) |
| WRKY22, WRKY29 | Defence genes | Arabidopsis thaliana | Huang and Hwang (2020) |
| FRK1, and PR1 | Defence genes | Arabidopsis thaliana | Huang and Hwang (2020) |
Concluding remark
The remarkable abilities of Agrobacterium never cease to attract plant biologists. The bacterium captures immense attention because of its pathogenesis as well as its biotechnological significance. The unique action of Agrobacterium is determined by virulence and host proteins. Research on plant–Agrobacterium interaction provides an insight into the molecular communication between these two organisms. The more we learn about host–pathogen interactions, the more interested we become in the next step in their path. There is still much to be explored in terms of plant–Agrobacterium interaction. For instance, VirE2 was recently shown to enter the plant cells via clathrin-mediated endocytosis, however, it remains unclear how T-DNA and other virulence effector proteins enter via the cell membrane. Furthermore, the T-complex enters the nucleus via VirE2-mediated cytoplasmic trafficking, but it is unclear how other virulence proteins, such as VirF and VirD5, enter the plant nucleus. The exact mechanism of T-DNA integration is still elusive.
Future prospects
As we gain an understanding of layered immunity during plant–Agrobacterium interaction, we can utilise this information in plant breeding to control crown gall. By elevating and stacking the extracellular immunity receptors, it might be possible to broaden the disease resistance. In addition, modulating the downstream signal transduction pathway to promote the expression of defence-related genes would enhance disease resistance. Recent genetic editing techniques, such as CRISPR/Cas9, allow the targeted modification of extracellular immune receptors and might be useful in exploring the downstream pathway in plants during Agrobacterium infection.
Author contribution statement
MT conceived the idea and wrote the article. DC and AKM reviewed the article. All authors are agreeing with the final draft.
Acknowledgements
The authors would like to acknowledge the Director CSIR-NBRI for the facilities to carry out this work. MT is indebted to Banaras Hindu University for the registration (Sept.2014/271) and UGC for providing fellowship. The review article bears the NBRI manuscript number; CSIR-NBRI_MS/2022/03/14
Data availability
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abu-Arish A, Frenkiel-Krispin D, Fricke T, Tzfira T, Citovsky V, Wolf SG, Elbaum M. Three-dimensional reconstruction of Agrobacterium VirE2 protein with single-stranded DNA. J Biol Chem. 2004;279(24):25359–25363. doi: 10.1074/jbc.M401804200. [DOI] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 2007;26(10):2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6(10):973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Baker CJ, Smith J, Rice C. Apoplast redox metabolism: Effect of acetovanillone (apocynin) and acetosyringone, on their co-oxidation and redox properties. Physiol Mol Plant Pathol. 2020 doi: 10.1016/j.pmpp.2020.101481. [DOI] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI) Mol Plant. 2015;8(4):521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Binns AN, Zhao J. The MexE/MexF/AmeC efflux pump of Agrobacterium tumefaciens and its role in Ti plasmid virulence gene expression. J Bacteriol. 2020 doi: 10.1128/JB.00609-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourras S, Rouxel T, Meyer M. Agrobacterium tumefaciens gene transfer: how a plant pathogen hacks the nuclei of plant and nonplant organisms. Phytopathology. 2015;105(10):1288–1301. doi: 10.1094/PHYTO-12-14-0380-RVW. [DOI] [PubMed] [Google Scholar]
- Bourras S, Meyer M, Grandaubert J, Lapalu N, Fudal I, Linglin J, Ollivier B, Blaise F, Balesdent M-H, Rouxel T (2012) Incidence of genome structure, DNA asymmetry, and cell physiology on T-DNA integration in chromosomes of the phytopathogenic fungus Leptosphaeria maculans. G3: Genes, Genomes, Genetics 2 (8):891–904 [DOI] [PMC free article] [PubMed]
- Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14(13):3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi GA, Ankenbauer RG, Nester EW. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci. 1990;87(17):6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi GA, Martinetti G, Leigh JA, Lee CC, Thienes C, Nester EW. Role of Agrobacterium tumefaciens ChvA Protein in Export of β-1, 2-Glucan. J Bacteriol. 2007;189(18):6742. doi: 10.1128/jb.171.3.1609-1615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A, Herrera-Estrella L, Inzé D, Van Haute E, Van Montagu M, Schell J, Zambryski P (2019) Introduction of genetic material into plant cells. In: Biotechnology and biological frontiers. Routledge, pp 480–493 [DOI] [PubMed]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Wong ML, Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc Natl Acad Sci. 1989;86(4):1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Kapelnikov A, Oliel S, Zakai N, Rojas MR, Gilbertson RL, Tzfira T, Loyter A. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J Biol Chem. 2004;279(28):29528–29533. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BP. Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol. 2015;53:541–563. doi: 10.1146/annurev-phyto-080614-120114. [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16(9):537. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Levy A, Tzfira T. The ongoing saga of Agrobacterium–host interactions. Trends Plant Sci. 2008;13(3):102–105. doi: 10.1016/j.tplants.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Dallo SF, Zhang B, Denno J, Hong S, Tsai A, Haskins W, Ye JY, Weitao T (2012) Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. Scient World J [DOI] [PMC free article] [PubMed]
- Das A. Identification of a Carboxy-Terminal Glutamine-Rich Domain in Agrobacterium tumefaciens Coupling Protein VirD4 Required for Recognition of T-Strand DNA and Not VirE2 as a Substrate for Transfer to Plant Cells. Mol Plant Microbe Interact. 2020;33(2):166–172. doi: 10.1094/MPMI-04-19-0099-R. [DOI] [PubMed] [Google Scholar]
- De Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16(9):839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- Ditt RF, Nester EW, Comai L. Plant gene expression response to Agrobacterium tumefaciens. Proc Natl Acad Sci. 2001;98(19):10954–10959. doi: 10.1073/pnas.191383498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditt RF, Kerr KF, de Figueiredo P, Delrow J, Comai L, Nester EW. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant Microbe Interact. 2006;19(6):665–681. doi: 10.1094/MPMI-19-0665. [DOI] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science. 2007;318(5849):453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet. 2006;38(2):258–263. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- Duprey A, Reverchon S, Nasser W. Bacterial virulence and Fis: adapting regulatory networks to the host environment. Trends Microbiol. 2014;22(2):92–99. doi: 10.1016/j.tim.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18(3):265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Sanders PR, Flick JS, Adams SP, Bittner ML, Brand LA, Fink CL, Fry JS. Expression of bacterial genes in plant cells. Proc Natl Acad Sci. 1983;80(15):4803–4807. doi: 10.1073/pnas.80.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book Am Soc Plant Biol. 2011 doi: 10.1199/tab.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst U, Zeng Y, Albert M, Witte AK, Fliegmann J, Felix G. Perception of Agrobacterium tumefaciens flagellin by FLS2 XL confers resistance to crown gall disease. Nature Plants. 2020;6(1):22–27. doi: 10.1038/s41477-019-0578-6. [DOI] [PubMed] [Google Scholar]
- Gan HM, Savka MA (2018) One more decade of Agrobacterium taxonomy. In: Agrobacterium Biology. Springer, pp 1–14 [DOI] [PubMed]
- Gao R, Lynn DG. Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J Bacteriol. 2005;187(6):2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cano E, Hak H, Magori S, Lazarowitz SG, Citovsky V. The Agrobacterium F-box protein effector VirF destabilizes the Arabidopsis GLABROUS1 enhancer/binding protein-like transcription factor VFP4, a transcriptional activator of defense response genes. Mol Plant Microbe Interact. 2018;31(5):576–586. doi: 10.1094/MPMI-07-17-0188-FI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67(1):16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol. 2010;48:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front Plant Sci. 2012;3:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. Integration of Agrobacterium T-DNA into the plant genome. Annu Rev Genet. 2017;51:195–217. doi: 10.1146/annurev-genet-120215-035320. [DOI] [PubMed] [Google Scholar]
- Guo M, Gao D, Jin Y. Progress in the formation and transfer of Agrobacterium T-complex. Prog Biochem Biophys. 2009;36(11):1408–1414. [Google Scholar]
- Guo M, Huang Z, Yang J. Is there any crosstalk between the chemotaxis and virulence induction signaling in Agrobacterium tumefaciens? Biotechnol Adv. 2017;35(4):505–511. doi: 10.1016/j.biotechadv.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Guo M, Ye J, Gao D, Xu N, Yang J. Agrobacterium-mediated horizontal gene transfer: Mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol Adv. 2019;37(1):259–270. doi: 10.1016/j.biotechadv.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Gust AA, Pruitt R, Nürnberger T. Sensing danger: key to activating plant immunity. Trends Plant Sci. 2017;22(9):779–791. doi: 10.1016/j.tplants.2017.07.005. [DOI] [PubMed] [Google Scholar]
- He F, Nair GR, Soto CS, Chang Y, Hsu L, Ronzone E, DeGrado WF, Binns AN. Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens. J Bacteriol. 2009;191(18):5802–5813. doi: 10.1128/JB.00451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel BC, Tomlinson AD, Morton ER, Choi J-H, Fuqua C. Agrobacterium tumefaciens exoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J Bacteriol. 2014;196(18):3221–3233. doi: 10.1128/JB.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hj S, Wang HH, Stefanato FL, Craze M, Bowden S, Wallington E, Zipfel C, Ridout CJ. Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 2015;206(2):606–613. doi: 10.1111/nph.13356. [DOI] [PubMed] [Google Scholar]
- Hou W, Singh RK, Zhao P, Martins V, Aguilar E, Canto T, Tenllado F, Dias ACP. Transgenic expression of Hyp-1 gene from Hypericum perforatum L. alters expression of defense-related genes and modulates recalcitrance to Agrobacterium tumefaciens. Planta. 2020;251(1):13. doi: 10.1007/s00425-019-03310-3. [DOI] [PubMed] [Google Scholar]
- Hu X, Zhao J, DeGrado WF, Binns AN. Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc Natl Acad Sci. 2013;110(2):678–683. doi: 10.1073/pnas.1215033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F-C, Hwang H-H. Arabidopsis reticulon-like4 (RTNLB4) Protein participates in agrobacterium infection and VirB2 peptide-induced plant defense response. Int J Mol Sci. 2020;21(5):1722. doi: 10.3390/ijms21051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhou Q, Sun P, Yang J, Guo M. Two Agrobacterium tumefaciens CheW proteins are incorporated into one chemosensory pathway with different efficiencies. Mol Plant Microbe Interact. 2018;31(4):460–470. doi: 10.1094/MPMI-10-17-0255-R. [DOI] [PubMed] [Google Scholar]
- Huang FC, Chi SF, Chien PR, Liu YT, Chang HN, Lin CS, Hwang HH. Arabidopsis RAB8A, RAB8B and RAB8D Proteins Interact with Several RTNLB Proteins and are Involved in the Agrobacterium tumefaciens Infection Process. Plant Cell Physiol. 2021;62(10):1572–1588. doi: 10.1093/pcp/pcab112. [DOI] [PubMed] [Google Scholar]
- Hwang H-H, Gelvin SB. Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell. 2004;16(11):3148–3167. doi: 10.1105/tpc.104.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A. The Arabidopsis G-protein β-subunit is required for defense response against Agrobacterium tumefaciens. Biosci Biotechnol Biochem. 2009;73(1):47–52. doi: 10.1271/bbb.80449. [DOI] [PubMed] [Google Scholar]
- Janda M, Lamparová L, Zubíková A, Burketová L, Martinec J, Krčková Z. Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol Plant Pathol. 2019;20(7):1005–1012. doi: 10.1111/mpp.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karppinen K, Derzsó E, Jaakola L, Hohtola A. Molecular cloning and expression analysis of hyp-1 type PR-10 family genes in Hypericum perforatum. Front Plant Sci. 2016;7:526. doi: 10.3389/fpls.2016.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Pour N, Condemine G, Hugouvieux-Cotte-Pattat N. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics. 2004;4(10):3177–3186. doi: 10.1002/pmic.200300814. [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Appelhagen I, Viehoever P, Li Y, Weisshaar B. The structural features of thousands of T-DNA insertion sites are consistent with a double-strand break repair-based insertion mechanism. Mol Plant. 2015;8(11):1651–1664. doi: 10.1016/j.molp.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Kopertekh L, Schiemann J. Transient production of recombinant pharmaceutical proteins in plants: evolution and perspectives. Curr Med Chem. 2019;26(3):365–380. doi: 10.2174/0929867324666170718114724. [DOI] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, Van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol. 2010;28(4):365–369. doi: 10.1038/nbt.1613. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium's endless hunt for new partners. Trends Genet. 2006;22(1):29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Citovsky V. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int J Develop Biol. 2013;57:467–481. doi: 10.1387/ijdb.130199bl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Citovsky V (2018) Beyond Agrobacterium-mediated transformation: horizontal gene transfer from bacteria to eukaryotes. In: Agrobacterium Biology. Springer, pp 443–462 [DOI] [PMC free article] [PubMed]
- Lapham RA, Lee LY, Xhako E, Gómez EG, Nivya VM, Gelvin SB. Agrobacterium VirE2 protein modulates plant gene expression and mediates transformation from its location outside the nucleus. Front Plant Sci. 2021;12:1051. doi: 10.3389/fpls.2021.684192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pan SQ. Agrobacterium delivers VirE2 protein into host cells via clathrin-mediated endocytosis. Sci Adv. 2017;3(3):e1601528. doi: 10.1126/sciadv.1601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jia Y, Hou Q, Charles TC, Nester EW, Pan SQ. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc Natl Acad Sci. 2002;99(19):12369–12374. doi: 10.1073/pnas.192439499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tang Y, Lin J, Cai W. Methods for genetic transformation of filamentous fungi. Microb Cell Fact. 2017;16(1):168. doi: 10.1186/s12934-017-0785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Christie PJ (2018) The Agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. In: Agrobacterium Biology. Springer, pp 233–260 [DOI] [PMC free article] [PubMed]
- Lin Y-H, Pierce BD, Fang F, Wise A, Binns AN, Lynn DG. Role of the VirA histidine autokinase of Agrobacterium tumefaciens in the initial steps of pathogenesis. Front Plant Sci. 2014;5:195. doi: 10.3389/fpls.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Wang H, Wang S, Jiang W, Shan C, Li B, Yang J, Zhang S, Sun W. Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR. J Integr Plant Biol. 2015;57(7):641–652. doi: 10.1111/jipb.12306. [DOI] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury S, Delaunay A, Mesnard F, Cronier D, Chabbert B, Geoffroy P, Legrand M. O-methyltransferase (s)-suppressed plants produce lower amounts of phenolic vir inducers and are less susceptible to Agrobacterium tumefaciens infection. Planta. 2010;232(4):975–986. doi: 10.1007/s00425-010-1230-x. [DOI] [PubMed] [Google Scholar]
- Meyer T, Renoud S, Vigouroux A, Miomandre A, Gaillard V, Kerzaon I, Prigent-Combaret C, Comte G, Moréra S, Vial L. Regulation of hydroxycinnamic acid degradation drives Agrobacterium fabrum lifestyles. Mol Plant Microbe Interact. 2018;31(8):814–822. doi: 10.1094/MPMI-10-17-0236-R. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Ram AF, Hooykaas PJ, van den Hondel CA. Agrobacterium-mediated transformation of Aspergillus awamori in the absence of full-length VirD2, VirC2, or VirE2 leads to insertion of aberrant T-DNA structures. J Bacteriol. 2004;186(7):2038–2045. doi: 10.1128/JB.186.7.2038-2045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamilarasan M, Prasad M. Plant innate immunity: an updated insight into defense mechanism. J Biosci. 2013;38(2):433–449. doi: 10.1007/s12038-013-9302-2. [DOI] [PubMed] [Google Scholar]
- Nair GR, Lai X, Wise AA, Rhee BW, Jacobs M, Binns AN. The integrity of the periplasmic domain of the VirA sensor kinase is critical for optimal coordination of the virulence signal response in Agrobacterium tumefaciens. J Bacteriol. 2011;193(6):1436–1448. doi: 10.1128/JB.01227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nester EW. Agrobacterium: nature’s genetic engineer. Front Plant Sci. 2015;5:730. doi: 10.3389/fpls.2014.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa Yokoi A, Saika H, Hara N, Lee LY, Toki GSB. Agrobacterium T-DNA integration in somatic cells does not require the activity of DNA polymerase θ. New Phytol. 2021;229(5):2859–2872. doi: 10.1111/nph.17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman A, Aqeel M, Lou Y. PRRs and NB-LRRs: from signal perception to activation of plant innate immunity. Int J Mol Sci. 2019;20(8):1888. doi: 10.3390/ijms20081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormeno-Orrillo E, Servín-Garcidueñas LE, Rogel MA, González V, Peralta H, Mora J, Martínez-Romero J, Martínez-Romero E. Taxonomy of rhizobia and agrobacteria from the Rhizobiaceae family in light of genomics. Syst Appl Microbiol. 2015;38(4):287–291. doi: 10.1016/j.syapm.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Park SY, Vaghchhipawala Z, Vasudevan B, Lee LY, Shen Y, Singer K, Waterworth WM, Zhang ZJ, West CE, Mysore KS. Agrobacterium T-DNA integration into the plant genome can occur without the activity of key non-homologous end-joining proteins. Plant J. 2015;81(6):934–946. doi: 10.1111/tpj.12779. [DOI] [PubMed] [Google Scholar]
- Pfeilmeier S, George J, Morel A, Roy S, Smoker M, Stransfeld L, Downie JA, Peeters N, Malone JG, Zipfel C. Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol J. 2019;17(3):569–579. doi: 10.1111/pbi.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinweha N, Asvarak T, Viboonjun U, Narangajavana J. Involvement of miR160/miR393 and their targets in cassava responses to anthracnose disease. J Plant Physiol. 2015;174:26–35. doi: 10.1016/j.jplph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Teige M, Hirt H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci. 2009;106(43):18414–18419. doi: 10.1073/pnas.0905599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EPi, Yasuda S, Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 2018;93(4):592–613. doi: 10.1111/tpj.13808. [DOI] [PubMed] [Google Scholar]
- El Sarraf N, Gurel F, Tufan F, McGuffin LJ. Characterisation of HvVIP1 and expression profile analysis of stress response regulators in barley under Agrobacterium and Fusarium infections. PloS one. 2019;14(6):e0218120. doi: 10.1371/journal.pone.0218120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, Canlas PE, Daudi A, Petzold CJ, Singan VR. Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 2015;11(3):e1004809. doi: 10.1371/journal.ppat.1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams M, Vial L, Chapulliot D, Nesme X, Lavire C. Rapid and accurate species and genomic species identification and exhaustive population diversity assessment of Agrobacterium spp. using recA-based PCR. System Appl Microbiol. 2013;36(5):351–358. doi: 10.1016/j.syapm.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lee LY, Gelvin SB. Is VIP1 important for A grobacterium-mediated transformation? Plant J. 2014;79(5):848–860. doi: 10.1111/tpj.12596. [DOI] [PubMed] [Google Scholar]
- Shilo S, Tripathi P, Melamed-Bessudo C, Tzfadia O, Muth TR, Levy AA. T-DNA-genome junctions form early after infection and are influenced by the chromatin state of the host genome. PLoS Genet. 2017;13(7):e1006875. doi: 10.1371/journal.pgen.1006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol. 2009;191(8):2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Townsend CO. A plant-tumor of bacterial origin. Science. 1907;25(643):671–673. doi: 10.1126/science.25.643.671. [DOI] [PubMed] [Google Scholar]
- Song G-q, Prieto H, Orbovic V (2019) Agrobacterium-Mediated Transformation of Tree Fruit Crops: Methods, Progress, and Challenges. Frontiers in plant science 10 [DOI] [PMC free article] [PubMed]
- Subramoni S, Nathoo N, Klimov E, Yuan Z-C. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci. 2014;5:322. doi: 10.3389/fpls.2014.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou J-M. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell. 2017;29(4):618–637. doi: 10.1105/tpc.16.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M, Gautam N, Indoliya Y, Kidwai M, Mishra AK, Chakrabarty D. A tau class GST, OsGSTU5, interacts with VirE2 and modulates the Agrobacterium-mediated transformation in rice. Plant Cell Rep. 2022 doi: 10.1007/s00299-021-02824-z. [DOI] [PubMed] [Google Scholar]
- Torres M, Jiquel A, Jeanne E, Naquin D, Dessaux Y, Faure D. Agrobacterium tumefaciens fitness genes involved in the colonization of plant tumors and roots. New Phytol. 2022;233(2):905–918. doi: 10.1111/nph.17810. [DOI] [PubMed] [Google Scholar]
- Trdá L, Fernandez O, Boutrot F, Héloir MC, Kelloniemi J, Daire X, Adrian M, Clément C, Zipfel C, Dorey S. The grapevine flagellin receptor Vv FLS 2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 2014;201(4):1371–1384. doi: 10.1111/nph.12592. [DOI] [PubMed] [Google Scholar]
- Tu H, Li X, Yang Q, Peng L, Pan SQ (2018) Real-time trafficking of agrobacterium virulence protein VirE2 inside host cells. In: Agrobacterium Biology. Springer, pp 261–286 [DOI] [PubMed]
- Tzfira T, Citovsky V. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 2002;12(3):121–129. doi: 10.1016/s0962-8924(01)02229-2. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20(13):3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol. 2006;17(2):147–54. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Valentini M, Gonzalez D, Mavridou DA, Filloux A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr Opin Microbiol. 2018;41:15–20. doi: 10.1016/j.mib.2017.11.006. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, Hooykaas PJ. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003;31(14):4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kregten M, de Pater S, Romeijn R, van Schendel R, Hooykaas PJ, Tijsterman M. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nature Plants. 2016;2(11):1–6. doi: 10.1038/nplants.2016.164. [DOI] [PubMed] [Google Scholar]
- Veena JH, Doerge R, Gelvin SB. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 2003;35(2):219–236. doi: 10.1046/j.1365-313x.2003.01796.x. [DOI] [PubMed] [Google Scholar]
- Wan W-L, Fröhlich K, Pruitt RN, Nürnberger T, Zhang L. Plant cell surface immune receptor complex signaling. Curr Opin Plant Biol. 2019;50:18–28. doi: 10.1016/j.pbi.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Peng W, Zhou X, Huang F, Shao L, Luo M. The putative Agrobacterium transcriptional activator-like virulence protein VirD5 may target T-complex to prevent the degradation of coat proteins in the plant cell nucleus. New Phytol. 2014;203(4):1266–1281. doi: 10.1111/nph.12866. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Yu M, Shih P-Y, Wu H-Y, Lai E-M. Stable pH Suppresses Defense Signaling and is the Key to Enhance Agrobacterium-Mediated Transient Expression in Arabidopsis Seedlings. Sci Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-34949-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang S, Huang F, Zhou X, Chen Z, Peng W, Luo M. VirD5 is required for efficient Agrobacterium infection and interacts with Arabidopsis VIP2. New Phytol. 2018;217(2):726–738. doi: 10.1111/nph.14854. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang W, Wang M, Cheng Q. Cloning and characterization of the PtVIP1 gene in Populus. J Fores Resz. 2019;30(6):2259–2266. [Google Scholar]
- Watt SA, Wilke A, Patschkowski T, Niehaus K. Comprehensive analysis of the extracellular proteins from Xanthomonas campestris pv. campestris B100. Proteomics. 2005;5(1):153–167. doi: 10.1002/pmic.200400905. [DOI] [PubMed] [Google Scholar]
- Wetzel ME, Olsen GJ, Chakravartty V, Farrand SK. The repABC plasmids with quorum-regulated transfer systems in members of the Rhizobiales divide into two structurally and separately evolving groups. Genome Biol Evol. 2015;7(12):3337–3357. doi: 10.1093/gbe/evv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AA, Binns AN. The receiver of the Agrobacterium tumefaciens VirA histidine kinase forms a stable interaction with VirG to activate virulence gene expression. Front Microbiol. 2016;6:1546. doi: 10.3389/fmicb.2015.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik AM, Nodine MD, Gaj MD. miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front Plant Sci. 2017;8:2024. doi: 10.3389/fpls.2017.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-F, Lin J-S, Shaw G-C, Lai E-M. Acid-induced type VI secretion system is regulated by ExoR-ChvG/ChvI signaling cascade in Agrobacterium tumefaciens. PLoS pathogens. 2012;8(9):e1002938. doi: 10.1371/journal.ppat.1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S. A chemical genetic approach demonstrates that MPK 3/MPK 6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 2014;77(2):222–234. doi: 10.1111/tpj.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li X, Tu H, Pan SQ. Agrobacterium-delivered virulence protein VirE2 is trafficked inside host cells via a myosin XI-K–powered ER/actin network. Proc Natl Acad Sci. 2017;114(11):2982–2987. doi: 10.1073/pnas.1612098114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZC, Haudecoeur E, Faure D, Kerr KF, Nester EW. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and γ-amino butyric acid reveals signalling cross-talk and Agrobacterium–plant co-evolution. Cell Microbiol. 2008;10(11):2339–2354. doi: 10.1111/j.1462-5822.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe. 2010;7(3):197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Lacroix B, Gafni Y, Citovsky V. Disassembly of synthetic Agrobacterium T-DNA–protein complexes via the host SCFVBF ubiquitin–ligase complex pathway. Proc Natl Acad Sci. 2013;110(1):169–174. doi: 10.1073/pnas.1210921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Boone L, Kocz R, Zhang C, Binns AN, Lynn DG. At the maize/Agrobacterium interface: natural factors limiting host transformation. Chem Biol. 2000;7(8):611–621. doi: 10.1016/s1074-5521(00)00007-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lee C-W, Wehner N, Imdahl F, Svetlana V, Weiste C, Dröge-Laser W, Deeken R. Regulation of oncogene expression in T-DNA-transformed host plant cells. PLoS pathogens. 2015;11(1):e1004620. doi: 10.1371/journal.ppat.1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.