ABSTRACT
Wildlife play a role in the acquisition, maintenance, and dissemination of antimicrobial resistance (AMR). This is especially true at the human-domestic animal-wildlife interface, like urbanized areas, where interactions occur that can promote the cross-over of AMR bacteria and genes. We conducted a 2-year fecal surveillance (n = 783) of a white-tailed deer (WTD) herd from an urban park system in Ohio to identify and characterize cephalosporin-resistant and carbapenemase-producing bacteria using selective enrichment. Using generalized linear mixed models we found that older (OR = 2.3, P < 0.001), male (OR = 1.8, P = 0.001) deer from urbanized habitats (OR = 1.4, P = 0.001) were more likely to harbor extended-spectrum cephalosporin-resistant Enterobacterales. In addition, we isolated two carbapenemase-producing Enterobacterales (CPE), a Klebsiella quasipneumoniae harboring blaKPC-2 and an Escherichia coli harboring blaNDM-5, from two deer from urban habitats. The genetic landscape of the plasmid carrying blaKPC-2 was unique, not clustering with other reported plasmids encoding KPC-2, and only sharing 78% of its sequence with its nearest match. While the plasmid carrying blaNDM-5 shared sequence similarity with other reported plasmids encoding NDM-5, the intact IS26 mobile genetic elements surrounding multiple drug resistant regions, including the blaNDM-5, has been reported infrequently. Both carbapenemase genes were successfully conjugated to a J53 recipient conferring a carbapenem-resistant phenotype. Our findings highlight that urban environments play a significant role on the transmission of AMR bacteria and genes to wildlife and suggest WTD may play a role in the dissemination of clinically and epidemiologically relevant antimicrobial resistant bacteria.
IMPORTANCE The role of wildlife in the spread of antimicrobial resistance is not fully characterized. Some wildlife, including white-tailed deer (WTD), can thrive in suburban and urban environments. This may result in the exchange of antimicrobial resistant bacteria and resistance genes between humans and wildlife, and lead to their spread in the environment. We found that WTD living in an urban park system carried antimicrobial resistant bacteria that were important to human health and resistant to antibiotics used to treat serious bacterial infections. This included two deer that carried bacteria resistant to carbapenem antibiotics which are critically important for treatment of life-threatening infections. These two bacteria had the ability to transfer their AMR resistance genes to other bacteria, making them a threat to public health. Our results suggest that WTD may contribute to the spread of antimicrobial resistant bacteria in the environment.
KEYWORDS: Escherichia coli, Klebsiella quasipneumoniae, white-tailed deer, antimicrobial resistance, carbapenemase, cephalosporinase, plasmids, wildlife
INTRODUCTION
Antimicrobial resistance (AMR) is a global health issue that if left unchecked could lead to 10 million deaths per year globally and cause economic crisis (1). In the United States alone, there are more than 2.8 million antibiotic-resistant infections per year that result in more than 35,000 annual deaths, costing the U.S. economy $4.6 billion annually in health care costs (2). Among resistant bacteria causing disease, extended-spectrum β-lactamase (ESBL) producers, those that confer resistance to cephalosporins, are considered a serious public health threat because infections have been steadily increasing since 2012 and they limit treatment options for serious illnesses like invasive Gram-negative infections (2). In these situations, carbapenem antibiotics are often prescribed, however, carbapenem-resistant infections are increasing in the U.S., particularly because of two epidemic carbapenem-resistant genotypes, blaKPC and blaNDM (3). Gram-negative infections with bacteria resistant to carbapenem antibiotics are considered an urgent public health threat that limit patient treatment options to drug classes with significant side effects like polymyxins (2, 4).
AMR determinants and bacteria can disseminate outside healthcare setting to communities and the natural environment (5, 6). Moreover, recent studies have identified increased or emerging resistance of cephalosporin- and carbapenem-resistant genotypes in diverse settings suggesting widespread dissemination (5, 7, 8). While the zoonotic transmission of antimicrobial resistant bacteria is well-characterized in domestic animals and humans, their dissemination in ecosystems and wildlife populations is underreported (9). Free-ranging, wild deer may serve as sentinels to evaluate environmental contamination and the epidemiology of AMR as they are intimately associated with their ecosystem and human activities (10). Moreover, like most urban species, they can adapt to anthropogenic environments and influences allowing their populations to expand beyond the ecosystem carrying capacity (11, 12).
We previously reported a temporal trend in β-lactam resistance among fecal Escherichia coli isolated from urban white-tailed deer (Odocoileus virginianus) in Northeast Ohio over a 10-year cross-sectional study (13). The objective of this study was to investigate and characterize phenotypic and genotypic cephalosporin- and carbapenem-resistant enteric bacteria in a wild, free-ranging white-tailed deer (WTD) herd. In addition, we aimed to characterize demographic and environmental risk factors that were associated with AMR colonization. We also describe the presence of carbapenem-resistant Klebsiella quasipneumoniae and E. coli harboring blaKPC-2 and blaNDM-5, respectively, from two WTD.
RESULTS
WTD sample demographics.
A total of 783 individual WTD fecal samples were collected between 2018 and 2019 (Table 1). Among the WTD for which a sample was collected, the distribution of sex was nearly equal with 416 (53.1%) females and 367 (46.9%) males. The age distribution was skewed with a majority of the sample consisting of adults (432; 55.2%) with fewer fawns (199; 25.4%) and yearlings (152; 19.4%). The number of WTD harvested from individual regions of the Metropark was quite variable ranging from 14 (1.8%) from region 1 to 185 (23.6%) from region 4. When regions were dichotomized into urban and suburban, a majority of the sample resided in urban (561; 71.7%) compared to suburban regions (222; 28.4%). The number of deer sampled across the 2-year time period was nearly equal (2018: 370 (47.3%); 2019: 413 (52.8%).
TABLE 1.
Demographics of 783 white-tailed deer sampled at an urban park located in Northeastern Ohio
| Demographics | WTD sampled | Total sampled | Percent (%) |
|---|---|---|---|
| Sex | 783 | ||
| Female | 416 | 53.1 | |
| Male | 367 | 46.9 | |
| Age | 783 | ||
| Fawn | 199 | 25.4 | |
| Yearling | 152 | 19.4 | |
| Adult | 432 | 55.2 | |
| Region | 783 | ||
| 1a | 14 | 1.7 | |
| 2a | 56 | 7.2 | |
| 3a | 140 | 17.9 | |
| 4a | 185 | 23.6 | |
| 5a | 95 | 12.1 | |
| 6a | 71 | 9.1 | |
| 7b | 42 | 5.4 | |
| 8b | 15 | 1.9 | |
| 9b | 113 | 14.4 | |
| 10b | 52 | 6.6 | |
| Yr | 783 | ||
| 2018 | 370 | 42.3 | |
| 2019 | 413 | 52.8 |
Indicates the region was classified as an urban.
Indicates the region was classified as suburban.
Prevalence of cephalosporin-resistant Enterobacterales and conferring genotypes.
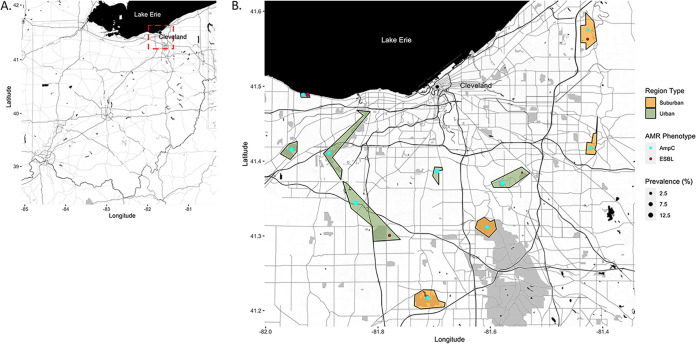
Seventy-nine (10.1%) WTD fecal samples had a cephalosporin resistant Enterobacterales (CeRE) phenotype (Table 2). The AmpC-like phenotype, as defined by reduced susceptibility to cefoxitin was observed across all sampled regions and was present in 76 (9.7%) of the fecal samples. This phenotype was conferred by the CMY genotype in 61 (7.8%) of deer sampled (Fig. 1). The ESBL phenotype, as defined by reduced susceptibility to cefepime, was found in nine (1.2%) of the fecal samples recovered from four of the 10 sampled regions. This phenotype was conferred by the CTX-M genotype in all cases of the ESBL phenotype. Gene sequencing identified that all isolates maintained the CTX-M-15 allele variation. Six samples (0.8%) had isolates that displayed both an AmpC-like and ESBL phenotype, but only three (0.3%) fecal samples harbored isolates with the CTX-M and CMY gene.
TABLE 2.
The phenotypic and genotypic prevalence of cephalosporin-resistant Enterobacterales (CeRE) recovered from 783 white-tailed deer sampled at an urban park located in Northeastern Ohio
| Classification | No. positive | No. tested | Prevalence (%) |
|---|---|---|---|
| Phenotype | |||
| CeRE | 79 | 783 | 10.1 |
| AmpC-like | 76 | 783 | 9.7 |
| ESBL | 9 | 783 | 1.2 |
| Genotype | |||
| CMY-2-like | 61 | 783 | 7.8 |
| CTX-Ma | 9 | 783 | 1.2 |
All CTX-M genes were the blaCTX-M-15 allelic variant.
FIG 1.
Map of the study location in Northeast Ohio. (A) Map of the Ohio region. Red dash box indicates a zoomed-region of the study sample area exemplified in B. (B) The study area centered in Northeast Ohio. Individual regions where deer were harvested are highlighted. Regions are colored to delineate urban versus suburban classifications. Dot plots within regions represent prevalence of WTD AmpC-like and ESBL phenotypes. The maps were generated using the ggmap and ggplot2 packages in R as described in Materials and Methods.
Association of demographic attributes and cephalosporin resistance.
Multivariable generalized linear mixed models were constructed to assess the relationship between demographic variables and CeRE (Table 3). Of the WTD harboring any CeRE, males were at 1.77 times greater odds (95% CI = 1.24 to 2.50; P = 0.001) compared with females after adjusting for age class, urbanization and year collected. Age was a predictor of CeRE recovery (P < 0.001) and differed by age group with yearling and adults being at 3.3 (95% CI = 2.45 to 4.45; P < 0.001) times and 2.26 (95% CI = 1.72 to 2.97, P < 0.001) times the odds of harboring a CeRE, respectively. Interestingly, the odds of being colonized with CeRE was reduced by 32% (95% CI = 0.57 to 0.82; P < 0.001) when adults were compared to yearlings. WTD from urban habitats were at 1.4 (95% CI: 1.15, 1.70; P = 0.001) times greater odds of having CeRE isolated compared with suburban deer. Interestingly, CeRE recovery decreased across the 2-year sampling period from 12.9% (48/370) to 7.5% (31/413). WTD from 2019 were at 42% lesser odds of maintaining a CeRE isolate compared with WTD sampled in 2018.
TABLE 3.
Epidemiologic risk factors for CeRE colonization among 783 white-tailed deer sampled at an urban park located in Northeastern Ohio assessed using multivariable linear mixed models
| Risk factor | CeRE prevalence | Odds ratio | 95% confidence interval | P-valuea |
|---|---|---|---|---|
| Sex | 0.008 | |||
| Female (Referent) | 33 (7.9) | |||
| Male | 46 (12.5) | 1.77 | (1.24 to 2.50) | 0.001 |
| Age | <0.001 | |||
| Fawn (Referent) | 11 (5.5) | |||
| Yearling | 23 (15.1) | 3.30 | (2.45 to 4.45) | <0.001 |
| Adult | 45 (10.4) | 2.26 | (1.72 to 2.97) | <0.001 |
| Adult vs. Yearling | 0.68 | (0.57 to 0.82) | <0.001 | |
| Urbanization | 0.0131 | |||
| Suburban (Referent) | 18 (8.1) | |||
| Urban | 61 (10.9) | 1.40 | (1.15 to 1.70) | 0.001 |
| Yr | <0.001 | |||
| 2018 (Referent) | 48 (13.0) | |||
| 2019 | 31 (7.5) | 0.58 | (0.45 to 0.73) | <0.001 |
P-values from univariate analysis of each individual factor (sex, age, urbanization, and year) included in the final model are presented. Odds ratios, 95% confidence intervals, and P-values are denoted for risk factors included in the multivariable analysis. Referent variable were the reference group used to calculate odds ratios.
Phenotypic and genomic characterization of two carbapenem-resistant isolates.
Two isolates recovered from two separate deer carried the epidemic carbapenemase genes blaKPC and blaNDM (Table 4). One isolate was recovered from an adult doe that was harvested from an urban area in 2018. This isolate was initially identified as a Klebsiella pneumoniae by MALDI-TOF, but was further characterized as a K. quasipneumoniae following genome analysis. The MLST scheme for K. pneumoniae identified this isolate as ST2712. This isolate carried two plasmids, a 170,848 bp IncFIB(K) plasmid that maintained the following acquired AMR genes: dfrA12, sul2, aph(3”)-Ib, aph(6)-Id, blaTEM-1, blaCTX-M-15, aac(3)-IIa, qnrB1, tet(A), aac(6’)-Ib-cr5, blaOXA-1, and catB3, and a 104,872-bp IncFIIK plasmid harboring the epidemic carbapenemase, blaKPC-2.
TABLE 4.
The plasmidome and associated resistome found in isolates recovered from white-tailed deer harboring the epidemic carbapenemase genes blaKPC-2 and blaNDM-5
| Species | Contig ID | Length | Plasmidome | Resistome |
|---|---|---|---|---|
| Klebsiella quasipneumoniae | Chromosome | 5,151,098 | ||
| pCMP243-1 | 170,848 | IncFIB(K) | dfrA14, sul2, aph(3”)-Ib, aph(6)-Id, blaTEM-1, blaCTX-M-15, aac(3)-IIa, qnrB1, tet(A), aac(6’)-Ib-cr5, blaOXA-1, catB3 | |
| pCMP243-2 | 104,872 | IncFII | blaKPC-2 | |
| Escherichia coli | Chromosome | 4,853,432 | blaEC | |
| pCMP42-1 | 120,124 | IncFII, IncFIA | mph(A), tet(A), dfrA12, aadA2, qacEdelta1, sul1, ble, blaNDM-5 | |
| pCMP42-2 | 51,705 | IncI1 | blaCMY-141 | |
| pCMP42-3 | 25,171 | |||
| pCMP42-4 | 6,647 | Col | ||
| pCMP42-5 | 5,411 | Col | ||
| pCMP42-6 | 4,072 | |||
| pCMP42-7 | 3,230 | Col |
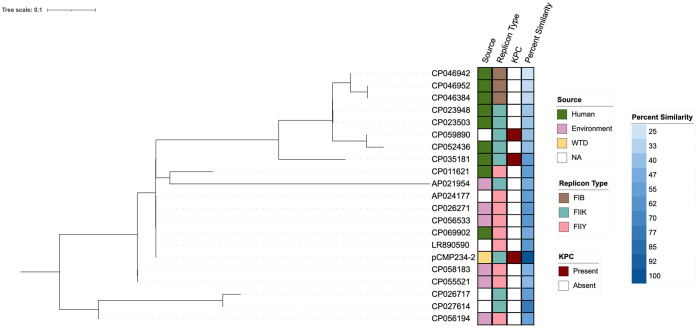
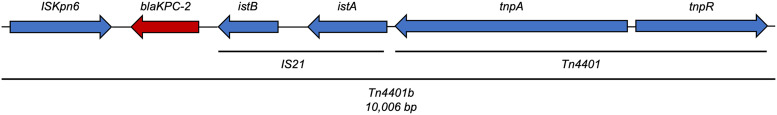
The IncFIIK plasmid pCMP234-2 was a novel blaKPC-2 harboring plasmid, sharing only 78% of its genome with its nearest match (CP027614) (Fig. 2). Using the top 20 BLAST queries, less than 1.5% of pCMP234-2 genome was used for whole-genome single nucleotide polymorphism (SNP) analysis suggesting great genetic heterogeneity among the top results. pCMP234-2 showed the highest genome percent similarity with isolates from environmental sources like wastewater and clustered with plasmids of replicon type IncFIIY. Interestingly, the plasmid clustered around and showed highest genomic similarity to other plasmids that did not harbor blaKPC. Closer examination of the genomic landscape adjacent to the KPC-2 gene showed the typical Tn4401b insertional element was responsible for blaKPC-2 mobilization (Fig. 3).
FIG 2.
De novo SNP phylogenetic analysis of pCMP243-2 compared with the top 20 NCBI BLAST results and the associated metadata including isolate source, plasmid replicon type, presence of blaKPC on the plasmid, and percent similarity to pCMP243-2. Phylogenetic SNP analysis was based on <1.5% of pCMP243-2’s genome because of genomic heterogeneity among the top 20 NCBI BLAST matches.
FIG 3.
Genetic map of the mobile genetic element carrying blaKPC-2 on pCMP243-2. The gene is mobilized on a Tn4401b transposable element that shares 100% coverage and identity with greater than 50 other isolates.
The second isolate was from the feces of a yearling buck that was harvested from an urban area in 2019. The isolate was identified as E. coli by MALDI-TOF and confirmed as an E. coli ST4450. This isolate maintained seven plasmids that ranged in size from 120,124 bp to 3,230 bp (Table 4). The first plasmid pCMP42-1 was identified as an IncFII plasmid and harbored the following resistance genes: mph(A), tet(A), dfrA12, aadA2, qacEdelta1, sul1, ble, and the epidemic carbapenemase, blaNDM-5. The second plasmid was a 51,705 bp IncI1 that harbored a blaCMY-141. The remaining plasmids did not harbor acquired resistance genes.
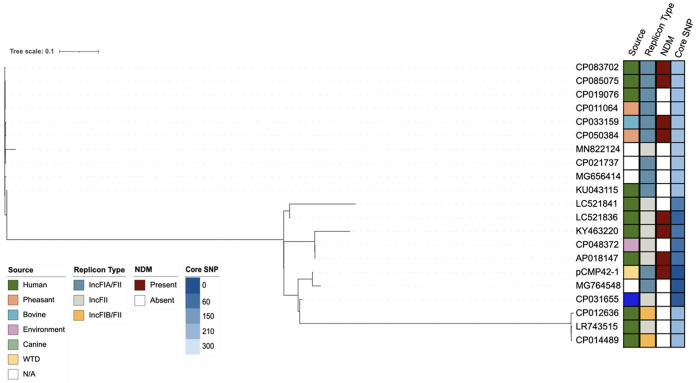
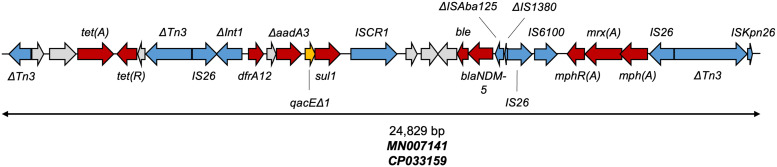
pCMP42-1 shared greater than 90% similarity with six of the top 20 NCBI BLAST plasmid sequences (Fig. 4). However, percent similarity ranged from 46% to 98% and phylogenetic analysis could only identify 43,343 nucleotide positions (36.1% of the pCMP42-1’s genome size) among the 21 genomes that could be used for core SNP analysis. The core SNPs distances ranged from 12 to 298 with an average of 205 SNP (95% CI = 158.0 to 252.4). pCMP42-1 clustered near plasmids in isolates from a diverse sources including humans, a canine, and the environment. Not surprisingly, pCMP42-1 clustered with plasmids of similar replicon type, but with variability among NDM maintenance. The NDM-5 gene was located within a 24-kb collection of truncated and intact insertional elements and resistance genes (Fig. 5). Within this region, IS26 insertional elements flanked a macrolide resistance element as well as an element that consisted of blaNDM-5 and a truncated class I integron harboring the intact sul1 and dfrA12 resistances genes as well as the truncated aadA3 aminoglycoside resistance gene. This element shared 100% coverage and identity, but not alignment, with two other isolates from the GenBank database (MN007141 and CP033159).
FIG 4.
SNP phylogenetic analysis of pCMP42-1 and the top 20 NCBI BLAST hits and associated metadata including isolate source, plasmid replicon type, presence of blaNDM within the plasmid sequence and SNP differences.
FIG 5.
The mobile and antimicrobial resistance genetic landscape surrounding blaNDM-5 on pCMP42-1. The nearly 25-Kb consists of two resistance regions, the blaNDM-5 region and the truncated class I integron region, both of which are flanked by IS26 elements. This element shares 100% coverage and identity, but not alignment, with two GenBank isolates: MN007141 and CP033159. Gene colors categorize genes as mobile genetic elements (blue), antimicrobial resistance (red), antiseptic resistance (yellow), and others (gray). Truncated genes are indicated with a Δ.
Phenotypic characterization identified that both isolates were multidrug resistant, with resistance to most of the drug classes tested (Table 5). The NDM-harboring E. coli required higher concentrations of all the tested carbapenems compared to the KPC-harboring K. quasipneumoniae. Nonetheless, both exemplified MICs indicating clinical carbapenem resistance.
TABLE 5.
Phenotypic AMR profilesa of the Klebsiella quasipneumoniae and Escherichia coli recovered from White-tailed deer and their respective transconjugantsb
| Antimicrobial | Klebsiella quasipneumoniae | J53-KPC-2 | E. coli | J53-NDM-5 | J53 |
|---|---|---|---|---|---|
| Amoxicillin/Clavulanic Acid | >32 | >32 | >32 | >32 | 4 |
| Ampicillin | >32 | >32 | >32 | >32 | 2 |
| Azithromycin | >16 | 4 | >16 | >16 | 2 |
| Aztreonam | >16 | 16 | >16 | >16 | ≤2 |
| Cefepime | >16 | 4 | >16 | >16 | ≤1 |
| Cefotaxime | >32 | 4 | >32 | >32 | ≤0.25 |
| Cefoxitin | 32 | 8 | >32 | >32 | 2 |
| Cefazolin | >16 | >16 | >16 | >16 | ≤8 |
| Ceftiofur | >8 | 8 | >8 | >8 | 0.25 |
| Ceftriaxone | >64 | 8 | >64 | >64 | ≤0.25 |
| Doripenem | 1 | 1 | >2 | >2 | ≤0.12 |
| Ertapenem | 2 | 1 | >4 | >4 | ≤0.25 |
| Imipenem | 4 | 2 | >8 | >8 | ≤0.5 |
| Meropenem | ≤1 | ≤1 | >8 | >8 | ≤1 |
| Piperacillin Tazobactam | >64 | 32 | >64 | >64 | ≤4 |
| Ticarcillin Clavulanic Acid | >128 | >128 | >128 | >128 | ≤16 |
| Chloramphenicol | 32 | 8 | 8 | 4 | 8 |
| Nalidixic Acid | 32 | 2 | >32 | 2 | 2 |
| Ciprofloxacin | >4 | ≤0.015 | >4 | ≤0.015 | ≤0.015 |
| Levofloxacin | 4 | ≤1 | >8 | ≤1 | ≤1 |
| Doxycycline | >16 | ≤2 | 16 | 8 | ≤2 |
| Minocycline | >16 | ≤2 | 4 | ≤2 | ≤2 |
| Tetracycline | >32 | ≤4 | >32 | >32 | ≤4 |
| Tigecycline | 4 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Colistin | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Polymyxin B | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Gentamicin | >16 | 1 | 1 | 0.5 | 0.5 |
| Streptomycin | >64 | 8 | 64 | >64 | 16 |
| Tobramycin | >8 | 2 | ≤1 | ≤1 | ≤1 |
| Amikacin | ≤4 | ≤4 | ≤4 | ≤4 | ≤4 |
| Sulfisoxazole | >256 | ≤16 | >256 | >256 | ≤16 |
| Trimethoprim Sulfamethoxazole | >4 | ≤0.12 | >4 | >4 | ≤0.12 |
Minimum inhibitory concentrations (μg/mL).
Phenotypic AMR of the recipient J53 isolate is included as reference.
Mobilization of carbapenem-resistance genes and confirmation of phenotypic resistance.
Conjugation experiments with both the K. quasipneumoniae with blaKPC-2 and the E. coli with blaNDM-5 successfully mobilized both carbapenemase genes to a J53 recipient strain and were confirmed by PCR. Moreover, phenotypic testing showed that these mobilized carbapenem resistance genes conferred phenotypic carbapenem resistance. In both cases, the transconjugant required markedly higher carbapenem concentrations for inhibition of growth compared with the recipient J53 strain (Table 5).
DISCUSSION
Here, we report a CeRE colonization prevalence of 10.1% in a wild, free-ranging deer population. Other studies investigating cephalosporin resistance prevalence among deer using methods similar to those in this study range are notably lower, near 2% (13, 14). However, our reported prevalence is quite low compared with fecal carriage in healthy companion animals and livestock, but relatively consistent with levels collected from humans (15, 16). It should be noted, however, that our observed prevalence estimated using selective media cannot be directly compared with national AMR surveillance data utilizing isolates recovered using media without selective pressure from antimicrobials.
One potential source of CeRE for deer exposure is surface water that receives treated wastewater effluent. Despite treatment, wastewater effluent can carry a host of resistant bacteria, some of which are resistant to drugs of last resort like carbapenems and colistin (5, 6, 17). In 2009, an estimated 4.5 billion gallons of untreated wastewater was discharged into Lake Erie. In addition, as of 2019 combined sewer overflows discharged an estimated 2.6 billion gallons of untreated effluent (18, 19), some into primary headwaters and streams within the Cleveland Metroparks as it flows to Lake Erie.
Contaminated surface waters can create an environmental reservoir for the exposure of deer and other wildlife to antimicrobial resistant bacteria. Another potential source of antimicrobial resistant bacteria contamination of waterways is via runoff from large farming operations where animals are intensively managed and antimicrobial use is common. In Northeast Ohio, farms, particularly intensive farming operations are rare and unlikely to contribute significantly to environmental dissemination of AMR bacteria (20). Lastly, companion animal feces are frequently found in these areas. Healthy companion animals frequently maintain CeRE bacteria in their gut and these bacteria and their determinants are frequently found contaminating soil and plants (15, 21). These areas can serve as hot spots for domestic animal-wildlife interface and can further contribute to the resistome of urban storm water runoff (22).
The allelic variation of the CeRE isolates was largely constrained to two main types, the CMY-2-like for the AmpC phenotype and CTX-M-15 for the ESBL phenotype. Livestock were previously considered to be the main reservoir of blaCMY-2, but global dissemination of this genotype via zoonotic foodborne transmission and other routes have contributed to human and companion animal colonization (23). This dissemination into multiple reservoirs has led to broad environmental dissemination (24). Conversely, the CTX-M-15 genotype has historically been identified as a variant causing global dissemination and disease among humans (25). Similar to blaCMY, blaCTX-M is globally disseminated and found in contaminated environments, surface waters, and domestic animals (26).
Two carbapenemase-producing Enterobacterales (CPE), one with blaKPC and one with blaNDM were isolated from two separate WTD from the same urban region during 2018 and 2019. Carbapenemase-producing bacteria are considered an urgent public health threat, and blaKPC-2 is considered a hospital epidemic allele most prevalent in K. pneumoniae and Enterobacter spp. (2, 27). The spread of blaKPC beyond hospitals occurs via municipal wastewater flows and treated wastewater effluent that contain CPE and the KPC genetic determinant (5, 6). Isolates with blaNDM only account for sporadic outbreaks in hospital setting in North America and Europe, but their ease of dissemination between Enterobacterales species facilitates their widespread presence within the community (28). Similar to blaKPC, blaNDM can be disseminated through wastewater, but it appears at lower frequencies (5). Dissemination via sources other than surface waters with wastewater inputs is extremely rare in the United States, as blaKPC and blaNDM are rarely reported in livestock, companion animals, and meat products (7, 8, 29–31).
Epidemic carbapenemases like KPC and NDM have been reported in birds, and recently blaOXA was reported in wild boars in Spain (14, 32). In the United States and Canada, there have been recent reports of blaKPC, blaIMP, and blaIMI isolated from corvids (33, 34). When found, major epidemic carbapenemase genotypes are often associated with urbanized habitats or proximity to wastewater treatment plants (32–35). The prevalence of carbapenemase-producing bacteria in many nonavian species has not been reported. Here, we report the novel findings that WTD can become colonized by enteric bacteria expressing these epidemic carbapenemase genes in the United States.
Mobilization of carbapenemase genes is an important contributor to the widespread CPE dissemination beyond human hospitals to communities, natural environments, and domestic and wild animals. The blaNDM-5 we recovered from a WTD was mobilized on an IncFII/FIA plasmid with a somewhat unique genetic backbone that only shared >90% similarity with six other plasmids in the GenBank curation. Interestingly, the epidemiologic dissemination of blaNDM-5 through members of Enterobacterales is partially a function of the self-transmissible IncX3 plasmid that drives their spread within and beyond hospitals into the community (36, 37). However, the genetic element immediately surrounding the NDM-5 reported in this study has been disseminated on both IncX and IncF plasmids (38). Here, we report a rare mobile genetic element variant that consists of three IS26 elements flanking two antimicrobial resistance regions, blaNDM-5 and the truncated class I integron. One recent publication highlighted the importance of flanking IS26 elements in transmitting multidrug-resistance regions that often contain blaNDM-5 (39). Moreover, these IS26 elements may promote interplasmid transfer as they can form circular intermediates and serve as scaffolds for plasmid fusions (39).
The blaKPC-2 we recovered from a WTD is on the Tn4401b transposon, an isoform of the Tn4401 transposable element that is responsible for epidemic expansion and dissemination of blaKPC-2 (40). This insertional element is nested within the mobilizable IncFII plasmid that facilitates its conjugal transfer. IncFII plasmids are known to carry blaKPC-2 in human clinical settings and can spread widely among Enterobacteriaceae (41). However, the IncFII(K) plasmid reported in this study seems to harbor a unique plasmid genetic background, sharing only 78% of its genome with its nearest match. In addition, this plasmid variant did not cluster or share genomic similarity with other blaKPC-2 carrying plasmids, suggesting a unique KPC-2-IncFII(K) combination variant and highlights broad reach of the mobile genetic element. Co-occurrence of other AMR genes on the same or additional plasmids suggest that both CPE isolates have experienced selection pressure in the past and acquired numerous determinants for a selective advantage. Our mobilization experiments show that both carbapenemase genes are easily conjugated to recipient bacteria and confer similar carbapenem-resistant phenotype in those recipients. Conjugal transfer can promote widespread dissemination between and within reservoir species.
Older, male deer were at greater odds of having CeRE present in the fecal microbiome relative to younger, female deer. Deer foraging habits are related to body size, with larger deer foraging more often to meet energy demands (42). Increased foraging efforts may increase the risk for exposure to contaminated environments. In addition, older male deer have larger home ranges compared with younger female deer (43). This increased home range allows for exposure to further diverse environments, some of which may be contaminated. Because of increased deer density, sexually dimorphic feeding areas may not be present. Female and young deer are less likely to consume forage when male deer are present (44). Interestingly, yearlings were at greater odds of harboring an antimicrobial resistant isolate. In some scenarios, yearlings can have larger home ranges than adults, allowing them more contact with different environments (43). Moreover, yearlings select high-protein forage to meet growing energy demands (45). Cultivated plants typically have higher crude protein ratios compared with wild plants promoting more yearling-human interactions.
Urbanization increased the odds of WTD having a CeRE by 1.4 times. The human-domestic animal-wildlife interface represent critical points for antimicrobial resistance crossover (46). Previous studies identified that proximity to domestic livestock and humans in farm settings increased antimicrobial resistance among birds and small mammals (47). Moreover, wildlife interaction and proximity to human waste management sites and wastewater treatment plants are considered a significant risk factor for AMR crossover (48). Recent studies in wildlife identified increased human populations and urbanization were risk factors for AMR bacteria colonization in mesopredators and birds (49, 50). However, among omnivores like primates, adaptation to humans was seen as a more significant risk factor compared to proximity (51).
We observed that WTD are a potentially important wildlife reservoir of cephalosporin-resistant bacteria. In addition, we report the first isolation of the carbapenemase-encoding blaKPC-2 and blaNDM-5 in wild WTD. This is a likely a result of contaminated environments in close proximity to humans, especially surface waters that receive treated or untreated wastewater.
MATERIALS AND METHODS
Study site and sample collection.
Fecal sample collection took place in a metropolitan park system in Northeastern Ohio, USA (Fig. 1A and B). WTD were harvested nightly between January 2018 to March 2018 during the 2018 sampling season and January 2019 to March 2019 during the 2019 sampling season. Harvesting was conducted as part of a deer population management program that was approved by the Ohio Department of Natural Resources. The use of the deer fecal samples for scientific research following their collection for other purposes was reviewed by The Ohio State University’s IACUC staff and determined to be exempt from mandated IACUC review. Ten regions within the park system were assigned for deer management efforts; six urban regions numbered 1 to 6 and four suburban regions numbered 7 to 10. Regions were classified as suburban or urban based on the 2019 ACS 5-year estimate census data (https://data.census.gov/) of the number of households within a half mile buffer. Regions were classified as urban if there was greater than 1 unit per acre and suburban if they contained less than 1 unit per acre (52).
Authorized sharpshooters conducted all harvesting efforts from roadside and tree stands overlooking bait stations. Harvested deer were transported to a centralized processing station. During processing, deer were weighed, aged, sexed, and field dressed. WTD age was estimated using tooth replacement and wear characteristics (53). Deer were divided into three age groups; fawns (<1 year of age), yearling (1 to 2 years), and adults (>2 years). During field dressing, an approximately 10-g fecal sample was sterilely collected from the distal large intestine and placed at 4°C until sample processing.
Sample processing and isolation.
A 4-g fecal sample was combined with 36 mL of MacConkey agar modified with 2 μg/mL cefotaxime and incubated overnight at 37°C. An inoculate of the MacConkey broth was then streaked onto three MacConkey agar plates that were modified with either 8 μg/mL of cefoxitin for isolation of Enterobacterales expressing the AmpC β-lactamase phenotype, 4 μg/mL of cefepime for isolation of Enterobacterales expressing the ESBL phenotype, or 0.5 μg/mL meropenem and 70 μg/mL zinc sulfate heptahydrate for the isolation of Enterobacterales expressing the carbapenemase-producing phenotype.
We selected a single lactose-fermenting isolate expressing the AmpC β-lactamase-producing phenotype and another expressing the ESBL-producing phenotype for further characterization. These isolates were tested for tryptophan utilization using the indole production assay. Isolates were then genotypically characterized for the presence of either blaCMY or blaCTX-M by PCR using previously reported primers (54, 55). PCR products of the expected molecular weight were cleaned and bidirectionally sequenced using a 3730 DNA analyzer (Applied Biosystems) and analyzed for allelic variation using the basic local alignment search tool (BLAST).
We also selected up to three morphologically distinct colonies with a carbapenemase-producing phenotype, giving preference to lactose fermenting isolates. We confirmed carbapenemase production using the CarbaNP test and then determined bacterial genus and species using MALDI-TOF (56). Isolates representing bacterial species not expected to have intrinsic carbapenemase production were tested for the presence of blaKPC, blaNDM, blaIMP, and blaVIM by PCR using previously reported primers (57–60).
Phenotypic and genotypic characterization of carbapenemase-producing isolates.
Two CPE isolate with transmissible carbapenemase genes underwent whole-genome sequencing using a long-read (PacBio; Pacific Biosciences, Menlo Park, CA) platform. DNA libraries were prepared using a 10-Kb template preparation protocol with the PacBio SMRTbell template prep kit v. 1.0. The reads were assembled using PacBio Hierarchical Genome Assembly Process 4.0 and contigs were circularized by Circlator (61, 62). Assembled sequences were assessed for MLST, acquired antimicrobial resistance genes, and plasmid content using PubMLST, ResFinder, AMRFinderPlus and PlasmidFinder, respectively (63–66).
Plasmids that harbored carbapenemase genes were assessed for genome similarity using NCBI BLAST. The top 20 BLAST hits for each plasmid were curated for phylogenetic analysis. Phylogenetic trees were assembled using the CSIPhylogeny pipeline using default parameters and the appropriate carbapenemase-encoding plasmid as the reference sequence (67). Constructed trees were visualized using the Interactive Tree of Life (iTOL) online tool (68).
The genetic landscape surrounding the carbapenemases genes for both CPE isolates was annotated using the Prokka and PGAP pipelines (69, 70). Mobile genetic elements within this genetic context were characterized using ISFinder (71). The similarity of these genetic elements with previous curated genomes were assessed using NCBI BLAST and the query coverage and percent identity were resulted.
An antimicrobial susceptibility profile was also generated for the isolate with transmissible carbapenemase genes using the Sensititre semiautomated broth microdilution system (NARMS CMV3AGNF, ESB1F and GNX2F panels; Thermo Fisher Scientific, Oakwood Village, OH) following Clinical and Laboratory Standards Institute (CLSI) guidelines (72). E. coli strains ATCC 25922 and 35218 were used as quality controls.
Conjugation experiments.
Overnight cultures of the two CPE isolates and a lactose-negative, sodium azide-resistant E. coli J53 strain were grown on LB agar with either 0.5 μg/mL meropenem and 70 μg/mL zinc sulfate heptahydrate or 100 μg/mL of sodium azide. A 1:1 mixture of CPE and the recipient E. coli were mixed in fresh, antibiotic-free nutrient broth and allowed to grow overnight at 37°C under static conditions. Broth mixtures were then serially diluted to 10−5 concentrations. A 100 μL aliquot from each dilution was plated on MacConkey agar modified with 100 μg/mL of sodium azide and 0.5 μg/mL meropenem and 70 μg/mL zinc sulfate heptahydrate. Up to three lactose negative isolates from each plate were selected. Isolates were tested for the presence of either the blaKPC or blaNDM using PCR as previously described. One PCR positive blaKPC and blaNDM representative isolate was tested for phenotypic resistance as previously described.
Data analysis.
Generalized linear mixed models were constructed to identify the association between phenotypic antimicrobial resistance and WTD sex, age class and weight, region classification of origin (urban versus suburban), and year of collection. A forward building model approach was employed and all independent variables with a P-value >0.2 were included in the multivariable model. Dependent factors with a P-value >0.05 and that did not alter the odds ratio by 20% upon removal from or addition to the model were dropped from the final multivariable analysis. All models were controlled for within-region clustering by including individual region as a random effect. Robust standard errors were estimated using the Huber-White sandwich methods. Adjusted odds ratios, their 95% confidence intervals and P-values were reported for all variables included in the final model. All statistical analysis was conducted using STATA v. 15 (StataCorp LLC, College Station, TX, USA).
Geographical maps were visualized using the ggmap package in R to import publicly available, open-source Stamen maps (available at https://stamen.com/open-source/). Resistant phenotype and study region data layers were overlaid on each map using the ggplot2 package in R.
Data availability.
Whole-genome sequence of K. quasipneumoniae and E. coli chromosomal and plasmid DNA were deposited in GenBank with biosample numbers SAMN23219801 and SAMN23219802, respectively.
ACKNOWLEDGMENTS
We thank the White-tailed deer management team and Erik Shaffer for managing, processing, and collecting deer samples as part of the deer management program. This study was funded in part by Cleveland Metroparks, Cleveland Metroparks Zoo and the USDA National Institute of Food and Agriculture, Grant/Award Number: 2013-68003-21282.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. Government.
Contributor Information
Thomas E. Wittum, Email: Wittum.1@osu.edu.
Christopher A. Elkins, Centers for Disease Control and Prevention
REFERENCES
- 1.World Health Organization. 2019. No time to wait: securing the future from drug-resistant infections. Report to the Secretary-General of the United Nations. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Center for Disease Control. 2019. Antibiotic resistant threats report, 2019. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 3.Center for Disease Control. 2020. Carbapenem-resistant Enterobacteriaceae. https://arpsp.cdc.gov/profile/arln/cre.
- 4.Kubin CJ, Ellman TM, Phadke V, Haynes LJ, Calfee DP, Yin MT. 2012. Incidence and predictors of acute kidney injury associated with intravenous polymyxin B therapy. J Infect 65:80–87. 10.1016/j.jinf.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Mathys DA, Mollenkopf DF, Feicht SM, Adams RJ, Albers AL, Stuever DM, Grooters SV, Ballash GA, Daniels JB, Wittum TE. 2019. Carbapenemase-producing Enterobacteriaceae and Aeromonas spp. present in wastewater treatment plant effluent and nearby surface waters in the US. PLoS One 14:e0218650. 10.1371/journal.pone.0218650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballash GA, Lee S, Mollenkopf DF, Mathys DA, Albers AL, Sechrist E, Feicht SM, Van Balen Rubio JC, Sullivan SMP, Lee J, Wittum TE. 2020. Pulsed electric field application reduces carbapenem- and colistin-resistant microbiota and blaKPC spread in urban wastewater. J Environ Manage 265:110529. 10.1016/j.jenvman.2020.110529. [DOI] [PubMed] [Google Scholar]
- 7.Mollenkopf DF, Mathys DA, Feicht SM, Stull JW, Bowman AS, Daniels JB, Wittum TE. 2018. Maintenance of carbapenemase-producing Enterobacteriaceae in a farrow-to-finish swine production system. Foodborne Pathog Dis 15:372–376. 10.1089/fpd.2017.2355. [DOI] [PubMed] [Google Scholar]
- 8.Daniels JB, Chen L, Grooters SV, Mollenkopf DF, Mathys DA, Pancholi P, Kreiswirth BN, Wittum TE. 2018. Enterobacter cloacae complex sequence type 171 isolates expressing KPC-4 carbapenemase recovered from canine patients in Ohio. Antimicrob Agents Chemother 62. 10.1128/AAC.01161-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold KE, Williams NJ, Bennett M. 2016. ‘Disperse abroad in the land': the role of wildlife in the dissemination of antimicrobial resistance. Biol Lett 12:20160137. 10.1098/rsbl.2016.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mo SS, Urdahl AM, Madslien K, Sunde M, Nesse LL, Slettemeås JS, Norström M. 2018. What does the fox say? Monitoring antimicrobial resistance in the environment using wild red foxes as an indicator. PLoS One 13:e0198019. 10.1371/journal.pone.0198019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith S, Wang J, Fanning S, McMahon BJ. 2014. Antimicrobial resistant bacteria in wild mammals and birds: a coincidence or cause for concern? Ir Vet J 67:8. 10.1186/2046-0481-67-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo T, Fidino M, Lehrer EW, Magle SB. 2017. Mammal diversity and metacommunity dynamics in urban green spaces: implications for urban wildlife conservation. Ecol Appl 27:2330–2341. 10.1002/eap.1611. [DOI] [PubMed] [Google Scholar]
- 13.Ballash GA, Munoz-Vargas L, Albers A, Dennis PM, LeJeune JT, Mollenkopf DF, Wittum TE. 2021. Temporal trends in antimicrobial resistance of fecal Escherichia coli from deer. Ecohealth 18:288–296. 10.1007/s10393-021-01559-3. [DOI] [PubMed] [Google Scholar]
- 14.Darwich L, Vidal A, Seminati C, Albamonte A, Casado A, López F, Molina-López RA, Migura-Garcia L. 2019. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS One 14:e0210686. 10.1371/journal.pone.0210686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathys DA, Mollenkopf DF, Bremer CA, Daniels JB, Wittum TE. 2017. Prevalence of AmpC- and extended-spectrum β-Lactamase-Harbouring Enterobacteriaceae in faecal flora of a healthy domestic canine population. Zoonoses Public Health 64:554–560. 10.1111/zph.12341. [DOI] [PubMed] [Google Scholar]
- 16.Landers TF, Mollenkopf DF, Faubel RL, Dent A, Pancholi P, Daniels JB, Wittum TE. 2017. Extended-spectrum β-lactam resistance in the enteric flora of patients at a tertiary care medical centre. Zoonoses Public Health 64:161–164. 10.1111/zph.12293. [DOI] [PubMed] [Google Scholar]
- 17.Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. 2013. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 18.Northeast Ohio Regional Sewer District. 2018. About project clean lake. https://www.neorsd.org/community/about-the-project-clean-lake-program/.
- 19.Northeast Ohio Regional Sewer District (NEORSD). 2020. Combined sewer overflow (CSO) long-term control plan consent decree: semi-annual progress report No. 17. https://www.neorsd.org/I_Library.php?SOURCE=library/Semi-Annual_Progress_Report_17.pdf&a=download_file&LIBRARY_RECORD_ID=7596.
- 20.United States Department of Agriculture. 2018. Ohio Agricultural statistics 2017–2018 annual bulletin. https://www.nass.usda.gov/Statistics_by_State/Ohio/Publications/Annual_Statistical_Bulletin/Ohio%20bulletin%202017-2018.pdf.
- 21.Gekenidis MT, Rigotti S, Hummerjohann J, Walsh F, Drissner D. 2020. Long-term persistence of blaCTX-M-15 in soil and lettuce after introducing extended-spectrum β-lactamase (ESBL)-producing Escherichia coli via manure or water. Microorganisms 8:1646. 10.3390/microorganisms8111646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Suits M, Wituszynski D, Winston R, Martin J, Lee J. 2020. Residential urban stormwater runoff: a comprehensive profile of microbiome and antibiotic resistance. Sci Total Environ 723:138033. 10.1016/j.scitotenv.2020.138033. [DOI] [PubMed] [Google Scholar]
- 23.Mollenkopf DF, Weeman MF, Daniels JB, Abley MJ, Mathews JL, Gebreyes WA, Wittum TE. 2012. Variable within- and between-herd diversity of CTX-M cephalosporinase-bearing Escherichia coli isolates from dairy cattle. Appl Environ Microbiol 78:4552–4560. 10.1128/AEM.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heider LC, Hoet AE, Wittum TE, Khaitsa ML, Love BC, Huston CL, Morley PS, Funk JA, Gebreyes WA. 2009. Genetic and phenotypic characterization of the bla(CMY) gene from Escherichia coli and Salmonella enterica isolated from food-producing animals, humans, the environment, and retail meat. Foodborne Pathog Dis 6:1235–1240. 10.1089/fpd.2009.0294. [DOI] [PubMed] [Google Scholar]
- 25.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 26.Cho S, Nguyen HAT, McDonald JM, Woodley TA, Hiott LM, Barrett JB, Jackson CR, Frye JG. 2019. Genetic characterization of antimicrobial-resistant Escherichia coli isolated from a mixed-use watershed in Northeast Georgia, USA. IJERPH 16:3761. 10.3390/ijerph16193761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson BM, El Chakhtoura NG, Patel S, Saade E, Donskey CJ, Bonomo RA, Perez F. 2017. Carbapenem-resistant Enterobacter cloacae in patients from the US Veterans Health Administration, 2006–2015. Emerg Infect Dis 23:878–880. 10.3201/eid2305.162034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vikram A, Schmidt JW. 2018. Functional blaKPC-2 sequences are present in U.S. beef cattle feces regardless of antibiotic use. Foodborne Pathog Dis 15:444–448. 10.1089/fpd.2017.2406. [DOI] [PubMed] [Google Scholar]
- 30.Cole SD, Peak L, Tyson GH, Reimschuessel R, Ceric O, Rankin SC. 2020. New Delhi Metallo-β-Lactamase-5-producing Escherichia coli in companion animals, United States. Emerg Infect Dis 26:381–383. 10.3201/eid2602.191221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballash GA, Albers AB, Mollenkopf DF, Sechrist E, Adams RJ, Wittum TE. 2021. Antimicrobial resistant bacteria recovered from retail ground meat products in the US include a Raoultella ornithinolytica co-harboring blaKPC-2 and blaNDM-5. Sci Rep 11:14041. 10.1038/s41598-021-93362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köck R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S, Jurke A. 2018. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect 24:1241–1250. 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Kutilova I, Janecko N, Cejkova D, Literak I, Papagiannitsis CC, Dolejska M. 2018. Characterization of blaKPC-3-positive plasmids from an Enterobacter aerogenes isolated from a corvid in Canada. J Antimicrob Chemother 73:2573–2575. 10.1093/jac/dky199. [DOI] [PubMed] [Google Scholar]
- 34.Kutilova I, Valcek A, Papagiannitsis CC, Cejkova D, Masarikova M, Paskova V, Davidova-Gerzova L, Videnska P, Hrabak J, Literak I, Dolejska M. 2020. Carbapenemase-producing Gram-negative bacteria from American crows in the United States. Antimicrob Agents Chemother 65:e00586-20. 10.1128/AAC.00586-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarabai H, Wyrsch ER, Bitar I, Dolejska M, Djordjevic SP. 2021. Epidemic HI2 plasmids mobilising the carbapenemase gene blaIMP-4 in Australian clinical samples identified in multiple sublineages of Escherichia coli ST216 colonising silver gulls. Microorganisms 9:567. 10.3390/microorganisms9030567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He T, Wei R, Zhang L, Sun L, Pang M, Wang R, Wang Y. 2017. Characterization of NDM-5-positive extensively resistant Escherichia coli isolates from dairy cows. Vet Microbiol 207:153–158. 10.1016/j.vetmic.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Tian D, Wang B, Zhang H, Pan F, Wang C, Shi Y, Sun Y. 2020. Dissemination of the blaNDM-5 gene via IncX3-type plasmid among. Enterobacteriaceae in Children mSphere 5:e00699-19. 10.1128/mSphere.00699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou H, Jia X, Liu H, Li S, Wu X, Huang S. 2020. Emergence of NDM-5-porducing Escherichia coli in a teaching hospital in Chongqing, China: IncF-type plasmids may contribute to the prevalence of blaNDM-5. Front Microbiol 11:334. 10.3389/fmicb.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao QY, Zhu JH, Cai RM, Zheng SR, Zhang LJ, Chang MX, Lu YW, Fang LX, Sun J, Jiang HX. 2021. IS26 is responsible for the evolution and transmission of blaNDM-5-harboring plasmids in Escherichia coli of poultry origin in China. mSystems 6:e0064621. 10.1128/mSystems.00646-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2013. Complete sequence of a bla(KPC-2)-harboring IncFII(K1) plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother 57:1542–1545. 10.1128/AAC.02332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luna RS, Duarte A, Weckerly FW. 2013. Influence of body size on dietary nutrition of white-tailed deer Odocoileus virginianus. J Fish Wildlife Manag 4:53–62. 10.3996/092012-JFWM-085. [DOI] [Google Scholar]
- 43.Miller K, Muller L, Demarais S. 2003. White-tailed deer, p 906–930. In Feldhamer GA, Thompson BC, Chapman JA. (ed), Wild mammals of North America, 2nd ed. John Hopkins University Press, Baltimore, MD, USA. [Google Scholar]
- 44.Cherry MJ, Conner M, Warren RJ. 2015. Effects of predation risk and group dynamics on white-tailed deer foraging behavior in a longleaf pine savanna. Beheco 26:1091–1099. 10.1093/beheco/arv054. [DOI] [Google Scholar]
- 45.Dostaler S, Ouellet JP, Therrien JF, Cote SD. 2011. Are feeding preferences of white-tailed deer related to plant constituents? J Wild Manage 75:913–918. 10.1002/jwmg.118. [DOI] [Google Scholar]
- 46.Hassell JM, Begon M, Ward MJ, Fèvre EM. 2017. Urbanization and disease emergence: dynamics at the wildlife-livestock-human interface. Trends Ecol Evol 32:55–67. 10.1016/j.tree.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathys DA, Mathys BA, Mollenkopf DF, Daniels JB, Wittum TE. 2017. Enterobacteriaceae harboring AmpC (blaCMY) and ESBL (blaCTX-M) in migratory and nonmigratory wild songbird populations on Ohio dairies. Vector Borne Zoonotic Dis 17:254–259. 10.1089/vbz.2016.2038. [DOI] [PubMed] [Google Scholar]
- 48.Greig J, Rajic A, Young I, Mascarenhas M, Waddell L, LeJeune J. 2015. A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses Public Health 62:269–284. 10.1111/zph.12147. [DOI] [PubMed] [Google Scholar]
- 49.Worsley-Tonks KEL, Miller EA, Anchor CL, Bender JB, Gehrt SD, McKenzie SC, Singer RS, Johnson TJ, Craft ME. 2021. Importance of anthropogenic sources at shaping the antimicrobial resistance profile of a peri-urban mesocarnivore. Sci Total Environ 764:144166. 10.1016/j.scitotenv.2020.144166. [DOI] [PubMed] [Google Scholar]
- 50.Ahlstrom CA, Bonnedahl J, Woksepp H, Hernandez J, Olsen B, Ramey AM. 2018. Acquisition and dissemination of cephalosporin-resistant E. coli in migratory birds sampled at an Alaska landfill as inferred through genomic analysis. Sci Rep 8:7361. 10.1038/s41598-018-25474-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsons MB, Travis DA, Lonsdorf EV, Lipende I, Elchoufi D, Gilagiza B, Collins A, Kamenya S, Tauxe RV, Gillespie TR. 2021. Antimicrobial resistance creates threat to chimpanzee health and conservation in the wild. Pathogens 10:477. 10.3390/pathogens10040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theobald DM. 2001. Land-use dyanmics beyond the American urban fringe. Geogr Rev 91:544–564. 10.2307/3594740. [DOI] [Google Scholar]
- 53.Severinghaus CW. 1949. Tooth development and weas as criteria of age in white-tailed deer. J Wildlife Management 13:195–216. 10.2307/3796089. [DOI] [Google Scholar]
- 54.Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother 45:2716–2722. 10.1128/AAC.45.10.2716-2722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wittum TE, Mollenkopf DF, Daniels JB, Parkinson AE, Mathews JL, Fry PR, Abley MJ, Gebreyes WA. 2010. CTX-M-type extended-spectrum β-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog Dis 7:1575–1579. 10.1089/fpd.2010.0615. [DOI] [PubMed] [Google Scholar]
- 56.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. 1996. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J Clin Microbiol 34:2909–2913. 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart TJ, Hossain A, Johnson JA, Goering RV, Thomson KS. 2003. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J Antimicrob Chemother 51:711–714. 10.1093/jac/dkg124. [DOI] [PubMed] [Google Scholar]
- 59.Gröbner S, Linke D, Schütz W, Fladerer C, Madlung J, Autenrieth IB, Witte W, Pfeifer Y. 2009. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J Med Microbiol 58:912–922. 10.1099/jmm.0.005850-0. [DOI] [PubMed] [Google Scholar]
- 60.Peirano G, Ahmed-Bentley J, Woodford N, Pitout JD. 2011. New Delhi metallo-beta-lactamase from traveler returning to Canada. Emerg Infect Dis 17:242–244. 10.3201/eid1702.101313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 62.Hunt M, De Silva N, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circulator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carattoli A, Zankari E, GarcÌa-Fern ·Ndez A, Voldby Larsen M, Lund O, Villa L, Ller Aarestrup M, Hasman ¯FH. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Letunic I, Bork P. 2019. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 70.Tatusova T, Ciufo S, Federhen S, Fedorov B, McVeigh R, O'Neill K, Tolstoy I, Zaslavsky L. 2015. Update on RefSeq microbial genomes resources. Nucleic Acids Res 43:D599–605. 10.1093/nar/gku1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-genome sequence of K. quasipneumoniae and E. coli chromosomal and plasmid DNA were deposited in GenBank with biosample numbers SAMN23219801 and SAMN23219802, respectively.