Abstract
Prevalence, antibiotic susceptibility, and genetic diversity were determined for Escherichia coli O157:H7 isolated over 11 months from four beef cattle feedlots in southwest Kansas. From the fecal pat (17,050) and environmental (7,134) samples collected, 57 isolates of E. coli O157:H7 were identified by use of bacterial culture and latex agglutination (C/LA). PCR showed that 26 isolates were eaeA gene positive. Escherichia coli O157:H7 was identified in at least one of the four feedlots in 14 of the 16 collections by C/LA and in 9 of 16 collections by PCR, but consecutive positive collections at a single feedlot were rare. Overall prevalence in fecal pat samples was low (0.26% by C/LA, and 0.08% by PCR). No detectable differences in prevalence or antibiotic resistance were found between isolates collected from home pens and those from hospital pens, where antibiotic use is high. Resistant isolates were found for six of the eight antibiotics that could be used to treat E. coli infections in food animals, but few isolates were multidrug resistant. The high diversity of isolates as measured by random amplification of polymorphic DNA and other characteristics indicates that the majority of isolates were unique and did not persist at a feedlot, but probably originated from incoming cattle. The most surprising finding was the low frequency of virulence markers among E. coli isolates identified initially by C/LA as E. coli O157:H7. These results demonstrate that better ways of screening and confirming E. coli O157:H7 isolates are required for accurate determination of prevalence.
The low infectious dose and high virulence of Escherichia coli O157:H7 make human infections particularly severe and life threatening (7, 28). Shiga toxins produced by E. coli O157:H7 are the principal virulence factors responsible for hemorrhagic colitis and hemolytic uremic syndrome in humans (14, 15). Antibiotic treatment is contraindicated for human E. coli O157:H7 infections, because certain antibiotics, such as fluoroquinolones, induce Shiga toxin-encoding bacteriophages in vivo and lead to increased expression of Shiga toxin genes (35). Antibiotics also may cause bacterial lysis, which could increase free Shiga toxin in the intestinal tract (13, 32, 33; L. E. Wolf, D. W. Acheson, L. L. Lincicome, and G. T. Keusch, VTEC '97: 3rd Int. Symp. Workshop Shiga Toxin (Verocytotoxin)-Producing E. coli Infect., abstr. V145/III, p. 60, 1997). Even though antibiotics should not be used therapeutically for E. coli O157:H7 infections, some evidence indicates that antibiotic resistance in isolates is increasing (17). Because many human E. coli O157:H7 infections are acquired from eating undercooked contaminated beef, it is important to determine if the organism develops resistance to antibiotics during food animal production. In addition, it is important to determine if resistant E. coli O157:H7 is a possible reservoir for spread of resistance factors to other microorganisms. The Food and Drug Administration, Centers for Disease Control and Prevention, and others believe that agricultural use of antibiotics accounts for the majority of increases in antibiotic-resistant human isolates (30).
Antibiotics typically are used at cattle feedlots therapeutically, for disease prophylaxes, and for growth promotion. Antibiotics also may be used to treat horses, pets, other livestock, and crops at feedlots. If an animal becomes sick at a feedlot, usually it is removed from its resident (home) pen and relocated to a hospital pen, where it is treated with antibiotics. When the animal is healthy, usually from 3 to 60 days later, it is returned to its home pen. While in the hospital pen, animals commingle with other animals receiving antibiotics for a variety of conditions. Therefore, one would expect to find a greater number of resistant bacterial isolates in hospital pens than in home pens.
Knowing the genetic diversity of E. coli O157:H7 collected over time could help determine if contamination at a feedlot is due to bacteria that are transient or indigenous (resident). Transient bacteria can be introduced into the feedlot on arriving cattle; in ingredients for cattle rations; from contaminated water sources; or by other means, such as other animals (wild or domestic), vehicles, and employees. Escherichia coli O157:H7 shed by animals might persist for long durations in contaminated soil, water, manure, and feed and be spread to other uninfected animals (9). Manure management, feed bunk and water trough sanitation, and feed management all contribute to preventing transient bacteria from becoming resident. Therefore, if bacteria collected over time are genetically similar, the feedlot could be contaminated with a resident strain, whereas a greater genetic diversity could indicate that strains are arriving from a variety of sources. However, LeClerc et al. (21) reported that E. coli O157:H7 strains may become hypermutable, thereby making them more adaptable to changing environments and perhaps increasing the organism's genetic diversity.
Genetic fingerprinting is a means by which epidemiologists have traced back infections to their probable sources. It also has been used to understand the ecology of E. coli O157:H7 and might be used as part of hazard analysis and critical control point (HACCP) programs for producers to reduce on-farm pathogens. Using genetic fingerprinting techniques, Kudva et al. (20) found multiple strains of E. coli O157:H7 in a single flock of sheep and showed that a single animal shed multiple strains simultaneously and that strains shed by individuals changed over time. Escherichia coli O157:H7 has been isolated from animal drinking water, animal feed, flies, and a pigeon at dairy farms in Wisconsin (27). The majority of isolates collected at these farms had the same genetic fingerprint. Although various methods are available for genetic characterization of bacterial isolates, random amplification of polymorphic DNA (RAPD) has been used successfully in the past for E. coli O157:H7 (4, 24, 31) and is less costly and time-consuming than other methods.
Escherichia coli O157:H7 strains typically contain genes that encode Shiga toxins (stx1 and/or stx2), attaching-and-effacing proteins such as intimin (eaeA), and hemolysins (hly) (2, 6, 12, 34). The presence of a plasmid-encoded enterohemolysin (E-Hly) in E. coli O157:H7 was first demonstrated in 1989 by Beutin et al. (3). Despite the presence of common virulence factors, strains of E. coli O157 may differ in their degree of pathogenicity. Baker et al. (1) found that strains of human origin were more virulent in gnotobiotic pigs than strains of bovine origin.
Cattle infected with E. coli O157:H7 show no signs of disease, and shedding of the organism in feces is sporadic and difficult to detect. One approach to assess prevalence is by sampling many feedlots once (USDA:APHIS:VS:Centers for Epidemiology and Animal Health: Escherichia coli O157:H7 shedding by feedlot cattle, 1995; Factors associated with Escherichia coli O157:H7 in feces of feedlot cattle, 1997 [http: //www.aphis.usda.gov / vs / ceah / cahm / Beef_Feedlot / bffeed.htm]). Because other studies have found E. coli O157:H7 prevalence to be low at livestock operations, many samples must be collected to obtain a sufficient number of isolates for analysis.
The major objectives of this study were to determine the prevalence of E. coli O157:H7 at four beef cattle feedlots by sampling repeatedly and intensively over an 11-month period and to determine if a difference in antibiotic susceptibility could be detected between isolates from home pens and those from hospital pens, where antibiotic use is high. An additional objective was to determine, through use of RAPD genetic fingerprinting, if the E. coli O157:H7 isolates were primarily resident or transient.
MATERIALS AND METHODS
Sample collection.
Four beef cattle feedlots (designated M, S, R, and V) in southwest Kansas were studied. Each feedlot was chosen for inclusion because it was large-scale (>35,000 animal capacity) and well managed and because pens in the feedlot were used continuously throughout the year. The cattle in the feedlots came from a variety of sources. Animals at each feedlot are kept in a pen with up to 250 animals for approximately 145 days until they are shipped to slaughter and another lot of cattle is placed in the pen. During the period of study, these feedlots reported using antibiotics therapeutically, but not prophylactically or for growth promotion. Samples were collected at each feedlot every 3 weeks throughout an 11-month period (16 collections). Typically, 30 home pens and all hospital and buller pens (pens with steers removed from their home pen because they were mounted repeatedly by other steers) were sampled at each feedlot during each of the 16 collections. One teaspoon of feces was collected from each of five different fresh fecal pats on the pen floor and placed together in a new zip-lock plastic bag. Six bags were collected from each study pen. Drinking water (50 ml) and rations (50 g) from each pen were sampled at each collection. Standing lagoons at each feedlot and individual ration components at each feedlot's feed mill also were sampled. Samples were placed in a cooler with frozen cool packs. At the end of a sampling day, new frozen cool packs were placed in the cooler, and the cooler was sealed and shipped by next-day delivery to the laboratory.
Selective enrichment and isolation of E. coli O157:H7.
On arrival at the laboratory, each composite fecal sample was mixed, and 1 g of feces or feed or 1 ml of water was removed and put into 10 ml of universal pre-enrichment broth (Difco, Inc., Detroit, Mich.) with a 15-μg/ml final concentration of novobiocin (Sigma-Aldrich, St. Louis, Mo.) and incubated at 42°C for 18 to 24 h. Then, the tube was vortexed, and a swab sample was plated onto sorbitol-MacConkey agar. The plate was streaked for isolation and incubated at 42°C for 18 h.
Identification by C/LA.
Identification by the combination of bacterial culture and latex agglutination (C/LA) was performed as follows. After incubation, up to five suspected E. coli O157:H7 colonies (gray-white) were picked and inoculated into triple-sugar iron agar slants and incubated at 42°C for 18 to 24 h. After incubation, suspected E. coli isolates (yellow slant and butt) were plated onto O157 Rainbow agar (Biolog, Inc., Hayward, Calif.) and incubated at 42°C for 18 to 24 h. Suspected O157 isolates (gray-black) were picked and streaked onto blood agar plates (BAP), which were incubated at 42°C for 18 to 24 h. Then, isolated colonies were checked for O157 LA (Remel, Lenexa, Kans.). Any isolates that agglutinated with the O157 latex were checked for H7 agglutination (Remel). If the isolate did not agglutinate with the H7 latex, the isolate was transferred to another BAP, incubated at 42°C for 18 to 24 h, and then rechecked for H7 agglutination. If the isolate again did not agglutinate with the H7 latex, the isolate was cultured and checked a third time.
Biochemical confirmation.
Isolates that agglutinated with the O157 latex were inoculated into an API20E strip (bioMerieux Vitek, Hazelwood, Mo.) for identification according to the manufacturer's specifications.
Storage.
If the API strip identified the isolate as E. coli, then the isolate was inoculated onto BAP and incubated at 42°C for 18 to 24 h. The isolate was inoculated onto Protect beads (Key Scientific Products, Round Rock, Tex.) according to the manufacturer's specifications and frozen at −70°C for long-term storage.
Antibiotic susceptibility testing.
Isolates were streaked for isolation onto BAP and incubated overnight at 37°C. Following incubation, an autoinoculator (Sensititre/Alamar AccuMed, Westlake, Ohio) was used to inoculate the wells of a 96-well plate with bacteria to a standard cell density. Isolates were inoculated into both a breakpoint panel with 17 antibiotics and a broth microdilution panel with 8 antibiotics. The antibiotics selected for the broth microdilution panel are used commonly at feedlots and are included on panels used by the National Antimicrobial Resistance Monitoring System (29). The breakpoint panel was a standard panel distributed by the manufacturer to test human isolates. The concentration of each antibiotic was selected by the manufacturer (Sensititre/Alamar) on the basis of National Committee for Clinical Laboratory Standards recommendations (23). After inoculation, the plates were incubated overnight at 37°C. Following incubation, the plates were read with an automated reader (Sensititre/Alamar AccuMed), and results were generated by use of SAMS software (Sensititre/Alamar AccuMed).
RAPD primers.
To narrow the search for arbitrary primers that would be suitable for this study, six primers that had been discriminatory in E. coli studies by others (22, 31) were used for RAPD. From these, the two primers that had shown the greatest ability to discriminate among E. coli O157:H7 isolates in preliminary testing were selected. All primers were purchased from Integrated DNA, Inc., Coralville, Iowa. The 10-mer primers used were 1254 (5′CCG CAG CCA A 3′) and 1283 (5′ GCG ATC CCC A 3′). The four primers that were not discriminatory were HLWL85, OPB17, 1247, and 1290. The E. coli O157:H7 isolates were analyzed with each of the two primers in at least three independent reactions, and bands that were clearly and consistently detected were considered to establish a fingerprint for that isolate.
RAPD procedure.
Isolates were grown for 18 to 24 h at 42°C in tubes with 5 ml of brain heart infusion broth. After incubation, the broth suspension was checked for optical density. Isolates were diluted with broth to standardize optical density and centrifuged. Total DNA (chromosomal and plasmid) was extracted by resuspending the cell pellet in 100 μl of Tris EDTA containing 20% Chelex. The suspension was incubated at 55°C for 15 min and then boiled for 7 min. Cell debris and Chelex were removed by centrifugation. For RAPD profiles of plasmid DNA, isolates were grown for 18 to 24 h at 42°C in brain heart infusion broth, and plasmids were extracted with a Qiagen (Valencia, Calif.) plasmid isolation kit. According to the manufacturer's specifications, this kit can isolate plasmids that range from 1 to 100 MDa in size. The DNA concentrations were about 5 ng/μl, as determined by the DNA dipstick method (Invitrogen, Carlsbad, Calif.). Each PCR mixture consisted of 1× reaction buffer, 1.5 mM MgCl2, 2 μM primer, 200 μM each deoxynucleoside triphosphates (dNTPs), 2.5 U of Taq polymerase (Fisher, St. Louis, Mo.); and 5 μl of DNA in a total reaction volume of 50 μl. Amplification was done with a thermal cycler (model PTC-100; MJ Research,Watertown, Mass.), which was set for four cycles at 94°C for 4 min, 37°C for 4 min, and 72°C for 4 min; followed by 30 cycles at 94°C for 1 min, 37°C for 1 min, and 72°C for 1 min; followed by 1 cycle at 72°C for 10 min.
After the reaction, the amplified DNA was electrophoresed on a 1.5% agarose gel for 90 min at 94 mA with ethidium bromide in the gel. After electrophoresis, the gel was placed on a Transilluminator, and photographs were taken. Observational gel analysis was done to compare the number and size of bands in each sample.
Virulence genes.
The following primers (New England Medical Center Hospital, Boston, Mass.) were used to analyze the isolates for the presence of virulence genes: stx1 forward primer (AAA TCG CCA TTC GTT GAC TAC TTC T), stx1 reverse primer (TGT CCA TTC TGG CAA CTC GCG ATG CA), stx2 forward primer (CAG TCG TCA CTC ACT GGT TTC ATC A), stx2 reverse primer (GGA TAT TCT CCC CAC TCT GAC ACC), eaeA forward primer (CAG GTC GTC GTG TCT GCT AAA), eaeA reverse primer (TCA GCG TGG TTG GAT CAA CCT), hlyA forward primer (GCA TCA TCA AGC GTA CGT TCC), and hlyA reverse primer (AAT GAG CCA AGC TGG TTA AGC T). The annealing temperatures were 63°C for the stx1 and stx2 primers and 59°C for the eaeA and hlyA genes. Amplification was done with a 94°C denaturation step for 1 min and an annealing step for 1 min, followed by 72°C for 1 min, for 40 cycles, followed by 1 cycle at 72°C for 10 min. The PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
Data analysis.
All data were entered into computer spreadsheets (Microsoft Excel, Bellevue, Wash.) and then converted to SAS (26) data sets, and tables were constructed to report prevalence data. For this study, the sampling unit was the pen and the population of inference comprised pens at large-scale beef cattle feedlots. Prevalence was computed as the number of pens with E. coli O157:H7 divided by the number of pens sampled for collection and feedlot combinations.
RESULTS
Prevalence.
From 24,184 samples collected, 45 samples were positive (0.19%). Multiple colonies were taken from each sample, and 57 total isolates of E. coli O157:H7 were identified by use of C/LA (Table 1). Forty-four of the positive samples were fecal pat samples (Table 2), and one was a water trough sample (3,186 collected). Of 2,871 ration samples, 581 ingredient samples, 312 lagoon water samples, 24 horse fecal samples, and 160 fecal pat samples from temporary holding pens at feedlot R, none tested positive for E. coli O157:H7.
TABLE 1.
Characterization of 57 E. coli O157:H7 isolates from fresh fecal pats collected from southwest Kansas beef cattle feedlotsa
| Collection | Feedlot | Pen ID | Pen type | PCR characterization
|
Lane | RAPD pattern
|
Feedlot strain no. | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| eaeA | hlyA | stx1 | stx2 | T | P | ||||||
| 3 | M | 128 | Home | − | − | − | − | 32 | 1 | 2 | 14 |
| 7 | M | 40 | Home | + | + | + | + | 31 | 1 | 14 | 42 |
| 13 | M | 114 | Home | − | − | − | − | 29 | 15 | 12 | 5 |
| 13 | M | 1022 | Hospital | − | − | − | − | 30b | 14 | 13 | 13 |
| 1 | R | 113 | Home | + | + | + | + | 45 | 2 | 2 | |
| 1 | R | 300 | Home | + | + | − | + | 44 | 1 | 14 | 34 |
| 1 | R | 505c | Home | + | + | − | + | 49 | 1 | 2 | 7 |
| 1 | R | 505c | Home | + | + | − | + | 48 | 2 | 16 | 20 |
| 1 | R | 47 | Hospital | + | + | + | − | 43 | 2 | 2 | 8 |
| 1 | R | 47c | Hospital | − | − | − | − | 47 | 1 | 2 | 22 |
| 1 | R | 47c | Hospital | + | + | + | + | 46 | 11 | 15 | 37 |
| 8 | R | 402 | Home | − | − | − | − | 44 | |||
| 8 | R | 81c | Home | − | − | − | − | 41 | 13 | 2 | 16 |
| 8 | R | 81c | Home | − | − | − | − | 42 | 12 | 2 | 48 |
| 11 | R | 45c | Hospital | + | + | + | + | 39 | 2 | 10 | 24 |
| 11 | R | 45c | Hospital | − | − | − | − | 40 | 1 | 2 | 26 |
| 12 | R | 256 | Hospital | − | − | − | − | 38 | 2 | 2 | 11 |
| 15 | R | 113 | Home | − | − | − | − | 36 | 2 | 3 | 46 |
| 16 | R | 97 | Buller | + | + | − | + | 34 | 1 | 2 | 38 |
| 16 | R | 107 | Home | + | + | + | + | 33 | 14 | 2 | 29 |
| 16 | R | 43 | Hospital | − | − | − | − | 35 | 10 | 3 | 10 |
| 1 | S | 409 | Home | + | + | − | + | 27 | 1 | 2 | 39 |
| 2 | S | 223 | Home | + | + | − | + | 26 | 1 | 10 | 18 |
| 5 | S | 303 | Home | + | + | − | + | 23 | 1 | 2 | 21 |
| 8 | S | 301 | Home | − | − | − | − | 15 | 1 | 8 | 4 |
| 8 | S | 228c | Home | + | + | + | + | 20 | 1 | 11 | 23 |
| 8 | S | 228c | Home | + | + | − | + | 21 | 1 | 2 | 27 |
| 8 | S | 228c | Home | + | + | + | + | 22 | 1 | 3 | 33 |
| 8 | S | 228c | Home | + | + | − | + | 18 | 1 | 10 | 54 |
| 8 | S | 228c | Home | + | + | − | + | 19b | 1 | 10 | |
| 8 | S | 409c | Home (W) | + | + | + | + | 17 | 3 | 9 | 1 |
| 8 | S | 409c | Home (W) | − | − | − | − | 16 | 4 | 2 | 43 |
| 13 | S | 110 | Home | − | − | − | − | 14 | 5 | 7 | 2 |
| 13 | S | 15 | Hospital | − | − | − | − | 12b | 6 | 5 | 32 |
| 13 | S | 19 | Hospital | − | − | − | − | 13b | 1 | 6 | 15 |
| 14 | S | 18 | Hospital | − | − | − | − | 11b | 7 | 5 | 47 |
| S | 20 | Buller | − | − | − | − | 28b | 16 | 2 | 51 | |
| 2 | V | 306 | Buller | + | + | + | + | 6 | 2 | 3 | 25 |
| 2 | V | 53 | Home | + | + | − | + | 7 | 2 | 2 | 52 |
| 2 | V | 75 | Home | − | − | − | − | 8 | 1 | 3 | 36 |
| 2 | V | 61c | Home | + | + | − | + | 9 | 1 | 4 | 28 |
| 2 | V | 61c | Home | − | − | − | − | 10 | 2 | 2 | 30 |
| 4 | V | 306c | Buller | + | + | + | + | 24 | 2 | 10 | 9 |
| 4 | V | 306c | Buller | + | + | − | + | 25 | 2 | 2 | 35 |
| 4 | V | 533 | Home | − | − | − | − | 5 | 7 | 2 | 17 |
| 7 | V | 83 | Home | + | + | − | + | 4b | 2 | 3 | 19 |
| 10 | V | 306 | Buller | − | − | − | − | 49 | |||
| 10 | V | 16 | Home | − | − | − | − | 3 | 8 | 2 | 40 |
| 10 | V | 71 | Home | − | − | − | − | 53 | |||
| 10 | V | 24c | Home | − | − | − | − | 1 | 9 | 1 | 3 |
| 10 | V | 24c | Home | − | + | − | − | 2 | 1 | 2 | 12 |
| 12 | V | 43 | Home | + | + | − | + | 54 | 1 | 2 | 31 |
| 13 | V | 433 | Home | − | − | − | − | 53 | 2 | 2 | 45 |
| 14 | V | 52 | Home | − | − | − | − | 52 | 2 | 2 | 50 |
| 15 | V | 29 | Home | − | − | − | − | 37 | 2 | 3 | 6 |
| 15 | V | 52 | Home | − | − | − | − | 51 | 2 | 2 | 41 |
| 16 | V | 43 | Home | − | − | − | − | 50b | 10 | 2 | |
Isolates were collected from pen floors at four southwest Kansas beef cattle feedlots (V, S, M, and R) every 3 weeks for 11 months, resulting in 16 collections. Data are sorted by feedlot and collection within feedlot. All isolates were from fecal pat samples, except for two from a water (W) sample. Lane numbers correspond to lanes in Fig. 1 in left-to-right, top-to-bottom order. RAPD T is the pattern identified from total DNA; RAPD P is the pattern identified from plasmids of the isolate. The genes amplified by PCR were eaeA (intimin), hlyA (hemolysin), stx1 (Shiga toxin I), and stx2 (Shiga toxin II). + and −, presence or absence, respectively, of the gene. Three isolates were not characterized by RAPD. Blank spaces indicate missing data. Feedlot strain number can be used to cross-reference additional information about these isolates (19).
Strain was resistant to five or more antibiotics.
Multiple isolates from the same sample.
TABLE 2.
Percentage of fecal pats positive for E. coli O157:H7a
| Feedlot | % Positive (n samples)
|
|||
|---|---|---|---|---|
| Home | Hospital | Buller | Total | |
| All isolates | ||||
| V | 0.43 (3,237) | 0.00 (541) | 1.90 (159) | 0.43 (3,937) |
| R | 0.21 (3,363) | 0.49 (1,013) | 0.32 (316) | 0.28 (4,692) |
| S | 0.20 (2,958) | 0.49 (610) | 0.27 (365) | 0.25 (3,933) |
| M | 0.09 (3,160) | 0.10 (980) | 0.00 (348) | 0.09 (4,488) |
| Total | 0.24 (12,718) | 0.29 (3,144) | 0.42 (1,188) | 0.26 (17,050) |
| PCR confirmed | ||||
| V | 0.12 | 0.00 | 1.26 | 0.15 |
| R | 0.12 | 0.30 | 0.32 | 0.17 |
| S | 0.17 | 0.00 | 0.27 | 0.15 |
| M | 0.03 | 0.00 | 0.00 | 0.02 |
| Total | 0.11 | 0.10 | 0.25 | 0.12 |
Results represent percentage of fecal pats positive for each type of pen (home, hospital, and buller), the pen type total percent, and the total percent for each feedlot for all isolates and for PCR-confirmed isolates. Multiple picks from a single sample were counted once. Sample sizes for PCR-confirmed isolates were identical to sample sizes for all isolates.
At a collection when E. coli O157:H7 was isolated, it was found in fewer than four samples at a yard or in a pen. E. coli O157:H7 was found in at least one feedlot at 14 of 16 collections (Table 1). It was isolated from significantly fewer collections at feedlot M (18.8%) than from the other feedlots (V, 56.3%; S, 37.5%; and R, 37.5%). Rarely was E. coli O157:H7 found from consecutive collections at a single feedlot, but it was isolated from five consecutive collections at feedlot V.
For isolates from fecal pat samples, no significant differences were detected among pen types, although the trend was that prevalence overall was higher in hospital and buller pens than in home pens (Table 2). Three home pens were positive twice, but one of these housed different animals at each of the two collections. One buller pen had E. coli O157:H7 in three collections, two of which were about 126 days apart, a sufficient length of time for the pen to contain different animals. No hospital pens tested positive twice. Only in one pen (a hospital pen) was E. coli O157:H7 found in more than one composite fecal sample on a sample collection day.
In home pens, more positive samples were found in spring (15 samples) and summer (14 samples) than in fall (8 samples) or winter (8 samples). In hospital pens, all positive samples except one were collected between January and April. For buller pens, positive samples were collected in spring, summer, and winter.
Antibiotic susceptibility.
Some isolates were resistant to antibiotics not approved for feedlot animals (Table 3). Resistant isolates were found for six of the eight (86%) antibiotics that could be used to treat E. coli infections in feedlot animals. All 57 isolates had intermediate resistance to the other two approved antibiotics. All isolates were resistant to tilmicosin, an antibiotic that is used in feed at many cattle feedlots. All but one isolate was susceptible to gentamicin that has been used for cattle. However, its use in feedlot cattle is now prohibited in the United States, and there is a voluntary moratorium on its extra-label use in cattle. Most isolates were susceptible to other antibiotics used at feedlots, such as trimethoprim-sulfamethoxazole and ceftiofur. All isolates were susceptible to ciprofloxacin, an antibiotic used widely in human medicine and similar to enrofloxacin, which recently has been approved for use in cattle. All isolates were resistant to antibiotics that do not target E. coli, except tylosin, to which all 57 isolates had intermediate susceptibility.
TABLE 3.
Antimicrobial susceptibility of all isolates and PCR-confirmed E. coli O157:H7 isolatesa
| Antibiotic (concn [μg/ml]) | No. of isolates with susceptibility
|
|||||
|---|---|---|---|---|---|---|
| All (n = 57)
|
PCR confirmed (n = 26)
|
|||||
| S | I | R | S | I | R | |
| Not approved for feedlot animals | ||||||
| E. coli targeting | ||||||
| Imipenem (4, 8) | 57 | 0 | 0 | 26 | 0 | 0 |
| Ciprofloxacin (1, 2) | 57 | 0 | 0 | 26 | 0 | 0 |
| Norfloxacin (4, 8) | 57 | 0 | 0 | 26 | 0 | 0 |
| Nitrofurantoin (32) | 57 | 0 | 0 | 26 | 0 | 0 |
| Gentamicin (4, 8) | 56 | 1 | 0 | 26 | 0 | 0 |
| Cefazolin (8, 16) | 48 | 0 | 9 | 24 | 0 | 2 |
| Cephalothin (8, 16) | 33 | 14 | 10 | 18 | 5 | 3 |
| Non-E. coli targeting | ||||||
| Oxacillin (2) | 0 | 0 | 57 | 0 | 0 | 26 |
| Vancomycin (4, 16) | 0 | 0 | 57 | 0 | 0 | 26 |
| Approved for feedlot animals | ||||||
| E. coli targeting | ||||||
| Trimethoprim-sulfamethoxazole (2/38) | 53 | 0 | 4 | 26 | 0 | 0 |
| Ceftiofur (0.015–32)b | 50 | 0 | 7 | 22 | 0 | 4 |
| Ampicillin (8) (0.015–32)b | 47 (48) | 0 | 10 (9) | 24 (21) | 0 | 2 (5) |
| Tetracycline (4, 8) (0.015–32)b | 31 (39) | 3 (0) | 23 (18) | 15 (18) | 1 (0) | 10 (8) |
| Amoxicillin/clavulanic acid (4/2) | 30 | 0 | 27 | 16 | 0 | 10 |
| Florfenicol (0.015–32)b | 0 | 57 | 0 | 0 | 26 | 0 |
| Spectinomycin (0.25–32)b | 0 | 57 | 0 | 0 | 26 | 0 |
| Rifampin (1, 2) | 0 | 2 | 55 | 0 | 1 | 25 |
| Non-E. coli targeting | ||||||
| Tylosin (0.312–20)b | 0 | 57 | 0 | 0 | 26 | 0 |
| Penicillin (0.03, 0.12, 8) | 0 | 0 | 57 | 0 | 0 | 26 |
| Clindamycin (0.5, 2) | 0 | 0 | 57 | 0 | 0 | 26 |
| Erythromycin (0.5, 4) (0.015–32)b | 0 | 0 | 57 | 0 | 0 | 26 |
| Tilmicosin (0.015–32)b | 0 | 0 | 57 | 0 | 0 | 26 |
Analysis includes multiple isolates from a single sample. S, susceptible; I, intermediate susceptibility; R, resistant. Concentrations of antibiotics are in parentheses after the antibiotic name and are expressed in micrograms per milliliter when reconstituted in 50 μl of broth. If there are two sets of antibiotic concentrations, the second set is for the broth microdilution method. When both methods were used and the number of isolates differed between the methods, the number of isolates in parentheses was determined by the broth microdilution method.
Susceptibility determined by broth microdilution instead of the breakpoint methods.
Isolates with multiple resistance.
Most isolates were not multiresistant (Table 4). Some isolates with multiple resistance were resistant to antibiotics that are not approved for use in feedlot animals. Of the isolates that were resistant to five or more antibiotics, four were from hospital pens, three were from home pens, and one was from a buller pen; none were from the same sample. Three of the eight multiply-resistant isolates were from one collection time, but from two feedlots. All were collected in spring from three feedlots.
TABLE 4.
Percentage of E. coli O157:H7 isolates not resistant or resistant to one or multiple antibiotics for all isolates and PCR-confirmed isolatesa
| No. of antibiotics | % of isolates resistant
|
|||
|---|---|---|---|---|
| All (n = 57)
|
PCR confirmed (n = 26)
|
|||
| Microdilution | Breakpoint | Microdilution | Breakpoint | |
| 0 | 63.2 | 1.8 | 65.4 | 3.8 |
| 1 | 24.6 | 35.1 | 19.2 | 42.3 |
| 2 | 0 | 29.8 | 0 | 26.9 |
| 3 | 12.3 | 17.5 | 7.0 | 19.2 |
| 4 | 0 | 1.8 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 |
| 6 | 0 | 8.8 | 0 | 7.7 |
| 7 | 0 | 3.5 | 0 | 0 |
| 8 | 0 | 1.8 | 0 | 0 |
Results represent the percentage of isolates not resistant or resistant in terms of all isolates (n = 57) and PCR-confirmed isolates (n = 26). The analysis includes multiple isolates from a single sample. Susceptibility testing for 8 of the 22 antibiotics was done by the broth microdilution method. Antibiotics that did not target E. coli (Table 3) were omitted from the analysis.
RAPD profiles of total DNA.
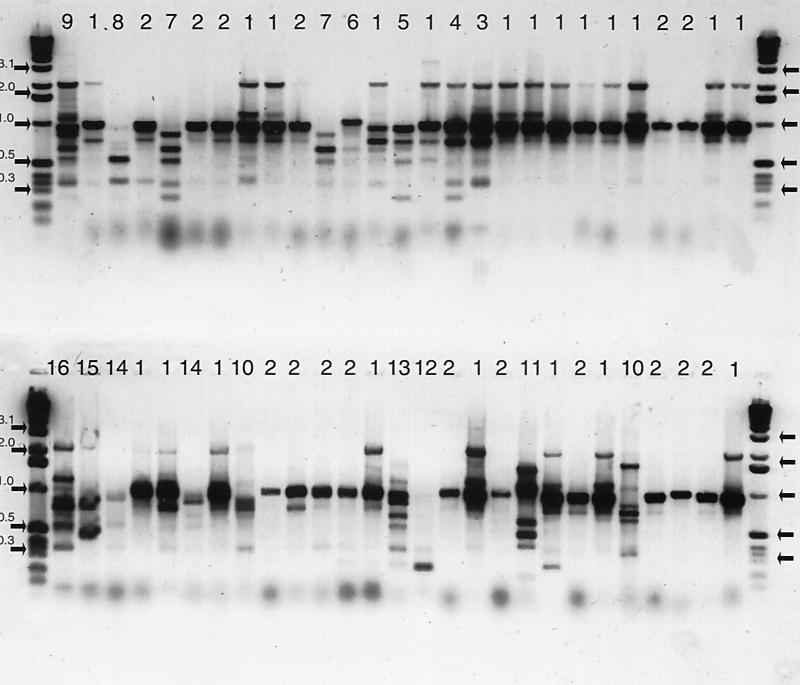
Of the 57 isolates, 54 were characterized by RAPD profiles of total DNA, and 16 unique patterns were identified (Fig. 1 and Table 1). Most isolates were either RAPD pattern 1 or RAPD pattern 2 (Table 5) and were found at all four feedlots. The remaining 14 RAPD patterns were identified only once. Clusters of unique RAPD patterns distinguished the feedlots from one another.
FIG. 1.
RAPD profiles of total DNA (negative image of horizontal gel electrophoreses) (primer 1254) of 54 of 57 E. coli O157: H7 isolates from fecal pats collected at four southwest Kansas beef cattle feedlots collected every 3 weeks for 11 months. Lanes that share the same number were judged to be identical. The number above each lane is the RAPD pattern number. The first and last lanes of each row are a 1-kb ladder standard with key fragment sizes indicated in kilobases and marked with arrows.
TABLE 5.
Number of E. coli O157:H7 isolates with each RAPD pattern of total DNA at southwest Kansas beef cattle feedlots
| RAPD pattern | No. of isolates with pattern
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All
|
PCR confirmed
|
|||||||||
| S | R | V | M | Total | S | R | V | M | Total | |
| 1 | 10 | 5 | 4 | 2 | 21 | 8 | 3 | 2 | 1 | 14 |
| 2 | 6 | 10 | 16 | 4 | 5 | 9 | ||||
| 3 | 1 | 1 | 1 | 1 | ||||||
| 4 | 1 | 1 | ||||||||
| 5 | 1 | 1 | ||||||||
| 6 | 1 | 1 | ||||||||
| 7 | 1 | 1 | 2 | |||||||
| 8 | 1 | 1 | ||||||||
| 9 | 1 | 1 | ||||||||
| 10 | 1 | 1 | 2 | |||||||
| 11 | 1 | 1 | 1 | 1 | ||||||
| 12 | 1 | 1 | ||||||||
| 13 | 1 | 1 | ||||||||
| 14 | 1 | 1 | 2 | 1 | 1 | |||||
| 15 | 1 | 1 | ||||||||
| 16 | 1 | 1 | ||||||||
| Total no. of patterns | 7 | 7 | 6 | 3 | 2 | 4 | 2 | 1 | ||
| Total no. of isolates | 16 | 16 | 18 | 4 | 54 | 9 | 9 | 7 | 1 | 26 |
Results represent four southwest Kansas beef cattle feedlots (S, R, V, and M) and were obtained from all isolates (n = 54) and PCR-confirmed isolates (n = 26). The analysis includes multiple isolates from a single sample. A blank space indicates the pattern was not identified at the feedlot.
At feedlot V, where E. coli O157:H7 was isolated from five consecutive collections, three isolates had the same RAPD pattern. Two of the three isolates that had the same pattern were collected from the same pen 3 weeks apart. This was the only pen at any feedlot where isolates from multiple collections had the same RAPD pattern (Table 1 and Fig. 1).
Multiple isolates from the same sample rarely had the same RAPD pattern. Of the nine samples with multiple picks (Table 1), only two had the same RAPD pattern (Fig. 1).
RAPD profiles of plasmid DNA.
Similar diversity (16 patterns) was found in the RAPD profiles of plasmid DNA isolated from the 54 E. coli O157:H7 isolates tested (Table 1).
Virulence genes.
The PCR showed that more than half the isolates did not have the virulence genes stx1, stx2, eaeA, and hlyA (Table 1). Of the 26 isolates that had at least one of the stx genes, 38.5% had both genes, and 57.7% had only stx2. One isolate (3.8%) had only stx1. All 26 isolates contained both the eaeA gene and the hlyA gene.
Overall diversity.
When all combinations of virulence factors, DNA patterns, and antibiotic resistance were considered, even greater diversity was found among the isolates. From samples in which two isolates were picked, neither isolate had the same RAPD patterns of total DNA and plasmid DNA. One sample had five isolates that were characterized. Two of these had the same RAPD patterns of total DNA and plasmid DNA and the same virulence genes, but were different in their antibiotic profiles. When all measures that were used to characterize the diversity of the isolates were considered, all but 8 of the 57 isolates were unique.
Comparison of all isolates with PCR-confirmed isolates.
We found that 31 of the 57 isolates did not have either the eaeA gene or one or both of the stx genes. This afforded the opportunity to compare non-type-specific E. coli with E. coli O157:H7. The salient difference between the two confirmation methods was a general halving of the culture prevalence rates.
When only the PCR-confirmed isolates were considered, overall prevalence was reduced to 0.08%. One isolate from the single positive (by C/LA) water sample was confirmed by PCR. The trend of hospital pens having a higher prevalence than home pens was not detectable when only PCR-confirmed isolates were analyzed (Table 2). The frequency of finding at least one positive feedlot at a collection dropped from 14 to 9 of 16 collections. Although feedlot prevalence was lower at each feedlot, the feedlot prevalence of PCR-confirmed E. coli O157:H7 was still lower at feedlot M (6.3%) than at the other feedlots (V, 18.8%; R, 18.8%; and S, 37.5%). Only at feedlot S was E. coli O157:H7 found from consecutive collections, where it was isolated from two consecutive collections 3 weeks apart.
Although the number was fewer, the PCR-confirmed isolates were resistant to the same antibiotics as all isolates (Table 3). Only two of the nine colonies that were resistant to five or more antibiotics had the eaeA gene (Table 4).
When only the 26 isolates with the eaeA gene were considered (Table 1), five RAPD patterns of total DNA were identified (Table 5). These PCR-confirmed isolates included multiple picks from a total of 20 samples. Of those samples with multiple isolates, none had the same combinations of RAPD patterns, virulence genes, or antimicrobial resistance pattern.
DISCUSSION
Prevalence.
Prevalence of E. coli O157:H7 was low at southwest Kansas cattle feedlots sampled intensively over time. Had only one isolate instead of up to five been selected from each culture plate, the prevalence would have been considerably lower, illustrating the importance of selecting multiple isolates from a plate. Improved methodology, such as immunomagnetic separation (5), might have increased our prevalence estimates, but it is costly and time-consuming and increases sensitivity only when the organism is present in very low concentrations. However, overestimation of prevalence (false positives) can result from using current methods such as polyclonal LA.
Others have reported similar low prevalence rates in cattle (8, 10, 11, 25, 28), but higher prevalence rates have been reported recently by Keen and Elder (16). That study used a new antigen capture test to screen multiple isolates from a sample before further testing, effectively increasing the probability of selecting a positive isolate.
Antibiotic susceptibility.
We expected to observe a difference in the antibiograms of isolates collected from home pens versus hospital pens, where convalescent animals had been treated recently with antibiotics. Because we had so few isolates, we were not able to detect a difference statistically.
We expected that most isolates would be resistant to antibiotics that are used heavily at feedlots, but that was not the case. For example, less than half of the isolates were resistant to tetracycline, which was one of the most heavily used antibiotics on feedlots. No isolate was susceptible to newer drugs (e.g., florfenicol, spectinomycin, and rifampin) that are used at feedlots. Prudent use of these newer antibiotics will be necessary to retain their efficacy and for resistant bacterial populations to revert to a susceptible phenotype.
We expected isolates from cattle feedlots to be susceptible to antibiotics not approved for feedlot animals. However, some isolates were resistant to cefazolin and cephalothin. Perhaps the organisms had acquired resistance from other organisms, were coselected through genetic linkage of resistance factors, or were resistant to antibiotics in the same class. The resistant organisms also could have been of human origin or could be naturally resistant to the antibiotics.
When reporting resistance patterns, researchers need to consider whether the antibiotic targets the organism. If it does not, resistance would not be associated with use or misuse of the antibiotic. Current concern about the increase of antibiotic resistance can be mitigated partially when the antimicrobial agent-bacterium combination is considered. For example, the data reported here could be interpreted to mean that E. coli O157:H7 is a public health concern because it is highly resistant to antibiotics such as vancomycin. Because this antibiotic does not target this organism and never would be used as treatment for an E. coli O157:H7 infection, this interpretation is incorrect. Before broad statements are made about an increase in antibiotic resistance because of antibiotic use, the antibiotics considered should target the organism, and evidence should be provided that use of antibiotics has caused this increase in antibiotic resistance.
The combination of resistance and whether the antibiotic targets the organism also must be addressed when reporting that organisms are multidrug resistant. Isolates in this study with multiple resistances were resistant to all but two antibiotics (cefazolin and gentamicin) that target E. coli and are approved for use in feedlot animals.
Why are isolates resistant when cattle are not treated for an E. coli infection? Even though antibiotic treatment is not used for E. coli O157:H7 infections in humans or food animals, we found that a third of the isolates were resistant to one or more antibiotics. One likely explanation for this resistance is the relative ease with which resistance factors are exchanged among promiscuous bacteria (21). Another possibility is that cattle are being treated with antibiotics for other conditions, thereby selecting for resistant populations of E. coli.
Genetic diversity.
The surprisingly high degree of genetic diversity indicates that E. coli O157:H7 probably was brought into these feedlots on the hide or in the feces of incoming cattle. Although some RAPD patterns of total DNA were unique to a feedlot, their infrequent occurrence does not support the hypothesis that strains are resident, but rather indicates that they were unique to incoming cattle. Some patterns were present in isolates at all feedlots and occurred with higher frequency, suggesting that they were resident strains. However, because of our sampling intensity, if they were resident strains, they would have been recovered at more collections. Isolates with common RAPD patterns of total DNA did not have the same RAPD profiles of plasmid DNA or virulence markers, further indicating the diversity of the organism and that the isolates were not resident strains.
The high diversity observed may make epidemiological traceback investigations of human infections to feedlots difficult. The organism's rare and sporadic occurrence makes it a poor choice as a sentinel organism for HACCP monitoring programs.
Virulence genes.
The presence of virulence markers has been used extensively in hybridization and PCR-based methods of detecting E. coli O157:H7. Some investigators have reported differences in virulence between E. coli O157:H7 strains recovered from humans and strains isolated from animals (18). If not all virulence markers are tested, then the molecular methods of detection might fail to identify the organism or to distinguish diversity among strains. Production of Shiga toxin may be higher in isolates derived from hospital pens in our study, and this testing is under way. Studies that do not test for virulence markers may overreport E. coli O157:H7 prevalence. For instance, commonly used polyclonal antibody tests may result in false positives (19). Conversely, some detection methods that require an isolate may underreport E. coli O157:H7 prevalence because of a lack of sensitivity or specificity.
Comparison of all isolates with PCR-confirmed isolates.
Research laboratories typically have used C/LA as the means of identifying E. coli O157:H7 when an isolate is required for further characterization, such as antibiotic susceptibility testing. The results presented here show that this methodology results in more than 50% false positives. Even when the same isolates are compared by two different PCR methods, different results can occur. The results from our PCR-confirmed isolates do not agree precisely (82% agreement) with those for the same isolates tested by Kimura et al. (19). Possible reasons for this discrepancy are the different primers used for each test, the difference in test types, and the laboratory-to-laboratory variation. These results illustrate the variability that can occur in results when different methods and laboratories are used to identify and characterize E. coli O157:H7. Nevertheless, genetic confirmation of isolates is highly recommended to increase the accuracy of all future prevalence studies.
ACKNOWLEDGMENTS
United States Department of Agriculture Cooperative State Research Education, and Extension Service grant 95–37201-2127, Food and Drug Administration grant FD-U-001574, and National Institutes of Health grant AI 39067 supported this work.
We thank Lori Helmle, Christy Davison, and Matthew Selee for valuable field collection support. We also thank Mike Hornback, Aaron Carman, David Stuever, Dusty Woods, Thao Ngo, Ramona Chitrakar, and Christa Irwin for skillful technical assistance.
Footnotes
This is contribution no. 01-108-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Baker D R, Moxley R A, Francis D H. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv Exp Med Biol. 1997;412:53–58. doi: 10.1007/978-1-4899-1828-4_6. [DOI] [PubMed] [Google Scholar]
- 2.Beebakhee G, Louie M, De Azavedo J, Brunton J. Cloning and nucleotide sequence of the eae gene homologue from enterohemorrhagic Escherichia coli serotype O157:H7. FEMS Microbiol Lett. 1992;91:63–68. doi: 10.1016/0378-1097(92)90563-4. [DOI] [PubMed] [Google Scholar]
- 3.Beutin L, Montenegro M A, Orskov I, Orskov F, Prada J, Zimmermann S, Stephan R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J Clin Microbiol. 1989;27:2559–2564. doi: 10.1128/jcm.27.11.2559-2564.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birch M, Denning D W, Law D. Rapid genotyping of Escherichia coli O157 isolates by random amplification of polymorphic DNA. Eur J Clin Microbiol Infect Dis. 1996;15:297–302. doi: 10.1007/BF01695661. [DOI] [PubMed] [Google Scholar]
- 5.Chapman P A, Cerdan Malo A T, Siddons C A, Harkin M. Use of commercial enzyme immunoassays and immunomagnetic separation systems for detecting Escherichia coli O157 in bovine fecal samples. Appl Environ Microbiol. 1997;63:2549–2553. doi: 10.1128/aem.63.7.2549-2553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenber M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin P M, Bell B P, Cieslak P R, Tuttle J, Barrett T J, Doyle M P, McNamara A M, Shefer A M, Wells J G. Large outbreak of Escherichia coli O157:H7 infections in the western United States: the big picture. In: Karmali MA, Goglio AG, editors. Recent advances in verocytotoxin-producing Escherichia coli infections. New York, N.Y: Elsevier; 1994. pp. 7–12. [Google Scholar]
- 8.Hancock D D, Besser T E, Kinsel M L, Tarr P I, Rice D H, Paros M A. The prevalence of Escherichia coli O157 in dairy and beef cattle in Washington state. Epidemiol Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock D D, Besser T E, Rice D H. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices. In: Kaper JB, O'Brien AD, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: American Society for Microbiology; 1998. pp. 85–91. [Google Scholar]
- 10.Hancock D D, Rice D H, Thomas L A, Dargatz D A, Besser T E. Epidemiology of Escherichia coli O157:H7 in feedlot cattle. J Food Prot. 1997;60:462–465. doi: 10.4315/0362-028X-60.5.462. [DOI] [PubMed] [Google Scholar]
- 11.Hancock D D, Besser T E, Rice D H, Herriott D E, Tarr P I. Longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol Infect. 1997;118:193–195. doi: 10.1017/s0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis K G, Kaper J B. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun. 1996;64:4826–4829. doi: 10.1128/iai.64.11.4826-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karch, Stockbine H N, O'Brien A. Growth of Escherichia coli in the presence of trimethoprim-sulfamethoxazole facilitates detection of Shiga-like toxin producing strains by colony blot assay. FEMS Microbiol Lett. 1986;35:141–145. [Google Scholar]
- 14.Karmali M A, Steele B T, Petric M, Lim C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet. 1983;i:619–620. doi: 10.1016/s0140-6736(83)91795-6. [DOI] [PubMed] [Google Scholar]
- 15.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 16.Keen J E, Elder R O. Proceedings of the 9th Symposium of the International Society for Veterinary Epidemiology and Economics. Breckenridge, Colo. 2000. High but variable enterohemorrhagic E. coli (EHEC) O157 fecal shedding in pens of slaughter-ready Kansas and Nebraska beef feedlot cattle; p. 453. [Google Scholar]
- 17.Kim H H, Samadpour M, Grimm L, Clausen C R, Besser T E, Baylor M, Kobayashi J M, Neill M A, Shoenknecht F D, Tarr P I. Characteristics of antibiotic-resistant Escherichia coli O157:H7 in Washington State, 1984–1991. J Infect Dis. 1994;170:1606–1609. doi: 10.1093/infdis/170.6.1606. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Nietfeldt J, Benson A K. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA. 1999;96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura R, Mandrell R E, Galland J C, Hyatt D R, Riley L W. Restriction-site-specific PCR as a rapid test to detect enterohemorrhagic E. coli O157:H7 strains in environmental samples. Appl Environ Microbiol. 2000;66:2513–2519. doi: 10.1128/aem.66.6.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudva I T, Hunt C W, Williams C J, Nance U M, Hovde C J. Evaluation of dietary influences on Escherichia coli O157:H7 shedding by sheep. Appl Environ Microbiol. 1997;63:3878–3886. doi: 10.1128/aem.63.10.3878-3886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 22.Madico G, Akopyants N S, Berg D E. Arbitrarily primed PCR DNA fingerprinting of Escherichia coli O157:H7 strains by using templates from boiled cultures. J Clin Microbiol. 1995;33:1534–1536. doi: 10.1128/jcm.33.6.1534-1536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1996. [Google Scholar]
- 24.Pacheco A B F, Guth B E C, Sores K C C, de Almeida D F, Ferreira L C S. Clonal relationships among Escherichia coli serogroup O6 isolates based on RAPD. FEMS Microbiol Lett. 1997;148:255–260. doi: 10.1111/j.1574-6968.1997.tb10297.x. [DOI] [PubMed] [Google Scholar]
- 25.Rahn K, Renwich S A, Johnson R P, Wilson J B, Clark R C, Alves D, McEwen S, Lior H, Spika J. Persistence of Escherichia coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiol Infect. 1997;119:251–259. doi: 10.1017/s0950268897007929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SAS Institute, Inc. SAS/STAT user's guide, release 6.03 edition. Cary, N.C: SAS Institute, Inc.; 1988. [Google Scholar]
- 27.Shere J A, Bartlett K J, Kaspar C W. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl Environ Microbiol. 1998;64:1390–1399. doi: 10.1128/aem.64.4.1390-1399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilden J, Young W, McNamara A, Custer C, Boesel B, Lambert-Fair M, Majokowski J, Vugia D, Werner S B, Hollingsworth J, Morris J G. A new route of transmission for Escherichia coli: infection from dry fermented salami. Am J Public Health. 1996;86:1142–1145. doi: 10.2105/ajph.86.8_pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tollefson L. FDA reveals plans for antimicrobial susceptibility monitoring. J Am Vet Med Assoc. 1996;208:459. [PubMed] [Google Scholar]
- 30.Tollefson L, Fedorka-Cray P J, Angulo F J. Public health aspects of antibiotic resistance monitoring in the USA. Acta Vet Scand Suppl. 1999;92:67–75. [PubMed] [Google Scholar]
- 31.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterspiel J, Ashkenazi S, Morrow A, Cleary T G. Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin 1. Infection. 1992;20:25–29. doi: 10.1007/BF01704889. [DOI] [PubMed] [Google Scholar]
- 33.Wong C S, Jelacic S, Habeeb R L, Watkins S L, Tarr P I. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342:1930–1936. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, McDaniel A D, Wolf L E, Keusch G T, Waldor M K, Acheson D W. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis. 2000;181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]