Abstract
The underlying mechanisms by which severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) leads to acute and long-term neurological manifestations remains obscure. We aimed to characterize the neuropathological changes in patients with coronavirus disease 2019 and determine the underlying pathophysiological mechanisms.
In this autopsy study of the brain, we characterized the vascular pathology, the neuroinflammatory changes and cellular and humoral immune responses by immunohistochemistry.
All patients died during the first wave of the pandemic from March to July 2020. All patients were adults who died after a short duration of the infection, some had died suddenly with minimal respiratory involvement. Infection with SARS-CoV-2 was confirmed on ante-mortem or post-mortem testing. Descriptive analysis of the pathological changes and quantitative analyses of the infiltrates and vascular changes were performed.
All patients had multifocal vascular damage as determined by leakage of serum proteins into the brain parenchyma. This was accompanied by widespread endothelial cell activation. Platelet aggregates and microthrombi were found adherent to the endothelial cells along vascular lumina. Immune complexes with activation of the classical complement pathway were found on the endothelial cells and platelets. Perivascular infiltrates consisted of predominantly macrophages and some CD8+ T cells. Only rare CD4+ T cells and CD20+ B cells were present. Astrogliosis was also prominent in the perivascular regions. Microglial nodules were predominant in the hindbrain, which were associated with focal neuronal loss and neuronophagia.
Antibody-mediated cytotoxicity directed against the endothelial cells is the most likely initiating event that leads to vascular leakage, platelet aggregation, neuroinflammation and neuronal injury. Therapeutic modalities directed against immune complexes should be considered.
Keywords: COVID-19, SARS-CoV-2, neurovascular injury, complement deposition, neuroinflammation
Lee et al. examine microvascular pathology in the brains of patients who died from COVID-19. They show that antibody-mediated cytotoxicity directed against brain endothelial cells is likely to be the initiating event that leads to vascular leakage, platelet aggregation, neuroinflammation and neuronal injury.
Introduction
Patients with severe acute respiratory syndrome with coronavirus-2 (SARS-CoV-2) infection can develop a wide variety of neurological complications. In severely ill patients, an encephalopathy may occur and has been attributed to hypoxia or multi-organ dysfunction although the pathophysiology remains unclear.1,2 Some may develop ischaemic or haemorrhagic strokes as well as a variety of post-viral immune-mediated syndromes.3,4 Encephalitis is rare; however, acute haemorrhagic encephalomyelitis5 or an acute disseminated encephalomyelitis including transverse myelitis have been reported.6,7 Microvascular pathology may occur in the brain and other organs; however, the underlying mechanisms are unknown.8 It remains unclear whether viral infection of brain cells can develop in these patients, or whether these syndromes are secondary to immune-mediated phenomena. SARS-CoV-2 has rarely been found in the CSF of patients with CNS symptoms,9,10 and autopsy studies have either failed to find the virus or the virus has been found in the brain at only low copy numbers without associated inflammation, which cannot explain the widespread pathology.11–14 Additionally, many patients complain of persistent symptoms of cognitive difficulties, extreme fatigue, sleep and autonomic dysfunction lasting several months after recovery from the acute infection suggesting a post-viral CNS syndrome that has been termed long-COVID or post-acute sequelae of SARS-CoV-2 infection, which resembles myalgic encephalomyelitis/chronic fatigue syndrome.15
To explore these possibilities, we examined autopsy brain tissue from patients who had died with coronavirus disease-19 (COVID-19) and performed detailed virological and immunohistopathological analysis.
Materials and methods
Patients
Nine patients (seven males and two females; age 24–73 years) were studied who had died during the first wave of the pandemic (March to July 2020). Co-morbidities included diabetes (n = 2, 22%), hypertension (n = 1, 11%), hypertension and diabetes (n = 1, 11%) and substance use disorder (n = 2, 22%). Five patients (56%) had died suddenly. Four (44%) were found dead at home, one (11%) in a subway. The remaining patients died within days to weeks after onset of symptoms. All patients had evidence of lung involvement at time of autopsy but only one patient required ICU admission. All patients had nasal swabs that were polymerase chain reaction positive for SARS-CoV-2 either before or after death. Preliminary findings from these patients were previously published.16 Patients in this series represent a subset of patients who showed microvascular abnormalities on post-mortem MRI. The control group consisted of nine males and one female; age 43–74 years. They were diagnosed with pulmonary infections (n = 2, 20%), systemic infections (n = 2, 20%), hypertension (n = 4, 40%), bipolar disorder (n = 1, 10%) and substance use disorder (n = 1, 10%). There was no statistically significant difference in demographic characteristics between groups. Fixation duration in 10% neutral buffered formalin was 2–7 weeks in both groups. Characteristics of COVID- 19 patients and non-COVID-19 controls are shown in Supplementary Table 1.
Immunohistochemistry
Chromogenic immunohistochemistry
Immunohistochemical analysis was performed on autopsied brain tissues including the olfactory bulb, frontal and temporal lobes, basal ganglia, hippocampus, thalamus, midbrain, pons, medulla oblongata and cerebellum (Supplementary Table 2). Brains were dissected in the coronal plane and selected regions were fixed in 10% neutral buffered formalin. The samples were then embedded in paraffin, cut into 5-μm thick sections and mounted on SuperFrost® Plus (Thermo Fisher Scientific). The sections were processed as follows: the slides were deparaffinized and rehydrated using xylene and graded ethanol. For general morphological evaluation, haematoxylin and eosin staining was performed. Immunohistochemical staining was performed either on a Leica Bond-MAX automated slide stainer (Leica Biosystems) using protocols from the manufacturer or manually. For manual staining, antigen retrieval was done by heat-induced antigen retrieval methods in 10 mM citrate buffer at pH 6.0 or 10 mM Tris-EDTA buffer at pH 9.0; or enzyme-induced antigen retrieval method with proteinase K. Peroxidase was blocked by incubating tissues in 0.3 to 3.0% hydrogen peroxide for 10 min at room temperature and protein blocking was done with protein block solution (Agilent) for 30 min at room temperature. Sections were incubated with primary antibodies overnight at room temperature. After washing with 1× Tris-buffered saline containing 0.05% Triton X-100, sections were incubated with PowerVision polymeric horseradish peroxidase anti-mouse IgG (Leica Biosystems) or PowerVision poly-HRP anti-rabbit IgG for 2 h at room temperature and developed with 3,3′-diaminobenzidine (DAB; Vector Laboratories). Sections were counterstained with haematoxylin (Agilent). Antibodies used for immunohistochemistry are listed in Supplementary Table 3.
Immunohistochemistry image analysis
Protein levels were determined by immunohistochemistry. Immunostained slides were scanned using a slide scanner (Aperio AT2, Leica Biosystems) and random images per group were captured from the whole-slide images scanned and quantified in a blinded manner using unbiased analysis software. The extent of fibrinogen deposition was quantified by mapping areas of strong and weak immunostaining using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Then, the percentage per total area was calculated. Data are expressed as percentage of fibrinogen deposition over the total area. The image analysing steps are illustrated in Supplementary Fig. 4.
For quantification of CD61 platelet aggregates, the number of vessels with CD61+ platelet aggregates was evaluated using Aperio ImageScope software (Leica Biosystems). Then, 15–20 regions of each stained brain section were randomly selected. Next, CD61+ vessels were counted in each region. Each area was 1.2 mm2. Data are expressed as the number of vessels with CD61+ platelet aggregates per mm2.
Platelet endothelial cell adhesion molecule 1 (PECAM-1) levels were assessed using ImageScope software (Leica Biosystems). From each stained brain section, 15–30 small blood vessels seen in cross section per 10 selected regions were selected. The area of the entire vessel and lumen area were measured by tracing the contours of the entire vessel and lumen within the vessel. Area of PECAM-1 immunostaining was determined by subtracting luminal area from entire vessel area. To exclude the effect of vessel size, the PECAM-1 area was normalized by dividing it by the blood vessel area. Data are presented as mean areas of PECAM-1.
Quantification of CD68, CD3, CD4, CD8, CD20 and TMEM-119+ cells was performed using Gen5 Image Prime software (BioTek, Winooski, VT, USA). Fifty regions of each stained brain section were randomly selected, and the number of positive cells was counted. Each area was 0.4 mm2. Values represent the average number of cells per area (cells/mm2). Foci of microglia (immunostaining for CD68) that were surrounding the neurons were counted and considered to represent foci of neuronophagia. Data are expressed the average number of foci of neuronophagia per area (n/mm2).
Multiplex fluorescence immunohistochemistry
Multiplex fluorescence immunohistochemistry was performed on 5-µm-thick formalin-fixed paraffin-embedded sections that were deparaffinized. Antigen was unmasked using the heat-induced antigen retrieval method. The sections were incubated with Human BD Fc Blocking solution (BD Biosciences) and then incubated in True Black Reagent (Biotium) to quench tissue autofluorescence. The sections were immunoreacted for 1 h at room temperature using 1–5 μg/ml cocktail mixtures of immunocompatible antibodies (Supplementary Table 3), followed by washing in PBS with 1 mg/ml bovine serum albumin and staining the sections for 1 h at room temperature using a 1 µg/ml cocktail mixture of appropriately cross-adsorbed secondary antibodies (Thermo Fisher; Jackson ImmunoResearch; Li-Cor Biosciences) conjugated to one of the following spectrally compatible fluorophores: Alexa Fluor 430, Alexa Fluor 488, Alexa Fluor 546, Alexa Fluor 594, Alexa Fluor 647, IRDye 680LT or IRDye 800CW. After washing, the sections were counterstained using 1 µg/ml DAPI (Thermo Fisher) for visualization of cell nuclei. Slides were then coverslipped and imaged using a multispectral epifluorescence microscope.
Multiplex fluorescence image acquisition
Images were acquired from whole specimen sections using the Axio Imager.Z2 slide scanning fluorescence microscope (Carl Zeiss AG) equipped with a ×20/0.8 Plan-Apochromat (Phase-2) objective (Carl Zeiss AG), an ORCA-Flash4.0 sCMOS digital camera (Hamamatsu), a 200W X-Cite 200DC broad band lamp source (Excelitas Technologies) and eight customized filter sets (Semrock) to detect the following fluorophores: DAPI, Alexa Fluor 430, Alexa Fluor 488, Alexa Fluor 546, Alexa Fluor 594, Alexa Fluor 647, IRDye 680LT and IRDye 800CW. Image tiles (600 × 600 µm viewing area) were individually captured at 0.325 µm/pixel spatial resolution, and the tiles seamlessly stitched into whole specimen images using the ZEN 2 image acquisition and analysis program (Carl Zeiss AG). A colour table was applied to each image channel to either match its emission spectrum or to set a distinguishing colour balance. Pseudo-coloured stitched images were exported to Adobe Photoshop, spatially aligned at a pixel level of resolution, and overlaid as individual layers to create multicoloured merged composites.
RNAscope
RNAscope in situ hybridization was performed according to the manufacturer’s instructions (Advanced Cell Diagnostics). In brief, formalin-fixed, paraffin-embedded section slides were dried for 1 h at 60°C, deparaffinized in fresh xylene and fresh 100% ethanol. RNAscope® hydrogen peroxide was added to the sections and incubate for 10 min at room temperature. For purposes of antigen retrieval, the slides were boiled in RNAscope® 1× Target Retrieval Reagent for 15 min and then incubated in RNAscope® Protease Plus for 30 min at 40°C. After washing, the slides were covered with drops of probes targeting specific genes that are interested and incubated for 2 h at 40°C. After hybridizing with the probe, the signals were amplified sequentially with amplifiers and labelled with a label probe using the 2.5 HD Detection Kit (as per the manufacturer’s procedure), at 40°C or room temperature. To reveal the signal, slides were incubated in a red working solution for 10 min at room temperature. Slides were then counterstained with 50% haematoxylin and mounted with EcoMount mounting medium. Images were captured using a whole-slide scanner (Aperio AT2). Probes used in RNAscope in situ hybridization are listed in Supplementary Table 4.
Digital spatial profiling data generation
Gene expression profiles in the brainstem of three COVID-19 patients and two non-COVID-19 controls were investigated using the NanoString GeoMX™ digital spatial profiling (DSP) platform with the whole Transcriptome Atlas for 18 704 human transcripts (NanoString). The DSP workflow was carried out by NanoString Technologies. Briefly, formalin-fixed, paraffine-embedded tissue slides were deparaffinized and rehydrated using xylene and graded ethanol. Antigen retrieval was performed in 10 mM citrate buffer at pH 6.0 for 15 min at 100°C. A multiplexed cocktail of antibodies, each with a unique ultraviolet-photocleavable indexing oligo, mouse anti-GFAP antibody clone GA5 (Thermo Fisher Scientific) conjugated to Alexa Fluor 488, rabbit anti-CD45 antibody clone D9M81 (Cell Signaling Technology) conjugated to Alexa Fluor 594, mouse anti-CD31 (PECAM-1) antibody clone JC/70A (Abcam) conjugated to Alexa Fluor 647 and Syto83 nucleic acid stain (Thermo Fisher Scientific), was applied to a slide-mounted brainstem tissue section. After staining, slides were scanned using a GeoMX™ DSP instrument to generate digital fluorescent images. Twelve regions of interest at a 500-µm circular diameter for each immune antigen on each slide were then selected based on the fluorescence imaging. Ultraviolet light illuminated regions of interest to photocleave antibodies and release oligos from region of interest. The released oligos were collected and deposited into designated wells on a microtitre plate. The oligos were then quantified by next-generation sequencing. Reads were processed into digital counts and mapped back to each region of interest, generating a map of transcript activity within the tissue architecture. The workflow of DSP data generation and representative regions of interest from each sample are illustrated in Supplementary Fig. 21.
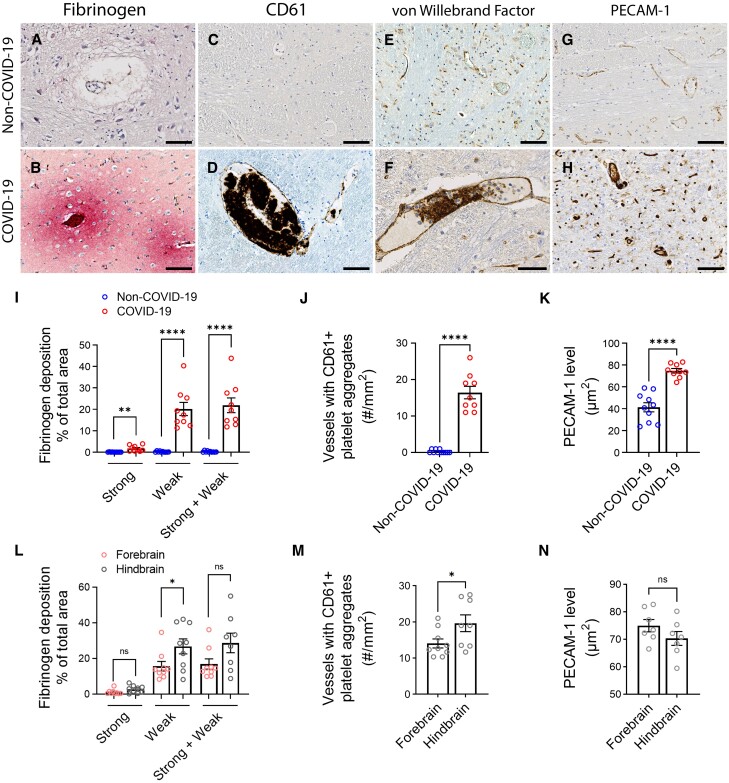
Figure 1.
Microvascular injury and thrombus formation. Immunostaining for (A and B) fibrinogen, (C and D) CD61 for activated platelets, (E and F) vWF and (G and H) PECAM-1 shows absence or minimal staining in the non-COVID-19 brain but in the COVID-19 patients, there was (B) perivascular leakage of fibrinogen, (D) platelet aggregates, (F) thrombi and (H) increased PECAM-1 on endothelial cells. Significantly greater (I) leakage of fibrinogen (**P = 0.0011, ****P < 0.0001) and (J) blood vessels with platelet aggregates were present in the COVID-19 brains (****P < 0.0001). (K) PECAM-1 was significantly increased in the brains of COVID-19 patients compared to non-COVID-19 controls (****P < 0.0001). (L–N) Dots represent the average of individual values in the brain regions that make up the forebrain or hindbrain of a patient. The forebrain includes the cerebrum, basal ganglia and thalamus. The hindbrain contains the pons, medulla and cerebellum. (L) Strong fibrinogen deposition was equally distributed in different regions of the brain, while weak deposition was more prevalent in the hindbrain compared to the forebrain (*P = 0.04). (M) There were more thrombi in the hindbrain compared to the forebrain of the COVID-19 patients (*P = 0.04). (N) PECAM-1 immunostaining was equally distributed in different brain regions of the COVID-19 patients. Data represents mean ± SEM. Scale bars = 100 µm.
Digital spatial profiling data analysis
Third quartile normalization
Normalization of raw distal counts were originally processed using the NanoString third quartile (Q3) normalization procedure (NanoString, Seattle, WA, USA). For Q3 normalization, raw counts were normalized against the 75th percentile from their own regions of interest. Regions of interest were scaled so that they all have the same value for their Q3 value. All the genes were divided by regions of interest for their respective Q3 count and all the genes in all regions of interest were multiplied by the geometric mean of Q3 counts for all regions of interest.
Quality control and regularized log transformation
Quality control was performed by generating relative log expression (RLE) plots from the Q3 and median of ratios normalized counts using the Exploratory Data Analysis and Normalization for RNA-Seq (EDASeq) package in R (v.4.1.1) scripting language (R Foundation for Statistical Computing). RLE plots were generated using EDASeq’s ‘plotRLE’ function. RLE plots can indicate unwanted sources of variation as deviations from the median expression line. RLE plots were generated using the Q3 normalization from NanoString, median of ratios normalization from the DESeq2 package in R,17 and the raw counts. Rlog transformation was performed using the DESeq2 package for certain analyses. Briefly, rlog transforms the raw counts to the log2 scale, minimizing inter-sample differences with respect to transcripts with small counts, and then normalizes counts to library size. Factors of unwanted variance (explained next) and sex were accounted for as batch effects using the removeBatchEffect function in the limma package in R18 after rlog transformation and median of ratios normalization and before plotting.
Removal of unwanted variance
Recent papers have identified improved methods for addressing unwanted variance in DSP data compared to the default NanoString analysis.19 First, lowly expressed genes (i.e. those with five or fewer transcripts across all samples) were filtered. To account for sources of unwanted variance in the DSP data we identified three housekeeping genes, C1orf43, EMC7 and PSMB420 to use as controls. Removal of unknown sources of variation was performed using the ‘RUVg’ function in the remove unwanted variation from RNA-Seq Data (RUVSeq) package in R.21 Briefly, RUVg uses housekeeping genes to estimate unwanted variation via factor analysis.
Differential expression analysis
Differentially expressed genes between groups were identified using the DESeq2 package in R.17 Un-normalized counts were used as input to DESeq2 and the design formula included sex and three of the factors of unknown variation identified by RUVSeq. Estimated log2 fold change (LFC) values were shrunk using adaptive shrinkage from the ashr package in R22 and plotted using the Enhanced Volcano package in R.23 Genes with a false discovery rate adjusted P-value <0.05 and | LFCs | ≥1 were considered significant. A positive LFC indicates greater expression of a gene in patients compared to controls and a negative LFC indicate less expression of that gene in patients compared to controls.
Principal component analysis and heat map
Rlog transformed counts were used for principal component analysis (PCA) and heat maps. PCA was performed using the ‘prcomp’ function in R and were plotted using the ‘autoplot’ function of the ggfortify package in R.24 Loading plots were generated from the PCA loadings and plotted using the ggplot2 package in R.25 Heat maps were generated to identify broad patterns in expression between samples among the transcripts that explained the most variance. The median absolute deviation statistic was used to select the top 1000 transcripts based on the amount of variability each gene explained across samples. These genes were then plotted in a heat map using the ‘heatmap.2’ function of the gplots package in R. The dendrograms and sample order were determined using complete-linkage agglomerative hierarchical clustering with a Euclidean distance function. Z-score values were centred and scaled along the row direction. Rows represent the median absolute deviation 1000 genes and columns represent samples.
Pathway analysis
The top 1550 genes in terms of raw P-value from DESeq2 were imported into the Qiagen Ingenuity Pathway Analysis (IPA) program for core analyses.26 The IPA analyses were exported to R for plotting in ggplot2. A Z-score <2 indicates significant upregulation of that canonical pathway and a Z-score >2 indicates significant downregulation of that canonical pathway.
Statistical analysis
Descriptive analyses were performed to provide information on the general characteristics of the study population. The Kolmogorov–Smirnov test was used to evaluate whether the distribution of numerical variables was normal. Accordingly, two independent sample t-tests and one-way ANOVA were used to compare between/among groups. The numeric variables were presented as the mean and standard errors. Distribution of data in dot-plots is provided, as well as bars to indicate mean values in the figures. A Pearson’s correlation coefficient was used to measure the statistical relationship between variables. A P-value < 0.05 was considered significant. Analyses were performed using GraphPad Prism (GraphPad Software, v.9.1.2, La Jolla, CA, USA).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material. Derived data supporting the findings of this study are available from the corresponding author on request.
Results
Microvascular injury and activation of endothelial cells
Fibrinogen is a large abundant protein in blood but is unable to cross the intact blood–brain barrier. Immunostaining for this protein showed areas of multifocal staining throughout the brain. Strong diffuse immunostaining was present in the perivascular regions with a gradient of weaker staining at increasing distance from the blood vessels (Fig. 1A and B and Supplementary Fig. 1). Fibrinogen leakage was evident in all COVID-19 cases (Supplementary Fig. 2). The non-COVID cases showed none or only minimal weak immunostaining (mean ± SEM = 1.73 ± 0.45 versus 0.03 ± 0.02% strong deposition, P = 0.0011; mean ± SEM = 20.14 ± 3.09 versus 0.18 ± 0.08% weak deposition, P < 0.0001; mean ± SEM = 21.88 ± 3.43 versus 0.22 ± 0.10% total deposition, P < 0.0001) (Fig. 1I). In some regions of COVID-19 patients, neurons and glial cells stained for fibrinogen, presumably representing uptake from the parenchyma (Supplementary Fig. 3). The area involved varied from 5–65% of the section, with strong immunostaining in <10% of the section (Supplementary Fig. 5). Strong staining of fibrinogen deposits showed no significant difference between brain regions, but weak staining was more in the hindbrain than in the forebrain (mean ± SEM = 26.87 ± 4.19 versus 15.75 ± 2.66% weak deposition, P = 0.04) (Fig. 1L and Supplementary Fig. 5). One patient for whom the olfactory bulb was available showed strong fibrinogen staining in 22% of the area (Supplementary Fig. 5). Thus, there was widespread disruption of the blood–brain barrier in all patients. Leakage of plasma proteins from blood vessels into the brain parenchyma was also confirmed by IgM staining (Supplementary Fig. 6). The pattern is similar to that of fibrinogen leakage, although the fibrinogen leakage was much more prominent, probably because its molecular mass is smaller and hence can diffuse further into the parenchyma.
To identify activated platelets, immunostaining was performed for integrin β3 or CD61, which is a cell adhesion molecule.27–29 Platelet aggregates were present in all brain regions of the COVID-19 patients (Supplementary Fig. 1). Some small blood vessels were occluded by the platelets, in others they lined the blood vessels as focal aggregates adhered to the endothelial cells or were present in the intraluminal space (Fig. 1C and D and Supplementary Fig. 7). Platelet aggregates were also found in the meningeal and choroid plexus blood vessels (Supplementary Fig. 7). Rare platelet aggregates were seen in the non-COVID-19 patients (mean ± SEM = 16 ± 2 versus 0.3 ± 0.1 vessels with aggregates/mm2, P < 0.0001) (Fig. 1C, D and J). All COVID-19 patients had platelet aggregates but were greater in the hindbrain compared to the forebrain (mean ± SEM = 20 ± 2 versus 14 ± 1 vessels with aggregates/mm2, P = 0.04) (Fig. 1M and Supplementary Fig. 8). Cases 3, 4 and 6 had extensive platelet aggregation in all brain regions (Supplementary Fig. 8), but microinfarcts were rarely seen. Immunostaining for von Willebrand factor followed the pattern of CD61, confirming the presence of thrombi in the blood vessels (Fig. 1E and F and Supplementary Fig. 1).
Immunostaining for PECAM-1 showed increased staining in the endothelial cells of the patients compared to controls (mean ± SEM = 74.71 ± 2.06 versus 41.29 ± 4.16 mm2, P < 0.0001) (Fig. 1G, H and K). Staining showed increased intensity and surface area involvement of the blood vessels in all brain regions with no significant difference between the different regions in COVID-19 patients (Fig. 1N and Supplementary Fig. 9). This supports the previous observation of increased platelets adherent to the endothelial cells.
Complement activation and immune complexes
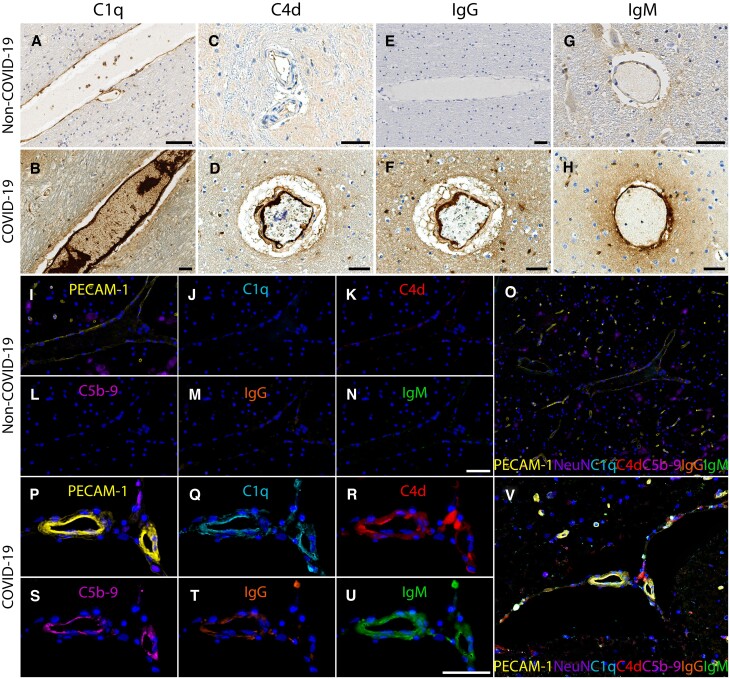
To determine whether there was activation of the complement cascade, we immunostained for several complement molecules. Complement components 1q and 4d (C1q, C4d) were present on endothelial cells and platelets. This was associated with deposition of IgG and IgM. Only minimal staining was seen in the non-COVID-19 controls (Fig. 2A–H and Supplementary Fig. 10). Multiplex immunostaining showed co-localization of PECAM-1 on endothelial cells with C1q, C4d and complement component complex (C5b-9) along with IgG and IgM (Fig. 2I–V and Supplementary Fig. 11). Deposition of immune complexes was seen in all regions of the brain. C1q, IgG and IgM were also present in the perivascular regions. They were predominantly in the extracellular matrix but were also present in some glial cells and neurons. Focal areas of C1q deposits were also seen on the myelin sheaths of axons (Supplementary Fig. 12). RNA in situ hybridization for C1QA and C3 showed that some endothelial cells and macrophages expressed the complement genes, suggesting that these cells may contribute to the complement cascade (Supplementary Figs 13 and 14).
Figure 2.
Complement activation and immune complexes. Immunostaining for (A and B) C1q, (C and D) C4d, (E and F) IgG, and (G and H) IgM shows minimal staining in the brains of non-COVID-19 controls and extensive deposition on endothelial cells in the blood vessels of COVID-19 patients. (I–V) Multiplex immunostaining (I) minimal PECAM-1 on endothelial cells and lack of deposition of (J) C1q, (K) C4d, (L) C5b-9, (M) IgG, and (N) IgM in a non-COVID-19 brain tissue. (O) is a composite of each of the markers. In a COVID-19 tissue, there is (P) increased PECAM-1 in endothelial cells and deposition of (Q) C1q, (R) C4d, (S) C5b-9, (T) IgG and (U) IgM. (V) shows co-localization of these markers on the endothelial cells. Scale bars = 50 µm.
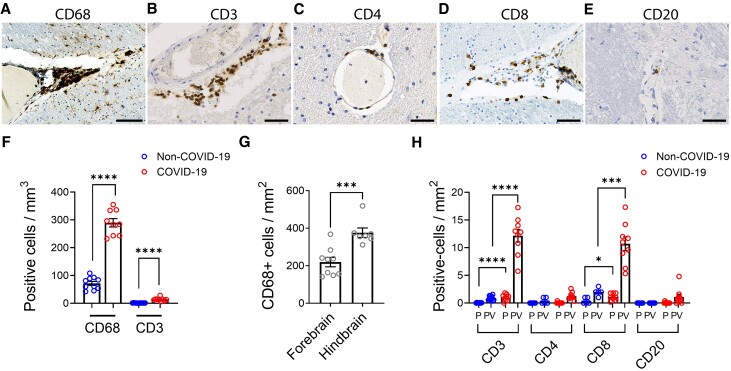
Figure 3.
Characterization of inflammatory infiltrates. The perivascular region shows infiltration of (A) CD68+ macrophages, (B) CD3+ T cells, (C) CD4+ T cells, (D) CD8+ T cells and (E) a few CD20+ B cells. (F) Increased amounts of CD68+ and CD3+ cells were present in COVID-19 cases compared to non-COVID-19 controls (****P < 0.0001). The predominant inflammatory response is mediated by macrophages with a lesser contribution from T-cell populations (nearly 20-fold difference). (G) CD68+ cells were present in greater numbers in the hindbrain compared to the forebrain of the COVID-19 patients (***P = 0.0007). Dots represent the average of individual values in the brain regions that make up the forebrain or hindbrain of a patient. The forebrain includes the cerebrum, basal ganglia and thalamus. The hindbrain contains the pons, medulla and cerebellum. (H) CD3+ and CD8+ T cells were significantly increased in COVID-19 cases compared to non-COVID-19 controls (*P = 0.04, ***P = 0.0003, ****P < 0.0001). CD8+ T cells were present in relatively higher numbers than CD4+ T cells. Only rare CD4+ T cells and CD20+ B cells were present. All lymphatic cell types were predominantly in the perivascular (PV) regions with few cells in the parenchyma (PC). Data represents mean ± SEM. Scale bars = 100 µm.
Inflammatory infiltrates
Macrophage infiltration, as identified by CD68 immunostaining, was a predominant cell type in the perivascular inflammatory infiltrate. CD68+ macrophages were present in all COVID-19 cases and there was no significant difference between them although they were in much greater numbers compared to non-COVID cases (mean ± SEM = 289.55 ± 14.99 versus 71.70 ± 7.04 cells/mm2, P < 0.0001) (Fig. 3A and F). Macrophages were present throughout the brain with higher infiltration in the hindbrain compared to the forebrain (mean ± SEM = 376.00 ± 25.00 versus 219.25 ± 25.11 cells/mm2, P = 0.0007) (Fig. 3G and Supplementary Figs 15 and 16). In some areas, aggregates of CD68+ cells formed microglial nodules (Supplementary Fig. 17).
Figure 5.
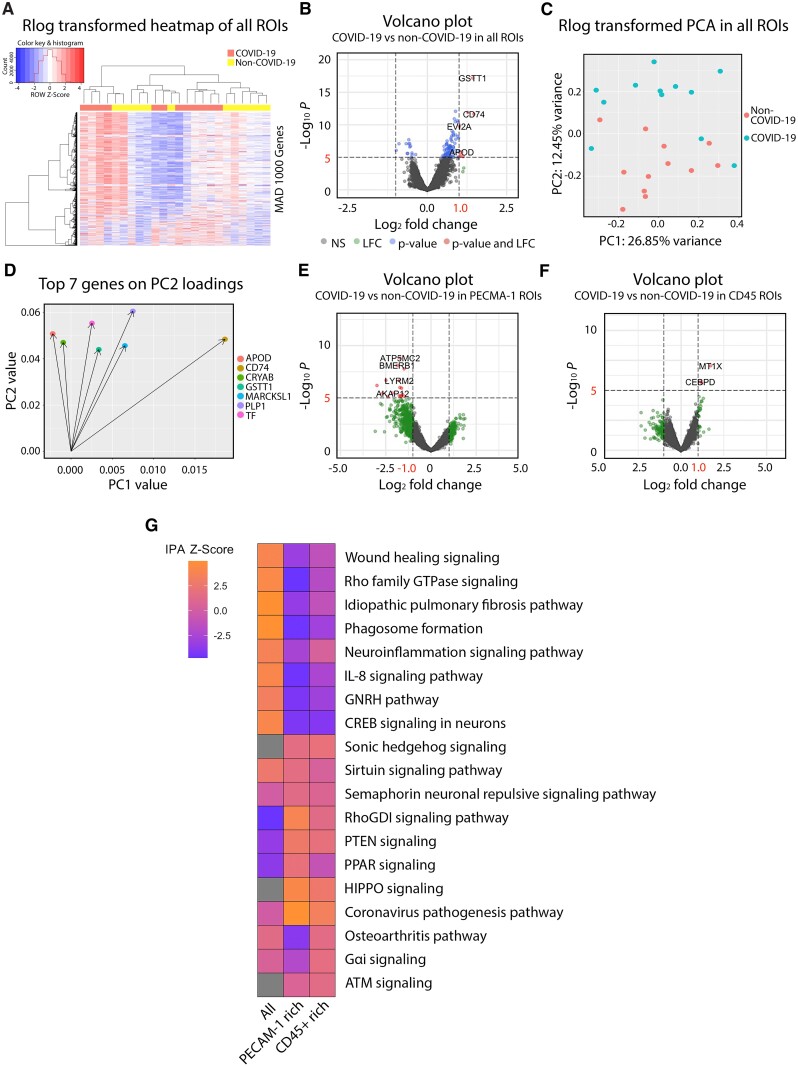
Spatial gene expression profiling. Gene expression data of the brainstem obtained with the NanoString GeoMx platform was analysed by DEA, PCA and IPA. (A–D) Gene expression profiling performed on all regions of interest (ROIs) was analysed. (A) Heat map of the top 1000 genes in terms of median absolute deviation across samples. Complete-linkage hierarchical clustering with a Euclidean distance function was used to generate dendrograms. Column values were annotated based on COVID patient (red) or control (yellow) status. Z-scores were scaled and centred based on row values and the histogram at the top left indicates no significant outliers or overly influential genes. (B) The volcano plot shows the relationship between shrunken LFCs and P-values from our DEA of patients versus controls. Red dots indicate differentially expressed genes with an LFC > 1 and P < 0.05. (C) PCA plot of regularized log (rlog) transformed counts highlighting patient and control samples. (D) Loading plot of the top seven features in terms of loadings from principal component 2, which separated patients from controls. Gene expression profiling performed on (E) PECAM-1 rich and (F) CD45-rich regions of interest were analysed and plotted in volcano plots. (G) IPA output table shows a list of differentially regulated pathways in the all, PECAM-1-rich and CD45-rich regions of interest of the COVID-19 brainstem. Upregulation of a pathway is represented by shades of orange and downregulation in blue.
Anti-CD3 antibody was used as a pan-T cell marker. The pattern of T cell infiltration was similar to that of the macrophages, except that there were fewer T cells compared to the extent of macrophage infiltrates (mean ± SEM = 14.00 ± 1.97 versus 289.55 ± 14.99 cells/mm2, P < 0.0001) (Fig. 3B and F). Subtyping of the T cells in the perivascular region showed that CD8+ cells were the predominant cell type compared to CD4+ cells (mean ± SEM = 10.91 ± 1.24 versus 1.26 ± 0.22 cells/mm2, P < 0.0001) (Fig. 3C, D and H and Supplementary Figs 15 and 16). T cells were localized within perivascular spaces with very few cells in the parenchyma (Fig. 3H and Supplementary Fig. 15). In some blood vessels, the T cells were adherent to the endothelial cells on the luminal side (Supplementary Fig. 15). Like macrophages, the T cells were also present in all regions of the brain and in all COVID-19 cases (Supplementary Figs 15 and 16). Very few CD20+ B cells were present (Fig. 3E and H and Supplementary Figs 15 and 16).
Microglial cell activation and neuronal injury
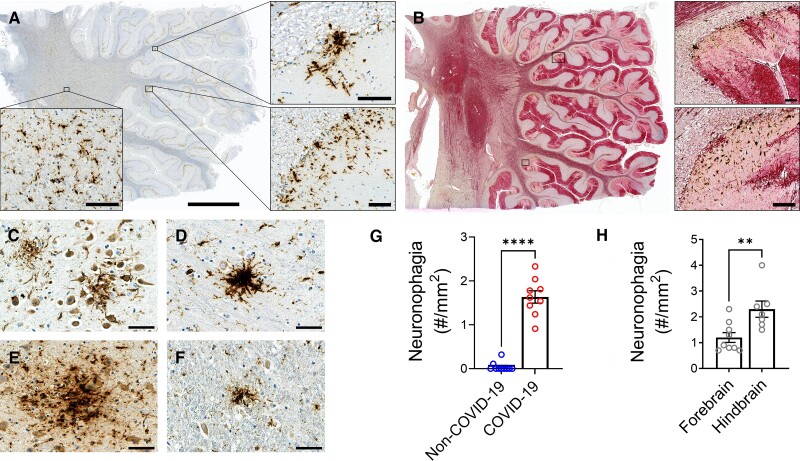
Microglia clusters associated with neuronal cell loss were prominent in the grey matter of the cerebellum and brainstem nuclei (Fig. 4 and Supplementary Figs. 18 and 19). Multifocal areas of loss of Purkinje cells, molecular cells and granule cells were seen in the cerebellum. To further delineate the pattern of Purkinje cell injury, the neurons were stained for calbindin. Loss of neuronal processes resulted in a stripped pattern in the molecular layer (Fig. 4B and Supplementary Fig. 18). The number of microglia clusters surrounding neurons were counted and considered to represent neuronophagia. Neuronophagia was present in all COVID-19 cases and in much greater numbers compared to non-COVID cases (mean ± SEM = 1.6 ± 0.0 versus 0.0 ± 0.0/mm2, P < 0.0001) (Fig. 4G). There were more foci of neuronophagia in the hindbrain compared to the forebrain (mean ± SEM = 2.3 ± 0.3 versus 1.2 ± 0.2/mm2, P = 0.007) (Fig. 4H and Supplementary Fig. 18).
Figure 4.
Microglial nodules and neuronal injury. Microglial nodules, clusters of microglia, surrounding neurons in the grey matter were present. (A) CD68+ cells were found in clusters in the cerebellum of case 9. (B) Double labelling of CD68 (brown) and calbindin (red) in the cerebellum of Case 9 shows multifocal loss of neuronal processes. (C–F) Clusters of CD68+ microglia surrounding neurons were present in the (C) hippocampal CA1 region of Case 2, (D) thalamus of Case 8, (E) pons of Case 7 and (F) solitary nucleus of the medulla. (G) The number of foci of neuronophagia was significantly increased in the brains of COVID-19 patients compared to non-COVID-19 controls (****P < 0.0001). (H) There were significantly more foci of neuronophagia in the hindbrain compared to the forebrain of the COVID-19 patients (**P = 0.0071). Dots represent the average of individual values in the brain regions that make up the forebrain or hindbrain of a patient. The forebrain includes the cerebrum, basal ganglia and thalamus. The hindbrain contains the pons, medulla and cerebellum. Data represents mean ± SEM. Scale bars = 50 µm.
Immunostaining of resident microglia with anti-TMEM119 antibody showed that activated TMEM119+ cells were present in all COVID-19 case and there was no significant difference amongst them although they were in much greater numbers compared to non-COVID cases (mean ± SEM = 230.85 ± 30.57 versus 41.20 ± 9.35 cells/mm2, P < 0.0001). Activated microglia were present in all regions of the brain but were most prominent in the hindbrain compared to the forebrain (mean ± SEM = 285.88 ± 34.28 versus 170.79 ± 30.48 cells/mm2, P = 0.027), where it was also associated with neuronophagia (Supplementary Fig. 19).
Reactive astrocytosis
GFAP immunostaining was used as a marker of astrocyte activation. Intense astrocyte activation was present in the perivascular regions including the walls of the blood vessels. In the cerebellum, Bergmann astrocytosis was present in areas of neuronal loss. Mild to moderate diffuse astrocytosis was seen in the white matter of the brain (Supplementary Fig. 20).
Spatial transcriptomics of the brainstem
DSP data generated from multiple region of interest comparable glial activation, immune cell infiltration and vascular niche identified using the morphological markers GFAP, CD45 and PECAM-1 in 2 non-COVID-19 and 3 COVID-19 brainstems were analysed (Supplementary Fig. 21).
We sought to determine the most appropriate method of normalization/transformation since we observed unwanted variation in the Q3 normalized counts using RLE plots (Supplementary Fig. 22). Based on previously published work on the analysis of DSP data, we used RUVSeq and DESeq2 to successfully address the unwanted variation.19 RLE plots of median of ratios normalized data in all regions of interest indicated successful normalization (Supplementary Fig. 22C). PCA plots that used the rlog transformed counts and accounted for unwanted variation showed separation of patients from controls (Supplementary Fig. 22F).
Differentially expressed genes between COVID-19 patients and controls were then identified (absolute LFC > 1; P-adj < 0.05). Significantly differentially expressed genes identified by differential expression analysis (DEA) in all regions of interest, PECAM-1 rich regions and CD45 rich regions of COVID-19 patients are listed in Supplementary Table 6. The heat map indicated some separation by patient/control status and hierarchical clustering suggested possible patient-enriched clusters on the basis of transcript expression patterns (Fig. 5A). A total of 900 genes, including 727 decreased in patients and 173 increased in patients, were significantly differentially expressed by adjusted P-value in all regions of COVID-19 patients compared to controls (Fig. 5B). The PCA demonstrated relatively good separation between groups (Fig. 5C). PCA loading plots were used to assess which features influenced relevant principal components and, therefore, separation of patients from controls (Fig. 5D). Based both on DEA and PCA, we highlight the importance of APOD (encoding apolipoprotein D), CD74 (encoding an MHC-II protein), GSTT1 (encoding glutathione S-transferase theta 1) and TF (encoding coagulation factor III) transcripts as key players in the pathophysiology of neuro-COVID-19 (Fig. 5D).
In addition, we identified 353 genes decreased in patients and six genes increased in patients in PECAM-1 rich regions and three genes increased in patients in CD45 rich regions of COVID-19 patients compared to controls (Fig. 5E and F). Of note, ATP synthase and anchoring genes (ATP5MC2 and AKAP12) were decreased in the PECAM-1 rich regions, and a metal ion transporter gene (MT1X) was increased in patients in the CD45 rich regions.
Subsequent IPA revealed that altered gene expression in all regions of interest of COVID-19 patients was associated with upregulation of wound healing signalling pathway, signalling by Rho family GTPases, pulmonary fibrosis idiopathic signalling pathway, phagosome formation, neuroinflammation signalling, IL-8 signalling, GNRH signalling and CREB signalling in neurons (Fig. 5G). This supports the hypothesis that COVID-19 patients exhibit robust innate immune responses in the brain. Transcriptional changes in both PECAM-1-rich regions of interest and CD45-rich regions of interest were uniquely associated with differential regulation of sonic hedgehog, RhoGDI, PTEN, HIPPO and coronavirus pathogenesis pathways (Fig. 5G). PECAM-1 rich regions were associated with the greatest degree of upregulation in the coronavirus pathogenesis pathway.
Correlation between fibrinogen leakage and inflammatory infiltrates
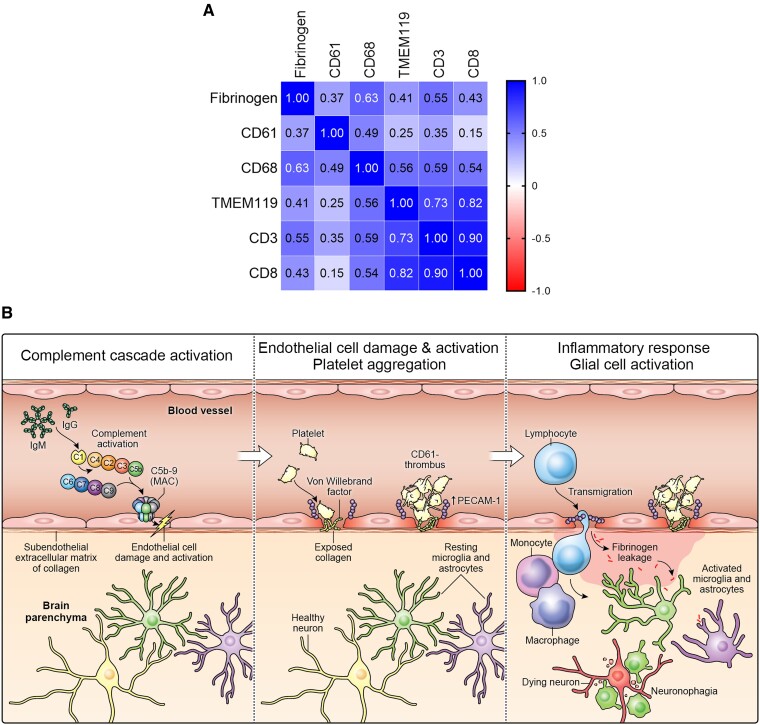
We determined the relationship between fibrinogen leakage (percentage/total area) and inflammatory markers (cells/mm2), or CD61+ platelet aggregates and inflammatory markers (CD61+ vessels/mm2). Fibrinogen deposition was positively correlated with the amount of CD68+ cells (r = 0.63, P < 0.0001), TMEM119+ cells (r = 0.41, P = 0.0034), CD3+ cells (r = 0.55, P < 0.0001) and CD8+ cells (r = 0.43, P = 0.0031). CD61+ platelet aggregates were also positively correlated with the amount of CD68+ cells (r = 0.49, P = 0.0003) and CD3+ cells (r = 0.35, P = 0.0109). A positive relationship was also found between fibrinogen deposition and CD61+ platelet aggregates (r = 0.37, P = 0.0084) (Fig. 6A and Supplementary Fig. 23).
Figure 6.
Neurovascular injury with complement activation and inflammation in COVID-19. (A) Correlation matrix of the various cell type markers. Pearson’s correlation coefficient (r) between two pairs of variables is shown in the heat map. The correlation coefficients are represented in terms of the changes of the intensity of red/blue colour, as shown in the colour bar. (B) The proposed cascade of events to explain the neuropathological findings is as follows: C1 binds to the IgG and IgM antibodies and activates the classic complement pathway. The end product of this cascade, C5b-9 binds to endothelial cells and causes endothelial cell damage. This leads to activation of endothelial cells and increased PECAM-1 and vWF release, resulting in platelet aggregation and thrombus formation. Simultaneously, there is leakage of serum proteins into the perivascular space, which leads to an influx of monocytes and T lymphocytes into the parenchyma. Monocytes differentiate to macrophages and there is activation of microglia and astrocytes in the brain parenchyma. This leads to neuronal injury and neuronophagia.
Detection of SARS-CoV-2 in brain
We used multiple techniques to detect SARS-CoV-2 in the brain. Polymerase chain reaction was performed using multiple primer sets to different regions of the virus on frozen and fixed tissues. A hybridization assay was used to capture any viral transcripts and was followed by sequencing. Fixed tissues were subjected to RNA in situ hybridization using RNAscope. Probes to detect SARS-CoV-2 spike (S) gene were validated in human embryonic kidney 293T cells transfected with the SARS-CoV-2 S plasmid. The cells were fixed in formalin and embedded in paraffin. All techniques failed to detect any virus in the brain, including regions where there were obvious signs of inflammation (Supplementary Fig. 24).
Discussion
Vascular pathology has been commonly described in SARS-CoV-2-infected patients. 1–5% of hospitalized patients with COVID-19 develop strokes, some without underlying risk factors.30 Ischaemic and haemorrhagic strokes may occur.31,32 Fibrin clots have also been found in small blood vessels in the brain.33 The cause of a hypercoagulable state is unclear but a generalized hyperinflammatory state11 and anti-phospholipid antibodies have been implicated.34 Rare cases of acute necrotizing haemorrhagic encephalopathy have also been described.5 Microcerebral haemorrhages may occur in critically ill patients.33 Using post-mortem high-resolution MRI, we previously found that most patients had widespread multifocal microvascular disease which corelated with vascular leakage and injury.16 In the present study, we characterized the pattern, mechanism and consequences of microvascular injury in patients with COVID-19. The loss of vascular integrity was evident by the presence of several large proteins in the perivascular regions that normally do not cross the blood–brain barrier. These included fibrinogen, C1q, IgG and IgM. Fibrinogen was present in high concentrations around the blood vessels with a gradual decrease in concentrations at greater distances from the vasculature. This suggests a leaky blood–brain barrier. All markers of vascular injury were more common in the hindbrain. Similar deposition of fibrinogen in the lumen of the blood vessels has been described in active inflammatory lesions of multiple sclerosis. But even in these patients there was no perivascular leakage of fibrinogen as described in our patients.35
We investigated whether the vascular compromise could be related to endothelial cell dysfunction. Previous studies have shown extensive endothelial cell injury in the lungs.36 Another study showed damage to endothelial cells in the brain resulting in empty basement membranes known as string blood vessels.37 We found increased levels of PECAM-1 on endothelial cells. In contrast, an in vitro study showed a decrease in PECAM-1 on endothelial cells following treatment with recombinant spike protein of SARS-CoV-2.38 Thus, the mechanism of increased PECAM-1 levels in the endothelial cells remains unclear. However, this molecule can serve as an adhesion molecule for platelets and platelet aggregates that were adhered to the endothelial cells was a prominent observation in this study. The platelets were activated and, in some instances, caused occlusion of the small blood vessels. These observations were supported by spatial transcriptomics data. The primary genes driving separation between CODIV-19 patients and controls, CD74 and TF, both contribute to thrombosis formation. In addition to its role in immune function, CD74 is known to be involved in thrombosis formation by contributing to the platelet cytoplasmic Ca2+ signalling pathway.39TF contributes to thrombosis formation by encoding coagulation factor III (TF, tissue factor) to initiate the coagulation cascade and has been reported to be increased in COVID-19.40 The semaphore signalling and the RhoGDI signalling pathways associated with vascular permeability were enriched in COVID-19 patients compared to controls. This may explain the microinfarcts seen in some COVID-19 patients on MRI.31 One study found that the lungs of patients with COVID-19 have unique vascular features consisting of endothelial damage, microthrombosis and intussusception angiogenesis.36 They found that out of 323 genes, 69 angiogenesis-related genes were differentially regulated in the lungs of COVID-19 patients and the amount of new vessels was 2.7 times as high compared to lungs of patients with influenza. Interestingly, we found 25 angiogenesis-related genes that were differentially expressed in brain of the COVID-19 patients, which overlapped with the previous study in lungs. We further found that signalling pathways involved in angiogenesis were dysregulated. These vascular gene dysregulations are consistent with the histopathological observations in our study.
Several makers of endothelial cell function have been studied in COVID-19 patients. Meta-analysis showed that higher plasma levels of vWF antigen, tissue-type plasminogen activator, plasminogen activator inhibitor-1 antigen and soluble thrombomodulin were associated with poor outcome.41 In the liver, strong vWF staining in sinusoidal endothelial cells was associated with increased platelet adhesion.42 Consistent with these observations, we found increased immunostaining for vWF in the microthrombi. Two patients had a history of drug abuse; however, there was nothing unusual about their vascular pathology compared to the other patients.
To determine whether the compromise of the endothelial cells maybe an immune-mediated phenomenon, we looked for the deposition of immunoglobulins. Aggregates of IgG and IgM were found on endothelial cells and platelet aggregates that co-localized with several members of the complement cascade. The presence of C1q, C4d and C5b-9 suggests activation of the classical complement pathway. We also found deposition of C1q and C3 in macrophages and endothelial cells, which has been shown to be induced by SARS-CoV-2 spike protein.43 Deposition of complement cascade and immunoglobulins suggests an immune-mediated injury to the endothelial cells. The antigen against which this immune response is targeted remains unknown. Possibly, the antibodies are directed against an antigen on the endothelial cells, e.g. anti-idiotypic antibodies against the spike protein would bind to the ACE-2 receptor on endothelial cells.44 Alternatively, immune complexes formed by the antibodies and spike protein that may bind to the ACE-2 receptor on the endothelial cells. The spike protein has been shown to compromise the blood–brain barrier in vitro.45 Elevated levels of factors involved in the classical complement pathway have been found in the plasma and autoantibodies that cross-react with brain antigens and the spike protein have been described in the CSF of patients with neuro-COVID-19.46 Critically ill patients with neurological manifestations have also been found to have autoantibodies in plasma and CSF against a number of CNS antigens including endothelium of blood vessels.47 Thus, while the damage to endothelial cells may not be unique to the CNS, the consequences of the breakdown of the blood–brain barrier are unique to the CNS.
We found the cellular infiltrates of macrophages, CD4+ T cells and CD8+ T cells in COVID-19 patients using immunohistochemistry, which was consistent with other studies.11,16 Mirroring these observations, the results of our spatial transcriptomics data demonstrated that the signalling pathways involved in the migration or trafficking of these cells were enhanced in regions rich in PECAM-1+ cells or CD45+ cells. These included RhoGDI, PTEN and Gαi signalling pathways. The cellular infiltrates were predominantly in the perivascular region and largely composed of macrophages. There were 10-fold more macrophages compared to T cells. Although there were few T cells, CD8+ cells out-numbered CD4+ cells and there were only rare B cells. This suggests that the inflammatory infiltrate was secondary to the leakage of serum proteins into the perivascular region as macrophages act as scavengers and help with the repair process. This is consistent with other studies that have found activated monocytes and macrophage markers in CSF48,49 and the brain.11 In one study, the CD8+ T cells were further characterized to show that they had both cytotoxic and exhaustion markers.50 We observed that the serum proteins such as fibrinogen and complement were taken up by glia and neurons. Similarly, astrocytosis was also most prominent in the perivascular regions suggesting that this was secondary to the vascular injury.
We also found multifocal loss of neurons in the hind brain including the cerebellum. This pattern of neuronal loss cannot be attributed to hypoxia where a more diffuse pattern of injury would be expected. The damaged neurons were often in close vicinity of the activated macrophages or microglia and there was evidence of neuronophagia suggesting that the neuronal injury was secondary to glial cell activation. Neuronophagia in the brainstem of patients with COVID-19 has been described previously.13 The underlying pathophysiological mechanisms of COVID-19-induced neuronal injury are unclear but may be explained by metabolic dysregulation and oxidative stress and DNA double-strand damage. Spatial transcriptomics of the brainstem, where neuronal injury was evident, indicated dysregulation of genes such as APOD, GSTT1, ATP5MC2 and MT1x and signalling pathways including PTEN signalling and PPAR signalling. Taken together, these findings are consistent with metabolic dysregulation. For example, APOD has been shown to be upregulated in the brain following HCoV-OC43 infection and has a neuroprotective effect by controlling the levels of peroxidated lipids.51 We also found genes and signalling pathways associated with oxidative stress and DNA damage were upregulated in COVID-19. These included the genes ATP5MC2 and MT1x, Sirtuin, and the HIPPO and ATM signalling pathways. While the dysregulation of these genes and pathways may explain some of the pathological observations in this study, they may also represent potential therapeutic targets.
Interestingly, several of the pathological findings were more prominent in the hindbrain, which is consistent with other studies although the cause remains unclear.4 It has been hypothesized that the virus may reach the brainstem via the olfactory pathways or the vagus nerve that innervates the respiratory and gastrointestinal tracts.52 However, we were unable to confirm the infection in the brainstem. Involvement of the brainstem could have dire consequences since many vital functions are controlled by this region. It may also explain many of the acute and persistent manifestations seen in patients with COVID-19.53,54 Importantly, five patients in our study died suddenly, most while sleeping, hence the possibility of central apnoea needs to be considered although cardiac arrythmia or dysautonomia could be contributory.
We and others have failed to detect the SARS-CoV-2 virus using a variety of highly sensitive techniques.16,55 However, a previous study showed that human brain endothelial cells can be infected with the virus in vitro.37 Another study found small amounts of detectable SARS-CoV-2 RNA in a few individuals in the olfactory bulb, medulla oblongata and the cerebellum and viral protein was found in areas of acute infarcts.52 A subsequent study found double stranded RNA in pericytes of brain blood vessels of patients with COVID-19 and implicated it in the breakdown of the blood–brain barrier.56 Very low levels of SARS-CoV-2 were also detected in other studies by polymerase chain reaction only, and not by RNA in situ hybridization or by immunohistochemistry. They concluded that the neuropathological observations did not result from direct viral infection of brain parenchyma but were more probably due to systemic inflammation.13 Our spatial transcriptomics data supports this in that most of the pathways that we identified as differentially regulated are related to the immune response.
On the basis of these observations, we propose the following cascade of events. Infection with SARS-CoV-2 triggers the formation of immune complexes activating the classical complement pathway. The mechanism by which the viral infection causes the formation of immune complexes is not clear as we were unable to find the virus or viral proteins in the tissues. One possibility is that anti-idiotypic antibodies against the spike protein might bind to the ACE-2 receptor on endothelial cells triggering the cascade of events.44 These immune complexes may cause multifocal areas of endothelial cell activation resulting in platelet activation, aggregation and formation of thrombi. Injury to the endothelial cells may result in leakage of serum proteins into perivascular regions. This may set up several reparative cellular processes, which include infiltration of monocytes that differentiate to macrophages with phagocytic activity to clean up the proteins. The serum proteins are also taken up by glial cells and neurons. The inflammatory process results in microglia activation causing neuronal injury and neuronophagia (Fig. 5B). Since several of the patients in our series died suddenly with very minor lung involvement, we believe that had these patients survived they would probably have progressed to develop long-COVID. Hence the pathological findings here are relevant to this population as well.
Our study has several limitations. Since several patients were found dead, medical histories and post-mortem intervals were not available. Even though we conducted extensive studies for detection of SARS-CoV-2, it is possible that our inability to detect the virus could be a technical or sampling artefact. Although the age groups of our patients were not perfectly matched, the controls were older and hence would have been expected to have more vascular pathology. Four of the controls had respiratory or systemic infections but did not demonstrate the pathological findings seen in the COVID-19 patients. We did not include any controls with microvascular disease hence we cannot be certain if the microvascular changes seen in the COVID-19 patients are specific for the infection.
Conclusions
Injury to the microvasculature by immune complexes with complement activation is the key central event that results in breakdown of the blood–brain barrier, microthromboses, perivascular inflammation and neuronal injury. We postulate that these events are central to the development of the neurological manifestations seen in acute COVID-19 and possibly in long-COVID. Importantly, these studies suggest that therapeutic approaches targeted against the development of immune complexes should be considered.
Supplementary Material
Acknowledgements
We thank NanoString Technologies for help with GeoMx DSP assay. We also thank Dr Farinaz Safavi for useful comments and suggestions. Dr Martha Quezado from National Cancer Institute at NIH provided the control brain tissues. Dr Zu Xi Yu at National Heart Lung and Blood Institute at NIH provided technical assistance. The views expressed in this article are those of the authors and do not reflect the official policy of the Department of Defense (DoD) or the US Government. The identification of specific products or instrumentation is considered an integral part of the scientific endeavour and does not constitute endorsement or implied endorsement on the part of the author, DoD or any component agency
Abbreviations
- C1q =
complement component 1q
- C4d =
complement component 4d
- COVID-19 =
coronavirus disease 2019
- DSP =
digital spatial profiling
- IPA =
Ingenuity Pathway Analysis
- LFC =
log2 fold change
- PCA =
principal component analysis
- RLE =
relative log expression
- rlog =
regularized log
- SARS-CoV-2 =
severe acute respiratory syndrome coronavirus 2
Contributor Information
Myoung Hwa Lee, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Daniel P Perl, Department of Pathology, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Joseph Steiner, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Nicholas Pasternack, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Wenxue Li, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Dragan Maric, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Farinaz Safavi, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Iren Horkayne-Szakaly, The Joint Pathology Center, Defense Health Agency, Silver Spring, MD 20910, USA.
Robert Jones, The Joint Pathology Center, Defense Health Agency, Silver Spring, MD 20910, USA.
Michelle N Stram, Office of Chief Medical Examiner, and Department of Forensic Medicine, New York University School of Medicine, New York, NY 10016, USA.
Joel T Moncur, The Joint Pathology Center, Defense Health Agency, Silver Spring, MD 20910, USA.
Marco Hefti, Department of Pathology, University of Iowa Roy J. and Lucille A. Carver College of Medicine, Iowa City, IA 52242, USA.
Rebecca D Folkerth, Office of Chief Medical Examiner, and Department of Forensic Medicine, New York University School of Medicine, New York, NY 10016, USA.
Avindra Nath, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Funding
Division of Intramural Research (NS003130), and K23NS109284 from NINDS, NIH; Roy J. Carver Foundation and the Iowa Neuroscience Institute.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Liotta EM, Batra A, Clark JR, et al. . Frequent neurologic manifestations and encephalopathy-associated morbidity in COVID-19 patients. Ann Clin Transl Neurol. 2020;7:2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frontera JA, Sabadia S, Lalchan R, et al. . A prospective study of neurologic disorders in hospitalized patients with COVID-19 in New York City. Neurology. 2021;96:e575–e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varatharaj A, Thomas N, Ellul MA, et al. . Neurological and neuropsychiatric complications of COVID-19 in 153 patients: a UK-wide surveillance study. Lancet Psychiatry. 2020;7:875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ross Russell AL, Hardwick M, Jeyanantham A, et al. . Spectrum, risk factors and outcomes of neurological and psychiatric complications of COVID-19: a UK-wide cross-sectional surveillance study. Brain Commun. 2021;3:fcab168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: Imaging features. Radiology. 2020;296:E119–E120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reichard RR, Kashani KB, Boire NA, Constantopoulos E, Guo Y, Lucchinetti CF. Neuropathology of COVID-19: a spectrum of vascular and acute disseminated encephalomyelitis (ADEM)-like pathology. Acta Neuropathol. 2020;140:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis A, Jain R, Frontera J, et al. . COVID-19 associated brain/spinal cord lesions and leptomeningeal enhancement: A meta-analysis of the relationship to CSF SARS-CoV-2. J Neuroimaging. 2021;31:826–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greuel S, Ihlow J, Dragomir MP, et al. . COVID-19: Autopsy findings in six patients between 26 and 46 years of age. Int J Infect Dis. 2021;108:274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Placantonakis DG, Aguero-Rosenfeld M, Flaifel A, et al. . SARS-CoV-2 is not detected in the cerebrospinal fluid of encephalopathic COVID-19 patients. Front Neurol. 2020;11:587384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis A, Frontera J, Placantonakis DG, Galetta S, Balcer L, Melmed KR. Cerebrospinal fluid from COVID-19 patients with olfactory/gustatory dysfunction: A review. Clin Neurol Neurosurg. 2021;207:106760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matschke J, Lütgehetmann M, Hagel C, et al. Neuropathology of patients with COVID-19 in Germany: A post-mortem case series. Lancet Neurol. 2020;19:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paniz-Mondolfi A, Bryce C, Grimes Z, et al. . Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J Med Virol. 2020;92:699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thakur KT, Miller EH, Glendinning MD, et al. . COVID-19 neuropathology at Columbia University Irving Medical Center/New York Presbyterian Hospital. Brain. 2021;144:2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solomon IH, Normandin E, Bhattacharyya S, et al. . Neuropathological features of COVID-19. N Engl J Med. 2020;383:989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nath A. Long-haul COVID. Neurology. 2020;95:559–560. [DOI] [PubMed] [Google Scholar]
- 16. Lee MH, Perl DP, Nair G, et al. . Microvascular injury in the brains of patients with COVID-19. N Engl J Med. 2021;384:481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ritchie ME, Phipson B, Wu D, et al. . limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 (7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhattacharya A, Hamilton AM, Furberg H, et al. . An approach for normalization and quality control for NanoString RNA expression data. Brief Bioinform. 2021;22:bbaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. [DOI] [PubMed] [Google Scholar]
- 21. Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stephens M. False discovery rates: a new deal. Biostatistics. 2017;18:275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blighe K, Rana S; Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1120. 2021.
- 24. Tang Y, Horikoshi M, Li W. ggfortify: Unified interface to visualize statistical result of popular R packages. The R Journal. 2016;8:474–485. [Google Scholar]
- 25. Wickham H. ggplot2: Elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 26. Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mamdouh Z, Chen X, Pierini LM, Maxfield FR, Muller WA. Targeted recycling of PECAM from endothelial surface-connected compartments during diapedesis. Nature. 2003;421:748–753. [DOI] [PubMed] [Google Scholar]
- 28. Newman PJ, Berndt MC, Gorski J, et al. . PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science. 1990;247:1219–1222. [DOI] [PubMed] [Google Scholar]
- 29. Simmons DL, Walker C, Power C, Pigott R. Molecular cloning of CD31, a putative intercellular adhesion molecule closely related to carcinoembryonic antigen. J Exp Med. 1990;171:2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shahjouei S, Tsivgoulis G, Farahmand G, et al. . SARS-CoV-2 and stroke characteristics: A report from the multinational COVID-19 stroke study group. Stroke. 2021;52:e117–e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keller E, Brandi G, Winklhofer S, et al. . Large and small cerebral vessel involvement in severe COVID-19: Detailed clinical workup of a case series. Stroke. 2020;51:3719–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merkler AE, Parikh NS, Mir S, et al. . Risk of Ischemic stroke in patients with Coronavirus Disease 2019 (COVID-19) vs patients with influenza. JAMA Neurol. 2020;77:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kirschenbaum D, Imbach LL, Rushing EJ, et al. . Intracerebral endotheliitis and microbleeds are neuropathological features of COVID-19. Neuropathol Appl Neurobiol. 2021;47:454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuo Y, Estes SK, Ali RA, et al. . Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Transl Med. 2020;12:eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wakefield AJ, More LJ, Difford J, McLaughlin JE. Immunohistochemical study of vascular injury in acute multiple sclerosis. J Clin Pathol. 1994;47:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ackermann M, Verleden SE, Kuehnel M, et al. . Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N Engl J Med. 2020;383:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wenzel J, Lampe J, Muller-Fielitz H, et al. . The SARS-CoV-2 main protease M(pro) causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat Neurosci. 2021;24:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raghavan S, Kenchappa DB, Leo MD. SARS-CoV-2 spike protein induces degradation of junctional proteins that maintain endothelial barrier integrity. Front Cardiovasc Med. 2021;8:687783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su H, Na N, Zhang X, Zhao Y. The biological function and significance of CD74 in immune diseases. Inflamm Res. 2017;66(3):209–216. [DOI] [PubMed] [Google Scholar]
- 40. Hottz ED, Azevedo-Quintanilha IG, Palhinha L, et al. . Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andrianto Al-Farabi MJ, Nugraha RA, Marsudi BA, Azmi Y. Biomarkers of endothelial dysfunction and outcomes in coronavirus disease 2019 (COVID-19) patients: A systematic review and meta-analysis. Microvasc Res. 2021;138:104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kondo R, Kawaguchi N, McConnell MJ, et al. . Pathological characteristics of liver sinusoidal thrombosis in COVID-19 patients: A series of 43 cases. Hepatol Res. 2021;51:1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaneko N, Satta S, Komuro Y, et al. . Flow-mediated susceptibility and molecular response of cerebral endothelia to SARS-CoV-2 infection. Stroke. 2021;52:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arthur JM, Forrest JC, Boehme KW, et al. . Development of ACE2 autoantibodies after SARS-CoV-2 infection. PLoS ONE. 2021;16:e0257016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buzhdygan TP, DeOre BJ, Baldwin-Leclair A, et al. . The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol Dis. 2020;146:105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song E, Bartley CM, Chow RD, et al. . Divergent and self-reactive immune responses in the CNS of COVID-19 patients with neurological symptoms. Cell Rep Med. 2021;2:100288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Franke C, Ferse C, Kreye J, et al. . High frequency of cerebrospinal fluid autoantibodies in COVID-19 patients with neurological symptoms. Brain Behav Immun. 2021;93:415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heming M, Li X, Rauber S, et al. . Neurological manifestations of COVID-19 feature T cell exhaustion and dedifferentiated monocytes in cerebrospinal fluid. Immunity. 2021;54:164–175.e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farhadian S, Glick LR, Vogels CBF, et al. . Acute encephalopathy with elevated CSF inflammatory markers as the initial presentation of COVID-19. BMC Neurology. 2020;20:248. [DOI] [PMC free article] [PubMed]
- 50. Schwabenland M, Salie H, Tanevski J, et al. . Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia-T-cell interactions. Immunity. 2021;54(7):1594–1610.e1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Do Carmo S, Jacomy H, Talbot PJ, Rassart E. Neuroprotective effect of apolipoprotein D against human coronavirus OC43-induced encephalitis in mice. J Neurosci. 2008;28:10330–10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meinhardt J, Radke J, Dittmayer C, et al. . Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci. 2021;24:168–175. [DOI] [PubMed] [Google Scholar]
- 53. Balcom EF, Nath A, Power C. Acute and chronic neurological disorders in COVID-19: potential mechanisms of disease. Brain. 2021;144:3576–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Frontera JA, Yang D, Lewis A, et al. . A prospective study of long-term outcomes among hospitalized COVID-19 patients with and without neurological complications. J Neurol Sci. 2021;426:117486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang AC, Kern F, Losada PM, et al. . Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bocci M, Oudenaarden C, Saenz-Sarda X, et al. . Infection of brain pericytes underlying neuropathology of COVID-19 patients. Int J Mol Sci. 2021;22:11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material. Derived data supporting the findings of this study are available from the corresponding author on request.