Abstract
Recessive variants in GBA1 cause Gaucher disease, a prevalent form of lysosome storage disease. GBA1 encodes a lysosomal enzyme that hydrolyzes glucosylceramide (GlcCer) into glucose and ceramide. Its loss causes lysosomal dysfunction and increased levels of GlcCer. We generated a null allele of the Drosophila ortholog Gba1b by inserting the Gal4 using CRISPR-Cas9. Here, we show that Gba1b is expressed in glia but not in neurons. Glial-specific knockdown recapitulates the defects found in Gba1b mutants, and these can be rescued by glial expression of human GBA1. We show that GlcCer is synthesized upon neuronal activity, and it is transported from neurons to glia through exosomes. Furthermore, we found that glial TGF-β/BMP induces the transfer of GlcCer from neurons to glia and that the White protein, an ABCG transporter, promotes GlcCer trafficking to glial lysosomes for degradation.
Glucosylceramide is synthesized upon neuronal activity and transported to glia via exosomes for degradation by Gba1b and White.
INTRODUCTION
The cell membrane of eukaryotic cells contains three major classes of lipids: phospholipids (mostly phosphatidylethanolamine and phosphatidylcholines), cholesterol, and sphingolipids. Sphingolipids serve as structural membrane components of cells, and many sphingolipids and their metabolites function in modulating diverse cellular processes (1). Although sphingolipids correspond to a relatively small fraction of lipid species, they are far more diverse than phospholipids (2).
The most elementary sphingolipid backbone, ceramide, is synthesized in the endoplasmic reticulum from acyl-CoA and serine. After synthesis, it is transported to the Golgi apparatus (Golgi) by the ceramide transfer protein (CERT) for further modification into glycosphingolipids and sphingomyelins (3, 4), which are then transported from the Golgi to the plasma membrane. These lipids modulate the stiffness of the plasma membrane (3). A severe reduction in ceramides, as observed upon the loss of CERT in Drosophila, increases the fluidity of plasma membranes and the susceptibility to reactive oxygen species (5) and severely reduces life span (6). In contrast, excessive levels of ceramides form gel-like membrane domains due to aggregation with other phospholipids (7, 8). This leads to reduced membrane fluidity and impaired membrane curvature (9). In Drosophila, ceramide accumulation in mitochondria negatively affects the mitochondrial function in a Parkinson’s disease (PD) model (10). Also, increased levels of ceramides impair membrane endocytosis in aged flies and lead to a reduction of life span (11). Moreover, a disequilibrium between ceramide and sphingomyelin due to disruption of ceramidases affects vesicle fusion, trafficking, and synaptic transmission at presynaptic terminals (12). Hence, either increased or decreased levels of ceramides affect neuronal homeostasis by regulating membrane fluidity.
Compared to the diversity of ceramides, glycosphingolipids are even more complex. There are hundreds of different forms of glycosphingolipids that are most prevalent in lipid rafts where they play important roles in cell signaling. Glucosylceramide (GlcCer) and galactosylceramide are the two major categories of glycosphingolipids (2, 4). The precise biological function of the vast majority of these lipids is ill-defined, yet knockout of many glycosphingolipid synthases in mice has revealed important functions for the different glycosphingolipids (13). Moreover, dysregulation of several glycosphingolipids is found in patients with severe childhood-onset diseases (14, 15). Autosomal recessive variants in Glucosylceramides Beta (GBA1) cause Gaucher disease (GD), the most prevalent lysosomal storage disease. GBA1 encodes a lysosomal hydrolase, β-glucosylceramidase, that hydrolyzes GlcCer into glucose and ceramide. Loss of GBA1 can manifest in infants (type II GD), leading to severe neurological symptoms and death before the age of two (16, 17). Many GD-associated GBA1 variants have been linked to PD (14, 15, 18–20). These findings indicate a close relationship between glycosphingolipid metabolism and rare as well as common neurological disorders.
Studies in flies (21–23), fish (24), and mice (25–27) have shown that loss of GBA1 orthologs leads to GlcCer accumulation, lysosomal dysfunction, ubiquitinated protein aggregation, mitochondrial dysfunction, loss of dopaminergic neurons, progressive locomotor defects, and a shorter life span. However, despite the association between elevated GlcCer levels and neurodegeneration, many key questions remain unanswered. These include the source and dynamics of GlcCer production and degradation at the cellular level. The precise role of GlcCer in GD pathophysiology is also not fully understood.
There are two orthologs of human GBA1 in Drosophila: Gba1a and Gba1b. Expression data from modENCODE (28) and FlyAtlas (29) indicate that Gba1a is predominantly expressed in the midgut, whereas Gba1b is expressed in the brain. In this study, we focus on Gba1b. We show that Gba1b is expressed and required in glia, but not in neurons, and that glial-specific expression of human GBA1 rescues systemic loss of Gba1b in flies. We also demonstrate that neuronal activity triggers the synthesis of GlcCer in neurons, which is then secreted from neurons via exosomes. GlcCer released from neurons is then taken up by glia for lysosomal degradation. Loss of the white gene, which encodes an ABCG transporter abundantly expressed in pigment glia (30, 31), severely affects the glial lysosomal degradation of GlcCer. Last, we present compelling evidence that the mechanism of transport of GlcCer from neurons to glia via exosomes is conserved in vertebrate neuronal cells and depends on transforming growth factor–β (TGF-β)/bone morphogenetic protein (BMP) signals produced in glia.
RESULTS
Gba1b is expressed in glial cells but not in neurons
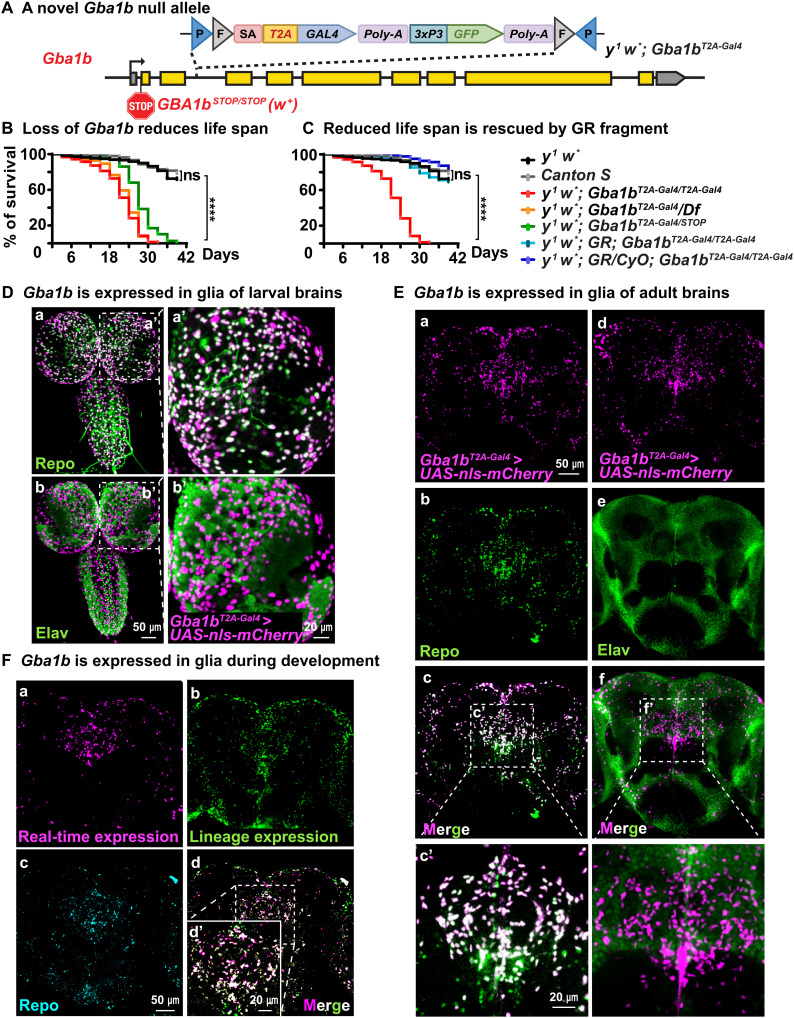
To determine the expression pattern of Gba1b in the central nervous system (CNS), we used the CRIMIC technology (32), as there is no antibody available for Drosophila Gba1b. We inserted a CRIMIC cassette containing SA-T2A-GAL4-PolyA into the third intron of Gba1b (Fig. 1A). This artificial exon drives the expression of GAL4 protein under the control of endogenous Gba1b regulatory elements while arresting transcription because of the presence of the Poly-A sequence (33). Reverse transcription polymerase chain reaction results show that this y1 w*; Gba1bT2A-Gal4 allele is a null allele (fig. S1A). Other alleles include w1118; dGBA1b−/− (abbreviated as Gba1b STOP), a severe loss-of-function allele that carries the mini-white+ gene (22), and w1118; Df(3R)BSC490/TM6C, Sb1 cu1 (Df for short), which uncovers Gba1b and several other genes (34). Flies that lack Gba1b (y1 w*; Gba1bT2A-Gal/T2A-Gal4, y1 w*; Gba1bT2A-Gal4/Df, and y1 w*; Gba1bT2A-Gal4/STOP) show a severely reduced life span when compared with y1 w* and Canton S flies (Fig. 1B). The reduced life span caused by the loss of Gba1b is fully rescued by the introduction of one or two copies of a 20-kb P{acman; CH322-118C10} (35), a genomic bacterial artificial chromosome (BAC) that contains the Gba1b and the mini-white+ gene as a dominant marker (Fig. 1C). The total GlcCer levels are increased 16-fold in y1 w*; Gba1bT2A-Gal/T2A-Gal4 mutant brains when compared to y1 w* controls (fig. S1B), consistent with what has been documented for the Gba1b STOP allele (22).
Fig. 1. Gba1b is expressed in glia.
(A) y1 w*; Gba1bT2A-Gal/T2A-Gal4: A construct containing attp-FRT-SA-T2A-GAL4-PolyA-3xP3GFP-FRT-attp was inserted in Gba1b using CRIMIC technology (32). This insertion creates a null allele as shown in fig. S1. It also leads to the production of GAL4 protein in the same temporal and spatial expression pattern as Gba1b. (B and C) Reduced life span of flies that lack Gba1b: These include (i) y1 w*; Gba1bT2A-Gal4/T2A-Gal4, (ii) y1 w*; Gba1bT2A-Gal4/Df, and (iii) y1 w*; Gba1bT2A-Gal/STOP (this is a red-eyed fly due to the presence of the Gba1b STOP allele). The Gba1bSTOP/STOP allele contains an early stop codon and an insertion of the mini-white+ gene (22). Reduced life span can be fully rescued by one or two copies of a genomic fragment (GR) that contains the Gba1b gene and the white+ gene (N > 200). ****P < 0.0001. (D and E) Gba1b is expressed in glia in larval and adult brains. The GAL4 protein produced by the y1 w*; Gba1bT2A-Gal4 insertion is used to drive the nls::mCherry protein (magenta). On the basis of costaining with Repo (green) present in the nuclei of glia, Gba1b is only expressed in glia. Moreover, there is no obvious colocalization between Elav (green), which marks neuronal nuclei, and nls::mCherry, which reports Gba1b expression, indicating that Gba1b is not expressed in neurons (N = 3). (F) Magenta and green signals represent real-time and lineage expression of Gba1b, respectively (see fig. S1, B and C). Again, Gba1b is only expressed in glia. Repo (canyon) labels the glial nuclei (N = 3).
To identify the cells that express Gba1b in the CNS, we used y1 w*; Gba1bT2A-Gal4 to drive UAS-nls (nuclear localization signal)::mCherry (32). The mCherry-positive nuclei represent cells that express Gba1b, and costaining with an antibody against Elav (a neuronal nuclear marker) (36) or Repo (a glial nuclear marker) (37) shows that Gba1b is expressed in glia of third instar larval (Fig. 1D) and adult brains (Fig. 1E). To determine whether neurons transiently express Gba1b during development, we used G-TRACE (38). It reports the historical/lineage expression of a gene (fig. S1C). G-TRACE of y1 w*; Gba1bT2A-Gal4 costaining with Repo or Elav indicates that Gba1b is not expressed in neurons during development (Fig. 1F and fig. S1D). Hence, these data demonstrate that Gba1b is expressed in glia in the CNS of Drosophila.
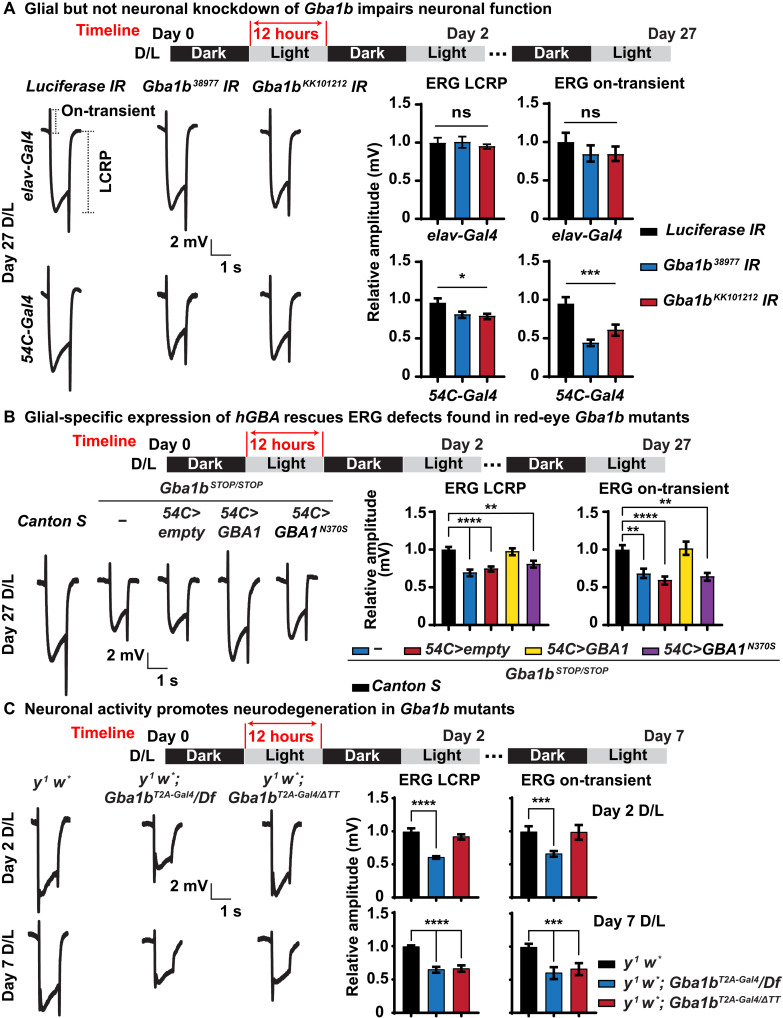
To explore the function of GBA1b in glia, we used the Drosophila visual system as our experimental model. The fly eye is composed of ~750 units called ommatidia. Each ommatidium contains eight photoreceptor neurons surrounded by pigment and cone cells, which function as glia (39, 40). We performed knockdowns of Gba1b in photoreceptor neurons using the elav-Gal4 driver (41) or in pigment glia using the 54C-Gal4 driver (39, 42). Flies were raised under constant darkness conditions until the third day after eclosion (day 0) and were aged for 27 days (day 27) in a 12-hour dark/12-hour light (D/L) cycle at 2500 lux. To assess neuronal function in adults, we performed electroretinogram (ERG) recordings. ERGs assess the neuronal function of the photoreceptors and synaptic transduction in the visual system (43). Upon light exposure, the amplitude of light coincident receptor potential (LCRP) measures the light-dependent phototransduction, whereas the amplitude of the on/off transient is a measure of synaptic transmission between photoreceptors and lamina neurons. As shown in Fig. 2A, glial-specific knockdown of Gba1b using RNA interference (RNAi) leads to a reduction of the LCRPs and on-transient. However, no defect is observed upon neuronal knockdown, consistent with the glial-specific expression of Gba1b. This indicates that Gba1b is required in glial cells to support proper eye function and that its loss affects the phototransduction cascade and synaptic transmission.
Fig. 2. Glial Gba1b is necessary and sufficient to support neuronal function.
(A) Gba1b is required in glia to support neuronal function. ERG recordings of flies of the indicated genotypes after 27 days of D/L cycles. Gba1b is specifically knocked down in neurons driven by elav-Gal4 (top) and in glia driven by 54C-Gal4 (bottom), respectively. Glial knockdown but not neuronal knockdown of Gba1b leads to reduction of LCRPs and on-transient. The ERG LCRP and on-transient amplitudes are quantified on the right. Error bars represent SEM (n ≥ 6); *P < 0.05 and ***P < 0.001. (B) Glial-specific expression of human GBA1 but not GBA1N370S rescues ERG phenotypes in Gba1b mutants. ERG recordings of flies of the indicated genotypes after 27 days of D/L cycles. Gba1bSTOP/STOP shows reduced ERG LCRP and on-transient, which can be fully rescued by glial expression of GBA1 driven by 54C-Gal4. The ERG LCRP and on-transient amplitudes are quantified on the right. Error bars represent SEM (n ≥ 6); **P < 0.01 and ****P < 0.0001. Flies that are tested in this experiment are red eyed because of the transgenes. (C) Neuronal activity promotes neurodegeneration in Gba1b mutants. ERG recordings of flies of the indicated genotypes after 2 and 7 days of D/L cycles. y1 w*; Gba1bΔTT is a partial loss-of-function allele with 40% remaining enzymatic activity (21). At day 2 (top), loss of Gba1b (y1 w*; Gba1bT2A-Gal4/Df) causes reduced ERG LCRP and on-transient amplitudes, whereas the partial loss-of-function allele (y1 w*; Gba1bT2A-Gal4/ΔTT) does not obviously affect the ERGs. At day 7 (bottom), both allelic combinations exhibit reduced LCRP and on-transient amplitudes. The data are quantified on the right. Error bars represent SEM (n ≥ 6); ***P < 0.001 and ****P < 0.0001. Flies that are tested in this experiment are phenotypically white.
To assess whether similar phenotypes were observed in Gba1b null mutants, we first recorded ERGs in Gba1bSTOP/STOP mutants and observed similar reduced LCRPs and on-transient (Fig. 2B). Expression of the human GBA1 reference complementary DNA (cDNA) driven specifically in glia by 54C-Gal4 rescues the ERG defects of Gba1bSTOP/STOP animals (Fig. 2B). However, glial expression of human GBA1N370S, a missense variant found in patients with GD that retains about 30% of β-glucosylceramidase enzymatic activity (44), fails to rescue the ERG defects (Fig. 2B). These data demonstrate that human GBA1 and fly Gba1b functions are evolutionarily conserved.
To determine the phenotype associated with the other Gba1b loss-of-function alleles, we performed ERGs on homozygous y1 w*; Gba1bT2A-Gal4/Gba1bT2A-Gal4 and transheterozygous y1 w*; Gba1bT2A-Gal4/Df. We also tested a partial loss-of-function combination, y1 w*; Gba1bT2A-Gal4/ΔTT. The w1118; GBA1ΔTT allele is a hypomorphic allele that retains about 40% of β-glucosylceramidase activity in fly brains (21). We first raised the mutant flies in constant darkness (D/D). These flies only show a very mild impairment in LCRPs and no defect in on-transient at day 7 (fig. S2A). To determine whether light-induced neuronal activity is required to cause ERG defects, we shifted the flies to the D/L condition. A complete loss of Gba1b impairs neuronal activity at day 2 (Fig. 2C), indicating that neuronal activity stimulated by light is required to induce a strong loss-of-function phenotype. However, the partial loss of Gba1b does not show ERG defects before day 7 (Fig. 2C). These data show that prolonged light-induced neuronal activity is required to trigger ERG defects when residual enzymatic activity is present. The ERG defects observed in Gba1b null flies are fully rescued by glial expression of human GBA1 reference cDNA but not by GBA1N370S (fig. S2B). These data show that Gba1b/GBA1 is required in glia and is sufficient to support normal function.
The above data raise an issue as to why some phenotypes are observed at day 27 whereas other assays reveal phenotype at a much earlier time point at day 2. The assays that reveal phenotypes in young flies (Fig. 2C) were all performed in flies that have white eyes (white− background), whereas those shown in Fig. 2 (A and B) were performed in red-eye flies (white+ background). Note that the ERG amplitudes of LCRP and on-transient are very similar in y1 w* (white− background) and Canton S (white+ background) flies after 7 days of a D/L cycle (fig. S2C), suggesting that the ERG defects shown in Fig. 2C are due to the lack of Gba1b but not loss of pigmentation in the fly eyes. Moreover, as shown in fig. S2C, y1 w* flies exhibit reduced ERG LCRP and on-transient amplitude at day 27 compared with Canton S flies, indicating that the white gene may play a role in this process. We will address this issue below. In summary, all the data presented so far indicate that Gba1b is required in glia to support neuronal function.
Loss of Gba1b leads to abnormal glia morphology and progressive neurodegeneration
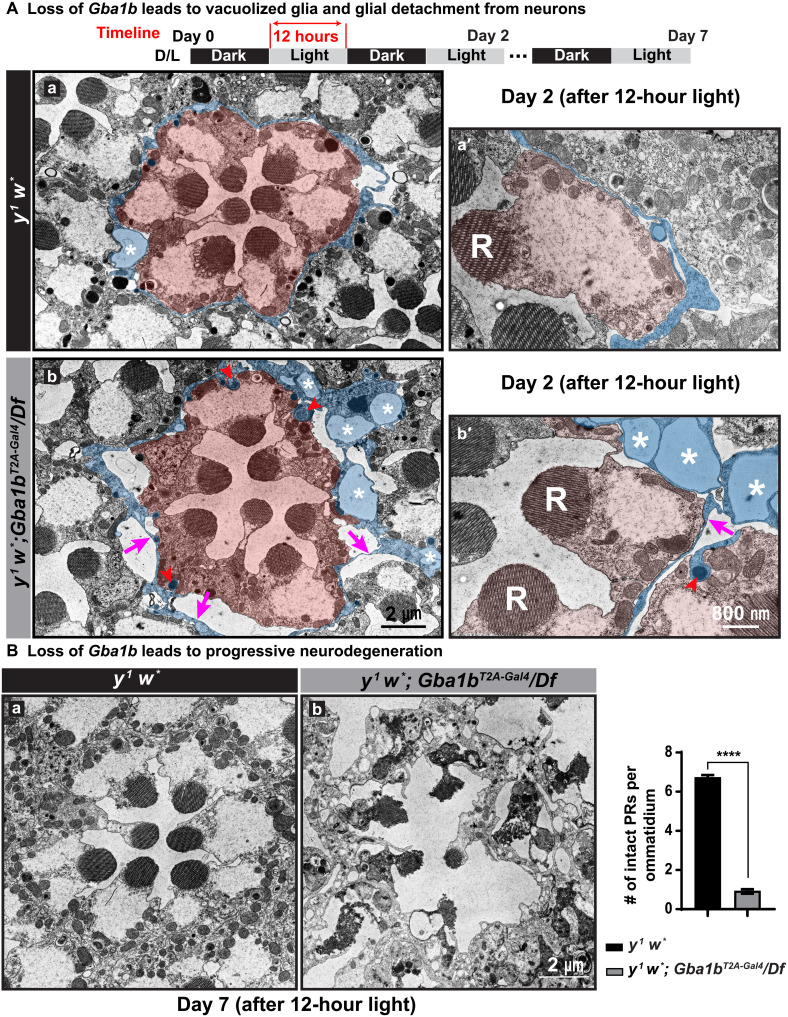
To assess ultrastructural defects in the fly retinas, we performed transmission electronic microscopy (TEM) at days 2 and 7 for flies kept in D/L cycles. All flies used in these experiments were in a white− background (y1 w*; Gba1bT2A-GAL4/Df or y1 w* control). After 2 days of D/L exposure, glia in the Gba1b null mutants exhibit large vacuoles (~1.5 μm2) that are not or rarely observed in y1 w* control animals [Fig. 3A (b and b′, white asterisks) and fig. S3A]. Glia in y1 w* flies are tightly associated with photoreceptor neurons (Fig. 3A, a and a′, highlighted in blue), whereas many glia in mutant retina are detached from the photoreceptor neurons [Fig. 3A (b and b′, magenta arrow) and fig. S3B]. We also observe a vast increase in the number of lysosomes in glia of mutant animals when compared to y1 w* flies at day 2 [Fig. 3A (b and b′, red arrowheads) and fig. S3C]. In contrast, we did not observe obvious morphological defects in photoreceptor neurons, nor did we observe an increase in lysosomes in these cells (Fig. 3A and fig. S3C). These data show that the primary morphological lesions originate in glia.
Fig. 3. Loss of Gba1b impairs glial morphology, which precedes neuronal loss.
(A) Loss of Gba1b leads to vacuolized glia. TEM images of fly retina of the indicated genotypes after 2 days of D/L cycles. Photoreceptor neurons are highlighted in orange, and pigment cells are highlighted in blue (a, a′, b, and b′). R, rhabdomeres. Red arrowheads point to lysosomes in pigment cells. y1 w*; Gba1bT2A-Gal4/Df null mutants exhibit an increased number of lysosomes in glia when compared to glia of y1 w* flies (b and b′). Magenta arrows indicate glial detachment, which is commonly seen in the retina of y1 w*; Gba1bT2A-Gal4/Df null mutants. Glial vacuoles are frequently seen in the retina of null mutant flies, and asterisks mark vacuoles in glia, which are rarely observed in y1 w* flies. (B) TEM images of fly retina of the indicated genotypes upon 7 days of D/L cycles. The overall morphology of the retina is severely affected in y1 w*; Gba1bT2A-Gal4/Df flies, whereas the retinas of y1 w* flies do not show obvious defects. The number of intact photoreceptors (PRs) per ommatidium is quantified on the right. Error bars represent SEM (n = 3); ****P < 0.0001. Flies that are tested in this experiment are phenotypically white.
As shown in Fig. 3B, TEM after 7 days of D/L cycles shows that the photoreceptor morphology is severely affected in Gba1b null mutants (Fig. 3B, b), consistent with the severe ERG defects documented in Fig. 2C. In contrast, the neurons and glia in y1 w*controls show no or very minor defects at this stage (Fig. 3B, a). The data clearly show that loss of Gba1b causes defects in glia that precede the defects in photoreceptor neurons. Moreover, Gba1b is required for neuronal maintenance.
Loss of Gba1b causes light/activity-dependent GlcCer accumulation in neurons and glia
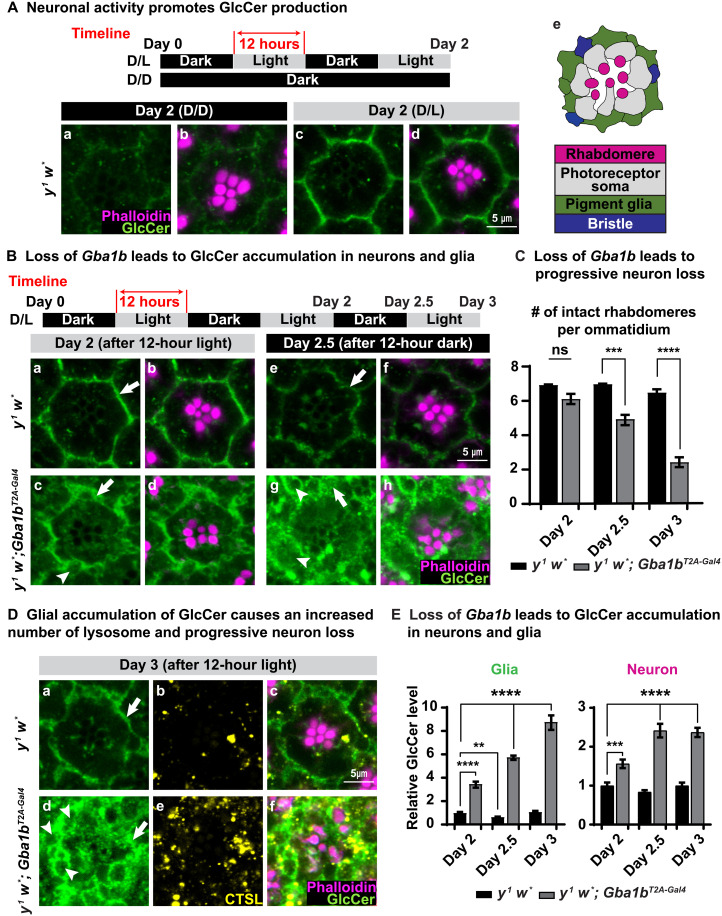
To determine how loss of Gba1b leads to functional (Fig. 2 and fig. S2) and morphological (Fig. 3) defects in the nervous system, we measured the distribution and levels of GlcCer, the substrate of Gba1b protein. As shown in Fig. 4A (a and b) and fig. S4A (a and b), both y1 w* and y1 w*; Gba1bT2A-Gal4 (homozygous y1 w*; Gba1bT2A-Gal4/T2A-Gal4 alleles are labeled y1 w*; Gba1bT2A-Gal4 hereafter) flies that are kept in constant darkness (D/D) do not show obvious GlcCer signals based on immunostaining (the green signals in Fig. 4A and fig. S4A). Upon 12 hours of light stimulation, we observed GlcCer accumulation in neurons and glia in the retinas of both y1 w* and y1 w*; Gba1bT2A-Gal4 flies [Fig. 4A (c and d) and fig. S4A (c and d)]. Hence, light-induced activity of photoreceptor neurons promotes the synthesis of GlcCer.
Fig. 4. Loss of Gba1b leads to GlcCer accumulation.
(A) Immunofluorescent images of fly retina of the indicated genotypes and the conditions under which the flies were raised: antibody against GlcCer in green, whereas phalloidin labels rhabdomeres in magenta. (a and b) Flies kept in the dark show very little GlcCer in the ommatidia of y1 w* (basal level). (c and d) After 12 hours of light exposure, GlcCer is synthesized and accumulates in retina of y w flies (n ≥ 9). (e) A cartoon image illustrating the structure of a fly ommatidium. (B) (a, b, e, and f) GlcCer accumulation in the glia of y1 w* flies is reduced after 12 hours of inactivation of neurons in the darkness. (a to d) After 2 days of D/L cycles, similar to y w flies, GlcCer accumulates in the glia of y1 w*; Gba1bT2A-Gal4 flies. However, GlcCer accumulates more in the neurons of y1 w*; Gba1bT2A-Gal4 flies. (c, d, g, and h) The glial accumulation of GlcCer fails to be degraded upon exposure to 12 hours of darkness in retina of y1 w*; Gba1bT2A-Gal4 flies (n ≥ 9). (C) Loss of Gba1b leads to progressive neurodegeneration. Numbers of intact PRs per ommatidium in (B) and (D) were quantified. Error bars represent SEM (n ≥ 9); ***P < 0.001 and ****P < 0.0001. (D) (a, c, d, and f) GlcCer progressively accumulates in the retina of y1 w*; Gba1bT2A-Gal4 flies, which is highly enriched in the glial region. (b and e) CTSL (yellow) represents lysosomes. The number of lysosomes is increased in y1 w*; Gba1bT2A-Gal4 flies compared with y1 w* flies (n ≥ 9). (E) Quantification of relative GlcCer levels in (B) and (D). Error bars represent SEM (n ≥ 9); **P < 0.01, ***P < 0.001, and ****P < 0.0001. All flies that are tested in this experiment are phenotypically white.
To determine whether neural activity triggers GlcCer synthesis besides the visual system, we overexpressed trpA1 in fly neurons driven by elav-GAL4 (45). trpA1 encodes a thermosensitive transient receptor potential channel (TRP channel). It is a cation channel that is not permeable to Ca2+ and Na+ ions at 18°C but is highly permeable to these ions at 29°C (46). Transgenic flies (elav-GAL4 >dTrpA1) and negative control flies (elav-GAL4 >empty) were raised at 18°C till 3 days after eclosion. They were then exposed at 29°C for 60 min to promote neuronal activity (fig. S4B). We performed immunostaining on paraffin brain sections using the GlcCer antibody. As shown in fig. S4B, a significant increase in GlcCer levels was observed in many brain areas of the fly that overexpressed dTrpA1 in the neurons, indicating that neuronal activity promotes GlcCer synthesis in the CNS.
To further assess the effect of light exposure on GlcCer accumulation and neurodegeneration, we exposed the y1 w* and y1 w*; Gba1bT2A-Gal4 flies for 12 hours to light [day 2; Fig. 4, B (a to d) and C] and then transferred them into the dark for 12 hours [day 2.5; Fig. 4, B (e to h) and C]. As shown in Fig. 4B (e and f), compared with Fig. 4B (a and b), the GlcCer levels in the y1 w* control flies are reduced upon dark exposure. In contrast, when compared with Fig. 4B (c and d), the levels of GlcCer in the y1 w*; Gba1bT2A-Gal4 flies are not reduced but rather accumulate in glia and photoreceptor neurons (Fig. 4B, g and h) upon dark exposure. Furthermore, the accumulation of GlcCer is exacerbated when the y1 w*; Gba1bT2A-Gal4 flies were further kept for a second period of 12 hours in light (day 3; Fig. 4D, a, c, d, and f). Moreover, we observed a robust expansion in size of the pigment glia (white arrows in Fig. 4, B and D) and an increase in vacuoles in these glia (white arrowheads in Fig. 4, B and D). To monitor the morphology of photoreceptor neurons, we used phalloidin to label the rhabdomeres (47). We do not observe neurodegeneration at day 2 [phalloidin in Fig. 4, B (b and d) and C]. This is consistent with previous TEM data at day 2 (Fig. 3A). However, we observe a progressive loss of photoreceptor neurons in the mutants at day 2.5 [phalloidin in Fig. 4, B (f and h) and C] and day 3 [phalloidin in Fig. 4, D (c and f) and C]. Note that loss of Gba1b causes morphological defects in the pigment glia before neurodegeneration, consistent with our TEM data (Fig. 3). In summary, the data show that light stimulation promotes the elevation of GlcCer in both neurons and pigment glia, and loss of Gba1b causes severe morphological defects in pigment glia followed by the loss of the photoreceptor neurons.
GlcCer transport to glia for lysosomal degradation by Gba1b
To explore how GlcCer accumulates in glia, we measured the relative elevation of GlcCer intensity in y1 w* and y1 w*; Gba1bT2A-Gal4 flies raised under different light stimulation paradigms (Fig. 4E). Upon light stimulation at day 2, we found that GlcCer accumulates more in both neurons and glia of y1 w*; Gba1bT2A-Gal4 mutants than in y1 w* flies (first two columns in the neuron and glia plots of Fig. 4E). When we compared the GlcCer levels between pigment glia of y1 w* flies at days 2, 2.5, and 3, we observed a fluctuation in levels that correlates with light exposure. This fluctuation of GlcCer in the pigment glia of y1 w* flies is consistent with the model that light/activity stimulates an increase in GlcCer levels, and darkness permits elevated GlcCer levels to return to a lower baseline (black columns in the glia plot of Fig. 4E). In contrast, in the absence of this light/activity, there is an accumulation with time in y1 w*; Gba1bT2A-Gal4 (gray columns in the glia plot of Fig. 4E). These data suggest that GlcCer is produced in active neurons and accumulates in both neurons and glia when Gba1b is lost.
Given that Gba1b is a lysosomal hydrolase, we assessed the presence of lysosomes in y1 w* and y1 w*; Gba1bT2A-Gal4 flies at day 3 using immunofluorescence staining of Cathepsin L (CTSL), a lysosomal enzyme (Fig. 4D, b, c, e, and f) (48). The y1 w*; Gba1bT2A-Gal4 mutant ommatidia show an increased number of lysosomes when compared with y1 w* (fig. S4D). Most of these lysosomes are localized in glia (Fig. 4D, b, c, e, and f), indicating that loss of Gba1b leads to a progressive glial GlcCer accumulation that impairs glial lysosomes.
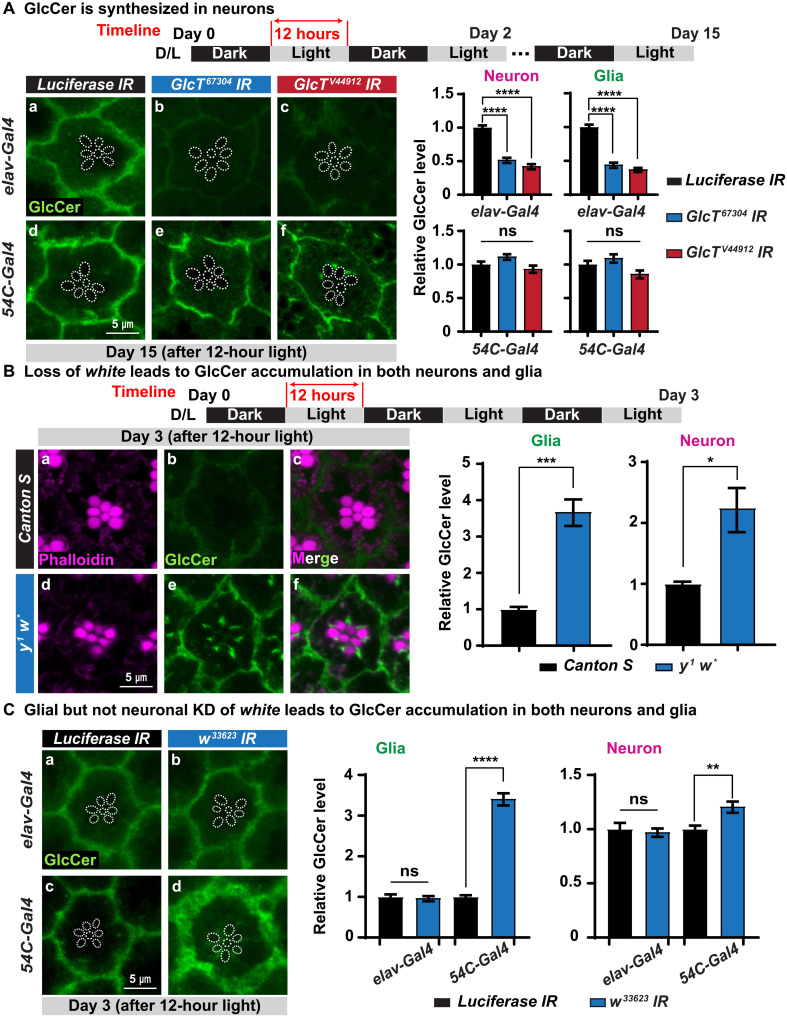
As GlcCer is produced upon neuronal activity and progressively accumulates in glia of y1 w*; Gba1bT2A-Gal4 mutants, we hypothesized that GlcCer is synthesized in neurons and subsequently transported to glia for lysosomal degradation by Gba1b. To test this hypothesis, we knocked down Glucosylceramide synthase (GlcT), the gene encoding the ceramide glucosyltransferase that produces GlcCer, either in neurons (driven by elav-Gal4) or in pigment glia (driven by 54C-Gal4) in wild-type flies. Neuronal (Fig. 5A, a to c) but not glial (Fig. 5A, d to f) knockdown of GlcT significantly reduces GlcCer levels, suggesting that GlcCer is indeed produced in neurons. The latest fly single-cell sequencing database shows that GlcT is highly expressed in neurons including the photoreceptor neurons (49).
Fig. 5. Loss of white in glia causes GlcCer accumulation.
(A) GlcCer is produced in neurons. Immunofluorescent images of fly ommatidia of the indicated genotypes after 15 days of D/L cycles. Glucosylceramide synthase (GlcT) was knocked down using two different RNAi constructs in neurons driven by elav-Gal4 (top) and in glia driven by 54C-Gal4 (bottom), respectively. Neuronal but not glial knockdown of GlcT leads to a reduction of GlcCer in both neurons and glia. Dashed circles outline rhabdomeres. Relative GlcCer levels are quantified on the right. Error bars represent SEM (n ≥ 11), ****P < 0.0001. Flies that are tested in this experiment are red eyed because of the transgene. (B) Loss of white causes GlcCer accumulation. Immunofluorescent images of fly retina of the indicated genotypes after 3 days of D/L cycles. GlcCer accumulates in the ommatidia of y w but not in Canton S flies. Relative GlcCer levels are shown on the right. Error bars represent SEM (n ≥ 7), *P < 0.05 and ***P < 0.001. (C) Glial but not neuronal knockdown (KD) of white causes GlcCer accumulation. Immunofluorescent images of fly retina of the indicated genotypes after 3 days of D/L cycles. The white mRNA was knocked down using a UAS-RNAi construct expressed in neurons by elav-Gal4 (top) and in glia driven by 54C-Gal4 (bottom), respectively. Knockdown of white in glia leads to an accumulation of GlcCer in both neurons and glia, whereas neuronal knockdown in neurons does not cause a phenotype. Relative GlcCer levels are quantified on the right. Error bars represent SEM (n = 11), **P < 0.01 and ****P < 0.0001. Flies that are tested in this experiment are red eyed because of the transgenes.
In summary, our data show that GlcCer is synthesized in neurons by GlcT upon light stimulation and is somehow transported to pigment glia for lysosomal degradation by Gba1b. Upon loss of Gba1b, GlcCer accumulates in the photoreceptor neurons and pigment glia, causing glial expansion and vacuoles in glia. This accumulation is toxic to both cell types, although the glial impairment precedes neuronal loss.
Loss of white exacerbates GlcCer accumulation and neurodegeneration
The y1 w*; Gba1bT2A-Gal4 allele was generated in a white− background using 3XP3-GFP as a dominant marker (32), whereas the Gba1bSTOP/STOP and all RNAi lines carry the mini-white as a dominant marker. As documented before (Fig. 2, B and C, and fig. S2B), y1 w*; Gba1bT2A-Gal4 mutants exhibit defects much earlier than Gba1bSTOP/STOP mutants, while both alleles are thought to be strong loss-of-function alleles (22). The white gene encodes an ABC [adenosine 5′-triphosphate (ATP)–binding cassette] transporter that is known to transport molecules that make up the pigment granule, a lysosomal-like organelle (30, 31). The fly White protein is conserved with human ABCG family transporters that are known to be lipid transporters (50). Considering that white has been found to affect light-induced photoreceptor degeneration and life span (51) similar to Gba1b, we assessed whether the white gene might affect GlcCer trafficking and participate in GlcCer metabolism.
To explore the impact of white on GlcCer metabolism and neurodegeneration, we performed immunostaining for GlcCer and compared its levels in neurons and glia of Canton S (white+) and y1 w* (white−) flies at day 3. As shown in Fig. 5B, GlcCer levels are significantly increased in both neurons and glia of y1 w* flies when compared with Canton S flies. Hence, loss of white is sufficient to cause GlcCer accumulation.
The White protein has been shown to be abundant in pigment glia and present at much lower levels in photoreceptor neurons (30, 31). Unfortunately, the antibody used to perform these studies is no longer available. To determine which type of cells requires the White protein, we knocked down the white gene in glia driven by 54C-Gal4 and in neurons using elav-Gal4. As shown in fig. S5A, neuronal knockdown does not cause an obvious alteration in pigmentation, whereas glial knockdown reduces or abolishes pigmentation in females and males, respectively. GlcCer does not accumulate in glia or neurons when white is knocked down in neuron (Fig. 5C, a and b). However, glial knockdown leads to an accumulation of GlcCer and an expansion of the glial cells (Fig. 5C, c and d). The data show that loss of white in glial cells impairs GlcCer degradation.
To assess how White may modulate GlcCer trafficking, we first determined the subcellular localization of ectopic expressed White protein in Drosophila Schneider 2 (S2) cells. As shown in fig. S5B, White::HA is colocalized with the multivesicular body (MVB) marker hepatocyte growth factor–regulated tyrosine kinase substrate (Hrs) (52) (fig. S5B, m to o), but not mitochondrial (Atp5a; fig. S5B, a to c), lysosomal (CTSL; fig. S5B, d to f), early endosomal (Rab5; fig. S5B, g to i), or late endosomal (Rab7; fig. S5B, j to l) markers. This indicates that White may modulate GlcCer trafficking by regulating the endolysosomal pathway in MVBs. We then exposed the S2 cells to Biotin-GlcCer and determined the White::HA subcellular localization (fig. S5C). White-HA remains associated with MVBs upon Biotin-GlcCer exposure (fig. S5C, a to f), but a substantial fraction of White::HA becomes colocalized with the lysosomal marker CTSL (fig. S5C, g to l), suggesting that White is associated with MVB that traffics to lysosomes upon GlcCer exposure.
GlcCer is transported from neurons to glia in mammalian cell coculture assays
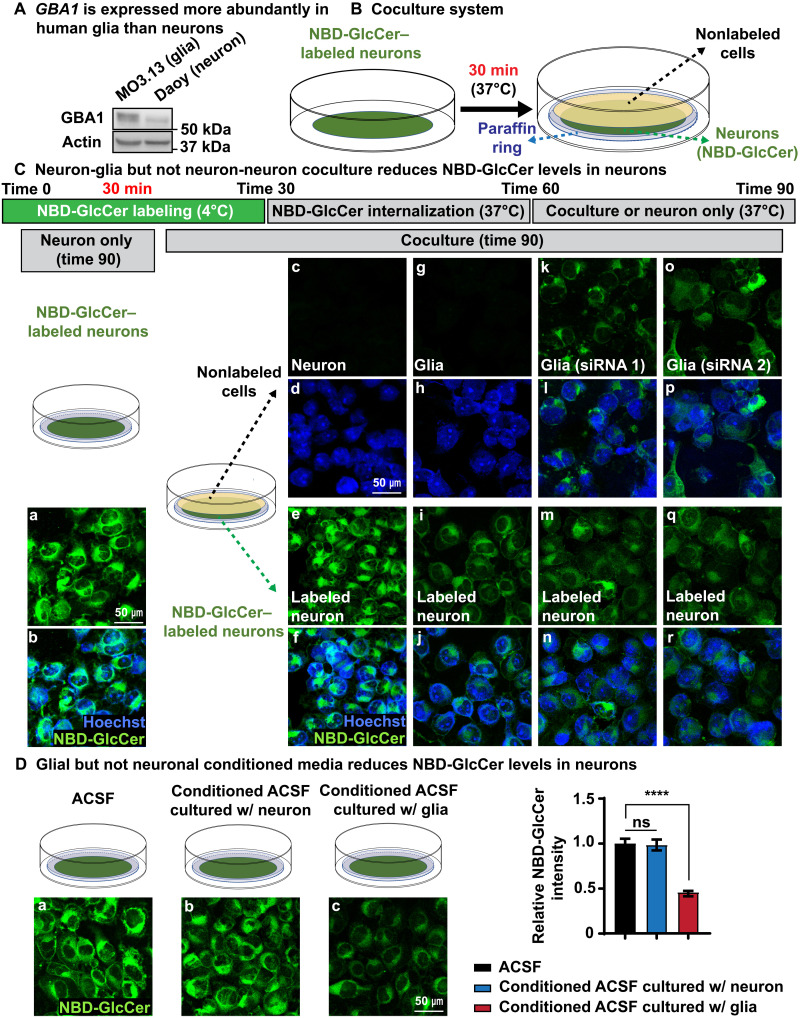
Our data in flies suggest that Gba1b plays an important role in GlcCer metabolism in glia. Human transcriptome data show that human GBA1 is expressed at higher levels in astrocytes than in neurons (53). The Brain RNA-Seq database (Brain RNA-Seq) shows that Gba, the mouse homolog of GBA1, is expressed more in microglia, oligodendrocytes, and astrocytes than in neurons (54). These results are consistent with a potential role of glia in GlcCer metabolism in our fly model. We therefore compared human GBA1 protein levels in a human medulloblastoma cell line with neuronal properties (Daoy cells; we will call them neurons hereafter) (55) and a human oligodendrocyte cell line (MO3.13 oligodendrocytes; we will call them glia hereafter) (56). The level of GBA1 is indeed significantly higher in these human glial cells than in Daoy neurons (Fig. 6A). We then established a coculture system to test whether the transport of GlcCer from neurons to glia occurs in mammalian cells and assess whether the role of GBA1 in glia during this process is evolutionarily conserved.
Fig. 6. Neuron-to-glia GlcCer transport is evolutionarily conserved in human cells.
(A) A human oligodendrocyte cell line, MO3.13, expresses more GBA1 than human Daoy neurons. (B) A cartoon illustrating the coculture system. Daoy cells are labeled with C6-NBD-GlcCer (green, NBD-GlcCer). A paraffin ring is then placed on top of the labeled neuron. Other cells, Daoy neurons, or glia are cultured on a coverslip. The latter are inverted so that they bathe in the same fluid as the labeled neurons. Hence, the two groups of cells face each other but are separated by the paraffin ring. (C) (a and b) After labeling at 4°C and 30 min at 37°C, NBD-GlcCer is partially internalized into the cytosol of labeled neurons. They are then cultured in ACSF for an additional 30 min. The NBD-GlcCer remains in the cells for the entire period. (c to f) Neuron-neuron coculture does not reduce the NBD-GlcCer levels in labeled neurons. Little to no NBD-GlcCer signal is detected in nonlabeled neurons. (g to j) Neuron-glia coculture leads to a decrease in NBD-GlcCer levels in labeled neurons. A faint signal is observed in the nonlabeled MO3.13 glia (g). (k to r) Labeled neurons are cocultured with nonlabeled MO3.13 glia in which GBA1 is knocked down using two different siRNAs. Again, the NBD-GlcCer level is reduced in the labeled neurons under (GBA1 knocked down) coculture conditions. NBD-GlcCer is retained in the nonlabeled MO3.13 glia. (D) Glial but not neuronal conditional medium promotes NBD-GlcCer release from labeled neurons. NBD-GlcCer–labeled neurons were incubated with three different conditional ACSF media as shown in (a) to (c) for 30 min, respectively. Only ACSF that was used to culture glia (c) is able to trigger NBD-GlcCer release from labeled neurons. Relative NBD-GlcCer intensity is quantified on the right. ****P < 0.0001.
To label the GlcCer, we incubated Daoy neurons with fluorescent C6-nitrobenzoxadiazole–tagged GlcCer (NBD-GlcCer for short) for 30 min at 4°C. This labels the plasma membrane of neurons (fig. S6A, a and b). Upon shifting the cells to 37°C for 30 min, the NBD-GlcCer is partially internalized into the cytosol (time 60; fig. S6A, c and d), and a similar signal is observed 30 min later (time 90; fig. S6A, e to g). To monitor whether NBD-GlcCer can be transferred to other cells, the labeled Daoy neurons were cocultured with four different types of cells: Daoy neurons, MO3.13 glia, and two independently derived MO3.13 glial cells in which GBA1 was knocked down by small interfering RNAs (siRNAs) (fig. S6C). In these assays, the labeled neurons are placed at the bottom, whereas the cell layer of the four different cell types is cultured on a coverslip that is placed above. The top layer of cells does not come into direct contact with the cells at the bottom as a ring of paraffin film separates both cell layers. Labeled neurons that were not cocultured with other cells (Fig. 6C, a and b) or labeled neurons that are overlaid with nonlabeled neurons do not show a reduction in NBD-GlcCer levels after 30 min [Fig. 6C (e and f) and fig. S6B]. Also, no to little NBD-GlcCer signal is detected in the nonlabeled neurons placed on top in this assay (Fig. 6C, c and d), indicating that NBD-GlcCer is not transported between neurons. However, when the labeled neurons were cocultured with glia, we observed a significant reduction of the levels of NBD-GlcCer in the labeled neurons [Fig. 6C (i and j) and fig. S6B]. Moreover, only a very faint NBD-GlcCer signal is now detected in glia. This suggests that glial cells trigger the transport of NBD-GlcCer from labeled neurons and that they may be able to uptake GlcCer for degradation [Fig. 6C (g and h) and fig. S6B].
If glial cells are able to take up the NBD-GlcCer released from labeled neurons, reduction of GBA1 in glia should lead to an accumulation of GlcCer. We therefore knocked down GBA1 in glia using two independent siRNAs (fig. S6C). We then overlaid the labeled neuronal cells with these glial cells to determine whether they accumulate GlcCer that is taken up from the medium. As shown in Fig. 6C, we observed a robust accumulation of NBD-GlcCer in these glial cells [Fig. 6C (k, l, o, and p) and fig. S6D], providing compelling evidence that NBD-GlcCer from labeled neurons is indeed transported to glia and that GBA1 plays a role in the degradation process. In summary, these data show that GlcCer is transported from neurons to glia and that this process is evolutionally conserved between flies and human cells.
TGF-β/BMP promotes the transfer of GlcCer from neurons to glia via exosomes
Given that GlcCer is transported from neurons to glia but not between neurons, we hypothesized that glia may release a signal that stimulates neurons to secrete GlcCer. We therefore collected conditioned medium from neuronal and glial cell cultures. When NBD-GlcCer–labeled neurons were exposed to conditioned artificial cerebrospinal fluid (ACSF) from neurons for 30 min, no obvious changes were observed at NBD-GlcCer levels (Fig. 6D, a and b). In contrast, conditioned ACSF medium from glia induced a severe reduction in NBD-GlcCer signal in labeled neurons (Fig. 6D, a and c). These data indicate that glia secrete a factor that induces the release NBD-GlcCer from labeled neurons.
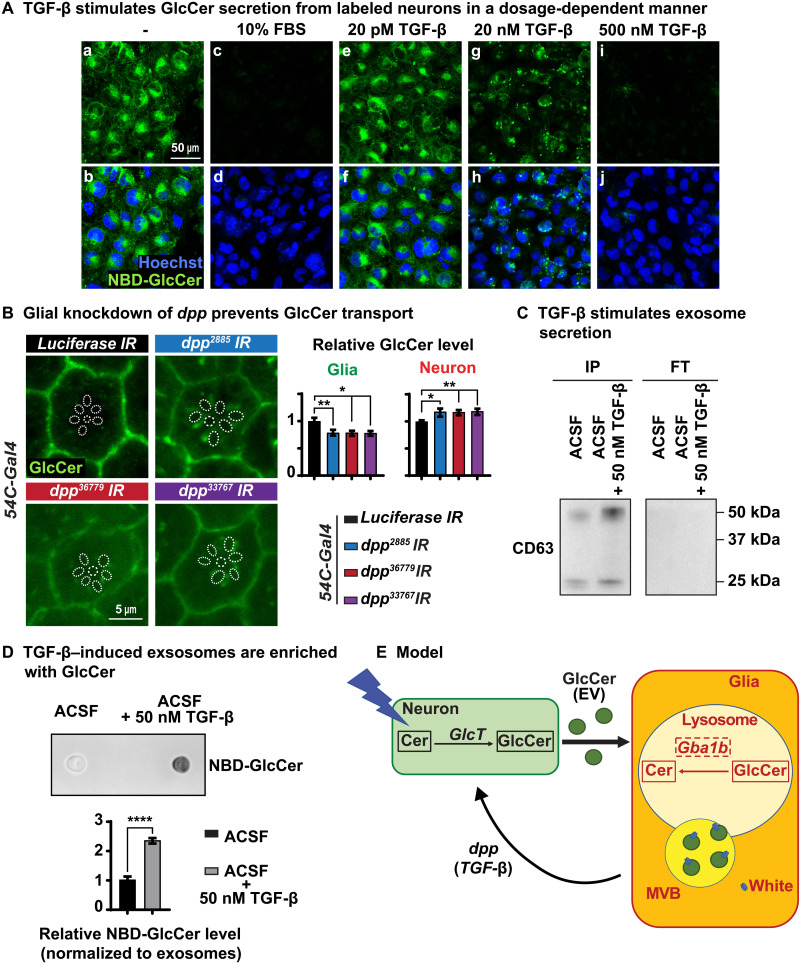
In all previous experiments, we used a defined medium: ACSF, which contains salts and glucose but no serum or growth factors. To explore whether cell signaling molecules present in other media are able to promote the release of GlcCer from neurons, we added 10% fetal bovine serum (FBS) to ACSF. To our surprise, ACSF containing 10% FBS led to a near-complete release of NBD-GlcCer from labeled neurons upon a 30-min exposure (Fig. 7A, a to d), suggesting that FBS contains key factors that can promote the release of NBD-GlcCer from neurons. FBS contains various growth factors including Insulin-like growth factor–binding protein 2 (IGFBP2), Transforming growth factor–β (TGF-β), and Glial growth factor (GGF), among other growth factors (57). In the Drosophila visual system, the cone and pigment cells that function as glial cells (39, 40) express elevated levels of dpp and daw, two genes encoding ligands of the TGF-β/BMP superfamily (fig. S7A) (49). Moreover, it has been reported that mammalian glia produce TGF-β1, which activates TGF-β signaling in adult mouse neurons (58). We therefore explored whether TGF-β can promote the transfer of GlcCer from neurons to glia.
Fig. 7. TGF-β triggers GlcCer secretion in the form of exosomes.
(A) Growth factor TGF-β stimulates the release of NBD-GlcCer of labeled neurons. NBD-GlcCer–labeled neurons were incubated with five different conditional ACSF media as indicated for 30 min, respectively. (a to d) ACSF with 10% FBS promotes the complete release of NBD-GlcCer from labeled neurons. (a, b, and e to j) ACSF with TGF-β triggers the release of NBD-GlcCer from labeled neurons in a dosage-dependent manner. (B) Glial knockdown of dpp causes elevated GlcCer in the photoreceptor neurons and reduced GlcCer level in the pigment glia. Relative GlcCer intensity is quantified on the right. Error bars represent SEM (n = 11), *P < 0.05 and **P < 0.01. (C) NBD-GlcCer–labeled neurons were incubated with two conditional ACSF media: ACSF only and ACSF with 50 nM TGF-β for 1 hour. Immunoprecipitation (IP) of CD63-positive exosomes from two conditional ACSF media as indicated. FT, flow-through. ACSF with 50 nM TGF-β contains significantly more exosomes from NBD-GlcCer–labeled neurons compared with ACSF only. (D) Exosomes released from NBD-GlcCer–labeled neurons are enriched with GlcCer. A dot blot was performed to determine the levels of NBD-GlcCer in exosomes isolated from the two conditional ACSF media in (C). Hence, ACSF with 50 nM TGF-β contains many more exosomes, and these exosomes are enriched with NBD-GlcCer. (E) Working model. GlcCer is synthesized in neurons upon neuronal activity. It is then transported to glia via exosomes for lysosomal degradation by Gba1b. TGF-β/Dpp secreted from glia is sufficient to stimulate neuronal release of exosomes, which are enriched with GlcCer. white (ABCG) plays a major role in glia in the degradation of GlcCer and is associated with MVBs, and loss of both white and Gba1b synergizes and exacerbates the accumulation of GlcCer, suggesting that both participate independently in the degradation of GlcCer.
We first tested whether human TGF-β was able to trigger the release of GlcCer from labeled neurons in our cell culture system. TGF-β (20 pM) did not affect the release of NBD-GlcCer (Fig. 7A, e and f), whereas 500 nM TGF-β led to a near-complete loss of NBD-GlcCer from labeled neurons (Fig. 7A, i and j). At lower doses of 20 nM, a dose in the physiological range (59), we observe a redistribution and a reduction of NBD-GlcCer in neurons (Fig. 7A, g and h). Note that these assays were performed after a 30-min exposure. Hence, these data show that the addition of TGF-β is sufficient to promote the release of GlcCer from labeled neurons in a short time frame.
To determine whether glial TGF-β signaling can stimulate GlcCer secretion in flies, we specifically reduced the level of expression of dpp in the pigment glia using three different RNAi lines including a documented RNAi line (60). As shown in Fig. 7B, glial knockdown of dpp causes a mild but significant elevation of GlcCer in photoreceptor neurons as well as a mild but significant reduction of GlcCer level in pigment glia. These data suggest that a glial-expressed dpp is involved in GlcCer release from neurons in Drosophila. In summary, the data indicate that TGF-β/BMP signals produced in glia promote the release of GlcCer produced in neurons.
To determine the mechanism by which GlcCer is transported from neurons to glia, we first explored the role of two apolipoproteins encoded by Neuronal Lazarillo and Glial Lazarillo (39, 61). We previously showed that Glial Lazarillo plays a critical role in the transport of neutral lipids from neurons to glia in the fly visual system (61). However, as shown in fig. S7B, a severe loss of these proteins induced by RNAi did not affect the levels of GlcCer in neurons and glia, suggesting that GlcCer transport from neurons to glia may not rely on lipoprotein particles.
Previous work has shown increased numbers of extracellular vesicles (EVs) in heads of Gba1b mutant flies (62), suggesting that increased levels of GlcCer lead to an increase in EVs. Moreover, Corrigan et al. (63) presented compelling data that Drosophila male reproductive glands secrete exosomes under the regulation of BMPs, members of TGF-β superfamily. Hence, GlcCer may be transported from neurons to glia via exosomes. To determine whether this is the case, we isolated CD63-positive exosomes (64) from ACSF media that were used to culture NBD-GlcCer–labeled neurons. As shown in Fig. 7C, Western blots show an increase in CD63-positive exosomes in ACSF upon the addition of 50 nM TGF-β when compared to ACSF without TGF-β. Dot blots of these CD63-positive exosomes exposed to TGF-β are highly enriched in NBD-GlcCer (Fig. 7D). Together, these data support the idea that TGF-β triggers GlcCer release from neurons to glia via exosomes.
To determine whether TGF-β promotes GlcCer-enriched exosome secretion, we purified exosomes from media of cells that were treated with and without 50 nM TGF-β for 1 hour and performed TEM. TEM images showed that vesicles purified from media of neurons that were treated with TGF-β have a diameter of around 80 nm (fig. S7C, b). However, no vesicles were detected from media of neurons that were not treated with TGF-β (fig. S7C, a). These data further support the notion that TGF-β triggers exosome secretion. Using the same experimental paradigm, we performed lipidomics on the neuronal media. As shown in fig. S7D, we observed significantly increased levels of ceramide and GlcCer in the cell culture media treated with TGF-β when compared to the nontreated cell culture media, indicating that TGF-β promotes ceramide and GlcCer secretion. No significant difference was observed in the cell pellets for ceramide and GlcCer, although they showed a consistent decrease for both (fig. S7D). In the context of the observations with the CD63-positive vesicles and the TEM data, we conclude that GlcCer-containing exosomes are secreted upon a TGF-β signal.
Given that exosomes are released by the fusion of MVBs with the plasma membrane and given that large MVBs are often observed by TEM in Drosophila photoreceptors, we performed RNAi experiments to reduce the levels of three ESCORT complex proteins in photoreceptor neurons: CHMP2B, Mvb12, and lsn. We assessed the levels of GlcCer in glia and observed no accumulation of GlcCer after 15 days of D/L cycles (fig. S7E). Hence, these data are consistent with the above observations that GlcCer is transported via exosomes from neurons to glia. Note that the levels of GlcCer in neurons are also decreased when we reduced the levels of these ESCORT complex proteins specifically in neurons. This is also consistent with the previous observations that ESCORT complex affects GlcCer synthesis (65, 66).
DISCUSSION
Here, we show that GlcCer, the substrate of Gba1b in flies and GBA1 in humans, is produced in neurons upon neuronal activity in flies and is transported from neurons to glia via exosomes. Upon exposure to TGF-β/BMP signals produced in glia, GlcCer-enriched exosomes are released from neurons, and these exosomes are taken up by glia. We propose that GlcCer is then endocytosed by glia where it is degraded in the lysosomes through the action of Gba1b/GBA1. We also show that Drosophila Gba1b is expressed in glia in the CNS during all developmental stages as well as in adults. Similarly, GBA1 protein levels are also much higher in human glial cells than neurons. The ABCG transporter, White, plays an important role in the degradation of GlcCer in pigment glia in flies and is present in MVBs (Fig. 7E).
Gba1b is necessary and sufficient in Drosophila glia to support neuronal function
We show that Gba1b is required in glia to support neuronal function and that the human GBA1, when expressed in glial cells, rescues the null mutant phenotype. However, the mutant GBA1N370S variant associated with GD and PD (20) fails to rescue Gba1b mutant phenotypes (21). Our data argue that expression of Gba1b in glia is necessary and sufficient to maintain proper neuronal activity.
Ultrastructural TEM images demonstrate that loss of Gba1b leads to impaired and vacuolized pigment glia before neuronal degeneration. These glial cells accumulate elevated levels of GlcCer and exhibit lysosomal expansion based on costaining with the lysosomal marker CTSL. Glial GlcCer accumulation impairs the morphology of glia first and is followed by the demise of neurons. Although most studies on GBA1 and GD focus on defects in neurons, a study by Keatinge et al. (24) reported that activation of microglia precedes neuronal demise in zebrafish. Knocking out Gba in both neurons and glia of mice driven by Nestin-Cre leads to paralysis 21 days after birth (27), whereas a knockdown of Gba in mice dopaminergic neurons causes robust microglial activation but does not affect motor behavior and does not cause dopaminergic neuron loss (67). These results again suggest that GBA1 may play a more important role in glia than in neurons in the pathogenesis of neurodegenerative diseases. However, we show that human GBA1 (Fig. 6) is present in neurons at relatively low levels when compared to glia. In addition, neuronal functions of GBA1 in some neurons have previously been reported (68–70). However, our data and these vertebrate studies show that GBA1 is present in glial cells as well (68–70). In summary, our data show that Gba1b plays an essential role in glia and is required for the maintenance of neurons in Drosophila.
Neuronal activity promotes GlcCer production
UDP-glucose:ceramide glucosyltransferase (UGCG, GlcT in flies) catalyzes the synthesis of GlcCer, and its loss leads to embryonic lethality (71). The conditional and selective removal of Ugcg in mouse neurons and glia causes severe neural abnormalities after birth and lethality at weaning (72). The synthesis of GlcCer has been shown to be triggered by endotoxin lipopolysaccharides as they induce Ugcg mRNA transcription (73). Increased levels of Ugcg mRNA have been shown in tumor cells (74). However, GlcCer synthesis has not been linked to neuronal activity to our knowledge. Our data show that y1 w* or y1 w*; Gba1bT2A-Gal4 mutant flies that are kept in the dark do not accumulate GlcCer in photoreceptor neurons or pigment glia. However, upon light exposure, y1 w* ommatidia show an accumulation of GlcCer in neurons and glia, but the accumulation of GlcCer is obviously greater in y1 w*; Gba1bT2A-Gal4 than in y1 w* ommatidia. When we trigger neuronal activity by activating Drosophila TrpA1 channel in the fly brain for 60 min, we observe a significant increase in GlcCer levels in most areas of the fly brain. Hence, these data show that neuronal activity promotes the production of GlcCer. Our data strongly indicate that neuronal activity promotes GlcCer synthesis. However, they do not exclude the possibility that neuronal activity inhibits GlcCer degradation.
The loss of the synthesis of GlcCer in glia, unlike the loss of GlcCer in neurons, is not associated with a phenotype. Similarly, loss of Ugcg in oligodendrocytes does not cause obvious phenotypes in mice (75), suggesting that GlcCer is produced by other cells and transported to glia (76). Consistently, we find that loss of GlcT, the fly homolog of UGCG, in glia does not affect the levels of GlcCer, whereas loss of GlcT in neurons strongly reduced the levels of GlcCer in neurons and glia. Neuronal activity leads to an increased level of GlcCer in neurons and glia. However, how the transport of GlcCer is regulated and what triggers the transport are unknown.
Glia secrete a TGF-β/BMP signal to promote the release of GlcCer-enriched exosomes
We found that only neuron-glia but not neuron-neuron coculture stimulates NBD-GlcCer release from labeled neurons. Moreover, conditional ACSF from glia but not from neurons promotes NBD-GlcCer release from neurons. These results implicate that molecular cues that are secreted from glia trigger GlcCer release. Our data show that TGF-β is sufficient to promote the release of NBD-GlcCer from neurons. After exposing the labeled neurons to TGF-β, we observed a very significant increase in CD63-positive exosomes and an increase in NBD-GlcCer in the exosomal fraction. Similarly, when neurons are not labeled but stimulated with TGF-β, we observe an increase in GlcCer in the culture media based on lipidomic assays. Given that lysosomal enzymes have been previously shown to be shared among nearby cells (77) and that Gba1b can be transported between cells via exosomes in flies (78), we also assessed whether GBA1 was present in the media or the exosomes produced by the neurons. However, we did not detect GBA1 proteins in exosomes or culture media. Hence, a significant fraction of neuronal GlcCer must be transported through exosomes to glia upon TGF-β exposure.
dpp and daw, two homologs of TGF-β/MBP superfamily, are expressed in the pigment glia but not in photoreceptors (fig. S7A) (49). A reduction of the expression of dpp in pigment glia leads to an increase in GlcCer in neurons, consistent with the data observed with cultured neurons. daw may also play a role in stimulating GlcCer secretion, and the observed defects may be enhanced by the loss of daw. In summary, our data argue that the transfer for exosomes containing GlcCer is an evolutionarily conserved process that is based on a TGF-β/BMP signaling.
Increased numbers of EVs were reported in heads of Gba1b mutant flies (62), suggesting that increased levels of GlcCer lead to an increase in EVs. Exosomes secreted by neurons have been shown to facilitate the clearance of β-amyloid. Moreover, several studies suggest that there is a positive correlation between the levels of GlcCer and α-synuclein aggregation (20, 79), and loss of GBA1 enhances α-synuclein transmission between cells (80). In addition, neuron-to-glia transport of α-synuclein has been reported, but the precise mechanisms are yet to be elucidated (81). In Drosophila, overexpression of α-synuclein in photoreceptor neurons causes increased numbers of MVBs (82). These data indicate that neuron-to-glia transport of α-synuclein may share the same exosomes as those that transport GlcCer, and given the elevated levels of GlcCer in glia, aggregation of α-synuclein in glia may be favored. It has been argued that glial cells play an important role in the progression of PD. α-Synuclein accumulates in astrocytes and has been associated with the recruitment of phagocytic microglia that engulf selected neurons in restricted brain regions possibly causing the clinical symptoms associated with PD (83).
White promotes GlcCer transport to lysosomes in glia
white encodes a transmembrane ABCG transporter (84). Its cellular and molecular functions are not well characterized, although it is the most commonly used marker in fly research. white is expressed in Drosophila pigment glia and at much lower levels in photoreceptor neurons (30, 31). White is required for the formation of pigment granules in pigment glia and photoreceptors. RNAi-mediated knockdown of white in pigment glia results in a severe loss of eye pigmentation (fig. S4). Loss of white also reduces life span by about 10%, affects climbing ability starting at day 30, and causes a progressive retinal degeneration (51).
A reduction of white expression using RNAi in glia alone leads to glial expansion and accumulation of GlcCer after 3 days of a D/L cycle. However, this phenotype is strongly exacerbated by the loss of Gba1b. These data suggest that white is required for the degradation of GlcCer in glia and that its loss synergizes with the loss of Gba1b, arguing that both proteins have separate functions that converge on GlcCer degradation. The White protein colocalizes with the MVB marker Hrs but not with the lysosomal marker CTSL, indicating that White is associated with membrane trafficking in the MVBs. Moreover, White proteins together with MVBs are relocated to lysosomes after GlcCer treatment (fig. S5B), suggesting that White modulates GlcCer trafficking to lysosome through a pathway that relies on MVBs. We propose that both white and Gba1b are required for the degradation of GlcCer and that in the absence of one or the other, some degradation occurs but that in the absence of both, no degradation of GlcCer occurs, leading to a highly toxic combination that will quickly affect neuronal function and strongly accelerate the phenotypes. This also raises the possibility that other ABCG transporters may participate in GlcCer degradation in other glial cells. In summary, we propose that active neurons produce GlcCer. Glial cells provide a TGF-β/BMP signal that stimulates GlcCer secretion of exosomes from neurons that are then endocytosed by glial cells for degradation by Gba1b and White.
MATERIALS AND METHODS
Reagents and resources
Experimental model and subject details
Drosophila
Flies were raised on molasses-based food at 25°C in constant dark unless otherwise noted. The full list of genotypes of fly stocks used can be found in the Key Resources Table.
Mammalian cell culture
Daoy and MO3.13 cells were used in this study. Cells were maintained in Dulbecco’s modified Eagle’s medium with GlutaMAX medium supplemented with 10% FBS, 1× pyruvate, and 1× penicillin and streptomycin.
Insect cell culture
S2 cells were used in this study. Cells were maintained in Schneider’s Drosophila Medium supplemented with 10% FBS and 1× penicillin and streptomycin (Tables 1 to 5).
Table 1. Experimental models: Organisms/strains.
BDSC, Bloomington Drosophila Stock Center; VDRC, Vienna Drosophila Resource Center; N/A, not applicable.
| Source | Identifier | |
|
y1 w*; TI{GFP[3xP3.cLa]=CRIMIC.TG4.0}Gba1bCR00541-TG4.0/ TM3, Sb1 Ser1 |
BDSC | FBti0199405 |
| w*; P{w[+mC]=UAS-mCherry.NLS}3 | BDSC | FBst0038424 |
| w1118; dGBA1b−/−, we renamed it to Gba1bSTOP/STOP | N/A | Gift from K. Kinghorn (22) |
|
w*; P{w[+mC]=UAS-RedStinger}4, P{w[+mC]=UAS- FLP.D}JD1, P{w[+mC]=Ubi-p63E(FRT.STOP) Stinger}9F6/CyO |
BDSC | G-TRACE, FBst0028280 |
| y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.JF01355}attP2 | BDSC | Luciferase RNAi, FBti0130444 |
|
y1 sc* v1 sev21; P{y[+t7.7] v[+t1.8]=TRiP.HMS01893} attP40 |
BDSC | Gba1b RNAi, FBst0038977 |
| P{KK107189}VIE-260B | VDRC | Gba1b RNAi, FBst0473085 |
| y1 w*; P{w[+m*]=GAL4}54C | BDSC | 54C-Gal4, FBti0100692 |
| y1 w*; Pbac{UAS-GBA1}/SM6a | This study | N/A |
| w*; Pbac{UAS-GBA1N370S}/CyO, Gal80; TM6,Tb/+ | N/A | Gift from L. Pallanck (21) |
| w1118; Df(3R)BSC490/TM6C, Sb1 cu1 | BDSC | Deficiency line for Gba1b, FBab0045306 |
|
y1 sc* v1 sev21; P{y[+t7.7] v[+t1.8]=TRiP.HMC06408} attP40 |
BDSC | GlcT RNAi, FBti0185552 |
| w1118; P{GD2142}v44912 | VDRC | GlcT RNAi, FBst0465833 |
| y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.HMS00017}attP2 | BDSC | w RNAi, FBti0140096 |
|
y1 sc* v1 sev21; P{y[+t7.7] v[+t1.8]=TRiP.HMC06329} attP40 |
BDSC | Glaz RNAi, FBti0185474 |
| P{KK107553}VIE-260B | VDRC | Nlaz RNAi, FBst0473194 |
| y1 w*; PBac{CH322-118C10}VK00037 | This study | 20-kb BAC genomic rescue fragment for Gba1b |
| y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.HMS01844}attP40 | BDSC | CHMP2B RNAi, FBti0149590 |
| y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.HMS02004}attP40 | BDSC | Mvb12 RNAi, FBti0149746 |
|
y1 sc* v1 sev21; P{y[+t7.7] v[+t1.8]=TRiP.HMS01747} attP40 |
BDSC | lns RNAi, FBti0149494 |
| UAS-dTrpA1 | N/A | Gift from P. Garrity (46) |
| w*; UAS-dpp-RNAi-1+2-homo/CyO;TM6b (2885) | N/A | dpp RNAi, gift from K. Wharton (60) |
| y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.JF02455}attP2 | BDSC | dpp RNAi, FBti0146826 |
| y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.JF02794}attP2 | BDSC | dpp RNAi, FBti0141012 |
Table 5. Primers and recombinant DNA and siRNA.
| Source | Identifier | |
|---|---|---|
| GTGCTCCCGACTGGCAGTTGC | This study | Gba1b PP1 forward primer |
| ACAGGCTTTGGGGAATGTTGG | This study | Gba1b PP1 reverse primer |
| GTACACTTCATGAGCATGGGCTG | This study | Gba1b PP2 forward primer |
| TGCAGGACTCCGTGTTCAATAGC | This study | Gba1b PP2 reverse primer |
| Plasmid: pUASTattb_white-HA | This study | N/A |
| Plasmid: pUASTattb_GBA1 | This study | N/A |
| Glucosylceramidase beta siRNA #1 | Thermo Fisher Scientific | s501314 |
| Glucosylceramidase beta siRNA #2 | Horizon Inspired Cell Solutions | J-006366-07-0005 |
Table 2. Experimental models: Cell lines.
ATCC, American Type Culture Collection.
| Source | Identifier | |
|---|---|---|
|
Drosophila melanogaster: Cell line S2: S2-DRSC |
Laboratory of N. Perrimon |
CVCL_Z232; FlyBase: FBtc0000181 |
| MO3.13 | Cedarlane Labs | cat#CLU301 |
| Daoy | ATCC | cat#ATCC-HTB-186 |
Table 3. Antibodies.
| Source | Identifier | |
|---|---|---|
| Rat anti-elav | DSHB | AB_528218 |
| Mouse anti-Repo | DSHB | AB_528448 |
| Rabbit anti-Glc-Cer | Glycobiotech | cat#RAS_0010 |
| Phalloidin 488 nm | Thermo Fisher Scientific | AB_2315147 |
| Mouse anti-ATP5α | Abcam | AN_301447 |
| DAPI | Thermo Fisher Scientific | AB_2629482 |
| Alexa 488–conjugated secondary antibodies |
Jackson ImmunoResearch Labs |
AB_2338059 |
| Alexa Cy3–conjugated secondary antibodies |
Jackson ImmunoResearch Labs |
AB_2338013 |
| Alexa Cy5–conjugated secondary antibodies |
Bioss Inc. | AB_11117143 |
| Rabbit anti-Rab5 | Abcam | ab31261 |
| Mouse anti-CTSL | R&D Systems | MAB22591 |
| Mouse anti-Rab7 | DSHB | AB_2722471 |
| Guinea pig anti–FL-Hrs | Bellen laboratory | GP30 |
| Mouse monoclonal anti-HA |
Covance | AB_10064220 |
| Rabbit anti-HA | Sigma-Aldrich | H6908 |
| C6-NBD-Glucosylceramide | Cayman Chemical | 23209 |
| C6-Biotin-Glucosylceramide | Cayman Chemical | 23208 |
| Hoechst 33342 | Invitrogen | H3570 |
| Mouse anti-CD63 | Invitrogen | Ts63 |
Table 4. Chemicals.
EMS, Electron Microscopy Sciences; Embed 812, EMBED 812 RESIN; NMA, (methyl-5-norbornene-2,3-dicarboxylic anhydride); DDSA, dodecenyl succinic anhydride specially distilled; DMP-30, DMP30.
| Source | Identifier | |
|---|---|---|
| VECTASHIELD | Vector Labs | cat#H-1000 |
| RapiClear | SunJin Lab Co. | N/A |
| Pierce Protease Inhibitor Tablets, EDTA-free |
Thermo Fisher Scientific |
cat#88266 |
| Schneider’s Drosophila medium | Thermo Fisher Scientific |
cat#21720 |
| Fetal bovine serum, heat- inactivated |
Sigma-Aldrich | cat#F4135 |
| Penicillin streptomycin | Thermo Fisher Scientific |
cat#15070063 |
| Cacodylic acid, trihydrate sodium 100 g |
EMS | cat#12300 |
| EM-grade glutaraldehyde, 25% Aq solution |
EMS | cat#16221 |
| Osmium tetroxide 4% Aq solution |
EMS | cat#19191 |
| Paraformaldehyde 16% Aq solution |
EMS | cat#15711 |
| Propylene oxide | EMS | cat#20411 |
| Koptec 200 Proof 100% ethanol anhydrous |
VWR | cat#89125-186 |
| Embed-812 | EMS | cat#14901 |
| NMA | EMS | cat#19001 |
| DDSA | EMS | cat#13711 |
| DMP-30 | EMS | cat#13600 |
| Uranyl acetate | EMS | cat#RT22400 |
| Lead nitrate | EMS | cat#RT17900-25 |
| Western Lightning Plus-ECL | PerkinElmer | cat#NEL105001EA |
| Recombinant human TGF-beta 3 protein |
R&D Systems | 243-B3 |
Method details
Life span
All flies of genotypes indicated in Fig. 1B were collected at eclosion and divided into multiple vials (20 flies per vial). They were raised at 25°C in a 12-hour dark/12-hour light cycle. All flies were transferred to fresh vials, and the number of dead flies was counted every 3 days. Survival rates were quantified from the total population.
Immunostaining
For immunostaining of fly retina, heads were dissected in cold 1× phosphate-buffered saline (PBS) and fixed in 3.7% formaldehyde diluted in PBS overnight at 4°C. On the second day, the retinae were dissected in cold PBS and fixed for an additional 30 min in 3.7% formaldehyde diluted in 1× PBS. Subsequently, the samples were washed with 0.1% Triton X-100 diluted in 1× PBS (0.1% PBST afterward). Washed samples were blocked in 5% natural goat serum (NGS) diluted in 0.1% PBST (5% NGST afterward) for 1 hour at room temperature (RT). After blocking, the samples were incubated overnight at 4°C with the following primary antibodies: rabbit anti-GlcCer (1:250; RAS_0010, Glycobiotech) and mouse anti-CTSL (1:1000; MAB22591, R&D Systems). On the third day, the samples were washed with 0.1% PBST, followed by incubation with Alexa 488–, Cy3-, or Cy5-conjugated secondary antibodies (111-545-144, 111-585-003, and 111-175-144, Jackson ImmunoResearch Labs and bs-2673R-Cy5.5, Bioss Inc.) and phalloidin 488 (AB_2315147, Thermo Fisher Scientific) for 1 hour at RT. After incubation with secondary antibodies, the samples were washed with 0.1% PBST. Last, the samples were mounted with VECTASHIELD (H-1000, Vector Laboratories) and kept at −20°C before imaging under a confocal microscope. All the confocal images were acquired with a Leica SP8 confocal microscope. All retinal confocal images were taken 5 μm below the surface. The fluorescent signals were separately quantified by outlining the neuronal or glial area, respectively. Confocal images were processed using Fuji (ImageJ) and Photoshop (Adobe).
For immunostaining of fly brains, larval and adult brains were dissected in cold 1× PBS and fixed in 4% paraformaldehyde diluted in 1× PBS overnight at 4°C. On the second day, the fixed brains were permeabilized with 2% Triton X-100 diluted in 1× PBS overnight at 4°C. The samples were blocked in 5% NGST under vacuum at RT for 2 hours. After blocking, the samples were incubated overnight at 4°C with the following primary antibodies: rat anti-elav [1:250; Rat-elav-7E8A10, DSHB (Developmental Studies Hybridoma Bank)] and mouse anti-Repo (1:50; Mouse-Repo-8D12, DSHB). Then, the samples were washed with 0.1% PBST, followed by incubation with Alexa 488–, Cy3-, or Cy5-conjugated secondary antibodies (111-545-144, 111-585-003, and 111-175-144, Jackson ImmunoResearch Labs and bs-2673R-Cy5.5, Bioss Inc.) for 1 hour at RT. Last, the samples were mounted with RapiClear (SunJin Lab Co.) and kept at −20°C before imaging under a confocal microscope. All the confocal images were acquired with a Leica SP8 confocal microscope. Confocal images were processed using Fuji (ImageJ) and Photoshop (Adobe).
For immunostaining of S2 cells, S2 cells were cultured on poly-d-lysine (PDL)–treated coverslips and fixed in 4% paraformaldehyde diluted in 1× PBS for 10 min at RT. Fixed cells were rinsed with 1× PBS followed by permeabilizing with 0.2% Triton X-100 diluted in 1× PBS for 10 min at RT. Subsequently, the cells were blocked with 1% bovine serum albumin diluted with 1× PBS for 1 hour at RT. After blocking, the samples were incubated overnight at 4°C with the following primary antibodies: mouse anti-Atp5α (1:250; Mouse-Repo-α5, DSHB), mouse anti-CTSL (1:1000; MAB22591, R&D Systems), rabbit anti-Rba5 (1:250; ab31261, Abcam), mouse anti-Rab7 (1:250; AB_2722471, DSHB), and guinea pig anti–FL-Hrs (1:250; GP30). Then, the samples were washed with 1× PBST, followed by incubation with Alexa 488–, Cy3-, or Cy5-conjugated secondary antibodies (111-545-144, 111-585-003, and 111-175-144, Jackson ImmunoResearch Labs and bs-2673R-Cy5.5, Bioss Inc.) for 1 hour at RT. Last, the samples were mounted with 4′,6-diamidino-2-phenylindole (DAPI) Fluoromount-G (SouthernBiotech) and kept at −20°C before imaging under a confocal microscope. All the confocal images were acquired with a Leica SP8 confocal microscope. Confocal images were processed using Fuji (ImageJ) and Photoshop (Adobe).
ERG recording
ERG recordings were performed by following the protocol documented in (85). Flies, as indicated in the manuscript, were glued on glass slides. Two electrodes were filled with 0.1 M NaCl. One of them was probed into the fly thorax, and the other one was placed on the fly eye. A 1-s pulse of light stimulation interval was shined. The ERG traces were recorded and analyzed by LabChart 8 Reader.
Transmission electron microscopy
The ultrastructure of the Drosophila visual system was processed following standard electron microscopy procedures as documented in (11). Adult heads were dissected and fixed in fixative [2% paraformaldehyde, 2.5% glutaraldehyde, 0.1 M sodium cacodylate, and 0.005% CaCl2 (pH 7.2)] at 4°C overnight and then postfixed in 1% OsO4. The fixed samples were dehydrated in ethanol and propylene oxide and then embedded in Embed-812 resin (Electron Microscopy Sciences) under vacuum. Photoreceptors were then sectioned and stained in 1% uranyl acetate and saturated lead nitrate. TEM images of photoreceptor sections were taken using a JEOL JEM 1010 transmission electron microscope at 80 kV with an AMT XR-16 mid-mount 16-megapixel digital camera.
Coculture assay
Cells were split and cultured on PDL-coated coverslips in 24-well plates at a density of 5 × 104 the day before the experiment. On the day of the assay, cells were washed with cold Hanks’ balanced salt solution (HBSS; Gibco). The cells were then incubated with 4 μM C6-NBD-GlcCer (Cayman) at 4°C for 30 min. C6-NBD-GlcCer is dissolved in cold HBSS by vortexing to create micelles. At 4°C, the plasma membrane of cells is labeled with C6-NBD-GlcCer. After labeling, the cells are rinsed with warm HBSS (37°C) to remove unlabeled C6-NBD-GlcCer. Subsequently, the cells were transferred in a cell culture incubator (37°C and 5% CO2) to avoid light for 30 min and allow C6-NBD-GlcCer to be taken up by cells. After C6-NBD-GlcCer labeling, a paraffin ring was placed on top of the labeled cells followed by placing nonlabeled cells on top of the paraffin ring. The two coverslips with labeled cells (beneath the paraffin ring) and nonlabeled cells (above the paraffin ring) were face-to-face with each other and cocultured in the warm (37°C) ACSF [125 mM NaCl, 5 mM KCl, 2 mM MgSO4, 24 mM NaHCO3, 1.25 mM NaH2PO4, 2 mM CaCl2, and 10 mM glucose (pH 7.4), filtered with a 0.22-nm filter] in a cell culture incubator (37°C and 5% CO2) to avoid light for 30 min. After coculture, cells were mounted in warm HBSS with Hoechst, and live images were taken immediately with a Leica SP8 confocal microscope.
Conditional ACSF media incubation assay
Cells were cultured in 15-cm dishes and reached confluency in normal cell culture media as mentioned above in the “Mammalian cell culture” section. Cells were then washed with warm ACSF twice and incubated with ACSF in a cell culture incubator (37°C and 5% CO2) overnight the day before the experiment. On the day of the assay, conditional ACSF media were harvested and filtered with 0.45-nm filters to remove cellular debris. Cells were labeled with C6-NBD-GlcCer as mentioned above. The labeled cells were then incubated with conditional ACSF media, as documented in Results, in a cell culture incubator (37°C and 5% CO2) to avoid light for 30 min. After conditional ACSF media incubation, cells were mounted in warm HBSS with Hoechst, and live images were taken immediately with a Leica SP8 confocal microscope.
FBS (10%) and TGF-β treatment assay
Cells were labeled with C6-NBD-GlcCer as mentioned above. Subsequently, labeled cells were incubated with 10% FBS or different concentrations of TGF-β diluted in warm ACSF in a cell culture incubator (37°C and 5% CO2) to avoid light for 30 min. After treatment, cells were mounted in warm HBSS with Hoechst, and live images were taken immediately with a Leica SP8 confocal microscope.
Exosome isolation assay
Cells were cultured on 10-cm plates to confluency (five plates per experimental condition). They were then labeled with C6-NBD-GlcCer. Subsequently, labeled cells were incubated with or without 50 nM TGF-β diluted in warm ACSF in a cell culture incubator (37°C and 5% CO2) to avoid light for 1 hour. After treatment, ACSF media were harvested, and total exosomes from the media were purified by using Total Exosome Isolation Reagent (Invitrogen). Then, CD63-positive exosomes were isolated from total exosomes by using Exosome-Human CD63 Isolation/Detection Reagent (Invitrogen).
Lipid extraction
Lipids were extracted as described previously (86). For hexosylceramide analysis from fly heads, deuterated GlcCer (d18:1/16:0 d3-GlcCer) was added to 1000 fly heads as an internal standard with 500 μl of 1-butanol:methanol (1:1, v/v). For multiplexed ceramides, hexosylceramides, and hexosylsphingosine analysis from Daoy cells and media, deuterated GlcCer (d18:1/16:0 d3-GlcCer), ceramide (d18:1 d7/18:0-Cer), and lyso globotriaosylsphingosine (lyso Gb3 d7) were added. Homogenates were sonicated for 60 min at RT and centrifuged at 13,000g for 10 min. The supernatant was dried using a vacuum centrifuge and reconstituted in chloroform:methanol (1:4, v/v) and centrifuged at 13,000g for 5 min to remove any insoluble salts. The supernatant was collected and injected for liquid chromatography–tandem mass spectrometry (LC-MS/MS) for lipid analysis.
Targeted analysis of lipids using LC-MS/MS
Vanquish Horizon UHPLC (Thermo Fisher Scientific, Waltham, MA) was coupled to an Orbitrap Fusion Tribrid ID-X mass spectrometer (Thermo Fisher Scientific, Waltham, MA) for analysis of lipids. Two targeted methods of lipids were developed using Hypersil Gold Vanquish C18 UHPLC column (2.1 mm × 15 cm, 1.9 μm), one for hexosylceramides and another for ceramides, hexosylceramides, sphingosine, and hexosylsphingosine. For both methods, a binary gradient of mobile phase A (water:acetonitrile, 6:4, v/v) and mobile phase B (isopropanol:methanol:acetonitrile, 8:1:1, v/v/v) with 0.1% formic acid and 10 mM ammonium formate at the flow rate of 300 μl/min was applied to separate lipids. Full-scan MS spectra at a resolution of 60,000 [mass/charge ratio (m/z) 200] and MS/MS spectra at a resolution of 15,000 (m/z 200) in scheduled parallel reaction monitoring mode were obtained with a column temperature of 50°C and a spray voltage of 3.5 kV in positive ion mode. Because of differences in target lipids between the two methods, slightly different parameters of LC-MS/MS including LC gradient, full-scan MS range, and collision energy in higher-energy collisional dissociation were applied.
For targeted analysis of hexosylceramides, mobile phase B was increased from 50 to 95% over 7 min, 95% for 6 min, 50% over 0.1 min, and 50% B for 4 min. A primary MS scan of 400 to 850 m/z was carried out to cover the range of hexosylceramides, and higher-energy collisional dissociation was set at 40%. For targeted analysis of multiplexed ceramides, hexosylceramides, and hexosylsphingosine, mobile phase B was increased from 20 to 60% over 2 min, 75% over 2.5 min, 85% over 1.5 min, 95% over 1 min, maintained at 95% for 6 min, 20% for 0.1 min, and equilibrated for 4 min. A full-scan MS of 250 to 900 m/z was acquired, and collision energy was set at 35% for ceramides and hexosylsphingosine and 40% for dihydroceramides and hexosylceramides.
Peak areas of lipids were calculated using Skyline (MacCoss Lab Software), normalized to the peak area of the deuterated internal standard, and corrected by the protein amount measured from the bicinchoninic acid assay. The total level of lipid for each class was calculated by adding the normalized peak areas of individual species.
Quantification and statistical analysis
All datasets were organized and analyzed in Microsoft Excel (version 2019) and GraphPad Prism (version 9.2.0) using two-tailed Student’s t test or one-way analysis of variance (ANOVA). For fly experiments, sample sizes are stated in the figure legends. All crosses were performed at least twice. For cell experiments, all studies were conducted for a minimum of three biological replicates. Error bars are shown as SEM. The criteria for significance are as follows: ns (not significant), P > 0.05; *P < 0.05; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We thank J. Shulman, S. Yamamoto, M. Wang, C. Wilson, H. Zoghbi, K. Kinghorn, and O. Kanca for insightful comments. We thank L. Pallanck, K. Kinghorn, K. Wharton, and P. Garrity for sharing fly stocks. We thank P. Marcogliese for cloning pUASTattb_GBA1 plasmid and M. Ma for cloning pUASTattb_white-HA plasmid. We thank H. Pan for injections to create transgenic flies. We thank the Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center for providing stocks and reagents. We acknowledge support from HHMI, the Jan and Dan Duncan Neurological Research Institute, and the Huffington Foundation.
Funding: This work was supported by the Huffington Foundation.
Author contributions: Conceptualization: L.W., G.L., and H.J.B. Methodology: L.W., G.L., Z.Z., Y.L., S.K.B., and A.P. Investigation: L.W., Z.Z., Y.L., S.K.B., and A.P. Visualization: L.W. and Z.Z. Supervision: G.L. and H.J.B. Writing—original draft: L.W. and H.J.B. Writing—review and editing: L.W., G.L., and H.J.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
References
REFERENCES AND NOTES
- 1.Chaurasia B., Summers S. A., Ceramides in metabolism: Key lipotoxic players. Annu. Rev. Physiol. 83, 1–28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraft M. L., Sphingolipid organization in the plasma membrane and the mechanisms that influence it. Front. Cell Dev. Biology. 4, 154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslow D. K., Weissman J. S., Membranes in balance: Mechanisms of sphingolipid homeostasis. Mol. Cell 40, 267–279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gault C. R., Obeid L. M., Hannun Y. A., An overview of sphingolipid metabolism: From synthesis to breakdown. Adv. Exp. Med. Biol. 688, 1–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kudo N., Kumagai K., Tomishige N., Yamaji T., Wakatsuki S., Nishijima M., Hanada K., Kato R., Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc. Natl. Acad. Sci. U.S.A. 105, 488–493 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao R. P., Yuan C., Allegood J. C., Rawat S. S., Edwards M. B., Wang X., Merrill A. H., Acharya U., Acharya J. K., Ceramide transfer protein function is essential for normal oxidative stress response and lifespan. Proc. Natl. Acad. Sci. U.S.A. 104, 11364–11369 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goñi F. M., Alonso A., Effects of ceramide and other simple sphingolipids on membrane lateral structure. Biochim. Biophys. Acta B Biomembr. 1788, 169–177 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Wang T.-Y., Silvius J. R., Sphingolipid partitioning into ordered domains in cholesterol-free and cholesterol-containing lipid bilayers. Biophys. J. 84, 367–378 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiantia S., Kahya N., Ries J., Schwille P., Effects of ceramide on liquid-ordered domains investigated by simultaneous AFM and FCS. Biophys. J. 90, 4500–4508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vos M., Dulovic-Mahlow M., Mandik F., Frese L., Kanana Y., Diaw S. H., Depperschmidt J., Böhm C., Rohr J., Lohnau T., König I. R., Klein C., Ceramide accumulation induces mitophagy and impairs β-oxidation in PINK1 deficiency. Proc. Natl. Acad. Sci. U.S.A. 118, e2025347118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin G., Lee P.-T., Chen K., Mao D., Tan K. L., Zuo Z., Lin W.-W., Wang L., Bellen H. J., Phospholipase PLA2G6, a parkinsonism-associated gene, affects Vps26 and Vps35, retromer function, and ceramide levels, similar to α-synuclein gain. Cell Metab. 28, 605–618.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Rohrbough J., Rushton E., Palanker L., Woodruff E., Matthies H. J. G., Acharya U., Acharya J. K., Broadie K., Ceramidase regulates synaptic vesicle exocytosis and trafficking. J. Neurosci. 24, 7789–7803 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allende M. L., Proia R. L., Simplifying complexity: Genetically resculpting glycosphingolipid synthesis pathways in mice to reveal function. Glycoconj. J. 31, 613–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robak L. A., Jansen I. E., van Rooij J., Uitterlinden A. G., Kraaij R., Jankovic J., Heutink P., Shulman J. M.; International Parkinson’s Disease Genomics Consortium (IPDGC), Nalls M. A., Plagnol V., Hernandez D. G., Sharma M., Sheerin U.-M., Saad M., S.-Sánchez J., Schulte C., Lesage S., Sveinbjörnsdóttir S., Arepalli S., Barker R., Ben Y., Berendse H. W., Berg D., Bhatia K., de Bie R. M. A., Biffi A., Bloem B., Bochdanovits Z., Bonin M., Bras J. M., Brockmann K., Brooks J., Burn D. J., Majounie E., Charlesworth G., Lungu C., Chen H., Chinnery P. F., Chong S., Clarke C. E., Cookson M. R., Cooper J. M., Corvol J. C., Counsell C., Damier P., Dartigues J.-F., Deloukas P., Deuschl G., Dexter D. T., van Dijk K. D., Dillman A., Durif F., Dürr A., Edkins S., Evans J. R., Foltynie T., Dong J., Gardner M., Gibbs J. R., Goate A., Gray E., Guerreiro R., Harris C., van Hilten J. J., Hofman A., Hollenbeck A., Holton J., Hu M., Huang X., Wurster I., Mätzler W., Hudson G., Hunt S. E., Huttenlocher J., Illig T., Jónsson P. V., Lambert J.-C., Langford C., Lees A., Lichtner P., Limousin P., Lopez G., Lorenz D., McNeill A., Moorby C., Moore M., Morris H. R., Morrison K. E., Escott-Price V., Mudanohwo E., O’Sullivan S. S., Pearson J., Perlmutter J. S., Pétursson H., Pollak P., Post B., Potter S., Ravina B., Revesz T., Riess O., Rivadeneira F., Rizzu P., Ryten M., Sawcer S., Schapira A., Scheffer H., Shaw K., Shoulson I., Shulman J., Sidransky E., Smith C., Spencer C. C. A., Stefánsson H., Bettella F., Stockton J. D., Strange A., Talbot K., Tanner C. M., Tashakkori-Ghanbaria A., Tison F., Trabzuni D., Traynor B. J., Uitterlinden A. G., Velseboer D., Vidailhet M., Walker R., van de Warrenburg B., Wickremaratchi M., Williams N., Williams-Gray C. H., Winder-Rhodes S., Stefánsson K., Martinez M., Wood N. W., Hardy J., Heutink P., Brice A., Gasser T., Singleton A. B., Excessive burden of lysosomal storage disorder gene variants in Parkinson’s disease. Brain 140, 3191–3203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin G., Wang L., Marcogliese P. C., Bellen H. J., Sphingolipids in the pathogenesis of Parkinson’s disease and parkinsonism. Trends Endocrinol. Metab. 30, 106–117 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Sidransky E., Gaucher disease: Complexity in a “simple” disorder. Mol. Genet. Metab. 83, 6–15 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Wong K., Sidransky E., Verma A., Mixon T., Sandberg G. D., Wakefield L. K., Morrison A., Lwin A., Colegial C., Allman J. M., Schiffmann R., Neuropathology provides clues to the pathophysiology of Gaucher disease. Mol. Genet. Metab. 82, 192–207 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Chang D., Nalls M. A., Hallgrímsdóttir I. B., Hunkapiller J., van der Brug M., Cai F.; International Parkinson’s Disease Genomics Consortium, 23andMe Research Team, Kerchner G. A., Ayalon G., Bingol B., Sheng M., Hinds D., Behrens T. W., Singleton A. B., Bhangale T. R., Graham R. R., A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidransky E., Nalls M. A., Aasly J. O., Aharon-Peretz J., Annesi G., Barbosa E. R., Bar-Shira A., Berg D., Bras J., Brice A., Chen C.-M., Clark L. N., Condroyer C., Marco E. V. D., Dürr A., Eblan M. J., Fahn S., Farrer M. J., Fung H.-C., Gan-Or Z., Gasser T., Gershoni-Baruch R., Giladi N., Griffith A., Gurevich T., Januario C., Kropp P., Lang A. E., Lee-Chen G.-J., Lesage S., Marder K., Mata I. F., Mirelman A., Mitsui J., Mizuta I., Nicoletti G., Oliveira C., Ottman R., Orr-Urtreger A., Pereira L. V., Quattrone A., Rogaeva E., Rolfs A., Rosenbaum H., Rozenberg R., Samii A., Samaddar T., Schulte C., Sharma M., Singleton A., Spitz M., Tan E.-K., Tayebi N., Toda T., Troiano A. R., Tsuji S., Wittstock M., Wolfsberg T. G., Wu Y.-R., Zabetian C. P., Zhao Y., Ziegler S. G., Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 361, 1651–1661 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Do J., McKinney C., Sharma P., Sidransky E., Glucocerebrosidase and its relevance to Parkinson disease. Mol. Neurodegener. 14, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis M. Y., Trinh K., Thomas R. E., Yu S., Germanos A. A., Whitley B. N., Sardi S. P., Montine T. J., Pallanck L. J., Glucocerebrosidase deficiency in drosophila results in α-synuclein-independent protein aggregation and neurodegeneration. PLOS Genet. 12, e1005944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinghorn K. J., Grönke S., Castillo-Quan J. I., Woodling N. S., Li L., Sirka E., Gegg M., Mills K., Hardy J., Bjedov I., Partridge L., A drosophila model of neuronopathic gaucher disease demonstrates lysosomal-autophagic defects and altered mTOR signalling and is functionally rescued by rapamycin. J. Neurosci. 36, 11654–11670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabasso O., Paul S., Dorot O., Maor G., Krivoruk O., Pasmanik-Chor M., Mirzaian M., Ferraz M., Aerts J., Horowitz M., Drosophila melanogaster mutated in its GBA1b ortholog recapitulates neuronopathic gaucher disease. J. Clin. Med. 8, 1420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keatinge M., Bui H., Menke A., Chen Y.-C., Sokol A. M., Bai Q., Ellett F., Costa M. D., Burke D., Gegg M., Trollope L., Payne T., McTighe A., Mortiboys H., de Jager S., Nuthall H., Kuo M.-S., Fleming A., Schapira A. H. V., Renshaw S. A., Highley J. R., Chacinska A., Panula P., Burton E. A., O’Neill M. J., Bandmann O., Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum. Mol. Genet. 24, 6640–6652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinclair G. B., Jevon G., Colobong K. E., Randall D. R., Choy F. Y. M., Clarke L. A., Generation of a conditional knockout of murine glucocerebrosidase: Utility for the study of Gaucher disease. Mol. Genet. Metab. 90, 148–156 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Osellame L. D., Rahim A. A., Hargreaves I. P., Gegg M. E., Richard-Londt A., Brandner S., Waddington S. N., Schapira A. H. V., Duchen M. R., Mitochondria and quality control defects in a mouse model of gaucher disease—Links to Parkinson’s disease. Cell Metab. 17, 941–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enquist I. B., Bianco C. L., Ooka A., Nilsson E., Månsson J.-E., Ehinger M., Richter J., Brady R. O., Kirik D., Karlsson S., Murine models of acute neuronopathic Gaucher disease. Proc. Natl. Acad. Sci. U.S.A. 104, 17483–17488 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.modENCODE Consortium, Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., Washietl S., Arshinoff B. I., Ay F., Meyer P. E., Robine N., Washington N. L., Stefano L. D., Berezikov E., Brown C. D., Candeias R., Carlson J. W., Carr A., Jungreis I., Marbach D., Sealfon R., Tolstorukov M. Y., Will S., Alekseyenko A. A., Artieri C., Booth B. W., Brooks A. N., Dai Q., Davis C. A., Duff M. O., Feng X., Gorchakov A. A., Gu T., Henikoff J. G., Kapranov P., Li R., MacAlpine H. K., Malone J., Minoda A., Nordman J., Okamura K., Perry M., Powell S. K., Riddle N. C., Sakai A., Samsonova A., Sandler J. E., Schwartz Y. B., Sher N., Spokony R., Sturgill D., van Baren M., Wan K. H., Yang L., Yu C., Feingold E., Good P., Guyer M., Lowdon R., Ahmad K., Andrews J., Berger B., Brenner S. E., Brent M. R., Cherbas L., Elgin S. C. R., Gingeras T. R., Grossman R., Hoskins R. A., Kaufman T. C., Kent W., Kuroda M. I., Orr-Weaver T., Perrimon N., Pirrotta V., Posakony J. W., Ren B., Russell S., Cherbas P., Graveley B. R., Lewis S., Micklem G., Oliver B., Park P. J., Celniker S. E., Henikoff S., Karpen G. H., Lai E. C., MacAlpine D. M., Stein L. D., White K. P., Kellis M., Acevedo D., Auburn R., Barber G., Bellen H. J., Bishop E. P., Bryson T. D., Chateigner A., Chen J., Clawson H., Comstock C. L. G., Contrino S., DeNapoli L. C., Ding Q., Dobin A., Domanus M. H., Drenkow J., Dudoit S., Dumais J., Eng T., Fagegaltier D., Gadel S. E., Ghosh S., Guillier F., Hanley D., Hannon G. J., Hansen K. D., Heinz E., Hinrichs A. S., Hirst M., Jha S., Jiang L., Jung Y. L., Kashevsky H., Kennedy C. D., Kephart E. T., Langton L., Lee O.-K., Li S., Li Z., Lin W., Linder-Basso D., Lloyd P., Lyne R., Marchetti S. E., Marra M., Mattiuzzo N. R., McKay S., Meyer F., Miller D., Miller S. W., Moore R. A., Morrison C. A., Prinz J. A., Rooks M., Moore R., Rutherford K. M., Ruzanov P., Scheftner D. A., Senderowicz L., Shah P. K., Shanower G., Smith R., Stinson E. O., Suchy S., Tenney A. E., Tian F., Venken K. J. T., Wang H., White R., Wilkening J., Willingham A. T., Zaleski C., Zha Z., Zhang D., Zhao Y., Zieba J., Identification of functional elements and regulatory circuits by drosophila modENCODE. Science 330, 1787–1797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chintapalli V. R., Wang J., Dow J. A. T., Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Borycz J., Borycz J. A., Kubów A., Lloyd V., Meinertzhagen I. A., Drosophila ABC transporter mutants white, brown and scarlet have altered contents and distribution of biogenic amines in the brain. J. Exp. Biol. 211, 3454–3466 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie S. M., Howells A. J., Cox G. B., Ewart G. D., Sub-cellular localisation of the white/scarlet ABC transporter to pigment granule membranes within the compound eye of Drosophila melanogaster. Genetica 108, 239–252 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Lee P.-T., Zirin J., Kanca O., Lin W.-W., Schulze K. L., Li-Kroeger D., Tao R., Devereaux C., Hu Y., Chung V., Fang Y., He Y., Pan H., Ge M., Zuo Z., Housden B. E., Mohr S. E., Yamamoto S., Levis R. W., Spradling A. C., Perrimon N., Bellen H. J., A gene-specific T2A-GAL4 library for Drosophila. eLife 7, e35574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diao F., White B. H., A novel approach for directing transgene expression in drosophila: T2A-Gal4 in-frame fusion. Genetics 190, 1139–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryder E., Ashburner M., Bautista-Llacer R., Drummond J., Webster J., Johnson G., Morley T., Chan Y. S., Blows F., Coulson D., Reuter G., Baisch H., Apelt C., Kauk A., Rudolph T., Kube M., Klimm M., Nickel C., Szidonya J., Maróy P., Pal M., Rasmuson-Lestander Å., Ekström K., Stocker H., Hugentobler C., Hafen E., Gubb D., Pflugfelder G., Dorner C., Mechler B., Schenkel H., Marhold J., Serras F., Corominas M., Punset A., Roote J., Russell S., The DrosDel deletion collection: A Drosophila genomewide chromosomal deficiency resource. Genetics 177, 615–629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venken K. J. T., Carlson J. W., Schulze K. L., Pan H., He Y., Spokony R., Wan K. H., Koriabine M., de Jong P. J., White K. P., Bellen H. J., Hoskins R. A., Versatile P(acman) BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao K., White K., Neural specificity of elav expression: Defining a Drosophila promoter for directing expression to the nervous system. J. Neurochem. 63, 41–51 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Stork T., Bernardos R., Freeman M. R., Analysis of glial cell development and function in Drosophila. Cold Spring Harb. Protoc. 2012, 1–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans C. J., Olson J. M., Ngo K. T., Kim E., Lee N. E., Kuoy E., Patananan A. N., Sitz D., Tran P., Do M.-T., Yackle K., Cespedes A., Hartenstein V., Call G. B., Banerjee U., G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 6, 603–605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., Sanz E., Li Z., Hui J., Graham B. H., Quintana A., Bellen H. J., Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlton-Perkins M. A., Sendler E. D., Buschbeck E. K., Cook T. A., Multifunctional glial support by Semper cells in the Drosophila retina. PLOS Genet. 13, e1006782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Nagaraj R., Banerjee U., Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development 134, 825–831 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Dolph P., Nair A., Raghu P., Electroretinogram recordings of Drosophila. Cold Spring Harb. Protoc. 2011, pdb.prot5550 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Alcalay R. N., Levy O. A., Waters C. H., Fahn S., Ford B., Kuo S.-H., Mazzoni P., Pauciulo M. W., Nichols W. C., Gan-Or Z., Rouleau G. A., Chung W. K., Wolf P., Oliva P., Keutzer J., Marder K., Zhang X., Glucocerebrosidase activity in Parkinson’s disease with and without GBA mutations. Brain 138, 2648–2658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roessingh S., Stanewsky R., The Drosophila TRPA1 channel and neuronal circuits controlling rhythmic behaviours and sleep in response to environmental temperature. Int. J. Mol. Sci. 18, 2028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X., Rahman R., Guo F., Rosbash M., Genome-wide identification of neuronal activity-regulated genes in Drosophila. eLife 5, e19942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verderame M., Alcorta D., Egnor M., Smith K., Pollack R., Cytoskeletal F-actin patterns quantitated with fluorescein isothiocyanate-phalloidin in normal and transformed cells. Proc. Natl. Acad. Sci. U.S.A. 77, 6624–6628 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spira D., Stypmann J., Tobin D. J., Petermann I., Mayer C., Hagemann S., Vasiljeva O., Günther T., Schüle R., Peters C., Reinheckel T., Cell type-specific functions of the lysosomal protease cathepsin L in the heart*. J. Biol. Chem. 282, 37045–37052 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Li H., Janssens J., Waegeneer M. D., Kolluru S. S., Davie K., Gardeux V., Saelens W., David F. P. A., Brbić M., Spanier K., Leskovec J., McLaughlin C. N., Xie Q., Jones R. C., Brueckner K., Shim J., Tattikota S. G., Schnorrer F., Rust K., Nystul T. G., Carvalho-Santos Z., Ribeiro C., Pal S., Mahadevaraju S., Przytycka T. M., Allen A. M., Goodwin S. F., Berry C. W., Fuller M. T., White-Cooper H., Matunis E. L., DiNardo S., Galenza A., O’Brien L. E.; J. A. T. Dow, F. C. A. Consortium§, Jasper H., Oliver B., Perrimon N., Deplancke B., Quake S. R., Luo L., Aerts S., Agarwal D., Ahmed-Braimah Y., Arbeitman M., Ariss M. M., Augsburger J., Ayush K., Baker C. C., Banisch T., Birker K., Bodmer R., Bolival B., Brantley S. E., Brill J. A., Brown N. C., Buehner N. A., Cai X. T., Cardoso-Figueiredo R., Casares F., Chang A., Clandinin T. R., Crasta S., Desplan C., Detweiler A. M., Dhakan D. B., Donà E., Engert S., Floc’hlay S., George N., González-Segarra A. J., Groves A. K., Gumbin S., Guo Y., Harris D. E., Heifetz Y., Holtz S. L., Horns F., Hudry B., Hung R.-J., Jan Y. N., Jaszczak J. S., Jefferis G. S. X. E., Karkanias J., Karr T. L., Katheder N. S., Kezos J., Kim A. A., Kim S. K., Kockel L., Konstantinides N., Kornberg T. B., Krause H. M., Labott A. T., Laturney M., Lehmann R., Leinwand S., Li J., Li J. S. S., Li K., Li K., Li L., Li T., Litovchenko M., Liu H.-H., Liu Y., Lu T.-C., Manning J., Mase A., Matera-Vatnick M., Matias N. R., McDonough-Goldstein C. E., McGeever A., McLachlan A. D., Moreno-Roman P., Neff N., Neville M., Ngo S., Nielsen T., O’Brien C. E., Osumi-Sutherland D., Özel M. N., Papatheodorou I., Petkovic M., Pilgrim C., Pisco A. O., Reisenman C., Sanders E. N., dos Santos G., Scott K., Sherlekar A., Shiu P., Sims D., Sit R. V., Slaidina M., Smith H. E., Sterne G., Su Y.-H., Sutton D., Tamayo M., Tan M., Tastekin I., Treiber C., Vacek D., Vogler G., Waddell S., Wang W., Wilson R. I., Wolfner M. F., Wong Y.-C. E., Xie A., Xu J., Yamamoto S., Yan J., Yao Z., Yoda K., Zhu R., Zinzen R. P., Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375, eabk2432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarr P. T., Tarling E. J., Bojanic D. D., Edwards P. A., Baldán Á., Emerging new paradigms for ABCG transporters. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 584–593 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreiro M. J., Pérez C., Marchesano M., Ruiz S., Caputi A., Aguilera P., Barrio R., Cantera R., Drosophila melanogaster white mutant w1118 undergo retinal degeneration. Front. Neurosci. 11, 732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lloyd T. E., Atkinson R., Wu M. N., Zhou Y., Pennetta G., Bellen H. J., Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108, 261–269 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Booth H. D. E., Hirst W. D., Wade-Martins R., The role of astrocyte dysfunction in Parkinson’s disease pathogenesis. Trends Neurosci. 40, 358–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O’Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., Barres B. A., Wu J. Q., An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casciati A., Tanori M., Manczak R., Saada S., Tanno B., Giardullo P., Porcù E., Rampazzo E., Persano L., Viola G., Dalmay C., Lalloué F., Pothier A., Merla C., Mancuso M., Human medulloblastoma cell lines: Investigating on cancer stem cell-like phenotype. Cancer 12, 226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boscia F., D’Avanzo C., Pannaccione A., Secondo A., Casamassa A., Formisano L., Guida N., Sokolow S., Herchuelz A., Annunziato L., Silencing or knocking out the Na+/Ca2+ exchanger-3 (NCX3) impairs oligodendrocyte differentiation. Cell Death Differ. 19, 562–572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng X., Baker H., Hancock W. S., Fawaz F., McCaman M., Pungor E., Proteomic analysis for the assessment of different lots of fetal bovine serum as a raw material for cell culture. Part IV. Application of proteomics to the manufacture of biological drugs. Biotechnol. Prog. 22, 1294–1300 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Luo J., Ho P. P., Buckwalter M. S., Hsu T., Lee L. Y., Zhang H., Kim D.-K., Kim S.-J., Gambhir S. S., Steinman L., Wyss-Coray T., Glia-dependent TGF-β signaling, acting independently of the TH17 pathway, is critical for initiation of murine autoimmune encephalomyelitis. J. Clin. Invest. 117, 3306–3315 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox D. A., Transforming growth factor-beta 3. Cell Biol. Int. 19, 357–371 (1995). [DOI] [PubMed] [Google Scholar]
- 60.Haley B., Hendrix D., Trang V., Levine M., A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 321, 482–490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu L., MacKenzie K. R., Putluri N., Maletić-Savatić M., Bellen H. J., The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 26, 719–737.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas R. E., Vincow E. S., Merrihew G. E., MacCoss M. J., Davis M. Y., Pallanck L. J., Glucocerebrosidase deficiency promotes protein aggregation through dysregulation of extracellular vesicles. PLOS Genet. 14, e1007694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corrigan L., Redhai S., Leiblich A., Fan S.-J., Perera S. M. W., Patel R., Gandy C., Wainwright S. M., Morris J. F., Hamdy F., Goberdhan D. C. I., Wilson C., BMP-regulated exosomes from Drosophila male reproductive glands reprogram female behavior. J. Cell Biol. 206, 671–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Logozzi M., Milito A. D., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M. L., Federici C., Iessi E., Brambilla D., Venturi G., Lozupone F., Santinami M., Huber V., Maio M., Rivoltini L., Fais S., High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLOS ONE 4, e5219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt O., Weyer Y., Baumann V., Widerin M. A., Eising S., Angelova M., Schleiffer A., Kremser L., Lindner H., Peter M., Fröhlich F., Teis D., Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. EMBO J. 38, e101433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt O., Weyer Y., Sprenger S., Widerin M. A., Eising S., Baumann V., Angelova M., Loewith R., Stefan C. J., Hess M. W., Fröhlich F., Teis D., TOR complex 2 (TORC2) signaling and the ESCRT machinery cooperate in the protection of plasma membrane integrity in yeast. J. Biol. Chem. 295, 12028–12044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soria F. N., Engeln M., Martinez-Vicente M., Glangetas C., López-González M. J., Dovero S., Dehay B., Normand E., Vila M., Favereaux A., Georges F., Bianco C. L., Bezard E., Fernagut P.-O., Glucocerebrosidase deficiency in dopaminergic neurons induces microglial activation without neurodegeneration. Hum. Mol. Genet. 26, 2603–2615 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Henderson M. X., Sedor S., McGeary I., Cornblath E. J., Peng C., Riddle D. M., Li H. L., Zhang B., Brown H. J., Olufemi M. F., Bassett D. S., Trojanowski J. Q., Lee V. M. Y., Glucocerebrosidase activity modulates neuronal susceptibility to pathological α-synuclein insult. Neuron 105, 822–836.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calandri I. L., Hawkes M. A., Marrodan M., Ameriso S. F., Correale J., Allegri R. F., The impact of an early strict nationwide lockdown on the pattern of consultation for neurological diseases. J. Neurol. Sci. 418, 117084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chao D. H. M., Kallemeijn W. W., Marques A. R. A., Orre M., Ottenhoff R., van Roomen C., Foppen E., Renner M. C., Moeton M., van Eijk M., Boot R. G., Kamphuis W., Hol E. M., Aten J., Overkleeft H. S., Kalsbeek A., Aerts J. M. F. G., Visualization of active glucocerebrosidase in rodent brain with high spatial resolution following in situ labeling with fluorescent activity based probes. PLOS ONE 10, e0138107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamashita T., Wada R., Sasaki T., Deng C., Bierfreund U., Sandhoff K., Proia R. L., A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. U.S.A. 96, 9142–9147 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jennemann R., Sandhoff R., Wang S., Kiss E., Gretz N., Zuliani C., Martin-Villalba A., Jäger R., Schorle H., Kenzelmann M., Bonrouhi M., Wiegandt H., Gröne H.-J., Cell-specific deletion of glucosylceramide synthase in brain leads to severe neural defects after birth. Proc. Natl. Acad. Sci. U.S.A. 102, 12459–12464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brennan P. J., Tatituri R. V. V., Brigl M., Kim E. Y., Tuli A., Sanderson J. P., Gadola S. D., Hsu F.-F., Besra G. S., Brenner M. B., Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat. Immunol. 12, 1202–1211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y.-Y., Patwardhan G. A., Xie P., Gu X., Giuliano A. E., Cabot M. C., Glucosylceramide synthase, a factor in modulating drug resistance, is overexpressed in metastatic breast carcinoma. Int. J. Oncol. 39, 425–431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saadat L., Dupree J. L., Kilkus J., Han X., Traka M., Proia R. L., Dawson G., Popko B., Absence of oligodendroglial glucosylceramide synthesis does not result in CNS myelin abnormalities or alter the dysmyelinating phenotype of CGT-deficient mice. Glia 58, 391–398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Radin N. S., Glucosylceramide in the nervous system—A mini-review. Neurochem. Res. 19, 533–540 (1994). [DOI] [PubMed] [Google Scholar]
- 77.Mikulka C. R., Dearborn J. T., Benitez B. A., Strickland A., Liu L., Milbrandt J., Sands M. S., Cell-autonomous expression of the acid hydrolase galactocerebrosidase. Proc. Natl. Acad. Sci. U.S.A. 117, 9032–9041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jewett K. A., Thomas R. E., Phan C. Q., Lin B., Milstein G., Yu S., Bettcher L. F., Neto F. C., Djukovic D., Raftery D., Pallanck L. J., Davis M. Y., Glucocerebrosidase reduces the spread of protein aggregation in a Drosophila melanogaster model of neurodegeneration by regulating proteins trafficked by extracellular vesicles. PLOS Genet. 17, e1008859 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gegg M. E., Schapira A. H. V., The role of glucocerebrosidase in Parkinson disease pathogenesis. FEBS J. 285, 3591–3603 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Bae E.-J., Yang N.-Y., Song M., Lee C. S., Lee J. S., Jung B. C., Lee H.-J., Kim S., Masliah E., Sardi S. P., Lee S.-J., Glucocerebrosidase depletion enhances cell-to-cell transmission of α-synuclein. Nat. Commun. 5, 4755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reyes J. F., Rey N. L., Bousset L., Melki R., Brundin P., Angot E., Alpha-synuclein transfers from neurons to oligodendrocytes. Glia 62, 387–398 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Chouhan A. K., Guo C., Hsieh Y.-C., Ye H., Senturk M., Zuo Z., Li Y., Chatterjee S., Botas J., Jackson G. R., Bellen H. J., Shulman J. M., Uncoupling neuronal death and dysfunction in Drosophila models of neurodegenerative disease. Acta Neuropathol. Commun. 4, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halliday G. M., Stevens C. H., Glia: Initiators and progressors of pathology in Parkinson’s disease. Mov. Disord. 26, 6–17 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Mackenzie S. M., Brooker M. R., Gill T. R., Cox G. B., Howells A. J., Ewart G. D., Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim. Biophys. Acta B Biomembr. 1419, 173–185 (1999). [DOI] [PubMed] [Google Scholar]
- 85.Wang S., Tan K. L., Agosto M. A., Xiong B., Yamamoto S., Sandoval H., Jaiswal M., Bayat V., Zhang K., Charng W.-L., David G., Duraine L., Venkatachalam K., Wensel T. G., Bellen H. J., The retromer complex is required for rhodopsin recycling and its loss leads to photoreceptor degeneration. PLoS Biol. 12, e1001847 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alshehry Z. H., Barlow C. K., Weir J. M., Zhou Y., McConville M. J., Meikle P. J., An efficient single phase method for the extraction of plasma lipids. Metabolites 5, 389–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strauss O., The retinal pigment epithelium in visual function. Physiol. Rev. 85, 845–881 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
References