Abstract
The transmembrane (TM) channel-like 1 (TMC1) and TMC2 proteins play a central role in auditory transduction, forming ion channels that convert sound into electrical signals. However, the molecular mechanism of their gating remains unknown. Here, using predicted structural models as a guide, we probed the effects of 12 mutations on the mechanical gating of the transduction currents in native hair cells of Tmc1/2-null mice expressing virally introduced TMC1 variants. Whole-cell electrophysiological recordings revealed that mutations within the pore-lining TM4 and TM6 helices modified gating, reducing the force sensitivity or shifting the open probability of the channels, or both. For some of the mutants, these changes were accompanied by a change in single-channel conductance. Our observations are in line with a model wherein conformational changes in the TM4 and TM6 helices are involved in the mechanical gating of the transduction channel.
Structural modeling, mutagenesis, and hair-cell physiology elucidate aspects of inner-ear mechanosensory channel gating.
INTRODUCTION
Our sense of hearing relies on the conversion of sound-induced mechanical stimuli into neural signals by cochlear hair cells of the inner ear. This process hinges on the ability of sensory transduction channels in hair cells to respond to mechanical stimuli by opening and closing, thereby generating fluctuating receptor currents (1). Despite decades of research, it was difficult to determine the molecular identity of the transduction channel.
Experiments with spontaneous and engineered mutations of the transmembrane (TM) channel-like 1 (TMC1) protein have now indicated that TMC1 (and likely TMC2) is a pore-forming protein of the mechanotransduction complex (2, 3). TMCs can assemble as dimers in solution and are structurally related to the TMEM16 (transmembrane protein 16) and TMEM63 / OSCA (hyperosmolality-gated calcium-permeable channel) proteins (3–7). Each subunit of the dimer is thought to contain 10 TM helices and encompass a peripheral ion-conduction pathway (fig. S1) (3, 5, 8). Channel activity of TMC homologs expressed in heterologous cell lines (9) is also consistent with the idea that TMC1 and TMC2 constitute the transduction channel pore.
These experiments did not, however, shed light on gating: on the mechanically evoked conformational changes that open and close the pore. Conformational states associated with conduction have been suggested for the related ion channels and lipid scramblases of the TMEM16 family. Specifically, in TMEM16 chloride channels and lipid scramblases, Ca2+-activated opening is thought to involve an initial rearrangement of the “gating helix” TM6 (10, 11). Channel opening further requires the disruption of interactions between TM4 and TM6 (12, 13).
In this study, we investigate the relationship between the structure of TMC1 and the mechanical activation of the hair-cell transduction channel. We used structure-prediction algorithms to further explore potential conformations of TMC1. On the basis of these, we designed 12 distinct mutations in TM4 and TM6 that might affect gating (table S1). We used an AAV (adeno-associated virus) gene delivery approach (3) to express mutant TMC1 channels in hair cells of Tmc1/2 double-knockout (DKO) mice. We then recorded currents from cochlear hair cells while simultaneously stimulating them with probes to deflect stereocilia (14). Comparing the mechanical activation curves of mutated TMC1 channels with those of wild-type (WT)–TMC1, we provide evidence for a model of transduction wherein conformational changes involving TM4 and TM6 of TMC1 are critical for channel opening induced by hair bundle deflection.
RESULTS
Predicted TMC1 structures
To identify residues that may influence gating, we began with our previous homology model for TMC1 (3). We then generated more refined models using the deep learning–based modeling algorithms, transform-restrained (tr) Rosetta (15) and RoseTTAFold (16) (see Materials and Methods; Fig. 1A). These methods rely on patterns in protein sequences and on amino acid interactions suggested by coevolution and use energy minimization techniques (17) to predict protein structures. Without relying on structures of related proteins, both trRosetta and RoseTTAFold predict an overall “TMEM16-like” fold for TMC1 that is very similar to that previously predicted from homology to TMEM16s (Fig. 1A) (3).
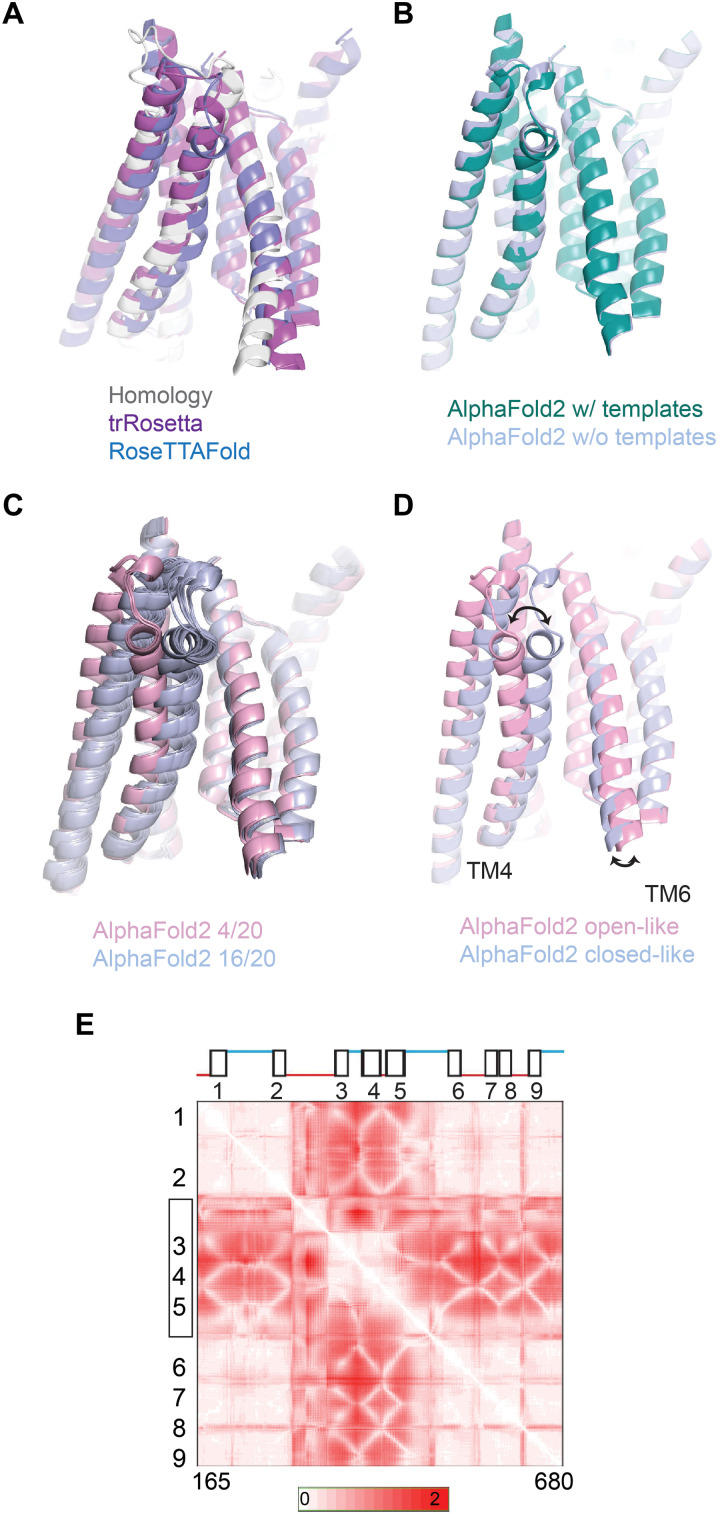
Fig. 1. Similarity in TMC1 structural predictions.
(A) Structural models for mouse TMC1. Shown in foreground are TM3, TM4, TM6, and TM8 viewed from within the membrane plane. Gray represents an I-TASSER model based on homology to TMEM16 channels. Purple shows a prediction by trRosetta. Blue is a template-free prediction by RoseTTAFold. The three models are nearly identical. (B) Structural model for TMC1 from AlphaFold2 is shown in green. The template-free AlphaFold 2 model is shown in blue. In the pore region, they are the same within 1- to 2-Å root mean square deviation. (C) Multiple structural models from AlphaFold2, generated with multiple seed parameters to achieve diversity. In 20 model iterations, 16 grouped in one conformation [blue; similar to that in (B)] and 4 grouped in a different conformation (pink). (D) Representative models from the two groups of conformations in (C). The conduction pathway for ions is between TM4 and TM6. In the more open conformation (pink), TM4 is more distant from TM6 by about 5 Å at the constricted region of the pore. (E) SD maps for all residue-residue distances for residues 165 to 680. For each residue pair, more intense red represents a larger change in distance between the two conformations. TM helices are numbered. The N terminus, C terminus, and TM10 were not included in the analysis.
We then performed predictions of the TMC1 structure using the AlphaFold2 program developed by DeepMind (18), (19) (Fig. 1B). The predictions with or without using a structural template were nearly identical and were very similar to previous models, especially regarding the overall fold of the pore region (Fig. 1B). AlphaFold2 reports high confidence scores for this region (fig. S2, A and B). There is now an experimentally determined x-ray crystal structure capturing TMC1’s TM2-TM3 loop together with CIB3 (calcium and integrin binding family member 3) (20), which is in line with what AlphaFold2 predicts for the unique fold of the TM2-TM3 loop domain in TMC1, further corroborating our confidence in the modeling (fig. 2C).
The models converge on a pore region in each subunit of the TMC1 dimer that is formed by TM4 to TM7 and exhibits an overall negative electrostatic potential, which is consistent with the cation selectivity of the channel (21–23). Several conserved residues within TM6 to 7 of mTMC1 contribute to this negative surface charge, including E514, E520, D528, D540, D557, E559, D569, and E567. Mutations of some of these residues, including D528 and D569, have been linked to hearing loss (24, 25).
Running the AlphaFold2 network multiple times using different seeds, we were able to increase the diversity of the predicted structural states (Fig. 1C). We generated 20 models (four random seeds and 5 models from each) and then aligned and clustered these structures using Chimera (29). By calculating the root mean square deviation values for every pairwise comparison between the 20 models, we found the structures clustered into two groups (16 structures in one group and 4 in the other). Representative structures from each group (shown in Fig. 1D) suggest a movement of TM4 relative to TM6, allowing the pore to widen or narrow (movie S1). In the first group, TM3 and TM4 were close to TM6 (“closed-like”; Fig. 1D). In the second, TM3 and TM4 maintained a close association with each other but were further from TM6 (“open-like”). The distance between the C-α atoms of residues 409 (on TM4) and 531 (on TM6) within the pore region is larger by ~5 Å in the open-like structures. The HOLE software (26) showed a water-filled pore conformation with a large conduction pathway for the open-like, but not closed-like, predicted structures (fig. 3A). The narrowest part of the pathway in this model occurs where N404 in TM4 approaches the positively charged R523 and the hydrophobic L524 in TM6. A thin cross section of the protein parallel to the membrane near N404 and L524 displays the opening clearly (fig. S3B). Notably, the change between the predicted states seems to be driven primarily not only by a ~10° tilting and translation of TM4 together with TM3 but also by smaller movements in TM6 (Fig. 1D). These predicted open-like structures are different from both the intermediate and open states of the TMEM16 scramblases (fig. S3D) (11, 13, 27) and display a pore diameter of ~6 Å in the narrowest dimension, which is smaller than that estimated for this channel on the basis of the sizes of the permeating molecules (5, 28).
We then generated 160 structures (32 random but distinct seeds and 5 models each) and mapped residue-residue distances to compare the AlphaFold2-generated structures (29). These maps showed that across the 160 structures, there was a large variability in the position of the TM3 to 5 helices with respect to the rest of the protein (TM1 to TM2 and TM6 to TM9; Fig. 1E). Most of the intracellular TM2-TM3 loop shows variability with the rest of the protein, as does the first half of the TM5-TM6 extracellular loop. However, these two loops also show variability with TM3 to 5. If the differences between the open-like and closed-like structures predicted by AlphaFold2 in fact represent a gating transition, then there may be three distinct domain movements associated with gating.
We also generated contact maps by EVcouplings software (30) (see Materials and Methods) and mapped them onto the open-like and closed-like structure predictions. Evolutionarily coupled residue pairs (ECs) are found in close proximity in the models, as expected (fig. S3C). However, there are notably more contacts (not just ECs) in the closed-like state (fig. S3C).
While these predicted structures will need to be validated in detail with experimental techniques such as x-ray crystallography or cryo–electron microscopy, they suggest a model wherein TMC1 pore helices TM4 and TM6, as in the structurally similar proteins of the TMEM16 and TMEM63/OSCA families, separate during channel gating (11, 31, 32).
Activation curves of virally expressed TMC1 in cochlear hair cells of Tmc1/2-DKO animals
If the pore helices TM4 and TM6 undergo a global rearrangement during gating, then mutations of critical residues within these helices may change the activation of the channel. We therefore designed and assessed the effects of mutations in this region of TMC1 (Fig. 2, A and B, and table S1). We generated AAV9-PHP.B viral vectors carrying a sequence encoding either WT or mutated TMC1 (Fig. 2C). Viral vectors were individually injected into the inner ears of Tmc1/2-DKO mice at postnatal day 1 (P1), using the round window approach (see Materials and Methods; Fig. 2C). Cochlear dissections were performed at P4 to P6. The tissue was placed in culture for an additional 3 to 7 days (Fig. 2C, bottom). The mid-basal section was used for electrophysiology, and the mid-apical section was used to assess viral transduction efficiency and functional rescue using an FM1-43 dye loading assay (see Materials and Methods). At this stage of cochlear development, Tmc2 is mostly down-regulated, but we used Tmc1/2-DKO animals to avoid the possibility of any contribution of TMC2 to transduction currents.
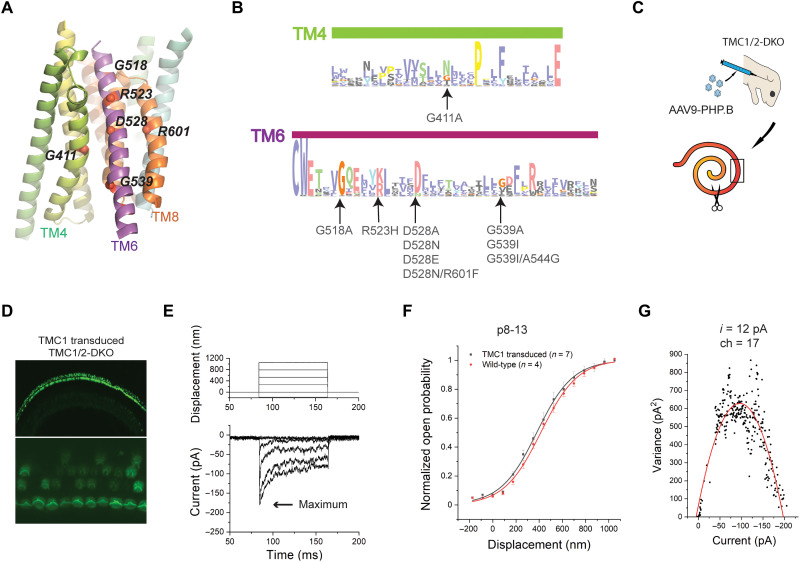
Fig. 2. Structure-based mutations of TMC1 and functional assays.
(A) Locations of the mutation sites in a view of the pore region from the plane of the membrane. (B) A sequence logo generated from multiple sequence alignments for TM4 and TM6 regions of TMCs; mutations in this study were indicated. The residues are colored according to the ClustalX coloring scheme. (C) Cartoon demonstrating AAV injection of neonatal Tmc1/2-DKO animals (top) and separation of the dissected cochlear explants into two sections (bottom). The scissors indicate where the tissue was split. The black box represents the mid-base region, used for electrophysiological recordings of transduction currents. (D) FM1-43 loading of cochlear HCs from Tmc1/2-DKO mice expressing virally expressed WT-TMC1 channels. The bottom panel is a magnified view at the plane of the bundles. (E) Representative transduction currents in response to −175- to 1050-nm bundle deflections (top) recorded from DKO IHCs expressing virally encoded WT-TMC1. (F) Activation curves. Normalized open probability curves are shown as a function of stimulus displacement (nanometers), in WT mice and Tmc1/2-DKO mice expressing WT-TMC1. The curves are fitted with a Boltzmann equation. (G) Single-channel current estimate (i = 12 pA) and number of channels (ch = 17) based on nonstationary noise analysis from a hair cell expressing viral WT-TMC1. Variance is plotted as a function of current and fitted with a parabolic equation.
Consistent with previous findings, we observed that cochlear hair cells of Tmc1/2-DKO mice do not take up FM1-43, indicating that they lack functional transduction channels (33, 34). In contrast, Tmc1/2-DKO hair cells transduced with AAV9-PHP.B-Tmc1 encoding WT-TMC1 showed FM1-43 uptake, indicating rescue of transduction activity (Fig. 2D). Specifically, the FM1-43 assay indicated that ~90% of inner hair cells (IHCs) expressed functional TMC1 channels. This is in line with recent studies demonstrating the use of AAV9-PHP.B vectors with cytomegalovirus (CMV) promoters as highly efficient for driving expression in hair cells (35, 36).
Following FM1-43 screening, electrophysiological recordings were performed using a stiff glass probe to stimulate hair bundles and whole-cell patch clamping to record currents, as previously described (3). First, we recorded currents from IHCs of WT mice at P8 to P13. Maximum current amplitudes of 660 ± 80 pA (n = 4) were obtained. In contrast, DKO hair cells expressing virally expressed WT-TMC1 channels yielded receptor currents with maximal amplitudes of 202 ± 33 pA, n = 7 (Fig. 2E). The amplitude and variability in the whole-cell currents were consistent with previous reports (3, 35).
To obtain activation curves for TMC1 channels, we measured IHC currents evoked by a series of 15 bundle step deflections ranging from −175 to 1050 nm (see Materials and Methods). Because the shape and slope of the activation curves reflect properties of individual channels, we normalized the currents for each cell to allow comparison to other cells (Fig. 2F).
We fitted the normalized activation curves of the channels with a Boltzmann equation
Here, Z represents the apparent mechanical sensitivity of the channel; it determines the slope of the activation curve (37). X0 represents the bundle deflection at which the channel open probability is 0.5. kBT is thermal energy (4.1 pN·nm at room temperature). The normalized activation curves that we obtained from IHCs of TMC1-injected Tmc1/2-DKO mice were consistent with previous observations using the same stimulation technique (Fig. 2F) (2, 3). The fits of the activation curves of virally transduced WT-TMC1 in IHCs of Tmc1/2-DKO animals yielded Z values of 0.027 ± 0.003 pN and X0 values of 380 ± 32 nm. Using nonstationary noise analysis of whole-cell currents (38, 39), we estimated single-channel currents as 12.5 ± 2.5 pA (Fig. 2G), which are also in line with our previous measurements (3).
Mutation of a glycine residue in TM4 alters the activation curve
We first targeted glycines within TM4 and TM6 because glycines within TM α helices of ion channels tend to act as hinge points for a gating conformational change (40). There is one glycine within TM4 (G411) that is highly conserved across TMCs (Figs. 2B and 3A); mutations of this residue have been associated with deafness (24). We mutated this residue to a helix-stabilizing alanine (41) to assess whether reducing the conformational flexibility of TM4 would affect gating.
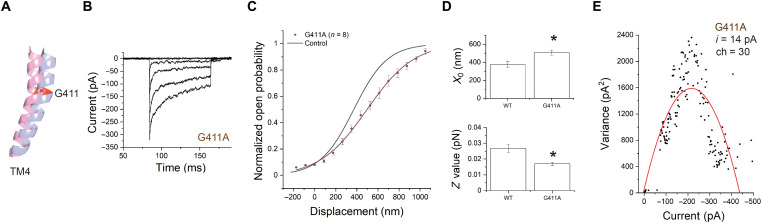
Fig. 3. Mutation of G411 on TM4 shifts the activation curve and changes the slope.
(A) Representation of TM4 showing G411A mutation site. Pink and blue represent the open- and closed-like configurations, respectively. (B) Representative transduction currents in response to −175- to 1050-nm deflections from IHCs expressing TMC1-G411A. (C) Activation curves from TMC1-G411A (red) and WT-TMC1 (black) expressed in DKO mice. (D) Fitting parameters X0 (position of half-activation) and Z (slope factor). Significance in differences in X0 (P = 0.01) and Z values (P = 0.01) is indicated as asterisks above the bars. (E) Nonstationary noise analysis for a hair cell expressing TMC1-G411A. Conductance did not change significantly.
Recordings from IHCs expressing TMC1 channels bearing the G411A mutation yielded maximum currents of 291 ± 38 pA (n = 8), which are comparable to those obtained from WT-TMC1–injected IHCs (Fig. 3B). However, the activation curves of TMC1-G411A were shifted rightward (X0 = 507 ± 28 nm) compared to WT-TMC1 (X0 = 380 ± 32 nm) (Fig. 3C). The slopes of the curves, represented by the Z values, were also substantially decreased—from 0.027 to 0.017 pN (table S2). Both these changes were significant with P < 0.05 (Fig. 3D). The single-channel current estimated from noise analysis was 14 ± 3.5 pA (Fig. 3E), which is close to the estimate for WT-TMC1 (3). Together, these findings are consistent with the idea that the G411A mutation influences the mechanical gating, but not the unitary conductance, of the channel.
Mutations of glycine residues on TM6 shift the activation curves
We then turned to the glycine residues in TM6. There are two: G518 is close to the extracellular side of the helix, and G539 is closer to the intracellular side (Figs. 2, A and B, and 4A). From IHCs expressing TMC1-G518A, we obtained maximum currents of 211 ± 11 pA (n = 8) that are comparable to those from WT-TMC1 (Fig. 4B and fig. S4A). TMC1-G518A activation curves were shifted to the right by ~110 nm (X0 = 493 ± 16 nm) compared to WT-TMC1 (X0 = 380 ± 32 nm) (Fig. 4, C and D, and table S2). From IHCs expressing TMC1-G539A, we also obtained typical maximum currents of 214 ± 38 pA (n = 14), and the activation curves were shifted rightward by ~140 nm (X0 = 518 ± 30 nm) (Fig. 4C). For both TMC1-G518A and TMC1-G539A, the difference in X0 compared to WT-TMC1 is statistically significant, with P < 0.05 (Fig. 4D).
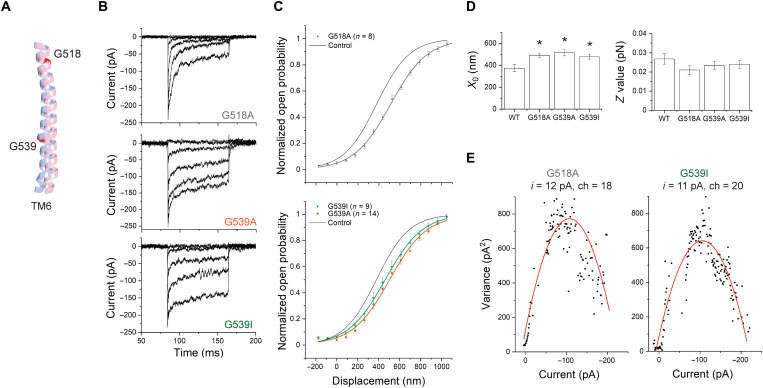
Fig. 4. Mutation of G518 or G539 on TM6 shifts the activation curves without changing the slope.
(A) Relative position of TM6 in open-like (pink) and closed-like (blue) structural models generated by AlphaFold2, showing G518 and G539. The predicted structures were aligned globally. (B) Representative transduction currents in IHCs expressing TMC1-G518A (top), TMC1-G539A (middle), and TMC1-G539I (bottom). (C) Activation curves from TMC1-G518A (top), TMC1-G539A (bottom), and TMC1-G539I (bottom). In both, the control WT-TMC1 activation curve is shown in black. (D) Fitting parameters X0 and Z. All three mutations shifted the activation curves (with P = 0.03, 0.01, and 0.02) without significantly changing slope (with P = 0.08, 0.44, and 0.13, respectively). (E) Nonstationary noise analysis for single hair cells expressing G518A (left) or G539I (right). Conductance did not significantly change from WT.
For both these mutations in TM6, the Z values were not significantly different from those for WT-TMC1 (Fig. 4D). A change in X0 without a change in Z suggests that these mutations in TM6 alter the open probability of the channel without changing the way it is coupled to the mechanical stimulus. Using a simple two-state model (see Materials and Methods), these changes in X0 can be interpreted as an increase in the energy of the open state compared to the closed state of ~0.5 kBT, thus favoring the closed state. Last, neither TMC1-G518A nor TMC1-G539A showed a significant effect on single-channel currents, with 12 ± 2.5 pA for G518A and 14 ± 2.5 pA for G539A (Fig. 4E).
The glycine at position 518 is highly conserved: 98% of the TMC sequences in our multiple sequence alignment show a glycine at this position. However, only 50% of the sequences have a glycine at position 539. A significant portion of the remaining sequences (41%) have an isoleucine or a valine at this position (table S3). These sequences include the closely related mouse TMC3, in which the amino acid corresponding to G539 is an isoleucine (fig. S4B). We found that the effect of a G539I mutation in TMC1 was similar to that of G539A (Fig. 4, B to D): IHCs expressing TMC1-G539I showed normal transduction current amplitude of 190 ± 14 pA (n = 9) and single-channel currents (14 ± 3 pA) but a ~100-nm rightward shift in the activation curves (X0 = 478 ± 25 nm) compared to WT-TMC1 (Fig. 4, C and D). Again, no significant change was observed in the Z value, implying that this mutation, similar to TMC1-G539A, does not affect the mechanical sensitivity of the channel (Fig. 4, C and D).
TMC3 has a glycine four residues toward the intracellular side of the protein, equivalent to position 544 in TMC1 (Fig. 2B and fig. S4B). We wondered whether a glycine anywhere in this region would provide sufficient flexibility and so reintroduced a glycine at position 544 in TMC1 lacking the glycine at position 539 (TMC1-G539I/A544G). However, we found that a glycine at position 544 did not substitute well for one at position 539, and the activation curve of TMC1-G539I/A544G was not significantly different from TMC1-G539I alone (fig. S4, C to E).
We conclude that mutation of either of the glycine residues at positions 518 and G539 on TM6 reduces the resting open probability, as indicated by the rightward shift in X0, even though they do not significantly contribute to the mechanical sensitivity of the channel as measured by the slope Z or to ion permeation. This is consistent with the idea that flexibility of TM6 is important for channel opening.
D528 on TM6 is critical for mechanical sensitivity and ion conduction
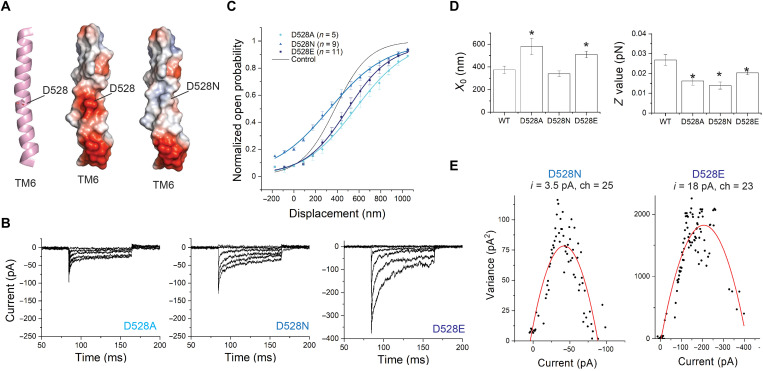
TM6 contains a central and highly conserved aspartate, D528 (Fig. 2, A and B). Tmc1 p.D528N is a recessive deafness mutation in mice (24). The corresponding residue within the Ca2 +-activated Cl− channel TMEM16A is a lysine (K645 in mouse TMEM16A), which has been linked to the voltage, Ca2+, and anion dependence of the channel activation (31). In the structural models for TMC1, D528 faces the channel pore (3, 5, 24) and significantly contributes to the negative surface charge (Fig. 5A and fig. S5A). To investigate whether D528 plays a role in mechanosensitive gating, we made the following mutations: D528A, D528N, and D528E.
Fig. 5. Mutations of D528 shift the activation curve and change the slope and can change single-channel currents.
(A) Position of D528 within TM6 of TMC1. TM6 is shown cartoon representation (left) with the side chain of D528 highlighted in stick. The right panels show TM6 in electrostatic surface representation. Electrostatic surface potentials are colored red and blue for negative and positive charges, respectively, and white color represents neutral residues. (B) Transduction currents from TMC1 bearing D528A (left), D528N (middle), or D528E (right) mutations. (C) Activation curves from IHCs expressing TMC1-D528A (cyan), D528N (blue), or D528E (navy). The fit for WT-TMC1 is shown in black as reference. (D) X0 and Z for each mutation compared to WT. All three mutations reduced the slope (P < 0.01), whereas the midpoints of curves for D528A and D528E (P = 0.01 and 0.01), but not D528N (P = 0.33), were shifted. (E) Nonstationary noise analysis for TMC1-D528N (left) and TMC1-D528E (right). Preservation of charge in the D528E mutant preserved single-channel conductance.
These mutations are also expected to change the surface electrostatics within the pore cavity (fig. S5A). We had previously found that IHCs expressing TMC1-D528C show significantly reduced transduction currents (~30% relative to WT-TMC1) (3). The currents were further diminished—irreversibly—with application of MTSET [(2-(trimethylammonium)ethyl)MethaneThioSulfonate] reagents (down to ~2% relative to WT), confirming their location within the pore (3).
Here, we first mutated D528 to an alanine to assess the effect of removing the aspartate side chain. We obtained average maximum currents of 80 ± 15 pA (n = 5), which are significantly smaller than that for other mutations that we examined (Fig. 5B and fig. S6). We found a 40% reduction in the slope of the TMC1-D528A activation curve compared to WT-TMC1 (Fig. 5, C and D, and table S2). In addition, the activation curve was shifted to the right by nearly 190 nm (566 ± 70 nm compared to 380 ± 32 nm). This apparent shift in the half-maximum location can be accounted for by the marked change in the slope; the resting open probability was not altered.
We then mutated D528 to asparagine (D528N), which is similar to aspartate in size but lacks the negative charge. The average maximum current was near normal at 148 ± 18 pA, n = 9 (Fig. 5B and fig. S6). We found that the D528N mutation also reduced the slope of the activation curve compared to WT-TMC1 (Fig. 5, C and D, and table S2). Reduction in slope in both D528A and D528N suggests that the aspartate side chain at this position contributes to the mechanical sensitivity of the channel. Furthermore, there was a leftward shift of the activation curve (X0 = 340 nm), indicating an increase in the resting open probability of TMC1-D528N (~20%) compared to WT-TMC1 (~6%) (Fig. 5C).
As D528 is both negatively charged and situated at a central location within the pore, we reasoned that the loss of charge in these mutants would reduce cation permeation. Consistent with another recent study, we found a notable decrease in TMC1-D528N single-channel currents: 3.5 ± 0.5 pA compared to 12.5 ± 2.5 pA for WT-TMC1 (Fig. 5E) (3, 24, 42). We therefore mutated D528 to glutamate, expecting that substitution with a negatively charged residue may have milder effects. IHCs expressing TMC1-D528E showed maximum average transduction currents of 332 ± 40 pA (n = 11). The large single-channel current for this mutant (17 ± 2.1 pA) suggests that the negative charge of the amino acid at this position is critical for attracting cations to the pore and so contributes to high conductance (Fig. 5E). However, the charge is not the sole determinant of D528’s contribution to channel activation: We found that the TMC1-D528E mutant had a lower slope than WT-TMC1 (Fig. 5, B to D, and table S2), suggesting that the size of the side chain of D528 matters for gating and that the role of D528 in gating is more than providing a negative charge. The fact that there is no glutamate at this position in a significant fraction of all the TMC sequences in the multiple sequence alignment is also consistent with this observation (table S4). Whether the amino acid size is changed by one carbon (D-to-E), or the size was kept but the charge removed (D-to-N), the mechanical sensitivity is affected.
It is possible that the observed effects of D528 mutations may be indirect through a reduction in Ca2+ permeation and its effect on the resting open probability. A recent study indeed reported a reduction in Ca2+ permeation in hair cells from Tmc1 p.D528N mice (24). However, a previously characterized deafness mutation M412K (“Beethoven”) and an M412C mutation also lead to a reduction in Ca2+ permeation (25, 43) but no change in the mechanical sensitivity of the channel as represented by the slope of the activation curves (2, 3), so it is more likely that D528 is directly involved in channel gating.
Mutations of a positively charged residue near D528 does not significantly alter TMC1’s mechanical sensitivity
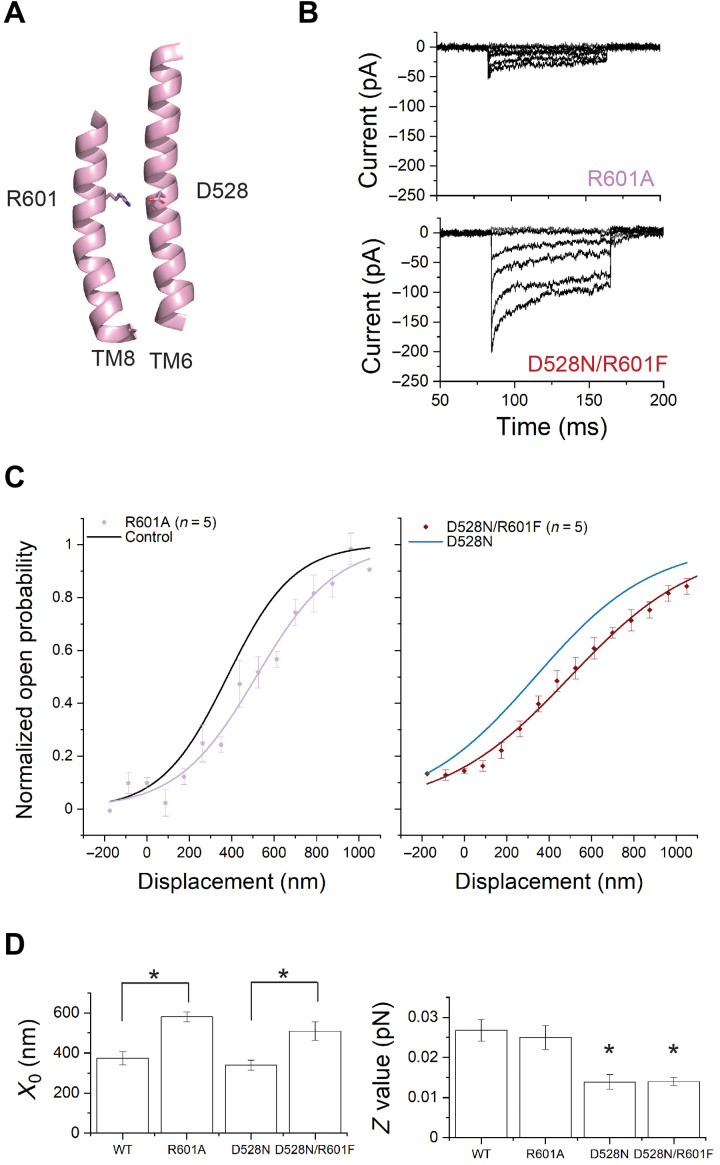
A positively charged residue near D528 is R601 on TM8. Evolutionary coupling analysis revealed that D528 on TM6 strongly coevolves with R601 (3). This is the second strongest coupling out of all possible analyzed pairs. Within a multiple sequence alignment of ~5000 TMC sequences, 90% of sequences have an aspartate at the position equivalent to 528. When it is an aspartate, the amino acid at the position equivalent to 601 is positively charged—either an arginine or a lysine. When 528 is not an aspartate (11%), 601 loses charge and is either a phenylalanine (F) or, less frequently, an isoleucine (I)—amino acids with hydrophobic chains (table S4).
The proximity of D528 on TM6 to R601 on TM8 in the structural models generated by AlphaFold2 suggests that they can form a salt bridge (Fig. 6A and fig. S5B). Similarly, in the structurally related hyperosmolality-gated OSCA calcium channels, interactions among TM6, TM7, and TM8 have been implicated in the gating of the channels (44). In the calcium-gated TMEM16A chloride channels, calcium ions bind among acidic residues in TM6, TM7, and TM8 to activate the channels (10). To assess the influence of the arginine at 601 on TMC1 gating, we mutated R601 in WT-TMC1 and in TMC1-D528N. TMC1-R601A yielded very low whole-cell currents (23 ± 8 pA, n = 5), while TMC1-D528N/R601F showed currents (166 ± 40 pA, n = 5) comparable to WT-TMC1 (Fig. 6B and fig. S6). In both WT-TMC1 and TMC1-D528N, mutation of R601 shifted the activation curves rightward, apparently stabilizing the closed state (R601A: X0 = 580 ± 25 nm; D528N/R601F: X0 = 510 ± 46 nm) (Fig. 6, C and D, and table S2). The shift in X0 is statistically significant for both (Fig. 6D). The Z values, representing mechanical sensitivity of the channel, were not significantly affected by these arginine mutations (Fig. 6D).
Fig. 6. Mutation of R601 shifts the activation curve without changing slope.
(A) Representation of TM8 showing R601. (B) Transduction currents from TMC1-R601A (top) and TMC1-D528N/R601F mutation (bottom). (C) Activation curves from IHCs expressing TMC1-R601A (C, lilac) or TMC1-D528N/R601F (D, crimson). The fits for WT-TMC1 (C, black) and TMC1-D528N (D, blue) are shown as reference. (D) X0 and Z values. R601A shifts the curve (P < 0.01) from WT without significantly changing slope (P = 0.1) and shifts the curve from D528N (P < 0.01) without further changing slope (P = 0.3).
Thus, removing a positive charge from TM8 that is likely involved in a salt bridge to TM6 changes the intrinsic energy difference between the open and closed states, without changing the efficiency with which the mechanical stimulus is coupled to channel gating. This implies further that the marked change in the mechanical sensitivity of the channel following mutation of D528, as measured by the slope, cannot be explained by electrostatic interaction between D528 and R601 but is caused by interaction between D528 and other residues or lipids.
DISCUSSION
Hearing relies on a number of proteins assembled at the tips of cochlear stereocilia; these include TMC1 and TMC2, PCDH15, LHFPL5, TMIE, and CIB2 (2, 45–47). These proteins are likely components of a mechanotransduction complex, which converts mechanical stimuli provided by sound waves into channel opening (48). Similar to other mechanosensitive ion channels, the hair-cell mechanotransduction complex is thought to contain pore-forming and force-sensitive gating elements. However, which proteins within the complex fulfill these roles has not been clear.
We previously used cysteine mutagenesis and cysteine-modifying reagents to implicate TMC1 TM domains 4 to 7 in the ion permeation pathway (3). Other studies involving mutations in these domains, including M412K (2, 43, 49), D569N (25), T416K, W554L, and D528N (25), are consistent with this finding, as are detailed molecular dynamic simulations of ion permeation using predicted TMC1 structures (8). The pore opening itself, however, is less well understood.
In this study, we used viral gene delivery of mutant TMC1 to Tmc1/2-DKO hair cells to study channel gating. To guide mutations, we first turned to machine learning–based, template-free structure prediction programs, including AlphaFold2 (18). Running AlphaFold2 multiple times with different starting conditions suggested structural diversity, with notable differences in the pore region of TMC1. These predicted conformations suggest that TM3 and TM4 may together move away from TM6 to open the channel (movie S1). Similarly, distancing of TM4 and TM6 enhances permeation in molecular dynamics simulations of the homology models (8).
For TMC1, the idea that conformational changes in TM4 and TM6 might bring their more extracellular portions in close contact and close off the permeation pathway is consistent with a gate for the hair-cell transduction channels that is located outside the binding site for charged blockers (50). Previous experiments demonstrated that M412 and D569 sites, which face the more intracellular part of the pore, are accessible from the extracellular solution only when the transduction channels are open but not when they are closed (3); they are also consistent with a gate near the outer part of the pore (fig. S5C).
The open-like conformations predicted by AlphaFold2 show a pore that is, in its narrowest dimension, not larger than about 6 Å in diameter. Careful physiology has suggested a larger open pore diameter, close to 12 Å (5, 28). It may be that the pore can expand further than suggested in these models, or it may be that large permeant ions can extend partially into the lipid as they pass through the pore (3).
On the basis of the predicted structures, we prepared a total of 12 mutations at seven distinct sites in TMC1. We found that mutations of a set of glycines within the TM6 helix led to rightward shifts in the activation curve without a change in slope. These glycine mutations lower the resting open probability of the channel, suggesting that they reduce the energy of the closed state without changing the way that force produced by tip link tension is conveyed to the gate. In the AlphaFold2-predicted structures, only small rearrangements are observed for TM6 (movie S1). Further functional, structural, and computational studies are needed to test the gating models that the predicted structures suggest.
On the other hand, mutations of the pore-lining residues G411 in TM4 and D528 in TM6 not only shifted the activation curve but also changed its slope. A reduction in the mechanical sensitivity of the transduction channel, as represented by the slope of the activation curve, suggests that the force applied by the tip link is less efficiently coupled to the relative energies of the open and closed states. According to the classical gating spring model (51), mechanical force is coupled to the channel through an elastic element known as the gating spring, which has not been molecularly identified. In this model, the slope factor Z of the activation curve (also known as the single-channel gating force) is defined as
where γ is the geometric gain between bundle deflection and gating spring extension, kGS is the gating spring stiffness, and b is the swing of the gate. Compared to mechanically tight hair bundles such as those from bullfrog, bundles in mammalian cochlea are difficult to stimulate with uniform deflection (52, 53). Uneven contact of hair cells with the stimulus probe results in underestimation of the Z value, by as much as 85% for IHCs (52, 53). Still, it is possible to compare the Z values from hair cells expressing mutated TMC1 channels and to determine whether Z has been altered by the mutations.
In the simple biophysical model, a change in Z may be due to a change in the gating spring stiffness or in the swing of the gate. It is difficult to imagine how the mutation of a single residue in TMC1 could reduce the stiffness of the gating spring; whatever it is, the gating spring must be able to stretch many tens of nanometers, and the structure of TMC1 is not conducive to such a huge rearrangement. From biophysical studies of bullfrog hair bundles, the gating swing has been estimated as 4 nm or more (54, 55), a distance much larger than the ~0.5-nm movement suggested by structural modeling that may close the outer part of the pore. Thus, it is likely that a large movement of one part of the channel propagates to the gate region to open the pore itself. Reduction in Z by mutation could also come about in how efficiently the force produced by the large gating movement changes channel open probability.
We have explored the possibility that D528 is coupled to the mechanical stimulus via salt bridge interactions with nearby positive charges with the protein itself. However, we find that mutations of R601 led to rightward shifts in the activation curves without a change in slope. This suggested that the proposed salt bridge between D528 and R601 may be important for stabilizing the closed channel conformation but does not seem to play a role in the way the gate is coupled to the mechanical stimulus.
Is TMC1 the force-sensing component of the mechanotransduction complex? It is possible that tip-link tension is conveyed to a distinct force-sensing protein within the complex and that a conformational change of that protein then promotes opening of the TMC pore (56). The block of gating compliance by pore-blocking compounds suggests that the pore is intimately associated with the conformational change produced by tension (54, 57). By changing the slope factor Z with single residue mutations in TM4 and TM6, this study specifically implicates TMC1 as part of the molecular sensor of tension. On the basis of this result, on ab inito structural modeling, and on gating movements in related channels, we propose also that the gating transition in TMC1 involves a separation of TM4 from TM6, to open the permeation pathway.
The classical gating spring model (1, 54) laid out a biophysical framework to explain the gating of the hair cells’ transduction channels. According to the model, the mechanical stimulus is transmitted to individual channel gates via molecular springs. Here, we explore in native cochlear hair cells the molecular correlate of the channel gate and determine a structural domain within TMC1 involved in channel opening in native cochlear hair cells.
Our findings here corroborate the overall similarities between TMC1 and the TMEM16 family members in that TM4 and TM6 helices are involved in both the permeation and the gating of the channel. Future experiments will now need to aim at understanding the unique features of TMC1. How is force transmitted from tip-link proteins to TMC1? What roles do lipids and other proteins of the transduction complex play in the mechanical gating of TMC1? Establishment of functional assays in heterologous cells lines, and technologies which can mechanically activate the channels, could greatly accelerate these efforts.
MATERIALS AND METHODS
Structure predictions
We compared our previously reported homology model for TMC1 (3) with the newly generated and more refined models by the trRosetta (15), RoseTTAFold (16), and AlphaFold2 (18). The template-free predictions with AlphaFold2 were performed in a Colab notebook setting (Google Research) (58). We selected the fast MMseq2 (Many against Many sequence-searching) method for generating multiple sequence alignments, which were prefiltered to 50% minimum coverage and 70% minimum identity to query sequence.
To obtain multiple conformations, we ran the network multiple times with different seeds (num_samples), corresponding to different multiple–sequence alignment cluster centers (58). We typically investigated the top 10 of the ranked structures. Structures were aligned in and visualized by PyMOL (version 2.4, Schrödinger). We aligned and clustered the structures using the program Chimera (29). The permeation pathways were modeled by the software HOLE (26).
Sequence alignment and evolutionary coupling analysis
Using EVcouplings software (59) (beta version of v0.1.1), which uses the Markov model-based sequence-search tool JackHMMER (60), we obtained a multiple sequence alignment of 5380 sequences. Alignments were built from the Uniref100 (61) dataset downloaded on 24 July 2020, using a maximum of five JackHMMER iterations. For further processing in the EVcouplings pipeline, we imposed a fragment requirement for each sequence to align to at least 70% of the query length, and analysis was restricted to just residue positions that contained at least 70% nongaps. We used the top 100 covarying residue pairs (evolutionary couplings) for further analysis.
AAV virus preparations
Mutations were introduced to specific locations in the mouse AAV-Tmc1ex1 vector, carrying a CMV promoter (3). We generated TMC1 constructs bearing the following amino acid substitutions: G518A, G539A, G539I, G539I/A544G, D528A, D528N, D528E, 601A, G411A, R601F/D528N, and A544G (GenScript, Piscataway NJ, USA). Constructs were screened with Sma I digest to check for inverted terminal repeat (ITR) integrity before packaging into AAV vectors. Viral vectors were packaged with AAV2 ITRs into the AAV9 capsid serotype PHP.B. Virus were generated and purified by the Viral Core at Boston Children’s Hospital with genomic titers (measured with human growth hormone primers) between 1 × 1013 and 5 × 1014 gc/ml. Vectors were aliquoted and stored at −80°C.
AAV virus injections
Neonatal Tmc1Δ/Δ/Tmc2Δ/Δ 1C57BL/6 mice (33) typically born on embryonic day 20 (E20) were placed with CD1 foster mothers. All animals were housed and bred in the animal facility at Harvard Medical School. Male and female pups were randomly chosen for the study. All animal protocols were approved by the Institutional Animal Care and Use Committee at Harvard Medical School. Before injection, mice were anesthetized using hypothermia. A postauricular incision was made near the left ear, and a cotton ball was inserted to spread the tissue. For round window membrane (RWM) injections, an injection needle made from a 1.4-mm glass capillary and pulled to a ~10-μm-diameter tip was positioned just above the RWM to confirm that viral suspension was released when the injection was started. Once confirmed, the needle was inserted through the RWM, and 1.2 μl was injected using a Nanoliter 2000 Injector (World Precision Instruments) at a rate of 65 nl/min. Following the procedure, the surgical incision was closed with sutures. The pups were then put on a 37°C heating board to recover and returned to their cages. Usually, three animals were injected in a session, and we typically performed three injection sessions per mutation.
Cochlear dissections
At P4 to P6, pups were euthanized by rapid decapitation, temporal bones were dissected, and the membranous labyrinth was isolated under a dissection microscope. Reissner’s membrane was peeled back, and the tectorial membrane and stria vascularis were mechanically removed. Organs of Corti were excised and cultured in medium containing Dulbecco’s Modified Eagle Medium supplemented with 1% fetal bovine serum, 10 mM Hepes, and carbenicillin (0.05 mg/ml) at 37°C in 5% CO2. Organ of Corti cultures were pinned flat beneath a pair of thin glass fibers glued at one end with Sylgard to an 18-mm round glass coverslip. The tissue was placed in culture for 3 to 7 days before electrophysiological studies.
FM1-43 uptake assays
Coverslips with adherent cochlear cultures were placed on a glass-bottomed chamber. The culture media was washed away with Leibovitz’s L15 medium three times and then incubated with 2 μM FM1-43 in L-15 for 1 min followed by incubation with 2 mM SCAS (4-Sulfonato calix[8]arene, sodium salt) in L-15 for 3 min and washed with L-15 two additional times before imaging.
Confocal imaging
Imaging was performed on an Olympus FV1000 confocal microscope. FM1-43 fluorescence (excitation at 488 nm with ~5 to 12% intensity) was measured on mounted cultures with a 60× (1.1 numerical aperture) water-immersion objective.
Electrophysiology
Transduction currents were recorded from IHCs using a Nikon Eclipse FN1 microscope with a 60× water-immersion objective and differential interference contrast optics and an Axopatch 200B patch clamp with a Digidata 1440 digitizer controlled by pCLAMP 10 software (Molecular Devices). The whole-cell voltage-clamp configuration was used for recordings. Currents were filtered at 5 kHz with a low-pass eight-pole Bessel filter. For hair-bundle stimulation, custom glass probes were made and polished to a diameter of ~4 μm to match the shape of the IHC bundles. The probe was attached to the probe holder with wax and shielded with grounded aluminum foil. The holder was moved by a piezo stack (Physik Instruments) driven by a custom high-voltage piezo driver amplifier. Bundles were displaced for 80 ms with 15-step displacements ranging from −175 to 1050 nm at 88-nm increments. For recordings, 1.5-mm outer diameter R-6 (8350) glass pipettes were pulled with a Narishige PC-10 puller and coated with prewarmed wax before use. These patch pipettes were filled with an internal solution containing 137 mM CsCl, 5 mM EGTA, 10 mM Hepes, 2.5 mM Na2-ATP, 0.1 mM CaCl2, and 3.5 mM MgCl2 and adjusted to pH 7.4 with CsOH, ~290 mmol/kg. The tissues were bathed in external solution containing 137 mM NaCl, 5.8 mM KCl, 0.7 mM NaH2PO4, 10 mM Hepes, 1.3 mM CaCl2, 0.9 mM MgCl2, and 5.6 mM glucose, vitamins, and essential amino acids, adjusted to pH 7.4 with NaOH, ~310 mmol/kg. Cells were held at a −80-mV potential, and a separate pipet flowed extracellular solution onto their apical surfaces.
Electrophysiology analysis
Data were analyzed with Clampfit (Molecular Devices) and ORIGIN (OriginLab). The average maximum current amplitudes as a function of probe displacement were plotted for each hair cell and fitted with a Boltzmann equation using ORIGIN as follows
The data were then normalized for each cell, such that A1 and A2 range from 0 to 1 to allow comparison across cells. Z values were derived from the relationship 1/dx = Z/kBT. For each TMC1 variant, the average X0 and Z values were calculated as the means ± SEM of all cells. The number of cells (n) is indicated in the text and figures. Student’s t test was used to compare the means, and P values of <0.05 are marked as significant. For mutations with no significant change in slope, the corresponding change in intrinsic energy difference was estimated as ∆X0/dx in units of kBT. For nonstationary noise analysis, the variance of the responses was plotted against the mean for each cell. As before, we averaged the data within 1-pA intervals (3) and fitted with a parabola to derive the relevant values
where σ2 is the variance, i is the single-channel current, I is the whole-cell current, and N is the number of channels (38). Cells were included in the analysis only if the plot of σ2 versus I yielded a parabolic dataset that was fit well by the equation above. Statistics are based on at least three cells for each condition.
Acknowledgments
We thank I. Anishchenko, A. Indzhykulian, C. Phillips, M. Sotomayor, and J. Lottheimer for helpful discussions; Y. Zhang for producing AAV vectors; B. Derfler for assistance with molecular preparations; and A. Griffith and J. Holt for providing Tmc1Δ/Δ/Tmc2 Δ/Δ mice.
Funding: This work was supported by National Institutes of Health grant R21 DC018631 (N.A.) and National Institutes of Health grant R01 DC000304 (D.P.C.).
Author contributions: N.A., B.P., K.D.K., and D.P.C. designed the experiments. B.P. acquired electrophysiology data. K.D.K. assembled instrumentation and performed FM1-43 assays and confocal imaging. P.I.T. performed cochlear dissections. Y.L. bred and maintained mice and performed cochlear injections. K.P.B. and D.S.M. identified and mapped evolutionarily coupled residue pairs onto structures. N.A., K.D.K., B.P., and D.P.C. wrote the manuscript.
Competing interests: D.S.M. is an advisor for Dyno Therapeutics, Octant, Juro Bio, Tectonic Therapeutics, and Genentech and a cofounder of Seismic. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The Tmc1Δ/Δ/Tmc2 Δ/Δ mice were provided under a material transfer agreement by A. J. Griffith (University of Tennessee College of Medicine). The Supplementary Materials include figs. S1 to S6, tables S1 to S4, and movie S1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S4
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Corey D. P., Hudspeth A. J., Kinetics of the receptor current in bullfrog saccular hair cells. J. Neurosci. 3, 962–976 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan B., Geleoc G. S., Asai Y., Horwitz G. C., Kurima K., Ishikawa K., Kawashima Y., Griffith A. J., Holt J. R., TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan B., Akyuz N., Liu X. P., Asai Y., Nist-Lund C., Kurima K., Derfler B. H., Gyorgy B., Limapichat W., Walujkar S., Wimalasena L. N., Sotomayor M., Corey D. P., Holt J. R., TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99, 736–753.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medrano-Soto A., Moreno-Hagelsieb G., McLaughlin D., Ye Z. S., Hendargo K. J., Saier M. H. Jr., Bioinformatic characterization of the anoctamin superfamily of Ca2+-activated ion channels and lipid scramblases. PLOS ONE 13, e0192851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballesteros A., Fenollar-Ferrer C., Swartz K. J., Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. eLife 7, e38433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn Y., Kim D. S., Pastan I. H., Lee B., Anoctamin and transmembrane channel-like proteins are evolutionarily related. Int. J. Mol. Med. 24, 51–55 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunzelmann K., Cabrita I., Wanitchakool P., Ousingsawat J., Sirianant L., Benedetto R., Schreiber R., Modulating Ca2+ signals: A common theme for TMEM16, Ist2, and TMC. Pflugers Arch. 468, 475–490 (2016). [DOI] [PubMed] [Google Scholar]
- 8.S. Walujkar, J. M. Lotthammer, C. R. Nisler, J. C. Sudar, A. Ballesteros, M. Sotomayor, In silico electrophysiology of inner-ear mechanotransduction channel TMC1 models. bioRxiv 2021.09.17.460860 [Preprint]. 10.1101/2021.09.17.460860 September 2021. https://doi.org/10.1101/2021.09.17.460860. [DOI]
- 9.Jia Y., Zhao Y., Kusakizako T., Wang Y., Pan C., Zhang Y., Nureki O., Hattori M., Yan Z., TMC1 and TMC2 proteins are pore-forming subunits of mechanosensitive ion channels. Neuron 105, 310–321.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Paulino C., Kalienkova V., Lam A. K. M., Neldner Y., Dutzler R., Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552, 421–425 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Falzone M. E., Rheinberger J., Lee B. C., Peyear T., Sasset L., Raczkowski A. M., Eng E. T., Di Lorenzo A., Andersen O. S., Nimigean C. M., Accardi A., Structural basis of Ca2+-dependent activation and lipid transport by a TMEM16 scramblase. eLife 8, e43229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam A. K. M., Dutzler R., Mechanism of pore opening in the calcium-activated chloride channel TMEM16A. Nat. Commun. 12, 786 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khelashvili G., Falzone M. E., Cheng X., Lee B. C., Accardi A., Weinstein H., Dynamic modulation of the lipid translocation groove generates a conductive ion channel in Ca2+-bound nhTMEM16. Nat. Commun. 10, 4972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. A. Indzhykulian, S. L. Johnson, G. S. Géléoc, Electrophysiological recordings of voltage-dependent and mechanosensitive currents in sensory hair cells of the auditory and vestibular organs of the mouse, in Developmental, Physiological, and Functional Neurobiology of the Inner Ear, A. K. Groves, Ed. (Springer, New York, 2022), pp. 221–264. [Google Scholar]
- 15.Yang H., Baker S. F., Gonzalez M. E., Topham D. J., Martinez-Sobrido L., Zand M., Holden-Wiltse J., Wu H., An improved method for estimating antibody titers in microneutralization assay using green fluorescent protein. J. Biopharm. Stat. 26, 409–420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G. R., Wang J., Cong Q., Kinch L. N., Schaeffer R. D., Millan C., Park H., Adams C., Glassman C. R., DeGiovanni A., Pereira J. H., Rodrigues A. V., van Dijk A. A., Ebrecht A. C., Opperman D. J., Sagmeister T., Buhlheller C., Pavkov-Keller T., Rathinaswamy M. K., Dalwadi U., Yip C. K., Burke J. E., Garcia K. C., Grishin N. V., Adams P. D., Read R. J., Baker D., Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–879 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohl C. A., Strauss C. E., Misura K. M., Baker D., Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.J. F. Moult, A. Kryshtafovych, T. Schwede, M. Topf, vol. CASP 14 Abstract Book.
- 20.Liang X., Qiu X., Dionne G., Cunningham C. L., Pucak M. L., Peng G., Kim Y. H., Lauer A., Shapiro L., Muller U., CIB2 and CIB3 are auxiliary subunits of the mechanotransduction channel of hair cells. Neuron 109, 2131–2149.e15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corey D. P., Hudspeth A. J., Ionic basis of the receptor potential in a vertebrate hair cell. Nature 281, 675–677 (1979). [DOI] [PubMed] [Google Scholar]
- 22.Kros C. J., Rusch A., Richardson G. P., Mechano-electrical transducer currents in hair cells of the cultured neonatal mouse cochlea. Proc. Biol. Sci. 249, 185–193 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Ricci A. J., Fettiplace R., Calcium permeation of the turtle hair cell mechanotransducer channel and its relation to the composition of endolymph. J. Physiol. 506, 159–173 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beurg M., Schimmenti L. A., Koleilat A., Amr S. S., Oza A., Barlow A. J., Ballesteros A., Fettiplace R., New Tmc1 deafness mutations impact mechanotransduction in auditory hair cells. J. Neurosci. 41, 4378–4391 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beurg M., Barlow A., Furness D. N., Fettiplace R., A Tmc1 mutation reduces calcium permeability and expression of mechanoelectrical transduction channels in cochlear hair cells. Proc. Natl. Acad. Sci. U.S.A. 116, 20743–20749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Kalienkova V., Clerico Mosina V., Bryner L., Oostergetel G. T., Dutzler R., Paulino C., Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM. eLife 8, e44364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farris H. E., LeBlanc C. L., Goswami J., Ricci A. J., Probing the pore of the auditory hair cell mechanotransducer channel in turtle. J. Physiol. 558, 769–792 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J. E., Huang C. C., Ferrin T. E., RRDistMaps: A UCSF Chimera tool for viewing and comparing protein distance maps. Bioinformatics 31, 1484–1486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopf T. A., Scharfe C. P., Rodrigues J. P., Green A. G., Kohlbacher O., Sander C., Bonvin A. M., Marks D. S., Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife 3, e03430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters C. J., Gilchrist J. M., Tien J., Bethel N. P., Qi L., Chen T., Wang L., Jan Y. N., Grabe M., Jan L. Y., The sixth transmembrane segment Is a major gating component of the TMEM16A calcium-activated chloride channel. Neuron 97, 1063–1077.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulino C., Neldner Y., Lam A. K., Kalienkova V., Brunner J. D., Schenck S., Dutzler R., Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. eLife 6, e26232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawashima Y., Geleoc G. S., Kurima K., Labay V., Lelli A., Asai Y., Makishima T., Wu D. K., Della Santina C. C., Holt J. R., Griffith A. J., Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 121, 4796–4809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Askew C., Rochat C., Pan B., Asai Y., Ahmed H., Child E., Schneider B. L., Aebischer P., Holt J. R., Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med. 7, 295ra108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J., Nist-Lund C., Solanes P., Goldberg H., Wu J., Pan B., Schneider B. L., Holt J. R., Efficient viral transduction in mouse inner ear hair cells with utricle injection and AAV9-PHP.B. Hear. Res. 394, 107882 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Ivanchenko M. V., Hanlon K. S., Devine M. K., Tenneson K., Emond F., Lafond J. F., Kenna M. A., Corey D. P., Maguire C. A., Preclinical testing of AAV9-PHP.B for transgene expression in the non-human primate cochlea. Hear. Res. 394, 107930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hudspeth A. J., Choe Y., Mehta A. D., Martin P., Putting ion channels to work: mechanoelectrical transduction, adaptation, and amplification by hair cells. Proc. Natl. Acad. Sci. U.S.A. 97, 11765–11772 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigworth F. J., The variance of sodium current fluctuations at the node of Ranvier. J. Physiol. 307, 97–129 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holton T., Hudspeth A. J., The transduction channel of hair cells from the bull-frog characterized by noise analysis. J. Physiol. 375, 195–227 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding S., Ingleby L., Ahern C. A., Horn R., Investigating the putative glycine hinge in Shaker potassium channel. J. Gen. Physiol. 126, 213–226 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luque I., Mayorga O. L., Freire E., Structure-based thermodynamic scale of alpha-helix propensities in amino acids. Biochemistry 35, 13681–13688 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Beurg M., Nam J. H., Fettiplace R., The speed of the hair cell mechanotransducer channel revealed by fluctuation analysis. J. Gen. Physiol. 153, e202112959 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corns L. F., Johnson S. L., Kros C. J., Marcotti W., Tmc1 point mutation affects Ca2+ sensitivity and block by dihydrostreptomycin of the mechanoelectrical transducer current of mouse outer hair cells. J. Neurosci. 36, 336–349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jojoa-Cruz S., Saotome K., Murthy S. E., Tsui C. C. A., Sansom M. S., Patapoutian A., Ward A. B., Cryo-EM structure of the mechanically activated ion channel OSCA1.2. eLife 7, e41845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B., Wu Z., Grillet N., Yan L., Xiong W., Harkins-Perry S., Muller U., TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron 84, 954–967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giese A. P. J., Tang Y. Q., Sinha G. P., Bowl M. R., Goldring A. C., Parker A., Freeman M. J., Brown S. D. M., Riazuddin S., Fettiplace R., Schafer W. R., Frolenkov G. I., Ahmed Z. M., CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat. Commun. 8, 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beurg M., Xiong W., Zhao B., Muller U., Fettiplace R., Subunit determination of the conductance of hair-cell mechanotransducer channels. Proc. Natl. Acad. Sci. U.S.A. 112, 1589–1594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudspeth A. J., How the ear’s works work. Nature 341, 397–404 (1989). [DOI] [PubMed] [Google Scholar]
- 49.Beurg M., Goldring A. C., Fettiplace R., The effects of Tmc1 Beethoven mutation on mechanotransducer channel function in cochlear hair cells. J. Gen. Physiol. 146, 233–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcotti W., van Netten S. M., Kros C. J., The aminoglycoside antibiotic dihydrostreptomycin rapidly enters mouse outer hair cells through the mechano-electrical transducer channels. J. Physiol. 567, 505–521 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markin V. S., Hudspeth A. J., Gating-spring models of mechanoelectrical transduction by hair cells of the internal ear. Annu. Rev. Biophys. Biomol. Struct. 24, 59–83 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Nam J. H., Peng A. W., Ricci A. J., Underestimated sensitivity of mammalian cochlear hair cells due to splay between stereociliary columns. Biophys. J. 108, 2633–2647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karavitaki K. D., Corey D. P., Sliding adhesion confers coherent motion to hair cell stereocilia and parallel gating to transduction channels. J. Neurosci. 30, 9051–9063 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howard J., Hudspeth A. J., Compliance of the hair bundle associated with gating of mechanoelectrical transduction channels in the bullfrog’s saccular hair cell. Neuron 1, 189–199 (1988). [DOI] [PubMed] [Google Scholar]
- 55.Cheung E. L., Corey D. P., Ca2+ changes the force sensitivity of the hair-cell transduction channel. Biophys. J. 90, 124–139 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christensen A. P., Corey D. P., TRP channels in mechanosensation: Direct or indirect activation? Nat. Rev. Neurosci. 8, 510–521 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Martin P., Bozovic D., Choe Y., Hudspeth A. J., Spontaneous oscillation by hair bundles of the bullfrog’s sacculus. J. Neurosci. 23, 4533–4548 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Mirdita, S. Ovchinnikov, M. Steinegger, ColabFold - Making protein folding accessible to all. bioRxiv 10.1101/2021.08.15.456425 [Preprint]. 15 August 2021. 10.1101/2021.08.15.456425. [DOI] [PMC free article] [PubMed]
- 59.Marks D. S., Colwell L. J., Sheridan R., Hopf T. A., Pagnani A., Zecchina R., Sander C., Protein 3D structure computed from evolutionary sequence variation. PLOS ONE 6, e28766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson L. S., Eddy S. R., Portugaly E., Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H., UniProt C., UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S4
References
Movie S1