Abstract
The metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “Syntrophus aciditrophicus” in cocultures with hydrogen-using microorganisms was studied. Cyclohexane carboxylate, cyclohex-1-ene carboxylate, pimelate, and glutarate (or their coenzyme A [CoA] derivatives) transiently accumulated during growth with benzoate. Identification was based on comparison of retention times and mass spectra of trimethylsilyl derivatives to the retention times and mass spectra of authentic chemical standards. 13C nuclear magnetic resonance spectroscopy confirmed that cyclohexane carboxylate and cyclohex-1-ene carboxylate were produced from [ring-13C6]benzoate. None of the metabolites mentioned above was detected in non-substrate-amended or heat-killed controls. Cyclohexane carboxylic acid accumulated to a concentration of 260 μM, accounting for about 18% of the initial benzoate added. This compound was not detected in culture extracts of Rhodopseudomonas palustris grown phototrophically or Thauera aromatica grown under nitrate-reducing conditions. Cocultures of “S. aciditrophicus” and Methanospirillum hungatei readily metabolized cyclohexane carboxylate and cyclohex-1-ene carboxylate at a rate slightly faster than the rate of benzoate metabolism. In addition to cyclohexane carboxylate, pimelate, and glutarate, 2-hydroxycyclohexane carboxylate was detected in trace amounts in cocultures grown with cyclohex-1-ene carboxylate. Cyclohex-1-ene carboxylate, pimelate, and glutarate were detected in cocultures grown with cyclohexane carboxylate at levels similar to those found in benzoate-grown cocultures. Cell extracts of “S. aciditrophicus” grown in a coculture with Desulfovibrio sp. strain G11 with benzoate or in a pure culture with crotonate contained the following enzyme activities: an ATP-dependent benzoyl-CoA ligase, cyclohex-1-ene carboxyl-CoA hydratase, and 2-hydroxycyclohexane carboxyl-CoA dehydrogenase, as well as pimelyl-CoA dehydrogenase, glutaryl-CoA dehydrogenase, and the enzymes required for conversion of crotonyl-CoA to acetate. 2-Ketocyclohexane carboxyl-CoA hydrolase activity was detected in cell extracts of “S. aciditrophicus”-Desulfovibrio sp. strain G11 benzoate-grown cocultures but not in crotonate-grown pure cultures of “S. aciditrophicus”. These results are consistent with the hypothesis that ring reduction during syntrophic benzoate metabolism involves a four- or six-electron reduction step and that once cyclohex-1-ene carboxyl-CoA is made, it is metabolized in a manner similar to that in R. palustris.
Biodegradation of aromatic compounds is an important component of the carbon cycle in various anoxic environments. Despite the large number of natural and synthetic homocyclic aromatic compounds, anaerobic microorganisms initially channel all aromatic substrates into a few central intermediates prior to ring cleavage (20). Benzoyl coenzyme A (CoA) is the most important of these intermediates since a large number of compounds, such as chloro-, nitro-, and aminobenzoates, aromatic hydrocarbons, and phenolic compounds, are initially converted to benzoyl-CoA prior to ring reduction and cleavage (18). The central pathways for benzoate and benzoyl-CoA metabolism under anaerobic conditions have been studied primarily in two microorganisms, the phototrophic, purple, nonsulfur bacterium Rhodopseudomonas palustris and the nitrate-reducing bacterium Thauera aromatica (18, 19). After activation of benzoate to benzoyl-CoA (1, 17), benzoyl-CoA is reduced to cyclohex-1,5-diene carboxyl-CoA by a benzoyl-CoA reductase, which has been purified from T. aromatica (8, 9, 26). Based on DNA sequence homology, it is believed that a similar reductive reaction occurs in R. palustris (14). After ring reduction, the pathways diverge in the two organisms. In T. aromatica, cyclohex-1,5-diene carboxyl-CoA is hydrated to 6-hydroxycyclohex-1-ene carboxyl-CoA (28). The latter compound is oxidized to 6-ketocyclohex-1-ene carboxyl-CoA, which is then hydrolytically cleaved to 3-hydroxypimelyl-CoA (29). The pathway in R. palustris is similar except that cyclohex-1,5-diene carboxyl-CoA is most probably reduced to cyclohex-1-ene carboxyl-CoA. The latter compound is metabolized to 2-ketocyclohexane carboxyl carboxyl-CoA, which is hydrolytically cleaved to pimelyl-CoA. The C7 ring cleavage products then undergo β-oxidation, which yields three molecules of acetate and one molecule of CO2.
Benzoate degradation also occurs under methanogenic conditions (37, 49). Tarvin and Buswell (49) observed degradation of benzoate in anoxic sediments with production of carbon dioxide and methane as the final end products. The discovery that methanogenic benzoate degradation to carbon dioxide and methane is mediated by a consortium of a fermentative (syntrophic) microorganism and hydrogen- and acetate-utilizing methanogens (15) and the subsequent isolation of the syntrophic partners (36) provided the opportunity to study the pathway for benzoate degradation under methanogenic conditions. So far, three species that syntrophically metabolize benzoate have been isolated (22, 36, 51), and all of these species belong to the genus Syntrophus. Benzoate degradation under syntrophic conditions has not been investigated as thoroughly as benzoate degradation under nitrate-reducing and phototrophic conditions due to the relatively slow growth rates and low cell yields of these organisms (3). However, recent studies have shown that benzoate is activated to benzoyl-CoA by an ATP-dependent ligase as the first step in benzoate metabolism (4, 46). Also, enzyme activities for glutaryl-CoA metabolism to acetate and CO2 have been detected in cell extracts of Syntrophus gentianae (46), and glutaryl-CoA dehydrogenase and the enzyme activities responsible for crotonyl-CoA metabolism to acetate have been detected in Syntrophus buswellii GA (2). The ring reduction and cleavage steps required for syntrophic benzoyl-CoA metabolism have not been investigated yet.
In this study, we investigated the pathway for syntrophic benzoate metabolism in “Syntrophus aciditrophicus” strain SB by identifying and quantifying metabolites produced during growth of this organism with benzoate, cyclohexane carboxylate, and cyclohex-1-ene carboxylate in cocultures with hydrogen-utilizing partners and by measuring the key enzyme activities postulated to be involved in benzoate metabolism. Below, we describe transient production of cyclohex-1-ene carboxylate and relatively larger amounts of cyclohexane carboxylate during benzoate degradation. This is consistent with the hypothesis that benzoyl-CoA reduction during syntrophic benzoate metabolism may involve a four- or six-electron reduction (46, 47). We hypothesize that the difference in benzoyl-CoA metabolism from that observed in R. palustris and T. aromatica may be due to the energetic constraints imposed by syntrophic metabolism of aromatic substrates.
MATERIALS AND METHODS
Microorganisms and media.
“S. aciditrophicus” SBT (= ATCC 700169T) was isolated from a sewage treatment plant in Norman, Okla. (22). Methanospirillum hungatei JF1 and Desulfovibrio sp. strain G11 were obtained from the culture collection of M. P. Bryant (Urbana, Ill.). All media and stock solutions were prepared anaerobically by the techniques described by Balch and Wolfe (6). The organisms were grown in a basal medium (33) lacking rumen fluid. To grow “S. aciditrophicus” in pure culture, crotonate (40 mM) was added to the basal medium (7) and the headspace was pressurized to 172 kPa with an 80% N2–20% CO2 gas mixture. M. hungatei and Desulfovibrio sp. strain G11 were grown in the basal medium containing 2 mM sodium acetate in the presence of 243 kPa of 80% H2–20% CO2. Sodium sulfate (15 mM) was included in the medium when Desulfovibrio sp. strain G11 was present. M. hungatei and Desulfovibrio sp. strain G11 cultures were incubated in a shaking incubator (100 rpm). Cocultures of “S. aciditrophicus” and M. hungatei or “S. aciditrophicus” and Desulfovibrio sp. strain G11 were established by adding a 15 to 20% (vol/vol) inoculum of each microorganism to the basal medium containing 1.2 to 1.5 mM sodium benzoate, sodium cyclohexane carboxylate, or sodium cyclohex-1-ene carboxylate as the substrate in the presence of a headspace containing 80% N2 and 20% CO2 (172 kPa). All inoculations were performed by using sterile disposable plastic syringes and needles that were degassed with oxygen-free nitrogen gas. All cultures were incubated at 37°C. T. aromatica DSM 6984 was obtained from the Deutsche Sammlung von Mikroorganismen (Braunschweig, Germany) and was cultured anaerobically at 28°C in a benzoate-nitrate medium (50). R. palustris CGA009 was kindly provided by Caroline S. Harwood and cultured as previously described (17).
Detection and quantification of metabolites by GC-MS.
Cocultures of “S. aciditrophicus” and either M. hungatei or Desulfovibrio sp. strain G11 were grown in 600 ml of basal medium with 1.4 mM sodium benzoate to detect metabolites of benzoate metabolism. Samples (60 ml) were withdrawn from the cultures at various times. The pH of each sample was brought to more than 12 for 30 min by stepwise addition of 1 N NaOH to hydrolyze putative thioester bonds. Each sample was then acidified to a pH of less than 2 with 12 N HCl. The samples were then extracted three times with 25-ml aliquots of ethyl acetate. The ethyl acetate extracts were filtered through anhydrous sodium sulfate to remove water, combined, and then concentrated to volumes of 2 to 3 ml under a vacuum. The concentrated ethyl acetate extract was then quantitatively transferred to 6-ml vials and evaporated to dryness under a stream of nitrogen gas. The dried ethyl acetate extract was then redissolved in 0.3 ml of ethyl acetate and derivatized with N,O-bis-(trimethylsilyl)triflouroacetamide (Pierce Chemicals, Rockford, Ill.). Each concentrated and derivatized extract was analyzed with a Hewlett-Packard 5890 series II gas chromatograph (GC) equipped with a Hewlett-Packard 5970 series mass spectrometer (MS) and a 30-m DB-5 fused silica capillary column (J & W Scientific, Folson, Calif.). Helium was used as the carrier gas at a flow rate of 0.8 ml/min. The oven temperature was held at 70°C for 5 min, increased at a rate of 10°C/min to 220°C, and then held at 220°C for 5 min. The controls for this experiment included heat-killed cocultures containing “S. aciditrophicus” and non-substrate-amended cocultures to differentiate between metabolites formed due to benzoate metabolism and metabolites present in the inoculum of crotonate-grown “S. aciditrophicus” cultures or H2-grown M. hungatei and Desulfovibrio sp. strain G11 cultures. All treatments were performed in triplicate. The metabolites were identified by comparing their retention times and mass spectral profiles with the retention times and mass spectral profiles of trimethylsilyl (TMS)-derivatized chemical standards and were quantified by comparison to standard curves constructed with the TMS derivatives of the compounds of interest. The detection limits ranged from 0.07 μM (pimelic acid) to 0.14 μM (cyclohexane carboxylic acid) under the experimental conditions used. The benzoate concentrations calculated by GC-MS analysis at different times were within ±10% of the benzoate concentrations determined by high-performance liquid chromatography analysis. R. palustris and T. aromatica cultures were grown with similar benzoate concentrations, and the samples were processed as described above. A similar protocol was used for detection of metabolites in cyclohexane carboxylate- or cyclohex-1-ene carboxylate-grown “S. aciditrophicus”-M. hungatei cocultures.
NMR spectroscopy.
Cocultures of “S. aciditrophicus” and M. hungatei were grown with 1.5 mM [ring-13C6] benzoate. Samples (100 ml) were withdrawn from the cultures at various times and acidified to a pH of less than 2 by dropwise addition of 12 N HCl. Each sample was extracted three times with ethyl acetate, concentrated under a vacuum, and dried under an N2 atmosphere as described above. The dried samples were then dissolved in deuterated chloroform (CDCl3). Non-substrate-amended and heat-killed controls were included as described above. A coculture of “S. aciditrophicus” and M. hungatei with unlabeled benzoate was used to ensure that none of the peaks observed in the nuclear magnetic resonance (NMR) spectra were due to 13C impurities in the solvents used. The NMR spectra of the organic solvent-extracted samples were obtained with a Unity INOVA 400-MHz NMR spectrometer (Varian) with a 13C resonance frequency of 100.573 MHz. The 13C spectra were obtained at 30°C by using a standard inverse-gated pulse sequence. The following experimental parameters were used: sweep width, 24,140 Hz; acquisition time, 1.00 s; and recycle delay, 8 s. The number of scans ranged from 500 to 36,000 depending on the sample concentration. The data were processed with 1-Hz line broadening.
Other analytical procedures.
Protein was determined as described previously (10). Growth was monitored by measuring absorbance at 600 nm. Benzoate was analyzed by high-performance liquid chromatography as described previously (21). Methane was analyzed by GC (23), and sulfate was analyzed by ion chromatography (32). Enzyme activities were determined with either a Beckman DU-64 spectrophotometer or a Shimadzu 2101-PC dual-beam spectrophotometer.
Preparation of cell extracts.
Cells were harvested by centrifugation (12,000 × g, 20 min, 4°C) and were washed by resuspending and recentrifuging the cell pellets three times in anoxic 100 mM Tris-HCl buffer (pH 7.8). The final cell pellet was suspended in the same buffer (0.2 g of cells/ml) containing 1 mM MgCl2, 2 mM dithiothreitol (DTT), and 0.2 mg of DNase per ml. Cells were broken under anaerobic conditions by two passages through a chilled French pressure cell at 110,400 kPa. Unbroken cells and cell debris were removed by centrifugation at 27,200 × g for 20 min at 4°C. The resulting supernatant, termed the cell extract, was used immediately in enzyme assays or was stored anaerobically in liquid nitrogen until it was used.
Enzyme assays.
Acyl-CoA ligase activity was assayed by measuring the amount of AMP formed in the CoA ligase reaction by a coupled enzyme assay (4). The reaction was initiated by adding 10 to 50 μl of cell extract, and then oxidation of NADH was monitored at 340 nm. Formation of 1 mol of AMP corresponded to oxidation of 2 mol of NADH. The substrates tested included benzoate, 2-, 3-, and 4- chlorobenzoates, 2-, 3-, and 4-fluorobenzoates, 4-hydroxybenzoate, picolinic acid, phenyl acetate, crotonate, n-butyrate, isobutyrate, heptanoate, and hexanoate. Formation of benzoyl-CoA from benzoate and CoA in cell extracts was confirmed by using [phenyl-14C] benzoate (56.9 mCi/mmol) as the substrate and a previously described procedure (17). The reaction was stopped after 2 min, and the assay mixture was extracted twice with ethyl acetate.
CoA-transferase activity was measured by using a procedure modified from that of Scherf and Buckel (43). Benzoyl-CoA was used as the CoA donor, and acetate was used as the CoA acceptor. The reaction mixture contained 100 mM phosphate buffer (pH 7.0), 0.2 M sodium acetate, 1 mM oxaloacetate, 1 mM 5,5′-dithio-bis-(2-nitrobenzoate), 0.1 mM benzoyl-CoA, and 4 U of citrate synthase in a total volume of 1 ml. The reaction was initiated by adding 5 to 50 μl of cell extract. The CoA liberated by citrate synthase activity reacted with 5,5′-dithio-bis-(2-nitrobenzoate) to form a yellow thiophenolate anion. The initial rates were determined by measuring the formation of this anion at 412 nm.
Cyclohex-1-ene carboxyl-CoA hydratase and 2-hydroxycyclohexane carboxyl-CoA dehydrogenase activities were assayed as previously described (40). Cyclohex-1-enecarboxyl-CoA hydratase activity was determined as the combined cyclohex-1-ene carboxyl-CoA hydratase and 2-hydroxycyclohexane carboxyl-CoA dehydrogenase activities assayed in the forward direction by using cyclohex-1-ene carboxyl-CoA as the substrate. 2-Hydroxycyclohexane carboxyl-CoA dehydrogenase activity was assayed in the reverse direction by using 2-ketocyclohexane carboxyl-CoA as the substrate. Both reactions were initiated by adding 1 to 5 μl of cell extract and were monitored by measuring oxidation of NADH at 340 nm. 2-Ketocyclohexane carboxyl-CoA hydrolase activity was assayed by using a procedure modified from the procedure of Perrota and Harwood (40). The reaction mixture contained 100 mM Tris-HCl buffer (pH 7.0), 100 mM MgCl2, 1 mM DTT, and 1 mM 2-ketocyclohexane carboxyl-CoA in a total reaction volume of 50 μl. The reaction was initiated by adding 1 to 5 μl of cell extract and was monitored by measuring the decrease in absorbance of the magnesium enolate complex at 314 nm.
Acyl-CoA dehydrogenase activity was assayed by the ferricenium hexafluorophosphate method (30). The reaction mixture contained 100 mM Tris-HCl buffer (pH 7.5), 0.1 mM ferricenium hexafluorophosphate, and 0.05 mM substrate in a total reaction volume of 1 ml. The reaction was initiated by adding 5 to 50 μl of cell extract. The enzyme activity was determined by monitoring the initial decrease in absorbance at 300 nm upon reduction of the ferricenium ion. For oxidation of 1 mol of an acyl-CoA substrate, 2 mol of ferricenium ions was required. The substrates examined included glutaryl-CoA, pimelyl-CoA, butyryl-CoA, octanoyl-CoA, and palmitoyl-CoA.
Enoyl-CoA hydratase activity was assayed indirectly by using a coupled assay with a mixture containing l-(+)-3-hydroxyacyl-CoA dehydrogenase (52). The reaction was initiated by adding crotonyl-CoA and was monitored by measuring reduction of NAD. l-(+)-3-Hydroxyacyl-CoA dehydrogenase activity was determined by measuring the oxidation of NADH coupled to reduction of S-acetoacetyl-CoA to 3-hydroxybutyryl-CoA (52). The reaction was initiated by adding S-acetoacetyl-CoA. 3-Ketoacyl-CoA thiolase activity was determined by monitoring CoA-dependent acetoacetyl-CoA cleavage (52). The reaction was initiated by adding CoA and was monitored by measuring the decrease in absorbance at 303 nm.
Phosphotransacetylase activity was assayed by measuring the formation of acetyl-CoA from acetyl-phosphate (52). The reaction was initiated by adding 10 to 50 μl of cell extract and was monitored by measuring the appearance of the thioester bond at 233 nm. Acetate kinase activity was assayed by the hydroxamate method (41). The reaction mixture contained 770 mM sodium acetate, 50 mM Tris-HCl buffer (pH 7.4), 1 mM MgCl2, 10 mM ATP, 10% hydroxylamine hydrochloride, and cell extract in a total volume of 1 ml. After incubation for 2 min at room temperature, the reaction was stopped by adding 1 ml of 10% trichloroacetic acid. The absorbance at 540 nm was measured by using a blank that contained all of the reagents except ATP.
Enzyme assays were performed aerobically at room temperature unless otherwise stated. The activities were corrected for the endogenous activities present in the cell extracts and were proportional to protein concentrations. Controls containing boiled extracts or lacking the substrate were included for each assay. Enzyme assays were performed by using cell extracts prepared from at least two or three different cell batches, and the coefficients of variation between replicate cultures were less than 15%.
Chemical synthesis.
2-Hydroxycyclohexane carboxylic acid was synthesized by reducing ethyl 2-cyclohexanone carboxylate with sodium borohydride in 95% ethanol as previously described (40). 2-Ketocyclohexane carboxylic acid was also synthesized from ethyl-2-cyclohexanone (12, 40). 2-Ketocyclohexane carboxyl-CoA, cyclohex-1-ene carboxyl-CoA, pimelyl-CoA, and glutaconyl-CoA were synthesized by reacting 2-ketocyclohexane carboxylic acid, cyclohex-1-ene carboxylic acid, pimelic acid, and glutaconic acid, respectively, with free CoA by using a procedure modified from the procedures of Merckel et al. (35) and Gallus and Schink (16). Crude preparations of the CoA thioesters were purified by using C18 reversed-phase cartridges (Sep-Pak Plus; Millipore Corp., Midford, Mass.) as previously described (40). Ferricenium hexafluorophosphate was synthesized from ferrocine and sodium hexafluorophosphate (30).
Chemicals.
Sodium benzoate was purchased from Sigma Chemical Co. (St. Louis, Mo.), cyclohexane carboxylic acid was purchased from Acros Organics (Fair Lawn, N.J.), and cyclohex-1-ene carboxylic acid was purchased from Aldrich Chemical Co. (Milwaukee, Wis.). [ring-13C6]benzoate and CDCl3 were purchased from Cambridge Isotope Laboratories (Andover, Mass.). S-Acetoacetyl-CoA, benzoyl-CoA, glutaryl-CoA, butyryl-CoA, octanoyl-CoA, CoA (sodium salt), ferrocine, sodium hexafluorophosphate, phosphoenolpyruvate, pyruvate kinase, myokinase, lactate dehydrogenase, citrate synthase, NADH, NAD+, crotonase, l-(+)-3-hydroxyacyl-CoA dehydrogenase, acetyl phosphate, and ATP were purchased from Sigma Chemical Co. All other chemicals used in this study were obtained from Sigma, Aldrich, or Fluka (Milwaukee, Wis.).
RESULTS
Metabolites produced during benzoate degradation.
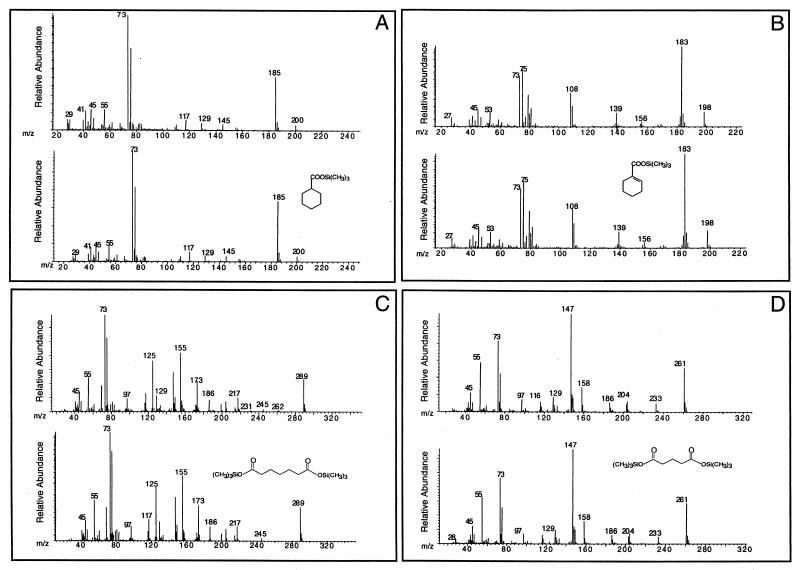
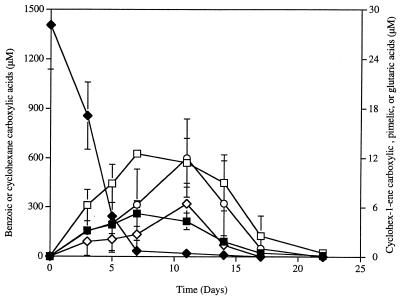
During growth of the “S. aciditrophicus”-M. hungatei cocultures with benzoate, cyclohexane carboxylate, cyclohex-1-ene carboxylate, pimelate, and glutarate were detected as their TMS derivatives. The TMS derivative of each compound had the same retention time and mass spectral profile as the the TMS derivative of the authentic chemical standard (Fig. 1). None of these compounds were detected in non-substrate-amended or heat-killed controls. Cyclohexane carboxylate (Fig. 1A) was transiently produced and accumulated to a maximum concentration of 260 μM in culture fluids (Fig. 2). The maximum concentration of cyclohexane carboxylate occurred when about 97.5% of the benzoate was consumed (Fig. 2). Cyclohex-1-ene carboxylate, pimelate, and glutarate (Fig. 1B to D) were also transiently produced and consumed, but these compounds were detected at much lower concentrations (Fig. 2). The maximum concentrations observed were 12.5, 6.4, and 11.3 μM for cyclohex-1-ene-carboxylate, pimelate, and glutarate, respectively, which represented 0.45 to 0.9% of the initial amount of benzoate. These four compounds were detected at similar concentrations regardless of whether the alkaline hydrolysis step was included and whether the entire culture or the cell-free culture fluid was analyzed.
FIG. 1.
Mass spectra of TMS derivatives of metabolites detected in “S. aciditrophicus”-M. hungatei benzoate-grown cocultures (upper spectra) and TMS derivatives of authentic chemical standards (lower spectra). (A) cyclohexane carboxylate-TMS; (B) cyclohex-1-ene carboxylate-TMS; (C) pimelate-TMS; (D) glutarate-TMS.
FIG. 2.
Transient detection of cyclohexane carboxylate (■), cyclohex-1-ene carboxylate (□), pimelate (◊), and glutarate (○) in “S. aciditrophicus”-M. hungatei cocultures grown with benzoate (⧫). Each compound was detected and quantified as its TMS derivative.
In addition, another metabolite with the same GC retention time and mass spectrum as the TMS derivative of 3-hydroxybutyrate was identified in “S. aciditrophicus”-M. hungatei culture fluids. However, this compound was also detected in non-substrate-amended and heat-killed controls, suggesting that 3-hydroxybutyrate was present in the cells used as the inoculum. 3-Hydroxybutyrate is an intermediate in crotonate metabolism by syntrophic bacteria (34, 52).
Cyclohexane carboxylate, cyclohex-1-ene carboxylate, pimelate, and glutarate were detected as their TMS derivatives in cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 at concentrations similar to those in the methanogenic cocultures (data not shown). Thus, the identities and quantities of metabolites produced during syntrophic growth on benzoate were independent of the hydrogen-utilizing partner.
Time course experiments relating methane production to benzoate consumption in “S. aciditrophicus”-M. hungatei cocultures indicated that the amount of methane produced per mole of benzoate consumed was much less than theoretically predicted (0.75 mol of methane per mol of benzoate) during the initial stages of benzoate metabolism. After 3 days, 72.7% of the initial benzoate present was consumed, and the ratio of amount of methane produced to amount of benzoate consumed was 0.3. After 5 days, 96% of the benzoate was consumed, and the methane-to-benzoate ratio was 0.49. Only after 100% of the benzoate was consumed (day 11) was the methane-to-benzoate ratio (0.76) close to the theoretical value, 0.75. Thus, it appears that some of the electrons produced during benzoate metabolism were used to reduce benzoyl-CoA to cyclohexane carboxylate (or its CoA derivative).
13C NMR analysis of culture extracts provided further evidence that cyclohexane carboxylate and cyclohex-1-ene carboxylate (or their CoA derivatives) were intermediates in benzoate metabolism by “S. aciditrophicus.” The 13C NMR spectrum of the zero-time sample from “S. aciditrophicus”-M. hungatei cocultures contained two peak clusters centered at 133.5 and 129.5 ppm, which resulted from C-1 and C-2 through C-6, respectively, of [ring-13C6]benzoic acid. This was confirmed by comparison to the [ring-13C6]benzoic acid standard in CDCl3. The 13C-labeled compounds detected during the experiment are listed in Table 1. Cyclohexane carboxylic acid was produced at the highest concentration of all compounds detected. The concentration of this compound slowly decreased over the experimental period, and it was no longer detectable by day 11. Coupling constants could not be determined for the C-3, C-4, and C-5 atoms of cyclohexane carboxylic acid since the differences in their resonance frequencies were less than the carbon-carbon coupling constant frequency (Jcc); thus, the n+1 coupling rule no longer was applicable (48). Benzoic acid was detected at low concentrations up to day 9. Signals from cyclohex-1-ene carboxylic acid were detected from day 3 through day 9. The concentration of acetic acid increased over the 11-day incubation period, and acetate was the only detectable compound by day 11. Both of the peaks for the methyl and carboxyl groups of acetate were doublets overlaid with a singlet (data not shown). The singlet at 20.8 ppm represented 36 to 40% of the total acetic acid detected in the methyl group and resulted from acetic acid molecules that were 13C labeled only at the methyl group. The presence of a carboxyl singlet indicated that some of the acetic acid molecules had 13C-labeled atoms only at the carboxyl position.
TABLE 1.
NMR data for compounds detected in “S. aciditrophicus”-M. hungatei cocultures grown on [ring-13C]benzoic acid
| Compound | Position | δ 13C (ppm) | Jcc(Hz) |
|---|---|---|---|
| Acetic acida | Carboxyl | 177.0 | 55.7 |
| Methyl | 20.7 | 56.9 | |
| Benzoic acida | C-1 | 133.5 | 53.6, 10.5 |
| C-2–C-6 | 127.7–130.7 | ||
| Cyclohexane carboxylic acidb | C-1 | 42.8 | 32.0 |
| C-2, C-6 | 28.7 | 31.9 | |
| C-3, C-5 | 25.7 | ||
| C-4 | 25.4 | ||
| Cyclohex-1-ene carboxylic acidc | C-1 | 142.6 | 70.3, 41.0 |
| C-2 | 147.5 | 69.8, 41.1 | |
| C-3–C-6 | 18.0–25.0 |
Time course experiments were conducted to identify and quantify metabolites resulting from benzoate metabolism by R. palustris and T. aromatica grown with a similar benzoate concentration (about 1.5 mM); similar sample volumes (about 60 ml) were analyzed. In both cases, none of the compounds mentioned above or any other potential intermediate of benzoate metabolism was detected. This indicated that the intermediates of benzoate degradation in R. palustris and T. aromatica were produced at much lower concentrations than was the case with “S. aciditrophicus.”
Metabolism of cyclohexane carboxylate and cyclohex-1-ene carboxylate by “S. aciditrophicus”-M. hungatei cocultures.
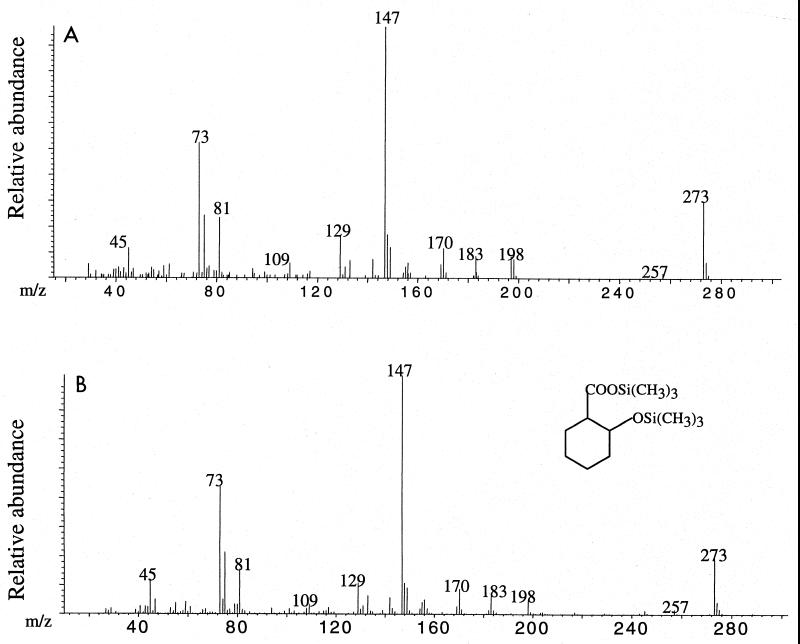
Cyclohex-1-ene carboxylate and cyclohexane carboxylate were metabolized without a lag by “S. aciditrophicus”-M. hungatei cocultures at rates slightly faster than the rate observed for benzoate (Table 2). Cocultures of “S. aciditrophicus” and M. hungatei grown with cyclohex-1-ene carboxylate and cyclohexane carboxylate were analyzed by GC-MS as described above. In cyclohex-1-ene carboxylate-grown cocultures, pimelate and glutarate were detected as their TMS derivatives at levels comparable to those found in benzoate-grown cultures; the maximum concentrations measured were 6.6 and 20.7 μM, respectively. Although cyclohexane carboxylate was detected (as its TMS derivative) in cyclohex-1-ene carboxylate-grown cultures, it was not produced at as high a concentration as observed in benzoate-grown cocultures. Instead, it was detected at levels comparable to the levels of pimelate and glutarate (e.g., at a maximum concentration of 4.2 μM). In addition, 2-hydroxycyclohexane carboxylate (Fig. 3) was detected as its TMS derivative in trace amounts (<0.5 μM). In cyclohexane carboxylate-grown cocultures, cyclohex-1-ene carboxylate, pimelate, and glutarate were detected. The maximum concentrations of these compounds in cyclohexane carboxylate-grown cultures were also comparable to those measured in benzoate-grown cocultures, (8.9, 6.1, and 15.7 μM for cyclohex-1-ene carboxylate, pimelate, and glutarate, respectively). None of the compounds mentioned above were detected in heat-killed or non-substrate-amended controls.
TABLE 2.
Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “S. aciditrophicus”-M. hungatei cocultures
| Compound | Amt of substrate added (μmol)a | Amt of methane produced (μmol)a | Ratio of amt of methane to amt of substrateb | Degradation rate (day−1) |
|---|---|---|---|---|
| Benzoate | 108.5 | 88.9 | 0.82 | 0.26 |
| Cyclohex-1-ene carboxylate | 115.5 | 166.2 | 1.44 | 0.35 |
| Cyclohexane carboxylate | 116.6 | 193.8 | 1.66 | 0.35 |
The values are averages based on duplicates.
The theoretical values for the ratio of amount of methane produced to amount of substrate degraded are 0.75 for benzoate, 1.25 for cyclohex-1-ene carboxylate, and 1.5 for cyclohexane carboxylate.
FIG. 3.
Mass specta of a TMS derivative of a metabolite detected in “S. aciditrophicus”-M. hungatei cyclohex-1-ene carboxylate-grown cocultures (A) and a synthesized TMS-derivatized 2-hydroxycyclohexane carboxylic acid standard (B).
Enzyme activities detected in cell extracts of “S. aciditrophicus.”
Cell extracts of benzoate-grown cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 contained an ATP-dependent benzoyl-CoA ligase activity (Table 3). Formation of benzoyl-CoA from benzoate was confirmed by the isotopic assay by using [phenyl-14C]benzoate (17). The results showed that 84% of the [14C]benzoate added was converted to benzoyl-CoA after 2 min. In addition to benzoate, 2-, 3-, and 4-fluorobenzoates, as well as 3-hydroxybenzoate, also served as substrates. No activity was detected with 4-hydroxybenzoate, 2-, 3-, or 4-chlorobenzoate, picolinic acid, phenylacetate, 4-hydroxyphenylacetate, crotonate, n-butyrate, isobutyrate, heptanoate, or hexanoate. No acyl-CoA ligase activity was detected in cell extracts of pure cultures of Desulfovibrio sp. strain G11 (data not shown). Similar levels of acyl-CoA activities were detected in crotonate-grown pure cultures of “S. aciditrophicus,” suggesting that this activity was constitutively present in “S. aciditrophicus.” Cell extracts of benzoate-grown cocultures contained very low levels of a benzoyl-CoA transferase activity. This activity was not detected in cell extracts of Desulfovibrio sp. strain G11 or pure cultures of “S. aciditrophicus.”
TABLE 3.
Enzyme activities detected in cell extracts of benzoate-grown cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 and in crotonate-grown pure cultures of “S. aciditrophicus”
| Enzymea | Sp act (nmol/min/mg of protein)
|
|
|---|---|---|
| Benzoate-grown coculture | Crotonate-grown pure culture | |
| Acyl-CoA ligases | ||
| Benzoate | 141 | 103 |
| 2-Fluorobenzoate | 70 | 107 |
| 3-Fluorobenzoate | 28 | 108 |
| 4-Fluorobenzoate | 102 | 129 |
| 3-Hydroxybenzoate | 36 | 6 |
| Benzoyl-CoA:acetate-CoA transferase | 3 | NDb |
| Cyclohex-1-ene 1-carboxyl-CoA hydratase | 57 | 2 |
| 2-Hydroxycyclohexane carboxyl-CoA dehydrogenase | 317 | 41 |
| 2-Ketocyclohexane carboxyl-CoA hydrolase | 391 | ND |
| Acyl-CoA dehydrogenases | ||
| Pimelyl-CoA | 1,144 | 581 |
| Glutaryl-CoA | 630 | 395 |
| Butyryl-CoA | NDc | 257 |
| Octanoyl-CoA | ND | 622 |
| Palmitoyl-CoA | ND | 256 |
| Enoyl-CoA hydratase | 18,780 | 23,880 |
| l-3-Hydroxyacyl-CoA dehydrogenase | 1,244 | 1,488 |
| 3-Ketoacyl-CoA thiolase | 287 | 323 |
| Phosphotransacetylase | 128 | ND |
| Acetate kinased | 21 | 10 |
Unless otherwise noted, activity was not detected in cell extracts of a pure culture of Desulfovibrio sp. strain G11.
ND, not detected.
Activity in a pure culture of Desulfovibrio sp. strain G11 was not determined.
The specific activity was 41 nmol min−1 mg of protein−1 in cell extract of a pure culture of Desulfovibrio sp. strain G11.
Cell extracts of benzoate-grown cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 contained many of the enzyme activities required for conversion of cyclohex-1-ene carboxyl-CoA to pimelyl-CoA (Table 3). The activity of cyclohex-1-ene carboxyl-CoA hydratase was measured by determining the combined cyclohex-1-ene carboxyl-CoA hydratase and 2-hydroxycyclohexane carboxyl-CoA dehydrogenase activities. The enzyme activity was present at levels similar to that reported for R. palustris (40). The hydratase activity was not detected in cell extracts of Desulfovibrio sp. strain G11, but low levels of activity were detected in crotonate-grown cultures of “S. aciditrophicus.” The activity was 28-fold higher in benzoate-grown cells than in crotonate-grown cultures of “S. aciditrophicus.” The activity of 2-hydroxycyclohexane carboxyl-CoA dehydrogenase was measured in the reverse direction by using 2-ketocyclohexane carboxyl-CoA as the substrate. Benzoate-grown “S. aciditrophicus”-Desulfovibrio sp. strain G11 cocultures contained high levels of the dehydrogenase activity. The specific activity of this enzyme was severalfold higher than the specific activity of cyclohex-1-ene carboxyl-CoA hydratase, a pattern similar to that reported for R. palustris (40). This enzyme activity was not detected in pure cultures of Desulfovibrio sp. strain G11 and was eightfold higher in benzoate-grown cocultures than in crotonate-grown pure cultures of “S. aciditrophicus” (Table 3). Cell extracts of benzoate-grown cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 also contained 2-ketocyclohexane carboxyl-CoA hydrolase at levels similar to that of 2-hydroxycyclohexane carboxyl-CoA dehydrogenase. This activity was not detected in pure cultures of Desulfovibrio sp. strain G11. The hydrolase activity was present at a level severalfold lower than that reported for R. palustris (40). Addition of 1 mM exogenous CoA did not stimulate the hydrolase activity, which is consistent with the ring cleavage reaction being hydrolytic rather than thiolytic. Unlike the activity in R. palustris cultures, the 2-ketocyclohexane carboxyl-CoA hydrolase activity in benzoate-grown cocultures was detectable only when DTT was included in the reaction mixture or when the reaction mixtures were processed in the anaerobic chamber. This suggested that the 2-ketocyclohexane carboxyl-CoA hydrolase in “S. aciditrophicus” may be oxygen sensitive. Also, no hydrolase activity was detected in crotonate-grown pure cultures of “S. aciditrophicus,” which suggested that the 2-ketocyclohexane carboxyl-CoA hydrolase activity in “S. aciditrophicus” was induced by growth on benzoate.
Cell extracts of benzoate-grown cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 contained high levels of pimelyl-CoA dehydrogenase, the first enzyme in the reaction sequence leading to the formation of glutaryl-CoA from pimelyl-CoA. The specific activity of this enzyme was twofold higher in benzoate-grown cocultures than in crotonate-grown pure cultures of “S. acidotrophicus” (Table 3). No activity was detected in cell extracts of pure cultures of Desulfovibrio sp. strain G11. Benzoate-grown cocultures of “S. aciditrophicus” and Desulfovibrio sp. strain G11 did not contain detectable acyl-CoA dehydrogenase activity when butyryl-CoA, octanoyl-CoA, or palmitoyl-CoA was the substrate. Also, glutaryl-CoA dehydrogenase activity was detected in cocultures as well as in crotonate-grown pure cultures of “S. aciditrophicus.” This activity was twofold higher in benzoate-grown cocultures than in crotonate-grown pure cultures. In addition to pimelyl-CoA dehydrogenase and glutaryl-CoA dehydrogenase activities, cell extracts of crotonate-grown pure cultures of “S. aciditrophicus” contained high levels of butyryl-CoA dehydrogenase, crotonyl-CoA dehydrogenase, and palmitoyl-CoA dehydrogenase activities (Table 3).
All of the enzymes required for conversion of crotonyl-CoA to acetyl-CoA and then to acetate were present in cell extracts of benzoate-grown “S. aciditrophicus”-Desulfovibrio sp. strain G11 cocultures, as well as in crotonate-grown pure cultures of “S. aciditrophicus.” The specific activities were more or less the same regardless of the substrate used for growth (Table 3).
DISCUSSION
Using GC-MS analysis, we identified cyclohexane carboxylate, cyclohex-1-ene carboxylate, pimelate, and glutarate in benzoate-grown cocultures of “S. aciditrophicus” and M. hungatei based on a comparison of the retention times and mass spectra of their TMS derivatives to the retention times and mass spectra of authentic chemical standards (Fig. 1 and 2). In addition to pimelate and glutarate, we also detected cyclohexane carboxylate and 2-hydroxycyclohexane carboxylate (Fig. 3) in cyclohex-1-ene carboxylate-grown cocultures and cyclohex-1-ene carboxylate in cyclohexane carboxylate-grown cocultures. These compounds were transiently produced and were not detected in heat-killed or non-substrate-amended controls, providing strong evidence that these compounds, probably as their CoA derivatives, are intermediates in the metabolism of benzoate, cyclohexane carboxylate, and cyclohex-1-ene carboxylate. 13C NMR spectroscopy confirmed that cyclohexane carboxylate and cyclohex-1-ene carboxylate were produced from benzoate. The fact that cyclohexane carboxylate, cyclohex-1-ene carboxylate, pimelate, and glutarate were detected when the alkaline hydrolysis step was omitted and were found in whole-culture broth (cells plus medium) and in cell-free culture broth suggests that “S. aciditrophicus” may excrete potential intermediates of benzoate and alicyclic acid metabolism. Several studies have detected free acids corresponding to potential intermediates in phenol (5) and benzoate (13) metabolism. Since “S. aciditrophicus” cocultures and pure cultures contain enzymatic activities that metabolize the various alicyclic and aliphatic CoA intermediates of benzoate metabolism (Table 3), this finding suggests that the CoA derivatives of the compounds mentioned above and not the free acids are the functional intermediates involved in benzoate and alicyclic acid metabolism.
Cyclohexane carboxylate was originally thought to be an intermediate in benzoate metabolism by R. palustris (13). However, subsequent investigations of cyclohexane carboxylate metabolism in this microorganism (27), as well as a better understanding of the biochemistry and genetics of benzoyl-CoA metabolism in R. palustris (38–40) and T. aromatica (28, 29), led to exclusion of cyclohexane carboxylate from the benzoyl-CoA pathway in both microorganisms. It was suggested that the transient production of cyclohexane carboxylate in R. palustris is a physiological response to excess reducing equivalents or is a mechanism to maintain appropriate intracellular levels of free CoA (27). Some of the earlier studies on benzoate degradation by methanogenic consortia from sewage sludge, rumen fluid, or anaerobic mud (15, 24, 37) also suggested that cyclohexane carboxylate is a benzoate degradation intermediate based on cochromatography data and the ability of the consortia to degrade cyclohexane carboxylate without a lag period. Given the number of different organisms that are present in such consortia, it was difficult to reach definitive conclusions regarding potential intermediates involved in the degradation of benzoate in these studies. However, we clearly show that relatively large amounts of cyclohexane carboxylate, accounting for about 18% of the benzoate carbon, are formed and consumed by “S. aciditrophicus” during growth with benzoate. These data suggest that cyclohexane carboxylate may serve as a repository for reducing equivalents generated during benzoate metabolism.
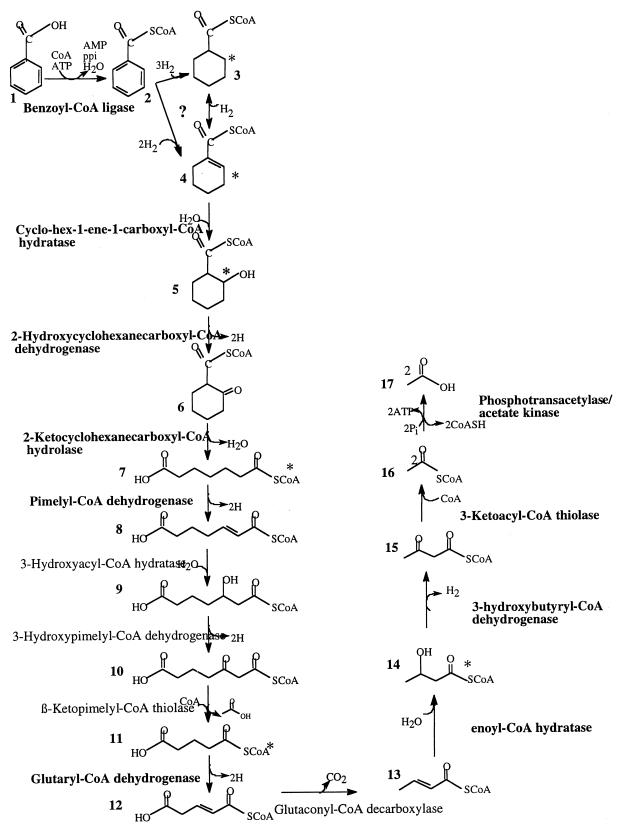
Detection of cyclohexane carboxylate, as well as cyclohex-1-ene carboxylate, pimelate, and glutarate, at levels not encountered in R. palustris or T. aromatica suggests that the pathway for benzoate metabolism in syntrophic bacteria is different from that in phototrophs and nitrate reducers. This difference is not surprising for two reasons. First, members of the genus Syntrophus, which belong to the δ subgroup of the class Proteobacteria, T. aromatica, which belongs to the β subgroup of the Proteobacteria, and R. palustris, which belongs to the α subgroup of the Proteobacteria, are phylogenetically distinct. Second, the energy constraints encountered in syntrophic aromatic metabolism (45–47) are not encountered by organisms that can obtain energy from respiration and photosynthesis. It is obvious that syntrophic microorganisms can obtain energy for growth on benzoate, yet it is not clear how net ATP production occurs if benzoate activation and ring reduction during syntrophic benzoate metabolism occur, as found in benzoate-degrading phototrophs and denitrifiers (18, 46, 47). This study and other studies (4, 46) demonstrated that benzoate activation proceeds by a benzoyl-CoA ligase reaction which uses two energy-rich bonds. We also detected a benzoate:acetyl-CoA transferase reaction that is similar to the succinyl-CoA:benzylsuccinate transferase reaction that has been suggested for benzylsuccinate activation in T. aromatica (31), which used only the equivalent of one energy-rich bond for benzoate activation. However, the level of activity of this enzyme in benzoate-grown cocultures is very low and is probably not sufficient to account for benzoate metabolism. While our evidence to date cannot exclude the possibility that there is a two-electron reduction of benzoyl-CoA to cyclohex-1,5-diene carboxylate, the transient production of large amounts of cyclohexane carboxylate suggests that ring reduction may differ in syntrophic metabolism. Thermodynamic calculations indicate that if the product of benzoyl-CoA reduction is cyclohex-1-ene carboxyl-CoA or cyclohexane carboxyl-CoA instead of cyclohex-1,5-diene carboxyl-CoA, the product observed in T. aromatica (9), then the reaction could proceed without ATP investment (44, 46). The detection of cyclohexane carboxylate and cyclohex-1-ene carboxylate in cell extracts is consistent with a four- or six-electron reduction step rather than a two-electron reduction step.
Although our work demonstrates that cyclohexane carboxylate is produced and consumed by benzoate-grown cultures of “S. aciditrophicus,” the role of this compound (or its CoA derivative) in syntrophic benzoate degradation is unclear. Cyclohexane carboxyl-CoA might be the product formed by a six-electron reduction of benzoyl-CoA. Cyclohexane carboxyl-CoA could then be oxidized to cyclohex-1-ene carboxyl-CoA by cyclohexane carboxyl-CoA dehydrogenase, and the latter compound could be metabolized in a manner similar to that observed in R. palustris (Fig. 4). However, this does not explain why cyclohexane carboxylate is produced at a level far higher than the levels of other metabolites. Another possible explanation is that cyclohexane carboxylate is produced in a dismutation reaction in which the reducing equivalents produced during oxidation of one benzoyl-CoA molecule are used to reduce another benzoyl-CoA molecule to cyclohexane carboxyl-CoA (Fig. 4). This scheme would require two distinct enzymes to reduce benzoyl-CoA, one enzyme to reduce benzoyl-CoA to cyclohex-1-ene carboxyl-CoA and another enzyme to reduce benzoyl-CoA to cyclohexane carboxyl-CoA. It has recently been suggested that benzoate dismutation (simultaneous oxidation and reduction of benzoate) is an alternative mechanism for hydrogen removal in methanogenic, benzoate-degrading enrichments in which the electron flow to methanogenesis is inhibited by bromoethanesulfonic acid (25). However, if such a reaction occurs in “S. aciditrophicus,” it is not clear what factors determine whether the reducing equivalents produced during benzoate metabolism form hydrogen or are used to reduce benzoyl-CoA to cyclohexane carboxyl-CoA. A third possibility is that benzoyl-CoA is reduced to cyclohex-1-ene carboxyl-CoA and then a dismutation reaction occurs in which oxidation of one molecule of cyclohex-1-ene carboxyl-CoA is coupled to reduction of three molecules of cyclohex-1-ene carboxyl-CoA (Fig. 4). However, the fact that cyclohex-1-ene carboxylate-grown cocultures do not accumulate cyclohexane carboxylate at levels comparable to those in benzoate-grown cultures argues against this possibility.
FIG. 4.
Proposed pathway for benzoate metabolism in “S. aciditrophicus.” Enzyme activities detected in “S. aciditrophicus” cell extracts are indicated by boldface type. The asterisks indicate compounds detected as their TMS derivatives in “S. aciditrophicus”-M. hungatei coculture extracts. 1, Benzoate; 2, benzoyl-CoA; 3, cyclohexane carboxyl-CoA; 4, cyclohex-1-ene carboxyl-CoA; 5, 2-hydroxycyclohexane carboxyl-CoA; 6, 2-ketocyclohexane carboxyl-CoA; 7, pimelyl-CoA; 8, 2-heptenedioc acid-1-CoA; 9, 3-hydroxypimelyl-CoA; 10, 3-ketopimelyl-CoA; 11, glutaryl-CoA; 12, glutaconyl-CoA; 13, crotonyl-CoA; 14, 3-hydroxybutyryl-CoA; 15, acetoacetyl-CoA; 16, acetyl-CoA; 17, acetate. The question mark indicates that the exact benzene ring reduction mechanism in “S. aciditrophicus” is not completely understood yet.
Enzyme studies in which “S. aciditrophicus” cell extracts were used indicated that the organism metabolizes cyclohex-1-ene carboxyl-CoA by a pathway similar to that observed in R. palustris. It could be argued that the same enzymes could also catalyze cyclohex-1,5-diene transformation to 6-hydroxycyclohex-1-ene carboxylate, 2-keto-6-hydroxycyclohex-1-ene carboxyl-CoA, and 3-hydroxypimelyl-CoA (19) since the latter substrates were not tested in our enzyme assays because they were not available commercially. However, detection of cyclohex-1-ene carboxylic acid, 2-hydroxycyclohexane carboxylic acid, and pimelic acid in culture extracts is consistent with a pathway similar to the R. palustris pathway. Also, the twofold-higher level of pimelyl-CoA dehydrogenase in benzoate-grown cells than in crotonate-grown cells is consistent with production of pimelate as the ring cleavage product (40). This activity was specific for pimelyl-CoA since benzoate-grown cells of “S. aciditrophicus” lacked detectable activity when butyryl-CoA, octanoyl-CoA, and palmitoyl-CoA were used as substrates. Thus, it is unlikely that the enzyme activity detected in benzoate-grown cells was due to the presence of a nonspecific acyl-CoA dehydrogenase. The pimelyl-CoA dehydrogenase activity, which was demonstrated for the first time in a benzoate-grown anaerobic microorganism, was not detected in a nitrate-reducing isolate when it was grown with benzoate (16), probably because benzoate degradation in the isolate proceeds via transformation of cyclohex-1,5-diene carboxyl-CoA to 3-hydroxypimelate without the formation of pimelyl-CoA. The dehydrogenase activity was detected, however, when the same isolate was grown on pimelate (16). It could also be argued that the observed cyclohex-1-ene carboxyl-CoA hydratase activity is due to the action of an enoyl-CoA hydratase, which acts primarily on short-chain unsaturated fatty acids (28). However, detection of cyclohex-1-ene carboxyl-CoA hydratase at 28-fold-higher levels when the organism was grown with benzoate than when it was grown with crotonate and detection of enoyl-CoA hydratase at similar levels in benzoate- and crotonate-grown cells indicate that a specific cyclohex-1-ene carboxyl-CoA hydratase activity is present in “S. aciditrophicus.”
An intriguing observation is the presence of both 12C and 13C carbons in the carboxyl and methyl moieties of acetate. Acetate molecules with 13C only in the methyl group most likely arose from pimelate in which one of the carboxyl groups was the original unlabeled carboxyl group of benzoate. The presence of acetate molecules that have 13C only in the carboxyl group is less easily understood since the use of a ring-13C6-labeled compound should result in acetate molecules with both carbons 13C labeled, except for the situation discussed above. It is possible that the glutaconyl-CoA decarboxylation reaction is in equilibrium with the bicarbonate pool and that this reaction allows an exchange of 12C and 13C atoms to occur.
In conclusion, although syntrophic benzoate metabolism follows the main themes observed in nitrate reducers and phototrophs, it appears that a third variant of the benzoyl-CoA pathway in which cyclohexane carboxylic acid is produced from benzoate may operate in microorganisms that syntrophically degrade benzoate (Fig. 4). The variation is probably imposed by the strict energy constraints of syntrophic metabolism. Detailed investigations are still required to determine the exact function of cyclohexane carboxylic acid in syntrophic benzoate metabolism and to determine how syntrophic microorganisms are able to obtain net ATP production to support growth on benzoate. The fact that three variants of the benzoyl-CoA pathway have been detected in the three groups of microorganisms that have been studied so far raises the question of how similar or different benzoate metabolism is in other physiological groups of microorganisms, such as sulfate-reducing and iron-reducing bacteria that have yet to be studied.
ACKNOWLEDGMENTS
This work was supported by DOE grant DE-FG03-96-ER-20214/A003. Funds for the Oklahoma Statewide Shared NMR Facility were provided by National Science Foundation grant BIR-9512269, by the National Science Foundation EPSCoR, by the Oklahoma State Regents for Higher Education, by the W. M. Keck Foundation, and by Conoco Inc.
We thank C. S. Harwood for technical assistance throughout this work and Jesus Maza for assistance in obtaining the NMR spectra.
REFERENCES
- 1.Altenschmidt U, Oswald B, Fuchs G. Purification and characterization of benzoate-coenzyme A and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J Bacteriol. 1991;173:5494–5501. doi: 10.1128/jb.173.17.5494-5501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auburger G, Winter J. Activation and degradation of benzoate, 3-phenylpropionate and crotonate by Syntrophus buswellii strain GA. Evidence for electron-transport phosphorylation during crotonate respiration. Appl Microbiol Biotechnol. 1996;44:807–815. doi: 10.1007/BF00178623. [DOI] [PubMed] [Google Scholar]
- 3.Auburger G, Winter J. Isolation and physiological characterization of Syntrophus buswellii strain GA from a syntrophic benzoate-degrading, strictly anaerobic coculture. Appl Microbiol Biotechnol. 1995;44:241–248. [Google Scholar]
- 4.Auburger G, Winter J. Purification and characterization of benzoyl-CoA ligase from a syntrophic, benzoate-degrading, anaerobic mixed-culture. Appl Microbiol Biotechnol. 1992;37:789–795. doi: 10.1007/BF00174847. [DOI] [PubMed] [Google Scholar]
- 5.Bakker G. Anaerobic degradation of aromatic compounds in the presence of nitrate. FEMS Lett. 1977;1:103–108. [Google Scholar]
- 6.Balch W E, Wolfe R S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaty P S, McInerney M J. Growth of Syntrophomonas wolfei in pure culture on crotonate. Arch Microbiol. 1987;147:389–393. [Google Scholar]
- 8.Boll M, Albracht S S P, Fuchs G. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. A study of adenosine triphosphate activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur J Biochem. 1997;244:840–851. doi: 10.1111/j.1432-1033.1997.00840.x. [DOI] [PubMed] [Google Scholar]
- 9.Boll M, Fuchs G. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Dean J A. Lange's handbook of chemistry. Vol. 7. New York, N.Y: McGraw-Hill; 1999. pp. 7.108–7.109. [Google Scholar]
- 12.Diekman W. Ueber cyklische B-Ketocarbonsauerester. Liebigs Ann Chem. 1901;317:98–109. [Google Scholar]
- 13.Dutton P L, Evans W C. The metabolism of aromatic compounds by Rhodopseudomonas palustris. A new, reductive method of aromatic ring metabolism. Biochem J. 1969;113:525–536. doi: 10.1042/bj1130525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egland P G, Pelletier D A, Dispensa M, Gibson J, Harwood C S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferry J G, Wolfe R S. Anaerobic degradation of benzoate to methane by a microbial consortium. Arch Microbiol. 1976;107:33–40. doi: 10.1007/BF00427864. [DOI] [PubMed] [Google Scholar]
- 16.Gallus C, Schink B. Anaerobic degradation of pimelate by newly isolated denitrifying bacteria. Microbiology. 1994;140:409–416. doi: 10.1099/13500872-140-2-409. [DOI] [PubMed] [Google Scholar]
- 17.Geissler J F, Harwood C S, Gibson J. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol. 1988;170:1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1999;22:439–458. [Google Scholar]
- 19.Harwood C S, Gibson J. Shedding light on anerobic benzene ring degradation: a process unique to prokaryotes? J Bacteriol. 1997;179:301–309. doi: 10.1128/jb.179.2.301-309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heider J, Fuchs G. Anaerobic metabolism of aromatic compounds. Eur J Biochem. 1997;243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins B T, McInerney M J, Warikoo V. Evidence for an anaerobic syntrophic benzoate degradation threshold and isolation of the syntrophic benzoate degrader. Appl Environ Microbiol. 1995;61:526–530. doi: 10.1128/aem.61.2.526-530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson B E, Bhupathiraju V K, Tanner R S, Woese C R, McInerney M J. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol. 1999;171:107–114. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- 23.Jenneman G E, McInerney M J, Knapp R M. Effect of nitrate on biogenic sulfide production. Appl Environ Microbiol. 1986;56:1205–1211. doi: 10.1128/aem.51.6.1205-1211.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keith C L, Bridges R L, Fina L R, Iverson K L, Claron J A. The anaerobic decomposition of benzoic acid during methane fermentation. IV. Dearomatization of the ring and volatile fatty acids formed on ring rupture. Arch Microbiol. 1978;118:173–176. doi: 10.1007/BF00415726. [DOI] [PubMed] [Google Scholar]
- 25.Kleerebezem R, Hulshoff L W, Lettinga G. Energetics of product formation during anaerobic degradation of phthalate isomers and benzoate. FEMS Microbiol Ecol. 1999;29:273–282. [Google Scholar]
- 26.Koch J, Eisenreich W, Bacher A, Fuchs G. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur J Biochem. 1993;211:649–661. doi: 10.1111/j.1432-1033.1993.tb17593.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuver J, Xu Y, Gibson J. Metabolism of cyclohexane carboxylic acid by the photosynthetic bacterium Rhodopseudomonas palustris. Arch Microbiol. 1995;159:337–345. doi: 10.1007/BF02529980. [DOI] [PubMed] [Google Scholar]
- 28.Laempe D, Eisenreich W, Bacher A, Fuchs G. Cyclohexa-1,5-diene-1-carboxyl-CoA hydratase, an enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1998;255:618–627. doi: 10.1046/j.1432-1327.1998.2550618.x. [DOI] [PubMed] [Google Scholar]
- 29.Laempe D, Jahn M, Fuchs G. 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1999;263:420–429. doi: 10.1046/j.1432-1327.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 30.Lehman T C, De H, Bhala A, Thorpe C. An acyl-coenzyme A dehydrogenase assay utilizing ferricenium ion. Anal Biochem. 1990;186:280–284. doi: 10.1016/0003-2697(90)90080-s. [DOI] [PubMed] [Google Scholar]
- 31.Leutwein C, Heider J. Anaerobic toluene-catabolic pathway in denitrifying Thauera aromatica: activation and beta-oxidation of the first intermediate, (R)-(+)-benzylsuccinate. Microbiology. 1999;145:3265–3271. doi: 10.1099/00221287-145-11-3265. [DOI] [PubMed] [Google Scholar]
- 32.Londry K L, Fedorak P M, Suflita J M. Anaerobic degradation of m-cresol by a sulfate-reducing bacterium. Appl Environ Microbiol. 1997;63:3170–3175. doi: 10.1128/aem.63.8.3170-3175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McInerney M J, Bryant M P, Pfenning N. Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens. Arch Microbiol. 1979;122:129–135. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- 34.McInerney M J, Wofford N Q. Enzymes involved in crotonate metabolism in Syntrophomonas wolfei. Arch Microbiol. 1992;158:344–349. [Google Scholar]
- 35.Merkel S M, Eberhard A E, Gibson J, Harwood C S. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rheudopseudomonas palustris. J Bacteriol. 1989;171:1–7. doi: 10.1128/jb.171.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mountfort D O, Bryant M P. Isolation and characterization of an anaerobic benzoate-degrading bacterium from sewage sludge. Arch Microbiol. 1982;133:249–256. [Google Scholar]
- 37.Nottingham P M, Hungate R E. Methanogenic fermentation of benzoate. J Bacteriol. 1969;98:1170–1172. doi: 10.1128/jb.98.3.1170-1172.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelletier D A, Harwood C S. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J Bacteriol. 2000;182:2753–2760. doi: 10.1128/jb.182.10.2753-2760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelletier D A, Harwood C S. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J Bacteriol. 1998;180:2330–2336. doi: 10.1128/jb.180.9.2330-2336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrotta J A, Harwood C S. Anaerobic metabolism of cyclohex-1-ene-1-carboxylate, a proposed intermediate of benzoate degradation, by Rhodopseudomonas palustris. Appl Environ Microbiol. 1994;60:1775–1782. doi: 10.1128/aem.60.6.1775-1782.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose I A. Acetate kinase of bacteria. Methods Enzymol. 1955;1:591–595. [Google Scholar]
- 42.Sadtler Research Laboratories. Standard spectra, carbon-13 NMR spectra. Philadelphia, Pa: Stadtler Research Laboratories; 1984. [Google Scholar]
- 43.Scherf U, Buckel W. Purification and properties of 4-hydroxybutyrate coenzyme A transferase from Clostridium aminobutyricum. Appl Environ Microbiol. 1991;57:2699–2702. doi: 10.1128/aem.57.9.2699-2702.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schöcke L, Schink B. Energetics of methanogenic benzoate degradation by Syntrophus gentianae in syntrophic coculture. Microbiology. 1997;143:2345–2351. doi: 10.1099/00221287-143-7-2345. [DOI] [PubMed] [Google Scholar]
- 46.Schöcke L, Schink B. Bichemistry of benzoate degradation by S. gentinae. Arch Microbiol. 1999;77:100–110. [Google Scholar]
- 47.Schöcke L, Schink B. Membrane-bound proton-translocating pyrophosphatase of Syntrophus gentianae, a syntrophically benzoate-degrading fermenting bacterium. Eur J Biochem. 1998;256:589–594. doi: 10.1046/j.1432-1327.1998.2560589.x. [DOI] [PubMed] [Google Scholar]
- 48.Stoecker M. Carbon-13–carbon-13 coupling constants of cycloalkane carboxylic acids. J Chem Res Synop. 1981;1981:52–53. [Google Scholar]
- 49.Tarvin D, Buswell A M. The methane fermentation of organic acids and carbohydrates. J Am Chem Soc. 1934;56:1751–1755. [Google Scholar]
- 50.Tschech A, Fuchs G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol. 1987;148:213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- 51.Wallrabenstein C, Gorny N, Springer N, Ludwig W, Schink B. Pure cultures of Syntrophus buswellii, definition of its phylogenetic status, and description of Syntrophus gentianae sp. nov. Syst Appl Microbiol. 1995;18:62–66. [Google Scholar]
- 52.Wofford N Q, Beaty P S, McInerney M J. Preparation of cell-free extracts and the enzymes involved in fatty acid metabolism in Syntrophomonas wolfei. J Bacteriol. 1986;167:136–142. doi: 10.1128/jb.167.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]