Abstract
Amyloid precursor protein (APP) is cleaved and processed sequentially by γ-secretase yielding amyloid β (Aβ) peptides of different lengths. Longer Aβ peptides are associated with the formation of neurotoxic plaques related to Alzheimer’s disease. Based on the APP substrate-bound structure of γ-secretase, we investigated the enzyme-substrate interaction using molecular dynamics simulations and generated model structures that represent the sequentially cleaved intermediates during the processing reaction. The simulations indicated an internal docking site providing strong enzyme-substrate packing interaction. In the enzyme-substrate complex, it is located close to the region where the helical conformation of the substrate is interrupted and continues toward the active site in an extended conformation. The internal docking site consists of two non-polar pockets that are preferentially filled by large hydrophobic or aromatic substrate side chains to stabilize binding. Placement of smaller residues such as glycine can trigger a shift in the cleavage pattern during the simulations or results in destabilization of substrate binding. The reduced packing by smaller residues also influences the hydration of the active site and the formation of a catalytically active state. The simulations on processed substrate intermediates and a substrate G33I mutation offer an explanation of the experimentally observed relative increase of short Aβ fragment production for this mutation. In addition, studies on a substrate K28A mutation indicate that the internal docking site opposes the tendency of substrate dissociation due to a hydrophobic mismatch at the membrane boundary caused by K28 during processing and substrate movement toward the enzyme active site. The proposed internal docking site could also be useful for the specific design of new γ-secretase modulators.
Significance
Proteolytic processing of amyloid precursor protein (APP) by γ-secretase is an important process involved in Alzheimer’s disease (AD). The sequential cleavage of the substrate along a binding channel depends on the balance between attractive and repulsive forces that determine the stability of the enzyme-substrate complex. Many familial AD mutations in APP can alter this equilibrium. Molecular dynamics simulations have been employed to identify an interaction region in the presenilin-1 subunit of γ-secretase where the substrate conformation switches from a helical to an extended conformation. This internal docking site provides strong substrate-enzyme attraction if filled with large hydrophobicand can effectively oppose dissociation. Simulations on substrate mutations support the model and explain the effect of several mutations on substrate processivity.
Introduction
γ-Secretase is an intramembrane protease that cleaves over 140 type-I membrane peptides. The catalytic center involves two aspartic acid residues located on transmembrane domain (TMD) 6 and 7 of the catalytic subunit presenilin (PS1 or PS2) (1, 2, 3). Among the known substrates, the amyloid precursor protein (APP) is the most studied substrate because of its potential involvement in the pathology of Alzheimer’s disease (AD) (4). According to the amyloid hypothesis, the cerebral plaques composed of amyloid β (Aβ) are the neurotoxic substance, inducing neuro-inflammation that eventually leads to cell death (5,6). Prior to the γ-secretase cleavage, the extracellular domain of APP is first shed by the soluble protease β-secretase to produce a transmembrane peptide termed APP CTFβ or C99 (7). C99 is subsequently recognized and cleaved in its TMD by γ-secretase, releasing N-terminal Aβ peptides and the C-terminal APP intracellular domain (AICD) (8, 9, 10, 11). The major Aβ peptides contain between 37 (Aβ37) and 43 (Aβ43) amino acids. The longer Aβ peptides, such as Aβ42 and Aβ43, are predominantly generated in cases of familial AD (FAD) due to mutations in PS1 or APP (12,13), and are highly aggregation prone and deposited in plaques (14,15).
Secretion of Aβ follows mainly two production lines starting by cleaving the amide bond between L49 and V50 (ε49) or between T48 and L49 (ε48) (8). After the initial cleavage, γ-secretase trims Aβ peptides in a three-residue stepwise proteolysis, following the sequence Aβ49-Aβ46-Aβ43-Aβ40-Aβ37 or Aβ48-Aβ45-Aβ42-Aβ38 (16). In addition, Aβ38 can also be derived from Aβ43, generating the VVIAT peptide (17). Modulators (GSM) and inhibitors (GSI) specific to desired substrates of γ-secretase are of significant medical interest because the latter compounds especially can selectively enhance the Aβ processivity and may reduce generation of long Aβ, which can form toxic aggregates (18, 19, 20).
To understand how APP is cleaved into Aβ peptides, biochemical photo-crosslinking assays have been used to study the process of C99 recruitment (21). In addition, substrate-based chemical probes have been employed to understand how C99 binds to γ-secretase (22, 23, 24, 25, 26). According to these studies, following interactions with exosites in the nicastrin (NCT) and presenilin enhancer-2 (PEN-2) subunits of γ-secretase, C99 binds to the γ-secretase exosite in the N-terminal fragment of PS1 (PS1-NTF) before it reaches the catalytic center between PS1-NTF and PS1-CTF (21). It has also been found that FAD mutations in APP and PS1 weaken the binding between γ-secretase and Aβ intermediates (17,27). Moreover, PS1 FAD mutants mis-position the C99 cleavage site domain by altering the interactions between substrate and enzyme (21,28). However, despite the extensive research on PS1 mutations, Aβ variants and AICD species (29, 30, 31, 32, 33, 34, 35), and binding of modulators and inhibitors (36, 37, 38), the molecular mechanism of how FAD mutations and GSMs modulate the generation of secreted Aβ species is still not completely resolved and fully understood.
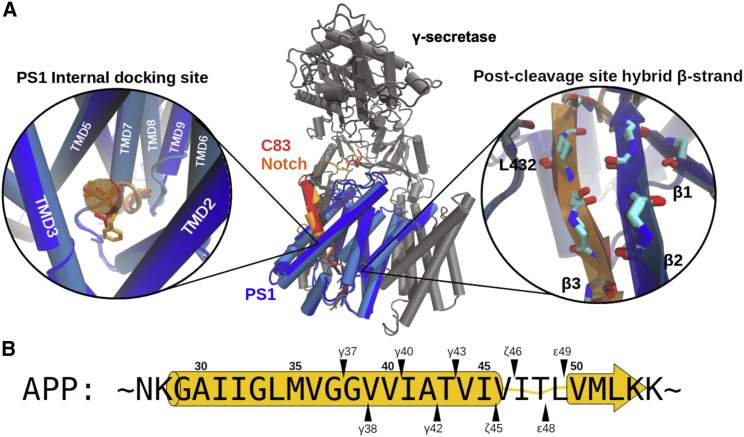
The first three-dimensional (3D) structures of substrate-bound γ-secretase complexes were solved in early 2019 (39,40). These structures show in molecular detail how C83 (a shortened C99), and Notch, another γ-secretase substrate regulating cell differentiation, bind to γ-secretase (39,40). Both substrates are bound in a very similar fashion in the space between TMD2 and TMD3 of PS1 (Fig. 1 A). In addition, both substrates adopt the same helix-loop-strand secondary structure profile. The cleavage site is located at the connection between the loop domain and the extended strand domain, which was predicted and later confirmed as a binding site of transition state analogs (TSAs) (41,42). Notably, several FAD mutations are found at the connection between the helical and loop segment of the Aβ substrate (over 12 FAD mutations were found within A42–V46; Fig. S1 A and B). The β strand at the C-terminal side of the substrate scissile bond, termed β3, is stabilized by forming a β sheet with β1, β2 strands and L432 of PS1 (Fig. 1 A, right panel). The disruption of the hybrid β sheet has been shown to reduce the secretion of AICD, suggesting its crucial role in γ-secretase proteolysis (39,40).
Figure 1.
Cryo-EM structure of substrate-bound γ-secretase and APP cleavage sites. (A) Superposition of the Notch1-bound (PDB: 6IDF) and C83-bound (PDB: 6IYC) γ-secretase structures resolved by cryo-EM (39,40) with γ-secretase cartoon model colored in gray and the catalytic subunit PS1 in dark blue (from PDB: 6IDF) or light blue (from PDB: 6IDF). The backbone RMSD of PS1, including L73-T291 and E376-I467, between two structures is 0.85 Å. Substrates are shown in red (C83) and orange (Notch1). (Left zoom-in) The PS1 internal docking site with V44-I45 of C83 (red) and F1748-F1749 of Notch1 (orange) aligned. (Right zoom-in) The hybrid β sheet formed downstream of the substrate cleavage site. (B) Sequence of APP and its consecutive cleavage sites targeted by γ-secretase. The two production lines are depicted above and below the APP sequence. The yellow cartoon representation in the background indicates the range of the helix-loop-strand conformation of the APP TMD in the bound state. To see this figure in color, go online.
Another characteristic interaction feature shared by the two substrates is a hydrophobic well-packed contact between substrate residues P5, P6 (measured relative to the initial cleavage site) and the TMD2–TMD5 of PS1 around the transition between the substrate loop segment and the helical part (Fig. 1 A, left panel). Interestingly, in contrast to the helical part of the bound substrate, the cryoelectron microscopy (cryo-EM) density of this substrate region is very well defined for the γ-secretase-substrate complexes (39,40).
The interaction region contributes significantly to substrate binding since designed helical substrate-derived inhibitors of γ-secretase that include residues beyond the helical part (but not reaching beyond the active site to form a β sheet), termed D-13, inhibit much more strongly to PS1 than just the helical segment of the substrate (D-10 (25)). In the following we term the contact site, distinct from the active site, the PS1 internal docking site to distinguish it from the previously mentioned external exosite (21). In a recent inhibitor design study, the helix-loop-strand feature of the C83 and Notch1 substrates was adopted by connecting a helical peptide inhibitor (HPI) and a TSA motif with a ω-aminoalkanoyl linker. The resulting inhibitor achieved a very low (subnanomolar) half maximal inhibitory concentration (IC50) potency, fivefold more potent than the TSA alone, suggesting it binds both at the PS1 internal docking site and at the active site (26).
In the present study, we used molecular dynamics (MD) and homology modeling approaches to investigate the non-polar contact between the substrate helical domain and the PS1 internal docking site and how APP mutations attenuate or promote the E-S stability. We constructed 23 substrate-bound γ-secretase complex variants by introducing APP mutations and modeling shorter Aβ peptides in silico and studied the MD in a 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) lipid bilayer.
The simulations indicate that the packing at the internal docking site is an essential element for stabilizing the helical domain of the substrate. Moreover, occupying the cavity between PS1 TMD3–TMD5 prevents excessive water molecules from disturbing the catalytic geometry. We modeled the putative binding poses of C99, Aβ49, Aβ46, Aβ43, and Aβ40, and identified competing forces that determine whether the Aβ peptide stays or leaves γ-secretase.
As the substrate is trimmed shorter, the membrane-anchoring residue K28, which also interacts with the NCT domain (43), pulls down the peripheral lipid molecules as the helical part of the substrate proceeds to the next cleavage pose. It attenuates the local membrane thickness, and imposes a force to pull the substrate out of the binding channel. The binding of especially hydrophobic residues at the internal docking site stabilizes binding and substrate processing.
To further verify the model, we simulated the Aβ40-γ-secretase complexes with the Aβ37/Aβ38-promoting APP mutation K28A (44,45) and G33I (46,47), which presumably weakens the membrane up-pulling force (meaning in the following toward the extracellular region) and enhances the substrate-binding affinity, respectively. Our model provides a molecular explanation of how mutations around V44 and I45 may alter the ε-cleavage and how APP mutations at K28 and G33 facilitate the secretion of shorter Aβ products. We offer a framework to elucidate how Aβ peptides are bound, trimmed, processed, and secreted by γ-secretase, and anticipate that our work will facilitate the structure-based design of more potent GSMs.
Materials and methods
Starting structures representing the unbound state of γ-secretase are generated by removing the ligand from the DAPT-bound (PDB: 5FN2 (48)) and C83-bound (PDB: 6IYC (40)) structures. In the Notch1-bound γ-secretase structure (PDB: 6IDF (39)) the substrate is slightly shifted compared with the arrangement in the C83-bound structure. Hence, simulations starting from the Notch1-bound γ-secretase structure (PDB: 6IDF (39)) could not successfully restore the hydrogen bond between the catalytic aspartate and G1753 (Fig. S2) that corresponds to the initial cleavage site. We reconstructed the Notch1-bound complex that can form the catalytic geometry at the expected substrate residue using the C83-bound γ-secretase complex as the template (PDB: 6IYC) and appropriate Notch1 substrate alignment (Table 1) with the MODELLER comparative modeling software (49). The artificially introduced D385A from the C83-bound structure (PDB: 6IYC) was restored using Ambertools18 (50). Residues between T291 and R377 of PS1 are not resolved in the γ-secretase structures and were not included in the simulations. Simulation boxes are prepared using CHARMM-GUI (51) and PPM online servers (52) and solvated with 500 POPC and TIP3P water molecules (53) and 0.15 M KCl salt. All simulations were performed using the Amber18 package and the ff14SB (54) force field for the proteins and the lipid17 (55) force field for the membrane lipids.
Table 1.
Residues of γ-secretase substrate with the corresponding cleavage position denoted by the subscript
| Notch1 | C99ε49 | C99ε48 | Aβ49ζ46 | Aβ46γ43 | Aβ43γ40 | Aβ40γ37 | |
|---|---|---|---|---|---|---|---|
| P22 | A1732 | K28∗ | N27 | G25 | E22 | F19 | K16 |
| P21 | Q1733 | G29 | K28∗ | S26 | D23 | F20 | L17 |
| P20 | L1734 | A30 | G29 | N27 | V24 | A21 | V18 |
| P19 | H1735 | I31 | A30 | K28∗ | G25 | E22 | F19 |
| P18 | F1736 | I32 | I31 | G29 | S26 | D23 | F20 |
| P17 | M1737 | G33 | I32 | A30 | N27 | V24 | A21 |
| P16 | Y1738 | L34 | G33 | I31 | K28∗ | G25 | E22 |
| P15 | V1739 | M35 | L34 | I32 | G29 | S26 | D23 |
| P14 | A1740 | V36 | M35 | G33 | A30 | N27 | V24 |
| P13 | A1741 | G37 | V36 | L34 | I31 | K28∗ | G25 |
| P12 | A1742 | G38 | G37 | M35 | I32 | G29 | S26 |
| P11 | A1743 | V39 | G38 | V36 | G33 | A30 | N27 |
| P10 | F1744 | V40 | V39 | G37 | L34 | I31 | K28∗ |
| P9 | V1745 | I41 | V40 | G38 | M35 | I32 | G29 |
| P8 | L1746 | A42 | I41 | V39 | V36 | G33 | A30 |
| P7 | L1747 | T43 | A42 | V40 | G37 | L34 | I31 |
| P6# | F1748# | V44# | T43# | I41# | G38# | M35# | I32# |
| P5# | F1749# | I45# | V44# | A42# | V39# | V36# | G33# |
| P4 | V1750 | V46 | I45 | T43 | V40 | G37 | L34 |
| P3 | G1751 | I47 | V46 | V44 | I41 | G38 | M35 |
| P2 | C1752 | T48 | I47 | I45 | A42 | V39 | V36 |
| P1 | G1753 | L49 | T48 | V46 | T43 | V40 | G37 |
| P1′ | V1754 | V50 | L49 | I47 | V44 | I41 | G38 |
| P2′ | L1755 | M51 | V50 | T48 | I45 | A42 | V39 |
| P3′ | L1756 | L52 | M51 | L49 | V46 | T43 | V40 |
The membrane-anchoring residue K28 is marked with an asterisk (∗), P5 and P6 are marked with a hash symbol (#).
For the enzyme-substrate complexes, two different protonation states of the active site with either the D257 or D385 protonated were considered. Start structures for γ-secretase in complex with the C99-Aβ49-Aβ46-Aβ43-Aβ40-Aβ37 substrates representing consecutive cleavages are constructed using MODELLER comparative modeling (with the last frame of the simulation at the prior cleavage binding pose and solvated in simulation boxes as described above). The total number of atoms for each simulation system is listed in Table 2.
Table 2.
Conducted simulations in the study
| aApo (5FN2) | aApo (6IYC) | Notch1S3 | C99ε49 | C99ε49,V44G | |
|---|---|---|---|---|---|
| Protonation | D385H | D385H | D257H,D385H | D257H,D385H | D257H,D385H |
| Atom # | 284,455 | 292,134 | 300,408 | 293,748 | 293,739 |
| C99ε49,I45G | C99ε49,GG | bC99ε48-1 | bC99ε48-2 | Aβ49ζ46 | |
| Protonation | D257H,D385H | D385H | D385H | D385H | D257H,D385H |
| Atom # | 293,736 | 293,727 | 301,518 | 294,453 | 293,523 |
| Aβ46γ43 | Aβ43γ40 | Aβ40γ37 | Aβ40γ37,G33I | Aβ40γ37,K28A | |
| Protonation | D257H,D385H | D257H,D385H | D257H,D385H | D257H,D385H | D257H,D385H |
| Atom # | 287,745 | 259,525 | 305,198 | 305,210 | 305,255 |
Each system is simulated for 600 ns twice with the same initial structure but different randomly generated initial velocity.
Three simulations of the each apo-form γ-secretase with initial structures taken from PDB 5FN2 and PDB 6IYC are performed with the D385-protonated PS1.
Only one trajectory with D385-protonated PS1 was simulated and analyzed in each of the C99ε48-bound γ-secretase complexes.
In a first step, each simulation system was minimized with maximal 70,000 steps with 10 kcal·mol−1 Å−2 positional restraint on protein using the MPI version of the pmemd program in Amber18 (50), followed by equilibration with gradually releasing positional restraint, from 10 to 0.1 kcal·mol−1 Å−2 on protein and 2.5 to 0 kcal·mol−1 Å−2 on membrane, for 400 ps at 303.15 K using the Cuda version of pmemd. The equilibrated systems are submitted to 600-ns production runs at a temperature of 303.15 K using the Langevin thermostat (56) and a pressure of 1 bar by Berendsen barostat (57). Employing the hydrogen mass repartitioning method (58) allowed a time step of 4 fs. For each of the unbound structures, three simulation (600 ns each) were performed. Also, in the case of the model with substrate placement at the ε48 cleavage site (D385 protonated), one production simulations was conducted. For all other cases (Table 1), two independent trajectories (with different starting velocities) are generated and both possible protonation states of D257 or D385 are considered. Hence, four simulations are conducted in each E-S complex. Cutoff distances of 9 Å and 12 Å for non-bonded interactions were tested on the C99-bound structure and exhibit similar root-mean-square fluctuations (RMSFs), root-mean-square deviation (RMSD) versus time, catalytic hydrogen bond formation, and similar substrate secondary structure during the simulations (Fig. S3, see also Figs. S4 and S5) and we used a real space cutoff of 9 Å for all simulations. In total, 52 trajectories were generated. The RMSD versus time for all simulations of γ-secretase, PS1, and the substrates are shown in Figs. S4 and S5 to illustrate the stability of the simulations.
Secondary structure was analyzed using the DSSP method (59) with cpptraj (60). Secondary structures of Alpha, 3-10, and Pi are classified as Helix and only Anti is classified as β strand in the main text. Water accessibilities of each substrate residue are calculated by counting the number of waters within 5 Å of the sidechain of the corresponding residue. The mean binding energy of the substrate is computed using the molecular mechanics energies combined with the Poisson-Boltzmann and surface area continuum solvation (MM/PBSA) method (61). Only the last 500 frames of each trajectory, in total 100 ns, are submitted for the energy evaluation. Dielectric constants are set to εlipid = 2 representing the membrane region between z = −18.5Å and z = +18.5Å, εprotein = 1 within the protein interior, and εwater = 80 otherwise to represent the aqueous environment. Noticing that K28 creates an uneven membrane thickness, only the substrate TMDs are taken for calculation to avoid energy divergence (62). Note that this approach approximates the whole membrane region with the implicit regions and therefore interactions involving water molecules, such as the scissile bond at the catalytic center, cannot be precisely calculated.
Results
PS1 internal docking site is stabilized by substrate binding
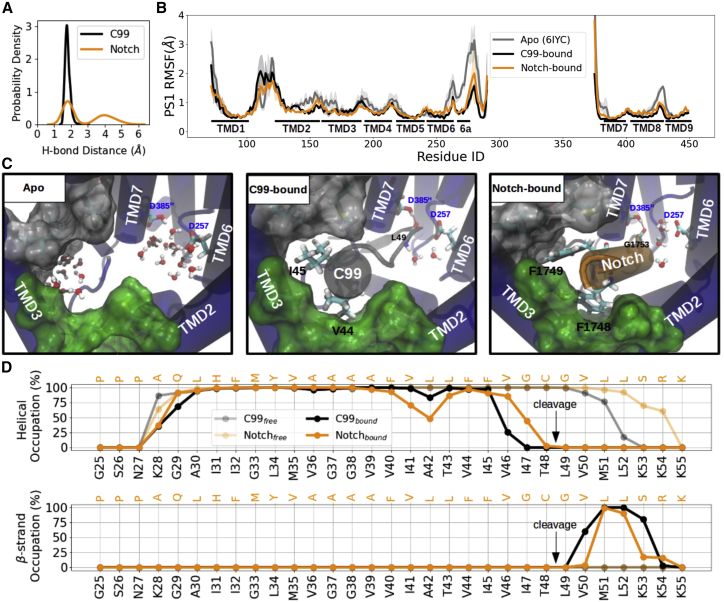
As a first step, we compared the dynamics of γ-secretase with and without the C99 or Notch (Notch1) substrates using MD simulations. To execute the intramembrane proteolysis, either D257 or D385 of PS1 needs to be protonated. In a previous publication, we identified D385 as the more likely protonated Asp residue based on a higher predicted pKA than D257 (33). Nevertheless, because of the uncertainty concerning the protonation state, most of our simulations are performed employing both the D385-protonated and the D257-protonated states of PS1. In the main text, results on the D385-protonated complexes are presented, and the results using the D257-protonated complex are provided in the supporting material (or discussed explicitly for comparison). The labeling of C99 residues is adapted to the Aβ numbering. During the simulations, the catalytic hydrogen bond, characterized by the distance between the protonated aspartate in PS1 and the substrate scissile bond, was observed in both complexes (Figs. 2 A and S6 A). Since the strong and covalent hydrogen bond that forms during catalysis with donor-acceptor distances of 2.2–2.5Å (63) is not accessible during classical MD simulations (requires expensive quantum mechanical calculations), a moderate hydrogen bond with donor-acceptor distances of 2.5–3.2Å (effective O-H distance of 1.5–2.2Å) stabilized by electrostatic interaction (63) is considered as an indicator of a productive catalytic geometry.
Figure 2.
MD simulations of unbound γ-secretase and bound to C99 and Notch substrates. (A) Probability density distribution of the catalytic hydrogen bond distance. (B) Residue-wise root-mean-square fluctuation (RMSF) of the γ-secretase catalytic subunit PS1. The unbound γ-secretase starting structures were used based on PDB 6IYC (gray). (C) View into the PS1 internal docking site in the unbound form (left), C99-bound (middle), and Notch-bound (right) γ-secretase complexes. PS1 is shown in blue cartoon representation, C99 in gray, and Notch in orange. The sub-pocket formed between TMD2 and TMD3 is shown as green surface, and the sub-pocket formed by TMD3-TMD5 and TMD7 is indicated as light gray surface. All residues defining these two pockets are listed in Table 2. Water molecules are shown in van der Waals (vdw) + bond representation. V44, I45, and the backbone of L49 of C99 and F1748, F1749, and the backbone of G1753 of Notch are shown in the licorice representation. The catalytic hydrogen bond is indicated as red dashed line between the substrate scissile bond and the protonated aspartic acid. (D) Secondary structure analysis of C99 (black) and Notch (orange) in γ-secretase bound form (solid line) and unbound (mostly helical) form (embedded in a membrane, transparent line). Helical and β strand occupancies are calculated as averages over the whole trajectories. To see this figure in color, go online.
The calculated RMSF profiles obtained from the comparative simulations indicate that substrate binding affects the flexibility pattern of enzyme and substrate (Fig. 2 B). The regions with significant changes in RMSF are mostly solvent accessible in the substrate-free (Apo) state and can be roughly assigned to two PS1 regions (Fig. 2 C). The first reduction in flexibility includes the residues at the C terminus of PS1-NTF, N terminus of TMD7, and the loop connecting TMD8 and TMD9, which respectively correspond to β1, β2, and the LPAL motif that form a sheet with the β3 strand of the substrate β3 (Fig. S7). The hybrid β sheet has been suggested as crucial element for γ-secretase with as-yet unknown mechanism (39,40). This region surrounds the enzyme active site and is not at the focus of the current study.
Another spatially distinct region with reduced RMSF upon substrate binding covers the residues from the C-terminal part of TMD2 to the N-terminal part of TMD5. Residues within this regime are mostly hydrophobic and in close contact with the P6 and P5 residues of the substrate, namely V44 and I45 of C99, and F1748 and F1749 of Notch-1 (Fig. 2 C; Tables 1 and 3). Higher RMSF differences in these two regions were also found in the simulations starting from another Apo-state cryo-EM structure with PDB 5FN2 (Fig. S8). Pocket detection on the Apo-state simulation with the program Mdpocket (64) shows that this region is also connected to the active site (Fig. S9 A and B). The volume of the pocket cleft fluctuates around a mean volume of ∼900 ± 400 Å3 (Fig. S9 C) in the Apo-state. Furthermore, a large ratio of apolar/polar surface area (∼380 Å2/60 Å2) indicates a preference toward non-polar or hydrophobic contacts (Fig. S9 D and E).
Table 3.
PS1 residues surrounding substrates P5 and P6 in the Aβ49-Aβ46-Aβ43-Aβ40-Aβ37 production line and their related FAD mutations
| Residues surrounding P5 | Residues surrounding P6 | |
|---|---|---|
| PS1 residues | S169, S170, L173, I213, L226, I229, M233, L286, I287, L383, I387 | M146, L150, I162, W165 |
| Related FAD mutations | S169P, ΔS169, S170F, L173W, I213L, I229F, M233T, M233L, L286V | M146L, L150P, W165G |
Note that several residues involved in this hydrophobic contact are reported FAD hotspots (https://alzforum.org), many of which are shown to increase the Aβ42/Aβ40 ratio and decrease the γ-secretase activity (Table 3; Fig. S1) (31). Likewise, residues near V44 and I45 of C99, including T43–V46, are known to have a direct impact on the ε-cleavage site (35,65). The information gives us a hint that the interaction around this region might be another critical E-S interaction, in addition to the hybrid β sheet C terminal to the substrate scissile bond.
Compared with the simulations of the unbound form of the substrates (embedded in the membrane), C99 and Notch undergo a significant conformational change from a full α helix into a helix-loop-strand conformation when bound at the PS1 binding channel (Fig. 2 D). The TMDs of C99 and Notch are composed of mostly hydrophobic residues. However, the N-terminal side of TMDs are rich in helix-destabilizing residues such as proline, glycine, serine, and asparagine, and the C-terminal side is capped by positively charged amino acid such as lysine and arginine. As expected, the β strand was formed and stayed firmly across substrates P1′ to P3′, especially on P2′, when binding to γ-secretase. During the MD simulations, the conformation of C99 residues T43 to I45 turn into a 310 helix in the second half of the trajectory (Fig. S10). In this conformation A42 points in the same direction as I45, hence allowing it in principle to proceed to the next cleavage pose by pure translational movement along the channel between TMD2 and TMD3 of PS1 (Fig. S11). Unwinding P8 and the 310 helix forming between P5 and P7 were also observed in the Notch-bound structure (Fig. S10). It is worth noting that A42 and I45 are substituted into helix-forming (especially 310 helix (66, 67, 68, 69)) α-aminoisobutyric acids (Aibs) in design of the γ-secretase HPI, suggesting that HPI binds similarly to the helical domain of C99/Notch (22,26).
By substituting the explicit solvent with the implicit membrane continuum, we calculated the binding energy contribution of each residue on the substrate separately for backbone and side chains (see section “materials and methods” and Fig. S12). The strongest backbone interacting residues are P1′–P3′, which form hydrogen bonds in the post-cleavage site hybrid β sheet. The contribution from sidechains in both substrates come predominantly from the C-terminal half of substrate TMDs. In the case of C99, residues on the N-terminal side of V44 and I45 contribute overall less to the interaction with the enzyme compared with C-terminal residues of the substrate. In particular, residues V44 and I45 in close contact with PS1 TMD2–TMD5 and TMD7 serve to anchor the substrate at the internal docking site. A similar trend was also observed in the Notch-bound complex, with F1748 and F1749 together contributing a binding energy around −6 kcal/mol. Similar results were obtained when the protonated aspartate was switched from D385 to D257 of PS1 (Fig. S5).
Modeling the ε48 binding pose of C99
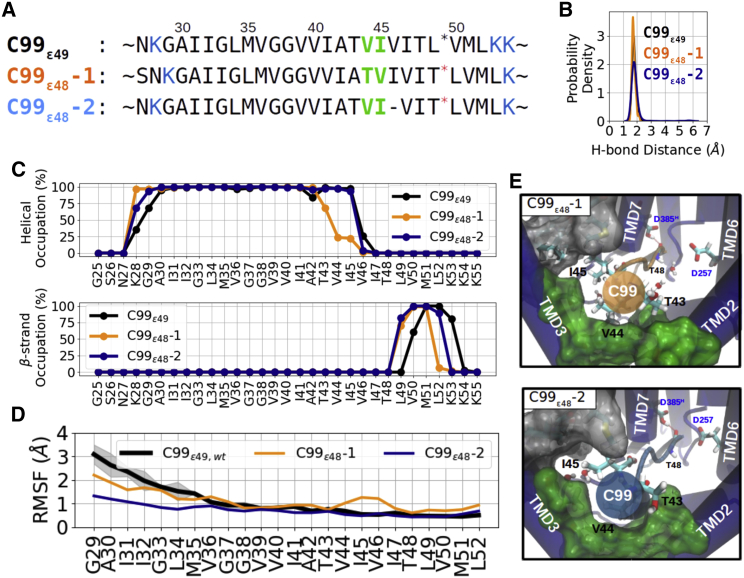
Besides the dominant ε49 cleavage, γ-secretase can alternatively also cleave C99 at the amide bond between T48 and L49, generating a C-terminal peptide (AICD51) and N-terminal Aβ48. The ε48 cleavage is the first cleavage of the production line producing the plaque-forming Aβ42 (17). Hence, it is of both biological and pathological interest to distinguish the structural difference between the ε49 and ε48 binding pose. We modeled two C99-γ-secretase putative binding models at the ε48 cleavage using comparative modeling with two different sequence alignment strategies (Fig. 3 A) The first model is generated by shifting the whole sequence by one residue toward the C-terminal side (C99ε48-1), whereas the second model keeps V44 and I45 at the same position and exposes the scissile bond by inserting an alignment gap between I45 and V46 (C99ε48-2).
Figure 3.
MD simulations of γ-secretase in complex with C99 shifted for cleavage at ε48 site. (A) Sequence alignments of the two models termed C99ε48-1 (orange), and C99ε48-2 (dark blue) relative to the residue positions in C99ε49 (black). Membrane-anchoring residues K28, K53, and K54 are marked in blue and residues residing in the hydrophobic pockets are marked in green. (B) Sampled probability density of the catalytic hydrogen bond distance in the MD simulation. (C) Secondary structure analysis of simulation on C99ε49 (black), C99ε48-1 (orange), C99ε48-2 (dark blue) bound to γ-secretase complexes. (D) RMSF of the substrates. (E) View (from the extracellular side) into the PS1 internal docking site of the two models C99ε48-1 (upper) and C99ε48-2 (lower) (representation same as in Fig. 2C). To see this figure in color, go online.
During the MD simulations of both reconstructed models, the catalytic hydrogen bond between the protonated aspartate and the carbonyl group of C99 T48 was found in over 90% of the trajectory frames, except for the first few nanoseconds while the systems were still considered undergoing the equilibration (Fig. 3 B). In addition, the β strand conformation is preserved at position P2′ and P3′ (Fig. 3 C). Despite the formed catalytic hydrogen bond, larger fluctuations were observed around substrate V44–V46 (Fig. 3 D) and a rotational substrate motion at the internal docking site was observed in the first reconstructed complex C99ε48-1. With the T43 rotated out of its starting position, the internal docking site was again filled up with V44 and I45 after ∼100 ns (Fig. S13; Video S1). As shown in the evolution of the secondary structure, the rotation was facilitated by the unwinding of the helix between T43 and I45 (Figs. 3 C and S10). In contrast, V44 and I45 stayed firmly in the PS1 internal docking site in the second reconstructed complex C99ε48-2. Consistent with the C99ε49 complex, residues T43–I45 exhibit a 310 helix while A42 was slightly disordered (Figs. 3 C and S10). Notably, while V46 stays mostly in the turn-and-bend conformation, calculated by the DSSP method (59), in the C99ε49 complex, it is classified into the none state in the C99ε48 complex, where the peptide bond is stretched so that the carbonyl group of T48 can be exposed to the catalytic aspartates (Fig. S10).
Importantly, our simulation result suggests that the V44 and I45 are recognized by the PS1 internal docking site and sterically allow both a cleavage at ε49 or ε48 position (a complete shift of the entire substrate sequence is sterically not required). Even the model with T43 intentionally placed in the P6 pocket of the internal docking site resulted in a rapid readjustment with V44 and I45 bound to the P5, P6 binding sites (Fig. 3 E). Hence, which ε-cleavage is executed depends mainly on the conformation of the residues of V46–T48 (30,35,70). This explains why mutations on residues remotely N terminal to V44 such as A21–D23 (30) and K28–A30 (71) do not alter the ε-cleavage preference. However, the docking pose present in the study might be altered when residues around V44 and I45 are mutated. Indeed, although some APP mutations around this region, such as T43I, V44A, and V44F, have been shown to decrease the ε49/ε48 cleavage propensity, other mutations such as I45F increase it (30,35,70).
Weakening the interaction at PS1 internal docking site leads to readjustment of substrate helical interface
Presumably, along the consecutive trimming process, C99 needs to move “downward” (from the extracellular toward the intracellular side of PS1) in order to expose the upstream scissile bonds to the catalytic center. Our simulations suggest that, due to the 310 helix formed within residues T43–I45, a translational movement without altering the substrate orientation is sufficient for the substrate proceeding to the next cleavage binding pose.
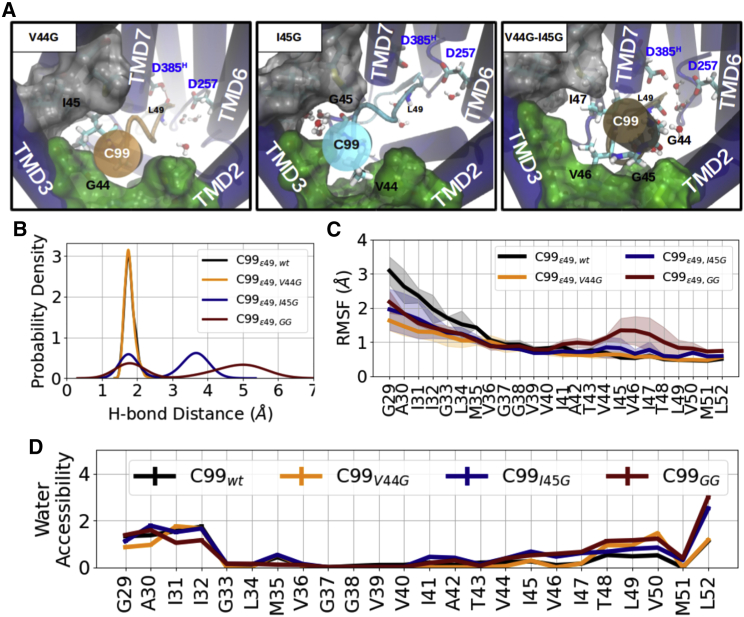
However, sequence alignment along the Aβ49-Aβ46-Aβ43-Aβ40-Aβ37 production line shows that, for this process, small residues such as A42, G38, and G33 are sometimes placed in the internal docking site (see Table 1). According to our hypothesis, such small residues may fail to stabilize the substrate at the PS1 internal docking site. Indeed, in support of our model, the experimental double mutation V44G-I45G on C99 was shown to significantly decrease the substrate cleavability and skewing the ε-cleavage site to generate longer AICD such as AICD49-99, AICD46-99, and AICD45-99 (65). To study these scenarios, we introduced in silico glycine substitutions of V44 or/and I45 on C99 to generate the complexes of C99V44G, C99I45G, and C99GG with γ-secretase and study the effect in MD simulations (Fig. 4 A).
Figure 4.
Glycine mutations at the internal docking site disturb the E-S interaction. (A) Top-view at the PS1 internal docking site of C99V44G (left), C99I45G (middle), and C99GG bound γ-secretase complexes (representation same as in Fig. 2C). (B) Probability density of the catalytic hydrogen bond distance during simulations. (C) RMSF of the substrate TMD and (D) residue-wise water accessibility of C99wt (black), C99V44G (orange), C99I45G (blue), and C99GG (brown) during simulations in complex with γ-secretase. To see this figure in color, go online.
Among the three C99 mutants, only C99V44G was able to largely preserve formation of the catalytic hydrogen bond (Fig. 4 B) and fluctuated the least (Fig. 4 C). In contrast, I45G and V44G-I45G mutants formed less frequent catalytic hydrogen bond (Fig. 4 B) and showed respectively mildly and largely more fluctuations at around V44–V46 of the substrates (Fig. 4 C). By computing the residue-wise water accessibility, we found that while I41–I47 remain in a dry environment in C99V44G and C99wt models, they are more exposed to the aqueous environment in the case of bound I45G and double-glycine substrate variants (Fig. 4 D).
As depicted in Fig. 2 C, in fact, in the simulations of substrate-free γ-secretase, the PS1 internal docking site is filled up with water molecules coming from the intracellular side of the membrane, in particular in the cavity between TMD3 and TMD5. When C99V44G or C99wt binds to the PS1 interior, the water-filling cavity is occupied by C99 I45 sidechain with a calculated binding energy contribution of ∼ −4.95 kcal/mol (Figs. 2 C, 4 A, and S14 A). However, when the water-blocking I45 is mutated to glycine, contributing a binding energy of only ∼ −0.61 kcal/mol, the cavity is again filled with water (Figs. 4 A and S14 A). In the case of the double substitution of V44G and I45G, the substrate helical domain rotates counterclockwise and V46 and I47 move into the PS1 internal docking site in one of the simulations (Fig. S15; Video S2). We note that none of the C99 glycine mutants perturbed the post-cleavage site hybrid β sheet (Fig. 2 C), but they all altered the local helicity around V44–V46 (Figs. S14 B and S10). The C99I45G and C99GG variants enrich the helical occupation on V46, by which V46 is drawn spatially closer to the water-filling cavity formed between TMD3 and TMD5 (similar results were observed with the D257H protonation state with a dissociation of the post-cleavage site hybrid β strand observed in the I45G mutant; Figs. S16 and S17).
Local hydrophobic mismatch induced by K28 in Aβn-γ-secretase binding complexes
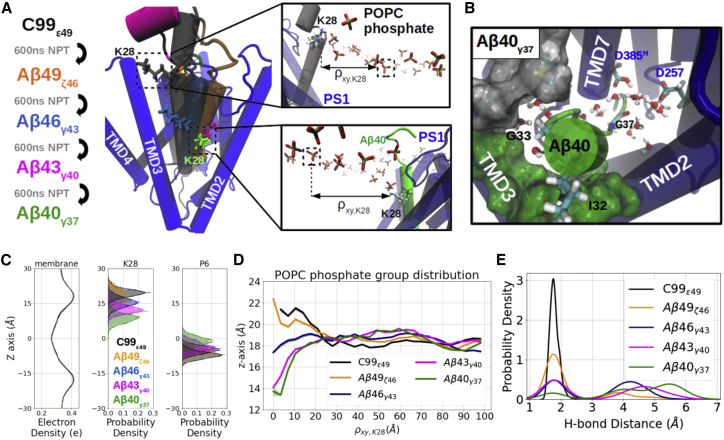
The substrate processing by PS1 in γ-secretase leads to a consecutive cleavage typically along the Aβ49-Aβ46-Aβ43-Aβ40-Aβ37 production line. It leads to the placement of substrate residues of different sizes into the internal docking site (see Table 1). In addition, the stepwise cleavage requires movement of the substrate toward the interior of the enzyme and toward the cytosolic side of the cell. On the N-terminal side at the entrance to the substrate-binding channel of PS1, the C99 substrate includes a K28 residue that is considered a membrane-anchoring residue for the substrate (44). K28 was also shown to interact with residue 241 on the NCT domain of γ-secretase (43). Mutation of K28 to a negatively charged residue or neutral residue enhances processivity of cleavage, leading to a relative increase of shorter secreted Aβ products (43,44,46,72). It is of interest to investigate the dynamics of the intermediate states that have to form during substrate processing and how binding to the internal docking site correlates with the movement of the K28 anchor residue. We consecutively constructed Aβn-γ-secretase complex models using the known full substrate complex as template for shifting the substrate sequence (see section “materials and methods” and the schematics in Fig. 5 A). Each product is named by its sequence length and cleavage site. For instance, Aβ49ζ46 stands for the docking pose of Aβ49 with V46 placed at the catalytic active site and ready for the ζ46 cleavage (Fig. S18).
Figure 5.
Comparative modeling and simulations of Aβn-γ-secretase complexes. (A) Schematic views of the modeling workflow and the top views (from the extracellular domain) of the binding poses of C99ε49 (black), Aβ49ζ46 (orange), Aβ46γ43 (blue), Aβ43γ40 (magenta), and Aβ40γ37 (green). Membrane-anchoring residue K28 of each substrate is shown explicitly in the licorice representation. Zoom-in views of the C99ε49 (upper) and Aβ40γ37 (lower) show how the position of K28 influences the local distribution of the POPC phosphate groups with radial distance from K28 on the xy plane denoted by ρxy,K28. (B) View into the PS1 internal docking site in the Aβ40γ37-bound γ-secretase (representation same as in Fig. 2C) (C). Distribution of the membrane electron density (left), membrane-anchoring residue K28 (middle), and substrate P6 (right) along the z axis in different Aβn-γ-secretase complexes. (D) Average z axis of the POPC phosphate on the extracellular side distributed along the radial distance ρxy,K28. (E) Probability density of the catalytic hydrogen bond distance. To see this figure in color, go online.
In the case of the simulations of the complexes with bound Aβ49ζ46, Aβ46γ43, Aβ43γ40 models the corresponding P5 and P6 residues (Table 1) all remaining at the initial placement in the PS1 internal docking site (Fig. S18). However, in model for the last cleavage pose Aβ40γ37 the residues P5 and P6 dissociated from the internal docking site during the simulations (Fig. 5 B; Video S3). It is likely that, besides the stabilization interaction at the PS1 internal docking site, the K28 anchor plays a role during this dissociation process.
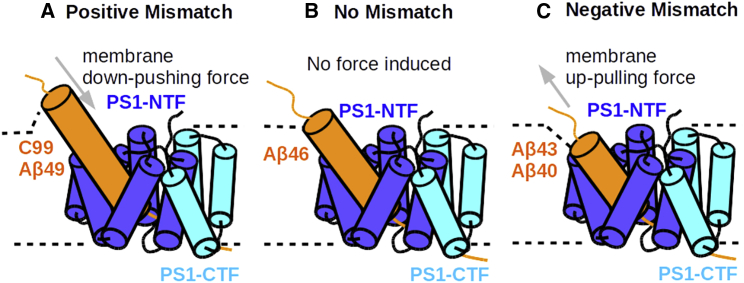
To study the influence of the membrane-anchoring effect of K28, we analyzed the dynamics of the lipid molecules as well as the Aβn-γ-secretase complex at the juxtamembrane region. The electron density of a lipid bilayer typically shows two peaks at around z = +18.5Å and z = −18.5Å, representing the choline groups of POPC of the two membrane leaves. The K28 and the P6 residues do not always stay at the same position on z axis relative to the center of the membrane for the different Aβ-bound complexes (Fig. 5 C). In the case of the Aβ49ζ46, Aβ46γ43, Aβ43γ40 models, the K28 moved deeper into the membrane as the substrate proceeds toward the shorter cleavage poses (Fig. 5 A, with residues at the internal docking site staying in place). Only in the case of the Aβ40γ37 complex residue P6 moved toward the exterior membrane side (Fig. 5 B). The time plot along the simulations shows that K28 of Aβ40 quickly approached the membrane upper leaf within the first 20ns followed by the pulling-up motion of P6 (Fig. S19). K28, carrying a positive charge, interacts with the negatively charged lipid phosphate groups and alters the peripheral membrane thickness (Fig. 5 A). With the strongest deformation close to K28, the membrane adapts slowly to its average thickness and reaches its equilibrium at the radial distance of 22.5 Å away from K28 on the xy plane, denoted as ρxy,K28 (Fig. 5 A and D). According to the mattress model, a hydrophobic mismatch in an uneven membrane may induce an elastic deformation free energy against the membrane deformation (73). We estimated the mismatch amplitude by fitting the curves with a simple function zPOPC = α e−βρ cos(γρ) + z(ρ = ∞), with α the mismatch amplitude, β the radial decaying rate, γ the membrane curvature, and z(ρ = ∞) the equilibrium height of the membrane upper leaf (Fig. S20). In the first two cleavages, namely C99ε49 and Aβ49ζ46, K28 stayed above the upper membrane leaf, inducing a positive mismatch, which promotes the substrate to move downward toward the next binding pose (α ∼ +3 Å; Figs. 5 A and 6 A). When proceeding to the Aβ46γ43 binding pose, K28 stayed at a similar height to the membrane extracellular surface (α ∼ 0 Å; Fig. 6 B). However, K28 no longer matches the average membrane height and creates a negative mismatch in the Aβ43γ40 and Aβ40γ37 docking poses; hence, the substrates in this case experience a membrane up-pulling force (α ∼ −4.75 Å; Figs. 5 A and 6 C).
Figure 6.
Illustration of the membrane elastic force induced by local hydrophobic mismatch based on the mattress model (73). (A) Positive mismatch as observed in C99ε49 and Aβ49ζ46 substrate-bound poses is induced when the substrate protrudes through the membrane bilayer. The membrane elastic potential trying to restore the optimal membrane thickness causes a force pushing the substrate toward the PS1 active site (termed “down-pushing force”). (B) No hydrophobic mismatch was observed in the Aβ46γ43-binding pose (elastic potential is zero or close to zero). (C) Negative mismatch observed in the Aβ43γ40 and Aβ40γ37 substrate-bound cases is induced when the substrate tends to contract the membrane bilayer. The membrane elastic potential keeping the membrane thickness causes an “up-pulling” force on the substrate. To see this figure in color, go online.
On the C-terminal side of the TMD helix are P5 and P6 residues sitting in the PS1 internal docking site (Fig. S18). Residue-wise binding energy analysis shows that the strongest binding affinity from the substrate helical domain are contributed by these two residues, and their binding strengths correlate with their size and hydrophobicity (Fig. S21 A and C). Notably, when a small residue is placed at position P5, such as A42 from the Aβ49ζ46 binding pose or G33 from the Aβ40γ37 binding pose, larger fluctuations were observed in the substrate TMD, in particularly around P5 (Fig. S22 A). In contrast, when a small residue is placed at P6, such as G38 from the Aβ46γ43 binding pose, only a slight increase in the fluctuation of substrate TMD was observed (Fig. S22 A). We note that the different effects upon placing a small residue at substrate P5 or P6 display a consistent behavior as observed in the simulations with C99 mutants V44G and/or I45G (Fig. 4 C).
With the negative hydrophobic mismatch inducing an up-pulling force at the juxtamembrane region and the weak helical binding force at the PS1 internal docking site, we hypothesize that the P6 movement observed for Aβ40γ37 is due to an imbalance between the two competing forces. For example, in the case of Aβ43γ40 residue, M35 and V36 placed in the internal docking site contribute a calculated binding energy of −6.5 ± 0.7 kcal/mol, whereas residues G33 and I32 placed in the internal docking site for Aβ40γ37 contribute only −2.8 ± 0.4 kcal/mol (Fig. S21 C). In the latter case, this small interaction energy cannot outbalance the hydrophobic mismatch, and the residues moved out of the pocket even though they experience a similar membrane mismatch amplitude (Fig. 5 B). On the other hand, although G38 and V39 of Aβ46γ43 contribute a relatively weak binding energy at the internal docking site (−4.05 ± 0.4 kcal/mol; Fig. S21 C), the absence of the hydrophobic mismatch makes Aβ46 stay steadily with the enzyme (Fig. 5 D; Video S3).
The gradual movement of the K28 during processing has also an effect on the helicity and water accessibility of the substrate TMD. With the local POPC molecules adjusting their positions to match K28, the local water-membrane interface moved toward the intracellular side, and residues around K28 are thus surrounded by water molecules (Fig. S22 B). The water molecules weaken the intramolecular hydrogen bonds required in helix formation, and the helical part of the substrate becomes shorter and shorter along the processive cleavage with residues at the N-terminal side of K28 turning into disordered conformations (Fig. S22 B and C, time evolution of the secondary structures are shown in Fig. S23). Notably, substrates P2–P5 are more exposed to water molecules in the Aβ40γ37 docking pose than other Aβ docking poses, agreeing with the C99I45G simulation where a glycine was also present at the P5 position (Fig. S22 B). The catalytic hydrogen bonds are stable in the first two cleavage poses, while they are seldom formed in the later three complexes, although still not totally vanished (Fig. 5 E). As a result of the local hydrophobic mismatch, P5 and P6 binding energies and the trend of catalytic hydrogen bonds along the consecutive binding process were similarly reproduced in the D257-protonated PS1 complexes, except that the substrate was pulled out from the internal docking site in both the Aβ43γ40 and Aβ40γ37 docking poses (Figs. S18, S24–S27).
APP mutations K28A and G33I stabilize substrate binding and catalytic geometry of Aβ40
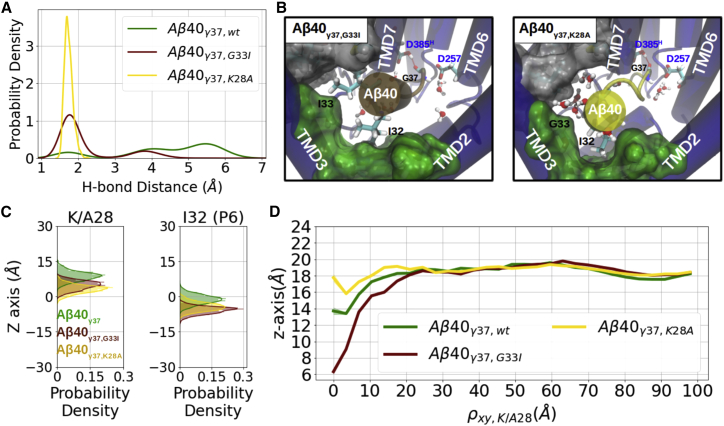
In the study of putative Aβn-γ-secretase complexes within the lipid bilayer, we observed a pair of competing forces that characterizes the decisive factors of γ-secretase holding or releasing the substrate, the membrane up-pulling force toward the extracellular membrane surface, and the E-S stabilization force at the PS1 internal docking site. This model provides a straightforward way to explain why certain APP mutations on K28 (44,45) and G33 (46,47) generate shorter Aβ products. It is due to reducing the membrane-anchoring feature at position K28 and/or enhancing the E-S binding affinity at the internal docking site. To verify the hypothesis, simulations of Aβ40-γ-secretase complexes with APP mutations K28A and G33I were conducted and analyzed.
Both APP mutations G33I and K28A result in an increased fraction of sampled catalytic geometries and stable I32 placement in the PS1 internal docking site (Fig. 7 A–C; Video S4). Notably, in the K28A mutant, water was observed accessing the pocket between TMD3 and TMD5, similarly to what was seen in the C99 I45G mutant at the ε49 cleavage pose (Fig. 4 A). G33I, in contrast, is expected to have a reverse influence compared with I45G in C99. Indeed, the introduced isoleucine with a larger hydrophobic side chain fills the pocket and prevents further water access (Fig. 7 B). In this case, side chains of P5 and P6 together contributed ΔEI32 + ΔEI33 = −8.2 ± 0.6 kcal/mol of E-S binding energy, around 5.4 kcal/mol stronger than estimated for the wild-type Aβ40 binding (Fig. S21 B and C).
Figure 7.
Influence of the APP mutations G33I and K28A on the Aβ40γ37 binding pose. (A) Probability density of the catalytic hydrogen bond distance. (B) Top-view at the PS1 internal docking site of Aβ40γ37,G33I-bound (left) and Aβ40γ37,K28A-bound (right) γ-secretase complexes. Atomic representation is the same as in Fig. 2C. (C) Distribution of the membrane-anchoring residue K28 (middle) and substrate P6 (right) along the z axis. (D) Average z axis of the POPC phosphate on the extracellular side distributed along the radial distance ρxy,K/A28. To see this figure in color, go online.
Indeed, the more favorable binding energy at the PS1 internal docking site in this case allowed K28 to move deeper into the membrane and to maintain and balance a much more negative hydrophobic mismatch (α approximately −12.7Å; Figs. 7 D, S20, and S28). Mutation K28A, on the other hand, alleviates the difference between the local and global membrane thickness (α ∼ +1.02; Figs. 7 D and S12) and, thus, mitigates the elastic potential stored in the membrane layer. With the G33 and I32 remaining in contact with the PS1 internal docking site, the local binding energy was observed to be approximately 2 kcal/mol more favorable than the wild-type Aβ40, mainly because of the enhanced interaction of I32 (Fig. S20 B and C). Although less pronounced, the same findings were also observed in the corresponding simulations with the D257-protonated γ-secretase complexes (Figs. S25, S26, S29, and S30).
Discussion
Intramembrane proteases are involved in several cell signaling pathways and a variety of diseases (74, 75, 76, 77, 78). γ-Secretase is an intramembrane aspartate protease cleaving its substrates within the lipid bilayer with the catalytic subunit presenilin. Once a APP substrate is bound to PS1 γ-secretase, Aβ peptides are sequentially produced and can dissociate. Longer products, such as Aβ42 and Aβ43, are prone to aggregate into AD-related fibrils. Several FAD mutations located on PS1 and APP are found to decrease γ-secretase activity and promote the secretion of longer Aβ peptides.
We have previously suggested that PS1 mutations located on TMD6, TMD7, and TMD9 can directly influence the ability of the enzyme recruiting the scissile bond to the catalytic center by free-energy calculation (33). Other groups have reported that mutations on PS1 and APP consistently destabilize the E-S complexes and, thus, tend to release longer Aβ peptides rather than bringing it to the next cleavage site (17,27,79). Since Aβ42 and Aβ43 are respectively generated from the Aβ48-Aβ45-Aβ42-Aβ38 and Aβ49-Aβ46-Aβ43-Aβ40-Aβ37 production lines, understanding how γ-secretase decides the first cleavage site and releases Aβ peptide at a certain step are of great biological and therapeutic interest.
In the present study, we investigate the role of an internal docking site PS1 that shows a well-packed interaction with substrate residues P5 and P6 shared by both C99 and Notch1 (Table 1). Our simulations indicate that this region interacts strongly with the C terminus of the substrate helical segment. In support of the critical role of the internal docking site, cross-linking experiments with the substituted cysteine accessibility method (SCAM) demonstrated that the pocket undergoes conformational changes in FAD-linked PS1 mutations and upon the binding of GSM-1 (80). Furthermore, a photo-affinity labeling experiment showed that V44 is the strongest binding residue of C99 (21). Compared with the substrate-free γ-secretase, we find that the water access to this pocket from the intracellular side is blocked upon substrate binding.
During the simulations, the substrate P5–P7 residues situated at the C terminus of the helical domain frequently adopted a 3-10 helix geometry, which well aligns the upstream i + 3 residues to the same interface. Hence, only a translational movement toward the next i + 3 cleavage pose is required during substrate processing (and no substrate rotation). The simulations on readjusted APP substrates support the view that both ε49 and ε48 cleavages are possible with the placement of V44 and I45 in the PS1 internal docking site (the cleavage then depends on the conformation between V46 and V50).
During the modeling of the docking poses following the initial cleavage, we identified the gradual change in membrane thickness centered at the APP membrane-anchoring residue K28 to oppose the substrate movement toward the PS1 active site. K28 interacts with the membrane but, in addition, also with other residues in the PS1 and nicastrin subunits (43). While the docking pose of Aβ46γ43 induced the least membrane hydrophobic mismatch in POPC membrane, positive mismatch was observed in the C99ε49 and Aβ49ζ46 complexes, and negative mismatches were observed in Aβ43γ40- and Aβ40γ37-bound complexes. The positive hydrophobic mismatch is beneficial for substrate processing because it causes a force to promote further substrate processing, whereas negative mismatch promotes release of the substrate to the extracellular space. Unlike Aβ43γ40 having large hydrophobic residues M35 and V36 located in the PS1 internal docking site, Aβ40γ37 with I32 and G33 in the internal docking site is indeed dragged out of the enzyme during simulations and the catalytic geometry at the active site was hardly formed. This indicates that the E-S interaction at the PS1 internal docking site on the one hand and the membrane up-pulling force caused by negative hydrophobic mismatch on the other hand are two critical factors determining whether the substrate can proceed to the next cleavage position. Our tug-of-war model between membrane and γ-secretase provides a useful explanation on why many mutations around K28 and G33 have an impact on C99 processivity. Indeed, mutations G29K and A30K, which both interact more strongly with the membrane phosphate, can induce an even stronger hydrophobic mismatch around K28 and have been shown to decrease the processivity (45). On the contrary, K28E, which repels the membrane phosphate groups, might further push the substrate to the next cleavage steps. Indeed, this mutation and K28A, K28Q, and K28L, which all alleviate the K28-lipid interaction, are found to produce even shorter peptides, such as Aβ33 and Aβ34 (44,45,72). Furthermore, mutating G33 to larger amino acids, such as G33A and G33I, which in our model enhance the enzyme binding force, also promote the Aβ40→Aβ37 processing in experiments (46,47). Among these mutations, we simulated the two well-reported mutations K28A and G33I, which address the balance between the forces in two different ways. Both mutations indeed stabilized the E-S geometry such that correct formation of a stable catalytic geometry at the active site became possible.
Interestingly, the results obtained for these two APP mutations suggest a new strategy on how the APP processivity could potentially be enhanced by ligands that can modulate the membrane thickness or enhance the Aβ40-γ-secretase interaction. Using the Aβ40-bound γ-secretase and a pocket search program (Fpocket (81)) we found three potential ligand docking pockets, namely D1, D2, and D3, at the Aβ40-γ-secretase interface (Fig. S31). While D1 and D2 located at the extracellular membrane could potentially modulate the membrane dynamics or enhance the E-S interaction at the juxta-membrane domain, D3 located at the PS1 internal docking site might allow stronger binding of the Aβ peptides to the PS1 internal docking site. Note that D1 is close to the binding pocket of E2012, an Aβ37- and Aβ38-promoting GSM, recently resolved by Yang et al. (42).
To sum up, our work provides a mechanistic explanation of how the balance between the interaction at the proposed PS1 internal docking site and negative hydrophobic mismatch at the substrate juxtamembrane domain plays an important role for the processivity of Aβ. Furthermore, our simulations suggest that the PS1 internal docking site, spatially distinct from the catalytic center (but forming an FAD hot spot), is an essential element for substrate positioning and stabilization. The local hydrophobic mismatch induced by APP K28, on the other hand, imposes a force on the substrate against the substrate binding. Based on this model, we successfully demonstrated how Aβ species can be modulated by enhancing the binding affinity at the PS1 internal docking site and weakening the hydrophobic mismatch. Our model not only paves a way for explaining how mutations on APP or PS1 alter the ε49/ε48 or Aβ43/Aβ40 ratio but also provides a potential target site for the development of small molecules that promote the secretion of shorter Aβ peptides.
Author contributions
S.-Y.C. performed the computational work, analyzed the data, and wrote the manuscript. M.Z. planned the research, interpreted the results, and wrote the manuscript.
Acknowledgments
We thank Manual Hitzenberger, Lukas Feilen, and Harald Steiner for useful discussion. This work is financially supported by Deutsche Forschungsgemeinschaft (DFG, 263531414/FOR 2290/project P7: Understanding Intramembrane Proteolysis).
Declaration of interests
The authors declare no competing interests.
Editor: Elizabeth Rhoades.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.05.023.
Supporting material
References
- 1.Wolfe M.S., Xia W., et al. Selkoe D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenthaler S.F., Haass C., Steiner H. Regulated intramembrane proteolysis – lessons from amyloid precursor protein processing. J. Neurochem. 2011;117:779–796. doi: 10.1111/j.1471-4159.2011.07248.x. [DOI] [PubMed] [Google Scholar]
- 3.Güner G., Lichtenthaler S.F. The substrate repertoire of γ-secretase/presenilin. Semin. Cell Dev. Biol. 2020;105:27–42. doi: 10.1016/j.semcdb.2020.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe M.S. Dysfunctional γ-secretase in familial Alzheimer’s disease. Neurochem. Res. 2019;44:5–11. doi: 10.1007/s11064-018-2511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy J.A., Higgins G.A. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 6.Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venugopal C., Demos C., et al. Sambamurti K. Beta-secretase: structure, function, and evolution. CNS Neurol. Disord. Drug Targets. 2008;7:278–294. doi: 10.2174/187152708784936626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Y., Misonou H., et al. Ihara Y. Distinct intramembrane cleavage of the β-amyloid precursor protein family resembling γ-Secretase-like cleavage of Notch. J. Biol. Chem. 2001;276:35235–35238. doi: 10.1074/jbc.c100357200. [DOI] [PubMed] [Google Scholar]
- 9.Sastre M., Steiner H., et al. Haass C. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C., Kim S.-H., et al. Sisodia S.S. Characterization of a Presenilin-mediated amyloid precursor protein carboxyl-terminal fragment: evidence for distinct mechanisms involved in γ-secretase processing of the App and Notch1 transmembrane domains. J. Biol. Chem. 2001;276:43756–43760. doi: 10.1074/jbc.c100410200. [DOI] [PubMed] [Google Scholar]
- 11.Weidemann A., Eggert S., et al. Evin G. A novel ε-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry. 2002;41:2825–2835. doi: 10.1021/bi015794o. [DOI] [PubMed] [Google Scholar]
- 12.Scheuner D., Eckman C., et al. Younkin S. Secreted amyloid β–protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat. Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 13.Tanzi R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2:a006296. doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwatsubo T., Odaka A., et al. Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 15.Lemere C.A., Lopera F., et al. Arango V J.C. The E280A presenilin 1 Alzheimer mutation produces increased Aβ42 deposition and severe cerebellar pathology. Nat. Med. 1996;2:1146–1150. doi: 10.1038/nm1096-1146. [DOI] [PubMed] [Google Scholar]
- 16.Takami M., Nagashima Y., et al. Ihara Y. γ-Secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of β-carboxyl terminal fragment. J. Neurosci. 2009;29:13042–13052. doi: 10.1523/jneurosci.2362-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okochi M., Tagami S., et al. Takeda M. γ-Secretase modulators and presenilin 1 mutants act differently on presenilin/γ-secretase function to cleave Aβ42 and Aβ43. Cell Rep. 2013;3:42–51. doi: 10.1016/j.celrep.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Bateman R.J., Siemers E.R., et al. Holtzman D.M. A gamma-secretase inhibitor decreases amyloid-beta production in the central nervous system. Ann. Neurol. 2009;66:48–54. doi: 10.1002/ana.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dovey H.F., John V., et al. Audia J.E. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J. Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe M.S. γ-secretase inhibitors and modulators for Alzheimer’s disease. J. Neurochem. 2012;120:89–98. doi: 10.1111/j.1471-4159.2011.07501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukumori A., Steiner H. Substrate recruitment of γ-secretase and mechanism of clinical presenilin mutations revealed by photoaffinity mapping. EMBO J. 2016;35:1628–1643. doi: 10.15252/embj.201694151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philip A.T., Devkota S., et al. Wolfe M.S. Designed helical peptides as functional probes for γ-secretase. Biochemistry. 2019;58:4398–4407. doi: 10.1021/acs.biochem.9b00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfe M.S. Probing mechanisms and therapeutic potential of γ-secretase in Alzheimer’s disease. Molecules. 2021;26:388. doi: 10.3390/molecules26020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bihel F., Das C., et al. Wolfe M.S. Discovery of a Subnanomolar helical D-tridecapeptide inhibitor of gamma-secretase. J. Med. Chem. 2004;47:3931–3933. doi: 10.1021/jm049788c. [DOI] [PubMed] [Google Scholar]
- 25.Kornilova A.Y., Bihel F., et al. Wolfe M.S. The initial substrate-binding site of gamma-secretase is located on presenilin near the active site. Proc. Natl. Acad. Sci. USA. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattarai S., Devkota S., et al. Wolfe M.S. Design of substrate transmembrane mimetics as structural probes for γ-secretase. J. Am. Chem. Soc. 2020;142:3351–3355. doi: 10.1021/jacs.9b13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szaruga M., Munteanu B., et al. Chávez-Gutiérrez L. Alzheimer’s-causing mutations shift Aβ length by destabilizing γ-secretase-Aβn interactions. Cell. 2017;170:443–456.e14. doi: 10.1016/j.cell.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Trambauer J., Rodríguez Sarmiento R.M., et al. Steiner H. Aβ43-producing PS1 FAD mutants cause altered substrate interactions and respond to γ-secretase modulation. EMBO Rep. 2019;21:e47996. doi: 10.15252/embr.201947996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chau D.-M., Crump C.J., et al. Li Y.-M. Familial Alzheimer disease presenilin-1 mutations alter the active site conformation of γ-secretase. J. Biol. Chem. 2012;287:17288–17296. doi: 10.1074/jbc.m111.300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimitrov M., Alattia J.-R., et al. Fraering P.C. Alzheimer’s disease mutations in APP but not γ-secretase modulators affect epsilon-cleavage-dependent AICD production. Nat. Commun. 2013;4:2246. doi: 10.1038/ncomms3246. [DOI] [PubMed] [Google Scholar]
- 31.Sun L., Zhou R., et al. Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc. Natl. Acad. Sci. USA. 2017;114:E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanbouch L., Schaack B., et al. Potier M.-C. Mutations in the juxtamembrane segment of the cholesterol-binding site of APP alter its processing and promotes production of shorter, less toxic Aβ peptides. bioRxiv. 2020 doi: 10.1101/2020.11.16.384891. Preprint at. [DOI] [Google Scholar]
- 33.Chen S.-Y., Zacharias M. How mutations perturb γ-secretase active site studied by free energy simulations. ACS Chem. Neurosci. 2020;11:3321–3332. doi: 10.1021/acschemneuro.0c00440. [DOI] [PubMed] [Google Scholar]
- 34.Xu T.-H., Yan Y., et al. Xu H.E. Alzheimer’s disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio. Cell Discov. 2016;2:16026. doi: 10.1038/celldisc.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devkota S., Williams T.D., Wolfe M.S. Familial Alzheimer’s disease mutations in amyloid protein precursor alter proteolysis by γ-secretase to increase amyloid β-peptides of ≥45 residues. J. Biol. Chem. 2021;296:100281. doi: 10.1016/j.jbc.2021.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beel A.J., Barrett P., et al. Jordan J.B. Nonspecificity of binding of γ-secretase modulators to the amyloid precursor protein. Biochemistry. 2009;48:11837–11839. doi: 10.1021/bi901839d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gertsik N., Am Ende C.W., et al. Li Y.-M. Mapping the binding site of BMS-708163 on γ-secretase with cleavable photoprobes. Cell Chem. Biol. 2017;24:3–8. doi: 10.1016/j.chembiol.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pozdnyakov N., Murrey H.E., et al. Johnson D.S. γ-Secretase modulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin. J. Biol. Chem. 2013;288:9710–9720. doi: 10.1074/jbc.m112.398602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang G., Zhou R., et al. Shi Y. Structural basis of Notch recognition by human γ-secretase. Nature. 2019;565:192–197. doi: 10.1038/s41586-018-0813-8. [DOI] [PubMed] [Google Scholar]
- 40.Zhou R., Yang G., et al. Shi Y. Recognition of the amyloid precursor protein by human γ-secretase. Science. 2019;363:eaaw0930. doi: 10.2210/pdb6iyc/pdb. [DOI] [PubMed] [Google Scholar]
- 41.Hitzenberger M., Zacharias M. Uncovering the binding mode of γ-secretase inhibitors. bioRxiv. 2019 doi: 10.1101/611194. Preprint at. [DOI] [PubMed] [Google Scholar]
- 42.Yang G., Zhou R., et al. Shi Y. Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Cell. 2021;184:521–533.e14. doi: 10.1016/j.cell.2020.11.049. [DOI] [PubMed] [Google Scholar]
- 43.Petit D., Hitzenberger M., et al. Chávez-Gutiérrez L. Extracellular interface between APP and Nicastrin regulates Aβ length and response to γ-secretase modulators. EMBO J. 2019;38:e101494. doi: 10.15252/embj.2019101494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kukar T.L., Ladd T.B., et al. Golde T.E. Lysine 624 of the amyloid precursor protein (APP) is a critical determinant of amyloid β peptide length. J. Biol. Chem. 2011;286:39804–39812. doi: 10.1074/jbc.m111.274696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung J.I., Ran Y., et al. Golde T.E. Complex relationships between substrate sequence and sensitivity to alterations in γ-secretase processivity induced by γ-secretase modulators. Biochemistry. 2014;53:1947–1957. doi: 10.1021/bi401521t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Page R.M., Gutsmiedl A., et al. Steiner H. β-Amyloid precursor protein mutants respond to γ-secretase modulators. J. Biol. Chem. 2010;285:17798–17810. doi: 10.1074/jbc.m110.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munter L.-M., Voigt P., et al. Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai X.C., Rajendra E., et al. Scheres S.H. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4:e11182. doi: 10.7554/elife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eswar N., Webb B., et al. Sali A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case D.A., Ben-Shalom I.Y., Kollman P.A., et al. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- 51.Lee J., Cheng X., et al. Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;110:405–413. doi: 10.1016/j.bpj.2015.11.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lomize M.A., Pogozheva I.D., et al. Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 54.Maier J.A., Martinez C., et al. Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickson C.J., Walker R.C., Gould I.R. Lipid21: Complex Lipid Membrane Simulations with AMBER. J. Chem. Theory Comput. 2022;3:1726–1736. doi: 10.1021/acs.jctc.1c01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goga N., Rzepiela A.J., et al. Berendsen H.J.C. Efficient algorithms for Langevin and DPD dynamics. J. Chem. Theor. Comput. 2012;8:3637–3649. doi: 10.1021/ct3000876. [DOI] [PubMed] [Google Scholar]
- 57.Berendsen H.J.C., Postma J.P.M., et al. Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 58.Balusek C., Hwang H., et al. Gumbart J.C. Accelerating membrane simulations with hydrogen mass repartitioning. J. Chem. Theory Comput. 2019;15:4673–4686. doi: 10.1021/acs.jctc.9b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 60.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 61.Miller B.R., McGee T.D., et al. Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 62.Hitzenberger M., Zacharias M. Structural modeling of γ -secretase Aβn complex formation and substrate processing. ACS Chem. Neurosci. 2019;10:1826–1840. doi: 10.1021/acschemneuro.8b00725. [DOI] [PubMed] [Google Scholar]
- 63.Jeffrey G.A. Oxford University Press; 1997. An Introduction to Hydrogen Bonding. [Google Scholar]
- 64.Schmidtke P., Bidon-Chanal A., et al. Barril X. MDpocket : open source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics. 2011;27:3276–3285. doi: 10.1093/bioinformatics/btr550. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez M.A., Biette K.M., et al. Wolfe M.S. Transmembrane substrate determinants for γ-secretase processing of APP CTFβ. Biochemistry. 2016;55:5675–5688. doi: 10.1021/acs.biochem.6b00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pike S.J., Boddaert T., et al. Clayden J. Participation of non-aminoisobutyric acid (Aib) residues in the 310 helical conformation of Aib-rich foldamers: a solid state study. New J. Chem. 2015;39:3288–3294. doi: 10.1039/c4nj01547a. [DOI] [Google Scholar]
- 67.Zhang L., Hermans J. 310 helix versus .alpha.-Helix: a molecular dynamics study of conformational preferences of aib and alanine. J. Am. Chem. Soc. 1994;116:11915–11921. doi: 10.1021/ja00105a034. [DOI] [Google Scholar]
- 68.Toniolo C., Bonora G.M., et al. Pedone C. Conformation of pleionomers of .alpha.-aminoisobutyric acid. Macromolecules. 1985;18:895–902. doi: 10.1021/ma00147a013. [DOI] [Google Scholar]
- 69.Bavoso A., Benedetti E., et al. Bonora G.M. Long polypeptide 310-helices at atomic resolution. Proc. Natl. Acad. Sci. U S A. 1986;83:1988–1992. doi: 10.1073/pnas.83.7.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolduc D.M., Montagna D.R., et al. Selkoe D.J. The amyloid-beta forming tripeptide cleavage mechanism of γ-secretase. Elife. 2016;5:e17578. doi: 10.7554/elife.17578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jung J.I., Premraj S., et al. Ran Y. Independent relationship between amyloid precursor protein (APP) dimerization and γ-secretase processivity. PLoS One. 2014;9:e111553. doi: 10.1371/journal.pone.0111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ousson S., Saric A., et al. Beher D. Substrate determinants in the C99 juxtamembrane domains differentially affect γ-secretase cleavage specificity and modulator pharmacology. J. Neurochem. 2013;125:610–619. doi: 10.1111/jnc.12129. [DOI] [PubMed] [Google Scholar]
- 73.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/s0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kühnle N., Dederer V., Lemberg M.K. Intramembrane proteolysis at a glance: from signalling to protein degradation. J. Cell Sci. 2019;132 doi: 10.1242/jcs.217745. [DOI] [PubMed] [Google Scholar]
- 75.Düsterhöft S., Künzel U., Freeman M. Rhomboid proteases in human disease: mechanisms and future prospects. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2200–2209. doi: 10.1016/j.bbamcr.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 76.Verhelst S.H.L. Intramembrane proteases as drug targets. FEBS J. 2017;284:1489–1502. doi: 10.1111/febs.13979. [DOI] [PubMed] [Google Scholar]
- 77.Drag M., Salvesen G.S. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turk B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 79.Ratovitski T., Slunt H.H., et al. Borchelt D.R. Endoproteolytic processing and stabilization of wild-type and mutant presenilin. J. Biol. Chem. 1997;272:24536–24541. doi: 10.1074/jbc.272.39.24536. [DOI] [PubMed] [Google Scholar]
- 80.Tominaga A., Cai T., et al. Tomita T. Conformational changes in transmembrane domain 4 of presenilin 1 are associated with altered amyloid- 42 production. J. Neurosci. 2016;36:1362–1372. doi: 10.1523/jneurosci.5090-14.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.