Abstract
Aim
To explore the quality control and implementation of the quantitative detection of liver fibrosis biomarkers, laminin (LN), collagen IV (Col Ⅳ), procollagen III amino‐terminal propeptide (PⅢNP), hyaluronic acid (HA), and cholyglycine (CG), in China.
Methods
Two quality control products were measured in different laboratories using different measurement methods and reagents, and the acquired results were subjected to analysis. The quantitative detection technique was based on the conventional assessment criteria, with a target value ±30% being employed.
Results
Hundred labs were involved in the External Quality Assessment with 88 laboratories completing the assessment, and the pass rates were 84%, 80.2%, 67.5%, 77.3%, and 58.3% for HA, LN, PⅢNP, Col Ⅳ, and CG, respectively. Chemiluminescence immunoassay was used most for HA (90.1%), LN (90.1%), PⅢNP (87.9%), and Col Ⅳ (82.9%) determination, whereas the chemiluminescence immunoassay (31.6%), latex‐enhanced immunoturbidimetry (36.7%), and homogeneous enzyme immunoassay (26.7%) were used for CG determination. The coefficients of variation for HA, LN, PⅢNP, Col Ⅳ, and CG in different laboratories were 3.3%–19.49%, 1.74%–38.81%, 1.97%–41.29%, 2.85%–41.69%, and 2.71%–41.8%, respectively.
Conclusion
The clinical quantitative detection of liver fibrosis biomarkers is highly performed in China. The existing problems are that there are many manufacturers producing reagents and instruments, the quality of reagents is uneven, the specificity and sensitivity of reagents are greatly different, the comparability of results of various systems is poor, and the accuracy and consistency between different systems are lacking. All above underscores the critical importance of EQA in improving and monitoring the identification of biomarkers for liver fibrosis.
Keywords: external quality assessment, internal quality control, liver fibrosis biomarkers, quality control products, quantitative detection and analysis
First, a questionnaire survey was conducted to investigate the current status of quantitative detection of liver fibrosis markers in clinical laboratories. Then, external quality assessment is carried out by detecting quality control products.
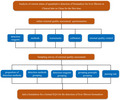
1. INTRODUCTION
The characteristics of chronic liver disease include increasing fibrosis, which may result in portal hypertension, cirrhosis, liver synthesis damage, and hepatocellular carcinoma. 1 , 2 Cholestatic and hepatotoxic chronic liver injuries are the two most prevalent forms of chronic liver injury. 3 Hepatotoxic injury is induced by persistent liver cell damage, such as HBV and/or HCV infection, and non‐alcoholic or alcoholic steatohepatitis. 4 , 5 More than 240 million individuals worldwide are infected with chronic HBV, with China being one of the countries with a high incidence of HBV. 6 , 7 Therefore, a large number of people with chronic HBV infection develop liver fibrosis in China.
Liver fibrosis is defined as the production of fibrous scarring caused by the accumulation of extracellular matrix (ECM) proteins (mainly type I and III crosslinked collagen) that substitute injured normal tissue. 1 , 3 To be frank, the stage of liver fibrosis could only be determined via the biopsy of the liver at present. Nevertheless, a biopsy of the liver can cause serious problems to patients in up to 3%, as well as mortality occurring in 0.03% of the cases. 5 For monitoring chronic liver disease progression, assessing fibrosis staging is critical, which determines prognosis and optimal treatment time, monitors treatment response, and assesses disease progression to reduce morbidity and mortality associated with cirrhosis sequelae. Thus, accurate non‐invasive tests for liver fibrosis diagnosis and staging are required. Non‐invasive fibrosis testing of fibrosis is gaining traction in clinical practice to diagnose liver fibrosis. For fibrosis assessment criteria, blood‐based indicators provide considerable benefits than standard biopsy of the liver, such as accessibility, cost, and safety. Serum markers estimate fibrosis using markers that measure ECM degradation/fibrogenesis. An extensive spectrum of chronic liver disorders, such as non‐alcoholic and alcoholic fatty liver disease, as well as chronic viral hepatitis, has been studied and verified with the aid of this technique.
Currently, five major liver fibrosis indicators are utilized in clinical laboratories, namely laminin (LN), hyaluronic acid (HA), procollagen III's N‐terminal peptide (PIIINP), cholyglycine (CG), and collagen IV (Col IV). 8 , 9 , 10 , 11 , 12 , 13 However, none of these biomarkers was included in the formal External Quality Assessment (EQA) plan. This means it is impossible to assess whether the measurement is accurate. The reliability and utility of disease diagnosis and treatment are determined by the accuracy of the clinical outcomes. The comparability and accuracy of findings across various methods employed in different laboratories, as a consequence, are critically essential considerations. EQA is a method in which the same samples are examined by various labs, and the findings are obtained, analyzed, and subjected to a comparison made by an external, independent organization. The detection and calibration capabilities of clinical laboratories are determined by the National Center for Clinical Laboratories (NCCL) via interlaboratory comparisons, and the laboratory progress is monitored. As a consequence, EQA contributes to the guarantee of comparability and accuracy of outcomes. Now that clinical laboratories are providing detection services for liver fibrosis biomarkers, it is necessary for the NCCL to conduct EQA services for them to ensure the accuracy and comparability of test results.
2. MATERIAL AND METHODS
2.1. Respondents
In the EQA survey for the quantitative identification of liver fibrosis biomarkers, the responders comprised clinical labs that had consented to be involved in it.
2.2. Questionnaire survey
In response to the query on "Investigations on the Quantitative Detection of Biomarkers of Liver Fibrosis," the clinical laboratories provided responses using the EQA system established by NCCL. In addition to the essential data, the questionnaire comprised of eight queries, including the quantitative identification of liver fibrosis biomarkers, the type of the sample, method of detection, the brand of the reagent, the brand of the instrument, the unit of the concentration used by the laboratory, and their readiness to be involved in the EQA survey (serum matrix) for the quantitative identification of liver fibrosis biomarkers (Table S1). After the data were obtained, a summary of the current state of the quantitative detection of liver fibrosis indicators in clinical labs was prepared.
2.3. EQA survey
The NCCL handled the notification of the EQA investigation and the submission of the application. The quality control items were provided to the labs that had submitted their applications. 202011 and 202012 were the two samples (comprising HA, LN, PⅢNP, Col Ⅳ, and CG biomarkers) with different concentrations (cut‐off values and high values).
2.4. Criteria of EQA evaluation
For the purpose of conducting the present research, the statistically robust mean ± 30% was used as the evaluation criterion. The detection result was considered as “qualified” if the deviation of the two samples was within the set range.
2.5. Statistical analysis
Based on the response and submitted findings, the accumulated number and the proportion of the questionnaire responses were calculated. The quantitative identification outcomes of the liver fibrosis biomarkers were classified in accordance with the reagents and evaluation made. Moreover, every reagent manufacturing company was named as reagents 1, 2, 3, and so on (Table S2). The coefficient of variation (CV), mean value, and bias between the target and the mean values of the reagent groups were derived when no less than two laboratories were in a single group.
3. RESULTS
3.1. The proportion of detection projects of liver fibrosis biomarkers
In the questionnaire, the proportion of the various detection items was evaluated. As shown in Table 1, 49.6% (112/226) of the laboratories quantitatively detected four liver fibrosis biomarkers (HA, LN, PⅢNP, and Col Ⅳ); 39.8% (90/226) of the laboratories detected five liver fibrosis biomarkers (HA, LN, PⅢNP, Col Ⅳ, and CG); 10.6% (24/226) of the laboratories detected only the CG biomarker. We assessed the proportion of distinct detection techniques with the aid of a questionnaire. The results revealed that the proportion of chemiluminescence immunoassay (CLIA) for HA, LN, PⅢNP, and Col Ⅳ detection was 86.6%. Other detection methods included latex‐enhanced immunoturbidimetry, up‐conversion luminescence immunoassay, (ULIA), electrochemiluminescence, (ECLIA), and radioimmunoassay (RIA), accounting for approximately 13.4% of the total detection methods. The common methods for CG detection were CLIA, latex‐enhanced immunoturbidimetry, and homogeneous enzyme immunoassay (HEI), accounting for 28.1%, 37.7%, and 28.9%, respectively. The other methods including ECLIA and RIA represented around 5.3% of the overall estimate.
TABLE 1.
Summary of questionnaire information
| Project | Amount | Proportion |
|---|---|---|
| HA, LN, PⅢNP, Col Ⅳ | 112 | 49.6% |
| HA, LN, PⅢNP, Col Ⅳ, and CG | 90 | 39.8% |
| CG | 24 | 10.6% |
| Method | Amount | Proportion (%) | |
|---|---|---|---|
| HA, LN, PⅢNP, Col Ⅳ | Chemiluminescence immunoassay, CLIA | 175 | 86.6 |
| Latex‐enhanced immunoturbidimetry | 17 | 8.4 | |
| Up‐conversion luminescence immunoassay, ULIA | 5 | 2.5 | |
| Electrochemiluminescence, ECLIA | 3 | 1.5 | |
| Radioimmunoassay, RIA | 2 | 1.0 | |
| CG | Chemiluminescence immunoassay, CLIA | 32 | 28.1 |
| Electrochemiluminescence, ECLIA | 3 | 2.6 | |
| Radioimmunoassay, RIA | 1 | 0.9 | |
| Latex‐enhanced immunoturbidimetry | 43 | 37.7 | |
| Homogeneous enzyme immunoassay, HEI | 33 | 28.9 | |
| Others | 2 | 1.8 |
The proportion of detection projects of liver fibrosis biomarkers and composition ratios of detection methods.
3.2. Group statistics on the basis of the methods of detection
For the quality control materials that were distributed, two concentrations (cut‐off value and high value) were established. For the purpose of collecting more data, the quality control materials were distributed to the labs that quantitatively detected four or five biomarkers. Moreover, the clinical labs that were involved in the present research employed a variety of methods, procedures, and reagents. “Other category” was used to classify laboratories that did not describe their detection techniques in their application. As illustrated in Table 2, robust CV, standard uncertainty and the robust standard deviation are all in accordance with the ISO13528 standards. The results revealed significant differences in the robust mean values among the various identification methods of the laboratories, indicating the necessity for grouped statistics. For HA detection using RIA, the quality control substance concentration increased with an increase in robust CV (24.1% vs 70.8%), which can be attributed to identification procedures defects and small trial size. With increased concentration, robust CV of chemiluminescence immunoassay‐HRP for HA, LN, PⅢNP, and Col Ⅳ showed a downward trend, while the chemiluminescence immunoassay‐Acridinium Ester showed an opposite trend. We tried to explain that maybe CLIA‐HRP is prone to be stable in HA, LN, Col Ⅳ, and PⅢNP detection when the sample concentration is increased, while CLIA‐AE is more suitable for detection at lower concentrations. Conversely, for Col IV detection, the robust CV of CLIA‐AE and microplate decreased with an increase in concentration. However, the detection of PⅢNP, HA, LN, and Col Ⅳ via ULIA was relatively stable. For CG detection, the robust CV of CLIA decreased with increased sample concentration. The robust CV of immunoturbidimetry increased significantly. We suspect that CLIA may have high reliability in detecting high concentrations.
TABLE 2.
The result of grouped statistics according to detection methods
| Batch number | Group | Total number | Robust mean | Robust SD | Standard uncertainty | Robust CV (%) | |
|---|---|---|---|---|---|---|---|
| HA | 202011 | All | 81 | 119.54 | 23.3 | 3.236 | 19.49 |
| Chemiluminescence immunoassay‐AMPPD | 8 | 97.2 | 26.66 | 12.594 | 27.42 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 128.39 | 9.97 | 2.657 | 7.77 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 118.36 | 21.7 | 7.524 | 18.34 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 121.61 | 14.58 | 3.645 | 11.99 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 54.55 | 4.1 | 2.958 | 7.51 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 102.74 | 5.24 | 3.782 | 5.1 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 2 | 137.1 | 33.04 | 29.201 | 24.1 | ||
| Latex‐enhanced immunoturbidimetry | 1 | – | – | – | – | ||
| Others | 1 | – | – | – | – | ||
| 202012 | All | 81 | 303.35 | 66.8 | 9.278 | 22.02 | |
| Chemiluminescence immunoassay‐AMPPD | 8 | 339.91 | 44.37 | 20.965 | 13.06 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 343.84 | 20.89 | 5.568 | 6.08 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 360.37 | 113.51 | 39.353 | 31.5 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 288.06 | 23.93 | 5.982 | 8.31 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 87.69 | 13.33 | 9.618 | 15.2 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 126.98 | 1.44 | 1.04 | 1.14 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 2 | 341.9 | 239.6 | 211.774 | 70.08 | ||
| Latex‐enhanced immunoturbidimetry | 1 | – | – | – | – | ||
| Others | 1 | – | – | – | – | ||
| LN | 202011 | All | 81 | 146.94 | 57.03 | 7.921 | 38.81 |
| Chemiluminescence immunoassay‐AMPPD | 8 | 120.43 | 40.49 | 19.127 | 33.62 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 124.14 | 8.98 | 2.394 | 7.24 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 115.95 | 57.95 | 20.091 | 49.98 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 189.48 | 7.04 | 1.761 | 3.72 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 131.99 | 17.45 | 12.59 | 13.22 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 81.91 | 1.98 | 1.432 | 2.42 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 2 | 326 | 49.72 | 43.942 | 15.25 | ||
| Latex‐enhanced immunoturbidimetry | 1 | – | – | – | – | ||
| Others | 1 | – | – | – | – | ||
| 202012 | All | 81 | 457.63 | 178.68 | 24.816 | 39.04 | |
| Chemiluminescence immunoassay‐AMPPD | 8 | 404.71 | 54.62 | 25.804 | 13.5 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 395.11 | 23.49 | 6.261 | 5.95 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 363.45 | 266.71 | 92.464 | 73.38 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 607.1 | 34.2 | 8.55 | 5.63 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 334.05 | 19.9 | 14.359 | 5.96 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 73.31 | 2.81 | 2.026 | 3.83 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 2 | 1213.85 | 39.53 | 34.942 | 3.26 | ||
| Latex‐enhanced immunoturbidimetry | 1 | – | – | – | – | ||
| Others | 1 | – | – | – | – | ||
| PⅢNP | 202011 | All | 83 | 12.36 | 4.98 | 0.683 | 40.28 |
| Chemiluminescence immunoassay‐AMPPD | 8 | 10.32 | 3.92 | 1.852 | 37.99 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 10.86 | 1.49 | 0.397 | 13.73 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 4.3 | 1.29 | 0.448 | 30.06 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 14.57 | 1.08 | 0.269 | 7.38 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 156.64 | 31.6 | 22.802 | 20.17 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 61.96 | 4.87 | 3.512 | 7.86 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 1 | – | – | – | – | ||
| Latex‐enhanced immunoturbidimetry | 3 | 4.19 | 3.01 | 2.175 | 71.99 | ||
| Others | 1 | – | – | – | – | ||
| 202012 | All | 83 | 30.85 | 12.74 | 1.747 | 41.29 | |
| Chemiluminescence immunoassay‐AMPPD | 8 | 23.96 | 9.27 | 4.378 | 38.67 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 35.22 | 3.87 | 1.032 | 11 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 10.23 | 4.86 | 1.686 | 47.55 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 29.37 | 2.68 | 0.671 | 9.13 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 475.43 | 355.83 | 256.801 | 74.84 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 59.72 | 4.49 | 3.242 | 7.52 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 1 | – | – | – | – | ||
| Latex‐enhanced immunoturbidimetry | 3 | 10.23 | 0.69 | 0.5 | 6.77 | ||
| Others | 1 | – | – | – | – | ||
| Col Ⅳ | 202011 | All | 88 | 78.34 | 32.66 | 4.352 | 41.69 |
| Chemiluminescence immunoassay‐AMPPD | 8 | 144.65 | 10.95 | 5.173 | 7.57 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 96.01 | 5.4 | 1.438 | 5.62 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 74.29 | 26 | 9.014 | 25 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 48.77 | 3.51 | 0.895 | 7.19 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 98.79 | 23.82 | 17.188 | 24.11 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 129.65 | 6.18 | 4.458 | 4.76 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 2 | 57.15 | 7.78 | 6.875 | 13.61 | ||
| Latex‐enhanced immunoturbidimetry | 8 | 61.82 | 25.69 | 10.704 | 41.55 | ||
| Others | 1 | – | – | – | – | ||
| 202012 | All | 88 | 306.87 | 110.22 | 14.686 | 35.92 | |
| Chemiluminescence immunoassay‐AMPPD | 8 | 454.64 | 189.92 | 89.731 | 41.78 | ||
| Chemiluminescence immunoassay‐HRP | 22 | 387.81 | 17.78 | 4.739 | 4.59 | ||
| Chemiluminescence immunoassay‐Acridinium Ester | 13 | 282.14 | 71.14 | 24.665 | 25.22 | ||
| Chemiluminescence immunoassay‐ABEI | 25 | 258.77 | 28.29 | 7.219 | 10.93 | ||
| Chemiluminescence immunoassay‐Microplate | 5 | 426.51 | 40.5 | 29.231 | 9.5 | ||
| Up‐conversion luminescence immunoassay, ULIA | 3 | 327.88 | 9.73 | 7.024 | 2.97 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 2 | 100.1 | 13.95 | 12.332 | 13.94 | ||
| Latex‐enhanced immunoturbidimetry | 8 | 135.41 | 73.86 | 30.776 | 54.55 | ||
| Others | 1 | – | – | – | – | ||
| CG | 202011 | All | 60 | 2.47 | 0.65 | 0.106 | 26.46 |
| Chemiluminescence immunoassay‐AMPPD | 2 | 2.02 | 1.14 | 1.007 | 56.39 | ||
| Chemiluminescence immunoassay‐HRP | 2 | 2.15 | 0.15 | 0.134 | 7.05 | ||
| Chemiluminescence immunoassay‐ABEI | 15 | 2.7 | 0.2 | 0.067 | 7.46 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 1 | – | – | – | – | ||
| Immunoturbidimetry | 7 | 2.28 | 0.5 | 0.238 | 22.1 | ||
| Latex‐enhanced immunoturbidimetry | 15 | 2.03 | 0.23 | 0.074 | 11.3 | ||
| Homogeneous enzyme immunoassay, HEI | 16 | 2.46 | 0.53 | 0.167 | 21.72 | ||
| Others | 1 | – | – | – | – | ||
| 202012 | All | 60 | 14.12 | 5.9 | 0.952 | 41.8 | |
| Chemiluminescence immunoassay‐AMPPD | 2 | 10.2 | 3.37 | 2.977 | 33.02 | ||
| Chemiluminescence immunoassay‐HRP | 2 | 17.16 | 1.24 | 1.099 | 7.25 | ||
| Chemiluminescence immunoassay‐ABEI | 15 | 12.89 | 0.31 | 0.103 | 2.38 | ||
| Electrochemiluminescence, ECLIA | 1 | – | – | – | – | ||
| Radioimmunoassay, RIA | 1 | – | – | – | – | ||
| Immunoturbidimetry | 7 | 14.84 | 8.22 | 3.885 | 55.42 | ||
| Latex‐enhanced immunoturbidimetry | 15 | 9.69 | 2.3 | 0.743 | 23.76 | ||
| Homogeneous enzyme immunoassay, HEI | 16 | 17.76 | 7.32 | 2.287 | 41.22 | ||
| Others | 1 | – | – | – | – |
If there is only one laboratory, robust standard deviation, standard uncertainty, and robust CV cannot be calculated.
3.3. Group statistics premised on detection reagents
Besides the detection techniques, the responses were classified according to the detection reagents used in view of the fact that the same detection technique can use different reagents; therefore, the classification by reagents increases accuracy. As shown in Table 3, reagents 1 and 13 represented the majority of the market share, indicating that they are frequently employed in most Chinese clinical labs for the quantitative identification of liver fibrosis biomarkers. Furthermore, the pass rates of the reagents 1 and 13 groups (100%) were higher than those of the other groups, suggesting that the larger sample size could increase the statistical accuracy. Although the reagent 1, 7, 10, 13, and 14 groups used CLIA for the detections of Col Ⅳ, LN, HA, and PⅢNP, the results varied, thereby suggesting the need for grouping based on the use of reagents and improving the commutability of quality control materials. For CG detection, reagents 1, 13, and 26 were commonly used in most Chinese clinical laboratories. Reagents 1 and 13 were used in the CLIA method, and reagent 26 was used in the HEI method. With increased sample concentration, CLIA’s robust CV decreased, whereas that of HEI was elevated, which was consistent with the findings of grouping by detection methods (Table 2).
TABLE 3.
The result of grouped statistics according to detection reagents
| Batch number | Group | Total number | Robust mean | Robust SD | Standard uncertainty | Robust CV (%) | |
|---|---|---|---|---|---|---|---|
| HA | 202011 | All | 81 | 119.54 | 23.3 | 3.236 | 19.49 |
| Reagent 1 | 22 | 131.94 | 7.81 | 2.131 | 5.92 | ||
| Reagent 2 | 2 | 137.1 | 33.04 | 29.201 | 24.1 | ||
| Reagent 5 | 3 | 102.74 | 5.24 | 3.782 | 5.1 | ||
| Reagent 7 | 5 | 42.17 | 1.39 | 0.778 | 3.3 | ||
| Reagent 8 | 3 | 63.91 | 0.8 | 0.709 | 1.25 | ||
| Reagent 9 | 3 | 106.87 | 9.92 | 8.767 | 9.28 | ||
| Reagent 10 | 5 | 101.61 | 5.36 | 2.995 | 5.27 | ||
| Reagent 11 | 3 | 99.39 | 4.9 | 3.533 | 4.93 | ||
| Reagent 13 | 25 | 122.45 | 13.49 | 3.372 | 11.01 | ||
| Reagent 14 | 5 | 265.2 | 11.58 | 6.473 | 4.37 | ||
| 202012 | All | 81 | 303.35 | 66.8 | 9.278 | 22.02 | |
| Reagent 1 | 22 | 349.89 | 16.54 | 4.511 | 4.73 | ||
| Reagent 2 | 2 | 341.9 | 239.6 | 211.774 | 70.08 | ||
| Reagent 5 | 3 | 126.98 | 1.44 | 1.04 | 1.14 | ||
| Reagent 7 | 5 | 63.93 | 9.96 | 5.567 | 15.58 | ||
| Reagent 8 | 3 | 140.03 | 1.6 | 1.418 | 1.15 | ||
| Reagent 9 | 3 | 230.33 | 33.53 | 29.64 | 14.56 | ||
| Reagent 10 | 5 | 311.77 | 16.08 | 8.988 | 5.16 | ||
| Reagent 11 | 3 | 375.08 | 6.61 | 4.773 | 1.76 | ||
| Reagent 13 | 25 | 289.44 | 22.98 | 5.744 | 7.94 | ||
| Reagent 14 | 5 | 705.18 | 42.11 | 23.541 | 5.97 | ||
| LN | 202011 | All | 81 | 146.94 | 57.03 | 7.921 | 38.81 |
| Reagent 1 | 22 | 123.44 | 7.72 | 2.107 | 6.26 | ||
| Reagent 2 | 2 | 326 | 49.72 | 43.942 | 15.25 | ||
| Reagent 5 | 3 | 81.91 | 1.98 | 1.432 | 2.42 | ||
| Reagent 7 | 5 | 127.6 | 2.23 | 1.244 | 1.74 | ||
| Reagent 8 | 3 | 251.22 | 130.25 | 115.122 | 51.85 | ||
| Reagent 9 | 3 | 91.58 | 3.39 | 2.998 | 3.7 | ||
| Reagent 10 | 5 | 63.27 | 3.26 | 1.822 | 5.15 | ||
| Reagent 11 | 3 | 87.38 | 1.76 | 1.269 | 2.01 | ||
| Reagent 13 | 25 | 189.18 | 7.18 | 1.794 | 3.79 | ||
| Reagent 14 | 5 | 235.67 | 15.85 | 8.86 | 6.73 | ||
| 202012 | All | 81 | 457.63 | 178.68 | 24.816 | 39.04 | |
| Reagent 1 | 22 | 394.83 | 18.76 | 5.118 | 4.75 | ||
| Reagent 2 | 2 | 1213.85 | 39.53 | 34.942 | 3.26 | ||
| Reagent 5 | 3 | 73.31 | 2.81 | 2.026 | 3.83 | ||
| Reagent 7 | 5 | 360.99 | 35.81 | 20.016 | 9.92 | ||
| Reagent 8 | 3 | 707.88 | 372.38 | 329.136 | 52.61 | ||
| Reagent 9 | 3 | 266.35 | 3.79 | 3.345 | 1.42 | ||
| Reagent 10 | 5 | 117.53 | 6.56 | 3.668 | 5.58 | ||
| Reagent 11 | 3 | 392.16 | 17.59 | 12.697 | 4.49 | ||
| Reagent 13 | 25 | 608.39 | 32.44 | 8.111 | 5.33 | ||
| Reagent 14 | 5 | 624.26 | 25.49 | 14.252 | 4.08 | ||
| PⅢNP | 202011 | All | 83 | 12.36 | 4.98 | 0.683 | 40.28 |
| Reagent 1 | 22 | 10.54 | 1.18 | 0.321 | 11.17 | ||
| Reagent 2 | 2 | 233.95 | 26.38 | 23.318 | 11.28 | ||
| Reagent 5 | 3 | 61.96 | 4.87 | 3.512 | 7.86 | ||
| Reagent 7 | 5 | 182.71 | 31.28 | 17.488 | 17.12 | ||
| Reagent 8 | 3 | 16.42 | 9.85 | 8.704 | 59.97 | ||
| Reagent 9 | 3 | 10.07 | 0.1 | 0.085 | 0.95 | ||
| Reagent 10 | 5 | 3.21 | 0.16 | 0.089 | 4.96 | ||
| Reagent 11 | 3 | 7.36 | 0.4 | 0.287 | 5.39 | ||
| Reagent 13 | 25 | 14.78 | 0.99 | 0.247 | 6.69 | ||
| Reagent 14 | 5 | 4.2 | 0.24 | 0.132 | 5.65 | ||
| Reagent 16 | 2 | 2.69 | 0.92 | 0.66 | 34.02 | ||
| 202012 | All | 83 | 30.85 | 12.74 | 1.747 | 41.29 | |
| Reagent 1 | 22 | 34.76 | 3.13 | 0.854 | 9 | ||
| Reagent 2 | 2 | 1858.1 | 1460.67 | 1291.059 | 78.61 | ||
| Reagent 5 | 3 | 59.72 | 4.49 | 3.242 | 7.52 | ||
| Reagent 7 | 5 | 618.64 | 108.46 | 60.633 | 17.53 | ||
| Reagent 8 | 3 | 29.43 | 14.28 | 12.623 | 48.53 | ||
| Reagent 9 | 3 | 17.24 | 0.64 | 0.567 | 3.72 | ||
| Reagent 10 | 5 | 6.21 | 0.25 | 0.139 | 3.99 | ||
| Reagent 11 | 3 | 16.8 | 0.94 | 0.675 | 5.57 | ||
| Reagent 13 | 25 | 29.38 | 2.14 | 0.536 | 7.29 | ||
| Reagent 14 | 5 | 9.38 | 0.19 | 0.103 | 1.97 | ||
| Reagent 16 | 2 | 9.56 | 0.69 | 0.5 | 7.25 | ||
| Col Ⅳ | 202011 | All | 88 | 78.34 | 32.66 | 4.352 | 41.69 |
| Reagent 1 | 22 | 94.82 | 4.01 | 1.094 | 4.23 | ||
| Reagent 2 | 2 | 57.15 | 7.78 | 6.875 | 13.61 | ||
| Reagent 5 | 3 | 129.65 | 6.18 | 4.458 | 4.76 | ||
| Reagent 7 | 5 | 100 | 5.45 | 3.048 | 5.45 | ||
| Reagent 8 | 3 | 161.97 | 9.82 | 8.682 | 6.07 | ||
| Reagent 9 | 3 | 70.49 | 23.37 | 20.66 | 33.16 | ||
| Reagent 10 | 5 | 107.02 | 12.58 | 7.035 | 11.76 | ||
| Reagent 11 | 3 | 151.01 | 2.14 | 1.546 | 1.42 | ||
| Reagent 13 | 25 | 47.65 | 4.12 | 1.051 | 8.65 | ||
| Reagent 14 | 5 | 56.4 | 4.83 | 2.702 | 8.57 | ||
| Reagent 17 | 7 | 64 | 17.15 | 8.75 | 26.79 | ||
| 202012 | All | 88 | 306.87 | 110.22 | 14.686 | 35.92 | |
| Reagent 1 | 22 | 385.22 | 17.66 | 4.818 | 4.59 | ||
| Reagent 2 | 2 | 100.1 | 13.95 | 12.332 | 13.94 | ||
| Reagent 5 | 3 | 327.88 | 9.73 | 7.024 | 2.97 | ||
| Reagent 7 | 5 | 401.93 | 52.77 | 29.498 | 13.13 | ||
| Reagent 8 | 3 | 404.9 | 38.82 | 34.311 | 9.59 | ||
| Reagent 9 | 3 | 200.4 | 44.48 | 39.314 | 22.2 | ||
| Reagent 10 | 5 | 408.7 | 30.17 | 16.864 | 7.38 | ||
| Reagent 11 | 3 | 637.21 | 6.36 | 4.593 | 1 | ||
| Reagent 13 | 25 | 254.99 | 23.07 | 5.886 | 9.05 | ||
| Reagent 14 | 5 | 235.62 | 6.72 | 3.758 | 2.85 | ||
| Reagent 17 | 6 | 146.89 | 38.78 | 19.791 | 26.4 | ||
| CG | 202011 | All | 60 | 2.47 | 0.65 | 0.106 | 26.46 |
| Reagent 1 | 5 | 3.44 | 0.71 | 0.395 | 20.58 | ||
| Reagent 7 | 4 | 1.3 | 0.02 | 0.021 | 1.85 | ||
| Reagent 13 | 15 | 2.74 | 0.18 | 0.056 | 6.4 | ||
| Reagent 17 | 3 | 2.2 | 1.06 | 0.935 | 48.09 | ||
| Reagent 19 | 2 | 1.88 | 0.29 | 0.255 | 15.37 | ||
| Reagent 24 | 4 | 1.94 | 0.07 | 0.046 | 3.77 | ||
| Reagent 25 | 4 | 4.06 | 0.1 | 0.085 | 2.37 | ||
| Reagent 26 | 10 | 2.46 | 0.19 | 0.074 | 7.61 | ||
| Reagent 29 | 3 | 2.1 | 0.13 | 0.095 | 6.24 | ||
| 202012 | All | 60 | 14.12 | 5.9 | 0.952 | 41.8 | |
| Reagent 1 | 5 | 24.02 | 4.17 | 2.333 | 17.37 | ||
| Reagent 7 | 4 | 7.82 | 0.45 | 0.397 | 5.74 | ||
| Reagent 13 | 15 | 12.93 | 0.35 | 0.113 | 2.71 | ||
| Reagent 17 | 3 | 29.4 | 25.68 | 22.695 | 87.33 | ||
| Reagent 19 | 2 | 11.96 | 7.1 | 6.272 | 59.36 | ||
| Reagent 24 | 4 | 8.21 | 1.14 | 0.714 | 13.91 | ||
| Reagent 25 | 4 | 27.78 | 2.92 | 2.58 | 10.51 | ||
| Reagent 26 | 10 | 17.28 | 3.04 | 1.2 | 17.58 | ||
| Reagent 29 | 3 | 8.71 | 0.54 | 0.392 | 6.24 |
There was only one case of reagent 3, 4, 6, 12, 15 for detection of HA, LN, PⅢNP, and Col Ⅳ, so statistical calculation could not be carried out.
There was only one case of reagent 2, 4, 12, 18, 20, 21, 22, 23, 27, 28 for detection of CG, so statistical calculation could not be carried out.
3.4. Group statistics premised on the principle of grouping
According to the statistical data shown in Tables 2 and 3, it is reasonable to hypothesize that the detection reagents and techniques were employed in their entirety. The reagent produced by a particular manufacturer is typically matched to the detection technique used by that manufacturer. Regardless of the data for grouping based on equipment, procedures, or reagents, grouping was conducted according to the number of labs involved: greater than or equal to 18 or 12 were in one group under ISO 13528. The “other” category was formed if they did not fit into any of the other groupings. According to ISO 13528, the findings were examined using the robust mean, robust standard deviation, and robust CV of the involved labs. However, owing to the limited number of participating labs, the grouping criterion was unsuitable. Therefore, the participating laboratories were classified into separate groups based on the number of laboratories using the same reagent ≥ 5 (Table 4). For the detection of PⅢNP, the lowest values were obtained using reagent 7 and the highest using reagent 14. However, for the detection of LN, the results obtained on using reagent 7 and reagent 14 were opposite. Furthermore, reagents 7 and 14 were both used in the CLIA method but labeled with different substrates. This can be attributed to the different sensitivity of the various reagents for the detection items. When the EQA of the quantitative detection of liver fibrosis biomarkers is properly performed, the variability of the results may be minimized by expanding the number of labs that were included in the present research.
TABLE 4.
The result and passing rates of grouped statistics according to the grouping principle
| Batch number | Group | Total number | Robust mean | Robust SD | Standard uncertainty | Robust CV | Total number | Number of passing labs | Pass rate (%) | |
|---|---|---|---|---|---|---|---|---|---|---|
| HA | 202011 | All | 81 | 119.54 | 23.3 | 3.236 | 19.49 | 19 | 14 | 70 |
| Reagent 1 | 22 | 131.94 | 7.812 | 2.131 | 5.92 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 42.17 | 1.391 | 0.778 | 3.3 | 5 | 5 | 100 | ||
| Reagent 10 | 5 | 101.61 | 5.358 | 2.995 | 5.27 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 122.45 | 13.486 | 3.372 | 11.01 | 25 | 24 | 96 | ||
| Reagent 14 | 5 | 265.2 | 11.58 | 6.473 | 4.37 | 5 | 5 | 100 | ||
| 202012 | All | 81 | 303.35 | 66.799 | 9.278 | 22.02 | 19 | 10 | 50 | |
| Reagent 1 | 22 | 349.88 | 16.536 | 4.511 | 4.73 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 63.93 | 9.959 | 5.567 | 15.58 | 5 | 4 | 80 | ||
| Reagent 10 | 5 | 311.77 | 16.079 | 8.988 | 5.16 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 289.44 | 22.975 | 5.744 | 7.94 | 25 | 25 | 100 | ||
| Reagent 14 | 5 | 705.18 | 42.111 | 23.541 | 5.97 | 5 | 5 | 100 | ||
| LN | 202011 | All | 81 | 146.94 | 57.032 | 7.921 | 38.81 | 19 | 6 | 30 |
| Reagent 1 | 22 | 123.44 | 7.723 | 2.107 | 6.26 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 127.6 | 2.226 | 1.244 | 1.74 | 5 | 5 | 100 | ||
| Reagent 10 | 5 | 63.27 | 3.26 | 1.822 | 5.15 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 189.18 | 7.175 | 1.794 | 3.79 | 25 | 25 | 100 | ||
| Reagent 14 | 5 | 235.67 | 15.85 | 8.86 | 6.73 | 5 | 5 | 100 | ||
| 202012 | All | 81 | 457.63 | 178.677 | 24.816 | 39.04 | 19 | 8 | 40 | |
| Reagent 1 | 22 | 394.83 | 18.764 | 5.118 | 4.75 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 360.99 | 35.805 | 20.016 | 9.92 | 5 | 5 | 100 | ||
| Reagent 10 | 5 | 117.53 | 6.562 | 3.668 | 5.58 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 608.39 | 32.442 | 8.111 | 5.33 | 25 | 25 | 100 | ||
| Reagent 14 | 5 | 624.26 | 25.494 | 14.252 | 4.08 | 5 | 5 | 100 | ||
| PⅢNP | 202011 | All | 83 | 12.36 | 4.978 | 0.683 | 40.27 | 21 | 7 | 31.8 |
| Reagent 1 | 21 | 10.54 | 1.178 | 0.321 | 11.17 | 22 | 20 | 95.2 | ||
| Reagent 7 | 5 | 182.71 | 31.284 | 17.488 | 17.12 | 5 | 5 | 100 | ||
| Reagent 10 | 5 | 3.21 | 0.159 | 0.089 | 4.96 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 14.78 | 0.989 | 0.247 | 6.69 | 25 | 25 | 100 | ||
| Reagent 14 | 5 | 4.2 | 0.237 | 0.132 | 5.65 | 5 | 4 | 80 | ||
| 202012 | All | 83 | 30.85 | 12.736 | 1.747 | 41.29 | 21 | 4 | 18.2 | |
| Reagent 1 | 21 | 34.76 | 3.129 | 0.854 | 9 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 618.64 | 108.464 | 60.633 | 17.53 | 5 | 5 | 100 | ||
| Reagent 10 | 5 | 6.21 | 0.248 | 0.139 | 3.99 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 29.38 | 2.142 | 0.536 | 7.29 | 25 | 20 | 80 | ||
| Reagent 14 | 5 | 9.38 | 0.185 | 0.103 | 1.97 | 5 | 4 | 80 | ||
| Col Ⅳ | 202011 | All | 88 | 78.33 | 32.66 | 4.352 | 41.69 | 22 | 8 | 36.4 |
| Reagent 1 | 22 | 94.82 | 4.011 | 1.094 | 4.23 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 100 | 5.452 | 3.048 | 5.45 | 6 | 5 | 83.3 | ||
| Reagent 10 | 5 | 107.02 | 12.584 | 7.035 | 11.76 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 47.65 | 4.121 | 1.051 | 8.65 | 5 | 5 | 100 | ||
| Reagent 14 | 5 | 56.4 | 4.834 | 2.702 | 8.57 | 25 | 23 | 95.8 | ||
| Reagent 17 | 6 | 64 | 17.146 | 8.75 | 26.79 | 5 | 5 | 100 | ||
| 202012 | All | 88 | 306.87 | 110.217 | 14.686 | 35.92 | 22 | 10 | 45.5 | |
| Reagent 1 | 22 | 385.22 | 17.664 | 4.818 | 4.59 | 22 | 21 | 100 | ||
| Reagent 7 | 5 | 401.93 | 52.768 | 29.498 | 13.13 | 6 | 4 | 66.7 | ||
| Reagent 10 | 5 | 408.7 | 30.168 | 16.864 | 7.38 | 5 | 5 | 100 | ||
| Reagent 13 | 25 | 254.99 | 23.069 | 5.886 | 9.05 | 5 | 5 | 100 | ||
| Reagent 14 | 5 | 235.62 | 6.722 | 3.758 | 2.85 | 25 | 24 | 100 | ||
| Reagent 17 | 6 | 146.89 | 38.783 | 19.791 | 26.4 | 5 | 5 | 100 | ||
| CG | 202011 | All | 60 | 2.47 | 0.654 | 0.106 | 26.46 | 30 | 19 | 63.3 |
| Reagent 1 | 5 | 3.44 | 0.707 | 0.395 | 20.58 | 5 | 4 | 80 | ||
| Reagent 13 | 15 | 2.74 | 0.175 | 0.056 | 6.4 | 15 | 15 | 100 | ||
| Reagent 26 | 10 | 2.46 | 0.187 | 0.074 | 7.61 | 10 | 10 | 100 | ||
| 202012 | All | 60 | 14.12 | 5.9 | 0.952 | 41.8 | 30 | 8 | 26.7 | |
| Reagent 1 | 5 | 24.02 | 4.173 | 2.333 | 17.37 | 5 | 5 | 100 | ||
| Reagent 13 | 15 | 12.93 | 0.35 | 0.113 | 2.71 | 15 | 15 | 100 | ||
| Reagent 26 | 10 | 17.28 | 3.037 | 1.2 | 17.58 | 10 | 9 | 90 |
All included labs were divided into groups and subjected to analyses according to the reagents they employed, with the findings being reviewed in relation to the target value ±30%. The desired outcome is a robust average value, with a dispersion of ±30% being the maximum permitted for the degree of dispersion of the data. A satisfactory result was judged to be achieved if the detection value fell within a certain range (Table 4). In the 202011 sample detection, the pass rates of HA, LN, PⅢNP, Col Ⅳ, and CG in the “other group” were 70%, 30%, 31.8%, 36.45%, and 63.3%, whereas those of reagents 1, 7, 10, 13, and 14 were nearly 100%. Therefore, if laboratories using the same reagent can be individually grouped, the pass rates are relatively high. This situation can also be seen in the detection of high‐concentration samples. The pass rate of the separate group was remarkably higher than that of the mixed group. The coefficient of variation for HA, LN, PⅢNP, Col Ⅳ, and CG in different laboratories were 3.3%–19.49%, 1.74%–38.81%, 1.97%–41.29%, 2.85%–41.69%, and 2.71%–41.8%, respectively. Generally, the lower CV appeared in the separate group, while the higher CV appeared in the mixed group. Therefore, these results signify the importance of grouping. With the increase in concentration, the pass rates of HA, LN, PⅢNP, Col Ⅳ, and CG, apart from the other group, showed no significant difference. The pass rates of HA, PⅢNP, and CG decreased with increasing concentrations, whereas that of LN and Col Ⅳ increased but not by much.
3.5. Total pass rates
After calculating the pass rate of each detection project, the total pass rates of all projects were calculated. The total pass rates of HA, LN, PⅢNP, Col Ⅳ, and CG were 84.0%, 80.2%, 67.5%, 77.3%, and 58.3%, respectively (ie pass for both low and high concentrations). Therefore, the pass rates of PⅢNP, Col Ⅳ, and CG need improvement (Table 5).
TABLE 5.
The total passing rates
| Project | Total number | Number of passing laboratories | Pass rate (%) |
|---|---|---|---|
| HA | 81 | 68 | 84.0 |
| LN | 81 | 65 | 80.2 |
| PⅢNP | 83 | 56 | 67.5 |
| Col Ⅳ | 88 | 68 | 77.3 |
| CG | 60 | 35 | 58.3 |
4. DISCUSSION
EQA (referred to as “proficiency testing”) is employed for the purpose of assessing the capacity of a laboratory to perform tests. It is an essential tool for identifying problems in a clinical laboratory and designing the appropriate solutions. EQA is an essential external monitoring technique for quality assurance, especially with the lack of a reference method and reference material. Several novel protein detection markers have recently been introduced into the clinical testing environment. This contributes to China's fast expansion of the medical diagnostic industry; however, it also highlights the significance of conducting the necessary interlaboratory quality assessment method. Therefore, the NCCL carried out an EQA survey for the quantitative identification of liver fibrosis biomarkers, which contributes to the preparation for a formal EQA program for the identification of liver fibrosis indicators.
There are three accepted methods for determining the evaluation criteria of EQA. The first method is based on clinical use‐value, which is based on the experience of clinicians (eg glycosylated hemoglobin). The second method is based on biological variation comprising a few indices that have been studied and can be found on the Westgard (https://www.westgard.com/minimum‐biodatabase1.htm) and EFLM (https://biologicalvariation.eu/) websites (eg cholesterol). The third method is based on existing detection technology. 14 , 15 , 16 , 17 , 18 The detection of liver fibrosis biomarkers uses the third method. For the data collected, the pass rates according to the target values of ±10%, 15%, 20%, 25%, and 30% were calculated. These results help to determine the appropriate criteria for existing detection capabilities.
According to the results of the EQA study conducted in China, the CLIA technique accounted for the greatest proportion of the quantitative detection of HA, LN, PⅢNP, and Col Ⅳ, representing around 90%, whereas, for the identification of CG, the CLIA, HEI, and latex‐enhanced immunoturbidimetry methods accounted for approximately one‐third each (Table 1). This suggested that despite the fact that there are multiple detection techniques, they are considerably concentrated. On analyzing the reported data, the results revealed significant differences based on the detection techniques employed, even in different orders of magnitude (Table 2). Even with the similar detection techniques but varied reagents, the detection values varied 3–100‐fold (Tables 3 and 4; reagents 1, 7, 10, 13, and 14). Although using a similar reagent and procedure, the numerical variance across various labs differed by more than twofold. These differences could be attributed to the following reasons. The reagents used were different, which adopt different methodologies (some manufacturers use the competition method, whereas others use the sandwich method). Additionally, the binding sites and affinity for quality control products along with sensitivity differ as different reagent manufacturers select different antibodies. The nature of the raw materials of quality control products (natural antigen or recombinant antigen) also contributes to the varied results. The quality control products employed in the present research were obtained from human serum, which might include a number of different antibody proteins in addition to the indicators for liver fibrosis. Hence, it is reasonable to hypothesize that the significant disparities in test findings across labs are due to changes in reagents, procedures, and characteristics, such as anti‐interference effects and specificity. In accordance with the existing regulations for medical device registration, the detection system that is formed of the reagents and their calibration products provided by the reagent manufacturers, as well as the “suitable equipment” listed on the kit's specifications, can serve as a supporting system, which may be defined as the “open” supporting system. The reasons for the substantial differences in outcomes between the “closed” and “open” matching methods are complicated. The development of various analytical systems, such as the data reading techniques, configuration of absorbance wavelengths, and built‐in calibrations, could be a possibility. Besides, plenty of invalid data were removed (such as wrong filling, missing file, and unit error) when analyzing the results, suggesting that the data reporting behavior of laboratory staff needs to be standardized. Therefore, the human factor in the data submission process cannot be ignored. According to the present EQA data, it is impossible to isolate a specific factor.
A large number of labs engaged in the quantitative identification of liver fibrosis biomarkers in China. Although the pass rate for HA and LN was relatively high in this survey, it does not suggest that the quality of this marker satisfied the clinical standards (Tables 4 and 5) in view of the fact that the evaluation criterion of target value set at ±30% is not very strict. At the same time, we found that the pass rate of the individual group (grouped according to ISO 13528) was substantially greater than the one of the other groups. This not only shows the necessity for grouping but also suggests that reagents with a small market share are at a disadvantage in grouping (Table 4). In addition to objective factors, many subjective factors also affect the results including data input error, reagent or method selection error, unit error, incorrect sample sequence use, internal quality control, and personnel operation error. All these factors restrict the accuracy of EQA results.
5. CONCLUSION
The present research is the first report to reveal the current state of the quantitative identification of liver fibrosis biomarkers in China, as far as we know. Despite the small number of labs involved in the present research, our findings indicate that there are a few drawbacks in the quantitative detection of liver fibrosis biomarkers. Commercially accessible quantitative detection kits need methodological study, and quality control procedures must be improved for the purpose of better understanding the reasons for differences in detection findings. Overall, the present research establishes the basis for the development of a formal EQA for the quantitative identification of liver fibrosis biomarkers. At the same time, it also provides guidance for the selection of instruments, methods, and reagents for laboratory liver fibrosis marker detection.
AUTHOR CONTRIBUTIONS
Chao Zhang was involved in acquisition and analysis of data. Chao Zhang and Chuanbao Zhang participated in management of data. Chao Zhang designed the study and drafted the manuscript.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
Table S1‐S2
ACKNOWLEDGMENTS
We thank Autobio for providing quality control products.
Zhang C, Zhang C. Analysis of current status of quantitative detection of biomarkers for liver fibrosis in Clinical labs in China. J Clin Lab Anal. 2022;36:e24490. doi: 10.1002/jcla.24490
Funding information
This work was supported by the National Natural Science Foundation of China (no. 82003809) and Beijing Hospital Nova Project (BJ‐2020‐087).
Contributor Information
Chao Zhang, Email: cbzhang@nccl.org.cn.
Chuanbao Zhang, Email: zc_mdy@163.com, Email: cbzhang@nccl.org.cn.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Adams LA. Biomarkers of liver fibrosis. J Gastroenterol Hepatol. 2011;26:802‐809. [DOI] [PubMed] [Google Scholar]
- 2. Caballeria L, Toran P, Caballeria J. Markers of hepatic fibrosis. Med Clin (Barc). 2018;150:310‐316. [DOI] [PubMed] [Google Scholar]
- 3. Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151‐166. [DOI] [PubMed] [Google Scholar]
- 4. Dhar D, Baglieri J, Kisseleva T, Brenner DA. Mechanisms of liver fibrosis and its role in liver cancer. Exp Biol Med (Maywood). 2020;245:96‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masuzaki R, Kanda T, Sasaki R, et al. Noninvasive assessment of liver fibrosis: current and future clinical and molecular perspectives. Int J Mol Sci. 2020;21(14):4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4‐S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tang LSY, Covert E, Wilson E, Kottilil S. Chronic hepatitis B infection: a review. JAMA. 2018;319:1802‐1813. [DOI] [PubMed] [Google Scholar]
- 8. El‐Mezayen HA, Habib S, Marzok HF, Saad MH. Diagnostic performance of collagen IV and laminin for the prediction of fibrosis and cirrhosis in chronic hepatitis C patients: a multicenter study. Eur J Gastroenterol Hepatol. 2015;27:378‐385. [DOI] [PubMed] [Google Scholar]
- 9. Enomoto H, Bando Y, Nakamura H, Nishiguchi S, Koga M. Liver fibrosis markers of nonalcoholic steatohepatitis. World J Gastroenterol. 2015;21:7427‐7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neuman MG, Cohen LB, Nanau RM. Hyaluronic acid as a non‐invasive biomarker of liver fibrosis. Clin Biochem. 2016;49:302‐315. [DOI] [PubMed] [Google Scholar]
- 11. Mak KM, Mei R. Basement membrane type IV collagen and laminin: an overview of their biology and value as fibrosis biomarkers of liver disease. Anat Rec (Hoboken). 2017;300:1371‐1390. [DOI] [PubMed] [Google Scholar]
- 12. Adams LA, Wang Z, Liddle C, et al. Bile acids associate with specific gut microbiota, low‐level alcohol consumption and liver fibrosis in patients with non‐alcoholic fatty liver disease. Liver Int. 2020;40:1356‐1365. [DOI] [PubMed] [Google Scholar]
- 13. Loomba R, Adams LA. Advances in non‐invasive assessment of hepatic fibrosis. Gut. 2020;69:1343‐1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kenny D, Fraser CG, Hyltoft Petersen P, Kallner A. Consensus agreement. Scand J Clin Lab Invest. 1999;59(7):585. [PubMed] [Google Scholar]
- 15. Kallner A, McQueen M, Heuck C. The Stockholm Consensus Conference on quality specifications in laboratory medicine, 25‐26 April 1999. Scand J Clin Lab Invest. 1999;59(7):475. [DOI] [PubMed] [Google Scholar]
- 16. Ceriotti F, Fernandez‐Calle P, Klee GG, et al. Criteria for assigning laboratory measurands to models for analytical performance specifications defined in the 1st EFLM Strategic Conference. Clin Chem Lab Med. 2017;55:189‐194. [DOI] [PubMed] [Google Scholar]
- 17. Kallner A. Quality specifications based on the uncertainty of measurement. Scand J Clin Lab Invest. 1999;59:513‐516. [DOI] [PubMed] [Google Scholar]
- 18. Sandberg S, Thue G. Quality specifications derived from objective analyses based upon clinical needs. Scand J Clin Lab Invest. 1999;59:531‐534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S2
Data Availability Statement
Not applicable.


