Significance
Mendelian randomization (MR) is a valuable tool for inferring the causal relationship between an exposure and an outcome. Great efforts have been made to relax MR assumptions to account for confounding due to pleiotropy. However, causal effects are often falsely detected between exposures and outcomes, even in the absence of genetic correlation. Here, we show that sample structure is a major confounding factor that is largely ignored by existing summary-level MR methods. To detect causal effects with well-calibrated statistical inference, we propose MR-APSS to account for pleiotropy and sample structure simultaneously by leveraging genome-wide information. Real data-analysis results suggest that MR-APSS not only avoids many false-positive findings, but also improves the statistical power of detecting causal effects.
Keywords: causal inference, Mendelian randomization, pleiotropy, sample structure, selection bias
Abstract
Mendelian randomization (MR) is a valuable tool for inferring causal relationships among a wide range of traits using summary statistics from genome-wide association studies (GWASs). Existing summary-level MR methods often rely on strong assumptions, resulting in many false-positive findings. To relax MR assumptions, ongoing research has been primarily focused on accounting for confounding due to pleiotropy. Here, we show that sample structure is another major confounding factor, including population stratification, cryptic relatedness, and sample overlap. We propose a unified MR approach, MR-APSS, which 1) accounts for pleiotropy and sample structure simultaneously by leveraging genome-wide information; and 2) allows the inclusion of more genetic variants with moderate effects as instrument variables (IVs) to improve statistical power without inflating type I errors. We first evaluated MR-APSS using comprehensive simulations and negative controls and then applied MR-APSS to study the causal relationships among a collection of diverse complex traits. The results suggest that MR-APSS can better identify plausible causal relationships with high reliability. In particular, MR-APSS can perform well for highly polygenic traits, where the IV strengths tend to be relatively weak and existing summary-level MR methods for causal inference are vulnerable to confounding effects.
Inferring the causal relationship between a risk factor (exposure) and a phenotype of interest (outcome) is essential in biomedical research and social science (1). Although randomized controlled trials (RCTs) are the gold standard for causal inference, RCTs can be very costly and sometimes even infeasible or unethical (e.g., random allocation to prenatal smoking) (2). Mendelian randomization (MR) was introduced to mimic RCTs for causal inference in observational studies (3, 4). Recently, MR analysis has drawn increasing attention (5) because it can take summary statistics from genome-wide association studies (GWASs) as input, including single-nucleotide polymorphism (SNP) effect-size estimates and their SEs, to investigate causal relationships among human complex traits.
MR is an instrumental variable (IV) method to infer the causal relationship between an exposure and an outcome, where genetic variants—e.g., SNPs—serve as IVs of the exposure (6, 7). To eliminate the influence of confounding factors, conventional MR methods rely on strong assumptions, including (A-I) IVs are associated with the exposure; (A-II) IVs are independent of confounding factors; and (A-III) IVs only affect the outcome through the exposure. However, assumptions (A-II) and (A-III) are often not satisfied in practice, due to confounding factors hidden in GWAS summary statistics, leading to false-positive findings (5, 8). To perform causal inference with genetic data, it is indispensable to distinguish two major confounding factors: pleiotropy (8) and sample structure (9, 10).
First, SNPs exhibit pervasive pleiotropic effects. Pleiotropy occurs when a genetic variant directly affects both exposure and outcome traits or indirectly through an intermediate phenotype (11). Pleiotropy can induce trait association or genetic correlation in the absence of causality (11). Due to the polygenicity of complex traits and linkage disequilibrium (LD) in the human genome, pleiotropic effects can widely spread across the whole genome (12). Therefore, a substantial proportion of SNPs can carry pleiotropic effects, and they fail to satisfy (A-II) and (A-III) on IVs in conventional MR methods.
Second, sample structure can lead to bias in SNP effect-size estimates and introduce spurious trait associations. Here, sample structure encompasses population stratification, cryptic relatedness, and sample overlap in GWASs of the exposure and outcome traits. In the presence of population stratification and cryptic relatedness, SNPs can affect the outcome through sample structure, and, thus, they violate assumptions (A-II) and (A-III) on IVs. Without correcting for sample structure, SNP effect-size estimates can be severely biased, which may lead to misinterpretation on trait association and, thus, many false-positive discoveries in causal inference. Sample overlap can also lead to spurious trait associations (13). Although principal component analysis (PCA) (14) and linear mixed models (LMMs) (15) are widely used to account for sample structure in GWASs, the results from LDSC (16) show that sample structure is often unsatisfactorily corrected in publicly available GWAS summary statistics.
To maximize the usage of publicly available GWAS summary statistics for causal inference, a number of summary-level MR methods have been developed, including Inverse Variance Weighted regression (IVW) (17), Egger (18), RAPS (19), dIVW (20), Weighted-median (21), Weighted-mode (22), Mendelian Randomization Analysis Using Mixture Models (MRMix) (23), CML-MA (24), and CAUSE (25). Despite these efforts, there are two major limitations in existing summary-level MR methods. First, most of them only use a small subset of SNPs passing the genome-wide significance (P ) for causal inference. To account for pleiotropy [including correlated pleiotropy and uncorrelated pleiotropy (25)], it is challenging to fit a flexible model with limited information from genome-wide significant SNPs. Second, existing summary-level MR methods presume that PCA- or LMM-based approaches have satisfactorily accounted for sample structure, and, thus, they largely ignore the influence of sample structure in GWAS summary statistics. Due to the complexity of human genetics, sample structure driven by socioeconomic status (26) or geographic structure (27) may not be fully corrected by routine adjustment, and it may remain as a major confounding factor hidden in GWAS summary statistics.
In this paper, we develop MR-APSS, a unified approach to MR Accounting for Pleiotropy and Sample Structure simultaneously. Specifically, we propose a foreground–background model to decompose the observed SNP effect sizes, where the background model accounts for confounding factors hidden in GWAS summary statistics, including correlated pleiotropy and sample structure, and the foreground model performs causal inference while accounting for uncorrelated pleiotropy. MR-APSS differs from existing methods in the following aspects. First, under the assumptions of LD score regression (LDSC) (16), the background model accounts for pleiotropy and sample structure using genome-wide summary statistics. In contrast, most summary-level MR methods only use SNPs passing the genome-wide significance (P ). Second, MR-APSS allows us to include more SNPs without achieving the genome-wide significance as IVs to improve statistical power. With the pre-estimated background model, MR-APSS can inform whether an SNP belongs to the background component or the foreground component. Even in the presence of many invalid IVs, the type I error will not be inflated because only the foreground signals are used for causal inference. As more SNPs are included, the increasing amount of the foreground signal can improve the statistical power.
To demonstrate the effectiveness of MR-APSS, we have performed a comprehensive simulation study and analyzed 640 pairs of exposure and outcome traits from 26 GWASs. In the simulation study, we showed that MR-APSS still had satisfactory performance when the assumptions of IVs were violated. We examined MR-APSS on a wide spectrum of complex traits using GWAS summary statistics, including psychiatric/neurological disorders, social traits, anthropometric traits, cardiovascular traits, metabolic traits, and immune-related traits. Real data results indicate that pleiotropy and sample structure are two major confounding factors. By rigorous statistical modeling of these confounding factors, MR-APSS not only avoids many false-positive findings, but also improves the statistical power of MR. When inferring causal relationships among highly polygenic traits, such as psychiatric disorders and social traits, the strengths of IVs tend to be relatively weak, and causal inference is vulnerable to confounding effects. Thus, existing MR methods will suffer from either low statistical power or inflated type I errors. The empirical results indicate that MR-APSS is particularly useful in this scenario because it accounts for confounding factors and allows for incorporating many IVs with moderate effects, demonstrating its advantage over existing MR methods.
Results
Overview of MR-APSS.
Causality, pleiotropy, and sample structure are three major sources to induce correlation between GWAS estimates of exposure-outcome traits. To distinguish causality from correlation, it is indispensable to eliminate the possibility that correlation is induced by confounding factors, such as pleiotropy and sample structure (including population stratification, cryptic relatedness, and sample overlap).
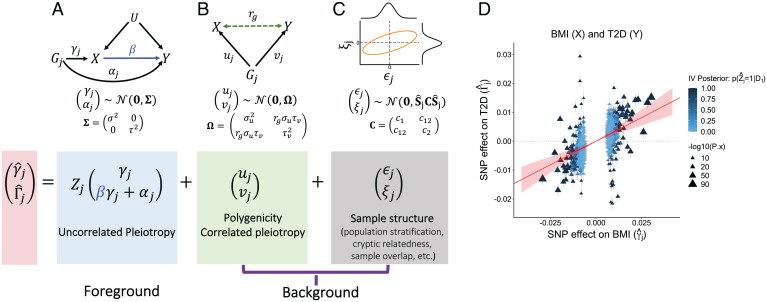
MR-APSS takes GWAS summary statistics of exposure and outcome traits as its input and performs causal inference based on a proposed foreground–background model (see an overview in Fig. 1 and details in Materials and Methods). Under the assumptions of LDSC (16) (see details in SI Appendix, section 1.1), the background model can effectively account for confounding factors by disentangling pleiotropy (Fig. 1B) and sample structure (Fig. 1C). This is because the pleiotropic effects can be tagged by LD, and the influence of sample structure is uncorrelated with LD (16). In addition to the LDSC assumptions in the background model, we have made two key assumptions for causal inference. First, we assume that the correlated pleiotropy effects can be approximately characterized by the genetic correlation, which can be estimated from genome-wide summary statistics. Second, we assume that the direct effect is independent of the instrument strength in our foreground model (known as the InSIDE condition). This is reasonable because correlated pleiotropy effects have been accounted for by using genome-wide genetic correlation. By further accounting for selection bias (28) due to selection of IVs (Materials and Methods), the foreground model can use the classical causal diagram to perform causal inference (Fig. 1A). In summary, our method requires the LDSC assumptions for the background model and the InSIDE condition for the foreground model to relax assumptions (A-II) and (A-III).
Fig. 1.
The MR-APSS approach. To infer the causal effect β between exposure X and outcome Y, MR-APSS uses a foreground–background model to characterize the estimated effects of SNPs Gj on X and Y ( and ) with SEs (), where the background model accounts for polygenicity, correlated pleiotropy (B) and sample structure (C), and the foreground model (A) aims to identify informative instruments and account for uncorrelated pleiotropy to perform causal inference. (D) We consider inferring the causal relationship between body mass index (BMI) and T2D as an illustrative example of MR-APSS. The estimated causal effect is indicated by a red line with its 95% CI indicated by the shaded area in transparent red color. Triangles indicate the observed SNP effect sizes ( and ). The color of triangles indicates the posterior of a valid IV, i.e., the posterior of an IV carrying the foreground signal (Zj = 1; dark blue) or not (Zj = 0; light blue).
Compared Methods.
Because MR-APSS uses the GWAS summary statistics as its input, we mainly compared MR-APSS with nine summary-level MR methods and grouped them (including MR-APSS) into three groups based on their assumptions, including IVW from group 1; Egger, RAPS, and dIVW from group 2; and Weighted-median, Weighted-mode, MRMix, CML-MA, CAUSE, and MR-APSS from group 3 (Table 1). We provide a review of them in SI Appendix, sections 2.1 and 2.2. We show theoretically that the IVW estimator and the dIVW estimator can be biased in the presence of pleiotropy and sample structure (SI Appendix, section 2.6). To establish a better connection with causal literature, we also provide a review of individual-level MR methods in SI Appendix, section 2.3 and Table S1. We conducted comparisons between summary-level MR methods and individual-level MR methods. Detailed results are provided in SI Appendix, sections 3.3 and 4.4 and Figs. S1, S6, and S16–S21.
Table 1.
Summary of 10 summary-level MR methods
| Method | (A-II) | (A-III) | Key assumptions | Sample structure | Selection bias |
|---|---|---|---|---|---|
| IVW (17) | ✓ | ✓ | ; All IVs are valid; NOME. | × | × |
| Egger (18) | ✓ | × | ; InSIDE (); Directional pleiotropy (); NOME. | × | × |
| RAPS (19) | ✓ | × | ; InSIDE (); Balanced pleiotropy (). | × | × |
| dIVW (20) | ✓ | × | ; InSIDE (); Balanced pleiotropy (). | × | ✓ |
| Weighted-median (21) | × | × | Majority valid; NOME. | × | × |
| Weighted-mode (22) | × | × | Plurality valid. | × | × |
| MRMix (23) | × | × | Plurality valid. | × | × |
| cML-MA (24) | × | × | Plurality valid. | × | × |
| CAUSE (25) | × | × | All IVs can be invalid; majority of IVs not be affected by correlated pleiotropy. | Sample overlap | × |
| MR-APSS | × | × | All IVs can be invalid; assumptions of LDSC (16) in the background model; InSIDE in the foreground model. | ✓ | ✓ |
Three IV assumptions: (A-I) IVs are associated with the exposure; (A-II) IVs are independent of confounders; and (A-III) IVs only affect the outcome through the exposure. NOME, the no-measurement error assumption. InSIDE, the instrument strength is independent of the direct effect. Majority valid, more than 50% of the IVs should be valid. Plurality valid, out of all groups of IVs having the same asymptotic ratio estimates of the causal effect, the largest group is the group of valid IVs.
Simulation Studies.
To evaluate MR-APSS in various scenarios and compare it with nine MR methods in Table 1, we first performed simulation studies under the MR-APSS model. After that, we investigated the robustness of MR-APSS in the presence of model misspecification.
For exposure and outcome traits, we used 47,049 SNPs on chromosomes 1 and 2 of 20,000 individuals of White British ancestry randomly drawn from the UK BioBank (UKBB). SNP effect sizes () were generated from the relationship shown in Fig. 1 and Eq. 1 in Materials and Methods. Based on real genotype data and simulated SNP effect sizes, we generated both traits and obtained summary statistics (see details in SI Appendix, section 3.1). The relationship shown in Fig. 1 is composed of the background signal and the foreground signal. For the background signal, polygenic effects (uj, vj) of all SNPs were normally distributed with variance components , such that the heritabilities of both exposure X and outcome Y were specified at 0.5. The magnitudes of the error terms were determined by the fixed sample sizes of 20,000. For the foreground signal, we randomly assigned 500 out of 47,049 SNPs as IVs. As the instrument strength (γj) and the magnitude of the direct effect (αj) are given by variance components and (Fig. 1), we specified to mimic real data scenarios. We set , so the magnitude of the direct effects in the foreground model is the same as that of the polygenic effects.
We compared MR-APSS with nine MR methods, including IVW, dIVW, RAPS, MRMix, cML-MA, Egger, CAUSE, Weighted-median, and Weighted-mode. Note that the performance of MR methods depends on the selected IVs. Using a stringent criterion, fewer SNPs will be selected, as IVs and MR methods tend to have lower power of detecting the causal effect and a lower false-positive rate. When more SNPs are included using a loose criterion, MR methods tend to have higher power, but a higher false-positive rate, because their model assumptions are more likely to be violated. To evaluate the performance of MR methods under null (β = 0), we used a stringent criterion (IV threshold ) to select IVs for IVW, dIVW, RAPS, MRMix, cML-MA, Egger, Weighted-median, and Weighted-mode. For CAUSE, we used its default threshold to include IVs. For MR-APSS, we used . For all nine MR methods, we applied LD pruning () to the selected IVs to ensure that they were nearly independent.
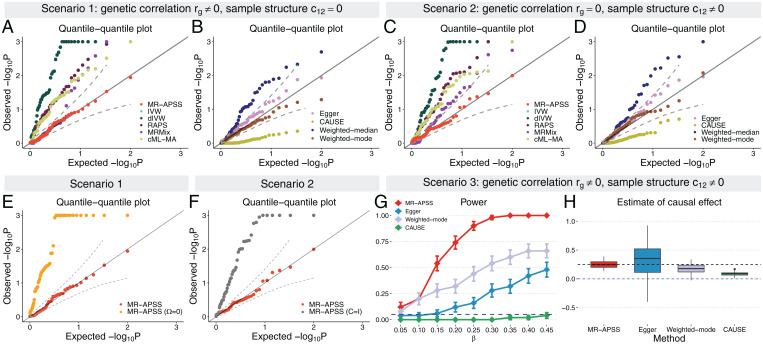
We first examined type I error control of different MR methods under null (β = 0) in the presence of genetic correlation induced by pleiotropy. We simulated data with genetic correlation, but without correlation in estimation errors. Quantile–quantile (QQ) plots of different MR methods are shown in Fig. 2 A, B, and E for genetic correlation (more results for different genetic correlations are given in SI Appendix, Fig. S2). Clearly, MR-APSS is the only method that produces well-calibrated P values. To better examine how MR-APSS accounted for polygenicity and pleiotropy, we manually set the variance component of MR-APSS to zero, i.e., . We denote this version of MR-APSS as MR-APSS (). As shown in Fig. 2E, MR-APSS produced well-calibrated P values, while MR-APSS () produced overly inflated P values. This suggests that variance component plays a critical role in accounting for polygenicity and pleiotropy. We also noticed different performance of alternative MR methods (Fig. 2 A and B). In the presence of nonzero genetic correlation, MR methods, such as IVW, dIVW, RAPS, MRMix, cML-MA, Weighted-median, and MR-APSS (), tended to produce inflated P values. Different from other MR methods, CAUSE produced very deflated P values, and, thus, CAUSE was very conservative in identifying causal effects.
Fig. 2.
Comparison of 10 summary-level MR methods on simulated data. (A–F) QQ plots of values from different methods under null simulations in the absence of causal effect (β = 0). Null simulations were performed under different scenarios. (A, B, and E) Null simulations with genetic correlation () induced by pleiotropy, but without correlation in estimation errors (). (C, D, and F) Null simulations in the presence of correlation in estimation errors () due to sample structure, but in the absence of nonzero genetic correlation (rg = 0). Based on results in A–D, MR-APSS, Egger, Weighted-mode, and CAUSE do not provide overly inflated P values. (G and H) Comparison of MR-APSS, Egger, Weighted-mode, and CAUSE under alternative simulations (). (G) The power under the settings that the causal effect size β varied from 0.05 to 0.45. (H) Estimates of causal effect under the alternative simulations (). The results were summarized from 50 replications.
Next, we examined the type I error control under null (β = 0) in the presence of correlation between estimation errors due to sample structure. Specifically, we set genetic correlation rg = 0 and simply generated correlation of estimation errors using 10,000 overlapped samples in exposure and outcome studies (more results for different c12 are given in SI Appendix, Fig. S3). We notice that correlation between estimation errors can also be induced by population stratification and cryptic relatedness. To avoid unrealistic simulation of population stratification, we investigated this issue when we performed real data analysis. The QQ plots of different MR methods are shown in Fig. 2 C, D, and F. IVW, dIVW, RAPS, MRMix, cML-MA, and Weighted-median produced overly inflated P values. These results indicate that correlation between estimator errors can be a major confounding factor, leading to false-positive findings. Again, CAUSE produced very deflated P values. To see how MR-APSS accounts for correlation between estimation errors, we set , i.e., and . In such a way, MR-APSS was forced to ignore the correlation between estimation errors. We denote this version of MR-APSS as MR-APSS (). As shown in Fig. 2F, MR-APSS () produced inflated P values. In contrast, MR-APSS produced well-calibrated P values. These results suggest that MR-APSS can satisfactorily account for correlation between estimation errors due to sample structure.
Finally, we examined the power of MR methods. As shown above, IVW, dIVW, RAPS, MRMix, cML-MA, and Weighted-median often produced overly inflated type I errors in the presence of either pleiotropy or sample structure. Hence, we only compared MR-APSS with Egger, Weighted-mode, and CAUSE. We simulated data with both genetic correlation and correlation between estimation error (). We varied the causal effect size β from 0.05 to 0.45. MR-APSS was the overall winner in terms of power (Fig. 2G). We further compared the estimation accuracy of the causal effects using MR-APSS, Egger, Weighted-mode, and CAUSE (Fig. 2H). Consistent with the literature (29), we observed that Egger had a very large estimation error. As discussed in SI Appendix, section 2.4, CAUSE often misinterprets the causal effect as correlated pleiotropy, leading to underestimation of the true causal effect. Consistently, we observed that the estimate of Weighted-mode and CAUSE was biased to the null (β = 0). In the above simulations, the foreground–background variance ratio was fixed at . We provide more results with different foreground–background variance ratios () in SI Appendix, Figs. S4 and S5.
To evaluate the robustness of MR-APSS in the presence of model misspecification, we also conducted simulations with the CAUSE model. The main patterns of the performance of the 10 MR methods largely remained the same. We provide details in SI Appendix, section 3.2 and Figs. S6–S8.
Real Data Analysis: Negative Control Outcomes.
To fairly examine the type I errors of MR methods, we use the negative control outcomes proposed by Sanderson et al. (9), where confounding factors (e.g., pleiotropy and sample structure) naturally exist. The traits that can serve as ideal negative control outcomes should satisfy two conditions. First, they should not be causally affected by any of the exposures considered. Second, the exposure and outcome traits could be affected by some unmeasured confounders, e.g., population stratification. Following the same way of Sanderson et al. (9) to choose negative control outcomes, we considered natural hair colors before graying (Hair color: black; Hair color: blonde; Hair color: light brown; and Hair color: dark brown) and skin tanning ability (Tanning) from UKBB because they are largely determined at birth, and they could be affected by sample structure.
We considered 26 exposure traits from UKBB and Genomics Consortiums (details for the GWAS sources are given in SI Appendix, Table S2). These traits can be roughly divided into five categories, including psychiatric/neurological disorders, social traits, anthropometric traits, cardiometabolic traits, and immune-related traits. The data-preprocessing steps for GWAS summary statistics are described in SI Appendix, section 4.1. The sample sizes of those GWASs range from 114,244 to 385,603, with a minimum of 15,954 for autism spectrum disorder (ASD) and a maximum of 898,130 for type 2 diabetes (T2D). Given the large sample sizes of GWASs, we used the genome-wide significance threshold as the IV threshold for IVW, dIVW, RAPS, Egger, MRMix, CML-MA, Weighted-median, and Weighted-mode in real data analysis. This stringent criterion helps to exclude invalid IVs for these methods and, thus, reduce their false-positive rates. Due to the stringent IV selection, we were not able to find enough SNPs (> 4) as IVs for four exposure traits, i.e., major depressive disorder (MDD), ASD, subject well-being, and the number of children ever born. For CAUSE (25), we used its default P value threshold to select IVs. For MR-APSS, we used as the default IV threshold.
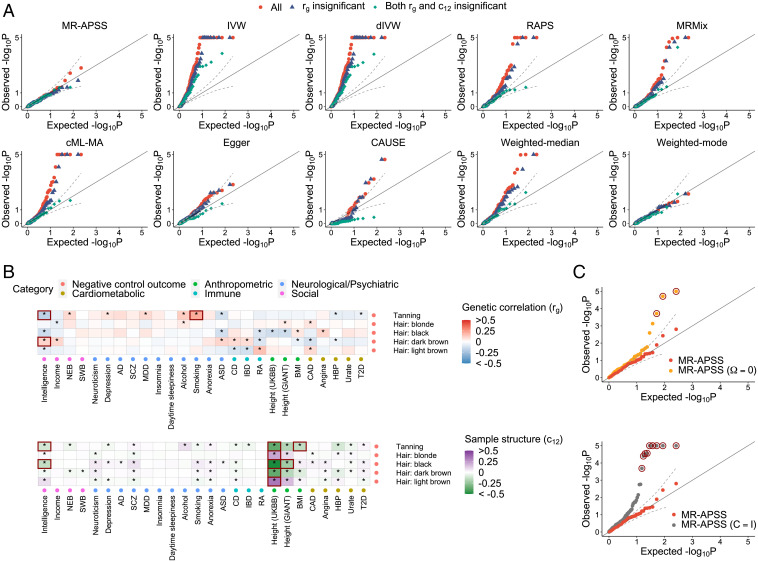
First, we applied MR-APSS and the nine summary-level MR methods to infer the causal effects between these 26 exposure traits and 5 negative control outcomes. To make the comparison fair, we focused on the results for 110 pairs, where each method had sufficient IVs for MR analysis. Ideally, these P values should be uniformly distributed between zero and one under the null (β = 0). Fig. 3A shows the QQ plots of values of the six methods (red dots). Clearly, MR-APSS and Weighted-mode produced well-calibrated P values. IVW, dIVW, RAPS, MRMix, cML-MA, and Weighted-median produced overly inflated P values, while Egger produced slightly inflated P values. CAUSE produced deflated P values in the beginning, but inflated P values later. We investigated the reasons why the five MR methods performed unsatisfactorily. As shown in Fig. 3B, we examined the estimates of two key parameters, rg and c12, of our background model, where rg is the genetic correlation capturing the overall correlated pleiotropic effects and c12 captures the correlation of estimation errors due to sample structure (e.g., population stratification, cryptic relatedness, and sample overlap). Among the 110 exposure–outcome trait pairs, 81 trait pairs had nearly zero genetic correlation, and 29 trait pairs had nonzero genetic correlation at the nominal level of 0.05 (marked by *). We also examined the correlation of estimation errors due to sample structure. Among the 110 trait pairs, 63 pairs had significant nonzero at the nominal level 0.05 (marked by *). To identify the major reason for the inflated P values produced by the nine MR methods, we restricted ourselves to the 81 trait pairs whose genetic correlation was nearly zero. For these 81 pairs, we generated the QQ plots of values of the 10 MR methods (blue triangles in Fig. 3A). Clearly, IVW, dIVW, RAPS, MRMix, cML-MA, and Weighted-median still produced overly inflated P values. Egger produced slightly better calibrated P values. CAUSE produced deflated P values in the beginning, but inflated P values later. We further restricted ourselves to trait pairs whose genetic correlation and correlation of estimation errors were both nearly zero. For these trait pairs (green diamond), MR-APSS, Weighted-mode RAPS, MRMix, cML-MA, Weighted-median, and Egger produced well-calibrated P values. IVW and dIVW still produced inflated P values. CAUSE produced very conservative P values. These results suggest that sample structure is another major confounding factor, in addition to pleiotropy.
Fig. 3.
Evaluation of the type I error control of MR methods using negative control outcomes. (A) QQ plots of values from 10 summary-level MR methods for causal inference between complex traits and negative control outcome. Red dots represent all 110 trait pairs tested by each method. Blue triangles represent the 81 trait pairs with insignificant genetic correlation at the nominal level of 0.05. Green diamonds represent the 29 trait pairs whose genetic correlations rg and c12 are both insignificant at the nominal level of 0.05. (B) Estimates of rg and c12 for trait pairs between 26 complex traits and 5 negative control outcomes. (C) QQ plots of values from MR-APSS, MR-APSS (), and MR-APSS () for trait pairs between 26 complex traits and 5 negative control outcomes. The circled P values correspond to the trait pairs marked by squares in (B), which are largely confounded by pleiotropy and sample structure.
It is worthwhile to mention that nonzero c12 can be induced by either population stratification or sample overlap. To see this, let us consider the relationship between Height (GIANT) (30) and Tanning from UKBB. Recall that parameters c1 and c2 capture the bias in estimation errors (), and parameter c12 captures their correlation (Fig. 1). By applying LDSC to estimate our background model, we obtained (SE ) for Height (GIANT) and (SE ) for Tanning, respectively. These results indicate that the publicly released GWAS summary statistics are affected by confounding factors, such as population stratification. By applying LDSC, we obtained (SE ). As we know, the samples from GIANT do not overlap with UKBB (31). Therefore, the nonzero value should be mainly attributed to population stratification. As a comparison, we also considered Height (UKBB) (32) and Tanning from UKBB. By applying LDSC, we obtained (SE 0.040) for Height (UKBB), suggesting that the released GWAS summary statistics of Height (UKBB) might potentially suffer from population stratification. By applying LDSC, we obtained (SE ) for Height (UKBB) and Tanning (UKBB). Such a nonzero value could be attributed to both population stratification and sample overlap.
To better examine the role of MR-APSS in accounting for pleiotropy or sample structure, we applied MR-APSS, but fixed and , respectively. We denote the two variations as MR-APSS () and MR-APSS (), where MR-APSS () does not account for pleiotropy, and MR-APSS () does not account for sample structure. As shown in Fig. 3 C–D, both MR-APSS () and MR-APSS () reported inflated P values. For example, based on Bonferroni correction, several trait pairs (marked with black circles in Fig. 3 C–D) were falsely detected as causal by MR-APSS () and MR-APSS (). As shown in Fig. 3B (marked by squares), their corresponding and values were significantly different from zero. By using negative control outcomes, we show that MR-APSS can produce well-calibrated P values by accounting for pleiotropy and sample structure.
Inferring Causal Relationships among Complex Traits.
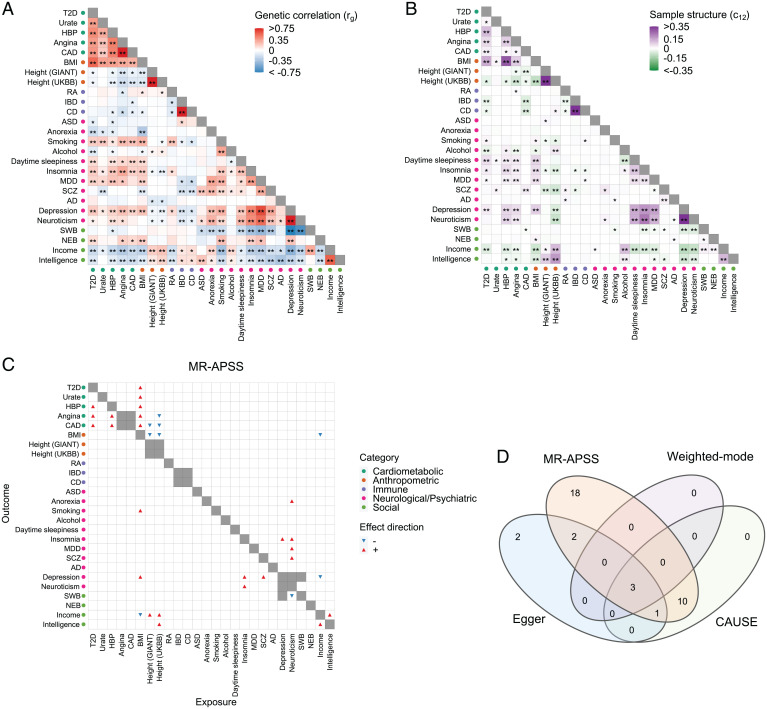
To perform causal inference, we considered 26 complex traits from 5 categories, including psychiatric/neurological disorders, social traits, anthropometric traits, cardiometabolic traits, and immune-related traits. Before applying MR methods, we examined the estimates of rg and c12 in the background model of MR-APSS for all 325 pairwise combinations of the 26 traits. We found that genetic correlation (rg) of 198 pairs significantly differed from zero at the nominal level of 0.05 (marked by in Fig. 4A). Among them, genetic correlation of 130 pairs remained significant after Bonferroni correction with (marked by in Fig. 4A). For the estimates of c12, 126 pairs had significant nonzero at the nominal level of 0.05 (marked by in Fig. 4B), and 76 pairs of them remained significantly different from zero after Bonferroni correction (marked by in Fig. 4B). Of note, 56 pairs of traits had significantly nonzero estimates of both and after Bonferroni correction. The above results suggest that both pleiotropy and sample structure are presented as major confounding factors for causal inference.
Fig. 4.
Application of MR-APSS to infer causal relationships between 26 complex traits. (A) Estimates of genetic correlation between 26 complex traits. Positive and negative estimates of genetic correlation are indicated in red and blue, respectively. Trait pairs with significant at the nominal level of 0.05 are marked by *. Trait pairs that remain to be significant after Bonferroni correction with are marked by **. (B) Estimates of c12 between 26 complex traits. Positive and negative estimates of c12 are shown in purple and green, respectively. Trait pairs with significant at the nominal level of 0.05 are marked by *. Trait pairs remain to be significant after Bonferroni correction with are marked by **. (C) Causal relationships detected by MR-APSS. The positive and negative estimates of causal effects of the exposure on the outcome are indicated by red up-pointing triangles and blue down-pointing triangles, respectively. (D) The Venn diagram shows the causal effects detected by MR-APSS, CAUSE, Egger, and Weighted-mode after Bonferroni correction.
We considered inferring the causal relationship between traits X and Y in both directions, i.e., (X as exposure and Y as outcome) and (Y as exposure and X as outcome). To avoid causal inference between two very similar phenotypes (e.g., Angina and coronary artery disease [CAD]), we excluded several trait pairs, which are marked in gray as nondiagonal cells in Fig. 4C. Therefore, 640 trait pairs remained for MR tests in total. We applied MR-APSS to these trait pairs using IV threshold and identified 34 significant causal relationships after Bonferroni correction (Fig. 4C, marked by triangles). As shown in Fig. 4A, many traits in social or neurological/psychiatric categories were observed to be genetically correlated with a wide range of complex traits from different categories. After accounting for pleiotropy and sample structure, the results from MR-APSS indicate that genetic correlation of many trait pairs should not be attributed to the causal effects. An example is Depression, which was also genetically correlated with 18 complex traits from different categories, such as body mass index (BMI) () from the Anthropometric category; and Insomnia () and schizophrenia (SCZ) () from the neurological/psychiatric category. MR-APSS only confirmed the causal effect of Depression on Insomnia (, P ). Clearly, MR-APSS can serve as an effective tool to distinguish causality from genetic correlation.
As a comparison, we also applied the nine compared methods to infer the causal relationships for the 640 trait pairs. We used as the IV selection threshold for IVW, dIVW, RAPS, Egger, MRMix, CML-MA, Weighted-median, and Weighted-mode and for CAUSE. For MR methods, including IVW, dIVW, RAPS, Egger, MRMix, CML-MA, Weighted-median, and Weighted-mode, only 541 trait pairs were tested because 99 trait pairs had less than four SNPs as IVs. For CAUSE, all 640 trait pairs were included. A summary of the causal relationships detected by the nine compared methods is given in SI Appendix, Figs. S22–S30. RAPS reported 58 trait pairs with significant causal effects after Bonferroni correction. Among them, 24 trait pairs were considered insignificant by MR-APSS after Bonferroni correction. Notably, RAPS made a similar assumption with the foreground model of MR-APSS; however, it has no background model to account for pleiotropy and sample structure. To better understand the difference between RAPS and MR-APSS, we applied MR-APSS () or MR-APSS () to those trait pairs. The testing P values of 18 trait pairs became significant based on Bonferroni correction. An example was BMI and Insomnia (SI Appendix, Table S3) with (SE 0.025) and (SE 0.010). RAPS produced with P . Without accounting for pleiotropy or sample structure, MR-APSS () and MR-APSS () reported with P and with P , respectively. After accounting for both pleiotropy and sample structure, MR-APSS estimated causal effect between BMI and Insomnia as with P . The results indicate that RAPS was likely affected by pleiotropy and sample structure.
Since IVW, dIVW, RAPS, MRMix, cML-MA, and Weighted-median tended to have higher type I errors than the nominal level, we mainly compared statistical power of MR-APSS with Egger, CAUSE, and Weighted-mode (Fig. 4D). A complete list of causal relationships among these traits detected by MR-APSS, Egger, CAUSE, and Weighted-mode are summarized in SI Appendix, Table S4. Based on Bonferroni correction, MR-APSS detected 18 significant causal effects that were not reported by CAUSE, Egger, and Weighted-mode, showing higher statistical power of MR-APSS. For example, MR-APSS detected significant causal effects of BMI on eight traits. Five of them were supported with evidence of causality from previous literature, including T2D (33), serum urate (Urate) (34), and three cardiovascular diseases (CVDs; high blood pressure [HBP], Angina, and CAD) (35). For these five supported trait pairs, Egger only detected three significant causal relationships (BMI on CAD, T2D, and HBP), and CAUSE only detected three significant causal relationships (BMI on Urate, HBP, and T2D), and, further, Weighted-mode detected two significant causal relationships (BMI on T2D and BMI on HBP). In addition to the confirmed findings, MR-APSS detected significant causal effects of BMI on Depression (, P ), ever smoked regularly (Smoking) (, P ), and Income (, P ). Those findings are consistent with results from previous MR studies (36–38), suggesting that being overweight not only increases the risk of depression and tobacco dependence, but also suffers from reduced income. Our results also revealed Neuroticism as an important health indicator, especially for human psychiatric health. Neuroticism is one of the Big Five personality traits, characterized by negative emotional states, including sadness, moodiness, and emotional instability. Higher neuroticism is associated with premature mortality and a wide range of mental illnesses or psychiatric disorders (31, 39). There is growing evidence that neuroticism plays a causal role in psychiatric disorders, such as SCZ (40) and MDD (41). Evidence from MR-APSS also supported the significant causal effect of Neuroticism on SCZ (, P ) and MDD (, P ). None of the three methods, CAUSE, Egger, and Weighted-mode, detected significant causal effects of Neuroticism on MDD or SCZ. MR-APSS also revealed that Neuroticism could be causally linked to Insomnia (, P ) and Anorexia (, P ). Weighted-mode and Egger did not report these two cases, and CAUSE only detected a significant causal effect between Neuroticism and Insomnia (, P ).
Type I Error Control and Statistical Power with Different IV Thresholds.
Existing summary-level MR methods select IVs based on a P value threshold (or an equivalent t value). In this section, we would like to highlight the advantages of our method. Regarding the type I error control, our method is insensitive to the choice of threshold. Regarding the improvement of statistical power, our method prefers a loose threshold, and we use P as the default setting in real applications. More details regarding the default IV threshold in real applications are given in SI Appendix, section 4.3.
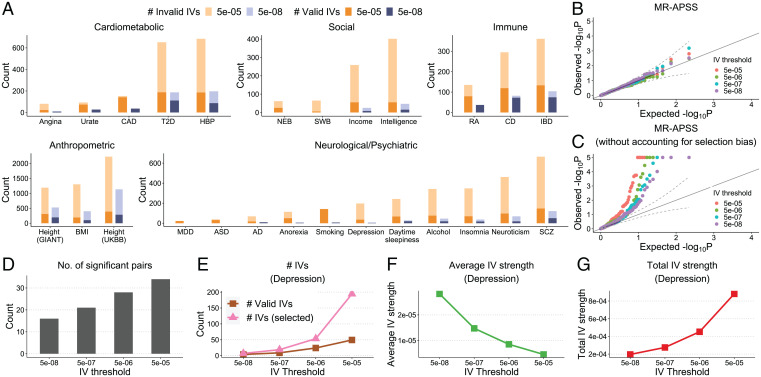
To examine the type I error control of MR-APSS when varying the IV thresholds, we varied the IV threshold from to when applying MR-APSS to infer the causal relationships between 26 complex traits and the 5 negative control outcomes. As more IVs are involved with a looser IV threshold, the number of invalid IVs increases because they are prone to the violation of MR assumptions. However, most of the IVs were detected by MR-APSS as invalid IVs (Fig. 5A). Since MR-APSS only uses the valid instrument strength in the foreground model for causal inference (Zj = 1), the type I error will not be inflated when more invalid IVs are included. As shown in Fig. 5B, the P values from MR-APSS for trait pairs between 26 complex traits and 5 negative control outcomes remain well-calibrated at different IV thresholds. These results confirm that the type I error of MR-APSS is insensitive to the IV threshold. It is important to note that correction of the selection bias is a critical step to control type I errors in MR-APSS. Without accounting for the selection bias, the magnitude of the true effect of a selected SNP is largely overestimated, and it tends to falsely contribute to the foreground signal (Zj = 1) for causal inference, thus producing false positives. To verify this, we modified MR-APSS to ignore selection bias and applied this modified version to the same trait pairs with negative control outcomes. Without accounting for the selection bias, the P values produced by the MR-APSS model given in Eq. 6 become inflated (Fig. 5C). When the threshold varies from to , the inflation of P values becomes more severe because more SNPs will falsely contribute to the foreground signal. As a comparison, we ran other summary-level MR methods to the same trait pairs. The QQ plots are shown in SI Appendix, Fig. S32. Clearly, P values produced by most summary-level MR methods (except Weighted-mode) become more inflated when the IV threshold becomes less stringent.
Fig. 5.
Evaluation of the performance of MR-APSS under different IV selection thresholds. (A) The average estimated number of valid IVs (dark color) and invalid IVs (light color) for traits from each category using IV thresholds and . (B and C) QQ plots of values from MR-APSS (B) and MR-APSS without accounting for selection bias (C) when applied to between 26 complex traits and 5 negative control outcomes. (D) The number of significant trait pairs between 26 complex traits identified by MR-APSS with different IV thresholds. (E–G) An illustrative examples of exposure: Depression. (E) The number of selected IVs Mt at threshold t and the estimated number of valid IVs. (F and G) The estimated average and total IV strengths.
As P values of MR-APSS are well-calibrated when the IV threshold varies from to , we can examine the statistical power of MR-APSS with different IV thresholds. We applied MR-APSS to infer the causal relationships among 26 complex traits by varying the IV threshold at , and . In general, we find that the average IV strength (defined in Eq. 11) decreases with the IV threshold becomes looser, and the total IV strength (defined in Eq. 12) increases as more IVs are included in the analysis. We provide two concrete examples to illustrate these points (see details in SI Appendix, section 4.2 and Fig. S14). As a result, the statistical power of MR-APSS can be improved by including SNPs with moderate effects. These results are confirmed in Fig. 5D, where the number of significant pairs identified by MR-APSS increases from 16 to 34 when the IV threshold becomes looser from to .
When investigating the causal relationship among 26 complex traits, the number of valid IVs, as well as the total IV strength, increased a lot by changing the IV threshold from to (Fig. 5A). We found that the social and neurological/psychiatric traits can benefit a lot from this property. Despite the large sample sizes for these traits, the number of IVs is too small to perform powerful MR analysis when using the IV threshold . For example, Depression only had a very small number of IVs using a stringent IV threshold . When the IV thresholds became looser, the number of selected IVs and the number of valid IVs increased a lot (Fig. 5E). Although the average IV strength decreased as the IV threshold became looser (Fig. 5F), the total IV strength increased dramatically (Fig. 5G). We also observed that, due to the limited number of IVs using a stringent IV threshold , MR-APSS could not detect a significant causal effect of Depression on Insomnia (, P ). By using a looser IV threshold, MR-APSS detected a significant causal relationship between Depression and Insomnia (, P ).
Discussion
In this paper, we have developed a summary-level MR method—namely, MR-APSS—to perform causal inference. To account for the confounding bias due to pleiotropy and sample structure, the background model of MR-APSS inherits the assumptions of LDSC. MR-APSS also assumes the InSIDE condition in the foreground model to infer the causal effect, i.e., . In other words, we assume that the association between the exposure and the outcome should be induced by their causal relationship, rather than rf, after accounting for confounding factors (e.g., correlated pleiotropy and sample structure) in the background model. Although our method relies on this assumption to infer the causal effect, we can empirically check the influence of this assumption via the following sensitivity analysis. Specifically, we can evaluate how the estimated causal effect changes when varies. In this way, users can obtain useful information about their inferred causal relationship under the perturbation of assumptions. We provide more details on sensitivity analysis in SI Appendix, section 1.5 and Fig. S13.
Besides the development of summary-level MR methods, we are aware of recent developments of individual-level MR methods, including sisVIVE (42), Two-Stage Hard Thresholding (43), GENIUS (44), GENIUS-MAWII (45), and Mendelian Randomization Mixed-Scale Treatment Effect Robust Identification (46). We believe that summary-level MR methods and individual-level MR methods are complementary to each other. On the one hand, summary-level methods relying on linear models only require marginal estimates and their SEs. Therefore, they are widely applicable to screen causal relationships between an exposure and an outcome. This is important because the access to individual-level data may be restricted due to privacy protection (47). On the other hand, individual-level methods can be more powerful than summary-level MR methods when individual-level data are accessible. First, individual-level MR methods can allow for a more flexible model to handle nonlinearity in causal inference. We are aware of several nonlinear MR methods using individual-level data (48, 49). Unlike linear MR methods, which approximate a population-averaged causal effect, the nonlinear MR methods estimate the localized average causal effects in each stratum of population using individual-level data. For example, a very recent MR study applies a nonlinear MR method to investigate whether a nonlinear model is a better fit for the relationship between diastolic blood pressure (DBP) and CVD (50). Second, individual-level MR methods can utilize more information, which is only available in individual-level GWAS datasets. For example, the individual-level methods GENIUS (44) and GENIUS-MAWII (45) require heteroscedasticity of the exposure, but this kind of information is not available in GWAS summary statistics. We find that GENIUS and GENIUS-MAWII are robust in the presence of pleiotropy and sample structure. The estimation efficiency of GENIUS and GENIUS-MAWII depends on their IV strengths, which are related to the heteroscedasticity of the exposure. In this regard, GENIUS and GENIUS-MAWII relax classical MR assumptions by requiring heteroscedasticity of the exposure, while MR-APSS relaxes classical MR assumptions by imposing the LDSC assumptions in its background model and the InSIDE condition in its foreground model. Through simulation studies and real data analyses, we find that GENIUS, GENIUS-MAWII, and MR-APSS are quite complementary to each other. We provide more detailed results in SI Appendix, sections 2.3, 3.3, and 4.4. In summary, we believe that summary-level methods and individual-level MR methods are complementary to each other, and they jointly contribute to the MR literature for causal inference. Summary-level MR methods are often preferred for large-scale screening of causal relationships, and individual-level MR methods can provide a closer examination for causal relationships of interest.
Similar to existing summary-level MR-methods, we consider linear models to perform causal inference, even for binary traits. To have better interpretation of the causal-effect estimates for binary traits, we show that the output from the observed 0–1 scale based on linear models can be transformed to the liability scale based on the probit models. We provide the details in SI Appendix, section 1.7.
Despite the improvement of MR-APSS over many existing MR methods, more research is needed for causal inference with genetic data. First, the background model is proposed to account for pleiotropy and sample structure hidden in GWASs of complex traits. The direct application of this model in some other contexts may not be suitable. For example, it is of great interest to infer the causal relationship between gene expression and complex diseases based on transcriptome-wide MR. However, it remains unclear what kind of signals should be considered as the background signals. The development of new statistical methods for transcriptome-wide MR is highly desirable. Second, multivariate MR is drawing more and more attention (51, 52). As some risk factors are known to be related to a certain type of disease, it is more interesting to ask what other risk factors can be inferred, conditioning on the known ones. We hope that MR-APSS can motivate more researchers to uncover more reliable causal relationships using rich genetic data resources.
Materials and Methods
The MR-APSS Approach.
MR-APSS takes GWAS summary statistics as input to perform causal inference, where and are the estimated j-th SNP’s effects on exposure X and outcome Y, respectively, and and are their SEs; is the selection criterion to ensure that SNP j is associated with X; and Mt is the number of SNPs selected as IVs using a threshold t of z values. To infer the causal effect β of exposure X on outcome Y, we propose to decompose the observed SNP effect sizes into background and foreground signals (Fig. 1):
| [1] |
where uj and vj are the polygenic effects of SNP j on X and Y, ϵj and ξj are the estimation errors of SNP effect sizes, γj is the remaining SNP effect on exposure X as the instrument strength, αj is the direct SNP effect on outcome Y, and Zj is a Bernoulli variable indicating whether SNP j has a foreground component (Zj = 1) or not (Zj = 0).
The Background Model of MR-APSS.
To model polygenic effects and their correlation induced by pleiotropy (Fig. 1B), we assume a variance component model
| [2] |
where (uj, vj) are random effects from a bivariate normal distribution with mean vector and covariance matrix , rg is the genetic correlation induced by pleiotropic effects between X and Y, and and are the variance of polygenic effects on X and Y, respectively. To account for bias and correlation in estimation errors due to sample structure, we consider the following model:
| [3] |
where , and the parameters c1 and c2 are used to adjust the bias in estimator errors, and c12 accounts for the correlation between the estimation errors. In the presence of population stratification and cryptic relatedness, c1 and c2 will deviate from one (typically larger than one). Moreover, either population stratification or sample overlap can induce covariance between the estimation errors, resulting in nonzero c12.
Under the assumptions of LDSC (16), we can exploit the LD structure of the human genome to account for confounding factors in the background model. Let be the LD score of SNP j, where rjk is the correlation between SNP j and SNP k. The key idea to adjust LD effects is based on the fact that the true genetic effects are tagged by LD, while the influence of sample structure is uncorrelated with LD. Then, we show that our background model (Zj = 0) can be written as (SI Appendix, section 1.1)
| [4] |
where pleiotropy and sample structure are captured by the first-order and zero-order terms of the LD score, respectively. Therefore, both and C in the background model are pre-estimated by LDSC using genome-wide summary statistics (SI Appendix, section 1.4.1). As observed in real data analysis, pleiotropy and sample structure are two major confounding factors for causal inference. We provide more discussion about the asymptotic distribution of summary statistics after principal component adjustment in SI Appendix, section 1.9.
The Foreground Model of MR-APSS.
By accounting for confounding factors using the background model, we only need three mild assumptions on instrument strength γj and direct effect αj to infer causal effect β, as shown in Fig. 1A. First, there exist some nonzero values in . Second, the strengths of instruments are independent of confounding factors. Third, the instrument strengths are independent of the direct effects (InSIDE condition), i.e., . Although our assumptions seem similar to those of existing methods, they are only imposed to the foreground signal, and, thus, they are much weaker than existing MR methods. Specifically, we assume that γj and αj are normally distributed and independent of each other:
| [5] |
The Foreground–Background Model of MR-APSS.
Now, we combine the background model and the foreground model to characterize the observed SNP effect sizes . Let be the probability that SNP j carries the foreground signal. Combining Eqs. 1, 2, 3, and 5 and integrating out γj, αj, uj, vj, ϵj, ξj, and Zj, we have the following probabilistic model:
| [6] |
where . A detailed derivation for Eq. 6 is given in SI Appendix, section 1.2. The theoretical justification of the uniformity of the approximated distribution for () in Eq. 6 for is given in SI Appendix, section 1.8.
Accounting for Selection Bias in MR-APSS.
Recall that SNPs are selected based on a P value threshold, or, equivalently, a threshold t of z score, i.e., . This selection process introduces nonignorable bias, i.e., , which has been known as the winner’s curse in GWAS (28, 53). To correct the selection bias in MR, we further take into account the selection condition . After some derivations (SI Appendix, section 1.3), model [6] becomes a mixture of truncated normal distributions:
| [7] |
where is the probability that the j-th SNP carries the foreground signal after selection.
Parameter Estimation and Statistical Inference.
In MR-APSS, the parameters of and in the background model are estimated by LDSC using genome-wide summary statistics. Given and , the log-likelihood function of the observed data can be written as:
| [8] |
To obtain the maximum likelihood estimate of model parameters , we then derive an efficient expectation-maximization algorithm (see details in SI Appendix, section 1.4.2). As a by-product, we can estimate the numbers of valid IVs and invalid IVs as and , respectively. Real data results of the estimated numbers of valid and invalid IVs are shown in Fig. 5A. The posterior of SNP j serving as a valid IV can be estimated as , as shown in dark blue in Fig. 1D. The likelihood ratio test can be conducted to examine the existence of the causal effect. Considering the following hypothesis test:
| [9] |
the likelihood-ratio test statistic is given by
| [10] |
where and are the parameter estimates obtained under hypotheses H1 and H0, respectively. Under the null hypothesis H0, the test statistic T is asymptotically distributed as , and its P value can be obtained accordingly.
IV Strength.
The performance of MR methods depend on the instrument strength. For MR-APSS, we define
| [11] |
| [12] |
which measure the average/total IV strength for those Mt SNPs with the selection criterion (). Given the observed summary statistics and the selection criterion t, we can use MR-APSS to obtain the posterior distributions of . Therefore, we can obtain the estimates of average IV strength and total IV strength defined in Eq. 11 and Eq. 12. According to the above definitions, the average and total IV strengths depend on both the IV threshold and sample size. In general, we find that the average IV strength decreases when the IV threshold becomes looser, and the total IV strength increases as more IVs are included in the analysis. Our definitions of IV strengths for the MR-APSS model are closely connected to the IV strengths defined in the MR literature (see details in SI Appendix, section 2.5).
Supplementary Material
Acknowledgments
We thank the editor and two anonymous reviewers for their very detailed and constructive comments, which have greatly helped to improve our manuscript. We also thank Prof. Lin S. Chen, Prof. Lan Wang, Prof. Baolin Wu, Prof. Zhigang Bao, and Prof. Dong Xia for their helpful comments and insightful discussions. This work is supported in part by Chinese Key-Area Research and Development Program of Guangdong Province Grant 2020B0101350001; Hong Kong Research Grant Council Grants 16307818, 16301419, 16308120, 12303618, 24301419, 14301120, and 16307221; Hong Kong Innovation and Technology Fund Grant PRP/029/19FX; Hong Kong University of Science and Technology Startup Grants R9405 and Z0428 from the Big Data Institute; Chinese University of Hong Kong Direct Grants 4053360 and 4053423; Chinese University of Hong Kong Startup Grant 4930181; the Chinese University of Hong Kong’s Project Impact Enhancement Fund and Science Faculty’s Collaborative Research Impact Matching Scheme; National Science Foundation of China Grant 12026610; Open Research Fund from Shenzhen Research Institute of Big Data Grant 2019ORF01004; and the Guangdong Provincial Key Laboratory of Big Data Computing, The Chinese University of Hong Kong, Shenzhen. The computational task for this work was performed by using the X-GPU cluster supported by the Research Grants Council Collaborative Research Fund Grant C6021-19EF.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106858119/-/DCSupplemental.
Data and Code Availability
All the GWAS summary statistics used in this paper are publicly available. The URLs for downloading the datasets are summarized in SI Appendix, Table S2. All study data are included in the article and/or supporting information. The MR-APSS software, the datasets, and sources codes for replicating the real data analysis are available at GitHub (https://github.com/YangLabHKUST/MR-APSS).
References
- 1.Rothman K. J., Greenland S., Causation and causal inference in epidemiology. Am. J. Public Health 95 (suppl. 1), S144–S150 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Bondemark L., Ruf S., Randomized controlled trial: The gold standard or an unobtainable fallacy? Eur. J. Orthod. 37, 457–461 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Smith G. D., Ebrahim S., ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Davey Smith G., Hemani G., Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23 (R1), R89–R98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pingault J. B., et al., Using genetic data to strengthen causal inference in observational research. Nat. Rev. Genet. 19, 566–580 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Burgess S., Butterworth A., Thompson S. G., Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baiocchi M., Cheng J., Small D. S., Instrumental variable methods for causal inference. Stat. Med. 33, 2297–2340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemani G., Bowden J., Davey Smith G., Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 27 (R2), R195–R208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanderson E., Richardson T. G., Hemani G., Davey Smith G., The use of negative control outcomes in Mendelian randomization to detect potential population stratification. Int. J. Epidemiol. 50, 1350–1361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang H. M., et al., Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solovieff N., Cotsapas C., Lee P. H., Purcell S. M., Smoller J. W., Pleiotropy in complex traits: Challenges and strategies. Nat. Rev. Genet. 14, 483–495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan D. M., Verbanck M., Do R., The landscape of pervasive horizontal pleiotropy in human genetic variation is driven by extreme polygenicity of human traits and diseases. SSRN [Preprint] (1 June 2018). https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3188410. [DOI] [PMC free article] [PubMed]
- 13.Burgess S., Davies N. M., Thompson S. G., Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 40, 597–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price A. L., et al., Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Loh P. R., et al., Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan B. K., et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium, LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didelez V., Sheehan N., Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 16, 309–330 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Bowden J., Davey Smith G., Burgess S., Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Q., Wang J., Hemani G., Bowden J., Small D. S., Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 48, 1742–1769 (2020). [Google Scholar]
- 20.Ye T., Shao J., Kang H., Debiased inverse-variance weighted estimator in two-sample summary-data Mendelian randomization. Ann. Stat. 49, 2079–2100 (2021). [Google Scholar]
- 21.Bowden J., Davey Smith G., Haycock P. C., Burgess S., Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwig F. P., Davey Smith G., Bowden J., Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi G., Chatterjee N., Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nat Commun 10, 1941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue H., Shen X., Pan W., Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am. J. Hum. Genet. 108, 1251–1269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison J., Knoblauch N., Marcus J. H., Stephens M., He X., Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat. Genet. 52, 740–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdellaoui A., et al., Genetic correlates of social stratification in Great Britain. Nat. Hum. Behav. 3, 1332–1342 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Haworth S., et al., Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat. Commun. 10, 333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zollner S., Pritchard J. K., Overcoming the winner’s curse: Estimating penetrance parameters from case-control data. Am. J. Hum. Genet. 80, 605–615 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S., Thompson S. G., Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood A. R., et al.; Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study, Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yengo L., et al.; GIANT Consortium, Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe K., et al., A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Ganz M. L., et al., The association of body mass index with the risk of type 2 diabetes: A case-control study nested in an electronic health records system in the United States. Diabetol. Metab. Syndr. 6, 50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka K., et al.; Osaka Twin Research Group, The relationship between body mass index and uric acid: A study on Japanese adult twins. Environ. Health Prev. Med. 20, 347–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan S. S., et al., Association of body mass index with lifetime risk of cardiovascular disease and compression of morbidity. JAMA Cardiol. 3, 280–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor A. E., et al., The effect of body mass index on smoking behaviour and nicotine metabolism: A Mendelian randomization study. Hum. Mol. Genet. 28, 1322–1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyrrell J., et al., Using genetics to understand the causal influence of higher BMI on depression. Int. J. Epidemiol. 48, 834–848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyrrell J., et al., Height, body mass index, and socioeconomic status: Mendelian randomisation study in UK Biobank. BMJ 352, i582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahey B. B., Public health significance of neuroticism. Am. Psychol. 64, 241–256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Os J., Jones P. B., Neuroticism as a risk factor for schizophrenia. Psychol. Med. 31, 1129–1134 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Farmer A., et al., Neuroticism, extraversion, life events and depression. The Cardiff Depression Study. Br. J. Psychiatry 181, 118–122 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Kang H., Zhang A., Cai T. T., Small D. S., Instrumental variables estimation with some invalid instruments and its application to Mendelian randomization. J. Am. Stat. Assoc. 111, 132–144 (2016). [Google Scholar]
- 43.Guo Z., Kang H., Tony Cai T., Small D. S., Confidence intervals for causal effects with invalid instruments by using two-stage hard thresholding with voting. J. Royal Stat. Soc. Ser. B. Stat. Methodol. 80, 793–815 (2018). [Google Scholar]
- 44.Tchetgen E. T., Sun B., Walter S., The GENIUS approach to robust Mendelian randomization inference. Stat. Sci. 36, 443–464 (2021). [Google Scholar]
- 45.Ye T., Liu Z., Sun B., Tchetgen E. T., GENIUS-MAWII: For robust Mendelian randomization with many weak invalid instruments. arXiv [Preprint] (2021). https://arxiv.org/abs/2107.06238. Accessed 13 July 2021.
- 46.Liu Z., Ye T., Sun B., Schooling M., Tchetgen E. T., On Mendelian Randomization Mixed-Scale Treatment Effect Robust Identification (MR MiSTERI) and estimation for causal inference. arXiv [Preprint] (2020). https://arxiv.org/abs/2009.14484. Accessed 30 September 2021. [DOI] [PubMed]
- 47.Zhu X., Stephens M., Bayesian large-scale multiple regression with summary statistics from genome-wide association studies. Ann. Appl. Stat. 11, 1561–1592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess S., Davies N. M., Thompson S. G.; EPIC-InterAct Consortium, Instrumental variable analysis with a nonlinear exposure-outcome relationship. Epidemiology 25, 877–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staley J. R., Burgess S., Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet. Epidemiol. 41, 341–352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arvanitis M., et al., Linear and nonlinear mendelian randomization analyses of the association between diastolic blood pressure and cardiovascular events: The J-curve revisited. Circulation 143, 895–906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanderson E., Davey Smith G., Windmeijer F., Bowden J., An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int. J. Epidemiol. 48, 713–727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuber V., Colijn J. M., Klaver C., Burgess S., Selecting likely causal risk factors from high-throughput experiments using multivariable Mendelian randomization. Nat. Commun. 11, 29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferguson J. P., Cho J. H., Yang C., Zhao H., Empirical Bayes correction for the Winner’s Curse in genetic association studies. Genet. Epidemiol. 37, 60–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the GWAS summary statistics used in this paper are publicly available. The URLs for downloading the datasets are summarized in SI Appendix, Table S2. All study data are included in the article and/or supporting information. The MR-APSS software, the datasets, and sources codes for replicating the real data analysis are available at GitHub (https://github.com/YangLabHKUST/MR-APSS).