Significance
An influential proposal for the evolution of intelligence in behaviorally sophisticated vertebrates is that increased diversity of synaptic proteins enabled enhanced neural plasticity. We recently completed an improved transcriptome for the nervous system of a simple neurobiological model, the marine mollusk Aplysia. A comparison of proteins that form synaptic scaffolds in Aplysia, in Octopus (a mollusk with an elaborate brain and complex behaviors), and in vertebrates revealed that several families of synaptic scaffold proteins in mollusks are absent in the vertebrate lineage. Despite dramatic differences in cognitive capacity, Octopus and Aplysia have very similar synaptic proteins. This suggests that another factor, such as increases in neural circuit complexity, may be the major contributor to the evolution of intelligence.
Keywords: neural plasticity, synaptic plasticity, evolution, neuromodulation, Aplysia
Abstract
The gastropod mollusk Aplysia is an important model for cellular and molecular neurobiological studies, particularly for investigations of molecular mechanisms of learning and memory. We developed an optimized assembly pipeline to generate an improved Aplysia nervous system transcriptome. This improved transcriptome enabled us to explore the evolution of cognitive capacity at the molecular level. Were there evolutionary expansions of neuronal genes between this relatively simple gastropod Aplysia (20,000 neurons) and Octopus (500 million neurons), the invertebrate with the most elaborate neuronal circuitry and greatest behavioral complexity? Are the tremendous advances in cognitive power in vertebrates explained by expansion of the synaptic proteome that resulted from multiple rounds of whole genome duplication in this clade? Overall, the complement of genes linked to neuronal function is similar between Octopus and Aplysia. As expected, a number of synaptic scaffold proteins have more isoforms in humans than in Aplysia or Octopus. However, several scaffold families present in mollusks and other protostomes are absent in vertebrates, including the Fifes, Lev10s, SOLs, and a NETO family. Thus, whereas vertebrates have more scaffold isoforms from select families, invertebrates have additional scaffold protein families not found in vertebrates. This analysis provides insights into the evolution of the synaptic proteome. Both synaptic proteins and synaptic plasticity evolved gradually, yet the last deuterostome-protostome common ancestor already possessed an elaborate suite of genes associated with synaptic function, and critical for synaptic plasticity.
During the course of animal evolution, the appearance of the nervous system was the transformative process that enabled the emergence of complex behaviors, including social interactions, behavioral plasticity, and learning. Indeed, the evolution of the nervous system beginning ∼600 million years ago was a key innovation driving the radiation of highly diverse animal species. A fundamental challenge in neuroscience and evolutionary biology is to identify the characteristics that account for the vast disparity in cognitive sophistication across animal groups, such as nematodes, frogs, and humans. What evolutionary innovations explain the remarkable cognitive sophistication of species such as octopus, corvids, and primates? Most explorations of this issue focus primarily on comparisons among mammals and on human exceptionalism (1, 2) and thus are evolutionarily narrow in scope. An alternative approach that compares nervous systems at the molecular level, applied by Emes, Grant, and colleagues, enables much broader evolutionary comparisons (3, 4). Their molecular analysis has the attractive feature of not being limited to anatomical comparisons of specific regions of the vertebrate brain, which exclude data on diverse invertebrate nervous systems. According to their hypothesis, the evolution of higher cognition in mammals required increased molecular diversity at the synapse (3, 4), resulting, in part, from the two rounds of whole-genome duplication in vertebrates. These whole-genome duplications produced multiple copies of every gene, including neuronal genes, which subsequently underwent either secondary loss or subfunctionalization. This “Synaptic Molecular Complexity Hypothesis” proposes that the nervous systems of cognitively sophisticated vertebrates benefited from the substantial expansions of families of synaptic proteins, including scaffold proteins (3, 4). Synaptic scaffold proteins mediate the tight colocalization of presynaptic Ca2+ channels, exocytosis proteins, and synaptic vesicles, the clustering of postsynaptic receptors, and the precise, coordinate organization of presynaptic exocytosis sites and postsynaptic receptors (5, 6). An alternative to this hypothesis is that increased cognitive capacity results from more elaborate neuronal circuitry in the central nervous system (CNS), which may be mediated by expansion of molecules that mediate axon guidance and wiring specificity.
To rigorously explore these evolutionary hypotheses, it is important to examine non-mammalian models of nervous system function at the molecular level. Adaptability of the nervous system, known as neural plasticity, is a fundamental characteristic of bilaterians that has dramatically contributed to their success (7–9). Central to the Synaptic Molecular Complexity Hypothesis is the assumption that the evolutionary expansion of scaffold proteins enhances the variety of synaptic plasticity possible, which, in turn, is required to support greater cognitive capacity (4). These multidomain scaffold proteins provide molecular substrates for synaptic modifications during neural plasticity and learning.
Among invertebrate species, one model system where neural plasticity has been extensively studied is the marine mollusk Aplysia californica (10–12). Through extensive research on signaling pathways, it is known that Aplysia neurons employ many of the same molecular cascades responsible for synaptic plasticity as do mammalian neurons (10–12). We have recently completed a high-coverage transcriptome assembly for the Aplysia CNS. This work follows an early Aplysia expressed sequence tag (EST) assembly (13) and a subsequent CNS transcriptome assembly generated in 2012 (SI Appendix, Table S1), and also a study of transcripts localized to Aplysia neuronal processes (14). The current improved transcriptome assembly is particularly advantageous for the accurate identification of the larger scaffold proteins, which are challenging to successfully predict from genomic sequences that are frequently fragmented in assemblies, due to long introns. Using this new transcriptome assembly, we have now asked two comparative questions: Are scaffold proteins at molluscan synapses less diverse than at mammalian synapses? Do simpler mollusks, such as Aplysia, have fewer sets of synaptic scaffold proteins than behaviorally sophisticated cephalopods, such as Octopus? Although it displays a relatively narrow range of behaviors, Aplysia is capable of robust behavioral plasticity, including both nonassociative and associative learning, which lasts days or even a few weeks (10–12). Octopus, in contrast, exhibits remarkable behavioral flexibility, including problem solving (15, 16). If increased cognitive capacity requires substantial expansions of families of synaptic proteins, we should observe such a difference between Aplysia and Octopus.
Our analysis of synaptic scaffold proteins has led to several interesting insights. Only in one specific instance did we identify a greater diversity of scaffold-protein family members in Octopus than in Aplysia: in a subfamily of transmembrane AMPA receptor-regulatory proteins (TARPs). In general, due to multiple rounds of genome duplication in vertebrates, there are more closely related scaffold proteins in vertebrate genomes. In contrast, a number of bilaterian families of scaffold proteins have either been lost in the chordate lineage or diversified in protostomes. Overall, our results challenge the hypothesis that cognitive capacity is determined by the molecular complexity of synapses. We argue that the increased synaptic protein complexity reported in mammals is, in part, due to a vertebrate-centric bias: The counting of proteins diversified in vertebrates; overlooking synaptic proteins present in protostomes that were lost in vertebrates; and ignoring protein-family expansions that occurred specifically in invertebrates.
We also found evidence that the evolution of synaptic transmission was a gradual rather than a unitary event. Protosynaptic proteins appeared progressively in early holozoans that lack neurons and synapses, such as single-celled choanoflagellates, sponges, and Trichoplax; these proteins were subsequently incorporated into synapses when neurons evolved. Moreover, neuromodulation and synaptic plasticity apparently evolved concurrently with synaptic transmission. Elaboration of both sets of processes had progressed dramatically by the last common bilaterian ancestor.
Results
An Improved Transcriptome Enables Examination of Synaptic Molecular Complexity.
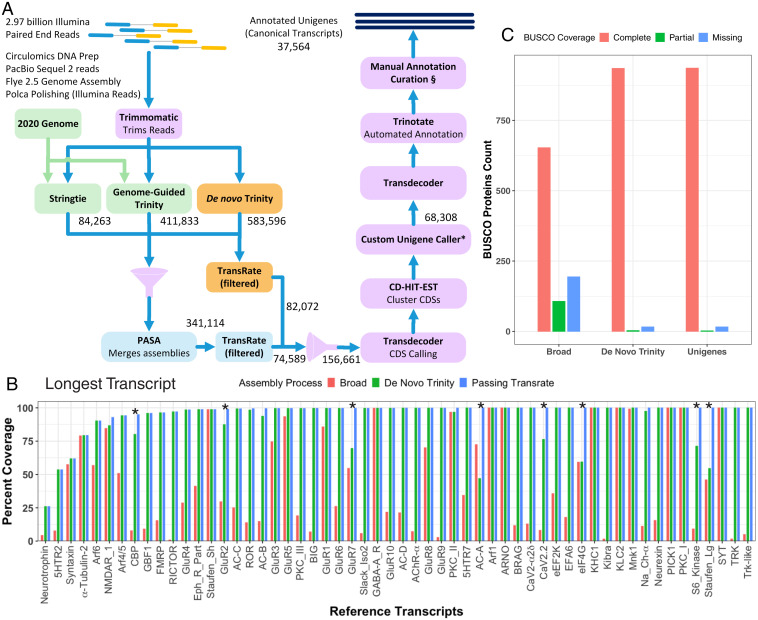
A major problem with generating complete transcriptome and genome assemblies of Aplysia has been the extensive fragmentation of these sequences, presumably due to an abundance of small repeats. One advantage of Aplysia is that due to extensive analysis of signaling pathways and neuronal genes, a number of messenger RNAs (mRNAs) have been sequenced and verified, many of which contain difficult-to-sequence and assemble GC-rich regions and repeats. We used a collection of previously cloned Reference Transcripts to assess the improvement of the transcriptome assembly with modified shearing times, increased read depth, and optimization of the assembly pipeline. Examples of fragmentation can be observed for specific Aplysia transcripts for both an earlier genome (SI Appendix, Fig. S1) and an earlier Trinity transcriptome assembly (SI Appendix, Fig. S2 and Table S1). We compared assemblies from libraries generated with a range of shearing times (from 1 to 6 min) and found that a reduction from 6 min to 200 s improved the assembled transcripts (SI Appendix, Fig. S3A). Deeper read coverage also improved the transcriptome assembly (SI Appendix, Fig. S3B). Nevertheless, even with reduced shearing time and more extensive reads, some fragmentation of transcripts persisted, as is quantitatively evident from the much greater reference transcript coverage by the total combined contigs versus the single longest contigs corresponding to individual Reference Transcripts (SI Appendix, Fig. S3 A and B).
An important step that increased the completeness of assemblies was to combine sets of contigs from genome-guided assemblers, frequently generating longer transcripts, with independent assemblies of transcripts generated by the de novo Trinity (DN-Trinity) assembler, which avoided frameshifts that were introduced by the genome-guided assembler StringTie (Fig. 1A, SI Appendix, Tables S2 and S3, Methods, and SI Appendix, Methods). Fig. 1B shows the increase in reference transcript coverage with this pipeline (see transcripts with asterisks in Fig. 1B; see SI Appendix, Fig. S4 for examples of improved Reference Protein coverage); this is a more sensitive measure of transcriptome completeness than BUSCO proteins (Fig. 1C). Several Reference Transcripts with incomplete coverage still remain in the final assembly; some of them (e.g., syntaxin, 5-HT2 receptor, eIF4G) are difficult to assemble due to highly repetitive sequences either in the coding region or the 3′ untranslated region. A few transcripts have low expression levels in the nervous system (e.g., neurotrophin) (Fig. 1B) but exhibit complete assembly in some peripheral tissues. It should be noted that Aplysia transcripts available on National Center for Biotechnology Information (NCBI) often show improvement compared with the two Broad assemblies on which they are based, because these are generated by a hybrid pipeline that incorporates large additional read sets and ESTs; however, we have observed that this NCBI hybrid pipeline sometimes introduces errors. We should emphasize that one cannot assess assemblies using nucleotide BLAST on NCBI, as the NCBI hybrid assembly for Aplysia includes individual sequences posted by Aplysia researchers; therefore, nearly all of our Reference Transcript sequences are correct on NCBI.
Fig. 1.
Assembly of transcriptome. (A) Assembly and annotation pipeline. Illumina paired-end reads (n = 2.97 billion) were trimmed with Trimmomatic and processed through three assemblers: DN-Trinity, which was used in all pilot assemblies; genome-guided de novo Trinity (GG-Trinity); and StringTie. Three assemblies were combined using PASA, and both the PASA contigs (n = 341,114) and the DN-Trinity contigs (n = 583,596) were evaluated with TransRate and filtered using custom combined TransRate contig scores (see SI Appendix, Methods). The combined 159,559 contigs were clustered based on ORFs with CD-HIT, and the longest ORF per cluster was identified as the unigene, yielding 71,104, which reduced to 39,486 unigenes with a minimum ORF length of 400 bp. Annotation was performed by Trinotate, supplemented with manual annotation. (*Custom Unigene Caller and §Manual annotation curation are described in SI Appendix, Methods) (B) A set of 57 Reference Transcripts was used to assess each set of contigs to obtain a more complete transcriptome assembly. This plot shows the percent coverage of Reference Transcripts by longest contig per transcript from the Broad 2013 Trinity transcriptome assembly, the final DN-Trinity assembly, and the combined set of contigs from PASA and DN-Trinity, after filtering through TransRate. 5HTR, serotonin receptor; AC, adenylyl cyclase; CBP, CREB-binding protein; CaV2.2, voltage-gated calcium channel type 2; eEF2K, eukaryotic elongation factor 2 kinase; eIF4G, eukaryotic initiation factor 4G; Eph_R_Part, ephrin receptor partial; FMRP, fragile X mental retardation protein; KHC, kinesin heavy chain; KIBRA, kidney and brain adapter protein; NaCh, sodium channel; RICTOR, rapamycin-insensitive companion of mammalian target of rapamycin; PKC, protein kinase C; TRK, tropomycin receptor kinase. Asterisks indicate substantial improvement of Reference Transcript sequences with the inclusion of contigs from genome-guided assemblers, relative to DN-Trinity contigs. (C) BUSCO scores for the 978 metazoan conserved proteins for predicted protein sequences from the 2013 Broad assembly, the DN-Trinity assembly, and the final combined set of unigenes.
Postsynaptic Scaffold Proteins.
With the benefit of this improved transcriptome assembly, we analyzed the synaptic scaffold proteins expressed in Aplysia CNS and compared the diversity of synaptic scaffold proteins in Aplysia with those in Octopus and in vertebrates. Both the presynaptic and postsynaptic scaffolds are important for synaptic complexity, neuromodulation, and activity-dependent plasticity. On the postsynaptic side, we have focused on both the transmembrane accessory proteins important for linking ligand-gated ion channels to intracellular scaffolds and the protein constituents of these scaffolds (SI Appendix, Fig. S5). In order to understand the expansion of scaffold-protein families, we examined orthologs in non-bilaterian phyla, as well as in other mollusks, arthropods, nematodes, and invertebrate deuterostomes.
TARPs.
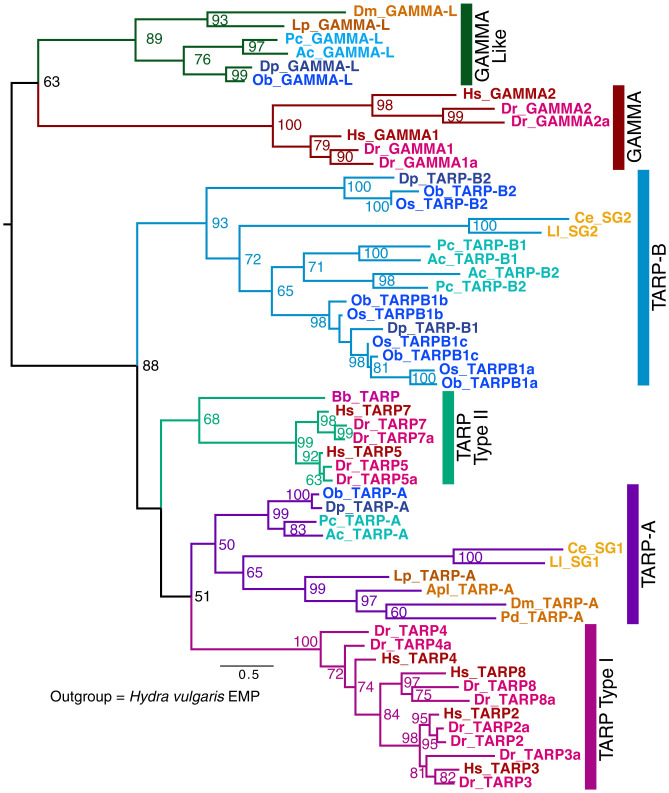
One of the most important families of scaffold proteins for vertebrate AMPA receptors is the TARPs, which are strongly implicated in the plasticity of synapses (17). Initially discovered through the mouse mutant stargazer, there are eight TARP proteins in vertebrates. Two of these (TARPs 1 and 6) function as the gamma subunits of calcium channels (indeed, in annotations, TARPs are often called gamma subunits). In our analysis, there are six distinguishable families (Fig. 2). These include three families in deuterostomes (18): type I TARPs (TARPs 2, 3, 4, and 8), type II TARPs (TARP 5 and 7), and gamma subunits (TARPs 1 and 6). In protostomes, there are also three TARP families: TARP-A, TARP-B, and gamma-like proteins. In dendrograms, protostome TARP-As and TARP-Bs and vertebrate type I TARPs and type II TARPs cluster together as a well-supported TARP family. Although the protostome and deuterostome gamma subunits sometimes cluster together, we are agnostic as to 1) whether these protostome proteins actually act as Ca2+ channel subunits and 2) whether the protostome gamma proteins and the vertebrate skeletal-muscle channel gamma subunits had a common ancestor. More complex trees that include other related proteins, such as the vertebrate claudins and the distinct invertebrate claudins (which are important for tight junctions and septate junctions, respectively), confirm the clustering of protostome and deuterostome TARPs (SI Appendix, Fig. S6).
Fig. 2.
Evolution of TARPs and related proteins. A dendrogram from a RAxML analysis (see Methods and SI Appendix, Table S2) with bootstrap values shown for values ≥50%. Hydra vulgaris EMP is the outgroup (not shown). Species names are shortened to two letters: Ac, Aplysia californica; Apl, Agrilus planipennis; Bb, Branchiostoma belcheri; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dp, Doryteuthis pealeii; Dr, Danio rerio; Hs, Homo sapiens; Lp, Limulus polyphemus; Ll, Loa loa; Ob, Octopus bimaculoides; Os, Octopus sinensis; Pc, Pomacea canaliculata; Pd, Polistes dominula. All species abbreviations from dendrograms are also defined in Dataset S1, which lists NCBI identifiers for dendrogram sequences. Vertebrate genes are colored in red shades, ecdysozoan genes are in orange shades, and mollusks are in blue shades. TARP-A and TARP-B are newly described clades of protostome TARPs identified in this dendrogram. We retained the original terms for the C. elegans TARPs (i.e., Stargazin, STG1, and STG2) to reflect that the function of these proteins was previously described. When a species has multiple isoforms of a TARP-B, they are numbered TARP-B1 and TARP-B2, and when one of these proteins has been expanded, it has been indicated as TARP-B1a and TARP-B1b. Vertebrate TARPs 1 and 6 are renamed as GAMMA1 and GAMMA2, respectively, to reflect the fact that they are not part of the larger TARP family. The closest protostome family to these gammas is called Gamma-like (GAMMA-L).
Protostome TARP-A members, including STG-1 in nematodes, often, but not always, cluster with type I TARPs (Fig. 2). These families share a common PDZ binding motif (TTPV) at their C terminus, known to interact with the PDZ domains of discs large (DLG) proteins, thereby contributing to anchoring of AMPA receptors to the post-synaptic density (PSD) (19). Protostome TARP-A members from Apis, Drosophila, and Caenorhabditis elegans regulate AMPA receptors (20, 21). Interestingly, some insect TARPs in the type-A group have isoforms with and without the C terminus TTPV motif due to alternative splicing, or they entirely lack the TTPV sequence, particularly in dipterans such as Drosophila. Both the protostome TARP-Bs and the deuterostome type II TARPs (e.g., TARPs 5 and 7) lack the TTPV motif and cluster separately from type I TARPs and TARP-As. These TARPs are also implicated in AMPA receptor regulation: TARP 5 and TARP 7 regulate AMPA receptors in mammals (22) and STG-2 regulates AMPA receptors in C. elegans (21). It is likely that these TARP groups diverged in the bilaterian ancestor, although we cannot rule out independent formations of type 1 and type 2 TARPs in deuterostomes and TARP-A and TARP-B in protostomes from a single founding TARP member in the bilaterian ancestor. There are no pre-bilaterian members of the TARP family, despite the presence of AMPA receptors and, presumably, glutamate neurotransmission in cnidarians. The four type I TARPs and the two type II TARPs found in mammals reflect the vertebrate genome duplication events; twice as many TARPs are present in Danio (eight type I and four type II TARPs, respectively), the result of an additional whole-genome duplication event. Many protostomes (e.g., nematodes, brachiopods) have single members of both the TARP-A and TARP-B families. In contrast, arthropods have a single TARP-A and apparently have lost TARP-B. Notably, several molluscan lineages have undergone independent expansions of the TARP-B family. In particular, octopuses have one TARP-A and four TARP-B members. Three of these TARP-B members appeared after the divergence of Octopus from squid. Aplysia and other gastropods have two TARP-B members from an independent duplication, as well as a TARP-A (i.e., three TARPs in Aplysia). It will be interesting to determine if these expansions are linked to the independent expansion of AMPA-like receptors in these species (23–25).
Vertebrate-Specific AMPA Receptor–Associated Proteins.
There are several other protein families with evidence supporting their role as AMPA receptor regulators, including GSG1, Dispanins, and Shisas (26). In these cases, it is not clear if there are orthologs in the Aplysia transcriptome or in other protostomes. We identified 21 transcripts in the Aplysia transcriptome that contain a domain characteristic of GSG1 (Pfam domain PF0082), but many of these do not demonstrate overt orthology with deuterostome sequences. Dispanins, such as SYNDIG1, have a conserved domain defined by PF04505 with 14 members in the Aplysia transcriptome, many with no clear deuterostome ortholog. From a chordate perspective, counting the recently evolved AMPA receptor-regulatory proteins leads to many more AMPA-receptor regulatory proteins in chordates (26), but unless similar efforts are made to uncover functionally related proteins in a protostome model, it will be impossible to know how many recently evolved protostome-specific AMPA receptor-regulatory proteins exist. Given the recent expansion of AMPA receptors in mollusks (23–25), it seems likely that additional AMPA receptor-regulatory proteins could be found.
NETOs.
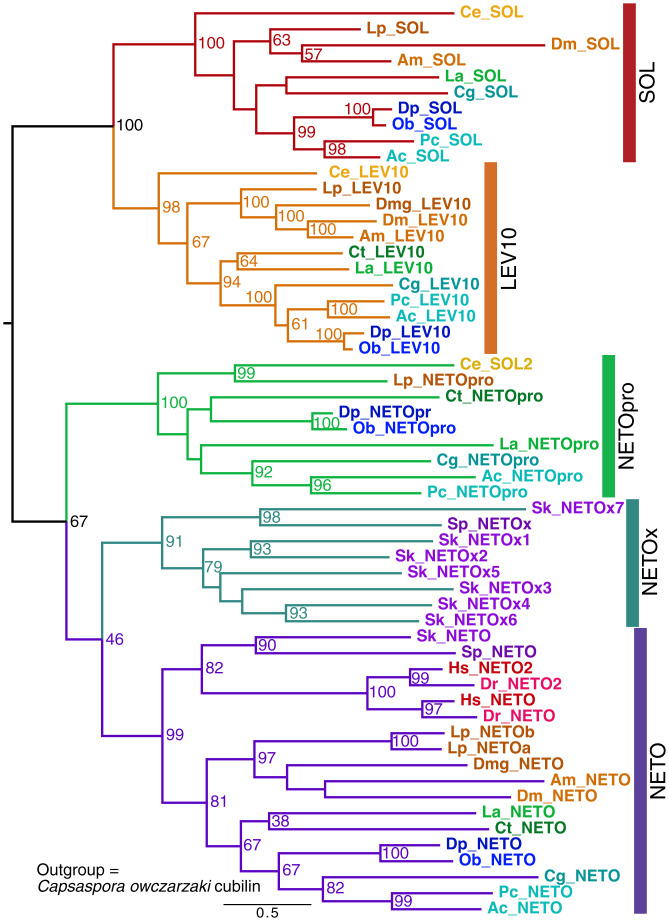
The NETO proteins have been shown to scaffold kainate receptors, but not AMPA receptors, in vertebrates. However, in C. elegans, SOL-1, a protein with cubilin domains related to NETO (SI Appendix, Fig. S5), does regulate AMPA receptors (27). We examined the expanded family of cubilin-containing proteins implicated in functions of ligand-gated neurotransmitter receptors (Fig. 3). We identified four Aplysia transcripts: one ortholog of vertebrate NETO; one protostome-specific NETO related to the C. elegans SOL-2, which is known to regulate AMPA receptors (26, 28); and representatives of two additional protostome-specific families, the SOL-1 family and the related LEV10 family [which scaffolds ligand-gated acetylcholine (ACh) receptors in C. elegans (29)]. Strikingly, chordates have lost three of these protostome families, retaining a single NETO family. In the hemichordate Saccoglossus, there was a large expansion of a NETO-related family, but only a single NETO is found in early branching chordates, with two members present after the genome duplication in vertebrates.
Fig. 3.
Evolution of NETO and related proteins. A dendrogram from a RAxML analysis with bootstrap values ≥50% shown. Species abbreviations for species not defined in the Fig. 1 legend: Am, Apis melifera; Cg, Crassostrea (Magallana) gigas; Ct, Capitella teleta; La, Lingula anatina; Sk, Saccoglossus kowalevskii; Sp, Strongylocentrotus purpuratus. The outgroup was Capsaspora owczarzaki cubilin (not shown). Vertebrate genes are colored in red shades, ambulacrarian genes are in purple shades, ecdysozoan genes are in orange shades, molluscan genes are in blue shades, and nonmolluscan spiralian genes are in green shades. We identified a new protostome-only branch of the family, NETO-Pro. LEV-10 (identified in a screen for resistance to the cholinergic agonist levamisole) and suppressor of lurcher (SOL) were named after the founding C. elegans proteins. SOL2 in C. elegans is a member of the NETOpro family. Note that the NETOx group of genes is specific for early-branching deuterostomes.
Additional Postsynaptic Scaffold Proteins.
We analyzed several additional families of postsynaptic scaffold proteins, described in the SI Appendix: the AMPA receptor–associated Cornichons, as well as the intracellular scaffolds, DLG, Homer, Shank, and GRIP (SI Appendix, Figs. S7–S11). For these five protein families, comparing numbers of genes across species, we observed a similar pattern in which Octopus and Aplysia have the same number of genes, humans have more gene isoforms than these mollusks, and Danio typically has more gene isoforms than humans, with the exception of Cornichons (SI Appendix, Table S4).
Presynaptic Scaffold Proteins.
The presynaptic active zone contains a dense cytoskeletal matrix formed by a number of multidomain scaffold proteins. We have analyzed several of these in the Aplysia CNS transcriptome, including the RIM superfamily, RIM binding protein (RIM-BP), CAST/ELKS, CASK, and unc13 (SI Appendix, Fig. S12)
RIM/Piccolo/Fife/Chalumeau Superfamily.
The scaffold proteins in the greater RIM family contain a FYVE zinc finger domain at the N terminus, followed by a Rab3A-binding α helix, a PDZ domain, and two C2 domains (SI Appendix, Fig. S12A). RIM interactions with unc13 regulate vesicle priming and/or release probability, and this effect is conserved from C. elegans to mouse (6, 30–32). The C terminus of the CaV2 Ca2+ channel binds to the PDZ domain of RIM, which contributes to localizing Ca2+ channels at active zones and positioning of synaptic vesicles proximal to sites of Ca2+ influx (33); RIM also participates in vesicle priming (6). In vertebrates, the RIM superfamily includes RIM1 and RIM2 and a pair of additional, larger, RIM-related scaffold proteins, Piccolo (also known as Aczonin) and Bassoon, with the same four domains (34, 35). These two closely related, large, presynaptic scaffold proteins were initially believed to be exclusive to vertebrates, as Drosophila lacks these proteins (36). However, a previous study found that Piccolo is an important kinesin transport cargo in Aplysia, where this scaffold protein contributes to synaptic strengthening during long-term facilitation in a learning model (37). Subsequently, Fife was identified in Drosophila as a novel, smaller, Piccolo-like protein, which is also found in bees (36) and in beetles (Dataset S1). In Drosophila, both Fife and RIM are important for homeostatic synaptic plasticity and contribute to the tight spatial coupling of synaptic vesicles to Ca2+ channels (38, 39), as does RIM in mammals (40). Clarinet, discovered more recently in C. elegans, is a highly divergent member of this superfamily, but appears to play a similar role in regulating transmitter release (41).
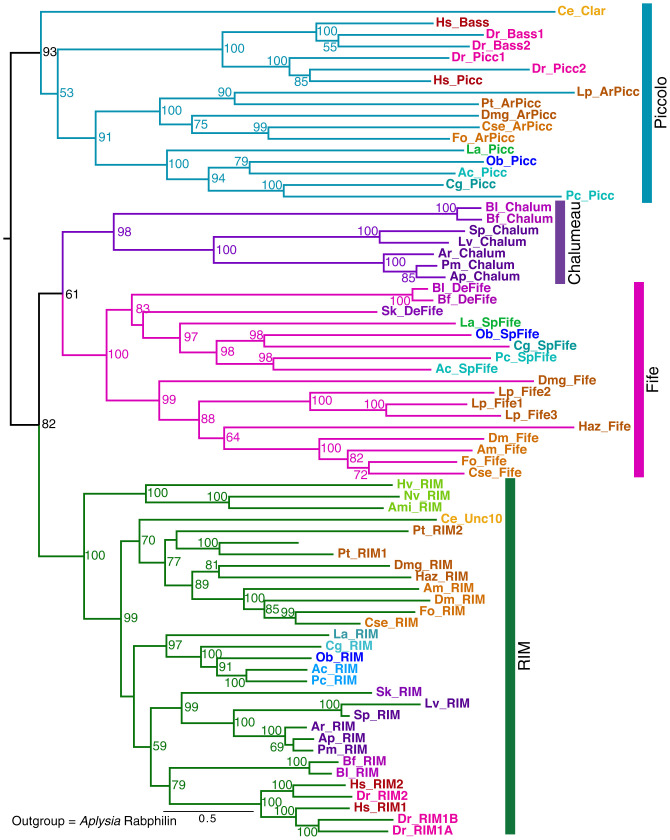
We identified three RIM-related proteins in the Aplysia CNS transcriptome assembly: a RIM ortholog, Aplysia Piccolo, and surprisingly, a Fife-like sequence, as Fifes had been previously thought to be specific to insects (36) or arthropods. Moreover, there are Fife-like sequences in two early diverging deuterostomes, Saccoglossus and Branchiostoma. The phylogenetic analysis revealed yet another RIM-like protein that is deuterostome specific, present in sea urchins and starfish, and also in Branchiostoma (Fig. 4). We named this small RIM superfamily member “Chalumeau.”
Fig. 4.
Evolution of RIM superfamily proteins across phylogenetic groups and domain structure of Aplysia unc13 proteins. Dendrogram for RIM, Piccolo, Bassoon, Fife, and Chalumeau family members from a RAxML analysis (with alignment trimmed with TrimAI; Methods and SI Appendix, Table S2). Bootstrap values ≥50% shown. Additional arthropods and echinoderms were added in this analysis to clarify the representation of Fifes, Piccolos, and Chalumeaus. Species abbreviations for species not defined in previous figure legends: Ami, Acropora millepora; Hv, Hydra vulgaris; Nv, Nematostella vectensis (cnidarians); Cse, Cryptotermes secundus; Haz, Hyalella azteca; Fo, Frankliniella occidentalis; Pt, Parasteatoda tepidariorum (arthropods); Aj, Anneissia japonica; Ar, Asterias rubens; Ap, Acanthaster planci; Lv, Lytechinus variegatus; Pv, Patiria miniata (echinoderms), Bf, Branchiostoma floridae (cephalochordate). For two genes characterized in C. elegans, we use the original gene names, clairnet, Clar, for a Piccolo family member, and Unc10 for RIM in this nematode. The outgroup was Aplysia rabphilin (not shown). Several unanticipated family members are indicated with specific names: ArPicc, arthropod Piccolo; SpFife, spiralian Fife; DeFife, deuterostome Fife. Note: Limulus has three Fife genes, and Bassoon is recently derived from Piccolo in vertebrates.
Because the presence of Piccolo appeared to be widespread among invertebrate bilaterians, we searched for related genes in arthropods. Contrary to what was previously reported, a number of arthropod groups have Piccolo as well as Fife, including the chelicerates Limulus and spiders, crustaceans (e.g., Daphnia and the amphipod Hyalea), and some insects (e.g., thrips, termites, and cockroaches). Therefore, unlike flies, bees, and nematodes, many invertebrate groups express both a Piccolo and a Fife. Chordates lost the Fife family after lancelets diverged.
To summarize, phylogenetic analysis suggests that Piccolo and Fife were both present before the deuterostome–protostome divergence (Fig. 4), whereas Chalumeau is present only in early deuterostomes. Piccolo and the closely related Bassoon diverged as a consequence of the vertebrate genome duplication. We did not detect any bilaterian species with all three proteins—Fife, Piccolo, and Chalumeau—as deuterostome genomes with Chalumeau do not have a Piccolo. Many species have lost one or more members of this family; for example, many insect groups lack Piccolo, as do echinoderms and lancelets. Alternatively, in these deuterostome groups, it is possible that Chalumeau evolved from Piccolo, though this relationship was not supported by the dendrograms Fig. 4.
In contrast to the other family members, RIM is present in all bilaterian clades. Two RIM-like proteins are present both in ctenophores and in placazoans (42). Our phylogenetic maximum-likelihood dendrogram analyses did not permit definitive delineation of the evolutionary relationships between these early RIMs (not included in the dendrogram) and the RIM families present in bilaterians.
The highly conserved RIM PDZ domain binds CaV2 at a C-terminal motif DDWC (33) that is nearly identical from cnidarians to humans (42), whereas other RIM-like family members have changes in the PDZ domain residues involved in ligand recognition. Divergent RIM-like sequences are found in earlier-branching metazoan phyla, including ctenophores, poriferans, and placazoans; however, neither the DDWC site on CaV2 nor the conserved residues in the PDZ domain that confer ligand specificity are found in these earlier-branching species (42). Thus, the interaction between RIM and CaV2 channels, which is important in localizing Ca2+ channels to the presynaptic release sites at bilaterian synapses (33), presumably evolved in the common ancestor of bilaterians and cnidarians; alternatively it is possible that the ctenophore RIM PDZ domain binds a putative PDZ ligand at the ctenophore CaV2 C terminus, as proposed by Piekut et al. (42).
Unc13.
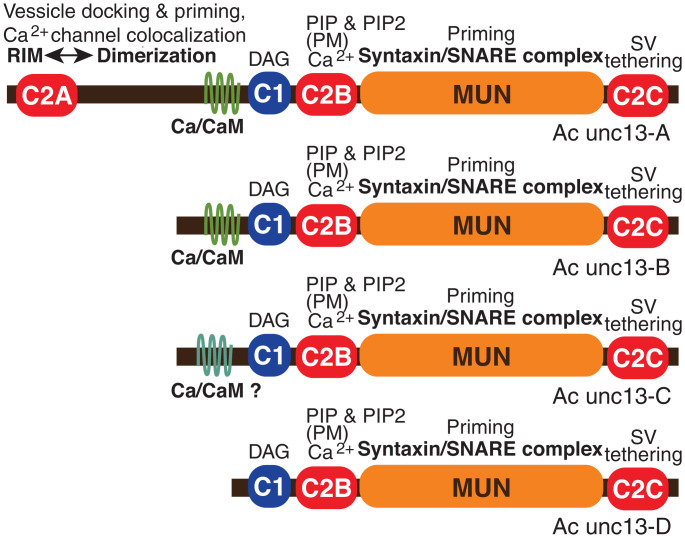
Unc13 exemplifies the large, multidomain scaffold proteins at synapses that have a series of interacting partners. Unc13 has been identified in early-branching animal clades, including sponges and Trichoplax, and the expansion of the unc13 protein and the expression of increased numbers of isoforms is believed to contribute to increased complexity of synaptic regulation across multiple bilaterian phyla. Many of these interactions are conserved from C. elegans and Drosophila to mammals (43). The essential core function of unc13 at the synapse (Munc13 in mammals) is mediated by the MUN domain, which interacts directly with the SNARE complex and is required for priming of synaptic vesicle release (43). In neurons in bilaterian species, all unc13 isoforms contain a series of four characteristic central and C-terminal domains (Fig. 5). The MUN domain is located near the C-terminal C2C domain, which interacts with synatpic vesicles. A pair of regulatory domains is located centrally: the C1 domain, which binds diacylglycerol (DAG), and, immediately adjacent, the C2B domain, which binds Ca2+ and the phospholipids PIP and PIP2 in the plasma membrane. When activated by Ca2+ and DAG, this combined C1–C2B regulatory module substantially enhances MUN domain function and vesicle priming. The phospholipid binding is also responsible for one form of Ca2+-gated, short-term plasticity, facilitating the initial interaction of synaptic vesicles with the phospholipid bilayer (44). The prototypic full-length form of unc13/Munc13 found in many bilaterians also contains an additional pair of modulatory sites: an N-terminal C2A domain with an adjacent α helix and a calmodulin (CaM)-binding, amphipathic α helix situated centrally, N-terminally to the C1 domain (Fig. 5). The C2A domain interacts with RIM (6), and indirectly with Rab3A, and mediates superpriming of vesicle release involving precise refinement of vesicle localization (30). There are related MUN-containing family members conserved in evolution and involved in release of dense core vesicles (e.g. unc13-D and BAI1-AP) (43); these diverged from unc13 in the last common metazoan ancestor (SI Appendix, Fig. S13) (43) and are not considered here.
Fig. 5.
Domains in alternative forms of unc13 expressed from alternative start sites from the Aplysia unc13 gene. The heterogeneity of isoforms is similar to the four Munc13 isoforms expressed in mammalian neurons: Munc13-1, bMunc13-2, ubMunc13-2, and Munc13-3. Unc13 interactions illustrated include C2A domain binding to RIM, which relieves inhibitory homodimerization; Ca2+/CaM binding site; C1 domain, which binds DAG; C2B domain, which binds Ca2+ and also binds PIP and PIP2, mediating localization to plasma membrane (PM); MUN domain, which facilitates a critical conformational change in syntaxin and ensures correct assembly of the SNARE complex; and the C terminus, including C2C, which tethers synaptic vesicles (SVs). DAG, Ca2+, and PIP/PIP2 binding to the C1-C2B module triggers activation of the MUN domain (43, 83). Note: Ac unc13-C contains a divergent, putative CaM-binding sequence (SI Appendix, Fig. S14); whether it actually binds Ca2+/CaM is not known (as indicated by “Ca/CaM-?”).
At mammalian synapses, there are three Munc13 genes expressed (SI Appendix, Fig. S13). The Munc13-2 gene is expressed as two distinct isoforms: a ubiquitous form (ubMunc13-2) and a brain-specific form (bMunc13-2). Of these four isoforms, bMunc13-2 and Munc13-3 lack the N-terminal C2A domain. C. elegans and Drosophila have a single gene, which is expressed in two and more than five isoforms, respectively. Evidence from C. elegans, Drosophila (45, 46), and mice (30, 47) demonstrates that unc13 isoforms that differ in these modulatory domains shift the balance between different types of short-term synaptic plasticity and sculpt the basal release properties of synapses in complex ways (48). For example, in mammals, ubMunc13-2 promotes frequency facilitation and augmentation, whereas Munc13-1 promotes short-term depression. Both Munc13-1 and ubMunc13-2 contain a characteristic Munc13-specific CaM-binding motif with an additional hydrophobic residue at position 26. This motif was initially believed to be unique to mammals, but is present in Drosophila and gastropods, including Aplysia (SI Appendix, Fig. S14). Both bMunc13-2 and Munc13-3 have divergent CaM-binding sequences that alter facilitation with repeated activity (47).
Analysis of the Aplysia transcriptome revealed that there is a single unc13 gene that is expressed in the CNS in four alternative forms, only one of which contains the N-terminal C2A domain (Fig. 5). These different isoforms are generated from different transcriptional start sites, determined by a family of alternative promoters located in introns of the unc13 gene. One of the Aplysia isoforms is truncated such that it lacks the CaM-binding site. Two of the isoforms, Ac-unc13-A and Ac-unc13-B, have a CaM-binding motif very similar to that described in Munc13-1 and ubMunc13-2, with a 1-5-8-10-(12)-26 pattern of hydrophobic residues (49) (SI Appendix, Fig. S14). Ac-unc13-C has a different pattern of hydrophobic residues. Whether this more divergent region in Ac-unc13-C actually binds Ca2+/CaM will need to be tested empirically.
Apparently, unc13 isoforms lacking the C2A domain have evolved multiple times in bilaterians; bMunc13-2 in mammals is an alternative product of a C2A-containing gene, and the C2A domain is found in some unc13 isoforms in arachnids but is missing from insects. The selection of unc13 isoforms expressed in individual Aplysia neurons, among the several available, may influence the short-term plasticity expressed at specific synapses. Differences in the expression of short-term synaptic plasticity, in turn, impact the types of long-term plasticity that will be induced at these synapses. Thus, the selection of unc13 isoforms expressed presynaptically sculpts both short-term and long-term synaptic plasticity, including the recruitment of postsynaptic forms of plasticity. Our analysis revealed that the diversity of unc13 isoforms in Aplysia (Fig. 5) is comparable to that in vertebrates. A preliminary analysis revealed substantial differences in expression levels of the four Aplysia unc13 isoforms, suggesting that most of the unc13 variants are expressed in a neuron-specific pattern in Aplysia CNS. The parallel evolution of multiple unc13 isoforms that differ in the precise CaM-binding site and in the presence of the C2A domain suggests there are fundamental roles for the diversity of this protein family in relatively simple nervous systems, as well as in more elaborate brains capable of sophisticated behaviors. Unc13, with all of the domains involved in regulating synaptic vesicle priming and mediating presynaptic synaptic plasticity, was already present early in animal evolution before the last common bilaterian ancestor (SI Appendix, Fig. S13) (43). As discussed later, this provides evidence for the co-evolution of synapses and the molecular substrates for synaptic plasticity.
Additional Presynaptic Scaffold Proteins.
We analyzed three additional families of presynaptic scaffold proteins, described in the SI Appendix: RIM-BP, CAST/ELKS, and CASK (SI Appendix, Figs. S15–S17). A comparison of gene content among species for these three scaffold proteins demonstrated that Octopus and Aplysia have the same number of genes; humans have more gene isoforms than these mollusks for CAST/ELKS and RIM-BP, though not for CASK; and Danio has more gene isoforms than humans, with the exception of RIM-BP (SI Appendix, Table S4).
Pfams.
One approach for assessing the diversity of protein family members is to tabulate the numbers of transcripts with nervous system–related domains as identified by Pfam IDs. Other than the previously described expansions in protocadherins and C2H2 zinc finger transcription factors, the Octopus proteome generally contained similar Pfam group content relative to Aplysia. Most importantly, we did not observe notable additional Pfam group expansions related to the synaptic proteome in Octopus (SI Appendix, Table S5 and Dataset S2). Although there are increased Pfams for most scaffold and signaling molecules in humans compared to Aplysia and Octopus, there are similar increases in Pfam numbers when comparing zebrafish to humans (SI Appendix, Table S5 and Dataset S2), suggesting this is linked to genome duplication, not cognitive capacity.
Discussion
We have generated an improved transcriptome assembly for Aplysia CNS using an optimized pipeline we developed and iterative evaluation of each version of the transcriptome assembly based on comparison with a set of Reference Transcripts, which had been previously cloned and characterized. Key steps in this improvement process were: 1) optimization of complementary DNA shearing time; 2) use of multiple RNA-seq assemblers, one de novo assembler and two genome-guided assemblers, followed by merging these assemblies using PASA; 3) removal of spurious contigs, including chimeras, with filtering using a modified TransRate score; and 4) clustering with CD-HIT of closely related transcripts (Fig. 1A). In approximately two-thirds of the transcript clusters, the longest open reading frames (ORFs) were in DN Trinity contigs, mainly due to frameshifts introduced by genome-guided assembly. In approximately one-third of the clusters, genome-guided assemblies improved the transcripts. If a set of predicted transcripts is to be used to assess representation of neuronal genes in distant phylogenetic groups, the completeness of the transcript assembly is critical. In a number of cases, we were able to stitch together fragmented transcript sequences in other mollusks based on orthologous Aplysia sequences. Thus, conclusions must always be considered tentative as to the absence of a gene in an invertebrate genome or transcriptome where assembly and gene prediction have not benefited from a closely related model system.
This newly assembled CNS transcriptome provides key insights into the Aplysia synaptic proteome and served as a resource to evaluate the hypothesis that transformative evolutionary advances in cognitive capacity and behavioral flexibility were a consequence of increases in synaptic complexity resulting from the increased gene numbers due to genome duplications in the vertebrate lineage (4). Here, we consider 1) this proposed Synaptic Molecular Complexity Hypothesis; 2) insights that emerged from our analysis concerning the progressive evolution of synaptic proteins and neuroplasticity; and 3) an alternative evolutionary mechanism for increased cognitive capacity and flexibility in both mammals and cephalopods.
Complexity of the Synaptic Proteome during Evolution.
A central concept of the Synaptic Molecular Complexity Hypothesis is that “multiplicative increases in the complexity of this protosynapse machinery secondary to genome duplications drove synaptic, neuronal, and behavioral novelty in vertebrates” (4). A critical flaw of this hypothesis is that it was based on an analysis that was highly vertebrate-centric, focusing exclusively on mammalian synaptic proteins; invertebrate families of synaptic proteins that are absent in vertebrates were not considered. Moreover, there is substantial, and perhaps unjustified, emphasis on the vertebrate-specific, whole-genome duplications (the two duplication events for most vertebrates and a third for bony fish). There have been large expansions of gene families in deuterostomes that led to functional diversity, which often have occurred independently of genome duplications [e.g., expansions of claudins (50), of immunoglobulin-related genes (51), and of olfactory receptors (52)].
Certainly, corvids, parrots, and many mammals, including primates, dolphins, and elephants, demonstrate particularly sophisticated cognitive capacity. Similarly, Octopus has much higher cognitive capacity than Aplysia. Do these increases in cognitive sophistication result from increased molecular complexity at individual synaptic sites? Does synaptic plasticity become more elaborate to support higher intelligence? The complete CNS transcriptome from Aplysia allows us to explore these questions.
Emes and Grant (4) emphasized expansions of synaptic scaffold proteins, receptors, and signaling enzymes found in mammals. A consequence of whole-genome duplications is that mammalian genomes encode more isoforms of signaling proteins than do those of invertebrates, such as C. elegans, Drosophila, gastropods, or cephalopods. However, previous examinations of synaptic signaling pathways have revealed a number of instances where families of proteins important for nervous system function in protostomes are not present in chordates, including: neuropeptide-gated ion channels (53); ACh receptors (e.g., inhibitory ACh-gated chloride channels); metabotropic glutamate receptors (26); octopamine receptors (54); classical calpains (55); and vesicular glutamate transporters (24). Counts of genes with specific Pfam domains provide a wholistic, if very general, index of expansions of protein families. Using Pfam counts, we also do not observe any striking expansions in important receptors or signaling molecules when comparing mollusks to vertebrates (SI Appendix, Table S5 and Dataset S2). Indeed, there are a greater number of neurotransmitter-gated ionotropic receptors in mollusks than in humans, due to substantial expansions in ionotropic glutamate receptors (23–25) and ligand-gated ACh receptors (56) (SI Appendix, Table S5 and Dataset S2).
We particularly focused on synaptic scaffold proteins that are implicated in synaptic plasticity and the precise spatial organization of synaptic sites. When comparing synaptic scaffold proteins between mollusks and vertebrates, we are in agreement with Emes et al. (3) that most scaffold proteins have more isoforms in vertebrates than in mollusks (SI Appendix, Table S4). However, there are families of scaffold proteins that are present in mollusks and other protostomes, but are absent in vertebrates; these include three NETO-related families implicated in receptor clustering and the RIM-related Fife family. Some of the presumed vertebrate-specific families (e.g., Piccolo), although absent in Drosophila, are, in fact, present in most protostomes but were specifically lost during insect evolution. Another important possibility is that heterogenous forms of scaffold proteins can be achieved through alternative transcripts from a single gene. We analyzed alternative transcripts only in the case of unc13, where we observed that different isoforms may achieve diversity comparable to the Munc13 isoforms in mammalian CNS (see also refs. 31 and 46).
It is also important to emphasize that diverse forms of scaffold proteins within a family in mammals may have distinct molecular interactions, such that they mediate the localization of different synaptic proteins that may contribute to different cognitive processes. This is clearly the case for the DLG family members such as SAP97 and PSD95 (57). Although specific behavioral deficits with knockout of a recently duplicated scaffold protein (57) are consistent with the hypothesis of Emes and Grant (4, 57), there are alternative possible explanations; specifically, there is not yet evidence that an individual DLG isoform plays a unique role in a specific form of synaptic plasticity involved in learning, rather than being expressed selectively at a critical site in a circuit. Moreover, given that Aplysia and Octopus have the same number of DLG genes, expansion of the family is certainly not necessary for increased cognitive capacity. As with unc13, alternative transcripts from a single gene in Aplysia, Octopus, and other invertebrate species may achieve increased diversity in synaptic plasticity.
If cognitive complexity were correlated with increased complexity of the synaptic proteome, we would have also expected to see substantial expansions of synaptic scaffold isoforms in Octopus compared with Aplysia. Although the Octopus genome demonstrates dramatic expansions in the protocadherin family and in zinc finger transcription factors (58), the Aplysia and Octopus transcriptomes have similar numbers of synaptic genes (SI Appendix, Table S5 and Dataset S2). Also, contrary to what would be predicted from the Synaptic Molecular Complexity Hypothesis, zebrafish often has more isoforms of synaptic scaffold proteins than do humans (59) (SI Appendix, Table S4), because teleost fish have undergone an additional round of genome duplication. This would suggest that more complex nervous systems are neither a direct consequence of increased synaptic protein diversity (comparing zebrafish and humans) nor is increased synaptic protein diversity required for a more complex nervous system (comparing Octopus and Aplysia) (SI Appendix, Table S5). Although the increase in synaptic proteins in zebrafish was not reflected by increased complexity of the zebrafish PSD proteome (59), it is difficult to exclude the possibility that PSD fractionation conditions developed for rodents might result in suboptimal retention of loosely associated PSD proteins in fish.
The proposal that increases in synaptic complexity drove cognitive capacity would imply that cognitively sophisticated animals have many more specialized synapses that differ based on increased complexity of specialized scaffolds. The mammalian “synaptomes” for excitatory synapses have been grouped into 37 types (60). However, there is little evidence that these subtypes, mainly categorized by the shapes of puncta of PSD scaffold proteins, are due to the diversity of the synaptic proteome. Moreover, it is possible that a similar approach examining synapses in organisms with simpler nervous systems would reveal a similar level of synaptic diversity. There are many specialized invertebrate synapses, which presumably also have specialized synaptic scaffolds [e.g., Drosophila photoreceptor synapses with a very precise molecular organization (61)].
One important caveat is that our analysis is limited to comparisons of predicted protein sequences from molluscan transcriptomes. In contrast, arguments favoring synaptic protein complexity in mammals are founded on proteomic analyses of the synaptome, as well as on the numbers of synaptic genes (3). Although both the Aplysia and Octopus transcriptomes that we analyzed were derived from CNS mRNA, we have no independent evidence that these scaffold proteins are localized to synapses. It should be emphasized that it is difficult to compare proteomic analyses from non-mammalian species, based either on pulldowns or on subcellular fractionation, as these protocols were generally optimized for mammalian synapses and key mammalian synaptic genes. For example, Emes et al. (3) compared synaptic complexity between Drosophila and mice, based on proteomics of pulldowns using the C terminus of NR2 and the Drosophila NR2 ortholog. However, Drosophila NR2 may not play a central role in the organization of insect synapses comparable to the important role of NR2 at mouse synapses. This does not necessarily mean that mammalian synapses are more complex, as Drosophila synapses may be organized around distinct protein networks.
Evolution of Synapses and Synaptic Plasticity.
One justification for a relationship between cognitive capacity and synaptic diversity is the need for an increased capacity for activity-dependent synaptic plasticity and neuromodulation. However, neural plasticity is also critical for adaptive success in quite simple nervous systems; indeed, even jellyfish exhibit behavioral flexibility (7). Neuromodulation and activity-dependent plasticity involve a complex set of processes that enables the nervous system to alter its output based on external inputs, behavioral state, and prior experience of the organism. One major contributing mechanism, particularly for neuromodulatory changes, is slow neurotransmission, whereby the release of neurotransmitters acting mainly through G protein–coupled receptors (GPCRs) allows the same neuronal circuit to produce different outputs, depending on the motivational state of the organism (e.g., injury, hunger, fear, sleep). Comparisons of Octopus with Aplysia and humans with zebrafish do not support the concept that the diverse molecular mechanisms that mediate slow neurotransmission have expanded in parallel with cognitive complexity (SI Appendix, Table S5 and Dataset S2). The transmitters that mediate slow neurotransmission (i.e., biogenic amines, ACh, and neuropeptides) have not expanded in animals with more sophisticated cognition, nor has there been a substantial expansion of the GPCRs that mediate the slow neuromodulatory responses. Signaling downstream of GPCRs is ancient, whether via intracellular messengers, or via membrane-delimited G proteins, via small G proteins such as Ras, or via protein kinases; these pathways have also not expanded in concert with cognitive capacity (SI Appendix, Table S5). Well-studied invertebrate neuromodulatory systems, such as the lobster stomatogastric system (62) or the Aplysia nervous system (63, 64) involve, if anything, more modulatory complexity than most vertebrate systems. For example, neuropeptides select among alternative motor patterns in the rhythmic chewing behavior of crustaceans (62). Repetition priming of central pattern generators is highly developed in the feeding (63) and locomotion circuitry (65) of Aplysia. Activity-dependent plasticity, also termed frequency-dependent plasticity, is present at all bilaterian synapses studied. Unc13 plays important roles in presynaptic activity-dependent plasticity, roles that are highly conserved from C. elegans and flies to mouse (43).
Learning is mediated by synaptic changes that are maintained over long periods of time. The molecular cascades important for inducing and maintaining memory are highly conserved over evolution, with little expansion of these molecules in vertebrates (66). Persistently active protein kinases that contribute to maintenance of memory were initially described in Aplysia (67). The translation of persistently active PKM ζ from an alternative PKC transcript (68) is a chordate innovation (69). However, the role of PKMs in memory maintenance is highly conserved and may be more diversified in Aplysia, where PKMs derived from multiple PKCs play roles in maintenance of specific forms of long-term facilitation (70). In summary, there is not evidence for a greater diversity of synaptic signaling pathways involved in induction or maintenance of long-term memory in mammals relative to other bilaterians.
It is worth considering some of the key innovations involved in the gradual adaptive evolution of more complex nervous systems. Importantly, many synaptic proteins such as Shank, DLG, Homer, and neuronal SNARES are present in single-celled or colonial choanoflagellates, which lack synapses. Ctenophores and cnidarians have markedly distinct molecular components of synapses (24, 71, 72), but whether this represents a divergence from a primitive synapse in their last common ancestor or the parallel, independent evolution of synapses is still unclear (73). Unlike cnidarians, ctenophores lack synaptotagmin (24), which is the primary Ca2+ sensor for fast synaptic transmission in bilaterians. In contrast, some important synaptic molecules are shared in ctenophores, cnidarians, and bilaterians, such as SNAREs, DLG, Homer, Shank, RIM, and unc13 (SI Appendix, Fig. S18). Ferlins, which are found in cnidarians and bilaterians, are also expressed in ctenophore neurons (72). Otoferlins are the major Ca2+ sensor for fast transmitter release in auditory and vestibular hair cells in mammals, and a related Ferlin may function as the Ca2+ sensor in comb jellies (24).
Whether or not neurons evolved independently in ctenophores and bilaterians, our analysis reveals a scenario where key molecular components of the synapse were acquired gradually, initially through exaptation and subsequently through addition of novel synaptic genes. The concept of exaptation of previously existing genes from organisms that lack neurons to explain the origin of some synaptic genes (e.g., Shank, Homer, DLG) was introduced by Srivastava et al. (74) and subsequently discussed by several authors (4, 24, 75, 76). Our analysis reveals that synaptic components were acquired at key nodes in animal evolution (SI Appendix, Figs. S18 and S19). The MUN domain is evolutionarily ancient (43). However, unc13 proteins with the domain structure characteristic of bilaterians appeared in the early-diverging metazoans, as they are present in ctenophores, poriferans, and Trichoplax. Both of the Ca2+ channels CaV1 and CaV2 were present in the last common ancestor of poriferans and Trichoplax (76), but the well-conserved C-terminal PDZ recognition motif that mediates CaV2 binding to RIM was added later (42) in the last common ancestor of cnidarians and bilaterians. Ligand-gated glutamate receptors and vesicular glutamate transmission exist in cnidarians, but receptor-associated proteins, such as TARPs and NETOs, evolved in bilaterians prior to the protostome-deuterostome divergence. Members of the RIM superfamily of scaffold proteins had evolved by the earliest metazoans (i.e., ctenophores, poriferans, and placozoans) (42); this family then diversified substantially in bilaterians. The gradual evolution of synaptic signaling and plasticity continued in protostomes and deuterostomes, but many of the important adaptations had occurred before the divergence of these two major bilaterian clades. From this perspective, we can understand that many of the genes integral to the highly structured bilaterian synapses have their origins across the evolutionary tree: in the protosynaptic genes found in single-celled holozoans, poriferans, and Trichoplax; in some molecular components found in the distinct synapses of ctenophores; and in molecular elements of cnidarian synapses (SI Appendix, Figs. S18 and S19). Thus, the evolution of the bilaterian synapse required many independent innovations. Whereas gradual adaptive evolution of synaptic signaling and plasticity continued in both protostomes and deuterostomes, most innovations had occurred in, or before, the last bilaterian common ancestor. Importantly, there is little evidence that later molecular synaptic adaptations are central to increased cognitive capacity in specific clades.
Cell Adhesion Molecules, Circuit Complexity, and Cognitive Capacity.
Broadly speaking, refinement of behavior and increased behavioral complexity involve both neuromodulatory mechanisms and increased complexity of neural circuitry. However, as suggested earlier, neuromodulation is present in the simplest nervous systems. Moreover, neural plasticity enhanced survival and evolutionary success across the animal phyla (8, 9), even in cnidarians (7). At the molecular level, increased complexity of the synaptic proteome enables more complex neuromodulation and activity-dependent plasticity; importantly, much of this molecular complexity that contributes to synaptic plasticity had evolved substantially before the divergence of the major bilaterian groups. In gastropod mollusks, as discussed above, synaptic plasticity involves highly elaborated molecular pathways, and the forms of synaptic plasticity are as diverse as those in the mammalian CNS (77–79).
We argue that increased behavioral complexity could be more easily explained by increased neuron number in conjunction with the evolution of circuitry linked to behavioral complexity, in the absence of substantial increases in synaptic complexity. Certainly, there is much more complex neuronal circuitry in Octopus than in Aplysia and in humans than in zebrafish, yet we found little evidence for increased complexity of the synaptic proteome paralleling enhanced cognitive sophistication. In contrast, large expansions in the protocadherin family of cell adhesion molecules have been described in both Octopus (58, 80) and vertebrates (81). These independent expansions suggest a molecular parallel underlying the evolution of complex nervous systems at the level of complexity of wiring rather than at the level of the synaptic proteome. Elaboration of associative cortexes in mammals and the appearance of specialized circuitry underlying language in humans are examples of links between the evolution of new circuitry and behavioral complexity. In Octopus, the circuitry underlying its rich behavioral repertoire is still being characterized. These innovations in circuitry may require novel molecular pathways or subtle alterations in neurogenesis and subsequent wiring, as has been described in mammals (82). Nevertheless, the formation of more complex circuitry, as found in Octopus, corvids, and primates, and the evolution of cognitive capacity, did not primarily involve substantial increases in the molecular complexity of synapses.
Methods
Processing of RNA from Aplysia CNS, phylogenetic analysis, and other methodological details are described in SI Appendix, Methods. The reads from each tissue were processed separately. For the CNS, 2.97 billion Illumina HiSeq reads were first trimmed and filtered (Fig. 1A and SI Appendix, Methods). These filtered reads were then assembled using three different approaches: de novo with Trinity; genome-guided de novo, also with Trinity; and pure genome-guided assembly with StringTie. (Software details are provided in SI Appendix, Table S2). These separate assemblies were then merged using PASA. We performed quality filtering of both the PASA transcripts and the original set of DN-Trinity transcripts using TransRate, adjusting the cumulative score calculation as described in SI Appendix, Methods. The longest coding sequence called per transcript from both TransRate-filtered PASA transcripts and TransRate-filtered DN-Trinity transcripts were clustered using CD-HIT–EST. The TransRate-passing transcript with the longest coding sequence within each cluster was selected. For a number of genes (∼69%), DN-Trinity provided more complete predicted transcripts (Fig. 1B and SI Appendix, Fig. S4). (Note that truncated transcripts sometimes resulted from frameshifts in genome-guided assemblies). We therefore retained the TransRate-passing DN-Trinity transcripts for a subsequent comparison step.
We refer to this set of 68,308 transcripts as “unigenes,” or 38,125 transcripts after filtering for those shorter than 400 bp (Fig. 1A). To track improvements to the quality of the transcriptome assembly as we developed this pipeline, the transcripts produced by each step were aligned to a set of 57 previously characterized Reference Transcripts from CNS, and coverage and completeness were evaluated.
Supplementary Material
Acknowledgments
We thank Weizhong Li for his help with CD-HIT and Bruce Krueger for his thoughtful comments on this manuscript. Adriano Senatore and Joseph Ryan generously provided recent ctenophore assemblies. This work was supported by NSF EAGER Award IOS-1255695 and NIH grant R01 MH 55880 grant to T.W.A.; by a Natural Sciences and Engineering Research Council of Canada Discovery grant and Canadian Institutes of Health Research project grant 340328 to W.S.; by funding from the HHMI to E.R.K.; and by a Hibbitt Early Career Fellowship to C.A. W.S. is James McGill Professor at McGill University.
Footnotes
Reviewers: J.B., University of Texas Health Science Center at Houston; E.C., Icahn School of Medicine at Mount Sinai; and S.V.P., The Scripps Research Institute.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122301119/-/DCSupplemental.
Data Availability
All Aplysia transcriptome and genome sequences are available at http://aplysiatools.org. Transcriptome sequences have been posted on NCBI (GenBank: GJYY00000000) (84). Dataset 1 provides Genbank IDs for all sequences analyzed in dendrograms, except for Doryteuthis; Doryteuthis peptide sequences were submitted to NCBI, but Genbank IDs are not yet available. These Doryteuthis peptide sequences are included in Dataset 1.
References
- 1.Cornélio A. M., de Bittencourt-Navarrete R. E., de Bittencourt Brum R., Queiroz C. M., Costa M. R., Human brain expansion during evolution is independent of fire control and cooking. Front. Neurosci. 10, 167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLean E. L., et al. , The evolution of self-control. Proc. Natl. Acad. Sci. U.S.A. 111, E2140–E2148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emes R. D., et al. , Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci. 11, 799–806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emes R. D., Grant S. G., Evolution of synapse complexity and diversity. Annu. Rev. Neurosci. 35, 111–131 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Biederer T., Kaeser P. S., Blanpied T. A., Transcellular nanoalignment of synaptic function. Neuron 96, 680–696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockmann M. M., et al. , A trio of active zone proteins comprised of RIM-BPs, RIMs, and Munc13s governs neurotransmitter release. Cell Rep. 32, 107960 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Dawson M. N., Hamner W. M., Rapid evolutionary radiation of marine zooplankton in peripheral environments. Proc. Natl. Acad. Sci. U.S.A. 102, 9235–9240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrot-Minnot M. J., Banchetry L., Cézilly F., Anxiety-like behaviour increases safety from fish predation in an amphipod crustacea. R. Soc. Open Sci. 4, 171558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters E. T., Nociceptive biology of molluscs and arthropods: Evolutionary clues about functions and mechanisms potentially related to pain. Front. Physiol. 9, 1049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkins R. D., Kandel E. R., Bailey C. H., Molecular mechanisms of memory storage in Aplysia. Biol. Bull. 210, 174–191 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Byrne J. H., Hawkins R. D., Nonassociative learning in invertebrates. Cold Spring Harb. Perspect. Biol. 7, a021675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins R. D., Byrne J. H., Associative learning in invertebrates. Cold Spring Harb. Perspect. Biol. 7, a021709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moroz L. L., et al. , Neuronal transcriptome of Aplysia: Neuronal compartments and circuitry. Cell 127, 1453–1467 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puthanveettil S. V., et al. , A strategy to capture and characterize the synaptic transcriptome. Proc. Natl. Acad. Sci. U.S.A. 110, 7464–7469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grasso F. W., Basil J. A., The evolution of flexible behavioral repertoires in cephalopod molluscs. Brain Behav. Evol. 74, 231–245 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Richter J. N., Hochner B., Kuba M. J., Pull or push? Octopuses solve a puzzle problem. PLoS One 11, e0152048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díaz-Alonso J., Nicoll R. A., AMPA receptor trafficking and LTP: Carboxy-termini, amino-termini and TARPs. Neuropharmacology 197, 108710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson A. C., Nicoll R. A., The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dakoji S., Tomita S., Karimzadegan S., Nicoll R. A., Bredt D. S., Interaction of transmembrane AMPA receptor regulatory proteins with multiple membrane associated guanylate kinases. Neuropharmacology 45, 849–856 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Walker C. S., et al. , Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 10781–10786 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R., et al. , Evolutionary conserved role for TARPs in the gating of glutamate receptors and tuning of synaptic function. Neuron 59, 997–1008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cull-Candy S. G., Farrant M., Ca2+ -permeable AMPA receptors and their auxiliary subunits in synaptic plasticity and disease. J. Physiol. 599, 2655–2671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greer J. B., Khuri S., Fieber L. A., Phylogenetic analysis of ionotropic l-glutamate receptor genes in the Bilateria, with special notes on Aplysia californica. BMC Evol. Biol. 17, 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abrams T. W., Sossin W. S., “Invertebrate genomics provide insights into the origin of synaptic transmission” in Oxford Handbook of Invertebrate Neurobiology, Byrne J. H., Ed. (Oxford University Press, Oxford, UK, 2018), pp. 123–150. [Google Scholar]
- 25.Moroz L. L., Nikitin M. A., Poličar P. G., Kohn A. B., Romanova D. Y., Evolution of glutamatergic signaling and synapses. Neuropharmacology 199, 108740 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos-Vicente D., Grant S. G., Bayés À., Metazoan evolution and diversity of glutamate receptors and their auxiliary subunits. Neuropharmacology 195, 108640 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y., Mellem J. E., Brockie P. J., Madsen D. M., Maricq A. V., SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature 427, 451–457 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Wang R., et al. , The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron 75, 838–850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gally C., Eimer S., Richmond J. E., Bessereau J. L., A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature 431, 578–582 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camacho M., et al. , Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat. Commun. 8, 15293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z., Tong X. J., Kaplan J. M., UNC-13L, UNC-13S, and Tomosyn form a protein code for fast and slow neurotransmitter release in Caenorhabditis elegans. eLife 2, e00967 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H., et al. , Heterodimerization of UNC-13/RIM regulates synaptic vesicle release probability but not priming in C. elegans. eLife 8, e40585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaeser P. S., et al. , RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144, 282–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., et al. , Aczonin, a 550-kD putative scaffolding protein of presynaptic active zones, shares homology regions with Rim and Bassoon and binds profilin. J. Cell Biol. 147, 151–162 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waites C. L., et al. , Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation. EMBO J. 32, 954–969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruckner J. J., et al. , Fife, a Drosophila Piccolo-RIM homolog, promotes active zone organization and neurotransmitter release. J. Neurosci. 32, 17048–17058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puthanveettil S. V., et al. , A new component in synaptic plasticity: Upregulation of kinesin in the neurons of the gill-withdrawal reflex. Cell 135, 960–973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruckner J. J., et al. , Fife organizes synaptic vesicles and calcium channels for high-probability neurotransmitter release. J. Cell Biol. 216, 231–246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller M., Liu K. S., Sigrist S. J., Davis G. W., RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J. Neurosci. 32, 16574–16585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaeser P. S., Deng L., Fan M., Südhof T. C., RIM genes differentially contribute to organizing presynaptic release sites. Proc. Natl. Acad. Sci. U.S.A. 109, 11830–11835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xuan Z., et al. , Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. eLife 6, e29276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piekut T., et al. , Early metazoan origin and multiple losses of a novel clade of RIM Presynaptic calcium channel scaffolding protein homologs. Genome Biol. Evol. 12, 1217–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dittman J. S., Unc13: A multifunctional synaptic marvel. Curr. Opin. Neurobiol. 57, 17–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipstein N., et al. , Munc13-1 is a Ca2+-phospholipid-dependent vesicle priming hub that shapes synaptic short-term plasticity and enables sustained neurotransmission. Neuron 109, 3980–4000.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Böhme M. A., et al. , Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca(2+) channel-vesicle coupling. Nat. Neurosci. 19, 1311–1320 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Pooryasin A., et al. , Unc13A and Unc13B contribute to the decoding of distinct sensory information in Drosophila. Nat. Commun. 12, 1932 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipstein N., et al. , Nonconserved Ca(2+)/calmodulin binding sites in Munc13s differentially control synaptic short-term plasticity. Mol. Cell. Biol. 32, 4628–4641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenmund C., et al. , Differential control of vesicle priming and short-term plasticity by Munc13 isoforms. Neuron 33, 411–424 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Castañeda F., et al. , Modular architecture of Munc13/calmodulin complexes: Dual regulation by Ca2+ and possible function in short-term synaptic plasticity. EMBO J. 29, 680–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukendi C., et al. , Evolution of the vertebrate claudin gene family: Insights from a basal vertebrate, the sea lamprey. Int. J. Dev. Biol. 60, 39–51 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Dornburg A., Yoder J. A., On the relationship between extant innate immune receptors and the evolutionary origins of jawed vertebrate adaptive immunity. Immunogenetics 74, 111–128 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Niimura Y., Nei M., Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. U.S.A. 102, 6039–6044 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Assmann M., Kuhn A., Dürrnagel S., Holstein T. W., Gründer S., The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biol. 12, 84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauknecht P., Jékely G., Ancient coexistence of norepinephrine, tyramine, and octopamine signaling in bilaterians. BMC Biol. 15, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hastings M. H., et al. , Novel calpain families and novel mechanisms for calpain regulation in Aplysia. PLoS One 12, e0186646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiao Y., Cao Y., Zheng Z., Liu M., Guo X., Massive expansion and diversity of nicotinic acetylcholine receptors in lophotrochozoans. BMC Genomics 20, 937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nithianantharajah J., et al. , Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat. Neurosci. 16, 16–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albertin C. B., et al. , The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayés À., et al. , Evolution of complexity in the zebrafish synapse proteome. Nat. Commun. 8, 14613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu F., et al. , Architecture of the mouse brain synaptome. Neuron 99, 781–799.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamanaka Y., Meinertzhagen I. A., Immunocytochemical localization of synaptic proteins to photoreceptor synapses of Drosophila melanogaster. J. Comp. Neurol. 518, 1133–1155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marder E., Bucher D., Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 69, 291–316 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Cropper E. C., Jing J., Perkins M. H., Weiss K. R., Use of the Aplysia feeding network to study repetition priming of an episodic behavior. J. Neurophysiol. 118, 1861–1870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mozzachiodi R., Lorenzetti F. D., Baxter D. A., Byrne J. H., Changes in neuronal excitability serve as a mechanism of long-term memory for operant conditioning. Nat. Neurosci. 11, 1146–1148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bruno A. M., Frost W. N., Humphries M. D., Modular deconstruction reveals the dynamical and physical building blocks of a locomotion motor program. Neuron 86, 304–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kandel E. R., The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 5, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz J. H., Cognitive kinases. Proc. Natl. Acad. Sci. U.S.A. 90, 8310–8313 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sacktor T. C., How does PKMζ maintain long-term memory? Nat. Rev. Neurosci. 12, 9–15 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Bougie J. K., et al. , The atypical protein kinase C in Aplysia can form a protein kinase M by cleavage. J. Neurochem. 109, 1129–1143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu J., et al. , Cell-specific PKM isoforms contribute to the maintenance of different forms of persistent long-term synaptic plasticity. J. Neurosci. 37, 2746–2763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moroz L. L., et al. , The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sachkova M. Y., et al. , Neuropeptide repertoire and 3D anatomy of the ctenophore nervous system. Curr. Biol. 31, 5274–5285.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Ryan J. F., Did the ctenophore nervous system evolve independently? Zoology (Jena) 117, 225–226 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Srivastava M., et al. , The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burkhardt P., The origin and evolution of synaptic proteins - choanoflagellates lead the way. J. Exp. Biol. 218, 506–514 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Moran Y., Zakon H. H., The evolution of the four subunits of voltage-gated calcium channels: ancient roots, increasing complexity, and multiple losses. Genome Biol. Evol. 6, 2210–2217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferguson L., et al. , Isoform specificity of PKMs during long-term facilitation in Aplysia is mediated through stabilization by KIBRA. J. Neurosci. 39, 8632–8644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin I., et al. , Spontaneous transmitter release recruits postsynaptic mechanisms of long-term and intermediate-term facilitation in Aplysia. Proc. Natl. Acad. Sci. U.S.A. 109, 9137–9142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wan Q., et al. , Protein kinase C acts as a molecular detector of firing patterns to mediate sensory gating in Aplysia. Nat. Neurosci. 15, 1144–1152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z. Y., Ragsdale C. W., Cadherin genes and evolutionary novelties in the octopus. Semin. Cell Dev. Biol. 69, 151–157 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Rubinstein R., Goodman K. M., Maniatis T., Shapiro L., Honig B., Structural origins of clustered protocadherin-mediated neuronal barcoding. Semin. Cell Dev. Biol. 69, 140–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franchini L. F., Genetic mechanisms underlying cortical evolution in mammals. Front. Cell Dev. Biol. 9, 591017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lipstein N., et al. , Munc13-1 is a Ca2+-phospholipid-dependent vesicle priming hub that shapes synaptic short-term plasticity and enables sustained neurotransmission. Neuron 109, 3980–4000.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Orvis J., et al., TSA: Aplysia californica isolate field caught, aquaculture reared, transcriptome shotgun assembly. Accession # GJYY00000000.1. NCBI GenBank. https://www.ncbi.nlm.nih.gov/nuccore/GJYY00000000. Deposited 10 June 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All Aplysia transcriptome and genome sequences are available at http://aplysiatools.org. Transcriptome sequences have been posted on NCBI (GenBank: GJYY00000000) (84). Dataset 1 provides Genbank IDs for all sequences analyzed in dendrograms, except for Doryteuthis; Doryteuthis peptide sequences were submitted to NCBI, but Genbank IDs are not yet available. These Doryteuthis peptide sequences are included in Dataset 1.