Abstract
Aging is characterized by the accumulation of damage to macromolecules and cell architecture that triggers a proinflammatory state in blood and solid tissues, termed inflammaging. Inflammaging has been implicated in the pathogenesis of many age-associated chronic diseases as well as loss of physical and cognitive function. The search for mechanisms that underlie inflammaging focused initially on the hallmarks of aging, but it is rapidly expanding in multiple directions. Here, we discuss the threads connecting cellular senescence and mitochondrial dysfunction to impaired mitophagy and DNA damage, which may act as a hub for inflammaging. We explore the emerging multi-omics efforts that aspire to define the complexity of inflammaging — and identify molecular signatures and novel targets for interventions aimed at counteracting excessive inflammation and its deleterious consequences while preserving the physiological immune response. Finally, we review the emerging evidence that inflammation is involved in brain aging and neurodegenerative diseases. Our goal is to broaden the research agenda for inflammaging with an eye on new therapeutic opportunities.
Aging has been conceptualized as a continuous duel between damage accumulation — due to a combination of environmental and endogenous processes — and resilience mechanisms that cope with such stressors and resolve damage (1). With aging, resilience mechanisms become less effective at repairing or removing damage and preventing its deleterious effects on health (2). Persistent molecular and cellular damage due to exhausted resilience is ultimately expressed as phenotypes of aging, including inflammaging, susceptibility to chronic diseases, physical and cognitive impairments, and, ultimately, frailty and death.
Atop the hierarchy of resilience is the immune system, the aggregate of cells, mediators, and signaling pathways that continuously patrol for pathogens or structural perturbations revealed as “unusual” molecular motifs. The immune system reacts to a variety of threats, such as symbiotic commensal and pathogenic microorganisms, pathogen-associated molecular patterns (PAMPs), and damage-associated molecular patterns (DAMPs) from endogenous and exogenous sources, and orchestrates defense responses aimed at eliminating the specific threat while minimizing damage to the host. While inflammation is important for tissue repair and regeneration, when abnormally intense or persistent, it can drive degeneration and chronic diseases.
The immune system undergoes numerous and profound changes with aging, which are extensively reviewed elsewhere (3–5). Hallmarks of immune aging are (a) a state of proinflammatory activation characterized by high circulating levels of proinflammatory cytokines — such as IL-6 and TNF-α — and localized tissue inflammation, and (b) an aberrant response to antigens and pathogens that could either be blunted, such as in flu vaccination, or excessive, such as in response to SARS-CoV-2 (6).
Considerable research in both animal models and humans has examined the causes and consequences of inflammaging (4). Although increased levels of inflammatory mediators (mostly IL-1, IL-6, TNF-α, and its receptors) are detected in all elderly individuals, higher levels of these biomarkers are associated with increased risk for many chronic conditions, including dementia, disability, and physical frailty. Inflammation’s causal role in cardiovascular disease was established by the CANTOS trial (Canakinumab Anti-Inflammatory Thrombosis Outcomes Study), which demonstrated that IL-1β inhibition reduced the risk of cardiovascular events versus the placebo, particularly in participants whose IL-6 levels were initially elevated (7).
Mechanisms identified as hallmarks of aging biology and immune cell dysfunction have all been hypothesized as causes of inflammation (8). Aging researchers now recognize that measuring a few cytokines in circulation fails to capture the complexity and potential ramifications of inflammaging. Immune cells in tissues, particularly lymphocytes and resident macrophages, show tissue-specific age-related changes likely connected to specific pathological processes (9). By measuring hundreds or thousands of molecules in a few drops of blood, scientists are attempting to identify (a) signatures of accelerated aging that are both informative of the complexity and diversity of the response and predictive of health outcomes and (b) key molecules and molecular mechanisms that can be targeted for intervention (10).
Given the extreme complexity of inflammaging, we focus herein on a few topics that have attracted considerable attention and controversy in the field. First, we discuss cellular senescence as a source of local and systemic inflammation. We highlight evidence that mitochondrial dysfunction is a nexus that binds impaired mitophagy with DNA damage and cellular senescence to ultimately foster a chronic inflammatory state. We then summarize efforts to identify circulating signatures of inflammation through “omics.” Finally, we review emerging data indicating that inflammation is involved in brain aging and dementia. Our intent is to discuss the causes and consequences of inflammaging and to enrich the research agenda toward the development of new therapeutic strategies.
Senescent cells in the inflammation-aging axis
Senescent cells and the senescence-associated secretory phenotype (SASP), now widely acknowledged as drivers of aging and age-related diseases, have emerged as key players in inflammaging. Cellular senescence can be initiated by multiple intrinsic and extrinsic nonlethal stresses, including genotoxic, oncogenic, mitochondrial, oxidative, inflammatory, paracrine signaling, and others (11). Hallmarks of cellular senescence include growth and replication arrest, resistance to apoptosis, chromatin remodeling, metabolic reprogramming, and morphological changes, among others (11–13). While highly specific criteria for defining cellular senescence are still debated, it is generally believed that all senescent cells secrete a set of diverse bioactive molecules, both soluble and within extracellular vesicles, known as the SASP. The SASP includes proteases, growth factors, and, notably, a collection of cytokines, chemokines, and other inflammatory mediators (14, 15). Via the SASP, senescent cells contribute to multiple chronic health conditions associated with inflammaging, e.g., atherosclerosis (16), osteoarthritis (17), cancer (18), myeloid skewing in the bone marrow (19), pulmonary fibrosis (20), Alzheimer’s disease and Parkinson’s disease (21–23), and insulin resistance (24). We detail next the contribution of senescent cells to inflammaging, how the aging immune system may contribute to senescent cell accumulation in peripheral tissues, and how assessing the burden of senescent cells in different tissues through “omics” may aid in the development of strategies to alleviate diseases associated with inflammaging.
Emerging role of senescence in the immune system.
Cells of the immune system can undergo cellular senescence or enter a senescent-like state (25–29). Yousefzadeh and colleagues recently suggested a causative link between a senescent immune system and nonlymphoid tissue aging (26). Deletion of ERCC1, a crucial DNA repair protein, in mouse hematopoietic cells leads to accelerated senescence of the immune system. Accordingly, mice lacking ERCC1 in hematopoietic cells, as well as aged mice, express elevated markers of senescence (p16, p21) in many immune cells with concomitant impairment of immune function. Strikingly, transfer of splenocytes from aged wild-type mice or mice lacking ERCC1 into young animals increased the senescence burden in nonlymphoid organs, suggesting that a senescent immune system contributes to aging phenotypic features in immune cells and in solid, nonlymphoid tissues. Thus, senescent immune cells may be central upstream targets for treating inflammaging-associated phenotypes and reducing systemic senescent cell burden.
Knowing whether senescent immune cells induce cellular senescence in peripheral tissues is central to designing strategies of prevention and treatment. T cell populations exhibiting multiple senescence markers and lacking proliferative capacity, in addition to B cells (30) and monocytes (31) that carry senescent features, have been described (32) and implicated in immunopathology. Accessible in circulation, senescent immune cells may prove easier to therapeutically eliminate than tissue-resident ones. Furthermore, owing to their proximity to blood, factors secreted into plasma by circulating senescent cells are more likely to be detectable and measurable in biomarker studies and be therapeutically targeted. Unfortunately, non-senescent immune cells can express and secrete, usually to a lesser degree, some of the same markers used to identify senescent cells, and research is needed to identify markers with the best discriminative value. For example, certain “senescent-like” macrophages showing high expression of p16INK4a and senescence-associated β-galactosidase (SA-β-gal) activity accumulate during aging, yet immunomodulatory agents can reverse their expression of these senescence markers (27, 28). Isolated p16INK4a-expressing cells from in vivo, however, express a range of senescence and SASP markers (29). These findings suggest that the beneficial effects of eliminating p16-positive cells are due solely to elimination of senescent cells, non-senescent macrophages, or some combination thereof.
Circulating SASP mediates senescent cell–immune system interactions.
The production of SASP inflammatory factors is primarily driven by NF-κB, a rapid-acting transcription factor that orchestrates the response to a range of cellular stressors (33). A p21/Rb pathway also produces a p21-activated secretory phenotype (PASP) that directs a CXCL14-mediated mechanism of senescent cell immunosurveillance (34). Many of the SASP components can activate and recruit cells of the immune system and drive inflammation, including the prototypical markers of chronic sterile inflammaging, i.e., IL-6, IL-1, and TNF-α (14, 35). Since a variety of circulating SASP factors increase with age, likely leaked from tissue-resident senescent cells, a subset of SASP factors have been proposed as biomarker candidates of aging and age-related diseases: growth differentiation factor 15 (GDF15), stanniocalcin 1 (STC1), matrix metalloproteinase 1 (MMP-1), inhibin subunit βA (also known as activin A, INHBA), TNF receptor superfamily member 1A (TNFRSF1A), and serpin family E member 1 (SERPINE1) (15, 35). Epidemiological studies indeed indicate that proteomic SASP panels predict chronological age, biological age, mortality, morbidity, and adverse health outcomes (35–37). High senescent cell burden also drives hematological phenotypes in mice, including blood clotting, which can be reversed upon elimination of senescent cells (38). Additionally, senescence occurs at sites of atherosclerotic plaques, where it participates in multiple stages of atherogenesis (16, 39) and, through a paracrine SASP mechanism, accelerates the atherosclerotic process. These observations suggest that the SASP is secreted into the blood at sufficient levels to contribute to systemic inflammaging and for detection. Consistent with this notion, the administration of dasatinib and quercetin (D+Q), a well-studied senolytic cocktail, in subjects with diabetic kidney disease reduced senescent cell numbers and circulating levels of SASP factors, such as IL-1α and IL-6 (20, 40–45). These initial findings are promising, but further studies are necessary (a) to confirm that the SASP in human biofluids correlates with senescent cell burden in tissues, (b) to identify the subset of SASP factors that are most representative of senescence and disease burden in various tissues, and (c) to identify features of the secretory phenotype that are potentially contributed by sources other than senescent cells.
The SASP directly affects the innate and adaptive immune systems (Figure 1) by promoting recruitment, differentiation, and/or activation of natural killer cells, macrophages, monocytes, neutrophils, and T lymphocytes (46). For instance, the SASP factors granulocyte colony-stimulating factor (G-CSF), macrophage colony-stimulating factor (M-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF) can stimulate the production and activity of monocytes and their differentiation into macrophages. The SASP also connects aging to immunometabolism, promoting the proliferation of the CD38+ population of macrophages that contribute to the age-associated decline of NAD+ (47). Moreover, SASP monocyte chemoattractant proteins (MCPs) and other cytokines/chemokines stimulate the activation and tissue infiltration of monocytes and macrophages. For example, injection of alginate-encapsulated senescent cells into mice causes the recruitment of M2-like resident peritoneal macrophages surrounding the capsules (27). Additionally, senescent fibroblasts in aged skin recruit proinflammatory monocytes by secreting MCP-1 (48). SASP factors may also blunt T cell responses in aged individuals by promoting the secretion of T cell–suppressing prostaglandin E2 (PGE2), implicating the SASP in the age-related decline of adaptive immunity (48).
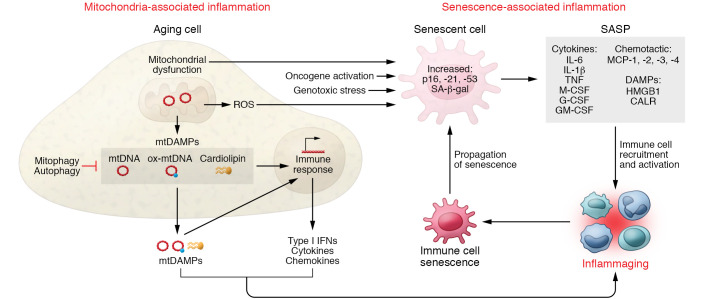
Figure 1. Interplay between cellular senescence, mitochondrial dysfunction, and the aging immune system.
During aging, increasing dysfunctional mitochondria contribute to inflammation through several pathways and interact with other hallmarks of aging such as cellular senescence and autophagy. Mitochondrial-derived damage-associated molecular patterns (mtDAMPs) accumulate in the cytosol, including mitochondrial DNA (mtDNA), oxidized mtDNA (ox-mtDNA), and cardiolipin. Accumulation of mtDAMPs leads to downstream production of inflammatory cytokines, type I interferons (IFNs), and chemokines via intracellular pattern recognition receptors to activate an immune response. The collective secretion of mtDAMPs, IFNs, cytokines, and chemokines into the extracellular environment contributes to inflammaging. With age, an increasing mitochondrial dysfunction and production of ROS lead to increased levels of senescent cells, which accumulate naturally with age and in response to multiple stimuli. The accumulation of senescent cells and the SASP increases the release of bioactive molecules, including cytokines, recruitment factors, and DAMPs, into circulation, thereby driving inflammaging. In turn, the accumulation of senescent lymphocytes in the aging immune system further propagates cellular senescence in peripheral tissues.
Senescent cells, including senescent fibroblasts, secrete DAMPs, which constitute endogenous molecules released from cells in response to internal and external stressors (15, 49). DAMPs that may trigger chronic sterile inflammation during aging and age-related pathologies include high-mobility group box 1 (HMGB1) and calreticulin (CALR) (50).
Translation to humans.
Massive efforts are under way to develop senotherapies that selectively eliminate senescent cells (senolytics) and/or modulate the secretory phenotype (senomorphics) as a strategy to mitigate inflammaging and its consequences. Several senolytics reduce or improve inflammatory or age-related phenotypes in preclinical models, specifically atherosclerosis (D+Q, navitoclax) (16, 41), osteoarthritis (navitoclax) (17), idiopathic pulmonary fibrosis (D+Q), and insulin resistance (23), among others, as well as rejuvenating hematopoietic stem cells in aged mice (navitoclax) (19). Cellular senescence is a driver of both physiologically beneficial phenotypes and age-related pathologies owing to the heterogeneous phenotypes of senescent cells that inhabit an organism or tissue, i.e., “senotype,” which is dependent on biological context and environmental factors. Achieving senotype specificity, or tailoring senolytics to harmful subsets of senescent cells while sparing potentially beneficial ones, presents a challenge to be addressed by future generations of senolytics (51, 52).
An aforementioned complexity of senotherapies is that cellular senescence has beneficial roles in wound healing, tumor suppression, and embryonic development, while also driving inflammaging (53). Removal of some senescent cell types can adversely affect wound healing and fibrosis of liver and perivascular tissue (54, 55). Attempts to distinguish between beneficial and pathological cell senescence at the proteome (15), transcriptome (56, 57), and functional levels have not been successful to date. Studies that capture the complex and dynamic nature of senescent cells by profiling across heterogeneous conditions are needed to identify precise senotype-specific biomarkers and to track the efficacy of interventions. In recent years, unbiased and comprehensive proteomic studies have uncovered an expanded list of factors that may help develop a signature of “senescence burden” in human plasma encompassing inflammatory factors (CXCLs, HMGB1, PTX3, etc.), growth factors (GDF15, IGFBPs), extracellular matrix–associated (ECM-associated) or ECM-modifying components (MMPs, TIMPs, CD44), and tailored sets of robustly expressed age-associated proteins (15). Looking forward, senolytic trials that precisely assess senescence burden with multi-omic and senotype-specific biomarker signatures are needed to identify the most effective senolytic drugs to combat inflammaging-associated pathologies.
Age-related mitochondrial dysfunction and inflammaging
Mitochondria provide the energy essential for all cellular activities, including the fueling of resilience mechanisms that counteract macromolecule damage accumulation with aging (1). Balancing between a cell’s energy/metabolic requirements and energy availability, mitochondria trigger adaptive responses and stress signals, including ROS production and activation of innate immunity. Mitochondrial function declines with aging in model organisms and humans (58). Impaired mitochondrial function, which involves excessive ROS and reduced ATP production, has been associated with visceral obesity, insulin resistance, higher circulating levels of proinflammatory markers, and loss of mobility, phenotypes that are characteristic of aging (59–64). Physical activity improves mitochondrial function in multiple tissues and reverses many features of aging, indicating the importance of efficient mitochondrial activity in preserving health and longevity (65).
Inflammation driven by impaired mitochondrial function occurs in the context of dysfunction in other aging hallmarks. Genomic instability, defective autophagy/mitophagy, and cellular senescence, three major hallmarks of aging, are intertwined with mitochondria-related inflammatory responses (Figure 1) (66). DNA damage accumulates with age and contributes to age-related phenotypic changes (67). DNA of mammal cells experiences over 105 modifications per day, many of which are ROS-related single-strand breaks that are detected by the DNA-break sensor poly(ADP-ribose) polymerase 1 (PARP1), and ultimately resolved by defined repair mechanisms (68). Persistently activated PARP1, however, consumes large amounts of NAD+, an essential coenzyme of sirtuins (SIRTs) (69). Depletion of NAD+ causes loss of efficient sirtuin activity, resulting in impaired functionality of mitochondrial SIRT3, leading to dysregulation in mitochondrial antioxidant systems, mitochondrial DNA (mtDNA) repair, and mitochondrial quality control and biogenesis (SIRT1) pathways, and may also promote senescence (70). Reduced NAD+ levels also impair SIRT2 activity, which normally deacetylates and inactivates the NLR family pyrin domain containing 3 (NLRP3) inflammasome, thereby counteracting inflammation (71). Indeed, NAD+ supplementation mitigates premature aging phenotypes associated with defects in DNA repair by restoring mitochondrial function and mitophagy (72, 73). Conversely, abnormal autophagy and mitophagy, often observed in chronic disease and aging, fail to remove dysfunctional mitochondria that release DAMPs and ROS, activating NLRP3 and mitochondria-related inflammation (74).
In physiological conditions, ROS operate as signaling molecules for diverse cellular functions and are rapidly quenched by antioxidants, such as thioredoxin and glutathione (75). However, impairments in oxidative phosphorylation increase the release of ROS, causing oxidative damage to macromolecules and ultimately cell dysfunction and death (76). Impaired mitochondria release DAMPs, composed of mostly oxidatively modified mtDNA and cardiolipin, into the cytosol through apoptosis-activated BAK/BAX macropore assemblies (77). mtDNA harbors conserved unmethylated CpG motifs characteristic of bacterial DNA, and when it enters the cytosol, it is sensed as “non-self” by the innate immune system via pattern recognition receptors, such as TLR9 and NLRs (78). Cardiolipin — a phospholipid that is normally segregated to the inner mitochondrial membrane and is essential to the maintenance of mitochondrial architecture and function — when herniated outside a damaged mitochondrial membrane, stimulates mitophagy, a process that eliminates dysfunctional mitochondria and promotes the formation of new mitochondria (79). In most severe cases, cardiolipin stimulates the release of cytochrome c and precipitates apoptosis (80). When ROS-damaged mtDNA and cardiolipin are released into the cytosol, various inflammatory pathways are activated. Responses that have been best studied involve the NLRP3 inflammasome, the DNA-sensing cGAS/STING pathway, and NF-κB, which are described in further detail in Figure 2.
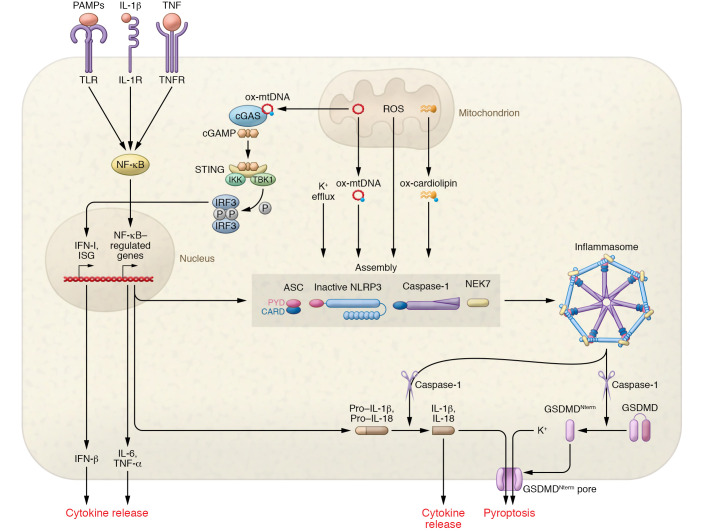
Figure 2. Mitochondria are hubs of inflammation.
Mitochondria are at the center of three inflammatory mechanisms: the NLRP3 inflammasome, cGAS/STING pathway, and NF-κB pathway. NLRP3 inflammasome activation is considered a two-step process: First, priming requires activation of NF-κB by PAMPs or DAMPs binding to pattern recognition receptors including TLRs or by cytokines, such as TNF-α or IL-1β, via their receptors. NF-κB stimulates the transcription and posttranslational stabilization of inactive forms of NLRP3, precursors of cytokines pro–IL-1β and pro–IL-18, and other NF-κB–related cytokines such as IL-6. Second, during activation, a multitude of DAMPs, such as ATP, cholesterol crystals, and urate crystals, promote inflammasome assembly, involving NIMA-related kinase 7 (NEK7), a sensor of K+ efflux (180). The activation likely requires K+ efflux, ROS production, and relocalization of cardiolipin to the outer mitochondrial membrane. It has been proposed that all stimuli that trigger the NRLP3 inflammasome converge on mitochondria, possibly via release of newly synthesized oxidized mtDNA, but K+ efflux may also be critical (181, 182). NLRP3 activation causes activation of caspase-1 and cleavage of pro–IL-1β and pro–IL-18 to their activated forms. Caspase-1 also cleaves gasdermin D (GSDMD), which oligomerizes to form pores in the cell membrane, which may lead to extracellular release of IL-1 and IL-18, K+ efflux, and massive swelling with membrane rupture (pyroptosis). The DNA sensor cGAS binds to mtDNA released into the cytosol and catalyzes production of the secondary messenger cyclic GMP-AMP (cGAMP) from ATP and GTP. cAGMP binds to the ER membrane adaptor STING, which is displaced to the perinuclear endosome and activates TANK binding kinase 1 (TBK1), which, in turn, phosphorylates interferon regulatory factor 3 (IRF3) that enters the nucleus and enhances transcription of type I IFN and IFN-stimulated genes (ISGs), while also activating NF-κB. In cell culture, cGAS overexpression induces IFN-β, whereas cGAS knockdown erases IFN-β induction by DNA transfection (183). cGAS/STING activation also enhances autophagy (184) and induces cellular senescence (185). NF-κB is activated by diverse stress signals and pathways related to defense and survival (89, 186). Multiple stress signals activate the NF-κB dimer (composed of p65/p50 subunits) that translocates to the nucleus, where it binds to consensus sequences in regulatory regions of target genes (187). NF-κB activation drives multiple pleiotropic effects, including enhancing the transcription of proinflammatory mediators, such as TNF-α and IL-6, that regulate both innate and adaptive immunity (95). CARD, caspase recruitment domain; GSDMDNterm, GSDMD amino-terminal cell death domain; LRR, leucine-rich repeat; PYD, pyrin domain.
Studies have demonstrated important roles of the NLRP3 inflammasome in aging and age-related diseases, such as atherosclerosis metabolic syndrome, type 2 diabetes, and Alzheimer’s disease (81–83). Autophagy can clear mitochondria-related DAMPs, as well as pro–IL-1β and ubiquitinated inflammasome complexes, thereby reducing the proinflammatory environment (84). Indeed, autophagy inhibition increases free mtDNA in the cytosol and the production of IL-1β (85). Ablation of the NLRP3 inflammasome protects mice from age-related increases in innate immune activation, alterations in the CNS transcriptome, and astrogliosis, while improving glycemic control and motor performance (86). Loss-of-function mutations in the STING gene are protective from aging-related diseases, especially among smokers affected by chronic pulmonary diseases, who typically have a proinflammatory state (87). Conversely, rare STING gain-of-function mutations are associated with massive inflammation especially in lungs (88). NF-κB complexes have been found inside mitochondria, juxtaposed to the inner membrane, and there is evidence that the NF-κB pathway is activated during mitochondrial dysfunction and oxidative stress, although the mechanism of activation is not defined (89). The canonical NF-κB pathway is triggered by both endogenous factors and environmental factors, including diet and air pollution, mostly via stimulation of proinflammatory receptors, such as the TNF receptor superfamily and the IL-1 receptor (90). NF-κB activation has been observed in numerous diseases associated with aging (91), including sarcopenia (92), osteoporosis (93), and neurodegenerative diseases (94).
Application of multi-omics approaches to study inflammaging
Progress has been made in understanding the complex relationship between aging and acute and chronic inflammation mostly due to the expansion of tools available to measure and quantify immune function and biomarkers. In vivo characterization of inflammation in humans has been particularly useful in revealing the nuances of biological responses to different stimuli (95). The emerging picture is that inflammation is orchestrated by a large number of cell types and mediators and takes on a wide range of forms to best address the specific challenge (96). Inflammation covers a wide spectrum of biological manifestations that are dictated by the inflammatory stimulus, the stage in the immune response, and a variety of person-specific host factors. The challenge is using biomarkers to distinguish between different nuances of inflammation and to determine how responses change with aging so as to pinpoint more precise therapeutic avenues that effectively reduce the burden of age-related disease.
Efforts to characterize inflammation in humans have relied on one or a few inflammatory proteins, with IL-6, TNF-α, and CRP being the most widely used (5). The blood and tissue level of these proteins increases with age (97) and, to varying degrees, has been associated with adverse age-related outcomes, including physical frailty (98), cardiovascular disease (7, 99), dementia (100), and mortality (101). It is now understood that these proteins represent only a component of inflammaging (102). Because of the complexity and pleiotropic nature of inflammatory signaling cascades, and the dynamic expression of inflammatory proteins following immune stressors, measuring only a single or a small number of inflammatory proteins increases the likelihood that contradictory results will be obtained from otherwise comparable studies. Multi-omics and multiplexing technology have provided investigators with necessary tools to characterize inflammation with a higher degree of resolution.
Multiplex platforms that assess an array of inflammatory indicators are increasingly used to characterize the multidimensional nature of inflammation. By pairing measures from multiple platforms, including transcripts, epigenetic modulators, proteins, and metabolites, with supervised or unsupervised dimension reduction approaches, investigators are beginning to make inferences about distinct components of the inflammatory network in different pathological settings using circulating biomarkers. For example, with a panel of 19 proteins measured in the blood of older adults enrolled in a community-based study, Morrisette-Thomas and colleagues used a principal component analysis to identify two distinct components of immune signaling, both of which were associated with current and future chronic age-related disease: (a) a component characterized by simultaneous coexpression of pro- and antiinflammatory proteins that showed a strong positive correlation with age (top proteins include sTNF-R1, sTNR-R2, IL-6, and TNF-α) and (b) a component characterized by an innate immune response that was not correlated with age (top proteins include MCP, IL-12, IL-8, and MIP) (103). Similar studies have also observed the separation of inflammation into three components (102, 104). With advances in multiplexing, investigators can now measure hundreds of immune proteins and transcripts in various matrices (105, 106). A recent analysis that used bulk RNA sequencing at ten time points in 17 tissues across the mouse lifespan found that genes linked to immune response pathways were among the most differentially expressed with age across organ systems. Two clusters of immune response genes, which included immunoglobulin J chain, β2-microglobulin, and complement C1q A chain (C1QA), were upregulated well before other gene clusters during the transition from middle to late life. Changes in the expression of these immune gene networks were proposed to originate from the infiltration of T and B cells into a diverse set of organ systems, most prominently in white adipose tissue (107).
Several studies have characterized changes in multiple biomarkers following an initial immune challenge. An early study that examined the transcriptional expression response to bacterial endotoxin revealed that different functional networks emerge during the response time course (108). For example, some proinflammatory cytokines (TNF-α and IL-1β) and chemokines (CCL2 and CCL10) showed the strongest expression at 2 to 4 hours, whereas the peak expression of antiinflammatory markers occurred at the 4- to 6-hour mark. A downregulation of transcriptional modules linked to mitochondrial bioenergetics and protein synthesis was also noted, highlighting the breadth of biological changes that coincidentally occur with inflammation. The study of temporal dynamics of the inflammatory response to a challenge may uncover a proinflammatory diathesis prior to the development of full-fledged inflammaging.
Single-cell RNA sequencing studies have revealed complex patterns of common and organ-specific immune cell changes with aging in mice and humans (109–111). In particular, leukocytes in advanced age acquire a proinflammatory profile (IL-1β, CD14, TNFRSF12A) and show decreased expression of antiinflammatory markers (e.g., CD9, CD88) (110). Additionally, monocytes and effector memory CD4+ T cells become more abundant, whereas plasmacytoid dendritic cells, naive CD4+ T cells, and CD8+ T cells decline with aging (111–113). An age-related increase in GZMK+ CD8+ T cells, i.e., a clonal, exhausted-like, granzyme K–expressing T cell, was recently identified as a prominent feature of inflammaging that contributes to increased inflammatory cytokines by promoting the SASP (112).
Developing immune age scores is an essential step to defining inflammaging and its relationship with adverse health outcomes. A recent study of adults used proteins, transcripts, and immune cell counting to construct an immune age (IMM-AGE) score that explained a significant portion of the interindividual cytokine response variability (114). The authors suggested that the biological age of the immune system is a primary determinant of overall inflammatory signaling. The IMM-AGE score was a stronger predictor of mortality than a DNA methylation clock in the same cohort. An inflammatory age score derived from levels of 73 inflammatory plasma and CSF proteins was used to study the relationship between age-related inflammation and dementia risk (115). Proteins involved in the response to cytokine stimulus (e.g., TNFSF14) and chemotaxis (e.g., CCL3, CXCL9) were ranked highest in the inflammatory age score, which accounted for approximately 40% of the variance in chronological age. A higher inflammatory age score was associated with Alzheimer’s disease (AD) dementia diagnosis, AD pathology, and lower cognition, indicating that a well-designed immune age score may have diagnostic and prognostic utility.
Genetics has long been considered one potential determinant of inflammatory aging that operates independent of exogenous factors such as infection and environmental exposures. For example, one large-scale GWAS found a median heritability of 37% for a broad set of immune cell types and immune traits, with some phenotypes showing heritability as high as 96% (116, 117). The heritability for many serum cytokines/chemokines ranges from 50% to 90% in healthy twins between 8 and 82 years of age, with IL-6 and IL-12p40 levels being especially driven by genetics. However, monozygotic twins show age-related divergence of inflammatory biomarker levels, suggesting that the nonheritable influence on inflammation increases with time (118). Like basal cytokine levels, the stimulus-induced cytokine production in response to bacterial, fungal, viral, or a nonmicrobial challenge can also be highly influenced by genetic variation. However, the degree to which genetics contributes to cytokine responses varies considerably according to the inflammatory stimulus, suggesting a genetic influence on trigger-specific responses (119–121). For example, the proportion of variance in TNF-α levels explained by genome-wide SNP data was more than 70% after ex vivo stimulation with fungi and more than 50% after ex vivo stimulation with LPS. While genetic factors explained little of the variation seen in TNF-α levels after stimulation with bacteria, such as E. coli, a large proportion of variation in IL-6 and IL-22 levels after challenge with bacterial stimuli was determined by genetics (119). Thus, while some aspects of inflammaging are likely driven by genotype, there is also evidence for the role of gene-environment interactions.
The application of multi-omics technology and the emergence of systems immunology has improved our understanding of inflammation and its interaction with the aging process. Many questions remain unsolved; perhaps the most important is how to identify and validate biomarkers for distinct inflammation subtypes (e.g., acute vs. chronic vs. tissue remodeling) and how to specify aspects of inflammation that can be selectively targeted to prevent age-related diseases.
New proteomic technologies for studying biomarker signatures.
Proteomic technologies, including mass spectrometry–based proteomics, aptamer-based arrays (SomaLogic), and proximity extension assay technology (Olink), are now available for the large-scale quantification and validation of thousands of proteins in human biological fluids or tissues (122–127). Although the advances in aptamer and proximity extension assay technology have significantly expanded access to large-scale proteomics, there remain certain limitations to these approaches, including platform-specific measurement variation, incomplete target validation, a bias toward measurement of secreted proteins, and a limited breadth of protein measurement (128). A full characterization of protein variability also remains a major technical challenge, since the approximately 20,300 human coding genes can produce hundreds of thousands of protein variants through alternative splicing and posttranslational modification (PTM) (129, 130). Though many of these proteins and proteoforms are likely relevant mediators of inflammation, most are not quantified by current large-scale proteomic approaches. Emerging mass spectrometry–based workflows with new computational pipelines as well as recently developed PTM analyses are starting to address this knowledge gap (131, 132). A recent landmark study introduced the Blood Proteoform Atlas, a reference map of proteoforms in 21 cell types in human blood and bone marrow (133). This platform has uncovered that proteoforms have higher cell type specificity than protein-level measurements, laying the groundwork for possible dissection of the different arms of the inflammatory response. By leveraging the specificity of the top-down proteoform with the robustness of bottom-up proteomic approaches (134), a new generation of biomarker signatures will likely emerge that permit accurate profiling and quantification of complex biological phenomena such as aging and inflammaging.
Inflammation and the aging brain
The brain is an immune-privileged organ with highly regulated innate and adaptive immune processes designed to maintain homeostasis and quickly clear pathogens while limiting exposure to peripheral immune challenges. However, in the context of inflammaging and dysregulation of the neuro-immune axis, the brain becomes increasingly vulnerable. Immune activation, both within and outside of the CNS, has been observed in many age-related neurological diseases. Mechanisms by which age-related inflammation and neuroimmune activation jointly affect brain health may be targets for disease prevention.
The neuro-immune axis.
Inflammaging can influence homeostatic neurobiological processes and precipitate or perpetuate the development of neuropathology. The blood-brain barrier (BBB) prevents blood molecules from freely entering the CNS and is a key conduit through which inflammatory factors interact with brain targets. The BBB in the brain microvasculature is composed of a continuous endothelial cell layer connected by tight junctions and sheathed by pericytes, basal membranes, and perivascular astrocytes (135). By shielding the brain from pathogens, neurotoxins, and immune molecules, the BBB helps to protect the brain from chemical and biological stressors. BBB permeability becomes less selective with aging because of a loss of tight junctions and a shift from tightly regulated receptor-mediated transcytosis toward nonspecific transcytosis of plasma proteins (136, 137). Age-related inflammatory factors in blood, such as TNF-α, can further increase BBB permeability by suppressing the expression of tight junctions and enhancing brain endothelial cell adhesion, ultimately increasing neuroinflammation (138–140).
Circulating TNF-α, IL-1β, and IL-6 bind to receptors in endothelial cells and induce the expression of cellular adhesion proteins (e.g., VCAM1) that further promote inflammation by triggering NF-κB signaling (Figure 3) (136, 140, 141). This process facilitates tethering of circulating myeloid cells to the brain endothelia, augmenting the intravascular inflammatory state and promoting the activation of microglia, the brain’s innate immune cell (140). In parallel, the entry of inflammatory cytokines and chemokines occurs through active transport or nonspecific caveolar transcytosis (137, 142). Inflammatory proteins may enter the brain through other channels as well, including via the choroid plexus and circumventricular organs (143, 144).
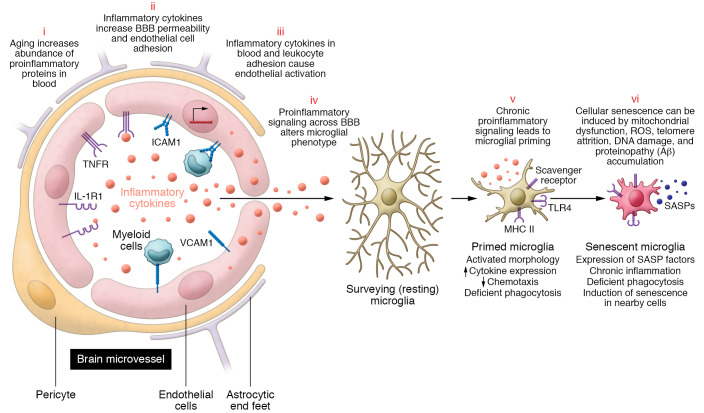
Figure 3. The role of inflammation in neurovascular and brain aging.
(i) Aging is associated with tissue-specific increases in proinflammatory protein expression, resulting in greater levels of inflammatory mediators (cytokines and chemokines) in peripheral circulation (blood). (ii) Inflammatory cytokines in blood signal cytokine receptors expressed in brain endothelial cells. This results in increased blood-brain barrier permeability and an upregulation of cellular adhesion molecules such as VCAM1. (iii) Inflammatory cytokines signal cytokine receptors on the luminal side of brain endothelial cells, leading to proinflammatory activation and leukocyte adhesion to vascular adhesion molecules, both of which promote proinflammatory endothelial activation. Together, these processes lead to an increased expression of inflammatory proteins in the brain parenchyma. (iv) Inflammatory proteins in the brain parenchyma can influence microglia and astrocyte phenotypes. For example, initial exposure of microglia to inflammatory mediators causes a transition from a surveilling homeostatic phenotype to an intermediate or activated phenotype. (v) Prolonged exposure to inflammatory signaling is believed to cause long-term microglial priming, characterized by exaggerated cytokine expression, reduced chemotaxis, and deficient phagocytosis. Primed microglia are more prone to aberrant expression in the context of disease. (vi) DNA damage, mitochondrial dysfunction, exposure to proteinaceous aggregates, and other forms of cellular stress can promote senescence of cells within the central nervous system, including brain endothelial cells and microglia. GFAP, glial fibrillary acidic protein; ICAM1, intercellular adhesion molecule 1; IL-1R1, IL-1 receptor type 1.
Peripheral inflammation, brain aging, and disease.
Many inflammatory mediators known to increase with aging (IL-1β, IL-18, sTNF-R1) are upregulated to an even greater extent in neurodegenerative conditions such as AD and Parkinson’s disease (PD) (100, 145–147). Studies have demonstrated that a proinflammatory state (148–150), inflammatory disease (e.g., rheumatoid arthritis) (151), and immune challenges (e.g., acute infection) (152, 153) all increase the risk for dementia over multiple decades. Genetic studies further support the role of immunity and inflammation in conditions such as AD, PD, and amyotrophic lateral sclerosis (ALS). For example, many polymorphisms associated with late-onset sporadic AD are in or near genes important for immunity, such as TREM2, CD33, CR1, GRN, and IL1RAP, and/or are expressed in brain immune cells including microglia (154). Counterintuitively, higher levels of IFN-γ and IL-12 (Th1 cytokines) were recently associated with slower cognitive decline in older adults, suggesting that the contribution of inflammatory signaling to neurodegenerative disease is complex and likely dependent on the inflammatory mediator in question (153).
Neuroinflammation, aging, and disease.
Neuroinflammation, a CNS-specific process characterized by proinflammatory cellular and molecular changes to microglia, astrocytes, and the BBB, is considered a core feature of aging and age-related neurological conditions, most notably AD, PD, and ALS (155). Age itself is associated with phenotypic changes to microglia, including deramified morphology, increased cytokine and chemokine expression, and an upregulation of MHC II and TLRs (156, 157). These changes, often referred to as microglial priming or sensitization, result in an amplified and prolonged glial inflammatory response after both central and peripheral immune challenges (158, 159). Astrocytes also demonstrate similar age-related changes, including elevated glial fibrillary acidic protein (GFAP) expression and hypertrophic morphology (160), both of which suggest a transition toward a proinflammatory state.
Aged glial cells respond less efficiently to antigens, including amyloid-β (Aβ) and α-synuclein, and have reduced phagocytic and antiinflammatory capacity (161–163). Delayed resolution of inflammation makes the brain particularly vulnerable to aberrant microglial responses to acute stressors, such as infection or tissue injury (164, 165). Microglia and astrocytes can also be primed and activated by age-related neuropathological processes, such as cerebrovascular lesions and protein aggregates (165, 166). Cellular senescence is yet another factor that can augment glial function and accelerate brain aging. Microglia, astrocytes, brain endothelial cells, and other CNS cell types show evidence of senescence and increased expression of senescence factors (167, 168). The expression of senescent factors, SASPs in particular, creates an inflammatory milieu within the brain believed to modulate neuroimmune processes while also compromising the integrity of the BBB (169, 170). The aggregation of misfolded proteins, including Aβ plaques and tau neurofibrillary tangles, may also promote neuronal and glial senescent phenotypes (171), as do other common facets of neurodegeneration, such as mitochondrial dysfunction and abnormal proteostasis (22, 172, 173). Although less direct, cellular senescence within the periphery and its associated SASP may also prime microglia and affect neurodegenerative processes through the immune-brain axis. Though each of these biological processes has been linked to neurodegeneration and cognitive decline, a causal role for cellular senescence in neurodegenerative disease has yet to be established. Still, many contend that removal of senescent cells within the CNS may be neuroprotective in older adults, particularly in the context of AD and PD (23, 174).
The extent to which neuroinflammation is either protective or pathogenic in the context of age-related changes is unknown and likely dependent on disease etiology and disease stage. For example, in AD and frontotemporal dementia, neuroinflammation and microglia activation potentiate the spreading of pathological tau neurofibrillary tangles (83, 175). In AD, this relationship may be mediated by NLRP3 inflammasome activation in microglia, a process that activates tau kinases (e.g., GSK-3β and CaMKII-α) and results in tau hyperphosphorylation (83). However, microglial activation may also be protective in these neurodegenerative conditions, for example by enhancing microglia phagocytosis of amyloid plaques in the context of AD (176–178). Much remains to be learned about how inflammaging and neuroinflammation interact to influence brain health and neurodegenerative disease.
Conclusions
Understanding the causes and consequences of inflammaging is one of the most active areas of research in aging and chronic diseases and one with powerful potential for translation. Work on biomarkers searching for signatures that discriminate between different aspects and triggers of inflammation is currently ongoing. Despite tremendous progress, the robustness and generalizability of developed signatures are not ready for clinical applications (179). Refining and validating these signatures in both animal models and humans and connecting them with specific biological mechanisms, age-related chronic diseases, and health outcomes remains a central research goal. An important question is whether signatures of circulating biomarkers will identify inflammatory processes that occur in different tissues and from different pathogenetic mechanisms. Progress in this field will require technological improvements in the measurement and analysis of multi-omics and rapid cycles of translation from model organisms to humans and vice versa to identify new promising therapeutic targets.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) and the Intramural Research Program, National Institute on Aging (NIA). NB was supported by Longevity Impetus Grants and the Office of Dietary Supplements Scholars Program. We thank Brigit Sullivan, NIH Library Editing Service.
Version 1. 07/15/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Walker et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2022;132(14):e158448. https://doi.org/10.1172/JCI158448.
Contributor Information
Keenan A. Walker, Email: keenan.walker@nih.gov.
Nathan Basisty, Email: nathan.basisty@nih.gov.
David M. Wilson, III, Email: dmwilson3@outlook.com.
Luigi Ferrucci, Email: ferruccilu@grc.nia.nih.gov.
References
- 1.Ferrucci L, et al. Measuring biological aging in humans: a quest. Aging Cell. 2020;19(2):e13080. doi: 10.1111/acel.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrucci L, et al. Time and the metrics of aging. Circ Res. 2018;123(7):740–744. doi: 10.1161/CIRCRESAHA.118.312816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franceschi C, et al. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14(10):576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 4.Ferrucci L, Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol. 2018;15(9):505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furman D, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25(12):1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bektas A, et al. A public health perspective of aging: do hyper-inflammatory syndromes such as COVID-19, SARS, ARDS, cytokine storm syndrome, and post-ICU syndrome accelerate short- and long-term inflammaging? Immun Ageing. 2020;17:23. doi: 10.1186/s12979-020-00196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridker PM, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolich-Zugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol. 2018;19(1):10–19. doi: 10.1038/s41590-017-0006-x. [DOI] [PubMed] [Google Scholar]
- 10.Williams SA, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25(12):1851–1857. doi: 10.1038/s41591-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorgoulis V, et al. Cellular senescence: defining a path forward. Cell. 2019;179(4):813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Segura A, et al. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28(6):436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. 2018;128(4):1238–1246. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppe JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basisty N, et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18(1):e3000599. doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Childs BG, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354(6311):472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23(6):775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demaria M, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7(2):165–176. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schafer MJ, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chinta SJ, et al. Cellular senescence is induced by the environmental neurotoxin paraquat and contributes to neuropathology linked to Parkinson’s Disease. Cell Rep. 2018;22(4):930–940. doi: 10.1016/j.celrep.2017.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bussian TJ, et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature. 2018;562(7728):578–582. doi: 10.1038/s41586-018-0543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat Neurosci. 2019;22(5):719–728. doi: 10.1038/s41593-019-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer AK, et al. Targeting senescent cells alleviates obesity-induced metabolic dysfunction. Aging Cell. 2019;18(3):e12950. doi: 10.1111/acel.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Zglinicki T, et al. Senescence in post-mitotic cells: a driver of aging? Antioxid Redox Signal. 2021;34(4):308–323. doi: 10.1089/ars.2020.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yousefzadeh MJ, et al. An aged immune system drives senescence and ageing of solid organs. Nature. 2021;594(7861):100–105. doi: 10.1038/s41586-021-03547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall BM, et al. Aging of mice is associated with p16(Ink4a)- and β-galactosidase-positive macrophage accumulation that can be induced in young mice by senescent cells. Aging (Albany NY) 2016;8(7):1294–1315. doi: 10.18632/aging.100991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall BM, et al. p16(Ink4a) and senescence-associated β-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging (Albany NY) 2017;9(8):1867–1884. doi: 10.18632/aging.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JY, et al. Cells exhibiting strong p16 INK4a promoter activation in vivo display features of senescence. Proc Natl Acad Sci U S A. 2019;116(7):2603–2611. doi: 10.1073/pnas.1818313116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frasca D. Senescent B cells in aging and age-related diseases: their role in the regulation of antibody responses. Exp Gerontol. 2018;107:55–58. doi: 10.1016/j.exger.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ong SM, et al. The pro-inflammatory phenotype of the human non-classical monocyte subset is attributed to senescence. Cell Death Dis. 2018;9(3):266. doi: 10.1038/s41419-018-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Covre LP, et al. The role of senescent T cells in immunopathology. Aging Cell. 2020;19(12):e13272. doi: 10.1111/acel.13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturmlechner I, et al. p21 produces a bioactive secretome that places stressed cells under immunosurveillance. Science. 2021;374(6567):eabb3420. doi: 10.1126/science.abb3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafer MJ, et al. The senescence-associated secretome as an indicator of age and medical risk. JCI Insight. 2020;5(12):e133668. doi: 10.1172/jci.insight.133668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka T, et al. Plasma proteomic signature of age in healthy humans. Aging Cell. 2018;17(5):e12799. doi: 10.1111/acel.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka T, et al. Plasma proteomic biomarker signature of age predicts health and life span. Elife. 2020;9:e61073. doi: 10.7554/eLife.61073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiley CD, et al. SILAC analysis reveals increased secretion of hemostasis-related factors by senescent cells. Cell Rep. 2019;28(13):3329–3337. doi: 10.1016/j.celrep.2019.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stojanović SD, et al. Senescence-induced inflammation: an important player and key therapeutic target in atherosclerosis. Eur Heart J. 2020;41(31):2983–2996. doi: 10.1093/eurheartj/ehz919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14(4):644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roos CM, et al. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell. 2016;15(5):973–977. doi: 10.1111/acel.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farr JN, et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat Med. 2017;23(9):1072–1079. doi: 10.1038/nm.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogrodnik M, et al. Cellular senescence drives age-dependent hepatic steatosis. Nat Commun. 2017;8:15691. doi: 10.1038/ncomms15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novais EJ, et al. Long-term treatment with senolytic drugs Dasatinib and Quercetin ameliorates age-dependent intervertebral disc degeneration in mice. Nat Commun. 2021;12(1):5213. doi: 10.1038/s41467-021-25453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hickson LJ, et al. Senolytics decrease senescent cells in humans: preliminary report from a clinical trial of Dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine. 2019;47:446–456. doi: 10.1016/j.ebiom.2019.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagiv A, Krizhanovsky V. Immunosurveillance of senescent cells: the bright side of the senescence program. Biogerontology. 2013;14(6):617–628. doi: 10.1007/s10522-013-9473-0. [DOI] [PubMed] [Google Scholar]
- 47.Covarrubias AJ, et al. Senescent cells promote tissue NAD(+) decline during ageing via the activation of CD38(+) macrophages. Nat Metab. 2020;2(11):1265–1283. doi: 10.1038/s42255-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers ES, et al. Recruitment of inflammatory monocytes by senescent fibroblasts inhibits antigen-specific tissue immunity during human aging. Nat Aging. 2021;1(1):101–113. doi: 10.1038/s43587-020-00010-6. [DOI] [PubMed] [Google Scholar]
- 49.Davalos AR, et al. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J Cell Biol. 2013;201(4):613–629. doi: 10.1083/jcb.201206006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman N, et al. DAMPs as mediators of sterile inflammation in aging-related pathologies. Ageing Res Rev. 2015;24(pt a):29–39. doi: 10.1016/j.arr.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Kirkland JL, et al. The clinical potential of senolytic drugs. J Am Geriatr Soc. 2017;65(10):2297–2301. doi: 10.1111/jgs.14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beerman I, et al. Short-term senolytic treatment: a paradigm to promote fracture repair during aging. J Clin Invest. 2022;132(8): a paradigm to promote fracture repair during aging. doi: 10.1172/JCI158871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burton DG, Krizhanovsky V. Physiological and pathological consequences of cellular senescence. Cell Mol Life Sci. 2014;71(22):4373–4386. doi: 10.1007/s00018-014-1691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grosse L, et al. Defined p16(High) senescent cell types are indispensable for mouse healthspan. Cell Metab. 2020;32(1):87–99. doi: 10.1016/j.cmet.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Casella G, et al. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019;47(21):11476. doi: 10.1093/nar/gkz879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernandez-Segura A, et al. Unmasking transcriptional heterogeneity in senescent cells. Curr Biol. 2017;27(17):2652–2660. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123(3):951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabbri E, et al. Insulin resistance is associated with reduced mitochondrial oxidative capacity measured by 31P-magnetic resonance spectroscopy in participants without diabetes from the Baltimore Longitudinal Study of Aging. Diabetes. 2017;66(1):170–176. doi: 10.2337/db16-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zampino M, et al. Poor mitochondrial health and systemic inflammation? Test of a classic hypothesis in the Baltimore Longitudinal Study of Aging. Geroscience. 2020;42(4):1175–1182. doi: 10.1007/s11357-020-00208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zampino M, et al. Cardiovascular health and mitochondrial function: testing an association. J Gerontol A Biol Sci Med Sci. 2021;76(2):361–367. doi: 10.1093/gerona/glaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi S, et al. 31P magnetic resonance spectroscopy assessment of muscle bioenergetics as a predictor of gait speed in the Baltimore Longitudinal Study of Aging. J Gerontol A Biol Sci Med Sci. 2016;71(12):1638–1645. doi: 10.1093/gerona/glw059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zane AC, et al. Muscle strength mediates the relationship between mitochondrial energetics and walking performance. Aging Cell. 2017;16(3):461–468. doi: 10.1111/acel.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian Q, et al. Muscle mitochondrial energetics predicts mobility decline in well-functioning older adults: the baltimore longitudinal study of aging. Aging Cell. 2022;21(2):e13552. doi: 10.1111/acel.13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rebelo-Marques A, et al. Aging hallmarks: the benefits of physical exercise. Front Endocrinol (Lausanne) 2018;9:258. doi: 10.3389/fendo.2018.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fakouri NB, et al. Toward understanding genomic instability, mitochondrial dysfunction and aging. FEBS J. 2019;286(6):1058–1073. doi: 10.1111/febs.14663. [DOI] [PubMed] [Google Scholar]
- 67.Schumacher B, et al. The central role of DNA damage in the ageing process. Nature. 2021;592(7856):695–703. doi: 10.1038/s41586-021-03307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abbotts R, et al. Coordination of DNA single strand break repair. Free Radic Biol Med. 2017;107:228–244. doi: 10.1016/j.freeradbiomed.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 70.Wiley CD, et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016;23(2):303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He M, et al. An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 2020;31(3):580–591. doi: 10.1016/j.cmet.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerutti R, et al. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 2014;19(6):1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang EF, et al. NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24(4):566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14(2):243–251. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rhee SG, et al. Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid Redox Signal. 2005;7(5-6):619–626. doi: 10.1089/ars.2005.7.619. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, et al. Mitochondria, oxidative stress and innate immunity. Front Physiol. 2018;9:1487. doi: 10.3389/fphys.2018.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galluzzi L, Vanpouille-Box C. BAX and BAK at the gates of innate immunity. Trends Cell Biol. 2018;28(5):343–345. doi: 10.1016/j.tcb.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 78.Jang JY, et al. The role of mitochondria in aging. J Clin Invest. 2018;128(9):3662–3670. doi: 10.1172/JCI120842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu CT, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M, et al. Surface-binding to cardiolipin nanodomains triggers cytochrome c pro-apoptotic peroxidase activity via localized dynamics. Structure. 2019;27(5):806–815. doi: 10.1016/j.str.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mastrocola R, et al. Metaflammation: tissue-specific alterations of the NLRP3 inflammasome platform in metabolic syndrome. Curr Med Chem. 2018;25(11):1294–1310. doi: 10.2174/0929867324666170407123522. [DOI] [PubMed] [Google Scholar]
- 82.Dixit VD. Nlrp3 inflammasome activation in type 2 diabetes: is it clinically relevant? Diabetes. 2013;62(1):22–24. doi: 10.2337/db12-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ising C, et al. NLRP3 inflammasome activation drives tau pathology. Nature. 2019;575(7784):669–673. doi: 10.1038/s41586-019-1769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun Q, et al. Inflammasome and autophagy regulation — a two-way street. Mol Med. 2017;23:188–195. doi: 10.2119/molmed.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13(3):255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Youm YH, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18(4):519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hamann L, et al. STING SNP R293Q is associated with a decreased risk of aging-related diseases. Gerontology. 2019;65(2):145–154. doi: 10.1159/000492972. [DOI] [PubMed] [Google Scholar]
- 88.Jeremiah N, et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest. 2014;124(12):5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Albensi BC. What is nuclear factor kappa B (NF-κB) doing in and to the mitochondrion? Front Cell Dev Biol. 2019;7:154. doi: 10.3389/fcell.2019.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bektas A, et al. Aging, inflammation and the environment. Exp Gerontol. 2018;105:10–18. doi: 10.1016/j.exger.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tilstra JS, et al. NF-κB in aging and disease. Aging Dis. 2011;2(6):449–465. [PMC free article] [PubMed] [Google Scholar]
- 92.Cai D, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119(2):285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 93.Chen Q, et al. DNA damage drives accelerated bone aging via an NF-κB-dependent mechanism. J Bone Miner Res. 2013;28(5):1214–1228. doi: 10.1002/jbmr.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steinman L. Nuanced roles of cytokines in three major human brain disorders. J Clin Invest. 2008;118(11):3557–3563. doi: 10.1172/JCI36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Medzhitov R. The spectrum of inflammatory responses. Science. 2021;374(6571):1070–1075. doi: 10.1126/science.abi5200. [DOI] [PubMed] [Google Scholar]
- 97.Parker D, et al. Age-related adverse inflammatory and metabolic changes begin early in adulthood. J Gerontol A Biol Sci Med Sci. 2019;74(3):283–289. doi: 10.1093/gerona/gly121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walker KA, et al. Midlife systemic inflammation is associated with frailty in later life: the ARIC Study. J Gerontol A Biol Sci Med Sci. 2019;74(3):343–349. doi: 10.1093/gerona/gly045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ridker PM, et al. Novel risk factors for systemic atherosclerosis: a comparison of C-reactive protein, fibrinogen, homocysteine, lipoprotein(a), and standard cholesterol screening as predictors of peripheral arterial disease. JAMA. 2001;285(19):2481–2485. doi: 10.1001/jama.285.19.2481. [DOI] [PubMed] [Google Scholar]
- 100.Shen XN, et al. Inflammatory markers in Alzheimer’s disease and mild cognitive impairment: a meta-analysis and systematic review of 170 studies. J Neurol Neurosurg Psychiatry. 2019;90(5):590–598. doi: 10.1136/jnnp-2018-319148. [DOI] [PubMed] [Google Scholar]
- 101.Varadhan R, et al. Simple biologically informed inflammatory index of two serum cytokines predicts 10 year all-cause mortality in older adults. J Gerontol A Biol Sci Med Sci. 2014;69(2):165–173. doi: 10.1093/gerona/glt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bandeen-Roche K, et al. Measuring systemic inflammatory regulation in older adults: evidence and utility. Rejuvenation Res. 2009;12(6):403–410. doi: 10.1089/rej.2009.0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morrisette-Thomas V, et al. Inflamm-aging does not simply reflect increases in pro-inflammatory markers. Mech Ageing Dev. 2014;139:49–57. doi: 10.1016/j.mad.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kip KE, et al. Global inflammation predicts cardiovascular risk in women: a report from the Women’s Ischemia Syndrome Evaluation (WISE) study. Am Heart J. 2005;150(5):900–906. doi: 10.1016/j.ahj.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 105.Kim CH, et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep. 2018;8(1):8382. doi: 10.1038/s41598-018-26640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Niewczas MA, et al. A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med. 2019;25(5):805–813. doi: 10.1038/s41591-019-0415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaum N, et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature. 2020;583(7817):596–602. doi: 10.1038/s41586-020-2499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 109.Kimmel JC, et al. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 2019;29(12):2088–2103. doi: 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tabula Muris C. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583(7817):590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mogilenko DA, et al. Immune ageing at single-cell resolution. Nat Rev Immunol. doi: 10.1038/s41577-021-00646-4. [published online November 23, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mogilenko DA, et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity. 2021;54(1):99–115. doi: 10.1016/j.immuni.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 113.Li M, et al. Age related human T cell subset evolution and senescence. Immun Ageing. 2019;16:24. doi: 10.1186/s12979-019-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alpert A, et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat Med. 2019;25(3):487–495. doi: 10.1038/s41591-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cullen NC, et al. Accelerated inflammatory aging in Alzheimer’s disease and its relation to amyloid, tau, and cognition. Sci Rep. 2021;11(1):1965. doi: 10.1038/s41598-021-81705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Orru V, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52(10):1036–1045. doi: 10.1038/s41588-020-0684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roederer M, et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161(2):387–403. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160(1-2):37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Y, et al. A functional genomics approach to understand variation in cytokine production in humans. Cell. 2016;167(4):1099–1110. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 120.Sliz E, et al. Genome-wide association study identifies seven novel loci associating with circulating cytokines and cell adhesion molecules in Finns. J Med Genet. 2019;56(9):607–616. doi: 10.1136/jmedgenet-2018-105965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ahola-Olli AV, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100(1):40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11(6):O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rosenberger G, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Collins BC, et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat Commun. 2017;8(1):291. doi: 10.1038/s41467-017-00249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rice SJ, et al. Absolute quantification of all identified plasma proteins from SWATH data for biomarker discovery. Proteomics. 2019;19(3):e1800135. doi: 10.1002/pmic.201800135. [DOI] [PubMed] [Google Scholar]
- 126.Lundberg M, et al. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39(15):e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pietzner M, et al. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat Commun. 2021;12(1):6822. doi: 10.1038/s41467-021-27164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith LM, et al. The Human Proteoform Project: defining the human proteome. Sci Adv. 2021;7(46):eabk0734. doi: 10.1126/sciadv.abk0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aebersold R, et al. How many human proteoforms are there? Nat Chem Biol. 2018;14(3):206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Basisty N, et al. Simultaneous quantification of the acetylome and succinylome by ‘one-pot’ affinity enrichment. Proteomics. 2018;18(17):e1800123. doi: 10.1002/pmic.201800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10(7):634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Melani RD, et al. The Blood Proteoform Atlas: a reference map of proteoforms in human hematopoietic cells. Science. 2022;375(6579):411–418. doi: 10.1126/science.aaz5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ntai I, et al. Integrated bottom-up and top-down proteomics of patient-derived breast tumor xenografts. Mol Cell Proteomics. 2016;15(1):45–56. doi: 10.1074/mcp.M114.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Erdő F, et al. Age-associated physiological and pathological changes at the blood-brain barrier: a review. J Cereb Blood Flow Metab. 2017;37(1):4–24. doi: 10.1177/0271678X16679420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Elahy M, et al. Blood-brain barrier dysfunction developed during normal aging is associated with inflammation and loss of tight junctions but not with leukocyte recruitment. Immun Ageing. 2015;12:2. doi: 10.1186/s12979-015-0029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang AC, et al. Physiological blood-brain transport is impaired with age by a shift in transcytosis. Nature. 2020;583(7816):425–430. doi: 10.1038/s41586-020-2453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lv S, et al. Tumour necrosis factor-alpha affects blood-brain barrier permeability and tight junction-associated occludin in acute liver failure. Liver Int. 2010;30(8):1198–1210. doi: 10.1111/j.1478-3231.2010.02211.x. [DOI] [PubMed] [Google Scholar]
- 139.Wong D, et al. Adhesion and migration of polymorphonuclear leukocytes across human brain microvessel endothelial cells are differentially regulated by endothelial cell adhesion molecules and modulate monolayer permeability. J Neuroimmunol. 2007;184(1-2):136–148. doi: 10.1016/j.jneuroim.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 140.Yousef H, et al. Aged blood impairs hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1. Nat Med. 2019;25(6):988–1000. doi: 10.1038/s41591-019-0440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang J, et al. Regulation of endothelial cell adhesion molecule expression by mast cells, macrophages, and neutrophils. PLoS One. 2011;6(1):e14525. doi: 10.1371/journal.pone.0014525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Licinio J, Wong ML. Pathways and mechanisms for cytokine signaling of the central nervous system. J Clin Invest. 1997;100(12):2941–2947. doi: 10.1172/JCI119846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fleischer V, et al. Translational value of choroid plexus imaging for tracking neuroinflammation in mice and humans. Proc Natl Acad Sci U S A. 2021;118(36):e2025000118. doi: 10.1073/pnas.2025000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sternberg EM. Neural-immune interactions in health and disease. J Clin Invest. 1997;100(11):2641–2647. doi: 10.1172/JCI119807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brosseron F, et al. Body fluid cytokine levels in mild cognitive impairment and Alzheimer’s disease: a comparative overview. Mol Neurobiol. 2014;50(2):534–544. doi: 10.1007/s12035-014-8657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lai KSP, et al. Peripheral inflammatory markers in Alzheimer’s disease: a systematic review and meta-analysis of 175 studies. J Neurol Neurosurg Psychiatry. 2017;88(10):876–882. doi: 10.1136/jnnp-2017-316201. [DOI] [PubMed] [Google Scholar]
- 147.Brosseron F, et al. Multicenter Alzheimer’s and Parkinson’s disease immune biomarker verification study. Alzheimers Dement. 2020;16(2):292–304. doi: 10.1016/j.jalz.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 148.Darweesh SKL, et al. Inflammatory markers and the risk of dementia and Alzheimer’s disease: a meta-analysis. Alzheimers Dement. 2018;14(11):1450–1459. doi: 10.1016/j.jalz.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 149.Gross AL, et al. Plasma markers of inflammation linked to clinical progression and decline during preclinical AD. Front Aging Neurosci. 2019;11:229. doi: 10.3389/fnagi.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walker KA, et al. Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat Aging. 2021;1(5):473–489. doi: 10.1038/s43587-021-00064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhou M, et al. Tumor necrosis factor (TNF) blocking agents are associated with lower risk for Alzheimer’s disease in patients with rheumatoid arthritis and psoriasis. PLoS One. 2020;15(3):e0229819. doi: 10.1371/journal.pone.0229819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sipilä PN, et al. Hospital-treated infectious diseases and the risk of dementia: a large, multicohort, observational study with a replication cohort. Lancet Infect Dis. 2021;21(11):1557–1567. doi: 10.1016/S1473-3099(21)00144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang HS, et al. Plasma IL-12/IFN-γ axis predicts cognitive trajectories in cognitively unimpaired older adults. Alzheimers Dement. 2021;18(4):645–653. doi: 10.1002/alz.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Takatori S, et al. Genetic risk factors for Alzheimer disease: emerging roles of microglia in disease pathomechanisms. Adv Exp Med Biol. 2019;1118:83–116. doi: 10.1007/978-3-030-05542-4_5. [DOI] [PubMed] [Google Scholar]
- 155.Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 156.Letiembre M, et al. Innate immune receptor expression in normal brain aging. Neuroscience. 2007;146(1):248–254. doi: 10.1016/j.neuroscience.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 157.Edler MK, et al. Microglia in aging and Alzheimer’s disease: a comparative species review. Cells. 2021;10(5):1138. doi: 10.3390/cells10051138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cunningham C, et al. Central and systemic endotoxin challenges exacerbate the local inflammatory response and increase neuronal death during chronic neurodegeneration. J Neurosci. 2005;25(40):9275–9284. doi: 10.1523/JNEUROSCI.2614-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cunningham C. Microglia and neurodegeneration: the role of systemic inflammation. Glia. 2013;61(1):71–90. doi: 10.1002/glia.22350. [DOI] [PubMed] [Google Scholar]
- 160.Norden DM, Godbout JP. Review: Microglia of the aged brain: primed to be activated and resistant to regulation. Neuropathol Appl Neurobiol. 2013;39(1):19–34. doi: 10.1111/j.1365-2990.2012.01306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hickman SE, et al. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci. 2008;28(33):8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Labzin LI, et al. Innate immunity and neurodegeneration. Annu Rev Med. 2018;69:437–449. doi: 10.1146/annurev-med-050715-104343. [DOI] [PubMed] [Google Scholar]
- 163.Wynne AM, et al. Protracted downregulation of CX3CR1 on microglia of aged mice after lipopolysaccharide challenge. Brain Behav Immun. 2010;24(7):1190–1201. doi: 10.1016/j.bbi.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cunningham C, Hennessy E. Co-morbidity and systemic inflammation as drivers of cognitive decline: new experimental models adopting a broader paradigm in dementia research. Alzheimers Res Ther. 2015;7(1):33. doi: 10.1186/s13195-015-0117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hoeijmakers L, et al. Microglial priming and Alzheimer’s disease: a possible role for (early) immune challenges and epigenetics? Front Hum Neurosci. 2016;10:398. doi: 10.3389/fnhum.2016.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haley MJ, et al. Microglial priming as trained immunity in the brain. Neuroscience. 2019;405:47–54. doi: 10.1016/j.neuroscience.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 167.Baker DJ, Petersen RC. Cellular senescence in brain aging and neurodegenerative diseases: evidence and perspectives. J Clin Invest. 2018;128(4):1208–1216. doi: 10.1172/JCI95145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yu HM, et al. Repeated lipopolysaccharide stimulation induces cellular senescence in BV2 cells. Neuroimmunomodulation. 2012;19(2):131–136. doi: 10.1159/000330254. [DOI] [PubMed] [Google Scholar]
- 169.Graves SI, Baker DJ. Implicating endothelial cell senescence to dysfunction in the ageing and diseased brain. Basic Clin Pharmacol Toxicol. 2020;127(2):102–110. doi: 10.1111/bcpt.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sahu MR, et al. Cellular senescence in the aging brain: a promising target for neurodegenerative diseases. Mech Ageing Dev. 2022;204:111675. doi: 10.1016/j.mad.2022.111675. [DOI] [PubMed] [Google Scholar]
- 171.Magini A, et al. Abnormal cortical lysosomal β-hexosaminidase and β-galactosidase activity at post-synaptic sites during Alzheimer’s disease progression. Int J Biochem Cell Biol. 2015;58:62–70. doi: 10.1016/j.biocel.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 172.Ashleigh T, et al. The role of mitochondrial dysfunction in Alzheimer’s disease pathogenesis. Alzheimers Dement. doi: 10.1002/alz.12683. [published online May 6, 2022]. [DOI] [PubMed] [Google Scholar]
- 173.Rai M, et al. Contribution of proteases to the hallmarks of aging and to age-related neurodegeneration. Aging Cell. 2022;21(5):e13603. doi: 10.1111/acel.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gonzales MM, et al. Senolytic therapy to modulate the progression of Alzheimer’s Disease (SToMP-AD): a pilot clinical trial. J Prev Alzheimers Dis. 2022;9(1):22–29. doi: 10.14283/jpad.2021.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pascoal TA, et al. Microglial activation and tau propagate jointly across Braak stages. Nat Med. 2021;27(9):1592–1599. doi: 10.1038/s41591-021-01456-w. [DOI] [PubMed] [Google Scholar]
- 176.Ewers M, et al. Increased soluble TREM2 in cerebrospinal fluid is associated with reduced cognitive and clinical decline in Alzheimer’s disease. Sci Transl Med. 2019;11(507):eaav6221. doi: 10.1126/scitranslmed.aav6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ewers M, et al. Higher CSF sTREM2 and microglia activation are associated with slower rates of beta-amyloid accumulation. EMBO Mol Med. 2020;12(9):e12308. doi: 10.15252/emmm.202012308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reifschneider A, et al. Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J. 2022;41(4):e109108. doi: 10.15252/embj.2021109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Moaddel R, et al. Proteomics in aging research: a roadmap to clinical, translational research. Aging Cell. 2021;20(4):e13325. doi: 10.1111/acel.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.He Y, et al. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530(7590):354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Munoz-Planillo R, et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38(6):1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhong Z, et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560(7717):198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sun L, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gui X, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yang H, et al. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A. 2017;114(23):E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lingappan K. NF-κB in oxidative stress. Curr Opin Toxicol. 2018;7:81–86. doi: 10.1016/j.cotox.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Barnabei L, et al. NF-κB: at the borders of autoimmunity and inflammation. Front Immunol. 2021;12:716469. doi: 10.3389/fimmu.2021.716469. [DOI] [PMC free article] [PubMed] [Google Scholar]