SUMMARY
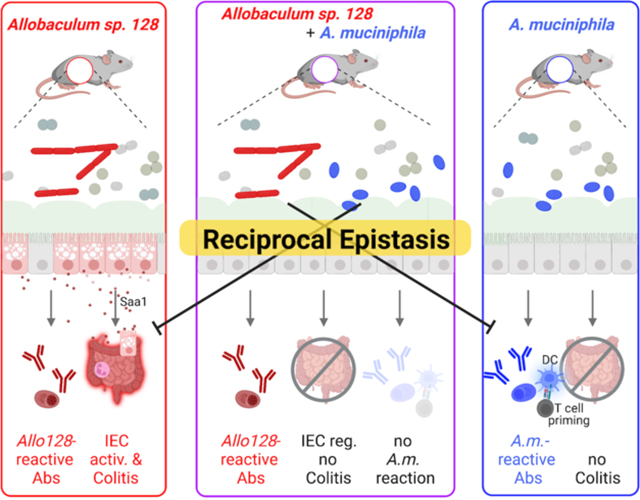
The impacts of individual commensal microbes on immunity and disease can differ dramatically depending on the surrounding microbial context, yet the specific bacterial combinations that dictate divergent immunological outcomes remain largely undefined. Here, we characterize an immunostimulatory Allobaculum species from an inflammatory bowel disease patient that exacerbates colitis in gnotobiotic mice. Allobaculum inversely associates with the taxonomically-divergent immunostimulatory species Akkermansia muciniphila in human microbiota-associated mice and human cohorts. Co-colonization with A. muciniphila ameliorates Allobaculum-induced intestinal epithelial cell activation and colitis in mice, while Allobaculum blunts the A. muciniphila-specific systemic antibody response and reprograms the immunological milieu in mesenteric lymph nodes by blocking A. muciniphila-induced dendritic cell activation and T cell expansion. These studies thus identify a pairwise reciprocal interaction between human gut bacteria that dictates divergent immunological outcomes. Furthermore, they establish a generalizable framework to define the contextual cues contributing to the ‘incomplete penetrance’ of microbial impacts on human disease.
Keywords: Gut microbiota, immunostimulatory commensals, mucosal immunity, inflammatory bowel disease, human gut bacteria, reciprocal epistasis, mesenteric lymph nodes, intestinal epithelium
Graphical Abstract
eTOC blurb
Microbial community context can critically alter commensal-induced immune responses. Here, Rice et al. describe a reciprocal interaction between a colitogenic human Allobaculum species and Akkermansia muciniphila that uniquely reprograms the immune responses elicited by either microbe alone, revealing non-linear impacts of interspecies interactions on host-immunity.
INTRODUCTION
Individual gut microbiota strains can influence diverse human phenotypes, but our understanding of the particular commensals that play causal roles in human health and disease remains limited. We and others have leveraged antigen-specific mucosal and systemic antibody responses to the commensal microbiota to identify specific commensal taxa that induce adaptive immune responses and critically exacerbate or ameliorate susceptibility to mouse models of human diseases (Kau et al., 2015; Palm & de Zoete et al., 2014; Viladomiu et al. 2017). For example, ‘pathogenic’ immunostimulatory bacteria can play potentially causal roles in inflammatory bowel disease (IBD), autoimmunity, and malnutrition, while ‘beneficial’ immunostimulatory species have been employed to treat metabolic syndrome and to augment cancer immunotherapy (Atarashi et al., 2015; Atarashi et al., 2017; Brown et al., 2015; Kau et al., 2015; Plovier et al., 2017; Routy et al., 2018; Zegarra-Ruiz et al., 2019). Nonetheless, potentially disease-driving bacteria are also found in apparently healthy individuals, and the effects of putative beneficial strains vary widely between subjects (Buffie et al., 2015; Ji et al., 2020; McDonald et al., 2018). Thus, the predictive power of strain carriage alone remains poor even for microbes with well-characterized disease-modulating activities, which severely hampers the accuracy of microbiome-based prognostics and constrains the efficacy of existing and emerging live bio-therapeutics.
One potentially key contributor to the ‘incomplete penetrance’ of microbial impacts on disease is that the effects of individual strains on host immunity can differ dramatically depending on the surrounding microbial community context (Belkaid et al., 2017; Buffie et al., 2013; Externest et al., 2000; Gould et al. 2018). However, the specific rules dictating these differential outcomes in humans remain almost entirely unclear. Here, by studying a unique colitogenic taxon isolated from IBD patients, we uncovered a reciprocal interaction between two phylogenetically distinct immunostimulatory taxa. Notably, co-colonization of gnotobiotic mice with these two unique immunogenic strains dramatically altered the immune responses elicited by each strain on its own. Borrowing from concepts in genetics, we refer to the relationship between these microbes as ‘epistatic’ to encapsulate the non-linear impacts of this inter-species interaction on host phenotypes. Thus, these studies begin to decode the specific contextual cues that may underlie interindividual variation in responses to immunostimulatory strains and establish a generalizable framework for identifying specific microbial combinations that dictate the context-dependent impacts of commensal microbes on human health and disease.
RESULTS
Immunostimulatory Allobaculum strains from IBD patients exacerbate colitis in simplified gnotobiotic mouse models
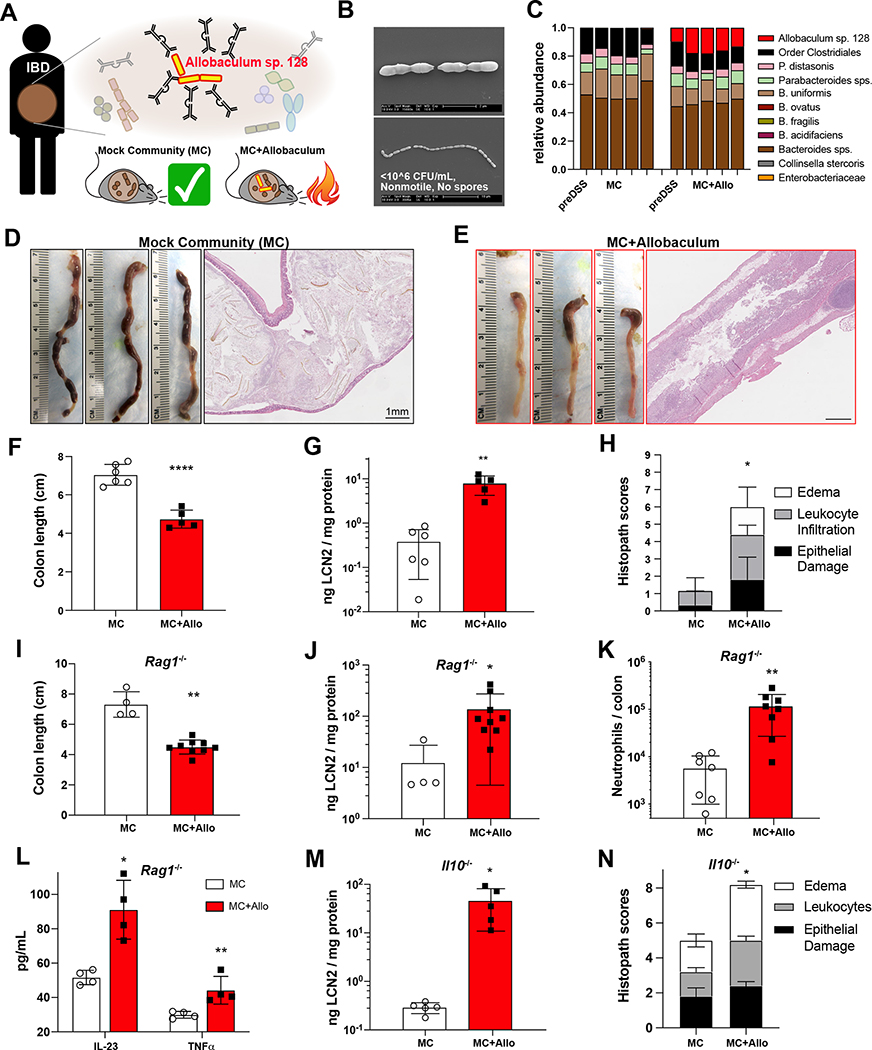
We previously identified and isolated a highly immunoglobulin A (IgA)-coated strain from the genus Allobaculum from the gut microbiota of an ulcerative colitis (UC) patient (Fig. 1A; Palm & de Zoete et al., 2014). This isolate, hereafter referred to as Allobaculum sp. 128, is culturable under strict anaerobic conditions, and is nonmotile and non-spore-forming (Fig. 1B). Based on full-length 16S rRNA gene sequence similarity and whole-genome sequencing, this strain is a member of an unnamed species from the genus Allobaculum and the prevalent, yet poorly characterized, family Erysipelotrichaceae (Greetham et al., 2004; Ha et al., 2020; Miyauchi et al., 2020). Because we found previously that IgA coating marks potentially colitogenic strains in human IBD patients, we set out to establish a reductionist gnotobiotic mouse model to examine the individual impact of this unique immunogenic strain on immunity and colitis. We colonized germ-free wild-type (WT) C57BL/6 mice with a non-colitogenic mock community (MC) of nine human gut bacteria representing three major phyla in the human gut microbiota (Palm & de Zoete et al., 2014) with or without Allobaculum sp. 128, and induced colitis by administering dextran sodium sulfate (DSS). We found that WT mice colonized with MC and Allobaculum sp. 128 exhibited severe, sometimes lethal, inflammation characterized by reduced colon length, elevated fecal lipocalin, and increased leukocyte infiltration, while mice colonized with MC alone showed only limited disease (Fig. 1C–1H). Notably, Allobaculum sp. 128 abundance remained stable during colitis, suggesting that Allobaculum sp. 128 does not “bloom” in response to inflammation (Fig. S1A–S1B). Allobaculum sp. 128 colonization also exacerbated DSS colitis in gnotobiotic Rag1−/− mice, demonstrating that the colitogenic activities of Allobaculum do not strictly require T or B cells in the acute DSS model (Fig. 1I–1L and S1C). Nonetheless, Allobaculum sp. 128 colonization also drove spontaneous disease development and expansion of Th17 cells in gnotobiotic Il10−/− mice (Fig. 1M–1N) (Fig. S1D–S1E). Finally, monocolonization with Allobaculum sp. 128 was sufficient to exacerbate acute DSS colitis as compared to a non-immunogenic strain from the family Erysipelotrichaceae (Fig. S1F–S1G).
Figure 1. An Allobaculum species from an ulcerative colitis patient exacerbates acute and chronic colitis in gnotobiotic mice.
(A) Identification and Isolation of IgA-coated Allobaculum sp. 128 from an ulcerative colitis patient. (B) Scanning electron micrographs of Allobaculum sp. 128 in vitro. Scale bars, 2μm (top), 10μm (bottom). (C-H) Germ-free WT mice were gavaged and colonized for seven days before treatment with 2% DSS-H2O ad libitum. (C) Fecal microbiota on d0 (first bar) and d7 (bars 2–5) of DSS. (D-E) Colons on d7 and representative H&E sections. Scale bars, 1mm. (F) Colon length on d7. (G) Fecal lipocalin (LCN2) on d2. (H) Histopathology scores. (I-L) Acute DSS colitis in Rag1−/− gnotobiotic mice: colon length (I), d2 fecal lipocalin (J), lamina propria CD45+Ly6G+ neutrophils (K), and colon explant cytokines (L). (M-N) Spontaneous colitis in Il10−/− gnotobiotic mice: fecal LCN2 (M) and colon histopathology scores (N). Welch’s t-test was used to compare microbiota groups. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. Error bars show mean ± SEM. (C-H) show one of N=6 independent experiments, n=5–6 mice per group. (I-L) show one of N=4 independent experiments, n=4–9 mice/group. (M-N) show one of N=3 independent experiments, n=5 mice/group.
To test whether other immunostimulatory strains from the genus Allobaculum also exhibit colitogenic activities, we identified a related Allobaculum strain in a UC patient from a separate IBD cohort and isolated this strain via high-throughput anaerobic culturomics and massively-parallel 16S rRNA gene sequencing. The full-length 16S rRNA sequence of this unique Allobaculum isolate was 95.2% similar to Allobaculum sp. 128, with a genome-level pairwise average nucleotide identity of 0.80, indicating that these are two different species within genus Allobaculum (Fig. S1H). Nonetheless, similar to the initial Allobaculum isolate, this second isolate also exacerbated DSS colitis in a simplified gnotobiotic mouse model (Fig. S1I–S1K).
Allobaculum sp. 128 elicits mucosal and systemic antibody responses at steady state
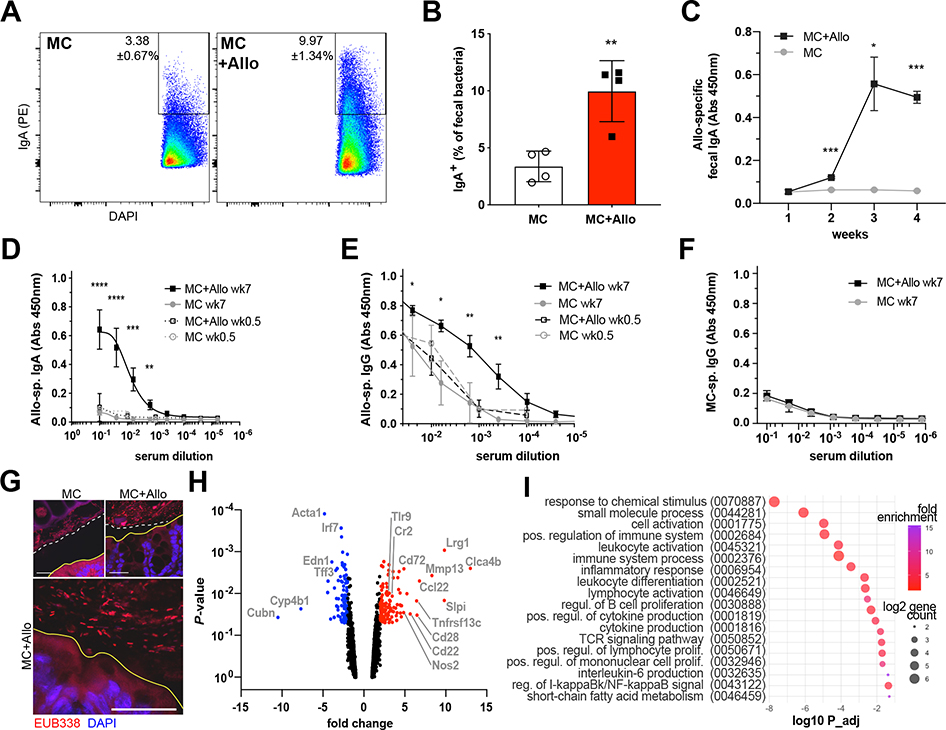
WT gnotobiotic mice colonized with Allobaculum sp. 128 showed no apparent intestinal inflammation in the absence of DSS treatment up to 12 weeks after colonization (Fig. S2A–S2B). However, because this strain was identified based on high levels of IgA coating in humans, we sought to directly interrogate the ability of Allobaculum to induce antigen-specific antibody responses in gnotobiotic mice in the absence of overt pathology. As expected, we found that WT gnotobiotic mice colonized with MC plus Allobaculum sp. 128 mounted a potent Allobaculum sp. 128-specific secretory IgA response (Fig. 2A–2C).
Figure 2. Allobaculum sp. 128 elicits mucosal and systemic immunity at steady state.
(A-B) GF WT mice colonized with MC or MC+Allobaculum sp. 128 were analyzed for fecal bacteria IgA coating (n=4 per group). (C) Allobaculum sp. 128-specific fecal water IgA and (D-E) serum IgA & IgG. (F) MC-specific serum IgG (n=3–6 mice per group). Welch’s t-test was used to compare microbiota groups at week 7. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. Data shown in (A-F) are representative of N=2 independent experiments. (G) Colon sections stained with bacterial FISH probe EUB338 and DAPI. Scale bars, 25μm. Solid yellow line: epithelial brush border; white dashed line: inner mucus layer. N=2 independent experiments with n=2 mice/group. (H) Bulk colon RNAseq at 4wks colonization (n=2–3 per group). Volcano plot of significantly differentially expressed genes and (I) gene ontology pathway enrichment for MC+Allo vs MC. Data shown in (H-I) is from N=1 experiment.
Select highly IgA-coated commensal strains, including gamma-Proteobacteria and Akkermansia muciniphila, also induce systemic IgA and IgG responses at steady state (Ansaldo et al., 2019; Castro-Dopico et al. 2019; Wilmore et al., 2018; Zeng et al., 2016). Similarly, Allobaculum sp. 128 colonization induced robust Allobaculum sp. 128-specific serum IgA and IgG responses at steady state (Fig. 2D–2E). However, systemic antibody responses to other strains in the mock community remained unchanged in the presence or absence of Allobaculum sp. 128 (Fig. 2F), as did total serum Ig titers and gut barrier permeability (Fig. S2C–S2E). Nonetheless, we observed a slight increase in Allobaculum sp. 128-specific fecal IgG and a modest decrease in colonic mucus thickness in Allobaculum sp. 128 colonized mice (Fig. S2F–S2G). Finally, consistent with the ability of Allobaculum to induce antigen-specific antibody responses, we also observed the presence of bacteria closely opposed to the colonic epithelium in MC + Allobaculum sp. 128 colonized mice (Fig. 2G).
To examine the Allobaculum sp. 128-induced mucosal response in an unbiased manner, we performed bulk RNA-seq on colon tissue isolated from gnotobiotic mice colonized with MC or MC + Allobaculum sp. 128. Gene ontology analysis of differentially expressed genes revealed an enrichment of genes involved in cytokine production, adaptive immune activation, and leukocyte proliferation in mice colonized with Allobaculum sp. 128 (Fig. 2H–2I), underscoring the immunostimulatory effects of this strain. Together, these data demonstrate that Allobaculum sp. 128 evokes antigen-specific mucosal and systemic antibody responses, as well as low-level intestinal inflammation, but is insufficient to trigger a wholesale disruption of the epithelial barrier or cause overt intestinal pathology on its own in wild-type mice.
Allobaculum is inversely correlated with the phylogenetically-divergent immunostimulatory commensal Akkermansia muciniphila
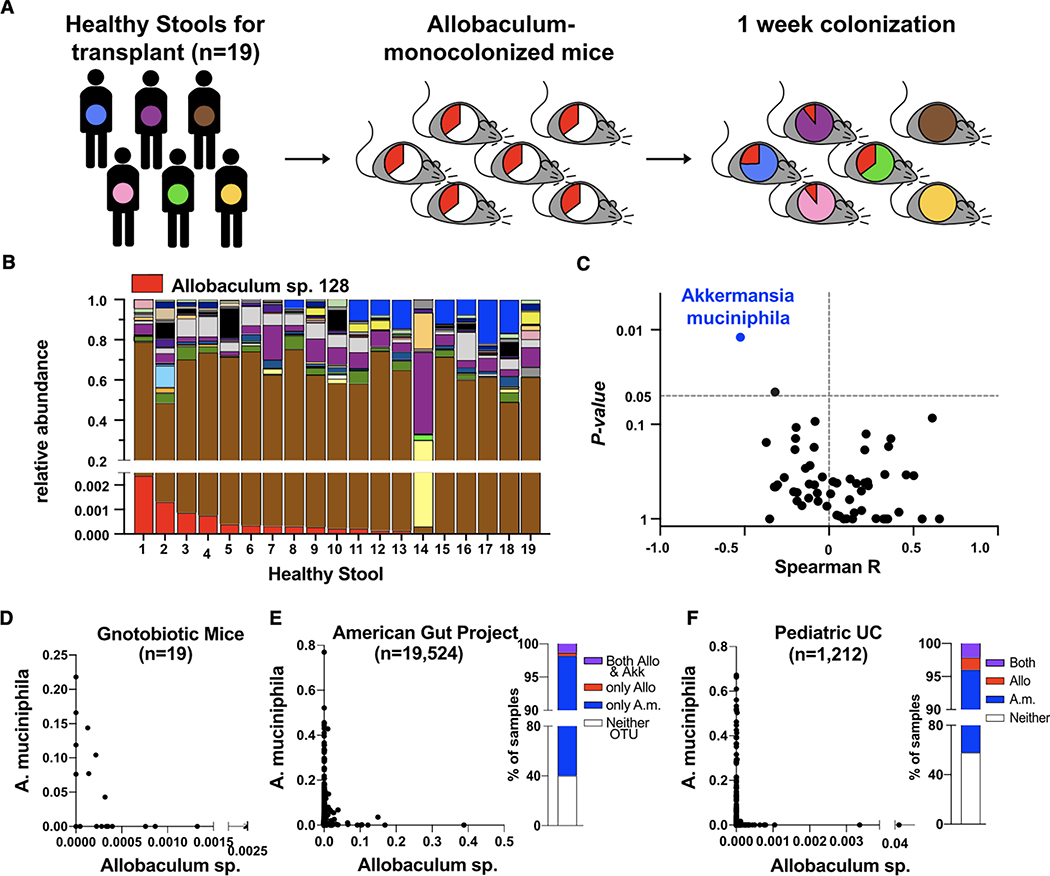
Our gnotobiotic mouse data suggest that immunostimulatory Allobaculum strains may play potentially causal roles in IBD. However, we also detected related Allobaculum strains in a meta-analysis of microbiome data from ostensibly healthy humans (American Gut Project) (McDonald et al., 2018; Table S1). One potential explanation for this observation is that specific microbial taxa present in healthy humans may protect against the colitogenic effects of Allobaculum. To begin to examine this hypothesis, we established a human microbiota-associated gnotobiotic mouse-based screen to reveal potential relationships between Allobaculum and diverse bacterial taxa from the human gut microbiota. Briefly, we mono-colonized individually housed germ-free mice with Allobaculum sp. 128 for 24 hours before gavaging each mono-colonized mouse with one of 19 different healthy human stool samples. After seven days, we evaluated microbial community composition in all mice via 16S rRNA gene sequencing (Fig. 3A–3B). As expected, mice colonized with different human samples harbored distinct microbial communities. Furthermore, we observed a range of Allobaculum sp. 128 colonization levels across these 19 unique community contexts (Fig. 3B). This variation in Allobaculum sp. 128 abundance was not due to variation in overall microbial diversity as there were no significant differences in richness or evenness between samples containing Allobaculum sp. 128 and those lacking Allobaculum sp. 128 (Fig. S3A). Thus, we hypothesized that specific microbial taxa may impact Allobaculum sp. 128 carriage or abundance. To identify such taxa, we calculated Spearman correlation coefficients for all genus-level OTUs paired with Allobaculum sp. 128 abundance and tabulated log likelihood ratios for each pairing. Remarkably, the well-known immunogenic mucinophile Akkermansia muciniphila (OTU_363731) exhibited the lowest Spearman coefficient (R = −0.52) and the most significant likelihood ratio (Fig. 3B–3D, S3B, and Table S2). To test whether this relationship between Allobaculum sp. 128 and A. muciniphila is generalizable to humans with naturally acquired microbiomes, we assessed the relative abundance of these two taxa in publicly available large-cohort studies of pediatric ulcerative colitis patients (n = 1,212) and healthy human volunteers (n = 19,524) (McDonald et al., 2018; Schirmer et al., 2018; Table S1). We found that A. muciniphila or Allobaculum exhibited a broadly similar anticorrelation to what we observed in our human microbiota-associated gnotobiotic mice (Fig. 3E–3F). Overall, these data reveal an inverse relationship between two phylogenetically distinct immunostimulatory commensal taxa and raise the possibility that A. muciniphila may influence Allobaculum-induced immune responses.
Figure 3. Allobaculum sp. 128 is inversely correlated with Akkermansia muciniphila in human microbiota-associated gnotobiotic mice.
(A) Experimental workflow. (B) GF WT mice were gavaged with Allobaculum sp. 128, then with healthy human stool 24h later. Fecal pellets were collected for microbiota profiling (n=19). Bacterial OTU legend shown in Figure S4B. (C) Spearman correlation of each genus OTU to Allobaculum sp. 128 abundance, and log-likelihood ratio P-values. (D) XY plot of data in (B). (E-F) Meta-analysis of human microbiome datasets (see Table S1): American Gut Project (McDonald, et al. 2018) and pediatric ulcerative colitis (UC) (Schirmer, et al. 2018). Data shown in (A-D) are from N=1 experiment.
A. muciniphila protects against Allobaculum-mediated exacerbation of DSS colitis
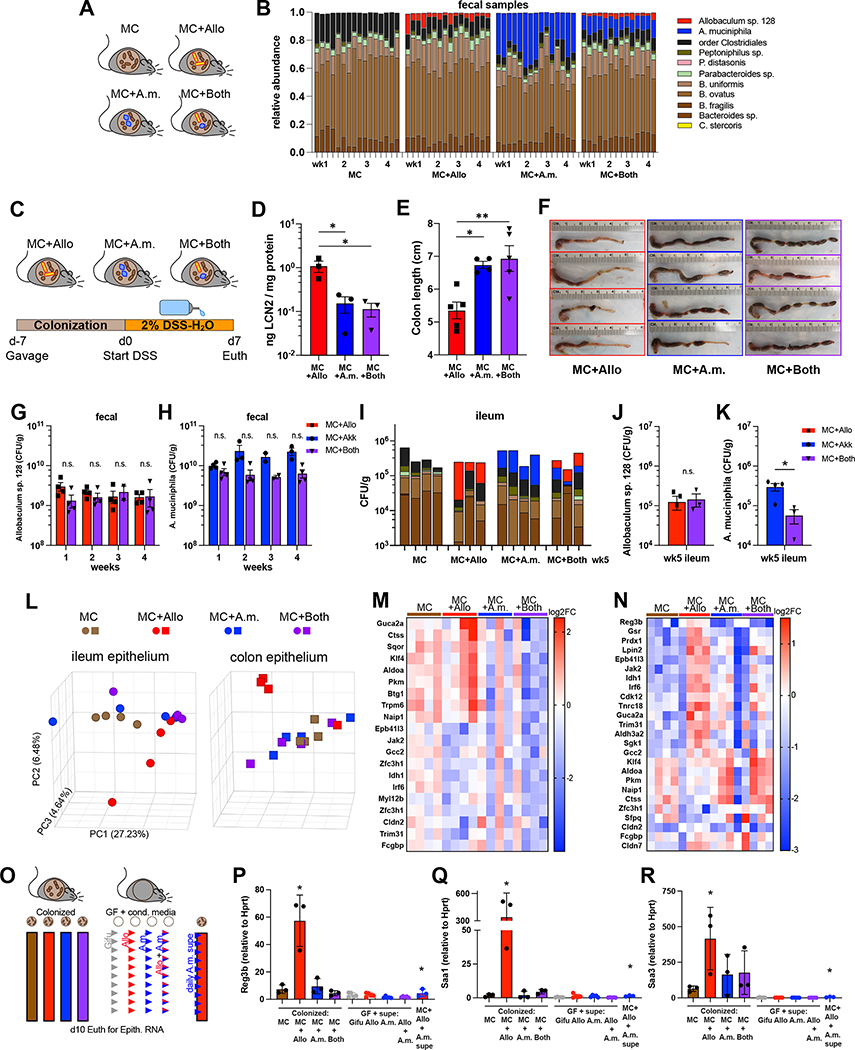
To test the potential effects of A. muciniphila on Allobaculum-induced immune responses, we colonized groups of WT gnotobiotic mice with either Allobaculum sp. 128, A. muciniphila (in-house human isolate 2G4), or both Allobaculum sp. 128 and A. muciniphila in the MC background and monitored their fecal microbiomes to ensure appropriate colonization (Fig. 4A–4B). Although we observed an anti-correlation between Allobaculum and A. muciniphila abundance across diverse community settings in both human microbiota-associated mice and humans (Fig. 3D–3F), we found that both species durably co-colonized gnotobiotic mice in the MC setting (Fig 4B). This is consistent with the observation that a subset of humans are co-colonized with A. muciniphila and Allobaculum, suggesting that these two species can co-exist in select settings. As expected, Allobaculum-colonized mice exhibited severe colitis after DSS treatment, as measured by fecal lipocalin and gross colon pathology. However, both A. muciniphila- and co-colonized mice displayed significantly lower levels of intestinal inflammation (Fig. 4C–4F).
Figure 4. A. muciniphila attenuates Allobaculum sp. 128-mediated intestinal epithelial cell activation and colitis.
(A) Experimental schematic. (B) Fecal microbiota composition profiled over four weeks (n=3–4 mice/group). (C) Experimental schematic for DSS colitis. (D) d2 fecal lipocalin and (E-F) colon length at euthanasia. (G-K) Absolute quantitative microbial profiling. (L) Principal component analysis (PCA) of gene expression in intestinal epithelial cells (IEC) after 2 weeks of colonization. (M-N) Log2 fold change of the top differentially expressed genes. (O) Experimental schematic: colonization with live commensal microbes or daily gavage with sterile culture supernatant for 10 days before RNA harvest from IEC. (P-R) Ileal IEC expression of key Allobaculum-induced genes. Error bars show mean ± SEM. Welch’s t-test was used to compare across MC+Allo+A.m.supe vs MC+Allo; * P<0.05, ** P<0.01. Data shown in (D-F) are one representative of N=2 experiments. Data shown in (A), (G-K), (L-N), and (O-R) are each from separate N=1 experiments.
To examine whether this protection was mediated by a reduction in Allobaculum sp. 128 abundance or localization, we assessed absolute Allobaculum sp. 128 abundance in the feces and ileal mucosa via absolute quantitative amplicon sequencing and FISH. We observed that co-colonization had no appreciable effect on the absolute abundance of Allobaculum sp. 128 in the feces or ileal mucosa, and a slight reduction in A. muciniphila CFUs (Fig. 4H, 4K). Nonetheless, we note that both Allobaculum sp. 128 and A. muciniphila durably co-colonized the ileal mucosa (Fig. 4G–4K and Fig. S3C). Furthermore, co-colonization also had only modest impacts on Allobaculum sp. 128 and A. muciniphila gene expression as measured by bacterial transcriptomics (Fig. S3D–S3F). Together, these data suggest that the impacts of co-colonization on colitis are not due to alterations in Allobaculum density or direct inter-bacterial interactions; however, we cannot completely rule out a role for direct niche competition in A. muciniphila-mediated amelioration of Allobaculum-induced colitis.
We next tested the effects of Allobaculum sp. 128, A. muciniphila, or co-colonization on IEC activation and observed that Allobaculum sp. 128 colonization elicited increased inflammatory gene expression in both ileal and colonic IECs, but this effect was blocked by co-colonization with A. muciniphila (Fig. 4L–4N). Daily gavage of germ-free mice with sterile Allobaculum sp. 128 culture supernatant for 10 days was insufficient to induce key inflammatory genes in IECs (Fig. 4O–4R). However, gavage with A. muciniphila supernatant profoundly suppressed the expression of Allobaculum-induced genes in MC+Allo colonized mice (Fig. 4O–4R).
A. muciniphila protects against Allobaculum-induced colitis in gnotobiotic mice colonized with a complete human gut microbial community and A. muciniphila-mediated protection is consistent across multiple A. muciniphila strains
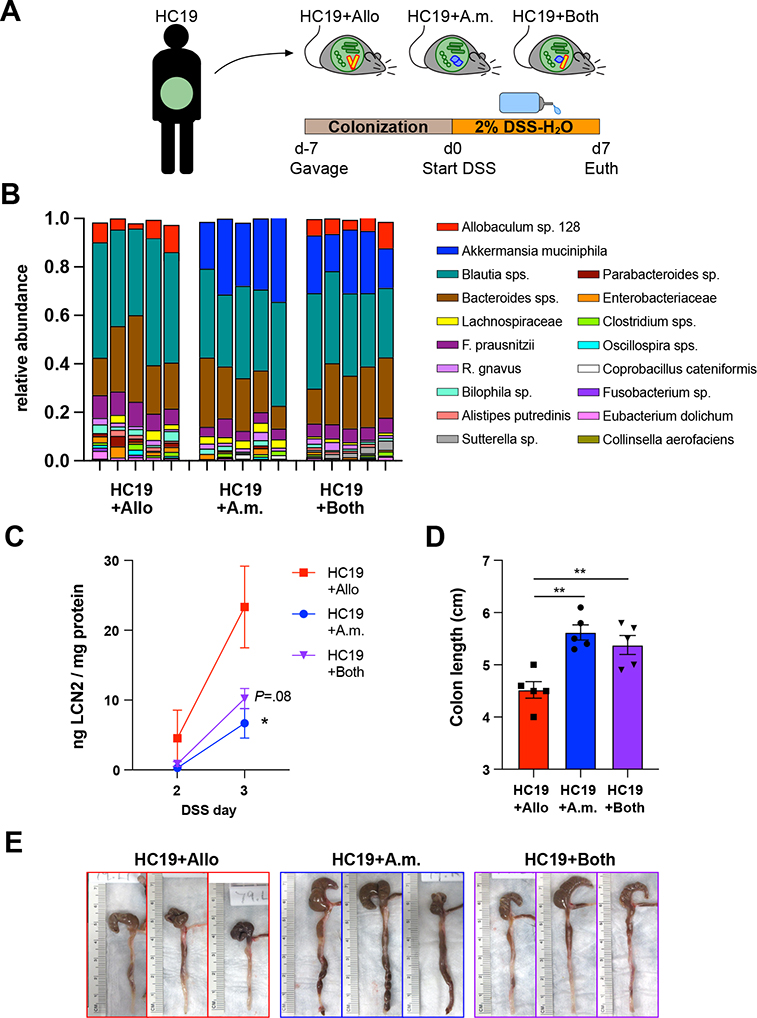
We next tested whether A. muciniphila could protect against Allobaculum-induced colitis in the context of a complex human gut microbial community. We colonized germ-free mice with homogenized stool from a healthy human donor plus either Allobaculum sp. 128, A. muciniphila, or both immunogenic strains and then induced colitis using DSS (Fig. 5A–5B). Consistent with what we observed in the context of a simplified mock community, co-colonized mice exhibited significantly less severe colitis as compared to mice colonized with Allobaculum sp. 128 in the absence of A. muciniphila (Fig. 5A–5E). Finally, to test whether the protective effects of A. muciniphila are consistent across strains, we assessed the impacts of type strain A. muciniphila (ATCC BAA-835) on Allobaculum-induced colitis and observed significant A. muciniphila-mediated amelioration of Allobaculum-mediated disease (Fig. S4A–S4E). Together, these data demonstrate that A. muciniphila ameliorates pathological intestinal immune responses incited by Allobaculum sp. 128 in multiple ecological contexts and across multiple independent strains.
Figure 5. A. muciniphila colonization protects against Allobaculum sp. 128-induced colitis in the context of a complex human microbiota.
(A) Experimental schematic using human fecal sample HC19. (B) Fecal microbiota composition on d3 of colitis. (C) Fecal lipocalin assessed on d2–3. (D-E) Colon length at d7 euthanasia. Error bars show mean ± SEM. Welch’s t-test was used to compare microbiota groups. * P<0.05, ** P<0.01. Data shown are from N=1 experiment.
Allobaculum sp. 128 blunts antigen-specific systemic antibody responses to A. muciniphila and oral vaccination
In addition to testing the effects of co-colonization on local intestinal inflammation, we also performed immunophenotyping of the mesenteric lymph nodes (MLN) after DSS treatment to assess potential inflammatory signatures outside the gut lamina propria (Fig. S4F–S4G). However, unlike in the colon, where A. muciniphila ameliorated Allobaculum-induced responses, co-colonization blunted putative A. muciniphila-induced immune responses. For example, MLNs from mice colonized with MC + A. muciniphila contained elevated levels of dendritic cells (DC) compared to those colonized by MC + Allobaculum sp. 128, and this effect was abrogated in mice co-colonized with both Allobaculum sp. 128 and A. muciniphila (Fig. S4H). These data imply that Allobaculum sp. 128 colonization may alter A. muciniphila-induced immune responses outside the colon. Based on this observation, we hypothesized that co-colonization may affect the development of Allobaculum sp. 128-specific and A. muciniphila-specific immune responses in a bi-directional manner.
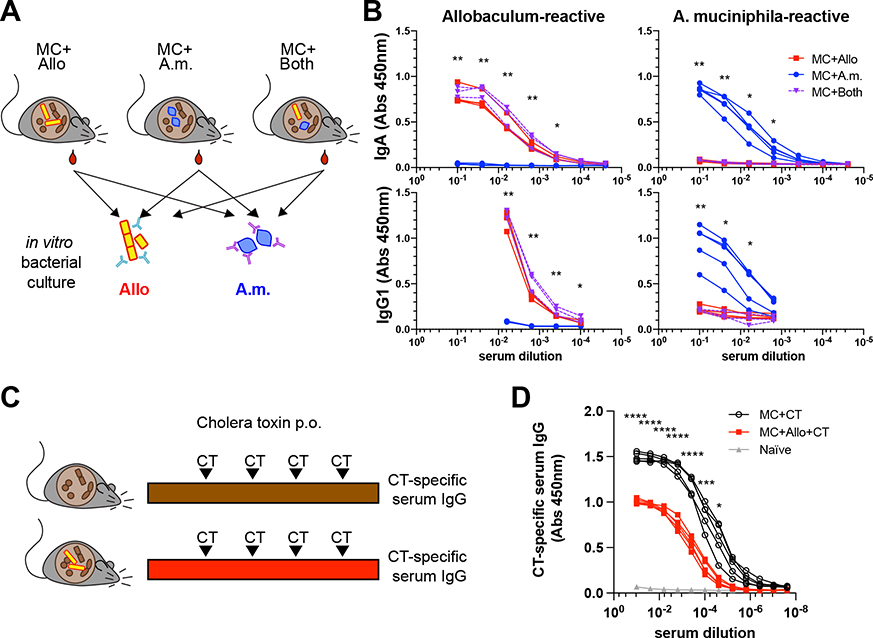
Because both Allobaculum sp. 128 and A. muciniphila induce potent systemic IgG responses, we next examined the effects of co-colonization on systemic antibody responses at steady state (Fig. 6A). As expected, colonization with A. muciniphila or Allobaculum sp. 128 in the MC background elicited potent bacterial-specific serum IgG and IgA responses (Fig. 6B). Despite the protective effects of A. muciniphila on Allobaculum-induced colitis, Allobaculum-specific antibody responses were unaltered after co-colonization. By contrast, Allobaculum sp. 128 co-colonization almost completely blocked the induction of A. muciniphila-specific serum IgA and IgG1 responses (Fig. 6B).Thus, co-colonization with Allobaculum sp. 128 and A. muciniphila reciprocally alters the immune responses elicited by each organism in isolation. We observed that co-colonization modestly reduced the absolute abundance of A. muciniphila in the terminal ileum (Fig. 4K), which could potentially explain why Allobaculum inhibits A. muciniphila-induced systemic IgG responses. However, colonization with Allobaculum sp. 128 also significantly decreased systemic IgG responses to oral vaccination with cholera toxin (CT), suggesting that the impacts of Allobaculum on A. muciniphila-induced responses are not simply due to reduced A. muciniphila colonization (Fig. 6C–6D). Overall, these data show that Allobaculum sp. 128 blunts systemic antibody responses to both endogenous A. muciniphila antigens and an exogenous vaccine antigen.
Figure 6. Allobaculum sp. 128 blunts antigen-specific serum antibody responses to A. muciniphila and oral vaccination.
(A) Experimental schematic. (B) Dilution curves of A. muciniphila-reactive and Allobaculum sp. 128-reactive serum IgA & IgG1 (n=3–5). (C-D) Cholera toxin (CT)-specific serum IgG at 5 weeks. Error bars show mean ± SEM. Welch’s t-test was used to compare MC+A.m. to MC+Both at each dilution. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. Data shown in (B) are representative from one of N=4 independent experiments. Data shown in (D) are from N=1 experiment.
Allobaculum sp. 128 and A. muciniphila elicit unique alterations in the immunological landscape in mucosal lymphoid organs, which are reciprocally reprogrammed by co-colonization
To further explore the individual and combined impacts of Allobaculum sp. 128 and A. muciniphila on host immune responses, we performed single-cell RNA sequencing (scRNA-seq) and simultaneous repertoire sequencing on mesenteric lymph node (MLN) and Peyer’s patch (PP) cells from gnotobiotic mice colonized for four weeks with either MC alone, MC with Allobaculum sp. 128 or A. muciniphila, or MC with both Allobaculum sp. 128 and A. muciniphila. We captured 4,391–10,306 cells per microbiota group, with 77.3–93.6% cells passing quality filters set to retain only viable cells with high-quality transcriptomes (Fig. S5A–S5B). After data scaling, dimensionality reduction, and manual annotation of clusters based on conserved marker genes, we observed significant microbiome-dependent alterations in the relative abundance of diverse immune cell populations (Fig. 7A–B and S6A).
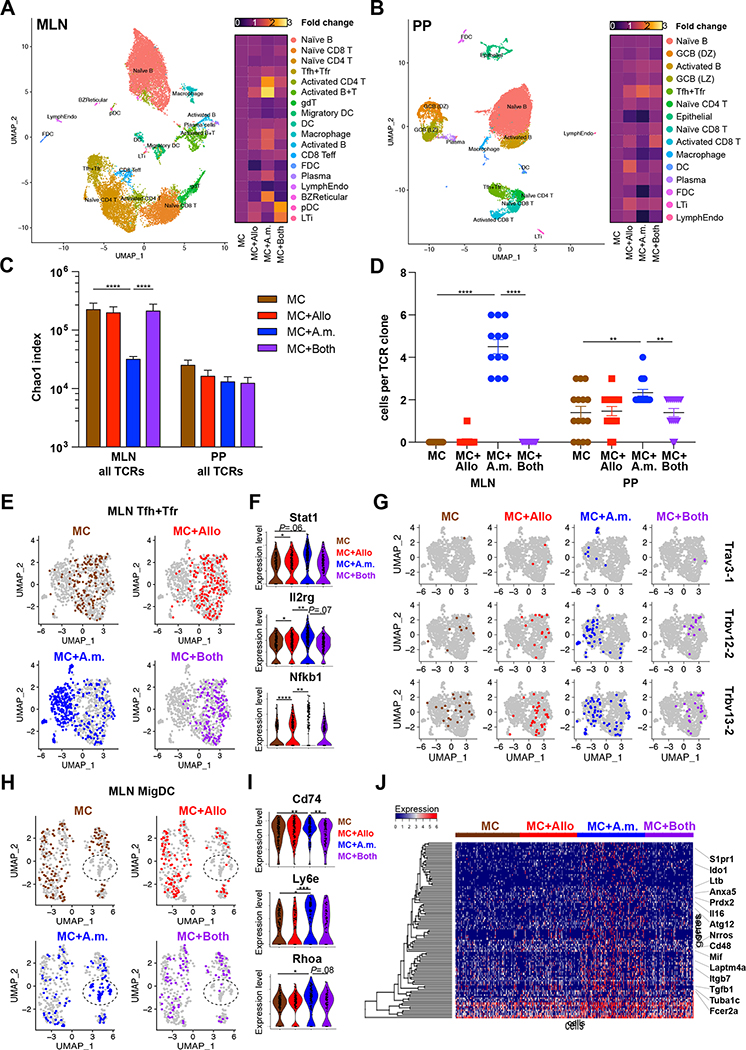
Figure 7. Allobaculum sp. 128 and A. muciniphila induce context-dependent transcriptomic reprogramming in mucosal lymphoid tissues.
(A-B) Annotated dimensionality reduction plots of single-cell gene expression libraries, pooled by tissue (A, MLN; B, PP). Right, heatmap of cell lineage relative frequency normalized to MC. (C) TCR repertoire diversity. (D) Top 12 most expanded clonotypes in MC+A.m. mice. (E,H) MLN Tfh+Tfr and MigDC were re-clustered and highlighted by microbiome. (F) Expression of key genes within Tfh+Tfr. (G) Prominent TCR clonotypes within MLN Tfh+Tfr induced by MC+A.m. (H) MLN MigDC UMAP clustering. (I) Expression of key MigDC antigen presentation genes. (J) Top 100 differentially expressed genes within MLN MigDC transcriptomes. Error bars show mean ± SEM. Welch’s t-test was used to compare gene expression across microbiome groups. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. Data shown from N=1 experiment with n=3 mice/group, pooled.
At baseline, Allobaculum sp. 128 colonization induced only subtle alterations in the immunological milieu in PP and MLN compared to mice colonized with MC alone, including slight increases in activated B and T cells, plasmacytoid dendritic cells, and lymphoid tissue inducer (LTi) cells in the MLN, and increased Tfh/Tfr, dendritic cell, and LTi in the PP (Fig. 7A–7B and S7A). However, A. muciniphila colonization induced an even more dramatic immunological restructuring, particularly in the MLN. This reprogramming was characterized by increases in activated CD4+ T cells and B cells, as well as increases in plasma cells, macrophages, and B cell zone reticular cells. Remarkably, most A. muciniphila-induced changes in the MLN were severely blunted upon co-colonization with Allobaculum sp. 128, while Allobaculum-induced alterations were either unaltered or enhanced upon co-colonization (Fig. 7A and S7B–S7E). A. muciniphila- and Allobaculum-induced alterations in PP cellularity were less dramatic overall and were characterized mainly by an increase in Tfh/Tfr cells, which was unaltered by co-colonization. Together, these data suggest that co-colonization with Allobaculum sp. 128 and A. muciniphila reprograms the immune responses elicited by each organism on its own.
Finally, we leveraged our scRNA-seq and TCR repertoire sequencing data to dissect the cellular mechanisms by which co-colonization with Allobaculum sp. 128 blunts the A. muciniphila-induced systemic antibody response. As expected, we observed that A. muciniphila-induced alterations in B cell clusters in the MLN were similarly blunted by co-colonization (Fig. S7C–S7E, clusters 6, 11, & 18). Since the systemic antibody response to A. muciniphila is T cell-dependent (Ansaldo et al., 2019), we next examined the activation and clonal expansion of T cells in individually colonized and co-colonized mice, with a specific focus on T follicular helper (Tfh) cells. A. muciniphila colonization alone was associated with the expansion of global TCR repertoire clonality, emergence of specific clonotypes in both the MLN and PP, and the appearance of a unique population of Tfh cells in the MLN (Fig. 7C–7G). However, these responses were nearly completely blocked by co-colonization with Allobaculum (Fig. 7C–7G). These data suggest that Allobaculum may prevent the initial priming of A. muciniphila-specific T cells in the MLN, for example by blocking A. muciniphila-induced activation or migration of professional antigen-presenting cells such as dendritic cells (DCs). Indeed, we found that A. muciniphila colonization elicited a unique population of migratory DCs (MigDC) in the MLN that exhibited enhanced expression of transcripts encoding antigen presentation machinery and activation markers, and the appearance of these cells was completely abrogated by co-colonization with Allobaculum (Fig. 7H–7J; cluster 10 in Fig. S6 & S7C–S7E). Overall, these data suggest that Allobaculum may block A. muciniphila-specific adaptive immune responses by preventing A. muciniphila-induced activation of intestinal dendritic cells.
DISCUSSION
Accumulating evidence suggests that individual immunostimulatory strains can play potentially causal roles in human health and disease, but the full spectrum of human gut microbes that shape human immunity remains to be defined. Furthermore, the presence or absence of such ‘causal’ strains alone remains a poor prognostic marker for phenotypic outcomes in humans, suggesting that additional factors may critically alter the responses elicited by specific microbial strains in different individuals. Here, by uncovering a pairwise reciprocal epistatic interaction between two immunostimulatory gut commensals, we begin to decode the microbiota-dependent contextual cues that dictate divergent immune responses to colonization with individual gut commensals. Overall, these studies support a model whereby the impacts of specific immunostimulatory strains in each individual are determined by the presence or absence of other immunomodulatory taxa.
We identified two independent Allobaculum strains from IBD patients that exacerbated colitis in gnotobiotic mouse models, implying that immunogenic Allobaculum species may play causal roles in disease in a subset of IBD patients. Notably, recent studies in mice and humans have identified phylogenetically related taxa as potential drivers of autoimmunity in mice and Crohn’s disease in humans. A mouse-associated strain of Allobaculum drove intestinal T cell responses and facilitated the induction of autoimmunity (Miyauchi et al., 2020), while Erysipleotrichaceae strains were enriched in creeping fat in Crohn’s disease patients (Ha et al., 2020). Although the specific mechanisms by which Allobaculum and its relatives induce intestinal inflammation remain unclear, an ability to invade the mucus layer and trigger inflammatory responses in IECs, including the production of serum amyloid A, may contribute to the pathogenic impacts of this genus (Atarashi et al., 2015; Lee et al., 2020; Miyauchi et al., 2020; van Muijlwijk et al., 2021). Notably, IECs from Allobaculum sp. 128-colonized mice displayed similar gene expression patterns to IECs from human ulcerative colitis patients, including changes in serum amyloid A, guanylate cyclase, cathepsins, and claudins (Parikh et al., 2019). Overall, Allobaculum and related taxa may be important drivers of pathological intestinal inflammation in humans and potential therapeutic targets for the treatment of inflammatory disease.
We and others have previously demonstrated that IgA coating can be used as a marker to identify potentially pathogenic immunostimulatory strains in IBD (Palm & de Zoete et al., 2014; Viladomiu et al., 2017). Indeed, we originally identified Allobaculum sp. 128 as a putative disease-driving microbe in IBD based on its high level of coating with secretory immunoglobulin IgA. However, highly IgA-coated taxa can also exhibit beneficial and immunoregulatory effects (Peterson et al., 2007; Kawamoto et al., 2014; Kubinak et al., 2015; Donaldson et al., 2018). For example, highly IgA-coated bacteria from healthy humans can protect against the pathogenic effects of IgA-coated taxa from undernourished children (Kau et al., 2015). Furthermore, A. muciniphila, which can protect against diet-induced obesity and is associated with enhanced responses to immunotherapy, is the most prevalent highly IgA-coated taxon in healthy humans (Png et al., 2010; Everard et al., 2013; Palm & de Zoete et al., 2014; Bajer et al., 2017; Routy et al., 2018). High IgA-coating thus marks microbes that elicit diverse adaptive immune responses at steady state. Our prior studies of IgA coating of the human gut microbiota suggested that each individual harbors only a handful of potent immunostimulatory strains (less than ten), which may compete for a limited number of unique immunogenic niches. These niches likely share specific features that facilitate active sampling by the immune system and thus initiation of innate and adaptive immune responses.
A growing body of work demonstrates that the specific immune responses induced by individual immunostimulatory commensal species are often highly dependent on the inflammatory and microbial context. For example, murine Helicobacter species that drive colitis in mouse models of IBD primarily trigger Treg responses in healthy animals, but the same microbial antigens elicit pathogenic effector T cells upon induction of colitis (Chai et al., 2017; Xu et al., 2018). Furthermore, A. muciniphila elicits Tfh responses in mice colonized with Altered Schaedler flora (ASF), but induces a mixture of Th cell types, including Th1, Th17, and Tregs, in the context of a complex microbiota (Ansaldo et al., 2019). The magnitude of the antigen-specific IgG response to A. muciniphila was also highly variable in the presence of a complex microbiota and some animals even lacked detectable A. muciniphila-induced T cell responses in these settings (Ansaldo et al., 2019). Our studies suggest that Allobaculum or other phylogenetically or functionally related taxa may explain this context-dependence of the adaptive immune response to A. muciniphila.
Conversely, while A. muciniphila co-colonization had no appreciable effect on Allobaculum-induced systemic IgG responses, both co-colonization and feeding with A. muciniphila supernatants ameliorated Allobaculum-induced IEC activation and colitis. Additional studies will be necessary to determine if these activities are conserved across diverse A. muciniphila strains and additional microbial contexts, as well as whether they are mediated by previously described A. muciniphila-derived immunomodulatory products (Plovier et al., 2017; Becken et al., 2021; Liu et al., 2021). Nonetheless, these observations may at least partially explain the relatively common detection of putative disease-driving taxa such as Allobaculum sp. in ostensibly healthy human subjects. In this model, the pathogenic effects of disease-driving taxa may be blunted by ‘epistatic’ interactions with phylogenetically divergent immunostimulatory taxa such as A. muciniphila. Notably, A. muciniphila is significantly more prevalent and abundant in healthy humans as compared to IBD patients, and carriage of Allobaculum alone is rare among healthy humans. Taken together, our data underscore the importance of microbial context in dictating immune responses elicited by individual commensal organisms and suggest that immunostimulatory strains provide critical contextual cues that alter the magnitude, specificity, or polarization of intestinal immune responses. Thus, the composite effects of the specific immunostimulatory strains present in each person (the immunostimulatory gut microbiota composition code) may determine individual immunological outcomes and susceptibility to immune-related diseases. Borrowing from concepts in genetics, we refer to the non-linear impacts of these interactions on the host as ‘epistatic’ since they uniquely alter the phenotypes elicited by either microbe on its own and can potentially be mediated by either direct or indirect inter-species interactions.
Using scRNA-seq, we found that both Allobaculum sp. 128 and A. muciniphila dramatically reshaped the immunological milieu in the PP and MLN at homeostasis and that co-colonization reprogrammed the immune responses elicited by each microbe on its own. These data thus demonstrate that immunostimulatory commensals critically shape the immunological environment in the gut, which may also impact the initiation or polarization of immune responses to intestinal antigens beyond the commensal microbiota, including food- and self-antigens, as well as mucosal vaccines. Indeed, we found that Allobaculum colonization also blunted systemic IgG responses to oral CT immunization. Although the precise molecular mechanisms by which A. muciniphila and Allobaculum sp. 128 reshape steady-state and pathophysiological immune responses remain unclear, our data suggest that impacts on critical cell types such as IECs and dendritic cells may explain at least some of the epistatic interactions between these gut commensals.
These proof-of-concept studies represent a critical first step toward establishing a microbiota composition code that more accurately predicts the impacts of immunogenic species/strains on human physiological trajectories. Furthermore, they provide a generalizable experimental framework to define the specific microbes and microbial interactions that dictate these divergent outcomes. The ability to predict the epistatic impacts of immunostimulatory taxa has potentially profound therapeutic implications, even beyond improving microbiome-based diagnostics and prognostics. For example, the approaches described here can potentially be used to identify specific microbial taxa that neutralize the pathogenic activities of detrimental immunostimulatory species/strains. These “precision probiotics” may be particularly useful for treating or preventing disease in individuals harboring “matched” disease-driving taxa. Conversely, carriage of specific taxa that blunt the beneficial effects of particular strains may predict non-responsiveness to live bio-therapeutics (e.g., proposed microbial adjuncts for cancer immunotherapy, including A. muciniphila), which may be useful as a gating strategy for patient selection or to identify patients that would benefit from a specific pre-treatment regimen (e.g., antibiotic pre-treatment prior to fecal microbiota transplantation). Overall, these studies begin to uncover the key contextual features that contribute to the incomplete penetrance of strain-specific microbial impacts on human disease and may eventually enable the development of improved microbiota-targeted interventions tailored to the specific microbial context of each individual.
LIMITATIONS OF THE STUDY
These studies reveal one example of pairwise reciprocal epistasis between two unique immunomodulatory taxa. Additional studies will be necessary to determine the broader relevance of reciprocal epistasis across diverse taxa and microbiota compositions. While our data suggest that the epistatic interactions described here are not primarily mediated by bacterial competition, we cannot completely rule out a role for direct inter-species interactions, and the relative importance of direct versus indirect interactions may vary across biological and experimental contexts. Furthermore, because we focused our efforts on understanding the immunological mechanisms of microbial epistasis, the specific bacterial molecules or behaviors that mediate these effects remain to be defined. Finally, determining the broader therapeutic potential of these observations in IBD will require an in-depth examination of the kinetics of epistasis-mediated protection both before, during, and after the onset of disease.
STAR★METHODS
Detailed Methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Noah W. Palm (noah.palm@yale.edu)
Materials availability.
Further information and requests for resources and reagents should be directed to the Lead Contact, Noah W. Palm (noah.palm@yale.edu).
Data and code availability.
Whole genome sequences, fecal 16S microbiota profiles, and bulk RNAseq data are available at NCBI Bioproject PRJNA739762. Single-cell RNAseq data are available at NCBI GEO GSE179165.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains.
Frozen stocks of each strain were streaked on Gut Microbiota Media agar (Goodman, et al. 2011) or Gifu Anaerobic Media agar (HyServe #05422) and incubated 48h at 37°C. Unless otherwise noted, all bacteria were grown in anaerobic conditions (gas composition: 4% H2, 10% CO2, 86% N2). Single colonies were picked into sterile broth and grown overnight at 37°C without shaking. 10μl aliquots of overnight broths were removed for alkaline lysis with boiling to extract genomic DNA, then identities of these monocultures were confirmed by PCR amplification of the 16S rRNA gene V4 region and Sanger sequencing (V4_F: GTGCCAGCMGCCGCGGTAA, V4_R: GGACTACHVGGGTWTCTAAT; or full-length 16S rRNA gene, using published primer sequences 8F and 1391R). Sequences were queried against NCBI and RDP databases.
Human fecal samples.
Human study protocols were approved by the Institutional Review Board (HIC #1607018104) of Yale School of Medicine. Informed consent was obtained from all participants and/or their legal guardians and all methods were performed according to relevant guidelines and regulations. Healthy subjects were recruited via advertisements on the Yale medical campus and in the New Haven Public Library. All fecal samples in this study were collected at home and stored on ice packs at −20°C before either overnight shipment or direct laboratory drop-off the day following collection in an insulated container. Samples were then stored at −80°C until use.
Animal experiments.
Germ-free mice (C57Bl/6, Rag1−/−, Il10−/−) were maintained in flexible film isolators (CBC) with all bedding, chow (Teklad 2018S), and water being autoclaved before import. All germ-free breeding isolators were regularly monitored for the presence of bacteria (both culture-dependent and -independent techniques). All experiments were conducted by transferring mice to positive pressure ventilated microisolator cages (Techniplast #ISO72P), and inoculating each mouse by oral gavage immediately upon transfer. Inocula were previously prepared in anaerobic culture and frozen at −80°C in media + 20% glycerol in gasket-sealed airtight glass vials (Wheaton). The day of inoculation, Wheaton vials were thawed to 25°C and 0.1mL gavaged per mouse. All animal protocols were approved by Yale University Institutional Animal Care and Use Committee (IACUC Protocol 2018-11513). All animal experiments were replicated in both male and female mice of 6–8 weeks of age. Dextran Sodium Sulfate (DSS; TdB Labs) was dissolved in sterile H2O to 2% w/v and passed through a 0.2μm vacuum filter before ad libitum administration. To assess gut permeability, mice were fasted for 4h before gavage with 600 mg/kg FITC-Dextran (Sigma Aldrich #46944). For oral vaccinations, 10μg of cholera toxin (List Biological Laboratories #100B) was administered by gavage weekly. Serum was collected under isoflurane anesthesia, by retro-orbital puncture.
METHOD DETAILS
Fecal sample processing.
Freshly defecated fecal samples were collected into sterile 2mL screw-cap tubes and rehydrated in 1mL sterile PBS, disrupted by 10sec bead beating (Lysing matrix D beads, MP Biomedicals) in a Biospec bead beater, then centrifuged 5min at 50×g to gently pellet large debris. Bacterial cell suspension was then transferred to sterile 2mL deep-well plates for downstream processing. Fecal bacteria were pelleted at 10,000×g for 10min, and clarified fecal water was removed for evaluation of Lipocalin-2 content by ELISA (R&D Systems DY1857). Bacterial pellet was resuspended in Qiagen PowerBead buffer, sonicated for 5min in sonicating water bath, lysis buffer was added, then complete lysis achieved by 0.1mm bead beating followed by genomic DNA isolation (Qiagen DNeasy Ultraclean Microbial; cat #12224). For absolute quantification of microbial strains in vivo, ZymoBIOMICS Spike-in Control I reagent was spiked into all sample wells before DNA extraction, according to manufacturer’s instructions (Zymo Research #D6320).
Microbiota profiling.
The 16S rRNA gene V4 region was amplified from each bacterial gDNA sample by PCR according to a dual-index multiplexing strategy (Kozich, et al. 2013), then amplicons were normalized and cleaned (Agencourt AMPure XP purification beads; Beckman Coulter #A63881). Samples were pooled and libraries were quantified by qPCR (KAPA Biosystems KK4835; Applied Biosystems QuantStudio 6 Flex instrument) then sequenced on an Illumina Miseq in 2×250 PE configuration, using a 500 cycle V2 reagent kit (#MS-102-2003).
Whole genome sequencing.
Overnight bacterial cultures were harvested by centrifugation, cells were lysed for high molecular weight gDNA extraction (Quick-DNA HMW MagBead Kit; Zymo Research #D6060). Genomic DNA was used to prepare two different types of sequencing libraries. Illumina’s Nextera XT kit (#FC-131-1024) was used to prepare short-read libraries, which were sequenced on Illumina Miseq (2×250), while Oxford Nanopore Technologies Ligation Sequencing kit (#SQK-LSK109) was used to prepare long-read libraries, which were sequenced using ONT MinION (Flow cell R9.4.1; #FLO-MIN106D).
Histology.
Whole mouse colons were placed in plastic histology cassettes and immersed in Bouin’s fixative fluid for 24h before transfer to 70% ethanol, paraffin embedding, sectioning, and H&E staining. Blinded slides were scored by a board-certified pathologist. For assessment of colonic mucus thickness, colon tissues were fixed in Carnoy’s solution for four hours, embedded in paraffin, sectioned, and stained with periodic acid Schiff (PAS). The thickness of the inner mucus layer) was quantified using ImageJ (Johansson et al. 2008; Kamphuis et al., 2017).
RNA-seq.
Colon tissues were opened longitudinally and washed thoroughly in sterile PBS until no visible fecal debris remained, then finely minced with a razor blade and transferred to 2mL screw-cap tubes with 1mL ice-cold TRI Reagent (Sigma Aldrich #T9424) and nuclease-free 0.1mm glass beads, thoroughly bead beating for 20sec *3, resting on ice in between. Bulk RNA samples were cleaned using Qiagen RNeasy Mini columns, DNase I digested, and quality checked on an Agilent Bioanalyzer RNA 6000 Nano Kit (#5067-1511). Colon RNAseq libraries were prepared by Yale Center for Genome Analysis staff and run using Illumina Hiseq 2×75 chemistry. Intestinal epithelial cell and bacterial RNAseq libraries were prepared using 60ng total RNA input into Illumina Total Stranded RNA Prep Kit with Ribo-zero Plus (#20040529) and sequenced using Illumina NovaSeq (2×150).
Fluorescence in situ hybridization.
1cm segments of mouse tissue were excised and fixed in Carnoy’s solution (1 Acetic Acid : 3 Chloroform : 6 Ethanol) for no more than 2 hours. Fixed tissues were embedded in paraffin for 5μm cryosectioning. Slides were deparaffinized in xylenes, rinsed in ethanol, and dried thoroughly before hybridization. Bacterial probe EUB-338 ([Cy3]-5’–GCTGCCTCCCGTAGGAGT-3’-[Cy3]) and VP403 ([biotin]-5’-CGAAGACCTTATCCTCCACG-3’-[biotin]) were used for staining at 1μg/mL in hybridization buffer (0.9M NaCl + 0.02M Tris, pH 7.5 + 20% Formamide + 0.05% SDS) in a humidified chamber for 2h at 46°C. After washing, slides were counterstained with DAPI and mounted in ProlongGold Antifade mounting media with overnight curing. Images were acquired on a Leica SP8 confocal microscope running LAS-X software version 3.1.5.
Bacterial flow cytometry.
Fecal bacterial cell suspensions were transferred to sterile LB+20% Glycerol and frozen at −80°C until further analysis. Bacteria were thawed on ice, then aliquoted 10^4 – 10^5 CFU per well of 2mL 96-deep-well plate (pellet not visible)(Moor, et al. 2016). Each staining reaction was blocked with normal rat serum for 15min, then washed in sterile PBS/0.1%BSA. Staining for endogenous coating by mouse IgA was performed at 1:100 with PE-conjugated eBioscience clone mA-6E1 (Thermo Fisher #12420482). Cells were washed three times in 500μl PBS, then transferred to 1.1mL microdilution tubes (VWR 20901-013) for analysis on a BD FACS Calibur instrument, including control tubes for sterile buffer (log FSC, log SSC), unstained cells, and secondary only-stained cells to set appropriate gates. A minimum of 50,000 events/sample were collected and analyzed using FlowJo v9.
Bacterial ELISAs.
Overnight broth cultures of bacterial strains of interest were washed three times in sterile PBS, then normalized to an OD600 of 0.1. Many 100μl aliquots were prepared and snap frozen in liquid nitrogen. To prepare ELISA plates, bacterial aliquots were thawed on ice, diluted further 1:10 in PBS, then coated 50μl/well of Nunc Maxisorp Immunoplates overnight at 4°C. The next day plates were spun 15min at 5000×g before discarding supernatant and confirming bacterial adhesion by phase contrast microscopy. Plates were blocked with 1%BSA in PBS before serially diluting serum or fecal water. After 2h incubation at RT, plates were washed four times with TBS-T, then mouse IgG was detected using HRP-conj. Goat anti-Ms-IgG (Thermo Fisher Scientific #31430; 1:6,000 dilution), or mouse IgA using HRP-conj. Goat anti-Ms-IgA (Sigma Aldrich A4789; 1:6,000 dilution). Plates were washed four times before detection with TMB (Pierce), stopped with 2N H2SO4, and read at Abs 450nm (Molecular Devices SpectraMax i3x).
Intestinal epithelial and lamina propria cell isolation.
Ileum and colon tissue was harvested into 25°C complete RPMI 1640 medium (supplemented with 10% FBS, Pen-Strep, L-Glutamine, HEPES). After thorough cleaning in sterile PBS to remove all visible fecal debris, tissues were shaken in strip buffer (HBSS + 1.5mM EDTA + 0.145 mg/mL DTT) at 37°C 225rpm for 20min to remove mucus and epithelial layers. Epithelial cells were filtered through stainless steel mesh, then centrifuged 10min 400×g, and resuspended in Trizol for RNA extraction. Remaining lamina propria tissue was shaken in strip buffer a second time, then minced and transferred to cRPMI + 0.5mg/mL DNase + 1mg/mL Collagenase D for 45min shaken at the same speed. Then cells were filtered twice through stainless steel mesh and lymphocytes enriched in a 40%−70% Percoll interface (20min at 600×g, brake off). Cells were aliquoted to round-bottom polystyrene microplates for Fc Blocking, fluor-conjugated antibody staining (see Table 1), and washing. Ex vivo cell restimulations were performed for 4h with 50ng/mL PMA + 1μM ionomycin, in the presence of brefeldin A (GolgiStop reagent, BD #554724), before surface staining, fixation, permeabilization, and intracellular staining.
MLN & PP cell isolation.
Mucosal lymphoid tissues were dissected and gently washed in sterile PBS, transferred to digestion media (serum-free RPMI 1640 supplemented with, Pen-Strep, L-Glutamine, HEPES, 2-mercaptoethanol, NEAA, Sodium Pyruvate, DNase I, and Collagenase D) in 30mL beaker with a small magnetic stir bar and stirred at 400rpm in 5%CO2 incubator for 15min. After stirring, beakers were transferred to ice and triturated with media containing 3% FBS, filtered through stainless steel mesh, centrifuged 350×g 10min 4°C. Cells were washed twice more in media to remove large debris chunks, then resuspended in PBS + 0.04%BSA and filtered again through 40μm nylon.
Single-cell RNA sequencing.
Single cell suspensions were counted by hemacytometer and normalized to 1e6/mL for submission to Yale Center for Genome Analysis staff for droplet generation and gel bead encapsulation using 10X Genomics Controller. Cell lysis, barcoding, and reverse transcription were performed using Chromium 5’ V2 chemistry according to manufacturer’s instructions. PCR-amplified gene expression libraries were quantified and evaluated for QC by Agilent Bioanalyzer, and sequenced on Illumina NovaSeq 6000 at a depth of 175M read pairs per library.
Dendritic cell–T cell co-cultures.
MLNs were digested to generate single cell suspensions as described above and MLN DCs were isolated from each mouse using EasySep Mouse Pan-DC Enrichment Kit (Stem Cell Technologies #19763), counted by hemacytometer, normalized for cell concentration, and plated at 7.5e4 cells/well of a round-bottom TC plate. OT-II cells were isolated from pooled spleens and peripheral lymph nodes of OT-II mice using EasySep Mouse Naïve CD4+ T cell isolation kit (Stem Cell Tech #19765), labeled for 20min in 5μM CellTraceViolet (Biolegend #425101), and plated at 2e5 cells/well with 100ng/mL OVA. On day 3 of co-culture, T cell proliferation was assessed by flow cytometry.
Bioinformatic analyses.
Phylogenetic analysis of bacterial taxa belonging to family Erysipelotrichaceae: 16S rRNA gene sequences from NCBI Genbank were aligned using Clustal Omega and alignments imported into MEGA v10.2.6. Phylogenetic trees were constructed using both neighbor-joining method and maximum likelihood estimation method, in each case bootstrapping for 1,000 replicates, both of which resulted in the same overall phylogeny. Trees were visualized using interactive Tree of Life (Letunic and Bork 2021). Whole genome assemblies: long-read fastq files were passed to Flye v2.6 for assembly (Kolmogorov et al., 2019) and short-read fastq files were used to finish assembling remaining contigs using Unicycler v0.4.9b (Wick et al., 2017). Microbiota profiling: 16S rRNA amplicon sequencing data were processed and analyzed using QIIME (v1.9), including rarefaction to 1000 reads/sample, elimination of reads below a frequency of 0.0001, open reference OTU picking, and filtering out contaminating OTUs known to originate from water control PCRs (Caporaso, et al. 2010; Lozupone, et al. 2012; McDonald, et al. 2012). Bulk RNAseq sequencing data were trimmed, aligned to mouse genome or bacterial genomes, and gene counts quantified using Partek Flow (v6.0). Gene lists were analyzed for GO enrichment using Panther v14 available at http://geneontology.org (Mi, et al. 2019). Single cell sequencing data were demultiplexed then processed using 10X Genomics cellranger count. Count matrices were imported into Seurat (v3.2.1) within R (v4.0.3) (Butler, et al. 2018), paired with microbiome metadata, filtered for nFeature_RNAs <500 & <5000 and percent mitochondrial genes <8%. Clusters were generated by UMAP with resolution = 0.8, manually annotated based on expression of conserved marker genes, then analyzed for differential expression across microbiome groups using FindMarkers. MLN cell clusters were analyzed for pairwise enrichment in microbiome conditions via MELD, using default parameters (Burkhardt et al., 2021). TCR repertoires were analyzed using Immunarch (Immunomind Team 2019).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was conducted in GraphPad Prism v9. Unless otherwise noted, data are plotted as mean ± SEM. Each figure legend describes the sample sizes of the data shown in that figure, as well as the specific statistical tests applied.
Supplementary Material
Figure S1. Allobaculum sp. 128 does not bloom during inflammation, and a second UC patient Allobaculum isolate is colitogenic in gnotobiotic mice, Related to Figure 1. (A) Time course of fecal microbiota profiling of MC+Allo colonized mice across 7 days of 2% DSS administration (n=4 mice/group). (B) Time course of fecal lipocalin (LCN-2) overlaid with Allobaculum sp. 128 abundance. (C) H&E-stained colon sections from Rag1−/− mice on d7 of DSS administration. Scale bars, 200μm. (D-E) Colon lamina propria CD4+IL-17A+ T cells from Il10−/− mice after 8 weeks colonization (n=5 mice/group). (F-G) GF mice were monocolonized with either Ery47 (control bacteria) or Allobaculum sp. 128 for 7 days before the induction of acute colitis via 2% DSS administration. Colonic inflammation was assessed by fecal lipocalin on d3 (F) and colon length on d7 (G) (n=5 mice/group). (H) Phylogenetic tree constructed using 16S rRNA gene sequences from members of family Erysipelotrichaceae, using maximum likelihood estimation, bootstrapped (BS) to 1,000 replicates. BS values are shown along branches. (I) Experimental schematic. (J) Second Allobaculum isolate (Allo2) was used for acute colitis model in WT gnotobiotic mice as in Figure 1C–1H. Three of six mice colonized with Allo2 were found dead (3 F.D.) prior to endpoint. (K) Fecal lipocalin, LCN2, was measured longitudinally by ELISA. Error bars show Mean ± SEM. Data shown in (A-B) from N=1 experiment. Data in (D-E) fron one of N=2 experiments. Data in (F-G) from N=1 experiment. Data shown in (J-K) from one of N=2 independent experiments, n=5–6 mice per group.
Figure S2. Unremarkable histopathology and total serum Ig of untreated Allobaculum sp. 128-colonized WT mice, Related to Figure 1. (A) Bouin’s-fixed H&E-stained colon sections from WT mice colonized with MC bacteria or MC+Allobaculum sp. 128, euthanized 12 weeks later. Scale bars, 200μm. (B) Blinded scoring for colitis. One experiment shown is representative of N=2 independent experiments. (C-D) Total serum IgA and IgG content at 7 weeks (n=3–4 mice/group). (E) FITC-dextran concentration in serum 1h after oral gavage (n=4–8 mice/group). (F) Allobaculum sp. 128-specific IgG in fecal water at 7 weeks post-colonization with MC or MC+Allo (n=5 mice/group). (G) Thickness of the colonic inner mucus layer after 2 weeks of colonization (tissues fixed in Carnoy’s solution to preserved the inner mucus layer; n=10–12 mice/group). Welch’s t-test was used to compare microbiota groups at each time point. ** P<0.01, * P<0.05. Data shown in (A-B) are from one representative of N=3 experiments. Data shown in (C-D) show one of N=2 experiments. Data in (E) are from N=1 experiments. Data in (F) show one of N=2 experiments. Data in (G) are compiled from N=2 experiments.
Figure S3. Microbial diversity cannot explain the relationship between Allobaculum and A. muciniphila, A. muciniphila and Allobaculum co-colonize the ileal mucosa, and co-colonization has minimal impacts on A. muciniphila and Allobaculum gene expression, Related to Figures 3 & 4. (A) Genus-level richness, Simpson’s diversity index, Shannon’s diversity index, and evenness of each microbiome that contained Allobaculum (n=14) or lacked Allobaculum sp. 128 (n=5). (B) Legend accompanying Fig. 4B. Data shown from N=1 experiment. (C) GF WT mice were mono- or bi-colonized as shown for 10 days. Terminal ilea were fixed and sections stained with bacterial FISH probes EUB338 (to stain Allobaculum) and VP403 (to stain A. muciniphila). Scale bars, 10μm. (D) In vivo bacterial transcriptomes from the ileum and colon were compared for differential expression of ORFs across single colonization or co-colonization conditions (MC+Allo vs MC+Both, and MC+Akk vs. MC+Both). (E) Allobaculum sp. 128 and (F) A. muciniphila ORFs that were differentially expressed in the ileum, along with their fold changes, P-values, and annotations (Prokka). Data are from N=1 experiment.
Figure S4. Type strain A. muciniphila attenuates Allobaculum sp. 128-mediated colitis and Allobaculum sp. 128 blunts A. muciniphila-induced dendritic cell responses in MLN, Related to Figure 5. (A) Experimental schematic for acute DSS colitis in WT gnotobiotic mice colonized with MC, MC+Allobaculum sp. 128, MC+A. muciniphilaT (type strain ATCC BAA-835), or MC+Allobaculum sp. 128+A. muciniphilaT (ATCC BAA-835) (n=4–6 mice/group). (B) Fecal microbiota profiling, (C) colon length, (D) d2 fecal lipocalin (LCN2), and (E) gross colon pathology. (F) Representative gating strategy for analysis of MLN cells performed in FlowJo after ≥100,000 events per sample were collected. (G) Immunophenotyping of MLN cell populations (% of viable cells) (n=4–6 mice/group). (H) Quantification of DCs (Live B220−TCRb−CD11b+CD11c+MHCII+). Welch’s t-test was used to compare microbiota groups. *P<0.05. Data shown in (A-E) are from N=1 experiment. Data shown in (F-H) are from one of N=2 independent experiments.
Figure S5. Approach for profiling microbiota-dependent mucosal immune landscape using single cell RNAseq, Related to Figure 7. (A) Schematic depicting single cell RNAseq (scRNAseq) experiment. (B) Quality control metrics used for filtration of scRNAseq data before proceeding to clustering and differential expression analyses. Only cells with 500–5000 RNA features were retained (between blue dashed lines), as well as cells with <8% genes of mitochondrial (mt) origin. Welch’s t-test was used to compare each cell lineage across microbiota groups; * P<0.05, ** P<0.01, *** P<0.001. N=1 experiment.
Figure S6. Expression of marker genes mapped to MLN cell clusters, Related to Figure 7. (A) Violin plots showing expression of marker genes across MLN cell clusters, numbered to match clusters shown in Figure S7.
Figure S7. Epistatic reversal of A. muciniphila-induced MLN immune cell clusters by co-colonization with Allobaculum sp. 128, and direct assessment of MLN DC function in co-colonized gnotobiotic mice, Related to Figure 7 and Figure S6. (A-E) Graph-based probabilistic analysis of MLN scRNAseq data, comparing two microbiota groups at a time. (A-B) MC+Allo relative to MC+Both or relative to MC. (C-E) MC+Akk relative to MC+Both reveals strong reversal (high MELD score) of cell clusters induced by A. muciniphila after co-colonization with Allobaculum sp. 128. The marker genes that define each cluster are displayed in Figure S6. (F-G) Ex vivo co-cultures of purified MLN DCs from gnotobiotic mice were examined for their capacity to prime CellTrace Violet (CTV)-labeled naïve OT-II T cells, by proliferation and dilution of CTV. Data shown are from N=1 experiment.
Table S1. Allobaculum and A. muciniphila abundance in two human cohorts broadly reflects an inverse relationship, Related to Figure 3. (Sheet 1. American Gut Project) Enumeration of sequencing data from American Gut Project (McDonald, et al. 2018; QIITA study IDs: 48742, 51570, 52698, 53379, & 54454). (Sheet 2. Schirmer_UC) Relative abundance data from pediatric ulcerative colitis (Schirmer, et al. 2018).
Table S2. Human microbiota-associated gnotobiotic mouse screen reveals inverse correlation between Allobaculum sp. 128 and A. muciniphila, Related to Figure 3. (Sheet 1. Relative Abundance) fecal microbiota profiles across n=19 gnotobiotic mice. (Sheet 2. Correlation Metrics, Sorted) Compiled statistics evaluating correlation and logistic regression between each pair of microbiota OTUs. (Sheet 3. Pearson R) Calculations of Pearson correlation. (Sheet 4. Spearman R) Calculations of Spearman correlation. (Sheet 5. Diversity Calcs) Calculations of alpha-diversity, beta-diversity, and evenness.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Ms Ly6G (FITC) | BD | Cat#551460; clone 1A8; Lot#6033810 |
| Rat anti-Ms CD4 (FITC) | BD | Cat#553729; clone GK1.5; Lot#5191688 |
| Hamster anti-Ms TCRbeta (BUV737) | BD | Cat#612821; clone H57-597; Lot#9331214 |
| Rat anti-Ms CD45 (APC-Cy7) | BD | Cat#557659; clone 30-F11; Lot#5357842 |
| Hamster anti-Ms CD3e (Pacific Blue) | BD | Cat#558214; clone 500-A2; Lot#5009871 |
| Rat anti-Ms IL-17A (AF647) | BD | Cat#560184; clone TC11-18H10; Lot#7104724 |
| Rat anti-Ms I-A/I-E (BV605) | BD | Cat#563413; clone M5/114.15.2; Lot#5170799 |
| Rat anti-CD11b (BUV395) | BD | Cat#565976; RRID AB_2738276 |
| Rat anti-Ms/Hu CD45R/B220 (BV711) | BioLegend | Cat#103255; clone RA3-6B2; Lot#B223589 |
| Goat anti-Ms IgG (AF647) | BioLegend | Cat#405322; RRID AB_2563045; Lot#B246161 |
| Rat anti-Ms IgA (PE) | eBioscience / Thermo Fisher Scientific | Cat#12420482; clone mA-6E1; Lot#2173312 |
| Rat anti-Ms CD16/32 | eBioscience / Thermo Fisher Scientific | Cat#14016186; clone 93; Lot#4333612 |
| Hamster anti-Ms/Hu CD11c (AF700) | eBioscience / Thermo Fisher Scientific | Cat#56011480; clone N418; Lot#E089601632 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit, for UV excitation | Thermo Fisher Scientific | Cat#L23105 |
| Goat anti-Ms IgG (H+L) (HRP) | Thermo Fisher Scientific | Cat#31430; Lot#UB278606 |
| Goat anti-Ms IgA (HRP) | Sigma Aldrich | Cat#A4789 |
| Bacterial and Virus Strains | ||
| Bacteroides sp. | Palm & de Zoete, et al. Cell 2014. | NWP_0582 |
| Parabacteroides distasonis | Palm & de Zoete, et al. Cell 2014. | NWP_0583 |
| Peptoniphilus sp. | Palm & de Zoete, et al. Cell 2014. | NWP_0584 |
| Bacteroides ovatus | Palm & de Zoete, et al. Cell 2014. | NWP_0585 |
| order Clostridiales UC | Palm & de Zoete, et al. Cell 2014. | NWP_0586 |
| family Lachnospiraceae UC | Palm & de Zoete, et al. Cell 2014. | NWP_0587 |
| Collinsella stercoris | Palm & de Zoete, et al. Cell 2014. | NWP_0588 |
| Bacteroides uniformis | Palm & de Zoete, et al. Cell 2014. | NWP_0589 |
| Parabacteroides sp. | Palm & de Zoete, et al. Cell 2014. | NWP_0590 |
| Allobaculum sp. 128 | Palm & de Zoete, et al. Cell 2014. | NWP_0324 |
| Allobaculum sp. Allo2 | this paper | NWP_0593 |
| Akkermansia muciniphila | Derrien, et al. 2004 | ATCC BAA-835 |
| Akkermansia muciniphila 2G4 | this paper | NWP_0598 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gifu Anaerobic Media | HyServe | Cat#05422 |
| Gut Microbiota Media (GMM) | Goodman, et al. 2011 | N/A |
| Dextran Sodium Sulfate (DSS) | TdB Labs | Cat#DB001 |
| Bouin’s Fixative Solution | Fisher Scientific | Cat#11201 |
| TRI Reagent | Sigma Aldrich | Cat#T9424 |
| TMB Substrate Kit | Thermo Fisher Scientific | Cat#34021 |
| Sulfuric Acid, 2.0 N | Avantor | Cat#H381-05 |
| RPMI 1640 | Thermo Fisher Scientific | Cat#11875-119 |
| DNase I, bovine pancreas | Sigma Aldrich | Cat#10104159001 |
| Collagenase D (type IV) | Sigma Aldrich | Cat#11088882001 |
| Percoll | VWR | Cat#89428-524 |
| Phorbol-12-myristate-13-acetate (PMA) | Sigma Aldrich | Cat#P1585-1MG |
| Ionomycin Calcium salt, ready made | Sigma Aldrich | Cat#I3909-1ML |
| GolgiStop Protein Transport Inhibitor | BD | Cat#554724 |
| Critical Commercial Assays | ||
| MagAttract Microbial DNA Kit (384) | Qiagen | Cat#27200-4 |
| PCR Purification Kit, Agencourt AMPure XP | Beckman Coulter | Cat#A63881 |
| NGS Library Quantification Complete kit (ABI Prism) | KAPA Biosystems | Cat#KK4835 |
| MiSeq Reagent Kit v2 (500 cycles) | Illumina | Cat#MS-102-2003 |
| Mouse Lipocalin-2/NGAL DuoSet ELISA | R&D Systems | Cat#DY1857 |
| Bioanalyzer RNA 6000 Nano Kit | Agilent | Cat#5067-1511 |
| ZymoBIOMICS Spike-in Control I (High Microbial Load) | Zymo Research | Cat#D6320 |
| Stranded Total RNA Prep kit for RNAseq libraries | Illumina | Cat#20040529 |
| Wet-to-Digital H&E histology services | Histowiz | N/A |
| Deposited Data | ||
| Allobaculum sp. 128 whole genome sequence | NCBI Genbank | CP078089 |
| Allobaculum sp. 'Allo2' whole genome sequence | NCBI Genbank | CP078088 |
| Bulk colon RNAseq | NCBI Bioproject | PRJNA739762 |
| Bulk intestinal epithelial RNAseq | NCBI Bioproject | PRJNA739762 |
| Bulk microbial RNAseq | NCBI Bioproject | PRJNA739762 |
| MLN & PP single cell RNAseq | NCBI GEO | GSE179165 |
| Experimental Models: Organisms/Strains | ||
| Germ-free C57BL/6 mice | University of Chicago Animal Resources Center | N/A |
| Germ-free Il10−/− mice | University of Michigan Gnotobiotics | N/A |
| Germ-free Rag1-/- | University of Michigan Gnotobiotics | N/A |
| Oligonucleotides | ||
| EUB-338 (FISH) | Canny, et al. 2006. | [Cy3]-5’-GCTGCCTCCCGTAGGAGT-3’-[Cy3] |
| VP403 (FISH) | Arnds, et al. 2010 | [biotin]-5’-CGAAGACCTTATCCTCCACG-3’-[biotin] |
| Hprt_qpcr_F | this paper | TCAGTCAACGGGGGACATAAA |
| Hprt_qpcr_R | this paper | GGGGCTGTACTGCTTAACCAG |
| Saa1_qpcr_F | this paper | TCTGGAGTTTTCCCAAGGGTG |
| Saa1_qpcr_R | this paper | GGTGAGTAGCTTCATCCTCTGTC |
| Saa3_qpcr_F | this paper | TGAAAGAAGCTGGTCAAGGGTC |
| Saa3_qpcr_R | this paper | TCCCCCGAGCATGGAAGTAT |
| Reg3b_qpcr_F | this paper | ATACCCTCCGCACGCATTAG |
| Reg3b_qpcr_R | this paper | TCTTTTGGCAGGCCAGTTCT |
| Software and Algorithms | ||
| GraphPad Prism | https://www.graphpad.com/scientific-software/prism/ | v9 |
| QIIME | http://qiime.org/ | v1.9 |
| MEGA | https://www.megasoftware.net/ | v10.2.6 |
| FlowJo | Treestar | v10 |
| Partek Flow | https://www.partek.com/partek-flow/ | v6 |
| Panther | http://geneontology.org/ | v14 |
| Seurat | https://satijalab.org/seurat/index.html | v3.2.1 |
| Immunarch | https://immunarch.com/ | v0.6.6 |
| R | https://www.r-project.org/ | v4.0.3 |
| Other | ||
| Anaerobic Culture Chambers | Coy | N/A |
| Flexible Film Gnotobiotic Isolators | Class Biologically Clean | N/A |
| Isocage P Microisolator Caging System | Techniplast | Cat#ISO72P |
| Bead Beater | Biospec | N/A |
| Spectramax i3x plate reader | Molecular Devices | Cat#i3x |
| QuantStudio 6 Flex Real-Time PCR Instrument | Applied Biosystems | Cat#4485699 |
Highlights for CHM title page:
Allobaculum (Allo) isolates from ulcerative colitis patients exacerbate colitis in mice
Immunostimulatory Allo. sps. inversely correlate with Akkermansia muciniphila (A. muc)
Co-colonization uniquely alters immune responses elicited by Allo. or A. muc. alone
Reciprocal “epistatic” inter-species interactions non-linearly impact host immunity
ACKNOWLEDGEMENTS
The authors acknowledge Dr. Xinran Liu & Morven Graham for their expertise with electron microscopy, Shrikant Mane, Chris Castaldi, Guilin Wang, and other Yale Center for Genome Analysis staff for library preparation and sequencing services, Yale Animal Resource Center staff for veterinary services, Dr. Stephanie Eisenbarth, Dr. Biyan Zhang, Dr. Xiangyun Yin and other lab members for IgG subtyping, DC expertise, and many helpful discussions, Dr. Ruaidhri Jackson for guidance with colon lamina propria cell preparations, Daniel Waizman for providing OT-II mice and T cell expertise, Dr. Andrew Wang and lab members for critical discussions of the manuscript, Dr. Guus van Muijlwijk for critical discussions of the manuscript, and all members for the Palm lab for their key observations and discussions.
FUNDING SOURCES
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers K22AI123477 and R21AI137935, the Kenneth Rainin Foundation, the Leona M. and Henry B. Helmsley Charitable Trust (3083), a Young Investigator Grant for Probiotics Research, and a sponsored research agreement with Artizan Biosciences. Approximately 20% of the funding for this research project (~$400,000) was financed with NIH funds; the remainder was financed by nongovernmental sources. NWP also gratefully acknowledges support from the Common Fund of the National Institutes of Health (DP2DK125119), Chan Zuckerberg Initiative, Aligning Science Across Parkinson’s, Emory University, the Ludwig Family and Mathers Foundations, the Pew Charitable Trust, the NIA and NIGMS (R01AG068863 and RM1GM141649), F. Hoffmann-La Roche Ltd, and the Yale Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
T.A.R. and N.W.P. are inventors on a provisional patent entitled “Compositions and Methods for Treating and Preventing Diseases or Disorders using Inter-Species Interactions” that is licensed to Artizan Biosciences. R.A.F., M.R.d.Z., and N.W.P. are co-founders of and consultants for Artizan Biosciences and N.W.P. is a co-founder of Design Biosciences.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, et al. (2019). Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnds J, Knittel K, Buck U, Winkel M, and Amann R (2010). Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Syst Appl Microbiol 33 (3), 139–148. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. (2017). Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J, and Drastich P (2017). Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 23, 4548–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becken B, Davey L, Middleton DR, Mueller KD, Sharma A, Holmes ZC, Dallow E, Remick B, Barton GM, David LA, et al. (2021). Genotypic and Phenotypic Diversity among Human Isolates of Akkermansia muciniphila. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Harrison OJ (2017). Homeostatic Immunity and the Microbiota. Immunity 46, 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, Wlodarska M, Willing BP, Vonaesch P, Han J, Reynolds LA, Arrieta MC, Uhrig M, Scholz R, Partida O, et al. (2015). Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nat Commun 6, 7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt DB, Stanley JS 3rd, Tong A, Perdigoto AL, Gigante SA, Herold KC, Wolf G, Giraldez AJ, van Dijk D, and Krishnaswamy S (2021). Quantifying the effect of experimental perturbations at single-cell resolution. Nat Biotechnol 39, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butto LF, Schaubeck M, and Haller D (2015). Mechanisms of Microbe-Host Interaction in Crohn’s Disease: Dysbiosis vs. Pathobiont Selection. Front Immunol 6, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny G, Swidsinski A, and McCormick BA (2006). Interactions of intestinal epithelial cells with bacteria and immune cells: methods to characterize microflora and functional consequences. Methods Mol. Biol. 341, 17–35. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, et al. (2019). Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis. Immunity 50, 1099–1114 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z, Perry JSA, Knoop KA, Tanoue T, Narushima S, et al. (2017). Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, and de Vos WM (2004). Akkermansia muciniphila gen. nov., sp nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Micr 54, 1469–1476. [DOI] [PubMed] [Google Scholar]
- Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. (2018). Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Lee SM, and Mazmanian SK (2016). Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110, 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Externest D, Meckelein B, Schmidt MA, and Frey A (2000). Correlations between antibody immune responses at different mucosal effector sites are controlled by antigen type and dosage. Infect Immun 68, 3830–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, and Gordon JI (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proceedings of the National Academy of Sciences of the United States of America 108, 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, Gavryushkin A, Carlson JM, Beerenwinkel N, and Ludington WB (2018). Microbiome interactions shape host fitness. Proc Natl Acad Sci U S A 115, E11951–E11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greetham HL, Gibson GR, Giffard C, Hippe H, Merkhoffer B, Steiner U, Falsen E, and Collins MD (2004). Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe 10, 301–307. [DOI] [PubMed] [Google Scholar]
- Ha CWY, Martin A, Sepich-Poore GD, Shi B, Wang Y, Gouin K, Humphrey G, Sanders K, Ratnayake Y, Chan KSL, et al. (2020). Translocation of Viable Gut Microbiota to Mesenteric Adipose Drives Formation of Creeping Fat in Humans. Cell 183, 666–683 e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji BW, Sheth RU, Dixit PD, Tchourine K, and Vitkup D (2020). Macroecological dynamics of gut microbiota. Nature Microbiology 5, 768-+. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, and Hansson GC (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105, 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis JBJ, Mercier-Bonin M, Eutamene H, and Theodorou V (2017). Mucus organisation is shaped by colonic content; a new view. Sci Rep 7, 8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, et al. (2015). Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 7, 276ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, Hattori M, and Fagarasan S (2014). Foxp3+ T Cells Regulate Immunoglobulin A Selection and Facilitate Diversification of Bacterial Species Responsible for Immune Homeostasis. Immunity 41, 152–165. [DOI] [PubMed] [Google Scholar]
- Kiner E, Willie E, Vijaykumar B, Chowdhary K, Schmutz H, Chandler J, Schnell A, Thakore PI, LeGros G, Mostafavi S, et al. (2021). Gut CD4(+) T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat Immunol 22, 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, and Pevzner PA (2019). Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37, 540–546. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, and Schloss PD (2013). Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl Environ Microb 79, 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak Jason L., Petersen C, Stephens WZ, Soto R, Bake E, O’Connell Ryan M., and Round June L. (2015). MyD88 Signaling in T Cells Directs IgA-Mediated Control of the Microbiota to Promote Health. Cell Host & Microbe 17, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, et al. (2020). Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 180, 79–91 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengfelder I, Sava IG, Hansen JJ, Kleigrewe K, Herzog J, Neuhaus K, Hofmann T, Sartor RB, and Haller D (2019). Complex Bacterial Consortia Reprogram the Colitogenic Activity of Enterococcus faecalis in a Gnotobiotic Mouse Model of Chronic, Immune-Mediated Colitis. Front Immunol 10, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation, Nucleic Acids Res; gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Lu W, Tian F, Zhao J, Zhang H, Hong K, and Yu L (2021). Akkermansia muciniphila Exerts Strain-Specific Effects on DSS-Induced Ulcerative Colitis in Mice. Front Cell Infect Microbiol 11, 698914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, and Knight R (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen YF, et al. (2018). American Gut: an Open Platform for Citizen Science Microbiome Research. Msystems 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, and Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme Journal 6, 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, and Foster KR (2016). Host Selection of Microbiota via Differential Adhesion. Cell Host Microbe 19, 550–559. [DOI] [PubMed] [Google Scholar]
- Mi HY, Muruganujan A, Ebert D, Huang XS, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, and Ohno H (2020). Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 585, 102–106. [DOI] [PubMed] [Google Scholar]
- Moor K, Fadlallah J, Toska A, Sterlin D, Balmer ML, Macpherson AJ, Gorochov G, Larsen M, and Slack E (2016). Analysis of bacterial-surface-specific antibodies in body fluids using bacterial flow cytometry. Nat Protoc 11, 1531–1553. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, et al. (2019). Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, and Gordon JI (2007). IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339. [DOI] [PubMed] [Google Scholar]
- Plichta DR, Juncker AS, Bertalan M, Rettedal E, Gautier L, Varela E, Manichanh C, Fouqueray C, Levenez F, Nielsen T, et al. (2016). Transcriptional interactions suggest niche segregation among microorganisms in the human gut. Nat Microbiol 1, 16152. [DOI] [PubMed] [Google Scholar]
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, et al. (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23, 107–113. [DOI] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, and Florin TH (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105, 2420–2428. [DOI] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Schirmer M, Denson L, Vlamakis H, Franzosa EA, Thomas S, Gotman NM, Rufo P, Baker SS, Sauer C, Markowitz J, et al. (2018). Compositional and Temporal Changes in the Gut Microbiome of Pediatric Ulcerative Colitis Patients Are Linked to Disease Course. Cell Host Microbe 24, 600–610 e604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Muijlwijk GH, van Mierlo G, Jansen P, Vermeulen M, Bleumink-Pluym NMC, Palm NW, van Putten JPM, and de Zoete MR (2021). Identification of Allobaculum mucolyticum as a novel human intestinal mucin degrader. Gut Microbes 13, 1966278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, et al. (2017). IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, and Holt KE (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. Plos Comput Biol 13, e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmore JR, Gaudette BT, Gomez Atria D, Hashemi T, Jones DD, Gardner CA, Cole SD, Misic AM, Beiting DP, and Allman D (2018). Commensal Microbes Induce Serum IgA Responses that Protect against Polymicrobial Sepsis. Cell Host Microbe 23, 302–311 e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegarra-Ruiz DF, El Beidaq A, Iñiguez AJ, Di Ricco ML, Vieira SM, Ruff WE, Mubiru D, Fine RL, Sterpka J, Greiling TM, Dehner C, and Kriegel MA (2019). A Diet-Sensitive Commensal Lactobacillus Strain Mediates TLR7-Dependent Systemic Autoimmunity. Cell Host & Microbe 25, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, and Nunez G (2016). Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 44, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Li P, Wu X, Lu G, Marcella C, Ji X, Ji G, and Zhang F (2020). Alterations of Akkermansia muciniphila in the inflammatory bowel disease patients with washed microbiota transplantation. Appl Microbiol Biotechnol 104, 10203–10215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Allobaculum sp. 128 does not bloom during inflammation, and a second UC patient Allobaculum isolate is colitogenic in gnotobiotic mice, Related to Figure 1. (A) Time course of fecal microbiota profiling of MC+Allo colonized mice across 7 days of 2% DSS administration (n=4 mice/group). (B) Time course of fecal lipocalin (LCN-2) overlaid with Allobaculum sp. 128 abundance. (C) H&E-stained colon sections from Rag1−/− mice on d7 of DSS administration. Scale bars, 200μm. (D-E) Colon lamina propria CD4+IL-17A+ T cells from Il10−/− mice after 8 weeks colonization (n=5 mice/group). (F-G) GF mice were monocolonized with either Ery47 (control bacteria) or Allobaculum sp. 128 for 7 days before the induction of acute colitis via 2% DSS administration. Colonic inflammation was assessed by fecal lipocalin on d3 (F) and colon length on d7 (G) (n=5 mice/group). (H) Phylogenetic tree constructed using 16S rRNA gene sequences from members of family Erysipelotrichaceae, using maximum likelihood estimation, bootstrapped (BS) to 1,000 replicates. BS values are shown along branches. (I) Experimental schematic. (J) Second Allobaculum isolate (Allo2) was used for acute colitis model in WT gnotobiotic mice as in Figure 1C–1H. Three of six mice colonized with Allo2 were found dead (3 F.D.) prior to endpoint. (K) Fecal lipocalin, LCN2, was measured longitudinally by ELISA. Error bars show Mean ± SEM. Data shown in (A-B) from N=1 experiment. Data in (D-E) fron one of N=2 experiments. Data in (F-G) from N=1 experiment. Data shown in (J-K) from one of N=2 independent experiments, n=5–6 mice per group.
Figure S2. Unremarkable histopathology and total serum Ig of untreated Allobaculum sp. 128-colonized WT mice, Related to Figure 1. (A) Bouin’s-fixed H&E-stained colon sections from WT mice colonized with MC bacteria or MC+Allobaculum sp. 128, euthanized 12 weeks later. Scale bars, 200μm. (B) Blinded scoring for colitis. One experiment shown is representative of N=2 independent experiments. (C-D) Total serum IgA and IgG content at 7 weeks (n=3–4 mice/group). (E) FITC-dextran concentration in serum 1h after oral gavage (n=4–8 mice/group). (F) Allobaculum sp. 128-specific IgG in fecal water at 7 weeks post-colonization with MC or MC+Allo (n=5 mice/group). (G) Thickness of the colonic inner mucus layer after 2 weeks of colonization (tissues fixed in Carnoy’s solution to preserved the inner mucus layer; n=10–12 mice/group). Welch’s t-test was used to compare microbiota groups at each time point. ** P<0.01, * P<0.05. Data shown in (A-B) are from one representative of N=3 experiments. Data shown in (C-D) show one of N=2 experiments. Data in (E) are from N=1 experiments. Data in (F) show one of N=2 experiments. Data in (G) are compiled from N=2 experiments.
Figure S3. Microbial diversity cannot explain the relationship between Allobaculum and A. muciniphila, A. muciniphila and Allobaculum co-colonize the ileal mucosa, and co-colonization has minimal impacts on A. muciniphila and Allobaculum gene expression, Related to Figures 3 & 4. (A) Genus-level richness, Simpson’s diversity index, Shannon’s diversity index, and evenness of each microbiome that contained Allobaculum (n=14) or lacked Allobaculum sp. 128 (n=5). (B) Legend accompanying Fig. 4B. Data shown from N=1 experiment. (C) GF WT mice were mono- or bi-colonized as shown for 10 days. Terminal ilea were fixed and sections stained with bacterial FISH probes EUB338 (to stain Allobaculum) and VP403 (to stain A. muciniphila). Scale bars, 10μm. (D) In vivo bacterial transcriptomes from the ileum and colon were compared for differential expression of ORFs across single colonization or co-colonization conditions (MC+Allo vs MC+Both, and MC+Akk vs. MC+Both). (E) Allobaculum sp. 128 and (F) A. muciniphila ORFs that were differentially expressed in the ileum, along with their fold changes, P-values, and annotations (Prokka). Data are from N=1 experiment.
Figure S4. Type strain A. muciniphila attenuates Allobaculum sp. 128-mediated colitis and Allobaculum sp. 128 blunts A. muciniphila-induced dendritic cell responses in MLN, Related to Figure 5. (A) Experimental schematic for acute DSS colitis in WT gnotobiotic mice colonized with MC, MC+Allobaculum sp. 128, MC+A. muciniphilaT (type strain ATCC BAA-835), or MC+Allobaculum sp. 128+A. muciniphilaT (ATCC BAA-835) (n=4–6 mice/group). (B) Fecal microbiota profiling, (C) colon length, (D) d2 fecal lipocalin (LCN2), and (E) gross colon pathology. (F) Representative gating strategy for analysis of MLN cells performed in FlowJo after ≥100,000 events per sample were collected. (G) Immunophenotyping of MLN cell populations (% of viable cells) (n=4–6 mice/group). (H) Quantification of DCs (Live B220−TCRb−CD11b+CD11c+MHCII+). Welch’s t-test was used to compare microbiota groups. *P<0.05. Data shown in (A-E) are from N=1 experiment. Data shown in (F-H) are from one of N=2 independent experiments.
Figure S5. Approach for profiling microbiota-dependent mucosal immune landscape using single cell RNAseq, Related to Figure 7. (A) Schematic depicting single cell RNAseq (scRNAseq) experiment. (B) Quality control metrics used for filtration of scRNAseq data before proceeding to clustering and differential expression analyses. Only cells with 500–5000 RNA features were retained (between blue dashed lines), as well as cells with <8% genes of mitochondrial (mt) origin. Welch’s t-test was used to compare each cell lineage across microbiota groups; * P<0.05, ** P<0.01, *** P<0.001. N=1 experiment.
Figure S6. Expression of marker genes mapped to MLN cell clusters, Related to Figure 7. (A) Violin plots showing expression of marker genes across MLN cell clusters, numbered to match clusters shown in Figure S7.
Figure S7. Epistatic reversal of A. muciniphila-induced MLN immune cell clusters by co-colonization with Allobaculum sp. 128, and direct assessment of MLN DC function in co-colonized gnotobiotic mice, Related to Figure 7 and Figure S6. (A-E) Graph-based probabilistic analysis of MLN scRNAseq data, comparing two microbiota groups at a time. (A-B) MC+Allo relative to MC+Both or relative to MC. (C-E) MC+Akk relative to MC+Both reveals strong reversal (high MELD score) of cell clusters induced by A. muciniphila after co-colonization with Allobaculum sp. 128. The marker genes that define each cluster are displayed in Figure S6. (F-G) Ex vivo co-cultures of purified MLN DCs from gnotobiotic mice were examined for their capacity to prime CellTrace Violet (CTV)-labeled naïve OT-II T cells, by proliferation and dilution of CTV. Data shown are from N=1 experiment.
Table S1. Allobaculum and A. muciniphila abundance in two human cohorts broadly reflects an inverse relationship, Related to Figure 3. (Sheet 1. American Gut Project) Enumeration of sequencing data from American Gut Project (McDonald, et al. 2018; QIITA study IDs: 48742, 51570, 52698, 53379, & 54454). (Sheet 2. Schirmer_UC) Relative abundance data from pediatric ulcerative colitis (Schirmer, et al. 2018).
Table S2. Human microbiota-associated gnotobiotic mouse screen reveals inverse correlation between Allobaculum sp. 128 and A. muciniphila, Related to Figure 3. (Sheet 1. Relative Abundance) fecal microbiota profiles across n=19 gnotobiotic mice. (Sheet 2. Correlation Metrics, Sorted) Compiled statistics evaluating correlation and logistic regression between each pair of microbiota OTUs. (Sheet 3. Pearson R) Calculations of Pearson correlation. (Sheet 4. Spearman R) Calculations of Spearman correlation. (Sheet 5. Diversity Calcs) Calculations of alpha-diversity, beta-diversity, and evenness.
Data Availability Statement
Whole genome sequences, fecal 16S microbiota profiles, and bulk RNAseq data are available at NCBI Bioproject PRJNA739762. Single-cell RNAseq data are available at NCBI GEO GSE179165.