Abstract
The identity of Frankia strains from nodules of Myrica gale, Alnus incana subsp. rugosa, and Shepherdia canadensis was determined for a natural stand on a lake shore sand dune in Wisconsin, where the three actinorhizal plant species were growing in close proximity, and from two additional stands with M. gale as the sole actinorhizal component. Unisolated strains were compared by their 16S ribosomal DNA (rDNA) restriction patterns using a direct PCR amplification protocol on nodules. Phylogenetic relationships among nodular Frankia strains were analyzed by comparing complete 16S rDNA sequences of study and reference strains. Where the three actinorhizal species occurred together, each host species was nodulated by a different phylogenetic group of Frankia strains. M. gale strains from all three sites belonged to an Alnus-Casuarina group, closely related to Frankia alni representative strains, and were low in diversity for a host genus considered promiscuous with respect to Frankia microsymbiont genotype. Frankia strains from A. incana nodules were also within the Alnus-Casuarina cluster, distinct from Frankia strains of M. gale nodules at the mixed actinorhizal site but not from Frankia strains from two M. gale nodules at a second site in Wisconsin. Frankia strains from nodules of S. canadensis belonged to a divergent subset of a cluster of Elaeagnaceae-infective strains and exhibited a high degree of diversity. The three closely related local Frankia populations in Myrica nodules could be distinguished from one another using our approach. In addition to geographic separation and host selectivity for Frankia microsymbionts, edaphic factors such as soil moisture and organic matter content, which varied among locales, may account for differences in Frankia populations found in Myrica nodules.
The nitrogen-fixing actinomycete Frankia establishes a symbiosis with actinorhizal plants, leading to root nodule formation. The host plants, distributed among eight different families and more than 200 species (9), typically colonize N-deficient and disturbed environments such as bogs, sandy coastal dunes, arctic tundra, mine spoils, and volcanic soils. Many actinorhizal species occur early after disturbances and are pioneer species.
Among the actinorhizal genera, Myrica, belonging to the Myricaceae family, has the widest geographic distribution. This genus is divided into 35 species, with 28 reported to be actinorhizal (9). They are small trees or shrubs common to nearly all major landmasses and with species occurring from tropical to temperate areas. According to morphological features (17), fossil and pollen records (26, 39), and a recent molecular study based on the rbcL gene (35), the Myricaceae family is considered the most primitive actinorhizal family.
Since the first confirmed isolation of the microsymbiont genus Frankia (11), a variety of methods have been used to obtain a coherent classification of the bacterial symbiont at the species level: soluble protein patterns (5, 6, 25), isoenzyme patterns (24), fatty acids (58), serology (4), DNA-DNA relatedness (1, 2, 23), and genome (19) and plasmid restriction analyses (52). These studies concluded that Frankia strains were heterogeneous and clustered within two main groups: the Alnus-Myrica-Casuarina group and the Elaeagnaceae group. Strains that infect Myrica spp. often grouped with Alnus-infective strains. However, some studies based on proteins (25, 55), host specificity (3), and genetic characterization (13, 60) pointed out the existence of strains isolated from or identified on Myrica spp. nodules having close relationships with Elaeagnaceae-infective strains. These studies led researchers to consider Myrica spp. promiscuous. Maggia and Bousquet (35) supported this hypothesis and linked it to the evolutionary position of the host plant. These authors have proposed that host plant evolution has proceeded toward more specificity, and therefore the most primitive actinorhizal plants, such as Myrica spp., should have a broad host range.
However, studies of strains isolated from Myrica spp. have some experimental limitations: (i) pure cultures tested for their infectivity often appeared to be atypical because they nodulated Elaeagnaceae but not their original host plant, (ii) some of the pure cultures were obtained after passage through an intermediate host, generally an Alnus species, before isolation, and (iii) to avoid the bias due to the isolation and culture steps, some authors identified the strains using direct detection in plant nodules, but the approaches used, even when based on the rrs gene or on the 16S-23S intergenic spacer, never yielded total 16S sequences allowing precise phylogenetic positioning.
The purpose of this study was to further elucidate phylogenetic and ecological relationships of Myrica-infective strains, Alnus-infective strains, and Elaeagnaceae-infective strains. We studied nodular Frankia strains from a site on which three different actinorhizal plants belonging to different infectivity groups (Myrica gale, Alnus incana subsp. rugosa, and Shepherdia canadensis) were growing in close proximity. Using PCR-restriction fragment length polymorphism (RFLP) analysis of the whole rrs gene, we analyzed the level of heterogeneity of M. gale-nodulating strains and the specificity of their genetic patterns compared with those of Alnus- and Shepherdia-infective strains. M. gale nodules were also sampled from two additional sites in order to detect relationships between site factors and Frankia strain diversity. The total rrs gene sequences from several Myrica isolates and field nodules were determined to assess the phylogenetic position of Myrica-infective strains with greater precision.
MATERIALS AND METHODS
Nodules and sites.
A total of 75 nodules were sampled at three sites (Table 1). Most of the nodules were collected at sites 1 and 2 in Wisconsin. Site 1 had pH values of 7 to 7.5 and organic matter percentages of 0.7 to 2.8. Site 2 had a pH value of 6.1 and an organic matter percentage of 9.6. Site 3 was in Glasgow, United Kingdom (U.K.), and consisted of an upland acid peat from the Trossachs with a pH range of 3.5 to 4.0 transferred 7 years previously along with M. gale transplants to a pit in a loam soil derived from glacial boulder clay in the Glasgow Botanic Gardens. The nodules came from roots close to the root collar and were probably derived from the upland acid peat soil. Several Alnus species grew near this site in the Gardens but no Myrica spp. M. gale occurred on all the sites, while S. canadensis and A. incana subsp. rugosa occurred only on site 1. On site 1 there is a steep moisture gradient on a lake shore sand dune, and M. gale, S. canadensis, and A. incana subsp. rugosa grow in close proximity. A. incana subsp. rugosa and M. gale occupy the moist lower dunes near interdunal ponds, while S. canadensis occurs on the drier upper slopes. Entire plants were excavated, and their root systems were removed intact before cleaning and sampling in order to ascertain the host origin of nodules.
TABLE 1.
Nodules and isolates used in this study
| Site no. | Nodule or straina | Designationb | Original host plant | Geographic origin | Source or reference |
|---|---|---|---|---|---|
| 1 | Alnus nodules (An)a | 1-1 to 1-7 | A. incana subsp. rugosa | Door County, Wis. | This study |
| 2-1 to 2-9 | |||||
| 3-1 to 3-8 | |||||
| Shepherdia nodules (Sn) | 1-1 to 1-4 | S. canadensis | This study | ||
| 2 | |||||
| 3-1 to 3-4 | |||||
| 4-1 to 4-3 | |||||
| 5-1 to 5-8 | |||||
| Myrica nodules | |||||
| MMn | 1-1 to 1-13 | M. gale | This study | ||
| 2 | |||||
| 3-1 to 3-6 | |||||
| 2 | MRn | 1-1 to 1-4 | M. gale | This study | |
| 2-1 to 2-5 | |||||
| 3 | MWnc | 1-1 to 1-2 | M. gale | Botanical Garden, Glasgow, U.K. | This study |
| Myrica isolatesd | |||||
| M16386 | M. californica | Westport-Legget, Calif. | 31 | ||
| M16464 | M. pensylvanica | Harvey Cedars, N.J. | 7 | ||
| M16467 | M. pensylvanica | Tinton Falls, N.J. | 7 | ||
| MgI5 | M. gale | Tupper Lake, N.Y. | 31 | ||
| MpI1 | M. pensylvanica | Nantucket, Mass. | 33 |
The designation used in phylogenetic trees is shown.
The first digit represents the plant number, and the second digit represents the nodule number on this plant.
Kindly provided by C. T. Wheeler (University of Glasgow, Glasgow, U.K.).
Kindly provided by D. Labeda (U.S. Department of Agriculture [USDA]). The accession numbers in the USDA collection are NRRL B-16386, NRRL B-16464, NRRL B-16467, NRRL B-16404, and NRRL B-16317, respectively, and the registration numbers are LLR 160401, RBR 162013, RBR 162021, DDB 16110210, and LLR 162001, respectively.
Bacterial strains.
Five Frankia strains isolated from three different Myrica species were included in this study (Table 1). They were grown in BuCT medium (28) at 28°C in the dark with weekly manual agitation.
DNA extraction.
DNA from root nodules was directly extracted according to Rouvier et al. (49). After washing the nodules with water, a single lobe was selected, and the outer layers were removed. Each lobe was crushed in 300 μl of TCP extraction buffer (100 mM Tris-HCl [pH 7], 0.5 M NaCl, 50 mM EDTA [pH 8], 2% [wt/vol] cethyltrimethylammonium bromide [Sigma, St. Louis, Mo.], 1% [wt/vol] polyvinylpolypyrrolidone [Sigma]). The homogenate was incubated at 65°C for 1 h and centrifuged twice at 6,000 × g for 5 min. The supernatant was extracted with an equal volume of chloroform-isoamyl alcohol (24:1, vol/vol) and centrifuged at 13,000 × g for 20 min. DNA from the aqueous phase was precipitated in ethanol for at least 2 h at −20°C. The sample was then centrifuged at 13,000 × g for 30 min, and the resulting DNA pellet was washed with 70% (vol/vol) ethanol, air dried, and dissolved in 10 μl of Tris-EDTA (TE) buffer (pH 7.5).
For isolated Frankia strains, hyphae were fragmented by repeatedly passing colonies through a 1.2-mm-diameter needle and were then washed three times in 500 μl of TE buffer (pH 7.5). DNA was extracted by sonication using a Cup Horn probe (Bioblock Scientific, Illkirch, France) for 1 to 2 min (level 2 and 50% active cycles). These samples were then kept at −20°C.
PCR amplification.
Prior to the PCR, DNA extracted from nodules was digested with NruI (Boehringer Mannheim, Meylan, France) to avoid amplification of the 16S ribosomal DNA (rDNA) of the chloroplasts present in the root cells (44). Amplification of the whole 16S rDNA was performed using primers FGPS-1509′-153 (5′-AAGGAGGGGATCCAGCCGCA-3′) and FGPS4-281bis (5′-ATGGA[G/A]AG[T/C]TTGATCCTGGCTCA-3′) (43) in a GeneAmp PCR system 2400 thermocycler (Perkin-Elmer Instruments, Norwalk, Conn.) under the following conditions: initial denaturation for 3 min at 94°C, 35 cycles of denaturation (1 min at 94°C), annealing (45 s at 60°C), and extension (1 min at 72°C), and a final extension for 7 min at 72°C. The reaction volume was 50 μl, containing 5 μl of diluted DNA, 1× PCR buffer (Gibco-BRL, Cergy-Pontoise, France), 1.5 mM MgCl2, W-1% detergent (0.05%, vol/vol) (Gibco-BRL), 0.5 μM each primer, and 2.5 U of Taq polymerase (Gibco-BRL). When amplification was not sufficient, T4 gene 32 protein (Boehringer Mannheim) was used to improve its yield. The amplification of DNA was checked by agarose gel electrophoresis (1%, wt/vol) with 5 μl of PCR product, and the gel was stained with ethidium bromide (0.5 μg/ml) (Sigma).
RFLP.
PCR products were digested for 2 h at the optimal conditions recommended by the manufacturer. The following restriction endonucleases were used: 10 U of CfoI, HaeIII, and MspI and 5 U of NdeII (all from Boehringer Mannheim). Restriction fragments were separated by electrophoresis on a 2.5% (wt/vol) Metaphor agarose gel (FMC Bioproducts, Rockland, Maine) containing ethidium bromide (0.5 μg. ml−1) and photographed with Ilford FP4 film. The combination of the restriction patterns obtained with the four endonucleases used allowed the determination of PCR-RFLP groups (or types) and the assignment of each strain to one of these groups. Nine reference strains for which the rrs genes had already been sequenced were also used, and their RFLP patterns were simulated using the MacVector software (International Biotechnology Kodak, Rochester, N.Y.). These strains, from different actinorhizal families, allowed us to clearly identify the nonisolated strains. A dendrogram was obtained by calculating the Dice coefficient and by using the UPGMA algorithm (53).
Sequencing.
The rrs gene of the five Frankia strains isolated from Myrica spp. and three nonisolated strains (nodules MRn2-2, Sn4-3, and Sn5-8) was sequenced by Génome Express S.A. (Grenoble, France). Before sequencing, the amplificon was purified with the QIAquick kit (Qiagen S.A., Les Ulis, France). Sequences were deposited in EMBL, and their accession numbers are given in Fig. 2. The sequences were aligned by using ClustalX software (57) and compared to reference sequences (from isolated and nonisolated Frankia strains and closely related bacteria) by using the MASE software (21). The distances were calculated according to Kimura's two-parameter method (30), and a dendrogram was obtained by using the neighbor-joining algorithm (50). The bootstrap values were determined from 1,000 replicates (22). The reference sequences were chosen in order to have a representative range of infectivity groups and the infectivity and effectivity capacities of the strains.
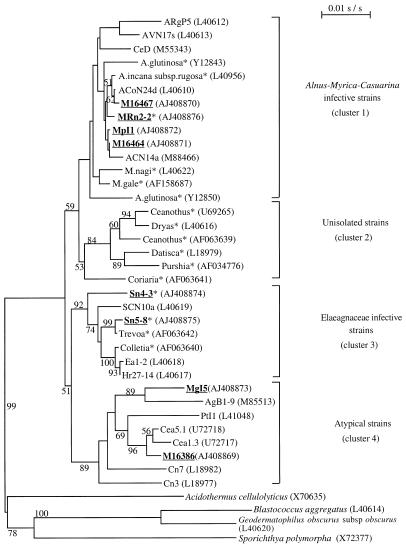
FIG. 2.
Phylogenetic tree based on rrs gene sequences obtained by the neighbor-joining algorithm (50). The numbers on the tree branches are bootstrap values. Only values larger than 50% are shown. GenBank accession numbers are shown in parentheses. Strains characterized in the present study are underlined and in bold. ∗, unisolated strains. Clusters are defined in reference 44.
RESULTS
DNA extraction and PCR amplification.
DNA extraction and rrs gene amplification were conducted for 40 nodules. The high concentration of plant molecules (tannins, polyphenols, polysaccharides, etc.) indicated by the dark color of the nodule extracts, together with the large size of the amplified fragment, may explain the low amplicon yields of some samples. The PCR products generated consisted of a unique amplicon of the expected size (approximatively 1,500 bp) for isolated and nonisolated strains.
RFLP analysis.
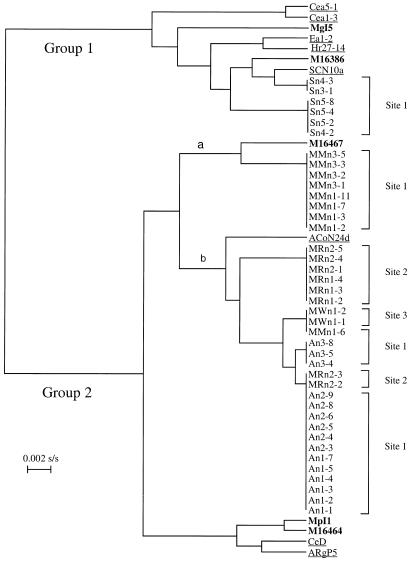
Because of the detection threshold of the agarose gel electrophoresis, fragments shorter than 80 bp were not considered for the analysis. The restriction patterns obtained with the four enzymes used are shown in Table 2. Depending on the restriction enzyme used, from 2 (NdeII) to 11 (HaeIII) different patterns were generated. Combining all the patterns obtained allowed the determination of 20 PCR-RFLP types. Seven types were obtained from amplified DNA of unisolated, nodular strains of M. gale, A. incana subsp. rugosa, and S. canadensis. Five types were from Frankia strains isolated from Myrica spp., and there were eight reference strain types. DNA from Elaeagnaceae strains, including both reference strains and unisolated (nodules) strains, was characterized by M2 and N2 patterns. Two Myrica spp. isolates together with two Ceanothus spp. isolates also harbored the N2 RFLP pattern. MspI restriction pattern analysis appears to be a useful tool for distinguishing between the two groups, which include strains that nodulate the Elaeagnaceae on the one hand, and Alnus, Myrica, and Casuarina on the other hand. The dendrogram obtained by the UPGMA algorithm is presented in Fig. 1. Frankia strains studied are divided into two groups. Group 1 includes Elaeagnaceae strains (reference strains and Shepherdia unisolated strains), the atypical strains Cea5.1 and Cea1.3, and strains MgI5 and M16386, isolated from M. gale and Myrica californica, respectively. Shepherdia unisolated strains are grouped with the only Shepherdia isolate known to be clearly separated phylogenetically from the Elaeagnus-Hippophaë branch. Group 2 includes strains infective on Alnus, Myrica, and Casuarina. The DNA of Frankia strains in Myrica nodules and Frankia DNA detected in Alnus nodules differed with the exception of Frankia DNA in nodules MRn2-2 and MRn2-3. The Frankia strains detected in Myrica nodules are phylogenetically related but clearly separated according to their site of origin.
TABLE 2.
Restriction patterns of amplified rrs gene from nodular Frankia strains and reference strains
| Nodule or strain | Restriction enzyme patternsa
|
Typesb | |||
|---|---|---|---|---|---|
| CfoI | HaeIII | MspI | NdeII | ||
| Alnus nodules | |||||
| An 1-1 | C1 | H1 | M1 | N1 | I |
| An 1-2 | C1 | H1 | M1 | N1 | I |
| An 1-3 | C1 | H1 | M1 | N1 | I |
| An 1-4 | C1 | H1 | M1 | N1 | I |
| An 1-5 | C1 | H1 | M1 | N1 | I |
| An 1-7 | C1 | H1 | M1 | N1 | I |
| An 2-3 | C1 | H1 | M1 | N1 | I |
| An 2-4 | C1 | H1 | M1 | N1 | I |
| An 2-5 | C1 | H1 | M1 | N1 | I |
| An 2-6 | C1 | H1 | M1 | N1 | I |
| An 2-8 | C1 | H1 | M1 | N1 | I |
| An 2-9 | C1 | H1 | M1 | N1 | I |
| An 3-4 | C2 | H1 | M1 | N1 | II |
| An 3-5 | C2 | H1 | M1 | N1 | II |
| An 3-8 | C2 | H1 | M1 | N1 | II |
| Shepherdia nodules | |||||
| Sn 3-1 | C3 | H2 | M2 | N2 | III |
| Sn 4-2 | C3 | H1 | M2 | N2 | IV |
| Sn 4-3 | C3 | H2 | M2 | N2 | III |
| Sn 5-2 | C3 | H1 | M2 | N2 | IV |
| Sn 5-4 | C3 | H1 | M2 | N2 | IV |
| Sn 5-8 | C3 | H1 | M2 | N2 | IV |
| Myrica nodules | |||||
| MMn 1-2 | C2 | H3 | M3 | N1 | V |
| MMn 1-3 | C2 | H3 | M3 | N1 | V |
| MMn 1-6 | C1 | H1 | M4 | N1 | VI |
| MMn 1-7 | C2 | H3 | M3 | N1 | V |
| MMn 1-11 | C2 | H3 | M3 | N1 | V |
| MMn 3-1 | C2 | H3 | M3 | N1 | V |
| MMn 3-2 | C2 | H3 | M3 | N1 | V |
| MMn 3-3 | C2 | H3 | M3 | N1 | V |
| MMn 3-5 | C2 | H3 | M3 | N1 | V |
| MRn 1-2 | C1 | H4 | M1 | N1 | VII |
| MRn 1-3 | C1 | H4 | M1 | N1 | VII |
| MRn 1-4 | C1 | H4 | M1 | N1 | VII |
| MRn 2-1 | C1 | H4 | M1 | N1 | VII |
| MRn 2-2 | C1 | H1 | M1 | N1 | I |
| MRn 2-3 | C1 | H1 | M1 | N1 | I |
| MRn 2-4 | C1 | H4 | M1 | N1 | VII |
| MRn 2-5 | C1 | H4 | M1 | N1 | VII |
| MWn 1-1 | C1 | H1 | M4 | N1 | VI |
| MWn 1-2 | C1 | H1 | M4 | N1 | VI |
| Myrica isolates | |||||
| M16386 | C3 | H2 | M5 | N2 | VIII |
| M16464 | C4 | H5 | M6 | N1 | IX |
| M16467 | C4 | H6 | M4 | N1 | X |
| MgI5 | C5 | H7 | M7 | N2 | XI |
| MpI1 | C4 | H8 | M6 | N1 | XII |
| Alnus strainsc | |||||
| ACoN24d | C4 | H5 | M1 | N1 | XIII |
| ARgP5 | C6 | H5 | M8 | N1 | XIV |
| Elaeagnaceae strainsc | |||||
| SCN10a | C3 | H5 | M2 | N2 | XV |
| Hr27-14 | C6 | H5 | M2 | N2 | XVI |
| Ea1-2 | C6 | H9 | M2 | N2 | XVII |
| Other strainsc | |||||
| Cea1.3 | C7 | H10 | M9 | N2 | XVIII |
| Cea5.1 | C8 | H10 | M9 | N2 | IXX |
| CeD | C6 | H11 | M6 | N1 | XX |
The letters designate the restriction enzyme used, and the number designates the pattern obtained with this enzyme.
The types represent the combination of patterns obtained with the four enzymes tested on the rrs gene.
Reference strains. Theoretical restriction patterns of the rrs gene using MacVector software.
FIG. 1.
Phylogenetic tree based on PCR-RFLP patterns analyzed with the UPGMA algorithm (53). Strains isolated from Myrica spp. are in bold. Reference strains (GenBank accession numbers, from top to bottom: U72718, U72717, L40618, L40617, L40619, L40610, M55343, and L40612) are underlined.
Sequencing.
The phylogenetic tree based on the 16S rDNA sequencing results and obtained by the neighbor–joining algorithm is shown in Fig. 2. In agreement with PCR-RFLP results, strains infective on Myrica spp. are divided into two groups. Isolated strains MpI1, M16464, and M16467 and unisolated strain MRn2-2 have as the closest phylogenetic neighbor strain ACN14a and belong to the Alnus-Myrica-Casuarina infectivity group. Isolates MgI5 and M16386, previously grouped with the Elaeagnaceae-infective strains (Fig. 1), have for their closest phylogenetic neighbor strain Cea5.1, isolated from a Ceanothus sp. (Rhamnaceae family), and form a coherent cluster, according to bootstrap values, with strains known to be atypical for their infectivity and/or effectivity phenotype. Unisolated Shepherdia strains Sn4-3 and Sn5-8 are closely related to strain FE39, isolated from Trevoa trinervis (Rhamnaceae family) and belong to the group of strains infective on members of the Elaeagnaceae and Rhamnaceae families.
DISCUSSION
M. gale-nodulating strains from three sites on two continents were genetically uniform (Fig. 1, group 2), grouping together with strains infecting Alnus and clustering with strain ACoN24d, representative of Frankia alni species. Nevertheless, specific genotypes apparently occurred at each site with few exceptions, leading to the conclusion that low diversity is present in each stand. This local dominance has been reported previously for A. incana and Myrica pensylvanica (15). Soil composition parameters that frequently correlated with the level of Frankia population diversity include soil moisture (18, 46, 48), pH (27, 29), and organic matter content (10). In this study, site 1 (neutral, low in organic matter, not flooded) and sites 2 and 3 (acidic, high in organic matter, flooded or moist) represent contrasting ecological environments that possibly support different types of Myrica-infective Frankia strains. It will be necessary to study more sites before drawing general conclusions.
All Elaeagnaceae strains, including Shepherdia-infective strains of site 1, were grouped in a distinct and deeply diverging branch of the phylogenetic tree (Fig. 1, group 1). In our study, all of the M. gale nodules sampled contained Alnus-like genotypes, and none contained Elaeagnaceae (or closely related)-infective Frankia strains. Other authors (13, 60) found “Elaeagnaceae-like genotypes” but on two other Myrica species nodules, M. cerifera and M. pensylvanica. Among the isolated strains tested, strain Mg15 isolated from M. gale was found to group with Elaeagnaceae strains by PCR-RFLP but not with the complete rrs gene sequence. However, this strain is atypical, since no nodulation was obtained on its original host plant, M. gale, but was obtained on Elaeagnaceae species (3, 31). Thus it can be concluded that even if Myrica is often considered a promiscuous actinorhizal genus, our results show that M. gale does not exhibit such promiscuity in its natural environment. The limited diversity of strains infective on M. gale could be correlated with the fact that this Myrica species typically grows in a narrow and possibly restrictive range of wet acidic and organic soils. Such a possibility has also been proposed by Clawson and Benson (13).
Results from site 1, where the three actinorhizal genera grow in close proximity, showed that each plant species is nodulated by a distinct Frankia genotype. Although Myrica- and Alnus-infective strains belong to the same host specificity group (HSG), distinct sets of closely related genotypes were associated with these species with only two exceptions. Surprisingly, in spite of the genetic proximity of Alnus- and Myrica-infective strains, Myrica strains from site 2 (Fig. 1, group 2b) were more closely related to Alnus strains from site 1 (Fig. 1, group 2b) than are Myrica strains from the same site 1 (Fig. 1, group 2a). Thus, strain selection in the soil by the host plant may entail a complex interaction between elements in the soil ecosystem and root-Frankia molecular dialog.
Phylogenetic positioning of the different groups of strains studied was achieved by sequencing the complete rrs gene of representative strains. This approach was chosen instead of the more rapid partial rrs sequencing. Indeed, the variable and conserved zones of the rrs gene, even if generally located in the same regions, were shown to exhibit variable positioning within a genus, which could result in misclassifications (12, 34, 45, 59). In the case of Frankia, the use of partial sequences leads to the separation of Casuarina- from Alnus-infective strains, while strains nodulating Elaeagnaceae become closer to strains nodulating Alnus (data not shown). This grouping is not consistent with most of the results reported on Frankia strains, including biochemical, physiological, and phylogenetic approaches (for a review, see reference 32). On the contrary, the phylogenetic tree based on complete rrs sequences (Fig. 2) confirms the results obtained using RFLP data (Fig. 1), demonstrating that the data used in the RFLP approach were sufficiently informative and discriminating even if only a few restriction enzymes were used. Some of them, such as NdeII, produced banding patterns that are highly specific for the different phylogenetic groups and are phylogenetically and taxonomically useful (37, 38). The bootstrap values shown in Fig. 2, even if not very high, appear to be significant. The topology of this tree is congruent with various phenotypic data, such as infection pathways (36), serology (4), cell wall 2-O-methyl-d-mannose abundance (54), cellular fatty acid composition (51), susceptibility to antibiotics (20), and more recent phylogenetic studies using the glnII gene (16).
For Myrica spp., rrs genes of five reference strains and one unisolated strain were sequenced. The isolates MpI1, M16464, and M16467, all isolated from M. pensylvanica, and the unisolated strain MRn2-2, from M. gale, are closely related to F. alni species and to unisolated strains nodulating Myrica and Alnus according to PCR-RFLP and sequencing data. This result confirms previous studies including these three isolates (5, 7, 8, 31, 42), where they belong to the cluster of Alnus-infective strains (cluster 1 [44]) and appear to be Nod+ Fix+ on their original host plant. According to Bloom and al. (7, 8), the Frankia strains isolated from M. pensylvanica are heterogeneous, which was confirmed by Clawson and Benson (13), who described this species as promiscuous in the field. The isolates MgI5 and M16386 clustered with isolated strains from various host plants. These strains have a common feature, lack of infectivity and/or effectivity on their original host plant, and group with the atypical strains in the phylogenetic tree (cluster 4 [44]). This positioning confirms what is already known about MgI5. Indeed, this strain is Nod− Fix− on its original host plant, M. gale (31), belongs to HSG 4, which includes strains infective only on the Eleaegnaceae species according to Baker (3), and grouped together with atypical strains in the study of Nittayajarn and al. (42). We therefore agree with the proposal that these marginal strains are not the real symbionts of the actinorhizal plants (47) but were present in their rhizosphere or at the nodule surface and were selected by the culture method. This highlights the necessity of working directly with nodular Frankia strains to avoid biases of isolation and culture.
Few studies examined the diversity and phylogenetic positioning of Shepherdia-infective strains. This study shows that Shepherdia-infective strains present more diversity than strains that nodulate Alnus or Myrica at site 1. In the phylogenetic tree based on rrs sequences, unisolated strains Sn4-3 and Sn5-8 grouped with SCN10a, close to a strain isolated from Trevoa trinervis (Rhamnaceae family) in the Elaeagnaceae-infective strain cluster (cluster 3 [44]). All the Shepherdia nodular DNA identified by the PCR-RFLP approach belongs to this group. The phylogenetic proximity of Elaeagnaceae- and Rhamnaceae-infective strains has been attributed to the common phyletic origin of these families (14, 56). Thus, the set of Shepherdia-infective strains identified in this study provides additional evidence for the congruence of host plant taxa and associated Frankia strain phylogenies. The group of strains that nodulate Ceanothus (Rhamnaceae family) appear to be an exception because they cluster with Rosaceae-infective strains in comparisons of nearly full length rrs sequences (14).
It now seems obvious that the apparent heterogeneity of Myrica isolates is partly due to the consideration of atypical isolated strains that probably originate from the nodule surface as an artifact of incomplete nodule surface sterilization during the isolation process. Such an isolation bias observed on Casuarina-infective strains has led to erroneous classification of strains and still hinders biodiversity studies (23, 41, 49). Because the Myricaceae family is the most primitive actinorhizal family, widely distributed and universally nodulated in its natural environment, it was suspected to harbor a large diversity of strains able to nodulate other introduced actinorhizal plants. Our study indicates that this is not the case for M. gale. Genetic differences in nodular Frankia strains among Myrica host species, soil types, and geographic sources remain to be more fully elucidated in the context of evolution, ecology, and conservation (40), particularly by sampling more sites with varied ecological conditions.
ACKNOWLEDGMENTS
We thank Joëlle Maréchal for useful advice and D. Labeda and C. T. Wheeler for providing biological samples.
This work was supported by a grant from the Ministère de la Recherche et de l'Education.
REFERENCES
- 1.Akimov V N, Dobritsa S V. Grouping of Frankia strains on the basis of DNA relatedness. Syst Appl Microbiol. 1992;15:372–379. [Google Scholar]
- 2.An S C, Wills J W, Riggsby W S, Mullin B C. Relationships of Frankia isolates based on deoxyribonucleic acid homology studies. Int J Syst Bacteriol. 1985;35:140–146. [Google Scholar]
- 3.Baker D. Relationships among pure cultured strains of Frankia based on host specificity. Physiol Plant. 1987;70:245–248. [Google Scholar]
- 4.Baker D, Pengelly W L, Torrey J G. Immunochemichal analysis of relationships among the isolated frankiae (Actinomycetales) Int J Syst Bacteriol. 1981;31:148–151. [Google Scholar]
- 5.Benson D R, Buchholz S E, Hanna D G. Identification of Frankia strains by two-dimensional polyacrylamide gel electrophoresis. Appl Environ Microbiol. 1984;47:489–494. doi: 10.1128/aem.47.3.489-494.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson D R, Hanna D. Frankia diversity in an alder stand as estimated by SDS-PAGE of whole cell protein. Can J Bot. 1983;61:2919–2923. [Google Scholar]
- 7.Bloom R A, Lechevalier M P, Tate R L. Physiological, chemical, morphological, and plant infectivity characteristics of Frankia isolates from Myrica pensylvanica: correlation to DNA restriction patterns. Appl Environ Microbiol. 1989;55:2161–2166. doi: 10.1128/aem.55.9.2161-2166.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom R A, Mullin B C, Tate R L. DNA restriction patterns and DNA-DNA solution hybridization studies of Frankia isolates from Myrica pensylvanica (bayberry) Appl Environ Microbiol. 1989;55:2155–2160. doi: 10.1128/aem.55.9.2155-2160.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bond G. Taxonomy and distribution of non-legume nitrogen-fixing systems. In: Gordon J C, Wheeler C T, editors. Biological nitrogen fixation in forest ecosystems: foundations and applications. The Hague, the Netherlands: Martinus Nijhoff; 1983. pp. 55–87. [Google Scholar]
- 10.Burleigh S H, Dawson J O. Occurrence of Myrica-nodulating Frankia in Hawaiian volcanic soils. Plant Soil. 1994;164:283–289. [Google Scholar]
- 11.Callaham D, Del Tredici P, Torrey J G. Isolation and cultivation in vitro of the actinomycete causing root nodulation in Comptonia. Science. 1978;199:899–902. doi: 10.1126/science.199.4331.899. [DOI] [PubMed] [Google Scholar]
- 12.Cilia V, Lafay B, Christen R. Sequence heterogeneity among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 13.Clawson M L, Benson D R. Natural diversity of Frankia in actinorhizal root nodules from promiscuous hosts in the Myricaceae. Appl Environ Microbiol. 1999;65:4521–4527. doi: 10.1128/aem.65.10.4521-4527.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clawson M L, Carù M, Benson D R. Diversity of Frankia strains in root nodules of plants from the families Elaeagnaceae and Rhamnaceae. Appl Environ Microbiol. 1998;64:3539–3543. doi: 10.1128/aem.64.9.3539-3543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clawson M L, Gawronski J, Benson D R. Dominance of Frankia strains in stands of Alnus incana subsp rugosa and Myrica pensylvanica. Can J Bot. 1999;77:1203–1207. [Google Scholar]
- 16.Cournoyer B, Lavire C. Analysis of Frankia evolutionnary radiation using glnII sequences. FEMS Microbiol Lett. 1999;177:29–34. doi: 10.1111/j.1574-6968.1999.tb13709.x. [DOI] [PubMed] [Google Scholar]
- 17.Cronquist A. The evolution and classification of flowering plants. 2nd ed. New York, N.Y: The New York Botanical Garden; 1988. [Google Scholar]
- 18.Dawson J O, Kowalski D G, Dart P J. Variation with soil depth, topographic position and host species in the capacity of soils from an Australian locale to nodulate Casuarina and Allocasuarina seedlings. Plant Soil. 1989;118:1–13. [Google Scholar]
- 19.Dobritsa S V. Restriction analysis of the Frankia spp. genome. FEMS Microbiol Lett. 1985;29:123–128. [Google Scholar]
- 20.Dobritsa S V. Grouping of Frankia strains on the basis of susceptibility to antibiotics, pigment production, and host specificity. Int J Syst Bacteriol. 1998;48:1265–1275. [Google Scholar]
- 21.Faulkner D V, Jurka J. Multiple aligned sequence editor (MASE) Trends Biochem Sci. 1988;13:321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez M P, Meugnier H, Grimont P A D, Bardin R. Deoxyribonucleic acid relatedness among members of the genus Frankia. Int J Syst Bacteriol. 1989;39:424–429. [Google Scholar]
- 24.Gardes M, Bousquet J, Lalonde M. Isozyme variation among 40 Frankia strains. Appl Environ Microbiol. 1987;53:1596–1603. doi: 10.1128/aem.53.7.1596-1603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardes M, Lalonde M. Identification and subgrouping of Frankia strains using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Physiol Plant. 1987;70:237–244. [Google Scholar]
- 26.Gladkova A N. Fragments of the history of the Myricaceae family. Pollen Spores. 1962;4:345. [Google Scholar]
- 27.Griffiths A P, McCormick L H. Effects of soil acidity on nodulation of Alnus glutinosa and viability of Frankia. Plant Soil. 1984;79:429–434. [Google Scholar]
- 28.Hooker J E, Wheeler C T. The effectivity of Frankia for nodulation and nitrogen fixation in Alnus rubra and A. glutinosa. Physiol Plant. 1987;70:333–341. [Google Scholar]
- 29.Jamann S, Fernandez M P, Moiroud A. Genetic diversity of Elaeagnaceae-infective Frankia strains isolated from various soils. Acta Oecol. 1992;13:395–405. [Google Scholar]
- 30.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 31.Lechevalier M P. Catalog of Frankia strains. Actinomycete. 1986;19:131–162. [Google Scholar]
- 32.Lechevalier M P. Minireview: taxonomy of the genus Frankia (Actinomycetales) Int J Syst Bacteriol. 1994;44:1–8. [Google Scholar]
- 33.Lechevalier M P, Lechevalier H A. The taxonomic position of the actinomycetic endophytes. In: Gordon J C, Wheeler C T, Perry D A, editors. Symbiotic nitrogen fixation in the managment of temperate forests. Corvallis, Oreg: Forest Research Laboratory, Oregon State University; 1979. pp. 111–122. [Google Scholar]
- 34.Ludwig W, Schleifer K H. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 35.Maggia L, Bousquet J. Molecular phylogeny of the actinorhizal Hamamelidae and relationships with host promiscuity towards Frankia. Mol Ecol. 1994;3:459–467. [Google Scholar]
- 36.Miller I M, Baker D D. Nodulation of actinorhizal plants by Frankia strains capable of both root hair infection and intercellular penetration. Protoplasma. 1986;131:82–91. [Google Scholar]
- 37.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution: analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller J. Fossil pollen records of extant angiosperms. Bot Rev. 1981;47:1–142. [Google Scholar]
- 40.Navarro E, Jaffre T, Gauthier D, Gourbiere F, Rinaudo G, Simonet P, Normand P. Distribution of Gymnostoma spp. microsymbiotic Frankia strains in New Caledonia is related to soil type and to host-plant species. Mol Ecol. 1999;8:1781–1788. doi: 10.1046/j.1365-294x.1999.00742.x. [DOI] [PubMed] [Google Scholar]
- 41.Nazaret S, Simonet P, Normand P, Bardin R. Genetic diversity among Frankia isolated from Casuarina nodules. Plant Soil. 1989;118:241–247. [Google Scholar]
- 42.Nittayajarn A, Mullin B C, Baker D D. Screening of symbiotic Frankiae for host specificity by restriction fragment length polymorphism analysis. Appl Environ Microbiol. 1990;56:1172–1174. doi: 10.1128/aem.56.4.1172-1174.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Normand P, Chapelon C. Direct characterization of Frankia and of close phyletic neighbors from an Alnus viridis rhizosphere. Physiol Plant. 1997;99:722–731. [Google Scholar]
- 44.Normand P, Orso S, Cournoyer B, Jeannin P, Chapelon C, Dawson J, Evtushenko L, Misra A K. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol. 1996;46:1–9. doi: 10.1099/00207713-46-1-1. [DOI] [PubMed] [Google Scholar]
- 45.Nour S M, Cleyet-Marel J C, Normand P, Fernandez M P. Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov. Int J Syst Bacteriol. 1995;45:640–648. doi: 10.1099/00207713-45-4-640. [DOI] [PubMed] [Google Scholar]
- 46.Pratt S D, Konopka A S, Murry M A, Ewers F W, Davis S D. Influence of soil moisture on the nodulation of post-fire seedlings of Ceanothus spp. growing in the Santa Monica Mountains of Southern California. Physiol Plant. 1997;99:673–679. [Google Scholar]
- 47.Ramirez-Saad H, Janse J D, Akkermans A D L. Root nodules of Ceanothus caerulus contain both the N2-fixing Frankia endophyte and a phylogenetically related Nod−/Fix− actinomycete. Can J Microbiol. 1998;44:140–148. [Google Scholar]
- 48.Righetti T L, Chard C H, Backhause R A. Soil and environmental factors related to nodulation in Cowania and Purshia. Plant Soil. 1986;91:147–160. [Google Scholar]
- 49.Rouvier C, Prin Y, Reddell P, Normand P, Simonet P. Genetic diversity among Frankia strains nodulating members of the family Casuarinaceae in Australia revealed by PCR and fragment length polymorphism analysis with crushed root nodules. Appl Environ Microbiol. 1996;62:979–985. doi: 10.1128/aem.62.3.979-985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;2:112–118. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Simon L, Jabaji-Hare S, Bousquet J, Lalonde M. Confirmation of Frankia species using cellular fatty acids analysis. Syst Appl Microbiol. 1989;11:229–235. [Google Scholar]
- 52.Simonet P, Normand P, Moiroud A, Lalonde M. Restriction enzyme digestion patterns of Frankia plasmids. Plant Soil. 1985;87:49–60. [Google Scholar]
- 53.Sokal R R, Sneath P H A. Principles of numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1963. [Google Scholar]
- 54.St.-Laurent L, Bousquet J, Simon L, Lalonde M. Separation of various Frankia strains in the Alnus and Elaeagnus host specificity group using sugar analysis. Can J Microbiol. 1987;33:764–772. [Google Scholar]
- 55.St-Laurent L, Lalonde M. Isolation and characterization of Frankia strains isolated from Myrica gale. Can J Bot. 1987;65:1356–1363. [Google Scholar]
- 56.Swensen S. The evolution of actinorhizal symbioses: evidence for multiple origins of the symbiotic association. Am J Bot. 1996;83:1503–1512. [Google Scholar]
- 57.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler C T, Hooker J E, Crose A, Berry A M M. The improvement and utilization in forestry of nitrogen fixation by actinorhizal plants with special reference to Alnus in Scotland. Plant Soil. 1986;90:393–406. [Google Scholar]
- 59.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- 60.Zimpfer J F, Kennedy G J, Smyth C A, Hamelin J, Navarro E, Dawson J O. Localization of Casuarina-infective Frankia near Casuarina cunninghamiana trees in Jamaica. Can J Bot. 1999;77:1248–1256. [Google Scholar]