Abstract
Degradative strains of fast-growing Mycobacterium spp. are commonly isolated from polycyclic aromatic hydrocarbon (PAH)-contaminated soils. Little is known, however, about the ecology and diversity of indigenous populations of these fast-growing mycobacteria in contaminated environments. In the present study 16S rRNA genes were PCR amplified using Mycobacterium-specific primers and separated by temperature gradient gel electrophoresis (TGGE), and prominent bands were sequenced to compare the indigenous Mycobacterium community structures in four pairs of soil samples taken from heavily contaminated and less contaminated areas at four different sites. Overall, TGGE profiles obtained from heavily contaminated soils were less diverse than those from less contaminated soils. This decrease in diversity may be due to toxicity, since significantly fewer Mycobacterium phylotypes were detected in soils determined to be toxic by the Microtox assay than in nontoxic soils. Sequencing and phylogenetic analysis of prominent TGGE bands indicated that novel strains dominated the soil Mycobacterium community. Mineralization studies using [14C]pyrene added to four petroleum-contaminated soils, with and without the addition of the known pyrene degrader Mycobacterium sp. strain RJGII-135, indicated that inoculation increased the level of degradation in three of the four soils. Mineralization results obtained from a sterilized soil inoculated with strain RJGII-135 suggested that competition with indigenous microorganisms may be a significant factor affecting biodegradation of PAHs. Pyrene-amended soils, with and without inoculation with strain RJGII-135, experienced both increases and decreases in the population sizes of the inoculated strain and indigenous Mycobacterium populations during incubation.
Polycyclic aromatic hydrocarbons (PAHs) consist of a class of chemicals with two or more fused benzene rings in linear, angular, or cluster arrangements. PAHs are ubiquitous: they are produced during fossil fuel combustion, waste incineration, or as by-products of industrial processes, such as coal gasification and petroleum refining, and are often released in large quantities into the environment (28, 29). High-molecular-weight PAHs are important constituents of petroleum as they are recalcitrant pollutants and because several of them are known mutagens or carcinogens. For example, the four-ring pyrene is mutagenic, whereas the five-ring benzo[a]pyrene is both mutagenic and carcinogenic (7).
There has been growing interest in mycobacteria due to their potential for PAH degradation. Recently described mycobacteria, such as Mycobacterium sp. strains RJGII-135 (hereafter called strain 135) and PYR-1, were isolated from petroleum-contaminated soils and shown to be degraders of high-molecular weight-PAHs such as pyrene and benzo[a]pyrene (15, 20, 45). Most of the described PAH-degrading mycobacteria are fast-growing species within the genus (4, 11, 16, 17, 18, 21, 22, 23, 27, 30, 31, 34), a clade distinct from the slow-growing group, which contains most of the known pathogenic species (14). It is likely that many other Mycobacterium species, including as-yet-uncultured strains, also possess the ability to degrade priority pollutants such as PAHs present in soil. However, little is known about the diversity and community structure of indigenous soil mycobacteria in either PAH-contaminated or pristine soils. The paucity of studies on mycobacterial ecology is partly due to their relatively low growth rate and hence susceptibility to overgrowth by faster-growing organisms in conventional methods of enrichment culture and isolation. Moreover, the selectivity of all culture media and the existence of uncultivable mycobacteria may cause further underestimation of the diversity of populations present in natural communities.
In the present study two culture-independent molecular techniques, PCR amplification of 16S rRNA genes and temperature gradient gel electrophoresis (TGGE), were used to compare the diversity and abundance of indigenous Mycobacterium populations among four different pairs of historically petroleum-contaminated soils. Similar to other molecular approaches, PCR-TGGE allows the detection of both culturable and nonculturable microorganisms and eliminates the problem of selectivity during culturing. PCR-TGGE allows multiple sample analyses on the same gel and provides a direct display of the community composition in both qualitative and semiquantitative ways. As many PAH compounds are both toxic and relatively recalcitrant to biodegradation, we hypothesized that heavily contaminated soils would contain less Mycobacterium diversity than their less contaminated counterparts. In addition, it has been suggested that the addition of degradative strains may stimulate the bioremediation of contaminated sites (5, 6, 13, 15, 20, 36, 39, 42, 45, 53). However, only in a few cases have the effects of the introduced strains on the microbial community structure been studied (35, 48). In the present study, we measured the mineralization of 14C-labeled pyrene in the four heavily contaminated soils, with and without inoculation of the pyrene degrader Mycobacterium sp. strain 135. These soil samples were further analyzed by the PCR-TGGE method to determine the relationship between inoculation, patterns of PAH degradation, and changes in the indigenous mycobacterial community structure in petroleum-contaminated soils.
MATERIALS AND METHODS
Soil samples.
Pairs of heavily contaminated soils (AT, CT, JT, and KT) and their less contaminated counterparts (AC, CC, JC, and KC) were obtained from four different petroleum-contaminated sites (sites A, C, J, and K). Sites A and C were petroleum refinery sites, whereas sites J and K were petroleum exploration and production sites. Each soil was chemically characterized; members of each pair were found to be similar in texture and other physical and chemical properties, and they do not contain unusually high levels of heavy metals (Table 1). Standard physical analyses were carried out by the Colorado Analytical Laboratory, Brighton. Soil texture was determined using the hydrometer method (12). Organic carbon content was determined by the Walkley-Black method using FeSO4 for titration (40). Soil pH was determined in a 1:1 ratio of soil and water. Cation exchange capacity was measured as described by Rhoades (43).
TABLE 1.
Physical characterization of soils
| Soil property (unit) | Less contaminated soils
|
Heavily contaminated soils
|
||||||
|---|---|---|---|---|---|---|---|---|
| AC | CC | JC | KC | AT | CT | JT | KT | |
| Texture | Silty clay loam | Silty clay | Loam | Loam | Silty clay loam | Silty clay | Loam | Loam |
| Clay (%) | 34 | 48 | 16 | 20 | 32 | 56 | 12 | 20 |
| Organic C (%) | 1.37 | 1.23 | 0.99 | 2.91 | 1.84 | 5.65 | 3.34 | 3.76 |
| pH | 6.5 | 7.8 | 7.3 | 5.5 | 7.7 | 7.6 | 7.7 | 6.8 |
| Cation exchange capacity (meq 100 g−1) | 17.0 | 18.7 | 6.3 | 19.3 | 17.4 | 20.3 | 7.8 | 10.2 |
| Cadmium (μg g−1) | NDa | ND | ND | ND | ND | ND | ND | ND |
| Chromium (μg g−1) | 16.4 | 26.3 | 5.0 | 10.8 | 14.2 | 118.5 | 5.0 | 9.8 |
| Lead (μg g−1) | 11.8 | 31.2 | 7.0 | 7.8 | 154 | 332 | 8.6 | 9.9 |
| Mercury (μg g−1) | ND | ND | ND | ND | ND | 0.455 | ND | ND |
| PAH (mg kg−1) | 0.220 | 15.0 | 37.0 | 0.0720 | 473 | 86.0 | 320 | 3.35 |
| Priority pollutant PAHs (mg kg−1) | 0.105 | 3.06 | 1.46 | 0.0170 | 27.1 | 10.5 | 18.2 | 0.262 |
| Total petroleum hydrocarbons (mg kg−1) | 21 | 54 | 3,050 | 18 | 4,650 | 2,450 | 27,500 | 6,250 |
| Mitrotox assay result | NAb | NA | NA | NA | Toxic | Nontoxic | Toxic | Nontoxic |
ND, none detected.
NA, not available.
PAH content was determined by the Arthur D. Little Laboratory (Cambridge, Mass.), using a gas chromatograph-mass spectrometer operated in the selective ion monitoring mode, after extraction from the soil by sonication with methylene chloride (Environmental Protection Agency [EPA] method 8270B). The same laboratory also determined the heavy metal content using EPA methods 6010A and 7471 and the total recoverable petroleum hydrocarbons using EPA method 9073. Microtox assays on the heavily contaminated soils were conducted by the Environmental and Water Resources Engineering Program at the University of Texas, Austin.
DNA extraction and purification.
Total bacterial DNA was extracted from soil samples by the procedure described by Zhou et al. (55), which combines both physical and chemical methods to maximize the recovery of DNA from soils of diverse composition. Briefly, 10 g of samples was ground in liquid nitrogen and thawed in a microwave oven for three cycles before the extraction of DNA with proteinase K and extended heating in a high-salt extraction buffer. To remove the humic acid coextracted with the DNA, the crude DNA extracts obtained were resuspended in distilled water (dH2O) and purified with Sepharose 4B spin columns (26).
PCR amplification of 16S ribosomal DNA (rDNA) from soil.
16S rRNA sequences of fast-growing Mycobacterium spp. commonly found in soil were obtained from GenBank and the Ribosomal Database Project (RDP), aligned, and used to design group-specific primers (3, 37). The sequence of the forward primer, MycF, was 5′-CGTGGGTGATCTGCCCT-3′ (Escherichia coli positions 121 to 137). The sequence of the reverse primer, MycR, was 5′-CGGCACGGATCCCAAGG-3′ (E. coli positions 858 to 844). MycR has a 40-base GC clamp (5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG-3′) linked to the 5′ end (25). Out of a total of 119 strains with the signature 16S rRNA sequence (E. coli positions 451 to 482) from the fast-growing clade of Mycobacterium in the RDP database, the designed Mycobacterium-specific primers exactly matched 80 of these strains. The remaining Mycobacterium strains in the database were mostly isolates from clinical specimens, which might not be commonly found in soil.
The PCR mixtures contained 5 ng of purified DNA extracts as templates, 200 μM deoxynucleoside triphosphates, 1 mM MgSO4, a 0.2-mg ml−1 final concentration of bovine serum albumin, 20 pmol of MycF, 20 pmol of MycR, and 5 μl of buffer A (Fisher Scientific, Pittsburgh, Pa.) in a final volume of 50 μl. After 5 min of denaturation at 94°C, 0.5 μl (2.5 U) of Taq DNA polymerase (Fisher Scientific) was added. Forty-one cycles of amplification were done under the following conditions: denaturation at 95°C for 1 min, annealing at 66°C for 1 min, and extension at 72°C for 2 min. The last cycle had an extension for 10 min. The amplified fragments are approximately 760 bp in size.
TGGE separation of rDNA.
PCR products were concentrated with a DNA SpeedVac (Savant, Farmingdale, N.Y.) and directly used for TGGE analysis with the DCode System (Bio-Rad, Hercules, Calif.). Six percent polyacrylamide gels (per 40 ml) were composed of 6 ml of 40% acrylamide-bisacrylamide (37.5:1), 1 ml of 50× Tris-acetate-EDTA buffer, 9 M urea, 40 μl of N,N,N′,N′-tetramethylethylethylenediamine, and 400 μl of 10% ammonium persulfate. Electrophoresis was performed at a constant voltage of 130 V and with a temperature gradient of 55 to 65°C for 16 h 40 min in 1.25× Tris-acetate-EDTA buffer. After electrophoresis, the gel was incubated for 40 min in SYBR Gold nucleic acid gel stain (Molecular Probes, Inc., Eugene, Oreg.). The number of bands per lane was determined visually and compared to those for PAH, priority pollutant PAH, and total petroleum hydrocarbon content using linear regression as implemented by Microsoft Excel.
PCR amplification of TGGE bands.
Several TGGE bands were excised, and the DNA was eluted with 10 μl of dH2O for 1 h before PCR amplification with MycF and eubacterial reverse primer 519R (5′-GA/TATTACCGCGGCG/TGCTG), which corresponds to E. coli positions 536 to 519. The reaction mixture (50 μl) contained 1 μl of a 104-fold dilution of the eluted DNA as the template, 200 μM deoxynucleoside triphosphates, 1 mM MgSO4, a 0.2-mg ml−1 final concentration of bovine serum albumin, 20 pmol of each primer, 5 μl of buffer A, and 0.5 μl of Taq DNA polymerase. The reaction conditions were similar to those described above except that 31 cycles were carried out. PCR products (approximately 400 bp in length) were purified with Wizard minicolumns (Promega, Madison, Wis.) before automated sequencing at the University of Cincinnati DNA Core Facility (Applied Biosystems model 377 or 373 sequencer; Perkin-Elmer, Norwalk, Conn.).
Sequencing and phylogenetic analysis of TGGE bands.
The DNA sequences obtained from TGGE bands were submitted to the ChimeraCheck program of the RDP at Michigan State University (37) to detect possible chimeras. No chimeras were found. A preliminary analysis of the 16S rRNA gene sequences was obtained by using the Advanced Blast Search program (1) available from GenBank (3) and the SequenceMatch program from the RDP (37). For phylogenetic analysis the sequences were initially aligned against other closely related Mycobacterium sequences using ClustalX version 1.8 (50), followed by manual alignment based on conserved features of primary and secondary structures. We conducted neighbor-joining, maximum-likelihood, and maximum-parsimony analyses as implemented by PAUP* version 4.0b2 (49). Nocardia farcinica was used as the outgroup to root the tree. Random stepwise addition of taxa was used.
Mineralization of PAH in soil samples.
To evaluate the effects of inoculation on PAH mineralization, Mycobacterium sp. strain 135 was grown (10% tryptic soy broth; 28°C for 5 days at 200 rpm) as an inoculum. Cells were harvested by centrifugation during the late exponential growth phase, washed, and diluted with dH2O before inoculation. A series of 50-ml serum bottles containing 5 g each of contaminated soil AT, CT, JT, or KT were divided into three groups and received the following treatments. Group 1 was inoculated with strain 135 (107 cells g of soil−1) and amended with 0.01 μCi of [14C]pyrene (34 pmol g of soil−1; specific activity, 58.7 mCi mmol−1) (Sigma, St. Louis, Mo.) dissolved in 50 μl of acetone, which was allowed to evaporate. Group 2 was amended with [14C]pyrene only. Group 3 was sterilized by autoclaving for a total of 2 h (1 h on each of two consecutive days) and then inoculated with strain 135 and amended with [14C]pyrene. Soils were brought up to 80% water-holding capacity, vortexed briefly to mix the contents, and then incubated in the dark at room temperature. The rate and extent of pyrene degradation during the period of incubation were measured by serum bottle radiorespirometry (32) using a Tri-Carb liquid scintillation analyzer (Packard Instrument Company, Downers Grove, Ill.). Cumulative mineralization was based on the total amount of labeled pyrene added to soils, which does not include degradation of indigenous pyrene. It is highly unlikely, however, that high levels of mineralization of indigenous pyrene would have occurred, as the bioavailability of large-ring PAHs in historically contaminated soils is generally low (19).
Changes in Mycobacterium community structure.
A series of beakers (250 ml) containing 100 g each of soils AT, CT, JT, and KT were divided into three groups and received the treatments as described above except that soils were amended with unlabeled pyrene (34 pmol g of soil−1) dissolved in acetone. After pyrene amendment, soils were well mixed and the acetone was allowed to evaporate. The beakers were then sealed with Parafilm, and the soil samples were incubated in the dark at room temperature. Ten grams of each soil was sampled at different time points during incubation for DNA extraction and TGGE analyses according to the methods described above.
Nucleotide sequence accession numbers.
The 16S rRNA sequences of mycobacterium phylotypes AT-3, CT-11, CT-12, JT-15, KT-19, KT-22, KT-23, AC-1, CT-9, CT-10, CT-24, CT-25, JC-13, JC-14, KT-26, KT-27, and CC-4 are available from GenBank under accession numbers AF220427 to AF220433, AF294742 to AF294750, and AF330695, respectively.
RESULTS
PCR-TGGE profiles of soils.
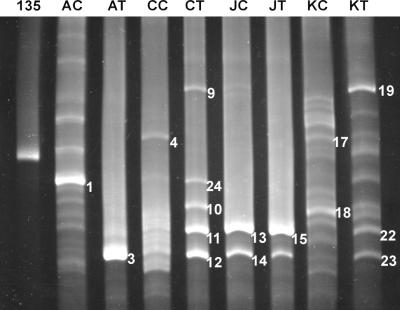
Community profiles of fast-growing mycobacteria in the eight soils are shown in Fig. 1, with each pair of soils giving distinct profiles. Each PCR-TGGE analysis was repeated at least three times, yielding identical profiles (data not shown). The reproducibility of the method is clearly shown in the similarity of the community profiles obtained for the four heavily contaminated soil samples in Fig. 1 and 4. Less contaminated soils AC, CC, and KC resulted in more TGGE bands (or phylotypes) than their heavily contaminated counterparts AT, CT, and KT; this was not true for the JC-JT soil pair, where only two distinct bands were seen in each soil. Soils AT and JT, which contain higher PAH contents and were shown as toxic in Microtox analysis, also gave fewer TGGE bands than did nontoxic soils CT and KT.
FIG. 1.
TGGE separation of the 16S rDNAs of fast-growing mycobacteria in eight soil samples. Lane 135, Mycobacterium sp. strain 135; lanes AC, CC, JC, and KC, less contaminated soil samples AC, CC, JC, and KC, respectively; lanes AT, CT, JT, and KT, heavily contaminated soil samples AT, CT, JT, and KT, respectively. TGGE bands 1, 3, 4, 9, 24, 10, 11, 12, 13, 14, 15, 17, 18, 19, 22, and 23 correspond to bands AC-1, AT-3, CC-4, CT-9, CT24, CT-10, CT-11, CT-12, JC-13, JC-14, JT-15, KC-17, KC-18, KT-19, KT-22, and KT-23, respectively. Electrophoresis was carried out with a temperature gradient of 55 to 65°C for 16 h 40 min in a 9 M urea–6% gel.
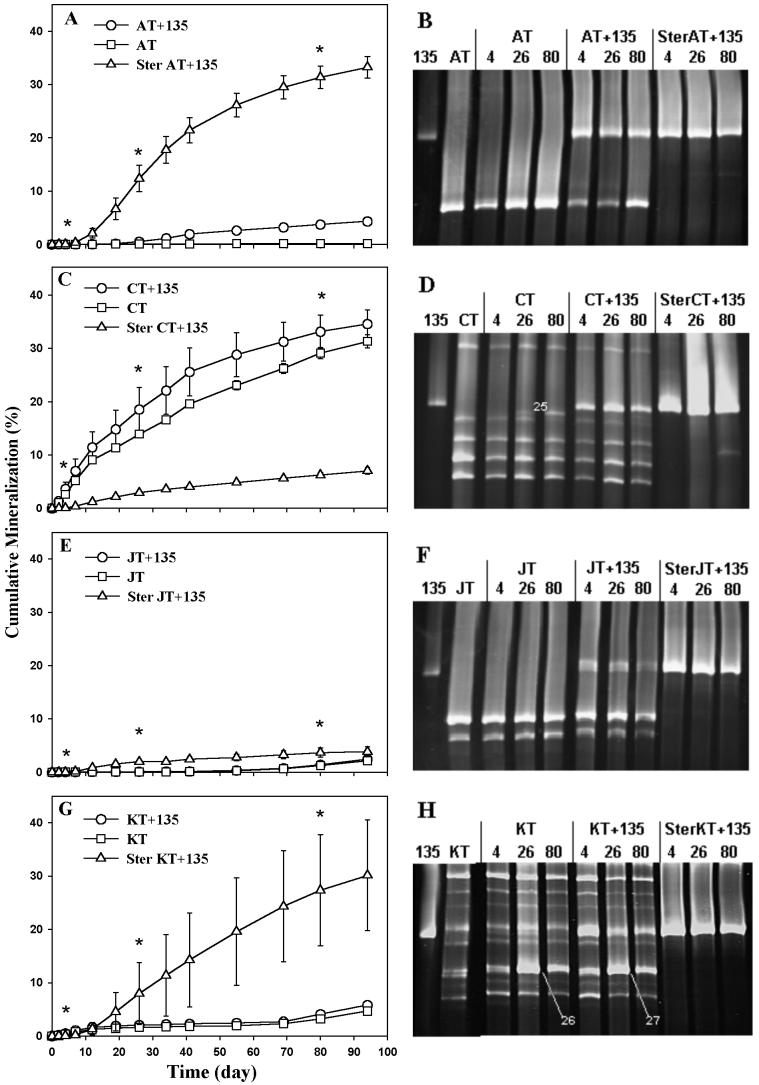
FIG. 4.
Pyrene mineralization curves and TGGE analysis of contaminated soils. (A, C, E, and G) Pyrene mineralization curves. AT, CT, JT, and KT are soils amended with pyrene only. AT+135, CT+135, JT+135, and KT+135 are soils amended with pyrene and Mycobacterium sp. strain 135. Ster AT+135, Ster CT+135, Ster JT+135, and Ster KT+135 are sterilized soils amended with pyrene and strain 135. Soils were sampled for TGGE analysis at days 4, 26, and 80 (asterisks). Error bars represent standard errors from three replicates. (B, D, F, and H) TGGE analysis. Lanes AT, CT, JT, and KT, unamended soil at day 0; lanes, 4, 26, and 80, pyrene-amended soils extracted at days 4, 26, and 80, respectively. Bands CT-25, KT-26, and KT-27 were excised for sequencing and phylogenetic analysis. Electrophoresis was carried out with a temperature gradient of 55 to 65°C for 16 h 40 min in a 9 M urea–6% gel.
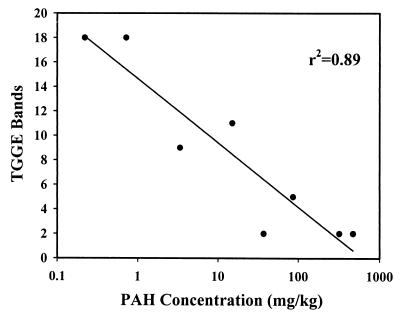
In comparisons among different pairs of soils, both members of the KC-KT pair have relatively low contamination levels and both members of the JC-JT pair have relatively high contamination levels (Table 1). In fact, soil KT had lower PAH contamination levels than soils CC and JC. This is reflected in the TGGE profiles, where soil KT had many more phylotypes than the other heavily contaminated soils and soil JC had only two prominent bands. When the PAH contents were compared across all eight soils, regardless of whether they are classified as less contaminated or heavily contaminated, a negative correlation between the number of TGGE bands and the log PAH content was observed, with an r2 value of 0.89 (Fig. 2). The priority pollutant PAH (r2 = 0.74) and total soil petroleum hydrocarbon (r2 = 0.81) contents also correlated with mycobacterial diversity, but to a lesser degree.
FIG. 2.
Relationship between PAH concentration and the number of TGGE bands recovered from individual soils. Data points represent the eight soils, including heavily contaminated and less contaminated soils.
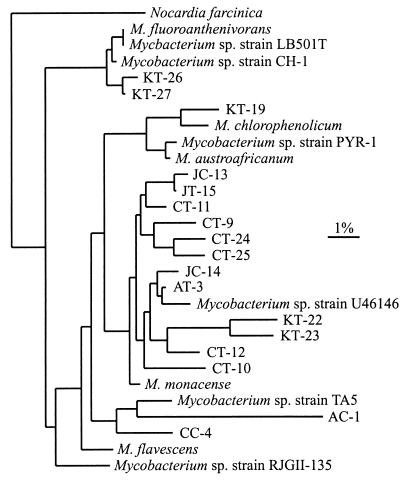
Several TGGE bands (AC-1, AT-3, CC-4, CT-9, CT-10, CT-11, CT-12, CT-24, JC-13, JC-14, JT-15, KC-17, KC-18, KT-19, KT-22, and KT-23 in Fig. 1), including both bright and faint bands, were excised, PCR amplified, and sequenced. Sequences obtained from bands KC-17 and KC-18 contained numerous ambiguous bases, presumably the result of overlapping bands, and were not analyzed further. Neighbor-joining analysis conducted with the available sequences in the RDP (37) and GenBank (3) databases indicates that the sequences from the TGGE bands are all related to other fast-growing strains of Mycobacterium but do not exactly match any known sequence (Fig. 3). Maximum-likelihood and maximum-parsimony analyses resulted in congruent trees (data not shown). The majority of the sequences fall into a clade which includes Mycobacterium monacense and Mycobacterium sp. strain U46146. The sequence from TGGE band KT-19 is most closely related to a clade containing Mycobacterium chlorophenolicum, while the sequences from bands AC-1 and CC-4, both isolated from less contaminated soils, are distantly related to Mycobacterium sp. strain TA5.
FIG. 3.
Neighbor-joining tree for sequences of 17 excised TGGE bands, strains of known Mycobacterium spp., and N. farcinica used as an outgroup. The scale bar corresponds to 0.01 estimated nucleotide substitution per sequence position. The 17 sequenced bands (GenBank accession numbers) are as follows: AC-1 (AF294742), AT-3 (AF220427), CC-4 (AF330695), CT-9 (AF294743), CT-10 (AF294744), CT-11 (AF220428), CT-12 (AF220429), CT-24 (AF294745), CT-25 (AF294746), JC-13 (AF294747), JC-14 (AF294748), JT-15 (AF220430), KT-19 (AF220431), KT-22 (AF220432), KT-23 (AF220433), KT-26 (AF294749), and KT-27 (AF294750). The 11 Mycobacterium spp. and the outgroup (accession numbers) are as follows: M. fluoroanthenivorans (AJ276274), Mycobacterium sp. strain LB501T (AJ245702), Mycobacterium sp. strain CH-1 (AF054278), Mycobacterium sp. strain RJGII-135 (U30661), M. flavescens (X52932), M. monacense (AF107039), Mycobacterium sp. strain U46146 (U46146), Mycobacterium sp. strain PYR-1 (U30662), M. austroafricanum (X93182), M. chlorophenolicum (X81926), Mycobacterium sp. strain TA5 (AB028483), and N. farcinica (X91041).
Mineralization of [14C]pyrene and changes in Mycobacterium community structure in soils.
Concurrent pyrene mineralization measurements and TGGE analyses of fast-growing Mycobacterium populations in heavily contaminated soil samples AT, CT, JT, and KT are shown in Fig. 4. In all sterilized soils inoculated with strain 135, mineralization occurred only after a lag period of approximately 7 days. In none of the treatments did the amount of pyrene mineralized to CO2 at the end of the experiment exceed 40% of that originally added. A band corresponding to strain 135 could be detected in all inoculated soils but not in uninoculated ones, indicating that the strain was not originally present in any of the soils (or that its number was below the detection limit of the PCR-TGGE method). The sequence of this band from inoculated soils was identical to that of strain 135.
Very little pyrene mineralization occurred in uninoculated toxic soil AT (Fig. 4A). Inoculation of the soil with strain 135 resulted in a significant, although small, increase in the level of pyrene mineralization in nonsterilized soil. Even higher levels of pyrene mineralization occurred when the soil was sterilized and then inoculated with strain 135. The intensity of the TGGE band for the major indigenous Mycobacterium phylotype in soil AT decreased at the beginning of the experiment but then recovered quickly as a bright band again in the day 26 and day 80 samples (Fig. 4B). Similarly, in inoculated soil AT, this band also decreased significantly in intensity initially, but it did not recover as a bright band until day 80, suggesting that this indigenous population was negatively affected by inoculation of strain 135 but could recover slowly over the course of the experiment. No change in the band intensity of strain 135 was observed in inoculated treatments of the soil, whether sterilized or not.
In contrast, considerable levels of pyrene mineralization occurred in the nontoxic soil CT, even when it was not inoculated (Fig. 4C). The inoculation of the soil with strain 135 increased the level of pyrene mineralization. However, sterilization and inoculation of the soil with strain 135 resulted in a significantly lower level of mineralization than that in either the native soil or the inoculated, nonsterilized soil. A new TGGE band (CT-25 in Fig. 4D) appeared in the uninoculated soil CT at day 26 (above band CT-24 in Fig. 1), with the band increasing in intensity by day 80. A new TGGE band at the same position also appeared in the day 80 sample of the inoculated soil, although the band was less intense than that in the uninoculated soil. Sequencing of band CT-25 and subsequent phylogenetic analysis indicated that it was most closely related to CT-24. With or without inoculation, the TGGE band intensities of other indigenous mycobacteria increased in day 26 samples but then decreased again in day 80 samples. This was also true for strain 135 in the inoculated soil. With inoculated, sterilized soil, strain 135 maintained a strong band throughout the period of incubation.
As with toxic soil AT, uninoculated soil JT exhibited very little pyrene mineralization (Fig. 4E). Inoculation of the soil with strain 135 did not result in any increase in pyrene mineralization. Only slightly higher levels of mineralization occurred when the soil was sterilized and then inoculated with strain 135. The TGGE profiles of soil JT did not show any change in the indigenous mycobacterial community structure over time in the uninoculated soil (Fig. 4F), but the band intensities of both indigenous mycobacteria and strain 135 decreased in the inoculated samples over the time of incubation. With sterilized soil, there appeared to be no significant change in the population size of strain 135 over time.
In uninoculated soil KT (Fig. 4G), pyrene mineralization was already detected by day 2, although it was maintained at a much lower level than in the other nontoxic soil, CT. This level of pyrene mineralization is higher than that occurring in toxic soils AT and JT. Inoculation of soil KT slightly increased mineralization, especially after day 70. However, much higher levels of pyrene mineralization occurred when the soil was sterilized and inoculated with strain 135. The brightness of the band for strain 135 was reduced after day 4 in inoculated soil KT but not in sterilized, inoculated soil (Fig. 4H).
Similar to the case for soil CT, new TGGE bands appeared in day 26 samples (KT-26 and KT-27 in Fig. 4H). The new bands appeared between the two bright, close bands seen on day 4 in Fig. 4H but were clearly resolved only under different TGGE conditions (data not shown). Bands KT-26 and KT-27 decreased slightly in intensity by day 80 in both uninoculated and inoculated soils. Phylogenetic analysis of band KT-26 and KT-27 sequences revealed the presence of Mycobacterium strains only distantly related to the previously sequenced bands (Fig. 3).
DISCUSSION
In this study, we used TGGE to examine the genetic diversity and population dynamics of fast-growing species of mycobacteria in petroleum-contaminated soil. TGGE profiles represent a minimum estimate of strain diversity, as only DNA fragments from the predominant species present in the community are displayed and different fragments with similar electrophoretic mobilities may overlap at the same position in the gel. Moreover, due to possible biases such as efficiency of cell lysis and PCR amplification of different templates, TGGE is only semiquantitative. Changes in the intensity of TGGE bands indicate changes in the population sizes of individual phylotypes, but comparisons of band intensity between different phylotypes are tenuous. Nevertheless, Henckel et al. (24) demonstrated that the major bands present do indeed represent the major components of the bacterial community.
The members of each of the four pairs of soil samples were similar in physical and chemical properties, making any differences in microbial community structure between the members of a pair of heavily contaminated and less contaminated soils attributable mainly to the presence of petroleum, although other human activities and vegetation may also play a role. Since soils AT and JT, both containing high levels of PAH and toxic by the Microtox assay, had less diverse TGGE profiles than those of the low-PAH, nontoxic soils CT and KT, Mycobacterium diversity may be reduced by the toxicity of PAHs. As shown in Fig. 2, mycobacterial communities were clearly less diverse in the soils containing large amounts of PAHs, although that may not be the direct cause of the reduction in diversity, as there are many other potentially toxic components present in petroleum.
Similar changes in microbial community structure and reduction in bacterial diversity in response to environmental stress and perturbation, e.g., contamination, have been well documented. Using phospholipid fatty acid analysis, MacNaughton et al. (35) demonstrated a community shift in a crude-oil-contaminated coastal site. Shi et al. (46) reported differences in the microbial community structures of uncontaminated and fuel-contaminated sand aquifers. Bååth et al. (2) also demonstrated by phospholipid fatty acid analysis that the species composition changed in soils amended with high levels of metal-rich sludge. Torsvik et al. (51) compared the total bacterial diversities in agricultural and forest soils and found that diversity in the agricultural soil was 2 to 5 times lower than that in the forest soil. A reduction in soil microbial diversity was also observed by Øvreås et al. (41) when they incubated agricultural soil with a mixture of methane and air.
In addition, a selection process in which the Mycobacterium community of the heavily contaminated soils had become dominated by one or a few populations may have occurred, since fewer TGGE bands showed up in the profiles of these soils. This is in accordance with other reports indicating that environmental stresses, including contamination, not only reduce the biodiversity of the original community but may also selectively enrich specific microorganisms that are more adapted to the new environment. Shi et al. (46) observed a proliferation of minor phylotypes within the fuel-contaminated aquifer upon toluene exposure. A study by Langworthy et al. (33) on a freshwater sedimentary microbial community demonstrated higher frequencies of PAH-degradative genes at contaminated sites. Studies on pristine soils and soils with a known history of PAH contamination revealed that pristine soils did not yield PAH degraders whereas contaminated soils harbored closely related PAH-degrading bacteria (39). Using denaturing gradient gel electrophoresis, Rooney-Varga et al. (44) also noted a selective enrichment of microorganisms in a petroleum-contaminated aquifer. Furthermore, Ferris et al. (10) reported that disturbance of a hot spring cyanobacterial mat community led to colonization by previously absent cyanobacterial populations in the disturbed areas.
Phylogenetic analysis of the DNA bands excised from the TGGE gels demonstrated the group specificity of the PCR primers used in this study. All phylotypes sequenced clearly fall within the fast-growing Mycobacterium group, which includes several known PAH degraders such as strains 135 and PYR-1 (14, 15, 16) and Mycobacterium flavescens (9). Moreover, Mycobacterium austroafricanum, isolated from a gasoline-contaminated site, can degrade many aliphatic and aromatic hydrocarbons (47), and M. chlorophenolicum is a pentachlorophenol degrader (38). Phylotypes KT-19, KT-26, KT-27, and AC-1 were all most closely related to known xenobiotic degraders such as M. chlorophenolicum and Mycobacterium sp. strains CH-1 and TA5 (8, 54). Interestingly, most of the dominant phylotypes were most closely related to M. monacense and Mycobacterium sp. strain U46146, two clinical isolates that contain the fast-growing Mycobacterium signature sequence (3). Although different pairs of soils had very different physical and chemical properties and extents of contamination, there was no clear association between soil type and the phylotypes present.
The low level of pyrene degradation in uninoculated soils AT and JT may be attributable to the low diversity of mycobacteria, and possibly other PAH-degrading microorganisms, in the soils, as few mycobacterial phylotypes were present. Similarly, the higher level of pyrene mineralization in soil CT may be a result of its higher diversity of mycobacteria. Soil KT, however, also contained many distinct mycobacterial populations, but only a low level of pyrene degradation occurred in the uninoculated soil, suggesting that many of these populations may not be pyrene degraders. The new TGGE bands that appeared after day 4 in soils CT and KT may represent minor degradative populations that increased in size during the period of incubation.
Strain 135 survived in all of the inoculated soils but over time appeared to decrease in number in nonsterile soils JT and KT, possibly explaining the enhancement of pyrene mineralization being greater in soils AT and CT. Higher mineralization rates in most of the sterilized soils suggest that competition with the indigenous microbiota affected the degradative activity of strain 135. This is supported by the TGGE profile of soil AT, where inoculation of the soil appeared to have severely decreased the size of the major indigenous Mycobacterium population in the soil. By day 80 partial recovery of this population occurred even though strain 135 maintained its population level in the soil.
Since sterilized and inoculated soil CT had a reduced level of pyrene mineralization compared to the uninoculated soil, indigenous microorganisms in the soil appear to be more effective than strain 135 alone for in situ pyrene mineralization. An alternative explanation is that sterilization of the soil might have produced toxic substances or otherwise altered chemical and physical conditions of the soil, thus decreasing mineralization (52).
Fast-growing mycobacteria are common saprophytes in soil, and many isolated strains are known PAH degraders. The mycolic acid-rich cell walls of these autochthonous soil bacteria may play a role in their utilization of hydrophobic substrates such as PAHs. This study provides information on the indigenous Mycobacterium community structure and its relation to petroleum contamination and PAH biodegradation in actual soils. A negative effect of the toxicity of contaminated soil on PAH degradation and community structure was demonstrated. Our studies also revealed that the introduction of degradative organisms may not be an appropriate choice for remediation of all contaminated soils. Thorough investigation and evaluation of site characteristics, both chemical and biological, are therefore necessary for identifying the most appropriate approach for bioremediation.
ACKNOWLEDGMENT
This work was supported by grant P42 ES 04908 from the National Institute of Environmental Health Sciences.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bååth E, Díaz-Raviña M, Frostegård Å, Campbell C D. Effect of metal-rich sludge amendments on the soil microbial community. Appl Environ Microbiol. 1998;64:283–245. doi: 10.1128/aem.64.1.238-245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D, Lipman D J, Ostell J. GenBank. Nucleic Acids Res. 1993;21:2963–2965. doi: 10.1093/nar/21.13.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodkorb T S, Legge R L. Enhanced biodegradation of phenanthrene in oil tar-contaminated soils supplemented with Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:3117–3121. doi: 10.1128/aem.58.9.3117-3121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bumpus J A. Biodegradation of polycyclic aromatic hydrocarbons by Phanerochaete chrysosporium. Appl Environ Microbiol. 1989;55:154–158. doi: 10.1128/aem.55.1.154-158.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 8.Churchill S A, Harper J P, Churchill P F. Isolation and characterization of a Mycobacterium species capable of degrading three- and four-ring aromatic and aliphatic hydrocarbons. Appl Environ Microbiol. 1999;65:549–552. doi: 10.1128/aem.65.2.549-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deanross D, Cerniglia C E. Degradation of pyrene by Mycobacterium flavescens. Appl Microbiol Biotechnol. 1996;46:307–312. doi: 10.1007/s002530050822. [DOI] [PubMed] [Google Scholar]
- 10.Ferris J M, Nold S C, Rersbech N P, Ward D M. Population structure and physiological changes within a hot spring microbial mat community following disturbance. Appl Environ Microbiol. 1997;63:1367–1374. doi: 10.1128/aem.63.4.1367-1374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritzsche C. Degradation of pyrene at low defined oxygen concentrations by a Mycobacterium sp. Appl Environ Microbiol. 1994;60:1687–1689. doi: 10.1128/aem.60.5.1687-1689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gee G W, Bauder J W. Particle-size analysis. In: Klute A, editor. Methods of soil analysis, part 1. Physical and mineralogical methods. Madison, Wis: American Society of Agronomy, Inc., and Soil Science Society of America, Inc.; 1986. pp. 383–411. [Google Scholar]
- 13.Goldstein R M, Mallory L M, Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985;50:977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govindaswami M, Feldhake D J, Kinkle B K, Mindell D P, Loper J C. Phylogenetic comparison of two polycyclic aromatic hydrocarbon-degrading mycobacteria. Appl Environ Microbiol. 1995;61:3221–3226. doi: 10.1128/aem.61.9.3221-3226.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosser R J, Warshawsky D, Vestal J R. Indigenous and enhanced mineralization of pyrene, benzo[a]pyrene, and carbazole in soils. Appl Environ Microbiol. 1991;57:3462–3469. doi: 10.1128/aem.57.12.3462-3469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosser R J, Warshawsky D, Vestal J R. Mineralization of polycyclic and N-heterocyclic aromatic compounds in hydrocarbon-contaminated soils. Environ Toxicol Chem. 1995;14:375–382. [Google Scholar]
- 17.Guerin W F, Jones G E. Mineralization of phenanthrene by a Mycobacterium sp. Appl Environ Microbiol. 1988;54:937–944. doi: 10.1128/aem.54.4.937-944.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haag F E. Die Saprophytischen Mykobakterien. Zentbl Bakteriol Abt 2. 1927;71:1–45. [Google Scholar]
- 19.Hatzinger P B, Alexander M. Effect of aging of chemicals in soil on their biodegradability and extractability. Environ Sci Technol. 1995;29:537–545. doi: 10.1021/es00002a033. [DOI] [PubMed] [Google Scholar]
- 20.Heitkamp A M, Cerniglia C E. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl Environ Microbiol. 1988;54:1612–1614. doi: 10.1128/aem.54.6.1612-1614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitkamp A M, Cerniglia C E. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl Environ Microbiol. 1989;55:1968–1973. doi: 10.1128/aem.55.8.1968-1973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heitkamp A M, Franklin W, Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol. 1988;54:2549–2555. doi: 10.1128/aem.54.10.2549-2555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heitkamp A M, Freeman J P, Miller D W, Cerniglia C E. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1988;54:2556–2565. doi: 10.1128/aem.54.10.2556-2565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1999. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson C R, Harper J P, Willoughby D, Roden E E, Churchill P F. A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl Environ Microbiol. 1997;63:4993–4995. doi: 10.1128/aem.63.12.4993-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jimenez I Y, Bartha R. Solvent-augmented mineralization of pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1996;62:2311–2316. doi: 10.1128/aem.62.7.2311-2316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones K C, Stratford J A, Waterhouse K S, Furlong E T, Giger W, Hites R A, Schaffner C, Johnston A E. Increases in the polynuclear aromatic hydrocarbon content of an agricultural soil over the last century. Environ Sci Technol. 1989;23:95–101. [Google Scholar]
- 29.Jones K C, Stratford J A, Tidridge P, Waterhouse K S. Polynuclear aromatic hydrocarbons in an agricultural soil: long-term changes in profile distribution. Environ Pollut. 1989;56:337–351. doi: 10.1016/0269-7491(89)90079-1. [DOI] [PubMed] [Google Scholar]
- 30.Kelley I, Freeman J P, Evans F E, Cerniglia C E. Identification of metabolites from the degradation of fluoranthene by Mycobacterium sp. strain PYR-1. Appl Environ Microbiol. 1993;59:800–806. doi: 10.1128/aem.59.3.800-806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleespies M, Kroppenstedt R M, Rainey F A, Webb L E, Stackebrandt E. Mycobacterium hodleri sp. nov., a new member of the fast-growing mycobacteria capable of degrading polycyclic aromatic hydrocarbons. Int J Syst Bacteriol. 1996;46:683–687. doi: 10.1099/00207713-46-3-683. [DOI] [PubMed] [Google Scholar]
- 32.Knaebel D B, Vestal J R. A comparison of double vial to serum bottle radiorespirometry to measure microbial mineralization in soils. J Microbiol Methods. 1988;7:309–317. [Google Scholar]
- 33.Langworthy D E, Stapleton R D, Sayler G S, Findlay R H. Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl Environ Microbiol. 1998;64:3422–3428. doi: 10.1128/aem.64.9.3422-3428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd-Jones G, Hunter D W F. Characterization of fluoranthene- and pyrene-degrading Mycobacterium-like strains by RAPD and SSU sequencing. FEMS Microbiol Lett. 1997;153:51–56. doi: 10.1111/j.1574-6968.1997.tb10462.x. [DOI] [PubMed] [Google Scholar]
- 35.MacNaughton S J, Stephen J R, Venosa A D, Davis G A, Chang Y-J, White D C. Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol. 1999;65:3566–3574. doi: 10.1128/aem.65.8.3566-3574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmood S K, Rama Rao P. Microbial abundance and degradation of polycyclic aromatic hydrocarbons in soil. Bull Environ Contam Toxicol. 1993;50:486–491. doi: 10.1007/BF00191235. [DOI] [PubMed] [Google Scholar]
- 37.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miethling R, Karlson U. Accelerated mineralization of pentachlorophenol in soil upon inoculation with Mycobacterium chlorophenolicum PCP1 and Sphingomonas chlorophenolica RA2. Appl Environ Microbiol. 1996;62:4361–4366. doi: 10.1128/aem.62.12.4361-4366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller J G, Lantz S E, Devereux R, Berg J D, Pritchard P H. Studies on the microbial ecology of polycyclic aromatic hydrocarbon biodegradation. In: Hinchee R E, Semprini L, Ong S K, editors. Bioremediation of chlorinated and PAH compounds. Boca Raton, Fla: Lewis Publishers; 1994. pp. 218–230. [Google Scholar]
- 40.Nelson D W, Sommers L E. Total carbon, organic carbon, and organic matter. In: Page A L, Miller R H, Keeney D R, editors. Methods of soil analysis, part 2. Chemical and microbiological properties. Madison, Wis: American Society of Agronomy, Inc., and Soil Science Society of America, Inc.; 1982. pp. 539–579. [Google Scholar]
- 41.Øvreås, Jensen L S, Daae F L, Torsvik V. Microbial community changes in a perturbed agricultural soil investigated by molecular and physiological approaches. Appl Environ Microbiol. 1998;64:2739–2742. doi: 10.1128/aem.64.7.2739-2742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramadan A M, El-Tayeb O M, Alexander M. Inoculum size as a factor limiting success of inoculation for biodegradation. Appl Environ Microbiol. 1990;56:1392–1396. doi: 10.1128/aem.56.5.1392-1396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhoades J D. Cation exchange capacity. In: Page A L, Miller R H, Keeney D R, editors. Methods of soil analysis, part 2. Chemical and microbiological properties. Madison, Wis: American Society of Agronomy, Inc., and Soil Science Society of America, Inc.; 1982. pp. 149–157. [Google Scholar]
- 44.Rooney-Varga J N, Anderson R T, Fraga J L, Ringelberg D, Lovley D R. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl Environ Microbiol. 1999;65:3056–3063. doi: 10.1128/aem.65.7.3056-3063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider J, Grosser R, Jayasimhulu K, Xue W, Warshawsky D. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl Environ Microbiol. 1996;62:13–19. doi: 10.1128/aem.62.1.13-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Zwolinski M D, Schreiber M E, Bahr J M, Sewell G W, Hickey W J. Molecular analysis of microbial community structures in pristine and contaminated aquifers: field and laboratory microcosm experiments. Appl Environ Microbiol. 1999;65:2143–2150. doi: 10.1128/aem.65.5.2143-2150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solano-Serena F, Marchal R, Casarégola S, Vasnier C, Lebeault J M, Vandecasteele J P. A Mycobacterium strain with extended capacities for degradation of gasoline hydrocarbons. Appl Environ Microbiol. 2000;66:2392–2399. doi: 10.1128/aem.66.6.2392-2399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephen J R, Chang Y-J, MacNaughton S J, Whitaker S L, Hicks C L, Leung K T, Flemming C A, White D C. Fate of metal-resistant inoculum in contaminated and pristine soils assessed by denaturing gradient gel electrophoresis. Environ Toxicol Chem. 1999;18:1118–1123. [Google Scholar]
- 49.Swofford D L. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b2. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 50.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torsvik V, Sørheim R, Goksøyr J. Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 52.Trevors J T. Sterilization and inhibition of microbial activity in soil. J Microbiol Methods. 1996;26:53–59. [Google Scholar]
- 53.Wagner-Döbler I, Bennasar A, Vancanneyt M, Strömpl C, Brümmer I, Eichner C, Grammel I, Moore E R B. Microcosm enrichment of biphenyl-degrading microbial communities from soils and sediments. Appl Environ Microbiol. 1998;64:3014–3022. doi: 10.1128/aem.64.8.3014-3022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yagi O, Hashimoto A, Iwasaki K, Nakajima M. Aerobic degradation of 1,1,1-trichloroethane by Mycobacterium spp. isolated from soil. Appl Environ Microbiol. 1999;65:4693–4696. doi: 10.1128/aem.65.10.4693-4696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J-Z, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]