Abstract
Cells assemble macromolecular complexes into scaffoldings that serve as substrates for catalytic processes. Years of molecular neurobiology research indicate that neurotransmission depends on such optimization strategies. However, the molecular topography of the presynaptic active zone (AZ), where transmitter is released upon synaptic vesicle (SV) fusion, remains to be visualized. Therefore, we implemented MINFLUX optical nanoscopy to resolve the AZ of rod photoreceptors. This was facilitated by a novel sample immobilization technique that we name heat-assisted rapid dehydration (HARD), wherein a thin layer of rod synaptic terminals (spherules) was transferred onto glass coverslips from fresh retinal slices. Rod ribbon AZs were readily immunolabeled and imaged in 3D with a precision of a few nanometers. Our 3D-MINFLUX results indicate that the SV release site in rods is a molecular complex of bassoon–RIM2–ubMunc13-2–Cav1.4, which repeats longitudinally on both sides of the ribbon.
3D-MINFLUX nanoscopy of heat-immobilized retinal sections reveals the molecular topography of rod photoreceptor active zones.
INTRODUCTION
The combination of multiscale structural approaches with molecular neurobiology and electrophysiology has given us a dynamic view of the active zone (AZ). The classic electron microscopic (EM) experiment that motivated intense interest in AZ structure arose from images of freeze-fractured frog neuromuscular junctions (1). That study resolved stimulation-dependent synaptic vesicle (SV) fusion sites that appeared as small indentations in the plasma membrane (PM). The fusion sites were arranged into two parallel rows that ran for several micrometers. Proximal to the fusion sites were intramembrane particles (IMPs) that could also been seen in the absence of stimulation, which suggested that they were residents of the AZ. Inspection of this AZ in smaller volumes using high-resolution EM tomography showed pairs of electron dense tethers connecting each SV to the PM (via a “rib”) and to the presumed IMPs (via “pegs”) (2, 3). The molecular identity of the tethers and IMPs has remained unknown. Efforts to resolve the physical positions of AZ proteins have involved the application of immuno-EM approaches to a variety of synapses (4, 5), which has put years of biochemical work into two-dimensional (2D) maps of the AZ with a resolution between 10 and 20 nm (6). A limitation of immuno-EM approaches is that labeling density is typically low, and imaging multiple proteins is challenging—especially if distinct epitopes are colabeled in a sample. For the last decade, superresolution light microscopy has offered a complementary perspective, with the advantage being the ability to image multiple proteins at a spatial resolution of ~25 nm in the xy plane (7–11). However, neither immuno-EM nor superresolution light microscopy offers suitable spatial resolution in 3D. Recent advances in fluorescence-based optical nanoscopy have overcome these limitations. For instance, the MINFLUX (minimal photon fluxes) method (12) combines single-molecule detection strategies used in Stochastical Optical Reconstruction Microscopy (STORM) microscopy with precise and rapid illumination strategies used in Stimulated Emission Depletion (STED) microscopy, which yields nanometer resolution in 3D (13).
The complex morphology of the rod photoreceptor (PR) synapse is an attractive target for high-resolution, 3D imaging. Rods send an individual synaptic terminal to a thin synaptic layer in the outer retina called the outer plexiform layer (OPL), creating the first synapses in the visual system, along with cones. The rod’s synaptic ending is spherical and fully envelopes the dendritic tips of postsynaptic rod bipolar and horizontal cells (rbc and hc, respectively) to create an extreme example of an “invaginating synapse” that is often referred to as the “rod spherule.” In mice, and most mammals, one very large ∩-shaped AZ is seated at the center of the rod spherule, and it is conspicuously marked by a presynaptic, electron-dense “synaptic ribbon” (see schematic in Fig. 1A). Synaptic ribbons are expressed in a subset of sensory cells in eyes and ears of mammals, and they differ in both their physical dimensions and the molecular composition of their AZs. Generally, synaptic ribbons emanate from the PM, they are covered in SVs, and the protein “ribeye” is essential for their formation (14, 15). The ribbon projects into the cytosol of the rod spherule and joins the PM where conventional AZ proteins are enriched. This arrangement is proposed to allow SVs to cycle through the ribbon by absorbing to and then translocating down the ribbon (16), and eventually fusing at the AZ in a Ca2+-dependent manner (for review see 17).
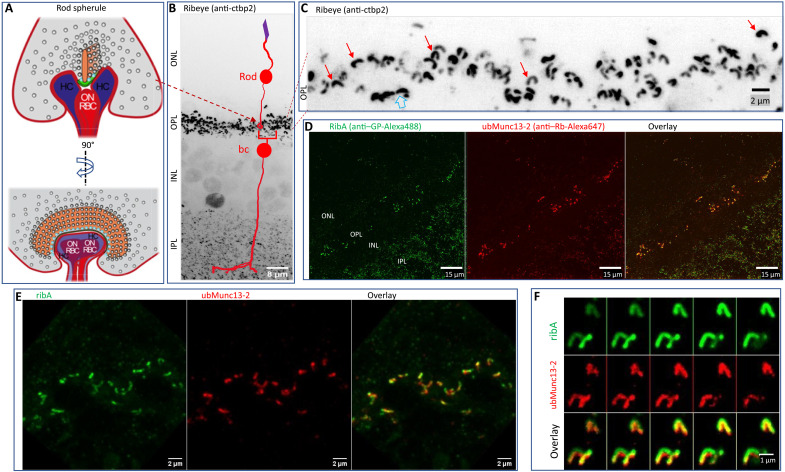
Fig. 1. Heat-assisted rapid dehydration (HARD) captures a thin layer of rod synaptic ribbons on a glass coverslip.
(A) Schematic of the rod ribbon synapse in two perspectives: front and side views (left and right). The rod spherule (gray) contains the following structures: the synaptic ribbon (orange), arciform density (AD) (green), synaptic vesicles (SVs) (gray spheres), and membrane proximal vesicles (cyan spheres); modified from (17). (B) Confocal image of a retinal slice processed for immunostaining with conventional fixation and permeabilization methods (see Materials and Methods). The anti-ctbp2 antibody was used to label the ribeye B domain. The intensely labeled OPL demarcates where PR ribbons are deposited. Smaller puncta in the inner retina [inner plexiform layer (IPL)] represent retinal bipolar cell synaptic ribbons. The outer and inner nuclear layers (ONL and INL, respectively) are also indicated with text, and although not purposefully targeted for immunolabeling, nuclei in the INL do express detectable levels of ctbp-2 (as a transcription factor) independent of ribeye. The image represents a maximal intensity Z projection summed over 40 optical sections, taken at 320-nm steps. (C) OPL shows the ∩-shaped morphology of rod ribbons, prepared and imaged as described in (A), but only 20 optical sections were used (movie S1). (D) A HARD sample stained for ribeye A domain (ribA; guinea pig polyclonal) and the active zone protein ubMunc13-2 (rabbit polyclonal). Only the OPL and IPL exhibit immunolabeling (movie S2). (E) Close-up of the OPL imaged in a HARD sample [illustrated as in (D)]. Ribbons were within 2 μm from the glass surface (10 sections at 200-nm increments) (movie S3). (F) Zoomed-in view of rod ribbons in a HARD sample shows the arrangement of ribeye around ubMunc13-2, which matches a previous description for ribeye and ubMunc13-2 colocalization in the OPL that was achieved with conventional fixation and staining procedures (25).
Whether the ribbon-type AZ can be divided into subregions is debated. For instance, are certain AZ protein expressed on each side of the ribbon (bilaterally), at or near the PM? In the case of voltage-dependent Ca2+ channels (Cav), they may not need to be expressed bilaterally if release is mediated by a large Ca2+ domain that extends well beyond the base of the ribbon. While studies on salamander rods indicate global Ca2+ (18, 19) and ribbon-independent release sites (20) support a substantial fraction of release, experiments on mouse rods indicate that the readily releasable pool (RRP) of SVs is ribbon dependent and mediated by a Ca2+ nanodomain (21). The latter implies a nanometer-scale proximity of Ca2+ channels and vesicular release sites (17). Despite recent progress (7, 11), assessing the precise topography of Ca2+ channels and candidate determinants of the vesicular release site such as Munc13 has remained difficult.
To address these questions, we imaged the positions of AZ proteins within mouse rod spherules. This entailed development of a method for immobilizing AZs on glass that we call heat-assisted rapid dehydration (HARD). Confocal images of immuno-labeled HARD samples exhibited the expected molecular morphology for a ribbon-AZ; in addition, essential ultrastructural features (EM results) were maintained. Evaluation of HARD samples with 3D-MINFLUX nanoscopy yielded single-molecule localization precision of <6 nm and in two colors. The results show that the AZ proteins bassoon, Rab3 Interacting Molecule-2 (RIM2), Cav1.4 channels, and ubMunc13-2 run along the length of the AZ and likely create two independent rows of release sites on both sides of the ribbon. We discuss the advantages of nanoscopic imaging of AZ structure in 3D, the implications of our results to rod physiology, and, more generally, the molecular architecture of SV release sites.
RESULTS
Immobilization of a thin layer of rod AZs through HARD
The initial attempt to implement MINFLUX for the study of rod ribbons began with freshly made retinal slices, as typically used for electrophysiological recordings (21). These slices were processed for immunofluorescence (IF) imaging using conventional fixation and permeabilization protocols (see Materials and Methods). Figure 1B presents a confocal image made from a retinal slice in which the protein ribeye was indirectly stained using an anti-ctbp2 primary antibody that targets the B domain of ribeye (see description of protein epitopes in fig. S1 and Table 1) (22, 23), followed by staining with a secondary antibody conjugated to the Alexa Fluor 647 fluorophore. Inspection of the ctbp2-labeled slices with confocal microscopy shows PR terminals in the OPL form ribeye-positive puncta that are larger and more intensely labeled than bipolar ribbons in the inner plexiform layer (IPL) (Fig. 1B). Cone ribbons are also deposited in the OPL, and they are readily distinguished from rod ribbons. First, a rod ribbon has a contour length approximately double that of a cone ribbon, 1.5 versus 0.8 μm. Second, each rod forms only a single ribbon (Fig. 1C and movie S1), while a cone terminal is populated with a cluster of ~13 ribbons (Fig. 1C, open arrow) (24). Our first attempts to image ribeye-labeled rod ribbons in slices with 2D-MINFLUX yielded an excessively high density of photon emissions that prohibited single-fluorophore localizations. Therefore, several approaches to reduce the density of photon emissions from rod synapses were attempted as laid out in greater detail in the Supplementary Materials.
Table 1. Antibody list.
aa, amino acids; Ms, mouse; ihcs, inner hair cells.
| Primary antibody |
Epitope, species (sequence database) |
Catalog no. and source; references to mouse ihc and rod ribbons |
| Rabbit anti– ubMunc13-2 |
aa 182–408 | B. Cooper, MPI-NAT, Goettingen; Ms rods (25) |
| Mouse anti-bassoon | aa 756–1001 mouse (CAA76598.1) |
SAP7F407, Abcam; Ms rods (51) |
| Rabbit anti-RIM2 | aa 909–1076 rat RIM2-4C (UniProt ID: Q9JIS1-3) |
140 303, Synaptic Systems; Ms rods (32) and Ms ihcs (52) |
| Mouse anti–ctbp2 | aa 361–445 ribeye (UniProt ID: O54855) |
612044, BD Biosciences; Ms rods (15) and ihcs (14) |
| Rabbit anti-Cav1.4, α1F subunit |
aa 1667–1985 mouse Cav1.4 (UniProt ID: Q7TNI3) |
365 003, Synaptic Systems; Ms rods (53) |
| Guinea pig anti– ribeye A-domain |
aa 95–207 rat ribeye (UniProt ID: Q9EQH5-2) |
192 104, Synaptic Systems; Ms rods (53) and Ms ihcs (54) |
| Rabbit anti-CAST | aa 655–670 rat ERC2 (UniProt id: Q8K3M6) |
143 103, Synaptic Systems; Ms rods (31) |
| Guinea pig anti-piccolo |
aa 2012–2351 rat piccolo (UniProt id: Q9JKS6) |
142 104, Synaptic Systems; Ms rods (55) and Ms ihcs (56) |
| Secondary antibodies |
Source | |
| Goat anti-rabbit, Alexa 488 |
A11008, Thermo Fisher Scientific |
|
| Donkey anti-mouse, Alexa 488 |
A21202, Thermo Fisher Scientific |
|
| Goat anti-Guinea pig, Alexa 488 |
A11073, Thermo Fisher Scientific |
|
| Goat anti-mouse, Alexa 546 |
A11003, Thermo Fisher Scientific |
|
| Goat anti-rabbit, Alexa 647 |
A21244, Thermo Fisher Scientific |
|
| Donkey anti-mouse, Alexa 647 |
A31571, Thermo Fisher Scientific |
|
| Anti-mouse, CF660C | Dye: Biotium; coupled in-house (Abberior) |
In short, what proved to be helpful was (i) a new approach for decreasing photon emission from out of focus rod ribbons and (ii) evaluating synaptic ribbon AZ proteins that were expressed at a lower copy number than ribeye. The new approach entailed placing a fresh, unfixed retinal slice on a glass coverslip warmed to 50°C and retracting it after ~30 s (fig. S2; Supplementary Materials for detailed method). A thin layer of retina remained on the glass and appeared as a translucent film for ~1 s before transitioning to a grayish color (dehydrated). After a few additional minutes of curing on the thermal plate, coverslips were processed for immunostaining. This involved rehydrating the sample in isotonic phosphate-buffered saline (PBS) and then incubation in blocking buffer [2% bovine serum albumin (BSA)] before applying primary antibodies, and neither chemical fixation nor permeabilization was used (see Materials and Methods). The labeled sample was shown to yield a thin layer of synaptic ribbons from the inner and outer retina (Fig. 1D), and a normal organization for rod ribbons was apparent in the OPL (Fig. 1D and movies S2 and S3). Furthermore, labeled ribbons were tightly colocalized with the vesicle-priming protein ubMunc13-2 (Fig. 1, D to F), which is in agreement with a study that used conventional fixation methods on mouse retinal slices (25).
We refer to this immobilization procedure as HARD, and we (26) note the following advantages of this sample preparation: (i) It does not require chemical fixation, (ii) does not require surface functionalization of the glass, (iii) makes the AZs readily accessible for immunolabeling without detergent permeabilization, and (iv) delivers a thin layer of retina on the glass (Fig. 1, D to F, and movies S2 and S3). Before further presenting the molecular architecture of the rod AZ in HARD samples, the physical composition of retinal material that transferred to the glass coverslip is described.
HARD samples maintain key structural features of rod PRs
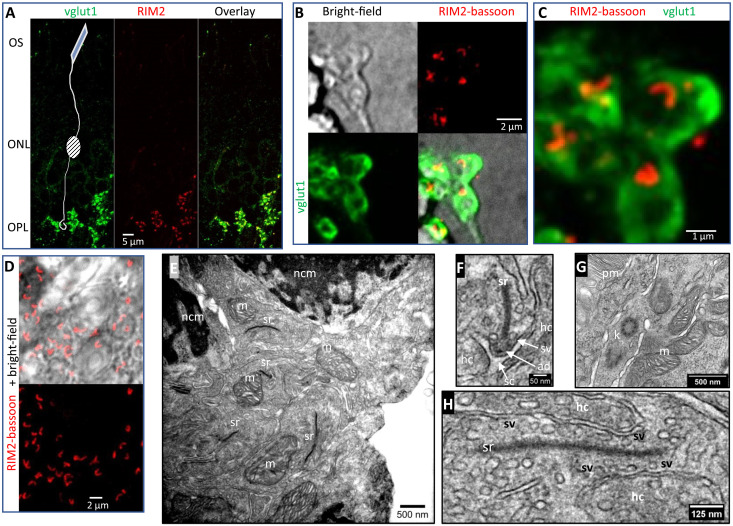
To further investigate what structures transferred to the coverslip, we made bright-field (BF) and IF images of HARD samples. To outline rod terminals in the OPL, an antibody against the vesicular glutamate transporter type 1 (vglut1) was used in combination with antibodies targeting AZ proteins RIM2 and bassoon (Fig. 2, A to C). Rod terminals could be found within ~1 to 2 μm from the surface of the glass coverslip (Fig. 2B). The corresponding IF image shows vglut1 signal in rod terminals, which surround the characteristically large rod ribbon AZ profiles (Fig. 2B). A zoomed-in view shows a cluster of vglut1-positive terminals along with their AZ profiles (Fig. 2C and movies S4 and S5). In a separate region of interest, the BF image shows PR nuclei accompanied by a thin layer of rod ribbons in the OPL (Fig. 2D and movie S6). Next, HARD samples that had been stored for a few days in a desiccated environment were rehydrated, then immersed in conventional aldehyde fixatives, and processed for EM. Ultrastructural features common to the OPL and outer PR segments were easily identified. First, PR nuclei positioned near large synaptic ribbons were identified as OPL territory (Fig. 2E). Some of the synaptic ribbons in the OPL were sectioned such that up to 1 μm of ribbon was visible, accompanied by a nearby mitochondrion, which is a characteristic of a rod synaptic terminal (Fig. 2E). Upon closer inspection, the rod ribbons were seen to tether SVs, and a subset of SVs at the base of the ribbon were docked at the presynaptic membrane (Fig. 2F). In other micrographs, the boundary between the PR outer and inner segments displayed organelles with ultrastructural features that were comparable to conventionally fixed retinal tissue. For example, PR substructures identified here include the following: kinocilia, photopigment-membrane discs, mitochondria in the ellipsoid body (Fig. 2H), and, lastly, a longitudinal section through a rod ribbon (Fig. 2H).
Fig. 2. HARD samples preserve important ultrastructural features of rod synaptic terminals.
(A) Overview of a HARD sample immuno-labeled for vglut1 (GP polyclonal; green) and RIM2 (Rb polyclonal; red) shows staining of PR terminals in the OPL. The location of the outer segments (OS) and ONL is indicated with text yet not purposefully labeled. (B) HARD sample visualized with BF optics shows rod terminals on the glass coverslip. IF signal for vglut1 (green) outlines rod terminals. Rod AZs are colabeled with RIM2 (anti–Rb-CF680) and BSN (anti–Ms-Alexa647) and presented as an aggregate red signal (see Table 1 for antibodies). (C) Higher-resolution image of IF signals are presented in (B) (movie S4: series of optical sections through terminals bounded by vglut1 in green, and in red bassoon + RIM2; movie S5: red channel alone with bassoon + RIM2). (D) Separate example of HARD sample labeled for bassoon + RIM2 (red) and combined with BF to show a thin layer of OPL on the glass (movie S6). (E to H) EM of HARD samples derived from sections made within a micrometer of the glass coverslip. In (E), a pair of rod soma is indicated in the top portions of the image by their electron dense, nuclear chromatin material (nmc). In the center of the image, several rod terminals are present, characterized by their synaptic ribbon (sr) paired closely to a single mitochondrion (m). (F) Zoomed-in view of a rod ribbon, which captures the height of the ribbon and associated synaptic structures: horizontal cell dendrites (hc), ribbon AD (ad), synaptic cleft (sc), and AZ docked synaptic vesicles (sv). (G) Other structures in the PRs at the juncture between PR ellipsoids and OS are indicated as follows: kinocillium (k), and OS pigment membrane (pm). (H) Longitudinal section through a rod ribbon.
Rod ribbon AZs in HARD samples maintain their molecular architecture at the level of confocal microscopy
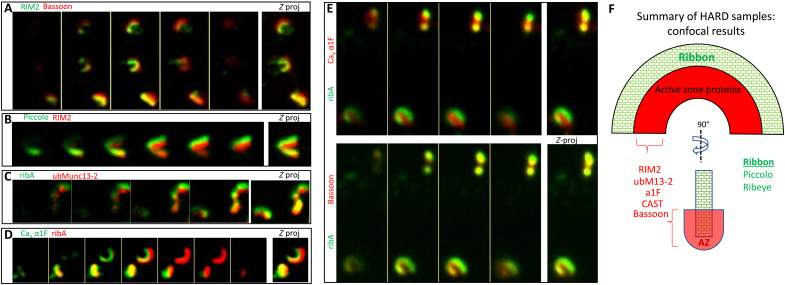
To compare immunolabeled HARD samples to published data that used conventional aldehyde fixation–based methods, we tested a variety of AZ protein antibodies. HARD samples imaged on an inverted, spinning disc confocal microscope showed the typical IF features for ribeye (using anti-ctbp2 to label the ribeye B domain or an anti–ribeye A domain antibody; fig. S1) and additional AZ proteins (Fig. 3, A to E, and fig. S3, A to F). Only the colabeling for ribeye and scaffolding AZ protein piccolo showed complete overlap at the confocal level (fig. S3E), in keeping with piccolo and ribeye demarcating the body of the ribbon (4, 27–29). In contrast, bassoon, another large AZ scaffolding protein, fills the inner face of the ribeye-labeled ribbon (Fig. 3F and fig. S3F). The inner face of the ribbon profile also shows RIM2 (costained with ribeye; fig. S3A) or piccolo (Fig. 3B and fig. S3D), which partially overlaps with bassoon (Fig. 3A and fig. S3B). Next, costaining of the Cav1.4 channel’s pore-forming α1F subunit with ribeye (Fig. 3, D and E, and fig. S3F) and bassoon (Fig. 3E and fig. S3F) shows that the IF from membrane-bound α1F subunit overlaps more with bassoon than ribeye. The other AZ proteins examined here are CAST (also referred to as ERC2) (30, 31) and ubMunc13-2 (25) that localize to the inner face of the ribbon (CAST: fig. S3C; ubMunc13-2: Figs. 1E and 3C). Overall, except for piccolo, the AZ proteins assessed in HARD samples with confocal microscopy are localized to the inner half of the body of the ribbon (see cartoon in Fig. 3F), which agrees with previous results derived from traditional IF approaches carried out on retinal slices.
Fig. 3. HARD samples preserve the molecular architecture of rod ribbons at the level of confocal microscopy.
(A and B) RIM2 overlaps with the interior aspects of bassoon and piccolo. (C and D) Costained for ribA (anti–GP-Alexa488) and active zone proteins (anti–Rb-Alexa647): ubMunc13-2 or α1F (rabbit polyclonal). Here, the ribbon (ribeye) surrounds these AZ proteins. Images in (E) represent a triple, colabeling of ribA (anti–GP-Alexa488) versus α1F (anti–Rb-Alexa647) and bassoon (anti–Ms-Alexa546). Here, both α1F and bassoon are interior to the body of the ribbon (ribA), but the α1F signal overlaps less with ribeye than bassoon, which suggests that α1F is interior to bassoon. (F) Summary of active zone protein localizations, all of which were interior to the body of the rod ribbon (ribeye and piccolo) and determined in this study with HARD samples and confocal microscopy. Only RIM2 and α1F were analyzed in combination with bassoon, and they overlap with bassoon. CAST and ubMunc13-2 were only costained with ribeye; hence, their positions relative other AZ proteins were not assessed here.
Survey of AZ proteins with 2D-MINLFUX nanoscopy
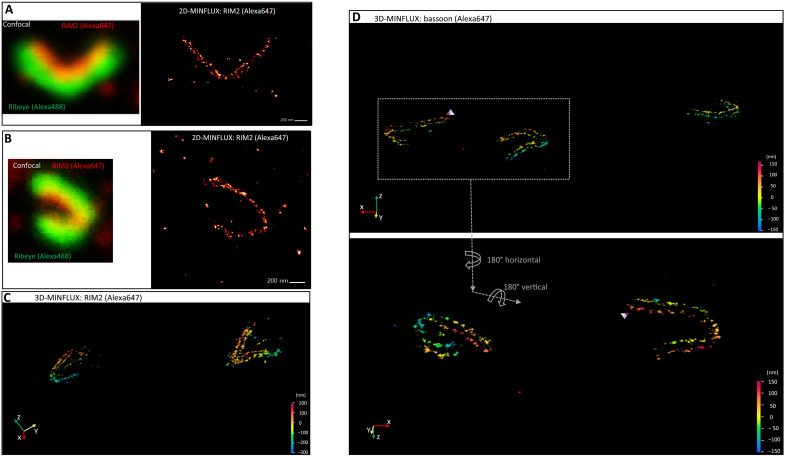
As stated earlier, our first approach using 2D-MINFLUX attempted to image ribeye-labeled rod ribbons in slice, but this gave an overwhelmingly high density of photon emissions. Therefore, labeling against RIM2 was used, because it gives lower signal intensity (presumably expressed at a lower copy number than ribeye), and HARD samples were used to reduce the density of photon emissions, including out-of-focus signals. This approach readily gave favorable results that showed that RIM2 localizations were composed of puncta that appear to fall into linear arrays (Fig. 4, A and B). Moreover, in some instances, the RIM2 patterns suggested an arrangement of two parallel rows (Fig. 4A).
Fig. 4. 3D-MINFLUX nanoscopy revolves the convoluted topography of the rod ribbon AZ.
(A and B) confocal image on the left shows a rod ribbon indirectly labeled for RIM2 and ribeye. On the right, the 2D-MINFLUX image of RIM2 is presented. (C) Image of RIM2 derived from 3D-MINFLUX nanoscopy. The ribbon on the right appears to have two parallel rows of RIM2 localizations. Color scale indicates distance in the z direction, and here, it ranges from +200 to −300 nm. X, y, and z axes are indicated in the bottom left corner. (D) 3D-MINFLUX images of three rod ribbons were labeled for bassoon, and each shows parallel rows of bassoon. The two ribbons on the left, in the top panel, are presented at higher magnification in the bottom panel, and the images have been flipped vertically and horizontally to illustrate the 3D nature of the ribbons. Distance scales in top and bottom panels range from +170 to −150 nm and +150 to −150 nm, respectively.
Localization patterns for piccolo and CAST were distinct from RIM2. 2D-MINFLUX images of piccolo shows a broader and denser labeling pattern than what was witnessed for RIM2 (fig. S4A). The thick band of piccolo has a width of ~200 nm in certain places (fig. S4A, i and ii), which is close to the height of the ribbon (fig. S5A), consistent with published data (29) and our confocal images showing that piccolo colocalizes with ribeye (figs. S3E and S4, A and B). Colabeling of CAST and ribeye showed far less overlap than any other AZ protein when examined with confocal microscopy (figs. S3C and S4, C and D), which is in line with earlier studies that have concluded CAST resides near or in the arciform density (AD) (30), a territory below the base of the ribbon (fig. S5A and Fig. 2F). The pattern of staining attained with 2D-MINFLUX reveals a subtle line of localizations that run the length of the interior of the ribbon. Occasional bright clusters of localizations appear within or outside the interior region of the ribbon (fig. S4C, i and ii). These hotspots may indicate a higher density of CAST, an artifactual aggregation of secondary antibody or sites where blinking is preferentially activated (hyperactive).
3D-MINFLUX: AZ proteins are expressed bilaterally along the length of the ribbon
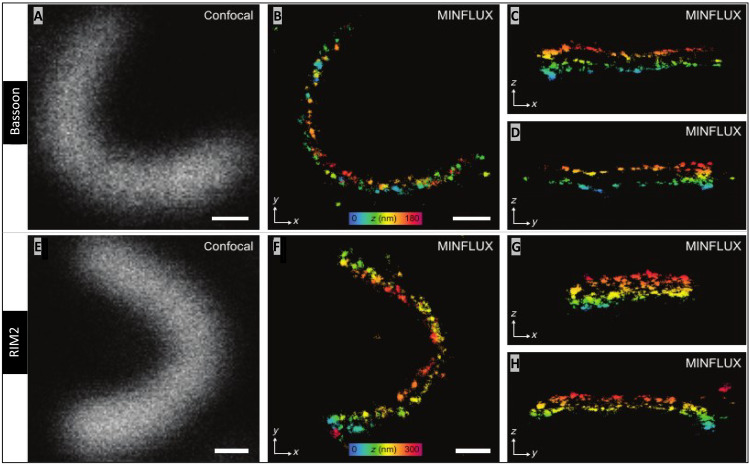
Confocal images of HARD samples show that rod ribbons are not all lying flat in the xy plane, but, instead, they have a complex 3D structure (Figs. 1E and 3, A to E, and movies S4 and S6). To overcome the limitations of rendering these 3D structures in 2D, we turned to 3D-MINFLUX nanoscopy. Figure 4 shows a field of view with multiple rod ribbon AZs, and, here, two parallel rows of RIM2 or bassoon can be seen twisting in space (depth is indicated on a color scale; Fig. 4, C and D). Examination of an individual bassoon-labeled AZ (Fig. 5A), at specific orientations, revealed a series of discrete localizations, which, on a small spatial scale, appear linearly arranged in single file (Fig. 5, B to D). This is most apparent when watching the ribbon about a fixed axis (Fig. 5B and movies S7 and S8). The same arrangement of two rows describes the pattern of RIM2 localizations when viewed at fixed angles (Fig. 5, E to H) or in rotation (movies S9 and S10). Quantitation of the spacing between rows gave the following average distances in nanometer (means ± SD; N ribbons) for bassoon and RIM2: 83.0 (7.9; 6) and 89.6 (8.3; 5), respectively. The separation of the RIM2 rows tended to be greater than that found for bassoon, although this did not reach statistical significance (P = 0.2100; Student’s t test). The localizations precision for each protein was between 4 and 6 nm (see Materials and Methods).
Fig. 5. 3D-MINFLUX reveals parallel rows of RIM2 and bassoon at rod AZs.
(A) Confocal image of a bassoon labeled rod ribbon. (B) Side view of the ribbon (z-projection), which masks the two rows of bassoon (movies S7 and S8: rotating bassoon labeled ribbon). (C and D) Top-down view of the central and end regions of the ribbon, such that the two rows of bassoon are apparent (C and D are y- and x-projections, respectively). Color distance scale (range: 0 to180 nm) presented in (B) indicates the height of the ribbon (distance in the z direction) when the ribbon lies in the zx- and zy-plane as in (C) and (D), respectively. (E to H) RIM2-labeled ribbon presented in the same manner as for bassoon in (A) to (C), and here, the color scale ranges from 0 to 300 nm (movies S9 and S10: rotating RIM2-labeled ribbon). White scale bars in (A), (B), (E), and (F) are 200 nm in length.
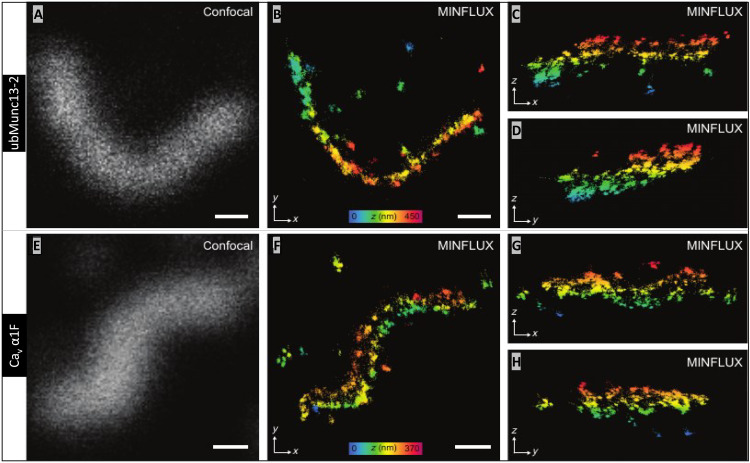
The other two AZ proteins examined in 3D-MINFLUX were ubMunc13-2 (Fig. 6, A to D) and the Cav1.4 α1F subunit (Fig. 6, E to H). The two rows of ubMunc13-2 are apparent at certain angles (Fig. 6B and movies S11 and S12), and the same can be said for the α1F subunit (Fig. 6F and movies S13 and S14). The distance (in nanometers) between the rows of ubMunc13-2 and α1F subunit were (means ± SD; N ribbons): 112.8 (12.7; 5) and 117.7 (11.0; 5), respectively. The distances separating the ubMunc13-2 and α1F subunit rows were statistically indistinguishable from each other (P = 0.532), but they were further apart than bassoon and RIM2. The two rows of α1F subunits are separated by 34.7 nm more than the rows of bassoon (P = 0.001), and the rows of ubMunc13-2 are spaced apart by 29.2 nm more than the rows of bassoon (P = 0.0002). The two rows of α1F are separated by 28.1 nm more than the rows of RIM2 (P = 0.0091), and the rows of ubMunc13-2 are separated by 23.4 nm more than the rows of RIM2 (P = 0.0019).
Fig. 6. Cav1.4 channels and ubMunc13-2 are bilaterally distributed about the rod ribbon AZ.
(A) Confocal image of ubMunc13-2 and, in (B), the corresponding 3D-MINFLUX image presented as a z-projection (movies S11 and S12: rotating ubMunc13-2–labeled rod ribbon). The same ribbon in (B) is presented as y- and x-projections in (C) and (D), respectively, partially revealing the two tracks of ubMunc13-2 in the central and end regions of the ribbon. (E to H) A ribbon labeled for Cav1.4 α1F subunits is presented in the same manner as ubMunc13-2 (movies S13 and S14: rotating α1F-labeled ribbon). Z direction, color scale bars in (B) and (F) range from 450 to 370 nm, respectively. White scale bars in (A), (B), (E), and (F) are 200 nm in length.
Colabeling of bassoon and RIM2 reveals their overlapping 3D distributions
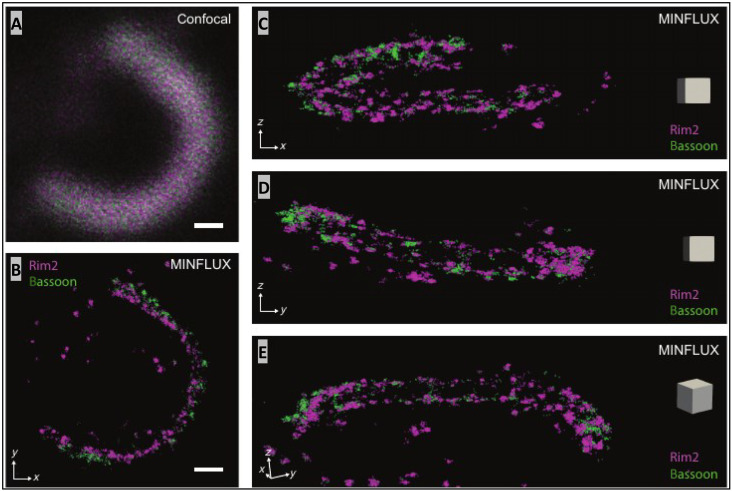
To better relate the 3D topographies of AZ proteins, we performed two-color 3D-MINLFUX imaging on samples labeled for bassoon with CF660c and RIM2 with Alexa647. Spectral unmixing of emissions from single-molecule localizations placed RIM2 slightly beneath the inner face of the bassoon profile (i.e., in the z dimension using the concavity of rod AZs as a feature) within ~20 nm (Fig. 7B). In addition, bassoon and RIM2 localizations overlapped with each other within each row along the length of the ribbon, which is consistent with the finding above that, on average, the distance between the two rows of proteins is not statistically different. The clear advantage of dual-probe versus single-probe 3D-MINFLUX is that we were able to determine that bassoon sat above RIM2 rather than below it and that the individual localizations overlap in the xy plane.
Fig. 7. Simultaneous imaging of bassoon and RIM2 with 3D-MINFLUX shows that they are intimately colocalized.
(A) Confocal image of a rod ribbon AZ stained for bassoon (CF660C) and RIM2 (Alexa 647), without spectral unmixing. (B) The corresponding two-color 3D-MINFLUX image, viewed from the side (z-projection) shows RIM2 slightly interior to bassoon. Scale bars in (A) and (B): 200 nm. Additional images of this ribbon are presented in (C) and (D) as y- and x-projections, respectively. In (E) the image is tilted to better reveal the arrangement of RIM2 and bassoon into two rows. The gray square box in each panel (C to E) has an edge with a length of 100 nm.
DISCUSSION
This 3D-MINFLUX study reports a highly efficient optical nanoscopy approach for elucidating the molecular architecture of the presynaptic AZ. The approach was enabled by the transfer of a thin layer of retinal tissue to the coverslip by means of HARD. Confidence in the structural preservation of synapses in the HARD samples was supported by results from standard confocal and EM. Analyzing the AZ of rod PRs in HARD samples with 3D-MINFLUX allowed for the localization of approximately a thousand single-molecule fluorescence events in 3D space, with 4- to 6-nm localization precision in less than 30 min in many instances. The results indicate that AZ proteins are organized in two parallel rows running on either side of the base of the rod ribbon. The continuity of the AZ over a contour of 1.5 μm suggests that SV release sites are formed with serial regularity, and these findings offer novel insight into the structural basis of the functional operation of the rod ribbon synapse. More generally, the combination of 3D-MINFLUX and tissue transfer by the HARD method provides a powerful approach for decipher the structure of synapses and other nanoscale functional units of cells in a near-native tissue context.
The 3D structure of the HARD immobilized rod synapses visualized in confocal IF (Fig. 1 to 3) are consistent with published images of immuno-labeled rod ribbons in slices (15, 21, 24, 26, 29). In addition, EM images of HARD samples demonstrated that synaptic structures in rod terminals (Fig. 2, E to G) and subcellular structures in the inner and outer segments of RPs were preserved (Fig. 2H). HARD samples presented substantially less unwanted extraneous signal than what is commonly encountered in conventionally fixed retinal sections (32). Unwanted signal can come from autofluorescence, uncoupled dye partitioning into membranes, or nonspecific adsorption of primary and/or secondary antibodies onto sticky surfaces (33). Aldehyde can exacerbate nonspecific signals (e.g., autofluorescence) and impair the binding of antibodies to their target epitopes (33), which has the effect of lowering the signal of interest while enhancing unwanted background noise. In addition, we did not use detergents, because they can cause protein extraction and thus lower the density of available epitopes (33). We posit that the high degree of labeling witnessed in the 3D images benefited from preservation of the AZ proteins and their epitopes (for instance, see Fig. 5, B to D). An additional source of unwanted signal arises from out-of-focus emissions that can interfere with single-molecule localization routines. It does not matter if the out-of-focus signal originates from specific labeling of AZ targets or nonspecific background, because when photon emissions reach excessively high rates, single-molecule localization is no longer possible (13). The thin layer of HARD sample greatly reduced signal redundancy in the z direction.
The AZ proteins that we studied in 3D were localized to two rows that ran continually for the length of the rod ribbon AZ. The separation of the two rows fits well with our general understanding of the physical dimensions at the base of the ribbon (fig. S5A). From EM studies, there are estimated to be ~40 SVs docked on either side of the ribbon, at its base, and running the length (~1.5 μm) of the ribbon (25, 32). Assuming that the upper estimate of the distance separating SV release sites on either side of the ribbon is approximated by the sum of the following components: (i) width of a rod ribbon, (ii) lengths of filaments attaching SVs to the ribbon body (“tethers”), and (iii) diameters of two SVs, and then a distance of 132 nm separates the release sites (fig. S5A). The epitopes of the AZ proteins that we studied fit within this upper estimate (fig. S5B). Rows of scaffolding protein epitopes for bassoon and RIM2 are separated by ~87 nm, and rows of Cav1.4 channel, (α1F subunit) and ubMunc13-2 are separated by distances of ~116 nm. The spacing of the rows of epitopes exceeds the width of the AD, which is a subregion that sits beneath the base of the ribbon. Assuming that RIM2 and bassoon extend tens of nanometers beyond where the antibody epitopes are localized (34), they may interact with proteins at the base of the ribbon or AD and, in addition, interact with laterally displaced release site partners at the PM (fig. S5C). In contrast, because ubMunc13-2 is a globular, well-structured protein (~180 kDa), its location is comparatively better approximated by the localizations of its epitope. In the case of the α1F subunit, it is the only transmembrane membrane protein (~230 kDa) that we studied. The antibody used here was directed against the cytosolic C terminus of α1F, which may extend only a few nanometers from the channel’s pore; thus, the epitope therein is likely a good estimate of the channel location. Notably, the spacing between the two rows of Cav channels and ubMunc13-2 is close to our upper estimate of the distances between release sites on either side of the ribbon, differing by 14 nm (118 nm versus 132 nm, respectively). Half of the 14-nm difference is what we suggest as the spacing between Cav channels and SV release site within an AZ on one side of the ribbon.
Functional estimates of the spatial coupling between vesicular release sites and Cav channels suggest that release from mouse rods is controlled by a Ca2+ nanodomain about the open channel (21). From electrophysiological results, nearly 90 SVs (45 SVs per side) are readily released from the mouse rod ribbon within 1 ms of the onset of a strong stimulation. This RRP of SVs was similar in size when low (0.5 mM EGTA) and high (10 mM EGTA) concentrations of intracellular Ca2+ buffering were used, suggesting that the releasable SVs were within ~10 nm of the Cav channel(s) (in other words, a Ca2+ nanodomain rather than a microdomain or global Ca2+) (21). This result is consistent with other studies carried out on auditory hair cells (35–37) and rbcs (15, 38) that also concluded a Ca2+ nanodomain controls release. Furthermore, the RRP of SVs at mouse rods was highly dependent on the presence of the ribbon, as ribbonless rods showed a 75% reduction in the size of their RRP (21), which is consistent with studies on ribbonless mouse rbcs (15). Incorporating the physiological results from mouse rods with our 3D-MINFLUX results suggests that the ~115-nm spacing between rows of AZ proteins (average of ubMunc13-2 and Cav1.4 spacings) is our best estimate of where SVs are primed and released of each side of the base of the ribbon (fig. S5B). Our 3D imaging results argue against the possibility that Cav1.4 channels are restricted to the recesses of the AD as previously suggested (31), because (i) the AD is too narrow and (ii) a Ca2+ nanodomain formed within the AD would be too far from the SV release sites to control release. Why would the rod ribbon create two parallel AZs?
AMPA receptor–bearing hc spines are positioned within 30 nm of the release sites (Fig. 2, F and G) (39), on either side of the ribbon (see schematic in Figs. 1A and 2, G and H). In mouse, an elongated, individual hc spine is assigned to one side of the ribbon [see schematic in Fig. 1A; (26)]. It is possible that an AZ on one side of the ribbon communicates independently with the spine it abuts. This is likely under circumstances that favor Ca2+ nanodomain control of release. The functional value of a one-to-one pairing of an AZ-to-spine may be that this allows an hc spine to specifically direct inhibitory feedback to the rod AZ that it is paired to. For example, in an individual cone terminal that expresses several synaptic ribbons, inhibitory feedback from an hc is circumscribed to the synaptic ribbon that it contacts (40). An entirely different scenario describes communication between rods and their postsynaptic rbcs. Each rod spherule contains a pair of rbc dendrites (26). They are thought to receive their glutamatergic input from the same rod AZ release sites as hcs (41), but the rbc dendrites are centrally located, displaced 200 nm from the rod ribbon AZs (26). This is assumed to allow rbc dendrites to monitor release from both sides of the ribbon, which doubles the number of release sites over which the release rate is averaged. In theory, this should enhance the ability of the synapse to signal single-photon events (41, 42).
Our 3D-MINFLUX results build on earlier studies that investigated the molecular architecture of mouse rod AZs and mechanisms of release. Previous studies using immuno-EM techniques determined the location of AZ proteins by analyzing 2D cross sections through the large, curved rod AZ(s). They reported that the AZ proteins (i.e., ubMunc13-2 and Cav1.4 channels) were positioned around the base of the ribbon, either in or near the AD, or laterally displaced from the ribbon where SVs are docked (4, 17, 25, 31). We find that these AZ proteins are positioned laterally, rather than in the AD, which we propose positions them in proximity (<10 nm) to SV docking sites (see fig. S5, A and B). We view this as a refinement of earlier models, which was facilitated by 3D imaging with a localization precision under 6 nm. Furthermore, the positioning of the AZ proteins concurs with our recent physiological study on wild-type and ribbonless mouse rods that suggested that a Ca2+ nanodomain controls the fusion of SVs docked at the base, and on both sides, of the ribbon (21). In that study, we also found that a fraction (25%) of release persisted in ribbonless rods, yet the coupling of Cav channels and SVs remained on a nanoscale. This is in contrast to measurements of evoked release from salamander rods, which represents the bulk of literature on this topic, that have reported Ca2+ nanodomain control of release in addition to a larger portion of release that is under the control of global Ca2+ (18, 19) and possibly independent of the ribbon (20). To explain the differences in the physiological behavior of mouse and salamander rods, with reference to Cav channel topographies, two divergent scenarios are given. First, the Cav channels in salamander terminals may be arranged into arrays distinct from the two rows of AZ proteins that we report for mouse rod ribbons. Alternatively, the same two rows of AZs may be formed about each salamander ribbon, but because salamander rods express five to seven ribbons per terminal (18), their ribbons may be positioned close enough to one another to combine Ca2+ domains (18). Applying 3D-MINFLUX to study Cav channels in salamander rods and other ribbon expressing cells will provide new structural and functional insight.
Our findings are consistent with several detailed descriptions of the molecular topography of AZ formed by other ribbon and conventional synapses. Several STED imaging studies on mouse cochlear inner hair cells (ihcs) have resolved highly ordered pre- and postsynaptic arrangements of synaptic proteins. In particular, linear strips of AZ proteins accurately describe the ihc ribbon-AZ (28, 43), and here, a nanodomain of Ca2+ has been imaged in living ihcs (8). A potential difference to rod AZs could be that the Cav1.3 channels of the ihc AZ cluster underneath the presynaptic density and feature a variable set of channels at the perimeter of the cluster that are tightly linked to SV release sites (44, 45). However, proper elucidation of Cav1.3 channel topography, in combination with RIM-binding protein (46), will require MINFLUX analysis or freeze-fracture immunolabeling EM (47, 48). Additional high-resolution detail comes from work at the fly neuromuscular junction where different forms of Unc13 are differentially localized at the AZ, to carry out unique functional roles (11). Similarly, Munc13-1 clusters have been indicated to form SV release sites at AZs of cultured hippocampal neurons (7) Some of the approaches used in those studies can be implemented to better elucidate the structural and functional organization of mouse rod ribbons. For instance, by using additional epitopes for a given protein, we should be able to better understand the orientation of AZ proteins about a release site. Even in the absence of such information in the current study, we were able to gain useful insight into mechanisms of release.
MATERIALS AND METHODS
Retinal slices and HARD
Animals were handled in accordance with the Max Planck Institute and German national animal care guidelines. Retinal slices freshly prepared from wild-type C57BL/6 mice were made with the same procedure used for electrophysiological recordings (26). Briefly, dissected portions of retina were absorbed onto pieces of nitrocellulose membrane, vitreal side down, and then sectioned with a tissue chopper (custom-made). Slicing was carried out in normal mouse extracellular solution (MES) with a low Ca2+ concentration that had the following composition: 135 mM NaCl, 2.5 mM KCl, 0.5 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 15 mM Hepes, pH adjusted to 7.35, with NaOH and an osmolarity of 295 mOsmol. Five minutes after slicing, the extracellular solution was exchanged to an MES with 2 mM CaCl2 at 20°C and incubated for 10 min before fixation. Glass coverslips used in these experiments were cleaned as follows: sonicated in deionized water for 20 min and then heat-dried in an oven at 50°C. To preserve retinal tissue for confocal, EM, and MINFLUX, we used HARD; see the Supplementary Materials for details on preparing HARD samples. Briefly, this entailed momentarily placing a 150- to 200-μm-thick fresh retinal slices (still attached to nitrocellulose membrane) directly onto glass coverslips that rested on a histological heat plate maintained at a temperature of 50°C. HARD samples were not treated with chemical fixatives or permeabilizing agents before or during immunostaining. The HARD samples could be stored in a desiccated chamber at 23°C for 2 weeks until needed, although, in most instances, the samples were immunostained immediately after preparation.
Immunohistochemistry
Immunohistochemistry of fixed retinal slices was performed as described (32). For indirect immunolabeling, HARD samples were first blocked for 60 min at 23°C with 2% BSA dissolved in PBS and then incubated with primary antibodies at a dilution of 1:1000 in 2% BSA-PBS for 12 to 16 hours at 4°C. Following incubation in the primary antibody, the HARD sample coverslips were washed on a rotating table for 2 to 4 hours, starting with 2% BSA-PBS for 30 min at 4°C, followed by three exchanges in 1× PBS for 30 min of wash time per exchange. Next, the HARD samples were incubated 1 hour with the secondary antibody at 23°C in 2% BSA-PBS. Samples were then washed two times in 2% BSA-PBS for 30 min at 23°C. In most instances, the ribbon was also labeled after labeling AZ proteins. This entailed a 1- to 2-hour incubation with 1:2000 dilution of anti-ctbp2 (mouse) or anti–ribeye A (guinea pig) at 23°C.
A list of primary and secondary antibodies is provided in Table 1, and references to published works using these antibodies can also be found therein. The epitopes used to make the antibodies are summarized in fig. S1. The Alexa Fluor 647 dye was used for MINFLUX. In addition, an anti-mouse secondary antibody coupled to the CF660C dye (Biotium), made in-house (Abberior Instruments), was used for two-color 3D-MINFLUX imaging in combination with an anti-rabbit Alexa 647 as the second fluorophore (Fig. 7). Stained samples were kept in PBS at 4°C until time of use (within 5 days of rehydration). For initial evaluation and documentation of the HARD sample procedure, immunolabeled samples were imaged with a spinning disc confocal Visiscope (Visitron Systems) at the Live-Cell Imaging Facility (Max Planck Institute for Multidisciplinary Sciences). Images were processed with ImageJ-Fiji software. Imaging results were derived from immunolabeling experiments that were performed in duplicate or triplicate, and multiple preparations were used to assess each primary antibody.
Sample mounting for MINFLUX microscopy
To enable sample stabilization during MINFLUX measurements, gold nanorods (Nanopartz Inc., A12-40-980-CTAB-DIH-1-25) were added as fiducials before mounting the samples in imaging buffer as described before (12). In brief, an undiluted dispersion of the nanorods was applied to the ready-made samples for 5 to 10 min. For MINFLUX imaging, samples were mounted in GLOX buffer [50 mM tris-HCl, 10 mM NaCl, 10% (w/v) glucose, and catalase (64 μg/ml), glucose oxidase (0.4 mg/ml), and 10 to 25 mM mercaptoethylamine (pH 8.0)] (12, 49). After mounting, samples were sealed with twinsil (picodent).
MINFLUX nanoscopy
For confocal and MINFLUX microscopy, an Abberior Instruments MINFLUX microscope equipped with a 642-nm (continuous wave) excitation laser, a 405-nm (continuous wave) activation laser, an spatial light modulator-based beam shaping module, and an electro-optical detector-based MINFLUX scanner was applied (13). Fluorescence photons emitted from the sample were counted using two avalanche photodiodes together with fluorescence filters (650 to 750 nm). In addition to the MINFLUX channels, the microscope has epifluorescence and 488-nm confocal imaging beam paths, which allow to identify structures based on ribeye–Alexa Fluor 488 IF-labeled structures. To enable measurements with molecular precision, the Abberior MINFLUX microscope is equipped with a reflection-based stabilization unit, based on a 980-nm laser. The MINFLUX microscope was calibrated daily as described previously (13). The precision of 3D-MINFLUX localizations is as follows [range of precision (nm), and the range of number of localizations]: Cav1.4 α1F: 5.01 to 5.53 nm, and 423 to 1550; bassoon: 4.29 to 4.34 nm and 359 to 614 ; ubMunc13-2: 4.65 to 5.58 nm, and 423 to 1283; RIM2: 4.94 to 5.27 nm, and 417 to 3751. The precision of the 2D-MINFLUX localizations is as follows [range of precision (nm) and number of localizations (N)]: CAST: 3.30 to 7.17 nm and 319 to 2776 localizations; piccolo: 3.34 to 3.91 nm and 2476 to 7809 N. To measure the distances between rows of AZ proteins, we used the ruler in ParaView (50) while rotating the image in the 3D viewer as shown in Fig. 5 (C and D). Five measurements of distance between rows were from each rod ribbon AZ, including the ends and central region. Data are presented as means ± SD. The measurements were from five to six rod AZs, and the unpaired Student’s t test was used to make statistical comparisons between AZ datasets.
Electron microscopy
HARD samples were rehydrated and subsequently immersed in freshly prepared and prewarmed 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.0) for 1 hour at room temperature. Samples were then kept in a cold room for 2 days. Subsequently, they were washed with double distilled water three times for 5 min each. The samples were dehydrated in a graded series of ethanol (30, 50, 75, and 100%, 5 min each) with a final dehydration in propylene oxide for 5 min at room temperature before resin infiltration with a 1:1 mixture of propylene oxide and EPON (EMbed 812; 14121, Electron Microscopy Sciences, Hatfield, PA, USA) for 1 hour and two fresh replacements of 100% EPON (first for 1 hour, and second overnight at 4°C). For the final embedding, the samples were covered with BEEM capsules (70,000; Electron Microscopy Sciences, Hatfield, PA, USA) filled with fresh EPON resin and cured in an oven at 60°C for 48 hours. Ultrathin (100-nm) sections were cut perpendicular to the coverslip and collected onto grids for transmission EM using a Thermo Fisher Scientific Talos L120C equipped with a Ceta 4K CMOS camera.
Acknowledgments
We thank B. Cooper and E. Neher for comments on the manuscript and A. Politi and P. Lenart for assistance with confocal microscopy.
Funding: This research project was funded by the Deutsche Forschungsgemeinschaft (DFG) for Quantitative Synaptology: CRC1286 (T.M.), Max-Planck-Society Fellowship (T.M.), the DFG under Germany’s Excellence Strategy—EXC 2067/1-390729940 (T.M.), and the German Ministry for Education and Research (BMBF) through grants to Abberior Instruments (grant 13N14122; LiveCell3DNanoscopy). In addition, this research is supported by Fondation Pour l’Audition (FPA RD-2020-10) to T.M.
Author contributions: Conceptualization: T.M. and C.P.G. Methodology: C.P.G. (HARD samples), C.A.W. and I.J. (MINIFLUX imaging), and T.W. and R.S. (MINFLUX Programming and Instrumentation). Investigation: C.P.G. (immunolabeling, confocal and MINFLUX imaging, and analysis), I.J. and C.A.W. (MINFLUX imaging and analysis), J.N. (analysis), and D.R. (electron microscopy). Visualization: C.P.G., I.J., C.A.W., and J.N. Supervision: C.P.G., C.A.W., and T.M. Writing—original draft: C.P.G. and T.M. Writing—review and editing: All authors.
Competing interests: The authors declare that they have no competing interests in the production and presentation of results. I.J., C.A.W., R.S., and T.W. work at Abberior Instruments that develops and manufacture superresolution fluorescence microscopes, including the 3D-MINFLUX system used here. Funding from the German Ministry for Education and Research (BMBF) to Abberior was used to facilitate the academic collaboration.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Supplemental Materials: HARD samples
HARD sample rationale and development
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S14
REFERENCES AND NOTES
- 1.Heuser J. E., Reese T. S., Dennis M. J., Jan Y., Jan L., Evans L., Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell Biol. 81, 275–300 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harlow M. L., Ress D., Stoschek A., Marshall R. M., McMahan U. J., The architecture of active zone material at the frog’s neuromuscular junction. Nature 409, 479–484 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Jung J. H., Szule J. A., Marshall R. M., McMahan U. J., Variable priming of a docked synaptic vesicle. Proc. Natl. Acad. Sci. U.S.A. 113, E1098–E1107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limbach C., Laue M. M., Wang X., Hu B., Thiede N., Hultqvist G., Kilimann M. W., Molecular in situ topology of Aczonin/Piccolo and associated proteins at the mammalian neurotransmitter release site. Proc. Natl. Acad. Sci. 108, E392–E401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holderith N., Lorincz A., Katona G., Rózsa B., Kulik A., Watanabe M., Nusser Z., Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat. Neurosci. 15, 988–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brose N., Brunger A., Cafiso D., Chapman E. R., Diao J., Hughson F. M., Jackson M. B., Jahn R., Lindau M., Ma C., Rizo J., Shin Y. K., Sollner T. H., Tamm L., Yoon T. Y., Zhang Y., Synaptic vesicle fusion: Today and beyond. Nat. Struct. Mol. Biol. 26, 663–668 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto H., Ariyoshi T., Kimpara N., Sugao K., Taiko I., Takikawa K., Asanuma D., Namiki S., Hirose K., Synaptic weight set by Munc13-1 supramolecular assemblies. Nat. Neurosci. 21, 41–49 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Neef J., Urban N. T., Ohn T.-L., Frank T., Jean P., Hell S. W., Willig K. I., Moser T., Quantitative optical nanophysiology of Ca2+ signaling at inner hair cell active zones. Nat. Commun. 9, 290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehmann N., van de Linde S., Alon A., Ljaschenko D., Keung X. Z., Holm T., Rings A., DiAntonio A., Hallermann S., Ashery U., Heckmann M., Sauer M., Kittel R. J., Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5, 4650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klevanski M., Herrmannsdoerfer F., Sass S., Venkataramani V., Heilemann M., Kuner T., Automated highly multiplexed super-resolution imaging of protein nano-architecture in cells and tissues. Nat. Commun. 11, 1552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Böhme M. A., Beis C., Reddy-Alla S., Reynolds E., Mampell M. M., Grasskamp A. T., Lützkendorf J., Bergeron D. D., Driller J. H., Babikir H., Göttfert F., Robinson I. M., O’Kane C. J., Hell S. W., Wahl M. C., Stelzl U., Loll B., Walter A. M., Sigrist S. J., Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca2+ channel–vesicle coupling. Nat. Neurosci. 19, 1311–1320 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Balzarotti F., Eilers Y., Gwosch K. C., Gynnå A. H., Westphal V., Stefani F. D., Elf J., Hell S. W., Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Schmidt R., Weihs T., Wurm C. A., Jansen I., Rehman J., Sahl S. J., Hell S. W., MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nat. Commun. 12, 1478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jean P., Lopez de la Morena D., Michanski S., Jaime Tobón L. M., Chakrabarti R., Picher M. M., Neef J., Jung S., Gültas M., Maxeiner S., Neef A., Wichmann C., Strenzke N., Grabner C., Moser T., The synaptic ribbon is critical for sound encoding at high rates and with temporal precision. eLife 7, e29275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxeiner S., Luo F., Tan A., Schmitz F., Südhof T. C., How to make a synaptic ribbon: RIBEYE deletion abolishes ribbons in retinal synapses and disrupts neurotransmitter release. EMBO J. 35, 1098–1114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaithianathan T., Henry D., Akmentin W., Matthews G., Nanoscale dynamics of synaptic vesicle trafficking and fusion at the presynaptic active zone. eLife 5, e13245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moser T., Grabner C. P., Schmitz F., Sensory processing at ribbon synapses in the retina and the cochlea. Physiol. Rev. 100, 103–144 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Thoreson W. B., Rabl K., Townes-Anderson E., Heidelberger R., A highly Ca2+-sensitive pool of vesicles contributes to linearity at the rod photoreceptor ribbon synapse. Neuron 42, 595–605 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rieke F., Schwartz E. A., Asynchronous transmitter release: Control of exocytosis and endocytosis at the salamander rod synapse. J. Physiol. 493, 1–8 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M., Hook M. J. V., Zenisek D., Thoreson W. B., Properties of ribbon and non-ribbon release from rod photoreceptors revealed by visualizing individual synaptic vesicles. J. Neurosci. 33, 2071–2086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabner C. P., Moser T., The mammalian rod synaptic ribbon is essential for Cav channel facilitation and ultrafast synaptic vesicle fusion. eLife 10, e63844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zenisek D., Horst N. K., Merrifield C., Sterling P., Matthews G., Visualizing synaptic ribbons in the living cell. J. Neurosci. 24, 9752–9759 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz F., Königstorfer A., Südhof T. C., RIBEYE, a component of synaptic ribbons: A protein’s journey through evolution provides insight into synaptic ribbon function. Neuron 28, 857–872 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Kerov V., Laird J. G., Joiner M., Knecht S., Soh D., Hagen J., Gardner S. H., Gutierrez W., Yoshimatsu T., Bhattarai S., Puthussery T., Artemyev N. O., Drack A. V., Wong R. O., Baker S. A., Lee A., α2δ-4 is required for the molecular and structural organization of rod and cone photoreceptor synapses. J. Neurosci. 38, 6145–6160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper B., Hemmerlein M., Ammermuller J., Imig C., Reim K., Lipstein N., Kalla S., Kawabe H., Brose N., Brandstatter J. H., Varoqueaux F., Munc13-independent vesicle priming at mouse photoreceptor ribbon synapses. J. Neurosci. 32, 8040–8052 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagiwara A., Kitahara Y., Grabner C. P., Vogl C., Abe M., Kitta R., Ohta K., Nakamura K., Sakimura K., Moser T., Nishi A., Ohtsuka T., Cytomatrix proteins CAST and ELKS regulate retinal photoreceptor development and maintenance. J. Cell Biol. 217, 3993–4006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khimich D., Nouvian R., Pujol R., tom Dieck S., Egner A., Gundelfinger E. D., Moser T., Hair cell synaptic ribbons are essential for synchronous auditory signalling. Nature 434, (2005). [DOI] [PubMed] [Google Scholar]
- 28.Michanski S., Smaluch K., Steyer A. M., Chakrabarti R., Setz C., Oestreicher D., Fischer C., Möbius W., Moser T., Vogl C., Wichmann C., Mapping developmental maturation of inner hair cell ribbon synapses in the apical mouse cochlea. Proc. Natl. Acad. Sci. U.S.A. 116, 6415–6424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller T. M., Gierke K., Joachimsthaler A., Sticht H., Izsvák Z., Hamra F. K., Fejtová A., Ackermann F., Garner C. C., Kremers J., Brandstätter J. H., Regus-Leidig H., A multiple piccolino-RIBEYE interaction supports plate-shaped synaptic ribbons in retinal neurons. J. Neurosci. 39, 2606–2619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deguchi-Tawarada M., Inoue E., Takao-Rikitsu E., Inoue M., Kitajima I., Ohtsuka T., Takai Y., Active zone protein CAST is a component of conventional and ribbon synapses in mouse retina. J. Comp. Neurol. 495, 480–496 (2006). [DOI] [PubMed] [Google Scholar]
- 31.tom Dieck S., Altrock W. D., Kessels M. M., Qualmann B., Regus H., Brauner D., Fejtová A., Bracko O., Gundelfinger E. D., Brandstätter J. H., Molecular dissection of the photoreceptor ribbon synapse physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J. Cell Biol. 168, 825–836 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabner C. P., Gandini M. A., Rehak R., Le Y., Zamponi G. W., Schmitz F., RIM1/2-mediated facilitation of Cav1.4 channel opening is required for Ca2+-stimulated release in mouse rod photoreceptors. J. Neurosci. 35, 13133–13147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stradleigh T. W., Ishida A. T., Fixation strategies for retinal immunohistochemistry. Prog. Retin. Eye Res. 48, 181–202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gundelfinger E. D., Reissner C., Garner C. C., Role of bassoon and piccolo in assembly and molecular organization of the active zone. Front. Synaptic Neurosci. 7, 19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandt A., Few CaV1.3 channels regulate the exocytosis of a synaptic vesicle at the hair cell ribbon synapse. J. Neurosci. 25, 11577–11585 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graydon C. W., Cho S., Li G.-L., Kachar B., von Gersdorff H., Sharp Ca2+ nanodomains beneath the ribbon promote highly synchronous multivesicular release at hair cell synapses. J. Neurosci. 31, 16637–16650 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts W. M., Jacobs R. A., Hudspeth A. J., Colocalization of ion channels involved in frequency selectivity and synaptic transmission at presynaptic active zones of hair cells. J. Neurosci. 10, 3664–3684 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarsky T., Tian M., Singer J. H., Nanodomain control of exocytosis is responsible for the signaling capability of a retinal ribbon synapse. J. Neurosci. 30, 11885–11895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao-Mirotznik R., Harkins A. B., Buchsbaum G., Sterling P., Mammalian rod terminal: Architecture of a binary synapse. Neuron 14, 561–569 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Beckwith-Cohen B., Holzhausen L. C., Wang T.-M., Rajappa R., Kramer R. H., Localizing proton-mediated inhibitory feedback at the retinal horizontal cell–cone synapse with genetically-encoded pH probes. J. Neurosci. 39, 651–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao-Mirotznik R., Buchsbaum G., Sterling P., Transmitter concentration at a three-dimensional synapse. J. Neurophysiol. 80, 3163–3172 (1998). [DOI] [PubMed] [Google Scholar]
- 42.van Rossum M. C., Smith R. G., Noise removal at the rod synapse of mammalian retina. Vis. Neurosci. 15, 809–821 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Chapochnikov N. M., Takago H., Huang C.-H., Pangršič T., Khimich D., Neef J., Auge E., Göttfert F., Hell S. W., Wichmann C., Wolf F., Moser T., Uniquantal release through a dynamic fusion pore is a candidate mechanism of hair cell exocytosis. Neuron 83, 1389–1403 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Wong A. B., Rutherford M. A., Gabrielaitis M., Pangršič T., Göttfert F., Frank T., Michanski S., Hell S., Wolf F., Wichmann C., Moser T., Developmental refinement of hair cell synapses tightens the coupling of Ca2+ influx to exocytosis. EMBO J. 33, 247–264 (2014), doi: 10.1002/embj.201387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pangršič T., Gabrielaitis M., Michanski S., Schwaller B., Wolf F., Strenzke N., Moser T., EF-hand protein Ca2+ buffers regulate Ca2+ influx and exocytosis in sensory hair cells. Proc. Natl. Acad. Sci. U.S.A. 112, E1028–E1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hibino H., Pironkova R., Onwumere O., Vologodskaia M., Hudspeth A. J., Lesage F., RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca2+ channels. Neuron 34, 411–423 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butola T., Alvanos T., Hintze A., Koppensteiner P., Kleindienst D., Shigemoto R., Wichmann C., Moser T., RIM-binding protein 2 organizes Ca2+ channel topography and regulates release probability and vesicle replenishment at a fast central synapse. J. Neurosci. 41, 7742–7767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura Y., Harada H., Kamasawa N., Matsui K., Rothman J. S., Shigemoto R., Silver R. A., Di Gregorio D. A., Takahashi T., Nanoscale distribution of presynaptic Ca2+ channels and its impact on vesicular release during development. Neuron 85, 145–158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gwosch K. C., Pape J. K., Balzarotti F., Hoess P., Ellenberg J., Ries J., Hell S. W., MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 17, 217–224 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Ahrens J., Geveci B., Law C., ParaView: An end-user tool for large data visualization. Vis. Handb. 717–731 (2005). [Google Scholar]
- 51.Dick O., tom Dieck S., Altrock W. D., Ammermüller J., Weiler R., Garner C. C., Gundelfinger E. D., Brandstätter J. H., The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron 37, 775–786 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Jung S., Oshima-Takago T., Chakrabarti R., Wong A. B., Jing Z., Yamanbaeva G., Picher M. M., Wojcik S. M., Göttfert F., Predoehl F., Michel K., Hell S. W., Schoch S., Strenzke N., Wichmann C., Moser T., Rab3-interacting molecules 2α and 2β promote the abundance of voltage-gated CaV1.3 Ca2+ channels at hair cell active zones. Proc. Natl. Acad. Sci. U.S.A. 112, E3141–E3149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryl M., Urbasik A., Gierke K., Babai N., Joachimsthaler A., Feigenspan A., Frischknecht R., Stallwitz N., Fejtová A., Kremers J., von Wittgenstein J., Brandstätter J. H., Genetic disruption of bassoon in two mutant mouse lines causes divergent retinal phenotypes. FASEB J. 35, e21520 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Klotz L., Wendler O., Frischknecht R., Shigemoto R., Schulze H., Enz R., Localization of group II and III metabotropic glutamate receptors at pre- and postsynaptic sites of inner hair cell ribbon synapses. FASEB J. 33, 13734–13746 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Gehlen J., Aretzweiler C., Mataruga A., Fahlke C., Müller F., Excitatory amino acid transporter EAAT5 improves temporal resolution in the retina. eNeuro 8, ENEURO.0406-21.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butola T., Wichmann C., Moser T., Piccolo promotes vesicle replenishment at a fast central auditory synapse. Front. Synaptic Neurosci. 9, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zampighi G. A., Schietroma C., Zampighi L. M., Woodruff M., Wright E. M., Brecha N. C., Conical tomography of a ribbon synapse: Structural evidence for vesicle fusion. PLOS ONE 6, e16944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dembla E., Dembla M., Maxeiner S., Schmitz F., Synaptic ribbons foster active zone stability and illumination-dependent active zone enrichment of RIM2 and Cav1.4 in photoreceptor synapses. Sci. Rep. 10, 5957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitz F., Bechmann M., Drenckhahn D., Purification of synaptic ribbons, structural components of the photoreceptor active zone complex. J. Neurosci. 16, 7109–7116 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Supplemental Materials: HARD samples
HARD sample rationale and development
References
Movies S1 to S14