Abstract
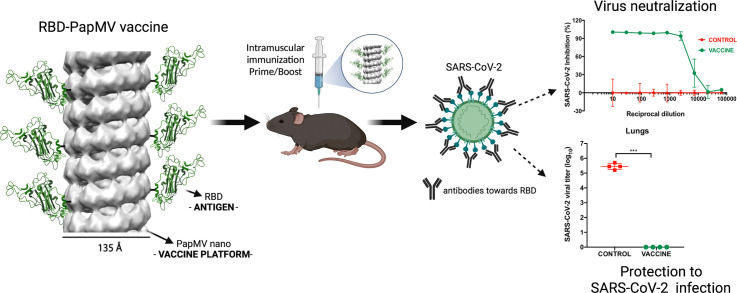
A vaccine candidate to SARS-CoV-2 was constructed by coupling the viral receptor binding domain (RBD) to the surface of the papaya mosaic virus (PapMV) nanoparticle (nano) to generate the RBD-PapMV vaccine. Immunization of mice with the coupled RBD-PapMV vaccine enhanced the antibody titers and the T-cell mediated immune response directed to the RBD antigen as compared to immunization with the non-coupled vaccine formulation (RBD + PapMV nano). Anti-RBD antibodies, generated in vaccinated animals, neutralized SARS-CoV-2 infection in vitro against the ancestral, Delta and the Omicron variants. At last, immunization of mice susceptible to the infection by SARS-CoV-2 (K18-hACE2 transgenic mice) with the RBD-PapMV vaccine induced protection to the ancestral SARS-CoV-2 infectious challenge. The induction of the broad neutralization against SARS-CoV-2 variants induced by the RBD-PapMV vaccine demonstrate the potential of the PapMV vaccine platform in the development of efficient vaccines against viral respiratory infections.
Keywords: Vaccine platform; Papaya mosaic virus (PapMV); Rod-shaped nanoparticle; SARS-CoV-2; Receptor binding domain (RBD), Sortase (SrtA), broad protection
Graphical abstract
The papaya mosaic virus nanoparticle (PapMV nano) vaccine platform is an inducer of the innate immune system and it is therefore an excellent tool for the development of vaccines. To produce the COVID-19 vaccine candidate, the surface of PapMV nano was couple to the SARS-CoV-2 receptor-binding domain (RBD) to generate the RBD-PapMV candidate vaccine. Immunization with the RBD-PapMV vaccine resulted in high levels of antibodies directed to RBD that neutralized the infection of the ancestral SARS-CoV-2, as well as the delta, and omicron variants in cell culture. The immune response elicited by RBD-PapMV also protected susceptible transgenic mice from SARS-CoV-2 infection in vivo.
Background
The ongoing COVID-19 pandemic, caused by the SARS-CoV-2 virus, has highlighted once more the critical role of vaccines in limiting the spread of infectious diseases1. Traditionally, vaccines against viral diseases are made of live-attenuated or inactivated pathogens that necessitate growing the pathogen in confinement facilities, which is time consuming and challenging to upscale2. The traditional approaches led to production of COVID-19 vaccines that were only partially efficient (around 50 % efficacy). On the other hand, the latest recombinant technologies were shown to be faster and more effective2., 3.. Novel vaccine platforms emerged allowing faster design, vaccine production, and shortening the time to enter clinical trials2., 3.. The fastest technologies are based on the utilization of nucleic acids (DNA or mRNA), including viral vectors, encoding the SARS-CoV-2 spike (S) protein as the vaccine antigen4. Following immunization, S is produced by the cells of the patients and then recognized by the immune system, leading to the desired protection through the induction of neutralizing antibodies to the S antigen2., 3.. The adenovirus vector5, used to transport the DNA encoding for the S antigen to the nucleus of the patient's cells was one of the first vaccine available against SARS-CoV-26. However, a slight increase in the rate of intracranial venous thrombosis (ICTV) and thrombocytopenia in adults >70 years were reported in patients receiving the ChAdOx-1-S vaccine based on an adenovirus of simian origin7., 8., 9.. No correlation was detected with mRNA-based vaccines, which are considered as a safer alternative10. The mRNA vaccines emerged as a fast and effective approach with a protection >90%11., 12., 13., 14., 15., 16. and is the favored vaccine in Europe and North America2., 17.. The mRNA-based vaccines are made of a modified mRNA encoding for the S protein of the SARS-CoV-2 ancestral strain that is embedded into a lipidic nanosphere used to transfect cells after injection for inducing production of the S protein11., 12. in vaccinated patients.
Despite being well positioned to respond to pandemic scenarios, the use of mRNA vaccines during the COVID-19 pandemic revealed some caveats, such as adverse reactions induced after immunization exceeding those induced by current seasonal vaccines, the need for deep freezing at −80 °C18., 19. for distribution, and a rapidly declining immunity after immunization20. Considering that SARS-CoV-2 infections will remain a recurring problem for years to come, pursuing the development of novel vaccine platforms is very important to continue improving the quality, stability and safety of the vaccines of the future21.
Vaccines based on the use of adjuvanted recombinant proteins6., 11., 12., 22., 23., 24. have also been developed and made available at a later stage of the pandemic. They will play an important role to better control the COVID-19 pandemic considering the shortage of SARS-CoV-2 vaccines2., 17.. They could also be used in a heterologous prime-boost immunization approach with mRNA-based vaccines that could potentially improve the protection against new emerging SARS-CoV-2 variants25., 26..
The use of protein-based nanoparticle vaccine platforms to enhance and modulate antigen immunogenicity has emerged as a promising approach for controlling pandemic outbreaks27., 28.. Nanoparticles harbor many advantages over other vaccine platforms, including (i) improved antigen diffusion to the lymph node, mostly through passive drainage29., 30.; (ii) enhancement of the immune response due to the triggering of innate immunity by intrinsic pathogen-associated molecular patterns31., 32., 33., 34.; and (iii) improved antigen stability when conjugated to the nanoparticle surface28., 35., 36., 37., 38., 39.. The use of viral nucleocapsid self-assembled into a nanoparticle has been successful in vaccine development37., 38., 39., 40., 41. and may play an important role in the vaccine of tomorrow.
Recently, the use of plant-virus-based nanoparticles as a vaccine platform42., 43., 44., 45., 46. has emerged as an interesting and effective alternative for the design of vaccines against coronavirus. Presentation of the protein antigen at the surface of the nanoparticle stabilized the antigen and improved the antigen-specific immune response38., 39., 40., 43., 44., 46..
In this report, we propose the use of a novel rod-shaped nanoparticle made of the papaya mosaic virus (PapMV) coat protein (CP) and a synthetic ssRNA named PapMV nano. This nanoparticle has great potential for the development of vaccines because it has been shown to (i) be safe in human47, (ii) trigger innate immunity through the stimulation of toll-like receptors (TLR) 7/834, (iii) be easily engineered with a vaccine antigen to enhance its immunogenincity32., 37., 38., 39., 40., 48., 49., 50., and (iiii) present vaccine antigens to immune cells in a repetitive and highly ordered manner that is ideal for activation of the B cells specific to the antigen51. Exploiting the PapMV nano technology, we designed a candidate vaccine to SARS-CoV-2 using the RBD antigen produced and purified from human cells as the vaccine antigen52., 53..
Methods
Production and purification of recombinant proteins
PapMV nano
The design, production and purification of PapMV CP containing four glycines at its N-terminus (Sortase A recognition signal) and a 6His-tag at its C-terminus was previously reported39.
Sortase A (SrtA)
SrtA was expressed in E. coli and purified by ion matrix affinity chromatography (IMAC) as previously reported37.
RBD
The RBD protein (aa 331–591, accession no. NC_045512.2), production and purification of the protein were previously reported52.
Coupling reaction with Sortase A transpeptidase
Coupling of recombinant RBD to the surface of PapMV nano using Sortase A was previously described37., 39.. The optimal ratio of RBD:Sortase:PapMV nano (in μM) for this reaction was 30:50:50. The uncoupled RBD and free SrtA were removed by dialysis. The final product was stored at 4 °C.
Analytical analysis of the RBD-PapMV vaccine
SDS-PAGE and assessment of coupling efficiency
The intensity of the protein bands as observed by SDS-PAGE, were quantified using the software ImageJ (version 1.49) (free software; https://imagej.nih.gov/ij/index.html). The coupled band (RBD-PapMV CP) has a molecular weight equivalent to the sum of the RBD and PapMV CP. The coupling efficiency is defined as a percentage calculated using the area under the curve (AUC) of coupled and non-coupled PapMV CP bands:
RBD mass represents 57 % of the coupled protein's weight (RBD-PapMV CP). Consequently, the amount of coupled RBD can be calculated as:
coupled RBD (μg) = (Total amount of PapMV nano (μg) * coupling efficiency) * 0.57.
Immunoblotting
Protein identity was confirmed by western blotting using an in-house anti-PapMV CP polyclonal antibody or an anti-SARS-CoV-2 Spike Subunit 1 polyclonal antibody (Thermo Fisher, USA). An anti-rabbit antibody coupled with alkaline phosphatase (Jackson Immunoresearch, USA,) was used as a secondary antibody. Bands were coloured using step 1 nitro blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) as a revealing reagent (ThermoFisher Scientific, Waltham, MA, USA).
Nanoparticle structural features
The size distribution of PapMV nanoparticles was assessed by dynamic light scattering (DLS) using a ZetaSizer Nano ZS (Malvern, UK). The nanoparticle shape was observed with using a FEI-TECNAI-Spirit transmission electron microscope (FEI, USA). Samples were stained with 3 % uranyl acetate and fixated on carbon formvar grids, as previously described by Thérien et al., 201737. The physical properties of PapMV nanoparticles were assessed before and after the coupling reaction.
Stability of RBD-PapMV
To monitor the quality and stability of RBD-PapMV vaccine candidate stored at 4 °C, the coupling efficiency, size, distribution, shape, and appearance of vaccine candidates were assessed weekly up to 30 days, using the previously described techniques.
Mice immunization and viral challenge
The immunogenicity of RBD-PapMV vaccine candidate was assessed in female BALB/c mice (Charles River, USA). Mice, 10/group, were immunized twice by intramuscular injection, 21 days apart, with RBD-PapMV (100 μg, including 5.1 μg of coupled RBD), RBD (5.1 μg) + PapMV nano (100 μg) or formulation buffer consisting of 10 mM Tris-HCl. Mice were bled before the first immunization for obtaining naïve sera. Twenty days after the first injection, 5 mice per group were euthanized, and blood was extracted by cardiac puncture for ELISA and neutralization assay. The remaining five mice/group were bled at day 39 for assessing the antibody titer, and were euthanized at day 42, when blood was extracted through cardiac puncture. The spleen was harvested for the ELISPOT assay.
Protection assay
Transgenic K18-hACE2 mice (Jackson Laboratory, USA), expressing the human ACE2 receptor, were used for the challenge. Mice, 8/group, were immunized following the schedule described above. Mice received either one or two doses of the RDB-PapMV vaccine (115 μg of nanoparticles coupled to 4.6 μg of RBD), PapMV nano alone (115 μg) twice, or formulation buffer (same as above). Additionally, five non-treated non-infected mice were included as a control. At day 42, mice were challenged by intranasal instillation with 104 TCID50 (50 % of tissue culture infectious dose) of the SARS-CoV-2 Ancestral strain, excluding the non-infected control. The virus strain was provided by the ‘Institut national de santé publique du Québec’(INSPQ). Body weight, clinical score and survival were recorded for 5 days using a blinded protocol. At days 2 and 5 post infection (p.i.) four mice per group were euthanized to assess the viral titer in the lung and nasal turbinates. Non-treated non-infected mice were euthanized only at day 5 p.i. Additionally, the lungs from 5 days p.i. were used for histopathology analysis and the detection of pro-inflammatory cytokines.
Antibody titration by ELISA
The antibody titers directed towards RBD in the sera of BALB/c mice were assessed by enzyme linked immunosorbent assay (ELISA) as described previously37. Results are expressed as antibody endpoint titers >3-fold OD450nm of the background value consisting of a pool of pre-immune sera.
IFN-γ detection by ELISPOT
The frequency of interferon gamma (IFN-γ)-secreting cells in the spleen of BALB/c mice was measured by enzyme-linked immunospot (ELISPOT) assay. ELISPOT assay was performed using an IFN-γ murine ELISPOT Kit (Abcam, UK) as described by the manufacturer. Splenocytes from immunized and control mice were stimulated with RBD (25 μg/ml), Concanavalin A (5 μg/ml, positive control - Invitrogen, USA), or culture medium in 96-well culture plates (Corning, USA) for 72 h at 37 °C, 5 % CO2. Then, activated cells were transferred to MultiScreen-IP filter plate (MilliporeSigma, USA) coated with IFN-γ capture antibody and incubated for 48 h at 37 °C, 5 % CO2. Finally, plates were revealed following the manufacturer's instructions.
Neutralization of SARS-CoV-2 variants
The capacity of mice sera to neutralize SARS-CoV-2 infection was assessed in vitro using a microneutralization assay (MNA) previously described54. The SARS-CoV-2 ancestral, Delta, and Omicron variants, as well as the permissive cell line Vero.E6 were kindly provided by the INSPQ. In brief, sera from immunized mice were heat inactivated at 56 °C for 1 h. Then, mice sera and a monoclonal antibody (MAb) against the Spike protein (Absolute Antibody, UK, 3 μg/ml), used as a positive control, were diluted 1/10 followed by 1/3 serial dilutions. Each dilution was incubated with 20 TCID50 of SARS-CoV-2 for 1 h at room temperature and used to infect Vero.E6 cell monolayers. Then, the infectious mixture was removed, and cells were incubated for 72 h with media containing sera or MAb at the same dilutions54. Cells not exposed to the virus were the negative controls (“no virus”), while cells treated with virus alone were the positive control of infection (“virus only”). Finally, the cell monolayers were fixed with formaldehyde 4 %. The amount of virus was assessed by immunostaining of the viral nucleoprotein (N) with an anti-N polyclonal antibody (Rockland) from rabbit, followed by an anti-rabbit IgG secondary antibody conjugated to peroxidase (Jackson Immunoresearch, USA). The optical density measured at 450 nm from samples and controls was used to calculate the percentage of infection using the following formula:
100 – ((ODx – Average of “no virus” wells) / (Average of “virus only wells” – Average of “no virus wells”) * 100), where X corresponds to the absorbance for each sample. Non-linear regression curve fit analysis was performed to calculate the ID50 values.
Assessment of viral titer in lungs and nasal turbinates
SARS-CoV-2 titers in the lung and nasal turbinate of infected K18-hACE2 transgenic mice were assessed by end-point titration as previously described55. Lungs and nasal turbinate were harvested at 2 and 5 d.p.i. and homogenized. The cytopathic effect for each sample was recorded and the TCID50 were computed using the Reed–Muench method56. Titers were expressed as log10 of TCID50/g55.
Assessment of pro-inflammatory cytokines in the lung
The levels of pro-inflammatory cytokines interleukin (IL)-6, tumor necrosis factor (TNF)-alpha and the chemokine KC/GRO (CXCL1) in the lungs of SARS-CoV-2 infected K18-hACE2 mice was measured using a V-Plex custom cytokine panel (Meso Scale Discovery - MSD, USA), according to the manufacturer's instructions. The concentration of cytokines in lung homogenate was calculated by interpolation of the calibrator's curves as described by the manufacturer.
Lung histopathology
For assessing the pulmonary tissue inflammatory response histologically after SARS-CoV-2 (ancestral strain) challenge, lungs of K18-hACE2 transgenic mice were extracted at 5 d.p.i. The left lung was fixed with 10 % buffered formalin, embedded in paraffin, sectioned at 4 μm thickness, and stained with hematoxylin-eosin. The histopathological inflammatory scores (IS) were determined by a pathologist, Dr. C. Couture, with experience in pulmonary pathology (see supplementary table 1 for the criteria used to assess the IS). A semi-quantitative scale was used to score bronchial/endobronchial, peribronchial, perivascular, interstitial, pleural and intra-alveolar inflammation. Capillary vascular congestion and pulmonary edema were also evaluated, and the inflammatory cellular infiltrate was characterized to determine if the inflammation was acute (neutrophilic) or chronic (lymphohistiocytic). The results were expressed as total pulmonary inflammatory scores.
Statistical analysis
Data was analyzed with Graph Pad PRISM 7.0b software (GraphPad Software, Inc., USA). One-way ANOVA test followed by Tukey's test was used to compare statistical differences among groups of mice regarding the antibody titers, the IFN-γ secretory response, and the viral titers in the lungs and nasal turbinate. Non-linear regression analysis using an inhibition curve fit model was performed to calculate the ID50 values of the MNA neutralization curves. The differences among groups regarding the percentage of viral inhibition at each dilution of the neutralization curves were assessed using two-way repeated measures ANOVA, with Tukey's as the post-test. Finally, a two-way ANOVA and Tukey's post-test were performed to compare the levels of each cytokine in lung homogenates, as well as the total inflammation scores derived from the histology analysis. Differences were considered significant for p < 0.05.
Results
Generation of RBD-PapMV vaccine
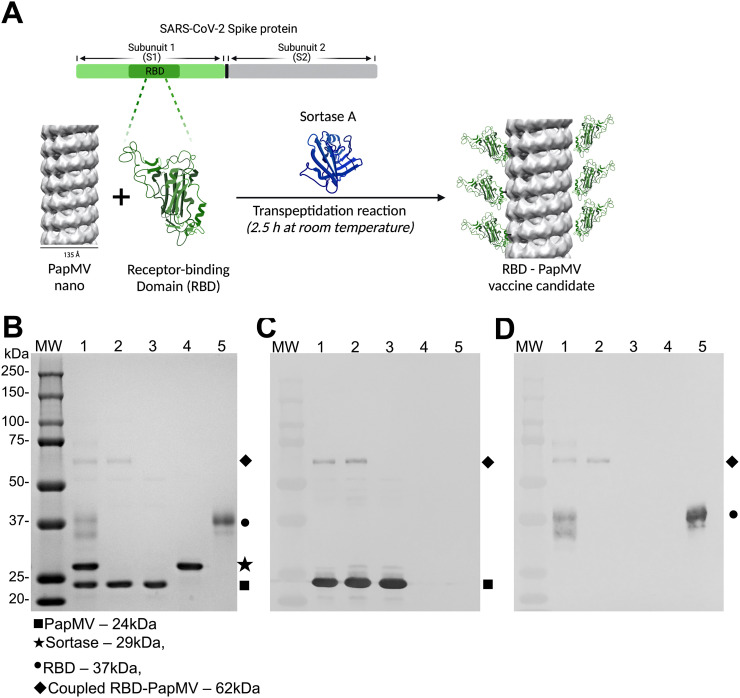
The coupling of the RBD antigen, from the SARS-CoV-2 ancestral strain, to the PapMV nanoparticle was performed using SrtA, a bacterial transpeptidase that induces the formation of a covalent link between two proteins through recognition of the LPETGG donor and G-repeat acceptor motifs57. Coupling of the RBD to the PapMV nano using SrtA results in a nanoparticle surface decorated with RBD protein (schematic in Fig. 1A). The resulting coupled protein comprised PapMV CP linked to the RBD protein, as shown by the appearance of a 62 kDa fusion protein on SDS-PAGE (Fig. 1B). Based on the intensity of the bands on the SDS-PAGE, we assessed that approximately 9 % of the PapMV CP subunits were coupled with the RBD (Fig. 1B lane 2). Immunoblotting confirmed that the RBD-PapMV coupled protein contains the PapMV CP (Fig. 1C) and the RBD (Fig. 1D).
Fig. 1.
Coupling of the RBD to PapMV nano. (A) Schematic representation of the coupling reaction of the RBD at the surface of the PapMV nano (RBD-PapMV), created with BioRender.com. (B) SDS-PAGE of the coupling reaction. Molecular weight markers are shown to the left. Lane 1: coupling reaction containing PapMV nano, Sortase A and the RBD; lane 2: purified RBD-PapMV vaccine; lane 3: PapMV CP; lane 4: Sortase A; and lane 5: RBD. (C, D) Western blotting using a polyclonal serum raised against PapMV CP (C) or RBD (D); a gel identical to panel B was blotted on the nitrocellulose membrane.
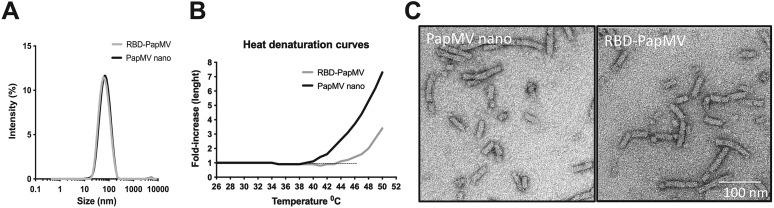
The mean length of the RBD-PapMV was measured by DLS. The average size of RBD-PapMV nanoparticles, 80 nm in length, was indistinguishable from PapMV nano (Fig. 2A). A heat denaturation curve was performed using the DLS to compare the stability of the PapMV nano with that of the RBD-PapMV vaccine candidate (Fig. 2B). Interestingly, the point of inflection leading to denaturation and aggregation of the nanoparticles was increased by 5 °C with the coupling of the RBD (RBD-PapMV; 45 °C) as compared to the free PapMV nano (40 °C). The transmission electron microscopy confirmed the rod-like structure of the RBD-PapMV nanoparticle, an average length of approximately 80 nm, and a width of 14 nm (Fig. 2C). The coupling of the RBD could not be observed by the EM probably because of the small size (32 kDa) of the protein. Additionally, RBD-PapMV was stable for 1 month at 4 ± 3 °C with no significant changes observed for the size of the nanoparticles, the rod-shape, or the coupling efficiency (Supplementary Fig. 2A, B and C).
Fig. 2.
Length and appearance of RBD-PapMV. (A) Dynamic light scattering (DLS) shows that the RBD-PapMV and PapMV nano have a similar average length of 80 nm. (B) Heat denaturation curve of the RBD-PapMV and the PapMV nano using the DLS. C) Transmission electron micrographs of PapMV nano and RBD-PapMV nanoparticles.
Assessment of the humoral response induced by the RBD-PapMV vaccine
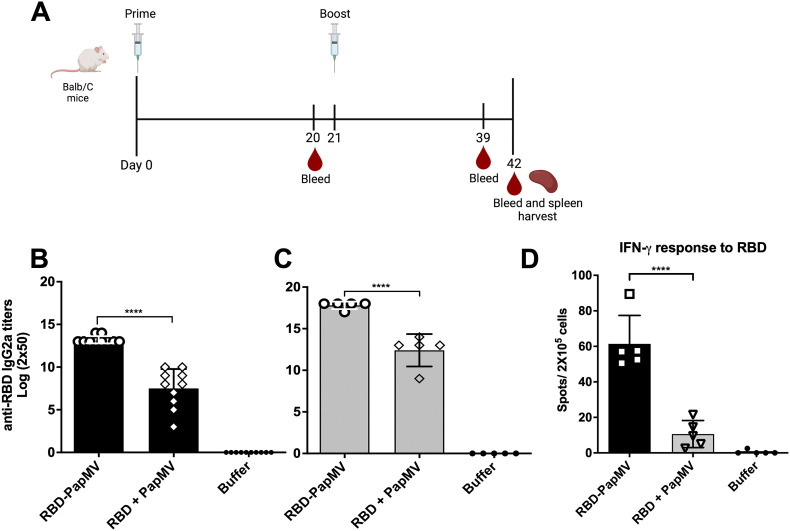
BALB/c mice were immunized twice intramuscularly with either RBD-PapMV, RBD + PapMV or formulation buffer. Mice sera was harvested at days 20 and 39 (schematic of the schedule in Fig. 3A). The levels of IgG1, related with T helper (Th) 2 response, and IgG2a, related with Th1 response58., 59., were assessed by ELISA against the RBD antigen. After one immunization, the IgG2a titers to RBD were 32-folds higher with the RBD-PapMV vaccine as compared to the RBD + PapMV uncoupled formulation (Fig. 3B). Likewise, the boost with the vaccine candidate was superior to the uncoupled control by 64-folds (Fig. 3C). For both conditions, the boost immunization led to higher IgG2a titers (Fig. 3B, C). The total IgG and the IgG1 titers against RBD also reflected significantly higher titers for RBD-PapMV after prime and boost when compared to uncoupled formulation (Supplementary Fig. 2). The mean ratio IgG2a/IgG1 after one or two doses of the RBD-PapMV vaccines were 1.39 and 1.33, respectively, indicating a preference towards the Th1 response.
Fig. 3.
Assessment of the humoral response induced by the RBD-PapMV vaccine. A) BALB/c mice were immunized twice, intramuscularly, at day 0 and 21, with RBD-PapMV, uncoupled RBD + PapMV or Buffer. Bleeding occurred at day 0, 20, 39 and 42. At day 42, animals were bled, sacrificed, and the spleens were harvested. IgG2a titers towards RBD assessed by ELISA using day 20 (B) or day 39 sera (C) are presented. D) T-cell mediated immune response elicited by the vaccine. Spleens of 5 mice per group were harvested at day 42. The T-cell mediated immune response was assessed by ELISPOT assay using the RBD to stimulate the splenocytes. ***p < 0.001, **** p < 0.0001. Schedule (A) created with BioRender.com
Assessment of the T cell mediated immune response induced by the RBD-PapMV vaccine
To assess the capacity of RBD-PapMV vaccine to elicit a T cell-mediated immune response, an ELISPOT assay was performed to measure the secretion of IFN-γ by the CD4+ and CD8+ T cells after stimulation with the RBD antigen (Fig. 3D). The results revealed that the coupled vaccine (RBD-PapMV) is 5.8 folds more efficient than the uncoupled formulation in inducing the proliferation of RBD-specific CD8+ and CD4+ T cells. It also confirmed that the RBD-PapMV vaccine triggers a Th1-type immune response.
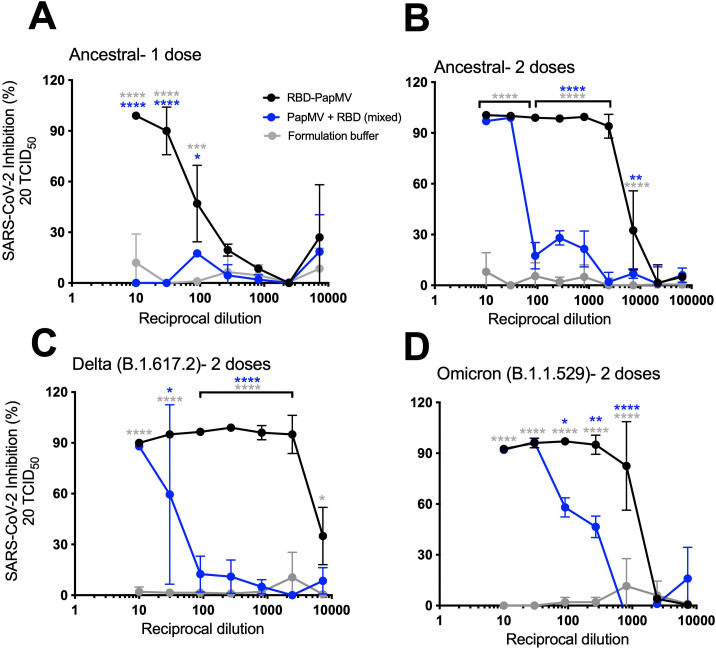
Microneutralization of SARS-CoV-2 variants
A microneutralization assay (MNA) was conducted to determine the capacity of sera from vaccinated mice to inhibit the infection of Vero.E6 cells by SARS-CoV-2. The MNAs were validated using a commercial monoclonal antibody directed to the RBD protein (Supplementary Fig. 3).
After only one immunization, a 1:10 sera dilution from mice immunized with the RBD-PapMV vaccine was able to completely neutralize SARS-CoV-2 infection (ancestral strain) (Fig. 4A). Then, the neutralization efficacy faded rapidly with the serum dilution. Sera from animals immunized once with the uncoupled formulation failed to neutralize infection. The sera of mice immunized twice with RBD-PapMV revealed a robust neutralization against the ancestral strain up to 1:2430-folds (Fig. 4B). The uncoupled formulation neutralized virus infection but it faded rapidly after only a 30-fold dilution.
Fig. 4.
Microneutralization assays against the variants. Graphs represent the percentage of SARS-CoV-2 inhibition of infection vs. the reciprocal dilution of the sera. The first dilution is 1/10, follow by a series of 1/3 dilution. Neutralization of SARS-CoV-2 Alpha (B1.1.7) infection using sera from mice immunized once (A) or twice (B) with RBD-PapMV, RBD + PapMV or Buffer. (C) MNA against the Delta variant. (D) MNA against the Omicron variant. Statistical differences between the RBD-PapMV and the buffer groups are showed in grey (*) while differences between RBD-PapMV and the RBD + PapMV are shown in blue (*). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To assess the potential of the vaccine to elicit broad neutralization, MNAs were performed with the Delta (B.1.617.2) or the Omicron (B.1.1.529) variants. Given the superior performance of the boost immunization, only these sera were used for the assay. The sera showed a similar trend for the neutralization of the Delta variant, with complete neutralization up to 2430-fold dilution for RBD-PapMV and 10-fold dilution for the uncoupled formulation (Fig. 4C). With Omicron, dilution of 270- and 810-fold of sera from mice vaccinated with RBD-PapMV generated 95 % and 85 % inhibition, respectively, after which, the efficacy decreased rapidly (Fig. 4D). The sera generated by the uncoupled formulation led to efficient inhibition only with sera diluted 10- and 30-fold.
The MNA assay provides a read-out of the efficacy of virus neutralization that is influenced by two factors: (i) the amount of antibodies in the sera and (ii) their affinity for RBD. Therefore, it is possible to determine a constant of affinity for such antibodies, called the neutralization score per unit of titer (NS), by dividing the neutralization efficacy by the antibody titer. The ID50 value of each curve, representing the inhibitory dilution at which 50 % of the viral neutralization is attained, constitutes a convenient indicator of neutralization efficacy. Using this value, the formula would be: ID50 / antibody titer = NS. High NS corresponds to serum with a higher neutralization efficacy per titer of antibodies (IgG2a titers). This is an indicator of effectiveness of the antibodies found in the sera to neutralize infection (Table 1 ). The NS ratio between one and two immunizations of RBD-PapMV is 321.5/5.7 = 56.4, which reveals that for the same amount of anti-RBD antibodies, the serum of mice immunized twice is 56-folds more efficient in neutralizing SARS-CoV-2 infection than those immunized once. RBD-PapMV injected twice is also (54-folds) more efficient in neutralization of the ancestral strain than the uncoupled formulation. With the Delta variant, the RBD-PapMV is (333/3) 111-folds more efficient in the neutralization than the uncoupled formulation, and (68/13) 5.2-folds with Omicron.
Table 1.
Calculation of the neutralization scores per unit of titer (NS) for each SARS-CoV-2 variant.
| Variant | Vaccine | Nb immunizations | ID50 | NS |
|---|---|---|---|---|
| Alpha | RBD-PapMV | 1 | 74.6 | 74.6/13.2 = 5.7 |
| RBD-PapMV | 2 | 5722 | 5722/17.8 = 321.5 | |
| RBD + PapMV | 2 | 72.9 | 72.9/12.4 = 5.9 | |
| Delta | RBD-PapMV | 2 | 5831 | 5931/17.8 = 333 |
| RBD + PapMV | 2 | 36.8 | 36.8/12.4 = 3 | |
| Omicron | RBD-PapMV | 2 | 1210 | 1210/17.8 = 68 |
| RBD + PapMV | 2 | 161.5 | 161.5/12.4 = 13 |
Nb: number, ID50: dilution for 50 % the viral neutralization.
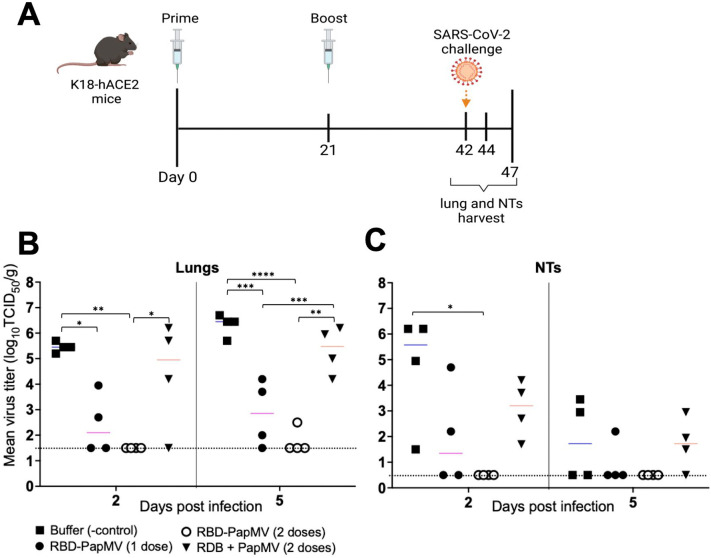
Protection from SARS-CoV-2 viral challenge
The protection efficacy of the RBD-PapMV vaccine was assessed using the hACE2 (K18) mice expressing the human ACE2 receptor that are susceptible to SARS-CoV-2 infection60. Vaccines and controls were administered intramuscularly once or twice with a 21-day interval. As control groups, mice were immunized twice with PapMV nano or the formulation buffer. Mice were challenged with the SARS-CoV-2 ancestral strain by intranasal instillation at day 42. On days 44 and 47, four mice per group were sacrificed to harvest the lungs and nasal turbinates (NT) and assess the viral titers (Fig. 5A). Viral titers were determined by end-point dilution of lung and NT homogenates in Vero.E6 cell monolayers (Fig. 5B). At 2 and 5 d.p.i., one dose of RBD-PapMV reduced the viral titer in the lungs as compared with the buffer control (Fig. 5B). Notably, two doses of the RBD-PapMV vaccine reduced the viral titers below the limit of detection for 4 out of 4 animals at day 2 p.i., and 3 out of 4 animals at day 5 p.i. (Fig. 5B). In the NTs, only the animals immunized twice with RBD-PapMV showed a decrease in the virus titers at day 2 p.i.
Fig. 5.
Challenge of vaccinated K18-hACE2 mice. (A) Schedule of the operations created with BioRender.com. K18-hACE2 mice were immunized intramuscularly once or twice with RBD-PapMV and twice with PapMV nano or formulation buffer. Mice were challenged with SARS-CoV-2 at day 42. The lungs and nasal turbinates (NTs) were harvested at 2 (day 44) and 5 (day 47) days post-challenge. (B) Viral titers in the lungs of animals harvested either at day 2 or 5 post challenge. (C) Virus titers in the nasal turbinates. *p < 0.05, ** < 0.01, ***p < 0.001, ****p < 0.0001.
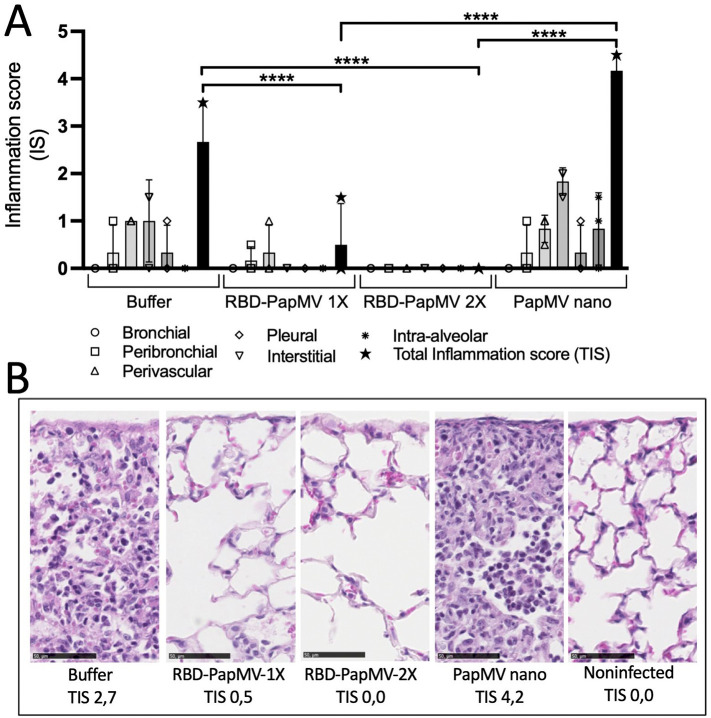
Lung histopathology and cytokine profile in lungs of infected animals
Lungs of infected mice with SARS-CoV-2 were harvested at day 5 post-infection. Inflammation, when present, was lymphohistiocytic (chronic) in nature and predominantly interstitial and perivascular in distribution. Mice immunized with the buffer or the naked PapMV had an average total inflammation score (TIS) of 2.7 and 4.2, respectively, which was significantly higher than mice immunized with one or two doses of the RBD-PapMV vaccine that showed a TIS 0.5 and 0, respectively (Fig. 6A, B). As expected, uninfected mice (negative controls) showed no sign of inflammation (TIS 0).
Fig. 6.
Histology of the lungs of the infected animals at day 5 post-inoculation with SARS-CoV-2. A) The average inflammation scores of 3 mice/group from the bronchial, peribronchial, perivascular, pleural, interstitial and intra-alveolar spaces were plotted. The total inflammation scores (TIS) were also shown for each group. B) Representative micrographs of the lungs for each group. From left to right: mice injected with buffer, mice vaccinated with either 1 or 2 doses of the RBD-PapMV, mice injected with 2 doses of PapMV nano alone and non-infected negative control mice. Total inflammation scores (TIS) represent averages of triplicates. Hematoxylin-Eosin (HE) stain. Scale bar is 50 μM. ****p < 0.0001.
Additionally, inflammation was assessed by measuring the levels of pro-inflammatory cytokines (IL-6, TNF-α and KC/Gro) in lung homogenates at 5 d.p.i. The groups immunized with one or two doses of the RBD-PapMV vaccine presented levels of IL-6 (<30 pg/mL) comparable to the non-infected animals, while the other infected groups (Buffer and PapMV) had high levels (80 and 165 pg/mL, respectively) of IL-6 (Supplementary Fig. 4).
Discussion
Pandemics constitute the most severe manifestation of a viral outbreak. With the increasing frequency of these events, there is a need to develop versatile vaccine platforms able to strongly stimulate the immune system in a safe manner. These platforms should be readily available, stable for easy transportation, and made with globally accessible technology. The development of PapMV nano technology is an attempt to respond to these needs.
In this study, we demonstrate that the coupling of the SARS-CoV-2 RBD to the surface of PapMV nano leads to a more effective protection as compared with the uncoupled formulation. Coupling to PapMV nano significantly increased the amount and quality of antibodies directed to the RBD antigen, as revealed by micro-neutralization assays. The coupling of RBD to PapMV nano increased the efficacy of neutralization of the ancestral and the Delta variant by >100-folds as compared with the non-coupled formulation. The engagement of the TLR7 by the RBD-PapMV vaccine may act as a co-stimulatory signal that enhances and accelerates the antibody class switch towards the production of the IgG2a subclass and the production of higher affinity antibodies to the RBD antigen34., 61.. PapMV nano was reported to accelerate germinal center formation, and to promote affinity/avidity maturation of specific IgG and isotype switching to IgG2a subclass62. Recently, these events were linked with the stimulation of TLR7, which is consistent with our data61.
To our knowledge, the PapMV nano is one of the few vaccine technologies available that are capable to stimulate the TLR7/8. Stimulation of TLR8 is a main advantage since these receptors are abundant in monocytes, macrophages and dendritic cells that are important players in the development of a strong and prolonged immune response63. Other TLR7/8 agonists are small dinucleotide analogs that can be only used topically due to their toxicity and are not suitable for the development of safe vaccines64. This feature also distinguishes the PapMV nano technology from the mRNA-based vaccines, since the later are poor activators of TLR7/8 because of the incorporation of modified nucleotides on the mRNA to optimize the translation efficacy65., 66..
The sera from animals immunized with the RBD-PapMV vaccine effectively neutralized the ancestral and Delta variants. However, its efficacy dropped by 4.7-fold with Omicron. The 16 mutations found in the RBD sequence, as compared with the ancestral SARS-CoV-267 could explain this drop in neutralization efficacy. The PapMV nano probably increased the broadness of the antibody response to the RBD, explaining the generation of antibodies capable of cross-neutralization as previously reported in another study by our group68.
Vaccines inducing production of antibodies with broad protection are needed considering that current SARS-CoV-2 mRNA vaccines offer only partial protection to the Omicron variant69. A recent study has demonstrated that multimerization of the RBD 24 times on a ferritin backbone generated a vaccine that showed a broad protection to several SARS-CoV-2variants70. In another study, the multimerization (20 times) of the RBD antigen on the surface of I50–53 nanoparticles lead to the development of an effective vaccine capable to trigger neutralizing antibodies to the ancestral strains of SARS-CoV-271. Those results, like for ours, suggest that the quality of the response to the RBD is improved by multimerization of the RBD antigen that facilitates the activation of the relevant B-cell lineage that are specific to the RBD antigen71. In contrast with the ferritin and the I53–50 technologies that are spheres of approximately 30 nm, the RBD-PapMV vaccine harbors a rod-shape nanoparticle of 80 nm that displays a longer surface to the B-cells to establish the contact with the B-cell receptor. In addition, PapMV nano carries a TLR7/8 stimulatory activity that might provide an advantage compared to these other systems.
The trigger of a strong T-cell response to RBD by the RBD-PapMV vaccine can also contribute to the development of broader protection against other SARS-CoV-2 variants since many of the MHC-class I and II epitopes found in the RBD domain are not affected by the mutations in the Omicron RBD. The coupling of RBD to PapMV nano increased the internalization of the RBD antigen by antigen-presenting cells as previously shown40, that will direct the RBD to cellular compartments involved in the presentation of RBD-derived CTL epitopes on MHC-class I and II. This is consistent with previous studies by our group that demonstrated presentation of an inserted CTL epitope in the PapMV nano on MHC-I through a proteasome-independent mechanism72., 73..
RBD-PapMV nanoparticles were shown to be stable at 4 ± 3 °C for 1 month without loss of integrity (supplementary Fig. 3). This stability of the vaccine preparation is critical to ensure its distribution, as it is easier to distribute a vaccine that does not need to be frozen, an advantage over mRNA-based vaccines.
Overall, the PapMV nano technology is very promising for the development of effective vaccines and it is distinct among the other protein-based nanoparticles based on its capacity to stimulate the TLR 7/833., 34., which is responsible for the trigger of a broad neutralizing immunity that is needed for protections against novel variants. We foresee this technology to be very useful in the future to enhance and broaden the immune response to the S protein by performing a prime-boost regimen with mRNA vaccines. Additionally, the production of this vaccine is entirely performed in recombinant systems, bacteria for the PapMV-nano and in mammal cell culture for the RBD, which facilitates the scale up of production and insures an affordable production cost.
Ethics statement
All animal work was previously approved by the “Comité de Protection des Animaux— CHUQ (CPA-CHUQ)” of the institution. The authorization numbers were CHU-20-684 and CHU-21-811.
Author contributions
The concept of the experiments was discussed and elaborated with DL, SMOU, PS, MB and MELG. The development of the methodology to design the experiments involved the contributions of DL, SMOU, MB, MELG, LJB, CG, MF, ID, LF, OF, XX, AK, MG, HR, MS, CC and MBaz. The formal analysis of the results was performed by SMOU, MB and DL. The main investigator providing the resources (funding) for the experiments and directing the personnel involved in the study is DL. The original draft was prepared by SMOU and DL. MB, PS, CC, LF and MBaz contributed to the review and editing of the manuscript.
Financial support
Thanks to the Canadian Institute of Health Research (CIHR-grant #440390), the Natural Sciences and Engineering Research Council (NSERC-grant# 2016-05852) of Canada and the CoVaRR-net network for funding this research program.
CRediT authorship contribution statement
Santa-Mariela Olivera-Ugarte: Methodology, Validation, Investigation, Writing – original draft. Marilène Bolduc: Methodology, Validation, Investigation, Supervision. Marie-Ève Laliberté-Gagné: Methodology, Investigation. Léa-Jeanne Blanchette: Methodology, Investigation. Caroline Garneau: Methodology, Investigation. Maude Fillion: Methodology, Investigation. Pierre Savard: Supervision. Isabelle Dubuc: Methodology, Investigation. Louis Flamand: Supervision. Omar Farnòs: Methodology, Investigation. Xingge Xu: Methodology, Investigation. Amine Kamen: Supervision, Funding acquisition. Mégan Gilbert: Methodology, Investigation. Henintsoa Rabezanahary: Methodology, Investigation. Martina Scarrone: Methodology, Investigation. Christian Couture: Supervision, Methodology, Investigation. Mariana Baz: Supervision. Denis Leclerc: Conceptualization, Supervision, Writing – original draft, Funding acquisition.
Declaration of competing interest
The authors declare no financial conflicts of interest.
Acknowledgments
We would like to thank Julie-Christine Lévesque of the “Plateforme de bio-imagerie du Centre de Recherche en Infectiologie” for transmission electron microscope service and the analysis of ELISPOT plates. Likewise, we thank Yann Breton for his assistance during the ELISPOT assay. We thank Helen Rothnie for the critical reading of the manuscript. Finally, we would like to thank the Canadian Institute of Health Research Canada for funding this research program (grant #440390).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nano.2022.102584.
Appendix A. Supplementary data
Supplementary material
References
- 1.Gebru A.A., et al. Global burden of COVID-19: situational analyis and review. Hum Antibodies. 2021;29:139–148. doi: 10.3233/hab-200420. [DOI] [PubMed] [Google Scholar]
- 2.Flanagan K.L., et al. Progress and pitfalls in the quest for effective SARS-CoV-2 (COVID-19) vaccines. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.579250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Constantin C., Pisani A., Bardi G., Neagu M. Nano-carriers of COVID-19 vaccines: the main pillars of efficacy. Nanomedicine (Lond) 2021;16:2377–2387. doi: 10.2217/nnm-2021-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho W., et al. Next-generation vaccines: nanoparticle-mediated DNA and mRNA delivery. Adv Healthc Mater. 2021;10 doi: 10.1002/adhm.202001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkashif A., Alhashimi M., Sayedahmed E.E., Sambhara S., Mittal S.K. Adenoviral vector-based platforms for developing effective vaccines to combat respiratory viral infections. Clin Transl Immunol. 2021;10 doi: 10.1002/cti2.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillus D., et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: a prospective cohort study. Lancet Respir Med. 2021;9:1255–1265. doi: 10.1016/S2213-2600(21)00357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteley W.N., et al. Association of COVID-19 vaccines ChAdOx1 and BNT162b2 with major venous, arterial, or thrombocytopenic events: a population-based cohort study of 46 million adults in England. PLoS Med. 2022;19 doi: 10.1371/journal.pmed.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayas A., et al. Bilateral superior ophthalmic vein thrombosis, ischaemic stroke, and immune thrombocytopenia after ChAdOx1 nCoV-19 vaccination. Lancet. 2021;397 doi: 10.1016/S0140-6736(21)00872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledford H. How could a COVID vaccine cause blood clots? Scientists race to investigate. Nature. 2021;592:334–335. doi: 10.1038/d41586-021-00940-0. [DOI] [PubMed] [Google Scholar]
- 10.Brazete C., Aguiar A., Furtado I., Duarte R. Thrombotic events and COVID-19 vaccines. Int J Tuberc Lung Dis. 2021;25:701–707. doi: 10.5588/ijtld.21.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brisse M., Vrba S.M., Kirk N., Liang Y., Ly H. Emerging concepts and technologies in vaccine development. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.583077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Q., et al. COVID-19 vaccines: current understanding on immunogenicity, safety, and further considerations. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.669339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munro A.P.S., et al. Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet. 2021;398:2258–2276. doi: 10.1016/S0140-6736(21)02717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh E.E., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prubeta B.M. Current state of the first COVID-19 vaccines. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soleimanpour S., Yaghoubi A. COVID-19 vaccine: where are we now and where should we go? Expert Rev Vaccines. 2021;20:23–44. doi: 10.1080/14760584.2021.1875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M., et al. COVID-19 vaccine development: milestones, lessons and prospects. Signal Transduct Target Ther. 2022;7:146. doi: 10.1038/s41392-022-00996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou M.T., et al. Safety and reactogenicity of 2 doses of SARS-CoV-2 vaccination in solid organ transplant recipients. Transplantation. 2021;105:2170–2174. doi: 10.1097/TP.0000000000003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy A., Alhatlani B. An overview of current COVID-19 vaccine platforms. Comput Struct Biotechnol J. 2021;19:2508–2517. doi: 10.1016/j.csbj.2021.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolgin E. COVID vaccine immunity is waning - how much does that matter? Nature. 2021;597:606–607. doi: 10.1038/d41586-021-02532-4. [DOI] [PubMed] [Google Scholar]
- 21.Dolgin E. How protein-based COVID vaccines could change the pandemic. Nature. 2021;599:359–360. doi: 10.1038/d41586-021-03025-0. [DOI] [PubMed] [Google Scholar]
- 22.Shinde V., et al. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capell T., et al. Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. 2020;25:635–643. doi: 10.1016/j.tplants.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung J.Y., Thone M.N., Kwon Y.J. COVID-19 vaccines: the status and perspectives in delivery points of view. Adv Drug Deliv Rev. 2021;170:1–25. doi: 10.1016/j.addr.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapkota B., et al. Heterologous prime-boost strategies for COVID-19 vaccines. J Travel Med. 2022;29 doi: 10.1093/jtm/taab191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R., et al. Immunogenicity of a heterologous prime-boost COVID-19 vaccination with mRNA and inactivated virus vaccines compared with homologous vaccination strategy against SARS-CoV-2 variants. Vaccines (Basel) 2022;10 doi: 10.3390/vaccines10010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X. Nanoparticle vaccines based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330. doi: 10.1016/j.immuni.2020.11.015. e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wibowo D., et al. Polymeric nanoparticle vaccines to combat emerging and pandemic threats. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li P., et al. Engineering biodegradable guanidyl-decorated PEG-PCL nanoparticles as robust exogenous activators of DCs and antigen cross-presentation. Nanoscale. 2017;9:13413–13418. doi: 10.1039/c7nr04470d. [DOI] [PubMed] [Google Scholar]
- 30.Manolova V., et al. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 31.Gause K.T., et al. Immunological principles guiding the rational Design of Particles for vaccine delivery. ACS Nano. 2017;11:54–68. doi: 10.1021/acsnano.6b07343. [DOI] [PubMed] [Google Scholar]
- 32.Bolduc M., et al. The quest for a nanoparticle-based vaccine inducing broad protection to influenza viruses. Nanomedicine. 2018;14:2563–2574. doi: 10.1016/j.nano.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Mathieu C., Rioux G., Dumas M.C., Leclerc D. Induction of innate immunity in lungs with virus-like nanoparticles leads to protection against influenza and Streptococcus pneumoniae challenge. Nanomedicine. 2013;9:839–848. doi: 10.1016/j.nano.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Lebel M., et al. Nanoparticle adjuvant sensing by TLR7 enhances CD8+ T cell-mediated protection from listeria monocytogenes infection. J Immunol. 2014;192:1071–1078. doi: 10.4049/jimmunol.1302030. [DOI] [PubMed] [Google Scholar]
- 35.Marcandalli J. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell. 2019;176:1420–1431. doi: 10.1016/j.cell.2019.01.046. e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grego E.A., et al. Polymeric nanoparticle-based vaccine adjuvants and delivery vehicles. Curr Top Microbiol Immunol. 2021;433:29–76. doi: 10.1007/82_2020_226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thérien A., et al. A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling. J Nanobiotechnology. 2017;15:54. doi: 10.1186/s12951-017-0289-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laliberté-Gagné M., et al. Increased immunogenicity of full-length protein antigens through sortase-mediated coupling on the PapMV vaccine platform. Vaccines (Basel) 2019;7 doi: 10.3390/vaccines7020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laliberté-Gagné M.E., et al. Modulation of antigen display on PapMV nanoparticles influences its immunogenicity. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carignan D., et al. Engineering of the PapMV vaccine platform with a shortened M2e peptide leads to an effective one dose influenza vaccine. Vaccine. 2015;33:7245–7253. doi: 10.1016/j.vaccine.2015.10.123. [DOI] [PubMed] [Google Scholar]
- 41.Kreimer A.R., et al. Evidence for single-dose protection by the bivalent HPV vaccine-review of the Costa Rica HPV vaccine trial and future research studies. Vaccine. 2018;36:4774–4782. doi: 10.1016/j.vaccine.2017.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tripathy S., Dassarma B., Bhattacharya M., Matsabisa M.G. Plant-based vaccine research development against viral diseases with emphasis on ebola virus disease: a review study. Curr Opin Pharmacol. 2021;60:261–267. doi: 10.1016/j.coph.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Santoni M., Zampieri R., Avesani L. Plant virus nanoparticles for vaccine applications. Curr Protein Pept Sci. 2020;21:344–356. doi: 10.2174/1389203721666200212100255. [DOI] [PubMed] [Google Scholar]
- 44.Hefferon K.L. Repurposing plant virus nanoparticles. Vaccines (Basel) 2018:6. doi: 10.3390/vaccines6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkataraman S., Hefferon K., Makhzoum A., Abouhaidar M. Combating human viral diseases: will plant-based vaccines be the answer? Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohsen M.O., et al. Neutralization of MERS coronavirus through a scalable nanoparticle vaccine. NPJ Vaccines. 2021;6:107. doi: 10.1038/s41541-021-00365-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langley J.M. 2744. A phase I randomized, observer-blind, controlled, dose escalation trial of the safety and tolerability of a single intramuscular dose of a PAL Adjuvant (Laboratory Code, FB-631) Co-administered with Seasonal TIV (2013–2014) to healthy adults ≥18–50 years of age. Open Forum Infect Dis. 2019;6 doi: 10.1093/ofid/ofz360.2421. [DOI] [Google Scholar]
- 48.Schneewind O., Missiakas D. Sortases, surface proteins, and their roles in Staphylococcus aureus disease and vaccine development. Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.PSIB-0004-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denis J., et al. Development of a universal influenza a vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26:3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 50.Leclerc D., Rivest M., Babin C., Lopez-Macias C., Savard P. A novel M2e based flu vaccine formulation for dogs. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachmann M.F., Jennings G.T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 52.Farnos O., et al. Rapid high-yield production of functional SARS-CoV-2 receptor binding domain by viral and non-viral transient expression for pre-clinical evaluation. Vaccines (Basel) 2020;8 doi: 10.3390/vaccines8040654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lainscek D., et al. A nanoscaffolded spike-RBD vaccine provides protection against SARS-CoV-2 with minimal anti-scaffold response. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9050431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amanat F., et al. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol. 2020;58 doi: 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baz M. Zika virus isolation, purification, and titration. Methods Mol Biol. 2020;2142:9–22. doi: 10.1007/978-1-0716-0581-3_2. [DOI] [PubMed] [Google Scholar]
- 56.Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 57.Jacobitz A.W., Kattke M.D., Wereszczynski J., Clubb R.T. Sortase transpeptidases: structural biology and catalytic mechanism. Adv Protein Chem Struct Biol. 2017;109:223–264. doi: 10.1016/bs.apcsb.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fornefett J., et al. Comparative analysis of humoral immune responses and pathologies of BALB/c and C57BL/6 wildtype mice experimentally infected with a highly virulent rodentibacter pneumotropicus (Pasteurella pneumotropica) strain. BMC Microbiol. 2018;18:45. doi: 10.1186/s12866-018-1186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuniga E.I., Macal M., Lewis G.M., Harker J.A. Innate and adaptive immune regulation during chronic viral infections. Annu Rev Virol. 2015;2:573–597. doi: 10.1146/annurev-virology-100114-055226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oladunni F.S., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang X., et al. TLR7 signaling shapes and maintains antibody diversity upon virus-like particle immunization. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.827256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rioux G., et al. Influence of PapMV nanoparticles on the kinetics of the antibody response to flu vaccine. J Nanobiotechnology. 2016;14:43. doi: 10.1186/s12951-016-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stogerer T., Stager S. Innate immune sensing by cells of the adaptive immune system. Front Immunol. 2020;11:1081. doi: 10.3389/fimmu.2020.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engel A.L., Holt G.E., Lu H. The pharmacokinetics of toll-like receptor agonists and the impact on the immune system. Expert Rev Clin Pharmacol. 2011;4:275–289. doi: 10.1586/ecp.11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carlile T.M., et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui Z. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871. doi: 10.1016/j.cell.2022.01.019. e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savard C., et al. Improvement of the trivalent inactivated flu vaccine using PapMV nanoparticles. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dejnirattisai W. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484. doi: 10.1016/j.cell.2021.12.046. e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saunders K.O., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brouwer P.J.M. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell. 2021;184:1188–1200. doi: 10.1016/j.cell.2021.01.035. e1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanafi L.A., et al. Two distinct chimeric potexviruses share antigenic cross-presentation properties of MHC class I epitopes. Vaccine. 2010;28:5617–5626. doi: 10.1016/j.vaccine.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 73.Leclerc D., et al. Proteasome-independent major histocompatibility complex class I cross-presentation mediated by papaya mosaic virus-like particles leads to expansion of specific human T cells. J Virol. 2007;81:1319–1326. doi: 10.1128/JVI.01720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material