Abstract
Metabolic plasticity is the ability of a biological system to adapt its metabolic phenotype to different environmental stressors. We used a whole-body and tissue-specific phenotypic, functional, proteomic, metabolomic and transcriptomic approach to systematically assess metabolic plasticity in diet-induced obese mice after a combined nutritional and exercise intervention. Although most obesity and overnutrition-related pathological features were successfully reverted, we observed a high degree of metabolic dysfunction in visceral white adipose tissue, characterized by abnormal mitochondrial morphology and functionality. Despite two sequential therapeutic interventions and an apparent global healthy phenotype, obesity triggered a cascade of events in visceral adipose tissue progressing from mitochondrial metabolic and proteostatic alterations to widespread cellular stress, which compromises its biosynthetic and recycling capacity. In humans, weight loss after bariatric surgery showed a transcriptional signature in visceral adipose tissue similar to our mouse model of obesity reversion. Overall, our data indicate that obesity prompts a lasting metabolic fingerprint that leads to a progressive breakdown of metabolic plasticity in visceral adipose tissue.
Keywords: Obesity, Metabolic plasticity, Visceral adipose tissue, Mitochondrial dysfunction, Exercise, Caloric restriction, Multi-organ approach, Human obesity, Two-steps bariatric surgery, Metabolic fingerprint
1. Introduction
Food availability, consumption surplus and sedentarism, predominant in most parts of the world, are the most significant environmental challenges that western societies are nowadays facing. According to the World Health Organization (WHO), 650 million adults were suffering from obesity in 2016, with a growing incidence that had become a significant concern in healthcare [62]. Moreover, obesity increases the risk for most common non-communicable diseases, including neurodegenerative and cardio-vascular diseases, cancer and type 2 diabetes (T2D). Tackling the biology underlying obesity-related T2D is a challenge, as it is a multi-organ disease, from both pathological and etiological perspectives. Although surgical, pharmacological and lifestyle-based interventions partially revert the obesity phenotype, are thwarted by a frequent failure to sustain body weight loss and health benefits, and frequent relapses [[1], [2], [3]].
The failure to reverse obesity and its comorbid diseases suggests that cellular and metabolic plasticity, may be impaired. Metabolic plasticity is interpreted as the ability of the cells to change and efficiently adapt their phenotypes to specific environmental cues. Such phenomenon takes place in chronic conditions and diseases as well as in aging [[4], [5], [6]]. However, whether obesity disrupts metabolic plasticity, notwithstanding therapeutic interventions, is still a conundrum. Hence, our major interest is to elucidate whether the metabolic changes in obesity and insulin resistance, originated at different levels, are reversible. Thus, deciphering how metabolic plasticity is orchestrated systemically.
In this context, the LiMa (Lifestyle Matters) project uses a comprehensive therapeutic approach focused on the systemic and tissue-specific impact of important environmental stressors (namely chronic overfeeding and physical inactivity). Elucidating how such obesity-induced metabolic perturbations originated in different organs are modulated after the implementation of a dietary and exercise intervention allows us to identify major specific vulnerabilities related to the disparity in disease progression and clinical outcomes. Here, we present an integrative multi-tiered approach to study the impact of such interventions on reverting obesity, in a mouse model of diet-induced obesity, and decipher whether this disease triggers a progressive breakdown of metabolic plasticity in key tissues.
Our study unveils a systemically unnoticed metabolic deterioration fingerprint in white adipose tissue (WAT), which is a decisive and critical player in obesity-related T2D progression. This tissue shows a persistent mitochondrial dysfunction that compromises tissue metabolic plasticity after obesity.
2. Results
2.1. Combined nutritional and exercise intervention impact on HFD-induced phenotype
We investigated whether diet-induced obesity and its related metabolic and tissue signature sequelae are reversible through modifications in the diet and activity patterns. Using mice, three experimental groups were defined: control group (Ctrl); HFD-induced obese and insulin-resistant group (HFD); and a third group (Int) that underwent a nutritional and exercise intervention (Fig. 1A). Concretely, the intervention combined a calorie restriction diet substitution (with poly- and mono-unsaturated rich oils and complex carbohydrates instead of saturated fats and simple sugars) with treadmill training, (see methods section for detailed group description). The HFD group showed increased body weight (BW) (Fig. 1B) and expanded fat volume (Fig. 1C and D). Moreover, HFD mice showed increased triglyceride content in liver and soleus skeletal muscle (Fig. 1E and F), along with disrupted metabolic flexibility, indicated by a narrower range of daily respiratory exchange ratio (RER) values (Fig. 1G and H, Fig. S1C), and parameters of impaired glucose metabolism (Fig. 1I) in comparison to the control group (Ctrl group). Glucose metabolism impairment was evidenced by both systemic insulin resistance (Fig. S1A) and the classical pancreatic overcompensation, i.e. increased pancreas size (Fig. 1K), glucose-stimulated insulin hypersecretion (GSIS) both in vivo and in vitro (Fig. 1J and L) and increased β cell mass (Figures 1M and N, Fig. S1B).
Fig. 1.
HFD- and combined intervention-induced phenotype. A: Experimental design scheme. B: Body weight for control (Ctrl, n = 80), high fat diet-fed (HFD, n = 70) and intervention (Int, n = 46) groups. C: Quantification of fat volume by NMR imaging and 7 T scan representative images per phenotype (Ctrl n = 4; HFD n = 4; Int n = 3). D: NMR representative images for each group for quantification in (C). E: Triglycerides (TG) levels in liver: Ctrl (n = 8), HFD (n = 7) and Int (n = 8). F: Triglycerides (TG) levels in soleus: Ctrl (n = 9), HFD (n = 8) and Int (n = 8). G: Group-averaged respiratory exchange ratio (RER) deviation from the 24h median for each animal for Ctrl (n = 11), HFD (n = 10) and Int (n = 6) for 48h in metabolic cages. H: Minimum and maximal values of deviation from median (RER variation, Figure E). I: Intraperitoneal glucose tolerance test (IGTT) for Ctrl (n = 54), HFD (n = 48) and Int (n = 12). The graph displayed combines data from different representative cohorts, so variation in n size is due to it and space limitations imposed by the treadmill for the intervention program (6 lanes, n = 12 maximum per cohort in order to avoid time differences). J: Basal (t = 0min) and 15min insulin levels during IGTT (panel H) for Ctrl (n = 15), HFD (n = 19) and Int (n = 13). K: Pancreas mean tissue weight for Ctrl (n = 6), HFD (n = 6) and Int (n = 4). L: Glucose-stimulated (2.8 and 16.7 mM) insulin secretion in vitro for pancreatic islets from animals in groups Ctrl (n = 10), HFD (n = 7) and Int (n = 7). M: Immunohistochemistry representative images of pancreatic islets (nuclei-blue, insulin-red, glucagon-green) from animals in Ctrl, HFD and Int. N: Normalized pancreatic islet insulin area by pancreas section area for Ctrl (n = 6), HFD (n = 6) and Int (n = 4). Ctrl: control; HFD: high fat diet-fed; Int: intervention. Data represented as box-and-whiskers plots (median, 25th to 75th percentiles and max/min values). ANOVA One-way and Post-hoc Tukey, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Combined nutritional and exercise intervention (Int group) reversed the observed altered parameters (Fig. 1B–I and 1K and L) except for persistent hyperinsulinemia (both fasting and after glucose stimulation) (Fig. 1J). This result was consistent with the maintained increase in β cell mass after the intervention (Figures 1M and N, Fig. S1B), even though in vitro GSIS was fully recovered (Figure 1L). Altogether, these results validated our diet-induced obesity animal model and the efficiency of the combined nutritional and exercise intervention to revert obesity and, partially, T2D phenotype.
2.2. Evaluation of tissue-specific responses to the combined intervention
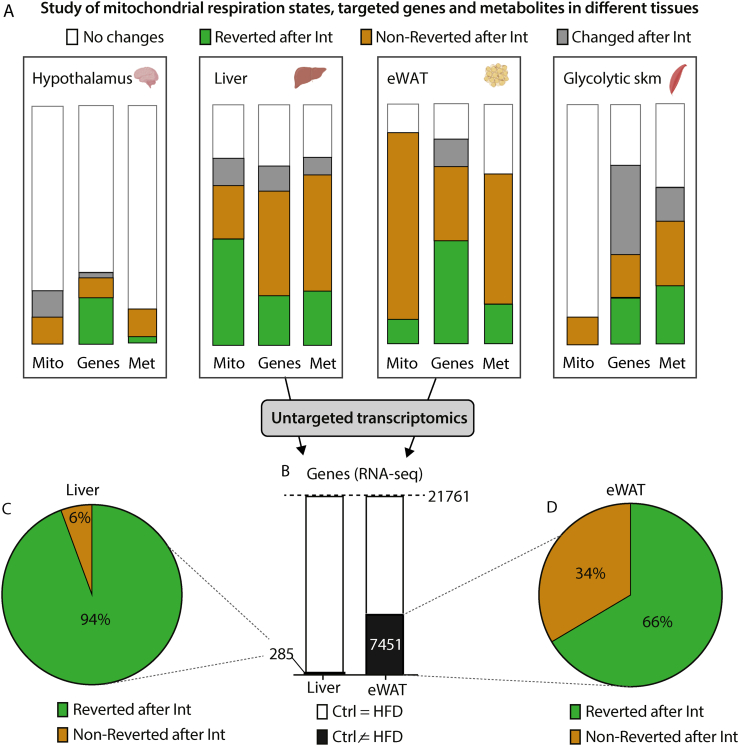
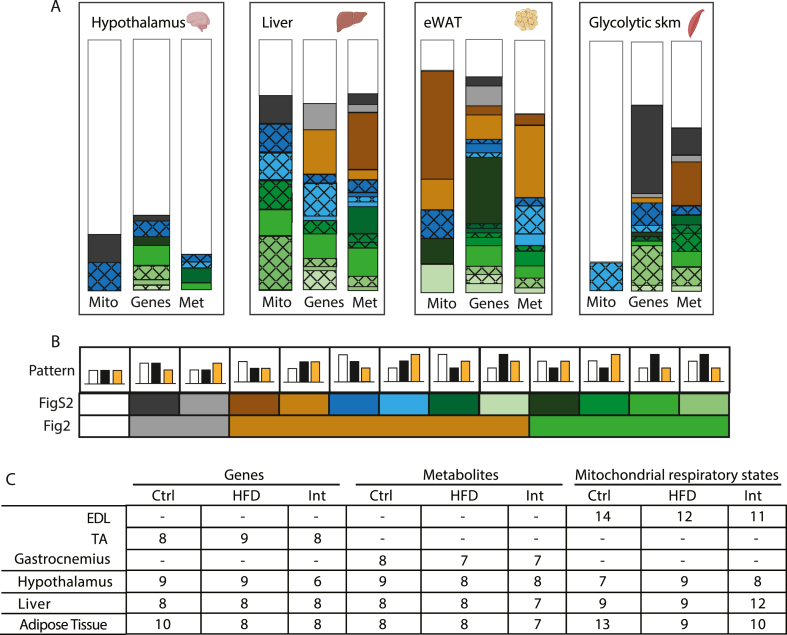
Next, we assessed the tissue-specific effect of the combined intervention on metabolic plasticity. We used a three-tiered approach to elucidate the pathophysiological impact of obesity-related insulin resistance in skeletal muscle, liver, epididymal WAT (eWAT), and hypothalamus. This approach consisted of evaluating, in these tissues, the reversibility patterns of different metabolic-related parameters: mitochondrial respiratory states (Mito); targeted expression profiles of genes related to glucose and fatty acid metabolism, mitochondrial biogenesis, function, and metabolic stress (Genes); and profiles of metabolites (Mets) (Fig. 2A for an illustrative simplification of reversibility patterns. Figs. S2A–C, check our research data for a more detailed profile information of parameters analysed and statistics).
Fig. 2.
Tissue-specific effects of HFD-feeding and nutritional and exercise intervention. A: Boxes highlight tissues analysed in this study (glycolytic skeletal muscle, hypothalamus, liver and epididymal white adipose tissue, eWAT), where we represent the changes induced by HFD and Intervention on mitochondrial respiratory states (Mito), gene expression (Genes) and metabolites (Met). Each bar corresponds to the 100% of respiratory states/genes/metabolites analysed. Bands of different colors represent fractions of respiratory states/genes/metabolites showing different patterns of change: white – no changed; green – significantly changed after HFD-feeding and reverted with intervention; red – significantly changed after HFD-feeding and not reverted with intervention; grey – significantly changed uniquely with intervention. See Fig. 2S and our research data for the original detailed representation, patterns definition, raw data and statistical details including n size. B: Bar graph for RNA-seq in liver and eWAT. The black area represents gene transcripts with significantly changed expression after HFD-feeding. C-D: Percentage of gene transcripts significantly reverted (green) or non-reverted (red) in liver (C) and eWAT (D) after the combined intervention. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The most prominent metabolic changes induced by HFD-feeding were observed in skeletal muscle, liver and eWAT (Fig. 2A combined green and dark orange bars). In the case of skeletal muscle, most of them were reverted after intervention (Fig. 2A green bars). However, the profound changes suffered by liver and eWAT exhibited a much higher degree of irreversibility across all three phenotypical tiers (Fig. 2A dark orange bars) despite the improvement in whole body metabolic phenotype promoted by the intervention. To broadly address the transcriptional signature of liver and eWAT, we performed next-generation RNA sequencing (RNA-seq) in these tissues before and after the intervention. Differential expression analysis revealed that, upon HFD-feeding, eWAT transcriptome was much more perturbed than the liver transcriptome (7,451 vs 285 altered genes, respectively) (Fig. 2B) and that upon intervention, the eWAT transcriptome exhibited a lesser degree of reversion (66% as compared to 94% in the liver) (Fig. 2C and D).
Collectively, these results argued for a tissue-specific heterogeneous susceptibility to obesity-induced consequences in metabolic plasticity. Concretely, eWAT showed the highest degree of alterations after a combined nutritional and exercise intervention.
2.3. Incomplete remodeling of eWAT after the combined intervention
To further characterize these wide obesity-related metabolic alterations in eWAT, we studied the transcriptional changes in more depth. We identified both downregulated (Fig. 3A) and upregulated genes (Fig. 3H) after HFD-feeding. Among the 3,869 downregulated genes with obesity, 1,036 recovered expression values similar to Ctrl animals, 1,464 partially recovered, but 1,369 remained downregulated after the intervention (Fig. 3A). Gene ontology enrichment analysis showed that non-reverted transcripts were related to biological processes such as biosynthesis, metabolism, response to oxidative stress and mitochondrial morphogenesis, organization and transport (Fig. 3B). In line with this, enrichment for cellular components identified genes with functions in the mitochondrial inner membrane as the top significantly enriched GO terms (Fig. S3A). The mitochondrial inner membrane is structured in cristae, whose density was significantly reduced with HFD-feeding and further decreased following the intervention (Fig. 3C–D, Fig. S3B). Decreased cristae density was observed irrespective of maintenance of the mitochondrial fractional area (mitochondrial content) and aspect ratio (Figs. S3C and S3D). Additionally, processing of OPA1, a protein located in the inner mitochondrial membrane and master regulator of cristae architecture, showed the same irreversible decrease with obesity and after the intervention (quantified as long/short isoforms ratio, Fig. 3E and F). High-resolution respirometry studies in permeabilized eWAT confirmed the functional relevance of defects in cristae density revealing impaired respiratory capacity (Fig. 3G).
Fig. 3.
Impairment of adipose tissue plasticity. A: Heatmap representing z-scores for RNA-seq cluster of transcripts downregulated after HFD-feeding (Ctrl n = 8, HFD n = 4, Int n = 4). B: Gene ontology biological process (GOBP) enrichment analysis for the 1369 transcripts downregulated in HFD and Int. C: Transmission electron microscopy (TEM) representative images of mitochondria cristae frequency, category 0, 1, 2 or 3 (see Methods section for category definition). D: Cristae density score according to categories detailed in C (Ctrl n = 3, HFD n = 4, Int n = 4). E: Normalized protein levels of OPA1 (represented as the long/short isoforms ratio) (Ctrl n = 7, HFD n = 9, Int n = 9). F: Representative western blott analysis of OPA1 expression; quantified in panel E. G: Mitochondrial respiratory states for eWAT homogenate (Ctrl n = 13, HFD n = 9, Int n = 10). Substrates; P: pyruvate, M: malate, G: glutamate and S: succinate. Pathways; N: NADH electron transfer-pathway, S: Succinate dehydrogenase-linked pathway and NS: NS-linked pathways. Respiratory rates; L: Leak-state; P: Oxidative phosphorylation (OXPHOS)-state; E: Electron transfer (ETS)-state [50]. H: Heatmap representing z-scores for RNA-seq cluster of transcripts upregulated after HFD-feeding (Ctrl n = 8, HFD n = 4, Int n = 4). I: Gene ontology biological process (GOBP) enrichment analysis for the 1129 transcripts upregulated in HFD and Int. J: Epididymal WAT weight (Ctrl n = 32, HFD n = 32, Int n = 26). K: Representative images for hematoxylin and eosin eWAT staining for each experimental group (Ctrl n = 3, HFD n = 4, Int n = 4). L: Average adipocyte area quantified from H&E staining images (Ctrl n = 3, HFD n = 4, Int n = 4). Ctrl: control; HFD: high fat diet-fed; Int: intervention. Data represented as box-and-whiskers plots (median, 25th to 75th percentiles and max/min values). ANOVA One-way and Post-hoc Tukey, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Among the 3,582 differentially upregulated genes after HFD-feeding (Figs. 3H), 1,064 achieved a complete reversion, 1,389 were partially reverted while the remaining 1,129 transcripts did not change after the intervention. Gene ontology enrichment analysis of these non-reverted transcripts revealed biological processes associated with the immune system and defense response (Fig. 3I). To further validate these findings, we investigated tissue immune cell infiltration in eWAT. The histological analysis showed hypertrophied adipocytes surrounded by crown-like structures (composed by immune cells) in the HFD group (Fig. 3K-L). Lastly, despite the improved systemic metabolic phenotype as well as the reduction of adipose tissue weight and adipocyte size promoted by the intervention (Fig. 3J and L), the histological analysis of eWAT in these animals showed a persistent alteration with the presence of dead adipocytes and a massive immune cell infiltration (Fig. 3K). These findings were corroborated by an increase in Caspase3-and Mac2-positive cells (Fig. 4D, Fig. S4D), markers of apoptosis and macrophages, respectively. Altogether, these results suggested that an active eWAT remodeling was occurring after the intervention, which could potentially explain the metabolic derangements observed so far.
Fig. 4.
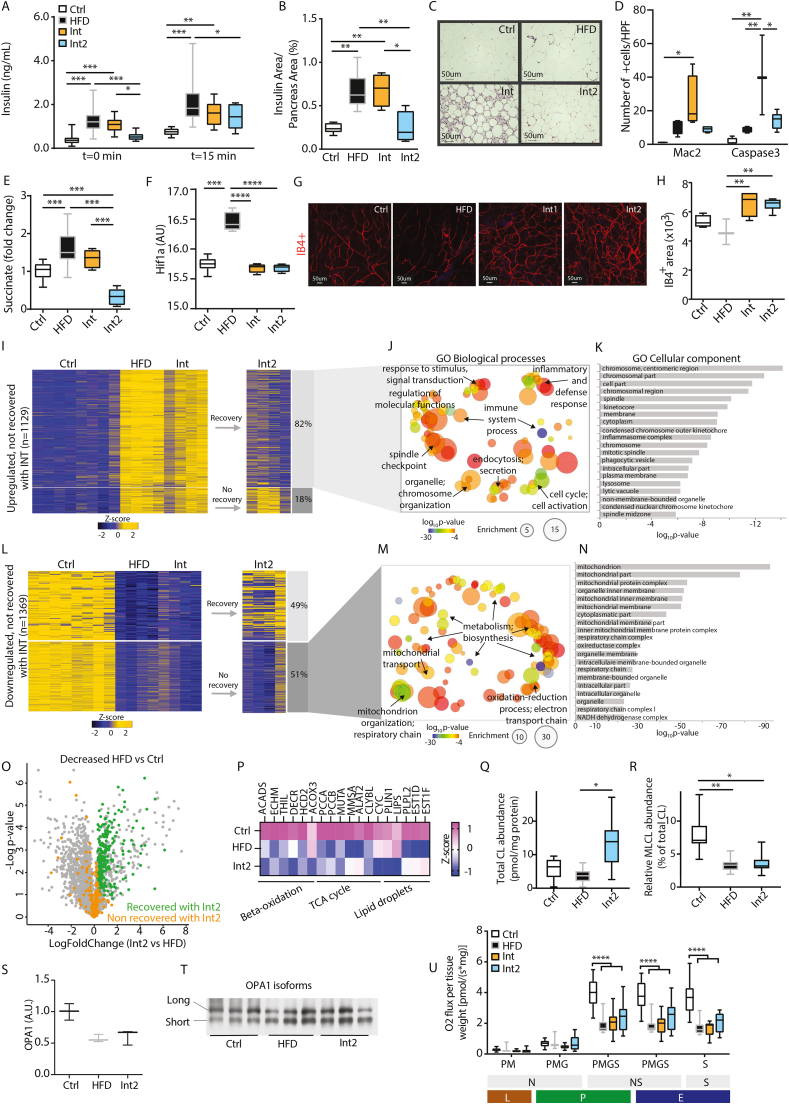
Mitochondrial deterioration in remodeled white adipose tissue. A: Basal (t = 0min) and 15min insulin levels during IGTT (Ctrl n = 28, HFD n = 30, Int n = 14, Int2 n = 10). B: Normalized pancreatic islet insulin area by total pancreas area for Ctrl n = 6, HFD n = 6, Int n = 4, Int2 n = 4. C: Representative images for hematoxylin and eosin in eWAT for each experimental group (Ctrl n = 3–4, HFD n = 4, Int n = 3–4, Int2 n = 4–5). D: Quantification for Caspase3-and Mac2-positive cells staining in eWAT for each experimental group (Fig. S4D) (Ctrl n = 3–4, HFD n = 4, Int n = 3–4, Int2 n = 4–5). E: Succinate metabolite levels in eWAT (Ctrl n = 15, HFD n = 13, Int n = 6, Int2 n = 6). F: Normalized Hif1-α gene expression in eWAT (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). G: Representative images of IB4-positive stained blood vessels in flat-mounted eWAT depots from each experimental group. H: IB4-positive area quantification (Ctrl n = 4, HFD n = 3, Int n = 4, Int2 n = 6). I: Heatmap representing z-scores for RNA-seq cluster of transcripts upregulated after HFD-feeding and not recovered after Int (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). J-K: Gene ontology biological processes (J) and cellular components (K) enrichment of the genes clustered in 4I that revert after Int2. L: Z-score for RNA-seq cluster of transcripts downregulated after HFD-feeding and not recovered after Int (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). M-N: Gene ontology biological processes (M) and cellular components (N) enrichment of the genes clustered in 4L that do not revert after Int2. O: Volcano plot displaying proteins decreased (HFD vs Ctrl) and their behavior after the second intervention (Int2 vs HFD) (n = 3/group). P: Heatmap showing Z-scored label free quantification (LFQ) intensities of proteins related to beta-oxidation, TCA cycle and lipid droplets dynamics differentially regulated in HFD and/or Int2 in comparison to Ctrl (n = 3/group). Q: Total cardiolipin (CL) abundance (Ctrl n = 5, HFD n = 10, Int2 n = 8). R: Relative monolysocardiolipin (MLCL) abundance (Ctrl n = 5, HFD n = 10, Int2 n = 8). S: Normalized protein levels of OPA1 (represented as the long/short isoforms ratio) (Ctrl n = 7, HFD n = 9, Int n = 9). T: Representative western blott analysis of OPA1 expression; quantified in panel S. U: Mitochondrial respiratory states for eWAT homogenate (Ctrl n = 13, HFD n = 9, Int n = 10, Int2 n = 13). Substrates; P: pyruvate, M: malate, G: glutamate and S: succinate. Pathways; N: NADH electron transfer-pathway, S: Succinate dehydrogenase-linked pathway and NS: NS-linked pathways. Respiratory rates; L: Leak-state; P: Oxidative phosphorylation (OXPHOS)-state; E: Electron transfer (ETS)-state. Ctrl: control; HFD: high fat diet-fed; Int: intervention; Int2: second intervention. Data represented as box-and-whiskers plots (median, 25th to 75th percentiles and max/min values). ANOVA One-way and Post-hoc Tukey, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
2.4. Mitochondrial dysregulation persisted after morphological eWAT remodeling
The negative energy balance induced by the combined intervention allowed for a global metabolic phenotypical recovery except for the maintained increase in pancreatic islets size (Figures 1L and M, Fig. 4SA) and the persistent eWAT mitochondrial dysfunction and immune cell infiltration (Figure 3, Figure 4SA). Due to the presumable transitory state in tissue remodeling following sustained weight loss and improved physical fitness, we hypothesized that an additional but milder intervention phase could facilitate the recovery of such irreversible features. To that end, we conducted this additional intervention phase for five more weeks in a separate group of mice with the same diet but undergoing pair-feeding and a less intense training program aiming at body weight maintenance (Int2 group). In addition, a subgroup of Ctrl animals fed with a chow diet were also extended for the whole intervention program to match Int2 animals’ age (Fig. S4A).
After this second intervention, body weight was maintained (Fig. S4B) and glucose tolerance was still comparable to the Int group (Fig. S4C). Furthermore, fasting insulin and glucose-stimulated insulin levels decreased (Fig. 4A), in line with reduced β cell mass (Fig. 4B), which argues for a recovery of the pancreas phenotype. The second intervention also improved eWAT morphology and immune cell infiltration (Fig. 4C, Fig. S4D), along with a progressive reduction of Caspase3-and Mac2-positive cells (Fig. 4D, Fig. S4D).
Adipose tissue remodeling and function can be influenced by tissue expandability as this leads to a limitation in oxygen and nutrient supply, often reflected by poor vascularization. In this sense, a limitation in oxygen triggers an increase in succinate levels, which stabilizes and activates HIF1-α, a master regulator of the cellular response to hypoxia and angiogenesis. To investigate whether the vasculature play a critical role in eWAT during the subsequent interventions, these parameters were assessed. On the one hand, a parallel increase in succinate levels as well as in HIF1- α expression was observed in obese animals (Fig. 4E and F), accompanied by a modest reduction in tissue vascularization revealed by confocal microscopy studies. On the other hand, however, same analysis evidenced the enhancement of eWAT vascularization after both interventions (Fig. 4G and H), as well as the normalization of these key mediators in angiogenic response; gene expression of Hif1α (Fig. 4F), Vegfα and Vegfr (Figs. S4E and S4F), and reduced levels of succinate (Fig. 4E).
Additionally, we evaluated whether this eWAT morphological recovery following the extended intervention was accompanied by the normalization of those non-reverted gene expression patterns previously observed. Among the 1,129 transcripts upregulated after HFD and Int (Figs. 3H), 82% had reverted to Ctrl values after Int2 (Fig. 4I). Gene ontology analysis of these now recovered genes revealed enrichment in biological processes linked to organelle and cytoskeleton organization, immune system, and inflammatory and defense response (Fig. 4J and K). Besides that, from the 1,369 transcripts downregulated after HFD and Int (Fig. 3A), only 49% were recovered (Figure 4L and Figs. S4G and S4H) after Int2 while 51% remained downregulated (Figure 4L). An enrichment in mitochondrial-related biological processes, such as electron transport chain, energy metabolism, biosynthesis, and mitochondrial organization and transport, was observed in these non-recovered set of genes (Figure 4M and N).
Mass spectrometry-based proteomics analysis of eWAT also displayed a similar dual pattern of recovery for those proteins downregulated after HFD feeding (Figure 4O, Fig. S4I). Among the 2,535 quantified proteins, 452 showed a significantly reduced levels in the HFD group in comparison to the Ctrl. As observed at a transcriptional level, a large fraction of these proteins (56%) was reverted with the implementation of the second combined intervention (Figure 4O, green dots). The Gene Ontology analysis of this recovered fraction of proteins highlighted a significant enrichment of the oxidative phosphorylation system (OXPHOS) and tricarboxylic acid cycle-related proteins, among others (Fig. S4J). A more specific mitochondrial proteome analysis further confirmed this recovery in the content of OXPHOS components, since respiratory chain-identified proteins were consistently decreased in the HFD group but remarkably increased following the extended intervention (Fig. S4K and S4L, purple dots). Interestingly, the same analysis also identified a consistent decrease of mitochondrial matrix-related proteins upon HFD conditions, but with a recovery pattern much less pronounced after intervention (Fig. S4K and S4L, orange dots), suggesting that many matrix-related proteins were among the remaining 199 proteins (44%) with reduced content in HFD animals that were still altered after the extended intervention (Fig. 4O, orange dots).
The refractory rearrangements of mitochondrial metabolism in eWAT primarily affected the pathways that harness fatty acids and amino acids to bioenergetics, since many direct and auxiliary enzymes in the early steps of β-oxidation (ACADS, ECHM, THIL, DECR, HCD2 or ACOX3), mitochondrial enzymes that replenish critical tricarboxylic acid cycle intermediates via the catabolism of branched-chain amino acids, cholesterol and odd-chain fatty acids (PCCA/B, MUTA, MMSA, ALAT2, CLYBL), as well as central components of the electron transport chain (CYC), remained strongly downregulated despite the intervention (Figure 4P). Moreover, we observed a decrease after the extended intervention of other proteins involved in relevant lipid metabolic processes, such as lipid droplets dynamics (PLIN1, LIPS, PLPL2, EST1D and EST1F) (Figure 4P).
Besides, and regarding the defects in cristae density observed previously, other aspects of mitochondrial membrane stability, such as the remodeling process of cardiolipins or OPA1 processing, were also evaluated after this extended intervention. On the one hand, even when total cardiolipin (CL) levels were already recovered (Figure 4Q), monolysocardiolipin (MLCL) relative abundance was decreased both in HFD and Int2 animals (Figure 4R), being indicative of decreased cardiolipin remodeling rates. The defective cardiolipin remodeling was potentially reinforced by an altered expression of genes related to the CL deacylation-reacylation cycle following the second intervention (i.e., Pla2g6, Lpgat1, Lclat1 or Hsd17b10) (Figs. S4M–P). In addition, the ratio between long and short OPA1 isoforms showed a similar decrease to the observed during obesity and the first intervention phase (Fig. 4S and T). Lastly, at a functional level, mitochondrial respiratory capacity also remained impaired after the extended intervention, as previously demonstrated in Int animals (Figure 4U). Altogether, these results demonstrate that the primary mitochondrial defect does not stem simply from the levels of OXPHOS subunits, and also reinforce its intrinsic persistence even after the resolution of tissue morphological remodeling, thus arguing for it to be a major determinant of a potential metabolic plasticity breakdown.
2.5. Enduring mitochondrial derangements drive eWAT metabolic plasticity breakdown
To obtain a more comprehensive understanding of the eWAT metabolic scenario, we studied the biological processes underlined by the downregulated genes following the subsequent experimental interventions. In doing so, we confirmed a progressive tissue deterioration at the transcriptional level (Fig. 5A). Three clusters of genes were defined, including 893, 601 and 678 downregulated genes following HFD-feeding, the first intervention and the extended intervention, respectively (Figs. 5A and S5A). The role of mitochondria as the primary organelle affected by HFD was confirmed after studying the cellular components and biological processes enriched in the three clusters (Figs. 5B and S5B). Furthermore, early alterations in peroxisomes were also revealed. Genes with functions in the Golgi and Endoplasmic Reticulum appeared downregulated after the first intervention, whereas genes within the proteasome complex were only decreased after the second intervention (Fig. 5B). Likewise, the respective enrichment of biological processes also displayed a spreading of early and cumulative defects in mitochondria-related metabolic and biosynthetic processes towards wider forms of mitochondrial and cellular stress, such as defects in mitochondrial translation, organization, protein transport and protein folding (Fig. S5B). Indeed, both cellular components and biological processes enriched within the downregulated genes after the different interventions aggregated and increased over time.
Fig. 5.
Metabolic plasticity breakdown in eWAT. A: Z-score for RNA-seq clusters of transcripts sequentially downregulated across the different experimental groups (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). B: Cumulative number of genes belonging to different enriched GOCC terms for each cluster in A. The same GOCC terms are indicated from HFD-cluster to Int2-cluster (y-axis), with increasing size (x-axis) denoting incremental number of genes for that specific term. C: Profile plot including the cluster of proteins that are differentially upregulated with HFD and maintained increased after the second intervention (Int2). Blue profiles show proteins specifically belonging to KEGG pathway Lysosome within this cluster. D: Heatmap showing Z-scored label free quantification (LFQ) intensities of metabolic stress-related proteins differentially regulated in Int2 in comparison to Ctrl (n = 3/group). E: Mitochondrial DNA (mtDNA) copy number in epididymal white adipose tissue (Ctrl n = 6, HFD n = 6, Int n = 6, Int2 n = 5, Ctrl + IF n = 8, HFD + Chow n = 4, HFD + IF n = 10, * indicate significant differences with Ctrl group); normalized by Ctrl mice refers to eWAT Ctrl group. Ctrl: control; HFD: high fat diet-fed; Int: intervention; Int2: second intervention; Ctrl + IF: control + dietary intervention; HFD + Chow: high fat diet-fed + chow Ad libitum; HFD + IF: high fat diet-fed + dietary intervention. F–H: Mitochondrial DNA (mtDNA) copy number in subcutaneous white adipose tissue (scWAT) (F), liver (G) and skeletal muscle gastrocnemius (H). Data represented as box-and-whiskers plots (median, 25th to 75th percentiles and max/min values). ANOVA One-way and Post-hoc Tukey, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We later investigated whether these transcriptionally driven assumptions also impact the proteome in Int2 animals. Some relevant patterns were underlined from this analysis. For instance, we identified a clear pattern of lysosomal proteins significantly enriched among those presenting an upregulation with obesity that persists following the second intervention (Fig. 5C). Besides the observed defective lipid metabolism (Figure 4P) we also further reinforced the early and cumulative mitochondrial and peroxisomal derangements observed since obesity (Fig. 4, Fig. 5B) by alterations in the content of proteins involved in reactive oxygen species (ROS) homeostasis, a process in which these organelles play a critical role. In this sense, several detoxifying proteins, including glutathione-related, were upregulated after the second intervention (GSHR, PRDX1, GLRX1, HMOX1, members of the aldose reductase superfamily ALD1 and ALD2, ABD12 or PLD3 among others) (Fig. 5D), which is indicative of high presence of oxidized species and increased glutathionylation activity. Despite this, several critical components of the intrinsic mitochondrial and peroxisomal ROS scavenging system, such as members of the glutathione peroxidase and S-transferase families (GPX1, GPX4, GPX7, GSTA3, MAAI, PEX5) and superoxide dismutase (SODC) were decreased following the second intervention phase (Fig. 5D).
Along with a marked state of cellular stress, Int2 animals also showed permanent deregulations in the content of proteins related to cellular and mitochondrial homeostatic response, which are important contributors to the organelle quality control system. For instance, we observed a significant decrease in the content of the mitochondrial chaperonines CH60 and CH10 (Fig. 5D), as well as other regulatory proteins of chaperonine activity, such as DNAJA3 (Fig. 5D), which also assists to mitochondrial DNA (mtDNA) maintenance. In this sense, protein levels of TFAM, a key player in mitochondrial genome replication and transcription, were also permanently decreased with obesity and the subsequent intervention (Fig. 5D).
A substantial reduction in mtDNA copy number in eWAT (Fig. 5E) supported the previous findings and further reinforced mitochondrial derangements as a key component in WAT metabolic deterioration. Contrarily, mtDNA in inguinal WAT depot (scWAT) and in other key metabolic tissues such as liver and skeletal muscle gastrocnemius remained unchanged (Fig. 5F–H). To elucidate the trigger for the decrease in mtDNA, we evaluated additional experimental groups following different dietary regimens to combat the diet-induced obesity (i.e. intermittent fasting or shift to chow diet). Mice placed on a HFD showed an eWAT mtDNA reduction similar to previously observed, regardless of the regime implemented afterwards (Fig. 5E). Conversely, such reduction was not present in animals that were not previously obese following the same dietary regime (Fig. 5E). Collectively, these results revealed an unresolved metabolic stress triggered by obesity that primarily impacts on mitochondrial performance.
2.6. Transcriptional signature in human visceral adipose tissue after weight loss
To address the translational value of our findings, we investigated the transcriptome of omental WAT samples from a unique human cohort of obese patients undergoing weight loss through a 2-step bariatric surgery procedure. These patients lost 41.6 ± 4.7 kg of BW between the first and second step, changing their BMI from 54.6 ± 2.0 to 39.7 ± 1.3 kg/m2 (p-value<0.001), and improved their glucose homeostasis parameters (fasting glucose pre: 6.32 ± 0.42 mmol/l, post: 5.54 ± 0.18 mmol/l, p-value = 0.06; HbA1c pre: 6.15 ± 0.16%, post: 5.51 ± 0.14%, p-value<0.001) (Fig. 6A). An unsupervised principal component analysis (PCA) of RNA-seq data revealed a highly significant difference (p < 0.001) between the pre and post conditions (before the first step and before the second step, respectively) in the second principal component, which accounts for 7% of the total transcriptional variability (Fig. 6B). A downstream supervised analysis between the two groups revealed the presence of 456 downregulated and 322 upregulated transcripts (FDR<0.1) in the post as compared to the pre-treatment condition (Fig. 6C). We explored the biological process enriched by these set of genes. Regarding the downregulated genes after the treatment the pattern was more diffuse and mainly related to the tissue structural organization and secretory activity (Fig. 6D), while among the upregulated genes we noticed a remarkable presence of metabolic and mitochondria-related terms (Fig. 6E). These later results would suggest a contraposition to the conclusions derived from our animal model but, due to the lack of healthy lean subjects as a reference, it is not possible to assess the extent of the transcriptional changes induced by obesity and, consequently, this analysis could be masking its real impact on metabolic plasticity.
Fig. 6.
Adipose tissue transcriptomics after weight loss interventions in humans. A: Table displaying the evolution of clinical parameters in 23 patients with obesity undergoing a 2-step bariatric surgery (pre-treatment: in the first surgery step – sleeve gastrectomy; post-treatment: in the second surgery step, Roux-en-Y gastric bypass). B: PCA displaying transcriptomics data from omental fat collected in those 23 patients during corresponding surgical procedures. C: Z-score for RNA-seq clusters of transcripts differentially expressed (post vs pre-treatment) from patients in A. D-E: Top-20 enriched Gene ontology biological process (GOBP) for the 456 transcripts downregulated (D) and 322 transcripts upregulated (E) after the treatment. F: Addressing the translational potential of the LiMa study. Bar graph showing the proportion of differentially expressed genes in comparison to all the identified genes for each of two representative GOBPs (mitochondrion organization and oxidation-reduction process). Green displays upregulated genes (recovered) after the weight loss intervention (light green: Int2 vs HFD in mice; dark green: post vs pre in humans). Red displays downregulated genes (non-recovered) after the weight loss intervention (light red: Int2 vs HFD in mice). For the mice comparison, only genes differentially downregulated HFD vs Ctrl were included. Of note, lack of lean controls in the human cohort (equivalent to Ctrl in mice) leaves the number of non-recovered genes unknown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Thus, we devised an alternative analysis focused on exploring the differences in the enrichment of biological processes between upregulated genes after bariatric surgery in humans and after the different interventions in our animal model, and thereby to compare the responsiveness to weight loss. For that purpose, we explored two common and general GO terms (oxidation-reduction process and mitochondrial organization) highlighted in both models (Fig. 6E for humans, Fig. S4G for mice) and compared the magnitude of changes. In both cases, the percentage of upregulated genes belonging to these GOterms was similar (<5%) but the degree of the remaining genes that were still downregulated after intervention (10–15%) was critical for the eWAT phenotype of our lean but formerly obese mice, and this incidence is still unknown in humans (Fig. 6F).
3. Discussion
Obesity-related T2D incidence is a leading healthcare challenge in most countries. Major efforts are devoted to develop new drugs and combinatorial therapies aimed to promote body weight loss and revert insulin resistance [2]. Lifestyle interventions are the most physiological and economical options [7]. However, their implementation, and the adherence of the obese population is not always successful in the long term. This is, in part, due to the metabolic, hormonal, and psychological changes associated with weight loss [1,3]. Given that conducting complex studies in human subjects is still difficult, appropriate animal models are still indispensable to unravel complex interactions between environmental stressors and genetic predisposition to obesity [8]. Our current approach, combining nutritional and exercise interventions, has proved successful in the recovery of nearly all phenotypical metabolic alterations in diet-induced obese animals, a model that resembles the early stages of human obesity-related T2D pathology [9,10]. Thus, we here provide valuable information that supports lifestyle interventions to overcome obesity, and its associated comorbidities.
Previous reports addressing tissue crosstalk in obesity and T2D have mainly focused on the direct relationship between two tissues (e.g., muscle-WAT, WAT-liver, hypothalamus-liver), as well as secreted factors governing such communication [11]. However, the implementation of multidisciplinary and more integrative approaches is promising, due to its potential contribution to broaden our understanding of the complexity of these diseases (https://www.diamap.eu/roadmap/domain/detail/3, [12]. By implementing this kind of experimental approach addressing the response of several key tissues in a context of a lifestyle-like intervention, we reveal in this study not only the impact of high fat diet feeding, but also how this metabolic fingerprint could influence the plasticity of such tissues after weight loss. This integrative analysis directly points at WAT as the most affected tissue as denoted by the magnitude of metabolic derangements occasioned during obesity, and the fact that many of these alterations did not normalize after the intervention. Importantly, our data confirm the previous observation that the obesogenic memory after weight loss is mainly conferred by the adipose tissue [13].
We then further confirmed this sustained fingerprint in epididymal WAT by a multi-tiered analysis, uncovering mitochondria as the primary affected organelle. Notably, mitochondrial derangements were strongly evidenced at multiple levels, from transcriptional to functional datasets. In particular, transcriptomics unveiled an extensive set of down-regulated genes, majorly framed within the mitochondrial compartment, which denoted a generalized impairment of metabolic processes. Proteomics data also evidenced a global impact on mitochondrial proteome, specially affecting the supportive role of amino acid and lipid catabolism, which let us to speculate on the plausible failure in the restoration of key pathways of core energy metabolism [14,15]. These results combined with the structural derangements manifested in the inner mitochondrial membrane, compellingly question the functionality of this organelle, despite a recovery in OXPHOS components at the protein level, [16]. Accordingly, the mitochondrial disturbances were translated into persistent defects in mitochondrial respiration at a functional level.
One of the most revealing insights is the fact that this metabolic disruption is not only present in phenotypically healthy animals, but also in an apparently functional adipose tissue. The plasticity of WAT in the face of nutrient oversupply under acute stress signals, such as hypoxia or inflammation, is well known [[17], [18], [19]]. However, the chronicity of such signals promoted by extended overfeeding periods can exert a maladaptive response. These results in persistent oxidative damage, severe hypoxic regions -accompanied by an inadequate angiogenic response-, adipocyte death, immune infiltration, and fibrosis [20]. More important, but far less understood, is how such perturbations impact the plasticity of the tissue upon the shrinkage promoted by weight-loss. Given the strong tissue atrophy that we observed after the first intervention period, we granted special attention to this factor. The massive tissue infiltration along with the inflammatory and immune transcriptional enrichment observed after the intervention share common traits with previously described early WAT responses to different weight loss strategies [13,[21], [22], [23]]. However, the mitochondrial phenotype was still present after the second intervention, which had resolved the inflammation and achieved complete tissue morphological remodeling. This demonstrates that mitochondrial defects were not just a consequence of a transitory state during tissue remodeling, but an intrinsic permanent tissue damage. Altogether, these data reinforce the claim of WAT metabolic plasticity breakdown.
Addressing the mechanisms leading to this persistent mitochondrial derangements is challenging due to multifactorial components occurring under physiological conditions. Nonetheless, omics data provided revealing insights about processes that could significantly contribute to this loss of metabolic plasticity. The decline of functional capabilities during the course of aging process is perhaps the most paradigmatic example of plasticity loss, and particularly their energy metabolism components. In this regard, most elements described in our model recapitulate many of the mitochondrial hallmarks described for age-related deterioration [4].
We then speculated about two major processes that could explain this hypothetical loss of tissue plasticity, based on this close parallelism between obesity-related T2D and aging; i) a sustained metabolic stress during obesity progression that could lead to such mitochondrial phenotype and ii) the inability of mitochondrial and cellular quality control mechanisms to overcome such derangements in an otherwise healthy tissue. Regarding the first option, the time frame of metabolic stress would be the critical determinant of this irreversible damage. In line with this, animal studies addressing HFD-induced transcriptional modulation of epididymal WAT in a time-resolved manner suggest a continuous exacerbation of mitochondrial defects as the most decisive event [24]. Of note, this particular deterioration reported in later-stages of obesity progression was still manifested in our model even when the obese phenotype was successfully resolved. This issue seems to be also a singularity of aging deterioration, most certainly proved by the recent comprehensive analysis of multi-tissue aging transcriptional dynamics, known as Tabula Muris Senis (or Mouse aging Cell Atlas, from The Tabula Muris Consortium) [25]. This work shares with our study two important findings: the first is a prominent susceptibility of gonadal WAT to age-related deterioration, as we observe in obesity conditions. The second is the strong detrimental similarities elicited by both obesity and aging. In addition, similar defects in gonadal WAT plasticity to those observed in our model were evidenced among the most remarkable traits of aged animals subjected to a calorie restriction-based intervention [6]. These results strengthen our thesis of an irreversible mitochondrial damage occurring during the course of the metabolic stress.
Additional experimental readouts also help to characterize this mitochondrial fingerprint left by obesity. For instance, the persistence in formerly obese mice of early mitochondrial and peroxisomal disorders, as denoted by alterations in the glutathione redox balance and the disruption of important scavenging systems, suggests a highly oxidative environment, which in turn is characteristic of an obesity-related WAT phenotype [26]. Moreover, the defective remodeling of cardiolipins and the role that pivotal enzymes, such as Pla2g6, Lpgat1, Lclat1, and Hsd17b10, play in this process add some insights in that direction. These results are in line with the reciprocal connection between pathological remodeling and oxidative stress, widely documented for several metabolic tissues [27], and various age-related degenerative conditions, including obesity-induced T2D [[28], [29], [30]]. This perpetuated oxidative insult is one of the plausible drivers of the obesity-driven mitochondrial damage, especially when it perturbs its physiological boundaries and favours the transition towards a pathological state of high ROS production [31]. The magnitude of reduction mtDNA copy number in this tissue is the most indicative sign of severe mitochondrial disturbances [32]. Indeed, mtDNA quantification in different tissues and experimental groups reinforced the distinctive vulnerability of epididymal WAT and pointed out unequivocally to high fat feeding as its driving force.
As raised before, the inability of mitochondrial and cellular quality control mechanisms to overcome aforesaid disorders, represents a second major process that could explain this hypothetical loss of WAT metabolic plasticity, Indeed, this inability also contributes to metabolic plasticity loss in aging models [5,33]. Mitochondrial import and folding deficiencies have been described as potential drivers of global proteostatic defects and the consequent cellular dysfunction spreading beyond this organelle in both aging [34] and obesity [35]. In this regard, we noticed that the dissemination of early metabolic defects into new biological processes related to protein transport and folding was paralleled by a reduction in the main mitochondrial chaperonines. Thus, the transcriptional defects initially confined to mitochondria were expanded to other cellular compartments such as Golgi, Endoplasmic Reticulum and ubiquitin and proteasome systems. Therefore, our results suggest that an unresolved dissemination of mitochondrial dysfunction is ultimately contributing to the unrecovered tissue phenotype.
Thus, the relevant question is: why such disrupted mitochondria or its defective components are not effectively recycled? A widely recognized acute response to remove defective mitochondria and recycle their components is an increase in lysosomal biogenesis and activity [36]. Of interest, impaired ability to foster these processes by the deregulation of lysosomal and proteosomal activities are pivotal events during aging progression [37,38]. In this respect, the global increase in lysosomal proteins observed in both HFD and Int2 animals seems to indicate an attempt to palliate the aggravated mitochondrial derangements. However, considering the transcriptional down-regulation of proteosomal activity after the second intervention, and the observed incapacity to re-establish proper functional mitochondria, we inferred that these signs (recycling and removal) are not translated into an effective response. Taken together, the current results provide robust signs of a defective mitochondrial clearance capacity as a plausible explanation for this perpetuated and accumulative tissue-specific mitochondrial disturbance.
We also investigated the translational value of our findings in a human cohort of patients with obesity undergoing weight loss after bariatric surgery. Despite the human cohort under evaluation represented a unique setting to study visceral adipose tissue dynamics, it was inevitably limited by the lack of a control group, since healthy lean individuals are not subjected to weight loss procedures and tissue sampling. Such limitations are frequent and heavily conditioned by ethical regulations when a surgical intervention is required to collect tissue samples. However, the few available human transcriptomic and proteomic data comparing lean and obese visceral WAT revealed strong similarities to the epididymal WAT phenotype that we observed in our HFD animals [[39], [40], [41], [42]]. Consistent with this, similarities were observed when studying the recovered fraction of genes in the post-intervention groups, both in the animal model and the human subjects. Therefore, the major relevance of our findings is also based on the fact that nearly all follow-up studies addressing the effects of weight loss have relied upon exploring adipose subcutaneous responses [43,44], and the study of visceral tissue in humans after weight loss or in healthy lean controls remains as a challenging endeavor. Thus, the impact of obesity in visceral WAT phenotype after weight loss remains critical for the irreversibility of certain aspects of the phenotype [13,45]. Further studies in human subjects should dissect whether “obesogenic memory” in visceral fat depots defines a distinct obesity sub phenotype and may link obesity to subsequent comorbidities and premature aging.
In summary, this study demonstrates that nutritional and exercise interventions, resembling a healthy lifestyle in humans, are able to revert the obesity-related T2D phenotype in mice. These benefits of a successful implementation of healthy lifestyle habits encourage additional research in both animal models and humans. However, the broad spectrum of data layers, analysed tissues and phenotypical characterization performed in this study revealed a silent disruption of mitochondrial homeostasis in visceral WAT. These lasting metabolic disturbances in visceral WAT suggest an initial and critical point of no return in obesity progression, which ultimately advocates for earlier and plausible weight loss interventions.
4. Materials and methods
4.1. Animals
C57BL/6JOlaHSD male mice purchased from Envigo (Indiana, IN, USA) were kept on a 12 h light/12 h dark cycle. Mice were assigned to two groups fed with different diets for 16 weeks: control group (Ctrl; fed with chow diet Teklad Global 14% Protein Rodent Maintenance Diet by Envigo; final inclusion criteria: body weight (BW) < 33 g, fasting normoglycemia, fasting normal insulin, glucose tolerance and insulin sensitivity); and a high-fat diet-fed group (HFD; D12451 by Research Diets, New Brunswick, NJ, USA), final inclusion criteria: BW > 37 g, fasting hyperglycemia, fasting hyperinsulinemia, glucose intolerance and insulin resistance). Comparative analysis between Ctrl and HFD animals (obesity-induced changes) was performed at this time-point. After 16 weeks on HFD, a subgroup of HFD mice was assigned to the intervention group (Int, fed with Preliminary Formula Rodent Diet with 45 kcal% Fat and Modification with Flaxseed and Olive Oil by Research Diets, New Brunswick, NJ, USA); and a further intervention was performed (Int2, fed with the same diet as the Int group), see intervention details below. All animal procedures were approved by the local ethics committee, Comitè Ètic d’Experimentació Animal at Universitat de Barcelona and the Departament d’Agricultura, Ramaderia, Pesca, Alimentació i Medi Natural at the Generalitat de Catalunya; complying with current Spanish and European legislation.
4.2. Nutritional and exercise intervention
Nutritional and exercise intervention (Int group) was initially for 5 weeks. The daily energy intake was different during the first week compared to the rest of weeks, being mice fed with 80% and then 100% in kcal of the daily energy intake determined for a Ctrl animal, respectively. Note that at the beginning of intervention it represents ∼60% of the caloric intake determined for an weight-matched animal (HFD), but this deficit is reduced as BW drops. Intervention diet aimed at maintaining energy content from fat (45%) and included several nutritional modifications: replacement of sucrose (simple carbohydrates) by corn starch (complex carbohydrates) and increase in mono- and polyunsaturated FA and omega-3 by changing oil origin (flaxseed and olive oil were used instead of lard). Intervention diet was combined with an exercise training program on a treadmill (Exer-6M Open Treadmill for Mice and Rats with Shocker and Software 2–102 M/m Columbus Instruments; Columbus, OH, USA). Acclimatization was performed already during the first week of intervention for 3 days prior 4 weeks of exercise training (5 days/week, 1 h/day). The protocol was designed to increase gradually the speed until reaching a final maximum speed of 20 m/min with a second level inclination (10°). To address new experimental hypothesis (Fig. 5), a second phase of intervention was implemented for 5 extra weeks (Int2 group). Int2 maintained the diet (100% in kcal content of the daily energy intake determined for a Ctrl animal) and the exercise program was adjusted to an alternate-day workout, 1 h/day with a fixed moderate speed (14–16 m/min) and inclination (5%).
4.3. Body composition
Mice were scanned using Magnetic Resonance Imaging in a 7.0 T BioSpec 70/30 horizontal (Bruker BioSpin, Ettlingen, Germany), equipped with an actively shielded gradient system (400 mT/m, 12 cm inner diameter). Animals were placed in a supine position in a Plexiglas holder with a nose cone for administering anesthetic (1.5% isoflurane in a mixture of 30% O2 and 70% N2O). Mice were scanned from below the head to the beginning of the tail, in 1.5 mm sections. A 3D reconstruction merging all the images was performed. Fat volume shown in white contrast was calculated merging the area values for each image. A Whole-body Composition Analyser (EchoMRI™, Houston, TX, USA) was used additionally.
4.4. Indirect calorimetry
TSE LabMaster (TSE Systems, Bad Homburg, Germany) was used as described previously [46,47]. Mice were acclimated for 24 h and then monitored for an additional 48 h. Data was collected every 30 min for O2 consumption and CO2 production to determine energy expenditure with standard analysis software provided with the calorimeter system. Heat production was calculated by the abbreviated Weir equation ([3.94(VO2) + 1.11(VCO2)] 1.44) [48]. Food and water intake, and locomotor activity were determined continuously for 48 h 30 min-RER values were averages for each animal in the same time point across 24 h. Median of these average for each animal was considered baseline, and the deviation to it was calculated, along with the maximal and minimum values.
4.5. Glucose homeostasis in vivo functional assays
Intraperitoneal Glucose Tolerance Test (IGTT) was performed after 16 h fasting (d-Glucose at 2 g/kg of mouse BW). Blood glucose was measured before and 15, 30, 60 and 120 min after glucose administration. Insulin levels at time 0 and 15 were measured by ELISA (90080 Crystal Chem Inc., Elk Grove Village, IL, USA) according to manufacturer's instructions. Insulin Tolerance Test (ITT) was performed after 4 h fasting (insulin Humulin R 100 UI/mL Regular by Eli Lilly and Company (Indiana, IN, USA), at 0.75 UI/kg of mouse BW). Blood glucose was measured using a glucometer before and 15, 30, 60 and 120 min after insulin administration.
4.6. In vitro glucose-stimulated insulin secretion (GSIS)
Animals were anaesthetized and 2–3 mL of fresh collagenase XI solution was injected in the clamped end of the bile duct inserted in the duodenum (Collagenase from Clostridium histolyticum, Type XI, 2–5 FALGPA units/mg solid, >1200 CDU/mg solid; C7657 Sigma, St. Louis, MI, USA) at 1000 U/mL in Hank's Balanced Salt Solution (HBSS without MgCl2 and CaCl2; Gibco, Thermo Fisher Scientific, Waltham, MA, USA). Pancreas tissue was then collected and incubated at 38 °C for 14 min before manual agitation, and after two washes with ice-cold HBSS containing CaCl2 (HBSS, Sigma, St. Louis, MI, USA), the pellet was resuspended in 7 mL and filtered (Cell Strainer 70 μm Nylon; BD Falcon, Bedford, MA, USA). The islets in the filter were washed twice with HBSS and plated in a RPMI solution by inverting the filter (RPMI, Gibco, Thermo Fisher Scientific, Waltham, MA, USA, supplemented with 2 mM l-glutamine, 1000 units/mL of both penicillin and streptomycin, Thermo Fisher Scientific, Waltham, MA, USA, and 10% FBS, Biosera, Manila, Philiphines). About 70 islets per animal were handpicked for each animal and cultured overnight in RPMI media, and then split in tubes of 15. To measure insulin secretion, islets were pre-incubated in agitation for 30 min in freshly prepared KRBH secretion solution (NaCl2 140 mM, KCl 4.5 mM, MgCl2·6H2O 1 mM, HEPES 20 mM, CaCl2 2.5 mM, 0.1% BSA, with 2.8 mM glucose) and then incubated in KRBH solution with 2.8 mM or 16.7 mM glucose for 1h. Supernatants were collected and for insulin content, remaining islets were lysed in glacial acetic (5.75% (v/v) in 0.1% BSA) following an overnight freezing cycle at −80 °C, warm them at 95 °C for 10 min, then centrifuging them at 4 °C and collecting the supernatant. Insulin levels were measured in supernatants from both insulin secretion and insulin content assays, by ELISA (Crystal Chem Inc., Elk Grove Village, IL, USA) according to manufacturer's instructions. Insulin secretion was normalized by insulin content.
4.7. Analytical measurements
For triglyceride measurements, lipids were extracted from tissue homogenates (100 mg of tissue in SDS 0.1%) using chloroform:methanol (2:1) to extract the organic phase pellet and resuspend it in ethanol. Triglyceride Reagent, Free Glycerol Reagent and Glycerol Standard Solution (T2449, F6428, G7793 Sigma, St. Louis, MI, USA) were used according to a modified protocol from the one provided by manufacturers in liver and skeletal muscle. Briefly, Free Glycerol Reagent was added to samples and standards in a 96-well microplate (NUNC, Denmark) incubated at 37 °C for 5 min. Absorbance at 540 nm was read before and after the addition of Triglyceride Reagent (Synergy HT Multi-Mode Microplate Reader, BioTek, Winooski, VT, USA).
4.8. Pancreas morphometry studies
Collected pancreas tissue was fixed overnight in formalin at 4 °C, dehydrated and embedded in parafin blocks (Embedding Center, Leica, Buffalo Grove, IL, USA), cut in 4 μm sections separated by 150 μm (Rotary Microtome, Leica, Buffalo Grove, IL, USA) and placed in Poly-l-Lysine treated microscope slides. Paraffin was removed, and the tissue was permeabilized in PBS1X+1%Triton, blocked with 3% BSA in PBS1 and incubated overnight at 4 °C with primary polyclonal antibodies for glucagon and insulin (Dako, Agilent, Santa Clara, CA, USA) (Research data repository). Secondary antibodies were conjugated with Alexa Fluor®488 and Alexa Fluor®555 (Life Technologies, Carlsbad, CA, USA) (Research data respository) and Hoechst was used for nuclei staining. An inverted-fluorescence microscope with camera (Leica, Buffalo Grove, IL, USA) and ImageJ were used to obtain and to measure the number and area of positive cells, respectively. The pancreas section was stained with tolonium chloride to measure the area and normalize the values.
4.9. Gene expression
50–100 mg of tissue were homogenized in 1 mL TRI Reagent (93289 Sigma, St. Louis, MI, USA) using magnetic beads (0.5 mm diameter ZrO2 for BulletBlender, NextAdvance, Troy, NY, USA) in a BulletBlender following manufacturer instructions for each tissue. For adipose tissue samples, a centrifugation was performed at this point to discard the surface layer. Supernatants were collected for RNA extraction according to manufacturer's instructions. cDNA was obtained using the High Capacity cDNA Reverse Transcription Kit (4368814 Applied Biosystems, Thermo Fisher Scientic, Waltham, MA, USA). cDNA dilution for each tissue was optimized. RT-PCR assays were performed in 384-well plates (Attendbio, Cerdanyola del Valles, Spain) using the 7900HT RT-PCR system (Thermo Fisher Scientific, Waltham, MA, USA) and the commercial reagents Premix Ex Taq™ (BN2001 TaKaRa Bio Inc., Kusatsu, Japan) according to recommended conditions. All Taqman® probes were obtained from Applied Biosystems (Research data repository). A standard curve of pooled samples was used for quantification.
4.10. Immunobloting
Tissues were homogenized with magnetic beads as described above and lysed using 300 μL of lysis buffer (Na2HPO4 10 mM, NaF 10 mM, Tris pH 7.5–8 50 mM, EDTA 5 mM, NaCl 150 mM and Triton X-100 150 mM) with the addition of a protease inhibitors cocktail (Roche), and mechanically disrupted through freeze-thaw cycles. Supernatants were collected for protein quantification and diluted at a given concentration in lysis buffer and Laemmli sample buffer 4X. Criterion XT Bis-Tris Gel (BioRad, Hercules, CA, USA) were used for electrophoresis. Transfer of the proteins to the PVDF membranes was performed using the Trans-Blot TurboTM Transfer System (BioRad, Hercules, CA, USA). Membrane was blocked for 1 h at RT (5% milk in TBS-T) and incubated overnight at 4 °C with primary antibodies (Research data repository). Secondary antibodies were incubated for 1 h at RT. PierceTM ECL Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA, USA) was used in an ImageQuant LAS 4000 (GE Healthcare, Chicago, IL, USA) for imaging of blots by chemiluminescence. ImageQuant TL Software (GE Healthcare, IL, USA) was used for the blot quantification.
4.11. High-resolution respirometry
Mitochondrial respiration was assessed by high-resolution respirometry (HRR) in glycolytic skeletal muscle, hypothalamus, liver and white adipose tissue. For sample preparation, each tissue was homogenized and permeabilized in fresh conditions as described in Ref. [49]. Experiments were performed in Oxygraph-2k system (Oroboros Instruments, Innsbruck, Austria) according to an established substrate-uncoupler-inhibitor titration (SUIT) HRR protocol (https://www.mitofit.org/index.php/SUIT-008), where the nomenclature used is defined (see further details in Ref. [50]). Briefly, LEAK respiration was measured in the presence of the NADH (N)-linked substrates Pyruvate (5 mM) and Malate (2 mM). Oxidative phosphorylation capacity was then determined by the addition of ADP (5 mM) at saturated concentrations, and cytochrome C (10 μM) was added to assess the integrity of the mitochondrial outer membrane before the addition of Glutamate (10 mM). Then, Succinate (10 mM) was added to stimulate the succinate (S)-linked pathway, allowing the convergent electron flow through both pathways simultaneously. Subsequent titration of the uncoupler carbonyl cyanide p-trifluoro-methoxyphenyl hydrazine (FCCP, 0.5 μM) was then performed to assess maximal non-coupled Electron Transfer (ET)-respiratory capacity mediated by NS-linked pathways. ET-respiratory capacity mediated uniquely by S-pathway was also evaluated inhibiting complex I by the addition of Rotenone (0.5 μM). Finally, residual oxygen consumption (ROX) was determined by the inhibition of complex III adding Antimycin A (2.5 μM) and this value was subtracted from O2 flux as a baseline for all respiratory states. The O2k-Software DatLab 7.4 was used for real-time data acquisition and analysis. Oxygen flux values were expressed relative to tissue wet weight per second (JO2, pmol mg-1 s-1).
4.12. Adipose tissue histology
Histological examination was done using 4 μm thick sections from formalin-fixed paraffin-embedded tissue samples, which were stained with hematoxylin and eosin (H&E) at the Pathology Department of the Hospital Clinic of Barcelona. Two-four μm thickness sections from paraffin embedded samples were used for immunohistochemistry at the Tumour Bank of the HCB-IDIBAPS Biobank (IDIBAPS, Barcelona) using the Leica Microsystems’ Bond-max™ automated immunostainer together with the Bond Polymer Refine Detection System (Leica Microsystems, Spain). Briefly, tissue sections were deparaffinized, and pretreated for antigen retrieval with citrate buffer, pH 6.0 20 min for Caspase 3 and Mac2. For macrophage immunostaining: primary monoclonal rat anti-murine antibody to Mac2 (Cedarlane Labs, Burlington, Ontario, Canada) (Research data repository) was used at 1:40,000 for 1 h, combined with a secondary rabbit anti-rat Ig at 1:3,000. For apoptosis: monoclonal rabbit anti-murine Caspase3 antibody (Cell Signaling Technology, Danvers, MA, USA) (Research data repository) was used at 1:500 for 1 h. Finally, samples were developed with diaminobenzidine and counterstained with hematoxylin.
4.13. Mitochondria electron microscopy
Anaesthetized animals were perfused transaortically with 50 mL heparinized saline (10 mg/L heparin in saline (0.9% NaCl)) followed by 100 mL of fixative solution (glutaraldehyde 2.5% + paraformaldehyde 2% in phosphate buffer 0.1 M pH = 7.4). Once perfused, a small piece of adipose tissue was collected and placed on vials with fixative solution for 24 h. Samples were rinsed four times with phosphate buffer, postfixed with 1% osmium tetroxide (EMS, Hatfield, PA, USA) for 2 h and rinsed with milliQ water. Samples were then dehydrated in an acetone series (50%, 70%, 90%, 96% and 100%, for 10 min each), and infiltrated and embedded in Epon resin (EMS, Hatfield, PA, USA). Ultrathin sections of 60 nm in thickness were obtained using a UC6 ultramicrotome (Leica, Buffalo Grove, IL, USA) and were stained with 2% uranyl acetate and lead citrate. Sections were observed in a Jeol 1010 (Gatan, Abitasu Yaesu, Japan) equipped with a tungsten cathode and Images were acquired at 80 kv with a CCD Megaview 1kx1k. Mitochondrial cristae density was evaluated distributing mitochondria in 4 categories: 0-not visible cristae; 1-very few cristae; 2-half of the mitochondria shows cristae, 3-equaly distributed cristae.
4.14. Whole mount adipose tissue immunostaining and vessel area quantification
Approximately 3 mm3 cubes of adipose tissue were cut from each sample and permeabilized for 1h with phosphate buffer saline (PBS) and 1% Triton X-100. Afterwards, tissues were blocked with blocking buffer (PBS + 0,3% Triton X-100 + 5% goat serum) for 2 h at room temperature. Primary antibody (Isolectin GS-B4 568, Invitrogen, Thermo Fisher Scientic, Waltham, MA, USA) (Research data repository) diluted (1:2000) in blocking buffer was incubated overnight at room temperature, with over day washings with PBS + 0,3% Triton-x 100. Afterwards, samples were clarified using 90% glycerol at 4 °C overnight. Samples were stored in ProLong™ Gold Antifade Reagent (P36930 Invitrogen, Thermo Fisher Scientic, Waltham, MA, USA) with DAPI. Images were taken with a Leica SP5 confocal microscope using 20X and 40X objectives (Leica, Buffalo Grove, IL, USA). All images are maximal z-stack projections. Images were processed using Volocity, Fiji and Adobe Photoshop CS5. Vessel density was quantified by measuring isolectin-B4 positive area, within a fixed square template, using the ImageJ software.
4.15. Phospholipid extraction
White adipose tissue (WAT) was homogenized in PBS of which 5% was removed to determine protein content via Bradford assay. Next, lipids were extracted twice from homogenate with 2:1 chloroform:methanol (Folch method) containing 0.5 μM CL(14:0)4 as internal standard and dried under N2 flow. Due to the high content of neutral lipids in WAT, phospholipids were separated by solid-phase extraction using Sep-Pak C18-columns (Waters Associates, Milford, MA, USA). Briefly, the lipid extract was re-suspended in 1 mL chloroform, applied on the column and washed thrice with 1 mL chloroform to elute neutral lipids. Phospholipids were eluted by quadruple addition of 2:1 chloroform:methanol and quadruple addition of methanol, collected and dried under N2 flow.
4.16. LC-MS/MS cardiolipins analysis
CL analysis was performed as described in Ref. [51]. Phospholipid extracts were dissolved in HPLC starting condition and subjected to HPLC-MS/MS analysis. Separation was achieved by reversed-phase HPLC with an Agilent Poroshell 120 EC-C8 2.7 μm 2.1 × 100mm column (Agilent Technologies, Santa Clara, CA, USA) on a Dionex Ultimate 3000 HPLC (Thermo Fisher Scientific, Waltham, MA, USA, 50 °C column oven, 0.4 mL/min flow) with running solvent A (60/40 Acetonitrile/H2O, 10 mM ammonium formate, 0.2% formic acid) and running solvent B (90/10 Isopropanol/Acetonitrile, 10 mM ammonium formate, 0.2% formic acid). Analytes were measured using a LTQ Velos MS (Thermo Fisher Scientific, Waltham, MA, USA) operated in negative ESI mode (3.8 kV, 275 °C capillary temperature, 460–1650 m/z) and data-dependent MS2 acquisition. Thermo raw data was converted to open-source MZML format and Peaks were integrated in MZmine2 [52]. Identification was based on a combination of accurate mass, (relative) retention times, and fragmentation spectra, compared to a library of standards. Data was corrected for internal standard, normalized on protein content, quantified using an external dilution series (CL(14:0)4, CL(18:1)4) and further analysed by an in-house pipeline in R. Significance was calculated using a one-way ANOVA with post hoc Tukey correction.
4.17. Quantification of mtDNA copy number
Mitochondrial DNA (mtDNA) content was determined by qPCR of total DNA extracted. NucleoSpin Tissue DNA purification kit (Macherey-Nagel, Dueren, Germany) was used to extract total DNA from adipose tissue according to manufacturer's protocol, and NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) to quantify DNA concentration. GoTaq® qPCR MasterMix (Promega, Madison, WI, USA) and 3 ng of DNA sample were combined to amplify both mtDNA and nuclear DNA by 7900HT RT-PCR system (Thermo Fisher Scientific, Waltham, MA, USA) using the following primers; cytochrome C subunit 2 (mt-Co2 Fw: CTACAAGACGCCACAT – mt-Co2 Rv: GAGAGGGGAGAGCAAT), 12S rRNA (Mt-Rnr1Fw: CTTCAGCAAACCCTAAAAAGG – Mt-Rnr1Rv: GCGTACTTCATTGCTCAATTC) and succinate dehydrogenase subunit A (Sdha Fw: TACTACAGCCCCAAGTCT - Sdha Rv: TGGACCCATCTTCTATGC). mtDNA content is referred to nuclear DNA (nDNA) as the copy number ratio of mtDNA to nDNA.
4.18. Metabolomics sample preparation
Pulverized liver tissue (50–100 mg) was mixed with ice-cold acetonitrile:water (1:1) and ultrasonicated, and the combined supernatant aqueous phases of 3 repetitions were frozen and lyophilized. The resultant pellet was dried and extracted with chloroform:methanol (2:1) by ultrasonication. The supernatant organic phase was collected and dried under N2 stream. Pulverized white adipose (30–70 mg), gastrocnemius (20–40 mg) and hypothalamus (10–25 mg) tissue was mixed with methanol and ultrasonicated before chloroform was added in two steps to final 1:1. Water was added 1:2:2 (water:methanol:chloroform) and the aqueous upper phase containing water and methanol and the organic lower phase were collected separately. An aqueous extract was dissolved in D2O (containing 0.67 mM trisilylpropionic, TSP) and the organic extract was reconstituted in CDCl3/CD3OD (2:1) solution (containing 1.18 mM tetramethylsilane, TMS). The supernatant of both phases was transferred into 5 mm NMR tubes.
4.19. Nuclear magnetic resonance (NMR) metabolomics analysis
1H NMR spectra were recorded at 310K on a Bruker Avance III 600 spectrometer (Bruker, Billerica, USA) operating at a proton frequency of 600.20 MHz using a 5 mm CPTCI triple resonance (1H, 13C, 31P). One-dimensional 1H pulse experiments of aqueous samples were performed. In order to suppress the residual water peak the experiments were recorded using the NOESY-presat sequence, with the mixing time set at 100 ms. A 75Hz-power irradiation was applied during recycling delay and mixing time to presaturate the solvent. A total of 256 transients were collected into 64 k data points for each 1H spectrum, being the spectral width set as 12 kHz (20 ppm). The exponential line broadening applied before Fourier transformation was of 0.3Hz. In order to remove the residual water moisture of deuterated methanol, lipidic samples were measured using a simple presaturation sequence. Thus, a 50Hz-power irradiation was used during recycling delay and mixing time to presaturate the solvent. Again, 256 transients were collected into 64 k data points for each 1H spectrum, being the spectral width set as 12 kHz (20 ppm). The exponential line broadening applied before Fourier transformation was also here of 0.3Hz. Of note, the frequency domain spectra were manually phased and baseline-corrected using TopSpin software (version 2.1, Bruker, Billerica, USA).
4.20. NMR data analysis
All the acquired 1H NMR spectra were phased, baseline-corrected, and referenced to the chemical shift of TSP or TMS signal at 0 ppm. For metabolic identification, references of pure compounds from the metabolic profiling AMIX spectra database (Bruker, Billerica, USA), human metabolome database (HMDB), and Chenomx databases were used (Chenomx Inc. Edmonton, Canada). After baseline correction, specific 1H NMR regions identified in the spectra were integrated for each extraction method entering the study using the AMIX 3.8 software package. Then, each integration region was normalized using the tissue weight used from each sample. Data (pre-) processing, data analysis, and statistical calculations were performed in RStudio (R version 3.0.2).
4.21. RNA-seq data processing and differential expression analysis
RNA samples from 20 WAT samples (Ctrl n = 8, HFD n = 4, Int n = 4 and Int2 n = 4) as well as 12 liver tissue samples (Ctrl n = 4, HFD n = 4 and Int n = 4) were sequenced by standard Illumina protocol to create raw sequence files (.fastq files), which underwent quality control analysis using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We aligned the quality checked reads to the Mouse Genome, version July 2007 (NCBI37/mm9) using TopHat version 2.1.1 allowing for unique alignments to the genome and up to two mismatches. The resulting alignments were summarized by Ensembl gene identifiers to evaluate number of uniquely aligned reads per transcript and per sample (raw read counts).
RNA-seq data were analysed using the limma package version 1.8.2 [53] available through the Bioconductor open source. The raw read counts were used as input to form a DGEList object combining the counts and associated annotation. Scale normalization was applied and the calculation of a normalized signal was performed by voom function of the limma package. This differential expression analysis was performed pair-wise between different experimental conditions, but per each tissue type separately. First, we identified genes either up- or downregulated in HFD conditions as compared to controls with a FDR<0.05. Then, we explored the expression modulation of these two gene sets after first intervention (Int). We could determine three distinct patterns: 1) genes fully recovered (with differential expression between HFD and Int but with no significant difference in expression between controls and Int); 2) genes partially recovered (not reaching the same expression level as in control conditions but showing the recovering tendency between HFD and Int); 3) genes which are not recovered (that continue to be differentially expressed between Int and controls, with no significant difference in expression between HFD and Int). In the case of WAT, for the third, not recovered category, we performed similar expression patterns analysis using the data of second intervention (Int2), defining the genes that are recovered or not after this additional experimental step.
4.22. Gene ontology analysis
For different gene sets, GO enrichments were performed using the GOstats R package [54], separately for biological processes and cell compartments related GO terms. Significant GO terms (p < 0.0001) were further summarized together by ReVigo software [55] in order to represent them as semantic-similarity scatter plot of the most significant GO terms, with a modification of the script to represent the GO enrichment values as dot size. To represent the cascade of GO terms affected in different steps of the experiment (HFD, Int and Int2, Fig. 5), we combined a biological and semantic approach, merging those GO terms that contained the same genes and that were related to the same biological process or cell compartment. In this analysis, general, ancestor GO terms (the highest level in GO terms tree; typically containing over 500 genes) as well as very specific GO terms (typically containing less than 5 genes) were excluded, in order to obtain a clearer view of the GO terms dynamics throughout three different experimental conditions.
4.23. Database and pattern data analysis
Gene expression (qPCR), metabolites content (metabolomics) and mitochondrial respiration states (high-resolution respirometry) patterns were analysed and are presented in Fig. 2. A Content Management System (CMS) dynamic database with a front-end user interface or a content management application (CMA) (idibaps.seeslab.net) was created along with a Content Delivery Application (CDA) that compiles all the information and updates the website. An entry for each mouse was generated including both phenotypical information (e.g., animal identification, group (Ctrl, HFD or Int), body weight, glucose levels (IGTT, ITT, etc) and specific-tissue data (e.g., mitochondrial respiratory values, gene expression data, protein content data, targeted metabolomics data, etc). Python scripts allowed correlation and comparison analysis of all the attributes entered in the database, which were structured in a pandas. DataFrame (http://pandas.pydata.org/) generated from the database. NumPy and SciPy were used for working with multi-dimensional arrays and matrices as well as for calculating high-level mathematical functions. Matplotlib was used for graphical representations (i.e., stacked bar plot). Pattern data analysis: to visually show the proportion of parameters that were reversed after the intervention we defined a 3-point vector for each parameter (e.g., gene expression, metabolite, mitochondrial respiration state) in which the 3 points correlated to the mean of this given parameter for 1) Ctrl group 2) HFD-fed group and 3) Int group (i.e., parameter=[Ctrl mean, HFD mean, Int mean]). Mann-Whitney comparisons were used to compare the groups in pairs. Once the significant differences were calculated, different patterns were defined: green for a reversion pattern after the intervention; red for no reversion in the intervention group, so the mean value in the HFD group, which is significantly different from the Ctrl group, is not different in comparison to the Int group; and grey for a difference in the Int group in comparison to the other two groups, between the ones there is no difference. A crossing pattern was added in the cases when there was no transitivity property. This happened in 3 scenarios: when Ctrl vs HFD showed no statistical difference, Ctrl vs Int showed no statistical difference, but HFD vs Int showed a statistical difference (Ctrl ≈ HFD, Ctrl ≈ Int, HFD≠Int); when Ctrl ≈ HFD, Ctrl≠Int, HFD ≈ Int; and when Ctrl≠HFD, Ctrl ≈ Int, HFD ≈ Int. Once these patterns were defined for gene expression, metabolites and mitochondrial respiration states in skeletal muscle, hypothalamus, liver and epididymal adipose tissue, stacked bar plots were used to visualize the relative percentage of all the genes/metabolites/respiratory states falling in the different patterns.
4.24. Proteomics eWAT sample preparation
Powdered fat pads were homogenized with ceramic beads (Percellys, 5500 rpm, 2 × 10 s, 30 s break) in the buffer containing 50 mM HEPES pH 7.4, 1% Triton X-100, 100 mM NaF, 10 mM sodium orthovanadate, 10 mM EDTA, 0.2% SDS, 100 mM NaCl. Homogenates were processed with Bioruptor (Diagenode SA; 10 min, cycle 30/30 s, at 4 C). Protein extracts were cleared from floating fat and tissue debris by centrifugation at 20000×g. 150 μg of proteins were precipitated with 4x volume of ice-cold acetone, incubated for 15 min at −80 C, followed by 120 min incubation at −20 C. Precipitated proteins were pelleted by 15 min centrifugation at 16,000×g and washed with ice-cold acetone twice. Air-dry pellets were resuspended in 8 M Urea/50 mM TAEB buffer supplemented with protease inhibitor cocktail (Sigma). 150 μg of the sample were reduced in 5 mM DTT at 25 C for 1 h, followed by treatment with 40 mM CAA for 30 min, in the dark, at room temperature. Lys-C protease was added at an enzyme:substrate ratio of 1:75 and incubated at 25 C for 4 h. The sample was diluted with 50 mM TEAB to a final concentration of 2 M Urea, and Trypsin was added at the ratio 1:75 followed by overnight digestion at 25 C. Sample was acidified with formic acid to a final concentration of 1%, and peptides were purified on the SDR-RP (C18) multi-stop-and-go-tip (Stage Tip) [56].
4.25. LCMS/MS proteomics eWAT analysis
LCMS/MS proteomics analysis was done as previously described [57]. Briefly, all samples were analysed on a Q Exactive Plus Orbitrap (Thermo Scientific) mass spectrometer that was coupled to an EASY nLC (Thermo Scientific). Peptides were loaded with solvent A (0.1% formic acid in water) onto an in-house packed analytical column (50 cm–75 μm I.D., filled with 2.7 μm Poroshell EC120C18, Agilent) and separated with 150 min gradients. The mass spectrometer was operated in data-dependent acquisition mode and MS1 survey scan was acquired from 300 to 1750 m/z at a resolution of 70,000. The top 10 most abundant peptides were isolated within a 1.8 Th window and subjected to HCD fragmentation using a collision energy of 27%. The AGC target was set to 5e5 charges, allowing a maximum injection time of 60 ms. Product ions were detected in the Orbitrap at a resolution of 17,500. Precursors were dynamically excluded for 20.0 s
All mass spectrometric raw data were processed with Maxquant (version 1.5.3.8) using default parameters. False discovery rates on protein and PSM level were estimated by the target-decoy approach to 1% (Protein FDR) and 1% (PSM FDR), respectively. The downstream comparative analysis was performed in Perseus (v1.6.1.1) [58] and the dataset was filtered for proteins quantified in all the samples in at least one experimental group. Missing values were imputed using a Gaussian distribution of random numbers (width: 30% of standard deviation, down-shift: 1.8 standard deviations). Comparative analysis between groups was performed using a t-test (FDR = 5% and S0 = 0.1). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (http://proteomecentral.proteomexchange.org) partner repository with the dataset identifier PXD025503.
4.26. Visceral adipose tissue samples from human cohort
Samples of omental visceral adipose tissue (AT) were obtained from 23 individuals (19 women, 4 men) in the context of the Leipzig obesity biobank and a 2-step bariatric surgery approach as described previously [13]. We included women and men with an age >18 years, a stable body weight 3 months prior to surgery (Mean ± SEM 152.5 ± 6.95 Kg; range: 92–236 Kg) without acute and severe infection, inflammatory or malignant diseases, with a BMI of 54.6 ± 2.00 (range: 37.1–78.9). All adipose tissue samples were collected during laparoscopic abdominal surgery as described previously [59]. Adipose tissue was immediately frozen in liquid nitrogen and stored at −80 °C. The study was approved by the Ethics Committee of the University of Leipzig (approval numbers: 363-10-13122010; 159-12-21052012), and performed in accordance to the declaration of Helsinki. All subjects gave written informed consent to use their data in anonymized form for research purposes before taking part in this study.
4.27. Human RNA-seq analysis
Human single-end and rRNA-depleted RNA-seq data were obtained thanks to the courtesy of Matthias Blüher lab (Helmholtz Zentrum München). Libraries were prepared with a SMARTseq protocol [60,61]. Briefly, RNA was enriched and reverse transcribed by Oligo(dT) and TSO primers and then, cDNA was amplified by ISPCR primers, tagmented with Tn5 using Nextera DNA Flex kit. Libraries were sequenced with Novaseq 6000. Adapters of raw reads were trimmed applying trimmomatic (v0.4) (10.1093/bioinformatics/btu170). Only reads which surpass a quality cut-off of 30 and a minimum read length of 36 nts were retained. Reads were mapped with the STAR alignment algorithm (10.1093/bioinformatics/bts635), allowing 100 multiple alignments per read. Standard pre- and post-mapping quality control was computed using FASTQC (v.0.11.4) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Gene count were conducted with featureCounts (v2.0.1) (10.1093/bioinformatics/btt656) where multiple mapped reads were fractionally counted. The human genome (GRCh38.p13) from GENCODE (10.1093/nar/gky955) served as reference. Data normalization (vst algorithm) and differential gene expression analysis (Wald algorithm) were computed using DESeq2 (v3.9) (10.18129/B9.bioc.DESeq2). As we observed high basal heterogeneity between individuals, we included sample pairing in the design to remove this batch effect. We therefore fitted an individual baseline for each patient in order to absorb patient-to-patient differences in expression before treatment. Genes with a FDR<0.1 were donated as differentially expressed. Gene ontology enrichments were performed applying the GOstats R-package (10.18129/B9.bioc.GOstats).
4.28. Statistical analysis
Results are expressed as mean ± SEM. The statistical significance among the three experimental groups was assessed using one-way ANOVA, and differences between means were subsequently tested by Tukey post-hoc test. Data curation involved the removal of values that were ±2 standard deviation (SD) in punctual situations. A p-value <0.05 was considered significant in all cases, meaning a confidence interval of 95% and setting significance level at α = 0.05. Tendencies with p-value between 0.05 and 0.07 are also indicated. All statistical analyses were performed using GraphPad 6 (GraphPad Software, Inc. La Jolla, CA, USA).
4.29. Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Pablo M. Garcia-Roves (pgarciaroves@ub.edu).
Research data
Research data corresponding to this manuscript are deposited in the following open-access repository: Mendeley Data, V1, doi: 10.17632/f38d6s33wm.1.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This work has been supported by Ministerio de Ciencia e Innovación (MICINN) Grant BFU2011- 24679 (P.M.G.-R.); Instituto de Salud Carlos III (ISCIII) Grant PI15/00701 (P.M.G.-R.) cofinanced by the European Regional Development Fund ‘‘A way to build Europe’‘; Government of Catalonia Support Grups de Recerca AGAUR 2017-SGR-204 (to J.C.P. and P.M.G-R.), 2017-SGR-736 (to J.I.M.-S.) and 2017-SGR-896 (to A.A.-M., M.S.-P., and R.G.). M.M. was recipient of a MINECO grant RTI2018-093864-B-I00. O.Y. was funded by MINECO grant BFU2017-87958. M.A.K. was funded by a Medical University of Innsbruck Start Grant. M.C. was funded by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement 725004). L. H. was funded by MINECO grants SAF2013-45887-R, SAF2017-83813-C3-1-R (granted to Dolors Serra (DS) and L.H.), co-funded by the European Regional Development Fund, the Centro de Investigación Biomédica en Red de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN) (Grant CB06/03/0001 to DS), the Government of Catalonia (2017SGR278 to DS), the Fundació La Marató de TV3 (201627–30 to DS), the European Foundation for the Study of Diabetes (EFSD)/Janssen-Rising Star and L’Oréal-UNESCO “For Women in Science” research fellowships. P.M.G.-R. was a recipient of a Ramon y Cajal contract (RYC-2009-05158) from MICINN. A.G-F. was a recipient of “Beca de Formació de Personal Investigador de l'IDIBAPS” fellowship and she is currently supported by an unconditional donation from the Novo Nordisk Foundation (NNF) to NNF Center for Basic Metabolic Research (NNF18CC0034900). P.G-P. was a recipient of predoctoral fellowship from the Universitat de Barcelona (APIF-UB). K.S was financed through “Advanced Postdoc Grant” from SFB 1218 (DFG, German Reuter Foundation, Projektnummer 269925409). J.M-B. was a recipient of predoctoral fellowship from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 675610. E.S-R. was recipient of a predoctoral fellowship from Ministerio de Economia y Empresa (MINECO) grant BES-2013-062796. This work was partially developed at the Esther Koplowitz Center (CEK, Barcelona, Spain). We want to acknowledge the following experts: Drs. Vidal-Puig A., Malagon M.M. and Raimundo N. for their advice and critical discussion of our work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102353.
Appendix ASupplementary data
The following are the Supplementary data to this article:
Fig. S1.
A: Insulin tolerance test in fasted mice (Ctrl n = 9; HFD n = 6; Int n = 6). B: Beta-cell size distribution per pancreas area (Ctrl n = 6; HFD n = 4; Int n = 4). Data represented as mean ± SEM or box-and-whiskers plots (median, 25th to 75th percentiles and max/min values) ANOVA One-way and Post-hoc Tukey, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Fig. S2.
A: Detailed representation of Fig. 2A uncovering sub-patterns within each group. Different colors indicate the different sub-patterns specified in B that correspond to patterns in Fig. 2A. B: Patterns are grouped in 4 different colors for Fig. 2A simplification. Mann-Whitney comparisons were used to compare the groups in pairs. Once the significant differences were calculated, different patterns were defined: green for a reversion pattern after the intervention; red for no reversion in the intervention group, so the mean value in the HFD group, which is significantly different from the Ctrl group, is not different in comparison to the Int group; and grey for a difference in the Int group in comparison to the other two groups, between the ones there is no difference. A crossing pattern was added in the cases when there was no transitivity property. This happened in 3 scenarios: when Ctrl vs HFD showed no statistical difference, Ctrl vs Int showed no statistical difference, but HFD vs Int showed a statistical difference (Ctrl≈HFD, Ctrl≈Int, HFD≠Int); when Ctrl≈HFD, Ctrl≠Int, HFD≈Int; and when Ctrl≠HFD, Ctrl≈Int, HFD≈Int. C: Table describing the n size used for the analysis of each experimental readout in every tissue under study.
Fig. S3.
A: Gene ontology cellular components (GOCC) enrichment analysis for the 1369 transcripts downregulated in HFD and Int. B: Percentage of mitochondria within the different mitochondrial cristae frequency categories according to Fig. 3C (Ctrl n = 3, HFD n = 4, Int n = 4). C: Mitochondrial fractional area calculated as mitochondrial area (μm2)/cytoplasm area (μm2) (Ctrl n = 3, HFD n = 4, Int n = 4). D: Mitochondrial aspect ratio calculated as the major axis/minor axis of the mitochondria ellipse shape (Ctrl n = 3, HFD n = 4, Int n = 4). Data represented as box-and-whiskers plots (median, 25th to 75th percentiles and max/min values).
Fig. S4.
A: Scheme with the 4 experimental groups indicating the non-reverted parameters after the first intervention (Int) and the rationale to then include a second intervention (Int2). B: Body weight evolution in the 4 experimental groups. Body weight of those Ctrl animals extended to match Int2 animals' age was recorded at final point (26–27 weeks), and dashed line indicates its stabilization along this period. C: Intraperitoneal glucose tolerance test (IGTT) (Ctrl n = 82, HFD n = 168, Int n = 32, Int2 n = 19). The graph displayed combines data from different representative cohorts, so variation in n size is due to it and space limitations imposed by the treadmill for the intervention program (6 lanes, n = 12 maximum per cohort in order to avoid time differences). D: Representative images for Caspase3-and Mac2-positive cells staining in eWAT for each experimental group (Ctrl n = 3–4, HFD n = 4, Int n = 3–4, Int2 n = 4–5). E-F: Vegfa (E) and Vegfr (F) normalized eWAT gene expression (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). G-H: Gene ontology biological processes (G) and cellular components (H) enrichment of the genes clustered in 4L that revert after Int2. I: PCA displaying the segregation of the adipose tissue proteome from the experimental groups Ctrl, HFD and Int2. J: KEGG pathways enriched in both recovered and not recovered proteins (Int2 vs HFD) showed in 4O. K-L: Volcano plot displaying mitochondrial-related proteins in the comparison HFD vs Ctrl (K) and Int2 vs HFD (L). Specific GOCC terms of mitochondrial locations/structures are highlighted in both graphs. (n = 3/group). M-P: Normalized gene expression of Pla2g6 (M), Lpgat1 (N), Lclat1 (O) and Hsd17b10 (P) in eWAT (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). Ctrl: control; HFD: high fat diet-fed; Int: intervention; Int2: second intervention. Data represented as mean ± SEM or as box-and-whiskers plots (median, 25th to 75th percentiles and max/min values). ANOVA One-way and Post-hoc Tukey, significant differences with respect to Ctrl *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Fig. S5.
A: Z-score for RNA-seq clusters of transcripts sequentially downregulated across the different experimental groups (Ctrl n = 8, HFD n = 4, Int n = 4, Int2 n = 4). B: Cumulative number of genes belonging to different enriched GOBP terms for each cluster in A. The same GOBP terms are indicated from HFD-cluster to Int2-cluster (y-axis), with increasing size (x-axis) denoting incremental number of genes for that specific term.
Fig. s6.
References
- 1.Bluher M. Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019;15:288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 2.Heymsfield S.B., Wadden T.A. Mechanisms, pathophysiology, and management of obesity. N. Engl. J. Med. 2017;376:1492. doi: 10.1056/NEJMc1701944. [DOI] [PubMed] [Google Scholar]
- 3.van Baak M.A., Mariman E.C.M. Mechanisms of weight regain after weight loss - the role of adipose tissue. Nat. Rev. Endocrinol. 2019;15:274–287. doi: 10.1038/s41574-018-0148-4. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gumeni S., Trougakos I.P. Cross talk of proteostasis and mitostasis in cellular homeodynamics, ageing, and disease. Oxid. Med. Cell. Longev. 2016 doi: 10.1155/2016/4587691. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn O., Drews L.F., Nguyen A., Tatsuta T., Gkioni L., Hendrich O., Zhang Q., Langer T., Pletcher S., Wakelam M.J.O., et al. A nutritional memory effect counteracts benefits of dietary restriction in old mice. Nat. Metab. 2019;1:1059–1073. doi: 10.1038/s42255-019-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saha S., Gerdtham U.G., Johansson P. Economic evaluation of lifestyle interventions for preventing diabetes and cardiovascular diseases. Int. J. Environ. Res. Publ. Health. 2010;7:3150–3195. doi: 10.3390/ijerph7083150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinert M., Clemmensen C., Hofmann S.M., Moore M.C., Renner S., Woods S.C., Huypens P., Beckers J., de Angelis M.H., Schurmann A., et al. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 2018;14:140–162. doi: 10.1038/nrendo.2017.161. [DOI] [PubMed] [Google Scholar]
- 9.Fonseca V.A. Defining and characterizing the progression of type 2 diabetes. Diabetes Care. 2009;32(Suppl 2):S151–S156. doi: 10.2337/dc09-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galgani J.E., Moro C., Ravussin E. Metabolic flexibility and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1009–E1017. doi: 10.1152/ajpendo.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priest C., Tontonoz P. Inter-organ cross-talk in metabolic syndrome. Nature Metabol. 2019;1:1177–1188. doi: 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- 12.Bäckhed F., Bugianesi E., Christofk H., Dikic I., Gupta R., Mair W.B., O'Neill L.A.J., Ralser M., Sabatini D.M., Tschöp M. The next decade of metabolism. Nature Metabol. 2019;1:2–4. doi: 10.1038/s42255-018-0022-7. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz J., Evers N., Awazawa M., Nicholls H.T., Bronneke H.S., Dietrich A., Mauer J., Bluher M., Bruning J.C. Obesogenic memory can confer long-term increases in adipose tissue but not liver inflammation and insulin resistance after weight loss. Mol. Metabol. 2016;5:328–339. doi: 10.1016/j.molmet.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrill J.S., Long E.K., Reilly B., Deng Y., Armitage I.M., Scherer P.E., Bernlohr D.A. Inflammation and ER stress regulate branched-chain amino acid uptake and metabolism in adipocytes. Mol. Endocrinol. 2015;29:411–420. doi: 10.1210/me.2014-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrada-Alcalde I., Tenorio-Guzman M.R., Tovar A.R., Salinas-Rubio D., Torre-Villalvazo I., Torres N., Noriega L.G. Metabolic fate of branched-chain amino acids during adipogenesis, in adipocytes from obese mice and C2C12 myotubes. J. Cell. Biochem. 2017;118:808–818. doi: 10.1002/jcb.25755. [DOI] [PubMed] [Google Scholar]
- 16.Brandt T., Mourier A., Tain L.S., Partridge L., Larsson N.G., Kuhlbrandt W. Changes of mitochondrial ultrastructure and function during ageing in mice and Drosophila. Elife. 2017;6 doi: 10.7554/eLife.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carobbio S., Pellegrinelli V., Vidal-Puig A. Adipose tissue function and expandability as determinants of lipotoxicity and the metabolic syndrome. Adv. Exp. Med. Biol. 2017;960:161–196. doi: 10.1007/978-3-319-48382-5_7. [DOI] [PubMed] [Google Scholar]
- 18.Sun K., Wernstedt Asterholm I., Kusminski C.M., Bueno A.C., Wang Z.V., Pollard J.W., Brekken R.A., Scherer P.E. Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5874–5879. doi: 10.1073/pnas.1200447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wernstedt Asterholm I., Tao C., Morley T.S., Wang Q.A., Delgado-Lopez F., Wang Z.V., Scherer P.E. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metabol. 2014;20:103–118. doi: 10.1016/j.cmet.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crewe C., An Y.A., Scherer P.E. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J. Clin. Invest. 2017;127:74–82. doi: 10.1172/JCI88883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung D.Y., Ko H.J., Lichtman E.I., Lee E., Lawton E., Ong H., Yu K., Azuma Y., Friedline R.H., Lee K.W., et al. Short-term weight loss attenuates local tissue inflammation and improves insulin sensitivity without affecting adipose inflammation in obese mice. Am. J. Physiol. Endocrinol. Metab. 2013;304:E964–E976. doi: 10.1152/ajpendo.00462.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller R.S., Becker K.G., Prabhu V., Cooke D.W. Adipocyte gene expression is altered in formerly obese mice and as a function of diet composition. J. Nutr. 2008;138:1033–1038. doi: 10.1093/jn/138.6.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamarron B.F., Mergian T.A., Cho K.W., Martinez-Santibanez G., Luan D., Singer K., DelProposto J.L., Geletka L.M., Muir L.A., Lumeng C.N. Macrophage proliferation sustains adipose tissue inflammation in formerly obese mice. Diabetes. 2017;66:392–406. doi: 10.2337/db16-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derous D., Kelder T., van Schothorst E.M., van Erk M., Voigt A., Klaus S., Keijer J., Radonjic M. Network-based integration of molecular and physiological data elucidates regulatory mechanisms underlying adaptation to high-fat diet. Genes Nutr. 2015;10:470. doi: 10.1007/s12263-015-0470-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaum N., Lehallier B., Hahn O., Palovics R., Hosseinzadeh S., Lee S.E., Sit R., Lee D.P., Losada P.M., Zardeneta M.E., et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature. 2020;583:596–602. doi: 10.1038/s41586-020-2499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jankovic A., Korac A., Buzadzic B., Otasevic V., Stancic A., Daiber A., Korac B. Redox implications in adipose tissue (dys)function--A new look at old acquaintances. Redox Biol. 2015;6:19–32. doi: 10.1016/j.redox.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chicco A.J., Sparagna G.C. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Romestaing C., Han X., Li Y., Hao X., Wu Y., Sun C., Liu X., Jefferson L.S., Xiong J., et al. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell Metabol. 2010;12:154–165. doi: 10.1016/j.cmet.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Ye B., Miller S., Yuan H., Zhang H., Tian L., Nie J., Imae R., Arai H., Li Y., et al. Ablation of ALCAT1 mitigates hypertrophic cardiomyopathy through effects on oxidative stress and mitophagy. Mol. Cell Biol. 2012;32:4493–4504. doi: 10.1128/MCB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song C., Zhang J., Qi S., Liu Z., Zhang X., Zheng Y., Andersen J.P., Zhang W., Strong R., Martinez P.A., et al. Cardiolipin remodeling by ALCAT1 links mitochondrial dysfunction to Parkinson's diseases. Aging Cell. 2019;18 doi: 10.1111/acel.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sies H. Oxidative eustress: on constant alert for redox homeostasis. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filograna R., Mennuni M., Alsina D., Larsson N.G. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. 2020 doi: 10.1002/1873-3468.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel S., Wanet A., De Pauw A., Rommelaere G., Arnould T., Renard P. Crosstalk between mitochondrial (dys)function and mitochondrial abundance. J. Cell. Physiol. 2012;227:2297–2310. doi: 10.1002/jcp.23021. [DOI] [PubMed] [Google Scholar]
- 34.Boos F., Labbadia J., Herrmann J.M. How the mitoprotein-induced stress response safeguards the cytosol: a unified view. Trends Cell Biol. 2020;30:241–254. doi: 10.1016/j.tcb.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 35.An Y.A., Crewe C., Asterholm I.W., Sun K., Chen S., Zhang F., Shao M., Funcke J.B., Zhang Z., Straub L., et al. Dysregulation of amyloid precursor protein impairs adipose tissue mitochondrial function and promotes obesity. Nat. Metab. 2019;1:1243–1257. doi: 10.1038/s42255-019-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deus C.M., Yambire K.F., Oliveira P.J., Raimundo N. Mitochondria-lysosome crosstalk: from physiology to neurodegeneration. Trends Mol. Med. 2020;26:71–88. doi: 10.1016/j.molmed.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Saez I., Vilchez D. The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genom. 2014;15:38–51. doi: 10.2174/138920291501140306113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terman A., Gustafsson B., Brunk U.T. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem. Biol. Interact. 2006;163:29–37. doi: 10.1016/j.cbi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Serrano M., Camafeita E., Lopez J.A., Rubio M.A., Breton I., Garcia-Consuegra I., Garcia-Santos E., Lago J., Sanchez-Pernaute A., Torres A., et al. Differential proteomic and oxidative profiles unveil dysfunctional protein import to adipocyte mitochondria in obesity-associated aging and diabetes. Redox Biol. 2017;11:415–428. doi: 10.1016/j.redox.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Perez R., Garcia-Santos E., Ortega-Delgado F.J., Lopez J.A., Camafeita E., Ricart W., Fernandez-Real J.M., Peral B. Attenuated metabolism is a hallmark of obesity as revealed by comparative proteomic analysis of human omental adipose tissue. J. Proteonomics. 2012;75:783–795. doi: 10.1016/j.jprot.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Perez R., Ortega-Delgado F.J., Garcia-Santos E., Lopez J.A., Camafeita E., Ricart W., Fernandez-Real J.M., Peral B. Differential proteomics of omental and subcutaneous adipose tissue reflects their unalike biochemical and metabolic properties. J. Proteome Res. 2009;8:1682–1693. doi: 10.1021/pr800942k. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Q., Fu Z., Gong Y., Seshachalam V.P., Li J., Ma Y., Liang H., Guan W., Lin S., Ghosh S., et al. Metabolic health status contributes to transcriptome alternation in human visceral adipose tissue during obesity. Obesity. 2020;28:2153–2162. doi: 10.1002/oby.22950. [DOI] [PubMed] [Google Scholar]
- 43.Burhans M.S., Hagman D.K., Kuzma J.N., Schmidt K.A., Kratz M. Contribution of adipose tissue inflammation to the development of type 2 diabetes mellitus. Compr. Physiol. 2018;9:1–58. doi: 10.1002/cphy.c170040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Kolk B.W., Muniandy M., Kaminska D., Alvarez M., Ko A., Miao Z., Valsesia A., Langin D., Vaittinen M., Paakkonen M., et al. Differential mitochondrial gene expression in adipose tissue following weight loss induced by diet or bariatric surgery. J. Clin. Endocrinol. Metab. 2021 doi: 10.1210/clinem/dgab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mardinoglu A., Heiker J.T., Gartner D., Bjornson E., Schon M.R., Flehmig G., Kloting N., Krohn K., Fasshauer M., Stumvoll M., et al. Extensive weight loss reveals distinct gene expression changes in human subcutaneous and visceral adipose tissue. Sci. Rep. 2015;5 doi: 10.1038/srep14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfluger P.T., Herranz D., Velasco-Miguel S., Serrano M., Tschop M.H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfluger P.T., Kirchner H., Gunnel S., Schrott B., Perez-Tilve D., Fu S., Benoit S.C., Horvath T., Joost H.G., Wortley K.E., et al. Simultaneous deletion of ghrelin and its receptor increases motor activity and energy expenditure. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G610–G618. doi: 10.1152/ajpgi.00321.2007. [DOI] [PubMed] [Google Scholar]
- 48.Weir J.B. New methods for calculating metabolic rate with special reference to protein metabolism. J. Physiol. 1949;109:1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canto C., Garcia-Roves P.M. High-resolution respirometry for mitochondrial characterization of Ex vivo mouse tissues. Curr. Protoc. Mol. Biol. 2015;5:135–153. doi: 10.1002/9780470942390.mo140061. [DOI] [PubMed] [Google Scholar]
- 50.Gnaiger E. Mitochondrial respiratory states and rates. MitoFit Preprint Arch. 2019;10 26124/mitofit:190001.v6. [Google Scholar]
- 51.Oemer G., Lackner K., Muigg K., Krumschnabel G., Watschinger K., Sailer S., Lindner H., Gnaiger E., Wortmann S.B., Werner E.R., et al. Molecular structural diversity of mitochondrial cardiolipins. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4158–4163. doi: 10.1073/pnas.1719407115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falcon S., Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 55.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 57.Szczepanowska K., Senft K., Heidler J., Herholz M., Kukat A., Hohne M.N., Hofsetz E., Becker C., Kaspar S., Giese H., et al. A salvage pathway maintains highly functional respiratory complex I. Nat. Commun. 2020;11:1643. doi: 10.1038/s41467-020-15467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyanova S., Cox J. Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 2018;1711:133–148. doi: 10.1007/978-1-4939-7493-1_7. [DOI] [PubMed] [Google Scholar]
- 59.Kloting N., Fasshauer M., Dietrich A., Kovacs P., Schon M.R., Kern M., Stumvoll M., Bluher M. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 2010;299:E506–E515. doi: 10.1152/ajpendo.00586.2009. [DOI] [PubMed] [Google Scholar]
- 60.Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 61.Song Y., Milon B., Ott S., Zhao X., Sadzewicz L., Shetty A., Boger E.T., Tallon L.J., Morell R.J., Mahurkar A., et al. A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genom. 2018;19:696. doi: 10.1186/s12864-018-5066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.WHO https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight.