Abstract
Satellite cells (SCs), the adult Pax7-expressing stem cells of skeletal muscle, are essential for muscle repair. However, in vitro investigations of SC function are challenging due to isolation-induced SC activation, loss of native quiescent state, and differentiation to myoblasts. In the present study, we optimized methods to deactivate in vitro expanded human myoblasts within a 3D culture environment of engineered human skeletal muscle tissues (“myobundles”). Immunostaining and gene expression analyses revealed that a fraction of myoblasts within myobundles adopted a quiescent phenotype (3D-SCs) characterized by increased Pax7 expression, cell cycle exit, and activation of Notch signaling. Similar to native SCs, 3D-SC quiescence is regulated by Notch and Wnt signaling while loss of quiescence and reactivation of 3D-SCs can be induced by growth factors including bFGF. Myobundle injury with a bee toxin, melittin, induces robust myofiber fragmentation, functional decline, and 3D-SC proliferation. By applying single cell RNA-sequencing (scRNA-seq), we discover the existence of two 3D-SC subpopulations (quiescent and activated), identify deactivation-associated gene signature using trajectory inference between 2D myoblasts and 3D-SCs, and characterize the transcriptomic changes within reactivated 3D-SCs in response to melittin-induced injury. These results demonstrate the ability of an in vitro engineered 3D human skeletal muscle environment to support the formation of a quiescent and heterogeneous SC population recapitulating several aspects of the native SC phenotype, and provide a platform for future studies of human muscle regeneration and disease-associated SC dysfunction.
Keywords: Engineered skeletal muscle, Human, Satellite cells, scRNA-seq, Quiescence, Activation
1. Introduction
Satellite cell function is an essential aspect of skeletal muscle growth and repair [1-3]. Mechanistic studies of SC behavior and function, such as the roles of Notch activation in maintaining SC quiescence and self-renewal [4,5], MAPK signaling in SC activation [6-8], and canonical Wnt signaling in SC differentiation [9,10] have relied on the use of mouse models, which although highly valuable, may not fully reflect SC function in human muscle, especially in disease and aging [11-13]. Recently, single-cell RNA-sequencing (scRNA-seq) studies of human muscle tissue samples have demonstrated a large diversity of resident muscle cells including the existence of two distinct SC populations expressing Pax7 [14], a transcription factor associated with regenerative potential in adult muscle [1]. While in vitro studies could provide additional insights in the human SC homeostasis and injury response, enzymatic muscle dissociation to isolate SCs induces their immediate exit from quiescence (activation), followed by loss of Pax7 expression, proliferation, differentiation [15,16], and, eventually, replicative senescence [17]. To be able to study human SCs in their native quiescent state in vitro, isolated activated SCs must be deactivated without inducing irreversible senescence. Critical to this objective is the ability to recapitulate the cellular, chemical, and physical aspects of the native SC microenvironment (niche) to which SCs are acutely sensitive [18].
Previously, culture of human SCs in a “quiescence media” on integrin-coated collagen I fibers that acted as a niche mimetic yielded reversible SC quiescence for only 3–4 days, likely because of the lack of key constituents of the native SC niche, such as the myofibers [19]. The presence of myotubes in 2D cultures enables the formation of Pax7+ ‘reserve cells’ (RCs) that possess certain characteristics of quiescent SCs [20,21]. RCs have been used in vitro to study the effects of glucose [22] and Notch [23] and Wnt [24] pathways in SC formation and quiescence. Additionally, recent advances in tissue engineering methodologies have enabled the development of biomimetic, 3D skeletal muscle tissues containing a pool of Pax7+ cells that allow studies of muscle injury response [25-32]. Several years ago, we optimized a method for engineering functional 3D human skeletal muscle tissues (‘myobundles’) made from primary myoblasts expanded in 2D culture [25,33-35]. By two weeks of 3D culture, the myobundles harbor a pool of Pax7+ SC-like cells abutting aligned differentiated myofibers and demonstrate physiological responses to electrical and acetylcholine stimulation [25], drugs [25,34], exercise [33], and inflammatory factors [35]. Considering that the formation of myobundles requires phenotypic transitions of 2D-expanded myoblasts into both functional myofibers and Pax7+ SC-like cells, we reasoned that this system would allow us to study in detail the process of human myoblast deactivation, whereby a quiescent SC phenotype is attained via cell cycle exit and programming into a homeostatic state, as well as transcriptional changes during SC activation in response to injury. Furthermore, while the function of native SC niche in homeostasis and muscle repair is supported by a number of resident non-muscle cells (e.g. vascular, neuronal, fibrogenic, immune cells) [36,37], we sought to reveal specific roles that muscle fibers, a critical niche constituent, have in human SC maintenance and injury-induced activation by utilizing human myobundles made from purified myogenic cells.
To enable these studies, we first optimized conditions to maximize the formation of myobundle-hosted Pax7+ cells (3D-SCs), and characterized their expression of myogenic regulatory factors, cell cycle, and Notch signaling genes relative to the starting population of 2D-expanded myoblasts. We then evaluated the functional roles of Notch and Wnt signaling in deactivation of 2D myoblasts into 3D-SCs and identified a robust injury method for 3D-SC re-activation. We subsequently applied scRNA-seq analysis - for the first time in engineered skeletal muscle tissues - to reveal 3D-SC heterogeneity reminiscent of that found in native human skeletal muscle [14], define and characterize the deactivation process from 2D myoblasts to 3D-SCs, and reveal transcriptional changes within the activated 3D-SC population reflective of native SC activation in vivo [37-40]. Together, our results provide a foundation for the future in vitro studies of human SC formation, quiescence, heterogeneity, and re-activation in various pathophysiological contexts.
2. Results
2.1. Optimizing 2D myoblast purity to increase Pax7+ cell density in 3D myobundles
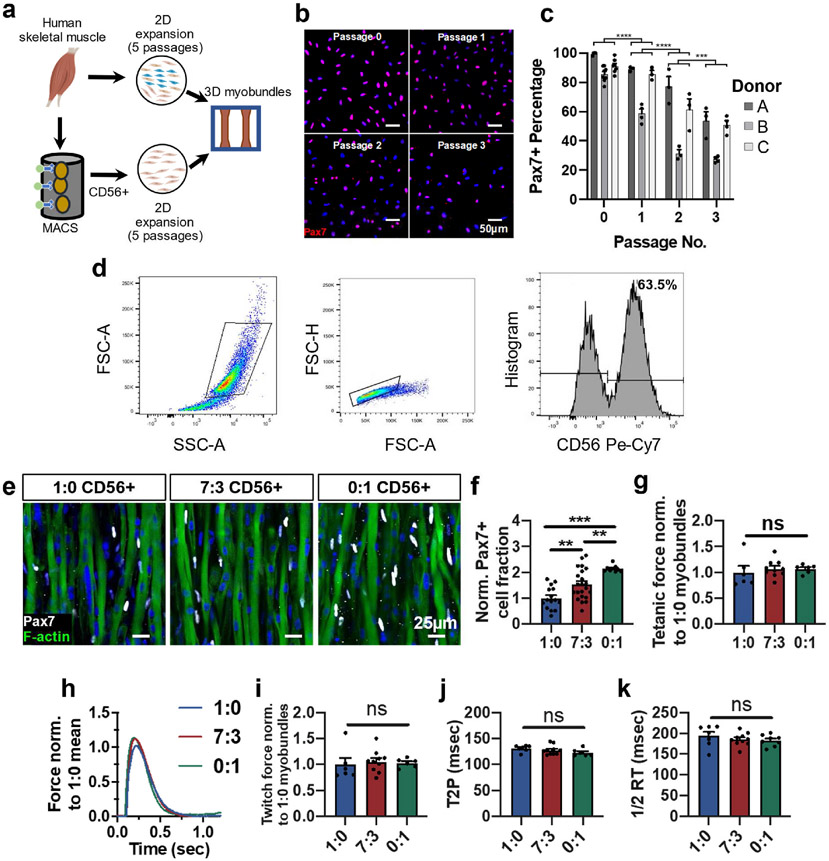
Traditionally, engineered 3D skeletal muscle tissues made from primary muscle cells are constructed using a heterogeneous cell population containing both myoblasts and non-myogenic cells [25,31,33]. While non-myogenic cells such as fibroblasts [41], endothelial cells [42], and fibro-adipogenic progenitors [43] play important roles in regulating the SC microenvironment in vivo, dissociated cells from different muscle donors contain varying proportions and makeups of non-myogenic cells, making it challenging to assess consequences on SC quiescence and function. Furthermore, in the context of in vitro myogenesis, the non-myogenic cells may interfere with fusion and early formation of myotubes that are necessary for homing of SCs [44]. We therefore sought to test the effects of using a pure population of CD56+ myogenic cells [45-47] on the formation of Pax7+ SCs within myobundles. Magnetic-activated cell sorting (MACS) of CD56+ myoblasts from muscle biopsies (Fig. 1a) yielded highly pure myogenic cells across multiple donors (91 ± 4.8% Pax7+, mean ± SEM) (Fig. 1b and c). Subsequent continuous expansion of these cells in 2D culture using standard growth media [25] significantly reduced Pax7+ cell fraction (Fig. 1b and c), as previously shown [17]. We then generated myobundles using CD56+ cells expanded for 5 passages and incorporated at 0%, 30%, or 100% of the starting cell mixture—a heterogeneous muscle population initially composed of approximately 66% CD56+ cells (Fig. 1d). This increase in starting CD56+ cell fraction yielded an increase in Pax7+ cells in 1-week differentiated myobundles (Fig. 1e and f) without effects on tetanic force generation (Fig. 1g, mean absolute force for all groups pooled: 3.87 ± 0.15 mN) or twitch amplitude, time-to-peak tension, and half-relaxation time (Fig. 1h-k). These results suggested that increasing the purity of the starting myoblast population increased the Pax7+ percentage in 3D myobundles and that the Pax7+ cell pool within myobundles could be formed in the absence of non-myogenic CD56−cells.
Fig. 1. CD56-based myoblast purification to increase formation of Pax7+ cells in myobundles.
a, Schematic of experimental flow from primary muscle cell isolation to 2D culture (unsorted or CD56+ MACS enrichment) and myobundle construction. b,c, Representative images of Pax7 staining in expanded CD56+ cells (nuclei stained with DAPI) at specified passages (b) and corresponding quantification (c, n = 3–6 coverslips per passage per donor, N = 3 donors). d, Representative flow cytometry analysis of muscle cell population expanded for 5 passages from muscle biopsy, highlighting the CD56+ cell fraction. e,f Representative whole bundle stains of 1-week differentiated myobundles with varying ratios (1:0, 7:3, and 0:1) of initial heterogeneous cells to CD56+ cells stained for Pax7 and F-actin (e) and corresponding quantification of Pax7 cell fraction normalized to 1:0 group (f, N = 1 donor, n = 9–22 myobundles per group). g, Tetanic force of 1-week differentiated myobundles normalized to 1:0 group (N = 1 donor, n = 4–10 myobundles per group). h, Representative twitch force trace from 1-week differentiated myobundles comprised with varying ratios (1:0, 7:3, and 0:1) of initial heterogeneous cells to CD56+ cells normalized to the mean twitch force of 1:0 myobundles and quantified twitch force (i), time to peak tension (T2P, j), and half-relaxation time (1/2 RT, k) (N = 1 donor, n = 6–10 myobundles per group). Data: mean ± SEM. *p < .05, **p < .01, ***p < .001; ns, not significant.
2.2. Optimizing 2D myoblast expansion to increase Pax7+ cell density in myobundles
To improve the expansion capacity of CD56+ myoblasts in long-term 2D culture (Supplementary Fig. 1a), we evaluated the treatment with 10 μM p38 inhibitor SB203580 (p38i), which was previously shown to increase PAX7 expression and engraftment potential of human myoblasts, but was only applied for up to 1 week in vitro [48]. We found that p38i-treated human myoblasts proliferated more rapidly than vehicle control (DMSO)-treated cells to undergo a 216-fold cell expansion between passages 3 and 5 (Supplementary Fig. 1b). Despite increased myoblast proliferation, p38i-treatment did not enhance PAX7 gene expression (Supplementary Fig. 1c), in contrast to previous findings in short-term culture [19]. To more accurately assess the gene expression of the Pax7+ cell pool in the myobundles, we enzymatically dissociated 1-week differentiated myobundles and strained the resulting cell lysate through a 30 μm filter to enrich for small mononuclear cells while removing differentiated myofibers. Relative to vehicle (DMSO) or p38i-expanded myoblasts, mononuclear cells isolated from corresponding 3D myobundles showed significantly enhanced PAX7 expression (Supplementary Fig. 1c). Additionally, compared to vehicle control, p38i treatment during 2D expansion significantly increased the fraction of Pax7+ cells in myobundles by 33 ± 0.07% (mean ± SEM, Supplementary Figs. 1d and e). Furthermore, after 1 week of 3D differentiation, myobundles derived from p38i-expanded myoblasts exhibited reduced force production, likely due to the inhibitory effects of p38i on differentiation during 2D expansion [49,50] that persisted during 3D differentiation in myobundles (Supplementary Fig. 1f). Collectively, these studies showed that a Pax7+ cell pool within engineered 3D myobundles can be amplified by chronic p38i treatment during 2D myoblast expansion.
2.3. Cell cycle arrest and myogenic regulatory factor expression during myobundle formation and differentiation
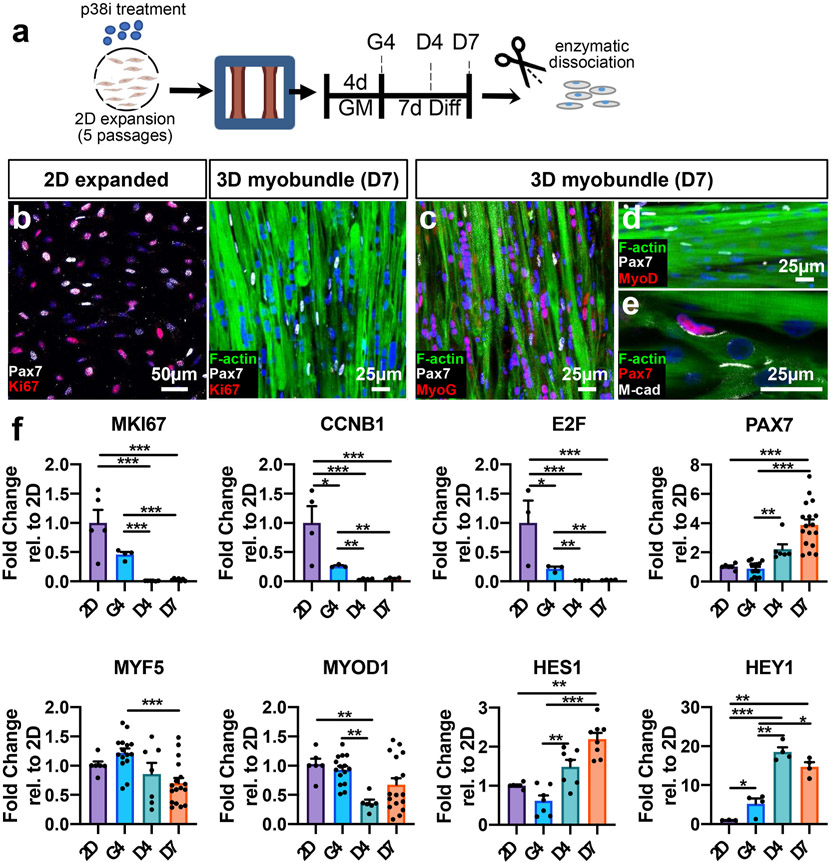
To further determine how 3D myobundle environment influences deactivation of p38i-expanded CD56+ myoblasts, we isolated mononuclear cells from myobundles at different time points of culture and characterized them for markers of cell cycle arrest, expression of myogenic regulatory factors, and Notch signaling-associated genes [51] (Fig. 2a). Immunostaining analyses revealed that similar to quiescent SCs in vivo and in contrast to 2D expanded cells, Pax7+ cells in 1-week differentiated myobundles were non-proliferative (Pax7+/Ki67−) (Fig. 2b) and negative for downstream markers of myogenic commitment, MyoD and MyoG (Fig. 2c-d). Furthermore, M-cadherin, a cell-cell adhesion protein, was expressed at the interface of Pax7+ cells and myofibers similar to observations in vivo [52] (Fig. 2e). We then assessed gene expression in 2D myoblasts and mononuclear cells isolated from 3D myobundles at 4 days of growth culture (G4), 4 days of differentiation (D4), and 7 days of differentiation (D7) (Fig. 2f). We found that expression of proliferation genes MKI67, E2F, and CCNB1 declined with the transition from 2D to 3D growth and differentiation (Fig. 2f), consistent with immunohistological findings (Fig. 2b). This exit from cell cycle was associated with a progressive increase in PAX7 gene expression in 3D mononuclear cells (Fig. 2f) as well as time-dependent changes in MYF5 and MYOD1 expression (Fig. 2f), suggestive of a heterogeneous and dynamic nature of the mononuclear cell population in myobundles. In addition, gene expression of downstream Notch signaling targets, HES1 and HEY1, was found to progressively increase (Fig. 2f). Collectively, these results suggested that upon transition to a 3D culture and switch to differentiation media, a subset of 2D-expanded, already activated myogenic cells did not undergo fusion and further differentiation into myofibers, but instead deactivated to acquire a quiescent Pax7+ SC-like phenotype, likely maintained by niche-mimetic interactions with abutting myofibers via M-cadherin junctions and upregulated Notch signaling. We therefore termed this pool of deactivated SC-like cells in myobundles as 3D-SCs.
Fig. 2. Cell cycle arrest and gene expression in transition from 2D expansion to 3D myobundle culture.
a, Schematic of experimental timeline (2D expansion for 5 passages with p38i followed by 3D culture). GM: 3D growth media, Diff: 3D differentiation media, G4 day 4 in growth media, D4,7, days 4,7 in differentiation media). b, Representative images of Pax7 and Ki67 staining in 2D-expanded passage-5 CD56+ myoblasts and D7 myobundles with myofibers stained with F-actin (DAPI-stained nuclei are blue). c-e, Representative images of D7 myobundles showing expression of transcription factors Pax7, MyoG, and MyoD, and cell adhesion molecule M-cadherin (M-cad). f, Gene expression in cells isolated from passage 5 expanded 2D cultures (2D) or myobundles (G4, D4, D7) normalized to RPL13 and shown relative to 2D group (N = 1 donor, n = 3–17 samples per group, 1 sample = cells from 4 to 8 myobundles or separate 2D cultures). Data: mean ± SEM. *p < .05; **p < .01; ***p < .001.
2.4. Notch and Wnt signaling in 3D-SC deactivation and maintenance of the quiescent state
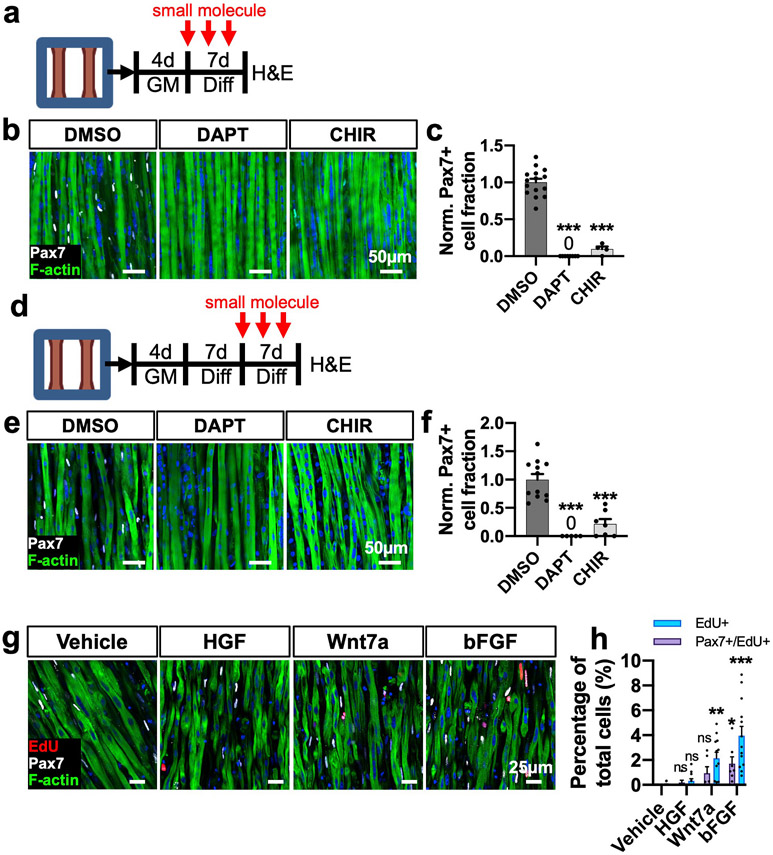
During both postnatal muscle development [9] and injury-induced regeneration [53], Notch and Wnt signaling pathways play determining roles in SC self-renewal vs. commitment [10,54]. Specifically, activation of Notch signaling is critical for preserving the SC pool [4,5], while constitutive activation of canonical Wnt signaling diminishes the SC pool during muscle regeneration due to precocious differentiation [53,55]. To determine if 3D-SC formation is similarly reduced by disruption of these pathways during myoblast deactivation, we applied 3 μM gamma secretase inhibitor (DAPT) to inhibit the Notch pathway or 3 μM GSK3 beta inhibitor (CHIR 99021) to stabilize beta-catenin and activate the canonical Wnt pathway during the first week of myobundle differentiation (Fig. 3a). Inhibiting Notch with DAPT or activating Wnt with CHIR significantly reduced 3D-SC density in myobundles (Fig. 3b and c). Assuming that the two drugs did not induce significant death to mononuclear cells, these findings suggest that disrupting the Notch or Wnt pathway prevents myoblast deactivation and acquisition of 3D-SC quiescence, leading instead to extensive myoblast fusion into myofibers. To further determine if perturbing these pathways would similarly impair the maintenance of 3D-SC quiescence, we treated 1-week differentiated myobundles with DAPT or CHIR during the second week of differentiation when 3D-SCs had been already established (Fig. 3d). Interestingly, each molecule significantly decreased the Pax7+ cell fraction (Fig. 3e and f), suggesting that similar to native SCs, quiescent 3D-SCs also rely on Notch activation to actively maintain quiescence [56,57] and remain sensitive to Wnt-induced commitment [9,55,57]. Collectively, these studies demonstrated critical roles of Notch and Wnt signaling in the formation and maintenance of a quiescent 3D-SC pool in myobundles.
Fig. 3. Notch and Wnt modulation on 3D SC quiescence and maintenance.
a, Schematic diagram of experimental design and treatment timing (GM: 3D growth media, Diff: 3D differentiation media). b, Representative images of whole-mount myobundles treated with DMSO (vehicle control), DAPT (3 μm), or CHIR (3 μm) during the first week of 3D differentiation stained for Pax7 and F-actin. c, Quantification of normalized Pax7+ cell fraction from whole myobundle stains (N = 1 donor, n = 4–15 myobundles per group). d, Schematic diagram of experimental design and treatment timing. e, Representative images of whole-mount myobundles treated with DMSO, CHIR, or DAPT during the second week of 3D differentiation stained for Pax7 and F-actin. f, Quantification of normalized Pax7 cell fraction from whole myobundle stains (N = 1 donors, n = 5–12 myobundles per group). g, Representative longitudinal sections of 14-day myobundles stained for EdU, Pax7, and F-actin after 1 week of treatment with specified growth factors. h, Quantification of EdU + percentage of nuclei (N = 2 donors, n = 6–13 myobundles per group). Data: mean ± SEM. *p < .05; **p < .01; ***p < .001; ns, not significant.
2.5. Growth factor response of 3D-SCs in myobundles
In addition to being quiescent, SCs need to remain responsive to extracellular cues to exit quiescence and activate. Hepatocyte growth factor (HGF), Wnt7a, and basic fibroblast growth factor (bFGF) can broadly stimulate SC activation and proliferation in vivo [58-61]. To test 3D-SC responsiveness to these mitogenic growth factors in vitro, myobundles were differentiated for 1 week to allow 3D-SC formation and cell cycle exit, followed by 1-week treatment every other day with 0.5 μM EdU and HGF (10 ng/ml) [62], Wnt7a (50 ng/ml) [63], or bFGF (20 ng/ml) [64]. Interestingly, HGF treatment did not significantly increase cellular EdU incorporation (Fig. 3g and h), which may be attributed to either sequestration of HGF by matricellular proteins and proteoglycans [65] inside myobundles or the need for injury-induced release of proteases to activate the HGF [66]. Unlike HGF, both Wnt7a and bFGF significantly increased the EdU incorporation in myobundles, which contained both activated 3D-SCs that co-expressed Pax7 and EdU as well as EdU+/Pax7-cells that presumably differentiated (and lost Pax7 expression) after initial activation (Fig. 3g and h). Together, these studies suggested that similar to native SCs in vivo, 3D-SCs within the myobundle environment remain sensitive to mitogenic growth factors and capable of re-entering the cell cycle.
2.6. Comparison of 3D-SCs with 2D culture-derived reserve cells
Reserve cells (RCs) form during myoblast differentiation in 2D culture and, similar to 3D-SCs, have been shown to exit the cell cycle, reduce MyoD expression, and increase Pax7 expression [21,67]. We thus preliminarily compared RCs and 3D-SCs generated from p5 myoblasts (Supplementary Fig. 2a) across several metrics including gene expression, ability to rapidly activate and proliferate, and capacity to generate functional muscle fibers. After 1 week of differentiation, RCs and 3D-SCs represented a significantly reduced proportion of Pax7+ cells compared to 2D p5 myoblasts and had withdrawn from the cell cycle, as shown by a lack of Pax7+/Ki67+ co-expression (Supplementary Figs. 2b-c). Compared to RCs, 3D-SCs showed increased expression of PAX7, SPRY1, and NOTCH3 and decreased expression of E2F gene (Fig. 3d), and, unlike RCs, had higher HEY1 expression than p5 myoblasts (Fig. 3d) [51,56]. Another key function of SCs in vivo is their ability to activate in response to tissue disruption [68]. To mimic this process, we isolated RCs and 3D-SCs from their respective tissue environments and plated them in growth media on culture plates. After 4 days in growth media, 3D-SCs expanded 3.8-fold vs. 2.1-fold-expanded RCs, suggesting that 3D-SCs more robustly re-activated in response to mitogenic stimulus (Supplementary Fig. 2e). We then utilized p5 2D-expanded myoblasts, RCs, and 3D-SCs to construct myobundles and assess their function. After one week of differentiation, the 3 myobundle groups exhibited similar cross-sectional areas (Supplementary Figs. 2f and g) with p5 myoblast-derived myobundles producing the greatest force (Supplementary Figs. 2h and i). Notably, 3D–SC–derived myobundles produced greater tetanic force than RC-derived myobundles (Supplementary Figs. 2h and i). Together, these preliminary comparisons suggest that 3D-SCs and RCs have functional differences which remain to be studied in more detail in the future.
2.7. scRNA-seq characterization of cellular heterogeneity during myoblast deactivation in myobundles
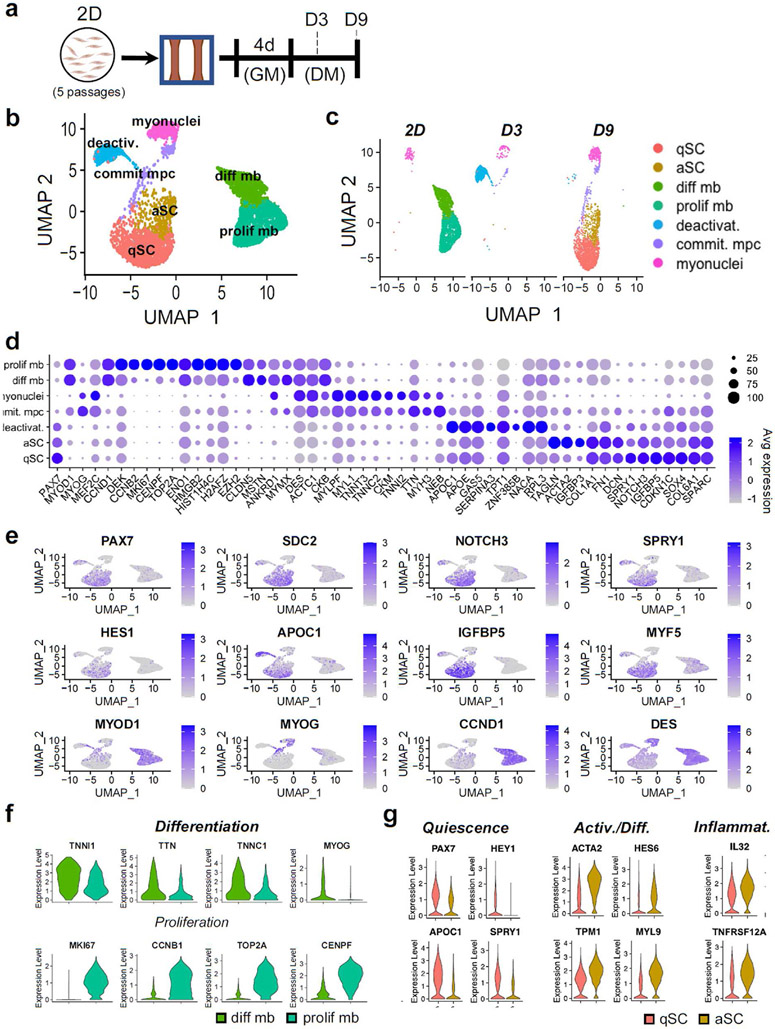
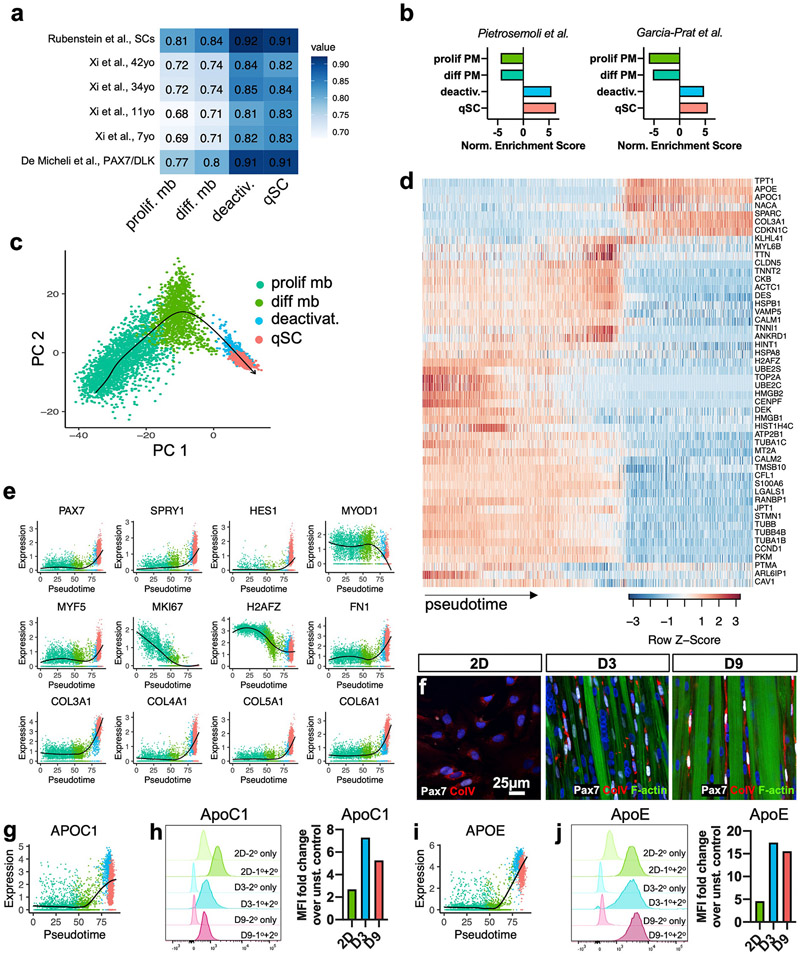
To better understand temporal changes in the transcriptome and cellular heterogeneity during the myoblast deactivation in myobundles, we performed scRNA-seq analysis of the following cell populations: passage 5 2D p38i-expanded CD56+ myoblasts used to make myobundles (2D), and mononuclear cells isolated after 3 and 9 days of myobundle differentiation (D3 and D9) (Fig. 4a). We used Seurat v3.2.2 [69] analysis package for count normalization and expression analysis and via the Louvain clustering method [69] identified 7 cell clusters corresponding to transcriptionally distinct states acquired during the myoblast deactivation (Fig. 4b-e). Specifically, the 2D myoblasts consisted of 2 dominant populations: 1) the differentiating population with higher expression of TNN11, TTN, TNNC1, and MYOG and 2) the proliferative population with higher expression of MK167, CCNB1, TOP2A, and CENPF (Fig. 4d,f). This finding is consistent with the presence of cycling Ki67+ and non-cycling Ki67-human myoblasts observed in 2D culture as well as similar distinction identified using scRNA-seq in primary mouse myoblasts [39]. The 2D myoblasts also contained a small non-proliferative myonuclei cluster characterized by no PAX7 and low MYOD1 expression paired with higher expression of MYOG, MEF2C, TNNT3, DES, CKM, and MYL1 (Fig. 4d and e), which likely originated from prematurely differentiated myoblasts.
Fig. 4. scRNA-seq clustering and marker analysis.
a, Schematic diagram of experimental layout. b, UMAP plot of cells isolated from all conditions. Each dot represents 1 cell, which is assigned to a distinctly colored cluster as identified by clustering analysis. c, UMAP plot split by each analyzed cell population (at different days of 2D and 3D culture) with cells colored by cluster. d, Dot plots displaying gene expression for each cluster with dot size representing percentage of cells expressing the gene and color representing average level of gene expression. e, UMAP plot with expression of selected genes associated with SC quiescence, activation, and myogenesis. f, Violin plots of the differentiating myoblast and proliferative myoblast clusters for differentiation and proliferation markers (padj < .01). g, Violin plots for the quiescent 3D-SC (qSC) and activated 3D-SC (aSC) cluster from the D9 condition for quiescence, activation, differentiation, and inflammatory markers (padj < .01).
In 3D myobundle culture, we also identified a distinct population of committed muscle progenitor cells (MPCs) lacking PAX7 and expressing MYOD1, MEF2C, and TNNT3 (Fig. 4c and d) as well as the myonuclei cluster identified in 2D myoblasts. Furthermore, the D3 experimental group revealed a critical intermediary cluster of deactivating cells which showed both: 1) upregulation of quiescence markers NOTCH3, SPRY1, APOE, and PAX7 and 2) downregulation of activation markers CCND1 and DES relative to the 2D differentiating and proliferative myoblast clusters (Fig. 4d and e).
Recent scRNA-seq studies have identified the existence of distinct SC subpopulations within both native homeostatic mouse [39] and human [70,71] muscles. Notably in uninjured human muscles, De Micheli et al. identified a “quiescent” and an “activated” population. In D9 myobundles, we similarly found clusters which based on their transcriptomic signature were termed quiescent (qSC) and activated (aSC) 3D-SCs. Consistent with descriptions of native human quiescent SCs, the myobundle-derived qSCs demonstrated strong expression of quiescence markers PAX7, SDC2 [14,72], FN1 [71], SPRY1 [56], and NOTCH3 [48] as well as low expression of MYOD1 and cell cycle genes (Fig. 4d,g). Compared to the qSC cluster, the aSC cluster exhibited reduced PAX7, FIEY1, APOC1, CDKN1C, and SPRY1 expression with increased expression of activation and differentiation markers ACTA2, ACTC1, TPM1, DES, and HES6 [73,74]. Similar to De Micheli et al. we also found relative upregulation of inflammatory markers IL32 and TNFRSF12A within the aSC population (Fig. 4d,g). To further determine if the distinction between myobundle D9 qSC versus aSC clusters was similar to the “quiescent” versus “activated” clusters identified De Micheli et al. [14], we compared the overlap in differentially expressed genes (padj < .05, ∣log2FC∣>0.25, minimum expressing cell percentage 10%). Of the 1588 genes identified in De Micheli’s “quiescent” versus “activated SCs [14] and 242 genes identified in D9 qSC versus aSC, a significant overlap of 63 genes was found (hypergeometric test, p~1.0e-4), suggesting that the 3D-SCs within myobundles recapitulate multiple aspects of native human SC heterogeneity.
2.8. Correlation of quiescent 3D-SCs in myobundles with native human SCs
To assess the similarity of the quiescent 3D-SCs with native human SCs, we integrated our dataset with previously published human scRNA-seq datasets [14,75,76] using Seurat v3 mutual nearest neighbors batch correction [69], and calculated the Spearman’s rank correlation for the top 2000 highly variable genes. We found that compared to 2D myoblast clusters, the 3D deactivating and qSC clusters demonstrated consistently higher correlation with native SCs (Fig. 5a). To further focus the analysis to genes known to be involved in SC quiescence, we used two previously described transcriptional signatures of quiescence identified in mouse SCs [77,78], mapped the genes to human homologs, and tested for enrichment of these signatures using gene set enrichment analysis (GSEA). While 2D myoblasts showed negative enrichment for these clusters, the deactivating cluster and qSCs in myobundles demonstrated progressive and significant enrichment for the SC quiescence signature (q < 0.05) (Fig. 5b). Interestingly, the deactivating cell cluster demonstrated correlation (Fig. 5a) and gene set enrichment (Fig. 5b) similar to the qSC cluster, suggesting the quiescence signature of 3D-SCs was established rapidly during early myobundle differentiation.
Fig. 5. Transcriptomic characterization of myoblast deactivation.
a, Heatmap of Spearman’s correlations between published human scRNA-seq clusters and proliferative myoblast, differentiating myoblast, deactivating cells, and qSC clusters. b, Normalized enrichment scores from GSEA of clusters using published mouse SC quiescence signatures by Pietrosemoli et al. [77] and Garcia-Prat et al. [78] as gene sets (q < .05). c, PCA plot for the proliferative myoblast, differentiating myoblast, deactivating cells, and qSC clusters from the 2D, D3, and D9 conditions with arrow delineating the trajectory generated by Slingshot. d, Heatmap of the top 50 important genes and their expression across pseudotime. e, Expression plots for selected genes across pseudotime with locally estimated scatterplot smoothing (LOESS) fit (black). f, Representative 2D, D3, and D9 cultures stained for Pax7, collagen V (Col V), and F-actin. g, APOC1 gene expression plot across pseudotime with LOESS fit. h, Flow cytometry fluorescence intensity histograms for ApoC1 protein with faint colors representing unstained controls (2° antibody only) and darker colors representing stained 2D, D3, and D9 samples (1°+2° antibody) and corresponding fold changes in the median fluorescence intensity (MFI) of stained vs. unstained samples. i-j, APOE gene and protein expression data analogous to that shown in g-h.
2.9. Trajectory inference on deactivation of 2D myoblasts to quiescent 3D-SCs
With the goal of further characterizing the deactivation process from 2D myoblasts to qSCs in myobundles, we performed pseudotime analysis of the 2D myoblast, D3 deactivating, and D9 qSC clusters using Slingshot [79]. The inferred trajectory bridged these clusters in the order: proliferative myoblasts, differentiating myoblasts, deactivating cells, and qSCs (Fig. 5c). Following the lineage assembly, pseudotime values were computed for each cell by projecting them onto principal curves [79]. To identify genes differentially expressed along this deactivation lineage, we applied the method used by Saelens et al. and constructed a random forest model to generate a pseudotime model [80]. The top 50 genes identified by the model’s feature importance metric were used to generate a heatmap (Fig. 5d). Forty-three of these genes were associated with activation, being expressed by proliferative and differentiating myoblasts, whereas 7 genes were associated with quiescence having high expression in deactivating cells and qSCs. When comparing these genes to descriptions of early activation and quiescence from in situ fixed mouse SCs [81], 21 of 43 activation genes and 3 of 7 quiescence genes were found to overlap. Genes highly expressed early in the pseudotime, corresponding to proliferative and differentiating myoblast clusters, spanned glycolytic metabolism (PKM [39]), proliferation (DEK, TOP2A, CENPF [82,83]), differentiation (CKB, DES, TNNI1, TNNT2, MYL6B, KLHL41 [84]), calcium-mediated signaling (CALM1 [85]) as well as cell motility (STMN1 [86,87]) and cytoskeletal structure (ACTC1, ANKRD1, TUBA1B, JPT1, S100A6 [88]), which are associated with SC activation [89,90]. In addition, genes coding heat shock proteins, which are associated with SC stress response and activation83, were reduced with deactivation. Further, the histone variant H2AFZ, associated with active transcription at the Myod locus in activated and differentiating mouse SCs84, 85, was downregulated across the deactivation trajectory (Fig. 5d and e). Interestingly, several ECM-related genes associated with skeletal muscle and the SC niche in vivo — FN1 [91], COL3A1 [92], COL4A1 [92], COL5A1 [93], and COL6A1 [94] — increased progressively during the deactivation process, suggesting qSCs may be remodeling their microenvironment and establishing a SC niche [51] (Fig. 5d-e). In immunostaining, collagen V expression appeared to increase across 2D, D3, and D9, but remained confined intracellularly (Fig. 5f), suggesting secretion, assembly, and remodeling of this protein may require extended time scales or additional microenvironmental factors. Late-stage deactivation was also associated with upregulation of SC markers APOE [51,95], APOC1 [14], and negative cell cycle regulators (CDKN1C and TPT1) [14]. To confirm if the differences in APOC1 and APOE gene expressions were maintained at the protein level, immunostaining intensity was compared via flow cytometry (Fig. 5g-j). Comparing the fold change in median fluorescence intensity (MFI) between stained (1°+2° antibody) and sample controls (2° antibody only) (Fig. 5h,j) revealed that both ApoE and ApoC1 were strongly increased in the D3 and D9 relative to 2D cells (Fig. 5h,j). Together, these studies revealed that myoblast deactivation in myobundles involved progressive transcriptomic changes reflective of cell cycle exit, metabolic downregulation, cytoskeletal and extracellular remodeling, and acquisition of quiescent SC markers.
2.10. Structural and functional characterization of myobundle injury by melittin
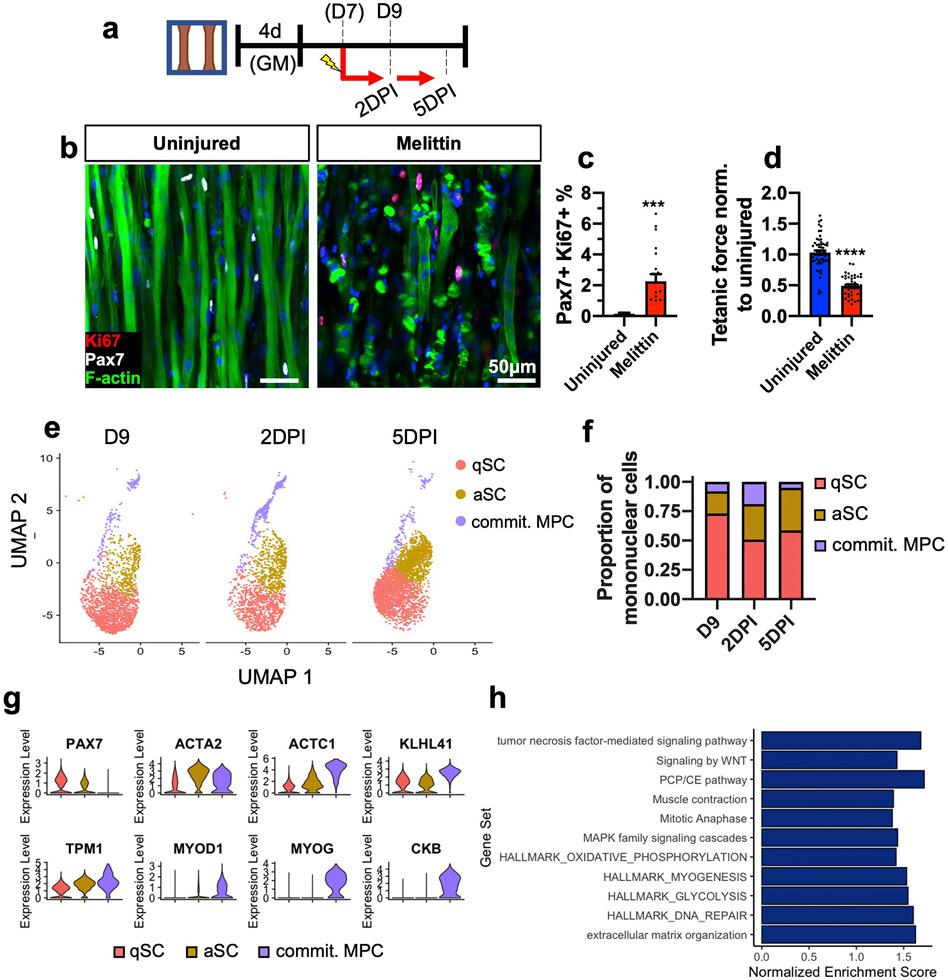
Rapid SC activation in response to injury is a key aspect of SC function and is critical for timely muscle regeneration in vivo [1]. Previous studies have reported activation of resident Pax7+ cells in 3D engineered rat and human muscles [26,27,31,32,96]. We evaluated the ability of bee venom (melittin, 10 μg/ml for 6 h), previously shown to induce skeletal muscle damage [97-99], to induce myobundle injury and 3D-SC activation (Fig 6a). Two days post injury (2DPI) by melittin, we consistently observed myofiber disruption and 3D-SC proliferation (Pax7+/Ki67+ cell increase, Fig. 6b and c), as well as deficits in twitch and tetanic contraction without effect on twitch kinetics (Fig. 6d, Supplementary Fig. 3). Additionally, we examined the 2-day response of myobundles to other injury-inducing agents, such as notexin (NTX), cardiotoxin (CTX), and the calcium ionophore A23187. While these treatments significantly reduced tetanic forces and slowed down twitch kinetics (Supplementary Figs. 4a-d), they exhibited a limited ability to induce myofiber disruption and 3D-SC proliferation in comparison to melittin (Supplementary Figs. 4e and f vs. Fig. 6c). We also found that by 5 and 7 days post melittin injury (5DPI and 7DPI), the percentage of proliferating Pax7+/Ki67+ cells was significantly reduced—which may indicate an early return to quiescence, 3D-SC death, or premature differentiation—and was not accompanied by recovery of tetanic force (Supplementary Figs. 5a-c). Taken together, the melittin-induced fiber fragmentation, muscle loss, and functional deficit in myobundles reproduced multiple aspects of muscle injury in vivo [100] and revealed that 3D-SCs are capable of injury-induced activation, cell cycle re-entry, and return to quiescence, similar to native SCs.
Fig. 6. Response of Myobundles and 3D SCs to injury.
a, Schematic diagram of experimental layout. DPI, days post-injury. b-d) Representative images of 1-week differentiated myobundles uninjured or injured by melittin and stained for Pax7, Ki67, and F-actin 2 days post-injury (b) and corresponding quantifications of percent Pax7+ Ki67+ nuclei (c, N = 2 donors, n = 20 myobundles per group) and tetanic force normalized to uninjured control (d, N = 3 donors, n = 44–60 myobundles per group). e,f UMAP plot of qSCs, aSCs, and committed MPCs at D9, 2DPI, and 5DPI (e) and corresponding proportions of each subpopulation within the total cell number (f). g, Violin plots of selected genes for the qSC and aSC clusters across all timepoints. h, GSEA of the 2DPI and 5DPI versus D9 aSC cluster with selected Gene Ontology and Reactome annotations (q < 0.05). Data: mean ± SEM. *p < .05; **p < .01; ***p < .001.
2.11. scRNA-seq characterization of the 3D-SC activation in response to myobundle injury
To further examine the transcriptomic signature of the 3D-SC activation in response to melittin, myobundles were injured on day 7 of differentiation and mononuclear cells were collected at 2DPI and 5DPI and analyzed by scRNA-seq (Fig. 6a). We found that in response to myobundle injury, the proportion of aSCs was increased (Fig 6e and f), either from a conversion of qSCs to aSCs and/or expansion of existing aSCs. Compared to aSCs and qSCs, a population of committed MPCs possessed higher expression of differentiation markers KLH41, MYOD1, MYOG, and CKB (Fig. 6g). Notably, the committed MPCs peaked at 2DPI and were diminished by 5DPI suggesting further differentiation to myotubes or apoptosis between these timepoints (Fig 6e and f). To further uncover the underlying mechanisms of the increased aSC population in response to injury, we applied gene set enrichment analysis (GSEA) to compare aSCs from D9 versus the combined injured timepoints (2DPI and 5DPI) using the Gene Ontology (biological processes and Hallmark) and Reactome Pathway annotations and illustrated both selected annotations (Fig 6h) and the full list of enriched terms (Supplementary Tables 4a-c) with q < 0.05. In comparison to the uninjured aSC cluster, the injured (2DPI and 5DPI) aSC clusters demonstrated significant enrichment for pathways consistent with those previously described for SC activation and muscle repair in mice, including: MAPK [6], FGF [8], Wnt [10], and TNF [38,101] signaling as well as glycolysis [39,40,102], proliferation [103], DNA repair [104], and myogenesis [103]. Together, these studies revealed transcriptomic underpinnings of the muscle-autonomous, injury-induced human 3D-SC activation, which would be difficult to study in native human muscle.
3. Discussion
In the presented work, we optimized the formation of quiescent 3D-SCs within tissue-engineered human myobundles, demonstrated the roles of Wnt and Notch signaling in 3D-SC formation and maintenance, and developed a protocol to activate 3D-SCs by injuring myobundles with a bee toxin. We also for the first time applied scRNA-seq analysis to tissue-engineered skeletal muscle to track and transcriptionally characterize the human myoblast deactivation process, formation of the quiescent 3D-SC pool, and injury-induced activation. Through these studies, we showed molecular evidence for phenotypic heterogeneity within the homeostatic 3D-SC pool resembling that reported for native human SCs in vivo [14]. As the starting population for these studies was CD56+ myogenic cells, our results provide new insights into muscle-autonomous regulation of human SC quiescence, deactivation, and re-activation.
While CD56+ cells have been used to engineer human muscle tissues [27,105-107], the effect of myogenic cell purity on the formation of quiescent Pax7+ cells, to our knowledge, has not been previously reported. We found that the Pax7+ 3D-SC formation in myobundles was increased with use of higher fraction of CD56+ cells (Fig. 1e and f), likely due to increased early-fusion events that permitted Pax7+ cell homing to myotubes, as we observed in engineered rat muscle [44]. Additionally, we found that p38 inhibition significantly accelerated human myoblast proliferation during long-term 2D expansion, while preserving Pax7 expression (Supplementary Figs. 1b and c), and increased the abundance of 3D-SCs (Pax7+ cell fraction) within myobundles (Supplementary Figs. 1d and e). Importantly, such obtained 3D-SCs exhibited key features of native quiescent SCs including elevated PAX7 expression and Notch signaling relative to 2D myoblasts (Fig. 2f), cell cycle arrest (Fig. 2b,f), potential to form functional myofibers (Supplementary Figs. 2h and i), and the ability to re-activate in response to growth factors (Fig. 3g and h) and muscle injury (Fig. 6b and c). Unlike a combination of biomaterial-based niche and multiple small molecules that maintained in vitro SC quiescence for up to 4 days [19], the human myobundle system contained a cell-based niche consisting of differentiated myofibers that supported the formation and maintenance of 3D-SC quiescence through Notch activation (Figs. 2f, 3a-f). These results not only confirmed the role of Notch signaling in the in vitro formation of quiescent SCs [23], but also suggested additional roles of Notch in human SC maintenance in vitro as observed in native mouse muscle [4,5]. Formation and maintenance of human 3D-SCs in myobundles were also disrupted by activation of canonical Wnt signaling, consistent with the studies in mice showing Wnt interference with Notch signaling [9] and SC depletion during regeneration [55].
Compared to RCs formed during 2D myoblast differentiation, 3D-SCs showed higher PAX7, SPRY1, and NOTCH3 expression (Supplementary Fig. 2d), faster re-activation and proliferation (Supplementary Fig. 2e), and greater functional output when used to form myobundles (Supplementary Figs. 2h and i). Interestingly, in our study, RCs did not show significantly higher PAX7 expression compared to 2D myoblasts, as previously reported [21], potentially due to use of P5 rather than P0–P1 myoblasts. In general, we expect that in vitro conditions can induce a spectrum of SC-like states that vary in similarity to the native SC phenotype observed in homeostatic muscle in vivo. It would be of interest to further compare RCs and 3D-SCs for their engraftment potential in vivo and for expression of miRNA repressors of myogenic progression in DDX6+ messenger ribonucleoprotein (mRNP) granules [108-110] to establish if 3D-SCs attain a phenotype more mimetic of native SCs.
Our RNA-seq analysis identified 7 distinct cell clusters including 2 populations of cultured myoblasts, proliferative and differentiating (Fig. 4f), which align with scRNA-seq-based description of proliferative and differentiated primary mouse myoblasts [39]. Previous scRNA-seq work has also identified several subpopulations of SCs in native mouse [39,111,112] and human [14,71] muscles. Although the overlap and relationship between the subpopulations across studies remains poorly understood, the qSC and aSC populations identified in the myobundle system respectively resembled the human PAX7+/DLK1+ (MuSC1) and PAX7low/MYF5+ (MuSC2) populations identified by De Micheli et al. [14]. Specifically, consistent with the differences in MuSC1 vs. MuSC2 [14], the myobundle-derived qSCs in our study expressed greater PAX7, SPRY1, CDKN1C, EGFR, DAG1, TCF4, COL4A1/2, and HEY1 relative to aSCs. Mirroring the MuSC2 population, aSCs expressed elevated levels of inflammatory markers (IL32 and TNFRSF12A), VAMP5, DES, EIF5A, and RAN. These results suggest that myobundle derived 3D-SCs are able of recapitulating important aspects of SC heterogeneity observed in native human muscle. We also noted high levels of FOS, JUN, and EGR1 in both qSCs and aSCs compared to 2D proliferating and differentiating myoblast populations (Supplementary Figs. 6a-c). Previously, high expression of these genes has been observed in SC clusters of other scRNA-seq datasets [70] and identified as artifacts of the enzymatic dissociation during sample preparation [81,113]. Future studies should examine various in vitro pre-fixation strategies [81] to limit activation of 3D-SCs induced by tissue dissociation.
An essential feature of muscle regeneration is SC deactivation and return to homeostatic quiescent state [114,115]; however, this process remains poorly understood despite being critical for preserving the SC pool and maintaining the regenerative potential of skeletal muscle between repeated injuries [103,116]. Our analysis identified a transcriptional trajectory of human myoblast deactivation in myobundles, reflective of a reduction in myoblast proliferation, metabolism, cytoskeletal reorganization, and differentiation (Fig. 5d and e), negative cell cycle regulation and ECM remodeling (Fig. 5d-f), and final acquisition of a homeostatic state characteristic of quiescent SCs in vivo [36,93,94,117]. Specifically, along the deactivation trajectory, we found increased PAX7 expression, decreased expression of the oncogene DEK [83], increased MYF5 expression suggestive of poised activation [108], increased expression of negative regulators of cell cycling (CDKN1C [51]) and FGF signaling (SPRY1 [118]), and increased Notch activation (HES1) [5]. The transcriptional changes identified here highlight the role of muscle-autonomous mechanisms in myoblast deactivation and may serve as a blueprint for future studies in mouse and human models of SC deactivation.
In mouse models, muscle regeneration studies traditionally apply toxins to damage the skeletal muscle and induce a regenerative response [119]. Here, we identified the bee venom melittin treatment to induce myobundle functional deficit, myofiber disruption, and 3D-SC activation, without functional recovery (Fig. 6b-d, Supp. Fig 5c). Previously, engineered human skeletal muscles have been injured using CTX without measurement of active force [26,96] or BaCl2 where tissues recovered functionally following injury [27], potentially due to a smaller tissue size or differential effects of BaCl2 vs. melittin. In our study, CTX, NTX, and A23187 injury all induced functional deficit, similar to melittin, but showed limited myofiber disruption and 3D-SC activation (Supplementary Figs. 4d-f vs. Fig. 6b-d). Importantly, in studying the melittin-induced injury response by scRNA-seq analysis, we observed an increasing proportion of aSCs relative to qSCs, and transcriptomic changes consistent with native mouse SC injury-induced activation including: increased MAPK signaling [6], glycolytic metabolism [39,40,102], and myogenesis [103]. Unexpectedly, the aSC cluster did not show strong expression of individual cell cycling genes (i.e. MKI67, CCND1) despite significant enrichment for proliferation-associated terms such as “Mitotic Anaphase” gene set (Fig. 6h) and Ki67 protein expression observed in immunostaining (Fig. 6b and c). This suggested that while proteins were still expressed at 2 days post-injury, proliferation-associated genes were already down-regulated. Accordingly, 3D-SC proliferation concluded by 5–7 days post-injury as determined by immunostaining for Ki67 (Supplementary Figs. 5a and b). This relatively short proliferation phase compared to SCs in vivo [112] corresponded with a lack of myobundle recovery (Supplementary Fig. 5c), and suggests that additional non-muscle cell types, such as macrophages and fibro-adipogenic progenitors (FAPs) [120-124], biochemical cues presented by muscle-specific ECM [125], and/or exogenous factors [126-128] may be necessary for robust regenerative response of human muscle. These intrinsic and extrinsic regulators of SC function [129], if incorporated in the 3D myobundle system, may also restore the native levels of Pax7 expression in deactivated myoblasts, known to rapidly decline during in vitro cell expansion [130].
In summary, we have shown that the 3D environment of engineered human myobundles provides a homeostatic niche for the formation of a Pax7+ cell pool that shares several characteristics with native human SCs, including expression of quiescence markers, Notch pathway utilization, and bi-population heterogeneity. Using this in vitro tissue-engineered system, we transcriptionally characterized muscle-autonomous processes of human myoblast deactivation, SC formation, and reactivation upon injury. This work lays a foundation for further exploration of human SCs at the epigenetic level and for investigations of SC function in models of muscle regeneration [32] and niche dysregulation [131,132].
4. Materials and methods
4.1. Human SC isolation and magnetic-activated cell sorting
Human skeletal muscle samples were obtained from surgical discards using protocols approved by the Duke University Institutional Review Board. Donor information, surgical discard source, and experiment attribution is described in Supplementary Table 1. Freshly harvested human muscle was immediately minced with scissors and digested in DMEM with 2 mg/ml collagenase (Worthington-Biochem, cat# LS11004174), 2% v/v dispase (Corning, cat #: 354235), and DNase I (Bio-rad, cat# 7326828) for ~90min at 37 °C on a rocker. The isolated cells were further sheared from fibers using a syringe and strained through 100 μm and 30 μm cell strainers. For heterogeneous cultures, these cells were directly plated for downstream use. For CD56+ cell enrichment, heterogenous suspensions were sorted using magnetic-activated cell sorting (MACS) for CD56+ per manufacturer instructions (Miltenyi Biotec, cat# 130-050-401). Briefly, dissociated cells were incubated in CD56+ MACS beads for 20min at 4 °C, then sorted on a MACS column, and washed 3 times with MACS buffer (0.2% BSA), yielding ~100-200 k cells per gram of muscle tissue. The sorted cells were subsequently plated at 10 cells/mm2 for downstream expansion and myobundle construction. Freshly plated cells were considered p0 with each round of replating after trypsinization adding to the passage number. During expansion, cells were cultured in 2D growth media (GM: low-glucose (1 g/L) DMEM (Sigma, cat# D6046), 10% FBS (Hyclone, cat# SH30071), dexamethasone (Sigma, cat# D4902), EGF (Prospecio, cat# CYT-217), and penicillin 64 mg/L (Sigma, cat# P7794)) without or supplemented with 10 μM p38 inhibitor (p38i) SB203580 (LC labs S-3400).
4.2. Fabrication and culture of tissue-engineered human myobundles
Myobundles were formed within polydimethylsiloxane (PDMS) molds containing two semi-cylindrical wells (7-mm long, 2-mm diameter), cast from 3D-machined Teflon masters, as previously described [25,33]. PDMS molds were coated with 2% (w/v) pluronic F-127 (Invitrogen) for 1 h at room temperature to prevent hydrogel adhesion. Laser-cut Cerex® frames positioned around the two wells were used to anchor the ends of the myobundles and facilitate handling. A cell solution (7.5 × 105 cells in 17.2 μL media per bundle + 2 μL of 50 U/mL thrombin in 1% BSA in PBS) and a gelling solution (3 μL media +10 μL Matrigel + 10 μL of 15 mg/mL fibrinogen in PBS) were prepared in separate vials on ice for up to eight myobundles per vial. Gelling solution was added to the cell solution, mixed thoroughly, and injected into the PDMS wells to polymerize at 37 °C for 30 min. Resulting myobundles were cultured dynamically on a rocker in 3D growth media (GM supplemented with 1.5 mg/mL 6-aminocaproic acid (ACA, Sigma)) for 4 days. The media was then switched to differentiation media (DM) consisting of a custom low-glucose, low amino acid (LAA) media to mimic physiological amino acid concentrations (as described in Ref. [133]), with 1x N2 serum supplement (Thermo Fisher), and 2 mg/mL ACA [134]. The media was exchanged every other day.
4.3. Isolation of mononuclear cells from myobundles
To isolate mononuclear cells (predominantly 3D-SCs), myobundles were minced with scissors in a 37 °C enzymatic digestion solution of 2% v/v dispase (Corning, cat #: 354235) and 15 mg/ml collagenase type II (Worthington-Biochem, cat# LS11004174) for 3–5 min. Subsequently, the solution was gently pipetted 5–6 times using a syringe with a 23G needle and incubated in the 37 °C water bath for 5–8 min for further digestion. This solution with enzymatically dissociated myobundles was further diluted in PBS and strained twice through 30 μm strainers to remove myofiber fragments and then the obtained cells were resuspended for further use. The total isolation time spanned approximately 30 min. For replating studies, isolated cells were seeded at 40,000 cells/well (6-well plate) in GM with p38 inhibitor at 10 μM for 4 days before quantifying fold change in cell numbers. For select experiments, replated cells were expanded for two passages before myobundle fabrication.
4.4. Reserve cell formation and isolation
To derive reserve cells (RCs), myoblasts were plated in 12-well or 6-well plates at P5 and expanded in growth media (same as 3D growth media) for 4 days to confluence, followed by differentiated for 5–7 days in differentiation media (LAA media with N2). To isolate the RCs, cultures were briefly trypsinized (0.025% for 3 min) to release single cells from the wells. To further reduce myotube contamination, cells were strained through a 100 μm and 30 μm strainers before downstream RNA isolation or continued culture. For replating studies, cells were seeded at 40,000 cells/well (6-well plate) in GM with p38 inhibitor at 10 μM for 4 days before quantifying fold change in cell numbers. For select experiments, replated RCs were expanded for two passages before myobundle fabrication.
4.5. Isometric force measurements in myobundles
Contractile properties of myobundles were measured using a custom, temperature-controlled force measurement setup described previously [25,33,135]. Briefly, single myobundles were anchored on one end to an immobilized PDMS block and on the opposite end to a suspended PDMS float connected to a force transducer. Tissues were stimulated using 40 V/cm, 5 ms electrical pulses via parallel platinum electrodes to generate twitch (single pulse) and tetanic (20 Hz for 1 s) contractions. A computer-controlled motorized linear actuator (Thor labs) connected to the PDMS float was used to stretch myobundles to a length (1.12 x culture length) that yielded the maximum generated force. Recorded force traces were analyzed for peak twitch or tetanus amplitude, time to peak twitch, and half-relaxation time using a custom MATLAB program [25,33,135].
4.6. Immunohistochemistry
Cell monolayers were fixed in 4% paraformaldehyde in PBS for 10 min and myobundles were fixed in 2% paraformaldehyde in PBS overnight at 4 °C [44]. Following fixation, samples were washed in PBS followed by blocking in 5% chick serum with 3% Triton-X 10. Primary and secondary antibodies (Supplementary Table 2) were applied overnight at 4 °C. Images were acquired using the Zeiss 880 inverted confocal microscope. For images of cryo-sectioned myobundles, fixed tissues were oriented in OCT relative to cutting plane either longitudinally (Fig. 3g) or cut in half and oriented perpendicularly for cross-sectional images (Supplementary Fig. 2f), frozen using liquid nitrogen, and cut in 10 μm sections.
4.7. Small molecule, growth factor, and injury administration to myobundles
For Wnt activation (CHIR99021 at 3 μM, Tocris Cat#:4423) and Notch inhibition (DAPT 3 μM, Cayman Cat #:13197), treatments were applied every other day with media changes during either the first or second week of differentiation. For growth factor treatments, HGF (10 ng/ml), Wnt7a (50 ng/ml), bFGF (10 ng/ml), or vehicle (1:2000 of 0.1% BSA) were applied every other day during the second week of differentiation. For EdU experiments, 0.5 μM EdU was co-administered with growth factors. For injury experiments, myobundles were treated for 6 h in 500 μl volume of media in 24-well dishes with melittin (10 μg/ml for 6 h, Sigma: M2272), cardiotoxin (0.4 μM for 6 h, latoxan: L8102), notexin (1 μg/ml for 6 h, latoxan: L8104), or A23187 (80μM for 1 h, Cayman: 11016).
4.8. RNA extraction and quantitative RT-PCR
RNA was isolated using RNeasy Plus Micro Kit (Qiagen), followed by RNA reverse-transcription using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR for muscle-related genes was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) according to manufacturer’s instructions and primers listed in Supplementary Table 3. The delta-delta (dd) Ct method was used for data quantification. dCt was determined by Ct (gene of interest) – Ct (housekeeping gene: RPL13). ddCt was calculated by dCt (condition) – dCt (control), with fold change being 2−ddct.
4.9. Single cell RNA-sequencing library construction
Cells were prepared for scRNA-seq analysis from a single continuous experiment (1 donor) during which samples were generated at the end of 2D culture before myobundle formation as well as at D3, D9, 2DPI, and 5DPI during myobundle culture. At each time point, cells were incubated in a freezing media (10% DMSO, 90% FBS) and stored in liquid nitrogen. At the end of the experiment, all samples were simultaneously processed and loaded onto the 10x Genomics Chromium Controller Single-Cell Instrument (10x Genomics) using the Chromium Single Cell 3’ Reagent V3 Kit. Cells were mixed with reverse transcription reagents, gel beads, and oil to generate single-cell gel beads in emulsions (GEM) for reverse transcription (RT). After RT, GEMs were broken and the single-stranded cDNA was purified with DynaBeads. cDNA was amplified by PCR and the cDNA product was purified with the SPRIselect Reagent Kit (Beckman Coulter). Sequencing libraries were constructed using the reagents provided in the Chromium Single-Cell 3′ Library Kit following the user guide (https://support.10xgenomics.com/single-cell-gene-expression/library-prep). scRNA libraries were sequenced with the DNBseq platform by BGI Genomics.
4.10. Single cell RNA-sequencing data processing and analysis
The Cell Ranger v3 software (10x Genomics) was used to demultiplex cellular barcodes to produce raw 3’ end read profiles for individual cells, and then to perform alignment against hg38 reference genome, filtering, barcode counting, and UMI (unique molecular identifier) counting, producing a feature-barcode matrix per sample. The downstream analysis with R package Seurat (v3.2.2) [69] was applied to perform quality control and subsequent analyses on the gene expression matrices produced by Cell Ranger. Specifically, we performed standard visualizations for number of genes per cell (nFeature_RNA), number of unique molecular identifiers per cell (nCount_RNA), and percent mitochondrial transcripts (Supplementary Fig. 6) before filtering, data clustering, and differential gene expression analysis. Cells were first filtered to retain those with >1000 UMIs, <5% mapping to mitochondrial genes, and >200 genes per cell. After standard normalization, variable features were identified using the “vst” method to select the top 2000 genes. Using cells from all conditions, clustering in Seurat (Louvain) was conducted with resolution 0.5. Cells from two clusters exhibiting both low nFeature_RNA (genes per cell) and nCount_RNA (number of molecules per cell), indicative of dead/dying cells, were filtered out. Differential gene expression between clusters was identified using the “Find-Markers” function with default parameters. Due to limited sizes of acquired muscle biopsies, we were not able to isolate enough human SCs to both use directly for scRNA-seq analysis and expand for downstream myobundle experiments. Therefore, to compare the 3D-SCs formed in myobundles with native, freshly isolated human SCs, we integrated our dataset using mutual nearest neighbors batch correction [69] with previously published human scRNA-seq datasets [14,75,76].
Trajectory analysis of scRNA-seq was performed using Slingshot [79]. From the clusters generated from Seurat, the 2D myoblast clusters, D3 transition cluster, and quiescent SC cluster were selected using cells only from the 2D, D3, and D9 timepoints. Next, Slingshot was run on first 4 principal components of PCA, and a lineage was generated using the quiescent SC cluster as the ending cluster. To illustrate trends in gene expression across pseudotime, we generated a heatmap and individual gene expression plots with locally estimated scatterplot smoothing (LOESS) lines. To identify important genes in the pseudotime analysis, a random forest model was trained on the top 300 highly variable genes (which captured 95.6% of the total variance) with a 90-10 train-test split. Given the pseudotime range (0–91) and median (57.71), the model showed strong performance on the held-out validation set with a coefficient of determination (R2) and mean absolute error (MAE) of 0.990 and 1.36.
Gene set enrichment analysis (GSEA) with FDR cutoff 0.05 was conducted within Clusterprofiler [136], including GO term analysis of the “Biological Process” sub-level and Reactome pathway databases. For the Hallmark gene set annotations, the msigdbr library [137] was analyzed within Clusterprofiler.
For analyzing enrichment of quiescence gene sets, gene lists were acquired from 2 previous studies [77,78] and tested for enrichment using Clusterprofiler. For Garcia-Prat et al. [78], genes were mapped to human orthologs using the biomaRt [138]. For the correlation heatmap, scRNA-seq from GSE130646 [75], GSE147457 [76], and GSE143704 [14] were processed, randomly downsampled to 400 cells per cluster, integrated using Seurat anchors (Seurat v3.2.2) [69], and the spearman’s correlation was calculated on the highly variable genes. For the comparisons to the De Micheli et al. dataset [14], differentially expressed genes (∣log2FC∣>0.25 and minimum expression in at least 10% of the cells) from the D9 timepoint between the qSC and aSC clusters were compared with De Micheli et al.’s MuSCl (Pax7+/DLK1+) and MuSC2 (PAX7low/MYF5+) clusters.
For the hypergeometric test, the significance in overlap was determined between 2 sets of differentially expressed genes: 1) D9 qSC versus D9 aSC and 2) De Micheli et al. MuSC1 (PAX7+DLK1+) versus MuSC2 (PAX7low MY5+) [14]. Differentially expressed genes were determined based on the cutoffs: ∣log2FC∣>0.25, minimum cell expressing percentage >10%, and padj < .05. The background number of genes (9290 genes) was restricted to include only transcripts expressed in at least 10% of cells.
4.11. Statistics
Data are expressed as mean ± SEM. For comparisons between 2 groups, Welch’s t-test was applied. For multiple comparisons, an ANOVA was used followed by post-hoc testing with Tukey’s honestly significant difference (HSD). Measurements related to contractile function (tetanic forces and twitch kinetics) represent individual myobundles. For histology, independent measurements indicate independent myobundles or separated 2D cultures. For qPCR analysis a single independent measurement was considered: 3D-SCs isolated from 4 to 8 myobundles, RCs isolated from 2 to 3 separated culture wells, or separated 2D cultures.
Supplementary Material
Acknowledgements
The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. This work was supported by NIH Grants AR065873, AR070543, and AR073470 from National Institute of Arthritis and Musculoskeletal and Skin Disease, UH3TR002142 from the NIH Common Fund for the Microphysiological Systems Initiative, and U01 EB028901 from the National Institutes of Biomedical Imaging and Bioengineering to NB; Startup fund from Duke University School of Medicine and the Regeneration Next Initiative to YD; and Duke Regeneration Next Initiative postdoctoral fellowship to YX.
Footnotes
CRediT authorship contribution statement
Jason Wang: Conceptualization, Writing – original draft, Investigation, Formal analysis. Torie Broer: Validation, Investigation, Formal analysis. Taylor Chavez: Validation, Formal analysis. Chris J. Zhou: Formal analysis, Investigation. Sabrina Tran: Formal analysis, Investigation. Yu Xiang: Formal analysis, Data curation, Resources. Alastair Khodabukus: Conceptualization, Supervision, Writing – review & editing. Yarui Diao: Supervision, Resources. Nenad Bursac: Conceptualization, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2022.121508.
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations. The sequencing data generated for this study will be deposited in the Gene Expression Omnibus under GSE188215.
References
- [1].Lepper C, Partridge TA, Fan CM, An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration, Development 138 (17) (2011) 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yablonka-Reuveni Z, The skeletal muscle satellite cell: still young and fascinating at 50, J. Histochem. Cytochem 59 (12) (2011) 1041–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Le Grand F, Rudnicki MA, Skeletal muscle satellite cells and adult myogenesis, Curr. Opin. Cell Biol 19 (6) (2007) 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA, Notch signaling is necessary to maintain quiescence in adult muscle stem cells, Stem Cell. 30 (2) (2012) 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, Tajbakhsh S, A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state, Stem Cell. 30 (2) (2012) 243–252. [DOI] [PubMed] [Google Scholar]
- [6].Jones NC, Tyner KJ, Nibarger L, Stanley HM, Cornelison DD, Fedorov YV, Olwin BB, The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell, J. Cell Biol 169 (1) (2005) 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jones NC, Fedorov YV, Rosenthal RS, Olwin BB, ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion, J. Cell. Physiol 186 (1) (2001) 104–115. [DOI] [PubMed] [Google Scholar]
- [8].Pawlikowski B, Vogler TO, Gadek K, Olwin BB, Regulation of skeletal muscle stem cells by fibroblast growth factors, Dev. Dynam 246 (5) (2017) 359–367. [DOI] [PubMed] [Google Scholar]
- [9].Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA, A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis, Cell Stem Cell 2 (1) (2008) 50–59. [DOI] [PubMed] [Google Scholar]
- [10].von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA, Wnt signaling in myogenesis, Trends Cell Biol. 22 (11) (2012) 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bareja A, Holt JA, Luo G, Chang C, Lin J, Hinken AC, Freudenberg JM, Kraus WE, Evans WJ, Billin AN, Human and mouse skeletal muscle stem cells: convergent and divergent mechanisms of myogenesis, PLoS One 9 (2) (2014), e90398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boldrin L, Muntoni F, Morgan JE, Are human and mouse satellite cells really the same? J. Histochem. Cytochem 58 (11) (2010) 941–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Maffioletti SM, Sarcar S, Henderson ABH, Mannhardt I, Pinton L, Moyle LA, Steele-Stallard H, Cappellari O, Wells KE, Ferrari G, Mitchell JS, Tyzack GE, Kotiadis VN, Khedr M, Ragazzi M, Wang W, Duchen MR, Patani R, Zammit PS, Wells DJ, Eschenhagen T, Tedesco FS, Three-Dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering, Cell Rep. 23 (3) (2018) 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].De Micheli AJ, Spector JA, Elemento O, Cosgrove BD, A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations, Skeletal Muscle 10 (1) (2020) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Blau HM, Webster C, Isolation and characterization of human muscle cells, Proc. Natl. Acad. Sci. U. S. A 78 (9) (1981) 5623–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, Di Santo JP, Buder-Browne GS, Wright WE, Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies, Aging Cell 6 (4) (2007) 515–523. [DOI] [PubMed] [Google Scholar]
- [17].Cooper RN, Thiesson D, Furling D, Di Santo JP, Butler-Browne GS, Mouly V, Extended amplification in vitro and replicative senescence: key factors implicated in the success of human myoblast transplantation, Hum. Gene Ther 14 (12) (2003) 1169–1179. [DOI] [PubMed] [Google Scholar]
- [18].Cosgrove BD, Sacco A, Gilbert PM, Blau HM, A home away from home: challenges and opportunities in engineering in vitro muscle satellite cell niches, Differentiation 78 (2–3) (2009) 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Quarta M, Brett JO, DiMarco R, De Morree A, Boutet SC, Chacon R, Gibbons MC, Garcia VA, Su J, Shrager JB, Heilshorn S, Rando TA, An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy, Nat. Biotechnol 34 (7) (2016) 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abou-Khalil R, Le Grand F, Chazaud B, Human and murine skeletal muscle reserve cells, Methods Mol. Biol 1035 (2013) 165–177. [DOI] [PubMed] [Google Scholar]
- [21].Laumonier T, Bermont F, Hoffmeyer P, Kindler V, Menetrey J, Human myogenic reserve cells are quiescent stem cells that contribute to muscle regeneration after intramuscular transplantation in immunodeficient mice, Sci. Rep 7 (1) (2017) 3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Furuichi Y, Kawabata Y, Aoki M, Mita Y, Fujii NL, Manabe Y, Excess glucose impedes the proliferation of skeletal muscle satellite cells under adherent culture conditions, Front. Cell Dev. Biol 9 (2021) 640399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kitzmann M, Bonnieu A, Duret C, Vernus B, Barro M, Laoudj-Chenivesse D, Verdi JM, Carnac G, Inhibition of Notch signaling induces myotube hypertrophy by recruiting a subpopulation of reserve cells, J. Cell. Physiol 208 (3) (2006) 538–548. [DOI] [PubMed] [Google Scholar]
- [24].Rochat A, Fernandez A, Vandromme M, Moles JP, Bouschet T, Carnac G, Lamb NJ, Insulin and wnt1 pathways cooperate to induce reserve cell activation in differentiation and myotube hypertrophy, Mol. Biol. Cell 15 (10) (2004) 4544–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N, Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs, Elife 4 (2015), e04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rajabian N, Shahini A, Asmani M, Vydiam K, Choudhury D, Nguyen T, Ikhapoh I, Zhao R, Lei P, Andreadis ST, Bioengineered Skeletal Muscle as a Model of Muscle Aging and Regeneration, Tissue Eng Part A, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fleming JW, Capel AJ, Rimington RP, Wheeler P, Leonard AN, Bishop NC, Davies OG, Lewis MP, Bioengineered human skeletal muscle capable of functional regeneration, BMC Biol. 18 (1) (2020) 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mills RJ, Parker BL, Monnot P, Needham EJ, Vivien CJ, Ferguson C, Parton RG, James DE, Porrello ER, Hudson JE, Development of a human skeletal micro muscle platform with pacing capabilities, Biomaterials 198 (2019) 217–227. [DOI] [PubMed] [Google Scholar]
- [29].Afshar ME, Abraha HY, Bakooshli MA, Davoudi S, Thavandiran N, Tung K, Ahn H, Ginsberg HJ, Zandstra PW, Gilbert PM, A 96-well culture platform enables longitudinal analyses of engineered human skeletal muscle microtissue strength, Sci. Rep 10 (1) (2020) 6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tiburcy M, Markov A, Kraemer LK, Christalla P, Rave-Fraenk M, Fischer HJ, Reichardt HM, Zimmermann WH, Regeneration competent satellite cell niches in rat engineered skeletal muscle, FASEB Bioadv 1 (12) (2019) 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Juhas M, Engelmayr GC Jr., Fontanella AN, Palmer GM, Bursac N, Biomimetic engineered muscle with capacity for vascular integration and functional maturation in vivo, Proc. Natl. Acad. Sci. U. S. A 111 (15) (2014) 5508–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Juhas M, Abutaleb N, Wang JT, Ye J, Shaikh Z, Sriworarat C, Qian Y, Bursac N, Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration, Nat Biomed Eng 2 (12) (2018) 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Khodabukus A, Madden L, Prabhu NK, Koves TR, Jackman CP, Muoio DM, Bursac N, Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle, Biomaterials 198 (2019) 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Khodabukus A, Kaza A, Wang J, Prabhu N, Goldstein R, Vaidya VS, Bursac N, Tissue-engineered human myobundle system as a platform for evaluation of skeletal muscle injury biomarkers, Toxicol. Sci 176 (1) (2020) 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen Z, Li B, Zhan RZ, Rao L, Bursac N, Exercise mimetics and JAK inhibition attenuate IFN-gamma-induced wasting in engineered human skeletal muscle, Sci. Adv 7 (4) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bentzinger CF, Wang YX, Dumont NA, Rudnicki MA, Cellular dynamics in the muscle satellite cell niche, EMBO Rep. 14 (12) (2013) 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yin H, Price F, Rudnicki MA, Satellite cells and the muscle stem cell niche, Physiol. Rev 93 (1) (2013) 23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen SE, Jin B, Li YP, TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK, Am. J. Physiol. Cell Physiol 292 (5) (2007) C1660–C1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dell’Orso S, Juan AH, Ko KD, Naz F, Perovanovic J, Gutierrez-Cruz G, Feng X, Sartorelli V, Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions, Development 146 (12) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ryall JG, Dell’Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, Sartorelli V, The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells, Cell Stem Cell 16 (2) (2015) 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G, Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration, Development 138 (17) (2011) 3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Verma M, Asakura Y, Murakonda BSR, Pengo T, Latroche C, Chazaud B, McLoon LK, Asakura A, Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and notch signaling, Cell Stem Cell 23 (4) (2018) 530–543 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Biferali B, Proietti D, Mozzetta C, Madaro L, Fibro-adipogenic progenitors cross-talk in skeletal muscle: the social network, Front. Physiol 10 (2019) 1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Juhas M, Bursac N, Roles of adherent myogenic cells and dynamic culture in engineered muscle function and maintenance of satellite cells, Biomaterials 35 (35) (2014) 9438–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Illa I, Leon-Monzon M, Dalakas MC, Regenerating and denervated human muscle fibers and satellite cells express neural cell adhesion molecule recognized by monoclonal antibodies to natural killer cells, Ann. Neurol 31 (1) (1992) 46–52. [DOI] [PubMed] [Google Scholar]
- [46].Agley CC, Rowlerson AM, Velloso CP, Lazarus NR, Harridge SD, Human skeletal muscle fibroblasts, but not myogenic cells, readily undergo adipogenic differentiation, J. Cell Sci 126 (Pt 24) (2013) 5610–5625. [DOI] [PubMed] [Google Scholar]
- [47].Agley CC, Rowlerson AM, Velloso CP, Lazarus NL, Harridge SD, Isolation and quantitative immunocytochemical characterization of primary myogenic cells and fibroblasts from human skeletal muscle, JoVE 95 (2015) 52049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Charville GW, Cheung TH, Yoo B, Santos PJ, Lee GK, Shrager JB, Rando TA, Ex vivo expansion and in vivo self-renewal of human muscle stem cells, Stem Cell Rep. 5 (4) (2015) 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM, Rejuvenation of the muscle stem cell population restores strength to injured aged muscles, Nat. Med 20 (3) (2014) 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB, p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice, Nat. Med 20 (3) (2014) 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S, Molecular signature of quiescent satellite cells in adult skeletal muscle, Stem Cell. 25 (10) (2007) 2448–2459. [DOI] [PubMed] [Google Scholar]
- [52].Irintchev A, Zeschnigk M, Starzinski-Powitz A, Wernig A, Expression pattern of M-cadherin in normal, denervated, and regenerating mouse muscles, Dev. Dynam 199 (4) (1994) 326–337. [DOI] [PubMed] [Google Scholar]
- [53].Jones AE, Price FD, Le Grand F, Soleimani VD, Dick SA, Megeney LA, Rudnicki MA, Wnt/beta-catenin controls follistatin signalling to regulate satellite cell myogenic potential, Skeletal Muscle 5 (2015) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Luo D, Renault VM, Rando TA, The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis, Semin. Cell Dev. Biol 16 (4–5) (2005) 612–622. [DOI] [PubMed] [Google Scholar]
- [55].Rudolf A, Schirwis E, Giordani L, Parisi A, Lepper C, Taketo MM, Le Grand F, Beta-catenin activation in muscle progenitor cells regulates tissue repair, Cell Rep. 15 (6) (2016) 1277–1290. [DOI] [PubMed] [Google Scholar]
- [56].Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, Brack AS, Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration, Cell Stem Cell 6 (2) (2010) 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Carlson ME, Silva HS, Conboy IM, Aging of signal transduction pathways, and pathology, Exp. Cell Res 314 (9) (2008) 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Greene EA, Allen RE, Growth factor regulation of bovine satellite cell growth in vitro, J. Anim. Sci 69 (1) (1991) 146–152. [DOI] [PubMed] [Google Scholar]
- [59].Cheng X, Huang H, Luo X, Shi B, Li J, Wnt7a induces satellite cell expansion, myofiber hyperplasia and hypertrophy in rat craniofacial muscle, Sci. Rep 8 (1) (2018) 10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Miller KJ, Thaloor D, Matteson S, Pavlath GK, Hepatocyte growth factor affects satellite cell activation and differentiation in regenerating skeletal muscle, Am. J. Physiol. Cell Physiol 278 (1) (2000) C174–C181. [DOI] [PubMed] [Google Scholar]
- [61].Allen RE, Sheehan SM, Taylor RG, Kendall TL, Rice GM, Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro, J. Cell. Physiol 165 (2) (1995) 307–312. [DOI] [PubMed] [Google Scholar]
- [62].Yamada M, Tatsumi R, Yamanouchi K, Hosoyama T, Shiratsuchi S, Sato A, Mizunoya W, Ikeuchi Y, Furuse M, Allen RE, High concentrations of HGF inhibit skeletal muscle satellite cell proliferation in vitro by inducing expression of myostatin: a possible mechanism for reestablishing satellite cell quiescence in vivo, Am. J. Physiol. Cell Physiol 298 (3) (2010) C465–C476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bentzinger CF, von Maltzahn J, Dumont NA, Stark DA, Wang YX, Nhan K, Frenette J, Cornelison DD, Rudnicki MA, Wnt7a stimulates myogenic stem cell motility and engraftment resulting in improved muscle strength, J. Cell Biol 205 (1) (2014) 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zeng C, Pesall JE, Gilkerson KK, McFarland DC, The effect of hepatocyte growth factor on Turkey satellite cell proliferation and differentiation, Poultry Sci. 81 (8) (2002) 1191–1198. [DOI] [PubMed] [Google Scholar]
- [65].Noriega-Guerra H, Freitas VM, Extracellular matrix influencing HGF/c-MET signaling pathway: impact on cancer progression, Int. J. Mol. Sci 19 (11) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tatsumi R, Allen RE, Active hepatocyte growth factor is present in skeletal muscle extracellular matrix, Muscle Nerve 30 (5) (2004) 654–658. [DOI] [PubMed] [Google Scholar]
- [67].Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y, Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ’reserve cells, J. Cell Sci 111 (Pt 6) (1998) 769–779. [DOI] [PubMed] [Google Scholar]
- [68].Li J, Han S, Cousin W, Conboy IM, Age-specific functional epigenetic changes in p21 and p16 in injury-activated satellite cells, Stem Cell. 33 (3) (2015) 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive integration of single-cell data, Cell 177 (7) (2019) 1888–1902, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].De Micheli AJ, Laurilliard EJ, Heinke CL, Ravichandran H, Fraczek P, Soueid-Baumgarten S, De Vlaminck I, Elemento O, Cosgrove BD, Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration, Cell Rep. 30 (10) (2020) 3583–3595 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Barruet E, Garcia SM, Striedinger K, Wu J, Lee S, Byrnes L, Wong A, Xuefeng S, Tamaki S, Brack AS, Pomerantz JH, Functionally heterogeneous human satellite cells identified by single cell RNA sequencing, Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Magli A, Incitti T, Kiley J, Swanson SA, Darabi R, Rinaldi F, Selvaraj S, Yamamoto A, Tolar J, Yuan C, Stewart R, Thomson JA, Perlingeiro RCR, PAX7 targets, CD54, integrin alpha9beta1, and SDC2, allow isolation of human ESC/iPSC-Derived myogenic progenitors, Cell Rep. 19 (13) (2017) 2867–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gao X, Chandra T, Gratton MO, Quelo I, Prud’homme J, Stifani S, St-Arnaud R, HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program, J. Cell Biol 154 (6) (2001) 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH, Hes6 regulates myogenic differentiation, Development 129 (9) (2002) 2195–2207. [DOI] [PubMed] [Google Scholar]
- [75].Rubenstein AB, Smith GR, Raue U, Begue G, Minchev K, Ruf-Zamojski F, D Nair V, Wang X, Zhou L, Zaslavsky E, Trappe TA, Trappe S, Sealfon SC, Single-cell transcriptional profiles in human skeletal muscle, Sci. Rep 10 (1) (2020) 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Xi H, Langerman J, Sabri S, Chien P, Young CS, Younesi S, Hicks M, Gonzalez K, Fujiwara W, Marzi J, Liebscher S, Spencer M, Van Handel B, Evseenko D, Schenke-Layland K, Plath K, Pyle AD, A human skeletal muscle atlas identifies the trajectories of stem and progenitor cells across development and from human pluripotent stem cells, Cell Stem Cell 27 (1) (2020) 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Pietrosemoli N, Mella S, Yennek S, Baghdadi MB, Sakai H, Sambasivan R, Pala F, Di Girolamo D, Tajbakhsh S, Comparison of multiple transcriptomes exposes unified and divergent features of quiescent and activated skeletal muscle stem cells, Skeletal Muscle 7 (1) (2017) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Garcia-Prat L, Perdiguero E, Alonso-Martin S, Dell’Orso S, Ravichandran S, Brooks SR, Juan AH, Campanario S, Jiang K, Hong X, Ortet L, Ruiz-Bonilla V, Flandez M, Moiseeva V, Rebollo E, Jardi M, Sun HW, Musaro A, Sandri M, Sol AD, Sartorelli V, Munoz-Canoves P, FoxO maintains a genuine muscle stem-cell quiescent state until geriatric age, Nat. Cell Biol 22 (11) (2020) 1307–1318. [DOI] [PubMed] [Google Scholar]
- [79].Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S, Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics, BMC Genom. 19 (1) (2018) 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Saelens W, Cannoodt R, Todorov H, Saeys Y, A comparison of single-cell trajectory inference methods, Nat. Biotechnol 37 (5) (2019) 547–554. [DOI] [PubMed] [Google Scholar]
- [81].Machado L, Esteves de Lima J, Fabre O, Proux C, Legendre R, Szegedi A, Varet H, Ingerslev LR, Barres R, Relaix F, Mourikis P, In situ fixation redefines quiescence and early activation of skeletal muscle stem cells, Cell Rep. 21 (7) (2017) 1982–1993. [DOI] [PubMed] [Google Scholar]
- [82].Yue L, Wan R, Luan S, Zeng W, Cheung TH, Dek modulates global intron retention during muscle stem cells quiescence exit, Dev. Cell 53 (6) (2020) 661–676 e6. [DOI] [PubMed] [Google Scholar]
- [83].Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA, Maintenance of muscle stem-cell quiescence by microRNA-489, Nature 482 (7386) (2012) 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gupta VA, Ravenscroft G, Shaheen R, Todd EJ, Swanson LC, Shiina M, Ogata K, Hsu C, Clarke NF, Darras BT, Farrar MA, Hashem A, Manton ND, Muntoni F, North KN, Sandaradura SA, Nishino I, Hayashi YK, Sewry CA, Thompson EM, Yau KS, Brownstein CA, Yu TW, Allcock RJ, Davis MR, Wallgren-Pettersson C, Matsumoto N, Alkuraya FS, Laing NG, Beggs AH, Identification of KLHL41 mutations implicates BTB-kelch-mediated ubiquitination as an alternate pathway to myofibrillar disruption in nemaline myopathy, Am. J. Hum. Genet 93 (6) (2013) 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tatsumi R, Wuollet AL, Tabata K, Nishimura S, Tabata S, Mizunoya W, Ikeuchi Y, Allen RE, A role for calcium-calmodulin in regulating nitric oxide production during skeletal muscle satellite cell activation, Am. J. Physiol. Cell Physiol 296 (4) (2009) C922–C929. [DOI] [PubMed] [Google Scholar]
- [86].Curmi PA, Nogues C, Lachkar S, Carelle N, Gonthier MP, Sobel A, Lidereau R, Bieche I, Overexpression of stathmin in breast carcinomas points out to highly proliferative tumours, Br. J. Cancer 82 (1) (2000) 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Balogh A, Mege RM, Sobel A, Growth and cell density-dependent expression of stathmin in C2 myoblasts in culture, Exp. Cell Res 224 (1) (1996) 8–15. [DOI] [PubMed] [Google Scholar]
- [88].Jurewicz E, Robaszkiewicz K, Moraczewska J, Filipek A, Binding of S100A6 to actin and the actin-tropomyosin complex, Sci. Rep 10 (1) (2020) 12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kawamura K, Takano K, Suetsugu S, Kurisu S, Yamazaki D, Miki H, Takenawa T, Endo T, N-WASP and WAVE2 acting downstream of phosphatidylinositol 3-kinase are required for myogenic cell migration induced by hepatocyte growth factor, J. Biol. Chem 279 (52) (2004) 54862–54871. [DOI] [PubMed] [Google Scholar]
- [90].Johnston AP, Baker J, Bellamy LM, McKay BR, De Lisio M, Parise G, Regulation of muscle satellite cell activation and chemotaxis by angiotensin II, PLoS One 5 (12) (2010), e15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA, Fibronectin regulates Wnt7a signaling and satellite cell expansion, Cell Stem Cell 12 (1) (2013) 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Thomas K, Engler AJ, Meyer GA, Extracellular matrix regulation in the muscle satellite cell niche, Connect. Tissue Res 56 (1) (2015) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, Tajbakhsh S, Mourikis P, Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche, Nature 557 (7707) (2018) 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, Vozzi G, Rando TA, Bonaldo P, Collagen VI regulates satellite cell self-renewal and muscle regeneration, Nat. Commun 4 (2013) 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Matsakas A, Barlow, Verpoorten S, Scully D, BS55 Regulation of Skeletal Muscle Stem Cell Function in a Mouse Model of Experimental Atherosclerosis and Hyperlipidaemia, Heart, 2019. [Google Scholar]
- [96].Trevisan C, Fallas MEA, Maghin E, Franzin C, Pavan P, Caccin P, Chiavegato A, Carraro E, Boso D, Boldrin F, Caicci F, Bertin E, Urbani L, Milan A, Biz C, Lazzari L, De Coppi P, Pozzobon M, Piccoli M, Generation of a functioning and self-renewing diaphragmatic muscle construct, Stem Cells Transl Med 8 (8) (2019) 858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Fletcher JE, Hubert M, Wieland SJ, Gong QH, Jiang MS, Similarities and differences in mechanisms of cardiotoxins, melittin and other myotoxins, Toxicon 34 (11–12) (1996) 1301–1311. [DOI] [PubMed] [Google Scholar]
- [98].Lee MT, Sun TL, Hung WC, Huang HW, Process of inducing pores in membranes by melittin, Proc. Natl. Acad. Sci. U. S. A 110 (35) (2013) 14243–14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lee JE, Shah VK, Lee EJ, Oh MS, Choi JJ, Melittin - a bee venom component - enhances muscle regeneration factors expression in a mouse model of skeletal muscle contusion, J. Pharmacol. Sci 140 (1) (2019) 26–32. [DOI] [PubMed] [Google Scholar]
- [100].Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS, Highly coordinated gene regulation in mouse skeletal muscle regeneration, J. Biol. Chem 278 (10) (2003) 8826–8836. [DOI] [PubMed] [Google Scholar]
- [101].Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, Sartorelli V, Puri PL, TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration, Cell Stem Cell 7 (4) (2010) 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Fu X, Zhu MJ, Dodson MV, Du M, AMP-activated protein kinase stimulates Warburg-like glycolysis and activation of satellite cells during muscle regeneration, J. Biol. Chem 290 (44) (2015) 26445–26456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Relaix F, Zammit PS, Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage, Development 139 (16) (2012) 2845–2856. [DOI] [PubMed] [Google Scholar]
- [104].Vahidi Ferdousi L, Rocheteau P, Chayot R, Montagne B, Chaker Z, Flamant P, Tajbakhsh S, Ricchetti M, More efficient repair of DNA double-strand breaks in skeletal muscle stem cells compared to their committed progeny, Stem Cell Res. 13 (3 Pt A) (2014) 492–507. [DOI] [PubMed] [Google Scholar]
- [105].Martin NR, Passey SL, Player DJ, Khodabukus A, Ferguson RA, Sharpies AP, Mudera V, Baar K, Lewis MP, Factors affecting the structure and maturation of human tissue engineered skeletal muscle, Biomaterials 34 (23) (2013) 5759–5765. [DOI] [PubMed] [Google Scholar]
- [106].Brady MA, Lewis MP, Mudera V, Synergy between myogenic and non-myogenic cells in a 3D tissue-engineered craniofacial skeletal muscle construct, J Tissue Eng Regen Med 2 (7) (2008) 408–417. [DOI] [PubMed] [Google Scholar]
- [107].Afshar Bakooshli M, Lippmann ES, Mulcahy B, Iyer N, Nguyen CT, Tung K, Stewart BA, van den Dorpel H, Fuehrmann T, Shoichet M, Bigot A, Pegoraro E, Ahn H, Ginsberg H, Zhen M, Ashton RS, Gilbert PM, A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction, Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Crist CG, Montarras D, Buckingham M, Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules, Cell Stem Cell 11 (1) (2012) 118–126. [DOI] [PubMed] [Google Scholar]
- [109].de Morree A, van Velthoven CTJ, Gan Q, Salvi JS, Klein JDD, Akimenko I, Quarta M, Biressi S, Rando TA, Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence, Proc. Natl. Acad. Sci. U. S. A 114 (43) (2017) E8996–E9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Sampath SC, Sampath SC, Ho ATV, Corbel SY, Millstone JD, Lamb J, Walker J, Kinzel B, Schmedt C, Blau HM, Induction of muscle stem cell quiescence by the secreted niche factor Oncostatin M, Nat. Commun 9 (1) (2018) 1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yartseva V, Goldstein LD, Rodman J, Kates L, Chen MZ, Chen YJ, Foreman O, Siebel CW, Modrusan Z, Peterson AS, Jovicic A, Heterogeneity of satellite cells implicates DELTA1/NOTCH2 signaling in self-renewal, Cell Rep. 30 (5) (2020) 1491–1503 e6. [DOI] [PubMed] [Google Scholar]
- [112].Oprescu SN, Yue F, Qiu J, Brito LF, Kuang S, Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration, iScience 23 (4) (2020) 100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].van Velthoven CTJ, de Morree A, Egner IM, Brett JO, Rando TA, Transcriptional profiling of quiescent muscle stem cells in vivo, Cell Rep. 21 (7) (2017) 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Motohashi N, Asakura A, Muscle satellite cell heterogeneity and self-renewal, Front. Cell Dev. Biol 2 (2014) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bazgir B, Fathi R, Rezazadeh Valojerdi M, Mozdziak P, Asgari A, Satellite cells contribution to exercise mediated muscle hypertrophy and repair, Cell J 18 (4) (2017) 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Jejurikar SS, Kuzon WM Jr., Satellite cell depletion in degenerative skeletal muscle, Apoptosis : an international journal on programmed cell death 8 (6) (2003) 573–578. [DOI] [PubMed] [Google Scholar]
- [117].Kuang S, Gillespie MA, Rudnicki MA, Niche regulation of muscle satellite cell self-renewal and differentiation, Cell Stem Cell 2 (1) (2008) 22–31. [DOI] [PubMed] [Google Scholar]
- [118].Chakkalakal JV, Jones KM, Basson MA, Brack AS, The aged niche disrupts muscle stem cell quiescence, Nature 490 (7420) (2012) 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hardy D, Besnard A, Latil M, Jouvion G, Briand D, Thepenier C, Pascal Q, Guguin A, Gayraud-Morel B, Cavaillon JM, Tajbakhsh S, Rocheteau P, Chretien F, Comparative study of injury models for studying muscle regeneration in mice, PLoS One 11 (1) (2016), e0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, Chawla A, Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration, Cell 153 (2) (2013) 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM, Muscle injury activates resident fibro/adipogenie progenitors that facilitate myogenesis, Nat. Cell Biol 12 (2) (2010) 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Segawa M, Fukada S, Yamamoto Y, Yahagi H, Kanematsu M, Sato M, Ito T, Uezumi A, Hayashi S, Miyagoe-Suzuki Y, Takeda S, Tsujikawa K, Yamamoto H, Suppression of macrophage functions impairs skeletal muscle regeneration with severe fibrosis, Exp. Cell Res 314 (17) (2008) 3232–3244. [DOI] [PubMed] [Google Scholar]
- [123].Mounier R, Theret M, Arnold L, Cuvellier S, Bultot L, Goransson O, Sanz N, Ferry A, Sakamoto K, Foretz M, Viollet B, Chazaud B, AMPKalpha1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration, Cell Metabol. 18 (2) (2013) 251–264. [DOI] [PubMed] [Google Scholar]
- [124].Mackey AL, Kjaer M, The breaking and making of healthy adult human skeletal muscle in vivo, Skeletal Muscle 7 (1) (2017) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Smoak MM, Hogan KJ, Grande-Allen KJ, Mikos AG, Bioinspired electrospun dECM scaffolds guide cell growth and control the formation of myotubes, Sci. Adv 7 (20) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Tidball JG, Villalta SA, Regulatory interactions between muscle and the immune system during muscle regeneration, Am. J. Physiol. Regul. Integr. Comp. Physiol 298 (5) (2010) R1173–R1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Borselli C, Storrie H, Benesch-Lee F, Shvartsman D, Cezar C, Lichtman JW, Vandenburgh HH, Mooney DJ, Functional muscle regeneration with combined delivery of angiogenesis and myogenesis factors, Proc. Natl. Acad. Sci. U. S. A 107 (8) (2010) 3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Fang J, Sia J, Soto J, Wang P, Li LK, Hsueh YY, Sun R, Faull KF, Tidball JG, Li S, Skeletal muscle regeneration via the chemical induction and expansion of myogenic stem cells in situ or in vitro, Nat Biomed Eng 5 (2021) 864–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Dumont NA, Wang YX, Rudnicki MA, Intrinsic and extrinsic mechanisms regulating satellite cell function, Development 142 (9) (2015) 1572–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Garcia SM, Tamaki S, Lee S, Wong A, Jose A, Dreux J, Kouklis G, Sbitany H, Seth R, Knott PD, Heaton C, Ryan WR, Kim EA, Hansen SL, Hoffman WY, Pomerantz JH, High-yield purification, preservation, and serial transplantation of human satellite cells, Stem Cell Rep. 10 (3) (2018) 1160–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Blau HM, Webster C, Pavlath GK, Defective myoblasts identified in Duchenne muscular dystrophy, Proc. Natl. Acad. Sci. U. S. A 80 (15) (1983) 4856–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Carlson ME, Conboy MJ, Hsu M, Barchas L, Jeong J, Agrawal A, Mikels AJ, Agrawal S, Schaffer DV, Conboy IM, Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses, Aging Cell 8 (6) (2009) 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kondash ME, Ananthakumar A, Khodabukus A, Bursac N, Truskey GA, Glucose Uptake and Insulin Response in Tissue-Engineered Human Skeletal Muscle, Tissue Eng Regen Med, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wang J, Zhou CJ, Khodabukus A, Tran S, Han SO, Carlson AL, Madden L, Kishnani PS, Koeberl DD, Bursac N, Three-dimensional tissue-engineered human skeletal muscle model of Pompe disease, Commun Biol 4 (1) (2021) 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Bian W, Bursac N, Soluble miniagrin enhances contractile function of engineered skeletal muscle, Faseb. J 26 (2) (2012) 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R package for comparing biological themes among gene clusters, OMICS 16 (5) (2012) 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection, Cell Syst 1 (6) (2015) 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W, BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis, Bioinformatics 21 (16) (2005) 3439–3440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations. The sequencing data generated for this study will be deposited in the Gene Expression Omnibus under GSE188215.