Abstract
αβ T cells are critical components of the adaptive immune system and are capable of inducing sterilizing immunity after pathogen infection and eliminating transformed tumor cells. The development and function of T cells is controlled through the T cell antigen receptor (TCR), which recognizes peptides displayed on major histocompatibility (MHC) molecules. Here, we review how T cells generate the ability to recognize self-peptide–bound MHC molecules and use signals derived from these interactions to instruct cellular development, activation thresholds, and functional specialization in the steady state and during immune responses. We argue that the basic tenants of T cell development and function follow Weber-Fetcher’s law of just noticeable differences and Wilder’s law of initial value. Together, these laws argue that the ability and quality of a response is scalable to the basal state of a system. Manifestation of these laws in T cells generates clone-specific activation thresholds that are based on perceivable differences between homeostasis and pathogen encounter (self versus nonself discrimination), as well as poised states for subsequent differentiation into specific effector cell lineages.
Introduction
As tradesmen, military generals, and economists have noted for millennia, thresholds based on the cost of acquisition control the fate of goods and borders. The quanta of cost, the associated value of an item or outcome, are relative; as Fechner noted in his studies of proportionality (1, 2), “the value of a moneypiece to an individual decreases with the amount of pieces which he already possesses.” Otherwise stated, how much is a franc worth to man with 2 francs as compared to a man with 20,000? With this in mind, during the 19th century, Ernst Weber and his student Gustav Fechner used studies of vision and weight to propose and advance theories on perception. Weber’s law of noticeable differences argues that there exists a minimum amount of change in stimulus intensity that is required to produce a noticeable variation in sensory experience (3). He proposed that the magnitude of a “just noticeable difference” is a constant proportion, based on a linear relationship with the original stimulus value (4):
where (ΔI) represents the change in stimulus, (I) represent the background stimulus, and k signifies the proportion or threshold value that is required for a stimulus to be noticed.
Fechner recognized that despite the inability to measure absolute values of a perceived sensation, one could measure the ability to identify changes in the intensity of a stimulus, that is, the sensitivity of a response (S). He argued that the sensitivity of a response is dependent upon individual thresholds (for example, a person with poor eyesight is less light-sensitive than one with proper eyesight), and proportional to the logarithm of the stimulus:
where k is a constant based on the context of the system and R is the quantity of the sensation. Logarithmic and power laws (5), and their derivatives, describe various human sensory systems, as well as cellular responses to stimulation.
Building upon these hypotheses, classical studies by Joseph Wilder of changes in blood pressure (BP) after injection of adrenaline demonstrated that sensitivity (or excitability to a stimulus) could provide both a positive or a negative response based on the initial value of the stimuli, that is, the basal state, and carried a time dimension. Furthermore, when the baseline BP was artificially increased to a higher level before the injection of adrenalin, the same original stimulus was no longer biologically active, thereby demonstrating signal antagonism. Thus, the initial value of the basal state directly influences whether a stimulus is capable of being biologically noticed (6). Together, Weber, Fechner, Wilder and other’s studies of human perception and biological response to stimuli addressed several fundamental problems in biology: what is the minimum stimulus that can be detected? What is the smallest difference that can distinguish between two stimuli? How does the basal state and antagonism relate to the intervals or scales that generate sensitivity thresholds?
Manifestation of these laws of perception on the αβ T cell repertoire is the presence of ligand recognition thresholds that instruct the development and survival of T cells, the ability to distinguish diseased from healthy tissue, and the generation of effector functions that orchestrate sterilizing immunity. These signals are derived from the T cell antigen receptor (TCR), co-receptors and adhesion molecules, as well as cytokines, metabolites, and growth factor receptors engaging their stimulatory ligands (7, 8). The initial value of T cell awareness, or the basal state of self-reactivity, is established during T cell development through recognition by the TCR of peptide (p) derived from self-protein and, in select cases lipids or metabolites, bound to host major histocompatibility complex (MHC) molecules. Noticeable differences in TCR signaling that arise during thymic selection are used to instruct the progression or arrest of the developmental process, as well as T cell lineage choice. As mature T cells migrate throughout the body, the basal state of self-reactivity in conjunction with cytokines sets naïve T cell frequencies, activation thresholds, and pre-immune biased differentiation programs. During infection, T-cell recognition of pathogen-derived pMHCs generates rapid TCR signal perturbations, a scalable process based on the affinity or half-life of the TCR-pMHC interaction, which drives cellular proliferation, acquisition of effector functions, differentiation, and memory formation (9–11). Fundamental to the ability of T cells to orchestrate pathogen clearance is the ability of lymphocytes to behave in a cell-autonomous manner, and to distinguish between self- and foreign pathogen–derived antigens.
Three cardinal features of immune perception are essential to solving the feat of broad pathogen-specificity and disease contextual T cell response patterns. First, at the earliest stages of T cell development, lineage precursor cells rearrange the hierarchy of their environmental sensing mechanisms and place TCR signaling at the apex (12). Second, recombinase activating gene (RAG)-mediated TCR recombination events combined with thymic selection processes export T cells that express TCRs with unique, clone-specific sequences (13–15). The functional capacity of T cells to discriminate between activating ligands is encompassed by the molecular sequence of the expressed TCR clonotype. An individual T cell expresses thousands of copies of the identical TCR on their cell surface, whereas other T cells within the repertoire express one of several million possible sequences within the antigen-binding site (16–18). These two attributes enable TCR-mediated recognition of pMHC molecules to be the gatekeeper of T cell activation and clonal expansion. The primacy of TCR signaling precludes naïve T cells from generically responding to the inflammatory milieu in the absence of cognate TCR ligand recognition (9).
Critical to the ability of the immune system to develop T cells and to provide disease-contextual T cell response pattern is a third cardinal feature: the establishment of scalable TCR signaling thresholds. Detection of activating ligands, leading to productive TCR signal transduction events, must be strong enough to overcome reversable steps and negative feedback processes embedded in the kinetic proofreading mechanisms that regulate TCR signaling (19–22). These mechanisms determine the minimum stimuli that can be recognized by T cells to propagate changes in gene transcription. Furthermore, pioneering studies in the 1990s reported that the TCR complex has a unique way to control signaling in mature T cells. By contrast to other receptors, the TCR is endowed with the ability to “interpret” very fine differences of the pMHC-TCR interactions and translate them into different outcomes (23, 24). Thus, TCR signaling is not a simple, threshold-based ON/OFF switch, able to either activate all T cell outcomes or none. Rather, the TCR is capable of sensing subtilities in the quantity and potency of the pMHC ligand being recognized and translating these interactions into scalable functional outcomes (23, 25). Indeed, fine differences in the quality of TCR stimuli distinguish T cell development from clonal elimination, and T cell activation and acquisition of effector functions from quiescence. The current consensus model argues that the activation of individual T cell clones is dependent upon the half-life or dwell-time of engagement of the TCR-pMHC interaction (24, 26, 27). More specifically, the half-lives of TCR-pMHC interactions are a composite of a normal distribution of individual binding events, and only the few rare, long–dwell time binding events (~10-fold greater than the average dwell-time) are capable of driving Nuclear Factor of Activated T cells (NFAT) translocation into the nucleus, thus providing a mechanistic basis for the observations that strong T cell activation is dependent upon antigen concentration as well as antigen quality (that is, the half-life of the TCR-pMHC interaction) (28).
The ability to track multiple T cell clonotypes during T cell development and immune responses has demonstrated that in vivo T cell behavior does not always match the expected outcome of in vitro–defined TCR-pMHC biophysical parameters. This understanding has led to the concept that homeostatic (basal-state) TCR signals derived from the engagement of self-pMHC ligands (self-reactivity), together with other environmental cues, are critical to establishing T cell triggering thresholds and effector lineage biases as well as pro-survival functions (29–33). During development, the strength (or quantity) of basal TCR signals regulates whether T cell maturation proceeds, and the lineage phenotype of the clone exported. During mature T cell homeostasis, the sensitivity of T cells to pathogens is modulated by the basal level of self-reactivity through altered amounts of enzymatically active, TCR signaling intermediaries and cytokine receptor abundance. Whereas, after infection, the magnitude of TCR and cytokine signals synergize with the initial cellular state of the clone to control the extent of proliferation, the induction of specific effector functions, and the differentiation into long-term memory T cells (11, 34–37). Thus, the concept that T cell responses are scalable, based on the potency and quantity of antigen, has to be viewed through the lens of the initial self-reactivity value of each clone and the environment for which it operates to explain how TCR-ligand recognition events can regulate the myriad of T cell development, activation, and differentiation processes.
Thymic acquisition of TCR signaling control of lymphocyte function
The design principle of early T cell development includes equipping thymocytes with an ability to use the quality of TCR-pMHC interactions to determine lineage development and function. This transition from stem cell to thymocyte includes altering the transcriptional and epigenetic landscape of the cell and is guided by classical cell-extrinsic factors (cytokine and growth factor signaling and cell surface receptor-ligand interactions) initiated and sustained within the unique thymic environment (38, 39). Many of the intermediary subsets do not express the cluster of differentiation (CD)4 or CD8 co-receptors and are thus named double-negative (DN) thymocytes (40–42). Thymic stroma–induced sensory inputs are initially translated by DN1 cells using the genetic architecture of stem cells to induce multiple rounds of proliferation and differentiation. As differentiation proceeds, the stem cell “legacy” transcription factor networks are rewired with the T cell lineage–committed genetic program, which focuses critical cellular activation and survival outcomes to the quality of TCR-generated signals (38).
The earliest thymic T cell precursors (ETPs), common lymphoid precursors and lymphoid-primed multipotent precursors, seed the thymus, and have the potential to differentiate into multiple cell lineages, including lymphocytes, natural killer (NK) cells, dendritic cells, and myeloid cells (Fig. 1) (43–47). In response to the thymic microenvironment, non-T cell lineages are largely blocked, and most thymic precursors are directed into the T cell lineage (38, 48). The expression of the receptor tyrosine kinase KIT, and the interleukin-7 receptor (IL-7R), on DN1 cells enables local expression of stem cell factor (SCF, the ligand for KIT) and IL-7 to provide survival and proliferation cues. Coincident with growth factor stimulation, crosslinking the receptor Notch1 on DN1 cells by Delta-like ligand 4 (DLL4) expressed on thymic stromal cells induces cleavage of the intracellular domain of Notch (Notch-IC), releasing Notch-IC to translocate to the nucleus. Notch-IC facilitates progression into the DN2 stages and commitment to the T cell lineage, in part, by inducing expression of genes encoding the transcriptional regulators Tcf7 (TCF1) and Gata3 (48–53). Regulation of gene expression by these molecules limits alternative cellular fate choices: modulation of PU.1 target genes limits myeloid and dendritic cell potential, the reduced abundance of FMS-like tyrosine kinase 3 (Flt3) stops B cell potential, and the induction of Bcl11b represses NK cell and type 2 innate lymphoid cells (ILC2) development (12, 54–59).
Fig. 1. Lineage relationships of αβ T cell development in the thymus.
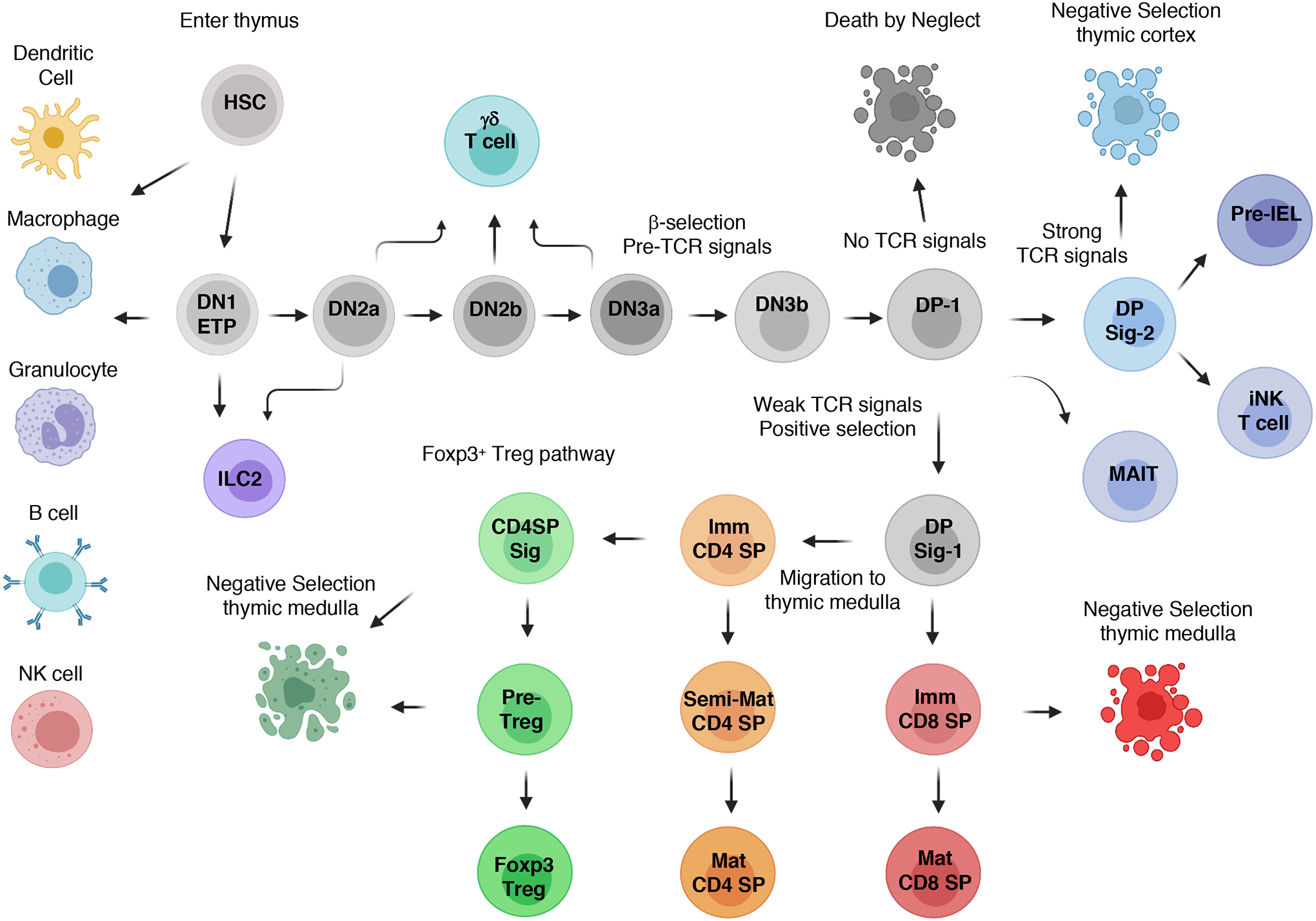
Fetal liver, followed by bone marrow-derived, hematopoietic stem cells (HSCs) enter the thymus, with most of the cells differentiating into early thymic progenitors (ETPs). Notch-1 signaling, in conjunction with proliferative and survival factors, induces progression to the double-negative 2 (DN2) stages and commitment to the T cell lineage, while limiting alternative cellular fates. TCF1 and other transcription factors induce the expression of genes encoding core TCR signaling components in DN2 thymocytes, whereas Rag1 and Rag2 expression enables the TRBV locus to be targeted for somatic recombination. Expression of a functional TCRβ chain allows DN3 thymocytes to progress through the β-selection checkpoint, which is followed by re-expression of RAG proteins to target the TRAV locus, leading to the generation of αβ TCR–expressing, resting double-positive 1 (DP-1) cells. The quality of TCR signaling derived from self-pMHC recognition at the DP-1 stage controls the initial stages of positive and negative selection, as well as the development of nonconventional T cells. CD4 and CD8 lineage choice occurs at the DP Sig-1 stage, which is followed by differentiation into immature CD4 or CD8 single-positive (SP) thymocytes. A second wave of negative selection can occur as immature CD4 and CD8 SP thymocytes migrate through the thymic medulla, with some immature CD4 SPs developing into Foxp3+ Treg cells. Note that only the major αβ thymocyte subsets are depicted.
TCF1 in DN2 thymocytes induces expression of genes encoding many of the core TCR signaling complex proteins, including CD3δ, the protein tyrosine kinase, Lck, and the adaptor protein linker of activation of T cells (Lat), as well as CD25 and Gata3 (Fig. 2) (52, 53). In addition, Notch-, E protein–, and Bc11b-dependent processes maximize the production of recombinase-activating gene 1 (Rag 1), Rag 2, CD3ε, as well as Pre-Tα (encoded by Ptrca) and terminal deoxynucleotide transferase (encoded by Dntt) (60–65). The generation of these, and additional TCR recombination and signaling molecules enable the TRBV locus to be targeted for somatic recombination, as well as the progression of thymocytes through the β-selection checkpoint. Thymic DN3 cells that generate an in-frame TCRβ chain form a surrogate TCR complex composed of the pre-Tα chain paired with the TCRβ chain, ζ-chains, and CD3 components (66–68).
Fig. 2. Canonical TCR signaling.
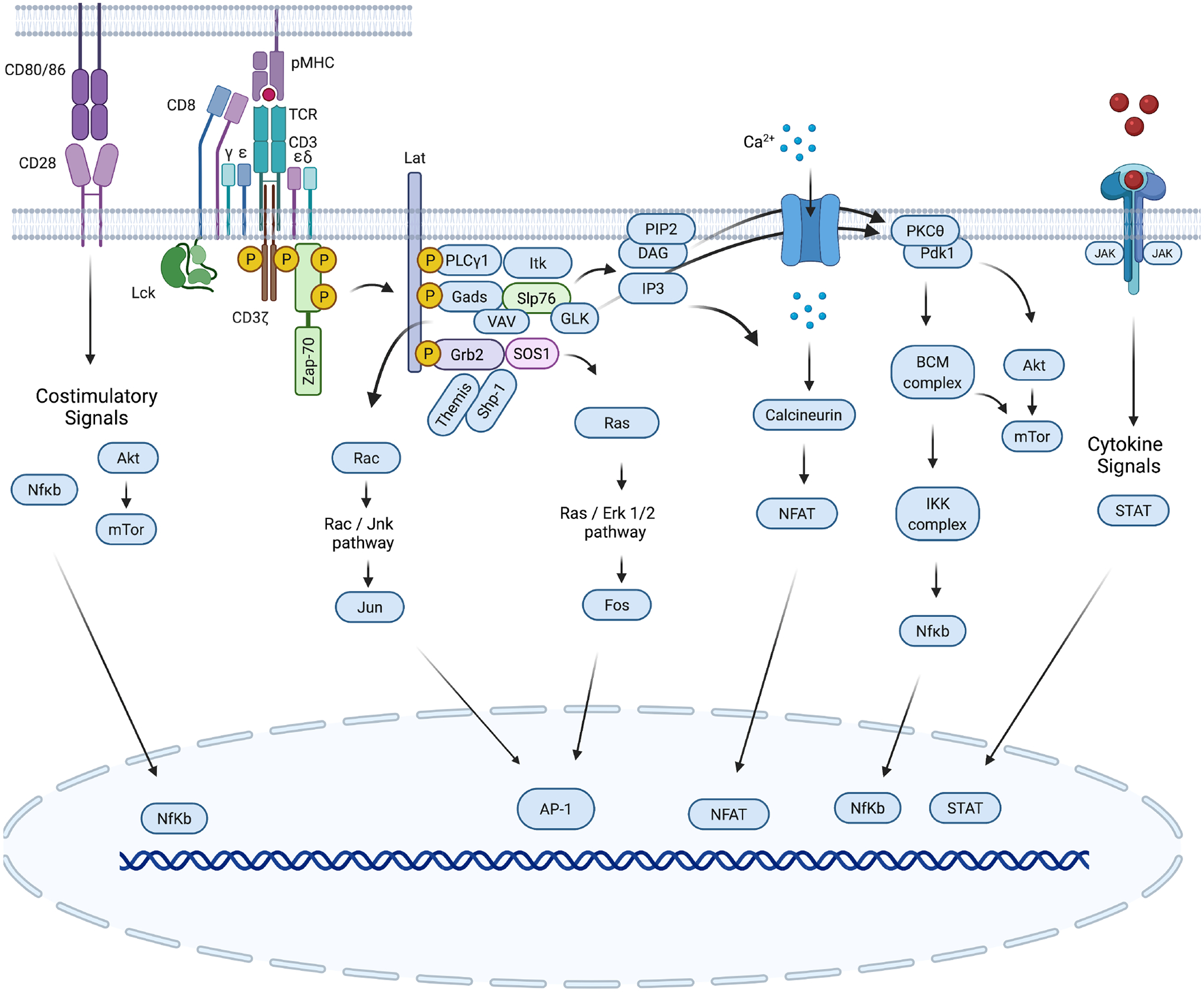
The TCR is composed of an αβ heterodimer that noncovalently associates with the CD3εδ and CD3εγ heterodimers and a ζ-chain homodimer (ζζ). Binding of the TCR to peptide-MHC complexes on the antigen-presenting cell (APC) stimulates TCR signaling. The co-receptor CD8 or CD4 interacts with the pMHC and approaches the TCR, enabling the kinase Lck to phosphorylate tyrosines within the ITAM motifs of the CD3 chain tails (10 in total). The kinase ZAP70 is then recruited to the TCR/CD3 complex through its association with phosphorylated ITAMs, and its phosphorylation by Lck causes its activation. Zap-70, in turn, phosphorylates the adapter proteins Slp76 and Lat, which form a signalosome that is a hub for other kinases and substrates to distribute and diversify the TCR signal. The enzyme PLCγ1 is activated by the kinase Itk and mediates the generation of diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). Both molecules act as second messengers for two important signaling cascades. IP3 initiates calcium (Ca2+) signaling, which leads to the activation of the transcription factor NFAT, whereas DAG and Ras-GRP enables activation of the Ras-ERK1/2 cascade (together with Grb-2–Sos). DAG, Slp76/Vav, and PI3K activity cooperate in the activation of the kinase PKCθ, which induces the formation of the CBM complex (Carma1/Bcl10/MALT1), which is followed by activation of the IKK complex (IKKα-IKKβ-IKKγ), leading to the nuclear translocation of the transcription factor NF-κB. The activity of the kinase PI3K, which is activated by TCR signals, can be amplified by costimulatory CD28 signals and enables the localization of the kinases PDK1 and Akt at the plasma membrane, where Pdk1 activates Akt. Subsequently, Akt regulates mTOR signaling, which modulates T cell metabolism. TCR engagement leads to an increase in the concentration of intracellular Ca2+, activation of the CBM complex and mitogen-activated protein kinase pathways (mediated by ERK and JNK), which ultimately results in the activation and nuclear translocation of the transcription factors NFAT, NF-κB, and AP-1.
Formation of the pre-TCR complex on the cell surface of thymocytes induces an autonomous signal, which requires proximal TCR signaling molecules, including CD3ε, Lat, Slp-76 and the kinases Lck, Syk and Zap70 (69–76). This process can be amplified through engagement of the pre-TCR with MHC ligands (77). In conjunction with Notch and IL-7 signaling, the pre-TCR signal activates phosphatidylinositol 3-kinases (PI3Ks), which promote cellular metabolism, proliferation, survival and differentiation (78–83). PI3K phosphorylates the membrane lipid phosphatidylinositol(4,5)bisphosphate (PIP2) to generate phosphatidylinositol(3,4,5)trisphosphate (PIP3), which recruits and activates Pdk1- and Akt-family kinases. Pdk1 is required for thymocyte proliferation and it activates Akt, which promotes glucose uptake, glycolysis, viability, and differentiation of DN3 and DN4 cells (84, 85). These processes trigger allelic exclusion of the second TRBV allele, initiation of TRAV gene-rearrangements at the early CD4+CD8+ ‘double-positive’ (DP) stage, expression of Rorgt and other factors, thereby promoting DP cell survival and differentiation into quiescent cells (Fig. 1) (86–89). Recombination of the TRAV locus facilitates the generation of millions of distinct αβTCRs, with each individual DP thymocyte expressing a unique αβ TCR clonotype (TCR sequence), which is then used for TCR signaling–dependent thymic selection (90).
Ligand recognition properties of TCRs expressed on pre-selection DP thymocytes
The central challenge of thymic development is to equip mature T cells with TCRs that have specificity for antigens presented by host MHC ligands; a prospect that is confounded by the immense diversity of classical MHC proteins, which are encoded by the most polymorphic genes in humans, and the enormous diversity of peptides capable of being presented by these molecules (91, 92). Correspondingly, TCR rearrangement generates exponentially greater diversity within the ligand-binding site: a mosaic of germline-encoded DNA sequences (CDR1 and CDR2), as well as the highly diverse CDR3s derived from the V and J gene segments, complimented with the random use of D gene segments and nontemplate nucleotide insertions and deletions. The result is the antigen-binding site of the TCR that localizes the V(D)J junctional amino acids within its center, which is surrounded by the CDR1 and CDR2 residues. When bound to ligands, there is a bias for the TCR CDR1 and CDR2 residues to engage the MHC, whereas the CDR3 residues often interact with the bound peptide (Fig. 3A) (14, 93, 94). Despite the enormous diversity of TCRs and of self-peptides and MHC-encoding alleles, approximately 10 to 20% of DP thymocytes have discernable reactivity with a given haplotype of MHC ligands (95–99). The underlying mechanisms that enable TCR rearrangement to generate antigen receptors with variegated values of self-reactivity and the ability of thymocytes to interpret different amounts of TCR signaling inputs to generate MHC-restricted T cell repertoires remain fundamental questions.
Fig. 3. The biochemical characteristics of amino acid residues within the V(D)J junctional portion of CDR3 calibrate the initial self-reactivity values of T cells.
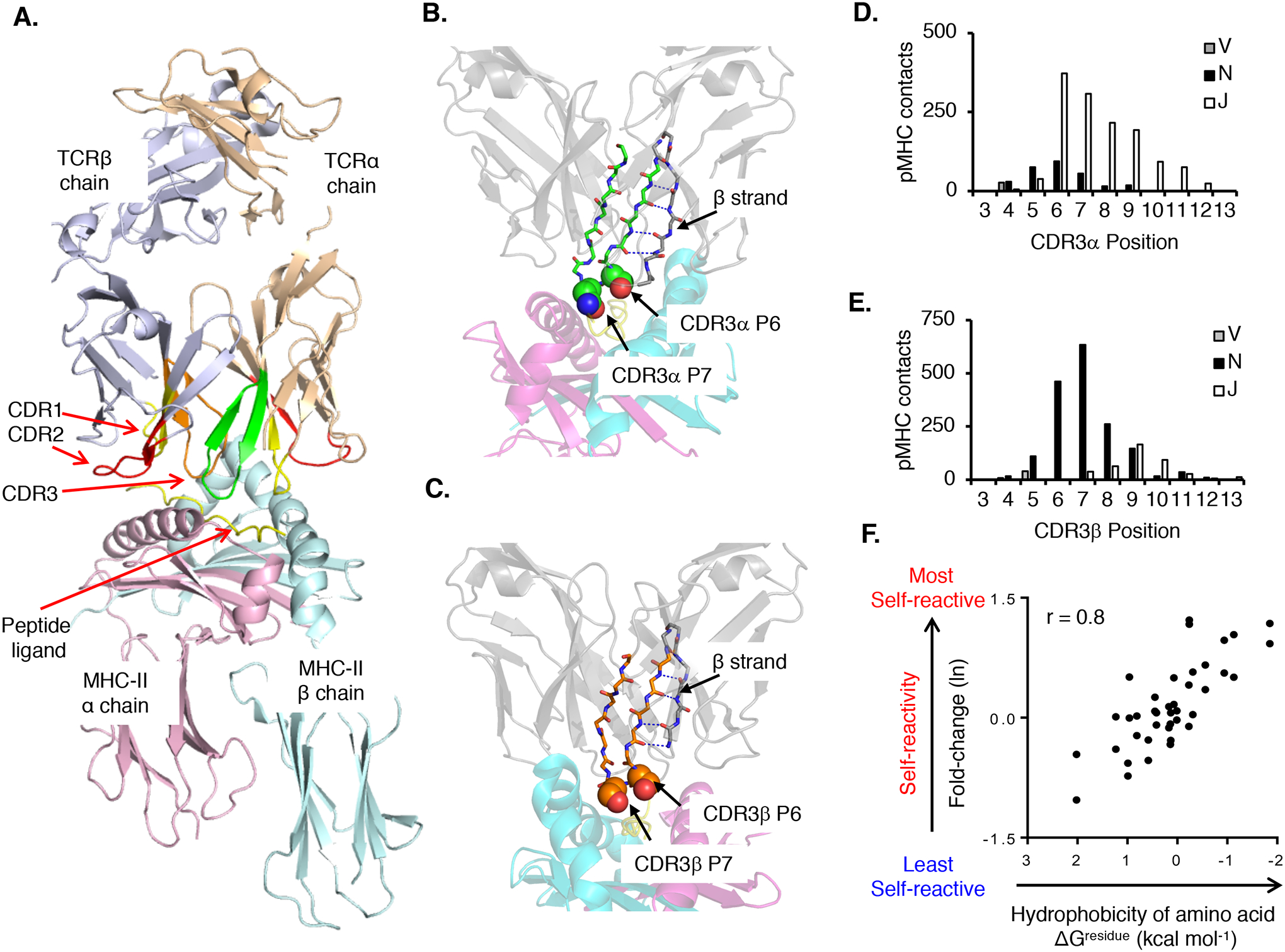
(A) Canonical orientation of an αβ TCR binding to pMHC. Highlighted are the TCRα chain (wheat) and TCRβ chain (purple), CDR1 (red), CDR2 (yellow), and CDR3α (green) and CDR3β (orange) binding to a pMHC-II ligand (PDB: 3C5Z). (B and C) A conserved β-strand within the TCRα (B) and the TCRβ V domains F-G loop (C) places CDR3 residues 6 (P6) and 7 (P7) at surface-exposed positions within the ligand-binding site. (D and E) CDR3α (D) and CDR3β (E) residues 6 and 7 form the most contacts with the pMHC. CDR3α residues contacting pMHC are primarily derived from Jα gene segments (~80%), whereas CDR3β residues contacting pMHC are primarily derived from N-region additions and Dβ gene segments (~80%). (F) Relative probability that a TCR will be self-pMHC reactive based on amino acid usage at CDR3 P6 or P7 as compared to the interfacial hydrophobicity value (ΔGresidue kcal mol−1) of each amino acid (98, 358). Each symbol represents an individual amino acid (n = 40). CDR3 contacts were assessed by using the following PDB accession codes: murine TCR–peptide–MHC class I: 1FO0, 1G6R, 1KJ2, 1LP9, 2CKB, 2O19, 2OL3, 3PQY, 3RGV, 3TF7, and 4MS8; murine TCR–peptide–MHC class II: 1D9K, 1U3H, 2PXY, 2Z31, 3C5Z, 3C6L, 3C60, 3QIB, 3QIU, 4P2Q, 4P2R, 4P5T, and 4P23; human TCR–peptide–MHC class I: 1AO7, 1BD2, 1MI5, 1OGA, 2AK4, 2BNQ, 2ESV, 2NX5, 3DXA, 3FFC, 3GSN, 3HG1, 3KPS, 3MV7, 3O4L, 3PWP, 3QDJ, 3SJV, 3UTS, 3UTT, and 4EUP; and human TCR–peptide–MHC class II: 1FTY, 1YMM, 1ZGL, 2IAM, 3O6F, 3PL6, 4E41, 4GRL, 4H1L, and 4P4K. Contacts were defined as two atoms localized within 4.0 Å, Ncont, CCP4 program suite 6.2.0.
The relatively high frequency at which DP thymocytes recognize host self-pMHC complexes that induces at least a basal level of signaling, argues that the specificities of TCR ligand-binding sites are not a random happenstance (95–101). Several characteristics of the TCR likely underlie their predilection for self-pMHC ligands. Macroscopically, the evolved specificity of TCRs for MHC ligands may involve the overall shape complementarity. The framework regions and protein fold of all TCRs are highly conserved. Across all TCR V genes, each CDR1 and CDR2 loop adopts one of a few possible canonical secondary structures, which limits major structural alterations derived from the combination of different TCR Vα and Vβ genes (102–104). However, the overall shape complementarity of TCRs with pMHC ligands is relatively poor (102), which enables minor sequence variations within the ligand-binding site of the TCR to have substantial influence on the affinity of pMHC binding. Furthermore, specific germline-encoded sequences within the CDRs modulate the frequency at which randomly generated TCRs bind to MHC ligands. Repetitive observation of particular CDR1 and CDR2 amino acid residues engaging MHC α-helices have supported the hypothesis that individual V gene residues have been evolutionarily selected for MHC binding. An example of this is a tyrosine residue at CDR2 position 48 of Vβ8.2 (Tyr48), and several other Vβs. Tyr48 is consistently observed contacting the main chain and side chains of MHC class I, MHC class II, and non-classical MHC molecules, albeit at different sites along the corresponding α-helix, and is often required for TCR recognition of pMHC ligands (105–111).
The biochemical characteristics of amino acid residues within the V(D)J junctional portion of the CDR3 synergize with germline-encoded features of the TCR to promote MHC ligand specificity. Analyses of TCR-pMHC crystal structures have found that a conserved β-strand within the TCR V domain F-G loop consistently places CDR3 residues 6 and 7 at surface-exposed positions within the ligand-binding site (Fig. 3, B and C) (98). For TCRα chains, these CDR3 residues are predominately derived from J gene segments, whereas for TCRβ chains, they are derived from N-region additions and D gene segments (Fig. 3, D and E). The positioning of these CDR3 residues at the focal point of the ligand-binding site enables their intrinsic biochemical properties to influence the binding properties of TCRs. Indeed, unbiased approaches have demonstrated that hydrophobic residues found at CDR3 positions 6 and position 7 increase the frequency and strength at which otherwise random TCRs engage pMHC complexes (Fig. 3F) (98, 112, 113). The ability of hydrophobic amino acids to promoting receptor-ligand binding likely occurs through a hydrophobic effect, which generates binding affinity through the exclusion of water molecules (114).
Boundary values of self-reactivity that control thymocyte development
Despite biases for MHC ligands, the self-reactivity value of most TCRs expressed on DP thymocyte is below perception (Fig. 1), which may portend an inability of the expressed TCR to recognize any pMHC ligand of the host, including ones that contain pathogen-derived peptides. At the opposite extreme, TCRs with very high self-reactivity values may subsequently have an inability to distinguish self-ligands from foreign ones, because recognition of both types of ligands could induce strong TCR signals. As an extension of this, inflammatory T cells that are unable to distinguish foreign- from self-ligands may cause a strong predisposition to autoimmune disease.
T cell development solves this Goldilocks-like conundrum by subjecting de novo thymocytes to positive and negative selection (115, 116). Positive selection requires a minimum level of TCR signaling derived from self-pMHC to induce thymocyte maturation, whereas thymocytes that receive TCR signals below this minimal threshold ”die by neglect” through cellular apoptosis (Fig. 4A). Similarly, thymocytes that receive strong TCR signals from self-pMHC, above that required for positive selection, often die by negative selection. However, the boundary values of self-reactivity that distinguish death by neglect, positive selection, and negative selection are not absolute, and mixed precursor-product relationships can ensue. For example, DP thymocytes that generate strong TCR signals, substantially above the threshold for positive selection, can be eliminated by negative selection or develop into nonconventional, ”innate-like” lymphocytes (117–121). Innate-like lymphocytes are often observed at mucosal sites, and, perhaps due to a deficiency in their ability to distinguish self- from foreign ligands, de-emphasize the use of TCR signals and use alternative environmental-sensing mechanisms to regulate lymphocyte function (122, 123).
Fig. 4. Model of αβ T cell development, lineage, and functional biases generated by basal self-reactivity values.
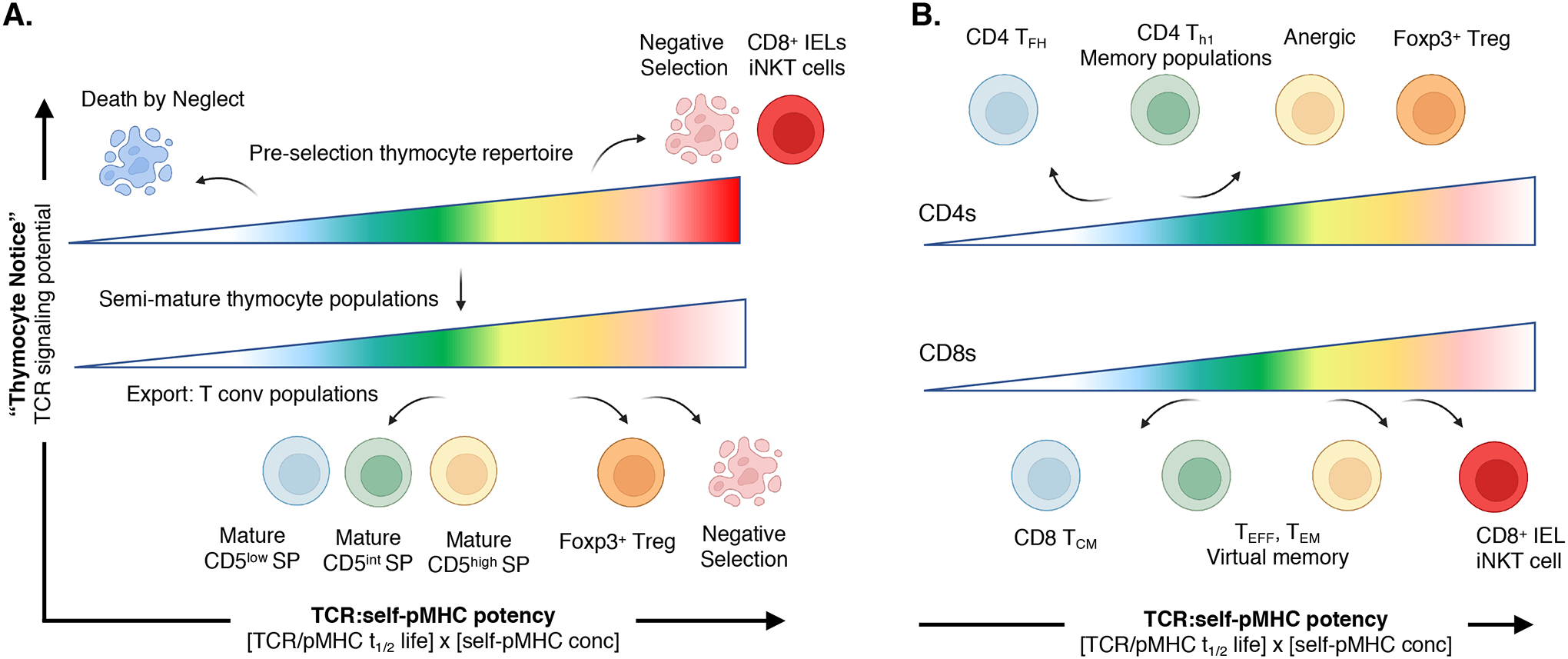
(A) Pre-selection DP thymocyte stage (top) requires TCR signaling thresholds to induce positive selection or death. Undetectable-to-weak signals results in thymocyte death by neglect, weak to moderate signals initiate positive selection, whereas stronger signals results in negative selection. CD5 expression further demarcates weak-to-strong, basal self-reactivity within the thymocyte lineages. Strong signals can also induce some MHC-I–restricted thymocytes to develop into “innate-like” CD8+ T cells. Semi-mature thymocyte populations (bottom) within the thymic medulla use continued weak TCR signaling to maintain the Tconv lineage development or stronger TCR signals to induce Foxp3-regulatory diversion or late-stage negative selection. (B) Basal self-reactivity predisposes CD4+ T cell (top) and CD8+ T cell (bottom) functional responses. Weak self-reactivity promotes CD4+ TFH and CD8+ TCM phenotypes. Increasing levels of self-reactivity bias the responding T cell to undergo stronger TEFF proliferative responses and form long-term memory as well as virtual memory, whereas the highest levels of self-reactivity are found in anergic, regulatory and innate-like T cells.
Precise quantification of the TCR–self-pMHC recognition properties that instruct thymocytes to develop into T cells have been enormously challenging; very few naturally occurring, positively selecting self-peptide ligands have been identified, and the affinities of such interactions are extremely weak (116, 124–126). To circumvent this bottleneck, stimulatory ligands that range from weak to strong potency for a particular T cell clonotype have been identified and tested for their ability to induce thymocyte selection of the corresponding thymocyte in vitro. These studies have revealed that pMHC ligands that promote positive selection have equilibrium affinities (KD) of ~ 500 μM to 1 mM (127, 128). These very weak affinity interactions largely preclude the ability to measure the half-life (t1/2) of the TCR-pMHC interaction with high confidence. Negative selection of DP thymocytes occurs when the self-pMHC interactions reach KD values of ~200 to 300 μM and t1/2 values of ~ 0.3 to 1 s, with MHC-I–restricted thymocytes requiring stronger stimuli to induce negative selection relative to MHC-II thymocytes. This discrepancy may be a consequence of CD4 binding more strongly to Lck than does CD8 (128, 129). There exists a sharp threshold between ligands that induce positive selection and those that drive negative selection, regardless of the concentration of the stimulatory ligand (127, 128, 130–132). Very few ligands are capable of initiating both positive and negative selection, depending on antigen concentration, with these interactions often promoting the development of innate like T cells (117–121, 133). Further enhancing the differences between positive and negative selectors are the development of “catch bonds” between the TCR and pMHC, which extend the dwell time during which the TCRs of the thymocytes engage stronger affinity ligand (134). It is suggested that negatively selecting ligands, but not positively selecting ligands, form these types of interactions. Whether the ability to form catch bonds is intrinsic to monomeric TCR and co-receptor/pMHC interactions or includes cellular components, for example, cell adhesion molecules that stabilize the TCR complex nanocluster interactions with the antigen-presenting cells (APC), remain debated.
Interpreting basal from excessive self-pMHC reactivity during thymocyte selection
Positive selection of thymocytes defines the minimum exogenous stimuli that can be detected through the TCR. Basing T cell maturation on a minimal signaling threshold, however, creates an essential problem. The diversity of the antigen-binding site of the TCR could, in an analogous fashion to antibodies, enable thymocytes to generate TCR signals after weak affinity engagement of any cell surface expressed protein. However, the function competency of αβ T cells is organized around their ability to uniquely recognize pMHC complexes. To ensure that thymic selection matures only MHC-restricted T cells, a critical protein tyrosine kinase, Lck, is sequestered from the TCR complex through direct binding to the cytoplasmic tails of the CD4 and CD8 co-receptors (128, 135–137). Lck is required for the initial phosphorylation of CD3 and ζ-chain components of the TCR complex that leads to downstream signaling (Fig. 2). Because the expression of Lck is limited, the amount of “co-receptor–free” Lck within the cytoplasm able to freely diffuse to the TCR complex without co-receptor is constrained (9). After antigen engagement, CD4 or CD8 co-receptors, together with associated Lck, are recruited to the TCR complex through the direct binding of CD8 and CD4 to the ectodomains of the MHC-I and MHC-II molecules, respectively (135). This molecular architecture ensures that the initial steps of TCR signaling, including the phosphorylation of CD3, Zap-70, Lat, Slp-76 and the activation of PI3K, calcium (Ca2+) flux, and other signaling pathways, require TCR engagement of ligand and cannot occur or are highly inefficient when derived from non-MHC ligands or when TCRs are bound in noncanonical orientations (137, 138).
The initial TCR signals that drive DP thymocyte selection manifest large changes in gene expression that are largely independent of the class or allele of the MHC being recognized. Layered into these initial changes in the transcriptional landscape are TCR signal quality–dependent alterations in gene expression and protein function, which set the interval between sufficient signaling for positive selection and excessive signaling that culminates in thymocyte apoptosis. The “common stem” of TCR-controlled gene networks include the canonical TCR transcriptional regulators EGR, Fos, and members of the NFAT, NF-κB, and Nur77 families (139, 140). Engagement of self-pMHC also induces expression of the genes encoding Id2 and Id3, transcriptional regulators that relieve a blockade on DP thymocyte differentiation mediated by E proteins and HEB (Tcf12). HEB and E2A (Tcf3) function as gatekeepers for further DP thymocyte differentiation by requiring that a functional αβTCR is expressed and that TCR-induced signals are generated (141–144). TCR signals promoting positive selection induce the production of anti-apoptotic Bcl2 family members, as well as IL-7R, both of which provide pro-survival cues (133, 140, 145, 146). Stronger TCR signals can induce development down the “agonist-selection” pathway (Fig. 1, Sig-2), which leads to the development of intraepithelial T cell precursors, iNK T cells, or clonal deletion through the increased production of the pro-apoptotic molecule Bim (Bcl2l11) and activation of Caspase 3, mediators of DP thymocyte cell death (146–149).
After the initial stages of positive selection, thymocytes destined to become conventional αβ T cells use asymmetrical TCR signaling to determine whether to differentiate into the CD4 or CD8 lineage. During lineage commitment, DP Sig-1 thymocytes reduce the abundance of CD8, which causes a cessation of TCR signaling in MHC-I–restricted, but not MHC-II–restricted, thymocytes (140, 150, 151). Two prototypic transcription factors that regulate CD4 and CD8 T cell differentiation are Thpok and Runx3d, respectively. TCR signal cessation induces the production of Runx3d, which in conjunction with TCF-1, silences CD4 gene expression in MHC-I–restricted thymocytes and induces CD8 re-expression (Fig. 1) (151–155). The molecular details of this process remain to be resolved and include a requirement for IL-7 and other intra-thymic cytokines to maintain survival of the intermediaries (156, 157). Runx3d further functions as a transcriptional silencer of Zbtb7b (which encodes Thpok) expression (158, 159). The re-establishment of TCR signals and the increased expression of Eomes and genes associated with granule- and interferon-γ-mediated cytotoxic functions induces differentiation into mature CD8 SP and cytotoxic lineage T cells (140, 151). In contrast, DP thymocytes that recognize MHC-II–presented self-peptides, thereby using CD4 as the co-receptor to recruit Lck to the TCR complex, maintain TCR signaling at the initial post-positive selection stage. Sustained TCR signaling enables Tcf-1 and Lef-1 to positively regulate ThPOK expression, which results in suppression of Runx3 expression and induces the expression of c-Myb, Gata3, and genes encoding other transcriptional regulators that promote the development of immature and then mature CD4+ T cells (140, 151, 157, 160).
Changing the basal state of self-reactivity: post selection thymocyte desensitize TCR signaling
Left unabated, manifestations of DP thymic selection pose a substantial predisposition to autoimmune diseases. Very weak TCR–self pMHC binding events would induce mature T cell lymphoproliferation, and overt T cell tolerance would extend only to those self-antigens that are presented within the thymic cortex during DP thymocyte selection. Counterbalancing these eventualities, post-selection CD4 and CD8 lineage–committed thymocytes alter their relative extent of self-reactivity by reducing the sensitivity of TCR signaling pathways. This process may also work bi-directionally for some clones with very weak, intrinsic self-reactivity (161). Thymocytes that maintain high self-reactivity despite this desensitization process or that recognize unique self-peptides presented within the medulla but not the cortex (162), are subject to late-stage negative selection or diversion into regulatory T cells after recognition of self-pMHC presented by thymic dendritic cells (tDCs) and medullary thymic epithelial cells (mTECs) (Fig. 4A). Thus, the maturation of T cells includes an intrinsic diminution of self-reactivity, which contributes to the establishment of graded TCR signaling thresholds; basal TCR signals derived from weak affinity self-pMHC interactions promote mature T cell survival, whereas lymphocyte activation and acquisition of effector functions require stimuli that are noticeably stronger, often derived from recognition of foreign-pMHC.
Post-selection thymocytes re-calibrate TCR signaling sensitivity through regulated gene transcriptional, translational, and posttranslational mechanisms. A set of thymocyte-intrinsic changes are digital in nature, whereby expression is present in DP thymocytes and absent in post-selection SP cells, or vice versa. Transcriptionally, the gene encoding the voltage-gated Na+ channel, VSGC, is exclusively expressed in DP thymocytes, which enables intracellular Ca2+ flux to occur after very weak self-pMHC interactions (161, 163). Similarly, thymocyte expressed, positive selection associated 1 (Tespa1) increases TCR sensitivity in DP thymocytes, whereas Themis inhibits the phosphatase activity of Shp-1 (164–166). DP thymocyte–specific sialylation patterns also increase the binding affinity of CD8 for MHC molecules (167). Reciprocally, post-selection thymocytes increase the abundance of multiple negative regulators of TCR signaling, including the phosphatases CD5 and CD45, which dephosphorylate TCR complex signaling proteins, and Cbl-b, which targets signaling molecules for degradation through its E3 ubiquitin ligase activity (168–171). The en bloc transcriptional changes leading to TCR desensitization occur in part to a reduction in the abundance of the microRNA Mir181a in post-selection thymocytes (126, 172). For several of the negative regulators, their abundance in post-selection thymocytes and T cells is dependent on and proportional to the magnitude of the TCR signals received during positive selection. This feature is thought to enable individual mature T cells to tune their TCR sensitivity in response to cues from the environment. Together, these processes contribute to a 10- to 30-fold reduction in TCR signaling sensitivity in post-selection thymocytes and limit autoimmunity (126, 173–175).
Late-stage thymocyte development: self-reactivity thresholds that distinguish Tconv cell development from late-stage negative selection and Foxp3+ Treg cell diversion
CD4 lineage– and CD8 lineage–committed thymocytes use chemokine gradients to migrate from the thymic cortex to the thymic medulla (176, 177). As thymocytes traverse the cortico-medullary junction, they interact with a network of tDCs and mTECs. Despite having already passed the first positive selection and lineage commitment checkpoints, immature CD4SP and CD8SP thymocytes that engage self-pMHC presented by mTECs or tDCs can continue along the Tconv cell differentiation process or undergo a second wave of deletion (Fig. 1). In addition, CD4SP thymocytes, and to a lesser extent CD8SP thymocytes, can be diverted into the thymic-derived regulatory (Treg) lineage (145, 162, 178–182). This “second wave of selection” occurs in part due to a set of novel self-peptides being presented to thymocytes, which are presented by migratory DCs that bring peripheral antigens to the thymus (183–185) and medullary-expressed antigens that are expressed due to the expression of Aire, Fezf2, or generated due to the presence of different proteosomes and proteases in TEC subsets (162, 186–188).
Detailed characteristics of thymocyte interactions with mTECs and tDCs that discriminate late-stage negative selection and thymic Foxp3+ Treg diversion remain outstanding, in part due to a knowledge gap about the self-antigens that drive these processes. Only a handful of self-antigens that instruct tTreg development have been identified (189–191). Nevertheless, TCR signaling reporters (117), TCRβ chain usage (98), the abundance of phosphatases such as CD5 (192), and certain TCR–self-antigen transgenic models (193–197) argue that tTreg cell differentiation occurs after the recognition of agonist self-pMHC ligands, which results in the tTreg repertoire having an increased self-reactivity relative to the CD4 Tconv cell repertoire (Fig. 4A). Agonist selection of tTreg cells, however, has not been uniformly observed, and the TCR repertoires of CD4+ Tconv cells and Foxp3+ Treg cells display some clonotype overlap (198–201). To explain these observations, it has been argued that the Foxp3 program results from altered or attenuated agonist TCR signals in conjunction with cytokine and co-stimulatory signals, which are insufficient to induce clonal deletion (162, 202–205).
TCR signals that distinguish continued Tconv cell development from Foxp3+ Treg diversion or negative selection could be ratcheted based on TCR-pMHC affinity and/or the concentration at which the self-pMHC complex is presented. Consistent with this hypothesis, a study has argued for a kinetic signaling model of Foxp3+ Treg cell development, with TCR-pMHC interactions that have a half-life (t1/2) between ~0.3 and 1 s being capable of inducing Foxp3+ Treg cell development, whereas longer interactions induce negative selection. TCR–self-pMHC interactions with shorter t1/2 values did not redirect thymocytes from becoming mature CD4 Tconv cells (191). This hypothesis dovetails with reports that TCR signal quality and downstream gene expression can be dependent upon TCR:pMHC binding kinetics (206–208). Noncanonical TCR docking on self-pMHC has also been proposed as a mechanism whereby T cells can be shuttled into the Foxp3+ Treg cell lineage. Alterations in the geometry of TCR-pMHC interactions can modify the activation of some TCR signaling cascades (209, 210), which may arise from an inability to form proper TCR-pMHC interactions or recruit co-stimulatory molecules (211). However, the relative contribution of noncanonical TCR:self-pMHC docking remains an open question because only two sets of Treg cell TCR structures have been solved bound to their self-pMHC ligands, with one report demonstrating that Treg cell TCRs can bind abnormally (212), whereas others found a highly conventional binding site orientation (191). After attenuated TCR signaling, the major developmental pathway for Foxp3+ Treg cells is thought to follow a two-step differentiation process, whereby immature CD4SP thymocytes increase the cell surface abundance of the high-affinity IL-2R, CD25, and TNFSF members (213–215). TCR signaling is thought to be important for increasing the abundance of the transcription factors NR4A1/3 and, with IL-2 signaling, initiating the Foxp3+ Treg cell epigenetic signature through the chromatin organizer SATB1 (216–219). Coincident CD28 co-stimulation is critical for NF-κB activation, enabling cRel to target the Foxp3 locus and to form an enhanceosome of transcription factors important for Foxp3 expression, which includes RelA, NFAT, Smad, and Creb (220–225). Furthermore, transient versus persistent TCR signals control Foxp3-dependent Treg cell differentiation through the Akt-mTOR axis (226).
T cell homeostasis and setting the activation threshold
After their export from the thymus, naïve αβ T cells recirculate between secondary lymphoid organs, blood, and lymph and sample antigenic peptides presented by MHC molecules on the surface of antigen-presenting cells (APCs). Most of the encounters between the TCR and self-pMHC are not prolonged enough to stimulate full activation of the naïve T cell. However, naïve T cells live for many weeks or even years in humans, relying on survival signals derived from weak TCR–self-pMHC interactions and the homeostatic cytokines IL-7 and IL-15 (133, 227–229). Once a naïve T cell engages its cognate ligand, full activation and differentiation requires TCR signaling molecules to undergo a series of reversable protein conformational changes and phosphorylation events. These events are part of the kinetic proofreading steps of TCR signal transduction, which act as a resistor of spurious TCR activation signals by including a time-delay on signaling (19–22). Thus, the commencement of TCR signaling is not instantaneous and requires TCRs to be bound by pMHC for a period of time, thereby generating a threshold that separates survival signals from those that induce immune responses.
The basal state of self-reactivity is T cell clone–specific, and T cells use these homeostatic TCR signal inputs to control their sensitivity to subsequent antigenic stimulation and to generate pre-immune T cell effector and memory lineage biases (Fig. 4B). Reflective of the law of initial value, basal TCR signaling influences the cellular state by regulating the abundance and phosphorylation status of key signaling molecules (230). Because the efficiency of TCR signaling is sensitive to the concentration of pre-formed signaling intermediates, this read-and-response mechanism enables T cells with low or high basal states of self-reactivity to be relatively quiescent during homeostasis (Fig. 5). However, the fine tuning of signaling molecule abundance and activity results in substantial functional heterogeneity; naïve T cells with high values of self-reactivity undergo stronger homeostatic proliferation and, after activation, antigen-specific expansion (31–33, 230–245) through improved sensitivity to the cytokine environment (32). As perhaps a direct test of the law of initial value, artificially lowering basal TCR input signals by transferring T cells into MHC-deficient mice is sufficient to lower T cell activation thresholds to an extent in which returning the T cells to an MHC-sufficient host leads to self-pMHC becoming autoimmune targets of what had been an immune tolerant T cell repertoire (246). Thus, in a reversable manner, basal state self-reactivity directly controls the cellular state of T cells and the ability of T cells to distinguish between self and foreign ligands.
Fig. 5. Potential mechanisms of how basal self-reactivity contributes to T cell sensitivity to pathogens.
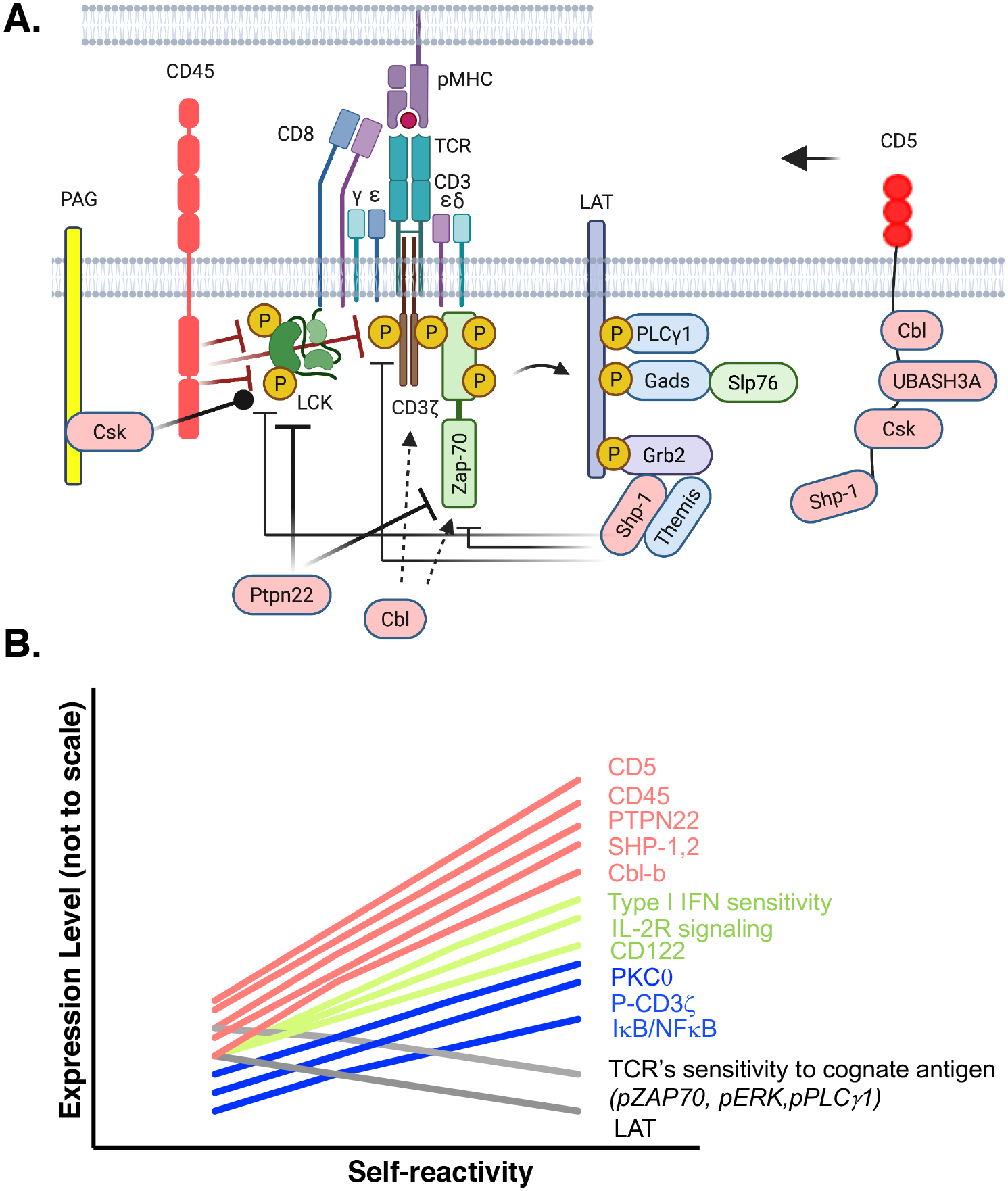
(A) High levels of self-reactivity increase the abundances of negative regulators of TCR signaling, including Shp-1, Shp-2, Cbl-b, CD5, Csk, Ptpn22, and CD45. CD45 dephosphorylates Lck and the CD3ζ chain to inhibit downstream signaling in the absence of antigen stimulation. The Lck kinase domain contains an activating tyrosine, Tyr394 and an inhibitory tyrosine, Tyr505. When Tyr394 is solely phosphorylated, Lck is fully active, whereas phosphorylation of Tyr505 inhibits Lck activity. By dephosphorylating both residues of Lck and Cd3ζ, CD45 limits, but does not abolish, Lck kinase function and counterbalances CD3ζ-chain phosphorylation. Csk is recruited to PAG and further limits Lck kinase activity by phosphorylating Lck Tyr505. Ptpn22 is a cytosolic phosphatase that once recruited to the plasma membrane can also inhibit Lck and Zap-70 activities. Shp-1 is recruited to the plasma membrane by different receptors, including CD5, and it inhibits TCR signaling by dephosphorylating Lck, Zap-70, and CD3ζ. Themis associates with LAT through Grb-2 and can inhibit SHP-1. c-Cbl and Cbl-b are E3 ubiquitin ligases that target ZAP-70 and CD3ζ for degradation and interrupt ZAP-70–CD3ζ interactions, respectively. CD5 is a membrane receptor that, when phosphorylated at cytosolic tyrosines by Lck, can recruit the inhibitory proteins c-Cbl, Cbl-b, UBASH3A, and Csk and bring them to the TCR complex. (B) Basal levels of self-reactivity influence TCR sensitivity through expression of negative regulators of TCR signals (Shp-1, Shp-2, Cbl, CD5, and CD45), which limit TCR signaling; phosphorylation of Zap-70, ERK, PLCγ1, and expression of LAT and IκB/NF-κB. Alterations further bias the T cell response to be receptive to cytokines, including IL-2 and type-1 IFN. The graph is for visualization purposes and relative differences in expression are not to scale.
Several TCR-centric mechanisms have been proposed to account for rheostat control of naïve T cell functionality based on basal self-reactivity values. During homeostasis, TCR signals induce a low level of continual TCR ζ-chain phosphorylation in the absence of cognate foreign antigen (29, 30). This is thought to increase the efficiency with which proximal TCR signaling events can be propagated. Basal TCR signals are also thought to promote TCR-Zap70 co-polarization in CD4 T cells after cognate antigen recognition and stimulate cell motility, enabling the T cell to scan the APC and form T cell–APC conjugates (29, 247). In addition, T cells with higher self-reactivity values increase the cell surface abundance of CD5 and the related molecule CD6. Signaling by CD5 (and CD6) induces Ca2+ flux, activates the PI3K, PKC, and CK2 signaling pathways, and induces the recruitment of Gads and SLP76 to the phosphorylated cytoplasmic domain of CD5 and CD6 (248–250). Paradoxically, CD5 and CD6 can also function as negative regulators of TCR signaling. CD5 can inhibit the TCR signaling kinase, Fyn (251), and associate with the phosphatase Shp-1, Ras-GAP, and the ubiquitin ligase Cbl (which targets CD3ζ and Zap-70 for degradation), as well as Csk (an inhibitor of Fyn and Lck) to impair T cell signaling (252). The context in which CD5 and CD6 function as either positive or negative regulators of TCR signaling may arise from the assembly of distinct signalosomes around LAT–SLP-76, depending on the strength of the self-pMHC interaction and the abundances of their ligands on APCs (253–255). Further clouding a simple TCR-centric model, the strength of basal TCR signals is inversely correlated with the abundance of CD8, which carries the kinase Lck, and the efficiency of generating proximal TCR signaling events (242, 244, 256), and influence T cell metabolism through Myc and Akt functions (257). Consistent with this, a report suggests that CD5hi naïve T cell have reduced amounts of LAT and increased amounts of the phosphatases Shp1, SHP2, Cbl-b, and CD45 compared to those of CD5lo T cells (Fig. 5) (256).
These studies raise the question as to how increases in the basal state of self-reactivity can enhance T cell responses while simultaneously increasing the threshold of TCR signaling. Alterations in cytokine responsiveness may solve this conundrum. The extent of self-reactivity directly influences the ability of IL-7, IL-15, and type I IFN, to transmit stimulatory and pro-survival signals through altering the abundance of cytokine receptors and their downstream signaling molecules (32, 256, 258). Although it is clear that naïve T cells integrate signals emanating from the TCR, cytokine receptors, and adhesion/co-stimulatory receptors during activation, the molecular details of this synergy are largely unknown, with a study suggesting that Themis may act as a crucial integrator in this process (259).
Layered into the overall reactionary aspects of the naïve T cell repertoire are ‘virtual memory’ and ‘innate memory’ clones that are foreign antigen–inexperienced T cells that yet carry a memory phenotype (260, 261). Consistent with the law of initial value, a commonality of theses anticipatory clones that have pre-formed effector functions is a high basal state of self-reactivity (262–264). Whereas innate memory T cells arise in the thymus and virtual memory T cells differentiate in the periphery, both subsets depend on IL-15 signals to induce production of the transcription factor Eomes, thereby providing these T cells an ability to secrete IFN-γ after antigen recognition (Fig. 4B) (260). Thus, the basal state self-reactivity influences the functionality and clonal complexity within the antigen-inexperienced repertoire, which is propagated through naïve, effector, and memory T cell subsets (Fig. 4B).
Primary T cell activation: outcomes based on the strength of TCR signals
The architecture of the TCR complex aids in the ability of T cells to distinguish homoeostatic from foreign- and diseased-self ligands and transmit signals of different quality. During homeostasis, the CD3ε and CD3ζ cytoplasmic domains of the TCR complex can bind to the inner leaflet of the plasma membrane, an arrangement that inserts the aromatic tyrosine of the immunoreceptor tyrosine-based activation motif (ITAM) within the plasma membrane (265–268). This process is a dynamic equilibrium that enables some Zap70 to be bound to phosphorylated CD3 components (269), although Zap70 remains in an inactivated state in the absence of Lck recruitment to the TCR complex and subsequent phosphorylation (270). This membrane-associated, biased equilibrium of TCR signaling components may limit spurious overt T cell activation, while enabling homeostatic signaling to be generated. Two ligand recognition properties contribute to overcoming this resistance to T cell activation: the half-life of the TCR-pMHC interaction (potency) and the local concentration at which the activating ligand is present (density). For many aspects of T cell biology, these two variables are not equivalent; growing evidence suggests that aspects of T cell activation and differentiation are a product of the summation of total signal (potency × density), whereas others require particular long-lived, potent TCR-pMHC engagements (206–208, 271, 272). Long-lived TCR-pMHC interactions, in particular, are thought to contribute to the initiation of productive TCR signals by enabling the ITAMs of the TCR complex to become maximally phosphorylated and subsequently induce NFAT translocation to the nucleus (28, 265–268).
The threshold for a T cell to enter into cell cycle and proliferate correlates with the quantity of Zap-70 activity that is reached (273). After TCR engagement by agonist pMHC ligands, the ITAMs of the TCR complex can be released from the membrane, potentially due to the shear force applied to the TCR and changes in the local environment, to become available for phosphorylation by co-receptor-bound Lck (265–268). Phosphorylation and reorganization of the TCR complex, including potential conformational changes within the CD3 and TCR components, enable the translation of extracellular TCR binding events into intracellular signaling (274–282). This process is scalable to an extent, because the number of phosphorylated ITAMs within the TCR complex is based on the potency and density of TCR-pMHC interactions (283, 284). The Zap-70 kinase binds to phosphorylated ITAMs within the TCR complex, even at a resting state (269). However, only long-lived and appropriately oriented TCR-pMHC interactions enable co-receptor-bound Lck to phosphorylate ITAM-associated Zap-70 (138), thereby stimulating its enzymatic activity (269). Built into this pathway is the inefficient ability of Zap-70 to phosphorylate its immediate downstream target, LAT, which controls the activation of multiple downstream signaling pathways. This enzymatic inefficiency of Zap-70 generates a TCR-proximal kinetic proofreading step that defines the minimal difference in stimuli that can distinguish homeostatic from T cell–activating signals (285).
For TCR signals that overcome the activation energy barrier, antigen potency and density, in coordination with cytokines, provide unique inputs that control a Myc-dependent cell division timer and an independent cell death timer, which regulates T cell proliferation and clonal expansion (286, 287). The magnitude of cell division (the accumulation of cells that enter into the cell cycle and the number of cell divisions) is programmed early by TCR and costimulatory signals, whereas the cytokine IL-2 governs the duration of cell division after T cell activation (288). Co-stimulation in the form of CD28-B7 interactions is crucial for naïve T cells to proliferate, because CD28 signaling reinforces several TCR signaling pathways, including PLCγ-Ca2+ mobilization, PI3K, NF-κB, and JNK (289). Synergistically, antigen potency and density influence the probability and rate at which T cells are activated and expand (273, 290), whereas ligand potency appears to be the dominant regulator of apoptosis after cell division (290). Initial T cell survival, the frequency at which daughter T cells survive versus undergo apoptosis after the first cell division, depends on the capacity to switch from a survival pathway that depends on a signaling pathway including IL-7, IL-7R, pSTAT5, and Bcl2, which regulates naïve T cell homeostasis, to a signaling pathway mediated by the TCR, Ca2+, ERK, and Bclxl/A1, which is determined by TCR signal strength (291). Together, these findings argue that the magnitude of TCR signals regulates T cell division, whereas antigen potency and cytokine signaling modulate T cell survival.
If TCR signal strength regulates T cell proliferation and survival, why do some weak-potency T cell responses occur in vivo? It is possible that synergies between high-density ligands and the amount of the cytokine IL-2 (or IL-12) enable naïve T cells to reach the threshold that triggers Myc production and cell survival (286). Consistent with this idea, IL-2 and co-stimulatory signals increase the ability of naïve T cells to proliferate in response to weak-potency ligands (292–294). The TCR-proximal signaling molecule Themis may also be critical in this process, because Themis-deficient mature CD8+ T cells show a defect in the expression of c-Myc and in proliferation in response to low-potency antigens and cytokines (259). Thus, for T cell clonal expansion, Myc appears to be the final arbiter that distinguishes pathogen recognition from homeostatic TCR signal inputs.
It is important to note that T cell clones do not function in isolation, and signals from neighboring T cells can help to achieve a coordinated, and hence, more effective response. Two mechanisms that help orchestrate T cell responses are regulatory T (Treg) cells (295, 296) and the influence of T cell quorum sensing. Quorum sensing is a process of cell-cell communication first described for bacteria in which the production and detection of extracellular chemicals are used to monitor cell population density and synchronize gene expression of the group, thereby enabling individuals to act in unison (297). Although still at an early stage of discovery, quorum sensing of T cell cytokine secretion and APC activation have been proposed as mechanisms to limit the ability of rare, potentially autoimmune clones from inducing pathology (298–300), as well as a control mechanism to coordinate CD8+ T cell responses (301).
T cell differentiation and memory formation
Once recruited into an immune response, naïve T cells have the potential to differentiate into a multitude of stable and metastable subsets described by the acquisition of specific effector functions. Activated CD4+ T cell subsets provide diverse aid to the clearance of pathogens. These include direct effector functions, providing help to CD8+ T cells and for B cell affinity maturation in the germinal center, as well as regulating the function of macrophages, DCs, and even other T cell subsets. CD8+ T cells, the sentries of the adaptive immune system, predominately acquire cytotoxic and type-1 effector functions (Tc1), including the ability to produce IFN-γ and directly lyse infected targets through the production of granzyme and perforin (302). The acquisition of cytotoxicity appears to be a default program, with ligand potency controlling how rapidly and uniformly T cells become activated at the population level (303, 304). However, for many CD4+ and CD8+ T cell effector functions, distinct calibrated levels of TCR signaling based on antigen density and potency are required.
How TCR signals synergize with the inflammatory and local tissue cytokine milieu to regulate CD4+ T cell effector differentiation continues to be a long-standing question in the field (305). CD4+ T cell differentiation is responsive to signal inputs from the TCR and cytokine receptors; independently, strong TCR stimulation in the absence of inflammatory cues can result in peripheral T cell anergy, deletion, or differentiation into regulatory lineages, an attractive therapeutic approach to limiting autoimmune disease (306–308). Reflective of need, naive CD4+ T cells can differentiate into CD4+ T helper 1 (TH1) cells, which are able to activate macrophages and dendritic cells; TH2 cells, which regulate eosinophils and mast cell degranulation; TH17 cells, which influence stromal cell activation and neutrophil recruitment; and T follicular helper (TFH) and germinal center (GC-TFH) cells, which provide B cell help. Within noninflammatory environments, self-, food-, and microbiota-specific CD4+ T cells can be induced to acquire anti-inflammatory regulatory properties (pTregs). Some T cell effector functions and differentiation fates closely align with TCR signal intensity (for TH1 and TFH cells), but the link for other cell types is less clear (TH2, TH17, and iTreg cells) (35–37, 207, 304, 309–313). TH1 and TFH cells require high levels of acute TCR signals (derived from high-affinity TCR-ligands and long dwell-time interactions or from high antigen density), which are required to fully phosphorylate all of the ζ-chain ITAMs and activate the signaling intermediates (Zap-70, Slp76, Shp1, and Cbl) necessary to support the downstream effector pathways mediated by ERK and Akt (36, 313, 314). Full activation of these signaling pathways is critical for the expression of the TFH and GC-TFH lineage transcription factor Bcl6 and the TH1 lineage transcription factors T-bet and Blimp-1 (36, 207, 253, 313, 315).
If both TH1 and TFH CD4+ T cells are induced by strong TCR signals, what are the distinguishing factors that direct the different effector lineage outcomes? Within the spectrum of TCR signals, the decision between TFH and TH1 cell differentiation first comes down to the abundance of the transcriptional regulator IRF4 that the TCR signal can support. IRF4 amounts are translated into preferential access to high- or low-affinity binding sites in the DNA regulatory elements that regulate the TFH cell versus TH1 cell transcriptional programs, respectively (206, 207). On a second step that further delineates the commitment towards TFH or TH1 cells, the extent of TCR signal regulates the input of IL-2 signals through the expression of IL-2R on the surface of effector T cells. Because IL-2 represses TFH cell differentiation (316, 317), strong TCR signals result in a higher frequency of cells that express the IL-2Rβ subunit and the high-affinity IL-2Rα chain, CD25, which further stimulates TH1 cell differentiation (315, 318, 319). However, layered into this process are the observations that increasing self-pMHC basal signaling inhibits early decisions towards TFH cell differentiation, while favoring a highly activated TH1 cell fate (Fig. 4B) (320). Mechanistically, how the initial self-reactivity value of a naïve T cell clone impinges on CD4+ T cell differentiation remains outstanding and may include metabolic as well as transcriptional regulation (311, 321–323).
CD8+ effector T cell differentiation is coupled with a metabolic shift into aerobic glycolysis and the preferential expression of the transcriptional regulators T-bet and Blimp-1 rather than Eomes and Bcl6 (324). Strong TCR signals control the relative abundances of IRF4, Blimp-1, and T-bet, thereby activating IFNγ and Gmzb promoters and sustaining effector function (325–328). Thus, a high TCR signaling threshold for cytotoxic CD8+ T cells, as well as for CD4+ TH1 and TFH cells, may be intentionally established to avoid spontaneous or overt responses that can lead to autoimmunity or cytokine storms in response to self-antigens. After clearance of an infection, most of the effector CD4+ and CD8+ T cells generated in an immune response die. Those that survive as memory T cells retain the antigen-specificity of the effector cells that participated in the response but, unlike them, are endowed with molecular properties that enable them to be long-lived and fast functional responders to re-infection (329). T cell memory is phenotypically and functional heterogenous, comprised of central memory (TCM), effector memory (TEM), peripheral memory (TPM), resident memory (TRM), and stem cell memory (TSCM) subsets (330). Each memory cell subset is endowed with certain overlapping and unique functional capacities (331). The TCR signaling threshold for both CD4+ and CD8+ T cell memory development is thought to be moderately low with weak signals capable of inducing the memory-associated transcriptional and metabolic memory programs (37, 315, 328, 332–335). Interestingly, virtual memory T cells preferentially develop into TCM cells after antigen recognition (336). These response features of the T cell repertoire may be used to maximize clonal diversity within the memory compartment.
Transcriptional control of T cell effector and memory development operates under a reciprocal balance between those transcriptional regulators associated with effector differentiation (for example, Blimp-1 and T-bet) and those associated with memory formation (for example, Eomes, TCF-1, and Bcl6) (37, 315, 325, 332, 337, 338). Weak-potency TCR ligands appear to have an inherent ability to induce the expression of genes encoding the memory-associated transcription factors Eomes and TCF-1, whereas they limit the expression of genes encoding transcription factors associated with effector cell differentiation and expansion (332, 339). However, it is not completely clear how these relationships are shaped at the TCR-proximal level. It has been postulated that weak-potency TCR signals regulate T cell memory through the assembly of signalosomes that are different from those assembled in response to strong TCR ligands (333, 340, 341). Consistent with this, experimental reduction of signals that are associated with strong TCR stimulation (including, Ca2+, ERK and mTOR) do not impair T cell memory (333, 340–342). Rather, signaling pathways mediated by NF-κB and Wnt, which are not distinctively associated with strong T cell activation, appear to have a more prominent role in directing T cell memory (343–346).
The hypothesis that weak T cell activation promotes memory cell formation is primarily drawn from studies of T cell central and effector memory cell differentiation (333, 341, 347, 348), with weak TCR signals favoring the development of TCM through the production of increased amounts of Eomes and Tcf-1 (315, 332). A similar conclusion has been reached for TRM cells in the lung, although whether this holds true for all tissues remains controversial (349, 350). In addition to antigenic signals, memory T cell differentiation requires pro-inflammatory signals from different cytokines. TCR signal strength regulates the abundance of pro-inflammatory cytokine receptors on T cells, and thus the mosaic of environmental inputs that the T cell receives (332, 351). It is believed that total overall stimulatory potential determines the extent of T effector cell and memory cell differentiation, with TSCM and TCM cells being less differentiated, whereas TEFF are fully differentiated (330) and have a lower survival potential. Yet, even though weak-potency TCR signals favor the development of memory T cells, memory T cells are also generated in response to strong TCR stimulation. Indeed, T cells that bear high-affinity TCRs are abundant in the memory repertoire because of their strong proliferative advantage (37, 352). Ensuring that immune responses to weak- and strong-potency antigens supports the generation of memory T cells and is consistent with the idea of guaranteeing T cell clonal diversity as the host loses thymic output and gains antigen experience in the memory pool with age (353). Undoubtedly, enabling the generation of memory T cells specific for a wider range of TCR ligands will increase the breadth of ligand reactivity, which may provide the host with an increased advantage to respond to rapidly evolving and new pathogens.
Concluding remarks
T cells, like many biological systems, are responsive to environmental stimuli to guide their development, calibrate their sensitivity to activation signals, and govern the magnitude and functional specialization during responses. Defining the first principles of T cell function has enabled the development of therapeutics to override tolerance mechanisms to unleash T cells to attack cancers and rebalance homeostasis in patients prone to autoimmunity. As the field develops, the principles behind the law of noticeable differences and the law of initial value should help guide our understanding of T cell behavior and answer a number of outstanding questions. How can vaccines be developed to generate T cells with a broad or narrow breadth of effector functions? Why are many highly self-reactive T cells eliminated, whereas others persist? Why do some T cell clones have autoimmune potential, whereas others are merely self-reactive? What is the nature of T cell clones that can be re-invigorated during chronic viral infections to eliminate the pathogen? How can designer chimeric antigen receptor (CAR) T cell therapies be tailored toward high-efficiency tumor eradication while minimizing off-target effects? Answers to these questions will likely involve characteristics of the interactions between the TCR and antigens, as well as the temporal and spatial constraints of when and where antigen is recognized. Indeed, reminiscent of the positive or negative regulation of blood pressure by adrenaline, proinflammatory T cells need defined periods of secession from TCR signaling to maintain peak function (354–356) and to limit their becoming non-functional or their differentiation into immunoregulatory cells (304, 311, 357).
Acknowledgments:
The authors thank M. Daniels, D. G. Pages, W.-L. Lo, R. Obst, and B. Stadinski for their critical review of the manuscript.
Funding:
E.S.H. is supported by the US National Institutes of Health (AI143976 and AR071269). E.T. is supported by the US National Institutes of Health (AI110420-06A1 and 1U01CA244314) and University of Missouri internal funds.
References and Notes
- 1.Fechner GT, Elemente der Psychophysik [Elements of psychophysics]. band 2. Leipzig: Breitkopf und Härtel., (1860). [Google Scholar]
- 2.Fechner GT, [First published .1860]. Howes DH; Boring EG (eds.). Elements of psychophysics [Elemente der Psychophysik]. volume 1. Translated by Adler HE. United States of America: Holt, Rinehart and Winston., (1966). [Google Scholar]
- 3.Weber EH, De pulsu, resorptione, audita et tactu.-Annotationes anatomicae et physiologicae-. Trs. by Ross HE, Academic Press, New York, 1978, (1834). [Google Scholar]
- 4.Ekman G, Weber’s Law and Related Functions. The Journal of Psychology 47, (1959). [Google Scholar]
- 5.MacKay DM, Psychophysics of perceived intensity: A theoretical basis for Fechner’s and Stevens’ laws. Science 139, 1213–1216 (1963).14019221 [Google Scholar]
- 6.WILDER J, THE LAW OF INITIAL VALUE IN NEUROLOGY AND PSYCHIATRY: Facts and Problems1. The Journal of nervous and mental disease 125, 73–86 (1957). [DOI] [PubMed] [Google Scholar]
- 7.Mueller DL, Jenkins MK, Schwartz RH, Clonal expansion versus functional clonal inactivation: a costimulatory signalling pathway determines the outcome of T cell antigen receptor occupancy. Annu Rev Immunol 7, 445–480 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Paul WE, Seder RA, Lymphocyte responses and cytokines. Cell 76, 241–251 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty AK, Weiss A, Insights into the initiation of TCR signaling. Nat Immunol 15, 798–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruterbusch M, Pruner KB, Shehata L, Pepper M, In Vivo CD4+ T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annual Review of Immunology 38, 705–725 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Jameson SC, Masopust D, Understanding Subset Diversity in T Cell Memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yui MA, Rothenberg EV, Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14, 529–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schatz DG, Oettinger MA, Baltimore D, The V(D)J recombination activating gene, RAG-1. Cell 59, 1035–1048 (1989). [DOI] [PubMed] [Google Scholar]
- 14.Davis MM, Bjorkman PJ, T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988). [DOI] [PubMed] [Google Scholar]
- 15.Oettinger MA, Schatz DG, Gorka C, Baltimore D, RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248, 1517–1523 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Arstila TP et al. , A direct estimate of the human alphabeta T cell receptor diversity. Science 286, 958–961 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Robins HS et al. , Overlap and effective size of the human CD8+ T cell receptor repertoire. Science translational medicine 2, 47ra64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins MK, Chu HH, McLachlan JB, Moon JJ, On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol 28, 275–294 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Hopfield JJ, Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A 71, 4135–4139 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ninio J, Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595 (1975). [DOI] [PubMed] [Google Scholar]
- 21.McKeithan TW, Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci U S A 92, 5042–5046 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lever M, Maini PK, van der Merwe PA, Dushek O, Phenotypic models of T cell activation. Nat Rev Immunol 14, 619–629 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Sloan-Lancaster J, Allen PM, Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu Rev Immunol 14, 1–27 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Siller-Farfan JA, Dushek O, Molecular mechanisms of T cell sensitivity to antigen. Immunol Rev 285, 194–205 (2018). [DOI] [PubMed] [Google Scholar]
- 25.De Magistris MT et al. , Antigen analog-major histocompatibility complexes act as antagonists of the T cell receptor. Cell 68, 625–634 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Tischer DK, Weiner OD, Light-based tuning of ligand half-life supports kinetic proofreading model of T cell signaling. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yousefi OS et al. , Optogenetic control shows that kinetic proofreading regulates the activity of the T cell receptor. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin JJY et al. , Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci Signal 12, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefanova I, Dorfman JR, Germain RN, Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 420, 429–434 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Surh CD, Sprent J, Homeostasis of naive and memory T cells. Immunity 29, 848–862 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Persaud SP, Parker CR, Lo WL, Weber KS, Allen PM, Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol 15, 266–274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fulton RB et al. , The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat Immunol 16, 107–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN, T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38, 263–274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabinowitz JD et al. , Altered T cell receptor ligands trigger a subset of early T cell signals. Immunity 5, 125–135 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Vanguri V, Govern CC, Smith R, Huseby ES, Viral antigen density and confinement time regulate the reactivity pattern of CD4 T-cell responses to vaccinia virus infection. Proc Natl Acad Sci U S A 110, 288–293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tubo NJ et al. , Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zehn D, Lee SY, Bevan MJ, Complete but curtailed T-cell response to very low-affinity antigen. Nature 458, 211–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosokawa H, Rothenberg EV, How transcription factors drive choice of the T cell fate. Nat Rev Immunol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah DK, Zuniga-Pflucker JC, An overview of the intrathymic intricacies of T cell development. J Immunol 192, 4017–4023 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Penit C, Vasseur F, Cell proliferation and differentiation in the fetal and early postnatal mouse thymus. J Immunol 142, 3369–3377 (1989). [PubMed] [Google Scholar]
- 41.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC, Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 26, 678–689 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Godfrey DI, Kennedy J, Suda T, Zlotnik A, A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol 150, 4244–4252 (1993). [PubMed] [Google Scholar]
- 43.Adolfsson J et al. , Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Serwold T, Ehrlich LI, Weissman IL, Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood 113, 807–815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlenner SM et al. , Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32, 426–436 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Luc S et al. , The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol 13, 412–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell JJ, Bhandoola A, The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452, 764–767 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC, Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol 178, 858–868 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Radtke F, MacDonald HR, Tacchini-Cottier F, Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol 13, 427–437 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Hosoya T et al. , GATA-3 is required for early T lineage progenitor development. J Exp Med 206, 2987–3000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visan I et al. , Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat Immunol 7, 634–643 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Germar K et al. , T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A 108, 20060–20065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber BN et al. , A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbauer F et al. , Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 38, 27–37 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Ojeda ME et al. , GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood 121, 1749–1759 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Ikawa T et al. , An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Li P et al. , Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Leid M, Rothenberg EV, An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Obaldia ME, Bhandoola A, Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol 33, 607–642 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Takeuchi A et al. , E2A and HEB activate the pre-TCR alpha promoter during immature T cell development. J Immunol 167, 2157–2163 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Ikawa T, Kawamoto H, Goldrath AW, Murre C, E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med 203, 1329–1342 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C, Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A 103, 9976–9981 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Georgescu C et al. , A gene regulatory network armature for T lymphocyte specification. Proc Natl Acad Sci U S A 105, 20100–20105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu W et al. , E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood 121, 1534–1542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welinder E et al. , The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci U S A 108, 17402–17407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Groettrup M et al. , A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell 75, 283–294 (1993). [DOI] [PubMed] [Google Scholar]
- 67.von Boehmer H, Fehling HJ, Structure and function of the pre-T cell receptor. Annu Rev Immunol 15, 433–452 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Yamasaki S, Saito T, Molecular basis for pre-TCR-mediated autonomous signaling. Trends Immunol 28, 39–43 (2007). [DOI] [PubMed] [Google Scholar]
- 69.DeJarnette JB et al. , Specific requirement for CD3epsilon in T cell development. Proc Natl Acad Sci U S A 95, 14909–14914 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malissen M et al. , Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J 14, 4641–4653 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W et al. , Essential role of LAT in T cell development. Immunity 10, 323–332 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Clements JL et al. , Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science 281, 416–419 (1998). [DOI] [PubMed] [Google Scholar]
- 73.Groves T et al. , Fyn can partially substitute for Lck in T lymphocyte development. Immunity 5, 417–428 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Pivniouk V et al. , Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell 94, 229–238 (1998). [DOI] [PubMed] [Google Scholar]
- 75.van Oers NS, Lowin-Kropf B, Finlay D, Connolly K, Weiss A, alpha beta T cell development is abolished in mice lacking both Lck and Fyn protein tyrosine kinases. Immunity 5, 429–436 (1996). [DOI] [PubMed] [Google Scholar]
- 76.Cheng AM et al. , The Syk and ZAP-70 SH2-containing tyrosine kinases are implicated in pre-T cell receptor signaling. Proc Natl Acad Sci U S A 94, 9797–9801 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X et al. , Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection. Science 371, 181–185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV, Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Kreslavsky T et al. , beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity 37, 840–853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tussiwand R et al. , The preTCR-dependent DN3 to DP transition requires Notch signaling, is improved by CXCL12 signaling and is inhibited by IL-7 signaling. Eur J Immunol 41, 3371–3380 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Boudil A et al. , IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte beta-selection. Nat Immunol 16, 397–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ciofani M, Zuniga-Pflucker JC, Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol 6, 881–888 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Hosokawa H, Rothenberg EV, Cytokines, Transcription Factors, and the Initiation of T-Cell Development. Cold Spring Harbor perspectives in biology 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Juntilla MM, Koretzky GA, Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol Lett 116, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fayard E, Moncayo G, Hemmings BA, Hollander GA, Phosphatidylinositol 3-kinase signaling in thymocytes: the need for stringent control. Sci Signal 3, re5 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Sun Z et al. , Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000). [DOI] [PubMed] [Google Scholar]
- 87.Guo J et al. , Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol 3, 469–476 (2002). [DOI] [PubMed] [Google Scholar]
- 88.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW, An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol 11, 240–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan J, Crittenden RB, Bender TP, c-Myb promotes the survival of CD4+CD8+ double-positive thymocytes through upregulation of Bcl-xL. J Immunol 184, 2793–2804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bradley P, Thomas PG, Using T Cell Receptor Repertoires to Understand the Principles of Adaptive Immune Recognition. Annu Rev Immunol 37, 547–570 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Robinson J et al. , The IMGT/HLA database. Nucleic Acids Res 37, D1013–1017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trowsdale J, Knight JC, Major histocompatibility complex genomics and human disease. Annual review of genomics and human genetics 14, 301–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garcia KC et al. , An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219 (1996). [DOI] [PubMed] [Google Scholar]
- 94.Rossjohn J et al. , T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Zerrahn J, Held W, Raulet DH, The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 88, 627–636 (1997). [DOI] [PubMed] [Google Scholar]
- 96.Merkenschlager M et al. , How many thymocytes audition for selection? J Exp Med 186, 1149–1158 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Meerwijk JP et al. , Quantitative impact of thymic clonal deletion on the T cell repertoire. J Exp Med 185, 377–383 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stadinski BD et al. , Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat Immunol 17, 946–955 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stritesky GL et al. , Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci U S A 110, 4679–4684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jerne NK, The somatic generation of immune recognition. Eur J Immunol 1, 1–9 (1971). [DOI] [PubMed] [Google Scholar]
- 101.Blackman M et al. , The T cell repertoire may be biased in favor of MHC recognition. Cell 47, 349–357 (1986). [DOI] [PubMed] [Google Scholar]
- 102.Ysern X, Li H, Mariuzza RA, Imperfect interfaces. Nature structural biology 5, 412–414 (1998). [DOI] [PubMed] [Google Scholar]
- 103.Housset D, Malissen B, What do TCR-pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol 24, 429–437 (2003). [DOI] [PubMed] [Google Scholar]
- 104.Al-Lazikani B, Lesk AM, Chothia C, Canonical structures for the hypervariable regions of T cell alphabeta receptors. J Mol Biol 295, 979–995 (2000). [DOI] [PubMed] [Google Scholar]
- 105.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC, Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol 8, 975–983 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Dai S et al. , Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity 28, 324–334 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garcia KC, Adams JJ, Feng D, Ely LK, The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol 10, 143–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Borg NA et al. , CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007). [DOI] [PubMed] [Google Scholar]
- 109.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P, Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature 458, 1043–1046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stadinski BD et al. , A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity 35, 694–704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yin L et al. , A Single T Cell Receptor Bound to Major Histocompatibility Complex Class I and Class II Glycoproteins Reveals Switchable TCR Conformers. Immunity 35, 23–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wirasinha RC et al. , alphabeta T-cell receptors with a central CDR3 cysteine are enriched in CD8alphaalpha intraepithelial lymphocytes and their thymic precursors. Immunol Cell Biol 96, 553–561 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Kasatskaya SA et al. , Functionally specialized human CD4(+) T-cell subsets express physicochemically distinct TCRs. eLife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chandler D, Interfaces and the driving force of hydrophobic assembly. Nature 437, 640–647 (2005). [DOI] [PubMed] [Google Scholar]
- 115.Mathis D, Benoist C, Back to central tolerance. Immunity 20, 509–516 (2004). [DOI] [PubMed] [Google Scholar]
- 116.Klein L, Kyewski B, Allen PM, Hogquist KA, Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moran AE et al. , T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 208, 1279–1289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McDonald BD, Bunker JJ, Ishizuka IE, Jabri B, Bendelac A, Elevated T cell receptor signaling identifies a thymic precursor to the TCRalphabeta(+)CD4(−)CD8beta(−) intraepithelial lymphocyte lineage. Immunity 41, 219–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mayans S et al. , alphabetaT cell receptors expressed by CD4(−)CD8alphabeta(−) intraepithelial T cells drive their fate into a unique lineage with unusual MHC reactivities. Immunity 41, 207–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chopp LB et al. , An Integrated Epigenomic and Transcriptomic Map of Mouse and Human alphabeta T Cell Development. Immunity 53, 1182–1201 e1188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kurd NS et al. , Factors that influence the thymic selection of CD8alphaalpha intraepithelial lymphocytes. Mucosal Immunol 14, 68–79 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Denning TL et al. , Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J Immunol 178, 4230–4239 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Bilate AM et al. , T Cell Receptor Is Required for Differentiation, but Not Maintenance, of Intestinal CD4(+) Intraepithelial Lymphocytes. Immunity 53, 1001–1014 e1020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hogquist KA et al. , Identification of a naturally occurring ligand for thymic positive selection. Immunity 6, 389–399 (1997). [DOI] [PubMed] [Google Scholar]
- 125.Lo WL et al. , An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells. Nat Immunol 10, 1155–1161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM, An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol 10, 1162–1169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Daniels MA et al. , Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 (2006). [DOI] [PubMed] [Google Scholar]
- 128.Stepanek O et al. , Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell 159, 333–345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wiest DL et al. , Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J Exp Med 178, 1701–1712 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hogquist KA et al. , T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 131.Alam SM et al. , T-cell-receptor affinity and thymocyte positive selection. Nature 381, 616–620 (1996). [DOI] [PubMed] [Google Scholar]
- 132.Gascoigne NR, Palmer E, Signaling in thymic selection. Curr Opin Immunol, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hogquist KA, Jameson SC, The self-obsession of T cells: how TCR signaling thresholds affect fate ‘decisions’ and effector function. Nat Immunol 15, 815–823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hong J et al. , A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat Immunol 19, 1379–1390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Germain RN, T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2, 309–322 (2002). [DOI] [PubMed] [Google Scholar]
- 136.Van Laethem F et al. , Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity 27, 735–750 (2007). [DOI] [PubMed] [Google Scholar]
- 137.Van Laethem F et al. , Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 154, 1326–1341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zareie P et al. , Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling. Science 372, (2021). [DOI] [PubMed] [Google Scholar]
- 139.Mingueneau M et al. , The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 14, 619–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chopp LB et al. , An integrated epigenomic and transcriptomic map of mouse and human αβ T cell development. Immunity This Issue, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Engel I, Murre C, E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J 23, 202–211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jones ME, Zhuang Y, Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27, 860–870 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jones-Mason ME et al. , E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity 36, 348–361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Emmanuel AO et al. , TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4(+)CD8(+) thymocytes. Nat Immunol 19, 1366–1378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Daley SR, Hu DY, Goodnow CC, Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-kappaB. J Exp Med 210, 269–285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yi J, Kawabe T, Sprent J, New insights on T-cell self-tolerance. Curr Opin Immunol 63, 14–20 (2020). [DOI] [PubMed] [Google Scholar]
- 147.Verstichel G et al. , The checkpoint for agonist selection precedes conventional selection in human thymus. Science immunology 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gangadharan D et al. , Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity 25, 631–641 (2006). [DOI] [PubMed] [Google Scholar]
- 149.McDonald BD, Bunker JJ, Erickson SA, Oh-Hora M, Bendelac A, Crossreactive alphabeta T Cell Receptors Are the Predominant Targets of Thymocyte Negative Selection. Immunity 43, 859–869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brugnera E et al. , Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity 13, 59–71 (2000). [DOI] [PubMed] [Google Scholar]
- 151.Taniuchi I, CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annu Rev Immunol 36, 579–601 (2018). [DOI] [PubMed] [Google Scholar]
- 152.Taniuchi I et al. , Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002). [DOI] [PubMed] [Google Scholar]
- 153.Woolf E et al. , Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 100, 7731–7736 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.He X et al. , The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005). [DOI] [PubMed] [Google Scholar]
- 155.Sun G et al. , The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol 6, 373–381 (2005). [DOI] [PubMed] [Google Scholar]
- 156.Park JH et al. , Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol 11, 257–264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Luckey MA et al. , The transcription factor ThPOK suppresses Runx3 and imposes CD4(+) lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat Immunol 15, 638–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.He X et al. , CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 (2008). [DOI] [PubMed] [Google Scholar]
- 159.Setoguchi R et al. , Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008). [DOI] [PubMed] [Google Scholar]
- 160.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J, GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19, 83–94 (2003). [DOI] [PubMed] [Google Scholar]
- 161.Lutes LK et al. , T cell self-reactivity during thymic development dictates the timing of positive selection. eLife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Klein L, Robey EA, Hsieh CS, Central CD4(+) T cell tolerance: deletion versus regulatory T cell differentiation. Nat Rev Immunol 19, 7–18 (2019). [DOI] [PubMed] [Google Scholar]
- 163.Lo WL, Donermeyer DL, Allen PM, A voltage-gated sodium channel is essential for the positive selection of CD4(+) T cells. Nat Immunol 13, 880–887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fu G et al. , Themis sets the signal threshold for positive and negative selection in T-cell development. Nature 504, 441–445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Plas DR et al. , Cutting edge: the tyrosine phosphatase SHP-1 regulates thymocyte positive selection. J Immunol 162, 5680–5684 (1999). [PubMed] [Google Scholar]
- 166.Wang D et al. , Tespa1 is involved in late thymocyte development through the regulation of TCR-mediated signaling. Nat Immunol 13, 560–568 (2012). [DOI] [PubMed] [Google Scholar]
- 167.Daniels MA et al. , CD8 binding to MHC class I molecules is influenced by T cell maturation and glycosylation. Immunity 15, 1051–1061 (2001). [DOI] [PubMed] [Google Scholar]
- 168.Tarakhovsky A et al. , A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science 269, 535–537 (1995). [DOI] [PubMed] [Google Scholar]
- 169.Azzam HS et al. , CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med 188, 2301–2311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Naramura M, Kole HK, Hu RJ, Gu H, Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc Natl Acad Sci U S A 95, 15547–15552 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McNeill L et al. , The differential regulation of Lck kinase phosphorylation sites by CD45 is critical for T cell receptor signaling responses. Immunity 27, 425–437 (2007). [DOI] [PubMed] [Google Scholar]
- 172.Li QJ et al. , miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129, 147–161 (2007). [DOI] [PubMed] [Google Scholar]
- 173.Yagi J, Janeway CA Jr., Ligand thresholds at different stages of T cell development. Int Immunol 2, 83–89 (1990). [DOI] [PubMed] [Google Scholar]
- 174.Davey GM et al. , Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J Exp Med 188, 1867–1874 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lucas B, Stefanova I, Yasutomo K, Dautigny N, Germain RN, Divergent changes in the sensitivity of maturing T cells to structurally related ligands underlies formation of a useful T cell repertoire. Immunity 10, 367–376 (1999). [DOI] [PubMed] [Google Scholar]
- 176.Takahama Y, Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol 6, 127–135 (2006). [DOI] [PubMed] [Google Scholar]
- 177.Lancaster JN, Li Y, Ehrlich LIR, Chemokine-Mediated Choreography of Thymocyte Development and Selection. Trends Immunol 39, 86–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cowan JE et al. , The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. J Exp Med 210, 675–681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kishimoto H, Sprent J, Negative selection in the thymus includes semimature T cells. J Exp Med 185, 263–271 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Weissler KA, Caton AJ, The role of T-cell receptor recognition of peptide:MHC complexes in the formation and activity of Foxp3(+) regulatory T cells. Immunol Rev 259, 11–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Malchow S et al. , Aire-dependent thymic development of tumor-associated regulatory T cells. Science 339, 1219–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Perry JSA et al. , Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41, 414–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huseby ES, Sather B, Huseby PG, Goverman J, Age-dependent T cell tolerance and autoimmunity to myelin basic protein. Immunity 14, 471–481 (2001). [DOI] [PubMed] [Google Scholar]
- 184.Bonasio R et al. , Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat Immunol 7, 1092–1100 (2006). [DOI] [PubMed] [Google Scholar]
- 185.Li J, Park J, Foss D, Goldschneider I, Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J Exp Med 206, 607–622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Anderson MS et al. , Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]
- 187.Takaba H et al. , Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell 163, 975–987 (2015). [DOI] [PubMed] [Google Scholar]
- 188.Murata S et al. , Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 (2007). [DOI] [PubMed] [Google Scholar]
- 189.Leonard JD et al. , Identification of Natural Regulatory T Cell Epitopes Reveals Convergence on a Dominant Autoantigen. Immunity 47, 107–117 e108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kieback E et al. , Thymus-Derived Regulatory T Cells Are Positively Selected on Natural Self-Antigen through Cognate Interactions of High Functional Avidity. Immunity 44, 1114–1126 (2016). [DOI] [PubMed] [Google Scholar]
- 191.Stadinski BD et al. , A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+ Treg cell development. Nat Immunol 20, 1046–1058 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Barthlott T, Kassiotis G, Stockinger B, T cell regulation as a side effect of homeostasis and competition. J Exp Med 197, 451–460 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Apostolou I, Sarukhan A, Klein L, von Boehmer H, Origin of regulatory T cells with known specificity for antigen. Nat Immunol 3, 756–763 (2002). [DOI] [PubMed] [Google Scholar]
- 194.Jordan MS et al. , Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol 2, 301–306 (2001). [DOI] [PubMed] [Google Scholar]
- 195.Aschenbrenner K et al. , Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol 8, 351–358 (2007). [DOI] [PubMed] [Google Scholar]
- 196.Legoux FP et al. , CD4+ T Cell Tolerance to Tissue-Restricted Self Antigens Is Mediated by Antigen-Specific Regulatory T Cells Rather Than Deletion. Immunity 43, 896–908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Malhotra D et al. , Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat Immunol 17, 187–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY, An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol 7, 401–410 (2006). [DOI] [PubMed] [Google Scholar]
- 199.van Santen HM, Benoist C, Mathis D, Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J Exp Med 200, 1221–1230 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pacholczyk R et al. , Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity 27, 493–504 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS, A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity 37, 475–486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bautista JL et al. , Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol 10, 610–617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Thiault N et al. , Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors. Nat Immunol 16, 628–634 (2015). [DOI] [PubMed] [Google Scholar]
- 204.Weist BM, Kurd N, Boussier J, Chan SW, Robey EA, Thymic regulatory T cell niche size is dictated by limiting IL-2 from antigen-bearing dendritic cells and feedback competition. Nat Immunol 16, 635–641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li MO, Rudensky AY, T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol 16, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Iwata A et al. , Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat Immunol 18, 563–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Krishnamoorthy V et al. , The IRF4 Gene Regulatory Module Functions as a Read-Write Integrator to Dynamically Coordinate T Helper Cell Fate. Immunity 47, 481–497 e487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Allison KA et al. , Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW, Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol 6, 490–496 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Adams JJ et al. , T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35, 681–693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schubert DA et al. , Self-reactive human CD4 T cell clones form unusual immunological synapses. J Exp Med 209, 335–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Beringer DX et al. , T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat Immunol, (2015). [DOI] [PubMed] [Google Scholar]
- 213.Burchill MA et al. , Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lio CW, Hsieh CS, A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mahmud SA et al. , Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat Immunol 15, 473–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kitagawa Y et al. , Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chorro L et al. , Interleukin 2 modulates thymic-derived regulatory T cell epigenetic landscape. Nature communications 9, 5368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sekiya T et al. , The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nature communications 2, 269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sekiya T et al. , Nr4a Receptors Regulate Development and Death of Labile Treg Precursors to Prevent Generation of Pathogenic Self-Reactive Cells. Cell reports 24, 1627–1638 e1626 (2018). [DOI] [PubMed] [Google Scholar]
- 220.Owen DL, Sjaastad LE, Farrar MA, Regulatory T Cell Development in the Thymus. J Immunol 203, 2031–2041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Isomura I et al. , c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med 206, 3001–3014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Vang KB et al. , Cutting edge: CD28 and c-Rel-dependent pathways initiate regulatory T cell development. J Immunol 184, 4074–4077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Long M, Park SG, Strickland I, Hayden MS, Ghosh S, Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity 31, 921–931 (2009). [DOI] [PubMed] [Google Scholar]
- 224.Zheng Y et al. , Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Oh H et al. , An NF-kappaB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function. Immunity 47, 450–465 e455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sauer S et al. , T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A 105, 7797–7802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Vrisekoop N et al. , Sparse production but preferential incorporation of recently produced naive T cells in the human peripheral pool. Proc Natl Acad Sci U S A 105, 6115–6120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Surh CD, Sprent J, Regulation of mature T cell homeostasis. Semin Immunol 17, 183–191 (2005). [DOI] [PubMed] [Google Scholar]
- 229.Jameson SC, Maintaining the norm: T-cell homeostasis. Nat Rev Immunol 2, 547–556 (2002). [DOI] [PubMed] [Google Scholar]
- 230.Grossman Z, Paul WE, Dynamic Tuning of Lymphocytes: Physiological Basis, Mechanisms, and Function. Annu Rev Immunol, (2015). [DOI] [PubMed] [Google Scholar]
- 231.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN, T Cell-Positive Selection Uses Self-Ligand Binding Strength to Optimize Repertoire Recognition of Foreign Antigens. Immunity 38, 263–274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Weber KS et al. , Distinct CD4+ helper T cells involved in primary and secondary responses to infection. Proc Natl Acad Sci U S A 109, 9511–9516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Myers DR, Zikherman J, Roose JP, Tonic Signals: Why Do Lymphocytes Bother? Trends Immunol 38, 844–857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jameson SC, Lee YJ, Hogquist KA, Innate memory T cells. Adv Immunol 126, 173–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kieper WC, Jameson SC, Homeostatic expansion and phenotypic conversion of naive T cells in response to self peptide/MHC ligands. Proceedings of the National Academy of Sciences of the United States of America 96, 13306–13311 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Smith K et al. , Sensory adaptation in naive peripheral CD4 T cells. J Exp Med 194, 1253–1261 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hawiger D, Masilamani RF, Bettelli E, Kuchroo VK, Nussenzweig MC, Immunological unresponsiveness characterized by increased expression of CD5 on peripheral T cells induced by dendritic cells in vivo. Immunity 20, 695–705 (2004). [DOI] [PubMed] [Google Scholar]
- 238.Dalloul A, CD5: a safeguard against autoimmunity and a shield for cancer cells. Autoimmun Rev 8, 349–353 (2009). [DOI] [PubMed] [Google Scholar]
- 239.Milam AAV et al. , Tuning T Cell Signaling Sensitivity Alters the Behavior of CD4(+) T Cells during an Immune Response. J Immunol 200, 3429–3437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lesourne R et al. , Themis, a T cell-specific protein important for late thymocyte development. Nat Immunol 10, 840–847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Palmer MJ, Mahajan VS, Chen J, Irvine DJ, Lauffenburger DA, Signaling thresholds govern heterogeneity in IL-7-receptor-mediated responses of naïve CD8+ T cells. Immunology and cell biology 89, 581–594 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Takada K, Jameson SC, Self-class I MHC molecules support survival of naive CD8 T cells, but depress their functional sensitivity through regulation of CD8 expression levels. J Exp Med 206, 2253–2269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Cho J-H et al. , An intense form of homeostatic proliferation of naive CD8+ cells driven by IL-2. J Exp Med 204, 1787–1801 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Park JH et al. , ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat Immunol 8, 1049–1059 (2007). [DOI] [PubMed] [Google Scholar]
- 245.Jameson SC, Fulton RB, Not all naive CD8 T cells are created equal. Immunol Cell Biol 89, 576–577 (2011). [DOI] [PubMed] [Google Scholar]
- 246.Bhandoola A et al. , Peripheral expression of self-MHC-II influences the reactivity and self-tolerance of mature CD4(+) T cells: evidence from a lymphopenic T cell model. Immunity 17, 425–436 (2002). [DOI] [PubMed] [Google Scholar]
- 247.Fischer UB et al. , MHC class II deprivation impairs CD4 T cell motility and responsiveness to antigen-bearing dendritic cells in vivo. Proc Natl Acad Sci U S A 104, 7181–7186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Soldevila G, Raman C, Lozano F, The immunomodulatory properties of the CD5 lymphocyte receptor in health and disease. Curr Opin Immunol 23, 310–318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Freitas CMT, Johnson DK, Weber KS, T Cell Calcium Signaling Regulation by the Co-Receptor CD5. Int J Mol Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Breuning J, Brown MH, T Cell Costimulation by CD6 Is Dependent on Bivalent Binding of a GADS/SLP-76 Complex. Mol Cell Biol 37, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bamberger M et al. , A new pathway of CD5 glycoprotein-mediated T cell inhibition dependent on inhibitory phosphorylation of Fyn kinase. J Biol Chem 286, 30324–30336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Voisinne G, Gonzalez de Peredo A, Roncagalli R, CD5, an Undercover Regulator of TCR Signaling. Front Immunol 9, 2900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Voisinne G et al. , Quantitative interactomics in primary T cells unveils TCR signal diversification extent and dynamics. Nat Immunol 20, 1530–1541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mori D et al. , The T cell CD6 receptor operates a multitask signalosome with opposite functions in T cell activation. J Exp Med 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Orta-Mascaro M et al. , CD6 modulates thymocyte selection and peripheral T cell homeostasis. J Exp Med 213, 1387–1397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cho JH et al. , CD45-mediated control of TCR tuning in naive and memory CD8(+) T cells. Nature communications 7, 13373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zhou X et al. , The deubiquitinase Otub1 controls the activation of CD8(+) T cells and NK cells by regulating IL-15-mediated priming. Nat Immunol 20, 879–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ju YJ et al. , Self-reactivity controls functional diversity of naive CD8(+) T cells by co-opting tonic type I interferon. Nature communications 12, 6059 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Brzostek J et al. , T cell receptor and cytokine signal integration in CD8(+) T cells is mediated by the protein Themis. Nat Immunol, (2020). [DOI] [PubMed] [Google Scholar]
- 260.White JT, Cross EW, Kedl RM, Antigen-inexperienced memory CD8(+) T cells: where they come from and why we need them. Nat Rev Immunol 17, 391–400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kawabe T, Zhu J, Sher A, Foreign antigen-independent memory-phenotype CD4(+) T cells: a new player in innate immunity? Nat Rev Immunol 18, 1 (2018). [DOI] [PubMed] [Google Scholar]
- 262.Miller CH et al. , Eomes identifies thymic precursors of self-specific memory-phenotype CD8(+) T cells. Nat Immunol 21, 567–577 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Drobek A et al. , Strong homeostatic TCR signals induce formation of self-tolerant virtual memory CD8 T cells. EMBO J 37, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.White JT et al. , Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nature communications 7, 11291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Aivazian D, Stern LJ, Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nature structural biology 7, 1023–1026 (2000). [DOI] [PubMed] [Google Scholar]
- 266.Xu C et al. , Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zhang H, Cordoba SP, Dushek O, van der Merwe PA, Basic residues in the T-cell receptor zeta cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci U S A 108, 19323–19328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Gagnon E, Schubert DA, Gordo S, Chu HH, Wucherpfennig KW, Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3epsilon cytoplasmic domain. J Exp Med 209, 2423–2439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.van Oers NS, Killeen N, Weiss A, ZAP-70 is constitutively associated with tyrosine-phosphorylated TCR zeta in murine thymocytes and lymph node T cells. Immunity 1, 675–685 (1994). [DOI] [PubMed] [Google Scholar]
- 270.Thill PA, Weiss A, Chakraborty AK, Phosphorylation of a Tyrosine Residue on Zap70 by Lck and Its Subsequent Binding via an SH2 Domain May Be a Key Gatekeeper of T Cell Receptor Signaling In Vivo. Molecular and cellular biology 36, 2396–2402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sloan-Lancaster J, Shaw AS, Rothbard JB, Allen PM, Partial T cell signaling: altered phospho-zeta and lack of zap70 recruitment in APL-induced T cell anergy. Cell 79, 913–922 (1994). [DOI] [PubMed] [Google Scholar]
- 272.Racioppi L, Ronchese F, Matis LA, Germain RN, Peptide-major histocompatibility complex class II complexes with mixed agonist/antagonist properties provide evidence for ligand-related differences in T cell receptor-dependent intracellular signaling. J Exp Med 177, 1047–1060 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Au-Yeung BB et al. , A sharp T-cell antigen receptor signaling threshold for T-cell proliferation. Proc Natl Acad Sci U S A 111, E3679–3688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B, Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 109, 901–912 (2002). [DOI] [PubMed] [Google Scholar]
- 275.Liu B, Chen W, Evavold BD, Zhu C, Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Depoil D, Dustin ML, Force and affinity in ligand discrimination by the TCR. Trends Immunol 35, 597–603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Robert P et al. , Kinetics and mechanics of two-dimensional interactions between T cell receptors and different activating ligands. Biophys J 102, 248–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Das DK et al. , Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci U S A 112, 1517–1522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC, Mechanosensing in T lymphocyte activation. Biophys J 102, L5–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kim ST et al. , The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem 284, 31028–31037 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Beddoe T et al. , Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity 30, 777–788 (2009). [DOI] [PubMed] [Google Scholar]
- 282.Brazin KN et al. , The T Cell Antigen Receptor alpha Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations. Immunity 49, 829–841 e826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Holst J et al. , Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat Immunol 9, 658–666 (2008). [DOI] [PubMed] [Google Scholar]
- 284.Love PE, Hayes SM, ITAM-mediated signaling by the T-cell antigen receptor. Cold Spring Harbor perspectives in biology 2, a002485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lo WL et al. , Slow phosphorylation of a tyrosine residue in LAT optimizes T cell ligand discrimination. Nat Immunol 20, 1481–1493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Heinzel S, Marchingo JM, Horton MB, Hodgkin PD, The regulation of lymphocyte activation and proliferation. Curr Opin Immunol 51, 32–38 (2018). [DOI] [PubMed] [Google Scholar]
- 287.Heinzel S et al. , A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat Immunol 18, 96–103 (2017). [DOI] [PubMed] [Google Scholar]
- 288.Marchingo JM et al. , T cell signaling. Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science 346, 1123–1127 (2014). [DOI] [PubMed] [Google Scholar]
- 289.Michel F, Attal-Bonnefoy G, Mangino G, Mise-Omata S, Acuto O, CD28 as a molecular amplifier extending TCR ligation and signaling capabilities. Immunity 15, 935–945 (2001). [DOI] [PubMed] [Google Scholar]
- 290.Hommel M, Hodgkin PD, TCR affinity promotes CD8+ T cell expansion by regulating survival. J Immunol 179, 2250–2260 (2007). [DOI] [PubMed] [Google Scholar]
- 291.Koenen P et al. , Mutually exclusive regulation of T cell survival by IL-7R and antigen receptor-induced signals. Nature communications 4, 1735 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Au-Yeung BB et al. , IL-2 Modulates the TCR Signaling Threshold for CD8 but Not CD4 T Cell Proliferation on a Single-Cell Level. J Immunol 198, 2445–2456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Deenick EK, Gett AV, Hodgkin PD, Stochastic model of T cell proliferation: a calculus revealing IL-2 regulation of precursor frequencies, cell cycle time, and survival. J Immunol 170, 4963–4972 (2003). [DOI] [PubMed] [Google Scholar]
- 294.van Gisbergen KPJM et al. , The costimulatory molecule CD27 maintains clonally diverse CD8(+) T cell responses of low antigen affinity to protect against viral variants. Immunity 35, 97–108 (2011). [DOI] [PubMed] [Google Scholar]
- 295.Ramsdell F, Rudensky AY, Foxp3: a genetic foundation for regulatory T cell differentiation and function. Nat Immunol 21, 708–709 (2020). [DOI] [PubMed] [Google Scholar]
- 296.Sakaguchi S et al. , Regulatory T Cells and Human Disease. Annu Rev Immunol 38, 541–566 (2020). [DOI] [PubMed] [Google Scholar]
- 297.Ng WL, Bassler BL, Bacterial quorum-sensing network architectures. Annu Rev Genet 43, 197–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Butler TC, Kardar M, Chakraborty AK, Quorum sensing allows T cells to discriminate between self and nonself. Proc Natl Acad Sci U S A 110, 11833–11838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Al-Yassin GA, Bretscher PA, Does T Cell Activation Require a Quorum of Lymphocytes? J Immunol 201, 2855–2861 (2018). [DOI] [PubMed] [Google Scholar]
- 300.Wong HS et al. , A local regulatory T cell feedback circuit maintains immune homeostasis by pruning self-activated T cells. Cell 184, 3981–3997 e3922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Zenke S et al. , Quorum Regulation via Nested Antagonistic Feedback Circuits Mediated by the Receptors CD28 and CTLA-4 Confers Robustness to T Cell Population Dynamics. Immunity 52, 313–327 e317 (2020). [DOI] [PubMed] [Google Scholar]
- 302.Zhang N, Bevan MJ, CD8(+) T cells: foot soldiers of the immune system. Immunity 35, 161–168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Richard AC et al. , T cell cytolytic capacity is independent of initial stimulation strength. Nat Immunol 19, 849–858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Trefzer A et al. , Dynamic adoption of anergy by antigen-exhausted CD4(+) T cells. Cell reports 34, 108748 (2021). [DOI] [PubMed] [Google Scholar]
- 305.Tubo NJ, Jenkins MK, TCR signal quantity and quality in CD4 T cell differentiation. Trends Immunol 35, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Schwartz RH, T cell anergy. Annu Rev Immunol 21, 305–334 (2003). [DOI] [PubMed] [Google Scholar]
- 307.Clemente-Casares X et al. , Expanding antigen-specific regulatory networks to treat autoimmunity. Nature 530, 434 (2016). [DOI] [PubMed] [Google Scholar]
- 308.Larche M, Wraith DC, Peptide-based therapeutic vaccines for allergic and autoimmune diseases. Nat Med 11, S69–76 (2005). [DOI] [PubMed] [Google Scholar]
- 309.Denton AE et al. , Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses. J Immunol 187, 5733–5744 (2011). [DOI] [PubMed] [Google Scholar]
- 310.Guy CS et al. , Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nature immunology 14, 262–270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Zinzow-Kramer WM, Weiss A, Au-Yeung BB, Adaptation by naive CD4(+) T cells to self-antigen-dependent TCR signaling induces functional heterogeneity and tolerance. Proc Natl Acad Sci U S A 116, 15160–15169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Keck S et al. , Antigen affinity and antigen dose exert distinct influences on CD4 T-cell differentiation. Proc Natl Acad Sci U S A 111, 14852–14857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Hwang S et al. , TCR ITAM multiplicity is required for the generation of follicular helper T-cells. Nature communications 6, 6982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kunzli M, Reuther P, Pinschewer DD, King CG, Opposing effects of T cell receptor signal strength on CD4 T cells responding to acute versus chronic viral infection. eLife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Snook JP, Kim C, Williams MA, TCR signal strength controls the differentiation of CD4(+) effector and memory T cells. Sci Immunol 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Pepper M, Pagán AJ, Igyártó BZ, Taylor JJ, Jenkins MK, Opposing signals from the bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ballesteros-Tato A et al. , Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36, 847–856 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Kotov DI et al. , TCR Affinity Biases Th Cell Differentiation by Regulating CD25, Eef1e1, and Gbp2. J Immunol 202, 2535–2545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Smith GA, Taunton J, Weiss A, IL-2Rbeta abundance differentially tunes IL-2 signaling dynamics in CD4(+) and CD8(+) T cells. Sci Signal 10, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Bartleson JM et al. , Strength of tonic T cell receptor signaling instructs T follicular helper cell-fate decisions. Nat Immunol 21, 1384–1396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Hwang SS et al. , mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 367, 1255–1260 (2020). [DOI] [PubMed] [Google Scholar]
- 322.Milam AAV et al. , Tonic TCR Signaling Inversely Regulates the Basal Metabolism of CD4(+) T Cells. Immunohorizons 4, 485–497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Myers DR, Wheeler B, Roose JP, mTOR and other effector kinase signals that impact T cell function and activity. Immunol Rev 291, 134–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Chen Y, Zander R, Khatun A, Schauder DM, Cui W, Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front Immunol 9, 2826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Intlekofer AM et al. , Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 326.Knudson KM, Goplen NP, Cunningham CA, Daniels MA, Teixeiro E, Low-affinity T cells are programmed to maintain normal primary responses but are impaired in their recall to low-affinity ligands. Cell reports 4, 554–565 (2013). [DOI] [PubMed] [Google Scholar]
- 327.Kallies A, Xin A, Belz GT, Nutt SL, Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 31, 283–295 (2009). [DOI] [PubMed] [Google Scholar]
- 328.Man K et al. , The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 329.Williams MA, Holmes BJ, Sun JC, Bevan MJ, Developing and maintaining protective CD8+ memory T cells. Immunol Rev 211, 146–153 (2006). [DOI] [PubMed] [Google Scholar]
- 330.Chung HK, McDonald B, Kaech SM, The architectural design of CD8+ T cell responses in acute and chronic infection: Parallel structures with divergent fates. J Exp Med 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Jameson SC, The Naming of Memory T-Cell Subsets. Cold Spring Harbor perspectives in biology 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Knudson KM, Goplen NP, Cunningham CA, Daniels MA, Teixeiro E, Low-Affinity T Cells Are Programmed to Maintain Normal Primary Responses but Are Impaired in Their Recall to Low-Affinity Ligands. Cell Reports 4, 554–565 (2013). [DOI] [PubMed] [Google Scholar]
- 333.Smith-Garvin JE et al. , T-cell receptor signals direct the composition and function of the memory CD8+ T-cell pool. Blood 116, 5548–5559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Solouki S et al. , TCR Signal Strength and Antigen Affinity Regulate CD8(+) Memory T Cells. J Immunol 205, 1217–1227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Man K et al. , Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 47, 1129–1141 e1125 (2017). [DOI] [PubMed] [Google Scholar]
- 336.Lee JY, Hamilton SE, Akue AD, Hogquist KA, Jameson SC, Virtual memory CD8 T cells display unique functional properties. Proc Natl Acad Sci U S A 110, 13498–13503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Banerjee A et al. , Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 185, 4988–4992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Kallies A, Xin A, Belz G, Nutt S, Blimp-1 Transcription Factor Is Required for the Differentiation of Effector CD8(+) T Cells and Memory Responses. Immunity, (2009). [DOI] [PubMed] [Google Scholar]
- 339.Nayar R et al. , Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol 192, 5881–5893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Ou-Yang CW, Zhu M, Sullivan SA, Fuller DM, Zhang W, The requirement of linker for activation of T cells in the primary and memory responses of CD8 T cells. J Immunol 190, 2938–2947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Knudson KM, Hamilton SE, Daniels MA, Jameson SC, Teixeiro E, Cutting edge: The signals for the generation of T cell memory are qualitatively different depending on TCR ligand strength. J Immunol 191, 5797–5801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Rao RR, Li Q, Odunsi K, Shrikant PA, The mTOR Kinase Determines Effector versus Memory CD8+ T Cell Fate by Regulating the Expression of Transcription Factors T-bet and Eomesodermin. Immunity, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Gattinoni L et al. , Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 15, 808–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Zhou X et al. , Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Knudson KM et al. , NFkappaB-Pim-1-Eomesodermin axis is critical for maintaining CD8 T-cell memory quality. Proc Natl Acad Sci U S A 114, E1659–E1667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Pearce E et al. , Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Huang F et al. , The tyrosine kinase Itk suppresses CD8+ memory T cell development in response to bacterial infection. Sci Rep 5, 7688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Roychoudhuri R et al. , BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 17, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Fiege JK et al. , The Impact of TCR Signal Strength on Resident Memory T Cell Formation during Influenza Virus Infection. J Immunol 203, 936–945 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Frost EL, Kersh AE, Evavold BD, Lukacher AE, Cutting Edge: Resident Memory CD8 T Cells Express High-Affinity TCRs. J Immunol 195, 3520–3524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.van Panhuys N, Klauschen F, Germain RN, T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization In Vivo. Immunity 41, 63–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Prlic M, Hernandez-Hoyos G, Bevan MJ, Duration of the initial TCR stimulus controls the magnitude but not functionality of the CD8+ T cell response. J Exp Med 203, 2135–2143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Hale JS, Boursalian TE, Turk GL, Fink PJ, Thymic output in aged mice. Proceedings of the National Academy of Sciences of the United States of America 103, 8447–8452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Hernandez-Lopez RA et al. , T cell circuits that sense antigen density with an ultrasensitive threshold. Science 371, 1166–1171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Jan M et al. , Reversible ON- and OFF-switch chimeric antigen receptors controlled by lenalidomide. Science translational medicine 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Weber EW et al. , Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science 372, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kalekar LA et al. , CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat Immunol 17, 304–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.White SH, Wimley WC, Membrane protein folding and stability: physical principles. Annual review of biophysics and biomolecular structure 28, 319–365 (1999). [DOI] [PubMed] [Google Scholar]


