Abstract
Denaturing gradient gel electrophoresis (DGGE) of DNA fragments generated by PCR with 16S ribosomal DNA-targeted group-specific primers was used to detect lactic acid bacteria (LAB) of the genera Lactobacillus, Pediococcus, Leuconostoc, and Weissella in human feces. Analysis of fecal samples of four subjects revealed individual profiles of DNA fragments originating not only from species that have been described as intestinal inhabitants but also from characteristically food-associated bacteria such as Lactobacillus sakei, Lactobacillus curvatus, Leuconostoc mesenteroides, and Pediococcus pentosaceus. Comparison of PCR-DGGE results with those of bacteriological culture showed that the food-associated species could not be cultured from the fecal samples by plating on Rogosa agar. On the other hand, all of the LAB species cultured from feces were detected in the DGGE profile. We also detected changes in the types of LAB present in human feces during consumption of a milk product containing the probiotic strain Lactobacillus rhamnosus DR20. The analysis of fecal samples from two subjects taken before, during, and after administration of the probiotic revealed that L. rhamnosus was detectable by PCR-DGGE during the test period in the feces of both subjects, whereas it was detectable by culture in only one of the subjects.
The distal human intestinal tract is colonized by numerous bacteria that form a complex community rich in bacterial species (22). This community is referred to as the intestinal microflora. Most current knowledge of the intestinal microflora has been obtained by using bacteriological culture techniques and microscopy. Bacteriological culture has been shown to have deficiencies in microbial ecological studies, particularly due to the presence of bacterial cells that are not detectable by culture methods (7). The application of recently developed culture-independent molecular techniques permits more detailed investigations of the intestinal microflora (16, 23). Most of the molecular techniques for the detection, identification, and classification of bacteria are based on the nucleotide sequence of 16S rRNA. Denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) of 16S ribosomal DNA (rDNA) amplicons have been demonstrated to be suitable tools for the analysis of microbial communities because they permit the detection of species and changes in community structure quickly and economically (14). Total bacterial DNA from the habitat of interest is extracted and a region with a hypervariable nucleotide base sequence of the 16S rRNA gene is amplified by PCR. The resulting mixture of 16S rDNA fragments is subjected to a denaturing gradient, established in a polyacrylamide gel with urea and formamide or increasing temperature, in order to separate the fragments and generate a “genetic fingerprint” of the community. The usefulness of PCR (universal primers) and DGGE and TGGE in the analysis of the biodiversity of gastrointestinal microfloras has already been clearly demonstrated (20, 24, 29).
Lactobacillus species are commonly present in human fecal samples (5). Some Lactobacillus species have also received considerable attention with respect to their putative healthful properties when ingested as probiotics (17, 25). The detection of particular bacterial species in human feces using single PCRs with species-specific primers is logistically daunting (9). PCR-DGGE has the potential to provide a more practical approach because lactobacilli can be detected and identified (or at least grouped) by comparison of the migration distance of their PCR amplicons in DGGE gels with those of reference strains. This approach has already been applied for the detection and identification of lactobacilli in the stomach contents of mice (26). In this ecosystem, the microbial community has a simple composition with large numbers of Lactobacillus cells compared to the microbial community human feces. In human feces, lactobacilli generally make up less than 1% of the community (19). The fecal Lactobacillus population cannot be detected by DGGE analysis of DNA fragments obtained from a PCR amplification with universal primers because this method detects the 90 to 99% most numerous species in the community (14, 29). There is thus a need for the derivation of specific primers to enable the monitoring of bacteria present as minority members of the microflora. The use of group-specific primers for actinomycetes (6) and cyanobacteria (15) in combination with DGGE has been reported for soil and lichens, respectively. Recently, PCR-DGGE with primers specific for the Lactobacillus reuteri phylogenetic group was applied to detect these lactobacilli in the feces of pigs (20).
In this paper, we describe the derivation and use of PCR primers specific for lactic acid bacteria (LAB) of the genera Lactobacillus, Pediococcus, Weissella, and Leuconostoc in conjunction with DGGE. This technique was used to detect these LAB in human feces. The bacterial species detected by this method were identified by sequencing of DNA excised from the gels or by comparison of the migration distances of the DNA amplicons with those of reference strains. We also compared the identity of LAB cultured from fecal samples with the results obtained by PCR-DGGE. Finally, changes in the LAB population during consumption of a product containing the probiotic strain Lactobacillus rhamnosus DR20 were investigated.
MATERIALS AND METHODS
Primer design.
The PROBE-DESIGN tool of the software package ARB (http://www.biol.chemie.tu-muenchen.de/pub/ARB/documentation/arb.ps) was used to derive 16S rDNA-targeted primers specific for lactobacilli. The potential target sequences for the deduced primers were compared with all sequences in the ARB database using the PROBE-MATCH tool. A 40-bp GC clamp (5′-CGCCCGGGGCGCGCCCCGGGCGGCCCGGGGGCACCGGGGG-3′) was attached to the reverse primer (Lac2) to obtain PCR fragments suitable for DGGE analysis (28). The melting behavior of the PCR fragments, as revealed by the software of the Poland server (http://www.biophys.uni-duesseldorf.de/POLAND/poland.html), served as a basis for primer optimization.
Bacterial strains and growth conditions.
The following bacteria were used in this study: Lactobacillus acidophilus DSM 20079T, L. brevis DSM 20054T, L. casei DSM 20011T, L. crispatus DSM 20584T, L. curvatus subsp. curvatus DSM 20019T, L. delbrueckii subsp. bulgaricus DSM 20081T, L. gasseri DSM 20243T, L. johnsonii DSM 10533T, L. plantarum DSM 20174T, L. paracasei subsp. paracasei LTH 2579, L. paracasei subsp. paracasei DSM 5622T, L. reuteri DSM 20016T, L. rhamnosus DSM 20021T, L. ruminis DSM 20403T, L. sakei DSM 20017T, L. salivarius subsp. salicinius DSM 20554T, L. sharpeae DSM 20505T, Bacteroides distasonis DSM 20701T, Bifidobacterium dentium ATCC 27678, Clostridium perfringens ATCC 13124, Enterococcus faecium DSM 20477T, Escherichia coli Shure (Stratagene), Eubacterium limosum DSM 20543T, Staphylococcus epidermidis DSM 20044T, Veillonella parvula DSM 2008T, and Weissella viridescens DSM 20410T.
Lactobacilli and W. viridescens were grown anaerobically in MRS medium (Difco) at 37°C. Enterococci and staphylococci were grown aerobically in M53 medium (Deutsche Sammlung von Mikroorganismen und Zellkulturen) at 37°C, and E. coli was grown under the same conditions in Luria-Bertani medium. All other bacteria were cultured anaerobically in brain heart infusion broth (Difco) at 37°C.
Fecal samples.
Samples from two studies were investigated. In the first study, fecal samples were obtained from four healthy subjects (A, B, C, D; two female, two male) aged 28 to 37 years. Freshly collected samples were diluted 10-fold in phosphate buffer (0.05 M [pH 7.0]) and stored at −82°C in 1-ml aliquots for later DNA extraction. To compare DGGE results with those of bacteriological culture, fresh fecal samples of subjects A, C, and D were homogenized and diluted 10-fold in prereduced diluent (containing 8.5 g of NaCl, 1.0 g of peptone, and 0.1 g of cysteine per liter [pH 7.0]) in an anaerobic chamber. Viable cell counts of lactobacilli were determined by plating on Rogosa SL agar (Difco) after incubation for 2 days. Eleven colonies were picked randomly from an agar plate containing 30 to 300 colonies. These subcultured isolates represented the predominant strains comprising the population selected on Rogosa agar (10). Finally, fecal samples of two subjects (C, D) were collected once a month for 6 months. The second study used fecal samples of two subjects that had been collected previously during a probiotic trial with L. rhamnosus DR20 (24).
DNA extraction.
Chromosomal DNA was isolated from an overnight culture of L. paracasei LTH 2579 as described previously (18). Total DNA of pure cultures was extracted as described previously (26). For extraction of the total DNA from fecal samples (1 ml), the frozen samples were thawed on ice and purified as described by Wang et al. (27) but, after washing of the cells, the pellet was resuspended in 100 μl of lysis buffer (6.7% sucrose, 50 mM Tris HCl [pH 8.0], 10 mM EDTA, 20 mg of lysozyme per ml, 1,000 U of mutanolysin per ml, 100 μg of RNaseA per ml). After incubation for 1 h at 37°C, 6 μl of sodium dodecyl sulfate (20%) and 5 μl of proteinase K solution (15 mg/ml) were added and the mixture was further incubated for ca. 15 min at 60°C until the cells lysed. After cooling on ice, 400 μl of Tris HCl (pH 8.0) was added and the mixture was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and twice with chloroform. After ethanol precipitation the DNA was dissolved in 100 μl of Tris HCl (pH 8.0).
PCR amplification.
Amplification was carried out using a GeneAmp 2400 Thermocycler (Perkin-Elmer) and the specific primers Lac1 and Lac2GC (Fig. 1). The reaction mixture (50 μl) contained 25-pmol amounts of each primer, 0.2 mM concentrations of each deoxyribonucleotide triphosphate, reaction buffer, 20 mM tetramethylammonium chloride, 25 μg of bovine serum albumin, 2.5 U of rTaq polymerase (Amersham Pharmacia Biotech), and 1 μl of DNA solution. The amplification program was 94°C for 2 min; 35 cycles of 94°C for 30 s, 61°C for 1 min, and 68°C for 1 min; and finally 68°C for 7 min. To determine the lower limit of PCR detection, a solution of chromosomal DNA (ca. 10 ng/μl) of L. paracasei LTH 2579 was serially diluted and subjected to PCR.
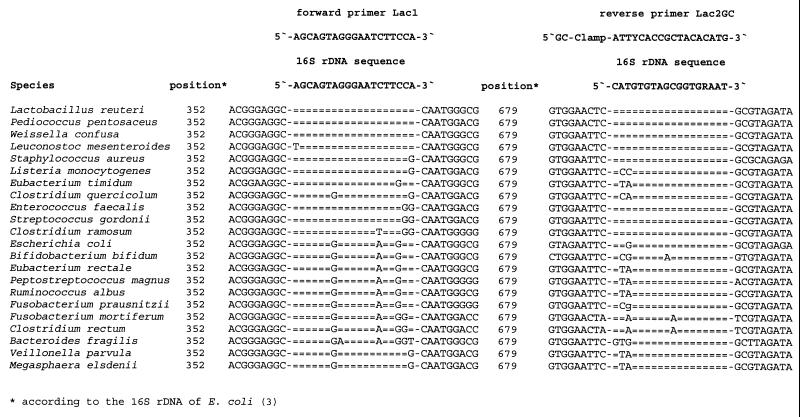
FIG. 1.
Primers for the specific amplification of 16S rDNA sequences of species of the Lactobacillus, Pediococcus, Leuconostoc, and Weissella group and alignment of the primer binding regions within the 16S rDNA sequences of related and nonrelated intestinal bacteria.
DGGE and excision of DNA fragments.
DGGE was performed as described previously (26), with the following modifications: the gel contained a 32.5 to 40% gradient of urea and formamide increasing in the direction of electrophoresis. Excision and purification of DNA fragments from DGGE gels were performed as described by Ben Omar and Ampe (2).
Bias determination in PCR-DGGE.
To determine whether DNA extraction and template annealing in the PCR amplification introduced bias to the results, cells of L. acidophilus DSM 20079T, L. paracasei subsp. paracasei DSM 5622T, L. reuteri DSM 20016T, L. salivarius subsp. salicinius DSM 20554T, and L. sharpae DSM 20505T were mixed to obtain final counts for each species of 5 × 107 and 5 × 108 cells/ml. DNA was extracted from a 1-ml aliquot of the mixtures, and PCR-DGGE was carried out as described for the fecal samples.
Sequencing and sequence analysis.
DNA sequences of PCR fragments obtained from pure cultures or from purified DGGE bands were determined by the dideoxy chain termination method using the AutoRead sequencing kit (Amersham Pharmacia Biotech), the LI-COR system (MWG Biotech), and the IRD 800 labeled primer Lac2Seq (5′-ATTTCACCGCTACACATG-3′). To determine the closest relatives of the partial 16S rDNA sequences, a search of the GenBank DNA database was conducted by using the BLAST algorithm (1). A similarity of >99% to 16S rDNA sequences of type strains was used as the criterion for identification.
RESULTS
Construction and evaluation of primers.
PCR primers Lac1 and Lac2 (Fig. 1) were derived for the amplification of 340 bp of the 16S rRNA gene (16S rDNA) of lactobacilli. In silico primer specificity analysis showed that the primers would not bind exclusively to the 16S rDNA of lactobacilli but would also anneal to that of Pediococcus spp., Leuconostoc spp., and Weissella spp. Alignment of amplified 16S rDNA sequences theoretically permitted the differentiation of Lactobacillus species with the exception of the L. casei group (L. casei, L. paracasei, L. rhamnosus, and L. zeae). For DGGE analysis, a GC clamp of 40 bases was linked to primer Lac2, resulting in primer Lac2GC. The PCR-amplified 16S rDNA fragments of various lactobacilli containing this GC clamp exhibited a melting behavior suitable for DGGE (data not shown).
The specificity of primers Lac1 and Lac2GC was evaluated using DNA extracted from all of the bacterial strains listed in Materials and Methods. A PCR product was only obtained when using DNA templates from the Lactobacillus species and from W. viridescens DSM 20410T. DGGE of the PCR amplicons from representative lactobacilli showed that most of the Lactobacillus species could be differentiated according to the migration distances of their respective 16S rDNA fragments. Examples are depicted in Fig. 2 (lanes 1 to 5). The amplicons from members of the L. casei group showed similar migration distances. The sensitivity of the PCR system was determined using chromosomal DNA of L. paracasei subsp. paracasei LTH 2579. A minimum of 100 fg of DNA was necessary to obtain a PCR product.
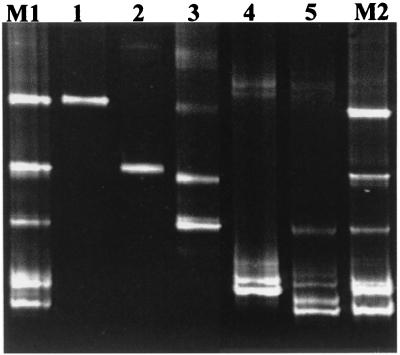
FIG. 2.
PCR-DGGE analysis of 16S rDNA fragments generated by PCR with the specific primers Lac1 and Lac2GC and DNA extracted from Lactobacillus species or mixed populations. Lanes: 1, L. sharpeae DSM 20505T; 2, L. acidophilus DSM 20079T; 3, L. salivarius subsp. salicinius DSM 20554T; 4, L. paracasei subsp. paracasei DSM 5622T; 5, L. reuteri DSM 20016T; M1 and M2, mixture of the Lactobacillus species containing 5 × 107 and 5 × 108 cells of each species per ml, respectively.
Detection of lactobacilli in fecal samples using DGGE.
An initial experiment with mixtures of Lactobacillus species prepared in vitro did not demonstrate difficulties with the detection of the various species. As shown in Fig. 2 (lanes M1 and M2), all five species were detected in the Lactobacillus population. Based on the intensity of the signals, a slight decrease was observed for the species L. acidophilus and L. salivarius subsp. salicinius in the mixture when compared to the other species in the mixed culture.
The usefulness of the Lac1 and Lac2GC primers in conjunction with DGGE in studying Lactobacillus populations in fecal samples of human subjects was tested. DGGE analysis of the PCR-amplified 16S rDNA fragments generated from the feces of four subjects (A to D) revealed different profiles in the case of each individual (Fig. 3). Sequencing of excised and purified DNA from gels showed that the fragments originated from species of the genera Lactobacillus, Pediococcus, Leuconostoc, and Weissella. This result was consistent with the prediction of the computer-based primer specificity analysis. Interestingly, DGGE revealed the presence of characteristically food-associated bacteria in the feces (e.g., L. sakei and L. curvatus).
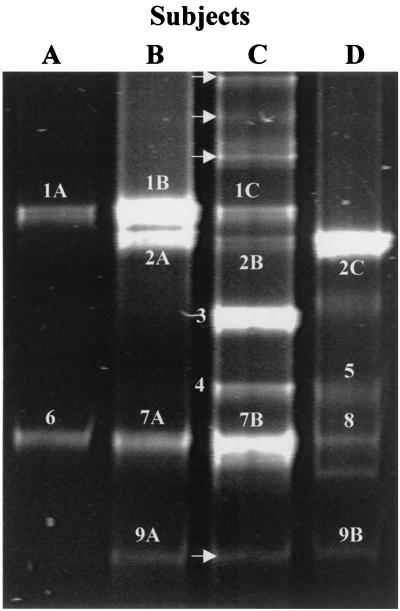
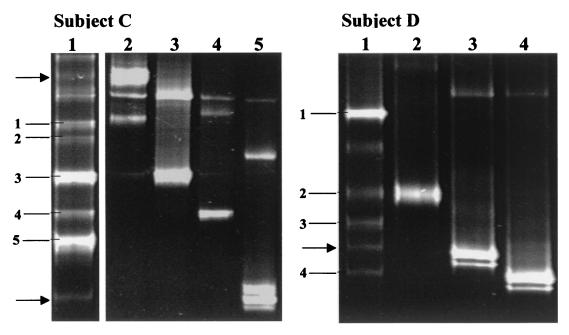
FIG. 3.
DGGE analysis of PCR-amplified 16S rDNA fragments obtained with primer pair Lac1 and Lac2GC and DNA isolated from human feces. The fragments were excised and sequenced. Based on sequence comparisons, the fragments were allotted to the following species: 1A to 1C, Lactobacillus sakei; 2A to 2C, L. curvatus; 3, L. acidophilus; 4, L. crispatus; 5, W. confusa; 6, P. pentosaceus; 7A and 7B, Leuconostoc mesenteroides; 8, L. fructivorans; 9A and 9B, L. casei group. Bands not resulting in sequences upon purification and sequencing are indicated by arrows.
Fecal samples from subjects C and D were collected monthly over a period of 6 months and the samples were subjected to DGGE analysis. As shown in Fig. 4, in the case of both subjects, fluctuations in the 16S rDNA fragment profiles were observed. Some fragments could be detected for a longer period (e.g., for both subjects fragments 1 and 2, and for subject C fragment 3) than others. Interestingly, these frequently detected fragments originated from Lactobacillus species typically found to be associated with foods and often used as starter organisms.
FIG. 4.
DGGE of PCR products obtained with primer pair Lac1 and Lac2 GC of fecal samples from subjects C and D taken each month over a period of 6 months. Sequence characterization of the excised fragments indicated the presence of 1A to 1D, L. sakei; 2A to 2D, L. curvatus; 3A, L. delbrueckii; and 4A, L. plantarum.
Comparison of results of DGGE analyses with those of bacteriological culture.
The fecal samples of subjects A, C, and D (Fig. 3) were additionally used for bacteriological culture on Rogosa agar. Although a DGGE profile could be obtained for subject A (Fig. 3, lane A), bacteria were not cultured from the fecal sample. For subjects C and D, the CFU/g values were 1.4 × 107 and 1.2 × 106, respectively. To obtain insight into the composition of the dominant Lactobacillus species of subjects C and D, 11 colonies were randomly picked and subcultured. DNA was extracted from the bacterial isolates and subjected to PCR amplification. DGGE analysis of these amplicons revealed different profiles for isolates from subjects C and D. These results are shown in Fig. 5, together with that of the corresponding fecal sample. The bacterial isolates were identified by sequencing as belonging to the species L. acidophilus, L. crispatus, L. plantarum, Weissella confusa, and Pediococcus pentosaceus and to the L. casei group.
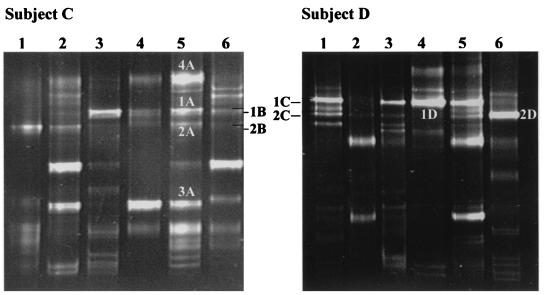
FIG. 5.
Comparison of PCR-DGGE and bacteriological culture to monitor the species composition of the Lactobacillus population in human feces. DGGE analyses were performed with PCR-amplified 16S rDNA fragments which were obtained with the primer pair Lac1 and Lac2GC and DNA extracted from fecal samples (lane 1) or the corresponding Lactobacillus isolates (subject C, lanes 2 to 5; subject D, lanes 2 to 4) cultured on Rogosa medium. The DNA fragments were allotted by sequence analysis to the following species. Isolates from subject C were as follows: lane 2, L. plantarum; lane 3, L. acidophilus; lane 4, L. crispatus; lane 5, L. casei group. DGGE profile of subject C revealed the following: fragment 1, L. sakei; fragment 2, L. curvatus; fragment 3, L. acidophilus; fragment 4, L. crispatus; fragment 5, Leuconostoc mesenteroides. Isolates from subject D were identified as follows: lane 2, W. confusa; lane 3, Pediococcus acidilactici; lane 4, L. casei group. DGGE profile of subject D identified the following: fragment 1, L. curvatus; fragment 2, W. confusa; fragment 3, Lactobacillus fructivorans; fragment 4, L. casei group. Fragments that did not result in sequences upon purification and sequencing are indicated by arrows.
Table 1 summarizes the comparisons of culture and PCR-DGGE results. All of the cultured species could be detected by PCR-DGGE. In three cases, faintly staining fragments present in the DGGE profiles of the fecal samples matched those obtained from the bacterial isolates (Fig. 5), but excision and sequencing were unsuccessful. DNA from several species (L. sakei, L. curvatus, and L. fructivorans, Leuconostoc mesenteroides, and Pediococcus pentosaceus) was detected by PCR-DGGE in the fecal samples of all of the subjects, but these species were not detected among the colonies cultured on Rogosa medium. These species are characteristically involved in food fermentations. They were present at least on the same order of magnitude as the cultured lactobacilli, as indicated by the intensity of staining of the DGGE fragments (e.g., L. mesenteroides in subject C [Fig. 5, subject C, lane 1, fragment 5] and L. curvatus in subject D [Fig. 5, subject D, lane 1, fragment 1]).
TABLE 1.
Bacterial counts of fecal samples from subjects A, C, and D and species detected by PCR-DGGE and by culture
| Subject | Cell count on Rogosa agar (CFU/g) | Bacterial speciesa | Detected by:
|
|
|---|---|---|---|---|
| PCR-DGGE | Culture (no. of colonies, out of 11) | |||
| A | <3 × 102 | L. sakei | + | − |
| P. pentosaceus | + | − | ||
| C | 1.4 × 107 | L. acidophilus | + | + (3) |
| L. crispatus | + | + (2) | ||
| L. casei group | +b | + (1) | ||
| L. plantarum | +b | + (5) | ||
| L. curvatus | + | − | ||
| L. sakei | + | − | ||
| L. mesenteroides | + | − | ||
| D | 1.2 × 106 | W. confusa | + | + (5) |
| P. acidilactici | +b | + (1) | ||
| L. casei group | + | + (5) | ||
| L. curvatus | + | − | ||
| L. fructivorans | + | − | ||
Species were identified by sequencing of the DNA fragment upon excision from the gel.
Visible fragment in the gel but sequencing was unsuccessful.
Investigation of fecal samples from probiotic trial.
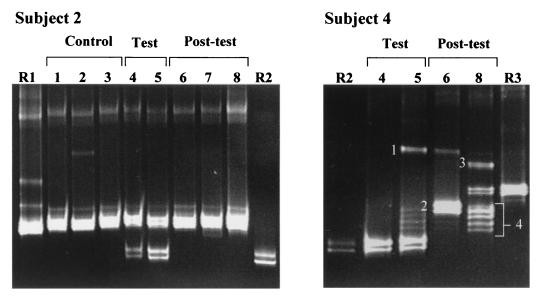
Fecal samples collected from two human subjects (subjects 2 and 4) who had consumed a milk product containing the probiotic strain L. rhamnosus DR20 (24) were investigated. The extracted DNAs of type strains of those species (L. rhamnosus, L. ruminis, and L. acidophilus) which had been cultured from these fecal samples on Rogosa agar during the probiotic trial were used as identification standards in DGGE gels. As summarized in Table 2 and shown in Fig. 6, PCR-DGGE using the group-specific primers Lac1 and Lac2GC detected the presence of L. rhamnosus only in the fecal samples of both subjects collected during the test period. As shown during the probiotic trial (24), L. rhamnosus was below detectable limits in the feces of subject 2 when using bacteriological culture, but was detectable using L. rhamnosus-specific PCR primers (Table 2). L. ruminis was cultured from all of the fecal samples collected from subject 2 and was also detected by PCR-DGGE. The feces of subject 4 harbored some species that were only detected by PCR-DGGE (Fig. 6, fragments 1 to 4). These species (L. sakei, L. delbrueckii, L. curvatus, and Leuconostoc mesenteroides) are considered to be food-associated bacteria. L. acidophilus was identified in the DGGE gel by reference to the migration distance of the fragment from the type strain (Fig. 6, R3 and sample 8). Detection of this species in the samples was in agreement with the culture result. PCR amplicons were not obtained from fecal samples 1 to 3 in the control period and sample 7 in the posttest period of subject 4. L. crispatus and L. acidophilus were, however, detected at levels of 3.2 × 105 and 1.6 × 105 CFU/g in samples 1 and 7, respectively (Table 2).
TABLE 2.
Bacterial counts of fecal samples from a probiotic trial and species detected by PCR-DGGE and by culture
| Subject no. and sampling period | Fecal sample no. | Cell counts on Rogosa agar (CFU/g) | Bacterial speciesa | Detected by:
|
|
|---|---|---|---|---|---|
| PCR-DGGE | Culture (no. of colonies, out of 10) | ||||
| Subject 2 | |||||
| Control | 1, 2, 3 | 3.2 × 108 to 4.0 × 109 | L. ruminis | + | + (10) |
| Test | 4, 5 | 4.0 × 108, 2.0 × 108 | L. ruminis | + | + (10) |
| L. casei group | + | − | |||
| Posttest | 6, 8 | 2.0 × 108, 2.5 × 108 | L. ruminis | + | + (10) |
| 7 | 2.0 × 108 | L. ruminis | + | + (9) | |
| L. casei group | − | + (1) | |||
| Subject 4 | |||||
| Control | 1 | 3.2 × 105 | L. crispatus | − | + (10) |
| 2, 3 | None | None | − | − | |
| Test | 4 | 1.6 × 105 | L. rhamnosus DR20 | +b | + (10) |
| 5 | 2.5 × 105 | L. rhamnosus DR20 | +b | + (10) | |
| L. sakei | +c | − | |||
| Posttest | 6 | None | L. delbrueckii | +c | − |
| L. sakei | + | − | |||
| 7 | 1.6 × 105 | L. acidophilus | − | + (9) | |
| 8 | 1.0 × 106 | L. acidophilus | + | + (10) | |
| L. curvatus | +c | − | |||
| Leuconostoc mesenteroides | +c | − | |||
Species were identified by comparison of DGGE profiles from fecal samples with those obtained from reference strains and from bacterial isolates as described by Tannock et al. (24).
Identified as belonging to the L. casei group.
Identified by sequencing of the DNA fragment upon excision from the gel.
FIG. 6.
DGGE analysis of 16S rDNA fragments obtained from the fecal samples of subjects 2 and 4 before, during, and after consumption of a probiotic product. Subject 2: R1, L. ruminis DSM 20403T; R2, L. rhamnosus DSM 20021T; 1 to 8, fecal samples taken during the control (1 to 3), test (4 and 5), and posttest period (6 to 8). Subject 4: R2, L. rhamnosus DSM 20021T; R3, L. acidophilus DSM 20079T; 4 to 8, fecal samples taken during the test (4 and 5) and posttest period (6 and 8). Samples from the control period (1 to 3) and sample 7 of the posttest period of subject 4 did not give PCR products. DNA fragments which did not match fragments of the reference strains were allotted upon sequencing to the following species: 1, L. sakei; 2, L. delbrueckii; 3, L. curvatus; and 4, Leuconostoc mesenteroides.
DISCUSSION
The presence of lactobacilli in fecal or intestinal samples has traditionally been demonstrated by bacteriological culture. This work has revealed that a relatively limited number of Lactobacillus species are regularly detectable in human feces (12). The PCR-DGGE system that we have described permits the detection in feces of not only Lactobacillus species considered to be intestinal inhabitants but also of LAB commonly associated with food and often used as starter organisms (21). At least half of the LAB that we detected in feces belonged to this category. The staining intensity of the DNA fragments in the gels indicated that the food-associated bacteria were present in the feces in similar numbers to the typical intestinal LAB. The food-associated species in the fecal samples were not cultivable on Rogosa agar. Either target DNA in dead cells, DNA released from cells that had lysed in the intestinal tract, or DNA from living cells in a noncultivable state was detected. Culture media other than Rogosa agar might support the growth of these latter cells, but we have not yet investigated this possibility. We believe that the detection of noncultivable LAB sequences in human feces was of interest because it made us wonder about the impact of these ingested bacteria on the consumer. It would be interesting to know, for example, in which part of the digestive tract the LAB change from cultivable to noncultivable and whether even in the noncultivable state they may have an impact on the immune system as they transit the gut in the digesta (4, 11). The irregular detection in human feces of noncultivable LAB originating in food strengthens the concept that only some of the Lactobacillus species detected in human studies are truly autochthonous to the intestinal ecosystem; even some of the cultivable species may be transient bacteria consumed in food (24).
LAB are difficult to detect in human feces when using universal PCR primers because these bacteria constitute only a minor part of the microflora. In the probiotic trial using L. rhamnosus DR20, the strain was not detected in human fecal samples using PCR-DGGE and universal primers HDA1GC and HDA2, although it was cultured from the feces of the subjects and represented the predominant Lactobacillus strain during the test period (24). The use of the Lac1 and Lac2GC primers lowered the detection limit so that L. rhamnosus could be detected in the fecal samples by PCR-DGGE. In general, Lactobacillus species present in numbers of >106 CFU/g of feces (wet weight) could be detected even in the presence of DNA from the predominant members of the fecal microflora (about 1011 cells/g) in the PCR.
PCR with primers Lac1 and Lac2GC amplified a 340-bp fragment of the V3 region of the 16S rRNA gene. This sequence permitted most of the Lactobacillus species to be differentiated. Members of the L. casei group have sequence variation in the V1 region between nucleotides 73 and 111 of the 16S rRNA gene (13) but none in the V3 region. Thus, they could not be differentiated in our system. Sequencing all of the fragments in a DGGE profile is laborious, and inaccurate identifications may occur because of the poor quality of some sequences deposited in databases. We found that it is easier to identify the bacteria by comparison of the PCR amplicon migration distances in DGGE gels with those of reference strains. It is noteworthy that the species comprising the L. acidophilus group cannot easily be identified by classical phenotypic tests (8) but can be easily differentiated by PCR-DGGE using the Lac1 and Lac2GC primers.
Our study has led to the derivation of PCR primers that amplify bacteria belonging to the Lactobacillus, Pediococcus, Leuconostoc, and Weisella group of LAB. The primers have been demonstrated to be useful in the analysis, at the species level, of these populations in human feces. This can be achieved simply and economically by a single PCR followed by DGGE. We think that the primers Lac1 and Lac2GC and PCR-DGGE have potential use in the following types of studies. Firstly, they could be used in studies to monitor the LAB content of human subjects in probiotic or other trials aimed at manipulating the composition of the intestinal microflora. Secondly, they could be used in large-scale microbial ecological investigations of the intestinal tract of humans and other animals in order to define the characteristic Lactobacillus microflora of particular hosts. Thirdly, they could be used to follow the bacterial successions that occur during the acquisition of the intestinal microflora. Finally, the results of our experiments suggest that there is potential for the derivation of group-specific PCR primers for the many phylogenetic groups of bacteria that comprise the intestinal microflora. The use of group-specific primers could provide information essential to an understanding of the dynamics, at the level of bacterial species, of the intestinal ecosystem.
ACKNOWLEDGMENTS
We thank M. Kranz for excellent technical assistance during sequencing. The participation of the subjects in this study is gratefully acknowledged.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ben Omar N, Ampe F. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl Environ Microbiol. 2000;66:3664–3673. doi: 10.1128/aem.66.9.3664-3673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Haller D, Bode C, Hammes W P, Pfeifer A M A, Schiffrin E J, Blum S. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammes W P, Vogel R F. The genus Lactobacillus. In: Wood B J B, Holzapfel W H, editors. The lactic acid bacteria. 2. The genera of lactic acid bacteria. London, United Kingdom: Blackie Academic and Professional; 1995. pp. 19–54. [Google Scholar]
- 6.Heuer H, Krsek M, Baker P, Smalla K, Wellingon E M H. Analyses of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein G, Pack A, Bonaparte C, Reuter G. Taxonomy and physiology of probiotic lactic acid bacteria. Int J Food Microbiol. 1998;41:103–125. doi: 10.1016/s0168-1605(98)00049-x. [DOI] [PubMed] [Google Scholar]
- 9.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCartney A L, Wang W, Tannock G W. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl Environ Microbiol. 1996;62:4608–4613. doi: 10.1128/aem.62.12.4608-4613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meydani S N, Ha W-K. Immunologic effects of yogurt. Am J Clin Nutr. 2000;71:861–72. doi: 10.1093/ajcn/71.4.861. [DOI] [PubMed] [Google Scholar]
- 12.Mitsuoka T. The human gastrointestinal tract. In: Wood B J B, editor. The lactic acid bacteria. 1. The lactic acid bacteria in health and disease. London, United Kingdom: Elsevier Applied Science; 1992. pp. 69–114. [Google Scholar]
- 13.Mori K, Yamazaki K, Ishiyama T, Katsumata M, Kobayashi K, Kawai Y, Inoue N, Shinano H. Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int J Syst Bacteriol. 1997;47:54–57. doi: 10.1099/00207713-47-1-54. [DOI] [PubMed] [Google Scholar]
- 14.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 15.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Sullivan D. Methods for analysis of the intestinal microflora. In: Tannock G W, editor. Probiotics: a critical review. Wymondham, United Kingdom: Horizon Scientific Press; 1999. pp. 23–44. [Google Scholar]
- 17.Reid G. The scientific basis for probiotic strains of Lactobacillus. Appl Environ Microbiol. 1999;65:3763–3766. doi: 10.1128/aem.65.9.3763-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt G, Hertel C, Hammes W P. Molecular characterisation of the dnaK operon of Lactobacillus sakei LTH681. Syst Appl Microbiol. 1999;22:321–328. doi: 10.1016/S0723-2020(99)80039-3. [DOI] [PubMed] [Google Scholar]
- 19.Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol. 2000;66:2263–2266. doi: 10.1128/aem.66.5.2263-2266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson J M, McCracken V J, Gaskins H R, Mackie R I. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain NM53. Appl Environ Microbiol. 2000;66:4705–4714. doi: 10.1128/aem.66.11.4705-4714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiles M E, Holzapfel W H. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 22.Tannock G W. Normal microflora. An introduction to microbes inhabiting the human body. London, United Kingdom: Chapman and Hall; 1995. [Google Scholar]
- 23.Tannock G W. Analysis of the intestinal microflora: a renaissance. Antonie Leeuwenhoek. 1999;76:265–278. [PubMed] [Google Scholar]
- 24.Tannock G W, Munro K, Harmsen H J M, Welling G W, Smart J, Gobal P K. Analyses of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. 2000;66:2578–2588. doi: 10.1128/aem.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughan E E, Mollet B, deVos W M. Functionality of probiotics and intestinal lactobacilli: light in the intestinal tract tunnel. Curr Opin Biotechnol. 1999;58:505–510. doi: 10.1016/s0958-1669(99)00018-x. [DOI] [PubMed] [Google Scholar]
- 26.Walter J, Tannock G W, Tilsala-Timisjarvi A, Rodtong S, Loach D M, Munro K, Alatossava T. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primer. Appl Environ Microbiol. 2000;66:297–303. doi: 10.1128/aem.66.1.297-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R-F, Cao W-W, Cerniglia C E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Hayes V M, Osinga J, Mulder I M, Looman M W G, Buys C H C M, Hofstra R M W. Improvement of fragment and primer selection for mutation detection by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1998;54:32–40. doi: 10.1093/nar/26.23.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoetendal E G, Akkermans A D L, de Vos W M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]