Abstract
Microtubules (MTs) and MT motor proteins form active 3D networks made of unstretchable cables with rod-like bending mechanics that provide cells with a dynamically changing structural scaffold. In this study, we report an antagonistic mechanical balance within the dynein–kinesin microtubular motor system. Dynein activity drives the microtubular network inward compaction, while isolated activity of kinesins bundles and expands MTs into giant circular bands that deform the cell cortex into discoids. Furthermore, we show that dyneins recruit MTs to sites of cell adhesion, increasing the topographic contact guidance of cells, while kinesins antagonize it via retraction of MTs from sites of cell adhesion. Actin-to-microtubule translocation of septin-9 enhances kinesin–MT interactions, outbalances the activity of kinesins over that of dyneins, and induces the discoid architecture of cells. These orthogonal mechanisms of MT network reorganization highlight the existence of an intricate mechanical balance between motor activities of kinesins and dyneins that controls cell 3D architecture, mechanics, and cell–microenvironment interactions.
Keywords: dynein, kinesin, microtubules, contact guidance, mechanobiology, motility
Microtubules (MTs) and MT motors have emerged as key drivers of dynamic changes to the cell cytoskeleton, controlling cell chirality,1,2 shape plasticity,3−7 3D architecture,8−11 motility signaling,12 mechanotransduction,13 and 3D migration (in immune cells).14 In cooperation with the integrin-based actomyosin adhesion system, MTs serve as a mechanosensor that integrates extra- and intracellular mechanical signals on a scale of the whole cell.15 Mechanistically, MTs form a cellular network of unstretchable rod-like cables that are embedded within sterically confining, laterally re-enforcing actomyosin networks providing cells with a means to resist mechanical deformations;16,17 e.g., in cardiac myocytes, MTs that are coaxially integrated into the myofibrillar network act as rod-like springs which mechanically oppose myofibril contractile shortening.18,19 In other cell types, MTs provide scaffolds that mechanically reinforce actively protruding pseudopodia, e.g., invadopodia,7,20−22 podosomes,23,24 invasive microtentacles,25,26 or dendritic cell protrusions that facilitate cell adhesion and spreading within soft fibrous 3D collagen matrices.27,28
At any given point in time, the 3D architecture of an individual cell is, ultimately, a result of a dynamic balance between the internal and external structural and biochemical and mechanical signals that are integrated by (at least in part) actomyosin and MT networks. For instance, cell body linearization is driven by an actomyosin-dependent inward collapse of the MT network into linear MT bundles,10,29 whereas integrin-dependent cell adhesion and spreading drives MT unbundling.30−32 Adaptations of cell shape to migration, e.g., dynamic interactions between the cell and the extracellular matrix (ECM), also require integration of biochemical and biomechanical signals between MTs and actomyosin which enables cells to sense, align with, and exhibit guided motility along nanotextured 3D microenvironments.32 While it is clear that MT and actomyosin networks play a crucial role in the control of cell shape and cell–microenvironment interactions, the precise nature of specific interactions between MTs and actomyosin, and the manner in which mechanical forces are transmitted and balanced between these two networks, remains elusive.
An accumulating body of evidence indicates that MT motor proteins actively control cell architecture and mechanics via regulating interactions between MTs and other cellular components like actomyosin and organelles as well as with the extracellular matrix (ECM). For example, recent findings demonstrate that dynein motors drive and coordinate spatial alignment of radially organized MT networks, centering microtubules organization centers (MTOCs) at the geometrical or mechanical center of the actomyosin network of the cell.33,34 Dyneins also control cell-type-specific changes in the cell architecture that drive downstream functional adaptations; i.e., in neurons, dyneins drive MT sliding and extension along the axonal axis,35−38 while in hematopoietic cell lineages, dyneins generate tensile forces in MT networks that surround the cell nucleus and ultimately result in the formation of nucleus-compressing 3D MT meshworks that favor mechanosensory activation of chromatin toward myeloid (and away from lymphoid) differentiation.39 Meanwhile, kinesins demonstrate similar mechanical and MT-restructuring effects, but in apparent opposition to the actions of dyneins. For example, kinesins have been shown to promote bundling and sliding of antiparallel, “telescopic” MT filaments that manifests as “looping” at the level of individual filaments40 and drive global reorganization of MTs at the level of the whole cell.41 These individual findings highlight the involvement of both kinesin and dynein motors in the control of cell structure and shape. However, very few studies, if any, have probed how or whether the synergy or antagonistic actions of both motors in tandem drive changes in cell shape, mechanics, or cell–microenvironment interactions, presumably due to challenges associated with cytotoxicity and the general lack of motor-specific inhibitors to study mesoscopic (whole cell) scale effects of kinesin and dynein balance.
Previous studies have linked the dynamics of actomyosin and MT networks to large-scale changes in cell structure9,42 using MT networks of enucleated mammalian platelets that have MTs organized into parallel, 2–4 μm ring-like structures known as marginal bands.43 During platelet activation, the action of dynein drives band expansion into the actomyosin platelet cortex,9,44 resulting in displacement of tension into the cortex, platelet discoid deformation, and eventually a disc-to-sphere transition.8,45 Thus, the 3D structure of platelets is dependent upon a delicate, mechanistic balance between antagonistic dynein-based MT forces and actomyosin-based tension within the platelet cortex that can be dynamically shifted via modulating dynein activity or that of other motors like myosins.8,9,45 As the direct mechanical antagonist of dyneins, kinesins oppose and prevent dynein-driven marginal bands overextension,44 likely via cross-linking/bundling MTs in the opposite direction of dynein-dependent MT antiparallel sliding.9,42 However, the exact mechanism of how marginal band homeostasis is maintained by the opposing actions of dynein–kinesin remains unresolved; i.e., whether the dyneins and kinesins drive the marginal band ring extension–compression balance via the indirect MT–actomyosin–MT9 or the direct MT–MT filament cross-linking and sliding.42 Moreover, the exact roles and mechanisms for dyneins and kinesins during platelets’ marginal band structural condensation around a singular multicoiled MT43 remain unknown.
Despite the significant progress in understanding the isolated effects for MT motor proteins on the organization of the MT network and MT–actomyosin interaction, the importance and the extent of mechanical antagonism between dyneins and kinesins at the mesoscopic whole cell level, e.g., for a cell with a regular size and unreduced structural complexity, remain elusive. It is also unclear which role the balance between kinesin and dynein motor activity plays in the cellular mechanobiology and cell–microenvironment interactions, e.g., structural adaptation and migration responsiveness to the microenvironment cues.
Results and Discussion
Inhibition of Dyneins and Myosins Induces Giant Microtubular Rings
We examine the morphological response of MDA-MB-231 triple negative human breast adenocarcinoma cells following the inhibition of dyneins and myosins. MDA-MB-231 cells are triple negative breast cancer cells and are chosen as our principal cell model due to our clinical interest in both (i) the role of microtubules and microtubular motors in the mesenchymal mode of cell migration and (ii) the microenvironment-directed contact guidance of cancer cells, one of the primary drivers of metastatic dissemination in triple negative breast cancer.32,46,47
For efficient and acute dynein inhibition, we utilize Dynapyrazole A (80 μM, 1 h, unless otherwise noted). Dynapyrazole A is a Ciliobrevin-derived inhibitor, which displays substantially higher stability and dynein-inhibition efficacy than Ciliobrevins.48 Similarly, we utilize a high dose of (−)-Blebbistatin (100 μM, 1 h) for acute suppression of actomyosin contractility.9,32
To capture the in-tissue complexity of cell adhesion faithfully, yet uniformly across large cell populations, we use an isotropic rectangular grid of collagen-1 “fibers” that support fibrillar cell spreading11 (Figure 1a). Collagen type-1, a major cell adhesion ligand in the ECM of physiological tissues, is organized into orthogonal sets of 1-μm-wide parallel lanes, spaced with a 15-μm-wide pitch to mimic collagen “fibers”. Micropatterned collagen grids prevent the excessive formation of the 2D lamellipodium.11,49 Abnormal 2D lamellipodium dominates cell adhesion and spreading scenarios on regular flat substrates, while a 1D “fiber” architecture induces a more natural quasi-3D modality of cell–ECM interactions.49 In order to closely recapitulate the average tissue mechanical rigidity, we microprint the collagen ECM grids on polyacrylamide hydrogel surfaces with a mechanical rigidity of G′ = 50 kPa.50,51
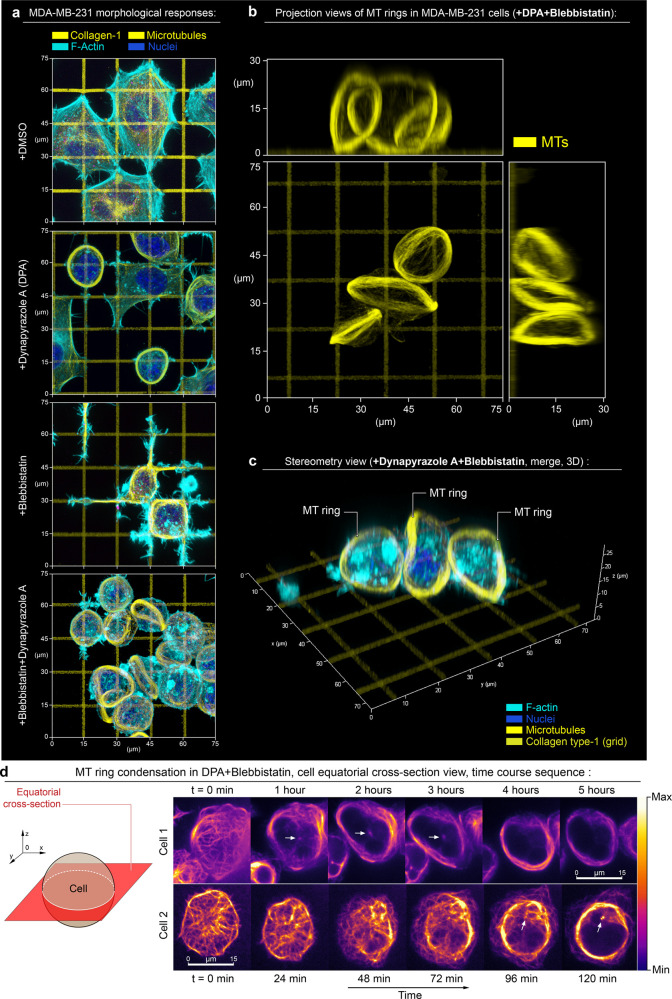
Figure 1.
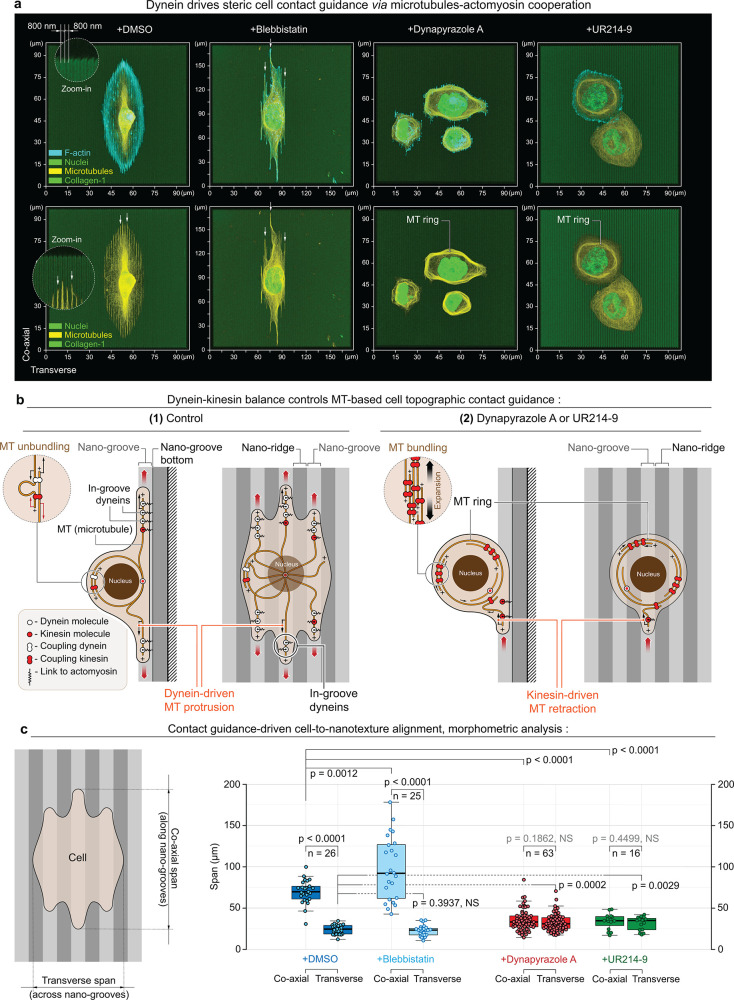
Top 3D overview of MDA-MB-231 cells during adhesion and spreading along the artificial extracellular matrix (orthogonal type-1 collagen grid, G′ = 50 kPa). (a) Micrographs present cell configurations in the control condition (+DMSO), during dynein inhibition (+Dynapyrazole A), in the presence of actomyosin contractility inhibitor (+Blebbistatin), and under combined dynein and actomyosin contractility suppression (+Blebbistatin+Dynapyrazole A). Upon biaxial spreading along the orthogonal collagen lanes in the control condition (+DMSO), cells develop a concave polygonal architecture with stress-fibers predominantly located at the mechanically tensed cell periphery—i.e., arching free cell edges. Suppression of the dynein motor activity (+Dynapyrazole A) induces mixed cell architectures, with 70% of cells partially developing MT rings along with cells having polygonal architecture. Inhibition of actomyosin contractility (+Blebbistatin) shifts cell organization toward the spreading network of dendritic protrusions that compliantly extend along the collagen grid. These protrusions feature the microtubule scaffold as their structural core. Notably, combined dynein and myosin II inhibition (+Blebbistatin+Dynapyrazole A) results in the reorganization of the MT network into MT ring in >90% cells. Similar to marginal bands in mammalian platelets, these rings deform cells into lenticular discoids. Separate channels are shown in Figure SI1. (b,c) 2D projection views (b) and stereometric 3D views with merged channels (c) of the MT rings, induced by +Dynapyrazole A+Blebbistatin treatment. (d) Time course sequence for MT network expansion and ring condensation in MDA-MB-231 cells (+ Dynapyrazole A+Blebbistatin; 130 μM and 50 μM respectively), equatorial confocal plane imaging, SiR-tubulin live microtubules labeling; see Movie 1. Note the MT network expansion-driven translocation throughout the cell volume toward the cell cortex, while MTOCs (arrows) remain separated from the ring within the cytosolic volume of cells. F-actin is labeled with Phalloidin-ATTO 647N. Chromatin is labeled with Hoechst.
Within 2 h, in control conditions (+DMSO), spreading cells acquire concave polygonal shapes with stress-fibers accumulating at the arching cell edges (Figures 1a and SI1, +DMSO). In this scenario, we observe a geometric configuration of adhesions, which presumably serves to induce high linear tension along the free cell edges.11,52 Suppression of actomyosin contractility with Blebbistatin results in polygonal cell shape collapse and “dendritic”-like cell spreading with cell protrusions growing along the collagen-1 grids (Figures 1a and SI1, +Blebbistatin), reflecting Arp2/3-driven low-contractility or “fluid-like” cell spreading, reported for 2D/3D ECM.11,27,28 Dynein inhibition with Dynapyrazole A induces global reorganization of the MT network into MT bundles with various stages of maturity for ∼70% of the total cell population and include a subset of circularized MT bundles (rings) in ∼50% of all cells (n = 80) (Figures 1a and SI1, Dynapyrazole A). Combined inhibition of dyneins and myosins drives substantially more prominent and uniform bundling of MTs across the entire cell population, resulting in the loss of cell spreading and MT reorganization into well-pronounced, 3D circular ring-like bundles (diameter ⌀ ≈ 20 μm) that are located around the cell peripheral circumference and deform the cell into the discoids (Figures 1a and SI1, +Blebbistatin+Dynapyrazole A), observed in ∼65% of the total cell population (n = 100). Notably, no cells with detectable MT rings are observed in +DMSO and +Blebbistatin treatment conditions (Figures 1a and SI1).
The analysis of 3D projections (X0Y, X0Z, Y0Z) for Blebbistatin+Dynapyrazole A-treatment (Figures 1b,c and SI2) indicates that MT rings induce the deformation of cells into lenticular discoids (⌀ ≈ 20–25 μm). This cell shape is indicative of an active deformation of the cell cortex from within the cell volume by mechanically expanding MT rings. Notably, Dynapyrazole A-induced discoid deformation is synergistically favored by the Blebbistatin cotreatment, suggesting that contractility of actomyosin within the cortex mechanically antagonizes cell shape plasticity and highlighting a relationship between F-actin cortex structural cross-linking and MT network pliability.53,54 Furthermore, an F-actin cytoskeleton lacking the contractile influence of myosin II would allow for both higher F-actin cortex viscous compliance, and deformability—meanwhile, the structural and mechanical integrity of F-actin-microtubular networks as a single system would still be maintained via their dynein-dependent interlinking33,55 and via steric interactions.16 Given the above, our interpretation is that an increase in F-actin network structural compliance in Blebbistatin is a predominant factor contributing toward the increased efficiency of MT spatial reorganization and ring formation in comparison to when cells are solely exposed to Dynapyrazole A.
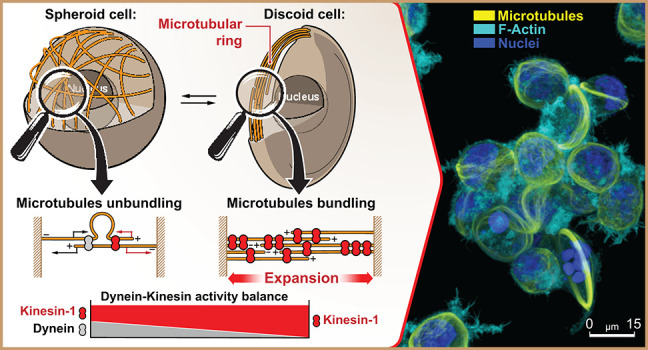
The cell discoids are structurally reminiscent of significantly smaller (⌀ ≈ 3 μm) MT marginal bands (MB) in highly specialized, enucleated, structural complexity-reduced mammalian platelets42 and in non-mammalian red blood cells.4 However, these structures were not previously reported for mesenchymal cells, e.g., for large cells with unreduced structural complexity. Dynamics of MT ring formation in +Dynapyrazole A+Blebbistatin shows that MTs, which are normally distributed roughly equally throughout the cell volume, translocate and redistribute almost entirely into the cortex, indicating the active spatial expansion of MT network (Figure 1d and Movies 1 and 2). Spatial expansion of MTs results in their spherical buckling into the MT ring, caused by the confining boundaries of the cell cortex (Figure 1c). This phenomenon, largely similar, but not fully identical with the MB formation in much smaller mammalian platelets,8,45 appears to highlight a mechanical antagonism that exists between the expanding MT ring and the cell cortex. Thus, we conclude that the cell lenticular discoid deformation during +Dynapyrazole A+Blebbistatin cotreatment is the result of the MT circular band extension into a relatively compliant cell cortex. Interestingly, MT band extension into a more contractile and rigid cell cortex leads to the persistence of the cell spheroid shape, accompanied by MT ring buckling.56
We observe that micropatterned collagen type-1 grids are more favorable for the MT ring formation and cell discoid deformation, as the excessive lamellipodium-driven cell spreading and flattening on the preliminary tested solid uncoated surfaces (used for super-resolution imaging) and collagen-coated surfaces (not shown) significantly slows down and effectively prevents structural reorganization of the MT network. These results suggest that the subcellular mechanisms that underpin cell adhesion and spreading processes interfere with and/or inhibit the formation of prominent MT ring, as the detachment of fully spread and flattened cells in response to the +Dynapyrazole A+Blebbistatin treatment is not effective (within the duration of the experiment) and is possibly hindered by the numerous nascent adhesions that, unlike mature focal adhesions, do not require robust cell contractility. Our general observations also indicate that the MT ring-like reorganization can occur in the spheroid cells prior to their spreading. However, it is important to highlight that MT ring formation is an extreme form of the MT cytoskeleton reorganization in response to the acute motor activity perturbations with high drug doses and that the absence of MT rings within a particular cell does not preclude the universality of the MT motor counterbalance mechanisms that we propose within this manuscript.
Notably, the positioning of the MTOCs within the cell volume is largely affected by suppression of dynein (Figures 1d, arrows, and SI3a,b); i.e., during +Dynapyrazole A ± Blebbistatin treatments the MTOC displays a loss of its canonical position at the center of the microtubular radial arrays, with the latter being drastically reorganized into the MT rings (Figure SI3). Indeed, the MT radial array formation in cells does not always mandate MTOCs as the primary nucleators of the radially organized microtubular filaments, but instead requires dyneins as the MT cross-linkers and MT-sliding motors for MTOC-independent self-organization.57−59 Thus, a more general mechanism for MT self-organization highlights microtubules’ minus- or plus-end collective spatial convergence, driven by the dominance of either MT minus- or plus-end-directed motor processivity.57−61 Currently, it is assumed that in certain cases MTOCs are the secondary structures at the centers of the MT radial arrays and are being transported to the self-organized MT array foci by the motors, e.g., dyneins.62,63 Our results are consistent with these findings and confirm that global microtubular network reorganization, driven by perturbations in MT motor balance, can also lead to both (i) the displacement of the MTOC from the canonical MT array foci and (ii) the structural detachment of the MTOC from the dominant MT network structures.
The robust trend of MT network reorganization into 3D MT rings during +Dynapyrazole A+Blebbistatin treatments is reversible, i.e., after the clear MT network translocation from the cell volume to the cell periphery, where MTs appear to condense into MT rings, the substitution of the +Dynapyrazole A+Blebbistatin incubation medium with freshly preheated (37 °C) and CO2-preincubated cell culture medium lacking inhibitors (washout) reverses inhibitor-dependent suppression of MDA-MB-231 cell spreading and also results in the rapid disassembly of MT rings; cells subsequently reacquire their spread polygonal architectures on the collagen type-1 grids (Figure SI4 and Movie 3). Thus, we conclude that cell 3D shape and gross MT network organization are governed at least in part by dynein–kinesin mechanical counterbalance and myosin activity status, and that this counterbalance is a reversible, dynamically adjustable mechanism that can be utilized for regulation of the key aspects of cell behavior, such as changing the MT network 3D architecture and the cell structure, and tuning the key aspects of cell mechanobiology.
Balance of Dynein and Kinesin Activities Controls Structure of Microtubule Network
We show that MDA-MB-231 cells develop giant MT rings in response to the suppression of dynein activity, while the formation of MT marginal bands in mammalian platelets, on the contrary, requires active dyneins yet inactive kinesins.9 We hypothesize that the reorganization of the isotropic MT network into circular bundles is likely driven by the disbalance of dynein and kinesin motor activities in either direction (Figure 2a-1). In this model, the action of dyneins and kinesins together creates an expansion–compaction balance of the MT network within the cell volume, as well as MT protrusion–retraction balance within cell adhesion sites (Figure 2a-1). Isolated activity of kinesins (e.g., +Dynapyrazole A) leads to both MT network expansion and kinesin-driven retraction of MTs from cell adhesion sites (Figures 2a-2 and SI5). Therefore, physical expansion of the MT network and retraction of MT filaments from peripheral adhesions each results from kinesin sliding along the MT filaments toward MT plus-ends, that if uncompensated for by the opposing dynein-dependent MT sliding toward minus-ends, lead to a progressing, antiparallel telescopic extension of MT filaments, as well as to the ejection of MTs by kinesin from the adhesion sites (Figures 2a-2 and SI5b).
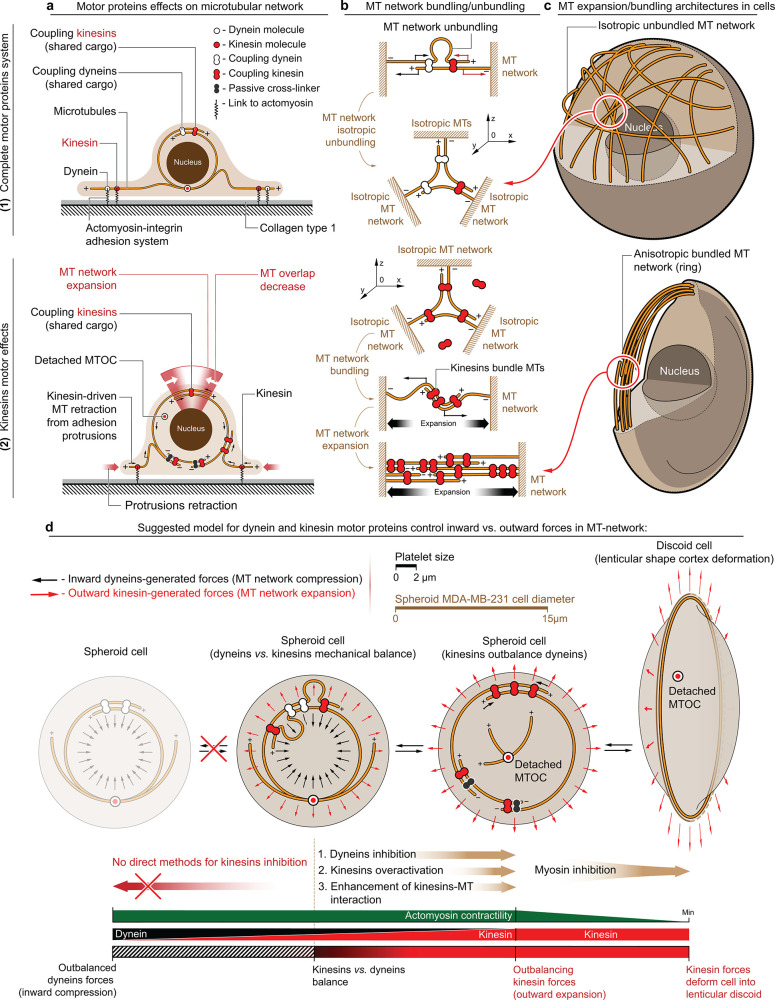
Figure 2.
Kinesin and dynein motor proteins counterbalance outward and inward-directed forces within MT network. (a) Schematic representation of dynein–kinesin superposition and sole kinesin-generated forces within the MT network: (1) Superpositioned dynein and kinesin forces counterbalance each other within both cell volume and at cell–substrate adhesion sites. (2) Kinesin motor proteins facilitate MT network expansion into the cell cortex within the cell volume via antiparallel telescopic MT sliding-out. Kinesin-driven MT sliding decreases MT overlap, extends overlapping antiparallel MT filaments (MT network expansion), and drives MT network retraction/expulsion from the cell-ECM adhesion sites. Note that dynein can act as the MTs’ passive cross-linker in +Dynapyrazole A+Blebbistatin that prevents MT network collapse into a radial MT array by kinesin-1-driven collective concentration of all MT plus-ends into a single focus. Active dynein also contributes as an antagonizing cross-linker during kinesin-1 overactivation, remaining relatively passive in comparison to kinesin’s enhanced MT processivity. (b) Key force-generating configurations of MT-motor proteins can effectively drive MT network compaction/expansion and MT bundling/unbundling. Note that dynein and kinesin motors both directly and indirectly bundle MTs and form coupled complexes, e.g., by cargo shared between two or more motors (coupled dynein and kinesin). (1) Simultaneous work of coupled dyneins and kinesins within antiparallel MT structures induce directionality conflict within sliding MTs, that results in MT filaments buckling and unbundling into scattered, misaligned, and isotropic MT networks. (2) Kinesin-only system (coupled kinesins) induces MT bundling and antiparallel MT “telescopic” sliding-out that drives 3D MT network collapse into the ring (circular bundle) and ring expansion into the cell cortex deforming cells into the discoids. (c) Nontreated spheroid cells with active kinesins and dyneins have unbundled MT network, scattered throughout cell volume. Isotropic MT network highlights dynamic mechanical balance of network compaction and expansion. +Dynapyrazole A+Blebbistatin-treated MDA-MB-231 cells develop kinesin-bundled MT rings that feature kinesin-driven ring expansion. MT ring expansion deforms individual cells into lenticular discoids. (d) Suggested mechanism describes mechanical and structural contribution of MT network into the cell cortex as a counterbalancing system of dynein and kinesin motors. Three alternative strategies are available for shifting the mechanical balance toward a higher contribution of kinesins—direct dynein inhibition (+Dynapyrazole A), kinesin-1 overactivation (+Kinesore), and MT network redecoration with the septin-9 that enhances kinesin-MT interactions (+UR214-9). Note that inhibition of cortex contractility, combined with outbalanced activity of kinesins, leads to the MT ring formation–expansion, and cell deformation into the lenticular discoid.
Notably, dyneins64 and kinesins65−68 each can cross-link and linearize MT networks into antiparallel bundles.40 These bundles are additionally enhanced through interaction with the actomyosin cytoskeleton, which serves in the mechanical transmission of dynein-driven forces between adjacent MT filaments.69 Our basic idea of mechanical antagonism between dyneins and kinesins is that these motors exhibit individual and combined structuring effects onto the bundling and unbundling of MT networks (Figure 2b). In the antiparallel MT bundles, the combined activity of dyneins and kinesins will lead to an MT sliding-in and sliding-out directionality conflict, consequent buckling of MTs which is followed by MT isotropic disarray,70,71 MT network unbundling and spatial misalignment (Figure 2b-1). Alternatively, in parallel (unidirectionally oriented) MT bundles, the principal lack of the antiparallel MT sliding activity will lead to low levels of MT reorganization and bundling dynamics for both dynein and kinesin motors. Thus, we neglect the contribution of the MT motor activity within the parallel MT bundles. Importantly, shifting the balance of dynein and kinesin motor activity via dominance of either motor enhances MT bundling due to directionally coherent sliding of MTs and due to a decrease of the MT sliding directionality conflict, as demonstrated on the example of kinesin-wise balance shift (Figure 2b-2), driving reorganization of the isotropic MT network into the single bundled structure (Figures 1a and SI1: +Dynapyrazole A and +Blebbistatin+Dynapyrazole A; Figures 1b and SI2). Since there is no technically reliable method for a detectable kinesin inhibition in nonplatelet cells, the reorganization of MTs due to shifting the dynein–kinesin balance toward dynein is depicted only hypothetically (Figure 2d).
The model of transition between isotropic and anisotropic MT networks fundamentally depends on the reorganization of cargo-coupled kinesin and dynein motors. For dynein motor-mediated MT-to-MT bundling and sliding activities, we consider the two most relevant, common, and abundant cargoes that are shared between dynein molecules positioned on the adjacent microtubules to act as their dynamic cross-linker: (1) the other adjacently localized microtubules64 and (2) F-actin, i.e., the actomyosin network that demonstrates a strong mechanical and structural linkage to the microtubules via the dynein–dynactin complex.33,34,55,72 Thus, in our model, F-actin and MT networks are the most abundant partners of the microtubule–dynein system that play the role of structural integrators—universally shared cargo between multiple dyneins, facilitating a universal medium for structural linking and transduction of dynein-generated mechanical forces between multiple adjacent microtubules. Similarly, kinesin remodels the microtubular architecture via its ability to slide microtubules along each other en masse and independently from the actomyosin network;41 i.e., in the kinesin-driven MT network reorganization, the microtubules participate both as the cargo of kinesins and as the actual trafficking tracks of kinesin cargos. In this scenario, kinesin effectively acts as a direct unmediated microtubule cross-linker,65 resulting in kinesin-driven MT bundling and sliding via their dual cargo-track role that orchestrates the formation of the elongating/expanding microtubular bundles and rings.
Based on the proposed model, we explain the formation of giant MT rings (a) and cell discoids (b) in Dynapyrazole A (±Blebbistatin) by the inhibition of dyneins and significant outbalance toward the kinesin-driven effects. These effects, such as MT bundling, extension of MT bundles, and their expansion into the cell cortex, result in the formation of the giant MT rings. These rings generate outward-directed forces sufficient for discoid deformation of the low-contractility actomyosin cell cortex during experiments with +Dynapyrazole A+Blebbistatin cotreatment (Figure 1a–c: +Dynapyrazole A+Blebbistatin; Figure 2d). Experimental characterization of the kinesin-driven expansion of MT networks into the MT rings during +Dynapyrazole A+Blebbistatin cotreatment demonstrates discoid deformation and identifies robust accumulation of total kinesins within the MT rings (Figures 3a and SI6), along with substantial MT ring diameter ⌀ dominance over the cells’ transverse width (Figures 3b and SI6a,b).
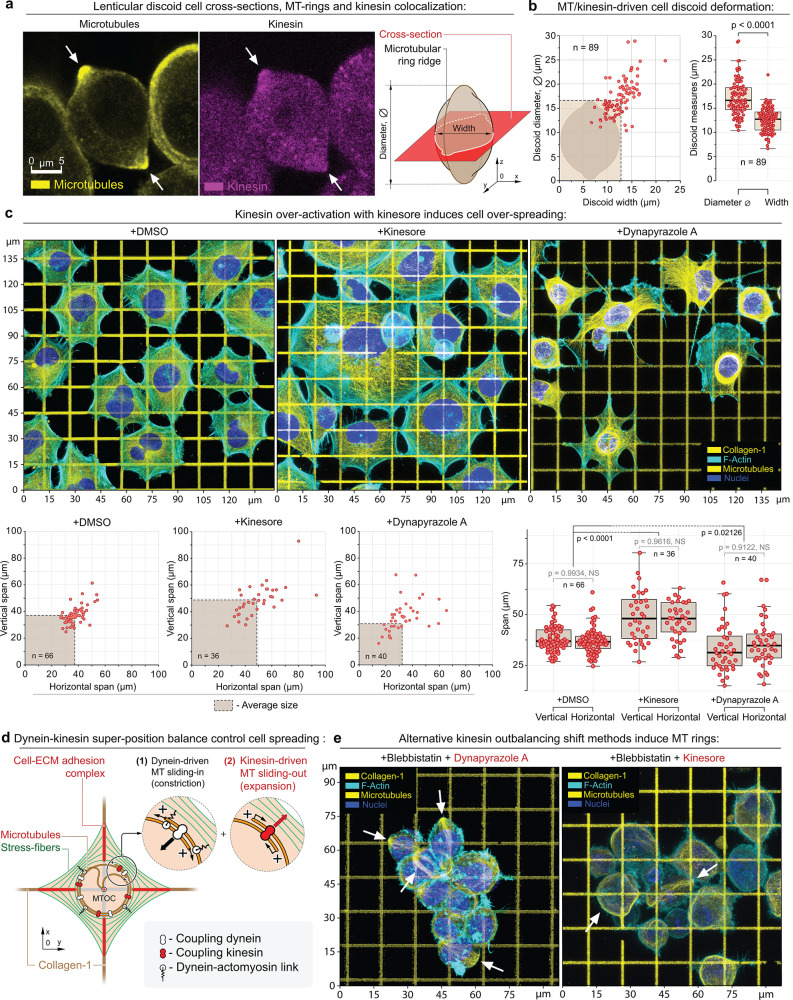
Figure 3.
Structural analysis of the MDA-MB-231 cells during alternative dynein–kinesin balance shift mechanisms. (a) MT ring median-plane cross-section in MDA-MB-231 cells (left) as shown in the schematic (right) in +Dynapyrazole A+Blebbistatin. Note kinesin accumulation at the MT rings (arrows); see also Figure SI3a–d. (b) MT rings expand and deform cells into the lenticular discoids (MT ring diameter > cell width). The plots depict the measurements of the cell discoid diameters (diameter of the MT ring) and the cells’ widths (transverse cell dimension). The dashed box depicts the averaged cell discoid diameter and width boundaries (means). (c) Top row – Adhesion and spreading of MDA-MB-231 cells on the collagen-1 grids (G′ = 50 kPa) in control condition (+DMSO) and during kinesin overactivation (+Kinesore), compared to the direct dynein inactivation (+Dynapyrazole A). Kinesin overactivation with +Kinesore does not inhibit the dynein activity, yet induces a significant increase of the biaxial spreading of cells in comparison to the control cells. Alternatively, dynein–kinesin balance shift toward the kinesin activity via the direct dynein inhibition (+Dynapyrazole A) induces a visible reduction of cell spreading and “dendritization” of cells that is similar to the loss of cell contractility shape during Blebbistatin-induced myosin II inhibition (Figure 1, +Blebbistatin). The loss of cell contractility in Dynapyrazole A is related to two main factors:74 the loss of MT-to-actomyosin mechanical cross-linking and force transmission as well as the loss of the dynein-driven mechanical contractility within the MT network (+Dynapyrazole A). Bottom row – Morphometric analysis of control (+DMSO) and +Kinesore- and +Dynapyrazole A-treated cells indicate a significant increase in cell biaxial spreading, driven by kinesin-mediated MT network expansion. (d) Suggested mechanism for the MT–actomyosin interaction in cells on the collagen grids. Dyneins and kinesins control the (1) MT network compaction and (2) MT network expansion, respectively. Dyneins also cross-link the MT network to the actomyosin cytoskeleton (stress-fibers), i.e., actomyosin, adhesion-bound matrix, adhesion complex, MTs, and MT motor protein subsystems are united into a single mechanical system. See also Figure SI4a. (e) Alternative methods for outbalancing the kinesin activity via +Dynapyrazole A+Blebbistatin and +Kinesore+Blebbistatin induce MT rings (arrows) and cell discoids. F-actin is labeled with Phalloidin-ATTO 647N. Chromatin is labeled with Hoechst. Pairwise one-way t test-derived p values are shown on the plots with corresponding n (size of individual cells measurement sets) for each condition, generated in triplicates.
In addition, we show that MT ring formation and discoid cell deformation can be induced across multiple cell types, e.g., in the MDA-MB-231 cells and in primary mesenchymal cells such as mouse embryonic fibroblasts (MEFs) (Figures SI7 and SI8). All tested cell types feature significantly larger scale and structural complexity compared to previously reported mammalian platelets (Figure 2d). Whether this phenomenon is universal across a wider range of large mammalian cells of unreduced complexity remains a subject of further investigation; however, favoring universality is the ubiquitous nature of the phenomena’s constituent components (MTs, MT motors, actin, myosin) and the relevant key dynamic properties that are maintained across a great number of mammalian cell types.
Control of Cell–ECM Adhesion and Spreading by Dynein–Kinesin Balance
We next proceed to examine the effect of the superposition of dynein and kinesin motor activities on cell adhesion and spreading on collagen ECMs. To do so, we modulate the dynein–kinesin balance via two alternative strategies: (a) kinesin-1 overactivation with Kinesore40 and (b) kinesin-wise shift via Dynapyrazole A-induced dynein suppression. We decided to focus on kinesin-1 as one of the most conventional and abundant members of the kinesin family among protozoa, fungi, and animals.73 Notably, Kinesore-driven kinesin-1 overactivation (50 μM, 1 h) enhances polygonal cell spreading along orthogonal collagen-1 lines compared to the controls (Figure 3c, +DMSO vs +Kinesore). Alternatively, a kinesin-wise balance shift via Dynapyrazole A-induced dynein inhibition leads to dendritization and poor polygonal spreading (Figure 1a and Figure 3c, +DMSO vs +Dynapyrazole A). Dendritization is analogous to the low cell contractility spreading mode seen during Blebbistatin treatment (Figures 1a and SI1, +Blebbistatin). We attribute these differences to the dynein activity status—i.e., the MT network contraction–expansion balance is controlled by the dynein–kinesin mechanical balance (Figures 3d and SI9a). However, dyneins facilitate mechanical cross-linking and mechanotransduction of the forces within the MT network (Figure SI9a) and the actomyosin cytoskeleton33,35,70,74−77 (Figure 3d). Thus, suppression of dyneins with Dynapyrazole A reduces both the MT network contractility (Figure SI9b) and the MT–actomyosin mechanotransduction leading to the partial loss of the overall cell–ECM contractility and impaired cell spreading. Therefore, we propose that a mechanical synergy between actomyosin and MT–dynein contractile machineries in the adhesion sites (Figures 3d and SI9a) facilitates the mechanosensory-stimulated cell adhesion and spreading11 (Figure SI9a-1). In this case, Kinesore-induced cell overspreading (Figure 3c, +Kinesore) is a result of the Kinesore-enhanced kinesin, i.e., kinesin-1 activity that expands the MT network yet preserves background contractility of the actomyosin–dynein–MT system, which sustains cell adhesion and spreading via mechanosensory stimulation, effectively resulting in maintaining of cell adhesion and enhanced cell spreading.
Nonspreading cell behavior, induced by the combination of kinesin overactivity and low actomyosin contractility, results in the formation of MT rings and discoid cells (∼35% of total cell population, n = 80) regardless of dynein’s MT-processing activity status (Figure 3e, +Kinesore+Blebbistatin, +Dynapyrazole A+Blebbistatin; Figure SI6e-f). These results indicate that MT rings are a structurally “self-locked” system that requires only MT-to-MT filament bundling and mechanical sliding, e.g., by kinesins65−68 (Figures 2 and 3a,b). That is, it does not require a dynein-mediated mechanical or structural integration with actomyosin for MT ring formation, as it does during cell adhesion-spreading on ECM. Furthermore, the dispensable role of dynein’s MT-processing activity during the kinesin-driven MT ring formation, both in +Kinesore+Blebbistatin and in +Dynapyrazole A+Blebbistatin conditions, indicates that MT rings are the extreme of kinesin outbalance within the existing range of dynein–kinesin mechanical balances, as both treatments induce the dynein–kinesin MT-sliding balance shift toward the kinesin activity via two principally different mechanisms.
Notably, the low-contractility dendritic cell spreading along the collagen type-1 grids (Figure 1a, +Blebbistatin; Figures SI1 and SI5a) reported for MDA-MB-23132 and MDA-MB-46811 cells and for MT rings are mutually exclusive (Figure SI5a), since MT ring formation is linked to the kinesin-wise motor balance shift that also drives an ejection of the microtubules from the dendritic protrusions (Figure SI5b).
Passive MT Cross-Linking by Dyneins Facilitates Microtubular Reorganization and Ring Formation
Dynapyrazole A targets MT-dependent ATPase activity of dyneins and specifically suppresses the dynein motor MT-processing function, while the dynein–MT binding affinity remains intact.48 One implication that follows from Dynapyrazole A-induced block of the dynein’s MT processivity is an increased lifetime of dynein–MT interactions, while the energy of dynein–MT interactions is likely to remain unchanged, since the ATP–dynein docking–hydrolysis cycle drives dynein-to-MT binding–unbinding and processivity. Nonprocessive but MT-binding dyneins could act as passive MT cross-linkers and principally antagonize the kinesin-driven MT–MT sliding. Thus, it cannot be ruled out that dynein’s MT–MT cross-linking and braking activity may have a significant role in the mechanical expansion of MT network and ring formation during +Dynapyrazole or +Dynapyrazole A+Blebbistatin treatments. However, during +Dynapyrazole A+Blebbistatin-induced MT ring formation, MT rings expand into the cell cortex and display circumference elongation and force generation with a magnitude large enough to deform cell cortex into a discoid shape (Figure 3a,b). The strong dynein-mediated passive MT–MT cross-linking into circular bundles should principally antagonize expansion of the ring. However, we routinely observe that expanding MT rings results in a discoid deformation of cells (discoid aspect ratio ∼1:1.4, Figure 3b) during +Dynapyrazole A+Blebbistatin treatments within 1-h-long experiments (Figure 1a). Thus, we favor the conclusion that passive dynein-mediated cross-linking of MT filaments is unlikely to serve or exert great influence as a decisive factor for MT ring formation, i.e., ring coalescence via lateral condensation (or bundling) of microtubules.
In addition to direct dynein-mediated MT–MT linkage,64 an alternative and more prominent mechanism of F-actin-mediated MT–MT cross-linking occurs via the dynein–F-actin molecular adaptor, dynactin, a universally constitutive multisubunit complex expressed in all eukaryotes.55,72,78,79 Dynein–dynactin complexes mechanically and structurally interlink MT and F-actin networks en masse, transmitting dynein-generated forces between individual MT filaments via F-actin networks.33 Thus, blocking the processivity of dyneins, if combined with retention of dynein’s passive cross-linking to the MTs, should structurally trap the MT network within the F-actin cortex, effectively preventing the spatial rearrangement of MT network into MT rings.
Since neither of the two complementary scenarios discussed above were observed—i.e., ring over-cross-linking by dyneins and dynein–dynactin-mediated MT retention in the actin cortex—we tested an alternative scenario of MT ring formation by Kinesore-induced kinesin-1 overactivation (+Kinesore+Blebbistatin). Treatment with +Kinesore+Blebbistatin induces the formation of expanding MT rings, indicating that the kinesin-mediated MT bundling activity and kinesin-driven MT–MT sliding65 could be the principal structural and mechanical factors responsible for MT ring formation and its expansion, respectively (Figure 3e). Moreover, kinesin activity is the only motor activity preserved across +Dynapyrazole A+Blebbistatin and +Kinesore+Blebbistatin treatments, indicating that kinesin-1 is the motor which actively drives expansion of MT rings into the cell cortex causing discoid cell deformation.
Interestingly, some synthetic biology systems display similar cytoskeletal filaments self-organized into the ringed structures.80,81 We find these reports on synthetic system behavior to be complementary to our findings, as in both the reported synthetic cells (giant unilamellar vesicles, i.e. - GUV) and in our cell model the filaments bundling into rings require an increase of the effective cross-linking of the constituent filaments. However, the reported F-actin self-organization into the rings within GUVs does not feature a balance of antagonistic motor activities and only requires an increase of the direct cross-linking between F-actin filaments. On the contrary, live cells form the MT rings and bundles in the context of the mechanical antagonism between several competing motor systems that results in the MTs bundling–unbundling dynamic balance.
Indeed, Dynapyrazole A inhibits only dynein’s MT-processing function. The effective dynein-mediated MT–MT cross-linking in Dynapyrazole A should remain largely unchanged. On the other hand, the Dynapyrazole A-mediated blockade of dynein processivity is experimentally sufficient to induce MT circular bundling (Figures 1a and SI1, +Dynapyrazole A). We propose that an increased effective MT bundling into the rings, that is observed during +Dynapyrazole A+Blebbistatin and +Kinesore+Blebbistatin treatments, mostly arises from the decrease of the active MT unbundling driven by antagonistic activity of dynein and kinesin motors. Therefore, and importantly, circular MT bundling occurs upon the balance shift in either direction (i.e., toward either dynein- or kinesin-dependent influence) and not solely from a direct increase of the MT–MT bundling by one of the cross-linkers. We have additionally verified that direct bundling of MTs, either by acute pharmacological MT stabilization with Paclitaxel (10 μM, 1 h) or more physiological overexpression of the microtubule-binding domain of ensconsin (EMTB-3xGFP, 48 h), is not sufficient to result in the formation of MT rings (Figure SI10).
Our results indicate that cells form MT bundles and rings in response to both +Dynapyrazole A ± Blebbistatin or +Kinesore ± Blebbistatin treatments. Thus, a selective suppression (+Dynapyrazole A ± Blebbistatin) or overactivation (+Kinesore ± Blebbistatin) of one microtubule motor subsystem increases the effective MT bundling. During this activity shift, MT bundling can outcompete MT unbundling normally supported by simultaneous dynein64 and kinesin (e.g., -kinesin-1)41,65 MT direct cross-linking and conflicting sliding activities coexisting within an entangled network of MTs (Figure 2b). Thus, intriguingly, our results demonstrate that the increase of the effective degree of microtubule cross-linking can be achieved not only by the direct increase of cross-linking molecules, e.g., by overexpression, but also via dynamic suppression of the mechanical antagonism between the conflicting MT motor subsystems that can occur upon dynein–kinesin balance shifting in either direction.
In order to directly evaluate the contribution of the dynein-based MT–MT passive cross-linking into circular bundling of microtubules, we utilized an alternative dynein inhibitor, Dynarrestin. As opposed to Dynapyrazole A, Dynarrestin suppresses dynein-to-MT binding affinity82 and prevents passive MT–MT cross-linking via nonprocessive dyneins. The result of +Dynarrestin+Blebbistatin treatment shows that suppression of the dynein-based MT bundling does not, in principle, prevent the MT ring formation (Figure SI11), as cells display a similar reorganization of MT network into the rings, although with a much lower efficiency (∼8% of total cell population; n = 100) compared to the more prominent effects of +Dynapyrazole A+Blebbistatin treatment (∼65% of total cell population, n = 100). Intriguingly, in addition to a lower frequency, the +Dynarrestin+Blebbistatin-induced MT rings on average demonstrate a poorer lateral condensation of individual MT bundles into rings in comparison to MT rings resulting from +Dynapyrazole A+Blebbistatin treatments. This result confirms a lower MT bundling upon Dynarrestin treatment and indicates a potential role for the dynein-based passive MT cross-linking activity in condensation of MT bundles into rings.
The direct comparison of +Dynarrestin+Blebbistatin and +Dynapyrazole A+Blebbistatin cotreatments highlights the importance of the dynein–MT binding affinity and passive MT–MT cross-linking for the MT bundling and ring formation. Key studies demonstrate that a motor balance shift toward either kinesin60 or dynein57,58,83,84 that is strong enough to render the opposing motor’s contribution as negligible leads to the microtubular reorganization into the radial star-like arrays, but not into the rings. Indeed, unidirectional sliding of MT motors toward either minus- or plus-ends along multiple microtubules leads to the collective microtubule motor-driven spatial convergence of the relevant MT ends at the single focus. Each motor (complex of motors) slides coupled microtubules along each other until their respective ends meet, inducing their clustering and formation of radial star-like MT arrays. A passive cross-linking would be important for preventing the collapse of microtubule networks into the radial single focus structures. Indeed, during +Dynapyrazole A+Blebbistatin and +Kinesore+Blebbistatin treatments, MT-binding activity of dynein is not directly affected, and cells in both treatments display a robust MT reorganization into the expanding rings. On the contrary, +Dynarrestin+Blebbistatin treated cells display a reduced efficiency of the MT reorganization. The majority of rings display a reduced lateral condensation. Because +Dynarrestin+Blebbistatin treatment drives the formation of the MT rings in ∼8% of cell population, we conclude that, along with dyneins, MTs in live cells are cross-linked by a multitude of other proteins, e.g., MAPs85 and Tau86,87 (and many others), that likely contribute, at least in part, to the MT reorganization into the rings. We concede that this interpretation would implicate the phenotype of circular MT reorganization as, potentially, cell-type specific, separating the phenomenology of MT ring formation in our system apart from the structurally reduced synthetic systems.
We conclude that the balance shift between antagonistic MT motor subsystems is the leading factor for the circular MT condensation and expansion of MT rings. We propose that a substantial motor disbalance, toward either of the motor subsystems, diminishes antagonistic motor-driven MT unbundling, which can increase effective MT–MT bundling during an acute experiment without increase in the expression of MT cross-linkers. We also propose that passive MT–MT cross-linking and braking of MT sliding are critical for preventing structural MT network collapse, e.g., into the radial star-like arrays.
Septins Blockade Stabilizes Microtubules and Activates Kinesin-1-Driven MT Ring Formation
Septins are nonpolar filament-forming GTPase proteins that serve as important cytoskeletal components regulating dynamics, structure, and mechanics of the plasma membrane, F-actin, and microtubules.88 Key studies indicate that in mammalian cells septin-7 and septin-9 regulate microtubule stability and consequently can control MT architecture via MT modification-mediated modulation of the MT motor activities. Namely, active septin-7 sustains HDAC6-driven MT deacetylation,89 while also preventing MAP4 from interaction with microtubules.90 Thus, a blockade of septin-7 leads to MT stabilization via redundantly duplicated pathways, i.e., through MAP4-MT adsorption and an increase of MT acetylation (Figure 4a). Direct interaction between septin-9 and microtubules stabilizes91 and cross-links92 microtubular networks. Notably, although septin-9 stabilizes microtubules, it also protects microtubular dynamics from a complete “freeze” by overstabilization with taxanes.93
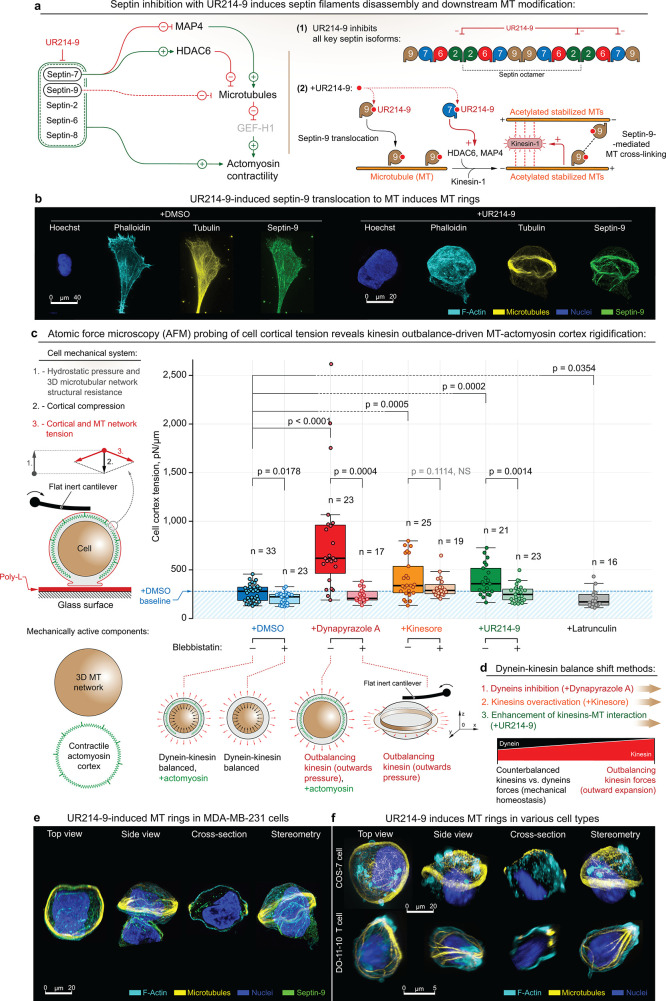
Figure 4.
Formation of MT ring via alternative dynein–kinesin balance shift mechanisms features cell rigidification as an outcome of MT network expansion. (a) Left – UR214-9-targeted septin blockade leads to the downstream stabilization of microtubules; i.e., septin-7 blockade leads to HDAC6 inactivation and the following microtubule acetylation and stabilization, coherent with the parallel effects of MAP4 release from septin-7 to the microtubular pool. In addition, total septin suppression leads to the loss of actomyosin contractile structures (e.g., stress-fibers) and to a total decrease of actomyosin contractility. (Right) Septin-9 blockade with UR214-9 leads to its relocation to the stabilized microtubules, where it increases microtubular cross-linking via direct septin-9 interactions and possibly via F-actin mediation. Stabilized acetylated MT activates kinesin-1, while combined with MT, cross-linking leads to the MT bundling and bundle extension. (b) Immunofluorescent visualization of stress-fiber-specific localization for septin-9 in MDA-MB-231 cells in control conditions (+DMSO) and septin-9 translocation onto the MT network, accompanied by MT ring formation during septin-2 inactivation (+UR214-9). See also Figure SI14a. For cells attached directly to the uncoated glass (for the purpose of super-resolution imaging), rings were formed in ∼10–15% of total cell population, n = 100 (estimates are made by visual examination). We attribute the quantitative differences with collagen gels to the nature of the substrates used in various experiments. (c) AFM analysis of the MDA-MB-231 cell rigidification upon kinesin-wise activity outbalance, induced by three alternative mechanisms. Left – Schematic of the cell mechanical system: both MT network and actomyosin represent mechanically active cell components that synergistically regulate cell mechanics. Right – Dynapyrazole A, Kinesore, and UR214-9 induce cell rigidification. Bottom – schematic representation of cell rigidification upon kinesin’s outbalancing activity: while +Dynapyrazole A+Blebbistatin, +Kinesore+Blebbistatin, and +UR214-9+Blebbistatin all induce MT rings, due to the rings’ anisotropic configuration their mechanical effects are nonreadable by AFM. (d) Schematic representation of the alternative mechanisms for inducing the kinesin’s outbalancing activity. Both dynein inhibition (+Dynapyrazole A), kinesin overactivation (+Kinesore), and kinesin–MT interaction enhancement via septin-9 F-actin-to-MT translocation (+UR214-9) unanimously induce cell rigidification via kinesin-driven MT network expansion. See also Figure SI9b. (e,f) UR214-9-induced kinesin overactivation results in MT ring formation in MDA-MB-231 cells (e), as well as in other tested cell lines, e.g., COS-7 (f, top, rings were formed in ∼10–20% of total cell population, n = 100, estimates are made by visual examination), and in DO-11-10 murine T cell line (f, bottom, rings were formed in ∼8–15% of total cell population, n = 100, estimates are made by visual examination). F-actin is labeled with Phalloidin-ATTO 647N. Chromatin is labeled with Hoechst. Pairwise one-way t test-derived p values are shown on the plots with corresponding n (size of individual cells measurement sets) for each condition, generated in triplicates.
For effective and abrupt septin blockade, we chose UR214-9 (Figures 4a, b and SI12a). It is an efficient low-molecular-weight cell-permeable non-cytotoxic mammalian septin inhibitor.94,95 UR214-9 was recently derived from the yeast septin inhibitor forchlorfenuron (FCF),96 with the latter being unsuitable for the complete and acute inhibition of mammalian septins due to its high cytotoxicity in high concentrations.
We demonstrate that UR214-9 can effectively inhibit all key septin isoforms, as we show by calculating UR214-9 key septin inhibition efficiency via in silico modeling of UR214-9 docking to septin-2, septin-7, and septin-9 and comparing its corresponding docking energies to the septin interaction energies with GTP and FCF (Figure SI12b and c). The computational in silico modeling data clearly indicates that UR214-9 is nearly equally effective at competition with GTP docking to septin-2, septin-7, and septin-9 and is clearly more effective than FCF.
UR214-9 treatment leads to disintegration of the septin filaments,97 as confirmed by the tests on MDA-MB-231 cells (Figure SI12c); i.e., UR214-9 (40 μM, 2 h) clearly induces septin-9 translocation from stress-fibers onto the MT network in MDA-MB-231 cells (Figure SI12c). Notably, in well-spread cells that were allowed to develop the polygonal architecture prior to the UR214-9 treatments, the septins (e.g., septin-9) accumulates preferentially at the peripheral free cell edge stress-fibers that feature the highest mechanical stress.52 Septin-9 translocation onto MT filaments upon addition of UR214-9 leads to MT reorganization into the buckles and rings of the subcellular size 2–5 μm rings (Figure SI12c) in ∼85–95% of total cell population, n = 150.
Adding UR214-9 to the MDA-MB-231 cells prior to their full spreading leads to the more complete MTs reorganization into the cell-sized MT rings (Figure 4b). Thus, for cells attached directly to the uncoated glass (for the purpose of super-resolution imaging), rings were formed in ∼10–15% of total cell population, n = 100 (estimates are made by visual examination), compared to ∼5–7% of total cell population, n = 75, +Dynapyrazole A ± Blebbistatin (estimates are made by visual examination), and ∼3–4% of total cell population, n = 75, +Kinesore+Blebbistatin (estimates are made by visual examination). We attribute the quantitative differences to the nature of the substrates used in various experiments. Examined rings are structurally similar to the rings induced by +Dynapyrazole A ± Blebbistatin (Figure 1a) and +Kinesore+Blebbistatin (Figure 3e) treatments.
Indeed, MT stabilization via the septin-7-HDAC6 signaling axis89 can directly contribute to the MT bundling and sliding via enhanced kinesin-1-to-MT binding affinity and enhanced kinesin-1 MT processivity.98,99 On the other hand, UR214-9-induced disintegration of the septin filaments leads to the release of monomeric septin-9, with its consequent accumulation on the microtubules, which also contributes to the stabilization of the MT, without freezing of their polymerization dynamics.93 Importantly, septin-9 deposition on microtubules additionally cross-links the MT network, contributing to their bundling. Intriguingly, we show that septin-9-enriched MT filaments are also enriched with F-actin (Figure SI12c, + UR214-9), indicating septin-9’s potential role as an MT-F-actin adaptor that can increase the MT cross-linking via numerous F-actin-related cross-linkers. However, our data on the well-spread polygonal MDA-MB-231 cells indicate that the MT bundling is not dictated by mere passive cross-linking of the stabilized MTs, as passively cross-linking MT bundles are usually formed throughout the entire cell volume, condensing at the cell’s spatial boundaries (cortex), while UR214-9 induced small, actively formed subcellular MT rings that condense upon MT circular buckling, routinely observed during kinesin-1 overactivation.40
UR214-9 treatment is able to induce MT network bundling-expansion into the MT rings via the septin-9 and septin-7-kinesin-1 signaling axis (Figure SI12b), and UR214-9 treated cells do not show signs of dynein suppression, as MT networks maintain their radial array architectures (Figure SI12c). Thus, it serves as an orthogonal strategy to confirm that the outbalance of kinesins over dyneins is a sufficient requirement for the MT network bundling and expansion. UR214-9-induced MT rings show that kinesin mechanical outbalance can induce rings regardless of the particular, and often redundant, molecular pathway affected (Figures SI13b and SI14b). The advantage of this approach is that UR214-9 allows alternative stimulation of kinesin-1 via MT modifications, e.g., regardless of the preexisting background cellular activity of kinesins and dyneins (Figures SI9b and SI13b,c).
Cell Mechanics Is Controlled by Dynein-Kinesin Balance
We summarize the previously described mechanistic findings into a cell mechanical model that predicts kinesin-driven cell rigidification upon kinesin-wise dynein–kinesin balance shift, followed by the MT network expansion (Figure 1d and Movies 1 and 2). That is, the MT network mechanically expands into the cell cortex, providing the cortex with the additional, outward-directed force-generating MT scaffold (Figures 2, 3a, and SI5b). This model-based prediction is strongly supported by cell discoid deformation with the expanding MT ring into the relaxed actomyosin cell cortex in Blebbistatin (Figures 3a,b and SI6a,b). We test the predicted kinesin-wise outbalance-induced cell rigidification with atomic force microscopy (AFM) live cell probing (Figure 4c). Indeed, we identify a significant cell rigidification (described by increase in surface cortical tension) across all three methods of the kinesin-wise MT motors balance shift; i.e., +Dynapyrazole A, +Kinesore, and +UR214-9 treatments unanimously induce the MDA-MB-231 cell rigidification (Figures 4c and SI9b). Their combinations with Blebbistatin reduce AFM rigidity readout to a lower level, due to actomyosin cortex relaxation (a) and MT network collapse into the spatially anisotropic ring (b), which is largely unavailable for AFM cantilever probing (Figure 4c). Thus, indeed all alternative and orthogonal methods of kinesin-wise MT motors balance shift induce MT scaffold structural expansion, mechanistically and structurally proving the existing dynein–kinesin mechanical balance that orchestrates MT network architecture and cell mechanics (Figures 4d and SI9b). Perhaps surprisingly, UR214-9-induced septin inhibition and its clear translocation from the contractile actomyosin (e.g., stress-fibers, Figure SI12c) still leads to the cell rigidification via mechanically active predominantly kinesin-1-driven MT network expansion, despite the loss of the septin-dependent actomyosin network integrity and contractility due to the loss of septins’ F-actin-stabilizing activity100 from UR214-9.
The universal character of changes in cell structural organization and its resulting mechanics, induced by three alternative mechanisms of dynein–kinesin activity balance shift (Figures 4c and SI9b) also extends into the universality of the described MT network structural reorganization in various cell types. That is, UR214-9-induced MT bundling-expansion into MT rings not only is observed in the principal MDA-MB-231 cell model (Figure 4e, ∼15%), but it appears to be universal for otherwise dissimilar cell types, such as COS-7 (∼20% of cell population, 2 h of UR214-9 incubation period) and DO-11-10 murine T cells (∼15% of cell population, 2 h of UR214-9 incubation period) (Figure 4f), highlighting the basic and universal importance of the septin–MT–motor protein signaling axis. Thus, the MT–motor proteins’ rebalancing effects are fundamental to cell mechanobiology regulation principles.
Dynein–Kinesin Balance Controls 2D Contact and “2.5D” Steric-Cell Guidance
Dynein motors regulate structural coherence between MT and actomyosin networks.33,36,101 On the other hand, the MT network mechanically (sterically) scaffolds cellular contact guidance on nanotextures.32 Therefore, we hypothesize that tunable coupling of MTs and actomyosin within the cell adhesion sites by balancing dynein–kinesin motor activity may play a significant role in cellular contact guidance at the adhesion sites (Figure 2a). According to this model, the sole kinesin-driven MT network “self-locking” into the MT ring and MT retraction from actomyosin cell adhesion interface can be in direct competition with the dyneins that facilitate mechanostructural integration of MT filaments into the cell actomyosin adhesion system33 and away from the giant MT ring structures (Figure 2a). Notably, this model is complementary to the previously reported indispensable scaffolding role of MT networks for cell adhesion and spreading20 by supporting low-contractility dendritic cell protrusions.11,27,28
We propose that dynein activity facilitates the integration of MT filaments as the contact guidance scaffold into the cell adhesion and into spreading actomyosin protrusions (Figure 5a, +DMSO). This activity of dyneins competes with antagonistic kinesin activity (Figure 5b-1), rendering dyneins as the key MT motor subsystem that controls cellular contact guidance via mechanical coupling of MTs and actomyosin and by pulling MT filaments into the nanogrooves as the cell spreading–elongation–directing scaffold. Indeed, Blebbistatin-induced suppression of actomyosin contractility does not affect cell contact guidance elongation along the nanotexture, as the MT in-groove alignment remains intact (Figure 5a,c, +DMSO vs +Blebbistatin). On the contrary, Dynapyrazole A-induced dynein inhibition completely abrogates insertion and elongation of MTs into and along nanogrooves on collagen-1 nanotextures, which is accompanied by the complete loss of cell alignment to the contact guidance cues of nanotextures (Figure 5a and c, +DMSO vs +Dynapyrazole A). Additionally, the kinesin activity outbalance induced by UR214-9 leads to identical results, such as the loss of contact guidance and formation of planar MT rings that are unaligned with underlying nanotextures (Figure 5a and c, +DMSO vs +UR214-9). Instead, MT network reorganizes into the “self-locked” bundled configurations with uncompensated kinesin activity that also retracts and expels the MTs from the actomyosin cytoskeleton in cell adhesion sites (Figure 5b-2). In summary, the dynein–kinesin balance controls cellular contact guidance in a sterically interactive microenvironment by recruitment of MTs to the sites of cell adhesion.
Figure 5.
Dynein motor-driven in-groove MT guidance is crucial for the contact guidance-induced alignment of the cell to the “2.5D” collagen nanotexture. (a) MDA-MB-231 cells (top) and their MT network (bottom) alignment to the sterically interactive collagen type-1-coated “2.5D” nanotextures (800/800 nm nanogrooves/nanoridges, depth: 600 nm). Left to right: (+DMSO), control conditions; (+Blebbistatin), during actomyosin low contractility state; (+Dynapyrazole), during dynein activity inhibition; and (+UR214-9), during septin-9 actin-to-MT translocation and kinesin overactivation. Note that both direct dynein inactivation (+Dynapyrazole A) and kinesin outbalance over dynein activity (+UR214-9) suppress MT network nanogroove alignment, induce planar MT rings, and consequently suppress cell contact guidance along the nanotextures. Note the MT-to-nanogroove alignment in active, balanced dynein cell states, +DMSO and +Blebbistatin (arrows), but the MT network reorganizes into rings with no alignment to the nanotexture during kinesin activity outbalance over dyneins, +Dynapyrazole A and +UR214-9. (b) Schematic views of dynein+MT-driven cell contact guidance along the nanotextures (1) and kinesin outbalance-driven loss of cell contact guidance (2) in either +Dynapyrazole A or +UR214-9. (c) Quantification of cell contact guidance as cell spreading spans measurement along vs across nanotextures shows no preferential response of cell spreading alignment to the nanotexture direction in both +Dynapyrazole A and +UR214-9. F-actin is labeled with Phalloidin-ATTO 647N. Chromatin is labeled with Hoechst. Pairwise one-way t test-derived p values are shown on the plots with corresponding n (size of individual cells measurement sets) for each condition, generated in triplicates.
Thus, mechanically and structurally antagonistic effects of dyneins and kinesins onto the MT network as the cell scaffold are emerging as an additional and important cell mechanobiological system that requires deeper insight. The inherent integration of the MT–actomyosin systems together via their motor proteins’ synergy appears as another dimension of the fundamental principles that regulate cell–microenvironment mechanical and structural integration and should find both theoretical and practical applications in cell biology, cell and tissue bioengineering, and medicine.
Conclusions
Pharmacologically induced MT rings serve as a striking visual indicator of a kinesin-wise motor disbalance within microtubule networks. We think that the specific phenomenon of MT ring formation in mesenchymal cells likely has only limited biological relevance. In our hands, the MT rings are simply an indicator of the most extreme form of a cell response to acute MT motor perturbation. However, the dynein–kinesin activity balance shift, that we propose within this manuscript to underpin the appearance of this extreme phenotype, may represent a key factor of large-scale cell mechanobiology that controls multiple key aspects of cell behavior, including adhesion, contact guidance, migration, and microenvironment mechanosensing.
Dynein actively cross-links the microtubules to actomyosin33 (e.g., via the dynactin complex55) and generates its own contractile forces.70,74 The loss of dynein activity may directly contribute to the partial loss of total cell contractility.38,74,102 Moreover, since MTs act as flexible but nonstretchable cables, their mechanical linkage to the structurally dynamic actomyosin cytoskeleton may directly contribute to long-distance mechanotransduction across the cell and tissues13,74,103 and to the stabilization of cell 3D architecture by compensating for structural (viscous) and mechanical energy dissipation within the dynamic actomyosin network.
Indeed, our data indicate that Dynapyrazole A-induced dynein suppression leads to the dendritization of cells, which is similar to the effect of Blebbistatin-induced decrease of actomyosin contractility (Figure 3c)28,104 and indicates a partial loss of cell contractility. Alternatively, kinesin-wise balance shift via Kinesore-induced kinesin-1 overactivation results in a greater cell spreading (Figure 3c), pointing out that the dynein–kinesin balance directly regulates the degree of MT network expansion, and with it the cell–microenvironment interaction and cell migratory properties.
Furthermore, dynein–kinesin balance regulates the topographic contact guidance of cells. Cell alignment to topographically anisotropic surfaces is mechanically and structurally enforced by the MT scaffold104 that provides steric interactions between cell microenvironment spatial guidance cues and the cytoskeleton. Dynein actively pulls the MT toward the adhesion interface. On the contrary, kinesin-wise motor balance shift results in MT retraction from adhesion sites which is accompanied by the loss of cell contact guidance (Figure 5).
The MT acetylation and MAP4 decoration are innate cell mechanisms for modulating the activities of kinesin and dynein motors, as well as microtubular reorganization.98,105 Septin-991 and Septin-789,90 are reported as the key regulators of microtubule post-translational modifications that can control the dynein–kinesin activity balance via septins’ downstream HDAC6 and MAP4 signaling pathways, linking dynein–kinesin balance to the state of the microtubular apparatus. Although we did not detect a strong and direct implication of microtubular post-translational modifications into acute pharmacologically induced dynein–kinesin balance shift or in the subsequent formation of MT rings (Figures SI7 and SI8), the role for changes in post-translational MT modifications could become more evident during subtle and prolonged dynein–kinesin activity balance shifts. However, it remains a subject of further investigation.
Materials and Methods
Cell Experiments
Human MDA-MB-231 cells (ATCC HTB-26), mouse embryonic fibroblasts (a gift from Dr. Robert S. Adelstein, NIH), and monkey COS-7 (ATCC CRL-1651) fibroblast cells were maintained in DMEM with 4.5 g/L d-glucose, l-glutamine, 110 mg/L sodium pyruvate (Corning Cellgro, Cat#10013CV), and 10% heat-inactivated FBS (HyClone, Cat#SH30071.03H) at 37 °C in 5% CO2. Mouse Do-11-10 thyoma cells (Sigma, Cat#85082301) were maintained in ImmunoCult-XF T Cell Expansion Medium (STEMCELL Technologies Inc., Cat#10981) with the addition of Human Recombinant Interleukin 2 (STEMCELL Technologies Inc., Cat#78036.3) at 37 °C in 5% CO2. We treated cells in glass-bottom 35 mm Petri dishes (MatTek Corp., Cat#P35G-1.5-14-C) using (−)-Blebbistatin enantiomer (Sigma, Cat#203391), Dynapyrazole A (Sigma, Cat#SML2127), Kinesore (Sigma, Cat#SML2361), Dynarrestin (Tocris, Cat#6526), Paclitaxel (Sigma, Cat#T7402), septin inhibitor UR214-9 (synthesized by Dr. Rakesh K. Singh), or dimethyl sulfoxide (Sigma, Cat#472301), as indicated in the main text.
Cells were seeded at high density onto micropatterned coverslips or unpatterned glass-bottom dishes (for super-resolution microscopy) in DMEM or Immunocult. After 30–60 min of spreading, media was replaced with DMEM or Immunocult medium containing indicated concentration of drugs. Cells were fixed with 4% ice-cold PFA solution after 1 h and used for immunofluorescence analysis.
MEF and COS-7 transfection with microtubule binding domain of ensconsin EMTB-3XGFP (Addgene Cat#26741) was performed in Opti-MEM (Gibco, Cat#31985-070) using SuperFect reagent (Qiagen, Cat#301307) according to the manufacturer’s protocol.
Synthesis of UR214-9
Equimolar mixture of 4-amino-2,6-dichloropyridine (Tokyo Chemical Industries, Cat#A2369) and 2-fluoro-3-(trifluoromethyl) phenyl isocyanate (Aldrich, Cat#472182-2G) in dimethylformamide (DMF) was stirred and heated at 65 °C in a sealed glass tube overnight. DMF was removed using a Buchi rotary evaporator. The crude reaction mixture was dissolved in a mixture of dichloromethane (DCM) + MeOH and purified by preparative thin-layer chromatography using Hexane:EtOAc (50:50) as eluent. The pure product band was scrapped off the glass plate and UR214-9 was stripped from the silica gel using DCM + MeOH through a sintered funnel. The solvent was evaporated using a Buchi rotary evaporator to obtain UR214-9 as an off-white powder. UR214-9 was dried overnight in a desiccator and analyzed by proton and carbon NMR and mass spectrometry. MS-APCI: [368.3, singlet, 100%; 370.2, doublet, 60%].
High-Precision Micropatterning
A detailed step-by-step protocol for polyacrylamide gel micron-scale micropatterning with collagen type-1 is described elsewhere.106 Briefly, microcontact soft lithography printing at the micron-scale precision is complicated by the van der Waals effects that provoke a collapse of the conventionally soft regular PDMS (rPDMS) microstamps onto the printed glass surface. In order to prevent the undesirable collapse, we substituted rPDMS microstamps with their composite analogues—i.e., soft, mechanically cushioning rPDMS blocks, veneered with a 0.5–0.8-mm-thick noncollapsing hard PDMS (hPDMS).104,107 For the detailed hPDMS preparation protocol, please see the hPDMS Formulation section. To cast the microprinting surfaces from the commercially fabricated and passivated molding matrix of desirable design (Minnesota Nano Center, University of Minnesota), we coated molds with a 0.5–0.8-mm-thick layer of hPDMS. The hPDMS layer was then cured at 70 °C for 30 min. Next, an ∼8-mm-thick layer of rPDMS is poured atop the cured hPDMS layer (rPDMS; 1:5 curing agent/base ratio, Sylgard-184, Dow Corning), and cured in the oven (at 70 °C for ∼1 h). The resulting composite microstamps were then carefully peeled off the molds and cut into 1 × 1 cm2 pieces as ready-to-use microstamps.
In order to fabricate the collagen micropatterns, we first microprinted α-collagen-1 rabbit pAb (AbCam, Cambridge, UK, Cat#ab34710; RRID: AB_731684), conjugated with biotin, ((+)-biotin N-hydroxysuccinimide ester, Sigma-Aldrich, Cat#H1759; as per the commercial protocol) and a fluorescent tag (Alexa Fluor succinimidyl esters, Invitrogen, Molecular Probes, Cat#A20000, Cat#A20003; as per supplied protocol) on a bare, thermally treated intermediate coverglass (FisherFinest Premium Cover Glass; #1, Cat#12-548-5 P; 450 °C, baked overnight in the furnace). For that, the microstamps were coated with the 0.2 mg/mL α-collagen-1 antibody PBS solution for 40 min at 37 °C in a dark humid chamber and then gently rinsed in deionized water and dried under a jet of argon or nitrogen immediately prior to the microcontact soft lithography procedure. Second, glass-bottom 35 mm cell culture dishes (MatTek Corp., Ashland, MA, Cat#P35G-1.0-20-C) were activated with 3-(trimethoxysilyl) propyl methacrylate (Sigma-Aldrich, Cat#6514, as per commercial protocol) to ensure the covalent cross-linking between the glass surface and PAA gels. Third, 5 μL of PAA premix (see the PAA Elastic Gel Premix section) of the projected rigidity G′ = 50 kPa50 with the addition of the 5% streptavidin–acrylamide (ThermoFisher, Cat#S21379) were, avoiding any air bubble formation, carefully “sandwiched” between the activated glass surface and the micropatterned intermediate glass surface promptly after adding a curing catalyst (aminopropyltriethoxysilane (APS)). The resulting “sandwiches” with cured PAA were then incubated in deionized water (1 h, 20 °C) for hypotonic PAA swelling that ensures gentle coverglass release from the PAA gel. The resulting dish-bound PAA gels were checked for quality of the transferred fluorescent α-collagen-1 Ab micropatterns on an epifluorescent microscope. Lastly, the α-collagen-1 Ab micropatterns on PAA gels were incubated with 1 mg/mL rat monomeric collagen-1 (Corning, NY, Cat#354249) in cold PBS (4 °C, 12 h) and then rinsed three times with cold PBS and utilized for experiments.
For making PAA collagen-functionalized nanogrooves, we used antibody-coated 2.5D stamps directly.106 Biotinylated antibodies on intermediate glass surfaces or stamps were cross-linked to polymerizing PAA gels. For that, we polymerized 7–10 μL of gel mix in the “sandwich” fashion between a patterned surface and glass-bottom 35 mm Petri dishes (MatTek Corp., Ashland, MA), that we activated with 3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich) in ethyl alcohol (Pharmco-Aaper) and acetic acid (Fisher Chemical) as per the commercial protocol. For preparation of G′ = 50 kPa gels, we mixed degassed 40% acrylamide, 15 μL; and 2% bis-acrylamide, 14.4 μL; with 2 mg/mL streptavidin–acrylamide, 3.33 μL; 10X PBS, 5 μL; deionized milli-Q water, 11.17 μL; TEMED, 0.1 μL; 10% APS, 1 μL.
hPDMS Formulation
Briefly, the hPDMS mixture was prepared as follows: 3.4 g of VDT-731 (Gelest, Inc., Cat#VDT-731), 18 μL of Pt catalyst (Platinum(0)-2,4,6,8-tetramethyl-2,4,6,8-tetravinylcyclotetrasiloxane complex solution) (Sigma-Aldrich, Cat#479543), and one drop of cross-linking modulator 2,4,6,8-tetramethyl-2,4,6,8-tetravinylcyclotetrasiloxane (Sigma-Aldrich, Cat#396281) were thoroughly mixed in a 50 mL conical bottom centrifuge tube on the vortex mixer for at least 30 s. Lastly, immediately before use, we added to the mixture 1 g of HMS-301 (Gelest, Inc., Cat#HMS-301), promptly mixed it for 30 s on a vortex mixer, and immediately used it for the mold coating. A detailed hard PDMS (hPDMS) formulation and related protocols are described elsewhere.107,108
PAA Elastic Gel Premix
For PAA premixes, we utilized 40% acrylamide (40% AA) base (BioRad, Cat#161-0140) and 2% bis-AA (BioRad, Cat#161-0142) cross-linker as described elsewhere.109,110 Additionally, streptavidin–acrylamide (Thermo Fisher, Cat#S21379) was added to the final concentration of 0.133 mg/mL to enable PAA gels cross-linking with biotinylated proteins of interest for the micropatterns transfer from the intermediate glass surface to the cured PAA gels. Briefly, for preparation of 50 μL of G′ = 50 kPa PAA gel premix, the components were mixed as follows: 40% AA, 15 μL; 2% bis-AA, 14.40 μL; 2 mg/mL streptavidin–acrylamide, 3.33 μL; 10X PBS, 5 μL; deionized milli-Q water, 11.17 μL; TEMED, 0.1 μL. The premix solutions were degassed and stored at 4 °C before use. To initiate curing, 1 μL of 10% APS was added to 50 μL of the PAA premix immediately before the PAA gel casting procedure.
Super- and High-Resolution and Confocal Imaging
We fixed cells with ice-cold (4 °C) DMEM containing 4% paraformaldehyde (Sigma, Cat#P6148) for the duration of 15 min. PFA-fixed samples were then rinsed in 1% BSA in PBS two or three times (5 min for each cycle), followed by 30-min-long blocking–permeabilization treatment in 0.1% Triton X-100 (X100, Sigma) solution with 10% BSA (Fisher, Cat#BP9704) in PBS (Thermo Fisher, Cat#10010023). For immunofluorescence sample staining, we utilized primary antibodies against β-tubulin (Sigma, Cat#T7816), detyrosinated α-tubulin (Abcam, Cat#ab43389), acetylated α-tubulin (Sigma, Cat#T6793), α-tubulin (Abcam, Cat#ab18251), γ-tubulin (Abcam, Cat#ab27074), dynein (EMD Millipore, Cat#MAB1618), kinesin (EMD Millipore, Cat#MAB1613), septin-9 (Sigma, Cat#HPA042564), and septin-2 (Clonentech, Cat#60075) antibodies, diluted in 1% BSA PBS. The duration of the incubation with any of the listed primary antibody solutions was 1 h at room temperature. Similarly, labeling with Alexa-Fluor-conjugated secondary antibodies (Thermo Fisher) was performed at their final concentration of 5 μg/mL for the duration of 1 h in 1% BSA PBS at room temperature. After washing out the excess secondary antibodies, F-actin was stained with fluorescent phalloidin (Phalloidin-ATTO 647N conjugate, Millipore-Sigma, Cat#65906; 10 U/mL). Chromatin was labeled with 1:1000 Hoechst solution (Tocris, Cat#5117). We mounted samples using 90% Glycerol (Sigma, Cat#G5516) in PBS.
High-resolution 3D and 2D imaging for cell morphometric analysis was performed on a Leica TCS SP8 laser scanning confocal microscope with LIAchroic Lightning system and LAS X Lightning Expert super-resolution capacity; 405, 488, 552, and 638 nm excitation diode lasers; with 40×/1.3 oil immersion objective (Leica, Germany). The scanning settings were optimized with Nyquist LAS X function with HyD2.0-SMD excitation sensors, at the regular pinhole size of 0.85 AU and the scanning frequency at 100 Hz. Each frequency channel was scanned in sequence in order to avoid the signal bleeding between the channels.
Alternatively, a Nikon TiE stand with an A1Rsi Confocal scan head, powered by NIS-Elements Confocal software (Nikon, Japan) used for cell imaging. Objectives used were PlanApo VC 20×/0.75 NA and PlanApo VC 60×WI/1.20NA, and excitation was provided sequentially by 405, 488, and 561 nm lasers. Fluorescence was collected through a 1.2 AU pinhole using emission filters of 425–475 nm, 500–550 nm, and 570–620 nm. Pixel size was adjusted to Nyquist sampling (voxel size x, y, z for the 20× objective; j, k, l for the 60× objective).
Morphometric analysis was performed automatically and/or manually utilizing LAS X (Leica, Germany) or NIS-Elements Advanced Research software (Nikon, Japan) as an integral part of the data analysis streamline “microscopy-to-measurement-to-analysis”. Video sequences were also analyzed with the ImageJ/FIJI “stacks” plug-in. Additionally, live cell imaging microscopy experiments were performed on an Olympus X81 (Olympus, Japan) microscopy inside a temperature (37 °C), CO2 (5%), and humidity-controlled chamber at 20× magnification. Brightfield and fluorescent images were obtained every 2 min using MetaMorph software (Molecular Devices, USA). Composite 2D/3D cells + micropattern images were reconstructed and assembled using NIS-Elements AR and linear image parametric adjustments. Figures were composed using unmodified NIS-Elements AR-generated TIFF images with Adobe Illustrator CC 2017 (Adobe).
Instant structured illumination microscopy (iSIM) was performed using the custom-built instant structured illumination microscope system (VisiTech Intl, Sunderland, UK) equipped with an Olympus UPLAPO-HR 100×/1.5 NA objective, two Flash-4 scientific CMOS cameras (Hamamatsu, Corp., Tokyo, Japan), an iSIM scan head (VisiTech Intl, Sunderland, UK), and a Nano-Drive piezo Z stage (Mad City Laboratories, Madison, WI). The iSIM scan head included the VT-Ingwaz optical destriping unit. Image acquisition and system control were done using MetaMorph Premiere software (Molecular Devices, LLC, San Jose, CA). Images were deconvolved with an iSIM-specific commercial plugin from Microvolution (Cupertino, CA) in FIJI.
Live Cell Imaging of Giant Microtubule Rings
Human MDA-MB-231 cells were transferred from confluent T25 flasks (CytoOne; Cat#CC7682-4325) into a 24-well plate (CytoOne; Cat#CC7682-7524) at 40% confluency. Cells were incubated overnight with 100 nM SiR-Tubulin (Cytoskeleton, Inc.; Cat#CY-SC002) and then seeded at high density onto micropatterned coverslips (same as for immunofluorescence; see above) in DMEM containing 100 nM SiR-Tubulin. After either 30 min or 1 h of spreading, media was replaced with DMEM containing 100 nM SiR-Tubulin, 130 μM Dynapyrazole A, and 100 μM Blebbistatin. Cells were imaged using a Visitech “instant” Structure Illumination Microscopy (iSIM) microscope, and images were captured either every 10 min or every 30 min in the far-red channel using a CMOS camera.
Atomic Force Microscopy
Suspended MDA-MB-231 triple negative breast cancer cells with passage number between P8 and P13 were plated on a glass-bottom dish (Willco Wells) precoated with 0.01% poly(l-lysine) (Sigma-Aldrich) with culture media solution (Life Technologies). Depending on the pharmacological drug treatment, cells were incubated with times between ∼15 and 75 min to allow them to weakly adhere to the glass-bottom dish surface and the drug to take maximum effect. AFM force spectroscopy experiments were performed using a Bruker BioScope Catalyst AFM system (Bruker) mounted on an inverted Axiovert 200 M microscope (Zeiss) equipped with a confocal laser scanning microscope 510 Meta (Zeiss) and a 40× objective lens (0.95 NA, Plan-Apochromat, Zeiss). The combined microscope instrument was placed on an acoustic isolation table (Kinetic Systems). During AFM experiments, cells were maintained at physiologically relevant temperature 37 °C using a heated stage (Bruker). A soft silicon nitride tipless AFM probe (HQ:CSC38/tipless/Cr–Au, MikroMasch) was used for gentle compression of spherical weakly adhered 231 cells. The AFM microcantilevers were precalibrated using the standard thermal noise fluctuations method, with estimated spring constants for microcantilevers used between 0.05 and 0.11 N/m. Immediately after probe precalibration, the AFM tipless probe was moved on top of a spherical MDA-MB-231 cell. Five to ten successive force curves were performed on each cell. The deflection set-point was set between 12 and 32 nm yielding applied forces between 0.95 and 2.7 nN.
All AFM force–distance curve measurements were analyzed using a custom-written MATLAB (The MathWorks) code to calculate the cellular surface cortical tension. For curve fitting, indentation depths between 0 and 600 nm were relatively consistent in yielding good fits (R2 > 0.75). Curves with poor fits R2 < 0.75 were discarded from the analysis. Additionally, we discarded noisy force curves and/or curves that presented jumps possibly due to cantilever and plasma membrane adhesion, slippage, or very weakly adherent moving cells.
Spherical MDA-MB-231 cells’ cellular surface cortical
tension
(T; pN/μm) was calculated by fitting each recorded
force–distance curve using the cortical tension method we previously
described that defines the force balance relating the applied cantilever
force with the pressure excess inside the rounded cells and the corresponding
actin cortex tension:  , where T is the cellular
surface tension, kc is the AFM cantilever
spring constant, Z is the Z-piezo
extension, and d is the cantilever mean deflection.53
, where T is the cellular
surface tension, kc is the AFM cantilever
spring constant, Z is the Z-piezo
extension, and d is the cantilever mean deflection.53
In Silico UR214-9-Septin Docking Energy Simulation
Swiss-Model111 was used to model the structures of septins. Missing residues and atoms were automatically completed by Swiss-Model in the modeling. Docking calculations were performed using MedusaDock,112,113 which models the flexibility of both the ligand and the receptor. Docking poses were evaluated using MedusaScore,114 a scoring function based on the physics-based force field accounting for the protein–ligand interaction energy. Starting from a low number of independent docking attempts, an ensemble of candidates was isolated that have the lowest binding energies to each receptor’s binding site. The maximum number of independent docking attempts was chosen as 1000, after analyzing the convergence of docking poses energies. By properly modeling the induced-fit phenomenon upon ligand binding,113 MedusaDock is not sensitive to the starting conformations of the receptors. Thus, the docking results were not biased by the starting conformations.
Statistical Analysis
Only pairwise comparisons as one-sided t tests between a control group and all other conditions are utilized to analyze the data, as well as between paired -Blebbistatin and + Blebbistatin cotreatment groups. Statistical analysis is performed using either KaleidaGraph 4.5.4 (Synergy Software) or Prism 7b (GraphPad Software, Inc.). The exact p values are indicated on the plots, unless the p < 0.0001, i.e., below cutoff lower limit for Kaleidagraph and Prism softwares. Sample size n for each comparison is reported in the corresponding plots (i.e., n reflects the number of measured individual cells). Number of replicates is 3, unless specified otherwise. Data are shown as box and whiskers diagrams: first quartile, median, third quartile, and 95% percent confidence interval.
Acknowledgments
E.D.T. and this work were supported by the Department of Pharmacology, Penn State College of Medicine via the startup funds. A.S.Z. and A.M. were supported by the FDA Intramural Research Program of the Center for Biologics Evaluation and Research. We thank Christian Combs and Daniela Malide for the Light Microscopy Core support at the National Heart, Lung, and Blood Institute, NIH. Work in the Burnette lab was supported by a MIRA to D.T.B. (R35 GM125028), a training grant to J.B.H. (T32 GM08320) and an American Heart Association fellowship to J.B.H. (AHA 836090). A.X.C.R. and C.S. were supported by the National Institutes of Health (NIH) Intramural Research Program in the National Institute of Biomedical Imaging and Bioengineering (NIH grant # ZIA EB000094) and by the NIH Distinguished Scholars Program. N.V.D. is supported by the NIH grant R35GM134864 and the Passan Foundation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c04435.
Detailed 3D micrographs of cell samples during key treatments; detailed 3D microimaging of the microtubular rings in discoid cells; MTOC position visualization in cell samples during key treatments; time course sequence of the microtubular rings formation and their post-washout dissolution; visualization of mutual exclusivity between MT rings and dendritic protrusions; kinesin visualization in discoid cells; post-translational modification of the microtubules in MT rings; analysis of detyrosinated MT filaments inclusion into the rings in MDA-MB-231 and MEF cells; schematic representation of cell mechanobiology regulation with MT motor balance; analysis of the effects of MT stabilization on the formation of the rings; MT ring formation during Dynarrestin+Blebbistatin treatment; in silico computational simulations of UR214-9 docking and interactions energies with Septin-2, -7, and -9; effects of UR214-9 treatment on Septin-9 localization in MDA-MB-231 cells; dynein and kinesin localization in MDA-MB-231 during DPA and DPA+Blebbistatin treatments; dynein, kinesin, and septin-9 localization in MDA-MB-231 cells during UR214-9 treatment (PDF)
Movie 1: MT network expansion and ring condensation in MDA-MB-231 cells (AVI)
Movie 2: Active spatial expansion of MT network (AVI)
Movie 3: Spread polygonal architectures on the collagen type-1 grids (AVI)
Author Contributions
■ E.D.T. observed MT ring induction and conceived, formulated, and theoretically conceptualized the project; E.D.T. and A.S.Z. developed the theoretical framework and designed the experimental strategies; A.X.C.R. developed the theoretical and experimental methods for dynamic live cell micromechanical probing; E.D.T., A.S.Z., and A.X.C.R. carried out the core mechanobiology, cell biology, and microscopy studies; E.D.T. microfabricated the artificial micropatterned ECM platforms for the MT rings study; A.M., C.S., and A.X.C.R. performed the AFM analysis; R.K.S. designed, synthesized, purified, chemically tested, and verified septin inhibitor UR214-9 and outlined septin-kinesin signaling axis framework; J.W. and N.V.D. developed and applied the high precision in silico molecular modeling for examining the UR214-9 septin-blocking efficiency; E.D.T., A.S.Z., J.B.H., D.T.B., A.M., and R.K.S. performed the UR214-9 experiments and data analysis; J.B.H. and D.T.B. performed the live cell MT ring formation video-microscopy; E.D.T., A.S.Z., J.W., N.V.D., and A.X.C.R. interpreted the data; all coauthors contributed to the manuscript editing; E.D.T. wrote the manuscript; E.D.T. oversaw all aspects of this study. A.M., C.S., and J.B.H. contributed equally.
The first version of this manuscript is also uploaded to the BioRxiv preprint service that was posted on May 26, 2021: Alexander S. Zhovmer, Alexis Manning, Chynna Smith, James. B. Hayes, Dylan T. Burnette, Alexander X. Cartagena-Rivera, Rakesh K. Singh, and Erdem D. Tabdanov. Mechanical Counterbalance of Kinesin and Dynein Motors in Microtubular Network Regulates Cell Mechanics, 3D Architecture, and Mechanosensing. 2021, doi: 10.1101/2021.05.25.445700, BioRxiv, https://www.biorxiv.org/content/10.1101/2021.05.25.445700v1 (Accessed 10-10-2021).
The authors declare no competing financial interest.
Notes
Data Availability: The authors declare that the data supporting the findings of this study are available within the paper or the Supporting Information, and any data can be made further available upon reasonable requests.
Supplementary Material
References
- Xu J.; Van Keymeulen A.; Wakida N. M.; Carlton P.; Berns M. W.; Bourne H. R. Polarity Reveals Intrinsic Cell Chirality. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (22), 9296–9300. 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak M.; Polak B.; Simunić J.; Boban Z.; Kuzmić B.; Thomae A. W.; Tolić I. M.; Pavin N. The Mitotic Spindle Is Chiral Due to Torques within Microtubule Bundles. Nat. Commun. 2018, 9 (1), 3571. 10.1038/s41467-018-06005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fygenson D. K.; Marko J. F.; Libchaber A. Mechanics of Microtubule-Based Membrane Extension. Phys. Rev. Lett. 1997, 79 (22), 4497–4500. 10.1103/PhysRevLett.79.4497. [DOI] [Google Scholar]
- Winckler B.; Solomon F. A Role for Microtubule Bundles in the Morphogenesis of Chicken Erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 1991, 88 (14), 6033–6037. 10.1073/pnas.88.14.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.; Naruse K.; Stamenović D.; Fredberg J. J.; Mijailovich S. M.; Tolić-Nørrelykke I. M.; Polte T.; Mannix R.; Ingber D. E. Mechanical Behavior in Living Cells Consistent with the Tensegrity Model. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (14), 7765–7770. 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E. M.; Tong Z.; Paul C. D.; Hung W.-C.; Stroka K. M.; Boggs A. E.; Martin S. S.; Konstantopoulos K. Physical Confinement Alters Tumor Cell Adhesion and Migration Phenotypes. FASEB J. 2012, 26 (10), 4045–4056. 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet B. P.; Noordstra I.; van Amersfoort M.; Katrukha E. A.; Ammon Y.-C.; ter Hoeve N. D.; Hodgson L.; Dogterom M.; Derksen P. W. B.; Akhmanova A. Mesenchymal Cell Invasion Requires Cooperative Regulation of Persistent Microtubule Growth by SLAIN2 and CLASP1. Dev. Cell 2016, 39 (6), 708–723. 10.1016/j.devcel.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieff S.; Alsina A.; Mathur A.; Nédélec F. J. Balance of Microtubule Stiffness and Cortical Tension Determines the Size of Blood Cells with Marginal Band across Species. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (17), 4418–4423. 10.1073/pnas.1618041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagouraga B.; Grichine A.; Fertin A.; Wang J.; Khochbin S.; Sadoul K. Motor-Driven Marginal Band Coiling Promotes Cell Shape Change during Platelet Activation. J. Cell Biol. 2014, 204 (2), 177–185. 10.1083/jcb.201306085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette D. T.; Ji L.; Schaefer A. W.; Medeiros N. A.; Danuser G.; Forscher P. Myosin II Activity Facilitates Microtubule Bundling in the Neuronal Growth Cone Neck. Dev. Cell 2008, 15 (1), 163–169. 10.1016/j.devcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabdanov E. D.; Puram V. V.; Win Z.; Alamgir A.; Alford P. W.; Provenzano P. P. Bimodal Sensing of Guidance Cues in Mechanically Distinct Microenvironments. Nat. Commun. 2018, 9 (1), 4891. 10.1038/s41467-018-07290-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf A.; Renkawitz J.; Hauschild R.; Girkontaite I.; Tedford K.; Merrin J.; Thorn-Seshold O.; Trauner D.; Häcker H.; Fischer K.-D.; Kiermaier E.; Sixt M. Microtubules Control Cellular Shape and Coherence in Amoeboid Migrating Cells. J. Cell Biol. 2020, 219 (6), 1. 10.1083/jcb.201907154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J. S.; Joca H. C.; Law R. A.; Williams K. M.; Kerr J. P.; Shi G.; Khairallah R. J.; Martin S. S.; Konstantopoulos K.; Ward C. W.; Stains J. P. Microtubules Tune Mechanotransduction through NOX2 and TRPV4 to Decrease Sclerostin Abundance in Osteocytes. Sci. Signaling 2017, 10 (506), 1. 10.1126/scisignal.aan5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabdanov E. D.; Rodríguez-Merced N. J.; Cartagena-Rivera A. X.; Puram V. V.; Callaway M. K.; Ensminger E. A.; Pomeroy E. J.; Yamamoto K.; Lahr W. S.; Webber B. R.; Moriarity B. S.; Zhovmer A. S.; Provenzano P. P. Engineering T Cells to Enhance 3D Migration through Structurally and Mechanically Complex Tumor Microenvironments. Nat. Commun. 2021, 12 (1), 1–17. 10.1038/s41467-021-22985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman S.; Vianay B.; Roca V.; De Pascalis C.. Microtubules Tune Mechanosensitive Cell Responses. 2020, 2020.07.22.205203, BioRxiv, 10.1101/2020.07.22.205203 (accessed on 10–10–2021),. [DOI] [PubMed] [Google Scholar]
- Brangwynne C. P.; MacKintosh F. C.; Kumar S.; Geisse N. A.; Talbot J.; Mahadevan L.; Parker K. K.; Ingber D. E.; Weitz D. A. Microtubules Can Bear Enhanced Compressive Loads in Living Cells Because of Lateral Reinforcement. J. Cell Biol. 2006, 173 (5), 733–741. 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez B. J.; Valentine M. T. Molecular Control of Stress Transmission in the Microtubule Cytoskeleton. Biochim. Biophys. Acta, Mol. Cell Res. 2015, 1853, 3015–3024. 10.1016/j.bbamcr.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Robison P.; Caporizzo M. A.; Ahmadzadeh H.; Bogush A. I.; Chen C. Y.; Margulies K. B.; Shenoy V. B.; Prosser B. L. Detyrosinated Microtubules Buckle and Bear Load in Contracting Cardiomyocytes. Science 2016, 352 (6284), aaf0659. 10.1126/science.aaf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y.; Caporizzo M. A.; Bedi K.; Vite A.; Bogush A. I.; Robison P.; Heffler J. G.; Salomon A. K.; Kelly N. A.; Babu A.; Morley M. P.; Margulies K. B.; Prosser B. L. Suppression of Detyrosinated Microtubules Improves Cardiomyocyte Function in Human Heart Failure. Nat. Med. 2018, 24 (8), 1225–1233. 10.1038/s41591-018-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet B. P.; Akhmanova A. Microtubules in 3D Cell Motility. J. Cell Sci. 2017, 130 (1), 39–50. 10.1242/jcs.189431. [DOI] [PubMed] [Google Scholar]
- Schoumacher M.; Goldman R. D.; Louvard D.; Vignjevic D. M. Actin, Microtubules, and Vimentin Intermediate Filaments Cooperate for Elongation of Invadopodia. J. Cell Biol. 2010, 189 (3), 541–556. 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino J.; Henriet E.; Ezzoukhry Z.; Goetz J. G.; Moreau V.; Saltel F. The Microenvironment Controls Invadosome Plasticity. J. Cell Sci. 2016, 129 (9), 1759–1768. 10.1242/jcs.182329. [DOI] [PubMed] [Google Scholar]
- Seano G.; Primo L. Podosomes and Invadopodia: Tools to Breach Vascular Basement Membrane. Cell Cycle 2015, 14 (9), 1370–1374. 10.1080/15384101.2015.1026523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maridonneau-Parini I. Control of Macrophage 3D Migration: A Therapeutic Challenge to Limit Tissue Infiltration. Immunol. Rev. 2014, 262 (1), 216–231. 10.1111/imr.12214. [DOI] [PubMed] [Google Scholar]
- Matrone M. A.; Whipple R. A.; Thompson K.; Cho E. H.; Vitolo M. I.; Balzer E. M.; Yoon J. R.; Ioffe O. B.; Tuttle K. C.; Tan M.; Martin S. S. Metastatic Breast Tumors Express Increased Tau, Which Promotes Microtentacle Formation and the Reattachment of Detached Breast Tumor Cells. Oncogene 2010, 29 (22), 3217–3227. 10.1038/onc.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killilea A. N.; Csencsits R.; Le E. B. N. T.; Patel A. M.; Kenny S. J.; Xu K.; Downing K. H. Cytoskeletal Organization in Microtentacles. Exp. Cell Res. 2017, 357 (2), 291–298. 10.1016/j.yexcr.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.; Jiang H.; Ho C.-H.; Grinnell F. Microtubule Function in Fibroblast Spreading Is Modulated According to the Tension State of Cell--Matrix Interactions. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (13), 5425–5430. 10.1073/pnas.0608030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba D.; Alisafaei F.; Savadipour A.; Rowe R. A.; Liu Z.; Pryse K. M.; Shenoy V. B.; Elson E. L.; Genin G. M. The Balance between Actomyosin Contractility and Microtubule Polymerization Regulates Hierarchical Protrusions That Govern Efficient Fibroblast–Collagen Interactions. ACS Nano 2020, 14 (7), 7868–7879. 10.1021/acsnano.9b09941. [DOI] [PubMed] [Google Scholar]
- Krendel M.; Gloushankova N. A.; Bonder E. M.; Feder H. H.; Vasiliev J. M.; Gelfand I. M. Myosin-Dependent Contractile Activity of the Actin Cytoskeleton Modulates the Spatial Organization of Cell-Cell Contacts in Cultured Epitheliocytes. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (17), 9666–9670. 10.1073/pnas.96.17.9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette D. T.; Shao L.; Ott C.; Pasapera A. M.; Fischer R. S.; Baird M. A.; Der Loughian C.; Delanoe-Ayari H.; Paszek M. J.; Davidson M. W.; Betzig E.; Lippincott-Schwartz J. A Contractile and Counterbalancing Adhesion System Controls the 3D Shape of Crawling Cells. J. Cell Biol. 2014, 205 (1), 83–96. 10.1083/jcb.201311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhn J. A. M.; Perez A. M.; Schneider I. C. Contact Guidance Diversity in Rotationally Aligned Collagen Matrices. Acta Biomater. 2018, 66, 248–257. 10.1016/j.actbio.2017.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabdanov E. D.; Puram V.; Zhovmer A.; Provenzano P. P. Microtubule-Actomyosin Mechanical Cooperation during Contact Guidance Sensing. Cell Rep. 2018, 25 (2), 328–338.e5. 10.1016/j.celrep.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A. J.; Schaeffer A.; De Pascalis C.; Letort G.; Vianay B.; Bornens M.; Piel M.; Blanchoin L.; Théry M. Acto-Myosin Network Geometry Defines Centrosome Position. Curr. Biol. 2021, 31, 1206. 10.1016/j.cub.2021.01.002. [DOI] [PubMed] [Google Scholar]
- Farina F.; Gaillard J.; Guérin C.; Couté Y.; Sillibourne J.; Blanchoin L.; Théry M. The Centrosome Is an Actin-Organizing Centre. Nat. Cell Biol. 2016, 18 (1), 65–75. 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A. N.; Patil A.; Black M. M.; Craig E. M.; Myers K. A.; Yeung H. T.; Baas P. W. Cytoplasmic Dynein Transports Axonal Microtubules in a Polarity-Sorting Manner. Cell Rep. 2017, 19 (11), 2210–2219. 10.1016/j.celrep.2017.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo U.; Winding M.; Lu W.; Gelfand V. I. Interplay between Kinesin-1 and Cortical Dynein during Axonal Outgrowth and Microtubule Organization in Drosophila Neurons. eLife 2015, 4, e10140 10.7554/eLife.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F. J.; Baas P. W. Microtubules Released from the Neuronal Centrosome Are Transported into the Axon. J. Cell Sci. 1995, 108, 2761–2769. 10.1242/jcs.108.8.2761. [DOI] [PubMed] [Google Scholar]
- Grabham P. W.; Seale G. E.; Bennecib M.; Goldberg D. J.; Vallee R. B. Cytoplasmic Dynein and LIS1 Are Required for Microtubule Advance during Growth Cone Remodeling and Fast Axonal Outgrowth. J. Neurosci. 2007, 27 (21), 5823–5834. 10.1523/JNEUROSCI.1135-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedzinski S.; Agsu G.; Vianay B.; Delord M.; Blanchoin L.; Larghero J.; Faivre L.; Théry M.; Brunet S. Microtubules Control Nuclear Shape and Gene Expression during Early Stages of Hematopoietic Differentiation. EMBO J. 2020, 39 (23), e103957 10.15252/embj.2019103957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall T. S.; Yip Y. Y.; Wallock-Richards D. J.; Pfisterer K.; Sanger A.; Ficek W.; Steiner R. A.; Beavil A. J.; Parsons M.; Dodding M. P. A Small-Molecule Activator of Kinesin-1 Drives Remodeling of the Microtubule Network. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (52), 13738–13743. 10.1073/pnas.1715115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly A. L.; Kim H.; Srinivasan D.; Lakonishok M.; Larson A. G.; Gelfand V. I. Kinesin-1 Heavy Chain Mediates Microtubule Sliding to Drive Changes in Cell Shape. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (27), 12151–12156. 10.1073/pnas.1004736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca-Zamora E. J.; Ferrer-Marín F.; Rivera J.; Teruel-Montoya R. Tubulin in Platelets: When the Shape Matters. Int. J. Mol. Sci. 2019, 20 (14), 3484. 10.3390/ijms20143484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-Hett S.; Richardson J. L.; Schulze H.; Drabek K.; Isaac N. A.; Hoffmeister K.; Shivdasani R. A.; Bulinski J. C.; Galjart N.; Hartwig J. H.; Italiano J. E. Jr. Visualization of Microtubule Growth in Living Platelets Reveals a Dynamic Marginal Band with Multiple Microtubules. Blood 2008, 111 (9), 4605–4616. 10.1182/blood-2007-10-118844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. G.Chapter 3: The Structure of Resting and Activated Platelets. In Platelets, 4th ed., Michelson A. D., Ed.; Academic Press: London, 2019; pp 47–77. [Google Scholar]
- Sadoul K. New Explanations for Old Observations: Marginal Band Coiling during Platelet Activation. J. Thromb. Haemostasis 2015, 13 (3), 333–346. 10.1111/jth.12819. [DOI] [PubMed] [Google Scholar]
- Ray A.; Morford R. K.; Ghaderi N.; Odde D. J.; Provenzano P. P. Dynamics of 3D Carcinoma Cell Invasion into Aligned Collagen. Integr. Biol. 2018, 10 (2), 100–112. 10.1039/C7IB00152E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A.; Lee O.; Win Z.; Edwards R. M.; Alford P. W.; Kim D.-H.; Provenzano P. P. Anisotropic Forces from Spatially Constrained Focal Adhesions Mediate Contact Guidance Directed Cell Migration. Nat. Commun. 2017, 8, 14923. 10.1038/ncomms14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman J. B.; Santarossa C. C.; Miller R. M.; Yu L. S.; Serpinskaya A. S.; Furukawa H.; Morimoto S.; Tanaka Y.; Nishitani M.; Asano M.; Zalyte R.; Ondrus A. E.; Johnson A. G.; Ye F.; Nachury M. V.; Fukase Y.; Aso K.; Foley M. A.; Gelfand V. I.; Chen Chemical Structure-Guided Design of Dynapyrazoles, Cell-Permeable Dynein Inhibitors with a Unique Mode of Action. eLife 2017, 6, 1. 10.7554/eLife.25174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A. D.; Wang F. W.; Matsumoto K.; Yamada K. M. One-Dimensional Topography Underlies Three-Dimensional Fibrillar Cell Migration. J. Cell Biol. 2009, 184 (4), 481–490. 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov S. V.; Sabass B.; Schwarz U. S.; Waterman C. M. High-Resolution Traction Force Microscopy. Methods Cell Biol. 2014, 123, 367–394. 10.1016/B978-0-12-420138-5.00020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.-H.; Frey M. T.; Burnham N. A.; Wang Y.-L. Substrate Rigidity Regulates the Formation and Maintenance of Tissues. Biophys. J. 2006, 90 (6), 2213–2220. 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M.; Pépin A.; Dressaire E.; Chen Y.; Bornens M. Cell Distribution of Stress Fibres in Response to the Geometry of the Adhesive Environment. Cell Motil. Cytoskeleton 2006, 63 (6), 341–355. 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- Cartagena-Rivera A. X.; Logue J. S.; Waterman C. M.; Chadwick R. S. Actomyosin Cortical Mechanical Properties in Nonadherent Cells Determined by Atomic Force Microscopy. Biophys. J. 2016, 110 (11), 2528–2539. 10.1016/j.bpj.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laevsky G.; Knecht D. A. Cross-Linking of Actin Filaments by Myosin II Is a Major Contributor to Cortical Integrity and Cell Motility in Restrictive Environments. J. Cell Sci. 2003, 116, 3761–3770. 10.1242/jcs.00684. [DOI] [PubMed] [Google Scholar]
- Fokin A. I.; David V.; Oguievetskaia K.; Derivery E.; Stone C. E.; Cao L.; Rocques N.; Molinie N.; Henriot V.; Aumont-Nicaise M.; Hinckelmann M.-V.; Saudou F.; Le Clainche C.; Carter A. P.; Romet-Lemonne G.; Gautreau A. M. The Arp1/11 minifilament of Dynactin Primes the Endosomal Arp2/3 Complex. Sci. Adv. 2021, 7 (3), 1. 10.1126/sciadv.abd5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieff S.; Alsina A.; Mathur A.; Nédélec F. J. Balance of Microtubule Stiffness and Cortical Tension Determines the Size of Blood Cells with Marginal Band across Species. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (17), 4418–4423. 10.1073/pnas.1618041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.; Malikov V.; Rodionov V. Self-Organization of a Radial Microtubule Array by Dynein-Dependent Nucleation of Microtubules. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (18), 10160–10165. 10.1073/pnas.181354198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikov V.; Cytrynbaum E. N.; Kashina A.; Mogilner A.; Rodionov V. Centering of a Radial Microtubule Array by Translocation along Microtubules Spontaneously Nucleated in the Cytoplasm. Nat. Cell Biol. 2005, 7 (12), 1213–1218. 10.1038/ncb1332. [DOI] [PubMed] [Google Scholar]
- Malikov V.; Kashina A.; Rodionov V. Cytoplasmic Dynein Nucleates Microtubules to Organize Them into Radial Arrays in Vivo. Mol. Biol. Cell 2004, 15 (6), 2742–2749. 10.1091/mbc.e03-10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec F. J.; Surrey T.; Maggs A. C.; Leibler S. Self-Organization of Microtubules and Motors. Nature 1997, 389 (6648), 305–308. 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- Surrey T.; Nédélec F.; Leibler S.; Karsenti E. Physical Properties Determining Self-Organization of Motors and Microtubules. Science 2001, 292, 1167. 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- Keating T. J.; Borisy G. G. Centrosomal and Non-Centrosomal Microtubules. Biol. Cell 1999, 91 (4–5), 321–329. 10.1111/j.1768-322X.1999.tb01090.x. [DOI] [PubMed] [Google Scholar]
- Cytrynbaum E. N.; Rodionov V.; Mogilner A. Computational Model of Dynein-Dependent Self-Organization of Microtubule Asters. J. Cell Sci. 2004, 117, 1381–1397. 10.1242/jcs.00919. [DOI] [PubMed] [Google Scholar]
- Tanenbaum M. E.; Vale R. D.; McKenney R. J. Cytoplasmic Dynein Crosslinks and Slides Anti-Parallel Microtubules Using Its Two Motor Domains. eLife 2013, 2, e00943 10.7554/eLife.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A.; Hause G.; Fink G.; Steinberg G. Conventional Kinesin Mediates Microtubule-Microtubule Interactions in Vivo. Mol. Biol. Cell 2006, 17 (2), 907–916. 10.1091/mbc.e05-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M. A.; Rice S. E. Microtubule-Associated Protein-Like Binding of the Kinesin-1 Tail to Microtubules. J. Biol. Chem. 2010, 285 (11), 8155–8162. 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.; Qiu W.; Gupta M. L. Jr; Pereira-Leal J. B.; Reck-Peterson S. L.; Pellman D. Mechanisms Underlying the Dual-Mode Regulation of Microtubule Dynamics by Kip3/Kinesin-8. Mol. Cell 2011, 43 (5), 751–763. 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M.; Drummond D. R.; Cross R. A.; McAinsh A. D. The Kinesin-14 Klp2 Organizes Microtubules into Parallel Bundles by an ATP-Dependent Sorting Mechanism. Nat. Cell Biol. 2009, 11 (6), 724–730. 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- Dogterom M.; Koenderink G. H. Actin-Microtubule Crosstalk in Cell Biology. Nat. Rev. Mol. Cell Biol. 2019, 20 (1), 38–54. 10.1038/s41580-018-0067-1. [DOI] [PubMed] [Google Scholar]
- Foster P. J.; Fürthauer S.; Shelley M. J.; Needleman D. J. Active Contraction of Microtubule Networks. eLife 2015, 4, 1. 10.7554/eLife.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte J. M.; Nédélec F. Large-Scale Microtubule Networks Contract Quite Well. eLife 2016, 5, 1. 10.7554/eLife.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimm M. L.; Henty-Ridilla J. L. New Twists in Actin-Microtubule Interactions. Mol. Biol. Cell 2021, 32 (3), 211–217. 10.1091/mbc.E19-09-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adio S.; Reth J.; Bathe F.; Woehlke G. Review: Regulation Mechanisms of Kinesin-1. J. Muscle Res. Cell Motil. 2006, 27 (2), 153–160. 10.1007/s10974-005-9054-1. [DOI] [PubMed] [Google Scholar]
- Kim D.; You E.; Rhee S. Dynein Regulates Cell Migration Depending on Substrate Rigidity. Int. J. Mol. Med. 2011, 29 (3), 440–446. 10.3892/ijmm.2011.867. [DOI] [PubMed] [Google Scholar]
- Wu J.; Misra G.; Russell R. J.; Ladd A. J. C.; Lele T. P.; Dickinson R. B. Effects of Dynein on Microtubule Mechanics and Centrosome Positioning. Mol. Biol. Cell 2011, 22 (24), 4834–4841. 10.1091/mbc.e11-07-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Lin F.; Wan Z.; Sun X.; Lu Y.; Huang J.; Wang F.; Zeng Y.; Chen Y.-H.; Shi Y.; Zheng W.; Li Z.; Xiong C.; Liu W. Profiling the Origin, Dynamics, and Function of Traction Force in B Cell Activation. Sci. Signaling 2018, 11 (542), 1. 10.1126/scisignal.aai9192. [DOI] [PubMed] [Google Scholar]
- Burakov A. V.; Nadezhdina E. S. Centering and Shifting of Centrosomes in Cells. Cells 2020, 9 (6), 1351. 10.3390/cells9061351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M. A.; Hoang H. T.; Urnavicius L.; Bullock S. L.; Carter A. P. In Vitro Reconstitution of a Highly Processive Recombinant Human Dynein Complex. EMBO J. 2014, 33 (17), 1855–1868. 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T. A. Dynactin. Annu. Rev. Cell Dev. Biol. 2004, 20, 759–779. 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Bashirzadeh Y.; Redford S. A.; Lorpaiboon C.; Groaz A.; Litschel T.; Schwille P.; Hocky G. M.; Dinner A. R.; Liu A. P. Actin Crosslinker Competition and Sorting Drive Emergent GUV Size-Dependent Actin Network Architecture. Commun. Biol. 2021, 4 (1), 1. 10.1038/s42003-021-02653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litschel T.; Kelley C. F.; Holz D.; Adeli Koudehi M.; Vogel S. K.; Burbaum L.; Mizuno N.; Vavylonis D.; Schwille P. Reconstitution of Contractile Actomyosin Rings in Vesicles. Nat. Commun. 2021, 12 (1), 2254. 10.1038/s41467-021-22422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höing S.; Yeh T.-Y.; Baumann M.; Martinez N. E.; Habenberger P.; Kremer L.; Drexler H. C. A.; Küchler P.; Reinhardt P.; Choidas A.; Zischinsky M.-L.; Zischinsky G.; Nandini S.; Ledray A. P.; Ketcham S. A.; Reinhardt L.; Abo-Rady M.; Glatza M.; King S. J.; Nussbaumer P.; et al. Dynarrestin, a Novel Inhibitor of Cytoplasmic Dynein. Cell Chem. Biol. 2018, 25 (4), 357–369.e6. 10.1016/j.chembiol.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy G. G.; Rodionov V. I. Lessons from the Melanophore. FASEB J. 1999, 13, S221–S224. 10.1096/fasebj.13.9002.S221. [DOI] [PubMed] [Google Scholar]
- Rodionov V. I.; Borisy G. G. Self-Centering in Cytoplasmic Fragments of Melanophores. Mol. Biol. Cell 1998, 9 (7), 1613–1615. 10.1091/mbc.9.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R.; John A. Microtubule-Associated Proteins as Direct Crosslinkers of Actin Filaments and Microtubules. IUBMB Life 2015, 67 (6), 395–403. 10.1002/iub.1384. [DOI] [PubMed] [Google Scholar]
- Cabrales Fontela Y.; Kadavath H.; Biernat J.; Riedel D.; Mandelkow E.; Zweckstetter M. Multivalent Cross-Linking of Actin Filaments and Microtubules through the Microtubule-Associated Protein Tau. Nat. Commun. 2017, 8 (1), 1–12. 10.1038/s41467-017-02230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cruz F.; Torres-Cruz F. M.; Monroy-Ramírez H. C.; Escobar-Herrera J.; Basurto-Islas G.; Avila J.; García-Sierra F. Fragmentation of the Golgi Apparatus in Neuroblastoma Cells Is Associated with Tau-Induced Ring-Shaped Microtubule Bundles. J. Alzheimer's Dis. 2018, 65 (4), 1185–1207. 10.3233/JAD-180547. [DOI] [PubMed] [Google Scholar]
- Spiliotis E. T.; Nakos K. Cellular Functions of Actin- and Microtubule-Associated Septins. Curr. Biol. 2021, 31 (10), R651–R666. 10.1016/j.cub.2021.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ageta-Ishihara N.; Miyata T.; Ohshima C.; Watanabe M.; Sato Y.; Hamamura Y.; Higashiyama T.; Mazitschek R.; Bito H.; Kinoshita M. Septins Promote Dendrite and Axon Development by Negatively Regulating Microtubule Stability via HDAC6-Mediated Deacetylation. Nat. Commun. 2013, 4 (1), 1–11. 10.1038/ncomms3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer B. E.; Haystead T.; Macara I. G. Mammalian Septins Regulate Microtubule Stability through Interaction with the Microtubule-Binding Protein MAP4. Mol. Biol. Cell 2005, 16 (10), 4648–4659. 10.1091/mbc.e05-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakos K.; Rosenberg M.; Spiliotis E. T. Regulation of Microtubule Plus End Dynamics by Septin 9.. Cytoskeleton 2019, 76 (1), 83–91. 10.1002/cm.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X.; Bowen J. R.; Knox T. K.; Zhou K.; Pendziwiat M.; Kuhlenbäumer G.; Sindelar C. V.; Spiliotis E. T. Novel Septin 9 Repeat Motifs Altered in Neuralgic Amyotrophy Bind and Bundle Microtubules. J. Cell Biol. 2013, 203 (6), 895–905. 10.1083/jcb.201308068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targa B.; Klipfel L.; Cantaloube I.; Salameh J.; Benoit B.; Poüs C.; Baillet A. Septin Filament Coalignment with Microtubules Depends on SEPT9_i1 and Tubulin Polyglutamylation, and Is an Early Feature of Acquired Cell Resistance to Paclitaxel. Cell Death Dis. 2019, 10 (2), 1–14. 10.1038/s41419-019-1318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. K.; Singh R. K.; Khazan N.; Kodza A.; Singh N. A.; Jones A.; Sivagnanalingam U.; Towner M.; Itamochi H.; Turner R.; Moore R. G. Development of Potent Forchlorfenuron Analogs and Their Cytotoxic Effect in Cancer Cell Lines. Sci. Rep. 2020, 10 (1), 3241. 10.1038/s41598-020-59824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K.; Kim K. K.; Khazan N.; Rowswell-Turner R. B.; Laggner C.; Jones A.; Srivastava P.; Hovanesian V.; Lamere L.; Conley T.; Pandita R.; Baker C.; Myers J. R.; Pritchett E.; Ahmad A.; Ruffolo L.; Jackson K.; Gerber S. A.; Ashton J.; Milano. et al., Septins Disruption Controls Tumor Growth and Enhances Efficacy of Herceptin. 2020, 2020.02.19.954529, bioRxiv, 10.1101/2020.02.19.954529 (accessed on 10–13–2021). [DOI] [Google Scholar]
- Hu Q.; Nelson W. J.; Spiliotis E. T. Forchlorfenuron Alters Mammalian Septin Assembly, Organization, and Dynamics. J. Biol. Chem. 2008, 283 (43), 29563–29571. 10.1074/jbc.M804962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems A. D.; Welf E. S.; Driscoll M. K.; Mazloom-Farsibaf H.; Chang B.-J.; Weiss B. G.; Chi J.; Dean K. M.; Fiolka R.; Danuser G.. Blebs Promote Cell Survival by Assembling Oncogenic Signaling Hubs. 2021, 04.23.441200, bioRxiv, 10.1101/2021.04.23.441200 (accessed on 08–11–2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanian L.; Berger C. L.; Hendricks A. G. Acetylated Microtubules Are Preferentially Bundled Leading to Enhanced Kinesin-1 Motility. Biophys. J. 2017, 113 (7), 1551–1560. 10.1016/j.bpj.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed N. A.; Cai D.; Lynne Blasius T.; Jih G. T.; Meyhofer E.; Gaertig J.; Verhey K. J. Microtubule Acetylation Promotes Kinesin-1 Binding and Transport. Curr. Biol. 2006, 16 (21), 2166–2172. 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Dolat L.; Hunyara J. L.; Bowen J. R.; Karasmanis E. P.; Elgawly M.; Galkin V. E.; Spiliotis E. T. Septins Promote Stress Fiber--Mediated Maturation of Focal Adhesions and Renal Epithelial Motility. J. Cell Biol. 2014, 207 (2), 225–235. 10.1083/jcb.201405050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R.; Cha B.-J.; Theurkauf W. E.; Saxton W. M. Dynein and the Actin Cytoskeleton Control Kinesin-Driven Cytoplasmic Streaming in Drosophila Oocytes. Development 2005, 132 (16), 3743–3752. 10.1242/dev.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R. B.; Seale G. E.; Tsai J.-W. Emerging Roles for Myosin II and Cytoplasmic Dynein in Migrating Neurons and Growth Cones. Trends Cell Biol. 2009, 19 (7), 347–355. 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. P.; Robison P.; Shi G.; Bogush A. I.; Kempema A. M.; Hexum J. K.; Becerra N.; Harki D. A.; Martin S. S.; Raiteri R.; Prosser B. L.; Ward C. W. Detyrosinated Microtubules Modulate Mechanotransduction in Heart and Skeletal Muscle. Nat. Commun. 2015, 6 (1), 1–14. 10.1038/ncomms9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabdanov E. D.; Puram V.; Zhovmer A.; Provenzano P. P. Microtubule-Actomyosin Mechanical Cooperation during Contact Guidance Sensing. Cell Rep. 2018, 25 (2), 328–338.e5. 10.1016/j.celrep.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper J. D.; Decker F.; Agana B.; Howard J. The Motility of Axonemal Dynein Is Regulated by the Tubulin Code. Biophys. J. 2014, 107 (12), 2872–2880. 10.1016/j.bpj.2014.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabdanov E. D.; Zhovmer A. S.; Puram V.; Provenzano P. P. Engineering Elastic Nano-and Micro-Patterns and Textures for Directed Cell Motility. STAR Protocols 2020, 1, 100013. 10.1016/j.xpro.2019.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid H.; Michel B. Siloxane Polymers for High-Resolution, High-Accuracy Soft Lithography. Macromolecules 2000, 33 (8), 3042–3049. 10.1021/ma982034l. [DOI] [Google Scholar]
- Odom T. W.; Love J. C.; Wolfe D. B.; Paul K. E.; Whitesides G. M. Improved Pattern Transfer in Soft Lithography Using Composite Stamps. Langmuir 2002, 18 (13), 5314–5320. 10.1021/la020169l. [DOI] [Google Scholar]
- Fischer R. S.; Myers K. A.; Gardel M. L.; Waterman C. M. Stiffness-Controlled Three-Dimensional Extracellular Matrices for High-Resolution Imaging of Cell Behavior. Nat. Protoc. 2012, 7 (11), 2056–2066. 10.1038/nprot.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov S. V.; Sabass B.; Schwarz U. S.; Waterman C. M. High-Resolution Traction Force Microscopy. Methods Cell Biol. 2014, 123, 367–394. 10.1016/B978-0-12-420138-5.00020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T.; Kopp J.; Guex N.; Peitsch M. C. SWISS-MODEL: An Automated Protein Homology-Modeling Server. Nucleic Acids Res. 2003, 31 (13), 3381–3385. 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Dokholyan N. V. MedusaDock 2.0: Efficient and Accurate Protein–Ligand Docking With Constraints. J. Chem. Inf. Model. 2019, 59 (6), 2509–2515. 10.1021/acs.jcim.8b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F.; Yin S.; Dokholyan N. V. Rapid Flexible Docking Using a Stochastic Rotamer Library of Ligands. J. Chem. Inf. Model. 2010, 50 (9), 1623–1632. 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S.; Biedermannova L.; Vondrasek J.; Dokholyan N. V. MedusaScore: An Accurate Force Field-Based Scoring Function for Virtual Drug Screening. J. Chem. Inf. Model. 2008, 48 (8), 1656–1662. 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.