Abstract
Studies over the past two decades have demonstrated that astrocytes are tightly associated with neurons and play pivotal roles in neural circuit development, operation, and adaptation in health and disease. Nevertheless, precisely how astrocytes integrate diverse neuronal signals, modulate neural circuit structure and function at multiple temporal and spatial scales, and influence animal behavior or disease through aberrant excitation and molecular output remains unclear. This Perspective discusses how new and state-of-the-art approaches, including fluorescence indicators, opto- and chemogenetic actuators, genetic targeting tools, quantitative behavioral assays, and computational methods, might help resolve these longstanding questions. It also addresses complicating factors in interpreting astrocytes’ role in neural circuit regulation and animal behavior, such as their heterogeneity, metabolism, and inter-glial communication. Research on these questions should provide a deeper mechanistic understanding of astrocyte-neuron assemblies’ role in neural circuit function, complex behaviors, and disease.
Keywords: actuator, astrocytes, behavior, computational approaches, genetic targeting, indicator, neural circuit
1 |. INTRODUCTION
“What is the function of glial cells in neural centers? The answer is still not known, and … it may remain unsolved for many years to come until physiologists find direct methods to attack it.” (Ramon y Cajal, 1911).
This prophecy turned out to be accurate. Astrocytes, glial cells of the central nervous system (CNS), are thought to regulate neural circuit formation, operation, and adaptation (Zuchero & Barres, 2015). During development, they help regulate neurogenesis, synapse number and function. In the adult CNS, they influence synaptic plasticity, neuronal excitability, network synchronization (e.g., brain state switching), rhythmic activity (e.g., in central pattern generating circuits), learning, and memory (Akther & Hirase, 2021; Allen & Eroglu, 2017; Araque et al., 2014; Di Castro & Volterra, 2021; Nimmerjahn & Bergles, 2015). Their molecular and functional heterogeneity suggests that astrocytes form regional networks, act in a neuronal cell type and circuit-specific manner, and—based on behavioral and machine learning approaches—may process information in ways complementary to neurons. Astrocytes also play essential roles in pathology. After CNS injuries and in patients with neurodegenerative disease, they adapt their structural and functional properties, thereby regulating neuronal health and survival (Pekny et al., 2016; Sofroniew, 2020). These critical advances in our understanding of astrocyte biology—from the subcellular to network level—are primarily due to recent technical progress, including new and improved fluorescent indicators, bidirectional actuators, sophisticated behavioral assays, recording techniques (e.g., wearable microscopes), computational methods, and theoretical frameworks allowing measurement, manipulation, and modeling of these cells’ in vivo structure and function across CNS regions (e.g., cortical, subcortical, brainstem, spinal cord).
Despite this progress, many longstanding questions remain (Box 1), including how astrocytes integrate and (nonlinearly) respond to diverse neuronal signals in their extracellular environment at the cellular and population level, how they convert this task- and animal state-dependent information into functional outputs that modulate neural circuit dynamics and animal behavior on various temporal and spatial scales, whether this influence on neurons—however sophisticated—primarily serves homeostatic functions (e.g., transmitter recycling, restoration of ionic balance, toxic substance removal, energy substrate delivery, synaptic scaling) or augments/complements neural circuit properties (e.g., increased computational power, flexibility, or learning and memory through coincidence detection-triggered excitatory and inhibitory synaptic gain modulation, extracellular K+-mediated changes in network excitability/synchrony, or temporally holding information about prediction error extent to update synaptic weights accordingly), and whether they perform similar computations in various brain and spinal cord regions that differ in cytoarchitecture and functional dynamics.
BOX 1.
Select unresolved questions
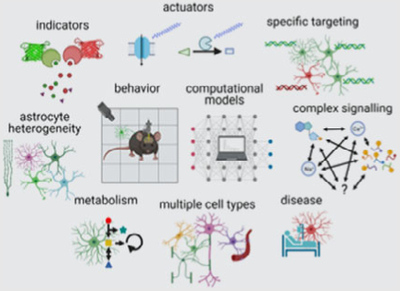
|
Overall
|
|
Indicators
|
|
Actuators
|
|
Genetic targeting approaches
|

|
Behavioral assays
|

|
Computational approaches
|
|
Astrocyte heterogeneity
|
|
Astrocyte metabolism
|
|
Contribution of other non-neuronal
cells
|
|
Disease
|
Filling this knowledge gap will require a systematic approach (1) measuring relevant neuronal signals in relation to astrocyte activity and morphology, (2) manipulating astrocyte activity and structural plasticity in physiologically relevant ways, (3) relating astrocytes’ functional and morphological changes to changes in neuronal dynamics and animal behavior, and (4) integrating the various data into a coherent picture using computational modeling and theoretical frameworks. Insights from these studies promise to transform our modern understanding of systems neuroscience and provide the conceptual framework for placing astrocytes in higher information processing.
In this perspective, we first provide an overview of state-of-the-art tools for measuring and manipulating astrocyte communication, including various indicators, actuators, and genetic targeting approaches. Next, we discuss how combining these interrogations with behavioral measurements and computational methods might help uncover astrocytes’ role in regulating neural circuit function and animal behavior. Finally, we consider how astrocyte heterogeneity and diversity and astrocytes’ interaction with other non-neuronal cells may complicate the picture. Given the wide range of topics covered in this perspective, the cited literature merely serves to exemplify where the field stands today, the challenges it faces, and tools and approaches that might help tackle them. For a more in-depth discussion of the various topics, we refer the reader to the cited and topical reviews in this Special Issue.
2 |. MEASURING, MANIPULATING, AND TARGETING ASTROCYTE FUNCTIONS
2.1 |. Indicators
The potential pathways by which neurons and astrocytes communicate with one another run the gamut of signaling mechanisms in the CNS, including chemical and electrical synaptic transmission and neuromodulation (Verkhratsky & Nedergaard, 2018). Critical for elucidating these molecular interactions are reagents that allow quantitative monitoring of corresponding signaling mechanisms in vivo. Most functional imaging is currently centered on a small number of mechanisms, particularly those that involve calcium excitation. To develop a deeper understanding of neuron-astrocyte communication, additional signaling mechanisms need to be studied. Fortunately, an increasingly broad and powerful toolbox of biosensors to track signaling between and within cells has recently become available (Table 1). This toolbox includes indicators for the readout of astrocyte input and output signals, as well as intracellular biochemical activity. Additionally, spectral variants of these indicators are beginning to allow the study of pathway interactions.
TABLE 1.
Genetically encoded indicators for measuring astrocyte functiona
| Available indicators for readout of neuron-astrocyte signaling | |||
|---|---|---|---|
| Indicator type | Indicator name(s) | Available color variants | References |
| Acetylcholine | iAChSnFR, GRABACh | Green | Borden et al., 2020; Jing et al., 2020 |
| Adenosine | GRABAdo | Green | Peng et al., 2020 |
| ATP | iATPSnFR, GRABATP | Green | Lobas et al., 2019; Wu, He, et al., 2021 |
| Dopamine | dLight, GRABDA | Green, yellow, red | Patriarchi et al., 2018; Patriarchi et al., 2020; Sun et al., 2018 |
| D-Serine | D-serFSb | Cyan-yellowb | Vongsouthi et al., 2021 |
| Endocannabinoid | GRABeCB | Green | Dong, He, et al., 2021 |
| GABA | iGABASnFR | Green | Marvin et al., 2019 |
| Glutamate | iGluSnFR | Blue, cyan, green, yellow | Marvin et al., 2018 |
| Glycine | GlyFSb | Cyan-yellowb | Zhang et al., 2018 |
| Nicotine | iNicSnFR | Green | Shivange et al., 2019 |
| Norepinephrine | nLight, GRABNE | Green | Feng et al., 2019; Oe et al., 2020 |
| Serotonin | sLight, iSeroSnFR, GRAB5–HT | Green | Patriarchi et al., 2018; Unger et al., 2020; Wan et al., 2021 |
| Available indicators for readout of intracellular signaling | |||
| ATP/ADP | Perceval | Green | Berg et al., 2009 |
| ATP | ATeamsb | Cyan-yellowb | Imamura et al., 2009 |
| Calcium | X-CaMP series, X-GECO series | Blue, green, yellow, red, near-infrared | Dalangin et al., 2020; Dana et al., 2016; Dana et al., 2019; Inoue et al., 2019; Qian et al., 2020; Shemetov et al., 2021; Shen et al., 2018 |
| cAMP | Flamindo series, R-FlincA, cAMPr | Yellow, red | Hackley et al., 2018; Harada et al., 2017; Kim, Shin, & Bae, 2021; Odaka et al., 2014; Ohta et al., 2018 |
| Chloride | SuperClomeleonb, ClopHensorNb | Cyan-yellowb, green-redb | Grimley et al., 2013; Raimondo et al., 2013 |
| ERK | RAB-EKARevb | Green-redb | Ding et al., 2015 |
| Glucose | iGlucoSnFR, FLIPglub, Green Glifons | Green, cyan-yellowb | Díaz-García et al., 2019; Fehr et al., 2003; Keller et al., 2021; Mita et al., 2019 |
| IP3 | IRIS-1b | Cyan-yellowb | Matsu-ura et al., 2006 |
| Lactate | eLACCO, Laconicb | Green, cyan-yellowb | Nasu et al., 2021; San Martín et al., 2013 |
| NADH/NAD+ | Peredoxb | Green-redb | Hung et al., 2011 |
| NADPH | iNapb | Cyan-yellowb | Tao et al., 2017 |
| pH | pHRedb, ClopHensorNb | Red, green-redb | Raimondo et al., 2013; Tantama et al., 2011 |
| PKA | ExRai-AKAR | Green | Zhang et al., 2021 |
| Potassium (K+) | GEPIIb, KIRINb, GINKO2, KRaION | Cyan-yellowb, green-redb, green | Bischof et al., 2017; Torres Cabán et al., 2021; Wu, Wen, et al., 2021 |
| ROS (H2O2) | FROG/Bb, HyPer7b, HyPerRed | Blue-greenb, greenb, red | Ermakova et al., 2014; Pak et al., 2020; Sugiura et al., 2020 |
| Available indicators for readout of astrocyte-neuron signaling | |||
| ATP, GABA, glutamate, etc. See above | See above | See above | |
Abbreviations: cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; IP3, inositol-trisphosphate; PKA, protein kinase A; ROS, reactive oxygen species.
This table does not cover existing synthetic indicators (e.g., CoroNa Green AM for sodium), which may be helpful in acute experiments. For additional genetically encoded indicators (e.g., for cAMP), see the cited reviews. For an overview of additional genetically encoded metabolite sensors, see Koveal et al., 2020; San Martin et al., 2014; Zhang et al., 2020; San Martin et al., 2022.
Ratiometric indicator.
2.1.1 |. Indicators for readout of astrocyte input
Indicators for neurotransmitters, neuromodulators, and neuropeptides will be crucial for determining precisely how animal behavior engages these pathways and shapes astrocytes’ excitation in a given region. The spatial and temporal heterogeneity of astrocytes’ calcium transients suggests that they are exposed to a continually varying cocktail of transmitters in their environment. How these complex time-varying patterns of neuronal (and non-neuronal) signals are then integrated by astrocytes, which calcium sources contribute to this process (e.g., plasma membrane calcium channels, ion exchangers, endoplasmic reticulum (ER), mitochondria, and potentially lysosomes; Heidemann et al., 2005; Semyanov et al., 2020), and what information the various astrocyte activity patterns encode remains largely unknown. Experimental evidence suggests that astrocytes in the CNS may use multiple parameters (e.g., transient onset, amplitude, duration, frequency, and spatial pattern) to encode information (Nelson et al., 2019; Nimmerjahn & Bergles, 2015). Direct measurement of the input signals together with astrocyte excitation in the context of quantifiable behavioral assays will be critical to address this question. These assays should allow control over the type of pathways being activated (e.g., dopamine) and their timing. Reward-based assays seem particularly well suited in this context (see below). While an increasing palette of indicators is starting to allow multiplex measurements, the number of signals that can be measured simultaneously will likely remain limited (e.g., due to indicator signal-to-noise ratio [SNR] or emission spectrum overlap). Therefore, the behavioral assays also need to be highly reproducible so that measurements with different indicator combinations obtained from different groups of animals can be compared.
2.1.2 |. Indicators for readout of astrocyte intracellular signaling
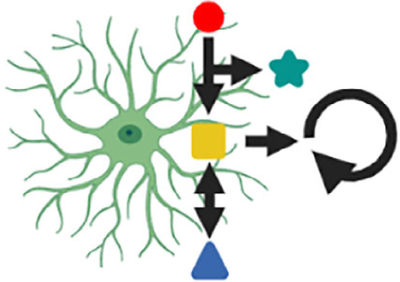
Indicators for the intracellular biochemical activity will be critical in determining its relationship to receptor activation and astrocyte responses. For example, astrocytes express several subtypes or levels of G-protein coupled receptors (GPCRs; Gs-, Gi-, and Gq-coupled). Activation of these receptors (e.g., by dopamine, norepinephrine, serotonin, acetylcholine) increases or decreases intermediate signals, such as inositol-trisphosphate (IP3), cyclic adenosine monophosphate (cAMP), reactive oxygen species (ROS), or inositol 1,4,5-triphosphate (IP3) receptor-binding protein released with IP3 (IRBIT) (Figure 1). These signals are then integrated spatially and temporally (e.g., at the mitochondrial or ER level), leading to calcium and protein kinase A (PKA) signals reflective of local and projection neuron activities. Elevations of calcium and PKA activity, in turn, are thought to jointly or individually modulate neuronal excitability and plasticity through several mechanisms and on various timescales (e.g., neuroactive substance release, extracellular potassium concentration changes, morphological alterations) (Dong et al., 2012; Horvat et al., 2016; Impey et al., 1998; Masmoudi-Kouki et al., 2007; Verkhratsky & Nedergaard, 2018). Nevertheless, precisely how local neural and projection neuron activities regulate these signals during animal behavior remains largely unknown. Measuring intracellular signaling in behaving animals will, therefore, provide vital clues as to how local and projection neuron activity is integrated within astrocytes, regulates astrocytes’ gene expression (e.g., through level or frequency of nuclear calcium transients), or regulates astrocyte-derived extracellular ion or neuroactive factor concentrations.
FIGURE 1.
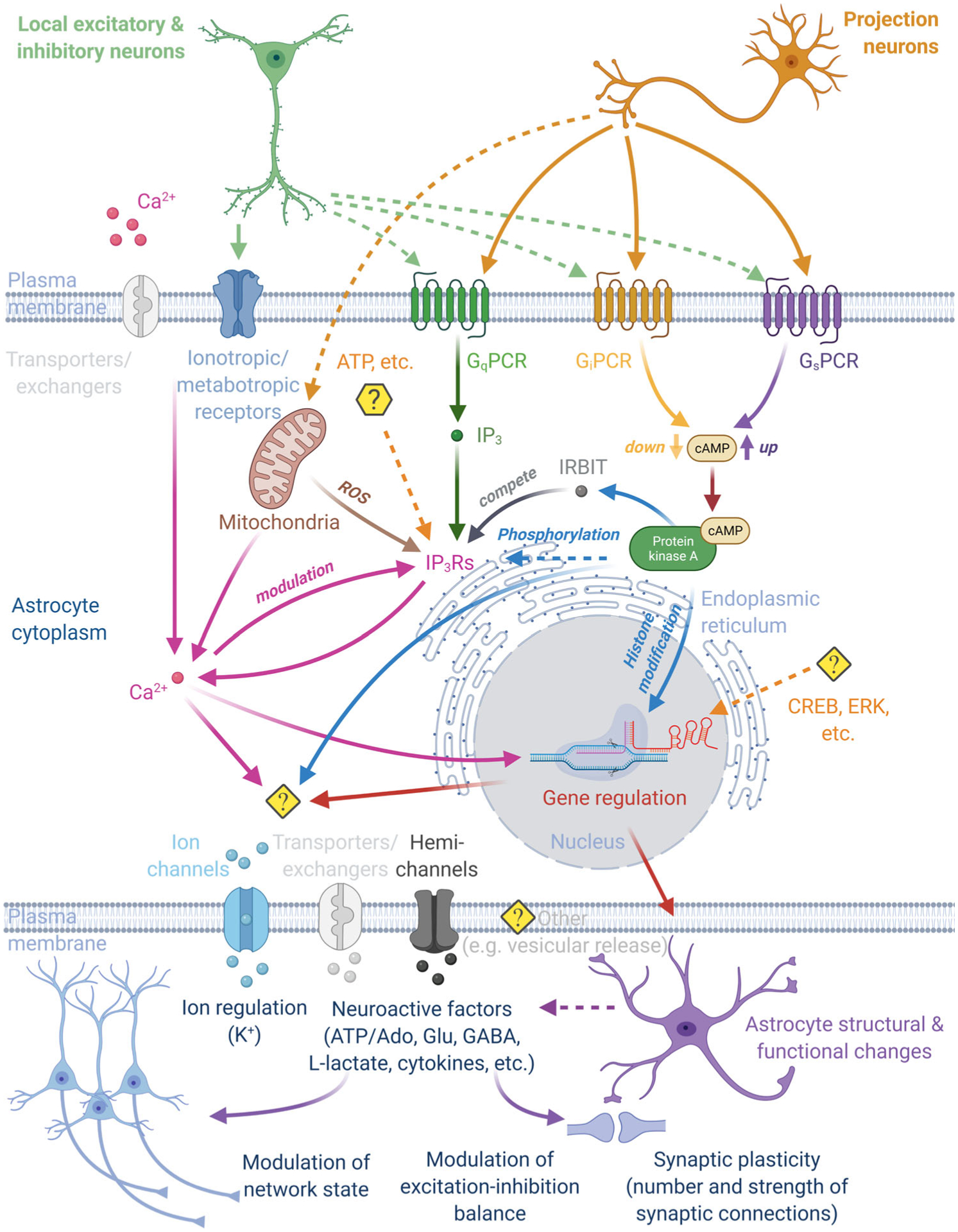
Astrocytes regulate and are controlled by neural circuits and animal behavior. Animal behavior activates a subset of local excitatory, inhibitory, and projection neurons, leading to the release of diverse molecular signals. Astrocytes are thought to spatially and temporally integrate these time-varying signals in their environment, with calcium and/or PKA playing central roles in this process. Signal integration also involves intermediate signals, such as reactive oxygen species (ROS), IP3, cAMP, and IRBIT. Astrocyte excitation, in turn, is thought to modulate neural circuit function (e.g., network state, excitation-inhibition balance, synaptic strength, or number) through different mechanisms (e.g., extracellular ion regulation, neuroactive factor release, perisynaptic process structure) and on various timescales (second to minutes) (see also Figures 2–3). Abbreviations: Ado, adenosine; cAMP, cyclic adenosine monophosphate; Glu, glutamate; IP3, inositol-trisphosphate; IRBIT, IP3 receptor-binding protein released with IP3; PKA, protein kinase A
2.1.3 |. Indicators for readout of astrocyte output
Indicators for soluble and contact-mediated signaling will be essential to relate changes in astrocyte excitation to neural circuit modulation. For example, proximity labeling and genomic profiling have revealed distinct adhesion molecules in perisynaptic astrocyte processes (PAPs) that modulate either excitatory or inhibitory synapses (Singh et al., 2016; Stogsdill et al., 2017). Whether there is specificity in the type of synapses being enveloped within an astrocyte’s territory and what the functional implications of such specificity might be (e.g., changes in excitation-inhibition balance) remains largely unknown. Indicators for molecules released by astrocytes (e.g., glutamate, GABA, ATP, D-serine, lactate) (Table 1) may also help resolve controversies and discrepancies regarding astrocyte-to-neuron signaling, including the conditions under which it occurs, its spatiotemporal dynamics, and the types of transmitters being released (Bonvento & Bolaños, 2021; Savtchouk & Volterra, 2018; Sherwood et al., 2021; Wolosker et al., 2016).
In summary, novel genetically encoded indicators are needed to shed light on the complex intra- and extracellular signaling utilized by astrocyte-neuron assemblies. This may require complementary targeting methods that restrict indicator expression to specific subcellular or intracellular compartments (Broussard et al., 2018; Henderson et al., 2015; Shemesh et al., 2020; Suzuki et al., 2014). However, close attention should be paid to the indicator expression level, as it can interfere with normal cell function (Armbruster et al., 2020). Control experiments (e.g., expression level titration, sensors with ablated target molecule binding) should be employed to rule out measurement artifacts whenever possible. Some signaling molecules may act on and be released by astrocytes (e.g., ATP), requiring additional interventions to distinguish between cause and effect. Additionally, some signaling molecules (e.g., K+, IP3) may spread and act intracellularly given astrocytes’ gap junctional coupling (including with other glial cells, such as oligodendrocytes [OLs]), a route that may need to be considered when interpreting physiological or actuator-evoked indicator signals and effects (see below). Nevertheless, increased capabilities to measure extra- and intracellular signaling promise to make a substantive difference in examining astrocyte function and their roles in brain circuits (Figure 1).
2.2 |. Actuators
Measurements with the functional and structural indicators mentioned above can reveal correlative relationships and constrain existing theoretical models of how molecular signaling, cellular dynamics, and animal behavior are linked. To corroborate these possible links and establish causal relationships, additional targeted manipulation of neuron-astrocyte communication will be necessary.
2.2.1 |. Actuators for astrocyte excitation
A wide variety of tools, including opto- and chemogenetic actuators, have been developed to tightly control the activity of genetically defined sets of neurons. All-optical approaches have provided intriguing insights into how the targeted neurons and their activity patterns encode and influence sensorimotor behavior (Carrillo-Reid et al., 2019; Robinson et al., 2020). The success of these approaches has raised hopes that a similar strategy might be possible for astrocytes. Indeed, by applying some of these tools to astrocytes, studies in mice and zebrafish have provided initial evidence for a causal link between these cells’ activity, neural circuit function, and animal behavior on both short and long timescales (Nagai, Yu, et al., 2021). These studies have also highlighted that astrocytes present unique challenges. Arguably, current actuators and the ways to control them (Table 2) are still a far cry from allowing us to manipulate astrocytes in ways that mimic their physiological operation. Neither the evoked intra- and extracellular dynamics (e.g., calcium, cAMP, potassium) nor their spatiotemporal pattern mimics experimentally observed astrocyte dynamics which, given their complexity, are near impossible to reproduce with current techniques (the same technical limitations apply to neuronal actuation). However, while the evoked activity may not be entirely physiological, it provides critical information regarding the aspects of neural circuit function astrocytes can control or how their targeted manipulation may be used for therapeutic applications.
TABLE 2.
Genetically encoded actuators for interrogating astrocyte functiona
| Available actuators for astrocyte “excitation” | |||
|---|---|---|---|
| Actuator type | Actuator name(s) | Mechanism | References |
| Calcium | ChR2, CatCh | Cation channel | Beppu et al., 2014; Figueiredo et al., 2014 |
| ArchT | Proton pump | Beppu et al., 2014; Poskanzer & Yuste, 2016 | |
| Opto-XRs | G-protein signalingb | Figueiredo et al., 2014 | |
| DREADDs | G-protein signalingb | Chai et al., 2017; Durkee et al., 2019 | |
| cAMP/cGMP | YFP-CaRhAC/YFP-CaRhGC | Enzyme activity (adenylyl/guanylyl cyclase) | Scheib et al., 2018 |
| Available actuators for astrocyte “inhibition” | |||
| Actuator type | Actuator name(s) | Mechanism | References |
| Calcium | CalEx | Reduces calcium signaling after expression of the plasma membrane calcium pump PMCA2 | Yu et al., 2018; Yu et al., 2021 |
| SpiCee | Reduces calcium signaling by calcium binding to a chimeric calmodulin- and ɑ-parvalbumin-based calcium buffer | Ros et al., 2020 | |
| iβARK | Attenuates Gq-GPCR-evoked calcium signaling by sequestering Gɑq-GTP | Nagai, Bellafard, et al., 2021 | |
| cGMP | SponGee | Reduces cGMP signaling by cGMP binding to a chimeric PKG1ɑ/PKG1β cGMP buffer | Ros et al., 2019 |
| IP3 | IP3 sponge | Competes with the endogenous IP3Rs for IP3 binding in a dose-dependent manner | Miyamoto & Mikoshiba, 2017; Uchiyama et al., 2002 |
Abbreviations: cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; GPCR, G-protein coupled receptor; IP3, inositol-trisphosphate.
While these actuators may target a specific signaling pathway, it is important to note that their effects can be broad, indirect, and divergent. For example, CalEx expression leads to numerous gene expression changes affecting multiple astrocyte functions at once (Nagai, Bellafard, et al., 2021). ChR2 and ArchT both elevate intracellular calcium levels but have opposing effects on glutamate release (Beppu et al., 2014). Calcium elevations can be partly due to autocrine signaling following stimulated transmitter release (e.g., ATP) (Figueiredo et al., 2014).
Unlike in neurons, “inhibitory” opsins and DREADDs typically lead to calcium excitation in astrocytes (Durkee et al., 2019; Poskanzer & Yuste, 2016).
2.2.2 |. Actuators for astrocyte inhibition
Aberrant astrocyte activity in the brain and spinal cord has been described in various disease models (Nelson et al., 2019; Nimmerjahn & Bergles, 2015). Tools to inhibit astrocytes’ excitation or transmission are, therefore, critical. For neurons, expression of tetanus toxin light chain (Kim et al., 2009), diphtheria toxin (Roselló-Díez et al., 2018), inhibitory opsins or DREADDs (Lerner et al., 2016; Rajasethupathy et al., 2016; Roth, 2016), and vesicular inhibitory amino acid transporter deletion (Hirrlinger et al., 2019; Rahman et al., 2015) have been used for silencing. Unfortunately, silencing astrocytes appears more difficult due to these cells’ distinct physiological properties. For example, opsins and DREADDs that inhibit neuronal firing (e.g., ArchT, hM4Di) typically mediate calcium excitation in astrocytes (Nagai et al., 2019; Poskanzer & Yuste, 2016). Genetically encoded chelators or pumps have been shown to attenuate intracellular signaling (e.g., calcium or cGMP increases) but currently provide limited temporal resolution or reversibility (Table 2) (Ros et al., 2019; Ros et al., 2020; Yu et al., 2018). Another challenge is compartment-specific manipulations (e.g., cell body, branches, or microdomains). Currently, only a few genetic targeting approaches exist for that purpose (e.g., H2B and Lck for nuclear and plasma membrane targeting, respectively).
Clearly, there is an urgent need for new and improved actuators that allow precise control over astrocytes’ spatial and temporal dynamics and specific signaling pathways. Thorough characterization, including transcriptomic and proteomic profiling, will be necessary to develop a mechanistic understanding of precisely how their activation influences astrocytes, neural circuits, and animal behavior, either directly or indirectly (Figueiredo et al., 2014; Octeau et al., 2019; Philtjens et al., 2021; Yu et al., 2020). Such experiments should also consider how the observed effects depend on stimulation parameters and the approach itself (e.g., light-induced heating-mediated effects; Courtney et al., 2021; Owen et al., 2019; Schmidt & Oheim, 2020). Together with established genetic interventions (e.g., CRISPR/Cas9-based approaches to genetically or epigenetically modify astrocytes’ ability to sense, process, or send signals), these new tools will be essential in mapping the interplay between astrocytes and neural circuits in behavior on both short and long timescales.
2.3 |. Genetic targeting approaches
While various genetic tools allow astrocyte targeting, differences exist in these approaches’ ability to control regional and temporal expression and target astrocyte subtypes. Astrocytes are a heterogeneous cell population. Their structural and functional properties depend on the CNS region. Even within a given area, astrocytes can show considerable heterogeneity. This suggests a complex interplay between these cells and their local environment and that astrocytes may modulate neural circuit function in a region-dependent manner. Therefore, genetic approaches for dissecting astrocytes’ role in neural circuits may need to match this diversity, as more widespread interventions (e.g., global gene knockout or signaling pathway activation) could obscure region- or subtype-specific effects. Genetic targeting of astrocytes with high specificity, efficiency, and in a region-dependent manner has turned out to be challenging (Hirbec et al., 2020). While studies to identify region- and subpopulation-specific promoters are ongoing, several existing approaches (either alone or in combination) can be used to target astrocytes with varying degrees of specificity.
Numerous transgenic mouse lines allow fluorescent indicator, actuator, Cre-DNA recombinase, or tet-system component expression in astrocytes using different promoters (e.g., GFAP, GLAST/SLC1A3, Aldh1l1) (Casper & McCarthy, 2006; Hirrlinger et al., 2006; Mori et al., 2006; Natsubori et al., 2015; Nolte et al., 2001; Pascual et al., 2005; Pfrieger & Slezak, 2012; Slezak et al., 2007; Tanaka et al., 2010; Winchenbach et al., 2016; Zhuo et al., 2001; see also https://networkglia.eu/en/animal_models). Each of these mouse lines has its advantages and disadvantages. None of them is entirely astrocyte-specific, though off-target expression varies by region, targets a homogenous subset of astrocytes in a given area or neural circuit, or captures all CNS astrocytes. Notably, germline recombination has been reported for several of these mouse lines (Luo et al., 2020; Zhang et al., 2013). While some of these issues can be overcome, at least in part, by temporally controlled transgene expression after postnatal development, corresponding data should be interpreted with the above limitations in mind.
Several strategies can be used to achieve more circuit-specific astrocyte targeting:
Viral vectors: Stereotactic delivery to the CNS region of interest allows spatially confined manipulations. Targeting is also achieved by leveraging viral tropism, astrocyte promoters (e.g., gfaABC1D), detargeting strategies (e.g., miR124), or injecting Cre-dependent vectors into astrocyte promoter-driven Cre-expressing mice. However, given the lack of truly pan-astrocyte or astrocyte sub-population-specific promoters (Hirbec et al., 2020), off-target expression in the investigated region should be quantified. Standard vector systems used to target astrocytes include adeno-associated viruses, adenoviruses, and lentiviruses. While it is beyond the scope of this Perspective to discuss these different systems in detail (see Delzor et al., 2013; Hirbec et al., 2020 for recent reviews), the following considerations determine which approach is most appropriate for a given application: packaging capacity, tropism, expression speed or level, spatial spread (viral particle size-dependent), immunogenicity, ability to infect dividing or nondividing cells, and genome integration. Direct stereotactic injections result in small lesions and corresponding glial responses. Some viral vectors allow systemic delivery, thereby preventing local injuries. However, these blood-tissue-barrier permeable vectors lead to off-target infection and other potential side effects (e.g., liver toxicity). In both cases, the immune response needs to be carefully considered.
Combinatorial genetics: This strategy allows targeting of cell populations not defined by the activity of a single promoter but the simultaneous or consecutive activity of two or more promoters (Table 3). However, while this approach has been successfully used for neurons, defining appropriate promoter combinations to target specific subsets of astrocytes or astrocytes in specific neural circuits is still an open question. Ongoing population-wide and single-cell sequencing studies promise to provide information about region-specific gene expression and promoters, thereby allowing future targeting of astrocyte subpopulations and interrogating their roles in regulating neural circuit function and animal behavior.
TABLE 3.
Genetic approaches for astrocyte targeting
| Approach | Concept | Properties | References |
|---|---|---|---|
| Split-Cre |
|
|
Beckervordersandforth et al., 2010; Hirrlinger et al., 2019; Hirrlinger, Scheller, et al., 2009; Jullien et al., 2003; Kim, Kolesnikov, et al., 2021 |
| Split-Cre-ERT2 |
|
|
Hirrlinger et al., 2019; Hirrlinger, Requardt, et al., 2009 |
| Split-Cre-Intein |
|
|
Luan et al., 2020; Salwig et al., 2019; Wang et al., 2012 |
| Combination of different DNA recombinases (e.g., Cre, Dre, Flp) |
|
|
Jensen & Dymecki, 2014; Liu et al., 2020; Madisen et al., 2015 |
| Light-activated DNA recombinases |
|
|
Weinberg et al., 2019; Yao et al., 2020 |
| Combination of DNA recombinases and the Tet-system |
|
|
Ahmadzadeh et al., 2020; Madisen et al., 2015; Rosellό-Díez et al., 2018 |
In summary, further development of genetic targeting approaches is needed to achieve astrocyte specificity, subtype, or circuit targeting.
3 |. RELATING ASTROCYTE FUNCTIONS TO NEURAL CIRCUIT OPERATION AND ANIMAL BEHAVIOR
3.1 |. Behavioral approaches
Evidence that astrocyte properties are modulated by sensory experience and motor behavior, and that astrocyte manipulation can affect neuronal properties and behavioral phenotype is a foundation for the hypothesis that these cells play crucial roles in regulating neural circuit function and animal behavior. Nevertheless, the diversity of experimental approaches and measurement conditions has led to a flood of data with sometimes seemingly contradictory results. Hence, despite the recent technical progress in measuring neuronal, astrocyte, and transmitter dynamics in behaving animals, precisely how the complex patterns of astrocyte excitation relate to neural circuit function and animal behavior remains unclear. Apart from the apparent model or technical differences, one possible explanation for this predicament is that task-related trials can generate unique behavioral and cellular activities, each influencing astrocytes’ dynamics in a deterministic yet complex nonlinear fashion. Hence, to link astrocyte excitation to neural circuit function and behavioral changes, it will be necessary to employ quantitative behavioral assays that allow precise control or readout of sensory input, internal state, and behavioral response, with or without complementary approaches (e.g., molecular genetic interventions).
3.1.1 |. Select insights from brain recordings
Studies in the brain have indicated that astrocytes’ complex intracellular signaling, most prominently calcium transients, is triggered by neurotransmitter and neuromodulator receptor activation on their surface; that receptor co-activation can lead to nonlinear responses (e.g., pairing sensory stimulation with motor-behavior evoked neuromodulator signaling); and that signal integration within astrocytes influences their subsequent activity (Bazargani & Attwell, 2017; Kastanenka et al., 2020; Nimmerjahn & Bergles, 2015). As a result, astrocyte signaling spans multiple spatial and temporal scales, from sub-second transients in single astrocytes to seconds- or even minutes-long transients in astrocytic networks. This complexity suggests that astrocytes may carry out computations on various timescales related to sensory processing, brain state modulation, and memory formation (Figure 2). For example, astrocytes can influence behavioral adaptation and spatial memory, as shown by genetic approaches to modulate astrocyte Gq GPCR signaling, either locally or across brain regions (Nagai, Bellafard, et al., 2021). However, as with any approach that globally manipulates astrocyte excitation (e.g., by opsin, DREADD, calcium pump, or chelator expression), it remains unclear to what extent these interventions mimic astrocytes’ varied responses and effects on neural circuits. Recent studies using reward-based behavioral assays and machine learning approaches have begun to link astrocyte excitation to information processing and behavioral variables. Using simultaneous imaging of astrocyte and neuronal calcium activity in the dorsal hippocampus (CA1 region) of head-fixed mice navigating a virtual environment, individual astrocytes’ activity was found to co-vary with virtual space location. The combined information about neuronal and astrocyte activity allowed the animal’s position to be reconstructed more faithfully in decoding algorithms than from neurons alone (Curreli et al., 2021). Measuring astrocyte calcium dynamics also in the hippocampal CA1 region but employing a different virtual environment, another group found that astrocytes seemingly encode reward location in an experience-dependent manner (i.e., in a familiar but not novel environment) (Doron et al., 2021). While the two studies disagreed regarding how astrocytes encode spatial information, both suggest that non-neuronal cells may contribute to information processing. Notably, astrocytes represented the spatial environment differently than neurons. However, the extent to which motor behavior itself influenced these studies’ astrocyte responses remains unclear. Astrocytes are known to exhibit widespread calcium excitation in response to locomotion (Ding et al., 2013; Nimmerjahn et al., 2009; Paukert et al., 2014). Run parameters, particularly the timing of astrocyte response onset relative to run onset and the period between runs, can strongly influence experimental results and interpretation. Addressing this concern head-on, a recent study in the mouse motor cortex showed that astrocyte population responses are linked to run onset, duration, and inter-run interval (Merten et al., 2021). Using a quantitative, reward-based visual detection task, statistical modeling, and machine learning approach, this study also showed that behavioral trial type and animal performance deterministically shaped astrocytes’ spatial and temporal response characteristics. Astrocyte population responses were found to encode the animals’ decision, reward, and sensory properties. Importantly, astrocyte responses seemed to carry behaviorally relevant information depending on and complementary to neuronal coding.
FIGURE 2.
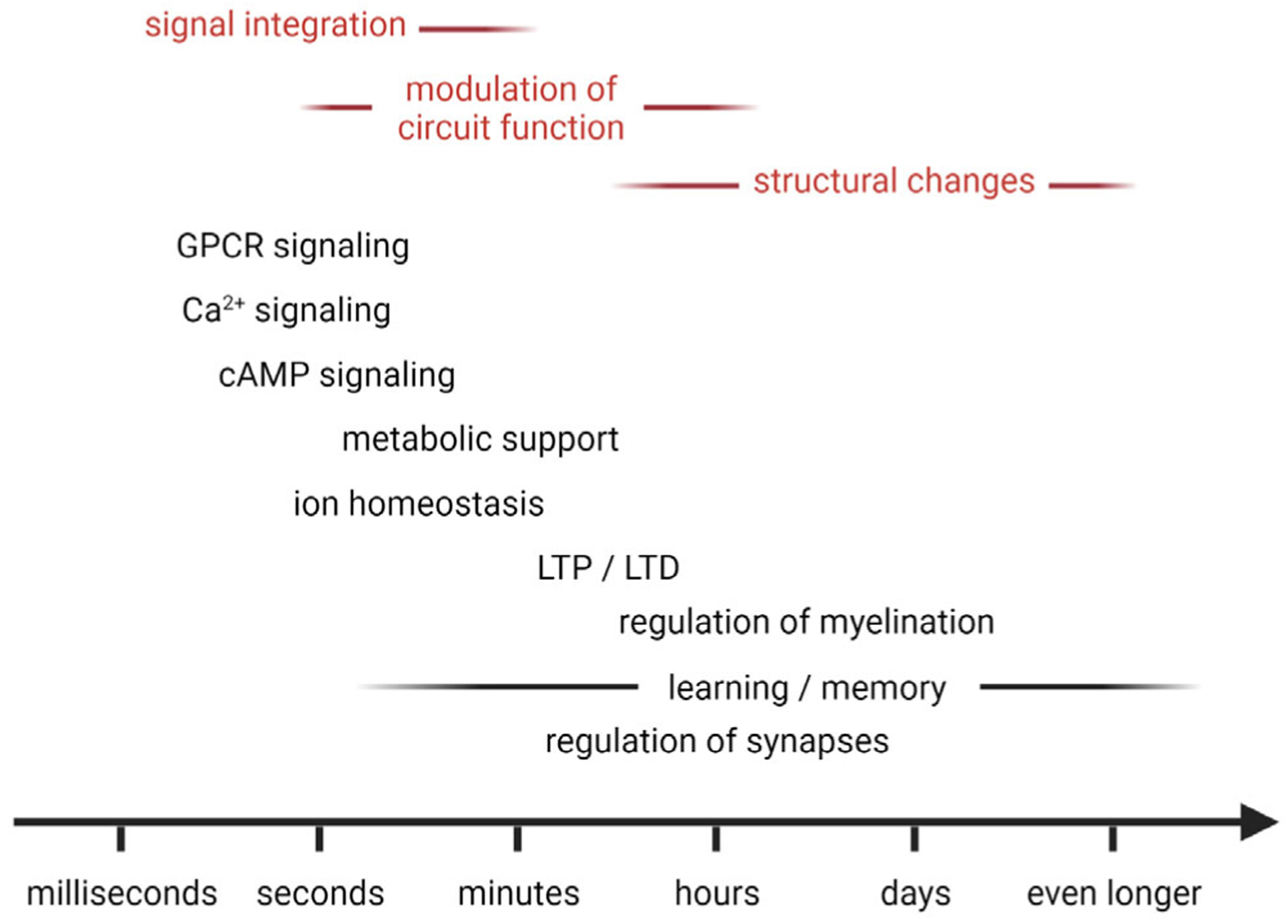
Astrocytes regulate neural circuits on various timescales. Astrocyte transients (e.g., calcium, cAMP) tend to be slow (sub-seconds to minutes), resulting from molecular signal integration in their environment. Similarly, astrocyte modulation of neural circuit function (e.g., through regulation of extracellular ion homeostasis, neuroactive factor release, or metabolic support) typically occurs on the seconds to minutes timescale, whereas morphological and gene expression-dependent changes (e.g., perisynaptic process structure, myelin regulation) influence neural circuit plasticity and function on the minutes to days or even longer timescale. Notably, the spatiotemporal dynamics of astrocytes’ functional signals, their spatial arrangement, gene expression, and coupling suggest that these cells serve complementary roles to neurons (see text for more details). Abbreviations: cAMP, cyclic adenosine monophosphate; GPCR, G-protein coupled receptor; LTD, long-term depression; LTP, long-term potentiation
3.1.2 |. Select insights from spinal cord recordings
To date, most spinal cord studies involving astrocytes have been performed in reduced preparations or anesthetized animals, mainly due to the technical difficulty of obtaining stable recordings from this CNS region during animal behavior, which typically results in large amplitude and nonlinear tissue displacements (Nelson et al., 2019). The few studies that have been conducted in awake mice demonstrate that anesthesia powerfully suppresses spinal neuron and astrocyte activity and that sensorimotor-driven astrocyte excitation is complex and regionally heterogeneous (Sekiguchi et al., 2016; Shekhtmeyster, Carey, et al., 2021; Shekhtmeyster, Duarte, et al., 2021). For example, using quantitative sensory and motor assays together with in vivo calcium imaging, superficial (i.e., lamina I–II) dorsal horn astrocytes were found to respond to innocuous sensory stimuli (tail pinch) with asynchronous increases in the frequency but not amplitude or duration of their microdomain transients. In contrast, noxious stimuli evoked concerted calcium transients throughout the superficial astrocyte syncytium within and across spinal segments, both in the presence and absence of a flight response (Sekiguchi et al., 2016; Shekhtmeyster, Duarte, et al., 2021). Using the same standardized behavioral assays, local neural activity was measured and compared to astrocyte excitation using separate groups of mice (Sekiguchi et al., 2016; Shekhtmeyster, Carey, et al., 2021). As expected, innocuous sensory stimuli of increasing intensity elevated the number and amplitude of responding excitatory neurons, suggesting that astrocytes’ microdomain transients are driven primarily by synaptic activity. Excitatory neural activity continued to rise monotonically for more intense and noxious stimuli, implying that the spatially and temporally distinct astrocyte syncytium responses in this stimulus regime are mediated not only by synaptic but also volume transmission (e.g., neuromodulator release from descending projection fibers).
Recent studies have also indicated that spinal cord astrocytes are regionally heterogeneous (Kronschläger et al., 2021; Molofsky et al., 2014; Shekhtmeyster, Carey, et al., 2021), that this heterogeneity is functionally relevant (Kelley et al., 2018; Kohro et al., 2020) and associated with distinct spatiotemporal excitation patterns. For example, using translaminar imaging in behaving mice with chronically implanted microprisms, astrocytes in sensory and premotor areas of the spinal dorsal horn were found to respond to noxious mechanical stimuli and locomotion in a lamina-specific manner (Shekhtmeyster, Carey, et al., 2021). In contrast, Tac1-expressing neurons, involved in pain processing, responded to the same noxious stimuli with calcium spiking in sensory regions but did not respond to locomotion, suggesting that lamina-specific astrocyte activity depends on the neuronal cell type. How, in turn, this astrocyte excitation may control neuronal activity on single-cell, circuit, and behavioral levels remains to be determined.
Studies focused on pain signaling also showed that Hes5-positive astrocytes in superficial spinal laminae gate descending noradrenergic commands that control mechanosensory behavior (Kohro et al., 2020). Additionally, dorsal horn astrocytes were shown to gate peripheral nociceptive signals. Aβ-fiber activation induced calcium excitation in dorsal horn astrocytes, in turn mediating long-term depression in neurokinin 1 receptor-positive projection neurons in an adenosine A1 receptor-dependent manner, thereby resulting in pain inhibition (Xu et al., 2021). Furthermore, dorsal horn astrocytes were shown to play essential roles in amplifying chronic itching through STAT3- and lipocalin-2-dependent mechanisms following their transition to a reactive state (Shiratori-Hayashi et al., 2015).
Similarly, ventral astrocytes seem to play essential roles in controlling motor behavior. Studies on various central pattern generator (CPG) circuits suggest that bi-directional communication between astrocytes and neurons constrains CPG output within an optimal operating range and in a state-dependent manner (Broadhead & Miles, 2021; Nelson et al., 2019). Multiple mechanisms (e.g., ATP, S100β, K+, transporter expression/activity) have been proposed to mediate this inhibitory feedback enabling motor actions to be tailored to the animal’s needs.
In summary, a deeper and more complete understanding of astrocytes’ contribution to neural circuit function and animal behavior may be achieved by systematically measuring, manipulating, quantifying, and modeling astrocytes’ functional status in the context of standardized and quantitative behavioral tasks. The use of such behavioral assays for relating molecular to cellular dynamics and animal behavior is all the more important given that the various dynamics may be challenging to measure simultaneously (e.g., due to a lack of appropriate indicator color variants). Nevertheless, stochastic changes in experimental conditions and behavioral state can create inherent variability across recordings and sessions, complicating the identification of relevant cellular and molecular mechanisms. Outwardly, identical behavioral states can differ in the internal state (e.g., attention), leading to differences in how local microcircuits and their constituents respond to sensory input (e.g., differences in tonic neuromodulator levels). One approach to deal with this intrinsic variability is to gather as much data as possible, including the animal’s internal state, external environment, and performance on the task, such that unbiased computational approaches can be used to identify similar trials that can then be clustered and compared. Another approach is to turn to statistical modeling and machine learning approaches capable of identifying or accounting for dependent and hidden variables.
3.2 |. Computational approaches
How do astrocytes integrate neuronal (and non-neuronal) signals? How do they regulate neural circuits on both short and long timescales? Computational analysis and modeling approaches are essential in answering these questions.
3.2.1 |. Computational analysis
Astrocyte signal (e.g., calcium) fluctuations occur in cell bodies, major branches, and distal processes/microdomains. Multiple types of events can occur within a single astrocyte due to intracellular signal compartmentalization. Different events can have diverse footprints and heterogeneous time courses. Signals can also propagate within or between astrocytes. This propagation can be heterogeneous in terms of initiating position, speed, direction, and spread, and the propagation patterns can be distinct for different events in the same region. Accurately quantifying events that change size or location across time, propagate within or across cells, and spatially overlap with other signals is difficult with the traditional region of interest (ROI)-based methods. Recently, probabilistically principled, unbiased, and data-driven frameworks, such as Astrocyte Quantification and Analysis (AQuA), have been developed (Table 4). These frameworks use advanced machine learning approaches and consider the specific properties of astrocyte signals, promising to more accurately and comprehensively quantify the full spectrum of astrocyte activity patterns in an unbiased manner. Similar approaches may allow quantification of extracellular signals in relation to astrocyte activity, ideally in all spatial dimensions. Recent studies have also suggested that ionic (e.g., calcium) baseline changes are physiologically relevant (King et al., 2020; Tufail et al., 2017). Quantifying and modeling these changes may be particularly important in various disease settings.
TABLE 4.
Computational tools for quantifying astrocyte or neuronal activitya
| Select tools for quantifying astrocyte excitation | ||
|---|---|---|
| Name | Properties | References |
| AQuA |
|
Wang et al., 2019 |
| CaSCaDe |
|
Agarwal et al., 2017 |
| GECIquant |
|
Venugopal et al., 2019 |
| N/A |
|
Bojarskaite et al., 2020 |
| Select tools for quantifying neuronal activity | ||
| CalmAn |
|
Giovannucci et al., 2019 |
| CASCADE |
|
Rupprecht et al., 2021 |
| EXTRACT |
|
Inan et al., 2021 |
| EZcalcium |
|
Cantu et al., 2020 |
| Minian |
|
Dong, Mau, et al., 2021 |
| MIN1PIPE |
|
Lu et al., 2018 |
| OnACID-E |
|
Friedrich et al., 2021 |
| Suite2p |
|
Pachitariu et al., 2017 |
This table does not cover denoising algorithms (e.g., Deep Interpolation) (Lecoq et al., 2021), which might enhance signal extraction from noisy data.
Complementary to the analysis of astrocytes’ complex spatiotemporal activity will be the accurate quantification of their three-dimensional structure (e.g., territory, arborization, microdomain structure) and intracellular organelle distribution (e.g., mitochondria, ER) with respect to their surrounding environment (e.g., excitatory and inhibitory synapses, neuromodulatory fibers, neuronal cell types). For example, astrocytes are in close contact with various types of neurons or neuronal structures and exhibit diverse morphologies across layers and tissue regions (Eilam et al., 2016; Lanjakornsiripan et al., 2018; Refaeli et al., 2021). The complex morphology and cellular environment place fundamental constraints on what kinds of signals astrocytes can sense, how they can be integrated locally and globally, and which types of commands they can transmit to neurons. The restricted intracellular space of astrocyte leaflets, for example, limits intracellular signal diffusion, enabling them to exert localized effects. Therefore, accurate anatomical data is crucial for model development as it provides fundamental geometric constraints on astrocytes’ functional dynamics and output.
3.2.2 |. Predictive modeling
Studies over the past decades have indicated that astrocytes actively respond to local and projection neuron activity, first by modulating cytosolic ion concentrations (e.g., calcium, sodium), then secreting signals, and ultimately modifying their gene expression pattern and morphology. Thus, while neurons are unarguably essential players in information processing, astrocytes need to be accounted for and integrated into circuit models to understand better how a given network operates or dysfunctions. Indeed, it is appealing to consider astrocytes and neurons as a unified circuit (“astrocyte-neuron assembly”) since they participate in information processing in complementary manners in terms of both temporal and spatial domains. Neurons communicate on the millisecond timescale locally and over long distances and at defined release sites. Astrocytes operate on the seconds or even longer timescale (Figure 2), transmitting signals more locally and diffusely. In silico approaches provide an opportunity to incorporate these multilevel and multiscale data into a coherent framework and make testable predictions. Among computational models that include astrocytes, four general types can be discerned (Manninen et al., 2018): single astrocyte models, astrocyte network models, neuron-astrocyte synapse models, and neuron-astrocyte network models. Over time, these models have become increasingly complex, some incorporating astrocytes’ intricate three-dimensional architecture and multiscale dynamics (Savtchenko et al., 2018).
In silico approaches offer many advantages. They allow investigation of questions difficult if not impossible to address in live animals in a tightly controlled manner. For example, guided by omic approaches, they can help assess how (patho-)physiological astrocyte variations affect various dynamic properties (e.g., how regional differences in IP3 receptor expression/distribution influence astrocyte calcium transients, or how disease-associated changes in K+ channel expression alter spatial buffering and neuronal excitability) (Denizot et al., 2019). Modeling also allows critical evaluation of experimental interventions (e.g., how different calcium indicator/exogenous buffer concentrations alter astrocyte compartment or network transients and, thereby, downstream signaling such as transmitter release) (Semyanov et al., 2020). Additionally, plausible mechanisms contributing to experimental observations can be explored (e.g., whether and how different astrocyte signaling pathways contribute to neuronal synchronization during attentional shifts or epileptic discharges). In silico approaches are also essential for generating new hypotheses or making experimentally testable predictions about astrocytes’ supportive or augmenting role in neural circuit function (e.g., their calcium elevations transiently store neuronal activity traces allowing them to determine optimal energy resource distribution or modulate specific synaptic connections upon recurrence or recall) (Gordleeva et al., 2019; Gordleeva et al., 2021; Kastanenka et al., 2020).
However, the power and accuracy of in silico models critically depend on information about dependent variables and experimental data, and while incorporating more variables can increase model flexibility, it also bears the risk of overfitting. Variables to consider may include:
The types and concentrations of neurotransmitters, neuromodulators, and neuropeptides (co-)released into the extracellular space (e.g., dopamine during rewarded behavioral task trials),
Their spatiotemporal characteristics (e.g., synaptic or volume transmission),
Their concentration-dependent action on relevant receptors (e.g., α1 or α2 receptor activation during high- or low-extracellular norepinephrine concentrations, respectively),
Nonlinear interactions between signaling pathways (e.g., glutamate and norepinephrine),
Constraints on astrocyte excitation (e.g., refractory duration to replenish intracellular stores following calcium depletion),
Astrocytes’ dynamic morphology and coupling (e.g., activity-dependent alterations),
Astrocyte heterogeneity (e.g., layer-specific differences in functional properties),
The potential influence of environmental or blood-borne factors (e.g., pO2 and pH),
Metabolic and circadian factors, and
Network state (e.g., asleep, awake, attentive).
Modeling these multilevel, multiscale interactions is, without a doubt, a daunting task and necessarily phenomenological at early stages. Perhaps the first step to tackle this challenge is to construct (or build on existing) simplified models that recapitulate defined sets of in vivo observations (e.g., astrocytes’ nonlinear calcium responses to neurotransmitter and neuromodulator receptor co-activation). As more in vivo data becomes available (e.g., from direct neuromodulator signaling measurements), these models will need to be iteratively updated. Models that recapitulate different aspects of neuron-astrocyte interaction will then need to be integrated and extended to larger cellular networks. Gradually, these models are expected to become more predictive and identify potential hidden factors that influence astrocyte excitation (e.g., blood-borne factors), thereby guiding future experiments. Measurements from different CNS areas or using behavioral task variations to interrogate a given circuit will help determine conserved or regionally unique mechanisms. Behavioral assays should allow control of signaling pathway recruitment (e.g., dopamine release) and quantitative readout of animal state, behavior, or the effect of targeted pathway alterations (e.g., by AAV-mediated interventions).
In summary, new and improved data analysis tools and mathematical models promise to provide the sorely needed quantitative description of how astrocytes spatially and temporally integrate molecular signals in their environment and how the intracellular dynamics that underlies this integration leads to functional outputs that modulate synaptic function or network state on various timescales.
4 |. CHALLENGES IN INTERPRETING ASTROCYTE FUNCTION
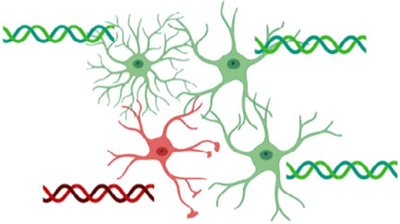
4.1 |. Astrocyte heterogeneity, interspecies diversity, and their relevance to neural circuit function and animal behavior
Astrocytes are a heterogeneous cell population. They differ in their structural and functional properties between, but also within CNS regions (Clarke et al., 2021; Emsley & Macklis, 2006; Farmer & Murai, 2017; Khakh & Deneen, 2019; Matias et al., 2019; Matyash & Kettenmann, 2010; Oberheim et al., 2012; Zhang & Barres, 2010). Various metrics have been defined to characterize cellular heterogeneity. Metrics commonly used to define neuronal subpopulations include the location (e.g., brain region or cell layer), morphology (e.g., local or projection neuron, axonal or dendritic arborization), gene expression (e.g., parvalbumin, somatostatin, vasoactive intestinal peptide), biochemical (e.g., neurotransmitter usage), and physiological properties (e.g., action potential firing properties). Similar metrics have been used to describe astrocyte heterogeneity (Table 5). Some overlap exists between the resulting cell populations.
TABLE 5.
Examples of astrocyte heterogeneity and diversity
| Heterogeneity/diversity metric | Example references |
|---|---|
| Location and morphology | |
|
Chai et al., 2017; Köhler et al., 2021; Lanjakornsiripan et al., 2018; Matyash & Kettenmann, 2010; Reichenbach & Wolburg, 2012; Somjen, 1988 |
| Gene expression profilea | |
|
Batiuk et al., 2020; Chai et al., 2017; Doyle et al., 2008; Gokce et al., 2016; John Lin et al., 2017; Lanjakornsiripan et al., 2018; Morel et al., 2017; Zeisel et al., 2015 |
| Biochemical and physiological properties | |
|
Chai et al., 2017; Fernández-Moncada et al., 2021; Herde et al., 2020; Hirrlinger et al., 2008; Kelley et al., 2018; Köhler et al., 2021; Köhler et al., 2018; Kronschläger et al., 2021; Miller et al., 2019; Oheim et al., 2018; Olsen et al., 2007; Theis & Giaume, 2012 |
| Species | |
|
Freeman & Rowitch, 2013; Nagai, Yu, et al., 2021 |
See Hirbec et al., 2020 for a recent overview on scRNAseq studies on glial cells.
Astrocyte heterogeneity is functionally relevant for the control of neural circuit function. For instance, gene expression differences across cortical layers include genes involved in synaptic regulation (e.g., Sparc, Mertk), neurotransmitter uptake (e.g., Slc1a3/GLAST), and astrocyte coupling (e.g., Gja1/Cx43) (Lanjakornsiripan et al., 2018). Intriguingly, these gene expression-defined “astrocyte layers” are distinct from cortical neuronal layers and differ between cortical areas (Bayraktar et al., 2020). Astrocyte populations support synaptogenesis in ways that depend on their gene expression profiles (John Lin et al., 2017). Hippocampal and striatal astrocytes differ in their K+ channel, gap junction protein (e.g., Gja1/Cx43), glutamine synthetase, calcium channel, and pump expression (Chai et al., 2017). The latter contributes to differences in calcium signaling (Chai et al., 2017). Given the central role of calcium excitation in astrocyte physiology, these disparities may be partly responsible for regional differences in gliotransmission, neuromodulation, structural dynamics, and blood flow regulation (Clarke et al., 2021; Khakh & Deneen, 2019; Semyanov et al., 2020). This heterogeneity may extend down to the subcellular level, with each astrocyte microdomain potentially performing different functions depending on its proximity to surrounding neurons or non-neuronal cells. Regional heterogeneity may also cause differences in injury responses, with the injury itself promoting astrocytes’ functional differentiation (Sofroniew, 2020). In summary, astrocyte heterogeneity will likely need to be considered in models of neural circuit function. Given the existing tools, determining exactly how local and regional astrocyte heterogeneity contributes to neural circuit function will be a significant experimental challenge.
The observation of astrocyte heterogeneity on different spatial scales raises the question about its origin. White matter astrocytes in the spinal cord are generated by distinct developmental domains and express specific transcription factor combinations that establish their positional identity (Hochstim et al., 2008). The regional identity of astrocytes in the brain is defined by region-specific transcriptional and epigenetic signatures, suggesting that they are derived from nucleus-specific progenitors (Herrero-Navarro et al., 2021). Additionally, the interaction of astrocytes with neighboring neurons contributes to their heterogeneity, for example, through sonic hedgehog signaling (Farmer et al., 2016; Lanjakornsiripan et al., 2018; Stogsdill et al., 2017). Neuronal activity (e.g., glutamatergic signaling) promotes astrocyte functional and morphological maturation (Morel et al., 2014). An instructive role for neuronal cues on astrocyte properties was also shown in Satb2 and Reeler mutant mice, in which changes in neuronal layering are accompanied by alterations in astrocyte layering (Bayraktar et al., 2020). Astrocyte heterogeneity, therefore, appears to result from developmental programs and local environmental adaptations (see Clarke et al., 2021 for an in-depth review) and may be relevant to disease.
In addition to astrocyte heterogeneity within a given organism’s CNS, astrocytes show diversity across species (see Nagai, Yu et al., 2021 for a comparison between Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, and Mus musculus). This raises the crucial yet experimentally challenging question of how astrocytes modulate neural circuit function and behavioral characteristics in humans. Human and experimental animal model astrocytes differ in many properties (Bedner et al., 2020; De Majo et al., 2020). Astrocyte processes cover larger volumes in the human brain and contact ~10–20 times as many synapses as in rodents (Oberheim et al., 2006). The astrocytic domain overlap is also more extensive in humans, and two unique types of astrocytes are present in the cortex (interlaminar and varicose projection astrocytes, potentially involved in long-distance communication) (Colombo & Reisin, 2004; Oberheim et al., 2009). Human astrocytes also show unique gene expression profiles in health and disease (Nagai, Yu, et al., 2021; Zhang et al., 2016). How these differences affect human neural circuit function, behavior, and disease remains to be determined.
One approach to studying human astrocyte diversity and its impact on brain circuit physiology has been to transplant stem or progenitor cell-derived human astrocytes into the rodent brain (Chen et al., 2015; de Majo et al., 2020; Goldman et al., 2015). A similar approach has been used to study primate-specific interlaminar astrocytes in mice (Padmashri et al., 2021). Strikingly, grafting human astrocytes into the mouse forebrain enhanced synaptic plasticity and learning (Han et al., 2013). Transferring human induced pluripotent stem cell (iPSC)-derived glial cells from schizophrenia patients into the mouse brain resulted in astrocytes with abnormal morphology, hypomyelination, and behavioral deficits resembling a schizophrenia phenotype. These findings suggest that human glial cells contribute to disease-specific circuit alterations (Windrem et al., 2017). Models that closely recapitulate the human tissue environment will be essential to understand better how astrocytes contribute to human brain and spinal cord function.
These examples of astrocyte heterogeneity and interspecies diversity highlight that we are just beginning to understand how the CNS’ complex cytoarchitecture and signaling control animal behavior. Vital questions that will likely drive research in the coming years include:
To what extent do current astrocyte classifications capture physiologically relevant sub-populations? Which other or additional parameters might help define functionally homogeneous populations to study their impact on neural circuits?
Which differences between astrocytes are crucial for controlling neural circuit function?
How does astrocyte heterogeneity due to cell-intrinsic properties or neuronal heterogeneity influence neural circuit function?
To what extent does astrocyte heterogeneity and diversity support neuronal function or serve a complementary role (e.g., control neural circuit activity or network state during animal behavior)?
How functionally heterogeneous are astrocytic microdomains? How does this heterogeneity relate to local neural circuits?
What are the conserved functions between human and model organism astrocytes? How are potential functional differences behaviorally relevant?
Tackling these questions will likely require developing novel tools and experimental approaches (see above). While it is generally accepted that brain or spinal cord function cannot be understood without taking neuronal diversity into account, this most likely applies to astrocytes as well.
4.2 |. Astrocyte metabolism and its relationship to animal behavior
Metabolic cooperation between neurons and astrocytes is perhaps one of the most intuitive examples of the functional interdependency of these cell types in health and disease. Various kinds of metabolic interactions have been identified, including the glutamate-glutamine cycle (Bak et al., 2006; Martinez-Hernandez et al., 1977), glutathione metabolism (Dringen et al., 2000; Hirrlinger & Dringen, 2010), and the astrocyte-neuron-lactate shuttle (ANLS) (Pellerin & Magistretti, 1994; see Bak & Walls, 2018; Barros & Weber, 2018 for a critical discussion of the ANLS). Recent evidence suggests that astrocyte metabolism does not only contribute to metabolic and energetic homeostasis of the tissue they reside in but plays a crucial role in behavioral regulation (Bonvento & Bolaños, 2021). Astrocyte glycogen metabolism, for example, is essential for memory consolidation in chickens (Gibbs, 2016; Gibbs et al., 2006). Cortical activity upon arousal induces astrocyte lactate release via adrenergic signaling in mice (Zuend et al., 2020). Astrocyte glycogenolysis and lactate transport from astrocytes to neurons, induced by stimulation of astrocytic β2-adrenergic receptors, is necessary for memory formation and consolidation in the rat hippocampus (Gao et al., 2016; Netzahualcoyotzi & Pellerin, 2020; Newman et al., 2011; Suzuki et al., 2011). Lactate modulates NMDA receptor-mediated signaling, plasticity-related gene expression, and learning-induced mRNA transcription in neurons (Descalzi et al., 2019; Yang et al., 2014). Notably, lactate can fulfill diverse roles in the target cells, including as an energy substrate or modulating cellular redox state, by directly activating (e.g., GPR81/HCAR1) or modulating receptors (e.g., NMDARs) and affecting various signaling (e.g., calcium) (Barros, 2013). Neuronal activity-related and animal stress-induced protein lactylation were also recently shown to depend on lactate availability (Hagihara et al., 2021), extending the range of lactate’s functions to epigenetic regulation. Additional roles for lactate (and other metabolites) are likely.
Mitochondrial reactive oxygen species (mtROS) also play critical roles in astrocyte metabolism and metabolic cooperation with neurons. Reduction of mtROS specifically in astrocytes leads to mouse behavioral changes in the open-field test and cognitive deficits in a novel object recognition test (Vicente-Gutierrez et al., 2019). Activation of type 1 cannabinoid receptors (CB1Rs) on astrocyte mitochondria results in reduced mtROS production, glycolysis, and lactate release, neuronal energy stress, and, consequently, impaired social behavior (Jimenez-Blasco et al., 2020), highlighting the powerful effects of these receptors on cell-type-specific metabolism, astrocyte-neuron assembly function, and animal behavior.
Another essential pathway involves the amino acid L-serine. This precursor of the NMDA receptor co-agonists D-serine and glycine is synthesized in astrocytes from the glycolysis intermediate 3-phosphoglycerate via the phosphorylated pathway. Inhibition of this pathway alters NMDA receptor signaling and LTP in the mouse hippocampus and spatial memory (Le Douce et al., 2020; Neame et al., 2019). Notably, L-serine and D-serine levels are reduced in a mouse model of Alzheimer’s disease, and deficits in synaptic plasticity and spatial memory were rescued by D-serine application (Le Douce et al., 2020). Astrocytic CB1Rs contribute to regulating the extracellular D-serine concentration, and deletion of these receptors in astrocytes results in impaired LTP and object recognition memory (Robin et al., 2018).
Recent evidence also suggests that astrocytes metabolize fatty acids via β-oxidation and that this pathway is involved in metabolic cooperation between astrocytes and neurons (Eraso-Pichot et al., 2018; Fecher et al., 2019; Ioannou et al., 2019; Konttinen et al., 2019; Schirmeier et al., 2021; Timper et al., 2020). Furthermore, saturated lipids have been identified as a neurotoxic factor released from astrocytes under pathological conditions (Guttenplan et al., 2021). Whether and how astrocytes’ fatty acid metabolism contributes to regulating neural circuit function and animal behavior remains to be investigated.
Together, these examples demonstrate that astrocyte metabolism and animal behavior mutually affect one another (Alberini et al., 2018; Bonvento & Bolaños, 2021). Astrocyte metabolism plays a vital role in learning, memory, and behavior. Its regulation occurs on many different and likely interconnected levels. Elucidating metabolic processes in astrocyte-neuron assemblies will be necessary for a deeper understanding of how they regulate neural circuit function and animal behavior.
4.3 |. Beyond astrocytes: The contribution of other glial cells to neural circuit function and animal behavior
Accumulating evidence indicates that astrocytes are not the only resident non-neuronal cells capable of modulating neural circuit function and animal behavior on various timescales. Microglia and oligodendroglial lineage cells (OLCs), either alone or in close coordination with astrocytes, can also modulate the structural and functional plasticity of neurons.
Microglia are the intrinsic immune sentinels of the CNS. Their highly motile processes continually survey the environment, allowing them to detect membrane-bound signals (e.g., phosphatidylserine) (Davalos et al., 2005; Fourgeaud et al., 2016; Huang et al., 2021; Nimmerjahn et al., 2005; Tufail et al., 2017). These physical cell–cell interactions are critical for shaping neural circuits and connections during development and disease (e.g., pruning immature, inactive, or dysfunctional synapses) (Hong et al., 2016; Li & Barres, 2018; Paolicelli et al., 2011; Wilton et al., 2019). They also contribute to learning-dependent synapse formation in the mature brain, thereby modulating animal behavior on a longer timescale (Parkhurst et al., 2013). Additionally, microglia communicate with neurons (and astrocytes) through soluble factors (e.g., purinergic signaling), allowing more rapid communication (Cserép et al., 2021). For example, microglia have recently been shown to control neuronal firing frequency and synchrony via adenosine, providing an important negative feedback loop for neural circuits (Badimon et al., 2020). Thus, microglial actions may need to be considered when investigating glial control of animal behavior.
Similarly, OLCs—which include oligodendroglia precursor cells (OPCs; also referred to as NG2 cells) and oligodendrocytes (OLs)—dynamically interact with neurons, regulating their fate, myelination, and circuit properties (Bonetto et al., 2021; Foster et al., 2019; Moore et al., 2020; Suminaite et al., 2019; see also the recent GLIA Special Issue on “Plasticity of Myelinating Glia”: Fields & Richardson, 2019). OPCs receive direct synaptic input from neighboring axons (Bergles et al., 2000), and glutamatergic signaling (synaptic and non-synaptic) affects OLCs on different levels (Kula et al., 2019). Developing OLs regulate myelin sheath elongation and stability in an activity-dependent manner (Baraban et al., 2018; Krasnow et al., 2018). Myelination of axons in the adult brain is also controlled by neuronal activity and has been shown to modulate motor learning, motor performance, sound localization, and memory persistence (Bacmeister et al., 2020; Ford et al., 2015; Gibson et al., 2014; McKenzie et al., 2014; Pan et al., 2020; Wang et al., 2020; Xiao et al., 2016). OLs adapt their metabolite supply according to axonal activity (Saab et al., 2016). Conversely, nutritional signals regulate OL proliferation, myelin regulatory factor-dependent perineuronal net remodeling, and tuning of the circuit that controls food intake and weight gain in the mediobasal hypothalamus (Kohnke et al., 2021). Adaptive myelination is also critical for spike-timing-dependent plasticity in neural circuits (Monje, 2018; Suminaite et al., 2019), and activity-dependent, brain region-specific oligodendrogenesis may be essential for episodic memory formation (Barboza et al., 2021).
While these examples show that microglia and OLCs dynamically modulate specific neural properties, the close interaction between non-neuronal cell types suggests an even more complex scenario (Figure 3). For example, astrocytes communicate with OLs (e.g., via gap junction-coupled pan-glial networks) (Griemsmann et al., 2015; Theis & Giaume, 2012) and contribute to myelination through lipids (e.g., cholesterol) and other signaling molecules (Camargo et al., 2017; Kıray et al., 2016; Molina-Gonzalez & Miron, 2019). Perinodal astrocytes control myelin thickness, nodal gap length, axonal excitability, and conduction velocity (Dutta et al., 2018; Lezmy et al., 2021). Microglia, too, communicate with OLs to affect myelination (Bar & Barak, 2019). Astrocytes closely interact and coordinate with microglia (e.g., synaptic pruning), and disruption of this signaling has been linked to various disease phenotypes (Jha et al., 2019; Liddelow et al., 2020). Microglia can modulate astrocyte gliotransmission by regulating VAMP2 proteins (Takata-Tsuji et al., 2021). Therefore, current evidence suggests that neurons, astrocytes, microglia, and OLCs co-operate multi-laterally to regulate neural circuit function. If and how other cell types (e.g., tanycytes, ependymal cells, and pericytes) also contribute to this process remains to be investigated. Establishing a comprehensive view of these complex interactions underlying precise control of neural circuit function on various timescales in health and disease will be a significant challenge for future studies.
FIGURE 3.
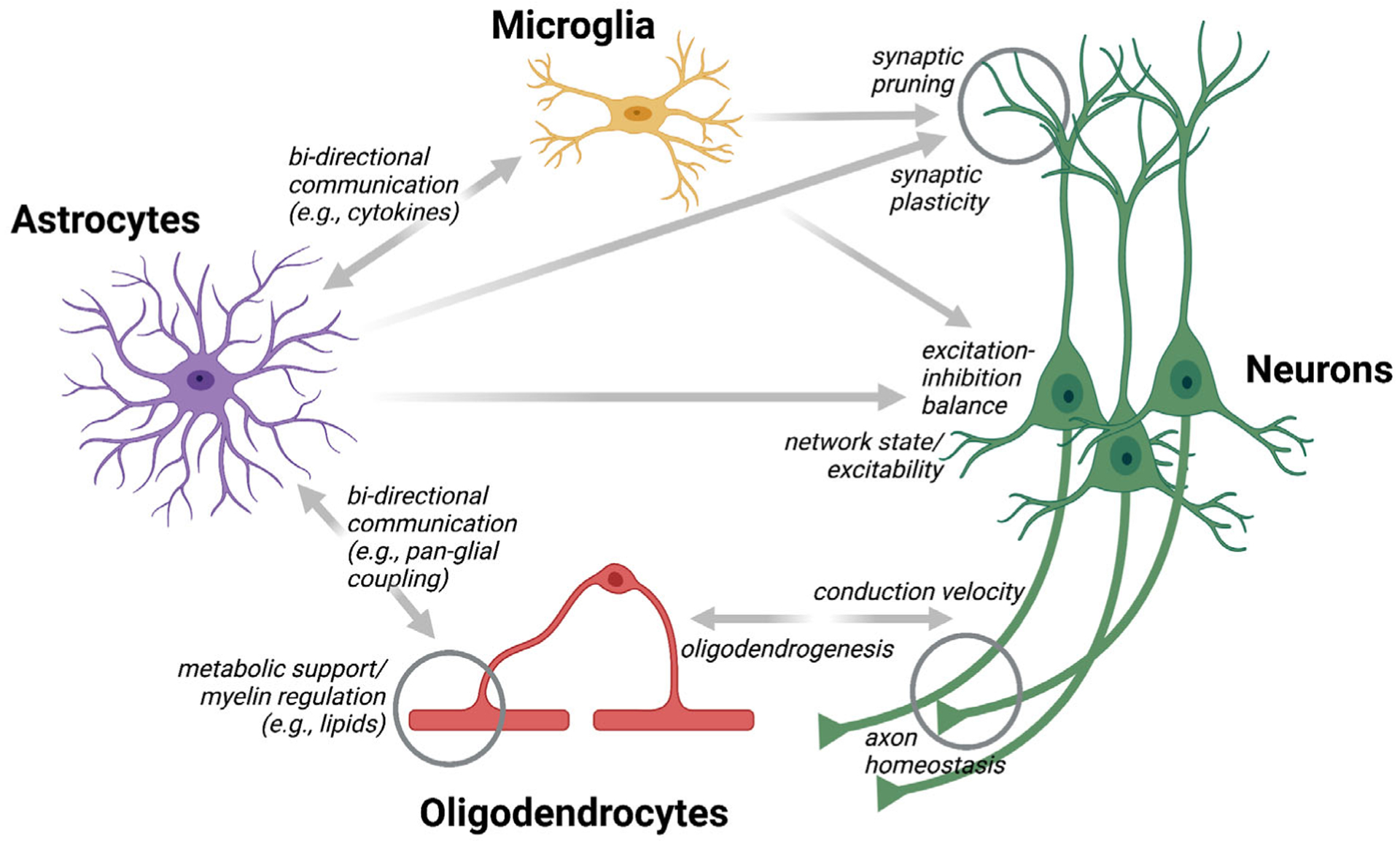
Astrocytes regulate neural circuits directly and indirectly. Astrocytes can modulate neural circuit function directly through different mechanisms (see Figure 1). Microglia have also been shown to control neuronal synapse number and activity on various timescales. Similarly, oligodendrocytes can regulate neurons’ axonal properties (e.g., conduction velocity) in an activity- and behavior-dependent manner. Astrocytes bidirectionally communicate with and regulate both microglia and oligodendrocytes (e.g., through diffusible messengers and physical interactions), allowing them to influence neural circuits indirectly and in a spatially and temporally distinct manner than their direct routes
5 |. CONCLUDING REMARKS
Astrocytes play essential roles in neural circuit function, including the control of extracellular ion homeostasis, transmitter recycling, metabolic supply, and synaptic plasticity. Yet, fundamental questions regarding their role in behavior-related neural circuit regulation remain unresolved. How do they integrate the diverse neuronal signals associated with animal behavior? How do they modulate neural circuit function and structure on behaviorally relevant timescales in response to these signals (Figure 1)? How does impairment of these functions contribute to disease? How can astrocytes’ beneficial roles be boosted in disease to protect CNS cells from damage and promote regeneration (Pekny et al., 2016)? Addressing these and other questions (Box 1) will likely require a multiscale approach, including functional, anatomical, molecular, and omic analyses and theoretical modeling. Novel indicators, actuators, genetic targeting approaches, quantitative behavioral assays, and computational methods are beginning to provide much-needed answers to these questions (Tables 1–4). However, integrating astrocytes into existing neural circuit models will not be easy. Astrocytes have unique properties presenting unique challenges. They regulate neural circuits on various spatial and temporal scales (from sub-micron to millimeters and from sub-seconds to days or more) (Figure 2), are functionally heterogeneous (Table 5), and closely coordinate with other non-neuronal cells (Figure 3). Tools to contend with astrocytes’ functional complexity and 3D arrangement are still in their infancy but will be critical for uncovering these cells’ sensitively orchestrated actions and adequately integrating them into predictive models.
Elucidating the full spectrum of astrocytes’ dynamic functions will also be vital for optimal disease treatment. Fundamental questions in this area include: How do astrocytes initiate or perpetuate pathophysiological CNS changes? How can these changes be controlled to minimize CNS damage and promote regeneration? How does astrocyte heterogeneity contribute to region-specific disease characteristics? How does the dynamic interplay of astrocytes with other non-neuronal cells, such as microglia, OLs, and circulating or infiltrating nonresident immune cells, influence various disease stages (Liddelow et al., 2020; Nutma et al., 2020)? Answering these fundamental questions will likely require a multidisciplinary approach involving researchers from neuroscience, immunology, pharmacology, clinical, and engineering disciplines.
In conclusion, uncovering the molecular, cellular, and circuit mechanisms by which astrocytes influence neural circuit dynamics promises to transform our understanding of how astrocyte-neuron assemblies shape brain and spinal cord computations and complex animal behaviors. Additionally, they will provide the basis for conceptually novel approaches to combat inflammatory and neurodegenerative conditions for which only limited or suboptimal treatment options exist.
ACKNOWLEDGMENTS
The authors wish to apologize to all colleagues whose important work was not directly cited due to the review period or space limitations, especially in the tables. Respective references and more details on the cited work can be found in the referenced topical reviews. We thank Guoqiang Yu for allowing the reuse and adaptation of Figure 1, produced as part of a joint U19 grant. All figures were created with Biorender.com. This work was funded, in part, by grants from the Deutsche Forschungsgemeinschaft (DFG) (HI1414/6-1, within priority program 1757, and HI1414/7-1 to Johannes Hirrlinger) and the National Institutes of Health (NIH) (U19 NS123719 and R01 NS108034 to Axel Nimmerjahn). Open access funding enabled and organized by Projekt DEAL.
Funding information
Deutsche Forschungsgemeinschaft, Grant/Award Numbers: Hi1414/6-1, Hi1414/7-1; National Institutes of Health, Grant/Award Numbers: R01 NS108034, U19 NS123719
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Agarwal A, Wu PH, Hughes EG, Fukaya M, Tischfield MA, Langseth AJ, Wirtz D, & Bergles DE (2017). Transient opening of the mitochondrial permeability transition pore induces microdomain calcium transients in astrocyte processes. Neuron, 93(3), 587–605.e7. 10.1016/j.neuron.2016.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzadeh E, Bayin NS, Qu X, Singh A, Madisen L, Stephen D, Zeng H, Joyner AL, & Rosello-Diez A (2020). A collection of genetic mouse lines and related tools for inducible and reversible intersectional misexpression. Development, 147(10), dev186650. 10.1242/dev.186650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akther S, & Hirase H (2021). Assessment of astrocytes as a mediator of memory and learning in rodents. Glia. 10.1002/glia.24099 [DOI] [PubMed] [Google Scholar]
- Alberini CM, Cruz E, Descalzi G, Bessières B, & Gao V (2018). Astrocyte glycogen and lactate: New insights into learning and memory mechanisms. Glia, 66(6), 1244–1262. 10.1002/glia.23250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ, & Eroglu C (2017). Cell biology of astrocyte-synapse interactions. Neuron, 96(3), 697–708. 10.1016/j.neuron.2017.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A, Carmignoto G, Haydon PG, Oliet SHR, Robitaille R, & Volterra A (2014). Gliotransmitters travel in time and space. Neuron, 81(4), 728–739. 10.1016/j.neuron.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster M, Dulla CG, & Diamond JS (2020). Effects of fluorescent glutamate indicators on neurotransmitter diffusion and uptake. eLife, 9, e54441. 10.7554/elife.54441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacmeister CM, Barr HJ, McClain CR, Thornton MA, Nettles D, Welle CG, & Hughes EG (2020). Motor learning promotes remyelination via new and surviving oligodendrocytes. Nature Neuroscience, 23(7), 819–831. 10.1038/s41593-020-0637-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badimon A, Strasburger HJ, Ayata P, Chen X, Nair A, Ikegami A, Hwang P, Chan AT, Graves SM, Uweru JO, Ledderose C, Kutlu MG, Wheeler MA, Kahan A, Ishikawa M, Wang Y, Loh YE, Jiang JX, Surmeier DJ, … Schaefer A (2020). Negative feedback control of neuronal activity by microglia. Nature, 586(7829), 417–423. 10.1038/s41586-020-2777-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak LK, Schousboe A, & Waagepetersen HS (2006). The glutamate/GABA-glutamine cycle: Aspects of transport, neurotransmitter homeostasis and ammonia transfer. Journal of Neurochemistry, 98(3), 641–653. 10.1111/j.1471-4159.2006.03913.x [DOI] [PubMed] [Google Scholar]
- Bak LK, & Walls AB (2018). Crosstalk opposing view: Lack of evidence supporting an astrocyte-to-neuron lactate shuttle coupling neuronal activity to glucose utilisation in the brain. Journal of Physiology, 596(3), 351–353. 10.1113/JP274945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar E, & Barak B (2019). Microglia roles in synaptic plasticity and myelination in homeostatic conditions and neurodevelopmental disorders. Glia, 67(11), 2125–2141. 10.1002/glia.23637 [DOI] [PubMed] [Google Scholar]
- Baraban M, Koudelka S, & Lyons DA (2018). Ca2+ activity signatures of myelin sheath formation and growth in vivo. Nature Neuroscience, 21(1), 19–23. 10.1038/s41593-017-0040-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza L, Bessieres B, Nazarzoda O & Alberini C (2021). Neuronal activity–driven oligodendrogenesis in selected brain regions is required for episodic memories. bioRxiv, 2021.12.10.472135. 10.1101/2021.12.10.472135 [DOI] [Google Scholar]
- Barros LF (2013). Metabolic signaling by lactate in the brain. Trends in Neurosciences, 36(7), 396–404. 10.1016/j.tins.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Barros LF, & Weber B (2018). Crosstalk proposal: An important astrocyte-to-neuron lactate shuttle couples neuronal activity to glucose utilisation in the brain. Journal of Physiology, 596(3), 347–350. 10.1113/JP274944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk MY, Martirosyan A, Wahis J, Vin F. d., Marneffe C, Kusserow C, Koeppen J, Viana JF, Oliveira JF, Voet T, Ponting CP, Belgard TG, & Holt MG (2020). Identification of region-specific astrocyte subtypes at single cell resolution. Nature Communications, 11(1), 1220. 10.1038/s41467-019-14198-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Bartels T, Holmqvist S, Kleshchevnikov V, Martirosyan A, Polioudakis D, Ben Haim L, Young AMH, Batiuk MY, Prakash K, Brown A, Roberts K, Paredes MF, Kawaguchi R, Stockley JH, Sabeur K, Chang SM, Huang E, Hutchinson P, … Rowitch DH (2020). Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nature Neuroscience, 23(4), 500–509. 10.1038/s41593-020-0602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargani N, & Attwell D (2017). Amines, astrocytes, and arousal. Neuron, 94(2), 228–231. 10.1016/j.neuron.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, Haslinger A, Lie DC, Beckers J, Yoder B, Irmler M, & Götz M (2010). In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell, 7(6), 744–758. 10.1016/j.stem.2010.11.017 [DOI] [PubMed] [Google Scholar]
- Bedner P, Jabs R, & Steinhäuser C (2020). Properties of human astrocytes and NG2 glia. Glia, 68(4), 756–767. 10.1002/glia.23725 [DOI] [PubMed] [Google Scholar]
- Beppu K, Sasaki T, Tanaka KF, Yamanaka A, Fukazawa Y, Shigemoto R, & Matsui K (2014). Optogenetic countering of glial acidosis suppresses glial glutamate release and ischemic brain damage. Neuron, 81(2), 314–320. 10.1016/j.neuron.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Berg J, Hung YP, & Yellen G (2009). A genetically encoded fluorescent reporter of ATP:ADP ratio. Nature Methods, 6(2), 161–166. 10.1038/nmeth.1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Roberts JD, Somogyi P, & Jahr CE (2000). Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature, 405(6783), 187–191. 10.1038/35012083 [DOI] [PubMed] [Google Scholar]
- Bischof H, Rehberg M, Stryeck S, Artinger K, Eroglu E, Waldeck-Weiermair M, Gottschalk B, Rost R, Deak AT, Niedrist T, Vujic N, Lindermuth H, Prassl R, Pelzmann B, Groschner K, Kratky D, Eller K, Rosenkranz AR, Madl T, … Malli R (2017). Novel genetically encoded fluorescent probes enable real-time detection of potassium in vitro and in vivo. Nature Communications, 8(1), 1422. 10.1038/s41467-017-01615-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojarskaite L, Bjørnstad DM, Pettersen KH, Cunen C, Hermansen GH, Åbjørsbråten KS, Chambers AR, Sprengel R, Vervaeke K, Tang W, Enger R, & Nagelhus EA (2020). Astrocytic Ca2+ signaling is reduced during sleep and is involved in the regulation of slow wave sleep. Nature Communications, 11(1), 3240. 10.1038/s41467-020-17062-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetto G, Belin D, & Káradóttir RT (2021). Myelin: A gatekeeper of activity-dependent circuit plasticity? Science, 374, eaba6905. 10.1126/science.aba6905 [DOI] [PubMed] [Google Scholar]
- Bonvento G, & Bolaños JP (2021). Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metabolism, 33(8), 1546–1564. 10.1016/j.cmet.2021.07.006 [DOI] [PubMed] [Google Scholar]
- Borden PM, Zhang P, Shivange AV, Marvin JS, Cichon J, Dan C, Podgorski K, Figueiredo A, Novak O, Tanimoto M, Shigetomi E, Lobas MA, Kim H, Zhu PK, Zhang Y, Zheng WS, Fan C, Wang G, Xiang B, … Looger LL (2020). A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. bioRxiv, 2020.02.07.939504. 10.1101/2020.02.07.939504 [DOI] [Google Scholar]
- Broadhead MJ, & Miles GB (2021). A common role for astrocytes in rhythmic behaviours? Progress in Neurobiology, 202, 102052. 10.1016/j.pneurobio.2021.102052 [DOI] [PubMed] [Google Scholar]
- Broussard GJ, Liang Y, Fridman M, Unger EK, Meng G, Xiao X, Ji N, Petreanu L, & Tian L (2018). In vivo measurement of afferent activity with axon-specific calcium imaging. Nature Neuroscience, 21(9), 1272–1280. 10.1038/s41593-018-0211-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo N, Goudriaan A, van Deijk A-LF, Otte WM, Brouwers JF, Lodder H, Gutmann DH, Nave K-A, Dijkhuizen RM, Mansvelder HD, Chrast R, Smit AB, & Verheijen MHG (2017). Oligodendroglial myelination requires astrocyte-derived lipids. PLoS Biology, 15(5), e1002605. 10.1371/journal.pbio.1002605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu DA, Wang B, Gongwer MW, He CX, Goel A, Suresh A, Kourdougli N, Arroyo ED, Zeiger W, & Portera-Cailliau C (2020). EZcalcium: Open-source toolbox for analysis of calcium imaging data. Frontiers in Neural Circuits, 14, 25. 10.3389/fncir.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reid L, Han S, Yang W, Akrouh A, & Yuste R (2019). Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell, 178(2), 447–457.e5. 10.1016/j.cell.2019.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper KB, & McCarthy KD (2006). GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Molecular and Cellular Neurosciences, 31(4), 676–684. 10.1016/j.mcn.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whitelegge JP, Coppola G, & Khakh BS (2017). Neural circuit-specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron, 95(3), 531–549.e9. 10.1016/j.neuron.2017.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Qian K, Chen W, Hu B, Blackbourn LW, Du Z, Ma L, Liu H, Knobel KM, Ayala M, & Zhang S-C (2015). Human-derived neural progenitors functionally replace astrocytes in adult mice. The Journal of Clinical Investigation, 125(3), 1033–1042. 10.1172/JCI69097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BE, Taha DM, Tyzack GE, & Patani R (2021). Regionally encoded functional heterogeneity of astrocytes in health and disease: A perspective. Glia, 69(1), 20–27. 10.1002/glia.23877 [DOI] [PubMed] [Google Scholar]
- Colombo JA, & Reisin HD (2004). Interlaminar astroglia of the cerebral cortex: A marker of the primate brain. Brain Research, 1006(1), 126–131. 10.1016/j.brainres.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Courtney CD, Sobieski C, Ramakrishnan C, Ingram RJ, Wojnowski NM, DeFazio RA, Deisseroth K & Christian-Hinman CA (2021). Evaluating the efficacy of Optoα1AR activation in astrocytes in modulating basal hippocampal synaptic excitation and inhibition. bioRxiv, 2021.01.06.425606. 10.1101/2021.01.06.425606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserép C, Pósfai B, & Dénes Á (2021). Shaping neuronal fate: Functional heterogeneity of direct microglia-neuron interactions. Neuron, 109(2), 222–240. 10.1016/j.neuron.2020.11.007 [DOI] [PubMed] [Google Scholar]
- Curreli S, Bonato J, Romanzi S, Panzeri S & Fellin T (2021). Glial place cells: Complementary encoding of spatial information in hippocampal astrocytes. bioRxiv, 2021.07.06.451296. 10.1101/2021.07.06.451296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalangin R, Drobizhev M, Molina RS, Aggarwal A, Patel R, Abdelfattah AS, Zhao Y, Wu J, Podgorski K, Schreiter ER, Hughes TE, Campbell RE & Shen Y (2020). Far-red fluorescent genetically encoded calcium ion indicators. bioRxiv, 2020.11.12.380089. 10.1101/2020.11.12.380089 [DOI] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, Patel R, Macklin JJ, Bargmann CI, Ahrens MB, Schreiter ER, Jayaraman V, Looger LL, Svoboda K, & Kim DS (2016). Sensitive red protein calcium indicators for imaging neural activity. eLife, 5, e12727. 10.7554/elife.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, Macklin JJ, Chen Y, Konnerth A, Jayaraman V, Looger LL, Schreiter ER, Svoboda K, & Kim DS (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature Methods, 16(7), 649–657. 10.1038/s41592-019-0435-6 [DOI] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, & Gan WB (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nature Neuroscience, 8(6), 752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- De Majo M, Koontz M, Rowitch D, & Ullian EM (2020). An update on human astrocytes and their role in development and disease. Glia, 68(4), 685–704. 10.1002/glia.23771 [DOI] [PubMed] [Google Scholar]
- Delzor A, Escartin C, & Déglon N (2013). Lentiviral vectors: A powerful tool to target astrocytes in vivo. Current Drug Targets, 14(11), 1336–1346. 10.2174/13894501113146660213 [DOI] [PubMed] [Google Scholar]
- Denizot A, Arizono M, Nägerl UV, Soula H, & Berry H (2019). Simulation of calcium signaling in fine astrocytic processes: Effect of spatial properties on spontaneous activity. PLoS Computational Biology, 15(8), e1006795. 10.1371/journal.pcbi.1006795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descalzi G, Gao V, Steinman MQ, Suzuki A, & Alberini CM (2019). Lactate from astrocytes fuels learning-induced mRNA translation in excitatory and inhibitory neurons. Communications Biology, 2, 247. 10.1038/s42003-019-0495-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Castro MA, & Volterra A (2021). Astrocyte control of the entorhinal cortex-dentate gyrus circuit: Relevance to cognitive processing and impairment in pathology. Glia. 10.1002/glia.24128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-García CM, Lahmann C, Martínez-François JR, Li B, Koveal D, Nathwani N, Rahman M, Keller JP, Marvin JS, Looger LL, & Yellen G (2019). Quantitative in vivo imaging of neuronal glucose concentrations with a genetically encoded fluorescence lifetime sensor. Journal of Neuroscience Research, 97(8), 946–960. 10.1002/jnr.24433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, O’Donnell J, Thrane AS, Zeppenfeld D, Kang H, Xie L, Wang F, & Nedergaard M (2013). α1-adrenergic receptors mediate coordinated Ca2+ signaling of cortical astrocytes in awake, behaving mice. Cell Calcium, 54(6), 387–394. 10.1016/j.ceca.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GCH, Zhang J, Quinn AM, Hughes TE, Maysinger D, Alford SC, Zhang Y, & Campbell RE (2015). Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nature Methods, 12(3), 195–198. 10.1038/nmeth.3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, He K, Dudok B, Farrell JS, Guan W, Liput DJ, Puhl HL, Cai R, Wang H, Duan J, Albarran E, Ding J, Lovinger DM, Li B, Soltesz I, & Li Y (2021). A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo. Nature Biotechnology. 10.1038/s41587-021-01074-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Chen X, Cui M, Yu X, Pang Q, & Sun J (2012). β2-adrenergic receptor and astrocyte glucose metabolism. Journal of Molecular Neuroscience, 48(2), 456–463. 10.1007/s12031-012-9742-4 [DOI] [PubMed] [Google Scholar]
- Dong Z, Mau W, Feng Y, Pennington ZT, Chen L, Zaki Y, Rajan K, Shuman T, Aharoni D & Cai DJ (2021). Minian: An open-source miniscope analysis pipeline. bioRxiv, 2021.05.03.442492. 10.1101/2021.05.03.442492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron A, Rubin A, Benmelech-Chovav A, Benaim N, Carmi T, Kreisel T, Ziv Y & Goshen I (2021). Hippocampal astrocytes encode reward location. bioRxiv, 2021.07.07.451434. 10.1101/2021.07.07.451434 [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, & Heintz N (2008). Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell, 135(4), 749–762. 10.1016/j.cell.2008.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dringen R, Gutterer M, & Hirrlinger J (2000). Glutathione metabolism in brain metabolic interaction between astrocytes and neurons in the defense against reactive oxygen species. European Journal of Biochemistry, 267(16), 4912–4916. 10.1046/j.1432-1327.2000.01597.x [DOI] [PubMed] [Google Scholar]
- Durkee CA, Covelo A, Lines J, Kofuji P, Aguilar J, & Araque A (2019). Gi/o protein-coupled receptors inhibit neurons but activate astrocytes and stimulate gliotransmission. Glia, 67(6), 1076–1093. 10.1002/glia.23589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta DJ, Woo DH, Lee PR, Pajevic S, Bukalo O, Huffman WC, Wake H, Basser PJ, SheikhBahaei S, Lazarevic V, Smith JC, & Fields RD (2018). Regulation of myelin structure and conduction velocity by perinodal astrocytes. Proceedings of the National Academy of Sciences, 115(46), 11832–11837. 10.1073/pnas.1811013115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam R, Aharoni R, Arnon R, & Malach R (2016). Astrocyte morphology is confined by cortical functional boundaries in mammals ranging from mice to human. eLife, 5, e15915. 10.7554/eLife.15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley JG, & Macklis JD (2006). Astroglial heterogeneity closely reflects the neuronal-defined anatomy of the adult murine CNS. Neuron Glia Biology, 2(3), 175–186. 10.1017/S1740925X06000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso-Pichot A, Brasó-Vives M, Golbano A, Menacho C, Claro E, Galea E, & Masgrau R (2018). GSEA of mouse and human mitochondriomes reveals fatty acid oxidation in astrocytes. Glia, 66(8), 1724–1735. 10.1002/glia.23330 [DOI] [PubMed] [Google Scholar]
- Ermakova YG, Bilan DS, Matlashov ME, Mishina NM, Markvicheva KN, Subach OM, Subach FV, Bogeski I, Hoth M, Enikolopov G, & Belousov VV (2014). Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nature Communications, 5(1), 5222. 10.1038/ncomms6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WT, Abrahamsson T, Chierzi S, Lui C, Zaelzer C, Jones EV, Bally BP, Chen GG, Theroux J-F, Peng J, Bourque CW, Charron F, Ernst C, Sjöström PJ, & Murai KK (2016). Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science, 351(6275), 849–854. 10.1126/science.aab3103 [DOI] [PubMed] [Google Scholar]
- Farmer WT, & Murai K (2017). Resolving astrocyte heterogeneity in the CNS. Frontiers in Cellular Neuroscience, 11, 1–7. 10.3389/fncel.2017.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecher C, Trovò L, Müller SA, Snaidero N, Wettmarshausen J, Heink S, Ortiz O, Wagner I, Kühn R, Hartmann J, Karl RM, Konnerth A, Korn T, Wurst W, Merkler D, Lichtenthaler SF, Perocchi F, & Misgeld T (2019). Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nature Neuroscience, 22(10), 1731–1742. 10.1038/s41593-019-0479-z [DOI] [PubMed] [Google Scholar]
- Fehr M, Lalonde S, Lager I, Wolff MW, & Frommer WB (2003). In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. Journal of Biological Chemistry, 278(21), 19127–19133. 10.1074/jbc.M301333200 [DOI] [PubMed] [Google Scholar]
- Feng J, Zhang C, Lischinsky JE, Jing M, Zhou J, Wang H, Zhang Y, Dong A, Wu Z, Wu H, Chen W, Zhang P, Zou J, Hires SA, Zhu JJ, Cui G, Lin D, Du J, & Li Y (2019). A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron, 102(4), 745–761.e8. 10.1016/j.neuron.2019.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Moncada I, Robles-Maldonado D, Castro P, Alegría K, Epp R, Ruminot I, & Barros LF (2021). Bidirectional astrocytic GLUT1 activation by elevated extracellular K+. Glia, 69(4), 1012–1021. 10.1002/glia.23944 [DOI] [PubMed] [Google Scholar]
- Fields RD, & Richardson WD (2019). Plasticity of myelinating glia. Glia, 67(11), 2005–2007. 10.1002/glia.23720 [DOI] [PubMed] [Google Scholar]
- Figueiredo M, Lane S, Stout RF, Liu B, Parpura V, Teschemacher AG, & Kasparov S (2014). Comparative analysis of optogenetic actuators in cultured astrocytes. Cell Calcium, 56(3), 208–214. 10.1016/j.ceca.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MC, Alexandrova O, Cossell L, Stange-Marten A, Sinclair J, Kopp-Scheinpflug C, Pecka M, Attwell D, & Grothe B (2015). Tuning of Ranvier node and internode properties in myelinated axons to adjust action potential timing. Nature Communications, 6, 8073. 10.1038/ncomms9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster AY, Bujalka H, & Emery B (2019). Axoglial interactions in myelin plasticity: Evaluating the relationship between neuronal activity and oligodendrocyte dynamics. Glia, 67(11), 2038–2049. 10.1002/glia.23629 [DOI] [PubMed] [Google Scholar]
- Fourgeaud L, Través PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagorska A, Rothlin CV, Nimmerjahn A, & Lemke G (2016). Tam receptors regulate multiple features of microglial physiology. Nature, 532(7598), 240–244. 10.1038/nature17630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, & Rowitch DH (2013). Evolving concepts of gliogenesis: A look way back and ahead to the next 25 years. Neuron, 80(3), 613–623. 10.1016/j.neuron.2013.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J, Giovannucci A, & Pnevmatikakis EA (2021). Online analysis of microendoscopic 1-photon calcium imaging data streams. PLoS Computational Biology, 17(1), e1008565. 10.1371/journal.pcbi.1008565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao V, Suzuki A, Magistretti PJ, Lengacher S, Pollonini G, Steinman MQ, & Alberini CM (2016). Astrocytic β2-adrenergic receptors mediate hippocampal long-term memory consolidation. Proceedings of the National Academy of Sciences, 113(30), 8526–8531. 10.1073/pnas.1605063113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs ME (2016). Role of glycogenolysis in memory and learning: Regulation by noradrenaline, serotonin and ATP. Frontiers in Integrative Neuroscience, 9, 70. 10.3389/fnint.2015.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs ME, Anderson DG, & Hertz L (2006). Inhibition of glycogenolysis in astrocytes interrupts memory consolidation in young chickens. Glia, 54(3), 214–222. 10.1002/glia.20377 [DOI] [PubMed] [Google Scholar]
- Gibson EM, Purger D, Mount CW, Goldstein AK, Lin GL, Wood LS, Inema I, Miller SE, Bieri G, Zuchero JB, Barres BA, Woo PJ, Vogel H, & Monje M (2014). Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science, 344(6183), 1252304. 10.1126/science.1252304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, Koay SA, Taxidis J, Najafi F, Gauthier JL, Zhou P, Khakh BS, Tank DW, Chklovskii DB, & Pnevmatikakis EA (2019). CaImAn an open source tool for scalable calcium imaging data analysis. eLife, 8, e38173. 10.7554/eLife.38173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhoff TC, & Quake SR (2016). Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-Seq. Cell Reports, 16(4), 1126–1137. 10.1016/j.celrep.2016.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SA, Nedergaard M, & Windrem MS (2015). Modeling cognition and disease using human glial chimeric mice. Glia, 63(8), 1483–1493. 10.1002/glia.22862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordleeva SY, Ermolaeva AV, Kastalskiy IA, & Kazantsev VB (2019). Astrocyte as spatiotemporal integrating detector of neuronal activity. Frontiers in Physiology, 10, 294. 10.3389/fphys.2019.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordleeva SY, Tsybina YA, Krivonosov MI, Ivanchenko MV, Zaikin AA, Kazantsev VB, & Gorban AN (2021). Modeling working memory in a spiking neuron network accompanied by astrocytes. Frontiers in Cellular Neuroscience, 15, 631485. 10.3389/fncel.2021.631485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griemsmann S, Höft SP, Bedner P, Zhang J, Staden E. v., Beinhauer A, Degen J, Dublin P, Cope DW, Richter N, Crunelli V, Jabs R, Willecke K, Theis M, Seifert G, Kettenmann H, & Steinhäuser C (2015). Characterization of Panglial gap junction networks in the thalamus, neocortex, and hippocampus reveals a unique population of glial cells. Cerebral Cortex, 25(10), 3420–3433. 10.1093/cercor/bhu157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimley JS, Li L, Wang W, Wen L, Beese LS, Hellinga HW, & Augustine GJ (2013). Visualization of synaptic inhibition with an optogenetic sensor developed by cell-free protein engineering automation. The Journal of Neuroscience, 33(41), 16297–16309. 10.1523/JNEUROSCI.4616-11.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan KA, Weigel MK, Prakash P, Wijewardhane PR, Hasel P, Rufen-Blanchette U, Münch AE, Blum JA, Fine J, Neal MC, Bruce KD, Gitler AD, Chopra G, Liddelow SA, & Barres BA (2021). Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature, 599(7883), 102–107. 10.1038/s41586-021-03960-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackley CR, Mazzoni EO, & Blau J (2018). cAMPr: A single-wavelength fluorescent sensor for cyclic AMP. Science Signaling, 11(520), eaah3738. 10.1126/scisignal.aah3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H, Shoji H, Otabi H, Toyoda A, Katoh K, Namihira M, & Miyakawa T (2021). Protein lactylation induced by neural excitation. Cell Reports, 37(2), 109820. 10.1016/j.celrep.2021.109820 [DOI] [PubMed] [Google Scholar]
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, Silva AJ, Takano T, Goldman SA, & Nedergaard M (2013). Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell, 12(3), 342–353. 10.1016/j.stem.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K, Ito M, Wang X, Tanaka M, Wongso D, Konno A, Hirai H, Hirase H, Tsuboi T, & Kitaguchi T (2017). Red fluorescent protein-based cAMP indicator applicable to optogenetics and in vivo imaging. Scientific Reports, 7(1), 7351. 10.1038/s41598-017-07820-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann AC, Schipke CG, & Kettenmann H (2005). Extracellular application of nicotinic acid adenine dinucleotide phosphate induces Ca2+ signaling in astrocytes in situ. Journal of Biological Chemistry, 280(42), 35630–35640. 10.1074/jbc.M507338200 [DOI] [PubMed] [Google Scholar]
- Henderson MJ, Baldwin HA, Werley CA, Boccardo S, Whitaker LR, Yan X, Holt GT, Schreiter ER, Looger LL, Cohen AE, Kim DS, & Harvey BK (2015). A low affinity GCaMP3 variant (GCaMPer) for imaging the endoplasmic reticulum calcium store. PLoS One, 10(10), e0139273. 10.1371/journal.pone.0139273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde MK, Bohmbach K, Domingos C, Vana N, Komorowska-Müller JA, Passlick S, Schwarz I, Jackson CJ, Dietrich D, Schwarz MK, & Henneberger C (2020). Local efficacy of glutamate uptake decreases with synapse size. Cell Reports, 32(12), 108182. 10.1016/j.celrep.2020.108182 [DOI] [PubMed] [Google Scholar]
- Herrero-Navarro Á, Puche-Aroca L, Moreno-Juan V, Sempere-Ferràndez A, Espinosa A, Susín R, Torres-Masjoan L, Leyva-Diaz E, Karow M, Figueres-Onate M, Lopez-Mascaraque L, Lopez-Atalaya JP, Berninger B, & López-Bendito G (2021). Astrocytes and neurons share region-specific transcriptional signatures that confer regional identity to neuronal reprogramming. Science Advances, 7(15), eabe8978. 10.1126/sciadv.abe8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H, Déglon N, Foo LC, Goshen I, Grutzendler J, Hangen E, Kreisel T, Linck N, Muffat J, Regio S, Rion S, & Escartin C (2020). Emerging technologies to study glial cells. Glia, 68(9), 1692–1728. 10.1002/glia.23780 [DOI] [PubMed] [Google Scholar]
- Hirrlinger J, & Dringen R (2010). The cytosolic redox state of astrocytes: Maintenance, regulation and functional implications for metabolite trafficking. Brain Research Reviews, 63(1–2), 177–188. 10.1016/j.brainresrev.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Hirrlinger J, Marx G, Besser S, Sicker M, Köhler S, Hirrlinger PG, Wojcik S, Eulenburg V, Winkler U, & Hülsmann S (2019). GABA-glycine Cotransmitting neurons in the ventrolateral medulla: Development and functional relevance for breathing. Frontiers in Cellular Neuroscience, 13, 517. 10.3389/fncel.2019.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirrlinger J, Requardt RP, Winkler U, Wilhelm F, Schulze C, & Hirrlinger PG (2009). Split-CreERT2: Temporal control of DNA recombination mediated by split-Cre protein fragment complementation. PLoS One, 4(12), e8354. 10.1371/journal.pone.0008354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang W, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, & Kirchhoff F (2009). Split-cre complementation indicates coincident activity of different genes in vivo. PLoS One, 4(1), e4286. 10.1371/journal.pone.0004286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirrlinger PG, Scheller A, Braun C, Hirrlinger J, & Kirchhoff F (2006). Temporal control of gene recombination in astrocytes by transgenic expression of the tamoxifen-inducible DNA recombinase variant CreERT2. Glia, 54(1), 11–20. 10.1002/glia.20342 [DOI] [PubMed] [Google Scholar]
- Hirrlinger PG, Wurm A, Hirrlinger J, Bringmann A, & Reichenbach A (2008). Osmotic swelling characteristics of glial cells in the murine hippocampus, cerebellum, and retina in situ. Journal of Neurochemistry, 105(4), 1405–1417. 10.1111/j.1471-4159.2008.05243.x [DOI] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, & Anderson DJ (2008). Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell, 133(3), 510–522. 10.1016/j.cell.2008.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Dissing-Olesen L, & Stevens B (2016). New insights on the role of microglia in synaptic pruning in health and disease. Current Opinion in Neurobiology, 36, 128–134. 10.1016/j.conb.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat A, Zorec R, & Vardjan N (2016). Adrenergic stimulation of single rat astrocytes results in distinct temporal changes in intracellular Ca2+ and cAMP-dependent PKA responses. Cell Calcium, 59(4), 156–163. 10.1016/j.ceca.2016.01.002 [DOI] [PubMed] [Google Scholar]
- Huang Y, Happonen KE, Burrola PG, O’Connor C, Hah N, Huang L, Nimmerjahn A, & Lemke G (2021). Microglia use TAM receptors to detect and engulf amyloid β plaques. Nature Immunology, 22(5), 586–594. 10.1038/s41590-021-00913-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, & Yellen G (2011). Imaging cytosolic NADH-NAD+ redox state with a genetically encoded fluorescent biosensor. Cell Metabolism, 14(4), 545–554. 10.1016/j.cmet.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, & Noji H (2009). Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proceedings of the National Academy of Sciences, 106(37), 15651–15656. 10.1073/pnas.0904764106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, Deloulme JC, Chan G, & Storm DR (1998). Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron, 21(4), 869–883. 10.1016/s0896-6273(00)80602-9 [DOI] [PubMed] [Google Scholar]
- Inan H, Schmuckermair C, Tasci T, Ahanonu BO, Hernandez O, Lecoq J, Dinç F, Wagner MJ, Erdogdu MA & Schnitzer MJ (2021). Fast and statistically robust cell extraction from large-scale neural calcium imaging datasets. bioRxiv, 2021.03.24.436279. 10.1101/2021.03.24.436279 [DOI] [Google Scholar]
- Inoue M, Takeuchi A, Manita S, Horigane S, Sakamoto M, Kawakami R, Yamaguchi K, Otomo K, Yokoyama H, Kim R, Yokoyama T, Takemoto-Kimura S, Abe M, Okamura M, Kondo Y, Quirin S, Ramakrishnan C, Imamura T, Sakimura K, … Bito H (2019). Rational engineering of XCaMPs, a multicolor GECI suite for in vivo imaging of complex brain circuit dynamics. Cell, 177(5), 1346–1360.e24. 10.1016/j.cell.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Ioannou MS, Jackson J, Sheu S-H, Chang C-L, Weigel AV, Liu H, Pasolli HA, Xu CS, Pang S, Matthies D, Hess HF, Lippincott-Schwartz J, & Liu Z (2019). Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell, 177(6), 1522–1535.e14. 10.1016/j.cell.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Jensen P, & Dymecki SM (2014). Essentials of recombinase-based genetic fate mapping in mice. Methods in Molecular Biology (Clifton, N.J.), 1092, 437–454. 10.1007/978-1-60327-292-6_26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha MK, Jo M, Kim J-H, & Suk K (2019). Microglia-astrocyte crosstalk: An intimate molecular conversation. The Neuroscientist, 25(3), 227–240. 10.1177/1073858418783959 [DOI] [PubMed] [Google Scholar]
- Jimenez-Blasco D, Busquets-Garcia A, Hebert-Chatelain E, Serrat R, Vicente-Gutierrez C, Ioannidou C, Gómez-Sotres P, Lopez-Fabuel I, Resch-Beusher M, Resel E, Arnouil D, Saraswat D, Varilh M, Cannich A, Julio-Kalajzic F, Bonilla-Del Río I, Almeida A, Puente N, … Marsicano G (2020). Glucose metabolism links astroglial mitochondria to cannabinoid effects. Nature, 583(7817), 603–608. 10.1038/s41586-020-2470-y [DOI] [PubMed] [Google Scholar]
- Jing M, Li Y, Zeng J, Huang P, Skirzewski M, Kljakic O, Peng W, Qian T, Tan K, Zou J, Trinh S, Wu T, Zhang S, Pan S, Hires SA, Xu M, Li H, Saksida LM, Prado VF, … Li Y (2020). An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nature Methods, 17(11), 1139–1146. 10.1038/s41592-020-0953-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Lin CC, Yu K, Hatcher A, Huang TW, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, Patel AJ, Arenkiel BR, Noebels JL, Creighton CJ, & Deneen B (2017). Identification of diverse astrocyte populations and their malignant analogs. Nature Neuroscience, 20(3), 396–405. 10.1038/nn.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien N, Sampieri F, Enjalbert A, & Herman JP (2003). Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Research, 31(21), e131. 10.1093/nar/gng131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastanenka KV, Moreno-Bote R, Pittà M. d., Perea G, Eraso-Pichot A, Masgrau R, Poskanzer KE, & Galea E (2020). A roadmap to integrate astrocytes into systems neuroscience. Glia, 68(1), 5–26. 10.1002/glia.23632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JP, Marvin JS, Lacin H, Lemon WC, Shea J, Kim S, Lee RT, Koyama M, Keller PJ, & Looger LL (2021). In vivo glucose imaging in multiple model organisms with an engineered single-wavelength sensor. Cell Reports, 35(12), 109284. 10.1016/j.celrep.2021.109284 [DOI] [PubMed] [Google Scholar]
- Kelley KW, Ben Haim L, Schirmer L, Tyzack GE, Tolman M, Miller JG, Tsai H-H, Chang SM, Molofsky AV, Yang Y, Patani R, Lakatos A, Ullian EM, & Rowitch DH (2018). Kir4.1-dependent astrocyte-fast motor neuron interactions are required for peak strength. Neuron, 98(2), 306–319.e7. 10.1016/j.neuron.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, & Deneen B (2019). The emerging nature of astrocyte diversity. Annual Review of Neuroscience, 42, 187–207. 10.1146/annurev-neuro-070918-050443 [DOI] [PubMed] [Google Scholar]
- Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, & Dymecki SM (2009). Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron, 63(3), 305–315. 10.1016/j.neuron.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Kolesnikov M, Peled-Hajaj S, Scheyltjens I, Xia Y, Trzebanski S, Haimon Z, Shemer A, Lubart A, Van Hove H, Chappell-Maor L, Boura-Halfon S, Movahedi K, Blinder P, & Jung S (2021). A binary Cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity, 54(1), 176–190. e7. 10.1016/j.immuni.2020.11.007 [DOI] [PubMed] [Google Scholar]
- Kim N, Shin S, & Bae SW (2021). cAMP biosensors based on genetically encoded fluorescent/luminescent proteins. Biosensors, 11(2), 39. 10.3390/bios11020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CM, Bohmbach K, Minge D, Delekate A, Zheng K, Reynolds J, Rakers C, Zeug A, Petzold GC, Rusakov DA, & Henneberger C (2020). Local resting Ca2+ controls the scale of astroglial Ca2+ signals. Cell Reports, 30(10), 3466–3477.e4. 10.1016/j.celrep.2020.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kıray H, Lindsay SL, Hosseinzadeh S, & Barnett SC (2016). The multifaceted role of astrocytes in regulating myelination. Experimental Neurology, 283(Pt B), 541–549. 10.1016/j.expneurol.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Winkler U, & Hirrlinger J (2021). Heterogeneity of astrocytes in Grey and White matter. Neurochemical Research, 46(1), 3–14. 10.1007/s11064-019-02926-x [DOI] [PubMed] [Google Scholar]
- Köhler S, Winkler U, Sicker M, & Hirrlinger J (2018). Nbce1 mediates the regulation of the NADH/NAD+ redox state in cortical astrocytes by neuronal signals. Glia, 66(10), 2233–2245. 10.1002/glia.23504 [DOI] [PubMed] [Google Scholar]
- Kohnke S, Buller S, Nuzzaci D, Ridley K, Lam B, Pivonkova H, Bentsen MA, Alonge KM, Zhao C, Tadross J, Holmqvist S, Shimizu T, Hathaway H, Li H, Macklin W, Schwartz MW, Richardson WD, Yeo GSH, Franklin RJM, … Blouet C (2021). Nutritional regulation of oligodendrocyte differentiation regulates perineuronal net remodeling in the median eminence. Cell Reports, 36(2), 109362. 10.1016/j.celrep.2021.109362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohro Y, Matsuda T, Yoshihara K, Kohno K, Koga K, Katsuragi R, Oka T, Tashima R, Muneta S, Yamane T, Okada S, Momokino K, Furusho A, Hamase K, Oti T, Sakamoto H, Hayashida K, Kobayashi R, Horii T, … Tsuda M (2020). Spinal astrocytes in superficial laminae gate brainstem descending control of mechanosensory hypersensitivity. Nature Neuroscience, 23(11), 1376–1387. 10.1038/s41593-020-00713-4 [DOI] [PubMed] [Google Scholar]
- Konttinen H, Gureviciene I, Oksanen M, Grubman A, Loppi S, Huuskonen MT, Korhonen P, Lampinen R, Keuters M, Belaya I, Tanila H, Kanninen KM, Goldsteins G, Landreth G, Koistinaho J, & Malm T (2019). Pparβ/δ-agonist GW0742 ameliorates dysfunction in fatty acid oxidation in PSEN1ΔE9 astrocytes. Glia, 67(1), 146–159. 10.1002/glia.23534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koveal D, Díaz-García CM, & Yellen G (2020). Fluorescent biosensors for neuronal metabolism and the challenges of quantitation. Current Opinion in Neurobiology, 63, 111–121. 10.1016/j.conb.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow AM, Ford MC, Valdivia LE, Wilson SW, & Attwell D (2018). Regulation of developing myelin sheath elongation by oligodendrocyte calcium transients in vivo. Nature Neuroscience, 21(1), 24–28. 10.1038/s41593-017-0031-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronschläger MT, Siegert ASM, Resch FJ, Rajendran PS, Khakh BS, & Sandkühler J (2021). Lamina-specific properties of spinal astrocytes. Glia, 69(7), 1749–1766. 10.1002/glia.23990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula B, Chen T-J, & Kukley M (2019). Glutamatergic signaling between neurons and oligodendrocyte lineage cells: Is it synaptic or non-synaptic? Glia, 67(11), 2071–2091. 10.1002/glia.23617 [DOI] [PubMed] [Google Scholar]
- Lanjakornsiripan D, Pior B-J, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, & Gotoh Y (2018). Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nature Communications, 9(1), 1623. 10.1038/s41467-018-03940-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douce J, Maugard M, Veran J, Matos M, Jégo P, Vigneron P-A, Faivre E, Toussay X, Vandenberghe M, Balbastre Y, Piquet J, Guiot E, Tran NT, Taverna M, Marinesco S, Koyanagi A, Furuya S, Gaudin-Guerif M, Goutal S, … Bonvento G (2020). Impairment of glycolysis-derived L-serine production in astrocytes contributes to cognitive deficits in Alzheimer’s disease. Cell Metabolism, 31(3), 503–517.e8. 10.1016/j.cmet.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Lecoq J, Oliver M, Siegle JH, Orlova N, Ledochowitsch P, & Koch C (2021). Removing independent noise in systems neuroscience data using deepInterpolation. Nature Methods, 18(11), 1401–1408. 10.1038/s41592-021-01285-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Ye L, & Deisseroth K (2016). Communication in neural circuits: Tools, opportunities, and challenges. Cell, 164(6), 1136–1150. 10.1016/j.cell.2016.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezmy J, Arancibia-Cárcamo IL, Quintela-López T, Sherman DL, Brophy PJ, & Attwell D (2021). Astrocyte Ca2+–evoked ATP release regulates myelinated axon excitability and conduction speed. Science, 374(6565), eabh2858. 10.1126/science.abh2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, & Barres BA (2018). Microglia and macrophages in brain homeostasis and disease. Nature Reviews. Immunology, 18(4), 225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Marsh SE, & Stevens B (2020). Microglia and astrocytes in disease: Dynamic duo or Partners in Crime? Trends in Immunology, 41(9), 820–835. 10.1016/j.it.2020.07.006 [DOI] [PubMed] [Google Scholar]
- Liu K, Jin H, & Zhou B (2020). Genetic lineage tracing with multiple DNA recombinases: A user’s guide for conducting more precise cell fate mapping studies. Journal of Biological Chemistry, 295(19), 6413–6424. 10.1074/jbc.REV120.011631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobas MA, Tao R, Nagai J, Kronschläger MT, Borden PM, Marvin JS, Looger LL, & Khakh BS (2019). A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nature Communications, 10(1), 711. 10.1038/s41467-019-08441-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li C, Singh-Alvarado J, Zhou ZC, Fröhlich F, Mooney R, & Wang F (2018). Min1pipe: A Miniscope 1-photon-based calcium imaging signal extraction pipeline. Cell Reports, 23(12), 3673–3684. 10.1016/j.celrep.2018.05.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Kuzin A, Odenwald WF, & White BH (2020). Cre-assisted fine-mapping of neural circuits using orthogonal split inteins. eLife, 9, e53041. 10.7554/eLife.53041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Ambrozkiewicz MC, Benseler F, Chen C, Dumontier E, Falkner S, Furlanis E, Gomez AM, Hoshina N, Huang W-H, Hutchinson MA, Itoh-Maruoka Y, Lavery LA, Li W, Maruo T, Motohashi J, Pai EL-L, Pelkey KA, Pereira A, … Craig AM (2020). Optimizing nervous system-specific gene targeting with Cre driver Lines: Prevalence of germline recombination and influencing factors. Neuron, 106(1), 37–65.e5. 10.1016/j.neuron.2020.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, Gu H, Mills M, Cheng A, Tasic B, Nguyen TN, Sunkin SM, Benucci A, Nagy A, Miyawaki A, … Zeng H (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron, 85(5), 942–958. 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manninen T, Havela R, & Linne M-L (2018). Computational models for calcium-mediated astrocyte functions. Frontiers in Computational Neuroscience, 12, 14. 10.3389/fncom.2018.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez A, Bell KP, & Norenberg MD (1977). Glutamine synthetase: Glial localization in brain. Science, 195(4284), 1356–1358. 10.1126/science.14400 [DOI] [PubMed] [Google Scholar]
- Marvin JS, Scholl B, Wilson DE, Podgorski K, Kazemipour A, Müller JA, Schoch S, Quiroz FJU, Rebola N, Bao H, Little JP, Tkachuk AN, Cai E, Hantman AW, Wang SS-H, DePiero VJ, Borghuis BG, Chapman ER, Dietrich D, … Looger LL (2018). Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nature Methods, 15(11), 936–939. 10.1038/s41592-018-0171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V, Leite M, Kawashima T, Jensen TP, Kolb I, Knott EL, Novak O, Podgorski K, Leidenheimer NJ, Rusakov DA, Ahrens MB, Kullmann DM, & Looger LL (2019). A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nature Methods, 16(8), 763–770. 10.1038/s41592-019-0471-2 [DOI] [PubMed] [Google Scholar]
- Masmoudi-Kouki O, Gandolfo P, Castel H, Leprince J, Fournier A, Dejda A, Vaudry H, & Tonon M-C (2007). Role of PACAP and VIP in astroglial functions. Peptides, 28(9), 1753–1760. 10.1016/j.peptides.2007.05.015 [DOI] [PubMed] [Google Scholar]
- Matias I, Morgado J, & Gomes FCA (2019). Astrocyte heterogeneity: Impact to brain aging and disease. Frontiers in Aging Neuroscience, 11, 59. 10.3389/fnagi.2019.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsu-ura T, Michikawa T, Inoue T, Miyawaki A, Yoshida M, & Mikoshiba K (2006). Cytosolic inositol 1,4,5-trisphosphate dynamics during intracellular calcium oscillations in living cells. The Journal of Cell Biology, 173(5), 755–765. 10.1083/jcb.200512141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, & Kettenmann H (2010). Heterogeneity in astrocyte morphology and physiology. Brain Research Reviews, 63(1–2), 2–10. 10.1016/j.brainresrev.2009.12.001 [DOI] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, Faria J. P. d., Emery B, Tohyama K, & Richardson WD (2014). Motor skill learning requires active central myelination. Science, 346(6207), 318–322. 10.1126/science.1254960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merten K, Folk RW, Duarte D & Nimmerjahn A (2021). Astrocytes encode complex behaviorally relevant information. bioRxiv, 2021.10.09.463784. 10.1101/2021.10.09.463784 [DOI] [Google Scholar]
- Miller SJ, Philips T, Kim N, Dastgheyb R, Chen Z, Hsieh Y-C, Daigle JG, Datta M, Chew J, Vidensky S, Pham JT, Hughes EG, Robinson MB, Sattler R, Tomer R, Suk JS, Bergles DE, Haughey N, Pletnikov M, Hanes J, & Rothstein JD (2019). Molecularly defined cortical astroglia subpopulation modulates neurons via secretion of Norrin. Nature Neuroscience, 22(5), 741–752. 10.1038/s41593-019-0366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita M, Ito M, Harada K, Sugawara I, Ueda H, Tsuboi T, & Kitaguchi T (2019). Green fluorescent protein-based glucose indicators report glucose dynamics in living cells. Analytical Chemistry, 91(7), 4821–4830. 10.1021/acs.analchem.9b00447 [DOI] [PubMed] [Google Scholar]
- Miyamoto A, & Mikoshiba K (2017). Probes for manipulating and monitoring IP3. Cell Calcium, 64, 57–64. 10.1016/j.ceca.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Molina-Gonzalez I, & Miron VE (2019). Astrocytes in myelination and remyelination. Neuroscience Letters, 713, 134532. 10.1016/j.neulet.2019.134532 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai H-H, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, & Rowitch DH (2014). Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature, 509(7499), 189–194. 10.1038/nature13161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M (2018). Myelin plasticity and nervous system function. Annual Review of Neuroscience, 41, 61–76. 10.1146/annurev-neuro-080317-061853 [DOI] [PubMed] [Google Scholar]
- Moore S, Meschkat M, Ruhwedel T, Trevisiol A, Tzvetanova ID, Battefeld A, Kusch K, Kole MHP, Strenzke N, Möbius W, de Hoz L, & Nave K-A (2020). A role of oligodendrocytes in information processing. Nature Communications, 11(1), 5497. 10.1038/s41467-020-19152-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L, Chiang MSR, Higashimori H, Shoneye T, Iyer LK, Yelick J, Tai A, & Yang Y (2017). Molecular and functional properties of regional astrocytes in the adult brain. Journal of Neuroscience, 37(36), 8706–8717. 10.1523/JNEUROSCI.3956-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L, Higashimori H, Tolman M, & Yang Y (2014). Vglut1+ neuronal glutamatergic signaling regulates postnatal developmental maturation of cortical protoplasmic astroglia. Journal of Neuroscience, 34(33), 10950–10962. 10.1523/JNEUROSCI.1167-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Tanaka K, Buffo A, Wurst W, Kühn R, & Götz M (2006). Inducible gene deletion in astroglia and radial glia—A valuable tool for functional and lineage analysis. Glia, 54(1), 21–34. 10.1002/glia.20350 [DOI] [PubMed] [Google Scholar]
- Nagai J, Bellafard A, Qu Z, Yu X, Ollivier M, Gangwani MR, Diaz-Castro B, Coppola G, Schumacher SM, Golshani P, Gradinaru V, & Khakh BS (2021). Specific and behaviorally consequential astrocyte Gq GPCR signaling attenuation in vivo with iβARK. Neuron, 109(14), 2256–2274.e9. 10.1016/j.neuron.2021.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Rajbhandari AK, Gangwani MR, Hachisuka A, Coppola G, Masmanidis SC, Fanselow MS, & Khakh BS (2019). Hyperactivity with disrupted attention by activation of an astrocyte synaptogenic cue. Cell, 177(5), 1280–1292.e20. 10.1016/j.cell.2019.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Yu X, Papouin T, Cheong E, Freeman MR, Monk KR, Hastings MH, Haydon PG, Rowitch D, Shaham S, & Khakh BS (2021). Behaviorally consequential astrocytic regulation of neural circuits. Neuron, 109(4), 576–596. 10.1016/j.neuron.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu Y, Murphy-Royal C, Wen Y, Haidey JN, Molina RS, Aggarwal A, Zhang S, Kamijo Y, Paquet M-E, Podgorski K, Drobizhev M, Bains JS, Lemieux MJ, Gordon GR, & Campbell RE (2021). A genetically encoded fluorescent biosensor for extracellular L-lactate. Nature Communications, 12(1), 7058. 10.1038/s41467-021-27332-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsubori A, Takata N, & Tanaka KF (2015). Observation and manipulation of glial cell function by virtue of sufficient probe expression. Frontiers in Cellular Neuroscience, 9, 176. 10.3389/fncel.2015.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neame S, Safory H, Radzishevsky I, Touitou A, Marchesani F, Marchetti M, Kellner S, Berlin S, Foltyn VN, Engelender S, Billard J-M, & Wolosker H (2019). The NMDA receptor activation by D-serine and glycine is controlled by an astrocytic Phgdh-dependent serine shuttle. Proceedings of the National Academy of Sciences, 116(41), 20736–20742. 10.1073/pnas.1909458116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson NA, Wang X, Cook D, Carey EM, & Nimmerjahn A (2019). Imaging spinal cord activity in behaving animals. Experimental Neurology, 320, 112974. 10.1016/j.expneurol.2019.112974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzahualcoyotzi C, & Pellerin L (2020). Neuronal and astroglial monocarboxylate transporters play key but distinct roles in hippocampus-dependent learning and memory formation. Progress in Neurobiology, 194, 101888. 10.1016/j.pneurobio.2020.101888 [DOI] [PubMed] [Google Scholar]
- Newman LA, Korol DL, & Gold PE (2011). Lactate produced by glycogenolysis in astrocytes regulates memory processing. PLoS One, 6(12), e28427. 10.1371/journal.pone.0028427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, & Bergles DE (2015). Large-scale recording of astrocyte activity. Current Opinion in Neurobiology, 32, 95–106. 10.1016/j.conb.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, & Helmchen F (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science, 308(5726), 1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Mukamel EA, & Schnitzer MJ (2009). Motor behavior activates Bergmann glial networks. Neuron, 62(3), 400–412. 10.1016/j.neuron.2009.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C, Matyash M, Pivneva T, Schipke CG, Ohlemeyer C, Hanisch UK, Kirchhoff F, & Kettenmann H (2001). GFAP promoter-controlled EGFP-expressing transgenic mice: A tool to visualize astrocytes and astrogliosis in living brain tissue. Glia, 33(1), 72–86. [PubMed] [Google Scholar]
- Nutma E, van Gent D, Amor S, & Peferoen LAN (2020). Astrocyte and oligodendrocyte cross-talk in the central nervous system. Cells, 9(3), 600. 10.3390/cells9030600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Goldman SA, & Nedergaard M (2012). Heterogeneity of astrocytic form and function. Methods in Molecular Biology, 814, 23–45. 10.1007/978-1-61779-452-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, Ransom BR, Goldman SA, & Nedergaard M (2009). Uniquely hominid features of adult human astrocytes. Journal of Neuroscience, 29(10), 3276–3287. 10.1523/JNEUROSCI.4707-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Wang X, Goldman S, & Nedergaard M (2006). Astrocytic complexity distinguishes the human brain. Trends in Neurosciences, 29(10), 547–553. 10.1016/j.tins.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Octeau JC, Gangwani MR, Allam SL, Tran D, Huang S, Hoang-Trong TM, Golshani P, Rumbell TH, Kozloski JR, & Khakh BS (2019). Transient, consequential increases in extracellular potassium ions accompany Channelrhodopsin2 excitation. Cell Reports, 27(8), 2249–2261.e7. 10.1016/j.celrep.2019.04.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaka H, Arai S, Inoue T, & Kitaguchi T (2014). Genetically-encoded yellow fluorescent cAMP indicator with an expanded dynamic range for dual-color imaging. PLoS One, 9(6), e100252. 10.1371/journal.pone.0100252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oe Y, Wang X, Patriarchi T, Konno A, Ozawa K, Yahagi K, Hirai H, Tsuboi T, Kitaguchi T, Tian L, McHugh TJ, & Hirase H (2020). Distinct temporal integration of noradrenaline signaling by astrocytic second messengers during vigilance. Nature Communications, 11(1), 471. 10.1038/s41467-020-14378-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oheim M, Schmidt E, & Hirrlinger J (2018). Local energy on demand: Are ‘spontaneous’ astrocytic Ca2+-microdomains the regulatory unit for astrocyte-neuron metabolic cooperation? Brain Research Bulletin, 136, 54–64. 10.1016/j.brainresbull.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Ohta Y, Furuta T, Nagai T, & Horikawa K (2018). Red fluorescent cAMP indicator with increased affinity and expanded dynamic range. Scientific Reports, 8(1), 1866. 10.1038/s41598-018-20251-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen ML, Campbell SL, & Sontheimer H (2007). Differential distribution of Kir4.1 in spinal cord astrocytes suggests regional differences in K+ homeostasis. Journal of Neurophysiology, 98(2), 786–793. 10.1152/jn.00340.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen SF, Liu MH, & Kreitzer AC (2019). Thermal constraints on in vivo optogenetic manipulations. Nature Neuroscience, 22(7), 1061–1065. 10.1038/s41593-019-0422-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C, Dipoppa M, Schröder S, Rossi LF, Dalgleish H, Carandini M & Harris KD (2017). Suite2p: Beyond 10,000 neurons with standard two-photon microscopy. bioRxiv, 061507. 10.1101/061507 [DOI] [Google Scholar]
- Padmashri R, Ren B, Oldham B, Jung Y, Gough R, & Dunaevsky A (2021). Modeling human-specific interlaminar astrocytes in the mouse cerebral cortex. Journal of Comparative Neurology, 529(4), 802–810. 10.1002/cne.24979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak VV, Ezerin¸a D, Lyublinskaya OG, Pedre B, Tyurin-Kuzmin PA, Mishina NM, Thauvin M, Young D, Wahni K, Gache SAM, Demidovich AD, Ermakova YG, Maslova YD, Shokhina AG, Eroglu E, Bilan DS, Bogeski I, Michel T, Vriz S, … Belousov VV (2020). Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metabolism, 31(3), 642–653.e6. 10.1016/j.cmet.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Mayoral SR, Choi HS, Chan JR, & Kheirbek MA (2020). Preservation of a remote fear memory requires new myelin formation. Nature Neuroscience, 23(4), 487–499. 10.1038/s41593-019-0582-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, & Gross CT (2011). Synaptic pruning by microglia is necessary for normal brain development. Science, 333(6048), 1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, Hempstead BL, Littman DR, & Gan W-B (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell, 155(7), 1596–1609. 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual O, Casper KB, Kubera C, Zhang J, Revilla-Sanchez R, Sul J-Y, Takano H, Moss SJ, McCarthy K, & Haydon PG (2005). Astrocytic purinergic signaling coordinates synaptic networks. Science, 310(5745), 113–116. 10.1126/science.1116916 [DOI] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong W-H, Folk RW, Broussard GJ, Liang R, Jang MJ, Zhong H, Dombeck D, von Zastrow M, Nimmerjahn A, Gradinaru V, Williams JT, & Tian L (2018). Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science, 360(6396), eaat4422. 10.1126/science.aat4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Mohebi A, Sun J, Marley A, Liang R, Dong C, Puhger K, Mizuno GO, Davis CM, Wiltgen B, von Zastrow M, Berke JD, & Tian L (2020). An expanded palette of dopamine sensors for multiplex imaging in vivo. Nature Methods, 17(11), 1147–1155. 10.1038/s41592-020-0936-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukert M, Agarwal A, Cha J, van Doze A, Kang JU, & Bergles DE (2014). Norepinephrine controls astroglial responsiveness to local circuit activity. Neuron, 82(6), 1263–1270. 10.1016/j.neuron.2014.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Pekna M, Messing A, Steinhäuser C, Lee J-M, Parpura V, Hol EM, Sofroniew MV, & Verkhratsky A (2016). Astrocytes: A central element in neurological diseases. Acta Neuropathologica, 131(3), 323–345. 10.1007/s00401-015-1513-1 [DOI] [PubMed] [Google Scholar]
- Pellerin L, & Magistretti PJ (1994). Glutamate uptake into astrocytes stimulates aerobic glycolysis: A mechanism coupling neuronal activity to glucose utilization. Proceedings of the National Academy of Sciences, 91(22), 10625–10629. 10.1073/pnas.91.22.10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Wu Z, Song K, Zhang S, Li Y, & Xu M (2020). Regulation of sleep homeostasis mediator adenosine by basal forebrain glutamatergic neurons. Science, 369(6508), eabb0556. 10.1126/science.abb0556 [DOI] [PubMed] [Google Scholar]
- Pfrieger FW, & Slezak M (2012). Genetic approaches to study glial cells in the rodent brain. Glia, 60(5), 681–701. 10.1002/glia.22283 [DOI] [PubMed] [Google Scholar]
- Philtjens S, Turnbull MT, Thedy BP, Moon Y & Kim J (2021). Single-cell resolution analysis of the crosstalk between chemogenically activated astrocytes and microglia. bioRxiv, 2020.04.27.064881. 10.1101/2020.04.27.064881 [DOI] [Google Scholar]
- Poskanzer KE, & Yuste R (2016). Astrocytes regulate cortical state switching in vivo. Proceedings of the National Academy of Sciences, 113(19), E2675–E2684. 10.1073/pnas.1520759113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Cosio DMO, Piatkevich KD, Aufmkolk S, Su W-C, Celiker OT, Schohl A, Murdock MH, Aggarwal A, Chang Y-F, Wiseman PW, Ruthazer ES, Boyden ES, & Campbell RE (2020). Improved genetically encoded near-infrared fluorescent calcium ion indicators for in vivo imaging. PLoS Biology, 18(11), e3000965. 10.1371/journal.pbio.3000965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman J, Besser S, Schnell C, Eulenburg V, Hirrlinger J, Wojcik SM, & Hülsmann S (2015). Genetic ablation of VIAAT in glycinergic neurons causes a severe respiratory phenotype and perinatal death. Brain Structure & Function, 220(5), 2835–2849. 10.1007/s00429-014-0829-2 [DOI] [PubMed] [Google Scholar]
- Raimondo JV, Joyce B, Kay L, Schlagheck T, Newey SE, Srinivas S, & Akerman CJ (2013). A genetically-encoded chloride and pH sensor for dissociating ion dynamics in the nervous system. Frontiers in Cellular Neuroscience, 7, 202. 10.3389/fncel.2013.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Ferenczi E, & Deisseroth K (2016). Targeting neural circuits. Cell, 165(3), 524–534. 10.1016/j.cell.2016.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y Cajal S (1911). Histologie du systeme nerveux de l’homme et des vertebres. Maloine Paris, A. (Ed.). [Google Scholar]
- Refaeli R, Doron A, Benmelech-Chovav A, Groysman M, Kreisel T, Loewenstein Y, & Goshen I (2021). Features of hippocampal astrocytic domains and their spatial relation to excitatory and inhibitory neurons. Glia, 69(10), 2378–2390. 10.1002/glia.24044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A, & Wolburg H (2012). Astrocytes and ependymal glia. In Kettenmann H & Ransom B (Eds.), Neuroglia (3rd ed., pp. 35–39). Oxford University Press. [Google Scholar]
- Robin LM, Oliveira da Cruz JF, Langlais VC, Martin-Fernandez M, Metna-Laurent M, Busquets-Garcia A, Bellocchio L, Soria-Gomez E, Papouin T, Varilh M, Sherwood MW, Belluomo I, Balcells G, Matias I, Bosier B, Drago F, Van Eeckhaut A, Smolders I, Georges F, … Marsicano G (2018). Astroglial CB1 receptors determine synaptic D-serine availability to enable recognition memory. Neuron, 98(5), 935–944.e5. 10.1016/j.neuron.2018.04.034 [DOI] [PubMed] [Google Scholar]
- Robinson NTM, Descamps LAL, Russell LE, Buchholz MO, Bicknell BA, Antonov GK, Lau JYN, Nutbrown R, Schmidt-Hieber C, & Häusser M (2020). Targeted activation of hippocampal place cells drives memory-guided spatial behavior. Cell, 183(6), 1586–1599.e10. 10.1016/j.cell.2020.09.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros O, Baudet S, Zagar Y, Loulier K, Roche F, Couvet S, Aghaie A, Atkins M, Louail A, Petit C, Metin C, Mechulam Y, & Nicol X (2020). SpiCee: A genetic tool for subcellular and cell-specific calcium manipulation. Cell Reports, 32(3), 107934. 10.1016/j.celrep.2020.107934 [DOI] [PubMed] [Google Scholar]
- Ros O, Zagar Y, Ribes S, Baudet S, Loulier K, Couvet S, Ladarre D, Aghaie A, Louail A, Petit C, Mechulam Y, Lenkei Z, & Nicol X (2019). SponGee: A genetic tool for subcellular and cell-specific cGMP manipulation. Cell Reports, 27(13), 4003–4012.e6. 10.1016/j.celrep.2019.05.102 [DOI] [PubMed] [Google Scholar]
- Roselló-Díez A, Madisen L, Bastide S, Zeng H, & Joyner AL (2018). Cell-nonautonomous local and systemic responses to cell arrest enable long-bone catch-up growth in developing mice. PLoS Biology, 16(6), e2005086. 10.1371/journal.pbio.2005086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL (2016). DREADDs for neuroscientists. Neuron, 89(4), 683–694. 10.1016/j.neuron.2016.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht P, Carta S, Hoffmann A, Echizen M, Blot A, Kwan AC, Dan Y, Hofer SB, Kitamura K, Helmchen F, & Friedrich RW (2021). A database and deep learning toolbox for noise-optimized, generalized spike inference from calcium imaging. Nature Neuroscience, 24(9), 1324–1337. 10.1038/s41593-021-00895-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab AS, Tzvetavona ID, Trevisiol A, Baltan S, Dibaj P, Kusch K, Möbius W, Goetze B, Jahn HM, Huang W, Steffens H, Schomburg ED, Perez-Samartin A, Perez-Cerda F, Bakhtiari D, Matute C, Löwel S, Griesinger C, Hirrlinger J, … Nave K-A (2016). Oligodendroglial NMDA receptors regulate glucose import and axonal energy metabolism. Neuron, 91(1), 119–132. 10.1016/j.neuron.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwig I, Spitznagel B, Vazquez-Armendariz AI, Khalooghi K, Guenther S, Herold S, Szibor M, & Braun T (2019). Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. The EMBO Journal, 38, (12), e102099. 10.15252/embj.2019102099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martín A, Arce-Molina R, Aburto C, Baeza-Lehnert F, Barros LF, Contreras-Baeza Y, Pinilla A, Ruminot I, Rauseo D, & Sandoval PY (2022). Visualizing physiological parameters in cells and tissues using genetically encoded indicators for metabolites. Free Radical Biology and Medicine, 182, 34–58. 10.1016/j.freeradbiomed.2022.02.012 [DOI] [PubMed] [Google Scholar]
- San Martín A, Ceballo S, Ruminot I, Lerchundi R, Frommer WB, & Barros LF (2013). A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS One, 8(2), e57712. 10.1371/journal.pone.0057712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin A, Sotelo-Hitschfeld T, Lerchundi R, Fernandez-Moncada I, Ceballo S, Valdebenito R, Baeza-Lehnert F, Alegria K, Contreras-Baeza Y, Garrido-Gerter P, Romero-Gómez I, & Barros LF (2014). Single-cell imaging tools for brain energy metabolism: A review. Neurophotonics, 1(1), 1–9. 10.1117/1.NPh.1.1.011004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savtchenko LP, Bard L, Jensen TP, Reynolds JP, Kraev I, Medvedev N, Stewart MG, Henneberger C, & Rusakov DA (2018). Disentangling astroglial physiology with a realistic cell model in silico. Nature Communications, 9(1), 3554. 10.1038/s41467-018-05896-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savtchouk I, & Volterra A (2018). Gliotransmission: Beyond black-and-white. The Journal of Neuroscience, 38(1), 14–25. 10.1523/JNEUROSCI.0017-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheib U, Broser M, Constantin OM, Yang S, Gao S, Mukherjee S, Stehfest K, Nagel G, Gee CE, & Hegemann P (2018). Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 Å structure of the adenylyl cyclase domain. Nature Communications, 9(1), 2046. 10.1038/s41467-018-04428-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmeier S, Hertenstein H, McMullen E, Deharde L & Brankatschk M (2021) Glycolytically impaired glial cells fuel neural metabolism via β-oxidation. ResearchSquare. 10.21203/rs.3.rs-251408/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E, & Oheim M (2020). Infrared excitation induces heating and calcium microdomain hyperactivity in cortical astrocytes. Biophysical Journal, 119(11), 2153–2165. 10.1016/j.bpj.2020.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi KJ, Shekhtmeyster P, Merten K, Arena A, Cook D, Hoffman E, Ngo A, & Nimmerjahn A (2016). Imaging large-scale cellular activity in spinal cord of freely behaving mice. Nature Communications, 7, 11450. 10.1038/ncomms11450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semyanov A, Henneberger C, & Agarwal A (2020). Making sense of astrocytic calcium signals–from acquisition to interpretation. Nature Reviews. Neuroscience, 21(10), 551–564. 10.1038/s41583-020-0361-8 [DOI] [PubMed] [Google Scholar]
- Shekhtmeyster P, Carey EM, Duarte D, Ngo A, Gao G, Nelson NA, Clark CL & Nimmerjahn A (2021) Multiplex, translaminar imaging in the spinal cord of behaving mice. bioRxiv, 2021.12.23.474039. 10.1101/2021.12.23.474039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhtmeyster P, Duarte D, Carey EM, Ngo A, Gao G, Olmstead JA, Nelson NA & Nimmerjahn A (2021) Trans-segmental imaging in the spinal cord of behaving mice. bioRxiv, 2021.12.23.474042. 10.1101/2021.12.23.474042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh OA, Linghu C, Piatkevich KD, Goodwin D, Celiker OT, Gritton HJ, Romano MF, Gau R, Yu C-C, Tseng H-A, Bensussen S, Narayan S, Yang C-T, Freifeld L, Siciliano CA, Gupta I, Wang J, Pak N, Yoon Y-G, … Boyden ES (2020). Precision calcium imaging of dense neural populations via a cell-body-targeted calcium indicator. Neuron, 107(3), 470–486.e11. 10.1016/j.neuron.2020.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemetov AA, Monakhov MV, Zhang Q, Canton-Josh JE, Kumar M, Chen M, Matlashov ME, Li X, Yang W, Nie L, Shcherbakova DM, Kozorovitskiy Y, Yao J, Ji N, & Verkhusha VV (2021). A near-infrared genetically encoded calcium indicator for in vivo imaging. Nature Biotechnology, 39(3), 368–377. 10.1038/s41587-020-0710-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dana H, Abdelfattah AS, Patel R, Shea J, Molina RS, Rawal B, Rancic V, Chang Y-F, Wu L, Chen Y, Qian Y, Wiens MD, Hambleton N, Ballanyi K, Hughes TE, Drobizhev M, Kim DS, Koyama M, … Campbell RE (2018). A genetically encoded Ca2+ indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biology, 16(1), 9. 10.1186/s12915-018-0480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood MW, Arizono M, Panatier A, Mikoshiba K, & Oliet SHR (2021). Astrocytic IP3Rs: Beyond IP3R2. Frontiers in Cellular Neuroscience, 15, 695817. 10.3389/fncel.2021.695817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori-Hayashi M, Koga K, Tozaki-Saitoh H, Kohro Y, Toyonaga H, Yamaguchi C, Hasegawa A, Nakahara T, Hachisuka J, Akira S, Okano H, Furue M, Inoue K, & Tsuda M (2015). Stat3-dependent reactive astrogliosis in the spinal dorsal horn underlies chronic itch. Nature Medicine, 21(8), 927–931. 10.1038/nm.3912 [DOI] [PubMed] [Google Scholar]
- Shivange AV, Borden PM, Muthusamy AK, Nichols AL, Bera K, Bao H, Bishara I, Jeon J, Mulcahy MJ, Cohen B, O’Riordan SL, Kim C, Dougherty DA, Chapman ER, Marvin JS, Looger LL, & Lester HA (2019). Determining the pharmacokinetics of nicotinic drugs in the endoplasmic reticulum using biosensors. Journal of General Physiology, 151(6), 738–757. 10.1085/jgp.201812201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz L-J, Kim IH, Manhaes AC, Rodrigues WS Jr., Pamukcu A, Enustun E, Ertuz Z, Scheiffele P, Soderling SH, Silver DL, Ji R-R, Medina AE, & Eroglu C (2016). Astrocytes assemble Thalamocortical synapses by bridging NRX1α and NL1 via Hevin. Cell, 164(1–2), 183–196. 10.1016/j.cell.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slezak M, Goritz C, Niemiec A, Frisen J, Chambon P, Metzger D, & Pfrieger FW (2007). Transgenic mice for conditional gene manipulation in astroglial cells. Glia, 55(15), 1565–1576. 10.1002/glia.20570 [DOI] [PubMed] [Google Scholar]
- Sofroniew MV (2020). Astrocyte reactivity: Subtypes, states, and functions in CNS innate immunity. Trends in Immunology, 41(9), 758–770. 10.1016/j.it.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somjen GG (1988). Nervenkitt: Notes on the history of the concept of neuroglia. Glia, 1(1), 2–9. 10.1002/glia.440010103 [DOI] [PubMed] [Google Scholar]
- Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji R-R, & Eroglu C (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature, 551(7679), 192–197. 10.1038/nature24638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K, Mihara S, Fu N, & Hisabori T (2020). Real-time monitoring of the in vivo redox state transition using the ratiometric redox state sensor protein FROG/B. Proceedings of the National Academy of Sciences, 117(27), 16019–16026. 10.1073/pnas.1918919117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suminaite D, Lyons DA, & Livesey MR (2019). Myelinated axon physiology and regulation of neural circuit function. Glia, 67(11), 2050–2062. 10.1002/glia.23665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zeng J, Jing M, Zhou J, Feng J, Owen SF, Luo Y, Li F, Wang H, Yamaguchi T, Yong Z, Gao Y, Peng W, Wang L, Zhang S, Du J, Lin D, Xu M, Kreitzer AC, … Li Y (2018). A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell, 174(2), 481–496.e19. 10.1016/j.cell.2018.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Stern SA, Bozdagi O, Huntley GW, Walker RH, Magistretti PJ, & Alberini CM (2011). Astrocyte-neuron lactate transport is required for long-term memory formation. Cell, 144(5), 810–823. 10.1016/j.cell.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Kanemaru K, Ishii K, Ohkura M, Okubo Y, & Iino M (2014). Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nature Communications, 5, 4153. 10.1038/ncomms5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata-Tsuji F, Chounlamountri N, Do L-D, Philippot C, Novion Ducassou J, Couté Y, Achour SB, Honnorat J, Place C, & Pascual O (2021). Microglia modulate gliotransmission through the regulation of VAMP2 proteins in astrocytes. Glia, 69(1), 61–72. 10.1002/glia.23884 [DOI] [PubMed] [Google Scholar]
- Tanaka KF, Ahmari SE, Leonardo ED, Richardson-Jones JW, Budreck EC, Scheiffele P, Sugio S, Inamura N, Ikenaka K, & Hen R (2010). Flexible accelerated STOP tetracycline operator-knockin (FAST): A versatile and efficient new gene modulating system. Biological Psychiatry, 67(8), 770–773. 10.1016/j.biopsych.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M, Hung YP, & Yellen G (2011). Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. Journal of the American Chemical Society, 133(26), 10034–10037. 10.1021/ja202902d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, Zou Y, Shi M, Liu R, Su N, Du J, Zhou H-M, Zhu L, Qian X, Liu H, Loscalzo J, & Yang Y (2017). Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nature Methods, 14(7), 720–728. 10.1038/nmeth.4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, & Giaume C (2012). Connexin-based intercellular communication and astrocyte heterogeneity. Brain Research, 1487, 88–98. 10.1016/j.brainres.2012.06.045 [DOI] [PubMed] [Google Scholar]
- Timper K, Del Río-Martín A, Cremer AL, Bremser S, Alber J, Giavalisco P, Varela L, Heilinger C, Nolte H, Trifunovic A, Horvath TL, Kloppenburg P, Backes H, & Brüning JC (2020). Glp-1 receptor signaling in astrocytes regulates fatty acid oxidation, mitochondrial integrity, and function. Cell Metabolism, 31(6), 1189–1205.e13. 10.1016/j.cmet.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Cabán CC, Yang M, Lai C, Yang L, Subach F, Smith BO, Piatkevich KD & Boyden ES (2021). Tuning the sensitivity of genetically encoded fluorescent potassium indicators through structure-guided and genome mining strategies. bioRxiv, 2021.10.07.463355. 10.1101/2021.10.07.463355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufail Y, Cook D, Fourgeaud L, Powers CJ, Merten K, Clark CL, Hoffman E, Ngo A, Sekiguchi KJ, O’Shea CC, Lemke G, & Nimmerjahn A (2017). Phosphatidylserine exposure controls viral innate immune responses by microglia. Neuron, 93(3), 574–586.e8. 10.1016/j.neuron.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Yoshikawa F, Hishida A, Furuichi T, & Mikoshiba K (2002). A novel recombinant hyperaffinity inositol 1,4,5-trisphosphate (IP3) absorbent traps IP3, resulting in specific inhibition of IP3-mediated calcium signaling. Journal of Biological Chemistry, 277(10), 8106–8113. 10.1074/jbc.M108337200 [DOI] [PubMed] [Google Scholar]
- Unger EK, Keller JP, Altermatt M, Liang R, Matsui A, Dong C, Hon OJ, Yao Z, Sun J, Banala S, Flanigan ME, Jaffe DA, Hartanto S, Carlen J, Mizuno GO, Borden PM, Shivange AV, Cameron LP, Sinning S, … Tian L (2020). Directed evolution of a selective and sensitive serotonin sensor via machine learning. Cell, 183(7), 1986–2002.e26. 10.1016/j.cell.2020.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal S, Srinivasan R, & Khakh BS (2019). GECIquant: Semi-automated detection and quantification of astrocyte intracellular Ca2+ signals monitored with GCaMP6f. In de Pittà M & Berry H (Eds.), Springer series in computational neuroscience. Computational Glioscience (pp. 455–470). Springer International Publishing. 10.1007/978-3-030-00817-8_17 [DOI] [Google Scholar]
- Verkhratsky A, & Nedergaard M (2018). Physiology of Astroglia. Physiological Reviews, 98(1), 239–389. 10.1152/physrev.00042.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Gutierrez C, Bonora N, Bobo-Jimenez V, Jimenez-Blasco D, Lopez-Fabuel I, Fernandez E, Josephine C, Bonvento G, Enriquez JA, Almeida A, & Bolaños JP (2019). Astrocytic mitochondrial ROS modulate brain metabolism and mouse behaviour. Nature Metabolism, 1(2), 201–211. 10.1038/s42255-018-0031-6 [DOI] [PubMed] [Google Scholar]
- Vongsouthi V, Whitfield JH, Unichenko P, Mitchell JA, Breithausen B, Khersonsky O, Kremers L, Janovjak H, Monai H, Hirase H, Fleishman SJ, Henneberger C, & Jackson CJ (2021). A rationally and computationally designed fluorescent biosensor for d-serine. ACS Sensors, 6(11), 4193–4205. 10.1021/acssensors.1c01803 [DOI] [PubMed] [Google Scholar]
- Wan J, Peng W, Li X, Qian T, Song K, Zeng J, Deng F, Hao S, Feng J, Zhang P, Zhang Y, Zou J, Pan S, Shin M, Venton BJ, Zhu JJ, Jing M, Xu M, & Li Y (2021). A genetically encoded sensor for measuring serotonin dynamics. Nature Neuroscience, 24(5), 746–752. 10.1038/s41593-021-00823-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Qi G, Zhou Y, Zhang Y, Wang B, Hu P, & Jin Y (2020). Single-cell ATP detection and content analyses in electrostimulus-induced apoptosis using functionalized glass nanopipettes. Chemical Communications (Cambridge, England), 56(10), 1561–1564. 10.1039/c9cc08889j [DOI] [PubMed] [Google Scholar]
- Wang P, Chen T, Sakurai K, Han B-X, He Z, Feng G, & Wang F (2012). Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Scientific Reports, 2, 497. 10.1038/srep00497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, DelRosso NV, Vaidyanathan TV, Cahill MK, Reitman ME, Pittolo S, Mi X, Yu G, & Poskanzer KE (2019). Accurate quantification of astrocyte and neurotransmitter fluorescence dynamics for single-cell and population-level physiology. Nature Neuroscience, 22(11), 1936–1944. 10.1038/s41593-019-0492-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg BH, Cho JH, Agarwal Y, Pham NTH, Caraballo LD, Walkosz M, Ortega C, Trexler M, Tague N, Law B, Benman WKJ, Letendre J, Beal J, & Wong WW (2019). High-performance chemical- and light-inducible recombinases in mammalian cells and mice. Nature Communications, 10(1), 4845. 10.1038/s41467-019-12800-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton DK, Dissing-Olesen L, & Stevens B (2019). Neuron-glia signaling in synapse elimination. Annual Review of Neuroscience, 42, 107–127. 10.1146/annurev-neuro-070918-050306 [DOI] [PubMed] [Google Scholar]
- Winchenbach J, Düking T, Berghoff SA, Stumpf SK, Hülsmann S, Nave K-A, & Saher G (2016). Inducible targeting of CNS astrocytes in Aldh1l1-CreERT2 BAC transgenic mice. F1000Research, 5, 2934. 10.12688/f1000research.10509.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windrem MS, Osipovitch M, Liu Z, Bates J, Chandler-Militello D, Zou L, Munir J, Schanz S, McCoy K, Miller RH, Wang S, Nedergaard M, Findling RL, Tesar PJ, & Goldman SA (2017). Human iPSC glial mouse chimeras reveal glial contributions to schizophrenia. Cell Stem Cell, 21(2), 195–208.e6. 10.1016/j.stem.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosker H, Balu DT, & Coyle JT (2016). The rise and fall of the D-serine-mediated gliotransmission hypothesis. Trends in Neurosciences, 39(11), 712–721. 10.1016/j.tins.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-Y, Wen Y, Serre NBC, Laursen CCH, Dietz AG, Taylor BR, Aggarwal A, Rancic V, Becker M, Ballanyi K, Podgorski K, Hirase H, Nedergaard M, Fendrych M, Lemieux MJ, Eberl DF, Kay AR, Campbell RE & Shen Y (2021). A sensitive and specific genetically encodable biosensor for potassium ions. bioRxiv, 2021.10.07.463410. 10.1101/2021.10.07.463410 [DOI] [Google Scholar]
- Wu Z, He K, Chen Y, Li H, Pan S, Li B, Liu T, Xi F, Deng F, Wang H, Du J, Jing M, Li Y (2022). A sensitive GRAB sensor for detecting extracellular ATP in vitro and in vivo. Neuron, 110, (5), 770.–782.e5. 10.1016/j.neuron.2021.11.027 [DOI] [PubMed] [Google Scholar]
- Xiao L, Ohayon D, McKenzie IA, Sinclair-Wilson A, Wright JL, Fudge AD, Emery B, Li H, & Richardson WD (2016). Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nature Neuroscience, 19(9), 1210–1217. 10.1038/nn.4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Ford NC, He S, Huang Q, Anderson M, Chen Z, Yang F, Crawford LK, Caterina MJ, Guan Y, & Dong X (2021). Astrocytes contribute to pain gating in the spinal cord. Science Advances, 7(45), eabi6287. 10.1126/sciadv.abi6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ruchti E, Petit JM, Jourdain P, Grenningloh G, Allaman I, & Magistretti PJ (2014). Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proceedings of the National Academy of Sciences, 111(33), 12228–12233. 10.1073/pnas.1322912111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Yuan P, Ouellette B, Zhou T, Mortrud M, Balaram P, Chatterjee S, Wang Y, Daigle TL, Tasic B, Kuang X, Gong H, Luo Q, Zeng S, Curtright A, Dhaka A, Kahan A, Gradinaru V, Chrapkiewicz R, … Cetin A (2020). RecV recombinase system for in vivo targeted optogenomic modifications of single cells or cell populations. Nature Methods, 17(4), 422–429. 10.1038/s41592-020-0774-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Xue L, Hiblot J, Griss R, Fabritz S, Roux C, Binz P-A, Haas D, Okun JG, & Johnsson K (2018). Semisynthetic sensor proteins enable metabolic assays at the point of care. Science, 361(6407), 1122–1126. 10.1126/science.aat7992 [DOI] [PubMed] [Google Scholar]
- Yu X, Moye SL, & Khakh BS (2021). Local and CNS-wide astrocyte intracellular calcium signaling attenuation in vivo with CalExflox mice. The Journal of Neuroscience, 41(21), 4556–4574. 10.1523/JNEUROSCI.0085-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Nagai J, Marti-Solano M, Soto JS, Coppola G, Babu MM, & Khakh BS (2020). Context-specific striatal astrocyte molecular responses are phenotypically exploitable. Neuron, 108(6), 1146–1162. e10. 10.1016/j.neuron.2020.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, & Linnarsson S (2015). Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science, 347(6226), 1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- Zhang J, Dublin P, Griemsmann S, Klein A, Brehm R, Bedner P, Fleischmann BK, Steinhäuser C, & Theis M (2013). Germ-line recombination activity of the widely used hGFAP-Cre and nestin-Cre transgenes. PLoS One, 8(12), e82818. 10.1371/journal.pone.0082818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-F, Liu B, Hong I, Mo A, Roth RH, Tenner B, Lin W, Zhang JZ, Molina RS, Drobizhev M, Hughes TE, Tian L, Huganir RL, Mehta S, & Zhang J (2021). An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nature Chemical Biology, 17(1), 39–46. 10.1038/s41589-020-00660-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Herde MK, Mitchell JA, Whitfield JH, Wulff AB, Vongsouthi V, Sanchez-Romero I, Gulakova PE, Minge D, Breithausen B, Schoch S, Janovjak H, Jackson CJ, & Henneberger C (2018). Monitoring hippocampal glycine with the computationally designed optical sensor GlyFS. Nature Chemical Biology, 14(9), 861–869. 10.1038/s41589-018-0108-2 [DOI] [PubMed] [Google Scholar]
- Zhang Y, & Barres BA (2010). Astrocyte heterogeneity: An underappreciated topic in neurobiology. Current Opinion in Neurobiology, 20(5), 588–594. 10.1016/j.conb.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MSB, Li G, Duncan III JA, Cheshier SH, Shuer LM, Chang EF, Grant GA, Hayden Gephart MG, & Barres BA (2016). Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron, 89(1), 37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cheng X, Zhao Y, & Yang Y (2020). Lighting up live-cell and in vivo central carbon metabolism with genetically encoded fluorescent sensors. Annual Review of Analytical Chemistry, 13(1), 293–314. 10.1146/annurev-anchem-091619-091306 [DOI] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, & Messing A (2001). Hgfap-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis, 31(2), 85–94. 10.1002/gene.10008 [DOI] [PubMed] [Google Scholar]
- Zuchero JB, & Barres BA (2015). Glia in mammalian development and disease. Development, 142(22), 3805–3809. 10.1242/dev.129304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuend M, Saab AS, Wyss MT, Ferrari KD, Hösli L, Looser ZJ, Stobart JL, Duran J, Guinovart JJ, Barros LF, & Weber B (2020). Arousal-induced cortical activity triggers lactate release from astrocytes. Nature Metabolism, 2(2), 179–191. 10.1038/s42255-020-0170-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


