Abstract
Altered mitochondrial DNA (mtDNA) occurs in neurodegenerative disorders like Alzheimer’s disease (AD); how mtDNA synthesis is linked to neurodegeneration is poorly understood. We previously discovered Nutrient-induced Mitochondrial Activity (NiMA), an inter-organelle signaling pathway where nutrient-stimulated lysosomal mTORC1 activity regulates mtDNA replication in neurons by a mechanism sensitive to amyloid-β oligomers (AβOs), a primary factor in AD pathogenesis (Norambuena et al., 2018). Using 5-ethynyl-2′-deoxyuridine (EdU) incorporation into mtDNA of cultured neurons, along with photoacoustic and mitochondrial metabolic imaging of cultured neurons and mouse brains, we show these effects being mediated by mTORC1-catalyzed T40 phosphorylation of superoxide dismutase 1 (SOD1). Mechanistically, tau, another key factor in AD pathogenesis and other tauopathies, reduced the lysosomal content of the tuberous sclerosis complex (TSC), thereby increasing NiMA and suppressing SOD1 activity and mtDNA synthesis. AβOs inhibited these actions. Dysregulation of mtDNA synthesis was observed in fibroblasts derived from tuberous sclerosis (TS) patients, who lack functional TSC and elevated SOD1 activity was also observed in human AD brain. Together, these findings imply that tau and SOD1 couple nutrient availability to mtDNA replication, linking mitochondrial dysfunction to AD.
Keywords: Alzheimer’s disease, Insulin, Amino acids, mTOR, Tau
1. Introduction
The human brain represents ~2% of the total body mass, but one fifth and one quarter of the total daily oxygen and glucose expenditure occurs there, respectively (Rolfe and Brown, 1997). Exquisite regulation of this costly oxygen and glucose budget is crucial to cognitive functions, especially in humans. The complexity of this daunting task multiplies when considering metabolic adjustments needed to support the activity of specific neuronal populations. It is not surprising, therefore, that much remains to be learned about how neurons orchestrate signaling pathways that control mitochondrial activity.
Reduction in brain glucose utilization is a well-known feature in cognitive disorders like Alzheimer’s disease (AD) (Camandola and Mattson, 2017). Fuoro-2-deoxyglucose positron-emission tomography (FDG-PET) studies of human brains have shown a larger decline in glucose utilization in the hippocampus and cortex in AD brain as compared to individuals without dementia (Crane et al., 2013; Croteau et al., 2018; Gordon et al., 2018; Kapogiannis and Mattson, 2011). At the organ level, PET studies tracking oxygen-15 showed that cerebral metabolic rate of oxygen is reduced in several AD brain regions and its decline positively correlates with severity of dementia (Collins et al., 2015; Ishii et al., 1996). As the main oxygen-consuming organelle, mitochondria not only are the central cellular source of nutrient-derived chemical energy in the form of ATP, but also are key contributor of the total cellular ROS (Balaban et al., 2005), which per se serve as physiological envoys as well as noxious molecules when in excess (Shadel and Horvath, 2015). Along these lines, mitochondrial dysfunction has been long linked to a variety of neurodegenerative disorders (Burté et al., 2014; Chen and Chan, 2009; Sheng and Cai, 2012), in particular AD (Calkins et al., 2011; DuBoff et al., 2013; Manczak et al., 2011). At the molecular level, deficient brain energy metabolism has been linked to both compromised expression of electron transport chain proteins (Adav et al., 2019; Cottrell et al., 2001; Valla et al., 2001) and mtDNA mutations in neurons (Mosconi et al., 2007). These observations not only highlight a maternally inherited risk factor in AD (Liang et al., 2008; Mosconi et al., 2007), but also suggest that dysregulation of mechanisms regulating the mitochondrial genome may account for brain energy metabolism deficiencies.
Mitochondria play a critical role in cell and body physiology by regulating the homeostasis of lipids, calcium and reactive oxygen species (ROS), cell death, inflammasome activation and others (Labbé et al., 2014; Nunnari and Suomalainen, 2012; Zhong et al., 2018). Mitochondrial dynamics and function are highly regulated by extracellular cues or global changes to enable timely adjustment on mitochondrial metabolism. This process also involves the coordinated expression of >1000 nuclear-encoded genes and those encoded by mtDNA (Nunnari and Suomalainen, 2012; Suomalainen and Battersby, 2017). Neurodegenerative disorders (Burté et al., 2014; DuBoff et al., 2013), type 2 diabetes (De Felice and Ferreira, 2014) and cancer (Vyas et al., 2016), have all been linked to defects in mtDNA maintenance (Nunnari and Suomalainen, 2012; Trifunovic et al., 2004). Although mutations in mtDNA have not been directly linked to AD, brain oxidative metabolism deficiencies do associate with mtDNA mutations that affect expression of energy metabolism genes in posterior cingulate neurons (Liang et al., 2008; Mosconi et al., 2007). Furthermore, diminished mitochondrial functioning in AD is associated with single nucleotide polymorphisms, germline variants, deletions affecting the expression of electron transport chain genes and reduced base-excision repair in mtDNA (Wang et al., 2020). Still, the mechanisms coordinating nutrient availability, cellular redox states and mtDNA maintenance are not fully understood.
Superoxide dismutases (SODs) are metallo-enzymes that catalyze removal of superoxide free radicals (Miao and Clair, 2009). Copper and Zinc SOD (CuZnSOD or SOD1), and Manganese SOD (MnSOD or SOD2) are conserved intracellular proteins (Valentine et al., 2005); SOD1 is the major SOD that is widely distributed throughout the cytosol, mitochondrial intermembrane space and the nucleus, whereas SOD2 exclusively resides in mitochondria(Weisiger and Fridovich, 1973). SOD1 activity restricts the superoxide accumulation during mitochondrial respiration, and along with peroxidases, directly controlling cellular ROS levels and oxidative damage. In addition, recent evidence suggests that SOD1 can directly modulate respiration by stabilizing casein kinase 1, a mitochondrial respiration repressor (Reddi and Culotta, 2013). SOD1 dysfunctions have been linked to ALS (Valentine et al., 2005) and seems to play a role in cancer (Che et al., 2016). Although the potential involvement of SOD1 in AD has been suggested before (Murakami et al., 2011; Yoon et al., 2009), mechanistic evidence remains elusive.
Aβ peptides and tau can work in coordination to disrupt neuronal function at different levels, including organelle functioning (Guo et al., 2017; Norambuena et al., 2018; Polanco et al., 2018). For example, we recently discovered an inter-organelle signaling pathway, NiMA (Nutrient-induced Mitochondrial Activity), in which activation of the lysosome-associated multi-subunit kinase, mechanistic target of rapamycin complex 1 (mTORC1) by insulin or amino acids, leads to inhibition of mtDNA synthesis and increase mitochondrial oxidative phosphorylation (OXPHOS) in vitro and in vivo (Norambuena et al., 2018). In addition, our group previously found that soluble AβOs trigger lipid raft-and Rac1dependent mTORC1 activity at the plasma membrane (PM), by a mechanism dependent on tau (Norambuena et al., 2017). As a consequence, AβOs trigger neuronal cell cycle re-entry, a harbinger of neuron death in AD (Arendt et al., 2010), and inhibit NiMA as well (Norambuena et al., 2018). The molecular mechanism regulating the NiMA signaling pathway is still not fully understood, but the recent addition of SOD1 to the expanding list of proteins regulated by mTORC1 (Tsang et al., 2018), opened the possibility that SOD1 is an upstream NiMA regulator and a mediator of its disruption by the AβO-tau axis in AD.
Using EdU incorporation into mtDNA, metabolic imaging at single-cell and tissue levels by 2-photon fluorescence lifetime microscopy (2P-FLIM) (Norambuena et al., 2018), along with multi-parametric photoacoustic microscopy (MP-PAM) (Ning et al., 2015), we now show that tau-dependent activation of lysosomal mTORC1 inhibits SOD1 activity in neurons by a mechanism that requires phosphorylation of SOD1 at T40. This allows mTORC1 to downregulate mtDNA synthesis in the presence of nutrients, a process that we found to be dysregulated by AβOs. SOD1 activity was stimulated by AβOs in cultured neurons and was elevated in human AD brain. Finally, pharmacologically inhibiting SOD1 activity with ATN-224 restrained mtDNA synthesis and inhibited respiration in live cells and in the mouse cortex. Thus, SOD1 is a part of a neuronal nutrient-sensing machinery that is based on tau and mTORC1, and functionally connects extracellular cues to mitochondrial functioning, enabling nutrients to sustain OXPHOS and prevent unwanted mtDNA synthesis. It is possible that disruption of NiMA contributes to oxidative damage and propagation of mtDNA mutations linked to diseases such as AD (Blanchard et al., 1993; Carrieri et al., 2001; Chagnon et al., 1999; Chen et al., 2016; Corral-Debrinski et al., 1994; Coskun et al., 2004; Hudson et al., 2012; Inczedy-Farkas et al., 2014; Lakatos et al., 2010; Van Der Walt et al., 2004), ALS (Wiedemann et al., 2002) and TS (Sakamoto et al., 2018).
2. Materials and methods
2.1. Cell culture and materials
Human embryonic kidney (HEK293) were grown in DMEM/F12 media (GIBCO) supplemented with HyClone cosmic calf serum (GE Healthcare) and 50 μg/ml gentamycin (GIBCO).
Human tuberous sclerosis fibroblasts with a polymorphisms in TSC1 gene (catalog number GM06149 and GM06121) were obtained from the Coriell Institute, and were cultured according to the vendor’s specifications in Eagle’s minimal essential medium (GIBCO) supplemented with 15% Optima fetal bovine serum (Atlanta Biologicals).
To prepare primary mouse neurons, brain cortices were dissected from E17/18 WT (C57/Bl6) or tau knockout mice (Dawson et al., 2001), and primary neuron cultures were prepared as described previously (Norambuena et al., 2018; Norambuena et al., 2017). Experiments were performed using cultures grown for at least 10 days in Neurobasal medium (GIBCO) supplemented with B27 (GIBCO) and 50 μg/ml gentamycin before being used for experiments, except that lentivirus transductions were performed 3 days in advance of all other experimental procedures.
Amyloid-β oligomers (AβOs) were prepared as follow. Lyophilized, synthetic Aβ1–42 (AnaSpec) was dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (Sigma-Aldrich) to ~1 mM and evaporated overnight at room temperature. The dried powder was resuspended for 5 min at room temperature in 40–50 μl dimethylsulfoxide to ~1 mM and sonicated for 10 min in a water bath. To prepare oligomers, the dissolved, monomeric peptide was diluted to ~400 μl (100 μM final concentration) in Neurobasal medium (GIBCO), incubated 48 h at 4 °C with rocking, and then centrifuged at 14,000g for 15 min to remove fibrils. For all experiments, AβOs were diluted into tissue culture medium to a final concentration of ~1.5 μM total Aβ1–42 (Supplemental Figs. 3D and 6B and C) or as indicated in Fig. 6A and Supplemental Fig. 5.
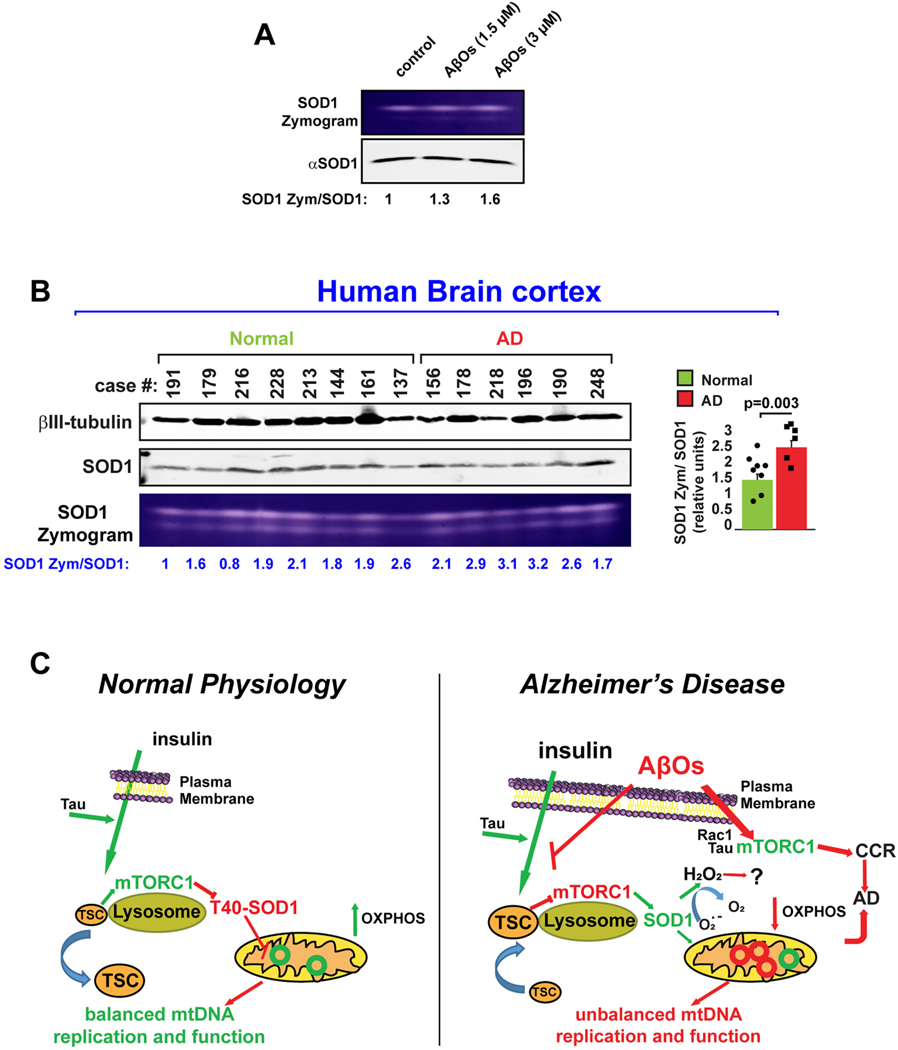
Fig. 6.
SOD1 activity is increased in AD.
(A) SOD1 activity was followed in WT mouse neurons cultures treated with different concentrations of AβO during 1 h. In-gel activity assays showed that AβO increased SOD1 activity in a dose-dependent manner. Data are representative of three independent assays. (B) SOD1 activity is higher in AD human brains. SOD1 activity assays were analyzed in human brain cell extracts taken from deceased patients affected by AD and compared to normal controls. Samples were analyzed by In-gel activity assays and the average SOD1 zymogen: SOD1 protein ratio in Normal and AD groups were analyzed by using student t-test. No significant outliers were detected on each group of patient-derived brain samples (Grubss’test; GraphPad software).
(C) Model. Under physiological conditions, nutrient stimulated and tau-dependent activation of lysosomal mTORC1 leads to phosphorylation of SOD1 at T40, directly regulating mitochondrial oxidative pathways and mtDNA synthesis by a mechanism independent of Ck1γ2, but likely involving substrates sensitive to oxidation by hydrogen peroxide. Insulin and amino acid-regulated mitochondrial functions are blocked by AβOs, which increased the lysosomal positioning of TSC, likely decreasing mTORC1 activity and stimulating it instead at the PM, where mTORC1 kinase activity triggers cell cycle re-entry (CCR), a frequent prelude to cortical neuron death in Alzheimer’s disease (AD). Thus, SOD1 is at the crossroads of regulating mitochondrial activity through the NiMA pathway and mediating its disruption in AD.
2.2. Lentivirus production and infection
Lentiviral particles for shRNA knockdowns and hTau 2N4R and 0N4R protein expression in mouse cortical neurons were prepared as follows. The expression plasmids, pCSC-SP-PW-NepX (also known as BOB-NepX) or pLKO.1 (Addgene plasmids 12,340 and 10,878, respectively), and the packaging vectors, pSPAX2 and pMD2.G (Addgene plasmids 12,260 and 12,259, respectively) were transfected using Lipofectamine 3000 (ThermoFisher) into HEK293T cells grown in 15 cm Petri dishes to ~80% confluence in DMEM (GIBCO) supplemented with 10% HyClone cosmic calf serum (GE Healthcare). Each transfection was with 15 μg total DNA at a 50%/37.5%/12.5% ratio of expression vector/pSPAX2/pMD2.G. Lentivirus-conditioned medium was collected 24 and 48 h after the start of transfection. Lentiviral particles were concentrated in a Beckman Coulter Optima LE-80 K ultracentrifuge for 2 h at 23,000 rpm (95,000 gav) at 4 °C in an SW28 rotor, resuspended in 400 μl Neurobasal medium and stored at −80 °C in 20 μl aliquots. Cultured neurons were transduced in Neurobasal/B27 medium and incubated for 72 h before assays were performed.
2.3. Human brain cell extracts
This study was approved by the Institutional Review Board for Health Sciences Research at the University of Virginia. Human frontal cortex brain samples were previously acquired via generous donation from the University of Virginia Brain Resource Facility. A total of 8 patients free from pathological evidence of Alzheimer’s disease (mean age 73.5 ± 3.9 years, 50% female, average post-mortem interval (PMI) 9.1 ± 1.6 h) were compared to 6 patients with Alzheimer’s pathology (mean age 76.8 ± 2.3 years, 50% female, average PMI 7.5 ± 1.8 h). There was no significant difference in age (p = 0.53) or PMI (p = 0.51) between normal and AD subjects. Portioned brain samples in 1.5 ml Eppendorf tubes were thawed on ice, and one scoop of 0.5 mm zirconium oxide beads was added along with 100–200 μl of lysis buffer to cover sample and beads. Lysis buffer consisted of RIPA buffer (Bioworld, #42020024–2); 1:100 HALT™ protease inhibitor cocktail and 1:100 EDTA (Thermo Scientific, #78430); 1:100 phosphatase inhibitor cocktail 2 (Sigma Aldrich, #P5726); and 1:100 phosphatase inhibitor cocktail 3 (Sigma Aldrich, #P0044). This mixture was placed for 5–15 min in a bullet blender at 4 °C until fully homogenized, after which homogenate was placed in a centrifuge at 10,000 RPM for 10 min at 4 °C. Finally, supernatant was transferred to a new tube and used as protein lysate for subsequent SOD1 activity assays (see below) and western blots. A similar procedure was performed to prepare mouse brain cell extracts.
2.4. Superoxide dismutase 1 activity, oxygen consumption and PTEN activity assays
Superoxide dismutase 1 activity was evaluated as described previously (Weydert and Cullen, 2010). This method detects SOD activity from freshly as well as frozen specimens (Weydert and Cullen, 2010). Briefly, mammalian cells were washed three times in phosphate-buffered saline (Fisher Scientific BP665), lysed in 50 mM phosphatase buffer (0.05 M KH2PO4, 0.05 M K2HPO4, pH 7.8) with a Sonicator (FisherScientific FB120) set at 40% power for 30s on ice. Cell lysates were diluted in native 2× sample buffer (Bio Rad 1,610,738). 20–30 uls of the protein samples were separated in 12% native PAGE gels, which were stained with nitro blue tetrazolium chloride (Abcam, catalog number ab146262), as described previously (Weydert and Cullen, 2010). The 12% native polyacrylamide gels used were optimized for separation of SOD-2 (88 kD) from SOD-1 (32 kD), and as we show in Fig. S8, cyanide completely abolished formazan deposition on gels of HEK293 cell extracts, as well as for SOD1-GFP, but not for the more slowly migrating bands that mark SOD2. To visualize SOD1 and SOD2 activities, gels were rinsed with water three times and placed on a light box for 20 min to 2 h. SOD1 and SOD2 bands were identified either by sodium cyanide treatment in the gel staining step as described by Weydert and Cullen (Weydert and Cullen, 2010; see also Fig. S8) or shRNA mediated knockdown of SOD1 in mouse neurons. In parallel, same amount of cell lysates were separated by SDS-PAGE followed by western blotting to detect SOD-1 protein and the housekeeping ribosomal S6 protein. Similar procedure was followed when SOD1 activity was evaluated from human brain cell extracts. In all cases, SOD1 enzyme activity was normalized to the amount of SOD1 protein detected by western blotting. A Li-Cor Odyssey imaging station that provides ~5 logs of linear response (~100,000 shades of linear intensity) was used for direct scanning and software-driven quantitation of fluorescently labeled western blots. The Odyssey software was also used to quantify zymogram TIFF images captured by a 16-bit (65,536 shades of linear intensity) transparency scanner attached to an iMac. Thus, the SOD1 zymogen:SOD1 protein ratios are independent of differences in protein loading between experimental conditions or between brain tissue samples.
For Oxygen consumption, WT mouse cortical neurons grown in 96 well plates (black plate, clear bottom, Costar catalog number 3603) were serum-starved in Hank’s balanced salt solution, as described earlier (Norambuena et al., 2018), then treated for 1 h with the SOD1 inhibitor and copper chelator, ATN-224 Cayman Chemical (cat no. 23553). Oxygen consumption was assayed using the Extracellular O2 Consumption Assay (Abcam catalog number ab197243).
To evaluate PTEN Oxidation, hydrogen peroxide-induced oxidation of PTEN was performed as described by Juarez et al. (Juarez et al., 2008) with few modifications. Briefly, WT neurons were serum starved in HBSS during 2 h, then treated with 500 nM hydrogen peroxide for 15 min. Neurons were washed in cold PBS on ice, lysed in RIPA buffer (1% Nonidet P40, 0.25% sodium deoxicolate, 150 mM NaCl, 50 mM Tris-HCl pH = 7.5, 200 μM sodium orthovanadate, 10 μM NaF, 100 nM okadaic acid) supplemented with 10 mM N-ethylmaleimide during 30 min at 4 °C. Samples were run on a nonreducing SDS-PAGE and transferred to nitrocellulose membranes (BioRad cat. No. 1620112). The same membranes were incubated with antibodies against PTEN and S6 ribosomal protein as a loading control. Oxidized PTEN runs faster than the reduced form on a SDS-PAGE (Juarez et al., 2008).
2.5. cDNA constructs and shRNA sequences
Mouse TSC2 knockdowns was described by us before (Norambuena et al., 2017). pcDNA3 LysoTorcar was a kind gift of Dr. Jin Zhang (University of California, San Diego). Human 2N4R tau vector (Norambuena et al., 2017) was amplified by PCR and transferred to pBOB-NepX between the AgeI and HpaI sites using the following primers: forward 5′ TTA ACC GGT ATG GCT GAG CCC CGC CAG 3′ and reverse 5′ ATT GTTAAC CTA CAA ACC CTG CTT GGC CAG 3′. The same strategy was use to transfer hTau 0N4R WT (see below) to the pBOB NepX lentiviral vector. To knockdown TSC2, a shTSC2-pLKO.1 construct was described previously (Norambuena et al., 2017).
The human SOD1 phospho-null (T40A) and phospho-mimetic (T40E) constructs were generated using the NEB Q5® Site-Directed Mutagenesis Kit (cat noE0554S) using as a template the human SOD1 construct pF146 pSOD1WTAcGFP1described below. Primers design and procedure was done following manufacturer recommendations (http://nebasechanger.neb.com/).
The following constructs were from Addgene:
pRK7-FLAG-TSC1 (plasmid 8995; gift from Dr. John Blenis).
pRK5-GFP-hTAU 0N4R WT (plasmid 46,904; gift from Dr. Karen Ashe).
pF146 pSOD1WTAcGFP1 (plasmid 26,407: gift from Elizabeth Fisher).
pSPAX2 and pMD2.G (plasmids 12,259 and 12,260; gift from Didier Trono).
The following plasmids were purchased from The RNAi Consortium of the Broad Institute; 1) Mouse Nprl3 (TRCN0000175195). 2) Mouse SOD1 (TRCN0000101046).
2.6. Immunoblotting
Samples were resolved by SDS-PAGE using either 10 or 12% acrylamide/bis-acrylamide gels and transferred to 0.22 μm nitrocellulose (Bio-Rad). Membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences), and were incubated with primary antibodies and secondary IRDye-labeled antibodies (see Supplemental Table 1) diluted into antibody buffer (Odyssey blocking buffer diluted 1:1 in PBS/0.1% Tween 20). All antibody incubations were for 1 h at room temperature or overnight at 4 °C, and 3 washes of 5 min each with PBS/0.1% Tween 20 were performed after each antibody step. Membranes were dried by placing them on sheets of filter paper prior to quantitative imaging with an Odyssey imaging station (LI-COR Biosciences).
2.7. NADH and NADPH measurement
2.7.1. Time-correlated single photon counting (TCSPC) Fluorescence Lifetime Imaging Microscopy (FLIM)
FLIM was recorded on a Zeiss LSM-780 NLO confocal/multiphoton microscopy system comprising an inverted Axio Observer Z1 microscope, an X-Cite 120PC Q mercury arc light source (Excelitas Technologies) for cell selection, a motorized stage for automated scanning, an IR Chameleon Vision-II ultrafast Ti:sapphire laser for multiphoton excitation (Coherent), a Zeiss 40 × 1.3 NA oil immersion Planapo objective, an environmental chamber (PeCon GmbH, Germany) that envelops the microscope stage to control temperature and CO2 level, and a 3-channel FLIM system based on three HPM-100–40 GaAsP-based hybrid detectors and 3 SPC-150 TCSPC boards (Becker & Hickl). The SPC-150 boards are synchronized with the 2-photon excitation laser and the Zeiss LSM-780 NLO scan head signal. Ex 740 nm; Em450/50 nm.
2.7.1.1. Imaging.
Mouse neurons, and HEK293 cells were grown in 35-mm glass-bottom dishes, and maintained at 37 °C in 5% CO2/95% air on the stage of the Zeiss LSM-780 NLO microscope. All cultures were serum- starved in Hank’s balanced salt solution (HBSS) for 2 h before addition of ATN 224 to 2 μM, and imaged before and 60 min after treatment. The laser was tuned to 740 nm with an average power of 7 mW at the specimen plane, and NAD(P)H fluorescence was collected using a 450–500 nm emission filter (Objective lens, 20×). For each experiment, 5–10 field of views were recorded in the descanned mode, and then each field of view was subjected to a 40-s acquisition in the non-descanned mode. The laser power and acquisition time were selected to ensure enough photons per pixel while avoiding photodamage to cells. Next, ROIs corresponding to mitochondria were selected from the NAD(P)H photon image for lifetime analysis. Both the lifetimes and fractions of free and enzyme-bound NAD(P)H were calculated on a per pixel basis, from which pseudo-color images of the bound NAD(P)H and histograms of the frequency distribution of a2% (fraction of bound NAD(P)H) were generated.
2.7.1.2. Processing.
FLIM images were processed with SPCImage software (v5; Becker & Hickl) with non-linear least square for optimal fitting. Data images were exported for analysis using SPCImage to establish the free and enzyme-bound fractions of NAD(P)H. The intensity images were used to document mitochondrial morphology and to generate ROIs with a custom plug-in for ImageJ (http://imagej.net/Welcome) to capture the discrete and heterogeneous nature of mitochondrial dynamics. A custom ImageJ macro extracted parameters of interest, most notably the fraction of enzyme-bound NAD(P)H. A custom macro in Microsoft Excel further processed the data by individual ROI and produced histograms, charts and statistics, followed by merging the different field of views for charting.
3. Other fluorescence microscopy procedures
3.1. Primary mouse neuron transfections
6–7 days old cultures of WT or Tau KO (Dawson et al., 2001) neurons growing on no. 1.5 thickness, 12 mm round glass coverslips in 24-well culture dishes were transfected with human SOD1-GFP WT, TA or TE using Lipofectamine 2000 (ThermoFisher). Each transfection was performed by diluting 0.5 μg of plasmid in 50 μls of plain Neurobasal medium. On a different tube, 1 μl of Lipofectamine 2000 (ThermoFisher) was diluted in 50 μls of the same medium. The final DNA/Lipofectamine complexes (100uls) were added to the cells maintained in 100 μls of conditioned medium. 5–6 h after transfection, 300 μls of previously saved conditioned medium was added back to the cultures. EdU uptake assay was performed 24 h later as described below. Cells were fixed with 3.7% para-formaldehyde in PBS, and the coverslips were sealed to glass slides using Fluormount G (ThermoFisher).
3.2. Human tuberous sclerosis fibroblasts
Cells growing on #1.5 thickness, 12 mm round glass coverslips in 24-well dishes were transfected with 0.5 μg of pRK7-FLAG-TSC1 using Lipofectamine 2000 (ThermoFisher) following the same procedure described in the previous paragraph. Expression of exogenously encoded TSC1 was monitored 24 h later by immunofluorescence. Cells were rinsed in PBS and fixed for 15 min in 3.7% para-formaldehyde. Next, they were washed and permeabilized in washing buffer (0.2% Tween 20 in PBS) 3 times for 5 min each and incubated for 60 min in blocking buffer (PBS version; LI-COR Biosciences). Fixed cells were then incubated for 1 h each with primary and secondary antibodies diluted into blocking buffer supplemented with Tween 20 to 0.2%, with several PBS washes after each antibody step. Finally, coverslips were mounted onto glass slides using Fluormount-G (ThermoFisher). Samples were imaged on a Nikon Eclipse Ti equipped with a Yokogawa CSU-X1 spinning disk head, 60 × 1.4 NA Planapo objective, a Hamamastu Flash 4.0 scientific CMOS camera, and 405 nm, 488 nm, 561 nm and 640 nm lasers.
3.3. Mitochondrial DNA (mtDNA) replication experiments
To visualize mtDNA replication by we used Click-it chemistry as described by others (Lewis et al., 2016). EdU is a thymidine analog that readily incorporates into active replicating DNA, which can be detected by a copper catalyzed reaction that covalently links (“clicks”) the alkyne group in EdU to the picolyl azide group in an Alexa Fluor® dye (Salic and Mitchison, 2008). Mitochondria were labeled with Mitotracker CMXRos (Invitrogen catalog number M7512), and AlexaFluor 647-EdU-tagged nucleoids were revealed using the Click-It chemistry (Salic and Mitchison, 2008). Briefly, WT mouse cortical neurons grown on glass coverslips were serum-starved in HBBS for 2 h in the presence or absence of AβOs (see below) and aphidicolin (7.5 μM) before cells were treated for 3 h with nutrients (1 μM insulin plus 0.8 mM L-leucine [Sigma Aldrich catalog numbers I5500 and L8000, respectively] and 0.4 mM L-arginine [Acros Organics catalog number 105001000)]). For immunofluorescence, cells were incubated with 100 nM Mitotracker Red CMXRos for 60 min before being assayed. The immunofluorescence labeling procedure was as described for human tuberous sclerosis fibroblasts. Automatic counting of Perinuclear mitochondrial nucleoids per neuron was performed by using the “Particle Analysis” plugging in ImageJ software (https://imagej.net/imaging/particle-analysis). Briefly, each channel on the RGB stack was separated and saved as an 8-bit white/black image. The corresponding EdU channel was analyzed for background immunofluorescence subtraction and a threshold range was manually set to isolate the object of interests (EdU foci). Finally, all the EdU foci in the field of view (fov) were automatically recorded using the “analyze particle” tool. To total EdU foci/cell shown on figures was finally calculated by dividing the number of EdU foci per fov for the total number of cells contained on each fov.
3.4. Co-localization assays
Co-localization of mTOR with LAMP1 or Mitotracker CMXRos was quantified by an ImageJ plug-in (http://rsbweb.nih.gov/ij/index.html) for the Manders coefficient, as described before(Norambuena et al., 2017), which is based on the Pearson Correlation Coefficient (Dunn et al., 2011). Briefly, images were taken by a Nikon eclipse inverted microscope equipped with a Yokagawa spinning disk confocal head, and a 40 × 1.4 NA lens with a ~ 200 nm resolution along the X and Y axes. Each micrograph was captured using a Hamamatsu Flash 4.0 CMOS camera and yielded ~3.2 million pixels, which is sufficient to produce images that are not pixel limited for the full theoretical resolution of the objective. Results are based on the quantitative analyses of dozens of neurons under each experimental condition. Each analysis was performed on a pixel-by-pixel basis by using Pearson’s coefficient correlation calculation with ImageJ software at 256 shades of gray/channel/pixel. This method is far more sensitive and accurate than can be inferred by simple visual inspection of digital images displayed on a computer screen.
3.5. Multiparametric Photoacoustic Microscopy (MP-PAM)
12–16 week-old male CD-1 mice were used in this study. All animal procedures were approved by the Institutional Animal Care and Use Committee at Washington University in St. Louis. Following hair removal, a surgical incision was made in the scalp. The exposed skull was cleaned. Then, the mouse skull region over a 3 × 3 mm2 ROI was carefully removed to expose the cortex for topical application of ATN-224. After craniotomy, the anesthetized mouse was transferred to a stereotaxic instrument. The animal body temperature was maintained at 37 °C via a heating pad and the local brain temperature was also maintained at 37 °C via a temperature-controlled water tank. Ultrasound gel was applied between the open-skull window and water tank for acoustic coupling. Following the baseline imaging, the ultrasound gel was gently removed and a solution of ATN-224 (4 μM) was applied topically. The exposed mouse cortex was treated with the SOD1 inhibitor solution for 80 min. Then, the ROI was covered again with ultrasound gel and subjected to post-treatment MP-PAM imaging (Cao et al., 2017). For 2P-FLIM imaging of the live mouse brain, the same pre-imaging procedure was used, but in this case for 12 week old male C57BL/6 mice. After surgery, the mice were transferred to a stereotaxic frame at the side of the Zeiss LSM-780 microscope. An objective converter was used to convert the inverted LSM-780 to upright configuration. A 20×/NA 0.85 water immersion objective lens was coupled to the converter and gently moved to the cranial window for FLIM imaging.
4. Statistics
The paired t-test was used for analyzing 2P-FLIM assays shown in Figs. 4A–C and Fig. S7B. Since there were thousands of ROIs obtained per image, the average of the ROIs for each field of view was calculated to reduce the sample size and thus the quantity of false positives. All data obtained follow the assumptions of normal distributions. All data displayed follow a frequency distribution relative to a2% bound to NADPH.
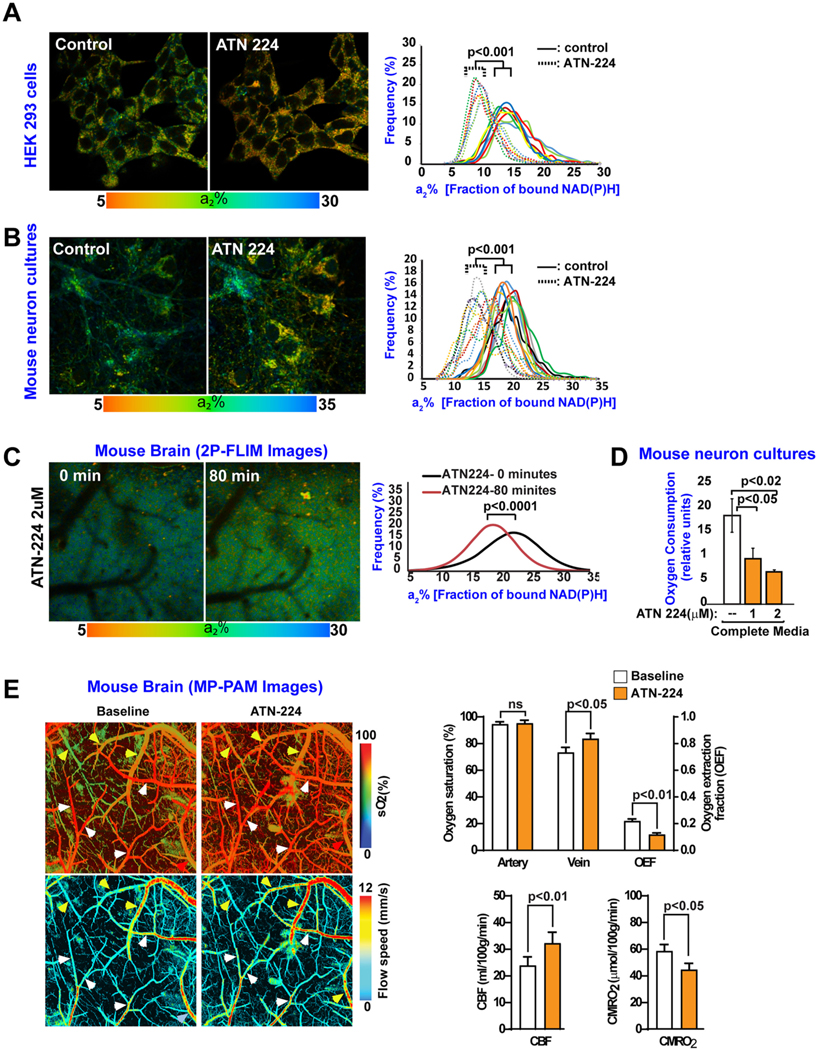
Fig. 4.
ATN-224 regulates mitochondrial activity in live cells and mouse brain.
(A-B) a2% values (fraction of bound NAD(P)H; baseline control, and 60 min after ATN-224 treatment) were recorded pixel by pixel for HEK293 cells (A) and mouse neurons (B). Each colored line in the panel graphs refers to a single field of view containing ~15 cells, and each pair of solid and dotted lines (same color) refers to the same field of view before and after ATN-224 treatment, respectively. All statistical analyses were performed using Student’s two-tailed paired t-tests. Data are representative of two independent assays.
(C) 2P-FLIM images (a2% values) of the live mouse brain cortex were recorded pixel by pixel before and after an 80 min treatment with ATN-224 through an open skull window. Statistical analyses were performed using Student’s two-tailed unpaired t-test. Data are representative of three independent assays.
(D) ATN-224 reduces oxygen consumption in mouse neurons in culture. Mouse neurons cultures were kept in complete medium in the absence or presence of the SOD1 inhibitor, ATN-224. One hour later, oxygen consumption was measured. The data were collected from three independent assays in which each experimental condition contained six replicates. Error bars represent mean ± s.e.m. Significance of differences were determined using an paired two-tailed Student’s t-test assuming equal variance.
(E) ATN-224 reduces oxygen consumption in live mouse brain. MP-PAM imaging of wild-type (WT) mouse cerebral cortex through an open-skull window 80 min. After topical application of ATN-224. An increase in blood oxygenation of the cortical vasculature was observed, indicating decreased oxygen extraction and consumption due to downregulation of mitochondrial activity. Data were obtained from three mice, each of which was measured once for O2 saturation, oxygen extraction fraction (OEF), and cerebral metabolic rate of oxygen (CMRO2). Error bars represent ± s.e.m. White arrows: Arteries; yellow arrows: Veins.
For MP-PAM assays, vessel segmentations and quantitative analysis were done following our established protocol (Cao et al., 2017). The paired t-test was used for comparing the cerebral hemodynamics and oxygen metabolism before and after application of the compounds, which is shown in Fig. 4E. A p-value of less than 0.05 was considered statistically significant.
Significance of differences in all SOD1 assays, Edu uptake, oxygen consumption and colocalization experiments were determined using an unpaired two-tailed Student’s t-test assuming equal variance. SOD1 activity in human brain samples were further analyzed for outliers using Grubbs’s test using GraphPad free software (https://www.graphpad.com/quickcalcs/Grubbs1.cfm ).
5. Results
5.1. Tau regulates lysosomal mTORC1 activity and mtDNA synthesis
To gain a better understanding of the molecular mechanism regulating mtDNA synthesis through the NiMA pathway, we first asked whether it is influenced by tau. We began by comparing the incorporation of 5-ethyl-2′-deoxyuridine (EdU) into mtDNA in mouse primary neuron cultures from WT or tau KO mice (Dawson et al., 2001). As shown previously by our group (Norambuena et al., 2018), WT mouse neurons readily incorporate EdU into mitochondrial nucleoids in a 3 h incubation period, mainly in neuronal perikarya. Under these conditions, an average of ~16 active EdU foci per neuronal perikaryon were detected (Fig. 1A,B). In agreement with our previous observations (Norambuena et al., 2018), we also observed that the number of actively replicating nucleoids was reduced by ~30% when lysosomal mTORC1 activity was stimulated with a mixture of amino acids (arginine and leucine, or R + L) plus insulin (Fig. 1A,B). By contrast, the number of nucleoids in tau KO mouse neurons was ~2-fold greater than in WT neurons and was insensitive to nutrient stimulation (Fig. 1A,B). Re-expression of full length 2N4R human tau in tau KO neurons reduced the basal levels of EdU uptake by ~30% compared to uninfected neurons and restored EdU uptake sensitivity to nutrient stimulation (Fig. 1C,D). These observations imply that tau expression promotes lysosomal mTORC1 activity, and by extension, suppresses mtDNA synthesis.
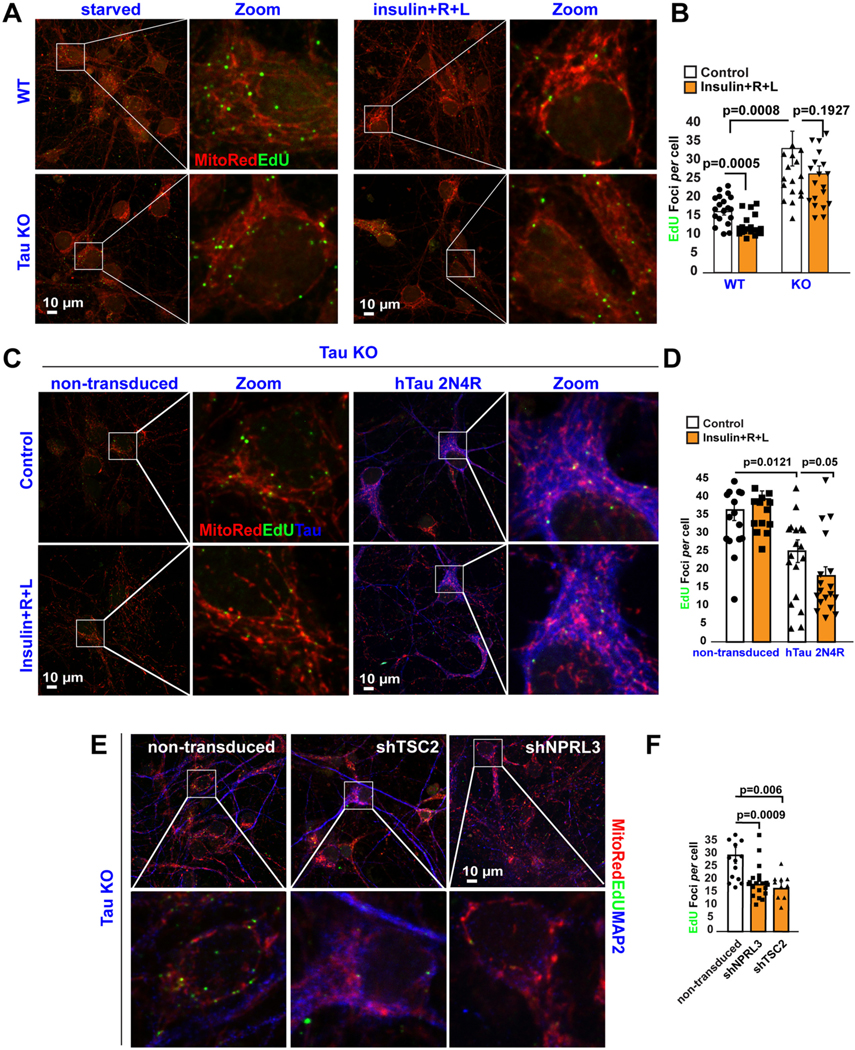
Fig. 1.
Tau regulates lysosomal mTORC1 signaling and mtDNA synthesis.
(A-B) DNA replication in mitochondrial nucleoids was detected in both WT and tau KO mouse cortical neurons after a 3-h pulse of the thymidine analog, EdU. Cells were labeled additionally with the mitochondrial marker, Mitotracker CMXRos. Quantification of EdU uptake into perinuclear nucleoids showed that nutrients inhibit mtDNA replication (EdU foci/neurons) by ~30% in WT (control: 16.35+/− 0.93; n = 271 cells vs nutrients:11.9±0.68 EdU foci/cell; n = 251 cells) but not in Tau KO neurons (control: 36.4+/− 2.9 EdU foci/cell; n = 123 cells versus nutrients: 26.4±2.16 EdU foci/cell; n = 120 cells). Each data point in the graph represents the average number of EdU foci/cell/field of view (FoV) recorded in this experiment and each FoV contained between 2 and 15 cells. Error bars represent mean ± s.e. m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of at least three independent assays.
(C-D) DNA replication in mitochondrial nucleoids was detected in non-transduced tau KO neurons or after re-expressing human Tau 2N4R construct. Neurons were serum-starved in HBSS for 2 h. Next, the cells were pulse-labeled for 3 h with EdU. Note that forcing hTau expression alone yielded an ~30% decrease in EdU incorporation into mtDNA in the absence of nutrients (36.48 ±2.9 EdU foci/cell; n = 123 cells vs 25.1 +/− 3.13 EdU foci/cell; n = 75 cells), further decreasing by 20% in the presence of nutrients (control: 25.1 +/− 3.13 EdU foci/cell; n = 75 cells vs nutrients: 18.27 +/− 2.5 EdU foci/cell; n = 87 cells). Each data point in the graph represents the average number of EdU foci/cell/FoV recorded in this experiment and each FoV contained between 2 and 15 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of at least three independent assays.
(E-F) Genetic activation of mTORC1 activity mimics nutrient-mediated inhibition of mtDNA synthesis in Tau KO neurons. Neurons were serum-starved in HBSS for 2 h. Next, the cells were pulse-labeled for 3 h with EdU (Non-transduced: 29.8±3.13 EdU foci/cell; n = 221 cells vs shNPRL3: 18.3 ±1.24 EdU foci/cell; n = 336 cells vs shTSC2: 16.8±2.5 EdU foci/cell; n = 201 cells). Each data point in the graph represents the average number of EdU foci/cell/FoV recorded in this experiment and each FoV contained between 2 and 10 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t- test assuming equal variance. Data are representative of three independent assays.
To test this idea directly, we monitored the effect of tau expression on lysosomal mTORC1 activity by expressing LysoTorcar, a biosensor of lysosomal mTORC1 activity (Zhou et al., 2015). LysoTorcar corresponds to the full-length 4EBP1 protein, a well-known mTORC1 substrate for phosphorylation at T37, T46 and S65, flanked by Cerulean and Ypet fluorescent proteins. Phosphorylation of those sites can be monitored by western blotting and signifies mTORC1 activation, which can be reversed by dephosphorylation. Finally, the N-terminal ~25% of LysoTorcar corresponds to LAMP1, which targets the fusion protein to lysosomes (Zhou et al., 2015) (Fig. S1A). Consistent with its ability to report mTORC1 activity, basal LysoTorcar phosphorylation in HEK293 cells was reduced by ~70% by the mTOR inhibitor, Torin 1 (Fig. S1A). To test if tau affects mTORC1 activity, HEK293 cells were co-transfected with LysoTorcar, and either GFP or GFP-tau (human 0N4R isoform) for 24 h. Then, cells were serum-starved for 1.5 h before adding 100 nM insulin and R + L for another 25 min. Under baseline, unstimulated conditions, phosphorylation of LysoTorcar on T37 and T46 and S65 was ~40% higher in cells expressing GFP-tau compared to GFP, and the level increased by an extra 50% when cells were stimulated with either insulin or R + L (Fig. S1B). These results suggest that dysregulation of mtDNA synthesis in tau KO neurons occurs as a consequence of lower basal levels of mTORC1 activity. In fact, when the activity of lysosomal mTORC1 was genetically increased in tau KO mouse neurons by shRNA-mediated reduction of either of two suppressors of mTORC1 kinase activity, the TS complex (TSC) or GATOR1 complex (using shTSC2 or shNPRL3, respectively) we observed a ~ 50% reduction in the number of replicating mitochondrial nucleoids (Fig. 1E,F), reaching values similar to those detected in WT neurons (Fig. 1A). As shown before by our group, this strategy increases mTORC1 activity by ~50% (Norambuena et al., 2017).
5.2. Tau regulates lysosomal positioning of the TSC
Activation of lysosomal mTORC1 by nutrients can be achieved by R + L-stimulated recruitment of mTORC1 from cytosol to the lysosomal membrane (Saxton and Sabatini, 2017) or insulin-induced detachment from the lysosomal surface of the TSC complex (Menon et al., 2014). As shown by our group previously (Norambuena et al., 2017), stimulation of WT primary neuron cultures for 30 min with a mixture of R + L and insulin increased the amount of mTOR on the lysosomal surface, as judged by an increase in its co-localization with the lysosomal marker, LAMP1 (Fig. 2A,C). A similar increase was also observed in tau KO neurons (Fig. 2A,C). Interestingly, nutrient stimulation reduced the lysosomal content of TSC2, a subunit of the TSC complex, in WT neurons, but not in tau KO neurons (Fig. 2B,D). Re-expressing 2N4R human tau in tau KO neurons (Western Blots in Fig. 2G) did not alter mTOR co-localization with LAMP1 under nutrient-deprived conditions but trended towards enabling increased co-localization after nutrient stimulation (Fig. 2E). On the other hand, tau re-expression in tau KO neurons led to reduced co-localization of TSC2 with LAMP1 in nutrient-deprived neurons, which decreased further after nutrient stimulation (Fig. 2F). These collective results reveal that tau controls lysosomal mTORC1 activity and mtDNA synthesis by regulating the lysosomal localization of the mTORC1 suppressor, TSC.
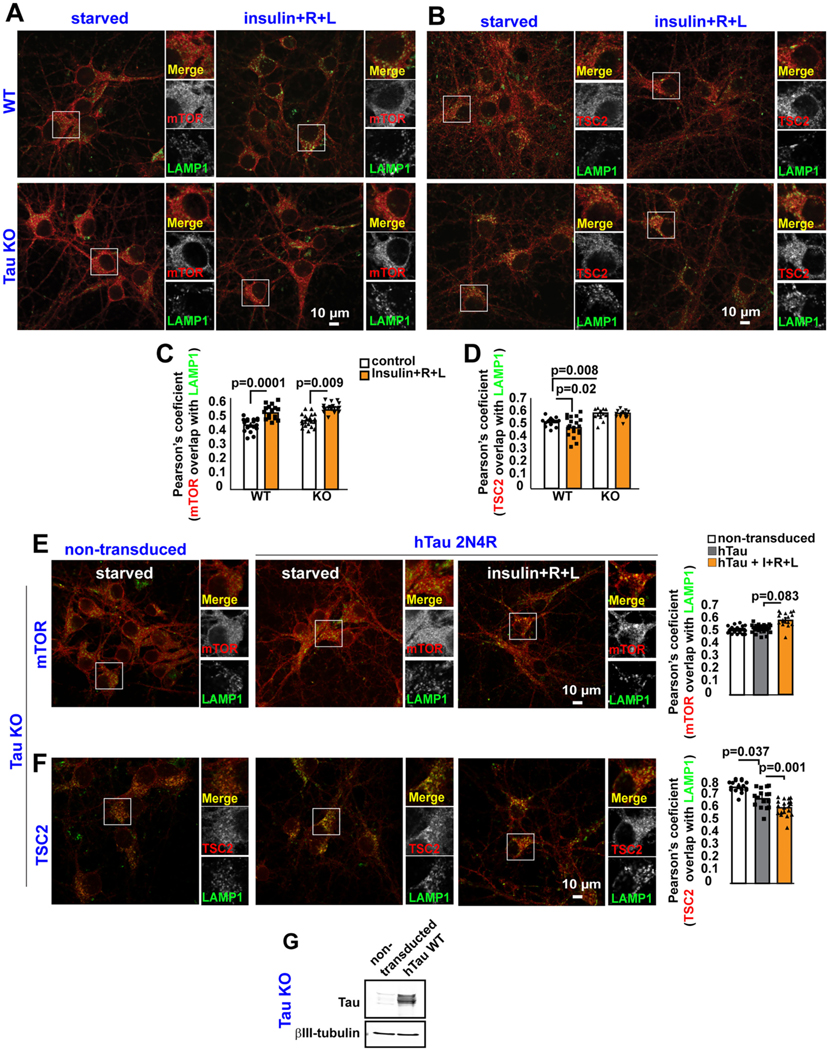
Fig. 2.
Tau Regulates TSC complex positioning on Lysosomes.
(A-B) WT or Tau KO cortical neurons were maintained in Hank’s saline buffer for 2 h and then treated with insulin, plus arginine and leucine (R + L) for 30 min. Neurons were fixed followed by double immunofluorescence for mTOR and LAMP1, or TSC2 and LAMP1. (C–D) Quantification of experiments shown in A-B. Each data point in the graph represents the average Pearson’s coefficient/FoV recorded in this experiment and each FoV contained between 2 and 15 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of at least three independent assays.
(E-F) Non-transduced or human 2N4R Tau-expressing Tau KO cortical neurons starved of insulin and amino acids for 2 h were treated with insulin, plus R + L for 30 min, and then were labeled by double immunofluorescence for mTOR and LAMP1, or TSC2 and LAMP1. Each data point in the graph represents the average Pearson’s coefficient value/FoV recorded in this experiment and each FoV contained between 2 and 15 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of at least three independent assays. (G) Representative western blot showing expression of human 2N4R Tau in Tau KO cortical neurons.
5.3. mtDNA synthesis is insensitive to nutrients in TS human fibroblasts
TS is a developmental disorder caused by loss-of-function mutations in TSC1 or TSC2 whose protein products, TSC1 and TSC2, along with TBC1D7, constitute the TSC (Gao et al., 2002). TS patients develop benign, tuber-like tumors in brain, heart, kidney and skin, and are prone to cognitive deficits (Prather and de Vries, 2004), autism (Smalley, 1998), epilepsy (Thiele, 2004) and other neurological symptoms (Orlova and Crino, 2010). Since TSC is a major regulator of mTORC1 activity, we evaluated mtDNA synthesis in human fibroblasts derived from patients affected by TSC. Mutations in TSC1 rendered mtDNA synthesis refractory to nutrient stimulation in human TSC fibroblasts (Fig. S2A). Re-expressing WT human TSC1 in these cells not only increased the number of nucleoids in starved fibroblasts but also restored sensitivity to nutrient-stimulation (Fig. S2B). These data reinforce our previous report (Norambuena et al., 2018) that malfunctioning of lysosomal mTORC1 causes mitochondria to be refractory to insulin and amino acids signaling in at least two diseases: in AD, in which AβOs triggers mTORC1 activation at the PM (Norambuena et al., 2017), and in tuberous sclerosis, due to loss of functional TSC complex.
5.4. Tau controls mtDNA synthesis through SOD1
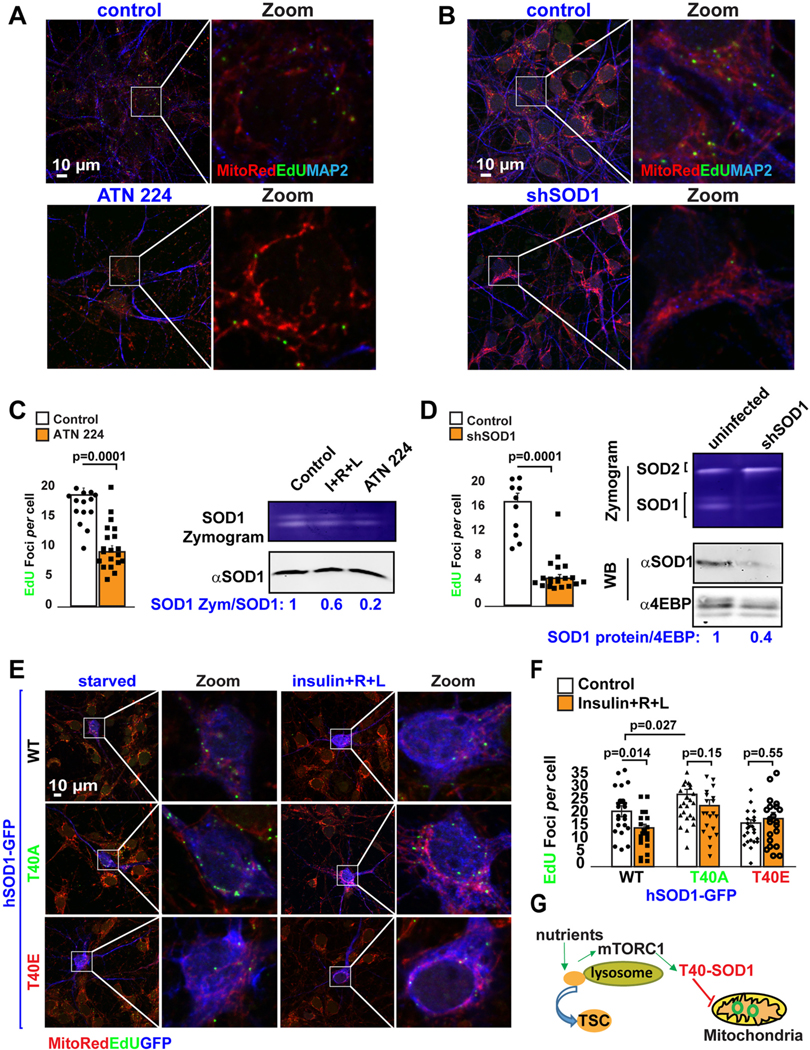
mTORC1 phosphorylates and regulates the activity of numerous substrates (Hsu et al., 2011; Yu et al., 2011). We previously showed that NiMA was independent of well-known mTORC1 substrates including S6K, 4EBP/eIF4E and BcL-xL (Norambuena et al., 2018). We did not, however, previously examine superoxide dismutase 1 (SOD1), which was recently shown to be an mTORC1 substrate (Tsang et al., 2018), and serves as the main regulator of the cytosolic redox state by consuming superoxide produced during mitochondrial respiration and generating the signaling molecule hydrogen peroxide (Veal et al., 2007). SOD1 thus emerged as a potential coordinator of extracellular cues and redox regulation with mitochondrial function through the NiMA pathway. To test whether SOD1 activity is part of the tau-mTORC1 signaling pathway, we first evaluated SOD1 activity by using zymograms or In-gel activity assays (Weydert and Cullen, 2010) in WT and tau KO neurons that were starved, or stimulated with insulin. SOD1 activity in WT neurons was reduced by ~60% after 30 min of treatment with nutrients (Fig. S3A). In contrast, SOD1 activity in tau KO neurons was reduced by insulin stimulation by only~10% (Fig. S3B). Conversely, re-expressing 0N4R human tau in tau KO mouse neurons decreased SOD1 activity by 40% in the absence of mTORC1 stimulation and decreased even more after nutrient treatment (Fig. S3C). Finally, SOD1 activity was ~20% higher in the brain of the Tau KO mouse compared to WT (Fig. S3E). These collective results imply that tau regulates SOD1 through its regulation of lysosomal mTORC1 activity, and by extension SOD1 regulates mtDNA synthesis. Consistent with that interpretation, the basal level of EdU-positive mitochondrial nucleoids in neurons was reduced by ~50% by the pharmacological SOD1 inhibitor, ATN 224, and by ~75% by shRNA-mediated reduction of SOD1 (Fig. 3A–D).
Fig. 3.
Threonine 40 on SOD1 regulates mtDNA synthesis through the NiMA pathway.
(A-B) DNA replication in mitochondrial nucleoids was detected in WT mouse cortical neurons treated with the SOD1 inhibitor, ATN-224 (A), or in SOD1 knockdown neurons (B). Neurons were serum-starved in HBSS for 2 h. Next, the cells were pulse-labeled for 3 h with EdU. (C–D) Quantification of EdU uptake into nucleoids showed that ATN-224 and SOD1 reduction inhibit mtDNA replication by ~60% (control: 18.7 ± 1.16 EdU foci/cell, n = 332 vs ATN224: 9.1 ± 1 EdU foci/cell, n = 150 cells) and ~ 70% (control: 16.9 ± 1.4 EdU foci/cell, n = 345 vs shSOD1: 4.4 ± 0.6 EdU foci/cell, n = 262), respectively. SOD1 activity under each of these conditions was evaluated using in-gel activity assays (n = 3). Each data point in the graphs represents the average number of EdU foci/cell/FoV recorded in this experiment and each FoV contained between 2 and 15 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of three independent assays.
(E) DNA replication in mitochondrial nucleoids was evaluated in WT mouse cortical neurons expressing hSOD1-GFP constructs before and after nutrient stimulation. Quantification of EdU uptake into nucleoids showed that cells expressing either hSOD1T40A-GFP or hSOD1T40E-GFP renders mtDNA synthesis insensitive to nutrient regulation (hSOD1-GFPWT control: 21.4±2.2 EdU foci/cell vs nutrients: 15+/− 1.4 EdU foci/cell; vs hSOD1-GFPT40E control:16.8±1.3 EdU foci/cell vs nutrients: 18.5±2.4 EdU foci/cell; vs hSOD1-GFPT40A control: 28±1.9 EdU foci/cell vs nutrients: 24+/− 2.3 EdU foci/cell. (G) Graphical summary of results presented in this figure. Each data point in the graph represents the average number of EdU foci/GFP-expressing cell/FoV recorded in this experiment and each FoV contained between 1 and 2 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of three independent assays.
mTORC1 phosphorylates human SOD1 at T40 (Tsang et al., 2018), thereby downregulating SOD1 activity. To test whether phosphorylation of SOD1 on T40 was required for nutrient-mediated regulation of mtDNA synthesis we introduced single amino acid substitutions into a GFP-tagged WT human SOD1 fusion protein to produce phosphomimetic (T40E) and phospho-null (T40A) mutants. Nutrient stimulation by insulin and R + L reduced the number of replicating nucleoids by 25% in mouse neurons expressing GFP-SOD1WT (Fig. 3E, F). In contrast, the number of nucleoids in neurons expressing GFP-SOD1T40A under starvation conditions was ~25% higher than in neurons expressing GFP- SOD1WT and was insensitive to nutrient stimulation (Fig. 3E, F). Conversely, expression of GFP-SOD1T40E reduced the base-line number of nucleoids by 24% compared to expression of GFP-SOD1WT, reaching levels similar to cells expressing GFP-SOD1WT after nutrient stimulation (Fig. 3E, F). In addition, the number of mitochondrial nucleoids in GFP-SOD1T40E expressing neurons was insensitive to nutrient stimulation (Fig. 3E, F). Finally, the number of nucleoids in tau KO neurons, which showed lower levels of mTORC1 activity and double the number of EdU foci per cell compared to WT neurons, was decreased by ~50% by expressing GFP-SOD1T40E (Fig. S4). Thus, phosphorylation at T40 in SOD1 regulates mtDNA synthesis through the tau-mTORC1 signaling pathway (Fig. 3G).
5.5. ATN 224 inhibits mitochondrial respiration in cultured cells and mouse brain
Having demonstrated that the tau-mTORC1-SOD1 signaling pathway regulates mtDNA synthesis, we then explored whether SOD1 activity is required for respiration in cultured HEK293 cells, mouse neurons, or in mouse brain. For the cultured cells we used 2-photon fluorescence lifetime imaging (2P-FLIM) to monitor NAD(P)H lifetimes and the degree to which these co-enzymes associate with partner enzymes in perinuclear mitochondria, as shown previously (Norambuena et al., 2018). The “bound fraction” of NAD(P)H, expressed as “a2%” (Lakowicz, 2006), positively correlates with mitochondrial respiration (Norambuena et al., 2018) and biosynthetic pathways (Blacker et al., 2014). Following serum starvation in Hank’s balanced salt solution (HBSS) for 2 h, individual fields of view were assayed by 2P-FLIM before and 60 min after stimulation of the cells with Bis(choline)tetrathiomolybdate (ATN-224), a copper chelator and SOD1 inhibitor (Chidambaram et al., 1984). ATN-224 caused a similar decrease in enzyme-bound NAD(P)H in both HEK293 and neurons, signifying a downregulation in oxidative phosphorylation (OXPHOS) activity (Fig. 4A,B). In addition, ATN-224 significantly reduced oxygen consumption in neuron cultures (Fig. 4D).
We next asked whether SOD1 activity also regulates mitochondrial respiration at the tissue level in vivo. First, we used 2P-FLIM to monitor changes in NAD(P)H lifetimes in live mouse brain. This approach revealed that topical application of ATN-224 for 80 min through a cranial window caused a substantial decrease in the fraction of enzyme-bound NAD(P)H (Fig. 4C). In parallel, we applied multiparametric photoacoustic microscopy (MP-PAM) to measure oxygen-metabolic responses in the live mouse brain (Cao et al., 2017). MP-PAM imaging through a cranial window enables simultaneous, high-resolution imaging of total hemoglobin concentration (CHb), oxygen saturation of hemoglobin (SO2), and cerebral blood flow (Ning et al., 2015). This approach revealed that ATN-224 caused significant increases in venous SO2, without changes in cerebral blood flow (CBF; Fig. 4E). These hemodynamic responses to ATN-224 were accompanied by decreases in both oxygen extraction fraction (OEF) and the cerebral metabolic rate of oxygen (CMRO2; Fig. 4E). In our previous study we provided evidence that NiMA regulate OXPHOS in neuronal and non-neuronal cells as well as in the live mouse brain by a mechanism mediated by mTORC1 (Norambuena et al., 2018). Considering the observations presented so far, our collective results suggest that mTORC1-mediated inhibition of SOD1, through phosphorylation of T40, inhibits mtDNA synthesis and stimulates mitochondrial respiration in both proliferating and non- proliferating cells. The inhibitory effect of ATN-224 on OXPHOS in live cells and mouse brain seems counterintuitive, but likely occurs as a consequence of inhibiting cytochrome-c oxidase through copper sequestration (Nair and Mason, 1966), rather than by SOD1 inhibition.
5.6. Tau controls nutrient-mediated suppression of mtDNA synthesis and its dysregulation by AβOs
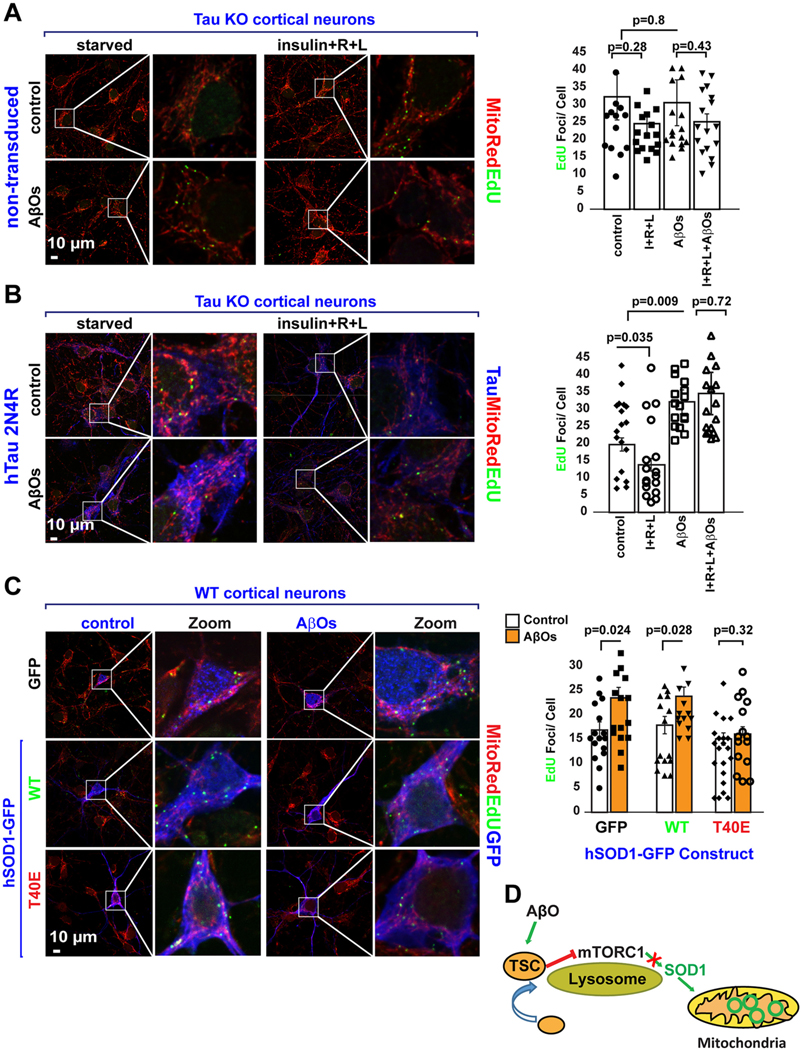
We previously reported that AβO-mediated disruption of NiMA requires tau expression (Norambuena et al., 2018). As tau expression in tau KO neurons restores normal lysosomal mTORC1 signaling and mtDNA synthesis in neurons (Fig. 1), we tested whether tau is also required for nutrient-induced inhibition of mtDNA synthesis, and the upregulation of mtDNA synthesis triggered by AβOs (Norambuena et al., 2018). Contrary to the inhibitory action of nutrients on mtDNA synthesis observed in WT mouse neurons (Fig. 1A), nutrients did not significantly decrease the nucleoid content of tau KO neurons (Fig. 5A). Likewise, the number of nucleoids in tau KO neurons were not affected by AβOs in either the absence or presence of nutrients (Fig. 5A). Tau re-expression not only reduced the basal level of nucleoids by 30%, but also restored inhibition of mtDNA synthesis by nutrient stimulation (Fig. 5B). Furthermore, tau re-expression in tau KO neurons re-established the ability of AβOs to increase mtDNA synthesis (Fig. 5B) and to abolish the inhibitory effect of nutrients (Fig. 5B). Having established that tau permits proper lysosomal mTORC1 signaling and its counteraction by AβOs, we then asked whether T40 phosphorylation on SOD1 mediates AβO dysregulation of mtDNA synthesis. AβO-treatment increased the number of nucleoids by ~30% in neurons expressing either GFP or GFP-SOD1WT, but not in GFP-SOD1T40E expressing neurons (Fig. 5C). AβOs therefore signal through the tau-mTORC1-SOD1 signaling pathway to dysregulate mtDNA synthesis (Fig. 5D).
Fig. 5.
AβO-mediated dysregulation of mtDNA synthesis requires tau and is mediated by SOD1 T40.
(A) DNA replication in mitochondrial nucleoids was detected in tau KO mouse neurons. Neurons were serum-starved in HBSS for 2 h. Next, the cells were pulse- labeled for 3 h with EdU in the absence or presence of nutrients (a mixture of insulin plus R + L, AβOs, or a combination of both). Quantification of EdU uptake into nucleoids showed that mtDNA replication was insensitive to either nutrient stimulation (control: 32 ± 6.7 EdU foci/cell, n = 80 vs nutrients: 25 ± 2.6 EdU foci/ cell; n = 102), AβO (31 ± 6.6 EdU foci/cell, n = 102) or AβOs plus nutrients (25.2 ± 2.2 EdU foci/cell, n = 71). Each data point in the graph represents the average number of EdU foci/cell/FoV recorded in this experiment and each FoV contained between 2 and 15 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of three independent assays.
(B) Cortical mouse neurons from tau KO mice were transduced with lentivirus to express hTau 2N4R, which not only rescued the ability of tau KO neurons to reduce mtDNA synthesis in the presence of nutrients (control: 19.8 ± 1.9 EdU foci/cell n = 69 cells vs nutrients: 13.9 ± 1.8 EdU foci/cell n = 65 cells) but also to increase it upon AβO treatment (control:19.8 ± 1.9 EdU foci/cell, n = 69 cells vs AβO: 32 ± 3.5 EdU foci/cell, n = 55 cells). Tau re-expression also rescued the ability of AβO to block nutrient-mediated inhibition of mtDNA synthesis (nutrients: 13.9 ± 1.8 EdU foci/cell n = 65 cells vs AβO + nutrients: 34.6 ± 6.2 EdU foci/cell n = 55 cells). Each data point in the graph represent the average number of EdU foci/GFP-expressing cell/FoV recorded in this experiment and each FoV contained between 1 and 2 GFP-expressing cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of three independent assays.
(C) DNA replication in mitochondrial nucleoids was detected in WT mouse cortical neurons expressing either GFP, hSOD1WT-GFP or hSOD1T40E-GFP. Note that AβO treatment increase mtDNA synthesis by ~25% in both GFP (control:16 ± 1.7 EdU foci/cell n = 30 cells vs AβO: 23.5 ± 2.2 EdU foci/cell, n = 35 cells) and hSOD1WT- GFP (control: 18 ± 1.8 EdU foci/cell n = 35 cells vs AβO: 24 ± 1.9 EdU foci/cell, n = 33 cells) but not in hSOD1T40E-GFP expressing neurons (control: 16.2 ± 1.4 EdU foci/cell n = 40 cells vs AβO: 17.41 ± 1.8 EdU foci/cell n = 33 cells).
(D) Graphical summary of results presented in this figure.
Each data point in the graph represents the average number of EdU foci/ GFP-expressing cell/FoV recorded in this experiment and each FoV contained between 1 and 2 cells. Error bars represent mean ± s.e.m. Significance of differences were determined using an unpaired two-tailed Student’s t-test assuming equal variance. Data are representative of three independent assays.
5.7. SOD1 activity is elevated in Alzheimer’s disease human brain
Previous work by our group and others has shown that AβOs impair insulin/IGF1 signaling (De Felice and Ferreira, 2014; Norambuena et al., 2017). Mechanistically these effects are partially explained by a reduction in AKT activity in the presence of AβOs (Fig. S5), which in turn increases lysosomal positioning of TSC and thereby suppresses mTORC1 kinase activity (Norambuena et al., 2017). As expected, AβO treatment increased SOD1 activity in HEK 293 cells (Fig. S6B) and mouse neuron cultures (Fig. 6A, Fig. S3D and Fig. S6C). In addition, an AβO-mediated increase in SOD1 activity was not observed in tau KO neurons (Fig. S3D). Again, tau expression in Tau KO neurons reduced SOD1 activity by ~40% and restored the ability of AβOs to increase SOD1 activity (Fig. S3D). Finally, we explored SOD1 activity in AD human brain. As shown in Fig. 6B, SOD1 activity was evaluated by using In-gel activity assays in brain samples taken from 8 controls and 6 CE donors, SOD1 activity was ~40% higher in AD compared to normal donors.
6. Discussion
Although considerable progress has been made in the past few decades of AD research, therapies proven to delay or prevent symptom onset, or slow disease progression are still lacking. Of particular relevance is the fact that ~50% of people with type 2 diabetes (T2D) are eventually affected by sporadic AD (Schrijvers et al., 2010). A key feature of T2D is multi-organ metabolic failure, which for brain manifests as reduced insulin sensitivity and increased mitochondrial dysfunction in neurons (De Felice and Ferreira, 2014). Thus, understanding the mechanistic details of the insulin signaling pathway in neurons is pivotal to understanding its pathophysiological effects in AD. Using live incorporation of EdU into mitochondrial nucleoids along with metabolic imaging in live cells and mouse brain, we now expand the mechanistic understanding of an inter-organelle signaling pathway that we recently discovered (Norambuena et al., 2018). We show here that activation of lysosomal mTORC1 by insulin and amino acids regulates mitochondrial respiration and mtDNA synthesis by a mechanism controlled by tau and likely, mTORC1-catalyzed phosphorylation of SOD1 at T40. Previous work have reported increased SOD activity in fibroblasts cells lines stablished from AD and normal patients (Zemlan et al., 1989) or no changes in brains from control subjects and AD patients (Marklund et al., 1985). Using In-gel activity assays, which allows measuring SOD1 activity independently of SOD2 (Weydert and Cullen, 2010), we found higher SOD1 activity in AD human brains, which is also triggered by AβOs in a tau-dependent manner in neuronal cultures, providing additional evidence for how disruption of this lysosome-to-mitochondria signaling pathway represents a seminal step in at least two pathological contexts, AD and tuberous sclerosis.
mTOR plays a major physiological role regulating cellular metabolism (Saxton and Sabatini, 2017). These functions require the precise regulation of multiple cellular pathways through serine and/or threonine phosphorylation of potentially >100 substrates (Hsu et al., 2011; Yu et al., 2011). Our discovery of the NiMA pathway not only demonstrates that mitochondria quickly react to changes in mTORC1 activity by a mechanism independent of protein synthesis and gene expression, but also established a novel mTORC1 function regulated by its recently discovered substrate, SOD1 (Tsang et al., 2018), independently of other well-known mTORC1 substrates, like Bcl-xl, S6 kinase and 4EBP/eIF4E (Norambuena et al., 2018). SOD1 is a main regulator of cytosolic redox states (Miao and Clair, 2009), but its functional role for mitochondria is not fully understood. We found that SOD1 is part of the neuronal nutrient sensing machinery that mechanistically connects nutrient-mediated activation of lysosomal mTORC1 to mtDNA synthesis and activity. Our observation that inhibition of SOD1 activity with ATN-224 reduced baseline OXPHOS in cells and mouse brains, suggests that interventions targeting SOD1 activity could be beneficial by ameliorating oxidative damage and propagation of oxidized or mutated mtDNA in AD. A note of caution however should be taken considering ATN-224’s pro-apoptotic and pro-oxidant effects in the treatment of cancer (Glasauer et al., 2014).
It is accepted that some cell types, such as hepatocytes or cancer cells grown under hypoxic conditions, can consume oxygen in a mitochondrion-independent way, a process known as “non-mitochondrial oxygen consumption (NMOC)”. It is believed that NMOC may contribute 20–30% of the total consumptions under those conditions (Banh et al., 2016). Although our assays do not differentiate between mitochondrial and NMOC, it is likely that between 70 and 80% correspond to mitochondrial utilization. This idea is also supported by the following observations: (i) mTOR is also a main regulator on mitochondrial oxygen consumption (Schieke et al., 2006) (ii) in our previous work, we demonstrated that insulin and amino acids regulate mitochondrial functioning as measured by analyzing oxygen consumption, mitochondrial ROS and ATP utilization through mTOR in neuron cultures; (iii) we also showed that oxygen consumption induced by amino acids in live mouse brains, was completely blocked by the mTORC1 inhibitor rapamycin (Norambuena et al., 2018). Thus, it is likely that mitochondria account for the vast majority of the oxygen consumed in neurons in culture and live mouse brains in the assays presented in this study.
We recently provided mechanistic evidence for how the AβO-tau axis pathologically orchestrates a signaling pathway that at least partially explains how brain insulin resistance may lead to AD by promoting ectopic neuronal cell cycle re-entry and mitochondrial dysfunction (Norambuena et al., 2018; Norambuena et al., 2017; Polanco and Götz, 2018). AβOs can induce an insulin resistant-like phenotype in neurons by sequestering insulin receptors from dendrites and reducing neuronal response to insulin (Zhao et al., 2007). AKT phosphorylation at both T308 and S473 leads to full activation (Yang et al., 2015), and we found that AβO treatment reduces the phosphate content on AKT, suggesting that AβOs inhibit AKT activity and its downstream targets. In fact, we observed that tau re-expression in tau KO neurons restored the lysosomal distribution of the TSC complex, a direct target of AKT phosphorylation by mTORC1 (Menon et al., 2014), its sensitivity to nutrient stimulation, and both mTORC1 and SOD1 activities. In addition, tau re-expression also rescued the ability of AβOs to pathologically increase mtDNA synthesis. This ability of tau both to restore “normal” signaling and to facilitate pathological signaling by AβOs is ironic, but possibly explained by the ability of tau to scaffold signaling pathways that became noxious in the presence of AβO, as for example by the activation mTORC1 (Norambuena et al., 2017) and Fyn (Ittner et al., 2010) at the PM. This paradoxically dual effect should be considered when designing interventions aimed at reducing tau expression in the context of AD and other tauopathies (DeVos et al., 2017). In this sense, it is interesting to note that reduction of mTOR genetic dosage or rapamycin treatment (a cellular phenocopy of tau reduction in neurons) ameliorates cognitive deficits in AD transgenic mice (Caccamo et al., 2014; Caccamo et al., 2010). However, rapamycin’s adverse side effects have precluded its broader use in humans (Li et al., 2014). Further work aimed at understanding the role of SOD1 on the NiMA pathway is necessary for developing more specific interventions. As discuss below, hydrogen peroxide treatment reduced mtDNA synthesis without affecting mitochondrial OXPHOS (Fig. S7A and B), suggesting that substrates sensitive to oxidation by peroxide are involved and may represent future therapeutic targets.
How does SOD1 regulate mitochondrial function? SOD1 activity has been shown to be required for repressing respiration in yeast and potentially in human cells (Reddi and Culotta, 2013), a process involving SOD1-mediated interaction and stabilization of casein kinase gamma 2 (Ck1γ2), a respiration repressor (Reddi and Culotta, 2013). However, we could not find evidence of such a regulatory mechanism in HEK293 cells or mouse primary neurons, as neither prolonged (24 h) treatment with ATN-224 nor exposure to AβOs, conditions that respectively abolish or stimulate SOD1 activity, modified Ck1γ2 expression (Fig. S6A, B and C). In line with the latter, we did not observe changes in Ck1γ2 expression in human AD brains, in which SOD1 activity was higher compared to non-AD controls (Fig. S6D and Fig. 6B, respectively). Thus, we conclude that in human cells, mouse neurons and human brain, Ck1γ2 expression is not regulated by SOD1 activity and by extension, does not controls mitochondrial function. However, we did observe that hydrogen peroxide (a by-product of SOD1 activity) treatment was able to reduce mtDNA synthesis (Fig. S7A), suggesting that nutrient-mediated inhibition of mtDNA synthesis is regulated by one or more substrates sensitive to oxidation by H2O2. In this respect, it is interesting to note that CaMKII and PKA, whose activity can be modified by H2O2 (Burgoyne et al., 2012), were previously shown to be dysregulated by AβOs (Norambuena et al., 2017; Seward et al., 2013). Future work is necessary to understand how nutrient-mediated activation of mTORC1, oxidant sensing machinery and kinases activity are integrated to orchestrate proper inter-organelle responses.
The almost exclusively neuronal expression of tau likely conferred neurons with advantages allowing them to perform critical functions during the human lifespan. These functions are not only related to its role as a regulator of cargo transport along microtubules (Dixit et al., 2008; Ebneth et al., 1998; Swanson et al., 2017; Trinczek et al., 1999; Vershinin et al., 2007), but as we show here, by allowing neurons to properly respond to insulin and nutrients, and to coordinate extracellular cues to cytosolic redox and mitochondrial functioning. Here, we show that tau expression favors insulin signaling, which in turn reduces the amount of tuberous sclerosis complex (TSC) on the lysosome. TSC is a constitutive lysosomal mTORC1 inhibitor whose abundance on the lysosomal surface is decreased by insulin signaling and amino acids (Demetriades et al., 2014; Menon et al., 2014). Thus, our new results mechanistically implicate tau in regulating mTORC1 by controlling the lysosomal content of TSC, although further details of the mechanism remain to be determined. The importance of these observations are exemplified by the fact that tau deletion leads to brain insulin resistance in mice (Marciniak et al., 2017). Its downstream consequences are also highlighted in this study by showing that tau sustains nutrient-mediated activity of mTORC1 and SOD1. Thus, tau mechanistically connects four key players in AD: insulin and nutrient signaling, mTORC1 function, SOD1 (redox regulation), and mitochondrial activity (De Felice and Ferreira, 2014). The results of this study, which are summarizes in Fig. 6C, allow us to propose that the tau-mTORC1-SOD1 signaling pathway mechanistically connects mtDNA synthesis and maintenance to nutrient signaling in neurons, opening the possibility that dysregulation of this signaling pathway contributes to mitochondrial oxidative damage and propagation of mutated mtDNA in neurodegeneration. This process potentially could reach pathological levels by changing the balance between normal and damaged mtDNA.
Finally, it is noteworthy that two of the proteins in the pathway described here are emblematic of neurodegeneration: tau in AD and numerous non-Alzheimer’s tauopathies, and SOD1 in ALS, which is an example of a non-Alzheimer’s tauopathy (Lee et al., 2001). We are proposing that a steady conversion of soluble tau into insoluble aggregates may lead to reduction of lysosomal mTORC1 activity, and by extension, increased SOD1 activity, cytosolic redox dysregulation, and mitochondrial dysfunction in AD.
7. Conclusions
Our studies suggests that AβOs cause Tau-dependent, mTORC1-mediated SOD1 dysregulation and by extension, mitochondrial dysfunction in AD. Perhaps agents other than AβOs similarly lead to Tau-dependent SOD1 dysregulation in non-Alzheimer’s tauopathies, including ALS.
Supplementary Material
Acknowledgments
We thanks Dr. Anthony Spano (Department of Biology, University of Virginia) for providing N-ethylmaleimide. Dr. Chengsan Sun (Department of Biology, University of Virginia) for mouse brain surgical assistance. We also thanks the Keck Center for Cellular Imaging (Department of Biology, University of Virginia) for making possible our live cell mitochondrial metabolic imaging.
Funding
We are grateful for financial support from NIH/NIA (grant R01AG067048 to AN) the Alzheimer’s and Related Diseases Research Award Fund (grant 17-5 to AN), University of Virginia’s Brain Institute and Virginia Alzheimer’s Disease Center Leadership Award (AP, AN and GSB), the Owens Family Foundation (GSB), NIH/NIA (grant RF1 AG051085 to GSB); NIH/Office of the Director for funds to purchase a Zeiss 780 microscope that was used in this study (OD016446 to AP), the Cure Alzheimer’s Fund (GSB), the Alzheimer’s Association (grants ZEN-16-363266 to GSB), Webb and Tate Wilson (GSB), The Virginia Chapter of the Lady’s Auxiliary of the Fraternal Order of Eagles (GSB), and the University of Virginia President’s Fund for Excellence (GSB).
Footnotes
CRediT authorship contribution statement
Andrés Norambuena: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. Xuehan Sun: Investigation, Writing – review & editing. Horst Wallrabe: Methodology, Data curation, Writing – review & editing. Ruofan Cao: Investigation, Writing – review & editing. Naidi Sun: Investigation, Writing – review & editing. Evelyn Pardo: Investigation, Writing – review & editing. Nutan Shivange: Investigation, Writing – review & editing. Dora Bigler Wang: Investigation. Lisa A. Post: Methodology, Writing – review & editing. Heather A. Ferris: Supervision, Writing – review & editing. Song Hu: Supervision, Writing – review & editing. Ammasi Periasamy: Supervision, Writing – review & editing. George S. Bloom: Supervision, Writing – review & editing.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbd.2022.105737.
References
- Adav SS, Park JE, Sze SK, 2019. Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer’s disease. Mol. Brain 10.1186/s13041-019-0430-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt T, Brückner MK, Mosch B, Lösche A, 2010. Selective cell death of hyperploid neurons in Alzheimer’s disease. Am. J. Pathol 177, 15–20. 10.2353/ajpath.2010.090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T, 2005. Mitochondria, oxidants, and aging. Cell 120, 483–495. 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Banh RS, Iorio C, Marcotte R, Xu Y, Cojocari D, Rahman AA, Pawling J, Zhang W, Sinha A, Rose CM, Isasa M, Zhang S, Wu R, Virtanen C, Hitomi T, Habu T, Sidhu SS, Koizumi A, Wilkins SE, Kislinger T, Gygi SP, Schofield CJ, Dennis JW, Wouters BG, Neel BG, 2016. PTP1B controls non- mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nat. Cell Biol 18, 803–813. 10.1038/ncb3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain AJ, Szabadkai G, Duchen MR, 2014. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun 5 10.1038/ncomms4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard BJ, Park T, Fripp WJ, Lermanv LS, Ingram VM, 1993. A mitochondrial DNA deletion in normally aging and in alzheimer brain tissue. Neuroreport. 10.1097/00001756-199306000-00051. [DOI] [PubMed] [Google Scholar]
- Burgoyne JR, Oka S, Ale-Agha N, Eaton P, 2012. Hydrogen peroxide sensing and signaling by protein kinases in the cardiovascular system. Antioxid. Redox Signal 18, 1042–1052. 10.1089/ars.2012.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burté F, Carelli V, Chinnery PF, Yu-Wai-Man P, 2014. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol 11, 11–24. 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Majumder S, Richardson A, Strong R, Oddo S, 2010. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and tau: effects on cognitive impairments. J. Biol. Chem 285, 13107–13120. 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, De Pinto V, Messina A, Branca C, Oddo S, 2014. Genetic reduction of mammalian target of rapamycin ameliorates Alzheimer’s disease-like cognitive and pathological deficits by restoring hippocampal gene expression signature. J. Neurosci 34, 7988–7998. 10.1523/JNEUROSCI.0777-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins MJ, Manczak M, Mao P, Shirendeb U, Reddy PH, 2011. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum. Mol. Genet 20, 4515–4529. 10.1093/hmg/ddr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camandola S, Mattson MP, 2017. Brain metabolism in health, aging, and neurodegeneration. EMBO J. 10.15252/embj.201695810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Li J, Ning B, Sun N, Wang T, Zuo Z, Hu S, 2017. Functional and oxygen-metabolic photoacoustic microscopy of the awake mouse brain. Neuroimage 150, 77–87. 10.1016/j.neuroimage.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri G, Bonafé M, De Luca M, Rose G, Varcasia O, Bruni A, Maletta R, Nacmias B, Sorbi S, Corsonello F, Feraco E, Andreev KF, Yashin AI, Franceschi C, De Benedictis G, 2001. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer’s disease. Hum. Genet 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D, 1999. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French- Canadian founder population. Am. J. Med. Genet . [DOI] [PubMed] [Google Scholar]
- Che M, Wang R, Li X, Wang HY, Zheng XFS, 2016. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov. Today 10.1016/j.drudis.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC, 2009. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet 18, R169–R176. 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu C, Parker WD, Chen H, Beach TG, Liu X, Serrano GE, Lu Y, Huang J, Yang K, Wang C, 2016. Mitochondrial DNA rearrangement Spectrum in brain tissue of Alzheimer’s disease: analysis of 13 cases. PLoS One. 10.1371/journal.pone.0154582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidambaram MV, Barnes G, Frieden E, 1984. Inhibition of ceruloplasmin and other copper oxidases by thiomolybdate. J. Inorg. Biochem 22, 231–239. 10.1016/0162-0134(84)85008-4. [DOI] [PubMed] [Google Scholar]
- Collins J-A, Rudenski A, Gibson J, Howard L, O’Driscoll R, 2015. Relating oxygen partial pressure, saturation and content: the haemoglobin-oxygen dissociation curve. Breathe (Sheffield, England) 11, 194–201. 10.1183/20734735.001415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, McKee AC, Beal MF, Graham BH, Wallace DC, 1994. Marked changes in mitochondrial dna deletion levels in alzheimer brains. Genomics. 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Beal MF, Wallace DC, 2004. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc. Natl. Acad. Sci. U. S. A 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell DA, Blakely EL, Johnson MA, Ince PG, Turnbull DM, 2001. Mitochondrial enzyme-deficient hippocampal neurons and choroidal cells in AD. Neurology. 10.1212/WNL.57.2.260. [DOI] [PubMed] [Google Scholar]
- Crane PK, Walker R, Hubbard RA, Li G, Nathan DM, Zheng H, Haneuse S, Craft S, Montine TJ, Kahn SE, McCormick W, McCurry SM, Bowen JD, Larson EB, 2013. Glucose levels and risk of dementia. N. Engl. J. Med 369, 540–548. 10.1056/NEJMoa1215740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau E, Castellano CA, Fortier M, Bocti C, Fulop T, Paquet N, Cunnane SC, 2018. A cross-sectional comparison of brain glucose and ketone metabolism in cognitively healthy older adults, mild cognitive impairment and early Alzheimer’s disease. Exp. Gerontol 10.1016/j.exger.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Dawson HN, Ferreira A, Eyster MV, Ghoshal N, Binder LI, Vitek MP, 2001. Inhibition of neuronal maturation in primary hippocampal neurons from τ deficient mice. J. Cell Sci 114, 1179–1187. 10.1242/jcs.114.6.1179. [DOI] [PubMed] [Google Scholar]
- De Felice FG, Ferreira ST, 2014. Inflammation, defective insulin signaling, and mitochondrial dysfunction as common molecular denominators connecting type 2 diabetes to Alzheimer disease. Diabetes. 10.2337/db13-1954. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA, 2014. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799. 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, Wegener AJ, Chen G, Shen T, Tran H, Nichols B, Zanardi TA, Kordasiewicz HB, Swayze EE, Bennett CF, Diamond MI, Miller TM, 2017. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med 9 10.1126/scitranslmed.aag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Ross JL, Goldman YE, Holzbaur ELF, 2008. Differential regulation of dynein and kinesin motor proteins by tau. Science (80-.) 319. 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBoff B, Feany M, Götz J, 2013. Why size matters - balancing mitochondrial dynamics in Alzheimer’s disease. Trends Neurosci. 10.1016/j.tins.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH, 2011. A practical guide to evaluating colocalization in biological microscopy. AJP Cell Physiol. 300, C723–C742. 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow EM, Mandelkow E, 1998. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer’s disease. J. Cell Biol 143 10.1083/jcb.143.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D, 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- Glasauer A, Sena LA, Diebold LP, Mazar AP, Chandel NS, 2014. Targeting SOD1 reduces experimental non–small-cell lung cancer. J. Clin. Invest 124, 117–128. 10.1172/JCI71714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BA, Blazey TM, Su Y, Hari-Raj A, Dincer A, Flores S, Christensen J, McDade E, Wang G, Xiong C, Cairns NJ, Hassenstab J, Marcus DS, Fagan AM, Jack CR, Hornbeck RC, Paumier KL, Ances BM, Berman SB, Brickman AM, Cash DM, Chhatwal JP, Correia S, Förster S, Fox NC, Graff-Radford NR, la Fougère C, Levin J, Masters CL, Rossor MN, Salloway S, Saykin AJ, Schofield PR, Thompson PM, Weiner MM, Holtzman DM, Raichle ME, Morris JC, Bateman RJ, Benzinger TLS, 2018. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer’s disease: a longitudinal study. Lancet Neurol. 10.1016/S1474-4422(18)30028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Tang P, Chen L, Liu P, Hou C, Zhang X, Liu Y, Chong L, Li X, Li R, 2017. Amyloid β-induced redistribution of transcriptional factor EB and lysosomal dysfunction in primary microglial cells. Front. Aging Neurosci 10.3389/fnagi.2017.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM, 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322. 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Sims R, Harold D, Chapman J, Hollingworth P, Gerrish A, Russo G, Hamshere M, Moskvina V, Jones N, Thomas C, Stretton A, Holmans PA, O’Donovan MC, Owen MJ, Williams J, Chinnery PF, 2012. No consistent evidence for association between mtDNA variants and Alzheimer disease. Neurology. 10.1212/WNL.0b013e31824e8f1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inczedy-Farkas G, Trampush JW, Perczel Forintos D, Beech D, Andrejkovics M, Varga Z, Remenyi V, Bereznai B, Gal A, Molnar MJ, 2014. Mitochondrial DNA mutations and cognition: a case-series report. Arch. Clin. Neuropsychol 10.1093/arclin/acu016. [DOI] [PubMed] [Google Scholar]
- Ishii K, Kitagaki H, Kono M, Mori E, 1996. Decreased medial temporal oxygen metabolism in Alzheimer’s disease shown by PET. J. Nucl. Med 37, 1159–1165. [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, Wölfing H, Chieng BC, Christie MJ, Napier IA, Eckert A, Staufenbiel M, Hardeman E, Götz J, 2010. Dendritic function of tau mediates amyloid-β toxicity in alzheimer’s disease mouse models. Cell 142. 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Juarez JC, Manuia M, Burnett ME, Betancourt O, Boivin B, Shaw DE, Tonks NK, Mazar AP, Doñate F, 2008. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. U. S. A 105, 7147–7152. 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapogiannis D, Mattson MP, 2011. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer’s disease. Lancet Neurol. 10, 187–198. 10.1016/S1474-4422(10)70277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé K, Murley A, Nunnari J, 2014. Determinants and functions of mitochondrial ´ behavior. Annu. Rev. Cell Dev. Biol 30, 357–391. 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- Lakatos A, Derbeneva O, Younes D, Keator D, Bakken T, Lvova M, Brandon M, Guffanti G, Reglodi D, Saykin A, Weiner M, Macciardi F, Schork N, Wallace DC, Potkin SG, 2010. Association between mitochondrial DNA variations and Alzheimer’s disease in the ADNI cohort. Neurobiol. Aging 10.1016/j.neurobiolaging.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz JR, 2006. Principles of Fluorescence Spectroscopy Principles of Fluorescence Spectroscopy, Principles of Fluorescence Spectroscopy, 3rd edn. Springer, New York, USA. 10.1007/978-0-387-46312-4. [DOI] [Google Scholar]
- Lee VM-Y, Goedert M, Trojanowski JQ, 2001. Neurodegenerative Tauopathies. Annu. Rev. Neurosci 24, 1121–1159. 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Lewis SC, Uchiyama LF, Nunnari J, 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science (80-.) 353, aaf5549. 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kim SG, Blenis J, 2014. Rapamycin: one drug, many effects. Cell Metab. 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA, 2008. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci. U. S. A 105, 4441–4446. 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Calkins MJ, Reddy PH, 2011. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum. Mol. Genet 20, 2495–2509. 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak E, Leboucher A, Caron E, Ahmed T, Tailleux A, Dumont J, Issad T, Gerhardt E, Pagesy P, Vileno M, Bournonville C, Hamdane M, Bantubungi K, Lancel S, Demeyer D, Eddarkaoui S, Vallez E, Vieau D, Humez S, Faivre E, Grenier-Boley B, Outeiro TF, Staels B, Amouyel P, Balschun D, Buee L, Blum D, 2017. Tau deletion promotes brain insulin resistance. J. Exp. Med 214 10.1084/jem.20161731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund SL, Adolfsson R, Gottfries CG, Winblad B, 1985. Superoxide dismutase isoenzymes in normal brains and in brains from patients with dementia of Alzheimer type. J. Neurol. Sci 67, 319–325. 10.1016/0022-510X(85)90156-X. [DOI] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD, 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mtorc1 at the lysosome. Cell 156, 1771–1785. 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Clair DK, 2009. Regulation of superoxide dismutase genes: implications in disease. Free Radic. Biol. Med 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Brys M, Switalski R, Mistur R, Glodzik L, Pirraglia E, Tsui W, De Santi S, de Leon MJ, 2007. Maternal family history of Alzheimer’s disease predisposes to reduced brain glucose metabolism. Proc. Natl. Acad. Sci 104, 19067–19072. 10.1073/pnas.0705036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Murata N, Noda Y, Tahara S, Kaneko T, Kinoshita N, Hatsuta H, Murayama S, Barnham KJ, Irie K, Shirasawa T, Shimizu T, 2011. SOD1 (copper/zinc superoxide dismutase) deficiency drives amyloid β protein oligomerization and memory loss in mouse model of Alzheimer disease. J. Biol. Chem 286, 44557–44568. 10.1074/jbc.M111.279208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair PM, Mason HS, 1966. Reconstitution of cytochrome c oxidase from an apo- enzyme and cu(I). Biochem. Biophys. Res. Commun 10.1016/0006-291X(66)90261-0. [DOI] [PubMed] [Google Scholar]
- Ning B, Sun N, Cao R, Chen R, Kirk Shung K, Hossack JA, Lee J-M, Zhou Q, Hu S, 2015. Ultrasound-aided multi-parametric photoacoustic microscopy of the mouse brain. Sci. Rep 5, 18775. 10.1038/srep18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena A, Wallrabe H, McMahon L, Silva A, Swanson E, Khan SSS, Baerthlein D, Kodis E, Oddo S, Mandell JWJW, Bloom GSGS, 2017. mTOR and neuronal cell cycle reentry: how impaired brain insulin signaling promotes Alzheimer’s disease. Alzheimers Dement. 13, 152–167. 10.1016/j.jalz.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena A, Wallrabe H, Cao R, Wang DB, Silva A, Svindrych Z, Periasamy A, Hu S, Tanzi RE, Kim DY, Bloom GS, 2018. A novel lysosome-to-mitochondria signaling pathway disrupted by amyloid-β oligomers. EMBO J. e100241 10.15252/embj.2018100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A, 2012. Mitochondria: in sickness and in health. Cell. 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova KA, Crino PB, 2010. The tuberous sclerosis complex. Ann. N. Y. Acad. Sci 1184, 87–105. 10.1111/j.1749-6632.2009.05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco JC, Götz J, 2018. Are you TORCing tau me? Amyloid-β blocks the conversation between lysosomes and mitochondria. EMBO J. 10.15252/embj.2018100839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco JC, Li C, Bodea LG, Martinez-Marmol R, Meunier FA, Götz J, 2018. Amyloid-β and tau complexity - Towards improved biomarkers and targeted therapies. Nat. Rev. Neurol 10.1038/nrneurol.2017.162. [DOI] [PubMed] [Google Scholar]
- Prather P, de Vries PJ, 2004. Behavioral and cognitive aspects of tuberous sclerosis complex. J. Child Neurol 19, 666–674. 10.1177/08830738040190090601. [DOI] [PubMed] [Google Scholar]
- Reddi AR, Culotta VC, 2013. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC, 1997. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev 77, 731–758. 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Yamasaki T, Sumiyoshi T, Utsunomiya N, Takeda M, Kamba T, Nakamura E, Ogawa O, 2018. A family case with germline TSC1 and mtDNA mutations developing bilateral eosinophilic chromophobe renal cell carcinomas without other typical phenotype of tuberous sclerosis. J. Clin. Pathol 71, 936–943. 10.1136/jclinpath-2018-205211. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ, 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A 105, 2415–2420. 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM, 2017. mTOR signaling in growth, metabolism, and disease. Cell. 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Aponte AM, Shen R-F, Balaban RS, Finkel T, 2006. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem 281, 27643–27652. 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Schrijvers EMC, Witteman JCM, Sijbrands EJG, Hofman A, Koudstaal PJ, Breteler MMB, 2010. Insulin metabolism and the risk of Alzheimer disease: the Rotterdam study. Neurology. 10.1212/WNL.0b013e3181ffe4f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seward MEME, Swanson E, Norambuena A, Reimann A, Cochran JN, Li R, Roberson EDED, Bloom GSGS, Nicholas Cochran J, Li R, Roberson EDED, Bloom GSGS, 2013. Amyloid-β signals through tau to drive ectopic neuronal cell cycle re-entry in Alzheimer’s disease. J. Cell Sci 126, 1278–1286. 10.1242/jcs.1125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Horvath TL, 2015. Mitochondrial ROS signaling in organismal homeostasis. Cell. 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z-H, Cai Q, 2012. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci 13, 77–93. 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, 1998. Autism and tuberous sclerosis. J. Autism Dev. Disord 10.1023/A:1026052421693. [DOI] [PubMed] [Google Scholar]
- Suomalainen A, Battersby BJ, 2017. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- Swanson E, Breckenridge L, McMahon L, Som S, McConnell I, Bloom GS, 2017. Extracellular tau oligomers induce invasion of endogenous tau into the Somatodendritic compartment and axonal transport dysfunction. J. Alzheimers Dis 58, 803–820. 10.3233/JAD-170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele EA, 2004. Managing epilepsy in tuberous sclerosis complex. J. Child Neurol. 19, 680–686. 10.1177/08830738040190090801. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, Törnell J, Jacobs HT, Larsson N-G, 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423. 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Trinczek B, Ebneth A, Mandelkow EM, Mandelkow E, 1999. Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J. Cell Sci 112. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Chen M, Cheng X, Qi Y, Chen Y, Das I, Li X, Vallat B, Fu LW, Qian CN, Wang HY, White E, Burley SK, Zheng XFS, 2018. SOD1 phosphorylation by mTORC1 couples nutrient sensing and redox regulation. Mol. Cell 10.1016/j.molcel.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JS, Doucette PA, Potter SZ, 2005. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- Valla J, Berndt JD, Gonzalez-Lima F, 2001. Energy hypometabolism in posterior cingulate cortex of Alzheimer’s patients: superficial laminar cytochrome oxidase associated with disease duration. J. Neurosci 10.1523/jneurosci.21-13-04923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA, 2004. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci. Lett 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA, 2007. Hydrogen Peroxide Sensing and Signaling. Mol. Cell 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP, 2007. Multiple-motor based transport and its regulation by tau. Proc. Natl. Acad. Sci. U. S. A 104 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Zaganjor E, Haigis MC, 2016. Mitochondria and Cancer. Cell. 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhao F, Ma X, Perry G, Zhu X, 2020. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener 10.1186/s13024-020-00376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisiger RA, Fridovich I, 1973. Mitochondrial superoxide dismutase. Site of synthesis and intramitochondrial localization. J. Biol. Chem 10.1016/S0021-9258(19)43735-6. [DOI] [PubMed] [Google Scholar]
- Weydert CJ, Cullen JJ, 2010. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc 5 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann FR, Manfredi G, Mawrin C, Beal MF, Schon EA, 2002. Mitochondrial DNA and respiratory chain function in spinal cords of ALS patients. J. Neurochem 80, 616–625. 10.1046/j.0022-3042.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- Yang G, Murashige DS, Humphrey SJ, James DE, 2015. A positive feedback loop between Akt and mTORC2 via SIN1 phosphorylation. Cell Rep. 10.1016/j.celrep.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Yoon EJ, Park H-J, Kim G-Y, Cho H, Choi J-H, Park H-Y, Jang J-Y, Rhim H, Kang S, 2009. Intracellular amyloid beta interacts with SOD1 and impairs the enzymatic activity of SOD1: implications for the pathogenesis of amyotrophic lateral sclerosis. Exp. Mol. Med 41, 611–617. 10.3858/emm.2009.41.9.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon S-O, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J, 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326. 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlan FP, Thienhaus OJ, Bosmann HB, 1989. Superoxide dismutase activity in Alzheimer’s disease: possible mechanism for paired helical filament formation. Brain Res. 476, 160–162. 10.1016/0006-8993(89)91550-3. [DOI] [PubMed] [Google Scholar]
- Zhao W-Q, De Felice FG, Fernandez S, Chen H, Lambert MP, Quon MJ, Krafft GA, Klein WL, 2007. Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J. 22, 246–260. 10.1096/fj.06-7703com. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin X, Wong J, Ding S, Seki E, Schnabl B, Hevener AL, Greenberg HB, Kisseleva T, Karin M, 2018. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Clister TL, Lowry PR, Seldin MM, Wong GW, Zhang J, 2015. Dynamic visualization of mTORC1 activity in living cells. Cell Rep. 10.1016/j.celrep.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.