Abstract
Inhibitory and activating immune receptors play a key role in modulating the amplitude and duration of immune responses during infection and in maintaining immune balance in homeostatic conditions. The CD200 Receptor (CD200R) gene family in humans encodes one inhibitory receptor, CD200R1, and one putative activating member, CD200R1 Like (CD200R1L). It is demonstrated that CD200R1L is endogenously expressed by human neutrophils and activates cellular functions such as reactive oxygen species (ROS) production via Syk, PI3Kβ, PI3Kδ, and Rac GTPase signaling. Phylogenetic analysis shows that CD200R1L is present in many species among vertebrates, ranging from birds to primates, suggesting that evolutionary conservation of this receptor is critical for protection against co‐evolving pathogens. The duplication event that generated CD200R1L from CD200R occurred several times throughout evolution, supporting convergent evolution of CD200R1L. In our phylogenetic trees, CD200R1L has longer branch lengths than CD200R1 in most species, suggesting that CD200R1L is evolving faster than CD200R1. It is proposed that CD200R1L represents a hitherto uncharacterized activating receptor on human neutrophils.
Keywords: NETosis, phylogeny, reactive oxygen species
Graphical Abstract
First study to report CD200R1L protein expression in humans; CD200R1L induces production of reactive oxygen species and IL‐8.
Abbreviations
- AUC
area under the curve
- CD
cluster of differentiation
- CD200R
CD200 receptor 1
- CD200R1L
CD200 receptor 1‐like
- cDNA
copy Deoxyribonucleic acid
- DAMP
Danger associated molecular patterns
- DAP12
DNAX‐activation protein 12
- TYRO
protein tyrosine kinase‐binding protein
- DPI
diphenyleneiodonium chloride
- eGFP (GFP)
enhanced green fluorescent protein
- FACS
fluorescence activated cell sorting
- FcγR
Fcγ receptor
- FMLP
N‐Formylmethionine‐leucyl‐phenylalanine
- GEF
Guanine exchange factor
- HEK293T
Human embryo kidney 293 T cells
- IL
interleukin
- ITAM
immunoreceptor tyrosine‐based activation motif
- LPS
lipopolysaccharide
- MFI
mean fluorescent intensity
- MPO
myeloperoxidase
- mRNA
messenger ribonucleic acid
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NET
Neutrophil extracellular trap
- PI3K
Phosphoinositide 3‐kinase
- PKC
protein Kinase C
- PLA2
phospholipase A2
- PLC
phospholipase C
- PMA
Phorbol 12‐myristate 13‐acetate
- qPCR
quantitative polymerase chain reaction
- ROS
reactive oxygen species
- Syk
spleen tyrosine kinase
1. INTRODUCTION
Inhibitory and activating immune receptors are crucial for the regulation of immune responses. These receptors can be present in pairs of highly related activating and inhibitory receptors such as the paired Cd200r gene family, which in humans includes one inhibitory, CD200R1, and one putative activating member, CD200R1L. The inhibitory CD200R1 (from here: CD200R) is expressed on a variety of myeloid cells, T cells and dendritic cells 1 , 2 and binds to the ubiquitously expressed glycoprotein CD200 and in mice also to the recently identified iSec1/2. 3 , 4 In mice, four CD200R1L‐related genes (termed mCd200rla‐d or Cd200rL1‐4) have been identified. 4 In contrast to CD200R1, the activating CD200RLa‐d/CD200RL1‐4 do not bind to CD200 and iSec1 ligand. 3
Inhibitory receptors signal via an inhibitory motif present in their cytoplasmic domain. In contrast, paired activating receptors have a short cytoplasmic domain that lacks a signaling motif. Instead, these receptors contain a charged amino acid residue in the transmembrane region, involved in association with adapter proteins that can relay activating signals through immunoreceptor tyrosine‐based activation motif (ITAM) or YxxM motifs. 4 Mouse Cd200rla and b associate with DAP12 and DAP12 association is necessary for proper CD200R1L surface expression. 4
Due to the inability to detect human CD200R1L protein expression, the signalling capacity and biological significance of the human activating CD200R1L are not known. cDNA for human CD200RL1 could be isolated from peripheral blood, however on the basis of differences in amino acid sequence with the mouse homologue and other genes in this group, it has been suggested that the gene does not encode for a functional protein. 4
However, a few reports have shown that CD200R1L gene expression is associated with the risk of developing psoriasis, atopic dermatitis and also helminth infection, 5 , 6 suggesting that the CD200R1L gene does encode a functional protein. In humans the predicted CD200R1L amino acid sequence contains a charged residue in the transmembrane region, suggesting human CD200R1L also could relay activating signals through recruitment of adaptor molecules.
Pathogenic pressure is thought to be an important driver of paired receptor evolution. 7 For the CD200R family a “counterbalance theory” was proposed 7 , 8 if a pathogen exploits the inhibitory receptor to down‐regulate responses against itself, it may, because of similarities in structure, also bind the activating receptor and induce an opposing activating signal. The activating receptors would then evolve to retain pathogen binding but lose the host ligand‐binding activity. Evidence that activating receptors evolved from the inhibitory ones relies on the fact that these receptors appeared later in evolution with greater variation in gene numbers. 9 Genetic divergence by gene duplication and mutations might be the mechanisms behind inhibitory to activating receptor transition. 10 , 11 The majority of inhibitory receptors are highly homologous to their paired activating receptors. 12 Despite similarity, the known ligands of inhibitory family members are usually not shared with the activating ones, as is the case for the CD200R family. 13
Here we sought to identify endogenous human CD200R1L expression, characterize what cellular responses it induces, and study how this paired receptor has evolved.
2. MATERIALS AND METHODS
2.1. Donors and cell isolation
Peripheral blood from healthy donors was collected in lithium‐heparin tubes (Greiner Bio‐One). Neutrophils were isolated by Ficoll‐Paque density gradient centrifugation (GE Healthcare). The pellet was resuspended in 50 ml ice‐cold ACK Lysis Buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.4) for red blood cell lysis. Afterward, neutrophils were resuspended in Dulbecco's Modified Eagle Medium (Gibco Life Technologies), enriched with 10% v/v FCS, PenStrep (Life Technologies; 50 U/ml), and l‐glutamine (Life Technologies; 2 mM). Purity of isolated neutrophils was analyzed using the CELL‐DYN Emerald (Abbott Diagnostics) and was > 90%. In order to exclude that contaminating cells present in our Ficoll isolated granulocytes were responsible for CD200R1L expression, FACS sorted neutrophils were isolated from whole blood based on FCS/SSC, and expression of CD16(+), CD14(‐), and CCR3(‐) expression (Supplementary Fig. S2B) using a BD FACSAria™ III Cell Sorter.
All experiments were performed in accordance with local guidelines and regulations approved by the Medical Ethical Committee of the University Medical Centre Utrecht and all blood donors gave informed consent.
2.2. Transfection and plasmids
To test the anti‐CD200R1L antibody specificity, 0.5 × 106 HEK923T cells were cultured for 24 h in a 6‐wells plate and subsequently transfected with plasmids encoding eGFP (empty vector), DAP12 (GFP‐tagged, kindly provided by Marion Brown, Oxford), CD200R (untagged), CD200R1L (Myc‐tagged, from Origene), or with a combination of DAP12 and CD200R1L. Untransfected cells (only treated with transfection agent) were used as control. Transfection mixes containing 1 μg of plasmid DNA, 2.5 μl FuGene transfection reagent (Promega) and 200 μl Opti‐MEM (Gibco) were incubated for 20 min. Afterward, 800 μl Opti‐MEM was added, the total mix was pipetted on the cells and cells were incubated for overnight transfection. The next day, the medium was replaced with RPMI medium (Gibco, with 10% v/v FCS, PenStrep (Life Technologies; 50 U/ml) and l‐glutamine (Life Technologies; 2 mM)). Cells were collected in ice‐cold PBS for flow cytometric analysis.
2.3. Flow cytometric analysis
HEK923T were washed with FACS Buffer (PBS supplemented with 1% BSA (w/v) and 0.01% NaN3) and stained with anti‐CD200R or anti‐CD200R1L antibodies for 20 min at 4°C. Cells were washed with FACS buffer and resuspended with the secondary antibody for 20 min. HEK293T were intracellularly stained with anti‐MYC antibody using FIX and PERM Cell Permeabilization Kit (Life Technologies (Invitrogen)). See Supplementary Fig. S1 for the gating strategy. For flow cytometric analysis of primary immune cells, whole blood was lysed for erythrocytes (155 mm NH4Cl, 10 mM KHCO3, 0.1 nM EDTA, pH7.2‐7.4) for 10–15 min, cells were washed with FACS Buffer and resuspended in 25 μl of antibody mix. After 20 min incubation at 4°C, cells were washed with FACS buffer and resuspended with the second antibody mix (for two‐step staining in case of biotinylated or unlabelled antibodies). Flow cytometric analysis was performed on a FACS Canto II machine or a LSRFortessa Cell Analyzer (both from BD Biosciences) and data was analysed using Flow Jo version 10.
Antibodies used for flow cytometric analysis can be found in Supplementary methods.
2.4. Western blot
For western blot analysis of CD200R1L, neutrophils (both isolated by Ficoll and FACS sorted) were lysed in boiling modified 2× Laemmli buffer (125 mM Tris‐HCl, pH 6.8, 4% SDS, 20% glycerol, 10% 2‐mercaptoethanol, 0.004% bromophenol blue) for 5 min. Samples were loaded under denaturing conditions on AnyKD gradient gels (BioRad). Protein was transferred to PVDF membranes (Immobilon‐P PVDF 45um, Merck Chemicals BV). Membranes were blocked in 5% fat‐free milk (Elk, Campina, The Netherlands) in TBS 0.05% Tween‐20 (TTBS), and incubated overnight with CD200R1L‐biotin antibody (Sino Biological), anti‐MYC‐biotin antibody (Cell Signaling) or CD200R‐biotin (Serotec) in 1% Elk TTBS. Blots were washed and HRP labelled secondary antibody or streptavidin‐HRP (bioLegend) was added for 1 h at 4 °C. The blots were imaged using SuperSignal West Femto (Thermo Fisher Scientific) on a Bio‐rad Chemidoc MP.
2.5. qPCR analysis
mRNA from purified Ficoll isolated neutrophils was isolated using QIAGEN mRNA micro isolation kits, followed by iScript (Biorad) cDNA synthesis according to the manufacturer protocol. qPCR for CD200R and CD200R1L was performed on a StepOnePlus Realtime PCR system (AB Instruments) with SYBR Select Master Mix (Life Technologies). Data are represented as 2(Ct(B2M)‐Ct(target)).
| Target | Forward | Reverse |
|---|---|---|
| CD200R1L | TAGTTCATGCATGGGTGGAAAG | AAAGCACAGCATTTATATCCATCAG |
| CD200R | GACCAGAGAGGGTCTCACCA | TTGAAGCGGCCACTAAGAAG |
| B2M | GATGAGTATGCCTGCCGTGT | TGCGGCATCTTCAAACCTCC |
2.6. Determination of ROS production
96‐well Microfluor 2 plates (Thermo Scientific) were coated overnight at 4°C with 1 μg/ml anti‐CD32 (clone IV.3; Stem Cell), anti‐CD200R1L (Sino Biological), IgG1 isotype control (eBioscience) or with PBS (Sigma‐Aldrich). The next day, wells were washed twice with PBS. Ficoll isolated neutrophils were resuspended in HEPES+++ buffer (20 mM HEPES, 132 mM NaCl, 1 mM MgSO4, 6 mM KCl, 1.2 mM KH2PO4, pH = 7.4, 0.5% BSA, 1 mM Ca2+, and 5 mM D‐glucose). 100 μl of HEPES+++ supplemented with 100 μM AmplexRed® (Thermo Fischer) and 2 U/ml HRP and 100 μl of cell suspension were added to each well. Fluorescence was measured each minute using a Fluoroscan Ascent machine (λexcitation = 545 nm, λemission = 590 nm) for 90 min. All conditions were performed in triplicates.
To screen for downstream proteins that are involved in CD200R1L‐induced ROS production, Ficoll isolated neutrophils were pre‐incubated for 30 min at 37°C in HEPES+++ with the following inhibitors: R406 (Syk inhibitor; 1 μM; Selleck Chemicals LLC), wortmannin (pan‐phosphoinositide 3‐kinase (PI3K) inhibitor; 1 μM; Sigma‐Aldrich), Akt VIII (Akt inhibitor; 2 μM; Cayman Chemical), BAPTA (Ca2+ chelator; 10 μM; Focus Biomolecules), EHT‐1864 (Rac GTPase inhibitor; 15 μM; Cayman Chemical), GSK 650394 (SGK inhibitor; 10 μM; Tocris), BYL719 (p110α inhibitor; 3 μM; Cayman Chemical), TGX‐221 (p110β inhibitor; 3 μM; Cayman Chemical), AS‐604850 (p110γ inhibitor; 10 μM; Cayman Chemical) or CAL‐101 (p110δ inhibitor; 3 μM; Cayman Chemical). For all PI3K/p110 inhibitors, solvents (DMSO or ethanol) reached a concentration of 0.2% v/v. ROS assay was carried out in the presence of the inhibitors.
To assess the role of Fc receptors in ROS production, neutrophils were incubated at 37°C for 15 min with or without FcR blocking agent (Miltenyi). To exclude possible interactions of our antibody with other receptors due to glycosylation patterns we heat denatured anti‐CD200R1L and isotype control antibodies for 5 min at 90C. Cells were washed and the ROS assay was performed as described above.
Area under the curve (AUC) for each stimulus was calculated after subtraction of background/PBS ROS production.
2.7. Enzyme‐linked immunisorbent assay
Neutrophils (both Ficoll isolated and FACS sorted) were stimulated with isotype control, anti‐ CD200R1L (1 μg/mL or 10 μg/mL) or LPS (100 ng/mL) for 24 h and cell‐free supernatant was harvested and stored at −20°C until assayed by ELISA according to the manufacturer's protocol (IL‐8 ELISA Life Technologies, cat number 88‐8086‐88). MAXIsorb plates (NUNC) were coated with 50μL capture antibody diluted in coating buffer for overnight at 4C. The plates were washed 3 times with 200μL PBS 0.05% Tween‐20, and blocked with 100μL blocking buffer provided with the kit. The standard was diluted as the manufacturer recommended, and both the standard curve samples and assay samples were incubated for 2h at room temperature while shaking. Plates were washed 3 times with 200μL PBS 0.05% Tween‐20, and incubated 1 hour at room temperature with 50μL detection antibody in blocking buffer, followed by 3 times 200μL washes and incubation with strep‐HRP in blocking buffer. Before development with TMB substrate, the plates were washed 5 times with 200μL PBS 0.05% Tween‐20. Optical densities (OD) were measured at 450 nM and 560 nM as control. With PRISM 8.3 we extrapolated from a 4‐parameter dose response curve based on the concentrations of the standard curve and their OD‐values.
2.8. Microscopic live imaging NET assay
For this study, we used a semi‐automatic approach to quantify NET release in a live imaging assay, which is based on previous described methods. 16 , 40 , 41 Briefly, Ficoll isolated neutrophils were incubated in RPMI 10% containing 20 μM Hoechst 33342 for 30 min at 37 °C and washed twice with RPMI 1640 (without phenol red) supplemented with 2% FBS, 50 U/ml Penicillin‐Streptomycin, and 10 mM HEPES (referred to as RPMI‐2% hereafter). Neutrophils were seeded on 0.001% poly‐L‐lysine (Sigma Aldrich) pre‐coated wells of a clear bottom 96‐wells plate (Ibidi) and stimulated with isotype control (10 μg/ml, eBioscience), CD200R1L (10 μg/mL, Sino Biological) or 25 ng/ml phorbol 12‐myristate 13‐acetate (PMA) (Sigma Aldrich) for 4h. All stimuli were resuspended in RPMI‐2% containing 4 nM Sytox Green (Life Technologies). After 4h cells were spun down and medium was gently discarded. Neutrophils were fixed with 2% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature (RT). NET release was measured using the Pathway 855 bioimaging system (BD Biosciences) with a 10x objective. In order to study the effect of ROS in CD200R1L induced NET release, the live imaging procedure described above was performed in the presence or absence of chemical NADPH oxidase inhibitor, Diphenyleneiodonium (DPI) (Sigma‐Aldrich). Data was analysed using FIJI software as previously described. 16 MPO stainings were performed using the IncuCyte ZOOM (Essen BioScience) microscope as previously described. 42 Briefly, neutrophils (2 × 10 6 cells/ml) were incubated for 5 min in the dark at room temperature with the membrane‐permeable NUCLEAR‐ID Red DNA dye to stain nuclei (Enzo Life Sciences). Cells were washed three times with RPMI medium to remove excess dye. Labelled neutrophils were plated at a cell density of 20.000 cells/100ul per well in a 96‐well flat clear bottom polystyrene tissue culture treated microplate (Costar 3596, Corning) to facilitate segmentation and analysis by the integrated imaging software. Sytox Green was added to the plates concomitantly with the stimuli for 4h. Neutrophils were fixed with 2% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature (RT). NET release was measured using the IncuCyte ZOOM (Essen BioScience) microscope with a 20x objective, using phase contrast, red (800 ms exposure) and green (400 ms exposure) channels. For the MPO staining, mouse anti‐human MPO‐FITC antibody (Abcam) was added with the stimuli instead of Sytox green dye, and imaged for 4h every 15 minutes with phage, red (1600 ms exposure) and green (800 ms exposure) channels using the incucyte microscope.
To assess the role of Fc receptors in NETosis induction, neutrophils were incubated at 37°C for 15 min with or without FcR blocking agent (Miltenyi). Cells were washed and the NETosis assay was performed as described above. Data was analysed as described above.
2.9. Phylogenetic analysis
A total of 128 sequences from 57 species were identified using BLAST (human CD200R was given as query, using a local BLAST installation). We did not explicitly search for the origins of these molecules (i.e., we did not make extra efforts to find the earliest CD200R and CD200R1L proteins). Instead we made use of a stringent threshold in BLAST searches to make sure that our analysis contains only the proteins, where the homology with CD200R and CD200R1L is very likely. The sequences are extracted from the reference sequence database, the sequence alignments were made using MAFFT software, 43 and the phylogeny was made using RAxML software 44 (see Supplementary Table 1 for reference sequences used).
2.10. Statistical analysis
All data was analysed using GraphPad Prism 7. Comparison of conditions was performed using Wilcoxon signed‐rank test (experiments were performed at least 6 times with different donors) or Man Whitney test. A p‐value < 0.05 was considered significant.
2.11. Data availability
No datasets were generated during the course of this study.
3. RESULTS
3.1. Human primary neutrophils express CD200R1L
mRNA expression of CD200R1L has been detected in human peripheral blood, but endogenous CD200R1L protein expression was never reported. 4 We revisited this issue as new antibodies directed against CD200R1L became commercially available. To validate that the monoclonal mouse antibody against human CD200R1L did not cross‐react with the inhibitory CD200R, we transiently transfected HEK293T cells with either CD200R‐ or CD200R1L‐Myc‐encoding vectors and stained the cells with anti‐CD200R1L for flow cytometric analysis (Fig. 1A, Supplementary Fig. S1A). We included co‐expression of DAP12‐GFP with CD200R1L‐Myc, since it has been reported that mouse CD200RLa and CD200RLb require DAP12 for stable surface expression. 14 Non‐transfected cells, cells transfected with an empty vector or only a DAP12‐encoding vector were used as controls. Only cells that were transfected with CD200R1L or DAP12/CD200R1L expression plasmids stained positive with the anti‐CD200R1L antibody (Fig. 1A). Importantly, the anti‐CD200R1L antibody did not bind to CD200R transfected cells. CD200R1L protein expression was also determined by Western blot. Samples were stained using anti‐MYC antibody (Fig. 1B, left side) or anti‐CD200R1L (Fig 1B, right side). We observed a band of around 45kDA in both blots, which is consistent with the reported molecular mass of rhCD200R1L (approximately 45–60 kDa due to glycosylation). CD200R1 and CD200R1L qPCR primers were validated in the same samples of transfected cells (Fig. 1C). Only samples transfected with CD200R1L alone or in combination with DAP12 showed amplification using our primers. Based on these results, we conclude that the anti‐CD200R1L antibody and the CD200R1L primers only bind human CD200R1L and do not cross‐react with CD200R1.
FIGURE 1.
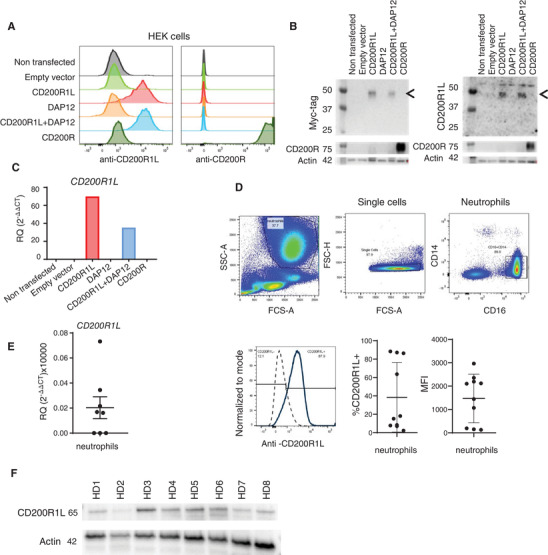
Anti‐CD200R1L antibody specificity and CD200R1L expression in human healthy donor neutrophils. (A–C) HEK293T transfected with DNA vectors expressing indicated proteins, and analysed by (A) flow cytometry, (B) Western blot, or (C) qPCR. (A) n = 3 from 3 independent experiments, one representative experiment is shown. Gating strategy available in Supplementary Fig. S1A. (B) Samples were stained using anti‐MYC antibody (left side) or anti‐CD200R1L (right side). Arrows depict CD200R1L bands. n = 2 from 2 independent experiments. (C) n = 2 from 2 independent experiments. (D–F) Expression of CD200R1L assessed by (D) flow cytometry, (E) qPCR, and (F) Western blot. (D) A representative FACS staining indicating CD200R1L expression in neutrophils from a healthy donor. Quantification indicate the % and MFI of CD200R1L positive neutrophils. 10 different donors were analysed in 5 independent experiments (E) CD200R1L expression in purified healthy donor neutrophils by qPCR analysis, n = 7 from 3 independent experiments (F) and by Western blot n = 8 from 4 independent experiments. Actin and CD200R1L were probed on the same membrane.
Full‐length blots are included in the Supplementary Information
We used these validated tools to study CD200R1L expression in human blood. We detected expression of CD200R1L on CD16+ neutrophils by flow cytometry (Fig. 1D), a cell type that was previously not studied for CD200R1L expression, and validated protein expression of CD200R1L by Western Blot as a protein of approximately 65 kDa (Fig. 1F). Although all donors tested expressed CD200R1L, the level of expression of CD200R1L was variable between donors. Neutrophils may express an alternatively glycosylated form of CD200R1L that is bigger than the one expressed by HEK cells. In addition to protein expression of CD200R1L, we also detected mRNA expression (Fig. 1E) in isolated neutrophils by qPCR. However, CD200R1L protein and mRNA expression was variable between donors: 3 out of 8 healthy donors didn't show detectable CD200R1L mRNA levels and 4 out of 10 donors had less than 10% CD200R1L expressing neutrophils, with MFI values ranging from 153 to 2965. Coherent with previous reports, 4 we did not detect CD200R1L mRNA in total PBMC (data not shown). In order to further validate CD200R1L expression in neutrophils we compared Ficoll isolated granulocytes (purity above 90%) with FACS sorted neutrophils (purity above 99%; Supplementary Figs S1B and S1C). We observed that CD16+CD14– neutrophils isolated by both methods expressed CD200R1L. We therefore concluded that CD200R1L mRNA and protein is expressed in human neutrophils.
3.2. CD200R1L elicits ROS and IL‐8 cytokine production
Neutrophils are one of the first cell types to be recruited to sites of infection, and can kill pathogens through a combination of different cytotoxic mechanisms. One of the hallmarks associated with the antimicrobial and inflammatory actions of neutrophils is the activation of a powerful burst of reactive oxygen species (ROS). Therefore, we tested whether ligation of CD200R1L induces ROS production by neutrophils. We observed that cross‐linking CD200R1L with immobilized antibodies resulted in significantly more ROS production compared to IgG1 isotype control (p = 0.0049; Fig. 2A, B). ROS production was not due to binding of the antibody to Fcγ‐receptors (FcγR), since CD200R1L‐induced ROS production was not affected by FcγR‐blockade, while FcγR‐blocking did decrease CD32/FcγRII‐induced ROS production (Fig. 2C). In conclusion, cross‐linking of CD200R1L with a monoclonal antibody elicits an FcγR‐independent extracellular oxidative burst in vitro, demonstrating that CD200R1L is a functional protein on primary human neutrophils. In addition to ROS, we observed that anti‐CD200R1L stimulated neutrophils produced IL‐8 (Figure 2D). We observed that CD200R1L stimulation led to increased IL‐8 production compared to isotype control treated samples and was in the same order of magnitude as after stimulation with 100 ng/mL of LPS. No differences in CD200R1L‐induced ROS and IL‐8 cytokine production were observed when Ficoll isolated granulocytes and FACS sorted neutrophils were compared (Supplementary Fig S2A and B respectively). This indicates that neutrophils were the main population responding to CD200R1L stimulation. Heat denaturation of anti‐CD200R1L and isotype control abrogated ROS production excluding possible interactions with other receptors due to glycosylation patterns (Supplementary Fig S2C).
FIGURE 2.
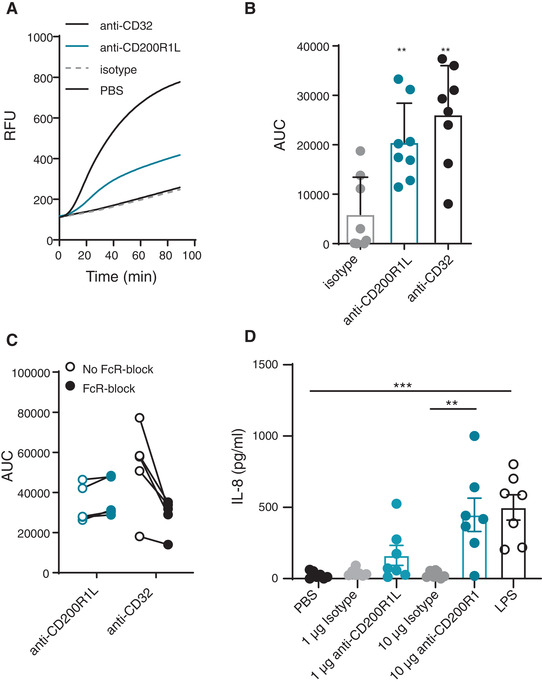
CD200R1L stimulation induces ROS and cytokine production by neutrophils. (A–C) ROS production by neutrophils after 90 minutes stimulation with isotype, anti‐CD200R1L, or anti‐CD32. (A) Representative ROS production in a healthy donor. RFU: relative fluorescent units. B) Quantification ROS production as the area under the curve (AUC) in multiple donors stimulated as in 2A. Wilcoxon signed‐rank test was performed to compare ROS production in isotype versus anti‐CD200R1L or anti‐CD32. **p < 0.01, 8 donors from 4 experiments. Bars represent means + SD. (C) Areas under the curve (AUC) of ROS production with and without FcR block with anti‐CD32 or anti‐CD200R1L stimulation (p = 0.05, n = 5 from 3 independent experiments). (D) IL‐8 production by neutrophils after PBS, isotype, anti‐CD200R1L and LPS stimulation for 24 h. one‐way ANOVA with Tukey multicomparison test. **p < 0.01, ***p < 0.001, n = 7 from 4 experiments
3.3. CD200R1L‐induced ROS production relies on Syk, PI3K and Rac GTPase signalling
Mouse CD200Ra‐b activates Spleen Tyrosine Kinase (Syk) through DAP12, followed by activation of PI3 kinase (PI3K), after which the signal diverges. 4 ROS production in neutrophils has been shown to be largely dependent on ITAM–mediated signalling pathways triggered by DAP12‐ and/or FcRγ‐ receptors. We pre‐treated neutrophils with small molecular inhibitors for Syk, PI3K or downstream effectors of PI3K, such as Akt, SGK or Rac GTPase, and the calcium chelator BAPTA to address the contribution of these pathways in human CD200R1L‐mediated ROS production. Inhibition of Syk, PI3K, Rac and Akt decreased CD200R1L‐induced ROS production, whereas inhibition of SGK, and chelating Ca2+ did not affect ROS production (Fig. 3A, B). The pooled data shown in Fig. 3B are corrected for background ROS production, which is probably induced by the isolation procedure. PI3K is involved in both CD200R1L‐mediated and spontaneous ROS production, as indicated by the negative value after background correction.
FIGURE 3.
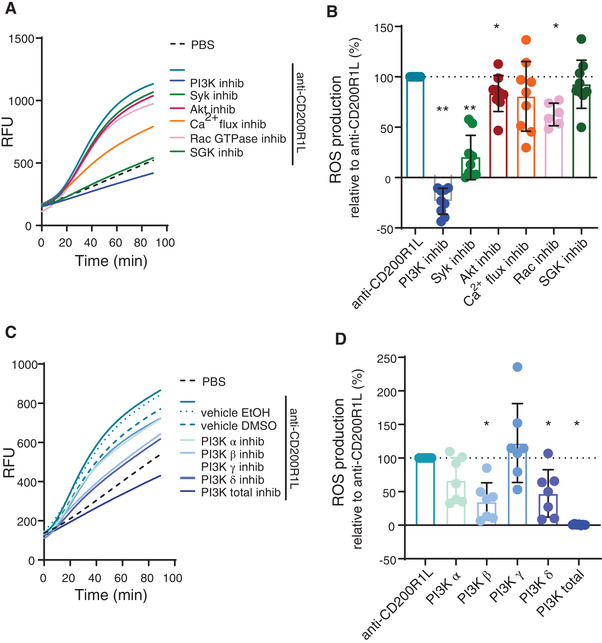
Inhibition of downstream effector molecules Syk, PI3K and Rac GTPase reduces CD200R1L‐induced ROS production. (A,B) anti‐CD200R1L induced ROS production after pre‐incubation with small molecule inhibitors. (A) example experiment and (B) quantification of the area under the curve normalized to anti‐CD200R1L. Syk, PI3K, Akt: n = 9; Rac GTPase: n = 6 donors in 3 independent experiments. (C, D) example experiment and (D) quantification of the ROS production in anti‐CD200R1L‐stimulated neutrophils treated with PI3K isoform inhibitors normalized to anti‐CD200R1L. n = 7 donors in 3 independent experiments. A Wilcoxon signed‐rank test was used to compare conditions to vehicle control. *p < 0.05, **p < 0.01. Bars represent mean + SD
To further dissect how PI3K is involved in CD200R1L‐induced ROS in neutrophils, we specifically inhibited catalytic subunits of class I PI3K isoforms, a subgroup of the PI3K family that is associated with induction of the oxidative burst. 15 Using small molecular inhibitors that target isoforms of the catalytic subunit p110 we found that inhibition of PI3Kβ and −δ decreased CD200R1L‐induced ROS production, indicating that these proteins are involved in the downstream CD200R1L signalling cascade. Inhibition of PI3Kα and ‐γ did not lead to a significant decrease in ROS production (Fig. 3C, D) at the concentrations we used. Together these data show that CD200R1L induces ROS dependent on Syk, PI3Kβ and −δ, Akt and Rac.
3.4. CD200R1L induces classical ROS‐dependent NETosis
Another important neutrophil function that can be induced by ROS production is the release of extracellular traps (NETs). In order to understand if CD200R1L triggering leads to NET‐release, we cross‐linked CD200R1L with the monoclonal antibody and measured NETosis after 4h using live imaging as described previously. 16 Activating CD200R1L on neutrophils led to a significant increase in NET release compared to isotype control treated neutrophils (Fig. 4A), however the CD200R1L‐mediated NETosis was small compared to PMA. The increase of NET release was still observed if we pre‐treat neutrophils with an Fc blocking agent demonstrating that CD200R1L‐induced NET release is independent of Fc receptor activation (Supplementary Fig. S3A). Characteristic of the NETosis process we observed that MPO stained CD200R1L‐induced NETs (Supplementary Fig. S3B and Supplementary video 1). Since it has been shown that ROS production precedes classical NADPH dependent NETosis, 17 we blocked NADPH activity by means of the pharmacological inhibitor diphenyleneiodonium chloride (DPI) and measured NET release after 4h. DPI significantly reduced NETosis after CD200R1L triggering and after stimulation with PMA as positive control (Fig. 4B) but did not affect isotype induced NET release. Taken together, our data show that CD200R1L can induce ROS and low amounts of ROS‐dependent NETosis.
FIGURE 4.
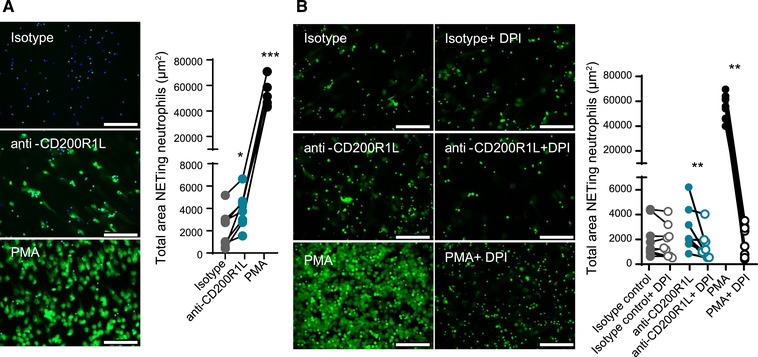
CD200R1L stimulation induces NET release. (A) NETosis induced after 4 h stimulation with isotype control, anti‐CD200R1L or PMA analyzed by SYTOX‐green staining. Representative images for each stimulation are shown. Data are presented as total area NETing neutrophils (μm2). Mann‐Whitney test comparing anti‐CD200R1L or PMA stimulation with isotype control. (B) Purified healthy donor neutrophils were stimulated with IgG1 isotype control, anti‐CD200R1L, PMA with or without DPI for 4 h. Representative images for each stimulation are shown and data was analyzed as in Fig. 4A). Wilcoxon test. (A, B) Scale 100μ. Data from 8 donors in 4 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
3.5. Evolution of CD200R‐CD200R1L paired receptor
Our data show that in human neutrophils, CD200R1L is expressed as a functional receptor. To explore the paired CD200R family dynamics through evolutionary history, we used phylogenetic analysis. We analysed 128 sequences from 57 different vertebrate species to construct a phylogenetic tree (Supplementary Fig. S4). Our analysis revealed that CD200R was present in the common ancestor of birds and of mammals (Supplementary Fig. S4, summarized in Fig. 5A), while CD200RL probably appeared later in evolution. The gene duplications that gave rise to CD200R1L genes occurred several times independently: for example, in the common ancestor of birds and in the common ancestor of primates. This suggests that the CD200R‐CD200R1L paired receptors have undergone convergent evolution. We hypothesize that similar selective pressures have led to the independent evolution of the activating CD200R family members in distantly related species. Since it is thought that pathogenic pressure drives faster evolution, we analysed the branch lengths in the CD200R family phylogenetic tree. Branch lengths depict how many mutations occurred in the evolutionary time between lineages. We found that activating CD200R family members have longer branch lengths than their inhibitory counterparts (Fig. 5B), suggesting that activating receptors accumulate mutations at a faster pace. This implies a stronger pathogenic pressure on the activating CD200R‐family members compared to their inhibitory counterparts. Importantly, these results are also coherent with the anti‐microbial functions that CD200R1L activates in neutrophils.
FIGURE 5.
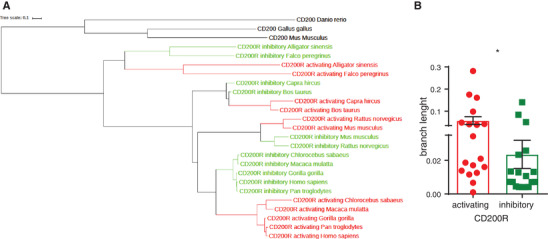
CD200R1L is evolutionary conserved and is evolving faster than CD200R . (A) A summarized phylogenetic tree based on analysis of 128 sequences from 57 species (see Supplementary Table 1 for sequence reference). Sequence alignments were made using MAFFT software, and the phylogeny was made using RAxML software. (see Supplementary Fig. S4 for extended tree). (B) Primate tree branch lengths of CD200R and CD200R1L is depicted
4. DISCUSSION
Neutrophils are the most abundant circulating leukocytes. However, the full spectrum of their biological functions is just starting to emerge. Here we show that the immune activating CD200R1L, in contrast to what was previously reported, 4 is endogenously expressed and functional in human neutrophils. Moreover, the CD200R/CD200R1L receptor pair is evolutionarily conserved and duplicated multiple times throughout evolution, suggestive of its importance in immune regulation. The faster evolution of activating family members agrees with the previously proposed theory 7 , 8 that under pathogen pressure, these receptors evolve quicker. These results highlight not only the characterization of a previously unidentified activating receptor on neutrophils but also its functional consequences.
Antibody‐mediated cross‐linking of CD200R1L on human neutrophils prompted ROS production. The NADPH oxidase is regulated by the collective action of several intracellular signalling pathways, including those driven by PI3K, 18 phospholipase C (PLC)/Ca2+/protein kinase C (PKC), 19 phospholipase D (PLD), 20 phospholipase A2 (PLA2), 21 and p38/Erk. 22 PI3Kγ and PI3Kβ isoforms are required for superoxide production after FMLP stimulation. 23 There is also evidence that class I PI3Ks can activate Rac2 through regulation of one of its guanine exchange factor (GEF), PRex1. 24 By making use of specific inhibitors we demonstrated that CD200R1L‐induced ROS production on human neutrophils relies on Syk, PI3K and Rac GTPase signalling. Surprisingly, we observed that inhibiting Rac GTPase only partly decreased ROS production. This might be due to insufficient Rac inhibition or the presence of compensatory mechanisms from the different Rac isoforms.
Notably, ROS can also increase the overall antimicrobial response of neutrophils by activating the release of granules, inducing NETs, and stimulating the production of pro‐inflammatory cytokines. 25 Two main major routes to induce NETosis have been proposed: the classical, ROS dependent and the non‐classical, ROS‐independent route. 16 , 26 , 27 Upon stimulation with PMA, anti‐LL37 28 and non‐opsonized Staphylococcus aureus, 29 neutrophils can undergo ROS‐dependent NETosis through the NADPH oxidase complex, as shown by the abrogation of NET release by using enzymatic inhibitors of this complex 29 , 30 or by studies with neutrophils from NADPH‐deficient patients/CGD. 16 , 31 , 32 In contrast, ROS‐independent NETosis is induced by for example opsonised S.aureus 33 and uric acid. 34 We observed that CD200R1L stimulation led to small but significant classical NADPH oxidase dependent NET release compared to isotype control antibody but much lower than PMA induced NETs. NETosis induced by stimuli such as opsonized bacteria is generally 3‐fold less than PMA induced netosis, 16 which is not in the same range as what we observe for CD200R1L which is 10 fold less than PMA. A direct comparison to other more physiologic NET inducing agents such as PSM peptides 35 or other antibodies 36 and/or immune complexes 37 would provide better insight on the levels of CD200R1L induced NETs. Moreover, further in vivo studies are necessary to address the biological relevance of the relatively low CD200R1L‐induced NET release in the context of infection.
Organisms at all developmental stages rely on the expression and function of genes to cope with the quickly evolving environmental and pathogenic pressure. The ability to sense and adapt to environmental and pathogenic changes is vital for organisms to maintain cellular functions and assure survival. Some genes may have also been lost through evolution if the pathogenic pressure ceased to exist. It is hypothesized that the efficacy of the adaptive immune system has allowed vertebrates to decrease the often‐impressive diversity of innate effector molecules that was available in earlier lineages. 38 For the evolution of the CD200R family a “counterbalance theory” has been proposed. 8 Our data support this theory because independent selection for activating receptors across species suggests that these receptors co‐evolved with pathogens as pathogen recognition receptors. Moreover, we found that CD200R1L has evolved in amphibians and mammals by convergent evolution, demonstrating CD200R1L was selected for in multiple occasions. Adding to this, activating CD200R family members are evolving faster than their inhibitory counterpart suggesting an ever‐present pathogenic pressure. It is also remarkable that the number of activating receptors is very variable between species implying that a particular selection of inhibitory and/or activating receptors may be of benefit in the ongoing battle between pathogens and host.
Although its name suggests otherwise, CD200R1L is not a receptor for CD200. 13 Studies have shown that several herpes viruses acquired host CD200 that can bind to the inhibitory CD200R but not to the activating ones. 39 The source and identity of the CD200R1L ligand remains elusive. However, given the genetic association with the risk of developing psoriasis, atopic dermatitis and helminth infection, 5 , 6 we hypothesise that bacteria and helminths as likely candidates. Furthermore, in addition to pathogens, circulating host‐derived molecules released from dying and damaged cells after hypoxia, trauma, and cell death (DAMPs) could potentially be CD200R1L ligands. CD200R family therefore, is a prototype gene family which offers the opportunity to study the “arms races” caused by host/pathogen interactions.
AUTHORSHIP
M.I.P.R., J.S., R.G., N.S., and G.H.A.W. conducted experiments, M.I.P.R., J.S., N.S. and CK analysed data; MIPR drafted manuscript; CK, MV and LM edited manuscript; MV and LM supervised the study and confirmed submission.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table 1: Antibodies used for flow cytometric analysis
Supplementary Figure S1: Gating strategy for CD200R1L expression.
A) Cells were gated based on the expression of Myc‐CD200R1L (stained with AlexaF700) and/or DAP12 (GFP) depending on the transfection condition. Surface expression of CD200R1L and CD200R was accessed on the colour coded gates. The same colour code was used for the histograms on Fig. 1. n = 3 from 3 independent experiments, one representative experiment is shown. See also Fig. 1.
B) Whole blood samples, FACS sorted neutrophils and Ficoll isolated granulocytes were stained for CD14, CD16, CD200R1L and isotype control. The purity of Ficoll isolated granulocytes was above 90% and sorted neutrophils reached 99% purity.
C) CD200R1L expression in FACS sorted healthy donor neutrophils by Western blot. n = 8 from 4 independent experiments. Actin and CD200R1L bands were probed on the same membrane. Full‐length blots are included in the Supplementary Information file.
Supplementary Figure 2‐ CD200R1L stimulation of Ficoll isolated granulocytes and sorted neutrophils induces comparable ROS and cytokine production.
A) Example plots and quantification of anti‐CD32, anti‐CD200R1l or isotype iduced ROS production by Ficoll isolated and Sorted neutrophils, with quantification. RFU: relative fluorescent units. For quantification the areas under the curves (AUC) were calculated. N = 7 donors from 4 independent experiments. Significance was tested by a Wilcoxon signed‐rank test. *: p < 0.05, **: p < 0.01.
B) IL‐8 production by Ficoll isolated granulocytes and sorted neutrophils after PBS, isotype, anti‐CD200R1L or LPS stimulation for 24h. IL‐8 production was measured by ELISA. Significance tested by one way anova with Tukey multicomparison test. n = 7 from 4 independent experiments. *: p < 0.05, **: p < 0.01.
C) Example plot and quantification of anti‐CD32, anti‐CD200R1L and isotype induced ROS production by Ficoll isolated granulocytes and sorted neutrophils. Isotype and anti‐CD200R1L were coated ‘native’ and heat denatured. 4 donors in 2 independent experiments. Significance was calculated by paired T test. *: p < 0.05, **: p < 0.01.
Supplementary Figure S3: CD200R1L stimulation induces NET release
A) Purified healthy donor neutrophils labelled with NUCLEAR‐ID Red dye (red) were pre‐stimulated with Fc blocking agent before Sytox green (green) and anti‐CD200R1L or isotype stimulation for 4h. Samples were fixed with PFA and NET release was measured using the IncuCyte ZOOM (Essen Biosciences) with a 20x objective. Representative images for each stimulation are shown. Data was analysed using IncuCyte software as previously described. 42 (anti‐CD200R1L versus isotype, p = 0.0313, Wilcoxon test). Scale 100μ. Data from 6 donors from 1 experiment.
B) Purified healthy donor neutrophils labelled with NUCLEAR‐ID Red dye (red) were stimulated with anti‐CD200R1L in the presence of MPO‐FITC for 4h. NET release was measured every 15 min using the IncuCyte ZOOM (Essen Biosciences) with a 20x objective. Representative images for the MPO staining from the CD200R1L stimulation are shown. Scale 100μ. Data from 4 donors from 2 independent experiments.
Supplementary Figure S4: Extended phylogenetic tree for CD200R1L and CD200R .
A total of 128 sequences from 57 species were identified using BLAST (human CD200R was given as query, using a local BLAST installation, see Supplementary Table 1 for sequence reference). Sequence alignments were made using MAFFT software, and the phylogeny was made using RAxML software. See also Fig. 5.
Supplement Material
Pascoal Ramos MI, Keşmir C, Stok JE, et al. CD200R1L is a functional evolutionary conserved activating receptor in human neutrophils. J Leukoc Biol. 2022;111:367–377. 10.1002/JLB.2A0520-334R
REFERENCES
- 1. Meyaard L, Adema GJ, Chang C, et al. LAIR‐1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity. 1997;7:283‐290. [DOI] [PubMed] [Google Scholar]
- 2. Meyaard L. LAIR‐1, a widely distributed human ITIM‐bearing receptor on hematopoietic cells. Curr Top Microbiol Immunol. 1999;244:151‐157. [DOI] [PubMed] [Google Scholar]
- 3. Kojima T, Tsuchiya K, Ikemizu S, et al. Novel CD200 homologues iSEC1 and iSEC2 are gastrointestinal secretory cell‐specific ligands of inhibitory receptor CD200R. Sci Rep. 2016;6:36457. 10.1038/srep36457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wright GJ, Cherwinski H, Foster‐Cuevas M, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J Immunol. 2003;171:3034‐3046. [DOI] [PubMed] [Google Scholar]
- 5. Baurecht H, Hotze M, Brand S, et al. Genome‐wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genet. 2015;96:104‐120. 10.1016/j.ajhg.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fumagalli M, Pozzoli U, Caglian R, et al. The landscape of human genes involved in the immune response to parasitic worms. BMC Evol Biol. 2010;10:264. 10.1186/1471-2148-10-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akkaya M, Barclay AN. How do pathogens drive the evolution of paired receptors?. Eur J Immunol. 2013;43:303‐313. 10.1002/eji.201242896. [DOI] [PubMed] [Google Scholar]
- 8. Barclay AN, Hatherley D. The counterbalance theory for evolution and function of paired receptors. Immunity. 2008;29:675‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abi‐Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin‐like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319‐1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217‐251.217‐51. [DOI] [PubMed] [Google Scholar]
- 11. Wilson MD, Cheung J, Martindale DW, Scherer SW, Koop BF. Comparative analysis of the paired immunoglobulin‐like receptor (PILR) locus in six mammalian genomes: duplication, conversion, and the birth of new genes. Physiol Genomics. 2006;27:201‐218. 10.1152/physiolgenomics.00284.2005. [DOI] [PubMed] [Google Scholar]
- 12. Lanier LL. Face off–the interplay between activating and inhibitory immune receptors. Curr Opin Immunol. 2001;13:326‐331. [DOI] [PubMed] [Google Scholar]
- 13. Hatherley D, Cherwinski H, Moshref M, Barclay AN. Recombinant CD200 protein does not bind activating proteins closely related to CD200 receptor. J Immunol. 2005;175:2469‐2474. [DOI] [PubMed] [Google Scholar]
- 14. Voehringer D, Rosen DB, Lanier LL, Locksley RM. CD200 receptor family members represent novel DAP12‐associated activating receptors on basophils and mast cells. J Biol Chem. 2004;279:54117‐54123. [DOI] [PubMed] [Google Scholar]
- 15. Condliffe AM, Davidson K, Anderson KE, et al. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432‐1440. 10.1182/blood-2005-03-0944. [DOI] [PubMed] [Google Scholar]
- 16. van der Linden M, Westerlaken GHA, van der Vlist M, van Montfrans J, Meyaard L. Differential signalling and kinetics of neutrophil extracellular trap release revealed by quantitative live imaging. Sci Rep. 2017;7:6529. 10.1038/s41598-017-06901-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kirchner T, Möller S, Klinger M, et al. The impact of various reactive oxygen species on the formation of neutrophil extracellular traps. Mediators Inflamm. 2012;2012:849136. 10.1155/2012/849136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao XP, Zhu X, Fu J, et al. Blockade of class IA phosphoinositide 3‐kinase in neutrophils prevents NADPH oxidase activation‐ and adhesion‐dependent inflammation. J Biol Chem. 2007;282:6116‐6125. 10.1074/jbc.M610248200. [DOI] [PubMed] [Google Scholar]
- 19. Watson F, Robinson J, Edwards SW. Protein kinase C‐dependent and ‐independent activation of the NADPH oxidase of human neutrophils. J Biol Chem. 1991;266:7432‐7439. [PubMed] [Google Scholar]
- 20. Usatyuk PV, Gorshkova IA, He D, et al. Phospholipase D‐mediated activation of IQGAP1 through Rac1 regulates hyperoxia‐induced p47phox translocation and reactive oxygen species generation in lung endothelial cells. J Biol Chem. 2009;284:15339‐15352. 10.1074/jbc.M109.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dana R, Leto TL, Malech HL, Levy R. Essential requirement of cytosolic phospholipase A2 for activation of the phagocyte NADPH oxidase. J Biol Chem. 1998;273:441‐445. [DOI] [PubMed] [Google Scholar]
- 22. Brown GE, Stewart MQ, Bissonnette SA, et al. Distinct ligand‐dependent roles for p38 MAPK in priming and activation of the neutrophil NADPH oxidase. J Biol Chem. 2004;279:27059‐27068. 10.1074/jbc.M314258200. [DOI] [PubMed] [Google Scholar]
- 23. Nguyen GT, Green ER, Mecsas J. Neutrophils to the ROScue: mechanisms of NADPH oxidase activation and bacterial resistance. Front Cell Infect Microbiol. 2017;7:373. 10.3389/fcimb.2017.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong X, Mo Z, Bokoch G, et al. P‐Rex1 is a primary Rac2 guanine nucleotide exchange factor in mouse neutrophils. Curr Biol. 2005;15:1874‐1879. 10.1016/j.cub.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 25. Rosales C. Neutrophil: a cell with many roles in inflammation or several cell types?. Front Physiol. 2018;9:113. 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rochael NC, Guimarães‐Costa AB, Nascimento MTC, et al. Classical ROS‐dependent and early/rapid ROS‐independent release of Neutrophil Extracellular Traps triggered by Leishmania parasites. Sci Rep. 2015;5:18302. 10.1038/srep18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Branzk N, Papayannopoulos V. Molecular mechanisms regulating NETosis in infection and disease. Semin Immunopathol. 2013;35:513‐530. 10.1007/s00281-013-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Avondt K, Fritsch‐Stork R, Derksen RH, Meyaard L. Ligation of signal inhibitory receptor on leukocytes‐1 suppresses the release of neutrophil extracellular traps in systemic lupus erythematosus. PLoS One. 2013;8:e78459. 10.1371/journal.pone.0078459. [doi];PONE‐D‐13‐21533 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuchs TA, Abed U, Goosmann C, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Remijsen Q, Berghe TV, Wirawan E, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baehner RL, Nathan DG. Leukocyte oxidase: defective activity in chronic granulomatous disease. Science. 1967;155:835‐836. [DOI] [PubMed] [Google Scholar]
- 32. Chiriaco M, Salfa I, Di Matteo G, Rossi P, Finocchi A. Chronic granulomatous disease: clinical, molecular, and therapeutic aspects. Pediatr Allergy Immunol. 2016;27:242‐253. 10.1111/pai.12527. [DOI] [PubMed] [Google Scholar]
- 33. Pilsczek FH, Salina D, Poon KKH, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol. 2010;185:7413‐7425. [DOI] [PubMed] [Google Scholar]
- 34. Arai Y, Nishinaka Y, Arai T, et al. Uric acid induces NADPH oxidase‐independent neutrophil extracellular trap formation. Biochem Biophys Res Commun. 2014;443:556‐561. 10.1016/j.bbrc.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 35. Bjornsdottir H, Rudin AD, Klose FP, et al. Phenol‐soluble modulin alpha peptide toxins from aggressive staphylococcus aureus induce rapid formation of neutrophil extracellular traps through a reactive oxygen species‐independent pathway. Front Immunol. 2017;8:257. 10.3389/fimmu.2017.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aleman OR, Mora N, Cortes‐Vieyra R, Uribe‐Querol E, Rosales C. Differential use of human neutrophil fc gamma receptors for inducing neutrophil extracellular trap formation. Journal of Immunology Research. 2016. doi:Artn 290803410.1155/2016/2908034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Behnen M, Leschczyk C, Möller S, et al. Immobilized immune complexes induce neutrophil extracellular trap release by human neutrophil granulocytes via FcgammaRIIIB and Mac‐1. J Immunol. 2014;193:1954‐1965. 10.4049/jimmunol.1400478. [DOI] [PubMed] [Google Scholar]
- 38. Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol. 2014;5:459. 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwong LS, Akkaya M, Barclay AN, Hatherley D. Herpesvirus orthologues of CD200 bind host CD200R but not related activating receptors. J Gen Virol. 2016;97:179‐184. 10.1099/jgv.0.000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoffmann JH, Schaekel K, Gaiser MR, Enk AH, Hadaschik EN. Interindividual variation of NETosis in healthy donors: introduction and application of a refined method for extracellular trap quantification. Exp Dermatol. 2016;25:895‐900. 10.1111/exd.13125. [DOI] [PubMed] [Google Scholar]
- 41. Brinkmann V, Goosmann C, Kuhn LI, Zychlinsky A. Automatic quantification of in vitro NET formation. Front Immunol. 2012;3:413. 10.3389/fimmu.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gupta S, Chan DW, Zaal KJ, Kaplan MJ. A high‐throughput real‐time imaging technique to quantify NETosis and distinguish mechanisms of cell death in human neutrophils. J Immunol. 2018;200:869‐879. 10.4049/jimmunol.1700905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059‐3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stamatakis AMKDDTFBMA. RAxML‐NG: a fast, scalable, and user‐friendly tool for maximum likelihood phylogenetic inference. BioRxiv. 2019:447110. 10.1101/447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: Antibodies used for flow cytometric analysis
Supplementary Figure S1: Gating strategy for CD200R1L expression.
A) Cells were gated based on the expression of Myc‐CD200R1L (stained with AlexaF700) and/or DAP12 (GFP) depending on the transfection condition. Surface expression of CD200R1L and CD200R was accessed on the colour coded gates. The same colour code was used for the histograms on Fig. 1. n = 3 from 3 independent experiments, one representative experiment is shown. See also Fig. 1.
B) Whole blood samples, FACS sorted neutrophils and Ficoll isolated granulocytes were stained for CD14, CD16, CD200R1L and isotype control. The purity of Ficoll isolated granulocytes was above 90% and sorted neutrophils reached 99% purity.
C) CD200R1L expression in FACS sorted healthy donor neutrophils by Western blot. n = 8 from 4 independent experiments. Actin and CD200R1L bands were probed on the same membrane. Full‐length blots are included in the Supplementary Information file.
Supplementary Figure 2‐ CD200R1L stimulation of Ficoll isolated granulocytes and sorted neutrophils induces comparable ROS and cytokine production.
A) Example plots and quantification of anti‐CD32, anti‐CD200R1l or isotype iduced ROS production by Ficoll isolated and Sorted neutrophils, with quantification. RFU: relative fluorescent units. For quantification the areas under the curves (AUC) were calculated. N = 7 donors from 4 independent experiments. Significance was tested by a Wilcoxon signed‐rank test. *: p < 0.05, **: p < 0.01.
B) IL‐8 production by Ficoll isolated granulocytes and sorted neutrophils after PBS, isotype, anti‐CD200R1L or LPS stimulation for 24h. IL‐8 production was measured by ELISA. Significance tested by one way anova with Tukey multicomparison test. n = 7 from 4 independent experiments. *: p < 0.05, **: p < 0.01.
C) Example plot and quantification of anti‐CD32, anti‐CD200R1L and isotype induced ROS production by Ficoll isolated granulocytes and sorted neutrophils. Isotype and anti‐CD200R1L were coated ‘native’ and heat denatured. 4 donors in 2 independent experiments. Significance was calculated by paired T test. *: p < 0.05, **: p < 0.01.
Supplementary Figure S3: CD200R1L stimulation induces NET release
A) Purified healthy donor neutrophils labelled with NUCLEAR‐ID Red dye (red) were pre‐stimulated with Fc blocking agent before Sytox green (green) and anti‐CD200R1L or isotype stimulation for 4h. Samples were fixed with PFA and NET release was measured using the IncuCyte ZOOM (Essen Biosciences) with a 20x objective. Representative images for each stimulation are shown. Data was analysed using IncuCyte software as previously described. 42 (anti‐CD200R1L versus isotype, p = 0.0313, Wilcoxon test). Scale 100μ. Data from 6 donors from 1 experiment.
B) Purified healthy donor neutrophils labelled with NUCLEAR‐ID Red dye (red) were stimulated with anti‐CD200R1L in the presence of MPO‐FITC for 4h. NET release was measured every 15 min using the IncuCyte ZOOM (Essen Biosciences) with a 20x objective. Representative images for the MPO staining from the CD200R1L stimulation are shown. Scale 100μ. Data from 4 donors from 2 independent experiments.
Supplementary Figure S4: Extended phylogenetic tree for CD200R1L and CD200R .
A total of 128 sequences from 57 species were identified using BLAST (human CD200R was given as query, using a local BLAST installation, see Supplementary Table 1 for sequence reference). Sequence alignments were made using MAFFT software, and the phylogeny was made using RAxML software. See also Fig. 5.
Supplement Material
Data Availability Statement
No datasets were generated during the course of this study.


