Abstract
Lipocalin 2 (LCN2) is an adipokine that accomplishes several functions in diverse organs. However, its importance in muscle and physical exercise is currently unknown. We observed that following acute high‐intensity exercise (“Gran Sasso d'Italia” vertical run), LCN2 serum levels were increased. The Wnt pathway antagonist, DKK1, was also increased after the run, positively correlating with LCN2, and the same was found for the cytokine Interleukin 6. We, therefore, investigated the involvement of LCN2 in muscle physiology employing an Lcn2 global knockout (Lcn2 −/−) mouse model. Lcn2 −/− mice presented with smaller muscle fibres but normal muscle performance (grip strength metre) and muscle weight. At variance with wild type (WT) mice, the inflammatory cytokine Interleukin 6 was undetectable in Lcn2 −/− mice at all ages. Intriguingly, Lcn2 −/− mice did not lose gastrocnemius and quadriceps muscle mass and muscle performance following hindlimb suspension, while at variance with WT, they lose soleus muscle mass. In vitro, LCN2 treatment reduced the myogenic differentiation of C2C12 and primary mouse myoblasts and influenced their gene expression. Treating myoblasts with LCN2 reduced myogenesis, suggesting that LCN2 may negatively affect muscle physiology when upregulated following high‐intensity exercise.
Keywords: adipokines, bone, exercise, lipocalin 2, muscle, myogenesis
Lipocalin2 (Lcn2) is an adipokine that exerts several physiological functions. We found it to be higher in sera following high‐intensity exercise in runners, and it directly correlated with DKK1 and IL6. Removing Lcn2 in mice causes no gross muscle alterations, but reduces muscle loss following hindlimb suspension in the largest muscles in the hindlimb, while it increases soleus muscle loss. In vitro treatment with rLcn2 reduces myogenesis and myogenic gene expression, therefore high Lcn2 following challenge could be detrimental for muscle.

1. INTRODUCTION
Lipocalin 2 (LCN2) is a multifunctional protein belonging to the lipocalin superfamily (Lögdberg & Wester, 2000) and involved in many pathological processes, such as inflammation (Abella et al., 2015), myocarditis (Ding et al., 2010), acute kidney injury (Mishra et al., 2003, 2006) and bone physiopathology (Capulli et al., 2009; Capulli et al., 2018; Costa et al., 2013; Mosialou et al., 2017; Rucci et al., 2015; Veeriah et al., 2016). From our investigations, LCN2 emerges as a complex player in bone biology, detrimental for bone health when upregulated in mechanical unloading (Rucci et al., 2015). Surprisingly, its absence causes osteopenia in basal conditions (Capulli et al., 2018), but this occurrence is associated with altered energy metabolism rather than with a direct effect of LCN2 on bone cells.
Although the role of LCN2 has been studied in several tissues and is still under scrutiny, its effect on muscle and exercise biology has been poorly investigated so far. Indeed, it has been recently demonstrated that the lack of LCN2 reduces satellite cell proliferation, impairing acute muscle damage repair (Rebalka et al., 2018). Moreover, a large‐scale analysis performed on longissimus dorsi muscles, explanted from mice subjected to space flight, showed an upregulation of LCN2 compared to earth gravity control (Gambara et al., 2017). Further reports claimed that LCN2 is a negative determinant of myocardial health (Ding et al., 2010; Marques et al., 2017; Sung et al., 2017; Xu et al., 2012).
In this study, we aimed at characterising LCN2 at the skeletal muscle level in healthy male human athletes and in mice. We report that serum LCN2 increases after acute high‐intensity exercise (“Gran Sasso d'Italia” vertical run) and correlates with the Wingless‐related integration site (Wnt) pathway inhibitor, Dikkopf‐1 (DKK1). Intriguingly, lack of LCN2 reduces muscle fibre size and influences gene expression in a mouse model, although it is dispensable for normal muscle function.
2. MATERIALS AND METHODS
2.1. Materials
Dulbecco's modified Eagle's Medium (DMEM), fetal bovine serum (FBS), penicillin, streptomycin and trypsin were from GIBCO. Basic fibroblast growth factor (bFGF) was from Peprotech while recombinant mouse (rm)LCN2 (cat#1857‐LC), anti‐LCN2 antibody (cat# AF1757, RRID:AB_354974), human (cat# DLCN20) and mouse (cat# MLCN20) LCN2 enzyme‐linked immunosorbent assay (ELISA) were from R&D. HRP‐conjugated secondary antibody was from Santa Cruz Biotechnology (Cat# sc‐2020, RRID:AB_631728). The fluorescent secondary antibody for FACS (Cat# A‐21085, RRID:AB_2535742) was from Thermo Fisher Scientific. Vacuette serum‐separator tubes were from Greiner Bio‐One. Sterile plasticware and syringes were from Falcon Becton‐Dickinson or Costar. TRIzol reagent, primers and reagents for reverse transcription polymerase chain reaction were from Invitrogen. The Sensimix SYBR Green QPCR master mix was from Bioline. RT2‐Array pathway finder was from Qiagen (Cat# PAMM‐014ZA). Masson's trichrome kit was from Bio Optica. Undercarboxylated Osteocalcin (ucOCN) ELISA kit was from Elabscience, Houston, TX (cat# E‐EL‐H5493), while human (cat# ab171580) and mouse (cat# ab210965) myoglobin, Dikkopf‐1 (DKK1, cat# ab100501) and interleukin‐6 (IL6) (cat# ab100712) ELISA kits were from Abcam. Human IL‐6 ELISA was from R&D (cat# D6050). C2C12 cells were obtained from the European Collection of Authenticated Cell Cultures (Cat# 91031101, RRID:CVCL0188) through Sigma Aldrich Co. All the other reagents were of the purest grade from Sigma Aldrich Co.
2.2. Animals
Lcn2 −/−mice (background C57BL6/J, IMSR Cat# JAX:000664, RRID:IMSR_JAX:000664) were generated and kindly provided by Dr. Tak Wah Mak (University Health Network) (Berger et al., 2006). They are vital, with an overall normal lifespan and fertility. All procedures involving animals and their care were conducted in conformity with national and international laws and policies (European Economic Community Council Directive 86/609, OJ L 358, 1, December 12, 1987; Italian Legislative Decree 4.03.2014, n.26, Gazzetta Ufficiale della Repubblica Italiana no. 61, March 4, 2014; ethical approval protocol N.365/2017‐PR) and the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. Mice were housed in the animal facility of the University of L'Aquila, Italy, at the following conditions: temperature (24°C); humidity (60%–65%); dark/light cycle (12/12 h). They had access to food and water ad libitum and were fed with a standard diet (Mucedola code: 4RF21) composed of 60.8% carbohydrates, 21% proteins, 3.45% fat, 6.8% fibres, 7.95% trace elements, and 12% humidity.
2.3. Hindlimb suspension
For the hindlimb suspension experiments, which were conducted according to Rucci et al., 2015, 8‐week‐old male mice (WT C57BL/6 or Lcn2 −/−) were used. A strip of medical tape was wrapped around the tail of the animals, leaving an overhang that was securely clipped to an overhead wire, the height of which was adjusted to maintain the mice suspended at an approximately 30° angle. The swivel apparatus allowed animals ready access to food and water and to move freely into the cage using their forelimbs. The day before the experiment, mice were acclimated into the experimental room, and hindlimb grip force was performed using a grip force metre equipped with a specifically designed force transducer (Ugo Basile, Varese, Italy). Grip force analyses were repeated after 7, 14, and 20 days on both normally caged mice (Normal Loading Condition, NLC) and hindlimb suspended mice (HLS) of both genotypes. Immediately after the grip force, mice were put back in their experimental condition (i.e., NLC or HLS). After 21 days, mice were euthanized by CO2 inhalation, serum and muscles were harvested and frozen or fixed as appropriate.
2.4. Cell line
The C2C12 mouse myoblast‐like cell line was cultured in DMEM plus 10% FBS for maintenance, while when treating with rmLCN2 C2C12 cells were kept in fusion medium (DMEM plus 5% horse serum) for 4 days in total, with two medium changes, before fixation or RNA extraction. Cells were supplied with penicillin/streptomycin and glutamine and kept in a humidified 37°C incubator with 95% air 5% CO2.
2.5. Primary myoblast cultures
Mouse primary myoblasts were cultured from the hindlimb muscles of 4–6‐week‐old CD1 mice using standard protocols (Keire et al., 2013; Springer et al., 2001). Briefly, mice were euthanised by CO2 inhalation, and muscles were collected from hindlimbs and forelimbs. Muscles were then digested with collagenase/dispase, strained through a 70μm mesh and plated. Fibroblasts were depleted by detaching cells by mechanical displacement in phosphate‐buffered saline (PBS) with no calcium and magnesium, and pre‐plating for 15 min on collagen‐coated dishes at each passage to allow selective attachment of fibroblasts. Cells were kept in 80% Ham's F10, 20% FBS, 25 μg/ml bFGF before complete fibroblast depletion, and 40% DMEM, 40% Ham's F10, 20% FBS, 25 μg/ml bFGF afterwards. When treating with rmLCN2, cells were kept in fusion medium (DMEM plus 5% horse serum) for 4 days in total, with two medium changes, before fixation or RNA extraction. Cells were supplied with penicillin/streptomycin and glutamine and kept in a humidified 37°C incubator with 95% air 5% CO2.
2.6. Vertical run
Vertical run is a high‐intensity acute exercise, taking place on the Gran Sasso mountain located in the Central Italy Apennines. Male runners from different European countries competed in a timed race which consisted of a 3.6 km long race with a 1.03 km vertical ascension (29.5% slope, Figure S1A), which is usually completed in 40–50 min (speed = 75–80 m/min). This was an outdoor run, and the weather was unremarkable, with no rain and 25–27°C temperature. Fifteen competitors (Table 1) chose to participate in our study and were given self‐administered informed consent in English or Italian depending on their preferred language.
Table 1.
Features of “Gran Sasso d'Italia” vertical run participants
| ID | Age | Sex | Hours of training/week | Professional (Y/N) |
|---|---|---|---|---|
| 1 | 17 | M | 5 | N |
| 2 | 18 | M | 7 | N |
| 3 | 29 | M | 3.5 | N |
| 4 | 30 | M | 3 | N |
| 5 | 32 | M | 3.5 | N |
| 6 | 33 | M | 10 | Y |
| 7 | 35 | M | 4 | Y |
| 8 | 38 | M | 6.5 | Y |
| 9 | 39 | M | 14 | Y |
| 10 | 44 | M | 6 | Y |
| 11 | 45 | M | 3.5 | Y |
| 12 | 46 | M | 5 | N |
| 13 | 48 | M | 12 | N |
| 14 | 51 | M | 7 | N |
| 15 | 54 | M | 10 | Y |
2.7. Human serum sampling
About 5 ml of venous blood were collected from participants 30 min before the race and no more than 30 min after the race. Blood was collected on‐site into serum‐separator tubes by a medical doctor and brought to the laboratory in controlled‐temperature boxes, before being centrifuged at 2000g for 10 min. Sera were then harvested in aseptic conditions and frozen at −80°C for storage. When needed for the ELISA or Reflotron tests, serum was thawed on ice and used following the manufacturer's instructions. The protocol was approved by Asl (Agenzia Sanitaria Locale) Ethical Committee Milan 1, Milan, Italy (protocol ID: MARC01).
2.8. Mouse serum sampling
Blood was collected by cardiac puncture from 1‐, 3‐, 6‐, 12‐month‐old WT C57/BL6/J (henceforth WT) or Lcn2 −/− mice after sacrifice by CO2 inhalation, allowed to clot for 15 min at room temperature in 1.5 ml tubes, centrifuged at 2000g for 10 min and stored at −80°C until use.
2.9. ELISA and reflotron tests
Sera from participants to the vertical run were used for ELISA detection of LCN2, undercarboxylated osteocalcin (ucOCN), myoglobin, DKK1 and IL‐6 according to the manufacturers' instructions. Reflotron PLUS reactive strips were used to evaluate creatine kinase as described in the instruction sheet.
For WT and Lcn2 −/− mice analyses, sera were used to detect LCN2, myoglobin or IL‐6 by ELISA, according to the manufacturers' instructions. Reflotron PLUS reactive strips were used to evaluate creatine kinase as described in the instruction sheet.
2.10. Western blot
Six‐month‐old WT or Lcn2 −/− mice were sacrificed, and proteins were extracted from diaphragm, quadriceps, soleus and Extensor Digitorum Longus (EDL) muscle tissues. RIPA buffer was used for the extraction, which was carried out using an IKA® ultra‐turrax homogeniser, before three cycles of freeze‐thaw. Protein concentration was quantified using the Bradford assay, and 50 μg of whole tissue lysate was loaded in a 12% poly‐acrylamide gel. After blotting on nitrocellulose, membranes were stained with Ponceau S to visualise protein bands. Blocking was performed with 5% nonfat dry milk in tris‐buffered saline plus 0.1% Tween‐20 (TBS‐T). Membranes were then incubated with the anti‐LCN2 antibody at a 1:300 dilution overnight at 4°C with gentle shaking, washed three times in TBS‐T, followed by HRP‐conjugated secondary antibody and ECL‐based chemiluminescence detection, which was carried out with a ChemiDOC‐XR cabinet (Biorad).
2.11. Comparative real‐time RT‐PCR
Total RNA was extracted from mouse diaphragm, quadriceps, soleus, EDL muscle tissues using the TRIzol® method. One μg of RNA was reverse‐transcribed into cDNA using Murine Moloney leukaemia virus reverse transcriptase and the equivalent of 0.1 μg was processed using The Sensimix SYBR Green QPCR master mix for real‐time PCR. Results, calculated as fold to WT average using the ∆∆Ct method, were normalised with the housekeeping gene β‐actin. Primer sequences are available in Table 2.
Table 2.
Sequences of PCR primers used in this study
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Il6 | GAGGATACCACTCCCAACAGACC | AAGTGCATCATCGTTGTTCATACA |
| Il1b | GCCCATCCTCTGTGACTCAT | AGGCCACAGGTATTTTGTCG |
| Tnfa | CTCCCTTTGCAGAACTCAGG | AGCCCCCAGTCTGTATCCTT |
| Myod1 | TACAGTGGCGACTCAGATGC | TAGTAGGCGGTGTCGTAGCC |
| Myog | CCAACCCAGGAGATCATTTG | ATATCCTCCACCGTGATGCT |
| Mef2c | GGGGACTATGGGGAGAAAAA | ATCTCACAGTCGCACAGCAC |
| Pax7 | TGGCCAAACTGCTGTTGATTAC | CGTCTCCAGAGGGTTTCCTG |
| Utrophin | GAACAGCAAAGGGCCACAAG | GGACTGGGAGGGGTCATAGT |
| Desmin | CAACTTCCGAGAAACCAGCC | AGACCACAAAGGGGTGATCG |
| Actnb | TGTTACCAACTGGGACGACA | TCTCAGCTGTGGTGGTGAAG |
Abbreviation: RT‐PCR, reverse transcription polymerase chain reaction.
2.12. Real‐time RT2‐arrays
Total RNA was extracted from mouse C2C12 cells following treatment with vehicle or 1 μg/ml rmLCN2 using the TRIzol® method. One 1 μg of total RNA was reverse‐transcribed as described above, and the whole cDNA was mixed with the appropriate amount of SYBR mix and water following the manufacturer's instructions. The mix was then equally distributed among the wells of a mouse RT2‐array pathway finder. A total of three experiments were ran and analysed using the manufacturer's online software, using an average of all available housekeeping genes as a reference to calculate fold regulations. Genes having C t > 30 in both control and treated samples were not considered for the analysis.
2.13. 3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium (MTT) assay
3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium (MTT) assay was performed by applying tetrazolium salt (5 mg/ml in PBS) directly to the cell cultures (final concentration 0.8 mg/ml) in a 96‐well plate. After incubating for 3 h, the supernatant was removed, and the water‐insoluble salts derived from the reaction were dissolved in dimethyl sulfoxide. Optical density was then read at 595 nm in an ELISA reader, and blank‐subtracted to obtain the experimental value.
2.14. Cytofluorimetric analysis
Diaphragms from 6‐month‐old WT mice were explanted, rinsed in PBS, and enzyme‐digested as described in the “Primary myoblast cultures” paragraph. Raw lysates were then filtered through a 40 μm strainer to select single cells rather than clumps or mature myotubes, centrifuged, and resuspended in FACS buffer (0.5% BSA plus 5 mM EDTA in PBS). Cells were then fixed in 2% neutral‐buffered formalin, permeabilized in 0.1% Tween‐FACS buffer and incubated with anti‐LCN2 antibody at a 1:10 dilution, followed by incubation with fluorescent secondary antibody at 1:100 dilution. Cells were then analysed using a BD FACSMelody (Becton Dickinson and Company) to assess the % of LCN2+ cells.
2.15. Skeletal muscle histology and histomorphometry
After the sacrifice of mice by CO2, hindlimbs were isolated, skin and fur removed and limbs immersed in 10% neutral buffered formalin. After 48 h of fixation, quadriceps were isolated from the limbs and processed for paraffin embedding. Five‐μm‐thick cross sections were obtained within 50 μm from the longitudinal mid‐section of the muscle, stained with haematoxylin‐eosin or Masson's trichrome staining, dehydrated and mounted with the permanent medium. For fibre size analyses, haematoxylin‐eosin‐stained quadriceps muscle fibres from the whole section were selected and their minimum Feret diameter was calculated using the image analysis software NIH ImageJ (RRID:SCR_003070) version 1.50i. This method was preferred compared to the fibre area analysis because less prone to error due to slight differences in sectioning angles (Dubach‐Powell, 2011). To calculate the % of intact fibres, we used the method described by Grounds et al. (Grounds, 2012). Briefly, after haematoxylin‐eosin
staining, the non‐centrally nucleated fibres were identified, counted and expressed as a percentage of the total number of fibres. To calculate the % of collagen, we used the software ImageJ to select aniline blue‐stained tissue in quadricep muscles stained with Masson's trichrome method, and calculated the % of collagen area (stained in blue) over the total area (Gutpel et al., 2015).
2.16. Statistics
Results were expressed as the mean ± SD of at least three independent experiments or ≥3 mice/group. Correlation analyses were performed using Pearson's correlation test. For the vertical run experiment, paired Student's t test was used to evaluate differences between before and after the race. In every other instance, unpaired Student's t test or Mann–Whitney test was used when comparing two groups, depending on whether the distribution was normal (D'Agostino‐Pearson omnibus test or Shapiro–Wilk normality test when N was too low to perform the former). To compare curves in longitudinal studies, GraphPad Prism (RRID:SCR_002798, version 7.0) was used to run curve fitting tests to evaluate whether one curve could efficiently fit the datasets compared. A p value less than .05 was considered statistically significant.
3. RESULTS
3.1. LCN2 increases after high‐intensity acute exercise and correlates with DKK1
Fifteen male amateur or professional runners (age 17–54 years) participating in the vertical run were enroled. This is a race in which participants run on a 29.5% slope for 3.6 km, ascending for more than 1000 metres in altitude in 40–50 min (Figure S1a), thus representing a very intense physical activity. We collected the blood right before and after the race to evaluate whether this exercise would influence serum levels of LCN2 and other markers. Indeed, the analyses showed that LCN2 levels were significantly increased after the race (Figure 1a) while no differences were observed in the basal conditions (e.g., before race) between professional runners and amateurs (Figure S1b). Serum levels of the muscle damage markers myoglobin (Figure 1b) and creatine kinase (Figure 1c) were also increased compared to those before the race, reflecting the high intensity of the workout. Since it is known that LCN2 influences bone biology (Capulli et al., 2018; Rucci et al., 2015), we also analysed the bone deposition marker ucOCN, finding it unremarkable (Figure 1d). Finally, we analysed the serum levels of the Wnt pathway inhibitor, DKK1, finding it significantly increased after the race (Figure 1e). Interestingly, also the inflammatory cytokine IL6, which has been already linked to LCN2 (Costa et al., 2013; Rucci et al., 2015; Sultan et al., 2012), was increased after the race (Figure 1f). Finally, serum levels of DKK1 (Figure 1g) and IL6 (Figure 1h) significantly correlated with LCN2 serum levels.
Figure 1.
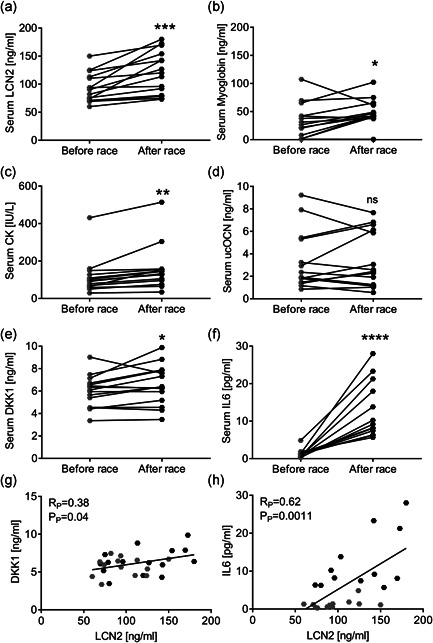
“Gran Sasso d'Italia” vertical run. Fifteen participants took part in the vertical run. Blood was collected right before and after the race. (a–f) Serum analyses showing levels of (a) LCN2, (b) myoglobin, (c) creatine kinase (CK), (d) undercarboxylated osteocalcin (ucOCN), (E) DKK1 and (F) IL6. (G,H) Pearson's correlation analyses between LCN2 and (g) DKK1 or (h) IL6 serum levels. (a–f) Paired Student's t test. *p < .05, **p < .01, ***p < .001 and ****p < .0001 after race vs before race
3.2. LCN2 is expressed in mouse muscles
Based on these results, we evaluated the role of LCN2 in mouse muscle biology. Among diaphragm, quadriceps, EDL and soleus of wild type (WT) male mice, the former showed the highest transcriptional expression of Lcn2 compared to the other muscles, with EDL showing the lowest relative expression (Figure 2a). To better understand the role of LCN2 in muscle, we then employed a global Lcn2 −/− mouse (Berger et al., 2006), which, as expected, showed undetectable levels of circulating and muscle expression of the LCN2 protein, demonstrated by ELISA (Figure 2b) and Western blot analysis (Figure 2c and Figure S2) respectively, compared to WT mice. These results also showed that WT diaphragm, quadricep and soleus expressed LCN2 at the protein level, while it was nearly undetectable in EDL (Figure 2c), thus confirming the differences observed at the transcriptional level. Finally, to gain further confirmation that LCN2 is expressed in the diaphragm, which presented with the highest LCN2 levels, we ran a cytofluorimetric analysis to assess the % of LCN2 positive cells among all mononuclear cells in this muscle, finding that about 3.5% of cells expressed LCN2 (Figure 2d).
Figure 2.
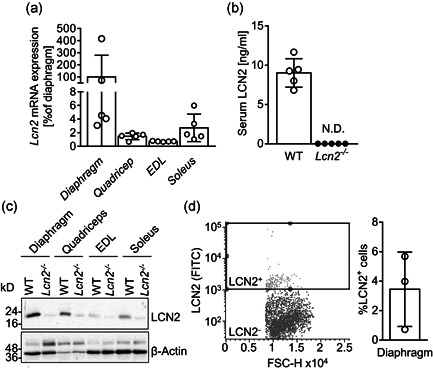
LCN2 expression in mouse muscle tissues. (a) Lcn2 mRNA expression analysed in muscle sections from diaphragm, quadriceps, EDL and soleus of WT male mice. Results are the % to the average of the diaphragm ΔC ts (N = 5 mice per group). (b) ELISA showing circulating levels of LCN2 in WT and Lcn2 −/− mice. (c) Western blot representative of three mice per group, showing muscle expression of LCN2 and β‐Actin as a loading control. (d) Cytofluorimetric analysis of mononuclear cells extracted from the diaphragm of 6‐month‐old WT mice and stained for LCN2. Left panel: representative dot plot with gating box; right panel: quantification of percent LCN2 positive cells. EDL, Extensor Digitorum Longus; ELISA, enzyme‐linked immunosorbent assay; mRNA, messenger RNA; WT, wild‐type
3.3. Lcn2 −/− mice have normal muscle performance, undamaged muscles, but have a smaller fibre diameter
We next assessed whether LCN2 could influence muscle performance by subjecting WT and Lcn2 −/− mice to the grip strength metre, finding no differences between the two genotypes at all ages evaluated (Figure 3a). Moreover, quadriceps (Figure S3a), EDL (Figure S3b), soleus Figure S3c), tibialis anterior (Figure S3d) and diaphragm (Figure S3e) weights were also unaffected by the lack of LCN2 during the lifespan. Consistently, when we analysed the sera of WT and Lcn2 −/− mice, we found no differences in the muscle damage markers, myoglobin (Figure 3b) and creatine kinase (Figure 3c). At variance with WT mice, IL6 was consistently undetectable in Lcn2 −/− mice (Figure 3d) during their lifespan. Finally, DKK1 was unremarkable between the two genotypes (Figure 3e).
Figure 3.
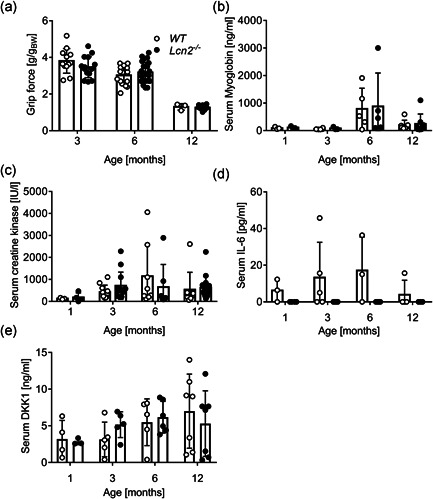
Effect of the lack of LCN2 on muscle phenotype. WT and Lcn2 ‐/− mice were subjected to (a) grip force analysis at the ages indicated in the abscissa. Results are expressed as grip force (g)/body weight(g). After sacrifice, sera were harvested and analysed for (b) myoglobin, (c) CK, (d) IL6 and (b, d, e) DKK1 by ELISA or (c) by the Reflotron assay. Curve fitting test. *p < .05 versus WT. CK, creatine kinase; ELISA, enzyme‐linked immunosorbent assay; IL6, interleukin‐6; WT, wild‐type
Histopathological evaluation of quadricep sections from WT and Lcn2 −/− mice of 3, 6, and 12 months of age showed no difference in the % of intact fibres (Figure 4a–c). Muscle collagen was also fully comparable between WT and Lcn2 −/− mice at all ages investigated (Figure 4d–f). However, when we analysed the minimum Feret diameter of quadricep fibres, we found that it was significantly smaller in Lcn2 −/− mice compared to WT (Figure 4g–i).
Figure 4.
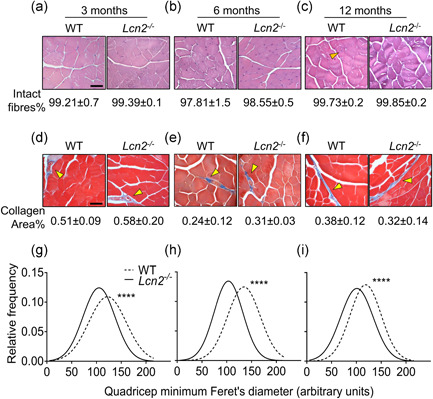
Histological analysis of WT andLcn2 −/− mice quadriceps. Quadriceps from WT and Lcn2 −/− mice were explanted, fixed, paraffin‐embedded and sectioned at 5 μm. (a–c) Haematoxylin‐eosin staining performed on histological sections from (a) 3‐, (b) 6‐, and (c) 12‐month‐old mice to quantify the % of intact fibres. (d–f) Masson's trichrome staining to evaluate the % collagen area on histological sections from (d) 3‐, (e) 6‐ and (f) 12‐month‐old mice. (g–i) After haematoxylin‐eosin staining, minimum Feret's diameter from quadricep fibres was assessed via software (NIH ImageJ, version 1.50i.) in 3‐, (h) 6‐, and (i) 12‐month‐old mice. Gaussian curves were interpolated to better represent the fibre size distributions. (a–f) N = 3–5 mice per group; (g–i) N > 1000 fibres per group, arising from N = 3–5 mice per group. Scale bar = 50 μm; orange arrowhead: example of non‐intact (centrally nucleated) fibre; yellow arrowhead: collagen area stained in blue. Student's t test. ****p < .0001 versus WT. WT, wild‐type
3.4. Effect of Lcn2 deletion on muscle gene expression
Next, we assessed the transcriptional profile of muscles from WT and Lcn2 −/− mice by focusing on gene expression of inflammatory cytokines (Il6, Il1b, and Tnfa), as well as muscle differentiation and function‐related genes, including myoblast determination protein (Myod)1, Myogenin (Myog), Myocyte‐specific enhancer factor (Mef)2c, Paired box protein (Pax7), Utrophin and Desmin. At 3 months of age, diaphragm from Lcn2 −/− mice present with higher levels of the foetal analogue of dystrophin, Utrophin (Fairclough et al., 2013; Song et al., 2019; Tinsley et al., 1996; Tinsley et al., 1998), while all the other genes were unaffected (Figure 5a). As for the quadriceps, Figure 5b shows that Lcn2 −/− mice have reduced expression of Il1b and a trend of increase of Myog (p = .07) and Myod1 (p = .07) compared to WT. Soleus of 3‐month‐old Lcn2 −/− mice showed increased Myog expression, along with higher Pax7, a marker of muscle satellite cells, whereas all other genes analysed were not modulated (Figure 5c). However, at 12 months of age, all genes were unremarkable in the diaphragm (Figure 5d) and quadriceps (Figure 5e), while showing a trend inversion in soleus, where Myog and Pax7 were both significantly reduced (Figure 5f) along with Tnfa in Lcn2 −/− mice compared to WT.
Figure 5.
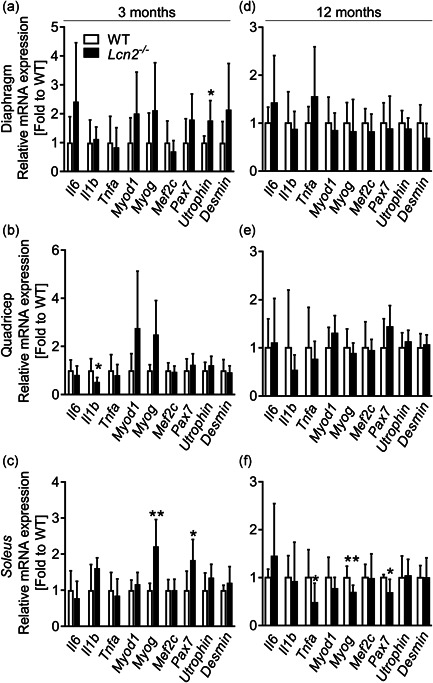
Transcriptional profiling of WT andLcn2 −/−mice muscles. RNA was extracted from (a–c) 3‐ or (d–f) 12‐month‐old WT and Lcn2 −/− mice muscles. One μg of total RNA was reverse‐transcribed into cDNA and used for transcriptional analyses. (a, d) Diaphragm, (b, e) quadriceps and (c, f) soleus. The genes analysed are indicated in the abscissa. N = 5–10. Student's t test or Mann–Whitney test. *p < .05; **p < .01 vs WT. cDNA, complmentary DNA; IL6, interleukin‐6; mRNA, messenger RNA; WT, wild‐type
3.5. Effect of Lcn2 genetic removal on muscle function loss under unloading conditions
To better elucidate whether Lcn2 played a role in muscle in non‐basal conditions, we challenged mice with hindlimb suspension, a well‐known in vivo model employed to mimic an unloading condition, eventually leading to muscle and bone loss. In these experiments, WT or Lcn2 −/− mice were hanged by the tail so that the hindlimb muscles were unloaded (HLS group), while control mice were kept in normal loading conditions (NLC). As expected, hindlimb muscle performance, evaluated by grip force test, was impaired in HLS mice over the timeframe of the experiment compared to NLC mice (Figure 6a). Intriguingly, muscle performance of Lcn2 −/− mice in HLS, was comparable to those in NLC (Figure 6b), although a direct comparison between WT and Lcn2 −/− HLS mice showed no significant differences. Moreover, creatine kinase levels were unremarkable in all conditions evaluated (Figure 6c).
Figure 6.
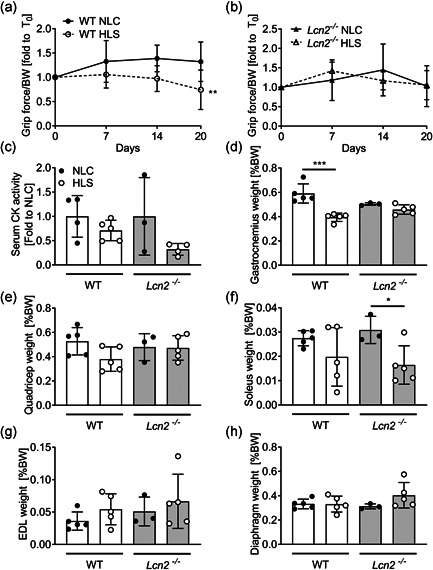
Effect of the lack of Lcn2 on muscle phenotype under mechanical unloading conditions. Eight‐week‐old WT and Lcn2 −/− mice were subjected to hindlimb suspension for 3 weeks. (a, b) Grip force was performed during the timeframe of the experiment in hindlimb suspended (HLS) or normal loading condition (NLC) (a) WT and (b)Lcn2 −/− mice. At sacrifice, serum was harvested to measure (c) CK serum activity. (d) Gastrocnemius, (e) quadriceps, (d) soleus, (g) EDL and (H) diaphragm muscles explanted from HLS and NLC mice were weighted and plotted as % of body weight. (a, b) Curve fitting test, N = 3–5. (c–h) Student's t test. *p < .05; ***p < .01; ***p < .001. CK, creatine kinase; EDL, Extensor Digitorum Longus; WT, wild‐type
At sacrifice, we weighted representative hindlimb muscles to confirm that mass was lost during the experiment. Indeed, this was the case for Gastrocnemius in HLS WT mice versus NLC, while Lcn2 −/− HLS mice did not lose muscle mass compared to Lcn2 −/− NLC (Figure 6d). In quadriceps, we observed a similar trend, although differences in weight between WT HLS and NLC were only close to significance (p = .06), while no differences were observed between NLC and HLS Lcn2 −/− mice (Figure 6e). In Soleus muscles, we noticed a different behaviour of muscle weight, which was not significantly affected in WT HLS mice versus NLC, while it was significantly reduced in Lcn2 −/− HLS versus NLC (Figure 6f). As for the EDL weight, this was not affected in either WT or Lcn2 −/− mice (Figure 6g), and the same was found for the diaphragm (Figure 6h). We next performed a gene profiling analysis on quadricep muscles, where we first checked for Lcn2 expression, which we found unremarkable between HLS and NLC WT, and as expected we did not detect any expression in Lcn2 −/− mice (Figure 7a). The muscle function/differentiation‐related genes Myod1 (Figure 7b), Myogenin (Figure 7c), Mef2c (Figure 7d), Pax7 (Figure 7e), and Desmin (Figure 7f) were not significantly affected in either genotype following hindlimb suspension. However, while a trend of decrease (p = .07) was found in Utrophin expression in WT mice following HLS, this was not the case in Lcn2 −/− mice (Figure 7g). As for the cytokine expression, Il6 was significantly lower in WT mice following HLS, while this was not observed in Lcn2 −/− mice (Figure 7h). Il1b was unremarkable in both genotypes after HLS, although we found it to be downregulated in NLC Lcn2 −/− mice vs NLC WT, as expected (Figure 7i), while Tnfa expression was unremarkable (Figure 7j).
Figure 7.
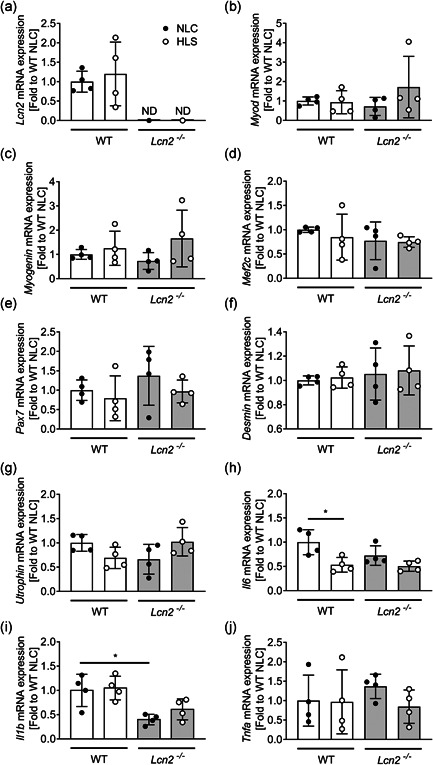
Effect of the lack of Lcn2 on muscle gene expression under mechanical unloading conditions. Eight‐week‐old WT and Lcn2 −/− mice were subjected to hindlimb suspension for 3 weeks. At sacrifice, quadriceps RNA was extracted, reverse‐transcribed and subjected to real‐time RT‐PCR to analyse the expression of (a) Lcn2, (b) Myod, (c) Myogenin, (d) Mef2C, (e) Pax7, (f) Desmin, (g) Utrophin, (H) Il6, (I) Il1b, and (J) Tnfa. Results are shown as fold to WT NLC. Student's t test. *p < .05. IL, interleukin; RT‐PCR, reverse transcrition polrmerase chain reaction
3.6. Recombinant LCN2 treatment reduces myoblast differentiation while increasing Bmp2 expression
To obtain some molecular insights into the effect of LCN2 on muscle cells, we used the C2C12 cell line, representing an in vitro model of murine myogenic progenitors (Burattini et al., 2004), along with primary mouse myoblasts. The results showed that treating C2C12 myoblasts under fusion medium with 1 μg/ml rmLCN2 decreased their differentiation into myotubes, as evaluated by phase‐contrast microscopy and 4′,6‐diamidino‐2‐phenylindole nuclear counterstaining, thus increasing the % of myoblast‐like, poorly differentiated, cells (Figure 8a). However, the number of nuclei/myotube was not affected, indicating that only the first phases of myogenesis were impaired, rather than the tube elongation after the first fusions. Furthermore, the cell metabolic activity, analysed by MTT assay, was not affected by LCN2 at 24–72 h, no matter if using differentiation (Figure 8b) or maintenance medium (Figure 8c). We ran similar experiments using mouse primary myoblasts, treated with 0.2 μg/ml rmLCN2, observing similar results both for differentiation (Figure 8d) and cell metabolic activity (Figure 8e,f). To verify whether the observed effect on myoblasts was due to reduction in muscle differentiation genes, we performed gene expression analyses for the Myod and Myogenin, finding them both downregulated by the treatment with rmLCN2 (Figure 8g).
Figure 8.
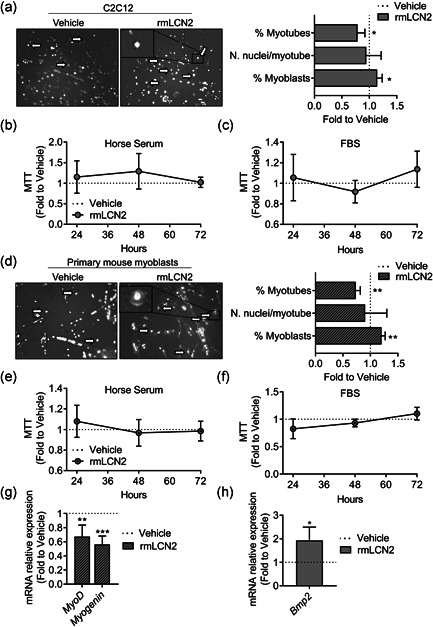
Effect of rmLCN2 on muscle cells in vitro. (a) C2C12 cells were treated with 1 µg/ml LCN2 under a myogenic differentiation medium (DMEM supplemented with 5% horse serum) for 4 days. Then cells were fixed and stained with DAPI to evidence nuclei. Merged phase‐contrast DAPI images were collected to analyse the % of myonuclei, the % of myoblasts and the number of nuclei/myotube. (b) C2C12 were treated with 1 µg/ml rmLCN2 for 24, 48, or 72 h under differentiation medium or (c) DMEM plus 10% FBS. MTT was carried out to analyse cell metabolic activity. (d) Primary CD1 mouse myoblasts were treated with 200 ng/ml rmLCN2 for 24, 48, or 72 h under differentiation medium, then cells were fixed and stained with DAPI to evidence nuclei. Merged phase‐contrast DAPI images were collected to analyse the % of myonuclei, the % of myoblasts and the number of nuclei/myotube. (e) Myoblasts were treated with 200 ng/ml rmLCN2 for 24, 48, or 72 h under differentiation medium or (f) DMEM/F10 plus 20% FBS and 2.5 ng/ml bFGF. MTT was carried out to analyse cell metabolic activity. (g) Myoblasts were treated for 4 days with 200 ng/ml rmLCN2 under differentiation medium, then RNA was extracted, reverse‐transcribed, and Myod and Myogenin expression was analysed. (h) C2C12 cells were treated as described in (a), then RNA was extracted, reverse‐transcribed and subjected to signal transduction pathway finder RT2‐array. After applying cut‐off criteria (threshold cycle [C t]<30, p‐value <.05) and validating the results with a separate set of primer, we found Bmp2 to be significantly upregulated in cells treated with rmLCN2. (a, d) Arrow and insets: myoblast‐like cells. Student's t test. *p < .05; **p < .01 versus vehicle. DMEM, Dulbecco's modified Eagle's medium; bFGF, basic fibroblast growth factor; DAPI, 4′,6‐diamidino‐2‐phenylindole; FBS, fetal bovine serum; MTT, 3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium
Since the signalling mechanism through which LCN2 acts is unclear and, to the best of our knowledge, it was never investigated in muscle cells, we performed RT2 signal transduction pathway finder array on C2C12 cells treated with 1 µg/ml rmLCN2 in fusion medium for 4 days. This assay included 86 genes related to different transduction pathways, such as tumor growth factor‐β, Wnt, nuclear factor‐κB, JAK/STAT, p53 signalling and more. We applied cut‐off criteria (threshold cycle [C t]<30, p‐value < .05) and validated the regulation of the resulting genes. This analysis eventually provided us with a single upregulated gene: Bone Morphogenic Protein 2 (Bmp2) (Figure 8g), a gene of the TFGβ superfamily that is known to prevent in vivo myogenic differentiation (Musgrave et al., 2001). Based on this finding, we assessed Bmp2 expression also in the diaphragm, quadriceps and soleus of WT and Lcn2 −/− finding no differences at all the ages evaluated (Figure S4).
4. DISCUSSION
LCN2 is an interesting multifunctional protein, and new functions emerge constantly. In this study, we investigated its role in exercise and muscle biology, finding that it is upregulated after high‐intensity exercise, and correlates with the Wnt pathway inhibitor DKK1 and the inflammatory cytokine IL6 in humans. In animal models, LCN2 is expressed in some muscles, especially diaphragm, quadriceps and soleus. Lcn2 −/− mice have normal grip force, muscle weight, muscle connective tissue and muscle fibres. However, the size of the muscle fibres was consistently smaller in Lcn2 −/− mice compared to WT at all ages evaluated, despite having normal muscle weight. Another interesting observation was that the proinflammatory cytokine IL6 was undetectable in the sera of Lcn2 −/− mice during their adult lives. A link between LCN2 and IL6 has already been described (Costa et al., 2013; Rucci et al., 2015; Sultan et al., 2012) and here we confirmed this observation, in line with the knowledge that LCN2 also plays a proinflammatory role, perhaps in partnership with IL6. Our transcriptional analyses on WT and Lcn2 −/− mice muscles at 3 and 12 months of age, evaluating both inflammatory molecules and muscle‐related genes, showed that, at 3 months of age, the absence of LCN2 increases Utrophin in the diaphragm, and Myog and Pax7 in soleus, while reducing Il1b, another well‐described pro‐inflammatory cytokine, in quadriceps. However, this is no longer true at 12 months of age, when most genes are not significantly affected. The only significant differences were observed in soleus, with a paradoxical reduction of Myog and Pax7. Soleus muscles are slow twitching, as opposed to quadriceps and diaphragm, which are mostly fast twitching in the mouse: this important difference may have a role in determining the differential gene expression observed and is worthy of further investigation, also considering that Lcn2 has a role in energy metabolism (Capulli et al., 2018; Mosialou et al., 2017; Paton et al., 2013), which is one of the key differences between fast‐ and slow‐twitch fibres (Barclay et al., 1993). Rebalka et al. (Rebalka et al., 2018) observed a low satellite cell number and activation in 5–7 months old Lcn2 −/− mice after acute muscle damage. We can then hypothesise that ageing is another factor that could impair the muscle satellite cell pool in Lcn2 −/− mice at least in a specific muscle, such as the soleus.
Treating muscle cells with rmLCN2 in vitro also led to a negative effect, with an impairment in myogenic differentiation. Consistently, rmLCN2 treatment led to the mRNA upregulation of Bmp2, as emerged from RT2‐array signal transduction pathway finder analyses (Figure 8h). This factor has been reported by multiple investigators to reduce stability and/or expression of myogenic factors, and to redirect the differentiation of mesenchymal progenitors from muscle‐lineage to osteoblast‐lineage, as it was demonstrated mainly in C2C12 and other cell models (Katagiri et al., 1994, 1997; Liu et al., 2009; Vinals & Ventura, 2004). However, the link between Lcn2 and Bmp2 is not found in the opposite condition, as investigating its expression in Lcn2 −/− muscles showed no differences versus WT.
Interestingly, we also found that DKK1, another inhibitor of the Wnt signalling reported to reduce myogenesis (Huraskin et al., 2016), positively correlated with the increased serum LCN2 levels in the vertical run participants. Consistently, we have previously found that DKK1 serum levels positively correlated with serum LCN2 levels in human healthy women, along with an indirect relationship between age and DKK1 when using LCN2 as a predictor. Altogether, our results strengthen the functional link between these two proteins. Nevertheless, similar to what we observed with Bmp2, the link between Lcn2 and Dkk1 is not found in the opposite condition, as investigating its levels in Lcn2 −/− sera showed no differences versus WT at all ages evaluated.
As a matter of fact, recent reports indicate that Lcn2 may also be involved in inflammatory myopathies, a heterogeneous class of muscle diseases characterised by muscle weakness and inflammation (Schmidt, 2018). Intriguingly, LCN2 was found to be upregulated in an experimental model of polymyositis, an inflammatory myopathy, at all timepoints analysed (Emmer et al., 2019), thus providing another link between muscle, inflammation and LCN2. However, whether this molecule plays an active role in the determination of sarcopenia remains to be clarified. With the aim to dissect this aspect, we ran hindlimb suspension experiments on WT and Lcn2 −/− mice, which allows to mimic a condition of muscle and bone loss due to reduced mechanical loading. We found that removing Lcn2 genetically significantly prevents weight loss of gastrocnemius; however, the opposite was found in soleus, where WT mice have no significant reduction of weight following HLS, while Lcn2 −/− did. Moreover, the hindlimb grip force test revealed that WT mice lost muscle strength over time versus NLC, while Lcn2 −/− HLS mice were comparable to their respective NLC. It is important to underline, however, that no statistical difference was observed upon a direct comparison between WT HLS and Lcn2 −/− HLS mice. Therefore, although the results are promising, the role of Lcn2 in muscle unloading will need to be further elucidated and confirmed with different models.
In conclusion (Figure S5), we showed that LCN2 is upregulated following acute high‐intensity exercise in humans,and positively correlates with DKK1. Furthermore, treatment with LCN2 reduces myogenic differentiation and upregulates BMP2 in C2C12 myoblasts. These results suggest that the role of LCN2 in physiological conditions in muscle is minor, but it might determine a reduction of myogenic differentiation in conditions in which its expression increases, such as following acute high‐intensity exercise.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
Research funded by AFM‐Téléthon (grant #20006) to Nadia Rucci and Agenzia Spaziale Italiana (ASI) to Nadia Rucci and AnnaTeti. Open access funding provided by Universita degli Studi dell'Aquila within the CRUI‐CARE Agreement.
Ponzetti, M. , Aielli, F. , Ucci, A. , Cappariello, A. , Lombardi, G. , Teti, A. , & Rucci, N. (2022). Lipocalin 2 increases after high‐intensity exercise in humans and influences muscle gene expression and differentiation in mice. J Cell Physiol, 237, 551–565. 10.1002/jcp.30501
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abella, V. , Scotece, M. , Conde, J. , Gómez, R. , Lois, A. , Pino, J. , Gómez‐Reino, J. J. , Lago, F. , Mobasheri, A. , & Gualillo, O. (2015). The potential of lipocalin‐2/NGAL as biomarker for inflammatory and metabolic diseases. Biomarkers, 20(8), 565–571. 10.3109/1354750X.2015.1123354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay, C. J. , Constable, J. K. , & Gibbs, C. L. (1993). Energetics of fast‐ and slow‐twitch muscles of the mouse. The Journal of Physiology, 472, 61–80. 10.1113/jphysiol.1993.sp019937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, T. , Togawa, A. , Duncan, G. S. , Elia, A. J. , You‐Ten, A. , Wakeham, A. , Fong, H. E. , Cheung, C. C. , & Mak, T. W. (2006). Lipocalin 2‐deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia‐reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America, 103(6), 1834–1839. 10.1073/pnas.0510847103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burattini, S. , Ferri, R. , Battistelli, M. , Curci, R. , Luchetti, F. , Falcieri, E. (2004). C2C12 murine myoblasts as a model of skeletal muscle development: Morpho‐functional characterization, European Journal of Histochemistry, 48(3), 223–233, 10.4081/891 [DOI] [PubMed] [Google Scholar]
- Capulli, M. , Ponzetti, M. , Maurizi, A. , Gemini‐Piperni, S. , Berger, T. , Mak, T. W. , Teti, A. , & Rucci, N. (2018). A Complex role for Lipocalin 2 in bone metabolism: Global ablation in mice induces osteopenia caused by an altered energy metabolism. Journal of Bone and Mineral Research, 33(6), 1141–1153. 10.1002/jbmr.3406 [DOI] [PubMed] [Google Scholar]
- Capulli, M. , Rufo, A. , Teti, A. , & Rucci, N. (2009). Global transcriptome analysis in mouse calvarial osteoblasts highlights sets of genes regulated by modeled microgravity and identifies A “mechanoresponsive osteoblast gene signature. Journal of Cellular Biochemistry, 107(2), 240–252. 10.1002/jcb.22120 [DOI] [PubMed] [Google Scholar]
- Costa, D. , Lazzarini, E. , Canciani, B. , Giuliani, A. , Spanò, R. , Marozzi, K. , Manescu, A. , Cancedda, R. , & Tavella, S. (2013). Altered bone development and turnover in transgenic mice over‐expressing Lipocalin‐2 in bone. Journal of Cellular Physiology, 228(11), 2210–2221. 10.1002/jcp.24391 [DOI] [PubMed] [Google Scholar]
- Ding, L. , Hanawa, H. , Ota, Y. , Hasegawa, G. , Hao, K. , Asami, F. , Watanabe, R. , Yoshida, T. , Toba, K. , Yoshida, K. , Ogura, M. , Kodama, M. , & Aizawa, Y. (2010). Lipocalin‐2/neutrophil gelatinase‐B associated lipocalin is strongly induced in hearts of rats with autoimmune myocarditis and in human myocarditis. Circulation Journal, 74(3), 523–530. 10.1253/circj.CJ-09-0485 [DOI] [PubMed] [Google Scholar]
- Dubach‐Powell, J. (2011). Quantitative determination of muscle fiber diameter (minimal Feret's diameter) and percentage of centralized nuclei. Treat‐NMD, 1–16. [Google Scholar]
- Emmer, A. , Abobarin‐Adeagbo, A. , Posa, A. , Jordan, B. , Delank, K. S. , Staege, M. S. , Surov, A. , Zierz, S. , & Kornhuber, M. E. (2019). Myositis in Lewis rats induced by the superantigen Staphylococcal enterotoxin A. Molecular Biology Reports, 46(4), 4085–4094. 10.1007/s11033-019-04858-9 [DOI] [PubMed] [Google Scholar]
- Fairclough, R. J. , Wood, M. J. , & Davies, K. E. (2013). Therapy for Duchenne muscular dystrophy: Renewed optimism from genetic approaches. Nature Reviews Genetics, 14(6), 373–378. 10.1038/nrg3460 [DOI] [PubMed] [Google Scholar]
- Gambara, G. , Salanova, M. , Ciciliot, S. , Furlan, S. , Gutsmann, M. , Schiffl, G. , Ungethuem, U. , Volpe, P. , Gunga, H. C. , & Blottner, D. (2017). Microgravity‐induced transcriptome adaptation in mouse paraspinal longissimus dorsi muscle highlights insulin resistance‐linked genes. Frontiers in Physiology, 5(8), 279. 10.3389/fphys.2017.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds, M. (2012). Quantification of histopathology in haemotoxylin and eosin stained muscle sections. TREAT‐NMD Neuromuscular Network, Id), 1–14. Retrieved from http://www.treat-nmd.eu/resources/research-resources/dmd-sops/ [Google Scholar]
- Gutpel, K. M. , Hrinivich, W. T. , & Hoffman, L. M. (2015). Skeletal muscle fibrosis in the mdx/utrn+/‐mouse validates its suitability as a murine model of Duchenne muscular dystrophy. PLOS One, 10(1), 10.1371/journal.pone.0117306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huraskin, D. , Eiber, N. , Reichel, M. , Zidek, L. M. , Kravic, B. , Bernkopf, D. , von Maltzahn, J. , Behrens, J. , & Hashemolhosseini, S. (2016). Wnt/β‐catenin signaling via Axin2 is required for myogenesis and, together with YAP/Taz and tead1, active in IIa/IIx muscle fibers. Development (Cambridge), 143(17), 3128–3142. 10.1242/dev.139907 [DOI] [PubMed] [Google Scholar]
- Katagiri, T. , Akiyama, S. , Namiki, M. , Komaki, M. , Yamaguchi, A. , Rosen, V. , Wozney, J. M. , Fujisawa‐Sehara, A. , & Suda, T. (1997). Bone morphogenetic protein‐2 inhibits terminal differentiation of myogenic cells by suppressing the transcriptional activity of MyoD and myogenin. Experimental Cell Research, 230(2), 342–351. 10.1006/excr.1996.3432 [DOI] [PubMed] [Google Scholar]
- Katagiri, T. , Yamaguchi, A. , Komaki, M. , Abe, E. , Takahashi, N. , Ikeda, T. , Rosen, V. , Wozney, J. M. , Fujisawa‐Sehara, A. , Suda, T. 1994). Bone morphogenetic protein‐2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. Journal of Cell Biology, 127(6 I), 1755–1766. 10.1083/jcb.127.6.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keire, P. , Shearer, A. , Shefer, G. , & Yablonka‐Reuveni, Z. (2013). Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods in Molecular Biology, 946, 431–468. 10.1007/978-1-62703-128-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , Ginn, S. L. , Lek, M. , North, K. N. , Alexander, I. E. , Little, D. G. , & Schindeler, A. (2009). Myoblast sensitivity and fibroblast insensitivity to osteogenic conversion by BMP‐2 correlates with the expression of Bmpr‐1a. BMC Musculoskeletal Disorders, 10, 51. 10.1186/1471-2474-10-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lögdberg, L. , & Wester, L. (2000). Immunocalins: A lipocalin subfamily that modulates immune and inflammatory responses. Biochimica et Biophysica Acta ‐ Protein Structure and Molecular Enzymology, 1482(1–2), 284–297. 10.1016/S0167-4838(00)00164-3 [DOI] [PubMed] [Google Scholar]
- Marques, F. Z. , Prestes, P. R. , Byars, S. G. , Ritchie, S. C. , Würtz, P. , Patel, S. K. , Booth, S. A. , Rana, I. , Minoda, Y. , Berzins, S. P. , Curl, C. L. , Bell, J. R. , Wai, B. , Srivastava, P. M. , Kangas, A. J. , Soininen, P. , Ruohonen, S. , Kähönen, M. , Lehtimäki, T. , … Charchar, F. J. (2017). Experimental and euman evidence for lipocalin‐2 (neutrophil gelatinase‐associated lipocalin [NGAL]) in the development of cardiac hypertrophy and heart failure. Journal of the American Heart Association, 6(6), 10.1161/JAHA.117.005971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, J. , Ma, Q. , Kelly, C. , Mitsnefes, M. , Mori, K. , Barasch, J. , & Devarajan, P. (2006). Kidney NGAL is a novel early marker of acute injury following transplantation. Pediatric Nephrology, 21(6), 856–863. 10.1007/s00467-006-0055-0 [DOI] [PubMed] [Google Scholar]
- Mishra, J. , Ma, Q. , Prada, A. , Mitsnefes, M. , Zahedi, K. , Yang, J. , Barasch, J. , & Devarajan, P. (2003). Identification of neutrophil gelatinase‐associated lipocalin as a novel early urinary biomarker for ischemic renal injury. Journal of the American Society of Nephrology, 14(10), 2534–2543. 10.1097/01.ASN.0000088027.54400.C6 [DOI] [PubMed] [Google Scholar]
- Mosialou, I. , Shikhel, S. , Liu, J. M. , Maurizi, A. , Luo, N. , He, Z. , Huang, Y. , Zong, H. , Friedman, R. A. , Barasch, J. , Lanzano, P. , Deng, L. , Leibel, R. L. , Rubin, M. , Nickolas, T. , Chung, W. , Zeltser, L. M. , Williams, K. W. , Pessin, J. E. , & Kousteni, S. (2017). MC4R‐dependent suppression of appetite by bone‐derived lipocalin 2. Nature, 543(7645), 385–390. 10.1038/nature21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrave, D. S. , Pruchnic, R. , Wright, V. , Bosch, P. , Ghivizzani, S. C. , Robbins, P. D. , & Huard, J. (2001). The effect of bone morphogenetic protein‐2 expression on the early fate of skeletal muscle‐derived cells. Bone, 28(5), 499–506. 10.1016/S8756-3282(01)00413-6 [DOI] [PubMed] [Google Scholar]
- Paton, C. M. , Rogowski, M. P. , Kozimor, A. L. , Stevenson, J. L. , Chang, H. , & Cooper, J. A. (2013). Lipocalin‐2 increases fat oxidation in vitro and is correlated with energy expenditure in normal weight but not obese women. Obesity, 21(12), 640–648. 10.1002/oby.20507 [DOI] [PubMed] [Google Scholar]
- Rebalka, I. A. , Monaco, C. , Varah, N. E. , Berger, T. , D'souza, D. M. , Zhou, S. , Mak, T. W. , & Hawke, T. J. (2018). Loss of the adipokine lipocalin‐2 impairs satellite cell activation and skeletal muscle regeneration. American Journal of Physiology‐Cell Physiology, 315(5), C714–C721. 10.1152/ajpcell.00195.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucci, N. , Capulli, M. , Piperni, S. G. , Cappariello, A. , Lau, P. , Frings‐Meuthen, P. , Heer, M. , & Teti, A. (2015). Lipocalin 2: A new mechanoresponding gene regulating bone homeostasis. Journal of Bone and Mineral Research, 30(2), 357–368. 10.1002/jbmr.2341 [DOI] [PubMed] [Google Scholar]
- Schmidt, J. (2018). Current classification and management of inflammatory myopathies. Journal of Neuromuscular Diseases, 5(2), 109–129. 10.3233/JND-180308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Morales, L. , Malik, A. S. , Mead, A. F. , Greer, C. D. , Mitchell, M. A. , Petrov, M. T. , Su, L. T. , Choi, M. E. , Rosenblum, S. T. , Lu, X. , VanBelzen, D. J. , Krishnankutty, R. K. , Balzer, F. J. , Loro, E. , French, R. , Propert, K. J. , Zhou, S. , Kozyak, B. W. , … Stedman, H. H. (2019). Non‐immunogenic utrophin gene therapy for the treatment of muscular dystrophy animal models. Nature Medicine, 25, 1505–1511. 10.1038/s41591-019-0594-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, M. L. , Rando, T. A. , & Blau, H. M. (2001). Gene delivery to muscle. Current Protocols in Human Genetics, 31, 13. 10.1002/0471142905.hg1304s31 [DOI] [PubMed] [Google Scholar]
- Sultan, S. , Pascucci, M. , Ahmad, S. , Malik, I. A. , Bianchi, A. , Ramadori, P. , Ahmad, G. , & Ramadori, G. (2012). LIPOCALIN‐2 is a major acute‐phase protein in a rat and mouse model of sterile abscess. Shock, 37(2), 191–196. 10.1097/SHK.0b013e31823918c2 [DOI] [PubMed] [Google Scholar]
- Sung, H. K. , Chan, Y. K. , Han, M. , Jahng, J. , Song, E. , Danielson, E. , Berger, T. , Mak, T. W. , & Sweeney, G. (2017). Lipocalin‐2 (NGAL) attenuates autophagy to exacerbate cardiac apoptosis induced by myocardial ischemia. Journal of Cellular Physiology, 232(8), 2125–2134. 10.1002/jcp.25672 [DOI] [PubMed] [Google Scholar]
- Tinsley, J. , Deconinck, N. , Fisher, R. , Kahn, D. , Phelps, S. , Gillis, J. M. , & Davies, K. (1998). Expression of full‐length utrophin prevents muscular dystrophy in mdx mice. Nature Medicine, 4(12), 1441–1444. 10.1038/4033 [DOI] [PubMed] [Google Scholar]
- Tinsley, J. M. , Potter, A. C. , Phelps, S. R. , Fisher, R. , Trickett, J. I. , & Davies, K. E. (1996). Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature, 384(6607), 349–353. 10.1038/384349a0 [DOI] [PubMed] [Google Scholar]
- Veeriah, V. , Zanniti, A. , Paone, R. , Chatterjee, S. , Rucci, N. , Teti, A. , & Capulli, M. (2016). Interleukin‐1β, lipocalin 2 and nitric oxide synthase 2 are mechano‐responsive mediators of mouse and human endothelial cell‐osteoblast crosstalk. Scientific Reports, 19(6), 29880. 10.1038/srep29880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinals, F. , & Ventura, F. (2004). Myogenin protein stability is decreased by BMP‐2 through a mechanism implicating Id1. Journal of Biological Chemistry, 279(44), 45766–45772. 10.1074/jbc.M408059200 [DOI] [PubMed] [Google Scholar]
- Xu, G. , Ahn, J. , Chang, S. , Eguchi, M. , Ogier, A. , Han, S. , Park, Y. , Shim, C. , Jang, Y. , Yang, B. , Xu, A. , Wang, Y. , & Sweeney, G. (2012). Lipocalin‐2 induces cardiomyocyte apoptosis by increasing intracellular iron accumulation. Journal of Biological Chemistry, 287(7), 4808–4817. 10.1074/jbc.M111.275719 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


