Abstract
Background and Purpose
The transient receptor potential vanilloid 4 (TRPV4) cation channel participates in multiple physiological processes and is also at the core of different diseases, making this channel an interesting pharmacological target with therapeutic potential. However, little is known about the structural elements governing its inhibition.
Experimental Approach
We have now combined in silico drug discovery and molecular dynamics simulation based on Xenopus tropicalis xTRPV4 structure with functional studies measuring cell Ca2+ influx mediated by human TRPV4 channel to characterize the binding site of known TRPV4 inhibitors and to identify novel small molecule channel modulators.
Key Results
We have found that the inhibitor HC067047 binds to a pocket conformed by residues from S2–S3 linker (xTRPV4‐D542), S4 (xTRPV4‐M583 and Y587 and S5 (xTRPV4‐D609 and F613). This pocket was also used for structure‐based virtual screening in the search of novel channel modulators. Forty potential hits were selected based on the lower docking scores (from ~250,000 compounds) and their effect upon TRPV4 functionally tested. Three were further analysed for stability using molecular dynamics simulation and functionally tested on TRPV4 channels carrying mutations in the binding pocket. Compound NSC151066, shown to require residue xTRPV4‐M583 for its inhibitory effect, presented an IC50 of 145 nM and demonstrated to be an effective antiviral against Zika virus with a potency similar to HC067047.
Conclusion and Implications
Together, we propose structural insights into the inhibition of TRPV4 and how this information can be used for the design of novel channel modulators.
LINKED ARTICLES
This article is part of a themed issue on Structure Guided Pharmacology of Membrane Proteins (BJP 75th Anniversary). To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v179.14/issuetoc
Keywords: antiviral, drug discovery, HC067047, in silico, inhibition, molecular dynamics, RN1734, structure, TRPV4
Abbreviations
- 4α‐PDD
4α‐phorbol 12,13‐didecanoate
- EETs
epoxyeicosatrienoic acids
- MD
molecular dynamics
- MM‐GBSA
Molecular Mechanics‐Generalized Born Surface Area
- PEI
polyethylenimine
- SI
selectivity index
- TRPV4
transient receptor potential vanilloid 4
What is already known
The TRPV4 channel is an interesting pharmacological target with therapeutic potential.
What this study adds
The structural basis for TRPV4 interaction with known inhibitors
The identification of novel inhibitors based on this binding site.
What is the clinical significance
The effectiveness of novel TRPV4 inhibitors on the infection caused by Zika virus n vitro.
1. INTRODUCTION
The transient receptor potential vanilloid 4 (TRPV4) is a widely expressed nonselective cation channel that is activated by physical stimuli such as hypotonicity (Arniges et al., 2004; Liedtke et al., 2000; Strotmann, Harteneck, Nunnenmacher, Schultz, & Plant, 2000), mechanical forces (Andrade et al., 2005), moderate heat (Garcia‐Elias et al., 2013; Güler et al., 2002; Watanabe, Davis, et al., 2002) or UVB radiation (Moore et al., 2013). Different ligands also modulate TRPV4 activity, including epoxyeicosatrienoic acids (EETs) (Berna‐Erro et al., 2017; Watanabe et al., 2003), bisandrographolide (Smith, Maloney, Pothen, Clardy, & Clapham, 2006), flavonoids (Shao et al., 2019; Zhang et al., 2019) and synthetic agonists such as 4α‐phorbol 12,13‐didecanoate (4α‐PDD) (Watanabe, Vriens, et al., 2002) or GSK1016790A (Thorneloe et al., 2008). TRPV4 participates in multiple physiological processes, including cell and systemic volume homeostasis (Arniges et al., 2004; Fernandez‐Fernandez et al., 2008; Liedtke & Friedman, 2003; Tian et al., 2009), endothelial function and angiogenesis (Vriens et al., 2005), epithelial hydroelectrolyte transport (Takayama, Shibasaki, Suzuki, Yamanaka, & Tominaga, 2014), nociception (Alessandri‐Haber, Dina, Joseph, Reichling, & Levine, 2006), bladder voiding (Gevaert et al., 2007), ciliary beat frequency regulation (Andrade et al., 2005; Jung et al., 2018; Lorenzo, Liedtke, Sanderson, & Valverde, 2008), innate immunity (Alpizar et al., 2017; Galindo‐Villegas et al., 2016), matrix stiffness (Rahaman et al., 2014) and osteoarticular maintenance (Muramatsu et al., 2007; O'Conor, Leddy, Benefield, Liedtke, & Guilak, 2014). TRPV4 is also involved in pathophysiological conditions ranging from genetic forms of skeletal and peripheral nervous diseases (Nilius & Voets, 2013) to viral infection (Doñate‐Macián et al., 2018) and airways disease (Zhu et al., 2009).
Considering the role of TRPV4 in different pathological conditions, it is not surprising the interest of researchers and pharmaceutical industry to find appropriate channel modulators. The first specific TRPV4 antagonists were found more than a decade ago, HC067047 (Everaerts et al., 2010) and RN1734 (Vincent et al., 2009), but exhibited limitations such as pharmacokinetics profile and toxicity that prevented their progression to the clinic. Recently, new channel modulators have been identified in the publication as compound 36, a quinazolin derivative and potent agonist (EC50 60 nM) with high aqueous solubility (Atobe et al., 2019), GSK2798745, a potent (IC50 16 nM) and orally active antagonist (Cheung et al., 2017) already in phase I clinical trials for the treatment of stable heart failure (Goyal et al., 2019) along with GSK3491943 and GSK3527497, which effectively blocked TRPV4‐induced pulmonary oedema and showed a suitable in vivo pharmacodynamics and toxicity (Brnardic et al., 2018; Cheung et al., 2017). However, despite the current availability of potent and specific novel TRPV4 antagonists, the mechanisms underlying their modulation of channel activity are not completely understood.
In the present study, we have combined in silico drug discovery and molecular dynamics (MD) with functional analysis to identify the HC067047 binding pocket within the structure of TRPV4. Moreover, we used the inhibitor docking site to screen and test novel small molecules with inhibitory activity on TRPV4.
2. METHODS
2.1. Cells and transfections
HeLa cells (ECACC93021013, CVCL_0030) were transiently transfected with human TRPV4 wild‐type (WT)/mutant cDNAs cloned in the pcDNA3.1 vector as previously described (Doñate‐Macián et al., 2018; Fernandes et al., 2008). In brief, HeLa cells were cultured in DMEM with 10% FBS at 37°C and 5% CO2. Cells were seeded onto poly‐l‐lysine‐coated coverslips in 24‐well plates and transiently transfected with human TRPV4‐WT/mutant constructs using polyethylenimine (PEI) reagent (ExGen500, Fermentas MBI). For each coverslip, 1 μg of the TRPV4 constructs and 4.8 μl of PEI (7 equivalents) were added. The human hepatocarcinoma cell line Huh7/Scr (Src transformed CVCL_E049, kindly provided by F. Chisari, The Scripps Research Institute, La Jolla, CA) was used for viral infection experiments and cultured as previously described (Doñate‐Macián et al., 2018).
2.2. Materials and plasmid generation
All reagents were obtained from Sigma, unless otherwise indicated. Fura‐2 AM was obtained from Invitrogen (UK). GSK1016790A, HC067047 and RN1734 were obtained from Tocris Bioscience (UK). Compounds used for the initial screening were purchased from National Cancer Institute Developmental Therapeutic Program (NCI‐DTP). TRPV4 mutants were generated from pCDNA3‐TRPV4‐WT plasmid (Doñate‐Macián et al., 2018; Fernandes et al., 2008). Mutations were introduced by site‐directed mutagenesis using the QuikChange kit (Stratagene). The primers used for mutagenesis were as follows: TRPV4‐D546A forward ATTCTCTCTTCATTGCTGGCTCCTTCCAG; TRPV4‐D546A reverse CTGGAAGGAGCCAGCAATGAAGAGAGAAT; TRPV4‐Y591A forward CTGGATGAATGCCCTTGCCTTCACCCGTGGGC; TRPV4‐Y591A reverse GCCCACGGGTGAAGGCAAGGGCATTCATCCAG; TRPV4‐Y591H forward GGATGAATGCCCTTCACTTCACCCGTGGG; TRPV4‐Y591H reverse CCCACGGGTGAAGTGAAGGGCATTCATCC; TRPV4‐M587A forward GTCCTGGGCTGGGCGAATGCCCTTTACTT; and TRPV4‐M587A reverse AAGTAAAGGGCATTCGCCCAGCCCAGGAC.
2.3. Binding site identification and druggability assessment of the HC067047
The SiteMap module in Schrödinger (Schrodinger, 2020) (SCR_016748) was used to assess the druggability of the HC067047 binding site based on the Xenopus TRPV4 PDB: 6BBJ (Deng et al., 2018) and on the rat TRPV1 PDB: 3J5R (Cao, Liao, Cheng, & Julius, 2013) electron microscopy structures. The preprocessed protein was submitted to SiteMap using default parameters to calculate properties such as the exposure, the volume of the pocket and the degree of hydrophobicity, generating contour maps (“site maps”), which were scored using Site Score values. For each site, receptor grids for docking were generated using cubic grids with a size of 20 Å. The grid centre was set at the centroid of each site map. Molecules were docked using Glide program (Friesner et al., 2006) in Schrödinger suite 2019‐2 using default settings (Schrodinger, 2020). The OPLS3 force field was used for the docking protocol. The ligand was drawn in 2D sketcher and prepared with the LigPrep suite (SCR_016746). The ligand was docked using standard precision (SP) mode by enabling flexible ligand sampling for docking procedures. The 10 best conformations for each ligand were saved. The default scoring function Glide Docking Score was calculated and used for sorting the poses. The conformations with lower docking scores were selected and this binding site was selected as the druggable site. The molecular docking was replicated to other chains and the MD simulations were accomplished. To perform all processes described before, a workflow from KNIME software (Schrodinger, 2020) was used. Mutations were performed through the Schrodinger maestro graphical interface.
2.4. Molecular dynamics (MD) simulations
We performed four independent MD simulations using the Desmond program (Bowers et al., 2006) in Schrödinger suite 2019‐2. Each protein–ligand complex was inserted in a POPC membrane of 140 × 140 Å (approximate) and solvated using explicit TIP3P water models and an orthorhombic box with periodic boundary conditions. All complexes were neutralized with 0.15 mol·L−1 of NaCl and parametrized with OPLSe force field. Each simulation was performed for a total of 500 ns with a recording interval of 100 ps. NPT ensemble at standard conditions of T = 310.15 K and P = 1 atm was used. Ligand interaction diagrams were performed using the “simulation interaction diagram program” module of Schrodinger maestro.
2.5. Free energy calculations
Molecular Mechanics‐Generalized Born Surface Area (MM‐GBSA) was used to estimate the binding free energy of the TRPV4 protein–ligand, for the WT protein and its mutants. For MM‐GBSA calculation, the final 100 ns were extracted from a total of 500‐ns production MD trajectories. The surface area, MM/GBSA approach, was used as implemented in the Prime module from Schrödinger suite 2019‐2 using the default settings (Greenidge, Kramer, Mozziconacci, & Wolf, 2013). The MM/GBSA analysis was performed on three subsets of each system: the protein alone, the ligand alone, and the complex (protein–ligand). The total free energy was calculated, including all the molecular mechanics contributions (bond, angle and dihedral energies, electrostatic and van der Waals energies).
2.6. Ratiometric Ca2+ concentration measurements and solutions
Cytosolic [Ca2+] signals were obtained from transfected HeLa cells loaded with 4.5‐μM fura‐2 AM (Invitrogen) as previously described (Arniges et al., 2004; Fernandes et al., 2008) and represented normalized relative to the fluorescence ratio (340/380) measured prior to cell stimulation or inhibitor addition. Cells were incubated in isotonic solutions containing fura‐2 AM for 30 min at room temperature. The cells were then washed thoroughly with isotonic solution. Measurements of intracellular Ca2+ concentrations were obtained by using an Olympus IX70 inverted microscope (Olympus, Hamburg, Germany). The excitation light (340 and 380 nm) was supplied by a Polychrome IV monochromator (Till Photonics, Martinsried, Germany) and directed towards the cells under study by a 505DR dichromatic mirror (Omega Optical, Brattleboro, VT). Fluorescence images were collected by a digital charge‐coupled device camera (Hamamatsu Photonics) after being passed through a 535DF emission filter (Omega Optical) by using AquaCosmos software (Hamamatsu Photonics). Isotonic bath solutions used for Ca2+ imaging experiments contained 140‐mM NaCl, 2.5‐mM KCl, 1.2‐mM CaCl2, 0.5‐mM MgCl2, 5‐mM glucose and 10‐mM HEPES (pH 7.3, adjusted with Tris). Hypotonic solution (30%) was obtained by reducing NaCl content and adjusting osmolarity to 220 mOsm·L−1. Experiments were double‐blinded and carried at room temperature (~24°C).
2.7. Data and statistical analysis
Data are expressed as mean ± SEM (unless otherwise indicated). Statistical analysis using SigmaPlot 12.5 software was assessed with Student's unpaired t‐test, Mann–Whitney U test, one‐way ANOVA followed by Bonferroni post hoc test, or Kruskal–Wallis test followed by Dunn post hoc test, as appropriate. Differences were considered significant if P value < 0.05 and depicted as *. The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology.
2.8. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY http://www.guidetopharmacology.org and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander et al., 2019).
3. RESULTS
We used computational modelling methods such as molecular docking and MD simulations for an atomistic insight into the interactions between the TRPV4 cation channel and its specific inhibitor HC067047, which was used as a prototypical compound to find the structural features of the binding pocket. The binding mode of the inhibitor was assessed in the WT channel complex and in the mutants generated. For the in silico analysis, we used the recently reported structure of Xenopus tropicalis TRPV4 (xTRPV4) (Deng et al., 2018), which shares 78% sequence identity with the human protein, whereas all functional analyses were run on the human protein (hTRPV4).
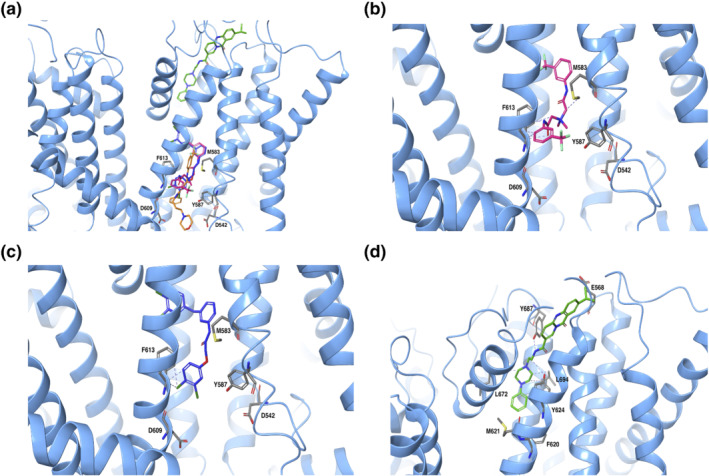
Docking poses clustered in four cavities in TRPV4, representing a unique binding site defined in the TRPV4 fourfold symmetry (Figure 1a,b). The structural information of the xTRPV4‐HC067047 complex over an MD trajectory of 500 ns was investigated to characterize crucial intermolecular interactions in the binding site. We used MM‐GBSA to estimate the binding energy of HC067047 to the TRPV4‐WT complex obtained from the last 100 ns of MD simulation of a total of 500 ns, which was −75.1 ± 7.4 kcal·mol−1 (mean ± SD; n = 4). Based on the most frequent interactions, that is, residues showing more than 50% of time occupancy (displayed in the magnified view in Figure 1a), we identified a HC067047 binding site in the xTRPV4‐WT system conformed by residues from S2–S3 linker (D542, corresponding to hTRPV4‐D546), S4 (M583 and Y587, corresponding to hTRPV4‐M587 and Y591, respectively) and S5 (D609 and F613, corresponding to hTRPV4‐D613 and F617) (Figure 1c). This binding pocket shows partial overlapping with the binding site proposed for 5′,6′‐EET (Berna‐Erro et al., 2017) and, less pronouncedly, with the 4α‐PDD channel agonist (Vriens, Owsianik, Janssens, Voets, & Nilius, 2007), which suggests that the site is a hotspot of channel regulation.
FIGURE 1.
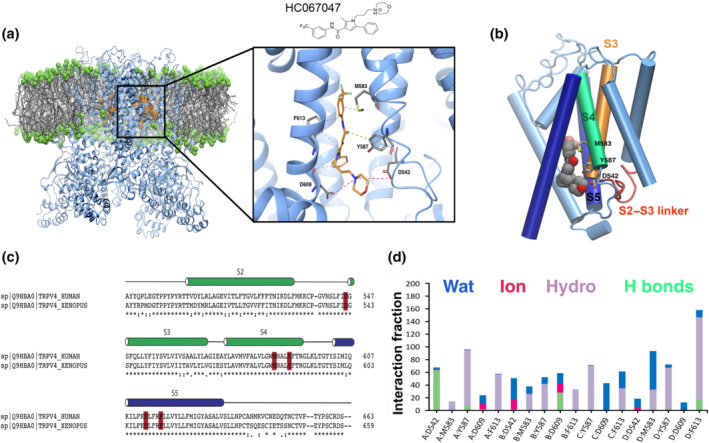
Predicted model of HC067047 interaction with TRPV4. (a) Location of HC067047 is shown on the tertiary structure of the Xenopus tropicalis TRPV4. The image corresponds the last conformation of HC067047 on the binding pocket after 500‐ns MD; right, a zoom‐in view of the HC067047 binding pocket. Residues important for interaction with HC067047 are coloured in grey. Hydrophobic interactions, H bonds and ionic interactions are shown as dashed lines in purple, green and fuchsia, respectively. (b) Interactions between HC067047 and residues D542, M583 and Y587 of a single subunit in the xTRPV4‐WT. (c) Sequence alignment of S2 to S5 from X. tropicalis and human TRPV4. Key residues within the HC067047 binding site marked in red. *Identical residues; conserved substitutions (same amino acid group); semi‐conserved substitution (similar shapes). (d) Interaction fraction plots depicting protein–HC067047 contacts during the course of 500‐ns MD simulation. Hydrophobic interactions, H bonds, ionic interactions and water bridges are shown in purple, green, fuchsia and blue, respectively. The stacked bar charts are normalized over the course of the trajectory, for example, a value of 0.7 suggests that 70% of the simulation time the specific interaction is maintained. Values over 1.0 are possible as protein residues may make multiple contacts of same subtype with the ligand
Analysis of the main contacts that HC067047 generated over the molecular simulation trajectory of 500 ns (Figure 1d) revealed that residues D542 and D609 engaged mainly in hydrogen bond interactions, direct or water‐mediated, as well as ionic interactions with the protonated amine group and oxygen of morpholine ring in HC067049, with an occupancy of over 50% during the time of the trajectory. The Y587, M583 and F613 also played an important role in the protein–ligand interactions, through hydrophobic contacts. Next, we computed the behaviour of xTRPV4 complexes in which two residues that showed high occupancy and stabilize HC067047 interaction with the TRPV4 were changed, residues D542 and Y587 (Figure 2). We generated TRPV4 complexes substituting these residues to alanine, D542A (Figure 2a,d) and Y587A (Figure 2b,e), or changing the charge of the side chain, Y587H (Figure 2c,f). MD of the xTRPV4‐D542A complex showed, first, an increase in the HC067047 binding energy (−69.7 ± 4 kcal·mol−1, n = 4) and, second, the loss of the interaction between A542 and HC067047, although the interaction of HC067047 with Y587 was strengthened through water‐mediated hydrogen bonds and hydrophobic interactions (Figure 2a,d). On the other hand, the Y587A mutant complex preserved the overall interactions presented in the WT complex, although the hydrophobic interaction with the amino acid A587 was lost (Figure 2b,e). However, the cation–π or hydrogen bond interactions between the protonated amine of HC067047 morpholine ring and D609 were strengthened with an occupancy of over 50% in the system. These small differences in protein–ligand interactions in the xTRPV4‐Y587A complex did not alter the HC067047 binding energy of the xTRPV4‐Y587A complex (−73.8 ± 11 kcal·mol−1, n = 4). We also generated a xTRPV4‐Y587H complex, which provided relevant data reinforcing the relevance of Y587 in the interaction with HC067047. We observed that in the xTRPV4‐Y587H complex, the interaction with the residue D609 is strongly affected by the mutation, although the hydrophobic and hydrogen bond interactions with residue H587 are evident yet, the occupancy is less than 100% compared to the complexes without mutation in this residue. These changes and the decrease in the time of occupancy of few interactions caused a loss of affinity of the ligand, so its stability at the binding site is affected and, consequently, the binding energy is increased (−68.8 ± 4 kcal·mol−1, n = 4).
FIGURE 2.
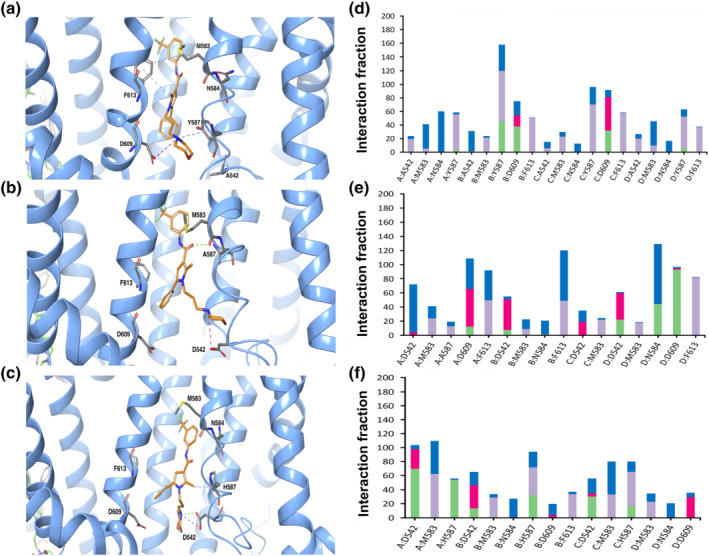
Predicted model of HC067047 interaction with mutant xTRPV4 systems. (a–c) Images corresponding to the last conformation of HC067047 on the binding pocket after 500‐ns MD in the xTRPV4‐D542A (a), xTRPV4‐Y587A (b) and xTRPV4‐Y587H (c) systems. (d–f) Interaction fraction plots depicting protein–HC067047 contacts during the course of 500‐ns MD simulation in the xTRPV4‐D542A (d), xTRPV4‐Y587A (e) and xTRPV4‐Y587H systems (f). Hydrophobic interactions, H bonds, ionic interactions and water bridges are shown in purple, green, fuchsia and blue, respectively. The stacked bar charts are normalized over the course of the trajectory: for example, a value of 0.7 suggests that 70% of the simulation time the specific interaction is maintained. Values over 1.0 are possible as some protein residue may make multiple contacts of same subtype with the ligand
The HC067047 binding site identified in silico was tested on human TRPV4 expressed in HeLa cells. The channel activity was evaluated by imaging the entry of calcium into hTRPV4‐transfected and fura‐2‐loaded HeLa cells exposed to the TRPV4 agonist GSK1016790A (Thorneloe et al., 2008). First, we showed that TRPV4‐WT was activated by GSK1016790A and inhibited by HC067047 in a dose‐dependent manner (Figure 3a). TRPV4‐D546A, like TRPV4‐WT, was activated by GSK1016790A but showed reduced inhibition by submaximal concentrations of HC067047 (Figure 3b). Analysis of TRPV4‐WT inhibition by HC067047 reported an IC50 of 2.7 ± 1 nM whereas the IC50 for TRPV4‐D546A was 499 ± 36 nM (Figure 3c). Channel inhibition by HC067047 was also tested in response to cell swelling in the presence of 30% hypotonic solutions. TRPV4‐WT channels showed a pronounced inhibition by 10‐nM HC067047 (Figure 3d) whereas no inhibition was detected in HeLa cells transfected with TRPV4‐D546A (Figure 3e).
FIGURE 3.
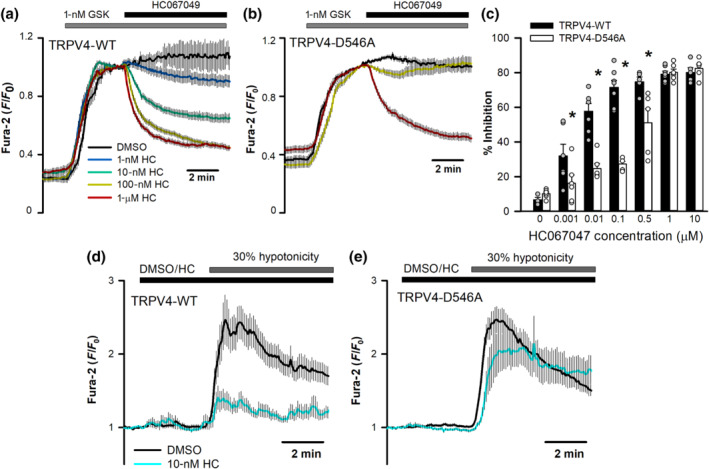
hTRPV4‐D546A mutation affects HC067047 inhibition. (a, b) Changes in intracellular [Ca2+] (indicated by normalized fura‐2 ratios) in HeLa cells transfected with hTRPV4‐WT (a) or hTRPV4‐D546A (b) cDNAs, after perfusion with 1‐nM GSK1016790A and addition of HC067047 (or vehicle) at the indicated concentrations at time t = 0. Traces are means ± SEM of 50 (DMSO), 71 (1 nM), 91 (10 nM), 95 (100 nM), 98 (500 nM), 94 (1 μM) and 63 (10 μM) cells expressing TRPV4‐WT and 59 (DMSO), 66 (1 nM), 93 (10 nM), 96 (100 nM), 44 (500 nM), 82 (1 μM) and 47 (10 μM) cells expressing TRPV4‐D546A. (c) Percentage inhibition of GSK1016790A‐induced increases in intracellular Ca2+ at different HC067047 concentrations. Mean ± SEM of 5 (DMSO), 6 (1 nM), 7 (10 nM), 7 (100 nM), 5 (500 nM), 7 (1 μM) and 5 (10 μM) independent experiments with TRPV4 and 6 (DMSO), 6 (1 nM), 6 (10 nM), 5 (100 nM), 5 (500 nM), 6 (1 μM) and 5 (10 μM) independent experiments with TRPV4‐D546A. (d, e) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐WT (d) or hTRPV4‐D546A (e) cDNAs, after perfusion with a 30% hypotonic solution in the presence (WT, n = 60; D546A, n = 75) or absence (WT, n = 69; D546A, n = 78) of 10‐nM HC067047
Next, we tested the relevance of residue Y591 on the binding of HC067047. This residue has been previously shown to participate in channel gating (Vriens et al., 2007). Thus, we first tested the activity of Y591 mutants. Neither hTRPV4‐Y591A nor hTRPV4‐Y591H responded to GSK1016790A (Figure 4a) but responded to 30% hypotonicity (Figure 4b,c). Using the hypotonicity‐induced activation of hTRPV4, we observed that hTRPV4‐Y591H, like hTRPV4‐D545A, was not inhibited by 10‐nM HC067047 (Figure 4b,d), whereas hTRPV4‐Y591A was fairly inhibited (Figure 4c,d). These results are in agreement with the in silico observations pointing to an altered binding of HC067047 to the channel carrying xTRPV4‐D542A and xTRPV4‐Y587H mutations, but normal TRPV4–HC067047 interaction in the case of the xTRPV4‐Y587A mutation.
FIGURE 4.
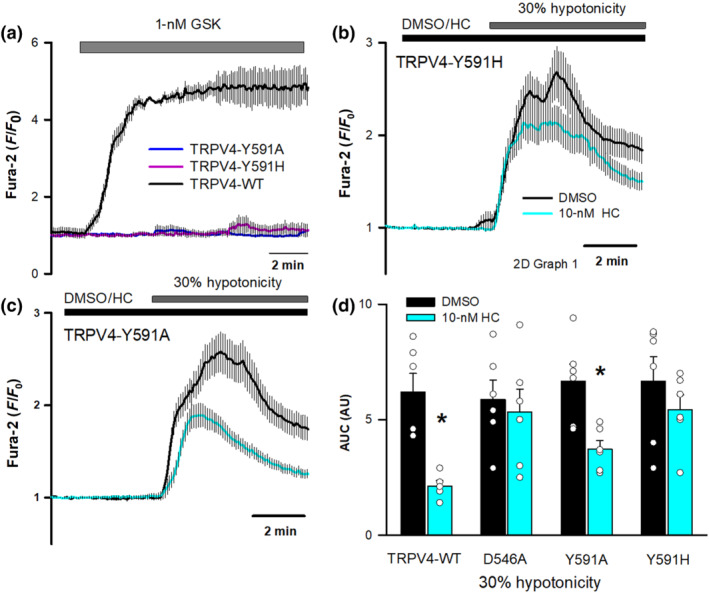
Effect of hTRPV4‐Y591 mutations on HC067047 inhibition. (a) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐WT (n = 30), hTRPV4‐Y591A (n = 31), or hTRPV4‐Y591H (n = 34) cDNAs after perfusion with 1‐nM GSK1016790A. (b, c) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐Y591H (b) or hTRPV4‐Y591A (c) cDNAs after perfusion with a 30% hypotonic solution in the presence (Y591H, n = 96; Y591A, n = 92) or absence (Y591H, n = 75; Y591A, n = 69) of 10‐nM HC067047. (d) Mean and individual dot plots of the AUC in HeLa cells transfected with the indicated cDNAs and exposed to 30% hypotonic solutions in the presence or absence of 10‐nM HC067047. N = 6 for all conditions except DMSO TRPV4‐WT (n = 7) and DMSO TRPV4‐D546A (n = 5)
We also addressed if other TRPV4 inhibitors such as RN1734 and ruthenium red, structurally unrelated to HC067047, may also use the HC067047 binding pocket. RN1734 (Vincent et al., 2009) dose‐dependently inhibited GSK1016790A activated hTRPV4‐WT (Figure 5a,c), whereas RN1734 hardly inhibited TRPV4‐D546A (Figure 5b,c). However, unlike HC056047 (Figure 4), RN1734 reduced both TRPV4‐Y591A and TRPV4‐Y591H responses to 30% hypotonicity, undistinguishable from the inhibition seen in the TRPV4‐WT channel (Figure 5d–f). As with the activation by GSK1016790A, TRPV4‐D546A channel activation by hypotonicity was not blocked by RN1734 (Figure 5f). Together, these results suggest that both HC067047 and RN1734 require residue D546 for their inhibitory effect on TRPV4, whereas residue Y591 only participates in the HC067047 interaction with and inhibition of TRPV4. On the other hand, the blocking effect of ruthenium red, a nonselective cation channel blocker (Voets et al., 2002), including TRPV4, that binds to the pore domain of TRP channels, was not affected by mutations on residues D546 (Figure 6a–d) or Y591 (Figure 6e,f), independently of the activating stimuli.
FIGURE 5.
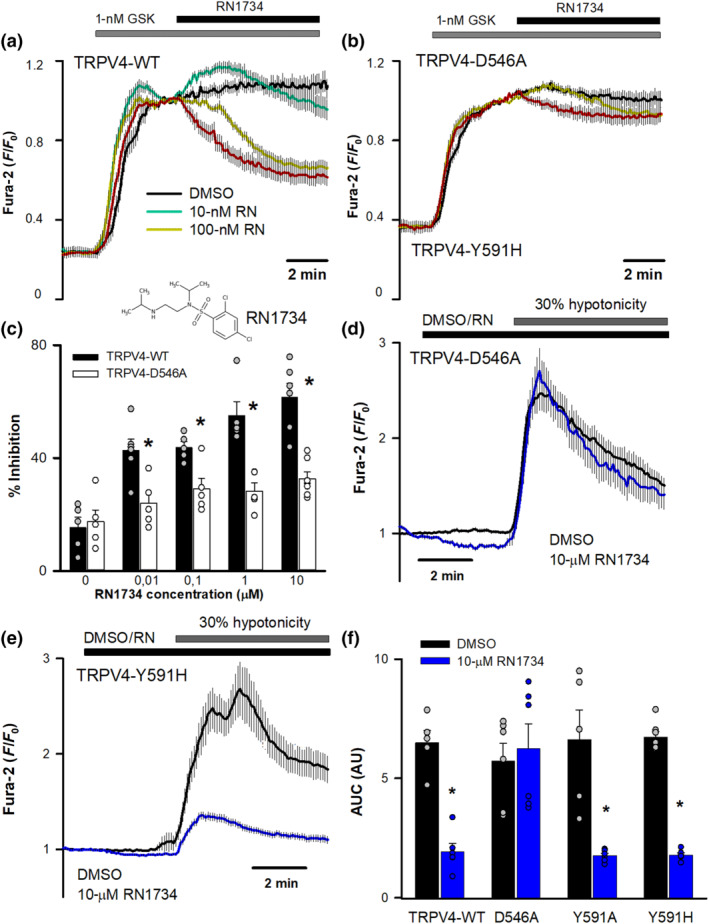
HC067047 binding pocket is partially shared by the inhibitor RN1734. (a, b) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐WT (a) or hTRPV4‐D546A (b) cDNAs, after perfusion with 1‐nM GSK1016790A and addition of RN1734 (or vehicle) at the indicated concentrations at time t = 0. Traces are means ± SEM of 47 (DMSO), 81 (10 nM), 78 (100 nM), 82 (1 μM) and 93 (10 μM) cells expressing TRPV4‐WT and 72 (DMSO), 75 (10 nM), 80 (100 nM), 78 (1 μM) and 60 (10 μM) cells expressing TRPV4‐D546A. (c) Percentage inhibition of GSK1016790A‐induced increases in intracellular Ca2+ at different RN1734 concentrations. N = 5 for all conditions except 10‐nM and 10‐μM HC067047 TRPV4‐WT (n = 6) and 10‐μM HC067047 TRPV4‐D546A (n = 7). (d, e) Changes in intracellular [Ca2+] (means ± SEM) in HeLa cells transfected with hTRPV4‐D546A (d) or hTRPV4‐Y591H (e) cDNAs, after perfusion with a 30% hypotonic solution in the presence (D546A, n = 83; Y591H, n = 88) or absence (D546A, n = 78; Y591H, n = 75) of 10‐μM RN1734. (f) AUC in HeLa cells transfected with the indicated cDNAs and exposed to 30% hypotonic solutions in the presence or absence of 10‐μM RN1734. N number of experiments from left to right: 5, 6, 5, 7, 6, 6, 6 and 6
FIGURE 6.
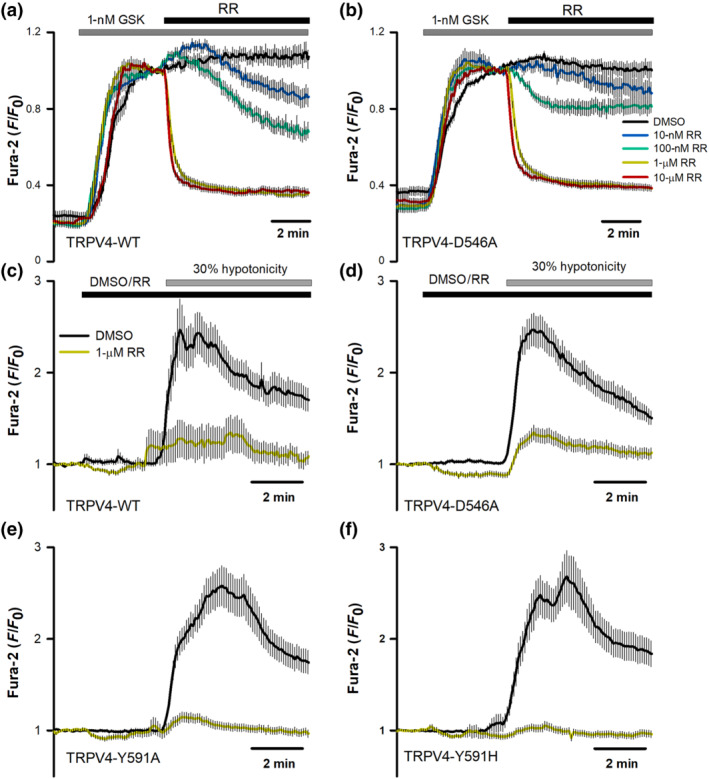
Mutations in hTRPV4 residues D546 and Y591 do not affect channel inhibition by ruthenium red. (a, b) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐WT (a) or hTRPV4‐D546A (b) cDNAs, after perfusion with 1‐nM GSK1016790A and addition of ruthenium red (or vehicle) at the indicated concentrations at time t = 0. Traces are means ± SEM of 48 (DMSO), 75 (10 nM), 67 (100 nM), 95 (1 μM) and 70 (10 μM) cells expressing TRPV4‐WT and 72 (DMSO), 53 (10 nM), 63 (100 nM), 73 (1 μM) and 80 (10 μM) cells expressing TRPV4‐D546A. (c–f) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐WT (c), D546A (d), Y591A (e), or Y591H (f) cDNAs, after perfusion with a 30% hypotonic solution in the presence or absence of 1‐μM ruthenium red (RR). N values for DMSO in WT, D546A, Y591A and Y591H are 23, 31, 27 and 35, respectively. N values for RR in WT, D546A, Y591A and Y591H are 30, 33, 31 and 34, respectively. Statistical analysis of the effect of RR on wild‐type versus mutant TRPV4 channels activated by GSK1916790A or hypotonicity reported a P > 0.05 for all conditions tested in panels (a)–(f), except for the comparison of the values obtained with 100‐nM RR in the presence of GSK (panels a and b) which was 0.04
Once the antagonist–protein interactions observed in silico were validated by mutagenesis studies, we run a structure‐based virtual screening using the National Cancer Institute (NCI) database (SCR_014886) to discover new TRPV4 inhibitor molecules. To perform the screening, we used the HC067047 binding pocket to do the grid box for docking. The same configuration protocol used for HC067047 was used for the virtual screening process. A KNIME workflow with Schrodinger software nodes was used to perform all procedures of TRPV4 inhibitor's choice and screening (Schrodinger suite, 2019). Of the ~250,000 compounds present in the open NCI database (https://cactus.nci.nih.gov/download/nci), we selected 40 potential hits based on the lower docking scores, which were tested using Ca2+ imaging in HeLa cells transfected with hTRPV4‐WT.
TRPV4‐WT was activated by 10‐nM GSK1016790A and each of the inhibitor candidates was added at a concentration of 1 μM (Figure 7a). TRPV4 inhibition percentage for each drug candidate was calculated as the decrease observed in GSK response following addition of the candidate drug (Figure 7a). Among the 40 candidates, 16 showed a statistically significant TRPV4 inhibition, of which we selected three candidates with an inhibitory potential similar to HC067047 and RN1734: NSC151066, NSC212069 and NSC600684 (Figure 7a,b). These three candidates were further tested at different concentrations (Figure 7c–f) and their IC50 obtained, which were 145 ± 47 nM for 151066, 69 ± 19 nM for 212069 and 363 ± 150 nM for 600684.
FIGURE 7.
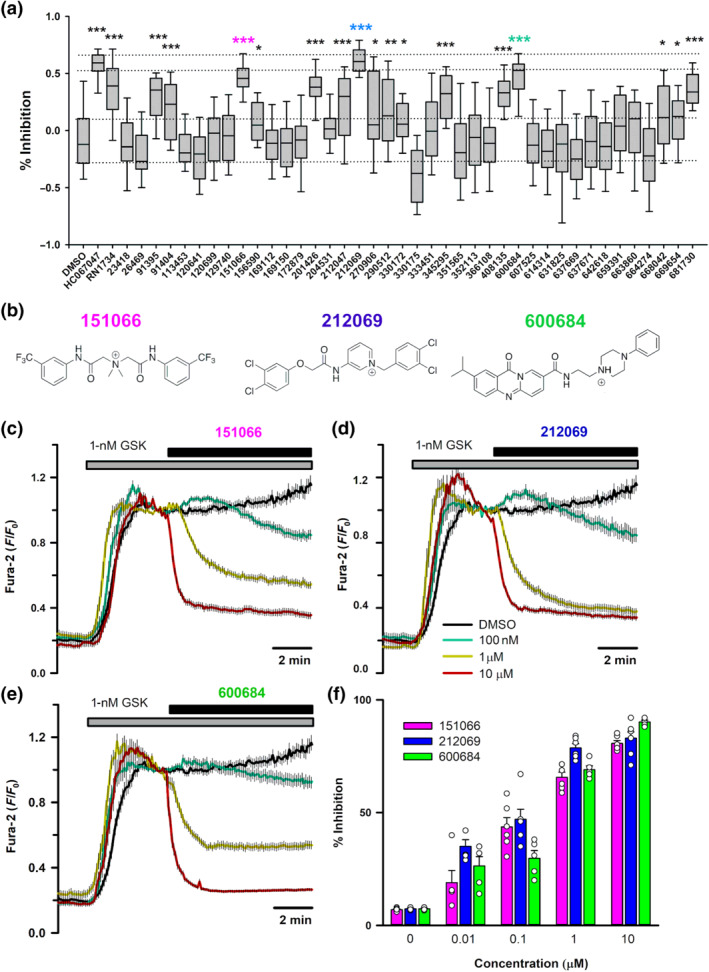
Effect of selected hits from the structure‐based screening on hTRPV4 channel activity. (a) Percentage of inhibition of hTRPV4‐WT transfected in HeLa cells and activated by 1‐nM GSK1016790A in the presence of different compounds and known channel inhibitors at 1 μM (n ≥ 26 for each compound). (b) Chemical structures of three hits selected for further analysis. (c–e) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐WT cDNA after perfusion with 1‐nM GSK1016790A and addition of 151066 (c), 212069 (d) and 600684 (e) at the indicated concentrations at time t = 0. Traces are means ± SEM of 52, 46, 57, 57 and 67 cells for compound 151066; 47, 39, 51, 46 and 67 cells for compound 212069; and 67, 39, 48, 57 and 50 cells for compound 600684. (f) Percentage inhibition of GSK1016790A‐induced increases in intracellular Ca2+ at different concentrations of 151066 (N = 5, 5, 6, 6 and 6), 212069 (N = 5, 5, 6, 6 and 7) and 600684 (N = 5 for all concentrations)
These three compounds were analysed for stability studies using MD simulation. The simulation studies revealed that 151066 (pink) and 212069 (blue) have good stability, similar to HC067047 (orange) within the pocket, while 600684 (green) was positioned close to the extracellular region of the protein, unlikely to interact with the key residues described in the HC067047 binding pocket (Figure 8a). Similar to the xTRPV4‐HC067047 complex, hydrophobic interactions with amino acids M583 and F613 were observed for xTRPV4‐151066 (Figure 8b) and xTRPV4‐212069 (Figure 8c) complexes, whereas the xTRPV4‐600684 complex (Figure 8d) showed no interactions with any of the HC067047 key binding residues. This prediction was also tested in HeLa cells transfected with different hTRPV4 mutants. All three compounds, at a concentration of 1 μM, significantly inhibited the hTRPV4‐D546A (Figure 9a,d) and hTRPV4‐Y591H channels (Figure 9b,e). However, hTRPV4‐M587A channel showed reduced inhibition by 212069 and 151066, compared to TRPV4‐WT and TRPV4‐D546A channels, and considerable inhibition by 600684 and HC067047 (Figure 9c,d), in agreement with the MD results that proposed residue M587 is important for the binding of 151066 and 212069 compounds.
FIGURE 8.
Interaction of new compounds with residues of the binding pocket. (a) Image corresponding to the last conformation of HC067047 (orange), 151066 (fuchsia), 212069 (blue) and 600684 (green) on the binding pocket of TRPV4 after 500‐ns MD. (b) Close up image of 151066 interactions. (c) Close up image of 212069 interactions. (d) Close up image of 600684 interactions. Hydrophobic interactions, H bonds and ionic interactions are shown in dashed lines in purple, fuchsia and green, respectively
FIGURE 9.
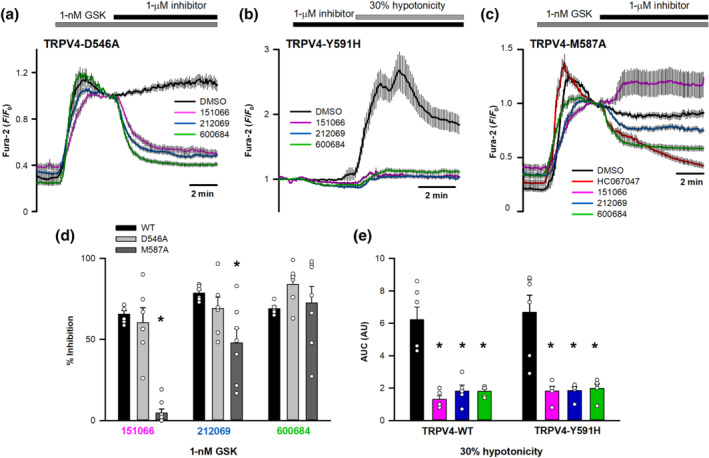
Effect of D546A, Y591H and M587A mutations on the inhibitory activity of the new inhibitors. (a) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐D546A (DMSO, n = 30; 151066, n = 39; 212069, n = 48; 600684, n = 54) after perfusion with 1‐nM GSK1016790A and addition of different inhibitors (or vehicle) at 1 μM. (b) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐Y591H after perfusion with a 30% hypotonic solution in the presence or absence of different inhibitors (DMSO, n = 40; 151066, n = 49; 212069, n = 52; 600684, n = 55) at 1 μM. (c) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV4‐M587A (DMSO, n = 63; 151066, n = 90; 212069, n = 84; 600684, n = 104) after perfusion with 1‐nM GSK1016790A and addition of different inhibitors. (d) Percentage inhibition of GSK1016790A‐induced increases in intracellular Ca2+ concentrations generated by TRPV4‐WT (151066, n = 6; 212069, n = 6; 600684, n = 5), TRPV4‐D546A (n = 6 for all compounds) and TRPV4‐M587A (151066, n = 8; 212069, n = 7; 600684, n = 7) channels using different compounds. (e) Mean and individual dot plots of the AUC in HeLa cells transfected with the indicated cDNAs and exposed to 30% hypotonic solutions in the absence (black) or the presence of 1 μM of 151066 (pink), 212069 (blue) or 600684 (green). N = 5 for all conditions except for both control conditions (n = 6)
Next, we tested the effect of these three new TRPV4 inhibitors on the activity of the closely related TRPV1 channel (Caterina et al., 1997). TRPV1 channel was activated with capsaicin followed by addition of the new compounds at a concentration of 1 μM (Figure 10a). While 151066 showed no inhibitory effect, both 212069 and 600684 showed significant inhibition (Figure 10a,b). The interaction of HC067047, 151066, 212069 and 600684 compounds with rat TRPV1 capsaicin binding site (Cao et al., 2013; Liao, Cao, Julius, & Cheng, 2013) was also studied in silico. Results obtained using a molecular docking approach confirmed that capsaicin (Figure 10c, yellow) stabilized at 3.06–4.55 Å from amino acids that modulate the TRPV1 vanilloid sensitivity (Y511, M547, T550 and L669) (Gao, Cao, Julius, & Cheng, 2016; Zhao et al., 2013). Compounds 212069 (blue) and 600684 (green) (Figure 10c) showed relevant proximity to amino acids involved in the vanilloid sensitivity of TRPV1 (particularly Y511), while HC067047 (orange) and compound 151066 (pink) showed a different structural conformation, losing the interaction with many of these residues, particularly Y511 (Figure 10d). Together, this set of experiments confirmed certain degree of TRPV4 specificity, versus TRPV1, for compound 151066.
FIGURE 10.
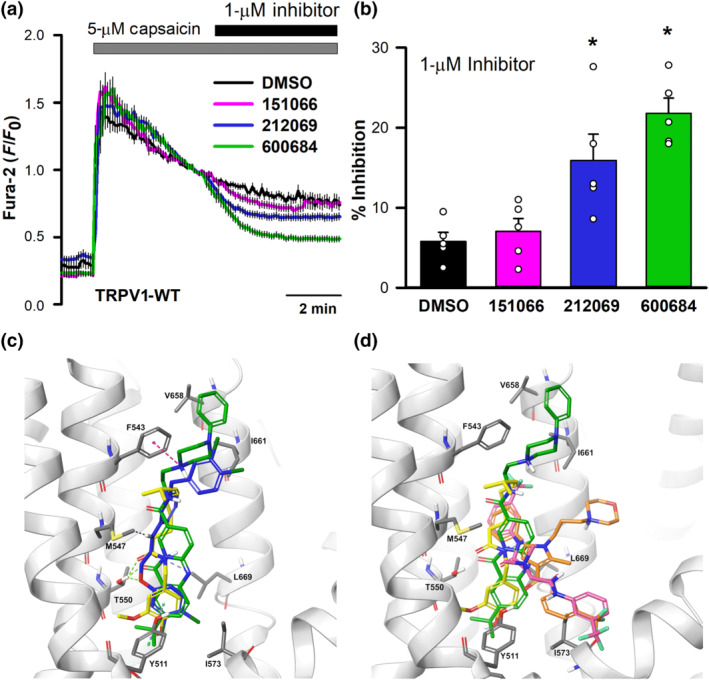
Interaction of new compounds with TRPV1. (a) Changes in intracellular [Ca2+] in HeLa cells transfected with hTRPV1 (DMSO, n = 40; 151066, n = 40; 212069, n = 43; 600684, n = 36 cells) after perfusion with 5‐μM capsaicin and addition of DMSO, or the indicated compounds at 1 μM. (b) Percentage inhibition of capsaicin‐induced increases in intracellular Ca2+ concentrations generated by TRPV1 (n = 5 for all conditions). (c) Image corresponding to the binding of capsaicin (yellow), 212069 (blue) and 600684 (green) on the binding capsaicin pocket of TRPV1. (d) Image similar to (c) with the addition of HC067047 (orange) and 151066 (fuchsia)
Finally, we addressed the potential use of 151066 in an experimental condition in which TRPV4 inhibitors have proven effective antivirals. We have recently reported the involvement of TRPV4 in Zika virus infectivity of Huh7 cells and that genetic or pharmacological inhibition of TRPV4 reduced viral infectivity (Doñate‐Macián et al., 2018). We first demonstrated that 151066, similar to HC067047, inhibited GSK1016790A‐mediated currents in human Huh7 cells endogenously expressing TRPV4 (Figure 11a). Second, Huh7 cells were exposed to Zika virus at 0.01 MOI and infectivity was assessed measuring the activity of a luciferase reporter inserted in the viral RNA genome. Compound 151066 reduced viral infectivity with an EC50 of 22 nM and 50% cytotoxic concentration (CC50) of 2.99 μM (Figure 11b), which gave a selectivity index (SI) calculated as CC50/EC50 of 135.9, in the same range of that previously reported for HC067047 (SI = 187) (Doñate‐Macián et al., 2018).
FIGURE 11.
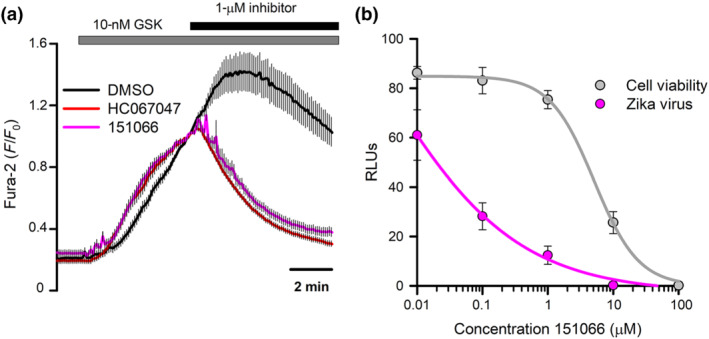
Inhibition of TRPV4 with 151066 reduces Zika virus infectivity. (a) Changes in cytosolic [Ca2+] measured in Huh7 cells after perfusion with 10‐nM GSK1016790A and addition of 1‐μM 151066 or HC067047. (b) Zika virus infection and cell viability in Huh7 cells treated with increasing concentrations of 151066. The data are presented as mean ± SEM of three independent experiments at each 151066 concentrations
4. DISCUSSION
Over the last decade, virtual screening has been successfully employed in the discovery of bioactive lead compounds; however, only a few TRPV4 modulators have been identified that can be used for the treatment of TRPV4‐related diseases. The combination of in silico MD with calcium imaging to evaluate TRPV4 channel activity has proven effective in the characterization of the structural basis of TRPV4 modulation. We have previously identified domains required for subunit tetramerization and trafficking (Garcia‐Elias et al., 2015) and demonstrated that the mechanotransducing and osmotransducing messenger 5′,6′‐EET (Fernandes et al., 2008; Vriens et al., 2004; Watanabe et al., 2003) gates TRPV4 by a direct action on a site formed by residues from the S2–S3 linker, S4 and S4–S5 linker (Berna‐Erro et al., 2017). We have now characterized the binding pocket of the TRPV4 inhibitor, HC067047 (Thorneloe et al., 2008), which is delimited by the S2–S3 linker, S4 and S5, partially overlapping with the EET binding site. Within this pocket, we identified hTRPV4‐D546 as a key residue in HC067047‐mediated inhibition, with also hTRPV4‐Y591 playing a prominent role, as mutations in both residues affect inhibition by HC067047. This binding pocket is not unique for HC067047 as another channel inhibitor, RN1734, also requires residue hTRPV4‐D546, to exert its inhibitory activity. That this binding pocket is a bona fide inhibitory site was further confirmed by our structure‐based virtual screening. In silico analysis of a database using HC067047 as a prototypical inhibitor and its binding pocket generated a short list of 40 hits, three of which were effective TRPV4 inhibitors and one of them, 151066, was shown to depend on the integrity of the binding site to inhibit TRPV4, as hTRPV4‐M587A became insensitive to 151066.
Our results locate the TRPV4 antagonist binding site at a structural location that appear to be a hotspot for modulator binding in TRP channels. TRPC6 channel binds its inhibitor AM1473 in a region delimited by transmembrane segments S1–S4 (Bai et al., 2020) and TRPV6 uses residue Y466 on S3 to bind its inhibitor 2‐aminoethoxydiphenyl borate (Singh, Saotome, Mcgoldrick, & Sobolevsky, 2018). Interaction with agonist has also pinpointed to this structural domain formed by S1–S4 transmembrane segments. In addition to the already mentioned binding of EET to TRPV4 (Berna‐Erro et al., 2017), icilin, a synthetic cooling agonist, binds to TRPM8 in this site (Yin et al., 2019) and TRPV1 binds capsaicin through residues located in S4 and S4–S5 linker (Yang et al., 2015).
Together, our structure‐based in silico approach has shown a good predictive ability, confirming the current trend for the use of this type of approach in the development of potent and specific modulators (Schaller et al., 2020), in addition to be a powerful tool to investigate the ligand‐dependent protein activity at the mechanistic level. Furthermore, MD‐based virtual screening presents interesting advantages:‐ (1) prioritization of compounds by virtual screening, compared to high‐throughput screenings, can reduce considerably the number of hits requiring experimental testing, (2) MD‐based conformations generate a more realistic dynamic ligand–protein interaction and may optimize the identification of hits (Carlson et al., 2000) and (3) offers valuable information, currently incomplete, on dynamic structures that can be fed into artificial intelligence approaches to drug discovery in the era of big data (Vamathevan et al., 2019).
In summary, we have revealed useful insights that can be exploited for the design future drugs to target different TRPV4‐linked pathologies and underline the emerging potential of the combination of structure‐based in silico simulations with in vitro functional studies in structural pharmacology.
AUTHOR CONTRIBUTIONS
P.D.‐M. and M.A.V. conceived and designed the study. P.D.‐M. and F.R.‐M. performed calcium imaging experiments and generated the required molecular biology tools. Y.D., J.C. and F.G.‐N. performed in silico analysis. J.D. and G.P.‐V. designed and performed viral infection assays. F.G.‐N. and J.D. contributed to the interpretation of the data. M.A.V. wrote the manuscript. All authors edited the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
5.
ACKNOWLEDGEMENTS
We thank Dr. Andres Merits (University of Tartu, Estonia) for kindly providing the plasmid encoding the ZIKV with NanoLuc, Spanish Ministry of Economy and Competitiveness through grants RTI2018‐099718 (to M.A.V.) and BFU2016‐80039‐R (to J.D.), an institutional “Unidad de Excelencia María de Maeztu” CEX2018‐000792‐M and FEDER funds. F.G.‐N. thanks Fondecyt Regular projects 1170733, the US Army of USA, W911NF‐14‐1‐0520 and The Centro Interdisciplinario de Neurociencia de Valparaíso is a Millennium Institute supported by the Millennium Scientific Initiative of the Ministerio de Economía, Fomento y Turismo P029‐022‐F.
Doñate‐Macian, P. , Duarte, Y. , Rubio‐Moscardo, F. , Pérez‐Vilaró, G. , Canan, J. , Díez, J. , González‐Nilo, F. , & Valverde, M. A. (2022). Structural determinants of TRPV4 inhibition and identification of new antagonists with antiviral activity. British Journal of Pharmacology, 179(14), 3576–3591. 10.1111/bph.15267
Pablo Doñate‐Macian and Yorley Duarte contributed equally to this work.
[Correction added on 9 November 2020, after first online publication: the copyright line has been corrected in this current version.]
Funding information Chilean Fondo Nacional de Desarrollo Tecnológico (FONDECYT), Grant/Award Number: 1170733; Spanish Ministerio de Ciencia, Innovación y Universidades, Grant/Award Numbers: BFU2016‐80039‐R, RTI2018‐099718; The Centro Interdisciplinario de Neurociencia de Valparaíso is a Millennium Institute supported by the Millennium Scientific Initiative of the Ministerio de Economía, Fomento y Turismo, Grant/Award Number: P029‐022‐F; US Army of USA, Grant/Award Number: W911NF‐14‐1‐0520; Fondecyt Regular; Unidad de Excelencia María de Maeztu, Grant/Award Number: CEX2018‐000792‐M
DATA AVAILABILITY STATEMENT
All data generated for this article is available in the main text.
REFERENCES
- Alessandri‐Haber, N. , Dina, O. A. , Joseph, E. K. , Reichling, D. , & Levine, J. D. (2006). A transient receptor potential vanilloid 4‐dependent mechanism of hyperalgesia is engaged by concerted action of inflammatory mediators. The Journal of Neuroscience, 26, 3864–3874. 10.1523/JNEUROSCI.5385-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , … Sharman, J. L. (2019). The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology, 176, S142–S228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpizar, Y. A. , Boonen, B. , Sanchez, A. , Jung, C. , López‐Requena, A. , Naert, R. , … Vanoirbeek, J. A. (2017). TRPV4 activation triggers protective responses to bacterial lipopolysaccharides in airway epithelial cells. Nature Communications, 8, 1059. 10.1038/s41467-017-01201-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, Y. N. , Fernandes, J. , Vazquez, E. , Fernandez‐Fernandez, J. M. , Arniges, M. , Sanchez, T. M. , … Valverde, M. A. (2005). TRPV4 channel is involved in the coupling of fluid viscosity changes to epithelial ciliary activity. The Journal of Cell Biology, 168, 869–874. 10.1083/jcb.200409070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arniges, M. , Vázquez, E. , Fernández‐Fernández, J. M. , Valverde, M. A. , Vazquez, E. , & Fernandez‐Fernandez, J. M. (2004). Swelling‐activated Ca2+ entry via TRPV4 channel is defective in cystic fibrosis airway epithelia. The Journal of Biological Chemistry, 279, 54062–54068. 10.1074/jbc.M409708200 [DOI] [PubMed] [Google Scholar]
- Atobe, M. , Nagami, T. , Muramatsu, S. , Ohno, T. , Kitagawa, M. , Suzuki, H. , … Kawanishi, M. (2019). Discovery of novel transient receptor potential vanilloid 4 (TRPV4) agonists as regulators of chondrogenic differentiation: Identification of quinazolin‐4(3H)‐ones and in vivo studies on a surgically induced rat model of osteoarthritis. Journal of Medicinal Chemistry, 62, 1468–1483. 10.1021/acs.jmedchem.8b01615 [DOI] [PubMed] [Google Scholar]
- Bai, Y. , Yu, X. , Chen, H. , Horne, D. , White, R. , Wu, X. , … Huang, X. (2020). Structural basis for pharmacological modulation of the TRPC6 channel. eLife, 9, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna‐Erro, A. , Izquierdo‐Serra, M. , Sepúlveda, R. V. , Rubio‐Moscardo, F. , Doñate‐Macián, P. , Serra, S. A. , … Valverde, M. A. (2017). Structural determinants of 5′,6′‐epoxyeicosatrienoic acid binding to and activation of TRPV4 channel. Scientific Reports, 7, 10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J. , Chow, D. E. , Xu, H. , Dror, R. O. , Eastwood, M. P. , Gregersen, B. A. , … & Salmon, J. K. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, p. 43.
- Brnardic, E. J. , Ye, G. , Brooks, C. , Donatelli, C. , Barton, L. , McAtee, J. , … Lawhorn, B. G. (2018). Discovery of pyrrolidine sulfonamides as selective and orally bioavailable antagonists of transient receptor potential vanilloid‐4 (TRPV4). Journal of Medicinal Chemistry, 61, 9738–9755. 10.1021/acs.jmedchem.8b01317 [DOI] [PubMed] [Google Scholar]
- Cao, E. , Liao, M. , Cheng, Y. , & Julius, D. (2013). TRPV1 structures in distinct conformations reveal activation mechanisms. Nature, 504, 113–118. 10.1038/nature12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, H. A. , Masukawa, K. M. , Rubins, K. , Bushman, F. D. , Jorgensen, W. L. , Lins, R. D. , … McCammon, J. (2000). Developing a dynamic pharmacophore model for HIV‐1 integrase. Journal of Medicinal Chemistry, 43, 2100–2114. 10.1021/jm990322h [DOI] [PubMed] [Google Scholar]
- Caterina, M. J. , Schumacher, M. A. , Tominaga, M. , Rosen, T. A. , Levine, J. D. , & Julius, D. (1997). The capsaicin receptor: A heat‐activated ion channel in the pain pathway. Nature, 389, 816–824. 10.1038/39807 [DOI] [PubMed] [Google Scholar]
- Cheung, M. , Bao, W. , Behm, D. J. , Brooks, C. A. , Bury, M. J. , Dowdell, S. E. , … Thorneloe, K. S. (2017). Discovery of GSK2193874: An orally active, potent, and selective blocker of transient receptor potential vanilloid 4. ACS Medicinal Chemistry Letters, 8, 549–554. 10.1021/acsmedchemlett.7b00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Z. , Paknejad, N. , Maksaev, G. , Sala‐Rabanal, M. , Nichols, C. G. , Hite, R. K. , & Yuan, P. (2018). Cryo‐EM and X‐ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nature Structural & Molecular Biology, 25, 252–260. 10.1038/s41594-018-0037-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doñate‐Macián, P. , Jungfleisch, J. , Pérez‐Vilaró, G. , Rubio‐Moscardo, F. , Perálvarez‐Marín, A. , Diez, J. , & Valverde, M. A. (2018). The TRPV4 channel links calcium influx to DDX3X activity and viral infectivity. Nature Communications, 9, 2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaerts, W. , Zhen, X. , Ghosh, D. , Vriens, J. , Gevaert, T. , Gilbert, J. P. , … Voets, T. (2010). Inhibition of the cation channel TRPV4 improves bladder function in mice and rats with cyclophosphamide‐induced cystitis. Proceedings of the National Academy of Sciences of the United States of America, 107, 19084–19089. 10.1073/pnas.1005333107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, J. , Lorenzo, I. M. , Andrade, Y. N. , Garcia‐Elias, A. , Serra, S. A. , Fernandez‐Fernandez, J. M. , & Valverde, M. A. (2008). IP3 sensitizes TRPV4 channel to the mechano‐ and osmotransducing messenger 5′‐6′‐epoxyeicosatrienoic acid. The Journal of Cell Biology, 181, 143–155. 10.1083/jcb.200712058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Fernandez, J. M. , Andrade, Y. N. , Arniges, M. , Fernandes, J. , Plata, C. , Rubio‐Moscardo, F. , … Valverde, M. A. (2008). Functional coupling of TRPV4 cationic channel and large conductance, calcium‐dependent potassium channel in human bronchial epithelial cell lines. Pflügers Archiv, 457, 149–159. 10.1007/s00424-008-0516-3 [DOI] [PubMed] [Google Scholar]
- Friesner, R. A. , Murphy, R. B. , Repasky, M. P. , Frye, L. L. , Greenwood, J. R. , Halgren, T. A. , … Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein‐ligand complexes. Journal of Medicinal Chemistry, 49, 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Galindo‐Villegas, J. , Montalban‐Arques, A. , Liarte, S. , de Oliveira, S. , Pardo‐Pastor, C. , Rubio‐Moscardo, F. , … Mulero, V. (2016). TRPV4‐mediated detection of hyposmotic stress by skin keratinocytes activates developmental immunity. Journal of Immunology, 196, 738–749. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Cao, E. , Julius, D. , & Cheng, Y. (2016). Article TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature, 534, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Elias, A. , Berna‐Erro, A. , Rubio‐Moscardo, F. , Pardo‐Pastor, C. , Mrkonjić, S. , Sepúlveda, R. V. , … Valverde, M. A. (2015). Interaction between the linker, pre‐S1, and TRP domains determines folding, assembly, and trafficking of TRPV channels. Structure, 23, 1404–1413. 10.1016/j.str.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Garcia‐Elias, A. , Mrkonjic, S. , Pardo‐Pastor, C. , Inada, H. , Hellmich, U. A. , Rubio‐Moscardó, F. , … Valverde, M. A. (2013). Phosphatidylinositol‐4,5‐biphosphate‐dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proceedings of the National Academy of Sciences of the United States of America, 110, 9553–9558. 10.1073/pnas.1220231110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert, T. , Vriens, J. , Segal, A. , Everaerts, W. , Roskams, T. , Talavera, K. , … Nilius, B. (2007). Deletion of the transient receptor potential cation channel TRPV4 impairs murine bladder voiding. Journal of Clinical Investigation, 117, 3453–3462. 10.1172/JCI31766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, N. , Skrdla, P. , Schroyer, R. , Kumar, S. , Fernando, D. , Oughton, A. , … Cheriyan, J. (2019). Clinical pharmacokinetics, safety, and tolerability of a novel, first‐in‐class TRPV4 ion channel inhibitor, GSK2798745, in healthy and heart failure subjects. American Journal of Cardiovascular Drugs, 19, 335–342. [DOI] [PubMed] [Google Scholar]
- Greenidge, P. A. , Kramer, C. , Mozziconacci, J. C. , & Wolf, R. M. (2013). MM/GBSA binding energy prediction on the PDBbind data set: Successes, failures, and directions for further improvement. Journal of Chemical Information and Modeling, 53, 201–209. 10.1021/ci300425v [DOI] [PubMed] [Google Scholar]
- Güler, A. D. , Lee, H. , Iida, T. , Shimizu, I. , Tominaga, M. , & Caterina, M. (2002). Heat‐evoked activation of the ion channel, TRPV4. The Journal of Neuroscience, 22, 6408–6414. 10.1523/jneurosci.22-15-06408.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, C. , Fernández‐Dueñas, V. , Plata, C. , Garcia‐Elias, A. , Ciruela, F. , Fernández‐Fernández, J. M. , & Valverde, M. A. (2018). Functional coupling of GABAA/B receptors and the channel TRPV4 mediates rapid progesterone signaling in the oviduct. Science Signaling, 11, eaam6558. 10.1126/scisignal.aam6558 [DOI] [PubMed] [Google Scholar]
- Liao, M. , Cao, E. , Julius, D. , & Cheng, Y. (2013). Structure of the TRPV1 ion channel determined by electron cryo‐microscopy. Nature, 504, 107–112. 10.1038/nature12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke, W. , Choe, Y. , Marti‐Renom, M. A. , Bell, A. M. , Denis, C. S. , Hudspeth, A. J. , … Heller, S. (2000). Vanilloid receptor‐related osmotically activated channel (VR‐OAC), a candidate vertebrate osmoreceptor. Cell, 103, 525–535. 10.1016/s0092-8674(00)00143-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke, W. , & Friedman, J. M. (2003). Abnormal osmotic regulation in trpv4 −/− mice. Proceedings of the National Academy of Sciences of the United States of America, 100, 13698–13703. 10.1073/pnas.1735416100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, I. M. , Liedtke, W. , Sanderson, M. J. , & Valverde, M. A. (2008). TRPV4 channel participates in receptor‐operated calcium entry and ciliary beat frequency regulation in mouse airway epithelial cells. Proceedings. National Academy of Sciences. United States of America, 105, 12611–12616. 10.1073/pnas.0803970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C. , Cevikbas, F. , Pasolli, H. A. , Chen, Y. , Kong, W. , Kempkes, C. , … Jokerst, N. M. (2013). UVB radiation generates sunburn pain and affects skin by activating epidermal TRPV4 ion channels and triggering endothelin‐1 signaling. Proceedings of the National Academy of Sciences, 110, E3225–E3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu, S. , Wakabayashi, M. , Ohno, T. , Amano, K. , Ooishi, R. , Sugahara, T. , … Matsuda, A. (2007). Functional gene screening system identified TRPV4 as a regulator of chondrogenic differentiation. The Journal of Biological Chemistry, 282, 32158–32167. 10.1074/jbc.M706158200 [DOI] [PubMed] [Google Scholar]
- Nilius, B. , & Voets, T. (2013). The puzzle of TRPV4 channelopathies. EMBO Reports, 14, 152–163. 10.1038/embor.2012.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Conor, C. J. , Leddy, H. A. , Benefield, H. C. , Liedtke, W. B. , & Guilak, F. (2014). TRPV4‐mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proceedings of the National Academy of Sciences of the United States of America, 111, 1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman, S. O. , Grove, L. M. , Paruchuri, S. , Southern, B. D. , Abraham, S. , Niese, K. A. , … Olman, M. A. (2014). TRPV4 mediates myofibroblast differentiation and pulmonary fibrosis in mice. The Journal of Clinical Investigation, 124, 5225–5238. 10.1172/JCI75331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, D. , Šribar, D. , Noonan, T. , Deng, L. , Nguyen, T. N. , Pach, S. , … Wolber, G. (2020). Next generation 3D pharmacophore modeling. Wiley Interdisciplinary Reviews: Computational Molecular Science, 10, e1468. [Google Scholar]
- Schrodinger . (2020). Schrodinger Release 2020–1: Schrodinger KNIME Extensions.
- Schrodinger suite . (2019). Protein Preparation Wizard; Epik version 2.2.
- Shao, J. , Han, J. , Zhu, Y. , Mao, A. , Wang, Z. , Zhang, K. , … Ma, X. (2019). Curcumin induces endothelium‐dependent relaxation by activating endothelial TRPV4 channels. Journal of Cardiovascular Translational Research, 12, 600–607. [DOI] [PubMed] [Google Scholar]
- Singh, A. K. , Saotome, K. , Mcgoldrick, L. L. , & Sobolevsky, A. I. (2018). Structural bases of TRP channel TRPV6 allosteric modulation by 2‐APB. Nature Communications, 9, 2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P. L. , Maloney, K. N. , Pothen, R. G. , Clardy, J. , & Clapham, D. E. (2006). Bisandrographolide from Andrographis paniculata activates TRPV4 channels. The Journal of Biological Chemistry, 281, 29897–29904. 10.1074/jbc.M605394200 [DOI] [PubMed] [Google Scholar]
- Strotmann, R. , Harteneck, C. , Nunnenmacher, K. , Schultz, G. , & Plant, T. D. (2000). OTRPC4, a nonselective cation channel that confers sensitivity to extracellular osmolarity. Nature Cell Biology, 2, 695–702. [DOI] [PubMed] [Google Scholar]
- Takayama, Y. , Shibasaki, K. , Suzuki, Y. , Yamanaka, A. , & Tominaga, M. (2014). Modulation of water efflux through functional interaction between TRPV4 and TMEM16A/anoctamin 1. The FASEB Journal, 28, 2238–2248. [DOI] [PubMed] [Google Scholar]
- Thorneloe, K. S. , Sulpizio, A. C. , Lin, Z. , Figueroa, D. J. , Clouse, A. K. , McCafferty, G. P. , … Misajet, B. A. (2008). N‐((1S)‐1‐{[4‐((2S)‐2‐{[(2,4‐Dichlorophenyl)sulfonyl]amino}‐3‐hydroxypropanoyl)‐1‐piperazinyl]carbonyl}‐3‐methylbutyl)‐1‐benzothiophene‐2‐carboxamide (GSK1016790A), a novel and potent transient receptor potential vanilloid 4 channel agonist induces urin. Pharmacology and Experimental Therapeutics, 326, 432–442. [DOI] [PubMed] [Google Scholar]
- Tian, W. , Fu, Y. , Garcia‐Elias, A. , Fernández‐Fernández, J. M. , Vicente, R. , Kramer, P. L. , … Cohen, D. M. (2009). A loss‐of‐function nonsynonymous polymorphism in the osmoregulatory TRPV4 gene is associated with human hyponatremia. Proceedings of the National Academy of Sciences of the United States of America, 106, 14034–14039. 10.1073/pnas.0904084106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamathevan, J. , Clark, D. , Czodrowski, P. , Dunham, I. , Ferran, E. , Lee, G. , … Zhao, S. (2019). Applications of machine learning in drug discovery and development. Nature Reviews. Drug Discovery, 18, 463–477. 10.1038/s41573-019-0024-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, F. , Acevedo, A. , Nguyen, M. T. , Dourado, M. , DeFalco, J. , Gustafson, A. , … Duncton, M. A. (2009). Identification and characterization of novel TRPV4 modulators. Biochemical and Biophysical Research Communications, 389, 490–494. 10.1016/j.bbrc.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Voets, T. , Prenen, J. , Vriens, J. , Watanabe, H. , Janssens, A. , Wissenbach, U. , … Nilius, B. (2002). Molecular determinants of permeation through the cation channel TRPV4. The Journal of Biological Chemistry, 277, 33704–33710. 10.1074/jbc.M204828200 [DOI] [PubMed] [Google Scholar]
- Vriens, J. , Owsianik, G. , Fisslthaler, B. , Suzuki, M. , Janssens, A. , Voets, T. , … Nilius, B. (2005). Modulation of the Ca2 permeable cation channel TRPV4 by cytochrome P450 epoxygenases in vascular endothelium. Circulation Research, 97, 908–915. 10.1161/01.RES.0000187474.47805.30 [DOI] [PubMed] [Google Scholar]
- Vriens, J. , Owsianik, G. , Janssens, A. , Voets, T. , & Nilius, B. (2007). Determinants of 4α‐phorbol sensitivity in transmembrane domains 3 and 4 of the cation channel TRPV4. The Journal of Biological Chemistry, 282, 12796–12803. 10.1074/jbc.M610485200 [DOI] [PubMed] [Google Scholar]
- Vriens, J. , Watanabe, H. , Janssens, A. , Droogmans, G. , Voets, T. , & Nilius, B. (2004). Cell swelling, heat, and chemical agonists use distinct pathways for the activation of the cation channel TRPV4. Proceedings of the National Academy of Sciences of the United States of America, 101, 396–401. 10.1073/pnas.0303329101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, H. , Davis, J. B. , Smart, D. , Jerman, J. C. , Smith, G. D. , Hayes, P. , … Nilius, B. (2002). Activation of TRPV4 channels (hVRL‐2/mTRP12) by phorbol derivatives. The Journal of Biological Chemistry, 277, 13569–13577. 10.1074/jbc.M200062200 [DOI] [PubMed] [Google Scholar]
- Watanabe, H. , Vriens, J. , Prenen, J. , Droogmans, G. , Voets, T. , & Nilius, B. (2003). Anandamide and arachidonic acid use epoxyeicosatrienoic acids to activate TRPV4 channels. Nature, 424, 434–438. 10.1038/nature01807 [DOI] [PubMed] [Google Scholar]
- Watanabe, H. , Vriens, J. , Suh, S. H. , Benham, C. D. , Droogmans, G. , & Nilius, B. (2002). Heat‐evoked activation of TRPV4 channels in a HEK293 cell expression system and in native mouse aorta endothelial cells. The Journal of Biological Chemistry, 277, 47044–47051. 10.1074/jbc.M208277200 [DOI] [PubMed] [Google Scholar]
- Yang, F. , Xiao, X. , Cheng, W. , Yang, W. , Yu, P. , Song, Z. , … Zheng, J. (2015). Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nature Chemical Biology, 11, 518–524. 10.1038/nchembio.1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y. , Le, S. C. , Hsu, A. L. , Borgnia, M. J. , Yang, H. , & Lee, S. Y. (2019). Structural basis of cooling agent and lipid sensing by the cold‐activated TRPM8 channel. Science (80‐.), 363, eaav9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Mao, A. , Xiao, W. , Zhang, P. , Han, X. , Zhou, T. , … Ma, X. (2019). Morin induces endothelium‐dependent relaxation by activating TRPV4 channels in rat mesenteric arteries. European Journal of Pharmacology, 859, 172561. 10.1016/j.ejphar.2019.172561 [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Zhu, L. , Zhu, Y. , Cao, J. , Li, S. , Huang, Q. , … Zhu, X. (2013). The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nature Cell Biology, 15, 1434–1444. 10.1038/ncb2880 [DOI] [PubMed] [Google Scholar]
- Zhu, G. , Gulsvik, A. , Bakke, P. , Ghatta, S. , Anderson, W. , Lomas, D. A. , … Pillai, S. G. (2009). Association of TRPV4 gene polymorphisms with chronic obstructive pulmonary disease. Human Molecular Genetics, 18, 2053–2062. 10.1093/hmg/ddp111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated for this article is available in the main text.


