Abstract
Target capture has emerged as an important tool for phylogenetics and population genetics in nonmodel taxa. Whereas developing taxon‐specific capture probes requires sustained efforts, available universal kits may have a lower power to reconstruct relationships at shallow phylogenetic scales and within rapidly radiating clades. We present here a newly developed target capture set for Bromeliaceae, a large and ecologically diverse plant family with highly variable diversification rates. The set targets 1776 coding regions, including genes putatively involved in key innovations, with the aim to empower testing of a wide range of evolutionary hypotheses. We compare the relative power of this taxon‐specific set, Bromeliad1776, to the universal Angiosperms353 kit. The taxon‐specific set results in higher enrichment success across the entire family; however, the overall performance of both kits to reconstruct phylogenetic trees is relatively comparable, highlighting the vast potential of universal kits for resolving evolutionary relationships. For more detailed phylogenetic or population genetic analyses, for example the exploration of gene tree concordance, nucleotide diversity or population structure, the taxon‐specific capture set presents clear benefits. We discuss the potential lessons that this comparative study provides for future phylogenetic and population genetic investigations, in particular for the study of evolutionary radiations.
Keywords: Bromeliaceae, phylogenomics, plant radiation, population structure, target capture, Tillandsia
Abstract
La captura selectiva de secuencias de ADN ha surgido como una herramienta importante para la filogenética y la genética de poblaciones en taxones no‐modelo. Mientras que el desarrollo de sondas de captura específicas para cada taxón requiere un esfuerzo sostenido, las colecciones de sondas universales disponibles pueden tener una potencia disminuida para la reconstrucción de relaciones filogenéticas poco profundas o de radiaciones rápidas. Presentamos aquí un conjunto de sondas para la captura selectiva desarrollado recientemente para Bromeliaceae, una familia de plantas extensa, ecológicamente diversa y con tasas de diversificación muy variables. El conjunto de sondas se centra en 1776 regiones de codificación, incluyendo genes supuestamente implicados en rasgos de innovación clave, con el objetivo de potenciar la comprobación de una amplia gama de hipótesis evolutivas. Comparamos la potencia relativa de este conjunto de sondas diseñado para un taxón específico, Bromeliad1776, con la colección universal Angiosperms353. El conjunto específico da lugar a un mayor éxito de captura en toda la familia. Sin embargo, el rendimiento global de ambos kits para reconstruir árboles filogenéticos es relativamente comparable, lo que pone de manifiesto el gran potencial de los kits universales para resolver las relaciones evolutivas. Para análisis filogenéticos o de genética de poblaciones más detallados, como por ejemplo la exploración de la congruencia de los árboles de genes, la diversidad de nucleótidos o la estructura de la población, el conjunto de captura específico para Bromeliaceae presenta claras ventajas. Discutimos las lecciones potenciales que este estudio comparativo proporciona para futuras investigaciones filogenéticas y de genética de poblaciones, en particular para el estudio de las radiaciones evolutivas.
1. INTRODUCTION
Targeted sequencing approaches have emerged as a promising tool for studying evolutionary relationships in nonmodel taxa, enabling researchers to retrieve large data sets while requiring few genomic resources (Bossert & Danforth, 2018; Escudero et al., 2020; McDonnell et al., 2021; Soto‐Gomez et al., 2019). Using custom baits, the method largely retrieves the same loci across a wide taxonomic scale, obtains comparable and mergeable data sets and may be combined with genome‐skimming (Lemmon & Lemmon, 2013; Weitemier et al., 2014). Pre‐existing knowledge of the targeted loci further provides opportunities to address specific questions on both deep and shallow timescales (Hale et al., 2020; Lemmon et al., 2012). Finally, the method does not necessarily require a reference genome, is highly cost‐effective and, with the ability to sequence herbarium samples, reduces the need for extensive sampling campaigns (Blaimer et al., 2016; Hale et al., 2020; Weitemier et al., 2014). Target capture has been successfully applied to resolve phylogenies in diverse groups, from arthropods such as bees (Xylocopa, Blaimer et al., 2016; Apidae, Bossert et al., 2019) and Araneae (Hexathelidae, Hedin et al., 2018) to mammals (Cetacea, McGowen et al., 2020), and in numerous plant groups (Heuchera, Folk et al., 2015; Gesneriaceae, Ogutcen et al., 2021; Zingiberales, Sass et al., 2016 to name a few). The method's utility for studies at microevolutionary scales has been to date marginally explored, but several studies have pointed to the ability to analyse genomic diversity and estimate population genomic parameters (Choquet et al., 2019; Christmas et al., 2017; Derrien & Ramos‐Onsins, 2020; de La Harpe et al., 2019; Sanderson et al., 2020). Nonetheless, the development of probes for target enrichment may pose several challenges: first, the need to identify regions conserved enough to ensure recovery, yet polymorphic enough to provide ample information (Soto‐Gomez et al., 2019; Villaverde et al., 2018). Second, probe design requires detecting regions without pervasive copy number polymorphism (Kadlec et al., 2017; Lemmon et al., 2012), a particular challenge for angiosperms and other groups, where duplication events are ubiquitous (Van de Peer et al., 2017).
In contrast, universal kits offer an attractive alternative that require reduced efforts to establish, and provide comparable data sets across wider ranges of taxa (Johnson et al., 2019; Kadlec et al., 2017). Such kits were designed to retrieve single‐copy markers, for example, in the broad scope of amphibians (Hime et al., 2021), anthozoans (Quattrini et al., 2018), vertebrates (Lemmon et al., 2012) or angiosperms (Johnson et al., 2019). In the latter example, the Angiosperms353 kit is designed to target 353 single‐copy genes across angiosperms. So far the kit has been employed successfully in resolving phylogenies, including but not limited to Nepenthes (Murphy et al., 2020), Schefflera (Shee et al., 2020) and the rapid radiations of Burmeistera (Bagley et al., 2020) and Veronica (Thomas et al., 2021), establishing the kit as an eminent tool in macroevolutionary research. Its utility at microevolutionary levels is yet to be fully realized, although several works have established its suitability to deliver informative signals at a lower taxonomic level (Beck et al., 2021) and in acquiring population genomics parameters (Slimp et al., 2021). The use of highly conserved markers in a universal kit may, however, limit resolution power. Generally, taxon‐specific baits are expected to deliver a higher information content and hence more accurate results (Kadlec et al., 2017), as enrichment success is known to drop with the level of divergence between sequences used for probe design and the targeted taxa (Liu et al., 2019). However, one study comparing the power of the universal Angiosperms353 kit and a taxon‐specific kit to resolve phylogenomic relationship in Cyperaceae reported surprisingly similar performance (Larridon et al., 2020) and similar findings were reported in Malinae (Ufimov et al., 2021) and in Ochnaceae (Shah et al., 2021). It remains to be established whether these findings apply to other taxa and other evolutionary scales, including at population level, where ample genomic variability is required to resolve intraspecific relationships and investigate patterns of genetic differentiation.
Until recently, the technology available to investigate evolutionary questions in rapidly evolving groups featuring high net diversification rates has presented major obstacles, in particular for nonmodel groups. Decreasing costs of sequencing coupled with an ever‐growing plethora of bioinformatic tools for data processing and downstream analysis has led to an increase in the use of methods like whole‐genome sequencing, RNA sequencing and restriction‐site associated DNA sequencing (RAD‐Seq) in lieu of traditional methods employing few conserved markers (de La Harpe et al., 2017; McKain et al., 2018; Weitemier et al., 2014; Zimmer & Wen, 2013). Whole‐genome sequencing however remains costly, posing barriers for research targeting large numbers of samples, organisms with large genomes and nonmodel organisms for which the availability of high‐quality genomic resources is often limited (Hollingsworth et al., 2016; Supple & Shapiro, 2018). While RAD‐seq is an affordable alternative and widely used in population genetics, the resulting data sets may fall short when screened for homologous sequences across distantly related lineages (but see, e.g., Heckenhauer et al., 2018). Additionally, RAD‐seq is less feasible when using degraded DNA from herbarium samples, and the use of short and inconsistently represented loci across phylogenetic sampling may result in low information content and difficulties in assessing paralogy (Jones & Good, 2016; Lemmon & Lemmon, 2013; McKain et al., 2018).
Rapid evolutionary radiations are key stages in the evolutionary history across the Tree of Life and highly recurrent, hence an essential part of biodiversity research (Gavrilets & Losos, 2009; Givnish et al., 2014; Hughes et al., 2015; Soltis et al., 2019; Soltis & Soltis, 2004). Fast evolving groups provide potent opportunities to investigate important questions in evolutionary biology, such as the interplay between ecological and evolutionary processes in shaping biodiversity. A few notable study systems are the cichlid fish (McGee et al., 2020; Salzburger, 2018), Heliconius butterflies (Dasmahapatra et al., 2012; Moest et al., 2020), Anolis lizards (McGlothlin et al., 2018; Stroud & Losos, 2020), Darwin's finches (Lamichhaney et al., 2015; Zink & Vázquez‐Miranda, 2019), white‐eyes birds (Moyle et al., 2009) and New World lupins (Nevado et al., 2016). Nevertheless, much remains unknown about the genomic basis underlying species diversification outside these intensively studied systems.
Research of rapidly diversifying lineages presents several challenges. First, a brief diversification period typically leads to imperfect reproductive barriers and incomplete lineage sorting, reflected in significant gene tree discordance and ambiguous relationships (Degnan & Rosenberg, 2009; Lamichhaney et al., 2015; Pease et al., 2016; Straub et al., 2014). In addition, understanding ‘speciation through time’ poses a methodological challenge and requires connecting two conceptual worlds: macroevolutionary investigations, concerned with spatial and ecological patterns over deeper timescales, and microevolutionary approaches, providing insight into the processes acting during population divergence and speciation (Bragg et al., 2016; de La Harpe et al., 2017). Resolving phylogenomic relationships and disentangling the contribution of different genomic processes through time typically require large‐scale genomic data sets and thorough taxon sampling efforts (Lemmon & Lemmon, 2013; Linder, 2008; Straub et al., 2012).
Here, we present Bromeliad1776, a new bait set for targeted sequencing, designed to address a wide range of evolutionary hypotheses in Bromeliaceae: from producing robust phylogenies to studying the interplay of genomic processes during speciation and the genetic basis of trait shifts, such as photosynthetic and pollination syndrome. This highly diverse Neotropical radiation provides an excellent research system for studying the drivers and constraints of rapid adaptive radiation (Benzing, 2000; Givnish et al., 2011; Loiseau et al., 2021; Mota et al., 2020; Palma‐Silva & Fay, 2020; Wöhrmann et al., 2020). Bromeliaceae as a whole is considered an adaptive radiation (Benzing, 2000; Givnish et al., 2011) and contains several rapidly radiating lineages, most notably within Bromelioideae (Aguirre‐Santoro et al., 2020) and Tillandsioideae (Loiseau et al., 2021). It is a species‐rich and charismatic monocot family, consisting of over 3000 species, including crops in the genus Ananas and other economically important species (Luther, 2008). Members of the family are characterized by a distinctive leaf rosette that often impounds rainwater in central tanks (phytotelmata). A diversity of arthropods and other animal species and microbes reside in bromeliad tanks, in some cases even leading to protocarnivory and other forms of nutrient acquisition (Givnish et al., 1984; Leroy et al., 2016). Bromeliads present a diversity of repeatedly evolving adaptive traits, which allowed them to occupy versatile habitats and ecological niches (Benzing, 2000). CAM photosynthesis, water‐absorbing trichomes, formation of tank habit, extensive rates of epiphytism and a diversity of pollination syndromes are some of the adaptations correlated with high rates of diversification within the family (Benzing, 2000; Crayn et al., 2004; Givnish et al., 2014; Kessler et al., 2020; Quezada & Gianoli, 2011).
To assess the utility of the Bromeliad1776 kit, we performed a comparison between our taxon‐specific kit and the universal Angiosperms353 kit using several methods across different evolutionary timescales. We present Bromeliad1776 in the light of methodological considerations on bait design, data handling, analyses and other practical considerations.
2. MATERIALS AND METHODS
2.1. Custom bait design
Whole‐genome sequences and gene models from Ananas comosus v.3 (Ming et al., 2015) were used to design a bait set aiming to target (i) single‐copy protein coding genes distributed across the whole genome, (ii) genes previously described as associated with key innovation traits in Bromeliaceae (see below), (iii) markers previously used for phylogenomic inference in Bromeliaceae and (iv) genes orthologous to those in the Angiosperms353 bait set. The 1776 selected genes are detailed in Table S1.
Genes in subset i were selected based on genetic diversity parameters calculated using whole‐genome sequence and RNAseq data previously published by de La Harpe et al. (2020); data publicly available online at SRA Bioproject (PRJNA649109) with the popgenome R package v.2.1.6 (Pfeifer et al., 2014). Genomic regions were retained in this category if they shared at least 70% identity between A. comosus and T. sphaerocephala, and if they had nucleotide diversity (π) values not exceeding the 90% quantile of the (π) distribution across genes for four Tillandsia species (Tillandsia australis, Tillandsia fasciculata, Tillandsia floribunda and T. sphaerocephala; data and analysis performed by de La Harpe et al. (2020)). We further excluded genes with a total exonic size smaller than 1100 bp, or individual exons smaller than 120 bp. Next, copy number variation was calculated based on clustering of A. comosus and Tillandsia transcriptome assemblies to generate three copy number categories—’single copy’, ‘low copy’ (i.e. less than five copies) and ‘high copy’ (i.e. five or more copies). We included only single‐copy genes in the design for bait subset i. Finally, we excluded genes that were located in genomic regions outside those assigned to linkage groups in the A. comosus reference (Ming et al., 2015). A total of 1243 genes were identified for this part.
The bait subset of genes associated with key innovative traits in Bromeliaceae (subset ii above) included (1) genes putatively under positive selection along branches relevant to C3/CAM shifts (de La Harpe et al., 2020), (2) genes that exhibit differential gene expression between CAM and C3 Tillandsia species (de La Harpe et al., 2020) and (3) genes putatively associated with photosynthetic and developmental functions, or with flavonoid and anthocyanin biosynthesis, according to the literature (e.g. Goolsby et al., 2018; Ming et al., 2015; Palma‐Silva et al., 2016; Wai et al., 2017). Ananas comosus genes with the highest match scores (calculated as lowest E‐score in BLASTP, Madden (2003)) against the sequences of genes from the literature were added to the bait set (see Table S2 for details). A total of 1612 genes underpinning innovative traits were included in the bait design, regardless of criteria used for subset i for size, similarity and duplication rate.
Markers previously used for phylogenomic inference in Bromeliaceae (subset iii) were obtained from the literature, spanning 13 genes (e.g. Barfuss et al., 2016; Machado et al., 2020; Schulte et al., 2009, see TS2 for full list). Genes orthologous to those in the Angiosperms353 bait set (Johnson et al., 2019) were identified using the orthologous gene models from A. comosus based on gene annotations (Ming et al., 2015) or using BLASTP (Madden, 2003), totalling 281 genes.
Finally, we used a draft genome of T. fasciculata (Jaqueline Hess, personal communication) to exclude from all candidates genes that exhibited multiple BLASTN hits, if they have not been previously described as duplicated within the genus (de La Harpe et al., 2020). Specifically, we excluded genes that matched another genomic sequence of at least 100 bp with high similarity score (>80%) and low E‐value (<10−5). In an additional round of filtering performed by the manufacturer of the final bait set, Arbor Biosciences (Ann Arbor, MI, USA), multicopy genes with sequences that are more than 95% identical were collapsed into a single sequence and baits with more than 70% GC content or containing at least 25% repeated sequences were excluded. In addition, targets including exons smaller than 80 bp were completed with regions flanking the exons according to the A. comosus reference genome. The final kit included 1776 genes: 801 genes in subset i, 681 genes associated with key innovative traits, 13 genes representing phylogenetic markers and 281 genes orthologous to the Angiosperms353 set. Probes were designed with 57,445 80‐mer baits tiling across targets in 2× coverage, targeting approximately 2.3 Mbp. The kit is subsequently referred to as the Bromeliad1776 bait set. Further specifications can be found in Tables S1 and S2 and in the github repository: https://github.com/giyany/Bromeliad1776/tree/main/MS_2021_scripts.
2.2. Plant material collection
We sampled a total of 70/72 Bromeliaceae samples (for Angiosperms353 and for Bromeliad1776, accordingly; Table S3), including 56 accessions from the Tillandsioideae subfamily and 16 representing the other subfamilies, except Navioideae. The divergence time between Tillandsioideae and subfamily Bromelioideae to which A. comosus belongs is estimated at 15 Mya (according to Givnish et al., 2014). Within Tillandsioideae, we sampled 38/40 individuals from five species of the Tillandsia subgenus Tillandsia (‘clade K’ in Barfuss et al., 2016; Sampling in Mexican populations illustrated in Figure S1).
2.3. Library preparation & enrichment
DNA extractions were performed using a modified CTAB protocol (Doyle & Doyle, 1987), purified using Nucleospin® gDNA cleanup kit from Macherey‐Nagel (Hudlow et al., 2011) following the supplier's instructions with a twofold elution step and finally quantified with Qubit® 3.0 Fluorometer (Life Technologies).
For each sample, 200 ng DNA was sheared using Bioruptor® Pico sonication device (Diagenode) aiming for an average insert size of 350 bp, dried in a speed vacuum Eppendorf concentrator 5301 (Eppendorf) and eluted in 30 L ddH2O. Genomic libraries were prepared using the NEBNext® Ultra TM II DNA Library Prep Kit for Illumina® (New England Biolabs) using reagents at half volumes following Hale et al. (2020) and using 11 PCR cycles, increased up to 13 cycled for libraries with low genomic output. Samples were double‐indexed with NEBNext® Multiplex Oligos for Illumina® (New England Biolabs). Fragment sizes were inspected with Agilent Bioanalyzer (Agilent Technologies), and concentrations were measured with Qubit® 3.0 Fluorometer. Subpools of 11–14 equimolar genomic libraries were prepared using phylogenetic proximity and DNA concentrations of the genomic libraries, which ranged from 2.62 to 118.0 ng/L, following Soto‐Gomez et al. (2019).
We used the Angiosperms353 and the Bromeliad1776 bait sets from Arbor Biosciences to enrich each subpool of genomic libraries independently with a single hybridization reaction of myBaits® target capture kits from Arbor Biosciences, following Hale et al. (2020). Average fragment size and DNA yield were estimated for each subpool using Agilent Bioanalyzer and Qubit® 3.0 Fluorometer. Subpools were then pooled in equimolar conditions and sequenced at Vienna BioCenter Core Facilities on Illumina® NextSeq™ 550 (2 × 150 bp, Illumina). Sequencing was conducted independently for either bait kit.
2.4. Data processing
The raw sequence data in BAM format were demultiplexed using deml v.1.1.3 (Renaud et al., 2015) and samtools view v.1.7 (Li et al., 2009), converted to fastq using bamtools v.2.4.0 (Barnett et al., 2011) and quality checked using fastqc v.0.11.7 (Andrews, 2010). Reads were then trimmed for adapter content and quality using trimgalore v.0.6.5 (Krueger, 2019), a wrapper tool around fastqc and cutadapt, using settings ‐‐fastqc ‐‐retain unpaired. Sequence quality and adapter removal were confirmed with FastQC reports.
Quality and adapter‐trimmed reads were aligned to A. comosus reference genome v.3 (Ming et al., 2015) using bowtie2 (Langmead & Salzberg, 2012) with the ‐‐very‐sensitive‐local option to increase sensitivity and accuracy. Samtools (Li et al., 2009) was then used to remove low‐quality mapping and sort alignments by position, and PCR duplicates were marked using MarkDuplicates from picardtools v.2.25 (Picard Toolkit, 2019). Summary statistics of the mapping step were generated using samtools stats. Variants were called using freebayes v1.3.2‐dirty (Garrison & Marth, 2012), and sites marked as MNP/complex were decomposed and normalized using the script ‘vcfallelicprimitives’ from vcflib (Garrison, 2012). Next, AN/AC field was calculated using bcftools v.1.7 (Li, 2011) and variant calls were filtered using vcflib (Garrison & Marth, 2012) and bcftools. Given that freebayes does not perform automatic variant filtering steps, we identified sets of parameters that generate reliable final SNP sets, based on two independent criteria: the highest transition/transversion ratios as reported by snpsift (SnpEff Cingolani et al., 2012) and the lowest π N/π S (see Section 2.7). After a detailed evaluation, we used the following criteria to generate two high‐quality SNP sets, one for each bait set: we considered genotype calls with per‐sample coverage below 10×—as missing (NA) and excluded variants (i) marked as indels or neighbouring indels within a distance of 3 bp, (ii) with depth of coverage at the SNP level lower than 500×, (iii) with less than 10 reads supporting the alternate allele at the SNP level or (iv) with more than 40% missing data. All genes in the Bromeliad1776 that passed the filtering criteria were included in the SNP set, regardless of their function. Summary statistics of the final SNP sets were generated using the script vcf2genocountsmatrix.py, namely the total number of SNPs, the proportion of on‐target SNPs and the proportion of SNPs in some specific genomic contexts, with A. comosus genome v.3 as a reference. The full data processing script align_and_trim.sh and the vcf2genocountsmatrix.py script are both available at https://github.com/giyany/Bromeliad1776.
2.5. Bait specificity and efficiency
To explore bait specificity, we calculated the percentage of high‐quality trimmed reads on‐target using samtools stats and bedtools intersect v2.25.0 (Quinlan & Hall, 2010) using the script calculat_bait_target_specifity.sh (available from https://github.com/giyany/Bromeliad1776). Targets for Bromeliad1776 were defined as the bait sequences plus their 500‐bp flanking regions. Targets for Angiosperms353 were defined using orthogroups to A. comosus: gene annotations from the bait set were used to assign genes to orthogroups using orthofinder (Emms & Kelly, 2019). When several orthogroups were found for a single Angiosperms353 gene, we included all, resulting in 559 A. comosus genes assigned to orthogroups. Within the orthogroups, targets were again defined as exonic regions plus their 500 bp flanking regions.
To provide insights into determinants of bait capture success, we calculated bait efficiency for all baits of Bromeliad1776. For each bait, efficiency was calculated as the number of high‐quality reads uniquely mapping to each bait target region, averaged over samples. We then tested for the correlation of capture efficiency to several bait characteristics (copy number, GC content, number and size of exons in targeted gene, size of baits and phylogenetic distance to A. comosus) with a generalized linear model or Kruskal–Wallis test in r v.4.0.3 (R Core Team, 2020) using a negative binomial family.
2.6. Phylogenomic analyses
We inferred phylogenomic relationships for all samples using two methods: a concatenation method, and a coalescent‐based species tree estimation. The latter method was included as concatenation methods do not account for gene tree incongruence, which may result in high support for an incorrect topology (Kubatko & Degnan, 2007), especially in the presence of notable incomplete lineage sorting. In addition, gene tree incongruence analysis provides insight into molecular genome evolution, including the extent of incomplete lineage sorting and other genomic processes such as hybridization and introgression (Galtier & Daubin, 2008; Wendel & Doyle, 1998).
We used the variant and nonvariant genotypes to create a phylip matrix with vcf2phylip v.2.0 (Ortiz, 2019) and constructed a maximum‐likelihood species tree for each bait set with raxml‐ng v.0.9.0 (Kozlov et al., 2019), using 250 bootstrap replicates and a GTR model with an automatic MRE‐based bootstrap convergence test. Next, we constructed a species tree using astral‐iii v.5.7.7 (hereafter: ASTRAL, Zhang et al., 2018). For both the Angiosperms353 and the Bromeliad1776 sets, we separated the matrix into independent genomic windows, defining each window as a gene according to the known exons and a 500‐bp flanking region. For Angiosperms353, we extracted the 559 genes (assigned to orthogroups as explained above) as genomic windows using bedtools intersect. For Bromeliad1776, genomic windows were extracted using the A. comosus gene sequences included in bait design. All loci and all accessions were included in species tree inference regardless of the percentage of missing data, since taxon completeness of individual gene trees is important for statistical consistency of this approach, and we expected only low levels of fragmentary sequences (Mirarab, 2019; Nute et al., 2018). After excluding genes with zero coverage, 269 genes and 1600 genes were included in species tree inference for Angiosperms353 and Bromeliad1776, respectively.
For each gene, a maximum‐likelihood gene tree was inferred using pargenes (Morel et al., 2019) with raxml‐ng (Kozlov et al., 2019), using a GTR model with an automatic MRE‐based bootstrap convergence test. Loci with insufficient signal may reduce the accuracy of species tree estimation (Mirarab, 2019), hence, in all gene trees, nodes with a bootstrap support smaller than 10 were collapsed using Newick utilities (Junier & Zdobnov, 2010). A species tree was then generated in ASTRAL with quartet support and posterior probability for each tree topology. The number of conflicting gene trees was calculated using phyparts and visualized using the script phypartspiecharts.py (available from https://github.com/mossmatters/MJPythonNotebooks).
2.7. Population structure and nucleotide diversity estimates
To explore the genetic structure within the Tillandsia species complex, we focused on five species from 15 localities (Table S3 and Figure S1). We first used plink v.1.9 (Chang et al., 2015) to filter out SNPs in linkage disequilibrium. Population structure was further explored through individual ancestry analysis, with identity‐by‐descent matrix calculated by plink and inference of population structure using admixture v.1.3. with K values ranging from one to ten, and 30 replicates for each K, using a block optimization method (Alexander & Lange, 2011). A summary of the admixture results was obtained and presented using pong (Behr et al., 2016). The set of LD‐pruned biallelic SNPs was further filtered to allow a maximum of 10% missing data and used to perform a principal components analysis (PCA) with snprelate v.1.20.1 (Zheng et al., 2012). Finally, for each Tillandsia species, we used the strategy of Leroy et al. (2021) to compute synonymous (π S) and nonsynonymous (π N) nucleotide diversities and Tajima's D, from fasta sequences using seq_stat_coding (Leroy et al., 2021).
3. RESULTS
3.1. Higher mapping rates and capture efficiency for taxon‐specific set
On average, 4,401,958 (803,464–12,693,516) paired‐end reads per accession were generated per Angiosperms353 library and 2,962,023 (1,282,762–6,298,880) per Bromeliad1776 library. Overall, the mapping rates to the A. comosus reference genome were higher for libraries enriched with Bromeliad1776, with an average mapping rate of 82.3% (61.8%–95.9%) and 42.8% (22.1%–77.9%), for Bromeliad1776 and Angiosperms353, respectively (Figure S2, Table S4). Higher mapping rates were recorded for subfamilies Bromelioideae and Puyoideae, as compared to Tillandsioideae, for both the Angiosperms353 and Bromeliad1776 sets (see Figures S3 and S4, respectively). This may reflect the effect of reference bias, and in the case of Bromeliad1776, it may be further amplified by our kit design based on A. comosus (subfamily Bromelioideae). Bait specificity was high for Bromeliad1776 with on average 90.4% reads on‐target (76.5%–94.2%), while for Angiosperms353 bait specificity was 14.0% (4.6%–30.1%; see Figure S2). Mapping rates and bait specificity were positively correlated for both bait sets (GLM, p < .01).
3.2. Bait efficiency depends on the genomic context
We investigated factors that may influence bait efficiency, starting with the contribution of gene copy number variation. We assumed three categories regarding the number of paralogs per orthogroup: single‐copy, low‐copy (i.e. less than five copies) and high‐copy (i.e. five or more copies). The number of gene copies had a significant effect on bait efficiency and post hoc Dunn's test supported significant differences in efficiency for comparisons between low‐copy and high‐copy, and between single‐copy and low‐copy (P = 2.8−44). Low‐copy genes exhibit the lowest enrichment success, suggesting that the bait efficiency is not simply correlated with the number of gene copies (Figure 1). We also recovered a significant effect of the intragenic GC content and GC content of the baits on bait efficiency (GLM, P = 1.5−68). Finally, we investigated the possible link between efficiency and gene structure. Average exon sizes (P < 2.0−16) and total number of exons per gene (P = 1.1−89) were also positively correlated with enrichment success. The size of the smallest exon for all targeted genes was however not correlated with bait efficiency. Sequence similarity, measured as per cent of identity between Tillandsia sequences and those of A. comosus, was positively correlated with capture success (P = 4.8−13; Figure 1).
FIGURE 1.
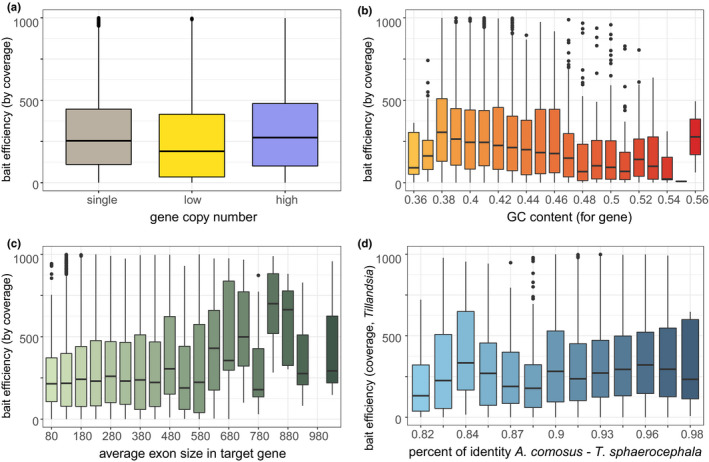
Effects of (a) putative gene copy number, (b) gene GC content, (c) average exon size, and (d) per cent of identity on bait efficiency in Bromeliad1776 bait set, measured as the number of high‐quality reads uniquely mapping to bait target region across samples. Continuous variable was binned and y‐values higher than 1000 excluded for visualization in b–d
3.3. Both kits provided a large number of SNPs
After variant calling and filtering, we identified 47,390 and 209,186 high‐quality SNPs for the Angiosperms353 and the Bromeliad1776 bait sets, respectively. On average, missing data represented 23.7% of genotype calls per individual in Angiosperms353, but only 6.3% for the Bromeliad1776 kit. The differences in amount of missing data are likely associated with the higher mean depth per site across the Bromeliad1776 kit (6602), as compared to Angiosperms353 (3437). Focusing on the subgenus Tillandsia, we identified 15,622 SNPs for Angiosperms353 (including a total of 18.9% missing data) compared to 65,473 polymorphic sites (2.9% missing data) for Bromeliad1776. In both full data sets and the subset including only Tillandsia samples, Bromeliad1776 recovered more variants in intronic regions compared with Angiosperms353. Angiosperms353 recovered a large proportion of off‐target SNPs, whereas in Bromeliad1776 approximately 15% of the SNPs were recovered from flanking regions (Table 1). We discuss ascertainment bias that may rise due to the nonrandom selection of markers in the supporting information.
TABLE 1.
Number and characteristics of the variants obtained for Angiosperms353 and Bromeliad1776
| indv Nr. | SNP Nr. | Site mean depth | SNPs in exonic regions | SNPs in intronic regions | SNPs in intergenic regions | On‐target SNPs | Flanking SNPs | Off‐target SNPs | |
|---|---|---|---|---|---|---|---|---|---|
| intragenic vcf | |||||||||
| Angiosperms353 | 70 | 47,390 | 3447 | 40,628 (85.7%) | 4376 (9.2%) | 2386 (5.1%) | 8424 (17.8%) | 3488 (7.4%) | 35,478 (74.8%) |
| Bromeliad1776 | 72 | 209,186 | 6601.7 | 170,893 (81.7%) | 35,790 (17.1%) | 2503 (1.2%) | 162,924 (77.9)% | 37,661 (18.0%) | 8601 (4.11%) |
| pop‐level vcf | |||||||||
| Angiosperms353 | 38 | 15,622 | 1837.8 | 13,345 (85.5%) | 1442 (9.2%) | 835 (5.3%) | 3032 (19.4%) | 1129 (7.22%) | 11,461 (73.4%) |
| Bromeliad1776 | 40 | 65,473 | 3914.9 | 54,636 (83.5%) | 9967 (15.2%) | 870 (1.3%) | 51,405 (78.5%) | 10,588 (16.2%) | 3480 (5.3%) |
3.4. Similar phylogenomic resolution in concatenation method, Bromeliad1776 outperforms Angiosperms353 for species tree reconstruction
The Angiosperms353 and Bromeliad1776‐based maximum‐likelihood phylogenetic trees recovered the same backbone phylogeny of Bromeliaceae, clustering subfamily Tillandsiaoedeae and the subgenus Tillandsia with high bootstrap values (Figure S5). Neither set obtained high support for interpopulation structure for Tillandsia gymnobotrya, but highly supported nodes separated T. fasciculata accessions from Mexico and from other locations, and the populations of T. punctulata for the Bromeliad1776 data set were similarly separated. The tree topologies were identical, with the notable exception of the placements of Tillandsia biflora and Racinaea ropalocarpa and the genus Deuterocohnia (Figure S5, purple arrow). Overall, internal nodes are strongly supported for both sets, except for Hechtia carlsoniae as sister to Tillandsioideae, which is poorly supported for both sets. While several internal nodes are slightly less supported for the Angiosperms353 set, overall these results demonstrate the efficacy of both kits in phylogenomic reconstruction using concatenation approaches, indicating that as few as 47 k SNPs within variable regions provide reliable information to resolve phylogenetic relationships within the recent evolutionary radiation of Tillandsia.
Species trees as inferred with ASTRAL for both data sets likewise provided an overall strong local posterior support (Figure 2, see also Figures S8 and S9). Several nodes however exhibit lower local posterior support values for the Angiosperms353 tree than for the Bromeliad1776 tree. The topology for the Bromeliad1776 ASTRAL tree was similar to the ML tree, but differed again by placing Deuterocohnia as sister taxa to Puyoideae only. In the Angiosperms353 tree, the topology differed from both ML trees and the ASTRAL Bromeliad1776 tree in several nodes. H. carlsoniae was placed as a sister taxa to all other subfamilies in the Angiosperm353 phylogeny. Notably, the placement of Catopsis and Glomeropitcrania differed, as well as the placement of Cipurosis subandinai, T. biflora and R. ropalocarpa. Several internal nodes were poorly supported, such as the node separating the tribe Catopsideae and core Tillandsioideae, and the nodes separating Tillandsioideae from all other subfamilies. The differences in topology between the Angiosperms353 ASTRAL tree to all other trees (ML trees and Bromeliad1776 ASTRAL tree) together with the low posterior support suggest lower resolution power and a poor fit of this data set for resolving a species tree.
FIGURE 2.
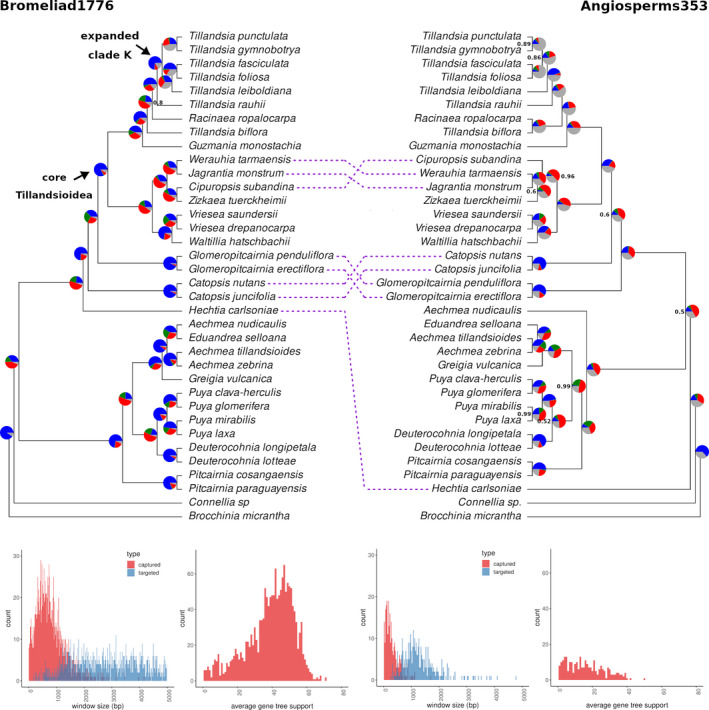
Coalescent‐based species trees generated ASTRAL‐III for samples enriched with Bromeliad1776 (left) and Angiosperms353 (right, flipped for mirroring), on 269 and 1600 genes for each set, respectively. Node values represent local posterior probabilities (pp) for the main topology and are equal to 1 unless noted otherwise. Pie charts at the nodes show levels of gene tree discordance: the percentages of concordant gene trees (blue), the top alternative bipartition (green), other conflicting topologies (red) and uninformative gene trees (grey). At bottom, length and average bootstrap support for gene trees from either data set, according to the design of the bait set used for enrichment: Angiosperms353 (right) and Bromeliad1776 (left). Each gene was considered a single genomic window
The length and average size of the input gene trees different among sets, with average window length of 304.6 bp and 819.9 bp and average gene tree support of 16.9 and 38.9 for Angiosperms353 and Bromeliad1776 bait sets, respectively (Figure 2). An examination of gene tree concordance constructed with Bromeliad1776 data set allowed us to identify variable levels of gene tree conflict among nodes (Figure 2). Gene tree discordance was especially high for the split between Tillandsioideae and other subfamilies, as well as for the split between Puyoideae and taxa assigned to Bromelioideae. Furthermore, gene tree discordance and the proportion of uninformative gene trees were especially high for splits among clades within the K.1 and K.2 clades of subgenus Tillandsia. A similar analysis with Angiosperms353 yielded evidence for gene tree discordance, but a considerable number of gene trees were reported to be noninformative (grey part of the pie charts), especially within subgenus Tillandsia (Figure 2).
3.5. Strong interspecific structure, but little evidence for within‐species population structure
After LD‐pruning and retaining maximum 10% missing data, 1025 and 32,941 biallelic SNPs were included for the Tillandsia PCA analysis of the Angiosperms353 and Bromeliad1776 data sets, respectively. Overall, both data sets provided evidence for interspecific structure, but not for population structure, with Bromeliad1776 resulting in border‐line higher resolution (slightly better separating T. foliosa from T. fasciculata). The percentage of explained variance was higher in the Bromeliad1776 set (19.3% and 16.5% for PC1 and PC2) as compared to the Angiosperms353 data set (14.5% and 11.8%, see Figures 3 and S6). Based on these two PCAs, we found no evidence for spatial genetic structure within each species, since accessions did not cluster by geographic origin on the two PCs presented, or any other PCs we investigated (see Figure S6).
FIGURE 3.
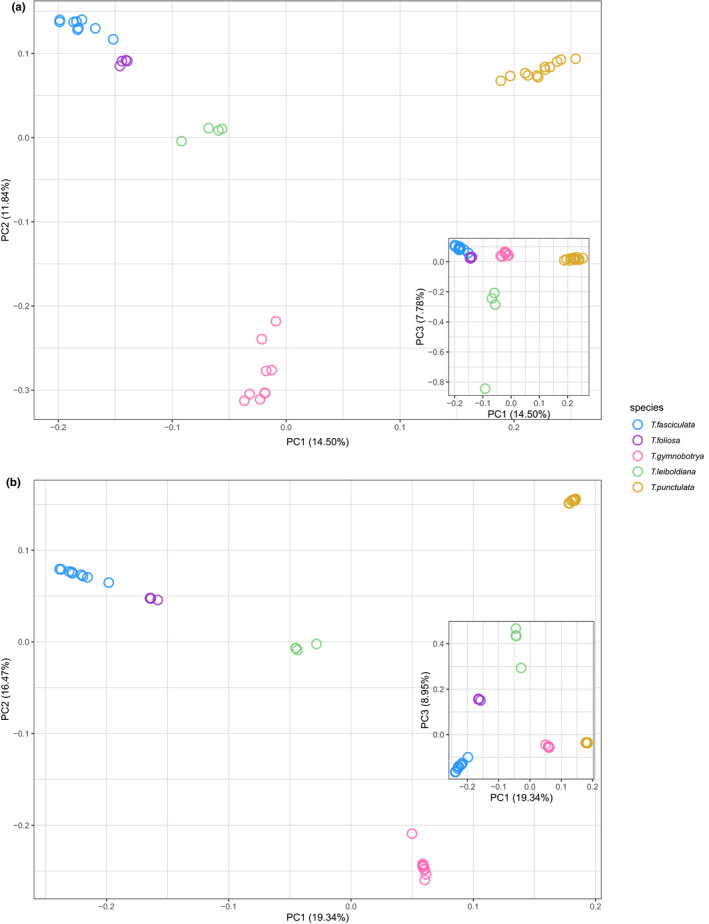
Principal component analysis (PCA) plot for samples of Tillandsia subgenus Tillandsia enriched with two bait sets: (a) Angiosperms353 (1025 variants); (b) Bromeliad1776 (32,941 variants). Colours indicate different species according to legend
In addition to PCA, we performed admixture analyses based on 9804 and 42,613 variants for the Angiosperms353 and Bromeliad1776 sets, respectively (Figure 4). We used a cross‐validation strategy to identify the best K and found clear support for K = 5 for the Bromeliad1776 set (Figure S7). In contrast, the CV pattern for the Angiosperms353 set varied widely, providing limited information about the best K. Lowest CV values were however observed for K = 9 with locally low values for K = 5 and K = 3 (Figure S7). We further investigated the admixture bar plots at different values of K. For K = 5, very similar patterns can be observed for both sets, with the recovered clusters reflecting the expected species boundaries. The main difference between the two data sets was the ability of the Bromeliad1776 set to reach a more consistent solution (‘consensus’) among 30 runs, especially at large K, as compared to the runs based on the Angiosperms353 bait set. The Bromeliad1776 was also able to distinguish between different sampling localities of T. punctulata and of T. fasciculata at K = 7–8 (Figure 4).
FIGURE 4.
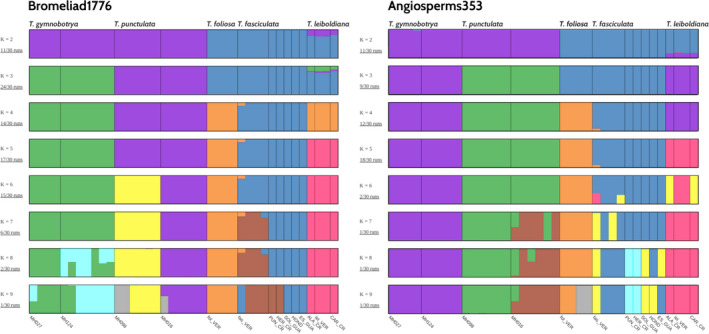
Population structure of 5 Tillandsia subgenus Tillandsia species from 14 sampling locations inferred with the ADMIXTURE software. Samples were enriched with either of two bait sets: Angiosperms353 (9804 variants after LD‐pruning) and Bromeliad1776 (42,613 variants after LD‐pruning), showing values of K = 2 to K = 9. Colours represent genetically differentiated groups, while each accession is represented by a vertical bar
3.6. Distinct diversities hint at different demographic processes
Nucleotide diversity estimates were calculated for the Bromeliad1776 data set only, due to difficulties obtaining a reliable SNP set with Angiosperms353 (see Section 2.4). Averaged levels of nucleotide diversity at synonymous sites π S greatly varied among species, from to for T. foliosa and T. fasciculata, respectively (Table S5; Figure 5). Given the recent divergence of these different species and their roughly similar life history traits, they are expected to share relatively similar mutation rates; hence, the observed differences in π S are expected to translate into differences of long‐term N e. Looking at the distribution of π S across genes, we found broader or narrower distributions depending on the species, which explains the observed differences in averaged π S, as typically represented by the median of the distribution (vertical bars, Figure 5). Most species exhibit distributions of Tajima's D (Figure 5) that are centred around zero, with the notable exception of T. punctulata. The distribution of this species is shifted towards positive Tajima's D values, therefore indicating a recent population contraction, suggesting that this species experienced a unique demographic trajectory as compared to the other species.
FIGURE 5.
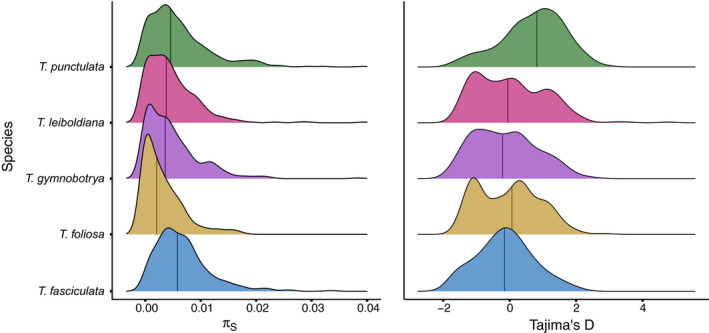
Distribution of Tajima's D and synonymous (π S) nucleotide diversity within each species for the Bromeliad1776 kit
4. DISCUSSION
4.1. A taxon‐specific bait set performs marginally better for phylogenomics
In this study, we compared the information content and performance of a taxon‐specific bait set and a universal bait set for addressing questions on evolutionary processes at different scales in a highly diverse Neotropical plant group, including recently radiated clades. We found that the taxon‐specific kit provided a greater number of segregating sites, yet contrary to our expectations, the abundance of information content did directly translate to a greater resolution power.
The universal and taxon‐specific sets performed comparably when investigating macroevolutionary patterns: the inferred species trees are remarkably consistent between the two bait sets (Figures 2 and S5). Notably, both sets were sufficiently informative to reconstruct the relationships among the fastest radiating clades. These results resonate with previous comparative works (e.g. in Burmeistera, Bagley et al., 2020; in Buddleja, Chau et al., 2018; and in Cyperus, Larridon et al., 2020), where taxon‐specific markers provided higher gene assembly success, but a comparable number of segregating sites for phylogenetic inference, indicating that universal bait sets are nearly as effective as taxon‐specific bait sets, even in fast evolving taxa. The main advantage of the bromeliad taxon‐specific set is its ability to provide additional resolution for deeper examination of gene tree incongruence (Figure 2), currently a fundamental tool in phylogenomic research (Edwards, 2009; Morales‐Briones et al., 2021; Pease et al., 2016).
The taxon‐specific bait set performed marginally better to address hypotheses at more recent evolutionary scales and provided arguably clearer evidence for inference of species genomic structure using clustering methods. In fact, genetic markers obtained from both data sets provided sufficient information to infer species but no geographic structure, suggesting that Tillandsia could be characterized by high gene dispersal among populations. Considering that the Angiosperms353 kit has shown potential to provide within‐species signal, as recently demonstrated by Beck et al. (2021) on Solidago ulmifolia, and to estimate demographic parameters from herbarium specimen (Slimp et al., 2021), we would expect the taxon‐specific set to accurately reveal a geographical genetic structure. However, the present study is generally based on small sample sizes per species (n = 4–8), mostly sampled within a limited geographic range, limiting our ability to draw robust conclusions on the levels of intraspecific population structure.
The Bromeliad1776 kit provided a substantially larger number of segregating sites (more than 200 k vs. 47 k in Angiosperms353; Table 1, Figure S2) due to higher enrichment success, following the expectation for higher sequence variation in custom‐made loci (Figure 1, see also Bragg et al., 2016; de La Harpe et al., 2019; Kadlec et al., 2017). We accordingly found that rates of molecular divergence are distinctly correlated with enrichment success in our sampling (Figure 1), following the expectation that a universal kit will provide fewer segregating sites.
However, the difference in resolution power between the kits cannot be ascribed solely to the different numbers of SNPs, but rather to the length and variability of the obtained regions. The topology obtained with the Angiosperm353 data set under the multispecies coalescent model was substantially different from all other inferred trees and the input gene trees provided a low power to detect patterns of gene tree discordance (Figure 2). We additionally observed that the highly conserved regions targeted by Angiosperms353 are shorter in comparison to Bromeliad1776 targets and thus result in shorter input windows for species tree inference (Figure 2). Hence, the patterns of gene tree discordance in the Angiosperms353 data set likely indicate incorrect gene tree estimation or other model misspecifications, rather than a biological signal. Specifically, coalescence‐based methods are sensitive to gene tree estimation error (Zhang et al., 2018) and perform better with gene trees estimated from unlinked loci long enough and variable enough to render sufficient signal per gene tree—this is especially true for data sets with many taxa. The high rates of uninformative genes trees, found in almost half of the intergenic nodes in the Angiosperms353 data set, are expected with increased levels of gene tree error, which in turn reduce the accuracy of ASTRAL (Mirarab, 2019; Sayyari & Mirarab, 2016). In contrast, the Bromeliad1776 ASTRAL tree (Figure 2, left and Figure S9) resolved phylogenetic relationships among taxa with high posterior probability and a topology similar to the ML tree. Gene tree discordance analysis revealed high incongruence around certain nodes, possibly reflecting rapid speciation events.
Since inference of phylogenetic relationships under the multispecies coalescent and exploration of gene tree discordance are both pivotal to phylogenomic research (Degnan & Rosenberg, 2009; Edwards et al., 2016; Pease et al., 2016), a taxon‐specific kit provides a clear advantage especially in recent rapid radiations, where gene tree conflict and incomplete lineage sorting are expected to be prevalent (Dornburg et al., 2019; Kubatko & Degnan, 2007; Roch & Warnow, 2015). In that regard, inference of the species tree with the Bromeliad1776 is a tool to drive further hypotheses concerning evolutionary and demographic processes in the evolution of Tillandsia. Moreover, the features of the loci targeted provide an important opportunity to study selection (see Section 4.3).
4.2. Insights on Bromeliaceae phylogeny and demographic processes in Tillandsia
Both bait sets resolved the phylogeny of Bromeliaceae, including the fastest evolving lineages of the subfamily Tillandsioideae. The results generally agreed with previous findings of the relationships among taxa (Givnish et al., 2011, 2014). Several findings that contrast with the expected known phylogeny may point at a complexity of genomic processes in the evolutionary history of Bromeliaceae subfamilies. Both the ML tree and species tree did not support a monophyly of the subfamily Pitcairnioideae, which was represented by four samples and two genera in our phylogeny: Deuterochonia and Pitcarnia. Rather, the genus Deuterochonia was sister to subfamily Puyoideae or sister to both Puyoideae and Bromelioideae subfamilies, inconsistent with the results of Barfuss et al. (2016) and Granados Mendoza et al. (2017). Interestingly, in a visualization of gene tree discordance we found high levels of incongruence and a high percentage of trees supporting an alternative topology in the node splitting the genera, indicating that several genomic processes such as hybridization and incomplete lineage sorting may have accompanied divergence in this group, contributing to the phylogenetic conflict and extending the challenges in resolving these evolutionary relationships. Within the core Tillandsioideae, the tribes Tillandsieae and Vrieseeae were found to be monophyletic, in accordance with previous work on the subfamily (Barfuss et al., 2016). Finally, within our focal group Tillandsia subgenus Tillandsia, clade K as suggested by Barfuss et al. (2016) and clades K.1 and K.2 as proposed by Granados Mendoza et al. (2017) were all well supported, further in agreement with their interpretation of Mexico and Central America as a centre of diversity for subgenus Tillandsia. Within Tillandsia, incongruence was prominent at the recent splits within clade K.1. and clade K.2 as expected in a recent rapid radiation, a result of high levels of incomplete lineage sorting, hybridization and introgression (Berner & Salzburger, 2015).
When applied to methods in population genetics, we obtained some evidence for a difference in demographic processes and in the level of genetic variation among species. This was especially true for the taxon‐specific bait set: for example, the bait set differentiated between populations of T. punctulata and T. fasciculata, but not T. gymnobotrya in a maximum‐likelihood tree and ancestry analysis (Figures 4 and S5), indicating differences in interpopulation genetic structure among species. The evidence for different demographic processes in these species extended to estimates of Tajima's D, where lower values may indicate a recent bottleneck. In addition, we found a unique distribution of nucleotide diversity for T. foliosa, possibly reflecting a low effective population size for this endemic species in contrast to the closely related, but widespread T. fasciculata. In all cases, our limited sampling given the large size of the family constrains our ability to draw conclusions of a ‘true’ phylogeny and to account for population structure. Our finding however suggests that nuclear markers obtained with a target capture technique can highlight genomic processes and be further applied to address questions in population genomics with a wider sampling scheme.
4.3. Future prospects and implications for research in Bromeliaceae and rapid radiations
Beyond the scope of this study, the availability of a bait set kit for Bromeliaceae provides a prime genetic resource for investigating several topical research questions on the origin and maintenance of Bromeliaceae diversity. Manifold studies of bromeliad phylogenomics set forth the challenges of resolving species‐level phylogenies with a small number of markers, particularly in young and speciose groups (Goetze et al., 2017; Granados Mendoza et al., 2017; Loiseau et al., 2021; Versieux et al., 2012). This particularly curated bait set allows highly efficient sequencing across taxa: within our study, we found high mapping success with 82.3% average read mapping. As expected, we documented a difference in enrichment success among taxa, explained by divergence time to the reference used for bait design (see Figure S4), suggesting possible deviations from the assumptions of nonrandomly distributed missing data that may mislead phylogenetic inference (Lemmon et al., 2009; Streicher et al., 2016; Xi et al., 2016). However, given the large enrichment success, downstream analysis with deliberate methodology can account for possible biases and provide robust inference with strict data filtering (Molloy & Warnow, 2018; Streicher et al., 2016). Hence, target enrichment with Bromeliad1776 can produce large data sets with consistent representation between taxa, allowing repeatability between studies and retaining the possibility for global synthesis by including sequence baits orthologous to the universal Angiosperms353 bait set. Moreover, with specific knowledge of the loci targeted in this set, the ability to obtain the same sequences across taxa and experiments and to differentiate genic regions with the use of A. comosus models, this bait set offers a broad utility for research in population genomics.
Another important feature in the Bromeliad1776 set is the inclusion of genes putatively associated with key innovative traits in Bromeliaceae with a focus on C3/CAM shifts. Little is known about the molecular basis of the CAM pathway, an adaptation to arid environments which evolved independently and repeatedly in over 36 plant families (Chen et al., 2020; Heyduk et al., 2019; Silvera et al., 2010). CAM phenotypes are considered key adaptations in Bromeliaceae, associated with expansion into novel ecological niches. In Tillandsia, C3/CAM shifts were found to be particularly associated with increased rates of diversification (Crayn et al., 2004; Givnish et al., 2014; de La Harpe et al., 2020). The Bromeliad1776 bait set offers opportunities to address specific questions on the relationship between rapid diversification and photosynthetic syndromes in this clade, including testing for gene sequence evolution. Additionally, the inclusion of multicopy genes, combined with newly developed pipelines for studying gene duplication and ploidy (Morales‐Briones et al., 2021; Viruel et al., 2019), is beneficial for studying the role of gene duplication and loss in driving diversification. With the increasing ubiquity of target baits as a genomic tool, we expect to see additional pipelines and applications emerging, further expanding the utility of target capture for both macro‐and microevolutionary research.
5. CONCLUSIONS
Even as whole‐genome sequencing becomes increasingly economically feasible, target capture is expected to remain popular due to its extensive applications in research. We found that evaluating the differences in resolution power between universal and taxon‐specific bait sets is far from a trivial task, and we attempted to lay out a methodological roadmap for researchers wishing to reconstruct the complex evolutionary history of rapidly diversifying lineages. While a taxon‐specific set offers exciting opportunities beyond phylogenomic and into research of molecular evolution, its development is highly time‐consuming, requires community‐based knowledge and may cost months of work when compared with out‐of‐the‐box universal kits. Our results suggest that universal kits can continue to be employed when aiming to reconstruct phylogenies, in particular as this may offer the possibility to use previously published data to generate larger data sets. However, for those wishing to deeply investigate evolutionary questions in certain lineages, a taxon‐specific kit offers certain benefits during data processing stages, where knowledge of the design scheme and gene models is extremely useful, and the possible return of costs is especially high for taxa emerging as model groups. We furthermore encourage groups designing taxon‐specific kits to include also universal probes, furthering the mission to complete the tree of life.
AUTHOR CONTRIBUTIONS
CL, MP and GY conceived the study. CL provided funding. TK coordinated sample collection, MdLH, VGJ and GY collected data. MHJB and WT identified species. GY designed bait kit, with guidance from JH and MP. CGC, JV, NR, MHJB and GY performed molecular work. GY and TL analysed the data using feedback from JV and OP. GY wrote the manuscript with significant input from all co‐authors.
OPEN RESEARCH BADGES
This article has earned an Open Data badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results.
Supporting information
Supplementary Material
Tables S1‐S5
ACKNOWLEDGEMENTS
This paper is dedicated to Christian Lexer, a wonderful mentor, friend and colleague. The research was supported with funding from the Christian Lexer professorship start‐up BE772002. The analyses benefited from the Vienna Scientific Cluster (VSC) and the Montpellier Bioinformatics Biodiversity (MBB) platform services. We thank Huiying Shang and Aram Drevekenin for help with code, Kelly Swarts, Claus Vogl and Matt Johnson for insightful discussions and advice. We thank the members of Swiss SNSF Sinergia project CRSII3_147630 for accession sampling and three anonymous reviewers for their comments on an earlier version of this manuscript.
Yardeni, G. , Viruel, J. , Paris, M. , Hess, J. , Groot Crego, C. , de La Harpe, M. , Rivera, N. , Barfuss, M. H. J. , Till, W. , Guzmán‐Jacob, V. , Krömer, T. , Lexer, C. , Paun, O. , & Leroy, T. (2022). Taxon‐specific or universal? Using target capture to study the evolutionary history of rapid radiations. Molecular Ecology Resources, 22, 927–945. 10.1111/1755-0998.13523
Christian Lexer, Ovidiu Paun, and Thibault Leroy shared last authorship.
DATA AVAILABILITY STATEMENT
Targeted sequencing reads generated for this project are available at NCBI‐SRA under BioProject PRJNA759878; for accession numbers, see Table S4. The probe set and the relevant supporting information are available in Dryad (https://doi.org/10.5061/dryad.mpg4f4r11). The bioinformatics scripts are available at https://github.com/giyany/Bromeliad1776/tree/main/MS_2021_scripts.
REFERENCES
- Aguirre‐Santoro, J. , Salinas, N. R. , & Michelangeli, F. A. (2020). The influence of floral variation and geographic disjunction on the evolutionary dynamics of Ronnbergia and Wittmackia (Bromeliaceae: Bromelioideae). Botanical Journal of the Linnean Society, 192(4), 609–624. 10.1093/botlinnean/boz087 [DOI] [Google Scholar]
- Alexander, D. H. , & Lange, K. (2011). Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics, 12(1), 246. 10.1186/1471-2105-12-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute. [Google Scholar]
- Bagley, J. C. , Uribe‐Convers, S. , Carlsen, M. M. , & Muchhala, N. (2020). Utility of targeted sequence capture for phylogenomics in rapid, recent angiosperm radiations: Neotropical Burmeistera bellflowers as a case study. Molecular Phylogenetics and Evolution, 152, 106769. 10.1016/j.ympev.2020.106769 [DOI] [PubMed] [Google Scholar]
- Barfuss, M. H. , Till, W. , Leme, E. M. , Pinzón, J. P. , Manzanares, J. M. , Halbritter, H. , Samuel, R. , & Brown, G. K. (2016). Taxonomic revision of Bromeliaceae subfam. Tillandsioideae based on a multi‐locus DNA sequence phylogeny and morphology. Phytotaxa, 279(1), 1–97. 10.11646/phytotaxa.279.1.1 [DOI] [Google Scholar]
- Barnett, D. W. , Garrison, E. K. , Quinlan, A. R. , Strömberg, M. P. , & Marth, G. T. (2011). BamTools: A C++ API and toolkit for analyzing and managing BAMfiles. Bioinformatics, 27(12), 1691–1692. 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, J. B. , Markley, M. L. , Zielke, M. G. , Thomas, J. R. , Hale, H. J. , Williams, L. D. , & Johnson, M. G. (2021). Is Palmer’s elm leaf goldenrod real? The Angiosperms353 kit provides within‐species signal in Solidago ulmifolia s.l. bioRxiv. 10.1101/2021.01.07.425781 [DOI] [Google Scholar]
- Behr, A. A. , Liu, K. Z. , Liu‐Fang, G. , Nakka, P. , & Ramachandran, S. (2016). Pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics, 32(18), 2817–2823. 10.1093/bioinformatics/btw327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing, D. H. (2000). Bromeliaceae: Profile of an adaptive radiation. Cambridge University Press. [Google Scholar]
- Berner, D. , & Salzburger, W. (2015). The genomics of organismal diversification illuminated by adaptive radiations. Trends in Genetics, 31(9), 491–499. 10.1016/j.tig.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Blaimer, B. B. , Lloyd, M. W. , Guillory, W. X. , & Brady, S. G. (2016). Sequence capture and phylogenetic utility of genomic ultraconserved elements obtained from pinned insect specimens. PLoS One, 11(8), e0161531. 10.1371/journal.pone.0161531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert, S. , & Danforth, B. N. (2018). On the universality of target‐enrichment baits for phylogenomic research. Methods in Ecology and Evolution, 9(6), 1453–1460. 10.1111/2041-210X.12988 [DOI] [Google Scholar]
- Bossert, S. , Murray, E. A. , Almeida, E. A. B. , Brady, S. G. , Blaimer, B. B. , & Danforth, B. N. (2019). Combining transcriptomes and ultraconserved elements to illuminate the phylogeny of Apidae. Molecular Phylogenetics and Evolution, 130, 121–131. 10.1016/j.ympev.2018.10.012 [DOI] [PubMed] [Google Scholar]
- Bragg, J. G. , Potter, S. , Bi, K. , & Moritz, C. (2016). Exon capture phylogenomics: Efficacy across scales of divergence. Molecular Ecology Resources, 16(5), 1059–1068. 10.1111/1755-0998.12449 [DOI] [PubMed] [Google Scholar]
- Chang, C. C. , Chow, C. C. , Tellier, L. C. , Vattikuti, S. , Purcell, S. M. , & Lee, J. J. (2015). Second‐generation PLINK: Rising to the challenge of larger and richer datasets. Giga‐Science, 4(1), 7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, J. H. , Rahfeldt, W. A. , & Olmstead, R. G. (2018). Comparison of taxon‐specific versus general locus sets for targeted sequence capture in plant phylogenomics. Applications in Plant Sciences, 6(3), e1032. 10.1002/aps3.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐Y. , Xin, Y. , Wai, C. M. , Liu, J. , & Ming, R. (2020). The role of cis ‐elements in the evolution of crassulacean acid metabolism photosynthesis. Horticulture Research, 7(1), 1–8. 10.1038/s41438-019-0229-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, M. , Smolina, I. , Dhanasiri, A. K. S. , Blanco‐Bercial, L. , Kopp, M. , Jueterbock, A. , Sundaram, A. Y. M. , & Hoarau, G. (2019). Towards population genomics in non‐model species with large genomes: A case study of the marine zooplankton Calanus finmarchicus . Royal Society Open Science, 6(2), 180608. 10.1098/rsos.180608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas, M. J. , Biffin, E. , Breed, M. F. , & Lowe, A. J. (2017). Targeted capture to assess neutral genomic variation in the narrow‐leaf hopbush across a continental biodiversity refugium. Scientific Reports, 7(1), 41367. 10.1038/srep41367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani, P. , Platts, A. , Wang, L. L. , Coon, M. , Nguyen, T. , Wang, L. , Land, S. J. , Lu, X. , & Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly, 6(2), 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crayn, D. M. , Winter, K. , & Smith, J. A. C. (2004). Multiple origins of crassulacean acid metabolism and the epiphytic habit in the Neotropical family Bromeliaceae. Proceedings of the National Academy of Sciences of the United States of America, 101(10), 3703–3708. 10.1073/pnas.0400366101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra, K. K. , Walters, J. R. , Briscoe, A. D. , Davey, J. W. , Whibley, A. , Nadeau, N. J. , … The Heliconius Genome Consortium (2012). Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature, 487(7405), 94–98. 10.1038/nature11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Harpe, M. , Hess, J. , Loiseau, O. , Salamin, N. , Lexer, C. , & Paris, M. (2019). A dedicated target capture approach reveals variable genetic markers across micro‐ and macroevolutionary time scales in palms. Molecular Ecology Resources, 19(1), 221–234. 10.1111/1755-0998.12945 [DOI] [PubMed] [Google Scholar]
- de La Harpe, M. , Paris, M. , Hess, J. , Barfuss, M. H. J. , Serrano‐Serrano, M. L. , Ghatak, A. , Chaturvedi, P. , Weckwerth, W. , Till, W. , Salamin, N. , Wai, C. M. , Ming, R. , & Lexer, C. (2020). Genomic footprints of repeated evolution of CAM photosynthesis in a Neotropical species radiation. Plant, Cell & Environment, 43(12), 2987–3001. 10.1111/pce.13847 [DOI] [PubMed] [Google Scholar]
- de La Harpe, M. , Paris, M. , Karger, D. N. , Rolland, J. , Kessler, M. , Salamin, N. , & Lexer, C. (2017). Molecular ecology studies of species radiations: Current research gaps, opportunities and challenges. Molecular Ecology, 26(10), 2608–2622. 10.1111/mec.14110 [DOI] [PubMed] [Google Scholar]
- Degnan, J. H. , & Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology & Evolution, 24(6), 332–340. 10.1016/j.tree.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Derrien, T. , & Ramos‐Onsins, S. (2020). Assessing a novel sequencing‐based approach for population genomics in non‐model species. Peer Community in Genomics, 1, 100002. 10.24072/pci.genomics.100002 [DOI] [Google Scholar]
- Dornburg, A. , Su, Z. , & Townsend, J. P. (2019). Optimal rates for phylogenetic inference and experimental design in the era of genome‐scale data sets. Systematic Biology, 68(1), 145–156. 10.1093/sysbio/syy047 [DOI] [PubMed] [Google Scholar]
- Doyle, J. J. , & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19(1), 11–15. [Google Scholar]
- Edwards, S. V. (2009). Is a new and general theory of molecular systematics emerging? Evolution, 63(1), 1–19. 10.1111/j.1558-5646.2008.00549.x [DOI] [PubMed] [Google Scholar]
- Edwards, S. V. , Xi, Z. , Janke, A. , Faircloth, B. C. , McCormack, J. E. , Glenn, T. C. , Zhong, B. , Wu, S. , Lemmon, E. M. , Lemmon, A. R. , Leaché, A. D. , Liu, L. , & Davis, C. C. (2016). Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics. Molecular Phylogenetics and Evolution, 94, 447–462. 10.1016/j.ympev.2015.10.027 [DOI] [PubMed] [Google Scholar]
- Emms, D. M. , & Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology, 20(1), 238. 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero, M. , Nieto‐Feliner, G. , Pokorny, L. , Spalink, D. , & Viruel, J. (2020). Editorial: Phylogenomic approaches to deal with particularly challenging plant lineages. Frontiers in Plant Science, 11, 591762. 10.3389/fpls.2020.591762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk, R. A. , Mandel, J. R. , & Freudenstein, J. V. (2015). A protocol for targeted enrichment of intron‐containing sequence markers for recent radiations: A phylogenomic example from Heuchera (Saxifragaceae). Applications in Plant Sciences, 3(8), 1500039. 10.3732/apps.1500039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier, N. , & Daubin, V. (2008). Dealing with incongruence in phylogenomic analyses. Philosophical Transactions of the Royal Society B: Biological Sciences, 363(1512), 4023–4029. 10.1098/rstb.2008.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E. (2012). Vcflib: A C++ library for parsing and manipulating VCF files. Github. https://github.com/ekg/vcflib [Google Scholar]
- Garrison, E. , & Marth, G. (2012). Haplotype‐based variant detection from short‐read sequencing. arXiv:1207.3907 [q‐bio]. [Google Scholar]
- Gavrilets, S. , & Losos, J. B. (2009). Adaptive radiation: Contrasting theory with data. Science, 323(5915), 732–737. 10.1126/science.1157966 [DOI] [PubMed] [Google Scholar]
- Givnish, T. J. , Barfuss, M. H. J. , Ee, B. V. , Riina, R. , Schulte, K. , Horres, R. , Gonsiska, P. A. , Jabaily, R. S. , Crayn, D. M. , Smith, J. A. C. , Winter, K. , Brown, G. K. , Evans, T. M. , Holst, B. K. , Luther, H. , Till, W. , Zizka, G. , Berry, P. E. , & Sytsma, K. J. (2014). Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Molecular Phylogenetics and Evolution, 71, 55–78. 10.1016/j.ympev.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Givnish, T. J. , Barfuss, M. H. J. , Van Ee, B. , Riina, R. , Schulte, K. , Horres, R. , Gonsiska, P. A. , Jabaily, R. S. , Crayn, D. M. , Smith, J. A. C. , Winter, K. , Brown, G. K. , Evans, T. M. , Holst, B. K. , Luther, H. , Till, W. , Zizka, G. , Berry, P. E. , & Sytsma, K. J. (2011). Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: Insights from an eight‐locus plastid phylogeny. American Journal of Botany, 98(5), 872–895. 10.3732/ajb.1000059 [DOI] [PubMed] [Google Scholar]
- Givnish, T. J. , Burkhardt, E. L. , Happel, R. E. , & Weintraub, J. D. (1984). Carnivory in the Bromeliad Brocchinia reducta, with a cost/benefit model for the general restriction of carnivorous plants to sunny, moist, nutrient‐poor habitats. The American Naturalist, 124(4), 479–497. 10.1086/284289 [DOI] [Google Scholar]
- Goetze, M. , Zanella, C. M. , Palma‐Silva, C. , Büttow, M. V. , & Bered, F. (2017). Incomplete lineage sorting and hybridization in the evolutionary history of closely related, endemic yellow‐flowered Aechmea species of subgenus Ortgiesia (Bromeliaceae). American Journal of Botany, 104(7), 1073–1087. 10.3732/ajb.1700103 [DOI] [PubMed] [Google Scholar]
- Goolsby, E. W. , Moore, A. J. , Hancock, L. P. , Vos, J. M. D. , & Edwards, E. J. (2018). Molecular evolution of key metabolic genes during transitions to C4 and CAM photosynthesis. American Journal of Botany, 105(3), 602–613. 10.1002/ajb2.1051 [DOI] [PubMed] [Google Scholar]
- Granados Mendoza, C. , Granados‐Aguilar, X. , Donadío, S. , Salazar, G. A. , Flores‐Cruz, M. , Hágsater, E. , Starr, J. R. , Ibarra‐Manríquez, G. , Fragoso‐Martínez, I. , & Magallón, S. (2017). Geographic structure in two highly diverse lineages of Tillandsia (Bromeliaceae). Botany‐Botanique, 95(7), 641–651. 10.1139/cjb-2016-0250 [DOI] [Google Scholar]
- Hale, H. , Gardner, E. M. , Viruel, J. , Pokorny, L. , & Johnson, M. G. (2020). Strategies for reducing per‐sample costs in target capture sequencing for phylogenomics and population genomics in plants. Applications in Plant Sciences, 8(4), e11337. 10.1002/aps3.11337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckenhauer, J. , Samuel, R. , Ashton, P. S. , Abu Salim, K. , & Paun, O. (2018). Phylogenomics resolves evolutionary relationships and provides insights into floral evolution in the tribe Shoreeae (Dipterocarpaceae). Molecular Phylogenetics and Evolution, 127, 1–13. 10.1016/j.ympev.2018.05.010 [DOI] [PubMed] [Google Scholar]
- Hedin, M. , Derkarabetian, S. , Ramírez, M. J. , Vink, C. , & Bond, J. E. (2018). Phylogenomic reclassification of the world’s most venomous spiders (Mygalomorphae, Atracinae), with implications for venom evolution. Scientific Reports, 8(1), 1636. 10.1038/s41598-018-19946-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk, K. , Moreno‐Villena, J. J. , Gilman, I. S. , Christin, P.‐A. , & Edwards, E. J. (2019). The genetics of convergent evolution: Insights from plant photosynthesis. Nature Reviews Genetics, 20(8), 485–493. 10.1038/s41576-019-0107-5 [DOI] [PubMed] [Google Scholar]
- Hime, P. M. , Lemmon, A. R. , Lemmon, E. C. M. , Prendini, E. , Brown, J. M. , Thomson, R. C. , Kratovil, J. D. , Noonan, B. P. , Pyron, R. A. , Peloso, P. L. V. , Kortyna, M. L. , Keogh, J. S. , Donnellan, S. C. , Mueller, R. L. , Raxworthy, C. J. , Kunte, K. , Ron, S. R. , Das, S. , Gaitonde, N. , … Weisrock, D. W. (2021). Phylogenomics reveals ancient gene tree discordance in the amphibian tree of life. Systematic Biology, 70(1), 49–66. 10.1093/sysbio/syaa034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, P. M. , Li, D.‐Z. , van der Bank, M. , & Twyford, A. D. (2016). Telling plant species apart with DNA: From barcodes to genomes. Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1702), 20150338. 10.1098/rstb.2015.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudlow, W. R. , Krieger, R. , Meusel, M. , Sehhat, J. C. , Timken, M. D. , & Buoncristiani, M. R. (2011). The NucleoSpin® DNA Clean‐up XS kit for the concentration and purification of genomic DNA extracts: An alternative to microdialysis filtration. Forensic Science International. Genetics, 5(3), 226–230. 10.1016/j.fsigen.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Hughes, C. E. , Nyffeler, R. , & Linder, H. P. (2015). Evolutionary plant radiations: Where, when, why and how? New Phytologist, 207(2), 249–253. 10.1111/nph.13523 [DOI] [PubMed] [Google Scholar]
- Johnson, M. G. , Pokorny, L. , Dodsworth, S. , Botigué, L. R. , Cowan, R. S. , Devault, A. , Eiserhardt, W. L. , Epitawalage, N. , Forest, F. , Kim, J. T. , Leebens‐Mack, J. H. , Leitch, I. J. , Maurin, O. , Soltis, D. E. , Soltis, P. S. , Wong, G.‐S. , Baker, W. J. , & Wickett, N. J. (2019). A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k‐medoids clustering. Systematic Biology, 68(4), 594–606. 10.1093/sysbio/syy086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. R. , & Good, J. M. (2016). Targeted capture in evolutionary and ecological genomics. Molecular Ecology, 25(1), 185–202. 10.1111/mec.13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier, T. , & Zdobnov, E. M. (2010). The Newick utilities: High‐throughput phylogenetic tree processing in the UNIX shell. Bioinformatics (Oxford, England), 26(13), 1669–1670. 10.1093/bioinformatics/btq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec, M. , Bellstedt, D. U. , Maitre, N. C. L. , & Pirie, M. D. (2017). Targeted NGS for species level phylogenomics: “made to measure” or “one size fits all”? PeerJ, 5, e3569. 10.7717/peerj.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, M. , Abrahamczyk, S. , & Krömer, T. (2020). The role of hummingbirds in the evolution and diversification of Bromeliaceae: Unsupported claims and untested hypotheses. Botanical Journal of the Linnean Society, 192(4), 592–608. 10.1093/botlinnean/boz100 [DOI] [Google Scholar]
- Kozlov, A. M. , Darriba, D. , Flouri, T. , Morel, B. , & Stamatakis, A. (2019). RAxML NG: A fast, scalable and user‐friendly tool for maximum likelihood phylogenetic inference. Bioinformatics, 35(21), 4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, F. (2019). Trim Galore: A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI‐digested RRBS‐type (Reduced Representation Bisulfite‐Seq) libraries. Babraham Institute, Babraham Bioinformatics. [Google Scholar]
- Kubatko, L. S. , & Degnan, J. H. (2007). Inconsistency of phylogenetic estimates from concatenated data under coalescence. Systematic Biology, 56(1), 17–24. 10.1080/10635150601146041 [DOI] [PubMed] [Google Scholar]
- Lamichhaney, S. , Berglund, J. , Almén, M. S. , Maqbool, K. , Grabherr, M. , Martinez‐Barrio, A. , Promerová, M. , Rubin, C.‐J. , Wang, C. , Zamani, N. , Grant, B. R. , Grant, P. R. , Webster, M. T. , & Andersson, L. (2015). Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature, 518(7539), 371–375. 10.1038/nature14181 [DOI] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larridon, I. , Villaverde, T. , Zuntini, A. R. , Pokorny, L. , Brewer, G. E. , Epitawalage, N. , Fairlie, I. , Hahn, M. , Kim, J. , Maguilla, E. , Maurin, O. , Xanthos, M. , Hipp, A. L. , Forest, F. , & Baker, W. J. (2020). Tackling rapid radiations with targeted sequencing. Frontiers in Plant Science, 10, 1655. 10.3389/fpls.2019.01655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, A. R. , Brown, J. M. , Stanger‐Hall, K. , & Lemmon, E. M. (2009). The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Systematic Biology, 58(1), 130–145. 10.1093/sysbio/syp017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, A. R. , Emme, S. A. , & Lemmon, E. M. (2012). Anchored hybrid enrichment for massively high‐throughput phylogenomics. Systematic Biology, 61(5), 727–744. 10.1093/sysbio/sys049 [DOI] [PubMed] [Google Scholar]
- Lemmon, E. M. , & Lemmon, A. R. (2013). High‐throughput genomic data in systematics and phylogenetics. Annual Review of Ecology, Evolution, and Systematics, 44(1), 99–121. 10.1146/annurev-ecolsys-110512-135822 [DOI] [Google Scholar]
- Leroy, C. , Carrias, J.‐F. , Céréghino, R. , & Corbara, B. (2016). The contribution of microorganisms and metazoans to mineral nutrition in bromeliads. Journal of Plant Ecology, 9(3), 241–255. 10.1093/jpe/rtv052 [DOI] [Google Scholar]
- Leroy, T. , Rousselle, M. , Tilak, M.‐K. , Caizergues, A. E. , Scornavacca, C. , Recuerda, M. , Fuchs, J. , Illera, J. C. , De Swardt, D. H. , Blanco, G. , Thébaud, C. , Milá, B. , & Nabholz, B. (2021). Island songbirds as windows into evolution in small populations. Current Biology, 31(6), 1303–1310.e4. 10.1016/j.cub.2020.12.040 [DOI] [PubMed] [Google Scholar]
- Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27(21), 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , & Durbin, R. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, H. P. (2008). Plant species radiations: Where, when, why? Philosophical Transactions of the Royal Society B: Biological Sciences, 363(1506), 3097–3105. 10.1098/rstb.2008.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Johnson, M. G. , Cox, C. J. , Medina, R. , Devos, N. , Vanderpoorten, A. , Hedenäs, L. , Bell, N. E. , Shevock, J. R. , Aguero, B. , Quandt, D. , Wickett, N. J. , Shaw, A. J. , & Goffinet, B. (2019). Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nature Communications, 10(1), 1485. 10.1038/s41467-019-09454-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau, O. , Mota Machado, T. , Paris, M. , Koubínová, D. , Dexter, K. G. , Versieux, L. M. , Lexer, C. , & Salamin, N. (2021). Genome skimming reveals widespread hybridization in a Neotropical flowering plant radiation. Frontiers in Ecology and Evolution, 9, 322. 10.3389/fevo.2021.668281 [DOI] [Google Scholar]
- Luther, H. E. (2008). An alphabetical list of bromeliad binomials (11th ed.). The Marie Selby Botanical Gardens Sarasota, The Bromeliad Society International. [Google Scholar]
- Machado, T. M. , Loiseau, O. , Paris, M. , Weigand, A. , Versieux, L. M. , Stehmann, J. R. , Lexer, C. , & Salamin, N. (2020). Systematics of Vriesea (Bromeliaceae): Phylogenetic relationships based on nuclear gene and partial plastome sequences. Botanical Journal of the Linnean Society, 192(4), 656–674. 10.1093/botlinnean/boz102 [DOI] [Google Scholar]
- Madden, T. (2003). The BLAST Sequence Analysis Tool. National Center for Biotechnology Information (US). [Google Scholar]
- McDonnell, A. J. , Baker, W. J. , Dodsworth, S. , Forest, F. , Graham, S. W. , Johnson, M. G. , Pokorny, L. , Tate, J. , Wicke, S. , & Wickett, N. J. (2021). Exploring Angiosperms353: Developing and applying a universal toolkit for flowering plant phylogenomics. Applications in Plant Sciences, 9(7), 11443. 10.1002/aps3.11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, M. D. , Borstein, S. R. , Meier, J. I. , Marques, D. A. , Mwaiko, S. , Taabu, A. , Kishe, M. A. , O’Meara, B. , Bruggmann, R. , Excoffier, L. , & Seehausen, O. (2020). The ecological and genomic basis of explosive adaptive radiation. Nature, 586(7827), 75–79. 10.1038/s41586-020-2652-7 [DOI] [PubMed] [Google Scholar]
- McGlothlin, J. W. , Kobiela, M. E. , Wright, H. V. , Mahler, D. L. , Kolbe, J. J. , Losos, J. B. , & Brodie, E. D. (2018). Adaptive radiation along a deeply conserved genetic line of least resistance in Anolis lizards. Evolution Letters, 2(4), 310–322. 10.1002/evl3.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowen, M. R. , Tsagkogeorga, G. , Álvarez‐Carretero, S. , dos Reis, M. , Struebig, M. , Deaville, R. , Jepson, P. D. , Jarman, S. , Polanowski, A. , Morin, P. A. , & Rossiter, S. J. (2020). Phylogenomic resolution of the Cetacean tree of life using target sequence capture. Systematic Biology, 69(3), 479–501. 10.1093/sysbio/syz068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain, M. R. , Johnson, M. G. , Uribe‐Convers, S. , Eaton, D. , & Yang, Y. (2018). Practical considerations for plant phylogenomics. Applications in Plant Sciences, 6(3), e1038. 10.1002/aps3.1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming, R. , VanBuren, R. , Wai, C. M. , Tang, H. , Schatz, M. C. , Bowers, J. E. , Lyons, E. , Wang, M.‐L. , Chen, J. , Biggers, E. , Zhang, J. , Huang, L. , Zhang, L. , Miao, W. , Zhang, J. , Ye, Z. , Miao, C. , Lin, Z. , Wang, H. , … Yu, Q. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics, 47(12), 1435–1442. 10.1038/ng.3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab, S. (2019). Species tree estimation using ASTRAL: Practical considerations. arXiv:1904.03826 [q‐bio]. [Google Scholar]
- Moest, M. , Van Belleghem, S. M. , James, J. E. , Salazar, C. , Martin, S. H. , Barker, S. L. , Moreira, G. R. P. , Mérot, C. , Joron, M. , Nadeau, N. J. , Steiner, F. M. , & Jiggins, C. D. (2020). Selective sweeps on novel and introgressed variation shape mimicry loci in a butterfly adaptive radiation. PLOS Biology, 18(2), e3000597. 10.1371/journal.pbio.3000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy, E. K. , & Warnow, T. (2018). To include or not to include: The impact of gene filtering on species tree estimation methods. Systematic Biology, 67(2), 285–303. 10.1093/sysbio/syx077 [DOI] [PubMed] [Google Scholar]
- Morales‐Briones, D. F. , Gehrke, B. , Huang, C.‐H. , Liston, A. , Ma, H. , Marx, H. E. , & Tank, D. C. , & Yang, Y. (2021). Analysis of paralogs in target enrichment data pinpoints multiple ancient polyploidy events in Alchemilla s.l. (Rosaceae). Systematic Biology, 1063–5157. 10.1101/2020.08.21.261925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, B. , Kozlov, A. M. , & Stamatakis, A. (2019). ParGenes: A tool for massively parallel model selection and phylogenetic tree inference on thousands of genes. Bioinformatics, 35(10), 1771–1773. 10.1093/bioinformatics/bty839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota, M. R. , Pinheiro, F. , Leal, B. S. D. S. , Sardelli, C. H. , Wendt, T. , & Palma‐Silva, C. (2020). From micro‐ to macroevolution: Insights from a Neotropical bromeliad with high population genetic structure adapted to rock outcrops. Heredity, 125(5), 353–370. 10.1038/s41437-020-0342-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle, R. G. , Filardi, C. E. , Smith, C. E. , & Diamond, J. (2009). Explosive Pleistocene diversification and hemispheric expansion of a “great speciator”. Proceedings of the National Academy of Sciences of the United States of America, 106(6), 1863–1868. 10.1073/pnas.0809861105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, B. , Forest, F. , Barraclough, T. , Rosindell, J. , Bellot, S. , Cowan, R. , Golos, M. , Jebb, M. , & Cheek, M. (2020). A phylogenomic analysis of Nepenthes (Nepenthaceae). Molecular Phylogenetics and Evolution, 144, 106668. 10.1016/j.ympev.2019.106668 [DOI] [PubMed] [Google Scholar]
- Nevado, B. , Atchison, G. W. , Hughes, C. E. , & Filatov, D. A. (2016). Widespread adaptive evolution during repeated evolutionary radiations in New World lupins. Nature Communications, 7(1), 12384. 10.1038/ncomms12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nute, M. , Chou, J. , Molloy, E. K. , & Warnow, T. (2018). The performance of coalescent based species tree estimation methods under models of missing data. BMC Genomics, 19(5), 286. 10.1186/s12864-018-4619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogutcen, E. , Christe, C. , Nishii, K. , Salamin, N. , Möller, M. , & Perret, M. (2021). Phylogenomics of Gesneriaceae using targeted capture of nuclear genes. Molecular Phylogenetics and Evolution, 157, 107068. 10.1016/j.ympev.2021.107068 [DOI] [PubMed] [Google Scholar]
- Ortiz, E. M. (2019). Vcf2phylip v2.0: Convert a VCF matrix into several matrix formats for phylogenetic analysis. Zenodo. 10.5281/zenodo.2540861 [DOI] [Google Scholar]
- Palma‐Silva, C. , & Fay, M. F. (2020). Bromeliaceae as a model group in understanding the evolution of Neotropical biota. Botanical Journal of the Linnean Society, 192(4), 569–586. 10.1093/botlinnean/boaa003 [DOI] [Google Scholar]
- Palma‐Silva, C. , Ferro, M. , Bacci, M. , & Turchetto‐Zolet, A. C. (2016). De novo assembly and characterization of leaf and floral transcriptomes of the hybridizing bromeliad species (Pitcairnia spp.) adapted to Neotropical Inselbergs. Molecular Ecology Resources, 16(4), 1012–1022. 10.1111/1755-0998.12504 [DOI] [PubMed] [Google Scholar]
- Pease, J. B. , Haak, D. C. , Hahn, M. W. , & Moyle, L. C. (2016). Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biology, 14(2), e1002379. 10.1371/journal.pbio.1002379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer, B. , Wittelsbürger, U. , Ramos‐Onsins, S. E. , & Lercher, M. J. (2014). PopGenome: An efficient Swiss army knife for population genomic analyses in R. Molecular Biology and Evolution, 31(7), 1929–1936. 10.1093/molbev/msu136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard toolkit (2019). Broad Institute, GitHub repository. http://broadinstitute.github.io/picard/ [Google Scholar]
- Quattrini, A. M. , Faircloth, B. C. , Dueñas, L. F. , Bridge, T. C. L. , Brugler, M. R. , Calixto‐Botía, I. F. , DeLeo, D. M. , Forêt, S. , Herrera, S. , Lee, S. M. Y. , Miller, D. J. , Prada, C. , Rádis‐Baptista, G. , Ramírez‐Portilla, C. , Sánchez, J. A. , Rodríguez, E. , & McFadden, C. S. (2018). Universal target‐enrichment baits for anthozoan (Cnidaria) phylogenomics: New approaches to long‐standing problems. Molecular Ecology Resources, 18(2), 281–295. 10.1111/1755-0998.12736 [DOI] [PubMed] [Google Scholar]
- Quezada, I. M. , & Gianoli, E. (2011). Crassulacean acid metabolism photosynthesis in Bromeliaceae: An evolutionary key innovation. Biological Journal of the Linnean Society, 104(2), 480–486. 10.1111/j.1095-8312.2011.01713.x [DOI] [Google Scholar]
- Quinlan, A. R. , & Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Renaud, G. , Stenzel, U. , Maricic, T. , Wiebe, V. , & Kelso, J. (2015). deML: Robust demultiplexing of Illumina sequences using a likelihood‐based approach. Bioinformatics (Oxford, England), 31(5), 770–772. 10.1093/bioinformatics/btu719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch, S. , & Warnow, T. (2015). On the robustness to gene tree estimation error (or lack thereof) of coalescent‐based species tree methods. Systematic Biology, 64(4), 663–676. 10.1093/sysbio/syv016 [DOI] [PubMed] [Google Scholar]
- Salzburger, W. (2018). Understanding explosive diversification through cichlid fish genomics. Nature Reviews Genetics, 19(11), 705–717. 10.1038/s41576-018-0043-9 [DOI] [PubMed] [Google Scholar]
- Sanderson, B. J. , DiFazio, S. P. , Cronk, Q. C. B. , Ma, T. , & Olson, M. S. (2020). A targeted sequence capture array for phylogenetics and population genomics in the Salicaceae. Applications in Plant Sciences, 8(10), e11394. 10.1002/aps3.11394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass, C. , Iles, W. J. D. , Barrett, C. F. , Smith, S. Y. , & Specht, C. D. (2016). Revisiting the Zingiberales: Using multiplexed exon capture to resolve ancient and recent phylogenetic splits in a charismatic plant lineage. PeerJ, 4, e1584. 10.7717/peerj.1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyari, E. , & Mirarab, S. (2016). Fast coalescent‐based computation of local branch support from Quartet Frequencies. Molecular Biology and Evolution, 33(7), 1654–1668. 10.1093/molbev/msw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte, K. , Barfuss, M. H. J. , & Zizka, G. (2009). Phylogeny of Bromelioideae (Bromeliaceae) inferred from nuclear and plastid DNA loci reveals the evolution of the tank habit within the subfamily. Molecular Phylogenetics and Evolution, 51(2), 327–339. 10.1016/j.ympev.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Shah, T. , Schneider, J. V. , Zizka, G. , Maurin, O. , Baker, W. , Forest, F. , Brewer, G. E. , Savolainen, V. , Darbyshire, I. , & Larridon, I. (2021). Joining forces in Ochnaceae phylogenomics: A tale of two targeted sequencing probe kits. American Journal of Botany, 108(7), 1201–1216. 10.1002/ajb2.1682 [DOI] [PubMed] [Google Scholar]
- Shee, Z. Q. , Frodin, D. G. , Cámara‐Leret, R. , & Pokorny, L. (2020). Reconstructing the complex evolutionary history of the Papuasian Schefflera radiation through Herbariomics. Frontiers in Plant Science, 11, 258. 10.3389/fpls.2020.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera, K. , Neubig, K. M. , Whitten, W. M. , Williams, N. H. , Winter, K. , & Cushman, J. C. (2010). Evolution along the crassulacean acid metabolism continuum. Functional Plant Biology, 37(11), 995–1010. 10.1071/FP10084 [DOI] [Google Scholar]
- Slimp, M. , Williams, L. D. , Hale, H. , & Johnson, M. G. (2021). On the potential of Angiosperms353 for population genomics. Applications in Plant Sciences, 9(7), e11419. 10.1002/aps3.11419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis, P. S. , Folk, R. A. , & Soltis, D. E. (2019). Darwin review: Angiosperm phylogeny and evolutionary radiations. Proceedings of the Royal Society B: Biological Sciences, 286(1899), 20190099. 10.1098/rspb.2019.0099 [DOI] [Google Scholar]
- Soltis, P. S. , & Soltis, D. E. (2004). The origin and diversification of angiosperms. American Journal of Botany, 91(10), 1614–1626. 10.3732/ajb.91.10.1614 [DOI] [PubMed] [Google Scholar]
- Soto‐Gomez, M. S. , Pokorny, L. , Kantar, M. B. , Forest, F. , Leitch, I. J. , Gravendeel, B. , & Viruel, J. (2019). A customized nuclear target enrichment approach for developing a phylogenomic baseline for Dioscorea yams (Dioscoreaceae). Applications in Plant Sciences, 7(6), e11254. 10.1002/aps3.11254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, S. C. K. , Moore, M. J. , Soltis, P. S. , Soltis, D. E. , Liston, A. , & Livshultz, T. (2014). Phylogenetic signal detection from an ancient rapid radiation: Effects of noise reduction, long‐branch attraction, and model selection in crown clade Apocynaceae. Molecular Phylogenetics and Evolution, 80, 169–185. 10.1016/j.ympev.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Straub, S. C. K. , Parks, M. , Weitemier, K. , Fishbein, M. , Cronn, R. C. , & Liston, A. (2012). Navigating the tip of the genomic iceberg: Next‐generation sequencing for plant systematics. American Journal of Botany, 99(2), 349–364. 10.3732/ajb.1100335 [DOI] [PubMed] [Google Scholar]
- Streicher, J. W. , Schulte, J. A. , & Wiens, J. J. (2016). How should genes and taxa be sampled for phylogenomic analyses with missing data? An empirical study in Iguanian Lizards. Systematic Biology, 65(1), 128–145. 10.1093/sysbio/syv058 [DOI] [PubMed] [Google Scholar]
- Stroud, J. T. , & Losos, J. B. (2020). Bridging the process‐pattern divide to understand the origins and early stages of adaptive radiation: A review of approaches with insights from studies of Anolis lizards. Journal of Heredity, 111(1), 33–42. 10.1093/jhered/esz055 [DOI] [PubMed] [Google Scholar]
- Supple, M. A. , & Shapiro, B. (2018). Conservation of biodiversity in the genomics era. Genome Biology, 19(1), 131. 10.1186/s13059-018-1520-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, A. E. , Igea, J. , Meudt, H. M. , Albach, D. C. , Lee, W. G. , & Tanentzap, A. J. (2021). Using target sequence capture to improve the phylogenetic resolution of a rapid radiation in New Zealand Veronica. American Journal of Botany, 108(7), 1289–1306. 10.1002/ajb2.1678 [DOI] [PubMed] [Google Scholar]
- Ufimov, R. , Zeisek, V. , Píšová, S. , Baker, W. J. , Fér, T. , Loo, M. , Dobeš, C. , & Schmickl, R. (2021). Relative performance of customized and universal probe sets in target enrichment: A case study in subtribe Malinae. Applications in Plant Sciences, 9(7), e11442. 10.1002/aps3.11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y. , Mizrachi, E. , & Marchal, K. (2017). The evolutionary significance of polyploidy. Nature Reviews Genetics, 18(7), 411–424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Versieux, L. M. , Barbará, T. , Wanderley, M. D. G. L. , Calvente, A. , Fay, M. F. , & Lexer, C. (2012). Molecular phylogenetics of the Brazilian giant bromeliads (Alcantarea, Bromeliaceae): Implications for morphological evolution and biogeography. Molecular Phylogenetics and Evolution, 64(1), 177–189. 10.1016/j.ympev.2012.03.015 [DOI] [PubMed] [Google Scholar]
- Villaverde, T. , Pokorny, L. , Olsson, S. , Rincón‐Barrado, M. , Johnson, M. G. , Gardner, E. M. , Wickett, N. J. , Molero, J. , Riina, R. , & Sanmartín, I. (2018). Bridging the micro‐ and macroevolutionary levels in phylogenomics: Hyb‐Seq solves relationships from populations to species and above. New Phytologist, 220(2), 636–650. 10.1111/nph.15312 [DOI] [PubMed] [Google Scholar]
- Viruel, J. , Conejero, M. , Hidalgo, O. , Pokorny, L. , Powell, R. F. , Forest, F. , Kantar, M. B. , Soto Gomez, M. , Graham, S. W. , Gravendeel, B. , Wilkin, P. , & Leitch, I. J. (2019). A target capture‐based method to estimate ploidy from Herbarium specimens. Frontiers in Plant Science, 10, 937. 10.3389/fpls.2019.00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai, C. M. , VanBuren, R. , Zhang, J. , Huang, L. , Miao, W. , Edger, P. P. , & Ming, R. (2017). Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple. The Plant Journal, 92(1), 19–30. 10.1111/tpj.13630 [DOI] [PubMed] [Google Scholar]
- Weitemier, K. , Straub, S. C. K. , Cronn, R. C. , Fishbein, M. , Schmickl, R. , McDonnell, A. , & Liston, A. (2014). Hyb‐Seq: Combining target enrichment and genome skimming for plant phylogenomics. Applications in Plant Sciences, 2(9), 1400042. 10.3732/apps.1400042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F. , & Doyle, J. J. (1998). Phylogenetic incongruence: Window into genome history and molecular evolution. In Soltis D. E., Soltis P. S., & Doyle J. J. (Eds.), Molecular systematics of plants II: DNA sequencing (pp. 265–296). Springer US. [Google Scholar]
- Wöhrmann, T. , Michalak, I. , Zizka, G. , & Weising, K. (2020). Strong genetic differentiation among populations of Fosterella rusbyi (Bromeliaceae) in Bolivia. Botanical Journal of the Linnean Society, 192(4), 744–759. 10.1093/botlinnean/boz096 [DOI] [Google Scholar]
- Xi, Z. , Liu, L. , & Davis, C. C. (2016). The impact of missing data on species tree estimation. Molecular Biology and Evolution, 33(3), 838–860. 10.1093/molbev/msv266 [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Rabiee, M. , Sayyari, E. , & Mirarab, S. (2018). ASTRAL‐III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics, 19(6), 153. 10.1186/s12859-018-2129-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Levine, D. , Shen, J. , Gogarten, S. M. , Laurie, C. , & Weir, B. S. (2012). A high‐performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics, 28(24), 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, E. A. , & Wen, J. (2013). Reprint of: Using nuclear gene data for plant phylogenetics: Progress and prospects. Molecular Phylogenetics and Evolution, 66(2), 539–550. 10.1016/j.ympev.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Zink, R. M. , & Vázquez‐Miranda, H. (2019). Species limits and phylogenomic relationships of Darwin’s Finches remain unresolved: Potential consequences of a volatile ecological setting. Systematic Biology, 68(2), 347–357. 10.1093/sysbio/syy073 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Tables S1‐S5
Data Availability Statement
Targeted sequencing reads generated for this project are available at NCBI‐SRA under BioProject PRJNA759878; for accession numbers, see Table S4. The probe set and the relevant supporting information are available in Dryad (https://doi.org/10.5061/dryad.mpg4f4r11). The bioinformatics scripts are available at https://github.com/giyany/Bromeliad1776/tree/main/MS_2021_scripts.


