SUMMARY
Reduction of crop yield due to iron (Fe) deficiency has always been a concern in agriculture. How Fe insufficiency in floral buds affects pollen development remains unexplored. Here, plants transferred to Fe‐deficient medium at the reproductive stage had reduced floral Fe content and viable pollen and showed a defective pollen outer wall, all restored by supplying floral buds with Fe. A comparison of differentially expressed genes (DEGs) in Fe‐deficient leaves, roots, and anthers suggested that changes in several cellular processes were unique to anthers, including increased lipid degradation. Co‐expression analysis revealed that ABORTED MICROSPORES (AMS), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1, and BASIC HELIX‐LOOP‐HELIX 089/091/010 encode key upstream transcription factors of Fe deficiency‐responsive DEGs involved in tapetum function and development, including tapetal ROS homeostasis, programmed cell death, and pollen outer wall formation‐related lipid metabolism. Analysis of RESPIRATORY‐BURST OXIDASE HOMOLOG E (RBOHE) gain‐ and loss‐of‐function under Fe deficiency indicated that RBOHE‐ and Fe‐dependent regulation cooperatively control anther reactive oxygen species levels and pollen development. Since DEGs in Fe‐deficient anthers were not significantly enriched in genes related to mitochondrial function, the changes in mitochondrial status under Fe deficiency, including respiration activity, density, and morphology, were probably because the Fe amount was insufficient to maintain proper mitochondrial protein function in anthers. To sum up, Fe deficiency in anthers may affect Fe‐dependent protein function and impact upstream transcription factors and their downstream genes, resulting in extensively impaired tapetum function and pollen development.
Keywords: Fe deficiency, tapetum, pollen development, reactive oxygen species, mitochondria, Arabidopsis thaliana
Significance Statement
Insufficient iron (Fe) in floral buds may be the primary cause of pollen development disruption, most likely affecting Fe‐dependent activities and upstream transcriptional regulation in the tapetum.
INTRODUCTION
Seed production and the majority of our food supply rely on the success of sexual reproduction, which requires viable pollen and ovules. Pollen development in Arabidopsis starts from a pollen mother cell (PMC) that undergoes meiosis to produce a tetrad of four haploid microspores. After release from the tetrad, the young microspores (YMs) expand and form vacuolated pollen (VP) with a large vacuole inside. Next, the VP undergoes mitosis to become binuclear (BN). Finally, the BN pollen undergoes further mitosis to form the tricellular (TC) pollen. The tapetum encloses the locule to provide space and nutrients for pollen development. In addition, the tapetum also synthesizes and secretes components such as callase to release the YM from the tetrad (Mepham and Lane, 1969) and sporopollenin for the formation of the outer pollen wall (also called exine) (Gu et al., 2014; Xu et al., 2014), as well as neutral lipids for the pollen coat formation. Eventually, the tapetum gradually degenerates through programmed cell death (PCD) (Luo et al., 2013) and disappears before anther dehiscence.
Several transcription factors have been reported to be critical for pollen development. DYSFUNCTIONAL TAPETUM 1 (DYT1) (Gu et al., 2014; Zhang et al., 2006), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION 1 (TDF1) (Gu et al., 2014), ABORTED MICROSPORE (AMS) (Ma et al., 2012; Xu et al., 2014, 2010), and MYB80 (also named MYB103 or Male Sterile 188) (Wang et al., 2018) form a signaling cascade. This gene regulatory network precisely regulates tapetum function and development, such as sporopollenin biosynthesis, reactive oxygen species (ROS) production, and PCD in the tapetum (Xie et al., 2014; Zhang et al., 2014). DYT1, TDF1, AMS, and MYB80 each have their specific target genes and are involved in diverse tapetum functions (Li et al., 2017). BASIC HELIX‐LOOP‐HELIX (bHLH) 089/091/010 interacts with DYT1 and AMS. It mediates their transcriptional activity on target genes (Lou et al., 2018; Wang et al., 2018; Xu et al., 2010). They also independently play essential roles in stress responses and hormone signaling (Zhu et al., 2015).
Iron (Fe) is an essential nutrient for plants because it plays a critical role in metabolic processes such as photosynthesis, mitochondrial respiration, and the synthesis of fatty acids. Fe deficiency has become the most common nutritional disorder in plants. The typical symptoms of Fe deficiency include chlorosis of young leaves and reduced crop yield and quality (Abadía et al., 2004; Àlvarez‐Fernàndez et al., 2006). The responses to Fe deficiency in vegetative tissues have been extensively studied (Lopez‐Millan et al., 2013; Mai and Bauer, 2016; Pan et al., 2015; Rodriguez‐Celma et al., 2013). Although it has been shown that Fe content is critical for flower development, especially male reproductive tissue (Ravet et al., 2009; Roschzttardtz et al., 2011; Schuler et al., 2012; Takahashi et al., 2003), the detailed effects of Fe deficiency on reproductive tissues are yet to be investigated. A previous study suggested that developing flowers receive Fe from the roots and, to a lesser extent, from senescent leaves (Morrissey and Guerinot, 2009). The FERRIC REDUCTASE DEFECTIVE (FRD) and YELLOW STRIPE1‐LIKE (YSL) transporters, as well as NICOTIANAMINE SYNTHASE (NAS), are necessary for long‐distance Fe transport from roots to leaves and flowers. Mutants with defects in these genes have higher rates of pollen abortion (Roschzttardtz et al., 2011; Schuler et al., 2012; Yokosho et al., 2016). These findings suggest that Fe homeostasis is vital for pollen development. However, since these mutants show defects in reproductive tissues and vegetative tissues, it is difficult to demonstrate whether impaired pollen development in these mutants is due to abnormal vegetative growth or changes in Fe homeostasis in reproductive tissues.
Tapetal cells are highly metabolically active as they synthesize and transfer components for pollen growth. They require an abundance of energy and intermediates and mainly rely on mitochondria to support their functions. Therefore, defects in mitochondrial function and dynamics result in impaired tapetum function and increased pollen abortion (Balk and Leaver, 2001; Chen et al., 2019; Luo et al., 2013). Fe is essential for mitochondrial respiratory activity because mitochondrial respiratory complexes require a Fe‐S cluster and heme to carry out electron transfer (Vigani, 2012). A rice (Oryza sativa) mutant with a defect in the mitochondrial Fe transporter (MIT) had reduced mitochondrial Fe content and fertility, but the mechanism was not characterized (Bashir et al., 2011).
The actual occurrence of tapetum PCD is essential for producing viable pollen. Maintaining proper anther ROS levels (Xie et al., 2014), mainly contributed by the tapetum and developing pollen (Hu et al., 2011), is critical for regulating tapetum PCD (De Storme and Geelen, 2014; Wilson and Zhang, 2009). Loss or abnormal expression of the RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) gene encoding tapetal NADPH oxidase affects anther ROS production and tapetum PCD (Xie et al., 2014). Fe is involved in ROS regulation in cells because it triggers the Fenton reaction and is required for the function of ROS‐producing and ROS‐scavenging enzymes (Inupakutika et al., 2016; Mittler, 2017). Fe has been recognized as a potential initiator catalyzing ROS production involved in animal PCD (Dixon and Stockwell, 2014). The ferritin mutant has high levels of Fe accumulation and oxidative stress in flowers, which result in male sterility (Ravet et al., 2009; Sudre et al., 2013). However, it is still unknown how Fe deficiency affects cellular processes in anthers, especially those involving mitochondria, ROS, and PCD in the tapetum.
We decreased Fe supply during reproductive growth and observed a reduction in Fe content in floral buds and an increase in pollen abortion; these phenotypes were restored by providing an exogenous supply of Fe to floral buds of plants growing in Fe‐deficient medium. Analysis of anther transcriptomes showed that the transcriptional changes in Fe‐deficient anthers might be correlated with several defects in anthers and pollen, including disrupted tapetum degeneration, anther ROS homeostasis, pollen outer wall formation, and lipid metabolism. In addition, we found that the tapetum‐enriched transcription factors AMS, TDF1, and bHLH089/091/010 might play essential roles in regulating a large portion of genes differentially expressed in response to Fe deficiency. Thus, the critical changes in molecular regulation in anthers seem to associate with cellular alterations under Fe deficiency.
RESULTS
Reduction of viable pollen and aberrant pollen morphology under Fe deficiency
To understand the influence of Fe deficiency specifically during pollen development, we grew plants hydroponically in a Fe‐sufficient medium until bolting (when primary inflorescences had one to two opening flowers; Figure S1a). Reproductive growth under Fe deficiency, with regard to floral organs and siliques, was comparable to that under Fe sufficiency (Figure S1b). Alexander staining revealed that most pollen from plants continuously grown in Fe‐sufficient medium had dark‐purple cytoplasm and a blue‐green pollen wall, which indicated that pollen was viable (Figure 1a). However, a portion of the pollen from plants grown under Fe deficiency had blue‐green pollen walls and lacked dark‐purple cytoplasm, which indicated that the pollen was non‐viable (Figure 1b). The quantification of viable pollen revealed that under Fe deficiency, the production of viable pollen was almost 40% lower than that of plants grown in Fe‐sufficient medium (Figure 1c). In addition, the in vitro germination rate of pollen developed under Fe deficiency was dramatically lower (60%) than that of plants grown under Fe sufficiency (Figure 1d and Figure S1c). These results indicated that Fe deficiency decreases the production of viable pollen.
Figure 1.
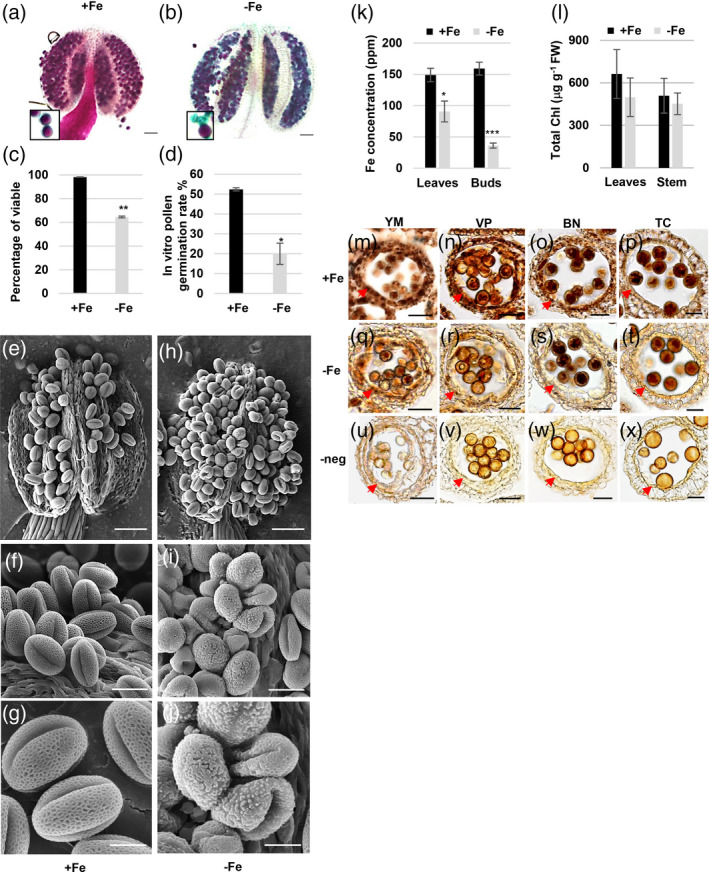
The changes in the viability and morphology of pollen, Fe content, and chlorophyll content under Fe deficiency.
(a, b) The viability of pollen in anthers of plants grown for 18 days after transplantation to Fe‐sufficient (+Fe, a) and ‐deficient (−Fe, b) conditions was analyzed using Alexander staining. Bars = 50 μm. The enlarged areas show viable pollen (purple) and non‐viable pollen (blue‐green). (c) The percentage of viable pollen was scored from anthers stained with Alexander stain. (d) The percentage of pollen that germinated was scored. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, **P < 0.01 between −Fe and +Fe conditions (Student’s t‐test). (e–j) Cryo‐SEM images of mature pollen in the dehiscent anthers of plants grown under +Fe (e–g) and −Fe (h–j) conditions. Bars = 50 μm in (e and h), 20 μm in (f and i), and 10 μm in (g and j). (k) Fe concentration in leaves and floral buds from plants grown under Fe‐sufficient (+Fe) and ‐deficient (−Fe) conditions was analyzed by ICP‐MS. (l) Chlorophyll (Chl) content in rosette leaves and inflorescence stems from plants grown under +Fe and −Fe conditions. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, ***P < 0.005 between −Fe and +Fe anthers (Student’s t‐test). (m–x) Sections of anthers from plants grown under +Fe (m–p) and −Fe (q–t) conditions were stained with Perls/DAB. Bars = 20 μm. (u–x) Sections of anthers from plants grown under +Fe conditions were stained with DAB alone as a negative control (‐neg). The arrows in panels (m) to (x) indicate the tapetum. YM, young microspore; VP, vacuolated pollen; BN, binucleate pollen; TC, tricellular pollen.
To investigate the defects in pollen formation under Fe deficiency, we used cryo‐scanning electron microscopy (cryo‐SEM) to investigate the morphology and structure of mature pollen (Figure 1e–j). The cryo‐SEM results showed that pollen grains from Fe‐sufficient plants had a reticule‐like outer wall structure and were folded inward at three apertures, forming an olive shape (Figure 1e–g). A subset of pollen grains from plants grown under Fe deficiency was collapsed and had an irregular outer wall structure (Figure 1h–j). Since the synthesis and secretion of the outer pollen wall precursor sporopollenin is conducted by the tapetum, the tapetum plays a dominant role in forming the outer pollen wall. These results imply that Fe deficiency might disrupt tapetum function.
Fe content in different tissues and Fe distribution in anthers under Fe deficiency
To reveal the changes in Fe content in unopened floral buds at all stages and also in mature rosette leaves under Fe‐deficient conditions, we measured Fe content in floral buds and leaves using inductively coupled plasma mass spectrometry (ICP‐MS). The results showed that Fe content was about 70% lower in floral buds and 40% lower in leaves of plants transferred to Fe‐deficient medium than in those assigned to Fe‐sufficient medium (Figure 1k). Fe is essential for chlorophyll biosynthesis and the function of photosynthesis complexes in leaves, associated with the supply of carbohydrates to sink organs such as floral buds. Therefore, we examined total chlorophyll content in rosette leaves and inflorescence stems and metabolites, including carbohydrates and several amino acids, in floral buds. The results showed that our Fe‐deficient condition did not significantly affect the chlorophyll contents in rosette leaves and inflorescence stems (Figure 1l), suggesting a negligible impact on chlorophyll biosynthesis and function. In addition, relative contents of soluble sugars, such as sucrose, glucose, and fructose, and amino acids in floral buds developed under Fe deficiency were comparable to or even higher than those in Fe‐sufficient buds (Table S1), implying the supply of carbon and nitrogen was probably not compromised.
Since the pollen defects and a significant reduction in Fe content were found in floral buds under Fe deficiency, we further analyzed the histochemical distribution of Fe in developing anthers at different stages (from YM to TC) using Perls/DAB stain, a standard stain to detect Fe in tissues (Roschzttardtz et al., 2013) (Figure 1m–x). The sections stained without potassium ferrocyanide were used as the negative control (Figure 1u–x). Our results revealed that Fe accumulated in the tapetum, microspores, and anther wall layers at the YM and VP stages under Fe‐sufficient conditions (Figure 1m,n). Furthermore, at the BN and TC stages, the intensity of the Fe signal detected by Perls/DAB staining was higher in the tapetum and pollen than in the surrounding cell layers under Fe‐sufficient conditions (Figure 1o,p). In contrast, under Fe deficiency, the intensity of the Fe signal at the YM to TC stages was lower, especially in the tapetum (Figure 1q–t), compared with that under Fe‐sufficient conditions (Figure 1m–p). The results indicated that when Fe supply is insufficient at the reproductive stage, Fe accumulation decreases in floral buds, especially in the tapetum.
Fe deficiency affects tapetum morphology and delays tapetum degeneration
Since Fe deficiency decreased Fe content in the tapetum and disrupted the outer pollen wall, which mainly relies on tapetum function, we made semi‐thin sections of anthers to investigate whether the morphology and structure of the tapetum had defects during different developmental stages under Fe deficiency (Figure 2a1–b6). There were no apparent differences in the tapetum between Fe‐sufficient and ‐deficient anthers from the tetrad to the VP stage (Figure 2a1–a3,b1–b3), but hypertrophy of the tapetum was observed at the BN stage (Figure 2b4) and the tapetum became vacuolated at the TC stage (Figure 2b5) under Fe deficiency. In addition, at the late TC stage, the tapetum disappeared when the septum was still thick under Fe‐sufficient conditions (Figure 2a6). Still, under Fe deficiency, the tapetum remained even when the septum became thin (seemed to be degenerating) (Figure 2b6).
Figure 2.
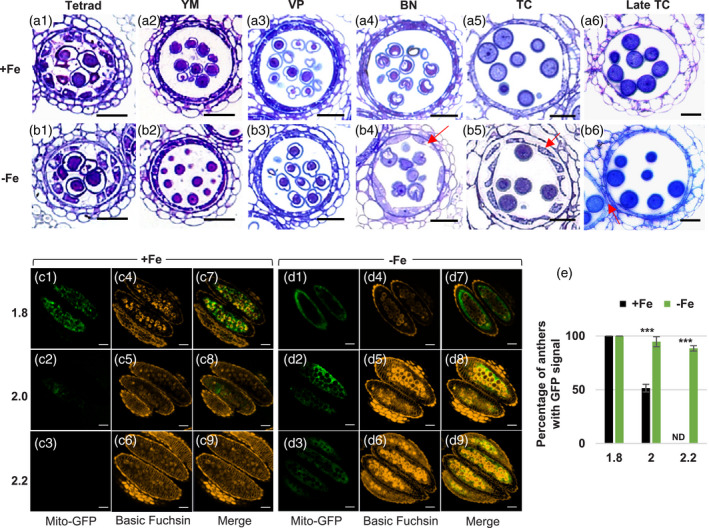
The morphology and degeneration of the tapetum under Fe deficiency.
(a, b) Semi‐thin sections of anthers from plants grown under Fe‐sufficient (a1–a6) and ‐deficient (b1–b6) conditions. The arrows in (b4 and b5) indicate abnormal tapetum morphology. The arrow in (b6) indicates the degenerating septum. Bars = 20 μm in (a1–b6). YM, young microspore; VP, vacuolated pollen; BN, binucleate pollen; TC, tricellular pollen. (c, d) Anthers showing tapetum‐specific expression of mito‐GFP from 1.8‐, 2.0‐, and 2.2‐mm buds of Fe‐sufficient (+Fe, c) and ‐deficient (−Fe, d) plants were examined. Basic Fuchsin was used to stain lignin to differentiate anther stage in (c4–c6 and d4–d6). (c7–c9 and d7–d9) are merged images of mito‐GFP and Basic Fuchsin. Bars = 50 μm. (e) Quantification of the percentage of anthers with GFP signals (n > 10). Data are shown as mean ± standard error of three independent experiments. ***P < 0.005 between −Fe and +Fe conditions (Student’s t‐test). ND, not detected.
To confirm whether Fe deficiency delays tapetum degeneration, we analyzed the stages of tapetum degeneration initiated using the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay to detect DNA fragmentation, a relatively early step in cell death. At stage 9, no DNA fragmentation was seen in the tapetum in either Fe‐sufficient or Fe‐deficient anthers. At stage 10, DNA fragmentation was detected in a few tapetal cells in the Fe‐sufficient anthers but not in the tapetal cells in the Fe‐deficient anthers. At stage 11, DNA fragmentation was seen in several tapetal cells in both Fe‐sufficient and ‐deficient anthers (Figure S2). These results seem to suggest that DNA fragmentation was slightly delayed in the tapeta grown under Fe deficiency. In addition, we used a transgenic line, which was generated in our previous study and expressed mitochondria‐targeted GFP (mito‐GFP) specifically in tapetal cells, to monitor the presence of the tapetum. Our previous study showed that the GFP signal is only present in the tapetum and not in the locule (Chen et al., 2019). After the TC stage, more and more lignin accumulates in the endothecium and epidermis layers of the anther wall following development (Mitsuda et al., 2005; Quilichini et al., 2014; Ursache et al., 2018). Based on the correlation between bud size and anther/pollen development (Chen et al., 2019), pollen is at the TC stage when bud size is 1.4 mm. In Figure 2c,d, we examined the presence of the tapetum in anthers from the middle to very late TC stage (from buds which had sizes from 1.8 to 2.2 mm) characterized by detecting lignin content stained with Basic Fuchsin. Under Fe‐sufficient conditions, tapetal cells, visualized by mito‐GFP, were detected in anthers from 1.8‐mm buds; the signal remained in anthers from 2.0‐mm buds; and no signal was seen in anthers from 2.2‐mm buds (Figure 2c1–c9). Under Fe deficiency, the tapetum was detected in anthers from 1.8‐ and 2.0‐mm buds and still remained in anthers from 2.2‐mm buds (Figure 2d1–d9). We quantified the percentage of anthers containing a GFP signal from 1.8‐, 2.0‐, and 2.2‐mm floral buds (Figure 2e). The results showed that under Fe‐sufficient conditions, the tapetum was present in all anthers from 1.8‐mm buds, absent in 50% of anthers from 2.0‐mm buds, and absent in 100% of anthers from 2.2‐mm buds. However, under Fe deficiency, approximately 90% of anthers from 2.2‐mm buds still contained the tapetum, indicating that tapetal PCD was delayed.
Restoration of floral Fe content and viable pollen by supplying Fe to floral buds
Since Fe deficiency usually accompanies alterations in the contents of other metals, especially Co, Cu, Mn, and Zn (Korshunova et al., 1999; Perea‐Garcia et al., 2013; Vert et al., 2002), we measured metal contents using ICP‐MS and observed higher Cu, Co, and Mg contents in floral buds under Fe‐deficient conditions (Table S2). Thus, even though we grew plants in a Fe‐deficient medium only after inflorescence initiation, we cannot exclude the possibility that the increase in non‐viable pollen might result from impaired vegetative tissues under Fe deficiency. To determine whether Fe insufficiency in floral buds was the primary cause of the reduction in viable pollen, we provided an exogenous supply of Fe by dropping a solution with Fe2‐EDTA (+Fe) or EDTA (mock) on the floral buds of plants growing in Fe‐deficient medium. Treatment with exogenous Fe2‐EDTA but not EDTA rescued the production of viable pollen (Figure 3a–e), the Fe contents of floral buds (as measured using the bathophenanthroline disulfonate [BPDS] method [Figure 3f]), and the outer wall and shape of mature pollen (Figure S3). These results indicated that insufficient Fe levels in the floral buds were the primary cause of the pollen defects observed under our Fe‐deficient conditions.
Figure 3.
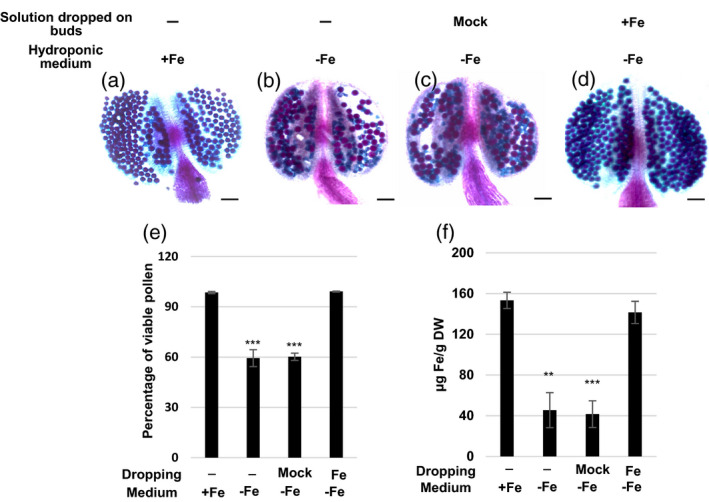
Pollen viability and Fe content in floral buds of plants grown in Fe‐deficient medium.
(a–d) Alexander staining of mature pollen from plants grown in Fe‐sufficient (+Fe, a and b) and Fe‐deficient (−Fe, c and d) hydroponic medium and treated without (a, b) and with 50 μm Na2‐EDTA (Mock, c) or 50 μm NaFe‐EDTA (Fe, d) by dropping the solution on floral buds. (e) Percentage of viable pollen calculated from Alexander staining of pollen. (f) Fe concentration of floral buds. Data are shown as mean ± standard error of three independent experiments. **P < 0.01, ***P < 0.001 compared with Fe‐sufficient plants (Student’s t‐test). Bars = 50 μm in (a–d).
Differentially expressed genes in anthers, leaves, and roots under Fe deficiency
To uncover transcriptional regulation in Fe‐deficient anthers and its link to pollen and tapetum defects, we analyzed the transcriptomes of anthers of plants grown under Fe‐sufficient and ‐deficient conditions using RNA sequencing (RNA‐seq). The processes of transcriptome analysis used in this study are shown in a flowchart (Figure S4). First of all, in our anther transcriptome, we detected the upregulation of several known Fe deficiency‐responsive (−Fe) genes (Hindt et al., 2017; Le Jean et al., 2005; Rodriguez‐Celma et al., 2019, 2013; Urzica et al., 2012), confirmed by quantitative real‐time PCR (qPCR) (Figure S5a). Thus, the results indicated that the Fe deficiency response occurs in anthers. A comparison of a set of Fe‐sufficient and ‐deficient root and leaf transcriptomes (Grillet et al., 2018) with our anther transcriptome showed that a total of 723, 639, and 1164 differentially expressed genes (DEGs) (adjusted P‐value [P.adj] < 0.05) were identified under Fe deficiency in anthers, leaves, and roots, respectively (Figure 4a). Nine genes (listed in Table S3) were commonly differentially expressed in the root, leaf, and anther transcriptomes under Fe deficiency. A comparison of the −Fe DEGs in three tissues showed that 86.4% (625/723) of anther DEGs were anther‐unique, 67.8% (433/639) of leaf DEGs were leaf‐unique, and 81.7% (951/1164) of root DEGs were root‐unique (Figure 4a), indicating that the majority of DEGs responding to Fe deficiency were tissue‐unique. Therefore, transcriptional alteration caused by Fe deficiency in anthers might differ from that in roots and leaves, and transcriptome analysis of Fe‐deficient anthers may facilitate the dissection of anther‐unique Fe deficiency responses.
Figure 4.
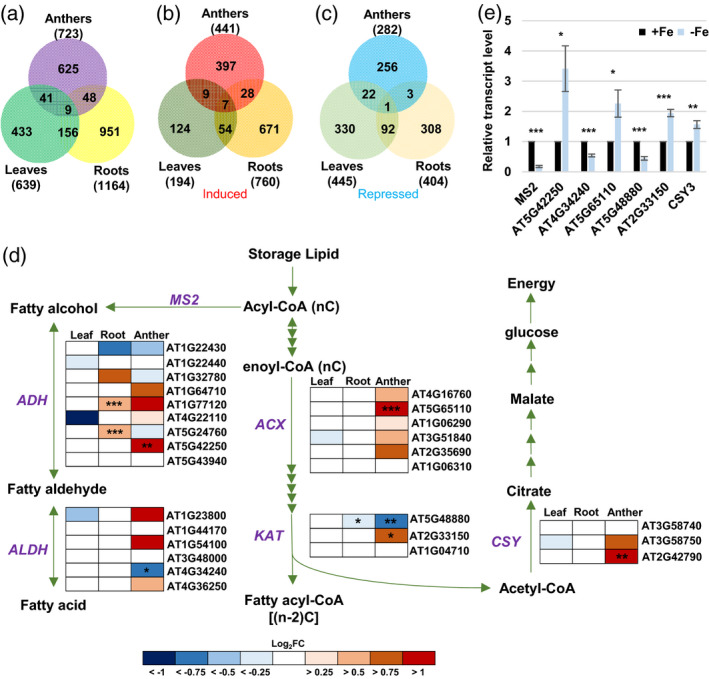
Analysis of Fe deficiency‐responsive genes and genes involved in the fatty acid degradation pathway.
(a–c) Venn diagrams of total (a), induced (b), and repressed (c) DEGs in Fe‐deficient anthers, leaves, and roots (P.adj < 0.05). (d) Changes in expression of genes involved in the fatty acid degradation pathway (P = 0.00377) under Fe deficiency. MS2, MALE STERILITY 2; ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; ACX, acyl‐CoA oxidase; KAT, 3‐ketoacyl‐CoA thiolase; CSY, citrate synthase. Enrichment analysis of anther DEGs was performed using EXPath 2.0. Boxes indicate relative expression changes in genes encoding enzymes. Log2FC, log2(fold change of expression levels) from RNA‐seq data in Fe‐deficient (−Fe) versus ‐sufficient (+Fe) tissues. *P.adj < 0.05, **P.adj < 0.01, ***P.adj < 0.001 between −Fe and +Fe conditions. (e) Comparison of transcript levels of genes from panel (d) between anthers from −Fe and +Fe plants as analyzed by qPCR. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 between −Fe and +Fe anthers (Student’s t‐test).
Pathway enrichment analysis of anther‐unique DEGs
To characterize the critical metabolic pathways altered explicitly in Fe‐deficient anthers, we performed pathway enrichment analysis for 653 (397 induced plus 256 repressed; Figure 4b,c) anther‐unique −Fe DEGs. The results showed that a total of 15 pathways were significantly (P < 0.05) enriched (Table S4). Among 15 enriched pathways, ‘starch and sucrose metabolism’, ‘nitrogen metabolism’, ‘nicotinate and nicotinamide metabolism’, and ‘alanine, aspartate, and glutamate metabolism’ might be linked to the changes in metabolites in Fe‐deficient anthers. ‘Phenylpropanoid biosynthesis’ and ‘fatty acid degradation’ were also enriched in anther‐unique −Fe DEGs. Nine anther‐unique −Fe DEGs were involved in the phenylpropanoid biosynthesis pathway, of which six were detected in the tapetum transcriptome (Li et al., 2017) and repressed under Fe deficiency (Table S5). The phenylpropanoid pathway occurs in the tapetum, and phenylpropanoid derivatives are essential components of sporopollenin, the outer pollen wall precursor (Xue et al., 2020). Thus, the misregulation of the phenylpropanoid biosynthesis pathway under Fe deficiency might be related to an impaired pollen outer wall.
It is known that the supply of fatty acids is essential for sporopollenin biosynthesis (de Azevedo Souza et al., 2009; Lallemand et al., 2013; Shi et al., 2015). Four anther‐unique genes, encoding an alcohol dehydrogenase (ADH, AT5G42250), an aldehyde dehydrogenase (ALDH3/ALDH3l1, AT4G34240), acyl‐CoA oxidase (ACX2, AT5G65110), and a 3‐ketoacyl‐CoA thiolase (KAT2, AT2G33150), were involved in the fatty acid degradation pathway. ADH and ALDH are critical enzymes for generating fatty alcohol and fatty aldehyde, intermediates of sporopollenin, from fatty acid (de Azevedo Souza et al., 2009). ACXs and KATs are involved in the first and last steps of fatty acid degradation and are critical for fatty acid degradation. Mutants with defects in ACXs failed to catabolize storage lipids (Pinfield‐Wells et al., 2005; Pracharoenwattana et al., 2005). In addition, MALE STERILITY 2 (MS2), which is involved in converting fatty acid to fatty alcohol (Chen et al., 2011), was repressed in Fe‐deficient anthers. Furthermore, fatty acid degradation was blocked in a mutant with a defect in peroxisomal citrate synthase (CSY). CSY is vital for the transfer of carbon from storage lipids to glucose by multiple processes (Figure 4d) (Eckardt, 2005; Pracharoenwattana et al., 2005), and csy loss‐of‐function mutants cannot metabolize triacylglycerol (TAG) (Pracharoenwattana et al., 2005), suggesting that CSY plays a role in facilitating fatty acid degradation. We found that the transcript levels of ACX2, KAT2, and CSY3 (AT2G42790) were upregulated and MS2 and KAT5 (AT5G48880) were downregulated in Fe‐deficient anthers (Figure 4d,e). Although KATs participate in fatty acid degradation, KAT5 is highly correlated with genes associated with flavonoid biosynthesis (Carrie et al., 2007; Yonekura‐Sakakibara et al., 2008), and KAT5 has been proposed to play a role in the regulatory network of carbon source to direct the carbon flux toward the biosynthesis of flavonoids (Perez de Souza et al., 2020), one of the principal components of sporopollenin (Shi et al., 2015). The upregulation of ACX2, KAT2, and CSY3 might imply an increase in fatty acid degradation, which is also suggested by a decrease in neutral lipids (a primary type of storage lipids) in the tapetum (Figure 5). The downregulation of KAT5 and MS2 might indicate a decrease in flavonoid and fatty alcohol biosynthesis, which might facilitate fatty acid degradation. The differential expression of MS2, AT5G42250, AT4G34240, AT5G65110, AT5G48880, and AT2G33150 was confirmed by qPCR (Figure 4e). The downregulation of genes involved in phenylpropanoid biosynthesis and upregulation of genes linked to fatty acid degradation suggests a reduction in components for the synthesis of sporopollenin.
Figure 5.
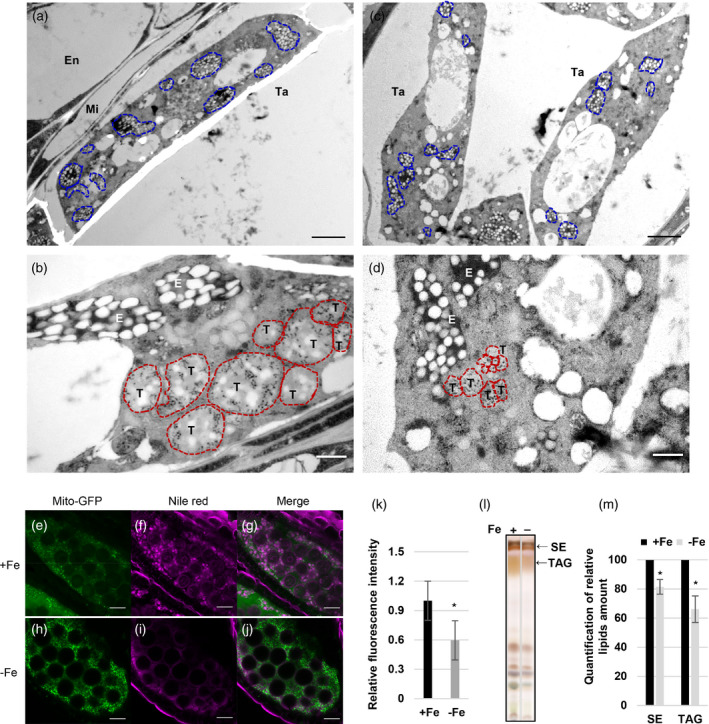
Tapetosomes, elaioplasts, and neutral lipid content in anthers under Fe deficiency.
(a–d) Fe‐sufficient (+Fe, a and b) and Fe‐deficient (−Fe, c and d) tapetal cells at the BN stage were observed using TEM. Blue (a and c) and red (b and d) dash lines indicate elaioplasts and tapetosomes, respectively. Bars = 2 μm (a and c) and 0.5 μm (b and d). Ta, tapetum; Mi, middle layer; En, endothecium; T, tapetosome; E, elaioplast. (e–j) Arabidopsis anthers showing tapetum‐specific expression of mito‐GFP from +Fe (e–g) and −Fe (h–j) plants were stained with Nile red (f, i) at the TC stage. (g and j) The merged images of mito‐GFP and Nile red. Bars = 20 μm. (k) Quantification of relative (versus WT under +Fe) Nile red fluorescence intensity of the tapetum (n > 10). (l) Lipids were extracted from anthers of Brassica napus and separated by TLC. SE, sterol ester; TAG, triacylglycerol. (m) Quantification of relative (versus WT under +Fe) amounts of SE and TAG in panel (l). Data are shown as mean ± standard error of three independent experiments. *P < 0.05, **P < 0.01 between −Fe and +Fe conditions (Student’s t‐test).
Since each of the five enzymes ADH, ALDH, ACX, KAT, and CSY is encoded by more than one gene (Figure 4d) in Arabidopsis and they might be regulated in different manners in tissues under Fe deficiency, we analyzed the changes in expression of 27 genes encoding five enzymes in the anther, leaf, and root transcriptomes in response to Fe deficiency (Figure 4d). Among the 27 genes, although the transcript levels of five genes were changed under Fe deficiency in the leaves, none of them showed a statistically significant change (P.adj < 0.05); in the roots, the transcript levels of five genes were changed, and three of them showed statistically significant differences. Of note, in anthers, the transcript levels of 20 genes were altered, and six of them showed statistically significant changes. These results suggest that although the five enzymes in the fatty acid degradation pathway are present in all three tissues, the genes encoding the five enzymes seem to be altered more obviously in anthers than in leaves and roots under Fe deficiency.
Fe deficiency affects neutral lipid accumulation in the tapetum
To examine whether fatty acid degradation was facilitated under Fe deficiency, we checked storage lipid levels in anthers under Fe deficiency. In Brassicaceae anthers, including Arabidopsis, neutral lipids are the primary type of storage lipid, most of which are stored in two organelles, the tapetosomes and elaioplasts, in the tapetum (Hsieh and Huang, 2007). At the BN stage, under Fe deficiency, tapetosomes and elaioplasts in the tapetum were fewer and smaller than those under Fe sufficiency (Figure 5a–d). We further used Nile red, a specific dye for neutral lipids, to analyze the neutral lipid content in anthers expressing tapetal mito‐GFP generated in our previous study (Chen et al., 2019). Under Fe sufficiency, neutral lipids (Nile red signal) were mainly present in the tapetum, marked by mito‐GFP (Figure 5e–g). Under Fe deficiency, the levels of neutral lipids in the tapetum were lower (Figure 5h–j). The quantification of Nile red intensity (neutral lipid content) in the tapetum (area showing mito‐GFP) revealed that the levels of neutral lipids in the tapetum were lower under Fe deficiency (Figure 5k).
The characteristics of tapetum cells in Brassica and Arabidopsis are highly similar (Chalhoub et al., 2014; Owen and Makaroff, 1995; Platt et al., 1998), especially with regard to lipid storage in tapetosomes and elaioplasts (Hsieh and Huang, 2007; Kim et al., 2005). To further demonstrate that this phenomenon occurs in Arabidopsis and other closely related species, we analyzed neutral lipids in Brassica anthers grown under Fe‐sufficient and ‐deficient conditions. Taking advantage of the large size of Brassica anthers and the established method for quantifying the relative amount of TAG and sterol esters (SEs) in Brassica anthers, we used TLC to separate SEs and TAG in anthers of Brassica napus, which contains large amounts of SEs and TAG in the tapetum (Hsieh and Huang, 2007) (Figure 5l). Quantification of the SEs and TAG revealed that their levels were lower under Fe deficiency (Figure 5m). These results indicated that Fe deficiency reduces the contents of neutral lipids, including TAG and SEs, and impairs tapetosome and elaioplast formation in the tapetum.
Identification of hub transcription factors in transcriptional alteration in the Fe‐deficient tapetum
Stress conditions can cause tapetal dysfunction by changing the expression of genes involved in tapetum and microspore development, signaling, and metabolic pathways (Jin et al., 2013; Parish et al., 2012). Since most tapetum‐specific genes are involved in the regulation of tapetum function and development, we compared our anther‐unique DEGs with published tapetum and mature pollen transcriptomes (Li et al., 2017; Rahmati Ishka et al., 2018) to identify ‘tapetum‐specific −Fe DEGs’, detected in the tapetum, but not the mature pollen transcriptome. Among anther‐unique DEGs, 37 of 397 induced genes and 114 of 256 repressed genes were tapetum‐specific (Figure 6a,b). We performed co‐expression analysis of 151 tapetum‐specific DEGs (37 induced plus 114 repressed genes) using ExPath2.0 to identify the hub genes involved in adjusting tapetum function under Fe deficiency. The co‐expression network of tapetum‐specific −Fe DEGs showed that AMS, which encodes a transcription factor essential for tapetum development and function (Lou et al., 2018; Ma et al., 2012; Wang et al., 2018; Xu et al., 2014) that acts as a master regulator of sporopollenin biosynthesis (Xu et al., 2014), was one of four genes with the largest number of correlations with other DEGs. TDF1/AtMYB35 had the second‐largest and bHLH089 and bHLH091 had the third‐largest number of correlations with other DEGs (Figure 6c). It has been shown that DYT1, TDF1, AMS, bHLH089/091/010, and MYB80 can form a genetic regulatory pathway to regulate tapetum function and development. Each has its specific target genes involved in controlling diverse tapetum functions (Cui et al., 2016; Li et al., 2017; Xu et al., 2010; Zhu et al., 2015). We examined the transcript levels of AMS, TDF1, bHLH089/091/010, DYT1, and MYB80 in anthers. The results showed that AMS, TDF1, and bHLH089/091/010 were downregulated, but DYT1 and MYB80 levels were not significantly (P > 0.05) changed under Fe deficiency (Figure 6d and Figure S5b). These results suggested that Fe deficiency affects AMS, TDF1, and bHLH089/091/010 but not DYT1 and MYB80 at the transcriptional level.
Figure 6.
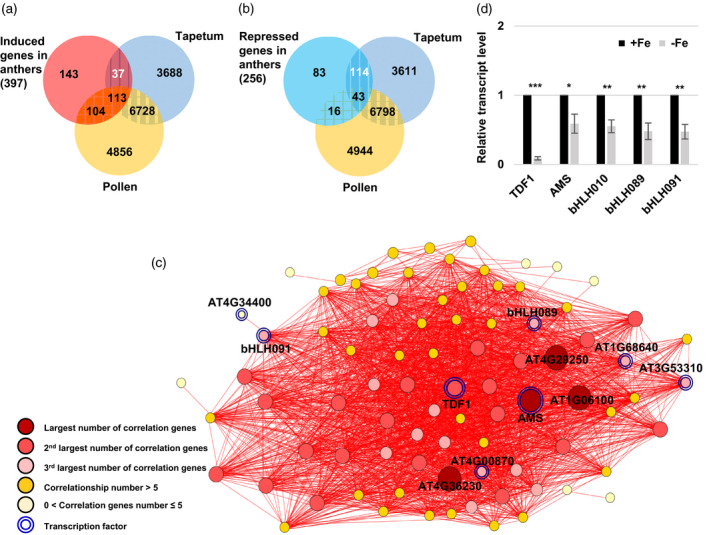
Identification and analysis of tapetum‐unique DEGs under Fe deficiency.
(a, b) Venn diagrams showing the overlap of induced (a) and repressed (b) DEGs (P.adj < 0.05) in Fe‐deficient anthers with genes detected in the tapetum and mature pollen transcriptomes. (c) Tapetum‐unique DEGs (37 upregulated plus 114 repressed DEGs in panels (a) and (b)) were selected for co‐expression analysis. Each node represents a gene. Two genes connected by a red line indicate a significant positive correlation (Pearson’s correlation coefficient > 0.80) across different conditions determined using EXPath 2.0. (d) Relative transcript levels of TDF1, AMS, bHLH089, bHLH091, and bHLH010 in anthers between −Fe and +Fe plants as determined by qPCR. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, ***P < 0.001 between −Fe and +Fe anthers (Student’s t‐test).
A large portion of tapetum‐expressed −Fe DEGs are also DEGs of the ams, tdf1, and bhlhs mutant transcriptomes
To reveal how transcriptional alterations of AMS, TDF1, and bHLH089/091/010 are related to the changes in tapetum function under Fe deficiency, we analyzed transcriptomes of the tdf1, ams, and triple bhlhs mutants (Li et al., 2017; Ma et al., 2012; Xu et al., 2014) and identified 2389, 1415, and 1081 DEGs (between mutant and wild type [WT]) for the ams, tdf1, and bhlhs mutants, respectively. To focus on DEGs of the ams, tdf1, and bhlhs mutants expressed in the tapetum, we selected the mutant DEGs which were expressed in the published tapetum transcriptome as tapetum‐expressed mutant DEGs and obtained 1324, 651, and 390 tapetum‐expressed mutant DEGs from 2389, 1415, and 1081 DEGs of the ams, tdf1, and bhlhs mutants, respectively (Figure S4). By comparing the tapetum‐expressed −Fe DEGs and tdf1, ams, and bhlhs mutant DEGs, we found that 50% (79 of 157 repressed genes in Figure 7a and 75 of 150 induced genes in Figure 7b) of the −Fe DEGs were also the DEGs in the ams, tdf1, and bhlhs mutants. Among the 154 (79 plus 75) −Fe DEGs, approximately 43, 31, and 20% of the repressed genes and 30, 7, and 34% of the induced genes were found to be DEGs in the ams, tdf1, and bhlhs mutants, respectively. This suggests that the downregulation of AMS and bHLHs might be associated with both repression and induction of tapetum‐expressed −Fe genes. However, the downregulation of TDF1 might be more related to the repression of tapetum‐expressed −Fe genes. In addition, some −Fe DEGs only belonged to the ams mutant transcriptome but not tdf1, which might be because of the biphasic regulation of AMS protein (Ferguson et al., 2017).
Figure 7.
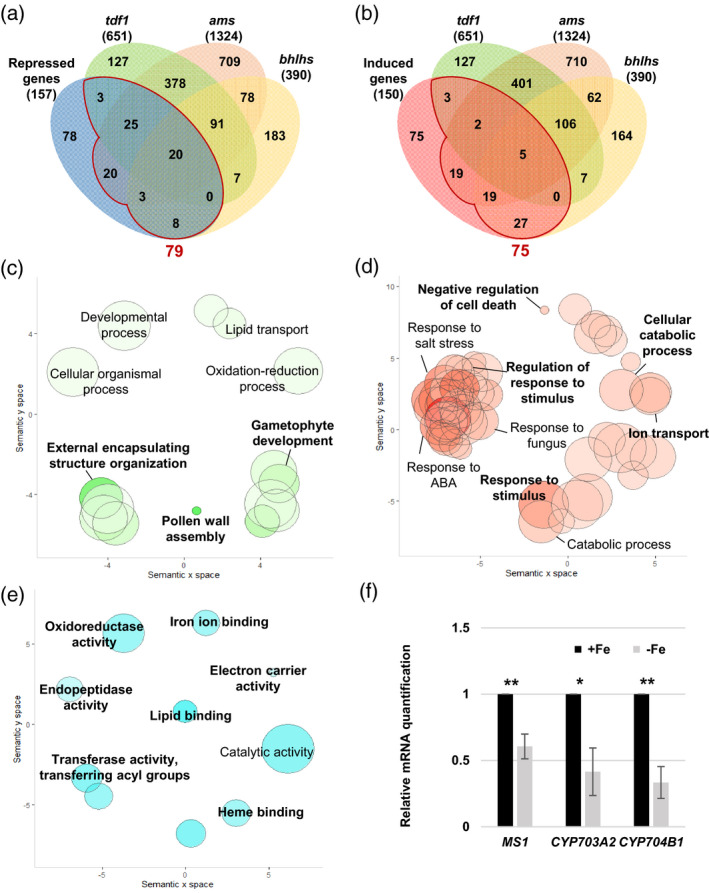
Analysis of tapetum‐expressed DEGs in Fe‐deficient anthers and in tdf1, ams, and bhlhs mutants.
(a, b) Venn diagrams showing the overlap of 157 repressed (a) and 150 induced (b) tapetum‐expressed genes (P.adj < 0.05) in Fe‐deficient anthers and 651, 1324, and 390 tapetum‐expressed DEGs in the tdf1, ams, and bhlhs mutant transcriptomes, respectively. A total of 79 repressed and 75 induced DEGs, i.e., the sum of gene numbers from the regions in panels (a) and (b) enclosed by red lines, in Fe‐deficient anthers overlapped with DEGs in the tdf1, ams, and bhlhs mutants. (c–e) GO enrichment analysis of the −Fe DEGs detected in tdf1, ams, or bhlhs. Enriched GO terms in the biological process category for the 79 Fe deficiency‐repressed (c) and 75 Fe deficiency‐induced (d) genes indicated in panels (a) and (b), respectively. (e) Enriched GO terms in the molecular function category for the 79 Fe deficiency‐repressed genes detected in tdf1, ams, or bhlhs. Semantic representation of significantly enriched GO categories. Color intensity reflects the significance of the enrichment test, with dark colors corresponding to lower P‐values (<0.05). Circle size indicates the relative frequency of the GO term in the EBI GOA database (Barrell et al., 2009). (f) Comparison of mRNA levels between anthers of −Fe and +Fe plants as determined by qPCR. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, **P < 0.01 between −Fe and +Fe anthers (Student’s t‐test).
With regard to the Gene Ontology (GO) terms in the biological process category, the 79 genes repressed under Fe deficiency, and also the DEGs in the tdf1, ams, and bhlhs mutants, were enriched in GO terms linked to pollen wall formation such as ‘pollen wall assembly’, ‘gametophyte development’, and ‘external encapsulating structure organization’ (Figure 7c and Figure S6). The GO terms of the 75 Fe deficiency‐induced genes, and also the DEGs in the tdf1, ams, and bhlhs mutants, were mainly related to responses to stress and stimuli as well as ‘negative regulation of cell death’, ‘catabolic process’, and ‘ion transport’ (Figure 7d). MS1, MS2, CYP704B1, CYP703A2, and TKPR2, which are associated to GO terms linked to pollen wall formation, are known to be required for sporopollenin biosynthesis. It has been reported that mutants with defects in these genes show abnormal pollen walls (Chen et al., 2011; Dobritsa et al., 2009; Grienenberger et al., 2010; Lallemand et al., 2013; Vizcay‐Barrena and Wilson, 2006; Xiong et al., 2016). qPCR confirmed the downregulation of MS1, MS2, CYP703A2, and CYP704B1 in Fe‐deficient anthers (Figure 7f; the MS2 result is shown in Figure 4e).
The enriched GO terms in the molecular function category of the 79 repressed genes were associated with Fe‐dependent activities, including ‘iron ion binding’, ‘heme binding’, ‘oxidoreductase activity’, and ‘electron carrier activity’, and lipid‐related activities that included ‘lipid binding’, ‘transferase activity of acyl group’, and ‘endopeptidase activity’ (Figure 7e). In addition, many genes annotated to ‘iron ion binding’ and ‘heme binding’ encoded proteins requiring Fe for their function, including RBOHE, CYP703A2, CYP704B1, CYP98A8, CYP86C4, PRX9, ATCB5‐B, and AT2G24800, encoding a putative peroxidase. Thus, these results suggested that Fe deficiency might repress a portion of genes encoding proteins with Fe‐dependent activities through affecting AMS, TDF1, and bHLHs.
Changes in RBOHE expression alter pollen defects under Fe deficiency
It has been shown that tapetum‐expressed RBOHs, RBOHE, and RBOHC (also called ROOT HAIR DEFECTIVE 2 [RHD2]), are necessary for anther ROS production and that proper expression of tapetum‐expressed RBOHs is critical for the precise occurrence of tapetum PCD and pollen development (Xie et al., 2014). Therefore, to check whether RBOHs are linked to pollen defects under Fe deficiency, we examined RBOHE and RBOHC expression in Fe‐sufficient and ‐deficient anthers by qPCR. The results showed that RBOHE expression was higher than RBOHC expression under Fe sufficiency, and RBOHE but not RBOHC was downregulated under Fe deficiency (Figure 8a). This result implies that RBOHE might play a dominant role in anthers under Fe‐sufficient conditions and seems to respond to Fe deficiency transcriptionally. Furthermore, we used H2DCFDA staining to investigate anther ROS levels and found that the anther ROS levels were dramatically lower at most stages under Fe deficiency compared with the same stages under Fe sufficiency (Figure 8b,c and Figure S7), which is opposite to the increase in ROS in Fe‐deficient roots and leaves (Tewari et al., 2013; Zhai et al., 2018). As ROS can serve as signaling molecules to impact cellular processes, the diverse ROS regulation in various tissues might be associated with their different responses to Fe deficiency.
Figure 8.
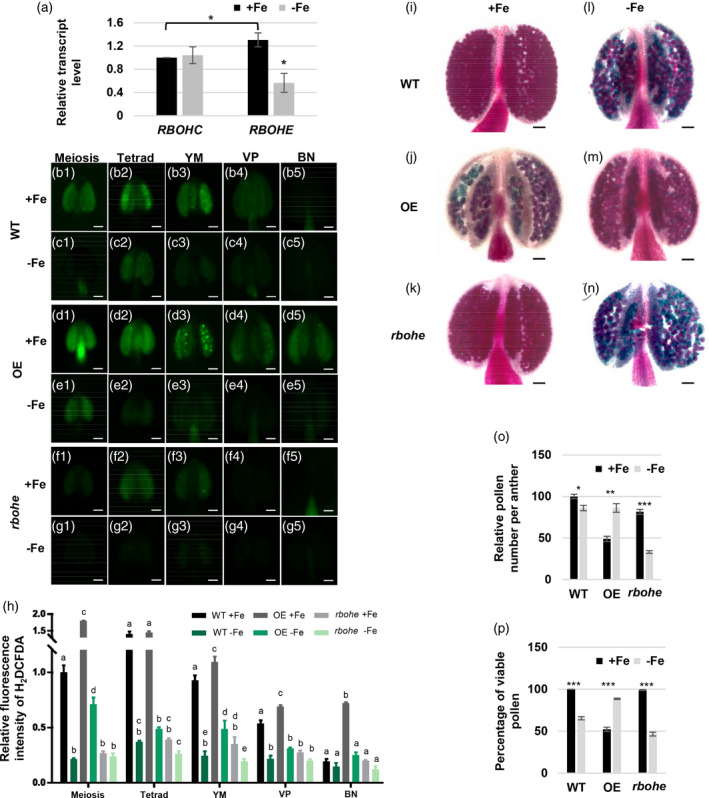
The expression level of RBOH in anthers and the effect of RBOHE level on pollen viability and anther ROS levels under Fe sufficiency and deficiency.
(a) The transcript levels of RBOHC and RBOHE in anthers grown under Fe‐sufficient (+Fe) and ‐deficient (−Fe) conditions were examined by qPCR. (b–g) ROS levels in different stage anthers of wild‐type (WT), Osg6b:RBOHE overexpression (OE), and rbohe mutant plants grown under +Fe and −Fe conditions were analyzed by H2DCFDA staining. (h) Quantification of H2DCFDA fluorescence intensity in anthers at different stages. Different letters represent statistically significant differences between +Fe and −Fe conditions and different lines in each stage according to two‐way anova with Tukey’s multiple comparisons test (P < 0.05) (Data S1). Pollen viability in anthers from WT, OE, and rbohe mutant plants grown under +Fe (i–k) and −Fe (l–n) conditions using Alexander staining. Viable pollen is purple and non‐viable pollen is blue‐green. (o) The quantification of relative pollen number per anther (versus WT under +Fe). (p) The quantification of the percentage of viable pollen. All of the pollen grains in anthers were counted. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005 between −Fe and +Fe conditions (Student’s t‐test). Bars = 50 μm. YM, young microspore; VP, vacuolated pollen; BN, binucleate pollen.
A previous study showed that Osg6B:RBOHE overexpression (OE) lines have higher transcript levels of RBOHE than WT and the rbohe null mutant has no detectable RBOHE transcripts (Xie et al., 2014). Therefore, to investigate whether the reduction of anther ROS levels under Fe deficiency was RBOHE‐dependent, we examined anther ROS levels in OE, rbohe, and WT anthers (Figure 8d–g and Figure S7). The quantification of anther ROS levels under Fe‐sufficient conditions showed that compared with WT, the OE line had higher anther ROS levels and the rbohe mutant had lower anther ROS levels at most analyzed stages (Figure 8b,d,f,h, Figure S7, Data S1). Under Fe‐deficient conditions, the WT and the OE line had lower anther ROS levels at most stages, and the rbohe mutant had lower anther ROS levels at the YM stage compared with anther ROS levels under Fe‐sufficient conditions (Figure 8c,e,g,h, Figure S7, Data S1). These results indicated that anther ROS are mainly generated in an RBOHE‐dependent manner, but Fe‐dependent and RBOHE‐independent ROS generation also contribute to ROS levels in anthers.
To reveal whether impaired pollen development is associated with the downregulation of RBOHE under Fe deficiency, we used Alexander staining to detect viable pollen in the WT, OE, and rbohe anthers just before dehiscence (Figure 8i–n) and quantified the production of viable pollen as the total pollen per anther and the percentage of viable pollen in anthers (Figure 8o,p). Under Fe sufficiency, the OE line and the rbohe mutant had approximately 50% and 20% less total pollen per anther than WT (Figure 8o), indicating that a portion of pollen degenerated early and did not remain in anthers before dehiscence. In addition, compared with the WT, the percentage of viable pollen was 50% lower in the OE line and unaltered in the rbohe mutant under Fe sufficiency (Figure 8p). Importantly, under Fe deficiency, the total pollen number per anther was slightly lower in the WT, restored in the OE line, and dramatically lower in the rbohe mutant (Figure 8o) than the number under Fe‐sufficient conditions. The production of viable pollen was lower in the WT, restored in the OE line, and dramatically lower in the rbohe mutant under Fe deficiency (Figure 8i–p). These results indicated that pollen defects caused by Fe deficiency were rescued by overexpression of RBOHE and pollen defects under Fe deficiency became more severe in anthers lacking RBOHE. To sum up, the expression status of RBOHE in anthers seems to be associated with the production of viable pollen under Fe deficiency.
Mitochondrial function and status were altered under Fe deficiency
The tapetum is highly metabolically active and requires a lot of energy. In addition, proper regulation of anther ROS levels is necessary to produce viable pollen (Xie et al., 2014). Mitochondria are the primary source of both energy and ROS through mitochondrial respiration. Fe is vital for the activities of mitochondrial respiratory complexes, and Fe deficiency affects mitochondrial respiration (Vigani, 2012). To investigate whether mitochondrial function, especially mitochondrial respiration, was influenced by Fe deficiency, we measured the oxygen consumption rate (OCR) of anthers under Fe sufficiency and deficiency. The results showed that anthers grown under Fe deficiency had an approximately 30% reduction in OCR compared to anthers grown under Fe sufficiency (Figure 9a). Since the alteration of overall mitochondrial respiration can result from changes in the activities of mitochondrial respiratory complexes, mitochondrial density, and mitochondrial morphology (Mishra and Chan, 2016; Sesso et al., 2012), we further examined how these mitochondrial characteristics were altered in anthers grown under Fe deficiency.
Figure 9.
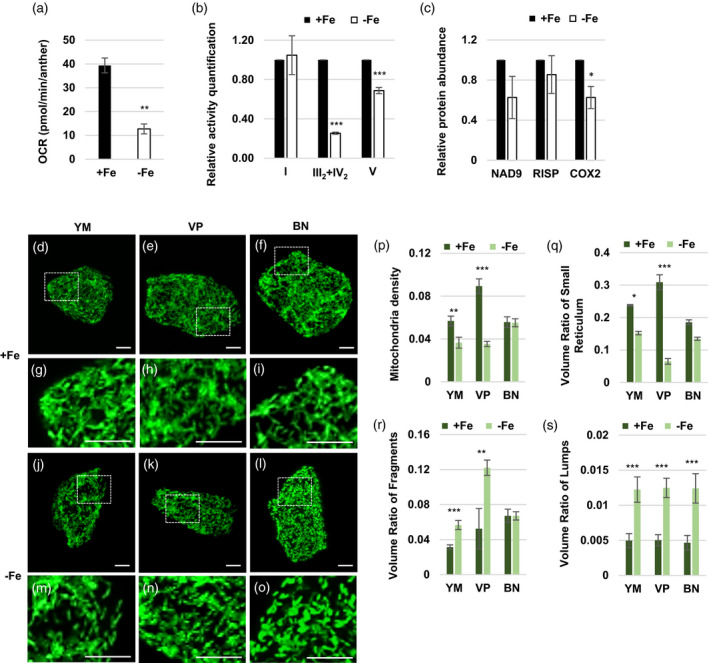
Mitochondrial function and status in anthers and tapeta under Fe deficiency.
(a) The average basal oxygen consumption rates (OCRs) of six anthers (two from each stage) collected at the YM, VP, and BN stages from plants grown under Fe‐sufficient (+Fe) and ‐deficient (−Fe) conditions. (b) The relative mitochondrial complex activity. The same volumes of crude mitochondrial extract from anthers of Brassica napus were separated by BN‐PAGE. Then, the in‐gel activity of mitochondrial complexes I, IV, and V was quantified. (c) Relative protein abundances of NAD9, RISP, and COX2 were determined by Western blot analysis. (d–o) Mitochondrial morphology in the tapetum at the YM, VP, and BN stages. Individual tapetal cells expressing mito‐GFP from plants grown under +Fe (d–i) and −Fe (j–o) conditions were photographed under a Zeiss LSM780. Projected mitochondrial (GFP) images from Z‐stacks are shown. Bars = 5 μm. (p–s) The morphological features of tapetal cell mitochondria were analyzed by MicroP 3D automatic quantification software. Data are shown as mean ± standard error of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005 between −Fe and +Fe conditions (Student’s t‐test). YM, young microspore; VP, vacuolated pollen; BN, binucleate pollen.
Using B. napus, we analyzed the activities of mitochondrial respiratory complexes in stage‐mixed anthers by a blue native (BN) gel‐based activity assay (Pineau et al., 2008). The results showed that compared with Fe‐sufficient conditions, mitochondrial complex I activity had no apparent change, and supercomplex III2+IV2 and complex V (F1) activities were lower under Fe deficiency (Figure 9b, Figure S8a–d). Furthermore, we quantified the abundance of cytochrome c oxidase subunit 2 (COX2), NADH dehydrogenase subunit 9 (NAD9), and Rieske iron‐sulfur protein (RISP) in B. napus anthers by Western blot analysis. The results showed COX2 abundance was slightly lower, but the protein abundances of NAD9 and RISP were not affected in anthers grown under Fe deficiency (Figure 9c, Figure S8e). These results suggested that Fe deficiency mainly reduced mitochondrial supercomplex III+IV and complex V activity in anthers through mechanisms other than decreasing the relative abundance of respiratory complexes in mitochondria.
Furthermore, we wanted to reveal whether mitochondrial density and morphology were altered under Fe deficiency. Since (i) tapetal cells and developing pollen contain a higher density of mitochondria per area compared with other anther layers (Lee and Warmke, 1979), (ii) there is an apparent reduction in Fe content in the tapetum in Fe‐deficient anthers, and (iii) mitochondrial density and morphology in tapetal cells at the YM to BN stages are more distinguishable compared with other stages (Chen et al., 2019), we analyzed the changes in mitochondrial density and morphology in the tapetum from the YM to BN stages under Fe deficiency using the approach established in our previous study (Chen et al., 2019). The whole population of mito‐GFP‐labeled mitochondria in isolated tapetal cells was imaged and analyzed (Figure S9a–d). Under Fe deficiency, tapetal mitochondria became more fragmented and swollen at the YM, VP, and BN stages than those under Fe‐sufficient conditions (Figure 9d–o). Therefore, the geometric and textural features of tapetal mitochondria at the YM, VP, and BN stages were further analyzed. The mitochondria in single tapetal cells at different stages were classified into six subtypes: fragments, lumps, simple tubules, branched tubules, small reticulum, and large reticulum (representative tapetal cells are shown in Figure S10). Quantification of mitochondrial features showed that under Fe deficiency, mitochondrial density and the volume ratio of the small reticulum were dramatically lower at the YM and VP stages (Figure 9p,q). The volume ratio of fragment‐type mitochondria, which have low respiratory activity (Mishra and Chan, 2016; Westermann, 2012), to all mitochondria was higher at the YM and VP stages (Figure 9r). The volume ratio of lump‐type mitochondria, which are swollen and usually associated with mitochondrial dysfunction (Crompton, 1999; Green and Reed, 1998; Zoratti and Szabò, 1995), to all mitochondria was higher at all three stages (Figure 9s). Our results indicated that Fe deficiency caused a reduction in mitochondrial density and an increase in fragmented and swollen mitochondria in the tapetum. Together, the decrease of mitochondrial respiratory efficiency (indicated by OCR) of anthers grown under Fe deficiency was probably caused by a decline in mitochondrial complex activity and mitochondrial density and an increase in fragmented mitochondria.
DISCUSSION
Fe deficiency in floral buds is the leading cause of defects in pollen development
Impaired reproductive growth, including defective pollen development, was detected in mutants with defects in Fe transport (Le Jean et al., 2005; Roschzttardtz et al., 2011; Yokosho et al., 2016) and in WT plants grown under Fe deficiency from the vegetative stage to the reproductive stage (Briat et al., 2015; El‐Jendoubi et al., 2014). However, the impaired reproductive growth may be caused by insufficient Fe levels in reproductive tissues and significant changes in vegetative tissues such as impaired photosynthesis. Therefore, it has been difficult to interpret whether Fe deficiency is the direct cause of defects in reproductive growth and how Fe insufficiency affects pollen development. Although the Fe content in leaves was also lower under our Fe‐deficient conditions, the total chlorophyll content in the leaves, an indicator of photosynthesis efficiency, was not significantly lower. In addition, the contents of the main carbohydrates and amino acids were not changed and were even slightly higher in floral buds when plants were grown in Fe‐deficient medium. Importantly, we revealed that the Fe content and pollen defects in plants grown under Fe‐deficient conditions were restored by supplying Fe to floral buds only. Our results indicate that Fe deficiency in floral buds directly impacts pollen development and suggest that directly spraying Fe solution on reproductive tissues may be an efficient approach for rescuing reproductive growth when there is insufficient Fe available in the soil. Under Fe deficiency, Fe content in anthers was lower in the tapetum layer, and further cellular and molecular studies showed that Fe deficiency affects pollen development mainly through impairing tapetum function. However, the possibility that Fe insufficiency in pollen also disrupts its cellular processes and contributes to reduced viable pollen cannot be excluded.
Transcriptional alteration of AMS, TDF1, and bHLH089/091/010 might be linked to several tapetal defects under Fe deficiency
Through analyzing how the anther transcriptome is affected by Fe insufficiency, we found that AMS, TDF1, and bHLH089/091/010 are critical upstream transcription factors, and their transcriptional alteration may affect their downstream genes such as RBOHE, MS1, MS2, CYP703A2, and CYP704B1, resulting in many defects in the tapetum under Fe deficiency. Although previous studies have shown that DYT1, TDF1, AMS, and MYB80 are critical for tapetum function and have their specific target genes involved in diverse tapetum functions (Li et al., 2017), it is unknown whether and how these transcription factors might be affected under Fe deficiency. This study showed that Fe deficiency affects AMS, TDF1, and bHLH089/091/010 (bHLHs), but not DYT1 and MYB80, at the transcriptional level. Since the function of DYT1 is influenced not only by transcriptional regulation but also by the regulation of protein subcellular localization and protein–protein interaction (Cui et al., 2016), we could not exclude the possibility of a change in protein levels or function of DYT1 (and probably also MYB80) under Fe deficiency. Notably, the expression of TDF1 was repressed more severely than the expression of AMS and bHLHs (Figure 6d). Whether the downregulation of TDF1 is the primary change causing the transcriptional alteration in Fe‐deficient anthers is a critical issue that needs to be further studied.
We also revealed that Fe deficiency changes transcript levels of several genes encoding Fe‐dependent proteins such as RBOHE, CYP703A2, CYP704B1, CYP98A8, CYP86C4, PRX9, AT2G24800, encoding a putative peroxidase, and ATCB5‐B. It has been shown that CYP98A8, CYP703A2, and CYP704B1 are direct targets of AMS (Xu et al., 2014), and thus their downregulation might result from the reduction in AMS transcript levels under Fe deficiency. Although −Fe DEGs were not enriched in GO terms related to mitochondrial function, our results show that mitochondrial respiratory complex activities were reduced under Fe deficiency, possibly because less Fe was available for the Fe‐dependent subunits of respiratory complexes (Figure 9b). These results suggest that Fe deficiency may reduce the activities and/or transcript levels of Fe‐dependent proteins in anthers. However, since only nuclear‐encoded transcripts were analyzed in our transcriptomes, we could not exclude the possibility that the reduction of mitochondrial respiratory activities was caused by a change in the transcript levels of genes encoded by the mitochondrial genome. To sum up, Fe deficiency might cause functional alterations in some Fe‐dependent proteins because of the lack of available Fe as their cofactor and also affect some Fe‐dependent proteins at the transcriptional level through alteration of their upstream transcription factors. However, how Fe deficiency induces alterations in the levels of upstream transcription factors in the tapetum and whether Fe deficiency directly or indirectly impacts transcription factors need to be further investigated.
Fe deficiency increases mitochondrial dysfunction and elimination in the tapetum
Mitochondria are very dynamic and frequently undergo fusion and fission to change their morphology. Mitochondrial status, including morphology and density in cells, is tightly linked to the demand for energy and metabolites in cells (Mishra and Chan, 2016; Pfluger et al., 2015; Skulachev, 2001). Although it was previously suggested that the number, shape, and ultrastructure of mitochondria are different in Fe‐deficient plant cells (Dell'Orto et al., 2002; Landsberg, 1986, 1994; Pascal and Douce, 1993; Vigani et al., 2009), it is not clear whether mitochondrial density changes or how the morphologies of mitochondria change under Fe deficiency. In this study, we observed a reduction in mitochondrial density and an increase in fragmented and swollen mitochondria in the Fe‐deficient tapetum. Mitochondrial swelling is mainly caused by a lack of selective permeability of the inner membrane, which usually causes a loss of mitochondrial membrane potential (Zancani et al., 2015).
Mitochondria lacking membrane potential are dysfunctional and tend to be removed by autophagy. Mitochondrial fragmentation can also facilitate the removal of dysfunctional mitochondria. In addition, impairment of mitochondrial respiratory function can inhibit mitochondrial fusion, leading to mitochondrial fragmentation (Sauvanet et al., 2010). Thus, the reduction of mitochondrial complex activity in Fe‐deficient anthers might cause mitochondrial fragmentation, promoting the elimination of dysfunctional (swollen) mitochondria. However, our transcriptome results show that Fe deficiency‐induced DEGs were not enriched in GO terms related to mitochondria in the biological process category. These results suggest that under Fe deficiency, the decline in mitochondrial density in tapetal cells might not result from the reduction in mitochondrial biogenesis but rather from an increase in the elimination of mitochondria.
Mitochondrial dysfunction might be linked to an increase in fatty acid degradation in Fe‐deficient tapeta
Mitochondrial function is critical for tapetum function and pollen development, which has been demonstrated, especially in cytoplasm male sterile lines, and is well described in several reviews (Hanson and Bentolila, 2004; Hu et al., 2014; Islam et al., 2014). The present study revealed that Fe deficiency increased the volume of dysfunctional mitochondria and reduced total mitochondrial density in tapetal cells, which might decrease mitochondrial respiration as suggested by a lower OCR and decrease mitochondrial energy production in the tapetum. To support pollen development, the tapetum is highly metabolically active and requires an ample energy supply. In addition, mitochondrial dysfunction may induce a retrograde signal to alter the transcriptional regulation of nuclear genes involved in metabolic and signaling pathways (Busi et al., 2011; Ng et al., 2013; Van Aken et al., 2016; Van Aken and Whelan, 2012). Therefore, mitochondrial dysfunction and the reduction in energy supply from mitochondria caused by Fe deficiency might be the driving forces impacting transcriptional readjustment and resulting in changes in metabolic pathways, including the alternative ways to generate energy in the tapetum. It has been suggested that CSY can use acetyl‐CoA, the product of fatty acid degradation, and oxaloacetate to form citrate; citrate can be converted to malate through the glyoxylate pathway; and then malate can be used to regenerate glucose, which can undergo glycolysis to produce energy (Eckardt, 2005) (Figure 4). Therefore, fatty acid degradation can provide carbon for glucose synthesis for energy production. The anther‐unique DEGs induced by Fe deficiency were enriched in GO terms related to the fatty acid degradation pathway. Its key enzymes, including ACX2 and KAT2, and CSY3 were upregulated in Fe‐deficient anthers, suggesting the enhancement of fatty acid degradation and citrate synthesis from acetyl‐CoA to regenerate glucose further. The increase in fatty acid degradation under Fe deficiency was also observed in Saccharomyces cerevisiae (Shakoury‐Elizeh et al., 2010). In that report the authors hypothesized that the changes in metabolite levels suggested upregulation of glycolytic activity for energy production, which is consistent with the loss of energy generated through respiration as a consequence of Fe deficiency.
A model of molecular and cellular responses to Fe deficiency in the tapetum
Based on our studies of the effects of Fe deficiency on pollen development, we propose that Fe insufficiency causes transcriptional alteration of genes encoding upstream transcription factors such as TDF1, AMS, and bHLH089/091/010 which subsequently contribute to many defects in the tapetum and pollen (Figure 10). The repression of AMS, which results from the downregulation of TDF1, might contribute to the downregulation of RBOHE to affect anther ROS levels and tapetum PCD. The repression of AMS combined with the downregulation of bHLH089/091/010 might repress the expression of sporopollenin biosynthesis enzymes, which are essential for the formation of the outer pollen wall. Notably, the depletion of Fe might decrease anther ROS production by affecting RBOHE‐dependent and ‐independent ROS generation and alter mitochondrial function and status, including the reduction of respiratory activities and mitochondrial density. In addition, Fe deficiency might facilitate fatty acid degradation probably to compensate for the energy deficit caused by changes in mitochondrial function and status. Therefore, Fe deficiency in anthers might alter protein function and trigger extensive transcriptome reprogramming, which might impair several tapetal cellular processes and eventually disrupt pollen development.
Figure 10.
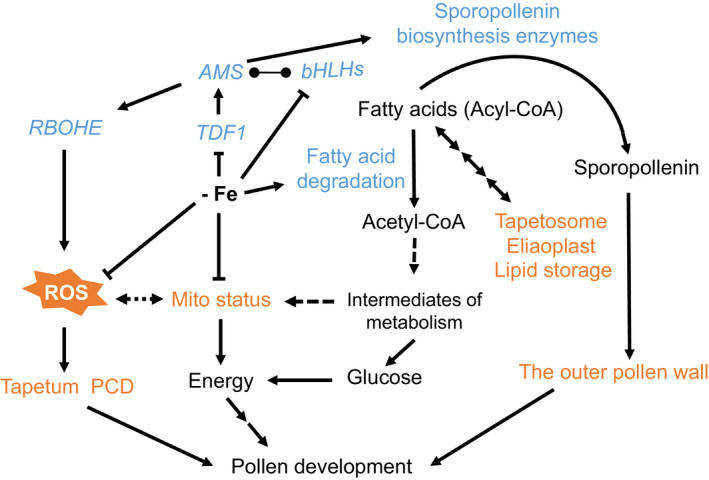
A model of molecular and cellular responses to Fe deficiency in the tapetum.
In Fe‐deficient anthers, TDF1, AMS, and bHLH089/091/010 are downregulated and lead to many defects in the tapetum and pollen. The downregulation of AMS might alter the expression of RBOHE and genes encoding sporopollenin biosynthesis enzymes. The downregulation of RBOHE reduces ROS production, which may affect tapetum PCD. The change in mitochondrial status possibly results from the decrease in Fe‐dependent mitochondrial protein function. Mitochondrial (Mito) status is crucial for tapetum function in pollen development, as shown in our previous study (Chen et al., 2019). Fe deficiency facilitates fatty acid degradation and fatty acid metabolic flux with reduced lipid storage, and the produced acetyl‐CoA can be used as an intermediate for carbon metabolism and as an energy source to compensate for mitochondrial dysfunction. Taken together, Fe deficiency in anthers triggers extensive transcriptome reprogramming, which might contribute to several tapetum defects, and Fe insufficiency might have impacts on Fe‐dependent proteins and mitochondrial status. These changes in molecular and cellular processes result in disrupted pollen development. The blue font represents genes/proteins involved in molecular regulation. The orange font indicates cellular processes that are defective under Fe deficiency. The line ending in circles represents protein interactions.
EXPERIMENTAL PROCEDURES
Plant growth and plant materials
Arabidopsis plants (Arabidopsis thaliana, ecotype Columbia‐0), including the Osg6B:RBOHE OE line, the rbohe null mutant, and transgenic Arabidopsis expressing mito‐GFP, and B. napus were grown at 22°C under approximately 110 μmol m−2 sec−1 light intensity and a 16 h light/8 h dark cycle. Arabidopsis seeds were surface sterilized and grown on agar plates for 10 days with half‐strength modified Hoagland’s medium (Millner and Kitt, 1992) containing 250 μm KH2PO4, 1% sucrose, and 1% bactoagar, adjusted to pH 5.7. Plants were then transferred to a hydroponic system consisting of half‐strength modified Hoagland’s medium without sucrose for another 18 days. Next, plants were transferred to Fe‐sufficient conditions (50 μm NaFe‐EDTA) or Fe‐deficient conditions (no NaFe‐EDTA added) and grown for 18 days. Then samples were collected for further analysis. For supplying exogenous Fe to floral buds, plants were grown in a Fe‐deficient hydroponic medium and 50 μm Na2‐EDTA or 50 μm NaFe‐EDTA was dropped onto the inflorescences once a day for 18 days.
To obtain enough fresh anthers to isolate crude mitochondrial extracts or lipids, we chose B. napus (B1128‐3, Tao Yuan #4) from the World Vegetable Center. Brassica napus seeds were surface sterilized and grown on agar plates with half‐strength modified Hoagland’s medium with 1% sucrose and 1% bactoagar for 7 days and transferred to the hydroponic system of half‐strength modified Hoagland’s medium without sucrose for another 4 weeks. Next, B. napus plants were transferred to Fe‐sufficient conditions (50 μm Fe) or Fe‐deficient conditions (no Fe added) and grown for 4 weeks, and then samples were collected for further analysis.
The transgenic line expressing mito‐GFP specifically in the tapetum was generated in our previous study (Chen et al., 2019). In brief, the mito‐GFP fragment in the mt‐gk construct obtained from Dr. Andreas Nebenführ at the University of Tennessee, USA, was recloned into the pCambia1304 plasmid under the control of the AT5G62080 tapetum‐specific promoter (Huang et al., 2013), a gift from Dr. Anthony H.C. Huang (University of California, Riverside) and Dr. Ming‐Der Huang (National Sun Yat‐sen University, Taiwan). Transgenic Arabidopsis lines expressing mito‐GFP specifically in the tapetum were generated by floral dipping and selected using antibiotics and the presence of a GFP signal in anthers.
Quantification of viable pollen and total pollen and in vitro pollen germination
To quantify viable pollen and total pollen number per anther, we collected unopened floral buds at anther stage 12–13, in which pollen was mature and anthers were still not dehiscent, and stained them using Alexander solution (Alexander, 2009) overnight at 4°C. The stained anthers were carefully isolated to maintain their integrity. The isolated anthers were put under a cover slide and pressed gently until the individual pollen could be observed, and anthers were still not broken and photographed using a Zeiss Imager Z1 to count viable pollen and total pollen number per anther. The pollen germination assay was performed following a previously described protocol (Li, 2011). The percentage of pollen that germinated was scored from ≥20 mature florals buds from at least five plants in one experiment. Three independent experiments were performed.
Cryo‐scanning electron microscopy analysis
Anthers from mature buds were loaded on a stub, frozen in a liquid nitrogen slush, and transferred to a sample preparation chamber at −160°C. After 5 min, the temperature was raised to −85°C, and the sample was sublimed for 15 min. After coating with platinum at −130°C, the samples were transferred to a cryo‐stage SEM chamber and observed at −160°C using a cryo‐scanning electron microscope (FEI Quanta 200 SEM/Quorum Cryo System PP2000TR FEI) set at 20 kV.
ICP‐MS
The metal contents of different tissues were analyzed by ICP‐MS (Agilent 7800, Agilent Technologies, Santa Clara, CA, USA). Fresh samples were dried at 70°C for 3 days. Approximately 5 mg dry weight per sample was transferred into a Teflon vessel and digested with 1 ml 65% HNO3 (Tracepur) and 0.5 ml H2O2 (Suprapur; Merck, Darmstadt, Germany) in a MarsXpress microwave digestion system (CEM; Matthews, NC, USA). Tomato (Solanum lycopersicum) leaves (SRM‐1573a) from the National Institute of Standards and Technology (Gaithersburg, MD, USA) were used as a standard. The volume was adjusted to 10 ml with H2O. The calibration solutions were prepared by diluting multi‐element stock standard solutions (ICP Multi‐Element Standard VI; Merck). Digested solutions were analyzed, and the results were inspected before sample analysis. Matrix‐induced background interference was corrected by subtraction of a reagent blank. The concentration of each element was measured from three independent replicates.
Chlorophyll measurements
Chlorophyll was extracted according to the procedure described in a previous study (Moran, 1982). Briefly, rosette leaves and stems were collected from plants (five per sample) grown under Fe‐sufficient and Fe‐deficient conditions. Approximately 0.1 g of tissue was ground in liquid N2 and mixed with 1 ml of 80% acetone for 12 h at 4°C in complete darkness. Samples were centrifuged at 16 000 g for 5 min, and the supernatants were collected in new Eppendorf tubes. Extraction was carried out two more times, and extracts were pooled in a 96‐well plate. Absorbance was measured using a spectrophotometer (EONC; Biotek Instruments, Winooski, VT, USA). Chlorophyll content was calculated using the following formula described in a previous study (Arnon, 1949):
where V = volume of the extract (ml) and W = weight of fresh tissues (g).
Fe staining with the Perls/DAB procedure
Since we established a correlation between the anther/pollen stage and the size of floral buds in our previous study (Chen et al., 2019), we are able to correctly identify the stages of anthers based on the size of floral buds and we do not need to open anthers to check the pollen stage. Arabidopsis buds at different stages were collected based on bud size (Chen et al., 2019), fixed in 4% glutaraldehyde and 4% dimethyl sulfoxide, 0.25% glutaraldehyde, 0.1% Tween 20, and 0.1% Triton X‐100, placed in a vacuum at 4°C overnight, and then washed in PBS for 10 min. Samples were dehydrated in an ethanol series and then processed by gradually decreasing the ratio of ethanol to pure Sub‐X (Leica, Vienna, Austria). Samples were then embedded by gradually decreasing the ratio of Sub‐X to pure Paraplast (Leica) at 60°C. Ten‐micrometer sections of the embedded floral buds were mounted on glass slides for Perls/DAB staining. Perls staining for Fe content in situ was performed as described previously (Sands et al., 2016). Briefly, sections were incubated in xylene for 3 min twice and then air‐dried. The sections were incubated with Perls stain containing 1% potassium ferrocyanide trihydrate and 5% PVP in 0.05 m HCl solution for 60 min. As a negative control, the sections were incubated in 0.05 m HCl solution without potassium ferrocyanide trihydrate. The sections were washed twice with Milli‐Q H2O (Millipore Co., MA, USA) followed by incubation in methanol containing 10 mm NaN3 and 0.3% H2O2 for 75 min. Sections were washed twice with PBS. Sections were then incubated for 40 min in 0.025% DAB (w/v) and 0.12% H2O2 (v/v) in 0.01 m Tris‐HCl, pH 7.4. The reaction was stopped by washing twice with PBS.
Metabolite profiling by GC‐TOF‐MS
Approximately 50 mg of floral buds was collected from Arabidopsis and extracted following a previously published procedure (Tsai et al., 2016). Metabolite analysis was performed using the Pegasus 4D GCxGC‐TOF‐MS system (LECO, St. Joseph, MI, USA) at the Metabolomics Core Facility, Agricultural Biotechnology Research Center, Academia Sinica, Taiwan. Three independent experiments (biological replicates) were conducted.
Fe content measurement
Fe concentration in floral buds (Figure 3f) was measured using the BPDS method (Pan et al., 2015; Tsai et al., 2018). Flowers from 10 plants were dried at 60°C for 2 days, and dry weight was recorded. Then, 225 μl of 65% (v/v) HNO3 was added to the dried samples, and the samples were incubated at 96°C for 6 h. Next, 150 μl of 30% (v/v) H2O2 was added, and samples were incubated at 56°C for 2 h. Next, 225 μl of sterile water was added, and 5 μl of the sample solution was mixed in a solution containing 1 mm BPDS, 0.6 m sodium acetate, and 0.48 m hydroxylamine hydrochloride. The concentration of Fe2+‐BPDS3 complexes was determined by measuring the absorbance at 535 nm in a PowerWave XS2 microplate spectrophotometer (BioTek; Power Wave XS2). Fe concentrations were determined against a standard curve made with FeCl3 that was treated using the same procedure as described above.
TUNEL assay
Buds were fixed in 4% paraformaldehyde, 4% dimethyl sulfoxide, 0.25% glutaraldehyde, 0.1% Tween 20, and 0.1% Triton X‐100 and placed in a vacuum at 4°C for 16 h. Buds were dehydrated in an ethanol series and then processed by gradually decreasing the ratio of ethanol to pure Sub‐X (Leica). Buds were then embedded by gradually reducing the proportion of Sub‐X to pure Paraplast (Leica) at 60°C. Then, 10‐mm sections were assessed with a TUNEL kit (DeadEnd Fluorometric TUNEL System; Promega, Madison, WI, USA). Samples were imaged using a Zeiss LSM 780 confocal laser‐scanning microscope.
Sample preparation for semi‐thin and ultra‐thin cross‐sectioning
Arabidopsis buds at different stages were fixed in 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 m sodium phosphate buffer, pH 7.0, at room temperature for 4 h followed by three 20‐min rinses. Samples were post‐fixed in 1% OsO4 in the same buffer at room temperature for 4 h, followed by three 20‐min rinses. Samples were dehydrated in an acetone series and embedded in Spurr’s resin. Anther stages were categorized into meiosis, tetrad, YM, VP, BP, and TC based on cytological observations with a light microscope as described previously (Sanders et al., 1999). Semi‐thin sections (1–2 μm) were cut using glass knives, stained with 0.1% Toluidine blue, and observed and photographed with a Zeiss Imager Z1 microscope. Ultra‐thin sections (70–90 nm) were obtained using a Reichert Ultracut S or Leica EM UC6 (Leica) and collected using 100 mesh copper grids. The sections were stained with 5% uranyl acetate in 50% methanol for 20 min and 0.4% lead citrate for 6 min. Sections were observed using an FEI Tecnai G2 Spirit TWIN Transmission Electron Microscope at 80 kV, and images were taken with a Gatan Orius CCD camera.
Detection of the tapetum in late‐stage anthers
Based on observation of the structure of the tapetum in a previous study, the tapetum degenerates at later stages, with its cytoplasmic contents released in the locule following cell death (Quilichini et al., 2014). Transgenic Arabidopsis plants specifically expressing GFP in the tapetum were used to monitor the tapetum. Anthers were stained with 0.2% Basic Fuchsin (CAS no. 58969‐01‐0; Sigma, Darmstadt, Germany) (Ursache et al., 2018) in PBS for 5 h and then washed twice with PBS. Anthers were imaged using a Zeiss LSM 780 confocal laser‐scanning microscope. The same GFP fluorescence parameters were utilized for unbiased quantification between samples. At least 12 anthers from more than three plants in one experiment were scored to calculate the percentage of anthers with GFP signal. Three independent experiments were performed.
RNA extraction and transcriptome sequencing
Arabidopsis anthers at different stages (from the PMC to the TC stage) were collected from three independent experiments, and total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) as described in the manufacturer’s instructions and processed with RNase free DNase I (Qiagen). RNA quality and concentration were determined using a NanoDrop‐1000 Spectrophotometer, an RNA Nano Chip on a Bioanalyzer, and Qubit. The cDNA libraries were built using the Illumina TruSeq RNA Sample Preparation v.2 kit (Illumina, San Diego, CA, USA) and sequenced in the single‐end format using the Illumina MiSeq System (MiSeq 150 cycles v.3, SR150) at the High Throughput Sequencing Core at the Biodiversity Research Center at Academia Sinica.
Transcriptome analysis
Approximately 20 million reads were collected from each anther library. Raw reads from leaf, root (GSE87760), pollen (SRP110833), and tapetum (SRS2426392 and SRS2426393) transcriptomes were downloaded from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra). Our anther RNA‐seq reads and downloaded raw reads were examined for quality using FastQC (v.0.11.5; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure that the library preparation and sequencing data were suitable for analysis and further processed by Trimmomatic (v.0.35) to remove adapter sequences and remove low‐quality reads (Bolger et al., 2014). To perform mapping and quantify transcript abundance, we performed quasi‐alignment using Salmon (v.0.12.0) (Patro et al., 2017), which required a reference index from the Arabidopsis reference genome (v.TAIR10) (https://www.arabidopsis.org/). Counts per gene from Salmon were summarized using the R package tximport and further normalized by DEseq2 (Love et al., 2014; Soneson et al., 2015). Benjamini and Hochberg’s method was used to adjust the P‐values to correct for the false discovery rate, and genes with an adjusted P‐value (P.adj) of <0.05 were considered DEGs (Hochberg, 1995). Venn diagrams were produced using jvenn (Philippe Bardou et al., 2014). For the pollen and tapetum transcriptomes, we used transcripts per million from Salmon, which were converted by zFPKM normalization (Hart et al., 2013). We defined the threshold as normalized zFPKM ≥ 0, a rather stringent criterion for determining expressed genes. Microarray data for tdf1 (GSE102528) were downloaded and analyzed using Genespring GX software (Agilent, Palo Alto, CA, USA). DEGs of tdf1 were identified with a cut‐off of fourfold change in expression compared with WT and statistical significance of the t‐test against zero (P < 0.05); multiple testing correction was performed using Storey with bootstrapping (q < 0.1). DEGs of the ams and bhlhs triple mutants were directly obtained from the published data (Li et al., 2017; Ma et al., 2012; Xu et al., 2014). The DEGs of ams were combined from several published datasets (Li et al., 2017; Ma et al., 2012; Xu et al., 2014; Zhu et al., 2015).
Gene Ontology, pathway enrichment, and co‐expression analysis
The Singular Enrichment Analysis module computes GO enrichment in a group of target genes by comparing them to the Arabidopsis genome (TAIR10) using AgriGO v.2.0 (Tian et al., 2017; Trapnell et al., 2012). The Benjamini–Yekutieli method was used to adjust the P‐values for the multiple testing correction using default parameters in AgriGO v.2.0. The GO terms with an adjusted P‐value of <0.05 were defined as significantly enriched terms. REVIGO was used to summarize and visualize the GO terms using SimRel semantic similarity measurements (dispensability < 0.4) (Supek et al., 2011; Xie et al., 2014). To obtain essential GO terms in the biological process category, we chose the GO terms based on dispensability <0.3 and log10(P‐value) <−3.5 from the list in Table S6 (dispensability <0.4); they are shown in Figure 7d. Pathway enrichment analysis and co‐expression analysis were performed using the EXPath 2.0 microarray database (Chien et al., 2015). Pathways with a cumulative probability of P < 0.05 of hypergeometric distribution were considered significantly enriched pathways of a group of input genes. A co‐expression correlation network was built using EXPath 2.0 with a threshold Pearson correlation coefficient of >0.8 and with ‘all conditions’ selected from the microarray database.
Quantitative real‐time PCR
One microgram of total RNA was used as the template for reverse transcription with SuperScript III (Invitrogen, Carlsbad, CA, USA), and the cDNA was diluted 20‐fold. Diluted cDNA (5 μl) was used as the template for qPCR in a 20‐μl reaction containing SYBR Green PCR Master Mix (Applied Biosystems, Vilnius, Lithuania) performed on a 7500 Fast RealTime PCR System (Applied Biosystems) using the following program: 20 sec at 95°C, followed by 40 cycles of 3 sec at 95°C and 30 sec at 60°C, with an additional melt curve stage consisting of 15 sec at 95°C, 1 min at 60°C, and 15 sec at 95°C. The ACT8 expression level was used as an internal control. The expression values of target genes were normalized to the corresponding value of the WT. Three independent experiments were performed. The primers used are listed in Table S7.
Detection of anther ROS
Anthers of different stages were stained with 10 μm of the fluorescent dye H2DCFDA (Invitrogen, Eugene, OR, USA) in PBS. For staining, anthers were placed in H2DCFDA solution in a vacuum for three minutes, kept in darkness at room temperature for 2 h, and washed with PBS twice. The stained samples were imaged by a Zeiss Imager Z1 using the same parameter settings and without overexposure to perform comparative quantification using AxioVision SE64 Rel. 4.9.1.
Nile red staining
A 100 μg ml−1 (w/v) stock solution of Nile red (Sigma; N3013) in acetone was diluted to 1 μg ml−1 in 25% glycerol, vortexed, and added directly to the anthers before the coverslip was put in place for 3.5 h. The images of samples presented in Figure 5e–j were acquired using a Zeiss LSM 780 confocal laser‐scanning microscope. Z‐stacks of the Nile red signal were acquired by a Zeiss Imager Z1, and parameters for fluorescence images were set the same without overexposure for comparative quantification using AxioVision SE64 Rel. 4.9.1 (Figure 5k).
Lipid extraction and TLC
Lipids were extracted as described in a previous study (Bligh and Dyer, 1959). Briefly, 100 mg of pooled anthers at the PMC to TC stages from Fe‐sufficient and ‐deficient B. napus was homogenized in liquid nitrogen. Then 100 μl ice‐cold chloroform and 200 μl ice‐cold methanol were added to the homogenate, and samples were mixed by vortexing for 2 min and centrifuged at 845 g for 5 min at 4°C. The supernatant was saved in another tube. Then, 100 μl chloroform, 200 μl methanol, and 80 μl of 0.45% KCl were added to the pellet. The samples were mixed by vortexing for 2 min and then centrifuged at 845 g for 5 min at 4°C. The supernatants were mixed, 200 μl chloroform and 120 μl of 0.45% KCl were added, and the samples were centrifuged at 845 g for 5 min at 4°C. The upper layer was carefully removed. The lower layer was dried at 70°C and resuspended in 100 μl chloroform:methanol (2:1), and 30 μl was used for separation on TLC plates (Knittelfelder and Kohlwein, 2017), which were developed in hexane:diethyl ether:acetic acid (70:30:1, v/v/v). Neutral lipids were stained by incubating for 10 sec in a MnCl2 solution containing 0.63 g MnCl2·4H2O, 60 ml methanol, 4 ml sulfuric acid, and 60 ml H2O. The plates were dried for 15 min, heated at 170°C for about 2 min, and then scanned (Epson Expression 12000XL) for densitometric quantification. ImageJ was used to distinguish the peaks of the bands from the densitographic curves and the area of each peak was calculated in relative units. The amount of lipids under Fe deficiency relative to that under Fe sufficiency (100%) is shown.
Anther respiration rate measurements
Anther respiration was measured using a Seahorse XF analyzer (Seahorse Bioscience, Billerica, MA, USA) following a published method (Sew et al., 2013) and the manufacturer’s instructions. Briefly, the eight‐well sensor cartridge was hydrated in 200 μl of XF Calibrant Solution (Seahorse Bioscience) per well for 2 h at room temperature before the assay. Anthers at the YM, VP, and BN stages were fixed at the bottom of wells with 1 μl tissue adhesive glue (3 M Vetbond Tissue Adhesive, Neuss, Germany). After 2 min, 180 μl of respiration buffer (2 mm HEPES, 2 mm MES, and 0.4 mm CaCl2, pH 7.6) was added to the wells, followed by eight cycles of mixing (2 min), waiting (3 min), and measurement (5 min). The OCRs of anthers were recorded by Seahorse XF Acquisition and Analysis Software (v.2.6; Seahorse Bioscience).
Preparation of crude mitochondrial extracts
Crude mitochondrial extracts were isolated as previously described (Pineau et al., 2008). Approximately 200 mg of fresh anthers from B. napus was ground using a pestle and mortar in 2 ml ice‐cold extraction buffer containing 0.6 m sucrose, 0.2% polyvinylpyrrolidone 40, 8 mm cysteine, 4 mm EDTA, 0.2% bovine serum albumin, and 75 mm MOPS‐KOH, pH 7.6. The lysate was centrifuged at 1300 g for 4 min twice, and the supernatant was centrifuged at 21 130 g for 25 min at 4°C. Next, the pellet was resuspended in 200 μl of 0.3 m sucrose, 10 mm MOPS‐KOH, pH 7.2. The suspensions were washed with 600 μl water, centrifuged at 21 130 g for 25 min, and resuspended in 150 μl buffer containing 50 mm Bis‐Tris‐HCl, pH 7, 0.5 m 6‐aminohexanoic acid, 1 mm EDTA, 0.3 m sucrose, and protease inhibitor cocktail (Invitrogen). Next, 30 μl of 10% β‐dodecyl maltoside was added, and the samples were incubated at 4°C for 15 min and then centrifuged at 21 130 g for 10 min. Then 16 μl 5% Coomassie Blue G‐250 (in 0.020 m Bis‐Tris‐HCl, pH 7, 0.5 m 6‐aminohexanoic acid) was added. Next, an aliquot equivalent to 16 mg (fresh weight) of anthers was loaded onto a BN gel as described previously (Pineau et al., 2008, 2005; Wittig et al., 2006). After electrophoresis, the gels were stained with Coomassie Blue or mitochondrial activity staining was performed in‐gel.
Mitochondria in‐gel activity staining
In‐gel staining for measuring mitochondrial complex activities was carried out according to the protocol established by Schertl and Braun (2015). Briefly, gels were washed twice for 5 min in H2O. For complex I activity, gel strips were incubated in 0.1 m Tris‐HCl, pH 7.4, 0.14 mm NADH, and 0.1% NBT. For complex IV activity, gel strips were stained in 50 mm phosphate buffer, pH 7.4, 0.1% DAB, and 0.1% cytochrome c. Complex V activity staining was carried out in 35 mm Tris‐HCl, 270 mm glycine, pH 8.3 for 2 h, and then the gels were incubated in assay buffer (35 mm Tris‐HCl, 270 mm glycine, 14 mm MgCl2, 0.2% Pb(NO3)2, 8 mm ATP, pH 8.3). The reactions were stopped by fixing the gels in 45% methanol and 10% acetic acid; for complex V, 45% methanol only was used. Gels were transferred to water and scanned (Epson Expression 12000XL) for densitometric quantification of the bands (VisionWorks Life Science). The band density is given in relative units. The value of each band was normalized by the amount of protein on gels stained with Coomassie Blue, and the intensity under Fe deficiency relative to that under Fe sufficiency (100%) is shown.
Western blot analysis
Crude mitochondrial extract from anthers of B. napus (equivalent to 10 mg fresh weight) was mixed with 3× SDS sample buffer (187.5 mm Tris‐HCl, pH 6.8, 15% β‐mercaptoethanol, 6% SDS, 30% glycerol, 0.03% bromophenol blue) for 10–15 min at 100°C and separated by 15% SDS‐PAGE in running buffer (25 mm Tris, 192 mm glycine, 0.1% SDS). After the protein markers were separated, proteins were transferred to a PVDF membrane in 1× transfer buffer (0.02 m Tris‐base, 0.15 m glycine, 10% methanol). Next, the PVDF membrane was stained with Ponceau S and photographed. The PVDF membrane was washed three times with TBS‐T, incubated in 1× TBS‐T with 5% milk powder for 1 h, and then incubated with primary antibodies (anti‐NAD9 (Lamattina et al., 1993), anti‐RISP (Carrie et al., 2010), and anti‐COX2 [Agrisera, AS04 054A]) overnight at 4°C. The membrane was washed three times with TBS‐T and incubated with secondary antibody for 2 h. Protein bands were visualized by enhanced chemiluminescence (Clarity Western ECL Substrate, Bio‐Rad; Biospectrum imaging system, CA, USA). The pictures were used for densitographic analysis of the bands. ImageJ was used to distinguish the peaks of the bands from the densitographic curves, and the area of each peak was calculated, which was quantified in relative units. The peak area of each band was normalized by the amount of protein on the membrane stained using Ponceau S. The amount of protein under Fe deficiency was normalized to that under Fe sufficiency (100%).
Quantification of tapetal volume and mitochondrial morphology
Anthers were fixed in 4% paraformaldehyde (EMS, Cat. 15710) in a vacuum for 30 min and washed twice with 1× PBS buffer for 10 min. Tapetal cells were pressed out from fixed anthers and separated from the anther wall on the slide. Tapetal cells on the slide were stained with 12 mm propidium iodide for 20 min and washed with 1× PBS. For the quantification of mitochondria, Z‐stack images of whole tapetal cells were acquired by a Zeiss LSM 780 confocal microscope with a 63×/1.4 oil objective, 0.22 μm × 0.22 μm × 0.25 μm scaling, 1024 × 1024 pixel resolution, 16‐bit image format, and 0.6 scanner zoom. For analysis of mitochondrial characteristics, we followed our previously established procedure using the homemade MicroP 3D automatic quantification software (Chen et al., 2019).
Statistical analysis
Statistical analyses were carried out between two groups using a two‐tailed Student’s t‐test to identify statistically significant differences. In addition, comparisons among multiple groups were performed by two‐way analysis of variance (anova) with Tukey’s multiple comparisons test using GraphPad Prism software (v. 7; GraphPad Software, San Diego, CA, USA; Data S1). A P‐value of <0.05 was considered statistically significant. Data are presented as mean ± standard error of three independent biological analyses.
ACCESSION NUMBERS
RNA‐seq data from this article can be found in GEO under accession number GSE153588. The genes mentioned in this article are as follows: RBOHE, AT1G19230; AMS, AT2G16910; bHLH010, AT2G31220; bHLH089, AT1G06170; bHLH091, AT2G31210; TDF1/AtMYB35, AT3G28470.
AUTHOR CONTRIBUTIONS
THH and DFS designed the study; THH performed the experiments; THH and DFS analyzed results; THH and DFS wrote the original article; THH and DFS reviewed and edited the article. All the authors read and discussed the manuscript and gave approval for the final version to be published.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Figure S1. The morphology of plants and in vitro germination of pollen grown under Fe‐sufficient and ‐deficient conditions.
Figure S2. DNA fragmentation in Fe‐sufficient and ‐deficient anthers.
Figure S3. Cryo‐SEM images of pollen in dehiscent anthers.
Figure S4. The workflow of Fe‐deficient anther transcriptome analysis.
Figure S5. The changes in transcript levels of known −Fe genes, DYT1 and MYB80, in anthers under Fe deficiency.
Figure S6. Analysis of the expression of genes that are repressed in anther tissue upon Fe deficiency in the tapetal transcriptomes of ams, tdf1, and bhlhs mutants.
Figure S7. The effect of RBOHE expression on anther ROS levels under Fe sufficiency and deficiency.
Figure S8. The activities and protein abundance of mitochondrial respiratory complexes in developing anthers under Fe deficiency.
Figure S9. Mitochondria labeled with mito‐GFP in isolated tapetal cells.
Figure S10. 3D numerical analysis of mitochondrial morphology.
Table S1. Metabolite profiling in floral buds.
Table S2. Element contents in leaves and floral buds.
Table S3. The DEGs in anthers, leaves, and roots under Fe deficiency.
Table S4. Pathway enrichment analysis of anther‐unique DEGs under Fe deficiency.
Table S5. Anther‐unique DEGs under Fe deficiency in the phenylpropanoid biosynthesis pathway.
Table S6. GO enrichment analysis of tapetum‐expressed genes induced by Fe deficiency and also differentially expressed in the tdf1, ams, and bhlhs mutants.
Table S7. Primers used for qPCR.
Data S1. Statistical analysis of H2DCFDA fluorescence intensity in Fe‐sufficient and ‐deficient anthers at each developmental stage in different lines.
ACKNOWLEDGMENTS
We thank all lab members. We also thank Dr. Yan Zhang for providing the Osg6B:RBOHE and rbohe null mutant seeds; Dr. Kuo‐Chen Yeh, Dr. Jin‐Chi Lo, and Ms. I‐Chien Tang for help with ICP‐MS; Dr. Ho‐Ming Chen and Mr. Bo‐Han Hou for help with transcriptome analysis; Dr. Chung‐Chih Lin for assistance with the 3D automatic quantification software; the Plant Cell Biology Core Lab for help with tissue sectioning and sample processing for SEM and TEM; Dr. Jim Whelan for providing the anti‐RISP antibody; and Dr. Jean Michel Grienenberger and Dr. Géraldine Bonnard for providing the anti‐NAD9 antibody. In addition, we thank the staff of the High Throughput Sequencing Core in the Biodiversity Research Center at Academia Sinica for performing the NGS experiments and the staff of the Metabolomics Core Facility, Agricultural Biotechnology Research Center, Academia Sinica, Taiwan for metabolite analysis. We used the confocal microscope at the Scientific Instrument Center of Academia Sinica with the assistance of Ms. Shu‐Chen Shen. This work was supported by grants from Academia Sinica, Taiwan (034007), and the Ministry of Science Technology, Taiwan (107‐2311‐B‐001‐011‐MY3). We thank Academia Sinica, Taiwan, and the Ministry of Science Technology, Taiwan, for funding support.
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
References
- Abadía, J. , Álvarez‐Fernández, A. , Rombolá, A.D. , Sanz, M. , Tagliavini, M. & Abadía, A. (2004) Technologies for the diagnosis and remediation of Fe deficiency. Soil Science. Plant Nutrition, 50, 965–971. [Google Scholar]
- Alexander, M.P. (2009) Differential staining of aborted and nonaborted pollen. Stain Technology, 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Àlvarez‐Fernàndez, A. , Abadía, J. & Abadía, A. (2006) Iron deficiency, fruit yield and fruit quality. In: Barton L.L. & Abadia J. (Eds.) Iron nutrition in plants and rhizospheric microorganisms. Dordrecht: Springer, pp. 85–101. [Google Scholar]
- Arnon, D.I. (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris . Plant Physiology, 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J. & Leaver, C.J. (2001) The PET1‐CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. The Plant Cell, 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou, P. , Mariette, J. , Escudié, F. , Djemiel, C. & Klopp, C. (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinformatics, 15(1), 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell, D. , Dimmer, E. , Huntley, R.P. , Binns, D. , O'Donovan, C. & Apweiler, R. (2009) The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Research. 37(Database issue): D396–D403. 10.1093/nar/gkn803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir, K. , Ishimaru, Y. , Shimo, H. , Nagasaka, S. , Fujimoto, M. , Takanashi, H. et al. (2011) The rice mitochondrial iron transporter is essential for plant growth. Nature Communications, 2, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh, E.G. & Dyer, W.J. (1959) A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology, 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. & Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat, J.F. , Dubos, C. & Gaymard, F. (2015) Iron nutrition, biomass production, and plant product quality. Trends in Plant Science, 20, 33–40. [DOI] [PubMed] [Google Scholar]
- Busi, M.V. , Gomez‐Lobato, M.E. , Rius, S.P. , Turowski, V.R. , Casati, P. , Zabaleta, E.J. et al. (2011) Effect of mitochondrial dysfunction on carbon metabolism and gene expression in flower tissues of Arabidopsis thaliana . Molecular Plant, 4, 127–143. [DOI] [PubMed] [Google Scholar]
- Carrie, C. , Giraud, E. , Duncan, O. , Xu, L. , Wang, Y. , Huang, S. et al. (2010) Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. Journal of Biological Chemistry, 285, 36138–36148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie, C. , Murcha, M.W. , Millar, A.H. , Smith, S.M. & Whelan, J. (2007) Nine 3‐ketoacyl‐CoA thiolases (KATs) and acetoacetyl‐CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Molecular Biology, 63, 97–108. [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A.P. , Tang, H. , Wang, X. et al. (2014) Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chen, P.Y. , Wu, C.C. , Lin, C.C. , Jane, W.N. & Suen, D.F. (2019) 3D imaging of tapetal mitochondria suggests the importance of mitochondrial fission in pollen growth. Plant Physiology, 180, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Yu, X.H. , Zhang, K. , Shi, J. , De Oliveira, S. , Schreiber, L. et al. (2011) Male Sterile2 encodes a plastid‐localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiology, 157, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, C.H. , Chow, C.N. , Wu, N.Y. , Chiang‐Hsieh, Y.F. , Hou, P.F. & Chang, W.C. (2015) EXPath: a database of comparative expression analysis inferring metabolic pathways for plants. BMC Genomics, 16(Suppl 2), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton, M. (1999) The mitochondrial permeability transition pore and its role in cell death. The Biochemical Journal, 341(Pt 2), 233–249. [PMC free article] [PubMed] [Google Scholar]
- Cui, J. , You, C. , Zhu, E. , Huang, Q. , Ma, H. & Chang, F. (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. The Plant cell, 28, 1078–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza, C. , Kim, S.S. , Koch, S. , Kienow, L. , Schneider, K. , McKim, S.M. et al. (2009) A novel fatty Acyl‐CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. The Plant Cell, 21, 507–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme, N. & Geelen, D. (2014) The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant, Cell and Environment, 37, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Orto, M. , Pirovano, L. , Villalba, J.M. , González‐Reyes, J.A. & Zocchi, G. (2002) Localization of the plasma membrane H+‐ATPase in Fe‐deficient cucumber roots by immunodetection. Plant and Soil, 241, 11–17. [Google Scholar]
- Dixon, S.J. & Stockwell, B.R. (2014) The role of iron and reactive oxygen species in cell death. Nature Chemical Biology, 10, 9–17. [DOI] [PubMed] [Google Scholar]
- Dobritsa, A.A. , Shrestha, J. , Morant, M. , Pinot, F. , Matsuno, M. , Swanson, R. et al. (2009) CYP704B1 is a long‐chain fatty acid omega‐hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiology, 151, 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt, N.A. (2005) Peroxisomal citrate synthase provides exit route from fatty acid metabolism in oilseeds. The Plant Cell, 17, 1863–1865. [Google Scholar]
- El‐Jendoubi, H. , Vazquez, S. , Calatayud, A. , Vavpetic, P. , Vogel‐Mikus, K. , Pelicon, P. et al. (2014) The effects of foliar fertilization with iron sulfate in chlorotic leaves are limited to the treated area. A study with peach trees (Prunus persica L. Batsch) grown in the field and sugar beet (Beta vulgaris L.) grown in hydroponics. Frontiers in Plant Science, 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, A.C. , Pearce, S. , Band, L.R. , Yang, C. , Ferjentsikova, I. , King, J. et al. (2017) Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytologist, 213, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R. & Reed, J.C. (1998) Mitochondria and Apoptosis. Science, 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Grienenberger, E. , Kim, S.S. , Lallemand, B. , Geoffroy, P. , Heintz, D. , Souza Cde, A. et al. (2010) Analysis of TETRAKETIDE alpha‐PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. The Plant Cell, 22, 4067–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet, L. , Lan, P. , Li, W. , Mokkapati, G. & Schmidt, W. (2018) IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nature Plants, 4, 953–963. [DOI] [PubMed] [Google Scholar]
- Gu, J.N. , Zhu, J. , Yu, Y. , Teng, X.D. , Lou, Y. , Xu, X.F. et al. (2014) DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. The Plant Journal, 80, 1005–1013. [DOI] [PubMed] [Google Scholar]
- Hanson, M.R. & Bentolila, S. (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. The Plant Cell, 16(Suppl), S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, T. , Komori, H. , LaMere, S. , Podshivalova, K. & Salomon, D.R. (2013) Finding the active genes in deep RNA‐seq gene expression studies. BMC Genomics, 14, 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt, M.N. , Akmakjian, G.Z. , Pivarski, K.L. , Punshon, T. , Baxter, I. , Salt, D.E. et al. (2017) BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana . Metallomics, 9, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg, Y.B.A.Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 57, 289–300. [Google Scholar]
- Hsieh, K. & Huang, A.H.C. (2007) Tapetosomes in brassica tapetum accumulate endoplasmic reticulum‐derived flavonoids and alkanes for delivery to the pollen surface. The Plant Cell, 19, 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Huang, W. , Huang, Q. , Qin, X. , Yu, C. , Wang, L. et al. (2014) Mitochondria and cytoplasmic male sterility in plants. Mitochondrion, 19, 282–288. 10.1016/j.mito.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Hu, L. , Liang, W. , Yin, C. , Cui, X. , Zong, J. , Wang, X. et al. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. The Plant Cell, 23, 515–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M.D. , Chen, T.L. & Huang, A.H. (2013) Abundant type III lipid transfer proteins in Arabidopsis tapetum are secreted to the locule and become a constituent of the pollen exine. Plant Physiology, 163, 1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inupakutika, M.A. , Sengupta, S. , Devireddy, A.R. , Azad, R.K. & Mittler, R. (2016) The evolution of reactive oxygen species metabolism. Journal of Experimental Botany, 67, 5933–5943. [DOI] [PubMed] [Google Scholar]
- Islam, M.S. , Studer, B. , Møller, I.M. , Asp, T. & Staub, J. (2014) Genetics and biology of cytoplasmic male sterility and its applications in forage and turf grass breeding. Plant Breeding, 133, 299–312. [Google Scholar]
- Jin, Y. , Yang, H. , Wei, Z. , Ma, H. & Ge, X. (2013) Rice male development under drought stress: phenotypic changes and stage‐dependent transcriptomic reprogramming. Molecular Plant, 6, 1630–1645. [DOI] [PubMed] [Google Scholar]
- Kim, H.U. , Li, Y. & Huang, A.H.C. (2005) Ubiquitous and endoplasmic reticulum‐located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. The Plant Cell, 17, 1073–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittelfelder, O.L. & Kohlwein, S.D. (2017) Thin‐layer chromatography to separate phospholipids and neutral lipids from yeast. Cold Spring Harbor Protocols, 2017(5), 10.1101/pdb.prot085456. [DOI] [PubMed] [Google Scholar]
- Korshunova, Y.O. , Eide, D. , Clark, W.G. , Guerinot, M.L. and Pakrasi, H.B. (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Molecular Biology, 40, 37–44. [DOI] [PubMed] [Google Scholar]
- Lallemand, B. , Erhardt, M. , Heitz, T. & Legrand, M. (2013) Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiology, 162, 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina, L. , Gonzalez, D. , Gualberto, J. & Grienenberger, J.‐M. (1993) Higher plant mitochondria encode an homologue of the nuclear‐encoded 30‐ kDa subunit of bovine mitochondrial complex I. European Journal of Biochemistry, 217, 831–838. [DOI] [PubMed] [Google Scholar]
- Landsberg, E.‐C. (1986) Function of rhizodermal transfer cells in the Fe stress response mechanism of Capsicum annuum L. Plant Physiology, 82, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg, E.‐C. (1994) Transfer cell formation in sugar beet roots induced by latent Fe deficiency. Plant and Soil, 165, 197–205. [Google Scholar]
- Le Jean, M. , Schikora, A. , Mari, S. , Briat, J.F. & Curie, C. (2005) A loss‐of‐function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. The Plant Journal, 44, 769–782. [DOI] [PubMed] [Google Scholar]
- Lee, S.L. & Warmke, H.E. (1979) Organelle size and number in fertile and cytoplasmic male‐sterile corn. American Journal of Botany, 66, 141–148. [Google Scholar]
- Li, D.D. , Xue, J.S. , Zhu, J. & Yang, Z.N. (2017) Gene regulatory network for tapetum development in Arabidopsis thaliana . Frontiers in Plant Science, 8, 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. (2011) Arabidopsis pollen tube germination. Bio‐Protocol Bio, 101, e73. [Google Scholar]
- Lopez‐Millan, A.F. , Grusak, M.A. , Abadia, A. & Abadia, J. (2013) Iron deficiency in plants: an insight from proteomic approaches. Frontiers in Plant Science, 4, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, Y. , Zhou, H.‐S. , Han, Y. , Zeng, Q.‐Y. , Zhu, J. & Yang, Z.‐N. (2018) Positive regulation of AMS by TDF1 and the formation of a TDF1‐AMS complex are required for anther development in Arabidopsis thaliana . New Phytologist, 217, 378–391. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D. , Xu, H. , Liu, Z. , Guo, J. , Li, H. , Chen, L. et al. (2013) A detrimental mitochondrial‐nuclear interaction causes cytoplasmic male sterility in rice. Nature Genetics, 45, 573–577. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Feng, B. & Ma, H. (2012) AMS‐dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes. BMC Plant Biology, 12, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, H.‐J. & Bauer, P. (2016) From the proteomic point of view: Integration of adaptive changes to iron deficiency in plants. Current Plant Biology, 5, 45–56. [Google Scholar]
- Mepham, R.H. & Lane, G.R. (1969) Role of the Tapetum in the development of tradescantia pollen. Nature, 221, 282–284. [Google Scholar]
- Millner, P.D. & Kitt, D.G. (1992) The Beltsville method for soilless production of vesicular‐arbuscular mycorrhizal fungi. Mycorrhiza, 2, 9–15. [Google Scholar]
- Mishra, P. & Chan, D.C. (2016) Metabolic regulation of mitochondrial dynamics. Journal of Cell Biology, 212, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N. , Seki, M. , Shinozaki, K. & Ohme‐Takagi, M. (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell, 17, 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. (2017) ROS are good. Trends in Plant Science, 22, 11–19. [DOI] [PubMed] [Google Scholar]
- Moran, R. (1982) Formulae for determination of chlorophyllous pigments extracted with N,N‐Dimethylformamide. Plant Physiology, 62, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey, J. & Guerinot, M.L. (2009) Iron uptake and transport in plants: the good, the bad, and the ionome. Chemical Reviews, 109, 4553–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, S. , Ivanova, A. , Duncan, O. , Law, S.R. , Van Aken, O. , De Clercq, I. et al. (2013) A membrane‐bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. The Plant Cell, 25, 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, H.A. & Makaroff, C.A. (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma, 185, 7–21. [Google Scholar]
- Pan, I.C. , Tsai, H.H. , Cheng, Y.T. , Wen, T.N. , Buckhout, T.J. & Schmidt, W. (2015) Post‐transcriptional coordination of the Arabidopsis iron deficiency response is partially dependent on the E3 ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2). Molecular & Cellular Proteomics: MCP, 14, 2733–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish, R.W. , Phan, H.A. , Iacuone, S. & Li, S.F. (2012) Tapetal development and abiotic stress: a centre of vulnerability. Functional Plant Biology, 39(7), 553. [DOI] [PubMed] [Google Scholar]
- Pascal, N. & Douce, R. (1993) Effect of iron deficiency on the respiration of Sycamore (Acer pseudoplatanus L.) cells. Plant Physiology, 103, 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R. , Duggal, G. , Love, M.I. , Irizarry, R.A. & Kingsford, C. (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nature Methods, 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea‐Garcia, A. , Garcia‐Molina, A. , Andres‐Colas, N. , Vera‐Sirera, F. , Perez‐Amador, M.A. , Puig, S. et al. (2013) Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low‐phosphate signaling. Plant Physiology, 162, 180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Souza, L. , Garbowicz, K. , Brotman, Y. , Tohge, T. & Fernie, A.R. (2020) The acetate pathway supports flavonoid and lipid biosynthesis in Arabidopsis. Plant Physiology, 182, 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger, P.T. , Kabra, D.G. , Aichler, M. , Schriever, S.C. , Pfuhlmann, K. , Garcia, V.C. et al. (2015) Calcineurin links mitochondrial elongation with energy metabolism. Cell Metabolism, 22, 838–850. [DOI] [PubMed] [Google Scholar]
- Pineau, B. , Layoune, O. , Danon, A. & De Paepe, R. (2008) L‐galactono‐1,4‐lactone dehydrogenase is required for the accumulation of plant respiratory complex I. Journal of Biological Chemistry, 283, 32500–32505. [DOI] [PubMed] [Google Scholar]
- Pineau, B. , Mathieu, C. , Gerard‐Hirne, C. , De Paepe, R. & Chetrit, P. (2005) Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. Journal of Biological Chemistry, 280, 25994–26001. [DOI] [PubMed] [Google Scholar]
- Pinfield‐Wells, H. , Rylott, E.L. , Gilday, A.D. , Graham, S. , Job, K. , Larson, T.R. et al. (2005) Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. The Plant Journal, 43, 861–872. [DOI] [PubMed] [Google Scholar]
- Platt, K.A. , Huang, A.H.C. & Thomson, W.W. (1998) Ultrastructural study of lipid accumulation in tapetal cells of Brassica napus L. Cv. Westar during microsporogenesis. International Journal of Plant Sciences, 159, 724–737. [Google Scholar]
- Pracharoenwattana, I. , Cornah, J.E. & Smith, S.M. (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. The Plant Cell, 17, 2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini, T.D. , Douglas, C.J. & Samuels, A.L. (2014) New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana . Annals of Botany, 114, 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati Ishka, M. , Brown, E. , Weigand, C. , Tillett, R.L. , Schlauch, K.A. , Miller, G. et al. (2018) A comparison of heat‐stress transcriptome changes between wild‐type Arabidopsis pollen and a heat‐sensitive mutant harboring a knockout of cyclic nucleotide‐gated cation channel 16 (cngc16). BMC Genomics, 19, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet, K. , Touraine, B. , Boucherez, J. , Briat, J.F. , Gaymard, F. & Cellier, F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. The Plant Journal, 57, 400–412. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Celma, J. , Connorton, J.M. , Kruse, I. , Green, R.T. , Franceschetti, M. , Chen, Y.T. et al. (2019) Arabidopsis BRUTUS‐LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proceedings of the National Academy of Sciences, 116, 17584–17591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Celma, J. , Pan, I.C. , Li, W. , Lan, P. , Buckhout, T.J. & Schmidt, W. (2013) The transcriptional response of Arabidopsis leaves to Fe deficiency. Frontiers in Plant Science, 4, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz, H. , Conejero, G. , Divol, F. , Alcon, C. , Verdeil, J.L. , Curie, C. et al. (2013) New insights into Fe localization in plant tissues. Frontiers in Plant Science, 4, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz, H. , Seguela‐Arnaud, M. , Briat, J.F. , Vert, G. & Curie, C. (2011) The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. The Plant Cell, 23, 2725–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M. , Bui, A.Q. , Weterings, K. , McIntire, K.N. , Hsu, Y.‐C. , Lee, P.Y. et al. (1999) Anther developmental defects in Arabidopsis thaliana male‐sterile mutants. Sexual Plant Reproduction, 11, 297–322. [Google Scholar]
- Sands, S.A. , Leung‐Toung, R. , Wang, Y. , Connelly, J. & LeVine, S.M. (2016) Enhanced histochemical detection of iron in paraffin sections of mouse central nervous system tissue: application in the APP/PS1 mouse model of Alzheimer's disease. ASN Neuro, 8, 10.1177/1759091416670978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvanet, C. , Duvezin‐Caubet, S. , di Rago, J.P. & Rojo, M. (2010) Energetic requirements and bioenergetic modulation of mitochondrial morphology and dynamics. Seminars in Cell & Developmental Biology, 21, 558–565. [DOI] [PubMed] [Google Scholar]
- Schertl, P. & Braun, H.‐P. (2015) Activity measurements of mitochondrial enzymes in native gels. Methods in Molecular Biology, 1305, 131–138. [DOI] [PubMed] [Google Scholar]
- Schuler, M. , Rellán‐Álvarez, R. , Fink‐Straube, C. , Abadía, J. & Bauer, P. (2012) Nicotianamine functions in the Phloem‐based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. The Plant cell, 24, 2380–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesso, A. , Belizario, J.E. , Marques, M.M. , Higuchi, M.L. , Schumacher, R.I. , Colquhoun, A. et al. (2012) Mitochondrial swelling and incipient outer membrane rupture in preapoptotic and apoptotic cells. Anatomical Record (Hoboken), 295, 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sew, Y.S. , Stroher, E. , Holzmann, C. , Huang, S. , Taylor, N.L. , Jordana, X. et al. (2013) Multiplex micro‐respiratory measurements of Arabidopsis tissues. New Phytologist, 200, 922–932. [DOI] [PubMed] [Google Scholar]
- Shakoury‐Elizeh, M. , Protchenko, O. , Berger, A. , Cox, J. , Gable, K. , Dunn, T.M. et al. (2010) Metabolic response to iron deficiency in Saccharomyces cerevisiae . Journal of Biological Chemistry, 285, 14823–14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Cui, M. , Yang, L. , Kim, Y.J. & Zhang, D. (2015) Genetic and biochemical mechanisms of pollen wall development. Trends in Plant Science, 20, 741–753. [DOI] [PubMed] [Google Scholar]
- Skulachev, V.P. (2001) Mitochondrial filaments and clusters as intracellular power‐transmitting cables. Trends in Biochemical Sciences, 26, 23–29. [DOI] [PubMed] [Google Scholar]
- Soneson, C. , Love, M.I. & Robinson, M.D. (2015) Differential analyses for RNA‐seq: transcript‐level estimates improve gene‐level inferences. F1000Research, 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre, D. , Gutierrez‐Carbonell, E. , Lattanzio, G. , Rellan‐Alvarez, R. , Gaymard, F. , Wohlgemuth, G. et al. (2013) Iron‐dependent modifications of the flower transcriptome, proteome, metabolome, and hormonal content in an Arabidopsis ferritin mutant. Journal of Experimental Botany, 64, 2665–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F. , Bosnjak, M. , Skunca, N. & Smuc, T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M. , Terada, Y. , Nakai, I. , Nakanishi, H. , Yoshimura, E. , Mori, S. et al. (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. The Plant Cell, 15, 1263–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari, R.K. , Hadacek, F. , Sassmann, S. & Lang, I. (2013) Iron deprivation‐induced reactive oxygen species generation leads to non‐autolytic PCD in Brassica napus leaves. Environmental and Experimental Botany, 91, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, T. , Liu, Y. , Yan, H. , You, Q. , Yi, X. , Du, Z. et al. (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research, 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D.R. et al. (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.H. , Rodriguez‐Celma, J. , Lan, P. , Wu, Y.C. , Velez‐Bermudez, I.C. & Schmidt, W. (2018) Scopoletin 8‐hydroxylase‐mediated fraxetin production is crucial for iron mobilization. Plant Physiology, 177, 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K.J. , Lin, C.Y. , Ting, C.Y. & Shih, M.C. (2016) Ethylene‐regulated glutamate dehydrogenase fine‐tunes metabolism during anoxia‐reoxygenation. Plant Physiology, 172, 1548–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache, R. , Andersen, T.G. , Marhavy, P. & Geldner, N. (2018) A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. The Plant Journal, 93, 399–412. [DOI] [PubMed] [Google Scholar]
- Urzica, E.I. , Casero, D. , Yamasaki, H. , Hsieh, S.I. , Adler, L.N. , Karpowicz, S.J. et al. (2012) Systems and trans‐system level analysis identifies conserved iron deficiency responses in the plant lineage. The Plant Cell, 24, 3921–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken, O. , Ford, E. , Lister, R. , Huang, S. & Millar, A.H. (2016) Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. The Plant Journal, 88, 542–558. [DOI] [PubMed] [Google Scholar]
- Van Aken, O. & Whelan, J. (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Frontiers in Plant Science, 3, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G. , Grotz, N. , Dedaldechamp, F. , Gaymard, F. , Guerinot, M.L. , Briat, J.F. et al. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell, 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani, G. (2012) Discovering the role of mitochondria in the iron deficiency‐induced metabolic responses of plants. Journal of Plant Physiology, 169, 1–11. [DOI] [PubMed] [Google Scholar]
- Vigani, G. , Maffi, D. & Zocchi, G. (2009) Iron availability affects the function of mitochondria in cucumber roots. New Phytologist, 182, 127–136. [DOI] [PubMed] [Google Scholar]
- Vizcay‐Barrena, G. & Wilson, Z.A. (2006) Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. Journal of Experimental Botany, 57, 2709–2717. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Guo, Z.L. , Zhou, W.T. , Zhang, C. , Zhang, Z.Y. , Lou, Y. et al. (2018) The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiology, 178, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, B. (2012) Bioenergetic role of mitochondrial fusion and fission. Biochimica et Biophysica Acta (BBA) ‐ Bioenergetics, 1817(10), 1833–1838. [DOI] [PubMed] [Google Scholar]
- Wilson, Z.A. & Zhang, D.B. (2009) From Arabidopsis to rice: pathways in pollen development. Journal of Experimental Botany, 60, 1479–1492. [DOI] [PubMed] [Google Scholar]
- Wittig, I. , Braun, H.P. & Schagger, H. (2006) Blue native PAGE. Nature Protocols, 1, 418–428. [DOI] [PubMed] [Google Scholar]
- Xie, H.T. , Wan, Z.Y. , Li, S. & Zhang, Y. (2014) Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. The Plant Cell, 26, 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, S.X. , Lu, J.Y. , Lou, Y. , Teng, X.D. , Gu, J.N. , Zhang, C. et al. (2016) The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana . The Plant Journal, 88, 936–946. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Ding, Z. , Vizcay‐Barrena, G. , Shi, J. , Liang, W. , Yuan, Z. et al. (2014) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. The Plant Cell, 26, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Yang, C. , Yuan, Z. , Zhang, D. , Gondwe, M.Y. , Ding, Z. et al. (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana . The Plant Cell, 22, 91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J.S. , Zhang, B. , Zhan, H. , Lv, Y.L. , Jia, X.L. , Wang, T. et al. (2020) Phenylpropanoid derivatives are essential components of sporopollenin in vascular plants. Molecular Plant, 13, 1644–1653. [DOI] [PubMed] [Google Scholar]
- Yokosho, K. , Yamaji, N. & Ma, J.F. (2016) OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. Journal of Experimental Botany, 67, 5485–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura‐Sakakibara, K. , Tohge, T. , Matsuda, F. , Nakabayashi, R. , Takayama, H. , Niida, R. et al. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene‐metabolite correlations in Arabidopsis. The Plant Cell, 20, 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancani, M. , Casolo, V. , Petrussa, E. , Peresson, C. , Patui, S. , Bertolini, A. et al. (2015) The permeability transition in plant mitochondria: the missing link. Frontiers in Plant Science, 6, 1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, L. , Sun, C. , Feng, Y. , Li, D. , Chai, X. , Wang, L. et al. (2018) AtROP6 is involved in reactive oxygen species signaling in response to iron‐deficiency stress in Arabidopsis thaliana . FEBS Letters, 592, 3446–3459. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Liu, D. , Lv, X. , Wang, Y. , Xun, Z. , Liu, Z. et al. (2014) The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. The Plant Cell, 26, 2939–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Sun, Y. , Timofejeva, L. , Chen, C. , Grossniklaus, U. & Ma, H. (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development, 133, 3085–3095. [DOI] [PubMed] [Google Scholar]
- Zhu, E. , You, C. , Wang, S. , Cui, J. , Niu, B. , Wang, Y. et al. (2015) The DYT1‐interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. The Plant Journal, 83, 976–990. [DOI] [PubMed] [Google Scholar]
- Zoratti, M. & Szabò, I. (1995) The mitochondrial permeability transition. Biochimica et Biophysica Acta (BBA) ‐ Reviews on Biomembranes, 1241(2), 139–176. 10.1016/0304-4157(95)00003-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The morphology of plants and in vitro germination of pollen grown under Fe‐sufficient and ‐deficient conditions.
Figure S2. DNA fragmentation in Fe‐sufficient and ‐deficient anthers.
Figure S3. Cryo‐SEM images of pollen in dehiscent anthers.
Figure S4. The workflow of Fe‐deficient anther transcriptome analysis.
Figure S5. The changes in transcript levels of known −Fe genes, DYT1 and MYB80, in anthers under Fe deficiency.
Figure S6. Analysis of the expression of genes that are repressed in anther tissue upon Fe deficiency in the tapetal transcriptomes of ams, tdf1, and bhlhs mutants.
Figure S7. The effect of RBOHE expression on anther ROS levels under Fe sufficiency and deficiency.
Figure S8. The activities and protein abundance of mitochondrial respiratory complexes in developing anthers under Fe deficiency.
Figure S9. Mitochondria labeled with mito‐GFP in isolated tapetal cells.
Figure S10. 3D numerical analysis of mitochondrial morphology.
Table S1. Metabolite profiling in floral buds.
Table S2. Element contents in leaves and floral buds.
Table S3. The DEGs in anthers, leaves, and roots under Fe deficiency.
Table S4. Pathway enrichment analysis of anther‐unique DEGs under Fe deficiency.
Table S5. Anther‐unique DEGs under Fe deficiency in the phenylpropanoid biosynthesis pathway.
Table S6. GO enrichment analysis of tapetum‐expressed genes induced by Fe deficiency and also differentially expressed in the tdf1, ams, and bhlhs mutants.
Table S7. Primers used for qPCR.
Data S1. Statistical analysis of H2DCFDA fluorescence intensity in Fe‐sufficient and ‐deficient anthers at each developmental stage in different lines.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.


