Abstract
A previous study in 356 healthy Finnish volunteers showed that glycochenodeoxycholate 3‐O‐glucuronide (GCDCA‐3G) and glycodeoxycholate 3‐O‐glucuronide (GDCA‐3G) are promising biomarkers of organic anion transporting polypeptide 1B1 (OATP1B1). In the same cohort, we now evaluated the performances of two other OATP1B1 biomarkers, coproporphyrin I (CPI) and III (CPIII), and compared them with GCDCA‐3G and GDCA‐3G. Based on decreased (*5 and *15) and increased (*14 and *20) function SLCO1B1 haplotypes, we stratified the participants to poor, decreased, normal, increased, and highly increased OATP1B1 function groups. Fasting plasma CPI concentration was 68% higher in the poor (95% confidence interval, 44%, 97%; P = 1.74 × 10−10), 7% higher in the decreased (0%, 15%; P = 0.0385), 10% lower in the increased (3%, 18%; P = 0.0087), and 23% lower in the highly increased (1%, 40%; P = 0.0387) function group than in the normal function group. CPIII concentration was 27% higher (7%, 51%; P = 0.0071) in the poor function group than in the normal function group. CPI and CPIII detected poor OATP1B1 function with areas under the precision‐recall curve (AUPRC) of 0.388 (95% confidence interval, 0.197, 0.689) and 0.0798 (0.0485, 0.203), and receiver operating characteristic curve (AUROC) of 0.888 (0.851, 0.919) and 0.731 (0.682, 0.776). The AUPRC and AUROC of GCDCA‐3G were, however, 0.389 (0.258, 0.563) and 0.100 (−0.0046, 0.204; P = 0.0610) larger than those of CPI, and 0.697 (0.555, 0.831) and 0.257 (0.141, 0.373; P < 0.0001) larger than those of CPIII. In conclusion, these data indicate that plasma CPI outperforms CPIII in detecting altered OATP1B1 function, but GCDCA‐3G is an even more sensitive OATP1B1 biomarker than CPI.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Coproporphyrin I (CPI) and coproporphyrin III (CPIII) are promising biomarkers to measure organic anion transporting polypeptide (OATP) 1B activity in humans. In vitro data suggest that CPI and CPIII are better substrates for OATP1B1 than for OATP1B3.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ Are CPI and CPIII sensitive OATP1B1 biomarkers in humans? What is the performance of CPI and CPIII compared with the highly specific and sensitive OATP1B1 endogenous probes glycochenodeoxycholate 3‐O‐glucuronide (GCDCA‐3G) and glycodeoxycholate 3‐O‐glucuronide (GDCA‐3G)?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ CPI clearly outperformed CPIII in distinguishing the genetically poor, decreased, increased, and highly increased OATP1B1 function groups, indicating higher sensitivity as an OATP1B1 biomarker. However, GCDCA‐3G showed an even better sensitivity than CPI. Careful standardization of plasma CP handling and storing is needed for reliable quantification.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ CPI may be a useful endogenous probe substrate along with the more sensitive GCDCA‐3G for measuring OATP1B1 activity during drug development and in clinical practice.
Organic anion transporting polypeptides (OATPs) 1B1 and 1B3 are influx transporters located on the sinusoidal membrane of human hepatocytes. 1 , 2 , 3 , 4 , 5 They mediate the hepatic uptake of a diverse array of endogenous compounds and xenobiotics, including drugs from several important therapeutic classes such as statins, HIV protease inhibitors, and angiotensin II receptor antagonists. Inhibition of OATP1B1 and OATP1B3 by concurrent medication and genetic variability affecting OATP1B1 activity are recognized risk factors for clinically relevant adverse drug reactions, such as statin‐induced myopathy. 6 , 7 , 8 , 9 , 10 Therefore, early identification of OATP1B‐mediated drug–drug interaction (DDI) risk is of high importance during drug development. 11
In the past few years, the research of OATP1B endogenous probe substrates as early signals for DDIs have shown significant progress. Circulating OATP1B substrates, such as coproporphyrins (CPs), hexadecanedioate, tetradecanedioate, direct bilirubin, and many glucuronidated and sulfated bile acids, have demonstrated high potential for measuring OATP1B activity. 12 , 13 , 14 Regarding CPs as OATP1B biomarkers, CPI and CPIII have been shown to detect strong dose‐dependent OATP1B inhibition by rifampin, and also moderate or mild inhibition by other OATP1B inhibitors in healthy volunteers. 15 , 16 , 17 , 18 , 19 , 20 Furthermore, a recent study demonstrated the usefulness of CPI and CPIII along with glucuronidated and sulfated bile acids in identifying OATP1B inhibition in cancer patients. 21 Moreover, a common genetic variant in the SLCO1B1 gene, c.521T>C, reduces the function of OATP1B1 and associates with increased CPI levels, 8 , 22 , 23 , 24 supporting the utility of CPI as an endogenous OATP1B probe.
CPI and CPIII, are porphyrin metabolites arising from heme biosynthesis that are excreted unchanged into the bile and urine. Both are substrates of OATP1B1 and OATP1B3, but only CPIII is also a substrate of OATP2B1. 16 , 25 In addition, the hepatic efflux transporters multidrug resistance–associated protein (MRP) 2, MRP3, and MRP4 transport CPI and CPIII. 20 , 26 , 27 Moreover, Rotor syndrome, a rare inherited complete deficiency of OATP1B1 and OATP1B3, leads to decreased hepatic uptake and increased urinary excretion of CPs. 28
Using a genetic association study, we recently demonstrated that plasma glycochenodeoxycholate 3‐O‐glucuronide (GCDCA‐3G) and glycodeoxycholate 3‐O‐glucuronide (GDCA‐3G) are highly sensitive and specific biomarkers for OATP1B1 activity. 29 Up until recently, CPs have been suggested as the most sensitive OATP1B endogenous probes; 30 however, there are no studies comparing the specificities of CPI and CPIII as OATP1B1 biomarkers with GCDCA‐3G and GDCA‐3G. Moreover, the effects of increased function SLCO1B1 variants on plasma CP concentrations have not been studied previously. Therefore, the aim of the present study was to investigate the impact of increased and decreased function variants of SLCO1B1 on the plasma concentrations of CPI and CPIII. The study was carried out in the same set of individuals in whom we previously investigated the impact of SLCO1B1 variants on GCDCA‐3G and GDCA‐3G concentrations, thus allowing direct comparison of the performance of these compounds as OATP1B1 biomarkers.
METHODS
Study participants and samples
A total of 356 healthy unrelated White Finnish volunteers (183 women, 173 men) participated in the study after giving a written informed consent. The mean ± standard deviation (SD) age of the participants was 24.1 ± 4.1 years, weight 69.7 ± 12.1 kg, and body mass index 22.9 ± 2.7. Participants were not on any continuous medication and were not tobacco smokers. Blood samples for measuring endogenous compounds were collected as part of a previously published 31 and a yet unpublished single dose pharmacokinetic study (Trial 1, European Union Drug Regulating Authorities Clinical Trials Database (EudraCT) number 2011‐004645‐40 and Trial 2, EudraCT number 2015‐000540‐41). Trial 1 was carried out between January 2012 and October 2014 and Trial 2 between May 2015 and October 2017. Following an overnight fast at 7–8 a.m. and before the study drug administration, a 10‐mL blood sample was collected from each participant into a light‐protected ethylenediaminetetraacetic acid–containing tube. The tube was placed on ice immediately after sampling and plasma was separated within 30 minutes. The plasma samples were stored at −80°C until analysis. The study protocols were approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (Helsinki, Finland).
Chemicals
CPI and CPIII, and stable isotopically labeled CPI 15N4 and CPIII 15N4 were purchased from Toronto Research Chemicals (Toronto, ON, Canada). Other chemicals and organic solvents were of commercially available analytical grade.
Quantification of plasma CPI and CPIII
The plasma samples were prepared by mixed‐mode anion exchange extraction on 96‐well plates (Oasis MAX μElution, Waters, Milford, MA) using a Freedom EVO automated liquid handling system (Tecan Group, Männedorf, Switzerland). Prior to extraction, 150 µL aliquot of plasma, 150 µL of freshly prepared 12% phosphoric acid, and 20 µL of stable isotope‐labeled internal standard solution (25 ng/mL in water each) were mixed and loaded into the plate. Samples were washed with 200 μL of 5% ammonium hydroxide and 200 μL of methanol, and eluted twice with 30 μL of 5% formic acid in methanol‐acetonitrile (60:40 volume/volume). The sample extracts were dried using a GeneVac centrifugal evaporator (Thermo Fisher Scientific, Waltham, MA), followed by reconstitution in 40 µL of 25% methanol containing 1% formic acid. The measurement of CPI and CPIII were carried out using a Sciex 6500 QTRAP+ liquid chromatography–tandem mass spectrometer (Sciex, Toronto, ON, Canada). The chromatographic separation was achieved on Acquity UPLC BEH C18 column (2.1 × 100 mm I.D.; 1.7 μm particle size) (Waters) using a mobile phase gradient of 0.1% formic acid in water and 0.1% formic acid in acetonitrile as previously described. 32 The mass spectrometer was operated in positive multiple reaction monitoring mode using the singly and doubly charged ion transitions of 655 to 596 and 328 to 238 for CPI and CPIII, and 659 to 600 and 330 to 240 for the isotopically labeled internal standards. The lower limits of quantification were 0.02 ng/mL, and the interday coefficient of variations were below 15% at relevant concentrations for both analytes.
Genotyping
Genomic DNA was extracted from ethylenediaminetetraacetic acid anticoagulated blood samples using the Maxwell 16 LEV Blood DNA Kit on a Maxwell 16 Research automated nucleic acid extraction system (Promega, Madison, WI). The participants were genotyped using either the Illumina Infinium Core Exome (n = 183) or Global Screening Array (n = 173) microchips at the Technology Centre of the Institute for Molecular Medicine Finland (Helsinki, Finland). Only the single‐nucleotide variants (SNVs), which were available on both chips and had minor allele frequencies over 0.01 were included in the analyses. Success rate of 97% and Hardy‐Weinberg equilibrium P < 10−5 were employed as quality thresholds for including genotype data in statistical analysis. In addition, the participants were genotyped for the SLCO1B1 c.388A>G (rs2306283, p.N130D), c.463C>A (rs11045819, p.P155T), c.521T>C, and c.1929A>C (rs34671512, p.L643F) SNVs with TaqMan genotyping assays on a QuantStudio 12K Flex Real‐Time PCR System (Thermo Fisher Scientific). The SLCO1B1 haplotypes *1 (c.388A‐c.463C‐c.521T‐c.1929A, previously known as *1A), *5 (c.388A‐c.463C‐c.521C‐c.1929A), *14 (c.388G‐c.463A‐c.521T‐c.1929A), *15 (c.388G‐c.463C‐c.521C‐c.1929A), *20 (c.388G‐c.463C‐c.521T‐c.1929C, previously known as *35), and *37 (c.388G‐c.463C‐c.521T‐c.1929A, previously known as *1B) were computed with PHASE v2.1.1. 33 , 34 , 35 , 36 , 37 SLCO1B1*14 and *20 alleles were considered to be increased function alleles, *5 and *15 to be decreased function alleles, and *1 and *37 to be normal function alleles. The subjects were divided into the following genotype‐predicted phenotype groups: normal function (*1 and *37 homozygotes and compound heterozygotes), decreased function (carriers of one decreased function allele together with a normal or increased function allele), poor function (carriers of two decreased function alleles), increased function (carriers of one increased function allele together with a normal function allele), and highly increased function (carriers of two increased function alleles). For comparison, the results were also calculated by considering *37 to be an increased function allele (Table S1 ).
Statistical analysis
The data were analyzed with the statistical programs JMP Genomics 8.0 (SAS Institute, Inc., Cary, NC) and IBM SPSS 25 for Windows (Armonk, NY). The concentrations of CPI and CPIII were logarithmically transformed before analysis. Possible differences in CPI and CPIII concentrations between the two clinical trials in which the samples were collected, and effects of demographic covariates (sex and logarithmically transformed bodyweight and blood hemoglobin concentration) were investigated using a forward stepwise linear regression analysis. P value thresholds of 0.05 for entry and 0.10 for removal were employed as the stepping method criteria. Genome‐wide association analyses were carried out using linear regression analysis with additive coding and the significant demographic covariates set as fixed factors. A P value of below 5 × 10−8 was considered genome‐wide significant. For genome‐wide significant SNVs, SLCO1B1 haplotypes and genotype‐predicted phenotype groups, analysis of variance adjusting for significant demographic covariates was carried out with pairwise comparisons with the Fisher’s least significant difference method. A P value of below 0.05 was considered statistically significant. Precision‐recall (PR) and receiver operating characteristic (ROC) analyses were carried out and areas under the precision‐recall (AUPRC) and receiver operating characteristic (AUROC) curves were calculated with MedCalc Statistical Software version 19.7 (MedCalc Software bv, Ostend, Belgium) using 1,000 bootstrap iterations for confidence intervals (CIs).
RESULTS
Fasting plasma CPI and CPIII concentrations in healthy volunteers
The geometric fasting plasma mean CPI and CPIII concentrations were 0.57 ng/mL (95% CI, 0.55–0.59 ng/mL; range 0.10–1.6 ng/mL) and 0.079 ng/mL (95% CI, 0.076–0.082 ng/mL; range 0.019–0.19 ng/mL), showing moderate variability between individuals. In a forward stepwise linear regression analysis, sex and clinical trial number (Trial 1 or Trial 2) were significantly associated with plasma CPI concentration, and sex, clinical trial number, and body weight with CPIII concentration. Women had on average 26% and 28% lower (P = 1.23 × 10−19 and P = 5.22 × 10−14) CPI and CPIII concentrations than men. The concentration of CPI was 21% and that of CPIII was 15% lower (P = 2.21 × 10−12 and P = 1.88 × 10−6) in Trial 2 than in Trial 1. CPIII concentration was 3% higher per 10% increase in body weight (P = 0.0137). The association of body weight with CPI concentration in the linear regression analysis was borderline significant (1% increase per 10% increase in body weight, P = 0.0511). Therefore, sex, trial number, and body weight were set as covariates in the genetic association analyses. In the final linear regression model, blood hemoglobin concentration showed no association with CPI or CPIII concentration (P = 0.365 and P = 0.509). Associations of CPI and CPIII concentrations with these covariates are shown in Figure S1 .
Genome‐wide association study for plasma CPI and CPIII
In a genome‐wide association analysis, only the SLCO1B1 rs4149056 (c.521T>C, p. Val174Ala) SNV showed significant association with the plasma concentration of CPI (P = 6.16 × 10−10) (Figure 1 ). When adjusting for the effect of rs4149056, no other SNVs showed genome‐wide significant associations with CPI concentration. Plasma CPIII concentration was not associated with any SNV at the genome‐wide significance level.
Figure 1.
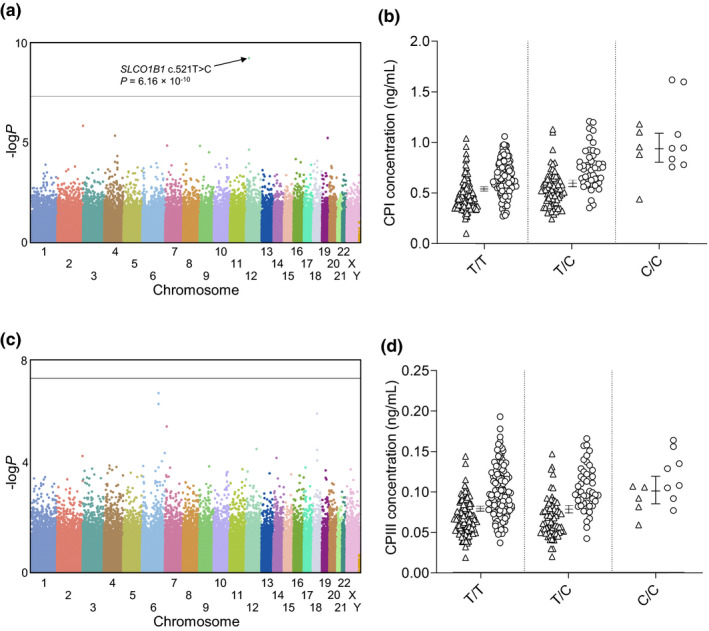
Manhattan plots of the fasting plasma (a) CPI and (c) CPIII and the respective effects of SLCO1B1 c.521T>C on plasma (b) CPI and (d) CPIII levels. Horizontal line in a and c represents the genome‐wide significance level of 5 × 10−8. Horizontal lines in b and d indicate geometric marginal mean values and whiskers indicate 95% confidence intervals. Triangles, women; circles, men (b and d). CP, coproporphyrin.
Effects of SLCO1B1 genotypes on CPI and CPIII concentrations
In individuals that were homozygous variant or heterozygous for the SLCO1B1 c.521T>C SNV, the mean plasma concentration of CPI was 74% (P = 1.42 × 10−11) or 11% (P = 0.0012) higher than in individuals with the T/T genotype (Figure 1 ). For the plasma CPIII concentrations, only individuals with the C/C genotype differed significantly from the T/T homozygotes (28% higher concentration; P = 0.0052). Of SLCO1B1 haplotypes, the highest CPI concentrations were seen in individuals homozygous or compound heterozygous for the SLCO1B1*5 or *15 alleles, and the lowest plasma concentrations in those with different combinations of the *14 and *20 alleles (Table 1 and Figure 2 ). This trend was less clear for CPIII concentrations.
Table 1.
Effects of SLCO1B1 haplotypes on the plasma concentrations of CPI and CPIII in 356 healthy volunteers
| Genotype (n) | Geometric mean ng/mL (95% CI) |
Ratio to *1/*1 (95% CI) |
P |
|---|---|---|---|
| CPI | |||
| *14/*14 (1) | 0.341 (0.199, 0.583) | 0.616 (0.359, 1.06) | |
| *14/*37 (6) | 0.368 (0.295, 0.459) | 0.666 (0.532, 0.834) | 4.32 × 10−4 |
| *5/*14 (3) | 0.420 (0.308, 0.574) | 0.760 (0.554, 1.04) | 0.0891 |
| *5/*20 (1) | 0.450 (0.263, 0.771) | 0.814 (0.474, 1.39) | |
| *14/*20 (4) | 0.455 (0.348, 0.595) | 0.823 (0.626, 1.08) | 0.160 |
| *1/*20 (12) | 0.480 (0.412, 0.561) | 0.870 (0.739, 1.02) | 0.0921 |
| *20/*37 (7) | 0.513 (0.419, 0.629) | 0.928 (0.753, 1.14) | 0.485 |
| *1/*14 (34) | 0.529 (0.482, 0.580) | 0.957 (0.861, 1.06) | 0.407 |
| *37/*37 (3) | 0.547 (0.400, 0.748) | 0.990 (0.722, 1.36) | 0.951 |
| *14/*15 (11) | 0.548 (0.466, 0.644) | 0.992 (0.837, 1.17) | 0.921 |
| *1/*1 (116) | 0.553 (0.526, 0.581) | 1 | |
| *1/*37 (47) | 0.564 (0.521, 0.610) | 1.02 (0.929, 1.12) | 0.669 |
| *15/*20 (6) | 0.574 (0.460, 0.716) | 1.04 (0.828, 1.30) | 0.744 |
| *15/*37 (17) | 0.609 (0.534, 0.693) | 1.10 (0.958, 1.27) | 0.175 |
| *1/*5 (6) | 0.611 (0.490, 0.761) | 1.11 (0.882, 1.39) | 0.385 |
| *1/*15 (69) | 0.615 (0.577, 0.656) | 1.11 (1.03, 1.21) | 0.0108 |
| *15/*15 (11) | 0.912 (0.776, 1.07) | 1.65 (1.39, 1.96) | 1.37 × 10−8 |
| *5/*15 (2) | 1.08 (0.737, 1.57) | 1.95 (1.33, 2.86) | 6.84 × 10−4 |
| CPIII | |||
| *14/*14 (1) | 0.0530 (0.0288, 0.0975) | 0.669 (0.363, 1.23) | |
| *5/*14 (3) | 0.0565 (0.0397, 0.0804) | 0.713 (0.498, 1.02) | 0.0636 |
| *14/*37 (6) | 0.0606 (0.0473, 0.0778) | 0.765 (0.593, 0.987) | 0.0395 |
| *5/*20 (1) | 0.0633 (0.0344, 0.117) | 0.799 (0.433, 1.47) | |
| *1/*5 (6) | 0.0719 (0.0560, 0.0922) | 0.907 (0.702, 1.17) | 0.455 |
| *1/*20 (12) | 0.0741 (0.0622, 0.0884) | 0.935 (0.778, 1.12) | 0.477 |
| *20/*37 (7) | 0.0782 (0.0622, 0.0985) | 0.988 (0.779, 1.25) | 0.918 |
| *1/*37 (47) | 0.0785 (0.0717, 0.0858) | 0.990 (0.890, 1.10) | 0.852 |
| *1/*15 (69) | 0.0786 (0.0730, 0.0846) | 0.992 (0.904, 1.09) | 0.863 |
| *15/*20 (6) | 0.0789 (0.0615, 0.102) | 0.997 (0.771, 1.29) | 0.979 |
| *1/*1 (116) | 0.0793 (0.0749, 0.0839) | 1 | |
| *15/*37 (17) | 0.0796 (0.0686, 0.0922) | 1.00 (0.857, 1.18) | 0.963 |
| *1/*14 (34) | 0.0819 (0.0738, 0.0909) | 1.03 (0.918, 1.16) | 0.587 |
| *14/*15 (11) | 0.0832 (0.0692, 0.0999) | 1.05 (0.865, 1.27) | 0.626 |
| *37/*37 (3) | 0.0842 (0.0591, 0.120) | 1.06 (0.742, 1.52) | 0.741 |
| *14/*20 (4) | 0.0843 (0.0622, 0.114) | 1.06 (0.781, 1.45) | 0.692 |
| *15/*15 (11) | 0.0999 (0.0831, 0.120) | 1.26 (1.04, 1.53) | 0.0184 |
| *5/*15 (2) | 0.105 (0.0683, 0.162) | 1.33 (0.859, 2.05) | 0.202 |
Data are adjusted for sex, body weight, and clinical trial. CI, Confidence interval; CPI, coproporphyrin I; CPIII, coproporphyrin III.
Figure 2.
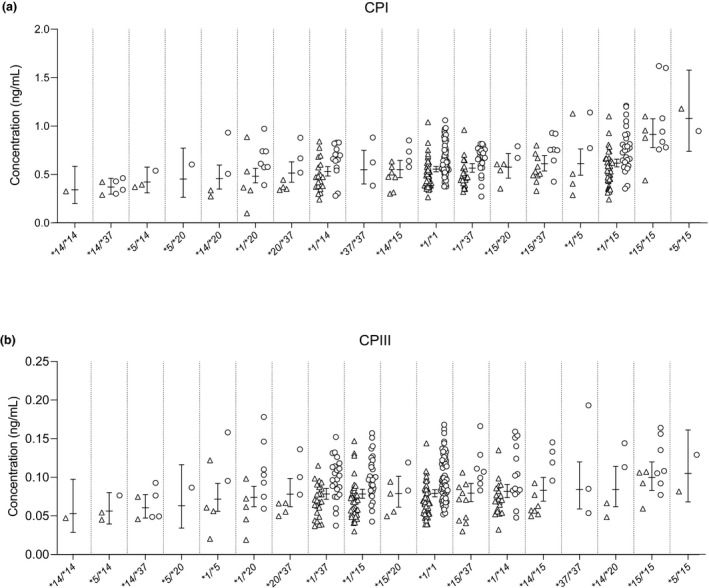
Effects of SLCO1B1 haplotypes on the plasma concentrations of (a) CPI and (b) CPIII. Horizontal lines indicate geometric estimated marginal mean values and whiskers indicate 95% confidence intervals. Triangles, women; circles, men. CP, coproporphyrin.
We then stratified the participants on the basis of decreased (*5 and *15) and increased (*14 and *20) function SLCO1B1 haplotypes to poor, decreased, normal, increased, and highly increased OATP1B1 function groups (Table 2 and Figure 3 ). Fasting plasma CPI concentration was 68% higher in the poor (95% CI, 44%, 97%; P = 1.74 × 10−10), 7% higher in the decreased (0%, 15%; P = 0.0385), 10% lower in the increased (3%, 18%; P = 0.0087), and 23% lower in the highly increased (1%, 40%; P = 0.0387) function group than in the normal function group. CPIII concentration was 27% higher (7%, 51%; P = 0.0071) in the poor function group than in the normal function group, but no significant difference was found between the decreased, increased, and highly increased function groups vs. the normal function group. When considering SLCO1B1*37 as an increased function allele, the difference in CPI concentration between the increased and normal function groups was reduced to a nonsignificant 2% (−1%, 9%; P = 0.539) (Table S1 ).
Table 2.
Effect of OATP1B1 phenotype on plasma biomarker concentrations in healthy volunteers
| Phenotype (n) | Geometric mean ng/mL (95% CI) | Ratio to normal function (95% CI) | P |
|---|---|---|---|
| CPI | |||
| Highly increased function (5) | 0.429 (0.337, 0.546) | 0.772 (0.603, 0.986) | 0.0387 |
| Increased function (59) | 0.498 (0.464, 0.534) | 0.896 (0.825, 0.972) | 0.00873 |
| Normal function (166) | 0.556 (0.533, 0.580) | 1 | |
| Decreased function (113) | 0.597 (0.567, 0.629) | 1.07 (1.00, 1.15) | 0.0385 |
| Poor function (13) | 0.937 (0.806, 1.09) | 1.68 (1.44, 1.97) | 1.74 × 10−10 |
| CPIII | |||
| Highly increased function (5) | 0.0769 (0.0586, 0.101) | 0.972 (0.738, 1.28) | 0.839 |
| Increased function (59) | 0.0774 (0.0715, 0.0838) | 0.978 (0.892, 1.07) | 0.642 |
| Normal function (166) | 0.0791 (0.0754, 0.0829) | 1 | |
| Decreased function (113) | 0.0780 (0.0736, 0.0826) | 0.986 (0.914, 1.06) | 0.707 |
| Poor function (13) | 0.101 (0.0850, 0.119) | 1.27 (1.07, 1.51) | 0.00711 |
| GCDCA‐3G a | |||
| Highly increased function (5) | 11.2 (6.61, 19.0) | 0.403 (0.235, 0.689) | 9.46 × 10−4 |
| Increased function (59) | 22.4 (19.2, 26.2) | 0.805 (0.673, 0.963) | 0.0178 |
| Normal function (166) | 27.9 (25.4, 30.5) | 1 | |
| Decreased function (113) | 44.5 (39.8, 49.8) | 1.60 (1.38, 1.85) | 6.43 × 10−10 |
| Poor function (13) | 239 (172, 331) | 8.57 (6.10, 12.0) | 1.46 × 10−29 |
| GDCA‐3G a | |||
| Highly increased function (5) | 43.0 (20.4, 90.6) | 0.675 (0.317, 1.44) | 0.307 |
| Increased function (59) | 42.6 (34.3, 52.9) | 0.668 (0.518, 0.860) | 0.00179 |
| Normal function (166) | 63.8 (56.0, 72.6) | 1 | |
| Decreased function (113) | 105 (90.1, 123) | 1.65 (1.35, 2.03) | 2.04 × 10−6 |
| Poor function (13) | 367 (231, 583) | 5.76 (3.56, 9.30) | 4.33 × 10−12 |
CPI and CPIII data are adjusted for sex, body weight, and clinical trial. GCDCA‐3G and GDCA‐3G data are adjusted for sex. Phenotypes classified by SLCO1B1 allele pairs: highly increased function phenotype, two increased function (*14 or *20) alleles; increased function phenotype, one normal function (*1 or *37) allele with one increased function allele; normal function phenotype, two normal function alleles; decreased function phenotype, one normal function allele or one increased function allele with one decreased function (*5 or *15) allele; poor function phenotype, two decreased function alleles. CI, confidence interval; CPI, coproporphyrin I; CPIII, coproporphyrin III; GCDCA‐3G, glycochenodeoxycholate 3‐O‐glucuronide; GDCA‐3G, glycodeoxycholate 3‐O‐glucuronide; OATP1B1, organic anion transporting polypeptide 1B1.
These data were reanalyzed from a previously published study. 29
Figure 3.
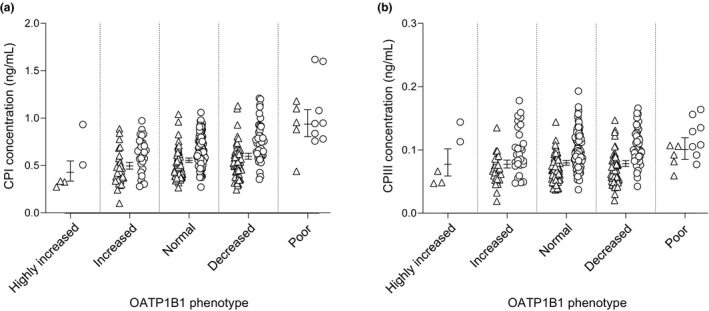
Plasma (a) CPI and (b) CPIII concentrations in predicted OATP1B1 phenotypes. Horizontal lines indicate geometric marginal mean values and whiskers indicate 95% confidence intervals. Triangles, women; circles, men. CP, coproporphyrin. OATP1B1, organic anion transporting polypeptide 1B1.
Comparison of biomarker performance
In the entire cohort, CPI and CPIII distinguished SLCO1B1 c.521C/C homozygotes from the T/C and T/T genotypes with AUPRC (95% CI) values of 0.388 (0.197, 0.689) and 0.0798 (0.0485, 0.203), and AUROC values of 0.888 (P < 0.0001) and 0.731 (P = 0.0002) (Table 3 and Figure 4 ). In men, CPI and CPIII detected C/C homozygotes with AUPRC values of 0.414 (0.143, 0.788) and 0.0883 (0.0451, 0.273), and AUROC values of 0.885 (P < 0.0001) and 0.683 (P = 0.0538). In women, the respective values for CPI and CPIII were 0.458 (0.104, 0.917) and 0.0836 (0.0335, 0.213), and 0.866 (P = 0.0021) and 0.788 (P = 0.0067).
Table 3.
Comparison of plasma biomarker performances in detecting SLCO1B1 c.521T>C variant homozygous genotype
| OATP1B1 biomarker |
Mean AUPRC (95% CI) |
F1max | Mean difference from GCDCA‐3G (95% CI) | Mean AUROC (95% CI) | Difference from GCDCA‐3G | |
|---|---|---|---|---|---|---|
| Mean (95% CI) | P value | |||||
| Entire cohort (n = 356) baseline AUPRC = 0.037 and baseline AUROC = 0.5 | ||||||
| GCDCA‐3G a |
0.777 (0.584, 0.934) |
0.692 | — |
0.988 (0.970, 0.996) |
— | — |
| GDCA‐3G a |
0.605 (0.353, 0.868) |
0.621 |
−0.172 (−0.0820, −0.301) |
0.876 (0.837, 0.908) |
−0.112 (0.022, −0.247) |
0.1021 |
| CPI |
0.388 (0.197, 0.689) |
0.432 |
−0.389 (−0.258, −0.563) |
0.888 (0.851, 0.919) |
−0.100 (0.00462, −0.204) |
0.0610 |
| CPIII |
0.0798 (0.0485, 0.203) |
0.167 |
−0.697 (−0.555, −0.831) |
0.731 (0.682, 0.776) |
−0.257 (−0.141, −0.373) |
<0.0001 |
| Men (n = 173) baseline AUPRC = 0.046 and baseline AUROC = 0.5 | ||||||
| GCDCA‐3G a |
0.935 (0.704, 1.00) |
0.857 | — |
0.996 (0.972, 1.000) |
— | — |
| GDCA‐3G a |
0.698 (0.318, 0.944) |
0.714 |
−0.237 (−0.0682, −0.515) |
0.875 (0.817, 0.921) |
−0.121 (0.0587, −0.300) |
0.1871 |
| CPI |
0.414 (0.143, 0.788) |
0.400 |
−0.521 (−0.277, −0.810) |
0.885 (0.828, 0.928) |
−0.111 (−0.0219, −0.201) |
0.0147 |
| CPIII |
0.0883 (0.0451, 0.273) |
0.211 |
−0.847 (−0.730, −0.937) |
0.683 (0.608, 0.752) |
−0.313 (−0.126, −0.499) |
0.0010 |
| Women (n = 183) baseline AUPRC = 0.027 and baseline AUROC = 0.5 | ||||||
| GCDCA‐3G a |
0.616 (0.297, 1.00) |
0.833 | — |
0.993 (0.968, 1.000) |
— | — |
| GDCA‐3G a |
0.524 (0.110, 0.938) |
0.600 |
−0.0923 (0.282, −0.520) |
0.853 (0.794, 0.901) |
−0.140 (0.125, −0.405) |
0.3010 |
| CPI |
0.458 (0.104, 0.917) |
0.533 |
−0.158 (0.248, −0.633) |
0.866 (0.808, 0.912) |
−0.128 (0.103, −0.358) |
0.2784 |
| CPIII |
0.0836 (0.0335, 0.213) |
0.235 |
−0.533 (−0.291, −0.944) |
0.788 (0.721, 0.845) |
−0.206 (0.000483, −0.412) |
0.0505 |
AUPRC, area under precision‐recall curve; AUROC, area under receiver operating characteristic curve; CI, Confidence interval; CPI, coproporphyrin I; CPIII, coproporphyrin III; F1max, maximum harmonic mean of the precision and recall over all measurement levels; GCDCA‐3G, glycochenodeoxycholate 3‐O‐glucuronide; GDCA‐3G, glycodeoxycholate 3‐O‐glucuronide; OATP1B1, organic anion transporting polypeptide 1B1; — , not applicable.
These data were reanalyzed from a previously published study. 29
Figure 4.
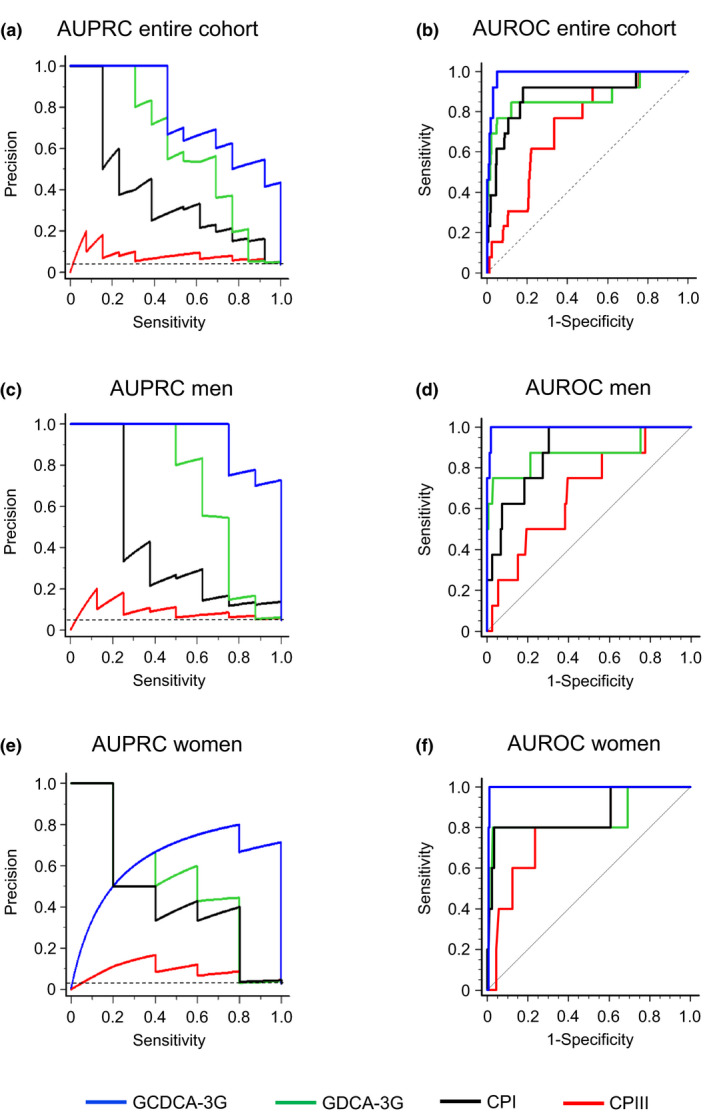
Performance of plasma biomarkers in detecting SLCO1B1 c.521C/C genotype in (a, b) the entire cohort of 356 healthy volunteers, in (c, d) 173 men and in (e, f) 183 women. AUPRCs represent the trade‐off of precision and sensitivity for every possible cutoff value. Horizontal dashed lines (a, c, e) represent the baseline AUPRC values of 0.046, 0.027, and 0.037, respectively, which correspond to the proportion of c.521C/C genotypes. AUROCs show relationships between sensitivity and specificity for every possible cutoff value. Dashed lines (b, d, f) represent random classifier. CP, coproporphyrin. AUPRC, area under the precision‐recall curve; AUROC, area under the receiver operating characteristic curve; GCDCA‐3G, glycochenodeoxycholate 3‐O‐glucuronide; GDCA‐3G, glycodeoxycholate 3‐O‐glucuronide.
We then compared the performances of CPI, CPIII, and the previously analyzed GCDCA‐3G and GDCA‐3G (Table 3 and Table S2 ). GCDCA‐3G outperformed the other OATP1B1 biomarkers in detecting the c.521C/C genotype and was chosen as a reference in PR and ROC analyses. In the entire cohort, the AUPRCs of GDCA‐3G, CPI, and CPIII were significantly smaller than the AUPRC of GCDCA‐3G, by 0.172 (95% CI, 0.0820, 0.301), 0.389 (0.258, 0.563), and 0.697 (0.555, 0.831), respectively. In men, the respective AUPRC differences were similar to the differences within the entire cohort, whereas in women only the AUPRC of CPIII was significantly lower than the AUPRC of GCDCA‐3G. Regarding AUROCs as a measure of performance, only CPI and CPIII in men (by 0.111; P = 0.0147, and 0.313; P = 0.0010) and CPIII in the whole cohort (by 0.257; P < 0.0001) were significantly smaller than the AUROCs of GCDCA‐3G. Moreover, when comparing the biomarkers’ sensitivities in detecting the different genotype‐predicted OATP1B1 function groups vs. the normal function group, GCDCA‐3G showed the best overall performance (Table 2 , Tables S1 and S2).
DISCUSSION
Plasma CPI and CPIII are among the most promising endogenous OATP1B biomarkers in humans. While previous preclinical data have shown that these compounds are better substrates for OATP1B1 than for OATP1B3, 25 the specificity of CPI and CPIII as OATP1B1 biomarkers in humans has not been directly compared with other candidate biomarkers. In this study, we investigated the effects of decreased and increased function SLCO1B1 variants on plasma CPI and CPIII levels in 356 healthy white Finnish volunteers. We then compared the performance of these compounds with GCDCA‐3G and GDCA‐3G to further evaluate the suitability of CPI and CPIII as OATP1B1 biomarkers. Moreover, we carried out a genome‐wide association study to identify other genes potentially associated with CPI and CPIII concentrations.
In the genome‐wide analysis, only the decreased function SLCO1B1 c.521T>C variant showed a genome‐wide significant association with plasma CPI levels, whereas no genome‐wide significant associations were observed for CPIII. These data indicate that common variability in genes other than SLCO1B1 is unlikely to be an important determinant of the fasting plasma concentrations of CPI and CPIII. Of note, no signals were detected for the efflux transporters MRP2, 3, or 4, which are known to transport CPI and CPIII. Plasma CPI and CPIII concentrations were 1.7‐fold and 1.3‐fold higher in individuals with the homozygous SLCO1B1 c.521C/C genotype than in noncarriers of the variant. In comparison, the fasting plasma concentrations of GCDCA‐3G and GDCA‐3G were 9.2‐fold and 6.4‐fold higher in the c.521C/C homozygotes than in noncarriers in our previous study in the same study cohort. 29 As the SLCO1B1 c.521C/C genotype results in a nearly complete OATP1B1 deficiency, 8 , 29 these results suggest that OATP1B1 plays a more important role in the hepatic uptake of GCDCA‐3G and GDCA‐3G than in the uptake of CPI and CPIII. The data further suggest that OATP1B1‐mediated transport is less important for the kinetics of CPIII than for CPI. This could be explained by the 6‐fold higher renal clearance of CPIII compared with CPI 16 and that CPIII is also a substrate for OATP2B1. 25
To gain more insight into the sensitivity of CPI and CPIII as OATP1B1 biomarkers, we then investigated the effects of increased and decreased function SLCO1B1 haplotypes on plasma CP levels. Low CPI concentrations were more common among individuals carrying the SLCO1B1*14 or *20 increased function alleles and the highest concentrations were seen in participants carrying the *5 and *15 decreased function alleles. A similar, but less clear trend was observed for CPIII levels. We stratified the study participants on the basis of these alleles to poor, decreased, normal, increased, and highly increased function genotype groups. Plasma CPI concentration was significantly higher in both the poor and decreased function genotype groups, and significantly lower in the increased and highly increased function groups than in the normal function group. These findings support both the role of CPI as an OATP1B1 biomarker and that *14 and *20 are associated with increased OATP1B1 activity. 29 , 34
A previous study in 391 Japanese individuals demonstrated that the SLCO1B1*15 allele is associated with increased plasma concentrations of CPI. 24 There appear to be no previous studies on the effects of SLCO1B1 variants on the plasma concentrations of CPIII or on the effects of increased function SLCO1B1 alleles on plasma CPI concentrations. In the Japanese population, the increased function alleles SLCO1B1*14 and *20 are very rare, 38 whereas *37 is the most common allele. Therefore, the SLCO1B1*37/*37 genotype was used as a reference in the Japanese study. In that study, the plasma CPI concentration seemed to be slightly higher in *1/*37 heterozygotes and in *1/*1 homozygotes than in *37/*37 homozygotes, but the differences between these genotype groups were not statistically significant. Similarly in our study, no clear effect of the SLCO1B1*37 allele on CPI concentration could be observed, although individuals compound heterozygous for *37 and *14 showed significantly lower plasma CPI concentration than individuals with the SLCO1B1*1/*1 genotype. When considering SLCO1B1*37 as an increased function allele, the increased function SLCO1B1 genotype group was no longer significantly different from the normal function group (Table S1 ). These data suggest that SLCO1B1*37 may be associated with a slightly elevated OATP1B1 activity compared with SLCO1B1*1, but the effect is so small that the allele could be considered a normal function allele.
Consistent with previous data regarding CPI, 22 we saw on average 30% higher plasma CPI and CPIII concentrations in men than in women. These sex‐related differences in plasma CP levels have been suggested to be at least partly due to a higher heme synthesis rate in men. 22 , 39 In one study, blood hemoglobin level was associated with plasma CPI levels, but no independent association between hemoglobin and CPI concentrations was found after adjusting for sex. 39 Similarly, in our study, blood hemoglobin concentration showed no independent association with CPI or CPIII concentration in the final linear regression model including sex as an independent variable. On the other hand, the concentrations of some other endogenous OATP1B1 substrates have also shown similar sex‐related differences. For example, in our previous study, plasma GCDCA‐3G and GDCA‐3G were higher in men than in women. 29 While the sex differences in GCDCA‐3G and GDCA‐3G concentrations could be partly due to a lower bile acid synthesis rate in women, the finding that also CPI and CPIII levels are lower in women suggests that OATP1B1 activity could be higher in women. The potential sex‐related difference in OATP1B1 activity may have important implications for drug safety, e.g., for evaluating DDIs and their clinical significance. A previous study found no sex difference in the liver expression of OATP1B1 in humans, 35 and further studies are required to verify whether OATP1B1 activity differs between the sexes and its underlying mechanisms.
One of the advantages of the present study was that we were able to compare the performances of CPs with GCDCA‐3G and GDCA‐3G as OATP1B1 probes. Such comparison was particularly interesting as GCDCA‐3G and GDCA‐3G have demonstrated strong potential as OATP1B1 biomarkers with exceptionally high sensitivities in detecting OATP1B1 deficiency. 29 To estimate the relative performance of each of the four endogenous probes, we quantified their efficiencies in discriminating the poor OATP1B1 function genotype using AUPRC and AUROC values.
AUPRC shows the relationship of the precision (positive predictive value) against the recall (sensitivity) for different thresholds, whereas AUROC displays the relationship between the sensitivity and the specificity. GCDCA‐3G demonstrated the highest efficiency in all PR and ROC analyses, whereas CPIII showed the weakest performance. CPI and GDCA‐3G performed similarly well, with the PR data favoring GDCA‐3G and ROC data favoring CPI. While the interpretation of the ROC curve alone may result in too optimistic values in heavily imbalanced data sets, 40 here both PR and ROC tests showed a relatively good potential for CPI to act as an OATP1B1 biomarker in healthy volunteers. Taken together, these data suggest that CPI displays an almost as good potential as GDCA‐3G for detecting OATP1B1 deficiency, but it underperforms compared with GCDCA‐3G.
A previous study investigated the effects of the nonselective OATP1B inhibitor rifampin on the plasma concentrations of GCDCA‐3G, GDCA‐3G, and CPI in healthy volunteers. 17 In that study, a single 600‐mg oral dose of rifampin increased the AUC of these OATP1B biomarkers 14.9‐fold, 4.0‐fold, and 3.7‐fold, respectively. Our previous 29 and present data show a similar rank order in the effects of the SLCO1B1 c.521C/C genotype on mean plasma concentrations of GCDCA‐3G (9.2‐fold), GDCA‐3G (6.4‐fold) and CPI (1.7‐fold) as compared with the T/T reference genotype. The smaller relative effect of SLCO1B1 genotype on CPI compared with the effect of rifampin suggests that OATP1B3 may play a more important role in the hepatic uptake of CPI than in that of GCDCA‐3G or GDCA‐3G. 25
In our study, plasma CPI levels showed much lower interindividual variability than GCDCA‐3G or GDCA‐3G. For example the coefficients of variation among participants with the SLCO1B1*1/*1 genotype were 31% for CPI, 79% for GCDCA‐3G, and 107% for GDCA‐3G. 29 A previous study in Japanese individuals showed a similarly low interindividual variability in plasma CPI concentrations. 24 Moreover, another study demonstrated that CPI shows less diurnal variation than GCDCA‐3G and GDCA‐3G. 17 As a consequence of lower variability, CPI performed equally well with GDCA‐3G as an OATP1B1 biomarker in the ROC and PR analyses despite a smaller effect of SLCO1B1 genotype on CPI than GDCA‐3G concentration.
In the present study, fasting plasma CP concentrations were measured from samples collected in two previous clinical trials, carried out during 2012–2014 and 2015–2017, respectively. To our surprise, we observed slightly higher mean CPI and CPIII levels in the samples from the earlier trial than in those from the newer trial. In both clinical trials, the samples were collected according to the same protocol and stored at −80°C. However, the type of light‐protected tube used for sample storage differed between the trials. Several previous studies have reported coproporphyrins to be photosensitive. 32 , 41 In our study, all the samples were processed using the same automated liquid handling protocol, with the total benchtop processing time being less than the recommended maximum of 2 hours. 41 Therefore, we consider it unlikely that photodegradation could explain the observed differences. On the other hand, as CPs exist in plasma at low concentrations (below 1 ng/mL), they could be relatively sensitive to also other physical factors such as surface adsorption into storage tubes. Of note, GCDCA‐3G and GDCA‐3G, which were previously measured from the same studies, exist in plasma at >100 times higher levels and no such concentration differences in the samples from the two trials with different storage tubes were detected. Nevertheless, this incidental observation highlights the importance of careful standardization of CP sample handling and storing. Such detailed protocols are especially crucial in multicenter trials, in which the samples are collected from several study sites.
To conclude, CPI and CPIII plasma concentrations are significantly increased among individuals with poor OATP1B1 function. Our findings support the previous data that CPI, in particular, is a sensitive biomarker to investigate OATP1B1 activity. However, in the light of the present study and the previous findings within the same study population, both CPI and CPIII are less sensitive OATP1B1 biomarkers than GCDCA‐3G. 29
Funding Information
This study was supported by grants from the European Research Council (Grant agreement 282109), State Funding for University‐Level Health Research (Finland), and the Sigrid Jusélius Foundation (Helsinki, Finland).
Conflict of Interest
The authors declared no competing interests for this work.
Author Contributions
M. Neuvonen and M. Niemi wrote the manuscript. M. Niemi designed the research. M. Neuvonen, A.T., P.H., J.T.B., and M. Niemi performed the research. M. Neuvonen and M. Niemi analyzed the data.
Supporting information
Fig S1
Table S1
Table S2
Acknowledgments
We thank Eija Mäkinen‐Pulli and Lisbet Partanen for skillful technical assistance.
References
- 1. Hsiang, B. et al. A novel human hepatic organic anion transporting polypeptide (OATP2). Identification of a liver‐specific human organic anion transporting polypeptide and identification of rat and human hydroxymethylglutaryl‐CoA reductase inhibitor transporters. J. Biol. Chem. 274, 37161–37168 (1999). [DOI] [PubMed] [Google Scholar]
- 2. König, J. , Cui, Y. , Nies, A.T. & Keppler, D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am. J. Physiol. Gastrointest. Liver Physiol. 278, G156–G164 (2000). [DOI] [PubMed] [Google Scholar]
- 3. Abe, T. et al. Identification of a novel gene family encoding human liver‐specific organic anion transporter LST‐1. J. Biol. Chem. 274, 17159–17163 (1999). [DOI] [PubMed] [Google Scholar]
- 4. König, J. , Cui, Y. , Nies, A.T. & Keppler, D. Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. J. Biol. Chem. 275, 23161–23168 (2000). [DOI] [PubMed] [Google Scholar]
- 5. Kalliokoski, A. & Niemi, M. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol. 158, 693–705 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neuvonen, P.J. , Niemi, M. & Backman, J.T. Drug interactions with lipid‐lowering drugs: mechanisms and clinical relevance. Clin. Pharmacol. Ther. 80, 565–581 (2006). [DOI] [PubMed] [Google Scholar]
- 7. Pasanen, M.K. , Neuvonen, M. , Neuvonen, P.J. & Niemi, M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet. Genomics 16, 873–879 (2006). [DOI] [PubMed] [Google Scholar]
- 8. Niemi, M. , Pasanen, M.K. & Neuvonen, P.J. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol. Rev. 63, 157–181 (2011). [DOI] [PubMed] [Google Scholar]
- 9. SEARCH Collaborative Group et al. SLCO1B1 variants and statin‐induced myopathy–a genomewide study. N. Engl. J. Med. 359, 789–799 (2008). [DOI] [PubMed] [Google Scholar]
- 10. Ramsey, L.B. et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin‐induced myopathy: 2014 update. Clin. Pharmacol. Ther. 96, 423–428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. International Transporter Consortium et al. Membrane transporters in drug development. Nat. Rev. Drug Discov. 9, 215–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu, X. et al. Clinical probes and endogenous biomarkers as substrates for transporter drug‐drug interaction evaluation: perspectives from the International Transporter Consortium. Clin. Pharmacol. Ther. 104, 836–864 (2018). [DOI] [PubMed] [Google Scholar]
- 13. Yee, S.W. et al. Metabolomic and genome‐wide association studies reveal potential endogenous biomarkers for OATP1B1. Clin. Pharmacol. Ther. 100, 524–536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Müller, F. , Sharma, A. , König, J. & Fromm, M.F. Biomarkers for in vivo assessment of transporter function. Pharmacol. Rev. 70, 246–277 (2018). [DOI] [PubMed] [Google Scholar]
- 15. Takehara, I. et al. Comparative study of the dose‐dependence of OATP1B inhibition by rifampicin using probe drugs and endogenous substrates in healthy volunteers. Pharm. Res. 35, 138 (2018). [DOI] [PubMed] [Google Scholar]
- 16. Lai, Y. et al. Coproporphyrins in plasma and urine can be appropriate clinical biomarkers to recapitulate drug‐drug interactions mediated by organic anion transporting polypeptide inhibition. J. Pharmacol. Exp. Ther. 358, 397–404 (2016). [DOI] [PubMed] [Google Scholar]
- 17. Mori, D. et al. Dose‐dependent inhibition of OATP1B by rifampicin in healthy volunteers: comprehensive evaluation of candidate biomarkers and OATP1B probe drugs. Clin. Pharmacol. Ther. 107, 1004–1013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen, H. et al. Further studies to support the use of coproporphyrin I and III as novel clinical biomarkers for evaluating the potential for organic anion transporting polypeptide 1B1 and OATP1B3 inhibition. Drug Metab. Dispos. 46, 1075–1082 (2018). [DOI] [PubMed] [Google Scholar]
- 19. Zhang, Y. et al. Detection of weak organic anion‐transporting polypeptide 1B inhibition by probenecid with plasma‐based coproporphyrin in humans. Drug Metab. Dispos. 48, 841–848 (2020). [DOI] [PubMed] [Google Scholar]
- 20. Kunze, A. , Ediage, E.N. , Dillen, L. , Monshouwer, M. & Snoeys, J. Clinical investigation of coproporphyrins as sensitive biomarkers to predict mild to strong OATP1B‐mediated drug‐drug interactions. Clin. Pharmacokinet. 57, 1559–1570 (2018). [DOI] [PubMed] [Google Scholar]
- 21. Mori, D. et al. Alteration in the plasma concentrations of endogenous organic anion‐transporting polypeptide 1B Biomarkers in patients with non‐small cell lung cancer treated with paclitaxel. Drug Metab. Dispos. 48, 387–394 (2020). [DOI] [PubMed] [Google Scholar]
- 22. Mori, D. et al. Effect of OATP1B1 genotypes on plasma concentrations of endogenous OATP1B1 substrates and drugs, and their association in healthy volunteers. Drug Metab. Pharmacokinet. 34, 78–86 (2019). [DOI] [PubMed] [Google Scholar]
- 23. Yee, S.W. et al. Organic anion transporter polypeptide 1B1 polymorphism modulates the extent of drug‐drug interaction and associated biomarker levels in healthy volunteers. Clin. Transl. Sci. 12, 388–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuki, Y. et al. Substantially increased plasma coproporphyrin‐I concentrations associated with OATP1B1*15 allele in Japanese general population. Clin. Transl, Sci. 14, 382–388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bednarczyk, D. & Boiselle, C. Organic anion transporting polypeptide (OATP)‐mediated transport of coproporphyrins I and III. Xenobiotica 46, 457–466 (2016). [DOI] [PubMed] [Google Scholar]
- 26. Gilibili, R.R. et al. Coproporphyrin‐I: a fluorescent, endogenous optimal probe substrate for ABCC2 (MRP2) suitable for vesicle‐based MRP2 inhibition assay. Drug Metab. Dispos. 45, 604–611 (2017). [DOI] [PubMed] [Google Scholar]
- 27. Chatterjee, S. et al. Transporter activity changes in nonalcoholic steatohepatitis: assessment with plasma coproporphyrin I and III. J. Pharmacol. Exp. Ther. 376, 29–39 (2021). [DOI] [PubMed] [Google Scholar]
- 28. Wolkoff, A.W. , Wolpert, E. , Pascasio, F.N. & Arias, I.M. Rotor's syndrome. A distinct inheritable pathophysiologic entity. Am. J. Med. 60, 173–179 (1976). [DOI] [PubMed] [Google Scholar]
- 29. Neuvonen, M. et al. Identification of glycochenodeoxycholate 3‐O‐glucuronide and glycodeoxycholate 3‐O‐glucuronide as highly sensitive and specific OATP1B1 biomarkers. Clin. Pharmacol. Ther. 109, 646–657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen, H. et al. Comparative evaluation of plasma bile acids, dehydroepiandrosterone sulfate, hexadecanedioate, and tetradecanedioate with coproporphyrins I and III as markers of OATP inhibition in healthy subjects. Drug Metab. Dispos. 45, 908–919 (2017). [DOI] [PubMed] [Google Scholar]
- 31. Hirvensalo, P. et al. Enantiospecific pharmacogenomics of fluvastatin. Clin. Pharmacol. Ther. 106, 668–680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramanathan, R. , King‐Ahmad, A.J. , Holliman, C.L. & Rodrigues, A.D. A highly selective and sensitive LC‐MS/HRMS assay for quantifying coproporphyrins as organic anion‐transporting peptide biomarkers. Bioanalysis 9, 1787–1806 (2017). [DOI] [PubMed] [Google Scholar]
- 33. Giacomini, K.M. et al. International Transporter Consortium commentary on clinically important transporter polymorphisms. Clin. Pharmacol. Ther. 94, 23–26 (2013). [DOI] [PubMed] [Google Scholar]
- 34. Ramsey, L.B. et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 22, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nies, A.T. et al. Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med. 5, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stephens, M. & Donnelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stephens, M. , Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pasanen, M.K. , Neuvonen, P.J. & Niemi, M. Global analysis of genetic variation in SLCO1B1. Pharmacogenomics 9, 19–33 (2008). [DOI] [PubMed] [Google Scholar]
- 39. Suzuki, Y. et al. Relationship of hemoglobin level and plasma coproporphyrin‐I concentrations as an endogenous probe for phenotyping OATP1B. Clin. Transl. Sci. 14, 1403–1411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saito, T. & Rehmsmeier, M. The precision‐recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 10, e0118432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Njumbe Ediage, E. et al. Development of an LC‐MS method to quantify coproporphyrin I and III as endogenous biomarkers for drug transporter‐mediated drug‐drug interactions. J. Chromatogr. B. Analyt. Technol. Biomed. Life. Sci. 1073, 80–89 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2


