Abstract
Background
Prostate‐specific membrane antigen (PSMA) is highly expressed in poorly differentiated, metastatic, and castration‐resistant prostate cancers. Recently, 68Ga‐PSMA positron emission tomography/computed tomography has been successfully developed as an effective diagnostic tool for prostate cancer. However, the pathophysiological functions of PSMA in prostate tumors remain unclear.
Methods
We examined the protein expression of PSMA in tumor endothelial cells in human prostate tumors by immunohistochemistry. Prostate cancer tissues were resected by robotic surgery in 2019 at Ehime University from patients with prostate cancer. In vitro, we prepared conditioned medium (CM) derived from a PSMA‐positive human prostate cancer cell line, LNCaP, cultured on collagen I gels. We then examined PSMA expression in human umbilical vascular endothelial cells (HUVECs) cultured with the CM. We assessed angiogenic activities by treatment of HUVECs with LNCaP‐derived CM using a tube formation assay that mimics angiogenesis.
Results
Immunohistochemistry of PSMA and CD31, a marker of endothelial cells, and PSMA‐expressing tumor endothelial cells were observed in 4 of 33 prostate cancer patients (12.1%). We also found that the 10,000g pellet fraction of the LNCaP‐derived CM containing PSMA‐positive membranes, such as microvesicles transformed HUVECs “PSMA‐negative” into “PSMA‐positive.” Furthermore, treatment of HUVECs with the 10,000g pellet fraction of the LNCaP‐derived CM significantly promoted tube formation, mimicking angiogenesis in a PSMA‐dependent manner.
Conclusions
Our findings revealed the existence of PSMA‐positive tumor endothelial cells in human prostate tumors, which enhances tumor angiogenesis in prostate cancer tissues.
Keywords: human umbilical vascular endothelial cells (HUVECs), LNCaP, prostate cancer specimen, prostate‐specific membrane antigen (PSMA), tube formation, tumor endothelial cells
1. INTRODUCTION
Prostate‐specific membrane antigen (PSMA) is highly expressed in poorly differentiated, metastatic, and castration‐resistant human prostate cancers. 1 , 2 , 3 In contrast, normal prostate epithelial cells express less PSMA. 4 , 5 Clinically, 68Ga‐PSMA PET/CT has been successfully established as an effective diagnostic tool for prostate cancer. 6 , 7 , 8 , 9 , 10 , 11 A Lutetium‐177 labeled‐anti‐PSMA antibody and PSMA ligands have been developed as targeted molecular radiotherapy for castration‐resistant prostate cancer patients. 12 , 13 , 14
From a molecular standpoint, PSMA is a type II transmembrane protein that possesses both glutamate carboxypeptidase II and folate hydrolase enzyme activities. 15 , 16 , 17 Although normal human endothelial cells express lower levels of PSMA, 4 , 5 tumor endothelial cells in various solid tumor tissues (e.g., thyroid cancer, glioma, breast cancer, nonsmall cell lung cancer, colorectal cancer, renal cell carcinoma) express PSMA at high levels. 18 , 19 , 20 , 21 , 22 , 23 , 24 The tumor angiogenic activities in mice lacking PSMA were drastically reduced. 18 Mechanistically, PSMA regulates angiogenesis by modulating integrin signaling. 19 In the case of glioblastoma, PSMA promotes angiogenesis by regulating the nuclear factor kappa B signaling pathway. 20 These data suggest that PSMA enhances the angiogenic activity of tumor endothelial cells in various solid tumors. However, in human prostate cancers, the pathophysiological significance of PSMA in prostate cancer tissues, including tumor angiogenesis, remains unclear.
Here, we performed immunohistochemistry of PSMA in human prostate cancer tissues and found that PSMA‐expressing tumor endothelial cells were observed in 4 of 33 prostate cancer patients (12.1%). In vitro, the 10,000g pellet fraction of conditioned medium (CM) derived from PSMA‐positive prostate cancer cell line (LNCaP cells) transformed normal endothelial cells “PSMA‐negative” into “PSMA‐positive.” Treatment of endothelial cells with the 10,000g pellet fraction of CM derived from LNCaP cells significantly promoted tube formation, mimicking angiogenesis. We also showed that the 10,000g pellet fraction of CM derived from LNCaP cells included PSMA proteins and membranes. Our results suggest that PSMA‐expressing prostate cancer cells release PSMA‐positive membranes such as microvesicles, which transform neighboring endothelial cells from PSMA‐negative into PSMA‐positive, promoting tumor angiogenesis.
2. MATERIALS AND METHODS
2.1. Antibodies
The following antibodies were purchased from the manufacturers as indicated: rabbit anti‐CD31 antibody (ab28364, dilution 1:1000; Abcam), mouse anti‐PSMA antibody (Clone 3E6, M3620, dilution 1:1000 for immunohistochemistry; Dako), donkey Alexa488‐conjugated anti‐rabbit immunoglobulin G (IgG) antibody (A21206, dilution 1:500; Life Technologies Corporation), donkey Alexa594‐conjugated anti‐mouse IgG antibody (A21203, dilution 1:500; Life Technologies Corporation), rabbit anti‐PSMA antibody (12702S, dilution 1:1000 for western blotting and immunofluorescence; Cell Signaling Technology), mouse anti‐glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) antibody (5A12, dilution 1:6000; Wako), goat Cy3‐conjugated anti‐rabbit IgG antibody (A10520, dilution 1:2000; Molecular Probes), goat Alexa488‐conjugated anti‐mouse IgG antibody (A11001, dilution 1:2000; Molecular Probes), horseradish peroxidase (HRP)‐conjugated anti‐rabbit IgG antibody (W4011, dilution 1:2000; Promega), and HRP‐conjugated anti‐mouse IgG antibody (W4021, dilution 1:2000; Promega).
2.2. Plasmids
PSMA was amplified with the Halo‐PSMA vector (FHC9160; Promega) using the following primer pairs: Myc‐PSMA‐F: ATGGAACAAAAACTCATCTCAGAAGAGGATCTGATGTGGAATCTCCTTCACGA and PSMA‐R: TTAGGCTACTTCACTCAAAG, PSMA‐F: ATGTGGAATCTCCTTCACGA and PSMA‐myc‐R: TTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGGCTACTTCACTCAAAGTCT. The PCR products were introduced into the blunt ends of the CSII‐CMV‐MCS‐IRES2‐Bsd vector (a kind gift from Dr. Hiroyuki Miyoshi, RIKEN).
2.3. Cell culture
LNCaP, PC3, and DU145 cells were purchased from ATCC and maintained at 37°C with 5% CO2 in RPMI (Wako) supplemented with 10% fetal bovine serum (FBS), 20 U/ml penicillin, and 100 µg/ml streptomycin. HEK293T cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle medium (Wako) supplemented with 10% FBS, 20 U/ml penicillin, and 100 µg/ml streptomycin. Human umbilical vascular endothelial cells (HUVECs) were purchased from Lonza. HUVECs were maintained at 37°C with 5% CO2 in EBM‐2 (Lonza) according to the manufacturer's instructions. HUVECs at passages 2–4 were used for the experiments. HUVECs cultured on collagen I gels were treated with the CM. PC3 cells that stably express Myc‐PSMA or PSMA‐myc were established by selection with 50 µg/ml blasticidin S (Wako) after infection with lentivirus carrying the Myc‐PSMA or PSMA‐myc gene.
2.4. Transfection
For transfection of plasmids into HEK293T cells, GeneJuice (Millipore) was used according to the manufacturer's instructions. At 48 h posttransfection, the cells were subjected to subsequent experiments. Transfection of prostate cancer cells with small interfering RNAs (siRNAs) (10 nM) was performed using RNAimax (Invitrogen) according to the manufacturer's instructions. Subsequent experiments were performed 72 h posttransfection. The following validated siRNA duplex oligomers were purchased and used for knockdown experiments: GGGCGAUCUAGUGUAUGUUAACUAU (siPSMA; Invitrogen). Control siRNA was purchased from Sigma‐Aldrich (SIC‐001).
2.5. Lentiviral expression
Lentiviruses carrying Myc‐PSMA or PSMA‐myc were generated as described previously (Watanabe, 2020). The CSII‐CMV‐MCS‐IRES2‐Bsd, pCAG‐HIVgp, and pCMV‐VSVG‐RSV‐Rev vectors were kind gifts from Dr. Hiroyuki Miyoshi (RIKEN).
2.6. Western blotting
Western blotting was performed as described previously. 21
2.7. RT‐PCR
Real‐time PCR was performed as previously described. 22
The following pairs of primers were used:
5′‐CAGCTGGAAATATCCTAAATCTGA‐3′ (PSMA sense primer), 5′‐TTGGATGAACAGGAATACTTGGAA‐3′ (PSMA antisense primer), 5′‐TGCACCACCAACTGCTTAGC‐3′ (GAPDH sense primer) and 5′‐GGCATGGACTGTGGTCATGAG‐3′ (GAPDH antisense primer).
2.8. Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 30 min at room temperature and permeabilized with 0.1% Triton X‐100 in PBS for 15 min at room temperature. For staining of endogenous PSMA, cells were fixed with 10% trichloroacetic acid in PBS for 15 min at 4°C and permeabilized with 0.05% saponin in PBS for 5 min at room temperature. After blocking with 3% bovine serum albumin in PBS for 30 min at room temperature, the cells were incubated with primary antibodies and then with fluorophore‐conjugated secondary antibodies. To stain the nuclei, fixed cells were treated with Hoechst 33342 (dilution 1:2000; Molecular Probes) at room temperature for 1 h.
2.9. Confocal microscopy
Confocal microscopy was performed using the A1R laser confocal microscope (Nikon) with a ×60 1.27 Plan‐Apochromat water immersion lens. Images were analyzed using the ImageJ or FIJI software (NIH).
2.10. Preparation of CM
The 1.5 × 105 cells/ml of prostate cancer cells were seeded on 6‐well plates coated with 500 µl of collagen I gel in each well. Three days later, the culture medium was removed and replaced with 2 ml of fresh RPMI medium in each well. The cells were incubated for 3 days, after which the media were collected and centrifuged at 1000g for 5 min. The supernatants were saved and defined as the conditioned media. For the concentration of a 10,000g pellet fraction of CM, 10 ml of CM was centrifuged at 10,000g, and the supernatants were removed. The pellet was resuspended in 1 ml of EBM‐2.
2.11. Tube formation assay on collagen I gels
Tube formation assays were performed as previously described. 23 Tube length was measured using ImageJ or FIJI (NIH).
2.12. Centrifugation
The CM was centrifuged at 1000g for 10 min at 4°C to pellet the debris. The supernatant media was subjected to centrifugation at 10,000g for 10 min at 4°C, and the pellets were designated as 10,000g pellets. The supernatant was transferred to an ultracentrifuge tube for further centrifugation at 100,000g for 60 min at 4°C (Optima XL80K; Beckman Coulter Inc.), and the pellets were defined as 100,000g pellets. The collected supernatant was passed through 0.35 µm filter to remove the larger protein aggregates or vesicles.
2.13. Immunohistochemistry
Immunohistochemistry was performed on 10% neutral buffered formalin‐fiixed and paraffin‐embedded tissue samples, which were cut on a microtome (3–5 µm thick) and stained according to standard protocols. The antibodies used in these studies were a mouse monoclonal to PSMA (M3620; Dako) and a rabbit polyclonal against CD31 (ab28364; Abcam). Human prostate tumor samples were prepared from 10% neutral buffered formalin‐fiixed surgical samples. All samples were judged by pathologists of Ehime University. Sampling was approved by the local institutional review board and ethics committee of Ehime University Hospital (Approval No. 1812008).
2.14. Membrane labeling
The membrane labeling assay was performed using a PKH26 Red Fluorescent Cell Linker Mini Kit for General Cell Membrane Labeling (MINI26‐1 kit; Sigma‐Aldrich) according to the manufacturer's instructions. Briefly, 10,000g of the pellet was suspended in 1 ml Diluent C and mixed with 2× dye Solution; 1 ml of diluent with 4 µl of the PKH26 dye solution. The pellets were then washed with PBS and centrifuged three times at 10,000g. Pellets (10,000g) were suspended in 1 ml of EBM‐2.
2.15. Statistical analysis
Statistical comparisons were made using the two‐tailed the Student t test or one‐way analysis of variance, followed by Tukey's post hoc test.
3. RESULTS
3.1. PSMA is expressed in tumor endothelial cells as well as prostate epithelial cells in human prostate tumors
We performed immunohistochemical evaluations of the expression of PSMA protein in the tumor endothelial cells of human primary prostate cancer tissues that were surgically resected at Ehime University Hospital by immunohistochemistry. Unexpectedly, we found that CD31‐positive tumor endothelial cells showed slight expression of PSMA in 4 of 33 cases (12.1%), whereas all prostate cancer cells highly expressed PSMA (Figure 1A and Table 1). Since normal endothelial cells in normal prostate tissues do not express PSMA (Figure 1B), 5 these data suggest that PSMA‐positive prostate cancer cells may possess the ability to transform PSMA‐negative endothelial cells into PSMA‐positive endothelial cells.
Figure 1.
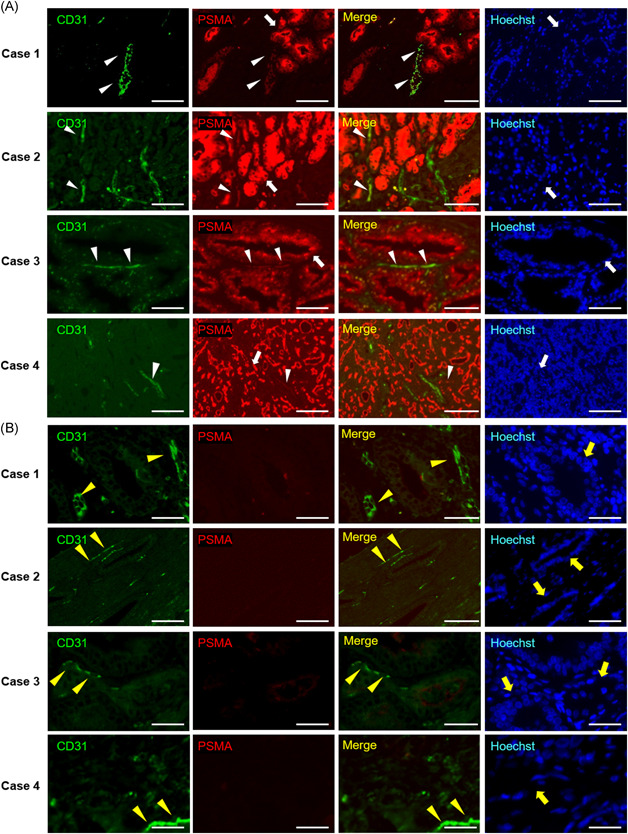
Expression of PSMA protein in human prostate cancer tissues and adjacent benign prostate tissues (n = 33). Representative images of immunohistochemical staining for PSMA and CD31 (a marker of endothelial cells) in human prostate cancers (A) and their adjacent benign prostate tissues (B) are shown. The neighboring sections of human prostate cancer tissues were subjected to fluorescence immunohistochemistry for PSMA (red) and CD31 (green). The PSMA‐positive tumor endothelial cells are indicated by arrowheads. White arrows indicate PSMA‐positive prostate cancer cells. The PSMA‐negative normal endothelial cells are indicated by yellow arrowheads. Yellow arrows indicate PSMA‐negative normal prostate cells. Bars: 100 µm. See also Table 1. Case 1. 67‐year‐old male PSA 9.65 ng/ml, Gleason score 4 + 5, pT2c EPE0, RM0, ly1, v0, n0, and sv0. Case 2. 68‐year‐old male PSA 5.5 ng/ml, Gleason score 4 + 5, pT2c EPE0, RM1, ly1, v0, n1, sv0. Case 3. 69‐year‐old male PSA 11.49 ng/ml, Gleason score 4 + 5, pT2a EPE0, RM0, ly1, v0, n1, sv0. Case 4. 68‐year‐old male PSA 13.4 ng/ml, Gleason score 4 + 4, pT3a EPE1, RM0, ly1, v1, n1, and sv0. PSMA, prostate‐specific membrane antigen [Color figure can be viewed at wileyonlinelibrary.com]
Table 1.
Clinical information of human prostate cancer tissues containing PSMA‐positive or PSMA‐negative tumor endothelial cells
| Prostate tumors with PSMA‐positive tumor endothelial cells (n = 4) | Prostate tumors with PSMA‐negative tumor endothelial cells (n = 29) | |
|---|---|---|
| Median age, years (range) | 68.25 (67–69) | 69.66 (51–80) |
| Gleason score | ||
| ≥8 | 4 | 10 |
| <8 | 0 | 19 |
| Median preoperative PSA, ng/ml (range) | 11.49 (5.5–13.4) | 6.346 (3.8–24.0) |
| EPE+ | 1 (25%) | 5 (17.24%) |
| RM+ | 1 (25%) | 5 (17.24%) |
| ly+ | 4 (100%) | 14 (48.27%) |
| v+ | 1 (25%) | 0 (0%) |
| n+ | 4 (100%) | 21 (72.41%) |
| sv+ | 0 (0%) | 3 (10.34%) |
Note: A total of 33 prostate cancer specimens were resected in robotic surgeries at Ehime University Hospital. The number of PSMA‐positive or PSMA‐negative tumor endothelial cells neighboring human prostate epithelial cancer cells in prostate tumor tissues were counted. We defined CD31‐positive endothelial cells as being PSMA‐positive when PSMA staining signals exceeded the threshold of 80
Abbreviations: EPE, extraprostatic extension; ly, lymph invasion; n, lymph node metastasis; PSA, prostate specific antigen; PSMA, prostate‐specific membrane antigen; RM, resection margin; sv, seminal vesicle invasion; v, venous invasion.
3.2. CM derived from LNCaP cells induce PSMA expression in HUVECs
Since tumor endothelial cells neighboring PSMA‐positive prostate cancers express PSMA (Figure 1A and Table 1), we speculated that PSMA may be able to transform PSMA‐negative endothelial cells into PSMA‐positive tumor endothelial cells. To address these mechanisms, we produced CM derived from various human prostate cancer cell lines (PC3, DU145, and LNCaP cells) cultured on plastic dishes (condition 1) or collagen I gels (condition 2) (Figure 2A). We found that, among the prostate cancer cells tested here, LNCaP cells expressed the PSMA protein (Figure 2A). LNCaP cells cultured on collagen I gels formed spheres (Figure 2A). HUVECs seeded on collagen I gels were then cultured with CM obtained under conditions 1 or 2. As shown in Figure 2B, HUVECs cultured with CM derived from LNCaP cells in condition 2 expressed PSMA protein, although control HUVECs cultured with EBM‐2 medium showed undetectable levels of PSMA protein. CM derived from LNCaP cells in condition 1 had a relatively weaker ability to induce PSMA expression in HUVECs compared to CM derived from LNCaP cells in condition 2 (Figure 2B). We then prepared and used CM derived from prostate cancer cells in condition 2. In contrast to the CM derived from LNCaP cells, PSMA protein was not detected in HUVECs cultured with CM derived from PC3 or DU145 in condition 2 (Figure 2C). Biochemically, we confirmed that CM derived from LNCaP cells, but not PC3 or DU145 cells, in condition 2 induced the protein expression of PSMA in HUVECs (Figure 2D). The messenger RNA (mRNA) expression of PSMA was also significantly induced by treatment of HUVECs with CM derived from LNCaP cells in condition 2 (Figure 2E). These data suggest that a PSMA‐positive prostate cancer cell line, LNCaP, releases factors in its CM that can transform PSMA‐negative HUVECs into PSMA‐positive HUVECs.
Figure 2.

PSMA was detected in HUVECs cultured with the conditioned medium derived from LNCaP cells. (A) Confocal images of prostate cancer cells (PC3, DU145, LNCaP cells) cultured in condition 1 or condition 2. The scheme of each condition by which each conditioned medium (CM) was prepared is shown on the right. To prepare the CM, 1.5 × 105 cells of prostate cancer cells were seeded on 6‐well plastic dishes (condition 1) or collagen I gels (condition 2). Three days later, the media were replaced with fresh media. The cells were then incubated for another 3 days, then the media were collected as CM. Bars: 100 µm. (B, C) Confocal images of HUVECs cultured with the CM derived from LNCaP cells (B) or PC3 and DU145 cells (C). The CM was diluted to half of its concentration with EBM‐2. The resulting solution was added to the HUVECs seeded on the collagen I gels. Seventy‐two hours later, cells were subjected to immunofluorescence staining for PSMA. Bars: 100 µm. (D) Western blots of HUVEC lysates cultured with the CM derived from prostate cancer cells in condition 2 for 72 h. The CM was diluted to half of its concentration with EBM‐2. The resulting solution was added to the HUVECs seeded on the collagen I gels. The lysates from LNCaP cells were used as a positive control of PSMA expression. (E) The mRNA expression of PSMA in HUVECs cultured with the CM derived from LNCaP cells in condition 2 for 72 h. The CM was diluted to half of its concentration with EBM‐2. The resulting solution was added to the HUVECs seeded on the collagen I gels. Data are mean ± SEM from three independent experiments. **p < .01. HUVEC, human umbilical vascular endothelial cell; mRNA, messenger RNA; PSMA, prostate‐specific membrane antigen [Color figure can be viewed at wileyonlinelibrary.com]
A previous study has shown that CM derived from PSMA‐positive prostate cancer cell lines (LNCaP or MDA‐PCa‐2b cells) cultured on plastic bottom dishes rarely induced PSMA expression in HUVECs. 24 In contrast, CM derived from LNCaP cells cultured on collagen I gels induced PSMA expression in HUVECs cultured on collagen I gel (Figure 2A,B). Collectively, collagen I gels are critical for LNCaP cells to release factors that transform PSMA‐negative HUVECs into PSMA‐positive HUVECs.
3.3. The 10,000g pellet fractions of CM derived from LNCaP cells can transform PSMA‐negative HUVECs into PSMA‐positive HUVECs
We then sought to identify the fractions in the LNCaP‐derived CM from LNCaP cells, which can transform PSMA‐negative HUVECs into PSMA‐positive HUVECs. We fractionated the CM from LNCaP cells into 10,000g pellets, 100,000g pellets, and 100,000g supernatant by centrifugation. Half of the 100,000g supernatant was filtered to remove large microsomes. HUVECs were then cultured with the CM fractions from LNCaP cells. As shown in Figure 3A, HUVECs cultured with the 10,000g pellet fraction expressed the PSMA protein in their cytosol. The other fractions did not induce the same transformation of HUVECs from PSMA‐negative into PSMA‐positive (Figure 3A). Since the 10,000g pellet fraction includes comparatively large vesicles such as microvesicles, 25 , 26 , 27 , 28 it is likely that microvesicles released from LNCaP cells may be able to induce this transformation. To examine the possibility that cell membranes derived from LNCaP cells were endocytosed into or fused to HUVECs, we labeled the 10,000g pellet fraction of the CM derived from LNCaP cells with a membrane dye. This was then added to the medium containing HUVECs. We found that the membrane dye partially colocalized with PSMA in HUVECs (Figure 3B). The 10,000g pellet fraction of CM derived from LNCaP cells included PSMA proteins (Figure 3C). The CMs derived from PC3 cells stably overexpressing Myc‐PSMA or PSMA‐myc transformed PSMA‐negative HUVECs into PSMA‐positive HUVECs (Figure 3D). Taken together, these data suggest that PSMA‐positive membranes released from PSMA‐expressing prostate cancer cells could transform HUVECs from a PSMA‐negative to a PSMA‐positive state.
Figure 3.

Fractionation of the CM derived from LNCaP cells. (A) Confocal images of HUVECs cultured with each fraction of CM derived from LNCaP cells. Each fraction was diluted to half of its concentration with EBM‐2 or suspended with EBM‐2, and added to the HUVECs seeded on the collagen I gels. Seventy‐two hours later, cells were subjected to immunofluorescence staining for PSMA. The PSMA‐positive HUVECs were shown by arrowheads. Bars: 100 µm. The representative images from three independent experiments were shown. (B) Confocal images of HUVECs cultured with 10,000g pellet fractions of CM derived from LNCaP cells. The 10,000g pellet fraction was labeled with a membrane marker dye, before the suspension in EBM‐2 medium. The labeled 10,000g pellet was cultured with HUVECs for 6 h, and cells were subjected to immunofluorescence staining for PSMA. Bars: 10 µm (left) and 2 µm (right; magnified images of squares in left images). The representative images from three independent experiments were shown. (C) Western blots of 10,000g pellet fraction of CM derived from LNCaP cells. The lysates from HUVECs and LNCaP cells were used as negative and positive controls of PSMA expression, respectively. The representative blot data from three independent experiments were shown. (D) Confocal images of HUVECs cultured with the CM derived from PC3 cells that stably express Myc‐PSMA or PSMA‐myc. The CM was diluted in half with EBM‐2, and added to the HUVECs seeded on the collagen I gels. Seventy‐two hours later, cells were subjected to immunofluorescence staining for PSMA. Bars: 100 µm. The representative images from three independent experiments were shown. CM, conditioned medium; HUVEC, human umbilical vascular endothelial cell; mRNA, messenger RNA; PSMA, prostate‐specific membrane antigen [Color figure can be viewed at wileyonlinelibrary.com]
3.4. The 10,000g pellet fractions of CM derived from PSMA‐expressing prostate cancer cells promote angiogenesis in vitro
Finally, we examined the effects of the 10,000g pellet fraction of CM derived from PSMA‐expressing prostate cancer cells on endothelial function. To this end, we performed a tube formation assay of HUVECs to mimic angiogenesis. The 10,000g pellet fraction of CM derived from LNCaP cells significantly enhanced tube formation in HUVECs (Figure 4A,B). PSMA knockdown significantly suppressed the increase in tube length of HUVECs (Figure 4A–C). We also prepared a 10,000g pellet fraction of the CM derived from PC3 cells that exogenously overexpressed PSMA (Figure 4D). The 10,000g pellet fraction of the PSMA‐expressing PC3 cell‐derived CM significantly promoted tube formation in HUVECs compared to that derived from parental PC3 cells, which do not endogenously express PSMA (Figure 4E,F). These data suggest that PSMA‐positive membranes released from PSMA‐expressing prostate cancer cells enhance angiogenesis in vitro.
Figure 4.

The tube formation assay of HUVECs cultured with 10,000g pellet fraction of CM derived from prostate cancer cells. (A) Representative images of tube formation. HUVECs seeded on collagen I gel were treated with the 10,000g pellet fraction of CM derived from LNCaP cells for 6 h, and packed on collagen I followed by VEGF‐A stimulation for 66 h. HUVECs were stained with Calcein‐AM before acquisition of images. To examine the PSMA dependency, the CM was prepared from LNCaP cells depleted of PSMA. Bars: 100 µm. (B) The quantitation of (A). Total tube lengths from three independent experiments were measured and normalized to those of cells cultured with normal EBM‐2. Data are the means ± SEM. *p < .05; n.s., not significant. (C) Western blots of LNCaP cell lysates, 72 h posttransfection with the indicated siRNAs. (D) Confocal images of PC3 cells stably expressing Myc‐PSMA or PSMA‐myc. Bars: 100 µm. (E) Representative images of tube formation. HUVECs seeded on collagen I gel were treated with the 10,000g pellet fraction of CM derived from PC3 cells for 6 h, and packed in collagen I followed by VEGF‐A stimulation for 66 h. HUVECs were stained with Calcein‐AM before acquisition of images. Bars: 100 µm. (F) The quantitation of (E). Total tube lengths from three independent experiments were measured and normalized to those of PC3 (parental). Data are the means ± SEM. *p < .05; **p < .01. CM, conditioned medium; HUVEC, human umbilical vascular endothelial cell; mRNA, messenger RNA; PSMA, prostate‐specific membrane antigen [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
PSMA is highly expressed in prostate cancer tissues, and its expression increases with tumor aggressiveness, metastatic disease, and disease recurrence. 1 , 2 , 3 PSMA is also expressed in several normal tissues such as the salivary glands, kidney, epididymis, ovary, ileum‐jejunum, and in astrocytes, while normal prostate tissues express less PSMA. 4 , 5 Recently, it has been reported that tumor endothelial cells of various solid malignant cancer tissues express PSMA, although normal endothelial cells do not express PSMA. 29 , 30 , 31 , 32 , 33 , 34 , 35 In the case of prostate tumors, a previous study showed that tumor endothelial cells neighboring human prostate cancer cells did not express PSMA. 5 Our present results indicate that, although the percentage of prostate cancer tumors containing PSMA‐positive tumor endothelial cells is low (12.1%), PSMA‐positive tumor endothelial cells exist in human prostate tumors (Figure 1A and Table 1). One remaining issue is the low PSMA positivity in tumor endothelial cells in prostate tumors in which prostate tumor epithelial cells highly express PSMA. We found that, in vitro, the tissue culture of both LNCaP cells (PSMA‐positive) and HUVECs (PSMA‐negative) on collagen I gels is critical to transform PSMA‐negative HUVECs into PSMA‐positive HUVECs (Figure 2B) as well as the promotion of angiogenesis (Figure 4A). The CM derived from LNCaP cells cultured on plastic dishes did not induce tube formation of HUVECs, 36 suggesting the necessity of collagen I gels to release angiogenic factors in 10,000g pellet fractions (such as microvesicles) from LNCaP cells. It is likely that the potency of PSMA‐positive prostate cancer cells to transform PSMA‐negative endothelial cells into PSMA‐positive endothelial cells could be influenced by their environments (e.g., extracellular matrix) in prostate tumors in vivo.
In this study, we found that the 10,000g pellet fraction of CM derived from LNCaP cells transformed PSMA‐negative endothelial cells to PSMA‐positive endothelial cells. We also confirmed that PSMA‐positive HUVECs promoted tube formation compared with PSMA‐negative HUVECs. Immunohistochemistry revealed that the tumor endothelial cells neighboring the epithelial prostate cancer cells in the resected specimen expressed PSMA. It is likely that PSMA‐positive microvesicles are endocytosed into or fused to endothelial cells in prostate tumors (Figure 5). Immunohistochemistry showed that PSMA expression in tumor endothelial cells was detected in some, but not all cases of prostate cancer. There were no correlations between the presence of PSMA‐positive tumor endothelial cells and malignancy indices such as Gleason score, serum PSA level, vascular invasion, or metastatic state. The clinical significance of the presence of PSMA‐positive tumor endothelial cells should be investigated in the future.
Figure 5.
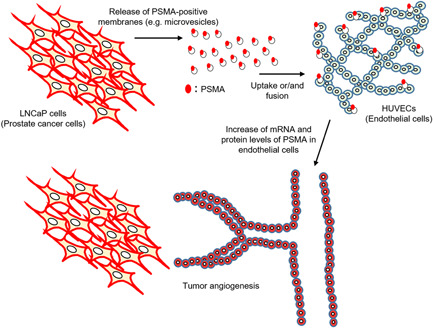
Scheme of this study. PSMA‐positive prostate cancer cells release PSMA‐positive membranes, such as microvesicles. The PSMA‐positive membranes are endocytosed in or fused to endothelial cells, followed by transformation into “PSMA‐positive” endothelial cells. PSMA expression in endothelial cells promotes angiogenesis PSMA‐expressing epithelial cells in human prostate tumors may contribute to active tumor angiogenesis through the transformation of PSMA‐negative endothelial cells into PSMA‐positive tumor endothelial cells in human prostate cancer tissues. PSMA, prostate‐specific membrane antigen [Color figure can be viewed at wileyonlinelibrary.com]
Treatment of 10,000g pellet fractions of the CM derived from LNCaP cells, which endogenously express PSMA, not only transformed PSMA‐negative HUVECs into PSMA‐positive HUVECs but also induced the mRNA expression of PSMA in HUVECs. PSMA‐positive membranes released from LNCaP cells, such as microvesicles, can induce transcription of PSMA. A previous study showed that a low molecular weight fraction (10–50 kDa) in CM prepared from SK‐RC‐13 (a renal cell carcinoma cell line), HCT‐15 (a colorectal cancer cell line) or MDA‐MB‐231 cell (a breast cancer cell line) induces PSMA expression in HUVECs. 24 The microvesicles released from LNCaP cells cultured on collagen I gels may include components in the low molecular weight fraction (10–50 kDa). Further analysis is needed to investigate the characteristics of the 10,000g pellet fraction (e.g., microvesicles) CM derived from LNCaP cells.
The molecular mechanisms underlying the release of LNCaP cell‐derived PSMA‐positive membranes from LNCaP cells and their uptake into HUVECs are still unknown. The signaling pathway through PSMA‐positive membranes, which induce angiogenesis of endothelial cells, has not yet been dissolved. The endocytic or fusion process of the PSMA‐positive membranes in normal endothelial cells may be attractive targets for the development of new antiangiogenic drugs, which could inhibit the transformation of normal endothelial cells into PSMA‐positive tumor endothelial cells.
5. CONCLUSIONS
We conclude that PSMA‐positive membranes released from PSMA‐expressing prostate cancer cells transform PSMA‐negative endothelial cells into PSMA‐positive endothelial cells. PSMA‐endothelial cells acquire high angiogenic activity. PSMA‐expressing epithelial prostate cancer cells in human prostate tumors may contribute to tumor angiogenesis through the transformation of PSMA‐negative endothelial cells into PSMA‐positive tumor endothelial cells in human prostate cancer tissues.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Ryuta Watanabe: designed and performed the experiments, analyzed the data, interpreted the results, and wrote the paper. Masashi Maekawa: designed and performed the experiments, analyzed the data, interpreted the results, and wrote the paper. Mie Kurata: validated the immunohistochemistry as a pathologist. Takeshi Kiyoi: performed immunohistochemistry. Noriyoshi Miura: designed the experiments and interpreted the results. Tadahiko Kikugawa: designed the experiments and interpreted the results. Shigeki Higashiyama: designed the experiments, interpreted the results and wrote the paper. Takashi Saika: designed the experiments, interpreted the results and wrote the paper.
ACKNOWLEDGMENTS
We thank Ms. Mami Chosei, Dr. Tomohisa Sakaue, and Ms. Izumi Tanimoto (Ehime University) for providing technical assistance. This work was supported by: JSPS KAKENHI Grant Number 19K18613 to RW, JSPS KAKENHI Grant Number 17K11142 to TK, JSPS KAKENHI Grant Number 18K09137 to NM.
Watanabe R, Maekawa M, Kiyoi T, et al. PSMA‐positive membranes secreted from prostate cancer cells have potency to transform vascular endothelial cells into an angiogenic state. The Prostate. 2021;81:1390‐1401. 10.1002/pros.24237
Ryuta Watanabe and Masashi Maekawa equally contributed to this work.
Contributor Information
Ryuta Watanabe, Email: watanabe.ryuta.gk@ehime-u.ac.jp.
Takashi Saika, Email: saika.takashi.ol@ehime-u.ac.jp.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Heston WDW. Characterization and glutamyl preferring carboxypeptidase function of prostate specific membrane antigen: a novel folate hydrolase. Urology. 1997;49(3A Suppl):104‐112. [DOI] [PubMed] [Google Scholar]
- 2. Israeli RS, Powell CT, Fair WR, Heston WDW. Molecular cloning of a complementary DNA encoding a prostate‐specific membrane antigen. Cancer Res. 1993;53(2):227‐230. [PubMed] [Google Scholar]
- 3. Pinto JT, Suffoletto BP, Berzin TM, et al. Prostate‐specific membrane antigen: a novel folate hydrolase in human prostatic carcinoma cells. Clin Cancer Res. 1996;2(9):1445‐1451. [PubMed] [Google Scholar]
- 4. Perner S, Hofer MD, Kim R, et al. Prostate‐specific membrane antigen expression as a predictor of prostate cancer progression. Hum Pathol. 2007;38(5):696‐701. [DOI] [PubMed] [Google Scholar]
- 5. Silver DA, Pellicer I, Fair WR, Heston WD, Cordon‐Cardo C. Prostate‐specific membrane antigen expression in normal and malignant human tissues. Clin Cancer Res. 1997;3(1):81‐85. [PubMed] [Google Scholar]
- 6. Hijazi S, Meller B, Leitsmann C, et al. Pelvic lymph node dissection for nodal oligometastatic prostate cancer detected by 68Ga‐PSMA‐positron emission tomography/computerized tomography. Prostate. 2015;75(16):1934‐1940. [DOI] [PubMed] [Google Scholar]
- 7. Ceci F, Uprimny C, Nilica B, et al. 68Ga‐PSMA PET/CT for restaging recurrent prostate cancer: which factors are associated with PET/CT detection rate? Eur J Nucl Med Mol Imaging. 2015;42(8):1284‐1294. [DOI] [PubMed] [Google Scholar]
- 8. Morigi JJ, Stricker PD, van Leeuwen PJ, et al. Prospective comparison of 18F‐fluoromethylcholine versus 68Ga‐PSMA PET/CT in prostate cancer patients who have rising PSA after curative treatment and are being considered for targeted therapy. J Nucl Med. 2015;56(8):1185‐1190. [DOI] [PubMed] [Google Scholar]
- 9. Eiber M, Maurer T, Souvatzoglou M, et al. Evaluation of hybrid 68Ga‐PSMA ligand PET/CT in 248 patients with biochemical recurrence after radical prostatectomy. J Nucl Med. 2015;56(5):668‐674. [DOI] [PubMed] [Google Scholar]
- 10. Afshar‐Oromieh A, Avtzi E, Giesel FL, et al. The diagnostic value of PET/CT imaging with the 68Ga‐labelled PSMA ligand HBED‐CC in the diagnosis of recurrent prostate cancer. Eur J Nucl Med Mol Imaging. 2015;42(2):197‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Afshar‐Oromieh A, Malcher A, Eder M, et al. PET imaging with a [68Ga]gallium‐labelled PSMA ligand for the diagnosis of prostate cancer: biodistribution in humans and first evaluation of tumour lesions. Eur J Nucl Med Mol Imaging. 2013;40(4):486‐495. [DOI] [PubMed] [Google Scholar]
- 12. Baum RP, Kulkarni HR, Schuchardt C, et al. 177Lu‐Labeled prostate‐specific membrane antigen radioligand therapy of metastatic castration‐resistant prostate cancer: safety and efficacy. J Nucl Med. 2016;57(7):1006‐1013. [DOI] [PubMed] [Google Scholar]
- 13. Zechmann CM, Afshar‐Oromieh A, Armor T, et al. Radiation dosimetry and first therapy results with a (124)I/(131)I‐labeled small molecule (MIP‐1095) targeting PSMA for prostate cancer therapy. Eur J Nucl Med Mol Imaging. 2014;41(7):1280‐1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kratochwil C, Giesel FL, Stefanova M, et al. PSMA‐targeted radionuclide therapy of metastatic castration‐resistant prostate cancer with 177Lu‐labeled PSMA‐617. J Nucl Med. 2016;57(8):1170‐1176. [DOI] [PubMed] [Google Scholar]
- 15. Zhu ZY, Zhong CP, Xu WF, et al. PSMA mimotope isolated from phage displayed peptide library can induce PSMA specific immune response. Cell Res. 1999;9(4):271‐280. [DOI] [PubMed] [Google Scholar]
- 16. Aggarwal S, Singh P, Topaloglu O, Isaacs JT, Denmeade SR. A dimeric peptide that binds selectively to prostate‐specific membrane antigen and inhibits its enzymatic activity. Cancer Res. 2006;66(18):9171‐9177. [DOI] [PubMed] [Google Scholar]
- 17. Shen D, Xie F, Edwards WB. Evaluation of phage display discovered peptides as ligands for prostate‐specific membrane antigen (PSMA). PLoS One. 2013;8(7):e68339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caromile LA, Dortche K, Rahman MM, et al. PSMA redirects cell survival signaling from the MAPK to the PI3K‐AKT pathways to promote the progression of prostate cancer. Sci Signal. 2017;10(470):eaag3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Conway RE, Petrovic N, Li Z, Heston W, Wu D, Shapiro LH. Prostate‐specific membrane antigen regulates angiogenesis by modulating integrin signal transduction. Mol Cell Biol. 2006;26(14):5310‐5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao Y, Zheng H, Li L, et al. Prostate‐specific membrane antigen (PSMA) promotes angiogenesis of glioblastoma through interacting with ITGB4 and regulating NF‐kappaB signaling pathway. Front Cell Dev Biol. 2021;9:598377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maekawa M, Tanigawa K, Sakaue T, et al. Cullin‐3 and its adaptor protein ANKFY1 determine the surface level of integrin beta1 in endothelial cells. Biol Open. 2017;6(11):1707‐1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tezuka‐Kagajo M, Maekawa M, Ogawa A, et al. Development of human CBF1‐targeting single‐stranded DNA aptamers with antiangiogenic activity in vitro. Nucleic Acid Ther. 2020;30(6):365‐378. [DOI] [PubMed] [Google Scholar]
- 23. Tanigawa K, Maekawa M, Kiyoi T, et al. SNX9 determines the surface levels of integrin beta1 in vascular endothelial cells: implication in poor prognosis of human colorectal cancers overexpressing SNX9. J Cell Physiol. 2019;234(10):17280‐17294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen DP, Xiong PL, Liu H, et al. Induction of PSMA and internalization of an anti‐PSMA mAb in the vascular compartment. Mol Cancer Res. 2016;14(11):1045‐1053. [DOI] [PubMed] [Google Scholar]
- 25. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 26. Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2.1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raposo G, Nijman HW, Stoorvogel W, et al. Affiliations expand B lymphocytes secrete antigen‐presenting vesicles. J Exp Med. 1996;1 183(3):1161‐1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szatanek R, Baran J, Siedlar M, Baj‐Krzyworzeka M. Isolation of extracellular vesicles: determining the correct approach (Review). Int J Mol Med. 2015;36(1):11‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bychkov A, Vutrapongwatana U, Tepmongkol S, Keelawat S. PSMA expression by microvasculature of thyroid tumors—Potential implications for PSMA theranostics. Sci Rep. 2017;7(1):5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nomura N, Pastorino S, Jiang P, et al. Prostate specific membrane antigen (PSMA) expression in primary gliomas and breast cancer brain metastases. Cancer Cell Int. 2014;14(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kasoha M, Unger C, Solomayer EF, et al. Prostate‐specific membrane antigen (PSMA) expression in breast cancer and its metastases. Clin Exp Metastasis. 2017;34(8):479‐490. [DOI] [PubMed] [Google Scholar]
- 32. Schmidt LH, Heitkötter B, Schulze AB, et al. Prostate specific membrane antigen (PSMA) expression in non‐small cell lung cancer. PLoS One. 2017;12(10):e0186280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wernicke AG, Edgar MA, Lavi E, et al. Prostate‐specific membrane antigen as a potential novel vascular target for treatment of glioblastoma multiforme. Arch Pathol Lab Med. 2011;135(11):1486‐1489. [DOI] [PubMed] [Google Scholar]
- 34. Milowsky MI, Nanus DM, Kostakoglu L, et al. Vascular targeted therapy with anti‐prostate‐specific membrane antigen monoclonal antibody J591 in advanced solid tumors. J Clin Oncol. 2007;25(5):540‐547. [DOI] [PubMed] [Google Scholar]
- 35. Chang SS, Reuter VE, Heston WD, Bander NH, Grauer LS, Gaudin PB. Five different anti‐prostate‐specific membrane antigen (PSMA) antibodies confirm PSMA expression in tumor‐associated neovasculature. Cancer Res. 1999; 59(13):3192‐3198. [PubMed] [Google Scholar]
- 36. Liu T, Jabbes M, Nedrow‐Byers JR, Wu LY, Bryan JN, Berkman CE. Detection of prostate‐specific membrane antigen on HUVECs in response to breast tumor‐conditioned medium. Int J Oncol. 2011;38(5):1349‐1355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


