Abstract
Purpose: To assess colony morphology of Staphylococcus aureus isolates for target shape (T1) and its utility in the identification of methicillin-resistant S. aureus (MRSA).
Methods: Staphylococcus species isolated from blood cultures were studied for colony morphology characteristics. A polymerase chain reaction (PCR) test was performed on positive blood culture bottles for the detection of S. aureus and methicillin resistance. Colony morphology was read at 24 and 48 hours and defined as follows: target shaped (T1) - an elevated colony center encircled by a pale zone, which is surrounded by a single ring of peripheral enhancement giving a ‘target’ appearance; dome-shaped (T2) with an elevated center lacking the ‘target’ appearance.
Results: At 48 hours, 73.7% of MRSA and 59.5% of coagulase-negative staphylococci (CoNS) showed T1 morphology. T1 morphology has a sensitivity of 73.68% and specificity of 93.55% amongst S. aureus for identification of methicillin resistance and a high positive predictive value (95.45%) at 48 hours.
Conclusion: T1 morphology has a modest sensitivity with specificity and positive predictive value amongst S. aureus for identification of methicillin resistance at 48 hours. It can be potentially used for the identification of MRSA, especially in resource-limited settings and wherein a molecular test is not repeated if PCR testing has already identified methicillin-sensitive S. aureus (MSSA) on a recent specimen on the same patient.
Keywords: mrsa, target shaped, blood agar, colony morphology, methicillin resistant staphylococcus aureus, staphylococcus aureus
Introduction
Staphylococcus aureus is a gram-positive coccus that can colonize nares and is an opportunistic pathogen leading to diseases including pneumonia, endocarditis, and bacteremia [1]. The colonies show characteristic yellow, round, large (1-3 mm), convex, and opaque colonies with beta hemolysis. Based on oxacillin resistance they are characterized as methicillin-resistant S. aureus (MRSA) or methicillin-sensitive S. aureus (MSSA). MRSA can produce non-hemolytic and white colonies, which resemble coagulase-negative staphylococci (CoNS). MRSA is a multi-drug resistant pathogen with resistance to penicillins and cephalosporins [2]. In this study, we reviewed S. aureus isolates for a novel colony morphology characteristic, which can aid in the identification of MRSA.
The article was presented as a poster at World Microbe Forum, 20-24 June 2021.
Materials and methods
This is a prospective study in which Staphylococcus species isolated from blood cultures were included. No identifiable information was collected during the study period and consent was waived by the institutional review board of the University of Texas Health, San Antonio (UTHSA), Texas, United States.
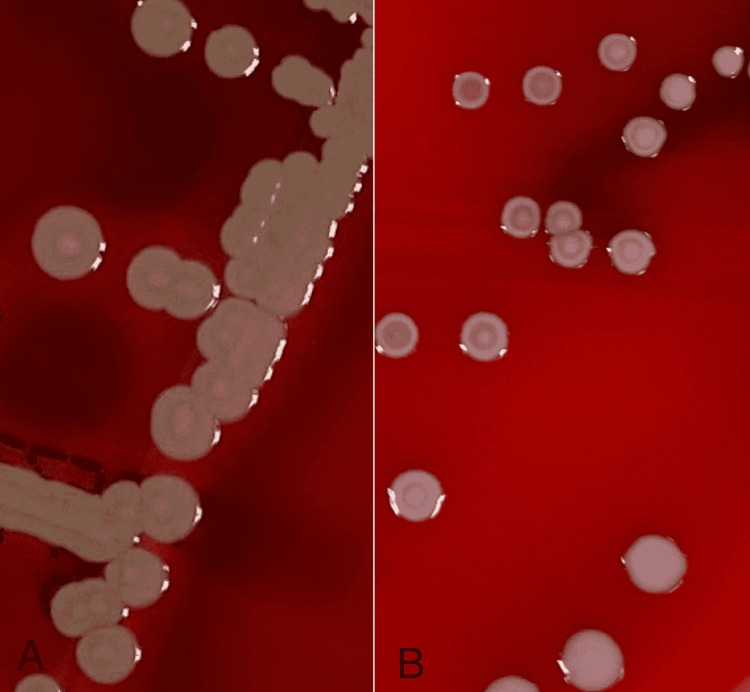
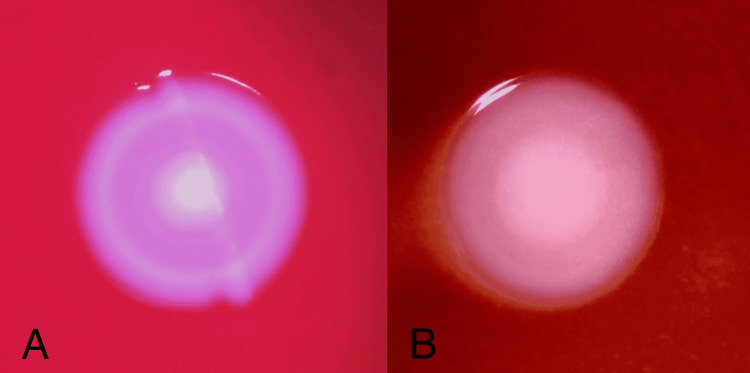
Inclusion criteria were well-isolated colonies of S. aureus on blood agar plates and exclusion criteria were small colonies and confluent colonies on blood agar plates. Blood agar plates were incubated for 24 and 48 hours at 35 °C with 5% CO2. The plates were read with the naked eye. If needed, a magnifying glass was used for assistance in visualizing the colonies. Colony shapes were classified as target-shaped (T1) or dome-shaped (T2). T1 is an elevated colony center encircled by a pale zone, which is surrounded by a single ring of peripheral enhancement giving a ‘target’ appearance (Figure 1). Magnified images of T1 colonies in CoNS and MRSA under dissecting microscope are depicted in Figure 2A and Figure 2B, respectively. T2 colony is with an elevated center and lacks the ‘target’ appearance (Figure 3).
Figure 1. Target-shaped (T1) colony morphology: (A) in a MRSA isolate; (B) in a CoNS isolate.
MRSA: methicillin-resistant Staphylococcus aureus; CoNS: coagulase-negative staphylococci
Figure 2. (A) Colony of CoNS observed under the dissecting microscope showing target shape (T1); (B) Colony of MRSA observed under the dissecting microscope showing target shape (T1).
MRSA: methicillin-resistant Staphylococcus aureus; CoNS: coagulase-negative staphylococci
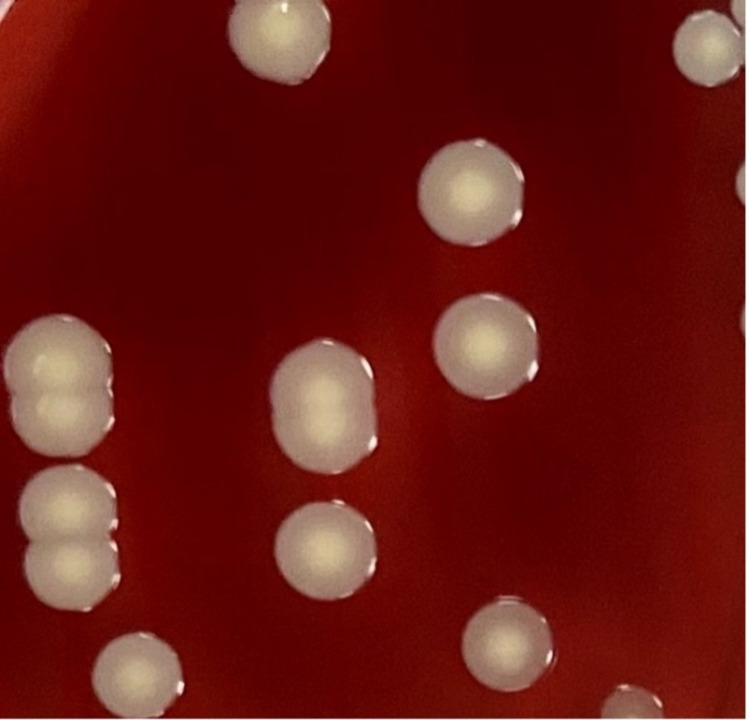
Figure 3. Dome-shaped (T2) colony morphology in Staphylococcus aureus lacking the target shape.
A commercially available PCR test (Cepheid® Xpert MRSA/SA Blood Culture test; Cepheid, Sunnyvale, California, United States) was performed on all first positive S. aureus blood culture bottles for the confirmation of S. aureus and detection of methicillin resistance.
Results
A total of 239 cases were screened for the study. Forty isolates were excluded from the study as the colonies were tiny or confluent, which prohibited the evaluation of colony morphology. After the exclusion of cases, a total of 199 Staphylococcus isolates were included in the study. Of the Staphylococcus isolates, 111 isolates were CoNS species. Of the remaining 88, 57 were MRSA and 31 were MSSA.
The colony morphologies recorded at 24 hours and 48 hours for target shape were as follows: At 24 hours, 19.3% of MRSA, 0.06% of MSSA, and 5.4% of CoNS had T1 colony morphology. At 48 hours, 73.7% of MRSA and 59.5% of CoNS showed T1 morphology as depicted in Table 1. T1 colony morphology has a sensitivity of 73.68% and specificity of 93.55% amongst S. aureus to detect MRSA strains at 48 hours with a positive predictive value of 95.45% (95%CI 84.49-98.78%).
Table 1. Number of isolates showing target-shaped (T1) and dome-shaped (T2) colony morphology of Staphylococcus species at 48 hours.
MRSA: methicillin-resistant Staphylococcus aureus; MSSA: methicillin-sensitive Staphylococcus aureus; CoNS: coagulase-negative staphylococci
| Organism (n) | Target-shaped (T1) | Dome-shaped (T2) | Neither T1 or T2 |
| MRSA (57) | 42 | 13 | 2 |
| MSSA (31) | 2 | 27 | 2 |
| CoNS (111) | 66 | 0 | 45 |
Discussion
The rapid identification of MRSA can be performed by fluorescence in situ hybridization (ISH), microarray, multiplex PCR, real-time PCR, and mass spectrometry. Identification can also be performed from positive blood culture bottles [3-12]. These tests may be cost-prohibitive in some settings. Traditional, less expensive but slower identification techniques include culture and subsequent confirmation of MRSA by disk diffusion and broth microdilution susceptibility testing.
A commonly used phenotypic method is screening isolated colonies by disk diffusion. Oxacillin and cefoxitin disks are most often used for screening. However, cefoxitin is recommended for screening as it can induce mecA and mecC better than oxacillin [13,14]. Chromogenic agar plates may be used to detect MRSA. The sensitivity of tests varied amongst different studies. In a study by Cesur et al., oxacillin resistance screen agar base (ORSAB) and CHROMagar™ MRSA media (Kanto Chemical Co. Inc., Tokyo, Japan) were compared to the oxacillin disk diffusion test reference method [15]. ORSAB had a sensitivity, specificity, and positive predictive value of 97.7%, 40%, and 36.5%, respectively. CHROMagar MRSA had a sensitivity, specificity, and positive predictive value of 95.5%, 37.6%, and 35.7% respectively. In a study by Onze-Lieve-Vrouwziekenhuis et al., MRSA II agar had a sensitivity and specificity of 98.9% and 89.4%, respectively, at 48 hours whereas MRSA ID had a sensitivity and specificity of 98.2% and 84.7%, respectively, at 48 hours respectively [16].
Other alternative methods have been studied for the identification of MRSA in histopathologic sections. These include immunohistochemistry (IHC) and ISH studies. IHC studies utilize the species-specific antigens of S. aureus and Penicillin-Binding Protein 2 (PBP2′) for identification [6]. MRSA can also be identified with ISH by demonstration of both the species-specific gene and mecA. However, these studies were predominantly performed on nonarchival sections such as unfixed frozen sections or paraformaldehyde-FFSs [17].
A literature search for T1 colony morphology on blood agar plates for the identification of MRSA was conducted. However, no similar studies were identified.
Limitations
First, care should be taken to read the morphology in well-isolated colonies. Confluent colonies may be falsely read as target shaped (T1) colony. Second, colony morphology at 48 hours should be considered for final results as the sensitivity increased from 19.3% to 73.68% for detection of MRSA at 48 hours; MSSA can, in rare cases, show a T1 morphology at 24 hrs. For example, on further incubation to 48 hours, a double zone of enhancement can be observed (deviating from the defined T1 shape colony) as in Figure 4.
Figure 4. Colony of MSSA showing two zones of enhancement with an elevated colony center at 48 hours (deviating from defined target shape).
MSSA: methicillin-sensitive Staphylococcus aureus
Conclusions
T1 morphology has a modest sensitivity with a specificity and positive predictive value amongst S. aureus for identification of methicillin resistance at 48 hours. T1 colony morphology at 48 hours can be potentially used for the identification of MRSA, especially in resource-limited settings as it does not require additional reagents, antibiotic disks, or media for detection.
T1 morphology can also be utilized in a setting where a molecular test is not repeated if PCR testing has already identified MSSA on a recent specimen on the same patient. Isolates can be further incubated for 24 hours to detect T1 colony morphology, which is suggestive of MRSA. Thereby, only cases showing T1 colonies at end of 48 hours can be further tested for confirmation by additional methods such as PCR or latex agglutination, improving patient care. The T1 colony morphology adds to the colony morphologic characteristics of MRSA.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
The authors have declared that no competing interests exist.
Human Ethics
Consent was obtained or waived by all participants in this study
Animal Ethics
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
References
- 1.A pursuit of Staphylococcus aureus continues: a role of persister cells. Chang J, Lee RE, Lee W. Arch Pharm Res. 2020;43:630–638. doi: 10.1007/s12272-020-01246-x. [DOI] [PubMed] [Google Scholar]
- 2.Detection of methicillin resistance in Staphylococcus aureus by polymerase chain reaction and conventional methods: a comparative study. Pillai MM, Latha R, Sarkar G. J Lab Physicians. 2012;4:83–88. doi: 10.4103/0974-2727.105587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapid methods for detection of MRSA in Clinical Specimens. Palavecino EL. Methods Mol Biol. 2020;2069:29–45. doi: 10.1007/978-1-4939-9849-4_2. [DOI] [PubMed] [Google Scholar]
- 4.Blood culture-based diagnosis of bacteraemia: state of the art. Opota O, Croxatto A, Prod'hom G, Greub G. Clin Microbiol Infect. 2015;21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Multicenter evaluation of the Staphylococcus QuickFISH method for simultaneous identification of Staphylococcus aureus and coagulase-negative staphylococci directly from blood culture bottles in less than 30 minutes. Deck MK, Anderson ES, Buckner RJ, et al. J Clin Microbiol. 2012;50:1994–1998. doi: 10.1128/JCM.00225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PNA FISH: present and future impact on patient management. Forrest GN. Expert Rev Mol Diagn. 2007;7:231–236. doi: 10.1586/14737159.7.3.231. [DOI] [PubMed] [Google Scholar]
- 7.Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. Buchan BW, Ginocchio CC, Manii R, et al. PLoS Med. 2013;10:0. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapid detection of Gram-positive organisms by use of the Verigene Gram-positive blood culture nucleic acid test and the BacT/Alert Pediatric FAN system in a multicenter pediatric evaluation. Sullivan KV, Turner NN, Roundtree SS, et al. J Clin Microbiol. 2013;51:3579–3584. doi: 10.1128/JCM.01224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comparison between MALDI-TOF MS and FilmArray Blood Culture Identification panel for rapid identification of yeast from positive blood culture. Paolucci M, Foschi C, Tamburini MV, Ambretti S, Lazzarotto T, Landini MP. J Microbiol Methods. 2014;104:92–93. doi: 10.1016/j.mimet.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 10.Interest of real-time PCR Xpert MRSA/SA on GeneXpert(®) DX System in the investigation of staphylococcal bacteremia (Article in French) Scanvic A, Courdavault L, Sollet JP, Le Turdu F. Pathol Biol (Paris) 2011;59:67–72. doi: 10.1016/j.patbio.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Stevenson LG, Drake SK, Murray PR. J Clin Microbiol. 2010;48:444–447. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A differential centrifugation protocol and validation criterion for enhancing mass spectrometry (MALDI-TOF) results in microbial identification using blood culture growth bottles. March-Rosselló GA, Muñoz-Moreno MF, García-Loygorri-Jordán de Urriés MC, Bratos-Pérez MA. Eur J Clin Microbiol Infect Dis. 2013;32:699–704. doi: 10.1007/s10096-012-1797-1. [DOI] [PubMed] [Google Scholar]
- 13.M100-S23. Performance Standards for Antimicrobial Susceptibility Testing, 30th edition. Wayne, Pennsylvania, United States: Clinical and Laboratory Standards Institute; 2020. Clinical and Laboratory Standards Institute. M100-S23. Performance Standards for Antimicrobial Susceptibility Testing; Thirtieth Edition. [Google Scholar]
- 14.Methicillin-resistant Staphylococcus aureus. Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A, Harbarth S. Nat Rev Dis Primers. 2018;4:18033. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 15.Evaluation of oxacillin resistance screening agar and chromogenic MRSA agar media for the detection of methicillin resistance in Staphylococcus aureus clinical isolates (Article in Turkish) Cesur S, Yildiz E, Irmak H, Aygün Z, Karakoç E, Kinikli S, Demiröz AP. https://pubmed.ncbi.nlm.nih.gov/20549963/ Mikrobiyol Bul. 2010;44:279–284. [PubMed] [Google Scholar]
- 16.Performance of a new chromogenic medium, BBL CHROMagar MRSA II (BD), for detection of methicillin-resistant Staphylococcus aureus in screening samples. Van Vaerenbergh K, Cartuyvels R, Coppens G, Frans J, Van den Abeele AM, De Beenhouwer H. J Clin Microbiol. 2010;48:1450–1451. doi: 10.1128/JCM.02103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Histochemical identification of methicillin-resistant Staphylococcus aureus: contribution to preventing nosocomial infection. Shimomura R, Tsutsumi Y. Semin Diagn Pathol. 2007;24:217–226. doi: 10.1053/j.semdp.2007.07.004. [DOI] [PubMed] [Google Scholar]