Abstract
NF-κB essential modulator (NEMO) regulates NF-κB signaling by acting as a scaffold for the kinase IKKβ and by directing its activity towards the NF-κB inhibitor, IκBα. Here, we show that a highly conserved central region of NEMO termed the Intervening Domain (IVD, aa 112–195) plays a key role in NEMO function. We determined a structural model of full-length NEMO by small-angle X-ray scattering (SAXS), and show that full-length, wild-type NEMO becomes more compact upon binding of a peptide including the NEMO-binding domain of IKKβ (aa 701–745). Mutation of conserved IVD residues (9SG-NEMO) disrupts this conformational change in NEMO and abolishes the ability of NEMO to propagate NF-κB signaling in cells, although the affinity of 9SG-NEMO for IKKβ compared to that of wild-type is unchanged. Based on these results, we propose a model in which the IVD is required for a conformational change in NEMO that is necessary for its ability to direct phosphorylation of IκBα by IKKβ. Our findings suggest a molecular explanation for certain disease-associated mutations within the IVD and provide insight into the role of conformational change in mammalian signaling scaffold proteins.
Keywords: NEMO, IκB kinase, scaffold protein, small-angle X-ray scattering (SAXS), conformational change, NF-kappaB (NF-κB), signal transduction, protein domain, protein structure
INTRODUCTION
Signaling scaffold proteins facilitate the temporal, spatial, orientational, and contextual interactions between intracellular signaling proteins1. They may bind and regulate two or more proteins of the same pathway or may coordinate signaling between pathways2. The most basic function of scaffolds is to provide a binding platform to co-localize signaling kinases and/or phosphatases with their substrates, thereby directing and regulating phosphorylation events3,4. This spatial and temporal coordination affords greater signaling specificity, by decreasing the likelihood of background and non-specific interactions5 while potentially increasing the rate of the desired reaction. Scaffolds can play a more active role than simple co-localization, for example by allosterically regulating the activity of their binding partners6. In some cases, scaffolds exert complex allosteric control to regulate the interactions between signaling components7. Because of their ability to facilitate complex signaling events, scaffold proteins have been engineered in order to modify dose-response to inputs as well as signaling outcomes7.
Recent work has suggested links between conformational dynamics and scaffold function. A pH-induced conformational change in the scaffold protein IscU has been suggested to play a role in iron-sulfur cluster biosynthesis8. Structural and kinetic studies have shown that Kinase Suppressor of Ras (KSR) undergoes an allosteric transition upon B-Raf binding, stimulating phosphorylation of Mitogen-Activated Protein-Kinase Kinase9. Deletion mapping and a partial crystal structure of the yeast scaffold Ste5 have been used to generate a model of autoinhibition, in which a conformational change in Ste5 acts as a gate to present the binding surface for Fus3, which then propagates a mating-specific signal10. Nonetheless, the extent to which particular scaffold proteins act as rigid binding templates, flexible platforms that allow interaction of their binding partners through stochastic motions, or dynamic structures that regulate signaling through ligand-induced conformational change, remains poorly understood.
The scaffold protein Nuclear Factor κB essential modulator (NEMO; UniProtKB entry Q9Y6K9) plays a central regulatory role in the canonical Nuclear Factor κB (NF-κB) signaling pathway. NF-κB comprises a family of related transcription factors that regulate the expression of hundreds of genes involved in diverse cellular and organismal processes11. Misregulation of the NF-κB pathway, often due to gene mutation of signaling components, contributes to numerous human diseases, especially those associated with chronic inflammation, immunodeficiency, and cancer11. Mutations in the human NEMO gene/protein, in particular, are associated with a variety of diseases, many having a B-cell immunodeficiency component12.
In unstimulated cells, NF-κB is usually maintained in an inactive state in the cytoplasm in complex with one of a family of inhibitory proteins known as Inhibitors of κB (IκB). The IκB protein most commonly studied in conjunction with NEMO function and canonical NF-κB signaling is IκBα13,14. A key regulator of the canonical NF-κB pathway is the IκB Kinase (IKK) complex, which comprises two kinase-containing subunits, IKKα or IKKβ bound to a dimer of NEMO (also known as IKKγ). Studies with IKK knockout mice indicate that IKKβ is the primary functional partner for NEMO15. Upon pathway activation, for example by stimulation of cells with tumor necrosis factor α (TNFα), interleukin-1β (IL-1β), or lipopolysaccharide (LPS), the IKK complex phosphorylates IκBα, triggering its ubiquitination and degradation by the proteasome, and thus allowing NF-κB to translocate to the nucleus where it activates NF-κB-responsive genes
It is not known whether NEMO serves simply as a passive docking site for IKK proteins or whether it exerts allosteric control over signaling events. However, the latter possibility is supported by biophysical measurements using intrinsic tryptophan fluorescence and 8-anilinonapthalene-1-sulfonic acid (ANS) fluorescence, which showed that NEMO undergoes a conformational change upon ubiquitin binding that is correlated with tighter binding to N- and C-terminal ligands and localization near the plasma membrane16. Additionally, Hauenstein et al. provided evidence that residues 112–150 are a site of negative regulation of NEMO, which maintains it in an auto-inhibited state until binding of poly-ubiquitin induces activation and enables IKKβ binding17.
Human NEMO is a 419 aa protein that exists as a constitutive dimer18,19. There is no X-ray crystal structure of full-length NEMO, but structures exist for several fragments or domains, in some cases bound to protein ligands (Figure 1). These structures include the following: a fragment of NEMO comprising residues 44–111 bound in a 2:2 complex with IKKβ(701–745)20; the Helix 2 (HLX2) domain of a NEMO dimer in complex with the viral FLICE inhibitory protein21; the combined coiled-coil 2 and leucine zipper (CoZi) domains unliganded22, bound to di-ubiquitin23,24, or in complex with other natural or artificial protein ligands25,26; and an NMR structure of the zinc finger (ZF) domain27. However, two regions of NEMO have not yet been structurally characterized: the region spanning residues 111–195, which connects the IKKβ binding domain and HLX2, and the 49 residues from aa 345–393, which connect the LZ domain with the C-terminal ZF (Figure 1). We do not include residues 1–43 among the structurally uncharacterized parts of NEMO, as this region is unstructured20,28.
Figure 1.
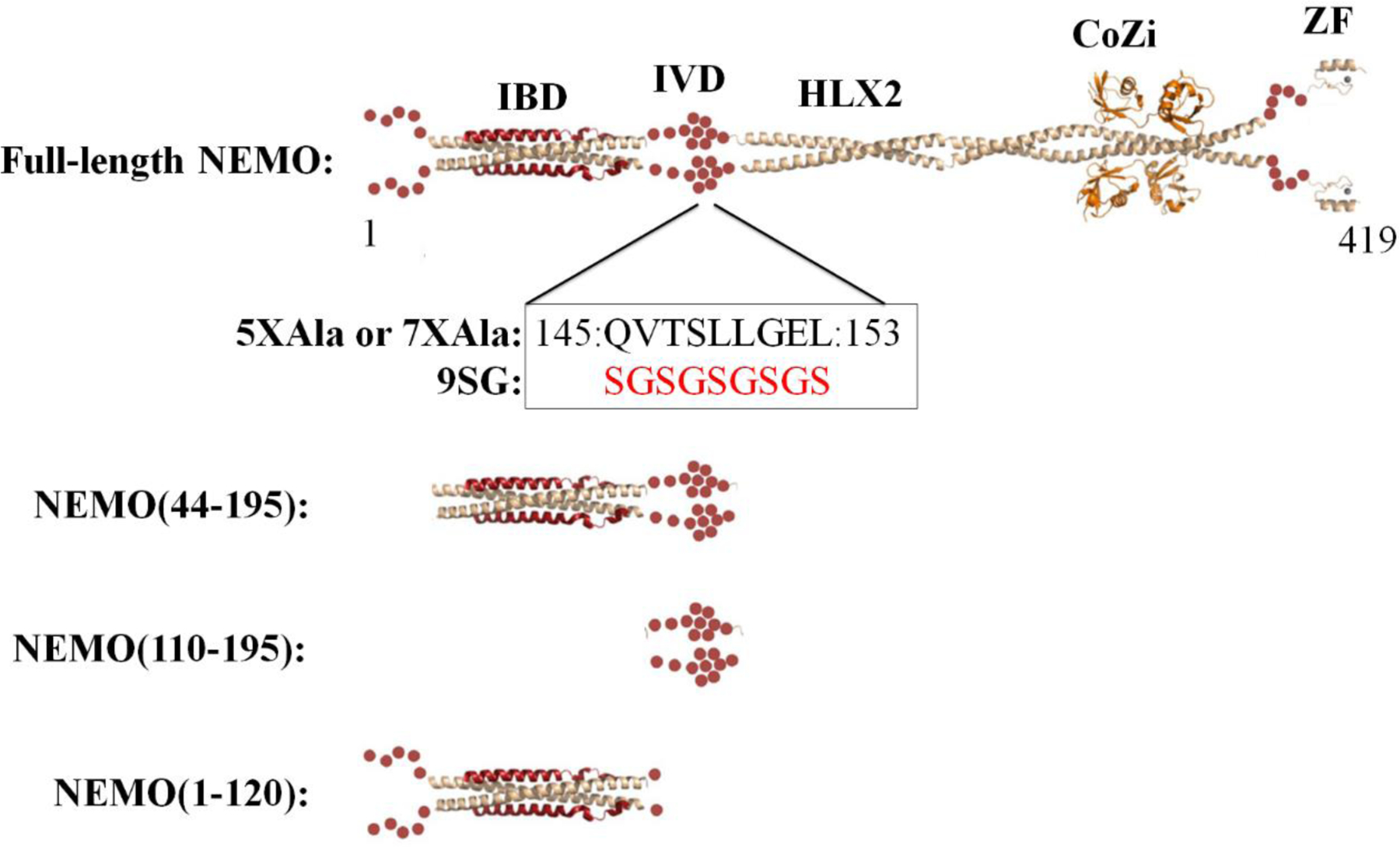
Domain map of NEMO with existing experimentally determined structures of individual NEMO domains and constructs used in this study. NEMO in beige ribbon; IKKβ(701–745) in red ribbon; linear di-ubiquitin in orange ribbon; red spheres represent structurally uncharacterized regions of NEMO. IBD, IKKβ binding domain; IVD, intervening domain; HLX2, coiled-coil 2; CoZi, leucine zipper; ZF, zinc finger.
The minimal NEMO-binding motif on IKKβ comprises 11 residues (aa 735–745)29, however, binding affinity equal to that of full-length IKKβ requires up to 45 residues20,23. It has been reported that NEMO also directly binds IκBα, through an interaction involving the C-terminal ZF domain of NEMO30. Analytical ultracentrifugation experiments using a NEMO construct encompassing residues 1–355 indicate that this major portion of NEMO has an extended, rod-like structure18. However, the mechanism by which NEMO directs the activity of IKKβ toward IκBα is not known. We hypothesize that NEMO undergoes a conformational change to bring the kinase active site of IKKβ into proximity with the phosphorylation sites on IκBα. Given the relatively simple coiled-coil structure that has been established for regions of NEMO extending from aa 44–111 and from aa 195–345, the structural mechanism underlying such a conformational change is unclear.
In this report, we have investigated the role in NEMO function of the previously uncharacterized region that connects the IKKβ binding domain (IBD) to the HLX2 domain. We have termed this region (aa 112–195) the Intervening Domain (IVD). The IVD is the largest contiguous region of NEMO that remains structurally uncharacterized, and includes sites of several disease-associated mutations31. We show that the IVD is highly conserved, is required for high affinity binding to IKKβ, confers structural stability on NEMO, and is required for activation of IKK in cells. Moreover, we use Small Angle X-ray Scattering (SAXS) to determine a structural model of unliganded full-length NEMO, and to show that the IVD appears to be a site of conformational regulation, mediating IKKβ-induced conformational changes that we propose play a role in NEMO and IKK complex function.
MATERIALS AND METHODS
Plasmid preparation for bacterial protein expression
NEMO(1–120) was subcloned into the pDEST-His-SUMO vector containing an N-terminal His6-SUMO tag (Invitrogen) as previously described28. Wild-type NEMO (WT-NEMO), 5XAla-NEMO, and 7XAla-NEMO were subcloned as described previously 19,28 into the pET24b(+) vector (Novagen) via NdeI and XhoI sites, with WT-NEMO transformed into Rosetta 2(DE3)pLysS competent cells (Novagen) and 5XAla-NEMO and 7XAla-NEMO transformed into T7 Express Iq Competent E. coli cells (New England Biolabs). NEMO(44–195) and NEMO(110–195) were subcloned out of a codon-optimized template (GenScript) by polymerase chain reaction, and ligated into a SUMOstar vector (LifeSensors) via BsaI and XhoI sites. These constructs were transformed into T7 Express competent cells (New England Biolabs) to take advantage of the codon optimization. 9SG was subcloned from the native, non-codon optimized 7XAla-NEMO template using the NEB Q5 Site-Directed Mutagenesis Kit (New England Biolabs), and transformed into Rosetta 2(DE3)pLysS competent cells (Novagen).
Protein expression and purification
WT-NEMO, 5XAla-NEMO, 7XAla-NEMO, 9SG-NEMO and SUMO-tagged NEMO(1–120) were expressed in bacterial cells and purified as described previously19,28. NEMO(44–195) and NEMO(110–195) were expressed identically to 5XAla-NEMO and 7XAla-NEMO, with a slight alteration during purification to remove a SUMO purification tag. Briefly, all NEMO-transformed cells were grown with shaking at 250 rpm at 37 °C overnight and used to inoculate fresh media at a 1:200 dilution. This culture was grown at 37 °C until OD600 reached between 0.4 and 0.6. Protein expression was induced by the addition of 1 mM isopropyl β-D-thiogalactoside (IPTG; GoldBio) and incubated for 4 h. Cells were pelleted by centrifugation and stored at −20 °C. The pellets were lysed by microfluidizer (Microfluidics Model #M110P) and incubated with 0.1 μL/mL Universal Nuclease (Thermo Fisher Pierce) and 10 μL/mL Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific) for 20 min. Urea (8 M) was then added to the lysate to prevent chaperones and other contaminating proteins from co-purifying with NEMO. The lysate was clarified by ultracentrifugation at 38,000 rpm for 30 min, sonicated to shear remaining large DNA fragments, and filtered through a 0.8 μm filter. The solubilization of SUMO-tagged NEMO(1–120) was slightly amended, with the lysate immediately centrifuged, the supernatant discarded, and the cell pellet washed and suspended with buffer supplemented with 0.5% Triton X-100, to which urea (8 M) was then added. These proteins were then purified as described previously19. Briefly, for WT-NEMO, 5XAla, 7XAla, 9SG-NEMO, and SUMO-tagged NEMO(1–120) lysate was applied to a 5 mL HisTrap High Performance column (GE Healthcare, Piscataway, NJ), and washed with 5 column volumes of 20 mM sodium phosphate, 500 mM NaCl, 40 mM imidazole, and 6 M urea (pH 7.4). The column-bound protein was refolded by applying a urea gradient from 6 to 0 M (20 column volumes). The folded protein was eluted with increasing imidazole concentration up to 500 mM imidazole in 20 mM sodium phosphate, 500 mM NaCl (pH 7.4). For WT-NEMO, lysate and buffers were supplemented with 5 mM TCEP to reduce misfolding and aggregation. Refolded NEMO from the nickel-NTA column was concentrated and applied to a HiPrep 26/60 Sephacryl S-300 HR column (GE Healthcare) equilibrated with 20 mM sodium phosphate and 500 mM NaCl (pH 7.4). For the NEMO(44–195) and NEMO(110–195) constructs, purification omitted the final gel filtration step. Instead, the protein was concentrated to 1 mL and incubated with SUMOstar Protease 1 (LifeSensors) for 2 h at room temperature, and then 4 °C overnight. This mixture was then applied to a 5 mL HisTrap High Performance column for subtractive affinity chromatography; His-tagged SUMOstar tag and SUMOstar Protease 1 bound the column, while the untagged protein flowed through. Purified proteins were pooled and concentrated to < 3 mL. WT-NEMO, 5XAla-NEMO, 7XAla-NEMO, and 9SG-NEMO protein concentrations were calculated from the absorbance at 280 nm (NanoDrop, Thermo Fisher Scientific) using an extinction coefficient of 14400 M−1cm−1, and 11460 M−1cm−1 for SUMO-tagged NEMO(1–120) (uncleaved). NEMO(44–195) and NEMO(110–195) contain no tryptophan residues, so protein concentration was calculated by Lowry protein assay (Folin & Ciocalteu Reagent, Thermo-Fischer Scientific) and densitometry via SDS-PAGE in comparison to protein standards. Yield of protein was approximately 10 mg per 2.5 g cell pellet.
Fluorescence anisotropy
The fluorescence anisotropy assay used herein has been described previously19,32. Briefly, assays were carried out in black 96-well polypropylene plates with a total volume of 200 μL. The assay buffer contained 50 mM Tris pH 7.4, 200 mM NaCl, 0.01% Triton X-100, and 1 mM DTT. NEMO concentrations were varied from 10 pM to 1 μM while keeping FITC-IKKβ(701–745) peptide (tracer probe; Genscript) constant at 15 nM. Plates were incubated at 25 °C for 1 h prior to reading. Fluorescence anisotropy was read using a SpectraMax M5 plate reader using λ = 488/520 ex/em. Anisotropy was calculated as described previously19.
Circular dichroism spectroscopy
NEMO constructs were diluted to 8.6 μM in a buffer containing 20 mM sodium phosphate and 500 mM NaCl at pH 7.4. Spectra were obtained using a 1 mm 300 μL quartz cuvette in an Applied Photophysics Chirascan CD spectrometer. Buffer-subtracted spectra from 180 to 260 nm with a step of 1 nm were measured at 10 °C and mean residue ellipticity (in units of deg.cm2/dmol) was calculated from the raw signal (in units of millidegrees) 33. The presence of coiled-coil was detected from the ratio of ellipticity at 222/208 nm35. For thermal denaturation experiments34, mean molar residue ellipticity was calculated from the signal at 222 nm, and traced as the sample temperature was ramped at 1 °C /min from 10 °C to 70 °C. Slowing the ramping rate to 0.5 °C /min had no effect on the thermal denaturation. Melting temperatures (Tm) were determined as the maximum of a plot of the first derivative of the thermal denaturation curve, against temperature. For experiments in which 2,2,2-trifluoroethanol (TFE) was included, NEMO was concentrated to 2.5 mg/mL before 10-fold dilution with TFE to yield a 90% TFE solution. Spectra were then obtained as described above. The percentage α-helix was calculated as described previously by Zhou and colleagues28, the ratio between Θ222 in aqueous buffer and that in TFE-containing buffer (Equation 1):
| (1) |
Small-angle X-ray scattering data collection, processing, and analysis
SAXS data for 5XAla-NEMO were collected on beamline X9 of the National Synchrotron Light Source, 9SG samples were collected on beamline 16-ID-LiX of the National Synchrotron Light Source II, at Brookhaven National Laboratories (Upton, NY), and all other samples were collected on beamline 4–2 of the Stanford Synchrotron Radiation Light Source, at the SLAC National Accelerator Laboratory, (Menlo Park, CA). Samples were transported frozen and, after thawing, were filtered with a 0.22 μm filter, centrifuged to remove any particulates. All experiments were conducted in 20 mM sodium phosphate, 500 mM NaCl, pH 7.4. Protein concentrations were ~1.6 mg/mL and ~0.8 mg/mL for 9SG-NEMO and 1 and 4.4 mg/mL for 5XAla-NEMO. For solution SAXS of WT-NEMO and NEMO(44–195), concentrations of 3.0, 2.3, 1.5, and 0.75 mg/mL protein were used. For NEMO(44–195), the difference in scattering intensity with increasing concentration showed that at the higher protein concentration the sample was approaching the solubility limit, such that the actual protein concentration present in solution was slightly lower than expected. For these data sets, the protein concentration in solution was therefore estimated directly from the scattering intensity (see figure legends S4 and S6). Samples were loaded into 8-tube PCR strips at ambient temperature and aspirated into a thin-walled 1.5-mm diameter sample tube using an automated system as described previously36.
Size exclusion small angle X-ray scattering (SEC-SAXS) was conducted on beamline 4–2 at the Stanford Synchrotron Radiation Lightsource equipped with an online Superdex 200 Increase 5/150 GL column (GE Healthcare) using a protein concentration of 5.8 mg/mL for WT-NEMO and 15 mg/mL for NEMO(44–195) and a flow rate of 0.05 mL/min.
At X9, scattered intensities were determined from q = 0.005 to 1.99 Å−1 using an X-ray wavelength of 0.9184 Å and detected via a Mar 165 CCD detector located 3.4 m from the sample. At LiX, scattered intensities were determined from q = 0.005 to 2.47 Å−1 using an X-ray wavelength of 0.9184 Å and detected via a Pilatus3 1M SAXS detector and two Pilatus3 300K WAXS detectors. At 4–2, scattered intensities were determined from q = 0.005 to 1.5 Å−1 using an X-ray wavelength of 1.127 Å and detected with a Pilatus3 X 1M detector. Three replicate data sets were collected. Data processing was performed using the X9, LiX, or 4–2 software packages to produce radially averaged intensity profiles extending over the entire scattering range. For solution SAXS of WT, 5XAla, and 9SG the data from two concentrations were checked to be consistent at low q and merged using PRIMUS37. For SEC-SAXS data, 500 to 600 frames were recorded, with an exposure taken every 5 s and, the first 100 frames that were part of the column void volume averaged and used for buffer background subtraction. Data through the elution profile were averaged every 5 frames, analyzed for sample aggregation, and used for subsequent analysis.
X-ray scattering patterns were initially checked for aggregation using the ATSAS suite37. The relative intensity versus inverse scattering angle, Kratky, and Porod plots were generated using Microsoft Excel and plotted with GraphPad Prism. The Rg and the P(r) function were calculated using GNOM38 with the data range indicated in Table S1. The P(r) was calculated by starting with autoGNOM in GNOM and manually adjusting the Dmax value to optimize the total quality estimate value while optimizing curve smoothness and monitoring its fit to the data. The alpha value was slightly increased to improve the curve smoothness while still keeping a good fit to the data. AllosMod-FoXS39,40 was used to validate data analysis quality. Input included the sequence of 5XAla-NEMO, unliganded crystal structures of NEMO(44–111), NEMO(197–249), and NEMO(280–321), and the experimental scattering curve for 5XAla-NEMO. For 5XAla-NEMO analysis with IKKβ(701–745), the IKKβ-bound crystal structure of NEMO(44–111) was used in place of the unliganded structure.
Following P(r) function determination by GNOM38, this information was used in DAMMIN41 or DAMMIF42 to generate 10 ab initio models of the shape of each construct of NEMO using default parameters. The 10 models were averaged using DAMAVER43. The final output of DAMAVER was then re-run through DAMMIN for final refinement, using default parameters. The goodness of fit of this final model was determined by FoXS39.40 in CHIMERA by χ-value calculation. The volume envelope of the model was generated using the “Fit in Map” feature of UCSF CHIMERA44. CHIMERA was also used to align the NEMO(44–195) volume envelope with that of the 5XAla-NEMO model for direct comparison, as well as to align each unliganded construct to its liganded counterpart. Fitting of the envelope with previously determined X-ray crystal structures of individual NEMO domains IBD, HLX2, and CoZi was accomplished by aligning the structures using BUNCH45 and superimposing on the envelope using CHIMERA44. This atomistic modeling fit well to the original scattering data (total quality estimate = 0.74 theoretical scattering versus experimental) but should be considered provisional due to SAXS resolution limits.
Mammalian cell culture and transfection
HEK 293, NEMO-deficient 293T, and NEMO-deficient mouse fibroblast (NIH3T3) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Biologos) as described previously19,46,47. NEMO-deficient 293T cells were generated by CRISPR/Cas9 disruption of a short sequence near the transcription start site of the NEMO gene48; elimination of NEMO protein expression in this cell line is shown in Fig. 6B. The creation of pcDNA-FLAG and pBABE-puro vectors for the expression of WT-NEMO and 7XAla-NEMO has been described previously19,46. The 9SG mutation in NEMO was created using the NEB Q5 Site-Directed Mutagenesis Kit (New England Biolabs). The pDNA-FLAG and pBABE vectors for the 9SG variant were created by standard recombinant DNA techniques. Transfection of 293T cells and retroviral transduction of mouse NEMO knockout cells were performed as described previously19. Virally transduced NEMO-knockout fibroblasts were selected using 2.5 μg/mL puromycin (Sigma) for approximately 1−2 weeks49.
Figure 6.
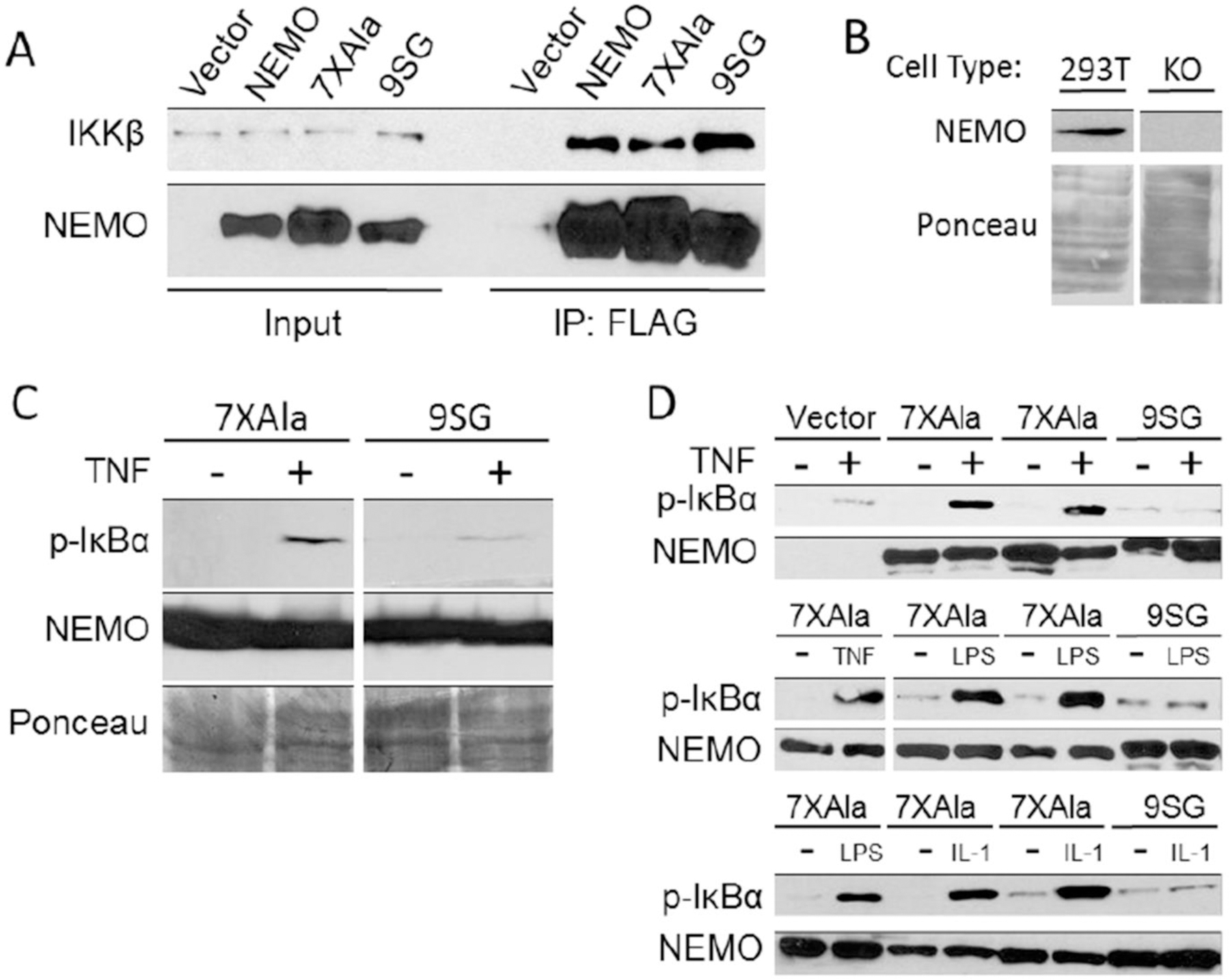
The 9SG mutation abolishes the ability of NEMO to function in signal-induced activation of IκB kinase. (A) The indicated FLAG-tagged NEMO constructs were transfected into 293T cells, and extracts were immunoprecipitated (IP) with anti-FLAG beads. Immunoprecipitates were then analyzed by anti-FLAG (bottom) and anti-IKKβ (top) Western blotting. In the Input lanes, 4% of the extract used in the immunoprecipitations was analyzed by Western blotting. (B) Whole cell extracts from wild-type and NEMO-knockout 293T cells were analyzed by anti-NEMO Western blotting (top) or Ponceau staining for total protein (bottom). (C) 293T NEMO knockout cells were transfected with plasmids for the expression of 7XAla- or 9SG-NEMO. Cells that were either untreated (−) or treated with TNFα (+) were analyzed by Western blotting for phospho-IκBα or NEMO or by Ponceau staining. (D) Mouse NEMO knockout fibroblasts were stably transduced with retroviral vectors for the indicated NEMO proteins. Stable cell lines were then untreated (−) or treated (+) with the indicated compounds. Extracts were analyzed by Western blotting for the indicated proteins.
Western blotting and co-immunoprecipitation
Western blotting was performed as described previously46. For analyzing phosphorylation of IκBα, cells were first treated with 20 ng/mL TNFα (R&D Systems) for 10 min, 2 ng/mL of IL-1 for 6 min, or 100 ng/mL of LPS for 12 min. Cells were then lysed directly in 2X SDS sample buffer. In all cases, samples containing approximately equal amounts of protein were separated on SDS-polyacrylamide gels, proteins were transferred to nitrocellulose membranes, and filters were incubated overnight at 4 °C with NEMO antiserum (cat. #2685, Cell Signaling Technology; 1:1000 dilution) or with phospho-IκBα antiserum (cat. #9246, Cell Signaling Technology; 1:1000 dilution). Horseradish peroxidase-conjugated secondary antiserum was added, and immunoreactive proteins were detected by SuperSignal Dura West chemiluminescence (Thermo Fisher Scientific). For co-immunoprecipitation experiments, transfected 293 cells in 100-mm tissue culture dishes were lysed in 500 μL of AT buffer [20 mM HEPES (pH 7.9), 1 mM EDTA, 1 mM EGTA, 20 mM Na4P2O7, 1 mM DTT, 1% (v/v) Triton X-100, 20% (w/v) glycerol, 1 mM Na3VO4, 1 μg/mL PMSF, 1 μg/mL leupeptin, and 1 μg/mL pepstatin]. An aliquot (20 μL) was saved as the input sample. To the remainder of the lysate was added 30 μL of anti-FLAG agarose beads (Sigma catalog no. A2220), and samples were incubated overnight with rocking at 4 °C. The beads were washed several times with AT buffer, and then bound proteins were removed by heating the samples at 90 °C in SDS sample buffer containing β-mercaptoethanol. Samples (input or immunoprecipitates) were then analyzed by reducing SDS-PAGE followed by Western blotting with the IKKβ antiserum (sc-7607, Santa Cruz Biotechnology), NEMO antiserum (catalog no. #2685; Cell Signaling Technology), or FLAG antiserum (catalog no. #2368; Cell Signaling Technology) as described previously19.
RESULTS
Design and Preparation of Recombinant NEMO Constructs for in vitro Studies
To enable investigation of the structure and function of the NEMO IVD through biochemical and biophysical studies in vitro, we created bacterial expression vectors for a variety of NEMO constructs. We previously showed that NEMO proteins with five (5XAla-NEMO; C11/76/95/131/167A) or seven (7XAla-NEMO; C11/54/76/95/131/167/347A) Cys-to-Ala mutations show increased solubility compared to WT-NEMO when expressed in bacteria, and retain signaling function when expressed in mammalian cells19 (and see Table S2 for comparative biochemical, biophysical, and biological evidence). For the current studies, recombinant full-length 5XAla-NEMO was purified as previously described (see Materials and Methods)19. The 9SG-NEMO variant (described below) was constructed in the 7XAla-NEMO background. Full-length versions of WT, 5XAla-, 7XAla and 9SG-NEMO proteins include a C-terminal His6 tag for affinity purification and the affinity tag was not removed for the described studies. A previously described truncated NEMO(1–120)28 lacking the full IVD (aa 112–195) was expressed with an N-terminal His6 tag preceding a fused SUMO fusion partner, which do not affect the protein function in vitro and were not removed. We also expressed a truncated NEMO construct, comprising residues 44–195, which includes the IBD (aa 44–111) plus the adjacent IVD (aa 112–195) and a NEMO construct comprising residues 110–195, which contains only the IVD. NEMO(44–195), and a NEMO(110–195) construct comprising the IVD alone, were each subcloned into a SUMOstar vector that contained a His6 tag N-terminal to a SUMOstar fusion partner, fused at the N terminus of the NEMO fragment, which was removed by proteolysis after purification. Note that NEMO(44–195) contains an inter-chain disulfide at residue 54, and so is constitutively dimeric. Although cysteine is the native residue at position 54, WT-NEMO is likely fully reduced in cells46. WT-NEMO is constitutively dimeric in the absence of any disulfides, and the presence or absence of a disulfide at position 54 has no measurable effect on the binding affinity of NEMO for IKKβ.19 A disulfide at position 54 can, however, help stabilize short NEMO fragments containing only the IKKβ binding domain by stabilizing the coiled-coil structure of this region as a covalent dimer, thereby preserving a conformation that resembles the structure of this region of NEMO as it exists in full-length NEMO28.
Analysis of NEMO Solution Structure by SAXS
SAXS reveals information about macromolecular structure and dynamics in solution. The raw X-ray scattering data are radially averaged about the beam center, and then buffer-subtracted to obtain the specific scattering intensity of the protein as a function of scattering angle. This information can be analyzed to reveal features of a protein’s structure, including the generation of a three-dimensional shape reconstruction with a resolution on the order of 10–50 Å50,51. The solution structure of NEMO in complex with MBP-tagged herpesvirus FLICE inhibitory protein (vFLIP) has been determined by SAXS17. To probe the structure of unliganded NEMO, we analyzed 5XAla-NEMO using SAXS. The key SAXS results from our study are summarized in Table S1. The scattering data are shown in Figure 2A as scattering intensity (I) plotted against the modulus of the scattering vector (q, Å−1). We note that the Guinier plot (ln(I) versus q2) for this data set (Figure 2B) is not linear below q2 = 0.0006 Å−2. However, unlike the situation for globular proteins, Guinier plots for extended macromolecules are not necessarily linear in this region50,51. To assess any effects from aggregation, a concentration series was performed (Figure 2E–F), which shows a similar dependency of ln(I) on q2 across concentrations (0.75 – 3.0 mg/mL) in the Guinier region below q2 = 0.0008. If interfering aggregation were an issue, we would expect a discrepancy in the curvature of the plots at low q2, which would be concentration dependent; this did not occur in the concentration range tested. Similarly, the experimental scattering data plotted as (log(I(q)) vs q and Kratky plots (q2 I(q) vs q) gave similar curves at all concentrations tested (Figure S6).
Figure 2.

SAXS model of 5XAla-NEMO. (A) Scattering curve for 5XAla-NEMO (blue), with theoretical scattering (red) calculated from the model shown in panel D. Inset: the error-weighted residual difference plot, Δ/σ = [I exp(q) − cI mod(q)]/σ(q) versus q for model fitting to experimental scattering. (B) Guinier plot. Linearity at low q is not necessarily expected for a partially flexible, extended protein50,51. (C) Distance-distribution function generated by GNOM. (D) Shape reconstruction generated by DAMMIN (gray envelope) with superposed NEMO model (blue) generated from three NEMO domains by BUNCH. “N-term” and “C-term” labels denote NEMO N terminus and C terminus, respectively; the Intervening Domain is denoted “IVD.” (E) Guinier plots showing lack of aggregation with increasing protein concentration for WT-NEMO.
The scattering data can be used to calculate a distance-distribution function, P(r), which can be plotted as a histogram of all possible pairwise atom distances. The form of the P(r) plot is characteristic of the three-dimensional shape of the molecule, and the maximum x-value equals the maximum dimension of the scattering particle50,51. Because of the observed non-linearity in the Guinier plot at low q, the values for radius of gyration (Rg) throughout this analysis were obtained from the P(r) plot rather than by using the Guinier approximation. The P(r) plot for 5XAla-NEMO, calculated using GNOM, is shown in Figure 2C. GNOM assigns an error-weighted scoring function, total quality estimate, a global parameter minimized in fitting that is used to evaluate the difference between the scattering profile computed from the model and the measured scattering profile38. Analysis of the data for 5XAla-NEMO resulted in a total quality estimate of 0.583. For comparison, analysis of a SAXS curve calculated by FoXS for a theoretical, extended coiled-coil protein with approximately the same overall length as NEMO (280 Å) resulted in a χ value of 0.601 (Figure S1). The P(r) plot indicates that 5XAla-NEMO has a highly extended structure, with the greatest number of vectors at short interatomic distances and a long tail extending to a maximum particle dimension, Dmax, of 315 Å. SAXS Analysis of WT-NEMO yielded similar values to 5XAla-NEMO, with a Dmax of 305 Å (total quality estimate of 0.807. These values are similar to the Dmax observed by Hauenstein et al.17 for the NEMO-MBP-vFLIP complex (320 Å), which they argue is in the same conformation as unliganded NEMO as determined by gel filtration analysis. Fragments of NEMO encompassing residues 44–11120, 197–24921, and 260–33322, have been crystallized and shown to be dimeric coiled coils (Figure 1). These three segments sum to 186 residues, corresponding to ~280 Å of extended α-helical structure. Comparing that value to our value of Dmax of 315 Å for 5XAla-NEMO, indicates that the remaining 233 residues cannot all be in an extended coiled-coil conformation, and must be disordered or assume more compact forms.
The SAXS data were further analyzed to generate low-resolution structures based on the P(r) data, by modeling the protein as beads on a fixed lattice using the software package DAMMIN41. Ten models were generated, averaged, and refined; prior to averaging, their average normalized spatial discrepancy was 0.58 ± 0.3. This unitless value reflects variability among DAMMIN models, with values c.a.1 indicating acceptable convergence on a final structure. The final, highest scoring model was fit into a volume map using UCSF CHIMERA44, revealing the shape of the average scattering particle in solution (Figure 2D). FoXS was used to calculate a theoretical scattering curve from the final DAMMIN model39.40, demonstrating how well the model represents the original scattering data (Figure 2A). The quality of the fit calculated by FoXS (χ = 0.6) indicates a good fit to the original experimental scattering data39.
The SAXS-generated structural model for 5XAla-NEMO reveals an extended structure containing several kinks. The extended, coiled-coil segments of three published X-ray crystal structures of the IBD, HLX2 and CoZi domains of NEMO were modeled together using BUNCH45 and superposed onto the SAXS envelope as shown in Figure 2D. The results indicate that the C-terminal ZF domains of NEMO are not visible in the SAXS structure, suggesting that the ZF domains, which are connected to the CoZi domain by a 50 aa linker of unknown structure, are not in a fixed position relative to the remainder of NEMO. Similarly, N-terminal residues 1–43 appear to be absent from the SAXS envelope, consistent with previous results indicating that this region is structurally disordered20,28. The small projection visible at the center of the extended structure may reflect residual electron density resulting from a small fraction of dimer-dimer contacts involving this region of the molecule, as has been proposed18,51. Notably, the region of the modeled structure between the IBD and HLX2 domains does not appear to have sufficient space to accommodate the 84 residues that connect these two domains (aa 112–195) as an extended coiled-coil, and appears somewhat globular, suggesting the IVD has a more compact structure.
Sequence Analysis of the IVD Region of NEMO
Functionally important regions of proteins are often highly conserved through evolution. To determine whether the sequence of the IVD is conserved across species, we performed a multiple sequence alignment (MSA) using annotated NEMO sequences obtained from BLASTp (National Center for Biotechnology Information). Redundant sequences, splice variants, and hypothetical sequences were removed, as were fragmentary sequences with fewer than 50 residues and sequences for the functionally distinct NEMO-like protein optineurin. Of the resulting 38 unique NEMO sequences, 27 were from mammals, 10 from fish or amphibians, and one from Drosophila. Among these 38 species, core regions of functionally important domains, such as the IBD (IKKβ binding), the HLX2 domain (vFLIP binding), the CoZi domains (ubiquitin binding), and the ZF (IκBα binding) are particularly highly conserved, with lower conservation towards the borders of these domains and in the segments linking them (Figure S2). The IVD also shows high sequence conservation in its core segment (aa 134–161), including a stretch of nine residues (145-QV/ATSLLGEL-153) that, aside from a Val/Ala polymorphism at position 146, is identical among all mammalian and non-mammalian vertebrate species (Figure 3). The finding that the IVD is as highly conserved as domains of NEMO that are known to have functional roles supports the hypothesis that the IVD constitutes a functionally important domain.
Figure 3.

Alignment of the core sequence of the NEMO Intervening Domain. Shown is the region that spans residues 141 to 161 of human NEMO. Residues in blue are identical to the corresponding position in human NEMO. The full alignment is shown in Figure S2.
Influence of the IVD on Binding Affinity for IKKβ in vitro
Previous studies have shown that 5XAla-NEMO binds a 45-mer peptide comprising the NEMO-interacting region of IKKβ with an affinity of 2.8 ± 1.1 nM, similar to the affinities observed for WT NEMO and the 7XAla variant19 (see Table S2 for collated data). In contrast, several covalently or noncovalently dimeric NEMO fragments containing only the IBD bind IKKβ peptide with a substantially reduced affinity of 70–560 nM28,53. The lower affinity seen with IBD-only constructs has been attributed to subtle differences in structure or flexibility compared to WT-NEMO28,53. To determine whether the adjacent IVD region influences the binding affinity of NEMO for IKKβ, we tested a NEMO construct comprising residues 44–195 which spans both the IBD and the IVD (aa 112–195).
The binding of NEMO(44–195) to IKKβ peptide was measured using a fluorescence anisotropy (FA) binding assay described previously19,32. The results of the FA assay (Figure 4A) show that, consistent with previous results, WT-NEMO bound the peptide with KD = 2.3 ± 0.7 nM (n = 3), 5XAla-NEMO bound the peptide with KD = 3.2 ± 0.5 nM (n = 3), and 7XAla bound the peptide with KD = 4.9 ± 1.1 nM (n = 3). NEMO(44–195) also bound IKKβ peptide with high affinity (KD = 10.6 ± 2.3 nM, n = 3), comparable to the result for the three full-length NEMO constructs but ~10-fold higher affinity than the value for NEMO(1–120) (KD = 78 ± 10 nM) measured in this study and consistent with previously measured value KD = 70–120 nM28) (Figure 4D). These results show that the presence of the full IVD increases the IKKβ binding affinity of the adjacent IBD to the value measured for full-length, WT-NEMO. A control construct, NEMO(110–195), comprising the IVD alone, showed no detectable binding to the IKKβ peptide (Figure 4A and D).
Figure 4.
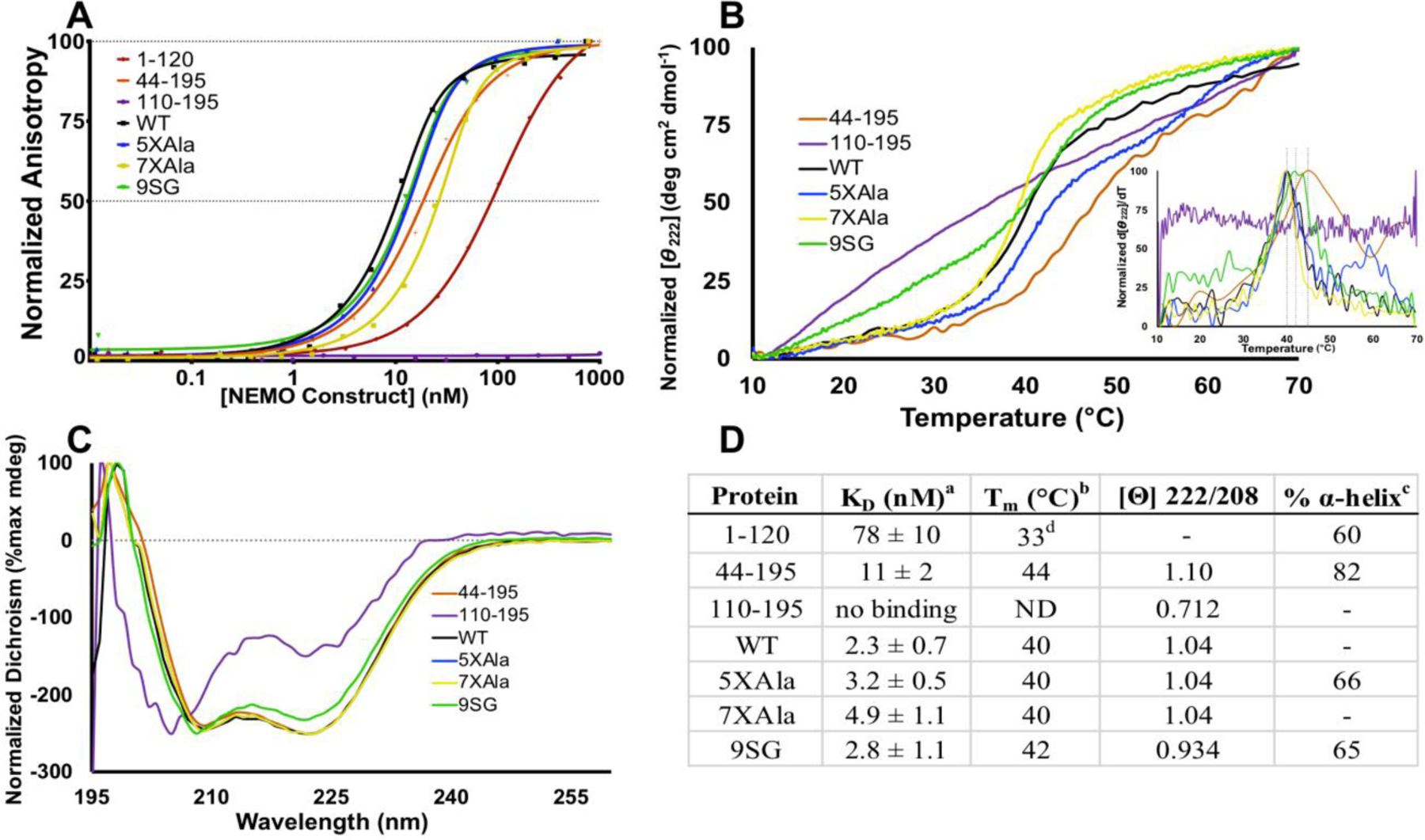
The IVD contributes to NEMO-IKKβ binding affinity and NEMO thermal stability. (A) Fluorescence anisotropy binding assays. The indicated NEMO proteins were titrated from 0.01 nM – 1000 nM, with FITC-labeled IKKβ(701–745) kept constant at 15 nM. Results are representative of three independent experiments performed with triplicate samples. (B) The indicated NEMO proteins were subjected to thermal denaturation, as monitored by CD. An increase in signal at Θ = 222 nm corresponds to loss of secondary structure. Inset: first derivative of melting curve used to identify Tm value. (C) CD spectra of NEMO constructs determined at 10 °C. The negative peak with minima at 208 and 222 nm is indicative of α-helical content. (D) Table of key results. aKD for binding FITC-IKKβ(701–745) measured using the FA binding assay. bDetermined by CD, monitoring change in Θ222 as temperature was increased at 1 °C/min. c The % α-helical content was determined from the CD spectrum in aqueous buffer compared with that in 90% TFE.d The Tm for NEMO(1–120) was reported previously, and is shown here for reference28.
As a second test of the influence of the IVD on IKKβ binding, we made a NEMO construct in which the highly conserved sequence (145-QVTSLLGEL-153) at the core of the IVD was replaced with a flexible Ser-Gly repeat sequence (i.e., SGSGSGSGS). This NEMO variant, termed 9SG-NEMO, was constructed in the 7XAla background. In the FA binding assay 9SG-NEMO had a binding affinity for IKKβ peptide of KD = 2.7 ± 1.1 nM (n = 3) (Figure 4A). Thus, 9SG-NEMO binds IKKβ with an affinity similar to that of WT-NEMO, 5XAla-NEMO, 7XAla-NEMO, and NEMO(44–195), and displays a higher affinity for IKKβ than that seen for the IBD-only construct NEMO(1–120) (Figure 4D). These results show that full binding affinity of NEMO for IKKβ in vitro requires the IVD region, consistent with close structural communication between the IBD and the IVD, but that IKKβ binding is tolerant to mutations in the IVD that affect the biological function of the protein (vide infra).
Effect of the IVD on the Structural Stability of the NEMO IKK Binding Domain (IBD)
To further investigate possible structural and functional communication between the IVD and the IBD we tested whether the IVD affects the thermal stability of the IBD, using circular dichroism spectroscopy (CD) to monitor thermal unfolding through the associated loss of secondary structure. The results showed that 5XAla-NEMO had a melting temperature of Tm = 40 °C (Figure 4B). We previously showed that the covalently dimeric IBD-only construct NEMO(1–120) is less stable, melting at a temperature of 33 °C 28. Notably, NEMO(44–195) showed a single cooperative melting transition with Tm = 44 °C, comparable to the Tm for 5XAla-NEMO. The ability of the IVD to stabilize NEMO into a single, cooperatively unfolding unit indicates that the IBD and IVD are structurally interconnected. The 9SG-NEMO variant also showed similar structural stability to 5XAla-NEMO, melting with Tm = 42°C (Figure 4B and D). This result suggests that the 9SG mutation does not disrupt the overall structural integrity of NEMO, consistent with the finding, described above, that the 9SG mutation does not decrease the affinity of the IBD for IKKβ peptide binding.
To determine whether the 9SG mutation causes a substantial disruption of NEMO secondary structure, we quantified helical content by comparing the CD spectra in aqueous buffer with those measured in 90% 2,2,2-trifluoroethanol (TFE), a solvent that induces adoption of α-helical structure regardless of the protein primary structure54,55. Using the spectrum obtained in 90% TFE as representative of a 100% α-helical structure, the ratio of the molar ellipticity at 222 nm measured in aqueous buffer to the corresponding value in 90% TFE gives an estimate of the α-helix content of a protein in aqueous conditions. These measurements showed that, in aqueous buffer, 9SG-NEMO was 65% α-helix, approximately the same as 7XAla-NEMO (66% α-helix) (Figures 4D and S3). This result shows that the 9SG mutation does not significantly disrupt the overall secondary structure of NEMO. Using the same approach, NEMO(44–195) was found to be 82% α-helix. To verify that 90% TFE was sufficient to induce full α-helicity, NEMO(44–195) was also evaluated in 97% TFE, yielding an essentially identical result (81% α-helix; data not shown). This high α-helical content suggests that the IVD is primarily folded with α-helical secondary structure. The presence of dimeric, fully folded coiled coils was then assessed from the ratio [θ] 222/208 taken from the CD spectra. A value ≥1 indicates fully-folded coiled-coils, with ≤0.9 indicating single-stranded alpha helices. WT-NEMO, 5XAla-NEMO, 7XAla-NEMO, and NEMO(44–195) all display values greater than 1 (Figure 4D), indicating dimeric coiled-coil, whereas for 9SG-NEMO [θ] 222/208 = 0.93, which indicates disruption of coiled-coil, but not of overall alpha-helical content. NEMO(110–195) has [θ] 222/208 = 0.71, indicating significant disruption of coiled-coil19. However, unlike full-length NEMO constructs, which are constitutive noncovalent homodimers19, and NEMO(1–120) which is covalently dimeric via an interchain disulfide bond through Cys5428, NEMO(110–195) is likely monomeric under the conditions tested. Therefore, the low α-helical content and failure to observe a cooperative melting transition seen for NEMO(110–195), as well as the reduction in coiled-coil content, may not be reflective of the structure of this region of the protein as it exists in the context of full-length NEMO.
Analysis of IVD Solution Structure and IKKβ-Induced Conformational Change by SAXS
To probe the structure of NEMO(44–195) in solution, SAXS was employed as described above for 5XAla-NEMO. Experimental scattering is shown in Figure 5A, with overlaid theoretical scattering calculated from the shape reconstruction model. The fit is reasonable, with χ = 0.3. Although there is no evidence for aggregation, the Guinier plot (Figure 5B) is not linear over q2 = 0.0008 Å−2. As stated (vide infra) a linear Guinier region is not necessarily expected for extended proteins. The P(r) function for NEMO(44–195) is shown in Figure 5C (total quality estimate of 0.807), and the shape reconstruction in Figure 5D. The notable consistency between the shape reconstruction of the NEMO(44–195) fragment and the corresponding region of full-length 5XAla-NEMO (Figure 5E) shows that the conformation of the IBD-IVD region within full-length NEMO is well represented by the shorter construct NEMO(44–195).
Figure 5.

5XAla-NEMO and WT-NEMO become more compact upon IKKβ-binding. (A) SAXS scattering curves for 5XAla-NEMO, WT-NEMO, and NEMO(44–195), with and without IKKβ(701–745). Data for unliganded NEMO(44–195) and WT and NEMO(44–195), complexed with IKKβ in a 1:1.2 ratio and subjected to SEC-SAXS. Theoretical scattering curves calculated from the resulting structural models for the unbound proteins are overlaid in red. (B) Guinier plots for the data sets shown in (A). (C) Distance-distribution functions generated by GNOM for the data sets shown in (A). (D) Shape reconstruction for unbound NEMO(44–195) generated by DAMMIF. (E) Shape reconstruction for NEMO(44–195) in orange superposed on the shape reconstruction for 5XAla-NEMO (gray), from Figure 2.
A previous study16 showed changes in both the intrinsic tryptophan fluorescence of NEMO (via Trp6) and in ANS fluorescence intensity upon addition of an 11-mer peptide comprising the minimal NEMO Binding Domain (NBD) of IKKβ, leading the authors to suggest that NEMO undergoes a ligand-induced conformational change upon binding IKKβ. To further investigate this proposed conformational change, we used SAXS to determine whether the size or shape of WT-NEMO, 5XAla-NEMO, or of NEMO(44–195) changes upon addition of the IKKβ(701–745) peptide, which comprises the region of IKKβ required for high binding affinity to NEMO (the native IKKβ oligomeric state is dimeric56). The scattering curves, Guinier plots, and P(r) functions for WT-NEMO, 5XAla-NEMO, and NEMO(44–195) with and without IKKβ(701–745) are shown in Figure 5A–C, and corresponding concentration studies for complexes of WT-NEMO and NEMO(44–195) with IKKβ(701–745) are shown in Figure S4. Notably, the P(r) function for 5XAla-NEMO (with IKKβ, total quality estimate of 0.580) indicates that upon binding IKKβ NEMO condenses in structure. Specifically, Dmax for 5XAla-NEMO decreases from 315 to 255 Å. For NEMO(44–195), (with IKKβ, total quality estimate of 0.591) Dmax shows no significant change in the absence and presence of IKKβ (93 and 95 Å, respectively). Importantly Dmax represents the maximum distance between atoms in the scattering mass, and thus reflects a property of the largest scattering particle. Therefore, addition of a smaller peptide should not affect Dmax except by inducing a change in the conformation of NEMO. Although one might expect the Rg value to change with addition of a small peptide in excess, the Rg of the unstructured peptide is much smaller than the Rg of the NEMO construct, such that the scattering from the unbound peptide acts as a nearly flat background in the region of q used to estimate the Rg of the protein. To ensure that that the results were not being affected by a high molar ratio of free IKKβ in solution, WT-NEMO was incubated with a 1.2 molar excess of IKKβ(701–745), followed by separation via online SEC-SAXS to eliminate any free IKKβ as well as removing any aggregates. This treatment yielded a narrow peak upon UV absorption detection. The Dmax decreased from 305 to 256 Å for WT-NEMO in the absence and presence of IKKβ peptide. In addition, the Rg (averaged square root distance of each scatterer to the center of mass50,51) calculated from the P(r) plot decreased significantly from 101 to 82 Å from the addition of the IKKβ peptide to WT-NEMO. The resolution of SAXS is not sufficient to allow conclusions about the detailed atomic structure of the complex. However, these solution scattering results clearly indicate that binding of IKKβ peptide causes NEMO to adopt a conformation that is more compact than that seen for unbound NEMO as demonstrated by a reduction in the Rg and length of the maximum distance vector in the scattering mass.
Effect of IVD Mutations on Downstream Signaling in Cells
To assess the importance of the IVD for NEMO activity in vivo, we first compared the ability of WT-, 7XAla-, and 9SG-NEMO to interact with endogenous IKKβ in cells. The 7XAla-NEMO was included because 9SG-NEMO was created in the 7XAla-NEMO background. The 5XAla-NEMO was used in the described biophysical experiments; however, 7XAla NEMO has been shown19 to behave identically in terms of binding and cell signaling activity (see Table S2). Human 293T cells were transfected with overexpression vectors for FLAG-tagged versions of WT-NEMO, 7XAla-NEMO, or 9SG-NEMO, and the binding of these proteins to endogenous IKKβ was evaluated in anti-FLAG co-immunoprecipitation (Co-IP) assays. Similar amounts of IKKβ were co-precipitated with all three NEMO proteins (Figure 6A). No IKKβ was co-precipitated using extracts from cells transfected with the empty vector control. The ability of 9SG-NEMO to pull down IKKβ to a similar extent as WT-NEMO is consistent with the observation that the 9SG variant protein retained the ability to bind IKKβ in vitro.
As a first test of whether the IVD plays a role in the ability of NEMO to mediate NF-κB pathway signaling, we assessed the ability of transiently transfected 9SG-NEMO to restore TNFα-induced phosphorylation of IκBα in a 293T cell line in which NEMO expression had been disrupted by CRISPR/Cas9 targeting (Figure 6B). As compared to 7XAla-NEMO, 9SG-NEMO had a reduced ability to restore TNFα-dependent phosphorylation of IκBα (Figure 6C).
As a more extensive test of 9SG-NEMO function, we used retroviral transduction to stably reconstitute mouse NEMO knockout cells with 7XAla- or 9SG-NEMO. We have previously shown that this method expresses biologically relevant levels of NEMO protein and can be used to test NEMO activity19,46. Knockout cells reconstituted with 7XAla-NEMO showed easily detectable phosphorylation of IκBα upon treatment with three known NF-κB pathway activators, TNFα, LPS, or IL-1β (Figure 6D). In contrast, there was little to no induction of IκBα phosphorylation in cells reconstituted with 9SG-NEMO or the empty vector control. Similar levels of NEMO were expressed in all cells. To ensure that the lack of activity of the 9SG variant was not due to the 7XAla background, we also created the 9SG mutation in the WT-NEMO background. As shown in Figure S5, the 9SG variant in the WT-NEMO background was also defective for supporting TNFα-induced phosphorylation of IκBα in mouse NEMO knockout cells, whereas WT-NEMO restored TNFα-dependent phosphorylation of IκBα in these cells.
Taken together, these results show that disruption of the highly conserved core IVD sequence by the 9SG mutation does not abolish the ability of NEMO to interact with IKKβ in vitro or in vivo, but does disrupt the ability of NEMO to support activation of IKK in human and mouse cells in response to a variety of upstream inducers of NF-κB signaling.
Effect of the IVD Mutation on IKKβ-Induced Conformational Change Measured by SAXS
Our cell-based reconstitution experiments showed that the IVD is required for NEMO-dependent activation of IKK in vivo. Although the in vitro binding affinity of the 9SG variant of NEMO for IKKβ is unchanged, there is a loss of signaling function in vivo. To investigate whether the signaling defect in 9SG-NEMO might be due to an inability to undergo the proposed ligand-induced conformational change, we analyzed the 9SG-NEMO variant by SAXS in the presence and absence of the IKKβ(701–745) peptide. The scattering curves, Guinier plots, and P(r) plots for these conditions are shown in Figure 7. Unlike 5XAla-NEMO and WT-NEMO, 9SG-NEMO solution scattering is not consistent with conversion to a more compact form upon binding the IKKβ peptide, as shown by the P(r) functions (total quality estimate = 0.529 and 0.620, respectively). In fact, 9SG-NEMO appears to elongate upon IKKβ peptide binding, from Dmax = 250 to 280 Å. The apparent lengthening of 9SG-NEMO upon IKKβ peptide binding may reflect an overall rigidification of 9SG-NEMO, which increases order and thereby allows more protein mass to be resolved against the solvent. In any case, upon binding of IKKβ peptide, 9SG-NEMO fails to take on the more compact form observed with both WT- and 5XAla-NEMO, suggesting that the 9SG mutation in the IVD domain eliminates a ligand-induced conformational change.
Figure 7.
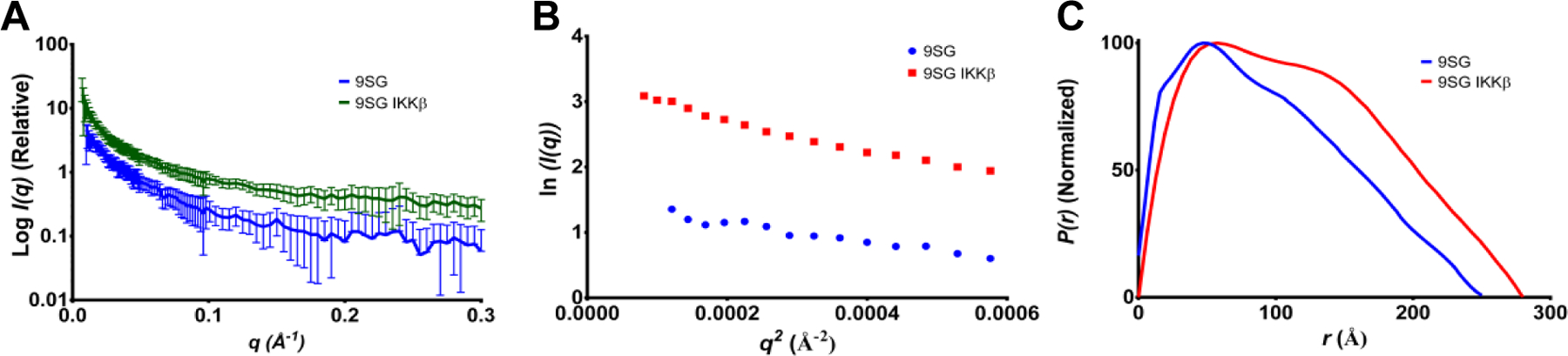
The IVD-region mutations present in 9SG-NEMO abolish the ability of IKKβ(701–745) binding to induce a more compact conformation. (A) SAXS scattering curve for 9SG-NEMO with (green) and without (blue) IKKβ(701–745). (B) Guinier plot for 9SG-NEMO. Linearity at low q is not necessarily expected for a partially flexible, extended protein50,51. (C) Distance-distribution function generated by GNOM.
DISCUSSION
Here, we show that a previously uncharacterized central region of NEMO, which we term the Intervening Domain (IVD), is required for high-affinity binding to IKKβ, confers thermal stability on NEMO, and is necessary for downstream activation of NF-κB signaling in vivo. Moreover, we show that introduction of the 9SG mutation in a highly conserved core region of the IVD sequence disrupts both the induced conformational change in vitro and NEMO’s ability to support phosphorylation of IκBα by IKKβ in vivo. Taken together, these results lead us to propose that the IVD plays a role in a signal-induced conformational change in NEMO that is required for downstream signaling. Therefore, our results support the notion that NEMO is an allosterically regulated scaffold protein.
A functional role for the IVD is supported by the finding that several mutations associated with genetic diseases occur in or near the IVD, including D113N, R123W, L153R, R173G, R175P and Q183H11,31. These mutations do not significantly reduce NEMO expression57–61, and thus likely exert their effects by altering NEMO function. The last residue of the targeted region in the 9SG variant (i.e., L153) is the site of an EDA-ID (ectodermal dysplasia with immunodeficiency)-associated mutation (L153R) which has been shown to render NEMO defective for NF-κB signaling59. Among the other disease-associated mutations within the IVD, D113N, R123W, L170P, R173G, R182P and Q183H are associated with Incontinentia Pigmenti (IP), Q157P with anhidrotic ectodermal dysplasia with immunodeficiency, osteopetrosis, and lymphedema (OL-EDA-ID), and A169P and R175P with EDA-ID31.
Several previous studies have tested the ability of disease-associated NEMO mutants to propagate NF-κB signaling12,62,63. Many of these disease-associated NEMO variants remain functional in response to at least one stimulus of canonical NF-κB signaling. In this study, we show that substitution of a strictly conserved nine-residue segment at the core of the IVD with a series of nine serine and glycine residues (i.e., the 9SG mutation) abolishes the ability of NEMO to function in canonical signaling in response to TNFα, LPS or IL-1β. Therefore, it is likely that the 9SG mutation disrupts a fundamental property of NEMO, such as the ability to undergo a productive ligand-induced conformational change. In support of this hypothesis, using SAXS we identified a ligand-induced conformational change that occurs in WT-NEMO but not 9SG-NEMO. The conformational change in NEMO may serve to bring IKKβ, which binds towards the N-terminal end of NEMO, into close proximity with IκBα, which binds at the C-terminal end30, thereby directing the kinase activity of IKKβ toward its specific substrate. Previously, the allosteric activation of binding partners by scaffolds has been shown by demonstrating an increase in activity6,9,64. Here, we present structural evidence suggesting how scaffold function occurs in NEMO.
There is no reported X-ray crystal structure of any fragment of NEMO that includes the IVD region. Thus, the structural details of how the IVD modulates IKKβ binding affinity and mediates the proposed ligand-dependent conformational change remain unclear. However, the observation that NEMO(44–195) – comprising only the IKKβ binding domain plus the adjacent IVD – shows a single cooperative melting transition indicates the existence of an intimate structural connection between the IVD and IBD. This structural inter-dependence is consistent with the existence of allosteric communication between these domains. The 9SG mutation has no effect on IKKβ binding in vitro or in co-IP experiments using human cell extracts. Thus, we propose that the abolition of the signaling function in cells resulting from the 9SG mutation is because the IVD is required for a critical conformational change in NEMO.
A previous study reported evidence, based on intrinsic tryptophan fluorescence and ANS fluorescence, that NEMO undergoes a ligand-induced conformational change16. Our results suggest that the IVD plays an important role in this conformational change, and in the overall signaling function of NEMO. We propose a model in which the IVD is required for a conformational change in NEMO that is necessary for its ability to direct phosphorylation of IκBα by IKKβ. The concept of a conformational change requirement for signaling propagation by a scaffold protein has precedence. For example, the yeast scaffold Ste5 uses a conformational change as a signaling gate to expose the binding surface on Fus3 to propagate a mating-specific signal10. However, whereas Ste5 requires cofactor binding to relieve an auto-inhibitory interaction and reveal a binding surface on Fus3 that leads to propagation of signaling, the binding surface of NEMO is constitutively available for the catalytic component of the IKK complex, IKKβ. In the case of NEMO, the binding of IKKβ triggers a decrease in the maximum length of the protein, as measured by SAXS, consistent with the need to bring the N-terminal region of NEMO with bound IKKβ close to its C-terminal region where IκB binds. Other NEMO ligands may also affect the conformation of NEMO. In particular, it has been reported that noncovalent binding of poly-ubiquitin affects the rate of a ligand-induced conformational change of NEMO16.
A recent study has suggested that the region comprising residues 112–150, encompassing part of the IVD, is a site of negative regulation of NEMO17. The authors proposed that this region can participate in an intramolecular interaction with the HLX2 region to maintain NEMO in an auto-inhibited, compact state prior to poly-ubiquitin binding. Their model is based, in part, on in vitro measurements showing that, whereas di-ubiquitin bound to truncated NEMO constructs lacking the 112–150 region, it did not bind to full-length NEMO. However, M1-linked poly-ubiquitin was able to bind to full-length NEMO, suggesting that this more extended form of poly-ubiquitin can outcompete the self-inhibition and convert NEMO to an open, active conformation. The authors also determined a SAXS model for full-length NEMO bound to an MBP-tagged herpesvirus FLICE inhibitory protein (vFLIP) that, like our unliganded full-length model, shows a shorter total length (c.a. 320 Å) than expected for a fully extended coiled-coil protein. These authors accounted for the shorter than expected length of their NEMO/MBP-vFLIP SAXS structure by postulating that, in the unbound state, NEMO is folded back on itself, consistent with their proposed autoinhibitory intramolecular self-association model. However, the ability to directly observe such a shape in their SAXS structure was affected by the presence of the large MBP-vFLIP ligand, a polypeptide of over 500 aa, which gives the overall molecular envelope a comma shape, and makes it difficult to assess the shape of NEMO itself within this large envelope.
The SAXS structure of unbound, full length NEMO that we describe here is not consistent with NEMO being extensively folded back on itself, as the molecular envelope is wide enough to accommodate only a single coiled-coil for most of its length. Instead, we attribute the shorter than expected length of NEMO to the adoption of a compact, folded structure in the IVD region (as opposed to a fully-extended coiled-coil). Moreover, our results indicate that the function of the IVD is clearly not limited to auto-inhibition; if this were the case then mutations introduced to the IVD would result in a gain of function, whereas our results show that the 9SG mutation in the IVD results in NEMO loss of function. There are multiple events required for full NEMO scaffolding activity, including IKKβ binding, poly-ubiquitin binding, and phosphorylation. It is possible, therefore, that the IVD is involved in auto-inhibition as well as in mediating a conformational change that serves other important functions, such as bringing the IKKβ bound near the N terminus of NEMO into proximity of the IκBα bound near the C terminus.
In summary, we have shown that the highly conserved Intervening Domain of NEMO is required for downstream activation of IKKβ, which is required for canonical NF-κB signaling. An approach analogous to the one taken herein may be useful for identifying regulatory regions of other scaffold proteins. The study of undruggable targets may benefit from this approach, by enabling the identification of regions that can be targeted with small molecules to modulate function by affecting the target’s ability to undergo an essential conformational change. SAXS is a powerful tool to probe these ligand-induced conformational changes. A high-throughput application of SAXS might be useful in identifying therapeutics that alter either unliganded- or liganded-conformational states of a target protein. Scaffold proteins are a class of target proteins that could benefit from this approach, as direct functional assays are often difficult to develop for scaffolds due to their lack of intrinsic catalytic activity.
Supplementary Material
Scheme 1.

Simplified scheme illustrating how NEMO mediates the phosphorylation of IκB by IKKα/β in response to activation of an upstream receptor (in this example, TNFR1), leading to ubiquitination and degradation of IκB, releasing NF-κB to translocate to the nucleus where it stimulates expression of target genes. Abbreviations: TNF, tumor necrosis factor; TNFR1, p55 TNF receptor; TRAF6, TNF receptor associated factor 6; RIP, receptor-interacting serine/threonine-protein kinase 1; TAK1, TGF β activated kinase 1, a.k.a. mitogen-activated protein kinase kinase kinase 7; p50/p65, a dimer comprising one form of the transcription factor NF-κB; IκB, inhibitor of κB; NEMO, NF-κB essential modulator; IKKα/β, inhibitor of κB kinase α and β; Ub, ubiquitin. The red asterisks represent sites of phosphorylation.
ACKNOWLEDGMENTS
This research was supported by NIH grant GM117350 to A.W., T.D.G., and K.N.A. L.K., Y.L., and M.B. received funding from the Boston University Undergraduate Research Opportunities Program. S.P. was supported by NSF-REU awards BIO-1262934 and CHE-1156666. The CD spectrometer was purchased under NSF grant CHE1126545. We thank the staff at the X9 and LiX beamlines at Brookhaven National Laboratory (BNL) and at 4–2 at Stanford Synchrotron Radiation Light Source (SSRL) for assistance with SAXS experiments. Use of SSRL is supported by US Department of Energy (DOE) Contract No. DE-AC02–76SF00515. Use of BNL is supported by NIH grant P41GM111244, DOE Office of Biological and Environmental Research (KP1605010), DOE Contact No. DE-SC0012704 and DE-AC02–98CH10886.
Abbreviations
- aa
amino-acid residues
- CD
circular dichroism
- DTT
dithiothreitol
- FA
fluorescence anisotropy
- FITC
fluorescein isothiocyanate
- FITC-IKKβ
IKKβ(701−745)-derived peptide labeled with fluorescein isothiocyanate
- HLX2
second coiled coil domain of NEMO
- IκB
inhibitor of κB
- IKK
inhibitor of κB kinase
- IBD
IKKβ binding domain
- IVD
intervening domain
- CoZi
leucine zipper
- NEMO
NF-κB essential modulator
- NF-κB
nuclear factor κB
- RANKL
RANK ligand
- TNFα
tumor necrosis factor α
- TCEP
tris(2-carboxyethyl)phosphine
- ZF
zinc finger
Footnotes
Supporting Information.
Supplemental Table S1 showing summary SAXS results, supplemental Table S2 comparing full-length NEMO constructs, and supplementary figures showing theoretical SAXS profiles, multiple sequence alignment of NEMO orthologs, CD spectra of NEMO constructs, additional plots for SAXS concentration series, comparison of SAXS models generated by alternative algorithms, and comparison of ability of WT-NEMO and 9SG-NEMO in the wild-type background to promote activation of IKKβ.
REFERENCES
- (1).Shaw AS, and Filbert EL (2009) Scaffold proteins and immune-cell signalling. Nat Rev Immunol 9, 47–56. [DOI] [PubMed] [Google Scholar]
- (2).Cortese MS, Uversky VN, and Keith Dunker A (2008) Intrinsic disorder in scaffold proteins: Getting more from less. Prog Biophys Mol Biol 98, 85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pan CQ, Sudol M, Sheetz M, and Low BC (2012) Modularity and functional plasticity of scaffold proteins as p(l)acemakers in cell signaling. Cell Signal 24, 2143–2165. [DOI] [PubMed] [Google Scholar]
- (4).Dhanasekaran DN, Kashef K, Lee CM, Xu H, and Reddy EP (2007) Scaffold proteins of MAP-kinase modules. Oncogene 26, 3185–3202. [DOI] [PubMed] [Google Scholar]
- (5).Vaquero J, Nguyen Ho-Bouldoires TH, Clapéron A, and Fouassier L (2017) Role of the PDZ-scaffold protein NHERF1/EBP50 in cancer biology: from signaling regulation to clinical relevance. Oncogene 3067–3079. [DOI] [PubMed] [Google Scholar]
- (6).Pawson T (1995) Protein modules and signalling networks. Nature 373, 573–580. [DOI] [PubMed] [Google Scholar]
- (7).Good MC, Zalatan JG, and Lim WA (2011) Scaffold Proteins: Hubs for Controlling the Flow of Cellular Information. Science 332, 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dai Z, Kim JH, Tonelli M, Ali IK, and Markley JL (2014) pH-Induced Conformational Change of IscU at Low pH Correlates with Protonation/Deprotonation of Two Conserved Histidine Residues. Biochemistry 5290–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Brennan DF, Dar AC, Hertz NT, Chao WCH, Burlingame AL, Shokat KM, and Barford D (2011) A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369. [DOI] [PubMed] [Google Scholar]
- (10).Zalatan JG, Coyle SM, Rajan S, Sidhu S, and Lim WA (2012) Conformational Control of the Ste5 Scaffold Protein Insulates Against MAP Kinase Misactivation. Science 337, 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Courtois G, and Gilmore TD (2006) Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene 25, 6831–6843. [DOI] [PubMed] [Google Scholar]
- (12).Maubach G, Schmädicke A-C, and Naumann M (2017) NEMO Links Nuclear Factor-κB to Human Diseases. Trends Mol Med 23, 1138–1155.29128367 [Google Scholar]
- (13).Hayden MS, and Ghosh S (2008) Shared Principles in NF-κB Signaling. Cell 132, 344–362. [DOI] [PubMed] [Google Scholar]
- (14).Ghosh G, Wang VY-F, Huang D-B, and Fusco A (2012) NF-κB regulation: lessons from structures. Immunol Rev 246, 36–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rudolph D, Yeh W-C, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, and Mak TW (2000) Severe liver degeneration and lack of NF-kB activation in NEMO/IKKg-deficient mice. Genes Dev 14, 854–862. [PMC free article] [PubMed] [Google Scholar]
- (16).Catici DAM, Horne JE, Cooper GE, and Pudney CR (2015) Poly-ubiquitin drives the molecular interactions of NF-κB essential modulator by allosteric regulation. J. Biol Chem 290, 14130–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hauenstein AV, Xu G, Kabaleeswaran V, and Wu H (2017) Evidence for M1-Linked Polyubiquitin-Mediated Conformational Change in NEMO. J Mol Biol 429, 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ivins FJ, Montgomery MG, Smith SJM, Morris‑Davies AC, Taylor IA, and Rittinger K (2009) NEMO oligomerization and its ubiquitin-binding properties. Biochem J 421, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cote SM, Gilmore TD, Shaffer R, Weber U, Bollam R, Golden MS, Glover K, Herscovitch M, Ennis T, Allen KN, and Whitty A (2013) Mutation of Nonessential Cysteines Shows That the NF-κB Essential Modulator Forms a Constitutive Noncovalent Dimer That Binds IκB Kinase-β with High Affinity. Biochemistry 52, 9141–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Rushe M, Silvian L, Bixler S, Chen LL, Cheung A, Bowes S, Cuervo H, Berkowitz S, Zheng T, Guckian K, Pellegrini M, and Lugovskoy A (2008) Structure of a NEMO/IKK-Associating Domain Reveals Architecture of the Interaction Site. Structure 16, 798–808. [DOI] [PubMed] [Google Scholar]
- (21).Bagnéris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, Waksman G, and Barrett T (2008) Crystal Structure of a vFlip-IKKγ Complex: Insights into Viral Activation of the IKK Signalosome. Mol Cell 30, 620–631. [DOI] [PubMed] [Google Scholar]
- (22).Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, and Dikic I (2009) Specific Recognition of Linear Ubiquitin Chains by NEMO Is Important for NF-κB Activation. Cell 136, 1098–1109. [DOI] [PubMed] [Google Scholar]
- (23).Lo Y-C, Lin S-C, Rospigliosi CC, Conze DB, Wu C-J, Ashwell JD, Eliezer D, and Wu H (2009) Structural Basis for Recognition of Diubiquitins by NEMO. Mol Cell 33, 602–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, and Fukai S (2009) Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett 583, 3317–3322. [DOI] [PubMed] [Google Scholar]
- (25).Grubisha O, Kaminska M, Duquerroy S, Fontan E, Cordier F, Haouz A, Raynal B, Chiaravalli J, Delepierre M, Israël A, Véron M, and Agou F (2010) DARPin-Assisted Crystallography of the CC2-LZ Domain of NEMO Reveals a Coupling between Dimerization and Ubiquitin Binding. J Mol Biol 395, 89–104. [DOI] [PubMed] [Google Scholar]
- (26).Fujita H, Rahighi S, Akita M, Kato R, Sasaki Y, Wakatsuki S, and Iwai K (2014) Mechanism Underlying I B Kinase Activation Mediated by the Linear Ubiquitin Chain Assembly Complex. Mol Cell Biol 34, 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cordier F, Vinolo E, Véron M, Delepierre M, and Agou F (2008) Solution Structure of NEMO Zinc Finger and Impact of an Anhidrotic Ectodermal Dysplasia with Immunodeficiency-related Point Mutation. J Mol Biol 377, 1419–1432. [DOI] [PubMed] [Google Scholar]
- (28).Zhou L, Yeo AT, Ballarano C, Weber U, Allen KN, Gilmore TD, and Whitty A (2014) Disulfide-Mediated Stabilization of the IκB Kinase Binding Domain of NF-κB Essential Modulator (NEMO). Biochemistry 53, 7929–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).May MJ, D’Acquisto F, Madge LA, Glockner J, Pober JS, and Ghosh S (2000) Selective Inhibition of NF-kB Activation by a Peptide That Blocks the Interaction of NEMO with the IκB Kinase Complex. Science 289, 1550–1554. [DOI] [PubMed] [Google Scholar]
- (30).Schröfelbauer B, Polley S, Behar M, Ghosh G, and Hoffmann A (2012) NEMO Ensures Signaling Specificity of the Pleiotropic IKKβ by Directing Its Kinase Activity toward IκBα. Mol. Cell 47, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Senegas A, Gautheron J, Maurin AGD, and Courtois G (2015) IKK-related genetic diseases: probing NF-κB functions in humans and other matters. Cell. Mol. Life Sci 72, 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Golden MS, Cote SM, Sayeg M, Zerbe BS, Villar EA, Beglov D, Sazinsky SL, Georgiadis RM, Vajda S, Kozakov D, and Whitty A (2013) Comprehensive Experimental and Computational Analysis of Binding Energy Hot Spots at the NF-κB Essential Modulator/IKKβ Protein–Protein Interface. J. Am. Chem. Soc 135, 6242–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Greenfield NJ (2007) Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc 1, 2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Greenfield NJ (2007) Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc 1, 2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kwok SC, and Hodges RS (2004) Stabilizing and Destabilizing Clusters in the Hydrophobic Core of Long Two-stranded α-Helical Coiled-coils. J. Biol. Chem 279, 21576–21588. [DOI] [PubMed] [Google Scholar]
- (36).Allaire M, and Yang L (2011) Biomolecular solution X-ray scattering at the National Synchrotron Light Source. J Synchrotron Radiat 18, 41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, and Svergun DI (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr 45, 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25, 493–503. [Google Scholar]
- (39).Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2013) Accurate SAXS Profile Computation and its Assessment by Contrast Variation Experiments. Biophys J 105, 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2016) FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res 44, W424–W429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Svergun DI (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Franke D, and Svergun DI (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr 42, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Volkov VV, and Svergun DI (2003) Uniqueness of ab-initio shape determination in small-angle scattering. J Appl Crystallogr 36, 860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (45).Petoukhov MV, and Svergun DI (2005) Global Rigid Body Modeling of Macromolecular Complexes against Small-Angle Scattering Data. Biophys J 89, 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Herscovitch M, Comb W, Ennis T, Coleman K, Yong S, Armstead B, Kalaitzidis D, Chandani S, and Gilmore TD (2008) Intermolecular disulfide bond formation in the NEMO dimer requires Cys54 and Cys347. Biochem Biophys Res Commun 367, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Liang M-C, Bardhan S, Pace EA, Rosman D, Beutler JA, Porco JA, and Gilmore TD (2006) Inhibition of transcription factor NF-κB signaling proteins IKKβ and p65 through specific cysteine residues by epoxyquinone A monomer: Correlation with its anti-cancer cell growth activity. Biochem Pharmacol 71, 634–645. [DOI] [PubMed] [Google Scholar]
- (48).Babaei M, Liu Y, Wuerzberger-Davis SM, Yeo AT, Kagermazova L, McCaslin EZ, Miyamoto S, and Gilmorea TD (2018) CRISPR/Cas9-based Editing of a Sensitive Transcriptional Regulatory Element to Achieve Cell Type-Specific Knockdown of the NEMO Scaffold Protein. bioRxiv 450320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gapuzan M-ER, Pitoc GA, and Gilmore TD (2003) Mutations within a conserved protein kinase A recognition sequence confer temperature-sensitive and partially defective activities onto mouse c-Rel. Biochem Biophys Res Commun 307, 92–99. [DOI] [PubMed] [Google Scholar]
- (50).Putnam CD, Hammel M, Hura GL, and Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40, 191–285. [DOI] [PubMed] [Google Scholar]
- (51).Roig-Solvas B, Brooks D, and Makowski L (2019) A direct approach to estimate the anisotropy of protein structures from small-angle X-ray scattering. J Appl Cryst 52, 274–283. [Google Scholar]
- (52).Tegethoff S, Behlke J, and Scheidereit C (2003) Tetrameric Oligomerization of I B Kinase (IKKβ) Is Obligatory for IKK Complex Activity and NF- B Activation. Mol Cell Biol 23, 2029–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Guo B, Audu CO, Cochran JC, Mierke DF, and Pellegrini M (2014) Protein Engineering of the N-Terminus of NEMO: Structure Stabilization and Rescue of IKKβ Binding. Biochemistry 53, 6776–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Luidens MK, Figge J, Breese K, and Vajda S (1995) Predicted and Trifluoroethanol-Induced a-Helicity of Polypeptides. Biopolymers 39, 367–376. [DOI] [PubMed] [Google Scholar]
- (55).Luo P, and Baldwin RL (1997) Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry 36, 8413–8421. [DOI] [PubMed] [Google Scholar]
- (56).Liu S, Misquitta YR, Olland A, Johnson MA, Kelleher KS, Kriz R, Lin LL, Stahl M, and Mosyak L (2013) Crystal Structure of a Human IκB Kinase Asymmetric Dimer. J. Biol. Chem 288, 22758–22767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Salt BH, Niemela JE, Pandey R, Hanson EP, Deering RP, Quinones R, Jain A, Orange JS, and Gelfand EW (2008) IKBKG (nuclear factor-κB essential modulator) mutation can be associated with opportunistic infection without impairing Toll-like receptor function. J Allergy Clin Immunol 121, 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Fusco F (2004) Molecular analysis of the genetic defect in a large cohort of IP patients and identification of novel NEMO mutations interfering with NF-κB activation. Hum Mol Genet 13, 1763–1773. [DOI] [PubMed] [Google Scholar]
- (59).Hanson EP, Monaco-Shawver L, Solt LA, Madge LA, Banerjee PP, May MJ, and Orange JS (2008) Hypomorphic nuclear factor-κB essential modulator mutation database and reconstitution system identifies phenotypic and immunologic diversity. J Allergy Clin Immunol 122, 1169–1177.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ku C-L, Picard C, Erdos M, Jeurissen A, Bustamante J, Puel A, von Bernuth H, Filipe-Santos O, Chang H-H, Lawrence T, Raes M, Marodi L, Bossuyt X, and Casanova J-L (2006) IRAK4 and NEMO mutations in otherwise healthy children with recurrent invasive pneumococcal disease. J. Med. Genet 44, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Hsiao P-F, Lin S-P, Chiang S-S, Wu Y-H, Chen H-C, and Lin Y-C (2010) NEMO gene mutations in Chinese patients with incontinentia pigmenti. J Formos Med Assoc 109, 192–200. [DOI] [PubMed] [Google Scholar]
- (62).Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israël A, Heiss NS, Klauck SM, Kioschis P, Wiemann S, Poustka A, Esposito T, Bardaro T, Gianfrancesco F, Ciccodicola A, D’Urso M, Woffendin H, Jakins T, Donnai D, Stewart H, Kenwrick SJ, Aradhya S, Yamagata T, Levy M, Lewis RA, and Nelson DL (2000) Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature 405, 466–472. [DOI] [PubMed] [Google Scholar]
- (63).Haverkamp MH, Marciano BE, Frucht DM, Jain A, van de Vosse E, and Holland SM (2014) Correlating Interleukin-12 Stimulated Interferon-γ Production and the Absence of Ectodermal Dysplasia and Anhidrosis (EDA) in Patients with Mutations in NF-κB Essential Modulator (NEMO). J Clin Immunol 436–443. [DOI] [PubMed] [Google Scholar]
- (64).Jeong J, VanHouten JN, Kim W, Dann P, Sullivan C, Choi J, Sneddon WB, Friedman PA, and Wysolmerski JJ (2017) The scaffolding protein NHERF1 regulates the stability and activity of the tyrosine kinase HER2. J. Biol Chem 292, 6555–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


