Significance
Brain metastasis with current limited treatment options is a common complication in advanced cancer patients, and breast-to-brain metastasis (B2BM) is one of the major types. In this work, we report that brain metastasis oncogenic long noncoding RNA (BMOR) is a key brain-enriched long noncoding RNA for the development of B2BM. We demonstrate that BMOR allows B2BM cells to colonize the brain tissue by evading immune-mediated killing in the brain microenvironment. At the molecular level, BMOR binds and inactivates IRF3 in B2BM cells. Finally, BMOR silencer can effectively suppress the development of brain metastasis in vivo. Therefore, our findings reveal a way in which cancer cells evade immune-mediated killing in the brain microenvironment for brain metastasis development and establish therapeutic targets with potential targeted strategies against B2BM.
Keywords: brain metastasis, lncRNA, immune evasion, BMOR, IRF3
Abstract
Brain metastases, including prevalent breast-to-brain metastasis (B2BM), represent an urgent unmet medical need in the care of cancer due to a lack of effective therapies. Immune evasion is essential for cancer cells to metastasize to the brain tissue for brain metastasis. However, the intrinsic genetic circuits that enable cancer cells to avoid immune-mediated killing in the brain microenvironment remain poorly understood. Here, we report that a brain-enriched long noncoding RNA (BMOR) expressed in B2BM cells is required for brain metastasis development and is both necessary and sufficient to drive cancer cells to colonize the brain tissue. Mechanistically, BMOR enables cancer cells to evade immune-mediated killing in the brain microenvironment for the development of brain metastasis by binding and inactivating IRF3. In preclinical brain metastasis murine models, locked nucleic acid-BMOR, a designed silencer targeting BMOR, is effective in suppressing the metastatic colonization of cancer cells in the brain for brain metastasis. Taken together, our study reveals a mechanism underlying B2BM immune evasion during cancer cell metastatic colonization of brain tissue for brain metastasis, where B2BM cells evade immune-mediated killing in the brain microenvironment by acquiring a brain-enriched long noncoding RNA genetic feature.
Breast cancer, the most prevalent cancer type in humans, is one of the most common causes of brain metastases (1, 2). However, for patients with brain metastases, including breast-to-brain metastasis (B2BM), treatment options beyond radiotherapy and neurological surgery are limited (3). The estimated median survival time of patients with B2BM is only ∼4–6 mo (4). These issues underscore the need for research into the new mechanisms underlying B2BM to establish new therapeutic targets and new effective therapies against B2BM. However, the underlying biology and molecular basis of B2BM are far from being clearly understood. In particular, it is largely unknown how B2BM cells can avoid immune-mediated killing in the brain microenvironment to colonize the brain tissue for brain metastasis.
As B2BM has great clinical significance, quite a few protein-coding genes have been identified to contribute to the development of B2BM (5). In comparison to protein-coding genes, long noncoding RNAs (lncRNAs), defined as transcripts greater than 200 nt in length but without the potential to encode proteins, are much more abundant throughout the human genome (6). LncRNAs are becoming attractive potential therapeutic targets for cancer treatment due to their important roles in cancer development and progression (7). However, only countable lncRNAs, such as X-inactive-specific transcript (XIST) (8) and lncRNA associated with BCBM (lnc-BM) (9), have been shown to contribute to the development of B2BM via different mechanisms. Lnc-BM promotes B2BM by regulating the production of interleukin (IL)-6 and recruiting macrophages (9), and loss of lncRNA XIST promotes B2BM by activating MSN-c-Met and reprogramming microglia (8). Although limited, these previously characterized B2BM-related lncRNAs implicate the potential important roles of lncRNAs in B2BM. Tissue-specific genes or tissue-enriched genes are a group of genes that preferentially display enriched expression in one or several tissues and are thought to be important in mechanisms underlying human genetic traits and diseases (10). Several brain-specific protein-coding genes were found to be expressed in B2BM cells to promote brain-specific/preference metastasis, such as ST6GALNAC5 (11) and neuronal GluN2B (12). Although lncRNAs are found to be more likely to exhibit tissue-specific expression patterns than protein-coding genes (13), not a brain-specific or brain-enriched lncRNA has been identified to be acquired by metastatic cells for their brain metastasis behaviors. It is largely unknown whether and how the general tissue-specific expression and function of lncRNAs can be linked to the organ-specific tropism of metastasis, such as B2BM.
In this study, we uncovered a brain metastasis promoting lncRNA in B2BM cells, designated brain metastasis oncogenic lncRNA (BMOR). BMOR is a brain-enriched lncRNA exhibiting a specifically enriched expression pattern in normal intracranial tissues such as the brain versus extracranial tissues, including the breast. We demonstrate that BMOR is required for the development of brain metastasis and is both necessary and sufficient to promote cancer cells to colonize the brain tissue. Mechanistically, BMOR shields cancer cells from immune-mediated killing in the brain microenvironment by binding and inactivating IRF3. Finally, we show that our designed BMOR silencer, named locked nucleic acid (LNA)-BMOR, can effectively suppress the development of brain metastasis by B2BM cells in vivo. Taken together, this study provides a lncRNA-mediated mechanism underlying the immune evasion of B2BM cells for the development of B2BM and establishes therapeutic targets with potential intervention strategies against B2BM.
Results
BMOR Is a Brain-Enriched lncRNA Required for Brain Metastasis Development.
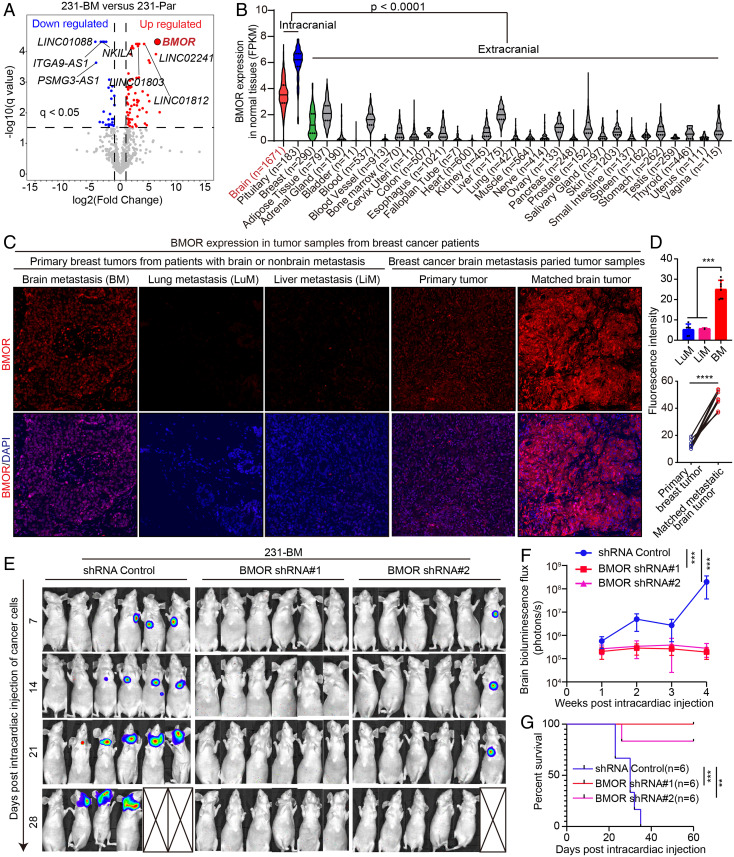
To identify driver lncRNAs for the development of B2BM, we profiled and compared lncRNA expression in B2BM cells (231-BM) and parental non-B2BM MDA-MB-231 cells (231-Par) via RNA-sequencing (RNA-seq). The results revealed a set of differentially expressed lncRNAs, including 277 up-regulated and 262 down-regulated lncRNAs in 231-BM versus 231-Par using the criteria |log2FC| >1 and q < 0.05 (Fig. 1A and Dataset S1), and the ten most differentially expressed lncRNAs were selected for validation by RT–qPCR (SI Appendix, Fig. S1A). The RNA-seq and RT–qPCR results consistently revealed a transcript of the intergenic gene ENSG00000203930, designated BMOR, to be one of the most up-regulated lncRNAs in 231-BM compared with 231-Par, indicating a potential important role of BMOR in brain metastasis development (Fig. 1A and SI Appendix, Fig. S1A and Dataset S1).
Fig. 1.
BMOR is a brain-enriched lncRNA that is required for the development of brain metastasis. (A) A volcano plot represents the expression changes of all lncRNA genes: significantly down- and up-regulated lncRNAs (231-BM vs. 231-Par: |log2 FC| >1 and q < 0.05) are colored blue and red, respectively; LncRNAs that do not show significant expression changes are colored gray; Selected differentially expressed lncRNAs are labeled with gene names, where BMOR as one of the most up-regulated lncRNAs is highlighted in red color. (B) A violin illustration shows BMOR expression in human normal tissues by retrieving and analyzing data generated by the Genotype-Tissue Expression (GTEx) project. (C and D) BMOR expression in tumor samples from breast cancer patients. The BMOR expression level was examined in 32 tumor samples from 24 breast cancer patients by RNA-FISH assays: 24 tumor samples from 16 patients diagnosed with brain metastasis, including 8 primary breast tumor samples from 8 patients and 16 matching primary breast and brain metastatic tumor samples from 8 other patients, and 8 primary breast tumor samples from 8 patients diagnosed with nonbrain metastasis, including 6 tumor samples from 6 patients diagnosed with lung metastasis and 2 tumor samples from 2 other patients diagnosed with liver metastasis. (C) Representative fluorescence images (200×) of RNA-FISH assays: BMOR (red) and nuclear DAPI (blue). (D) Quantification of BMOR expression levels. ***P < 0.001, ****P < 0.0001 (t test). (E–G) Impacts of BMOR on the development of brain metastasis using brain metastasis murine models generated by intracardiac injection of 231-BM cells with BMOR depletion (i.e., BMOR shRNA 1 and 2) compared with those without BMOR depletion (shRNA Control). (E) Bioluminescence images of mice with the indicated treatment over time. (F) Quantification of bioluminescence flux in mice with the indicated treatment over time. ***P < 0.001 (t test). (G) Overall survival of mice with the indicated treatment. Log-rank test: P = 0.0005 for shRNA control versus BMOR shRNA 1 and P = 0.0072 for shRNA control versus BMOR shRNA 2. **P < 0.01, ***P < 0.001.
It is noted that one of the distinctive features of lncRNAs is their highly tissue-specific or tissue-enriched expression patterns (14). Using both in silico and experimental methods, we confirmed that BMOR is a real lncRNA 1,257 nt in length without the potential to encode proteins and shows a predominant distribution in cell nuclei (SI Appendix, Fig. S1 B–G). Interestingly, when interrogating the genotype-issue expression (GTEx) benign tissue RNA-seq dataset (10), we found that in normal tissues, BMOR only displays substantial expression in intracranial tissues, including brain and pituitary, but not in breast and other extracranial tissues, suggesting that BMOR is a brain-enriched lncRNA (Fig. 1B). Impressively, in our collected breast cancer samples analyzed by RNA-fluorescence in situ hybridization (RNA-FISH), we found that BMOR shows markedly elevated expression in breast cancer metastasis to brain versus breast cancer metastasis to nonbrain tissues, including lung and liver (Fig. 1 C and D). Consistently, when we further analyzed the expression profiles of breast tumors with specific tissue metastasis information by retrieving all expression data of breast tumors from the Human Cancer Metastasis Database (15), the results showed that BMOR only displays substantial expression in breast cancer metastasis to the brain compared with breast cancer metastasis to lung, liver, and other nonbrain tissues (SI Appendix, Fig. S2). These data suggest that brain-enriched BMOR contributes to the brain-specific/preference metastasis of breast cancer. Then, we intended to ascertain whether and how BMOR mediates the development of B2BM. To investigate this point, we utilized a well-established breast cancer brain metastasis murine model generated by intracardiac injection of cancer cells into immune-deficient nude mice and examined brain metastasis-related oncogenic events, including the development of metastatic lesions in mouse brains and overall survival of mice. The results showed that in comparison with control cells, 231-BM cells with BMOR depleted by BMOR short hairpin RNAs (shRNAs) failed to develop into brain metastases (Fig. 1 E and F and SI Appendix, Fig. S3 A and C). The recipient mice bearing BMOR-depleted 231-BM cells showed significantly longer survival than the control mice (Fig. 1G and SI Appendix, Fig. S3B). These data suggest that BMOR is an oncogenic brain-enriched lncRNA that is essential for brain metastasis development.
BMOR Is Both Necessary and Sufficient for Cancer Cells to Colonize the Brain Tissue.
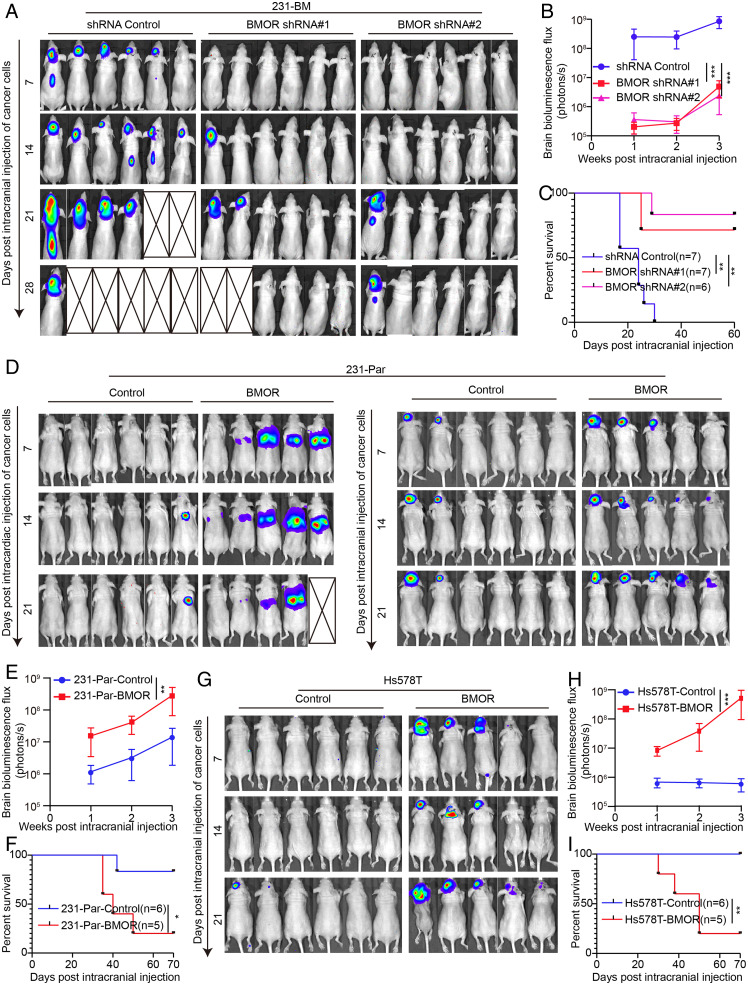
Although many parameters have been reported to contribute to brain metastasis, it remains a key unaddressed question of how cancer cells are able to colonize the brain tissue for brain metastasis. To examine whether BMOR plays a role in the metastatic colonization of cancer cells in the brain, in addition to a murine model of brain metastasis made by intracardiac injection of cancer cells, we used intracranial injection of cancer cells to generate a murine model of brain metastasis. In the intracranial injection brain metastasis murine model, we found that BMOR-depleted 231-BM cells versus control significantly reduced their abilities to colonize the brain tissue and develop into brain metastasis, resulting in significantly longer survival of the recipient mice (Fig. 2 A–C). Furthermore, intracranial, but not intracardiac, injection of 231-Par cells ectopically overexpressing BMOR compared with the control promoted the colonization of cancer cells in the brain and the development of brain metastasis (Fig. 2 D and E and SI Appendix, Fig. S4 A and C). In agreement with that of 231-Par cells, intracranial injection of Hs578T cells ectopically overexpressing BMOR versus control markedly enhanced the colonization of cancer cells in the brain and the development of brain metastasis (Fig. 2 G and H and SI Appendix, Fig. S4B). The recipient mice bearing intracranial injection of BMOR-overexpressing cancer cells, including both 231-Par and Hs578T cells, versus their corresponding controls showed significantly shorter overall survival times (Fig. 2 F and I). These results strongly suggest that brain-enriched lncRNA BMOR is both necessary and sufficient to promote cancer cells to colonize the brain tissue for brain metastasis.
Fig. 2.
BMOR is both necessary and sufficient to drive cancer cells to colonize the brain tissue for brain metastasis. (A–C) Impacts of BMOR on the metastatic colonization of cancer cells in the brain tissue for brain metastasis using intracranial injection of 231-BM cells with versus without BMOR depletion. (A) Bioluminescence images of mice with the indicated treatment over time. (B) Quantification of bioluminescence flux in mice with the indicated treatment over time. ***P < 0.001 (t test). (C) Overall survival of mice with the indicated treatment. Log-rank test: P = 0.0029 for shRNA control versus BMOR shRNA 1 and P = 0.0010 for shRNA control versus BMOR shRNA 2. **P < 0.01. (D–F) Impacts of BMOR on the metastatic colonization of cancer cells in the brain tissue for brain metastasis using brain metastasis murine models generated by intracardiac and intracranial injection of 231-Par cells with BMOR overexpression (BMOR) versus those without BMOR overexpression (Control). (D) Bioluminescence images of mice with the indicated treatment over time. (E) Quantification of bioluminescence flux in mice with the indicated treatment across time. **P < 0.01 (t test). (F) Overall survival of mice with the indicated treatment. Log-rank test: P = 0.0309. *P < 0.05. (G–I) Impacts of BMOR on the brain metastasis behaviors of cancer cells using brain metastasis murine models generated by intracranial injection of Hs578T cells with versus without BMOR overexpression. (G) Bioluminescence images of mice with the indicated treatment over time. (H) Quantification of bioluminescence flux in mice with the indicated treatment over time. ***P < 0.001 (t test). (I) Overall survival of mice with the indicated treatment. Log-rank test: P = 0.0081. **P < 0.01.
BMOR Shields Cancer Cells from Immune-Mediated Killing in the Brain Microenvironment.
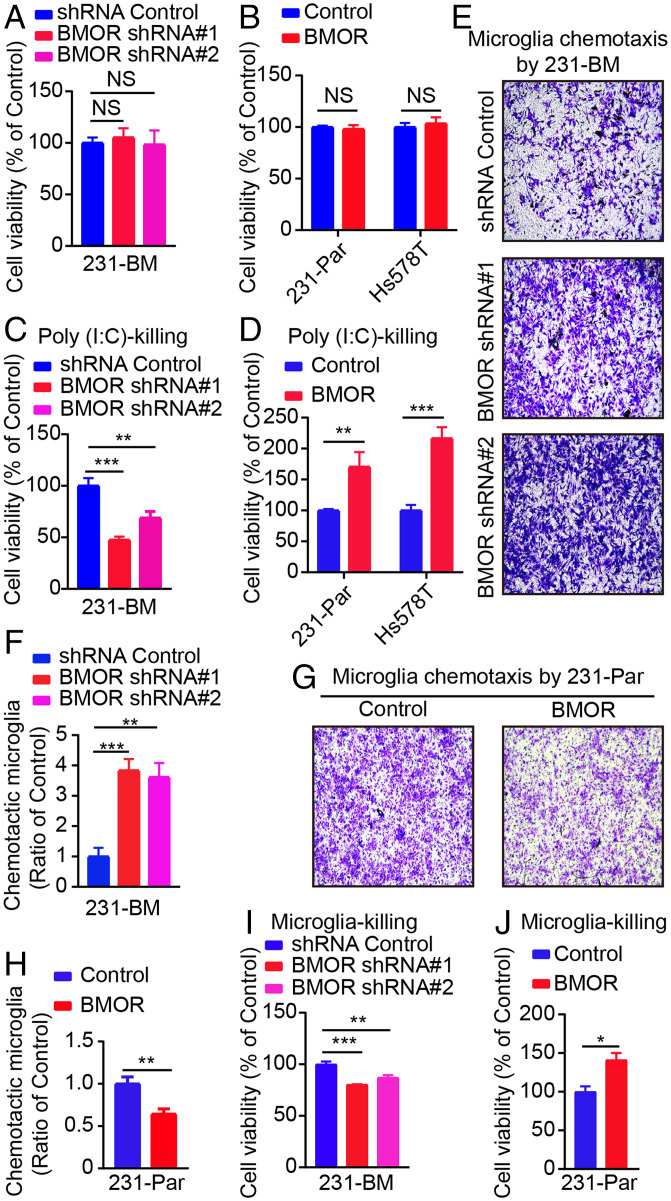
To gain clues about BMOR activity, we first performed RNA-seq transcriptome analysis on 231-BM cells with and without BMOR depletion. The results showed that depletion of BMOR results in reprogramming of the transcription network in 231-BM cells (Dataset S2). Further Gene Set Enrichment Analysis (GSEA) of the RNA-seq data revealed that depletion of BMOR in 231-BM cells versus the control down-regulated several immune response pathways important for inducing the cytotoxicity of cancer cells, such as the interferon (IFN) response, such as IFN-α and IFN-γ response, and tumor necrosis factor (TNF) signaling, such as the TNF-α signaling via nuclear factor κB (NF-κb) (SI Appendix, Fig. S5 A and B and Dataset S3). Consistently, IFN-α and IFN-γ responses as well as TNF-α signaling via NF-κb were also found to be down-regulated in 231-BM cells versus 231-Par cells as well as in breast cancer metastatic brain tumors compared with primary breast tumors in GSEA analysis of the related transcriptome data (SI Appendix, Fig. S5 C and D and Datasets S4 and S5). Furthermore, RT–qPCR confirmed that depletion of BMOR in 231-BM cells compared with the control significantly increased the expression of a set of genes related to the important immune response pathways for killing cancer cells, such as genes related to the IFN response, such as IFN-α and IFN-β, and genes related to TNF signaling, such as TNF-α and TNF-β (SI Appendix, Fig. S5E). Conversely, overexpression of BMOR in both 231-Par and Hs578T cells versus the corresponding controls significantly decreased the expression of these genes (SI Appendix, Fig. S5F). These data suggest that suppression of cytotoxic immune responses is critical in brain metastasis development, where BMOR plays an important role. Interestingly, without additional treatment, neither depletion of BMOR in 231-BM cells nor overexpression of BMOR in 231-Par and Hs578T cells versus the corresponding controls notably affected the viability of cancer cells (Fig. 3 A and B). In addition, migration and invasion assays showed that depletion of BMOR versus control did not decrease the number of migrated 231-BM cells, and overexpression of BMOR versus control cannot increase the number of migrated 231-Par cells (SI Appendix, Fig. S6 A–D). These data suggest that BMOR does not enhance the migration and growth potential of cancer cells. As described above, BMOR is sufficient to promote intracranially but not intracardially injected cancer cells to colonize the brain tissue, which contains brain-resident immune cells such as microglia (Fig. 2 A–I). Considering the inhibitory effects of BMOR on several immune response pathways that are important for inducing the cytotoxicity of cancer cells, our data suggest that BMOR drives cancer cells to colonize the brain tissue by protecting cancer cells from immune-mediated killing in the brain microenvironment. Then, we intended to further investigate this point. Poly(I:C) is a commonly used synthetic immunostimulant for promoting the immune-induced cytotoxicity of cancer cells (16). To confirm the protective effect of BMOR against the immune-mediated killing of cancer cells, we first examined whether and how modulating BMOR expression would impact the viability of cancer cells with Poly(I:C) treatment. The results showed that upon Poly(I:C) treatment, BMOR-depleted 231-BM cells versus control cells showed significantly decreased cell viability, and both 231-Par and Hs578T cells with BMOR overexpression compared with their corresponding controls exhibited significantly increased cell viability (Fig. 3 C and D). The lactate dehydrogenase (LDH) assay is a common method for determining cytotoxicity-induced cell death by measuring the activity of LDH released by damaged cells (17). The results of LDH assays showed that upon Poly(I:C) treatment but not without Poly(I:C) treatment, depletion of BMOR versus control significantly increased the cytotoxicity of 231-BM cells, and overexpression of BMOR significantly decreased the cytotoxicity of 231-Par and Hs578T cells compared with the corresponding controls (SI Appendix, Fig. S6 E–G). These data support a protective role of BMOR against the immune-mediated cytotoxicity of cancer cells. To further validate this point, we carried out cancer cell-immune cell coculture assays. Although the brain is known to contain various immune cells for innate and adaptive immunity, microglia are documented to be the brain’s primary resident immune cells for the immune-mediated elimination of nonbrain resident cells, including metastatic cancer cells (18, 19). The results showed that in the cancer cell-microglia coculture system, the chemotaxis of microglia was enhanced and decreased upon coculture with BMOR-depleted 231-BM and BMOR-overexpressing 231-Par cells versus the corresponding controls, respectively (Fig. 3 E–H). In addition, in line with the described findings in Poly (I:C) treatment, when cancer cells were cocultured with microglia, the depletion of BMOR in 231-BM cells compared with the control significantly decreased the viability of cancer cells, and the overexpression of BMOR in 231-Par cells versus the control significantly increased the viability of cancer cells (Fig. 3 I and J). These results suggest that the acquired expression of a brain-enriched lncRNA, BMOR, enables cancer cells to evade immune-mediated killing in the brain microenvironment to colonize the brain tissue for brain metastasis.
Fig. 3.
BMOR protects cancer cells from immune-mediated killing. (A) Impacts of BMOR depletion on the viability of 231-BM cells without additional treatment. NS for not significant (t test). (B) Impacts of BMOR overexpression on the viability of 231-Par and Hs578T cells without additional treatment. NS for not significant (t test). (C) Impacts of BMOR depletion on the viability of 231-BM cells with additional Poly(I:C) treatment for Poly(I:C)-killing effects. **P < 0.01, ***P < 0.001 (t test). (D) Impacts of BMOR overexpression on the viability of 231-Par and Hs578T cells with additional Poly(I:C) treatment for Poly(I:C)-killing effects. **P < 0.01, ***P < 0.001 (t test). (E and F) Impacts of BMOR depletion on the chemotaxis of microglia cocultured with 231-BM cells: (E) Representative images; (F) Quantification. **P < 0.01, ***P < 0.001 (t test). (G and H) Impacts of BMOR overexpression in 231-Par cells on the chemotaxis of microglia: (G) Representative images; (H) Quantification. **P < 0.01 (t test). (I) Impacts of BMOR depletion on the viability of 231-BM cells cocultured with microglia to assess microglia-killing effects. **P < 0.01, ***P < 0.001 (t test). (J) Impacts of BMOR overexpression on cell viability of 231-Par cells cocultured with microglia to assess microglia-killing effects. *P < 0.05 (t test).
BMOR Binds IRF3 to Inhibit IRF3 Activation.
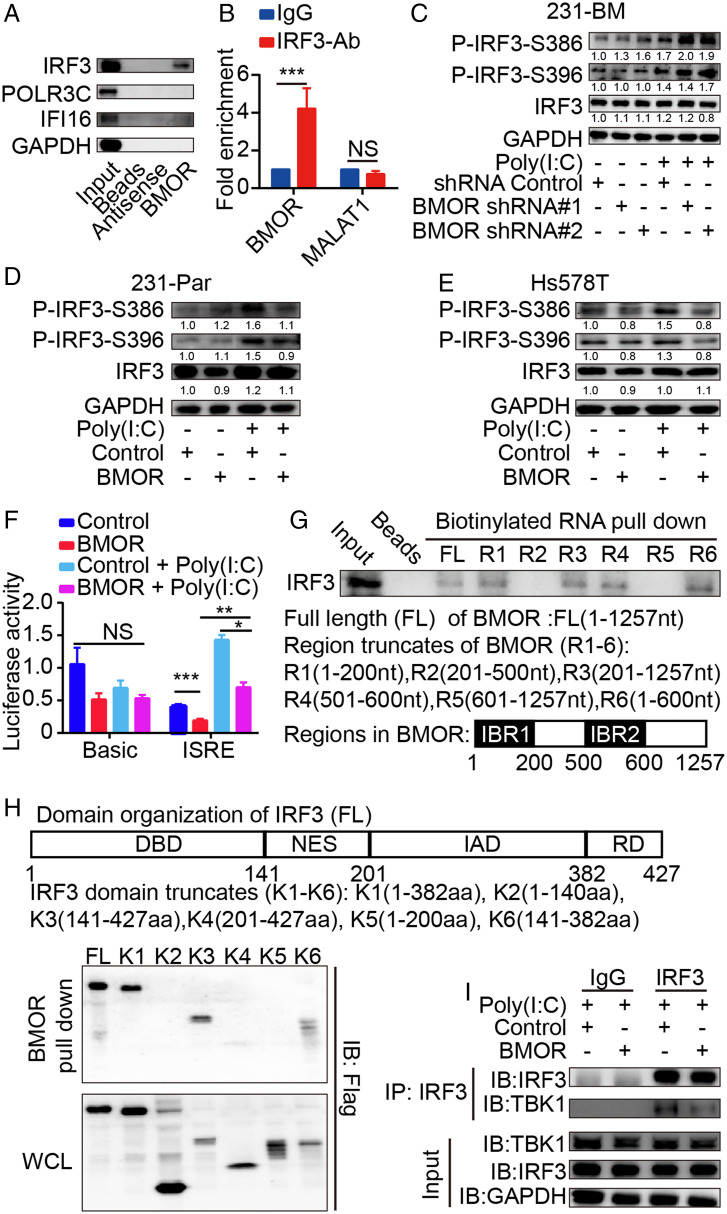
To ascertain the BMOR-mediated intrinsic circuits in cancer cells for their evasion of immune-mediated killing in the brain microenvironment, we searched for BMOR interacting molecules in brain metastatic cancer cells. IRF3, a key regulatory factor for immune-mediated killing of cancer cells by activating apoptotic factors such as BAX or by producing various IFNs or cytokines to induce cell cytotoxicity (20), was identified by RNA pull down-mass spectrometry (MS) and further confirmed to be a BMOR binding protein by RNA pull down-Western blot and RNA immunoprecipitation (RIP)-qPCR assays (Fig. 4 A and B and SI Appendix, Fig. S7A and Dataset S6). Previous studies have validated the potent capability of Poly(I:C) in activating IRF3 to kill cancer cells with increased IRF3-S386 and IRF3-S396 phosphorylation (i.e., P-IRF3-S386 and P-IRF3-S396) levels as established markers (21, 22). We found that depletion of BMOR in 231-BM cells markedly increased the levels of P-IRF3-S386 and P-IRF3-S396 but not IRF3 expression, particularly upon Poly(I:C) treatment (Fig. 4C and SI Appendix, Fig. S7B). Conversely, overexpression of BMOR in both 231-Par and Hs578T cells versus their corresponding controls markedly reduced the levels of P-IRF3-S386 and P-IRF3-S396 without notably affecting IRF3 expression (Fig. 4 D and E and SI Appendix, Fig. S7 C and D). Consistent results were observed in HEK293T cells (SI Appendix, Fig. S7E). These data suggest that BMOR binds IRF3 to antagonize IRF3 phosphorylation and activation. IRF3 activation for immune responses is characterized by transcriptional activation of various genes with IFN-stimulated response elements (ISREs), including many interferon-stimulated genes (ISGs), IFNs, and various cytokine genes (23, 24). As described above, BMOR can markedly inhibit the expression of ISGs, IFNs, and cytokine genes, including IFN-α and IFN-β, which are the most well-characterized target genes of IRF3 (25) (SI Appendix, Fig. S5 E and F). In agreement with these findings, luciferase reporter assays showed that overexpression of BMOR and depletion of BMOR versus the corresponding controls significantly decreased and increased the luciferase activity of the IRSE-containing promoter, respectively (Fig. 4F and SI Appendix, Fig. S7F). Furthermore, we carried out domain mapping assays and found that the NES plus IAD domain of IRF3 is responsible for IRF3 binding to the 1–200 region (i.e., IBR1) and 500–600 region (i.e., IBR2) of BMOR (Fig. 4 G and H and SI Appendix, Fig. S7 G and H). Interestingly, previous studies have revealed TBK1 to be an IRF3 master activator, where TBK1 binds the NES plus IAD domain of IRF3 to phosphorylate and activate IRF3 for both antitumoral and antiviral immune responses (26–28). Consistent with these findings, overexpression of BMOR and depletion of BMOR, respectively, reduced and increased TBK1 coprecipitation with IRF3 in co-immunoprecipitation (coIP) experiments, suggesting that the TBK1-IRF3 interaction is inhibited by BMOR (Fig. 4I and SI Appendix, Fig. S7I). These data suggest that BMOR, with an inhibitory role in IRF3 activation, suppresses IRF3-mediated immune responses, which are important in promoting the immune-induced cytotoxicity of cancer cells.
Fig. 4.
BMOR binds and inhibits IRF3. (A) Representative Western blot images of IRF3, POLR3C, and IFI16 in RNA pull down-Western blot assays. (B) Enrichment of BMOR and control lncRNA MALAT1 in RIP-qPCR assays. ***P < 0.001; NS, not significant (t test). (C–E) Impacts of BMOR on IRF3 phosphorylation by Western blotting with quantifications using GAPDH as the internal control. (C) Impacts of BMOR depletion on IRF3 phosphorylation in 231-BM cells. (D) Impacts of BMOR overexpression on IRF3 phosphorylation in 231-Par cells. (E) Impacts of BMOR overexpression on IRF3 phosphorylation in Hs578T cells. (F) Impacts of BMOR on the transcriptional activity of the ISRE-containing promoter by luciferase reporter assays. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant (t test). (G) Mapping the IRF3-binding region in BMOR using RNA pull down-Western blot assays. IBR1: IRF3-binding region 1. IBR2: IFR3-binding region 2. (H) Mapping the BMOR-binding domain in IRF3 using RNA pull down-Western blot assays. (I) Impacts of BMOR on TBK1 binding to IRF3 using coIP assays.
Targeting BMOR Is Effective against the Metastatic Colonization of Cancer Cells in the Brain.
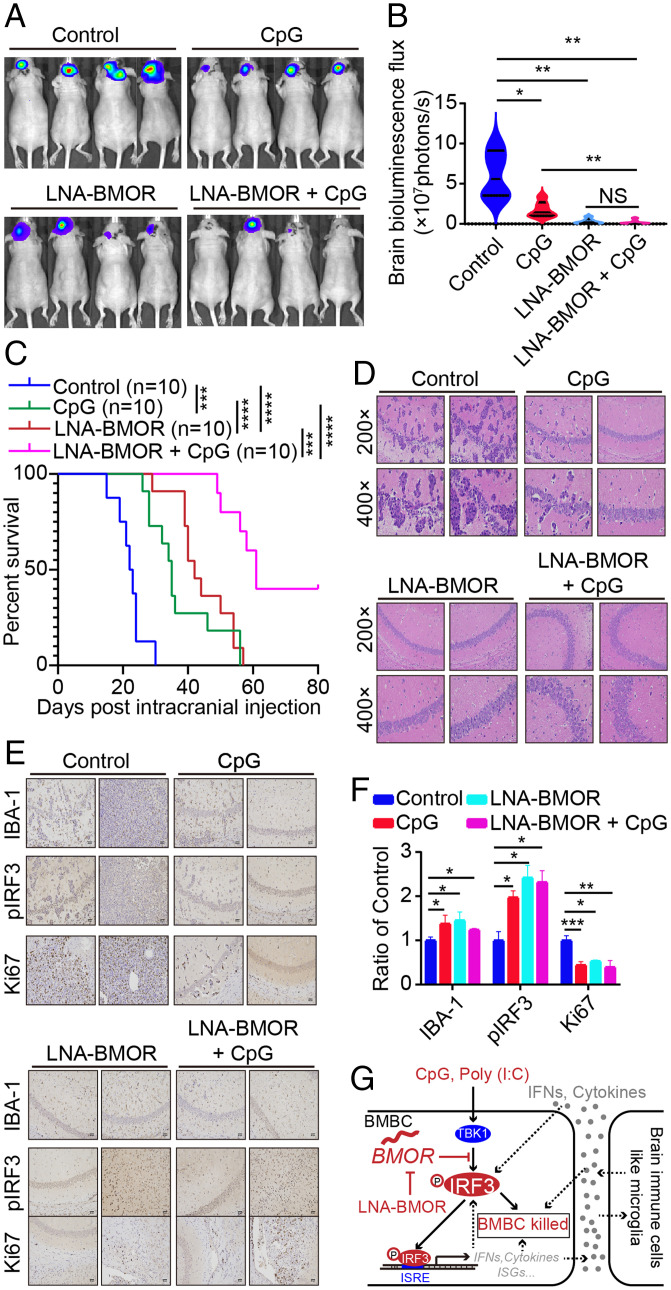
To further validate the critical role of BMOR-mediated mechanisms in brain metastasis and provide potential intervention strategies against brain metastasis, we designed a BMOR-silencer (LNA-BMOR) and used CpG, a Food and Drug Administration-approved immunostimulant with a well-characterized potent capacity to activate IRF3 (29). Although CpG has not been approved for the treatment of cancer patients, CpG has been investigated in many clinical trials for the treatment of malignancy including phase II clinical trials for the treatment of patients with brain tumors (https://clinicaltrials.gov/) (30). Impressively, compared with CpG, LNA-BMOR showed a better at least comparable effectiveness against the metastatic colonization of intracranially injected 231-BM cells in the mouse brain (Fig. 5 A and B). Furthermore, the mice treated with LNA-BMOR or CpG versus the control showed significantly longer survival (Fig. 5C). Strikingly, in these mice, more than 25% of mice undergoing cotreatment with LNA-BMOR and CpG were viable until the end of the experimental period, with their intracranial injection sites showing normal-like brain histology (Fig. 5 C and D). Then, we carried out immunohistochemical staining using antibodies against pIRF3 (marker for IRF3 activation), ki67 (marker for the colonization capability of metastatic cells), and IBA-1 (marker for the microglia-mediated killing of metastatic cells). In agreement with all our results described above, the results showed that both monotreatment and cotreatment with LNA-BMOR and CpG significantly increased pIRF3 and IBA-1 and reduced ki67 at the metastatic lesions generated by intracranial injection of 231-BM cells (Fig. 5 E and F). In agreement with the inhibitory role of BMOR in reducing the chemotaxis of microglia in the cancer cell-microglia coculture system as described above, the result of immunohistochemical staining using an antibody for F4/80 (a well-characterized microglial marker) showed that LNA-BMOR and CpG treatment compared with control can increase the recruitment of microglia in the brain metastatic lesions (Fig. 3 E–H and SI Appendix, Fig. S7 J and K). These data suggest that BMOR can play a certain role in shaping the brain tumor microenvironment (TME). Future work is needed to further investigate the specific role of BMOR in the formation of the TME in the brain to favor brain metastasis development. Nevertheless, in this study, by focusing on the role of BMOR in protecting cancer cells from immune-induced cytotoxicity in the brain microenvironment, we reveal that the lncRNA BMOR-mediated mechanism with potentially effective intervention strategies is important for the development of brain metastasis, where BMOR inhibits IRF3 activation to shield cancer cells from immune-mediated killing in the brain microenvironment (Fig. 5G).
Fig. 5.
Targeting BMOR-mediated evasion of immune-mediated killing is effective against the metastatic colonization of cancer cells in the brain for brain metastasis. (A–C) Effectiveness of LNA-BMOR and CpG treatment against the metastatic colonization of intracranially injected 231-BM cells in the brain. (A) Bioluminescence images of mice with the indicated treatment over time. (B) Quantification of bioluminescence flux in mice with the indicated treatment over time. *P < 0.05, **P < 0.01, NS, not significant (t test). (C) Overall survival of mice with the indicated treatment. Log-rank test: P = 0.0002 for CpG versus Control, P < 0.0001 for LNA-BMOR versus Control, P < 0.0001 for LNA-BMOR versus CpG, P = 0.0002 for LNA-BMOR versus LNA-BMOR + CpG, and P < 0.0001 for CpG versus LNA-BMOR + CpG. ***P < 0.001, ****P < 0.0001. (D–F) Pathological analysis of metastatic lesions in mouse brains generated by intracranially injected 231-BM cells with the indicated treatment. (D) Representative H&E staining images. (E) Representative immunohistochemical staining images using antibodies for the indicated proteins (200×). (F) Quantification of immunohistochemical staining images. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant (t test). (G) A schematic model for the druggable BMOR-mediated mechanism underlying cancer evasion of immune-mediated killing in the brain. BMBC, brain metastatic breast cancer cell.
Discussion
Brain metastases with B2BM, one of the most common types, have not yet been fully functionally or mechanistically resolved, and these patients show poor prognoses due to limited treatment options (2). Our study reports BMOR to be an oncogenic lncRNA for the development of B2BM. BMOR is a brain-enriched lncRNA expressed in B2BM cancer cells and is required for brain metastasis development. Functionally, BMOR is both necessary and sufficient to drive cancer cells to colonize the brain tissue by promoting cancer cell evasion of immune-mediated killing in the brain microenvironment for brain metastasis. At the molecular level, BMOR binds IRF3 and inhibits IRF3 activation to suppress the immune response-induced cytotoxicity of cancer cells. Targeting BMOR or BMOR-mediated mechanisms is effective against the metastatic colonization of cancer cells in the brain for brain metastasis in vivo.
In humans, less than 2% of the genome encodes proteins, and at least 75% is actively transcribed into noncoding RNAs, most of which are cataloged as lncRNAs (31). LncRNAs have emerged as key players in cancer development and progression (7). However, despite much more abundancy in the gene number of lncRNAs than that of protein-coding genes, limited lncRNAs have been functionally characterized. Given the generally tissue-specific or tissue-enriched expression and function of lncRNAs, lncRNAs are proposed to probably play important roles in the organ-specific tropism of cancer metastasis (32). However, there is little evidence to link the highly tissue-specific features of lncRNAs to the organ-specific/preference metastasis of cancer including brain metastasis of cancer, and not a brain-specific or brain-enriched lncRNA has been identified and characterized to contribute to cancer metastasis to the brain. Our study reports that BMOR is a brain-enriched lncRNA expressed in B2BM cells and is essential for the metastatic colonization of cancer cells in the brain for the development of brain metastasis. Furthermore, BMOR was found to be only substantially expressed in breast cancer metastasis to brain tissue versus breast cancer metastasis to nonbrain tissues. Thus, our data suggest that brain-enriched lncRNA BMOR contributes to the brain-preference metastasis of breast cancer, providing mechanistic insights into the formation and preference of brain metastasis.
Caner avoidance of immune destruction via immune evasion, previously segregated as one of the “emerging hallmarks,” is now considered to be a core hallmark of cancer (33). Immune evasion is essential for cancer cells to grow into tumors in both primary sites and distant metastatic organs, including the brain with unique features but not as “immunoprivileged” as previously thought (34). A huge research effort has revealed many immune-related functional and compositional changes for the development of cancer (35). However, due to the complexity of the immune system and cancer immunity, how cancer cells can avoid immune destruction is still not fully understood. Nevertheless, based on our current understandings of immune surveillance and cancer immune evasion, we can conclude that three major immune evasion modes are utilized by cancer cells to avoid immune destruction. Mode 1 is “escaping immune detection,” Mode 2 is “suppressing immune activation,” and Mode 3 is “avoiding immune-mediated killing.” Cancer cells likely orchestrate one but at least one of these three modes to avoid immune destruction to successfully develop into malignancy. Currently, we have a relatively better understanding of Modes 1 and 2. For Mode 1, there are quite a few studies to explain the underlying mechanisms, with loss of MHC I antigen presentation claimed to be an important mechanism for cancer cells to escape immune detection (36). For Mode 2, heavily focused studies in past decades have identified several important immune checkpoints inhibiting immune activation and established quite a few immune checkpoint inhibitors, such as anti-PD1 and anti-CTLA4 antibodies, for the clinical treatment of cancer (37). In comparison to Modes 1 and 2, Mode 3, by which cancer cells can avoid destruction by immune-induced cytotoxicity, is the last line of defense for cancer cells against the final killing effect of immune surveillance to successfully develop into malignancy. However, the mechanisms underlying Mode 3 remain poorly studied. Until recently, although not using cells or models related to brain metastasis, a study identified and reported a set of protein-coding genes expressed by cancer cells to promote cancer-intrinsic evasion of killing by T cells, where discrete functional modules controlling the IFN response-induced and TNF signaling-induced cytotoxicity were claimed to play critical roles (38). Interestingly, both the IFN response and TNF signaling are among the well-characterized activities of IRF3 activation for antitumoral and antiviral immune responses (25). Although lncRNA-mediated IRF3 signaling for cancer metastasis has not been reported, lncRNAs are claimed to play fundamental roles in cancer immune regulation by modulating various other immune-related signaling pathways in cancer metastasis (32, 39). Congruent with previous studies and current understandings of immune surveillance and cancer immune evasion, our study demonstrates that lncRNA BMOR promotes cancer-intrinsic evasion of immune-mediated killing for the development of brain metastasis, where BMOR binds IRF3 to inhibit IRF3 activation. Our work reveals an intrinsic mechanism in cancer cells mediated by a lncRNA to promote cancer immune evasion through an “avoiding immune-mediated killing” mode for brain metastasis, expanding our knowledge of the intrinsic genetic circuits that are critical for caner avoidance of immune destruction via immune evasion.
It is persistently updating on the concept of the TME, which dates back to as early as the 19th century, when Virchow proposed the relationship between inflammation and cancer in 1863 and Paget put forward the “seed and soil” theory in 1889 (40, 41). For brain metastasis, the brain TME is now emerging as a critical regulator (42). The TME, known to be a complicated ecosystem full of heterogeneity, is far from being fully functionally and mechanistically characterized. Nevertheless, increasing evidence strongly suggests that the immune TME plays important roles in both immune evasion of cancer and cancer immunotherapeutic efficacy (43). Interestingly, the recently identified protein-coding genes contributing to the evasion of immune-mediated killing were also highlighted to have important effects within the immune TME (38). Regarding cancer metastasis to the brain, it is perhaps not surprising that the immune TME in the brain will have unique features compared with the immune TME in nonbrain tissues due to unique brain-resident cells including immune cells such as microglia (42). Although not fully understood, microglia are believed to play a vital role in shaping the immune TME in the brain for brain metastasis and have recently been reported to be one of the major immune cell determinants of the brain TME (44). In agreement with these findings, we found that BMOR plays an inhibitory role in both the chemotaxis and activation of microglia, suggesting the important role of BMOR-inhibited microglia in the formation of an immunosuppressive TME in the brain for brain metastasis. Microglia in the TME are tumor-associated macrophages (TAMs), which are major players in the TME (45). In addition to microglia, TAMs in the brain TME also contain macrophages (also called tissue macrophages). A recent study detected type I IFN signaling to be enriched in both microglia and macrophages in the TME of brain metastases by carrying out leading-edge metagenes analysis (44). TBK1 is one of the IRF3 master activators that binds and phosphorylates IRF3 (27). TBK1-activated IRF3 is a key regulator that promotes type I IFNs to protect against cancer (46). Our results show that BMOR inhibits IRF3 activation by preventing IRF3 binding to its upstream activator TBK1 and repressing the expression of key genes (i.e., IFN-α and IFN-β) in type I IFN signaling. Type I IFNs play important regulatory roles in determining the multifarious function of both microglia and macrophages for the formation of an immunosuppressive TME (47). However, it is still largely unknown how cancer cells, microglia and macrophages cooperate to shape an immunosuppressive TME in the brain for brain metastasis. Future works are needed to further investigate whether and how brain-enriched BMOR can have specific or different impacts on microglia and macrophages in the brain TME. Nevertheless, our data, together with previous studies, support the notion that BMOR inhibits the antitumor functions of both microglia and macrophages to contribute to the formation of an immunosuppressive TME in the brain for brain metastasis, where BMOR-inhibited type I IFN signaling likely plays important roles.
Taken together, in comparison to the reported protein-coding genes and lncRNAs working models in brain metastasis and cancer immunity, we report that brain-enriched lncRNA-BMOR acquired by B2BM cancer cells is both necessary and sufficient to promote cancer cells to colonize the brain tissue by evading immune-mediated killing in the brain microenvironment for brain metastasis. This represents a way for cancer cells to avoid immune-mediated disruption. In addition to providing insights into cancer-intrinsic mechanisms underlying cancer immune evasion and brain metastasis, our findings also establish therapeutic targets with effective intervention strategies for the future treatment of B2BM patients.
Materials and Methods
Cell Lines and Culture Conditions.
Human breast cancer cell lines, including MDA-MB-231 (231-Par), brain metastatic cell line (231-BM), and Hs578T, were cultured in DMEM supplemented with 10% FBS (WISENT). The microglia cell line HMC3 was grown in MEM supplemented with 10% FBS. HEK293T cells were cultured in DMEM supplemented with 10% FBS. All cell lines were maintained in a humidified incubator with an atmosphere of 5% CO2 at 37 °C.
Total RNA Extraction and RNA-Seq.
In brief, total RNA was extracted using TRIzol Reagent (Life Technologies) following the manufacturer’s introductions. For RNA-seq, an Illumina high-throughput sequencer was used. Then, raw RNA-seq data were analyzed by using a commonly used FastQC-Tophat2-cufflinks workflow (48).
Patient Sample Collection and RNA-FISH Experiment.
Formalin-fixed paraffin-embedded (FFPE) tissue sections of breast tumors and metastases were obtained from breast cancer patients at Sun Yat-sen University Cancer Center with written informed consent. The clinical sample collection and experiment were carried out in accordance with guidelines and protocols approved by the Ethics and Scientific Committees of Sun Yat-sen University Cancer Center. RNA-FISH of FFPE tissue sections was used to examine BMOR expression in both breast cancer brain metastasis and nonbrain metastasis tumor samples. The sequence of the FISH probe against BMOR was 5′-AAACGTGCACAGGGCACGAA-3′ labeled with Cy5. RNA-FISH was performed and optimized according to previously described protocols (49). In brief, the tissue slides were treated with 1% pepsin (diluted in 10 mM HCl), followed by an incubation with 20 nM FISH probe in hybridization buffer (5 M NaCl, 1 M Tris × HCl, pH 8.0, 20% deionized formamide in 2× SSC) for 5 min at 83 °C. Then, hybridization was performed for 18 h at 42 °C, followed by a washing step. After that, the tissue slides were incubated with DAPI to counterstain the cell nuclei for 15 min. A Zeiss Image Scope System was used to capture the FISH images. All samples and images were reviewed and interpreted by experienced pathologists.
Brain Metastasis Mouse Model and Drug Treatment.
BALB/c nu/nu female mice (4- to 6-wk-old) were purchased from the Model Animal Research Center of Nanjing University. Intracardiac injection and intracranial injection of cancer cells were used to generate brain metastasis murine models. In brief, for the brain metastasis mouse model generated by intracardiac injection, mice underwent a left ventricular injection of 2 × 105 breast cancer cells resuspended in 100 µL PBS. For the brain metastasis mouse model generated by intracranial injection, mice were cerebrally injected with 2 × 104 breast cancer cells resuspended in 10 µL PBS . For drug treatment, scrambled LNA gapmers are used as LNA controls, and LNA-BMORs are LNA gapmers targeting BMORs. LNA control and LNA-BMOR containing phosphorothioate backbone modifications indicated by “*” in the following sequences: 5′-G*C*T*C*C*C*T*T*C*A*A*T*C*C*A*A-3′ for the LNA control and 5′-C*A*A*G*G*C*G*C*G*G*A*C*T*T*A -3′ for LNA-BMOR. In drug treatment experiments, mice were cerebrally injected with 2 × 104 cancer cells resuspended in 10 µL PBS, followed by cerebral injection of 0.9% saline-suspended drugs, including 120 pmol LNA control, 120 pmol LNA-BMOR, 20 µg CpG (InvivoGen), and a cotreatment with 120 pmol LNA-BMOR and 20 µg CpG. Mice were monitored using MRI or the IVIS Imaging System as indicated in the figures. All animal studies were carried out in compliance with the guidelines approved by the Animal Ethical and Welfare Committee of Nanjing Normal University.
RNA Pull Down.
All experiments were carried out by following the manufacturers’ protocols. In brief, BMOR or antisense RNA was transcribed in vitro from the vector PCDNA3.1 BMOR and PCDNA3.1 BMOR-Antisense using T7 RNA polymerase (Novoprotein). Then, the transcripts were biotin labeled with the Biotin RNA Labeling Mix (Roche). Whole-cell lysates were incubated with 6 µg biotinylated transcripts for 1 h at room temperature. The RNA–protein complexes were isolated with streptavidin magnetic beads (MedChemExpress) for further mass spectrometry (MS) identification or Western blot analysis.
RIP and CoIP.
Briefly, for RIP assays, cell lysates were prepared in RIP buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.1% Nonidet P-40 5% glycerol 0.5 mM DTT) supplemented with RNase inhibitor. IRF3 antibodies using immunoglobulin G control were incubated with cell lysates overnight at 4 °C. Then, 50 µL Protein A/G Magnetic Beads (MedChemExpress) were added and incubated with the mixture for 2 h. Coprecipitated RNAs were extracted and analyzed by RT–qPCR. The primers used are listed in SI Appendix, Table S1. For the coIP assay, cell lysates were prepared by using lysis buffer (50 mM Tris × HCl pH 7.0, 10 mM EDTA, 1% SDS, 5 µL/mL protease inhibitor). Then, the cell lysates were mixed with the primary antibody and Protein A/G Magnetic Beads (MedChemExpress) overnight at 4 °C. After that, the beads were washed three times using PBS, followed by Western blot analysis.
Luciferase Reporter Assay.
In brief, a promoter containing an ISRE was cloned into pGL3-Basic. Cells were seeded in 24-well plates, and plasmid transfection was performed when cells reached 80% confluency. Luciferase activity was measured after 48 h using the Dual-Luciferase reporter system (Promega) according to the manufacturer’s instructions.
Viability and Chemotaxis Assay.
Cell viability was measured using the CCK-8 kit according to the protocol recommended by the manufacturer (Dojindo Laboratories). Briefly, cells were seeded into 96-well plates. Cell viability was assessed by measuring the absorbance at 450 nm using a Bio-Rad iMark plate reader. For chemotaxis assays, briefly, a cancer cell-microglia coculture system was created by using transwell migration chambers (Merck Millipore), where microglia were placed in the upper chamber, and cancer cells were placed in the lower chambers. Chemotaxis microglia cells were fixed with methanol and stained with 0.5% crystal violet.
Immunohistochemistry and H&E Staining.
Immunohistochemistry and HE staining were performed following standard protocols. Briefly, tumor/tissue samples were fixed in 4% paraformaldehyde and then embedded in paraffin and sectioned. For immunohistochemistry staining, the sections of tumor samples were stained overnight with anti-pIRF3 antibody (Affinity Biosciences, AF2436), anti-ki67 antibody (Abcam, ab15580), anti-IBA-1 antibody (Abcam, ab5076) or anti-F4/80 antibody (Abcam, ab16911). Then, a SABC immunohistochemical staining kit (BOSTER) was used for secondary and tertiary antibody staining. H&E was used to stain the cell nucleus and cytoplasm, respectively. Images were collected using a Zeiss microscope.
Software and Statistical Analysis.
All data are expressed as the mean ± SD and analyzed using GraphPad Prism 7.0 software. For experiments consisting of only two groups, the data were evaluated by Student's t test (two-tail). For experiments involving multiple groups, the data were analyzed by one-way ANOVA. For survival analysis, log-rank P was used. When P < 0.05, statistical analysis considered statistical significance.
Supplementary Material
Acknowledgments
We thank Dr. Ceshi Chen, Dr. Hainin Tan, Dr. Yukun Cui, and Dr. Peng Sun for their help with the collection of breast cancer samples. We also thank Dr. Jean Paul Thiery for his helpful discussion. This work was supported by the National Natural Science Foundation of China (81974447 and 81902679), Natural Science Foundation of Jiangsu Province (SBK2020010058), Guangdong Natural Science Foundation (2021A1515010760), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200230119/-/DCSupplemental.
Data Availability
Additional procedures are described in SI Appendix, Supplementary Information Text. All study data are included in the article and SI Appendix. The original transcriptome sequencing data (RNA-seq) were deposited in the Short Reads Archive database of National Center for Biotechnology Information (BioProject: PRJNA777370) and the nucleotide sequence of BMOR was deposited in GenBank (OL351836).
References
- 1.Sung H., et al. , Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Achrol A. S., et al. , Brain metastases. Nat. Rev. Dis. Primers 5, 5 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Lin X., DeAngelis L. M., Treatment of brain metastases. J. Clin. Oncol. 33, 3475–3484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arslan U. Y., et al. , Breast cancer subtypes and outcomes of central nervous system metastases. Breast 20, 562–567 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Pedrosa R. M. S. M., Mustafa D. A., Soffietti R., Kros J. M., Breast cancer brain metastasis: Molecular mechanisms and directions for treatment. Neuro-oncol. 20, 1439–1449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kung J. T. Y., Colognori D., Lee J. T., Long noncoding RNAs: Past, present, and future. Genetics 193, 651–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M., The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Xing F., et al. , Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 78, 4316–4330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S., et al. , JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Invest. 127, 4498–4515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium G. T.; GTEx Consortium, The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bos P. D., et al. , Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Q., et al. , Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 573, 526–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward M., McEwan C., Mills J. D., Janitz M., Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 1, 2–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Statello L., Guo C. J., Chen L. L., Huarte M., Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng G., et al. , HCMDB: The human cancer metastasis database. Nucleic Acids Res. 46 (D1), D950–D955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchi F., Pretto S., Tagliabue E., Balsari A., Sfondrini L., Exploiting poly(I:C) to induce cancer cell apoptosis. Cancer Biol. Ther. 18, 747–756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar P., Nagarajan A., Uchil P. D., Analysis of cell viability by the lactate dehydrogenase assay. Cold Spring Harb. Protoc. 2018, 10.1101/pdb.prot095497 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Gutmann D. H., Kettenmann H., Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron 104, 442–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright-Jin E. C., Gutmann D. H., Microglia as dynamic cellular mediators of brain function. Trends Mol. Med. 25, 967–979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chattopadhyay S., et al. , Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 29, 1762–1773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B., et al. , Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3403–E3412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W., et al. , Contribution of Ser386 and Ser396 to activation of interferon regulatory factor 3. J. Mol. Biol. 379, 251–260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrilenas K. K., et al. , DNA-binding landscape of IRF3, IRF5 and IRF7 dimers: Implications for dimer-specific gene regulation. Nucleic Acids Res. 46, 2509–2520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura T., Yanai H., Savitsky D., Taniguchi T., The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Petro T. M., IFN regulatory factor 3 in health and disease. J. Immunol. 205, 1981–1989 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Chen F., et al. , Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 21, 498–510 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald K. A., et al. , IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y., Chen Z. J., STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oganesyan G., et al. , IRF3-dependent type I interferon response in B cells regulates CpG-mediated antibody production. J. Biol. Chem. 283, 802–808 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Ursu R., et al. , Intracerebral injection of CpG oligonucleotide for patients with de novo glioblastoma-A phase II multicentric, randomised study. Eur. J. Cancer 73, 30–37 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Iyer M. K., et al. , The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S. J., Dang H. X., Lim D. A., Feng F. Y., Maher C. A., Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 21, 446–460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanahan D., Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Louveau A., et al. , Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiam-Galvez K. J., Allen B. M., Spitzer M. H., Systemic immunity in cancer. Nat. Rev. Cancer 21, 345–359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhatchinamoorthy K., Colbert J. D., Rock K. L., Cancer immune evasion through loss of MHC class I antigen presentation. Front. Immunol. 12, 636568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pardoll D. M., The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawson K. A., et al. , Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 586, 120–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., et al. , Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 11, 1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin M. Z., Jin W. L., The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 5, 166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paget S., The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 8, 98–101 (1989). [PubMed] [Google Scholar]
- 42.Quail D. F., Joyce J. A., The microenvironmental landscape of brain tumors. Cancer Cell 31, 326–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binnewies M., et al. , Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klemm F., et al. , Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643–1660.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chanmee T., Ontong P., Konno K., Itano N., Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 6, 1670–1690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo S. R., et al. , STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aricò E., Castiello L., Capone I., Gabriele L., Belardelli F., Type I interferons and cancer: An evolving story demanding novel clinical applications. Cancers (Basel) 11, E1943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kukurba K. R., Montgomery S. B., RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015, 951–969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang K. C., et al. , MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat. Commun. 11, 6438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional procedures are described in SI Appendix, Supplementary Information Text. All study data are included in the article and SI Appendix. The original transcriptome sequencing data (RNA-seq) were deposited in the Short Reads Archive database of National Center for Biotechnology Information (BioProject: PRJNA777370) and the nucleotide sequence of BMOR was deposited in GenBank (OL351836).