Significance
Plastic individuals can buffer environmental changes, maintaining a stable performance across gradients. Plasticity is therefore thought to be particularly beneficial for the survival of wild populations that experience large environmental fluctuations, such as diel and seasonal temperature changes. Maintaining plasticity is widely assumed to be costly; however, empirical evidence demonstrating this cost is scarce. Here, we predict that if plasticity is costly, it would be readily lost in a stable environment, such as a laboratory. To test this, we measured a diverse range of phenotypic traits, spanning gene expression, physiology, and behavior, in wild and laboratory zebrafish acclimated to 15 temperatures. We show that laboratory fish have lost plasticity in many traits, demonstrating that maintaining plasticity carries a cost.
Keywords: plasticity, domestication, temperature, ectotherm
Abstract
Plasticity can allow organisms to maintain consistent performance across a wide range of environmental conditions. However, it remains largely unknown how costly plasticity is and whether a trade-off exists between plasticity and performance under optimal conditions. Biological rates generally increase with temperature, and to counter that effect, fish use physiological plasticity to adjust their biochemical and physiological functions. Zebrafish in the wild encounter large daily and seasonal temperature fluctuations, suggesting they should display high physiological plasticity. Conversely, laboratory zebrafish have been at optimal temperatures with low thermal fluctuations for over 150 generations. We treated this domestication as an evolution experiment and asked whether this has reduced the physiological plasticity of laboratory fish compared to their wild counterparts. We measured a diverse range of phenotypic traits, from gene expression through physiology to behavior, in wild and laboratory zebrafish acclimated to 15 temperatures from 10 °C to 38 °C. We show that adaptation to the laboratory environment has had major effects on all levels of biology. Laboratory fish show reduced plasticity and are thus less able to counter the direct effects of temperature on key traits like metabolic rates and thermal tolerance, and this difference is detectable down to gene expression level. Rapid selection for faster growth in stable laboratory environments appears to have carried with it a trade-off against physiological plasticity in captive zebrafish compared with their wild counterparts.
In ectotherms, body temperature is affected by the environmental temperature, and higher body temperatures increase biological rates (e.g., enzyme activity and metabolic rates) (1). Thermal performance curves illustrate this relationship, whereby performance under acute temperature exposure typically peaks at an optimal temperature (Topt) and then decreases at nonoptimal temperatures (2–4). However, over longer exposure to a temperature, individuals can remodel their physiology to restore homeostasis and counteract direct thermal effects (2, 5). Such thermal acclimation or compensation can improve performance in varying environments through physiological plasticity (5–7). Physiological plasticity describes the ability of an organism to adjust their physiology in different environments, in this case, to counteract the direct effect of temperature. Thermal performance breadth describes the range of temperatures over which performance is above or equal to 80% of the maximum (3), and this breadth can be extended through acclimation. Consequently, physiological plasticity has been hypothesized to be adaptive in heterogeneous environments (8) and would be reflected by broad thermal performance curves for a wide range of traits after acclimation (5). Maintaining high performance over a large thermal range may require large scale alterations in gene expression and therefore a greater capacity to modify transcriptional processes (9).
Having the ability to perform consistently across a wide range of temperatures could come with physiological or biochemical trade-offs. Such trade-offs could include investment in sensory and regulatory mechanisms, production costs (e.g., simultaneous expression of optimal and suboptimal enzyme isoforms, as each is optimized for a specific environmental condition), or could be genetically costly (e.g., if plastic genes are linked to genes for nonbeneficial/costly traits) (10, 11). If true, a trade-off could be predicted between physiological plasticity on one hand and performance at optimal temperatures on the other (12–14). Thermal performance curves with a narrower breadth and a higher peak could therefore be expected in populations that have evolved in the absence of environmental heterogeneity (15). In this case, selection should favor developmental pathways specific to the narrow environmental range experienced and select for peak performing nonplastic individuals (16). Despite this, little empirical evidence exists linking higher plasticity to decreased performance in a narrow range of stable environments (17, 18).
Here, we evaluate whether adaptation to homogenous environments reduces physiological plasticity over time using an approach comparing wild-caught versus laboratory zebrafish. We examine a suite of traits that span all levels of their biological organization in response to acclimation to temperatures across their entire thermal range. By using a multi- rather than single-trait approach, we aim to enhance our understanding of the role of plasticity at the whole organism level (19, 20).
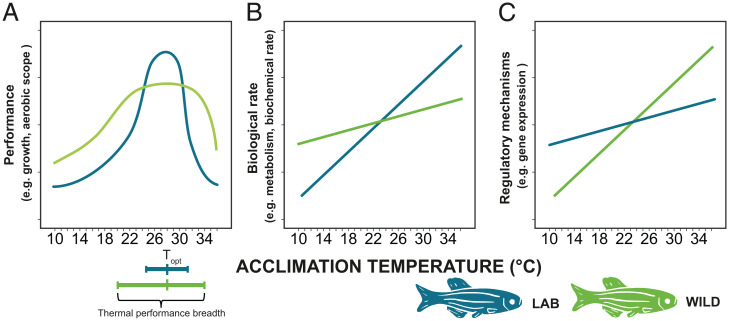
Zebrafish are one of the most commonly used experimental animals (21). In the wild, zebrafish experience large seasonal thermal fluctuations which can range from 17.3 up to 38.6 °C (22, 23). Biomedical research laboratories rear zebrafish at optimal temperature for growth and fecundity (28-28.5 °C) (24–26). The AB wild-type zebrafish line was originally brought into laboratories in the 1970s and has undergone a domestication process for more than 150 generations [our estimate based on Howe et al. (27); see SI Appendix, Supplementary Text]. Consequently, the line has adapted to life in small aquaria, high population densities, dry food, and to handling by humans (28, 29). One previously overlooked consequence of domestication is the potential adaptation to optimal temperature with low thermal variance and few thermal extremes (30). The domestication of zebrafish can thus be viewed as a long-term evolution experiment to optimal temperature with little thermal variation. We hypothesize that maintaining thermal plasticity comes at a cost, and adaptation to a stable thermal environment has thus reduced the physiological plasticity in domesticated zebrafish compared to wild zebrafish. We therefore predict that wild zebrafish should be able to acclimate and adjust their biochemistry and physiology through physiological plasticity to counteract direct thermal effects on biological rates to a higher degree than laboratory zebrafish. Wild zebrafish should thus be able to maintain close to normal function over a wider range of nonoptimal temperatures, while laboratory zebrafish should display high performance at a narrower thermal range and faster deterioration of performance when approaching nonoptimal temperatures (Fig. 1A). Additionally, as laboratory zebrafish might be adapted to life at optimal temperatures, we predict that the laboratory zebrafish should show higher performance in the measured traits at optimal temperatures (Fig. 1A). We also predict that wild fish have more consistent performance over different temperatures (i.e., shallower slope in the thermal reaction norm) (Fig. 1B). This would require the wild fish to have an enhanced regulatory capacity compared to the laboratory fish (Fig. 1C).
Fig. 1.
Predictions of how reduced physiological plasticity can be detected: (A) We predict that laboratory zebrafish (blue) will show higher performance in the measured traits at the optimal temperature (Topt) but have a narrower thermal performance breadth (≥80% performance) than wild zebrafish (light green). (B) We predict that wild zebrafish will show a higher capacity for adjusting their biochemistry and physiology (through acclimation or physiological plasticity) to counteract direct thermal effects on biological rates. This will be evident in wild zebrafish maintaining close to normal biological rates (i.e., metabolism) across different temperatures, post acclimation, while laboratory zebrafish will display more extreme rates as temperatures increase or decrease. (C) To allow a consistent performance after acclimation, we predict greater regulation in the underlying mechanisms in the wild zebrafish compared to the laboratory zebrafish.
To test these predictions, we exposed 300 juvenile laboratory zebrafish and 300 juvenile wild-caught F1 generation zebrafish to the full range of temperatures that zebrafish can survive at for 35 d (SI Appendix, Fig. S1). After 1 mo of thermal acclimation at 15 different temperatures (10–38 °C), both wild and laboratory fish were subjected to a range of phenotypic testing at their acclimation temperature. The phenotyping included behavior (swimming activity and alarm cue response), growth rate, metabolism (standard metabolic rate [SMR] and maximum metabolic rate [MMR]), startle response time, maximum swim speed, thermal tolerance, gene expression, RNA:DNA ratio, and red blood cell size (see Methods for further details). The 38 °C exposure caused elevated mortality in both the wild and laboratory zebrafish and was thus terminated.
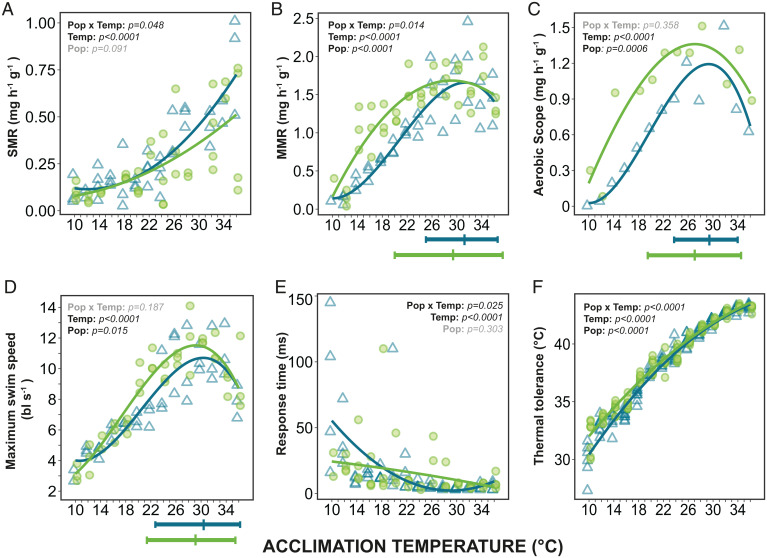
We found that laboratory fish had lower physiological plasticity in many of the phenotypic traits compared to wild fish, consistent with the hypothesis of loss of physiological plasticity following adaptation to stable environments (predicted in Fig. 1 A and B). SMR was higher in the laboratory fish at the highest temperatures (Fig. 2A), indicating a lower ability to compensate for the direct effect of temperature. This suggests that laboratory fish have a higher aerobic requirement to sustain baseline maintenance, which corresponded with the higher expression found in metabolic genes in all studied tissues at 36 °C (Fig. 3G and SI Appendix, Fig. S3). In addition, a reduced thermal performance breadth was detected in the MMR of the laboratory fish, where the breadth was 6.6 °C less than the wild fish (Fig. 2B and SI Appendix, Table S7).
Fig. 2.
Thermal performance curves of wild and laboratory zebrafish for metabolic rates and other physiological traits after acclimation to temperatures from 10 °C to 36 °C. (A) Standard metabolic rate (SMR), each point represents an individual fish. (B) Maximum metabolic rate (MMR), each point represents a group of six fish (measured together). (C) Aerobic scope, the difference between mean MMR and mean SMR. (D) Maximum swim speed (body lengths sec−1) where each point represents a group of six fish. (E) Response time (latency to respond to a stimulus), each point represents the median response time of an individual fish. (F) Thermal tolerance (using the critical thermal maxima method: CTmax) of individual fish. Optimal temperatures for performance and thermal performance breadths (80% performance) are illustrated beneath (B–D) and exact values as shown in SI Appendix, Table S7. The P values for the statistical models are indicated on each panel: Temp, effect of temperature on trait; Pop, difference between wild (light green circles) and laboratory (blue triangles) fish (intercept at 23 °C); and Pop × Temp, interaction (see SI Appendix, Tables S1 and S2).
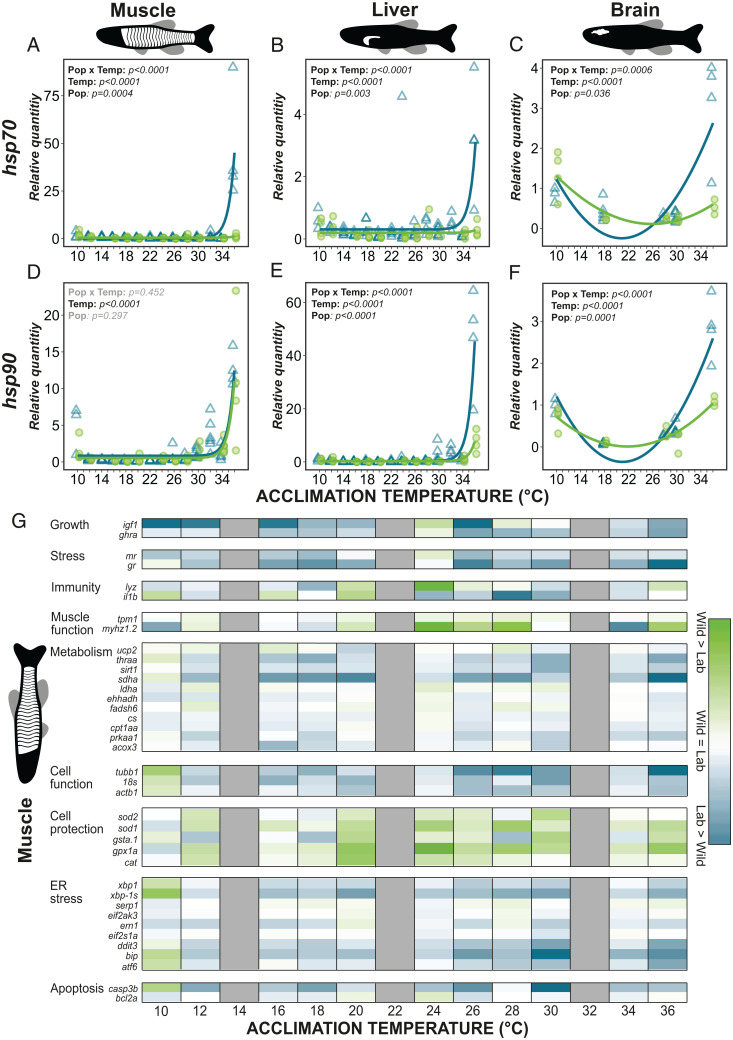
Fig. 3.
Comparison of gene expression in wild and laboratory zebrafish acclimated to temperatures from 10 °C to 36 °C. Heat shock protein (hsp) expression (A–F): hsp70 and hsp90 in the muscle (A and D), liver (B and E), and brain (C and F). Statistical model results indicated on each panel: Temp, effect of temperature on expression; Pop, difference between wild (light green circles) and laboratory (blue triangles) fish (at 23 °C); Pop × Temp, interaction. (G) Heatmap showing the difference in muscle gene expression (log transformed) between wild and laboratory zebrafish. Genes are grouped by functional groups and colors represent relative expression within these groups where green shows a higher expression in wild fish and blue a higher expression in laboratory fish. White illustrates no difference. Grey illustrates not analyzed. Full gene names and functions can be found in SI Appendix, Table S3.
As aerobic scope is the difference between MMR and SMR, the lower MMR and higher SMR results in a narrower thermal aerobic scope profile in the laboratory fish (Fig. 2C). This could in turn explain the differences in maximum swim speed, where the thermal performance breadth of laboratory fish was within 1 °C of the wild fish, but the peak performance was lower (Fig. 2D and SI Appendix, Tables S1, S2, and S7). Aerobic scope is indicative of the total aerobic capacity of the fish, and a narrower aerobic scope in the laboratory fish means simultaneous aerobic processes (e.g., growth, digestion, and swimming) can only occur at reduced rates at nonoptimal temperatures (31, 32).
Reductions in the physiological plasticity of laboratory fish were also detected in startle response time (Fig. 2E) and thermal tolerance (Fig. 2F). The laboratory fish had steeper thermal reaction norms than the wild fish and were thus less able to counteract the direct effect of temperature through acclimation. These steeper reaction norms were especially visible at the lower acclimation temperatures where laboratory fish had a longer response time and a lower thermal tolerance than wild fish. Consequently, laboratory fish appear less well adapted to respond to stimuli or cope with acute thermal challenges at lower acclimation temperatures. These findings indicate a loss of physiological plasticity in diverse physiological traits in the laboratory fish and reduced performance at suboptimal temperatures.
The loss of physiological plasticity in laboratory fish was also detectable at the gene level. This was apparent as a dramatic increase in heat shock protein expression (hsp70 and hsp90) at the highest acclimation temperatures in muscle, liver, and brain of laboratory fish (Fig. 3 A–F). Wild fish had a heat shock response at 36 °C but to a lesser extent than the laboratory fish. The higher heat shock expression in the laboratory fish could be due to failure of function (33–36), for example, a lack of heat tolerant isozymes (37) that may have been expressed at a higher degree in the wild fish (as predicted in Fig. 1C). This suggests higher continuous heat stress and heat injury (38) in the laboratory fish than in the wild fish. Additionally, there may have been a breakdown of regulation, such as a higher production of oxygen radicals due to leakier mitochondria (39), which could have led to more protein damage in the laboratory fish. The latter explanation is supported by the higher expression of genes relating to the induction of apoptosis and endoplasmic reticulum (ER) stress at the warmest temperatures in the laboratory fish across all three tissues sampled, suggesting that they experienced greater levels of cellular stress from misfolded proteins (Fig. 3G and SI Appendix, Fig. S3). The gene expression data therefore strengthen the evidence of reduced physiological plasticity in the laboratory fish and supports the suggestion that although the laboratory fish are coping, they are less able to adjust their biochemistry and physiology to nonoptimal temperatures than the wild fish.
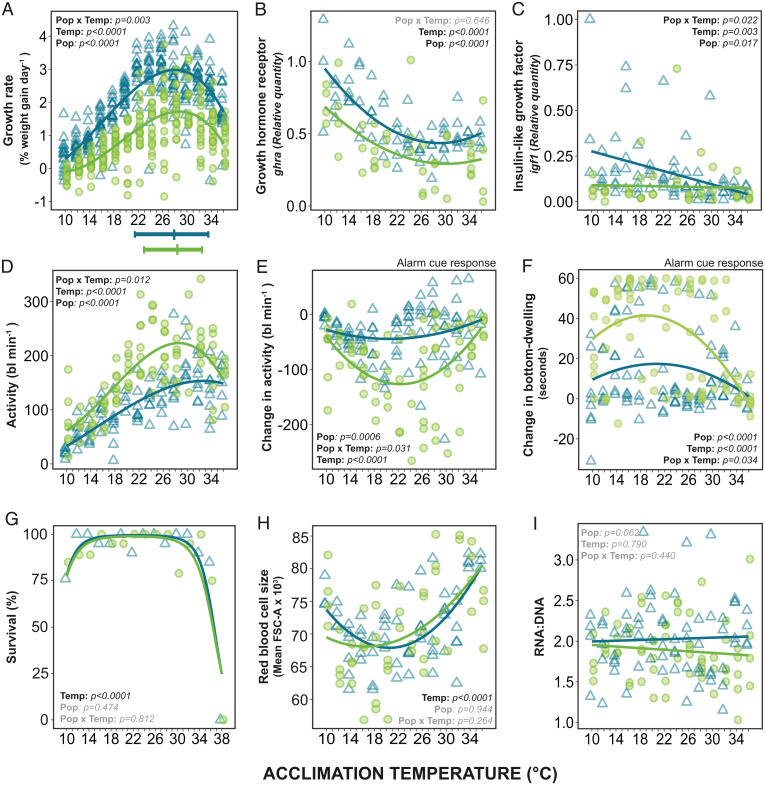
If maintaining plasticity is costly, laboratory fish should show reduced plasticity in combination with higher performance at optimal temperature. This prediction was confirmed, as growth rates of the laboratory fish were 40% (weight) and 35% (length) higher than wild fish at the optimal temperature (Topt) (Fig. 4A and SI Appendix, Fig. S2A). These higher growth rates in the laboratory fish could partly be due to reduced costs of physiological plasticity, but it is currently unclear what the main components of these costs are. They have been suggested to be genetic (10) or production costs (e.g., continuous production of thermally nonoptimal enzyme isoforms) (40), or investment in the sensory and regulatory mechanisms required for maintaining physiological plasticity (10, 41–43). This explanation is consistent with the high physiological plasticity in many traits, and low growth rates observed in the wild fish.
Fig. 4.
Growth, behavior, survival, and cell size in wild (green circles) and laboratory (blue triangles) zebrafish acclimated from 10 °C to 36 °C. Growth rate (A–C): (A) specific growth rate for weight for each individual fish; (B) relative quantity of growth hormone receptor α (ghra) in muscle tissue; and (C) relative quantity of insulin-like growth factor 1 (igf) in muscle tissue. Behavioral (D–F): (D) swimming activity (body lengths [bl] moved per minute) for individual fish prealarm cue; (E) change in activity in response to an alarm cue; and (F) change in time bottom-dwelling (sec min−1) in response to alarm cue, compared with prealarm cue. Survival (G): percentage survival of wild and laboratory fish after 35 d of acclimation to temperatures from 10 to 38 °C. (H) Red blood cell size (FSC-A). (I) RNA:DNA ratio (proxy of current growth). Maximum performance and upper and lower thermal performance breadths (80% performance) are illustrated beneath (A). Statistically significant differences indicated on each panel: Temp, significant effect of temperature on trait; Pop, significant difference between wild and laboratory fish (at 23 °C); Pop × Temp, significant interaction (see SI Appendix, Tables S1, S2, and S5).
Redundant genes that are not expressed can lose function due to neutral processes such as mutations or genetic drift (44). Physiological traits are often polygenic (45) and due to the likely complex genetic architecture of several of our measured traits, mutations would have to occur at a range of loci to cause functional changes. The slow mutational rate (thousands of generations) relative to the time the laboratory fish have spent in stable conditions (150 generations) (46, 47) makes it unlikely that this reduction of physiological plasticity in laboratory fish is due solely to mutation events. Changes in allele frequencies due to genetic drift, as well as directional selection for the stable laboratory environment, therefore, appear likely to be the mechanisms behind the phenotypic differences we observed. Furthermore, different laboratory strains of zebrafish that originate from different sources and collected from the wild at different times all have similar levels of heterozygosity and allele frequency patterns (48). This similarity suggests directional selection is the main driving force of adaptation to the laboratory environment rather than genetic drift. Differences in the regulation of gene expression could also be a mechanism contributing to the phenotypes we observed (49).
We predicted that laboratory fish should show higher growth only at optimal temperatures (Fig. 1A), but interestingly their growth was greater across all temperatures (Fig. 4A). Domestication generally increases growth rate in captivity in a range of organisms (50–52), and the elevated growth rate at all temperatures could be a spill-over effect from strong selection on growth performance at optimal temperature. In fish, higher activity in the growth hormone/IGF1 axis has been implicated in elevating growth after domestication (53), which matches the higher expression of the related genes ghra and igf1 in the muscle of laboratory fish (Fig. 4 B and C) in the present experiment. However, the growth hormone/IGF1 axis is only thought to be one of several mechanisms that contribute to the elevated growth of domesticated fish (54). Adaptation to the diet, high density, and aquaria are additional components of domestication that may have contributed to the higher growth of laboratory fish (55).
Meal digestion, assimilation, and growth are energetically costly and can reduce the aerobic scope available for other processes (32, 56). The lower aerobic scope despite higher growth rate, meaning less available residual aerobic scope, may contribute to the lower spontaneous activity and maximum swim speed seen in the laboratory fish (Figs. 2E and 4D). Laboratory fish also responded behaviorally less to an alarm cue (Fig. 4 E and F) across all temperatures than wild fish did. In the laboratory, such alarm behaviors are redundant as conditions are homogenous, food is abundant and there is no natural predation. A higher activity, maximum swim speed, and alarm cue response should be beneficial in the wild to escape predators, find food and gain information about the environment (57), but the resulting heightened alertness in the wild fish may be energetically demanding (58, 59) and could thus also contribute to the lower growth rate of wild fish. Conversely, by allocating fewer resources to growth, the wild fish may have been able to increase overall performance across a broader range of temperatures, highlighting a potential trade-off between physiological plasticity and growth.
At the coldest temperatures (10–12 °C), our results indicate that physiological plasticity is insufficient for normal function in both populations. Maximum swim speed, MMR, and alarm cue response showed a similar performance decrease in both wild and laboratory fish (Figs. 2 B and D and 4 E and F). Activity, growth and survival (Fig. 4 A, D, and G) were also reduced at these temperatures. The capacity that physiological plasticity has to restore high performance may be limited at these temperatures due to constraints in the underlying biochemical and physiological mechanisms of the traits (5, 37). These are temperatures that fall outside the range usually encountered in the wild (60), and the fish may therefore lack sufficiently cold-optimized enzyme isoforms (40, 61) or lipid regulation allowing sufficient membrane fluidity (62, 63). We have shown that while wild zebrafish are able to maintain high performance across a wider range of temperatures compared to a laboratory strain of zebrafish, there are environmental extremes where otherwise adaptive physiological plasticity fails to compensate for direct thermal effects.
Not all traits differed between the wild and laboratory zebrafish, for example, red blood cell (RBC) size increased similarly at high and low temperatures in both strains (Fig. 4H). An increase in RBC size may be due to RBC swelling caused by a physiological heat stress response and β-adrenergic stimulation (64–66). This may provide an explanation for the larger cell size at higher acclimation temperatures and corresponds to the increased expression of heat shock proteins we observed (Fig. 3 D–F). The increase at low temperatures may be an adaptive acclimation response, similar to the larger cells generally observed in both cold water-adapted and -acclimated fishes (67–70). Similarly, there were also no differences in RNA:DNA ratio across temperature or between the wild and laboratory fish (Fig. 4I) despite large differences in whole organism growth rate and gene expression (Fig. 4 A–C). RNA:DNA is a commonly used as a proxy for recent growth rate (71, 72), yet another study in zebrafish (73) also found a change in whole organism growth and gene expression but not RNA:DNA, suggesting that RNA:DNA may be a poor proxy for growth in zebrafish.
In this study, we show that laboratory zebrafish have lost a certain degree of physiological plasticity in many traits across all levels of biological organization: behavioral, physiological, and genetic. The lower plasticity and higher growth in the laboratory population is consistent with the hypothesis of a trade-off between plasticity and performance. A loss of physiological plasticity and improved growth rates in laboratory zebrafish is likely the result of selection for higher growth and adaption to stable and optimal temperatures in the laboratory in combination with other domestication effects, showing that physiological plasticity can be rapidly reduced in stable environments.
Methods
Experimental Animals.
The experiment was conducted November–December 2017 at the Norwegian University of Science and Technology (NTNU), Trondheim, Norway. A total of 300 wild zebrafish and 300 laboratory zebrafish were used in the experiment and acclimated to temperatures from 10 to 38 °C. The wild zebrafish were the offspring of wild-caught zebrafish collected from multiple sites in West Bengal, India and brought into the laboratory at NTNU in November 2016 (30, 74, 75). The wild fish were produced specifically for this experiment by random mating. The laboratory zebrafish were from the AB-wt strain, obtained from the Kavli Institute, Trondheim in October 2017. At the Kavli Institute the AB zebrafish line had been reared in an automated and controlled zebrafish rack system which minimized any fluctuations in water parameters, including temperature. Both wild and laboratory fish were produced using the same reproduction protocol as outlined in detail in Morgan et al. (75). All fish were held in similar holding tanks at 28 °C until the experiment commenced. The experiments were approved by the Norwegian Food Safety Authority (permit no. 8578).
Thermal Acclimation.
At the start of the experiment, all fish were anesthetized in buffered MS222 (conc. 110 mg/L); tagged using visible implant elastomers (VIE) (76); weighed (±0.001 g); and their standard length (±0.01 mm) was measured. Within each population, the fish were then haphazardly distributed between 15 aquaria with 20 fish per aquaria. Each aquarium was assigned to one temperature from 10 to 38 °C. The aquaria were 63-L, well aerated, and contained a filtration system and a plastic plant. Fish were fed ad libitum twice a day with dry fish flakes (Tetra pro), and water was changed regularly. To avoid acute thermal stress, the water temperatures were increased or decreased from 28 °C in a daily-stepwise manner using titanium heaters (TH-100, Aqua Medic) controlled by thermostats (ITC-306T, Inkbird), until final acclimation temperatures were reached (taking 0 to 9 days). Acclimation temperatures above 28 °C were increased at a rate of 1 °C day−1, whereas lower acclimation temperatures were reached by reducing the temperature by 2 °C day−1 until 12 °C and by 1 °C day−1 thereafter. Temperatures were monitored in real time and continuously recorded (Picotech TC-08) in each aquarium (SI Appendix, Fig. S1). Temperatures were kept constant at the desired temperatures for the duration of the experiment. The aquaria for both wild and laboratory fish were distributed between three climate-controlled rooms, one for the coldest (10–14 °C), one for intermediate (16–26 °C), and one for the warmest temperatures (28–38 °C). Acclimation temperatures and wild versus laboratory aquaria within each room were randomly distributed to reduce any within-room effects. The measurements of the fish traits were initiated 23 d after the start of the thermal ramping and continued until the final sampling on day 35.
Due to differing growth rates of wild and laboratory fish, the wild fish were 4–5 wk old at start of the experiment while the laboratory fish were 3–4 wk old. This enabled us to keep the size and life stage similar at the time of final sampling. In addition, due to the high number of fish and the many traits measured it was not possible to measure the wild and laboratory fish at the same time. The laboratory fish were therefore tagged, measured, and acclimated 12 d after the wild fish. Staggering the start time allowed phenotyping on both populations after the same acclimation time. The experimental dates for the wild fish were February 11, 2017, until July 12, 2017, and the laboratory fish from November 14, 2017, until December 19, 2017.
Quantification of Phenotypic Traits.
All phenotypic tests were performed at the water temperature at which fish were acclimated. Growth and survival were measured for all fish. We measured behavior, metabolic rates, response time, maximum swim speed and thermal tolerance in a subset of fish at each acclimation temperature. The same individuals had multiple traits quantified whenever possible. As some variables required longer duration measurements than other quicker tests, it was not possible to measure everything on all individuals and sample sizes therefore differ between variables. We minimized any differences that might occur due to sampling time by measuring two different temperatures each day and ensuring the temperature order of the wild and laboratory fish was the same. Organs were sampled for further analyses (e.g., biochemical and gene expression) from the fish that did not undergo any phenotyping. Phenotyping was not possible at 38 °C due to high mortality in both the wild and laboratory zebrafish resulting in these tanks being terminated.
Optimal Temperature and Thermal Performance Breadth.
For performances estimated to have a third order polynomial relationship with temperature, the performance breadth was defined and calculated as the difference between the two temperatures where performance was 80% of the maximum performance (3, 77). Performance breadth was excluded in cases where it went above 39 °C (i.e., activity) as this was outside the bounds of the experimental temperatures and exceeded long-term upper thermal tolerance (78). Optimal temperature was calculated as the peak of the polynomial curve, representing maximum performance.
Growth and Survival.
Weight (±0.001 g) and standard length (±0.01 mm) were measured at the start of the experiment during the tagging procedure and again at the end of the experiment. Specific growth rate (SGR) was then calculated for each fish as the percentage of weight gained per day (79). Mortality in the aquaria during the experiment was recorded and used to calculate the survival.
Behavior and Alarm Cue Response.
Behavior was measured individually for eight fish from each acclimation temperature in both wild and laboratory zebrafish, using a method similar to Vossen et al. (80). All behavioral trials within each strain (laboratory or wild) were completed within 3 d. The behavioral assay tanks measured 30 × 7 × 40 cm (length × breadth × height), with the water column at 25 cm, and with backs and sides painted opaque to minimize visual disturbance to the fish. Each tank was closed with a lid to maintain water temperature. The tanks were filmed at a front view using a USB camera (Kurokesu C1 IR). Eight assay tanks were filmed simultaneously spread evenly onto two shelves (SI Appendix, Fig. S8). The eight fish belonging to the same acclimation temperature were split in two and assayed on different days, thus each trial measured four fish from one acclimation temperature and four fish from another acclimation temperature.
Video recording was started once all eight fish had been carefully netted and put into their respective tanks. After 15 min, a conspecific cue (homogenized whole zebrafish) was added to the water via tubes connected to a timer-activated water pump, triggering an alarm cue response. Each tank’s tube ended in a pipette tip positioned above the water line and aimed against the tank’s wall to minimize water disturbance. The video was stopped, and the trial ended 10 min after the cue was added. After each trial the tanks were thoroughly cleaned to remove any chemical cues and were filled with fresh water.
Behavior was analyzed from videos using the automatic tracking software EthoVision XT12 (Noldus). Each assay tank was divided into three zones: surface (top 10% of water column), middle (83%), and bottom (7%; a zone small enough to identify bottom-dwelling behavior from random swimming) and the video analyzed separately precue (15 min) and postcue (2 min). The behaviors quantified from the videos were: total distance moved (body lengths per min; converted from cm), distance to the surface (cm), duration at the surface (seconds per minute), duration at the bottom (bottom dwelling; seconds per minute), and time spent freezing (total seconds per minute not moving). The alarm-cue response was calculated as the change in behavior post cue relative to the baseline precue behavior (by subtracting the precue behavior from the postcue behavior).
Metabolic Rates.
SMR.
SMR was measured using intermittent-flow respirometry (Loligo Systems) following best practices outlined in Clark et al. (31). Rates of oxygen consumption were recorded in four of the eight fish that had undergone the behavior trials and two SMR systems were run simultaneously for two different acclimation temperatures. All SMR measurements within each strain (wild or laboratory) were completed within 7 d. Each system contained four chambers (0.018 L), which were placed inside a temperature-controlled water bath with well-aerated water. Fish were tested individually with one fish per chamber. Each chamber was connected to a flush pump and a recirculation pump. When the flush pump was turned off the system was closed and the oxygen consumption (ṀO2) of the fish was recorded. The recirculation pump ensured there was constant mixing of the water within the chamber. The duration of the flush and measuring periods were adjusted for each acclimation temperature to prevent the dissolved oxygen falling below 80% air saturation, with warmer temperatures requiring shorter measurement periods. The oxygen concentration within the chamber was measured using fiber-optic leads focused on contactless sensor spots (Firesting O2, PyroScience) that were fixed on the inside of the chamber. The optodes were calibrated to 100% dissolved oxygen before adding the fish to the chamber and an external temperature sensor was used throughout. All fish had been starved for 24 h prior to beginning the SMR measurements. Each fish was carefully transferred to their chamber to minimize stress and measurements were started immediately once all fish were in place. Oxygen consumption was then continuously recorded overnight for 18 h to get resting values by allowing sufficient time for the fish to recover from the handling stress and the novel environment. At the end of the trial the fish were removed from their chambers, anesthetized, identified, and weighed, before entering the next performance test. After the fish had been removed from their chambers, background measurements were recorded for 10 min in all the chambers to account for bacterial respiration and algal photosynthesis. The system was then thoroughly bleached and rinsed before starting the next trial. All ṀO2 data were analyzed using the respR package in R (81) whereby SMR was calculated as the average of the lowest 10% of rates after outliers were removed, background respiration was subtracted, and the value converted into mg O2−1 h−1 g−1 by accounting for the volume of the respirometer and mass of the fish (31).
MMR.
Groups of five to seven fish were used to measure MMR using a method similar to Nilsson et al. (82). At the lowest acclimation temperatures, the size of the fish was very small and therefore oxygen consumption rates were very low. This meant measurements were not possible on individual fish as the volume of water in the swim tunnel was too large to accurately detect changes in the oxygen levels. In addition, zebrafish swim better in groups than individually. Three groups of fish were tested from each acclimation temperature. The fish were added to a customized 0.4 L circular glass food container (6 × 14 cm, 365+ IKEA). The chamber contained a raised stainless steel mesh platform with a magnetic stirring bar underneath creating a water flow. A piece of 4-cm diameter plastic pipe was added to the middle of the chamber to keep the fish swimming in the outer circumference of the chamber. Two holes were drilled into the lid, one that was connected to a flush pump for water exchange, and one with a chimney for water outlet and to allow an optode (Firesting O2, PyroScience) to be placed inside the chamber for measurements of oxygen consumption. Once the fish were added, the chamber was placed into a larger tank of water that contained plastic plants to minimize stress and kept the temperature constant. The tank lay on top of a magnetic stir plate and the chamber was sealed ensuring no air bubbles remained. The speed of the water was adjusted using the magnetic stir plate and the fish were first allowed to swim at a comfortable speed (i.e., the slowest speed at which the fish showed steady swimming) for 10 min. The speed was then increased for 5 min up until the fish could not maintain swimming. The speed was then adjusted down slightly (10%) so the fish were able to regain swimming and maintain their position in the water for at least 1 min. The fish were held at this speed for a maximum of 10 min during which time the oxygen consumption was measured. The rate of oxygen consumption during this time was used for the MMR calculations. MMR was analyzed using the respR package in R similar to the SMR. Since the measurement was taken on a group of fish, their individual masses were summed for use in the calculations, again accounting for the volume of the respirometer. The resulting MMR values were shown in mg O2−1 h−1 g−1.
Aerobic scope.
The aerobic scope was calculated by subtracting the mean SMR from the mean MMR at each acclimation temperature for the wild and laboratory zebrafish.
Maximum Swim Speed.
Group swim performance.
Maximum group swim speed was also obtained using a group swimming method similar to Usui et al. (83). The maximum swim speed was the speed at which the fish were able to maintain their position in the water column for at least 1 min. If two or more fish were not able to keep up with the group, then the speed was reduced. The arbitrary speeds of the stir plate were converted to cm s−1 by producing a calibration curve for the inner, middle, and outer part of the chambers. The fish mainly swam at the outer edge, and the water speed where the fish were positioned was calculated as the average of the middle and outer speeds. The speeds were then converted to body lengths per second. The mean length of each group of fish (measured at the end of the experiment) was used for calculation of body lengths per second.
Individual swim performance.
In addition to the group measurement of maximum swim speed, a swim tunnel respirometer (1.5 L) (Loligo Systems) was used to test the maximum swim speed of individual fish, similar to the methods in Killen et al. (84). Each fish was individually added to the swim chamber and left to swim at a low speed (i.e., the slowest speed at which the fish showed steady swimming) for 5 min to become accustomed to the chamber. The speed was then increased, and the fish were tested at two intermediate speeds for 5-min intervals. After this, the speed was gradually increased until the fish was not able to maintain its position in the water column. The speed was then adjusted down slightly, and the fish was left to swim for 30 s. This final speed was used as the maximum swimming speed. The arbitrary speeds of the motor were converted to cm s−1 by producing a calibration curve. Speeds were calibrated at four temperatures to check for a temperature effect on water speed, but there was no difference between them. The speeds were converted to body lengths per second using the length of the fish measured at the end of the experiment. This method of testing swim capacity was inferior to the group swimming in that many individuals appeared to show low motivation to swim, often letting themselves rest on the back grid at speeds much lower than their maximum. Testing groups of fish in species of shoaling fish can reduce stress and increase performance (85), so the group swimming was deemed the more reliable method. Oxygen consumption measurements from the single fish were too low to measure reliably at the lower temperatures and were therefore not included.
Response Time.
Response time, sometimes referred to as reaction latency, was measured in the same fish that had undergone the behavior and SMR tests (n = 2–4 per acclimation temperature and population) using methods similar to Gingins et al. (86). Each fish was tested individually three times. Similarly to the SMR measurements, all response time trials for each strain were completed within 7 d. The fish was placed into a 63-L (60 × 30 × 35 cm) aquarium with a reduced water level (10 cm depth; 18-L). The water temperature was controlled using a water-bath with heating and cooling elements to ensure the fish were tested at their acclimation temperature. The tank had a startle weight hanging above it, which was released by pulling a trigger string. The weight fell through a large diameter water PVC pipe without touching the sides. The pipe was placed so that it protruded 1 cm below the surface so that the fish could not see the weight before it hit the water. A mirror was placed under the aquarium at a 45° angle so that a high-speed camera (RX100 MkV, Sony) could record the fish from below. Once the fish was motionless and within the cameras field of view the weight was released. The event was recorded at 1000 frames s−1. Each fish was tested three times. However, if the fish did not respond it was tested up to five times. The response time was measured by counting the number of frames (each frame represented 1 ms) from when the weight hit the water until the fish first responded using VLC media player software (VLC, https://www.videolan.org/vlc/). The fish usually responded to the drop with a C-start, a common escape/startle reflex in fish. The response time is presented as the median time for each individual.
Thermal Tolerance.
Thermal tolerance was the final performance taken for the fish on the termination day of the experiment (day 35). The acute upper thermal tolerance was measured using the critical thermal maxima (CTmax) test following the protocol described by Morgan et al. (30, 75, 87). At each acclimation temperature, six to eight fish were tested together. Starting at their respective acclimation temperatures, the temperature was ramped up at a rate of 0.3 °C min−1 until the fish lost equilibrium and/or showed disorganized swimming (30, 87). At this point, they were removed from the tank, their individual CTmax was recorded, and the fish were euthanized before the final measurements (length and weight) were made and the termination of the experiment.
Molecular Analyses.
The fish that did not undergo the performance tests outlined above were euthanized, measured (weight and length), and sampled on the termination day (day 35). Within each population and at each acclimation temperature, three to six fish were used for molecular analyses. The brain, liver, and a muscle sample were dissected out of the fish on ice. These samples were stored in RNAlater (Invitrogen) and frozen at -80 °C until analysis.
Primers for 44 genes, suspected and previously reported to be related to thermal acclimation, were designed for real-time PCR with Primer3plus (https://www.primer3plus.com/) using GenBank (https://www.ncbi.nlm.nih.gov/genbank/) or were taken from previous publications, as indicated in SI Appendix, Table S3. Molecular analysis was performed at University of Greenwich, UK and MRC London Institute of Medical Sciences, UK. Total RNA from muscle, liver, and brain was extracted using the ReliaPrep RNA Cell Miniprep Kit (Promega), including a DNA wipe-out step with DNase I. For muscle samples this method was combined with a Tri-Reagent (Sigma-Aldrich) extraction prior to the RNA Kit step. RNA concentration was determined by Nanodrop ND-2000 (Thermo Fisher Scientific) and diluted to a common concentration of 50 ng µL−1 for muscle and liver, and 20 ng µL−1 for brain with nuclease free water (Promega). The GoScript Reverse Transcriptase (Promega) kit was used with Oligo(dT) primers to transcribe 500 ng of muscle RNA, 200 ng of liver RNA and 100 ng of brain RNA into cDNA. cDNA was stored at −20 °C until further use.
From all liver and muscle samples, plus a selection of brain samples, the expression of 44 genes (SI Appendix, Table S3) was analyzed with two technical replicates using the qPCR Biomark HD system (Fluidigm) based on 96.96 dynamic arrays (GE chips), as previously described in Miest et al. (88). A preamplification step was performed with a 500 nM pool of all primers in Preamp Master Mix (Fluidigm) and 1.25 µl cDNA per sample for 2 min at 95 °C followed by 10 cycles with 15 s at 95 °C and 4 min at 60 °C. Obtained preamp products were diluted 1:10 with low EDTA-TE buffer. The preamplified product was loaded onto the chip with SsoFast-EvaGreen Supermix Low Rox (Bio Rad Laboratories) and DNA-Binding Dye Sample Loading Reagent (Fluidigm). Primers were loaded onto the chip at a concentration of 50 µM in Assay Loading Reagent (Fluidigm) and low EDTA-TE Buffer. A dilution series was included to calculate primer efficiency and nontemplate controls were included to ensure samples were contamination free. The chip was run according to the Fluidigm 96.96 PCR protocol with a melting temperature (TM) of 60 °C. qBase+ 3.1 software (Biogazelle; http://www.qbaseplus.com) was used to select housekeeping (HK) genes and to verify stability of HK gene expression throughout analyzed samples (89). The following HKs were used for muscle: hif1ab, ef1a, cox4i1, and ndufs3 with a stability of M < 0.15; for liver ehhadh, cat, sod1, sod2, hif1ab, cox4i1, and ndufs3 were used with M < 0.15; and for brain hif1ab and ndufs3 were used with a stability of M < 0.15 (90). qBase+ was used to calculate compensated normalized relative quantity (CNRQ) in relation to a reference sample (i.e., laboratory zebrafish, 10 °C, sample 2) (90).
RNA:DNA Ratio Measurements.
RNA:DNA ratio can be used as a snapshot proxy for protein production and growth in fish and can be responsive to thermal acclimation (91). RNA:DNA measurements were carried out on the same fish as the molecular analyses. RNA and DNA were quantified according to (92, 93). A precooled stainless-steel ball was added to frozen muscle tissue in a 2 mL round-bottom Eppendorf tube. The tubes were placed in a precooled (−80 °C) homogenization block and homogenized (Tissuelyser, QIAgen) for 2 min (25 Hz). The tubes were moved to a homogenization block at room temperature, 100 μL sarcosil Tris-EDTA (STEB) buffer (0.5% vol/vol) was added, and homogenization was repeated. A total of 400 μL Tris-EDTA (pH 8.0) was added, and the tubes were centrifuged at 16 000 × g, 4 °C for 15 min. DNA and RNA was quantified from 10 to 20 μL of supernatant, using the Qubit (Life Technologies) RNA and dsDNA BR (broad range) assays, according to the supplier’s instructions. All measurements were carried out in duplicate.
Flow Cytometry.
Cell size has been connected to the performance in thermal tolerance in ectotherms (94). Here, we used flow cytometry as a quick method for red blood cell size estimates. Cell size was measured in the same fish as those used for RNA:DNA and molecular analyses. Blood was collected from the tail according to the protocol of Pedroso et al. (95). Collected blood (0.5–5.0 μL) was transferred to 0.9 × phosphate buffered saline (PBS, 1 mL) and centrifuged at 0.5 × g for 10 min. The supernatant was removed, and cells were resuspended in 0.9× PBS (0.5 mL). Samples were analyzed using a Novocyte flow cytometer (ACEA). To ensure that only single red blood cells (RBCs) were included in the analysis, two gates were created: one that selected events with low side scatter (SSC), and one that selected the events with both low forward scatter height (FSC-H) and low forward scatter area (FSC-A), which generally included the highest counts in the plot. Poor quality samples were excluded from the analysis. These included samples with low counts (<10,000 events), samples in which <4% of events were within the selected gate, and samples with unstable FSC-A signal over time. The mean of FSC-A from the gated cell population was used as a measure of RBC size.
Statistical Analyses.
Each phenotypic trait was analyzed separately, whereby the measured trait was the response variable and acclimation temperature, population (wild or laboratory) and their interaction (acclimation temperature × population) were the predictor variables. Weight was included as a covariate and was kept when it improved the model. This decision was based on an ANOVA which compared models with and without weight, including it when P < 0.05. Temperature was centered on the mean acclimation temperature (23 °C). For most phenotypic traits, model selection was used to test whether the effect of temperature was linear, quadratic (second order polynomial), or cubic (third order polynomial). The quality of the models were compared based on an ANOVA (significance criteria P < 0.05) and model parsimony using AIC (ΔAIC >2). When the ANOVA and the AIC-values showed different results, the simplest model was chosen. Only linear and quadratic temperature effects were tested when there was no biological reason to assume a cubic temperature effect or in those instances when the data were limited. Note that the same model (i.e., linear or quadratic temperature effect) was fitted to both populations for each trait to allow direct comparison. The significance of the predictor variables in the best model were estimated using a two-way ANOVA. Where the response variable was not normally distributed and/or the distribution was bound at the extremes (i.e., phenotypic traits: survival, bottom dwelling, time spent freezing and time spent at surface), the data were converted to proportions and GLMs were fitted with a binomial error distribution. Model selection was performed using parsimony (ΔAIC >2) and the significance of the predictor variables in the best model were estimated using a χ2 test (P < 0.05). When the response variable rapidly increased or decreased with temperature and was not bound (i.e., phenotypic traits: hsp70 and hsp90 in muscle and liver tissue), an exponential model was deemed the most appropriate model fit.
All figures in the main text show the raw data for ease of interpretation. Statistical outputs from ANOVAs are shown in SI Appendix, Tables S1 and S4 and model estimates in SI Appendix, Tables S2 and S6. Outputs from χ2 tests are shown in SI Appendix, Table S5. All analyses were carried out in R 3.4.3 (R Core Team, 2017) with effect sizes with P values less than 0.05 considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Research Council of Norway (Norges Forskningsråd Grant 62942 to F.J.), the Department of Biology, NTNU, and the Outstanding Academic Fellows Programme (OAFP) grant, NTNU. A.L. was supported by University of Greenwich VC studentship. Gene expression analysis was partly funded by the Fisheries Society of the British Isles Small Research Grant 2019 to J.J.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201919119/-/DCSupplemental.
Data Availability
Code and datasheets have been deposited in Figshare (DOI: 10.6084/m9.figshare.17121464).
References
- 1.Fry F. J., Responses of vertebrate poikilotherms to temperature. Thermobiology, 375–409 (1967). [Google Scholar]
- 2.Angilletta M. J., Niewiarowski P. H., Navas C. A., The evolution of thermal physiology in ectotherms. J. Therm. Biol. 27, 249–268 (2002). [Google Scholar]
- 3.Huey R. B., Stevenson R. D., Integrating thermal physiology and ecology of ectotherms: A discussion of approaches. Integr. Comp. Biol. 19, 357–366 (1979). [Google Scholar]
- 4.Schulte P. M., Healy T. M., Fangue N. A., Thermal performance curves, phenotypic plasticity, and the time scales of temperature exposure. Integr. Comp. Biol. 51, 691–702 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Seebacher F., White C. R., Franklin C. E., Physiological plasticity increases resilience of ectothermic animals to climate change. Nat. Clim. Chang. 5, 61–66 (2015). [Google Scholar]
- 6.Gunderson A. R., Stillman J. H., Plasticity in thermal tolerance has limited potential to buffer ectotherms from global warming. Proc. Biol. Sci. 282, 20150401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingsolver J. G., Huey R. B., Evolutionary analyses of morphological and physiological plasticity in thermally variable environments. Integr. Comp. Biol. 38, 545–560 (1998). [Google Scholar]
- 8.Garland T. Jr., Kelly S. A., Phenotypic plasticity and experimental evolution. J. Exp. Biol. 209, 2344–2361 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Somero G. N., The physiology of climate change: How potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J. Exp. Biol. 213, 912–920 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Dewitt T. J., Sih A., Wilson D. S., Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 13, 77–81 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Scheiner S. M., Barfield M., Holt R. D., The genetics of phenotypic plasticity. XI. Joint evolution of plasticity and dispersal rate. Ecol. Evol. 2, 2027–2039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevin L.-M., Lande R., Mace G. M., Adaptation, plasticity, and extinction in a changing environment: Towards a predictive theory. PLoS Biol. 8, e1000357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huey R. B., Kingsolver J. G., Evolution of thermal sensitivity of ectotherm performance. Trends Ecol. Evol. 4, 131–135 (1989). [DOI] [PubMed] [Google Scholar]
- 14.Richards C. L., Bossdorf O., Muth N. Z., Gurevitch J., Pigliucci M., Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9, 981–993 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Liefting M., Hoffmann A. A., Ellers J., Plasticity versus environmental canalization: Population differences in thermal responses along a latitudinal gradient in Drosophila serrata. Evolution 63, 1954–1963 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Snell-Rood E. C., Van Dyken J. D., Cruickshank T., Wade M. J., Moczek A. P., Toward a population genetic framework of developmental evolution: The costs, limits, and consequences of phenotypic plasticity. BioEssays 32, 71–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murren C. J., et al. , Constraints on the evolution of phenotypic plasticity: Limits and costs of phenotype and plasticity. Heredity 115, 293–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Relyea R. A., Costs of phenotypic plasticity. Am. Nat. 159, 272–282 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Forsman A., Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity 115, 276–284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen M. E., Papaj D. R., Why study plasticity in multiple traits? New hypotheses for how phenotypically plastic traits interact during development and selection. Evolution 10.1111/evo.14464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nüsslein-Volhard C., Dahm R., Zebrafish (OUP Oxford, 2002). [Google Scholar]
- 22.Mondal R., Bhat A., Temporal and environmental drivers of fish-community structure in tropical streams from two contrasting regions in India. PLoS One 15, e0227354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engeszer R. E., Patterson L. B., Rao A. A., Parichy D. M., Zebrafish in the wild: A review of natural history and new notes from the field. Zebrafish 4, 21–40 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Schaefer J., Ryan A., Developmental plasticity in the thermal tolerance of zebrafish Danio rerio. J. Fish Biol. 69, 722–734 (2006). [Google Scholar]
- 25.Westerfield M., The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) (University of Oregon Press, 1995). [Google Scholar]
- 26.Lawrence C., The husbandry of zebrafish (Danio rerio): A review. Aquaculture 269, 1–20 (2007). [Google Scholar]
- 27.Howe D. G., et al. , ZFIN, the zebrafish model organism database: Increased support for mutants and transgenics. Nucleic Acids Res. 41, D854–D860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsay J. M., et al. , Whole-body cortisol response of zebrafish to acute net handling stress. Aquaculture 297, 157–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright D., Ward A. J., Croft D. P., Krause J., Social organization, grouping, and domestication in fish. Zebrafish 3, 141–155 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Morgan R., et al. , Are model organisms representative for climate change research? Testing thermal tolerance in wild and laboratory zebrafish populations. Conserv. Physiol. 7, coz036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark T. D., Sandblom E., Jutfelt F., Aerobic scope measurements of fishes in an era of climate change: Respirometry, relevance and recommendations. J. Exp. Biol. 216, 2771–2782 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Jutfelt F., et al. , ‘Aerobic scope protection’ reduces ectotherm growth under warming. Funct. Ecol. 35, 1397–1407 (2021). [Google Scholar]
- 33.Queitsch C., Sangster T. A., Lindquist S., Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Rutherford S. L., Between genotype and phenotype: Protein chaperones and evolvability. Nat. Rev. Genet. 4, 263–274 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Rutherford S. L., From genotype to phenotype: Buffering mechanisms and the storage of genetic information. BioEssays 22, 1095–1105 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Rutherford S. L., Lindquist S., Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Somero G. N., Temperature adaptation of enzymes: Biological optimization through structure-function compromises. Annu. Rev. Ecol. Syst. 9, 1–29 (1978). [Google Scholar]
- 38.Jørgensen L. B., Malte H., Ørsted M., Klahn N. A., Overgaard J., A unifying model to estimate thermal tolerance limits in ectotherms across static, dynamic and fluctuating exposures to thermal stress. Sci. Rep. 11, 12840 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cadenas E., Davies K. J. A., Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–230 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Somero G. N., Proteins and temperature. Annu. Rev. Physiol. 57, 43–68 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Kussell E., Leibler S., Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Seebacher F., et al. , Plasticity of oxidative metabolism in variable climates: Molecular mechanisms. Physiol. Biochem. Zool. 83, 721–732 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Patapoutian A., Peier A. M., Story G. M., Viswanath V., ThermoTRP channels and beyond: Mechanisms of temperature sensation. Nat. Rev. Neurosci. 4, 529–539 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Albalat R., Cañestro C., Evolution by gene loss. Nat. Rev. Genet. 17, 379–391 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Crawford D. L., Schulte P. M., Whitehead A., Oleksiak M. F., Evolutionary physiology and genomics in the highly adaptable killifish (Fundulus heteroclitus). Compr. Physiol. 10, 637–671 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Lacy R. C., Loss of genetic diversity from managed populations: Interacting effects of drift, mutation, immigration, selection, and population subdivision. Conserv. Biol. 1, 143–158 (1987). [Google Scholar]
- 47.Masel J., King O. D., Maughan H., The loss of adaptive plasticity during long periods of environmental stasis. Am. Nat. 169, 38–46 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suurväli J., et al. , The laboratory domestication of zebrafish: From diverse populations to inbred substrains. Mol. Biol. Evol. 37, 1056–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaern M., Elston T. C., Blake W. J., Collins J. J., Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 6, 451–464 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Kimura T., Hamada Y., Growth of wild and laboratory born chimpanzees. Primates 37, 237–251 (1996). [Google Scholar]
- 51.Gjedrem T., Genetic improvement of cold-water fish species. Aquacult. Res. 31, 25–33 (2000). [Google Scholar]
- 52.Zuidhof M. J., Schneider B. L., Carney V. L., Korver D. R., Robinson F. E., Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 2005. Poult. Sci. 93, 2970–2982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleming I. A., Agustsson T., Finstad B., Johnsson J. I., Björnsson B. T., Effects of domestication on growth physiology and endocrinology of Atlantic salmon (Salmo salar). Can. J. Fish. Aquat. Sci. 59, 1323–1330 (2002). [Google Scholar]
- 54.Causey D. R., et al. , Proteomic comparison of selective breeding and growth hormone transgenesis in fish: Unique pathways to enhanced growth. J. Proteomics 192, 114–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robison B. D., Rowland W., A potential model system for studying the genetics of domestication: Behavioral variation among wild and domesticated strains of zebra danio (Danio rerio). Can. J. Fish. Aquat. Sci. 62, 2046–2054 (2005). [Google Scholar]
- 56.Secor S. M., Specific dynamic action: A review of the postprandial metabolic response. J. Comp. Physiol. B 179, 1–56 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Plaut I., Critical swimming speed: Its ecological relevance. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 131, 41–50 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Millidine K. J., Armstrong J. D., Metcalfe N. B., Presence of shelter reduces maintenance metabolism of juvenile salmon. Funct. Ecol. 20, 839–845 (2006). [Google Scholar]
- 59.Killen S. S., Reid D., Marras S., Domenici P., The interplay between aerobic metabolism and antipredator performance: Vigilance is related to recovery rate after exercise. Front. Physiol. 6, 111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spence R., et al. , The distribution and habitat preferences of the zebrafish in Bangladesh. J. Fish Biol. 69, 1435–1448 (2006). [Google Scholar]
- 61.Hochachka P. W., Somero G. N., Biochemical Adaptation: Mechanism and Process in Physiological Evolution (Oxford University Press, 2002). [Google Scholar]
- 62.Hazel J. R., Thermal adaptation in biological membranes: Is homeoviscous adaptation the explanation? Annu. Rev. Physiol. 57, 19–42 (1995). [DOI] [PubMed] [Google Scholar]
- 63.Hazel J. R., Williams E. E., The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 29, 167–227 (1990). [DOI] [PubMed] [Google Scholar]
- 64.Templeman N. M., LeBlanc S., Perry S. F., Currie S., Linking physiological and cellular responses to thermal stress: β-adrenergic blockade reduces the heat shock response in fish. J. Comp. Physiol. B 184, 719–728 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Nikinmaa M., Effects of adrenaline on red cell volume and concentration gradient of protons across the red cell membrane in the rainbow trout, Salmo gairdneri. Mol. Physiol. 2, 287–297 (1982). [Google Scholar]
- 66.Forster M. E., et al. , Catecholamine release in heat-stressed Antarctic fish causes proton extrusion by the red cells. J. Comp. Physiol. B 168, 345–352 (1998). [Google Scholar]
- 67.Verberk W. C. E. P., et al. , Shrinking body sizes in response to warming: Explanations for the temperature-size rule with special emphasis on the role of oxygen. Biol. Rev. Camb. Philos. Soc. 96, 247–268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van de Pol I. L. E., Hermaniuk A., Verberk W. C. E. P., Interacting effects of cell size and temperature on gene expression, growth, development and swimming performance in larval zebrafish. Front. Physiol. 12, 738804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hermaniuk A., van de Pol I. L. E., Verberk W. C. E. P., Are acute and acclimated thermal effects on metabolic rate modulated by cell size? A comparison between diploid and triploid zebrafish larvae. J. Exp. Biol. 224, jeb227124 (2021). [DOI] [PubMed] [Google Scholar]
- 70.Van Voorhies W. A., Bergmann size clines: A simple explanation for their occurrence in ectotherms. Evolution 50, 1259–1264 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Bulow F. J., RNA–DNA ratios as indicators of recent growth rates of a fish. J. Fish. Res. Board Can. 27, 2343–2349 (1970). [Google Scholar]
- 72.Buckley L., Caldarone E., Ong T.-L., RNA–DNA ratio and other nucleic acid-based indicators for growth and condition of marine fishes. Hydrobiologia 401, 265–277 (1999). [Google Scholar]
- 73.Masuda Y., Oku H., Okumura T., Nomura K., Kurokawa T., Feeding restriction alters expression of some ATP related genes more sensitively than the RNA/DNA ratio in zebrafish, Danio rerio. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 152, 287–291 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Sundin J., et al. , On the observation of wild zebrafish (Danio rerio) in India. Zebrafish 16, 546–553 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Morgan R., Finnøen M. H., Jensen H., Pélabon C., Jutfelt F., Low potential for evolutionary rescue from climate change in a tropical fish. Proc. Natl. Acad. Sci. U.S.A. 117, 33365–33372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hohn C., Petrie-Hanson L., Evaluation of visible implant elastomer tags in zebrafish (Danio rerio). Biol. Open 2, 1397–1401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phillips B. L., Llewelyn J., Hatcher A., Macdonald S., Moritz C., Do evolutionary constraints on thermal performance manifest at different organizational scales? J. Evol. Biol. 27, 2687–2694 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Åsheim E. R., Andreassen A. H., Morgan R., Jutfelt F., Rapid-warming tolerance correlates with tolerance to slow warming but not growth at non-optimal temperatures in zebrafish. J. Exp. Biol. 223, p.jeb229195 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Hopkins K. D., Reporting fish growth: A review of the basics1. J. World Aquacult. Soc. 23, 173–179 (1992). [Google Scholar]
- 80.Vossen L. E., et al. , Low concentrations of the benzodiazepine drug oxazepam induce anxiolytic effects in wild-caught but not in laboratory zebrafish. Sci. Total Environ. 703, 134701 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Harianto J., Carey N., Byrne M., respR—An R package for the manipulation and analysis of respirometry data. Methods Ecol. Evol. 10, 912–920. (2019). [Google Scholar]
- 82.Nilsson G. E., Östlund-Nilsson S., Penfold R., Grutter A. S., From record performance to hypoxia tolerance: Respiratory transition in damselfish larvae settling on a coral reef. Proc. Biol. Sci. 274, 79–85 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Usui T., et al. , The French press: A repeatable and high-throughput approach to exercising zebrafish (Danio rerio). PeerJ 6, e4292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Killen S. S., Croft D. P., Salin K., Darden S. K., Male sexually coercive behaviour drives increased swimming efficiency in female guppies. Funct. Ecol. 30, 576–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Culbert B. M., Gilmour K. M., Balshine S., Social buffering of stress in a group-living fish. Proc. Biol. Sci. 286, 20191626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gingins S., Roche D. G., Bshary R., Mutualistic cleaner fish maintains high escape performance despite privileged relationship with predators. Proc. Biol. Sci. 284, 20162469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morgan R., Finnøen M. H., Jutfelt F., CT max is repeatable and doesn’t reduce growth in zebrafish. Sci. Rep. 8, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miest J. J., Arndt C., Adamek M., Steinhagen D., Reusch T. B. H., Dietary β-glucan (MacroGard®) enhances survival of first feeding turbot (Scophthalmus maximus) larvae by altering immunity, metabolism and microbiota. Fish Shellfish Immunol. 48, 94–104 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Vandesompele J., et al. , Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J., qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Malzahn A. M., Clemmesen C., Rosenthal H., Temperature effects on growth and nucleic acids in laboratory-reared larval coregonid fish. Mar. Ecol. Prog. Ser. 259, 285–293 (2003). [Google Scholar]
- 92.Caldarone E., Wagner M., St. Onge-Burns J., Buckley L. J., Protocol and guide for estimating nucleic acids in larval fish using a fluorescence microplate reader. Northeast Fisheries Science Center (2001). https://repository.library.noaa.gov/view/noaa/5145. Accessed 3 April 2018.
- 93.Izzo C., Doubleday Z. A., Schultz A. G., Woodcock S. H., Gillanders B. M., Contribution of water chemistry and fish condition to otolith chemistry: Comparisons across salinity environments. J. Fish Biol. 86, 1680–1698 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Leiva F. P., Calosi P., Verberk W. C. E. P., Scaling of thermal tolerance with body mass and genome size in ectotherms: A comparison between water- and air-breathers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pedroso G. L., et al. , Blood collection for biochemical analysis in adult zebrafish. J. Vis. Exp. (63) e3865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and datasheets have been deposited in Figshare (DOI: 10.6084/m9.figshare.17121464).