Abstract
Background:
Congenital heart disease (CHD) is the most common anomaly at birth, with a prevalence of approximately 1%. While infants born to mothers with diabetes or obesity have a 2–3-fold increased incidence of CHD, the cause of the increase is unknown. Damaging de novo variants (DNV) in coding regions are more common among patients with CHD, but genome-wide rates of coding and noncoding DNVs associated with these prenatal exposures have not been studied in patients with CHD.
Methods:
DNV frequencies were determined for 1,812 patients with CHD who had whole genome sequencing and prenatal history data available from the Pediatric Cardiac Genomics Consortium’s CHD GENES study. The frequency of DNVs was compared between subgroups using t-test or linear model.
Results:
DNV frequencies were compared for 1,812 patients with CHD and prenatal history data who were recruited to the Pediatric Cardiac Genomics Consortium’s CHD GENES study. The number of DNVs per CHD patient was higher with exposure to maternal diabetes (76.5 vs 72.1, t-test p-value 3.03x10-11), but the difference was no longer significant after including parental ages in a linear model (paternal and maternal correction p-value 0.42). No interaction was observed between diabetes risk and parental age (paternal and maternal interaction p-values 0.80 and 0.68, respectively). No difference was seen in DNV count per patient based on maternal obesity (72.0 vs 72.2 for maternal BMI <25 vs maternal BMI >30, t-test p-value 0.86).
Conclusions:
After accounting for parental age, the offspring of diabetic or obese mothers have no increase in DNVs compared with other children with CHD. These results emphasize the role for other mechanisms in the etiology of CHD associated with these prenatal exposures.
Journal Subject Terms: Congenital Heart Disease, Genetics, Obesity, Pregnancy
Keywords: congenital heart disease, de novo variant, maternal diabetes, obesity, whole genome sequencing
Introduction
Congenital heart disease (CHD) is the most common anomaly at birth with a prevalence of approximately 6–13 in 1000 births1,2. CHD can be caused by a variety of genetic anomalies including aneuploidies, copy number variants (CNVs) and inherited or de novo single nucleotide or small insertion/deletion variants (DNVs)3–8. DNVs are also associated with important outcomes for CHD patients such as the risk of neurodevelopmental delay and postoperative recovery3,8–10.
Offspring of obese11,12, hypertensive13, and diabetic mothers12,14 are more likely to have CHD than other infants. When attributable causes were identified for 1565 infants with CHD, maternal obesity was the most common modifiable risk factor15. The magnitude of increased risk is generally lower with obesity exposure than with diabetes exposure, though the two conditions often overlap. In a study of the National Birth Defects Prevention Study, CHD risk was elevated among overweight mothers regardless of gestational diabetes status, but the odds ratio (OR) for CHD was higher among mothers who also had gestational diabetes16. The mechanism(s) by which these prenatal exposures confer an increased risk of CHD remain unclear. As the prevalence of obesity and diabetes have risen in the past decade17,18, defining the precise cause of increased CHD risk in affected pregnancies has taken on additional urgency. Genetic risk may play a role, as mothers of children with conotruncal heart defects were more likely to have a high polygenic risk for type II diabetes than fathers19.
Extensive whole genome sequence of >1800 CHD trios (proband and parents) provides an opportunity to assess the contribution of DNVs, mediated by a variety of prenatal exposures, to congenital heart disease. We and others have demonstrated that each child has approximately 75 DNVs, coding and noncoding, not carried by their parents20,21. We hypothesized that if a prenatal exposure, such as maternal diabetes or obesity, increased the frequency of de novo mutations in the child, this increase should be reflected in the whole genome sequence of the trio.
First, we compared the prevalence of maternal gestational diabetes in mothers of CHD probands in the PCGC (Pediatric Cardiovascular Genomics Consortium)22 cohort of >10,000 CHD families. We also assessed the association between extracardiac anomalies and prenatal exposure to maternal diabetes or obesity in this cohort. We then determined if any of these prenatal exposures is associated with an increase in de novo single nucleotide or small insertion/deletion variants, by comparing CHD probands' whole genome sequence (WGS) to their parents' WGS. Additionally, we examined the association of prenatal exposures with extracardiac anomalies and presence of loss-of-function variants in known CHD genes.
Methods
CHD participants were recruited to the Congenital Heart Disease Network Study of the Pediatric Cardiac Genomics Consortium (CHD GENES: ClinicalTrials.gov identifier NCT01196182) as previously described23. All participants or their parents provided written informed consent using protocols that were reviewed and approved by institutional review boards at participating institutions. Whole genome sequence data used in this study has been deposited in the National Institutes of Health dbGaP resource. Researchers trained in human subject confidentiality protocols may request access to this data at dbgap.ncbi.nlm.nih.gov. Full methods are available in Supplemental Materials.
Results
Prevalence of PCGC CHD probands who were born to diabetic or obese mothers
Of the 12,842 CHD patients enrolled in the PCGC22 with prenatal history, whole genome sequencing (WGS) data was available for 1,812 CHD trios (Supplemental Table I and Supplemental Data I). Maternal diabetes, pre-gestational or gestational, was reported in 188 pregnancies. Maternal BMI was reported as >30 kg/m2 for 1605 pregnancies (259 with WGS data), and <25 kg/m2 for 7066 pregnancies (1075 with WGS data). For the overall cohort, as well as for all maternal ages 20 and above at time of birth, there was a 1.7–2.7 fold increase in gestational diabetes (GDM) and a 2.9–8.7 fold increase in pre-gestational diabetes (PDM) among mothers in PCGC compared to age-matched US birth cohorts18,24 (Table 1). Gestational diabetes and maternal obesity were both associated with an increased odds (1.39–1.57 fold) of an extracardiac anomaly compared to children of mothers without these risk factors (Table 2). The increase in extracardiac anomalies was observed for infants born to mothers with pre-gestational diabetes was nominal. The increase in extracardiac anomalies with gestational diabetes or maternal obesity remained true among younger (ages 20–30) and older (ages 30–40) mothers (Supplemental Table II).
Table 1.
Increased maternal diabetes prevalence among CHD pregnancies.
| GDM | PCGC | US Birth Cohort | GDM OR (95% CI) | GDM Binomial p-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Maternal Age, years | Father Age, mean years (range) | GDM Expose d | Total | Proportion Exposed | GDM Exposed | Total | Proportion Exposed | ||
| 30.1 (13.0–55.3) | 32.6 (13.6–68.9) | 846 | 11930 | 0.071 | 3340594 | 124428672 | 0.027 | 2.7 (2.5–2.9) | 6.05E-141 |
| <20 | 21.5 (13.6–42.2) | 8 | 584 | 0.014 | 103849 | 15208765 | 0.007 | 2.0 (0.9–4.0) | 6.92E-02 |
| 20–24 | 26.3 (16.4–62.4) | 58 | 1905 | 0.030 | 471311 | 33393397 | 0.014 | 2.2 (1.6–2.8) | 1.96E-07 |
| 25–29 | 30.5 (18.1–55.8) | 188 | 3211 | 0.059 | 896603 | 36454907 | 0.025 | 2.4 (2.1–2.8) | 5.54E-25 |
| 30–34 | 34.4 (19.5–68.9) | 302 | 3774 | 0.080 | 1033307 | 26314660 | 0.039 | 2.0 (1.8–2.3) | 3.88E-27 |
| 35–39 | 38.5 (21.5–60.4) | 201 | 1909 | 0.105 | 649536 | 10854462 | 0.060 | 1.7 (1.5–2.0) | 7.59E-12 |
| >40 | 42.7 (25.0–66.7) | 89 | 547 | 0.163 | 185988 | 2202481 | 0.084 | 1.9 (1.5–2.4) | 1.74E-07 |
| PDM | PCGC | US Birth Cohort | PDM OR (95% CI) | Binomial p-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Maternal Age, years | Father Age, mean years (range) | PDM Expose d | Total | Proportion Exposed | PDM Exposed | Total | Proportion Exposed | ||
| 30.7 (16.3–46.3) | 33.5 (17.0–56.3) | 494 | 11578 | 0.043 | 34010 | 3932094 | 0.009 | 5.1 (4.7–5.6) | 7.10E-183 |
| <20 | 22.1 (17.0–31.9) | 16 | 592 | 0.027 | 847 | 211827 | 0.004 | 6.9 (3.9–11.4) | 4.35E-09 |
| 20–24 | 26.9 (18.8–52.3) | 81 | 1928 | 0.042 | 4016 | 803153 | 0.005 | 8.7 (6.9–10.9) | 1.74E-46 |
| 25–29 | 30.7 (19.8–47.8) | 111 | 3134 | 0.035 | 8036 | 1148057 | 0.007 | 5.2 (4.3–6.3) | 4.43E-42 |
| 30–34 | 35.1 (21.3–56.3) | 155 | 3627 | 0.043 | 11100 | 1110010 | 0.010 | 4.4 (3.7–5.2) | 3.59E-49 |
| 35–39 | 37.8 (24.5–54.7) | 103 | 1811 | 0.057 | 7658 | 546995 | 0.014 | 4.2 (3.4–5.2) | 9.86E-32 |
| >40 | 43.0 (30.6–54.5) | 28 | 486 | 0.058 | 2353 | 112052 | 0.021 | 2.9 (1.9–4.2) | 2.36E-06 |
Abbreviations: CI, confidence interval; GDM, gestational diabetes mellitus; OR, odds ratio; PCGC, Pediatric Cardiac Genomics Consortium; PDM, pre-gestational diabetes mellitus; US, United States. Bonferroni p-value threshold: 0.0035
Table 2.
Increased extracardiac anomalies among CHD patients exposed to gestational diabetes or maternal obesity.
| Exposure Status | Extracardiac Anomalies Present, Number | Extracardiac Anomalies Absent, Number | Fisher OR (95% CI, p-value) |
|---|---|---|---|
| PGD Exposed | 25 | 31 | 1.32 (1.03–3.19, 2.92E-02) |
| PGD Non-Exposed | 3518 | 7929 | - |
| GDM Exposed | 349 | 502 | 1.57 (1.35–1.81, 1.04E-09) |
| GDM Non-Exposed | 3518 | 7929 | - |
| Maternal BMI >30 | 594 | 1007 | 1.39 (1.24–1.56, 1.84E-08) |
| Maternal BMI <25 | 2100 | 4948 | - |
Abbreviations: CI, confidence interval; GDM, gestational diabetes mellitus; OR, odds ratio; PDM, pre-gestational diabetes mellitus. Bonferroni p-value threshold: 7.14E-03
DNV frequencies in CHD children of obese or diabetic mothers compared with the DNV frequency in CHD children of mothers without these risk factors.
There was no significant difference in DNVs among CHD patients born to mothers with obesity. By contrast there were significantly more DNVs (76.5, both coding and noncoding) in CHD patients with prenatal exposure to maternal diabetes than in CHD patients whose mothers did not have diabetes (72.1; Table 3). However, diabetic mothers were significantly older than non-diabetic mothers and increased parental (both paternal and maternal) age is correlated with increased numbers of DNVs in the child20,21,25,26. After including parental ages in the linear model, there was no significant difference in the numbers of DNVs found in CHD offspring of diabetic mothers compared to the numbers of DNVs was found in CHD offspring of non-diabetic mothers (Table 4). This remained true after excluding all probands with isolated atrial septal defect (ASD, Supplemental Table III). Further, correcting for parental ages and diabetes exposure in a pairwise fashion did not identify any interactions between parental age and diabetes effects on CHD patient DNVs (Supplemental Table IV). When patients exposed to gestational or pre-gestational diabetes were separately analyzed, results were similar.
Table 3.
De novo variant frequency differs by parental age and exposure.
| Exposure Category | WGS trios | Mean DNVs | Standard Deviation DNVs | DNVs T-test p-value | Mean Maternal Age, Years | Mean Paternal Age, Years |
|---|---|---|---|---|---|---|
| No Diabetes | 1605 | 72.1 | 15.9 | - | 30.9 | 33.2 |
| Diabetes | 188 | 76.5 | 16.6 | 3.03E-11 | 33.1 | 35.9 |
| GDM | 132 | 76 | 16.8 | 1.13E-02 | 33.1 | 36.0 |
| PGD | 56 | 77.6 | 16.4 | 1.71E-02 | 33.1 | 35.9 |
| BMI <25 | 1075 | 72 | 16.0 | - | 30.9 | 33.2 |
| BMI >30 | 259 | 72.2 | 16.2 | 8.62E-01 | 31.2 | 33.8 |
| Maternal Age 20–30yo without Diabetes | 620 | 63.6 | 12.2 | - | 26.0 | 28.8 |
| Maternal Age 20–30yo with Diabetes | 48 | 63.4 | 14.2 | 0.915** | 26.1 | 29.5 |
| Maternal Age 20–30yo with GDM | 36 | 64.3 | 14.5 | 0.775** | 26.3 | 30.1 |
| Maternal Age 20–30yo with PGD | 12 | 60.6 | 13.5 | 0.454** | 25.7 | 27.9 |
| Maternal Age 30–40yo with Diabetes | 120 | 79.3 | 14.3 | 4.15E-09 *** | 34.9 | 37.3 |
| Maternal Age 30–40yo with GDM | 82 | 77.5 | 14.6 | 6.11E-08 *** | 34.8 | 37.3 |
| Maternal Age 30–40yo with PGD | 38 | 80.6 | 13.7 | 2.00E-07 *** | 34.5 | 37.4 |
Abbreviations: BMI, body mass index; CI, confidence interval; DNV, de novo variant; GDM, gestational diabetes mellitus; PDM, pre-gestational diabetes mellitus.
BMI as continuous variable in GLM
compared to Maternal Age 20–30 without Diabetes
compared to Maternal Age 20–30 with Diabetes
Table 4.
De novo variant frequency is primarily driven by parental age.
| Exposure Category | Diabetes Poisson GLM P-Value (Parameter Esimate, Standard Error) | Diabetes Poisson GLM P-Value with Maternal Age (Parameter Estimate, Standard Error) | Diabetes Poisson GLM P-Value with Paternal Age (Parameter Estimate, Standard Error) | Diabetes Poisson GLM P-Value with Parental Ages (Parameter Estimate, Standard Error) |
|---|---|---|---|---|
| Any Diabetes | 3.0E-11 (0.06, 0.01) | 0.643 (4.1E-03, 8.9E-03) | 0.709 (−3.3E-3, 8.9E-03) | 0.424 (−7.2E-03, 9.0E-03) |
| GDM | 4.2E-07 (0.05, 0.01) | 0.899 (−1.3E-03, 5.2E-04) | 0.368 (−9.5E-03, 0.01) | 0.219 (−0.01, 0.01) |
| PGD | 2.3E-06 (0.07, 0.02) | 0.284 (0.02, 0.02) | 0.471 (0.01, 0.02) | 0.666 (0.01, 0.02) |
Abbreviations: GDM, gestational diabetes mellitus; GLM, general linear model; PDM, pre-gestational diabetes mellitus. GLM comparison Bonferroni p-value threshold 0.0038 (13 total comparisons including Main and Supplemental Tables)
Genomic risk score does not indicate contribution of common variants associated with diabetes to DNV frequency
Genomic risk scores (GRSs) for Type 2 diabetes27 and hypertension28 were calculated for mothers using published variant weights. In a Poisson linear model, maternal diabetes GRS was nominally correlated with DNV frequency, but not after consideration of maternal diabetes status and parental age (Table 5). Maternal hypertension GRS was not correlated with DNV frequency.
Table 5.
Genomic risk score for diabetes and hypertension not associated with DNV frequency.
| Maternal GRS | Poisson GLM P-Value (Parameter Estimate, Standard Error) | Poisson GLM P-Value with Diabetes Exposure as Covariate (Parameter Estimate, Standard Error) | Poisson GLM P-Value with Parental Ages and Diabetes Exposure as Covariates (Parameter Estimate, Standard Error) |
|---|---|---|---|
| Diabetes | 0.04 (−321900, 159300) | 0.05 (−312100, 159300) | 0.89 (−22590, 159200) |
| Hypertension | 0.47 (−566, 790) | 0.65 (−355, 791) | 0.55 (471, 793) |
Abbreviations: GRS, genomic risk score. GLM comparison Bonferroni p-value threshold 0.025 (2 total comparisons)
Pathogenic CHD variants
Rare heterozygous variants in at least 138 human CHD genes confer congenital heart disease risk4,29 (Supplemental Table V). Rare loss-of-function (LOF) variants in these 138 genes, identified from whole exome sequence (WES) data, have been described for 4,443 PCGC probands10, including 3,672 with prenatal history data (Supplemental Table VI). Overall, 6% (206/3,672) of the PCGC cohort with both WES data and documented prenatal history had a LOF CHD gene variant. There was no difference observed in the likelihood of having a LOF CHD gene variant based on exposure to maternal gestational diabetes (Supplemental Data II).
Discussion
Identifying modifiable risk factors for CHD could lead to significant improvement in neonatal health. Many non-genetic CHD risk factors are well established, such as in utero rubella infection, maternal alcohol consumption, and exposure to toxic compounds such as thalidomide30. Neighborhood-level factors and occupational exposures have also been associated with increased CHD risk31,32. This is the first study to characterize genome-wide DNV frequency in CHD offspring of mothers with diabetes or obesity. Our analysis of DNVs among CHD patients, stratified by these perinatal exposures, indicates that increased rates of DNVs are not a common mechanism for the observed increase in CHD risk (Figure 1). Though a higher number of DNVs were associated with maternal diabetes, the increase was accounted for by the associated difference in parental ages. While the increased number of DNVs would lead to a small increased risk of a CHD gene variant, no excess of LOF variants in dominant CHD genes were observed in CHD patients with these prenatal exposures. Our results suggest other factors such as inherited genetic variants, maternal metabolic influences on the developing heart, or environmental factors as important areas of future research to better understand their impact on CHD risk.
Figure 1.
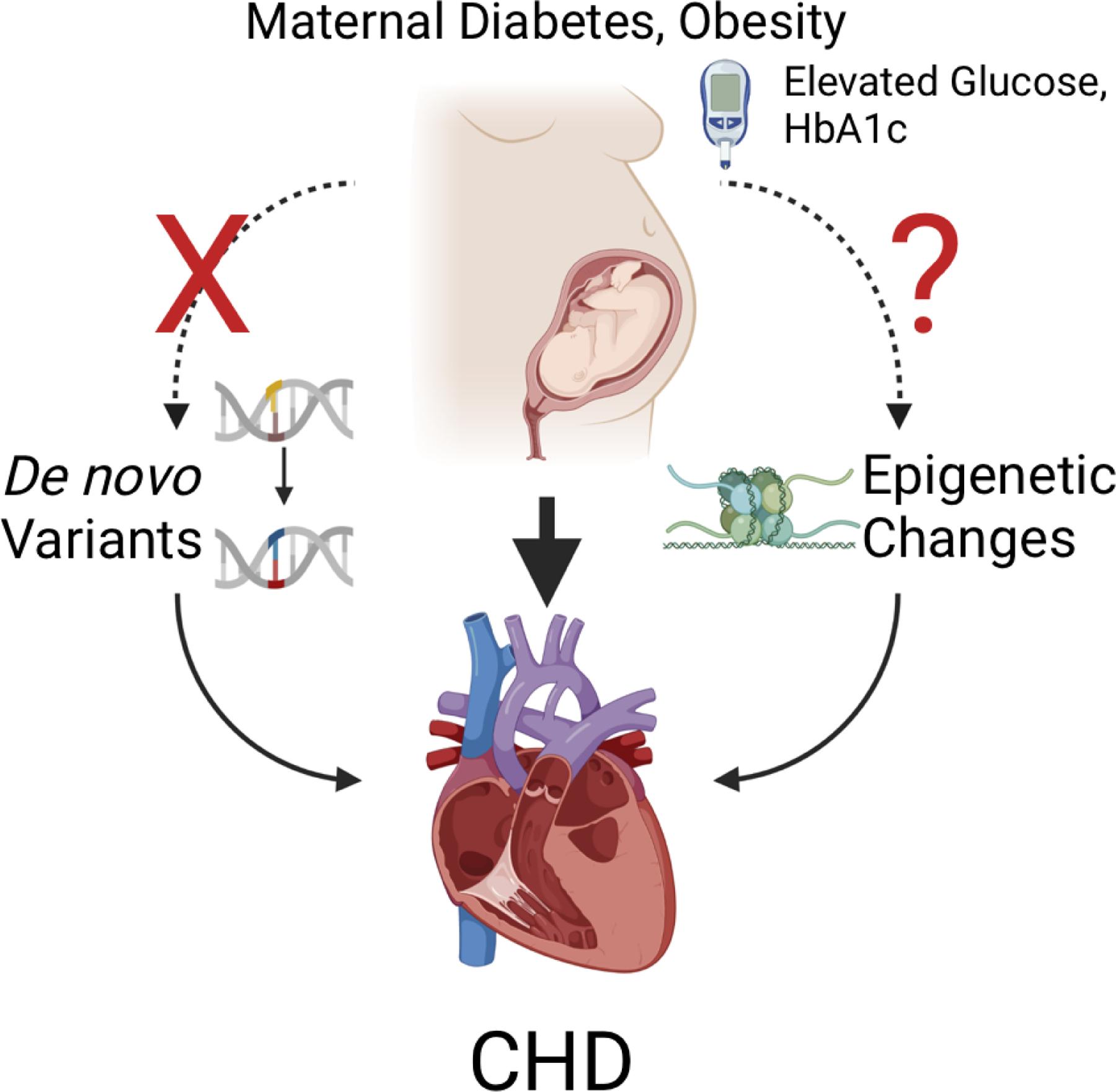
Maternal diabetes and obesity are risk factors for congenital heart disease (CHD). Potential mechanisms for risk include de novo variants (DNVs) and epigenetic changes, both of which are known to cause CHD. No increase in DNVs was associated with maternal diabetes or obesity, indicating that other mechanisms such as epigenetic changes are responsible for the increased CHD. Created with BioRender.
Other potential mechanisms for CHD risk
As stressors in the in utero environment are associated with epigenetic changes and many CHD genes also regulate chromatin state, we hypothesize that similar molecular pathways can be modified by environmental and genetic factors early in development. Elevated glucose and increased inflammation may contribute to CHD risk associated with maternal diabetes and obesity12. Maternal obesity and pre-gestational diabetes are both associated with alterations in glucose control, and exposure to hyperglycemia leads to abnormal gene expression in isolated cardiomyocytes33 as well as mouse models of development34. Maternal hyperglycemia leads to decreased chromatin accessibility at the eNOS locus and subsequent increase in Jarid2 expression in a mouse model of CHD sensitized by haploinsufficiency of Notch133. Supplementation of diabetic mice with cofactors for endothelial nitric oxide synthase (eNOS) during pregnancy reduces CHD, highlighting the potential role of endothelial dysfunction in CHD pathogenesis35. Glucose may be a dose-dependent teratogen, as higher hemoglobin A1c values are associated with greater risk of CHD36. Clinical severity of diabetes also correlates with CHD risk, as mothers with acute diabetes complications such as ketoacidosis during pregnancy were more likely to have an infant with CHD than those with uncomplicated diabetes37. Similarly, congenital malformations are more common with increasing severity of maternal obesity38. Potential protective factors include exercise39, first-trimester folic acid supplementation40, as nutritional deficiencies can be more prevalent among women with obesity, though the observed benefit has not been consistently observed41. Our data demonstrate that the biologic basis for these associations is not an increase in DNVs among CHD genes.
Shared risk with other developmental disorders
Extracardiac anomalies and neurodevelopmental impairments are commonly associated with CHD42,43. Maternal obesity and diabetes, alone and in combination, are also associated with an increased risk for neurodevelopmental disability among offspring without CHD44,45. Children born to mothers with both obesity and pre-gestational diabetes have a further elevated risk of neurodevelopmental disability44. The mechanism of risk may involve perturbations to early brain development, as BMI is negatively correlated with fronto-thalamic connectivity in the first month of life46. Consistent with previous studies, which have demonstrated larger odds ratios for pre-gestational diabetes association with multiple congenital anomalies than for isolated cases14, we also observed that maternal gestational diabetes and obesity were both associated with an increased likelihood of extracardiac anomalies (Table 2). Association of congenital anomalies with both pre-gestation diabetes as well as gestational diabetes, which is typically diagnosed at approximately 28 weeks of pregnancy, raises important questions about whether hyperglycemia and/or metabolic differences associated with obesity and insulin resistance is the primary teratogen responsible.
Limitations
Limitations include the use of questionnaires and review of patient medical records to determine maternal diabetes status. Neither the onset nor duration of gestational diabetes or information regarding maternal hypertension were available for our cohort. We recognize that maternal environmental exposures may modify DNV frequency; however these characteristics that were not measured in our cohort. We assessed DNVs that create single nucleotide polymorphisms and short insertions and deletions in coding and noncoding regions of the genome; future studies will consider other types of de novo variation (e.g., large insertions and deletions). Finally, it remains possible that subsets of diabetes or obesity exposures may influence de novo frequency, but further stratification was limited due to our sample size.
Future directions
These results emphasize the need to study mechanisms of CHD risk associated with modifiable risk factors. Genetic risk can be modified by environmental factors, and vice versa47. Abnormal DNA methylation profiles have been identified among CHD patients, but correlations with prenatal exposures are not reported48. Many pathogenic CHD variants demonstrate variable penetrance and expressivity, highlighting the possibility that environmental factors could also modify CHD severity. Mouse models of CHD have also demonstrated that penetrance of NOTCH1-related CHD is increased by exposure to gestational hypoxia49. Additional support for an interaction between genetic risk and prenatal exposures includes the finding that genetic associations with maternal hypertensive disorders were observed in both fetal and maternal genomes50, indicating that genetic as well as in utero environmental factors could contribute to increased CHD. Improved understanding of the basis of increased CHD associated with prenatal exposures could improve prenatal care to reduce the incidence of CHD and other congenital anomalies.
Supplementary Material
Acknowledgements
The authors would like to thank all participants and their families.
Funding Sources
The authors thank the participants of the PCGC and those in other human genetic studies that have enabled this research. This work was supported in part by grants from the Harvard Medical School Epigenetics & Gene Dynamics Award and American Heart Association Post-Doctoral Fellowship (S.U.M.), the Constance Goulandris Foundation (W.J.C.), the National Center for Research Resources (U01 HL098153), the National Center for Advancing Translational Sciences (UL1TR000003, 1TR002541) the National Institutes of Health (U01-HL098188, U01-HL098147, U01-HL098153, U01-HL098163, U01-HL098123 and U01-HL098162), the NIH Centers for Mendelian Genomics (5U54HG006504), and the Howard Hughes Medical Institute (R.P.L. and C.E.S.). Funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; nor the decision to submit the manuscript for publication. S.U.M. and C.E.S. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. S.U.M., J.G.S. and C.E.S conducted and are responsible for the data analysis.
Non-standard Abbreviation and Acronyms:
- ASD
atrial septal defect
- BMI
body mass index
- CHD
congenital heart disease
- CNV
copy number variant
- DNV
de novo variant
- GDM
gestational diabetes
- GRS
genomic risk score
- LOF
loss of function
- PCGC
Pediatric Cardiac Genomics Consortium
- PDM
pre-gestational diabetes
- WES
whole exome sequencing
- WGS
whole genome sequencing
Footnotes
Publisher's Disclaimer: Disclaimer: The manuscript and its contents are confidential, intended for journal review purposes only, and not to be further disclosed.
Clinical Trial Registration: https://clinicaltrials.gov/ct2/show/NCT01196182
Disclosures
None.
References
- 1.Leirgul E, Fomina T, Brodwall K, Greve G, Holmstrøm H, Vollset SE, Tell GS, Øyen N. Birth prevalence of congenital heart defects in Norway 1994–2009—A nationwide study. American Heart Journal 2014;168:956–964. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman MS, Smith AGC, Sable CA, Echko MM, Wilner LB, Olsen HE, Atalay HT, Awasthi A, Bhutta ZA, Boucher JLA, et al. Global, regional, and national burden of congenital heart disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Child and Adolescent Health 2020;4:185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015;350:1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature Genetics 2017;49:1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Chen W, Li W, Priest JR, Fu Y, Pang K, Ma B, Han B, Liu X, Hu S, et al. Exome-based case-control analysis highlights the pathogenic role of ciliary genes in transposition of the great arteries. Circulation Research 2020;126:811–821. [DOI] [PubMed] [Google Scholar]
- 6.Li AH, Hanchard NA, Furthner D, Fernbach S, Azamian M, Nicosia A, Rosenfeld J, Muzny D, D’Alessandro LCA, Morris S, et al. Whole exome sequencing in 342 congenital cardiac left sided lesion cases reveals extensive genetic heterogeneity and complex inheritance patterns. Genome Medicine 2017;9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Page DJ, Miossec MJ, Williams SG, Monaghan RM, Fotiou E, Cordell HJ, Sutcliffe L, Topf A, Bourgey M, Bourque G, et al. Whole Exome Sequencing Reveals the Major Genetic Contributors to Nonsyndromic Tetralogy of Fallot. Circulation Research 2019;124:553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SHA, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nature Genetics 2016;48:1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boskovski MT, Homsy J, Nathan M, Sleeper LA, Morton S, Manheimer KB, Tai A, Gorham J, Lewis M, Swartz M, et al. De novo Damaging Variants, Clinical Phenotypes and Post-Operative Outcomes in Congenital Heart Disease. Circulation: Genomic and Precision Medicine 2020;13:e002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morton SU, Shimamura A, Newburger PE, Opotowsky AR, Quiat D, Pereira AC, Jin SC, Gurvitz M, Brueckner M, Chung WK, et al. Association of Damaging Variants in Genes with Increased Cancer Risk among Patients with Congenital Heart Disease. JAMA Cardiology 2021;6:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson M, Razaz N, Edstedt Bonamy AK, Villamor E, Cnattingius S. Maternal Overweight and Obesity and Risk of Congenital Heart Defects. Journal of the American College of Cardiology 2019;73:44–53. [DOI] [PubMed] [Google Scholar]
- 12.Helle E, Priest JR. Maternal Obesity and Diabetes Mellitus as Risk Factors for Congenital Heart Disease in the Offspring. Journal of the American Heart Association 2020;9:e011541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanapo L, Donofrio MT, Ahmadzia HK, Gimovsky AC, Mohamed MA. The association of maternal hypertensive disorders with neonatal congenital heart disease: analysis of a United States cohort. Journal of Perinatology 2020;40:1617–1624. [DOI] [PubMed] [Google Scholar]
- 14.Tinker SC, Gilboa SM, Moore CA, Waller DK, Simeone RM, Kim SY, Jamieson DJ, Botto LD, Reefhuis J. Specific birth defects in pregnancies of women with diabetes: National Birth Defects Prevention Study, 1997–2011. American Journal of Obstetrics and Gynecology 2020;176:e1–176.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simeone RM, Tinker SC, Gilboa SM, Agopian AJ, Oster ME, Devine OJ, Honein MA. Proportion of selected congenital heart defects attributable to recognized risk factors. Annals of Epidemiology 2016;26:838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilboa SM, Correa A, Botto LD, Rasmussen SA, Waller DK, Hobbs CA, Cleves MA, Riehle-Colarusso TJ. Association between prepregnancy body mass index and congenital heart defects. American Journal of Obstetrics and Gynecology 2010;202:e1–10. [DOI] [PubMed] [Google Scholar]
- 17.Branum AM, Kirmeyer SE, Gregory ECW. Prepregnancy Body Mass Index by Maternal Characteristics and State: Data From the Birth Certificate, 2014. Natl Vital Stat Rep 2016;65:1–11. [PubMed] [Google Scholar]
- 18.Deputy NP, Kim SY, Conrey EJ, Bullard KM. Prevalence and Changes in Preexisting Diabetes and Gestational Diabetes Among Women Who Had a Live Birth — United States, 2012–2016. Morbidity and Mortality Weekly Report 2018;67:1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplinski M, Taylor D, Mitchell LE, Hammond DA, Goldmuntz E, Agopian AJ. The association of elevated maternal genetic risk scores for hypertension, type 2 diabetes and obesity and having a child with a congenital heart defect. PLoS ONE 2019;14:e0216477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter F, Morton SU, Kim SW, Kitaygorodsky A, Wasson LK, Chen KM, Zhou J, Qi H, Patel N, DePalma SR, et al. Genomic analyses implicate noncoding de novo variants in congenital heart disease. Nature Genetics 2020;52:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, Hardarson MT, Hjorleifsson KE, Eggertsson HP, Gudjonsson SA, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 2017;549:519–522. [DOI] [PubMed] [Google Scholar]
- 22.Hoang TT, Goldmuntz E, Roberts AE, Chung WK, Kline JK, Deanfield JE, Giardini A, Aleman A, Gelb BD, Neal M Mac, et al. The congenital heart disease genetic network study: Cohort description. PLoS ONE 2018;13:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, Pablo Kaski J, Kim R, Kline J, Mercer-Rosa L, Porter G, et al. The Congenital Heart Disease Genetic Network Study. Circulation Research 2013;112:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Ren X, He L, Li J, Zhang S, Chen W. Maternal age and the risk of gestational diabetes mellitus: A systematic review and meta-analysis of over 120 million participants. Diabetes Research and Clinical Practice 2020;162:108044. [DOI] [PubMed] [Google Scholar]
- 25.Girard SL, Bourassa CV, Lemieux Perreault L-P, Legault M-A, Barhdadi A, Ambalavanan A, Brendgen M, Vitaro F, Noreau A, Dionne G, et al. Paternal Age Explains a Major Portion of De Novo Germline Mutation Rate Variability in Healthy Individuals. PLoS ONE 2016;11:e0164212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldmann JM, Veltman JA, Gilissen C. De Novo Mutations Reflect Development and Aging of the Human Germline. Trends in Genetics 2019;35:828–839. [DOI] [PubMed] [Google Scholar]
- 27.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature Genetics 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars N, Benner C, Aguirre M, Venkataraman GR, Wainberg M, Ollila HM, Kiiskinen T, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nature Genetics 2021;53:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton SU, Quiat D, Seidman JG, Seidman CE. Genomic frontiers in congenital heart disease. Nature Reviews. Cardiology 2021; doi: 10.1038/s41569-021-00587-4. [DOI] [PMC free article] [PubMed]
- 30.Kalisch-Smith JI, Ved N, Sparrow DB. Environmental risk factors for congenital heart disease. Cold Spring Harbor Perspectives in Biology 2020;12:a037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Q, Dunn S, Wen SW, Lougheed J, Reszel J, Lavin Venegas C, Walker M. Neighbourhood maternal socioeconomic status indicators and risk of congenital heart disease. BMC Pregnancy and Childbirth 2021;21:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazekas-Pongor V, Fekete M, Csáky-Szunyogh M, Cseh K, Pénzes M. Parental occupational exposure and congenital heart diseases in a Hungarian case–control study. International Archives of Occupational and Environmental Health 2021;94:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu M, Zhu JY, LaHaye S, Majumdar U, Jiao K, Han Z, Garg V. Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease. JCI Insight 2017;2:e95085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano H, Fajardo VM, Nakano A. The role of glucose in physiological and pathological heart formation. Developmental Biology 2021;475:222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engineer A, Saiyin T, Lu X, Kucey AS, Urquhart BL, Drysdale TA, Norozi K, Feng Q. Sapropterin treatment prevents congenital heart defects induced by pre-gestational diabetes mellitus in mice. Journal of the American Heart Association 2018;7:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starikov R, Bohrer J, Goh W, Kuwahara M, Chien EK, Lopes V, Coustan D. Hemoglobin A1c in pre-gestational diabetic gravidas and the risk of congenital heart disease in the fetus. Pediatric Cardiology 2013;34:1716–1722. [DOI] [PubMed] [Google Scholar]
- 37.Øyen N, Diaz LJ, Leirgul E, Boyd HA, Priest J, Mathiesen ER, Quertermous T, Wohlfahrt J, Melbye M. Prepregnancy Diabetes and Offspring Risk of Congenital Heart Disease: A Nationwide Cohort Study. Circulation 2016;133:2243–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Block SR, Watkins SM, Salemi JL, Rutkowski R, Tanner JP, Correia JA, Kirby RS. Maternal pre-pregnancy body mass index and risk of selected birth defects: evidence of a dose-response relationship. Paediatric and Perinatal Epidemiology 2013;27:521–31. [DOI] [PubMed] [Google Scholar]
- 39.Schulkey CE, Regmi SD, Magnan RA, Danzo MT, Luther H, Hutchinson AK, Panzer AA, Grady MM, Wilson DB, Jay PY. The maternal-age-associated risk of congenital heart disease is modifiable. Nature 2015;520:230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu Y, Lin S, Zhuang J, Bloom MS, Smith M, Nie Z, Mai J, Ou Y, Wu Y, Gao X, et al. First-Trimester Maternal Folic Acid Supplementation Reduced Risks of Severe and Most Congenital Heart Diseases in Offspring: A Large Case-Control Study. Journal of the American Heart Association 2020;9:e015652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Øyen N, Olsen SF, Basit S, Leirgul E, Strøm M, Carstensen L, Granström C, Tell GS, Magnus P, Vollset SE, et al. Association Between Maternal Folic Acid Supplementation and Congenital Heart Defects in Offspring in Birth Cohorts From Denmark and Norway. Journal of the American Heart Association 2019;8:e011615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bellinger DC, Wypij D, Rivkin MJ, DeMaso DR, Robertson RL, Dunbar-Masterson C, Rappaport LA, Wernovsky G, Jonas RA, et al. Adolescents with d-transposition of the great arteries corrected with the arterial switch procedure: Neuropsychological assessment and structural brain imaging. Circulation 2011;124:1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nattel SN, Adrianzen L, Kessler EC, Andelfinger G, Dehaes M, Côté-Corriveau G, Trelles MP. Congenital Heart Disease and Neurodevelopment: Clinical Manifestations, Genetics, Mechanisms, and Implications. Canadian Journal of Cardiology 2017;33:1543–1555. [DOI] [PubMed] [Google Scholar]
- 44.Kong L, Norstedt G, Schalling M, Gissler M, Lavebratt C. The risk of offspring psychiatric disorders in the setting of maternal obesity and diabetes. Pediatrics 2018;142:e20180776. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Fallin MD, Riley A, Landa R, Walker SO, Silverstein M, Caruso D, Pearson C, Kiang S, Dahm JL, et al. The association of maternal obesity and diabetes with autism and other developmental disabilities. Pediatrics 2016;137:e20152206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spann MN, Scheinost D, Feng T, Barbato K, Lee S, Monk C, Peterson BS. Association of Maternal Prepregnancy Body Mass Index With Fetal Growth and Neonatal Thalamic Brain Connectivity Among Adolescent and Young Women. JAMA Network Open 2020;3:e2024661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gauderman WJ, Mukherjee B, Aschard H, Hsu L, Lewinger JP, Patel CJ, Witte JS, Amos C, Tai CG, Conti D, et al. Update on the State of the Science for Analytical Methods for Gene-Environment Interactions. American Journal of Epidemiology 2017;186:762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radhakrishna U, Vishweswaraiah S, Veerappa AM, Zafra R, Albayrak S, Sitharam PH, Saiyed NM, Mishra NK, Guda C, Bahado-Singh R. Newborn blood DNA epigenetic variations and signaling pathway genes associated with Tetralogy of Fallot (TOF). Plos One 2018;13:e0203893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapman G, Moreau JLM, I P E, Szot JO, Iyer KR, Shi H, Yam MX, O’Reilly VC, Enriquez A, Greasby JA, et al. Functional genomics and gene-environment interaction highlight the complexity of congenital heart disease caused by Notch pathway variants. Human Molecular Genetics 2020;29:566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinthorsdottir V, McGinnis R, Williams NO, Stefansdottir L, Thorleifsson G, Shooter S, Fadista J, Sigurdsson JK, Auro KM, Berezina G, et al. Genetic predisposition to hypertension is associated with preeclampsia in European and Central Asian women. Nature Communications 2020;11:5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics 2013:43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Research 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv.org. 2012:https://arxiv.org/abs/1207.3907.
- 59.Poplin R, Chang PC, Alexander D, Schwartz S, Colthurst T, Ku A, Newburger D, Dijamco J, Nguyen N, Afshar PT, et al. A universal snp and small-indel variant caller using deep neural networks. Nature Biotechnology 2018;36:983. [DOI] [PubMed] [Google Scholar]
- 60.Lambert SA, Gil L, Jupp S, Ritchie SC, Xu Y, Buniello A, McMahon A, Abraham G, Chapman M, Parkinson H, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nature Genetics 2021;53:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


