Abstract
The underlying molecular pathophysiology of feeding disorders, particularly in peripheral organs, is still largely unknown. A range of molecular factors encoded by appetite-regulating genes are already described to control feeding behaviour in the brain. However, the important role of the gastrointestinal tract in the regulation of appetite and feeding in connection to the brain has gained more attention in the recent years. An example of such inter-organ connection can be the signals mediated by leptin, a key regulator of body weight, food intake and metabolism, with conserved anorexigenic effects in vertebrates. Leptin signals functions through its receptor (lepr) in multiple organs, including the brain and the gastrointestinal tract. So far, the regulatory connections between leptin signal and other appetite-regulating genes remain unclear, particularly in the gastrointestinal system. In this study, we used a zebrafish mutant with impaired function of leptin receptor to explore gut expression patterns of appetite-regulating genes, under different feeding conditions (normal feeding, 7-day fasting, 2 and 6-hours refeeding). We provide evidence that most appetite-regulating genes are expressed in the zebrafish gut. On one hand, we did not observed significant differences in the expression of orexigenic genes (except for hcrt) after changes in the feeding condition. On the other hand, we found 8 anorexigenic genes in wild-types (cart2, cart3, dbi, oxt, nmu, nucb2a, pacap and pomc), as well as 4 genes in lepr mutants (cart3, kiss1, kiss1r and nucb2a), to be differentially expressed in the zebrafish gut after changes in feeding conditions. Most of these genes also showed significant differences in their expression between wild-type and lepr mutant. Finally, we observed that impaired leptin signalling influences potential regulatory connections between anorexigenic genes in zebrafish gut. Altogether, these transcriptional changes propose a potential role of leptin signal in the regulation of feeding through changes in expression of certain anorexigenic genes in the gastrointestinal tract of zebrafish.
Introduction
In recent years, the prevalence of eating disorders has steadily increased in human populations worldwide [1]. The critical roles of various endocrine and neural factors are demonstrated in the regulation of feeding behaviour during eating disorders [2]. However, the underlying molecular pathophysiology of these disorders, particularly in peripheral organs, is still largely unknown. Feeding behaviour is regulated by specific regions in the brain, which control hunger-driven activities, through endocrine signals originated from the brain itself and peripheral organs [3, 4]. Factors secreted by the brain, in particular the hypothalamic area, and in other potentially important organs, control feeding behaviour by inhibiting (anorexigenic) or stimulating (orexigenic) appetite. Those key factors or appetite-regulating genes, which encode a range of neuropeptides and their cognate receptors, can also be classified in orexigenic and anorexigenic categories based on their functions [5, 6]. While most studies have focused on appetite-regulating factors in the brain, it is becoming increasingly evident that the gastrointestinal tract plays an important role in appetite and feeding regulation as well, and that bidirectional signalling between the gastrointestinal tract and the brain exists [7–11]. In addition, various metabolic changes causing eating disorders are influenced by factors produced by both organs [12], by gut microbiota [13, 14], as well as by homeostatic metabolite signals and nutrients [15, 16]. Altogether, these external and internal factors initially communicate with molecular signals within the gastrointestinal tissues in order to exert their effects along the gut-brain axis. Hence, signals mediated through the gastrointestinal tract are among the major contributors of peripheral mechanisms regulating appetite in the brain [17]. These signals can affect hypothalamic circuitry to determine cessation of energy intake during meal ingestion and the return of appetite and hunger after fasting [17].
A prominent example of these molecular signals linking multiple organs and conveying the nutritional status and the size of energy stores to the brain is the hormone leptin [18]. Leptin inhibits food intake and functions as an anorexigen during appetite regulation in several vertebrates, including fish [19], and loss of function mutations in orthologous leptin genes in zebrafish and mammals increases body weight and promotes obesity [20–22]. The conserved metabolic function of leptin signalling across vertebrates provides an opportunity to use zebrafish as model organism to explore molecular mechanisms underlying more specific leptin functions in appetite regulation and feeding. Unlike mammals, zebrafish contains two leptin hormone genes, lepa and lepb, expressed in several peripheral organs [23]. In zebrafish, the main organs expressing leptin genes are liver and adipose tissue, and while short-term fasting causes decrease in hepatic lepb expression [23], lepa expression has been shown to increase directly after feeding and decrease after fasting in the whole body of wild-type zebrafish [24]. Also, long-term overfeeding had no particular effect on the leptin transcripts in the visceral adipose tissue of adult fish [25]. However, both leptin hormones mediate their signal through a single leptin receptor (lepr) gene, expressed in multiple brain regions, including the hypothalamus [26]. The regulatory effects of leptin signalling on appetite can be mediated through differential regulation of appetite regulating genes in the brain [27–30]. Nevertheless, leptin signal has also various effects on the gastrointestinal tract in relation to appetite and feeding [31, 32]. In mammals, leptin is produced in the stomach and its receptor is expressed in the intestine, mediating its effects on absorption of various nutrients in enterocytes, intestinal motility and cell proliferation, as well as secretion of different gastrointestinal hormones (reviewed in [32]). Subsequently, changes in intestinal conditions e.g. gut microbiota [14], trigger molecular signals, which are eventually sensed in specific brain regions. In zebrafish, both leptin and its receptor are expressed in the gastrointestinal tract and the enteric nervous system [33, 34]. The zebrafish intestinal expression of leptin and its receptor appeared to be responsive to fasting and feeding, indicating a potential regulatory role of leptin during changes of feeding conditions through the gut-brain axis [33, 34]. So far, studies addressing molecular mechanisms connecting the brain and gastrointestinal tissues under different feeding conditions are lacking in both fish and mammals. A first step to address this lack could be by generating information about transcriptional profiling of appetite regulating genes in the gut during changes in feeding conditions.
In this research, we assessed the expression levels of 37 appetite regulating genes in zebrafish gut (Table 1), which are already known to have also brain expression, under different feeding conditions [35]. The different feeding conditions to assess the expression dynamics of our target genes include normal feeding (control group), fasting and two refeeding steps (2 and 6 hours after refeeding). In addition, we investigated the expression of 11 genes with known gastrointestinal expression and function in zebrafish, as gut marker genes under the different conditions (Table 1). These marker genes were investigated to evaluate the effects of the different feeding conditions on gut functions. They include 4 genes for main anatomical compartments of gastrointestinal tract in zebrafish (apoa1a, apoa4a, aqp3a and ctsl1), 3 genes encoding important molecular transporters in gut (pept1, slc2a5 and sglt1), 2 genes encoding regulators of the release of digestive enzymes (ccka and cckar), a target of leptin signal (insa) and a gut expressed orexigenic gene (ghrl) (see Table 1). To explore the role of leptin signalling, we compared the expression levels of all these genes between gut samples from wild-type zebrafish and zebrafish with impaired leptin receptor function (lepr mutant). To do this, we first validated a suitable reference gene with stable expression and used its expression as normalization factor against the expression of our target genes. Through this study, not only we profiled expression changes of appetite regulating genes in zebrafish gut, but also, found genes whose transcriptional changes could be under the influence of leptin signalling. Finally, through pairwise expression correlation analysis, we propose potential loss and gain of transcriptional regulatory links, between the appetite regulating genes under impaired leptin signal. Our study provides for the first time information about the expression dynamics of appetite-regulating genes in zebrafish gut, under both different feeding conditions and impaired leptin signalling.
Table 1. Candidate target genes studied and their related function in fish.
| Genes | Description / Related function | Organism | References | |
|---|---|---|---|---|
| Gut marker genes | apoa1a apoa4a | Apolipoprotein A1 and A4 / Anterior intestine marker | Zebrafish | [36] |
| aqp3a | Aquaporin 3 / Posterior intestine marker | Zebrafish | [36] | |
| ccka cckar | Cholecystokinin and its receptor / Regulating the release of digestive enzymes | Goldfish Zebrafish | [30] | |
| ctsl1 | Cathepsin L1 / Posterior intestine marker | Zebrafish | [36] | |
| ghrl | Ghrelin / Orexigenic effects with intestinal expression | Goldfish Zebrafish | [37–39] | |
| insa | Insulin A / Intestinal target of leptin signal | Zebrafish | [40] | |
| pept1 | Peptide Transporter 1 / Intestinal hydrogen peptide cotransporter and responsive to leptin signal | Zebrafish | [41] | |
| slc2a5 | Solute carrier family 2 member 5 / Fructose transporter in small intestine | Zebrafish | [40] | |
| sglt1 | Sodium/glucose co-transporters 1 / Glucose and galactose transporter in small intestine | Goldfish | [42] | |
| Brain expressed appetite regulating genes | agrp (agrp1) | Agouti related neuropeptide / Orexigenic effects | Zebrafish | [43, 44] |
| apln, aplnr | Apelin, agtrl1 Ligand and its receptor / Orexigenic effects | Goldfish | [45, 46] | |
| cart1-4 | Cocaine and amphetamine regulated transcripts / Anorexigenic effects | Zebrafish | [47, 48] | |
| cnr1 | Cannabinoid receptor 1 / Orexigenic effects | Zebrafish | [47, 49] | |
| crh | Corticotropin-releasing hormone / Anorexigenic effects | Goldfish | [50, 51] | |
| dbi | Diazepam binding inhibitor, octadecaneuropeptide / Anorexigenic effects | Goldfish | [52, 53] | |
| galn, galr1,2 | Galanin/GMAP prepropeptide and its receptors / Orexigenic effects | Goldfish Zebrafish | [54–56] | |
| ghsr | Ghrelin receptor (growth hormone secretagogue receptor) / Orexigenic effects | Goldfish Zebrafish | [37–39] | |
| gnrh2,3 gnrhr1-4 | Gonadotropin releasing hormone 2 and 3, and their receptors / Anorexigenic effects | Goldfish Zebrafish | [57, 58] | |
| hcrt | Orexin, hypocretin neuropeptide precursor / Orexigenic effects | Goldfish Zebrafish | [59–61] | |
| kiss1, kiss1r | Prepro-Kisspeptin 1 and its receptor / Anorexigenic effects | Sea bass | [62] | |
| mc4r | Melanocortin 4 receptor / Anorexigenic effects | Goldfish | [63] | |
| mch, mchr1,2 | Pro-melanin-concentrating hormone and its receptors / Anorexigenic effects | Goldfish | [64, 65] | |
| nmu | Neuromedin U preproprotein / Anorexigenic effects | Goldfish | [66, 67] | |
| npy | Prepro-neuropeptide Y / Orexigenic effects | Goldfish Zebrafish | [68, 69] | |
| nucb2 | Nucleobindin 2/ Nesfatin 1 / Anorexigenic effects | Goldfish Zebrafish | [70–72] | |
| oxt | Oxytocin/neurophysin I prepropeptide / Anorexigenic effects | Zebrafish | [73] | |
| pacap | Pituitary adenylate cyclase activating polypeptide / Anorexigenic effects | Goldfish | [74] | |
| pomc | Pro-opiomelanocortin preproprotein / Anorexigenic effects | Goldfish Zebrafish | [75–77] | |
| pyy | Prepro-peptide YY / Anorexigenic effects | Goldfish | [78] | |
| scg2 | Secretogranin 2/ Secretoneurin / Orexigenic effects | Goldfish | [79, 80] | |
| spx | Spexin Hormone / Anorexigenic effects | Goldfish Zebrafish | [81, 82] | |
| trh | Thyrotropin-releasing hormone / Orexigenic effects | Goldfish | [83] |
Materials and methods
Fish breeding, fasting-refeeding scheme and sampling
Lepr sa12953 zebrafish were obtained from the European Zebrafish Resource Centre. The mutation was created by the Sanger Institute for the Zebrafish Mutation Project, replacing a thymine with an adenine on chromosome 6 and causing the formation of a premature codon stop and an incomplete translation of lepr mRNA, resulting in a premature stop codon and a shortened polypeptide [84]. The fish were kept in 3-liter recirculating tanks at 28.4°C under light:dark conditions of 14:10 h at the SciLife lab Genome Engineering Zebrafish facility at Uppsala University. Water quality was checked regularly by the facility. The fish were fed with dry pellets Zebrafeed® from Sparos® once in the morning and with Artemia twice a day (middays and evenings). The dry pellets consist of 63% of proteins, 14% of fats and oils, 1.8% of fibers, 12% of ash and 2.3% phosphorus [85]. At the start of the experiment, 20 homozygote lepr mutant zebrafish and 20 wild-type zebrafish of similar age and size were divided into four experimental groups of 5 fish each, including both males and females. Fish in the control groups were fed according to the above-mentioned feeding schedule for 7 days; in the fasting groups, fish were fasted for 7 days and in the two refeeding groups, fish were fasted for 7 days and sampled 2 hours and 6 hours after refeeding. The control fish were sampled 2 hours after feeding. A detailed description of the experimental set up can be found in Ahi et al., (2019a) [35]. Fish were euthanized using a tricaine solution (MS-222) of 0.4 mg/ml and immersion in ice. The viscera of zebrafish were dissected and the entire gastrointestinal tract of each fish was carefully removed, washed in saline buffer and transferred into RNAlater (Ambion Inc, Austin Texas) at 4°C for one day and then stored at -20°C. No significant differences in standard body length and net weight, as well as the hepato-somatic index (HSI) were shown between the genotypes (S1 Appendix). Fasting resulted in a 10%weight loss in both genotypes.
The fish handling procedures were approved by the Swedish Ethical Committee on Animal Research in Uppsala (permit C10/16). All methods were carried out in accordance with the guidelines and regulations of the Swedish Ethical Committee on Animal Research in Uppsala.
RNA extraction and cDNA synthesis
We extracted the RNA of sampled guts, following the Trizol (Ambion) method. In brief, the dissected guts were removed from RNAlater, dried and put into 1.5 ml Eppendorf tubes with 200 μl of Trizol. Then, each sample was carefully homogenized with a 25G Terumo needle and BD Plastipak 1 mL syringe. The steps included addition of chloroform (Sigma-Aldrich), incubations, centrifugations, upper phase separation, addition of glycoblue (Ambion) and isopropyl alcohol (Sigma-Aldrich). RNA precipitation and ethanol (VWR) washes were followed, according to already described procedures [35]. The RNA pellets were dissolved in 10 μl of nuclease-free water (Ambion), and genomic DNA removal steps were conducted using Turbo DNA-free kit (Ambion), according to the manufacturer´s instructions. The quality and quantity of RNA were assessed by NanoDrop 1000 v3.7 and 1000 ng of RNA input per sample was used for cDNA synthesis, with Superscript III First-Strand Synthesis System (Invitrogen). The cDNA synthesis protocol was performed in two steps, first by addition of 0.5 μl of random primers (50 ng/μl) and 0.5 dNTP (10 nM) to each RNA sample and incubation at 65°C for 5 minutes. Next, 2 μl of 5X First-Strand Buffer, 0.5 μl of DTT (0.1 M), 0.5 μl of RNase OUT (40 U/μl) and 0.5 μl of Superscript III RT (200 U/μl) were added to each RNA sample, followed by an incubation of 5 minutes at 25°C, 50 minutes at 50°C (RT enzyme reaction) and 15 minutes at 70°C (RT enzyme inhibition). The final volume of 10 μl of cDNA per sample was stored at -20°C until the qPCR step.
Gene selection, primer design and qPCR
To find gene(s) with stable expression (reference gene(s)) across the experimental groups and genotypes, we chose 8 candidate genes, based on previous studies on zebrafish, in which suitable references genes were validated across different developmental stages, experimental conditions and tissues [86–89]. We then selected 37 candidate target genes involved in appetite regulation in studies on goldfish and zebrafish (Cypriniformes), as well as 11 marker genes with known functions in the gut, as summarized in Table 1. To design primers for qPCR, we first retrieved the gene sequences from a zebrafish database, http://www.zfin.org [90]. The sequences were then imported into CLC Genomic Workbench (CLC Bio, Denmark) and the exon/exon boundaries were specified, using the annotated Danio rerio genome available in the Ensembl database, http://www.ensembl.org [91]. The Primer Express 3.0 (Applied Biosystems, CA, USA) and OligoAnalyzer 3.1 (Integrated DNA Technology) software were used to design primers, as described in our previous study [35] (S1 Appendix).
The preparation of qPCR reactions was done by mixing 1 μl of diluted cDNA (1:20) from each sample with 7.5 μl of qPCR Master mix, PowerUp SYBR Green Master Mix (Thermo Fischer Scientific), 0.3 μl of forward and reverse primers (10 μM) and 6.2 μl of nuclease-free water. The MxPro-3000 PCR machine (Stratagene, La Jolla, CA) was used for quantification, combined with the MxPro software (Stratagene) for data analysis. We performed two technical replicates for each biological replicate (per gene), following the common sample maximization method [92], in order to optimize our assay in each run. The qPCR reaction program was set to 2 minutes at 50°C, 2 minutes at 95°C, and 40 cycles of 15 second at 95°C and 1 minute at 62°C. At the end of the amplification step, we performed a dissociation run (60°C– 95°C) and the threshold cycles and efficiencies were calculated. To calculate efficiency values (E) for each primer pair, we generated standard curves, by making serial dilution of pooled cDNA of pooled samples. The detailed analysis of standard curves and efficiencies are described in our previous study [35] (S1 Appendix).
Gene expression analyses
In this study, we implemented three commonly used algorithms to validate the most suitable reference gene(s); BestKeeper [93], NormFinder [94] and geNorm [95]. These algorithms use different analysis methods to rank the most stably expressed reference genes, which their strengths and limitations are already discussed in details [96]. Next, the Cq value of the most stable reference gene, Cq reference, was used to normalize Cq values of target genes in each sample (ΔCq target = Cq target−Cq reference). Across all the samples, the biological replicate with the lowest expression for each gene was selected to calculate ΔΔCq values (ΔCq target− ΔCq calibrator). The relative expression quantities (RQ values) was estimated by 2−ΔΔCq, and their fold changes (logarithmic values of RQs) were used for statistical analysis [97]. For the direct comparisons of gene expression levels between the genotypes (within each experimental condition), the student t-test was applied. On the other hand, to compare the expression dynamic of a target gene across the different conditions (within each genotype), analysis of variance (ANOVA) test was applied, followed by Tukey’s HSD post hoc tests. Benjamini-Hochberg method was also applied to correct for the false positive rate in the multiple comparisons [98]. Pairwise Pearson correlation coefficients between the appetite regulating genes were calculated within each genotype, in order to identify possible similarities in expression patterns. A dendrogram hierarchical clustering, using expression levels of the appetite regulating genes as input, was constructed, to represent similarities between the experimental conditions and the genotypes. R project (http://www.r-project.org) was used for the statistical analyses [99]. Finally, using a zebrafish database for protein interactome, STRING v10 (http://string-db.org/) [100], we investigated potential interactions between the gene products.
Results
Reference gene validation
Validation of stably expressed gene(s) in every specific experimental conditions (so-called reference genes) is an essential step, in order to calculate the relative gene expression levels of target genes [101–103]. In this study, we assessed the expression levels of 8 candidate reference genes in the gut of lepr mutant and wild-type zebrafish, under different feeding conditions. The expression levels of the candidate reference genes showed considerable variations, with highest expression (lowest Cq, Cq≈19) for actb2, and lowest expression (highest Cq, Cq≈30) for tmem50a (S1 Fig). Interestingly, the variations in the expression levels of the candidate reference genes in the gut were very similar to their expression variations in the brain [35]. All the three algorithms, BestKeeper, geNorm and Normfinder, ranked tmem50a, as the most stable reference gene (Table 2). In further analyses, we considered the expression level of tmem50a gene as a normalization factor to calculate the expression levels of our target genes in each sample.
Table 2. Ranking of candidate reference genes in zebrafish gut in wild-type and lepr mutant adults, under different feeding conditions.
| BestKeeper | geNorm | NormFinder | |||||
|---|---|---|---|---|---|---|---|
| Ranking | r values | Ranking | SD values | Ranking | M values | Ranking | S values |
| tmem50a | 0.928 | ef1a | 0.461 | tmem50a | 0.623 | tmem50a | 0.186 |
| b2m | 0.892 | tmem50a | 0.624 | b2m | 0.674 | b2m | 0.271 |
| rpl13 | 0.882 | actb2 | 0.626 | ef1a | 0.707 | ef1a | 0.276 |
| ube2a | 0.857 | b2m | 0.647 | rpl13 | 0.732 | rpl13 | 0.298 |
| ef1a | 0.823 | rpl13 | 0.709 | actb2 | 0.733 | actb2 | 0.304 |
| rplp0 | 0.819 | p6gd | 0.717 | ube2a | 0.783 | p6gd | 0.315 |
| actb2 | 0.812 | ube2a | 0.799 | p6gd | 0.796 | ube2a | 0.359 |
| p6gd | 0.793 | rplp0 | 0.944 | rplp0 | 0.998 | rplp0 | 0.401 |
SD: standard deviation, r: Pearson product-moment correlation coefficient, SV: stability value, M: mean value of stability.
Expression analysis of orexigenic genes in zebrafish gut
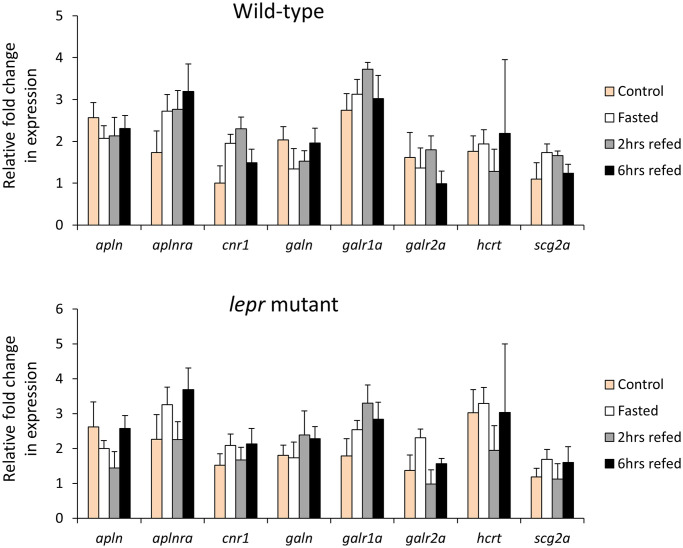
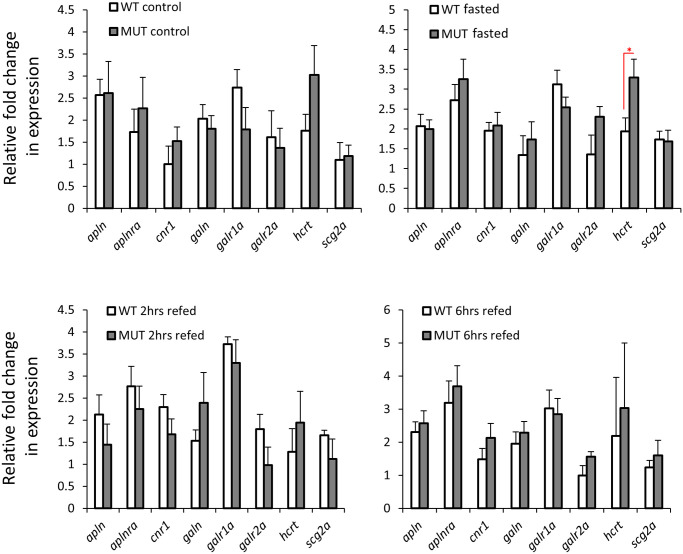
We assessed the expression levels of 12 orexigenic genes, already known to have brain expression, within wild-type or lepr mutant gut during the fasting-refeeding experiment [35]. We found 8 of these genes to be expressed in the gut as well (Fig 1), however, unlike the brain, none of these genes showed expression differences between the feeding conditions in both genotypes. These observations indicate that the orexigenic genes expressed in the gut might not have functional roles during changes in feeding conditions, under both normal and impaired leptin signalling. Next, we compared the expression levels of the 8 orexigenic genes, between lepr mutant and wild-type for each feeding condition, and found only one gene, hcrt, showing increased expression in the mutants in the fasting groups (Fig 2). This might indicate a functional role of hcrt in zebrafish gut during fasting, which is controlled by leptin signal. In general, these results suggest minimal or absence of functional roles for the gut expressed orexigenic genes, under different feeding conditions and leptin signal in zebrafish.
Fig 1. Expression dynamics of orexigenic genes in the gut of wild-type and lepr mutant zebrafish.
Means and standard errors of fold changes in expression of five biological replicates are shown for each experimental group. No significant differences between the experimental groups were detected in both genotypes.
Fig 2. Expression differences of orexigenic genes in the gut of wild-type versus lepr mutant zebrafish in each feeding group.
Means and standard errors of fold changes in expression of five biological replicates are shown for each experimental group. Significant differences between the lepr mutant and wild-type are delineated by asterisks (* P < 0.05).
Expression analysis of anorexigenic genes in zebrafish gut
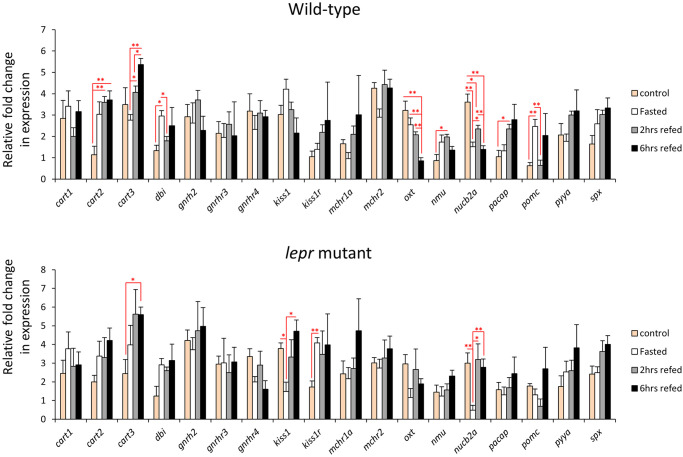
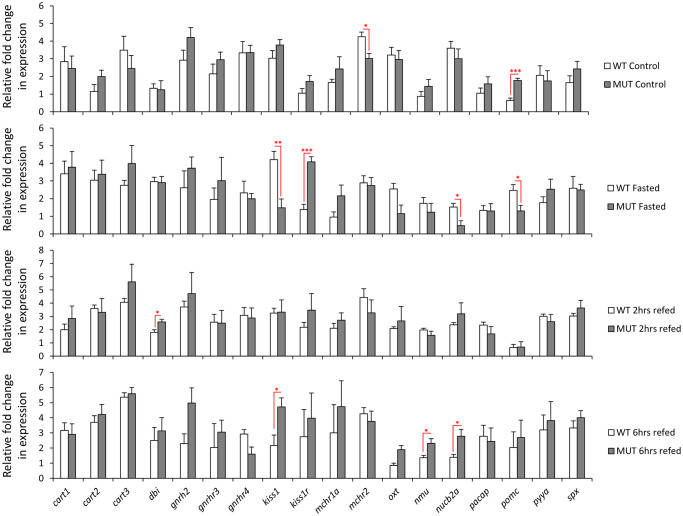
We assessed the expression levels of 25 anorexigenic genes, already known to have brain expression, in the gut of wild-type and lepr mutant zebrafish during the fasting-refeeding experiment [35]. We found 18 of these anorexigenic genes to be expressed in the gut as well (Fig 3). Among them, 8 genes showed expression differences between the feeding groups in the wild-type zebrafish (cart2, cart3, dbi, oxt, nmu, nucb2a, pacap, and pomc). This might indicate a functional role of the zebrafish gut in appetite control, through differential regulation of anorexigenic genes. The expression dynamics of these 8 genes showed also distinct variations, for example, dbi and pomc had increased expression during fasting, whereas cart2, cart3 and pacap showed increased expression in at least one of the refeeding groups. In the lepr mutants, only 4 genes showed expression differences between the feeding groups, i.e. cart3, kiss1 and kiss1r and nucb2a (Fig 3). This suggests that the expression differences of only 2 genes (cart3 and nucb2) are independent of leptin signal, and that the disappearance of differences in the other genes can possibly be linked to functional leptin signal. Moreover, kiss1 and kiss1r showed differences between the feeding groups only under the impaired leptin signal, indicating their potential leptin dependent function in the gut. In addition, we directly compared the expression of all the anorexigenic genes between the two genotypes, and observed expression differences for mchr2 and pomc in control groups, kiss1, kiss1r, nucb2a and pomc in fasting groups, dbi in 2 hrs refeeding, and kiss1, nmu and nucb2a in 6 hrs refeeding groups (Fig 4). Genes kiss1, nucb2a and pomc showed higher expression in the wild-types in the fasting groups, but opposite expression differences (higher in the mutant) were evident in the control groups for pomc and in the 6 hrs refeeding groups for kiss1 and nucb2a. Moreover, among the differentially expressed genes, kiss1 and kiss1r had the strongest expression difference between the two genotypes, but in opposite manner (in fasting groups). Altogether, these findings suggest potential functional roles for certain gut expressed anorexigenic genes under different feeding conditions and leptin signal activities in zebrafish.
Fig 3. Expression dynamics of anorexigenic genes in the gut of wild-type and lepr mutant zebrafish.
Means and standard errors of fold changes in expression of five biological replicates are shown for each experimental group. Significant differences between the experimental groups in each genotype are delineated by asterisks (* P < 0.05; ** P < 0.01).
Fig 4. Expression differences of anorexigenic genes in the gut of wild-type versus lepr mutant zebrafish in each feeding group.
Means and standard errors of fold changes in expression of five biological replicates are shown for each experimental group. Significant differences between the lepr mutant and wild-type are delineated by asterisks (* P < 0.05; ** P < 0.01; *** P < 0.001).
Expression analysis of gut marker genes in zebrafish
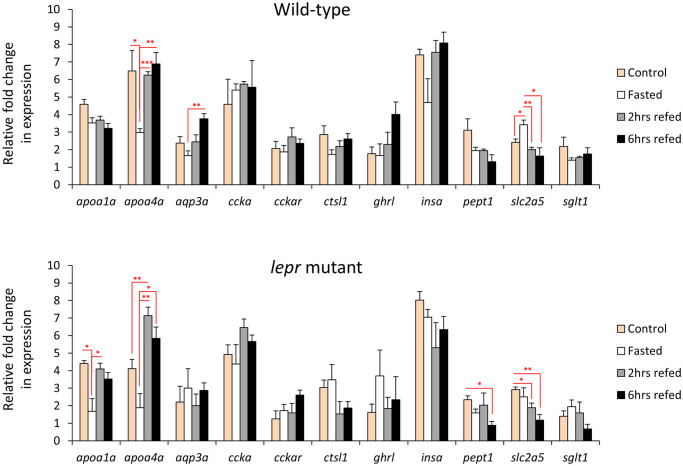
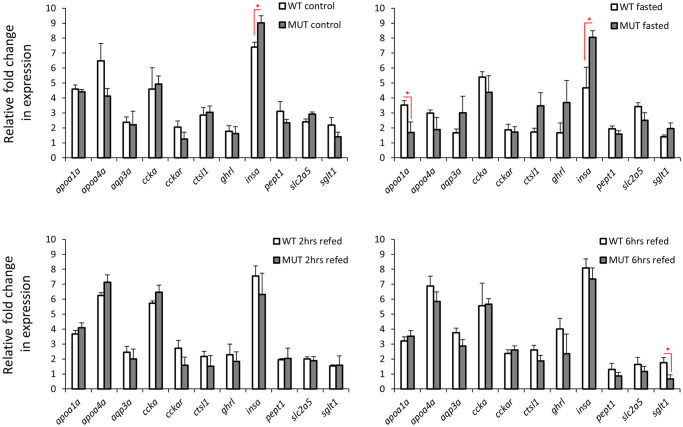
Finally, we tested the expression of a set of 11 target genes which are already known to be highly expressed in the gut and have different functions in the gastrointestinal system (Fig 5). We assessed the expression levels of these gut marker genes within wild-type and lepr mutant zebrafish, during the fasting-refeeding experiment. Among them, apoa4a and slc2a5 genes showed expression differences between the feeding groups in both genotypes, whereas aqp3a only in wild-types, and apoa1a and pept1 only in lepr mutants (Fig 5). The direct comparison between the two genotypes within each feeding group revealed expression differences only for insa in the control groups, insa and apoa1a in the fasting groups, and sglt1 in the 6 hrs refeeding groups (Fig 6). Unexpectedly, we did not find any expression differences for ghrl in both genotypes and between different feeding conditions, which is an orexigenic gene with known expression in gut and brain [38]. The findings demonstrate that changes in feeding conditions can lead to expression differences in certain gut marker genes, although expression changes in only few of these genes are affected by impaired leptin signal in zebrafish gut.
Fig 5. Expression dynamics of known gut-expressed genes in the gut of wild-type and lepr mutant zebrafish.
Means and standard errors of fold changes in expression of five biological replicates are shown for each experimental group. Significant differences between the experimental groups in each genotype are delineated by asterisks (* P < 0.05; ** P < 0.01; *** P < 0.001).
Fig 6. Expression differences of known marker genes in the gut of wild-type versus lepr mutant zebrafish in each feeding group.
Means and standard errors of fold changes in expression of five biological replicates are shown for each experimental group. Significant differences between the lepr mutant and wild-type are delineated by asterisks (* P < 0.05).
Expression correlation analyses of the appetite regulating genes in zebrafish gut
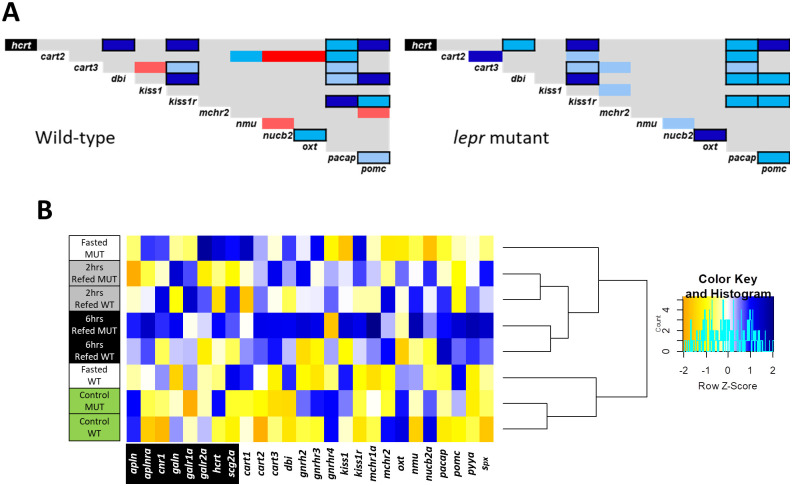
In order to predict regulatory connections between the genes within each genotype, we performed pairwise analysis of expression correlation for the appetite-regulating genes, which showed differential expression in previous steps (Fig 7A). Most of the identified correlations were positive in both genotypes. Negative expression correlations were also found, but only in wild-type zebrafish and between cart2/nucb2a, cart2/oxt, cart3/kiss1, mchr2/pomc and nmu/nucb2a, indicating leptin-dependent negative transcriptional regulatory connections in zebrafish gut. Most of the positive expression correlations appeared to be present in both genotypes, suggesting their positive transcriptional regulatory connections independent of leptin signal in zebrafish gut. Although, few positive expression correlations appeared to be gained under impaired leptin signal in gut, i.e. between cart2/kiss1r, cart3/mchr2, kiss1/mchr2 and nmu/nucb2a. Altogether, these results indicate transcriptional regulatory interactions between appetite-regulatory genes in the zebrafish gut. Moreover, these results revealed that impaired leptin signalling influences potential regulatory connections between appetite-regulating genes in the gut, particularly those connections, involving genes like cart2, cart3, kiss1, kiss1r, mchr2, nmu, nucb2a and oxt.
Fig 7. Connectedness of appetite-regulating genes and the feeding conditions based on the gene expression patterns.
(A) Pairwise expression correlations of appetite regulating genes in the gut of wild-type and lepr mutant zebrafish. The blue and red colours respectively indicate positive and negative Pearson correlation coefficients and their light to dark shadings show significant levels of P < 0.05, P < 0.01 and P < 0.001, respectively. The gene specified with black background is an orexigenic gene whereas the rest are anorexigenic genes. Pairwise correlations delineated with black borders are similar between the two genotypes. (B) Clustering of the experimental conditions in each genotype based on similarities in expression patterns of the appetite regulating genes (the blue and yellow colors indicate higher and lower expression level, respectively).
In addition, using the same differentially expressed genes as input in a zebrafish protein interactome prediction tool (STRING v10, http://string-db.org/), we revealed potential molecular interactions between products of hcrt, kiss1r, nmu, oxt, pacap and pomca, as well as their transcriptional connections to lepr in zebrafish (S1 Fig). It also appeared that the product of oxt gene had highest number of molecular interactions with the rest of the gene products, as well as potential transcriptional connection to lepr in zebrafish.
Using the combined expression patterns of all appetite regulating genes expressed in zebrafish gut, we conducted hierarchical clustering of the experimental groups in both genotypes (Fig 7A). This led to identification of the similarities and divergences between the feeding conditions, which might also be affected by impaired leptin signal. The analysis revealed that the refeeding groups were clustered distantly from the control feeding groups in both genotypes, while the 2hrs and 6hrs refeeding groups were closely branched together for both genotypes (Fig 7B). On the other hand, the most distant branching between the two genotypes was observed for the fasting groups, i.e. the wild-type fasting group was more similar to the control group, whereas the mutant fasting group was clustered with the refeeding groups (Fig 7B). This indicates that the gut expression of appetite-regulating genes represents the strongest divergence between wild-types and lepr mutants during fasting.
Discussion
The main aim of this study was to explore the expression dynamics of appetite-regulating genes in zebrafish gastrointestinal tract under different feeding conditions (Figs 1 and 3). We were also interested to investigate whether the impairment of leptin signal influences the expression of gut-expressed appetite regulating genes (Figs 2 and 4). Prior to addressing these aims, we first identified a reference gene, which is stably expressed in zebrafish gut under the different experimental conditions and assessed the expression of a handful of gut marker genes (Figs 5 and 6). In this study, however, we did not test the expression of marker genes involved in osmoregulation which is known to be affected by both fasting condition and leptin signal in fish [104, 105].
None of the orexigenic genes showed any significant changes in their expression dynamics under the different feeding conditions in both genotypes. This could imply an absence of functional roles of orexigenic genes in feeding regulation in the zebrafish gut. An unexpected result was observed for ghrl, a gut expressed orexigenic gene, which is shown to be transcriptionally induced in response to fasting and reduced in response to refeeding in zebrafish [38], but we did not find any changes in both genotypes between different feeding conditions. In the direct comparisons between the two genotypes only one orexigenic gene, hcrt or orexin, showed higher expression in lepr mutants in the fasting groups and a similar tendency was also observed in the normal feeding groups (Fig 2). This suggests a repressive effect of leptin signal on hcrt expression in zebrafish gut during fasting. We have not detected such a regulatory connection in our previous study in zebrafish brain, indicating thus tissue specificity of the transcriptional regulation of hcrt by the leptin signal [35]. Hypocretin (orexin) neuropeptide precursor gene, hcrt, encodes a hypothalamic precursor protein that, after proteolytic processing, gives rise to two neuropeptides, orexin A and orexin B. These neuropeptides play role in various physiological processes, such as regulation of sleep-wake cycle, reward-seeking, addiction, stress and feeding behaviour in zebrafish [106]. During fasting, hcrt, acts as a potent orexigenic neuropeptide in zebrafish brain [61]. In goldfish, leptin signal represses hcrt brain expression and thus inhibits its orexigenic effect on feeding behaviour [27].
Unlike the orexigenic genes, almost half of the gut-expressed anorexigenic genes (8 out of 18 genes) showed expression differences during changes in feeding conditions (Fig 3). This suggests that the potential regulatory effects of the gut on feeding regulation are mediated through anorexigenic genes in zebrafish. Among these differentially expressed anorexigenic genes, only two, cart3 and nucb2a, displayed changes in their expression dynamics in the gut of lepr mutants. This indicates that leptin signal plays important regulatory roles in transcriptional changes of the other 6 anorexigenic genes (cart2, dbi, oxt, nmu, pacap and pomc) in the zebrafish gut. For instance, induced expression of cart2, a member of cocaine and amphetamine regulated transcripts genes, in the refeeding groups, was only observed in the gut of wild-type zebrafish. This finding is opposite to their expression in the brain, where all members of cart genes showed reduced expression during fasting and refeeding in the wild-type zebrafish [35]. This also suggests that a recently found gene regulatory network (GRN) in zebrafish brain, consisting leptin signal and sp1 transcription factor as upstream regulators of cart genes, i.e. lepr-sp1-cart axis, does not exist in the gut [107]. Cart genes are shown to have conserved anorexigenic role in teleosts [30], including zebrafish [47, 48]. Although regulatory connections between cart genes and leptin are found in other species, such as goldfish [108], and mammals [109, 110], these connections are only reported in the brain.
Perhaps the most striking finding among the anorexigenic genes was the differential expression of prepro-kisspeptin 1, kiss1, and its receptor, kiss1r, between the different feeding conditions, only in the lepr mutant fish, as well as between the genotypes during fasting (Figs 3 and 4). Although, kiss1 and kiss1r are expressed in zebrafish gut and brain [111], so far their anorexigenic roles have only studied in sea bass (a Perciforme species) [62]. In mammals, the brain expression of kiss1 is induced by leptin administration [112], while fasting reduces its brain expression levels [113, 114]. Consistently, we have already observed reduced kiss1 expression after fasting in the brain of wild-type zebrafish but not in the lepr mutant, indicating its leptin-dependent transcriptional regulation during fasting in zebrafish [35]. In the gut, however, we found an opposite pattern; where kiss1 expression was reduced during fasting only in the lepr mutants and not in the wild-type fish. This suggests a potential opposite regulatory function of leptin signal on kiss1 expression between brain and gut in zebrafish. Interestingly, the kisspeptin receptor, kiss1r, which showed no expression difference between the genotypes in the brain [35], displayed an opposite expression pattern to its ligand gene (kiss1) in the gut, with increased expression during fasting. The regulatory connection between kiss1 (and its receptor) and leptin signal is not studied in the digestive system of vertebrates and these observations stress the necessity of future functional studies in the gut to unravel potential links. These results also suggest that the functional role of leptin signalling through regulation of kiss1/kiss1r during fasting/refeeding might be more important in the gut than in the brain of zebrafish.
Another interesting finding was the increased expression of pomc, pro-opiomelanocortin gene, during fasting in the gut of wild-type zebrafish and the absence of these changes in the lepr mutant fish. The basal gut expression level of pomc was higher in the lepr mutant under normal feeding condition, which was opposite to our previous findings in the brain [35]. During fasting, however, this pattern was reversed in the gut, with higher expression in wild-type, while in our previous study, no difference was observed between the two genotypes in the brain during fasting [35]. The implications of these changes are difficult to interpret, but it is obvious that there is a distinct leptin-dependent transcriptional regulation of pomc between the two organs under similar feeding conditions.
In zebrafish brain, almost all of the anorexigenic genes regulated by leptin signal showed reduced expression in the lepr mutant under normal feeding condition [35]. In the direct comparisons between the two genotypes in the gut, mchr2 was the only anorexigenic gene with reduced expression in the lepr mutants under normal feeding condition (Fig 4). Mchr2 encodes a melanin concentrating hormone (MCH) receptor gene and although the role of MCH signal in appetite-regulation has not been investigated in zebrafish, its anorexigenic role is already demonstrated in goldfish [64, 65]. In mice, leptin acts as an upstream regulator of MCH signal in the brain [115, 116]. Apart from the role of MCH signal in the regulation of immune responses, no studies have addressed other potential functions of this signal in zebrafish gut. Nucleobindin 2a or nesfatin 1, nucb2a, was another gene showing differences in the direct comparisons between the two genotypes, however, these differences were in opposite direction, i.e. reduced expression during fasting, but increased expression in 6 hrs refeeding in the gut of lepr mutant zebrafish. The anorexignenic role of nucb2a is already described in zebrafish, and interestingly, fasting results in decreased expression, both in the brain and the gut [72]. This may indicate that the decreased expression of nucb2a in both organs mitigates its anorexigenic effects during fasting in zebrafish. Our findings confirm the reduced expression of nucb2a during fasting in both genotypes, indicating a leptin independent regulation. However, the increase in nucb2a expression during refeeding, only in lepr mutants, might be linked to the function of leptin. Finally, dbi and nmu showed higher expression in the lepr mutants than wild-types in one of the refeeding groups. The octadecaneuropeptide or ODN is an endozepine peptide, which is generated by the cleavage of the product of dbi gene (diazepam-binding inhibitor) in the brain. In goldfish, brain injection of ODN has been shown to inhibits food intake and locomotor activity [52, 53]. These anorexigenic effects in goldfish are mediated through a mc4r and crh dependent signals [53], but we have not detected the expression of neither mc4r nor crh in the zebrafish gut. This suggests that the potential function of ODN is mediated through different signals in zebrafish gut.
Neuromedin U preproprotein gene, nmu, encodes a multifunctional neuropeptide, which controls the circulatory and digestive systems, energy homeostasis, and acts as anorexigenic factor in goldfish [66, 67]. mRNA transcripts of nmu were found in both the brain and the gut of goldfish [66], and in the brain of zebrafish [35]. Moreover, brain injection of Nmu causes suppression of food intake and locomotor activity in a dose-dependent manner in goldfish [66]. Although, nmu encoded protein is considered as a brain-gut neuropeptide, its exact role in feeding regulation through the digestive system remains unclear. The anorexigenic effects of nmu are mediated through crh or corticotropin-releasing hormone receptor in the goldfish brain [67], and we also found crh to be a downstream target of leptin signal in zebrafish brain [35]. Future studies are required to investigate if nmu expression in the gut plays also a similar anorexigenic role in zebrafish and, since crh is not expressed in gut, how its potential regulatory function is mediated in the gut. In mice and rats, the anorexigenic effects of nmu are independent of leptin signal in the brain [117], but in the present study we found that nmu expression in the zebrafish gut is linked to leptin signal during refeeding. Finally, similar to our previous observations in the brain [35], the strongest divergence in expression of appetite-regulating genes between the genotypes in the gut were found during fasting.
The hierarchical clustering of the experimental groups for both genotypes, based on the combined expression patterns of all appetite regulating genes tested in this study revealed that the strongest divergence between wild-types and lepr mutants happens during fasting in zebrafish intestine. While each of the other feeding groups clustered together regardless of their genotypes, the wild-type fasting group and the mutant fasting group branched distantly from each other (Fig 7B). In our previous study, we have found very similar results in the brain where the wild-type fasting group was clustered with control feeding group, whereas the mutant fasting group was clustered with refeeding groups [35]. This shows that both brain and intestine have similar divergent transcriptional in response to changes in feeding conditions for the appetite regulating genes and only during fasting the overall expression pattern for these genes is highly affected by leptin signal. Moreover, this might be an indicator of cross-talk between brain and intestine during changes in feeding condition through appetite regulating genes and their receptors in zebrafish. It is also important to note that since we could only breed limited number of fish for this study, due to low fertility rate of the lepr mutants [89], it was not possible to conduct other accurate tests of the feeding status, such as measuring blood levels of glucose, amino acids, lipids, etc., which require an entire fish to be used. However, we have shown in our previous study that the overall expression patterns of the appetite regulating genes and their receptors showed clear differences between the feeding conditions in zebrafish brain [35].
Among the gut marker genes tested in this study, apoa4a and slc2a5, showed changes in their expression dynamic in both wild-type and lepr mutant zebrafish. Apolipoprotein A4 gene (apoa4a) has conserved expression in villus-enterocyte of small intestine and hepatocytes of liver in zebrafish and mammals [36, 118]. Several functions have been attributed to apoa4a, such as regulating gluconeogenesis, lipid metabolism and transport, stomach emptying, acid secretion, and food intake (review [119]). In zebrafish, visceral upregulation of apoa4a is required for transition from endogenous to exogenous feeding, during mouth-opening larval stage [120]. The overexpression of apoa4a can lead to reduced food intake in both zebrafish and rats [121, 122]. In addition, it has been already demonstrated that extended fasting can reduce the expression of apoa4a in zebrafish liver [123] and its protein level in human body fluids [124]. Consistently, we found decreased expression of apoa4a in the gut in both genotypes, during fasting and this suggests its potential anorexigenic function in zebrafish. However, we found reduced expression of another apolipoprotein family member, apoa1a, only in the mutant group during fasting. Similarly, apoa1a is a conserved maker gene of the small intestine and liver in zebrafish and mammals [36, 125]. To our knowledge, there are no studies addressing transcriptional changes of apoa1a in response to fasting, its potential transcriptional regulation by leptin signal and its role in regulation of food intake in fish. Our findings of reduced apoa1a expression only in lepr mutant during fasting may imply on its leptin dependent role in lipid metabolism or food intake in zebrafish gut.
The second gene with differential expression dynamics during changes in feeding conditions in both genotypes, was slc2a5, but its expression pattern, unlike apoa4a, was different between the two genotypes. It displayed increased expression during fasting in the gut of wild-type zebrafish, whereas in lepr mutants, its expression was increased during normal feeding. Solute carrier family 2 member 5 or slc2a5 (previously named glut5) encodes a fructose transporter responsible for fructose uptake by the small intestine [126, 127]. The reduction in intestinal expression of slc2a5 during fasting, as well as its induced expression after refeeding, has been shown in rats [128]. Our finding show similar expression pattern for slc2a5 in the zebrafish gut during fasting and refeeding. Interestingly, leptin signal has regulatory effects on hepatic expression of slc2a5 in zebrafish [40]. In agreement with this study, we observed dysregulation of slc2a5 in lepr mutants, indicating leptin-dependent transcriptional response of slc2a5 to fasting and refeeding. The same study also showed increased visceral expression of an insulin gene (insa but not insb) in the absence of functional leptin signal [40]. Similarly, we also found higher expression of insa in the gut of lepr mutant zebrafish than wild-types in the control and fasting groups (Fig 6).
Two other marker genes showing differences in their expression dynamics between the genotypes were aqp3a, aquaglyceroporin-3, and pept1, intestinal H+/peptide cotransporter (also known as slc15a1) (Fig 5). Aqp3a showed increased expression levels at 6 hrs refeeding group compared to the fasting group, but only in wild-types. This indicates that this change might be dependent on leptin signal. Aqp3a encodes a conserved osmoregulatory channel protein on the membrane of epithelial cells in the large intestine in mammals and mucus cells of the posterior intestine of teleost fish [36, 129]. Thus, aqp3a is considered as a marker gene for the posterior part of intestine in vertebrates, which has an important role in water absorption, by increasing water permeability across cell membranes [36, 129, 130]. In mammals, leptin signal has been shown to control aqp3 expression in liver and adipose tissue [131, 132]. The leptin dependent transcriptional regulation of aqp3 is mediated through insulin and its downstream molecular cascade [131]. Such a regulatory connection between leptin signal and aqp3a has not been investigated in any tissues of teleost fish. Our finding suggests a regulatory link between leptin and aqp3a in zebrafish gut under different feeding conditions. The next gene, pept1 encodes a major peptide transporter expressed in the proximal part of zebrafish intestine, is localized to the brush border membrane of the epithelial cells and mediates the uptake of di- and tripeptides from the lumen into the enterocytes [133]. In a previous study in zebrafish, pept1 displayed increased expression during refeeding after short fasting [41], but we did not find such transcriptional changes. On the other hand, reduced intestinal expression of the mammalian orthologous of pept1 has been reported in the absence of leptin signal [134]. In the present study, we found reduced pept1 expression in 6 hrs refeeding group compared to the normal feeding group only in lepr mutant, which suggests potential regulatory connection between leptin and pept1 expression in zebrafish gut. Finally, in the direct comparison between the two genotypes, we found reduced expression of sglt1 in the mutants at 6 hrs refeeding group. The sodium/glucose co-transporter 1 gene or sglt1 (also known as slc5a1) encodes an integral membrane protein, which is the major mediator of dietary glucose and galactose uptakes from the intestine [135]. A complex regulatory effect of leptin signal on intestinal expression of sglt1 orthologue has been shown in mammals [136]. However, such a regulatory connection has not been investigated in fish.
Conclusions
The study revealed that most brain-expressed orexigenic and anorexigenic genes are also expressed in the zebrafish gut. Unlike in the brain, changes in feeding conditions do not affect the expression of orexigenic genes in the zebrafish gut, as only hcrt showed differential expression in the gut of zebrafish with impaired leptin signal. On the contrary, among the anorexigenic genes, several genes were differentially expressed in the gut of wild-type and lepr mutant zebrafish and significant differences were found under different feeding conditions. Taken together, based on the results of the present study, we propose a potential role of the gastrointestinal system in the regulation of feeding in zebrafish. However, future studies are required to investigate the expression patterns of these genes in different parts of the intestine to specify their changes in more anatomical detail. In addition, further functional studies can be very useful to unravel molecular cross-talks between brain and gut in regulation of feeding behaviour in zebrafish.
Supporting information
(XLSX)
(PDF)
Acknowledgments
The authors thank the Genome Engineering Zebrafish Facility at Uppsala University for its responsible management of our fish. Special thanks to Chrysoula Zalamitsou for her technical assistance through the study.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
The work was supported by the Carl Trygger Foundation for scientific research (CTS 16:413) and the grant was assigned to MS (the corresponding author). URL: https://www.carltryggersstiftelse.se/this-is-the-carl-trygger-foundation/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Galmiche M, Déchelotte P, Lambert G, Tavolacci MP. Prevalence of eating disorders over the 2000–2018 period: a systematic literature review. Am J Clin Nutr. 2019;109: 1402–1413. doi: 10.1093/ajcn/nqy342 [DOI] [PubMed] [Google Scholar]
- 2.Berner LA, Brown TA, Lavender JM, Lopez E, Wierenga CE, Kaye WH. Neuroendocrinology of reward in anorexia nervosa and bulimia nervosa: Beyond leptin and ghrelin. Mol Cell Endocrinol. 2018. doi: 10.1016/j.mce.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobrino Crespo C, Perianes Cachero A, Puebla Jiménez L, Barrios V, Arilla Ferreiro E. Peptides and Food Intake. Front Endocrinol (Lausanne). 2014;5: 58. doi: 10.3389/fendo.2014.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keen-Rhinehart E, Ondek K, Schneider JE. Neuroendocrine regulation of appetitive ingestive behavior. Front Neurosci. 2013;7: 213. doi: 10.3389/fnins.2013.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker JA, Bloom SR. Hypothalamic neuropeptides and the regulation of appetite. Neuropharmacology. 2012;63: 18–30. doi: 10.1016/j.neuropharm.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 6.Anubhuti Arora S. Role of neuropeptides in appetite regulation and obesity—A review. Neuropeptides. 2006;40: 375–401. doi: 10.1016/j.npep.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 7.Cegla J, Tan TM, Bloom SR. Gut–brain cross-talk in appetite regulation. Curr Opin Clin Nutr Metab Care. 2010;13: 588–593. doi: 10.1097/MCO.0b013e32833b64a3 [DOI] [PubMed] [Google Scholar]
- 8.Smeets PAM, Preissl H. Functional Neuroimaging of Appetite and Gut-Brain Interactions. Neuroendocrinology of Appetite. Chichester, UK: John Wiley & Sons, Ltd; 2016. pp. 174–200. [Google Scholar]
- 9.Zanchi D, Depoorter A, Egloff L, Haller S, Mählmann L, Lang UE, et al. The impact of gut hormones on the neural circuit of appetite and satiety: A systematic review. Neuroscience and Biobehavioral Reviews. Elsevier Ltd; 2017. pp. 457–475. doi: 10.1016/j.neubiorev.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 10.Clemmensen C, Müller TD, Woods SC, Berthoud HR, Seeley RJ, Tschöp MH. Gut-Brain Cross-Talk in Metabolic Control. Cell. Cell Press; 2017. pp. 758–774. doi: 10.1016/j.cell.2017.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weltens N, Iven J, Van Oudenhove L, Kano M. The gut-brain axis in health neuroscience: implications for functional gastrointestinal disorders and appetite regulation. Ann N Y Acad Sci. 2018;1428: 129–150. doi: 10.1111/nyas.13969 [DOI] [PubMed] [Google Scholar]
- 12.Buhmann H, le Roux CW, Bueter M. The gut–brain axis in obesity. Best Pract Res Clin Gastroenterol. 2014;28: 559–571. doi: 10.1016/j.bpg.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 13.Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. The microbiota–gut–brain axis in obesity. Lancet Gastroenterol Hepatol. 2017;2: 747–756. doi: 10.1016/S2468-1253(17)30147-4 [DOI] [PubMed] [Google Scholar]
- 14.Lutfi E, Basili D, Falcinelli S, Morillas L, Carnevali O, Capilla E, et al. The probiotic Lactobacillus rhamnosus mimics the dark-driven regulation of appetite markers and melatonin receptors’ expression in zebrafish (Danio rerio) larvae: Understanding the role of the gut microbiome. Comp Biochem Physiol Part B Biochem Mol Biol. 2021; 110634. doi: 10.1016/j.cbpb.2021.110634 [DOI] [PubMed] [Google Scholar]
- 15.Mithieux G. Crosstalk between gastrointestinal neurons and the brain in the control of food intake. Best Pract Res Clin Endocrinol Metab. 2014;28: 739–744. doi: 10.1016/j.beem.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 16.Mithieux G, Gautier-Stein A. Intestinal glucose metabolism revisited. Diabetes Res Clin Pract. 2014;105: 295–301. doi: 10.1016/j.diabres.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 17.Camilleri M. Peripheral Mechanisms in Appetite Regulation. Gastroenterology. 2015;148: 1219–1233. doi: 10.1053/j.gastro.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Londraville RL, Macotela Y, Duff RJ, Easterling MR, Liu Q, Crespi EJ. Comparative endocrinology of leptin: Assessing function in a phylogenetic context. Gen Comp Endocrinol. 2014;203: 146–157. doi: 10.1016/j.ygcen.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco AM, Soengas JL. Leptin signalling in teleost fish with emphasis in food intake regulation. Mol Cell Endocrinol. 2021;526: 111209. doi: 10.1016/j.mce.2021.111209 [DOI] [PubMed] [Google Scholar]
- 20.Broderick TL, Wang D, Jankowski M, Gutkowska J. Unexpected effects of voluntary exercise training on natriuretic peptide and receptor mRNA expression in the ob/ob mouse heart. Regul Pept. 2014;188: 52–59. doi: 10.1016/j.regpep.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 21.Wabitsch M, Funcke J-B, Lennerz B, Kuhnle-Krahl U, Lahr G, Debatin K-M, et al. Biologically Inactive Leptin and Early-Onset Extreme Obesity. N Engl J Med. 2015;372: 48–54. doi: 10.1056/NEJMoa1406653 [DOI] [PubMed] [Google Scholar]
- 22.Audira G, Sarasamma S, Chen J-R, Juniardi S, Sampurna BP, Liang S-T, et al. Zebrafish Mutants Carrying Leptin a (lepa) Gene Deficiency Display Obesity, Anxiety, Less Aggression and Fear, and Circadian Rhythm and Color Preference Dysregulation. Int J Mol Sci. 2018;19. doi: 10.3390/ijms19124038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorissen M, Bernier NJ, Nabuurs SB, Flik G, Huising MO. Two divergent leptin paralogues in zebrafish (Danio rerio) that originate early in teleostean evolution. J Endocrinol. 2009;201: 329–339. doi: 10.1677/JOE-09-0034 [DOI] [PubMed] [Google Scholar]
- 24.Tian J, He G, Mai K, Liu C. Effects of postprandial starvation on mRNA expression of endocrine-, amino acid and peptide transporter-, and metabolic enzyme-related genes in zebrafish (Danio rerio). Fish Physiol Biochem. 2015;41: 773–787. doi: 10.1007/s10695-015-0045-x [DOI] [PubMed] [Google Scholar]
- 25.Oka T, Nishimura Y, Zang L, Hirano M, Shimada Y, Wang Z, et al. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010;10: 21. doi: 10.1186/1472-6793-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q, Chen Y, Copeland D, Ball H, Duff RJ, Rockich B, et al. Expression of leptin receptor gene in developing and adult zebrafish. Gen Comp Endocrinol. 2010;166: 346–355. doi: 10.1016/j.ygcen.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volkoff H, Joy Eykelbosh A, Ector Peter R. Role of leptin in the control of feeding of goldfish Carassius auratus: interactions with cholecystokinin, neuropeptide Y and orexin A, and modulation by fasting. Brain Res. 2003;972: 90–109. doi: 10.1016/s0006-8993(03)02507-1 [DOI] [PubMed] [Google Scholar]
- 28.Yan A-F, Chen T, Chen S, Ren C-H, Hu C-Q, Cai Y-M, et al. Goldfish Leptin-AI and Leptin-AII: Function and Central Mechanism in Feeding Control. Int J Mol Sci. 2016;17: 783. doi: 10.3390/ijms17060783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imperatore R, D’Angelo L, Safari O, Motlagh HA, Piscitelli F, de Girolamo P, et al. Overlapping Distribution of Orexin and Endocannabinoid Receptors and Their Functional Interaction in the Brain of Adult Zebrafish. Front Neuroanat. 2018;12: 62. doi: 10.3389/fnana.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkoff H. The Neuroendocrine Regulation of Food Intake in Fish: A Review of Current Knowledge. Front Neurosci. 2016;10: 540. doi: 10.3389/fnins.2016.00540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen BA, Breen DM, Duca FA, Côté CD, Zadeh-Tahmasebi M, Filippi BM, et al. Jejunal Leptin-PI3K Signaling Lowers Glucose Production. Cell Metab. 2014;19: 155–161. doi: 10.1016/j.cmet.2013.11.014 [DOI] [PubMed] [Google Scholar]
- 32.Sáinz N, Barrenetxe J, Moreno-Aliaga MJ, Martínez JA. Leptin resistance and diet-induced obesity: central and peripheral actions of leptin. Metabolism. 2015;64: 35–46. doi: 10.1016/j.metabol.2014.10.015 [DOI] [PubMed] [Google Scholar]
- 33.Mania M, Maruccio L, Russo F, Abbate F, Castaldo L, D’Angelo L, et al. Expression and distribution of leptin and its receptors in the digestive tract of DIO (diet-induced obese) zebrafish. Ann Anat—Anat Anzeiger. 2017;212: 37–47. doi: 10.1016/j.aanat.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Suarez O, Cabo R, Abbate F, Randazzo B, Laurà R, Piccione G, et al. Presence and distribution of leptin and its receptor in the gut of adult zebrafish in response to feeding and fasting. Anat Histol Embryol. 2018;47: 456–465. doi: 10.1111/ahe.12384 [DOI] [PubMed] [Google Scholar]
- 35.Ahi EP, Brunel M, Tsakoumis E, Schmitz M. Transcriptional study of appetite regulating genes in the brain of zebrafish (Danio rerio) with impaired leptin signalling. Sci Rep. 2019;9: 20166. doi: 10.1038/s41598-019-56779-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Du J, Lam SH, Mathavan S, Matsudaira P, Gong Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics. 2010;11: 392. doi: 10.1186/1471-2164-11-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unniappan S, Lin X, Cervini L, Rivier J, Kaiya H, Kangawa K, et al. Goldfish Ghrelin: Molecular Characterization of the Complementary Deoxyribonucleic Acid, Partial Gene Structure and Evidence for Its Stimulatory Role in Food Intake. Endocrinology. 2002;143: 4143–4146. doi: 10.1210/en.2002-220644 [DOI] [PubMed] [Google Scholar]
- 38.Amole N, Unniappan S. Fasting induces preproghrelin mRNA expression in the brain and gut of zebrafish, Danio rerio. Gen Comp Endocrinol. 2009;161: 133–137. doi: 10.1016/j.ygcen.2008.11.002 [DOI] [PubMed] [Google Scholar]
- 39.Miura T, Maruyama K, Shimakura S-I, Kaiya H, Uchiyama M, Kangawa K, et al. Regulation of food intake in the goldfish by interaction between ghrelin and orexin. Peptides. 2007;28: 1207–1213. doi: 10.1016/j.peptides.2007.03.023 [DOI] [PubMed] [Google Scholar]
- 40.Michel M, Page-McCaw PS, Chen W, Cone RD. Leptin signaling regulates glucose homeostasis, but not adipostasis, in the zebrafish. Proc Natl Acad Sci U S A. 2016;113: 3084. doi: 10.1073/pnas.1513212113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koven W, Schulte P. The effect of fasting and refeeding on mRNA expression of PepT1 and gastrointestinal hormones regulating digestion and food intake in zebrafish (Danio rerio). Fish Physiol Biochem. 2012;38: 1565–1575. doi: 10.1007/s10695-012-9649-6 [DOI] [PubMed] [Google Scholar]
- 42.Blanco AM, Bertucci JI, Ramesh N, Delgado MJ, Valenciano AI, Unniappan S. Ghrelin Facilitates GLUT2-, SGLT1-and SGLT2-mediated Intestinal Glucose Transport in Goldfish (Carassius auratus). Sci Rep. 2017;7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y, Golling G, Thacker TL, Cone RD. Agouti-Related Protein (AGRP) Is Conserved and Regulated by Metabolic State in the Zebrafish, Danio rerio. Endocrine. 2003;22: 257–266. doi: 10.1385/ENDO:22:3:257 [DOI] [PubMed] [Google Scholar]
- 44.Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. 2007;21: 2042–2049. doi: 10.1096/fj.06-7503com [DOI] [PubMed] [Google Scholar]
- 45.Volkoff H, Wyatt JL. Apelin in goldfish (Carassius auratus): Cloning, distribution and role in appetite regulation. Peptides. 2009;30: 1434–1440. doi: 10.1016/j.peptides.2009.04.020 [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Sun P, Yang F, Kong T, Zhang R. Tributyltin disrupts feeding and energy metabolism in the goldfish (Carassius auratus). Chemosphere. 2016;152: 221–228. doi: 10.1016/j.chemosphere.2016.02.127 [DOI] [PubMed] [Google Scholar]
- 47.Nishio S-I, Gibert Y, Berekelya L, Bernard L, Brunet F, Guillot E, et al. Fasting Induces CART Down-Regulation in the Zebrafish Nervous System in a Cannabinoid Receptor 1-Dependent Manner. Mol Endocrinol. 2012;26: 1316–1326. doi: 10.1210/me.2011-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guillot R, Cortés R, Navarro S, Mischitelli M, García-Herranz V, Sánchez E, et al. Behind melanocortin antagonist overexpression in the zebrafish brain: A behavioral and transcriptomic approach. Horm Behav. 2016;82: 87–100. doi: 10.1016/j.yhbeh.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 49.Shimada Y, Hirano M, Nishimura Y, Tanaka T. A High-Throughput Fluorescence-Based Assay System for Appetite-Regulating Gene and Drug Screening. Gong Z, editor. PLoS One. 2012;7: e52549. doi: 10.1371/journal.pone.0052549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Pedro N, Alonso-Gómez AL, Gancedo B, Delgado MJ, Alonso-Bedate M. Role of corticotropin-releasing factor (CRF) as a food intake regulator in goldfish. Physiol Behav. 1993;53: 517–520. doi: 10.1016/0031-9384(93)90146-7 [DOI] [PubMed] [Google Scholar]
- 51.Bernier NJ. The corticotropin-releasing factor system as a mediator of the appetite-suppressing effects of stress in fish. Gen Comp Endocrinol. 2006;146: 45–55. doi: 10.1016/j.ygcen.2005.11.016 [DOI] [PubMed] [Google Scholar]
- 52.Matsuda K, Wada K, Miura T, Maruyama K, Shimakura SI, Uchiyama M, et al. Effect of the diazepam-binding inhibitor-derived peptide, octadecaneuropeptide, on food intake in goldfish. Neuroscience. 2007;150: 425–432. doi: 10.1016/j.neuroscience.2007.09.012 [DOI] [PubMed] [Google Scholar]
- 53.Matsuda K, Kojima K, Wada K, Maruyama K, Leprince J, Tonon M-C, et al. The Anorexigenic Action of the Octadecaneuropeptide (ODN) in Goldfish is Mediated Through the MC4R- and Subsequently the CRH Receptor-Signaling Pathways. J Mol Neurosci. 2010;42: 74–79. doi: 10.1007/s12031-010-9346-9 [DOI] [PubMed] [Google Scholar]
- 54.de Pedro N, Céspedes MV, Delgado MJ, Alonso-Bedate M. The galanin-induced feeding stimulation is mediated via α2-adrenergic receptors in goldfish. Regul Pept. 1995;57: 77–84. doi: 10.1016/0167-0115(95)91255-4 [DOI] [PubMed] [Google Scholar]
- 55.Volkoff H, Peter RE. Interactions between orexin A, NPY and galanin in the control of food intake of the goldfish, Carassius auratus. Regul Pept. 2001;101: 59–72. doi: 10.1016/s0167-0115(01)00261-0 [DOI] [PubMed] [Google Scholar]
- 56.Li L, Wei S, Huang Q, Feng D, Zhang S, Liu Z. A novel galanin receptor 1a gene in zebrafish: Tissue distribution, developmental expression roles in nutrition regulation. Comp Biochem Physiol Part B Biochem Mol Biol. 2013;164: 159–167. doi: 10.1016/j.cbpb.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 57.Nishiguchi R, Azuma M, Yokobori E, Uchiyama M, Matsuda K. Gonadotropin-releasing hormone 2 suppresses food intake in the zebrafish, Danio rerio. Front Endocrinol (Lausanne). 2012;3: 122. doi: 10.3389/fendo.2012.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoskins LJ, Xu M, Volkoff H. Interactions between gonadotropin-releasing hormone (GnRH) and orexin in the regulation of feeding and reproduction in goldfish (Carassius auratus). Horm Behav. 2008;54: 379–385. doi: 10.1016/j.yhbeh.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 59.Volkoff H, Bjorklund JM, Peter RE. Stimulation of feeding behavior and food consumption in the goldfish, Carassius auratus, by orexin-A and orexin-B. Brain Res. 1999;846: 204–9. doi: 10.1016/s0006-8993(99)02052-1 [DOI] [PubMed] [Google Scholar]
- 60.Nakamachi T, Matsuda K, Maruyama K, Miura T, Uchiyama M, Funahashi H, et al. Regulation by Orexin of Feeding Behaviour and Locomotor Activity in the Goldfish. J Neuroendocrinol. 2006;18: 290–297. doi: 10.1111/j.1365-2826.2006.01415.x [DOI] [PubMed] [Google Scholar]
- 61.Yokobori E, Kojima K, Azuma M, Kang KS, Maejima S, Uchiyama M, et al. Stimulatory effect of intracerebroventricular administration of orexin A on food intake in the zebrafish, Danio rerio. Peptides. 2011;32: 1357–1362. doi: 10.1016/j.peptides.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 62.Escobar S, Felip A, Zanuy S, Carrillo M. Is the kisspeptin system involved in responses to food restriction in order to preserve reproduction in pubertal male sea bass (Dicentrarchus labrax)? Comp Biochem Physiol Part A Mol Integr Physiol. 2016;199: 38–46. doi: 10.1016/j.cbpa.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 63.Cerdá-Reverter JM, Ringholm A, Schiöth HB, Peter RE. Molecular Cloning, Pharmacological Characterization, and Brain Mapping of the Melanocortin 4 Receptor in the Goldfish: Involvement in the Control of Food Intake. Endocrinology. 2003;144: 2336–2349. doi: 10.1210/en.2002-0213 [DOI] [PubMed] [Google Scholar]
- 64.Matsuda K, Shimakura S-I, Maruyama K, Miura T, Uchiyama M, Kawauchi H, et al. Central administration of melanin-concentrating hormone (MCH) suppresses food intake, but not locomotor activity, in the goldfish, Carassius auratus. Neurosci Lett. 2006;399: 259–263. doi: 10.1016/j.neulet.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 65.Matsuda K, Shimakura S-I, Miura T, Maruyama K, Uchiyama M, Kawauchi H, et al. Feeding-induced changes of melanin-concentrating hormone (MCH)-like immunoreactivity in goldfish brain. Cell Tissue Res. 2007;328: 375–382. doi: 10.1007/s00441-006-0347-5 [DOI] [PubMed] [Google Scholar]
- 66.Maruyama K, Konno N, Ishiguro K, Wakasugi T, Uchiyama M, Shioda S, et al. Isolation and characterisation of four cDNAs encoding neuromedin U (NMU) from the brain and gut of goldfish, and inhibitory effect of a deduced NMU on food intake and locomotor activity. J Neuroendocrinol. 2007;ja: 070927052250004. doi: 10.1111/j.1365-2826.2007.01615.x [DOI] [PubMed] [Google Scholar]
- 67.Maruyama K, Wada K, Ishiguro K, Shimakura S-I, Wakasugi T, Uchiyama M, et al. Neuromedin U-induced anorexigenic action is mediated by the corticotropin-releasing hormone receptor-signaling pathway in goldfish. Peptides. 2009;30: 2483–2486. doi: 10.1016/j.peptides.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 68.López-Patiño MA, Guijarro AI, Isorna E, Delgado MJ, Alonso-Bedate M, de Pedro N. Neuropeptide Y has a stimulatory action on feeding behavior in goldfish (Carassius auratus). Eur J Pharmacol. 1999;377: 147–153. doi: 10.1016/s0014-2999(99)00408-2 [DOI] [PubMed] [Google Scholar]
- 69.Yokobori E, Azuma M, Nishiguchi R, Kang KS, Kamijo M, Uchiyama M, et al. Neuropeptide Y Stimulates Food Intake in the Zebrafish, Danio rerio. J Neuroendocrinol. 2012;24: 766–773. doi: 10.1111/j.1365-2826.2012.02281.x [DOI] [PubMed] [Google Scholar]
- 70.Kerbel B, Unniappan S. Nesfatin-1 Suppresses Energy Intake, Co-localises Ghrelin in the Brain and Gut, and Alters Ghrelin, Cholecystokinin and Orexin mRNA Expression in Goldfish. J Neuroendocrinol. 2012;24: 366–377. doi: 10.1111/j.1365-2826.2011.02246.x [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez R, Kerbel B, Chun A, Unniappan S. Molecular, Cellular and Physiological Evidences for the Anorexigenic Actions of Nesfatin-1 in Goldfish. Roberts RG, editor. PLoS One. 2010;5: e15201. doi: 10.1371/journal.pone.0015201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatef A, Shajan S, Unniappan S. Nutrient status modulates the expression of nesfatin-1 encoding nucleobindin 2A and 2B mRNAs in zebrafish gut, liver and brain. Gen Comp Endocrinol. 2015;215: 51–60. doi: 10.1016/j.ygcen.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 73.Blechman J, Amir-Zilberstein L, Gutnick A, Ben-Dor S, Levkowitz G. The metabolic regulator PGC-1α directly controls the expression of the hypothalamic neuropeptide oxytocin. J Neurosci. 2011;31: 14835–40. doi: 10.1523/JNEUROSCI.1798-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuda K, Maruyama K, Nakamachi T, Miura T, Uchiyama M, Shioda S. Inhibitory effects of pituitary adenylate cyclase-activating polypeptide (PACAP) and vasoactive intestinal peptide (VIP) on food intake in the goldfish, Carassius auratus. Peptides. 2005;26: 1611–1616. doi: 10.1016/j.peptides.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 75.Liu S, Zhang C, Peng G. Effects of starvation on the expression of feeding related neuropeptides in the larval zebrafish hypothalamus. Yi chuan = Hered. 2016;38: 821–30. doi: 10.16288/j.yczz.16-087 [DOI] [PubMed] [Google Scholar]
- 76.Dalmolin C, Almeida DV, Figueiredo MA, Marins LF. Food intake and appetite control in a GH-transgenic zebrafish. Fish Physiol Biochem. 2015;41: 1131–1141. doi: 10.1007/s10695-015-0074-5 [DOI] [PubMed] [Google Scholar]
- 77.Cerdá-Reverter JM, Schiöth HB, Peter RE. The central melanocortin system regulates food intake in goldfish. Regul Pept. 2003;115: 101–113. doi: 10.1016/s0167-0115(03)00144-7 [DOI] [PubMed] [Google Scholar]
- 78.Gonzalez R, Unniappan S. Molecular characterization, appetite regulatory effects and feeding related changes of peptide YY in goldfish. Gen Comp Endocrinol. 2010;166: 273–279. doi: 10.1016/j.ygcen.2009.09.008 [DOI] [PubMed] [Google Scholar]
- 79.Trudeau VL, Martyniuk CJ, Zhao E, Hu H, Volkoff H, Decatur WA, et al. Is secretoneurin a new hormone? Gen Comp Endocrinol. 2012;175: 10–18. doi: 10.1016/j.ygcen.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 80.Mikwar M, Navarro-Martin L, Xing L, Volkoff H, Hu W, Trudeau VL. Stimulatory effect of the secretogranin-ll derived peptide secretoneurin on food intake and locomotion in female goldfish (Carassius auratus). Peptides. 2016;78: 42–50. doi: 10.1016/j.peptides.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 81.Zheng B, Li S, Liu Y, Li Y, Chen H, Tang H, et al. Spexin Suppress Food Intake in Zebrafish: Evidence from Gene Knockout Study. Sci Rep. 2017;7: 14643. doi: 10.1038/s41598-017-15138-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong MKH, Sze KH, Chen T, Cho CK, Law HCH, Chu IK, et al. Goldfish spexin: solution structure and novel function as a satiety factor in feeding control. Am J Physiol Metab. 2013;305: E348–E366. doi: 10.1152/ajpendo.00141.2013 [DOI] [PubMed] [Google Scholar]
- 83.Abbott M, Volkoff H. Thyrotropin Releasing Hormone (TRH) in goldfish (Carassius auratus): Role in the regulation of feeding and locomotor behaviors and interactions with the orexin system and cocaine- and amphetamine regulated transcript (CART). Horm Behav. 2011;59: 236–245. doi: 10.1016/j.yhbeh.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 84.Busch-Nentwich E, Kettleborough R, Dooley CM, Scahill C, Sealy I, White R, et al. Sanger Institute Zebrafish Mutation Project Mutant Data Submission. In: ZFIN Direct Data Submission. 2013. [Google Scholar]
- 85.Monteiro J, Martins S, Farias M, Costa T, Certal A. The Impact of Two Different Cold-Extruded Feeds and Feeding Regimens on Zebrafish Survival, Growth and Reproductive Performance. J Dev Biol. 2018;6: 15. doi: 10.3390/jdb6030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCurley AT, Callard G V. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9: 102. doi: 10.1186/1471-2199-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Casadei R, Pelleri MC, Vitale L, Facchin F, Lenzi L, Canaider S, et al. Identification of housekeeping genes suitable for gene expression analysis in the zebrafish. Gene Expr Patterns. 2011;11: 271–6. doi: 10.1016/j.gep.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 88.Xu H, Li C, Zeng Q, Agrawal I, Zhu X, Gong Z. Genome-wide identification of suitable zebrafish Danio rerio reference genes for normalization of gene expression data by RT-qPCR. J Fish Biol. 2016;88: 2095–2110. doi: 10.1111/jfb.12915 [DOI] [PubMed] [Google Scholar]
- 89.Tsakoumis E, Ahi EP, Schmitz M. Impaired leptin signaling causes subfertility in female zebrafish. Mol Cell Endocrinol. 2022;546: 111595. doi: 10.1016/j.mce.2022.111595 [DOI] [PubMed] [Google Scholar]
- 90.Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41: D854–60. doi: 10.1093/nar/gks938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, et al. Ensembl 2012. Nucleic Acids Res. 2012;40: D84–90. doi: 10.1093/nar/gkr991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8: R19. doi: 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26: 509–15. Available: http://www.ncbi.nlm.nih.gov/pubmed/15127793 doi: 10.1023/b:bile.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- 94.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64: 5245–50. doi: 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 95.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3: RESEARCH0034. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=126239&tool=pmcentrez&rendertype=abstract doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahi EP, Guðbrandsson J, Kapralova KH, Franzdóttir SR, Snorrason SS, Maier VH, et al. Validation of Reference Genes for Expression Studies during Craniofacial Development in Arctic Charr. PLoS One. 2013;8: e66389. doi: 10.1371/journal.pone.0066389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29: e45. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=55695&tool=pmcentrez&rendertype=abstract doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thissen D, Steinberg L, Kuang D, Thissen D, Steinberg L, Kuang D. Quick and Easy Implementation of the Benjamini-Hochberg Procedure for Controlling the False Positive Rate in Multiple Comparisons. J Educ Behav Stat. 2002;27: 77–83. [Google Scholar]
- 99.Team RDC. R: A Language and Environment for Statistical Computing. R Found Stat Comput Vienna, Austria. 2013.
- 100.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45: D362–D368. doi: 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27: 95–125. doi: 10.1016/j.mam.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 102.Ahi EP, Duenser A, Singh P, Gessl W, Sturmbauer C. Appetite regulating genes may contribute to herbivory versus carnivory trophic divergence in haplochromine cichlids. PeerJ. 2020;8: e8375. doi: 10.7717/peerj.8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ahi EP, Singh P, Duenser A, Gessl W, Sturmbauer C. Divergence in larval jaw gene expression reflects differential trophic adaptation in haplochromine cichlids prior to foraging. BMC Evol Biol. 2019;19: 150. doi: 10.1186/s12862-019-1483-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gorissen M, Flik G. Leptin in teleostean fish, towards the origins of leptin physiology. J Chem Neuroanat. 2014;61–62: 200–206. doi: 10.1016/j.jchemneu.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 105.Tseng YC, Hwang PP. Some insights into energy metabolism for osmoregulation in fish. Comparative Biochemistry and Physiology—C Toxicology and Pharmacology. Elsevier Inc.; 2008. pp. 419–429. doi: 10.1016/j.cbpc.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 106.Elbaz I, Levitas-Djerbi T, Appelbaum L. The Hypocretin/Orexin Neuronal networks in zebrafish. Current Topics in Behavioral Neurosciences. Springer Verlag; 2017. pp. 75–92. [DOI] [PubMed] [Google Scholar]
- 107.Ahi EP, Tsakoumis E, Brunel M, Schmitz M. Transcriptional study reveals a potential leptin-dependent gene regulatory network in zebrafish brain. Fish Physiol Biochem. 2021. doi: 10.1007/s10695-021-00967-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Volkoff H, Peter RE. Characterization of Two Forms of Cocaine- and Amphetamine-Regulated Transcript (CART) Peptide Precursors in Goldfish: Molecular Cloning and Distribution, Modulation of Expression by Nutritional Status, and Interactions with Leptin. Endocrinology. 2001;142: 5076–5088. doi: 10.1210/endo.142.12.8519 [DOI] [PubMed] [Google Scholar]
- 109.Kristensen P, Judge ME, Thim L, Ribel U, Christjansen KN, Wulff BS, et al. Hypothalamic CART is a new anorectic peptide regulated by leptin. Nature. 1998;393: 72–76. doi: 10.1038/29993 [DOI] [PubMed] [Google Scholar]
- 110.Lee SJ, Verma S, Simonds SE, Kirigiti MA, Kievit P, Lindsley SR, et al. Leptin stimulates neuropeptide Y and cocaine amphetamine-regulated transcript coexpressing neuronal activity in the dorsomedial hypothalamus in diet-induced obese mice. J Neurosci. 2013;33: 15306–17. doi: 10.1523/JNEUROSCI.0837-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Aerle R, Kille P, Lange A, Tyler CR. Evidence for the existence of a functional Kiss1/Kiss1 receptor pathway in fish. Peptides. 2008;29: 57–64. doi: 10.1016/j.peptides.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 112.Smith JT, Acohido B V., Clifton DK, Steiner RA. KiSS-1 neurones are direct targets for leptin in the ob/ob mouse. J Neuroendocrinol. 2006;18: 298–303. doi: 10.1111/j.1365-2826.2006.01417.x [DOI] [PubMed] [Google Scholar]
- 113.Matsuzaki T, Iwasa T, Kinouchi R, Yoshida S, Murakami M, Gereltsetseg G, et al. Fasting reduces the kiss1 mRNA levels in the caudal hypothalamus of gonadally intact adult female rats. Endocr J. 2011;58: 1003–1012. doi: 10.1507/endocrj.k11e-131 [DOI] [PubMed] [Google Scholar]
- 114.Wahab F, Ullah F, Chan YM, Seminara SB, Shahab M. Decrease in hypothalamic kiss1 and kiss1r expression: A potential mechanism for fasting-induced suppression of the HPG axis in the adult male rhesus monkey (Macaca mulatta). Horm Metab Res. 2011;43: 81–85. doi: 10.1055/s-0030-1269852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kokkotou EG, Tritos NA, Mastaitis JW, Slieker L, Maratos-Flier E. Melanin-Concentrating Hormone Receptor Is a Target of Leptin Action in the Mouse Brain. Endocrinology. 2001;142: 680–686. doi: 10.1210/endo.142.2.7981 [DOI] [PubMed] [Google Scholar]
- 116.Segal-Lieberman G, Bradley RL, Kokkotou E, Carlson M, Trombly DJ, Wang X, et al. Melanin-concentrating hormone is a critical mediator of the leptin-deficient phenotype. Proc Natl Acad Sci U S A. 2003;100: 10085–90. doi: 10.1073/pnas.1633636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hanada R, Teranishi H, Pearson JT, Kurokawa M, Hosoda H, Fukushima N, et al. Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat Med. 2004;10: 1067–1073. doi: 10.1038/nm1106 [DOI] [PubMed] [Google Scholar]
- 118.Sauvaget D, Chauffeton V, Citadelle D, Chatelet FP, Cywiner-Golenzer C, Chambaz J, et al. Restriction of apolipoprotein A-IV gene expression to the intestine villus depends on a hormone-responsive element and parallels differential expression of the hepatic nuclear factor 4α and γ isoforms. J Biol Chem. 2002;277: 34540–34548. doi: 10.1074/jbc.M206074200 [DOI] [PubMed] [Google Scholar]
- 119.Wang F, Kohan AB, Lo CM, Liu M, Howles P, Tso P. Apolipoprotein A-IV: A protein intimately involved in metabolism. Journal of Lipid Research. American Society for Biochemistry and Molecular Biology Inc.; 2015. pp. 1403–1418. doi: 10.1194/jlr.R052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu H, Liu E, Li Y, Li X, Ding C. Transcriptome analysis reveals increases in visceral lipogenesis and storage and activation of the antigen processing and presentation pathway during the mouth-opening stage in zebrafish larvae. Int J Mol Sci. 2017;18: 1634. doi: 10.3390/ijms18081634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fujimoto K, Fukagawa K, Sakata T, Tso P. Suppression of food intake by apolipoprotein A-IV is mediated through the central nervous system in rats. J Clin Invest. 1993;91: 1830–1833. doi: 10.1172/JCI116395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Otis JP, Zeituni EM, Thierer JH, Anderson JL, Brown AC, Boehm ED, et al. Zebrafish as a model for apolipoprotein biology: Comprehensive expression analysis and a role for ApoA-IV in regulating food intake. DMM Dis Model Mech. 2015;8: 295–309. doi: 10.1242/dmm.018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Drew RE, Rodnick KJ, Settles M, Wacyk J, Churchill E, Powell MS, et al. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio). Physiol Genomics. 2008;35: 283–295. doi: 10.1152/physiolgenomics.90213.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bouwman FG, De Roos B, Rubio-Aliaga I, Crosley LK, Duthie SJ, Mayer C, et al. 2D-electrophoresis and multiplex immunoassay proteomic analysis of different body fluids and cellular components reveal known and novel markers for extended fasting. BMC Med Genomics. 2011;4: 1–12. doi: 10.1186/1755-8794-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Elshourbagy NA, Boguski MS, Liao WSL, Jefferson LS, Gordon JI, Taylor JM. Expression of rat apolipoprotein A-IV and A-I genes: mRNA induction during development and in response to glucocorticoids and insulin. Proc Natl Acad Sci U S A. 1985;82: 8242–8246. doi: 10.1073/pnas.82.23.8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barone S, Fussell SL, Singh AK, Lucas F, Xu J, Kim C, et al. Slc2a5 (Glut5) is essential for the absorption of fructose in the intestine and generation of fructose-induced hypertension. J Biol Chem. 2009;284: 5056–5066. doi: 10.1074/jbc.M808128200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tseng YC, Chen RD, Lee JR, Liu ST, Lee SJ, Hwang PP. Specific expression and regulation of glucose transporters in zebrafish ionocytes. Am J Physiol—Regul Integr Comp Physiol. 2009;297: R275–R290. doi: 10.1152/ajpregu.00180.2009 [DOI] [PubMed] [Google Scholar]
- 128.Honma K, Masuda Y, Mochizuki K, Goda T. Re-feeding rats a high-sucrose diet after 3 days of starvation enhances histone H3 acetylation in transcribed region and expression of jejunal GLUT5 gene. Biosci Biotechnol Biochem. 2014;78: 1071–1073. doi: 10.1080/09168451.2014.912121 [DOI] [PubMed] [Google Scholar]
- 129.Ma T, Verkman AS. Aquaporin water channels in gastrointestinal physiology. Journal of Physiology. John Wiley & Sons, Ltd; 1999. pp. 317–326. doi: 10.1111/j.1469-7793.1999.0317t.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cutler CP, Martinez AS, Cramb G. The role of aquaporin 3 in teleost fish. Comparative Biochemistry and Physiology—A Molecular and Integrative Physiology. Elsevier Inc.; 2007. pp. 82–91. doi: 10.1016/j.cbpa.2006.09.022 [DOI] [PubMed] [Google Scholar]
- 131.Rodríguez A, Catalán V, Gómez-Ambrosi J, García-Navarro S, Rotellar F, Valentí V, et al. Insulin- and Leptin-Mediated Control of Aquaglyceroporins in Human Adipocytes and Hepatocytes Is Mediated via the PI3K/Akt/mTOR Signaling Cascade. J Clin Endocrinol Metab. 2011;96: E586–E597. doi: 10.1210/jc.2010-1408 [DOI] [PubMed] [Google Scholar]
- 132.Rodríguez A, Moreno NR, Balaguer I, Méndez-Giménez L, Becerril S, Catalán V, et al. Leptin administration restores the altered adipose and hepatic expression of aquaglyceroporins improving the non-alcoholic fatty liver of ob/ob mice. Sci Rep. 2015;5: 1–13. doi: 10.1038/srep12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Verri T, Kottra G, Romano A, Tiso N, Peric M, Maffia M, et al. Molecular and functional characterisation of the zebrafish (Danio rerio) PEPT1-type peptide transporter 1. FEBS Lett. 2003;549: 115–122. doi: 10.1016/s0014-5793(03)00759-2 [DOI] [PubMed] [Google Scholar]
- 134.Hindlet P, Bado A, Farinotti R, Buyse M. Long-term effect of leptin on H+-coupled peptide cotransporter 1 activity and expression in vivo: Evidence in leptin-deficient mice. J Pharmacol Exp Ther. 2007;323: 192–201. doi: 10.1124/jpet.107.125799 [DOI] [PubMed] [Google Scholar]
- 135.Röder P V., Geillinger KE, Zietek TS, Thorens B, Koepsell H, Daniel H. The role of SGLT1 and GLUT2 in intestinal glucose transport and sensing. PLoS One. 2014;9. doi: 10.1371/journal.pone.0089977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dominguez Rieg JA, Chirasani VR, Koepsell H, Senapati S, Mahata SK, Rieg T. Regulation of intestinal SGLT1 by catestatin in hyperleptinemic type 2 diabetic mice. Lab Investig. 2016;96: 98–111. doi: 10.1038/labinvest.2015.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting information files.