Here, Lin et al. investigated the mechanisms underlying self-renewal of spermatogonial stem cells and show that DOT1L, the sole H3K79 methyltransferase, is required for spermatogonial stem cell self-renewal. Their findings identify an essential function for DOT1L in adult stem cells and provide an epigenetic paradigm for regulation of spermatogonial stem cells.
Keywords: DOT1L, epigenetics, adult stem cell, self-renewal, spermatogonial stem cell, transplantation, histone methyltransferase
Abstract
Self-renewal of spermatogonial stem cells is vital to lifelong production of male gametes and thus fertility. However, the underlying mechanisms remain enigmatic. Here, we show that DOT1L, the sole H3K79 methyltransferase, is required for spermatogonial stem cell self-renewal. Mice lacking DOT1L fail to maintain spermatogonial stem cells, characterized by a sequential loss of germ cells from spermatogonia to spermatids and ultimately a Sertoli cell only syndrome. Inhibition of DOT1L reduces the stem cell activity after transplantation. DOT1L promotes expression of the fate-determining HoxC transcription factors in spermatogonial stem cells. Furthermore, H3K79me2 accumulates at HoxC9 and HoxC10 genes. Our findings identify an essential function for DOT1L in adult stem cells and provide an epigenetic paradigm for regulation of spermatogonial stem cells.
Males produce sperm continuously through adult life, driven by the presence of spermatogonial stem cells (SSCs) that continue to renew (Oatley and Brinster 2006; Brinster 2007; Law and Oatley 2020). SSCs are a subpopulation of undifferentiated spermatogonia that proliferate, differentiate, and eventually enter meiosis to become spermatocytes. After meiosis, spermatocytes become round spermatids (haploid germ cells), which undergo dramatic differentiation and morphogenesis to become elongated spermatids—spermatozoa with flagella. Failure in SSC self-renewal leads to a lack of sperm production and thus male infertility.
The presence of SSCs in testis was definitively demonstrated by SSC transplantation, in which donor germline progenitor cells were found to restore spermatogenesis when transplanted into germ cell-depleted testis in mice (Brinster and Avarbock 1994; Brinster and Zimmermann 1994). To date, SSC transplantation remains an unequivocal functional assay for SSCs (Oatley and Brinster 2006; Dobrinski 2008; Kubota and Brinster 2008). A second breakthrough in the field was the identification of in vitro culture conditions that permit indefinite expansion of SSCs on feeder cells with growth factors (Kanatsu-Shinohara et al. 2004; Kubota et al. 2004). These two technologies have spurred many functional studies of SSCs and have advanced translational applications of SSCs in animal transgenesis (Ciccarelli et al. 2020).
Adult tissues maintain homeostasis through a balance between adult stem cell renewal and differentiation. Although much is known about spermatogenesis, the regulation of SSC self-renewal remains poorly understood. Stem cell renewal requires both stem cell-intrinsic factors and external niche factors. To date, a number of SSC factors have been identified: PLZF, NANOS2, RB, GDNF, ETV5, BCL6B, and CARF. Their functions in SSC self-renewal have been identified through genetic studies. PLZF (also known as ZBTB16), a transcription factor, was the first identified germ cell-intrinsic SSC self-renewal factor (Buaas et al. 2004; Costoya et al. 2004). Historically, a spontaneous mutation in Plzf called luxoid was first reported in 1955 and exhibited pleiotropic defects in fertility, limb development, and skeleton development (Green 1955). Subsequent studies revealed that PLZF is expressed in spermatogonial progenitor cells and that Plzf mutant males exhibit a progressive loss of germ cells with age, demonstrating its role in SSC self-renewal. Mechanistically, PLZF binds to the promoters of thousands of genes and promotes SSC self-renewal through opposing the activity of mTORC1, a critical mediator of cell growth (Hobbs et al. 2010; Lovelace et al. 2016). NANOS2, an RNA-binding protein, and RB, the retinoblastoma tumor suppressor protein, are expressed in spermatogonia. Conditional deletion studies have shown that NANOS2 and RB are required for self-renewal of spermatogonial stem cells (Sada et al. 2009; Hu et al. 2013).
GDNF, ETV5, and CARF are expressed in Sertoli cells, the supporting somatic cells in the testis, and thus constitute SSC niche factors (Meng et al. 2000; Chen et al. 2005; Cui et al. 2020). GDNF, a cytokine, was identified as a key SSC niche factor, since mice heterozygous for the Gdnf deletion allele showed a depletion of spermatogonia, and mice overexpressing Gdnf had increased accumulation of undifferentiated spermatogonia in testis (Meng et al. 2000). GDNF stimulates SSC self-renewal through the cell surface coreceptors RET and GFRα1. Notably, the identification of GDNF as a Sertoli cell-secreted niche factor has enabled the in vitro culture of SSCs. When combined with bFGF, GDNF preserves stem cell activity and allows SSC expansion in vitro (Kanatsu-Shinohara et al. 2004; Kubota et al. 2004; Seandel et al. 2007). Bcl6b is a GDNF-responsive gene that functions in SSC renewal (Oatley et al. 2006). Mice deficient for the transcription factor ETV5 (previously known as ERM), which is expressed in Sertoli cells, exhibit an age-dependent depletion of undifferentiated spermatogonia (Chen et al. 2005). Interestingly, ETV5 is also expressed in SSCs in response to GDNF (Morrow et al. 2007; Oatley et al. 2007; Schmidt et al. 2009). Finally, loss of CARF (also known as CDKN2AIP) in Sertoli cells or spermatogonia leads to defects in SSC proliferation, which can be rescued by reactivation of the Wnt signaling pathway (Cui et al. 2020). Despite the identification of these key SSC self-renewal factors, the mechanistic basis of SSC self-renewal remains unclear. Moreover, PLZF does not appear to be regulated by GDNF, suggesting that additional unidentified factors remain and that their discovery will be critical for unraveling the mechanisms of SSC self-renewal.
DOT1L (disruptor of telomere silencing 1-like) is an evolutionarily conserved H3K79 methyltransferase that catalyzes H3K79 monomethylation, dimethylation, and trimethylation using SAM (S-adenosylmethionine) as the methyl donor (van Leeuwen et al. 2002). In general, H3K79 methylations are associated with active transcription (Vlaming and van Leeuwen 2016; Wood et al. 2018). Dot1, the yeast ortholog of DOT1L, is essential for telomere silencing, meiotic checkpoint control, and the DNA damage response (San-Segundo and Roeder 2000). Mouse DOT1L is essential for embryonic development, since Dot1l-null embryos die by E11.5 (Jones et al. 2008; Feng et al. 2010; Liao and Szabó 2020). Lineage-specific deletion studies show that DOT1L regulates neurogenesis and hematopoiesis by promoting proliferation of cerebral and hematopoietic progenitor cells (Jo et al. 2011; Franz et al. 2019). In MLL-rearranged leukemias, MLL1 translocation partners such as AF10 recruit DOT1L to cause aberrant H3K79 methylation and leukemogenesis (Okada et al. 2005; Deshpande et al. 2014). EPZ5676 is a potent DOT1L inhibitor and shows promising results in clinical trials for leukemia treatment (Sarno et al. 2020). Therefore, most mammalian DOT1L studies have focused on its role in leukemia. Due to the early embryonic lethality of the global Dot1l deletion, its role in male germ cell development has not been examined. However, maternal DOT1L is dispensable for embryonic development in mice (Liao and Szabó 2020). Here we unexpectedly discovered that DOT1L is a regulator of mouse SSC self-renewal. Mechanistically, DOT1L activates the transcription of the HoxC homeobox gene cluster in SSCs.
Results
DOT1L-dependent self-renewal of spermatogonial stem cells
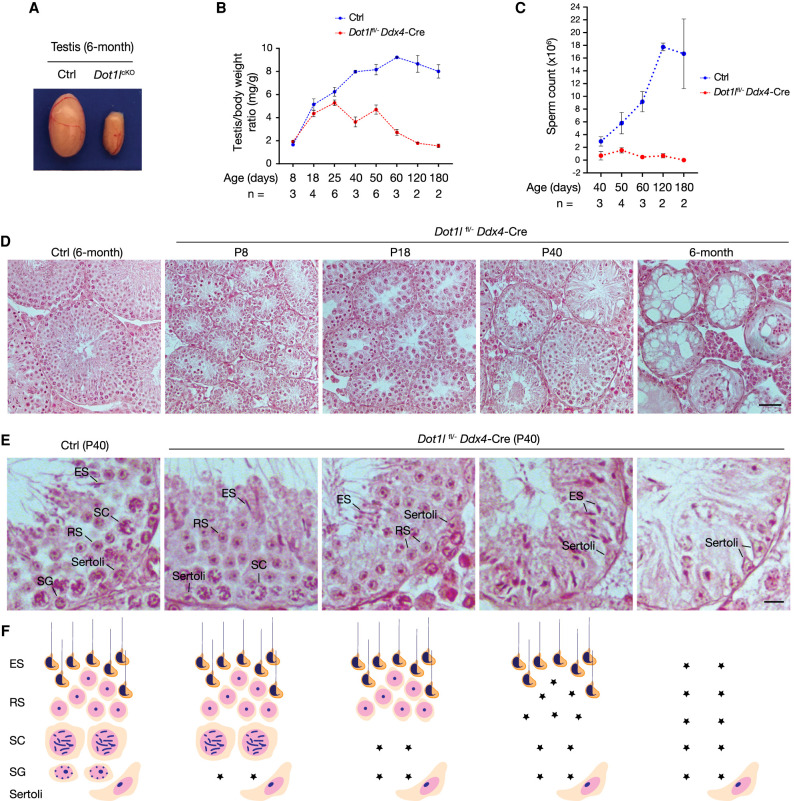
DOT1L is broadly expressed and its global loss leads to embryonic lethality (Jones et al. 2008; Feng et al. 2010). To investigate its role in mammalian spermatogenesis, we generated Dot1lfl/− Ddx4-Cre mice (referred to as Dot1lcKO) (Bernt et al. 2011). Ddx4-Cre is specifically expressed in germ cells beginning at embryonic day 15 and continuing after birth (Gallardo et al. 2007). The testis from 6-mo-old Dot1lcKO mice was dramatically smaller (Fig. 1A). However, the testis/body weight ratio of Dot1lcKO males was comparable with that of control males at postnatal day 8 (P8), P18, and P25, but sharply decreased at day 40 and beyond (Fig. 1B). Dot1lcKO males failed to sire offspring, as the sperm count of adult Dot1lcKO males was extremely low or zero (Fig. 1C). Together, these results show that DOT1L is essential for germ cell development in adult males.
Figure 1.
DOT1L is essential for SSC self-renewal in mice. (A) Testes from control (Dot1lfl/+ Ddx4-Cre) and Dot1lcKO male mice. (B,C) Testis/body weight ratio (B) and sperm count (C) of Dot1lcKO and control males at different ages. Control males: Dot1lfl/+, Dot1lfl/−, or Dot1lfl/+ Ddx4-Cre. (D) Histology of mouse testes at P8, P18, P40, and 6 mo. Scale bar, 50 µm. (Ctrl) Dot1lfl/+ Ddx4-Cre. (E,F) Representative seminiferous tubules showing sequential and progressive loss of germ cells from spermatogonia to elongated spermatids in Dot1lcKO testes at P40. (Ctrl) Dot1lfl/+ Ddx4-Cre. The diagram illustrates the presence and absence of different types of germ cells in the tubules above. Scale bar, 10 µm. (Asterisk) Loss of germ cells, (SG) spermatogonia, (SC) spermatocyte, (RS) round spermatid, (ES) elongated spermatid.
To investigate the cause of sperm production failure, we examined the first wave of spermatogenesis in juvenile testes. Embryonic gonocytes become spermatogonia after birth, a subset of which were thought to become spermatogonial stem cells (SSCs) (de Rooij 1998; Nakagawa et al. 2007; Law et al. 2019). The remaining spermatogonia proliferate, differentiate, enter meiosis, and initiate production of the first wave of sperm by day 35 in mice. Therefore, the first (juvenile) wave of spermatogenesis is SSC-independent. Notably, the testis weight was comparable between Dot1lcKO and control pups up to P25 (Fig. 1B). Histological analysis of Dot1lcKO testes showed the presence of spermatogonia (mitotic) at P8, spermatocytes (meiotic) at P18, and elongated spermatids (haploid) at P40 (Fig. 1D), at a time line expected in wild type. H3K79me2 was present in both germ cells and somatic cells in the control (Dot1lfl/+ Ddx4-Cre) testis (Supplemental Fig. S1). As expected, H3K79me2 was lost in germ cells but still present in somatic cells, including Sertoli cells, in all tubules from the Dot1lcKO testis (Supplemental Fig. S1). Together, these results demonstrate that the first wave of spermatogenesis occurs normally in Dot1lcKO males and that DOT1L is not required for survival of spermatogonia or for progression of meiosis.
We next closely examined 40-d-old testes, which contain adult waves of spermatogenesis (Fig. 1E). Adult waves of spermatogenesis are SSC-dependent: SSCs divide to preserve the stem cell population (self-renewal) and also produce spermatogonia that differentiate to initiate overlapping waves of spermatogenesis. The control seminiferous tubule consists of a complete cohort of spermatogonia, spermatocytes, round spermatids, and/or elongated spermatids (Fig. 1E, left panel). However, Dot1lcKO testes showed a variety of abnormal tubules with a loss of one or more types of germ cells. Dot1lcKO tubules showed a sequential loss of spermatogonia, spermatocytes, round spermatids, and finally elongated spermatids (Fig. 1E,F). The progressive germ cell loss phenotype beginning with spermatogonia is characteristic of a failure in SSC self-renewal, as observed in Gdnf, ETV5, Zbtb16, Rb, and Nanos2 mutant testes (Meng et al. 2000; Buaas et al. 2004; Costoya et al. 2004; Chen et al. 2005; Sada et al. 2009; Hu et al. 2013). As expected, the Dot1lcKO testis weight decreased progressively at day 50 and beyond (Fig. 1B). At 6 mo of age, all germ cells were lost in Dot1lcKO testes, resulting in a Sertoli cell only syndrome (Fig. 1D). These data strongly suggest that DOT1L is essential for SSC self-renewal. Furthermore, the SSC self-renewal defect in Dot1lcKO testes was much more severe than that in Zbtb16−/− testes, which still contain a significant fraction of tubules with germ cells even at 8 mo (Buaas et al. 2004). Absence of SALL4+ (a marker of spermatogonia) cells in P40 Dot1lcKO testes revealed a loss of spermatogonia in all tubules (Supplemental Fig. S2A,B; Hobbs et al. 2012; Gassei and Orwig 2013). Moreover, the number of SALL4+ spermatogonia was comparable between control and Dot1lcKO testes at P8, but significantly lower in Dot1lcKO testes at P18 and P25, providing further support for a defect in the SSC renewal (Supplemental Fig. S2C,D).
Requirement of DOT1L in SSC self-renewal in adulthood
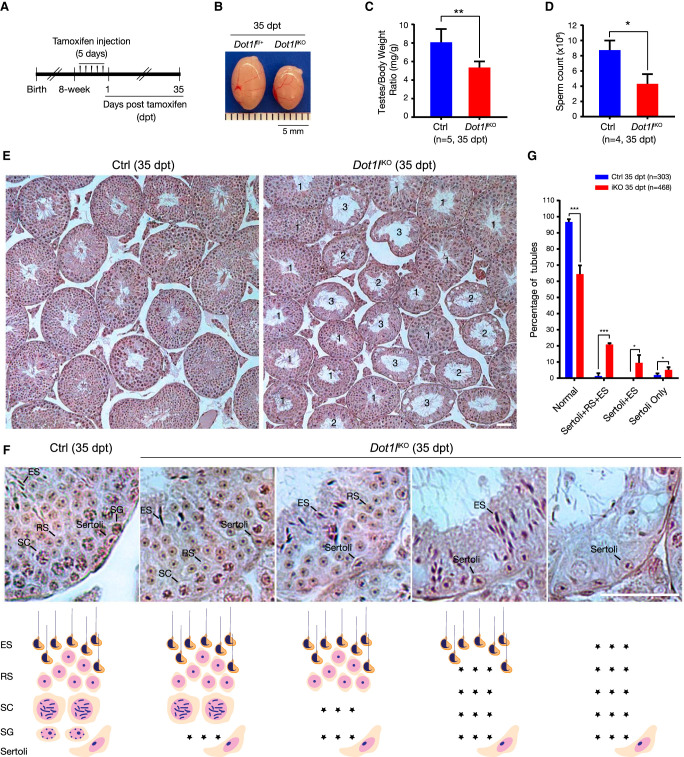
We next investigated the requirement of DOT1L in adult SSC self-renewal through tamoxifen-induced inactivation of Dot1l in 8-wk-old Dot1lfl/fl Ddx4-CreERT2 mice (referred to as Dot1liKO) (Fig. 2A). At 35 d posttamoxifen treatment (35 dpt), Dot1liKO males displayed a reduction in testis size and sperm count (Fig. 2B–D). Similar to P40 Dot1lcKO testes, adult Dot1liKO testes at 35 dpt exhibited a sequential loss of germ cells, starting from early germ cells (spermatogonia) to more differentiated germ cells (elongated spermatids) (Fig. 2E–G). The number of SALL4+ spermatogonia was sharply reduced in Dot1liKO testes at 35 dpt (Supplemental Fig. S3A,B,D). We looked earlier to determine how quickly the phenotype was observable. While Dot1liKO testes at 9 dpt showed no obvious histological defects, they already displayed a decrease in the number of SALL4+ spermatogonia (Supplemental Fig. S3B,C). The proliferation rate of SALL4+ spermatogonia was lower in 35-dpt Dot1liKO testes but was not statistically significant (Supplemental Fig. S3E,F). The number of TUNEL-positive germ cells was increased in 35-dpt Dot1liKO testes (Supplemental Fig. S3G,H). These results further support a requirement for DOT1L in SSC self-renewal in adult mice.
Figure 2.
Inducible deletion of Dot1l causes a failure in SSC self-renewal in adult mice. (A) Scheme of tamoxifen treatment in adult male mice. (B) Image of adult testes at 35 dpt. (Dot1liKO) Dot1lfl/fl Ddx4-CreERT2. (C) Testis/body weight ratio of control and Dot1liKO males. (**) P < 0.01, Student's t-test. (D) Epididymal sperm count of control and Dot1liKO males. (*) P < 0.05, Student's t-test. Control mice: Dot1lfl/+, Dot1lfl/fl, or Dot1lfl/+ Ddx4-CreERT2. (E) Histology of control and Dot1liKO testes at 35 dpt. Scale bar, 50 µm. Types of tubules in Dot1liKO testes: type 1, apparently normal; type 2, presence of Sertoli cells and round and elongated spermatids; and type 3, presence of Sertoli cells and elongated spermatids. (F) Progressive depletion of germ cells begins with spermatogonia in Dot1liKO testes at 35 dpt. The cartoon below depicts the germ cell composition of each tubule shown above. Scale bar, 50 µm. (Asterisk) Loss of germ cells, (SG) spermatogonia, (SC) spermatocyte, (RS) round spermatid, (ES) elongated spermatid. (G) Percentage of each type of seminiferous tubules in control and Dot1liKO testes at 35 dpt. Statistics: (*) P < 0.05, (***) P < 0.001, Student's t-test or Mann–Whitney U-test.
We examined the distribution of DOT1L and H3K79me2 in control and Dot1liKO testes at 35 dpt by immunofluorescence. In the control testis, the DOT1L signal was high in round spermatids but was present at low levels in other germ cells and somatic cells. In the Dot1liKO testis, DOT1L was depleted in germ cells in ∼75% of tubules at 35 dpt (Supplemental Fig. S4A). As expected, H3K79me2 was absent in germ cells but still present in somatic cells, including Sertoli cells, in 50% of the tubules from Dot1liKO testis (Supplemental Fig. S4B). The reduction of DOT1L and H3K79me2 in the Dot1liKO testis was confirmed by Western blotting analyses (Supplemental Fig. S4C–E).
DOT1L inhibition reduces SSC stemness after transplantation
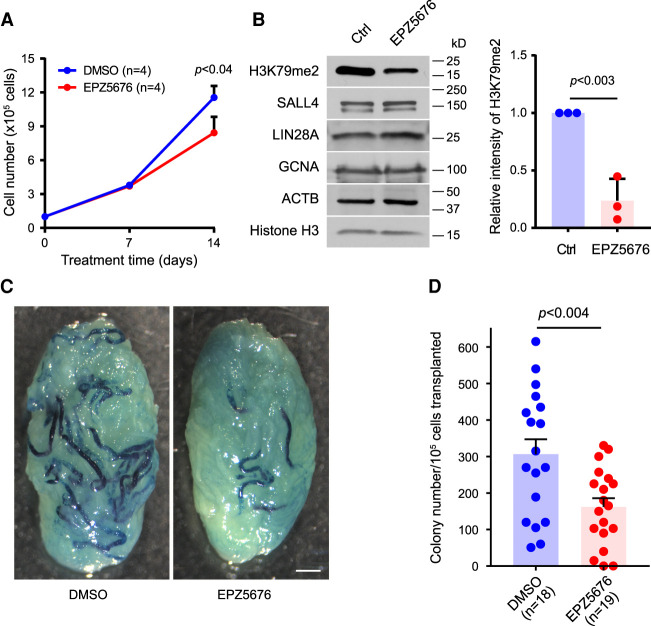
To further examine the role of DOT1L in SSC self-renewal, we treated SSCs with EPZ5676, a potent inhibitor of the DOT1L methyltransferase (Sarno et al. 2020), followed by SSC transplantation into testis. Mouse SSCs were derived from ROSA mice that express a LacZ reporter gene. SSCs, which form clumps in culture, were treated with EPZ5676 or DMSO as a vehicle control. The number of spermatogonia was comparable between the two groups at day 7 of culture, but at day 14 the number of spermatogonia in EPZ5676-treated cultures was 27% less than in DMSO cultures (Fig. 3A), suggesting that EPZ5676 reduces SSC proliferation and/or self-renewal. As expected, EPZ5676 treatment of SSCs reduced the abundance of H3K79me2 (Fig. 3B). After culturing with EPZ5676 or DMSO for 2 wk, clump-forming cells were transplanted into infertile mice to determine the stem cell activity (Brinster and Zimmermann 1994; Kubota and Brinster 2008). Two months after transplantation, the testes were stained with X-gal to quantify the number of LacZ-expressing SSCs in donor cell suspensions (Fig. 3C). EPZ5676 treatment of SSCs reduced the stem cell activity by 50% (Fig. 3D). These transplantation results further support a critical role for DOT1L in SSC self-renewal.
Figure 3.
Inhibition of DOT1L in mouse SSCs reduces stem cell activity. (A) Total number of spermatogonia in culture. (n) Number of experiments. Statistics: paired Student's t-test. (B) Down-regulation of H3K79me2 in mouse SSCs cultured with or without 2 µM EPZ5676 for 2 wk. The graph shows the relative intensity of H3K79me2 normalized to histone H3 from three experiments. (C) β-Gal staining of mouse testes at 2 mo after SSC transplantation. (D) Number of colonies derived from EPZ5676- or DMSO-treated SSCs after transplantation. (n) Number of recipient testes from four independent experiments. Statistics: unpaired Student's t-test.
DOT1L promotes the expression of the HoxC and Dlx3/4 transcription factors in SSCs
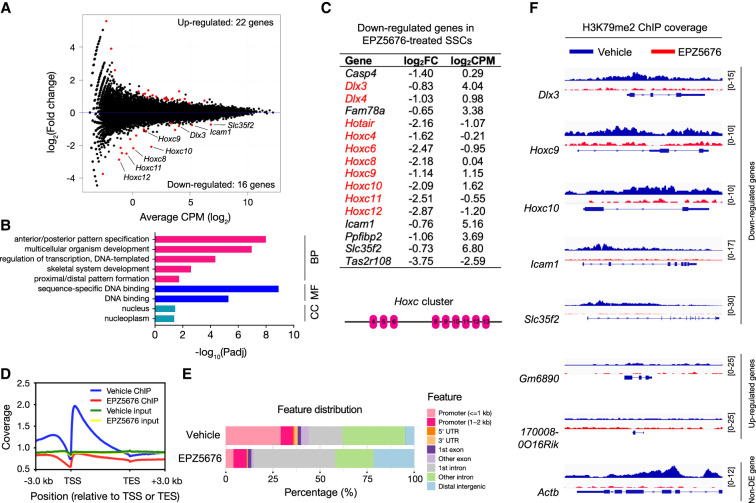
DOT1L catalyzes H3K79 methylation, and inactivation of DOT1L leads to a global loss of H3K79 methylation (Jones et al. 2008). H3K79 methylation marks are associated with active transcription (Vlaming and van Leeuwen 2016; Wood et al. 2018), suggesting that DOT1L may control SSC self-renewal through modulating transcription. To identify DOT1L-dependent genes in SSCs, we established SSC lines from DBA2 mice (Supplemental Fig. S5A; Kubota and Brinster 2008; Kanatsu-Shinohara et al. 2014). As expected, the SSCs expressed SALL4 and LIN28A, markers of undifferentiated spermatogonia (Supplemental Fig. S5B; West et al. 2009; Zheng et al. 2009; Hobbs et al. 2012). RNA-seq analysis was performed on SSCs collected at day 14 of inhibitor or mock treatment. With the FDR (false discovery rate) cutoff of 5%, we identified 38 differentially expressed (DE) genes in EPZ5676-treated SSCs: 22 up-regulated and 16 down-regulated (Fig. 4A; Supplemental Table S1). Gene ontology (GO) analysis of up-regulated DE genes did not reveal enrichment in any biological process or molecular function (Supplemental Fig. S5C). Strikingly, GO analysis of down-regulated DE genes revealed enrichment in “anterior/posterior pattern specification” and “sequence-specific DNA binding” (Fig. 4B), because nine out of 16 down-regulated DE genes encoded homeobox (Hox) transcription factors (Fig. 4C). Seven down-regulated genes belonged to the Hoxc gene cluster on chromosome 15: Hoxc4, Hoxc6, Hoxc8, Hoxc9, Hoxc10, Hoxc11, and Hoxc12, and two were Dlx genes (distal-less homeobox): Dlx3 and Dlx4 (Fig. 4C), which are adjacent genes on chromosome 11. Notably, Hotair, a lncRNA gene located between Hoxc11 and Hoxc12, was also down-regulated in EPZ5676-treated SSCs. The Hox family of transcription factors specifies segmental identity along the body axis in animals ranging from flies to humans (Mallo and Alonso 2013). As such, mutations in a particular Hox gene lead to homeotic transformation—fate switch of specific body segments. In mice, there are four Hox gene clusters located on four separate chromosomes: Hoxa, Hoxb, Hoxc, and Hoxd (Supplemental Fig. S5D). Notably, none of the members in the other three clusters (Hoxa, Hoxb, and Hoxd) were significantly down-regulated in EPZ5676-treated SSCs, indicating that DOT1L is specifically required for expression of the Hoxc cluster in SSCs (Supplemental Fig. S5D). Strikingly, Hoxc4 was identified as a GDNF-responsive gene in rat SSCs, and RNAi knockdown of Hoxc4 in vitro reduced the stem cell activity of rat SSCs in culture, as shown by transplantation (Schmidt et al. 2009). Differential expression of the Hoxc genes in EPZ5676-treated SSCs was confirmed by qRT-PCR (Supplemental Fig. S5E).
Figure 4.
Inhibition of DOT1L down-regulates Hoxc genes in SSCs. (A) MA plot of expression changes. Mouse SSCs were cultured with or without 2 µM EPZ5676 for 14 d. The DE genes are shown as red dots. (B) List of down-regulated DE genes in EPZ5676-treated SSCs. The diagram shows the organization of the Hoxc gene cluster on chromosome 15 (Mallo and Alonso 2013). (C) GO analysis of down-regulated DE genes in EPZ5676-treated SSCs. (BP) Biological process, (MF) molecular function, (CC) cellular component. (D) Position and enrichment of H3K79me2 ChIP-seq signals from vehicle (no inhibitor) and EPZ5676-treated SSCs relative to the TSS (transcription start site) and TES (transcription end site). Inputs are shown for comparison. Genes were binned to the same overall relative length. (E) Feature distribution of H3K79me2 ChIP-seq peaks in the mouse genome. (F) H3K79me2 profiles of selected DE genes and non-DE genes in vehicle- and EPZ5676-treated SSCs. Note that Actb is one of many non-DE genes with abundant H3K79me2. Numbers at the right indicate the signal range.
We derived Dot1l+/− and Dot1l−/− SSC lines (Supplemental Fig. S6A). Because Dot1l is essential for SSC self-renewal, the ability of Dot1l−/− spermatogonia to proliferate in culture seemed to be counterintuitive. As a precedent, Plzf−/− SSCs were successfully derived and maintained in long-term cultures (Hobbs et al. 2010). Western blot analyses confirmed the depletion of the DOT1L protein and H3K79me2 in Dot1l−/− SSCs (Supplemental Fig. S6B–D). By qRT-PCR analysis, we found that five Hoxc genes (Hoxc8, Hoxc9, Hoxc10, Hoxc11, and Hoxc12) were significantly down-regulated in Dot1l−/− SSC lines (Supplemental Fig. S6E), which was consistent with the RNA-seq results from EPZ5676-treated SSCs. Therefore, our RNA-seq analysis shows that DOT1L is required for transcription of the Hoxc genes in SSCs and thus implicates the Hoxc transcription factors in the regulation of SSC self-renewal.
Genome-wide distribution of H3K79me2 in mouse SSCs
We determined the distribution of H3K79me2 in SSCs treated with or without EPZ5676 by chromatin immunoprecipitation and next-generation sequencing (ChIP-seq). Consistent with prior reports (Vlaming and van Leeuwen 2016; Wood et al. 2018), we found that H3K79me2 accumulated significantly in the promoter regions or gene bodies with the highest levels immediately downstream from the transcription start sites (TSSs) (Fig. 4D; Supplemental Fig. S7A). Consistently, more than half of the H3K79me2 ChIP-seq reads mapped to the promoters, the first exons, and the first introns (Fig. 4E). As expected, the H3K79me2 signal was reduced substantially in EPZ5676-treated SSCs (Fig. 4D; Supplemental Fig. S7B). Six of 16 down-regulated genes such as Dlx3, Hoxc9, Hoxc10, Icam1, and Slc35f2 were occupied by H3K79me2, showing that they are DOT1L direct target genes (Fig. 4F; Supplemental Table S2). In contrast, all 22 up-regulated genes were devoid of H3K79me2 (Fig. 4F; Supplemental Table S2). While thousands of genes were associated with H3K79me2 in SSCs, the DOT1L inhibition only led to a small number of differentially expressed genes in SSCs. This discrepancy was also observed in other cells such as hematopoietic cells and neuronal progenitor cells (Bernt et al. 2011; Franz et al. 2019) and is in fact a hallmark of most epigenetic marks (Lenstra et al. 2011).
Discussion
DOT1L is the sole methyltransferase for H3K79 and is essential for development and thus viability (Jones et al. 2008). Our genetic and transplantation results demonstrate that DOT1L is essential for SSC self-renewal. Several stem cell-intrinsic factors, including RB, NANOS2, PLZF, ID4, and BCL6B, regulate SSC self-renewal. While depletion of RB or NANOS2 leads to a rapid loss of spermatogonial stem cells (Sada et al. 2009; Hu et al. 2013), mice lacking PLZF, ID4, or BCL6B exhibit milder defects in SSC self-renewal (Buaas et al. 2004; Costoya et al. 2004; Oatley et al. 2006, 2011). The defining characteristics in SSC self-renewal mouse mutants are a normal first wave of spermatogenesis and a sequential loss of germ cells beginning with spermatogonia in later waves of spermatogenesis. Notably, DMRT1 and PRAMEF12 are essential for the maintenance of spermatogonia (Takashima et al. 2013; Zhang et al. 2016; Wang et al. 2019). For factors essential for the survival of spermatogonia such as DMRT1 and PRAMEF12, it would be challenging to determine whether they also play a role in SSC self-renewal. Here we found that inactivation of DOT1L, like loss of RB, results in a dramatic defect in SSC self-renewal, showing that DOT1L is an essential germ cell-intrinsic regulator of SSC self-renewal. None of the known SSC self-renewal factors were down-regulated in EPZ5676-treated SSCs, suggesting that DOT1L may function in SSC self-renewal in parallel pathways.
DOT1L catalyzes H3K79 methylation, an epigenetic mark associated with actively transcribed genes. DOT1L interacts directly with the phosphorylated C-terminal domain of RNA polymerase II, which leads to its recruitment to transcribed genes (Kim et al. 2012). Our RNA-seq analysis revealed that seven HoxC cluster genes and two Dlx (homeobox) genes were down-regulated in EPZ5676-treated SSCs: HoxC4, HoxC6, HoxC8, HoxC9, HoxC10, HoxC11, HoxC12, Dlx3, and Dlx4. However, no genes from the other three Hox clusters (HoxA, HoxB, and HoxD) were down-regulated, suggesting that HoxC genes are specifically targeted in SSCs by DOT1L. In addition, Hoxc9, Hoxc10, Dlx3, and Dlx4 are marked by H3K79me2 (Fig. 4F; Supplemental Table S2). Six down-regulated Hox genes have been disrupted in mice (Supplemental Table S3). These mutants show homeotic transformation phenotypes—skeletal defects, as expected for Hox genes. All of the viable knockout mice were reported to be fertile, but fertility was not rigorously assessed, as the focus of these studies was on the role of Hox genes in skeletal development. Therefore, SSC self-renewal defects may have been missed. However, HoxC4 was identified as a GDNF-regulated gene in rat SSCs and was shown to be important for rat SSC self-renewal by siRNA knockdown and transplantation (Schmidt et al. 2009). In addition to the traditional homeotic skeletal function, Hox genes have important nontraditional roles. For example, HoxA9–13 are required for DOT1L-mediated malignant transformation of myeloid progenitor cells (Okada et al. 2005; Bernt et al. 2011; Deshpande et al. 2014). Therefore, future genetic studies are warranted to examine the role of the HoxC gene cluster and Dlx3/4 genes individually or collectively in SSC self-renewal.
Somatic cells can be reprogrammed into induced pluripotent stem (iPS) cells by defined factors (Takahashi and Yamanaka 2006). DOT1L functions as a barrier to somatic reprogramming into iPS cells. Specifically, inhibition of DOT1L enhances generation of iPS cells from somatic cells (Onder et al. 2012). This is also consistent with the dispensable role of DOT1L in ES cell self-renewal (Cao et al. 2020). In contrast, here we found that DOT1L is essential for self-renewal in SSCs, raising the possibility that DOT1L could also play a critical role in self-renewal of other adult stem cells. ES cells and iPS cells can be induced into primordial germ cell-like cells (PGCLCs) (Hayashi et al. 2011). To date, direct reprogramming of somatic cells into SSCs has not been achieved. Given its essential role in SSC self-renewal, DOT1L is expected to be required for reprogramming of somatic cells into SSCs in mammals, including humans.
Materials and methods
Generation of Dot1l conditional knockout mice
Dot1l floxed (Dot1lfl) mice were generated previously (Bernt et al. 2011). To inactivate Dot1l specifically in germ cells, Dot1l floxed mice were crossed with two Cre mouse strains: Ddx4-Cre (Jackson Laboratory 006954) and Ddx4-CreERT2 (Jackson Laboratory 024760) (Gallardo et al. 2007; John et al. 2008). For inducible deletion, tamoxifen (Sigma T5648) dissolved in corn oil (Sigma C8267) at a concentration of 20 mg/mL was intraperitoneally injected into 8-wk-old Dot1lfl/fl Ddx4-CreERT2 males (2 mg/30 g body weight) once per day for five consecutive days. Dot1lfl/+ Ddx4-CreERT2, Dot1lfl/+, or Dot1lfl/fl males injected with tamoxifen were used as controls. A toxic effect of EGFP-Cre in cultured SSCs was previously reported (Kanatsu-Shinohara et al. 2008). However, adult Dot1lfl/+ Ddx4-CreERT2 males treated with tamoxifen as described in this study showed no defects in SSC self-renewal or spermatogenesis. Testes were harvested for analysis at 9 or 35 d posttamoxifen treatment (dpt). PCR genotyping primers were as follows: the Dot1l floxed allele (418 bp): p2 (5′-CCCAAAAGGGTCTTTTCACA-3′) and p3 (5′-ATGGGATTTCATGGAAGCAA-3′); the Dot1l deletion allele (620 bp): p1 (5′-CTCACAGTCACATACTACCTCTGAC-3′) and p3 (5′-ATGGGATTTCATGGAAGCAA-3′); Ddx4-Cre (240 bp): VasaCre-1 (5′-CACGTGCAGCCGTTTAAGCCGCGT-3′) and VasaCre-2 (5′-TTCCCATTCTAAACAACACCCTGAA-3′); and Ddx4-CreERT2 (205 bp): KI-1 (5′-ATACCGGAGATCATGCAAGC-3′) and KI-2 (5′-GGCCAGGCTGTTCTTCTTA-3′). The University Laboratory Animal Resources (ULAR) at University of Pennsylvania provided veterinary care and husbandry for all of the mice, and the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania approved all procedures.
SSC culture and transplantation
Mouse SSC cultures were established as described previously (Kubota et al. 2004) with some modifications (Kubota and Kakiuchi 2020). In brief, testicular cells were prepared from 7-d-old B6.129S7-Gt(ROSA)26Sor/J mice (Jackson Laboratory) that expressed a LacZ (β-galactosidase) reporter gene in all tissues by enzymatic digestion using trypsin-EDTA (Thermo Fisher Scientific). Germ cells were cultured on SNL 76/7 feeders (from Allan Bradley, The Welcome Trust Sanger Institute) in a serum-free medium with 10 ng/mL GDNF (R&D Systems) and 0.5 ng/mL FGF2 (Corning) (Kubota and Kakiuchi 2020). After 2 mo in culture, 1 × 105 clump-forming cells were seeded on SNL feeder cells in 24-well plates with 2 µM EPZ5676 (Cayman Chemical) or DMSO (Sigma-Aldrich) as a vehicle control. Medium was changed every 2–3 d, and the cells were subcultured every 7 d. After culturing with EPZ5676 or DMSO for 2 wk, clump-forming cells were harvested using trypsin-EDTA and transplanted into recipient testes. For gene expression analysis, clumps were collected by gentle pipetting, and total RNA was extracted with TRIzol (Thermo Fisher Scientific) followed by RNeasy column purification (Qiagen). All cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Cultured SSC cells were transplanted into recipient C57BL/6 mouse testes via efferent duct injection as described (Kubota and Kakiuchi 2020). Recipient mice were treated with 44 mg/kg busulfan (Sigma-Aldrich) at least 6 wk prior to transplantation. Eight microliters of 8.3 × 105 cells/mL was transplanted into each testis. Two months after transplantation, the testes were stained with X-gal to quantify the number of SSCs in donor cell suspensions. Spermatogenic colonies consisting of X-gal-stained blue cells were counted and normalized to 1 × 105 cells transplanted into the recipient testes.
SSC culture for RNA-seq
Gelatin selection was used for enrichment of spermatogonia and removal of testicular somatic cells as described previously (Kanatsu-Shinohara et al. 2014). All cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2. For Western blotting and mRNA sequencing, SSCs were cultured on γ-irradiated MEFs prepared from E13.5 DR4 mouse embryos. Culture medium was based on IMDM (Sigma I3390) supplemented with 1% 10 μL/mL FBS (Hyclone SH30396.03), 5 mg/mL bovine albumin (MP Biomedicals 810661), 15 ng/mL rat GDNF (Peprotech 450-51), 10 ng/mL human FGF2 (Peprotech 100-18B), and other nutrients (Kanatsu-Shinohara et al. 2014). In brief (Supplemental Fig. S5A), one pair of testes from postnatal day 5–8 DBA/2J pups (Jackson Laboratory 000671) was harvested and dissociated with 1 mg/mL collagenase type IV (Sigma C5138) for 10 min, followed by digestion with 0.25% trypsin-EDTA (Gibco 25200-056) for 10 min. Single-cell suspension was plated on one 60-mm cell culture dish precoated with 0.1% gelatin and incubated overnight. Nonattached spermatogonia in the media were then transferred to MEF-coated plates. Medium was changed every 2–3 d. After 7–14 d of culture, SSCs formed clumps (Supplemental Fig. S5B) and were subcultured at a ratio of 1:3 to 1:6 every 5–7 d. To collect cells for RNA-seq and Western blotting, cell suspensions after digestion were plated on the 0.1% gelatin-coated cell culture dishes for 2 h to remove the adherent feeder cells.
DOT1L inhibitor treatment
EPZ5676 (Cayman Chemical 1380288-87-8) was dissolved in DMSO or 100% ethanol. During treatment, EPZ5676 solution was added to IMDM/FBS medium to achieve a final concentration of 2 μM in the experimental group. The same amount of solvent was added to the medium in the control group. Medium was changed every 2–3 d, and the cells were subcultured every 7 d.
RNA-seq analysis of SSCs
SSCs from two pups were established in culture and treated with EPZ5676 or solvent (triplicates per treatment) for 14 d as described above. Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific 15596026). Four micrograms of total RNA from each sample was used. Poly(A)+ mRNA was isolated with oligo(dT) magnetic beads and converted into cDNA to generate RNA-seq libraries using TruSeq stranded mRNA library preparation kit set A (Illumina RS-122-2101) by following the manufacturer's instructions. Six individual libraries (three EPZ5676-treated and three control samples) were pooled in equal amounts. Libraries were first test-sequenced on an Illumina MiSeq system (paired-end sequencing, 150 cycles, ∼1 million reads per library) and then on an Illumina NovaSeq 6000 system (single-end sequencing, 100 cycles, ∼25 million reads per sample) at the University of Pennsylvania Next-Generation Sequencing Core.
We mapped sequenced reads to the mouse (mm10) reference genome in the unique mapping mode. Uniquely mapped reads were used to evaluate expression of protein-coding genes. STAR was used for alignment with the default parameters. The annotation of genes was acquired from the UCSC RefSeq annotation. FeatureCount was used to determine the number of reads mapping to genes. Differential gene expression was called by using EdgeR with the FDR cutoff of 5%. The bulk RNA-seq data are available in GEO under accession number GSE181590.
qRT-PCR validation
The expression of nine down-regulated, two up-regulated, and nine nondifferentially expressed genes determined by RNA-seq was analyzed by quantitative real-time reverse transcription PCR (qRT-PCR). The same RNA samples for RNA sequencing and different RNA samples extracted from another four samples (two EPZ5676-treated and two ethanol-treated) were used. qRT-PCR primers are listed in Supplemental Table S4. Each reaction (in a final volume of 10 μL) consisted of 5 μL of 2× SYBR Green Master mix, 0.25 μL of each primer at 10 μM, 0.5 μL of cDNA, and 4.0 μL of H2O, except for Hoxc genes. For Hoxc genes, each reaction (in a final volume of 10 μL) consisted of 5 μL of 2× SYBR Green Master mix, 0.1 μL of each primer at 10 μM, 0.5 μL of cDNA, and 4.3 μL of H2O. Each sample was assayed in triplicates. The expression level was normalized to Gapdh using the 2−ΔΔCt method.
Chromatin immunoprecipitation and sequencing (ChIP-seq)
Mouse SSCs were cultured in the presence or absence of EPZ5676 for 7 d. SSCs were resuspended at 1 million/mL in PBS and cross-linked with formaldehyde (Insert) at a final concentration of 1% for 10 min at room temperature. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M for 5 min at room temperature. All of the following buffers were supplemented with protease inhibitors (Complete Mini, Roche, Halt, or Thermo Fisher Scientific). Cells were washed twice in ice-cold PBS and lysed in ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.1). Lysate of the equivalent of 1 million cells was transferred into microTUBE AFA fiber preslit snap cap tubes (Covaris PN 520045) and sonicated with a Covaris ME220 using the following settings: 130-sec duration, 75-W peak power, 20% duty factor, and 1000 cycles per burst. The resulting lysate was spun in a tabletop centrifuge at full speed for 10 min at 10°C. One percent of the supernatant was set aside as input control. The supernatant was diluted 1:10 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl at pH 8.1, 167 mM NaCl). H3K79me2 antibody (Abcam ab3594) was added at 2 µg per 1 million cells and incubated overnight at 4°C. Protein A+G-coated magnetic beads (Millipore 16663) were added for 3 h and then washed once with each of the following buffers: low-salt buffer (1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, 150 mM NaCl), high-salt buffer (1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% Igepal-CH 630 [NP-40], 1% deoxycholic acid sodium salt, 1 mM EDTA, 10 mM Tris-HCl at pH 8.1), and TE buffer (1 mM EDTA, 10 mM Tris-HCl at pH 8.1). Cross-linking was reversed in elution buffer (1% SDS, 0.1 M NaHCO3, 200 mM NaCl) overnight at 65°C. Beads were eluted by adding 20 µg/mL RNase A (Ambion AM2271) at 37°C and 100 µg/mL proteinase K (Invitrogen 43-337-93) for 1 h each at 55°C. DNA was purified using a PCR purification kit (Qiagen 28104). Eluted DNA was quantified using the Qubit assay (dsDNA HS assay kit, Invitrogen Q32851).
ChIP-seq libraries were prepared with the NEBNext Ultra II DNA library preparation kit for Illumina (NEB E7645S) by following the manufacturer's instructions. In brief, 1 ng of DNA from H3K79me2 ChIP was used. End repair, 5′ phosphorylation, and dA-tailing were performed in a thermocycler. The samples were incubated for 90 min at 20°C and subsequently for 30 min at 65°C. Adaptor (NEB E6440S) was diluted at 1:25 and ligated to the DNA sample. Adaptor-ligated DNA was cleaned up without size selection by using 0.8× sample volume of NEBNext sample purification beads. Purified adaptor-ligated DNA was labeled and enriched by PCR amplification with unique dual-index primer pairs (A11–H11; NEB E6440S). PCR reaction was cleaned up by using 0.9× sample volume of NEBNext sample purification beads. The libraries were analyzed on TapeStation, and the DNA concentration was measured using Qubit 4.0. Eight individual libraries (two vehicle ChIP, two EPZ5676-treated ChIP, two vehicle input, and two EPZ5676-treated input) were pooled in equal amounts. Libraries were sequenced with the NextSeq 1000/2000 P2 reagents v3 kit (Illumina 20046811) on an Illumina NextSeq 2000 system (single end, 100 cycles) at the University of Pennsylvania School of Veterinary Medicine Center for Host–Microbial Interactions Sequencing Core.
Fastq files of ChIP-seq were decoded by bcl2fastq2 (v2.20.0.422). The index and barcode in Fastq files were removed by Trimmomatic (v0.32). Rsubread (v 2.10.0) was used to map reads to the mouse reference genome (USCS GRCm38/mm10), and uniquely mapped reads were used for downstream analysis. After mapping, BAM files were sorted and indexed by SAMtools (v1.11). Duplicated reads were flagged by Picard (v1.96) and removed for downstream analysis. Peaks were called by MACS3 with FDR < 1%, P < 0.01, and broad parameters. Blacklist regions of ChIP-seq were downloaded from the ENCODE Project. Peaks were annotated by Chipseeker (v1.32.0) and ChIPpeakAnno (v3.30.0). ChIP-seq coverage was calculated with DeepTools (v3.4.3). Data were visualized with the IGV browser (v 2.12.3). The H3K79me2 ChIP-seq data are available in GEO under accession number GSE193575.
GO analysis
Gene ontology (GO) analysis of differentially expressed genes was performed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov).
Statistical analysis
Student's t-test or Mann–Whitney U-test were used for statistical analysis. Mean and standard deviation or standard error of mean were used to describe the average and distribution of the data.
Supplementary Material
Acknowledgments
We thank N. Wang and X. Zhang for help with SSC culture, B. Hermann for consultation on SSC studies, J. Schug for help with bulk RNA-seq, D.P. Beiting and C. Malekshahi for ChIP library sequencing, Prabu Reddi for anti-SP10 antibody, L.B. King for help with the manuscript, and Y. Guan and F. Yang for critical reading of the manuscript. Funding support was received from National Institute of Child Health and Human Development grant P50HD068157 (to M.S.B. and P.J.W.), National Natural Science Foundation of China grant 31771588 (to M.L.), a China Scholarship Council fellowship (to H.L.), and Japan Society for the Promotion of Science KAKENHI grant JP21K19194 (to H.K.).
Author contributions: P.J.W., M.L., and H.L. conceptualized the study. H.L. and K.C. carried out experiments and data analysis. H.K. and K.K. performed the SSC transplantation. K.C. and Y.L. performed bioinformatic analyses. S.S.R. and K.M.B. contributed to ChIP-seq. P.J.W., K.S., M.S.B., and M.L. analyzed data. P.J.W., H.L., H.K., K.C., and S.S.R. wrote the manuscript. All of the authors were involved in the discussion and commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349550.122.
Competing interest statement
The authors declare no competing interests.
References
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20: 66–78. 10.1016/j.ccr.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL. 2007. Male germline stem cells: from mice to men. Science 316: 404–405. 10.1126/science.1137741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Avarbock MR. 1994. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci 91: 11303–11307. 10.1073/pnas.91.24.11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. 1994. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci 91: 11298–11302. 10.1073/pnas.91.24.11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. 2004. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36: 647–652. 10.1038/ng1366 [DOI] [PubMed] [Google Scholar]
- Cao K, Ugarenko M, Ozark PA, Wang J, Marshall SA, Rendleman EJ, Liang K, Wang L, Zou L, Smith ER, et al. 2020. DOT1L-controlled cell-fate determination and transcription elongation are independent of H3K79 methylation. Proc Natl Acad Sci 117: 27365–27373. 10.1073/pnas.2001075117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, et al. 2005. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436: 1030–1034. 10.1038/nature03894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli M, Giassetti MI, Miao D, Oatley MJ, Robbins C, Lopez-Biladeau B, Waqas MS, Tibary A, Whitelaw B, Lillico S, et al. 2020. Donor-derived spermatogenesis following stem cell transplantation in sterile NANOS2 knockout males. Proc Natl Acad Sci 117: 24195–24204. 10.1073/pnas.2010102117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. 2004. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36: 653–659. 10.1038/ng1367 [DOI] [PubMed] [Google Scholar]
- Cui W, He X, Zhai X, Zhang H, Zhang Y, Jin F, Song X, Wu D, Shi Q, Li L. 2020. CARF promotes spermatogonial self-renewal and proliferation through Wnt signaling pathway. Cell Discov 6: 85. 10.1038/s41421-020-00212-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG. 1998. Stem cells in the testis. Int J Exp Pathol 79: 67–80. 10.1046/j.1365-2613.1998.00057.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AJ, Deshpande A, Sinha AU, Chen L, Chang J, Cihan A, Fazio M, Chen CW, Zhu N, Koche R, et al. 2014. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell 26: 896–908. 10.1016/j.ccell.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrinski I. 2008. Male germ cell transplantation. Reprod Domest Anim 43: 288–294. 10.1111/j.1439-0531.2008.01176.x [DOI] [PubMed] [Google Scholar]
- Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, Fields TA, Vivian JL, Fields PE. 2010. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 116: 4483–4491. 10.1182/blood-2010-03-276501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz H, Villarreal A, Heidrich S, Videm P, Kilpert F, Mestres I, Calegari F, Backofen R, Manke T, Vogel T. 2019. DOT1L promotes progenitor proliferation and primes neuronal layer identity in the developing cerebral cortex. Nucleic Acids Res 47: 168–183. 10.1093/nar/gky953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. 2007. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45: 413–417. 10.1002/dvg.20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K, Orwig KE. 2013. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One 8: e53976. 10.1371/journal.pone.0053976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MC. 1955. Luxoid, a new hereditary leg and foot abnormality in the house mouse. J Heredity 46: 91–99. 10.1093/oxfordjournals.jhered.a106545 [DOI] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. 2010. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 142: 468–479. 10.1016/j.cell.2010.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs RM, Fagoonee S, Papa A, Webster K, Altruda F, Nishinakamura R, Chai L, Pandolfi PP. 2012. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 10: 284–298. 10.1016/j.stem.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YC, de Rooij DG, Page DC. 2013. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci 110: 12685–12690. 10.1073/pnas.1311548110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. 2011. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 117: 4759–4768. 10.1182/blood-2010-12-327668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, Gallardo TD, Shirley LJ, Castrillon DH. 2008. Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev Biol 321: 197–204. 10.1016/j.ydbio.2008.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4: e1000190. 10.1371/journal.pgen.1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119: 1001–1012. 10.1016/j.cell.2004.11.011 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Takehashi M, Shinohara T. 2008. Brief history, pitfalls, and prospects of mammalian spermatogonial stem cell research. Cold Spring Harb Symp Quant Biol 73: 17–23. 10.1101/sqb.2008.73.033 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Matoba S, Morimoto H, Ogura A, Shinohara T. 2014. Improved serum- and feeder-free culture of mouse germline stem cells. Biol Reprod 91: 88. 10.1095/biolreprod.114.122317 [DOI] [PubMed] [Google Scholar]
- Kim SK, Jung I, Lee H, Kang K, Kim M, Jeong K, Kwon CS, Han YM, Kim YS, Kim D, et al. 2012. Human histone H3K79 methyltransferase DOT1L protein [corrected] binds actively transcribing RNA polymerase II to regulate gene expression. J Biol Chem 287: 39698–39709. 10.1074/jbc.M112.384057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Brinster RL. 2008. Culture of rodent spermatogonial stem cells, male germline stem cells of the postnatal animal. Methods Cell Biol 86: 59–84. 10.1016/S0091-679X(08)00004-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Kakiuchi K. 2020. Long-term ex vivo expansion of murine spermatogonial stem cells in a simple serum-free medium. Methods Mol Biol 2155: 165–182. 10.1007/978-1-0716-0655-1_14 [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. 2004. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci 101: 16489–16494. 10.1073/pnas.0407063101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law NC, Oatley JM. 2020. Developmental underpinnings of spermatogonial stem cell establishment. Andrology 8: 852–861. 10.1111/andr.12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law NC, Oatley MJ, Oatley JM. 2019. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat Commun 10: 2787. 10.1038/s41467-019-10596-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42: 536–549. 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Szabó PE. 2020. Maternal DOT1L is dispensable for mouse development. Sci Rep 10: 20636. 10.1038/s41598-020-77545-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan J, Hermann BP. 2016. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development 143: 1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M, Alonso CR. 2013. The regulation of Hox gene expression during animal development. Development 140: 3951–3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. 2000. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287: 1489–1493. 10.1126/science.287.5457.1489 [DOI] [PubMed] [Google Scholar]
- Morrow CM, Hostetler CE, Griswold MD, Hofmann MC, Murphy KM, Cooke PS, Hess RA. 2007. ETV5 is required for continuous spermatogenesis in adult mice and may mediate blood testes barrier function and testicular immune privilege. Ann N Y Acad Sci 1120: 144–151. 10.1196/annals.1411.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. 2007. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell 12: 195–206. 10.1016/j.devcel.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. 2006. Spermatogonial stem cells. Methods Enzymol 419: 259–282. 10.1016/S0076-6879(06)19011-4 [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. 2006. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci 103: 9524–9529. 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL. 2007. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 282: 25842–25851. 10.1074/jbc.M703474200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. 2011. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod 85: 347–356. 10.1095/biolreprod.111.091330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121: 167–178. 10.1016/j.cell.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Mancarci BO, Unternaehrer J, Gupta PB, et al. 2012. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483: 598–602. 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, Saga Y. 2009. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325: 1394–1398. 10.1126/science.1172645 [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. 2000. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell 11: 3601–3615. 10.1091/mbc.11.10.3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno F, Nebbioso A, Altucci L. 2020. DOT1L: a key target in normal chromatin remodelling and in mixed-lineage leukaemia treatment. Epigenetics 15: 439–453. 10.1080/15592294.2019.1699991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JA, Avarbock MR, Tobias JW, Brinster RL. 2009. Identification of GDNF-regulated genes important for spermatogonial stem cell self-renewal in the rat. Biol Reprod 81: 56–66. 10.1095/biolreprod.108.075358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, et al. 2007. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature 449: 346–350. 10.1038/nature06129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takashima S, Hirose M, Ogonuki N, Ebisuya M, Inoue K, Kanatsu-Shinohara M, Tanaka T, Nishida E, Ogura A, Shinohara T. 2013. Regulation of pluripotency in male germline stem cells by Dmrt1. Genes Dev 27: 1949–1958. 10.1101/gad.220194.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756. 10.1016/S0092-8674(02)00759-6 [DOI] [PubMed] [Google Scholar]
- Vlaming H, van Leeuwen F. 2016. The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma 125: 593–605. 10.1007/s00412-015-0570-5 [DOI] [PubMed] [Google Scholar]
- Wang Z, Xu X, Li JL, Palmer C, Maric D, Dean J. 2019. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat Commun 10: 5196. 10.1038/s41467-019-13193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. 2009. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460: 909–913. 10.1038/nature08210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K, Tellier M, Murphy S. 2018. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules 8: 11. 10.3390/biom8010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Oatley J, Bardwell VJ, Zarkower D. 2016. DMRT1 is required for mouse spermatogonial stem cell maintenance and replenishment. PLoS Genet 12: e1006293. 10.1371/journal.pgen.1006293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ. 2009. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol 9: 38. 10.1186/1471-213X-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.