In this study, Sudarshan et al. characterized the fusion protein produced by the EPC1-PHF1 translocations, which are highly recurrent chromosomal translocations in patients with endometrial stromal sarcomas (ESSs) and ossifying fibromyxoid tumors (OFMTs). Their results indicate that most chromosomal translocations linked to these sarcomas use the same molecular oncogenic mechanism through a physical merge of NuA4/TIP60 and PRC2 complexes, leading to mislocalization of histone marks and aberrant Polycomb target gene expression.
Keywords: EPC1, H3K27 methylation, H4 acetylation, HOXD, JAZF1, NuA4, PHF1, PRC2, SUZ12, TIP60
Abstract
Chromosomal translocations frequently promote carcinogenesis by producing gain-of-function fusion proteins. Recent studies have identified highly recurrent chromosomal translocations in patients with endometrial stromal sarcomas (ESSs) and ossifying fibromyxoid tumors (OFMTs), leading to an in-frame fusion of PHF1 (PCL1) to six different subunits of the NuA4/TIP60 complex. While NuA4/TIP60 is a coactivator that acetylates chromatin and loads the H2A.Z histone variant, PHF1 is part of the Polycomb repressive complex 2 (PRC2) linked to transcriptional repression of key developmental genes through methylation of histone H3 on lysine 27. In this study, we characterize the fusion protein produced by the EPC1-PHF1 translocation. The chimeric protein assembles a megacomplex harboring both NuA4/TIP60 and PRC2 activities and leads to mislocalization of chromatin marks in the genome, in particular over an entire topologically associating domain including part of the HOXD cluster. This is linked to aberrant gene expression—most notably increased expression of PRC2 target genes. Furthermore, we show that JAZF1—implicated with a PRC2 component in the most frequent translocation in ESSs, JAZF1-SUZ12—is a potent transcription activator that physically associates with NuA4/TIP60, its fusion creating outcomes similar to those of EPC1-PHF1. Importantly, the specific increased expression of PRC2 targets/HOX genes was also confirmed with ESS patient samples. Altogether, these results indicate that most chromosomal translocations linked to these sarcomas use the same molecular oncogenic mechanism through a physical merge of NuA4/TIP60 and PRC2 complexes, leading to mislocalization of histone marks and aberrant Polycomb target gene expression.
ATP-dependent remodelers and histone modifiers are key regulators of the structure and function of chromatin, essential for genome expression and stability, cell proliferation, development, and response to environmental cues. Post-translational modifications of specific residues on histones are also part of epigenetic mechanisms ensuring gene expression memory during cell divisions (Zentner and Henikoff 2013). Histone-modifying enzymes are often part of large multisubunit protein complexes. They are highly conserved and composed of various combinations of subunits like readers, writers, and erasers of histone marks; histone chaperones; and chromatin remodelers. The combination of different modules ensures specific localization and histone target selection, and enables epigenetic cross-talk and context-specific activity. The strictly coordinated activities of chromatin-modifying complexes ensure the proper functioning of the cell (Lalonde et al. 2014). Disruption of chromatin marks and their regulators can lead to various pathologies including cancer (Shen and Laird 2013).
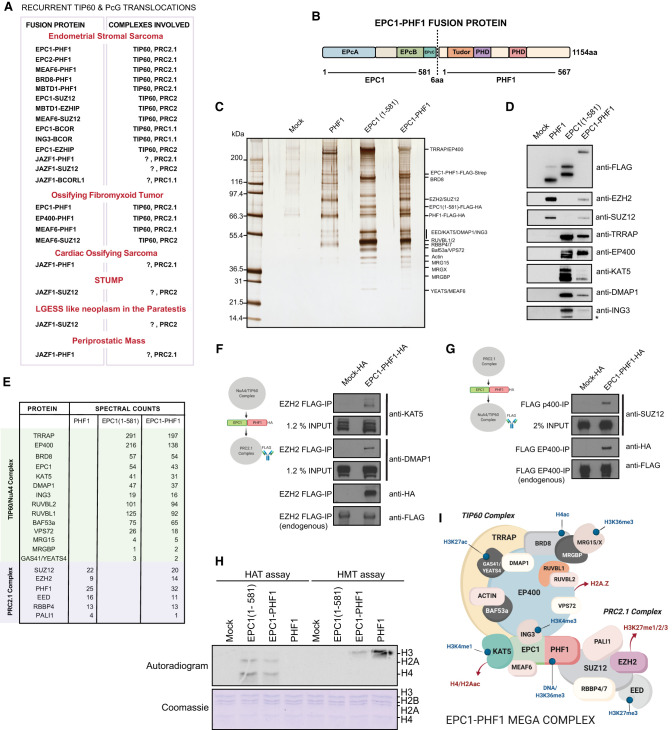
Recurrent chromosomal translocations producing oncogenic fusion proteins are common in many cancers and particularly prevalent in hematopoietic malignancies and sarcomas. These fusions frequently involve chromatin and transcription regulators and, in many cases, are thought to be the primary drivers of cancer. In recent years, disruption of chromatin dynamics has emerged as a consistent oncogenic mechanism used by these fusion proteins (for review, see Brien et al. 2019). Sarcomas are rare mesenchymal tissue cancers with distinct molecular profiles. One-third of all sarcomas harbor chromosomal translocations with otherwise normal karyotypes, showing robust clustering of gene expression profiles compared with normal tissues. Recurrent translocations are found in endometrial stromal sarcomas (ESSs) and ossifying fibromyxoid tumors (OFMTs) that potentially fuse subunits of distinct chromatin-modifying complexes with opposite functions in gene regulation; namely, the NuA4/TIP60 histone acetyltransferase (HAT) complex and Polycomb repressive complexes (Fig. 1A). Among the recurring chromosomal translocations that characterize ESSs, genes for five different subunits of the NuA4/TIP60 complex (EPC1/2, MBTD1, MEAF6, and BRD8) are repeatedly found fused to genes for different PRC2 components (PHF1/SUZ12/EZHIP) (Ferreira et al. 2018; Hoang et al. 2018; Momeni-Boroujeni et al. 2021). Interestingly, other soft tissue sarcomas such as OFMTs also harbor EPC1-PHF1, EP400-PHF1, and MEAF6-PHF1 fusions. OFMTs are rare cancers of uncertain cellular origin, and ∼50%–85% of cases show PHF1 translocations, with EP400-PHF1 being the most frequent (40%) (Schneider et al. 2016). JAZF1-PHF1 was reported in a case of cardiac ossifying sarcoma, and fusions of NuA4/TIP60 subunits with the PRC1.1 component BCOR have also been reported in ESSs, demonstrating that these types of fusion events represent a critical oncogenic mechanism in mesenchymal cancers (Fig. 1A; Schoolmeester et al. 2013).
Figure 1.
The EPC1-PHF1 fusion protein assembles a megacomplex merging NuA4/TIP60 and PRC2 complexes and their activities. (A) Table summarizing recurrent translocations found in soft tissue sarcomas that fuse NuA4/TIP60 complex proteins and Polycomb group proteins. (B) Schematic representation of the EPC1-PHF1 fusion protein. The numbers indicated are amino acids; protein domains retained in the fusion are indicated. (C) Silver-stained SDS-PAGE showing affinity-purified complexes. Labels at the right are proteins that were identified based on Western blotting and predicted molecular weights. (D) Western blots of selected NuA4/TIP60 and PRC2 complex subunits on the affinity-purified complexes shown in C. (E) Mass spectrometry analysis of the affinity-purified complexes shown in C. See also Supplemental Figure S2, A and B. (F,G) Affinity purification of endogenous PRC2 complex (EZH2 subunit) (F) and NuA4/TIP60 (EP400 subunit) (G), confirming formation of a merged megacomplex by EPC1-PHF1. (H) In vitro histone acetyltransferase (HAT) assay and histone methyltransferase (HMT) assay. (Top panel, lanes 2,3) KAT5 purified through EPC1(1–581) as well as EPC1-PHF1 fusion show HAT activity toward H2A and H4 on chromatin. (Top panel, lanes 7,8) EZH2 purified through PHF1 as well as EPC1-PHF1 fusion show HMT activity toward H3 on chromatin. Coomassie-stained SDS-PAGE gel was the loading control for nucleosomal histones. See also Supplemental Figure S2, C and D. (I) Schematic representation of the chimeric megacomplex assembled by the EPC1-PHF1 fusion protein.
NuA4/TIP60 is an evolutionarily conserved and multifunctional MYST family HAT complex with 17 distinct subunits (Steunou et al. 2014; Judes et al. 2015; Jacquet et al. 2016; Sheikh and Akhtar 2019). The catalytic subunit KAT5/Tip60 acetylates histones H4 and H2A, as well as variants H2A.Z and H2A.X, in the context of chromatin. In addition, the ATP-dependent remodeler subunit EP400 allows the exchange of canonical histone H2A in chromatin with variant histone H2A.Z (Billon and Côté 2013; Pradhan et al. 2016). There are also multiple subunits with different reader domains that allow context-dependent site-specific activity of the complex and epigenetic cross-talk (Steunou et al. 2014; Jacquet et al. 2016). The 836-amino-acid EPC1 protein is a noncatalytic scaffolding subunit of the TIP60 complex. The conserved N-terminal EPcA domain (amino acids 1–280) interacts with KAT5/Tip60, MEAF6, and ING3, thereby forming the human Piccolo NuA4 complex, enabling binding and acetylation of chromatin substrates (Boudreault et al. 2003; Doyon et al. 2004; Selleck et al. 2005; Lalonde et al. 2013; Xu et al. 2016). The C-terminal part of EPC1 is thought to associate with the rest of the NuA4/TIP60 complex based on data from the yeast NuA4 complex (Fig. 1B; Boudreault et al. 2003; Setiaputra et al. 2018; Wang et al. 2018). The NuA4/TIP60 complex is an important transcriptional coactivator that can be recruited to gene regulatory elements by several transcription factors. Through acetylation of histones as well as nonhistone substrates, it activates a multitude of gene expression programs, including proliferation, stress response, apoptosis, differentiation, and stem cell identity (Steunou et al. 2014; Judes et al. 2015; Sheikh and Akhtar 2019). The NuA4/TIP60 complex is also a key player in the response to DNA damage, assisting repair pathway choice as well as the repair process itself (Jacquet et al. 2016; Cheng et al. 2018 and references therein). All these functions are essential for cellular homeostasis; hence, NuA4/TIP60 subunits are often mutated or deregulated in cancers, and Tip60/KAT5 itself is a haploinsufficient tumor suppressor (Gorrini et al. 2007; Judes et al. 2015; Sheikh and Akhtar 2019).
Polycomb group proteins (PcG) are evolutionarily conserved proteins involved in development and transcriptional regulation, originally linked to HOX gene repression during Drosophila development (Schuettengruber et al. 2017). They are key regulators of mammalian development, differentiation, and cell fate decisions. PcG proteins assemble into multisubunit complexes, major ones being Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). These complexes cooperate with each other to create repressive chromatin regions through histone modifications and chromatin organization. While PRC1 catalyzes H2AK119 monoubiquitination and chromatin compaction, PRC2 catalyzes H3K27 methylation. The PRC2 complex is composed of the core components EZH2, SUZ12, EED, and RBBP4/7. Through a read–write mechanism, it can deposit H3K27me3 over large chromatin regions such as the HOX clusters. Other associated factors function to stabilize the PRC2 complex on chromatin and modulate its activity, defining distinct variant complexes PRC2.1 and PRC2.2. PRC2.1 contains one of the Polycomb-like (PCL) proteins PHF1, MTF2, or PHF19, as well as either PALI1/2 or EPOP (Laugesen et al. 2019; Loubiere et al. 2019). The 567-amino-acid PHF1 protein binds unmethylated CpG islands particularly at long linker DNA regions, stabilizes the binding of PRC2.1 complex on chromatin, and increases its activity (Cao et al. 2008; Sarma et al. 2008; Choi et al. 2017; Li et al. 2017). PHF1 is also known to bind the H3K36me3 mark through its Tudor domain (Musselman et al. 2012; Cai et al. 2013), restricting the catalytic activity of EZH2.
Many groups are working on the classification of ESSs using recurrent fusions as molecular markers. However, there is currently little molecular understanding about the consequences of these recurrent translocations and their primary role in tumorigenesis. To address this, we used biochemical and genomic approaches to study the product of a recurrent translocation found in ESSs and OFMTs that fuses the EPC1 subunit of the NuA4/TIP60 complex to the PHF1 subunit of the PRC2.1 complex (Fig. 1B; Micci et al. 2006; Antonescu et al. 2014). We investigated the molecular impact of the EPC1-PHF1 fusion protein by generating isogenic cell lines, used affinity purification followed by mass spectrometry to identify interactors, associated activities, and analyzed chromatin occupancy of the fusion protein. We also analyzed the effect of the fusion protein on global chromatin dynamics while correlating differential gene expression.
Results
To construct the fusion EPC1-PHF1 gene (Fig. 1B), we used a portion of the chimeric gene recovered by RT-PCR from total RNA isolated from surgically removed endometrial stromal sarcoma (a kind gift from Francesca Micci's group) (Micci et al. 2006). As necessary controls, we also subcloned full-length PHF1 and EPC1, as well as a portion of EPC1 corresponding to the fragment found in the fusion, referred to here as EPC1(1–581). We confirmed the expression of all constructs and then tested whether the EPC1-PHF1 fusion protein can act like an oncogenic driver by performing colony formation assay in HEK293T cells (Supplemental Fig. S1A,B). In contrast to full-length and truncated EPC1, which strongly inhibit growth, we observed that expression of EPC1-PHF1 leads to a greater number of colonies compared with controls (Supplemental Fig. S1B), supporting the idea that this gene fusion may be a driver event, giving a growth advantage to the expressing cells.
EPC1-PHF1 forms a megacomplex merging NuA4/TIP60 and PRC2.1
To isolate the native complex(es) formed by the EPC1-PHF1 fusion protein, we generated isogenic K562 cell lines by targeted integration of C-terminally TAP-tagged cDNAs into the AAVS1 safe harbor genomic locus (Dalvai et al. 2015). To circumvent issues related to protein overexpression, we used the moderately active PGK1 promoter, which was shown to achieve expression of NuA4/TIP60 and PRC2 subunits within 2.5-fold of the native levels (EP400, EPC1, MBTD1, and EZH2) (Dalvai et al. 2015). Our previous studies have demonstrated that this system enables purification of stable and stoichiometric protein complexes (Dalvai et al. 2015; Doyon and Côté 2016; Jacquet et al. 2016). We chose the K562 cell line because it is an ENCODE tier 1 cell line and can grow in suspension culture to high cell densities, ideal for biochemical and genomic studies.
Nuclear extracts from K562 cell lines expressing the fusion protein, individual fusion partners, or the empty tag (mock, C-terminal 3xFLAG-2xHA-2A-puromycin tag) were allowed to bind to anti-FLAG resin, and the bound material was eluted with 3xFLAG peptides. An alternate K562 cell line expressing EPC1-PHF1 with a C-terminal 3xFLAG-2xStrep tag was also used to improve yield and purity by binding the FLAG-eluted fraction to Streptactin beads followed by elution with biotin. SDS-PAGE and silver staining identified the components of the purified complexes (Fig. 1C). Mass spectrometry analysis identified all expected components of the NuA4/TIP60 and PRC2 complexes (Fig. 1D; Supplemental Fig. S2A,B). We further confirmed the complex subunits by Western blotting with NuA4/TIP60- and PRC2-specific antibodies (Fig. 1E). The PHF1 fraction contained the core subunits EZH2, EED, and SUZ12 of the PRC2 complex, as well as RBBP4 and the PRC2.1-specific PALI1. The EPC1(1–581) fraction contained all the subunits of the NuA4/TIP60 complex except MBDT1, which was expected since it associates through an interaction with the EPC1 C terminus (amino acids 644–672) (Jacquet et al. 2016; Zhang et al. 2020). Strikingly, the EPC1-PHF1 fraction contained subunits of both the TIP60 and PRC2.1 complexes. These results indicate that the EPC1-PHF1 fusion protein efficiently associates with both NuA4/TIP60 and PRC2.1. This was also supported by data in HEK293T cells (Supplemental Fig. S1C).
To determine whether the fusion protein's associations with the two complexes occur independently of each other or could occur simultaneously, we integrated 3xHA-tagged EPC1-PHF1 at the AAVS1 locus in K562 cell lines in which TALEN/CRISPR was used to introduce a 3xFLAG tag at endogenous EZH2 or EP400 genes, respectively (Dalvai et al. 2015). This allowed us to purify endogenous PRC2 and NuA4/TIP60 complexes and verify whether the fusion protein can physically bridge the two complexes. As shown in Figure 1, F and G, it is clear that expression of the EPC1-PHF1 fusion leads to the association of the PRC2 complex with NuA4/TIP60 and vice versa, demonstrating the formation of a megacomplex.
We then performed in vitro histone acetyltransferase (HAT) and histone methyltransferase (HMT) assays with purified fractions on native chromatin as substrate. We observed robust HAT activity toward histone H4 and H2A in both EPC1(1–581) and EPC1-PHF1 complexes, as expected for NuA4/TIP60 (Fig. 1H; Supplemental Fig. S2C). Simultaneously, we detected HMT activity toward histone H3 in PHF1 and EPC1-PHF1 complexes, as expected for PRC2 (Fig. 1H; Supplemental Fig. S2D). This demonstrates that the merged assembly maintains the enzymatic activities of both original complexes. Altogether, these results indicate that expression of the EPC1-PHF1 fusion protein in cells leads to the formation of a hybrid TIP60-PRC2 megacomplex that harbors opposite functions in terms of chromatin modifications and impact on gene regulation (Fig. 1I). It is important to note that we did not detect an effect of expressing the fusion protein or its partners on the bulk level of H3K27me3 (Supplemental Fig. S2E), in contrast to what has been previously proposed for the JAZF1-SUZ12 fusion (Ma et al. 2017).
The EPC1-PHF1 fusion complex is enriched at genomic loci bound by TIP60 and PRC2.1
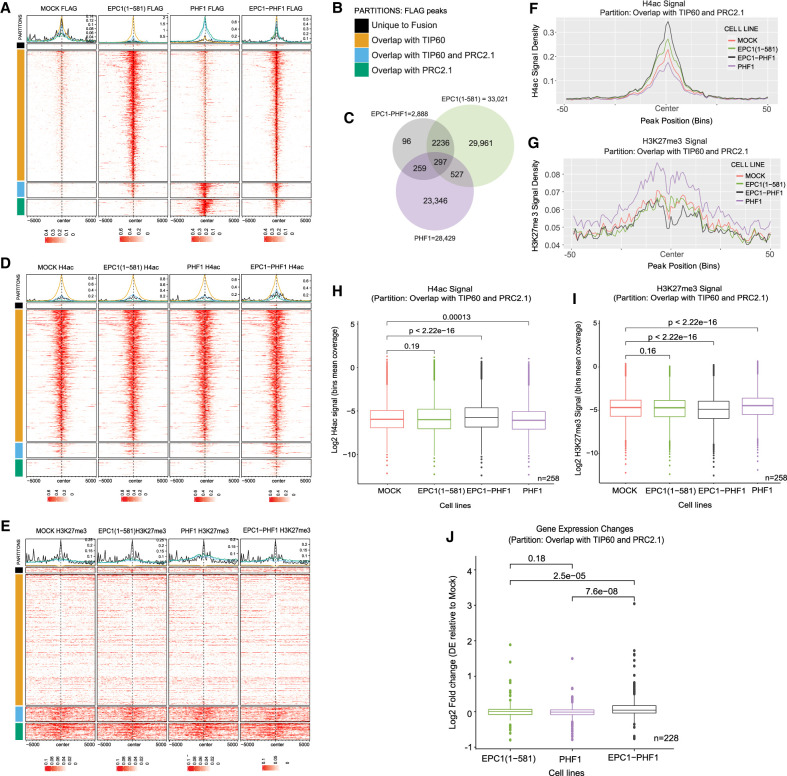
To determine the genomic locations where the EPC1-PHF1 megacomplex may act, we performed anti-FLAG chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) using the isogenic cell lines. We analyzed binding regions (significant peaks) of EPC1-PHF1, EPC1(1–581), and PHF1 in comparison with the empty vector cell line. While >20,000 bound regions were identified for each TIP60(EPC1) and PRC2.1(PHF1) complex, only 2888 peaks were called for the fusion (Fig. 2A–C), likely due to its lower expression (Fig. 1E; Supplemental Fig. S1C). Nevertheless, we focused on these locations bound by EPC1-PHF1 and determined overlaps with the regions bound by TIP60 or PRC2.1, or unique to the fusion. While most of the sites bound by the fusion overlap with TIP60-bound regions, a significant number also overlaps with PRC2.1 sites and with regions bound by both PRC2.1 and TIP60 (Fig. 2A–C; Supplemental Table S1). Only a small number of sites unique to the fusion was detected. These results indicate that the EPC1-PHF1 fusion protein is targeted to genomic locations normally bound by TIP60, PRC2.1, or both.
Figure 2.
The EPC1-PHF1 fusion complex induces global changes in histone marks and gene expression. (A) Enrichment of FLAG ChIP-seq signal in K562 cell lines. The heat maps show regions bound by EPC1-PHF1 (± 5000 bp to peak center), partitioned based on overlap with EPC1(1–581) and/or PHF1 FLAG peaks. (B) Partitions used for heat maps in A, D, and E. (TIP60) EPC1(1–581), (PRC2.1) PHF1. (C) Venn diagram showing the number FLAG ChIP-seq peaks and overlap between EPC1(1–581)-, PHF1-, and EPC1-PHF1-expressing K562 cell lines. (D) Enrichment of H4-penta-acetyl ChIP-seq signal at regions bound by EPC1-PHF1, partitioned based on overlap with EPC1(1–581) and/or PHF1 FLAG peaks. (E) Enrichment of H3K27me3 ChIP-seq signal in K562 cell lines at regions bound by EPC1-PHF1, partitioned based on overlap with EPC1(1–581) and/or PHF1 FLAG peaks. (F) Density plot of H4ac enrichment in K562 cell lines in a particular partition (EPC1-PHF1-bound regions and overlap with TIP60 and PRC2.1). Each bin corresponds to 100 bp. See Supplemental Figure S3 for other partitions. (G) Density plot of H3K27me3 enrichment in K562 cell lines in a particular partition (EPC1-PHF1-bound regions and overlap with TIP60 and PRC2.1). See Supplemental Figure S3 for other partitions. (H) Box plots showing H4ac enrichment level in K562 cell lines in a particular partition (EPC1-PHF1-bound regions and overlap with TIP60 and PRC2.1). See Supplemental Figure S3 for other partitions. Statistics were computed using bins; n = number of regions analyzed, where one region is 100 bins and the mean coverage over these 100 bins was used. P-value was calculated by Wilcoxon testing. (I) Box plots showing H3K27me3 enrichment level in K562 cell lines in a particular partition (EPC1-PHF1-bound regions and overlap with TIP60 and PRC2.1). See Supplemental Figure S3 for other partitions. Statistics were computed using bins; n = number of regions analyzed, where one region is 100 bins and the mean coverage over these 100 bins was used. P-value was calculated by Wilcoxon testing. (J) Gene expression changes at genes bound by EPC1-PHF1 in a particular partition (EPC1-PHF1-bound regions and overlap with TIP60 and PRC2.1). See Supplemental Figure S4 for other partitions. P-value was calculated by Wilcoxon testing.
Effect of EPC1-PHF1 on the chromatin and transcriptional landscape
Two major possible effects are expected from the presence of the fusion megacomplex at specific genomic loci: (1) the appearance of chromatin acetylation where PRC2.1 is normally bound on silenced regions of the genome, and (2) the appearance of H3K27 methylation where TIP60 is normally bound on expressed regions of the genome. We also speculated that the fusion protein may mislocalize TIP60-mediated H4 acetylation to poised bivalent chromatin regions, pushing them toward transcriptional activation. To test this, we performed H4 penta-acetyl (RRID: AB_310310) and H3K27me3 (RRID: AB_27932460) ChIP sequencing in our isogenic cell lines. Analysis of these histone marks in the four partitions of the sites bound by EPC1-PHF1 uncovered interesting significant changes. In regions that overlap with both TIP60 and PRC2.1, a significant increase of H4 acetylation and decrease of H3K27 methylation are seen in cells expressing the fusion (Fig. 2D–I; Supplemental Table S1). An increase of H4 acetylation is also detected on sites overlapping with PRC2.1 alone, while a decrease of H3K27me3 is less clear although obvious when only comparing with PHF1-expressing cells (Supplemental Fig. S3A–D). EPC1-PHF1-bound sites overlapping with TIP60 alone do not show a striking difference in terms of acetylation, while the drop of H3K27me3 is difficult to judge because of the very low starting level of this mark at these locations (Supplemental Fig. S3E–H). For the sites unique to the fusion, the low number makes it difficult to judge, but there seems to be an increase of acetylation (Supplemental Fig. S3I–L). Altogether, these results indicate that binding of the EPC1-PHF1 fusion alters the epigenome most strikingly by increasing H4 acetylation and decreasing H3K27 methylation at regions bound by both TIP60 and PRC2.1, as well as increasing acetylation at regions bound by PRC2.1 alone.
To understand the effect of EPC1-PHF1-induced changes in local histone modifications on transcription, we performed gene expression analysis of the isogenic cell lines (Supplemental Fig. S4A; Supplemental Table S1). Again focusing on the partition of the sites bound by EPC1-PHF1, genes neighboring regions that overlap with both TIP60 and PRC2.1 show significantly increased transcription in cells expressing the fusion (Fig. 2J). Genes neighboring regions that overlap with PRC2.1 also show a significant increase in expression, while changes at the other two partitions of EPC1-PHF1-bound sites are less striking (Supplemental Fig. S4B–D). Overall, the results indicate that the EPC1-PHF1-induced increase in H4 acetylation correlates with increased local gene expression. In parallel, we did not clearly detect locations bound by the fusion that show increased H3K27me3 and decreased transcription. On the contrary, the H3K27me3 mark deposited by PRC2 tends to decrease in regions with overlapping binding of TIP60 and PRC2.1, as local transcription increases. Interestingly, these regions could be linked to bivalent chromatin, as TIP60 is normally enriched in H3K4me3-containing regions (Fazzio et al. 2008; Ravens et al. 2015; Jacquet et al. 2016).
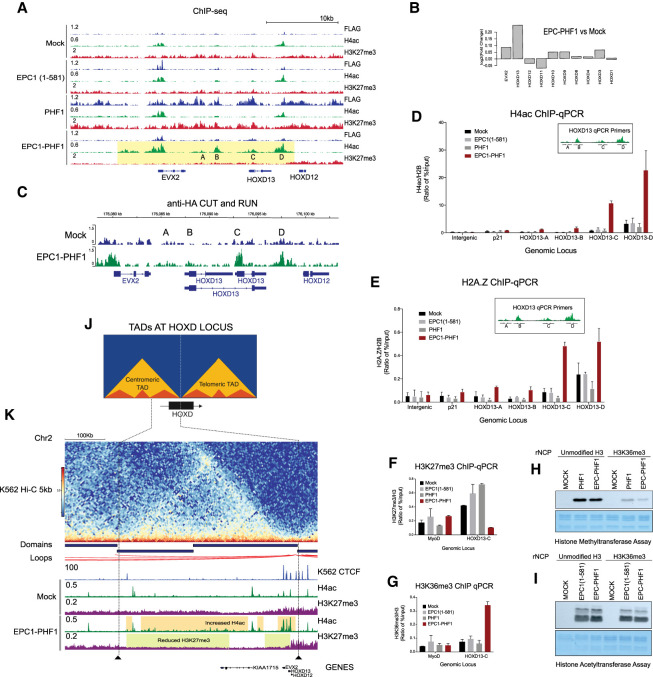
Since the increase of H4 acetylation signal at specific EPC1-PHF1-bound regions is predominant, this suggests that subverting NuA4/TIP60 activity to new targets by the fusion protein is the primary outcome. We found that the HOXD gene cluster in particular demonstrates this effect in a striking fashion. Representative ChIP-seq tracks of FLAG, H4 acetylation, and H3K27me3 at the posterior HOXD genes in the isogenic cells are shown in Figure 3A. Binding of EPC1(1–581) alone is detected in two precise locations coinciding with small pre-existing H4 acetylation islands at the end of the EVX2 gene and between HOXD12 and HOXD13. In contrast, PHF1 associates with the entire region, as expected from the H3K27me3 signal detected throughout the same region. Interestingly, while the H3K27me3 may be increased by the expression of exogenous PHF1, the two small H4 acetylation islands persist. Most strikingly, the EPC1-PHF1 cell line, while expressing much less fusion protein compared with EPC1(1–581) and PHF1 (Supplemental Fig. S6A), clearly demonstrates the appearance and mislocalization of H4 acetylation over a relatively large region (∼25 kb) (highlighted in yellow in Fig. 3A). Importantly, we could also observe a decrease in the levels of H3K27me3 in this same region, compared with the isogenic cell lines and also compared internally with the neighboring region (starting from the HOXD12 gene). This local increase in H4 acetylation correlates with a specific increase in expression of the EVX2 and HOXD13 genes located within the region (Fig. 3B). The FLAG signals reflecting EPC1-PHF1 binding in this region were too low to be called a peak but nevertheless showed higher mean reads over the increased acetylation peaks compared with mock cells (Supplemental Fig. S6G). To clearly demonstrate that EPC1-PHF1 is binding at these locations, we performed a CUT&RUN-seq experiment with an isogenic cell line expressing an HA-tagged fusion. We were able obtain more peaks through this analogous approach (Supplemental Table S1). Importantly, specific EPC1-PHF1 peaks were detected at the HOXD13 locations where new H4 acetylation appears (Fig. 3C). Moreover, 63% of EPC1-PHF1-FLAG ChIP-seq peaks overlap with the EPC1-PHF1-HA CUT&RUN peaks, validating that the peaks observed indeed correspond to the EPC1-PHF1 fusion (Supplemental Fig. S5A; Supplemental Table S1). Upon integrating CUT&RUN and ChIP-seq data, we observe that EPC1-PHF1 (HA CUT&RUN) predominantly binds regions occupied by TIP60, some PRC2.1, and some unique regions (Supplemental Fig. S5B,C). Validating our ChIP-seq analysis in Figure 2, we saw increased H4 acetylation and decreased H3K27 methylation signals at regions bound by EPC1-PHF1 overlapping both TIP60 and PRC2.1, as well as increased acetylation at regions bound by EPC1-PHF1 overlapping PRC2.1 alone (Supplemental Fig. S5D,E).
Figure 3.
EPC1-PHF1 induces changes in the chromatin landscape at the HOX gene clusters. (A) Representative ChIP-seq peaks at the HOXD gene cluster; the highlighted region in yellow shows H4 acetylation mislocalization (at regions labeled HOXD13-A, HOXD13-B, HOXD13-C, and HOXD13-D) and a reduction in H3K27me3 in the EPC1-PHF1-expressing K562 cell line. See Supplemental Figure S7 for similar effects at the other HOX clusters and Supplemental Figure S6D for similar effects in other genes. (B) Gene expression changes at the HOXD cluster in K562 cells expressing EPC1-PHF1 compared with cells expressing tag only (mock). See Supplemental Figure S4A for genome-wide gene expression changes. (C) Confirmation of EPC1-PHF1 localization at HOXD13 by anti-HA CUT&RUN sequencing. Representative peaks at the HOXD13 gene. See Supplemental Figure S5 for in-depth analysis. (D) Mislocalization of H4 acetylation determined by ChIP-qPCR. The EPC1-PHF1-expressing K562 cell line (bar graph colored maroon) shows increased H4 acetylation compared with control cell lines at HOXD13-A, HOXD13-B, HOXD13-C, and HOXD13-D regions, validating the results of ChIP-seq in A. Values are a ratio of percentage input of H4ac and H2B (n = 2). Error bars are range of the values. Intergenic (chromosome 12: 65,815,182–65,815,318) and p21 promoter are negative controls. (E) Mislocalization of variant histone H2A.Z determined by ChIP-qPCR. The EPC1-PHF1-expressing K562 cell line (bar graph colored maroon) shows increased H2A.Z occupancy compared with control cell lines at regions of H4 acetylation mislocalization (HOXD13-A, HOXD13-B, HOXD13-C, and HOXD13-D) shown in A. Values are a ratio of percentage input of H4ac and H2B. (n = 2). Error bars are range of the values. Intergenic (chroromosome12: 65,815,182–65,815,318) and p21 promoter are negative controls. (F,G) A decrease in H3K27me3 levels correlates with an increase in H3K36me3. (F) ChIP-qPCR with H3K27me3 antibody shows decreased level at the HOXD13-C region (gene body) only in the EPC1-PHF1-expressing K562 cell line (bar graph colored maroon), validating our ChIP-seq results in A. (G) ChIP-qPCR with H3K36me3 antibody shows increased level at the HOXD13-C region (gene body) only in the EPC1-PHF1-expressing K562 cell line (bar graph colored maroon). MyoD promoter is used as a negative control. Values are a ratio of percentage input of H4ac and H2B (n = 2). Error bars are range of the values. See also Supplemental Figure S6, B and C. (H,I) The H3K27me3 activity of EPC1-PHF1 is inhibited by the presence of H3K36me3. In vitro histone methyltransferase assay and histone acetyltransferase assay on recombinant nucleosomes (rNCP) with or without H3K36me3. Purified complexes were normalized using Western blotting for EZH2 and KAT5 (Supplemental Fig. S6E,F). Coomassie-stained SDS-PAGE gel is the loading control for recombinant nucleosomes. (J) Schematic representation of the topologically associated domains (TADs) and subdomains at the HOXD locus in mammalian cells. (K) Alignment of Hi-C data in K562 cells (Rao et al. 2014) with CTCF ChIP-seq in K562 cells (ENCODE group/Broad Institute histone marks) and ChIP-seq from this study (Fig. 4A). Highlighted regions indicate histone modification changes confined to a topological domain in the EPC1-PHF1-expressing cell line compared with the tag-only-expressing cell line (mock). See also Supplemental Figure S7.
To validate these observations, we performed ChIP-qPCR with primers at four sites of de novo H4 acetylation (HOXD13-A, HOXD13‐‐B, HOXD13-C, and HOXD13-D) (highlighted in Fig. 3A). Correcting for nucleosome occupancy with total H2B signal, we could confirm the specific increase in H4 acetylation at these locations in the cell line expressing EPC1-PHF1 (Fig. 3D).
Replacement of canonical H2A with H2A.Z in chromatin is another enzymatic activity of the NuA4/TIP60 complex through its EP400 subunit (Billon and Côté 2013; Pradhan et al. 2016). ChIP-qPCR with a H2A.Z-specific antibody revealed a similar mislocalization of this variant histone at the HOXD sites (Fig. 3E). These data clearly validate our hypothesis that the EPC1-PHF1 fusion complex can localize at genomic regions normally occupied by PRC2, mislocalizing the activities of the TIP60 complex to deregulate transcription. Other genes/genomic loci normally targeted by PRC2 and bound by the fusion show clear de novo H4 acetylation and a decrease of H3K27me3 (Supplemental Fig. S6D).
We were intrigued at the apparent reduction in the levels of H3K27me3 at the HOXD gene cluster in the EPC1-PHF1-expressing cells (Fig. 3A, highlighted in yellow). As shown above, we observed neither a global decrease in H3K27me3 levels (Supplemental Fig. S2E) nor destabilization of the PRC2 complex in our cell line (Fig. 1). Moreover, we could detect productive histone methylation by the EPC1-PHF1 megacomplex (Fig. 1H; Supplemental Fig. S2D). In a previous study (Musselman et al. 2012), we demonstrated that the Tudor domain of PHF1 binds to H3K36me3 and constrains PRC2-mediated H3K27me3 activity. We hypothesized that this could be part of the mechanism leading to a local decrease of H3K27me3. We checked for the levels of H3K27me3 and H3K36me3 at the HOXD13-C locus by ChIP-qPCR to confirm this possible cross-talk. A decrease in H3K27me3 was observed in EPC1-PHF1 cells compared with controls, validating our ChIP-seq results (Fig. 3F). We also saw an increase in H3K36me3 at the HOXD13-C locus in EPC1-PHF1 cells (Fig. 3G), likely linked to the increased transcription of the gene. This cross-talk was not observed at the NuA4/TIP60-bound and highly transcribed gene RPSA (Supplemental Fig. S6B; Jacquet et al. 2016) or at HOXA9, a repressed gene not occupied by the EPC1-PHF1 fusion complex (Supplemental Fig. S6C). We conclude that the mislocalization of H4 acetylation and the variant histone H2A.Z at the posterior HOXD gene locus in cells expressing EPC1-PHF1 leads to derepression and productive transcription. Subsequent deposition of H3K36me3, a histone mark linked to transcription elongation, blocks the spread of the repressive H3K27me3 mark, possibly in part through its recognition by the Tudor domain of PHF1 and direct effect on EZH2/PRC2 activity (Schmitges et al. 2011; Musselman et al. 2012; Jani et al. 2019; Finogenova et al. 2020). We confirmed this cross-talk in vitro with our purified fractions in HMT and HAT assays using recombinant nucleosomes carrying the H3K36me3 mark (Fig. 3H,I; Supplemental Fig. S6E,F). Both PHF1/PRC2.1 and EPC1-PHF1 complexes are clearly inhibited in their methyltransferase activity by the presence of H3K36me3, while TIP60 and EPC1-PHF1 complexes are not affected in their acetyltransferase activity.
The EPC1-PHF1-mediated effect on histone acetylation/methylation at HOXD is restricted to a specific topologically associating domain (TAD)
The HOXD gene cluster is involved in mammalian axial patterning, as well as limb and genital development. Spatiotemporal regulation of the HOXD locus depends on long-acting multiple enhancer sequences located in “gene deserts” outside the HOXD cluster. Mammalian cells organize such regulatory landscapes in structural units called topologically associating domains (TADs), which maintain genomic contacts even in the absence of transcription. These preformed 3D structures, delimited by CTCF and cohesin proteins, remain globally similar in cell types and are conserved from mice to humans (for reviews, see Bompadre and Andrey 2019; Szabo et al. 2019). The HOXD locus is positioned between two large TADs (Fig. 3J; Rodríguez-Carballo et al. 2017). During limb development, the telomeric TAD is activated and controls the transcription of early HOXD genes (forearm), while later the centromeric TAD controls the posterior HOXD genes (digits) (Andrey et al. 2013). A strong boundary limits the posterior HOX genes from getting activated aberrantly (Lonfat and Duboule 2015).
Chromatin modifications influence the dynamic mammalian TADs. The repressive H3K27me3 modification segregates chromatin into discrete subdomains, and studies suggest that these 3D clusters require PcG proteins and H3K27me3 (Szabo et al. 2019). The HOXD cluster is known to harbor an H3K27me3 dense region, and the 1.8-kb region between HOXD11 and HOXD12 (D11.12) may act as a putative mammalian Polycomb responsive element (PRE) (Woo et al. 2010). Recent studies have shown that this region acts as a nucleation hub for PRC2 to stably bind and spread H3K27me3 through intrachromosomal and interchromosomal interactions (Vieux-Rochas et al. 2015; Oksuz et al. 2018).
We observed that the Tip60/KAT5-mediated H4 acetylation and reduction of H3K27me3 at the HOXD locus in EPC1-PHF1 cells localized to the posterior HOXD genes. This led us to explore the possibility that a TAD boundary exists in this region. When we aligned the publicly available CTCF ChIP-seq data and Hi-C data set in K562, we observed that the chromatin changes induced by the EPC1-PHF1 fusion protein are restricted to a large portion of the centromeric TAD at the HOXD locus (Fig. 3J,K). Since Polycomb group protein-mediated long-range contacts are also at play at the HOXD locus, we looked for spreading of EPC1-PHF1-mediated H4 acetylation to other genomic loci that were shown to interact with the HOXD locus. Although existing Hi-C/5-C studies in K562 cells are not detailed enough to detect PcG-mediated TADs (Kundu et al. 2017) and many of the explored regions are not conserved from mice to humans (Vieux-Rochas et al. 2015; Oksuz et al. 2018), we could detect the presence of H4 acetylation at the HOXC locus (H3K27me3 spreading site), EVX1, HOXB13, CYP26B1, and LHX2 (H3K27me3 nucleation sites) (Supplemental Fig. S7). This observation suggests that the EPC1-PHF1 fusion protein could mislocalize activating histone marks through PcG-mediated chromatin contacts, activating an oncogenic transcriptional network. A similar mechanism was observed recently for an oncogenic EZH2 mutant that co-opted PcG-mediated long-range chromatin contacts to repress multiple tumor suppressors (Donaldson-Collier et al. 2019). Further work will be required to fully dissect the effect of the EPC1-PHF1 fusion complex on the structure and dynamics of TADs/Polycomb-mediated chromatin contacts.
JAZF1 is a stochiometric interaction partner of the NuA4/TIP60 complex
Various subunits of the NuA4/TIP60 complex can be fused to different subunits from Polycomb repressive complexes 1 and 2 in endometrial stromal sarcoma (Fig. 1A). However, the most frequently observed translocation is that of JAZF1-SUZ12, which occurs in >50% of all low-grade ESSs (LG-ESSs) (Ferreira et al. 2018). There are other recurrent translocations involving the JAZF1 protein in sarcomas; namely, JAZF1-PHF1 and JAZF1-BCORL1 (Fig. 1A; Micci et al. 2006; Schoolmeester et al. 2013; Allen et al. 2017). This connection led us to investigate whether JAZF1 interacts with the TIP60 complex.
The molecular function of the JAZF1 protein is largely unexplored. It is a transcription factor with three C2H2 zinc fingers, and studies have linked it to glucose production, lipid metabolism, ribosome biogenesis, metabolic disorders, and pancreatic cancer (Nakajima et al. 2004; Liao et al. 2019; Kobiita et al. 2020; Zhou et al. 2020). The JAZF1 protein has a homolog in S. cerevisiae called Sfp1 (Supplemental Fig. S8A), an essential transcription factor involved in regulation of ribosomal protein (RP), ribosomal biogenesis (RiBi), and cell cycle genes, responding to nutrients and stress through regulation by TORC1 (Marion et al. 2004; Lempiainen et al. 2009). Crucially, Sfp1 and the yeast NuA4 complex cooperate functionally to regulate RP and RiBi genes (Supplemental Fig. S8B; Reid et al. 2000; Loewith and Hall 2011; Rossetto et al. 2014).
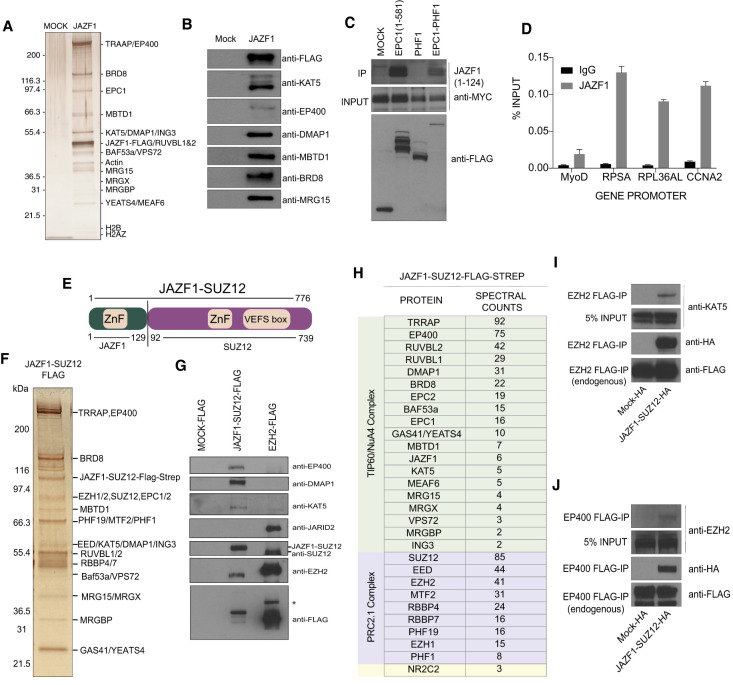
Since JAZF1 is not expressed in K562 cells (http://www.proteinatlas.org), we performed multiple cell compartment affinity purification coupled to tandem mass spectrometry (MCC-AP-MS/MS) in HEK293 as described in Lavallée-Adam et al. (2013). This technique allowed us to isolate protein–protein interaction occurring on the chromatin fraction. By purifying JAZF1, we recovered several NuA4/TIP60 core components with good reliability (Supplemental Fig. S8C). JAZF1 is highly expressed in the male and female reproductive tissues and thus may be involved in the tumorigenesis of LG-ESSs of all fusion subtypes. To confirm this interaction, we ectopically expressed JAZF1-3xFLAG-2xSTREP in K562 cells from the AAVS1 locus and tandem affinity-purified the protein from nuclear extracts. Silver staining of the purified fraction on gel, Western blot, and mass spectrometry analyses all confirm a very tight association of JAZF1 with the NuA4/TIP60 complex (Fig. 4A,B; Supplemental Fig. S8D), suggesting that JAZF1 can be a stable stoichiometric subunit. Accordingly, JAZF1 occupies the promoters of known TIP60-bound ribosomal protein genes RPSA and RPL36AL as determined by ChIP-qPCR (Fig. 4D; Jacquet et al. 2016).
Figure 4.
The JAZF1 protein stably associates with the NuA4/TIP60 complex. (A) Silver-stained SDS-PAGE showing tandem affinity-purified JAZF1. Labels at the right are proteins that were identified based on Western blotting and predicted molecular weights. The JAZF1 fraction shows all subunits of the NuA4/TIP60 complex. See also Supplemental Figure S8, C and D, for mass spectrometry analysis of JAZF1 purification from HEK293 and K562 cells, respectively. (B) Western blots of selected NuA4/TIP60 subunits on the affinity-purified fraction shown in A. (C) Immunoprecipitation of JAZF1 with the EPC1-PHF1 fusion. JAZF1(1–124) corresponding to the portion in the JAZF1-SUZ12 fusion protein that interacts with EPC1(1–581) and EPC1-PHF1. See Supplemental Figure S8E for data with full-length JAZF1 construct. (D) FLAG-JAZF1 ChIP-qPCR. Anti-FLAG ChIP-qPCR in HEK293 cells stably expressing FLAG-tagged JAZF1. Enrichment of JAZF1 can be seen at ribosomal protein gene promoters (RPSA and RPL36AL) regulated by NuA4/TIP60. CCNA2 is a positive control for JAZF1. Values are represented as percentage IP/input chromatin (n = 2). Error bars represent range of the values. (E) Schematic representation of the JAZF1-SUZ12 fusion protein. The numbers indicate are amino acids; protein domains retained in the fusion are indicated. (F) Silver-stained SDS-PAGE showing tandem affinity-purified JAZF1-SUZ12. Labels at the right are proteins that were identified based on Western blotting and predicted molecular weights. (G) Western blots of selected NuA4/TIP60 and PRC2 subunits on the affinity-purified fraction shown in F. Note the absence of JARID2 in the JAZF1-SUZ12 complex. (H) Mass spectrometry analysis of the affinity-purified JAZF1-SUZ12 complex shown in F. Note the absence of JARID2 and the presence of the three Polycomb-like paralogs PHF1, MTF2, and PHF19, identifying PRC2.1. (I,J) Formation of a TIP60-PRC2 megacomplex by JAZF1-SUZ12. (I) Immunoprecipitation of endogenously tagged EZH2. Tip60 can be detected only in the JAZF1-SUZ12-expressing cells. (J) Immunoprecipitation of endogenously tagged EP400. EZH2 can be detected only in the JAZF1-SUZ12-expressing cells.
To analyze this tight interaction in the context of the JAZF1-SUZ12 translocation, we performed coimmunoprecipitation experiments showing that the TIP60 complex still interacts strongly with the portion of JAZF1 (JAZF1[1–124]) that is fused to SUZ12 (Fig. 4C; Supplemental Fig. S8E). The same experiments showed that JAZF1 also still interacts with EPC1(1–581) and the EPC1-PHF1 megacomplex, but not with PHF1/PRC2.1. We then produced an isogenic cell line expressing the JAZF1-SUZ12 oncogenic fusion and purified the associated protein complex. Analysis of the purified fraction on gel by Western blot and mass spectrometry again showed the assembly of a chimeric megacomplex merging NuA4/TIP60 to PRC2.1 (Fig. 4E–H), confirmed again by fractionation of endogenous respective complexes (Fig. 4I,J). These conclusive data indicate that JAZF1 fusions could have the same effects on the epigenome as EPC1-PHF1.
Conserved oncogenic mechanism of EPC1-PHF1 and JAZF1-SUZ12 fusion proteins
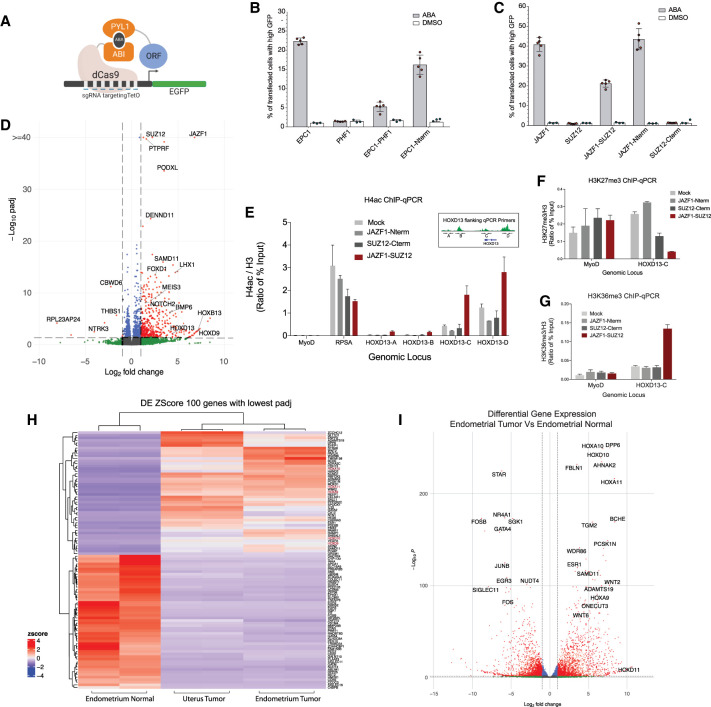
We have shown that the oncogenic EPC1-PHF1 fusion complex can lead to local transcription activation despite associating with a functional PRC2.1 complex. To further confirm the effect on transcription, we used a chemically inducible dCas9-based site-specific recruitment assay in HEK293T cells (Fig. 5A; Alerasool et al. 2022). In this system, we could detect specific transcriptional activation by the EPC1 protein, EPC1(1–581), and EPC1-PHF1, but not by PHF1 (Fig. 5B). Interestingly, we found that the JAZF1 protein is a very robust transcriptional activator in this reporter system. This is also the case for the JAZF1-SUZ12 fusion protein, as well as for the truncated JAZF1(1–124), but not SUZ12 by itself (Fig. 5C). ChIP-qPCR of H4 acetylation at the reporter gene also showed a specific increase with the two fusions but not with PHF1 (Supplemental Fig. S9A,B).
Figure 5.
The JAZF1-SUZ12 fusion protein has molecular impacts like EPC1-PHF1. (A) Model for the dCas9-based inducible reporter gene activation assay in B and C. (B,C) The transcription activation assay was quantified by flow cytometry analysis. (B) The percentage of cells transfected with EPC1, PHF1, EPC1-PHF1, or EPC1(1–581) expressing high GFP upon ABA treatment. EPC1-PHF1 is a transcriptional activator like EPC1 and EPC1 N terminus, whereas PHF1 is not. (C) The percentage of cells expressing high GFP when transfected with JAZF1, SUZ12, JAZF1-SUZ12, JAZF1(1–129), and SUZ12(93–740) and treated with ABA. JAZF1 (full length and N terminus) and JAZF1-SUZ12 act as transcriptional activators, whereas SUZ12 does not. At least 25,000 cells were analyzed for each replicate. Error bars represent SEM of five independent repeats. DMSO is used as a negative control. (D) Volcano plot of differential gene expression analysis of K562 cell line expressing JAZF1-SUZ12 fusion compared with an empty vector K562 cell line. (E) Mislocalization of H4 acetylation determined by ChIP-qPCR. The JAZF1-SUZ12-expressing K562 cell line (bar graph colored maroon) shows increased H4 acetylation compared with control cell lines at HOXD13-A, HOXD13-B, HOXD13-C, and HOXD13-D regions, like EPC1-PHF1 in Figure 4C. Values are represented as ratio of percentage of input chromatin of H4ac and H3 (n = 2). Error bars represent range of the values. RPSA promoter is a positive control for H4 acetylation, and MYOD promoter is a negative control. (F,G) A decrease in H3K27me3 levels at the HOXD locus correlates with increased H3K36me3 in JAZF1-SUZ12-expressing cells. (F) ChIP-qPCR with H3K27me3 antibody shows decreased level at the HOXD13-C region (gene body) in the JAZF1-SUZ12-expressing K562 cell line (bar graph colored maroon). MyoD promoter is a negative control. (G) ChIP-qPCR with H3K36me3 antibody shows increased level at the HOXD13-C region (gene body) only in the JAZF1-SUZ12-expressing K562 cell line (bar graph colored maroon). MyoD promoter is a negative control. Values are represented as ratio of percentage of input chromatin of H3K27me3 or H3K36me3 and H3 (n = 2). Error bars represent range of the values. See also Supplemental Figure S9, D and E. (H,I) RNA sequencing followed by differential gene expression analysis of two low-grade endometrial stromal sarcoma patient tissue samples with the presence of JAZF1-SUZ12 fusion. Pair-wise comparison was performed versus endometrial normal patient tissue (paired normal of endometrial tumor sample). (H) Heat map showing hierarchical clustering of the top 100 differentially expressed genes. (I) Volcano plot of differential gene expression analysis of endometrial tumor with JAZF1-SUZ12 fusion compared with adjacent normal endometrial tissue. See also Supplemental Figures S11 and S12.
The gene expression profiles of ESSs with different fusions (JAZF1 and TIP60) cluster together (Dewaele et al. 2014; Micci et al. 2016; Przybyl et al. 2018). We also performed an RNA-seq experiment with K562 cells expressing JAZF1-SUZ12 and again found specific increased expression of HOX genes/PRC2 targets, including HOXD13 (Fig. 5D; Supplemental Fig. S10; Supplemental Table S2). Recent interactome analyses of JAZF1-SUZ12 and JAZF1 also validate our results (Piunti et al. 2019; Procida et al. 2021). Altogether, these data confirm that JAZF1 associates with the TIP60 complex and that all of the currently detected fusion proteins in LG-ESSs bridge the TIP60 complex to Polycomb repressive complexes.
To confirm that the JAZF1-SUZ12 fusion generates the same molecular events on chromatin as EPC1-PHF1, we looked again at the HOXD locus by ChIP-qPCR. In JAZF1-SUZ12-expressing cells, we again detected an increase in H4 acetylation (Fig. 5E; Supplemental Fig. S9C), a decrease in H3K27 methylation (Fig. 5F; Supplemental Fig. S9D,E), and an increase in transcriptional elongation-associated H3K36me3 (Fig. 5G; Supplemental Fig. S9D,E) in the region flanking the HOXD13 gene. JAZF1-SUZ12 thus is able to mislocalize the TIP60 complex-associated histone mark through the JAZF1 protein, drawing parallels to the mislocalization mediated by EPC1-PHF1 (Fig. 3).
The loss of H3K27me3 in these data can be explained by the loss of the ZnB domain of SUZ12 in the JAZF1-SUZ12 fusion protein, which has been shown to reduce the binding of JARID2 and EPOP subunits (Chen et al. 2018a). SUZ12 acts as a platform to assemble the noncanonical subunits of the PRC2 complex, and a recent study shows that mutations in SUZ12 can modulate PRC2.1 versus PRC2.2 formation (Youmans et al. 2021). Furthermore, recent structural data suggest that the JARID2-containing PRC2.2 complex can partially override H3K36me3-mediated inhibition of EZH2 catalytic activity (Kasinath et al. 2021), whereas the PRC2.1 complex is known to be catalytically inhibited by the presence of H3K36me3 (Musselman et al. 2012; Finogenova et al. 2020). The loss of JARID2 interaction in JAZF1-SUZ12 skews it to associate with the PRC2.1 complex, as clearly shown by our purification (Fig. 4G,H), which is catalytically inhibited by the presence of the H3K36me3 modification.
Transcriptome analysis of human ESS tissue samples confirms up-regulation of PRC2 target genes
In order to support the mechanistic model proposed by studying the fusions in our heterologous model system, we performed RNA sequencing of two LG-ESS patient tissue samples with the JAZF1-SUZ12 translocation and one paired normal endometrial tissue (Fig. 5H,I; Supplemental Fig. S11A; Supplemental Table S3). Two pair-wise differential expression (DE) analyses were performed against the endometrial normal tissue sample. Pairwise differential expression analysis of endometrial tumor with the adjacent normal tissue is presented as a volcano plot in Figure 5I (the other ESS sample is shown in Supplemental Fig. S11A). Significantly up-regulated genes include many homeobox genes such as HOXA10, HOXD10, HOXA11, HOXD11, and HOXA9, showing that fusion proteins in LG-ESSs target the HOX gene cluster and drawing parallels to our results from K562 cell lines. Different pathway enrichment analyses clearly identified genes regulated by Polycomb group proteins and H3K27me3 as up-regulated in these endometrial tumors (Supplemental Figs. S11, S12; Supplemental Table S3), hinting that the JAZF1-SUZ12 fusion complex mistargets activating marks at these genes. Altogether, our data strongly support a model in which the mislocalization of TIP60 activities could be the predominant oncogenic mechanism of the recurrent fusion proteins found in endometrial stromal sarcomas and also other sarcomas.
Discussion
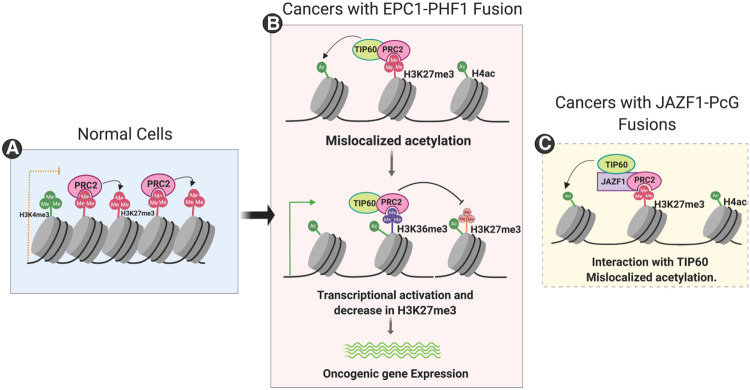
Our detailed biochemical work demonstrates that the EPC1-PHF1 and JAZF1-SUZ12 fusion proteins found in sarcomas assemble a megacomplex that merges two important chromatin-modifying activities with opposite functions in gene expression. Our results show that binding of this EPC1-PHF1/JAZF1-SUZ12 chimeric complex increases histone acetylation and H2A.Z incorporation within regions regulated by PRC2, notably within the HOXD gene cluster. This is correlated with increased gene expression, consistent with the notion that the fusions mistarget the HAT and histone exchange activities of NuA4/TIP60 to chromatin regions that are normally maintained in the silenced state (Fig. 6). Interestingly, mistargeting of histone acetylation at the HOXD cluster leads to loss of H3K27 trimethylation over an entire specific topologically associating domain (TAD) without affecting the neighboring ones, albeit also bound by PRC2 complexes. In theory, since the fusions are integrated in a megacomplex with PRC2.1, it could act at most PRC2.1 binding sites. We speculate that the presence of small pre-existing TIP60 binding sites within or in the vicinity of Polycomb/repressed regions—as we see at the HOXD cluster—could make a big difference. The TIP60 moiety of the EPC1-PHF1 megacomplex could be efficiently recruited there (by DNA-bound factors and/or small regions of H3K4me3 in bivalent chromatin), while the PRC2 moiety would allow it to spread along the region like PRC2 normally does, leading to de novo acetylation. This would lead to increased transcription, coupled with H3K36 trimethylation, which in turn inhibits the HMT activity of PRC2 (through recognition by the PHF1 Tudor domain), leading to a decrease of H3K27me3 over the region (Fig. 6, middle panel). It is important to note that H3K36 methylation has also been described as a potent boundary mark to block H3K27me3 spreading (Streubel et al. 2018). This mechanism would also explain why the clearer changes in histone marks and gene expression are found at EPC1-PHF1 mapped binding sites that overlap with both TIP60 and PRC2.1 (Fig. 2).
Figure 6.
Model for the oncogenic mechanism of EPC1-PHF1 fusion protein in soft tissue sarcomas. (A) In normal cells, the PRC2 complex occupies repressed or poised chromatin. (B) When EPC1-PHF1 fusion protein is expressed, it assembles a megacomplex combining NuA4/TIP60 and PRC2 complexes. The megacomplex occupies regions that have activating and repressive histone marks (such as the HOX clusters in K562) and mislocalizes NuA4/TIP60 activities (H4, H2A acetylation, and H2A.Z exchange). This tips the balance toward transcriptional activation. Levels of the transcription elongation-associated histone mark H3K36me3 increase and inhibit the deposition of the repressive H3K27me3 histone mark. Thus, the changes in chromatin landscape potentiate the expression of oncogenes. (C) Sarcomas with fusions of JAZF1 and PcG proteins also use a similar mechanism, since JAZF1 strongly interacts with the NuA4/TIP60 complex and mislocalizes its activities.
In addition, our study validates the protein JAZF1 as a potent transcription activator that stably associates with the NuA4/TIP60 complex. This information further integrates all of the reported translocations in low-grade endometrial stromal sarcomas (LG-ESSs) as producing a physical merge of TIP60 and PcG complexes, unifying the underlying molecular mechanism (Fig. 6, right panel). This is supported by the robust clustering of gene expression profiles of the LG-ESSs with various fusion proteins when compared with high-grade ESSs or tumors without fusion genes (Micci et al. 2016). Our findings further underline the importance of deregulated epigenetic modifiers in promoting oncogenesis, particularly in sarcomas (Nacev et al. 2020), in part through changes of the chromatin landscape over large genomic regions. The highly recurrent physical merger of TIP60 and PcG complexes in these sarcomas, through several distinct subunits, indicates that this event is the driving force in the oncogenic mechanism. It is somewhat reminiscent of the MLL fusion proteins in pediatric acute myeloid leukemia, in which the H3K4 HMT MLL is recurrently fused with diverse components of the superelongation complex, creating a large complex driving oncogenesis through altered transcription programs involving chromatin modifiers and readers as well as HOXA cluster misregulation (Mohan et al. 2010). A parallel can also be drawn to the discovery of various BRD4-linked Z4 complex proteins that are fused to NUT in NUT midline carcinoma (Shiota et al. 2018).
At the HOXD locus, the chromatin modification changes observed with EPC1-PHF1 spread over a large region that encompasses two domains within a larger TAD (Fig. 3J,K). This is especially interesting given recent studies demonstrating the nucleation and spreading of H3K27me3 through Polycomb domain contacts (Vieux-Rochas et al. 2015; Oksuz et al. 2018). Interestingly, EPC1-PHF1 localizes to the HOXD13/EVX2 nucleation region and mislocalizes H4 acetylation only in the centromeric TAD. We could also observe EPC1-PHF1 localization and H4 acetylation presence at “spreading” sites (Supplemental Fig. S7). A similar observation was made for the Ewing sarcoma fusion protein at the HOXD centromeric TAD leading to overexpression of HOXD13 as seen here (Svoboda et al. 2014; von Heyking et al. 2016). A question that emerges from our observation is whether mislocalization of activating marks will lead to disruption of these H3K27me3-centric repressive domains and disrupt the organization of chromatin domains.
Disruption of Polycomb-mediated repression by fusion proteins to activate stem cell-related gene expression programs has emerged as a common theme recently (Brien et al. 2019). Unlike in synovial sarcoma or NUT midline carcinoma, we did not observe extensive mislocalization/disruption in chromatin. This could be an artifact of our model system of study. Alternatively, the LG-ESS pathology is rare, milder, and not rapidly progressing and has limited metastasis. Hence, LG-ESSs may only deregulate few genes. Direct chromatin profiling in patient samples with the JAZF1-SUZ12 fusion will provide further evidence for this hypothesis.
Intriguingly, a newly described subunit of PRC2 complexes, EZHIP (CXORF67), was also found fused to the TIP60 subunit MBTD1 in LG-ESSs (Dewaele et al. 2014). EZHIP functions similarly to the oncogenic H3K27M mutant by binding to EZH2 and inhibiting its activity in trans (Hubner et al. 2019; Jain et al. 2019; Piunti et al. 2019). This mechanism seems similar to the trans inhibition of EZH2 through PHF1 Tudor-H3K36me3 binding and recently described direct inhibitory effect of H3K36me3 on EZH2 catalytic activity (Jani et al. 2019; Finogenova et al. 2020). The molecular characterization of MBTD1-EZHIP will further reveal whether parallels can be drawn with EPC1-PHF1/JAZF1-SUZ12.
Since endometrial stromal sarcomas and OFMTs are very rare, we were fortunate to get access to patient tissues. Although limited by sampling size, this is the first report of a gene expression comparison of a LG-ESS sample versus adjacent normal endometrial tissue. While the results clearly support the conclusions drawn from the model system, further analysis of more paired LG-ESS samples may shed light on the oncogenic signature of this rare cancer. Many pathways identified as enriched in our expression analysis of the two patient samples are implicated in oncogenesis of sarcomas (Supplemental Figs. S11, S12; Supplemental Table S3; Damerell et al. 2021).
The data presented in this study indicate that the mislocalization of NuA4/TIP60 leads to histone H4 acetylation and H2A.Z incorporation in chromatin, favoring local gene expression that can subsequently inhibit PRC2 activity. This succession of molecular events is likely the predominant oncogenic mechanism of the recurrent fusion proteins found in endometrial stromal sarcomas, ossifying fibromyxoid tumors, and other related sarcomas (Fig. 1A). Since the mutational burden of translocation-carrying cancers is relatively lower than other cancers (clonal homogeneity), targeted therapeutic approaches specific to the fusion protein are plausible (Taylor et al. 2011; Brien et al. 2019). According to our study, targeting TIP60 HAT activity with available small molecule inhibitors seems a viable approach (Brown et al. 2016). Alternatively, development of inhibitors designed to disrupt the interaction between JAZF1 and the TIP60 complex may also be a promising therapeutic approach.
Materials and methods
Cell culture
K562 cells were obtained from ATCC and maintained at 37°C under 5% CO2 in RPMI medium supplemented with 10% fetal bovine serum/newborn calf serum and GlutaMAX. HEPES-NaOH (25 mM at pH 7.4) was added during culture in Spinner flasks.
HEK293 cells were obtained from ATCC and maintained at 37°C under 5% CO2 in DMEM medium supplemented with 10% fetal bovine serum.
Construction of recombinant DNA
RT-PCR product from total RNA isolated from surgically removed endometrial stromal sarcoma tumors was a kind gift from Dr. Fransesca Micci (Micci et al. 2006). The cDNA contained the entire truncation of EPC1 and a fused portion of PHF1 up to base pair 139; the included portion of PHF1 contained a BglI restriction site, which was then used to reconstruct the full-length fusion gene using a clone of PHF1 obtained from Open Biosystems. This construct was then subcloned into the different expression plasmids. Human cDNA of JAZF1 was purchased from GE Healthcare. The open reading frame (ORF) of JAZF1 was subcloned into vectors and used to construct the truncation and the JAZF1-SUZ12 fusion by PCR. All constructs were verified by sequencing, and their expression was tested by transient transfection.
Cell line generation
EPC1-PHF1, JAZF1-SUZ12, and controls were targeted to the AAVS1 safe harbor locus in K562 cells using zinc finger nuclease (Hockemeyer et al. 2009) as described previously by Dalvai et al. (2015). For lentiviral overexpression, plasmids expressing proteins of interest were cotransfected with vesicular stomatitis virus G (VSV-G) plasmid DNA and Gag-Pol-Tet-Rev plasmid DNA into HEK293T cells using polyethyleneimine (PEI). Forty-eight hours to 72 h after transfection, supernatants containing lentivirus were harvested, and K562 cells were transduced in the presence of 8 µg/mL hexadimethrine bromide (polybrene; Sigma) at an MOI of 0.3. Two days after transduction, cells were selected with 0.5 µg/mL puromycin to get stable cell lines.
Purification of complexes
Native complexes were purified as described in detail before by Dalvai et al. (2015) and Doyon and Côté (2016). The purified complexes were loaded on NuPAGE 4%–12% Bis-Tris gels (Invitrogen) and visualized by silver staining. Fractions were then analyzed by mass spectrometry (details are in the Supplemental Material).
Affinity purification followed by immunoblotting
K562 cell lines were expanded to get 6 million to 8 million cells. Cells were collected, washed twice in 1× PBS (phosphate-buffered saline), and lysed for 30 min in 2× volume lysis buffer (450 mM NaCl, 10% glycerol, 50 mM Tris-HCl at pH 8, 1% Triton X-100, 2 mM MgCl2, 0.1 mM ZnCl2, 2 mM EDTA, 1 mM DTT, protease inhibitors), and the same volume of lysis buffer without salt was added to get a final salt concentration of 225 mM. Fractions were centrifuged to prepare whole-cell extracts. The extracts were incubated with FLAG-M2 agarose resin (Sigma) for 4 h at 4°C. The resin was centrifuged, washed in lysis buffer with 225 mM salt, and eluted with 3xFLAG peptide (Sigma). The eluted fraction was loaded onto 4%–15% gradient gels with input and immunoblotted with the appropriate antibodies.
ChIP and ChIP sequencing
FLAG ChIPs were performed according to the protocol described by Jacquet et al. (2016). Histone modification ChIPs were performed according to the protocol described by Lalonde et al. (2013). Libraries for sequencing were prepared as described (Jacquet et al. 2016). Samples were sequenced by 50-bp single reads on a HiSeq 2000 platform (Illumina). Reads from ChIP-seq experiments were obtained from the Genome Innovation Center at McGill University. Raw reads were trimmed using fastp v0.20.1 (Chen et al. 2018b). Quality check was performed on raw and trimmed data to ensure the quality of the reads using FastQC v0.11.7 (Andrews 2010) and MultiQC v1.5 (Ewels et al. 2016). Trimmed reads were aligned on the human genome (hg18) using bwa mem v0.7.17 (Li 2013) and SAMtools v1.13 (Li et al. 2009). Raw signal tracks and normalized tracks (in reads per million [RPM]) were produced from mapped reads using deepTools’ v2.17.0 bamCoverage tool (Ramírez et al. 2014) and BEDTools’ genomecov tool (Quinlan and Hall 2010), respectively. Tracks were converted to the bigwig format using bedGraphToBigWig v2.8 (Kent et al. 2010). MACS2 v2.2.1 software (Feng et al. 2012) was used to perform the peak calling. EPC1-PHF1 peak regions were separated into four partitions based on overlap with other factors (EPC1-PHF1 alone, EPC1-PHF1 and PHF1, EPC1-PHF1 and EPC1[1–581], and EPC1-PHF1, PHF1, and EPC1[1–581]). The peaks were annotated using the annotatePeak function in the ChipSeeker package v1.26.2 (Yu et al. 2015) in R v4.0.3 (https://www.r-project.org). Annotation was performed using the closest promoter to the EPC1-PHF1 binding site. Gene expression data corresponding to peak regions were extracted from the microarray data and then separated according to the previous partitions. Metaplots and graphical representations were produced with the ggplot2 v3.3.5 package (Wickham 2011). Box plot statistics were computed using bins; n = number of regions analyzed, where one region is 100 bins. Box plots and associated Wilcoxon tests were generated using the ggboxplot and stat_ compare_means functions, respectively, with the default settings of the ggpubr v0.4.0 package (https://CRAN.R-project.org/package=ggpubr). Heat maps were generated using ComplexHeatmap v2.6.2 (Gu et al. 2016) and EnrichedHeatmap v1.20.0 (Gu et al. 2018) packages.
Quantitative real-time PCRs were performed on a LightCycler 480 (Roche) with SYBR Green I (Roche) to confirm the specific enrichment at defined loci. The error bars represent standard errors based on two independent experiments. Quantitative real-time PCR primers used are listed in Supplemental Table S1.
CUT&RUN sequencing
The experiment was performed as detailed by Skene et al. (2018) with some modifications. K562 cells (0.5 million) were pelleted and resuspended in 1× PBS. Cells were cross-linked with 0.1% formaldehyde for 1 min. Cross-linked cells were washed at room temperature and bound to 10 μL of concanavalin A bead slurry. The cell and bead suspension was incubated in buffer containing 0.05% digitonin and 0.5 µg of anti-HA (Epicypher 13-2010) antibody overnight at 4°C. Afterward, the beads were washed with digitonin buffer, resuspended in buffer containing pAG-MNase ( 1:20 dilution; CUTANA EpiCypher) and digitonin, and incubated for 10 min with agitation. Beads were washed and resuspended in ice-cold digitonin buffer. CaCl2 (1 mM) was added and incubated for 2 h at 4°C. The reaction was stopped by addition of stop buffer containing EDTA and EGTA and incubated for 10 min at 37°C. Supernatant containing released chromatin was separated and decross-linked overnight at 55°C. DNA was purified with NEB Monarch PCR and DNA purification kit following the protocol enriching for short DNA fragments. DNA was quantified by Qubit HS DNA kit. The library was prepared following the manufacturer's instructions in NEBNext Ultra II DNA kit for low-input ChIP. One-hundred-base-pair paired-end sequencing was performed with the Illumina NovaSeq 6000 system. Raw reads were trimmed using fastp v0.21.0 (Chen et al. 2018b). Trimmed reads were aligned on the human genome (hg38) using bwa mem v0.7.17 (Li 2013) and SAMtools v1.13 (Li et al. 2009). Raw signal tracks and normalized tracks (in reads per million [RPM]) were produced from mapped reads using deepTools’ v2.17.0 bamCoverage tool (Ramírez et al. 2014) and BEDTools’ genomecov tool (Quinlan and Hall 2010), respectively. Tracks were converted to the bigwig format using bedGraphToBigWig v2.8 (Kent et al. 2010). MACS2 v2.2.1 software (Feng et al. 2012) was used to perform the peak calling with the following parameters: no λ, fragment size to 14, mfold from 5 to 50, and keeping all duplicated tags. The CUT&RUN files used in the heat map were lifted over to UCSC hg18 using UCSC lift over tool (Kuhn et al. 2013). Heat maps were generated using ComplexHeatmap v2.6.2 (Gu et al. 2016) and EnrichedHeatmap v1.20.0 packages (Gu et al. 2018), as described above for ChIP sequencing data.
HAT and HMT assays
Fractions of purified complexes were assayed for enzymatic activity on short oligonucleosomes and free histones isolated from HeLa S3 cells as described previously (Utley et al. 1996; Doyon et al. 2004; Musselman et al. 2012).
For the histone acetyltransferase (HAT) assays, 0.5 µg of the indicated substrates was incubated in a 15-µL reaction containing 50 mM Tris-HCl (pH 8.0), 5% glycerol, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate, and 0.125 µCi of [3H]-labeled acetyl-CoA (Perkin Elmer) or 0.15 mM unlabeled acetyl-CoA (Sigma) for 30 min at 30°C. Samples were either spotted on P81 membrane (GE Healthcare) for scintillation count or analyzed on 15% SDS-PAGE that was Coomassie-stained followed by treatment with Enhance (Perkin Elmer) and fluorography.
For the histone methyltransferase (HMT) assays, 0.5 µg of the indicated substrates was incubated in a 15-µL reaction containing 20 mM Tris-HCl (pH 8.0), 5% glycerol, 0.02 mM EDTA, 1 mM DTT, 1 mM PMSF, 50 mM KCl, and 0.55 µCi of [3H]AdoMet (S-adenosyl-L-[methyl-3H] methionine)/SAM (Perkin Elmer) or 0.05 mM unlabeled S-adenosyl methionine for 45 min at 30°C. Samples were either spotted on P81 membrane (GE Healthcare) for scintillation count or analyzed on 15% SDS-PAGE that was Coomassie-stained followed by treatment with Enhance (Perkin Elmer) and fluorography.
The histones acetyltransferase (HAT) assays and histones methyltransferase (HMT) assays with recombinant nucleosome core particles (rNCP) were performed in a volume of 15 µL as described above but using 0.5 µg of reconstituted/recombinant H3.3 mononucleosomes (unmodified or H3.3K36me3; Epicypher 16-0390). The amount of purified enzyme complex used was normalized by Western blotting to obtain similar amounts of TIP60 complex in HAT reactions or PRC2 complex in HMT reactions. For HAT, 2 µL of mock, 2 µL of EPC1-PHF1, and 0.635 µL of EPC1(1–581) were used. For HMT, 5 µL of mock, 5 µL of EPC1-PHF1, and 1.875 µL of PHF1 were used.
Recruitment activator assay
The assay was performed as described by Alerasool et al. (2022), Briefly, the HEK293T TRE3G-EGFP reporter cell line with ABI-dCas9 and a gRNA targeting seven tetO repeats in the TRE3G promoter was generated and a clone showing robust EGFP induction by a strong transcriptional activator VPR was selected for downstream assays. Ninety-six-well plates were seeded with 3 × 104 cells per well 1 d prior to transfection. One-hundred-fifty nanograms of each construct was transfected using polyethylemine (PEI). Transfected cells were induced 24 h after transfection by treatment with 100 µM abscisic acid. Forty-eight hours after induction, cells were dissociated and resuspended in flow buffer using a liquid handling robot and analyzed by LSRFortessa (BD). Flow cytometry data were analyzed using FlowJo by gating for positive gRNA (EBFP2), and then further for construct (TagRFP) expression. At least 25,000 cells were analyzed for each replicate.
Microarray
RNA samples were extracted using TRIzol reagent (Invitrogen) following the manufacturer's instructions. Duplicate RNA samples from EPC1-PHF1-expressing and control K562 cell lines were compared. We performed gene expression microarray experiments using the human Illumina HumanHT-12_V4 platform. We first log2-transformed and quantile-normalized the data before using them for the analyses presented here. All the data were preprocessed using the lumi Bioconductor package (Du et al. 2008).
Patient tissue samples
This study was approved by the Research Ethics Board of the University Health Network in Toronto, ON, Canada. Low-grade endometrial stromal sarcoma biobanked specimens (frozen) were obtained with broad consent. Fluorescence in situ hybridization (FISH) was performed to identify rearrangement involving JAZF1 and SUZ12.
mRNA sequencing
RNA from K562 cell lines was extracted using the Monarch total RNA purification kit (NEB) according to the manufacturer's instructions. RNA from frozen tissue samples were extracted using the miRNEasy micro kit (Qiagen). Only samples with RIN >7 were used for library preparation using the NEBNext Ultra II directional RNA library + NEBNext poly(A) mRNA dual-index kit. The sequencing run was performed on an Illumina NovaSeq 6000 system. One-hundred-base-pair paired-end reads were trimmed using fastp v0.20.1 (Chen et al. 2018b). Quality check was performed on raw and trimmed data to ensure the quality of the reads using FastQC v0.11.7 (Andrews 2010) and MultiQC v1.5 (Ewels et al. 2016). The quantification was performed with Kallisto v0.46.2 (Bray et al. 2016) against the human genome (hg38). The volcano graphical representations were produced with the Bioconductor package Enhanced Volcano (Blighe et al. 2018). Differential expression analysis was also performed using the DESeq2 v1.30.1 package (Love et al. 2014). All R analyses were done in R v4.0.3 (https://www.r-project.org). Fusion transcripts were identified using the STAR fusion pipeline (Haas et al. 2019). GSEA and enrichment were carried out using clusterProfiler version v4.2.2 (Padj cutoff value = 0.05).
Hi-C data alignment
Alignment of the ChIP sequencing data with previously published Hi-C data in K562 cells (Rao et al. 2014) was performed by converting the ChIP sequencing alignment from hg18 to hg19 using the CrossMap tool (Zhao et al. 2014). Hi-C data and ChIP sequencing data were then visualized using PyGenome Tracks tool V2.1 (Ramírez et al. 2018). The CTCF ChIP sequencing data used are from GEO: GSM733719 (Broad Institute/ENCODE group). The Hi-C data (heat map, domains, and loops) were downloaded from the data sets provided at Chorogenome (http://chorogenome.ie-freiburg.mpg.de/data_sources.html).
Data and software availability
NGS assays reported in this study are available at the GEO repository under the following accession numbers: ChIP-seq data and microarray expression analysis (GSE162544), CUT&RUN data (GSE196754), JAZF1-SUZ12 K562 RNA-seq (GSE196755), and patient sample RNA-seq (GSE196757).
All mass spectrometry files generated as part of this study were deposited at MassIVE (http://massive.ucsd.edu). The MassIVE IDs are MSV000083618, MSV000086476, and MSV000088749. The MassIVE FTP download links are ftp://massive.ucsd.edu/MSV000083618, ftp://massive.ucsd.edu/MSV000086476, and ftp://MSV000088749@massive.ucsd.edu.
Supplementary Material
Acknowledgments
We thank Céline Roques, Valérie Côté, and Philippe Cloutier for important technical support. We are very grateful to Professor Francesca Micci for providing the cDNA obtained from patient samples that covered the EPC1-PHF1 fusion. We thank Compute Canada for the use of supercomputers, McGill Genome Center for sequencing/expression microarray, and the Centre Hospitalier Universitaire de Québec-Université Laval proteomic platform. Models were created using Biorender.com. This work was supported by grants from the Canadian Institutes of Health Research (CIHR; FDN-143314) to J.C., the Government of Québec, Ministry of Economy and Innovation to B.C., and the Natural Sciences and Engineering Research Council (NSERC) of Canada to Y.D. (RGPIN-2014-059680) and J.-P.L. (RGPIN-2017-06124), and University of Toronto startup funds to M.T. D.S., M.-E.L., and K.J. were supported by PhD studentships from Fonds de la Recherche Québec-Santé (FRQS) and Fonds de la Recherche Québec-Nature/Technologie (FRQNT). N. Avvakumov was supported by a CIHR postdoctoral fellowship, and A.M. was supported by MSc studentships from NSERC and FRQNT. B.C. holds the Institut de Recherches Cliniques de Montréal Bell-Bombardier Research Chair. A.-C.G. holds the Canada Research Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics. J.-P.L. and Y.D. are Junior 2 FRQS scholars. J.C. holds the Canada Research Chair in Chromatin Biology and Molecular Epigenetics.
Author contributions: D.S., N. Avvakumov, M.-E.L., B.C., M.T., Y.D., and J.C. designed the experiments. D.S., N. Avvakumov, M.-E.L., N. Alerasool, A.M., K.J., C.L., J.-P.L., J.R., J.L., and Y.D. performed the experiments. C.J.-B., E.P., L.H., and S.T.S. analyzed the genomic data. M.Q.B. and M.R. provided the patient samples. J.-P.L., A.-C.G., B.C., M.T., Y.D., and J.C. supervised and secured funding. D.S. and J.C. wrote the manuscript with the help of coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348982.121.
Competing interest statement
The authors declare no competing interests.
References
- Alerasool N, Leng H, Lin ZY, Gingras AC, Taipale M. 2022. Identification and functional characterization of transcriptional activators in human cells. Mol Cell 82: 677–695.e7. 10.1016/j.molcel.2021.12.008 [DOI] [PubMed] [Google Scholar]
- Allen AJ, Ali SM, Gowen K, Elvin JA, Pejovic T. 2017. A recurrent endometrial stromal sarcoma harbors the novel fusion JAZF1-BCORL1. Gynecol Oncol Rep 20: 51–53. 10.1016/j.gore.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Sung YS, Chen CL, Zhang L, Chen HW, Singer S, Agaram NP, Sboner A, Fletcher CD. 2014. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors—molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer 53: 183–193. 10.1002/gcc.22132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Côté J. 2013. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta 1819: 290–302. 10.1016/j.bbagrm.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Blighe K, DeDionisio L, Christie KA, Chawes B, Shareef S, Kakouli-Duarte T, Chao-Shern C, Harding V, Kelly RS, Castellano L, et al. 2018. Gene editing in the context of an increasingly complex genome. BMC Genomics 19: 595. 10.1186/s12864-018-4963-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompadre O, Andrey G. 2019. Chromatin topology in development and disease. Curr Opin Genet Dev 55: 32–38. 10.1016/j.gde.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17: 1415–1428. 10.1101/gad.1056603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brien GL, Stegmaier K, Armstrong SA. 2019. Targeting chromatin complexes in fusion protein-driven malignancies. Nat Rev Cancer 19: 255–269. 10.1038/s41568-019-0132-x [DOI] [PubMed] [Google Scholar]
- Brown JA, Bourke E, Eriksson LA, Kerin MJ. 2016. Targeting cancer using KAT inhibitors to mimic lethal knockouts. Biochem Soc Trans 44: 979–986. 10.1042/BST20160081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, et al. 2013. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49: 571–582. 10.1016/j.molcel.2012.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. 2008. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 28: 1862–1872. 10.1128/MCB.01589-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jiao L, Shubbar M, Yang X, Liu X. 2018a. Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol Cell 69: 840–852.e5. 10.1016/j.molcel.2018.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018b. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Jobin-Robitaille O, Billon P, Buisson R, Niu H, Lacoste N, Abshiru N, Côté V, Thibault P, Kron SJ, et al. 2018. Phospho-dependent recruitment of the yeast NuA4 acetyltransferase complex by MRX at DNA breaks regulates RPA dynamics during resection. Proc Natl Acad Sci 115: 10028–10033. 10.1073/pnas.1806513115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Bachmann AL, Tauscher K, Benda C, Fierz B, Müller J. 2017. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat Struct Mol Biol 24: 1039–1047. 10.1038/nsmb.3488 [DOI] [PubMed] [Google Scholar]
- Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Côté J, Doyon Y. 2015. A Scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep 13: 621–633. 10.1016/j.celrep.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Damerell V, Pepper MS, Prince S. 2021. Molecular mechanisms underpinning sarcomas and implications for current and future therapy. Signal Transduc Target Ther 6: 246. 10.1038/s41392-021-00647-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele B, Przybyl J, Quattrone A, Finalet Ferreiro J, Vanspauwen V, Geerdens E, Gianfelici V, Kalender Z, Wozniak A, Moerman P, et al. 2014. Identification of a novel, recurrent MBTD1-CXorf67 fusion in low-grade endometrial stromal sarcoma. Int J cancer 134: 1112–1122. 10.1002/ijc.28440 [DOI] [PubMed] [Google Scholar]
- Donaldson-Collier MC, Sungalee S, Zufferey M, Tavernari D, Katanayeva N, Battistello E, Mina M, Douglass KM, Rey T, Raynaud F, et al. 2019. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet 51: 517–528. 10.1038/s41588-018-0338-y [DOI] [PubMed] [Google Scholar]
- Doyon Y, Côté J. 2016. Preparation and analysis of native chromatin-modifying complexes. Meth Enzymol 573: 303–318. 10.1016/bs.mie.2016.01.017 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Côté J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24: 1884–1896. 10.1128/MCB.24.5.1884-1896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. 2008. lumi: a pipeline for processing illumina microarray. Bioinformatics 24: 1547–1548. 10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174. 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. 10.1038/nprot.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Félix A, Lennerz JK, Oliva E. 2018. Recent advances in the histological and molecular classification of endometrial stromal neoplasms. Virchows Arch 473: 665–678. 10.1007/s00428-018-2470-6 [DOI] [PubMed] [Google Scholar]
- Finogenova K, Bonnet J, Poepsel S, Schäfer IB, Finkl K, Schmid K, Litz C, Strauss M, Benda C, Müller J. 2020. Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. Elife 9: e61964. 10.7554/eLife.61964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. 2007. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448: 1063–1067. 10.1038/nature06055 [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M, Ishaque N. 2018. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics 19: 234. 10.1186/s12864-018-4625-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Dobin A, Li B, Stransky N, Pochet N, Regev A. 2019. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol 20: 213. 10.1186/s13059-019-1842-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang L, Chiang S, Lee CH. 2018. Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology 50: 162–177. 10.1016/j.pathol.2017.11.086 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857. 10.1038/nbt.1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner JM, Muller T, Papageorgiou DN, Mauermann M, Krijgsveld J, Russell RB, Ellison DW, Pfister SM, Pajtler KW, Kool M. 2019. EZHIP / CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro-Oncology 21: 878–889. 10.1093/neuonc/noz058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, et al. 2016. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol Cell 62: 409–421. 10.1016/j.molcel.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, Bajic A, Juretic N, Deshmukh S, Venneti S, et al. 2019. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun 10: 2146. 10.1038/s41467-019-09981-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani KS, Jain SU, Ge EJ, Diehl KL, Lundgren SM, Müller MM, Lewis PW, Muir TW. 2019. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc Natl Acad Sci 116: 8295–8300. 10.1073/pnas.1819029116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judes G, Rifaï K, Ngollo M, Daures M, Bignon YJ, Penault-Llorca F, Bernard-Gallon D. 2015. A bivalent role of TIP60 histone acetyl transferase in human cancer. Epigenomics 7: 1351–1363. 10.2217/epi.15.76 [DOI] [PubMed] [Google Scholar]
- Kasinath V, Beck C, Sauer P, Poepsel S, Kosmatka J, Faini M, Toso D, Aebersold R, Nogales E. 2021. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371: eabc3393. 10.1126/science.abc3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiita A, Godbersen S, Araldi E, Ghoshdastider U, Schmid MW, Spinas G, Moch H, Stoffel M. 2020. The diabetes gene JAZF1 is essential for the homeostatic control of ribosome biogenesis and function in metabolic stress. Cell Rep 32: 107846. 10.1016/j.celrep.2020.107846 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. 2013. The UCSC genome browser and associated tools. Brief Bioinformatics 14: 144–161. 10.1093/bib/bbs038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Ji F, Sunwoo H, Jain G, Lee JT, Sadreyev RI, Dekker J, Kingston RE. 2017. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol Cell 65: 432–446.e5. 10.1016/j.molcel.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, et al. 2013. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev 27: 2009–2024. 10.1101/gad.223396.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde ME, Cheng X, Côté J. 2014. Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev 28: 1029–1041. 10.1101/gad.236331.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Højfeldt JW, Helin K. 2019. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell 74: 8–18. 10.1016/j.molcel.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallée-Adam M, Rousseau J, Domecq C, Bouchard A, Forget D, Faubert D, Blanchette M, Coulombe B. 2013. Discovery of cell compartment specific protein–protein interactions using affinity purification combined with tandem mass spectrometry. J Proteome Res 12: 272–281. 10.1021/pr300778b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, Shore D. 2009. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell 33: 704–716. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 10.48550/arXiv.1303.3997 [DOI] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al. 2017. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549: 287–291. 10.1038/nature23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao ZZ, Wang YD, Qi XY, Xiao XH. 2019. JAZF1, a relevant metabolic regulator in type 2 diabetes. Diabetes Metab Res Rev 35: e3148. 10.1002/dmrr.3148 [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonfat N, Duboule D. 2015. Structure, function and evolution of topologically associating domains (TADs) at HOX loci. FEBS Lett 589: 2869–2876. 10.1016/j.febslet.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Loubiere V, Martinez AM, Cavalli G. 2019. Cell fate and developmental regulation dynamics by polycomb proteins and 3D genome architecture. Bioessays 41: e1800222. 10.1002/bies.201800222 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Wang J, Wang J, Ma CX, Gao X, Patriub V, Sklar JL. 2017. The JAZF1-SUZ12 fusion protein disrupts PRC2 complexes and impairs chromatin repression during human endometrial stromal tumorogenesis. Oncotarget 8: 4062–4078. 10.18632/oncotarget.13270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci 101: 14315–14322. 10.1073/pnas.0405353101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci F, Panagopoulos I, Bjerkehagen B, Heim S. 2006. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res 66: 107–112. 10.1158/0008-5472.CAN-05-2485 [DOI] [PubMed] [Google Scholar]
- Micci F, Gorunova L, Agostini A, Johannessen LE, Brunetti M, Davidson B, Heim S, Panagopoulos I. 2016. Cytogenetic and molecular profile of endometrial stromal sarcoma. Genes Chromosomes Cancer 55: 834–846. 10.1002/gcc.22380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10: 721–728. 10.1038/nrc2915 [DOI] [PubMed] [Google Scholar]
- Momeni-Boroujeni A, Mohammad N, Wolber R, Yip S, Köbel M, Dickson BC, Hensley ML, Leitao MM Jr, Antonescu CR, Benayed R, et al. 2021. Targeted RNA expression profiling identifies high-grade endometrial stromal sarcoma as a clinically relevant molecular subtype of uterine sarcoma. Mod Pathol 34: 1008–1016. 10.1038/s41379-020-00705-6 [DOI] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff J, et al. 2012. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19: 1266–1272. 10.1038/nsmb.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacev BA, Jones KB, Intlekofer AM, Yu JSE, Allis CD, Tap WD, Ladanyi M, Nielsen TO. 2020. The epigenomics of sarcoma. Nat Rev Cancer 20: 608–623. 10.1038/s41568-020-0288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Fujino S, Nakanishi G, Kim YS, Jetten AM. 2004. TIP27: a novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acids Res 32: 4194–4204. 10.1093/nar/gkh741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. 2018. Capturing the onset of PRC2-mediated repressive domain formation. Mol Cell 70: 1149–1162.e5. 10.1016/j.molcel.2018.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Smith ER, Morgan MAJ, Ugarenko M, Khaltyan N, Helmin KA, Ryan CA, Murray DC, Rickels RA, Yilmaz BD, et al. 2019. CATACOMB: an endogenous inducible gene that antagonizes H3K27 methylation activity of polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv 5: eaax2887. 10.1126/sciadv.aax2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan SK, Su T, Yen L, Jacquet K, Huang C, Côté J, Kurdistani SK, Carey MF. 2016. EP400 deposits H3.3 into promoters and enhancers during gene activation. Mol Cell 61: 27–38. 10.1016/j.molcel.2015.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procida T, Friedrich T, Jack APM, Peritore M, Bönisch C, Eberl HC, Daus N, Kletenkov K, Nist A, Stiewe T, et al. 2021. JAZF1, a novel p400/TIP60/NuA4 complex member, regulates H2A.Z acetylation at regulatory regions. Int J Mol Sci 22: 678. 10.3390/ijms22020678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyl J, Kidzinski L, Hastie T, Debiec-Rychter M, Nusse R, van de Rijn M. 2018. Gene expression profiling of low-grade endometrial stromal sarcoma indicates fusion protein-mediated activation of the Wnt signaling pathway. Gynecol Oncol 149: 388–393. 10.1016/j.ygyno.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravens S, Yu C, Ye T, Stierle M, Tora L. 2015. Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells. Epigenetics Chromatin 8: 45. 10.1186/s13072-015-0039-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6: 1297–1307. 10.1016/S1097-2765(00)00128-3 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Carballo E, Lopez-Delisle L, Zhan Y, Fabre PJ, Beccari L, El-Idrissi I, Huynh THN, Ozadam H, Dekker J, Duboule D. 2017. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev 31: 2264–2281. 10.1101/gad.307769.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto D, Cramet M, Wang AY, Steunou AL, Lacoste N, Schulze JM, Côté V, Monnet-Saksouk J, Piquet S, Nourani A, et al. 2014. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J 33: 1397–1415. 10.15252/embj.201386433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. 2008. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 28: 2718–2731. 10.1128/MCB.02017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Schneider N, Fisher C, Thway K. 2016. Ossifying fibromyxoid tumor: morphology, genetics, and differential diagnosis. Ann Diagn Pathol 20: 52–58. 10.1016/j.anndiagpath.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Schoolmeester JK, Sukov WR, Maleszewski JJ, Bedroske PP, Folpe AL, Hodge JC. 2013. JAZF1 rearrangement in a mesenchymal tumor of nonendometrial stromal origin: report of an unusual ossifying sarcoma of the heart demonstrating JAZF1/PHF1 fusion. Am J Surg Pathol 37: 938–942. 10.1097/PAS.0b013e318282da9d [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. 2017. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171: 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. 2005. The Saccharomyces cerevisiae piccolo NuA4 histone acetyltransferase complex requires the enhancer of polycomb a domain and chromodomain to acetylate nucleosomes. Mol Cell Biol 25: 5535–5542. 10.1128/MCB.25.13.5535-5542.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, Ahmad S, Dalwadi U, Steunou AL, Lu S, Ross JD, Dong MQ, Côté J, Yip CK. 2018. Molecular architecture of the essential yeast histone acetyltransferase complex NuA4 redefines its multimodularity. Mol Cell Biol 38: e00570-17. 10.1128/MCB.00570-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh BN, Akhtar A. 2019. The many lives of KATs—detectors, integrators and modulators of the cellular environment. Nat Rev Genet 20: 7–23. 10.1038/s41576-018-0072-4 [DOI] [PubMed] [Google Scholar]
- Shen H, Laird PW. 2013. Interplay between the cancer genome and epigenome. Cell 153: 38–55. 10.1016/j.cell.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota H, Elya JE, Alekseyenko AA, Chou PM, Gorman SA, Barbash O, Becht K, Danga K, Kuroda MI, Nardi V, et al. 2018. ‘Z4’ complex member fusions in NUT carcinoma: implications for a novel oncogenic mechanism. Mol Cancer Res 16: 1826–1833. 10.1158/1541-7786.MCR-18-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, Henikoff S. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13: 1006–1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Steunou A-L, Rossetto D, Côté J. 2014. Regulating chromatin by histone acetylation. In Fundamentals of chromatin (ed. Workman JL, Abmayr SM), pp. 147–212. Springer, New York. [Google Scholar]
- Streubel G, Watson A, Jammula SG, Scelfo A, Fitzpatrick DJ, Oliviero G, McCole R, Conway E, Glancy E, Negri GL, et al. 2018. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell 70: 371–379.e5. 10.1016/j.molcel.2018.02.027 [DOI] [PubMed] [Google Scholar]
- Svoboda LK, Harris A, Bailey NJ, Schwentner R, Tomazou E, von Levetzow C, Magnuson B, Ljungman M, Kovar H, Lawlor ER. 2014. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics 9: 1613–1625. 10.4161/15592294.2014.988048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. 2019. Principles of genome folding into topologically associating domains. Sci Adv 5: eaaw1668. 10.1126/sciadv.aaw1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. 2011. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer 11: 541–557. 10.1038/nrc3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Owen-Hughes TA, Juan LJ, Côté J, Adams CC, Workman JL. 1996. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Meth Enzymol 274: 276–291. 10.1016/S0076-6879(96)74024-7 [DOI] [PubMed] [Google Scholar]
- Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. 2015. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci 112: 4672–4677. 10.1073/pnas.1504783112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heyking K, Roth L, Ertl M, Schmidt O, Calzada-Wack J, Neff F, Lawlor ER, Burdach S, Richter GH. 2016. The posterior HOXD locus: its contribution to phenotype and malignancy of Ewing sarcoma. Oncotarget 7: 41767–41780. 10.18632/oncotarget.9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ahmad S, Zhang Z, Côté J, Cai G. 2018. Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat Commun 9: 1147. 10.1038/s41467-018-03504-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2011. ggplot2. WIREs Comp Stat 3: 180–185. 10.1002/wics.147 [DOI] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110. 10.1016/j.cell.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J, Dai J, Zhu P, Chen Z. 2016. The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol Cell 63: 965–975. 10.1016/j.molcel.2016.07.024 [DOI] [PubMed] [Google Scholar]
- Youmans DT, Gooding AR, Dowell RD, Cech TR. 2021. Competition between PRC2.1 and 2.2 subcomplexes regulates PRC2 chromatin occupancy in human stem cells. Mol Cell 81: 488–501.e9. 10.1016/j.molcel.2020.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, He QY. 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. 2013. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20: 259–266. 10.1038/nsmb.2470 [DOI] [PubMed] [Google Scholar]
- Zhang H, Devoucoux M, Song X, Li L, Ayaz G, Cheng H, Tempel W, Dong C, Loppnau P, Côté J, et al. 2020. Structural basis for EPC1-mediated recruitment of MBTD1 into the NuA4/TIP60 acetyltransferase complex. Cell Rep 30: 3996–4002.e4. 10.1016/j.celrep.2020.03.003 [DOI] [PubMed] [Google Scholar]
- Zhao H, Sun Z, Wang J, Huang H, Kocher JP, Wang L. 2014. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30: 1006–1007. 10.1093/bioinformatics/btt730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Xu X, Wang H, Yang G, Yang M, Zhao X, Guo H, Song J, Zheng H, Zhu Z, et al. 2020. Effect of central JAZF1 on glucose production is regulated by the PI3K-Akt-AMPK pathway. FASEB J 34: 7058–7074. 10.1096/fj.201901836RR [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS assays reported in this study are available at the GEO repository under the following accession numbers: ChIP-seq data and microarray expression analysis (GSE162544), CUT&RUN data (GSE196754), JAZF1-SUZ12 K562 RNA-seq (GSE196755), and patient sample RNA-seq (GSE196757).
All mass spectrometry files generated as part of this study were deposited at MassIVE (http://massive.ucsd.edu). The MassIVE IDs are MSV000083618, MSV000086476, and MSV000088749. The MassIVE FTP download links are ftp://massive.ucsd.edu/MSV000083618, ftp://massive.ucsd.edu/MSV000086476, and ftp://MSV000088749@massive.ucsd.edu.