Abstract
Background
Real-time prediction is key to prevention and control of infections associated with health-care settings. Contacts enable spread of many infections, yet most risk prediction frameworks fail to account for their dynamics. We developed, tested, and internationally validated a real-time machine-learning framework, incorporating dynamic patient-contact networks to predict hospital-onset COVID-19 infections (HOCIs) at the individual level.
Methods
We report an international retrospective cohort study of our framework, which extracted patient-contact networks from routine hospital data and combined network-derived variables with clinical and contextual information to predict individual infection risk. We trained and tested the framework on HOCIs using the data from 51 157 hospital inpatients admitted to a UK National Health Service hospital group (Imperial College Healthcare NHS Trust) between April 1, 2020, and April 1, 2021, intersecting the first two COVID-19 surges. We validated the framework using data from a Swiss hospital group (Department of Rehabilitation, Geneva University Hospitals) during a COVID-19 surge (from March 1 to May 31, 2020; 40 057 inpatients) and from the same UK group after COVID-19 surges (from April 2 to Aug 13, 2021; 43 375 inpatients). All inpatients with a bed allocation during the study periods were included in the computation of network-derived and contextual variables. In predicting patient-level HOCI risk, only inpatients spending 3 or more days in hospital during the study period were examined for HOCI acquisition risk.
Findings
The framework was highly predictive across test data with all variable types (area under the curve [AUC]-receiver operating characteristic curve [ROC] 0·89 [95% CI 0·88–0·90]) and similarly predictive using only contact-network variables (0·88 [0·86–0·90]). Prediction was reduced when using only hospital contextual (AUC-ROC 0·82 [95% CI 0·80–0·84]) or patient clinical (0·64 [0·62–0·66]) variables. A model with only three variables (ie, network closeness, direct contacts with infectious patients [network derived], and hospital COVID-19 prevalence [hospital contextual]) achieved AUC-ROC 0·85 (95% CI 0·82–0·88). Incorporating contact-network variables improved performance across both validation datasets (AUC-ROC in the Geneva dataset increased from 0·84 [95% CI 0·82–0·86] to 0·88 [0·86–0·90]; AUC-ROC in the UK post-surge dataset increased from 0·49 [0·46–0·52] to 0·68 [0·64–0·70]).
Interpretation
Dynamic contact networks are robust predictors of individual patient risk of HOCIs. Their integration in clinical care could enhance individualised infection prevention and early diagnosis of COVID-19 and other nosocomial infections.
Funding
Medical Research Foundation, WHO, Engineering and Physical Sciences Research Council, National Institute for Health Research (NIHR), Swiss National Science Foundation, and German Research Foundation.
Introduction
Transmission of COVID-19 associated with health-care settings has been well documented across the pandemic.1 Hospital-onset COVID-19 infections (HOCIs) have been reported to account for 12·0–15·0% of all COVID-19 cases in health-care settings and up to 16·2% at the peaks of the pandemic.2 Although their effect is yet to be fully quantified, HOCIs amplify the pandemic by seeding further outbreaks.
Predicting which patients are at risk of health-care-associated infection (HCAI) can prevent onward transmission to patients and staff, also minimising workload during outbreaks. Traditionally, predicting HCAI has relied on identifying risk factors from combinations of patient clinical variables (eg, age, gender identity, and comorbidities) and hospital contextual variables (eg, colonisation pressure and patients’ length of stay).3 Although these approaches alone can perform reasonably well in identifying predictive risk factors of HCAIs, they overlook the fact that nosocomial spread of infection depends largely on the patient's contacts,4 which are heterogeneous5 and vary over time.6
Isolating and grouping patients who are infected, or suspected to be infected, to one area prevents onward spreading by interrupting transmission chains.7 Contact tracing of infected patients is effective at identifying disease super-spreaders,8 who are strong HOCI drivers,9, 10 and secondary cases and has played a pivotal role in national COVID-19 responses.11, 12, 13 However, exploiting the entire contact network, rather than direct contacts to individuals with known infection alone, provides greater information to characterise transmission.14 Indeed, early in the COVID-19 pandemic, population mobility and interactions guided national policy to reduce transmission.15 In health-care settings, the overall number of direct contacts of a patient is predictive of HCAI.16, 17, 18, 19 Yet, these studies16, 17, 18, 19 fail to use the full dynamic information of contacts.20
Research in context.
Evidence before this study
Throughout the COVID-19 pandemic, health-care facilities have had considerable numbers of hospital-onset COVID-19 infections (HOCIs). Despite substantially higher rates of COVID-19 morbidity and mortality among hospitalised patients, predictive models of HOCI are yet to be fully used in health-care settings. To address this gap, we have designed a machine-learning framework that integrates dynamic patient contact-networks with traditional patient clinical risk factors and contextual hospital variables. Patient contact networks are a natural approach to model the contact-mediated transmission of COVID-19 and other infectious diseases. Our study investigates the use of contact-network variables in predicting HOCIs at the patient level and their generalisability to various hospital settings. We performed two searches on PubMed (Sept 22, 2021) for English-language articles. Search one was on prediction of HOCIs, using the search terms “hospital-onset COVID-19 infections”, “nosocomial COVID-19”, “prediction”, and “forecasting”; search two was on the use of contact-networks for prediction of infections acquired in health-care settings, based on the search terms “healthcare-acquired infections”, “nosocomial infections”, “prediction”, “forecasting”, “contact networks”, and “dynamic contact networks”. Search one identified no studies performing a comprehensive investigation into risk factors of HOCI at the patient level. Although several works examined HOCI epidemiology, providing characterisation of contacts, these studies were performed at single hospital sites, with few patients, and did not include a risk-factor analysis. Other studies examined risk factors for predicting patient risk of COVID-19 on hospital admission; however, by definition, these studies target only community-onset COVID-19 infections and not HOCIs and thus do not capture the in-hospital sources of exposure risk. Search two identified studies that used the total number of patient contacts or the total number of contacts with infectious cases. However, no studies of infections acquired in health-care settings incorporated contact connectivity beyond a patient's immediate contacts to predict infection risk. Furthermore, the studies found in our searches did not use sophisticated network-theoretical measures or modelling techniques to predict individual patient risk, nor did they account for the time-varying nature of the contacts.
Added value of this study
To our knowledge, this is the first study to forecast HOCIs at the patient level by constructing contact-networks from routinely collected hospital bed records. To investigate the predictive use of patient contact-networks, we used a large multinational hospital dataset collected throughout extended periods of the COVID-19 pandemic in two hospital groups; one in London, UK, and one in Geneva, Switzerland. Using these datasets, we constructed and generalised models to predict HOCIs at the patient level both with and without measures of patient centrality calculated using the dynamic patient contact-networks. Our results show that variables extracted from patient contact-networks are strong predictors of HOCI in both testing and validation. Such network measures lead to improved prediction over standard risk-factor models on the basis of patient clinical data or hospital contextual variables. Most network-derived variables were significantly elevated in HOCIs, emphasising their importance as risk factors.
Implications of all the available evidence
This study shows that dynamic contact-networks provide novel sources of predictive power for respiratory infections acquired in health-care settings, improving the performance of traditional risk-factor prediction models for HOCIs. Contact-network-derived risk factors have the potential to enhance individualised infection prevention and early diagnosis. We designed a machine-learning framework to extract contact risk factors using routinely available bed administrative data and showed its novel and generalisable prediction power. The framework can be used in real time to generate daily risk predictions as part of a suite of surveillance tools in modern, data-driven infection prevention and control strategies.
In this study, we combine dynamic networks of patient contacts (based on bed allocation records) with clinical attributes and hospital contextual data into a novel forecasting framework to predict patient risk of HOCI acquisition for targeting preventive interventions. As a proof of principle, we perform a retrospective cohort study to assess the predictive power of risk factors that were extracted from patient-contact networks, constructed from routinely collected hospital data. We train and test models on a large London hospital dataset spanning the first two major UK surges of COVID-19 (ie, March 23–May 30, 2020 and Sept 7, 2020–April 24, 2021). We then validate the predictive gain from contact-network risk factors by applying the framework to an external dataset from a university-affiliated geriatric hospital in Geneva during surge one (ie, March 1–May 31, 2020) and to data from the same London hospital group after surge two (ie, after April 2–Aug 13, 2021) in the UK, when COVID-19 had become endemic.
Methods
Study design and participants
This international retrospective cohort study consists of a complete case analysis including all hospital inpatients with bed allocations. For training and testing we used data from a large London hospital group (Imperial College Healthcare NHS trust) (with approximately 1200 inpatient beds across five sites) from April 1, 2020, to April 1, 2021, capturing the UK's first surge (ie, March 23–May 30, 2020) and second surge (ie, Sept 7, 2020–April 24, 2021; appendix p 2). For validation, we applied the framework to a non-UK hospital group in the Department of Rehabilitation and Geriatrics, Geneva University Hospitals, Geneva, Switzerland (with approximately 600 inpatient beds across three sites), during Switzerland's first surge (ie, March 1–May 31, 2020), and to data from the same London hospital group after the second surge in the UK (ie, April 2–Aug 13, 2021). The infection prevention and control (IPC) measures are detailed in the appendix (p 2).
Patient data were extracted and de-identified by the business intelligence system (London), iCARE (London), and from in-house electronic health records (Geneva).
Inclusion criteria were any inpatient with an allocated bed during the study period. All inpatients were included in the formation of the dataset, whereas only patients spending 3 consecutive days or more in the hospital were used to predict HOCI acquisition (appendix p 2).
Individual patient consent was waived under the study's ethical approval covering non-consented pseudonymised patient data.
All analyses were approved by ethical committees (London: Imperial College London National Health Service [NHS] Trust service evaluations [Ref: 386,379,473] and ethics approval under 15\LO\0746; Geneva: Cantonal Ethics Committee [number CCER 2020–00827]).
Procedures
The consecutive days that a patient has spent in hospital before testing positive for SARS-CoV-2 reflects the likelihood of health-care acquisition because of the 2–14 days incubation period.21 Hence, we defined HOCIs in line with European and UK definitions, using the date of the first positive test for SARS-CoV-2 and symptom onset synonymously.22 We defined HOCI as infections in patients with a positive SARS-CoV-2 test sample 3 or more days after admission. We used this definition as a single category for HOCIs, which covers three types of HOCIs: indeterminate (ie, positive sample between 3 days and 6 days), probable (ie, positive sample between 7 days and 13 days), and definite (ie, positive sample after 14 days or more).22 These categories follow from genomic evidence,10 suggesting that a substantial proportion of low likelihood HOCIs (ie, patients who meet the definition of community-onset COVID-19 infection [COCI] and some who are allocated to intermediate HOCIs on the basis of days spent in hospital) are still hospital acquired, and the comprehensive admission screening policy in our study (appendix p 2). We defined COCI as infection in patients with a positive SARS-CoV-2 test sample up to 2 days after admission. We defined non-COVID-19 (ie, control) as patients with a negative SARS-CoV-2 test sample or patients who were not tested because of having had a positive test for SARS-CoV-2 in the past 90 days with no new symptoms or exposure to SARS-CoV-2.
Patient contacts were established by use of movement pathways from hospital electronic health records. We investigated three definitions of contact: patients coinciding on the same day in the same room, ward, and building, regardless of COVID-19 prevention measures, such as environmental ventilation (appendix p 2). The infectious period for patients with SARS-CoV-2 infection is defined as the 14 days before and 10 days after their first positive SARS-CoV-2 test result.21
Dynamic forecasting framework
We developed a framework to predict infections (appendix pp 2–6), enabling risk stratification, that combined dynamic patterns of contact, exposure to infection, and standard risk factors (R package). Fixed patient variables (eg, demographics) were collected, and dynamic, time-dependent variables (eg, contact-network graph-theoretical centrality for each patient and hospital contextual variables) were computed from a sliding time window to be used as model predictors over a forecasting horizon. In alignment with the maximum incubation period of COVID-19, we set the window length to 14 days21 and the forecasting horizon to 7 days.
For each time window, we extracted patient clinical variables, hospital contextual variables (relating to the hospital inpatient context), and contact-network variables (centrality measures) using network-theoretical analytics from each of the room, ward, and building contact networks derived from the data (panel ; appendix pp 3–4).
Panel. Model variable interpretation.
Patient clinical variable *
Age
Current age was specified in years.
Gender identity
Gender identity as recorded on hospital electronic health record at the time of study.
Patient type †
The specialities visited were cardiology, critical care, elderly care, gynaecology, haematology, infectious diseases, medicine (general), neurology, obstetrics, oncology, paediatrics, renal, respiratory, and surgery.
Hospital contextual variable ‡
Length of stay (total)
Total length of stay in hospital (days).
Length of stay (consecutive)
Total consecutive (ie, uninterrupted) days in hospital.
Length of stay (side rooms)
Total length of stay (days) in a side room (ie, isolation).
Background hospital COVID-19 prevalence
Background number of COVID-19 (hospital-onset COVID-19 infection and community-onset COVID-19 infection) cases in the hospital group.
Background prevalence of hospital-onset COVID-19 infection in hospital
Background number of hospital-onset COVID-19 infection cases in the hospital group.
Total hospital bed occupancy
Total number of hospitalised patients recorded in a room.
Bed, room, ward, and site moves
Number of times the patient moved between beds, rooms, wards, and sites.
Contact-network variable §
Infected degree
Total number of direct contacts with infectious patients.
Infected degree centrality
Days spent in direct contact with infectious patients, normalised by the total number of patients in hospital.
Infected closeness centrality
Measure of network distance relative to all infectious patients, normalised by the total number of patients in hospital.
Degree
Total number of direct contacts.
Degree centrality
Days spent in direct contact with other patients, normalised by the total number of patients in hospital.
Clustering coefficient
Measure of likelihood that a patient is also connected to the contacts of their immediate contacts.
Closeness centrality
Measure of network distance relative to all other patients, normalised by the total number of patients in hospital.
Betweenness centrality
Centrality of a patient with respect to the shortest paths through the contact network.
Page rank
Centrality measure of a patient in the contact network given by the importance of their neighbours in the network.
K-core number
Measure of how central a patient is to the most connected region of the graph.
Evaluation of machine learning models
We constructed and evaluated models to predict HOCI (appendix p 7), using a 70 to 30 training to testing data split (where 70% of patients were randomly selected and allocated to the training set, and the remaining 30% were allocated to the test set). Following an unbiased comparison (appendix p 7), we report results of the best machine learning model (eXtreme Gradient Boosting [XGBoost]).
Performance was measured by prediction on the test set, quantified by area under the receiver operating curve (AUC-ROC); balanced accuracy; sensitivity; specificity; and positive predictive values, negative predictive values, and positive and negative likelihood ratios, adjusted for multiple prediction bias (appendix p 7). To aid interpretation, we ranked variables by their predictive contribution using a recursive elimination strategy (appendix p 8).23
Two validation datasets were used: one external dataset from a non-UK hospital with three sites (ie, three sites of the Department of Rehabilitation and Geriatrics, Geneva University Hospitals, Geneva, Switzerland) collected during the first epidemic surge in Switzerland and one internal dataset from the same London hospital group from which data were initially collected after UK surges in COVID-19 was endemic. The same inclusion criteria were used for the training and testing dataset and the validation datasets (ie, all inpatients with a bed allocation during the dates of study were included in the formation of the dataset, whereas assignment of control and HOCI labels was restricted to patients who had spent 3 days or more in hospital).
To perform validation, we used the XGBoost model with hyper parameters optimised on the training data and then applied to the new data with available risk-factor variables. Due to the smaller size of the validation datasets compared with the test dataset, we report 5-fold cross validation performance.
Statistical analysis
Univariate variables analysis was performed to identify risk factors by comparing values between HOCI and control groups in patients who were in hospital for 3 days or more (appendix pp 2, 9). All inpatients with a bed allocation during the study periods were included in the computation of network-derived and contextual variables. In predicting patient-level HOCI risk, only inpatients spending 3 or more days in hospital during the study period were included. To establish significance, we used either Mann-Whitney U or χ2 tests and report p values adjusted for length of patient stay in hopsital (appendix p 5). Statistical analyses were done with R (version 4.0.4).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
A total of 51 157 patients were admitted to the London hospital group during the study's training and testing period (April 1, 2020–April 1, 2021). Of these patients, 3439 (6·7%) patients tested positive for SARS-CoV-2, including 2950 (5·8%) COCIs and 489 (1·0%) HOCIs (appendix p 9). Together, 21 576 (42·2%) patients had stayed at least 3 days in hospital and were included in the forecasting data (489 HOCIs and 21 087 non-HOCIs).
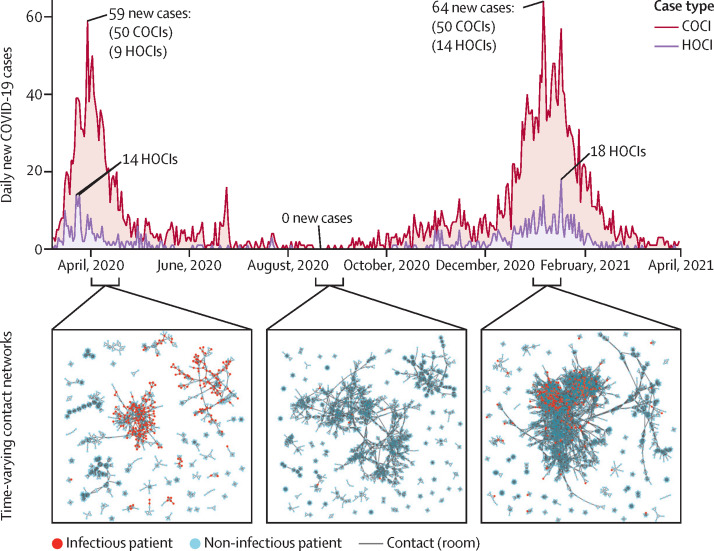
The prevalence of in-hospital COVID-19 cases had two surges congruent with national UK cases (figure 1 ). Surge one peaked on March 30, 2020 (ie, one day before the study period), at 59 new daily positive hospital cases (50 COCIs and nine HOCIs); surge two peaked on Jan 6, 2021, at 64 new daily cases (50 COCIs and 14 HOCIs). The two surges differed when analysing the time series (appendix p 10): the proportion of HOCIs was higher during surge two (17·8% HOCIs [406 of 2276 infections were HOCIs]) than during surge one (15·1% HOCIs [167 of 1107 infections were HOCIs]) and the correlation between HOCIs and COCIs was higher during surge two (R=0·79; p<0·0001) than during surge one (R=0·59; p<0·0001). The background variant makeup also varied between the UK surges, with the alpha (B.1.1.7) variant making up 59·3% and the delta (B.1.617.2) variant making up 1·1% of all nationally sequenced COVID-19 cases during surge two, whereas they were absent during surge one (appendix p 10).
Figure 1.
Background hospital infections and contact structure across the study period
Daily number of new patients who tested positive for COVID-19 within the hospital (COCI and HOCI) varied substantially across the study period. A peak of 59 cases was reached on March 30, 2020, and a peak of 64 cases was reached on Jan 6, 2021, dipping to zero new daily cases over days during July, August, September, and October. The patient-contact network also varied across the study period, with differences in connectivity and size of patient-contact clusters between each of the infection surges and during the summer period. COCI=community-onset COVID-19 infection. HOCI=hospital-onset COVID-19 infection.
The patient-contact network structure also varied throughout the pandemic (figure 1). The median number of contacts (degree) over networks across time was four in rooms (ie, the number of people sharing a room), 22 in wards (ie, the number of people sharing a ward), and 67 in buildings (ie, the number of people located in the same building at the same time), with an increasing trend over time (appendix p 12). Surge one had lower median degrees (three in rooms, 18 in wards, and 57 in buildings) than did surge two (four in rooms, 23 in wards, and 70 in buildings). Other network measures also varied over the study period (appendix p 12), with network metrics reflecting a denser contact-network in surge two than in surge one (figure 1).
Univariate analysis identified ten clinical variables that were differentially represented in patients with HOCI versus controls (table 1 ). Both age and gender identity were significantly different between patients with HOCI and controls, with HOCIs over-represented in older patients and those who identified as male. Regarding specialities, HOCIs were found in a higher proportion of patients in elderly care, general medicine, renal, and surgery compared with controls, and significantly lower proportions in patients from cardiology, gynaecology, obstetrics, and paediatrics.
Table 1.
Univariate analysis of variable sets for control versus HOCI data
| Control group (n=21 353) | HOCI group (n=465) | p value* | ||
|---|---|---|---|---|
| Patient clinical variables | ||||
| Age, years | 50·4 (27·3) | 69·2 (19·6) | <0·0001 | |
| Gender identity | ||||
| Female | 12 083 (57·3%) | 214 (43·8%) | <0·0001 | |
| Male | 9004 (42·7%) | 275 (56·2%) | <0·0001 | |
| Patient type | ||||
| Cardiology | 1476 (7·0%) | 13 (2·7%) | 0·0003 | |
| Critical care | 1497 (7·1%) | 44 (9·0%) | 0·12 | |
| Elderly care | 1645 (7·8%) | 76 (15·5%) | <0·0001 | |
| Gynaecology | 3037 (14·4%) | 14 (2·9%) | <0·0001 | |
| Haematology | 443 (2·1%) | 9 (1·8%) | 0·82 | |
| Infectious diseases | 232 (1·1%) | 4 (0·8%) | 0·77 | |
| Medicine (general) | 6136 (29·1%) | 217 (44·4%) | 0·0009 | |
| Neurology | 527 (2·5%) | 9 (1·8%) | 0·44 | |
| Obstetrics | 5208 (24·7%) | 12 (2·5%) | <0·0001 | |
| Oncology | 633 (3·0%) | 11 (2·2%) | 0·41 | |
| Paediatrics | 1097 (5·2%) | 8 (1·6%) | 0·0011 | |
| Renal | 1202 (5·7%) | 67 (13·7%) | <0·0001 | |
| Respiratory | 738 (3·5%) | 15 (3·1%) | 0·68 | |
| Surgery | 4829 (22·9%) | 142 (29·0%) | 0·0023 | |
| Hospital contextual variables | ||||
| Length of stay, days | 5·3 (2·6) | 7·3 (3·7) | <0·0001 | |
| Length of stay (consecutive), days | 3·8 (2·4) | 5·9 (3·6) | <0·0001 | |
| Length of stay (side rooms), days | 1·1 (2·6) | 2·7 (5·6) | <0·0001 | |
| Background hospital COVID-19 prevalence | 127 (174) | 372 (252) | <0·0001 | |
| Background hospital HOCI prevalence | 54·6 (35·6) | 19·1 (25·8) | <0·0001 | |
| Total hospital bed occupancy | 13 587 (3020) | 15 645 (2832) | <0·0001 | |
| Bed moves | 0·94 (0·81) | 1·00 (0·86) | 0·39 | |
| Room moves | 0·92 (0·79) | 0·96 (0·84) | 0·34 | |
| Ward moves | 0·66 (0·67) | 0·64 (0·66) | 0·57 | |
| Site moves | 0·04 (0·17) | 0·06 (0·23) | 0·11 | |
| Network variables | ||||
| Room-contact network | ||||
| Infected degree | 0·10 (0·55) | 0·74 (1·30) | <0·0001 | |
| Infected degree centrality | 0·00007 (0·00047) | 0·00063 (0·00130) | <0·0001 | |
| Infected closeness centrality | 0·0019 (0·0047) | 0·010 (0·010) | <0·0001 | |
| Degree | 5·4 (4·2) | 6·3 (4·2) | <0·0001 | |
| Degree centrality | 0·0020 (0·0016) | 0·0022 (0·0015) | 0·0001 | |
| Closeness centrality | 0·041 (0·034) | 0·064 (0·034) | 0·0002 | |
| Betweenness centrality | 0·0016 (0·0045) | 0·0018 (0·0035) | 0·16 | |
| PageRank | 0·00039 (0·00022) | 0·00039 (0·00024) | 0·58 | |
| Clustering coefficient | 0·073 (0·080) | 0·11 (0·10) | <0·0001 | |
| K-core number | 3·37 (2·34) | 3·71 (2·18) | 0·0010 | |
| Ward-contact network | ||||
| Infected degree | 1·4 (3·7) | 7·3 (8·3) | <0·0001 | |
| Infected degree centrality | 0·0010 (0·0031) | 0·0063 (0·0074) | <0·0001 | |
| Infected closeness centrality | 0·0088 (0·013) | 0·03010 (0·022) | <0·0001 | |
| Degree | 38 (25) | 41 (18) | 0·0080 | |
| Degree centrality | 0·012 (0·0080) | 0·012 (0·0050) | 0·54 | |
| Closeness centrality | 0·17 (0·050) | 0·20 (0·036) | <0·0001 | |
| Betweenness centrality | 0·0021 (0·0093) | 0·0017 (0·0040) | 0·022 | |
| PageRank | 0·00041 (0·00017) | 0·00040 (0·00017) | 0·10 | |
| Clustering coefficient | 0·10 (0·074) | 0·14 (0·093) | <0·0001 | |
| K-core number | 3·4 (2·3) | 3·7 (2·2) | 0·0011 | |
| Building-contact network | ||||
| Infected degree | 9·8 (24) | 43 (51) | 0·0009 | |
| Infected degree centrality | 0·0090 (0·026) | 0·042 (0·058) | <0·0001 | |
| Infected closeness centrality | 0·31 (0·062) | 0·34 (0·047) | <0·0001 | |
| Degree | 150 (130) | 210 (180) | <0·0001 | |
| Degree centrality | 0·046 (0·040) | 0·060 (0·050) | <0·0001 | |
| Closeness centrality | 0·31 (0·062) | 0·34 (0·047) | <0·0001 | |
| Betweenness centrality | 0·0014 (0·0047) | 0·0011 (0·0030) | 0·090 | |
| PageRank | 0·00041 (0·00019) | 0·00042 (0·00021) | 0·48 | |
| Clustering coefficient | 0·12 (0·064) | 0·14 (0·072) | 0·0019 | |
| K-core number | 85 (71) | 120 (88) | <0·0001 | |
Data are median (IQR) or n (%). Network, hospital contextual, and clinical variables were investigated for discriminatory power for HOCI (sample positive for SARS-CoV-2 at least 3 days after admission) versus control (sample not positive for SARS-CoV-2). Due to the sliding window, each patient can have multiple datapoints representing them on different days over the duration of their hospital stay. In addressment, patient variables are aggregated and averaged across time (appendix p 5). The significance test results show how the varying temporal profiles of patients could be used to classify HOCI versus control. Statistical analyses were performed using the Mann-Whitney U or the χ2 test. For clinical and contextual variables results are reported to 1 decimal point, whereas for network centralities results are given to 2 significant figures. HOCI=hospital-onset COVID-19 infection.
p values are adjusted for multiple testing as described in the the appendix (p 5).
Six of ten hospital contextual variables were significantly different between the HOCI and control groups (table 1). Relative to controls, patients with HOCI were associated with longer length of stay before testing positive and were in hospital during times of higher hospital-bed occupancy and during periods of increased background incidence of COVID-19. No significant difference between the HOCI and control groups was observed for variables related to movement rates (between beds, rooms, wards, and sites).
For network variables, 24 of 30 centrality measures were significantly higher in HOCI patients (eight of ten from each room-contact, ward-contact, and building-contact network; table 1). Network variables that were significantly higher in the HOCI group than in the control group across the three contact networks included measures accounting for infectious COVID-19 cases (ie, infected degree, infected degree centrality, and infected closeness centrality) and general network connectivity (ie, degree, closeness centrality, clustering coefficient, and K-core number).
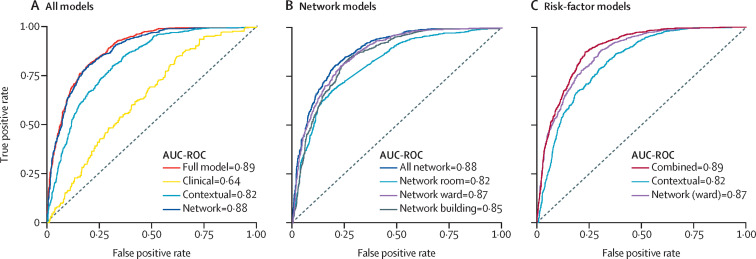
We trained different models on our London data using sets of variables of different types (panel). All models had high predictive power (table 2 ; figure 2 A, B). In particular, the model based solely on contact-network variables (AUC-ROC 0·88 [95% CI 0·86–0·90]) performed similarly to the model based on all variables (0·89 [0·88–0·90]) and yielded more predictive power than models using solely hospital context variables (0·82 [0·80–0·84]) or clinical variables (0·64 [0·62–0·66]). To ascertain the predictive power of different types of contacts, separate models were trained on variables from each of the three contact networks (ie, room, ward, and building). The model based on ward-contact network variables had the highest predictive power (0·87 [0·85–0·89]); yet building-contact (0·85 [0·83–0·87]) and room-contact (0·82 [0·80–0·84]) network models also yielded high performance.
Table 2.
Summary of test and validation set performance across variable groups
| AUC-ROC (95% CI) | Balanced accuracy | Sensitivity | Specificity | Positive predictive value | Negative predictive value | Positive likelihood ratio | Negative likelihood ratio | ||
|---|---|---|---|---|---|---|---|---|---|
| Test set performance models based on variable sets | |||||||||
| All types: patient clinical, hospital contextual, and network-derived | 0·89 (0·88–0·90) | 0·85 | 0·85 | 0·84 | 0·78 | 0·41 | 5·31 | 0·18 | |
| Patient clinical | 0·64 (0·62–0·66) | 0·61 | 0·46 | 0·75 | 0·55 | 0·44 | 1·84 | 0·72 | |
| Hospital contextual | 0·82 (0·80–0·84) | 0·80 | 0·87 | 0·73 | 0·68 | 0·37 | 3·22 | 0·18 | |
| Contact networks (all) | 0·88 (0·86–0·90) | 0·84 | 0·85 | 0·83 | 0·77 | 0·40 | 5·00 | 0·18 | |
| Room | 0·82 (0·80–0·84) | 0·80 | 0·77 | 0·82 | 0·74 | 0·41 | 4·28 | 0·28 | |
| Ward | 0·87 (0·85–0·89) | 0·85 | 0·90 | 0·80 | 0·75 | 0·39 | 4·50 | 0·13 | |
| Building | 0·85 (0·83–0·87) | 0·84 | 0·90 | 0·79 | 0·74 | 0·38 | 4·29 | 0·13 | |
| Test set risk-factor variable models | |||||||||
| Hospital contextual risk factors | 0·82 (0·80–0·84) | 0·80 | 0·89 | 0·70 | 0·66 | 0·36 | 2·97 | 0·16 | |
| Network (ward) risk factors | 0·87 (0·85–0·89) | 0·85 | 0·91 | 0·79 | 0·86 | 0·42 | 9·44 | 0·16 | |
| Combined (hospital contextual and network [ward]) risk factors | 0·89 (0·88–0·90) | 0·87 | 0·91 | 0·82 | 0·87 | 0·42 | 9·67 | 0·14 | |
| Validation set performance for models from surge 1 in Geneva hospital (epidemic)* | |||||||||
| Hospital contextual risk factors | 0·84 (0·82–0·86) | 0·82 | 0·97 | 0·66 | 0·68 | 0·31 | 2·85 | 0·05 | |
| Network (room) risk factors | 0·80 (0·77–0·83) | 0·80 | 0·76 | 0·84 | 0·78 | 0·39 | 4·75 | 0·29 | |
| Hospital contextual and network (room) risk factors | 0·88 (0·86–0·90) | 0·84 | 0·97 | 0·71 | 0·72 | 0·32 | 3·34 | 0·04 | |
| Validation set performance for London hospital group after surge 2 in the UK (endemic)† | |||||||||
| Hospital contextual risk factors | 0·49 (0·46–0·52) | 0·62 | 0·56 | 0·68 | 0·56 | 0·38 | 1·75 | 0·65 | |
| Network (ward) risk factors | 0·63 (0·60–0·66) | 0·71 | 0·66 | 0·76 | 0·67 | 0·39 | 2·75 | 0·45 | |
| Hospital contextual and network (ward) risk factors | 0·68 (0·64–0·70) | 0·74 | 0·70 | 0·78 | 0·70 | 0·39 | 3·18 | 0·38 | |
Performance is measured using AUC-ROC, balanced accuracy, sensitivity, specificity, positive predicted value, negative predicted value, the positive likelihood ratio, and the negative likelihood ratio, which operate on a collapsed confusion matrix to reduce bias (appendix p 7). AUC-ROC=area under the receiver operating characteristic curve.
For this non-UK hospital, contact-network risk factors are derived from the available room-contact network.
Contact-network variables were derived by use of the ward contact network (ie, the most predictive contact definition identified in training and testing).
Figure 2.
Model performance by variable set
(A) AUC-ROC (area under the curve [AUC]-receiver operating characteristic curve [ROC]) test set performance for models broken down by the major feature groups (ie, full, clinical, hospital contextual, and network). (B) A further network feature decomposition by network variables computing from all, room, ward, and building patient-contact networks. (C) Risk-factor model test set performance for the contextual risk-factor model, the network (ward) risk-factor model, and a combined model from both the contextual and network (ward) risk factors identified in table 2.
We then investigated models with fewer variables, by using only risk factors (ie, variables identified as significant; p<0·05 in table 1) among hospital contextual and ward-contact network variables. Clinical, room-contact network, and building-contact network variables were excluded due to comparably lower performance. Models based only on risk factors have equal performance to models including all variables (table 2; figure 2). Furthermore, the combined risk-factor model has the highest positive predictive value (0·87) and positive likelihood ratio (9·67) compared with all other variable-set models (table 2), in addition to high calibration (appendix p 14).
Using a stepwise-variable-elimination approach (appendix p 17), we ranked the combined set of risk factors (ie, hospital contextual plus ward-contact network). The hospital contextual variable “background hospital COVID-19 prevalence” was most predictive, followed by two ward-contact network variables: the infected contact network, which measures the network distance to all infectious cases, and the infected degree and degree centrality, which measures the direct contacts to infectious cases. A parsimonious model based on these three variables alone achieved AUC-ROC of 0·85 (95% CI 0·82–0·88), amounting to 95·5% of the combined model performance (appendix p 17). The same top three variables were also found when applying stepwise variable elimination to the entire variable set and to all the risk factors (table 2 and appendix p 17).
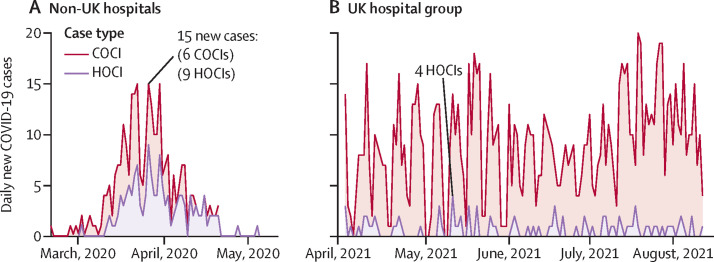
To validate the predictive power of contact-network variables, we applied our risk-factor models (without recalibrating the hyperparameters) to a Geneva-based geriatric hospital group during their first surge in cases (March 1–May 31, 2020). Over that period, 281 COVID-19 cases (138 COCIs and 143 HOCIs) were reported. Cases peaked on March 26, 2020, with 15 newly identified cases (nine HOCIs and six COCIs), reflecting the height of the early epidemic in Switzerland (figure 3 A). In this dataset, ward-level and building-level data were unavailable; hence, we constructed room-contact networks. On the basis of only hospital contextual risk factors, the model achieved a high prediction accuracy, but the inclusion of room-contact risk factors further increased performance (table 2).
Figure 3.
Epidemiology curves of study validation data
Newly identified COVID-19 cases are reported across time and are broken down by HOCI and COCI case types. (A) Non-UK (ie, Geneva) hospital caseload during an epidemic surge of cases. (B) UK hospital group after pandemic surges 1 and 2, when COVID-19 became endemic and non-surging. COCI=community-onset COVID-19 infection. HOCI=hospital-onset COVID-19 infection.
For further validation, we used additional data from the same London hospital group collected during an endemic period following surge two in the UK (April 2–Aug 10, 2021). During this time, 1·4 daily cases were reported on average, with no surging behaviour (figure 3B). Compared with UK surges 1 and 2, HOCIs constituted a lower percentage of all cases (186 [12·9%] of 1446 COVID-19 cases were HOCI compared with 167 [15·1%] of 1107 in UK surge one, and 406 [17·8%] of 2276 in UK surge two; appendix p 10). In this endemic setting, we found that the hospital contextual risk-factor model performed poorly with low sensitivity and specificity (table 2). The ward-contact network risk-factor model had substantially improved performance compared with the hospital contextual model. By further variable integration, performance was marginally improved with the combined risk-factor model and achieved higher AUC-ROC, sensitivity, and specificity as compared with the previous two models (table 2).
Discussion
We used network analysis in combination with machine learning to predict patient-level HOCI using routinely captured hospital data. To our knowledge, this is the first study to forecast individual patient HOCIs by extracting patient contact networks from bed records. Together with hospital contextual variables, we report patient contact-network centrality as a significant HOCI risk factor, able to increase predictive performance across all datasets analysed.
Transmission of SARS-CoV-2 in health-care settings has been associated with features such as limited isolation capacity, suboptimal individual infection prevention practices,24 physical distancing, presenteeism, environmental ventilation, and contaminated fomites, which can all be linked to particular patient groups.25 In our training and testing data, patients managed in elderly care, general medicine, renal, and surgical units were significantly over-represented in the HOCI group (table 1). Staffing levels and stress in critical care; complex pathways and excess movements, resulting in high contacts amongst surgery patients; and the strong community links in renal wards might have exacerbated transmission. Older patients and male gender identity being significantly over-represented in HOCIs reflects known features of the wider pandemic.26 Although IPC focuses on demographic and individual clinical risk variables,27, 28 our results show that such fixed variables are least predictive overall. Modern IPC might therefore improve management of outbreaks by including contextual and dynamic risk factors.
Behavioural factors, contact density, and ventilation between locations are known to affect risk of COVID-19 acquisition.29 These factors are consistent with the hospital contextual risk factors identified in our work. We found that background COVID-19 prevalence within the hospital group was the most predictive variable in our training and test data collected during pandemic surges. Although high case numbers increase transmission sources, background prevalence can also be a proxy for staffing stress and density changes, acting as potential exacerbators. Similarly, high HOCI risk from increased hospital-bed occupancy could be due to high patient loads, increased density, and staffing pressures, which make IPC challenging. Similar to other HCAIs, length of stay was significantly higher for HOCIs (table 1).3, 20 Length of stay and consecutive length of stay both being significantly longer in HOCIs than in controls also supports genomic analysis suggesting COVID-19 acquisition can be linked to previous admissions.10 Increased movement rates (ie, bed, room, ward, and site moves) were reported as a risk factor for HCAI locally,30 yet it was not significantly different for HOCIs in our data (table 1). The risk from movement rates alone is likely to be too general for HOCI, without specificity, and better captured via measures of contact-network centrality. Altogether, models based on hospital contextual variables showed strong predictive performance across epidemic surges. However, including network variables increased performance most notably in the endemic validation data (table 2).
Most contact-network variables (24 of 30 investigated, eight from each contact definition) were significantly higher in HOCIs (table 1), and the model based only on contact-network variables was as predictive as the model containing all variables (table 2; figure 2). The underlying network structure might, therefore, hold features exploitable for HOCI prediction with network mining tools.31 HOCIs were significantly more central in contact networks. Few studies have used contact data to investigate HCAI, and most have considered only direct contacts (ie, network degree).16, 17, 18, 19, 27 Similarly, COVID-19 transmission analysis outside hospital settings has been limited to direct contacts.12, 27 Consistent with these studies, our results show direct contacts as a strong risk factor of infection. Yet, the infected contact network (ward), measuring network connectedness to all known infections, was more predictive than direct infectious contacts (ie, infected degree), suggesting the presence of longer and indirect transmission chains that can affect contact tracing. Alternatively, disrupting underlying network connectivity by targeting patients with high centrality, together with screening and isolation based on risk factors, could be effective to reduce onward transmission.
To show generalisability, we applied our framework to data gathered from a hospital group that differed in both type (ie, geriatric vs long-term care) and country (ie, Switzerland vs the UK). Despite scarce contact data (ie, only room-level data were available), the framework was still highly predictive, and importantly, performance increased through the inclusion of contact-network risk factors. To further showcase its generalisability, we analysed data from the same London hospital group at a later date under differing epidemiological (ie, endemic) conditions (appendix p 10), changing IPC measures, newly emerging variants, and increasing vaccination rates. Although our framework achieved weaker performance on the endemic validation dataset, the inclusion of patient contact-network risk factors at the ward level substantially increased performance as compared with hospital-contextual risk factors, which did not have predictive capability (table 2).
The emergence of large databases with granular detail has allowed the construction and application of contact networks that can be integrated into routine IPC and public health policy. For instance, recorded movements within hospital (as studied here) or Bluetooth interactions of mobile users (eg, Corona-Warn-App in Germany) provide informative datasets that account for various underlying proxies in human interaction. The ubiquity of such data to construct contact networks is likely only to expand, with select hospitals introducing radiofrequency-identification tracking.32 Aimed at exploiting these emerging sources of data, our dynamic disease forecasting framework is designed to be portable to a range of settings and variables. The framework offers precise individual predictions of risk of infection acquisition and is thus amenable for risk stratification in real time, which can serve to guide dynamic IPC resource allocation for rapid screening, isolation, and grouping of patients at high risk of infection acquisition. By incorporating complex multimodal data sources into a single measure of predicted risk, our framework produces relevant and actionable outputs preventing disease acquisition.
Major challenges to effective IPC activity are low bed capacity and inadequate and overwhelmed isolation capacity, in addition to insufficient staffing and microbiological testing resources. These challenges to IPC were vastly exacerbated by the COVID-19 pandemic. We envisage the proposed framework to be used within a modern, data-driven IPC patient management system and able to assist optimal decisions in real-world scenarios. The predicted risk score for each patient can be used by clinicians to rank and prioritise (eg, identify patients at high risk for infection for isolation or grouping followed by targeted enhanced testing). In this way, HOCIs could be identified at the earliest opportunity, which in turn could optimise IPC measures and treatment. Patients at low risk of infection acquisition could also be potentially moved back to regular patient management faster, saving resources that are in demand. However, further work is needed to evaluate the direct implications (ie, clinical and economic) of identifying patients at high risk of infection. In addition to actionable clinical points, a key aspect of this framework is its dynamism and its ability to generate insight on demand. By aggregating complex data sources into single interpretable risk scores, a range of risk sources and their interactions are made accessible to hospital teams. Such data-driven insights, always integrated within human decision making, can enable hospital teams to become more flexible and responsive to complex, rapidly emerging disease threats.
Our study has several limitations. First, our contact definitions might not fully capture transmission (eg, connections via health-care workers);33, 34 indirect transmission over surfaces; non-room, ward, or building contact; or interactions from visitors. However, routinely collected patient bed allocations have been shown to capture implicitly non-patient interactions that align with organisational and speciality hospital structures.35 Staff and visitor contact data were not available in our data due to privacy restrictions, but such data should be investigated, in accordance with privacy preservations. Second, since our training and testing period occurred largely before the UK's vaccination rollout, we were unable to include vaccination status as a patient variable. With increasing levels of natural and induced immunity, inclusion of vaccination and recovery status might improve predictions; emerging new variants and incomplete vaccine coverage36 make the levels of susceptibility uncertain. Third, patient ethnicity was not available in our study. Due to its contextual complexities, and being a previously identified risk factor,37 ethnicity warrants specific and increased investigation in the future. Fourth, our data did not include ventilation or specific information about room arrangements (appendix p 2), which contribute to COVID-19 transmission.38 However, without accounting for ventilation, our models were highly predictive. Finally, various aspects of hospital organisation were altered across the pandemic, including changes in screening practice, personal protective equipment, or bed placement, which were not encoded here as variables.
Overall, our study emphasises that dynamic networks of patient contacts can aid personalised predictions of infection. Our study applies to respiratory virus transmission in hospital, using widely available patient bed records. Further work is needed to extend this framework to other infectious diseases, assessing the types of contact required for transmission, evaluating the implications of identifying a patient at high risk of infection acquisition, and understanding how it could be integrated into IPC more generally.
Data sharing
The processed anonymised training and testing dataset used in this study can be available on reasonable request to the corresponding authors. Patient pathways will not be provided as these are withheld by the corresponding authors’ organisation to preserve patient privacy. Data from the Imperial Clinical Analytics Research and Evaluation platform used in this study can be available to researchers on request. External validation data sources will not be provided as these are withheld by owners. Data regarding hospital COVID-19 admissions are freely available via the NHS COVID-19 hospital activity webpage (https://www.england.nhs.uk/statistics/statistical-work-areas/covid-19-hospital-activity). The code of the method is freely available as an R package (https://github.com/barahona-research-group/Dynamic-contact-infection-forecast) with examplar data sets.
Declaration of interests
After the analysis in this paper was completed, AM and MB received funding from UKRI Research England via the MedTech SuperConnector (awarded on June 1, 2022) for commercial viability testing of infection prevention models. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
We thank Imperial College London's NHS Infection Prevention and Control team and the iCARE team for supporting data access, cleaning, and interpretation. AM was supported in part by a scholarship from the Medical Research Foundation National PhD Training Programme in Antimicrobial Resistance Research (MRF-145–0004-TPG-AVISO) and by the NIHR Academy. RLP was funded by the Deutsche Forschungsgemeinschaft (Project-ID 424778381-TRR 295). MA and SH received funding from the Swiss National Science Foundation. AH is an NIHR Research Senior Investigator. AH is partly funded by the NIHR Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Infections in partnership with Public Health England, in collaboration with Imperial Healthcare Partners, University of Cambridge, and University of Warwick (NIHR grant code: NIHR200876). NZ was funded by the NIHR Health Protection Research Unit. AM, RLP, and MB acknowledge funding from the Engineering and Physical Sciences Research Council (grant EP/N014529/1) to MB, supporting the Engineering and Physical Sciences Research Council Centre for Mathematics of Precision Healthcare. AH and MB also received financial support from WHO for the investigation. The research was also supported by the NIHR Imperial Biomedical Research Centre and by the iCARE environment, and used the iCARE team and data resources. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health and Social Care, Public Health England, or WHO.
Contributors
AM, JRP, RLP, SM, and MB contributed to study concept and design. JRP, MA, SM, NZ, and FR contributed to data acquisition. AM, JRP, MA, and SM contributed to data analysis and accessed and verified the underlying data. AM, JRP, RLP, MA, AH, and MB contributed to the initial manuscript drafting. All authors contributed to data interpretation and final revisions of the manuscript. AH and MB contributed to study supervision. AM, JRP, RLP, MA, SM, SH, AH, and MB contributed to the discussion of the results and reviewed the data. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Footnotes
Variables attributable to individual patients. All variables were extracted from patient electronic health records at the time of the study.
Patients can be recorded as more than one patient type.
Length of stay variables are at a maximum of 14 days, and background prevalence of COVID-19 in hospital and hospital-onset COVID-19 infections capture the number of patients with COVID-19, or the total number of hospital patients, over the past 14 days. Each variable is computed over a given time window.
For each variable extracted from the contact network we provide the relative scale (ie, the spatial scale of the network considered by the variable during its calculation—eg, degree considers only its direct neighbours). A mathematical explanation of each variable is given in the appendix (pp 3–5). Each variable is also computed over a given time window.
Supplementary Material
References
- 1.Harapan H, Itoh N, Yufika A, et al. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13:667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barranco R, Vallega Bernucci Du Tremoul L, Ventura F. Hospital-acquired SARS-Cov-2 infections in patients: inevitable conditions or medical malpractice? Int J Environ Res Public Health. 2021;18:489. doi: 10.3390/ijerph18020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman J, McGowan JE., Jr Risk factors for nosocomial infection. J Infect Dis. 1978;138:811–819. doi: 10.1093/infdis/138.6.811. [DOI] [PubMed] [Google Scholar]
- 4.Cohen JE. Infectious diseases of humans: dynamics and control. JAMA. 1992;268 [Google Scholar]
- 5.Cevik M, Baral SD. Networks of SARS-CoV-2 transmission. Science. 2021;373:162–163. doi: 10.1126/science.abg0842. [DOI] [PubMed] [Google Scholar]
- 6.Holme P, Saramäki J. Temporal networks. Phys Rep. 2012;519:97–125. [Google Scholar]
- 7.Meyers L. Contact network epidemiology: bond percolation applied to infectious disease prediction and control. Bull Am Math Soc. 2007;44:63–86. [Google Scholar]
- 8.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Illingworth CJR, Hamilton WL, Warne B, et al. Superspreaders drive the largest outbreaks of hospital onset COVID-19 infections. eLife. 2021;10 doi: 10.7554/eLife.67308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lumley SF, Constantinides B, Sanderson N, et al. Epidemiological data and genome sequencing reveals that nosocomial transmission of SARS-CoV-2 is underestimated and mostly mediated by a small number of highly infectious individuals. J Infect. 2021;83:473–482. doi: 10.1016/j.jinf.2021.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge Y, Martinez L, Sun S, et al. COVID-19 transmission dynamics among close contacts of index patients with COVID-19: a population-based cohort study in Zhejiang province, China. JAMA Intern Med. 2021;181:1343–1350. doi: 10.1001/jamainternmed.2021.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferretti L, Wymant C, Kendall M, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368 doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kendall M, Milsom L, Abeler-Dörner L, et al. Epidemiological changes on the Isle of Wight after the launch of the NHS Test and Trace programme: a preliminary analysis. Lancet Digit Health. 2020;2:e658–e666. doi: 10.1016/S2589-7500(20)30241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman MEJ. Spread of epidemic disease on networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;66 doi: 10.1103/PhysRevE.66.016128. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Wang Z, Rader B, et al. Associations between changes in population mobility in response to the COVID-19 pandemic and socioeconomic factors at the city level in China and country level worldwide: a retrospective, observational study. Lancet Digit Health. 2021;3:e349–e359. doi: 10.1016/S2589-7500(21)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rewley J, Koehly L, Marcum CS, Reed-Tsochas F. A passive monitoring tool using hospital administrative data enables earlier specific detection of healthcare-acquired infections. J Hosp Infect. 2020;106:562–569. doi: 10.1016/j.jhin.2020.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamel M, Zoutman D, O'Callaghan C. Exposure to hospital roommates as a risk factor for health care-associated infection. Am J Infect Control. 2010;38:173–181. doi: 10.1016/j.ajic.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Shaughnessy MK, Micielli RL, DePestel DD, et al. Evaluation of hospital room assignment and acquisition of Clostridium difficile infection. Infect Control Hosp Epidemiol. 2011;32:201–206. doi: 10.1086/658669. [DOI] [PubMed] [Google Scholar]
- 19.Karan A, Klompas M, Tucker R, et al. The risk of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission from patients with undiagnosed coronavirus disease 2019 (COVID-19) to roommates in a large academic medical center. Clin Infect Dis. 2022;74:1097–1200. doi: 10.1093/cid/ciab564. [DOI] [PubMed] [Google Scholar]
- 20.Pastor-Satorras R, Vespignani A. Epidemic spreading in scale-free networks. Phys Rev Lett. 2001;86:3200–3203. doi: 10.1103/PhysRevLett.86.3200. [DOI] [PubMed] [Google Scholar]
- 21.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbas M, Zhu NJ, Mookerjee S, et al. Hospital-onset COVID-19 infection surveillance systems: a systematic review. J Hosp Infect. 2021;115:44–50. doi: 10.1016/j.jhin.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422. [Google Scholar]
- 24.Lanièce Delaunay C, Saeed S, Nguyen QD. Evaluation of testing frequency and sampling for severe acute respiratory syndrome coronavirus 2 surveillance strategies in long-term care facilities. J Am Med Dir Assoc. 2020;21:1574–1576. doi: 10.1016/j.jamda.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampf G, Brüggemann Y, Kaba HEJ, et al. Potential sources, modes of transmission and effectiveness of prevention measures against SARS-CoV-2. J Hosp Infect. 2020;106:678–697. doi: 10.1016/j.jhin.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenham C, Smith J, Morgan R. COVID-19: the gendered impacts of the outbreak. Lancet. 2020;395:846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Koh V, Marimuthu K, et al. Epidemiological and clinical predictors of COVID-19. Clin Infect Dis. 2020;71:786–792. doi: 10.1093/cid/ciaa322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soltan AAS, Kouchaki S, Zhu T, et al. Rapid triage for COVID-19 using routine clinical data for patients attending hospital: development and prospective validation of an artificial intelligence screening test. Lancet Digit Health. 2021;3:e78–e87. doi: 10.1016/S2589-7500(20)30274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Packer S, Hughes G, Edeghere O, Oliver I, Birney E. Using genomic concordance to estimate COVID-19 transmission risk across different community settings in England 2020/21. SSRN. 2021 https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3867682 published online June 15. (preprint). [Google Scholar]
- 30.Boncea EE, Expert P, Honeyford K, et al. Association between intrahospital transfer and hospital-acquired infection in the elderly: a retrospective case-control study in a UK hospital network. BMJ Qual Saf. 2021;30:457–466. doi: 10.1136/bmjqs-2020-012124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peach RL, Arnaudon A, Schmidt JA, et al. HCGA: highly comparative graph analysis for network phenotyping. Patterns (NY) 2021;2 doi: 10.1016/j.patter.2021.100227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho HJ, Zhang ZX, Huang Z, Aung AH, Lim W-Y, Chow A. Use of a real-time locating system for contact tracing of health care workers during the COVID-19 pandemic at an infectious disease center in Singapore: validation study. J Med Internet Res. 2020;22 doi: 10.2196/19437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbas M, Robalo Nunes T, Martischang R, et al. Nosocomial transmission and outbreaks of coronavirus disease 2019: the need to protect both patients and healthcare workers. Antimicrob Resist Infect Control. 2021;10:7. doi: 10.1186/s13756-020-00875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas M, Robalo Nunes T, Cori A, et al. Explosive nosocomial outbreak of SARS-CoV-2 in a rehabilitation clinic: the limits of genomics for outbreak reconstruction. J Hosp Infect. 2021;117:124–134. doi: 10.1016/j.jhin.2021.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myall AC, Peach RL, Weiße AY, et al. Network memory in the movement of hospital patients carrying antimicrobial-resistant bacteria. Appl Netw Sci. 2021;6:34. [Google Scholar]
- 36.Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sze S, Pan D, Nevill CR, et al. Ethnicity and clinical outcomes in COVID-19: a systematic review and meta-analysis. EClinicalMedicine. 2020;29 doi: 10.1016/j.eclinm.2020.100630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Gu J, Li K, et al. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg Infect Dis. 2020;26:1628–1631. doi: 10.3201/eid2607.200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed anonymised training and testing dataset used in this study can be available on reasonable request to the corresponding authors. Patient pathways will not be provided as these are withheld by the corresponding authors’ organisation to preserve patient privacy. Data from the Imperial Clinical Analytics Research and Evaluation platform used in this study can be available to researchers on request. External validation data sources will not be provided as these are withheld by owners. Data regarding hospital COVID-19 admissions are freely available via the NHS COVID-19 hospital activity webpage (https://www.england.nhs.uk/statistics/statistical-work-areas/covid-19-hospital-activity). The code of the method is freely available as an R package (https://github.com/barahona-research-group/Dynamic-contact-infection-forecast) with examplar data sets.