Abstract
The gut microbiota is in continuous interaction with the innermost layer of the gut, namely the epithelium. One of the various functions of the gut epithelium, is to keep the microbes at bay to avoid overstimulation of the underlying mucosa immune cells. To do so, the gut epithelia secrete a variety of antimicrobial peptides, such as chromogranin A (CgA) peptide catestatin (CST: hCgA352-372). As a defense mechanism, gut microbes have evolved antimicrobial resistance mechanisms to counteract the killing effect of the secreted peptides. To this end, we treated wild-type mice and CST knockout (CST-KO) mice (where only the 63 nucleotides encoding CST have been deleted) with CST for 15 consecutive days. CST treatment was associated with a shift in the diversity and composition of the microbiota in the CST-KO mice. This effect was less prominent in WT mice. Levels of the microbiota-produced short-chain fatty acids, in particular, butyrate and acetate were significantly increased in CST-treated CST-KO mice but not the WT group. Both CST-treated CST-KO and WT mice showed a significant increase in microbiota-harboring phosphoethanolamine transferase-encoding genes, which facilitate their antimicrobial resistance. Finally, we show that CST was degraded by Escherichia coli via an omptin-protease and that the abundance of this gene was significantly higher in metagenomic datasets collected from patients with Crohn’s disease but not with ulcerative colitis. Overall, this study illustrates how the endogenous antimicrobial peptide, CST, shapes the microbiota composition in the gut and primes further research to uncover the role of bacterial resistance to CST in disease states such as inflammatory bowel disease.
Subject terms: Antimicrobials, Microbial ecology
Introduction
The complex microbial community inhabiting the mammalian gut, and in particular their metabolic products, has a crucial effect on various health and disease states, such as inflammatory bowel disorders [1]. Gut microbiota, and its products, are in continuous interaction with the innermost layer of the gut, namely the gut epithelium. Part of the gut epithelial function is to keep the microbes at bay to avoid overstimulation of the underlying immune cells in the gut, and the subsequent inflammation [2, 3]. To do so, the gut epithelial cells secrete a wide variety of antimicrobial peptides.
CST, a proteolytically processed product of Chromogranin-A (CgA), acts as an antimicrobial peptide [4, 5]. The pro-protein CgA is abundantly expressed in endocrine and neuroendocrine cells including the epithelial enteroendocrine cells (EECs) [6–9]. CST consists of 21 amino acid residues and acts on a wide range of regulatory functions including immune, endocrine, metabolic, neurological, and cardiovascular functions [10–14]. For example, mice with a selective deletion of the CST-coding region of the Chga gene (CST-KO mice) displayed increased gut permeability, which was restored by CST treatment [15].
To date, a few studies have indicated associations between CST and the gut microbiota. Intrarectal administration of CST for 6 days, in mice, resulted in a shift in the gut microbiota composition, with a decrease of Firmicutes and an increase in the Bacteroidota phyla [16]. These findings suggest the role of CST as a modulator of the gut microbiota composition. This CST-modulatory effect could be mediated by its antimicrobial activity against a wide range of microorganisms. In fact, CST has been reported to have antimicrobial activity against pathogenic bacteria and filamentous fungi in in vitro studies [5]. In humans, CST showed antimicrobial effects against several skin microbes [17]. Nevertheless, the underlying mechanisms by which CST exerts its effects in vivo on the gut microbiota remain obscure. In this study, we show how CST plays a role in governing the colonization of the gut microbiota.
Results
Catestatin treatment affects the microbiota composition in CST-KO and WT mice
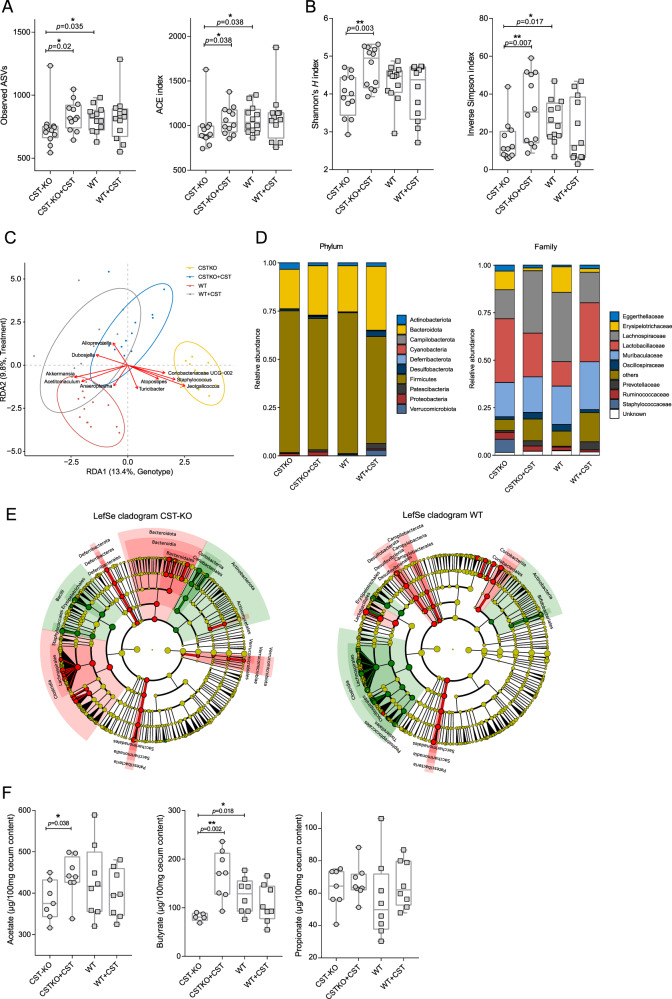
The composition of the gut microbiota is shaped by, among others, host-produced antimicrobial peptides [18, 19]. Here, we treated CST-KO and WT mice (n = 12) with 2 µg/g body weight/day CST by intraperitoneal injection for 15 consecutive days. Amplicon sequencing of the V3-V4 regions of the bacterial 16S rRNA gene was performed on the fecal samples of CST-KO and WT mice with and without CST treatment (n = 12). Microbial richness, assessed by the number of observed amplicon sequence variants (ASVs) and ACE index (Abundance-based Coverage Estimator), showed a significant decrease in CST-KO compared to WT mice (Fig. 1A). In contrast, the richness levels were restored upon treatment of CST-KO mice, but not in WT mice treated with CST. Next, the microbiota diversity was determined by Shannon’s H and inverted Simpson’s index, both indices are used to measure similar parameters of alpha diversity. Similar to the richness scores, the diversity index was significantly higher in the CST-treated CST-KO mice but not in the CST-treated WT group (Fig. 1B). The data highlight that the effect of the absence of CST in CST-KO mice on the microbiota diversity and richness could be restored upon treatment with CST, while in WT mice, with normal CST levels in their gut, the microbiota diversity and richness did not change with CST treatment.
Fig. 1. CST treatment affects the murine fecal microbiota.
A, B Alpha diversity was assessed using different metrics and is concordantly significantly increased upon CST treatment. A represents the species richness (observed ASVs, ACE) and B species diversity of the CST-KO and WT mice before and after CST treatment. Significance was tested with an unpaired Mann–Whitney test. Boxes represent the median with interquartile range, and whiskers represent the maxima and minima. C Genotype and treatment constrained redundancy analysis (RDA) on genus collapsed abundances. Arrows indicate the association of taxa with samples, with the length being a proxy for the strength of the association. Ellipses represent normal data generated by the R package ggplot2. Significant separation of clusters and contribution of the variables to the variance of the RDA was tested with Permutational Multivariate ANOVA (PERMANOVA) and revealed significant (p < 0.001) effects of CST administration, both in CST-KO and WT mice; gray – WT + CST, yellow – CST − KO, blue - CST − KO + CST. D Relative abundance of the present phyla and families. E Cladograms of a LefSe analysis (Linear discriminant analysis Effect Size) comparing microbiota changes upon CST treatment in CST-KO and WT mice. Significance was tested using a Kruskal–Wallis (KW) test and Linear Discriminant Analysis (LDA). A significant feature was considered when p < 0.01 and Log (LDA score) >3. The green color indicates the microbiota composition before CST treatment and the red color after CST treatment. F Concentrations of cecal acetate, butyrate, and propionate, of CST-treated (CST-KO, n = 7; WT, n = 8) and untreated animals (CST-KO, n = 8; WT, n = 8). Cecal acetate and butyrate were significantly increased in CST-treated CST-KO mice. Boxes represent the median with interquartile range, and whiskers represent the maxima and minima. Data were analyzed using a 2-tailed paired t-test, while 2 outliers were removed using ROUT in GraphPad Prism. Error bars are shown as mean ± SEM.
As a general exploratory analysis, principal component analysis (PCA) was performed and showed distinct clustering of the CST-KO, WT, and CST-treated groups (Supplementary Fig. 1A). Particularly, the CST-KO group clustered away from the other 3 groups, while the cluster of CST-treated WT mice showed a great spread and was overlapping with clusters of CST-treated CST-KO mice and WT mice.
To correct for any residual variation in the data, we further employed constrained RDA (redundancy analysis) to determine which bacterial taxa were associated with the different groups of mice at the genus level. The analysis was constrained to both, the mouse genotype and treatment (non-treated and CST-treated). Similar to the cluster separation detected by PCA analysis (Supplementary Fig. 1A), both constraints had a significant influence on the model (p < 0.001, determined by ANOVA-like permutation test), explaining 13.4% (genotype) and 9.8% (treatment) of the variation, respectively (Fig. 1C). Notably, the CST treated CST-KO group was shifted to the left side along RDA1, indicating a shift in “genotype” upon CST treatment, as opposed to WT, where the treatment group only moved along RDA2, which is associated with the treatment.
To assess the treatment effect exclusively on each genotype, we performed RDA constrained to untreated and CST-treated groups. On the genus level, Dubosiella and Romboutsia showed the strongest association with CST treatment along RDA1 (explaining ~31.2% of the variation) in the CST-KO mice (Supplementary Fig. 1B). In WT mice, RDA demonstrated an association of Faecalibaculum, Bifidobacterium, Romboutsia, and Anaeroplasma with the baseline along RDA1 (~17.2% explained variance), while Akkermansia was more associated with PC1 (~25.1% explained variance; residual variance). Alloprevotella and Candidatus Saccharimonas representing the strongest associations with CST treatment in WT (Supplementary Fig. 1B).
To further identify which bacterial taxa were affected by the CST treatment, pairwise comparisons of bacterial abundances were performed between CST-treated and untreated groups for each genotype. Focusing on the phylum level, Firmicutes decreased in relative abundance, while Bacteroidota, Patescibacteria, Desulfobacterota, and Proteobacteria increased with CST treatment in both CST-KO and WT groups. Verrucomicrobiota showed lower abundance in CST-KO but increased significantly in CST-treated groups (Fig. 1D). The changes depicted on the phyla level in CST-treated WT mice are consistent with findings from Rabbi et al, albeit different CST administration and dosage regimen [16]. These shifts in the main bacterial phyla upon CST treatment was further confirmed to be consistent in human microbiota by in vitro culturing of healthy human fecal samples with 10 µM CST (Supplementary Fig. 1C). On family level the most prominent changes were observed for Erysipelotrichaceae, which was consistently decreased in treated groups and Lachnospiraceae, which was increased in CST-KO, but decreased in WT in the CST treated groups. Further Lactobacillaceae was found to be decreased in CST-KO, but increased in WT CST treated groups (Fig. 1D). On the genus level, CST treatment reduced the abundance of Staphylococcus and Turicibacter in CST-KO and WT mice, while Alistipes, Akkermansia, and Roseburia, were significantly increased only in the CST-KO group (Supplementary Excel Sheet). Next, we used LEfSe (Linear discriminant analysis Effect Size; Segata et al., 2011) to complement our differential abundance analysis. The main discriminant feature separating the groups (untreated and CST-treated) in CST-KO mice were species from the genera Atopostipes, Jeotgalicoccus, Turicibacter, Staphylococcus, and Coriobacteriaceae UCG-002 family (Fig. 1E). In WT mice, the most discriminating genera were Lactobacillus, Candidatus Sacchararimonas, Turicibacter, and Enterococcus.
Changes in microbial composition are often accompanied by metabolic changes, in particular, the production of short-chain fatty acids (SCFAs) [20]. Thus, levels of SCFAs; acetate, butyrate, and propionate, were measured in the cecum of untreated and CST-treated CST-KO and WT mice (Fig. 1F). CST treatment significantly increased the levels of acetate and butyrate in CST-KO mice but not in the WT group. Specifically, butyrate was significantly lower in the CST-KO mice compared to their WT counterparts. Both observations are consistent with the observed significant increase in the SCFA-producing bacteria, Alistipes, Akkermansia, and Roseburia in the CST-KO group.
Overall, the results indicated a significant impact of CST treatment on the diversity and composition of the microbiota in the CST-KO mice, where CST is absent. This effect was much less prominent in WT mice, which have normal levels of CST.
CST treatment of CST-KO mice promotes the growth of core taxa present in WT mice
To investigate whether certain taxa exhibit similar behavior (increase or decrease in abundance) between genotypes when CST is administered, we focused on the core microbiota (common taxa between CST-treated groups), representing microbiota members, which directly (e.g., via resistance genes) or indirectly (e.g., via cross-feeding), resist the antimicrobial effect of CST. To do so, log-fold changes between CST treated and untreated groups for both WT and CST-KO mice were compared (Fig. 2). Almost all significant changes in core taxa (determined by unpaired Wilcoxon test) were exclusively higher in abundance in CST treated CST-KO mice (quadrants I and II), while, for WT, both significant increases and decreases in the abundance of core taxa were observed. These results coincide with the RDA analysis (cluster of CST-treated CST-KO group moving closer to WT) and indicate that the effect of CST treatment in CST-KO mice promotes the growth of core taxa associated with WT mice. Taxa present in quadrant II, which represent those common taxa between the untreated WT and treated CST-KO mice, are partially restored in their abundance in the latter group. Of note is, that Turicibacter was the only core taxon in both genotypes, exclusively negatively affected by CST treatment, exhibiting a significant decrease in abundance and therefore suggesting higher susceptibility to CST. To confirm this observation, we performed in vitro assays, to determine minimum inhibitory concentrations (MIC) for Turicibacter sanguinis. MIC was also performed for Bacteroides thetaiotaomicron, which increased in abundance upon CST treatment. The screening revealed low MIC for T. sanguinis (32 µM CST), while B. thetaiotaomicron had a higher MIC (>64 µM CST), thus confirming our in vivo observations. Reverse-phase HPLC analysis of spent culture supernatant after 24 h incubation with CST revealed degradation/uptake of CST from the medium, in the case of B. thetaiotaomicron, but seemingly less for T. sanguinis, hence explaining the higher MIC value in B. thetaiotaomicron (Supplementary Fig. 2). Taken together, the data imply that the changes in the gut microbiota composition associated with the CST treatment (Fig. 1) may be caused by the antimicrobial resistance of certain taxa to CST. However, the underlying mechanisms remain obscure.
Fig. 2. Taxa able to endure CST treatment benefit from its presence in the environment.
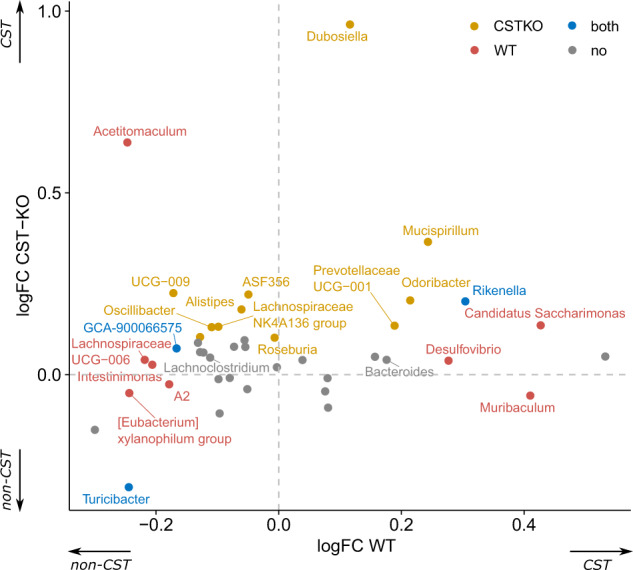
Scatter plot of log-fold changes, comparing abundance changes for significantly abundant genera between treated and untreated groups per genotype. Colors represent significance when testing for differential abundance between treated and untreated groups for each genotype.
Catestatin treatment promotes the abundance of the gut microbiota harboring antimicrobial resistance phosphoethanolamine transferase-encoding genes
To determine whether bacteria with an increase in abundance in CST-treated groups harbor antimicrobial resistance genes, a metagenomic prediction was performed using PiCRUSt2 on KO (KEGG Orthology) level. From a panel of known AMP resistance genes [21, 22], one KEGG Orthologue (K03760) was found to be significantly different in abundance between CST treated and untreated WT mice and close to significance in CST-KO, when corrected for multiple testing (Supplementary Excel Sheet).
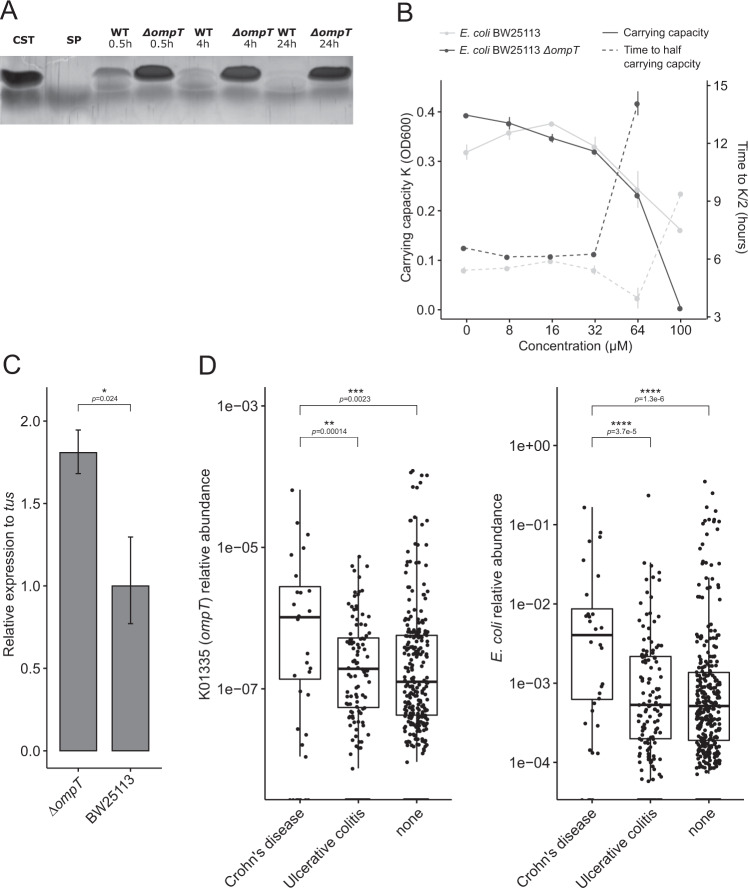
Sequences within this KEGG Orthogroup contain orthologs of EptA (E. coli) phosphoethanolamine transferase. As KEGG Orthologs only contain sequences from reference genomes with no specific gut metagenomic context, we sought to refine our search to investigate the taxonomical distribution of the EptA. To do so, the EptA protein sequence from E. coli BW25113 was used as a query to search the mouse gastrointestinal bacterial catalog (MGBC). Identified protein sequences were further filtered and a phylogenetic tree was constructed, which exhibited a distinct grouping of sequences without great dispersion and most of the sequences of the same phylum were clustered in separate clades. This showed a major subdivision between of the foremost sequences from Bacteroidota, Proteobacteria, and Campylobacterota (Fig. 3A).
Fig. 3. CST treatment promotes the abundance of the antimicrobial resistance gene, eptA.
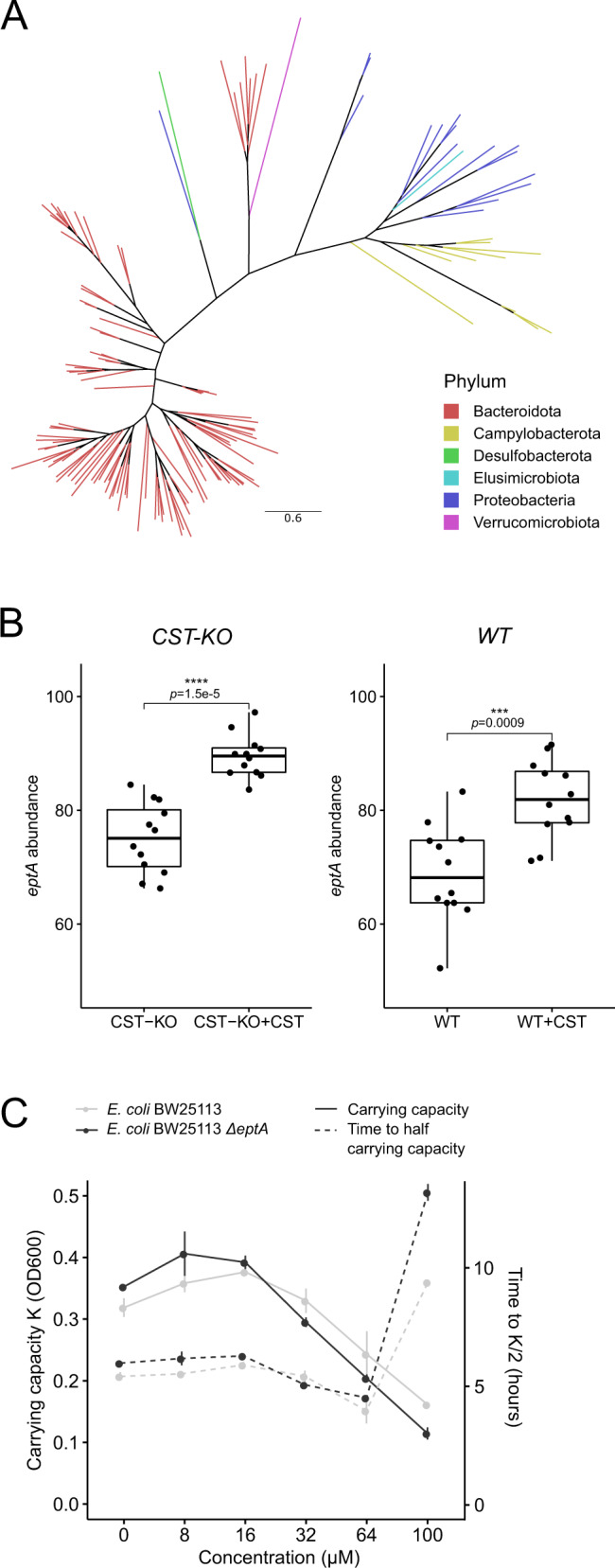
A Unrooted phylogenetic tree depicting the distribution of orthologous eptA-like genes throughout bacterial species in the mouse gut. The tree was pruned using Treemmer to aid visualization, while branches were colored by the phylum they belong to. B Boxplot showing a significant abundance increase of eptA harboring bacteria upon CST treatment, both in CST-KO and WT mice. The eptA abundance is defined as the sum of abundances from all taxa harboring an eptA gene. Boxes represent the median with interquartile range, and whiskers represent the maxima and minima. Significance was assessed by an unpaired t-test. C CST inhibits the growth of wild-type E. coli BW25113 and E. coli BW25113∆eptA strains, visualized by carrying capacity K (a measure of maximum OD600) and time taken to reach half the carrying capacity (inflection point of the growth curve at mid-log-phase) that depends on CST concentrations ranging from 8 to 100 µM.
Using the taxonomical distribution of eptA, we reanalyzed the abundance of genera harboring eptA upon CST treatment. Here, both CST-treated groups showed a significant increase in microbiota-harboring eptA, though the effect in the CST-KO group seemed to be more pronounced (Fig. 3B). To further support our findings that eptA-harboring core taxa increase in abundance in CST treated groups, we looked at individual log-fold changes of single taxa, as described above (Fig. 2). Indeed, eptA-harboring core taxa were higher in abundance in CST treated groups, regardless of the genotype (Supplementary Fig. 3A).
To further confirm the bioinformatics analysis outcome for the involvement of the eptA gene in CST resistance, wild-type E. coli BW25113 and an eptA knockout strain (∆eptA) were employed and were cultured in different concentrations (8–100 µM) of CST (Fig. 3C). The results indicate that the lack of eptA led to a decreased carrying capacity at higher CST concentrations (Fig. 3C), which is equivalent to the maximum population size of the culture, as well as a concentration-dependent prolonged lag-phase during the growth of the E. coli (Fig. 3C).
Similar to the majority of known antimicrobial peptides, CST is recognized by the PhoPQ system, which involves the induction of expression of the gene pmrD, followed by activation of pmrA, and eventually eptA expression [23]. To this end, we tested the expression of the above-mentioned transcription regulators in E. coli BW25113 strain. Most notably, phoP showed approximately 3-fold increase at 0.5 h after stimulation of E. coli BW25113 strain stimulated with 20 µM CST (sufficient concentration for detection without growth penalty), but only 2-fold and less prolonged in the control, before returning to baseline expression. The pmrA showed a later upregulation (after 1 h) compared to phoP in E. coli BW25113 CST-stimulated cultures, while pmrD expression spiked in the presence of CST, but was invariant between the different conditions (Supplementary Fig. 3B). Taken together, the results show a higher abundance of the bacteria-harboring eptA-like genes upon CST treatment, potentially providing these CST-resistant bacteria an advantage to colonize the gut and, in turn, indirectly affect the colonization of other bacterial taxa.
Catestatin peptide is degraded by E. coli omptin protease
The ability of E. coli BW25113∆eptA to still resist comparatively high CST concentrations lead us to test whether the bacterium harbors an additional resistance mechanism, such as degradation or uptake, similar to what we observed in B. thetaiotaomicron (Fig. 3D). We analyzed the spent culture supernatant by reverse-phase HPLC, which showed absence of CST-corresponding peaks after 24 h of incubation with E. coli BW25113∆eptA as well as E. coli BW25113 (Supplementary Fig. 4A). To test whether the enzyme involved in the CST degradation is excreted in the supernatant, sterile-filtered supernatants of non-stimulated E. coli BW25113 cultures were incubated with CST and subjected to tricine SDS-PAGE to visualize the degradation/uptake of CST. Indeed CST was cleaved by E. coli BW25113 after incubation for different time intervals from 30 min up till 24 h (Fig. 4A). This suggested the involvement of a secreted or membrane-anchored protease, which can also be present in secreted vesicles, in CST degradation. To identify which protease was involved, literature search was performed and revealed that E. coli harbors only 2 major outer membrane proteases, predicted to cleave CST [24]. Among those proteases is the omptin, an outer membrane protease, encoded by ompT. To confirm that omptin was responsible for the CST degradation, an E. coli ompT knockout strain (E. coli BW25113∆ompT) was employed and was cultured in different concentrations of CST. Compared to the wild-type and E. coli BW25113∆eptA strains, E. coli BW25113∆ompT showed no growth at 100 µM CST and an extensively prolonged lag-phase at 64 µM (Figs. 3D and 4B). Collectively, the results imply that wild-type E. coli BW25113 possesses the ability to cleave CST through its outer membrane protease omptin, providing the bacterium with another defense mechanism, together with EptA, to resist the antimicrobial action of CST.
Fig. 4. Omptin enzyme is responsible for the degradation of CST in E coli.
A Silver-stained tricine SDS-PAGE gels showing the cleavage of CST by wild-type E. coli BW25113, but not in E. coli BW25113∆ompT. Sterile non-stimulated culture supernatants were cultured with 100 µM CST and samples were collected at different time points; CST catestatin (2.3 kDa), SP substance P (1.3 kDa). B CST inhibits the growth of E. coli BW25113∆ompT, visualized by carrying capacity K (a measure of maximum OD600) and time taken to reach half the carrying capacity in CST concentrations ranging from 8 to 100 µM. C Baseline expression of eptA is increased in E. coli BW25113∆ompT compared and normalized to that of wild-type E. coli BW25113. tus was used as a housekeeping gene for normalization in both strains and statistical significance was calculated using a Wilcoxon test. D Relative abundance of omptin like genes or E. coli in patients with Crohn’s disease, ulcerative colitis, and healthy subjects. Axes are on log10-scale and significance was assessed using Wilcoxon test without stratification.
To further confirm the complementary function of EptA and omptin, we performed qPCR on control (unstimulated) and CST-stimulated cultures of wild-type E. coli BW25113 and E. coli BW25113∆ompT. The expression of eptA was upregulated about 1.7-fold in E. coli BW25113∆ompT without CST stimulation compared to the wild-type strain (Fig. 4C). When stimulated with 20 µM CST, an increase in the expression of ompT, but not eptA, was detected in wild-type E. coli BW25113 without stimulation and at 0.5 h after stimulation, while this effect was reversed at later time points (Supplementary Fig. 3B).
As E. coli BW25113 is considered a lab-strain, which might not be representative of naturally occurring E. coli strains, we also tested for the presence of ompT in two gut isolate strains, namely E. coli Nissle 1917 and E. coli DSM 11250. Both were found to be ompT carrying (Supplementary Fig. 4B). Taken together, the results indicate the capacity of E. coli strains harboring omptin to degrade CST, plausibly altering endogenous levels of the peptide in the gut in cases where the abundance of E. coli is significantly increased, such as in inflammatory bowel disease [25] To test this hypothesis, we investigated the abundance of ompT-like genes (KO term K01355) in different subtypes of inflammatory bowel diseases (IBD), which are also associated with altered levels of CST [15], using publicly available data from the MetaQuery database [26]. A significant increase in the abundance of ompT-like genes was found between non-IBD and Crohn’s disease patients. Similarly, there was a significant increase in the abundance of ompT-like genes between Crohn’s disease patients and patients with ulcerative colitis, but not between ulcerative colitis diseased and non-IBD subjects (Fig. 4D). This observation was consistent with the abundance of E. coli in those patients (Fig. 4D). Overall, the data indicate that CST degradation by bacteria-harboring omptin-like proteases may play a role in altered levels of the peptide previously detected in IBD patients, which in turn would result in an overgrowth of these bacteria and worsening of the disease situation.
Discussion
This study unravels the crucial role of CST on the microbiota composition in the fecal samples of WT and CST-KO mice. Specifically, there was a significant increase in the abundance of taxa that harbor the CST peptide-resistance enzyme, phosphoethanolamine transferase in CST-KO and WT mice treated with CST (Fig. 3B). The shifts observed in the microbial phyla in this study after treating WT mice with CST are in agreement with the findings of Rabbi et al., who also reported an increase in Bacteroidota and a decrease in Firmicutes, albeit a different route for the administration of CST and dosage [19]. Low abundance of Bacteroidota, especially Bacteroides/Prevotella have been linked to obesity, and CST-KO mice were described to be insulin resistant and obese besides having elevated blood pressure [13, 27]. In contrast, increased Bacteroidetes/Firmicutes ratio has been associated with IBD [28, 29], which is also characterized by elevated levels of CST [15]. Nevertheless, as CST is a multifunctional peptide, acting directly on the microbiota as well as on the host, any changes related to CST treatment should be viewed as a complex interplay and not direct causation originating from the microbiota [12]. Additionally, the lack of CST in CST-KO mice may have induced expression of other antimicrobial peptides, which, in turn, may contribute to the changes associated with the CST treatment.
CST treatment could, only to a small extent, restore the microbial composition in a CST-depleted environment, to the composition detected in an environment already primed with CST (WT mice) (Fig. 2). In general, the effect of CST treatment observed in the CST-KO group was significantly greater than that in the WT group (Fig. 1). This difference could be attributed to the presence of normal levels of CST in the WT mice; thus, the microbiota is already primed with the peptide and has evolved mechanisms to resist its antimicrobial effect. In contrast, the CST-KO group, in which CST is absent, may harbor bacteria that are susceptible to the CST antimicrobial activity but were eradicated upon CST treatment (Fig. 2). Especially the core taxa resisting CST treatment seem to benefit from their presence in the gut lumen environment, by exhibiting an increase in abundance. Introduction of CST to a CST-depleted environment did not necessarily promote exclusively health beneficial microbiota, but rather led to a more equilibrated composition, including also pathobiont taxa. For example, in CST-treated CST-KO, Oscillibacter and Mucispirillum (Fig. 2), both of which increased in relative abundance, have been found to increase in diet-induced obesity and might be considered as pathobionts [30, 31]. Similarly, Alistipes was shown to have a detrimental influence on colorectal cancer progression, despite contrasting evidence that it also has protective effects against colitis or cardiovascular diseases [32]. On the other side Odoribacter, a SCFA-producer, which increased in relative abundance in CST-treated WT mice (Fig. 2) has been linked to immunomodulatory effects and general health benefits presumably also in cooperation with Akkermansia [33].
So far, antimicrobial resistance mechanisms have been mostly investigated in pathogens due to their implication on antibiotic resistance. In in vitro studies, CST has been reported to have an inhibitory effect against the pathogenic gram-positive (Micrococcus luteus, Bacillus megaterium), gram-negative bacteria (Escherichia coli D22), and filamentous fungi (Neurospora crassa and Aspergillus fumigatus) [5]. Moreover, human CST showed antimicrobial effects against several skin microbes including the gram-positive (Staphylococcus aureus and Group A Streptococcus), gram-negative bacteria (E. coli O29, and Pseudomonas aeruginosa), yeast (Candida albicans), and filamentous fungi (Aspergillus niger, Aspergillus fumigatus, Trichophyton rubrum) [17]. Analogously, our data showed a significant decrease in the abundanace of Staphylococcus (Fig. 1), which is known to be potential pathogen in the gut, suggesting that CST can combat pathogens. However, as a defense mechanism, bacteria have evolved several systems to survive the effect of the antimicrobial peptides [21]. The ability to withstand these peptides provides a potential benefit in colonization over other members of the gut microbiota especially under pathological conditions [34]. Analogously, antimicrobial peptides are regularly used by the host to select for specific gut microbiota in homeostatic situations. For CST, the antimicrobial activity resides in the N-terminal of the peptide, which is highly cationic [5]. These positive charges in the peptide interact with the anionic components of the bacterial cell membrane, resulting in the permeabilization of microbial membranes, leading to cell lysis [7]. The present study suggests that CST may as well play a role in aiding the host to select for which microbes to colonize the gut. The abundance of bacteria encoding the phosphoethanolamine transferase genes increased upon CST treatment (Fig. 3A–E). This is not surprising since the E.coli phosphoethanolamine transferase eptA, for example, has been linked to colistin resistance, being a cyclic proteinogenic antibiotic [35]. EptA acts by modifying core lipopolysaccharide components with ethanolamine, increasing membrane charge and thus, repelling the antimicrobial agent. The fact that EptA-like proteins are abundant across a variety of different taxa present in the human gastrointestinal tract (Fig. 3A–C) implies that antimicrobial resistance is a mutualistic and important component in the selection of a specific microbiota composition by the host, through active modulation of antimicrobial peptide levels, such as CST. Indeed, this paradigm is supported by our findings that CST treatment induced a large-scale alteration in the microbiota composition not only in the CST-KO group but also in the WT group, both of which clustered closer to each other, and further away from the untreated groups (Fig. 1).
The induced expression of eptA gene in E. coli BW25113∆ompT (Fig. 4C) implies a complementary mechanism when other defense mechanisms are missing. Indeed, we uncovered the degradation of CST by wild-type E. coli, a member of the gut microbiota via omptin, an outer membrane protease, known to cleave other cationic antimicrobial peptides, such as protamine or LL-37 [36, 37]. Omptin proteases exhibit a strong requirement for arginine in the P1 position of the cleavage site, while P1’ can be either lysine, glycine, or valine [38]. Catestatin harbors one arginine – glycine site, thus omptin may potentially cleave at this position, yielding cateslytin and a hexapeptide. As cateslytin has been reported to have an antimicrobial activity [5], this cleavage might not completely ameliorate the effect of CST. Nonetheless, we observed a reduction of the inhibitory concentration in E. coli BW25113∆ompT (Fig. 4B). The presence of these fragments in the gut has its implications on the microbiota composition, as it may facilitate colonization of other CST-susceptible bacteria [39]. It remains elusive though, whether CST is taken up and accumulating inside the bacteria or is further degraded, and what the fate of the resulting cleavage fragments is, e.g., through uptake by other microbiota or interaction with the host. Analogously it remains unclear how other bacteria, such as B. thetaiotaomicron can degrade CST (Supplementary Fig. 2) and how widespread such a phenomenon is among the gut bacteria.
Levels of CST are increased in IBD patients, while E. coli was associated with the development and progression of different IBD types [25, 40]. The higher abundance of ompT-like genes in samples from Crohn’s disease patients, which coincided with a higher abundance of E.coli (Fig. 4D), suggests that bacteria harboring ompT-like genes, such as E. coli, may have a benefit to colonize the gut due to its ability to resist CST. In cases where the overall microbial composition is compromised, and the endogenous levels of CST are altered, like in IBD, such bacterial resistance may cause particular bacterial strains to be involved in worsening the gut inflammation. Overgrowth of gut bacteria with the capacity to resist CST may lead the host to secrete higher levels of CST as a defense mechanism. The levels of CST were reported to be higher in IBD patients compared to healthy subjects [15]. It remains unclear why only samples from Crohn’s disease patients but not ulcerative colitis showed a higher abundance of ompT as well as E. coli. One plausible explanation could be due to the different E. coli strains predominantly present in either Crohn’s disease or ulcerative colitis [25] (Fig. 4D).
A limitation of the present study is the relatively uncontrolled actual concentrations of CST that eventually reach the gut after intraperitoneal injection. This concentration variability could increase interindividual variability in the microbial composition. However, data on the physiological concentration of CST in the gut epithelia is sparse, except of a study reporting CST concentration in mice colonic tissue to be 0.35 nmol/mg of protein, compared to 0.863 nM circulating in plasma [15]. These data indicate that the CST concentrations are significantly higher in the colon compared to the plasma. Additionally, mice treated with CST intraperitonially, showed an increase in plasma CST from 0.86 nM to 1.72 nM in WT mice after 24 h, while CST-KO mice levels were elevated to 1.17 nM after 24 h [27].
Overall, the present study highlights the significant role of the CST peptide in determining the microbiota composition. The fact that gut bacteria with a capacity to resist the antimicrobial effect of CST become more abundant upon CST treatment, suggests that these bacteria may have gained an advantage over other microbiota members to survive the gut environment, which, indirectly shapes the colonization of the total community. Further studies should also explore the changes elicited by CST administration on the host side to gain a clearer picture of the direct and indirect effects of CST on the gut microbiota.
Materials and methods
Animals
All studies with mice were approved by the University of California San Diego (UCSD) and Veteran Affairs San Diego Healthcare System (VASDHS) Institutional Animal Care and Use Committees for the Mahata laboratory (UCSD: #S00048M; VA: #A13-002) and were performed in San Diego in adherence to the NIH Guide for the Care and Use of Laboratory Animals. Our experiment was based on a single cohort of mice which constisted of twelve mice per group each (control and CST-treatment, per genotype); out of those, twenty four male adult C57BL/6 J mice (age 20 weeks) were purchased from Jackson Laboratory (Bar Harbor, ME) and twenty four CST-KO mice (age 20-28 weeks) generated in the Mahata laboratory, were used. CST-KO mice have a deletion in the 63 bp CST domain from Exon VII of the Chga gene [27] in C57BL/6 background. Mice from WT and CST-KO groups were injected intraperitoneally once daily with CST (2 μg/g body weight; Genscript Biotech Corporation, Piscataway, NJ) for 15 days based on previous experimentation [15]. Mice were housed in 4 to 5 animals per cage and had free access to water and food (Normal Chow Diet, LabDiet 5001) in temperature and humidity-controlled rooms with a 12-hr light/dark cycle.
Determination of short-chain fatty acids (SCFAs) in cecal samples
Cecal samples (100 mg) were suspended in 1 mL of saturated NaCl (36%) solution. An internal standard (50 μL of 10.7 µM 2-methyl butyric acid in MQ water) was added and the samples were homogenized using glass beads. After the addition of 150 µL H2SO4 96%, SCFAs were extracted with 3 ml of ether. The ether layer was collected and dried with Na2SO4 (150 mg). The supernatant (0.5 µL) was analyzed using gas chromatography with flame ionization detection (Agilent, Santa Clara, California, USA). The system was equipped with a DB FFAP analytical column (30 m x 0.53 mm ID, 1.0 µm; Agilent) and helium GC grade (5.6) was used as carrier gas with a constant flow of 4.2 ml/min. The initial oven temperature was held at 100 °C for 3 min, ramped with 4 °C/min to 140 °C (isothermal for 5 min) and further with 40 °C/min to 235 °C (isothermal for 15 min). The resulting chromatograms were processed using ChemStation (Agilent Technologies).
DNA isolation and 16S rRNA gene sequencing
DNA isolation was performed on fecal samples that were collected from untreated and CST-treated WT and CST-KO mice. A phenol-chloroform-isoamyl alcohol procedure was used for DNA extraction [41]. Briefly, the pellet was resuspended in 1 ml of lysis buffer (940 µl TE buffer, 50 µl SDS 10% and 10 µl proteinase K 20 mg/ml) in a 2 ml screw cap micro-tube containing a mix of zirconium and glass beads. Then, samples were incubated at 58 °C for 1 hour and 150 µl of buffered phenol (Invitrogen, 15513-047) was added. To support lysis, samples were homogenized 3 × 30 s with 1-min intervals on ice in a mini bead-beater (Biospec, Bartlesville, USA). This was followed by the addition of 150 µl chloroform/isoamyl alcohol (24:1) and centrifugation at 16,000 × g for 10 min and 4 °C. The upper layer was carefully transferred to a clean tube, 300 µl of phenol-chloroform-isoamyl alcohol [25:24:1] was added, centrifuged and the process was repeated by adding 300 µl of chloroform-isoamyl alcohol [24:1]. To precipitate the DNA, the upper layer was transferred to a new tube and 1 volume of absolute isopropanol and 1/10 volume of 3 M sodium acetate was added. Samples were incubated overnight at −20 °C. Subsequently, samples were centrifuged at 16,000 × g for 20 min at 4 °C, the supernatant was discarded and the pellet was washed with 700 µl of 70% ethanol. After ethanol aspiration the DNA pellet was air-dried for 30 min and resuspended in 100 µl TE buffer. In each step, samples were vigorously mixed using a vortex.
Sequencing of the V3-V4 region of the bacterial 16S rRNA gene was carried out by Novogene Co. Ltd. Briefly, for sequencing library preparation, raw DNA extracts were diluted to 1 ng/µl in sterile water, and amplicons were generated by PCR (primers 341 F and 806 R) using a Phusion High-Fidelity PCR Master Mix (New England Biolabs). Amplification product quality was assessed by gel electrophoreses and samples were pooled in equimolar ratios. Libraries were generated with a NEBNext Ultra DNA Library Prep Kit for Illumina and sequencing was carried out on an MiSeq (Illumina) 250 bp paired-end platform. Initial processing of reads involved trimming of adapters and primers using a Novogene in-house pipeline (Novogene Co. Ltd, Cambridge, UK).
Microbiota analysis
Paired-end sequencing reads were filtered, denoised, merged, and classified with the dada2 package in the statistical programming language R while processing forward and reverse reads separately until merge [42, 43]. Briefly, reads were truncated to 220 bases and low-quality reads were filtered followed by dereplication. Error models were learned, while manually enforcing the monotonicity of the error function. Reads were denoised and merged with minimal overlap of 12 bp, while non-merging reads were concatenated. Singletons have been removed before performing bimera removal, followed by read classification using SILVA (V138) as a taxonomical reference database.
For downstream analysis, the phyloseq and microbiome packages were used, samples were rarefied to even depth and richness was determined by the number of observed amplicon sequence variants (ASVs), as well as ACE index and alpha diversity was assessed via Shannon’s H and inverted Simpson indexes [44, 45]. All ASVs were collapsed on genus level and cumulative sum scaling was applied using metagenomeSeq [46]. The resulting genus abundance table served as input for different types of ordinations such as principal component analysis (PCA) or redundancy analysis (RDA). The significance of constraints in constrained RDA was determined by permutation-like ANOVA. Differential abundance was assessed by unpaired Wilcoxon Rank Sum test using p < 0.05 as a significance threshold followed by FDR correction.
To generate the double log-fold change plot, first, the core microbiota were estimated for each group by filtering raw ASV counts to contain only more than 10 reads and be present in more than 10% of the samples. Next, only taxa intersecting between CST treated groups were selected and analyzed further. Differential abundance was assessed by a Wilcoxon Rank Sum test between treated and untreated groups per genotype and FDR corrected. Only taxa with an adjusted p < 0.05 were considered further. Their mean log-fold changes between groups were calculated and plotted against each other.
Further LefSe analysis was carried out, adjusting the alpha value for factorial Kruskal–Wallis test and the alpha value for pairwise Wilcoxon tests to 0.01. Additionally, the LDA score threshold was set to 3.0 [47].
For the functional analysis, the metagenomic prediction from 16S rRNA gene sequencing data was done using PiCRUSt2, using raw ASVs and unnormalized ASV abundances [48]. For this, ASVs were first filtered to have a minimal abundance of 10 reads and to be present in at least 1/10 of the samples. PiCRUSt2 was run with standard parameters and stratification enabled. The metagenomic data was further analyzed by using STAMP v2.1.3 [49]. For statistical analysis, White’s non-parametric t-test, two-sided, and Benjamini–Hochberg multiple test correction were used to reveal differences between groups.
Phylogenetic analysis
The amino acid sequence of the E. coli eptA gene (UniProt P30845) was searched with phmmer from the hmmer suite (v3.3.1; hmmer.org) against a non-redundant dataset of the mouse gastrointestinal bacterial catalog (MGBC) [50]. Hits were filtered on full sequence E-value, excluding all sequences with an E-value greater than 0.5× the median absolute deviation from the median. Additionally, only sequences originating from genomes with >90% completeness and <1 contamination were taken into account. Sequences were dereplicated, aligned with MAFFT (v7.453) and a phylogenetic tree was constructed using FastTree (v2.1.11) [51, 52]. The produced gene tree was visualized with FigTree (v1.4.4), to aid visibility the tree was pruned using Treemmer (v0.3; with parameter -RTL 0.95) [53].
Bacteria and growth inhibition assay
E. coli BW25113, BW25113∆ompT (JW0554) and BW25113∆eptA (JW5730) (Supplementary Table 1) were routinely grown aerobically in Difco Antibiotic Medium 3 (BD, Franklin Lakes, USA) at 37 °C degrees without agitation. Bacteria were inoculated from −80 °C stocks and grown overnight. Before the experiment, cultures were diluted at 1:100 in fresh medium from overnight cultures. Five different CST concentrations were tested in triplicates in a 96-well plate. For this, 10 µl of a 10× concentrated stock of CST were added to 90 µl of diluted culture to obtain final concentrations of 8, 16, 32, 64, and 100 µM. Bacterial growth was monitored for 24 h at 600 nm (OD600) with shaking for 5 s every 15 min using a BioTek ELx808 microplate reader (BioTek, Winooski, USA). Growth curves were fit using the growthcurver R package (v0.3.0).
Tricine SDS-PAGE
Overnight cultures of E. coli strains BW25113 and BW25113∆ompT (Supplementary Table 1) were centrifuged at 10,000 × g for 10 min at 4 °C and the supernatant was collected. The supernatant was further sterile filtered (0.2 µm filters) and to 90 µl of supernatant 10 µl of a 10× CST stock was added to reach a final concentration of 100 µM. Aliquots of 20 µl were taken at time points 0.5, 4, and 24 h.
To visualize CST degradation, culture supernatants were run on Tricine SDS-PAGE following a modified protocol [54]. Briefly, gels consisted out of a 4% stacking gel and a 16% resolving gel both with a cross-linker ratio of 19:1, and were prepared to standard procedure. For sample prep, 10 µl of each sample were mixed with 10 ml of reducing sample buffer (100 mM Tris-HCl, 1% SDS, 4% 2-mercaptoethanol, 0.02% Coomassie Brilliant Blue, 24% glycerol) and incubated at 70 °C for 5 min. Subsequently, 10 µl of the sample was loaded on the gel, first, run for ~30 min at 30 V, and then at 170 V for 90 min. Silver staining was applied to visualize proteins using various modifications in fixation to avoid loss of small proteins [55]. For this, gels were fixed (30% ethanol, 15% formalin, 5% acetic acid) for 60 min and quickly washed once with 50% ethanol and twice with MilliQ-filtered water. Gels were incubated for 40 min in 0.005% sodium thiosulfate, followed by incubation for 40 min in 0.1% silver nitrate solution containing 0.07% formalin. Washing with MilliQ-filtered water was performed and a 2% sodium carbonate solution containing 0.1% formalin was added. Gels were incubated for 1–2 min until bands became visible and development was stopped by removing the developer and adding 50 mM EDTA solution. Gels were then imaged in a ChemiDoc MP Imaging System (BioRad, Hercules, USA).
Expression analysis
Overnight cultures of E. coli BW25113, E. coli BW25113∆eptA, and E. coli BW25113∆ompT were resuspended 1:1000 in 6 ml of fresh Difco Antibiotic Medium 3 and grown until OD600 0.5 was reached. Aliquots of 1 ml were harvested, centrifuged at 10,000 × g for 10 min, and immediately stored at −80 °C. This and all subsequent centrifugation steps were done at 4 °C.
For culture stimulation with CST E. coli BW25113 or E. coli BW25113∆ompT was resuspended 1:1000 in 6 ml fresh Difco Antibiotic Medium 3 and grown until OD600 0.5. An aliquot of 5 ml was harvested and centrifuged at 3000 × g for 10 min. The pellet was resuspended in fresh medium containing 20 µM CST and gently mixed. Samples of 1 ml were taken at time points 0 h (before resuspension), 0.5 h, 1 h, 2 h, and 4 h, subsequently centrifuged at 10,000 × g for 10 min and immediately stored at −80 °C.
RNA was isolated using a phenol:chloroform:isoamyl alcohol procedure as previously described [56]. The harvested bacterial pellet was resuspended in 200 µl TE buffer and transferred to a tube containing 0.5 g of 0.1 mm zirconia/silica beads, 25 µl 10% SDS, and 200 µl phenol:chloroform:isoamyl alcohol solution. The subsequent procedure was performed as described and isolates were stored at −80 °C until further use.
qPCR was carried out according to standard procedures [57]. Briefly, 2 µg of RNA were reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit with RNAse Inhibitor (Applied Biosystems, Vilnius, Lithuania). Subsequently, 10 ng of cDNA were used for qPCR employing a PowerUp SYBR Green Master Mix (Applied Biosystems, Vilnius, Lithuania) and primers for genes eptA, ompT, phoP, pmrD, pmrA and tus. Reactions were performed in a clear 96-well plate using a CFX96 Real-Time PCR System (BioRad, Hercules, USA). Gene expression was assessed by normalizing to housekeeping gene tus using the R package qpcR (v1.4) for curve fitting and the pcr (v1.2.2) package for differential expression calculation employing the 2−∆∆Ct method as described in [58].
Statistical analysis
All statistical tests were performed using GraphPad Prism 7 or the statistical programming language R. Specific statistical tests are indicated in the text or figure legends. Normality was assessed by either D’Augustino-Pearson omnibus normality test or Shapiro–Wilk normality test. If normality was met a t-test was chosen, otherwise non-parametric Mann–Whitney was used. Outliers were assessed with GraphPads ROUT method (Q = 1).
Supplementary information
Acknowledgements
PG-D thanks the National Council of Science and Technology in Mexico (CONACyt) for the Ph.D. grant assigned to CVU 690069. We thank Dr. Matthias Heinemann of Department of Molecular Systems Biology, University of Groningen, the Netherlands, for providing us ΔeptA and ΔompT E. coli BW25113 mutant strain; Dr. Greet Vandermeulen of Department of chronic diseases and metabolism, Faculty of Medicine, KU Leuven, Belgium for the help with SCFAs analysis.
Author contributions
PG-D, MS, SKM, and SEA conceived and designed the study. PG-D, MS, BD, and SKM performed the experiments, and PG-D, MS, BD, and SEA analyzed the data. PG-D, MS, SEA wrote the original manuscript that was reviewed by AD, BD, KV, SKM, and SEA. Funding for these studies were acquired by SEA and SKM. All authors read and approved the final manuscript.
Funding
SKM is supported by a Merit Review Grant (I01 BX003934) from the Department of Veterans Affairs, USA. SEA is supported by a Rosalind Franklin Fellowship, co-funded by the European Union and the University of Groningen, The Netherlands.
Data availability
All data generated or analyzed during this study are included in this manuscript and its supplementary information files. The 16S rRNA gene amplicon sequence data were deposited under BioProject number PRJNA741992.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pamela González-Dávila, Markus Schwalbe.
Contributor Information
Sushil K. Mahata, Email: smahata@health.ucsd.edu
Sahar El Aidy, Email: sahar.elaidy@rug.nl.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01240-9.
References
- 1.Kåhrström CT, Pariente N, Weiss U. Intestinal microbiota in health and disease. Nature. 2016;535:47–47. doi: 10.1038/535047a. [DOI] [PubMed] [Google Scholar]
- 2.El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–79. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 3.Okumura R, Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med. 2017;49:e338–e338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahata SK, O’Connor DT, Mahata M, Yoo SH, Taupenot L, Wu H, et al. Novel autocrine feedback control of catecholamine release. A discrete chromogranin a fragment is a noncompetitive nicotinic cholinergic antagonist. J Clin Invest. 1997;100:1623–33. doi: 10.1172/JCI119686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briolat J, Wu SD, Mahata SK, Gonthier B, Bagnard D, Chasserot-Golaz S, et al. New antimicrobial activity for the catecholamine release-inhibitory peptide from chromogranin A. Cell Mol Life Sci. 2005;62:377–85. doi: 10.1007/s00018-004-4461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugardon K, Raffner R, Goumon Y, Corti A, Delmas A, Bulet P, et al. Antibacterial and antifungal activities of vasostatin-1, the N-terminal fragment of chromogranin A. J Biol Chem. 2000;275:10745–53. doi: 10.1074/jbc.275.15.10745. [DOI] [PubMed] [Google Scholar]
- 7.Aslam R, Atindehou M, Lavaux T, Haïkel Y, Schneider F, Metz-Boutigue M-H. Chromogranin A-derived peptides are involved in innate immunity. Curr Med Chem. 2012;19:4115–23. doi: 10.2174/092986712802430063. [DOI] [PubMed] [Google Scholar]
- 8.El-Salhy M, Patcharatrakul T, Hatlebakk JG, Hausken T, Gilja OH, Gonlachanvit S. Chromogranin A cell density in the large intestine of Asian and European patients with irritable bowel syndrome. Scand J Gastroenterol. 2017;52:691–7. doi: 10.1080/00365521.2017.1305123. [DOI] [PubMed] [Google Scholar]
- 9.Bartolomucci A, Possenti R, Mahata SK, Fischer-Colbrie R, Loh YP, Salton SR. The extended granin family: structure, function, and biomedical implications. Endocr Rev. 2011;32:755–97. doi: 10.1210/er.2010-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahata SK, Corti A. Chromogranin A and its fragments in cardiovascular, immunometabolic, and cancer regulation. Ann N Y Acad Sci. 2019;1455:34–58. doi: 10.1111/nyas.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corti A, Marcucci F, Bachetti T. Circulating chromogranin A and its fragments as diagnostic and prognostic disease markers. Pflugers Archiv Eur J Physiol. 2018;470:199–210. [DOI] [PubMed]
- 12.Mahata SK, Mahata M, Fung MM, O’Connor DT. Catestatin: a multifunctional peptide from chromogranin A. Regul Pept. 2010;162:33–43. doi: 10.1016/j.regpep.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying W, Mahata S, Bandyopadhyay GK, Zhou Z, Wollam J, Vu J, et al. Catestatin inhibits obesity-induced macrophage infiltration and inflammation in the liver and suppresses hepatic glucose production, leading to improved insulin sensitivity. Diabetes. 2018;67:841–8. doi: 10.2337/db17-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahata SK, Kiranmayi M, Mahapatra NR. Catestatin: a master regulator of cardiovascular functions. Curr Med Chem. 2018;25:1352–74. doi: 10.2174/0929867324666170425100416. [DOI] [PubMed] [Google Scholar]
- 15.Muntjewerff EM, Tang K, Lutter L, Christoffersson G, Nicolasen MJT, Gao H, et al. Chromogranin A regulates gut permeability via the antagonistic actions of its proteolytic peptides. Acta Physiol. 2021;232:e13655. [DOI] [PMC free article] [PubMed]
- 16.Rabbi MF, Munyaka PM, Eissa N, Metz-Boutigue MH, Khafipour E, Ghia JE. Human catestatin alters gut microbiota composition in mice. Front Microbiol. 2017;7:1–12. doi: 10.3389/fmicb.2016.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radek KA, Lopez-Garcia B, Hupe M, Niesman IR, Elias PM, Taupenot L, et al. The neuroendocrine peptide catestatin is a cutaneous antimicrobial and induced in the skin after injury. J Invest Dermatol. 2008;128:1525–34. doi: 10.1038/sj.jid.5701225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–68. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 19.Dupont A, Heinbockel L, Brandenburg K, Hornef MW. Antimicrobial peptides and the enteric mucus layer act in concert to protect the intestinal mucosa. Gut Microbes. 2014;5:761–5. doi: 10.4161/19490976.2014.972238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukuda N, Yahagi K, Hara T, Watanabe Y, Matsumoto H, Mori H, et al. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021;15:2574–90. doi: 10.1038/s41396-021-00937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuri R, Shprung T, Shai Y. Defensive remodeling: How bacterial surface properties and biofilm formation promote resistance to antimicrobial peptides. Biochim Biophys Acta Biomembr. 2015;1848:3089–100. doi: 10.1016/j.bbamem.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Jakobsson HE, Rodríguez‐Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16:164–77. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samantha A, Vrielink A. Lipid A Phosphoethanolamine Transferase: regulation, structure and immune response. J Mol Biol. 2020;432:5184–96. doi: 10.1016/j.jmb.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 25.Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev. 2019;32:1–16. doi: 10.1128/CMR.00060-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayfach S, Fischbach MA, Pollard KS. MetaQuery: a web server for rapid annotation and quantitative analysis of specific genes in the human gut microbiome. Bioinformatics. 2015;31:3368–70. doi: 10.1093/bioinformatics/btv382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying W, Tang K, Avolio E, Schilling JM, Pasqua T, Liu MA, et al. Immunosuppression of macrophages underlies the cardioprotective effects of CST (Catestatin) Hypertension. 2021;77:1670–82. doi: 10.1161/HYPERTENSIONAHA.120.16809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 2020;8:1–16. doi: 10.3390/microorganisms8111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indiani CMDSP, Rizzardi KF, Castelo PM, Ferraz LFC, Darrieux M, Parisotto TM. Childhood obesity and firmicutes/bacteroidetes ratio in the gut microbiota: a systematic review. Child Obes. 2018;14:501–9. doi: 10.1089/chi.2018.0040. [DOI] [PubMed] [Google Scholar]
- 30.Lam YY, Ha CWY, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herp S, Durai Raj AC, Salvado Silva M, Woelfel S, Stecher B. The human symbiont Mucispirillum schaedleri: causality in health and disease. Med Microbiol Immunol. 2021;210:173–9. doi: 10.1007/s00430-021-00702-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:1–15. doi: 10.3389/fimmu.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiippala K, Barreto G, Burrello C, Diaz-Basabe A, Suutarinen M, Kainulainen V, et al. Novel Odoribacter splanchnicus strain and its outer membrane vesicles exert immunoregulatory effects in vitro. Front Microbiol. 2020;11:1–14. doi: 10.3389/fmicb.2020.575455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPhee JB, Small CL, Reid-Yu SA, Brannon JR, Moual H LE, Coombes BK. Host defense peptide resistance contributes to colonization and maximal intestinal pathology by Crohn’s disease-associated adherent-invasive Escherichia coli. Infect Immun. 2014;82:3383–93. doi: 10.1128/IAI.01888-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Wei W, Lei S, Lin J, Srinivas S, Feng Y. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. MBio. 2018;9:1–18. doi: 10.3391/mbi.2018.9.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomassin JL, Brannon JR, Gibbs BF, Gruenheid S, Le Moual H. OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL-37. Infect Immun. 2012;80:483–92. doi: 10.1128/IAI.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desloges I, Taylor JA, Leclerc JM, Brannon JR, Portt A, Spencer JD, et al. Identification and characterization of OmpT-like proteases in uropathogenic Escherichia coli clinical isolates. Microbiologyopen. 2019;8:1–36. doi: 10.1002/mbo3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarter JD, Stephens D, Shoemaker K, Rosenberg S, Kirsch JF, Georgiou G. Substrate specificity of the Escherichia coli outer membrane protease OmpT. J Bacteriol. 2004;186:5919–25. doi: 10.1128/JB.186.17.5919-5925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkarni HM, Nagaraj R, Jagannadham MV. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol Res. 2015;181:1–7. doi: 10.1016/j.micres.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Muntjewerff EM, Dunkel G, Nicolasen MJT, Mahata SK, van den Bogaart G. Catestatin as a Target for Treatment of Inflammatory Diseases. Front Immunol. 2018;9:2199. [DOI] [PMC free article] [PubMed]
- 41.Santella RM. Approaches to DNA/RNA extraction and whole genome amplification: table 1. Cancer Epidemiol Biomark Prev. 2006;15:1585–7. doi: 10.1158/1055-9965.EPI-06-0631. [DOI] [PubMed] [Google Scholar]
- 42.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team. R: a language and environment for statistical computing. Vienna, Austria; 2019. https://www.r-project.org/.
- 44.Lahti L, Shetty S. microbiome R package. http://microbiome.github.io.
- 45.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed]
- 46.Paulson JN, Colin Stine O, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–2. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–8. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beresford-Jones BS, Forster SC, Stares MD, Notley G, Viciani E, Browne HP, et al. The Mouse Gastrointestinal Bacteria Catalogue enables translation between the mouse and human gut microbiotas via functional mapping. Cell Host Microbe. 2022;30:124–138.e8. doi: 10.1016/j.chom.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS, Arkin AP. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menardo F, Loiseau C, Brites D, Coscolla M, Gygli SM, Rutaihwa LK, et al. Treemmer: a tool to reduce large phylogenetic datasets with minimal loss of diversity. BMC Bioinforma. 2018;19:1–8. doi: 10.1186/s12859-018-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haider SR, Reid HJ, Sharp BL. Tricine-SDS-PAGE. In: Kurien B., Scofield R. editors. Protein electrophoresis. Methods in Molecular Biology (Methods and Protocols). Totowa, NJ: Humana Press; 2012. p. 81–91.
- 55.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 56.Zoetendal EG, Booijink CCGM, Klaassens ES, Heilig HGHJ, Kleerebezem M, Smidt H, et al. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1:954–9. doi: 10.1038/nprot.2006.143. [DOI] [PubMed] [Google Scholar]
- 57.Zhou K, Zhou L, Lim Q, Zou R, Stephanopoulos G, Too HP. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript and its supplementary information files. The 16S rRNA gene amplicon sequence data were deposited under BioProject number PRJNA741992.