Abstract
Objectives
To determine the reproducibility and replicability of studies that develop and validate segmentation methods for brain tumours on MRI and that follow established reproducibility criteria; and to evaluate whether the reporting guidelines are sufficient.
Methods
Two eligible validation studies of distinct deep learning (DL) methods were identified. We implemented the methods using published information and retraced the reported validation steps. We evaluated to what extent the description of the methods enabled reproduction of the results. We further attempted to replicate reported findings on a clinical set of images acquired at our institute consisting of high-grade and low-grade glioma (HGG, LGG), and meningioma (MNG) cases.
Results
We successfully reproduced one of the two tumour segmentation methods. Insufficient description of the preprocessing pipeline and our inability to replicate the pipeline resulted in failure to reproduce the second method. The replication of the first method showed promising results in terms of Dice similarity coefficient (DSC) and sensitivity (Sen) on HGG cases (DSC=0.77, Sen=0.88) and LGG cases (DSC=0.73, Sen=0.83), however, poorer performance was observed for MNG cases (DSC=0.61, Sen=0.71). Preprocessing errors were identified that contributed to low quantitative scores in some cases.
Conclusions
Established reproducibility criteria do not sufficiently emphasise description of the preprocessing pipeline. Discrepancies in preprocessing as a result of insufficient reporting are likely to influence segmentation outcomes and hinder clinical utilisation. A detailed description of the whole processing chain, including preprocessing, is thus necessary to obtain stronger evidence of the generalisability of DL-based brain tumour segmentation methods and to facilitate translation of the methods into clinical practice.
Keywords: magnetic resonance imaging, diagnostic radiology, neuroradiology, statistics & research methods
Strengths and limitations of this study.
This is an independent evaluation of the reproducibility of deep learning-based lesion segmentation studies that follow established reporting guidelines.
The clinical data set acquired at our institution was suitable for the replication part of the study.
This study did not aim to enable inferences about the clinical utility of the evaluated algorithms.
Introduction
The scientific community has directed substantial efforts at developing deep-learning (DL) methods for medical image analysis. DL methods have become the default choice under the claim of superior performance over classical algorithms.1–3 However, their outstanding performance comes at the cost of high complexity and inherent variability in model performance.3 Consequently, assessing which model design choices determine the empirical gains is challenging.3–5 Critics have also pointed out that scientific reporting of study designs has often been insufficient, and that the analysis of results tends to be biased towards authors’ desired outcomes.4 6 7 These issues present critical challenges to realising the potential of artificial intelligence and translating promising scientific algorithms into reliable and trusted clinical decision support tools.
In our previous work,5 we systematically explored the literature to identify whether prevalent brain lesion segmentation methods are a suitable basis for developing a tool that supports radiological brain tumour status assessment. Our findings corroborated the issues with reporting that may affect reproducibility.5 In particular, reporting of the preprocessing steps is inadequate in many instances.
The problem has been recognised by researchers, and efforts have been made to standardise reporting practices of DL validation studies. The checklist proposed by Pineau et al6 8 identifies a set of items to be reported pertaining to the presented models/algorithms, theoretical claims, data sets, code and experimental results. The reproducibility problem in relation to the specific field of medical image segmentation was highlighted by Renard et al in a literature review.3 The authors present recommendations for the framework description that provides specific context for medical image segmentation. Their recommended items to be reported3 are largely congruent with those proposed by Pineau.6 8 Renard et al,3 however, group their items by sources of variability in the model and evaluation framework, in contrast to grouping by scientific article section, as originally proposed by Pineau et al.6 8
Furthermore, Renard et al3 only identified three out of 29 studies included in their review to be sufficiently described according to their reproducibility recommendations. Two9 10 of the three were algorithms for brain tumour segmentation on MRI. To continue our pursuit of a technically validated DL brain tumour segmentation algorithm that is suitable for clinical validation, we attempted to reimplement the two methods.9 10
The two DL brain tumour segmentation methods were technically validated convolutional neural networks (CNNs). Kamnitsas et al9 developed a three-dimensional (3D) dual-pathway CNN with fully connected 3D conditional random fields.11 The method will be referred to as 3D dual-path CNN in this article. The authors made the method available for independent evaluation (https://github.com/deepmedic) but did not provide a trained model. The software came with a set of configurable network parameters and requirements for the input data. The input data requirements were: images in Neuroimaging Informatics Technology Initiative (NIfTI) file format12; images for each patient and reference labels with optional brain tissue masks (regions of interest—ROIs) had to be coregistered; all images fed to the network had to have the same voxel size; and for optimal performance, MRI signal intensities had to be standardised to have zero-mean and unit variance within each ROI.
Pereira et al developed a two-dimensional (2D) single-pathway CNN, referred to as 2D single-path CNN in this article. The authors published two network architectures (HGG—high-grade glioma and LGG—low-grade glioma) with trained weights.13 The preprocessing described in the original publication consisted of bias field correction with N4ITK,14 followed by intensity normalisation15 of each image. The input patch intensities were finally normalised with the mean and SD calculated from the training patches across each sequence. A roughly similar number of patches was extracted for each class (approximately 50 000 per class for HGG to match the number of patches extracted for training as stated in the original article). The segmentation result was further processed by removing clusters of voxels smaller than a predefined threshold of 10 000 mm3 and 3000 mm3 in HGG and LGG, respectively.
The aim of this study was therefore to determine the reproducibility and replicability of the two methods for brain tumour segmentation9 10 that Renard et al identified as adequately reported3; and to evaluate whether Renard’s and Pineau’s reproducibility recommendations are sufficient also for the task to segment an in-house clinical data set of brain tumours.
Material and methods
Overview
The study design is based on the assumption that the reproducibility items proposed by Renard et al are sufficient for reproduction and replication. We used the definitions of reproduction and replication from the National Academies of Sciences, Engineering and Medicine,16 which Pineau et al also refer to.6 Renard et al identified two methods for brain lesion segmentation9 10 as adequately reported,3 and we chose these two for the present study. Our goal was to implement the respective original methods with all processing steps and parameters and test them on the same data on which they were originally validated (reproducibility). As a measure of success, we compared quantitative results on segmentation accuracy to those reported in the original studies. We then attempted to replicate6 16 the findings: we performed an external validation on a clinically obtained data set from our institution.
Patient and public involvement
No patient involved.
Statistical analysis
We provide descriptive statistics (means and when possible SD) of segmentation evaluation metrics. The metrics we used are: Dice similarity coefficient—DSC, positive predictive value—PPV and sensitivity.
Reproducibility analysis
Evaluated segmentation algorithms
We implemented the two previously proposed DL algorithms for brain tumour segmentation: 3D dual-path CNN9 and 2D single-path CNN.10 In table 1, these algorithms are described in compliance with the reproducibility categories listed by Renard et al,3 together with libraries and computational parameters we used in our implementations. For our implementation, we used hyperparameters reported in the original articles. We trained the 3D dual-path CNN and tested both algorithms on a cluster with a Tesla V100 GPU (5120 cores; Nvidia, Santa Clara, California, USA), 32 GB RAM, and two 8-core Xeon Gold 6244 @ 3.60 GHz processors (Intel, Santa Clara, California, USA).
Table 1.
Description of the two algorithms implemented in the reproducibility analysis, 3D dual-path CNN9 and 2D single-path CNN,10 according to the reproducibility categories proposed by Renard et al3
| Main category | Subcategory | 3D dual-path CNN | 2D single-path CNN |
| Algorithm/model | Description of the DL architecture | Dual-path 3D CNN with a fully connected 3D CRF.11 | Single-path 2D CNN; two network architectures for HGG and LGG. |
| Dataset description | Image acquisition parameters | BraTS 2015 dataset18 | |
| Image size | |||
| Data set size | |||
| Link to the data set | |||
| Preprocessing description | Data excluded +reason | None | None |
| Augmentation transformation | Sagittal reflection of images | Rotation with multiples of 90° angles | |
| Final sample size | Not specified | ~1 800 000 for HGG ~1 340 000 for LGG |
|
| Training/validation/ testing split | Explanation if validation set not created | Training and testing sets provided by the BraTS challenge | |
| CV strategy +no of folds | Not specified | 5-fold CV on training set (n=274) | 1 subject in both HGG (n=220) and LGG (n=54) |
| Optimisation strategy | Optimisation algorithm +reference | RMSProp optimiser30and Nesterov’s momentum31 | Stochastic Gradient Descent and Nesterov’s momentum31 |
| Hyperparameters (learning rate a, batch size n, drop-out d) |
a=10-3 (halved when the convergence plateaus); n=10 d=50% (in the last 2 hidden layers) |
ainital=0.003 afinal=0.00003 n=128 dHGG=0.1 (in FC layers) dHGG=0.5 (in FC layers) |
|
| Hyperparameter selection strategy | CRF: 5-fold CV on a training subset HGG (n=44) and LGG (n=18) | Validation using 1 subject in both HGG (n=220) and LGG (n=54) | |
| Computing infrastructure | Name, class of the architecture, and memory size | NVIDIA GTX Titan X GPU using cuDNN V.5.0, 12 GB | GPU NVIDIA GeForce GTX 980 |
| Middleware | Toolbox used/in-house code +build version | Theano32 Python V.3.6.5, Tensorflow V.2.0.0/1.15.0, Nibabel V.3.0.2 Numpy V.1.18.2 |
Theano V.0.7.032 Lasagne V.0.1dev33 Python V.2.7.10 Numpy V.1.9.2 |
| Source code link +dependencies | https://github.com/deepmedic | http://dei-s2.dei.uminho.pt/pessoas/csilva/brats_cnn/ | |
| Evaluation | Metrics average +variations | Mean of DSC, Precision, and Sensitivity (calculated by the online evaluation system) | Boxplot and mean of DSC (calculated by the online evaluation system) |
| Our implementation middleware | |||
| Python version | 3.8.2 | 3.7.4 | |
| DL library | Tensorflow 2.2.1 | Theano (git version eb6a412), Lasagne (git version 5d3c63c) | |
| Numpy | 1.18.5 | 1.17.3 | |
| Nibabel | 3.0.2 | 3.2.1 | |
All the parameters and versions found in the first part of the table were specified in the original articles. The selection strategy of images to respective cross-validation folds was not specified. In the part ‘our implementation middleware’, we specify the Python version and libraries used for our implementations.
BraTS, Brain Tumour Segmentation Challenge; CNN, convolutional neural networks; CRF, conditional random field; CV, cross-validation; 2D, two dimensions; 3D, three dimensions; DL, deep learning; DSC, Dice similarity coefficient; FC, fully connected; HGG, high-grade glioma; LGG, low-grade glioma.
Image data set used for reproducibility analysis
Both algorithms were originally validated in the 2015 Brain Tumour Segmentation Challenge (BraTS),17 which consists of training and testing image sets of patients diagnosed with HGG and LGG. The training set contains 274 examinations (HGG n=220, LGG n=54). Each examination consists of T1-weighted (T1w) images before and after injection of contrast material (CM), T2w, and FLAIR (fluid-attenuated inversion recovery) images. The training data set additionally contains manual segmentations of tumour structures that serve as a criterion standard and delineate necrotic core, CE core, non-CE core and oedema. For the test set containing 110 examinations the criterion standard segmentations are not publicly available. Users can upload their segmentation results to an online system18 19 that internally compares the results with the hidden reference to determine per-case metrics (DSC, PPV, sensitivity and kappa). The system then returns summary measures (means and ranking position) to the user. Images in both sets are provided in.mha format and have been preprocessed with spatial normalisation,20 skull-stripping,21 and resampling to an isotropic resolution of 1 mm3 (linear interpolator).
Outcome parameters
We experimentally evaluated whether the two methods that Renard et al3 identified as reproducible according to their proposed criteria were possible to reproduce. Specifically, we examined whether enough information was given in the original articles or supplementary information for each processing step. If reimplementation did not reproduce the originally reported results, we contacted the authors directly to follow-up on any missing details and added this information to the results. Pereira et al13 supplied a pretrained model; for 3D dual-path CNN, we trained our reimplementation on the BraTS 2015 training data. Thereafter, we segmented the BraTS 2015 test set with both methods. We submitted the resulting segmentations to the online evaluation system18 and recorded the summary measures returned (mean DSC, mean sensitivity and mean PPV). Finally, we compared the summary measures with those available in the original publications.
Replication analysis
Evaluated segmentation algorithm
Only the 3D dual-path CNN was successfully reimplemented (cf. Results—Reproducibility study). External validation (replication analysis) on in-house clinical data was therefore carried out with this method. The segmentation models trained on the BraTS training data in the reproducibility analysis were applied to our dataset using a workstation with an Intel Core i7-6700HQ CPU @ 2.60 GHz processor and Nvidia GTX960M graphics card.
Image data set used for the replication analysis
The clinical in-house testing data set consisted of images from 27 cases (HGG n=12; LGG n=10; meningioma – MNG n=5). The set was selected for this study from a larger sample of image data. Data were anonymised and inclusion criteria were preoperative examinations, availability of manual expert reference segmentations, and imaging findings typical for the included types of pathology.
As in the BraTS data set, each MR examination included non-CM T1w, CM T1w, T2w, and FLAIR images. The images were provided in NIfTI12 format. Since we used a model trained on BraTS data to segment these images, we used the BraTS-Processor module from the BraTS Toolkit22 for preprocessing. Binary lesion segmentations had been prepared by trained raters and revised by a senior neurosurgeon (ASJ). Whole-tumour labels generated by delineation of T2/FLAIR hyperintensities were used for LGG. For HGG and MNG, the tumour core label was used, which had been delineated on CM T1w images and included CE tumour as well as any components enclosed by CE tumour. The reference segmentations were registered from the native space to the BraTS space following the transformation steps and using the registration matrices generated by the BraTS-Processor.22
Outcome parameters
The replicability of the 3D dual-path CNN was assessed by comparing DSC, sensitivity, and PPV derived from processing the clinical in-house data with those provided by the online system18 during the reproducibility analysis on the BraTS test set. We visually evaluated individual cases to determine causes of segmentation errors.
Based on findings from the reproducibility and the replication analysis we reviewed recommendations on reporting items proposed by Renard et al3 and Pineau et al.8 Challenges and failures in our attempts at reproduction and replication were documented and examined throughout the processes above. We then assessed and summarised these outcomes with suggested specific improvements to the reproducibility items for lesion segmentation on MRI for brain segmentation.
Results
Reproducibility study
3D dual-path CNN
BraTS data fulfilled most of the input requirements for the 3D dual-path CNN, apart from the format and the image intensity normalisation. To reproduce the study, all images were converted to NIfTI format, and MR signal intensities were normalised to have zero-mean and unit-variance within each ROI. We implemented these steps using SimpleITK for image conversion and an in-house python programme for signal intensity normalisation. Since the BraTS images are already skull-stripped, we generated brain masks for each patient by thresholding each image to include only non-zero voxels in order to reduce the runtime of the algorithm. The only changes we made in the 3D dual-path CNN configuration file were to set the number of input channels to all four available, as described in the original article (default in the source code was CE T1w and FLAIR), and to specify not to perform validation of the available samples, as the hyperparameters had already been defined for the model. Training the algorithm took approximately 27 hours, and testing took 14.5 min.
The quantitative evaluation shows that our reimplementation and testing of the 3D dual-path CNN on the BraTS 2015 data set achieved comparable results to those presented in the original study (table 2). We, therefore, deem the method reproducible.
Table 2.
Reproducibility results on BraTS 2015 presented in the original paper for the 3D dual-path CNN9 and for the 2D single-path CNN10 (original) and for our independent reproducibility analysis (this work)
| Dice similarity coefficient | Positive predictive value | Sensitivity | |||||||
| Whole tumour | Tumour core | CE tumour | Whole tumour | Tumour core | CE tumour | Whole tumour | Tumour core | CE tumour | |
| 3D dual-path CNN | |||||||||
| Original | 0.85 | 0.67 | 0.63 | 0.85 | 0.85 | 0.63 | 0.88 | 0.61 | 0.66 |
| This work | 0.85 | 0.68 | 0.64 | 0.85 | 0.83 | 0.62 | 0.88 | 0.64 | 0.70 |
| 2D single-path CNN | |||||||||
| Original | 0.78 | 0.65 | 0.75 | – | – | – | – | – | – |
| This work (HGG) | 0.36 | 0.25 | 0.17 | 0.36 | 0.21 | 0.29 | 0.54 | 0.58 | 0.17 |
| This work (LGG) | 0.25 | 0.14 | 0.13 | 0.40 | 0.51 | 0.37 | 0.25 | 0.10 | 0.10 |
Our analysis was carried out for HGG and LGG model parameters of the 2D single-path CNN. The results were congruent with the original analysis for the 3D dual-path CNN but they show an unsuccessful attempt to reproduce the 2D single-path CNN validation. The higher score in each column is emphasised in bold. Measures of dispersion or significance of differences were not available for the original method evaluation.
BraTS, Brain Tumour Segmentation Challenge; CE, contrast-enhanced; CNN, convolutional neural network; 2D, two dimensions; 3D, three dimensions; HGG, high-grade glioma; LGG, low-grade glioma.
2D single-path CNN
The preprocessing description by Pereira et al lacked certain parameters pertaining to the intensity normalisation: percentile points used to create a reference histogram for each sequence and glioma grade, and intensity parameters of the training patches. Furthermore, it was not specified which model architecture was used on the BraTS 2015 test set, where the data include both HGG and LGG. Despite the missing parameters, we made an attempt to reproduce the study. We used N4ITK bias field correction (as implemented in SimpleITK) with default parameters and a histogram normalisation procedure adapted from Reinhold et al.23 We decided on this implementation instead of the corresponding function in SimpleITK, because the latter requires a reference image or histogram, neither of which was available. For the final patch-normalisation step, the intensity parameters were not available, so we normalised each test image ROI to have zero-mean and unit variance. Finally, the results were postprocessed according to the procedure described by the authors. The testing time of the 2D single-path CNN was approximately 8 hours.
As the attempt was unsuccessful (results of the quantitative evaluation presented in table 2), we approached the lead author of the method and requested the missing information. The author generously provided information on the bias field correction as well as image histogram normalisation parameters.
Following this input, the N4ITK bias field correction was conducted using the implementation in Advanced Normalization Tools (ANTs)24 with the wrapper in Nipype25 with the following parameters specified: n_iterations=(20, 20, 20, 10), dimension=3, bspline_fitting_distance=200, shrink_factor=2, convergence_threshold=0. A visual inspection of the field inhomogeneity correction with ANTs/Nipype and the parameters given versus SimpleITK showed signal intensity differences in the tumour region (figure 1) that plausibly explained the failure to reproduce.
Figure 1.
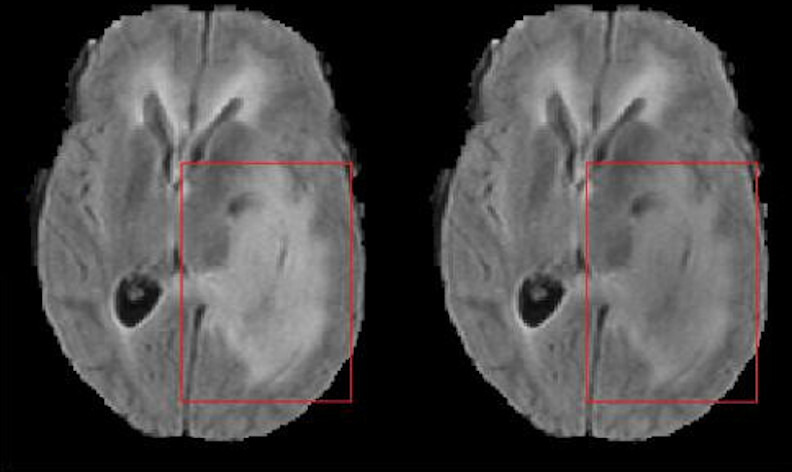
Comparison of the field inhomogeneity correction with ANTs/Nipype (left) and SimpleITK (right). Distinct differences in the FLAIR signal intensity of tumour tissue are visible (red squares). FLAIR, fluid-attenuated inversion recovery. ANTs, Advanced Normalization Tools.
The implementation of Nyul’s algorithm15 for intensity normalisation was developed in the lead author’s former lab, and the author was not at liberty to share the code. Instead, the author provided percentile points and corresponding intensity landmarks for each MR sequence used in their implementation. In the original study, however, the authors trained separate sets of parameters for LGG and HGG and could not retrieve the patch intensity parameters for patch normalisation. To compensate, we extracted the mean and SD from the training images by collecting intensity information of patches sampled from various brain regions to ensure class balance. We imposed a condition that for a given class, a certain percentage of patch pixels are labelled as that class. The values of mean and SD depended on the percentage value, and we did not succeed at finding a value that would improve the segmentation results. At this point, we decided not to pursue further efforts to reproduce the study.
Replication analysis
The replication analysis was conducted on the 3D dual-path CNN only. Quantitative results of the comparison of automatic segmented MRI collected in-house and expert delineations of the chosen tumour labels are presented in table 3.
Table 3.
3D dual-path CNN9 replication analysis results on in-house data for high-grade glioma (HGG) cases and meningioma (MNG) cases evaluated on the tumour core and for low-grade glioma (LGG) cases evaluated on the whole tumour label
| ID | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | Mean | SD |
| HGG cases tumour core | ||||||||||||||
| DSC | 0.88 | 0.85 | 0.80 | 0.85 | 0.89 | 0.85 | 0.57 | 0.89 | 0.86 | 0.81 | 0.87 | 0.14 | 0.77 | 0.22 |
| PPV | 0.84 | 0.86 | 0.72 | 0.84 | 0.85 | 0.79 | 0.41 | 0.85 | 0.80 | 0.73 | 0.80 | 0.08 | 0.72 | 0.23 |
| Sen | 0.93 | 0.85 | 0.89 | 0.87 | 0.92 | 0.91 | 0.89 | 0.93 | 0.93 | 0.91 | 0.96 | 0.61 | 0.88 | 0.09 |
| MNG cases tumour core | ||||||||||||||
| DSC | 0.84 | 0.80 | 0.56 | 0.09 | 0.77 | n.a. | 0.61 | 0.31 | ||||||
| PPV | 0.89 | 0.72 | 0.41 | 0.60 | 0.66 | 0.66 | 0.18 | |||||||
| Sen | 0.79 | 0.90 | 0.92 | 0.05 | 0.93 | 0.71 | 0.38 | |||||||
| LGG cases whole tumour | ||||||||||||||
| DSC | 0.35 | 0.70 | 0.89 | 0.58 | 0.93 | 0.85 | 0.83 | 0.85 | 0.54 | 0.77 | n.a | 0.73 | 0.18 | |
| PPV | 0.27 | 0.55 | 0.86 | 0.43 | 0.93 | 0.77 | 0.88 | 0.90 | 0.43 | 0.74 | n.a | 0.67 | 0.24 | |
| Sen | 0.52 | 0.93 | 0.92 | 0.89 | 0.93 | 0.95 | 0.78 | 0.80 | 0.75 | 0.80 | n.a | 0.83 | 0.13 | |
DSC, Dice similarity coefficient; n.a, not available; PPV, positive predictive value; Sen, sensitivity.
The average performance results of the replicability analysis using the in-house image set and the reproducibility results are compiled in table 4 for comparison.
Table 4.
Comparison of the mean results of the reproducibility (BraTS 2015 test set) and replicability (in-house image set) analysis of the 3D dual-path CNN9
| Data set: | In-house image set | BraTS 2015 test image set | ||
| Cases: | HGG | MNG | LGG+HGG | |
| Tumour core | DSC | 0.77 | 0.61 | 0.68 |
| PPV | 0.72 | 0.66 | 0.83 | |
| Sen | 0.88 | 0.71 | 0.64 | |
| Cases: | LGG | LGG+HGG | ||
| Whole tumour | DSC | 0.73 | 0.85 | |
| PPV | 0.83 | 0.85 | ||
| Sen | 0.67 | 0.88 | ||
BraTS, Brain Tumour Segmentation Challenge; CNN, convolutional neural network; 3D, three dimensions; DSC, Dice similarity coefficient; HGG, high-grade glioma; LGG, low-grade glioma; MNG, meningioma; PPV, positive predictive value; Sen, sensitivity.
The visual evaluation of individual cases revealed a variety of causes of poor performance. In HGG visual inspection of Case #07 results showed that the 3D dual-path CNN misclassified brain tissue voxels in the vicinity of the tumour core (figure 2, top row). A similar problem was observed in case #12 (figure 2, middle row). The algorithm failed to segment a tumour in MNG case #04 (figure 2, bottom row). While the tumour location and appearance (uncharacteristic for glioma) may be the reason for a poor result, we also note that the brain mask generated in the preprocessing by BraTS Processor failed to include a part of the reference label. For LGG the algorithm achieved relatively poor results for cases #01 and #09. The results obtained for LGG Case #01 revealed a segmentation error as a result of a preprocessing error: the brain mask included periocular tissue that was classified as tumour by the segmentation algorithm (figure 3, top row). In LGG Case #09, the 3D dual-path CNN labelled a substantial portion of the brain that was not included in the reference segmentation (figure 3, bottom row).
Figure 2.
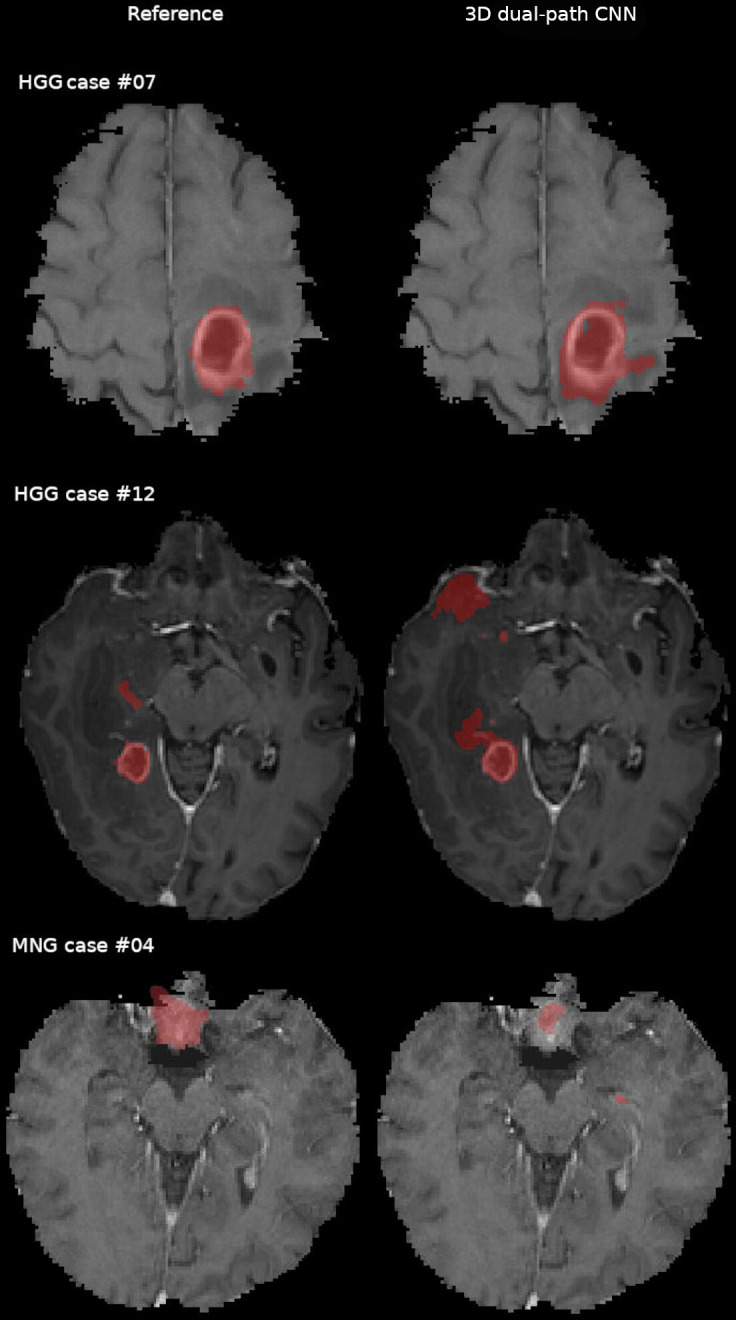
Comparison of the expert segmentation (reference) and the three-dimensional (3D) dual-path CNN tumour core segmentation in the in-house data for high-grade glioma (HGG) and meningioma (MNG) cases overlaid on contrast enhanced T1 weighted. Voxels misclassified by the 3D dual-path CNN are visible in HGG cases #07 and #12 (top and middle row). The 3D dual-path CNN failed to correctly outline the tumour and included normal brain structures in the left medial temporal lobe for MNG case #04 (bottom row). CNN, convolutional neural network.
Figure 3.
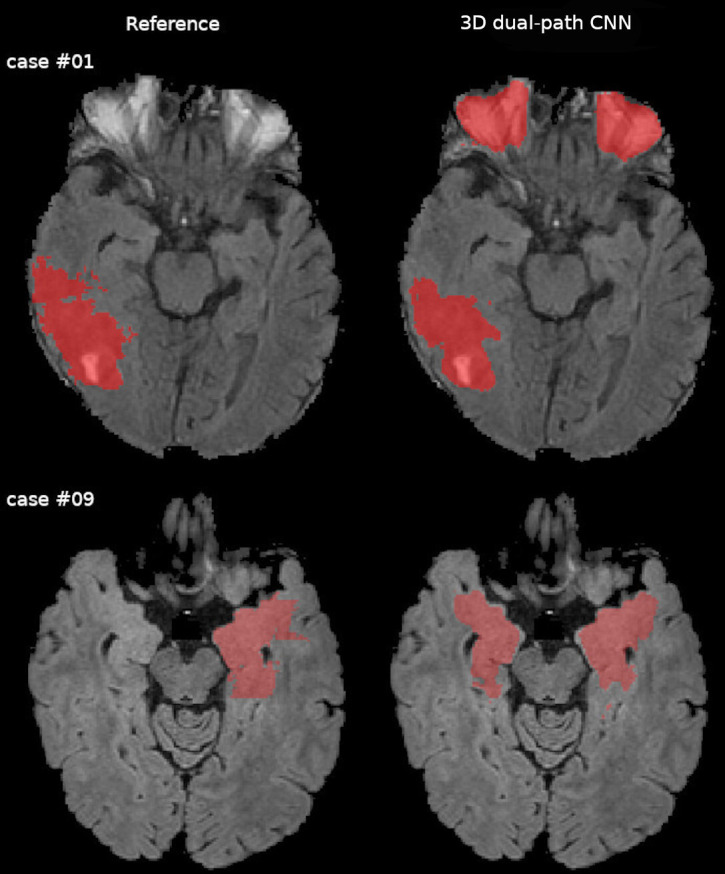
Comparison of the expert segmentation (reference) and the three-dimensional (3D) dual-path CNN whole tumour segmentation in the in-house data for low-grade glioma cases displayed overlaid on FLAIR images. Voxels misclassified by the 3D dual-path CNN are visible bilaterally in the orbit in case #01 (top row), which should have been excluded by the skull stripping procedure. In case #09 (middle row), the 3D dual-path CNN misclassified contralateral, sequence-depended FLAIR hyperintensities. CNN, convolutional neural network; FLAIR, fluid-attenuated inversion recovery.
Proposed updates to the checklist
From our results we deducted that insufficient description of the preprocessing was the main obstacle to reproducing Pereira’s et al10 results. We; therefore, present an updated reproducibility and replicability checklist for medical segmentation studies (table 5).
Table 5.
A suggested reproducibility and replicability checklist for automatic medical image segmentation studies
| Data set—description of the image data set used for model development and validation | |
|
□ □ □ □ |
| Data set preprocessing—description of the processing steps applied to the raw images before they can be fed to the segmentation model: | |
|
□ □ □ □ |
| Segmentation model—description of the model’s architecture used for the segmentation: | |
|
□ □ □ □ □ |
| Postprocessing—description of all processing steps and corresponding parameters applied to the output of the segmentation algorithm before evaluation: | |
|
□ □ □ |
| Model development—description of the training/validation and optimisation strategies: | |
|
□ □ □ □ □ □ □ □ |
| Computing infrastructure—description of the hardware used: | |
|
□ □ □ |
| Model evaluation—description of the model evaluation: | |
|
□ □ □ □ |
The update from the established checklists3 8 includes a new category data set preprocessing, and a new item in model evaluation category: Failed cases: number and reasons. We also regrouped the items into categories that provide a clearer structure for reporting in particular of reproducibility and replicability studies.
Discussion
Reproducibility and replicability of scientific results are the foundation of evidence-based medicine. In this work, we show that current guidelines for publishing validation studies on DL algorithms are incomplete. While attempting to reproduce the two studies on MR brain lesion segmentation that were identified as meeting current reproducibility recommendations,3 we found that only one of them was reproducible based on the published information. Remarkably, even after consultation with the authors of the second method, we were not able to obtain satisfactory segmentation results with their method. Our claims of reproducibility/non-reproducibility could not be supported with advanced statistical analysis; the online evaluation system18 (used to evaluate the segmentations in the original validation papers and our study) provides arithmetic means of the evaluation metrics without measures of dispersion. The small sample size of the in-house data along with the difference in tumour components segmented as a reference for HGG (tumour core) and LGG (whole tumour) further precludes a meaningful analysis of the statistical difference between the results obtained in the reproducibility and replicability analysis. We believe that our findings are nevertheless sufficient to support our conclusions.
We furthermore attempt to externally validate the findings reported for the 3D dual-path CNN on a set of own data. We found that the available preprocessing pipeline is not free from producing errors, which directly influences the segmentation outcome. Moreover, we observed a poorer performance of the algorithm in MNG cases. This is, however, a somewhat expected behaviour since the training set did not contain any MNG tumours. On the other hand, visual inspection also revealed that the 3D dual-path CNN segmentation errors may arise from preprocessing errors. Nonetheless, our results acquired with the BraTS-Processor and the 3D dual-path CNN are promising, and we have begun to explore the potential of this pipeline for clinical application. Unfortunately, the experience gained through this study suggests that the available algorithms are not, in their present form, ready to be implemented in clinical routines. This, despite their meeting the recommended criteria for reproducibility as outlined by Pineau et al6 8 and Renard et al.3 Improving the reproducibility of technical validation studies of DL segmentation methods will lay a foundation for producing strong evidence for what algorithms work best, when, and why. It will furthermore facilitate creating standardised evaluation frameworks and create a solid base for implementing DL tools in clinical routines.
Reproducibility criteria
The items that Renard et al3 identified as necessary to reproduce a DL methodology study are divided into information about hyperparameters (optimisation, learning rate, drop-out, batch size) and the data set used (training proportion, data augmentation and validation set). All these items are indeed included in the two studies we attempted to reproduce.9 10 The current recommendations do not, however, sufficiently stress the importance of thorough documentation of the image preprocessing chain.
The approach to preprocessing of the training and testing data is different between the two highlighted segmentation studies. The authors of the 3D dual-path CNN guarantee optimal performance of the algorithm on images prepared for the BraTS segmentation challenge (skull stripping, spatial normalisation, and resampling) with an additional intensity normalisation step. The 2D single-path CNN, on the other hand, achieved its reported high accuracy after more complex preprocessing had been applied. For our study, intensities of the whole images were corrected for field inhomogeneity, and histograms normalised across each sequence. The final preprocessing step involved patch normalisation. These procedures were not explicitly described. We requested the missing information from the authors, and while they were supportive in principle, they were unable to supply the patch intensity information. Unsurprisingly, the results show poor accuracy due to our inability to reproduce the intensity normalisation procedures conducted in the original study.
The problem of insufficient reporting of the preprocessing procedures has been recognised previously.5 While preprocessing may be less important in the context of segmentation challenges, evaluating the whole processing chain, from raw images to the final segmentation, is crucial in the context of application to independently collected data. Without the ability to reproduce the whole processing chain, meaningful method comparison and validation on external data becomes impossible.
Our findings prompt us to propose a significant modification to the previously reported reproducibility checklist by Pineau et al6 and Renard et al’s guidelines.3 We present this new checklist in table 5. First, we add what we conclude to be a necessary and sufficient description of the preprocessing. Second, we regroup the items to provide a clearer distinction between the various elements and aspects that are involved in the algorithm development versus the validation of the medical image segmentation tool: such a structure for providing a more transparent and easily implemented way of reporting is specifically designed to help those who seek to reproduce and replicate. More generally, these modifications are critical to improving the reproducibility and replicability of medical image segmentation methods. Since our updates are based on reproducibility and replicability of only two segmentation algorithms, we encourage researchers to comprehensively evaluate our checklist by including a broader selection of independently implemented algorithms for medical image segmentation.
Replication analysis
The external validation was conducted on locally acquired images. We cannot draw definitive conclusions regarding the 3D dual-path CNN’s performance in a clinical setting as statistical analysis would not be meaningful; in the in-house data, we evaluated separately tumour core label in HGG examinations and whole tumour label in LGG examinations. The BraTS evaluations for both tumour components are, on the other hand, done on a mix of HGG and LGG cases. Because of our small sample size, we also cannot make inferences about applying DL methods trained on glioma cases to other tumour cases. Our results, however, are promising. The analysis further highlighted how essential the preprocessing chain is for accurate brain tumour segmentation with the 3D dual-path CNN and likely with any other DL segmentation method.
In our pipeline, we used BraTS-Processor to take advantage of a tool that will automatically apply all the preprocessing steps that were also applied to the training set. Our analysis revealed segmentation errors that could be traced to errors in the preprocessing. Cases of errors in the skull stripping, which we observed in the in-house data, have been reported previously26 27 and will likely cause occasional problems in the future. Nonetheless, the processing pipeline generates segmentations that, even if erroneous in a few cases, will be easy to correct if the operator is equipped with a suitable interactive label editing tool. Developers of clinical tools should be aware of the issue and enable users to easily remove mislabelled regions.28
In addition to the noted preprocessing errors, we encountered another problem that likely influenced the results: the BraTS-Processor outputs images in the BraTS (MNI15229) space. To evaluate the automatic segmentations quantitatively, we had to transform the reference segmentations from the native space to the BraTS space as well. This resulted in visible distortions to the reference segmentations. Accordingly, the results we presented (table 5) likely underestimate the performance of the method (BraTS-Processor plus the 3D dual-path CNN) on externally acquired data. For a more accurate evaluation of a given processing pipeline, reference segmentations should be delineated on images in the BraTS space. While it may not be feasible in retrospective studies, it is a vital study design step for prospective studies.
Conclusions
Established reproducibility criteria for studies developing and validating DL lesion segmentation algorithms are not sufficient with regard to the preprocessing steps. The results of the reproducibility analysis led us to propose a new reproducibility checklist for medical image segmentation studies, especially if clinical utility of the algorithms is the goal. We further highlighted that even a fully reproducible preprocessing method is prone to errors on routine clinical images, which is likely to impair the segmentation outcome. We encourage researchers in the field of medical image segmentation to follow our modified checklist and assess it in terms of practical utility.
Supplementary Material
Acknowledgments
The computations were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at Chalmers Centre for Computational Science and Engineering (C3SE), partially funded by the Swedish Research Council through grant agreement no. 2018-05973. The authors would like to thank S. Pereira for providing us with details of his study, his support and interest in our work.
Footnotes
Contributors: EG conducted the study and led the writing of the article. RAH acts as the guarantor and was the main supervisor and consultant of the study progress and design choices. JS and IB-B were co-supervising the study progress at all stages. ASJ and TD provided us with the in-house collected images, reference segmentations, and design input for the external validation. All co-authors collaborated on manuscript composition and editing.
Funding: This work was supported by VGR InnovationsFonden (VGRINN-940050) and ALF funds (SU2018-03591 and ALFGBG-925851).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available in a public, open access repository. The code generated for this study is available from https://github.com/emiliagyska/repro_study.git.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the Swedish Ethical Review Authority, Dnr 702-18. The Swedish Ethical Review Authority waived the requirement of informed consent for this study (Dnr 702-18).
References
- 1.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 2.Park JE, Kickingereder P, Kim HS. Radiomics and deep learning from research to clinical workflow: neuro-oncologic imaging. Korean J Radiol 2020;21:1126–37. 10.3348/kjr.2019.0847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renard F, Guedria S, Palma ND, et al. Variability and reproducibility in deep learning for medical image segmentation. Sci Rep 2020;10:13724. 10.1038/s41598-020-69920-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipton ZC, Steinhardt J. Research for practice: troubling trends in machine-learning scholarship. Commun ACM 2019;62:45–53. 10.1145/3316774 [DOI] [Google Scholar]
- 5.Gryska E, Schneiderman J, Björkman-Burtscher I, et al. Automatic brain lesion segmentation on standard magnetic resonance images: a scoping review. BMJ Open 2021;11:e042660. 10.1136/bmjopen-2020-042660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pineau J, et al. Improving reproducibility in machine learning research (a report from the NeurIPS 2019 reproducibility program). ArXiv 2020. http://arxiv.org/abs/2003.12206 [Google Scholar]
- 7.Haibe-Kains B, Adam GA, Hosny A, et al. Transparency and reproducibility in artificial intelligence. Nature 2020;586:E14–16. 10.1038/s41586-020-2766-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pineau J. Machine learning reproducibility checklist. Available: https://www.cs.mcgill.ca/~jpineau/ReproducibilityChecklist.pdf [Accessed 01 Oct 2021].
- 9.Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78. 10.1016/j.media.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 10.Pereira S, Pinto A, Alves V, et al. Brain tumor segmentation using Convolutional neural networks in MRI images. IEEE Trans Med Imaging 2016;35:1240–51. 10.1109/TMI.2016.2538465 [DOI] [PubMed] [Google Scholar]
- 11.Krähenbühl P, Koltun V. Efficient inference in fully connected CRFs with Gaussian edge potentials. arXiv 2012:9. [Google Scholar]
- 12.Data Format Working Group, . NIfTI: — neuroimaging informatics technology initiative. Available: https://nifti.nimh.nih.gov/ [Accessed 28 Jan 2021].
- 13.Pereira S, Pinto A, Alves V. Brain tumor segmentation using Convolutional neural networks in MRI images. Available: http://dei-s2.dei.uminho.pt/pessoas/csilva/brats_cnn/ [DOI] [PubMed]
- 14.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29:1310–20. 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 2000;19:143–50. 10.1109/42.836373 [DOI] [PubMed] [Google Scholar]
- 16.E. National Academies of Sciences . Reproducibility and Replicability in science, 2019. [PubMed] [Google Scholar]
- 17.Menze BH, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993–2024. 10.1109/TMI.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BRATS - SICAS Medical Image Repository. Available: https://www.smir.ch/BRATS/Start2015 [Accessed 28 Jan 2021].
- 19.Kistler M, Bonaretti S, Pfahrer M, et al. The virtual skeleton database: an open access repository for biomedical research and collaboration. J Med Internet Res 2013;15, :e245. 10.2196/jmir.2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibanez L, et al. The ITK software guide. Kitware Inc, 2003: 2018. [Google Scholar]
- 21.Bauer S, Fejes T, Reyes M. A Skull-Stripping filter for ITK. Insight J 2012:859. [Google Scholar]
- 22.Kofler F, Berger C, Waldmannstetter D, et al. BraTS toolkit: translating BraTS brain tumor segmentation algorithms into clinical and scientific practice. Front Neurosci 2020;14:125. 10.3389/fnins.2020.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinhold JC, Dewey BE, Carass A, et al. Evaluating the impact of intensity normalization on MR image synthesis. Proc SPIE Int Soc Opt Eng 2019;10949. 10.1117/12.2513089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avants BB, Tustison NJ, Stauffer M, et al. The insight toolkit image registration framework. Front Neuroinform 2014;8:44. 10.3389/fninf.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorgolewski K, Burns CD, Madison C, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform 2011;5:13. 10.3389/fninf.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellner-Weldon F, Stippich C, Wiest R, et al. Comparison of perioperative automated versus manual two-dimensional tumor analysis in glioblastoma patients. Eur J Radiol 2017;95:75–81. 10.1016/j.ejrad.2017.07.028 [DOI] [PubMed] [Google Scholar]
- 27.Maier O, Wilms M, von der Gablentz J, et al. Extra tree forests for sub-acute ischemic stroke lesion segmentation in MR sequences. J Neurosci Methods 2015;240:89–100. 10.1016/j.jneumeth.2014.11.011 [DOI] [PubMed] [Google Scholar]
- 28.Dietvorst BJ, Simmons JP, Massey C. Overcoming algorithm aversion: people will use imperfect algorithms if they can (even slightly) modify them. Manage Sci 2018;64:1155–70. 10.1287/mnsc.2016.2643 [DOI] [Google Scholar]
- 29.Evans AC, Collins DL, Mills SR. 3D statistical neuroanatomical models from 305 MRI volumes. IEEE Conference Record Nuclear Science Symposium and Medical Imaging Conference 1993;3:1813–7. 10.1109/NSSMIC.1993.373602 [DOI] [Google Scholar]
- 30.Tieleman T, Hinton G. Lecture 6.5-rmsprop: divide the gradient by a running average of its recent magnitude. COURSERA: Neural Networks for Machine Learning 2012;4:26–31. [Google Scholar]
- 31.Sutskever I, Martens J, Dahl G. On the importance of initialization and momentum in deep learning. ICML 2013;3:1139–47. [Google Scholar]
- 32.Bastien F, et al. Theano: new features and speed improvements. ArXiv 2012. [Google Scholar]
- 33.Dieleman S, et al. Lasagne: first release 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a public, open access repository. The code generated for this study is available from https://github.com/emiliagyska/repro_study.git.


