Abstract
Environmental survival of Escherichia coli O157 may play an important role in the persistence and dissemination of this organism on farms. The survival of culturable and infectious E. coli O157 was studied using microcosms simulating cattle water troughs. Culturable E. coli O157 survived for at least 245 days in the microcosm sediments. Furthermore, E. coli O157 strains surviving more than 6 months in contaminated microcosms were infectious to a group of 10-week-old calves. Fecal excretion of E. coli O157 by these calves persisted for 87 days after challenge. Water trough sediments contaminated with feces from cattle excreting E. coli O157 may serve as a long-term reservoir of this organism on farms and a source of infection for cattle.
Escherichia coli O157 is an important human pathogen worldwide (12, 23). Most human infections are acquired from the consumption of foods and water contaminated directly or indirectly with bovine fecal material (2). Hence, lowering the amount of E. coli O157 excreted in the feces of individual cattle and minimizing the number of cattle excreting E. coli O157 are predicted to significantly reduce the incidence of human E. coli O157-related diseases (6, 17, 18, 20). Unfortunately, methods to effectively control E. coli O157 in cattle have yet to be identified.
Environmental persistence of E. coli O157 may play a key role in the epidemiology of this agent on farms. Cattle are considered the primary reservoir for this pathogen, but fecal excretion of E. coli O157 by cattle is only transient, typically lasting 3 to 4 weeks (14, 21, 26). In contrast, E. coli O157 can be repeatedly isolated from environmental sources on farms for periods lasting several years (14, 21, 24). The stability of molecular subtypes of E. coli O157 on farms despite the intermittent detection in cattle is consistent with the presence of nonbovine reservoirs for E. coli O157 on farms.
E. coli O157 shedding in cattle populations is spatially and temporally clustered, consistent with point sources of exposure to the organism, such as periodically contaminated feed or water (14, 21). Water offered to livestock is often of poor microbiological quality, and E. coli O157 is present in as many as 10% of troughs (9, 13). Although drinking water is recognized as an important vehicle in human E. coli O157 infections, it is not known whether cattle drinking from previously E. coli O157-contaminated water troughs are prone to colonization with this agent. If E. coli O157 persists and remains infectious in livestock water troughs, then farm management practices that target this environmental reservoir may ultimately aid in the control of E. coli O157 in cattle.
The goal of this study was to determine the significance of cattle water troughs as environmental reservoirs of E. coli O157. To that end, it was important to determine not only whether E. coli O157 could survive in this environment but also whether it would maintain the ability to infect cattle after undergoing the phenotypic, metabolic, and genetic changes often associated with prolonged environmental exposure (27, 28). It was also of interest to determine whether water chlorination, a common water treatment practice, would affect the survival of E. coli O157 in previously contaminated water troughs.
MATERIALS AND METHODS
Inoculation strain.
A bovine fecal isolate of E. coli O157 was identified by characteristic biochemical reactions, including lactose fermentation, the absence of sorbitol fermentation, and the absence of cleavage of 4-methylumbelliferyl glucuronide, by agglutination with a latex test for the O157 antigen (Oxoid, Basingstoke, Hampshire, United Kingdom), and by PCR detection of the stx2, eaeA, and fliCh7 genes (10, 30). A nalidixic acid-resistant strain of this isolate was selected by overnight culture in lauryl broth (Difco Laboratories, Detroit, Mich.) at 37°C followed by plating on MacConkey agar (Difco) containing 20 μg of nalidixic acid (MACNAL) (United States Biochemical Corporation, Cleveland, Ohio)/ml. A single colony (WSU2032) from this plating was further amplified by overnight culture in tryptic soy broth (TSB) (Difco) at 37°C.
A 12-week-old weaned Holstein calf was tested for fecal carriage of E. coli O157 by enrichment culture in TSB containing cefixime (50 ng/ml; Wyeth-Ayerst Laboratories, Pearl River, N.Y.) and vancomycin (40 μg/ml; Sigma Chemical Company, St. Louis, Mo.) followed by plating of serial 10-fold dilutions on sorbitol MacConkey agar (Difco) containing cefixime (50 ng/ml) and potassium tellurite (2.5 μg/ml; Sigma) (25). After three consecutive negative tests, the calf was orally challenged with 1010 CFU of an overnight TSB culture of WSU2032. Feces from this calf were collected from 2 to 7 days postchallenge and stored at room temperature. A portion of this fecal material was used in the subsequent experiments.
Experiment 1. (i) Survival of E. coli O157 in sediments of experimental microcosms.
Continuous-flow polypropylene chambers (80 liters, 57 by 47 by 30 cm) were used as experimental microcosms. To mimic recently contaminated water troughs, 12 kg of feces collected from the E. coli O157-challenged calf described above was combined with an equal weight of sediments freshly collected from feedlot water troughs. This mixture was equally divided among 12 microcosms. The initial concentration of E. coli O157 present in the sediments was 9 × 108 CFU/g, a total dose approximating that found in 10 kg of feces containing 107 CFU of E. coli O157/g. Clean water was pumped onto the surface of the narrow side of each microcosm at the rate of 160 liters/day. A drain was located directly opposite the input, 45 cm above the surface of the sediments. Troughs were loosely covered with lids and maintained in a secure fenced outdoor location from April to December 1999.
The microcosms were assigned to treatment groups, and the persistence of E. coli O157 was studied during two sequential periods comparing two different chlorine concentrations. In the first period (days 0 to 90), source water containing 0.15 ppm of free chlorine was allowed to flow from a header tank directly into six microcosms (chlorinated), whereas residual chlorine was removed by activated charcoal filtration from the input water of the remaining six microcosms (unchlorinated) (OMNIFilter model OB3 with GAC1 filter; Sta-Rite Industries Inc., Delavan, Wis.). During the second period (days 91 to 245), the chlorine level in the input water of the chlorinated microcosms was increased to 5 to 7 ppm by placing in the header tank of the chlorinated group a floating tablet chlorinator (HTH Floater; Arch Chemicals, Norwalk, Conn.) filled with 2.54-cm-diameter (14-g) trichloro-s-triazinetrione (stabilized chlorine) tablets (Arch Chemicals). The E. coli O157 concentrations in the microcosm sediments during these two periods were compared using a repeated-measure analysis of variance using the GLM ANOVA procedure of NCSS 2000 (NCSS, Kaysville, Utah). The type I error was set at 0.05 in two-tailed tests.
(ii) Detection of E. coli O157 in microcosms.
Sediment samples were collected in sterile sample bags at monthly intervals commencing on day 60. The samples settled at room temperature for at least 5 min before any water collected with the sample was decanted and discarded. Ten milliliters of sterile deionized distilled water was added to a 1-g (wet weight) aliquot of each sediment sample, and the diluted samples were mixed thoroughly (30 s, medium setting) (Stomacher 80; Seward Medical, London, United Kingdom). Additional 10-fold serial dilutions of the homogenized sample were made in sterile deionized distilled water, and 1-ml aliquots of each dilution were spread on 150-mm MACNAL plates. The plates were incubated overnight at 37°C, and lactose-positive colonies were enumerated. Ten lactose-positive colonies from each plate were further confirmed to be E. coli O157 using the criteria described in the previous section.
Experiment 2. (i) Calf challenge.
To determine whether E. coli O157 strains persisting in a microcosm for 6 months or longer remained infectious to cattle, four 10-week-old weaned male Holstein calves were challenged by sequential 14-day exposures to water from two randomly selected microcosms. Prior to this challenge, the animals were screened twice for fecal carriage of E. coli O157 as previously described. The calves were housed together in a large pen within a biocontainment facility and provided with free-choice hay and a calf starter grain ration typical of that fed to U.S. dairy calves.
Fresh water was added to the challenge microcosm to replace water consumed by the calves. Immediately after the microcosm was refilled, the concentration of E. coli O157 in the water column was determined by spread plating 1 ml of water on MACNAL plates and incubating them overnight at 37°C. Lactose-positive colonies were enumerated and subsequently identified as E. coli O157 as described above. An additional 20 ml of water was added to an equal volume of 2× concentrated TSB, incubated overnight at 37°C, and subsequently plated on MACNAL to identify E. coli O157 at concentrations below those detectable by the direct plating technique (<1 CFU/ml).
In order to differentiate between transient passage of E. coli O157 through the gastrointestinal tract of calves and proliferation and persistence of this organism in the calves (i.e., colonization), the experimental microcosms were replaced with a clean water source after the confirmation of E. coli O157 in calf feces. Initially, the microcosms were removed immediately following the detection of a single positive fecal sample and replaced following a culture-negative result. Subsequently, the calves were allowed to drink from the microcosms until at least two sequential fecal culture-positive results were obtained before the microcosms were removed from the calves' environment.
Calf fecal samples were cultured to detect E. coli O157 at 2 to 4-day intervals using overnight enrichment in TSB containing cefixime and vancomycin, as previously described, and spread plating of 1-ml aliquots of enriched broth onto MACNAL. Following the detection of the agent in two sequential samples from any single calf, subsequent fecal samples were analyzed quantitatively twice monthly until negative samples were obtained using the method described in the previous section for E. coli O157 in sediments. Fecal sampling was discontinued only when four sequential negative fecal cultures from each calf were obtained.
(ii) PFGE.
To confirm that the organisms recovered from the microcosms were of experimental origin and did not represent exogenous contamination of the experimental system, WSU2032 (the challenge organism) and three E. coli O157 isolates recovered from each experimental microcosm on days 183 and 245 of the study were examined by pulsed-field gel electrophoresis (PFGE) using XbaI digestion (7). Furthermore, three isolates collected on each sample day from the microcosms during the calf challenge experiment and three isolates collected from all calves on each sample day were evaluated by PFGE.
RESULTS
Experiment 1: survival of E. coli O157 in experimental microcosms.
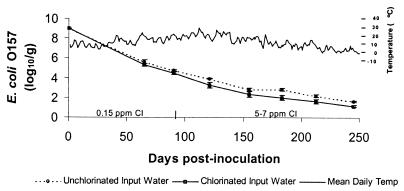
In the first study period (0 to 90 days postinoculation), the concentration of E. coli O157 in the microcosms decreased significantly with time (P < 0.01) (Fig. 1). Concentrations of E. coli O157 were significantly lower in microcosms receiving chlorinated water (0.15 ppm of free chlorine) than in those receiving unchlorinated source water (P = 0.04). There was no significant interaction between the effects of time and chlorination. In the second study period (days 91 to 245), during which a higher chlorination level was maintained in treated microcosms, the concentration of E. coli O157 in the sediments continued to decline in both groups (P < 0.01) and remained lower in the chlorinated microcosms (P = 0.02). Again, there was no significant interaction between the effects of time and chlorination. Culturable E. coli O157 strains remaining in the sediments of the microcosms at 183 and 245 days postinoculation averaged 102.82 and 101.62 CFU/g in the unchlorinated microcosms, respectively, and 102.00 and 101.16 CFU/g in the chlorinated microcosms, respectively.
FIG. 1.
Changes in log10 concentrations of E. coli O157 in sediments of microcosms simulating cattle water troughs. Chlorine concentration: prior to day 90, 0.15 ppm; after day 90, 5 to 7 ppm. Bars represent standard errors.
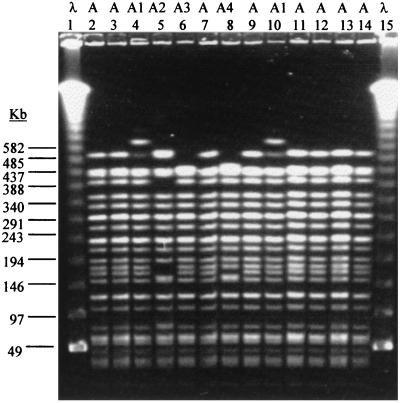
PFGE patterns of most isolates obtained from the microcosms at 183 and 245 days postinoculation were indistinguishable from that of the inoculum strain, WSU2032. Minor differences (Fig. 2) were observed in isolates obtained from two of the chlorinated microcosms on day 183 postinoculation (Fig. 2, patterns A1 and A2) and from a third chlorinated microcosm on day 245 postinoculation (Fig. 2, pattern A3). The microcosms from which these variants were obtained were not among those randomly selected for use in the second experiment.
FIG. 2.
PFGE patterns of recovered E. coli O157 isolates. Patterns, designated A (initial inoculum) and A1, A2, A3, and A4 (variants), are indicated at the top. Lanes 1 and 15, bacteriophage lambda DNA ladder standard for PFGE applications (Bio-Rad); lane 2, inoculum; lanes 3 to 6, microcosms; lanes 7 and 8, drinking water; lanes 9 to 13, calves; lane 14, inoculum. Molecular sizes are indicated at left.
Experiment 2: calf challenge.
For calf challenges, microcosms contaminated 183 days earlier were presented to the animals as the sole source of water. The first randomly selected microcosm had received unchlorinated input water and was presented to the calves from days 0 to 4 and again from days 7 to 14. Because E. coli O157 was detected in the feces of two calves on day 4, the microcosm water source was replaced with chlorinated water in a clean, automatically filling, 5-liter water bowl on days 5 and 6. The second randomly selected challenge microcosm had received chlorinated water and was the sole source of water available to the calves from days 15 to 28, by which time all exposed calves had become infected. The initial concentration of E. coli O157 in the water column of the first microcosm used for the challenge was 127 CFU/ml. However, the measured concentrations of suspended E. coli O157 strains in the microcosms during the challenge periods were variable and frequently below 1 CFU/ml (Table 1). Interestingly, the concentrations of E. coli O157 in the challenge microcosm increased on day 28 (that is, after the calves had become colonized), suggesting that recontamination may have occurred.
TABLE 1.
Water source and E. coli O157 concentrations in microcosms during the calf exposure experiment
| Microcosm | Concn (log10 CFU/ml) of E. coli O157 on daya:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 7 | 9 | 12 | 14 | 16 | 19 | 23 | 26 | 28 | >28 | |
| Unchlorinated water | 2.1 | 1.1 | 0.7 | 3 | E | E | ND | |||||||
| Chlorinated water | 0.8 | 0.6 | E | E | E | 1.4 | ||||||||
| Clean drinking water | NT | |||||||||||||
E, enrichment only, indicating that the agent was present at a concentration of <1 CFU/ml, the limit of quantitation; ND, none detected; NT, not tested.
All of the isolates recovered from the microcosms during the challenge experiment had PFGE banding patterns indistinguishable from or closely related to the test organism, WSU2032 (Fig. 2). Two isolates with identical minor differences (pattern A4; data not shown) from WSU2032 were collected on day 19 from the microcosms during the challenge experiment. No other pattern A4 isolates were obtained during the course of this study.
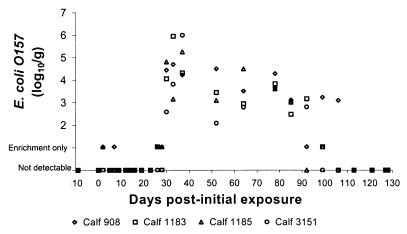
E. coli O157 was not detected in the feces of the calves prior to challenge with the microcosms. The calves remained alert, maintained normal appetites, and had no evidence of diarrhea throughout the duration of the experiment. The temporal pattern and magnitude of fecal detection of E. coli O157 in the challenged animals is displayed in Fig. 3. E. coli O157 was detected in calves 908 and 1185 on day 2 and again in calf 908 on day 7. All subsequent fecal cultures were negative until day 26. Between days 26 and 30 all animals became culture positive for E. coli O157 and remained so for up to an additional 87 days.
FIG. 3.
Temporal pattern of fecal excretion of E. coli O157 among calves drinking from microcosms contaminated with E. coli O157 6 months earlier.
During the entire period of fecal excretion, most calf isolates (107 of 108, 99%) matched exactly the PFGE pattern of the inoculum strain, WSU2032. The single isolate exhibiting minor PFGE differences from WSU2032 was isolated from calf 908 on day 99, and its pattern was indistinguishable from pattern A1.
DISCUSSION
The results from these studies demonstrate that cattle water troughs can serve as environmental reservoirs for E. coli O157 and as a long-term source for cattle infection. E. coli O157 remaining in the microcosms longer than 6 months maintained its ability to colonize cattle. Survival and proliferation of E. coli O157 in both clean drinking water and in filter-sterilized reconditioned meat-processing wastewater has been previously documented (22, 28, 29). Importantly, the experiments reported here differ from previous E. coli O157 aquatic survival studies in that they more closely represent natural contamination events in three significant ways: (i) the bacterial inoculum used was not a laboratory-grown isolate of E. coli O157 but rather was collected from feces of a calf actively excreting the organism and was representative of the most likely metabolic state and concentration of on-farm water trough contamination; (ii) the microcosms used were not treated in any way to remove competing microorganisms, so that the environmental persistence demonstrated here occurred in the presence of a natural bacterial flora present in cattle water troughs, including possible competitors; and (iii) the microcosms were not stagnant. This demonstrates that E. coli O157 remained part of the trough ecosystem despite a water retention period of only 0.5 days, a rate within the estimated range of water volume turnover in cattle water troughs.
The survival of E. coli O157 in these experiments parallels the observations of others that have found extended survival of other pathogenic bacteria in the natural aquatic environment (11). The survival of E. coli O157 in the microcosms was intended to mimic conditions present in naturally contaminated cattle water troughs. The survival of bacteria in aquatic systems is dependent upon many factors, notably exposure to sunlight, temperature, competition with and predation by other microflora, and nutrient availability (3). The influence each of these factors has on E. coli O157 survival in naturally contaminated troughs may be related to initial inoculum dose, trough design, location, and water trough management practices on farms and may differ from the results observed under experimental conditions.
The concentrations of E. coli O157 remaining in the microcosms may be viewed as underestimates because stressed bacteria may be sublethally injured and not proliferate on the selective media used in the assay (27). Although statistically significant differences between concentrations of E. coli O157 in chlorinated and unchlorinated systems were observed, the magnitude of the differences detected in the sediments of these experimental microcosms was small and considered unlikely to have major biological consequences, even when high levels of chlorine were added to the input water. However, chlorination may prove to be a useful adjunct in maintaining the microbiological quality of the water within livestock drinking troughs if practical methods to eliminate the accumulated sediments (a potential reservoir for bacterial persistence) are identified.
All isolates obtained from the experimental microcosms had PFGE banding patterns identical or closely similar to that of WSU2032. The isolates with minor differences from WSU2032 may have emerged during the infection of the calf used to generate the fecal inoculum, during laboratory passages, during environmental persistence in the microcosms, or following challenge. The emergence of novel, but closely related, PFGE clonal types during bovine colonization has been described previously (1). The closely related or identical PFGE patterns demonstrate the persistence of the initial inoculum in the microcosms for over 8 months.
Calf fecal shedding of E. coli O157 following exposure to challenge microcosms was characterized by an initial period of occasional single positive fecal samples followed by a period of sustained fecal E. coli O157 excretion. The intermittent detection of E. coli O157 in two calves during the first few days of this challenge experiment was consistent with the pattern of fecal shedding following one-time challenge of 107 CFU of E. coli O157 and most likely a result of passive shedding of the agent ingested with water, in the absence of significant replication or colonization of the gastrointestinal tract (8, 16). After day 28, calves excreted E. coli O157 in amounts and for durations similar to those reported by others following experimental challenges with higher doses of E. coli O157 (4, 5, 8).
Like the factors that govern the eventual clearance of this particular strain of E. coli from the gastrointestinal tract of cattle, the factors that contributed to the initial colonization of the calves remain undetermined. E. coli populations in the gastrointestinal tracts of young calves are in a continual state of fluctuation (19). Bacterial turnover associated with rumen development and interactions with other gastrointestinal flora may have resulted in the creation of a niche suitable for the colonization and proliferation of the E. coli O157 strain acquired from the drinking water microcosm (15, 31). Due to this complexity, calf infections following low-dose exposures may be uncommon stochastic events.
All four calves became colonized within a period of a few days. Based on the indistinguishable PFGE profiles of the isolates obtained from the calves and the drinking water, the water was the ultimate source of the colonizing organism. However, it is impossible to determine whether the water challenge was the direct source of the colonization of all four calves or whether a single calf initially infected from the water source subsequently transmitted the organism to the other three calves via a different route. Efficient calf-to-calf transmission in cohoused experimentally infected calves following low-dose challenges has been demonstrated (T. E. Besser et al., submitted for publication).
The genetic profile of the challenge organism, as determined by PFGE analysis, remained stable throughout the experiments. Likewise, no recognized changes in the management or appearance of either the microcosms or the calves occurred during the time of calf colonization. Nevertheless, cattle water troughs, the organism, the experimental calves, and their environments are complex ecosystems, and subtle changes with important effects on agent infectivity or host susceptibility are possible.
Observational studies have shown an association between the presence of E. coli O157 in cattle water troughs and the infection status of cattle drinking from these troughs (9, 26). While it is very likely that infected cattle frequently contaminate their water troughs with feces or saliva containing E. coli O157, the results of this study confirm that contaminated troughs can act as long-term reservoirs of the organism with a real potential for infection of cattle weeks or months later. Livestock water troughs contaminated with E. coli O157 and left without regular cleaning may serve as a reservoir of the agent on the farm for extended periods of time, such as during the cooler times of the year when E. coli O157 typically occurs at a very low prevalence in cattle. Since E. coli O157 can survive for extended periods within complex aquatic environments, caution should be used prior to the use of water for livestock or for human drinking and recreational purposes after a suspected contamination event.
REFERENCES
- 1.Akiba M, Sameshima T, Nakazawa M. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol Lett. 2000;184:79–83. doi: 10.1111/j.1574-6968.2000.tb08994.x. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong G L, Hollingsworth J, Morris J G., Jr Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 1996;18:29–51. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 3.Barcina I. Survival strategies of enteric bacteria in aquatic systems. Microbiologia. 1995;11:389–392. [PubMed] [Google Scholar]
- 4.Besser T E, Hancock D D, Pritchett L C, McRae E M, Rice D H, Tarr P I. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J Infect Dis. 1997;175:726–729. doi: 10.1093/infdis/175.3.726. [DOI] [PubMed] [Google Scholar]
- 5.Brown M, Gilbert P. Influence of the environment on the properties of vegetative microorganisms: an overview. In: Brown M, Gilbert P, editors. Microbiological quality assurance; a guide towards relevance and reproducibility of inocula. New York, N.Y: CRC Press; 1995. pp. 3–13. [Google Scholar]
- 6.Cassin M H, Lammerding A M, Todd E C, Ross W, McColl R S. Quantitative risk assessment for Escherichia coli O157:H7 in ground beef hamburgers. Int J Food Microbiol. 1998;41:21–44. doi: 10.1016/s0168-1605(98)00028-2. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. One-day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli serotype O157 by pulsed field gel electrophoresis (PFGE). Atlanta, Ga: PulseNet, the National Molecular Subtyping Network for Foodborne Disease Surveillance, Centers for Disease Control and Prevention; 1999. [Google Scholar]
- 8.Cray W C, Jr, Moon H W. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faith N G, Shere J A, Brosch R, Arnold K W, Ansay S E, Lee M-S, Luchansky J B, Kaspar C W. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl Environ Microbiol. 1996;62:1519–1525. doi: 10.1128/aem.62.5.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gannon V P, D'Souza S, Graham T, King R K. Specific identification of Escherichia coli O157:H7 using a multiplex PCR assay. Adv Exp Med Biol. 1997;412:81–82. doi: 10.1007/978-1-4899-1828-4_10. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez J. Modelling enteric bacteria survival in aquatic systems. Hydrobiologia. 1995;316:109–116. [Google Scholar]
- 12.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 13.Hancock D D, Besser T E, Rice D H, Ebel E D, Herriott D E, Carpenter L V. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev Vet Med. 1998;35:11–19. doi: 10.1016/s0167-5877(98)00050-6. [DOI] [PubMed] [Google Scholar]
- 14.Hancock D D, Besser T E, Rice D H, Herriott D E, Tarr P I. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol Infect. 1997;118:193–195. doi: 10.1017/s0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinton M, Linton A H, Hedges A J. The ecology of Escherichia coli in calves reared as dairy-cow replacements. J Appl Bacteriol. 1985;58:131–138. doi: 10.1111/j.1365-2672.1985.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R P, Cray W C, Jr, Johnson S T. Serum antibody responses of cattle following experimental infection with Escherichia coli O157:H7. Infect Immun. 1996;64:1879–1883. doi: 10.1128/iai.64.5.1879-1883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan D, McEwen S A, Lammerding A M, McNab W B, Wilson J B. Pre-slaughter control of Escherichia coli O157 in beef cattle: a simulation study. Prev Vet Med. 1999;41:55–74. doi: 10.1016/s0167-5877(99)00032-x. [DOI] [PubMed] [Google Scholar]
- 18.Jordan D, McEwen S A, Lammerding A M, McNab W B, Wilson J B. A simulation model for studying the role of pre-slaughter factors on the exposure of beef carcasses to human microbial hazards. Prev Vet Med. 1999;41:37–54. doi: 10.1016/s0167-5877(99)00033-1. [DOI] [PubMed] [Google Scholar]
- 19.Linton A H, Timoney J F. The ecology of chloramphenicol-resistance in Salmonella typhimurium and Escherichia coli in calves with endemic Salmonella infection. J Appl Bacteriol. 1981;50:115–129. doi: 10.1111/j.1365-2672.1981.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 20.Marks H M, Coleman M E, Lin C T, Roberts T. Topics in microbial risk assessment: dynamic flow tree process. Risk Analsis. 1998;18:309–328. doi: 10.1111/j.1539-6924.1998.tb01298.x. [DOI] [PubMed] [Google Scholar]
- 21.Mechie S C, Chapman P A, Siddons C A. A fifteen-month study of Escherichia coli O157:H7 in a dairy herd. Epidemiol Infect. 1997;118:17–25. doi: 10.1017/s0950268896007194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajkowski K T, Rice E W. Recovery and survival of Escherichia coli O157:H7 in reconditioned pork-processing wastewater. J Food Prot. 1999;62:731–734. doi: 10.4315/0362-028x-62.7.731. [DOI] [PubMed] [Google Scholar]
- 23.Reilly A. Prevention and control of enterohaemorrhagic Escherichia coli (EHEC) infections: memorandum from a WHO meeting. WHO Consultation on Prevention and Control of Enterohaemorrhagic Escherichia coli (EHEC) Infections. Bull W H O. 1998;76:245–255. [PMC free article] [PubMed] [Google Scholar]
- 24.Rice D H, McMenamin K M, Pritchett L C, Hancock D D, Besser T E. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol Infect. 1999;122:479–484. doi: 10.1017/s0950268899002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanderson M W, Gay J M, Hancock D D, Gay C C, Fox L K, Besser T E. Sensitivity of bacteriologic culture for detection of Escherichia coli O157:H7 in bovine feces. J Clin Microbiol. 1995;33:2616–2619. doi: 10.1128/jcm.33.10.2616-2619.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shere J A, Bartlett K J, Kaspar C W. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl Environ Microbiol. 1998;64:1390–1399. doi: 10.1128/aem.64.4.1390-1399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A, McFeters G. Injury of enteropathogenic bacteria in drinking water. In: McFeters G, editor. Drinking water microbiology. New York, N.Y: Springer-Verlag; 1990. pp. 368–379. [Google Scholar]
- 28.Wang G, Doyle M P. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J Food Prot. 1998;61:662–667. doi: 10.4315/0362-028x-61.6.662. [DOI] [PubMed] [Google Scholar]
- 29.Warburton D W, Austin J W, Harrison B H, Sanders G. Survival and recovery of Escherichia coli O157:H7 in inoculated bottled water. J Food Prot. 1998;61:948–952. doi: 10.4315/0362-028x-61.8.948. [DOI] [PubMed] [Google Scholar]
- 30.Wells J G, Shipman L D, Greene K D, Sowers E G, Green J H, Cameron D N, Downes F P, Martin M L, Griffin P M, Ostroff S M, et al. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J Clin Microbiol. 1991;29:985–989. doi: 10.1128/jcm.29.5.985-989.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao T, Doyle M P, Harmon B G, Brown C A, Mueller P O E, Parks A H. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J Clin Microbiol. 1998;36:641–647. doi: 10.1128/jcm.36.3.641-647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]