ABSTRACT
The microbiome influences the emotional and cognitive phenotype of its host, as well as the neurodevelopment and pathophysiology of various brain processes and disorders, via the well‐established microbiome–gut–brain axis. Rapidly accumulating data link the microbiome to severe neuropsychiatric disorders in humans, including schizophrenia, Alzheimer's and Parkinson's. Moreover, preclinical work has shown that perturbation of the microbiome is closely associated with social, cognitive and behavioural deficits. The potential of the microbiome as a diagnostic and therapeutic tool is currently undercut by a lack of clear mechanistic understanding of the microbiome–gut–brain axis. This review establishes the hypothesis that the mechanism by which this influence is carried out is synaptic plasticity – long‐term changes to the physical and functional neuronal structures that enable the brain to undertake learning, memory formation, emotional regulation and more. By examining the different constituents of the microbiome–gut–brain axis through the lens of synaptic plasticity, this review explores the diverse aspects by which the microbiome shapes the behaviour and mental wellbeing of the host. Key elements of this complex bi‐directional relationship include neurotransmitters, neuronal electrophysiology, immune mediators that engage with both the central and enteric nervous systems and signalling cascades that trigger long‐term potentiation of synapses. The importance of establishing mechanistic correlations along the microbiome–gut–brain axis cannot be overstated as they hold the potential for furthering current understanding regarding the vast fields of neuroscience and neuropsychiatry. This review strives to elucidate the promising theory of microbiome‐driven synaptic plasticity in the hope of enlightening current researchers and inspiring future ones.
Keywords: microbiome, central nervous system (CNS), enteric nervous system (ENS), neurons, synaptic plasticity, long‐term potentiation (LTP), neurotransmitters, immune system
I. INTRODUCTION
The microbiome–gut–brain axis refers to the way the resident gut microbiome communicates with and impacts the central nervous system. It has been firmly established (Ma, Forney & Ravel, 2012; Zarco, Vess & Ginsburg, 2012; Shreiner, Kao & Young, 2015; Tognini, 2017; Stiemsma & Michels, 2018; Altmäe, Franasiak & Mändar, 2019; Binyamin et al., 2020) that the microbiome is critical for normal physiological development and maintenance of homeostasis in all major multi‐organ systems of the body, and any significant perturbations to the microbiome may lead to a host of severe disorders (Shamriz et al., 2016; Seo, Boros & Holtzman, 2019; Goldberg et al., 2020; Shouval et al., 2020; Xavier et al., 2020; Zhang et al., 2020). Although the terms ‘microbiota’ and ‘microbiome’ are often used interchangeably, there is a small but important distinction. ‘Microbiota’ refers to the ecosystem of microorganisms in a given physiological environment, and includes colonizing entities such as bacteria, archaea, viruses, and eukaryotes, whereas ‘microbiome’ refers to the entirety of the microorganisms themselves along with their genetic material. This review focuses on the bacterial constituents of the gut microbiome (other constituents of the gut microbiome are not within the scope of this review).
Evidence links the microbiome to numerous phenomena (Leung & Thuret, 2015; Tognini, 2017; Strandwitz, 2018; Cryan et al., 2019) in the central nervous system (CNS), such as learning and memory, mental and emotional wellbeing and neuropsychiatric disorders, but the exact means by which these phenomena are shaped remains unclear. We propose that the microbiome affects the CNS via processes that contribute to synaptic plasticity in the CNS as well as the enteric nervous system (ENS), two systems that are engaged in constant bidirectional communication.
The ENS (Sasselli, Pachnis & Burns, 2012; Hyland & Cryan, 2016) is composed of two ganglionated plexi which facilitate the autonomic functionality of the gut. They do so independently of extrinsic sympathetic or parasympathetic inputs, earning the nickname ‘the second brain’. Although the ENS is a subsection of the peripheral nervous system, it is structurally similar to the CNS. Its various neuronal cell types are organized in a complex circuitry which orchestrates the integration of stimuli and induction of reflexes, enabling proper gut function. All of the major neurotransmitters that regulate neuronal processes in the brain also have vital enteric roles and are abundantly present in the gut (Mittal et al., 2017). Recent work has linked ENS dysfunction to the pathophysiology of diseases such as Parkinson's (Parashar & Udayabanu, 2017), Alzheimer's (Seo et al., 2019) and autism spectrum disorder (ASD) (Mulle, Sharp & Cubells, 2013), hitherto exclusively related to the CNS. The close association between the microbiome (Sasselli et al., 2012; Rao & Gershon, 2016) and the ENS suggests that the former plays a role in these devastating disorders.
Aside from regulating and maintaining the complex functionality of the gut, the enteric nervous system sends constant homeostatic feedback to the brain via the vagus nerve (Sasselli et al., 2012; Mittal et al., 2017). It is the longest parasympathetic nerve in the body. Not only does it enervate many organ systems, but it is also comprised mostly of afferent fibres carrying sensory information from the gut back to the brain. Vagal signalling, along with the hypothalamic–pituitary–adrenal axis (HPA) (Herman, 2018) and the immune system, constitute the bidirectional gut–brain axis. Since the microbiome has been shown to interact with and affect all three of these pathways (vagal signalling, HPA axis, immune system), it is well established that they are all crucial to the maintenance of the microbiome–gut–brain axis (Foster & McVey Neufeld, 2013; Rea, Dinan & Cryan, 2016; Strandwitz, 2018; Cryan et al., 2019; Frankiensztajn, Elliott & Koren, 2020).
Moreover, these three systems are tightly interconnected by several regulatory feedback mechanisms (Kabouridis & Pachnis, 2015; Obata & Pachnis, 2016; Fung, Olson & Hsiao, 2017; Yoo & Mazmanian, 2017; Kim, 2018). Vagal nerve endings present in the gut detect immunological mediators triggered by the microbiome and are then able to modulate the release of pro‐inflammatory cytokines by stimulating the release of acetylcholine (Ach). Additionally, vagal afferents are the conduit by which various microbial metabolites reach the brainstem solitary tract nucleus, which in turn projects to the hypothalamus and influences the HPA axis. The HPA axis is regarded as the body's primary stress‐response mechanism, whereby a cascade of signals between the hypothalamus, pituitary gland and adrenal cortex mediates glucocorticoid release, which initiates a metabolic and neuromodulatory stress response in the brain.
The mechanistic interface (Geuking et al., 2014; Kim, 2018; Zheng, Liwinski & Elinav, 2020) with the microbiome is distributed among the immune and enteric nervous systems and the HPA axis. Microbial metabolites engage the mucosal immune system and enteric nerve endings, both of which interact with one another, as well as the HPA axis. All of these, in turn, communicate information back to the CNS, which integrates the various stimuli and adapts accordingly. Moreover, there is growing evidence suggesting that the key to microbial involvement in host homeostasis lies within the microbial metabolites secreted into the gut and their interactions with various host pathways, such as those mentioned above.
A key aspect of neuronal tissue is its ability to adapt, physically as well as functionally, to a highly dynamic influx of stimuli and incorporate these adaptations into long‐lasting alterations in signal transduction and information retention. This is achieved by synaptic plasticity (Kotaleski & Blackwell, 2010; Bai & Suzuki, 2020), one of the most fundamental aspects of the nervous system, and seen in species ranging from flies to humans. It is worth mentioning that long‐term potentiation (LTP) and long‐term depression (LTD) are believed to only be cellular correlates of the natural processes we refer to as learning and memory and that the inductive properties are experimentally inferred. We believe that plasticity is an essential component of the microbiome–gut–brain axis because of its central role in modifying the CNS, ENS and HPA axis, as well as the immune system (Levy & Tasker, 2012; Cortés‐Mendoza et al., 2013; Maguire, 2014; Kabouridis & Pachnis, 2015).
Synaptic plasticity is the ability of neuronal tissue to undergo physical and functional changes at the level of individual connections between neurons (Citri & Malenka, 2008). Hebb was the pioneer behind the idea that resonating neuronal transmission among a transitory cellular repertoire lays the groundwork for memory formation. This theory assumes a fundamental adjustment in the firing rates of the participating synapses which takes place in direct correlation with the activity level in a given synapse (Hebb, 2002).
These changes, which are typically long‐lasting, occur in response to fluctuations in stimuli, allowing the host the ability to adapt to and thrive in dynamic environments. Quite simply, synaptic plasticity is cardinal to appropriate neuronal functionality. It is the cornerstone of learning, memory consolidation, emotional regulation, stimulus integration into behavioural adaptations and homeostatic maintenance of the brain and ENS. The various processes underlying these changes to synaptic architecture and transmission are induced primarily by LTP or LTD. Colloquially explained as ‘neurons that fire together wire together’, based on the ground‐breaking work of Hebb (2002), LTP refers to the strengthening of synapses, both physically and in terms of signal propagation capacity, as a result of high‐frequency stimuli. LTD (Collingridge et al., 2010) refers to a weakening of synaptic connectivity as a result of low‐frequency stimuli. LTP was initially observed in the hippocampus (Kumar, 2011), but observations have since been recorded in many other major areas of the brain. In studying synaptic plasticity, emphasis is given to areas of the brain that are known hubs of activity with projections to many other functional regions, especially structures that are implicated in learning and emotional regulation.
Synaptic plasticity (Glanzman, 2010; Kotaleski & Blackwell, 2010; Chater & Goda, 2014) is controlled by a combination of genes expressed via various signalling pathways, post‐translational modification of proteins, trafficking of receptors, and alterations to ion channel permeability. The revolutionary work of Kandel & Schwartz (1982) on the molecular mechanisms of synaptic plasticity corroborated Hebb's hypothesis and laid the groundwork for contemporary research.
Whilst the majority of work on the significance of synaptic plasticity has been conducted in the CNS, emerging evidence shows the importance of ENS plasticity. ENS‐mediated functions of the gut, including motility, hormone secretion and chemo‐sensitivity, have been shown to be altered following exposure to different nutrients and hormones, prolonged stress or inflammation. Although the exact mechanisms by which enteric neural tissue changes over the course of a lifetime remain to be elucidated, modulations in the ENS were shown to occur in response to specific nutrients, as well as peptide hormones secreted in the stomach and gut and sex hormones. Examples of these modulations include the hyper‐excitability of enteric neurons following an inflammatory response, altered action potential frequency across populations of neurons and lowered basal membrane potential (Mawe, Strong & Sharkey, 2009).
Given the increased importance attributed to ENS plasticity (Mawe et al., 2009; Schaefer, Van Ginneken & Copray, 2009), and in light of its implication in neurological disorders and the constant bidirectional communication with the CNS, we believe that the microbiome affects behaviour, cognition and emotional regulation by influencing plasticity in both the ENS and the CNS.
The aim of this review is to examine the microbiome–gut–brain axis in the context of synaptic plasticity, focusing on how the microbiome affects various plasticity‐inducing factors, including neurotransmitters, immune mediators and more.
The myriad ways in which the microbiome influences the ENS and CNS can be linked, to a great extent, to synaptic plasticity, both directly and indirectly (via mechanisms that contribute to synaptic plasticity). Therefore, we hypothesize that the most significant mechanistic relationship between the microbiome and the emotional, behavioural and cognitive phenotypes of the host is based on the microbiome's contribution to induction and maintenance of synaptic plasticity.
Our review provides initial support for this hypothesis and identifies mechanisms for further research through a discussion of potential research directions necessary to elucidate the connection between the microbiome and synaptic plasticity in the ENS and CNS. Understanding the mechanistic interaction between the microbiome and plasticity is of prime importance because of synaptic plasticity's involvement in severe neuropsychiatric disorders such as schizophrenia, ASD, bipolar disorder and depression. Relevant literature was sourced using the following search pattern: after searching for “synaptic plasticity” and becoming acquainted with the relevant terminology, each focal term was queried together with "microbiome", e.g “microbiome + synaptic plasticity”, “microbiome + LTP”, “microbiome + calcium signalling”, etc.
II. MECHANISMS OF SYNAPTIC PLASTICITY
An in‐depth review of the many aspects of synaptic plasticity is beyond the scope of this paper. Briefly, there are several types of plasticity associated with the frequency of stimuli received at the neuron, the brain region and the type of cells involved. Here we focus on plasticity‐associated mechanisms and signalling pathways that have been shown to be modulated by the microbiome, including neurotransmitters (De Vadder et al., 2018; Jadhav et al., 2018), calcium signalling (Engevik et al., 2019; Schalla & Stengel, 2020) and immune mediators (Schirmer et al., 2016; Fung et al., 2017; Kim, 2018; Xue et al., 2018; Cerovic, Forloni & Balducci, 2019). Additionally, we focus mainly on LTP because it is studied more robustly and is easily observed phenotypically, especially in terms of microbial involvement in its induction.
Plasticity is induced through neurotransmitters that are released from the presynaptic neuron and the postsynaptic cell (Goto, Yang & Otani, 2010; Korb & Finkbeiner, 2011; Lesch & Waider, 2012; Leal, Comprido & Duarte, 2014; Lu, Nagappan & Lu, 2014; Maguire, 2014; Pribiag & Stellwagen, 2014; Woolfrey & Dell'Acqua, 2015; Nanou & Catterall, 2018; Limanaqi et al., 2019). The terms ‘excitatory’ and ‘inhibitory’, as they pertain to neurotransmitters and synapses, refer to the alteration in the resting membrane potential of the postsynaptic cell and the subsequent probability of firing an action potential. Glutamate is the main excitatory neurotransmitter in the CNS, and its main target ionotropic receptors are the N‐methyl‐D‐aspartate receptor (NMDAR), α‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazolepropionic acid (AMPAR) and kainate. These receptors are crucial to plasticity processes, and neuropsychiatric disorders emerge in their absence (Rao & Finkbeiner, 2007; Zarate & Manji, 2008). Calcium, which enters the cell after activation of NMDARs, is fairly unique in that it triggers an elaborate signalling cascade that results in phosphorylation (and consequent increased expression) of AMPARs by Ca2+‐calmodulin‐dependent protein kinase 2 (CAMKII). Gamma‐aminobutyric acid (GABA) is the most abundant inhibitory neurotransmitter and has been shown to induce either LTP or LTD, according to the varying postsynaptic signalling cascades recruited by the different GABAergic receptor subunits (Jappy et al., 2016). Other neurotransmitters, such as serotonin (5‐HT) and dopamine, are also involved in plasticity‐related mechanisms, due to their wide distribution across the brain and their implication in a wide array of complex cognitive and emotional processes.
Contrary to the decades‐long belief that immune cells are only present in the brain during severe pathological situations (D'Agostino et al., 2012), it is now known that several immune mediators, under tightly controlled regulation, are necessary for homeostatic brain processes, including plasticity (Levin & Godukhin, 2017). The immune system is also functionally prominent in the ENS, where, in conjunction with all the neurotransmitters known to operate in the brain, it mediates many aspects of ENS functionality (Sasselli et al., 2012; Yoo & Mazmanian, 2017). The main branches of the gut–brain axis are comprised by numerous molecular and genetic pathways that are known to contribute to the induction and maintenance of synaptic plasticity.
III. EVIDENCE OF MICROBIAL EFFECTS ON PLASTICITY IN THE ENS AND CNS
Germ‐free (GF) animals are raised in a completely sterile environment, allowing them to serve as a platform for microbial transplants towards understanding causative effects of microbiome and host state on one another. While GF mice offer a blank slate for such studies, there is accumulating evidence that the GF condition is implicated in a range of health complications across the lifespan, which could make interpretation of findings more challenging (Dominguez‐Bello et al., 2019).
There is substantial evidence of altered behaviour, cognition and mood, in GF mice (Bercik et al., 2011; McVey Neufeld et al., 2015; Sarkar et al., 2020; Manca et al., 2020), indicating a microbial contribution to synaptic plasticity, as these phenotypic phenomena are rooted mechanistically in synaptic plasticity. GF mice also exhibit an altered neurophysiological transcription profile (Hölzel et al., 2010; Gareau et al., 2011; Heijtz et al., 2011; Ho, Lee & Martin, 2011; Lu et al., 2018), particularly in areas known to be highly involved in learning processes, such as the hippocampus and amygdala. Specifically, expression of immediate early genes (IEGs), which are directly involved in induction of plasticity, is markedly different in GF mice (Hoban et al., 2018). Evidence suggests that hippocampal plasticity in GF mice is regulated in a sex‐dependent manner (Darch et al., 2021). Due to the practical difficulty of conducting electrophysiological studies on GF mice, behavioural testing is often used as a proxy for altered plasticity (Medendorp et al., 2018). Young GF mice transplanted with the microbiome of aged mice exhibited a decrease in plasticity and in expression of neurotransmission‐related genes in the hippocampus, alongside marked cognitive deficits (D'Amato et al., 2020). The endocannabinoidome is a complex signalling pathway which has been linked to synaptic plasticity (Xu & Chen, 2015), learning and memory, behavioural adaptations and neurogenesis (Prenderville, Kelly & Downer, 2015). Manca et al. (2020) found the endocannabinoidome pathway to be impaired in GF mice, offering support to the hypothesis that it underlies some of the cognitive and behavioural differences in GF mice compared to specific‐pathogen‐free (SPF) mice. GF mice exhibit markedly altered behavioural and cognitive phenotypes, in part due to altered expression of brain‐derived neurotrophic factor (BDNF) and NMDARs, implicated in depression (BDNF) and neurogenesis and synaptic plasticity (NMDARs) (Neufeld et al., 2011). GF mice also have different levels of neurotransmitter secretion (Strandwitz, 2018), such as 5‐HT, dopamine and Ach, which were shown to influence plasticity in the brain. Several research groups have demonstrated the effects of antibiotics on behavioural phenotypes and expression profiles of key plasticity‐related compounds in the brain. Plasticity in the ENS is exemplified by increased levels of sensory sensitivity in the gut (Schaefer et al., 2009), induced by the microbiome (van Thiel et al., 2020). GF animals and those given antibiotics have altered gut neuronal transmission, coupled with a difference in membrane potential, either basally or following the firing of a neuronal signal (Carabotti et al., 2015). Calcium signalling is crucial in neuronal transmission and the induction of plasticity, and the microbiome has been shown to moderate the expression of calbindin, a calcium‐binding protein (McVey Neufeld et al., 2015).
Lactobacillus reuteri (Kunze et al., 2009) has been shown to alter the activity of calcium‐dependent channels in a subpopulation of enteric sensory neurons which are implicated in intestinal‐disorder‐derived pain. Rats fed an L. reuteri probiotic exhibited reduced levels of calcium‐dependent potassium channels, which lead to a lowered threshold for action‐potential firing, and consequently increased neuronal excitability. Another study (Perez‐Burgos et al., 2015) into the possible role of L. reuteri in amelioration of enteric pain sensations examined the effects of probiotic supplementation on capsaicin‐induced activation of transient receptor potential vanilloid 1 (TRPV1) channels. These channels are thought to mediate the transmission of nociceptive signals, and capsaicin is a known irritant which activates these channels in a manner leading to increased intracellular calcium currents. Mice treated with L. reuteri exhibited a reduced capsaicin‐induced excitatory response, accompanied by decreased TRPV1 expression.
There is thus a growing body of evidence that links the microbiome to the induction and maintenance of synaptic plasticity in both the ENS and CNS, suggesting that this crosstalk may be a key component of the microbiome–gut–brain axis.
IV. MICROBIOME–GUT–BRAIN AXIS – FROM THE GUT EPITHELIUM TO THE BRAIN AND BACK
The gut epithelium is the site of initial interaction between the resident microbiome and the host (Kabouridis et al., 2015; Okumura & Takeda, 2017; Solis et al., 2020). The physiological environment of the gut encompasses well‐developed neurological, immune and endocrine systems. The ENS is an autonomic neuronal network which communicates bi‐directionally with the CNS and autonomic nervous system (ANS) by way of both sympathetic and parasympathetic fibres synapsing in the gut. Enteric neurons express mechanoreceptors, chemo‐sensors and other signals which transmit a wide array of sensory, motor, molecular and vasodilator information back to the brain via the vagus nerve. The microbiome is thought to influence the neuronal activity of the ENS and CNS by secretion of neuromodulatory molecules (Fig. 1; Cryan & O'Mahony, 2011; Tognini, 2017; Strandwitz, 2018). Dopamine, noradrenaline (NA), 5‐HT, GABA and Ach are known to be produced by a wide array of bacteria (Strandwitz, 2018). Additionally, the gut microbiome was shown to play a significant role in the metabolism of tryptophan (Gao et al., 2020), which is the precursor of 5‐HT synthesis by the host. This is important because the majority of the body's 5‐HT is synthesised and utilized in the gut, with evidence of a constant feedback loop with 5‐HT synthesis in the brain.
Fig. 1.
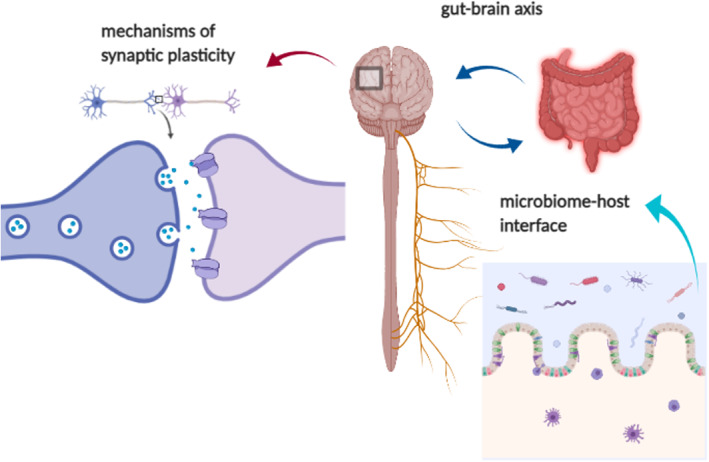
A general overview of the microbiome–gut–brain axis. The proposed mechanism by which the microbiome exerts its influence on the central nervous system is comprised of several elements. The first is interaction with the microbiome–host interface, the gut epithelium and enteric nervous and immune systems, which then synapse with the central nervous system via the vagus nerve and systemic immune interactions. The vagus is hypothesized to be a conduit for microbial metabolites directly to the brain, where, on a synaptic level, they influence the various mechanisms of synaptic plasticity.
Besides being the largest sensory organ, the gut is also the largest immune organ in the body, with the densest population of immune cells and molecular mediators (Yoo & Mazmanian, 2017). The mucosal immune system is highly specialized in its recognition of and response to pathogenic antigens (Carabotti et al., 2015; Rao & Gershon, 2016; Yoo & Mazmanian, 2017). Early‐life microbial colonization is thought to mediate maturation of the immune system, as shown in GF mice. Examples of microbiome–immune system interactions include microbial regulation of immune cell migration, differentiation and function of innate immunity constituents, induction of inflammation mediators (Geuking et al., 2014) and cytokine secretion modulation (Schirmer et al., 2016) by microbial short‐chain fatty acid production (Patel et al., 2012). Microbial perturbations are also linked to severe immunological disorders, including autoimmune diseases (Zhang et al., 2020).
The mucosal immune system and ENS are tightly intertwined. The mechanistic cooperation between these two systems begins in the early developmental stages – the functional constituents of the ENS mature concurrently with the immune system and with microbial colonization of the postnatal gut (Geuking et al., 2014; Kabouridis & Pachnis, 2015). Thus, the majority of enteric neurons and glial cells, as well as their connectivity, become fully formed under the direct influence of the developing microbiome. Peyer's patches, a vital element of the mucosal immune system, co‐mature with the ENS during this developmental period, under the same molecular signalling pathway (Patel et al., 2012). The immune system is thus highly involved in the many ENS‐modulated, CNS‐independent functions of the gut, including digestion and metabolism (Ma & Ma, 2019), gut motility (De Winter, 2010) and secretion of peptides and neurotransmitters (Khalil, Zhang & Engel, 2019).
The immune system is known to mediate processes in the CNS, with substantial evidence linking it to the induction of synaptic plasticity (Di Filippo et al., 2013). It is firmly established that increased systemic inflammation in later life contributes to decreased synaptic plasticity in the brain (Deleidi, Jãggle & Rubino, 2015), with pertinent evidence of an altered microbiome later in life (Nagpal et al., 2018) to support the hypothesis that the three phenomena are connected.
V. NEUROTRANSMITTERS
(1). Serotonin
The importance of neurotransmitters in the microbiome–gut–brain axis is compounded by the importance of neurotransmitters to a host of CNS (Lesch & Waider, 2012; Kraus et al., 2017) and ENS (Boesmans et al., 2008) processes and the ability of the microbiome to synthesise molecularly identical neurotransmitters that are biologically active in the host (Strandwitz, 2018). Additionally, the microbiome is known to mediate tryptophan metabolism by the host (O'Mahony et al., 2015). The neurotransmitter 5‐HT plays a vital role in both the CNS (Daubert & Condron, 2010) and ENS (Sasselli et al., 2012).
5‐HT is a functionally diverse neurotransmitter with extensive implications in neuroplasticity and modulation of a diverse range of functions (Daubert & Condron, 2010; Lesch & Waider, 2012; Lovelace et al., 2017). It is the most widely distributed transmitter in the brain, and its signalling pathways mediate not only homeostatic physiology, but also brain functionality, including sensory processing, cognitive control, emotional regulation, autonomic responses and motor activity, in a complex network of connections spanning the entire brain (Barton et al., 2008; Bennett & Maxwell, 2010; Lesch & Waider, 2012). Consequently, 5‐HT is a target of many physiological regulators, including modulators of gene transcription, neurotrophic peptides, and steroids, as well as psychotropic therapeutics, which impact the formation and activity of 5‐HT subsystems (Homberg et al., 2014). 5‐HT and BDNF are functionally linked. Examples of this functional interplay include the involvement of BDNF in the development and functionality of serotonergic neurons (Martinowich & Lu, 2008) and the co‐expression of BDNF and 5‐HT during heightened psychological arousal (Jiang et al., 2016).
The majority of the body's 5‐HT is contained within the gut (Yano et al., 2015); thus, the microbial communities colonizing the gut are hypothesized to exert an influence over its production and circulation, thereby affecting its downstream physiological roles (Clarke et al., 2013; Homberg et al., 2014; O'Mahony et al., 2015; Yano et al., 2015). 5‐HT is synthesized in two pathways. The first is from dietary tryptophan in enterochromaffin cells in the gut (Israelyan & Margolis, 2019), and the second is from albumin‐bound tryptophan that crosses the blood–brain barrier and is used for 5‐HT synthesis in the raphe nuclei of the brainstem (Lesch et al., 2012). The principal means of intraneuronal metabolism of 5‐HT is via oxidative deamination by monoamine oxidase resulting in formation of 5‐hydroxyindoleacetic acid (5‐HIAA) (Barton et al., 2008). Recent research supports microbial involvement in the metabolic processes pertaining to both of these metabolites (Waclawiková & El Aidy, 2018).
Preclinical studies have demonstrated that the GF condition is characterized by increased plasma tryptophan concentrations which can be normalized following microbial colonization of mice immediately post‐weaning (Clarke et al., 2013). Increased 5‐HT turnover, as indicated by the 5‐HIAA/5‐HT ratio, is also evident in the striatum of GF mice (Waclawiková & El Aidy, 2018). Other investigators have also reported elevations in plasma levels of both tryptophan and 5‐HT in GF mice compared to conventional animals (Wikoff et al., 2009). Interestingly, there may be a temporal effect of the colonization process. For example, one study reported that the elevated tryptophan concentrations in GF mice were reduced 4 days following the introduction of a microbiome but not at day 30 (El Aidy et al., 2012). Also of note is a recent study showing that GF rats have decreased hippocampal 5‐HT concentrations but exhibit stress‐induced elevation in both 5‐HT and 5‐HIAA, like conventional rats (Desbonnet et al., 2008). The impact of the gut microbiome on the CNS serotonergic system is not limited solely to microbiome‐deficient animals; administration of the probiotic Bifidobacterium infantis to rats resulted in reduced 5‐HIAA concentrations in the frontal cortex (El Aidy et al., 2012). Furthermore, there was also a marked increase in plasma concentrations of tryptophan and kynurenic acid in these animals. Additionally, the Bifidobacterium‐treated group exhibited reduced dihydrophenylacetic acid (DOPAC) and NA concentrations.
Clarke et al. (2013) showed that GF mice have impaired immune systems and heightened activity of the HPA axis but that sex differences exist in terms of BDNF and 5‐HT expression. BDNF expression was decreased in GF males, whereas 5‐HT, plasma tryptophan and 5‐HIAA levels were increased. Females did not exhibit significantly altered levels of these neurotransmitters. GF animals of both sexes exhibited a decreased kynurenine to tryptophan ratio (KYN/Trp) in plasma, which was restored to normal levels upon microbial colonization. The kynurenine pathway, which is heavily implicated in brain plasticity in health and disease, is responsible for catabolizing 95% of both CNS and peripheral tryptophan, with kynurenine being its primary catabolite. KYN/Trp is one of several ratios indicative of the pathway's functionality (Savitz, 2020). This neuromodulatory effect of Bifidobacterium is in line with reports of lowered NA levels in tricyclic antidepressant‐treated rats (Nakajima et al., 2012). Another study (Heijtz et al., 2011), which used behavioural testing to investigate the reduced anxiolytic behaviours of GF mice and the neurochemistry involved, found a significantly higher striatal turnover rate of NA, dopamine and 5‐HT in GF mice compared to SPF mice, but levels in the frontal cortex and hippocampus were unchanged.
In the ENS, 5‐HT secretion is mediated by the microbiome, and the activation of its 5‐HT4 receptor exerts neuro‐modulatory effects (Wood, 2001). De Vadder et al. (2018) showed that 5‐HT is necessary for adult ENS maintenance. Tryptophan hydroxylase (Tph) is the rate‐limiting enzyme in 5‐HT synthesis, and two different variants of this enzyme are used by enterochromaffin cells and neurons (Tph1 and Tph2, respectively), leading to 5‐HT synthesis in two distinct aggregations. De Vadder et al. (2018) also showed that Tph1‐deficient GF mice only exhibited a decrease in myenteric neuronal mass following colonization with a wild‐type microbiome, indicating that 5‐HT is crucial for healthy ENS development during early colonization of the gut. Additionally, conventional mice that were treated with irreversible Tph1 and Tph2 blockers showed decreased neuronal differentiation and less overall myenteric neuron functionality – factors which contribute to the efficacy of signal transduction and therefore induction of plasticity.
Selective serotonin‐reuptake inhibitors (SSRIs) are thought to exert their effects on gut 5‐HT in a manner that contributes to their anti‐depressant effects on the brain (Jones et al., 2020). Due to the role of the vagus nerve as a conduit between the gut, and consequently the microbiome, and the brain, McVey Neufeld et al. (2019) investigated the relationship between SSRI administration and vagal neuron firing rate, and the potential involvement of the microbiome in this bi‐directional pathway. Their results demonstrated that SSRI‐induced vagal neurostimulation is contingent upon signalling between the gut epithelium and afferent enteric neurons. Additionally, SSRI treatment altered the microbiome of the recipient mice, with the treatment group exhibiting reduced alpha diversity in comparison to the control group.
Taken together, these studies suggest the ability of the gut microbiome to influence the CNS and ENS serotonergic system profoundly, supporting our hypothesis that the gut microbiome is implicated in synaptic plasticity.
(2). Dopamine
Dopamine acts on both excitatory and inhibitory synapses distributed throughout the brain, but a high density of dopaminergic receptors in specific areas has led to the recognition of several major dopaminergic pathways implicated in a host of brain functions and neuropsychiatric disorders (Pignatelli & Bonci, 2015; Aarts et al., 2017; Parashar & Udayabanu, 2017). In the gut, several types of dopamine receptors mediate vasoactive and homeostatic effects.
A study examining the relationship between the microbiome and alcohol‐related addictive behaviour in mice found a strong correlation between the striatal expression of the dopamine receptor messenger RNA (mRNA) in the brains of addiction‐prone mice and specific microbial alterations in the gut after alcohol exposure followed by a three‐month abstinence period (Jadhav et al., 2018). Dopamine 1 receptors (D1Rs), comprising the direct pathway that mediates reinforcement learning and award‐seeking behaviour, were observed at higher levels, whereas the aversive‐learning modulator D2R was observed in lower abundance. Dopamine receptors (Chen, Hopf & Bonci, 2010; Goto et al., 2010; Nakano et al., 2010) have been shown to contribute to induction of stimulus‐dependent plasticity in areas of the brain responsible for emotional regulation. Jadhav et al. (2018) showed that changes in dopamine receptor expression were concomitant with microbial alterations in several taxa, including Lachnospiraceae, Syntrophococcus, Shuttleworthia, Gemella, Allobaculum, and Hydrogenoanaerobacterium.
Another study in humans with and without attention deficit hyperactivity disorder (ADHD) combined 16S sequencing with functional magnetic resonance imaging (fMRI) to connect microbial differences and dopaminergic‐transmission alteration in the striatum. The results showed increased Bifidobacterium levels in ADHD patients, coupled with deficient ventral striatal responses in a reward‐anticipation fMRI test (Aarts et al., 2017).
Dopamine signalling is widespread in the ENS and is necessary for gut motility (Rao & Gershon, 2016). Xue et al. (2018) compared dopamine synthesis, dopaminergic neuron viability and correlated immune activity in conventional and antibiotic‐treated mice. Microbially deficient mice exhibited markedly reduced tyrosine hydroxylase (TH; the rate‐limiting enzyme for dopamine synthesis) mRNA in the gut, resulting in increased interferon‐gamma production, and exacerbation of autoimmune hepatitis. They also showed that dopamine inhibits cytokine production via its D1R. These patterns can affect plasticity as demonstrated by the interconnectedness of the CNS and the immune system, especially as it pertains to the onset of pathophysiological situations (D'Agostino et al., 2012; Fung et al., 2017).
Emerging evidence links the microbiome to the pathophysiology of Parkinson's disease (PD) (Picconi, Piccoli & Calabresi, 2012), with dopamine playing a major role in this process. The pathophysiology of this devastating disease is well described as neurodegeneration of dopaminergic cells in the substantia nigra, associated with accumulation of α‐synuclein aggregates in the cellular bodies (synucleinopathy). Kim et al. (2019) demonstrated the vagus‐dependent spreading of α‐synuclein from the gut to the brain, correlating with a loss of dopaminergic neurons and characteristic PD symptoms. Interestingly, significant synucleinopathy is also present in the ENS. These findings, along with the fact that with the onset of PD, gastrointestinal symptoms often precede motor symptoms by several years (Elfil et al., 2020), may lead to a paradigm shift as to the true origin of PD. Helicobacter pylori has been shown to be overrepresented among PD patients and exacerbates symptoms by interfering with the metabolism of levodopa, the primary pharmaceutical administered to PD patients (Contin & Martinelli, 2010). Dysbiosis and small intestinal bacterial overgrowth (SIBO) are also characteristic of PD (Tan et al., 2014). PD patients’ bacterial profile indicates a reduction of Prevotellaceae, as well as a direct correlation between abundance of Enterobacteriaceae and severity of some motor symptoms (Parashar & Udayabanu, 2017).
In a murine model of PD (CDNF‐knockout mice), cerebral dopamine neurotrophic factor (CDNF) restored dopaminergic function in the nigrostriatal area of the brain (Lindahl et al., 2020). Lindahl et al. (2020) demonstrated that CDNF protects against enteric neuropathy occurring in old age, specifically in the submucosal plexus, which corresponds to enteric pathologies present in human PD patients. Interestingly, the CNS concentration of dopaminergic neurons in the CDNF‐knockout mice used for this study was unaltered.
VI. THE MICROBIOME AND NEURO‐IMMUNITY
The complex mechanisms of neuro‐immune regulation of gut physiology are numerous; therefore, a detailed exploration is beyond the scope of this review. We briefly review the main pathways by which neuro‐immune regulation is carried out, with evidence of microbial intervention on several aspects of this functional relationship.
The intestinal neuro‐immune interface encompasses (Yoo & Mazmanian, 2017) up to 80% of the body's immune cells, more than 100 million neurons and upwards of 100,000 synapses between enteric nerves and extrinsic (Uesaka et al., 2016) nerve endings from the vagus and pelvic nerves. While the ENS can function completely autonomously (Sasselli et al., 2012), the structural interface, with extrinsic sympathetic and parasympathetic nerves, allows the gut to send mechanical and chemosensory information back to the brain via the vagus nerve (Bonaz, Sinniger & Pellissier, 2017), and the brain to modulate gut functionality (Cryan et al., 2019) that requires nervous signal transmission, such as gut motility, contractility and vasodilation (Sasselli et al., 2012). Immunological structures (Yoo & Mazmanian, 2017) extend to the gut epithelium with the express purpose of sensing pathogenic material and enlisting an appropriate immune response. The myriad interactions between the microbiome and the enteric nervous system are clearly demonstrated in GF mice (Zheng et al., 2020). GF mice exhibit greatly reduced intra‐epithelial lymphocyte and immunoglobulin A (IgA) antibodies, an absence of T helper 17 (Th17) cells and an imbalance of Th1/Th2 cells (Kim, 2018). These phenomena indicate the involvement of the microbiome in both the innate and adaptive immune systems. The structural proximity of immune and nervous system structures gives rise to a functional overlap, compounded by molecular cross‐talk between the two systems (Khalil et al., 2019). Immune mediators are known to influence enteric neuronal activity, and neuroactive molecules affect the function of immune cells (Khalil et al., 2019). Macrophages, dendrites and T cells are activated by Ach, 5‐HT and vasoactive intestinal polypeptides secreted by enteric nerves, and cytokines, proteases and opioids released by the immune cells in turn modulate the sensitivity and transcriptional profile of neurons (Khalil et al., 2019). This is especially evident in gastrointestinal pathologies (Fung et al., 2017) which exhibit comorbidities of heightened immune responsivity and increased inflammation alongside electrical impairments in the gut and disturbances in perception of visceral pain. There is a growing body of evidence linking the microbiome to the onset and severity of inflammatory bowel disease (Ocansey et al., 2020). GF mice have a greater susceptibility for gastrointestinal dysfunction, and certain bacteria (Glassner, Abraham & Quigley, 2020) have been correlated to genetic mutations affecting epithelial defence against pathogens, mucosal barrier integrity, inflammatory gene expression and regulatory T‐cell differentiation (Schirmer et al., 2016; Kim, 2018; Lazar et al., 2018; Zheng et al., 2020).
Lactobacillus rhamnosus JB‐1 (JB‐1) (Liu et al., 2020) has been shown to require regulatory T cells for its role in mediating anxiolytic and depressive behaviours. Moreover, antibody depletion of CD25+ T regulatory (Treg) cells markedly reduced the beneficial behavioural effects of JB‐1. JB‐1 has been shown to exert mediating effects on anxiolytic and depressive behaviours in a vagus‐dependent manner (Perez‐Burgos et al., 2014), and it has also been shown to reduce GABA receptor expression (Bravo et al., 2011) in specific brain areas known to undergo extensive learning‐ and emotional‐regulation‐related plasticity. These multifaceted effects of JB‐1 exemplify the intricacy of the microbiome–gut–brain axis, with the multitude of constituents involved in its underlying mechanisms.
Additionally, the HPA axis also plays a vital role in the neuro‐immune regulation of the gut. Part of the limbic system, the HPA axis is the primary regulator of the stress response and is sensitive to a host of systemic molecules, including pro‐inflammatory cytokines released from the gut. In turn, corticotrophin releasing factor (CRF) from the hypothalamus triggers adrenal adrenocorticotropic hormone (ACTH) release and subsequent cortisol secretion. Aside from metabolically affecting most organ systems in the body, cortisol has been specifically implicated in mental disorders and stress‐related gastrointestinal and immune disorders (Wiley, Higgins & Athey, 2016). The microbiome has been shown to modulate the HPA axis extensively (Frankiensztajn et al., 2020). GF mice exhibit decreased anxiety and increased stress responsivity in conjunction with increased ACTH and cortisol levels. Bifidobacterium pseudocatenulatum (Moya‐Pérez et al., 2017) has been shown to prevent stress‐induced increases of corticosterone and hypothalamic catecholamines in mice, and also to cause a substantial decrease in stress‐induced inflammatory markers.
The vagus nerve (Bonaz et al., 2017), acting as a conduit between the ENS and the brain, synapses in the solitary tract nucleus, which sends projections to most regions of the brain, including the hypothalamus, further cementing the interconnectedness between the various branches of the microbiome–gut–brain axis. Evidence of vagus‐dependent microbial influence on the CNS includes (Fülling, Dinan & Cryan, 2019) Campylobacter jejuni's ability to increase anxiolytic behaviours, as well as Fos transcription factor (belonging to the IEG family) activity in vagal afferents and the solitary tract nucleus, the prevention of beneficial effects of L. rhamnosus JB‐1 in vagotomized mice (Liu et al., 2021) and the rescuing of social deficit by L. reuteri in animal models of ASD. The vagus nerve is also among the hypothesized channels by which bile acids (BAs) (Wu et al., 2020) and short‐chain fatty acids (Goswami, Iwasaki & Yada, 2018) exert their influence on the CNS. Short‐chain fatty acids are the product of microbial fermentation of dietary fibre and are thought to modulate a wide range of brain activities via immune, endocrine, vagal and other humoral pathways (Dalile et al., 2019). BAs are metabolically active hormonal mediators, known to be regulated by the microbiome (Monteiro‐Cardoso, Corlianò & Singaraja, 2021). BAs are synthesised in the liver and are systemically active, with BA receptors widely distributed in the brain as they are capable of crossing the blood–brain barrier. Hypothesized to act via the vagus nerve, BAs have been implicated in severe neuropsychiatric diseases (Sandhu et al., 2017).
VII. MICROBIAL EFFECTS ON ELECTROPHYSIOLOGY
The most direct evidence of the involvement of the microbiome in the regulation of synaptic plasticity comes from electrophysiological recordings of synaptic activity under different conditions of microbial treatment or after modulation of the host microbiome. Detecting changes in paired‐pulse facilitation is one mechanism to test effects on synaptic function.
Paired‐pulse facilitation (PPF) (Hu et al., 2013) refers to the effect wielded by a stimulated presynaptic cell on the magnitude of the second current emitted by a postsynaptic cell. This indicates a transient enhancement of transmission capacity following signal transduction in that specific synapse, a type of use‐dependent, short‐term plasticity. PPF is measured by plotting field excitatory postsynaptic potentials (fEPSPs) as a function of time elapsed between two consecutive action potential firings, and is hypothesized to be crucial for the induction of LTP (Tahmasebi et al., 2016).
One study (Davari et al., 2013) correlating microbial influence with EPSP transmission in the hippocampus studied diabetic and control rats after receiving probiotic treatment and undergoing a spatial learning test. The results showed that a probiotic cocktail composed of Lactobacillus acidophilus, Bifidobacterium lactis and Lactobacillus fermentum improved the learning task performance of both groups (diabetics and controls) and considerably rescued hippocampal EPSP response in the diabetic group. Also, the diabetic–probiotic group exhibited the greatest increase in LTP, which is significant due to the known phenomenon of diabetes‐induced learning and memory deficits (Zilliox et al., 2016).
Sgritta et al. (2019) utilized the AMPAR/NMDAR ratio in dopaminergic neurons to measure the effects of L. reuteri on social stimuli‐induced synaptic potentiation in a mouse model of ASD with known impairments to oxytocinergic transmission. AMPARs (Chater & Goda, 2014) play a crucial, dual role in the induction of synaptic plasticity. The opening of AMAPRs allows an influx of cations which triggers action potential firing and enlists calcium‐dependent signalling cascades (Nanou & Catterall, 2018; Kornijcuk et al., 2020) which catalyse long‐lasting changes in the structure and functionality of the neuron. NMDARs play an important role in AMPAR‐mediated plasticity (Moosmang, 2005; Hunt & Castillo, 2012), and the ratio of the two receptors’ expression levels mediates trends in neurotransmission that are linked to the induction and long‐term maintenance of plasticity (Rao & Finkbeiner, 2007). The results of this study showed that the AMPAR/NMDAR ratio, while similar at baseline levels between the ASD model and controls, failed to increase and induce LTP in the ASD mice in response to social stimuli alone. Additionally, L. reuteri administration rescued social deficits in the ASD mice in a vagus‐dependent manner, as confirmed by vagotomized controls. These findings suggest a direct effect of the microbiome on CNS transmission.
The major mechanisms through which the microbiome may affect LTP remain elusive. One possible mechanism is through the production of specific metabolites which can cross the blood–brain barrier and affect neuronal function and synaptic plasticity. Govindarajulu et al. (2020) set out to determine the potential effects on LTP induction by the microbiome‐dependent metabolite trimethylamine N‐oxide (TMAO). TMAO is a by‐product of microbial metabolism of nutrients, and has been implicated (Li et al., 2018) in age‐related cognitive impairments in mice. The rationale of this study was that a decline in LTP induction, influenced by increased levels of TMAO, is the mechanism behind old‐age cognitive deficits. The results of this study showed reduced fEPSP measurements and PPF in hippocampal slices incubated with TMAO, compared with controls. Western blot analysis revealed reduced expression of AMPAR subunits, which may account for the observed LTP deficits, as well as impaired basal synaptic transmission. This study provides evidence for a role of microbial metabolites in the regulation of synaptic plasticity.
The implications of direct microbial influence on electrophysiological events necessary for the induction and long‐term maintenance of synaptic plasticity are vastly important for understanding the mechanisms of the microbiome–gut–brain axis.
VIII. GENETIC EXPRESSION, SIGNALLING CASCADES AND NEUROTROPHIC FACTORS
One of the mechanisms responsible for synaptic plasticity is dynamically regulated gene expression and associated protein synthesis (Glanzman, 2010; Korb & Finkbeiner, 2011; Gipson, Kupchik & Kalivas, 2014; Berger et al., 2017). Patterns of gene expression vary widely, depending on the brain region undergoing the plasticity‐induced changes and the type of plasticity process taking shape, and these mechanisms have been studied extensively in humans and animal models. IEGs (Coppens et al., 2011) are heavily involved in plasticity induction due to their fast transcription that does not require de novo protein synthesis (Okuno, 2011). Prominent IEGs include the Arc, Fos and Egr gene families. Recently, researchers have begun to uncover connections between the gut microbiome and gene expression associated with plasticity (Dinan & Cryan, 2017).
The amygdala is a key element of the limbic system, responsible for the processing and integration of emotional‐related stimuli. Amygdala emotion‐induced learning and its consolidation into behavioural adaptations is realized via synaptic plasticity. Behavioural testing involving fear conditioning and socialization paradigms has become standard practice in animal studies investigating plasticity‐related functionality of the amygdala under different circumstances, especially in emotional disorders including post‐traumatic stress disorder, affective disorders and anxiety (Malter Cohen et al., 2013).
Hoban et al. (2018) conducted a preliminary examination of the brains of GF mice before subjecting them to a fear‐conditioning behavioural test. The researchers found that the mice exhibited a basally modified pattern of transcription, compared to conventionally raised animals, specifically as it pertains to the expression of IEGs including Fos, Egr2, Fosb and Arc. Such results support a neuronal basis for the well‐documented behavioural and emotional phenotypic alterations presented by GF animals. Also altered was the expression of genes necessary for neuronal functionality, synaptic transmission and developmental processes in the CNS. Following the behavioural test, GF mice did not appear to learn fear responses as well as their conventional counterparts, and subsequent genome‐wide transcriptome profiling revealed a total of 133 differentially expressed genes related to plasticity.
Another study (Stilling et al., 2018) focusing on the amygdala's role in social behaviour, and employing similar methods as above (investigation of brains of GF animals, behavioural testing and RNA sequencing), found significant differences in alternative splicing, intracellular signalling and phosphorylation, specifically in the mitogen‐activated protein kinase (MAPK) signalling pathway, which is heavily involved in regulation of synaptic plasticity (Falcicchia et al., 2020). These results suggest a baseline hyperactivity in the amygdalae of GF mice. GF mice did not perform as well, on average, in the behaviour test compared to GF mice colonized with a healthy microbiome at weaning or to conventional mice. Splicing activity was improved in GF mice following social interaction and was even more pronounced in ex‐GF mice, which displayed a behavioural response strongly resembling that of conventional mice. The expression profile of serotonergic receptor genes was not significantly different between GF and conventional animals of either sex. These results seem to suggest a mechanistic correlation between a healthy microbiome and functional integrity of the amygdala in inducing correct behavioural patterns in response to social stimuli.
Another study (Davari et al., 2013) examined the effects of probiotic treatment (L. acidophilus, Bi. lactis and L. fermentum) on the behavioural and electrophysiological aspects of diabetes‐induced learning and memory deficits in rats. Results of this study showed that diabetic animals that received probiotics exhibited markedly improved spatial memory, facilitated by an improved ability of the brain to perform LTP, a key element of synaptic plasticity.
A third study (Zolfaghari, Rabbani Khorasgani & Noorbakhshnia, 2020) measuring hippocampal CaMKII‐α and tumor necrosis factor (TNF‐α) expression showed that probiotic treatment with L. rhamnosus, L. reuteri or L. plantarum successfully impeded neuroinflammation‐induced memory deficits in mice. CaMKII‐α is one of the major factors responsible for induction of LTP. Lipopolysaccharide (LPS) is an endotoxic, neurotoxic and inflammatory molecule found in gram‐negative bacterial membranes, capable of stimulating the expression of TNF‐α and interleukins IL‐1β and IL‐6 (Zweigner, Schumann & Weber, 2006). The results of this study showed that L. rhamnosus ameliorated LPS‐induced impairment of hippocampal CaMKII‐α gene expression. Additional studies (Lim et al., 2017; Lee, Lim & Kim, 2018) highlight the ability of Lactobacillus to correct such impairments through administration of L. johnsonii CJLJ103 (LJ) to mice. LJ corrected two detrimental effects caused by LPS: nuclear factor kappa B (NF‐κB)‐induced hippocampal neuroinflammation and reduced BDNF expression.
Synaptic plasticity is a multi‐factorial process, enabled by differential gene expression and a consequent web of signalling cascades. Although some genes, like Arc and Fos, are directly linked to induction of plasticity, other markers of plasticity are activated indirectly, causing plasticity to occur downstream, for example via neurotransmitter release or BDNF (Leal et al., 2014; Falcicchia et al., 2020). Any functional modifications to the molecular machinery of the neuronal environment that lead to the induction of plasticity events can be traced back to altered gene expression and the ensuing protein synthesis, either of which can serve to indicate and quantify the processes taking place (Glanzman, 2010; Kotaleski & Blackwell, 2010).
BDNF is a growth factor that is abundantly expressed in all major brain regions and is implicated in a wide array of supportive and survival‐oriented functions, as well as higher cognitive functions such as learning, memory and emotional regulation (Cunha, Brambilla & Thomas, 2010; Lu et al., 2014; Loprinzi & Frith, 2019). Alterations in the expression or signalling of BDNF are implicated in the pathophysiology of mental disorders such as depression (Yu & Chen, 2011) and schizophrenia (Favalli et al., 2012), execution of motor functions (He et al., 2013) and LTP and consequent memory formation (Cunha et al., 2010; Loprinzi & Frith, 2019).
BDNF is crucial for the healthy development of the CNS, and its absence results in a multifaceted phenotype of cognitive and mental disorders, as evidenced from animal studies (Taliaz et al., 2010; Coppens et al., 2011; He et al., 2013; Jiang et al., 2016). Numerous studies examining the effects of the microbiome on CNS development show a clear correlation between a GF condition or antibiotic‐induced microbiome ablation with reduced BDNF levels in conjunction with aberrant cognitive and behavioural phenotypes. BDNF is tightly coupled with a range of processes occurring in synapses, particularly induction of plasticity, due to its involvement in many downstream signalling cascades and its presence in both inhibitory and excitatory synapses in all brain regions (Cunha et al., 2010; Favalli et al., 2012; Lu et al., 2014). Thus, it is probable that the microbiome plays an integral part in early developmental processes in the brain as it pertains to the expression of cardinal molecules such as BDNF.
Stilling et al. (2015) found enrichment of the transcription factors Fos, Egr2 and Nr4a1, as well as the plasticity‐related genes Arc and Homer1, in mice immediately following socialization in a behavioural test, indicating that plasticity activation is increased in brain areas that have been stimulated. The importance of examining the IEG expression profile concurrently with BDNF levels is that it was shown to be differentially expressed during BDNF‐dependent plasticity events (Coppens et al., 2011).
Also upregulated in socialized mice was the neuronal activity‐induced MAPK signalling pathway, paralleling the results of previous studies (Verma et al., 2016). A real‐time, quantitative, reverse transcription polymerase chain reaction (qRT‐PCR) was used to determine BDNF levels in GF mice as an indicator of cyclic AMP response element‐binding protein (CREB) expression. CREB is a crucial modulator of neuronal activity and is an important downstream effector of BDNF expression. Neufeld et al. (2011) showed similar results in a study that compared gene expression in the brains of GF and SPF mice, assisted by targeted neuronal stimulation provided by behavioural testing. GF mice exhibited anxiety‐like behaviour in the elevated plus maze, and their expression profile included decreased amygdaloid mRNA expression of NMDAR subunit NR2B, increased hippocampal BDNF levels and depleted levels of 5‐HT receptor 1A (5‐HT‐1A).
Correct gene expression in the CNS during neonatal development is critical for healthy maturation of countless brain functions, as evidenced in cases where the smallest alteration in gene expression results in complicated syndromes that affect an organism's entire well‐being (Veenstra‐VanderWeele et al., 2012; Zhan et al., 2014). Efforts to understand the impact of a healthy microbial transfer between mother and offspring on the latter's CNS development are aided by preclinical studies involving GF mice (Jenmalm, 2017; Torres et al., 2020). These studies can highlight the phenotypic differences in animals maturing with or without an intact microbiome. One such study (Lu et al., 2018) transferred the microbiome of preterm human infants to GF mice in order to pinpoint neuronal markers that were altered in congruence with the affected phenotype. Mice that received the preterm microbiome displayed poor development and a decreased expression profile of the neuronal markers NeuN and neurofilament‐L and myelination marker myelin basic protein (MBP). As is clinically observed in preterm infants, the ‘induced preterm’ mice exhibited increased neuroinflammation as measured by increased Nos1 and Igfbp3 brain mRNA expression, altered insulin‐like growth factor 1 (IGF‐1) signalling and decreased circulating insulin‐like growth factor binding protein 3 (IGFBP3). A different study (Chen et al., 2017) examined the microbial influence on hippocampal microRNA (miRNA) and mRNA expression. Utilizing behavioural tests to compare GF and SPF mice, followed by RNA sequencing, the researchers showed that the GF mice exhibited decreased anxiety‐like behaviour, supported by a deficient RNA expression profile, which was partially restored following colonization with SPF microbiome. The genetic significance of the RNA expression alterations lies in their importance in metabolic, cellular binding and guiding and molecular processes as they pertain to neuronal functionality. GF mice, repeatedly shown to exhibit reduced anxiolytic responses, were neurochemically examined in several major brain areas. In situ hybridization revealed reduced levels of NGFI‐A and BDNF mRNA. NGF is abundantly present in the hippocampus and takes part in HPA activity in an IEG‐inducible manner. NGFI‐A expression is implicated in situations related to stress, abnormal electrical activity such as seizures and following the induction of synaptic plasticity in the form of LTP (Olsson et al., 1994). This study also showed, by qPCR, a higher level of synaptophysin and postsynaptic density protein 95 (PSD‐95) (Ocansey et al., 2020) in the striata of GF mice (Olsson et al., 1994). Synaptophysin, expressed in most CNS neurons and particularly in neuroendocrine cells, is necessary for synaptic vesicle maturation and is used as a secondary indicator of developmental synaptogenesis (Hami et al., 2016). Also necessary for developmental processes in excitatory synapses is PSD‐95 (Prange et al., 2004). Microbial influence on proper CNS functionality is often caused by a perturbation to the normal microbiome as a result of infection or another microbial imbalance leading to an exacerbated presence of a certain pathogen. Gareau et al. (2011) introduced Citrobacter rodentium to GF and conventional mice and tested memory impairment in the presence or absence of an added stressor in the form of water avoidance stress (WAS). Following introduction to the aversive WAS stimulus the C. rodentium‐infected mice exhibited decreased hippocampal BDNF and c‐Fos levels, which were ameliorated by probiotic treatment. Interestingly, it was only the combination of C. rodentium and WAS that produced the alterations in BDNF and c‐Fos expression.
Due to its highly diverse functionality, BDNF is necessary for a number of brain processes, both homeostatic and plastic in nature. Evidence linking the modulation of BDNF to the microbiome signifies a step towards unravelling the mechanistic relationship between the microbiome and synaptic plasticity.
IX. SYNTHESIS
The existence of a microbiome–gut–brain axis is overwhelmingly supported, with correlations between the microbiome and a wide spectrum of cognitive, emotional and behavioural phenotypes, in health and disease, as well as homeostatic brain functionality. Currently, the vast potential of the microbiome as a diagnostic and therapeutic tool is undercut by a dearth in the mechanistic understanding of how the microbiome exerts its influence on the host's brain; however, some promising evidence is emerging. Probiotics (live, beneficial microorganisms introduced to the host), prebiotics (non‐digestible dietary fibre that serves as an energy source for the host's resident microbiome) and synbiotics (a combination of the two) have been increasingly used in a clinical setting by a variety of medical fields, exhibiting salubrious health effects, including normalization of neuropsychiatric symptoms (Moosmang, 2005; Kali, 2016; Cepeda, Katz & Blacketer, 2017; Mörkl et al., 2020; Morshedi, Saghafi‐Asl & Hosseinifard, 2020; Skott et al., 2020).
The microbiome interacts extensively with the mucosal immune system and the enteric nervous system, which operate synergistically with effects spanning many organ systems, particularly the brain. Microbial metabolites affect the brain through several branches of the microbiome–gut–brain axis via the parasympathetic vagus nerve that connects the ENS the CNS and the systemic immune system or the HPA axis which are interconnected as well. Synaptic plasticity is one of the most fundamental processes occurring in the brain, implicated in learning and memory consolidation. Dysfunctions in induction of synaptic plasticity are implicated in severe neuropsychiatric disorders. The mechanisms of plasticity are highly complex and include many types of receptors and their trafficking across membranes, signalling and cell adhesion molecules. The presynaptic cell is stimulated by a neurotransmitter which in turn activates ionic channels on the presynaptic cell, altering its membrane potential and ionic permeability, thereby affecting its ability to fire an action potential. ENS plasticity is emerging as a vastly important research target due to the complex interconnectedness between the CNS and ENS, as well as the latter's substantial involvement in various physiological and pathological phenomena. Moreover, the ENS shares many similarities with the CNS – from its neuronal circuitry to the neurotransmitters activating it.
The microbiome has been shown to influence several plasticity inducers as well as homeostatic maintenance, through various neurotransmitters, BDNF, and expression of relevant genes. While research into the effects of the microbiome on synaptic plasticity is shifting to the ENS, very few studies have investigated the interplay between plasticity in the ENS and the CNS, under the influence of the microbiome and its metabolites (Table 1). While conducting electrophysiological studies on GF mice is complicated by setting up adequate equipment within a sterile environment, we believe that incorporating such examinations into studies involving SPF or conventional mice is necessary. Electrophysiological studies of an animal's ENS as well as CNS are needed, together with measurements of plasticity‐related markers in both the CNS and the gut. Moreover, incorporating metabolomics analyses into microbiome studies aimed at unravelling the mechanistic connections between metabolites and synaptic plasticity will provide a more accurate picture of which metabolites are significantly altered, in conjunction with insights from electrophysiological studies. Additionally, we strongly believe in the big‐data approach of modelling such complex systems and interactions. There is evidence that computational‐oriented mapping of all the different molecular constituents of plasticity, the vast array of microbial metabolites and other physiological markers from the host, might yield a better understanding of how all these different elements fit together (Eetemadi et al., 2020; Mäki‐Marttunen et al., 2020).
Table 1.
Potential microbial effectors of synaptic plasticity
| Target nervous system | Hypothesized plasticity‐related mechanism affected | |
|---|---|---|
| Lactobacillus reuteri (LR) | ENS |
Calcium‐dependent channels in a subpopulation of enteric sensory neurons, implication in intestinal‐disorder derived pain (Kunze et al., 2009). LR probiotic‐treated mice exhibited reduced capsaicin‐induced excitatory response, accompanied by decreased expression of the nociceptive signal‐transmitting TRPV1 (Perez‐Burgos et al., 2015). Probiotic administration rescued social deficits in ASD mice in a vagus‐dependent manner. |
| Bifidobacterium infantis | CNS | Probiotic administration led to reduction of the serotonin intermediate 5‐HIAA in the frontal cortex, together with plasma increases of tryptophan, kynurenic acid, dihydroxyphenylacetic acid and noradrenaline (El Aidy et al., 2012). |
| Lachnospiraceae, Syntrophococcus, Shuttleworthia, Gemella, Allobaculum, and Hydrogenoanaerobacterium | CNS | Changes in dopamine receptor expression, known to contribute to induction of stimulus‐dependent plasticity in areas of the brain responsible for emotional regulation (Jadhav et al., 2018). |
| Bifidobacterium | CNS | Deficient ventral striatal responses in a reward anticipation fMRI test (Aarts et al., 2017). |
| Enterobacteriaceae | CNS | Correlated with severity of Parkinson's symptoms (Parashar & Udayabanu, 2017). |
| Lactobacillus rhamnosus JB‐1 (JB‐1) | CNS | Immune‐mediated modulation of anxiolytic and depressive behaviours (Liu et al., 2020). Affects anxiolytic and depressive behaviours in a vagus‐dependent manner (Perez‐Burgos et al., 2014), and has also been shown to reduce GABA receptor expression in specific brain areas known to undergo extensive learning and emotional‐regulation related plasticity (Bravo et al., 2011). L. rhamnosus ameliorated LPS‐induced impairment of hippocampal CaMKII‐α gene expression. |
| Bifidobacterium pseudocatenulatum | CNS | Prevention of stress‐induced HPA axis activation and consequent increases of corticosterone and hypothalamic catecholamines (Moya‐Pérez et al., 2017). |
| Campylobacter jejuni | Increase of anxiolytic behaviours, as well as Fos activity in vagal afferents and the solitary tract nucleus (Fülling et al., 2019). | |
| Lactobacillus acidophilus, Bifidobacterium lactis and/or Lactobacillus fermentum | CNS | Probiotic cocktail led to improvement in learning task performance, as well as rescue of hippocampal EPSP response and markedly improved spatial memory, facilitated by an improved ability of the brain to undergo LTP (Davari et al., 2013). |
| L. rhamnosus, L. reuteri and/or L. plantarum | CNS | Probiotic administration impeded neuroinflammation‐induced memory deficits in mice (Zolfaghari et al., 2020). |
| Lactobacillus johnsonii CJLJ103 | Probiotic administration of L. johnsonii CJLJ103 (LJ) to mice. LJ corrected two detrimental effects caused by LPS: NF‐κB‐induced hippocampal neuroinflammation and reduced BDNF expression (Lee et al., 2018) |
5‐HIAA, 5‐hydroxyindoleacetic acid; ASD, autism spectrum disorder; BDNF, brain‐derived neurotrophic factor; CaMKII‐α, calcium/calmodulin‐dependent protein kinase type II subunit alpha; CNS, central nervous system; ENS, enteric nervous system; EPSP, excitatory postsynaptic potential; fMRI, functional magnetic resonance imaging; Fos, transcription factor belonging to the immediate early gene family; GABA, gamma aminobutyric acid; HPA, hypothalamus–pituitary–adrenal axis; LPS, lipopolysaccharide; LR, Lactobacillus reuteri; LTP, long‐term potentiation; NF‐κB, nuclear factor kappa‐light‐chain‐enhancer of activated B cells; TRPV1, transient receptor potential vanilloid 1.
The objective of this review was to highlight the multi‐factorial involvement of the microbiome with the vastly important process of synaptic plasticity, and to emphasize the need for a research paradigm shift, where the pathophysiology of complex neuropsychiatric and neurodegenerative disorders is involved (Table 1). We believe that studying the microbiome–gut–brain axis should henceforth include a significant neuroscience‐driven research methodology. In‐depth explorations into the intricate processes involved with synaptic plasticity should put equal emphasis on the microbial and the neuronal–electrophysiological aspects of the process. Indeed, they should focus on the contribution of the microbiome and its metabolites, given their undisputed importance to cognitive, emotional and behavioural aspects of the host. Such processes have been shown to depend heavily on successful induction and maintenance of synaptic plasticity.
X. CONCLUSIONS
The microbiome has been unequivocally shown to affect the mental, emotional and cognitive functionality of the host, including both phenotypic manifestations and neurophysiological aspects.
Synaptic plasticity is a cardinal neurophysiological process occurring in both the CNS and ENS. It is responsible for the incorporation of long‐term functional changes to synaptic connectivity and structural integrity, and consequently, neuronal transmission. The consolidation of such plastic changes requires a host of molecular processes to occur, including alterations in gene expression and receptor trafficking, rapidly regulated signalling cascades, secretion of neurotransmitters and neurotrophic factors and more.
The microbiome has been shown to influence various different neurophysiological aspects of synaptic plasticity, with direct correlations established between microbial perturbations and neurotransmitter secretion, immediate early gene expression, neuronal surface receptor trafficking, neurophysiological properties affecting neuronal transmission and more. These diverse effects of the microbiome on the different mechanisms underlying synaptic plasticity suggest that the influence of the microbiome on the mental, cognitive and emotional wellbeing of the host is achieved by affecting synaptic plasticity directly and indirectly, thereby allowing for long‐lasting changes to take place.
XI. ACKNOWLEDGEMENTS
O.K. is supported by a European Research Council Consolidator grant (grant agreement no. 101001355).
REFERENCES
- Aarts, E. , Ederveen, T. H. A. , Naaijen, J. , Zwiers, M. P. , Boekhorst, J. , Timmerman, H. M. , Smeekens, S. P. , Netea, M. G. , Buitelaar, J. K. , Franke, B. , van Hijum, S. A. F. T. & Arias Vasquez, A. (2017). Gut microbiome in ADHD and its relation to neural reward anticipation. PLoS One 12, e0183509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmäe, S. , Franasiak, J. M. & Mändar, R. (2019). The seminal microbiome in health and disease. Nature Reviews Urology 16, 703–721. [DOI] [PubMed] [Google Scholar]
- Bai, Y. & Suzuki, T. (2020). Activity‐dependent synaptic plasticity in Drosophila melanogaster . Frontiers in Physiology 11, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D. A. , Esler, M. D. , Dawood, T. , Lambert, E. A. , Haikerwal, D. , Brenchley, C. , Socratous, F. , Hastings, J. , Guo, L. , Wiesner, G. , Kaye, D. M. , Bayles, R. , Schlaich, M. P. & Lambert, G. W. (2008). Elevated brain serotonin turnover in patients with depression. Archives of General Psychiatry 65, 38–46. [DOI] [PubMed] [Google Scholar]
- Bennett, A. & Maxwell, R. (2010). Synapse regression in depression: the role of 5‐HT receptors in modulating NMDA receptor function and synaptic plasticity. Australian & New Zealand Journal of Psychiatry 44, 301–308. [DOI] [PubMed] [Google Scholar]
- Bercik, P. , Denou, E. , Collins, J. , Jackson, W. , Lu, J. , Jury, J. , Deng, Y. , Blennerhassett, P. , Macri, J. , McCoy, K. D. , Verdu, E. F. & Collins, S. M. (2011). The intestinal microbiota affect central levels of brain‐derived neurotropic factor and behavior in mice. Gastroenterology 141, 599–609. [DOI] [PubMed] [Google Scholar]
- Berger, S. M. , Fernández‐Lamo, I. , Schönig, K. , Fernández Moya, S. M. , Ehses, J. , Schieweck, R. , Clementi, S. , Enkel, T. , Grothe, S. , von Bohlen und Halbach, O. , Segura, I. , Delgado‐García, J. M. , Gruart, A. , Kiebler, M. A. & Bartsch, D. (2017). Forebrain‐specific, conditional silencing of Staufen2 alters synaptic plasticity, learning, and memory in rats. Genome Biology 18, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binyamin, D. , Werbner, N. , Nuriel‐Ohayon, M. , Uzan, A. , Mor, H. , Abbas, A. , Ziv, O. , Teperino, R. , Gutman, R. & Koren, O. (2020). The aging mouse microbiome has obesogenic characteristics. Genome Medicine 12, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesmans, W. , Gomes, P. , Janssens, J. , Tack, J. & Berghe, P. V. (2008). Brain‐derived neurotrophic factor amplifies neurotransmitter responses and promotes synaptic communication in the enteric nervous system. Gut 57, 314–322. [DOI] [PubMed] [Google Scholar]
- Bonaz, B. , Sinniger, V. & Pellissier, S. (2017). The vagus nerve in the neuro‐immune axis: implications in the pathology of the gastrointestinal tract. Frontiers in Immunology 8, 1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo, J. A. , Forsythe, P. , Chew, M. V. , Escaravage, E. , Savignac, H. M. , Dinan, T. G. , Bienenstock, J. & Cryan, J. F. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proceedings of the National Academy of Sciences of the United States of America 108, 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabotti, M. , Scirocco, A. , Maselli, M. A. & Severi, C. (2015). The gut‐brain axis: interactions between enteric microbiota, central and enteric nervous systems. Annals of Gastroenterology 28, 203–209. [PMC free article] [PubMed] [Google Scholar]
- Cepeda, M. S. , Katz, E. G. & Blacketer, C. (2017). Microbiome‐gut‐brain axis: probiotics and their association with depression. The Journal of Neuropsychiatry and Clinical Neurosciences 29, 39–44. [DOI] [PubMed] [Google Scholar]
- Cerovic, M. , Forloni, G. & Balducci, C. (2019). Neuroinflammation and the gut microbiota: possible alternative therapeutic targets to counteract Alzheimer's disease? Frontiers in Aging Neuroscience 11, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater, T. E. & Goda, Y. (2014). The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Frontiers in Cellular Neuroscience 8, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. T. , Hopf, F. W. & Bonci, A. (2010). Synaptic plasticity in the mesolimbic system. Annals of the New York Academy of Sciences 1187, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Zeng, B. , Li, W. , Zhou, C. , Fan, S. , Cheng, K. , Zeng, L. , Zheng, P. , Fang, L. , Wei, H. & Xie, P. (2017). Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behavioural Brain Research 322, 34–41. [DOI] [PubMed] [Google Scholar]
- Citri, A. & Malenka, R. C. (2008). Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology 33, 18–41. [DOI] [PubMed] [Google Scholar]
- Clarke, G. , Grenham, S. , Scully, P. , Fitzgerald, P. , Moloney, R. D. , Shanahan, F. , Dinan, T. G. & Cryan, J. F. (2013). The microbiome‐gut‐brain axis during early life regulates the hippocampal serotonergic system in a sex‐dependent manner. Molecular Psychiatry 18, 666–673. [DOI] [PubMed] [Google Scholar]
- Collingridge, G. L. , Peineau, S. , Howland, J. G. & Wang, Y. T. (2010). Long‐term depression in the CNS. Nature Reviews Neuroscience 11, 459–473. [DOI] [PubMed] [Google Scholar]
- Contin, M. & Martinelli, P. (2010). Pharmacokinetics of levodopa. Journal of Neurology 257, 253–261. [DOI] [PubMed] [Google Scholar]
- Coppens, C. M. , Siripornmongcolchai, T. , Wibrand, K. , Alme, M. N. , Buwalda, B. , de Boer, S. F. , Koolhaas, J. M. & Bramham, C. R. (2011). Social defeat during adolescence and adulthood differentially induce BDNF‐regulated immediate early genes. Frontiers in Behavioral Neuroscience 5, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés‐Mendoza, J. , Díaz de León‐Guerrero, S. , Pedraza‐Alva, G. & Pérez‐Martínez, L. (2013). Shaping synaptic plasticity: the role of activity‐mediated epigenetic regulation on gene transcription. International Journal of Developmental Neuroscience 31, 359–369. [DOI] [PubMed] [Google Scholar]
- Cryan, J. F. & O'Mahony, S. M. (2011). The microbiome‐gut‐brain axis: from bowel to behavior. Neurogastroenterology & Motility 23, 187–192. [DOI] [PubMed] [Google Scholar]
- Cryan, J. F. , O'Riordan, K. J. , Cowan, C. S. M. , Sandhu, K. V. , Bastiaanssen, T. F. S. , Boehme, M. , Codagnone, M. G. , Cussotto, S. , Fulling, C. , Golubeva, A. V. , Guzzetta, K. E. , Jaggar, M. , Long‐Smith, C. M. , Lyte, J. M. , Martin, J. A. , et al. (2019). The microbiota‐gut‐brain axis. Physiological Reviews 99, 1877–2013. [DOI] [PubMed] [Google Scholar]
- Cunha, C. , Brambilla, R. & Thomas, K. L. (2010). A simple role for BDNF in learning and memory? Frontiers in Molecular Neuroscience 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, P. M. , Gottfried‐Blackmore, A. , Anandasabapathy, N. & Bulloch, K. (2012). Brain dendritic cells: biology and pathology. Acta Neuropathologica 124, 599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalile, B. , Van Oudenhove, L. , Vervliet, B. & Verbeke, K. (2019). The role of short‐chain fatty acids in microbiota–gut–brain communication. Nature Reviews Gastroenterology & Hepatology 16, 461–478. [DOI] [PubMed] [Google Scholar]
- D'Amato, A. , Di Cesare Mannelli, L. , Lucarini, E. , Man, A. L. , Le Gall, G. , Branca, J. J. V. , Ghelardini, C. , Amedei, A. , Bertelli, E. , Regoli, M. , Pacini, A. , Luciani, G. , Gallina, P. , Altera, A. , Narbad, A. , et al. (2020). Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity‐ and neurotransmission‐related proteins in young recipients. Microbiome 8(1), 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darch, H. T. , Collins, M. K. , O'Riordan, K. J. & Cryan, J. F. (2021). Microbial memories: sex‐dependent impact of the gut microbiome on hippocampal plasticity. European Journal of Neuroscience 54, 5235–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubert, E. A. & Condron, B. G. (2010). Serotonin: a regulator of neuronal morphology and circuitry. Trends in Neurosciences 33, 424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davari, S. , Talaei, S. A. , Alaei, H. & Salami, M. (2013). Probiotics treatment improves diabetes‐induced impairment of synaptic activity and cognitive function: behavioral and electrophysiological proofs for microbiome–gut–brain axis. Neuroscience 240, 287–296. [DOI] [PubMed] [Google Scholar]
- De Vadder, F. , Grasset, E. , Mannerås Holm, L. , Karsenty, G. , Macpherson, A. J. , Olofsson, L. E. & Bäckhed, F. (2018). Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proceedings of the National Academy of Sciences of the United States of America 115, 6458–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter, B. Y. (2010). Interplay between inflammation, immune system and neuronal pathways: effect on gastrointestinal motility. World Journal of Gastroenterology 16(44), 5523–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleidi, M. , Jãggle, M. & Rubino, G. (2015). Immune aging, dysmetabolism, and inflammation in neurological diseases. Frontiers in Neuroscience 9, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet, L. , Garrett, L. , Clarke, G. , Bienenstock, J. & Dinan, T. G. (2008). The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. Journal of Psychiatric Research 43, 164–174. [DOI] [PubMed] [Google Scholar]
- Di Filippo, M. , Chiasserini, D. , Gardoni, F. , Viviani, B. , Tozzi, A. , Giampà, C. , Costa, C. , Tantucci, M. , Zianni, E. , Boraso, M. , Siliquini, S. , de Iure, A. , Ghiglieri, V. , Colcelli, E. , Baker, D. , et al. (2013). Effects of central and peripheral inflammation on hippocampal synaptic plasticity. Neurobiology of Disease 52, 229–236. [DOI] [PubMed] [Google Scholar]
- Dinan, T. G. & Cryan, J. F. (2017). Brain‐gut‐microbiota axis and mental health. Psychosomatic Medicine 79, 920–926. [DOI] [PubMed] [Google Scholar]
- Dominguez‐Bello, M. G. , Godoy‐Vitorino, F. , Knight, R. & Blaser, M. J. (2019). Role of the microbiome in human development. Gut 68, 1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eetemadi, A. , Rai, N. , Pereira, B. M. P. , Kim, M. , Schmitz, H. & Tagkopoulos, I. (2020). The computational diet: a review of computational methods across diet, microbiome, and health. Frontiers in Microbiology 11, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy, S. , Kunze, W. , Bienenstock, J. & Kleerebezem, M. (2012). The microbiota and the gut‐brain axis: insights from the temporal and spatial mucosal alterations during colonisation of the germfree mouse intestine. Beneficial Microbes 3, 251–259. [DOI] [PubMed] [Google Scholar]
- Elfil, M. , Kamel, S. , Kandil, M. , Koo, B. B. & Schaefer, S. M. (2020). Implications of the gut microbiome in Parkinson's disease. Movement Disorders 35, 921–933. [DOI] [PubMed] [Google Scholar]
- Engevik, M. A. , Luk, B. , Chang‐Graham, A. L. , Hall, A. , Herrmann, B. , Ruan, W. , Endres, B. T. , Shi, Z. , Garey, K. W. , Hyser, J. M. & Versalovic, J. (2019). Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. mBio 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcicchia, C. , Tozzi, F. , Arancio, O. , Watterson, D. M. & Origlia, N. (2020). Involvement of p38 MAPK in synaptic function and dysfunction. International Journal of Molecular Sciences 21(16), 5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favalli, G. , Li, J. , Belmonte‐de‐Abreu, P. , Wong, A. H. C. & Daskalakis, Z. J. (2012). The role of BDNF in the pathophysiology and treatment of schizophrenia. Journal of Psychiatric Research 46, 1–11. [DOI] [PubMed] [Google Scholar]
- Foster, J. A. & McVey Neufeld, K.‐A. (2013). Gut–brain axis: how the microbiome influences anxiety and depression. Trends in Neurosciences 36, 305–312. [DOI] [PubMed] [Google Scholar]
- Frankiensztajn, L. M. , Elliott, E. & Koren, O. (2020). The microbiota and the hypothalamus‐pituitary‐adrenocortical (HPA) axis, implications for anxiety and stress disorders. Current Opinion in Neurobiology 62, 76–82. [DOI] [PubMed] [Google Scholar]
- Fülling, C. , Dinan, T. G. & Cryan, J. F. (2019). Gut microbe to brain signaling: what happens in vagus…. Neuron 101, 998–1002. [DOI] [PubMed] [Google Scholar]
- Fung, T. C. , Olson, C. A. & Hsiao, E. Y. (2017). Interactions between the microbiota, immune and nervous systems in health and disease. Nature Neuroscience 20, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, K. , Mu, C. , Farzi, A. & Zhu, W. (2020). Tryptophan metabolism: a link between the gut microbiota and brain. Advances in Nutrition 11, 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau, M. G. , Wine, E. , Rodrigues, D. M. , Cho, J. H. , Whary, M. T. , Philpott, D. J. , MacQueen, G. & Sherman, P. M. (2011). Bacterial infection causes stress‐induced memory dysfunction in mice. Gut 60, 307–317. [DOI] [PubMed] [Google Scholar]
- Geuking, M. B. , Köller, Y. , Rupp, S. & McCoy, K. D. (2014). The interplay between the gut microbiota and the immune system. Gut Microbes 5, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson, C. D. , Kupchik, Y. M. & Kalivas, P. W. (2014). Rapid, transient synaptic plasticity in addiction. Neuropharmacology 76, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzman, D. L. (2010). Common mechanisms of synaptic plasticity in vertebrates and invertebrates. Current Biology 20, R31–R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassner, K. L. , Abraham, B. P. & Quigley, E. M. M. (2020). The microbiome and inflammatory bowel disease. Journal of Allergy and Clinical Immunology 145, 16–27. [DOI] [PubMed] [Google Scholar]
- Goldberg, M. R. , Mor, H. , Magid Neriya, D. , Magzal, F. , Muller, E. , Appel, M. Y. , Nachshon, L. , Borenstein, E. , Tamir, S. , Louzoun, Y. , Youngster, I. , Elizur, A. & Koren, O. (2020). Microbial signature in IgE‐mediated food allergies. Genome Medicine 12, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, C. , Iwasaki, Y. & Yada, T. (2018). Short‐chain fatty acids suppress food intake by activating vagal afferent neurons. The Journal of Nutritional Biochemistry 57, 130–135. [DOI] [PubMed] [Google Scholar]
- Goto, Y. , Yang, C. R. & Otani, S. (2010). Functional and dysfunctional synaptic plasticity in prefrontal cortex: roles in psychiatric disorders. Biological Psychiatry 67, 199–207. [DOI] [PubMed] [Google Scholar]
- Govindarajulu, M. , Pinky, P. D. , Steinke, I. , Bloemer, J. , Ramesh, S. , Kariharan, T. , Rella, R. T. , Bhattacharya, S. , Dhanasekaran, M. , Suppiramaniam, V. & Amin, R. H. (2020). Gut metabolite TMAO induces synaptic plasticity deficits by promoting endoplasmic reticulum stress. Frontiers in Molecular Neuroscience 13, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hami, J. , Vafaei‐Nezhad, S. , Ivar, G. , Sadeghi, A. , Ghaemi, K. , Mostafavizadeh, M. & Hosseini, M. (2016). Altered expression and localization of synaptophysin in developing cerebellar cortex of neonatal rats due to maternal diabetes mellitus. Metabolic Brain Disease 31, 1369–1380. [DOI] [PubMed] [Google Scholar]
- He, Y.‐Y. , Zhang, X.‐Y. , Yung, W.‐H. , Zhu, J.‐N. & Wang, J.‐J. (2013). Role of BDNF in central motor structures and motor diseases. Molecular Neurobiology 48, 783–793. [DOI] [PubMed] [Google Scholar]
- Hebb, D. O. (2002). The Organization of Behavior: A Neuropsychological Theory. Mahwah, NJ: Lawrence Erlbaum Associates Inc. [Google Scholar]
- Heijtz, R. D. , Wang, S. , Anuar, F. , Qian, Y. , Bjorkholm, B. , Samuelsson, A. , Hibberd, M. L. , Forssberg, H. & Pettersson, S. (2011). Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, J. P. (2018). Regulation of hypothalamo‐pituitary‐adrenocortical responses to stressors by the nucleus of the solitary tract/dorsal vagal complex. Cellular and Molecular Neurobiology 38, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, V. M. , Lee, J.‐A. & Martin, K. C. (2011). The cell biology of synaptic plasticity. Science 334, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban, A. E. , Stilling, R. M. , Moloney, G. , Shanahan, F. , Dinan, T. G. , Clarke, G. & Cryan, J. F. (2018). The microbiome regulates amygdala‐dependent fear recall. Molecular Psychiatry 23, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzel, B. K. , Carmody, J. , Evans, K. C. , Hoge, E. A. , Dusek, J. A. , Morgan, L. , Pitman, R. K. & Lazar, S. W. (2010). Stress reduction correlates with structural changes in the amygdala. Social Cognitive and Affective Neuroscience 5, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberg, J. R. , Molteni, R. , Calabrese, F. & Riva, M. A. (2014). The serotonin–BDNF duo: developmental implications for the vulnerability to psychopathology. Neuroscience & Biobehavioral Reviews 43, 35–47. [DOI] [PubMed] [Google Scholar]
- Hu, S. G. , Liu, Y. , Chen, T. P. , Liu, Z. , Yu, Q. , Deng, L. J. , Yin, Y. & Hosaka, S. (2013). Emulating the paired‐pulse facilitation of a biological synapse with a NiOx‐based memristor. Applied Physics Letters 102, 183510. [Google Scholar]
- Hunt, D. L. & Castillo, P. E. (2012). Synaptic plasticity of NMDA receptors: mechanisms and functional implications. Current Opinion in Neurobiology 22, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland, N. P. & Cryan, J. F. (2016). Microbe‐host interactions: influence of the gut microbiota on the enteric nervous system. Developmental Biology 417, 182–187. [DOI] [PubMed] [Google Scholar]
- Israelyan, N. & Margolis, K. G. (2019). Reprint of: Serotonin as a link between the gut‐brain‐microbiome axis in autism spectrum disorders. Pharmacological Research 140, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav, K. S. , Peterson, V. L. , Halfon, O. , Ahern, G. , Fouhy, F. , Stanton, C. , Dinan, T. G. , Cryan, J. F. & Boutrel, B. (2018). Gut microbiome correlates with altered striatal dopamine receptor expression in a model of compulsive alcohol seeking. Neuropharmacology 141, 249–259. [DOI] [PubMed] [Google Scholar]
- Jappy, D. , Valiullina, F. , Draguhn, A. & Rozov, A. (2016). GABABR‐dependent long‐term depression at hippocampal synapses between CB1‐positive interneurons and CA1 pyramidal cells. Frontiers in Cellular Neuroscience 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenmalm, M. C. (2017). The mother‐offspring dyad: microbial transmission, immune interactions and allergy development. Journal of Internal Medicine 282, 484–495. [DOI] [PubMed] [Google Scholar]
- Jiang, D. , Jin, S. , Li, G. , Li, Q. , Li, Z. , Ma, H. , Zhuo, C. , Jiang, R. & Ye, M. (2016). Serotonin regulates brain‐derived neurotrophic factor expression in select brain regions during acute psychological stress. Neural Regeneration Research 11(9), 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L. A. , Sun, E. W. , Martin, A. M. & Keating, D. J. (2020). The ever‐changing roles of serotonin. The International Journal of Biochemistry & Cell Biology 125, 105776. [DOI] [PubMed] [Google Scholar]
- Kabouridis, P. S. , Lasrado, R. , McCallum, S. , Chng, S. H. , Snippert, H. J. , Clevers, H. , Pettersson, S. & Pachnis, V. (2015). The gut microbiota keeps enteric glial cells on the move; prospective roles of the gut epithelium and immune system. Gut Microbes 6, 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabouridis, P. S. & Pachnis, V. (2015). Emerging roles of gut microbiota and the immune system in the development of the enteric nervous system. Journal of Clinical Investigation 125, 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kali, A. (2016). Psychobiotics: an emerging probiotic in psychiatric practice. Biomedical Journal 39, 223–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel, E. & Schwartz, J. (1982). Molecular biology of learning: modulation of transmitter release. Science 218, 433–443. [DOI] [PubMed] [Google Scholar]
- Khalil, M. , Zhang, Z. & Engel, M. A. (2019). Neuro‐immune networks in gastrointestinal disorders. Visceral Medicine 35, 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. H. (2018). Immune regulation by microbiome metabolites. Immunology 154, 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Kwon, S.‐H. , Kam, T.‐I. , Panicker, N. , Karuppagounder, S. S. , Lee, S. , Lee, J. H. , Kim, W. R. , Kook, M. , Foss, C. A. , Shen, C. , Lee, H. , Kulkarni, S. , Pasricha, P. J. , Lee, G. , et al. (2019). Transneuronal propagation of pathologic α‐synuclein from the gut to the brain models Parkinson's disease. Neuron 103, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb, E. & Finkbeiner, S. (2011). Arc in synaptic plasticity: from gene to behavior. Trends in Neurosciences 34, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornijcuk, V. , Kim, D. , Kim, G. & Jeong, D. S. (2020). Simplified calcium signaling cascade for synaptic plasticity. Neural Networks 123, 38–51. [DOI] [PubMed] [Google Scholar]
- Kotaleski, J. H. & Blackwell, K. T. (2010). Modelling the molecular mechanisms of synaptic plasticity using systems biology approaches. Nature Reviews Neuroscience 11, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, C. , Castrén, E. , Kasper, S. & Lanzenberger, R. (2017). Serotonin and neuroplasticity – links between molecular, functional and structural pathophysiology in depression. Neuroscience & Biobehavioral Reviews 77, 317–326. [DOI] [PubMed] [Google Scholar]
- Kumar, A. (2011). Long‐term potentiation at CA3–CA1 hippocampal synapses with special emphasis on aging, disease, and stress. Frontiers in Aging Neuroscience 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, W. A. , Mao, Y.‐K. , Wang, B. , Huizinga, J. D. , Ma, X. , Forsythe, P. & Bienenstock, J. (2009). Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium‐dependent potassium channel opening. Journal of Cellular and Molecular Medicine 13, 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, V. , Ditu, L.‐M. , Pircalabioru, G. G. , Gheorghe, I. , Curutiu, C. , Holban, A. M. , Picu, A. , Petcu, L. & Chifiriuc, M. C. (2018). Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Frontiers in Immunology 9, 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal, G. , Comprido, D. & Duarte, C. B. (2014). BDNF‐induced local protein synthesis and synaptic plasticity. Neuropharmacology 76, 639–656. [DOI] [PubMed] [Google Scholar]
- Lee, H.‐J. , Lim, S.‐M. & Kim, D.‐H. (2018). Lactobacillus johnsonii CJLJ103 attenuates scopolamine‐induced memory impairment in mice by increasing BDNF expression and inhibiting NF‐kB activation. Journal of Microbiology and Biotechnology 28, 1443–1446. [DOI] [PubMed] [Google Scholar]
- Lesch, K.‐P. , Araragi, N. , Waider, J. , van den Hove, D. & Gutknecht, L. (2012). Targeting brain serotonin synthesis: insights into neurodevelopmental disorders with long‐term outcomes related to negative emotionality, aggression and antisocial behaviour. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 2426–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch, K.‐P. & Waider, J. (2012). Serotonin in the modulation of neural plasticity and networks: implications for neurodevelopmental disorders. Neuron 76, 175–191. [DOI] [PubMed] [Google Scholar]
- Leung, K. & Thuret, S. (2015). Gut microbiota: a modulator of brain plasticity and cognitive function in ageing. Healthcare 3, 898–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, S. G. & Godukhin, O. V. (2017). Modulating effect of cytokines on mechanisms of synaptic plasticity in the brain. Biochemistry (Moscow) 82, 264–274. [DOI] [PubMed] [Google Scholar]
- Levy, B. H. & Tasker, J. G. (2012). Synaptic regulation of the hypothalamic–pituitary–adrenal axis and its modulation by glucocorticoids and stress. Frontiers in Cellular Neuroscience 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Ke, Y. , Zhan, R. , Liu, C. , Zhao, M. , Zeng, A. , Shi, X. , Ji, L. , Cheng, S. , Pan, B. , Zheng, L. & Hong, H. (2018). Trimethylamine‐N‐oxide promotes brain aging and cognitive impairment in mice. Aging Cell 17, e12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.‐M. , Jang, H.‐M. , Jeong, J.‐J. , Han, M. J. & Kim, D.‐H. (2017). Lactobacillus johnsonii CJLJ103 attenuates colitis and memory impairment in mice by inhibiting gut microbiota lipopolysaccharide production and NF‐kB activation. Journal of Functional Foods 34, 359–368. [Google Scholar]
- Limanaqi, F. , Biagioni, F. , Busceti, C. L. , Ryskalin, L. , Soldani, P. , Frati, A. & Fornai, F. (2019). Cell clearing systems bridging neuro‐immunity and synaptic plasticity. International Journal of Molecular Sciences 20(9), 2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, M. , Chalazonitis, A. , Palm, E. , Pakarinen, E. , Danilova, T. , Pham, T. D. , Setlik, W. , Rao, M. , Võikar, V. , Huotari, J. , Kopra, J. , Andressoo, J.‐O. , Piepponen, P. T. , Airavaara, M. , Panhelainen, A. , et al. (2020). Cerebral dopamine neurotrophic factor–deficiency leads to degeneration of enteric neurons and altered brain dopamine neuronal function in mice. Neurobiology of Disease 134, 104696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Mian, M. F. , McVey Neufeld, K.‐A. & Forsythe, P. (2020). CD4+CD25+ T cells are essential for behavioral effects of Lactobacillus rhamnosus JB‐1 in male BALB/c mice. Brain, Behavior, and Immunity 88, 451–460. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Sanderson, D. , Mian, M. F. , McVey Neufeld, K.‐A. & Forsythe, P. (2021). Loss of vagal integrity disrupts immune components of the microbiota‐gut‐brain axis and inhibits the effect of Lactobacillus rhamnosus on behavior and the corticosterone stress response. Neuropharmacology 195, 108682. [DOI] [PubMed] [Google Scholar]
- Loprinzi, P. D. & Frith, E. (2019). A brief primer on the mediational role of BDNF in the exercise‐memory link. Clinical Physiology and Functional Imaging 39, 9–14. [DOI] [PubMed] [Google Scholar]
- Lovelace, M. D. , Varney, B. , Sundaram, G. , Lennon, M. J. , Lim, C. K. , Jacobs, K. , Guillemin, G. J. & Brew, B. J. (2017). Recent evidence for an expanded role of the kynurenine pathway of tryptophan metabolism in neurological diseases. Neuropharmacology 112, 373–388. [DOI] [PubMed] [Google Scholar]
- Lu, B. , Nagappan, G. & Lu, Y. (2014). BDNF and synaptic plasticity, cognitive function, and dysfunction. Handbook of Experimental Pharmacology 220, 223–250. [DOI] [PubMed] [Google Scholar]
- Lu, J. , Lu, L. , Yu, Y. , Cluette‐Brown, J. , Martin, C. R. & Claud, E. C. (2018). Effects of intestinal microbiota on brain development in humanized gnotobiotic mice. Scientific Reports 8, 5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Forney, L. J. & Ravel, J. (2012). Vaginal microbiome: rethinking health and disease. Annual Review of Microbiology 66, 371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N. & Ma, X. (2019). Dietary amino acids and the gut‐microbiome‐immune axis: physiological metabolism and therapeutic prospects. Comprehensive Reviews in Food Science and Food Safety 18, 221–242. [DOI] [PubMed] [Google Scholar]
- Maguire, J. (2014). Plasticity in the GABAergic regulation of the HPA axis (comment on DOI 10.1002/bies.201300178). BioEssays 36, 546–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäki‐Marttunen, T. , Iannella, N. , Edwards, A. G. , Einevoll, G. T. & Blackwell, K. T. (2020). A unified computational model for cortical post‐synaptic plasticity. eLife 9, e55714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter Cohen, M. , Jing, D. , Yang, R. R. , Tottenham, N. , Lee, F. S. & Casey, B. J. (2013). Early‐life stress has persistent effects on amygdala function and development in mice and humans. Proceedings of the National Academy of Sciences of the United States of America 110, 18274–18278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca, C. , Shen, M. , Boubertakh, B. , Martin, C. , Flamand, N. , Silvestri, C. & Di Marzo, V. (2020). Alterations of brain endocannabinoidome signaling in germ‐free mice. Biochimica et Biophysica Acta (BBA) ‐ Molecular and Cell Biology of Lipids 1865, 158786. [DOI] [PubMed] [Google Scholar]
- Martinowich, K. & Lu, B. (2008). Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology 33, 73–83. [DOI] [PubMed] [Google Scholar]
- Mawe, G. M. , Strong, D. S. & Sharkey, K. A. (2009). Plasticity of enteric nerve functions in the inflamed and postinflamed gut. Neurogastroenterology & Motility 21, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey Neufeld, K.‐A. , Bienenstock, J. , Bharwani, A. , Champagne‐Jorgensen, K. , Mao, Y. , West, C. , Liu, Y. , Surette, M. G. , Kunze, W. & Forsythe, P. (2019). Oral selective serotonin reuptake inhibitors activate vagus nerve dependent gut‐brain signalling. Scientific Reports 9, 14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey Neufeld, K. A. , Perez‐Burgos, A. , Mao, Y. K. , Bienenstock, J. & Kunze, W. A. (2015). The gut microbiome restores intrinsic and extrinsic nerve function in germ‐free mice accompanied by changes in calbindin. Neurogastroenterology & Motility 27, 627–636. [DOI] [PubMed] [Google Scholar]
- Medendorp, W. E. , Petersen, E. D. , Pal, A. , Wagner, L.‐M. , Myers, A. R. , Hochgeschwender, U. & Jenrow, K. A. (2018). Altered behavior in mice socially isolated during adolescence corresponds with immature dendritic spine morphology and impaired plasticity in the prefrontal cortex. Frontiers in Behavioral Neuroscience 12, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, R. , Debs, L. H. , Patel, A. P. , Nguyen, D. , Patel, K. , O'Connor, G. , Grati, M. , Mittal, J. , Yan, D. , Eshraghi, A. A. , Deo, S. K. , Daunert, S. & Liu, X. Z. (2017). Neurotransmitters: the critical modulators regulating gut‐brain axis. Journal of Cellular Physiology 232, 2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro‐Cardoso, V. F. , Corlianò, M. & Singaraja, R. R. (2021). Bile acids: a communication channel in the gut‐brain axis. NeuroMolecular Medicine 23, 99–117. [DOI] [PubMed] [Google Scholar]
- Moosmang, S. (2005). Role of hippocampal Cav1.2 Ca2+ channels in NMDA receptor‐independent synaptic plasticity and spatial memory. Journal of Neuroscience 25, 9883–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörkl, S. , Butler, M. I. , Holl, A. , Cryan, J. F. & Dinan, T. G. (2020). Probiotics and the microbiota‐gut‐brain axis: focus on psychiatry. Current Nutrition Reports 9, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshedi, M. , Saghafi‐Asl, M. & Hosseinifard, E.‐S. (2020). The potential therapeutic effects of the gut microbiome manipulation by synbiotic containing‐Lactobacillus plantarum on neuropsychological performance of diabetic rats. Journal of Translational Medicine 18(1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya‐Pérez, A. , Perez‐Villalba, A. , Benítez‐Páez, A. , Campillo, I. & Sanz, Y. (2017). Bifidobacterium CECT 7765 modulates early stress‐induced immune, neuroendocrine and behavioral alterations in mice. Brain, Behavior, and Immunity 65, 43–56. [DOI] [PubMed] [Google Scholar]
- Mulle, J. G. , Sharp, W. G. & Cubells, J. F. (2013). The gut microbiome: a new frontier in autism research. Current Psychiatry Reports 15, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, R. , Mainali, R. , Ahmadi, S. , Wang, S. , Singh, R. , Kavanagh, K. , Kitzman, D. W. , Kushugulova, A. , Marotta, F. & Yadav, H. (2018). Gut microbiome and aging: physiological and mechanistic insights. Nutrition and Healthy Aging 4, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K. , Obata, H. , Iriuchijima, N. & Saito, S. (2012). An increase in spinal cord noradrenaline is a major contributor to the antihyperalgesic effect of antidepressants after peripheral nerve injury in the rat. Pain 153, 990–997. [DOI] [PubMed] [Google Scholar]
- Nakano, T. , Doi, T. , Yoshimoto, J. & Doya, K. (2010). A kinetic model of dopamine‐ and calcium‐dependent striatal synaptic plasticity. PLoS Computational Biology 6, e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanou, E. & Catterall, W. A. (2018). Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron 98, 466–481. [DOI] [PubMed] [Google Scholar]
- Neufeld, K. M. , Kang, N. , Bienenstock, J. & Foster, J. A. (2011). Reduced anxiety‐like behavior and central neurochemical change in germ‐free mice. Neurogastroenterology & Motility 23, 255‐e119. [DOI] [PubMed] [Google Scholar]
- Obata, Y. & Pachnis, V. (2016). The effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology 151, 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocansey, D. K. W. , Zhang, L. , Wang, Y. , Yan, Y. , Qian, H. , Zhang, X. , Xu, W. & Mao, F. (2020). Exosome‐mediated effects and applications in inflammatory bowel disease. Biological Reviews 95, 1287–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura, R. & Takeda, K. (2017). Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Experimental & Molecular Medicine 49, e338–e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno, H. (2011). Regulation and function of immediate‐early genes in the brain: beyond neuronal activity markers. Neuroscience Research 69, 175–186. [DOI] [PubMed] [Google Scholar]
- Olsson, T. , Mohammed, A. H. , Donaldson, L. F. , Henriksson, B. G. & Seckl, J. R. (1994). Glucocorticoid receptor and NGFI‐A gene expression are induced in the hippocampus after environmental enrichment in adult rats. Molecular Brain Research 23, 349–353. [DOI] [PubMed] [Google Scholar]
- O'Mahony, S. M. , Clarke, G. , Borre, Y. E. , Dinan, T. G. & Cryan, J. F. (2015). Serotonin, tryptophan metabolism and the brain‐gut‐microbiome axis. Behavioural Brain Research 277, 32–48. [DOI] [PubMed] [Google Scholar]
- Parashar, A. & Udayabanu, M. (2017). Gut microbiota: implications in Parkinson's disease. Parkinsonism & Related Disorders 38, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A. , Harker, N. , Moreira‐Santos, L. , Ferreira, M. , Alden, K. , Timmis, J. , Foster, K. , Garefalaki, A. , Pachnis, P. , Andrews, P. , Enomoto, H. , Milbrandt, J. , Pachnis, V. , Coles, M. C. , Kioussis, D. , et al. (2012). Differential RET signaling pathways drive development of the enteric lymphoid and nervous systems. Science Signaling 5, ra55–ra55. [DOI] [PubMed] [Google Scholar]
- Perez‐Burgos, A. , Mao, Y. , Bienenstock, J. & Kunze, W. A. (2014). The gut‐brain axis rewired: adding a functional vagal nicotinic “sensory synapse”. The FASEB Journal 28, 3064–3074. [DOI] [PubMed] [Google Scholar]
- Perez‐Burgos, A. , Wang, L. , McVey Neufeld, K.‐A. , Mao, Y.‐K. , Ahmadzai, M. , Janssen, L. J. , Stanisz, A. M. , Bienenstock, J. & Kunze, W. A. (2015). The TRPV1 channel in rodents is a major target for antinociceptive effect of the probiotic Lactobacillus reuteri DSM 17938. The Journal of Physiology 593, 3943–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picconi, B. , Piccoli, G. & Calabresi, P. (2012). Synaptic dysfunction in Parkinson's disease. Advances in Experimental Medicine and Biology 970, 553–572. [DOI] [PubMed] [Google Scholar]
- Pignatelli, M. & Bonci, A. (2015). Role of dopamine neurons in reward and aversion: a synaptic plasticity perspective. Neuron 86, 1145–1157. [DOI] [PubMed] [Google Scholar]
- Prange, O. , Wong, T. P. , Gerrow, K. , Wang, Y. T. & El‐Husseini, A. (2004). A balance between excitatory and inhibitory synapses is controlled by PSD‐95 and neuroligin. Proceedings of the National Academy of Sciences of the United States of America 101, 13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenderville, J. A. , Kelly, Á. M. & Downer, E. J. (2015). The role of cannabinoids in adult neurogenesis. British Journal of Pharmacology 172, 3950–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribiag, H. & Stellwagen, D. (2014). Neuroimmune regulation of homeostatic synaptic plasticity. Neuropharmacology 78, 13–22. [DOI] [PubMed] [Google Scholar]
- Rao, M. & Gershon, M. D. (2016). The bowel and beyond: the enteric nervous system in neurological disorders. Nature Reviews Gastroenterology & Hepatology 13, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, V. R. & Finkbeiner, S. (2007). NMDA and AMPA receptors: old channels, new tricks. Trends in Neurosciences 30, 284–291. [DOI] [PubMed] [Google Scholar]
- Rea, K. , Dinan, T. G. & Cryan, J. F. (2016). The microbiome: a key regulator of stress and neuroinflammation. Neurobiology of Stress 4, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, K. V. , Sherwin, E. , Schellekens, H. , Stanton, C. , Dinan, T. G. & Cryan, J. F. (2017). Feeding the microbiota‐gut‐brain axis: diet, microbiome, and neuropsychiatry. Translational Research 179, 223–244. [DOI] [PubMed] [Google Scholar]
- Sarkar, A. , Harty, S. , Johnson, K. V.‐A. , Moeller, A. H. , Carmody, R. N. , Lehto, S. M. , Erdman, S. E. , Dunbar, R. I. M. & Burnet, P. W. J. (2020). The role of the microbiome in the neurobiology of social behaviour. Biological Reviews 95, 1131–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasselli, V. , Pachnis, V. & Burns, A. J. (2012). The enteric nervous system. Developmental Biology 366, 64–73. [DOI] [PubMed] [Google Scholar]
- Savitz, J. (2020). The kynurenine pathway: a finger in every pie. Molecular Psychiatry 25, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, K.‐H. , Van Ginneken, C. & Copray, S. (2009). Plasticity and neural stem cells in the enteric nervous system. The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 292, 1940–1952. [DOI] [PubMed] [Google Scholar]
- Schalla, M. A. & Stengel, A. (2020). Effects of microbiome changes on endocrine ghrelin signaling – a systematic review. Peptides 133, 170388. [DOI] [PubMed] [Google Scholar]
- Schirmer, M. , Smeekens, S. P. , Vlamakis, H. , Jaeger, M. , Oosting, M. , Franzosa, E. A. , ter Horst, R. , Jansen, T. , Jacobs, L. , Bonder, M. J. , Kurilshikov, A. , Fu, J. , Joosten, L. A. B. , Zhernakova, A. , Huttenhower, C. , et al. (2016). Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167, 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, D. , Boros, B. D. & Holtzman, D. M. (2019). The microbiome: a target for Alzheimer disease? Cell Research 29, 779–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgritta, M. , Dooling, S. W. , Buffington, S. A. , Momin, E. N. , Francis, M. B. , Britton, R. A. & Costa‐Mattioli, M. (2019). Mechanisms underlying microbial‐mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamriz, O. , Mizrahi, H. , Werbner, M. , Shoenfeld, Y. , Avni, O. & Koren, O. (2016). Microbiota at the crossroads of autoimmunity. Autoimmunity Reviews 15, 859–869. [DOI] [PubMed] [Google Scholar]
- Shouval, R. , Eshel, A. , Dubovski, B. , Kuperman, A. A. , Danylesko, I. , Fein, J. A. , Fried, S. , Geva, M. , Kouniavski, E. , Neuman, H. , Armon‐Omer, A. , Shahien, R. , Muller, E. , Noecker, C. , Borenstein, E. , et al. (2020). Patterns of salivary microbiota injury and oral mucositis in recipients of allogeneic hematopoietic stem cell transplantation. Blood Advances 4, 2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner, A. B. , Kao, J. Y. & Young, V. B. (2015). The gut microbiome in health and in disease. Current Opinion in Gastroenterology 31, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skott, E. , Yang, L. L. , Stiernborg, M. , Söderström, Å. , Rüegg, J. , Schalling, M. , Forsell, Y. , Giacobini, M. & Lavebratt, C. (2020). Effects of a synbiotic on symptoms, and daily functioning in attention deficit hyperactivity disorder – a double‐blind randomized controlled trial. Brain, Behavior, and Immunity 89, 9–19. [DOI] [PubMed] [Google Scholar]
- Solis, A. G. , Klapholz, M. , Zhao, J. & Levy, M. (2020). The bidirectional nature of microbiome‐epithelial cell interactions. Current Opinion in Microbiology 56, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiemsma, L. T. & Michels, K. B. (2018). The role of the microbiome in the developmental origins of health and disease. Pediatrics 141, e20172437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling, R. M. , Moloney, G. M. , Ryan, F. J. , Hoban, A. E. , Bastiaanssen, T. F. , Shanahan, F. , Clarke, G. , Claesson, M. J. , Dinan, T. G. & Cryan, J. F. (2018). Social interaction‐induced activation of RNA splicing in the amygdala of microbiome‐deficient mice. eLife 7, e33070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling, R. M. , Ryan, F. J. , Hoban, A. E. , Shanahan, F. , Clarke, G. , Claesson, M. J. , Dinan, T. G. & Cryan, J. F. (2015). Microbes and neurodevelopment – absence of microbiota during early life increases activity‐related transcriptional pathways in the amygdala. Brain, Behavior, and Immunity 50, 209–220. [DOI] [PubMed] [Google Scholar]
- Strandwitz, P. (2018). Neurotransmitter modulation by the gut microbiota. Brain Research 1693, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi, L. , Komaki, A. , Karamian, R. , Shahidi, S. , Sarihi, A. & Komaki, H. (2016). Interaction between paired‐pulse facilitation and long‐term potentiation during the stimulation of the cannabinoid and vanilloid systems in the dentate gyrus. Brain Research 1643, 27–34. [DOI] [PubMed] [Google Scholar]
- Taliaz, D. , Stall, N. , Dar, D. E. & Zangen, A. (2010). Knockdown of brain‐derived neurotrophic factor in specific brain sites precipitates behaviors associated with depression and reduces neurogenesis. Molecular Psychiatry 15, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, A. H. , Mahadeva, S. , Thalha, A. M. , Gibson, P. R. , Kiew, C. K. , Yeat, C. M. , Ng, S. W. , Ang, S. P. , Chow, S. K. , Tan, C. T. , Yong, H. S. , Marras, C. , Fox, S. H. & Lim, S.‐Y. (2014). Small intestinal bacterial overgrowth in Parkinson's disease. Parkinsonism & Related Disorders 20, 535–540. [DOI] [PubMed] [Google Scholar]
- Tognini, P. (2017). Gut microbiota: a potential regulator of neurodevelopment. Frontiers in Cellular Neuroscience 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, J. , Hu, J. , Seki, A. , Eisele, C. , Nair, N. , Huang, R. , Tarassishin, L. , Jharap, B. , Cote‐Daigneault, J. , Mao, Q. , Mogno, I. , Britton, G. J. , Uzzan, M. , Chen, C.‐L. , Kornbluth, A. , et al. (2020). Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ‐free mice. Gut 69, 42–51. [DOI] [PubMed] [Google Scholar]
- Uesaka, T. , Young, H. M. , Pachnis, V. & Enomoto, H. (2016). Development of the intrinsic and extrinsic innervation of the gut. Developmental Biology 417, 158–167. [DOI] [PubMed] [Google Scholar]
- van Thiel, I. A. M. , de Jonge, W. J. , Chiu, I. M. & van den Wijngaard, R. M. (2020). Microbiota‐neuroimmune cross talk in stress‐induced visceral hypersensitivity of the bowel. American Journal of Physiology‐Gastrointestinal and Liver Physiology 318, G1034–G1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra‐VanderWeele, J. , Muller, C. L. , Iwamoto, H. , Sauer, J. E. , Owens, W. A. , Shah, C. R. , Cohen, J. , Mannangatti, P. , Jessen, T. , Thompson, B. J. , Ye, R. , Kerr, T. M. , Carneiro, A. M. , Crawley, J. N. , Sanders‐Bush, E. , et al. (2012). Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proceedings of the National Academy of Sciences of the United States of America 109, 5469–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, R. , Harris, N. M. , Friedler, B. D. , Crapser, J. , Patel, A. R. , Venna, V. & McCullough, L. D. (2016). Reversal of the detrimental effects of post‐stroke social isolation by pair‐housing is mediated by activation of BDNF‐MAPK/ERK in aged mice. Scientific Reports 6, 25176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclawiková, B. & El Aidy, S. (2018). Role of microbiota and tryptophan metabolites in the remote effect of intestinal inflammation on brain and depression. Pharmaceuticals 11(3), 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff, W. R. , Anfora, A. T. , Liu, J. , Schultz, P. G. , Lesley, S. A. , Peters, E. C. & Siuzdak, G. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America 106, 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley, J. W. , Higgins, G. A. & Athey, B. D. (2016). Stress and glucocorticoid receptor transcriptional programming in time and space: implications for the brain‐gut axis. Neurogastroenterology & Motility 28, 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, J. D. (2001). Enteric nervous system, serotonin, and the irritable bowel syndrome. Current Opinion in Gastroenterology 17, 91–97. [DOI] [PubMed] [Google Scholar]
- Woolfrey, K. M. & Dell'Acqua, M. L. (2015). Coordination of protein phosphorylation and dephosphorylation in synaptic plasticity. Journal of Biological Chemistry 290, 28604–28612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Li, J.‐Y. , Lee, A. , Lu, Y.‐X. , Zhou, S.‐Y. & Owyang, C. (2020). Satiety induced by bile acids is mediated via vagal afferent pathways. JCI Insight 5(14), e132400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier, J. B. , Young, V. B. , Skufca, J. , Ginty, F. , Testerman, T. , Pearson, A. T. , Macklin, P. , Mitchell, A. , Shmulevich, I. , Xie, L. , Caporaso, J. G. , Crandall, K. A. , Simone, N. L. , Godoy‐Vitorino, F. , Griffin, T. J. , et al. (2020). The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends in Cancer 6, 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.‐Y. & Chen, C. (2015). Endocannabinoids in synaptic plasticity and neuroprotection. The Neuroscientist 21, 152–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, R. , Zhang, H. , Pan, J. , Du, Z. , Zhou, W. , Zhang, Z. , Tian, Z. , Zhou, R. & Bai, L. (2018). Peripheral dopamine controlled by gut microbes inhibits invariant natural killer T cell‐mediated hepatitis. Frontiers in Immunology 9, 2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, J. M. , Yu, K. , Donaldson, G. P. , Shastri, G. G. , Ann, P. , Ma, L. , Nagler, C. R. , Ismagilov, R. F. , Mazmanian, S. K. & Hsiao, E. Y. (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, B. B. & Mazmanian, S. K. (2017). The enteric network: interactions between the immune and nervous systems of the gut. Immunity 46, 910–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. & Chen, Z. (2011). The role of BDNF in depression on the basis of its location in the neural circuitry. Acta Pharmacologica Sinica 32, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate, C. A. & Manji, H. K. (2008). The role of AMPA receptor modulation in the treatment of neuropsychiatric diseases. Experimental Neurology 211, 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarco, M. , Vess, T. & Ginsburg, G. (2012). The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Diseases 18, 109–120. [DOI] [PubMed] [Google Scholar]
- Zhan, Y. , Paolicelli, R. C. , Sforazzini, F. , Weinhard, L. , Bolasco, G. , Pagani, F. , Vyssotski, A. L. , Bifone, A. , Gozzi, A. , Ragozzino, D. & Gross, C. T. (2014). Deficient neuron‐microglia signaling results in impaired functional brain connectivity and social behavior. Nature Neuroscience 17, 400–406. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Chen, B. , Zhao, L. & Li, H. (2020). The gut microbiota: emerging evidence in autoimmune diseases. Trends in Molecular Medicine 26, 862–873. [DOI] [PubMed] [Google Scholar]
- Zheng, D. , Liwinski, T. & Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Research 30, 492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilliox, L. A. , Chadrasekaran, K. , Kwan, J. Y. & Russell, J. W. (2016). Diabetes and cognitive impairment. Current Diabetes Reports 16, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari, S. I. , Rabbani Khorasgani, M. & Noorbakhshnia, M. (2020). The effects of lactobacilli (L. rhamnosus, L. reuteri, L. plantarum) on LPS‐induced memory impairment and changes in CaMKII‐α and TNF‐α genes expression in the hippocampus of rat. Physiology & Behavior 229, 113224. [DOI] [PubMed] [Google Scholar]
- Zweigner, J. , Schumann, R. R. & Weber, J. R. (2006). The role of lipopolysaccharide‐binding protein in modulating the innate immune response. Microbes and Infection 8, 946–952. [DOI] [PubMed] [Google Scholar]


