Abstract
The ability of RNA to catalyze RNA ligation is critical to its central role in many prebiotic model scenarios, in particular the copying of information during self‐replication. Prebiotically plausible ribozymes formed from short oligonucleotides can catalyze reversible RNA cleavage and ligation reactions, but harsh conditions or unusual scenarios are often required to promote folding and drive the reaction equilibrium towards ligation. Here, we demonstrate that ribozyme activity is greatly enhanced by charge‐mediated phase separation with poly‐L‐lysine, which shifts the reaction equilibrium from cleavage in solution to ligation in peptide‐RNA coaggregates and coacervates. This compartmentalization enables robust isothermal RNA assembly over a broad range of conditions, which can be leveraged to assemble long and complex RNAs from short fragments under mild conditions in the absence of exogenous activation chemistry, bridging the gap between pools of short oligomers and functional RNAs.
Keywords: catalysis, coacervates, ligases, peptides, ribozymes
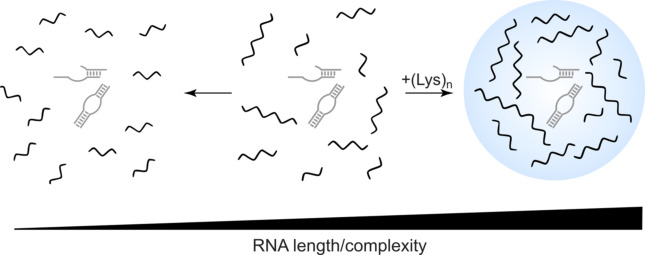
In solution, a prebiotically plausible ribozyme formed from short oligonucleotides reversibly cleaves an RNA substrate. However, in the presence of poly‐L‐lysine, phase separation occurs owing to charge interactions between the oppositely charged RNA and peptide. The resulting compartments greatly enhance ribozyme activity, driving the reaction equilibrium towards the ligation of long and complex RNA in mild conditions.
Introduction
The generation of RNA or other nucleic acid strands with sufficient length and sequence diversity to fold into functional catalysts and replicators is a key step in many models of early biology. Plausible pathways exist for the generation of activated ribonucleotides[ 1 , 2 , 3 ] and the formation of short oligomers by non‐enzymatic polymerization,[ 4 , 5 ] but the untemplated polymerization of long RNA oligomers containing all four nucleobases remains inefficient. [6] The formation of long and complex RNAs from pools of oligonucleotides can potentially bridge this gap, [7] as demonstrated by the assembly of a >200 nt RNA polymerase ribozyme from short oligomers by a fragmented version of the small nucleolytic hairpin ribozyme (HPz). [8]
HPz is a small self‐cleaving RNA motif that catalyzes the cleavage and ligation of specific RNA sequences.[ 9 , 10 ] The ability of HPz to fold into active conformations[ 11 , 12 ] is sensitive to environmental conditions such as ionic strength[ 13 , 14 ] and temperature. [15] In solution, the reaction equilibrium is shifted towards cleavage, whilst in dehydrating conditions (e.g. eutectic phases in ice, ethanol and drying) efficient ligation can be observed.[ 16 , 17 , 18 ] The ability of HPz to assemble from short oligonucleotides and to process a wide range of RNA junctions makes it an attractive model system for an early RNA catalyst.[ 8 , 16 ] The reversibility of the HPz‐catalyzed reaction allows for the possibility of recombination (Figure S1): First, RNA cleavage produces a strained 2′,3′‐cyclic phosphate functionalized substrate. Next, strand exchange then ligation can occur with other strands of a compatible sequence. Although relieving strain in the 2′,3′‐cyclic phosphate provides a limited enthalpic driving force for ligation, the associated entropic penalty results in cleavage being thermodynamically favored in solution for some systems. [19] These reactions are prebiotically appealing as they can occur independently from an exogenous RNA‐activation chemistry and have been shown to increase structural and informational diversity in pools of RNA oligomers. [20] Researchers have exploited recombination and ligation by the HPz ribozyme to assemble complex functional RNAs from short fragments,[ 8 , 21 ] to combine RNA enzymes and aptamers into larger, more complex aptazyme systems, [22] and to produce simple self‐replicators. [23] In combination with a polymerization chemistry capable of generating short oligomers, the realization of replicating and evolving systems may be possible. Ribozyme‐catalyzed recombination can require high magnesium concentrations to occur in solution, with minimal or fragmented HPz variants reliant on dehydration or freezing to drive ligation.[ 16 , 18 ] These requirements render recombination‐based RNA assembly incompatible with compartmentalization in some protocellular systems and narrow the range of viable environmental conditions. For example, high Mg2+ concentrations have a destabilizing effect on vesicles formed from single chain amphiphiles, [24] while phospholipid‐based vesicles are prone to fragmentation[ 25 , 26 ] and content loss [27] when exposed to freezing conditions.
Phase separation by oppositely charged polymers such as peptides and nucleic acids is a ubiquitous process that is both widely observed in modern biology[ 28 , 29 , 30 ] and exploited in synthetic biology.[ 31 , 32 ] Charged peptides are appealing components in origin of life scenarios, potentially supporting the function of early catalytic nucleic acids[ 33 , 34 ] and forming protocellular compartments such as coacervate droplets by phase separation. [35] Although liquid coacervate droplets are the most widely studied system within the context of the origin of life, other phases such as coaggregates[ 36 , 37 ] and gels [38] are also of interest. Spatial confinement, as well as the ability to localize reactions and concentrate key components, would have been valuable in early biology and is a prerequisite for open‐ended evolution. [39] The ability of coacervate phases to strongly concentrate divalent metal ions and RNA suggests a highly favorable environment for nucleic acid catalysis, [40] and interactions with polycationic peptides have been shown to enhance ribozyme activity in some cases.[ 41 , 42 ]
Despite this, catalysis by ribozymes has only recently been reconciled with coacervation,[ 43 , 44 ] and specific conditions are required to prevent loss of function due to misfolding. Previously reported enhancements in catalytic activity by condensed phases typically rely on the concentration and rescue of highly dilute systems below the ribozyme / substrate dissociation constant (K D), [44] and so far only ribozyme cleavage reactions have been demonstrated. [44] In order to realize complex, functional phase‐separated systems that may eventually be capable of open‐ended evolution, constructive processes such as the polymerization, ligation or recombination of nucleic acids must be supported.
In this work, we utilize poly‐L‐lysine ((Lys) n ) and a split HPz ribozyme as a model for interactions between early charged protopeptides and nucleic acid catalysts. Although cationic amino acids are crucial factors in protein‐nucleic acid interactions, and thus are essential components in any prebiotic scenario that leads to the emergence of protein synthesis, [45] basic cationic amino acids such as arginine and lysine only rarely emerge from model prebiotic syntheses.[ 46 , 47 , 48 , 49 ] However, ligation of these amino acids into peptides has been demonstrated in prebiotic conditions, [50] assuming sufficient feedstocks. The synthesis of non‐proteinaceous analogues such as ornithine and 2,4‐diamino‐butyric acid, potential precursors to extant cationic amino acids, [51] is more plausible.[ 51 , 52 , 53 ] Peptides composed of these analogues can act as functional substitutes to poly‐L‐lysine and have provided similar enhancements in ribozyme activity. [42] Depsipeptides, protopeptides which contain a mixture of ester and amide linkages, can be formed by dry‐wet cycles, [54] which selectively incorporate proteinaceous amino acids over non‐proteinaceous precursors, thus providing a potential mechanism for the enrichment of poorly abundant cationic amino acids in early polypeptides. [55] Unlike the model system used here, peptides or protopeptides emerging from prebiotic processes are likely not to have been homopeptides, or enantiomerically pure.
We show that charge‐based phase separation between (Lys) n and the HPz ribozyme RNA both enhances and modulates ribozyme activity, shifting the HPz reaction equilibrium from cleavage to ligation at ambient temperatures. This enables the production of both long and complex functional RNA from short fragments under isothermal conditions. Unlike under solution conditions, poly‐L‐lysine renders the HPz ribozyme functional over a broad range of temperatures and magnesium concentrations, even with varying lengths, charge ratios, and phase behaviors. These findings reconcile HPz‐catalyzed RNA recombination with a far broader range of environmental conditions, and furthermore suggest the ability of simple peptides or other more diverse polycations to support and enhance nucleic acid catalysis in heterogeneous prebiotic scenarios. [56]
Results
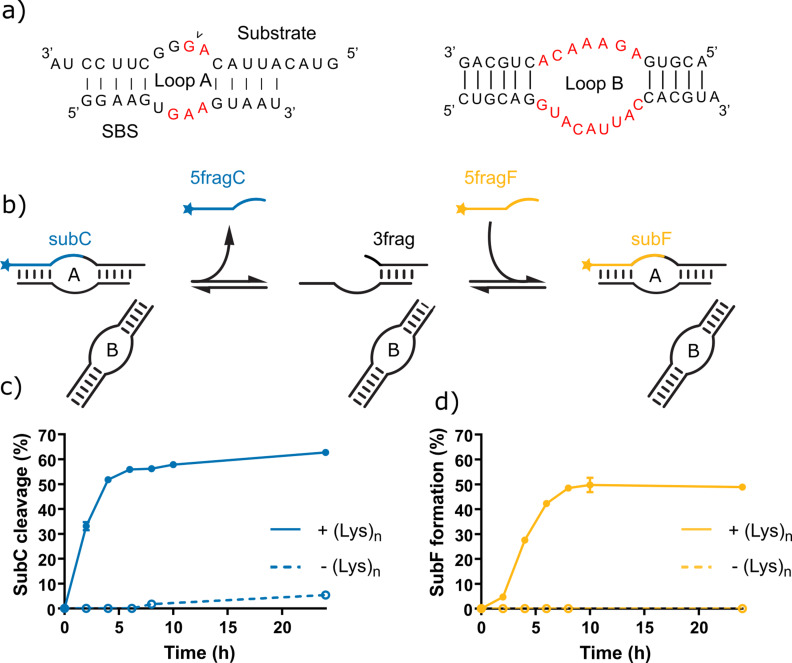
To investigate the recombination activity of the HPz ribozyme, we developed an assay that allows the independent characterization of both cleavage and ligation, based on a fragmented ribozyme system (Figure 1 a). [16] The ribozyme system comprises four strands forming two catalytic loops: the HPz loop B (two strands, 18 and 21 nt) and loop A, formed from a 14 nt substrate binding strand (SBS) and 3′‐Cy5 tagged 20 nt substrate (subC). Initially the ribozyme may cleave subC into two 10 nt products, one possessing the 3′‐Cy5 tag (5fragC), the other activated with a 2′,3′‐cyclic phosphate and capable of ligation (3frag). The reaction mixture also contains a 10 nt fragment identical in sequence to the 5fragC produced in the previous step but bearing a 3′‐FAM tag (5fragF). This fragment can compete for the substrate binding strand and forms a new FAM‐tagged 20 nt strand (subF) when ligated to the activated 3frag. The mechanism of recombination is described in Figure S1, and an example gel showing the activity of this system under various conditions is shown in Figure S2. In solution (Tris⋅HCl pH 8, 1 mM MgCl2, 30 °C), the cleavage of subC was slow, yielding only 5 % of 5fragC after 24 h (Figure 1 b, dashed line). No ligation of 5fragF to form subF was observed under these conditions (Figure 1 c, dashed line).
Figure 1.
The design and function of a simple split hairpin ribozyme recombination assay. a) The structure of the split HPz ribozyme, which is composed of loop A (substrate and substrate binding strand, or SBS) and loop B. The cleavage site is indicated with an arrow, and conserved bases are shown in red. b) Simplified diagram showing hairpin ribozyme‐catalyzed cleavage of a Cy5‐tagged 20 nt substrate (subC) (blue), strand exchange, and re‐ligation of a FAM‐tagged 10 nt fragment (5fragF) (yellow). c) Cleavage of the subC and d) ligation of the 5fragF over time by the HPz ribozyme with (Lys)19–72 (0.67:1 (Lys)19–72:RNA, 1 mM MgCl2, Tris⋅HCl pH 8, 30 °C) (solid line) and in solution (1 mM MgCl2, Tris⋅HCl pH 8, 30 °C) (dashed line).
We then investigated the effect of poly‐L‐lysine on the activity of the HPz system. Turbidity was observed after the addition of (Lys)19–72 (charge ratio=0.67:1 (Lys)19–72:RNA), as well as a drastic shift in ribozyme activity. Cleavage of subC proceeded rapidly compared to solution conditions, with approximately 60 % cleavage over 24 h (k cl=0.42±0.02 h−1). Strikingly, ligation of 5fragF, forming subF, was observed after just 2 h of reaction, with a final yield at 24 h of approximately 50 %. Whilst it is not possible to directly determine the rate of ligation in the recombination system due to the initial cleavage step followed by a presumably rate‐limiting strand exchange reaction, the ligation of a pre‐activated 3frag intermediate to 5fragF proceeded rapidly under these conditions, with an apparent ligation rate of k lig=0.076±0.004 min−1 (Figure S3).
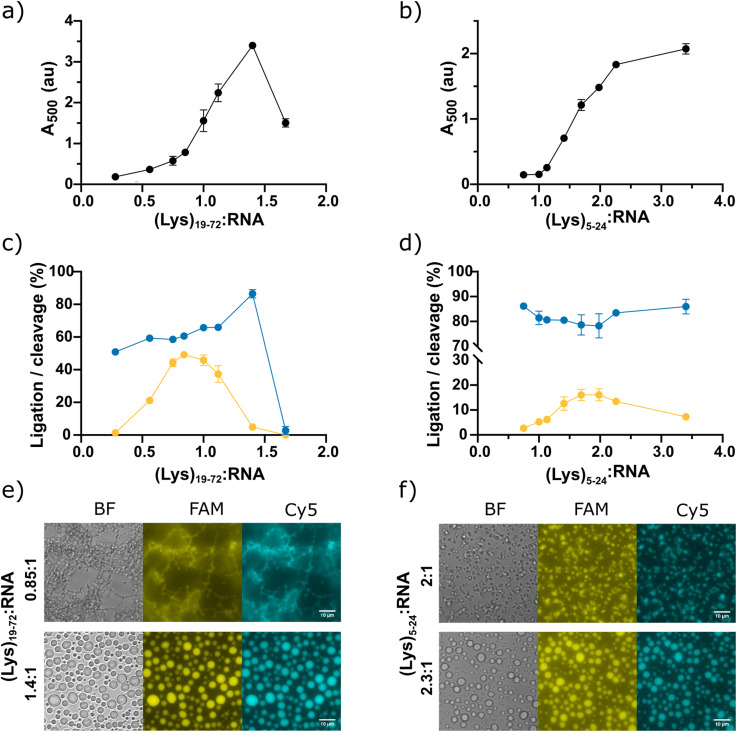
We then investigated the effect of polycation length and concentration on HPz ribozyme activity (Figure 2, Figure S4). Increasing concentrations of either (Lys)19–72 (Figure 2 a) or (Lys)5–24 (Figure 2 b) were titrated into a fixed concentration of RNA, and the degree of phase separation at the various (Lys) n :RNA ratios was measured by absorbance at 500 nm (Figure 2 a, b). The critical peptide concentrations for phase separation (CPs) were determined by the onset of turbidity. Phase separation occurred at a sub‐stoichiometric concentration of the longer peptide (CP19–72 ca. 0.7:1 (Lys)19–72:RNA), but not until a concentration ratio of near unity for the shorter peptide (CP5–24 ca. 1:1 (Lys)5–24:RNA). Activity measurements at t=24 h showed that (Lys)19–72 enhances both HPz cleavage and ligation at sub‐stochiometric ratios, but that both of these activities are suppressed at high ratios (Figure 2 c). The addition of (Lys)5–24 (Figure 2 d) led to high HPz cleavage activity and relatively weaker ligation compared to the longer (Lys)19–72 at all ratios tested, although the enhancement over solution conditions remained considerable. Optimal recombination of 5fragF to form subF was observed at charge ratios of 0.85:1 (Lys)19–72:RNA, and 2:1 (Lys)5–24:RNA. This enhancement of activity was found to be independent of the presence of the cationic Cy5 fluorophore on 5fragC (Figure S5). To determine the distribution of (Lys) n oligomers in our samples, we performed electrospray ionization mass spectrometry (ESI‐MS) (Figure S6). Surprisingly, the distributions were heavily biased towards shorter oligomers in both cases: for (Lys)5–24, only oligomers with n=3 to n=9 were detected, whilst for (Lys)19–72, only oligomers between n=16 to n=46 were observed (Figure S7). Whilst oligomers with lengths up to the specified upper limit are likely to be present in the samples, based on the manufacturer's characterization, these species appear to occur at such low concentrations that they fall below the detection limit of ESI‐MS.
Figure 2.
Influence of (Lys) n length and (Lys) n :RNA mixing ratio on phase separation and HPz activity. a) Variation in absorbance at 500 nm with varying ratios of (Lys)19–72:RNA. The minimal ratio for the onset of phase separation CP19‐72 ca. 0.7:1 (Lys)19–72:RNA. b) Variation in absorbance at 500 nm with varying ratios of (Lys)5–24. The minimal ratio for the onset of phase separation CP5‐24 ca. 1:1 (Lys)5–24:RNA. c) Endpoint cleavage and ligation activity of the HPz ribozyme with varying (Lys)19–72:RNA. d) Endpoint cleavage and ligation activity of the HPz ribozyme with varying (Lys)5–24:RNA. Endpoint ligation of 5fragF is shown in yellow, endpoint cleavage of subC is shown in blue. e) Example fluorescence microscopy images of (Lys)19–72:RNA condensates before (0.85:1 (Lys)19–72:RNA) and after (1.4:1 (Lys)19–72:RNA) the transition to liquid droplets. f) Example fluorescence microscopy images of (Lys)5–24:RNA condensates before (2:1 (Lys)5–24:RNA) and after (2.3:1 (Lys)5–24:RNA) the transition to liquid droplets. Brightfield imaging is shown in grey, FAM fluorescence in yellow and Cy5 fluorescence in cyan. Scale bars=10 μm. Full imaging of all datapoints is shown in Figures S8 and S9.
Brightfield and fluorescence microscopy of samples at each tested (Lys) n :RNA ratio revealed the formation of various separated phases with increasing charge ratio (Figures 2 e,f, S8 and S9). Increasing ratios of both (Lys)5–24 and (Lys)19–72:RNA led to the formation of particles that accumulated without coalescence into liquid droplets, and in which both labelled RNAs were strongly colocalized. This implies strong partitioning of the labelled substrate RNA into the condensed phase, which is likely to be responsible for the shift to ligation. At (Lys)19–72:RNA ratios near unity (0.85‐1:1), a web‐like structure was observed (Figures 2 e, Figure S9). For both poly‐L‐lysine lengths, a transition to liquid coacervate droplets was observed at excess charge ratios ((Lys)5–24:RNA ≥2.3:1, (Lys)19–72:RNA ≥1.1:1) (Figure 2 e,f). The phase separated particles initially formed as an unstable dispersion, eventually settling onto the bottom of the slide without wetting the passivated surface. Ligation activity was optimal at ratios immediately preceding the transition to coacervate droplets for both (Lys) n lengths ((Lys)5–24:RNA=2:1, (Lys)19–72:RNA=0.85:1). Although the formation of liquid droplets was associated with a reduction in activity, robust ligation activity was still observed in this phase at lower ratios (37.3 % yield at (Lys)19–72:RNA=1.1:1, 13.4 % yield at (Lys)5–24:RNA=2.3:1).
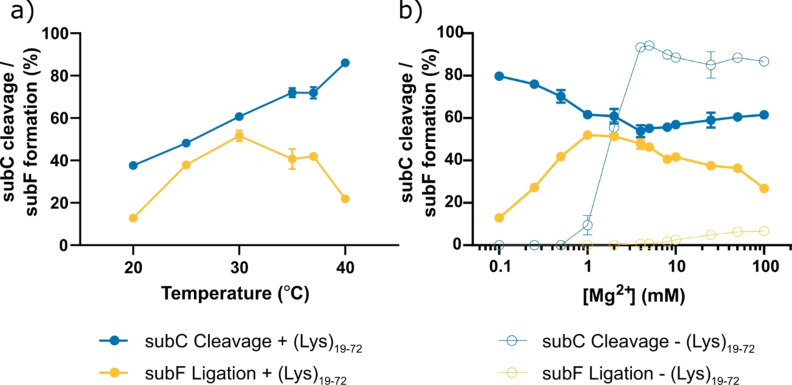
We sought to further characterize the system by studying endpoint recombination activity at the optimal (Lys)19–72:RNA ratio for ligation (0.75:1) at various temperatures and magnesium concentrations. The endpoint activities at various temperatures are shown in Figure 3 a. Optimal recombination yields were obtained at 30 °C (1 mM Mg2+, Tris⋅HCl pH 8, 52 % ligation after 24 h). Similar yields were observed at lower temperatures, although the time required to reach equilibrium increased (t 25 °C=96 h, t 20 °C=144 h). Increasing temperature shifted the reaction equilibrium towards cleavage, as expected due to the entropic cost of ligation inherent in the system. [57] Recombination activity was also supported across a broad range of Mg2+ concentrations (0.1–100 mM) in the presence of (Lys)19–72 (0.75:1), notably at concentrations as low as 0.1 mM Mg2+ (80 % subC cleavage, 13 % 5fragF ligation) (Figure 3 b). Optimal activity was observed between 1–5 mM Mg2+. Reduced 5fragF ligation yields above this point may be in part due to magnesium‐catalyzed hydrolysis of the 2′,3′‐cyclic phosphate, which is expected to slowly deplete the amount of activated 3frag in the system over time.
Figure 3.
Influence of temperature, magnesium concentration, and poly‐L‐lysine length and mixing ratio on recombination yield. a) The effect of reaction temperature on the endpoint recombination yield (0.75:1 (Lys)19–72:RNA, 1 mM MgCl2, Tris⋅HCl pH 8). The reaction was quenched at either 144 h (20 °C), 96 h (25 °C) or 24 h (30–40 °C). b) The effect of magnesium concentration on the HPz ribozyme recombination yield with (Lys) n (0.75:1 (Lys)19–72:RNA, 1 mM MgCl2, Tris⋅HCl pH 8, t=24 h) (solid circles) and in solution (Tris⋅HCl pH 8, 30 °C, t=24 h) (hollow circles). For both panels, cleavage of subC is shown in blue, whilst ligation of the 5fragF is shown in yellow.
To separate the effects of magnesium concentration [40] from other possible causes of recombination enhancement, we also measured activity under identical conditions but in the absence of peptide. Here, the reaction equilibrium was strongly shifted towards cleavage. No cleavage of subC was observed until 1 mM Mg2+, at which point the yield of 5fragC increased, reaching a maximum at 5 mM Mg2+ (94 % cleavage). The change in cleavage activity with magnesium concentration is sigmoidal and reminiscent of a ribozyme folding curve. Indeed, the measured midpoint of the cleavage data, 1.8 mM Mg2+, is similar to previously reported Mg2+ induced folding midpoints for the HPz ribozyme,[ 58 , 59 ] although differences in structure and fragmentation make a direct comparison difficult. In the absence of peptide, no ligation of 5fragF to form subF was observed until 4 mM Mg2+ (0.7 % yield), reaching a maximum yield of 7 % at 100 mM Mg2+ with a concomitant reduction in the measured proportion of 5fragC. This suggests that concentration of magnesium ions within the condensed phases does not alone account for the enhanced recombination activity observed in the presence of (Lys) n in this study.
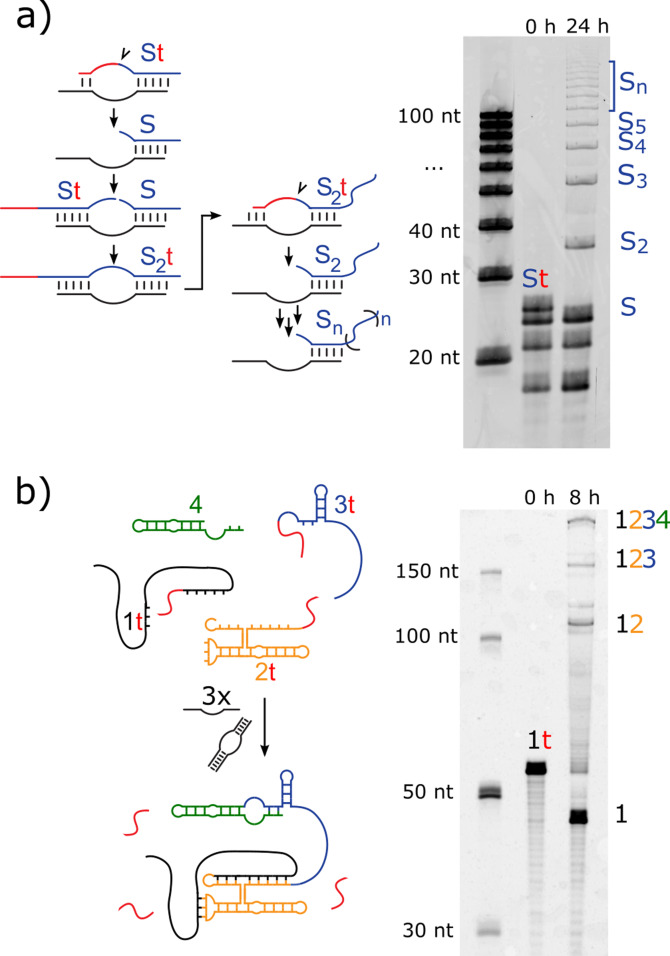
Having determined optimal conditions for ligation activity (Tris⋅HCl pH 8, 30 °C, 0.75:1 (Lys)19–72:RNA), we exploited the enhancing effect of (Lys)19–72 on the HPz ribozyme to form long RNA chains and complex structures from short fragments. The following ribozyme systems contain the same in trans loop B domain used in the previously described recombination assay (Figure 1 a), but combined with a range of substrate binding strands (SBSs) and substrates. First, we tested a recombination‐based RNA ladder system (Figure 4 a): the fragmented HPz ribozyme cleaves a short 3’‐tail from a 22 nt fragment (St), leaving a 16 nt 2′,3′‐cyclic phosphate functionalized fragment (S), which can then be concatenated. After 24 h incubation (Tris⋅HCl pH 8, 30 °C, 0.7:1 (Lys)19–72:RNA), the substrate tail was completely cleaved, and a ladder of concatenated products (Sn) was observed (longest product observed: n=15, 240 nt). The lack of uncleaved substrate is expected, as the cleaved 3′‐tail shares only three complementary bases with the substrate binding strand, and so should easily be displaced by the 5′ end of another substrate, making re‐ligation unfavorable. In addition, the 6 nt fragment may only be poorly concentrated in the condensed phase due to its short length. [43]
Figure 4.
Formation of long RNA chains and complex RNA by HPz recombination under optimized conditions. Example urea‐PAGE gels showing a) Concatenation of long RNA chains (>200 nt) by the HPz ribozyme with poly‐L‐lysine (0.75:1 (Lys)19–72:RNA, 1 mM MgCl2, Tris⋅HCl pH 8, t=24 h). The fragmented HPz ribozyme first binds the 22 nt substrate fragment (St), forming an A loop between substrate and binding strand. This docks with a B loop, then a 6 nt tail (t, shown in red) is cleaved from the substrate leaving a 2′,3′‐cyclic phosphate on the 16 nt reaction product (S). The cleavage site is indicated with an arrow. Another substrate (St or S) then displaces the cleaved tail, which has only three bases complementary to the substrate binding arm. In poly‐L‐lysine coacervates, or other concentrating conditions, the HPz ribozyme can then ligate the two fragments, forming a concatenated (S2) product. This process repeats, resulting in long chains of up to n>13 substrate fragments. The urea‐PAGE gel was imaged using SYBER Gold staining. b) Assembly of the RPR4 ribozyme (198 nt) from four fragments by HPz with poly‐L‐lysine (0.8:1 (Lys)19–72:RNA, 8 mM MgCl2, Tris⋅HCl pH 8, t=8 h, full length product yield=8 %). In this reaction, three different substrate binding strands bind pairs of oligomers from a set of four substrate strands. A short tail (t) is cleaved from fragments 1, 2 and 3, leaving 2′,3′‐cyclic phosphate functionalized fragments that be ligated together with fragment 4 to form the full‐length product. The assembly products are visualized using a fluorescent 5′‐FAM tag on fragment 1.
RNA polymerase ribozymes (RPRs), which catalyze the templated synthesis of RNA from nucleotide triphosphates, are considered analogues of an early RNA‐only replicator. [60] Although these ribozymes are capable of synthesizing long strands, their activity is not sufficient to quantitatively synthesize sequences with complex secondary structure beyond ca. 50 nt,[ 61 , 62 ] and as such self‐replication has not been demonstrated. These limitations may be overcome by stepwise modular assembly, in which the large and complex functional sequence is constructed from shorter fragments. As a proof of concept, we assembled the RPR4 ribozyme from short fragments in the presence of long poly‐L‐lysine (Figure 4 b), as previously demonstrated in‐ice. [8] Indeed, full length RPR4 was produced with a final yield of 7.8 % after 8 h (Tris⋅HCl pH 8, 30 °C, 0.8:1 (Lys)19–72 :RNA). The activity of the assembled product was not tested in this study. These yields are comparable to those observed in long‐term Mg2+‐free reactions driven by repeated 12 h freeze‐thaw cycles (10 % after 24 days). The similarity in the product yields observed in‐ice and in the condensed phase is both unexpected and notable: the absence of Mg2+ in previously reported freeze‐thaw cycle experiments allows the system to be driven towards the ligated state, as no cleavage reaction can occur during the thawing phase. In this study, the presence of Mg2+ means that the cleavage reaction is always active but is presumably counteracted by high RNA concentrations.
Discussion
In this study, we have shown that model ribozyme‐peptide interactions can drastically enhance the activity of a small nucleolytic ribozyme over a wide range of conditions. In many cases, phase separation with poly‐L‐lysine yielded activity in conditions where the ribozyme is otherwise completely inactive (<1 mM Mg2+ for cleavage and <4 mM Mg2+ for ligation). In conditions where the ribozyme was active in solution, the addition of (Lys)19–72 led to enhancements of up to 11‐fold for 5FragF ligation (4 mM Mg2+) and 65‐fold for subC cleavage (1 mM Mg2+). Importantly, phase separation shifted the equilibrium of the reversible transesterification reaction towards RNA ligation rather than cleavage, likely due to increased RNA concentration in the condensates. Both effects combined allow efficient and robust assembly of long RNA molecules under mild conditions in the absence of an exogenous activation chemistry. To the best of our knowledge, such a strong and rugged enhancement of ribozyme activity or shift in behavior by phase separation has not been previously reported.
Efficient recombination‐based RNA assembly by fragmented HPz systems is typically only observed in dehydrating environments, for example in eutectic ice, [16] alcoholic solutions, [63] or drying. [18] The shift from cleavage to ligation induced by (Lys) n suggests that interactions with cationic peptides could have greatly broadened the scope of recombination‐based RNA assembly on prebiotic Earth, perhaps providing comparable ligation yields to dehydrating environments with enhanced kinetics. Indeed, the rate of direct ligation with (Lys)19–72 was more than ten times faster than the rate of freezing‐induced ligation by a fragmented HPz ribozyme similar to that used here (k lig=0.006 min−1, T=−10 °C). [16] Poly‐L‐lysine‐supported HPz ribozyme catalysis is robust, with similar ligation yields observed across a range of temperatures below 35 °C. Similarly, the system is able to support ligation across a broad range of magnesium concentrations, even at levels far below that typically required for catalysis under solution conditions. This is especially relevant for fragmented ribozymes, which are prebiotically appealing due to their reduced complexity, but which require higher magnesium concentrations ([Mg2+]1/2=3 mM) for folding and activity than more complex species with additional stabilizing loops ([Mg2+]1/2=20–40 μM). [59]
In the present study, enhancements in ribozyme activity beyond typical solution behavior were observed across a range of charge ratios, with the greatest enhancements occurring before the formation of liquid droplets. Fluorescence microscopy showed strong concentration and colocalization of both tagged RNAs within all separated phases (Figures S8 and S9). This likely accounts for the observed enhancements in activity, analogous to hairpin ribozyme ligation in freezing and drying environments. [18] Assays performed in the absence of peptide suggest that increased Mg2+ concentrations, whilst probable in this system, [40] are alone insufficient to drive RNA ligation (Figure 3). Beyond simple concentration of reactants, increased hybridization, folding and thermal stability as a result of charge interactions with the peptide may contribute to the observed enhancements of activity. Interactions between lysine‐containing peptides and nucleic acids in amyloid gel fibers have previously been shown to promote the hybridization of nucleic acids below their K D, [37] and lysine containing copolymers have been reported to enhance the rate of DNA hybridization by over 200‐fold. [64] Even in the absence of phase separation, RNA polymerase ribozyme (RPR) holoenzyme assembly and activity is enhanced by interactions with simple lysine containing peptides and non‐proteinaceous analogues, [42] spermidine has been shown to stabilize the HPz ribozyme‐substrate complex and enhance substrate cleavage, [41] and cationic proto‐peptides have been shown to increase the thermal stability of folded RNA. [65] Such enhancements in hybridization as a result of charge interactions extend beyond peptides, and are a key aspect of the proposed role of clay surfaces in early molecular evolution. [66]
It is perhaps surprising that direct condensation of (Lys) n with catalytic RNA can support (let alone enhance) activity, given that ribozyme activity can be inhibited by strong polycation‐RNA interactions, which may induce misfolding.[ 45 , 67 ] It is also of interest that the transition to liquid coacervate droplets at high (Lys) n :RNA ratios was associated with a reduction in recombination activity. Similar phase transitions have been previously reported: poly‐L‐lysine has been shown to form precipitates when directly mixed with double‐stranded RNA, but liquid droplets with single stranded RNA. [68] We speculate that observed phase transition in the (Lys) n :HPz system may originate from reduced hybridization at excess charge ratios, which could also lead to a reduction in activity.
Previous studies have demonstrated that ribozyme‐catalyzed RNA cleavage is indeed supported when ribozymes are hosted within coacervate droplets.[ 43 , 44 ] Below the apparent ribozyme / substrate dissociation constant (K D), concentration by coacervation can rescue hammerhead and hairpin ribozyme activity, providing relative enhancements over dilute reactions in aqueous solution. [44] However, above the K D, hammerhead ribozyme activity in coacervate droplets is inhibited. [43] For RNA in coacervate droplets, this effect is most prominent in charge neutral conditions, or when the polycation is in excess. The activity of the hammerhead ribozyme when hosted in oligoarginine (R10)/ oligoaspartic acid (D10) coacervate droplets is inhibited at a 1:1 charge ratio of R10:D10. [44] However, in coacervate compositions with excess negative charge, activity can be enhanced over concentration‐limited solution conditions. [69] The suppression of activity at excess (Lys) n :RNA ratios reported here may be analogous, with strong lysine‐RNA interactions leading to misfolding when the peptide is in charge excess. Even so, the inhibition of activity in the coacervate phase is not total: strong recombination activity is observed in droplets at charge ratios near the transition point, suggesting that at least a portion of the RNA is able to remain folded and active. This explanation also accounts for the relative differences in activity suppression by (Lys)5–24 and (Lys)19–72 at excess peptide:RNA ratios, as longer polyions have a greater tendency to interact with their oppositely charged partners, and therefore may lead to greater misfolding and suppression of activity when in excess.[ 70 , 71 , 72 ] Peptide length dependent helicase‐like activity has been demonstrated for lysine/aspartic acid coacervates, in which droplets formed from shorter oligopeptides permitted hybridization of a short RNA duplex to a greater degree than droplets comprised of longer polyions. [73] Such effects have also been reported in extant biological systems, in particular Ddx4 protein coacervates, which melt double‐stranded DNA and stabilize single stranded species. [74] The suppression of activity with excess peptide may be overcome in the future by the use of heteropeptides or other homopeptides with varying charge density.
Conclusion
The observation that direct phase separation between short model peptides and ribozymes provides robust enhancements to catalytic activity under a broad range of conditions strengthens the case for both an early coevolution of RNA and peptides, and the argument that long and functional RNA emerged from pools of short oligomers via the action of small catalytic motifs. It is noteworthy that recombination is supported in this environment, as this implies that the key processes of strand release and reannealing also take place in addition to simply cleavage and ligation. Even the short poly‐L‐lysine used in this study, which is predominantly composed of oligomers of less than 9 residues, was able to phase separate and modulate the catalytic activity of the ribozyme. This suggests that short cationic protopeptides formed, for example, by dry‐wet cycling may also be capable of such enhancements. Further investigation into the interactions between heterogeneous proto‐peptides and catalytic nucleic acids is therefore of great interest. The greater enhancements provided by the longer model peptide perhaps emerged later, with the selection or synthesis of longer polycationic species.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
We wish to thank Martin Spitaler, Markus Oster, Giovanni Cardone, and the MPIB Imaging Core Facility for providing excellent guidance throughout the project, as well as subsidized equipment access. We would also like to thank Maria Victoria Sanchez Caballero, Barbara Agnes Steigenberger, Evelyn Stieger and the MPIB Mass Spectrometry Core Facility for their valuable assistance in characterizing the peptides used in this work. H.M. and E.Y.S. were supported by funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Project‐ID 364653263‐TRR 235). H.M. gratefully acknowledges support by the European Research Council (ERC) under the Horizon 2020 research and innovation programme (grant agreement ID: 802000, RiboLife). K.L.V., B.G., T.Y.D.T. and H.M. were supported by the Volkswagen Foundation with funding from the initiative “Life?—A Fresh Scientific Approach to the Basic Principles of Life” (grant number: 92772). Open access funding enabled and organized by Projekt DEAL. Open Access funding enabled and organized by Projekt DEAL.
K. Le Vay, E. Y. Song, B. Ghosh, T.-Y. D. Tang, H. Mutschler, Angew. Chem. Int. Ed. 2021, 60, 26096.
Contributor Information
Dr. Kristian Le Vay, Email: levay@biochem.mpg.de.
Dr. T.‐Y. Dora Tang, Email: tang@mpi-cbg.de.
Prof. Dr. Hannes Mutschler, Email: hannes.mutschler@tu-dortmund.de.
References
- 1. Becker S., Feldmann J., Wiedemann S., Okamura H., Schneider C., Iwan K., Crisp A., Rossa M., Amatov T., Carell T., Science 2019, 366, 76–82. [DOI] [PubMed] [Google Scholar]
- 2. Yadav M., Kumar R., Krishnamurthy R., Chem. Rev. 2020, 120, 4766–4805. [DOI] [PubMed] [Google Scholar]
- 3. Fialho D. M., Roche T. P., Hud N. V., Chem. Rev. 2020, 120, 4806–4830. [DOI] [PubMed] [Google Scholar]
- 4. Huang W., Ferris J. P., J. Am. Chem. Soc. 2006, 128, 8914–8919. [DOI] [PubMed] [Google Scholar]
- 5. Ferris J. P., Hill A. R., Liu R., Orgel L. E., Nature 1996, 381, 59–61. [DOI] [PubMed] [Google Scholar]
- 6. Monnard P. A., Kanavarioti A., Deamer D. W., J. Am. Chem. Soc. 2003, 125, 13734–13740. [DOI] [PubMed] [Google Scholar]
- 7. Briones C., Stich M., Manrubia S. C., RNA 2009, 15, 743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mutschler H., Wochner A., Holliger P., Nat. Chem. 2015, 7, 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferré-D'Amaré A. R., Biopolymers 2004, 73, 71–78. [DOI] [PubMed] [Google Scholar]
- 10. Shippy R., Lockner R., Farnsworth M., Hampel A., Appl. Biochem. Biotechnol. 1999, 12, 117–129. [DOI] [PubMed] [Google Scholar]
- 11. Chen S. J., Dill K. A., Proc. Natl. Acad. Sci. USA 2000, 97, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pressman A. D., Liu Z., Janzen E., Blanco C., Müller U. F., Joyce G. F., Pascal R., Chen I. A., J. Am. Chem. Soc. 2019, 141, 6213–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koculi E., Hyeon C., Thirumalai D., Woodson S. A., J. Am. Chem. Soc. 2007, 129, 2676–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen S. J., Annu. Rev. Biophys. 2008, 37, 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan T., Sosnick T. R., Fang X. W., Littrell K., Golden B. L., Shelton V., Thiyagarajan P., Proc. Natl. Acad. Sci. USA 2002, 98, 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vlassov A. V., Johnston B. H., Landweber L. F., Kazakov S. A., Nucleic Acids Res. 2004, 32, 2966–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vlassov A. V., Kazakov S. A., Johnston B. H., Landweber L. F., J. Mol. Evol. 2005, 61, 264–273. [DOI] [PubMed] [Google Scholar]
- 18. Kazakov S. A., Balatskaya S. V., Johnston B. H., RNA 2006, 12, 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hegg L. A., Fedor M. J., Biochemistry 1995, 34, 15813–15828. [DOI] [PubMed] [Google Scholar]
- 20. Mutschler H., Taylor A. I., Porebski B. T., Lightowlers A., Houlihan G., Abramov M., Herdewijn P., Holliger P., eLife 2018, 7, e43022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hieronymus R., Godehard S. P., Balke D., Müller S., Chem. Commun. 2016, 52, 4365–4368. [DOI] [PubMed] [Google Scholar]
- 22. Hieronymus R., Müller S., ChemSystemsChem 2021, 3, e2100003. [Google Scholar]
- 23. Gwiazda S., Salomon K., Appel B., Müller S., Biochimie 2012, 94, 1457–1463. [DOI] [PubMed] [Google Scholar]
- 24. Adamala K., Szostak J. W., Science 2013, 342, 1098–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Traïkia M., Warschawski D. E., Recouvreur M., Cartaud J., Devaux P. F., Eur. Biophys. J. 2000, 29, 184–195. [DOI] [PubMed] [Google Scholar]
- 26. MacDonald R. C., Jones F. D., Qui R., Biochim. Biophys. Acta Biomembr. 1994, 1191, 362–370. [DOI] [PubMed] [Google Scholar]
- 27. Litschel T., Ganzinger K. A., Movinkel T., Heymann M., Robinson T., Mutschler H., Schwille P., New J. Phys. 2018, 20, 055008. [Google Scholar]
- 28. Hyman A. A., Weber C. A., Jülicher F., Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [DOI] [PubMed] [Google Scholar]
- 29. Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A. A., Science 2009, 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- 30. Ukmar-Godec T., Hutten S., Grieshop M. P., Rezaei-Ghaleh N., Cima-Omori M. S., Biernat J., Mandelkow E., Söding J., Dormann D., Zweckstetter M., Nat. Commun. 2019, 10, 2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crosby J., Treadwell T., Hammerton M., Vasilakis K., Crump M. P., Williams D. S., Mann S., Chem. Commun. 2012, 48, 11832–11834. [DOI] [PubMed] [Google Scholar]
- 32. Tang T.-Y. D., Van Swaay D., DeMello A., Ross Anderson J. L., Mann S., Chem. Commun. 2015, 51, 11429–11432. [DOI] [PubMed] [Google Scholar]
- 33. Alva V., Söding J., Lupas A. N., eLife 2015, 4, e09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Das C., Frankel A. D., Biopolymers 2003, 70, 80–85. [DOI] [PubMed] [Google Scholar]
- 35. Oparin A. I., The Origin of Life and the Origin of Enzymes, Wiley, Hoboken, 1965. [Google Scholar]
- 36. Greenwald J., Kwiatkowski W., Riek R., J. Mol. Biol. 2018, 430, 3735–3750. [DOI] [PubMed] [Google Scholar]
- 37. Braun S., Humphreys C., Fraser E., Brancale A., Bochtler M., Dale T. C., PLoS ONE 2011, 6, e19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang D., Peng S., Hartman M. R., Gupton-Campolongo T., Rice E. J., Chang A. K., Gu Z., Lu G. Q., Luo D., Sci. Rep. 2013, 3, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adamski P., Eleveld M., Sood A., Kun Á., Szilágyi A., Czárán T., Szathmáry E., Otto S., Nat. Rev. Chem. 2020, 4, 386–403. [DOI] [PubMed] [Google Scholar]
- 40. Frankel E. A., Bevilacqua P. C., Keating C. D., Langmuir 2016, 32, 2041–2049. [DOI] [PubMed] [Google Scholar]
- 41. Chowrira B. M., Berzal-Herranz A., Burke J. M., Biochemistry 1993, 32, 1088–1095. [DOI] [PubMed] [Google Scholar]
- 42. Tagami S., Attwater J., Holliger P., Nat. Chem. 2017, 9, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drobot B., Iglesias-Artola J. M., Le Vay K., Mayr V., Kar M., Kreysing M., Mutschler H., Tang T.-Y. D., Nat. Commun. 2018, 9, 3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poudyal R. R., Guth-Metzler R. M., Veenis A. J., Frankel E. A., Keating C. D., Bevilacqua P. C., Nat. Commun. 2019, 10, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanco C., Bayas M., Yan F., Chen I. A., Curr. Biol. 2018, 28, 526–537. [DOI] [PubMed] [Google Scholar]
- 46. Hayatsu R., Studier M. H., Anders E., Geochim. Cosmochim. Acta 1971, 35, 939–951. [Google Scholar]
- 47. Yoshino D., Hayatsu K., Anders E., Geochim. Cosmochim. Acta 1971, 35, 927–938. [Google Scholar]
- 48. Plankensteiner K., Reiner H., Rode B. M., Mol. Diversity 2006, 10, 3–7. [DOI] [PubMed] [Google Scholar]
- 49. Patel B. H., Percivalle C., Ritson D. J., Duffy C. D., Sutherland J. D., Nat. Chem. 2015, 7, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Canavelli P., Islam S., Powner M. W., Nature 2019, 571, 546–549. [DOI] [PubMed] [Google Scholar]
- 51. Hartman H., Smith T., Life 2014, 4, 227–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Johnson A. P., Cleaves H. J., Dworkin J. P., Glavin D. P., Lazcano A., Bada J. L., Science 2008, 322, 404. [DOI] [PubMed] [Google Scholar]
- 53. Zaia D. A. M., Zaia C. T. B. V., De Santana H., Origins Life Evol. Biospheres 2008, 38, 469–488. [DOI] [PubMed] [Google Scholar]
- 54. Forsythe J. G., Yu S., Mamajanov I., Grover M. A., Krishnamurthy R., Fernández F. M., Hud N. V., Angew. Chem. Int. Ed. 2015, 54, 9871–9875; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 10009–10013. [Google Scholar]
- 55. Frenkel-Pinter M., Haynes J. W., C M., Petrov A. S., Burcar B. T., Krishnamurthy R., Hud N. V., Leman L. J., Williams L. D., Proc. Natl. Acad. Sci. USA 2019, 116, 16338–16346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Le Vay K., Mutschler H., Emerging Top. Life Sci. 2019, 3, 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nesbitt S. M., Erlacher H. A., Fedor M. J., J. Mol. Biol. 1999, 286, 1009–1024. [DOI] [PubMed] [Google Scholar]
- 58. Wilson T. J., Lilley D. M. J., RNA 2002, 8, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao Z. Y., Wilson T. J., Maxwell K., Lilley D. M. J., RNA 2000, 6, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnston W. K., Unrau P. J., Lawrence M. S., Glasner M. E., Bartel D. P., Science 2001, 292, 1319–1325. [DOI] [PubMed] [Google Scholar]
- 61. Attwater J., Wochner A., Holliger P., Nat. Chem. 2013, 5, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Horning D. P., Joyce G. F., Proc. Natl. Acad. Sci. USA 2016, 113, 9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vlassov A. V., Johnston B. H., Kazakov S. A., Oligonucleotides 2005, 15, 303–309. [DOI] [PubMed] [Google Scholar]
- 64. Wu L., Shimada N., Kano A., Maruyama A., Soft Matter 2008, 4, 744–747. [DOI] [PubMed] [Google Scholar]
- 65. Frenkel-Pinter M., Haynes J. W., Mohyeldin A. M., Martin C., Sargon A. B., Petrov A. S., Krishnamurthy R., Hud N. V., Williams L. D., Leman L. J., Nat. Commun. 2020, 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Franchi M., Ferris J. P., Gallori E., Orig Life Evol Biosph 2003, 33, 1–16. [DOI] [PubMed] [Google Scholar]
- 67. Chong P. A., Vernon R. M., Forman-Kay J. D., J. Mol. Biol. 2018, 430, 4650–4665. [DOI] [PubMed] [Google Scholar]
- 68. Vieregg J. R., Lueckheide M., Marciel A. B., Leon L., Bologna A. J., Rivera J. R., Tirrell M. V., J. Am. Chem. Soc. 2018, 140, 1632–1638. [DOI] [PubMed] [Google Scholar]
- 69. Poudyal R. R., Keating C. D., Bevilacqua P. C., ACS Chem. Biol. 2019, 14, 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Spruijt E., Westphal A. H., Borst J. W., Cohen Stuart M. A., Van Der Gucht J., Macromolecules 2010, 43, 6476–6484. [Google Scholar]
- 71. Li L., Srivastava S., Andreev M., Marciel A. B., De Pablo J. J., Tirrell M. V., Macromolecules 2018, 51, 2988–2995. [Google Scholar]
- 72. van der Gucht J., Spruijt E., Lemmers M., Cohen Stuart M. A., J. Colloid Interface Sci. 2011, 361, 407–422. [DOI] [PubMed] [Google Scholar]
- 73. Cakmak F. P., Choi S., Meyer M. C. O., Bevilacqua P. C., Keating C. D., Nat. Commun. 2020, 11, 5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nott T. J., Craggs T. D., Baldwin A. J., Nat. Chem. 2016, 8, 569–575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information