Abstract
Heparan sulfate (HS), a sulfated linear carbohydrate that decorates the cell surface and extracellular matrix, is ubiquitously distributed throughout the animal kingdom and represents a key regulator of biological processes and a largely untapped reservoir of potential therapeutic targets. The temporal and spatial variations in the HS structure underpin the concept of “heparanome” and a complex network of HS binding proteins. However, despite its widespread biological roles, the determination of direct structure-to-function correlations is impaired by HS chemical heterogeneity. Attempts to correlate substitution patterns (mostly at the level of sulfation) with a given biological activity have been made. Nonetheless, these do not generally consider higher-level conformational effects at the carbohydrate level. Here, the use of NMR chemical shift analysis, NOEs, and spin–spin coupling constants sheds new light on how different sulfation patterns affect the polysaccharide backbone geometry. Furthermore, the substitution of native O-glycosidic linkages to hydrolytically more stable S-glycosidic forms leads to observable conformational changes in model saccharides, suggesting that alternative chemical spaces can be accessed and explored using such mimetics. Employing a series of systematically modified heparin oligosaccharides (as a proxy for HS) and chemically synthesized O- and S-glycoside analogues, the chemical space occupied by such compounds is explored and described.
Introduction
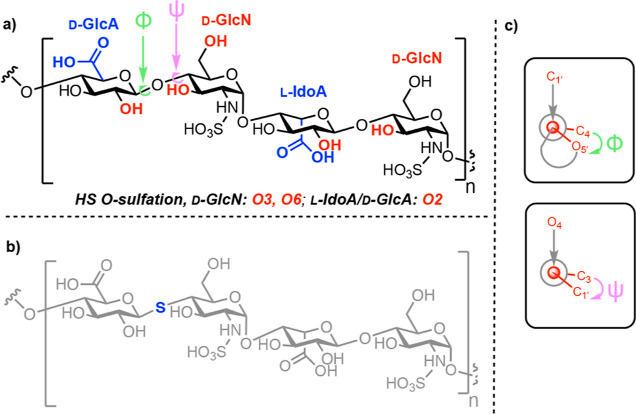
The cell surface is decorated with complex carbohydrates, which form a dense and structurally diverse layer, known as the glycocalyx. Among these carbohydrates, the glycosaminoglycan heparan sulfate (HS) is composed of a backbone consisting of β- (1,4)-linked d-glucuronic acid (GlcA) and α- (1,4)-linked N-acetyl-d-glucosamine (GlcNAc) residues. This repeating disaccharide is further modified in regional blocks, whereby N-acetylglucosamine is N-sulfated instead, GlcA undergoes epimerization at C5 to l-iduronic acid (IdoA), and O-sulfate groups are added at several positions1,2 (Figure 1a).
Figure 1.
(a) Example HS tetrasaccharide with complete N-sulfation and illustrating uronate C5 epimers alongside points for O-sulfation; free acid form shown; (b) replacement of glycosidic oxygen with sulfur to create an S-glycoside analogue; and (c) interglycosidic torsion angles represented by Newman projections: Phi (Φ) is the O5′–C1′–O4–C4 angle and Psi (ψ) is the C1′–O4–C4–C3 angle, and O4 is the linkage oxygen.
During the biosynthesis of HS, these structural modifications do not go to completion at all sites within the polysaccharide, giving rise to structural heterogeneity within the final HS chain.3,4 Consequently, HS can bind and regulate countless proteins of biological significance2,5−7 and represents an untapped reservoir of therapeutic potential.8 As a result of this biological ubiquity, studies to probe and understand HS structure-to-function effects with molecular precision are imperative. The most famous example in this context is the heparin/antithrombin interaction, but recent studies have unveiled new molecular fingerprints underpinning the biological function.9
Access to, and analysis of, structurally homogeneous HS oligosaccharide sequences has developed significantly in recent years,10−13 and the glycan toolbox of materials now available to deconvolute HS structural complexity is impressive. Such a toolbox, composed of chemically modified and synthetic HS saccharides, can be used to inform the HS structure-to-function effects at a molecular level.14,15 Within this context, unnatural sequences such as those obtained from the replacement of glycosidic linkage oxygen with sulfur (S-glycosides, Figure 1b) offer the exciting possibility of studying unique conformational preferences about the thioglycosidic and aglyconic bonds.16−18 Subsequent comparison to native sequences will improve our understanding and capability to perturb HS structure-to-function relationships as well as access the untapped carbohydrate chemical space.
Herein, to deconvolute and inform future HS biological structure-to-function studies, we have produced and utilized a series of systematically modified heparin oligosaccharides (as a proxy for HS), alongside O- and S-linked HS model disaccharides. NMR analysis is used to dissect the global and local conformational effects for distinct substituent/heteroatom patterns.
Materials and Methods
Heparin (UFH) from porcine mucosa was obtained from Bioiberica S.A. (Barcelona, Catalunya, Spain), while chemically modified heparin 6OH derivatives were prepared as described in ref (19). Briefly, N- and 6-O-desulfation was performed by heating heparin (200 mg, pyridinium salt) in a solution of DMSO containing 10% MeOH (25 mL) at 75 °C for 2 h. Re-N-sulfation was carried out by incubating the product of the latter reaction with the trimethyl-amine sulfur trioxide complex in a saturated sodium bicarbonate solution at 55 °C for 6 h. Oligosaccharides were isolated from heparinase digests of the polysaccharides separated on a Bio-Gel P-10 column (2.6 × 100 cm) at 0.5 mL/min in 0.25 M ammonium chloride previously calibrated with size-defined heparin oligosaccharides (Iduron, UK). Fractions (5 mL) were collected, and each peak was pooled separately, then desalted (HiPrep 26/10 Desalting column, GE Life Sciences), and freeze-dried. The peaks (Figure S1), corresponding to the size-defined heparin oligosaccharides, were detected by UV absorbance at 232 nm and had their structure confirmed by NMR (Figure S2).
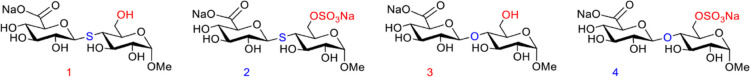
Synthesis of O- and S-HS Saccharide Analogues
O- and S-Disaccharides 1–4 were prepared as previously described.16,20 Briefly, S-disaccharides 1 and 2 were obtained by reacting (SN2) a suitable protected thiol derivative of GlcA (synthesized in four steps from d-glucurono-3,6-lactone) with a 4-triflate galactopyranoside precursor, followed by selective sulfation (for 2) and deprotection. O-Disaccharides 3 and 4 were prepared by glycosylation between a GlcA trichloroacetimidate donor and a 4-OH glucopyranoside derivative, followed by sulfation (for 4) and deprotection. Scheme 1 depicts the structures of 1–4.
Scheme 1. Chemical Structures of O- and S-HS Saccharide Analogues.
Nuclear Magnetic Resonance
Synthetic O- and S-HS saccharide analogues NMR experiments were performed at 300 K using an 800 MHz Bruker AVANCE III + spectrometer fitted with a CryoProbe. Unfractionated heparin, heparin oligosaccharides, and their chemically modified derivative NMR experiments were performed at 298, 310, or 343 K using a 500 MHz Bruker AVANCE NEO spectrometer fitted with a TXI probe. In addition to one-dimensional spectra, both homonuclear and heteronuclear two-dimensional spectra were collected. Total correlation spectroscopy (TOCSY, dipsi2gpph19) spectra were measured with a mixing time of 120–180 ms, while the mixing times for Nuclear Overhauser Effect Spectroscopy (NOESY, selnogpz.2) spectra varied between 120 and 300 ms. Heteronuclear Single Quantum Coherence (1H/13C HSQC, hsqcedetgpsp.3) spectra were collected using 32 scans, 2048 × 256 complex data points, spectral width in F1 5 KHz, in F2 11 KHz, and one-bond JCH = 145 Hz. All J-resolved heteronuclear multiple bond correlation (HMBC) spectra, Bruker pulse program hmbcetgpl3nd, were collected at 800 MHz with the JCH long-range value (cnst13) set to 3–8 Hz. The spectra were processed and assigned, and integration was performed using a Bruker TopSpin 4.1.4. Samples were prepared in deuterium oxide (>99.92%, Apollo Scientific) packed under argon to mitigate interferences arising from paramagnetic oxygen.
Results and Discussion
Distinct Sulfation Patterns Lead to Distant Conformational Effects in HS Oligosaccharides
Conventional NMR nuclei studies (mostly 1H and 13C) have been extensively used to provide details around conformational features of heparin/HS ligands.21 These studies have used natural, semisynthetic and fully synthetic materials with the same overall goal to extract structural information that can be correlated with biological function. NMR chemical shift analysis of HS polysaccharides has determined the extent to which different sulfation patterns (C2 of IdoA and C2 or C6 of GlcN) report on conformational changes throughout the polymer. For example, it was observed that sulfation at IdoA-C2 produces global conformation changes across the polymer backbone, whereas sulfation at C6 produces discrete changes, localized to the GlcN residue.22
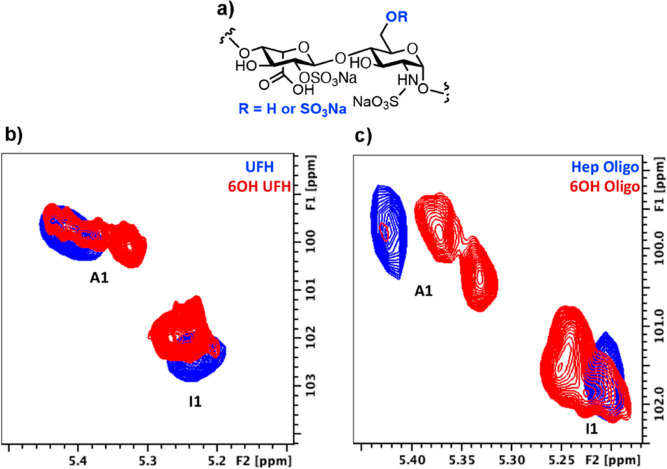
Here, HS oligosaccharides were produced using selective desulfation of heparin.19 These materials were analyzed using NMR to see whether the data obtained for these simpler, shorter oligosaccharides was comparable to that previously observed at the polymer level. The HSQC NMR spectra for this series of HS oligosaccharides are shown in Figure 2. Accordingly, anomeric chemical shift data are tabulated in Table 1. As expected, and agreeing with previously published results,22 minor changes were observed in anomeric position (both IdoA and GlcN) 1H/13C chemical shifts when unfractionated heparin (UFH) was selectively desulfated at C6 (6OH UFH) (Figure 2a). Equally, the values observed for a heparin decasaccharide (Hep Oligo) and its 6OH variant (6OH Oligo, Figure 2b) presented minor changes again, the most significant chemical shift change occurring at GlcN. Furthermore, as for UFH, desulfation at C6 led to little/no changes at C4 (Figure S3); chemical shifts, remaining at around δ = 79 ppm, imply that 6-O-sulfation had little influence on the backbone and overall saccharide conformation, as judged by chemical shift changes.
Figure 2.
(a) General HS sequence structure for UFH and Hep oligos examined, (b) HSQC NMR spectra for anomeric regions of UFH and its 6OH derivative, IdoA (I) and GlcN (A1) residues, and (c) HSQC NMR spectra for anomeric regions of Dp12 and its 6OH derivative, IdoA (I) and GlcN (A) residues. 1 denotes H1, anomeric position.
Table 1. Key 1H/13C Anomeric Chemical Shift Data.
| A1 (1H/13C ppm) | I1 (1H/13C ppm) | |
|---|---|---|
| UFH | 5.4/99.7 | 5.22/102.3 |
| 6OH UFH | 5.35/99.7 | 5.23/101.9 |
| Hep Oligo | 5.42/99.7 | 5.21/101.6 |
| 6OH Oligo | 5.35/100 | 5.23/101.6 |
Probing Iduronate Flexibility Using NOEs Within Model HS Oligosaccharides
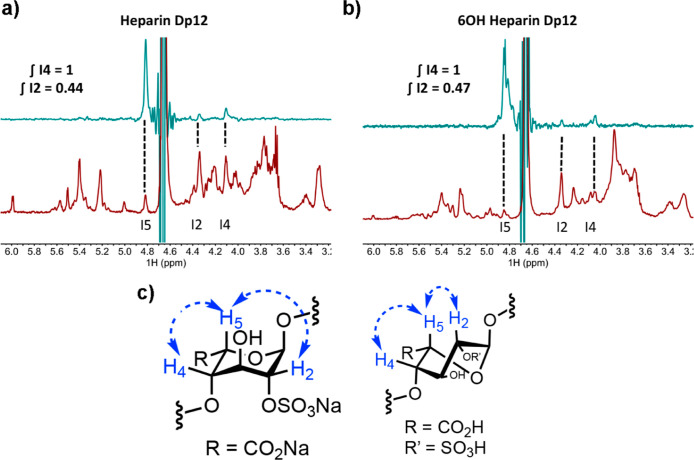
One of the key monosaccharide residues within HS, which bestows it with micro- and internal flexibility, is α-l-iduronic acid. Functionally, changes to the iduronate ring conformation seem to accommodate protein binding and lie at the heart of the prototypical interaction between heparin and AT.23 It is interesting to note, however, that the conformational properties of this residue, which involve fluidity through 4C1, 1C4, and 2S0 conformers, do not always lead to changes in overall chain orientation.24 Regardless, observations using model oligosaccharides may differ from those of polysaccharides. Building on the chemical shift analysis presented above, and since changes to the ring conformation alter the distance between vicinal hydrogen atoms, proton–proton NOEs were next used to probe iduronate ring conformation changes, correlating with distinct sulfation patterns of HS oligosaccharides. For this, selective irradiation of I5 was used, given its sensitivity to α-l-iduronic acid conformation and negligible overlap with signals from other protons (Figure SII).
Figure 3 shows selective 1D NOESY experiments for a series of chemically modified dodecasaccharides [degree of polymerization (Dp)12s]. The distribution of conformers was assessed by 1H–1H NOE analysis, which is particularly useful for the detection of the 2S0 conformer. In this conformer, the atomic distance between H2 and H5 (2.6 Å) compared with the 1C4 conformer (4.0 Å) can be easily extracted and inferred by measuring proton–proton NOEs: the shorter the distance, the stronger the NOEs,23 and any changes in equilibria between these would inevitably lead to changes in proton–proton NOE ratios. Sulfation at C6 of GlcN (Figure 3a,b) produced minor effects in the NOEs between I5 to I4 and I2 (judged by cross-peak integration), implying, once again, that such a modification position has a more localized effect in terms of saccharide conformation, which is confined around the GlcN residue. It is important to emphasize that this observation refers to the average conformation of all IdoA2S residues within a given chain since the resolution of NMR does not generally enable the differentiation between distinct residues positioned throughout the oligosaccharide. Furthermore, the dynamics of such conformer distributions is rather fast and, again, NMR does not provide a real-time picture of these events, rather, it provides the average over time.
Figure 3.
(a) Bottom, 1H NMR spectrum for heparin Dp12; top, selective 1D NOESY (irradiation of I5) with positions 5, 2, and 4 assigned; (b) bottom, 1H NMR spectrum for 6OH Dp12; top, selective 1D NOESY (irradiation of I5) with positions 5, 2, and 4 assigned; and (c) conformers 1C4 and 2S0 for IdoA2S residues which indicate distances between H2 and H5, and H4 and H5, displayed with dotted arrows.
Effect of C6 Sulfation within O- and S-Disaccharide Analogues of HS
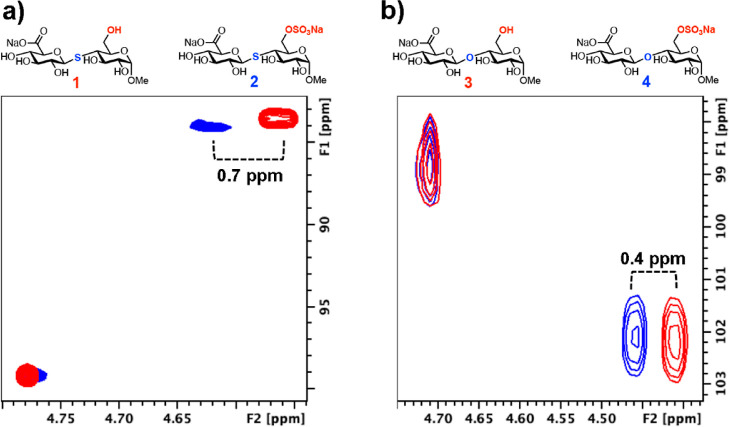
Utilizing O- and S-linked HS disaccharide analogues, bearing Glc in place of GlcN,25 the impact of C6 sulfation in shorter sequences was also investigated by NMR. Sulfation at C6 in S-glycoside 2 led to a significant change in 1H chemical shift of the adjacent uronic acid (0.7 ppm downfield change) when compared to the shift observed for its O-glycoside counterpart 4 (0.4 ppm) (Figure 4b). A small 13C chemical shift change at C1 (83.5 ppm for 1 to 84.05 ppm for 2) of sulfated S-glycoside uronate was also observed. These results suggest that instead of a localized change in the chemical environment in the case of O-glycoside 4, sulfation at C6 of S-glycoside 2 led to global changes in the overall saccharide chemical environment, as adjudged by the uronate chemical shift changes. These differences can be associated with changes in linkage geometry and changes to the uronate conformer (as evidenced by J coupling values, vide infra). Such an effect could be rationalized by the fact that sulfur is less electronegative than oxygen, increasing the shielded environment around the linkage, enabling the C6 sulfate to impart long-range conformational changes elsewhere in the chain.
Figure 4.
(a) HSQC NMR spectrum showing the effect of the 6-O-sulfation pattern upon S-glycosides and (b) HSQC NMR spectrum showing the effect of 6-O-sulfation pattern O-glycosides; black cross-peak = 6-OH compound and red cross-peak = 6-OSO3Na compound.
Spin–Spin Couplings Suggest Distinct Uronate Conformer Equilibria and Interglycosidic Dihedral Angles for O- and S-Linked HS Disaccharide Analogues
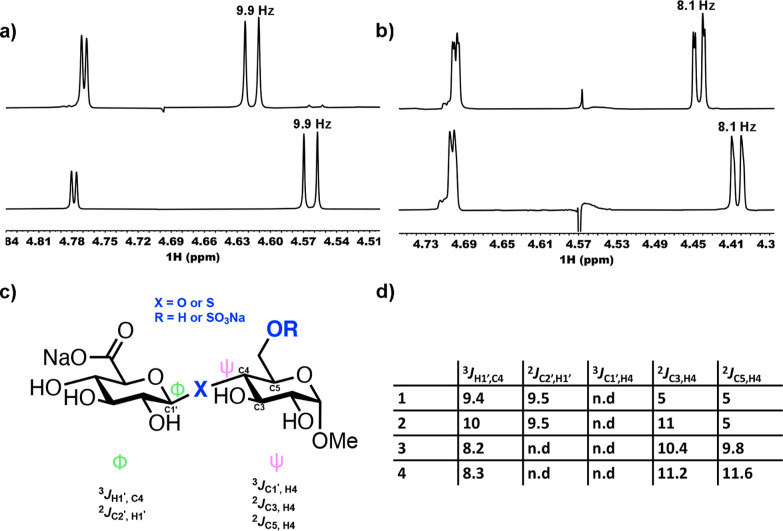
The conformational characteristics of glycans are, in part, influenced by the flexibility of their saccharide conformer equilibria and glycosidic linkages. 1H NMR analysis provides not only information around the constituent monosaccharides, based on the chemical shift, but also information around the dihedral angle between two coupled protons,26 for example, the non-equivalent protons on C1 and C2. Figure 5a,c shows the spin–spin coupling constants (3JH1,H2 of GlcA) for both O- and S-glycoside analogues of HS, 1–4. There were no observable differences resulting from sulfation at C6 for either 3 or 4 (3JH1,H2 = 8.1 Hz for 3 to 4 and 9.9 Hz for 1 to 2). However, switching from an O-glycosidic linkage to an S-glycoside led to an increase of 1.8 Hz (from 8.1 to 9.9 Hz) for this 3J coupling, suggesting a change in the solution-phase glucuronic acid conformation, certainly about the C1–C2 bond.
Figure 5.
(a) Anomeric region of 1H NMR spectra from 6-OH and 6-OSO3Na S-glycosides 1 and 2 (bottom to top), (b) anomeric region of 1H NMR spectra from 6-OH and 6-OSO3Na O-glycosides 3 and 4 (bottom to top), (c) scheme showing redundant J-couplings across the β (1 → 4) O- and S-glycosidic linkages, and (d) redundant J-couplings across the β (1 → 4) O- and S-glycosidic linkages values in Hz. n.d, not detectable.
Long-range heteronuclear coupling constant analysis using J-resolved NMR experiments were next employed to ascertain the effects of C6 sulfation and conversion of O- to S-glycosidic linkages upon glycosidic torsion angles. Dihedral angles across such linkages require four atoms linked by three consecutive bonds and represent the rotation angle between two planes around the middle bond (the saccharide units). Such angles can range from −180 to 180°, depending on the direction of rotation, are named phi (ϕ) and psi (ψ), and are illustrated in Figure 1c.27
As some of these constants are extremally small and at times undetectable (when dihedral angles are around 90°), resolution was sufficient at 800 MHz to enable measurement of only some of these. Yet, measuring Redundant J-couplings,28 which have been used to model conformational populations of O-glycosidic linkages, facilitated the further investigation into the effects of C6 sulfation and O- to S-glycosidic conversion upon glycosidic torsion angles (Figure 5c). Again, the O- to S-replacement led to changes in these coupling constants (Figure 5d), suggesting differences in linkage behavior. O-glycan sulfation at C6 did not induce changes in 3JH1’,C4 values (Figure 5d, 8.2 Hz for 3 and 8.3 Hz for 4), whereas a difference of 0.6 Hz was observed due to C6 sulfation in thioglycoside 2 (Figure 5d). Furthermore, there were significant changes to 2JC3,H4 in 1 and 2, which further suggest interglycosidic dihedral angle variations. Such changes may be explained by the shielding effect of sulfur versus oxygen (mentioned above) and longer C–S thioether bonds. Together, these results demonstrate that S-glycosides display conformational states that are unique from their natural O-glycosides.
Conclusions
The overall conformation of O- and S-glycoside analogues of HS is dependent on the substitution pattern within such molecules and their glycosidic linkage type. This observation agrees with previously described effects using systematically modified heparin polymers as a proxy for HS. Much of this also underpins many structure-to-function relationships and reinforces the idea that saccharides of similar backbones presenting different substitution patterns will display unique molecular architecture affecting their geometry, conformation, flexibility, and thus biological effect.
The conformational changes presented by S-glycoside analogues differ from those observed for their O-linked counterparts, highlighting that this heteroatom substitution enables access to previously untapped chemical space, which could be exploited as a powerful tool in investigating glycosaminoglycan–protein interactions and possibly inform the design of druggable glycan-based candidates.
Acknowledgments
Keele University is thanked for PhD studentship funding to L.N. and pump-priming funding to M.A.L. M.A.L. also thanks The Royal Society (IEC\NSFC\201116) and the Academy of Medical Sciences/Wellcome Trust (Springboard grant, SBF007\100054). G.J.M. thanks The U.K. Research and Innovation (UKRI, Future Leaders Fellowship, MR/T019522/1). FAPESP is thanked for project grant funding (17/14179-3, 19/19298-6, and 20/04899-1) to M.A.L., M.C.Z.M., and H.B.N., respectively.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02070.
Biogel P10 oligosaccharide fractionation; HSQC spectra for heparin and 6OH heparin oligosaccharide; HSQC spectra showing the C4 region of 6S (blue) and 6OH (red) heparin derivatives; and J-resolved HMBC spectra for S- and O-glycosides (PDF)
Author Present Address
The current address for TRR is Analytical and Biological Sciences, Research & Development, Science, Research and Innovation, Medicines & Healthcare Products Regulatory Agency, Blanche Lane, South Mimms, Hertfordshire, EN6 3QG, UK
Author Contributions
∇ M.C.Z.M., L.N., and C.O’S. are joint first authors. J.K., G.J.M., and M.A.L. are senior authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Leung A. W. Y.; Backstrom I.; Bally M. B. Sulfonation, an Underexploited Area: From Skeletal Development to Infectious Diseases and Cancer. Oncotarget 2016, 7, 55811–55827. 10.18632/oncotarget.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghetti M. C. Z.; Hughes A. J.; Rudd T. R.; Nader H. B.; Powell A. K.; Yates E. A.; Lima M. A. Heparan Sulfate and Heparin Interactions with Proteins. J. R. Soc. Interface 2015, 12, 20150589. 10.1098/rsif.2015.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghetti M. C. Z.; Deboni P.; Palomino C. M. V.; Braga L. P.; Cavalheiro R. P.; Viana G. M.; Yates E. A.; Nader H. B.; Lima M. A. ER-Golgi Dynamics of HS-Modifying Enzymes via Vesicular Trafficking Is a Critical Prerequisite for the Delineation of HS Biosynthesis. Carbohydr. Polym. 2021, 255, 117477. 10.1016/j.carbpol.2020.117477. [DOI] [PubMed] [Google Scholar]
- Meneghetti M. C. Z.; Gesteira Ferreira T.; Tashima A. K.; Chavante S. F.; Yates E. A.; Liu J.; Nader H. B.; Lima M. A. Insights into the role of 3-O-sulfotransferase in heparan sulfate biosynthesis. Org. Biomol. Chem. 2017, 15, 6792–6799. 10.1039/c7ob01533j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd T. R.; Preston M. D.; Yates E. A. The Nature of the Conserved Basic Amino Acid Sequences Found among 437 Heparin Binding Proteins Determined by Network Analysis. Mol. Biosyst. 2017, 13, 852–865. 10.1039/c6mb00857g. [DOI] [PubMed] [Google Scholar]
- Esko J. D.; Selleck S. B. Order out of Chaos: Assembly of Ligand Binding Sites in Heparan Sulfate. Annu. Rev. Biochem. 2002, 71, 435–471. 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Ori A.; Wilkinson M. C.; Fernig D. G. The Heparanome and Regulation of Cell Function: Structures, Functions and Challenges. Front. Biosci. 2008, 13, 4309–4338. 10.2741/3007. [DOI] [PubMed] [Google Scholar]
- Takemoto N.; Suehara T.; Frisco H. L.; Sato S.-i.; Sezaki T.; Kusamori K.; Kawazoe Y.; Park S. M.; Yamazoe S.; Mizuhata Y.; Inoue R.; Miller G. J.; Hansen S. U.; Jayson G. C.; Gardiner J. M.; Kanaya T.; Tokitoh N.; Ueda K.; Takakura Y.; Kioka N.; Nishikawa M.; Uesugi M. Small-Molecule-Induced Clustering of Heparan Sulfate Promotes Cell Adhesion. J. Am. Chem. Soc. 2013, 135, 11032–11039. 10.1021/ja4018682. [DOI] [PubMed] [Google Scholar]
- Jayson G. C.; Hansen S. U.; Miller G. J.; Cole C. L.; Rushton G.; Avizienyte E.; Gardiner J. M. Synthetic Heparan Sulfate Dodecasaccharides Reveal Single Sulfation Site Interconverts CXCL8 and CXCL12 Chemokine Biology. Chem. Commun. 2015, 51, 13846–13849. 10.1039/c5cc05222j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. J.; Hansen S. U.; Baráth M.; Johannessen C.; Blanch E. W.; Jayson G. C.; Gardiner J. M. Synthesis of a Heparin-Related GlcN-IdoA Sulfation-Site Variable Disaccharide Library and Analysis by Raman and ROA Spectroscopy. Carbohydr. Res. 2014, 400, 44–53. 10.1016/j.carres.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. J.; Gardiner J. M. Adaptable Synthesis of C-Glycosidic Multivalent Carbohydrates and Succinamide-Linked Derivatization. Org. Lett. 2010, 12, 5262–5265. 10.1021/ol102310x. [DOI] [PubMed] [Google Scholar]
- Hansen S. U.; Dalton C. E.; Baráth M.; Kwan G.; Raftery J.; Jayson G. C.; Miller G. J.; Gardiner J. M. Synthesis of L-Iduronic Acid Derivatives via [3.2.1] and [2.2.2] L-Iduronic Lactones from Bulk Glucose-Derived Cyanohydrin Hydrolysis: A Reversible Conformationally Switched Superdisarmed/Rearmed Lactone Route to Heparin Disaccharides. J. Org. Chem. 2015, 80, 3777–3789. 10.1021/jo502776f. [DOI] [PubMed] [Google Scholar]
- Baráth M.; Hansen S. U.; Dalton C. E.; Jayson G. C.; Miller G. J.; Gardiner J. M. Modular Synthesis of Heparin-Related Tetra-, Hexa- and Octasaccharides with Differential O-6 Protections: Programming for Regiodefined 6-O-Modifications. Molecules 2015, 20, 6167–6180. 10.3390/molecules20046167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongener I.; O’Shea C.; Wootton H.; Watkinson M.; Miller G. J. Developments in the Chemical Synthesis of Heparin and Heparan Sulfate. Chem. Rec. 2021, 21, 3238–3255. 10.1002/tcr.202100173. [DOI] [PubMed] [Google Scholar]
- Singh M.; Watkinson M.; Scanlan E. M.; Miller G. J. Illuminating glycoscience: synthetic strategies for FRET-enabled carbohydrate active enzyme probes. RSC Chem. Biol. 2020, 1, 352–368. 10.1039/d0cb00134a. [DOI] [PubMed] [Google Scholar]
- Koffi Teki D. S.-E.; Bil A.; Moreau V.; Chagnault V.; Fanté B.; Adjou A.; Kovensky J. Synthesis of Multivalent: S-Glycoside Analogs of a Heparan Sulfate Sequence. Org. Chem. Front. 2019, 6, 2718–2725. 10.1039/c9qo00581a. [DOI] [Google Scholar]
- Bundle D. R.; Rich J. R.; Jacques S.; Yu H. N.; Nitz M.; Ling C.-C. Thiooligosaccharide Conjugate Vaccines Evoke Antibodies Specific for Native Antigens. Angew. Chem. Int. Ed. 2005, 44, 7725–7729. 10.1002/anie.200502179. [DOI] [PubMed] [Google Scholar]
- Cao H.; Yu B. Synthesis of a S-Linked Heparan Sulfate Trisaccharide as the Substrate Mimic of Heparanase. Tetrahedron Lett. 2005, 46, 4337–4340. 10.1016/j.tetlet.2005.04.088. [DOI] [Google Scholar]
- Yates E. A.; Santini F.; Guerrini M.; Naggi A.; Torri G.; Casu B. 1H and 13C NMR Spectral Assignments of the Major Sequences of Twelve Systematically Modified Heparin Derivatives. Carbohydr. Res. 1996, 294, 15–27. 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- Koffi Teki D. S.-E.; Coulibaly B.; Bil A.; Vallin A.; Lesur D.; Fanté B.; Chagnault V.; Kovensky J. Synthesis of Novel S- and O-Disaccharides Analogs of Heparan Sulfate for Heparanase Inhibition. Org. Biomol. Chem. 2022, 20, 3528–3534. 10.1039/D2OB00250G. [DOI] [PubMed] [Google Scholar]
- Hughes A.; Meneghetti M.; Huang T.-Y.; Hung S.-C.; Elli S.; Guerrini M.; Rudd T.; Lima M.; Yates E. Investigating the Relationship between Temperature, Conformation and Calcium Binding in Heparin Model Oligosaccharides. Carbohydr. Res. 2017, 438, 58–64. 10.1016/j.carres.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Rudd T. R.; Yates E. A. Conformational Degeneracy Restricts the Effective Information Content of Heparan Sulfate. Mol. Biosyst. 2010, 6, 902–908. 10.1039/b923519a. [DOI] [PubMed] [Google Scholar]
- Hsieh P.-H.; Thieker D. F.; Guerrini M.; Woods R. J.; Liu J. Uncovering the Relationship between Sulphation Patterns and Conformation of Iduronic Acid in Heparan Sulphate. Sci. Rep. 2016, 6, 29602. 10.1038/srep29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov D.; Linhardt J. R.; Mayo H. K. NMR Solution Conformation of Heparin-Derived Hexasaccharide. Biochem. J. 1997, 328, 51–61. 10.1042/bj3280051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerduin P.; van Boeckel C. A. A.; Basten J. E. M.; Broekhoven M. A.; Lucas H.; Rood A.; van der Heijden H.; van Amsterdam R. G. M.; van Dinther T. G.; Meuleman D. G.; Visser A.; Vogel G. M. T.; Damm J. B. L.; Overklift G. T. Feasible Synthesis and Biological Properties of Six “non-Glycosamino” Glycan Analogues of the Antithrombin III Binding Heparin Pentasaccharide. Bioorg. Med. Chem. 1994, 2, 1267–1280. 10.1016/s0968-0896(00)82078-7. [DOI] [PubMed] [Google Scholar]
- Minch M. J. Orientational Dependence of Vicinal Proton-proton NMR Coupling Constants: The Karplus Relationship. Concepts Magn. Reson. 1994, 6, 41–56. 10.1002/cmr.1820060104. [DOI] [Google Scholar]
- Khan S.; Gor J.; Mulloy B.; Perkins S. J. Semi-Rigid Solution Structures of Heparin by Constrained X-Ray Scattering Modelling: New Insight into Heparin-Protein Complexes. J. Mol. Biol. 2010, 395, 504–521. 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Turney T.; Meredith R.; Pan Q.; Sernau L.; Wang X.; Hu X.; Woods R. J.; Carmichael I.; Serianni A. S. Conformational Populations of β-(1→4) O -Glycosidic Linkages Using Redundant NMR J -Couplings and Circular Statistics. J. Phys. Chem. B 2017, 121, 3042–3058. 10.1021/acs.jpcb.7b02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.